- 1Laboratory of Aquatic Nutrition and Feed, College of Fisheries, Guangdong Ocean University, Zhanjiang, China
- 2Aquatic Animals Precision Nutrition and High Efficiency Feed Engineering Research Center of Guangdong Province, Zhanjiang, China
- 3Key Laboratory of Aquatic, Livestock and Poultry Feed Science and Technology in South China, Ministry of Agriculture, Zhanjiang, China
The spleen is an important organ in the immune function of fish, and it is also important for hematogenesis and antibody and granulocyte production. However, the effect of oxidized fish oil on the spleen of hybrid grouper (♀ Epinephelus fuscoguttatus × ♂ Epinephelus lanceolatus) is unknown. In this study, hybrid groupers were fed with oxidized fish oil and the spleen index, antioxidant ability, histology and transcriptome were investigated. Oxidized fish oil did not affect the spleen index. Levels of superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GSH-Px) and malondialdehyde (MDA) in the spleen were significantly increased as the amount of oxidized fish oil in the diet increased, but the vitamin E concentration was significantly decreased. The morphological organization of the spleen was damaged with increased oxidative stress. And the spleen reacted to oxidative stress by platelet activation, FOXO and notch signaling pathways, which involved amyloid beta precursor protein binding family B member 1 interacting protein (APBB1IP) gene, glucose-6-phosphatase (G6PC) gene, histone acetyltransferase p300 (EP300) gene, insulin gene and notch 2 gene. In conclusion, the oxidized fish oil caused oxidative stress and damaged its structure. Additionally, oxidized fish oil changed the transcription profile of the spleen.
Introduction
Fish oil is an important source of lipids in aquaculture feed, being rich in unsaturated fatty acids such as eicosapentaenoic (EPA) and docosahexaenoic (DHA) acids (Long et al., 2021). However, during its production, storage and usage, fish oil is readily oxidized, producing a variety of primary and secondary metabolites including aldehydes, ketones, alcohols, esters, acids, and other substances (Yu et al., 2021). Fish oil oxidation in feed is a frequent occurrence in aquaculture and may induce oxidative stress in aquatic animals (Chen et al., 2019; Song et al., 2019).
The immune system of fish mainly comprises immune organs and tissues, immune cells and immune factors (Dalmo et al., 1997). The spleen has the functions of degrading and processing antigens and producing antibodies (Dalmo et al., 1997). Additionally, the spleen is also a major hematopoietic organ that can produce blood cells, endothelial cells, reticulocytes, macrophages and melanin macrophages (Graf and Schlüns, 1979). Research into fish spleen immunity mainly focuses on the effects of pathogens and environmental pollutants on the spleen. Pathogens including necrosis virus (Zhao H. et al., 2020), Yersinia ruckeri (Wang et al., 2021), Vibrio anguillarum (Song et al., 2021), and Aeromonas hydrophila (Xia et al., 2021) have been used to study the immune function of the spleen, as have environmental pollutants chlorpyrifos (Xing et al., 2019), cypermethrin and sulfamethoxazole (Zhao Z. et al., 2020) and heavy metals (Savassi et al., 2020). However, the response of the spleen to the toxic chemicals generated by oxidized fish oil is unknown.
The hybrid grouper (♀ Epinephelus fuscoguttatus × ♂ Epinephelus lanceolatus), a new species of grouper obtained through hybridization, has been noted as one of the most sought-after fish due to its excellent attributes such as high nutritional value, rapid growth and high disease resistance (Song S. G. et al., 2018). As a component of the fish immune system, the spleen is essential in resisting foreign antigens and regulating the immune response (Fu et al., 2015). In this study, the index, antioxidant ability and organizational structure of the hybrid grouper spleen were studied under oxidative stress. Additionally, the molecular mechanism of oxidative stress in the hybrid grouper spleen was investigated using transcriptome sequencing technology. In summary, the findings of this study will provide phenotypic information as well as a molecular foundation for further research into the mechanism of oxidative stress in the spleen.
Materials and Methods
Oxidized Fish Oil and Experimental Diets
Shandong Yuwang Pharmaceutical Co., Ltd. supplied fresh fish oil with a peroxide value (POV) of 1.15 mmol⋅kg–1. The technique of Chen et al. (2012) was used for preparing oxidized fish oil. This involved placing the bottle of fresh fish oil into a 1 L beaker and warming with a water bath at 55°C, during which the bottle was constantly injected with air using an air pump. A titration procedure (GB/T 5538–2005/ISO 3960:2001) was used to track the POV of the oxidized fish oil until it reached 231 mmol⋅kg–1, as described by Song C. et al. (2018). Finally, the oxidized fish oil refrigerated at –20°C for later use.
Protein sources in the diet were white fish meal, wheat gluten, corn gluten meal and soybean meal. Wheat flour was the primary source of sugar in the diet. Fresh and oxidized fish oil were the primary sources of oil in the diet. Four diets were prepared with fresh fish oil: oxidized fish oil ratios of 9:0 (R group), 6:3 (L group), 3:6 (M group), and 0:9 (H group). The composition and content of the experimental feed ingredients were shown in Table 1. The feed components were crushed, sieved using a 60-mesh sieve and weighed precisely according to the formula. After the various feed ingredients were mixed step by step, the corresponding oils were added. The raw materials were mixed evenly using a mixer, with an appropriate amount of water added to facilitate even mixing. A pelletizer was used to make 2.5 mm diameter pellets of the mix, which were dried at ambient temperature, packaged, and refrigerated at –20°C.
The analysis of experimental diets was as follows. The moisture content of the feed was determined by drying it at 105°C until a constant mass was reached (Zhao et al., 2015). The protein content was determined using a Kjeldahl nitrogen analyzer (FOSS KT8400; Li et al., 2021). The burning technique was used to determine the ash content (Nagy and Clair, 2000). The crude lipid content was determined using a lipid analyzer (ANKOM XT15; Purohit et al., 2016). High-performance liquid chromatography (GB/T 28717-2012) was used to evaluate the malondialdehyde (MDA) levels in the experimental diets (Long et al., 2022).
Experimental Fish and Feeding Trials
The experimental hybrid groupers, with the same genetic background and uniform specifications, were provided by a breeding hatchery (Zhanjiang, China). Before the experiment, the groupers were kept in an outdoor cement tank and fed commercial feed with 50% crude protein and 11% crude lipid (HaidaAquatic Diet Co. Ltd., Zhanjiang, China) for 28 days. The well-proportioned and robust hybrid groupers (average weight 30.34 ± 0.02 g) were split into four groups at random, each with three replicates and forty fish. The experiment was carried out in 1 m3 tempered barrels according to the indoor breeding system of the Donghai Island Breeding Base of Guangdong Ocean University. The aquaculture water was filtered by sponge and coral sand and then sterilized by ultraviolet light. For 65 days, fish were fed experimental diets twice a day, at 09:00 am and 16:00, until they were satiated. To ensure water quality, 50% of the water in the barrels was swapped daily. Throughout the test period, the water in the tempered glass barrels was continuously injected into air. The following physical and chemical characteristics of water were maintained: temperature 29 ± 2°C, dissolved O2 8.1 ± 0.41 mg⋅L–1 and ammonia 0.03 ± 0.01 mg⋅L–1.
Sample Collection
Fish were fasted for 24 h before being anesthetized with tricaine methanesulphonate (MS-222, 10 mg⋅L–1, Sigma-Aldrich) at the end of the experiment. Fourteen fish were randomly selected from each barrel. Of these, two fish were used to calculate the spleen index as follows: spleen index = spleen weight (g)/body weight (g) (Aghili et al., 2014). The spleens of four fish were mixed and stored in liquid nitrogen until enzyme activity was measured, the spleens of two fish were stored in 4% formaldehyde solution for section analysis and the spleens of six fish were mixed and stored in liquid nitrogen for transcriptome analysis.
Antioxidant Enzyme Ability
A spleen tissue sample (1 g) was placed in a test tube and phosphate buffer solution (9 mL) was added. After homogenization followed by centrifugation for 8 min (4°C, 6000 rpm), the supernatant was stored at 4°C. Test kits purchased from Nanjing Jiancheng Bioengineering Institute (China) were used to assess superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase (GSH-Px) activity, as well as vitamin E and MDA levels. The specific methods were the WST-1 method for SOD (Peskin and Winterbourn, 2017), the visible light method for CAT (Hadwan, 2018), the colorimetric method for GSH-Px (Shi et al., 2010) and vitamin E content (Jargar et al., 2012) and the TBA method for MDA (Dorsey and Jones, 2017).
Histopathology
The spleen fixed in paraformaldehyde was dehydrated in graded ethanol concentration, removed into xylene, then embedded in paraffin, sectioned, mounted with neutral gum and stained with hematoxylin and eosin. The morphological characteristics of the spleen were examined under a microscope and photographed as previously reported (Zhang Y. et al., 2019).
Transcriptome Profiling Analysis
Qualification and Quantification of RNA
Total RNA was extracted from the spleen using the Trizol method (Simms et al., 1993) and was subsequently processed with DNase-I. Agar gel electrophoresis was used to evaluate RNA integrity (Aranda et al., 2012) and the Nanodrop 2000 Ultra Micro Nucleic Acid Protein Analyzer was used to measure RNA concentration (Thermo Scientific, United States; Feng et al., 2018). Beijing Biomarker Bioinformatics Co., Ltd completed the library building and sequencing process.
