
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Mar. Sci., 21 October 2021
Sec. Marine Biology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.763470
 Kaidi Jin1†
Kaidi Jin1† Baolu Zhang2†
Baolu Zhang2† Qianqian Jin1
Qianqian Jin1 Zhongqiang Cai3
Zhongqiang Cai3 Lei Wei1
Lei Wei1 Xiaomei Wang3
Xiaomei Wang3 Yanxin Zheng3
Yanxin Zheng3 Baoyu Huang1
Baoyu Huang1 Meiwei Zhang1
Meiwei Zhang1 Yitao Qi4
Yitao Qi4 Yaqiong Liu1*
Yaqiong Liu1* Xiaotong Wang1*
Xiaotong Wang1*The Fujian oyster (Crassostrea angulate) is an important marine bivalve mollusk with high economic value. Gene function research and gene editing techniques have broad application prospects in oyster. The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) system has been widely used for genome engineering in many species. CRISPR-mediated gene editing has also been used successfully in the Pacific oyster through direct delivery of the CRISPR/Cas9 components into oyster embryos by microinjection. However, the low throughput and operational difficulties associated with microinjection is one of the factors limiting the widespread application of CRISPR/Cas9 in oysters. In this study, we attempted to deliver the CRISPR/Cas9-system into the embryos of C. angulate by electroporation. An all-in-one CRISPR/Cas9 vector plasmid was used as CRISPR/Cas9 system in this study. Electroporation was carried out using both eggs and blastula larvae. A large number of larvae became malformed or die after electroporation. A single base substitution mutation was detected in the D-larvae developed from electroporated eggs. Our results demonstrate that the CRISPR/Cas9 system can be delivered into embryos of C. angulate for gene editing by electroporation, which provides a reference and will further contribute to the future application of electroporation in mollusks.
In the past few decades, the marine mollusk farming industry has been developing rapidly. Various types of economic mollusks, including oysters, have been farmed on a large scale. However, some species decrease their reproductive efficiency and economic benefits after generations in the process of aquaculture, which adversely affects the development of molluscan aquaculture. Therefore, cultivating stable molluscan strains with fast growth, good quality, and strong stress resistance through artificial selection breeding is required. The development of genomic techniques, including CRISPR/Cas9 system-mediated genomic editing technology, can improve selection of breeding techniques for mollusks (Gutierrez et al., 2018; Hollenbeck and Johnston, 2018). In recent years, an increasing number of studies have focused on investigating the functions of different oyster genes. The discovery and better comprehension of the gene function will help in understanding the mechanism of oyster growth, development, and response to stress at the molecular level. In turn, it can lay a foundation for genome research and genetic engineering breeding of mollusks.
CRISPR/Cas9 is an adaptive immune defense system, which was formed during the long-term evolution of archaea and can resist virus invasion by specific cutting of foreign DNA. The modified CRISPR/Cas9 system uses gRNA to guide Cas9 protein to recognize and cut double-stranded DNA at target sites for gene knock-out or precise DNA editing (Jinek et al., 2012; Ran et al., 2013). The CRISPR/Cas9 system is a powerful tool for reverse genetics and gene function studies. Moreover, the discovery and development of the CRISPR system has presented new opportunities in breeding. After several years of development, original varieties can be improved by targeted mutations mediated by the CRISPR technology, increasing the efficiency and accuracy of breeding.
Given its successful application in model species (Bassett et al., 2013; Chang et al., 2013; Friedland et al., 2013; Sander and Joung, 2014; Gratz et al., 2015), the CRISPR/Cas9 system has been widely applied in many non-traditional model organisms, including Bombyx mori (Wang et al., 2013), cynomolgus monkeys (Niu et al., 2014), Daphnia magna (Nakanishi et al., 2014), and Oncopeltus fasciatus (Reding and Pick, 2020). In the recent years, the application of this technology has increased in some aquaculture species. In particular, the CRISPR/Cas9 system has been exploited for genetic engineering in the sea lamprey Petromyzon marinu, Oryzias latipes, and Oreochromis niloticus, which greatly facilitates the study of gene function in fish (Ansai and Kinoshita, 2014; Li et al., 2014; Square et al., 2015). In Exopalaemon carinicauda, a wide range of indels at the E. carinicauda chitinase 4 (EcChi4) gene locus in the genome was induced by the CRISPR/Cas9 system using a microinjection method (Gui et al., 2016). EcChi4 knockout prawns showed significantly higher death rates compared with wild-type prawns when challenged with Vibrio parahaemolyticus or Aeromonas hydrophila (Sun et al., 2017). The molt-inhibiting hormone (EcMIH), β, β-carotene 9′, and 10′-oxygenase (EcBCO2) genes, and the carotenoid isomerooxygenase gene EcNinaB-X1 were also successfully knocked out using CRISPR technology. These studies will contribute to a wide application of these genes in prawns breeding (Zhang et al., 2018; Sun et al., 2020a,b). In mollusks, transgenic embryos expressing mCherry protein were successfully created in Crepidula fornicate using the CRISPR system (Perry and Henry, 2015).
