
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 11 November 2021
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.774925
This article is part of the Research TopicMollusk Breeding and Genetic ImprovementView all 25 articles
Giant clams are found in a mutualistic association with Symbiodiniaceae dinoflagellates, however, the diversity of the giant clams, as well as the diversity and distribution of Symbiodiniaceae in different Tridacnine species remain relatively poorly studied in the South China Sea. In this study, a total of 100 giant clams belonging to small type’s giant clams, Tridacna maxima, T. crocea, and T. noae, were collected from Hainan and Xisha Islands. Based on mtDNA cytochrome c oxidase subunit 1 gene (COI) and 16S rRNA fragments, T. maxima and T. crocea showed a closer phylogenetic relationship than T. noae. All the three species of giant clams hosted Symbiodiniaceae including genera Symbiodinium (formerly Clade A) and Cladocopium (formerly Clade C). Geographically, symbionts in Cladocopium are restricted to Xisha Islands, probably because Cladocopium prefers to inhabit in waters with higher mean temperatures. The endosymbiont specificity among the three giant clam species was also detected. T. noae and T. crocea are found to harbor Symbiodinium preferentially, compared with Cladocopium. These results could provide important information to understand various endosymbionts occurring in giant clams in the South China Sea.
Giant clams (Tridacninae) inhabit tropical coral reefs throughout the Indo-Pacific oceans (Lucas, 2014). In the world, the Tridacninae subfamily includes two extant genera, Hippopus (two species) and Tridacna (10 extant species), and all these species host symbiotic zooxanthellae in the mantle tissues that plays important ecological roles in the coral reef ecosystem (Liu et al., 2021b). Recently, the natural resources of giant clams sharply decreased in many countries, including Australia, Indonesia, Singapore, Philippines, and Japan because of overfishing, habitat destruction, and global climate change (Copland and Lucas, 1988; Pringgenies et al., 1995; Neo and Todd, 2012; Neo et al., 2019). Therefore, all the giant clam species are listed in Appendix II of the Convention on International Trade in Endangered Species [United Nations Environment Programme-World Conservation Monitoring Center (UNEP-WCMC), 2007] and International Union for Conservation of Nature (IUCN) Red List of Threatened Species (Wells, 1997).
In China, giant clams are distributed mainly in the South China Sea, which can be divided into five geographical populations, Hainan Islands, Xisha Islands, Zhongsha Islands, Dongsha Islands, and Nansha Islands (Zhang et al., 2020). Eight giant clams have been reported in the South China Sea, including two large species Tridacna gigas and T. derasa; three middle species T. squamosa, H. hippopus, and H. porcellanus; and three small species T. maxima, T. crocea, and T. noae (Neo et al., 2017; Liu J. et al., 2020). Recent surveys found that large and middle type’s giant clams were in extremely low numbers, and small type’s giant clams became the dominant giant clam species (Liu et al., 2021a). However, the genetic and phenotypic variation in several giant clam species has been reported in many geographical locations (Nuryanto and Kochzius, 2009; Hui et al., 2016; Pappas et al., 2017), while there were only a few reports on the diversity of giant clams in the South China Sea (Lim et al., 2019; Liu J. et al., 2020).
Symbiodinium is found in endosymbiosis with marine invertebrates such as giant clams, corals, Porifera, and Foraminifera, which could provide their hosts with up to 100% of energy requirements (Stat et al., 2006; Venn et al., 2008; Reich et al., 2017). Since pediveliger larva stage, the symbiont between the giant clam and Symbiodinium is established (Liu C. et al., 2020; Zhang et al., 2020; Wang et al., 2021). The diversity and community structure of Symbiodiniaceae in the giant clam could hence likely affect the growth, reproduction, and photosynthetic efficiency of the host (DeBoer et al., 2012). According to the previous molecular taxonomic research, nine distinctive Symbiodinium clades (A to I) have been identified (Pochon and Gates, 2010). Each major clade could be further resolved into diverse genetic subclades through the use of highly variable DNA markers such as the nuclear ribosomal internal transcribed spacer (ITS) (Arif et al., 2014; Ikeda et al., 2017). Previous studies have shown that the diversity of endosymbiontic Symbiodiniaceae in host is influenced by numerous factors, such as host species, physiology of the hosts, and environmental factors (LaJeunesse et al., 2010, 2018; Hume et al., 2016; Pappas et al., 2017; Lim et al., 2019).
The aim of this study is to illustrate the diversity of three small type’s giant clams (T. maxima, T. crocea, and T. noae) using the mtDNA cytochrome c oxidase subunit 1 gene (CO1) and 16S rRNA gene in Hainan and Xisha Islands. Furthermore, the diversity of their endosymbiontic Symbiodiniaceae is also identified using DNA barcoding based on ITS1 region of rDNA.
All giant clams and experimental protocols used in this study were reviewed and approved by the committee and laboratory animal department of the Hainan University.
The small type’s giant clam samples (T. maxima, T. crocea, and T. noae) were collected from the two different sites, the lagoon of Zhaoshu, Xisha (112° 12′–112° 19′ E, 16° 57′–16° 59′ N) and the coastal waters of Sanya, Hainan (including Dongmao-Ximao Island, Luhuitou Peninsula, Yalong Bay and Wuzhizhou Island, 109° 21′–109°46′ E, 18° 11′–18°19′ N) in the South China Sea in June and July, and coded as Xisha01-Xisha77 and Sanya01-Sanya23, respectively. Briefly, about 1 cm2 mantle tissue was removed from each individual using sterile disposable razors and stored in 100% ethanol at 4°C.
A subsample of mantle tissue of size about 2 mm2 was used for the DNA extraction that was conducted using Tiangen DNA kit (Tiangen Biotech, Beijing, China) according to the instructions of the manufacturer.