Preparation of Libraries for Transcriptome Sequencing
For the RNA sample preparations, a total of 1 μg RNA per sample was utilized as input material. Following the manufacturer’s instructions, sequencing libraries were created using the NEBNext®UltraTM RNA Library Prep Kit for Illumina® (NEB, United States) and index codes were added to each sample’s sequences. mRNA was extracted from the total RNA using poly-Toligo-attached magnetic beads (Liu et al., 2019). In the NEBNext First Strand Synthesis Reaction Buffer, fragmentation was performed utilizing divalent cations at a high temperature (5X). M-MuLV reverse transcriptase and random hexamer primer were used to make first-strand cDNA. Next, DNA polymerase I and RNase H were used to synthesize second-strand cDNA. Exonuclease/polymerase was used to convert the remaining overhangs into blunt ends. To prepare for hybridization, NEBNext Adaptor, with a hairpin loop structure, was ligated after adenylation of the 3′ ends of DNA fragments. The library fragments were purified using AMPure XP to select cDNA fragments with a length preference of 240 bp (Beckman Coulter, Beverly, United States). This size-selected, adaptor-ligated cDNA was treated with 3 l USER enzyme (NEB, United States) for 15 min at 37°C followed by 5 min at 95°C. Then, PCR was carried out using Phusion High-Fidelity DNA polymerase, universal PCR primers and index (X) primer. Finally, the PCR products were purified using the AMPure XP system and the quality of the library was determined using the Agilent Bioanalyzer 2100 system.
Clustering and Sequencing
The index-coded samples were clustered using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina) on a cBot Cluster Generation System according to the manufacturer’s instructions. The library preparations were sequenced and paired-end reads were produced on an Illumina Hiseq 2000 platform after cluster formation (Liu et al., 2016).
Quality Control
The raw fastq readings were initially processed using in-house perl programs (Huang et al., 2020). Clean reads were produced in this phase by eliminating adapter-containing reads, ploy-N–containing reads and low-quality reads from the raw data (Zhang T. N. et al., 2019). The content of Q20, Q30 and GC in the clean data, as well as the sequence duplication level, were also computed (Zhang et al., 2014). All downstream analyses relied on high-quality, clean data.
Transcriptome Assembly
The left files from all libraries/samples were combined into a single large left.fq file and the right files (read2 files) were combined into a single large right.fq file (Yang et al., 2020). Trinity (Grabherr et al., 2011) was used to assemble the transcriptome based on left.fq and right.fq, with min kmer cov set to 2 and all other settings at their defaults.
Gene Functional Annotation
NR (NCBI non-redundant protein sequences), Pfam (protein families), KOG/COG/eggNOG (clusters of orthologous groups of proteins), Swiss-Prot (manually annotated and reviewed protein sequences), KEGG (Kyoto Encyclopedia of Genes and Genomes), GO (Gene Ontology), and TrEMBL (Atlas of Protein Sequence and Structure) databases were used to annotate gene function (Krause, 2008; Meng et al., 2020).
Quantification of Gene Expression Levels
For each sample, RSEM (Li and Dewey, 2011) was used to assess gene expression levels. Clean data was remapped onto the transcriptome that had been constructed. The mapping findings were used to calculate the read count for each gene.
Differential Expression Analysis
DESeq R (v. 1.10.1) was used for differential expression analysis of two conditions/groups (Hao and Feng, 2021). Using a model based on the negative binomial distribution, DESeq provides statistical procedures for detecting differential expression in digital gene expression data. The false discovery rate (FDR) was controlled by adjusting the P-values using Benjamini and Hochberg’s method (Jafari and Ansari-Pour, 2019). Genes identified by DESeq with an adjusted P-value of 0.05 were labeled as differentially expressed.
Gene Ontology Enrichment Analysis
The topGO R packages–based Kolmogorov–Smirnov test was used to perform GO enrichment analysis of differentially expressed genes (DEGs; Lin et al., 2020).
Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Analysis
Kyoto Encyclopedia of Genes and Genomes (Kanehisa et al., 2004) is a database resource for deriving information about the high-level functions and utilities of biological systems---such as cells, organisms and the ecosystem---from molecular-level data, particularly large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies1. To assess the statistical enrichment of differentially expressed genes in KEGG pathways, the KOBAS program (Xie et al., 2011) was utilized.
Data Analysis
SPSS 21.0 (Chicago, IL, United States) was used to handle experimental data and conduct the one-way analysis of variance (one-way ANOVA). The results were reported as mean ± standard error. When the test for homogeneity of variance had a P-value of greater than 0.05, multiple comparisons were performed. ANOVA using the Kruskal–Wallis one-way method was used to look for differences between groups with P-values of less than 0.05.
Results
Spleen Index of Hybrid Grouper Fed With Oxidized Fish Oil
The spleen index of the L, M, and H groups was not significantly different to that of the R group, but the spleen index of the H group was significantly smaller than that of the M group (Figure 1).
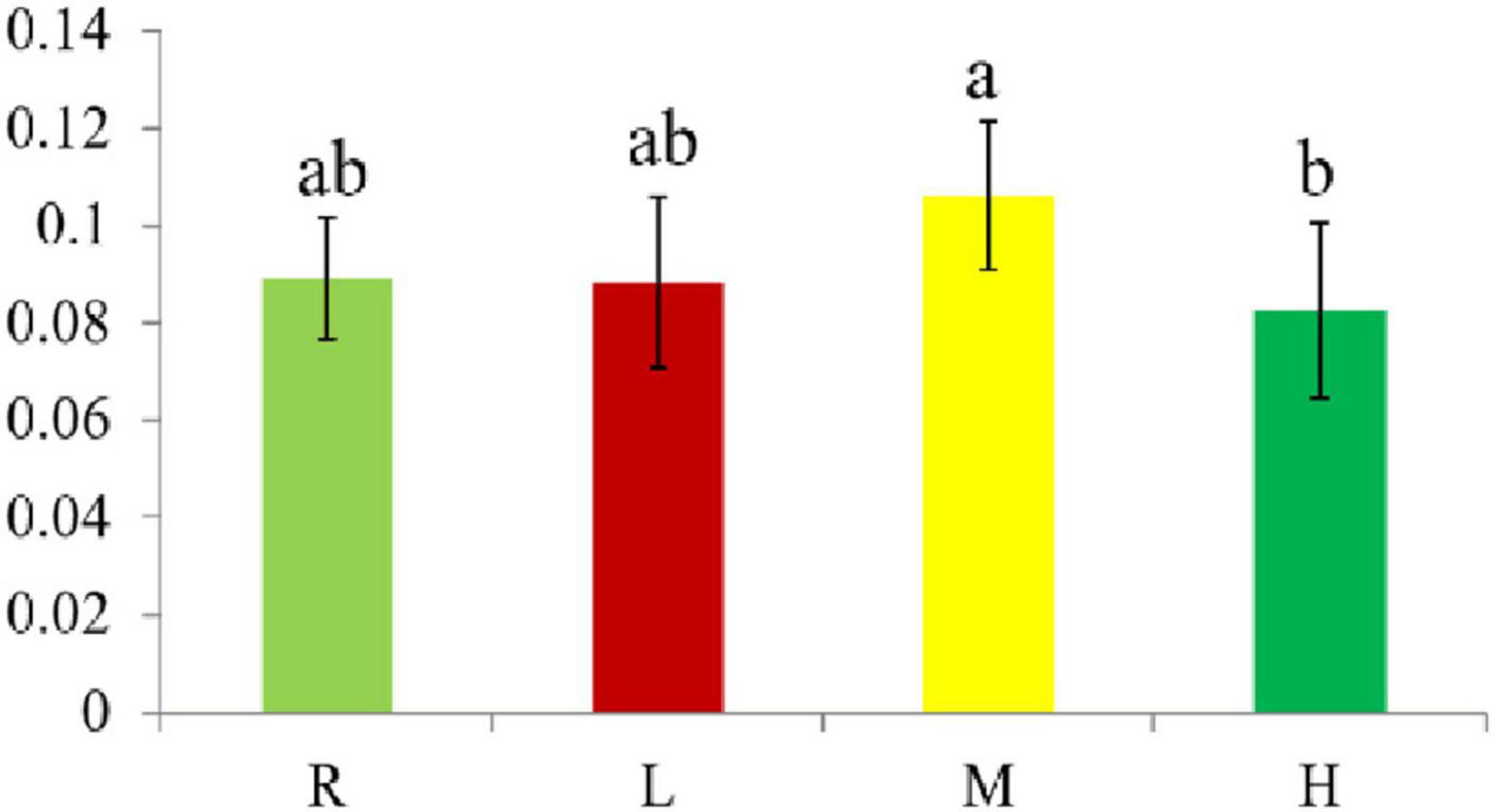
Figure 1. The Spleen index of hybrid grouper fed diets containing different oxidized fish oil level. Diverse little letters above histogram bars indicate significant differences (P < 0.05) in different dosage groups in Duncan’s multiple range tests, n = 6.
Antioxidant Performance of the Spleen
The SOD activity in the M and H groups was significantly higher than in the R and L groups (Figure 2A). CAT activity in the H group was significantly higher than in the R and L groups, but there was no significant difference between H and M groups (Figure 2B). GSH-Px activity significantly increased with increased dietary oxidized fish oil (Figure 2C). The vitamin E content in the M and H groups was significantly lower than in the R and L groups (Figure 2D). The MDA content in the H group was significantly higher than in the R and L groups but was not significantly different to the M group (Figure 2E).