Gene delivery is an important part of the genome-editing technology. Several methods, including physical, viral, and non-viral methods, are available for delivering the CRISPR/Cas9 system into the target cells (Chandrasekaran et al., 2018; Lino et al., 2018). Microinjection is a commonly used physical method of gene delivery in many animals. CRISPR/Cas9-mediated genomic editing has been successfully used in the embryonic cells of the Pacific oyster Crassostrea gigas through microinjection (Yu et al., 2019). Electroporation transfection is another commonly used physical method for gene delivery. Although electroporation is considered inefficient for gene delivery in zygote or embryonic cells, several studies have proposed technological solutions to improve the efficiency of electroporation for CRISPR delivery (Qin et al., 2015; Chen et al., 2016).
The Pitx gene is a transcription factor belonging to the highly evolutionarily conserved homeobox gene family in all animals, with a conserved function of regulating left-right asymmetry (Tran and Kioussi, 2021). In C. gigas, the expression level of the CgPitx gene was significantly higher in the right mantle, indicating that CgPitx can be associated with the asymmetric development of the left and right shells of Crassostrea gigas (Wei et al., 2018). The Fujian oyster (Crassostrea angulate) is an important marine biological resource as it is one of economic mollusks with large scale of cultivation on the southeast coast of China and even globally. In this study, we attempted to perform CRISPR-mediated gene editing by electroporation in the Fujian oyster (Crassostrea angulate), given that oyster sperm and egg cells are readily available, and high-throughput CRISPR delivery can be achieved using electroporation. The CaPitx gene was selected as the target gene for CRISPR/Cas9-mediated gene editing in the embryos of the Fujian oyster by electroporation. Our study aims to provide a reference for high-throughput CRISPR-mediated gene editing in oysters.
The Fujian oysters used in this study were collected from an aquaculture site in Xiamen, China. All oysters were acclimated in aerated seawater at 23°C for 3 days before the experiment. The oyster seedlings were grown in seawater at 23°C, micro-aerate was carried out 6 h after fertilization. After the D-larvae stage, the oyster seedlings were fed with chrysophyceae.
The gRNA target site of the CaPitx gene was designed using the online tool CCTop1 (Stemmer et al., 2015). The gRNA oligo “5′-CCCGAACTCGGGCTTCTGTG-3′” was selected for CaPitx. The pYSY-Cas9-gRNA-GFP vector plasmid, carrying a gRNA scaffold driven by a U6 promoter, a Cas9 expression cassette driven by a CMV promoter, and a GFP reporter gene driven by an EF1α promoter, was used as the express component of the all-in-one CRISPR/Cas9 system. The pYSY-Cas9-gRNA-GFP empty vector was linearized using BpiI. CaPitx-sgRNA was annealed and inserted into the empty vector using the In-Fusion cloning method to generate the CRISPR/Cas9 system in this study.
For electroporation, eggs or blastula larvae were collected and diluted to a concentration of 1 × 105 cells/mL. Then, the all-in-one CRISPR/Cas9 system (with a final concentration of 1 μg/mL) was added, and the mixture was incubated for 5 min at room temperature (21–25°C). Then, the mixture was transferred into a 0.4 cm electroporation cuvette (Bio-Rad Laboratories). Electroporation was performed in a Bio-Rad Gene Pulser Xcell system (Bio-Rad Laboratories) using a square wave pulse (100 V, with 15 ms pulse duration and four pulses separated by 100 ms pulse interval). After electroporation, the eggs or blastula larvae were washed in seawater and then fertilized with sperm or further developed in a bucket.
Fertilization rate was calculated by counting the percentage of 2-cell stage embryos 1 h after the egg and sperm were mixed. To calculate the survival rate, the living larvae were collected 24 h after fertilization; the number of living larvae was counted after the living larvae were killed by iodine tincture. The survival rate was calculated by dividing the number of living larvae by the total number of eggs. To calculate the malformation rate, iodine tincture was added to kill the living larvae collected 24 h after fertilization. Subsequently, the number of malformed larvae and normal D-larvae was counted. The malformation rate was calculated by dividing the number of malformed larvae by the total number of living larvae 24 h after fertilization.
For fluorescence analysis, D-larvae were collected and killed by iodine tincture 60 h after fertilization, and green fluorescence was observed using a fluorescence microscope. Fluorescent D-larvae were collected, and trace genomic DNA was extracted using the QIAamp DNA Micro Kit (Qiagen, United States). The target sequence was amplified by PCR, and the products were cloned into the pMD19-T Simple Vector (Takara) after purification using the AxyPrep DNA Gel Extraction Kit (Axygen). Sanger sequencing was performed to identify the mutations.