The COI, 16S, and ITS1 fragments of the giant clams and Symbiodiniaceae, respectively, were amplified by PCR, which were carried out in a total volume of 20 μl with 10 μl of 2 × Rapid Taq Master Mix (with Taq DNA polymerase, dNTP mix, MgCl2, and PCR buffer), 0.6 μl each of forward and reverse primers (10 μM), 1 μl of DNA diluted 1:5, and 7.8 μl of water. The primers were shown in Table 1. PCR conditions were set as follows: an initial denaturing step at 94°C for 5 min; 40 cycles of denaturing at 94°C for 30 s, annealing at 43–53°C for 30 s, and extension at 72°C for 90 s; and a final extension step at 72°C for 10 min. PCR products were sequenced at BGI (China).
The three fragments (COI, 16S, and ITS1) were aligned separately using Clustal W in MEGA X (Kumar et al., 2018). Sequences were converted into NEXUS format for phylogenetic analysis using DAMBE5 (Xia, 2013). Phylogenetic trees were reconstructed using Bayesian inference (BI; Huelsenbeck and Ronquist, 2001) in the CIPRES gateway (Miller et al., 2010). BI analyses were performed with MrBayes v.3.2.7 (Ronquist and Huelsenbeck, 2003), running four simultaneous Monte Carlo Markov chains (MCMC) for 10,000,000 generations, sampling every 1,000 generations and discarding the first 25% generations as burn-in. Two independent runs were performed to increase the chance of adequate mixing of the Markov chains and to increase the chance of detecting failure to converge, as determined by using Tracer v1.6. The effective sample size (ESS) of all the parameters was higher than 200. The resulting phylogenetic trees were visualized in FigTree v1.4.4.
Population genetic analyses were conducted based on the COI and 16S fragments of three giant clams. Genetic diversity indices, including haplotype diversity (Hd) and nucleotide diversity (π), were calculated in DNASP 5 (Librado and Rozas, 2009). To investigate the genetic relationships among haplotypes, a median-joining network was generated with PopART 1.7 (Clement et al., 2000). To infer if the three giant clams have experienced population expansion, Tajima’s D (Tajima, 1989) and Fu’s Fs statistics (Fu, 1997) of COI sequences were calculated in Arlequin 3.5 (Excoffier and Lischer, 2010). The significance levels were evaluated under 10,000 permutations.
A total of 88 COI and 89 16S sequences were obtained and submitted to Genbank (Supplementary Table 1). The phylogenetic relationships of three giant clams were reconstructed based on the aligned nucleotide sequences of COI (681 bp) and 16S (518 bp) fragments, respectively. Both BI analyses arrived at the identical topologies (Figures 1, 2).
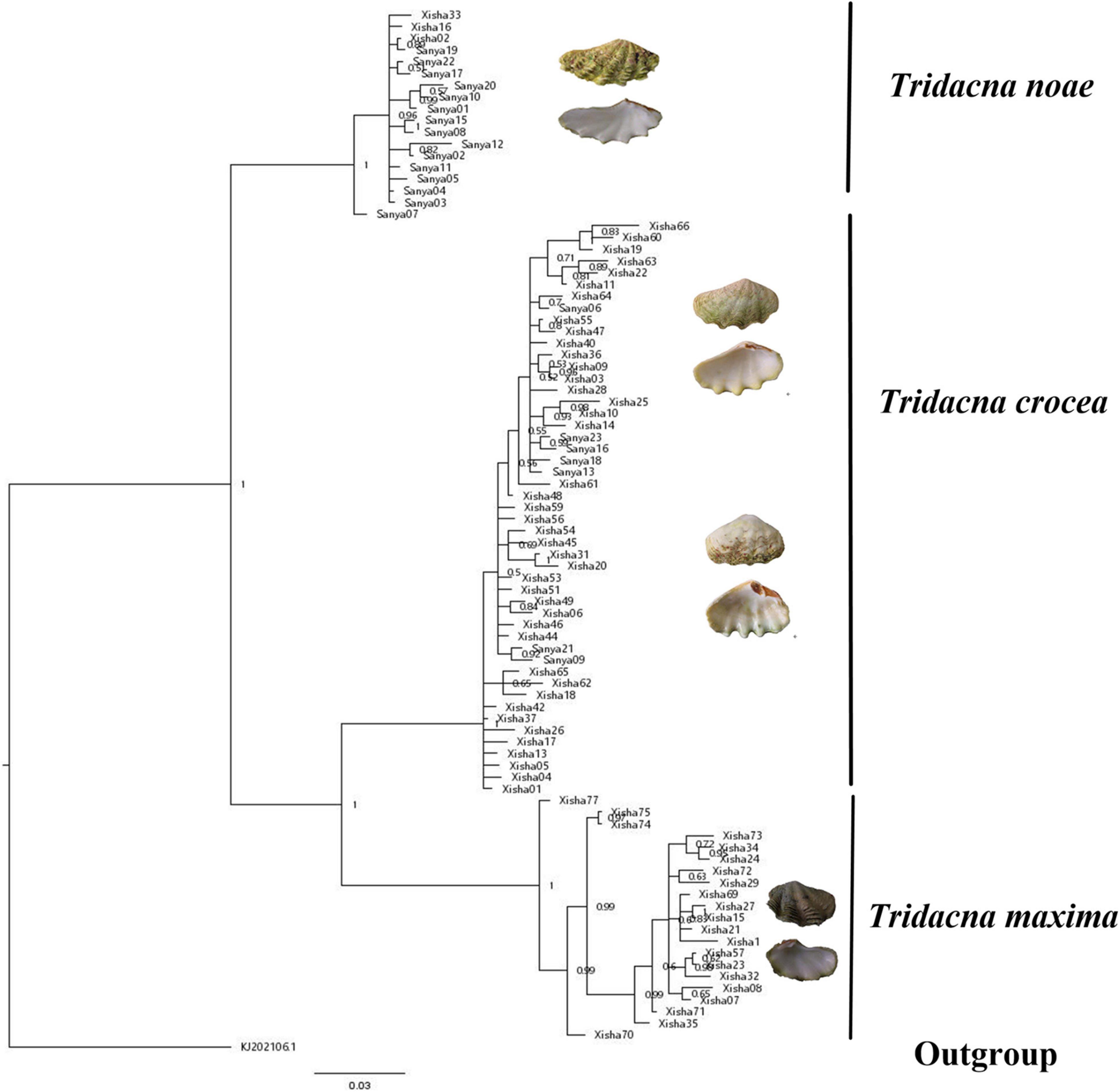
Figure 1. Binary indexed tree of the three giant clams based on the COI fragment. Hippopus hippopus (KJ202106) was used as outgroup. Numbers at nodes are posterior probabilities.