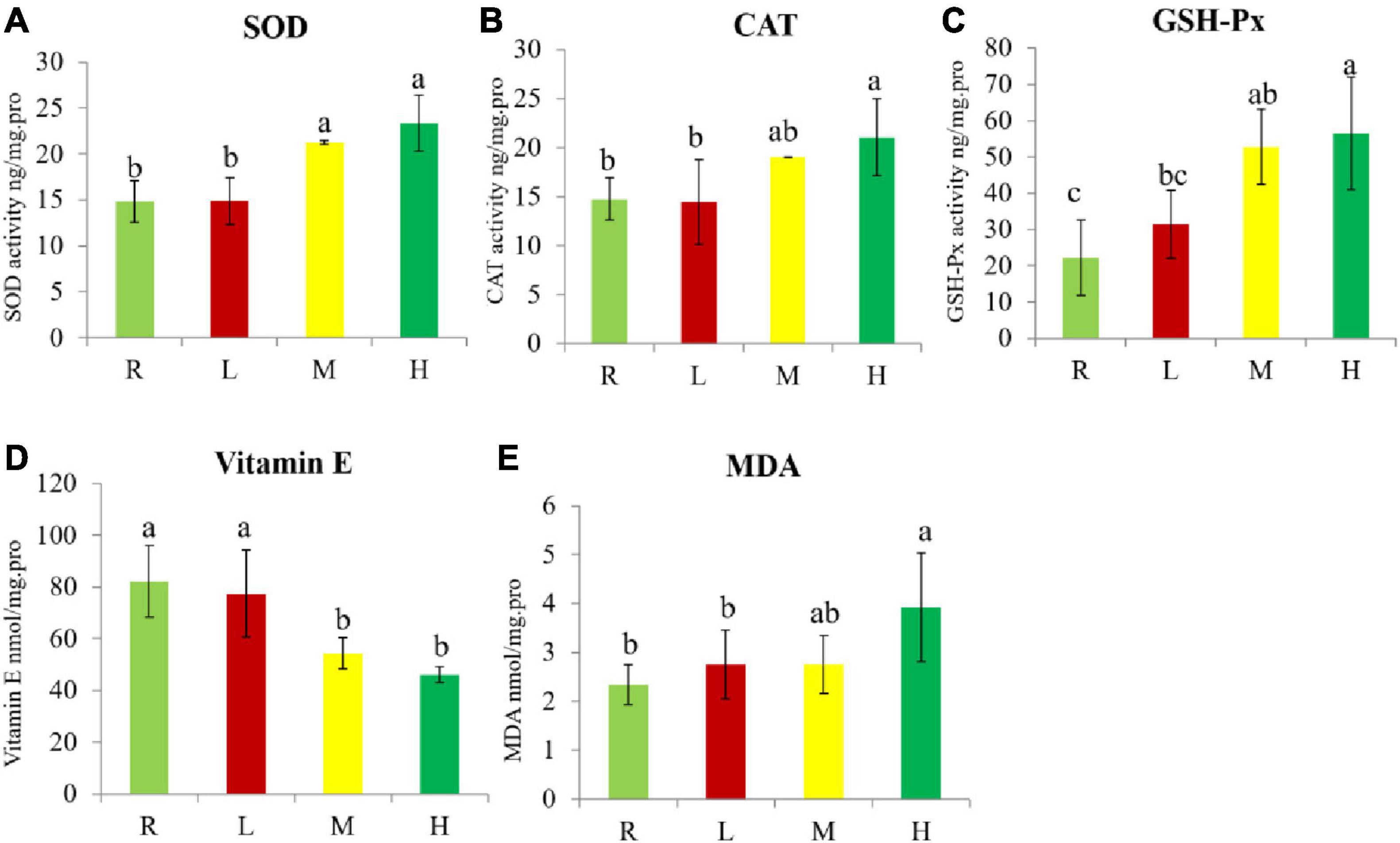
Figure 2. Antioxidant enzyme, Vitamin E, and MDA concentration in spleen of hybrid grouper. (A) Superoxide dismutase (SOD); (B) catalase (CAT); (C) glutathione peroxidase (GSH-Px); (D) vitamin E concentration; (E) malondialdehyde (MDA). Diverse little letters above histogram bars indicate significant differences (P < 0.05) in different dosage groups in Duncan’s multiple range tests, n = 3.
Spleen Histopathology
In the R group, the boundary between red pulp and white pulp was obvious, lymphocytes were numerous and dense, tiny arteries were clear and the sinusoid was full of blood cells (Figure 3A). In the L group, the boundary between red and white pulp was blurred, lymphocytes were numerous and dense, tiny arteries were clear and the sinusoid was full of blood cells (Figure 3B). In the M group, the boundary between red and white pulp was blurred, lymphocytes became scarce, the tiny artery was occluded and sinusoid blood cells started to decline (Figure 3C). In the H group, the boundary between red and white pulp was further blurred, lymphocytes became sparser, the tiny artery was also blocked and the number of blood cells in the sinusoid was further reduced (Figure 3D).
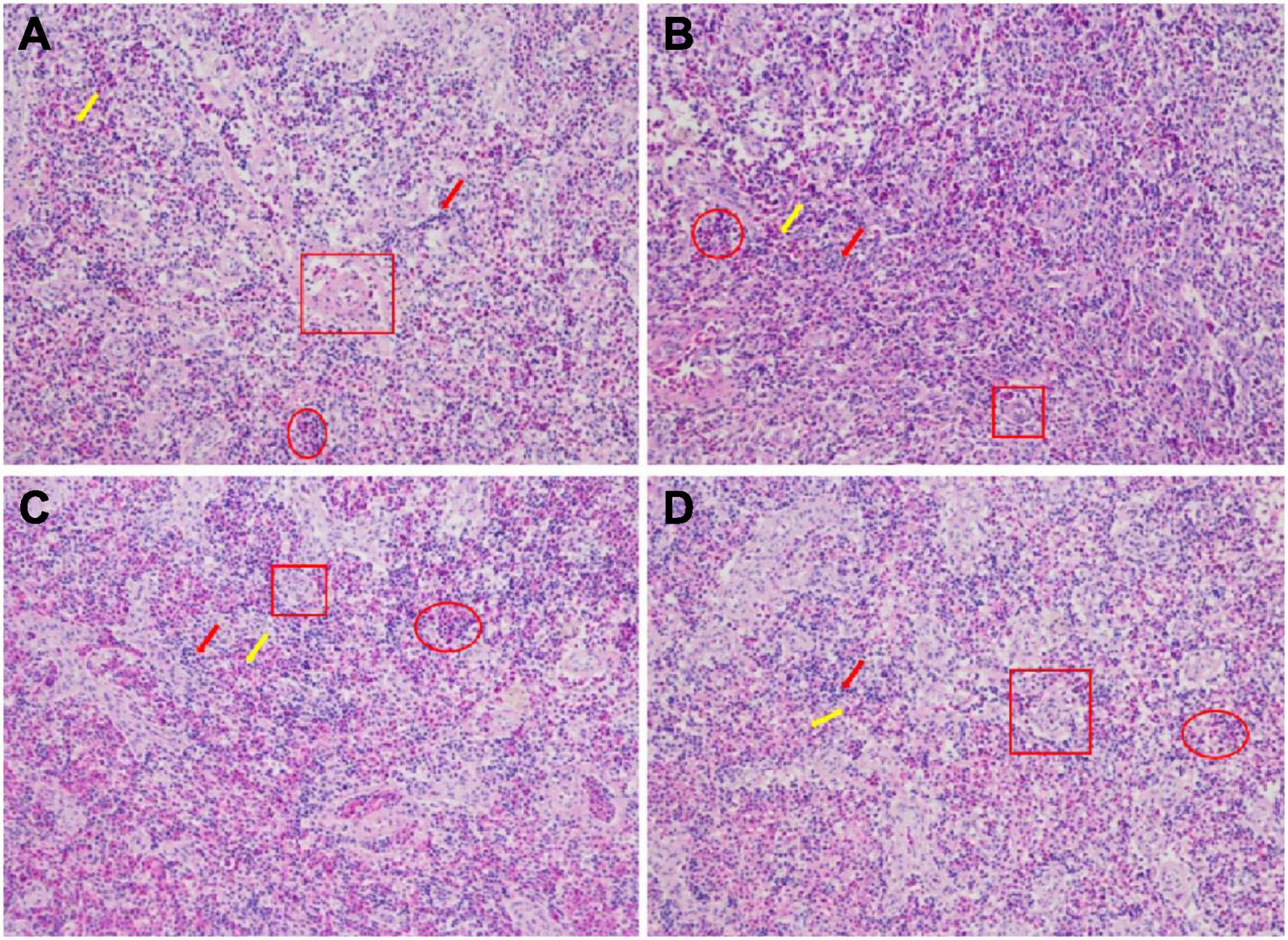
Figure 3. Histological examination images in spleen of hybrid grouper fed diets containing different oxidized fish oil level (magnification 200). R group (A), L group (B), M group (C), and H group (D). The red arrow represents the white pulp; the yellow arrow represents the red pulp; the red square represents the small arteries; the red ellipse represents the blood sinusoid.
Illumina Sequencing and de novo Assembly Between R and H Groups
In this study, three duplications in the R group and two duplications in the H group were used for transcription analysis. This was because one duplication was poorly correlated with the other two duplications in the H group. A total of 36.63 GB of clean data was collected from the spleen transcriptome sequencing results, with 5.99 GB of clean data for each sample and a Q30 base percentage of 93.76 per cent or higher. The sequencing quality was generally good, enabling subsequent research and analysis to be carried out. A total of 100,280 unigenes were recovered after assembly, with 22,604 of these measuring more than 1 kbp in length (Table 2). The data from the spleen transcriptome was been submitted to the Sequence Read Archive (GenBank accession number: PRJNA745324).
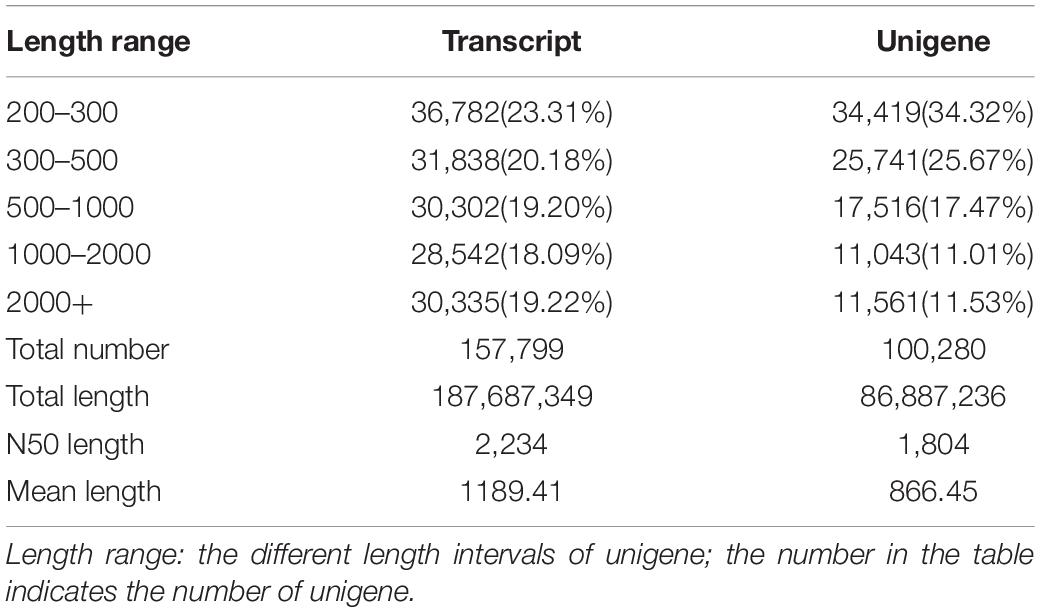
Table 2. Length distributions of the transcripts and unigenes in spleen of hybrid grouper fed fresh fish oil (R group) and highly oxidized fish oil (H group).