Pitx is a conserved gene that plays an important role in asymmetric development. In this study, the Hox domain of the C. angulata Pitx gene was used for gRNA design. sgRNA-CaPitx was fused into the pYSY-Cas-gRNA-GFP vector, which encodes a functional CRISPR/Cas9 system. The all-in-one fusion plasmid was then named CRISPR-CaPitx and was used as the genome-editing tool in this study.
The egg cells and blastula larvae were selected as gene-editing receptors. First, we attempted to introduce the CRISPR-CaPitx plasmid into egg cells by electroporation. Sperm and eggs were collected from the Fujian oysters with mature gonads (Figure 1A). The CRISPR-CaPitx plasmid was added to the egg suspension and mixed well, followed by a square-wave pulse under certain conditions (Figure 1A). After a short break, the egg cells were fertilized with the active sperms, and the mixture was continuously stirred in disorder for approximately 6 h until the larvae could swim. Normally developed blastula larvae have also been used as receptors for gene editing. The CRISPR-CaPitx plasmid was mixed with blastula larvae and then transferred by electroporation (Figure 1B). The oyster larvae not subjected to electroporation were used as blank controls. The oyster eggs or larvae electroporated without vector plasmids were used as negative controls.
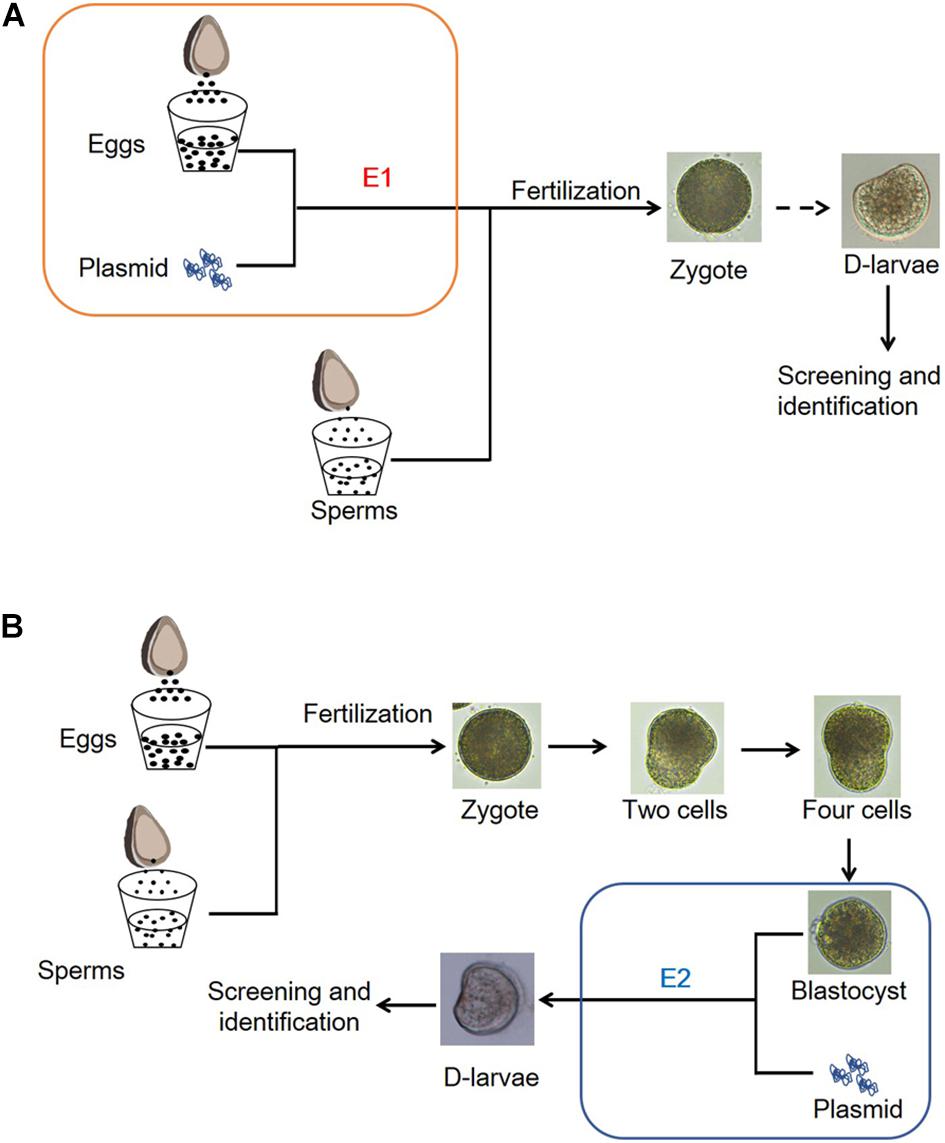
Figure 1. Schematic diagram of the mode of electroporation. (A) Eggs of the Fujian oyster were collected and then performed by electroporation before fertilization with sperms. (B) Electroporation was performed with blastula larvae.