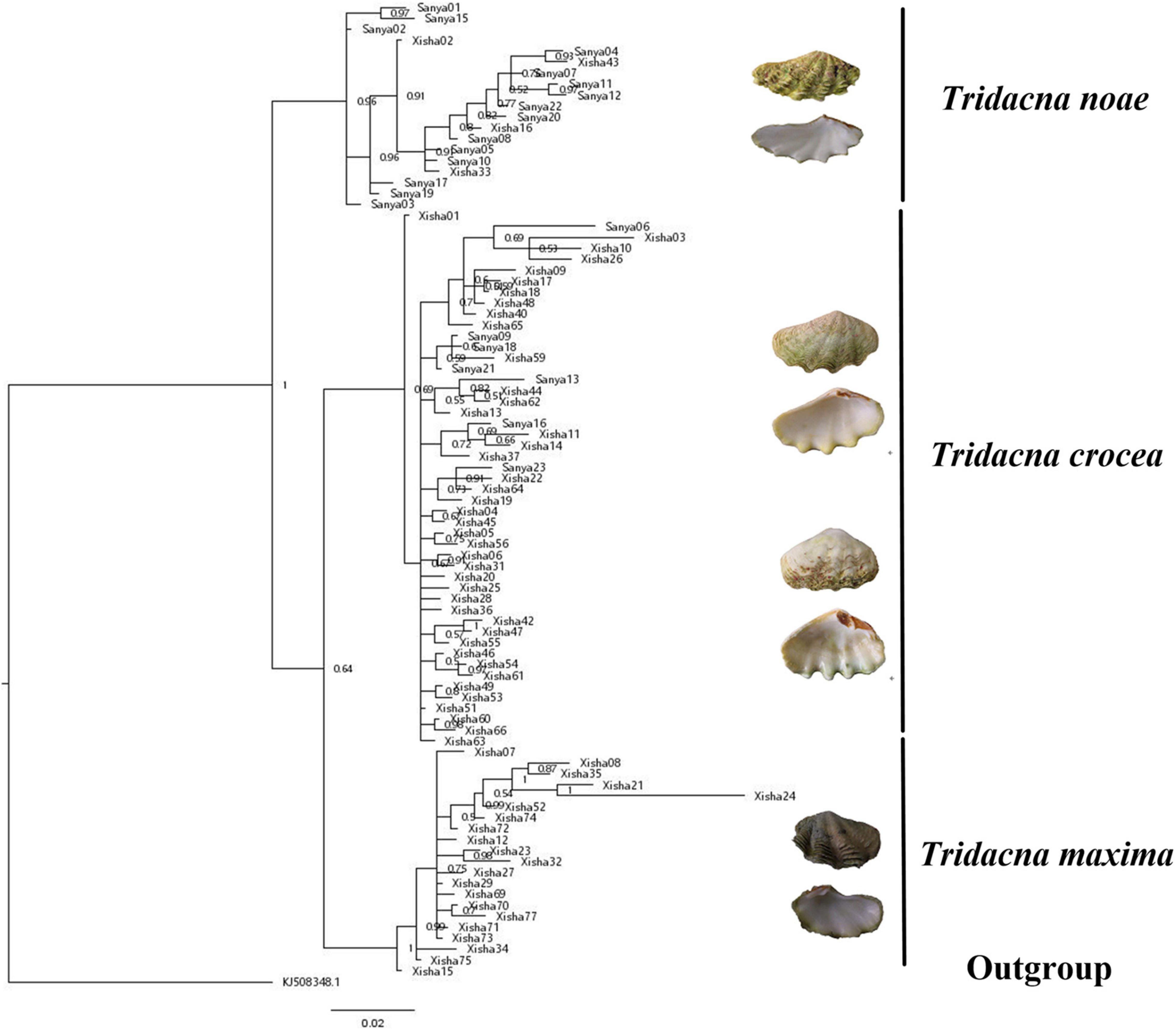
Figure 2. BI tree of three giant clams based on 16S fragment. Hippopus hippopus (KJ508348) was used as outgroup. Numbers at nodes are posterior probabilities.
According to the reconstructed phylogeny, T. maxima was recovered closer to T. crocea than to T. noae. Although the sister relationship between T. maxima and T. crocea was not highly supported in the 16S analysis (Figure 2), it arrived at a maximum support value in the COI phylogeny (Figure 1). Both the COI and 16S sequences supported the monophyly of T. crocea, however, high-intraspecific morphological diversity was found within this group, such as the values of shell width/shell length, byssal opening length/shell length, byssal opening width/shell length and number of hinge tooth (Supplementary Table 2).
The Hd and nucleotide diversity (π) across two sampling sites were presented in Table 2. The genetic diversity indices derived from COI were higher than those from 16S sequences. Compared with T. noae, T. crocea, and T. maxima showed relatively higher COI Hd and nucleotide diversity. Estimates of neutral tests for T. crocea and T. maxima of Xisha and T. noae of Sanya indicated population expansion by significant negative values of Fu’s Fs and Tajima’s D statistics (Table 2). The median-joining network analysis based on COI and 16S sequences revealed a star-like haplotype network (Figure 3).