Functional Annotation and Classification Between R and H Groups
There were 43,177 unigenes annotation results in the functional annotation across all of the databases. Among these, 13,830 (32%), 28,923 (67%), 26,284 (60.9%), 20,989 (48.6%), 28,094 (65.1%), 14,019 (32.6%), 31,728 (73.5%), 27,001 (62.5%), and 31,684 (73.4%) unigenes were annotated to the COG, GO, KEGG, KOG, Pfam, Swissprot, TrEMBL, eggnog, and NR databases, respectively (Table 3).
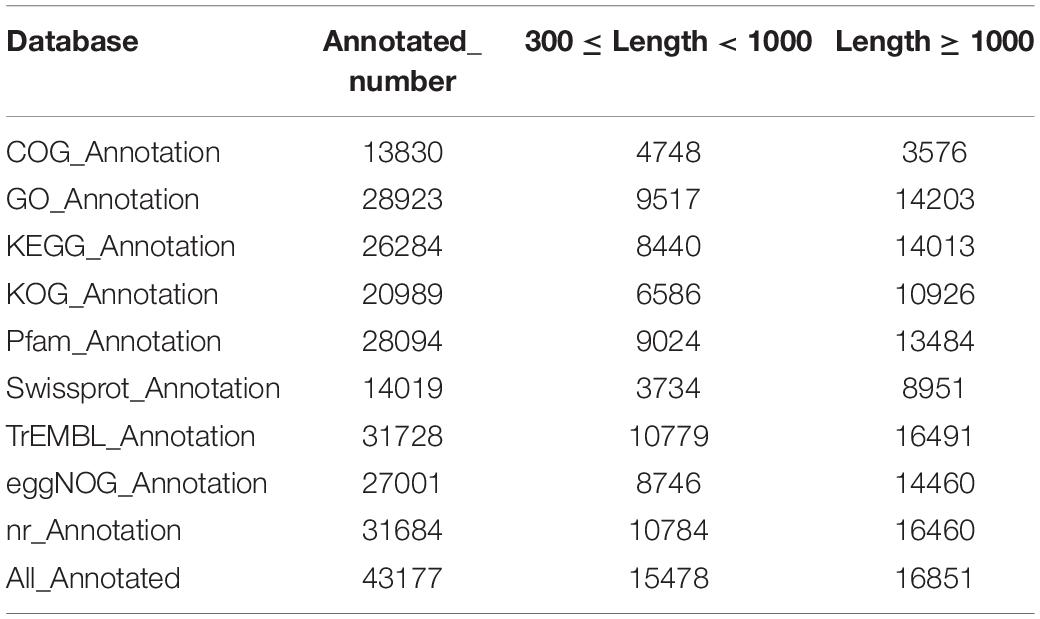
Table 3. The unigene’s annotation statistics in spleen of hybrid grouper fed fresh fish oil (R group) and highly oxidized fish oil (H group).
Identification of DEGs Between R and H Groups
The screening criteria included a false discovery rate (FDR) of less than 0.01 and a fold change (FC) of greater than or equal to 10. Between the R and H groups, there were 438 differentially expressed genes (DEGs), including 23 up-regulated genes and 415 down-regulated genes (Figure 4). According to substantial GO enrichment analyses, the enriched DEGs were mostly in biological regulation (Table 4). The KEGG pathway enrichment analysis revealed DEGs to be focused on platelet activation, the FOXO signaling and notch signaling pathway, and including amyloid beta precursor protein binding family B member 1 interacting protein (APBB1IP), glucose-6-phosphatase (G6PC), histone acetyltransferase p300 (EP300), insulin and notch 2 (Table 5).
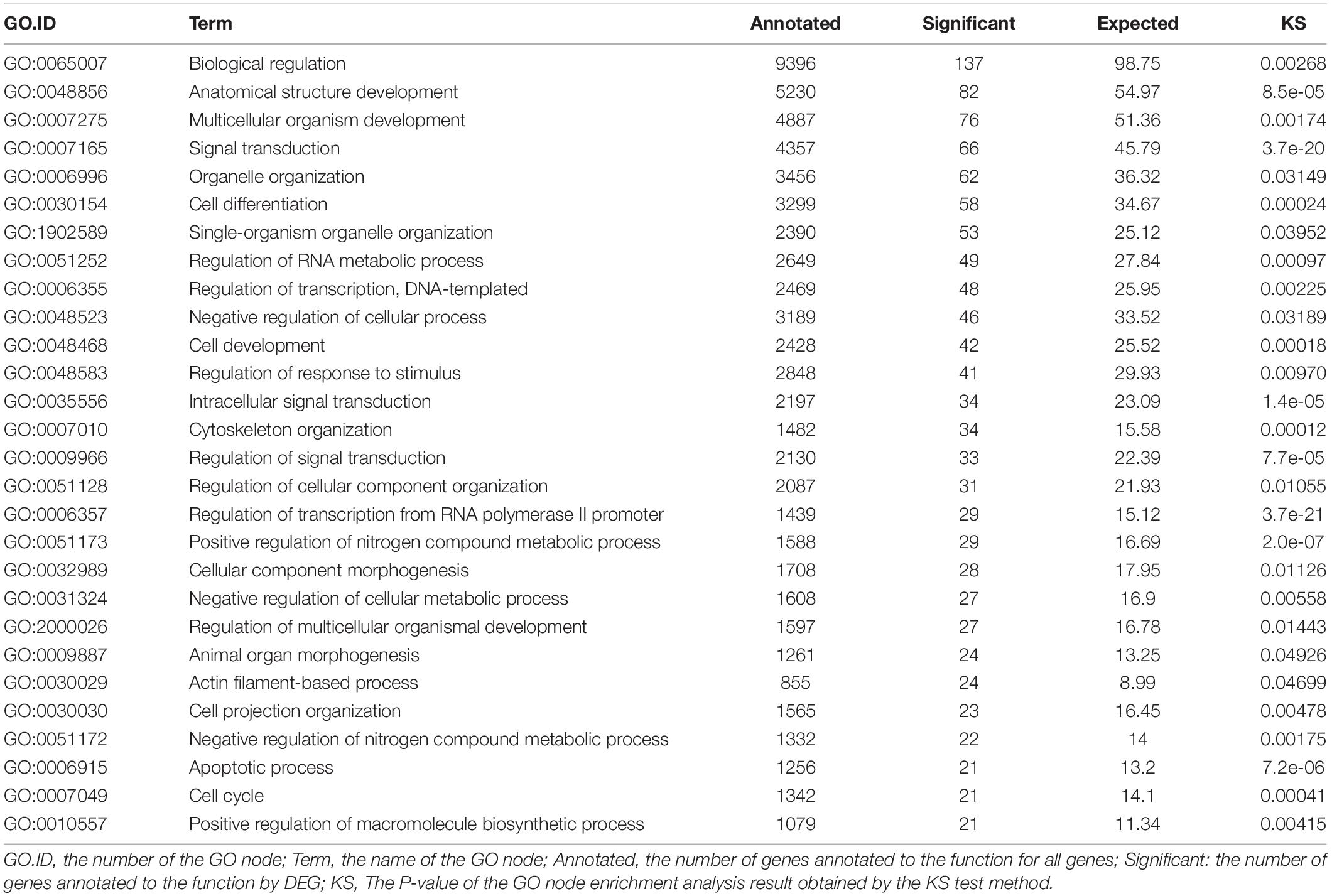
Table 4. Significant GO enrichment analysis of different expression unigenes in spleen of hybrid grouper fed fresh fish oil (R group) and highly oxidized fish oil (H group).
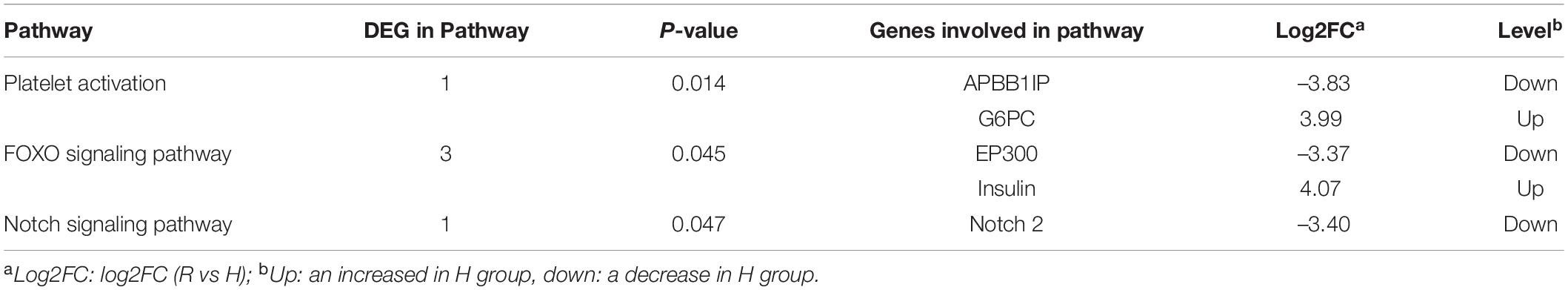
Table 5. Different expression unigene KEGG significant enrichment analysis in spleen of hybrid grouper fed fresh fish oil (R group) and highly oxidized fish oil (H group).