Fertilization rates were calculated 1 h after the addition of sperm. As shown in Figure 2A, 81 and 82% of eggs were fertilized successfully after electroporation with and without plasmid, respectively. Meanwhile, 96% of zygotes were cleaved in the blank control group. Most of the larvae that developed from the electroporated eggs died 24 h later. Thus, the survival rate was calculated 24 h after fertilization and 15% of the larvae in the blank control group were alive, while only 0.6 and 1% living larvae were observed in the electroporation with and without vector, respectively (Figure 2B). Moreover, a high malformation rate was evidenced after electroporation, with more than 80% of larvae not developing into normal D-larvae (Figure 2C). When electroporation was performed on blastula larvae, the survival rates of the transfected larvae were reduced to 5 and 9% (Figure 2B), and the malformation rate was reduced to 60 and 62% after transfection with or without plasmid (Figure 2C). These results suggest that electroporation severely affects larval development, thus, causing high mortality and malformation rates.
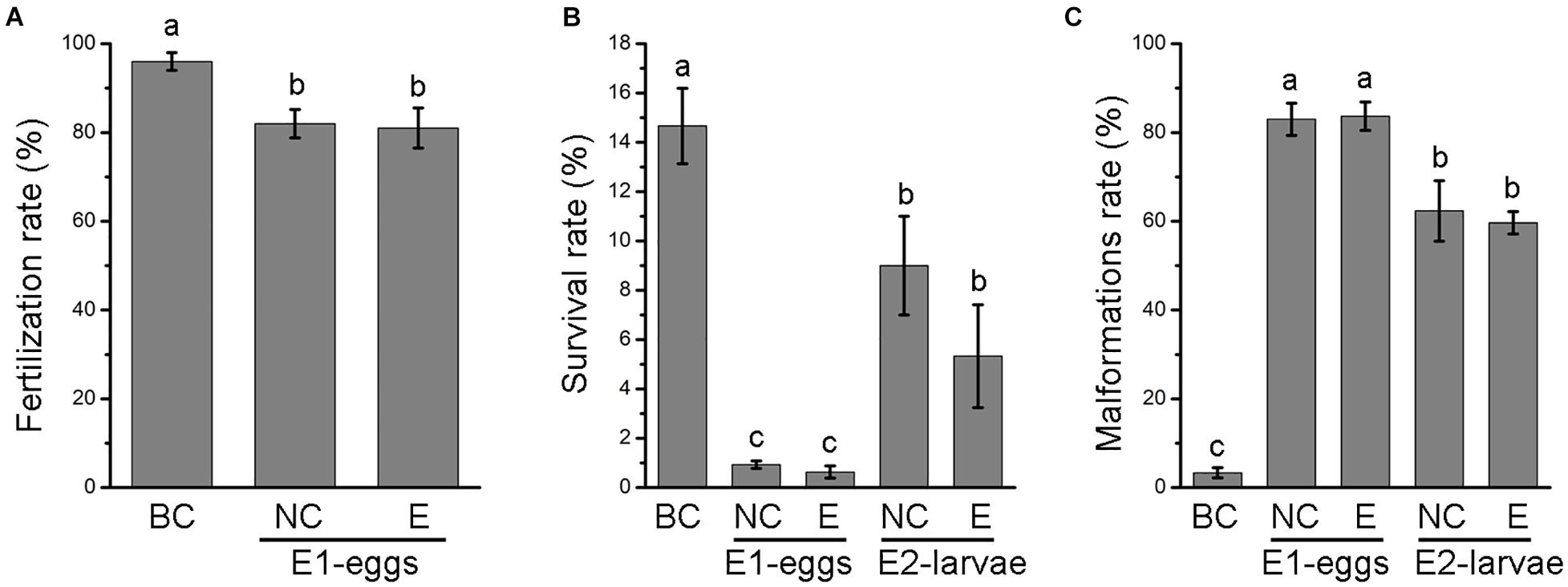
Figure 2. Fertilization rate (A), survival rate (B) and malformations rate (C) of oyster larvae after electroporation. BC, black control; NC, negative control; E, electroporation group. Error bars represent means ± SD of three independent repeats. One-way ANOVA were used for significance analysis (P < 0.05).
GFP was used as report gene to detect whether the plasmid vector was transferred into the larvae. Oyster larvae with GFP fluorescence were observed and screened by fluorescence microscopy 60 h after fertilization. Figure 3 shows that green fluorescence was detected in the larvae transfected by electroporation with the CRISPR-CaPitx plasmid, whereas no fluorescence was observed in the blank and negative control groups, indicating that the CRISPR-CaPitx plasmid was transferred into oyster larvae by electroporation. Then, D-larvae with green fluorescence were collected, and genomic DNA was extracted for amplification of the target fragment. After the purification, the target fragment was connected to a T-vector and transformed into E. coli for sequencing. Mutations were verified using Sanger sequencing. In D-larvae developed from electroporated eggs, DNA sequencing results revealed that a C to T single base substitution occurred near the Cas9 cut site (Figure 4). Although green fluorescence was observed, no mutations were detected in D-larvae developed from electroporated blastula. These results indicate that the CRISPR/Cas9 system can mediate mutagenesis in C. angulata by electroporation.
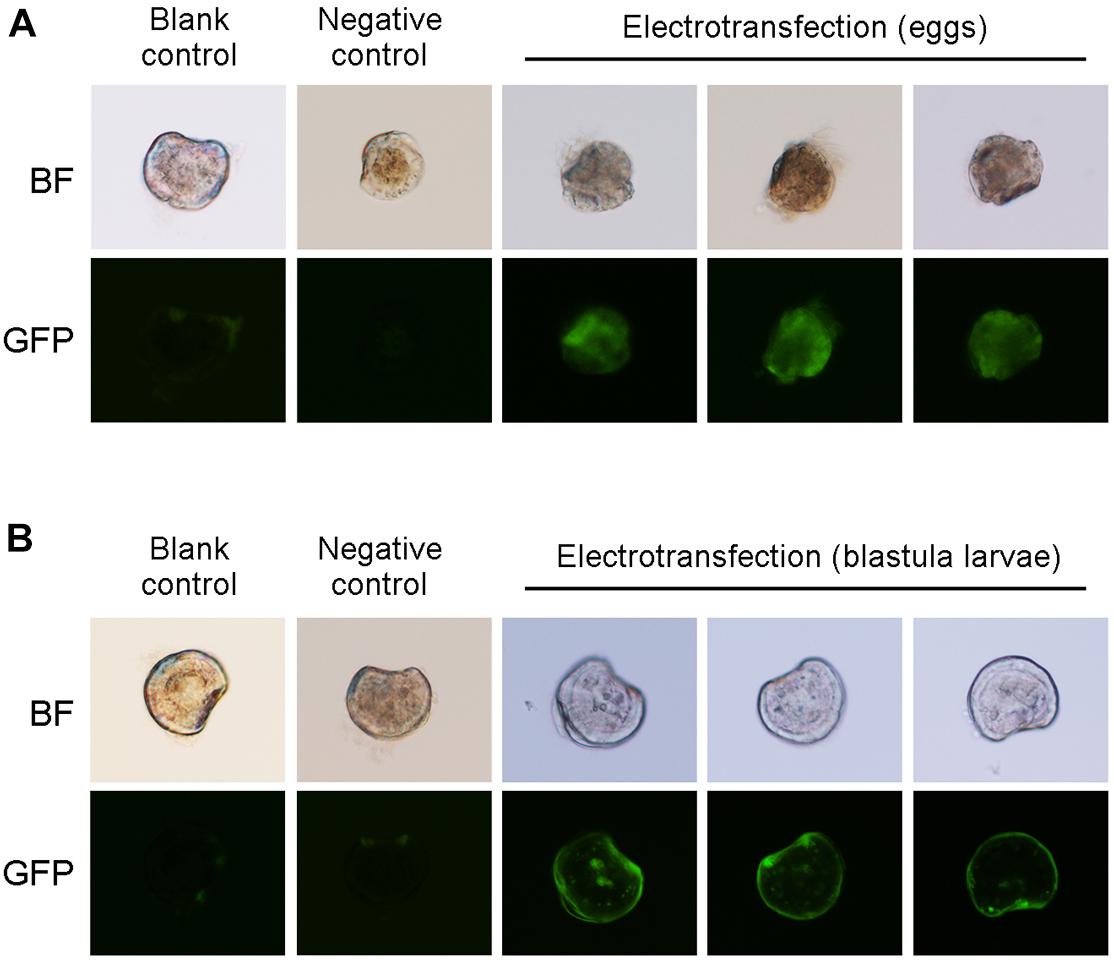
Figure 3. GFP fluorescence of the D-larvae development form electroporated eggs (A) and electroporated blastula larvae (B).
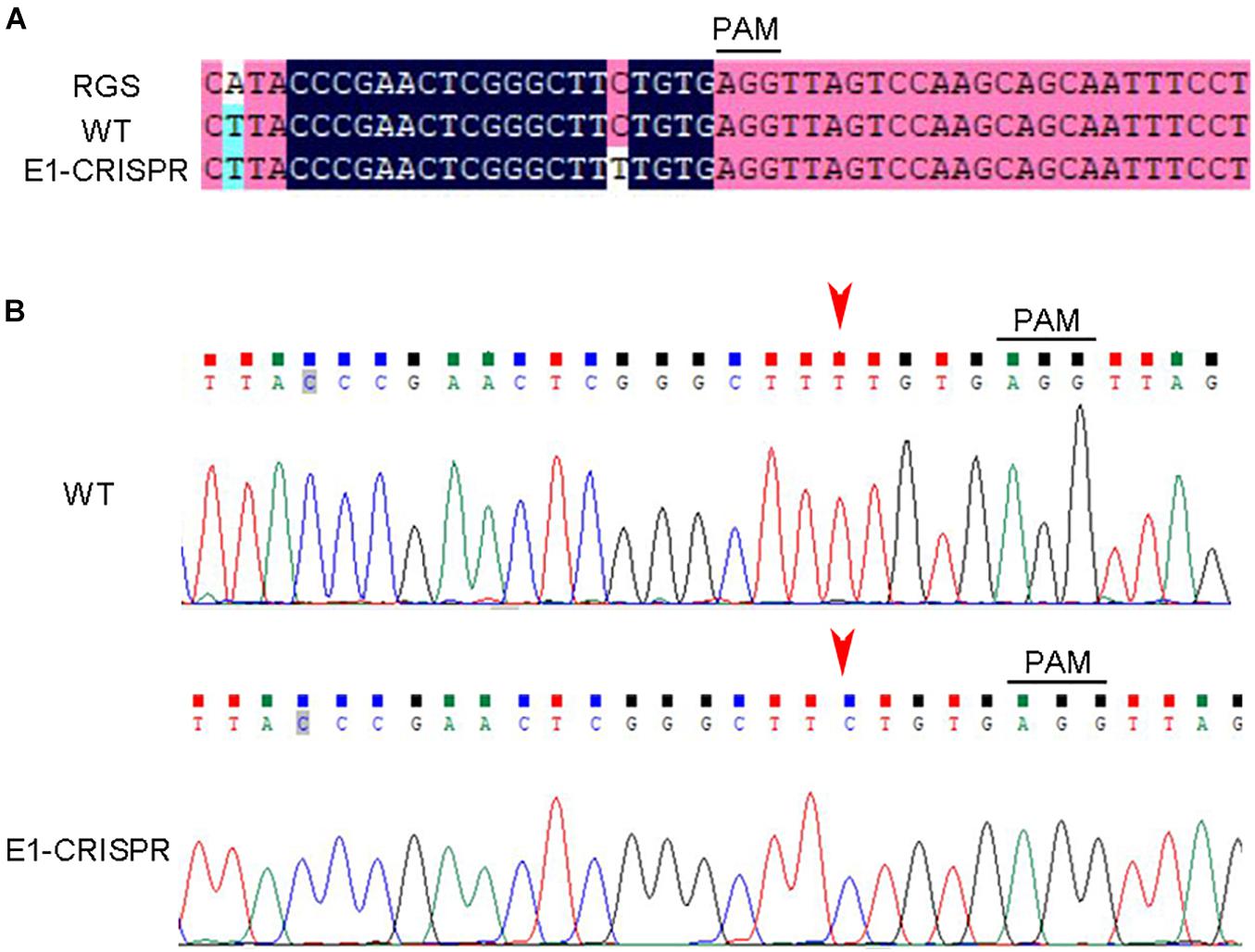
Figure 4. CRISPR/Cas9-mediated mutations in oyster D-larvae delivered by electroporation. (A) Comparison of sequencing results between wild type and edited larvae. (B) Sequencing traces of the target region of CaPitx. The PAM sequence is underlined by the black bar. The red arrow indicates the mutation site. PAM, protospacer adjacent motif. RGS, reference genome sequence. WT, wild-type. “E1-CRISPR” represent D-larvae developed from an electroporated egg.
Microinjection is a major option for delivering exogenous genetic material to many animals, including oysters (Gui et al., 2016; Chandrasekaran et al., 2018; Yu et al., 2019). Although microinjection is widely and effectively used, it has low throughput and high requirements for technology and equipment (especially for mollusks). The electroporation method was utilized to deliver the CRISPR/Cas9 system into oyster larvae in this study. First, we attempted gene editing in a single cell as much as possible. The CRISPR system was delivered into eggs by electroporation before mixing with sperm for fertilization (Figure 1A). Sixty hours later, green fluorescence was observed in whole D-larvae, suggesting that the CRISPR system was delivered into most cells of the D-larvae (Figure 3A). A single-base mutation was found and, further, confirmed near the Cas9 cleavage site (Figure 4). This finding suggests that although the CRISPR system was delivered into the D-larvae by electroporation, gene editing took place inefficiently in this study. Moreover, when electroporation was carried out on the blastula larvae, only some cells of the developed D-larvae could observe green fluorescence. In these D-larvae, no definite mutations were detected near the target set. We believe that because only a small number of the cells in the blastula larvae undergo electroporation, it is very difficult to identify even if editing occurs in some cells.
