
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 19 November 2021
Sec. Marine Evolutionary Biology, Biogeography and Species Diversity
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.772851
During the era of biodiversity loss, a complete species census and understanding where the different species occur is of high priority. Even though this knowledge has increased tremendously, mainly with expanded use of integrated taxonomic identification, there are groups where our knowledge is very limited, both in terms of diversity and distribution. Ctenophores are such a group. Due to a lack of identification literature, damage to specimens during net sampling and sample processing, difficulties with preservation and a considerably undescribed diversity within the phylum, this group is often hard to work with. A citizen science approach was applied during a mapping campaign on ctenophore diversity along the Norwegian coast in order to have a broad geographical coverage. This was achieved by a collaboration with five diving clubs along a south-north geographical gradient that were briefly introduced to ctenophore taxonomy and ecology and sampling techniques using Whatman® FTA® Cards. The data collected by the participating divers gave a broad spatial coverage and provided information on ctenophore diversity in these regions. The use of FTA® Cards in the sampling allowed successful species and genus level identification using DNA barcodes. However, small obstacles such as accurate morphological species identification and labor-intensive issues were identified that can impede the use of large-scale citizen science approaches to map ctenophore diversity and thus recommendations for future implications that address these issues are proposed here.
Science and international policies have declared understanding and conserving biodiversity as one of the priority areas for the coming decades, with a special focus on estimates and thorough quantification of biodiversity, the development of conservation tools, and measures to sustain biodiversity and ecosystem functions (IPBES, 2019). Actions to conserve biodiversity are an international goal, upheld by the Aichi Targets for 2020 by parties of the United Nations (UN) Convention on Biological Diversity (CBD) (Pereira et al., 2013). There are still groups of biota that remain insufficiently investigated, despite the significant increase in actions to understand biodiversity and achievements of several Aichi Biodiversity targets in the last 10–15 years (IPBES, 2019). Despite innovations in molecular technologies and integrated species identification that have fundamentally changed our understanding of marine biodiversity, our knowledge in marine community composition has been severely biased toward taxa that are relatively easy to sample, identify and quantify using traditional methods (e.g., Haddock, 2004; Pauly et al., 2008; McManus and Katz, 2009; Danovaro et al., 2016). Often, species that are of commercial and economical relevance have been prioritized. At the same time, other taxa have been excluded from monitoring programs such as taxa with overwhelming systematic complexity or numerous cryptic species, soft-bodied and fragile species, species with ontogenetic changes in their appearance or species which lack diagnostic characters (Verity and Smetacek, 1996). As a result, most of the existing long-term monitoring programs on marine plankton dynamics and diversity focus on spatial and temporal distribution patterns of phytoplankton or mesozooplankton groups that are easier to quantify and identify such as copepods or ichthyoplankton (Beaugrand, 2004; Hays et al., 2005; Wiltshire et al., 2010) while only a few time-series include groups that are difficult to sample or identify based on traditional methods such as protozooplankton or gelatinous zooplankton (Attrill et al., 2007; Harris, 2010; Hinder et al., 2012). This bias in biodiversity assessments thus limits the understanding of local-to-global patterns of biodiversity, challenging assessments on the ecological status of a system and hampering the predictions on future biodiversity change and species loss.
Citizen science (CS) approaches have a long history of involving the general public (both novices and experts) in, for example, bird watching programs which have contributed to observational avian data for centuries (Greenwood, 2007; Tulloch et al., 2013) and thus is not a new phenomenon., In fact, today the involvement of citizens from the non-scientific community in scientific research is a growing trend (e.g., Bonney et al., 2014; Pocock et al., 2014; Austen et al., 2016; Chandler et al., 2017; reviewed in Garcia-Soto et al., 2021). CS programs cover a wide taxonomic breadth of global biodiversity research (e.g., Greenwood, 2007; Theobald et al., 2015; Geijzendorffer et al., 2016; Chandler et al., 2017) have contributed to information on population dynamics, health and distribution of marine organisms, marine litter, and have also supported long-term monitoring programs of Marine Protected Areas (Deidun and Sciberras, 2017; Reviewed in Thiel et al., 2014 and in Garcia-Soto et al., 2021; Vohland et al., 2021). Recent improvements in CS have occurred through establishing the Citizen Science Global Partnership and the European Citizen Science Association as well as the research network of the COST Action CA15212 Citizen Science to Promote Creativity, Scientific Literacy, and Innovation throughout Europe to better collaborate on the shaping and development as well as using the different aspects of CS. Recently, it has been recognized that CS is not just a participatory way to contribute to scientific knowledge and to result in more data and information that would otherwise not be affordable (Tulloch et al., 2013), but also as a powerful tool for the generation and spread of scientific knowledge (Thiel et al., 2014). Including the public in data collection can increase and improve their knowledge and understanding of scientific research, thereby educating participants to a higher degree than they would with reading a finished scientific report. Tulloch et al. (2013) identified eight objectives for using volunteer-collected monitoring data, where both education and awareness were raised as main points. Thus, involvement in a CS project can contribute to the changing of attitudes and behaviors, bringing in environmental appreciation (Vohland et al., 2021).
The development of modern technologies has led to an advancement of CS approaches making it even easier for the public to get involved in state-of-the-art monitoring and biodiversity surveys (reviewed in Garcia-Soto et al., 2021). In marine jellyfish research, for example, different web-based and mobile apps have become popular tools to collect data through CS approaches, such as the Medjelly digital mobile app develop by the Institute of Marine Science (ICM-CSIC, Barcelona, Spain) which engages the public to contribute to real-time information on jellyfish species and distribution ranges. This app allows volunteers worldwide to collect and save data on jellyfish occurrences and environmental parameters directly on the Medjelly app while in the field, thus delivering up to date, daily jellyfish information along coastal areas, including predictive models for jellyfish blooms in the next 24 and 48 h (Marambio et al., 2016). Similarly, the recently launched Jelly Spotter app aims for a better localization of jellyfish in the Baltic Sea as well as identifying new invasive species. However, Martins et al. (2016) interviewed 110 marine users to understand beliefs about recording species in a CS project and found that apps are in general perceived as an acceptable tool but that some users were hesitant to use them due to technical design and the need for internet access. This shows that apps need to be user-friendly and easily accessible when applied to CS projects. Due to a rapidly growing technology today, there is reason to believe that apps and mobile devices can contribute substantially to monitoring programs, ecosystem surveillance and conservation. Thus, the public’s engagement can promote a higher acceptance and understanding of biodiversity research and help to support monitoring programs. However, if we wish to meet the future ambitions toward monitoring programs, there is a need to apply all possible sources for gathering data and information (Chandler et al., 2017).
Despite many advantages of CS approaches, several challenges remain. When discussing the use of CS in diversity monitoring, one of the main issues is that many researchers are not confident that the data obtained from volunteers is adequate and scientifically sound. This is especially the case when biological identification is concerned which is fundamental for biodiversity assessments. It raises questions concerning quality vs. quantity, since more data may not necessarily be more valuable if the data is misleading and biased (Balázs et al., 2021). However, during a species identification study on bumblebees (Austen et al., 2016), a group of insects of considerable conservation concern, the accuracy of identification was similar both for experts and non-experts at <60% for both. This study shows that experts and non-experts both make errors in species identification. It further demonstrates the importance of accurate species observations in ecological datasets and suggests that consideration should be given to possible inaccuracies, especially when such information is used to inform decision makers (Austen et al., 2016). Although there are issues with using CS, several studies show that with given the proper training, members of the public can collect data with high accuracy (Darwall and Dulvy, 1996; Fore et al., 2001; Boudreau and Yan, 2004). Accuracy of species identification can also be increased by combining morphological with molecular methods, also when using CS approaches. Examples of studies where the public is used to collect samples which are subsequently analyzed using DNA barcoding or eDNA techniques are multiple ranging from butterflies (Jisming-See et al., 2016) to full biodiversity assessments of marine parks (Marizzi et al., 2018) and ponds (Biggs et al., 2015; Buxton et al., 2018). Collecting data to be used for molecular analyses is a quick and technically straightforward method (Ficetola et al., 2008; Thomsen et al., 2012) and allows the validation and verification of the samples before being used.
