- 1School of Earth and Environmental Sciences, College of Natural Sciences, Seoul National University, Seoul, South Korea
- 2Research Institute of Oceanography, Seoul National University, Seoul, South Korea
- 3Department of Oceanography, Chonnam National University, Gwangju, South Korea
Dinoflagellates are a major component of marine ecosystems, and very cold and hot water may affect their survival. Global warming has amplified the magnitude of water temperature fluctuations. To investigate the molecular responses of dinoflagellates to very cold and hot water, we compared the differentially expressed genes of the phototrophic dinoflagellate Biecheleriopsis adriatica grown under optimal temperature and cold and heat stress. The number of genes upregulated or downregulated between optimal temperature and cold stress was twice than that between optimal temperature and heat stress. Moreover, the number of upregulated genes was greater than that of the downregulated genes under cold stress, whereas the number of upregulated genes was less than that of the downregulated genes under heat stress. Furthermore, among the differentially expressed genes, the number of genes upregulated under cold stress and with unchanged expression under heat stress was the highest, while the number of the genes downregulated under cold stress, but not under heat stress, was the second-highest. Facilitated trehalose transporter Tret1 and DnaJ-like subfamily B member 6-A were upregulated and downregulated, respectively, under cold stress; however, their expression remained unchanged under heat stress. In contrast, Apolipoprotein d lipocalin and Troponin C in skeletal muscle were upregulated and downregulated, respectively, under both cold and heat stress. This study provides insight into the genetic responses of dinoflagellates to climate change-driven large water temperature fluctuations.
Introduction
Dinoflagellates are ubiquitous and major components of marine ecosystems (Sherr and Sherr, 2007; Taylor et al., 2008; Kang et al., 2020b; Jeong et al., 2021a). They have all three trophic modes (i.e., autotrophy, mixotrophy, and heterotrophy) and play diverse roles as primary producers, prey, predators, symbionts, and parasites, which contribute to active biological interactions and material cycling in the ocean (Hansen, 1991; Coats, 1999; Jeong et al., 2010; Davy et al., 2012; Stoecker et al., 2017; Kang et al., 2018, 2019a, 2020b; Spilling et al., 2018; Eom et al., 2021). They sometimes cause red tides or harmful algal blooms, resulting in large-scale marine organism mortality and considerable economic loss to marine industries (Shumway, 1990; Landsberg, 2002; Flewelling et al., 2005; Jeong et al., 2021a; Sakamoto et al., 2021). Autotrophic/mixotrophic (phototrophic) dinoflagellates had the greatest annual integrated carbon retention in the eutrophicated Masan Bay, Korea, due to the relatively long bloom duration (Jeong et al., 2021b). Therefore, the abundance and growth rate of phototrophic dinoflagellates are important in marine ecosystems. Furthermore, diverse environmental changes, such as heat waves, cold waves, acidification, and eutrophication, affect the survival and abundance of phototrophic dinoflagellates (Gómez and Souissi, 2008; Heisler et al., 2008; Taucher et al., 2018).
Temperature is a major factor affecting dinoflagellate survival, growth rate, and abundance (Hinder et al., 2012; Kohli et al., 2014; Ok et al., 2019; Jang and Jeong, 2020; You et al., 2020). In particular, many dinoflagellates cannot survive in very cold or hot waters (Matsubara et al., 2007; Kibler et al., 2012; Xu et al., 2016; Lim et al., 2019; Kang et al., 2020a). Global warming, which is accelerating presently, has increased air and sea surface temperatures (Hansen et al., 2006; Smith et al., 2015). Furthermore, it amplifies the magnitude of water temperature fluctuations (Han and Lee, 2020). Thus, very cold periods in the winter and very hot periods in the summer are likely to affect the survival of dinoflagellates. An abnormal change in the ambient water temperature in coral reefs breaks the symbiotic relationship between the host and symbiotic dinoflagellates and subsequently causes coral bleaching (McClanahan et al., 2007; Veron et al., 2009; Graham and Nash, 2013). Thus, many studies on the effects of temperature on coral bleaching have focused on the changes in the eco-physiology and molecular responses of the host and symbiotic dinoflagellates (Jones et al., 2000; Saxby et al., 2003; Franklin et al., 2004; Leggat et al., 2011; Rosic et al., 2011; Bayer et al., 2012; Krueger et al., 2014; Levin et al., 2016; Gierz et al., 2017; Davies et al., 2018). However, very few studies have focused on understanding the effects of temperature on the molecular responses in dinoflagellates.
The temperature range for the survival of dinoflagellates is important for cell cycle, photosynthesis, distribution, and bloom dynamics (Iglesias-Prieto et al., 1992; Pospelova et al., 2008; Jeong et al., 2015; Jang and Jeong, 2020; Kang et al., 2020a). Dinoflagellates have experienced fluctuation in seawater temperature for a long time, and some species have evolved to adapt and survive in these environments (Brinkhuis et al., 1998; Crouch et al., 2001; Sluijs et al., 2005). Each dinoflagellate species has an optimal temperature for the maximum growth rate and the lowest and highest temperature limits for survival, which are different for each dinoflagellate species (Matsubara et al., 2007; Xu et al., 2010; Kibler et al., 2012; Jeong et al., 2018; Lim et al., 2019; Ok et al., 2019; Kang et al., 2020a; You et al., 2020). Some dinoflagellate species can survive at 4°C, while some can survive at 34°C (Morton et al., 1992; Band-Schmidt et al., 2003; Tong et al., 2010; Zheng et al., 2012; Xu et al., 2016). The survival of a dinoflagellate at a certain temperature may be affected by the presence and expression of temperature stress-regulating genes (Levin et al., 2016; Gierz et al., 2017; Davies et al., 2018).
To understand the responses of a dinoflagellate to a certain temperature that causes mortality, transcriptomes of the dinoflagellate grown under optimal temperature and cold and heat stress should be compared simultaneously. There have been several studies on comparative transcriptome analysis of dinoflagellates grown under optimal temperature and heat stress (Barshis et al., 2014; Fridey, 2015; Levin et al., 2016; Gierz et al., 2017; Davies et al., 2018; Bellantuono et al., 2019; Lin et al., 2019). However, only a few studies have compared the expression of a few target genes of dinoflagellates grown under optimal temperature and cold stress or optimal temperature and cold and heat stress (Deng et al., 2019; Kim et al., 2021; Wang et al., 2021).
The phototrophic dinoflagellate Biecheleriopsis adriatica, which belongs to the family Suessiaceae, was first described in 2009 (Moestrup et al., 2009). This dinoflagellate has been reported in the waters of many countries, including Korea, China, Greece, Japan, Palau Island, Philippines, and Singapore, as well as in the Adriatic Sea (Moestrup et al., 2009; Takahashi et al., 2014; Jang et al., 2015; Luo et al., 2015; Benico et al., 2019; Kang et al., 2019b; Tsipas, 2020). Furthermore, B. adriatica was reported to form a bloom with the phototrophic dinoflagellate Takayama sp., resulting in mass mortality of farmed milkfish in 2016 in Bolinao, Philippines (Benico et al., 2019). It is known that B. adriatica is consumed by many common heterotrophic dinoflagellates such as Aduncodinium glandula, Oxyrrhis marina, Gyrodinium dominans, Gyrodinium moestrupii, Luciella masanensis, Pfiesteria piscicida, and Oblea rotunda and the common naked ciliates Strombidinopsis sp. and Pelagostrobilidium sp. (Kang et al., 2019a). Thus, B. adriatica likely plays important ecological roles in marine ecosystems. Biecheleriopsis adriatica is positioned at the base of the clade consisting of the phototrophic dinoflagellate Ansanella granifera and species belonging to the family Symbiodiniaceae (LaJeunesse et al., 2018). Thus, B. adriatica is an important species for understanding the evolution of the order Suessiales. Cells of B. adriatica have been found in oceans worldwide at water temperatures of 17.7–32.0°C. The winter temperature of Korean waters is often below 5°C, whereas the summer temperature is above 30°C (Korea Hydrographic and Oceanographic Agency, 2021)1. Thus, in Korean waters, B. adriatica may produce cold-resistant and heat-resistant enzymes under cold and heat stress, respectively. Therefore, temperature-regulation-related genes may be differentially expressed under optimal temperature and cold and heat stress conditions.
This study explored differential gene expression of B. adriatica grown under optimal temperature and cold and heat stress. Furthermore, the differential expression pattern (upregulated, unchanged, and downregulated) of genes under cold stress, optimal temperature, and heat stress was investigated. The differential expression of genes under cold stress and optimal temperature and that under optimal temperature and heat stress yielded nine patterns. The results of this study provide a basis for understanding the molecular signs of responses of dinoflagellates to cold and heat stress and their adaption to global climate changes.
Materials and Methods
Preparation of Experimental Organism and Incubation at the Target Temperatures
Biecheleriopsis adriatica was isolated from plankton samples collected from surface waters off the coast of Tongyoung, southern Korea, using plankton samplers in August 2006, when the water temperature and salinity were 28.0°C and 31.0, respectively (Jang et al., 2015). The collected samples were filtered using a 154-μm Nitex mesh, and a clonal culture of B. adriatica BATY06 was established using two consecutive single-cell isolations (Jang et al., 2015). When B. adriatica concentration increased sufficiently, it was transferred to 32-, 270-, and 500-mL polycarbonate bottles containing f/2-Si medium (Guillard and Ryther, 1962). The bottles were placed on a shelf at 20°C for the cold stress experiment and 25°C for the optimal temperature and heat stress experiments under a 14 h:10 h light:dark cycle and illuminated with 50 μmol photons m–2 s–1 using cool white fluorescent light.
To determine the optimal, lowest, and highest water temperatures for survival, the growth rates of B. adriatica were measured at 14–34°C. The growth rates were almost zero at 14 and 32°C; thus, these two temperatures were used for cold and heat stress. The optimal temperature for maximum growth rate was 25°C.
For cold stress, a dense B. adriatica culture (approximately 100,000 cells mL–1) grown autotrophically at 20°C was distributed to three 800-mL tissue culture flasks (Falcon; Heidelberg, Germany) containing fresh f/2-Si medium. For heat stress and optimal temperature, a dense B. adriatica culture (approximately 100,000 cells mL–1) grown autotrophically at 25°C was distributed to six 800-mL tissue culture flasks containing fresh f/2-Si medium.
For RNA sequencing, the triplicate flasks for each target temperature were incubated in three temperature-controlled chambers set up at 14, 25, and 32°C for 11 days after gradual acclimation (Figure 1A). The temperature in each chamber was monitored using HOBO data loggers (HOBO MX2202 Pendant MX Temperature/Light Data Logger; Onset; Bourne, MA, United States).