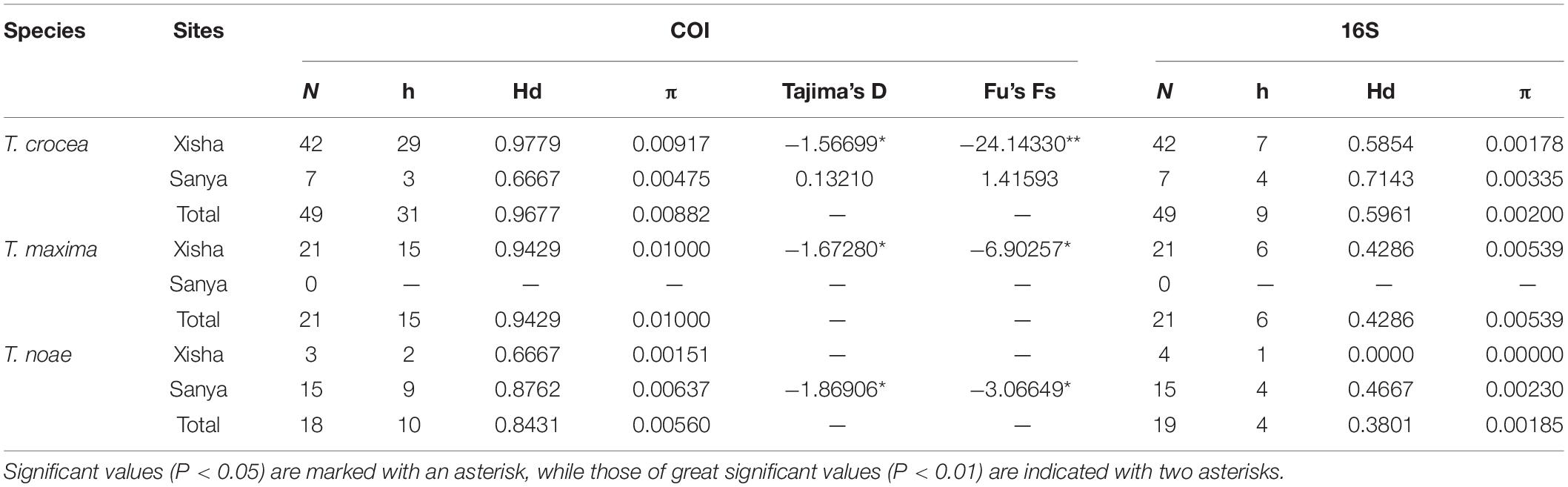
Table 2. Genetic diversity of the COI and 16S among different species and sites, and COI neutrality tests.
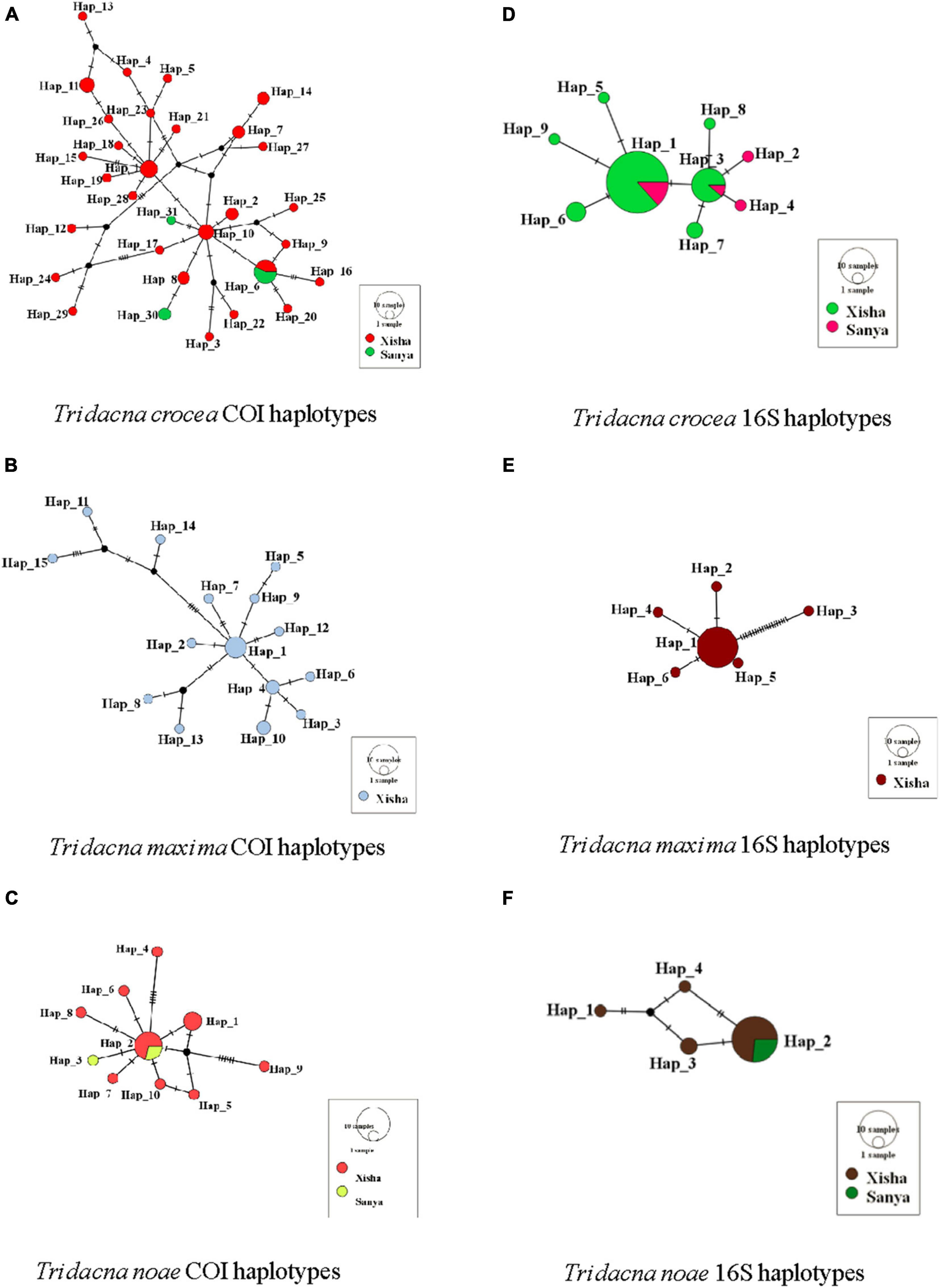
Figure 3. Network of three giant clams for COI and 16S data. (A) Tridacna crocea COI haplotypes; (B) Tridacna maxima COI haplotypes; (C) Tridacna noae COI haplotypes; (D) Tridacna crocea 16S haplotypes; (E) Tridacna maxima 16S haplotypes; (F) Tridacna noae 16S haplotypes. The color of the circle indicates the geographic region, and the size of the circle indicates the haplotype frequency.