Ctenophores are a good example of a marine group that is difficult to monitor and thus often neglected in long-term data series and monitoring efforts (Hay, 2006). In ecological samples, these extremely fragile specimens are often damaged when collected with traditional sampling nets or preserved with conventional fixatives (Podar et al., 2001; Thibault-Botha and Bowen, 2004; Hay, 2006; Mills and Haddock, 2007; Gorokhova and Lehtiniemi, 2010; Raskoff et al., 2010). Even though ctenophores are morphologically diverse, they possess several synapomorphies, such as biradial symmetry, eight rows of combs controlled by an apical organ, and specialized adhesive cells (e.g., Harbison, 1985; Podar et al., 2001; McConville, 2011) which limits the species level identification on damaged samples. Thus, the order or family of ctenophores can usually be identified while an identification to the correct species level is more demanding when only using traditional morphological identification procedures (e.g., McConville, 2011; Johansson et al., 2018; Majaneva et al., 2021; Shiganova and Abyzova, 2021). Scientific literature contains relatively few mentions of ctenophores from Norway, let alone specific species found in the region. So far, baseline data on species richness, diversity and distribution patterns of ctenophores are lacking (see e.g., Hosia and Falkenhaug, 2015; Johansson et al., 2018; Majaneva et al., 2021; Shiganova and Abyzova, 2021). The aim of this study was to target this knowledge gap by mapping the pelagic ctenophore diversity along the Norwegian coast from the North Sea to Svalbard. To increase the geographical coverage of the study area, a CS approach following the 9-step process for developing a citizen science project suggested by Bonney et al. (2009) together with DNA techniques was implemented. Due to major challenges in the ctenophore species identification, it was decided to use integrated species identification techniques. When molecular methods are applied, correct preservation of the sample is required to prevent DNA degradation, and samples are mainly preserved either by directly freezing at –20°C or by preserving in > 70% ethanol (EtOH) and subsequently freezing at –20°C. In addition, efficacy of common alternative products such as alcohol-based hand sanitizer and propylene and ethylene glycol-based automobile antifreeze have been tested with successful results (Steininger et al., 2015), but all are alcohol-based techniques. In Norway, > 60% alcohol is considered as narcotics and this limits the use of EtOH in CS projects. Thus FTA® Cards, a substrate based on Whatman FTA® technology that can store and protect DNA, and is thus suitable for collection of DNA samples, were tested. In this paper, the suitability of a CS approach and use of FTA® Cards were evaluated to map the morphologically challenging ctenophore taxa by addressing the following hypotheses:
H1: Difficulties in ctenophore identification can limit the quality of the data in the CS approach.
H2: Use of FTA® Cards is a simple, efficient method to increase species identification in the CS project.
To engage the public in current ctenophore diversity assessments, e-mails were sent to several diving clubs along the Norwegian coast in 2018. The clubs contacted were selected individually taking geographical aspects into account (Figure 1). In total, six diving clubs were selected based on their positive response and interest for the project: Kristiansand diving club, Mandal diving club, AEgir diving club, Levanger diving club, Steinkjer sports-diving club and Brønnøysund diving club. Each engaged diving club was visited in person during two trips: once in June 2018 (except Levanger and Steinkjer, who received all information via mail) and again in August 2018. The first visit aimed to provide detailed information on the research focus of the project, provide general information of the phylum and to get the divers acquainted with the use of FTA® Cards to be used for species collection. In addition, a simple identification manual, a sheet for data registration (date, location, temperature, visibility, species ID, photo ID, divers name) and FTA® Card user instructions was provided. Sampling strategies were decided by the diving clubs depending on their individual diving frequencies and locations. Divers involved were encouraged to take photographs of ctenophore specimens and to identify the individuals to the best of their knowledge using the identification manual before taking a body tissue sample of each specimen and smearing on the FTA® Card. In this way, data on both molecular and morphological species identification were collected. At the time of the first visits, an article on the project was published in Norway’s largest diving magazine “Dykking”1 to highlight the CS contribution and the importance of mapping the ctenophore diversity in Norway. The purpose of the second visit was to gather the samples, to talk about the diver’s experiences and to update them on the status of the project. Communication with the diving clubs was kept continuously through e-mail and social media. The divers involved in the sampling efforts received the results later, thereby creating enthusiasm among those involved.
After sampling through CS approaches, all FTA® Cards were collected, and DNA was extracted by using a modified protocol recommended by the manufacturer. DNA was extracted from the paper with a modified Chelex rapid-boiling procedure as explained in Granhag et al. (2012). Internal transcribed spacer 1 (ITS1) (app. < 300 bp) amplifications were performed on a MJ Research PTC 100 Thermal Cycler PCR with universal eukaryotic primers 1400F (5′ TGY ACACAC CGC CCG TC 3′) and 5′2Sr (5′ CCT AAG TTC AGC GGG TAG TCT CG 3′) (Podar et al., 2001; Chow et al., 2009). PCR mix consisted of 20 μL and contained 0.4 μL Phire® Hot Start II DNA Polymerase, 4 μL of Phire® 5x reaction buffer, 0.4 μL of each primer (final concentration 0.2 mmol), 1 μL of DNA template, 0.4 μL of dNTP 0.6 μL of 3% DMSO and 12.8 μL nuclease-free water (Podar et al., 2001) with cycle: 98°C–5 min (98°C–5 s, 64°C–5 s, 72°C–20 s) × 32, 72°C–2 min, 4°C–10 min. PCR products were verified in a 1.5% agarose gel in 1xTAE buffer. PCR products were purified using Illustra GFX PCR DNA and gel band purification kit following the cleaning procedure recommended by the manufacturer. Cycle sequencing of the PCR products was carried out by Macrogen Sequencing Service (Macrogen Inc., South Korea). The resulting nucleotide sequence electropherograms were checked by eye for poor base calls and sequence quality using Chromas Lite 2.1 (Technelysium Pty Ltd). The good-quality sequences were assembled using BioEdit software (Hall, 1999). To place the sequences phylogenetically, all available ITS1 sequences of Ctenophora and four Cnidaria sequences as an out-group from the NCBI Nucleotide database (accessed 28.08.2020) were used. Those sequences were combined with sequences from this study and aligned with the MAFFT online service (Katoh et al., 2019). The sequences were aligned using Q-INS-i strategy, which takes RNA secondary structure into account, and a gap-opening penalty of 1.53 and a gap extension penalty of 0.123. The alignments were visually checked, and identical sequences were excluded prior to the analyses. The alignments are available on request to the corresponding author. Bayesian phylogenetic analysis was performed with MrBayes 3.2.7a (Ronquist et al., 2012). Two independent runs with four Markov chains and 1,600,000 generations were carried out (average standard deviation of split frequencies 0.0094). No model was chosen prior to the analysis but sampled across the GTR model space with gamma-distributed rate variation across sites and a proportion of invariable sites. The resulting estimates (e.g., tree topology) were posterior probability weighted averages of the models. The sequences reported in this paper have been deposited in the European Nucleotide Archive with accession numbers: OU817762-OU817778).