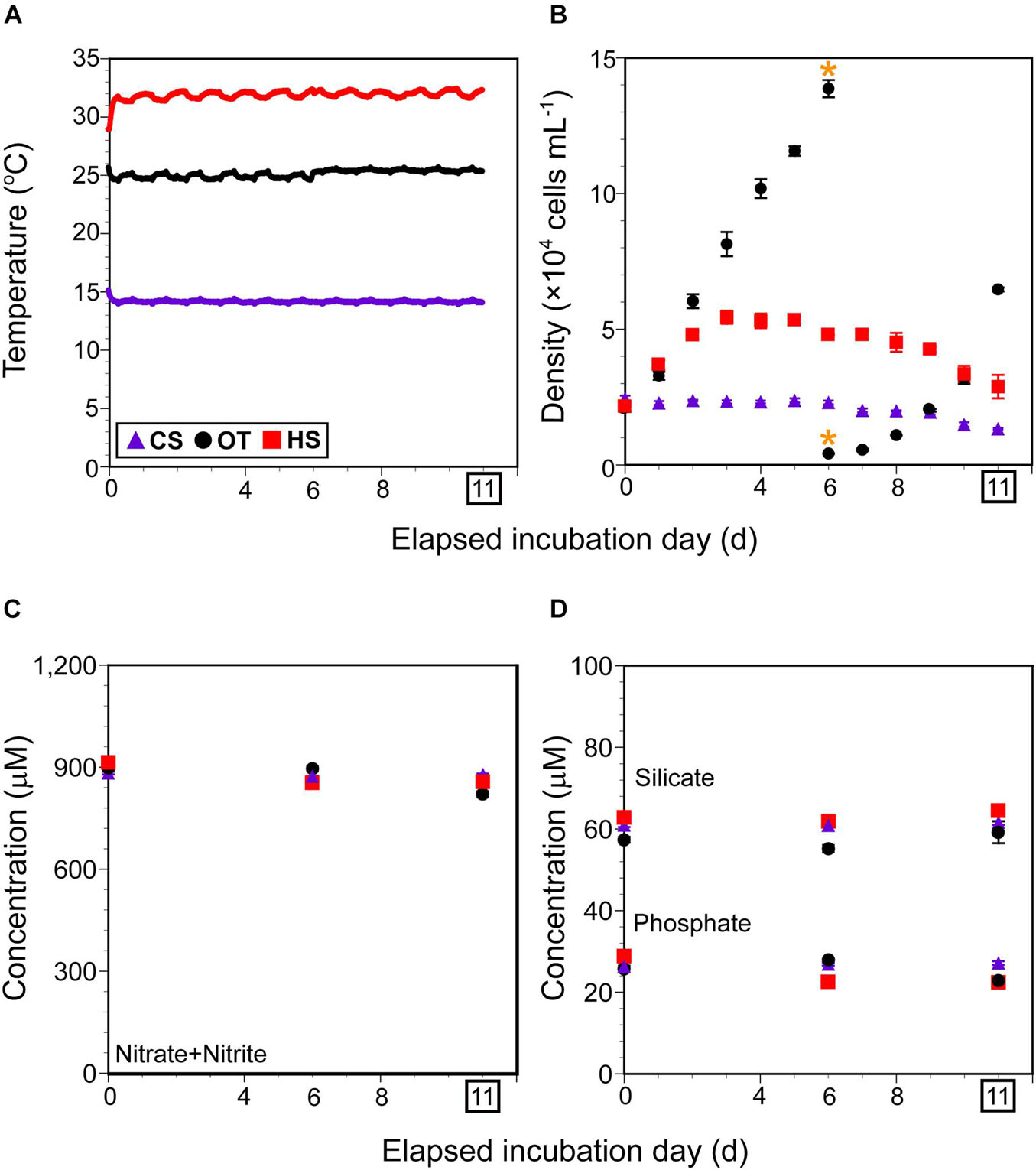
Figure 1. Incubation condition and cell density of Biecheleriopsis adriatica during the incubation. (A) Temperature (°C) as a function of elapsed incubation day in each temperature-controlled chamber monitored using HOBO data loggers for cold stress (CS), heat stress (HS), and optimal temperature (OT). (B) Average B. adriatica cell density (× 104 cells mL–1) in the triplicate flasks as a function of elapsed incubation day under CS, HS, and OT. Yellow asterisks indicate the day on which cultures were transferred into new flasks at OT. (C,D) Average concentration (μM) of nitrate plus nitrite, silicate, and phosphate in the triplicate flasks at days 0, 6, and 11 under each temperature condition. After incubating for 11 days under each condition, B. adriatica cells were harvested for RNA sequencing. Purple triangles, black circles, and red rectangles indicate CS, OT, and HS data points, respectively. Vertical symbols represent treatment means ± 1 SE.
To monitor B. adriatica cell density at each target temperature, a 5-mL aliquot was collected from each flask and fixed with 5% acidic Lugol’s solution daily. Aliquots were collected from the fixed samples, and ≥ 200 B. adriatica cells were enumerated using a Sedgewick-Rafter counting chamber to determine B. adriatica density. After incubating for 6 days at 25°C, B. adriatica reached the stationary phase and were transferred into new 800-mL flasks containing fresh f/2-Si medium.
To confirm that nutrient concentration did not affect B. adriatica growth, a 10-mL aliquot was collected from each flask on days 0, 6, and 11, gently filtered through GF/F filters (Whatman Inc., Floreham Park, NJ, United States), and stored at –20°C until nutrient concentration measurement. The concentrations of nitrate plus nitrite (hereafter NO3), phosphate (PO4), and silicate (SiO2) were measured using a nutrient auto-analyzer system (QuAAtro; Seal Analytical GmbH; Nordestedt, Germany). The samples diluted to 1/10 were used for the measurement to consider the limit concentration of the auto-analyzer system, and it was considered when the final nutrient concentration was calculated after the measurement.
Harvesting, RNA Extraction, and cDNA Library Construction
To harvest the similar cell number of B. adriatica (1 × 107 cells) after incubating for 11 days, 600, 200, and 300 mL aliquots were collected from each flask at 14, 25, and 32°C, respectively. The cells in the aliquots were collected by centrifugation at 2,808× g for 5 min (Labogene 1696R; Gyrozen Co., Gimpo, Korea), immediately frozen in liquid nitrogen, and stored at –80°C until RNA extraction.
Total RNA was extracted individually from each sample using the RNeasy Plant Mini Kit (Cat. No. 74903; Qiagen; Hilden, Germany) and treated with RNase-Free DNase set (Cat. No. 79254; Qiagen) to eliminate residual genomic DNA contaminants. The extracted RNA samples were purified using a RNeasy Kit (Cat. No. 74104; Qiagen), and the total RNA amount was assessed using Quant-IT RiboGreen (Cat. No. R11490; Invitrogen; Carlsbad, CA, United States). The integrity of the total RNA was evaluated using the TapeStation RNA Screen Tape (Cat. No. 5067-5576; Agilent Technologies; Santa Clara, CA, United States), and only RNA preparations with RNA integrity number (RIN) > 7.0 were used for RNA library construction.
The nine cDNA libraries were separately constructed using an Illumina TruSeq Stranded mRNA LT Sample Prep Kit (San Diego, CA, United States), and paired-end (2 × 100 bp) RNA of the pooled libraries was sequenced on an Illumina NovaSeq 6000 at Macrogen (Seoul, Korea). In the first step of this process, only eukaryotic polyadenylated mRNA was filtered to eliminate potential bacterial contaminants from the total RNA of B. adriatica.
The quality, total bases, total reads, GC content (%), and basic statistics of the sequenced raw reads of the nine libraries were calculated, and the raw reads of low-quality, adapter, contaminant DNA, or PCR duplicates were removed using Trimmomatic program (Bolger et al., 2014). The quality of the trimmed reads (clean reads) was checked using FastQC v0.11.72 before assembly.
Transcriptome Analyses
The clean reads for the nine libraries were assembled de novo using Trinity with the “–SS_lib_type FR” option, which is typically used for de novo transcriptome reconstruction (Grabherr et al., 2011). It combines the read sequences of a certain length of overlap to form long fragments without N gaps, called contigs. Only contigs longer than 200 bp were selected. The longest assembled contigs were filtered, and redundant contigs were removed (i.e., unigenes) using CD-HIT-EST, with a clustering identity of 0.98 (Li and Godzik, 2006).
For functional annotation, the unigenes were searched against NCBI non-redundant (NCBI nr) and Gene Ontology (GO) using BLASTx of DIAMOND software with an E-value default cutoff of 1.0E–5 (Buchfink et al., 2015).
Unigene expression levels were estimated using RSEM v1.3.1 (Li and Dewey, 2011). The clean reads from each sample were mapped back onto the assembled unigenes, and the read count value of each unigene was obtained from the mapping results. The unigene expression level (log2 fold change) between the samples under cold stress and optimal temperature or under heat stress and optimal temperature was calculated as fragments per kilobase of unigene per million mapped reads (FPKM). If more than one FPKM value was 0, it was excluded from the analysis to avoid reducing the power of the statistical hypothesis testing. After the addition of + 1 to each of the FPKM values from filtered unigenes, they were transformed to log2 scale and subjected to relative log expression (RLE) normalization. Statistically significant results of differential gene expression were obtained with |log2 fold change| ≥ 1 and p < 0.05, using the nbinomWaldTest of DESeq2 R library (Anders and Huber, 2010). Hierarchical clustering analysis of differentially expressed genes (DEGs) was performed using the Euclidean method and complete linkage, representing the similarity of expression patterns between unigenes and samples.
Based on the GO annotation results, functional unigene classification and enrichment were analyzed using Web Gene Ontology Annotation Plot (WEGO)3. The Pearson’s chi-square test was automatically carried out using WEGO to test whether the GO categories were significantly differentiated between the upregulated and downregulated gene datasets (Ye et al., 2006). Differences in GO categories between the two datasets were considered significant at a p-value < 0.05. In addition, the number of DEGs obtained with |log2 fold change| ≥ 1 and adjusted p-value (padj) < 0.05 was also determined (Benjamini and Hochberg, 1995).
Classification of Unigenes by the Trend in Expression Among the Conditions
The unigenes were classified into nine types based on the trends in expression under cold and heat stress against that under optimal temperature (Figure 2). Type 1 was composed of DEGs upregulated under both cold and heat stress; Type 2 consisted of unigenes upregulated under cold stress, but not significantly differentially expressed between optimal temperature and heat stress; Type 3 was composed of DEGs upregulated under cold stress but downregulated under heat stress; Type 4 was composed of unigenes not significantly differentially expressed between cold stress and optimal temperature but upregulated under heat stress; Type 5 consisted of unigenes not significantly differentially expressed between cold stress and optimal temperature and also between heat stress and optimal temperature; Type 6 consisted of unigenes not significantly differentially expressed between cold stress and optimal temperature but downregulated under heat stress; Type 7 was composed of DEGs downregulated under cold stress but upregulated under heat stress; Type 8 was composed of unigenes downregulated under cold stress but not significantly differentially expressed between optimal temperature and heat stress; Type 9 consisted of DEGs downregulated under both cold and heat stress. The number of unigenes in each type and the proportion of each type to all unigenes or DEGs was determined.
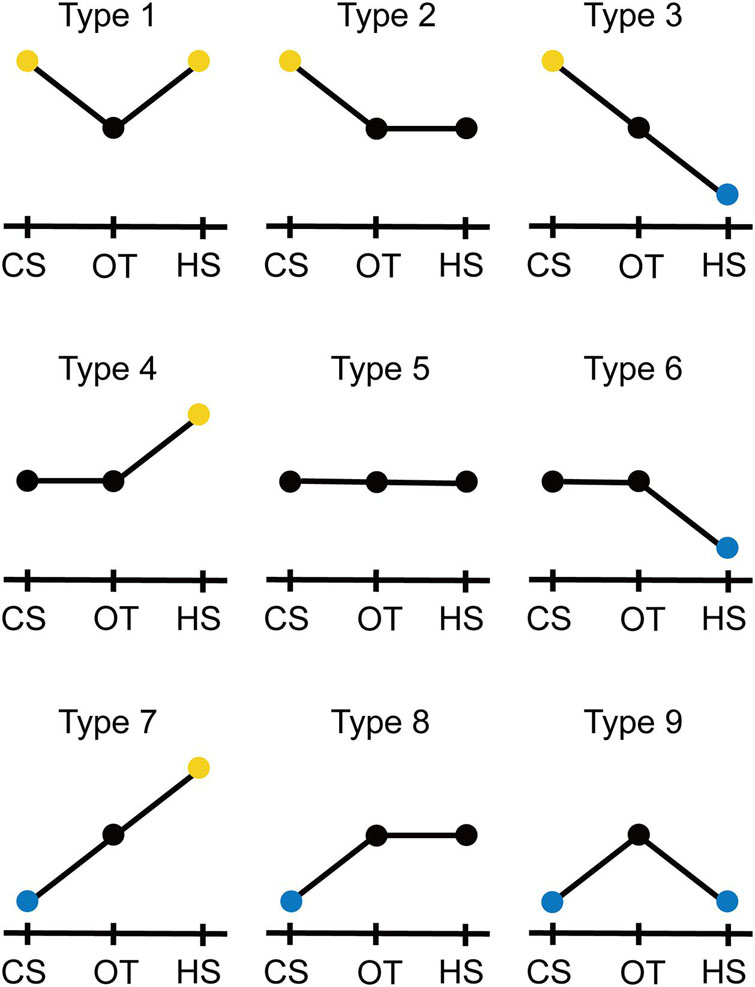
Figure 2. Standard of classification of the unigenes to nine types based on expression trend under cold stress (CS) and heat stress (HS) against optimal temperature (OT). Yellow and blue circles indicate the unigenes upregulated and downregulated, respectively, under CS or HS against OT. Black circles indicate the unigenes under the OT and with unchanged expression under CS or HS against OT. See details in “Materials and Methods” section.