A total of 59 ITS1 sequences were amplified and submitted to GenBank (Supplementary Table 1). The final 814 bp length alignment was used for phylogenetic reconstruction by BI inference (Figure 4). The phylogenetic tree was successfully separated into two clades (namely, Clade I and Clade II; Figure 4A) that were identified by blasting in NCBI. The results (not shown here) indicated that they corresponded to Symbiodinium and Cladocopium, respectively. Since considerable sequence divergence could impede accurate alignments between distantly related lineages (LaJeunesse, 2001), two phylogenetic analyses of Clade I and Clade II were further conducted, respectively. Clade I was not separated while Clade II could be divided into two subclades when they were considered individually (Figures 4B,C).
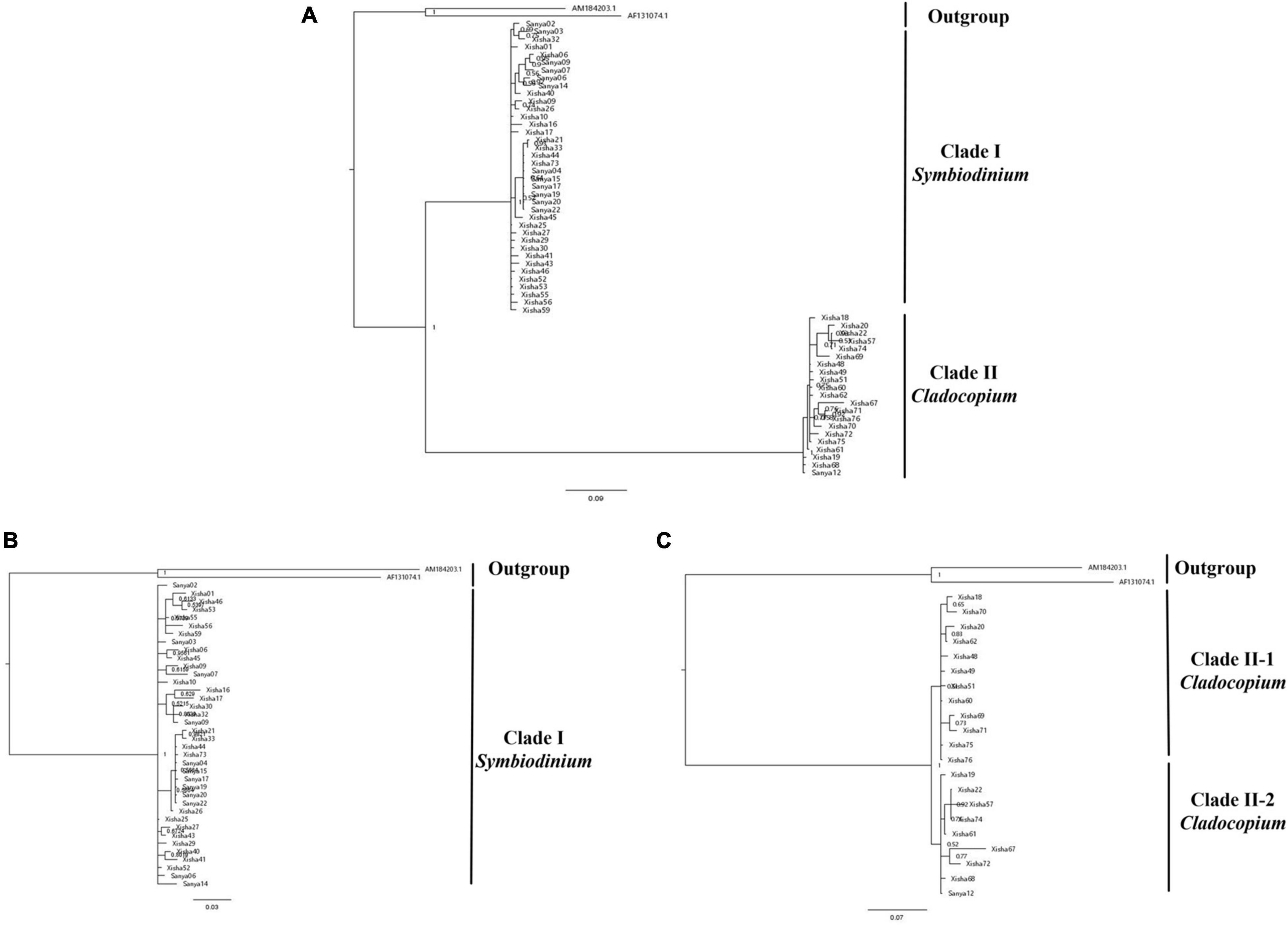
Figure 4. BI tree of zooxanthellae based on ITS1 fragment. Numbers at nodes are posterior probabilities. (A) Phylogenetic tree of all the zooxanthellae individuals using Gymnodinium (AM184203) and Gyrodinium (AF131074) as outgroup and (B) Phylogenetic tree of Clade I, and (C) Phylogenetic tree of Clade II, derived from the phylogeny of all zooxanthellae individuals, using the same outgroup.
The abundance of endosymbiont genus distribution, and their association to the three giant clam host species are shown in Table 3.
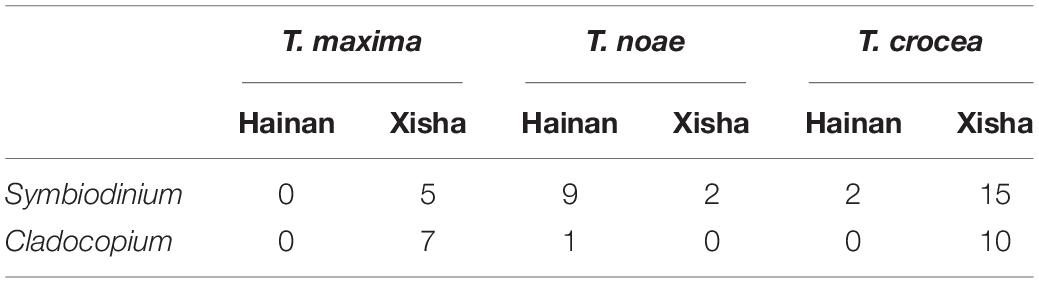
Table 3. Abundance of Tridacna clams with respective endosymbiont genus according to host species and sampling sites.