Five out of the six diving clubs that were committed to the CS efforts provided photographic material and tissue samples of ctenophore species (diving clubs in Kristiansand, Mandal, Stadt, Levanger and Steinkjer) (Figure 1 and Table 1). In total, 54 ctenophore specimens were collected and 42 of them included additional data, such as depth and water temperature. The CS campaign made it possible to have a broad geographical coverage, especially from the southern and western parts of Norway. Many of the participants reported low abundance of ctenophores during the sampling period, which can explain the relative low number of samples collected, but many participants continued to report observations and increased diversity and abundance after the CS campaign. In addition, as a positive result of the published article about the project in the diving magazine, a scuba diver provided information from the Oslofjord monthly (from September 2018 to February 2019) by sending updates regarding ctenophore diversity and density, water visibility, temperature and other additional biological data (not shown).
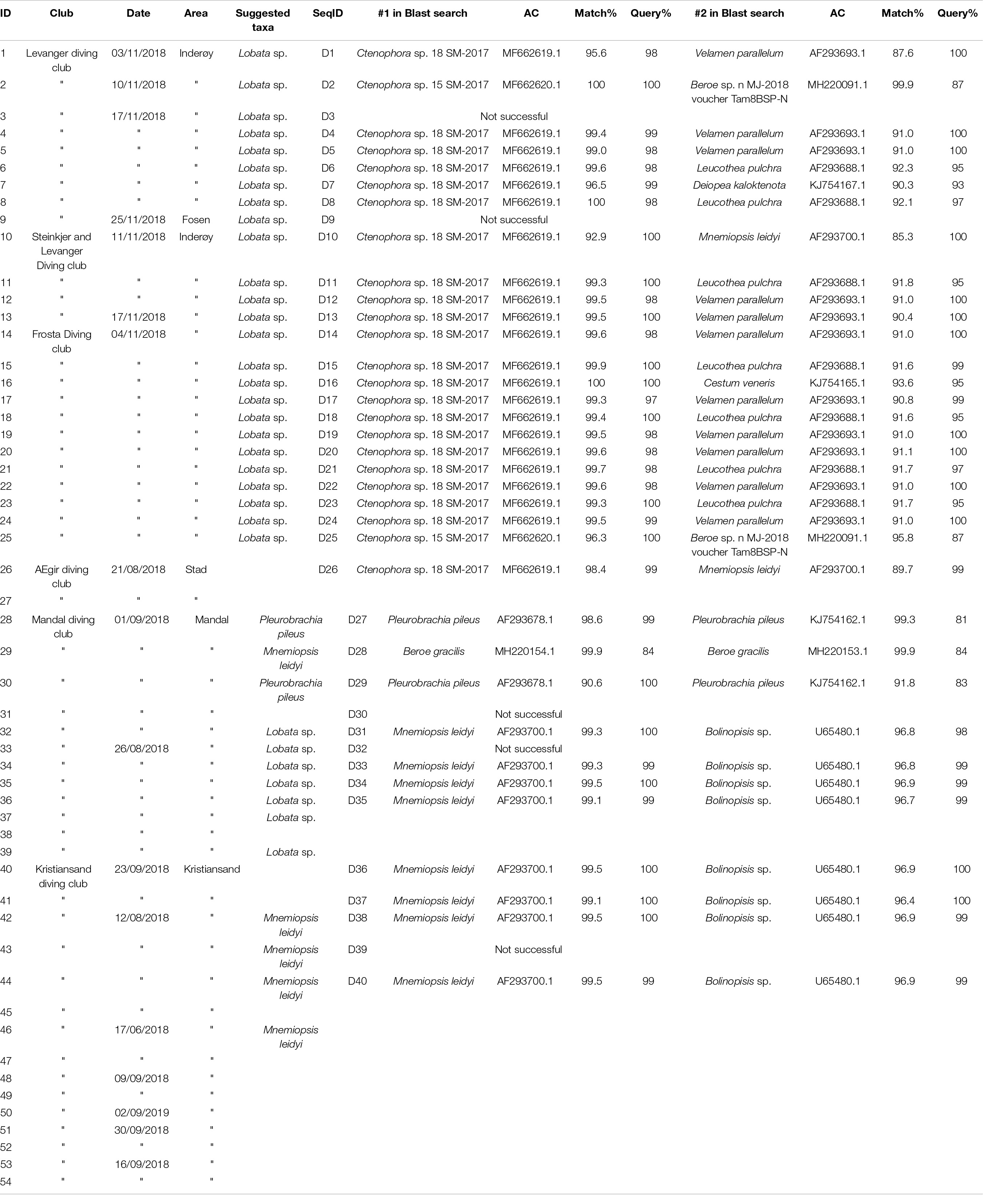
Table 1. Information on the ctenophore samples received from the participating diving clubs together with morphological and molecular species identification.
Information for 54 ctenophore specimens were received from the divers through the CS approach. Most specimens were morphologically identified by the CS participants to order and family level, while seven specimens representing two different species, Pleurobrachia pileus and Mnemiopsis leidyi, were identified to species level (Table 1). Good quality photos were received for 25 of the collected specimens. From which six ctenophore taxa could be morphologically identified: Bolinopsis infundibulum, M. leidyi, Euplokamis dunlapae, P. pileus, Beroe cucumis and undescribed cydippid ctenophore (“horned ctenophore”). The later (Figures 2D,F) was observed to have distinct appendages at the aboral end, resembling horns, a feature not observed in other taxa. The undescribed cydippid ctenophore had thick tentacles extending from the body, and a slender elongated body shape. The body shape resembles that of E. dunlapae, but with a narrower aboral end (Figure 2E). It was morphologically identified as a Lobata species (Figures 2C,G) and had lobes retracted and nested within the body. These individuals had a rounded aboral end and moderately rounded body shape. Divers from Kristiansand and Mandal were comfortable with identifying species within the Bolinopsidae family, describing to species level (e.g., M. leidyi). Specimens identified as B. cucumis had a high degree of branching between meridional canals which was a common trait observed by the divers for this specific species (Figure 2H). Euplokamis dunlapae was observed to have an elongated body, with long tentacles with little branching out from the tentacles (Figure 2E).