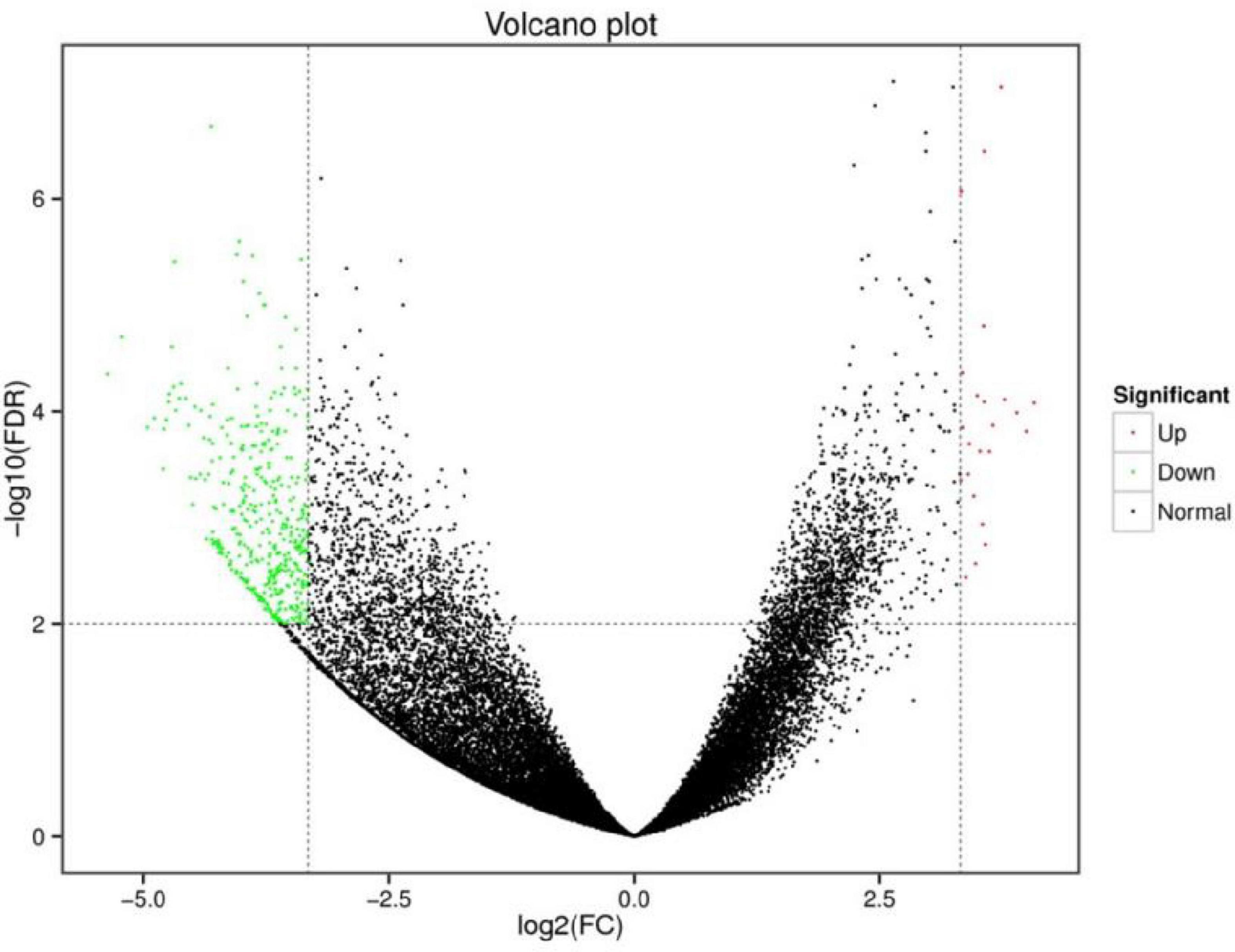
Figure 4. Volcano map of differentially expressed genes in spleen of hybrid grouper fed fresh fish oil (R group) and highly oxidized fish oil (H group).
Discussion
Oxidized Fish Oil Did Not Affect the Spleen Index
One of the most sensitive markers of poisoning is organ weight, which frequently precedes morphological alterations (Piao et al., 2013). The use of organ-to-body weight to evaluate treatment effects in toxicological research has emerged from the evaluation of organ weight changes in the context of body weight variations (Michael et al., 2007). In this study, the spleen index in the L, M, and H groups was not significantly different to the R group. This may have been because the dose of oxidized fish oil did not affect the spleen index.
Oxidized Fish Oil Caused Oxidative Stress in the Spleen
Oxidized fish oil has been proven in many studies to induce oxidative stress in fish (Song et al., 2019; Long et al., 2021). Antioxidant systems have evolved in reaction to oxidative stress, and include antioxidant enzymes such as SOD, CAT, GSH-Px, and non-enzymatic antioxidants including vitamin E, A, C, glutathione and uric acid (Nandi et al., 2019).
The initial line of defense against reactive oxygen species (ROS) is SOD, which catalyzes the dismutation of superoxide radicals (O2–) to molecular oxygen (O2) and hydrogen peroxide (H2O2) (Tosun et al., 2019). CAT can catalyze the decomposition of H2O2 into water and oxygen (Singh et al., 2018). As a selenium-dependent hydroperoxidase-reducing enzyme, GSH-Px is essential in lowering H2O2 and fatty acid hydroperoxides (Ursini and Maiorino, 2013). In the present study, the levels of SOD, CAT and GSH-Px in the spleen showed an increasing trend as the amount of oxidized fish oil in the diet increased, indicating that the spleen responded to increasing oxidative stress by continuously increasing antioxidant enzymes activity. However, previous studies found that there was a continuous decrease in the activity of antioxidant enzymes in the liver to cope with increasing oxidative stress (Long et al., 2021). Thus, the results of the present study were contrary to previous findings; this was because different organs have different resistance strategies in response to oxidative stress and different sensitivities to oxidized fish oil.
Vitamin E inhibits the formation of ROS when lipid undergoes oxidation (Pinto et al., 2020). In the present study, the vitamin E content in the spleen showed a decreasing trend as the amount of dietary oxidized fish oil increased, indicating that the spleen suppressed the production of ROS by continuously consuming vitamin E. Chen et al. (2012) also found that the vitamin E content in serum, the liver and muscle decreased in largemouth bass (Micropterus salmoides) as the amount of oxidized fish oil in the diet increased.
One of the end-products of lipid metabolism—and specifically polyunsaturated fatty acid peroxidation in cells—is MDA, which is used as a lipid peroxidation biomarker (Li, 2018). In the present study, the MDA content in the spleen gradually increased as the dietary oxidized fish oil increased, suggesting that an increase in the percentage of oxidized fish oil exacerbated splenic lipid peroxidation. This was in accordance with Rhynchocypris lagowski (Chen et al., 2019).
Oxidative Stress Damaged the Structure of the Spleen
Some studies have shown that oxidative stress can cause morphological changes to splenic tissue. Mycoplasma gallisepticum infection caused oxidative stress in chicken spleen, which led to histological abnormalities in the spleen (Hu et al., 2021). Flubendiamide and copper caused oxidative stress and changed the structure in rat spleen (Mandil et al., 2020). K2Cr2O7 caused oxidative stress and tissue lesions in rat spleen (García-Niño et al., 2015). The findings of the present study corroborated these previous findings. This phenomenon could be explained as follows. Oxidized fish oil induced excessive production of ROS in the spleen, with high quantities of ROS disrupting the normal redox state of cells and resulting in the production of peroxides and free radicals. These peroxides and free radicals have the potential to harm all cell components, including proteins, lipids and DNA.
Effect of Oxidative Stress on the Spleen Transcriptome Between R and H Groups
The mechanisms behind oxidative stress and splenic tissue damage induced by oxidized fish oil were unknown. Transcriptome analysis of the spleen samples from the R and H groups was undertaken to learn more about the effects of oxidative stress on the spleen.
Gene Ontology Enrichment Function Classification in Differentially Expressed Genes
In the GO enrichment function, the DEGs were mostly categorized as involved in biological regulation including signal transduction, regulation of response to stimulus and apoptotic processes, etc. Oxidized fish oil is a powerful stimulant that causes oxidative stress in the body (Yang et al., 2015; Zhang et al., 2021). In the present study, changes in antioxidant indices revealed that oxidized fish oil induced oxidative stress in the spleen. As a biological modulator and signal, oxidative stress not only has a deadly impact but also plays a critical role in controlling the function of cell membranes, which are essential for life (Betteridge, 2000). Oxidative stress alters the redox status of cells, resulting in the activation of protein kinases such as receptor and non-receptor tyrosine kinases, protein kinase C, and MAP kinase cascades, which cause a variety of physiological responses. These protein kinases are involved in cell activation, proliferation and differentiation, among other things. Furthermore, ROS and the resulting oxidative stress play important roles in apoptosis (Jarzaa̧b and Stryjecka-Zimmer, 2000). In general, the GO enrichment function of DEGs was in line with biological control caused by oxidative stress.
Effect of Oxidative Stress on Differentially Expressed Genes in Kyoto Encyclopedia of Genes and Genomes Pathways
Different gene products work together in organisms to accomplish biological tasks. Pathway annotation study of DEGs aids in the understanding of gene functions.
Effect of Oxidative Stress on Platelet Activation
Platelet activation is an essential element of thrombosis. The first event of activation is platelet shape change, which is followed by cytoskeleton rearrangement. Actin is the primary cytoskeletal component (Chen et al., 2017). In the present study, the actin filament–based process in the GO enrichment function of DEGs was consistent with previous research, which had shown that oxidative stress induced platelet activation by increasing the production of lipid peroxidation (Guo et al., 2019). Meanwhile, oxidative stress lowers platelet nitric oxide, which increases platelet activation and intracellular ROS generation (Manasa and Vani, 2016). In the present study, oxidative stress induced obstruction of splenic arterioles, which also showed from the side that platelet activation promoted the formation of thrombus. Furthermore, as a Rap1 binding protein, APBB1IP is primarily responsible for leukocyte recruitment and pathogen clearance through complement-mediated phagocytosis (Ge et al., 2021). The spleen is an essential immunological organ that can filter out cell debris, infections and aberrant cells, as well being as a source of red and white blood cells and immune cell subtypes (Lewis et al., 2007). In the present research, the APBB1IP gene involved in platelet activation was down-regulated under oxidative stress, indicating that oxidative stress impaired the immune function of the spleen.