The COI and 16S sequences generated identical topologies, supporting the separation of three major clades corresponding to three individual species T. crocea, T. maxima, and T. noae (Figures 1, 2). The latter two species are similar in general appearance, and they were even considered the same species (Rosewater, 1965). However, previous studies found that they could be distinguished by several morphological and ecological characteristics. For example, T. maxima was mainly distributed at the edges and crests of reefs, while T. noae was often found in the reef flat (Kubo and Iwai, 2007). In addition, T. noae has well-spaced rib scales, particularly on the upper third of the shell, while the ribs in T. maxima were closely crowded together (Su et al., 2014). Our phylogeny derived from COI and 16S fragments indicated a distant relationship between T. noae and T. maxima, shown as T. noae + (T. maxima and T. crocea), and therefore supported the validity of T. noae (Su et al., 2014). Although T. maxima has a short planktonic larvae duration (9 days; Lucas, 1988), it is widely distributed from the Red Sea to the central Pacific (Nuryanto and Kochzius, 2009). In our study, however, T. maxima was only found in Xisha, and this may probably be due to the low sample size of Sanya. High intraspecific morphological diversity was found within T. crocea, even though both the COI and 16S sequences supported the genetic monophyly of this species. Special attention should be paid during T. crocea identification since it shows high intraspecific diversity. Geographically, the sampling locations of T. crocea (Sanya and Xisha) in our study were restricted to the South China Sea, and corresponded to the western Pacific OTU (Liu J. et al., 2020).
A total of two clades (namely, Clade I and Clade II) were separated based on the ITS1 marker (Figure 4A), and they corresponded to Symbiodinium (formerly Clade A) and Cladocopium (formerly clade C; LaJeunesse et al., 2018), respectively. Until now, giant clam species have been reported to be associated with 30 Symbiodiniaceae phylotypes, all belonging to genera Symbiodinium, Cladocopium, and Durusdinium (not detected here) (Mies, 2019). In this study, Clade II could be further divided into two closely related lineages (Figure 4B), indicating the diversity of Cladocopium. Previous studies have also revealed that the diversity of Cladocopium was higher than Symbiodinium or Durusdinium in Dongsha Atoll (Lim et al., 2019), and this could be explained as the abundance of Cladocopium that is also species-rich among hosts in the South China Sea (Wong et al., 2016). Symbionts in Symbiodinium are widely distributed in the world (Baker, 2003; Tonk et al., 2013), and most members have been accustomed even to extreme conditions (Venn et al., 2008). For example, only Symbiodinium symbionts were found in the giant clams (Tridacna spp.; Pappas et al., 2017) in the Red Sea, which is characteristic of high heat and irradiance conditions. This could also be supported in our study, since the symbionts in Symbiodinium were found in both Xisha and Sanya, whereas most of the symbionts in Cladocopium were restricted to Xisha (Figure 4A), indicating that the former could be better adapted to the various conditions. This result may also be explained as that the clams with Cladocopium (formerly clade C) and Durusdinium (formerly clade D) were usually located in areas with higher mean temperatures as previous studies have revealed (DeBoer et al., 2012) since the climate of Xisha is warmer than Sanya at the same time.
This study reveals the endosymbiont specificity among the three giant clam species. At first, T. maxima associates slightly with Cladocopium (n = 7) over Symbiodinium (n = 5), but sample sizes are low. In the previous studies, both T. maxima host associations with Cladocopium and Symbiodinium endosymbionts have been reported. For example, in Dongsha (Lim et al., 2019), T. maxima associated preferentially with Cladocopium. In the Red Sea (Rossbach et al., 2021), however, strong T. maxima host associations with Symbiodinium endosymbionts were suggested. In French Polynesia, both Symbiodinium and Cladocopium were the dominant genera in T. maxima (Pochon et al., 2019). In order to better clarify T. maxima host associations in the South China Sea, future studies with broad sampling sites around this area are still needed. Different from T. maxima, the giant clams T. noae and T. crocea were found to associate with Symbiodinium over Cladocopium, and this host-endosymbiont specificity appears more obvious on T. noae. This result is supported by the study of Ikeda et al. (2017) showing that T. crocea had an apparent dominance of Symbiodinium (Clade A), although it is inconsistent with some previous studies which revealed T. noae and T. crocea host associations with Durusdinium (Lim et al., 2019) and Cladocopium (DeBoer et al., 2012), respectively. Within the stony coral Stylophora pistillata, Symbiodinium (Clade A) have been observed to be more related to individuals that inhabited shallow waters, whereas individuals in deeper waters were more associated with Cladocopium (Clade C) (Winters et al., 2009). In addition, Symbiodinium (Clade A) was found in other coral species inhabiting shallow waters (Rowan et al., 1997), and therefore, it is inferred that Symbiodinium (Clade A) tends to be more insensitive to the high irradiance and high temperature stresses of shallow waters (Ikeda et al., 2017). The habits of Symbiodinium (Clade A) may attribute to T. noae and T. crocea that inhabit shallow waters, associating preferentially with Symbiodinium.
In this study, the reconstructed phylogeny using the COI and 16S fragments indicated a relationship shown as T. noae + (T. maxima and T. crocea) and supported the validity of T. noae. A total of two clades of Symbiodiniaceae symbionts that corresponded to Symbiodinium (formerly Clade A) and Cladocopium (formerly clade C) were separated based on the ITS1 marker. The wide distribution of Symbiodinium may indicate that it could be better adapted to various conditions, while the restriction of Cladocopium may be explained as a temperature preference. This study also reveals the endosymbiont specificity that is related to the shared living habits between giant clams and dinoflagellates.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by The Committee and Laboratory Animal Department of Hainan University.