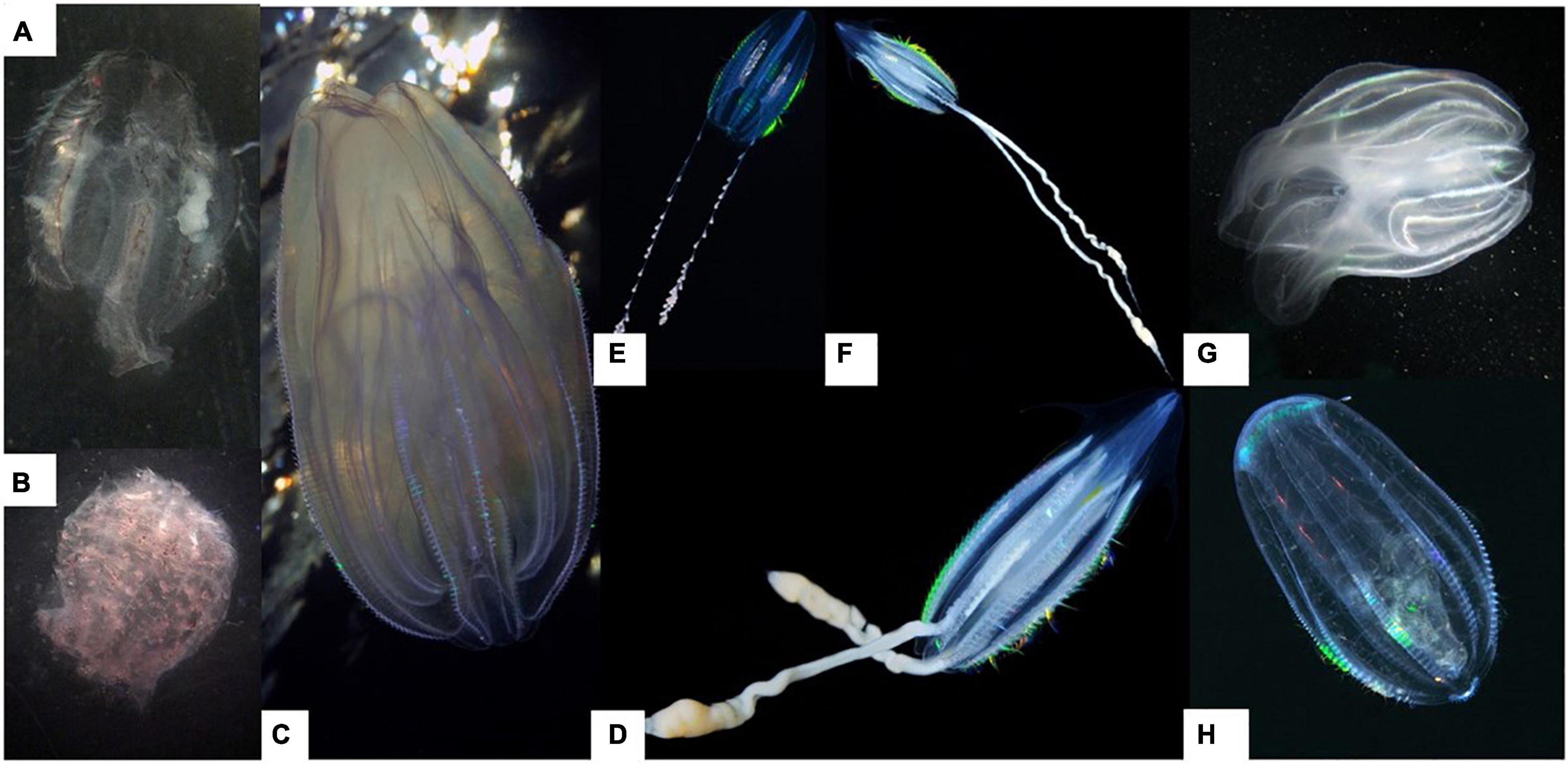
Figure 2. Ctenophore taxa identified along the Norwegian coast from Skagerrak to Svalbard: Typical example of net collected ctenophores (A) Mertensia ovum and (B) Beroe sp.; underwater images received from the citizen science participants (C) Bolinopsidae sp., (E) Euplokamis cf. dunlapae, (D,F) Undescribed cydippid ctenophore (same specimen), (G) Lobata spp. and (H) Beroe cucumis. Photos: (A,B) Sanna Majaneva, (C–F,H) Nils Aukan and (G) Vigdis Dahling.
In total, 34 of the specimens collected on DNA-paper were successfully sequenced for ITS1, of which three only in forward direction, resulting in five different ctenophore taxa. Specimens collected through the CS approach matched within the order of which the species had been morphologically identified (Table 1). Most of the specimens (22) morphologically identified as Lobata, matched with Ctenophora sp. 18 SM-2017 (MF662619) and two specimens with Ctenophora sp. 15 SM-2017 (MF662620) an unidentified cydippid larvae collected from Northern Norway (Halsband et al., 2018). These specimens also formed their own branch in the phylogenetic analysis (Figure 3). Four specimens morphologically identified as Lobata matched with M. leidyi (AF293700). Similarly, two individuals, morphologically identified as M. leidyi (D38 and D40) were confirmed as they matched with M. leidyi (AF293700) while comparing sequences through BLAST. The two tentaculate ctenophores identified by divers from Mandal diving club as P. pileus (D27 and D29), were confirmed by molecular methods to match P. pileus (AF293678) and nesting within the suggested Pleurobrachidae branch (Figure 3). Only one specimen identified as M. leidyi by the divers provided different results and was identified as a Beroe gracilis (MH220154) based on the molecular methods.
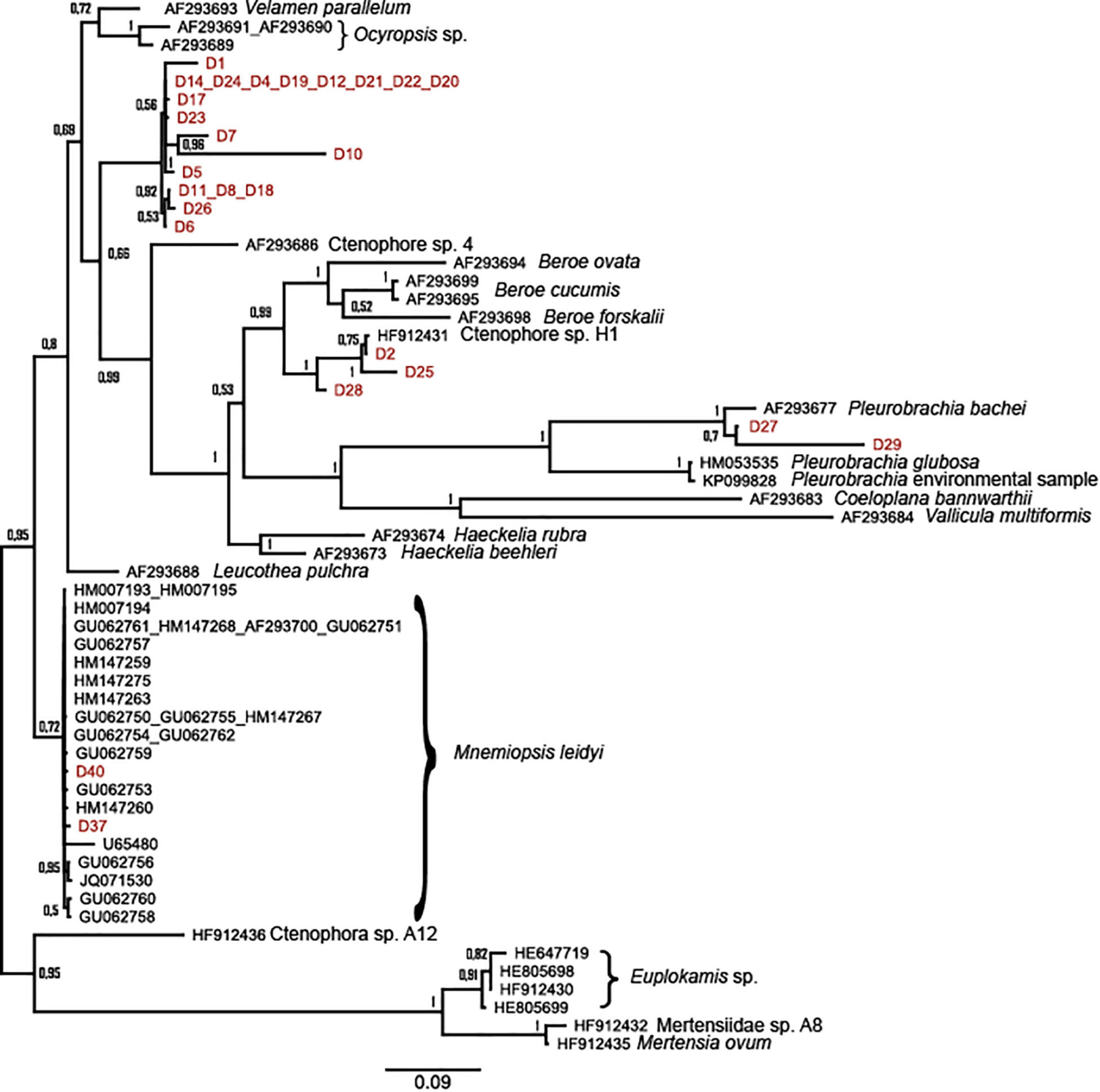
Figure 3. Maximum-likelihood tree for ITS1 of all ctenophore sequences in GenBank together with specimens collected in this study by the citizen science approach (marked red, D1-D40, see Table 1 for more information).
