
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Mar. Sci., 10 January 2022
Sec. Ocean Observation
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.767443
 Sophie Clayton1*
Sophie Clayton1* Harriet Alexander2*
Harriet Alexander2* Jason R. Graff3
Jason R. Graff3 Nicole J. Poulton4
Nicole J. Poulton4 Luke R. Thompson5,6
Luke R. Thompson5,6 Heather Benway7
Heather Benway7 Emmanuel Boss8
Emmanuel Boss8 Adam Martiny9,10
Adam Martiny9,10In this article, we present Bio-GO-SHIP, a new ocean observing program that will incorporate sustained and consistent global biological ocean observations into the Global Ocean Ship-based Hydrographic Investigations Program (GO-SHIP). The goal of Bio-GO-SHIP is to produce systematic and consistent biological observations during global ocean repeat hydrographic surveys, with a particular focus on the planktonic ecosystem. Ocean plankton are an essential component of the earth climate system, form the base of the oceanic food web and thereby play an important role in influencing food security and contributing to the Blue Economy. Despite its importance, ocean biology is largely under-sampled in time and space compared to physical and chemical properties. This lack of information hampers our ability to understand the role of plankton in regulating biogeochemical processes and fueling higher trophic levels, now and in future ocean conditions. Traditionally, many of the methods used to quantify biological and ecosystem essential ocean variables (EOVs), measures that provide valuable information on the ecosystem, have been expensive and labor- and time-intensive, limiting their large-scale deployment. In the last two decades, new technologies have been developed and matured, making it possible to greatly expand our biological ocean observing capacity. These technologies, including cell imaging, bio-optical sensors and 'omic tools, can be combined to provide overlapping measurements of key biological and ecosystem EOVs. New developments in data management and open sharing can facilitate meaningful synthesis and integration with concurrent physical and chemical data. Here we outline how Bio-GO-SHIP leverages these technological advances to greatly expand our knowledge and understanding of the constituents and function of the global ocean plankton ecosystem.
The physics, chemistry, and biology of the ocean system are irrevocably interlinked. Marine life and biological processes, particularly within the plankton, drive the global biogeochemical cycling of climatically-important elements (e.g., C, O, N, P, Si, Fe). Additionally, phytoplankton form the base of the oceanic food web accounting for roughly half of global primary productivity (Field et al., 1998) and underpinning an important part of the Blue Economy which is projected to be worth $3 trillion by 2030 (Rayner et al., 2019; Estes et al., 2021). The measurement and chronicling of physical (e.g., salinity and temperature) and chemical ocean properties (e.g., dissolved oxygen, the carbonate system and nutrients) have been broadly and systematically incorporated into global repeat surveys for many decades. By comparison, ocean life and biological processes, which directly contribute to sustaining valuable ocean fisheries, are chronically under-sampled in both time and space, with the notable exception of satellite-derived estimates of near-surface phytoplankton biomass (McClain, 2009; Siegel et al., 2013). As a result of this under-sampling, many important questions in oceanography remain unanswered, and our ability to observe and detect marine ecosystem responses to global climate change is restricted (Mieszkowska et al., 2014).
A globally consistent effort to quantify and study plankton communities across ocean basins will transform our understanding of plankton biogeography, marine food webs, and the biological regulation of elemental cycles. To date, the majority of survey programs have focused on “bio-discovery,” targeting unique ocean environments in order to generate a catalog of ocean life, leading to the discovery of new biodiversity by the Global Ocean Sampling Expedition (Rusch et al., 2007), Tara Oceans (Carradec et al., 2018), and Malaspina (Salazar et al., 2015; Acinas et al., 2019). It has been recognized that a central need exists to coordinate and merge observations of biology with concurrently measured physical and chemical properties across large spatial and temporal scales in order to study and model feedback loops between plankton ecosystems and chemical and physical ocean processes. Consistent and fully integrated observations will provide a beyond-baseline understanding of global plankton and pelagic organism biogeography, the biological regulation of particle composition and elemental stoichiometry, linking surface plankton diversity with the downward particle flux and C storage, and the regulation of deep ocean biodiversity. We also envisage that biological tracers such as microbial community composition in the deep ocean will help to uncover physical transport pathways that are not well-constrained by hydrographic observations alone. With sustained observations, we will be able to identify how characteristic shifts in ocean plankton communities may act as “biosensors” for ocean changes (Ustick et al., 2021).
In this paper, we outline a plan for Bio-GO-SHIP1, a globally consistent biological ocean observing program that integrates the latest observing technologies into an existing program (Sloyan et al., 2019). Integrating Bio-GO-SHIP within the existing Global Ocean Ship-based Hydrographic Investigation Program (GO-SHIP)2 naturally bridges physical, chemical, and biological measurements. This synergy has the potential to progressively inform our understanding of plankton biodiversity, the impacts of plankton community structure and activity on chemical inventories, and the physical connectivity between communities residing in apparently distinct oceanic provinces. By integrating existing mature observational technologies, Bio-GO-SHIP directly addresses the existing data gap in the measurement of biological and ecosystem EOVs which is hampering our ability to understand the role of the plankton ecosystem in the global climate system, the carbon cycle, and marine food webs. Below we highlight existing and new technologies that meet the demands of Bio-GO-SHIP and discuss aspects of data management and community access. Finally, we describe how repeat biological sections fit into and augment existing ocean observing systems, and how they will support future developments of autonomous observing platforms.
GO-SHIP is the most recent iteration in a series of global hydrography programs dating back to the Geochemical Ocean Sections Study in the 1970s (GEOSECS; Moore, 1984), followed in the late 1980s by the Joint Global Ocean Flux Study (JGOFS; Fasham et al., 2001), and the World Ocean Circulation Experiment in the 1990s (WOCE; Woods, 1985). The principal scientific objectives for long-term ship-based repeat hydrography programs have two closely linked components. Firstly, they aim to understand and document the large-scale distribution of ocean properties, their changes, and the drivers of those changes. Secondly, they assess the functioning of a warmer and more stratified ocean with increased dissolved inorganic carbon (DIC), lower pH, changes in circulation and ventilation processes, altered water cycle, and shrinking sea-ice. GO-SHIP organizes a global repeat-occupation effort, whereby most major ocean regions are sampled every decade in order to observe global changes. The program coordinates concurrent measurements of a suite of key physical and chemical essential ocean variables (EOVs) throughout the full ocean water column. While GO-SHIP is an international program, it is funded and executed nationally. Key to GO-SHIP's success is international agreement between the national partners on standard data collection and curation methods and protocols.
GO-SHIP's repeat decadal observations of ocean physics and chemistry have provided critical constraints on anthropogenic changes in ocean heat content (Roemmich et al., 2007; Purkey and Johnson, 2010; Waugh et al., 2013), penetration of carbon (Gruber et al., 2019), shoaling of the calcium carbonate saturation depth (Feely et al., 2004), and loss of oxygen (Schmidtko et al., 2017), all of which have direct and serious implications for ocean life. Unfortunately, routine measurements of EOVs to characterize life in the ocean—including its composition, abundance, and changes in distribution—which are fundamental to our understanding of marine ecosystems (Lombard et al., 2019; Boss et al., 2020) are missing. Integrating routine measurements of biological and ecosystem EOVs into global repeat sections represents an important step forward for developing both a holistic understanding of the functioning of marine ecosystems, and a baseline from which changes over the coming decades can be observed.
The increasing availability of mature technologies to measure key biological EOVs offers a unique opportunity for a truly integrated, global repeat sampling program. One example is the Atlantic Meridional Transect (AMT), which is a repeat transect spanning the Atlantic Ocean (Robins and Aiken, 1996). The AMT Program has incorporated routine measurements of biological EOVs since its inception in 1995 and serves as a model for integrating biological data collection into repeat surveys. Data from the AMT Program has revealed relationships between the phytoplankton size distribution and nutrient concentrations across large scale oceanic gradients (Cermeno et al., 2008). Another significant finding enabled by AMT data, was that scattering measured from space, contrary to expectations, is primarily caused by particles > 1μm (Organelli et al., 2018). These insights into the links between plankton communities, particle dynamics and biogeochemical cycles from the AMT were only possible thanks to the combination of in situ biological sampling and bio-optical measurements.
There have been a number of biological efforts in collaboration with the GO-SHIP program during the last 5 years on cruises to the Atlantic, Indian, and Pacific Oceans (Larkin et al., 2021). These GO-SHIP transects incorporating biology have allowed for a systematic analysis of large-scale gradients in plankton genomic diversity. The most abundant marine phytoplankton, Prochlorococcus, has been shown to adapt to nutrient availability through gene gains and losses (Coleman and Chisholm, 2010). This biological feature was applied as a living biosensor for the elemental type and severity of nutrient stress, and combined with complementary chemical and hydrographic data highlights the feedbacks between marine microbial metabolisms, hydrography and ocean biogeochemistry (Figure 1, Ustick et al. 2021). This analysis is supported by past nutrient-amendment bottle experiments and models (Moore et al., 2013), but also has uncovered many previously unrecognized regions of nutrient stress, suggesting that nutrient stress biogeography might be tied to shifts in vertical mixing and the aeolian supply of iron (Martiny et al., 2019). Genomic data has also been used to infer variations in C cycling strategies between ocean regions (Raes et al., 2021). Finally, plankton biodiversity patterns observed in these studies significantly diverged from current theoretical predictions suggesting that we still lack a fundamental understanding of the drivers of planktonic biodiversity (Raes et al., 2018).
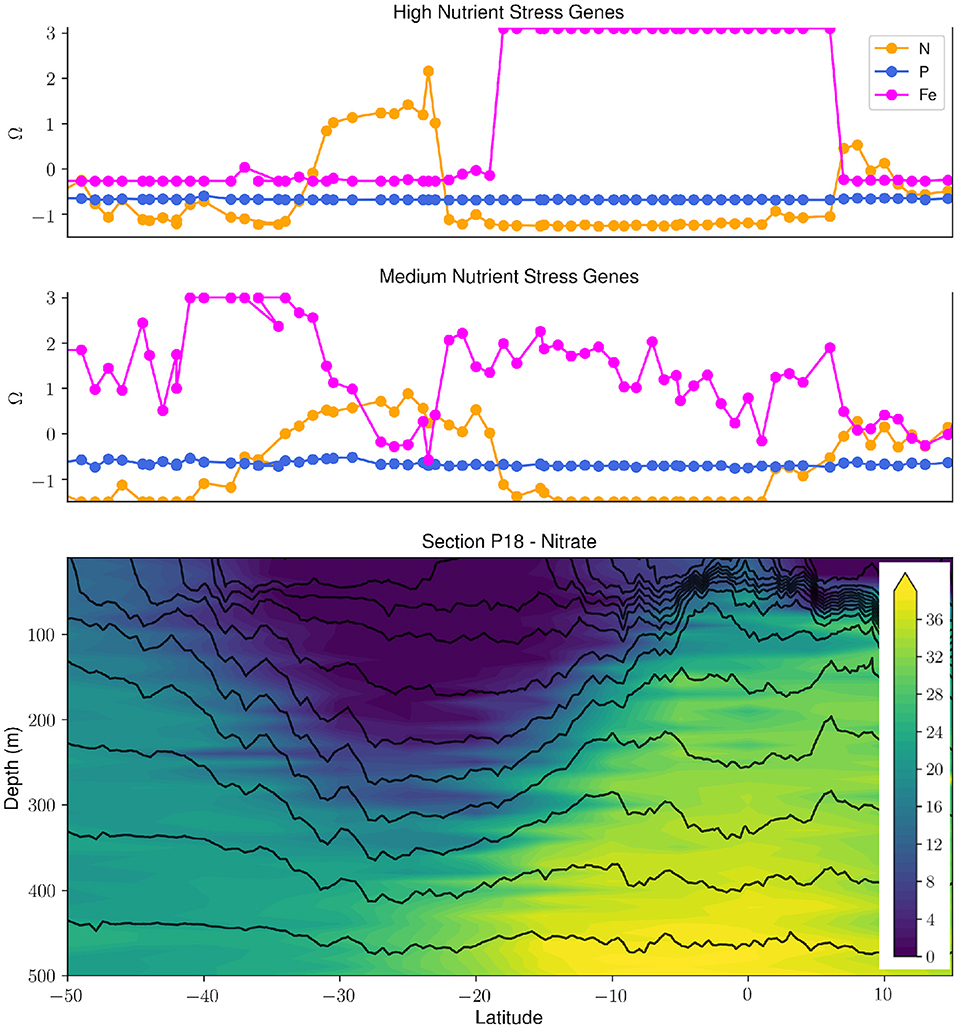
Figure 1. Synergy of biological, hydrographic and chemical data collected during the GO-SHIP P18 repeat section uncovers patterns and drivers of nutrient stress in Prochlorococcus. The upper two panels panel show the high and medium stress composite metrics (Ω) for N, Fe, and P (taken from Ustick et al., 2021). The bottom panel shows the nitrate concentration (μmol/kg) in the upper 500 m along the P18 transect with density contours (black contour lines). There is a clear relationship between the severity of N stress in Prochlorococcus and the depth of the nitracline. The P18 section is highlighted in red in Figure 2.
The elemental stoichiometry of ocean ecosystems is of fundamental importance to many biogeochemical processes such as the biological pump, nitrogen fixation, and the transfer of elements to higher trophic levels. Measurements of particulate organic matter on GO-SHIP transects demonstrated a clear latitudinal gradient in C:N:P (Garcia et al., 2018; Lee et al., 2021). Detailed hydrographic measurements from sections showed that ecosystems with a deep nutricline had elevated C:N and C:P. Furthermore, it was shown that shifts in genomic markers captured the impact of cellular nutrient limitation on C:N:P (Garcia et al., 2020). Finally, samples from GO-SHIP provided the first large-scale estimate of the carbon-to-oxygen remineralization ratio (Moreno et al., 2020). These efforts included using genomics to identify ecosystem functions and biodiversity patterns, the elemental stoichiometry of marine ecosystems, and the biogeography of the biological pump. Common to all these efforts are the clear linkages and integration between physical, chemical and biological observations.