To investigate the functions of representative DEGs belonging to each type, the unigenes annotated against eukaryotic organisms in the NCBI nr database with E-value cutoff E–30 were ranked by their expression level, and the functions of the high-ranking unigenes were examined. The hypothetical or predicted proteins were excluded from the identification and further screened using an NCBI conserved domain search. Unigenes with functions revealed previously were obtained. The rank of the unigenes was determined as follows: In the first step, the fold change values of the unigene expression under cold stress were arranged in the descending or ascending orders if they were upregulated or downregulated, respectively. In the second step, the unigenes were ranked. In the third step, the first and second steps were repeated for the fold change values of the same unigenes under heat stress. In the fourth step, the two rank numbers assigned to each unigene under cold and heat stress were added, and the unigenes were arranged according to the ascending order of the added number. If there was no differential expression of unigenes under cold or heat stress depending on the type, they were not calculated. Therefore, Type 5 was excluded from this calculation and examination of the function of the unigenes.
Results
Cell Density and Nutrient Concentration Under Three Different Temperatures
Under cold stress, the average B. adriatica cell density (± standard error) decreased slowly from 24,500 (± 1,030) cells mL–1 on day 0 to 13,330 (± 390) cells mL–1 on day 11, when they were harvested for RNA sequencing (Figure 1B). Under heat stress, the average B. adriatica cell density increased from 21,500 (± 1,410) cells mL–1 on day 0 to 54,400 (± 2,390) cells mL–1 on day 3 and then decreased slowly to 28,900 (± 4,320) cells mL–1 on day 11 (Figure 1B). Under optimal temperature, the average B. adriatica cell density increased from 20,500 (± 920) cells mL–1 on day 0 to 138,700 (± 3,180) cells mL–1 on day 6, entering the stationary phase. Thus, these cultures were transferred into new 800 mL flasks at an initial density of 4,200 (± 140) cells mL–1, and the B. adriatica cell density increased to 64,700 (± 770) cells mL–1 on day 11 (Figure 1B).
Under cold stress, the average (± standard error) NO3 concentration at day 0 was 882.8 (± 3.8) μM and remained unchanged on days 6 and 11 (Figure 1C). Similarly, the average PO4 and SiO2 concentration at day 0 were 26.9 (± 0.4) and 60.9 (± 0.5) μM, respectively, and remained unchanged on days 6 and 11 (Figure 1D).
The average NO3, PO4, and SiO2 concentrations on days 0, 6, and 11 under optimal temperature and heat stress were similar to those under cold stress (Figures 1C,D).
General Transcriptome Features of B. adriatica
Illumina sequencing of B. adriatica generated 130–202 million paired-end 100-bp clean reads per library (nine libraries = three triplicates at each temperature condition), with a trimming process for removing adapter sequences and poor-quality bases (Table 1). The GC content of the clean reads was between 54.30 and 54.92% and the ratio of bases with Phred quality scores ≥ 20 and 30 (Q20 and Q30) of the clean reads exceeded 99.14 and 96.70%, respectively (Table 1). A total of 198,703 unigenes were generated by a combined de novo assembly with an average 850 bp length and a 1,562 bp N50 (Table 1). Among the 198,703 unigenes, 35,370 (17.8%) and 65,580 (33.0%) were functionally annotated against the GO and NCBI nr databases, respectively, with an E-value default cutoff of 1.0E – 5.
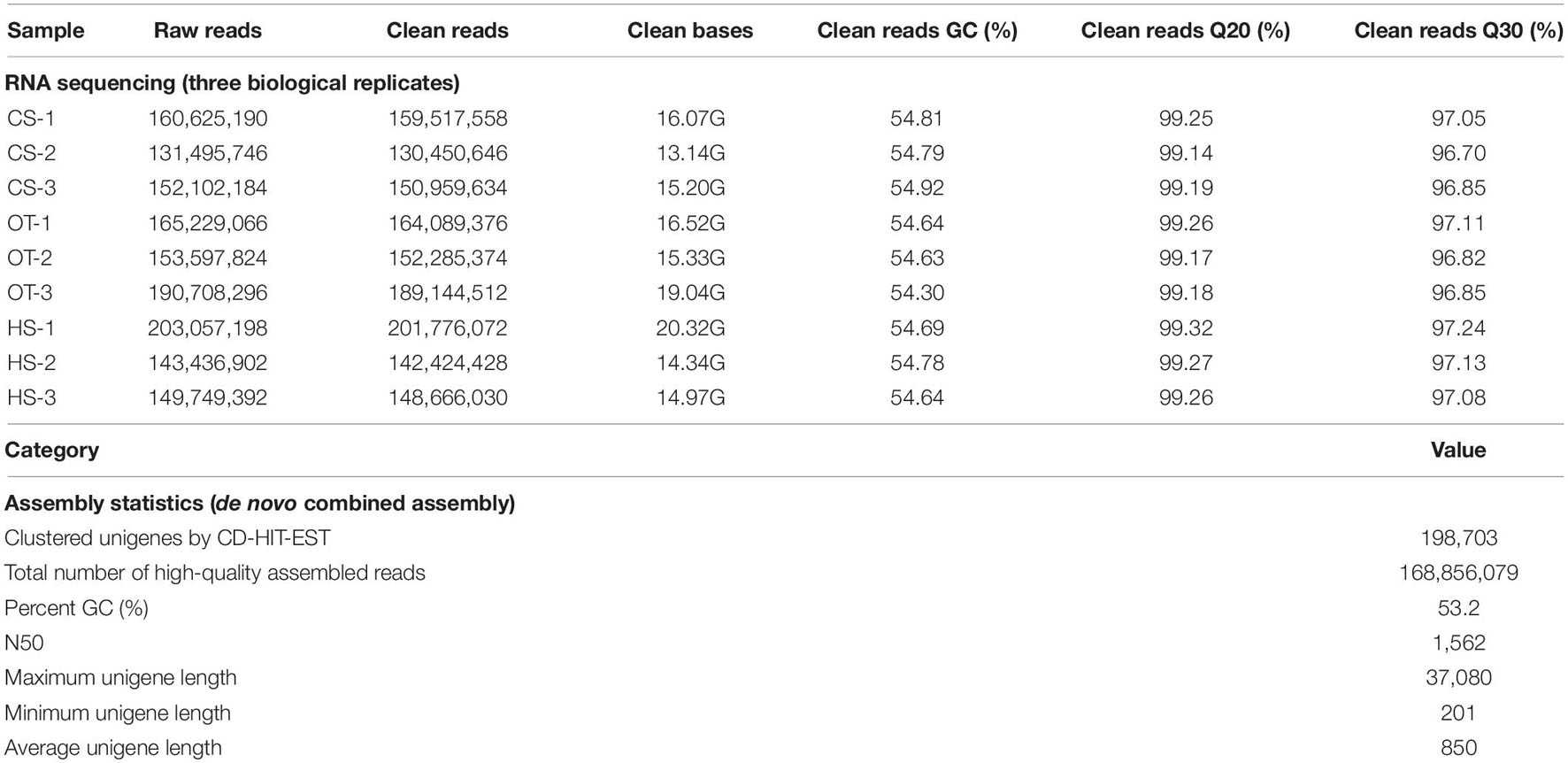
Table 1. Overall statistical results of the Illumina transcriptome sequencing and de novo assembly. CS, cold stress; OT, optimal temperature; HT, heat stress.
The number of DEGs in B. adriatica under cold and heat stress were 24,916 and 13,366, respectively, at the expression level of |log2 fold change| ≥ 1 and p < 0.05 (Figure 3). Under cold stress, 13,159 and 11,757 unigenes were upregulated and downregulated, respectively. In comparison, 6,345 upregulated and 7,021 downregulated unigenes were identified under heat stress (Figure 3). At the expression level of |log2 fold change| ≥ 1 and padj < 0.05, the number of DEGs in B. adriatica under cold and heat stress were 23,965 and 12,185, respectively (Supplementary Figure 1), and 12,812 and 11,153 genes were upregulated and downregulated, respectively, whereas 5,892 upregulated and 6,293 downregulated genes were identified under heat stress.
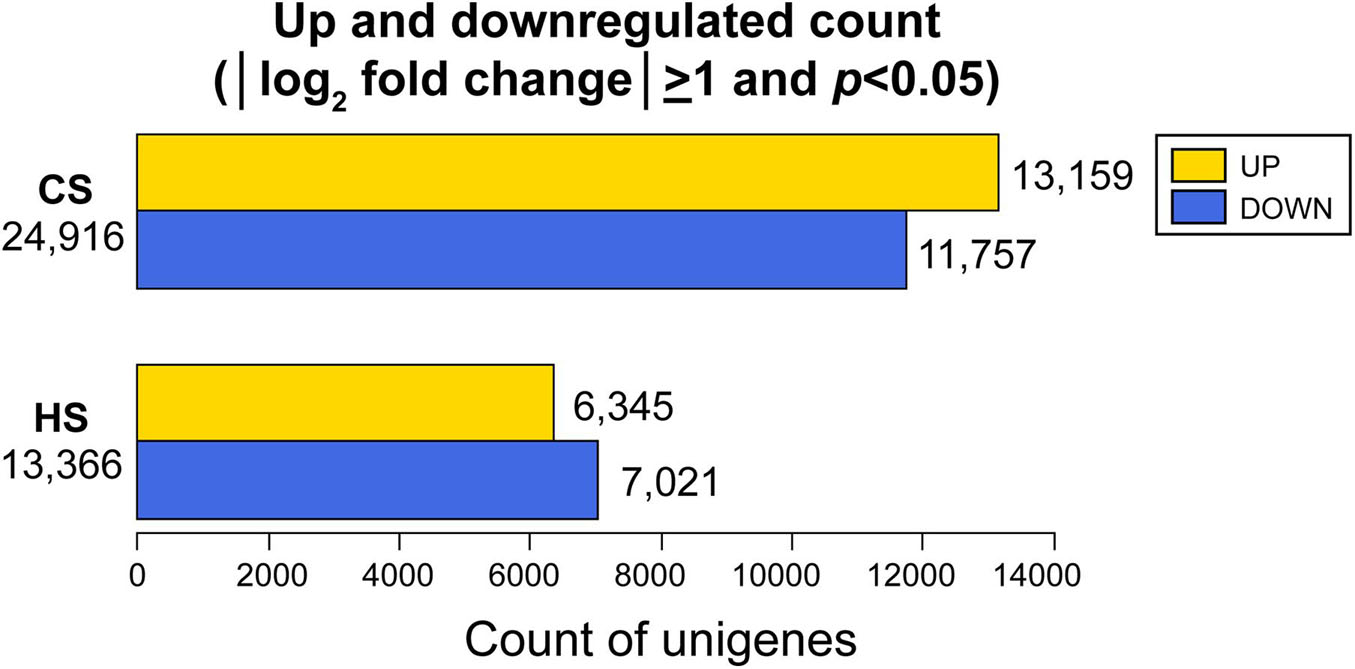
Figure 3. Upregulated and downregulated unigenes obtained with |log2 fold change| ≥ 1 and p < 0.05 under cold stress (CS) and heat stress (HS) against optimal temperature.
The heatmap for the DEGs depicting the result of hierarchical clustering analysis showed more similar patterns of DEGs between heat stress and optimal temperature than those between cold stress and optimal temperature (Figure 4).

Figure 4. Heat map of hierarchical clustering using the expression for 3,000 unigenes selected randomly (row; |log2 fold change| ≥ 1, p < 0.05) in nine samples (columns). The expression levels of each unigene across samples are shown as their row Z-scores. CS, cold stress; OT, optimal temperature; HS, heat stress.
Functional Classification of B. adriatica Differentially Expressed Genes Under Cold Stress
The biological function of the identified DEGs of B. adriatica under cold stress was investigated using GO analysis. The results of GO functional classification and enrichment were visualized and compared using WEGO (Figure 5).
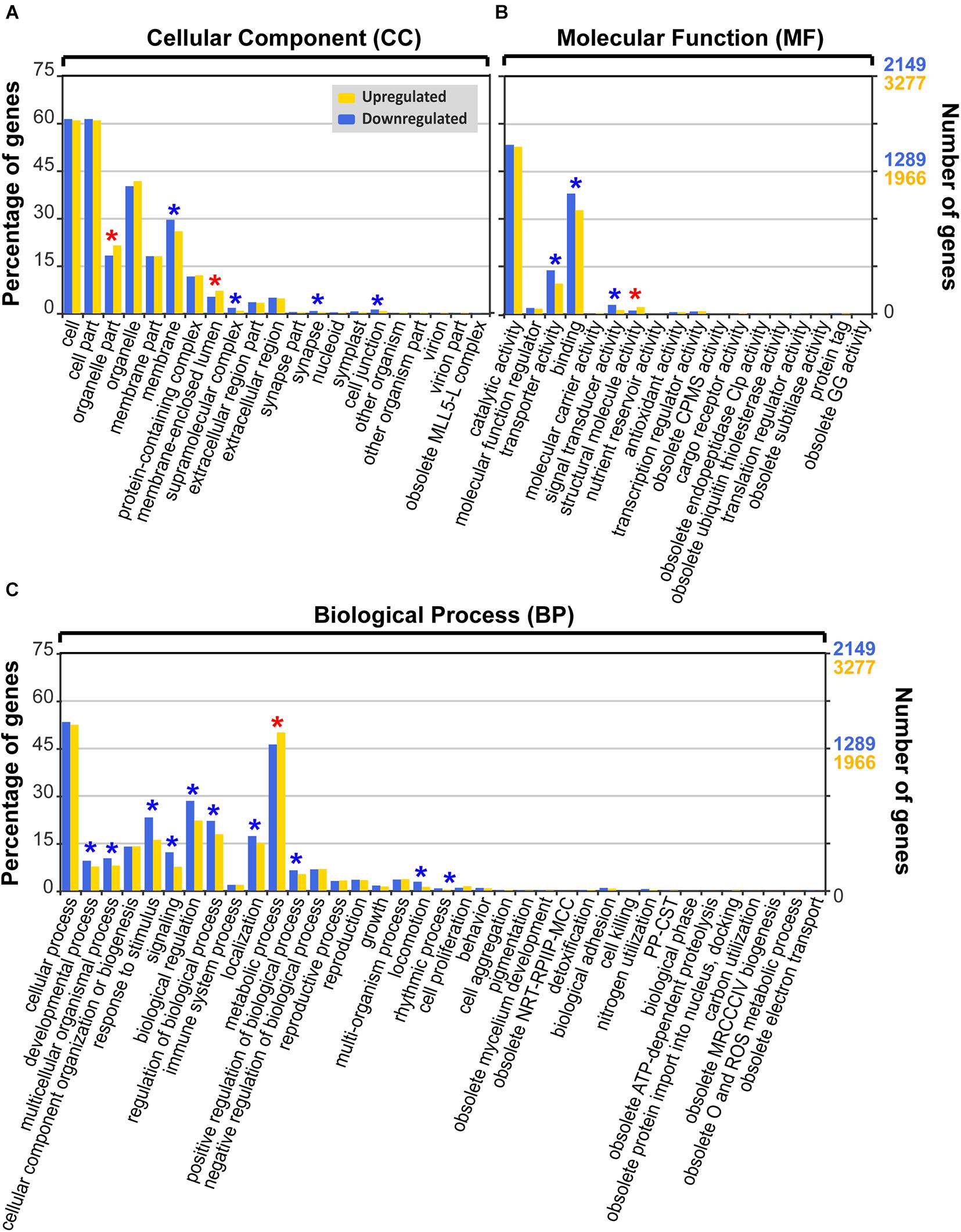
Figure 5. Gene Ontology (GO) functional classification (level 2) of differentially expressed genes (DEGs) of Biecheleriopsis adriatica under cold stress against optimal temperature. Category: (A) cellular component, (B) molecular function, and (C) biological process. The GO subcategories with significantly more upregulated genes than downregulated genes (p < 0.05, Pearson’s chi-squared test) are marked as red asterisks, whereas the GO subcategories with significantly more downregulated genes than upregulated genes are marked as blue asterisks. CPMS, cyclic pyranopterin monophosphate synthase; GG, gamma-glutamyltransferase; NRT-RPIIP-MCC, negative regulation of transcription from RNA polymerase II promoter during mitotic cell cycle; PP-CST, presynaptic process involved in chemical synaptic transmission; MRCCCIV, mitochondrial respiratory chain complex IV; O, oxygen; ROS, reactive oxygen species.
Under cold stress, 4,370 upregulated and 2,866 downregulated DEGs were annotated to the GO database (29.0% DEGs under cold stress), which were classified into 21, 18, and 37 GO subcategories in cellular component (CC), molecular function (MF), and biological process (BP) categories, respectively, in GO level 2 (Figure 5 and Supplementary Table 1).
In the CC category, “cell” and “cell part” subcategories possessed most DEGs, followed by “organelle” and “membrane” (Figure 5A and Supplementary Table 1). Six of the 21 subcategories had significantly different percentages of upregulated and downregulated DEGs (Pearson’s chi-squared test; p < 0.05) (Figure 5A, Table 2, and Supplementary Table 1). The percentage of upregulated DEGs was higher than that of downregulated DEGs in “membrane-enclosed lumen” and “organelle part” subcategories (Pearson’s chi-squared test; p = 0.001, p = 0.001), whereas the percentage of downregulated DEGs was higher than that of upregulated DEGs in “supramolecular complex,” “membrane,” “synapse,” and “cell junction” subcategories (Pearson’s chi-squared test; p < 0.001, p = 0.001, p = 0.013, p = 0.026; Figure 5A, Table 2, and Supplementary Table 1).
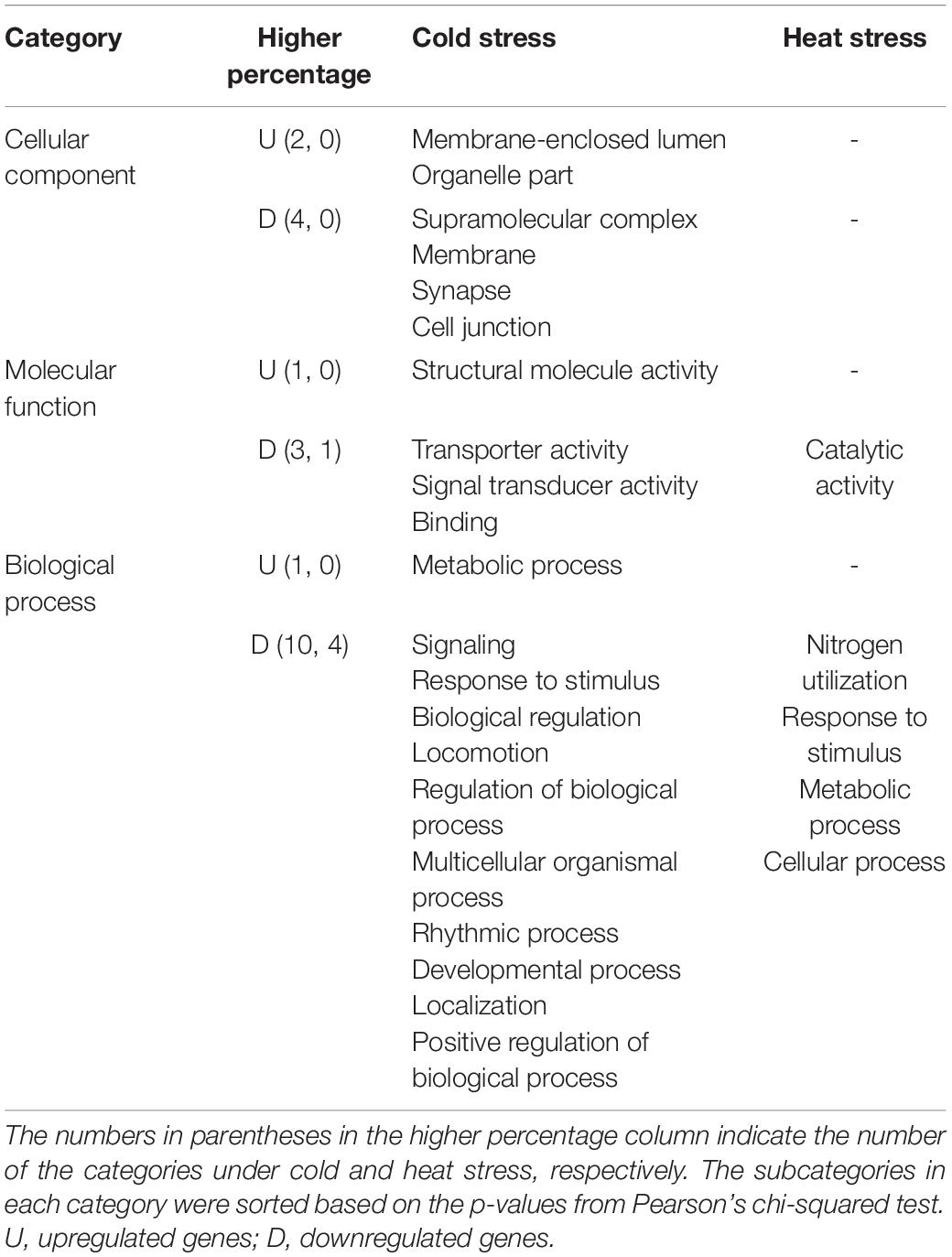
Table 2. Comparison of the subcategories with significant difference between the percentages of the upregulated and downregulated genes (Pearson’s chi-squared test; p < 0.05) in Biecheleriopsis adriatica under cold and heat stress in Gene Ontology (GO) functional classification level 2.
In the MF category, most DEGs belonged to “catalytic activity” and “binding” subcategories (Figure 5B and Supplementary Table 1). Four of the 18 subcategories had significantly different percentages of upregulated and downregulated DEGs (Pearson’s chi-squared test; p < 0.05; Figure 5B, Table 2, and Supplementary Table 1). The percentage of upregulated DEGs was higher than that of downregulated DEGs in the “structural molecule activity” subcategory (Pearson chi-squared test; p = 0.001), whereas the percentage of downregulated DEGs was higher than that of upregulated DEGs in “transporter activity,” “signal transducer activity,” and “binding” subcategories (Pearson’s chi-squared test; p < 0.001, p < 0.001, p < 0.001; Figure 5B, Table 2 and Supplementary Table 1).
In the BP category, “cellular process” subcategory had the highest percentage of DEGs, followed by “metabolic process” and “biological regulation” subcategories (Figure 5C and Supplementary Table 1). Eleven of the 37 subcategories had significantly different percentages of upregulated and downregulated DEGs (Pearson’s chi-squared test; p < 0.05; Figure 5C, Table 2, and Supplementary Table 1). Among them, the percentage of upregulated DEGs in “metabolic process” subcategory was higher than that of downregulated DEGs (Pearson’s chi-squared test; p = 0.001; Figure 5C, Table 2, and Supplementary Table 1).
At the expression level of |log2 fold change| ≥ 1 and padj < 0.05, there was no change in the trend of the results of GO analysis at level 2 under cold stress (Supplementary Figure 2).
In GO level 3, 77 subcategories had significant differences between the percentages of the upregulated and downregulated genes under cold stress (Pearson’s chi-squared test; p < 0.05; Supplementary Table 2). Among them, 23, 13, and 41 subcategories belonged to the CC, MF, and BP categories, respectively (Supplementary Table 2). Furthermore, the percentage of upregulated DEGs of 17 subcategories was higher than that of downregulated DEGs, whereas the remaining 60 subcategories had a higher percentage of downregulated DEGs than that of upregulated DEGs (Supplementary Table 2). The top 5 subcategories in GO level 3 with the largest differences between the percentage of the upregulated and downregulated genes significantly in B. adriatica under cold stress, based on the p-value, were “methylation,” “ion binding,” “cell communication,” “transmembrane transporter activity,” and “catalytic activity, acting on RNA” in sequence (Table 3).

Table 3. Comparison of the top five subcategories with the largest significant difference between the percentages of the upregulated and downregulated genes in Biecheleriopsis adriatica under cold and heat stress in Gene Ontology (GO) functional classification level 3.
Functional Classification of B. adriatica Differentially Expressed Genes Under Heat Stress
When the biological function of the identified B. adriatica DEGs under heat stress was investigated, 1,451 upregulated and 1,504 downregulated genes were annotated to the GO database (22.1% DEGs under heat stress), which were classified into 21, 12, and 33 GO subcategories in the CC, MF, and BP categories, respectively, at GO level 2 (Figure 6).
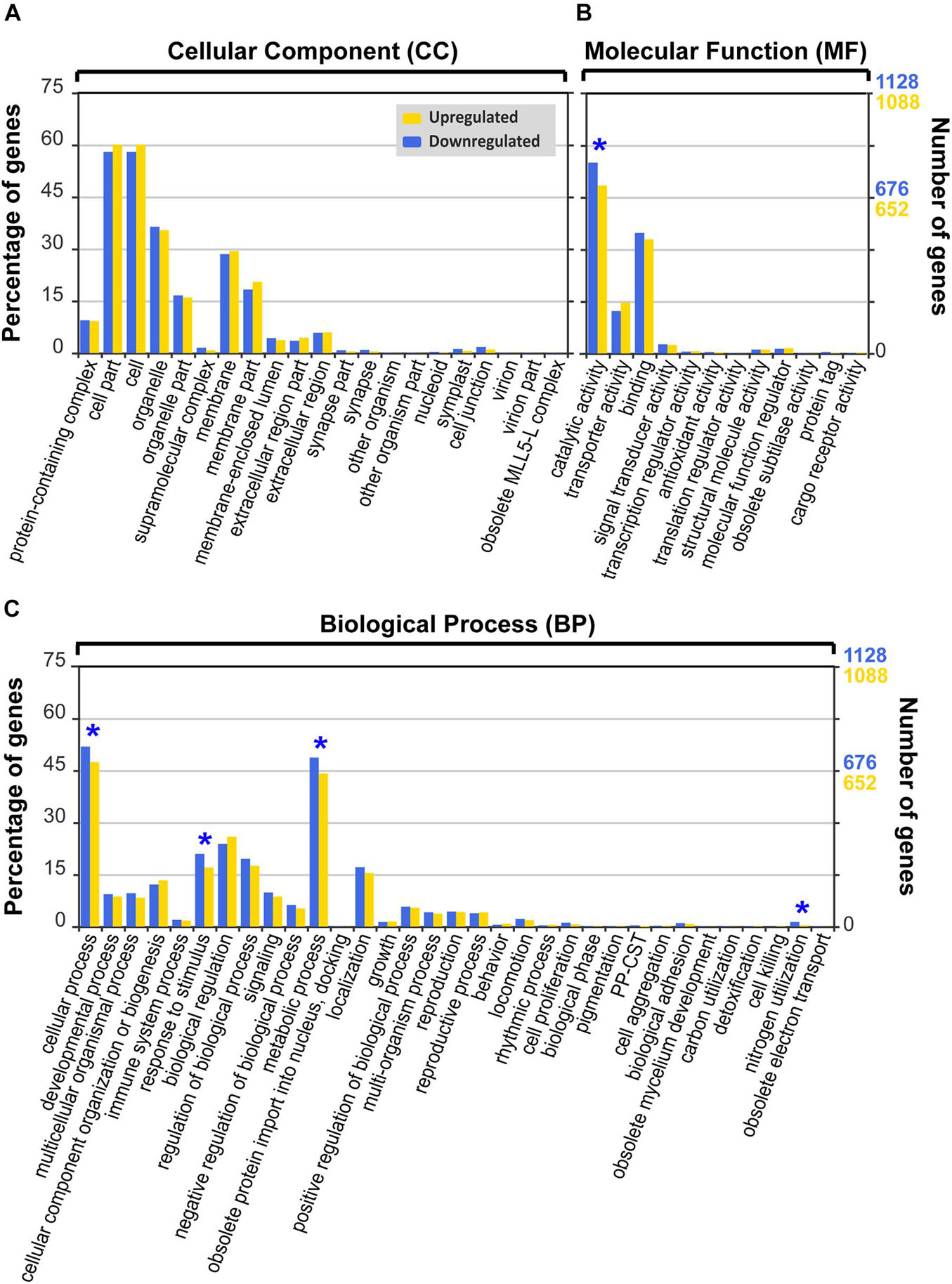
Figure 6. Gene Ontology (GO) functional classification (level 2) of differentially expressed genes (DEGs) of Biecheleriopsis adriatica under heat stress against optimal temperature. Category: (A) cellular component, (B) molecular function, and (C) biological process. The GO subcategories with significantly more downregulated genes than upregulated genes (p < 0.05, Pearson’s chi-squared test) are marked as blue asterisks. PP-CST: presynaptic process involved in chemical synaptic transmission.
In the CC category, “cell,” “cell part,” “organelle,” and “membrane” subcategories were dominant (Figure 6A and Supplementary Table 3). There were no subcategories with significant differences between the percentages of upregulated and downregulated genes (Figure 6A, Table 2, and Supplementary Table 3).
In the MF category, “catalytic activity” subcategory had the highest percentage of DEGs, followed by “binding” subcategory (Figure 6B and Supplementary Table 3). The “catalytic activity” subcategory had a higher percentage of downregulated genes than that of upregulated genes (Pearson’s chi-squared test; p < 0.001), but the percentage of downregulated genes in other subcategories was not significantly different from that of the upregulated genes (Figure 6B, Table 2, and Supplementary Table 3).
In the BP category, most DEGs were involved in the “cellular process” and “metabolic process” (Figure 6C and Supplementary Table 3). Four of the 33 subcategories had significantly different percentages of upregulated and downregulated DEGs (Pearson’s chi-squared test; p < 0.05; Figure 6C, Table 2, and Supplementary Table 3). The percentage of the downregulated genes in “nitrogen utilization,” “response to stimulus,” “metabolic process,” and “cellular process” subcategories were significantly higher than that of the upregulated genes (Pearson’s chi-squared test; p = 0.005, p = 0.006, p = 0.012, p = 0.016), whereas there were no significant differences between the percentage of the upregulated and downregulated genes in other subcategories (Figure 6C, Table 2, and Supplementary Table 3).
At the expression level of |log2 fold change| ≥ 1 and padj < 0.05, there was no change in the trend of the results of GO analysis under heat stress (Supplementary Figure 3).
In GO level 3, 29 subcategories had significant differences between the percentages of the upregulated and downregulated genes (Pearson’s chi-squared test; p < 0.05) under heat stress, with 9, 7, and 13 subcategories belonging to the CC, MF, and BP categories, respectively (Supplementary Table 4). Most subcategories had a higher percentage of downregulated genes than that of the upregulated genes (24 subcategories; Supplementary Table 4). The top 5 subcategories in GO level 3 with the largest differences between the percentage of the significantly upregulated and downregulated genes in B. adriatica under heat stress, based on the p-value, were “regulation of biological quality,” “small molecule metabolic process,” “response to abiotic stimulus,” “carbohydrate derivative binding,” and “ion binding” in sequence (Table 3).
Classification of Unigenes by the Trend in Expression Among the Temperature Conditions
When the unigenes were classified into nine types according to the expression trend, Type 5 had the greatest percentage of the number of the genes relative to that of all unigenes (83.4%; Figure 7A), followed by Type 2 (5.2%), Type 8 (4.8%), Type 6 (2.3%), Type 4 (1.8%), Type 1 (1.2%), Type 9 (1.0%), Type 3 (0.2%), and Type 7 (0.1%) (Figure 7A). Furthermore, Type 2 had the greatest percentage of the number of genes relative to that of the DEGs (31.0%; Figure 7B), followed by Type 8 (28.6%), Type 6 (13.6%), Type 4 (11.0%), Type 1 (7.4%), Type 9 (6.2%), Type 3 (1.4%), and Type 7 (0.8%) (Figure 7B). At the expression level of |log2 fold change| ≥ 1 and padj < 0.05, there was no change in this trend (Supplementary Figure 4).
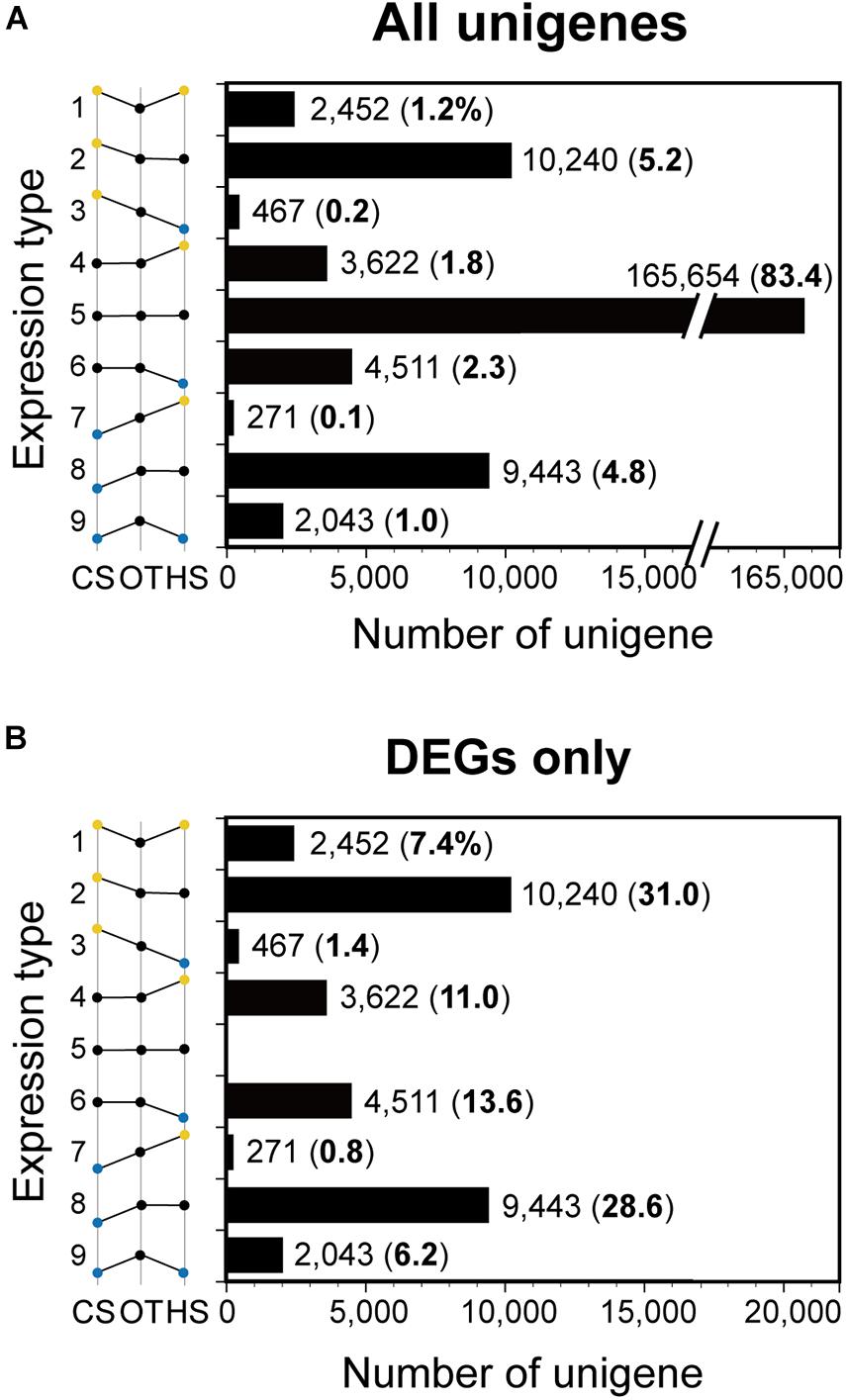
Figure 7. The number of unigenes of Biecheleriopsis adriatica in each expression type. The numbers in parenthesis indicate percentages of the number of the unigenes relative to that of (A) all unigenes and (B) differentially expressed genes (DEGs). CS, cold stress; OT, optimal temperature; HS, heat stress.
Among the unigenes ranked high by expression level, unigenes related to anti-stress, metabolic pathway, photosynthesis, and cell motility were detected in each type (Table 4). In Type 1, unigenes encoding “apolipoprotein d lipocalin” and “alpha-protein kinase vwkA” were detected; in Type 2, unigenes encoding “facilitated trehalose transporter Tret” and “glutathione S-transferase class-mu 28 kDa isozyme” were detected; in Type 3, unigenes encoding “fatty acid desaturase” and “fructose-bisphosphate aldolase” were detected; in Type 4, unigenes encoding “FAD-binding domain-containing protein” and “malate l-lactate dehydrogenase” were detected; in Type 6, unigenes encoding “proline/betaine transporter” and “aldo/keto reductase” were detected; in Type 7, unigenes encoding “chloroplast light harvesting complex protein, partial” and “intraflagellar transport protein 46-like” were detected; in Type 8, unigenes encoding “DnaJ-like subfamily B member 6-A” and “copia protein” were detected; in Type 9, unigenes encoding “troponin C, skeletal muscle” and “palmitoyl-monogalactosyldiacylglycerol delta-7 desaturase, chloroplastic” were detected (Table 4).
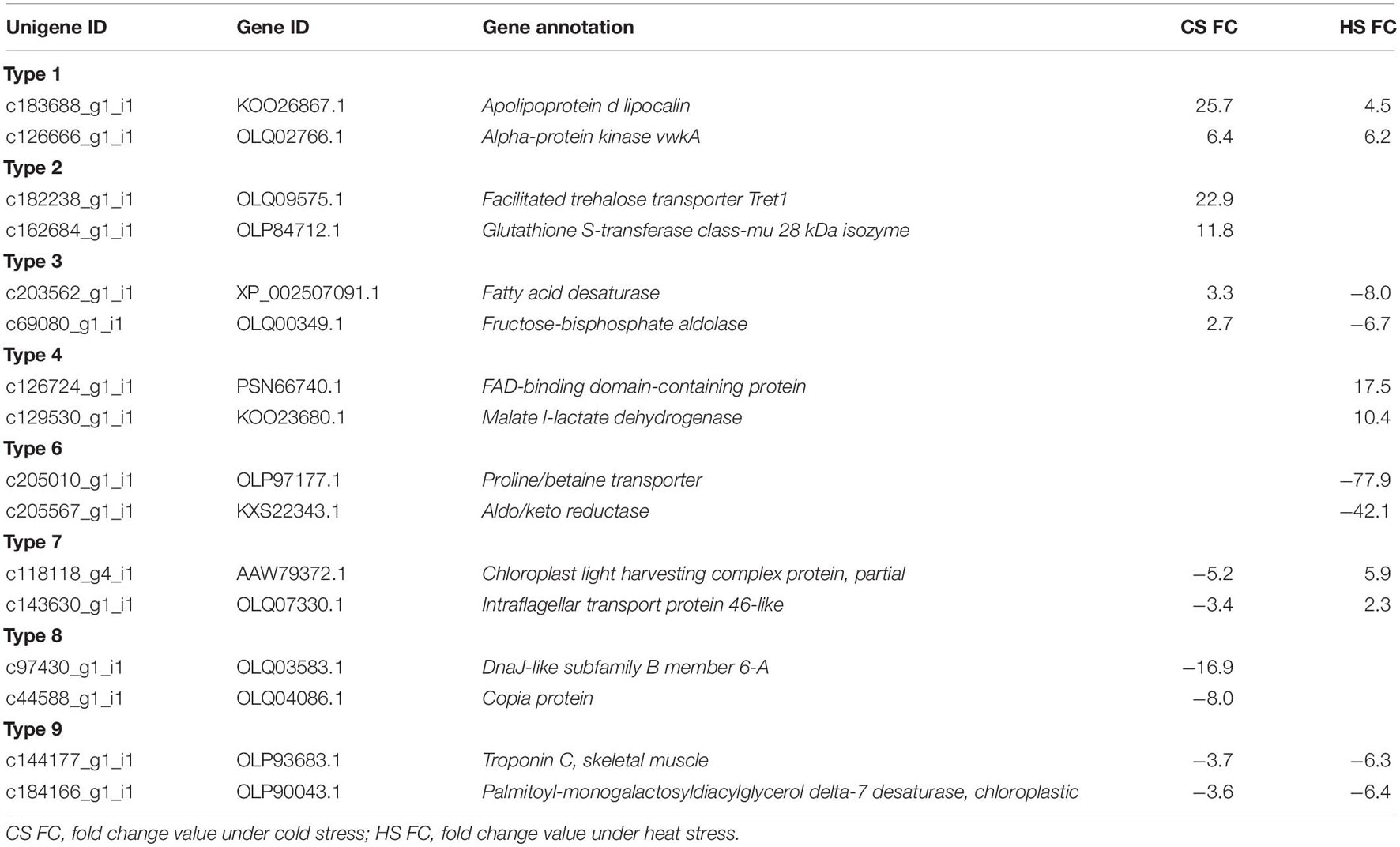
Table 4. Two differentially expressed unigenes of Biecheleriopsis adriatica belonged to each expression type.
Discussion
Dinoflagellates have a very large genome size ranging from 1.5 to 112 Gbp (Murray et al., 2016). In general, studies focus on the gene expression of dinoflagellates because it is difficult to determine their whole genome sequence (Murray et al., 2016). With advances in sequencing technology, complex transcriptional responses of dinoflagellates to diverse conditions have been reported, revealing a large number of unigenes (Murray et al., 2016; Caron et al., 2017). There have been many studies of de novo transcriptomics of dinoflagellates under different conditions of diverse environmental factors such as water temperature, nutrients, salinity, and light (e.g., Jaeckisch et al., 2011; Lowe et al., 2011; Morey et al., 2011; Bayer et al., 2012; Levin et al., 2016; Shi et al., 2017). The results of these studies provide a basis for better understanding of the responses of dinoflagellates to changes in environmental factors with regard to ecology and molecular biology.
Dinoflagellates experience both cold winter and hot summer in temperate regions (Selina et al., 2014; Kang et al., 2019b; Jang and Jeong, 2020). In temperate regions such as Korea, Japan, and China, sea waters become cold due to cold waves caused by the weakness of the stratospheric polar vortex by Arctic sea-ice loss or hot due to heat waves caused by the heat domes due to strengthening of the power of the North Pacific and Tibetan high pressures (Kim et al., 2014; Han and Lee, 2020; Lee et al., 2020; Inoue et al., 2021). Therefore, differences in growth and differential gene expression among very cold or hot and optimal temperatures should be understood to explore the survival of dinoflagellates in terms of eco-physiology and gene expression with respect to molecular biology. However, only three studies have explored the differential expression of two target genes of dinoflagellates grown under optimal temperature and cold stress or under optimal temperature and cold and heat stress (Deng et al., 2019; Kim et al., 2021; Wang et al., 2021). The expression of two cold shock domain protein genes in the mixotrophic dinoflagellate Prorocentrum cordatum exposed to low temperatures (4, 12, or 16°C) increased compared to that exposed to 20°C; the expression of saxitoxin biosynthesis gene sxtA4 increased in the toxic dinoflagellate Alexandrium catenella by cold shock (20→16°C), while that of sxtG increased by cold shock (20→16°C) and heat stress (12→20°C); heat shock protein 60 and 10 expressions were upregulated in the mixotrophic dinoflagellate Scrippsiella acuminata after exposure to lower (15, 10, and 5°C) and higher (25 and 30°C) temperatures compared to optimal temperature (20°C). This study showed that 5,233 genes were differentially expressed under both cold and heat stress. Furthermore, 19,683 genes were differentially expressed only under cold stress, while 8,133 genes were differentially expressed only under heat stress. Therefore, comparative transcriptome analysis of dinoflagellates grown under optimal temperature and cold and heat stress should be conducted simultaneously to understand the pattern of all expressed genes, and the present study provides a milestone for this topic.
Positive growth during the first 3 days under heat stress was observed. This likely reflected residual growth during pre-incubation. However, during the last 8 days, the abundance of B. adriatica continuously decreased, indicating that the cells were affected by heat stress when they were harvested.
The cultures used in this study were not completely axenic, although we attempted to minimize the effect of bacteria on the mRNA dataset of B. adriatica while preparing the experimental organisms and during RNA sequencing. It is generally very difficult for dinoflagellates to grow without bacteria; studies shown that many dinoflagellates have endosymbiotic relationships with bacteria or require bacteria for their survival (Alavi et al., 2001; Croft et al., 2005; Bolch et al., 2011; Cruz-López and Maske, 2016).
Nutrient concentrations during incubation under the three different temperature conditions were sufficiently high to enable dinoflagellate growth. Nutrient concentration is one of the major factors affecting dinoflagellate growth (Hwang and Lu, 2000; Nagasoe et al., 2010). Furthermore, nutrient limitations in ambient waters are known to affect gene expression in dinoflagellates (Morey et al., 2011; Lauritano et al., 2017).
General Transcriptome Features of B. adriatica
Some studies determined the number of DEGs of dinoflagellates at the expression level of p < 0.05 (e.g., Guo et al., 2016; Jang et al., 2019), whereas others determined that at the expression level of padj < 0.05 (e.g., Barshis et al., 2014; Bellantuono et al., 2019; Lin et al., 2019). The number of DEGs in B. adriatica at the expression level of p < 0.05 and padj < 0.05, were 33,049 and 30,877, respectively, indicating a 6.6% difference. The results of GO analysis and expression type at the expression level of p < 0.05, and padj < 0.05, did not change significantly. Furthermore, there was no change in the trends of any of the results.
At the expression level of p < 0.05, the larger number of upregulated DEGs than downregulated DEGs under cold stress indicates that B. adriatica expresses more genes in response to cold stress. In contrast, the higher number of downregulated DEGs than upregulated DEGs under heat stress suggests that B. adriatica restricts gene expression to save energy or that genes are damaged by heat shock. At the geological scale, the sea surface temperature during the mid- and late-Pleistocene (∼1.2 Ma) showed a cooling trend, although a glacial–interglacial cycle with large amplitudes of air temperature change was repeated (McClymont et al., 2013). The period of global warming in the last century was significantly shorter than that in the ice age. Thus, dinoflagellates may have acclimated to cold stress better than to heat stress. Furthermore, dinoflagellates conduct diurnal vertical migration, and some dinoflagellates pass through the thermocline and remain in cold water, although the surface water temperature increases significantly (Jeong et al., 2015). However, vertical migration may not provide a refuge when the surface water temperature decreases significantly. Thus, dinoflagellates may upregulate or downregulate gene expression under cold stress than under heat stress.
Functional Classification of B. adriatica Differentially Expressed Genes Under Cold and Heat Stress
The “structural molecule activity,” “membrane-enclosed lumen,” and “organelle part” subcategories in GO level 2 are related to cell structure and organelles. Cold stress may damage the protist cell structure. The morphology of the green alga Chlamydomonas reinhardtii changed significantly and its organelles were progressively disorganized and disrupted after decreasing the water temperature from 25 to 5°C (Valledor et al., 2013). After 24 h of water temperature decrease, the nucleolus of C. reinhardtii could not be distinguished; vacuoles and starch sheaths increased. Furthermore, the chloroplast changed its shape and flagella were shortened and disappeared after 120 h (Valledor et al., 2013). Similarly, under cold stress, B. adriatica might upregulate genes related to maintaining cell integrity instead of genes with other functions.
Under stress, microalgae tend to reserve extra metabolites by promoting metabolic processes or save energy by reducing metabolic processes (Levin et al., 2016; Sun et al., 2018). In this study, the “metabolic process” subcategory had a significantly higher percentage of upregulated DEGs than that of downregulated DEGs under cold stress. Therefore, B. adriatica may respond to cold stress by enhancing metabolic processes to reserve extra metabolites.
Under cold stress, the “signaling” subcategory had the greatest difference between the percentages of upregulated and downregulated DEGs, that is, the smallest p-value. This was followed by “response to stimulus” and “biological regulation.” The basic activities of B. adriatica, which perceive environmental change, transmit signals, and respond to changes by modulating biological processes or functions, seemed to be disturbed by cold stress.
Under cold stress, “methylation” had the highest difference between upregulated and downregulated genes in GO level 3. In organisms, methylation plays diverse roles, such as heavy metal modification, gene expression and protein function regulation, and RNA processing (Lee et al., 2010; Li et al., 2018). DNA methylation, which adds a methyl group to the DNA, can affect the gene expression of dinoflagellates, and temperature change can regulate DNA methylation levels (Bayer et al., 2012; Naydenov et al., 2015; Li et al., 2018). Therefore, cold stress may affect DNA methylation and gene expression in B. adriatica.
The number of subcategories showing a significant difference between the percentages of upregulated and downregulated DEGs under heat stress was lower than that under cold stress. The catalytic activity generally increases with temperature, but it decreases sharply due to enzyme denaturation if the temperature exceeds a certain threshold (Daniel and Danson, 2013). Therefore, enzyme denaturation in B. adriatica cells may occur at 32°C. Furthermore, nitrogen utilization efficiency decreases at non-optimal temperatures owing to the changes in cytoplasmic viscosity. Heat stress may also affect the cytoplasmic viscosity and nitrogen utilization efficiency in B. adriatica (Raven and Geider, 1988). Levin et al. (2016) reported that the downregulated DEGs of the thermo-tolerant dinoflagellate Cladocopium sp. under heat stress were distributed in more GO categories related to the metabolic and biosynthetic processes than those of thermo-sensitive Cladocopium sp. In some organisms, reduced metabolic and biosynthetic activity under stress is correlated with the increased survival time of organisms while saving energy (Hand and Hardewig, 1996). Therefore, B. adriatica may try to acclimate to high water temperatures with reduced energy consumption.
Under heat stress, only “regulation of biological quality” had a higher percentage of the upregulated DEGs than that of the downregulated DEGs among the top 5 subcategories. It includes many processes that modulate a qualitative or quantitative trait with a measurable attribute in an organism or part of an organism, such as size, mass, shape, and color (Gene Ontology). It was found that increasing the water temperature by 1°C caused an approximately 2.5% intra-specific decrease in protist cell volume (Atkinson et al., 2003). Furthermore, changes in cell morphology, such as cell shape, size, and color, due to heat stress have been observed in microalgae (Neustupa et al., 2008; Chokshi et al., 2015). The “responses to abiotic stimulus” subcategory had a higher percentage of downregulated genes than that of upregulated genes, suggesting that high temperature may not cause significant stress to B. adriatica or it adapts to high temperature rapidly.
Classification of Unigenes by the Expression Trend Among the Temperature Conditions
The number of unigenes differentially expressed under only cold or heat stress from that under optimal temperature (Types 2, 4, 6, and 8) was significantly more than that detected under both cold and heat stress (Types 1, 3, 7, and 9). This suggests that the mechanism of molecular responses of B. adriatica to cold stress is different from that to heat stress. Interestingly, the number of unigenes belonging to Types 1 (upregulated under both cold and heat stress) and 9 (downregulated under both cold and heat stress), Types 2 (upregulated under cold stress and unchanged under heat stress) and 8 (downregulated under cold stress and unchanged under heat stress), Types 3 (upregulated under cold stress and downregulated under heat stress) and 7 (downregulated under cold stress and upregulated under heat stress), and Types 4 (unchanged under cold stress and upregulated under heat stress) and 6 (unchanged under cold stress and downregulated under heat stress) were similar to a mirror image. Thus, B. adriatica may balance upregulation and downregulation of gene expression in response to temperature stress.
The “apolipoprotein d lipocalin (ApoD)” identified in Type 1 is an evolutionarily conserved anti-stress protein that enhances tolerance to freezing and oxidative stress in Arabidopsis thaliana. It was upregulated in the branching stony coral Acropora millepora under cool and warm treatments and the phototrophic dinoflagellate Prorocentrum donghaiense under phosphorus-depleted conditions (Charron et al., 2008; Ganfornina et al., 2008; Muffat and Walker, 2010; Wuitchik, 2018; Zhang et al., 2018). Furthermore, the unigene encoding “alpha-protein kinase vwkA” belonged to Type 1. The alpha-kinase family is involved in diverse cellular processes such as protein translation, Mg2+ homeostasis, intracellular transport, cell migration, adhesion, and proliferation (Middelbeek et al., 2010). Betapudi et al. (2005) reported vwkA involvement in regulating myosin II abundance and assembly behavior in the amoeba Dictyostelium discoideum. Myosin II is an actin-dependent molecular motor protein important for cell motility and cytokinesis (Maciver, 1996; Foth et al., 2006). Therefore, temperature stress may affect B. adriatica cell movement and division processes.
The unigenes encoding “facilitated trehalose transporter Tret1” and “glutathione S-transferase class-mu 28 kDa isozyme” belonged to Type 2. Trehalose improves insect and plant tolerance to cold stress (Kikawada et al., 2007; Iordachescu and Imai, 2008; Benoit et al., 2009). Therefore, trehalose may play the same role in B. adriatica as in insects and plants. Furthermore, glutathione S-transferase (GST) is an antioxidant; thus, it is considered to be a potential oxidative stress biomarker (Guo et al., 2014). In the dinoflagellate Fugacium sp., most of the 10 GST transcripts detected were upregulated under heat stress (Gierz et al., 2017). However, in this study, the B. adriatica GST gene was highly expressed under cold stress, but not heat stress. Therefore, this gene may be expressed by temperature stress, but whether the temperature stress is cold or heat stress is dependent on dinoflagellate species.
The unigenes encoding “fatty acid desaturase” and “fructose-bisphosphate aldolase (FBA)” belonged to Type 3. Temperature change affects the membrane fluidity of poikilothermic organisms such as bacteria, fungi, protozoa, plants, and animals (Murata and Los, 1997). Membrane fluidity perceives changes in environmental conditions and transduces signals to acclimate to these conditions (Murata and Los, 1997; Los and Murata, 2004). The cell membrane becomes rigid when exposed to low temperatures and becomes fluid at high temperatures (Murata and Los, 1997). The reduced membrane fluidity at low temperatures can be improved via membrane lipid desaturation by fatty acid desaturases (Sakamoto and Murata, 2002). Conversely, the increased membrane fluidity at high temperatures can be ameliorated by integrating de novo synthesized saturated fatty acids into membrane lipids and the presence of membrane-stabilizing proteins (Los et al., 2013). Therefore, the upregulation and downregulation of this gene under cold and heat stress, respectively, in this study corresponded with previous results. FBA is a key metabolic enzyme involved in glycolysis, gluconeogenesis, and the Calvin cycle (Flechner et al., 1999; Gross et al., 1999). Furthermore, it plays an important role in stress signaling in plants (Lu et al., 2012). Therefore, B. adriatica may be affected more by cold stress than that by heat stress.
The “FAD-binding domain-containing protein” belonging to Type 4 includes several genes playing key roles in many metabolic pathways, including the citric acid cycle, fatty acid beta-oxidation, and amino acid catabolism (Mansoorabadi et al., 2007; Kim and Winge, 2013). Moreover, “malate l-lactate dehydrogenase” belonging to Type 4 also functions in many metabolic pathways, including the tricarboxylic acid cycle, glyoxylate bypass, amino acid synthesis, and gluconeogenesis (Musrati et al., 1998; Minarik et al., 2002). They were upregulated only under heat stress, thus indicating that mechanisms for acclimation to temperature stress act differentially under cold and heat stress in B. adriatica metabolic pathways.
As “proline/betaine transporter” belonging to Type 6 is an amino acid transporter, its downregulation under heat stress suggests that the capacity to export amino acids in B. adriatica cell may reduce under heat stress (Li et al., 2021).
The unigenes encoding “chloroplast light harvesting complex (LHC) protein, partial” and “intraflagellar transport (IFT) protein 46-like” belonged to Type 7. LHC proteins function in light capture and photoprotection (Boldt et al., 2012). Three upregulated LHC genes were detected in Fugacium sp. under heat stress (Gierz et al., 2016). In contrast, under cold stress, LHC activity was reduced in C. reinhardtii and cereal grain Hordeum vulgare (Atienza et al., 2004; Valledor et al., 2013). Therefore, the results of this study are consistent with those of previous studies. The IFT system assembles and maintains eukaryotic cilia and flagella (Cole, 2003). The cilia and flagella are best known to play important roles as motile organelles, but they also function as sensory perception organelles that monitor the surrounding environment, such as chemicals, light, and temperature (Silflow and Lefebvre, 2001; Follit et al., 2006). Although B. adriatica speed was not measured during incubation, when observed under a microscope, B. adriatica cells swam more slowly under cold stress than under heat stress or optimal temperature.
The unigene encoding “DnaJ-like subfamily B member 6-A” was identified to belong to Type 8. Heat shock proteins (HSPs) are important chaperones that maintain cellular homeostasis under both favorable and unfavorable conditions (Wang et al., 2004). They are considered to be produced more under unfavorable conditions, such as temperature and light stress (Rajan and D’Silva, 2009). Unlike B. adriatica, which downregulates the unigene encoding DnaJ, DnaJ expression in other dinoflagellates was upregulated. The relative DnaJ transcription level in S. acuminata increased under both low and high temperatures than that under a moderate temperature; moreover, increased DnaJ expression in heat stress-exposed Cladocopium sp. and Fugacium kawagutii have been reported (Levin et al., 2016; Deng et al., 2019; Lin et al., 2019). In contrast, Gierz et al. (2017) reported that several transcripts encoding HSPs, including DnaJ, were identified in Fugacium sp. under heat stress, and the downregulated transcripts were more dominant than upregulated transcripts. Therefore, whether the expression of this gene is upregulated or downregulated is dependent on the dinoflagellate species.
The unigene encoding “troponin C, skeletal muscle” belonged to Type 9. Troponin C is a Ca2+ homeostasis protein of the calmodulin family (DeSalvo et al., 2010; Abassi et al., 2021). In the elkhorn coral Acropora palmata, this gene was downregulated under heat stress (DeSalvo et al., 2010). The downregulation of this gene under cold and heat stress in this study showed that Ca2+ homeostasis in B. adriatica may be affected by cold and heat stress. To elucidate this, more studies on the effect of temperature on the expression of this gene in dinoflagellates are required.
To better understand the response of dinoflagellate communities to diverse environmental changes, including cold waves, heat waves, ocean acidification, and local pollution, both eco-physiological and molecular characteristic changes should be explored together.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA756051).
Author Contributions
HK and HJ designed the study, drafted the manuscript, obtained the data, and performed the analyses with contribution from SP, JO, JY, SE, EP, SJ, and SL. All authors discussed the results and approved the final submitted manuscript.
Funding
This research was supported by the Useful Dinoflagellate Program of Korea Institute of Marine Science and Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (MOF) and the National Research Foundation (NRF) funded by the Ministry of Science and ICT (NRF-2020M3F6A1110582; NRF-2021M3I6A1091272; NRF-2021R1A2C1093379) award to HJ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank An Suk Lim and Eun Ji Kim for technical support. We also thank the editor and reviewers for their valuable comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.761095/full#supplementary-material
Supplementary Table 1 | Gene Ontology functional classification of differentially expressed genes of Biecheleriopsis adriatica under temperature stress conditions.
Supplementary Data Sheet 1 | Analysis results when differentially expressed genes of Biecheleriopsis adriatica were obtained with —log2 fold change— ≥ 1 and adjusted p-value < 0.05.
Footnotes
- ^ http://www.khoa.go.kr/oceangrid/gis/category/reference/distribution.do
- ^ http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- ^ http://wego.genomics.cn/
References
Abassi, S., Wang, H., and Ki, J. S. (2021). Characterization and Ca2+-induced expression of calmodulin (CaM) in marine dinoflagellates. Eur. J. Protistol. 77:125765. doi: 10.1016/j.ejop.2020.125765
Alavi, M., Miller, T., Erlandson, K., Schneider, R., and Belas, R. (2001). Bacterial community associated with Pfiesteria-like dinoflagellate cultures. Environ. Microbiol. 3, 380–396. doi: 10.1046/j.1462-2920.2001.00207.x
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1038/npre.2010.4282.1
Atienza, S. G., Faccioli, P., Perrotta, G., Dalfino, G., Zschiesche, W., Humbeck, K., et al. (2004). Large scale analysis of transcripts abundance in barley subjected to several single and combined abiotic stress conditions. Plant Sci. 167, 1359–1365. doi: 10.1016/j.plantsci.2004.07.006
Atkinson, D., Ciotti, B. J., and Montagnes, D. J. (2003). Protists decrease in size linearly with temperature: ca. 2.5% °C–1. Proc. R. Soc. Lond. Ser. B. Biol. Sci. 270, 2605–2611. doi: 10.1098/rspb.2003.2538
Band-Schmidt, C. J., Lechuga-Devéze, C. H., Kulis, D. M., and Anderson, D. M. (2003). Culture studies of Alexandrium affine (Dinophyceae), a non-toxic cyst forming dinoflagellate from Bahía Concepción Gulf of California. Bot. Mar. 46, 44–54. doi: 10.1515/BOT.2003.007
Barshis, D. J., Ladner, J. T., Oliver, T. A., and Palumbi, S. R. (2014). Lineage-specific transcriptional profiles of Symbiodinium spp. unaltered by heat stress in a coral host. Mol. Biol. Evol. 31, 1343–1352. doi: 10.1093/molbev/msu107
Bayer, T., Aranda, M., Sunagawa, S., Yum, L. K., DeSalvo, M. K., Lindquist, E., et al. (2012). Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PLoS One 7:e35269. doi: 10.1371/journal.pone.0035269
Bellantuono, A. J., Dougan, K. E., Granados-Cifuentes, C., and Rodriguez-Lanetty, M. (2019). Free-living and symbiotic lifestyles of a thermotolerant coral endosymbiont display profoundly distinct transcriptomes under both stable and heat stress conditions. Mol. Ecol. 28, 5265–5281. doi: 10.1111/mec.15300
Benico, G. A., Takahashi, K., Lum, W. M., Yñiguez, A. T., Azanza, R. V., Leong, S. C. Y., et al. (2019). First report of Biecheleriopsis adriatica in Bolinao, Northwestern Philippines and its wide distribution in Southeast Asia and adjacent waters. Philipp. J. Nat. Sci. 24, 34–41.
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Benoit, J. B., Lopez-Martinez, G., Elnitsky, M. A., Lee, R. E., and Denlinger, D. L. (2009). Dehydration-induced cross tolerance of Belgica antarctica larvae to cold and heat is facilitated by trehalose accumulation. Com. Biochem. Phys. A 152, 518–523. doi: 10.1016/j.cbpa.2008.12.009
Betapudi, V., Mason, C., Licate, L., and Egelhoff, T. T. (2005). Identification and characterization of a novel α-kinase with a von Willebrand factor A-like motif localized to the contractile vacuole and Golgi complex in Dictyostelium discoideum. Mol. Biol. Cell 16, 2248–2262. doi: 10.1091/mbc.e04-07-0639
Bolch, C. J., Subramanian, T. A., and Green, D. H. (2011). The toxic dinoflagellate Gymnodinium catenatum (Dinophyceae) requires marine bacteria for growth. J. Phycol. 47, 1009–1022. doi: 10.1111/j.1529-8817.2011.01043.x
Boldt, L., Yellowlees, D., and Leggat, W. (2012). Hyperdiversity of genes encoding integral light-harvesting proteins in the dinoflagellate Symbiodinium sp. PLoS One 7:e47456. doi: 10.1371/journal.pone.0047456
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brinkhuis, H., Bujak, J. P., Smit, J., Versteegh, G. J. M., and Visscher, H. (1998). Dinoflagellate-based sea surface temperature reconstructions across the Cretaceous–Tertiary boundary. Palaeogeogr. Palaeoclimatol. Palaeoecol. 141, 67–83. doi: 10.1016/S0031-0182(98)00004-2
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Caron, D. A., Alexander, H., Allen, A. E., Archibald, J. M., Armbrust, E. V., Bachy, C., et al. (2017). Probing the evolution, ecology and physiology of marine protists using transcriptomics. Nat. Rev. Microbiol. 15, 6–20. doi: 10.1038/nrmicro.2016.160
Charron, J. B. F., Ouellet, F., Houde, M., and Sarhan, F. (2008). The plant Apolipoprotein D ortholog protects Arabidopsis against oxidative stress. BMC Plant Biol. 8:86. doi: 10.1186/1471-2229-8-86
Chokshi, K., Pancha, I., Trivedi, K., George, B., Maurya, R., Ghosh, A., et al. (2015). Biofuel potential of the newly isolated microalgae Acutodesmus dimorphus under temperature induced oxidative stress conditions. Bioresour. Technol. 180, 162–171. doi: 10.1016/j.biortech.2014.12.102
Coats, D. W. (1999). Parasitic life styles of marine dinoflagellates. J. Eukaryot. Microbiol. 46, 402–409. doi: 10.1111/j.1550-7408.1999.tb04620.x
Cole, D. G. (2003). The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic 4, 435–442. doi: 10.1034/j.1600-0854.2003.t01-1-00103.x
Croft, M. T., Lawrence, A. D., Raux-Deery, E., Warren, M. J., and Smith, A. G. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93. doi: 10.1038/nature04056
Crouch, E. M., Heilmann-Clausen, C., Brinkhuis, H., Morgans, H. E., Rogers, K. M., Egger, H., et al. (2001). Global dinoflagellate event associated with the late Paleocene thermal maximum. Geology 29, 315–318. doi: 10.1130/0091-76132001029<0315:GDEAWT<2.0.CO;2
Cruz-López, R., and Maske, H. (2016). The vitamin B1 and B12 required by the marine dinoflagellate Lingulodinium polyedrum can be provided by its associated bacterial community in culture. Front. Microbiol. 7:560. doi: 10.3389/fmicb.2016.00560
Daniel, R. M., and Danson, M. J. (2013). Temperature and the catalytic activity of enzymes: a fresh understanding. FEBS Lett. 587, 2738–2743. doi: 10.1016/j.febslet.2013.06.027
Davies, S. W., Ries, J. B., Marchetti, A., and Castillo, K. D. (2018). Symbiodinium functional diversity in the coral Siderastrea siderea is influenced by thermal stress and reef environment, but not ocean acidification. Front. Mar. Sci. 5:150. doi: 10.3389/fmars.2018.00150
Davy, S. K., Allemand, D., and Weis, V. M. (2012). Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76, 229–261. doi: 10.1128/MMBR.05014-11
Deng, Y., Hu, Z., Chai, Z., and Tang, Y. Z. (2019). Molecular cloning of heat shock protein 60 (Hsp60) and 10 (Hsp10) genes from the cosmopolitan and harmful dinoflagellate Scrippsiella trochoidea and their differential transcriptions responding to temperature stress and alteration of life cycle. Mar. Biol. 166:7. doi: 10.1007/s00227-018-3455-3
DeSalvo, M. K., Sunagawa, S., Voolstra, C. R., and Medina, M. (2010). Transcriptomic responses to heat stress and bleaching in the elkhorn coral Acropora palmata. Mar. Ecol. Prog. Ser. 402, 97–113. doi: 10.3354/meps08372
Eom, S. H., Jeong, H. J., Ok, J. H., Park, S. A., Kang, H. C., You, J. H., et al. (2021). Interactions between common heterotrophic protists and the dinoflagellate Tripos furca: implication on the long duration of its red tides in the South Sea of Korea in 2020. Algae 36, 25–36. doi: 10.4490/algae.2021.36.2.22
Flechner, A., Gross, W., Martin, W. F., and Schnarrenberger, C. (1999). Chloroplast class I and class II aldolases are bifunctional for fructose-1, 6-biphosphate and sedoheptulose-1, 7-biphosphate cleavage in the Calvin cycle. FEBS Lett. 447, 200–202. doi: 10.1016/S0014-5793(99)00285-9
Flewelling, L. J., Naar, J. P., Abbott, J. P., Baden, D. G., Barros, N. B., Bossart, G. D., et al. (2005). Red tides and marine mammal mortalities. Nature 435, 755–756. doi: 10.1038/nature435755a
Follit, J. A., Tuft, R. A., Fogarty, K. E., and Pazour, G. J. (2006). The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell 17, 3781–3792. doi: 10.1091/mbc.e06-02-0133
Foth, B. J., Goedecke, M. C., and Soldati, D. (2006). New insights into myosin evolution and classification. Proc. Natl. Acad. Sci. U.S.A. 103, 3681–3686. doi: 10.1073/pnas.0506307103
Franklin, D. J., Hoegh-Guldberg, O., Jones, R. J., and Berges, J. A. (2004). Cell death and degeneration in the symbiotic dinoflagellates of the coral Stylophora pistillata during bleaching. Mar. Ecol. Prog. Ser. 272, 117–130. doi: 10.3354/meps272117
Fridey, K. (2015). Bioinformatics Approach to Determining Transcriptional and Translational Responses to Heat Stress in the Florida red Tide Dinoflagellate Karenia brevis. Master’s thesis. Charleston, SC: College of Charleston.
Ganfornina, M. D., Carmo, S. D., Lora, J. M., Torres-Schumann, S., Vogel, M., Allhorn, M., et al. (2008). Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell 7, 506–515. doi: 10.1111/j.1474-9726.2008.00395.x
Gierz, S. L., Forêt, S., and Leggat, W. (2017). Transcriptomic analysis of thermally stressed Symbiodinium reveals differential expression of stress and metabolism genes. Front. Plant Sci. 8:271. doi: 10.3389/fpls.2017.00271
Gierz, S. L., Gordon, B. R., and Leggat, W. (2016). Integral light-harvesting complex expression in Symbiodinium within the coral Acropora aspera under thermal stress. Sci. Rep. 6, 1–10. doi: 10.1038/srep25081
Gómez, F., and Souissi, S. (2008). The impact of the 2003 summer heat wave and the 2005 late cold wave on the phytoplankton in the north-eastern English Channel. C. R. Biol. 331, 678–685. doi: 10.1016/j.crvi.2008.06.005
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Graham, N. A. J., and Nash, K. L. (2013). The importance of structural complexity in coral reef ecosystems. Coral Reefs 32, 315–326. doi: 10.1007/s00338-012-0984-y
Gross, W., Lenze, D., Nowitzki, U., Weiske, J., and Schnarrenberger, C. (1999). Characterization, cloning, and evolutionary history of the chloroplast and cytosolic class I aldolases of the red alga Galdieria sulphuraria. Gene 230, 7–14. doi: 10.1016/S0378-1119(99)00059-1
Guillard, R. R., and Ryther, J. H. (1962). Studies of marine planktonic diatoms: I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can. J. Microbiol. 8, 229–239. doi: 10.1139/m62-029
Guo, R., Ebenezer, V., and Ki, J. S. (2014). PmMGST3, a novel microsomal glutathione S-transferase gene in the dinoflagellate Prorocentrum minimum, is a potential biomarker of oxidative stress. Gene 546, 378–385. doi: 10.1016/j.gene.2014.05.046
Guo, R., Lim, W. A., and Ki, J. S. (2016). Genome-wide analysis of transcription and photosynthesis inhibition in the harmful dinoflagellate Prorocentrum minimum in response to the biocide copper sulfate. Harmful Algae 57, 27–38. doi: 10.1016/j.hal.2016.05.004
Han, I. S., and Lee, J. S. (2020). Change the annual amplitude of sea surface temperature due to climate change in a recent decade around the Korean Peninsula. J. Korean Soc. Mar. Environ. Saf. 26, 233–241. doi: 10.7837/kosomes.2020.26.3.233
Hand, S. C., and Hardewig, I. (1996). Downregulation of cellular metabolism during environmental stress: mechanisms and implications. Annu. Rev. Physiol. 58, 539–563. doi: 10.1146/annurev.ph.58.030196.002543
Hansen, J., Sato, M., Ruedy, R., Lo, K., Lea, D. W., and Medina-Elizade, M. (2006). Global temperature change. Proc. Natl. