
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 04 February 2022
Sec. Marine Biology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.767024
This article is part of the Research TopicThe Philippine Seas: Biodiversity and Ecological Impacts of Natural and Anthropogenic stressors in Tropical Reef SystemsView all 14 articles
 Zabrina Bernice L. Malto1
Zabrina Bernice L. Malto1 Garry A. Benico1,2
Garry A. Benico1,2 Jeremiah D. Batucan1
Jeremiah D. Batucan1 James Dela Cruz3
James Dela Cruz3 Marc Lawrence J. Romero3
Marc Lawrence J. Romero3 Rhodora V. Azanza1
Rhodora V. Azanza1 Lilibeth A. Salvador-Reyes1*
Lilibeth A. Salvador-Reyes1*Surveillance and characterization of emerging marine toxins and toxigenic dinoflagellates are warranted to evaluate their associated health risks. Here, we report the occurrence of the ciguatera poisoning-causative dinoflagellate Gambierdiscus balechii in the Philippines. Toxin production and chemical diversity of secondary metabolites in G. balechii GtoxSAM092414, G. balechii Gtox112513, and the recently reported Gambierdiscus carpenteri Gam1BOL080513 were assessed using targeted and untargeted UPLC-MS/MS analysis and radioligand receptor-binding assay (RBA). 44-methylgambierone was produced by all three strains, albeit with different levels based on RBA and UPLC-HRMS/MS analysis. The fatty acid composition was similar in all strains, while subtle differences in monosaccharide content were observed, related to the collection site rather than the species. Molecular networking using the GNPS database identified 45 clusters belonging to at least ten compound classes, with terpene glycosides, carbohydrate conjugates, polyketides, and macrolides as major convergence points. Species-specific peptides and polyhydroxylated compounds were identified in G. balechii GtoxSAM092414 and G. carpenteri Gam1BOL080513, respectively. These provide a glimpse of the uncharacterized biosynthetic potential of benthic dinoflagellates and highlight the intricate and prolific machinery for secondary metabolites production in these organisms.
Ciguatera poisoning (CP) is one of the most common foodborne diseases associated with seafood consumption globally, having approximately 10,000–50,000 cases annually (Dickey and Plakas, 2010; Friedman et al., 2017; World Health Organization, 2020). Since 1988, 123 and 274 confirmed and suspected cases of CP, respectively, have been reported in the Philippines (Yñiguez et al., 2021). A survey of reef fishes from the Visayan and Sibuyan Seas in the Philippines showed that 4.46% were positive for ciguatoxins based on a mouse bioassay (Montojo et al., 2020). The limited surveillance of CP-causative organisms and associated toxins may contribute to underreporting of CP cases in the Philippines.
Benthic dinoflagellates belonging to the genera Gambierdiscus, Coolia, and Fukuyoa are recognized as the toxin producers linked to CP (Berdalet and Tester, 2018; Murray et al., 2020). Grazing by invertebrates and herbivorous fishes allows the benthic dinoflagellates and toxins to enter the food chain (Munday et al., 2017; Holmes et al., 2021). While fishes are the main vectors of CP, mollusks, crustaceans, and echinoderms are also linked to CP (Munday et al., 2017; Holmes et al., 2021).
The associated symptoms of CP, such as gastrointestinal, neurological, and cardiovascular symptoms and paradoxical dysesthesia (temperature reversal) (Bagnis et al., 1979), can be traced to the cellular effects of ladder-shaped polyether toxins collectively called ciguatoxins (CTX). CTX are lipophilic molecules that biotransform and bioaccumulate in fish. To date, more than 50 CTX analogs have been identified from fishes and benthic dinoflagellates, grouped as Pacific (P-CTX), Caribbean (C-CTX), and Indian (I-CTX) (Soliño and Costa, 2018; Chinain et al., 2020). CTX are potent activators of voltage-gated sodium channels and show significant toxicity in mice when given orally (Holmes et al., 2021).
Another class of toxins in benthic dinoflagellates is the water-soluble maitotoxins (MTX) that accumulate in the fishes’ digestive tract and liver. The presence of at least one sulfate moiety in the MTX backbone leads to significant hydrophilicity of these toxins and low bioaccumulation in fish flesh. MTX are the most potent marine toxins known to date, although MTX has lower toxicity compared to CTX via an oral route (Shmukler and Nikishin, 2017). The low oral bioavailability and bioaccumulation of MTX suggest that these compounds are not the main contributor to CP symptomatology (Holmes et al., 2021). To date, there are seven known MTX analogs, and their biological activity is varied (Estevez et al., 2020a,2021). MTX-1, MTX-2, and MTX-4 cause massive Ca2+ influx leading to cell death (Estevez et al., 2020a). MTX-3 causes a similar phenotype to CTX, although with lower potency (Boente-Juncal et al., 2019).
Among the MTX congeners, only MTX-1 have an assigned structure based on mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy. MTX-1 is the largest non-peptide toxin identified, with a molecular weight of 3425 Da, with 32 cyclic ether rings (Nonomura et al., 1996; Sasaki et al., 1996; Shmukler and Nikishin, 2017). Structural analysis of MTX-3 showed that it is smaller than MTX-1, with a mass of 1039 Da (Holmes and Lewis, 1994). Several groups undertook a targeted purification of MTX-3 to determine the structure. The purified material was analyzed by NMR and MS and indicated that MTX-3 is the 44-methyl analog of gambierone (Boente-Juncal et al., 2019; Murray et al., 2019). The difference in bioactivity, approximately 1,000-fold lower potency of 44-methylgambierone than the original data for partially purified MTX-3, may indicate a potential divergence in structure between MTX-3 and 44-methylgambierone (Holmes et al., 2021). Holmes and Lewis (1994) observed a potent toxic effect of partially purified MTX-3 when administered intraperitoneally in mice (Holmes and Lewis, 1994; Lewis et al., 1994). Purified 44-methylgambierone had lower toxicity, with LD50 of 20–38 mg/kg via intraperitoneal administration (Murray et al., 2020). Additional studies are necessary to ascertain the divergence in biological activity and structure between MTX-3 and 44-methylgambierone.
The complex structure of CTX and MTX and the limited biomass of dinoflagellate producers make the complete structural assignment and biological activity assessment of these toxins challenging. Several groups have implemented hyphenated liquid chromatography-mass spectrometry (LC-MS) to aid in toxin discovery. By using an LC-MS-based workflow, it is easier to screen extracts of benthic dinoflagellates for potential new congeners, and known CTX and MTX can be quantified and identified (Caillaud et al., 2010 and references cited therein; Chinain et al., 2010; Munday et al., 2017; Longo et al., 2019; Estevez et al., 2020a,2021; Murray et al., 2020; Tibiriçá et al., 2020; Gago-Martínez et al., 2021 and references cited therein; Mudge et al., 2021). UPLC-HRMS profiling of 252 marine microalgae belonging to 32 genera showed that 44-methylgambierone production is ubiquitous in eight Gambierdiscus species (Murray et al., 2020). The benthic dinoflagellate Coolia and Fukuyoa were likewise producers of 44-methylgambierone (Murray et al., 2020), with a putative new analog likely to be present in Coolia (Tibiriçá et al., 2020). While 44-methylgambierone has low toxicity via the oral route, UPLC-HRMS quantitation showed significant amounts in the producer dinoflagellate and snappers, suggesting potential contributions to CP. The intracellular concentration of 44-methylgambierone in Gambierdiscus sp. is 5.8–74 pg MTX-1 eq cell–1 (Longo et al., 2019), while bioaccumulation in snapper liver and muscles can increase the concentration (Kohli et al., 2014).
Apart from MTX and CTX, other compounds identified from CP-causative organisms include gambierone (Rodríguez et al., 2015), gambieroxide (Watanabe et al., 2013), gambierol (Satake et al., 1993), gambieric acids (Nagai et al., 1992), and the recently identified 29-methylgambierone (Mudge et al., 2021) and sulfo-gambierones (Yon et al., 2021). Gambierdiscus has the potential to produce compounds belonging to other molecular scaffolds, such as non-ribosomal peptide-polyketide hybrid compounds based on recent transcriptome profiling (Kohli et al., 2017; Van Dolah et al., 2020). The realization of the biosynthetic potential of Gambierdiscus has, however, yet to be established at the metabolome level.
Capitalizing on the improved methodology for mass spectrometry, untargeted metabolite analysis has become a mainstay tool to analyze chemical diversity in organisms. The Global Natural Products Social Molecular Networking Platform (GNPS) is based on spectral alignment to assess the similarities and relationships among molecules (Wang et al., 2016) and visualized using a molecular network. While GNPS is mainly used for biodiscovery of natural products (Teta et al., 2015; Naman et al., 2017; Ding et al., 2018; Via et al., 2018), it has been recently applied to dinoflagellate metabolites and demonstrated the unique metabolites from these organisms (Fiorini et al., 2020; Wu et al., 2020; Sibat et al., 2021). Molecular networking of five Dinophysis species identified the characteristic toxin profile for each strain and identified five new putative pectenotoxins (Sibat et al., 2021). Differences in the metabolites of Pseudo-nitzchia during the reproductive stages were evident from the GNPS-based analysis (Fiorini et al., 2020). Compounds with unprecedented chemical scaffolds in Pseudo-nitzchia extracts, not represented in reference databases, were observed (Fiorini et al., 2020). Improved qualitative and quantitative screening of okadaic acid and dinophysistoxins was achieved through molecular networking of extracts from Prorocentrum lima (Wu et al., 2020). New esters of okadaic acid and dinophysistoxins were also identified (Wu et al., 2020). A scientometric analysis on dinoflagellates research recommended using metabolite databases such as Dictionary of Natural Products, AntiBase, MassBank, and GNPS to share metabolites information to advance biomolecule discovery (Oliveira et al., 2020).
In this study, we looked at the biosynthetic potential of Gambierdiscus carpenteri and two Gambierdiscus balechii strains from the Philippines to produce toxins and other classes of secondary metabolites using high resolution mass spectrometry (HRMS) and the GNPS molecular networking platform. We probed the chemistry of the three Gambierdiscus strains and obtained insights into the production of ladder-shaped polyether toxins and other classes of compounds.
Three monoclonal cultures of Gambierdiscus spp. from The Marine Science Institute Red Tide Laboratory were used in this study (Table 1). Cultures were routinely maintained in filtered natural seawater (30 psu) supplemented with full strength IMK medium (WAKO, Tokyo, Japan) at 25 ± 2°C under 100 μmol photons m–2 s–1 of light using 40 W white fluorescent lamps, with a 12:12 h light:dark photoperiod. Large-scale 1-L cultures in Fernbach flasks were harvested after 21 days in the exponential phase by centrifugation at 4,000 × g for 15 min at 4°C (Thermo Fisher Scientific).
Cells of G. balechii GtoxSAM092414 and G. balechii Gtox112513 were observed with a Zeiss Axioskop 2 (Carl Zeiss, Göttingen, Germany) light microscope (LM). To visualize the thecal plates, cells were stained with Calcofluor white [5 μL, 10 × final concentration (Sigma-Aldrich)] and observed under a confocal laser scanning microscope (CLSM 710, Carl Zeiss, München Germany) at 420 nm wavelength. Autofluorescence of chloroplast was also observed using CLSM 710. Photos were taken with a Zeiss Axiocam MRm and processed using Zeiss Efficient Navigation (ZEN) software (Carl Zeiss, München Germany). Cell and thecal plate dimensions were measured from LM and CLSM micrographs. The modified Kofoidian tabulation system (Kofoid, 1909) described by Besada et al. (1982) was followed in naming the thecal plates of Gambierdiscus. These were compared to the previously identified G. carpenteri Gam1BOL080513 from Bolinao, Philippines (Vacarizas et al., 2018).
Genomic DNA was extracted from exponentially growing cultures using ISOLATE II Plant DNA Kit (Bioline, London, United Kingdom) following the manufacturer’s procedure. The D8–D10 region of large subunit ribosomal DNA (LSU rDNA) gene was amplified using the primers D8F and D10R (Litaker et al., 2009). Amplification was conducted in 50 μL reaction mix containing 45 μL of PCR supermix (Invitrogen, California, United States), 2 μL of each primer and 1 μL of template DNA. The thermal condition of PCR was as follows: initial denaturation step at 95°C for 2 min, followed by 35 cycles of 94°C for 30 s, 60°C for 2 min, and 72°C for 60 min, and finally an elongation step of 72°C for 5 min. Reaction was performed in a T100 Thermal cycler (Biorad, California, United States). Amplicons were purified using QIAquick Gel Purification Kit (Qiagen) and sent to 1st Base (Malaysia) for sequencing.
DNA sequences were aligned using MAFFT v7.110 (Katoh and Standley, 2013) with taxa downloaded from GenBank. The multiple sequences were manually edited and/or trimmed using BioEdit Sequence Alignment Editor v7.2.5 (Hall, 1999) with 58 selected taxa of Gambierdiscus comprising 17 species. For outgroups, four Fukuyoa spp., two Alexandrium spp., Akashiwo sanguinea and Prorocentrum micans were used. Maximum likelihood (ML) analysis was performed using PhyML (Guindon et al., 2010) with 500 bootstrap replicates. The best fitting substitution model for the ML tree as selected by the Smart Model Selection (SMS) program (Lefort et al., 2017) was general time reversible (GTR) with gamma distribution (G = 0.301) plus proportion of invariable sites (I = 0.881). Bayesian inference (BI) was computed via MrBayes v3.1.2 (Ronquist and Huelsenbeck, 2003) using a Metropolis-coupled Markov chain Monte Carlo run for 10 million generations with sampling at every 100 iterations. The best-fit substitution model for the BI tree, selected by jModelTest 2.1.10 (Darriba et al., 2012), was TIM3 + G (0.899) + I (0.3000). GenBank accession numbers are provided in the phylogenetic trees.
Gambierdiscus isolates were extracted according to the procedure of Murata et al. (1990). Lyophilized biomass from 1 L cultures were extracted using three volumes of acetone to yield the crude extracts. A portion of the crude extract (20.0 mg) was further fractionated on a Florisil® SPE-cartridge and eluted with 4:1 n-hexane:acetone (fraction A), 9:1 acetone:methanol (fraction B), and 1:1 acetone:methanol (fraction C). Fractions were dried and stored at –20°C until further analysis.
Dried crude extracts were resuspended in methanol and subjected to radioligand receptor-binding assay (RBA) based on the IAEA-Tecdoc-1729 for the detection of ciguatera toxins (IAEA, 2013). The test kit for RBA consisting of tritium-labeled brevetoxin-3 ([3H]PbTx-3), unlabeled PbTx-3 and porcine brain membrane was acquired from American Radiolabeled Chemicals. A 15 nM working solution of [3H]PbTx-3 was prepared by diluting with the assay buffer. Solutions of unlabeled PbTx-3, with final assay concentrations of 0.01 ng/mL to 1.0 μg/mL in half-log dilutions, were prepared for the CTX calibration curve. The assay was performed in a 96-well microtiter filter plate with FB glass fiber filter (0.65 μm pore size) by adding the following solutions in order: 35 μL assay buffer, 35 μL PbTx-3 standard solutions or extracts, 35 μL [3H]PbTx-3 working solution, and 105 μL porcine membrane homogenates. Plates were covered and incubated at 4°C for 1 h, filtered using a vacuum manifold and washed with 200 μL ice-cold assay buffer (2 ×). A 50 μL aliquot of the scintillation cocktail (OptiPhase, PerkinElmer) was added to each well and the reaction plate was further incubated for 1 h at room temperature. Plates were counted using a microplate scintillation counter (MicroBeta®, PerkinElmer). Curve fitting of the PbTx-3 standards was performed using a four-parameter logistic fit (Sigmoidal, 4PL) with variable slope. Limit of detection for the assay was 2.0 ×10–5 g PbTx-3 eq./g. Results are presented as pg PbTx-3 eq./cell by normalizing the acquired RBA values to the total cell counts of the cultures.
Gambierdiscus biomass was subjected to a two-stage sulfuric acid extraction according to the method of Templeton et al. (2012) to yield the total carbohydrates. The hydrolyzed carbohydrates were identified and quantified using the method of Schulze et al. (2017) with modifications. Monosaccharides were separated using an Acquity UPLC BEH Amide column, 1.7 μm, 2.1 × 50 mm (Waters) by a gradient program of acetonitrile/5 mM ammonium formate in water (both with 0.1% formic acid modifier): 90–75% acetonitrile for 8.5 min, and 75% acetonitrile for 4 min. Detection was done by multiple reaction monitoring (MRM) analysis (Shimadzu LCMS-8040). The optimized transitions for each standard are provided in Supplementary Table 1.
Data analysis was performed through manual peak integration using LabSolution (Shimadzu). Individual monosaccharide standards (Sigma) were prepared (0.156 μg/mL to 0.0098 μg/mL) in two-fold serial dilutions and injected three times for repeatability to generate the external calibration curves. Results are presented as % w/w (mg sugar/mg biomass), based on two biological replicates with three technical replicates each.
Solid-liquid extraction of the Gambierdiscus biomass was done according to the method of Bligh and Dyer (1959) to yield the chloroform soluble lipid extract. Fatty acid methyl esters (FAMEs) were prepared based on the AOAC official method 969.33 (AOAC, 2000b). Resulting FAMEs were separated and analyzed using gas chromatography with flame ionization detector (Shimadzu GC-2010) through the AOAC Official Method 963.22 (AOAC, 2000a). Chromatographic separation was done on a Supelco SP-2560 capillary column (0.25 μm, 100 m × 0.20 mm). Retention times of the eluted FAMEs were compared with known amounts of mixed reference standards composed of C6, C8, C10, C12, C14, C16, C18, C18:1, C18:2, C18:3, C20, C22, and C24. Peak areas were integrated using GCSolution (Shimadzu), and the response factors were calculated from the ratio between the peak area of the individual FAME and the internal standard (heneicosanoic acid methyl ester). Analysis was done using two biological replicates with three technical replicates each. Results are presented as % w/w (mg fatty acid/mg biomass).
UPLC-MS/MS analysis of all Florisil® fractions (1 mg/mL in acetonitrile) was performed using a Waters Acquity UPLC® H-Class System with a Xevo® G2-XS Quadrupole Time-of-Flight (QToF) high-resolution mass spectrometer. A 1 μL aliquot of each sample was injected in a Phenomenex Kinetex 2.6 μm C18 100Å column (50 × 2.1 mm) and eluted at 0.35 mL/min, using a gradient program of acetonitrile/water (0.1% formic acid modifier): 40–100% acetonitrile for 5.5 min, and 100% acetonitrile for 2 min. The mass spectrometer was set to observe at m/z 100–2,000 in positive ESI mode with an automated data dependent acquisition (DDA) MS/MS scan. Three DDA scans were acquired for each sample with increasing ramp collision energies: 15–25, 30–45, and 50–70 eV. DDA scan for the Pacific ciguatoxin 3C (P-CTX-3C, WAKO Chemicals) standard was acquired using the 15–25 eV ramp collision energy.
Chromatograms were converted to mzxml format using freely available MSConvert software1. A molecular network was created using the online workflow2 on the GNPS website3 (Wang et al., 2016). MS/MS data of the Florisil® fractions from each isolate were grouped to form the molecular network. The precursor ion mass tolerance was set to 2 Da and an MS/MS fragment ion tolerance of 0.1 Da. A network was created, with edges set to cosine score >0.7 and more than six matched peaks. Further, edges between two nodes were kept in the network if and only if each of the nodes appeared in each other’s respective top 10 most similar nodes. Finally, the maximum size of a molecular family was set to 100, and the lowest scoring edges were removed from the molecular families until the molecular family size was below this threshold. The spectra in the network were searched against the GNPS spectral libraries. All matches kept between network spectra, and library spectra were required to have a score >0.7 and at least six matched peaks.
Morphological characters (Figure 1) of the two Gambierdiscus strains (Gtox112513 and GtoxSAM092414) were identical to Gambierdiscus balechii (Fraga et al., 2016; Azanza et al., 2017). Cells were anterio-posteriorly compressed, measuring 53.6–63.5 μm (57.7 ± 2.7, n = 30) in depth (dorsoventral diameter) and 56.7–63.0 μm (60.1 ± 1.9, n = 30) in width. Cell shape was round to ellipsoid in apical view (Figures 1D,E). The epithecal and hypothecal surfaces were heavily areolated (Figures 1A,C, 2D). The presence of small and large accumulation bodies was observed (Figure 1B). The nucleus was large and elongated (Figure 1D). Cells had a rod-shaped, golden-brown chloroplast evenly distributed throughout the cell (Figure 1F). Thecal plates of the two strains as observed with fluorescence microscopy, especially the taxonomically informative plates of the genus such as the shape of the second apical plate (2′), third precingular plate (3″), and second antapical plate (2″″), support their identification as G. balechii. Thecal plate tabulation of the Gambierdiscus strain (Gtox112513) is only shown (Figure 2). The epitheca was composed of apical pore (Po), three apical plates and six precingular plates. In apical view, the first apical plate (1′) and sixth apical plate (6′) were barely visible (Figure 2A) but can be seen at ventral view (Figure 2C). Both plate 1′ and 6″ were the smallest among the apical and precingular plates, respectively. The second apical plate (2′) was the largest of the apical series and shaped like a hatchet, i.e., having a suture ratio of 2′/1″ and 2′/3″ = 0.60 (n = 30). The third precingular plate (3″) was asymmetrical (Figure 2A). The apical pore plate was oval with a fish-hook shaped slit (Figures 1A,D). The hypotheca was composed of two antapical plates (2″″), five postcingular plates (5″′), and a posterior sulcal plate (Sp). Among the five postcingular plates, the plate 4″′ was the largest. The second antapical plate (2″″) was narrow and pentagonal to trapezoidal (Figure 2B). Marginal borders of the plates in the epitheca and hypotheca were overlapping (Figures 2A,B).
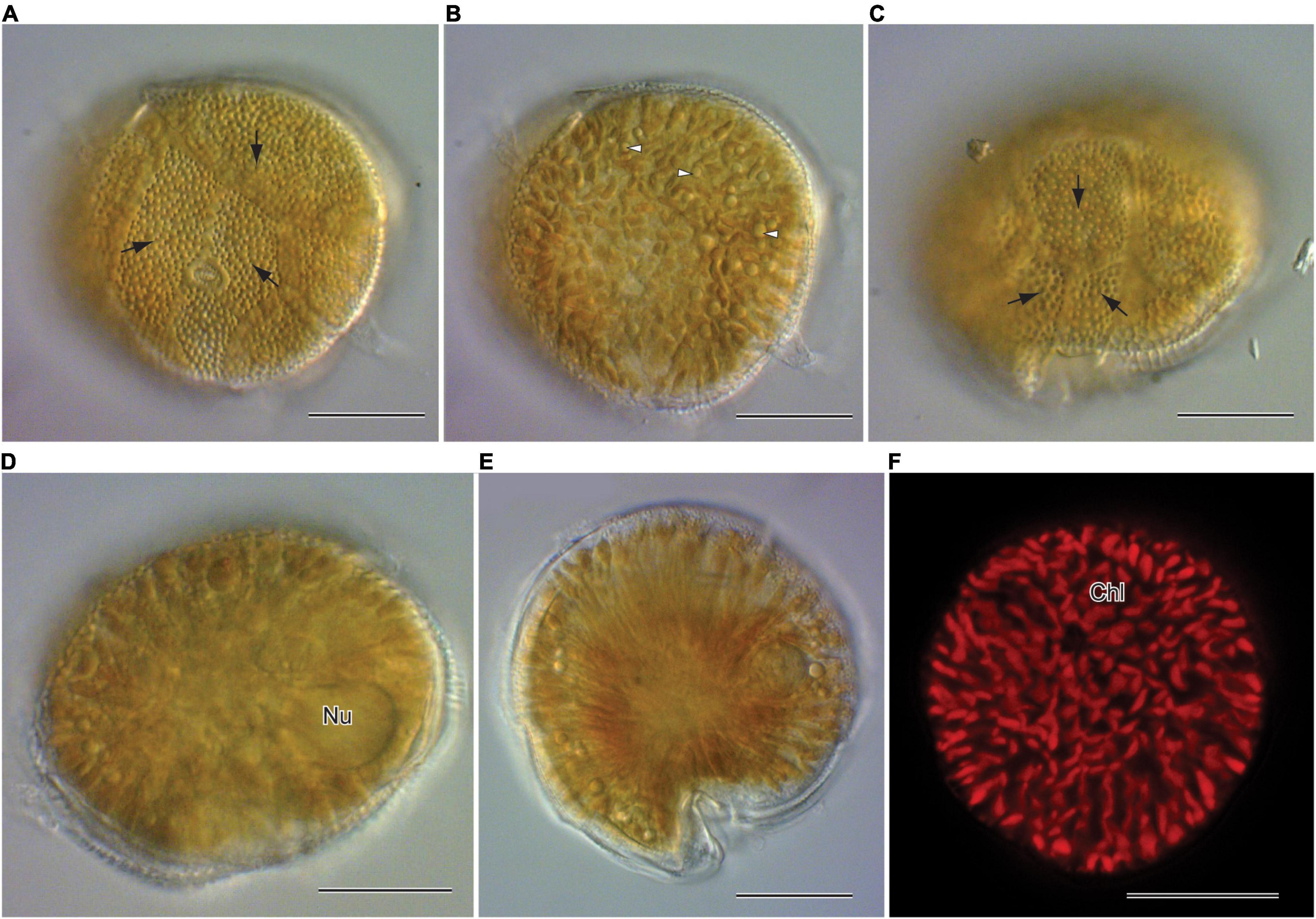
Figure 1. Light and fluorescence microscopy of Gambierdiscus balechii (A–D). Gtox112513; (E,F). GtoxSAM092414. (A) Surface focus from apical view showing heavy areolations (black arrows) (B) deeper focus from apical view showing the chloroplast and accumulation bodies (white arrowheads) (C) surface focus from antapical view showing heavy areolations (black arrows) (D) deeper focus from antapical view showing nucleus (Nu) (E) deeper focus from apical view showing the shape of the cell (F) fluorescence microscopy showing the chloroplast (Chl). Scale = 20 μm (A–F).
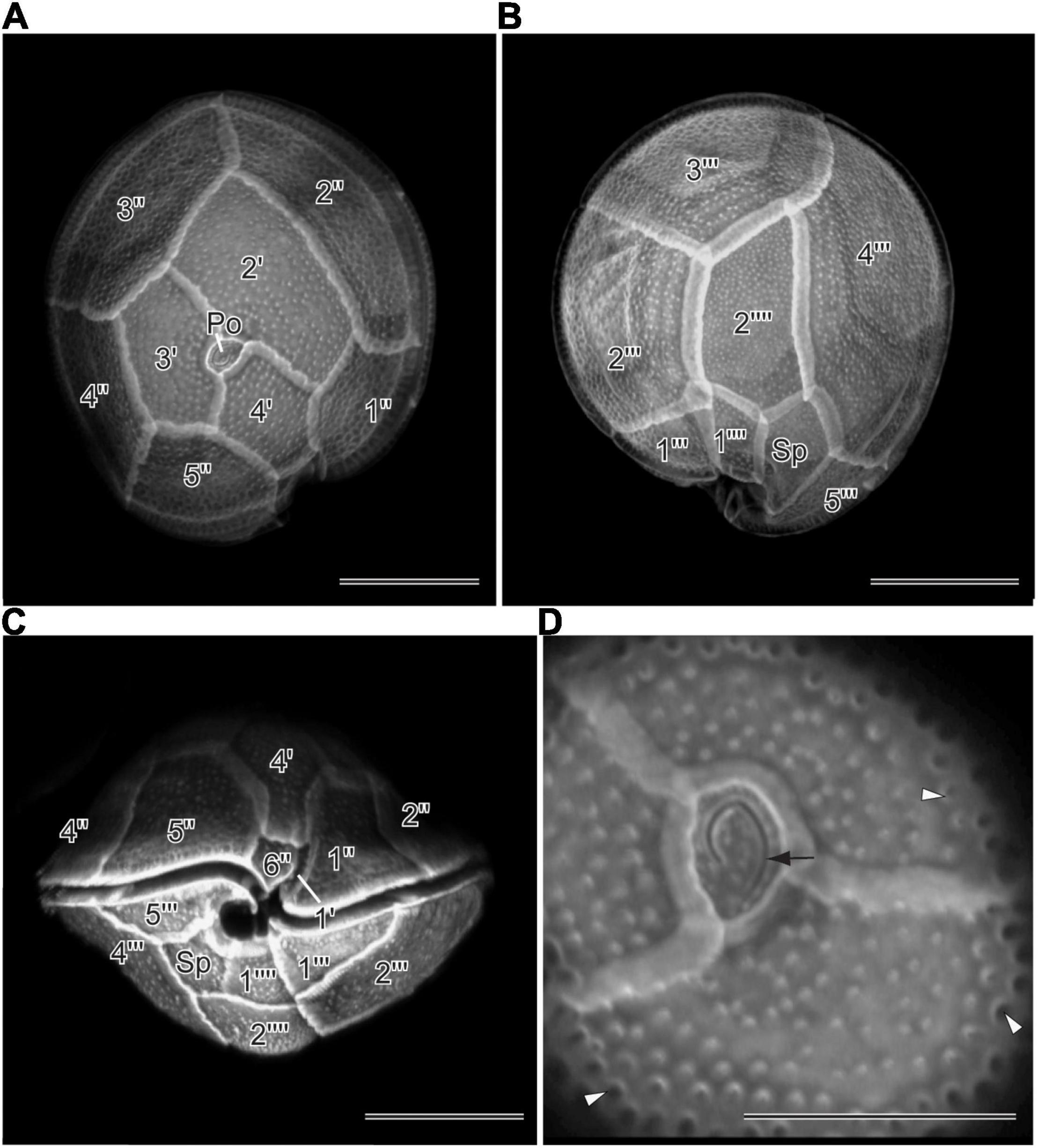
Figure 2. Confocal Laser Scanning Microscopy of calcofluor stained Gambierdiscus balechii (Gtox112513) (A) apical view (B) antapical view (C) ventral view (D). apical view. Apical pore plate showing the fishhook-shaped slit (black arrow) and pores (white arrow head). Scale = 20 μm (A–C), 10 μm (D). Apical pore plate (Po), apical plates (1′–4′), precingular plates (1′′–6′′), postcingular plates (1″′–5″′), and antapical plates (1″″ and 2″″).
Two new LSU rDNA (D8–D10) sequences were obtained from Gambierdiscus cultures established from Bolinao, Pangasinan (Gtox112513), and Guiuan, Eastern Samar, Philippines (GtoxSAM092414). The aligned sequences of 70 OTUs had 859 bps, of which 377 sites (43.9%) were variable, and 14 sites (1.6%) were parsimonious informative. Average base compositions were A = 1.05, C = 0.250, G = 0.250, and T = 1.60. The LSU sequences of the two strains differed at 14 positions (98.4% similarity). The maximum likelihood (ML) tree inferred from LSU rDNA is shown in Figure 3. Bootstrap support values derived from ML and posterior probability from Bayesian Inference (BI) analyses were given. The ML tree showed that Gtox112513 and GtoxSAM092414 clustered with previously reported G. balechii strains with strong support (BI/ML = 1.00/97%). This species formed a well-supported monophyletic clade (1.00/100%) comprising of G. pacificus, G. toxicus, G. cheloniae, G. lewisii, G. lapillus, G. scabrosus, G. belizeanus, G. honu, and two undescribed species. Gambierdiscus carpenteri (Gam1BOL080513) used in this study grouped with other reported G. carpenteri strains with strong support (0.96/94%) (Figure 3). This species formed a monophyletic clade composed of G. caribaeus and G. jejuensis with maximum support (1.00/100%).
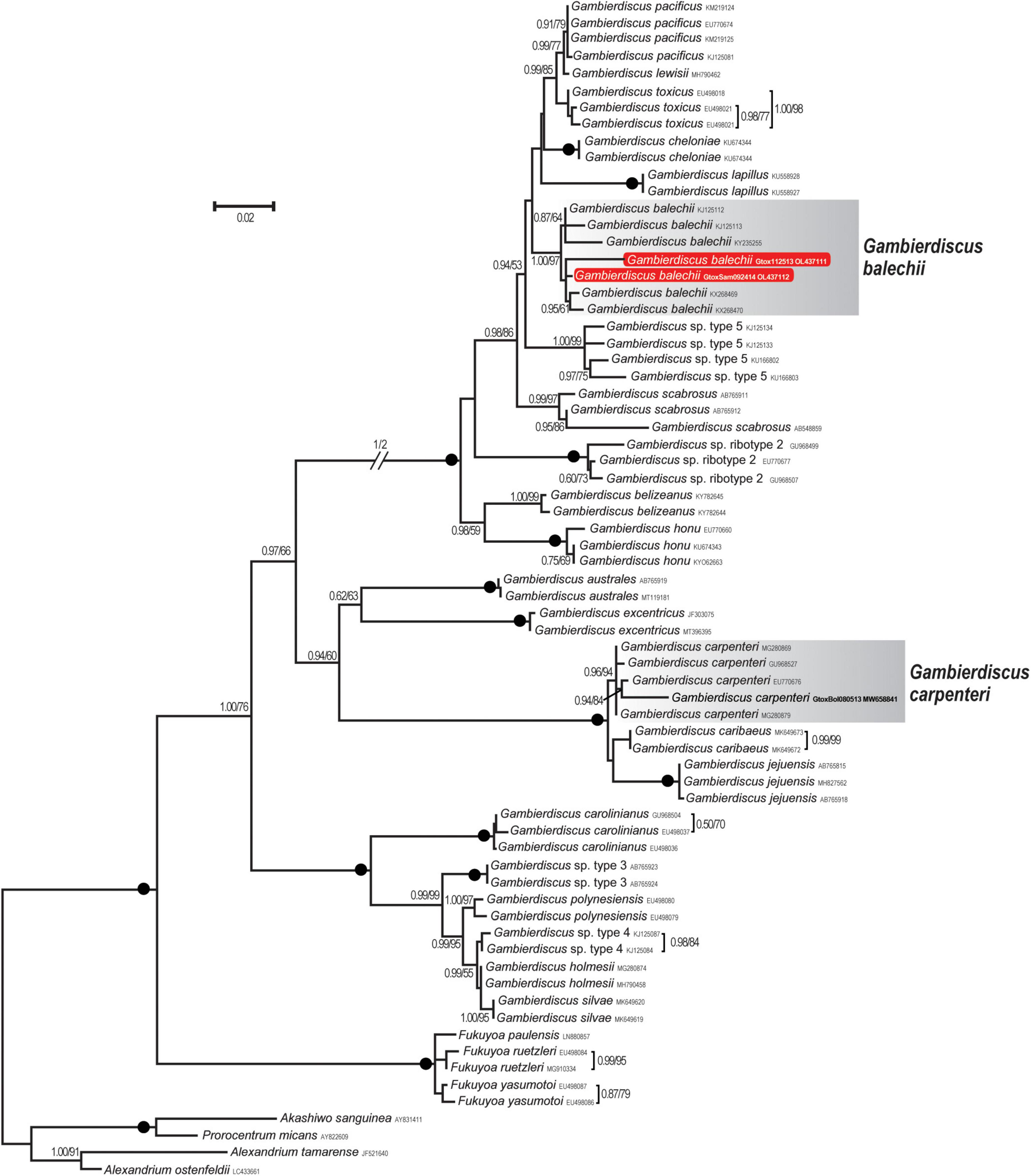
Figure 3. Maximum likelihood (ML) phylogeny of Gambierdiscus species inferred from D8-D10 LSU rDNA (859 bp) based on the GTR + I + G model. Posterior probabilities (PP, ≥ 0.7) of Bayesian inference and bootstrap support (BS, ≥ 50%) values of ML were indicated, and black dots on nodes indicate maximum supports for both BI and ML (PP/BS = 1.00/100). New DNA sequences analyzed in this study are highlighted in red box. Strains used in this study are highlighted in bold.
The toxin content of the three Gambierdiscus strains was assessed via RBA using the IAEA-Tecdoc-1729 to detect ciguatera toxins (IAEA, 2013). G. carpenteri Gam1BOL080513 gave the highest toxicity with 12.36 ± 4.38 pg PbTx-3 eq./cell (Figure 4). The G. balechii strains GtoxSAM092414 (0.80 ± 1.13 pg PbTx-3 eq./cell), and Gtox112513 (0.14 ± 0.05 pg PbTx-3 eq./cell) showed comparable toxicities. The G. balechii toxin content was approximately 15–91-fold lower than G. carpenteri Gam1BOL080513.
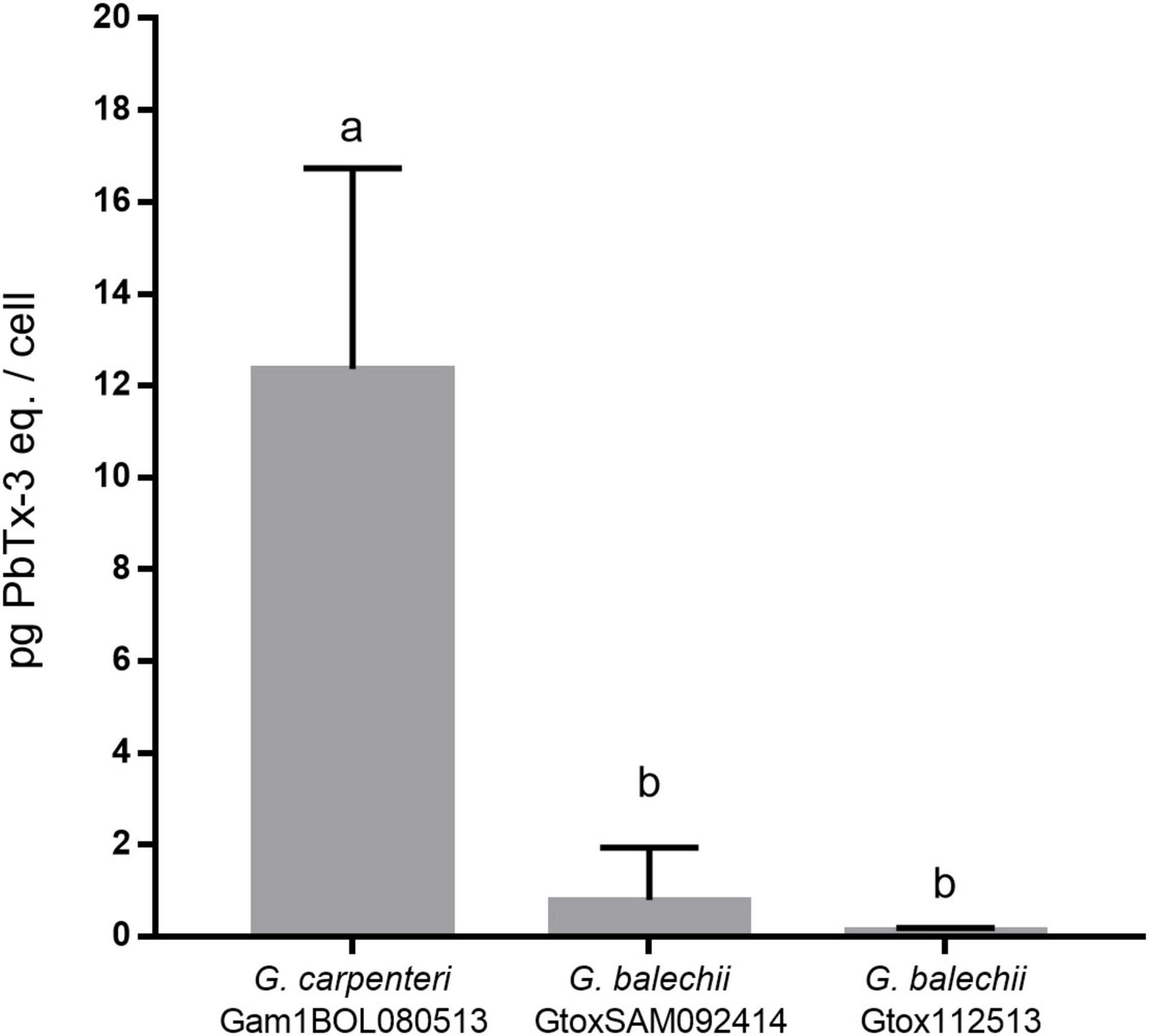
Figure 4. Toxin content of crude acetone extracts of G. carpenteri Gam1BOL080513, G. balechii GtoxSAM092414, and G. balechii Gtox112513 shown in pg PbTx-3 eq./cell. The two G. balechii strains had ∼15 to ∼91-fold lower toxin content compared to G. carpenteri. Results are presented mean + SD (n = 3). Different letters indicate significant difference after one-way ANOVA analysis (P < 0.05).
The total carbohydrate content of the three isolates was quantified using a colorimetric phenol-sulfuric acid assay (Masuko et al., 2005). G. carpenteri Gam1BOL080513 and G. balechii GtoxSAM092414 have comparable total carbohydrate content (Figure 5A). G. balechii Gtox112513 showed the highest total carbohydrate content with 12.66 ± 1.92% w/w.
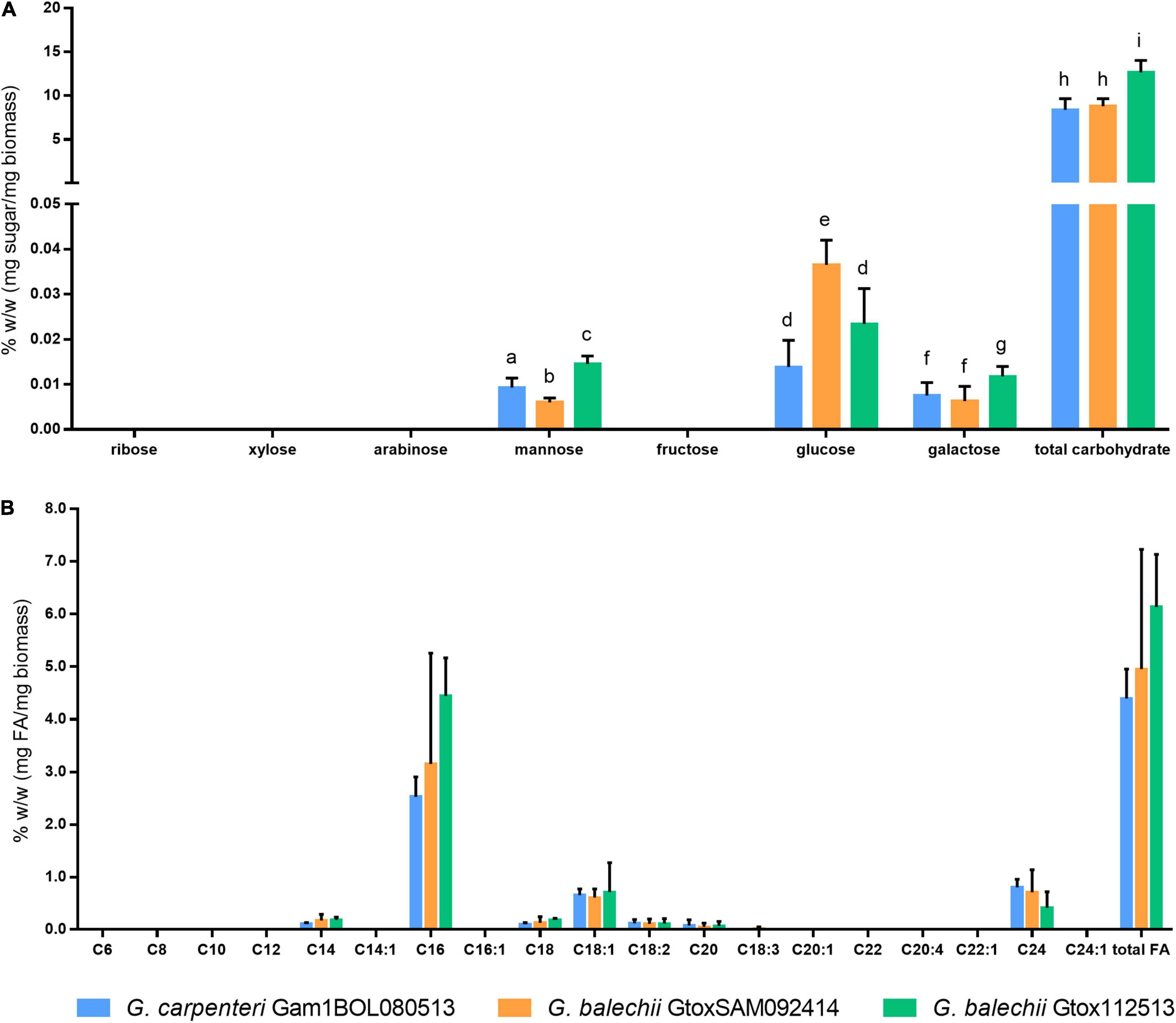
Figure 5. Monosaccharide (A) and fatty acid (B) content of crude carbohydrate and lipid extracts of G. carpenteri Gam1BOL080513, G. balechii GtoxSAM092414, and G. balechii Gtox112513 biomass. The three strains showed significant difference in the monosaccharide content but not in the fatty acid composition. Results are presented as mean + SD from two biological replicates with three technical replicates each. Different letters indicate significant differences after one-way ANOVA analysis (P < 0.05).
Carbohydrates were hydrolyzed according to the method of Schulze et al. (2017). Multiple reaction monitoring of the monosaccharides facilitated the identification and quantitation of the sugars. Galactose, mannose, and glucose were the major monosaccharides in the hydrolysates of the three Gambierdiscus isolates, with glucose being the principal monosaccharide (Figure 5A). G. balechii GtoxSAM092414 and G. balechii Gtox112513 gave the highest glucose and galactose content, respectively (Figure 5A). The mannose content significantly varied across the three isolates, with the highest amount in G. balechii Gtox112513 (Figure 5A).
Fatty acids from the biomass of the Gambierdiscus isolates were determined and quantified by converting the crude lipid extracts to fatty acid methyl esters (FAMEs) and detected using GC-FID based on the AOAC Official Method 963.22 (AOAC, 2000a). The total fatty acid content of the three Gambierdiscus strains was comparable (Figure 5B). The isolates also showed similar fatty acid profiles consisting of myristic (C14), palmitic (C16), stearic (C18), oleic (C18:1), linoleic (C18:2), arachidic (C20), and lignoceric (C24) acids (Figure 5B). Only G. balechii GtoxSAM092414 showed detectable levels of linolenic acid (C18:3) (Figure 5B). Principal fatty acids for all strains are C16 (2.53–4.55%), C18:1 (0.66–0.71%), and C24 (0.42–0.80%). The remaining fatty acids detected for all strains are saturated fatty acids, C14 (0.10–0.1%), and C20 (0.05–0.08%).
Untargeted metabolite profiling of the extracts was performed using reversed phase UPLC-MS and MSMS analysis and subsequently annotated using the GNPS database to identify the compounds and toxins produced by the three Gambierdiscus isolates. Figure 6 shows the molecular network generated from the extracts of the three strains of Gambierdiscus and P-CTX-3C (in lavender) as a seed compound with a known identity. The entire molecular network consists of 541 nodes with 666 connections and 45 clusters with 271 single nodes. Cosine scores ranged from 0.70 to 0.96, suggesting that the putative hits from the GNPS database and the metabolites in the Gambierdiscus extracts may not be identical but would likely have the same chemical scaffold. Hence, clusters were annotated based on their common functional groups. Based on this, 45 clusters were generated belonging to at least ten compound classes (Figure 6).
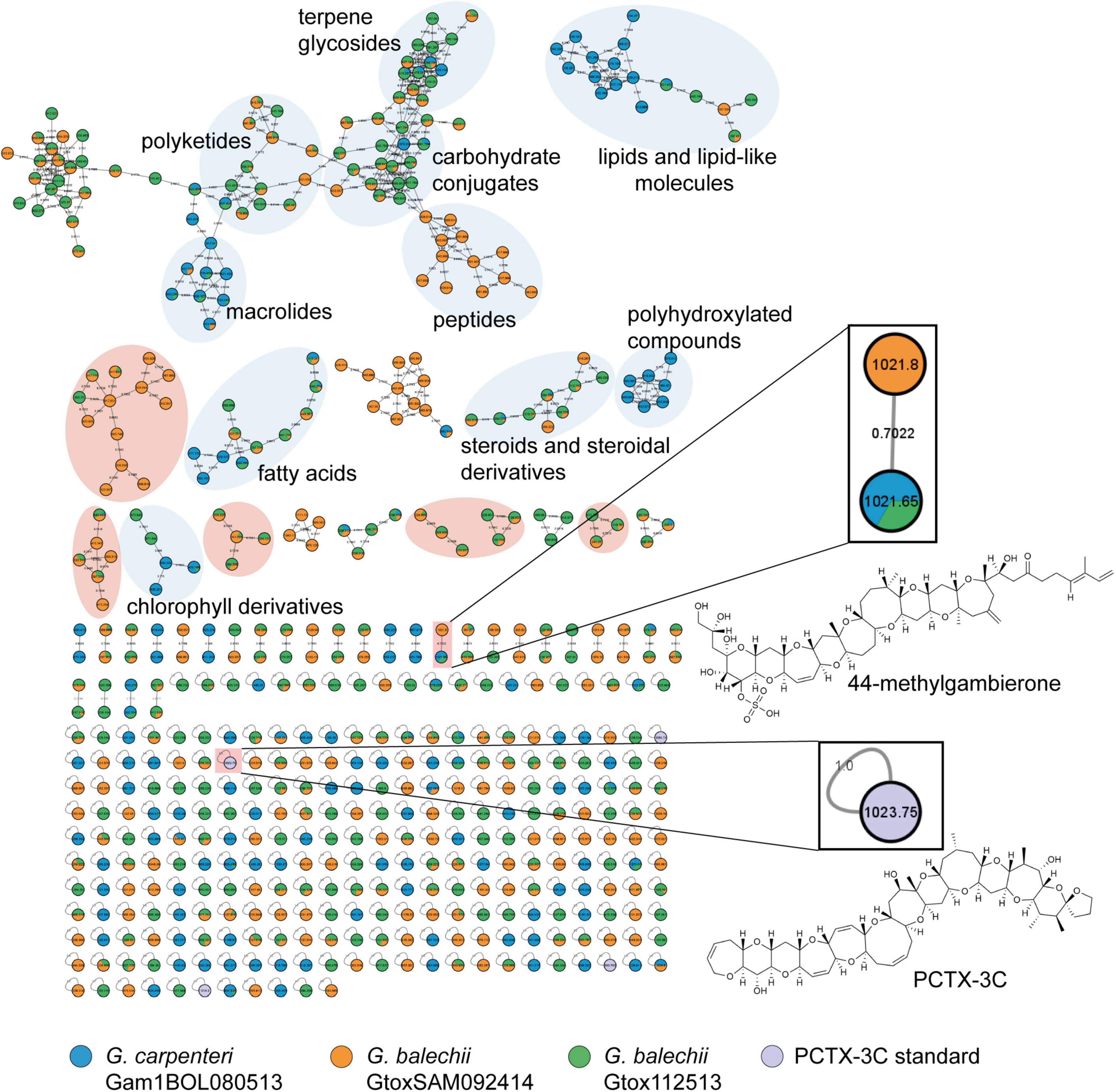
Figure 6. Molecular network generated from HRMS/MS spectra from the Florisil® fractions of three Gambierdiscus strains, and P-CTX-3C standard using Global Natural Product Social Molecular Networking (GNPS) tool. Each node corresponds to a compound in the given strain (indicated by the color/s of the node). The nodes were color-coded according to the strains: G. carpenteri Gam1BOL080513 in blue, G. balechii GtoxSAM092414 in orange, G. balechii Gtox112513 in green, and the P-CTX-3C standard in lavender. Similarity between nodes is represented by the gray curves labeled by the cosine score. Annotation of compound class or family was performed through matching of fragmentation patterns of known compounds available in the GNPS server. Enlarged inset are manually annotated toxins, authentic standard (P-CTX-3C) and 44-methylgambierone with their chemical structures. The three Gambierdiscus strains contain 44-methylgambierone and chemically diverse secondary metabolites. A high-resolution image of the molecular network is provided in the Supplementary Material (Supplementary Figure 1).
The largest cluster contains the most convergence of the compounds from the three strains, mainly consisting of terpene glycosides (2, 3), carbohydrate conjugates (4–6), polyketides, and macrolides (7–10) (Figure 6). Lipids and lipid-like molecules (11–13), fatty acids (14–16), and chlorophyll derivatives (17, 18) were present in all three strains. G. balechii strains also produced steroids and steroidal derivatives (19, 20). Strain-specific clusters were also observed, such as a peptide (21) cluster in the extract of G. balechii GtoxSAM092414 and polyhydroxylated (22, 23) compounds from G. carpenteri Gam1BOL080513 (Figure 6). The seed compound P-CTX-3C appears in the molecular network as a single node (Figure 6 inset). Manual annotation of the single nodes in the molecular network revealed two precursor masses, 1021.65 and 1021.8 Da, in the three extracts (Figure 6 inset) that clustered together. Examination of the chromatograms showed that these ions have identical retention times (Figure 7A) and MS1 profiles (Figure 7B). The observed masses matched the pseudomolecular ion corresponding to a water loss [M+H-H2O]+ of 44-methylgambierone (1). Further, a comparison of the MS/MS spectra to the published data (Boente-Juncal et al., 2019; Estevez et al., 2020b) for 44-methylgambierone (Table 2 and Supplementary Figure 2) showed similar fragmentation patterns, therefore, corroborating with the observed pseudomolecular ion. 44-methylgambierone was detected in fraction B of the G. balechii strains, and in fraction C of G. carpenteri Gam1BOL080513 (Supplementary Figures 3–5). The relative amount of 44-methylgambierone was semi-quantified by integrating the peak area corresponding to 44-methylgambierone (tR = 0.78–0.90 min) along with the solvent blank (Figure 7C). G. balechii Gtox112513 showed the highest amount of 44-methylgambierone compared to G. balechii GtoxSAM092414 and G. carpenteri Gam1BOL080513.
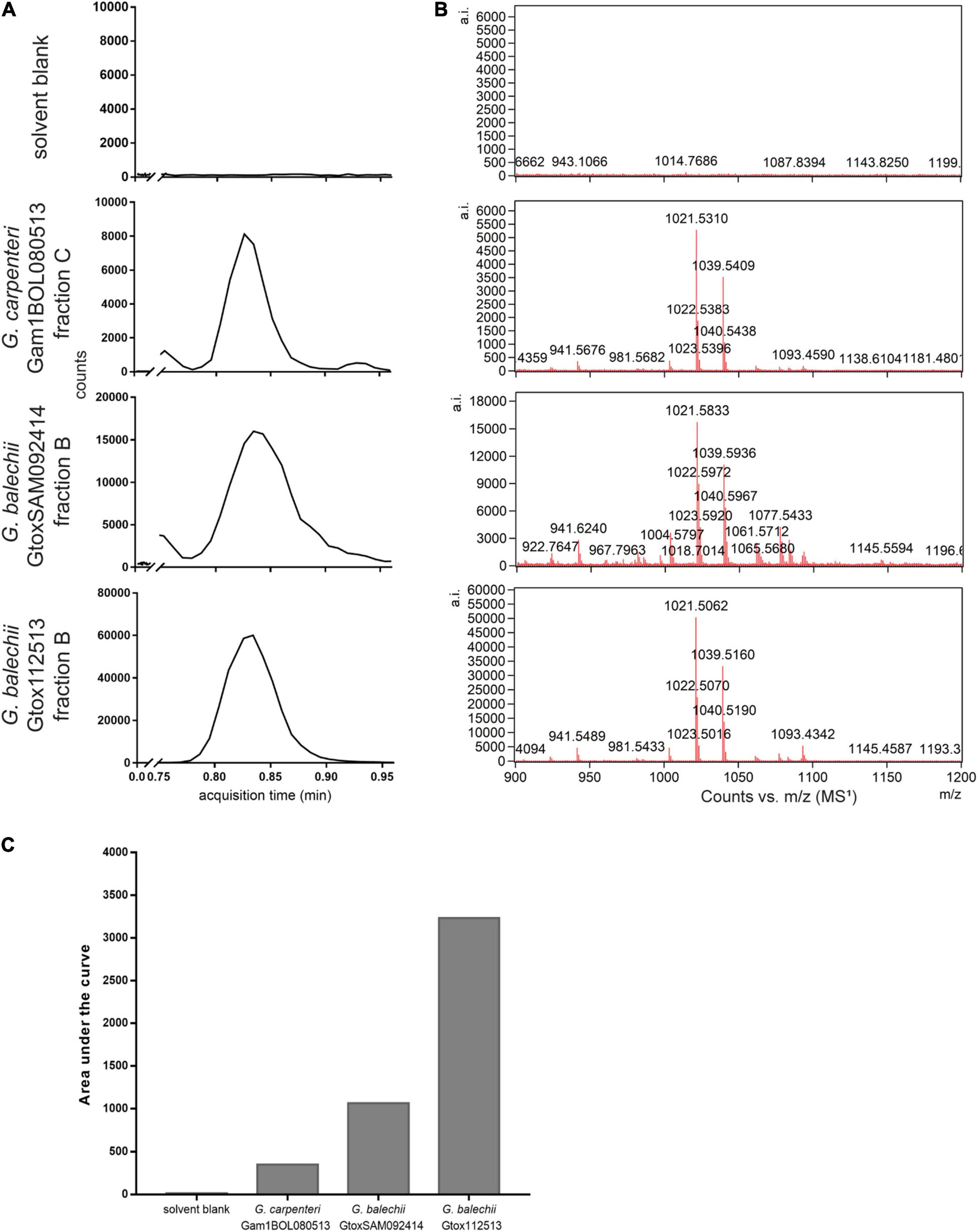
Figure 7. LC-HRMS chromatograms (A) of the Gambierdiscus Florisil® fractions (tR 0.75–0.95 min) and MS1 spectra corresponding to 44-methylgambierone (B). Relative amount of 44-methylgambierone was semi-quantified by integrating the area under the chromatogram peaks in (A) along with the solvent blank (C). G. balechii Gtox112513 contained the highest amount of 44-methylgambierone.
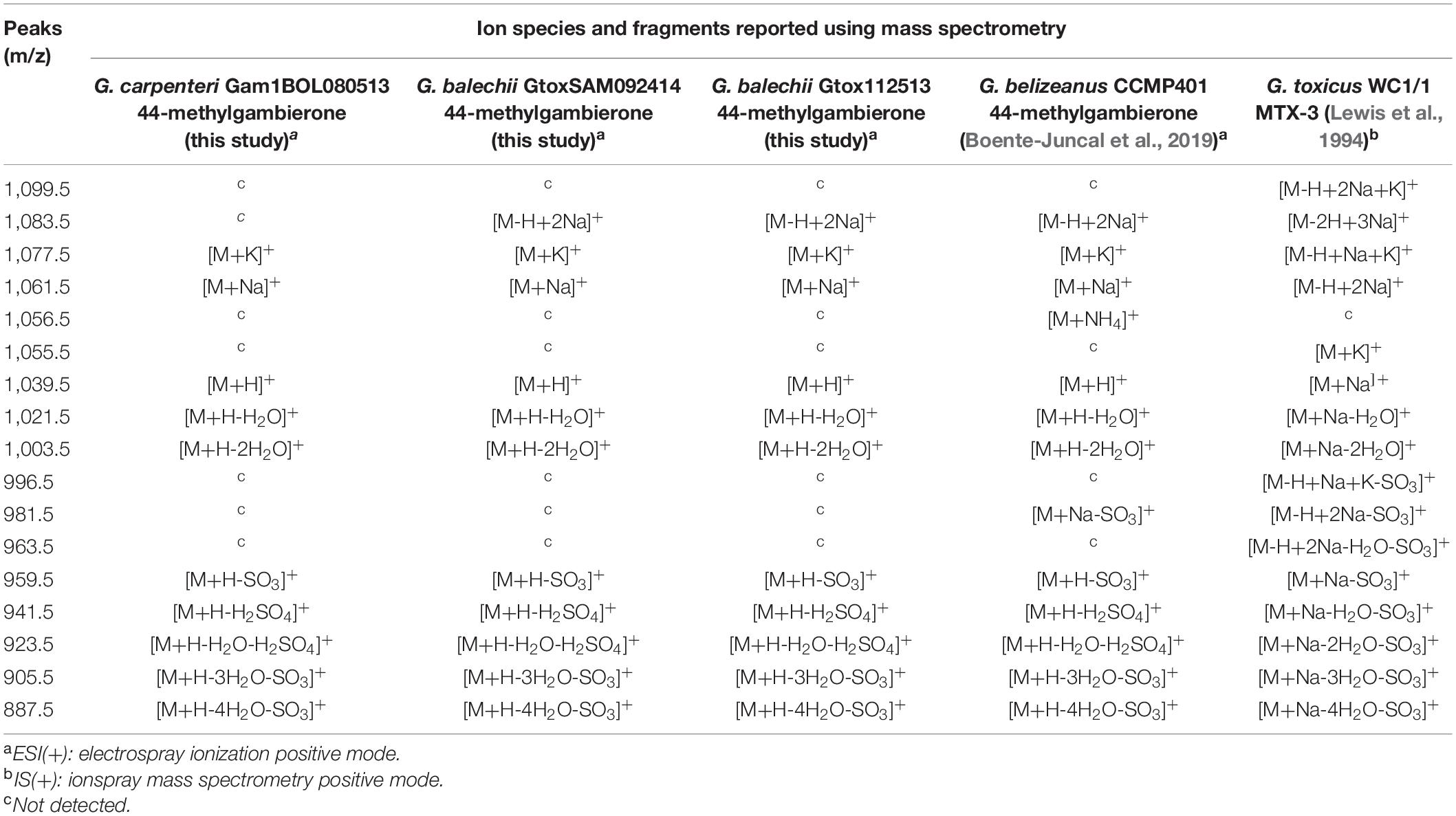
Table 2. Comparison between ions and fragments of 44-methylgambierone and MTX-3 from this study and literature values.
Identification of Gambierdiscus mainly relies on cell size, thecal morphological features, and molecular genetic data (Chinain et al., 1999; Litaker et al., 2009; Kretzschmar et al., 2017). In this study, two Gambierdiscus strains (Gtox112513, GtoxSAM092414) from Bolinao, Pangasinan, and Guiuan, Eastern Samar were characterized based on cellular and thecal plate morphology, and phylogeny inferred from D8–D10 LSU rDNA (Figures 1, 2). The morphology of these strains coincided with the key taxonomic features of G. balechii, as described by Fraga et al. (2016). Mainly, the cell size, thecal ornamentation, shape of second apical plate (2′), third precingular plate (3″) and second antapical plate (2″″) showed high resemblance to the original type material (Fraga et al., 2016). The Philippine strains possessed heavy areolation (reticulate-foveate) on their thecal plate surface, which is a character reported for G. balechii, G. belizeanus, G. cheloniae, G. lapillus, G. lewisii, G. scabrosus (Faust, 1995; Nishimura et al., 2014; Fraga et al., 2016; Smith et al., 2016; Kretzschmar et al., 2017, 2019). The shape of the taxonomically informative thecal plates i.e., hatchet shaped plate 2′, asymmetrical plate 3″ and narrow plate 2″″ of our strains was similar to G. balechii, G. cheloniae, and G. lewisii (Fraga et al., 2016; Smith et al., 2016; Kretzschmar et al., 2019). Finally, the cell dimension of the Philippine strains is nearly identical to Gambierdiscus balechii compared to other closely species (Supplementary Table 2; Fraga et al., 2016; Dai et al., 2017).
With the analyses of phylogenetic position inferred from D8-D10 LSU rDNA sequences, the morphological identification of the two Gambierdiscus strains as Gambierdiscus balechii was further resolved by forming a well-supported clade that includes the sequence of the type material of G. balechii from Celebes Sea, Manado, Indonesia (KX268470). This clade is also composed of strains from Rawa Island, Malaysia (KY235255) and another strain originally identified as Gamberidiscus type 6 from Marakei, Kiribati (KJ125112, KJ125113) but now designated as Gambierdiscus balechii (Dai et al., 2017). The identification of Gambierdiscus balechii adds to the report of this species in the tropical Pacific (Fraga et al., 2016; Zhang et al., 2016; Dai et al., 2017; Tester et al., 2020).
The toxicity of G. carpenteri Gam1BOL080513 (12.36 ± 4.38 pg PbTx-3 eq./cell) was comparable to the highest toxin content reported by Vacarizas et al. (2018) at 7.48 ± 0.49 pg PbTx-3 eq./cell. The difference may be attributed to the extraction procedure performed in this study, using acetone instead of methanol. Compared to other G. carpenteri strains in literature (Litaker et al., 2017; Pisapia et al., 2017; Díaz-Asencio et al., 2019), the Philippine strain showed relatively higher toxin content. Several Gambierdiscus species associated with macrophytes identified in Cuba, including G. carpenteri, have toxin content below the RBA limits of quantitation (Díaz-Asencio et al., 2019). G. carpenteri strains isolated from Hawaii (Pisapia et al., 2017), the Caribbean (Litaker et al., 2017), and Mexico (Litaker et al., 2017) showed femtogram levels of CTX-3C eq./cell using cell-based neuro-2a assay (CBA-N2a).
The toxin content in the Philippine strains (0.13–0.79 pg PbTx-3 eq./cell) of G. balechii was also higher than the literature values. G. balechii strains from Marekei, Kiribati have toxin values ranging from 1.1 to 19.9 fg P-CTX-1 eq./cell (Dai et al., 2017), while the Indonesian strain showed 3.4 ± 1.5 fg CTX-3C eq./cell (Pisapia et al., 2017) using CBA-N2a assay. However, a direct comparison cannot be established as the method for toxin determination, and the toxin standards used in these assays differed.
An integrated targeted and untargeted metabolite profiling approach was done to comprehensively characterize the chemistry of the three Gambierdiscus strains from the Philippines. Monosaccharides and fatty acids were profiled and quantified directly using HPLC-MS/MS and GC-FID, respectively. Secondary metabolites were assessed using unbiased UPLC-MS and MS/MS techniques. Florisil® fractionation of the crude extracts was performed to improve the detection of compounds produced at low amounts and, therefore, allowed for higher metabolite coverage in the UPLC-MS analysis. We expand and report the first chemical analysis of Gambierdiscus spp. using the GNPS platform. We focused on molecular clusters with generated GNPS annotation and manually annotated toxin clusters.
Metabolites from G. balechii strains were concentrated in the more lipophilic Florisil® fractions A and B, while G. carpenteri Gam1BOL080513 metabolites showed higher richness in the hydrophilic Florisil® fraction C. The molecular network suggests that the Gambierdiscus strains produce metabolites with high chemical diversity, including carbohydrates, lipids, and polyketides. Exploration of the molecular network (Figure 6) identified strain-specific clusters. For example, a cluster of polyhydroxylated compounds (22, 23, 38–42) was only observed in G. carpenteri Gam1BOL080513 (Figure 6). The two G. balechii strains showed related chemistries, evident from the clustering of their secondary metabolites (Figure 6 highlighted in orange) and consequently, high convergence. These species- and strain-specific clusters may be potential chemotaxonomic markers for identifying particular Gambierdiscus isolates. Further studies toward identifying these compounds and spatiotemporal variations on the production are needed to define the potential utility as chemotaxonomic markers.
The central cluster in the molecular network, which is a convergence of the three strains, contained annotated compounds belonging to terpene glycosides (2, 3) and carbohydrate conjugates (4–6). These are primarily carbohydrate-derived compounds connected to a lipophilic functional group. Based on the molecular network, these compounds are produced mainly by the two G. balechii strains.
Components of structural and storage polysaccharides were mainly detected through the targeted HPLC-MS-MRM approach (Figure 5A). The monosaccharides in the three Gambierdiscus strains are typical for microalgae (Templeton et al., 2012; Ortiz-Tena et al., 2016; Schulze et al., 2017). Glucose comprises the majority of the biomass since the main polysaccharide in dinoflagellate theca is cellulose (Okuda, 2002), and the primary storage polysaccharide for photosynthetic dinoflagellates is starch (Metting, 1996; Chen et al., 2013). Mannose and galactose were also reported as components of structural polysaccharides in dinoflagellates (Lewin et al., 1958; Brown, 1991). It is also the main organic component of mucilage found in the dinoflagellate Gonyaulax hyalina (MacKenzie et al., 2002) and a mixture of four phytoplanktonic taxa (Metaxatos et al., 2003). Benthic dinoflagellates, similar to Gambierdiscus, are known to produce mucilage as a means of attachment to their hosts (Heil et al., 1993; Rains and Parsons, 2015). In terms of the individual monosaccharide concentration, the glucose content was similar for the strains from Pangasinan, Philippines (Gam1BOL080513 and Gtox112513). A significant difference was observed for the strain from Eastern Samar, Philippines (GtoxSAM092414). In contrast, there was no evident relationship between the carbohydrate content and species suggesting spatial dependence. The carbohydrate content obtained in this study was significantly lower than other microalgae (Templeton et al., 2012; Ortiz-Tena et al., 2016; Schulze et al., 2017) and may be due to the partial hydrolysis of the complex carbohydrates using the sulfuric acid method.
Lipids and sterols are utilized as chemotaxonomic guides among dinoflagellates and eukaryotic microalgae, respectively (Leblond et al., 2006; Mooney et al., 2007; Desmond and Gribaldo, 2009). In particular, the 4-methyl sterol, dinosterol, has been used as a biomarker in marine sediments to determine past dinoflagellate blooms (Boon et al., 1979). Marine-derived terpenes are also metabolites of particular interest because of their wide range of bioactivity and structural diversity (Gross and König, 2006). Though most studies for these terpenes were in sponges and corals, symbiotic dinoflagellates particularly Symbiodinium sp. were identified as the actual producers of some of these compounds (Mydlarz et al., 2003).
Three lipid clusters were observed in the extracts of G. carpenteri and G. balechii (Figure 6). Sterols and terpenes (11–13) were putatively identified in the lipid and lipid-like molecules cluster. These compounds are common in all three strains. Other steroidal derivatives, mainly steroidal ketones (19, 20), also clustered together and were specific to the two G. balechii strains. Steroidal ketones have been recognized as possible intermediates in the diagenesis of sterols in sedimenting particulate matter and marine sediments (Gagosian and Smith, 1979; Gagosian et al., 1982).
Free fatty acids clustered together for the three strains (Figure 6) and corroborated with the GC-FAME analysis. The hydrolyzed fatty acids from lipids were similar across the three isolates (Figure 5B). One of the primary fatty acids detected was C18:1, a precursor of docosahexaenoic acid (DHA) and eicosapentanoic acid (EPA). The C18:1 fatty acid was previously suggested as fatty acid biomarker for dinoflagellates and classified as essential fatty acids to grazers (Joseph, 1975; Parrish et al., 2000). PUFAs, characteristic of benthic dinoflagellates (Usup et al., 2008), were barely detected in the FAME analysis because of the lack of standards. GNPS was able to annotate PUFAs 20-hydroxy-eicosatetraenoic acid (14, cosine score 0.84), 9-hydroxy-octadecatrienoic acid (15, cosine score 0.84), and DHA (16, cosine score 0.78) in the fatty acid cluster. Detection of C24:0 in all strains may also indicate the presence of very long chain PUFAs, which is hypothesized to be the precursor of C28 fatty acids octacosaheptaenoic [28:7(n-6)] and octacosaoctaenoic acid [28:8(n-3)] found in dinoflagellates (Leblond and Chapman, 2000). Very long chain PUFAs serve as a distinguishing feature of marine algae from higher plants (Harwood, 1998).
Since GNPS molecular networking is based on similarities among molecules, we used P-CTX-3C as a seed compound to cluster other structurally related molecules belonging to the CTX and MTX families of compounds. Surprisingly, P-CTX-3C and 44-methylgambierone appeared as single nodes (Figure 6 inset).
The 44-methylgambierone cluster (Figure 6 inset) showed the pseudomolecular ion [M+H-H2O]+ with a mass of 1021 Da. G. carpenteri Gam1BOL080513 and G. balechii Gtox112513 formed a single node for 44-methylgambierone with precursor mass of 1021.65 Da. G. balechii GtoxSAM092414 clustered with a separate node with precursor mass 1021.8 Da. The mass difference between 44-methylgambierone in the three Gambierdiscus species may be attributed to the mass error from the instrument rather than structural variation. Manual inspection of the MS and MS/MS spectra corresponding to these nodes showed similar m/z peaks (Figures 7A,B and Supplementary Figure 2) and retention time, indicating that the 44-methylgambierone in the three species are identical. Since 44-methylgambierone has an equivalent mass to 2,3-dihydroxyCTX-3C, the presence of a sulfate moiety is important to verify the maitotoxin backbone. The MS/MS fragmentation (Supplementary Figure 2) confirmed the presence of a sulfate moiety. 44-methylgambierone is the main MTX detected in this study presumably due to the high intracellular concentration of this compound in the three Gambierdiscus species.
However, the amount of 44-methylgambierone did not fully account for the observed toxicity in the RBA assay (Figure 7C) for G. carpenteri Gam1BOL080513. Since 44-methylgambierone has a low potency, the high ciguatoxicity in the RBA assay for G. carpenteri Gam1BOL080513 may suggest the production of additional toxins. Accordingly, the low RBA activity observed for G. balechii Gtox112513 and G. balechii GtoxSAM092414 may imply that 44-methylgambierone is the principal marine biotoxin in these two strains.
Detection of other MTX and CTX in G. carpenteri Gam1BOL080513 was attempted using multiple reaction monitoring (Supplementary Methods and Supplementary Table 3), as adapted from Estevez et al. (2021). Apart from 44-methylgambierone, other MTX and CTX analogs were not detected in G. carpenteri Gam1BOL080513 (Supplementary Figure 6). The targeted monitoring also validated the results from the untargeted metabolite analysis.
Other MTX and CTX congeners are usually present in very low amounts, limiting the detection of the parent mass under positive ionization. Enrichment of G. carpenteri Gam1BOL080513 through column fractionation and subsequent targeted analysis for CTX and MTX may aid in the detection of other marine biotoxins produced by this species. More sensitive methods for mass spectrometry detection, such as multiple reaction monitoring, have proved to be successful in detecting minor CTX and MTX in benthic dinoflagellate extracts. Molecular networking in the negative ionization mode may improve the detection limit by capitalizing on the presence of a sulfate moiety in MTX.
Pigments were also detected in the extracts of the three Gambierdiscus strains. Pheophorbide A (18) and pheophytin A (17), chlorophyll breakdown products, were putatively identified (cosine scores 0.80 and 0.87, respectively) in the chlorophyll derivatives cluster. Peridinin (10, cosine score 0.86) was also annotated in the polyketide cluster. Peridinin is an accessory pigment of several dinoflagellates, present as peridinin-chlorophyll-protein light harvesting complex used in photosynthesis (Song et al., 1976; Hofmann et al., 1996; Jiang et al., 2012). This pigment is also responsible for the characteristic orange to brown appearance of Gambierdiscus cells (Indelicato and Watson, 1986).
A cluster of polyhydroxylated compounds (22, 23, 38–42) specific to G. carpenteri Gam1BOL080513 was also observed. 26-(4′-carbamoyl-1,4′-bipiperidin-1′-yl)-2,15,17-trihydroxy-11-methoxy-3,7,12,14,16,18,22-heptamethyl-6,23,27,29-tetraoxo-8,30-dioxa-24-azatetracyclo[23.3.1.14,7.05,28]triaconta-1(28),2,4,9,19,21,25-heptaen-13-yl acetate (22, cosine score 0.79) was dereplicated from the GNPS spectral library. This putative compound is structurally related to linear polyhydroxylated compounds from dinoflagellates, such as the antifungal and hemolytic amphidinols, luteophanols, lingshuiol, and colopsinols produced by Amphidinium sp. (Echigoya et al., 2005). These compounds are cytotoxic through their membrane disrupting activity (Paul et al., 1997; Qi et al., 2007). Polyhydroxylated compounds are also highly valued because of their potent biological activity. Amphidinols, for example, are used as antifungal agents (Wakamiya et al., 2020). Related compounds may likewise have comparable bioactivity.
A strain-specific peptide cluster (21, 43–49) was putatively identified in the molecular network of G. balechii GtoxSAM092414. Peptide biosynthesis in dinoflagellates is understudied, principally due to the focused discovery of toxins synthesized by polyketide synthases (PKS). Non-ribosomal peptide synthetase (NRPS/PKS) are potentially widespread among dinoflagellates that are known producers of amine or amide-containing compounds such as Karenia brevis (Monroe and Van Dolah, 2008; López-Legentil et al., 2010; Van Dolah et al., 2017), and Ostreopsis spp. (Verma et al., 2019). NRPS/PKS hybrid sequences were also identified in the transcriptome of dinoflagellates which are not known producers of NRP-type metabolites, including Alexandrium (Vingiani et al., 2020) and Gambierdiscus (Kohli et al., 2017; Van Dolah et al., 2020). Brevisamide (Satake et al., 2008), ostreol A (Hwang et al., 2013), and alexandrolide (Satake et al., 2019) are NRPS-PKS hybrid compounds purified from dinoflagellates. Ostreol A is cytotoxic against brine shrimp at 0.9 μg/mL. Alexandrolide is cytotoxic against mouse lymphoid P388 cells at 4 μg/mL and inhibited the diatom Skeletonema costatum and Chattonella antiqua. NRPS-PKS hybrid compounds may serve as additional allelochemicals of dinoflagellates and could work in synergy with other toxins.
The targeted and untargeted chemical profiling and molecular networking of Philippine Gambierdiscus strains provided a glimpse of these organisms’ biosynthetic potential and chemical diversity. At the same time, we gained insights into the similarities and differences in the metabolite production in the three Gambierdiscus strains. While the production of fatty acids, carbohydrates and 44-methylgambierone are common among the three strains, we observed species- and strain-specific compounds. There is much more to marine toxins in the extracts of Gambierdiscus sp. The putative identification of polyhydroxylated and peptide-based compounds may suggest additional allelochemicals and potentially high value compounds from Gambierdiscus species. The putative identification from untargeted chemical profiling can guide targeted purification and chemical characterization of Gambierdiscus extracts.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm. nih.gov/genbank/, OL437111; OL437112 https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp, MSV000088378.
ZM: conceptualization, methodology, software, formal analysis, investigation, visualization, writing – original draft, review, and editing. GB: methodology, formal analysis, writing – original draft, review, and editing. JDC: formal analysis, resources, writing – review, and editing. JB: methodology, formal analysis, writing – review, and editing. MR and RA: resources, writing – review, and editing. LS-R: conceptualization, methodology, writing – original draft, review and editing, supervision, resources, project, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
This study was funded by the Department of Science and Technology—Philippine Council for Agriculture, Aquatic and Natural Resources Research and Development (DOST-PCAARRD) through the Hazard Detection and Mitigation Tools for Opportunistic Algal Blooms in a Changing Environment Research Program Project 3: Dynamics of Protein and Small Molecule Chemistry in HAB Causative Organisms (QMSR-MRRD-HAB-270-1275). Publication cost was supported by Upgrading Capacity, Infrastructure, and Assets for Marine Scientific Research in the Philippines (UPGRADE CIA) from the National Security Council.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank H. Junio and the Secondary Metabolites Profiling Laboratory of the Institute of Chemistry, University of the Philippines Diliman and K. B. Davis for assistance in the conduct of this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.767024/full#supplementary-material
AOAC (2000a). “Official method 963-22,” in Official Methods of Analysis of the Association of Official Analytical Chemists International, ed. W. Horwitz (Gaithersburg, MD: AOAC), 2200.
AOAC (2000b). “Official method 969-33,” in Official Methods of Analysis of the Association of Official Analytical Chemists International, ed. W. Horwitz (Gaithersburg, MD: AOAC), 2200.
Azanza, R. V., Benico, G. A., Iwataki, M., and Fukuyo, Y. (2017). Harmful Marine Dinoflagellates in the Philippines. Quezon City: The Marine Science Institute.
Bagnis, R., Kuberski, T., and Laugier, S. (1979). Clinical observations on 3,009 cases of ciguatera (fish poisoning) in the South Pacific. Am. J. Trop. Med. Hyg. 28, 1067–1073. doi: 10.4269/ajtmh.1979.28.1067
Berdalet, E., and Tester, P. A. (2018). “Key questions and recent research advances on harmful algal blooms in benthic systems,” in Global Ecology and Oceanography of Harmful Algal Blooms, eds P. M. Glibert, M. A. Burford, G. C. Pitcher, and M. M. Caldwell (Cham: Springer Nature), 261–286.
Besada, E. G., Loeblich, L. A., and Loeblich, A. R. III (1982). Observations on tropical, benthic dinoflagellates from ciguatera-endemic areas: Coolia, Gambierdiscus, and Ostreopsis. Bull. Mar. Sci. 32, 723–735.
Bligh, E. G., and Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol 37, 911–917. doi: 10.1139/cjz-2013-0052
Boente-Juncal, A., Álvarez, M., Antelo, Á, Rodríguez, I., Calabro, K., Vale, C., et al. (2019). Structure elucidation and biological evaluation of maitotoxin-3, a homologue of gambierone, from Gambierdiscus belizeanus. Toxins 11:79. doi: 10.3390/toxins11020079
Boon, J. J., Rijpstra, W. I. C., De Lange, F., De Leeuw, J. W., Yoshioka, M., and Shimizu, Y. (1979). Black Sea sterol—a molecular fossil for dinoflagellate blooms. Nature 277, 125–127. doi: 10.1038/277125a0
Brown, M. R. (1991). The amino-acid and sugar composition of 16 species of microalgae used in mariculture. J. Exp. Mar. Bio. Ecol. 145, 79–99. doi: 10.1016/0022-0981(91)90007-J
Caillaud, A., De La Iglesia, P., Darius, H. T., Pauillac, S., Aligizaki, K., Fraga, S., et al. (2010). Update on methodologies available for ciguatoxin determination: perspectives to confront the onset of ciguatera fish poisoning in Europe. Mar. Drugs 8, 1838–1907. doi: 10.3390/md8061838
Chen, C. Y., Zhao, X. Q., Yen, H. W., Ho, S. H., Cheng, C. L., Lee, D. J., et al. (2013). Microalgae-based carbohydrates for biofuel production. Biochem. Eng. J. 78, 1–10. doi: 10.1016/j.bej.2013.03.006
Chinain, M., Darius, H. T., Ung, A., Cruchet, P., Wang, Z., Ponton, D., et al. (2010). Growth and toxin production in the ciguatera-causing dinoflagellate Gambierdiscus polynesiensis (Dinophyceae) in culture. Toxicon 56, 739–750. doi: 10.1016/j.toxicon.2009.06.013
Chinain, M., Faust, M. A., and Pauillac, S. (1999). Morphology and molecular analyses of three toxic species of Gambierdiscus (Dinophyceae): G. pacificus, sp. nov., G. australes, sp. nov., and G. polynesiensis, sp. nov. J. Phycol. 35, 1282–1296. doi: 10.1046/j.1529-8817.1999.3561282.x
Chinain, M., Gatti, C. M., Roué, M., and Darius, H. T. (2020). “Ciguatera-causing dinoglagellates in the genera Gambierdiscus and Fukuyoa: distribution, ecophysiology and toxicology,” in Dinoflagellates: Morphology, Life History and Ecological Significance, ed. D. S. Rao (New York, NY: Nova Science Publishers), 405–457.
Dai, X., Mak, Y. L., Lu, C. K., Mei, H. H., Wu, J. J., Lee, W. H., et al. (2017). Taxonomic assignment of the benthic toxigenic dinoflagellate Gambierdiscus sp. type 6 as Gambierdiscus balechii (Dinophyceae), including its distribution and ciguatoxicity. Harmful Algae 67, 107–118. doi: 10.1016/j.hal.2017.07.002
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). JModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Desmond, E., and Gribaldo, S. (2009). Phylogenomics of sterol synthesis: insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biol. Evol. 1, 364–381. doi: 10.1093/gbe/evp036
Díaz-Asencio, L., Vandersea, M., Chomérat, N., Fraga, S., Clausing, R. J., Litaker, R. W., et al. (2019). Morphology, toxicity and molecular characterization of Gambierdiscus spp. towards risk assessment of ciguatera in south central Cuba. Harmful Algae 86, 119–127. doi: 10.1016/j.hal.2019.05.007
Dickey, R. W., and Plakas, S. M. (2010). Ciguatera: a public health perspective. Toxicon 56, 123–136. doi: 10.1016/j.toxicon.2009.09.008
Ding, C. Y. G., Pang, L. M., Liang, Z. X., Goh, K. K. K., Glukhov, E., Gerwick, W. H., et al. (2018). MS/MS-based molecular networking approach for the detection of aplysiatoxin-related compounds in environmental marine cyanobacteria. Mar. Drugs 16:505. doi: 10.3390/md16120505
Echigoya, R., Rhodes, L., Oshima, Y., and Satake, M. (2005). The structures of five new antifungal and hemolytic amphidinol analogs from Amphidinium carterae collected in New Zealand. Harmful Algae 4, 383–389. doi: 10.1016/j.hal.2004.07.004
Estevez, P., Castro, D., Le, M., Sibat, M., Tud, A., Dickey, R., et al. (2021). Toxicity screening of a Gambierdiscus australes strain from the Western Mediterranean Sea and identification of a novel maitotoxin analogue. Mar. Drugs 19:460.
Estevez, P., Rambla-alegre, M., Aligizaki, K., Gago-martinez, A., and Hess, P. (2020a). Use of mass spectrometry to determine the diversity of toxins produced by Gambierdiscus and Fukuyoa. Toxins 12:305.
Estevez, P., Sibat, M., Leão-Martins, J. M., Costa, P. R., Gago-Martínez, A., and Hess, P. (2020b). Liquid chromatography coupled to high-resolution mass spectrometry for the confirmation of Caribbean ciguatoxin-1 as the main toxin responsible for ciguatera poisoning caused by fish from European Atlantic coasts. Toxins 12:267. doi: 10.3390/toxins12040267
Faust, M. A. (1995). Observation of sand-dwelling toxic dinoflagellates (Dinophyceae) from widely differing sites, including two new species. J. Phycol. 31, 996–1003.
Fiorini, F., Borgonuovo, C., Ferrante, M. I., and Brönstrup, M. (2020). A metabolomics exploration of the sexual phase in the marine diatom Pseudo-nitzschia multistriata. Mar. Drugs 18, 313. doi: 10.3390/md18060313
Fraga, S., Rodriguez, F., Riobo, P., and Bravo, I. (2016). Gambierdiscus balechii sp . nov (Dinophyceae), a new benthic toxic dinoflagellate from the Celebes Sea (SW Pacific Ocean). Harmful Algae 58, 93–105. doi: 10.1016/j.hal.2016.06.004
Friedman, M. A., Fernandez, M., Backer, L. C., Dickey, R. W., Bernstein, J., Schrank, K., et al. (2017). An updated review of ciguatera fish poisoning: clinical, epidemiological, environmental, and public health management. Mar. Drugs 15:72. doi: 10.3390/md15030072
Gago-Martínez, A., Estevez, P., Castro, D., Barrios, C., Hess, P., and Sibat, M. L. (2021). “Characterisation of ciguatoxins,” in EFSA Supporting Publication 2021:EN-6649 (Vigo: University of Vigo), doi: 10.2903/sp.efsa.2021.EN-6649
Gagosian, R. B., and Smith, S. O. (1979). Steroids ketones in surface sediments from the south-west African shelf. Nature 277, 287–289. doi: 10.1038/277287a0
Gagosian, R. B., Smith, S. O., and Nigrelli, G. E. (1982). Vertical transport of steroid alcohols and ketones measured in a sediment trap experiment in the equatorial Atlantic Ocean. Geochim. Cosmochim. Acta 46, 1163–1172. doi: 10.1016/0016-7037(82)90002-3
Gross, H., and König, G. M. (2006). Terpenoids from marine organisms: unique structures and their pharmacological potential. Phytochem. Rev. 5, 115–141. doi: 10.1007/s11101-005-5464-3
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Harwood, J. L. (1998). “Membrane lipids in algae,” in Lipids in Photosynthesis: Structure, Function and Genetics. Advances in Photosynthesis and Respiration, eds S. Paul-André and M. Norio (Dordrecht: Springer), 53–64.
Heil, C. A., Maranda, L., and Shimizu, Y. (1993). “Mucus-associated dinoflagellates: large-scale culturing and estimation of growth rate,” in Toxic Phytoplankton Blooms in the Sea: Proceedings of the Fifth International Conference on Toxic Marine Phytoplankton, eds T. J. Smayda and Y. Shimizu (Amsterdam: Elsevier), 501–506.
Hofmann, E., Wrench, P. M., Sharples, F. P., Hiller, R. G., Welte, W., and Diederichs, K. (1996). Structural basis of light harvesting by carotenoids: peridinin-chlorophyll-protein from Amphidinium carterae. Science 272, 1788–1791.
Holmes, M. J., and Lewis, R. J. (1994). Purification and characterisation of large and small maitotoxins from cultured Gambierdiscus toxicus. Nat. Toxins 2, 64–72. doi: 10.1002/nt.2620020204
Holmes, M. J., Venables, B., and Lewis, R. J. (2021). Critical review and conceptual and quantitative models for the transfer and depuration of ciguatoxins in fishes. Toxins 13:515. doi: 10.3390/toxins13080515
Hwang, B. S., Yoon, E. Y., Kim, H. S., Yih, W., Park, J. Y., Jeong, H. J., et al. (2013). Ostreol A: a new cytotoxic compound isolated from the epiphytic dinoflagellate Ostreopsis cf. ovata from the coastal waters of Jeju Island, Korea. Bioorganic Med. Chem. Lett. 23, 3023–3027. doi: 10.1016/j.bmcl.2013.03.020
IAEA (2013). Detection of Harmful Algal Toxins Using the Radioligand Receptor Binding Assay A Manual of Methods. Monaco: IAEA.
Indelicato, S. R., and Watson, D. A. (1986). Identification of the photosynthetic pigments of the tropical benthic dinoflagellate Gambierdiscus toxicus. Mar. Fish. Rev. 48, 44–47.
Jiang, J., Zhang, H., Kang, Y., Bina, D., Lo, C. S., and Blankenship, R. E. (2012). Characterization of the peridinin-chlorophyll a-protein complex in the dinoflagellate Symbiodinium. Biochim. Biophys. Acta Bioenerg. 1817, 983–989. doi: 10.1016/j.bbabio.2012.03.027
Joseph, J. D. (1975). Identification of 3, 6, 9, 12, 15-octadecapentaenoic acid in laboratory-cultured photosynthetic dinoflagellates. Lipids 10, 395–403. doi: 10.1007/BF02532443
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kofoid, C. A. (1909). On Peridinium steini Jörgensen, with a note on the nomenclature of the skeleton of the Peridinidae. Arch. Für Protistenkd. 16, 25–47.
Kohli, G. S., Campbell, K., John, U., Smith, K. F., Fraga, S., Rhodes, L. L., et al. (2017). Role of modular polyketide synthases in the production of polyether ladder compounds in ciguatoxin-producing Gambierdiscus polynesiensis and G. excentricus (Dinophyceae). J. Eukaryot. Microbiol. 64, 691–706. doi: 10.1111/jeu.12405
Kohli, G. S., Papiol, G. G., Rhodes, L. L., Harwood, D. T., Selwood, A., Jerrett, A., et al. (2014). A feeding study to probe the uptake of Maitotoxin by snapper (Pagrus auratus). Harmful Algae 37, 125–132. doi: 10.1016/j.hal.2014.05.018
Kretzschmar, A. L., Larsson, M. E., Hoppenrath, M., Doblin, M. A., and Murray, S. A. (2019). Characterisation of two toxic Gambierdiscus spp. (Gonyaulacales, Dinophyceae) from the Great Barrier Reef (Australia): G. lewisii sp. nov. and G. holmesii sp. nov. Protist 170:125699. doi: 10.1016/j.protis.2019.125699
Kretzschmar, A. L., Verma, A., Harwood, T., Hoppenrath, M., and Murray, S. (2017). Characterization of Gambierdiscus lapillus sp. nov. (Gonyaulacales, Dinophyceae): a new toxic dinoflagellate from the Great Barrier Reef (Australia). J. Phycol. 53, 283–297. doi: 10.1111/jpy.12496
Leblond, J. D., Anderson, B., Kofink, D., Logares, R., Rengefors, K., and Kremp, A. (2006). Fatty acid and sterol composition of two evolutionarily closely related dinoflagellate morphospecies from cold Scandinavian brackish and freshwaters. Eur. J. Phycol. 41, 303–311. doi: 10.1080/09670260600804843
Leblond, J. D., and Chapman, P. J. (2000). Lipid class distribution of highly unsaturated long chain fatty acids in marine dinoflagellates. J. Phycol. 36, 1103–1108.
Lefort, V., Longueville, J.-E., and Gascuel, O. (2017). SMS: smart model selection in PhyML. Mol. Biol. Evol. 34, 2422–2424. doi: 10.1093/molbev/msx149
Lewin, J. C., Lewin, R. A., and Philpott, D. E. (1958). Observations on Phaeodactylum tricornutum. J. Gen. Microbiol. 18, 418–426. doi: 10.1016/j.tig.2019.05.007
Lewis, R. J., Holmes, M. J., Alewood, P. F., and Jones, A. (1994). Ionspray mass spectrometry of ciguatoxin-1, maitotoxin-2 and -3, and related marine polyether toxins. Nat. Toxins 2, 56–63.
Litaker, R. W., Holland, W. C., Hardison, D. R., Pisapia, F., Hess, P., Kibler, S. R., et al. (2017). Ciguatoxicity of Gambierdiscus and Fukuyoa species from the Caribbean and Gulf of Mexico. PLoS One 12:e0185776. doi: 10.1371/journal.pone.0185776
Litaker, R. W., Vandersea, M. W., Faust, M. A., Kibler, S. R., Chinain, M., Holmes, M. J., et al. (2009). Taxonomy of Gambierdiscus including four new species, Gambierdiscus caribaeus, Gambierdiscus carolinianus, Gambierdiscus carpenteri and Gambierdiscus ruetzleri (Gonyaulacales, Dinophyceae). Phycologia 48, 344–390. doi: 10.2216/07-15.1
Longo, S., Sibat, M., Viallon, J., Darius, H. T., Hess, P., and Chinain, M. (2019). Intraspecific variability in the toxin production and toxin profiles of in vitro cultures of Gambierdiscus polynesiensis (Dinophyceae) from French Polynesia. Toxins 11:735. doi: 10.3390/toxins11120735
López-Legentil, S., Song, B., DeTure, M., and Baden, D. G. (2010). Characterization and localization of a hybrid non-ribosomal peptide synthetase and polyketide synthase gene from the toxic dinoflagellate Karenia brevis. Mar. Biotechnol. 12, 32–41. doi: 10.1007/s10126-009-9197-y
MacKenzie, L., Sims, I., Beuzenberg, V., and Gillespie, P. (2002). Mass accumulation of mucilage caused by dinoflagellate polysaccharide exudates in Tasman Bay, New Zealand. Harmful Algae 1, 69–83. doi: 10.1016/S1568-9883(02)00006-9
Masuko, T., Minami, A., Iwasaki, N., Majima, T., Nishimura, S. I., and Lee, Y. C. (2005). Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal. Biochem. 339, 69–72. doi: 10.1016/j.ab.2004.12.001
Metaxatos, A., Panagiotopoulos, C., and Ignatiades, L. (2003). Monosaccharide and amino acid composition of mucilage material produced from a mixture of four phytoplanktonic taxa. J. Exp. Mar. Bio. Ecol. 294, 203–217. doi: 10.1016/S0022-0981(03)00269-7
Metting, F. B. (1996). Biodiversity and application of microalgae. J. Ind. Microbiol. Biotechnol. 17, 477–489. doi: 10.1007/bf01574779
Monroe, E. A., and Van Dolah, F. M. (2008). The toxic dinoflagellate Karenia brevis encodes novel type I-like polyketide synthases containing discrete catalytic domains. Protist 159, 471–482. doi: 10.1016/j.protis.2008.02.004
Montojo, U., Tanyag, B., Perelonia, K. B., Cambia, F., and Oshiro, N. (2020). Ciguatera in the Philippines: examining reef fish vectors and its causative benthic dinoflagellates in Visayan and Sibuyan Seas. Philipp. J. Fish. 27, 19–29. doi: 10.31398/tpjf/27.1.2019a0015
Mooney, B. D., Nichols, P. D., De Salas, M. F., and Hallegraeff, G. M. (2007). Lipid, fatty acid, and sterol composition of eight species of Kareniaceae (Dinophyta): chemotaxonomy and putative lipid phycotoxins. J. Phycol. 43, 101–111. doi: 10.1111/j.1529-8817.2006.00312.x
Mudge, E. M., Robertson, A., Leynse, A. K., McCarron, P., and Miles, C. O. (2021). Selective extraction of gambierone and related metabolites in Gambierdiscus silvae using m-aminophenylboronic acid–agarose gel and liquid chromatography–high-resolution mass spectrometric detection. J. Chromatogr. B 1188:123014. doi: 10.1016/j.jchromb.2021.123014
Munday, R., Murray, S., Rhodes, L. L., Larsson, M. E., and Harwood, D. T. (2017). Ciguatoxins and maitotoxins in extracts of sixteen Gambierdiscus isolates and one Fukuyoa isolate from the South Pacific and their toxicity to mice by intraperitoneal and oral administration. Mar. Drugs 15:208. doi: 10.3390/md15070208
Murata, M., Legrand, A. M., Ishibashi, Y., Fukui, M., and Yasumoto, T. (1990). Structures and configurations of ciguatoxin from the moray eel Gymnothorax javanicus. J. Am. Chem. Soc. 112:4380–4386. doi: 10.1021/ja00167a040
Murray, J. S., Nishimura, T., Finch, S. C., Rhodes, L. L., Puddick, J., Harwood, D. T., et al. (2020). The role of 44-methylgambierone in ciguatera fish poisoning: acute toxicity, production by marine microalgae and its potential as a biomarker for Gambierdiscus spp. Harmful Algae 97:101853. doi: 10.1016/j.hal.2020.101853
Murray, J. S., Selwood, A. I., Harwood, D. T., van Ginkel, R., Puddick, J., Rhodes, L. L., et al. (2019). 44-Methylgambierone, a new gambierone analogue isolated from Gambierdiscus australes. Tetrahedron Lett. 60, 621–625. doi: 10.1016/j.tetlet.2019.01.043
Mydlarz, L. D., Jacobs, R. S., Boehnlein, J., and Kerr, R. G. (2003). Pseudopterosin biosynthesis in Symbiodinium sp., the dinoflagellate symbiont of Pseudopterogorgia elisabethae. Chem. Biol. 10, 1051–1056. doi: 10.1016/j
Nagai, H., Murata, M., Torigoe, K., Satake, M., and Yasumoto, T. (1992). Gambieric acids, new potent antifungal substances with unprecedented polyether structures from a marine dinoflagellate Gambierdiscus toxicus. J. Org. Chem. 57, 5448–5453. doi: 10.1021/jo00046a029
Naman, C. B., Rattan, R., Nikoulina, S. E., Lee, J., Miller, B. W., Moss, N. A., et al. (2017). Integrating molecular networking and biological assays to target the isolation of a cytotoxic cyclic octapeptide, samoamide A, from an American Samoan marine cyanobacterium. J. Nat. Prod. 80, 625–633. doi: 10.1021/acs.jnatprod.6b00907
Nishimura, T., Sato, S., Tawong, W., Sakanari, H., Yamaguchi, H., and Adachi, M. (2014). Morphology of Gambierdiscus scabrosus sp. nov. (Gonyaulacales): a new epiphytic toxic dinoflagellate from coastal areas of Japan. J. Phycol. 50, 506–514. doi: 10.1111/jpy.12175
Nonomura, T., Sasaki, M., Matsumori, N., Murata, M., Tachibana, K., and Yasumoto, T. (1996). The complete structure of maitotoxin, Part II: configuration of the C135-C142 side chain and absolute configuration of the entire molecule. Angew. Chemie Int. Ed. 35, 1675–1678.
Okuda, K. (2002). Structure and phylogeny of cell coverings. J. Plant Res. 115, 283–288. doi: 10.1007/s10265-002-0034-x
Oliveira, C. Y. B., Oliveira, C. D. L., Müller, M. N., Santos, E. P., Dantas, D. M. M., and Gálvez, A. O. (2020). A scientometric overview of global dinoflagellate research. Publications 8, 1–18. doi: 10.3390/publications8040052
Ortiz-Tena, J. G., Rühmann, B., Schieder, D., and Sieber, V. (2016). Revealing the diversity of algal monosaccharides: fast carbohydrate fingerprinting of microalgae using crude biomass and showcasing sugar distribution in Chlorella vulgaris by biomass fractionation. Algal Res. 17, 227–235. doi: 10.1016/j.algal.2016.05.008
Parrish, C. C., Abrajano, T. A., Budge, S. M., Helleur, R. J., Hudson, E. D., Pulchan, K., et al. (2000). “Lipid and phenolic biomarkers in marine ecosystems: analysis and applications,” in Marine Chemistry. The Handbook of Environmental Chemistry, ed. P. J. Wangersky (Berlin: Springer), 193–223.
Paul, G. K., Matsumori, N., Konoki, K., Murata, M., and Tachibana, K. (1997). Chemical structures of amphidinols 5 and 6 isolated from marine dinoflagellate Amphidinium klebsii and their cholesterol-dependent membrane disruption. J. Mar. Biotechnol. 5, 124–128.
Pisapia, F., Holland, W. C., Hardison, D. R., Litaker, R. W., Fraga, S., Nishimura, T., et al. (2017). Toxicity screening of 13 Gambierdiscus strains using neuro-2a and erythrocyte lysis bioassays. Harmful Algae 63, 173–183. doi: 10.1016/j.hal.2017.02.005
Qi, X. M., Yu, B., Huang, X. C., Guo, Y. W., Zhai, Q., and Jin, R. (2007). The cytotoxicity of lingshuiol: a comparative study with amphidinol 2 on membrane permeabilizing activities. Toxicon 50, 278–282. doi: 10.1016/j.toxicon.2007.04.001
Rains, L. K., and Parsons, M. L. (2015). Gambierdiscus species exhibit different epiphytic behaviors toward a variety of macroalgal hosts. Harmful Algae 49, 29–39. doi: 10.1016/j.hal.2015.08.005
Rodríguez, I., Genta-Jouve, G., Alfonso, C., Calabro, K., Alonso, E., Sánchez, J. A., et al. (2015). Gambierone, a ladder-shaped polyether from the dinoflagellate Gambierdiscus belizeanus. Org. Lett. 17, 2392–2395. doi: 10.1021/acs.orglett.5b00902
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Sasaki, M., Matsumori, N., Nonomura, T., Murata, M., Tachibana, K., and Yasumoto, T. (1996). The complete structure of maitotoxin, Part I: configuration of the C1-C14 side chain. Angew. Chemie Int. Ed. 35, 1672–1675.
Satake, M., Bourdelais, A. J., Van Wagoner, R. M., Baden, D. G., and Wright, J. L. C. (2008). Brevisamide: an unprecedented monocyclic ether alkaloid from the dinoflagellate Karenia brevis that provides a potential model for ladder-frame initiation. Org. Lett. 10, 3465–3468. doi: 10.1021/ol801243n
Satake, M., Honma, D., Watanabe, R., and Oshima, Y. (2019). Alexandrolide, a diatom growth inhibitor isolated from the dinoflagellate Alexandrium catenella. Tetrahedron Lett. 60, 1341–1344. doi: 10.1016/j.tetlet.2019.04.019
Satake, M., Murata, M., and Yasumoto, T. (1993). Gambierol: a new toxic polyether compound isolated from the marine dinoflagellate Gambierdiscus toxicus. J. Am. Chem. Soc. 115, 361–362. doi: 10.1021/ja00054a061
Schulze, C., Strehle, A., Merdivan, S., and Mundt, S. (2017). Carbohydrates in microalgae: comparative determination by TLC, LC-MS without derivatization, and the photometric thymol-sulfuric acid method. Algal Res. 25, 372–380. doi: 10.1016/j.algal.2017.05.001
Shmukler, Y. B., and Nikishin, D. A. (2017). Ladder-shaped ion channel ligands: current state of knowledge. Mar. Drugs 15, 1–30. doi: 10.3390/md15070232
Sibat, M., Réveillon, D., Antoine, C., Carpentier, L., Rovillon, G. A., Sechet, V., et al. (2021). Molecular networking as a novel approach to unravel toxin diversity of four strains of the dominant Dinophysis species from French coastal waters. Harmful Algae 103:102026. doi: 10.1016/j.hal.2021.102026
Smith, K. F., Rhodes, L., Verma, A., Curley, B. G., Harwood, D. T., Kohli, G. S., et al. (2016). A new Gambierdiscus species (Dinophyceae) from Rarotonga, Cook Islands: Gambierdiscus cheloniae sp. nov. Harmful Algae 60, 45–56. doi: 10.1016/j.hal.2016.10.006
Soliño, L., and Costa, P. R. (2018). Differential toxin profiles of ciguatoxins in marine organisms: chemistry, fate and global distribution. Toxicon 150, 124–143. doi: 10.1016/j.toxicon.2018.05.005
Song, P.-S., Koka, P., Prézelin, B. B., and Haxo, F. T. (1976). Molecular topology of the photosynthetic light-harvesting pigment complex, peridinin-chlorophyll a-protein, from marine dinoflagellates. Biochemistry 15, 4422–4427. doi: 10.1021/bi00665a012
Templeton, D. W., Quinn, M., Van Wychen, S., Hyman, D., and Laurens, L. M. L. (2012). Separation and quantification of microalgal carbohydrates. J. Chromatogr. A 1270, 225–234. doi: 10.1016/j.chroma.2012.10.034
Tester, P., Wickliffe, L., Jossart, J., Rhodes, L., Enevoldsen, H., Adachi, M., et al. (2020). “Global distribution of genera Gambierdiscus and Fukuyoa,” in From Ecosystems to Socioecosystems. Proceedings of the 18th International Conference on Harmful Algae, ed. P. Hess (Nantes: International Society for the Study of Harmful Algae), 138–143.
Teta, R., Della Sala, G., Glukhov, E., Gerwick, L., Gerwick, W. H., Mangoni, A., et al. (2015). Combined LC-MS/MS and molecular networking approach reveals new cyanotoxins from the 2014 cyanobacterial bloom in Green Lake, Seattle. Environ. Sci. Technol. 49, 14301–14310. doi: 10.1021/acs.est.5b04415
Tibiriçá, C. E. J. D. A., Sibat, M., Fernandes, L. F., Bilien, G., Chomérat, N., Hess, P., et al. (2020). Diversity and toxicity of the genus Coolia Meunier in Brazil, and detection of 44-methyl gambierone in Coolia tropicalis. Toxins 12:327. doi: 10.3390/toxins12050327
Usup, G., Hamid, S. Z., Chiet, P. K., Wah, C. K., and Ahmad, A. (2008). Marked differences in fatty acid profiles of some planktonic and benthic marine dinoflagellates from Malaysian waters. Phycologia 47, 105–111. doi: 10.2216/07-55.1
Vacarizas, J., Benico, G., Austero, N., and Azanza, R. (2018). Taxonomy and toxin production of Gambierdiscus carpenteri (Dinophyceae) in a tropical marine ecosystem: the first record from the Philippines. Mar. Pollut. Bull. 137, 430–443. doi: 10.1016/j.marpolbul.2018.10.034
Van Dolah, F. M., Kohli, G. S., Morey, J. S., and Murray, S. A. (2017). Both modular and single-domain Type I polyketide synthases are expressed in the brevetoxin-producing dinoflagellate, Karenia brevis (Dinophyceae). J. Phycol. 53, 1325–1339. doi: 10.1111/jpy.12586
Van Dolah, F. M., Morey, J. S., Milne, S., Ung, A., Anderson, P. E., and Chinain, M. (2020). Transcriptomic analysis of polyketide synthases in a highly ciguatoxic dinoflagellate, Gambierdiscus polynesiensis and low toxicity Gambierdiscus pacificus, from French Polynesia. PLoS One 15:e0231400. doi: 10.1371/journal.pone.0231400
Verma, A., Kohli, G. S., Harwood, D. T., Ralph, P. J., and Murray, S. A. (2019). Transcriptomic investigation into polyketide toxin synthesis in Ostreopsis (Dinophyceae) species. Environ. Microbiol. 21, 4196–4211. doi: 10.1111/1462-2920.14780
Via, C. W., Glukhov, E., Costa, S., Zimba, P. V., Moeller, P. D. R., Gerwick, W. H., et al. (2018). The metabolome of a cyanobacterial bloom visualized by MS/MS-based molecular networking reveals new neurotoxic smenamide analogs (C, D, and E). Front. Chem. 6:316. doi: 10.3389/fchem.2018.00316
Vingiani, G. M., Stalberga, D., De Luca, P., Ianora, A., De Luca, D., and Lauritano, C. (2020). De novo transcriptome of the non-saxitoxin producing Alexandrium tamutum reveals new insights on harmful dinoflagellates. Mar. Drugs 18:386. doi: 10.3390/MD18080386
Wakamiya, Y., Ebine, M., Matsumori, N., and Oishi, T. (2020). Total synthesis of amphidinol 3: a general strategy for synthesizing amphidinol analogues and structure-activity relationship study. J. Am. Chem. Soc. 142, 3472–3478. doi: 10.1021/jacs.9b11789
Wang, M., Carver, J. J., Phelan, V. V., Sanchez, L. M., Garg, N., Peng, Y., et al. (2016). Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 34, 828–837. doi: 10.1038/nbt.3597
Watanabe, R., Uchida, H., Suzuki, T., Matsushima, R., Nagae, M., Toyohara, Y., et al. (2013). Gambieroxide, a novel epoxy polyether compound from the dinoflagellate Gambierdiscus toxicus GTP2 strain. Tetrahedron 69, 10299–10303. doi: 10.1016/j.tet.2013.10.022
World Health Organization (2020). World Health Organization Ciguatera Fish Poisoning. Available online at: https://www.who.int/foodsafety/areas_work/chemical-risks/CFP/en/ (accessed February 2, 2020).
Wu, H., Chen, J., Peng, J., Zhong, Y., Zheng, G., Guo, M., et al. (2020). Nontarget screening and toxicity evaluation of diol esters of okadaic acid and dinophysistoxins reveal intraspecies difference of Prorocentrum lima. Environ. Sci. Technol. 54, 12366–12375. doi: 10.1021/acs.est.0c03691
Yñiguez, A. T., Lim, P. T., Leaw, C. P., Jipanin, S. J., Iwataki, M., Benico, G., et al. (2021). Over 30 years of HABs in the Philippines and Malaysia: what have we learned? Harmful Algae 102:101776. doi: 10.1016/j.hal.2020.101776
Yon, T., Sibat, M., Robert, E., Lhaute, K., Holland, W. C., Litaker, R. W., et al. (2021). Sulfo-Gambierones, two new analogs of gambierone produced by Gambierdiscus excentricus. Mar. Drugs 19:657.
Keywords: Gambierdiscus carpenteri, Gambierdiscus balechii, ciguatera fish poisoning, marine toxins, dinoflagellates, secondary metabolites, 44-methylgambierone
Citation: Malto ZBL, Benico GA, Batucan JD, Dela Cruz J, Romero MLJ, Azanza RV and Salvador-Reyes LA (2022) Global Mass Spectrometric Analysis Reveals Chemical Diversity of Secondary Metabolites and 44-Methylgambierone Production in Philippine Gambierdiscus Strains. Front. Mar. Sci. 8:767024. doi: 10.3389/fmars.2021.767024
Received: 30 August 2021; Accepted: 13 December 2021;
Published: 04 February 2022.
Edited by:
Yehuda Benayahu, Tel Aviv University, IsraelReviewed by:
Richard J. Lewis, The University of Queensland, AustraliaCopyright © 2022 Malto, Benico, Batucan, Dela Cruz, Romero, Azanza and Salvador-Reyes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lilibeth A. Salvador-Reyes, bHNyZXllc0Btc2kudXBkLmVkdS5waA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.