QC was involved with the data curation, investigation, and writing the original draft. ZG worked with the software and supervision. AW was involved in the supervision and funding acquisition. CL was helped in the conceptualization, writing review and editing, supervision, and funding the acquisition. YY was helped in the data curation, formal analysis, methodology, and writing the original draft. All authors contributed to the article and approved the submitted version.
This research was supported financially by the National Key Research and Development Program of China (2019YFD0901301), the Key Research and Development Project of Hainan Province (ZDYF2019153), and the Starting Research Fund from the Hainan University (RZ2100003081).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.774925/full#supplementary-material
Arif, C., Daniels, C., Bayer, T., Banguera-Hinestroza, E., Barbrook, A., Howe, C. J., et al. (2014). Assessing Symbiodinium diversity in scleractinian corals via nextgeneration sequencing-based genotyping of the ITS2 rDNA region. Mol. Ecol. 23, 4418–4433. doi: 10.1111/mec.12869
Baker, A. C. (2003). Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 34, 661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417
Clement, M., Posada, D., and Crandall, K. A. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x
Copland, J. W. E., and Lucas, J. S. E. (1988). Canberra Australia Australian Centre for International Agricultural Research. Giant clams in Asia and the pacific. Monographs 9:274.
DeBoer, T. S., Baker, A. C., Erdmann, M. V., Jones, P. R., and Barber, P. H. (2012). Patterns of Symbiodinium distribution in three giant clam species across the biodiverse Bird’s Head region of Indonesia. Mar. Ecol. Prog. Ser. 444, 117–132. doi: 10.3354/meps09413
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fu, Y. (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925. doi: 10.1093/genetics/147.2.915
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755.
Hui, M., Agus, N., and Marc, K. (2016). Concordance of microsatellite and mitochondrial DNA markers in detecting genetic population structure in the boring giant clam Tridacna crocea across the Indo-Malay Archipelago. Mar. Ecol. 38:e12389.
Hume, B., Voolstra, C. R., Arif, C., D’ Angelo, C., Burt, J. A., Eyal, G., et al. (2016). Ancestral genetic diversity associated with the rapid spread of stress-tolerant coral symbionts in response to Holocene climate change. Proc. Natl. Acad. Sci. U. S. A. 113, 4416–4421. doi: 10.1073/pnas.1601910113
Ikeda, S., Yamashita, H., Kondo, S. N., Inoue, K., Morishima, S. Y., and Koike, K. (2017). Zooxanthellal genetic varieties in giant clams are partially determined by species-intrinsic and growth-related characteristics. PLoS One 12:e0172285. doi: 10.1371/journal.pone.0172285
Kubo, H., and Iwai, K. (2007). On sympatric two species within Tridacna “maxima”. Annu. Rep. Okinawa Fish. Ocean Res. Cent. 68, 205–210.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35:1547. doi: 10.1093/molbev/msy096
LaJeunesse, T. C. (2001). Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the ITS region: in search of a “species” level marker. J. Phycol. 37, 866–880. doi: 10.1046/j.1529-8817.2001.01031.x
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., et al. (2018). Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580. doi: 10.1016/j.cub.2018.07.008
LaJeunesse, T. C., Pettay, D. T., Sampayo, E. M., Phongsuwan, N., Brown, B., Obura, D. O., et al. (2010). Long-standing environmental conditions, geographic isolation and host-symbiont specificity influence the relative ecological dominance and genetic diversification of coral endosymbionts in the genus Symbiodinium. J. Biogeogr. 37, 785–800.
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lim, S. S. Q., Huang, D., Soong, K., and Neo, M. L. (2019). Diversity of endosymbiotic Symbiodiniaceae in giant clams at Dongsha Atoll, northern South China Sea. Symbiosis 78, 251–262.
Liu, C., Li, X., Wu, C., Wang, A., and Gu, Z. (2020). Effects of three light intensities on the survival, growth performance and biochemical composition of two size giant clams Tridacna crocea in the Southern China Sea. Aquaculture 528:735548.
Liu, C., Wu, C., Li, X., Liu, C., Wang, A., Gu, Z., et al. (2021a). Distribution and the influence of substrate on its mantle color of giant clam Tridacna noae in coastal waters of Sanya. Acta Hydrobiol. Sinica 45, 645–651.
Liu, C., Yang, X., Sun, Y., Yang, Y., Wang, A., He, L., et al. (2021b). Effects of the daily light/dark cycle on photosynthetic performance, oxidative stress and illumination-related genes in boring giant clam Tridacna crocea. Mar. Biol. 168:71.
Liu, J., Cui, D., Wang, H., Chen, J., Liu, H., and Zhang, H. (2020). Extensive cryptic diversity of giant clams (Cardiidae: Tridacninae) revealed by DNA-sequence-based species delimitation approaches with new data from Hainan Island, South China Sea. J. Mollus. Stud. 86, 56–63. doi: 10.1093/mollus/eyz033
Lucas, J. S. (1988). Giant clams: description, distribution and life history. Giant clams in Asia and the Pacific. ACIAR Monogr. 9, 21–33.
Marco, A., Lizano, D., and Santos, M. D. (2014). ARTICLE Updates on the status of giant clams Tridacna spp. and Hippopus hippopus in the Philippines using mitochondrial CO1 and 16S rRNA genes. Philipp. Sci. Lett. 7, 187–200.
Mies, M. (2019). Evolution, diversity, distribution and the endangered future of the giant clam–Symbiodiniaceae association. Coral Reefs 38, 1067–1084.
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in 2010 Gateway Computing Environments Workshop (GCE), (New Orleans: IEEE).
Neo, M. L., Lim, K. K., Yang, S., Soong, G. Y., Masucci, G. D., Biondi, P., et al. (2019). Status of giant clam resources around Okinawa-jima Island, Ryukyu Archipelago, Japan. Aquat. Conserv. 29, 1002–1011. doi: 10.1002/aqc.3033
Neo, M. L., and Todd, P. A. (2012). Giant clams (Mollusca: Bivalvia: Tridacninae) in Singapore: history, research and conservation. Raffles Bull. Zool. 25, 67–78.