Bio-GO-SHIP builds on the success of the previous efforts described above to establish an integrated biological observing program tied to the core GO-SHIP program. Crucial to this effort are the recommendations of the recently-convened SCOR Working Group 154 (“Integration of Plankton-Observing Sensor Systems to Existing Global Sampling Programs”; Boss et al. 2018) who assessed the feasibility of integrating different observational approaches for measuring biological and ecosystem EOVs into sustained global ocean observation programs, with a particular focus on GO-SHIP. Bio-GO-SHIP has been launched as a pilot project with funding from NOAA and NASA through the National Oceanographic Partnership Program. Although initially a US-based initiative, one of the goals of Bio-GO-SHIP is to develop and test standard community-approved protocols for global biological observations that can be applied broadly. Standard observational protocols, as already being employed in programs such as GeoTraces (Anderson et al., 2014), allow for the comparison and integration of data on biological EOVs across scientific cruises. This is currently not possible for many biological and ecosystem EOVs due to differing protocols. By leveraging the ship-time already committed for GO-SHIP sections, and by using sensors and instruments that can produce high-throughput underway data in conjunction with discrete sample collection, Bio-GO-SHIP is a cost-effective program that will likely add less than ~ 10% to the current operating costs of GO-SHIP and result in a large return on investment in terms of new scientific insights.
Detailed observations of the biological components of marine planktonic ecosystems have historically been restricted to targeted process studies of limited duration and spatial extent, such as the North Atlantic Aerosols and Marine Ecosystems Study (NAAMES; Behrenfeld et al., 2019; Penna and Gaube, 2019) and EXport Processes in the Ocean from Remote Sensing (EXPORTS; Siegel et al., 2016), or long term ocean time series including the Hawaii Ocean Timeseries (HOT; Karl and Church, 2014), the Bermuda Atlantic Time Series (BATS; Michaels and Knap, 1996), the CArbon Retention In A Colored Ocean Time-Series Program (CARIACO; Muller-Karger et al., 2019), and the Continuous Plankton Recorder Survey (Batten et al., 2019). Barriers to global-scale extended studies include the expense and labor involved with collecting, processing, and analyzing biological samples and associated properties. However, over the last two decades, many biological observational technologies have matured, been ground-tested and broadly applied, and are now capable of affordable and high-throughput sampling, making it feasible to mount a global biological sampling program in conjunction with existing repeat hydrographic survey programs. Additionally, by creating a consistent and community vetted protocols for global biological observations, we will be able to compare results across scientific cruises, which is currently not possible due to divergent observational protocols. For the first time, these observational tools coupled with consistent protocols and analytical pipelines, will make routine global repeat sections of biological and ecosystem EOVs a reality (Bojinski et al., 2014; Miloslavich et al., 2018).
Within the changing ocean environment, a baseline of sustained, consistent measurements is central to our ability to study, characterize, and monitor patterns in biodiversity and the downstream impacts of that diversity on ecosystem and biogeochemical processes. The GO-SHIP program currently includes the routine sampling of several biogeochemical EOVs, including dissolved oxygen, nutrients, dissolved organic carbon, and pCO2 (Sloyan et al., 2019). Here we provide a brief overview of the technologies and sampling strategies that will be included in the Bio-GO-SHIP program to study planktonic ecosystems, with a focus on biological EOVs central to pelagic ecosystems: specifically microbial, phytoplankton, zooplankton, and fish biomass and diversity (Miloslavich et al., 2018). By combining multiple sampling strategies, we plan to measure these variables in overlapping and complementary ways (Figure 2).
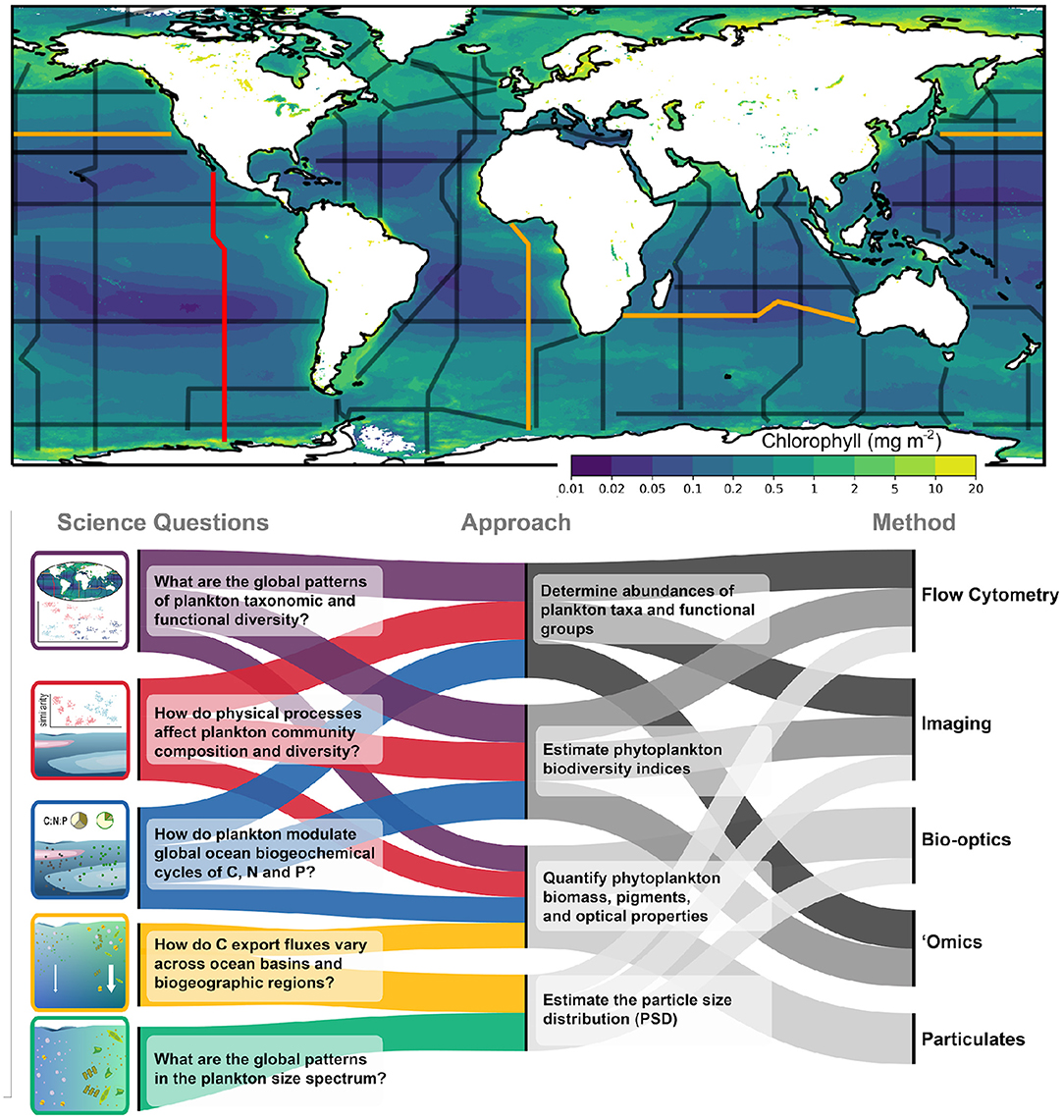
Figure 2. Biological sampling to be incorporated into GO-SHIP. The upper panel shows the GO-SHIP repeat sections overlain on global mean Chlorophyll (mg m−2) obtained from a MODIS mission composite spanning 2002–2021. Section P18 from which the data in Figure 1 was collected is highlighted in red. Highlighted in orange are sections I05, P02, and A13.5, which will be the first sections fully sampled as part of the Bio-GO-SHIP program in 2022–2023. The lower panel summarizes the key science questions that Bio-GO-SHIP addresses, the observational approaches and the range of different measurement types that will be used as part of the Bio-GO-SHIP program.
Microbial communities (comprised of prokaryotic and eukaryotic microbes and viruses) are centrally important to the functioning of the ocean. Using a combination of particulate, 'omic, and optical measures, we will be able to track their (relative) biomass and taxonomic composition. Flow cytometry will be used to target the smallest class of cells (0.5–10 μm), from which we can quantify cell numbers and extrapolate biomass, as well as coarse taxonomic composition. 'Omics are a well-developed and broadly used set of approaches to assess the diversity and function of microbial communities (Gilbert and Dupont, 2011; Sunagawa et al., 2015; Ustick et al., 2021). Metabarcoding, targeting conserved genes such as 16S or 18S, and shotgun metagenomics, which randomly samples DNA from the environment, shed light on the taxonomic composition of the microbial communities. Metatranscriptomics, which randomly samples the RNA from the environment, both provide information on the taxonomic and functional potential and a proxy for activity of the community.
Phytoplankton diversity, biomass and physiology will be assessed using a combination of 'omic, optical, and particulate measures. Phytoplankton span a wide range of size classes from < 1μm to > 2000μm (Finkel et al., 2009), necessitating the combination of multiple optical tools (flow cytometry, imaging flow cytometry, video imaging) to fully sample and estimate their contribution to biomass and cell size distribution (Lombard et al., 2019). By integrating bio-optical measures of fluorescence, absorption and backscatter at multiple wavelengths (e.g., SeaBird flbb, AC-S, and bb3) and Fast Repetition Rate Fluorometry (FRRF), we can also get information on the taxonomic composition of the community and their photo-physiological status (Vaillancourt, 2004; Suggett et al., 2009; Organelli et al., 2017). All of this Bio-GO-SHIP data will be critical to enable the testing of algorithms for biological (e.g., phytoplankton size distribution, PSD; phytoplankton function types, PFT) and biogeochemical (e.g., POC, primary productivity) proxies from remote sensing data, to refine them and to eventually produce better ones, particularly with the upcoming launch of the Plankton, Aerosol, Cloud, ocean Ecosystem (PACE) mission (Werdell et al., 2019). As with the heterotrophic microbial communities, 'omics provide a window into the fine-scale taxonomic and functional diversity of the eukaryotic plankton. In particular, 18S amplicon data has been successfully used to delineate protistan diversity within communities (de Vargas et al., 2015), while metatranscriptomics can reveal community function, activity and metabolism (Marchetti et al., 2012; Alexander et al., 2015; Carradec et al., 2018).
Pelagic consumers, fish and zooplankton, are central to supporting higher trophic levels, including humans. Given the increasing patchiness with organism size within the pelagic environment, common sampling techniques include extended trawling or net-based collections (e.g., MOCNESS). Such tools require a significant time investment and may not always be feasible on GO-SHIP or similar survey efforts. Rather, there are a variety of techniques that can be applied to derive proxies for abundance and taxonomic composition including imaging (e.g., Underwater Vision Profiler, UVP), acoustics and environmental DNA (eDNA). Active acoustics can be used to assess the abundance and community composition of large zooplankton and small fish (Howe et al., 2019). eDNA metabarcoding, a method which has become more commonly used in the conservation and ecological fields, amplifies marker genes to assess and estimate the presence and relative abundance of larger organisms (read: animals and multi-cellular plants) based on sloughed cells (Thomsen et al., 2012; Suter et al., 2020).
Each of the tools detailed above provide information on a particular property of the pelagic ecosystem that can be used to answer specific questions in oceanography: e.g., metabarcoding quantifies species composition, flow cytometry can illuminate shifts in community size structure over space and time, and bio-optics can used to infer rates of primary productivity underway. More so, it is in the combination and integration of multiple tools and approaches (and resulting combined datasets) that a more coherent understanding of the functioning of the ocean ecosystem can be built. As can be seen from the observational tools listed above, these distinct sampling approaches and technologies often result in overlap for quantities of interest (Figure 2). For instance, particle size data produced by multiple approaches or instruments [e.g., Laser In-Situ Scattering and Transmissometery (LISST), Flow Cytometry (FCM), Imaging Flow Cytobot (IFCB)] overlap across a portion of their datasets and provide a plankton size distribution ranging from microbes to large phytoplankton that could not be achieved from single instrument (Lombard et al., 2019). Multiple methods might be used to assess the taxonomic diversity of a community, with metabarcoding and metagenomics describing presence, metatranscriptomics highlighting active organisms, and high-throughput identifying the morphology. Similarly, 'omics targeting of nutrient-related genes with either metagenomics or metatranscriptomics can be combined with fast repetition rate fluorometry (FRRF) and particulate C:N:P to assess the dominant nutritional state of a community. Using complimentary techniques will provide a more accurate and comprehensive assessments of each EOV.
Advances in ocean technologies drive new discoveries, but maintaining pace with these advances is challenging and requires the attention of the community. The existing technologies that form the basis for the Bio-GO-SHIP program are currently mature, with a Technology Readiness level (TRL) between 7 and 9. However, although we are currently employing mature technologies, we are also anticipating the rapid development within areas of biological sampling. This race to keep pace with evolving technologies is particularly evident within the area of 'omics, as sequencing platforms rapidly shift and improve overall sequencing yield and potential contiguous read length (Levy and Myers, 2016). Looking only 10 years into the future, it is difficult to imagine what our potential capabilities might be within this sphere. As such, it is of paramount importance that we consider ways to ensure that the sequencing data we collect now is forward compatible with data we will collect 10 years from now. To ensure that this is possible, we must consider the best practices for intercalibration of these types of datasets. Additionally, there are great benefits to “biobanking” samples and “databanking” images for future analysis with the improved technology (Jarman et al., 2018). The collection and storage of samples and images in this way will be key to being able to detect future changes in the ocean ecosystem. We also expect that there will be improvements in bio-optics, imaging, cytometry and acoustics instrumentation and analysis, but these are likely to be more incremental given the relative maturity of those technologies.
Ship-based observations remain the “gold standard” in oceanographic exploration. However, these efforts are costly, and much effort has been put into developing autonomous observation technologies. Moored arrays and autonomous robotic instrumentation (e.g., Argo floats, gliders, and drifters) have enabled continuous remote observation of several physical oceanographic parameters (e.g., temperature, salinity, currents) and show great promise for the collection of chemical and biological data. Profiling Biogeochemical-Argo (BGC-Argo) floats instrumented with sensors that collect biogeochemically relevant measures including chlorophyll a, oxygen, nitrate, pH, backscatter, and multispectral downwelling irradiance as part of the Southern Ocean Carbon and Climate Observations and Modeling (SOCCOM) and Global Ocean Biogeochemistry Array (GO-BGC) programs, and whose data is comparable to those collected via shipboard observation, is a good example of the use of these technologies for biological investigations (Claustre et al., 2020). There is potential for developing remote and automated technologies to facilitate the collection of the biological data we detail in section 3.1 above. In particular, autonomous ecogenomic samplers, like the Environmental Sample Processor (ESP) (Scholin et al., 2017), have already demonstrated the possibility to sample DNA or RNA remotely (Ottesen et al., 2013). Additionally, new platforms that facilitate the fine-scale sampling of dissolved and particulate seawater biochemistry along vertical profiles, such as Clio (Breier et al., 2020), stand to expand the potential of automated collection of these parameters and decrease required ship time. Rigorous calibration and validation of such autonomous biological observing platforms will be key in their successful development and deployment. GO-SHIP cruises are currently used as a platform not only for the deployment of BGC-Argo profiling floats, but also provide key data for subsequent validation of the data provided by the on-board biogeochemical sensors and sampling (Bittig et al., 2019). We envision that Bio-GO-SHIP will provide similar synergies for deploying new automated biological sensors and, thus, accelerate the development and adoption of remote biological observational strategies.
Developing guidelines for the use of and consistent analysis of data produced by the technologies described above has been the focus of several working groups in recent years. This includes the SCOR WG 154 report (Boss et al., 2018, 2020), an Ocean Carbon Biogeochemistry (OCB)-sponsored small working group on Phytoplankton Taxonomy (“Data Standards and Practices for Taxon-Resolved Phytoplankton Observations,” Neeley et al., 2021), and an OCB-sponsored working group on Ocean Nucleic Acids 'Omics Intercalibration3. Although invaluable in building understanding, individual data types provide only a limited view of the whole system. Here, we outline an integrated approach to synthesizing physical and chemical oceanographic data (temperature, salinity, density, velocity, vertical profiles) with continuous (e.g., optics) or discrete (e.g., omics, FCM, particulate nutrients) biological measures. Beginning to holistically integrate these different data types will help us address fundamental questions in biological oceanography, inform the development of new biogeochemical models and enable their validation (Lombard et al., 2019). With increasing, high resolution biological measurements across the global ocean, the application of machine learning approaches to enable faster processing of data (e.g., the classification of images of plankton, Gonzalez et al., 2019), enable better extrapolation of processes in time and space (e.g. Flombaum et al., 2013; Selden et al., 2021) and ultimately allow for the interrogation of feedbacks between ocean physics, chemistry and biology.
Data sharing and dissemination is a challenging aspect of large-scale programs, like Bio-GO-SHIP, that collect high volumes of diverse data types. A first order issue is ensuring that all the data generated through the Bio-GO-SHIP program aligns with FAIR (Findable, Accessible, Interoperable, and Reusable) data management practices (Wilkinson et al., 2016). There is also a need to align this data with existing conventions in ocean and biological sciences (e.g., EOVs, ECVs, taxonomy). More so, ensuring that integrated, linked metadata with consistent vocabularies is present will be crucial. Additionally, as the program grows and develops, samples will be taken by many different individuals, so methodological documentation, data quality control, and intercalibration will be important.
There are many good models of data management and data sharing within the oceanographic community (e.g. Bowie and Tagliabue, 2018; Acinas et al., 2019). In particular, Tara Oceans implemented a highly successful system for data management and accessibility through a combination of robust collection of linked metadata and user-focused curated final data products made available alongside the raw data products (Villar et al., 2018). GO-SHIP's success in facilitating new scientific discoveries has been largely supported by a clear, consistent and well-documented set of data requirements and data submission timelines and policies4. GO-SHIP ranks data as being Level 1, 2, or 3, with Level 1 data being the highest priority for all GO-SHIP cruises. Level 2 data are highly desirable and should be collected when possible, and Level 3 data are considered ancillary and should only be collected if this can be done without interfering with the collection of the higher priority data. Biological measurements which are currently made as part of the GO-SHIP program (e.g., chlorophyll, HPLC pigments and primary production) are considered to be Level 3 measurements. It is envisaged that the Bio-GO-SHIP effort will raise some biological measurements to Level 2, and some eventually to Level 1. Bio-GO-SHIP will employ data management strategies used by these groups and integrate data-platforms to conform to the current best practices in data management and data requirements within the oceanographic community. Given the wide range of data types produced by the Bio-GO-SHIP program, the data will inevitably be curated within several data repositories (e.g., GenBank, BCO-DMO, CCHDO, NASA SeaBASS, OBIS). However, the Bio-GO-SHIP website provides a central repository that describes all of the data, lists its locations and DOIs and ensures redundancy for data discovery and access.
A mechanistic and coherent understanding of ecosystems and the capacity for marine organisms to evolve is crucial in light of climate change. While there are many well-developed physical-chemical ocean observing campaigns (e.g., GO-SHIP, GeoTraces, OSNAP, RAPID), few programs have incorporated the systematic collection of biological data across the global ocean. Tara Oceans, Malaspina, and the Global Ocean Survey represent a proof of concept and showcase the potential of global scale biological data collection. Still, a routine biological component of the global ocean observing system has yet to be established. The importance of biological measures at a global scale is undeniable and recognized as such by the Global Ocean Observing System (GOOS) EOVs. The maturation of sequencing, bio-optical, and high-throughput imaging technologies over the past two decades make global-scale systematic repeated biological observations tractable.
Ultimately, we envision that Bio-GO-SHIP will not be unique or operate in isolation and the more frequently these types of biological observations are incorporated into global-scale studies (e.g., BioGeoTraces, Biller et al., 2018) the better. The scientific community studying these highly complex and dynamic systems will benefit from greater volumes of consistent, high-quality data. More measurements, regardless of the program name, will ultimately accelerate discovery. A key component of the Bio-GO-SHIP program, as with GO-SHIP, is the goal to rapidly generate open access data for the community. Open data democratizes the scientific process and allows for greater participation in oceanography within and beyond the scientific community.
Bio-GO-SHIP augments the existing GO-SHIP program by collecting core biological measurements in a globally consistent and integrated program. Building on preceding biological survey programs and incorporating the use of high-throughput underway sampling, optical and imaging systems, Bio-GO-SHIP will generate data at higher spatial resolution than is typically achieved in other global scale sampling programs. The program is being developed in a highly interdisciplinary environment, working directly with physical and chemical oceanographers to scale up the biological observational strategies generally reserved for targeted process studies to global scale repeat sections. The technologies now exist to expand the core measurements of global hydrographic programs to build our understanding of interacting physical-chemical-biological processes and feedbacks that control and mediate the global biogeochemical cycles and link surface and deep ocean ecosystems. Although difficult to quantify, the benefits derived from making such consistent and sustained biological ocean observations will likely far outweigh the cost of establishing and maintaining the Bio-GO-SHIP program into the future (Rayner et al., 2019).
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
SC, HA, AM, NP, and JG contributed to conception of the manuscript. SC and HA drafted the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
The Bio-GO-SHIP pilot program was funded under the National Oceanographic Partnership Program as an inter-agency partnership between NOAA and NASA, with the US Integrated Ocean Observing System and NOAA's Global Ocean Monitoring and Observing program (HA, SC, JG, AM, and NP). HA was supported by a WHOI Independent Research and Development award. AM was supported by funding from NSF OCE-1848576 and 1948842 and NASA 80NSSC21K1654. JG was funded by NASA from grants 80NSSC17K0568 and NNX15AAF30G. LT was supported by award NA06OAR4320264 06111039 to the Northern Gulf Institute by NOAA's Office of Oceanic and Atmospheric Research, U.S. Department of Commerce.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acinas, S. G., Sanchez, P., Salazar, G., Cornejo-Castillo, F. M., Sebastian, M., Logares, R., et al. (2019). Metabolic architecture of the deep ocean microbiome. bioRxiv. doi: 10.1101/635680
Alexander, H., Rouco, M., Haley, S. T., Wilson, S. T., Karl, D. M., and Dyhrman, T. S. (2015). Functional group-specific traits drive phytoplankton dynamics in the oligotrophic ocean. Proc. Natl. Acad. Sci. U.S.A. 112, E5972–E5979. doi: 10.1073/pnas.1518165112
Anderson, R. F., Mawji, E., Cutter, G. A., Measures, C. I., and Jeandel, C. (2014). Geotraces: changing the way we explore ocean chemistry. Oceanography 27, 50–61. doi: 10.5670/oceanog.2014.07
Batten, S. D., Abu-Alhaija, R., Chiba, S., Edwards, M., Graham, G., Jyothibabu, R., et al. (2019). A global plankton diversity monitoring program. Front. Mar. Sci. 6:321. doi: 10.3389/fmars.2019.00321
Behrenfeld, M. J., Moore, R. H., Hostetler, C. A., Graff, J., Gaube, P., Russell, L. M., et al. (2019). The north atlantic aerosol and marine ecosystem study (NAAMES): science motive and mission overview. Front. Mar. Sci. 6:122. doi: 10.3389/fmars.2019.00122
Biller, S. J., Berube, P. M., Dooley, K., Williams, M., Satinsky, B. M., Hackl, T., et al. (2018). Marine microbial metagenomes sampled across space and time. Sci. Data. 5, 1–7. doi: 10.1038/sdata.2018.176
Bittig, H. C., Maurer, T. L., Plant, J. N., Schmechtig, C., Wong, A. P., Claustre, H., et al. (2019). A BGC-argo guide: planning, deployment, data handling and usage. Front. Mar. Sci. 6, 502. doi: 10.3389/fmars.2019.00502
Bojinski, S., Verstraete, M., Peterson, T. C., Richter, C., Simmons, A., and Zemp, M. (2014). The concept of essential climate variables in support of climate research, applications, and policy. Bull. Am. Meteorol. Soc. 95, 1431–1443. doi: 10.1175/BAMS-D-13-00047.1
Boss, E., Waite, A., Muller-Karger, F., Yamazaki, H., Wanninkhof, R., and Uitz, J. (2018), Beyond chlorophyll fluorescence: the time is right to expand biological measurements in ocean observing programs. Limnol. Ocean. Bulletin. 27, 89–90. doi: 10.1002/lob.10243.
Boss, E., Waite, A. M., Uitz, J., Acinas, S. G., Sosik, H. M., Fennel, K., et al. (2020). Recommendations for Plankton Measurements on the GO-SHIP Program With Relevance to Other Sea-Going Expeditions. SCOR Working Group 154 GO-SHIP Report. Scientific Committee on Oceanic Research.
Bowie, A., and Tagliabue, A. (2018). Geotraces data products: standardising and linking ocean trace element and isotope data at a global scale. Elements Int. Mag. Mineral. Geochem. Petrol. 14. 436–437. doi: 10.2138/gselements.14.6.436
Breier, J. A., Jakuba, M. V., Saito, M. A., Dick, G. J., Grim, S. L., Chan, E. W., et al. (2020). Revealing ocean-scale biochemical structure with a deep-diving vertical profiling autonomous vehicle. Sci. Robot. 5:eabc7104. doi: 10.1126/scirobotics.abc7104
Carradec, Q., Pelletier, E., Da Silva, C., Alberti, A., Seeleuthner, Y., Blanc-Mathieu, R., et al. (2018). A global ocean atlas of eukaryotic genes. Nat. Commun. 9:373. doi: 10.1038/s41467-017-02342-1
Cermeno, P., Maranin, E., Harbour, D., Figueira, F. G., Crespo, B. G., Huete-Ortega, M., et al. (2008). Resource levels, allometric scaling of population abundance, and marine phytoplankton diversity. Limnol. Oceanogr. 53, 312–318. doi: 10.4319/lo.2008.53.1.0312
Claustre, H., Johnson, K. S., and Takeshita, Y. (2020). Observing the global ocean with biogeochemical-argo. Annu. Rev. Mar. Sci. 12, 23–48. doi: 10.1146/annurev-marine-010419-010956
Coleman, M. L., and Chisholm, S. W. (2010). Ecosystem-specific selection pressures revealed through comparative population genomics. Proc. Natl. Acad. Sci. U.S.A. 107, 18634–18639. doi: 10.1073/pnas.1009480107
de Vargas, C., Audic, S., Henry, N., Decelle, J., Mahe, F., Logares, R., et al. (2015). Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605–1261605. doi: 10.1126/science.1261605
Estes, M., Anderson, C., Appeltans, W., Bax, N., Bednarsek, N., Canonico, G., et al. (2021). Enhanced monitoring of life in the sea is a critical component of conservation management and sustainable economic growth. Mar. Policy 132:104699. doi: 10.1016/j.marpol.2021.104699
Fasham, M. J., Balino, B. M., Bowles, M. C., Anderson, R., Archer, D., Bathmann, U., et al. (2001). A new vision of ocean biogeochemistry after a decade of the joint global ocean flux study (JGOFS). AMBIO J. Human Environ. 2001, 4–31.
Feely, R. A., Sabine, C. L., Lee, K., Berelson, W., Kleypas, J., Fabry, V. J., et al. (2004). Impact of anthropogenic Co2 on the CaCo3 system in the oceans. Science 305, 362–366. doi: 10.1126/science.1097329
Field, C. B., Behrenfeld, M. J., Randerson, J. T., and Falkowski, P. (1998). Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240. doi: 10.1126/science.281.5374.237
Finkel, Z. V., Beardall, J., Flynn, K. J., Quigg, A., Rees, T. A. V., and Raven, A. J. (2009). Phytoplankton in a changing world: cell size and elemental stoichiometry. J. Plankton Res. 32, 119–137. doi: 10.1093/plankt/fbp098
Flombaum, P., Gallegos, J. L., Gordillo, R. A., Rincon, J., Zabala, L. L., Jiao, N., et al. (2013). Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 110, 9824–9829. doi: 10.1073/pnas.1307701110
Garcia, C. A., Baer, S. E., Garcia, N. S., Rauschenberg, S., Twining, B. S., Lomas, M. W., et al. (2018). Nutrient supply controls particulate elemental concentrations and ratios in the low latitude Eastern Indian Ocean. Nat. Commun. 9, 1–10. doi: 10.1038/s41467-018-06892-w
Garcia, C. A., Hagstrom, G. I., Larkin, A. A., Ustick, L. J., Levin, S. A., Lomas, M. W., et al. (2020). Linking regional shifts in microbial genome adaptation with surface ocean biogeochemistry. Philos. Trans. R. Soc. B 375:20190254. doi: 10.1098/rstb.2019.0254
Gilbert, J. A., and Dupont, C. L. (2011). Microbial metagenomics: beyond the genome. Annu. Rev. Mar. Sci. 3, 347–371. doi: 10.1146/annurev-marine-120709-142811
Gonzalez, P., Castano, A., Peacock, E. E., Díez, J., Del Coz, J. J., and Sosik, M. H. (2019). Automatic plankton quantification using deep features. J. Plankton Res. 41, 449–463. doi: 10.1093/plankt/fbz023
Gruber, N., Clement, D., Carter, B. R., Feely, R. A., Van Heuven, S., Hoppema, M., et al. (2019). The oceanic sink for anthropogenic Co2 from 1994 to 2007. Science 363, 1193–1199. doi: 10.1126/science.aau5153
Howe, B. M., Miksis-Olds, J., Rehm, E., Sagen, H., Worcester, P. F., and Haralabus, G. (2019). Observing the oceans acoustically. Front. Mar. Sci. 6:426. doi: 10.3389/fmars.2019.00426
Jarman, S. N., Berry, O., and Bunce, M. (2018). The value of environmental DNA biobanking for long-term biomonitoring. Nat. Ecol. Evol. 2, 1192–1193. doi: 10.1038/s41559-018-0614-3
Karl, D. M., and Church, M. J. (2014). Microbial oceanography and the hawaii ocean time-series programme. Nat. Rev. Microbiol. 12, 699–713. doi: 10.1038/nrmicro3333
Larkin, A. A., Garcia, C. A., Garcia, N., Brock, M. L., Lee, J. A., Ustick, L. J., et al. (2021). High spatial resolution global ocean metagenomes from bio-GO-SHIP repeat hydrography transects. Sci. Data 8, 1–6. doi: 10.1038/s41597-021-00889-9
Lee, J. A., Garcia, C. A., Larkin, A. A., Carter, B. R., and Martiny, C. A. (2021). Linking a latitudinal gradient in ocean hydrography and elemental stoichiometry in the Eastern Pacific Ocean. Glob. Biogeochem. Cycles 35:e2020GB006622. doi: 10.1029/2020GB006622
Levy, S. E., and Myers, R. M. (2016). Advancements in next-generation sequencing. Annu. Rev. Genom. Hum. Genet. 17, 95–115. doi: 10.1146/annurev-genom-083115-022413
Lombard, F., Boss, E., Waite, A. M., Uitz, J., Stemmann, L., Sosik, H. M., et al. (2019). Globally consistent quantitative observations of planktonic ecosystems. Front. Mar. Sci. 6:196. doi: 10.3389/fmars.2019.00196
Marchetti, A., Schruth, D. M., Durkin, C. A., Parker, M. S., Kodner, R. B., Berthiaume, C. T., et al. (2012). Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc. Natl. Acad. Sci. U.S.A. 109, E317–E325. doi: 10.1073/pnas.1118408109
Martiny, A. C., Lomas, M. W., Fu, W., Boyd, P. W., Chen, L., Cutter, G. A., et al. (2019). Biogeochemical controls of surface ocean phosphate. Sci. Adv. 5:eaax0341. doi: 10.1126/sciadv.aax0341
McClain, C. R. (2009). A decade of satellite ocean color observations. Annu. Rev. Mar. Sci. 1, 19–42. doi: 10.1146/annurev.marine.010908.163650
Michaels, A. F., and Knap, A. H. (1996). Overview of the U.S. JGOFS Bermuda Atlantic time-series study and the hydrostation s program. Deep Sea Res. II Top. Stud. Oceanogr. 43, 157–198. doi: 10.1016/0967-0645(96)00004-5
Mieszkowska, N., Sugden, H., Firth, L. B., and Hawkins, J. S. (2014). The role of sustained observations in tracking impacts of environmental change on marine biodiversity and ecosystems. Philos. Trans. A Math. Phys. Eng. Sci. 372:20130339. doi: 10.1098/rsta.2013.0339
Miloslavich, P., Bax, N. J., Simmons, S. E., Klein, E., Appeltans, W., Aburto-Oropeza, O., et al. (2018). Essential ocean variables for global sustained observations of biodiversity and ecosystem changes. Glob. Change Biol. 24, 2416–2433. doi: 10.1111/gcb.14108
Moore, C. M., Mills, M. M., Arrigo, K. R., Berman-Frank, I., Bopp, L., Boyd, P. W., et al. (2013). Processes and patterns of oceanic nutrient limitation. sNat. Geosci. 6, 701–710. doi: 10.1038/ngeo1765
Moore, W. S. (1984). Review of the GEOSECS project. Nuclear Instrum. Methods Phys. Res. 223, 459–465. doi: 10.1016/0167-5087(84)90692-6
Moreno, A. R., Garcia, C. A., Larkin, A. A., Lee, J. A., Wang, W.-L., Moore, J. K., et al. (2020). Latitudinal gradient in the respiration quotient and the implications for ocean oxygen availability. Proc. Natl. Acad. Sci. U.S.A. 117, 22866–22872. doi: 10.1073/pnas.2004986117
Muller-Karger, F. E., Astor, Y. M., Benitez-Nelson, C. R., Buck, K. N., Fanning, K. A., Lorenzoni, L., et al. (2019). The scientific legacy of the cariaco ocean time-series program. Annu. Rev. Mar. Sci. 11, 413–437. doi: 10.1146/annurev-marine-010318-095150
Neeley, A., Beaulieu, S., Proctor, C., Cetinic, I., Futrelle, J., Ramos-Santos, I., et al. (2021). Standards and practices for reporting plankton and other particle observations from images. doi: 10.25607/OBP-1634
Organelli, E., Dall'Olmo, G., Brewin, R. J., Tarran, G. A., Boss, E., and Bricaud, A. (2018). The open-ocean missing backscattering is in the structural complexity of particles. Nat. Commun. 9, 1–11. doi: 10.1038/s41467-018-07814-6
Organelli, E., Nuccio, C., Lazzara, L., Uitz, J., Bricaud, A., and Massi, L. (2017). On the discrimination of multiple phytoplankton groups from light absorption spectra of assemblages with mixed taxonomic composition and variable light conditions. Appl. Opt. 56, 3952–3958. doi: 10.1364/AO.56.003952
Ottesen, E. A., Young, C. R., Eppley, J. M., Ryan, J. P., Chavez, F. P., Scholin, C. A., et al. (2013). Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc. Natl. Acad. Sci. U.S.A. 110, E488–E497. doi: 10.1073/pnas.1222099110
Penna, A. D., and Gaube, P. (2019). Overview of (sub)mesoscale ocean dynamics for the NAAMES field program. Front. Mar. Sci. 6:384. doi: 10.3389/fmars.2019.00384
Purkey, S. G., and Johnson, G. C. (2010). Antarctic bottom water warming between the 1990s and the 2000s: contributions to global heat and sea level rise budgets. J. Clim. 23, 6336–6351. doi: 10.1175/2010JCLI3682.1
Raes, E. J., Bodrossy, L., Van De Kamp, J., Bissett, A, Ostrowski, M., Brown, M. V., et al. (2018). Oceanographic boundaries constrain microbial diversity gradients in the south pacific ocean. Proc. Natl. Acad. Sci. U.S.A. 115, E8266–E8275. doi: 10.1073/pnas.1719335115
Raes, E. J., Karsh, K., Sow, S. L., Ostrowski, M., Brown, M. V., van de Kamp, J., et al. (2021). Metabolic pathways inferred from a bacterial marker gene illuminate ecological changes across south pacific frontal boundaries. Nat. Commun. 12, 1–12. doi: 10.1038/s41467-021-22409-4
Rayner, R., Jolly, C., and Gouldman, C. (2019). Ocean observing and the blue economy. Front. Mar. Sci. 6:0. doi: 10.3389/fmars.2019.00330
Robins, D., and Aiken, J. (1996). The Atlantic Meridional Transect: an oceanographic research programme to investigate physical, chemical, biological and optical variables of the Atlantic ocean. Underwater Technol. 21, 8–14. doi: 10.3723/175605496783328529
Roemmich, D., Gilson, J., Davies, R., Sutton, P., Wijffels, S., and Riser, S. (2007). Decadal spin up of the deep subtropical gyre in the South Pacific. J. Phys. Oceanogr. 37, 162–173. doi: 10.1175/JPO3004.1
Rusch, D. B., Halpern, A. L., Sutton, G., Heidelberg, K. B., Williamson, S., Yooseph, S., et al. (2007). The sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 5:e77. doi: 10.1371/journal.pbio.0050077
Salazar, G., Cornejo-Castillo, F. M., Benítez-Barrios, V., Fraile-Nuez, E., Alvarez-Salgado, X. A., Duarte, C. M., et al. (2015). Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J. 10, 596–608. doi: 10.1038/ismej.2015.137
Schmidtko, S., Stramma, L., and Visbeck, (2017). Decline in global oceanic oxygen content during the past five decades. Nature 542, 335–339. doi: 10.1038/nature21399
Scholin, C., Birch, J., Jensen, S., Marin, R. III, Massion, E., Pargett, D., et al. (2017). The quest to develop ecogenomic sensors: A 25-year history of the environmental sample processor (ESP) as a case study. Oceanography 30, 100–113. doi: 10.5670/oceanog.2017.427
Selden, C. R., Chappell, P. D., Clayton, S., Macías-Tapia, A., Bernhardt, P. W., and Mulholland, R. M. (2021). A coastal n2 fixation hotspot at the cape hatteras front: elucidating spatial heterogeneity in diazotroph activity via supervised machine learning. Limnol. Oceanogr. 66, 1832–1849. doi: 10.1002/lno.11727
Siegel, D., Behrenfeld, M. J., Maritorena, S., McClain, C. R., Antoine, D., Bailey, S. W., et al. (2013). Regional to global assessments of phytoplankton dynamics from the seawifs mission. Remote Sens. Environ. 135, 77–91. doi: 10.1016/j.rse.2013.03.025
Siegel, D. A., Buesseler, K. O., Behrenfeld, M. J., Benitez-Nelson, C. R., Boss, E., Brzezinski, M. A., et al. (2016). Prediction of the export and fate of global ocean net primary production: the EXPORTS science plan. Front. Mar. Sci. 3:22. doi: 10.3389/fmars.2016.00022
Sloyan, B. M., Wanninkhof, R., Kramp, M, Johnson, G. C., Talley, L. D., Tanhua, T., et al. (2019). The global ocean ship-based hydrographic investigations program (GO-SHIP): a platform for integrated multidisciplinary ocean science. Front. Mar. Sci. 6:445. doi: 10.3389/fmars.2019.00445
Suggett, D., Moore, C., Hickman, A., and Geider, R. (2009). Interpretation of fast repetition rate (FRR) fluorescence: signatures of phytoplankton community structure versus physiological state. Mar. Ecol. Prog. Ser. 376, 1–19. doi: 10.3354/meps07830
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., et al. (2015). Structure and function of the global ocean microbiome. Science 348, 1261359–1261359. doi: 10.1126/science.1261359
Suter, L., Polanowski, A. M., Clarke, L. J., Kitchener, J. A., and Deagle, E. B. (2020). Capturing open ocean biodiversity: comparing environmental DNA metabarcoding to the continuous plankton recorder. Mol Ecol. 30, 3140–3157. doi: 10.1111/mec.15587
Thomsen, P. F., Kielgast, J., Iversen, L. L., Müller, P. R., Rasmussen, M., and Willerslev, E. (2012). Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE 7:e41732. doi: 10.1371/journal.pone.0041732
Ustick, L. J., Larkin, A. A., Garcia, C. A., Garcia, N. S., Brock, M. L., Lee, J. A., et al. (2021). Metagenomic analysis reveals global-scale patterns of ocean nutrient limitation. Science 372, 287–291. doi: 10.1126/science.abe6301
Vaillancourt, R. D. (2004). Light backscattering properties of marine phytoplankton: relationships to cell size, chemical composition and taxonomy. J. Plankton Res. 26, 191–212. doi: 10.1093/plankt/fbh012
Villar, E., Vannier, T., Vernette, C., Lescot, M., Cuenca, M., Alexandre, A., et al. (2018). The ocean gene atlas: exploring the biogeography of plankton genes online. Nucleic Acids Res. 46, W289–W295. doi: 10.1093/nar/gky376
Waugh, D. W., Primeau, F., DeVries, T., and Holzer, M. (2013). Recent changes in the ventilation of the southern oceans. Science 339, 568–570. doi: 10.1126/science.1225411
Werdell, P. J., Behrenfeld, M. J., Bontempi, P. S., Boss, E., Cairns, B., Davis, G. T., et al. (2019). The plankton, aerosol, cloud, ocean ecosystem mission: status, science, advances. Bull. Am. Meteorol. Soc. 100, 1775–1794. doi: 10.1175/BAMS-D-18-0056.1
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., et al. (2016). The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3:160018. doi: 10.1038/sdata.2016.18
Keywords: biological oceanography, plankton ecosystems, ocean observing, repeat hydrography, GO-SHIP program
Citation: Clayton S, Alexander H, Graff JR, Poulton NJ, Thompson LR, Benway H, Boss E and Martiny A (2022) Bio-GO-SHIP: The Time Is Right to Establish Global Repeat Sections of Ocean Biology. Front. Mar. Sci. 8:767443. doi: 10.3389/fmars.2021.767443
Received: 30 August 2021; Accepted: 06 December 2021;
Published: 10 January 2022.
Edited by:
Laura Lorenzoni, National Aeronautics and Space Administration (NASA), United StatesReviewed by:
Lachlan McKinna, Go2Q Pty Ltd, AustraliaCopyright © 2022 Clayton, Alexander, Graff, Poulton, Thompson, Benway, Boss and Martiny. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sophie Clayton, c2NsYXl0b25Ab2R1LmVkdQ==; Harriet Alexander, aGFsZXhhbmRlckB3aG9pLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.