Effect of Oxidative Stress on the FOXO Signaling Pathway
Many cellular physiological processes, including apoptosis, cell-cycle regulation, glucose metabolism, oxidative stress resistance and lifespan, are influenced by the FOXO signaling pathway (Ge et al., 2020). Forkhead box O (FOXO) is the O-type subfamily of the forkhead transcription factor superfamily, which comprises FOXO1, FOXO3, FOXO4, and FOXO6 (Xie et al., 2012). FOXO activity is constantly altered to react to a variety of external stimuli of different types and intensities (Xie et al., 2012). Oxidative signals stimulate the FOXO transcription factor family, which controls cell growth and tolerance to oxidative damage (Gómez-Crisóstomo et al., 2014). Akasaki et al. (2014) found that decreased expression of FOXO transcription factors in chondrocytes increased their susceptibility to oxidative stress-induced death. The FOXO signaling pathway and its genes, including G6PC, EP300 and insulin, reacted to oxidative stress, indicating that oxidative stress destroyed the cell signaling pathways and affected cell function and expression (Chen et al., 2016). FOXO1 is glycosylated via the hexosamine glycosylation pathway under oxidative stress (Butt et al., 2011) and FOXO1 is O-glycosylated, resulting in increased production of G6PC and other gluconeogenic genes (Housley et al., 2008; Kuo et al., 2008). The G6PC gene was up-regulated in the spleen under oxidative stress in this research, indicating that the spleen reacted to oxidative stress through FOXO1 glycosylation as well.
EP300 is the essential enzyme for the acetylation of FOXOs (Van Der Heide and Smidt, 2005). Under oxidative stress, acetylation of FOXOs—like glycosylation—has been demonstrated to control transcriptional activity and mediate many biological activities of FOXOs (Essers et al., 2004). It has been observed that acetylation of FOXO factors reduces FOXO-mediated transcriptional activity by breaking the interface between FOXO factors and target DNA, as well as changing the role of FOXOs role from promoting cell cycle arrest to oxidative stress prevention to cell death (Van Der Heide and Smidt, 2005). However, the spleen responded to oxidative stress by reducing the expression of EP300 gene in this study.
Wang et al. (2007) demonstrated that oxidative stress increased SIRT2 expression in cells by binding to FOXO3a and decreasing its level of acetylation, which was similar to the results of the present study. This phenomenon may have been due to the spleen responding to oxidative stress via other mechanisms, with these still requiring further exploration. Furthermore, FOXO proteins have a key role in regulating the effects of insulin and growth hormones on a variety of physiological processes, including cell proliferation, apoptosis and metabolism (Barthel et al., 2005). FOXO protein, as a key target of insulin action, may suppress cell proliferation, induce apoptosis and improve cellular resilience to oxidative stress (Barthel et al., 2005). In the present study, the spleen synthesized insulin by enhancing the expression of insulin genes, thereby acting on FOXO protein to resist oxidative stress. Smith and Shanley (2013) found that stronger oxidative stress led to a short-term activation of insulin signaling, which was consistent with the up-regulation of insulin gene expression in the spleen under oxidative stress in the present study.
Effect of Oxidative Stress on the Notch Signaling Pathway
Notch signaling is an important pathway for communication between adjacent cells to regulate cell development. The three components of the notch signaling cascade are the notch receptor, notch ligand (DSL protein) and intracellular effector molecule (CSL-DNA binding protein) (Ehebauer et al., 2006). When their corresponding transmembrane ligands bind to the extracellular domains of notch 1, notch 2, notch 3, and notch 4 (a single-pass transmembrane receptor protein), the notch receptor is activated (Mu et al., 2013). In particular, notch 2 is generally involved in cell development and fate, as well as immune functions (Pan et al., 2013). Sakata-Yanagimoto and Chiba (2012) found that notch 2 was necessary for the development of B cells in the marginal zone of the spleen and regulated the differentiation of dendritic cells (DCs) of the spleen. Additionally, notch 2 regulates intestinal immune cells such as mast cells and DCs, and it plays an important role in the differentiation of helper T cells from CD4 T cells and the activation of cytotoxic T cells. Lewis et al. (2011) also found that DC-specific deletion of the notch 2 receptor resulted in a drop in DC numbers in the spleen, as well as the loss of CD11b + CD103 + DCs in the intestinal lamina propria and a concomitant decrease in IL-17–producing CD4 + T cells. In the present study, the notch 2 gene in the spleen was down-regulated under oxidative stress, showing that oxidative stress damaged the immune function of the spleen. The significant GO enrichment analysis of differentially expressed unigenes (DEGs), including cell differentiation, cell development, negative regulation of cellular processes, cytoskeleton organization, cellular component morphogenesis, cell projection organization, cell morphogenesis and apoptotic processes suggested that the immune cells of the spleen were negatively affected by oxidative stress. Additionally, histological analysis of the spleen showed a reduction in immune cells (T cells and B cells) arising from oxidative stress.
Conclusion
Oxidized fish oil caused oxidative stress in the spleen, which led to spleen tissue damage. More importantly, oxidative stress changed the spleen transcriptome profile, and the transcription profile of the spleen was altered through platelet activation, FOXO and the notch signaling pathway.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA745324.
Ethics Statement
The animal study was reviewed and approved by both animal studies were performed strictly based on the guidelines in the ‘Laboratory Animal Treatment and Usage Guide.’ Animal Ethics Committee of Guangdong Ocean University accepted the protocols (Zhanjiang, China).
Author Contributions
SL and ZL were in charge of breeding experiments, data analysis, and manuscript writing. XY and HL were in charge of breeding experiments. XD and BT were in charge of experimental guidance and manuscript revision. SZ was in charge of the purchase of experimental consumables. SP, TL, XS, and YY were in charge of maintenance of laboratory equipment and guidance of breeding experiments. All authors designated in this manuscript actively participated in this research.
Funding
This study was supported by the National Natural Science Fund (31972808), Guangdong Basic and Applied Basic Research Foundation (2021A1515011165), the Department of Education of Guangdong Province (2021ZDZX4005), the China Agriculture Research System (CARS-47), and Science and Technology Bureau of Zhanjiang (2020A03010 and 2020A05003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Thanks to the staffs of the Donghai Island Biological Research Base of Guangdong Ocean University for maintaining the breeding system. Meanwhile, thanks to the postgraduates of the Animal Nutrition and Feed Laboratory for their assistance in sample collection.
Footnotes
References
Aghili, T., Arshami, J., Tahmasbi, A. M., and Haghparast, A. R. (2014). Effects of Hypericum perforatum extract on IgG titer, leukocytes subset and spleen index in rats. Avicenna J. Phytomed. 4, 413–419. doi: 10.22038/ajp.2014.3166
Akasaki, Y., Alvarez-Garcia, O., Saito, M., Caramés, B., Iwamoto, Y., and Lotz, M. K. (2014). FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthr. Rheumatol. 66, 3349–3358. doi: 10.1002/art.38868
Aranda, P. S., Lajoie, D. M., and Jorcyk, C. L. (2012). Bleach gel: a simple agarose gel for analyzing RNA quality. Electrophoresis 33, 366–369. doi: 10.1002/elps.201100335
Barthel, A., Schmoll, D., and Unterman, T. G. (2005). FoxO proteins in insulin action and metabolism. Trends Endocrinol. Metab. 16, 183–189. doi: 10.1016/j.tem.2005.03.010
Butt, A., Kaleem, A., Iqbal, Z., Walker-Nasir, E., Saleem, M., Shakoori, A. R., et al. (2011). Functional regulation of DNA binding of FOXO1 by post translational modifications: in silico study. Pak. J. Zool. 43, 1167–1175.
Chen, H. Y., Chen, Y. M., Wu, J., Yang, F. C., Lv, Z., Xu, X. F., et al. (2016). Expression of FOXO6 is associated with oxidative stress level and predicts the prognosis in hepatocellular cancer. Medicine 95:e3708. doi: 10.1097/MD.0000000000003708
Chen, X., Fan, X., Tan, J., Shi, P., Wang, X., Wang, J., et al. (2017). Palladin is involved in platelet activation and arterial thrombosis. Thromb. Res. 149, 1–8.
Chen, X., Wang, Q., Guo, Z., Zhao, Y., Gao, Y., Yu, T., et al. (2019). Effects of dietary oxidized fish oil on growth performance and antioxidant defense mechanism of juvenile Rhynchocypris lagowski Dybowski. Aquaculture 512:734368. doi: 10.1016/j.aquaculture.2019.734368
Chen, Y. J., Liu, Y. J., Yang, H. J., Yuan, Y., Liu, F. J., Tian, L. X., et al. (2012). Effect of dietary oxidized fish oil on growth performance, body composition, antioxidant defence mechanism and liver histology of juvenile largemouth bass Micropterus salmoides. Aquac. Nutr. 18, 321–331. doi: 10.1111/j.1365-2095.2011.00900.x
Dalmo, R. A., Ingebrigtsen, K., and Bogwald, J. (1997). Non-specific defence mechanisms in fish, with particular reference to the reticuloendothelial system (RES). J. Fish Dis. 20, 241–273. doi: 10.1046/j.1365-2761.1997.00302.x
Dorsey, B. M., and Jones, M. A. (2017). Healthy Components of Coffee Processing by-Products. Amsterdam: Elsevier Inc.
Ehebauer, M., Hayward, P., and Martinez-Arias, A. (2006). Notch signaling pathway. Sci. STKE 2006:cm7. doi: 10.1126/stke.3642006cm7
Essers, M. A. G., Weijzen, S., De Vries-Smits, A. M. M., Saarloos, I., De Ruiter, N. D., Bos, J. L., et al. (2004). FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 23, 4802–4812. doi: 10.1038/sj.emboj.7600476
Feng, T., Li, S., Wang, S., and Pan, J. (2018). Cross priming amplification with nucleic acid test strip analysis of mutton in meat mixtures. Food Chem. 245, 641–645.
Fu, X., Li, N., Lin, Q., Guo, H., Liu, L., Huang, Z., et al. (2015). Early protein ORF086 is an effective vaccine candidate for infectious spleen and kidney necrosis virus in mandarin fish Siniperca chuatsi. Fish Shellfish Immunol. 46, 200–205. doi: 10.1016/j.fsi.2015.05.052
García-Niño, W. R., Zatarain-Barrón, Z. L., Hernández-Pando, R., Vega-García, C. C., Tapia, E., and Pedraza-Chaverri, J. (2015). Oxidative stress markers and histological analysis in diverse organs from rats treated with a hepatotoxic dose of Cr(VI): effect of curcumin. Biol. Trace Elem. Res. 167, 130–145. doi: 10.1007/s12011-015-0283-x
Ge, G., Long, Y., Shi, L., Ren, J., Yan, J., Li, C., et al. (2020). Transcriptomic profiling revealed key signaling pathways for cold tolerance and acclimation of two carp species. BMC Genomics 21:539. doi: 10.1186/s12864-020-06946-8
Ge, Q., Li, G., Chen, J., Song, J., Cai, G., He, Y., et al. (2021). Immunological role and prognostic value of APBB1IP in pan-cancer analysis. J. Cancer 12, 595–610. doi: 10.7150/jca.50785
Gómez-Crisóstomo, N. P., Rodríguez Martínez, E., and Rivas-Arancibia, S. (2014). Oxidative stress activates the transcription factors FoxO 1a and FoxO 3a in the hippocampus of rats exposed to low doses of ozone. Oxid. Med. Cell. Longev. 2014:805764. doi: 10.1155/2014/805764
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Graf, R., and Schlüns, J. (1979). Ultrastructural and histochemical investigation of the terminal capillaries in the spleen of the carp (Cyprinus carpio L.). Cell Tissue Res. 196, 289–306. doi: 10.1007/BF00240103
Guo, J., Wang, J., and Feng, J. (2019). Aspirin resistance mediated by oxidative stress-induced 8-Isoprostaglandin F2. J. Clin. Pharm. Ther. 44, 823–828. doi: 10.1111/jcpt.12838
Hadwan, M. H. (2018). Simple spectrophotometric assay for measuring catalase activity in biological tissues. BMC Biochem. 19:7. doi: 10.1186/s12858-018-0097-5
Hao, F., and Feng, Y. (2021). Cannabidiol (CBD) enhanced the hippocampal immune response and autophagy of APP/PS1 Alzheimer’s mice uncovered by RNA-seq. Life Sci. 264, 118624. doi: 10.1016/j.lfs.2020.118624
Housley, M. P., Rodgers, J. T., Udeshi, N. D., Kelly, T. J., Shabanowitz, J., Hunt, D. F., et al. (2008). O-GlcNAc regulates FoxO activation in response to glucose. J. Biol. Chem. 283, 16283–16292. doi: 10.1074/jbc.M802240200
Hu, W., Zhang, W., Shah, S. W. A., Ishfaq, M., and Li, J. (2021). Mycoplasma gallisepticum infection triggered histopathological changes, oxidative stress and apoptosis in chicken thymus and spleen. Dev. Comp. Immunol. 114:103832. doi: 10.1016/j.dci.2020.103832
Huang, J., Guo, Y., Hou, Q., Huang, M., and Zhou, X. (2020). Dynamic changes of the bacterial communities in roast chicken stored under normal and modified atmosphere packaging. J. Food Sci. 85, 1231–1239. doi: 10.1111/1750-3841.15038
Jafari, M., and Ansari-Pour, N. (2019). Why, when and how to adjust your P values? Cell J. 20, 604–607. doi: 10.22074/cellj.2019.5992
Jargar, J. G., Hattiwale, S. H., Das, S., Dhundasi, S. A., and Das, K. K. (2012). A modified simple method for determination of serum α-tocopherol (Vitamin E). J. Basic Clin. Physiol. Pharmacol. 23, 45–48. doi: 10.1515/jbcpp-2011-0033
Jarzaa̧b, A., and Stryjecka-Zimmer, M. (2000). Oxidative stress and apoptosis. Ann. Univ. Mar. Curie Sklodowska B 63, 67–71. doi: 10.2478/v10079-008-0010-6
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–D280. doi: 10.1093/nar/gkh063
Krause, A. (2008). Large scale protein sequence clustering - not solved but solvable. Curr. Bioinform. 1, 247–254. doi: 10.2174/157489306777011987
Kuo, M. S., Zilberfarb, V., Gangneux, N., Christeff, N., and Issad, T. (2008). O-GlcNAc modification of FoxO1 increases its transcriptional activity: a role in the glucotoxicity phenomenon? Biochimie 90, 679–685. doi: 10.1016/j.biochi.2008.03.005
Lewis, K. L., Caton, M. L., Bogunovic, M., Greter, M., Grajkowska, L. T., Ng, D., et al. (2011). Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity 35, 780–791. doi: 10.1016/j.immuni.2011.08.013
Lewis, S. M., Williams, A., and Eisenbarth, S. C. (2007). Structure and function of the immune system in vertebrates. Neonatal Immun. 6085, 1–55. doi: 10.1007/978-1-59259-825-0_1
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, X., Wang, Y., Li, H., Jiang, X., Ji, L., Liu, T., et al. (2021). Chemical and quality evaluation of Pacific white shrimp Litopenaeus vannamei: influence of strains on flesh nutrition. Food Sci. Nutr. 9, 5352–5360. doi: 10.1002/fsn3.2457
Lin, J., Ning, J., Lu, X., Chen, M., Cao, W., and Wang, C. (2020). Transcriptomic analysis and expression of C-type lectins in response to Vibrio parahaemolyticus challenge in Scapharca subcrenata. Fish Shellfish Immunol. 106, 365–373. doi: 10.1016/j.fsi.2020.08.016
Liu, J. J., Lu, L. Y., Liu, H. H., Li, Y. S., Su, X., Zhang, B. Z., et al. (2019). De novo assembly of the transcriptome for Greenbug (Schizaphis graminum Rondani) and analysis on insecticide resistance-related genes. Entomol. Res. 49, 363–373. doi: 10.1111/1748-5967.12383
Liu, Y., Mi, Y., Zhang, J., Li, Q., and Chen, L. (2016). Illumina-based transcriptomic profiling of Panax notoginseng in response to arsenic stress. Bot. Stud. 57:13. doi: 10.1186/s40529-016-0128-8
Long, S., Dong, X., Tan, B., Zhang, S., Chi, S., Yang, Q., et al. (2022). The antioxidant ability, histology, proximate, amino acid and fatty acid compositions, and transcriptome analysis of muscle in juvenile hybrid grouper (♀ Epinephelus fuscoguttatus ×♂ Epinephelus lanceolatus) fed with oxidized fish oil. Aquaculture 547:737510. doi: 10.1016/j.aquaculture.2021.737510
Long, S., Dong, X., Tan, B., Zhang, S., Xie, S., Yang, Q., et al. (2021). Growth performance, antioxidant ability, biochemical index in serum, liver histology and hepatic metabolomics analysis of juvenile hybrid grouper (♀ Epinephelus fuscoguttatus ×♂ Epinephelus lanceolatus) fed with oxidized fish oil. Aquaculture 545:737261.
Manasa, K., and Vani, R. (2016). Influence of oxidative stress on stored platelets. Adv. Hematol. 2016:4091461. doi: 10.1155/2016/4091461
Mandil, R., Prakash, A., Rahal, A., Singh, S. P., Sharma, D., Kumar, R., et al. (2020). In vitro and in vivo effects of flubendiamide and copper on cyto-genotoxicity, oxidative stress and spleen histology of rats and its modulation by resveratrol, catechin, curcumin and α-tocopherol. BMC Pharmacol. Toxicol. 21:29. doi: 10.1186/s40360-020-00405-6
Meng, R., Zhang, J. Y., Zhang, D. G., Nie, Z. L., and Meng, Y. (2020). Data of de novo assembly and annotation of transcriptome from Aspidistra fenghuangensis (Asparagaceae: Nolinoideae). Data Br. 31, 105738. doi: 10.1016/j.dib.2020.105738
Michael, B., Yano, B., Sellers, R. S., Perry, R., Morton, D., Roome, N., et al. (2007). Evaluation of organ weights for rodent and non-rodent toxicity studies: a review of regulatory guidelines and a survey of current practices. Toxicol. Pathol. 35, 742–750.
Mu, X., Isaac, C., Greco, N., Huard, J., and Weiss, K. (2013). Notch signaling is associated with ALDH activity and an aggressive metastatic phenotype in murine osteosarcoma cells. Front. Oncol. 3:143. doi: 10.3389/fonc.2013.00143
Nagy, T. R., and Clair, A. L. (2000). Precision and accuracy of dual-energy X-ray absorptiometry for determining in vivo body composition of mice. Obes. Res. 8, 392–398.
Nandi, A., Yan, L. J., Jana, C. K., and Das, N. (2019). Role of catalase in oxidative stress- and age-associated degenerative diseases. Oxid. Med. Cell. Longev. 2019:9613090. doi: 10.1155/2019/9613090
Pan, W. C., Kile, M. L., Seow, W. J., Lin, X., Quamruzzaman, Q., Rahman, M., et al. (2013). Genetic susceptible locus in NOTCH2 interacts with arsenic in drinking water on risk of type 2 diabetes. PLoS One 8:e0070792. doi: 10.1371/journal.pone.0070792
Peskin, A. V., and Winterbourn, C. C. (2017). Assay of superoxide dismutase activity in a plate assay using WST-1. Free Radic. Biol. Med. 103, 188–191.
Piao, Y., Liu, Y., and Xie, X. (2013). Change trends of organ weight background data in Sprague Dawley rats at different ages. J. Toxicol. Pathol. 26, 29–34. doi: 10.1293/tox.26.29
Pinto, M., Benfeito, S., Fernandes, C., and Borges, F. (2020). Antioxidant Therapy, Oxidative Stress, and Blood-Brain Barrier: The Road of Dietary Antioxidants. Amsterdam: Elsevier Inc.
Purohit, A. S., Reed, C., and Mohan, A. (2016). Development and evaluation of quail breakfast sausage. LWT Food Sci. Technol. 69, 447–453. doi: 10.1016/j.lwt.2016.01.058
Sakata-Yanagimoto, M., and Chiba, S. (2012). “Notch2 and immune function,” in Notch Regulation of the Immune System. Current Topics in Microbiology and Immunology, eds F. Radtke (Berlin, Heidelberg: Springer), 360. Available online at: https://doi.org/10.1007/82_2012_235
Savassi, L. A., Paschoalini, A. L., Arantes, F. P., Rizzo, E., and Bazzoli, N. (2020). Heavy metal contamination in a highly consumed Brazilian fish: immunohistochemical and histopathological assessments. Environ. Monit. Assess. 192:542. doi: 10.1007/s10661-020-08515-8
Shi, L. G., Yang, R. J., Yue, W. B., Xun, W. J., Zhang, C. X., Ren, Y. S., et al. (2010). Effect of elemental nano-selenium on semen quality, glutathione peroxidase activity, and testis ultrastructure in male Boer goats. Anim. Reprod. Sci. 118, 248–254.
Simms, D., Cizdziel, P., and Chomczynski, P. (1993). TRIzol: a new reagent for optimal single-step isolation of RNA. Focus 3, 99–102.
Singh, R., Singh, A., and Sachan, S. (2018). Enzymes Used in the Food Industry: Friends or Foes?. Amsterdam: Elsevier Inc.
Smith, G. R., and Shanley, D. P. (2013). Computational modelling of the regulation of Insulin signalling by oxidative stress. BMC Syst. Biol. 7:74. doi: 10.1186/1752-0509-7-41
Song, C., Liu, B., Xu, P., Ge, X., and Zhang, H. (2019). Emodin ameliorates metabolic and antioxidant capacity inhibited by dietary oxidized fish oil through PPARs and Nrf2-Keap1 signaling in Wuchang bream (Megalobrama amblycephala). Fish Shellfish Immunol. 94, 842–851. doi: 10.1016/j.fsi.2019.10.001
Song, C., Liu, B., Xu, P., Xie, J., Ge, X., Zhou, Q., et al. (2018). Oxidized fish oil injury stress in Megalobrama amblycephala: evaluated by growth, intestinal physiology, and transcriptome-based PI3K-Akt/NF-κB/TCR inflammatory signaling. Fish Shellfish Immunol. 81, 446–455. doi: 10.1016/j.fsi.2018.07.049
Song, L., Dong, X., and Hu, G. (2021). Transcriptome analysis of red sea bream (Pagrus major) head kidney and spleen infected by Vibrio anguillarum. Aquac. Rep. 21:100789. doi: 10.1016/jaqrep.2021.100789
Song, S. G., Chi, S. Y., Tan, B. P., Liang, G. L., Lu, B. Q., Dong, X. H., et al. (2018). Effects of fishmeal replacement by Tenebrio molitor meal on growth performance, antioxidant enzyme activities and disease resistance of the juvenile pearl gentian grouper (Epinephelus lanceolatus ♂× Epinephelus fuscoguttatus ♀). Aquac. Res. 49, 2210–2217. doi: 10.1111/are.13677
Tosun, M., Yağcı, R., and Erdurmuş, M. (2019). “Glaucoma and antioxidant status,” in Handbook of Nutrition, Diet and the Eye, eds V. R. Preedy and R. R. Watson (Cambridge, MA: Academic Press), 203–219.
Van Der Heide, L. P., and Smidt, M. P. (2005). Regulation of FoxO activity by CBP/p300-mediated acetylation. Trends Biochem. Sci. 30, 81–86. doi: 10.1016/j.tibs.2004.12.002
Wang, D., Sun, S., Li, S., Lu, T., and Shi, D. (2021). Transcriptome profiling of immune response to Yersinia ruckeri in spleen of rainbow trout (Oncorhynchus mykiss). BMC Genomics 22:292. doi: 10.1186/s12864-021-07611-4
Wang, F., Nguyen, M., Qin, F. X. F., and Tong, Q. (2007). SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell 6, 505–514.
Xia, X. H., Liu, G. P., Wu, X. L., Cui, S. S., Yang, C. H., Du, Q. Y., et al. (2021). Effects of Macleaya cordata extract on TLR20 and the proinflammatory cytokines in acute spleen injury of loach (Misgurnus anguillicaudatus) against Aeromonas hydrophila infection. Aquaculture 544:737105. doi: 10.1016/j.aquaculture.2021.737105
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, 316–322. doi: 10.1093/nar/gkr483
Xie, Q., Chen, J., and Yuan, Z. (2012). Post-translational regulation of FOXO. Acta Biochim. Biophys. Sin. 44, 897–901. doi: 10.1093/abbs/gms067
Xing, H., Chen, J., Peng, M., Wang, Z., Liu, F., Li, S., et al. (2019). Identification of signal pathways for immunotoxicity in the spleen of common carp exposed to chlorpyrifos. Ecotoxicol. Environ. Saf. 182:109464. doi: 10.1016/j.ecoenv.2019.109464
Yang, K., Xie, K., Zhu, Y. X., Huo, S. M., Hoffmann, A., and Hong, X. Y. (2020). Wolbachia dominate Spiroplasma in the co-infected spider mite Tetranychus truncatus. Insect Mol. Biol. 29, 19–37. doi: 10.1111/imb.12607
Yang, S. P., Liu, H. L., Wang, C. G., Yang, P., Sun, C. B., and Chan, S. M. (2015). Effect of oxidized fish oil on growth performance and oxidative stress of Litopenaeus vannamei. Aquac. Nutr. 21, 121–127. doi: 10.1111/anu.12143
Yu, H., Ren, Y., Wei, H., Xing, W., Xu, G., Li, T., et al. (2021). Dietary oxidized fish oil negatively affected the feed utilization, health status and fillet quality of juvenile Amur sturgeon, A. schrenckii. Aquaculture 546:737290. doi: 10.1016/j.aquaculture.2021.737290
Zhang, D. G., Zhao, T., Hogstrand, C., Ye, H. M., Xu, X. J., and Luo, Z. (2021). Oxidized fish oils increased lipid deposition via oxidative stress-mediated mitochondrial dysfunction and the CREB1-Bcl2-Beclin1 pathway in the liver tissues and hepatocytes of yellow catfish. Food Chem. 360:129814. doi: 10.1016/j.foodchem.2021.129814
Zhang, N., Zhang, H. J., Zhao, B., Sun, Q. Q., Cao, Y. Y., Li, R., et al. (2014). The RNA-seq approach to discriminate gene expression profiles in response to melatonin on cucumber lateral root formation. J. Pineal Res. 56, 39–50. doi: 10.1111/jpi.12095
Zhang, T. N., Yang, N., Goodwin, J. E., Mahrer, K., Li, D., Xia, J., et al. (2019). Characterization of circular RNA and microRNA profiles in septic myocardial depression: a lipopolysaccharide-induced rat septic shock model. Inflammation 42, 1990–2002. doi: 10.1007/s10753-019-01060-8
Zhang, Y., Li, Z., Kholodkevich, S., Sharov, A., Feng, Y., Ren, N., et al. (2019). Cadmium-induced oxidative stress, histopathology, and transcriptome changes in the hepatopancreas of freshwater crayfish (Procambarus clarkii). Sci. Total Environ. 666, 944–955.
Zhao, H., Wang, Y., Guo, M., Mu, M., Yu, H., and Xing, M. (2020). Grass carps co-exposed to environmentally relevant concentrations of cypermethrin and sulfamethoxazole bear immunodeficiency and are vulnerable to subsequent Aeromonas hydrophila infection. Environ. Pollut. 266:115156. doi: 10.1016/j.envpol.2020.115156
Zhao, X. H., Yang, Z. Q., Bao, L. B., Wang, C. Y., Zhou, S., Gong, J. M., et al. (2015). Daidzein enhances intramuscular fat deposition and improves meat quality in finishing steers. Exp. Biol. Med. 240, 1152–1157. doi: 10.1177/1535370214564755
Zhao, Z., Zhang, C., Lin, Q., Li, N. Q., Huang, Z. B., Zhao, M., et al. (2020). Single-walled carbon nanotubes as delivery vehicles enhance the immunoprotective effect of an immersion DNA vaccine against infectious spleen and kidney necrosis virus in mandarin fish. Fish Shellfish Immunol. 97, 432–439. doi: 10.1016/j.fsi.2019.12.072
Keywords: hybrid grouper (♀ Epinephelus fuscoguttatus × ♀E. lanceolatus), oxidized fish oil, spleen, oxidative stress, antioxidant ability, transcriptome
Citation: Long S, Li Z, Dong X, Yan X, Liu H, Tan B, Zhang S, Pan S, Li T, Suo X and Yang Y (2021) The Effect of Oxidized Fish Oil on the Spleen Index, Antioxidant Activity, Histology and Transcriptome in Juvenile Hybrid Grouper (♀ Epinephelus fuscoguttatus × ♂ Epinephelus lanceolatus). Front. Mar. Sci. 8:779305. doi: 10.3389/fmars.2021.779305
Received: 18 September 2021; Accepted: 01 November 2021;
Published: 23 November 2021.
Edited by:
Erchao Li, Hainan University, ChinaCopyright © 2021 Long, Li, Dong, Yan, Liu, Tan, Zhang, Pan, Li, Suo and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohui Dong, ZG9uZ3hpYW9odWkyMDAzQDE2My5jb20=
†These authors have contributed equally to this work
 Shuisheng Long
Shuisheng Long Zhihao Li1,2†
Zhihao Li1,2† Xiaohui Dong
Xiaohui Dong Hao Liu
Hao Liu Beiping Tan
Beiping Tan Shuang Zhang
Shuang Zhang