The citizen science approach conducted in the present study suggests that the involvement of citizens (both novices and experts) from the non-scientific community in collecting data can provide valuable data for monitoring and diversity assessments on larger spatial and temporal scales. Here, CS data was provided for a broad range of geographic locations along the Norwegian coast, allowing the project to create more even spatial distribution along Norway’s coastline from South to North when CS was combined with traditional research vessel field work. Our results suggest that FTA® Card-based CS approaches are suitable for mapping ctenophore diversity over wide geographical scales. However, some modifications such as selecting proper DNA regions used as the barcode for the protocol are required. The project also pinpointed some gray areas in the CS approach chosen and noted areas of improvement.
Citizen science has been described as a practical and efficient way to achieve the geographic extent required to document species distribution ranges, ecological patterns and to address ecological questions at relevant scales (reviewed in Tulloch et al., 2013). Results of this study highlight the significant value of applying CS approaches to increase geographical coverage of the study area. The participating diving clubs collected specimens from regions not accessible by traditional sampling efforts with research vessels, such as very shallow or narrow coastal areas. It also allowed samples to be collected almost simultaneously in different regions which would not have been possible with traditional research cruises or monitoring programs due to logistical issues. Comparing the sampling with divers vs. standard plankton sampling using nets, it can be considered as highly likely that divers cover a larger area in the upper surface layer. However, divers may to a greater extent overlook specimens when mapping an area as for example small specimens are hard to detect by the naked eye. Also, the level of consistency in sampling frequency between diving clubs varied; Mandal, Kristiansand, and AEgir arranged diving trips every weekend during Summer and Autumn while other diving clubs showed different frequencies.
Despite the above-mentioned benefits, CS can also have weaknesses in terms of contribution to species identification and biodiversity monitoring when both quantitative and qualitative assessments are needed. One of the factors causing lack of confidence in the scientific community toward CS data is the temporal resolution (Tulloch et al., 2013). For example, in many Bird Atlas surveys conducted by CS, the data may cover multiple years of surveys, but are typically treated as snapshots in time in the analysis and affect the temporal resolution. The present CS campaign focused solely on qualitative taxonomic assessments while quantitative estimates could not be achieved. Thus, the snapshot data provided in this study was suitable for the research question addressed. Similarly, accuracy on species identification has often been questioned in CS programs. In this study, we noted that most of the participants were not comfortable identifying down to species level but rather identified to order level, such as Lobata. Divers were, in some cases, comfortable with identifying Mnemiopsis leidyi and Pleurobrachia pileus to species. These identifications were also supported by the molecular identification. During diving, divers do not always have the best visibility nor the time to thoroughly study specimens, and gelatinous species no doubt add to the challenges. As ctenophores are generally known to be hard to identify, most of the scientific reports from the same region identify ctenophore species to the same level as participating divers e.g., to the order or family level or sometimes even as ctenophore indet (e.g., Johansson et al., 2018; Majaneva et al., 2021). It is also important to note that the participants of this CS study were divers, who often had many years of experience on marine life and photography, yet many of the participants had little knowledge on ctenophores (pers. com.). This is in-line with previous studies where it has been observed that the CS programs that contribute most to the Global Biodiversity Information Facility tend to serve existing and vibrant communities of expert amateurs and hobbyists (Lawrence, 2006; Bell et al., 2008; Lewandowski and Specht, 2015). In these cases, a relatively high species identification accuracy can be achieved (Austen et al., 2016), as the accuracy is dependent on the skills of each individual citizen. To increase the accuracy of species identification, multiple options to combine a CS approach with molecular identification exists. In this study the questions regarding species identification were resolved, since non-researchers only contributed with material for a molecular analysis and did not perform morphological identification themselves. However, when determining the benefits of CS approaches it is important to consider the taxa in question as well as the region. For ctenophores, the current knowledge is far more limited than birds, for example (as reviewed in Tulloch et al., 2013). Thus, the benefit of using CS when mapping ctenophore diversity can be considerably lower than for CS programs with Bird Atlases.
Reaching out to diving clubs along the Norwegian coast was a pleasant experience, and the response was overwhelming. Divers appeared to have a broad interest in the oceans and marine life, and a general knowledge on species and groups, even some knowledge on ctenophores which are a poorly studied group even in the scientific world. It seemed that the task challenged the divers but, on the other hand, was not too demanding as needed information was provided together with the DNA sample on the FTA® Cards. To gain larger datasets, the sampling period for the CS should have been longer as, in this study, the ctenophore abundance increased after the sampling period was over. This method also increases people’s awareness of ctenophores, as a great number of citizens were not aware of the phylum and were surprized to hear that they in fact inhabit Norwegian coastal waters and fjords. Divers often sent pictures asking about species identification and shared that they, after the project began, were a lot more observant of ctenophores than before. Previously, the accomplishments of CS programs have been attributed by behavioral psychologists to participants who appreciate the opportunity to build on their existing interests, try something new with little effort, and participate in projects associated with place and community (Lawrence, 2006; Bell et al., 2008). A CS approach can be used for exchanging knowledge and data, both within the scientific and the public community, and CS approaches can thus be considered as very valuable in terms of scientific outcome and outreach. Increased awareness, spread of education and ecological knowledge were some of the points mentioned during the many talks with participating divers.
The data collected in this study document the presence of two common ctenophore species along the Norwegian coast: Mnemiopsis leidyi and Pleurobrachia pileus. In addition, a minimum of three taxa were recorded which did not find a full match in the Blast search or group with published sequences in the phylogenetic analysis (Figure 3). Samples D2 and D25 grouped close to Ctenophora sp. H1 (HF912431; Majaneva and Majaneva, 2013), an unidentified ctenophore collected from Svalbard and potentially related to Beroidea. Most of the specimens collected by the CS participants matched with Ctenophora sp. 18 SM-2017 and Ctenophora sp. 15 SM-2017 (Halsband et al., 2018), an unidentified cydippid larva collected from Northern Norway. Cydippid larva is an early developmental stage, found in most ctenophores belonging to the orders Lobata and Cydippida (Podar et al., 2001). However, as divers are only able to collect specimens visible to the naked eye, the specimens collected here are most likely not in cydippid larval stage. The divers had identified these specimens as Lobata and thus only the species M. leidyi and Bolinopsis infundibulum would be possible options. As the specimens did not cluster with M. leidyi, and no good-quality sequences (only < 70 bp) exist for B. infundibulum it would make the latter one the potential species in question. Use of another gene region or combination of multiple gene regions, could provide better species identification for these specimens, but on the other hand would increase the workload and the costs.