Our results also illustrated that electroporation leads to massive larval death and deformity. Due to the large number of oyster eggs and sperm, it is simple to obtain a large number of embryos. Although the success rate is very low, a certain number of successfully transfected larvae can still be obtained on the premise of a large number. It is, therefore, still possible to achieve high-throughput gene editing by electroporation despite its low transfection efficiency and high mortality and malformation rate in oysters.
The application of CRISPR-mediated gene editing technology in marine mollusks still faces great challenges, either in functional studies after gene knockout or during genetic engineering breeding. Thus, gene editing in oysters may be more suitable for studying the function of related genes during early development nowadays. To conclude, this study provides experimental data for gene editing in oyster embryos via the electroporation delivery of CRISPR. Our results can provide useful reference for a widespread application of gene editing technology in mollusks in the future.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by Institutional Review Board of LuDong University (LDU-IRB20210916001).
XTW and YL designed the experiments. KJ, BZ, QJ, BH, MZ, YQ, and YL carried out the experiments. LW, ZC, XMW, and YZ contributed to the oyster collection and oyster seedling culture. YL and XTW wrote the manuscript. All authors reviewed the manuscript.
This research was supported by the National Key R&D Program of China (No. 2018YFD0901400), the Fine Agricultural Breeds Project of Shandong Province, China (2019LZGC020), the National Natural Science Foundation of China (Nos. 42076088, 41876193, and 41906088), the Special Funds for Taishan Scholars Project of Shandong Province, China (No. tsqn201812094), the Shandong Provincial Natural Science Foundation, China (No. ZR2019MC002), the Modern Agricultural Industry Technology System of Shandong Province, China (SDAIT-14- 03), and the Plan of Excellent Youth Innovation Team of Colleges and Universities in Shandong Province, China (2019KJF004).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Haigang Qi for providing the sequence of the CaPitx gene.
Ansai, S., and Kinoshita, M. (2014). Targeted mutagenesis using CRISPR/Cas system in medaka. Biol. Open 3, 362–371. doi: 10.1242/bio.20148177
Bassett, A. R., Tibbit, C., Ponting, C. P., and Liu, J. L. (2013). Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4, 220–228. doi: 10.1016/j.celrep.2013.06.020
Chandrasekaran, A. P., Song, M., Kim, K. S., and Ramakrishna, S. (2018). Different methods of delivering CRISPR/Cas9 into cells. Prog. Mol. Biol. Transl. Sci. 159, 157–176. doi: 10.1016/bs.pmbts.2018.05.001
Chang, N., Sun, C., Gao, L., Zhu, D., Xu, X., Zhu, X., et al. (2013). Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23, 465–472. doi: 10.1038/cr.2013.45
Chen, S., Lee, B., Lee, A. Y., Modzelewski, A. J., and He, L. (2016). Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation of zygotes. J. Biol. Chem. 291, 14457–14467. doi: 10.1074/jbc.m116.733154
Friedland, A. E., Tzur, Y. B., Esvelt, K. M., Colaiácovo, M. P., Church, G. M., and Calarco, J. A. (2013). Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10, 741–743.
Gratz, S. J., Rubinstein, C. D., Harrison, M. M., Wildonger, J., and O’connor-Giles, K. M. (2015). CRISPR-Cas9 genome editing in Drosophila. Curr. Protoc. Mol. Biol. 111, 31.2.1–31.2.20.
Gui, T., Zhang, J., Song, F., Sun, Y., Xie, S., Yu, K., et al. (2016). CRISPR/Cas9-mediated genome editing and mutagenesis of EcChi4 in Exopalaemon carinicauda. G3 6, 3757–3764. doi: 10.1534/g3.116.034082
Gutierrez, A. P., Matika, O., Bean, T. P., and Houston, R. D. (2018). Genomic selection for growth traits in Pacific Oyster (Crassostrea gigas): potential of low-density marker panels for breeding value prediction. Front. Genet. 9:391. doi: 10.3389/fgene.2018.00391
Hollenbeck, C. M., and Johnston, I. A. (2018). Genomic tools and selective breeding in Molluscs. Front. Genet. 9:253. doi: 10.3389/fgene.2018.00253
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Li, M., Yang, H., Zhao, J., Fang, L., Shi, H., Li, M., et al. (2014). Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics 197, 591–599. doi: 10.1534/genetics.114.163667
Lino, C. A., Harper, J. C., Carney, J. P., and Timlin, J. A. (2018). Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 25, 1234–1257. doi: 10.1080/10717544.2018.1474964
Nakanishi, T., Kato, Y., Matsuura, T., and Watanabe, H. (2014). CRISPR/Cas-mediated targeted mutagenesis in Daphnia magna. PLoS One 9:e98363. doi: 10.1371/journal.pone.0098363
Niu, Y., Shen, B., Cui, Y., Chen, Y., Wang, J., Wang, L., et al. (2014). Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156, 836–843. doi: 10.1016/j.cell.2014.01.027
Perry, K. J., and Henry, J. Q. (2015). CRISPR/Cas9-mediated genome modification in the mollusc, Crepidula fornicata. Genesis 53, 237–244. doi: 10.1002/dvg.22843
Qin, W., Dion, S. L., Kutny, P. M., Zhang, Y., Cheng, A. W., Jillette, N. L., et al. (2015). Efficient CRISPR/Cas9-mediated genome editing in mice by zygote electroporation of nuclease. Genetics 200, 423–430. doi: 10.1534/genetics.115.176594
Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., and Zhang, F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308.
Reding, K., and Pick, L. (2020). High-efficiency CRISPR/Cas9 mutagenesis of the white gene in the milkweed bug Oncopeltus fasciatus. Genetics 215, 1027–1037. doi: 10.1534/genetics.120.303269
Sander, J. D., and Joung, J. K. (2014). CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355. doi: 10.1038/nbt.2842
Square, T., Romasek, M., Jandzik, D., Cattell, M. V., Klymkowsky, M., and Medeiros, D. M. (2015). CRISPR/Cas9-mediated mutagenesis in the sea lamprey Petromyzon marinus: a powerful tool for understanding ancestral gene functions in vertebrates. Development 142, 4180–4187.
Stemmer, M., Thumberger, T., Del Sol, Keyer, M., Wittbrodt, J., and Mateo, J. L. (2015). CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10:e0124633.
Sun, Y., Liu, M., Yan, C., Yang, H., Wu, Z., Liu, Y., et al. (2020a). CRISPR/Cas9-mediated deletion of beta, beta-carotene 9′, 10′-oxygenase gene (EcBCO2) from Exopalaemon carinicauda. Int. J. Biol. Macromol. 151, 168–177. doi: 10.1016/j.ijbiomac.2020.02.073
Sun, Y., Yan, C., Liu, M., Liu, Y., Wang, W., Cheng, W., et al. (2020b). CRISPR/Cas9-mediated deletion of one carotenoid isomerooxygenase gene (EcNinaB-X1) from Exopalaemon carinicauda. Fish Shellfish Immunol. 97, 421–431. doi: 10.1016/j.fsi.2019.12.037
Sun, Y., Zhang, J., and Xiang, J. (2017). A CRISPR/Cas9-mediated mutation in chitinase changes immune response to bacteria in Exopalaemon carinicauda. Fish Shellfish Immunol. 71, 43–49. doi: 10.1016/j.fsi.2017.09.065
Tran, T. Q., and Kioussi, C. (2021). Pitx genes in development and disease. Cell Mol. Life Sci. 78, 4921–4938. doi: 10.1007/s00018-021-03833-7
Wang, Y., Li, Z., Xu, J., Zeng, B., Ling, L., You, L., et al. (2013). The CRISPR/Cas system mediates efficient genome engineering in Bombyx mori. Cell Res. 23, 1414–1416. doi: 10.1038/cr.2013.146
Wei, L., Xu, F., Wang, Y., Cai, Z., Yu, W., He, C., et al. (2018). The molecular differentiation of anatomically paired left and right mantles of the Pacific Oyster Crassostrea gigas. Mar. Biotechnol. 20, 425–435. doi: 10.1007/s10126-018-9806-8
Yu, H., Li, H., Li, Q., Xu, R., Yue, C., and Du, S. (2019). Targeted gene disruption in Pacific Oyster based on CRISPR/Cas9 ribonucleoprotein complexes. Mar Biotechnol. 21, 301–309. doi: 10.1007/s10126-019-09885-y
Keywords: CRISPR/Cas9, electroporation, Crassostrea angulate, gene editing, mollusk
Citation: Jin K, Zhang B, Jin Q, Cai Z, Wei L, Wang X, Zheng Y, Huang B, Zhang M, Qi Y, Liu Y and Wang X (2021) CRISPR/Cas9 System-Mediated Gene Editing in the Fujian Oysters (Crassostrea angulate) by Electroporation. Front. Mar. Sci. 8:763470. doi: 10.3389/fmars.2021.763470
Received: 24 August 2021; Accepted: 27 September 2021;
Published: 21 October 2021.
Edited by:
Menghong Hu, Shanghai Ocean University, ChinaCopyright © 2021 Jin, Zhang, Jin, Cai, Wei, Wang, Zheng, Huang, Zhang, Qi, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaqiong Liu, bHlxcGthQDE2My5jb20=; Xiaotong Wang, d2FuZ3hpYW90b25nOTk5QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.