Neo, M. L., Wabnitz, C. C., Braley, R. D., Heslinga, G. A., Fauvelot, C., Van Wynsberge, S., et al. (2017). Giant clams (Bivalvia: Cardiidae: Tridacninae): a comprehensive update of species and their distribution, current threats and conservation status. Oceanogr. Mar. Biol. 55, 87–388. doi: 10.1201/b21944-5
Nuryanto, A., Duryadi, D., Soedharma, D., and Blohm, D. (2007). Molecular phylogeny of giant clams based on mitochondrial DNA cytochrome coxidase I gene. Hayati 14, 162–166. doi: 10.11646/zootaxa.4007.2.1
Nuryanto, A., and Kochzius, M. (2009). Highly restricted gene flow and deep evolutionary lineages in the giant clam Tridacna maxima. Coral Reefs 28, 607–619.
Pappas, M. K., He, S., Hardenstine, R. S., Kanee, H., and Berumen, M. L. (2017). Genetic diversity of giant clams (Tridacna spp.) and their associated Symbiodinium in the central Red Sea. Mar. Biodivers. 47, 1209–1222. doi: 10.1007/s12526-017-0715-2
Pochon, X., and Gates, R. D. (2010). A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawaii. Mol. Phylogenet. Evol. 56, 492–497. doi: 10.1016/j.ympev.2010.03.040
Pochon, X., Wecker, P., Stat, M., Berteaux-Lecellier, V., and Lecellier, G. (2019). Towards an in-depth characterization of Symbiodiniaceae in tropical giant clams via metabarcoding of pooled multi-gene amplicons. PeerJ 7:e6898. doi: 10.7717/peerj.6898
Pringgenies, D., Suprihatin, J., and Lazo, L. (1995). Spatial and size distribution of giant clams in the Karimunjawa Islands, Indonesia. Phuket Mar. Biol. Cent. Spec. Publ. 15, 133–135.
Reich, H. G., Robertson, D. L., and Goodbody-Gringley, G. (2017). Do the shuffle: changes in Symbiodinium consortia throughout juvenile coral development. PLoS One 12:e0171768. doi: 10.1371/journal.pone.0171768
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Rossbach, S., Hume, B. C., Cárdenas, A., Perna, G., Voolstra, C. R., and Duarte, C. M. (2021). Flexibility in Red Sea Tridacna maxima-Symbiodiniaceae associations supports environmental niche adaptation. Ecol. Evol. 11, 3393–3406.
Rowan, R., Knowlton, N., Baker, A., and Jara, J. (1997). Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature 388, 265–269. doi: 10.1038/40843
Satoe, A., Tomura, T., Saitoh, S., Yokokura, R., Kawanishi, Y., Shinjo, R., et al. (2012). Soft Coral Sarcophyton (Cnidaria: Anthozoa: Octocorallia) Species Diversity and Chemotypes. PLoS One 7:e30410. doi: 10.1371/journal.pone.0030410
Stat, M., Carter, D., and Hoegh-Guldberg, O. (2006). The evolutionary history of Symbiodinium and scleractinian hosts-Symbiosis, diversity, and the effect of climate change. Perspect. Plant Ecol. 8, 23–43. doi: 10.1016/j.ppees.2006.04.001
Su, Y., Hung, J. H., Kubo, H., and Liu, L. L. (2014). Tridacna noae (Röding, 1798)–a valid giant clam species separated from T. maxima (Röding, 1798) by morphological and genetic data. Raffles Bull. Zool. 62, 124–135.
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. doi: 10.1093/genetics/123.3.585
Tonk, L., Sampayo, E. M., Weeks, S., Magno-Canto, M., and Hoegh-Guldberg, O. (2013). Host-specific interactions with environmental factors shape the distribution of Symbiodinium across the Great Barrier Reef. PLoS One 8:e68533. doi: 10.1371/journal.pone.0068533
United Nations Environment Programme-World Conservation Monitoring Center (UNEP-WCMC) (2007). UNEP-WCMC Species Database: CITES-listed Species. Cambridge: UNEP.
Venn, A. A., Loram, J. E., and Douglas, A. E. (2008). Photosynthetic symbioses in animals. J. Exp. Bot. 59, 1069–1080. doi: 10.1093/jxb/erm328
Wang, J., Zhou, Z., Ma, H., Li, J., Qin, Y., Wei, J., et al. (2021). Genetic Recombination of the Mantle Color Pattern of Two Boring Giant Clam (Tridacna crocea) Strains. Front. Mar. Sci. 8:657762. doi: 10.3389/fmars.2021.657762
Wells, S. (1997). Giant Clams: Status, Trade and Mariculture, and the Role of CITES Management. Gland: IUCN.
Winters, G., Beer, S., Zvi, B. B., Brickner, I., and Loya, Y. (2009). Spatial and temporal photoacclimation of Stylophora pistillata: zooxanthella size, pigmentation, location and clade. Mar. Ecol. Prog. Ser. 384, 107–119. doi: 10.3354/meps08036
Wong, J. C., Thompson, P., Xie, J. Y., Qiu, J. W., and Baker, D. M. (2016). Symbiodinium clade C generality among common scleractinian corals in subtropical Hong Kong. Reg. Stud. Mar. Sci. 8, 439–444. doi: 10.1016/j.rsma.2016.02.005
Xia, X. (2013). DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evol. 30, 1720–1728. doi: 10.1093/molbev/mst064
Keywords: Tridacna, Symbiodiniaceae, symbiont, COI, 16S rRNA, ITS1
Citation: Chao Q, Gu Z, Wang A, Liu C and Yang Y (2021) Diversity of Three Small Type’s Giant Clams and Their Associated Endosymbiotic Symbiodiniaceae at Hainan and Xisha Islands, South China Sea. Front. Mar. Sci. 8:774925. doi: 10.3389/fmars.2021.774925
Received: 13 September 2021; Accepted: 12 October 2021;
Published: 11 November 2021.
Edited by:
Liqiang Zhao, Guangdong Ocean University, ChinaReviewed by:
Gang Ni, Ocean University of China, ChinaCopyright © 2021 Chao, Gu, Wang, Liu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunsheng Liu, bGNzNTExM0AxNjMuY29t; Yi Yang, eWl5YW5nb3VjQG91dGxvb2suY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.