The classical morphology-based taxonomy of ctenophores (e.g., Harbison, 1985; Mills and Haddock, 2007) heavily relies on very detailed characteristics which are often unfamiliar to non-taxonomists, such as the branching structure of the gastrovascular system and an axial portion parallel to the ctenophore oral–aboral axis (mouth, pharynx, infundibulum). Among plankton researcher, other characteristics traditionally used in ctenophore systematics include the size, shape and position of organs (e.g., relative size of comb rows, position of tentacle roots, absence/presence of tentillae on the tentacle (Harbison, 1985; Majaneva, 2014). There is a common understanding that morphological identification of ctenophores can be challenging, also for experts. There is a lack of identification literature, considerable undescribed diversity within the phylum, specimens are often highly damaged (resulting from net sampling and sample processing, Figures 2A,B) and difficult to preserve (meaning that type specimens are generally not available for examination) (Haddock, 2004; Hay, 2006). To include molecular species identification into CS programs, the protocol in use should not only be handy to use but also provide accurate species identification, especially when considering CS approaches over larger geographical scales. FTA® Cards proved to be suitable for gelatinous DNA collection as high percentage (> 90%) of the FTA® Card data was correctly done and contained good-quality DNA to be analyzed. The next step in molecular species identification is choosing the correct DNA barcode. On the molecular species identification side, the small subunit (18S) ribosomal RNA gene is widely used, and several published protocols exits, thus 18S can serve as a useful marker for molecular identification (e.g., Gorokhova et al., 2009). However, despite the large number of publicly available ctenophore sequences in terms of species coverage as well as the number of specimens per species, this region is known to be highly conserved among ctenophores limiting the species level identification (Podar et al., 2001; Majaneva, 2014; Alamaru et al., 2017; Majaneva et al., 2021). The common barcoding gene mtCOI has shown to be promising for ctenophore species identification (Alamaru et al., 2017), but previously there have been limitations such as low number of sequences in reference libraries (see Christianson et al., 2021 for details). Furthermore, recently published protocols show that successful primers varied by taxon; whereas several combinations of primers successfully amplified the COI fragment for several species, some genera require customized primers (Christianson et al., 2021; Majaneva, unpublished data). For a few species, the variable ITS1 is sequenced (Majaneva and Majaneva, 2013; Simion et al., 2015) and, due to its short length and existing protocols, it is relatively easy to use. Despite the success rate being good, the limited reference library did not allow species level identification for most of the specimens, even though the ctenophore diversity of this regions is low.
The purpose of this study was to see how a small-scale citizen science approach could be integrated to increase the geographical coverage, as part of a mapping campaign on ctenophore diversity along the Norwegian coast. Accurate morphological species identification is challenging within the ctenophore groups and appears to be a common problem for both scientists and participants in the CS approach. Difficulties in morphological identification are mainly due to the fragile structure of ctenophores and the requirement that specimens need to be in good shape for accurate species identification. Using divers is therefore a positive contribution, as the specimens are in good condition and not harmed. However, time and visibility, in addition to other factors, can affect the diver’s ability to identify species. On the other hand, including divers through the CS approach contributed to gathering a great amount of data over a larger geographical area than would have been possible without the CS approach. Not only did the project gain from the CS approach, but the participants also increased their awareness and knowledge on the ctenophore species and improved their ecological understanding. Despite being time consuming and labor-intensive in terms of providing information, materials, planning trips, and collecting data from multiple different participants, the pros of integrating a CS approach clearly override the cons. There is, however, room for improvement of CS programs and methods on how to plan, carry out and follow up project where the citizens are involved. FTA® Cards proved to be a suitable method for CS projects as they were easy to use, gave good quality data, and, allowed verification of species identification using an integrated approach.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, OU817762-OU817778.
EJ and SM conceptualized and designed the study. EJ conducted the field work. EJ, SM, and M-AØ performed the molecular analyses. EJ and SM wrote the manuscript with contributions from M-AØ and NA. SM and NA provided funding acquisition, supervision, and guidance on data analysis, interpretation, and manuscript writing. All authors contributed to the article and approved the submitted version.
This work was supported by the Norwegian Taxonomy Initiative (Project No. 70184235/Ctenophores—native aliens in Norwegian waters) and the European Union’s Horizon 2020 Research and Innovation Programme (Grant Agreement No. 774499) as part of GoJelly (work package 2: “Driving mechanisms and predictions of jellyfish blooms”).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We wish to thank all the participants from the diving clubs for help with sampling and their enthusiasm as well as Nils Aukan and Vigdis Dahling for sharing their knowledge and underwater photographs.
Alamaru, A., Hoeksema, B. W., van der Meij, S. E., and Huchon, D. (2017). Molecular diversity of benthic ctenophores (Coeloplanidae). Sci. Rep. 7:6365. doi: 10.1038/s41598-017-06505-4
Attrill, M. J., Wright, J., and Edwards, M. (2007). Climate-related increases in jellyfish frequency suggest a more gelatinous future for the North Sea. Limnol. Oceanogr. 52, 480–485. doi: 10.4319/lo.2007.52.1.0480
Austen, G. E., Bindemann, M., Griffiths, R. A., and Roberts, D. L. (2016). Species identification by experts and non-experts: comparing images from field guides. Sci. Rep. 6:33634. doi: 10.1038/srep33634
Balázs, B., Mooney, P., Nováková, E., Bastin, L., and Arsanjani, J. J. (2021). “Data quality in citizen science,” in The Science of Citizen Science, eds K. Vohland, A. Land-Zandra, R. Lemmens, J. Perello, M. Ponti, R. Samson, et al. (Cham: Springer), 139.
Beaugrand, G. (2004). Monitoring marine plankton ecosystems. 1: description of an ecosystem approach based on plankton indicators. Mar. Ecol. Prog. Ser. 269, 69–81. doi: 10.3354/meps269069
Bell, S., Marzano, M., Cent, J., Kobierska, H., Podjed, D., Vandzinskaite, D., et al. (2008). What counts? Volunteers and their organisations in the recording and monitoring of biodiversity. Biodivers. Conserv. 17, 3443–3454. doi: 10.1007/s10531-008-9357-9
Biggs, J., Ewald, N., Valentini, A., Gaboriaud, C., Dejean, T., Griffiths, R. A., et al. (2015). Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus). Biodivers. Conserv. 183, 19–28. doi: 10.1016/j.biocon.2014.11.029
Bonney, R., Cooper, C. B., Dickinson, J., Kelling, S., Phillips, T. B., Rosenberg, K. V., et al. (2009). Citizen science: a developing tool for expanding science knowledge and scientific literacy. BioScience 59, 977–984. doi: 10.1525/bio.2009.59.11.9
Bonney, R., Shirk, J. L., Phillips, T. B., Wiggins, A., Ballard, H. L., Miller-Rushing, A. J., et al. (2014). Citizen science: next steps for citizen science. Science 343, 1436–1437. doi: 10.1126/science.1251554
Boudreau, S. A., and Yan, N. D. (2004). Auditing the accuracy of a volunteer-based surveillance program for an aquatic invader Bythotrephes. Environ. Monit. Assess. 91, 17–26. doi: 10.1023/B:EMAS.0000009228.09204.b7
Buxton, A. S., Groombridge, J. J., and Griffiths, R. A. (2018). Comparison of two citizen scientist methods for collecting pond water samples for environmental DNA studies. Citizen Sci. Theory Pract. 3:2. doi: 10.5334/cstp.151
Chandler, M., See, L., Copas, K., Bonde, A. M., López, B. C., Danielsen, F., et al. (2017). Contribution of citizen science towards international biodiversity monitoring. Biodivers. Conserv. 213, 280–294. doi: 10.1016/j.biocon.2016.09.004
Chow, S., Ueno, Y., Toyokawa, M., Oohara, I., and Takeyama, H. (2009). Preliminary analysis of length and gc content variation in the ribosomal first internal transcribed spacer (ITS1) of marine animals. Mar. Biotechnol. 11, 301–306. doi: 10.1007/s10126-008-9153-2
Christianson, L., Johnson, S., Schultz, D., and Haddock, S. (2021). Hidden diversity of Ctenophora revealed by new mitochondrial COI primers and sequences. Authorea [Preprints]. doi: 10.1111/1755-0998.13459
Danovaro, R., Carugati, L., Berzano, M., Cahill, A. E., Carvalho, S., Chenuil, A., et al. (2016). Implementing and innovating marine monitoring approaches for assessing marine environmental status. Front. Mar. Sci. 3:213. doi: 10.3389/fmars.2016.00213
Darwall, W. R., and Dulvy, N. K. (1996). An evaluation of the suitability of non-specialist volunteer researchers for coral reef fish surveys. Mafia Island, Tanzania—a case study. Biol. Conserv. 78, 223–231. doi: 10.1016/0006-3207(95)00147-6
Deidun, A., and Sciberras, A. (2017). Unearthing marine biodiversity through citizen science: the Spot the Jellyfish and the Spot the Alien Fish campaign case studies from the Maltese Islands (Central Mediterranean). Bull. Ent. Soc. Malta 9, 86–88.
Ficetola, G. F., Miaud, C., Pompanon, F., and Taberlet, P. (2008). Species detection using environmental DNA from water samples. Biol. Lett. 4, 423–425. doi: 10.1098/rsbl.2008.0118
Fore, L. S., Paulsen, K., and O’Laughlin, K. (2001). Assessing the performance of volunteers in monitoring streams. Freshw. Biol. 46, 109–123. doi: 10.1111/j.1365-2427.2001.00640.x
Garcia-Soto, C., Seys, J. J., Zielinski, O., Busch, J. A., Luna, S. I., Baez, J. C., et al. (2021). Marine Citizen Science: current state in Europe and new technological developments. Front. Mar. Sci. 8:241. doi: 10.3389/fmars.2021.621472
Geijzendorffer, I. R., Regan, E. C., Pereira, H. M., Brotons, L., Brummitt, N., Gavish, Y., et al. (2016). Bridging the gap between biodiversity data and policy reporting needs: an Essential Biodiversity Variables perspective. J. Appl. Ecol. 53, 1341–1350. doi: 10.1111/1365-2664.12417
Gorokhova, E., and Lehtiniemi, M. (2010). Reconsidering evidence for Mnemiopsis invasion in European waters. J. Plankton Res. 32, 93–95. doi: 10.1093/plankt/fbp105
Gorokhova, E., Lehtiniemi, M., Viitasalo-Frösen, S., and Haddock, S. H. (2009). Molecular evidence for the occurrence of ctenophore Mertensia ovum in the northern Baltic Sea and implications for the status of the Mnemiopsis leidyi invasion. Limnol. Oceanogr. 54, 2025–2033. doi: 10.4319/lo.2009.54.6.2025
Granhag, L., Majaneva, S., and Moller, L. F. (2012). First recordings of the ctenophore Euplokamis sp. (ctenophora, cydippida) in Swedish coastal waters and molecular identification of this genus. Aquat. Invasions 7, 455–463. doi: 10.3391/ai.2012.7.4.002
Greenwood, J. J. (2007). Citizens, science and bird conservation. J. Ornithol. 148, 77–124. doi: 10.1007/s10336-007-0239-9
Haddock, S. H. (2004). A golden age of gelata: past and future research on planktonic ctenophores and cnidarians. Hydrobiologia 530, 549–556. doi: 10.1007/s10750-004-2653-9
Hall, T. A. (1999). Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Nucleic Acids Symp. Ser. 41, 95–98.
Halsband, C., Majaneva, S., Hosia, A., Emaus, P. A., Gaardsted, F., Zhou, Q., et al. (2018). Jellyfish summer distribution, diversity and impact on fish farms in a Nordic fjord. Mar. Ecol. Prog. Ser. 591, 267–279. doi: 10.3354/meps12274
Harbison, G. R. (1985). “On the classification and evolution of the Ctenophora,” in The Origin and Relationships of Lower Invertebrates, The Systematic Association, Special, Vol. 28, eds S. C. Morris, J. D. George, R. Gibson, and H. M. Platt (Oxford: Claredon Press).
Harris, R. (2010). The L4 time-series: the first 20 years. J. Plankton Res. 32, 577–583. doi: 10.1093/plankt/fbq021
Hay, S. (2006). Marine ecology: gelatinous bells may ring change in marine ecosystems. Curr. Biol. 16, 679–682. doi: 10.1016/j.cub.2006.08.010
Hays, G. C., Richardson, A. J., and Robinson, C. (2005). Climate change and marine plankton. Trends Ecol. Evol. 20, 337–344. doi: 10.1016/j.tree.2005.03.004
Hinder, S. L., Manning, J. E., Gravenor, M. B., Edwards, M., Walne, A. W., Burkill, P. H., et al. (2012). Long-term changes in abundance and distribution of microzooplankton in the NE Atlantic and North Sea. J. Plankton Res. 34, 83–91. doi: 10.1093/plankt/fbr087
Hosia, A., and Falkenhaug, T. (2015). Invasive ctenophore Mnemiopsis leidyi in Norway. Mar. Biodivers. Rec. 8:e31.
IPBES (2019). in Global Assessment Report on Biodiversity and Ecosystem Services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services, eds E. S. Brondizio, J. Settele, S. Díaz, and H. T. Ngo, Bonn: IPBES Secretariat.
Jisming-See, S. W., Sing, K. W., and Wilson, J. J. (2016). DNA barcodes and citizen science provoke a diversity reappraisal for the “ring” butterflies of Peninsular Malaysia (Ypthima: satyrinae: nymphalidae: lepidoptera). Genome 59, 879–888. doi: 10.1139/gen-2015-0156
Johansson, M. L., Shiganova, T. A., Ringvold, H., Stupnikova, A. N., Heath, D. D., and MacIsaac, H. J. (2018). Molecular insights into the ctenophore genus Beroe in Europe: new species, spreading invaders. J. Hered. 109, 520–529. doi: 10.1093/jhered/esy026
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinformatics 20, 1160–1166. doi: 10.1093/bib/bbx108
Lawrence, A. (2006). No personal motive? Volunteers, biodiversity, and the false dichotomies of participation. Ethics Place Environ. 9, 279–298. doi: 10.1080/13668790600893319
Lewandowski, E., and Specht, H. (2015). Influence of volunteer and project characteristics on data quality of biological surveys. Biol. Conserv. 29, 713–723. doi: 10.1111/cobi.12481
Majaneva, S. (2014). Understanding the biodiversity and ecological importance of ctenophores – lessons from Arctic and Baltic Mertensia ovum. – W. & A. de Nottbeck Foundation. Sci. Rep. 41, 1–74.
Majaneva, S., and Majaneva, M. (2013). Cydippid ctenophores in the coastal waters of Svalbard: is it only Mertensia ovum? Polar Biol. 36, 1681–1686. doi: 10.1007/s00300-013-1377-6
Majaneva, S., Ringvold, H., Johansen, E., Østensen, M. A., and Hosia, A. (2021). Hiding in plain sight–Euplokamis dunlapae (Ctenophora) in Norwegian waters. J. Plankton Res. 43, 257–269.
Marambio, M., Lopez, L., Yahia, O. K. D., Yahia, N. D., Deidun, A., Piraino, S., et al. (2016). “The Medjelly App: a preventive and mitigation tool against jellyfish blooms involving a citizen science network,” in Proceedings of the 5th International Jellyfish Bloom Symposium, Barcelona.
Marizzi, C., Florio, A., Lee, M., Khalfan, M., Ghiban, C., Nash, B., et al. (2018). DNA barcoding: a first assessment of biodiversity in Marine Park by citizen scientists. PLoS One 13:e0199015. doi: 10.1371/journal.pone.0199015
Martins, V. Y., Christidis, L., Lloyd, D. J., and Pecl, G. (2016). Understanding drivers, barriers and information sourves for public participation in marine citizen science. J. Sci. Commun. 15:A02. doi: 10.22323/2.15020202
McConville, K. (2011). Towards a Comprehensive Taxonomic Key for the Phylum Ctenophora. Intern Papers, MBARI.
McManus, G. B., and Katz, L. A. (2009). Molecular and morphological methods for identifying plankton: what makes a successful marriage? J. Plankton Res. 31, 1119–1129. doi: 10.1093/plankt/fbp061
Mills, C. E., and Haddock, S. D. (2007). Ctenophores, Fourth Edn. Berkeley, CA: University of California press, 168–173.
Pauly, D., Graham, W., Libralato, S., Morissette, L., and Palomares, M. D. (2008). “Jellyfish in ecosystems, online databases, and ecosystem models,” in Jellyfish Blooms: Causes, Consequences, and Recent Advances, eds K. A. Pitt and J. E. Purcell (Dordrecht: Springer), 67–85. doi: 10.1007/978-1-4020-9749-2_5
Pereira, H. M., Ferrier, S., Walters, M., Geller, G. N., Jongman, R. H. G., Scholes, R. J., et al. (2013). Essential biodiversity variables. Science 339, 277–278. doi: 10.1126/science.1229931
Pocock, M. J. O., Chapman, D. S., Sheppard, L. J., and Roy, H. E. (2014). Choosing and Using Citizen Science: A Guide to When and How to Use Citizen Science to Monitor Biodiversity and the Environment. Wallingford: Centre for ecology and hydropogy, natural environment research council.
Podar, M., Haddock, S. H., Sogin, M. L., and Harbison, G. R. (2001). A molecular phylogenetic framework for the phylum Ctenophora using 18S rrna genes. Mol. Phylogenet. Evol. 21, 218–230. doi: 10.1006/mpev.2001.1036
Raskoff, K. A., Hopcroft, R. R., Kosobokova, K. N., Purcell, J. E., and Youngbluth, M. (2010). Jellies under ice: ROV observations from the Arctic 2005 Hidden Ocean Expedition. Deep Sea Res. Part II Top. Stud. Oceanogr. 57, 111–126. doi: 10.1016/j.dsr2.2009.08.010
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Shiganova, T. A., and Abyzova, G. A. (2021). Revision of Beroidae (Ctenophora) in the southern seas of Europe: systematics and distribution based on genetics and morphology. Zool. J. Linn. Soc. zlab021. doi: 10.1093/zoolinnean/zlab021
Simion, P., Bekkouche, N., Jager, M., Queinnec, E., and Manuel, M. (2015). Exploring the potential of small RNA subunit and ITS sequences for resolving phylogenetic relationships within the phylum Ctenophora. Zoology 118, 102–114. doi: 10.1016/j.zool.2014.06.004
Steininger, M. S., Hulcr, J., Šigut, M., and Lucky, A. (2015). Simple and efficient trap for bark and ambrosia beetles (Coleoptera: curculionidae) to facilitate invasive species monitoring and citizen involvement. J. Econ. Entomol. 108, 1115–1123. doi: 10.1093/jee/tov014
Theobald, E. J., Ettinger, A. K., Burgess, H. K., DeBey, L. B., Schmidt, N. R., Froehlich, H. E., et al. (2015). Global change and local solutions: tapping the unrealized potential of citizen science for biodiversity research. Biol. Conserv. 181, 236–244. doi: 10.1016/j.biocon.2014.10.021
Thibault-Botha, D., and Bowen, T. (2004). Impact of formalin preservation on Pleurobrachia bachei (Ctenophora). J. Exp. Mar. Biol. Ecol. 303, 11–17. doi: 10.1016/j.jembe.2003.10.017
Thiel, M., Penna-Díaz, M. A., Luna-Jorquera, G., Sala, S., Sellanes, J., and Stotz, W. (2014). Citizen scientists and marine research: volunteer participants, their contributions and projection for the future. Oceanogr. Mar. Biol. Annu. Rev. 52, 257–314.
Thomsen, P. F., Kielgast, J. O. S., Iversen, L. L., Wiuf, C., Rasmussen, M., Gilbert, M. T. P., et al. (2012). Monitoring endangered freshwater biodiversity using environmental DNA. Mol. Ecol. 21, 2565–2573. doi: 10.1111/j.1365-294X.2011.05418.x
Tulloch, A. I., Possingham, H. P., Joseph, L. N., Szabo, J., and Martin, T. G. (2013). Realising the full potential of citizen science monitoring programs. Biol. Conserv. 165, 128–138. doi: 10.1016/j.biocon.2013.05.025
Verity, P. G., and Smetacek, V. (1996). Organism life cycles, predation, and the structure of marine pelagic ecosystems. Mar. Ecol. Prog. Ser. 130, 277–293. doi: 10.3354/meps130277
Vohland, K., Land-Zandstra, A., Ceccaroni, L., Lemmens, R., Perelló, J., Ponti, M., et al. (2021). “The science of citizen science evolves. chapter 1,” in The Science of Citizen Science, eds K. Vohland, A. Land-Zandstra, L. Ceccaroni, R. Lemmens, J. Perelló, M. Ponti, et al. (Cham: Springer), 1–12. doi: 10.1007/978-3-030-58278-4_1
Keywords: DNA identification, Citizen science, Ctenophora, zooplankton, FTA cards®
Citation: Johansen E, Aberle N, Østensen M-A and Majaneva S (2021) Assessing the Value of a Citizen Science Approach for Ctenophore Identification. Front. Mar. Sci. 8:772851. doi: 10.3389/fmars.2021.772851
Received: 08 September 2021; Accepted: 25 October 2021;
Published: 19 November 2021.
Edited by:
Jin Sun, Ocean University of China, ChinaReviewed by:
Mattias Johansson, University of The Bahamas, BahamasCopyright © 2021 Johansen, Aberle, Østensen and Majaneva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sanna Majaneva, c2FubmFtYUBudG51Lm5v
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.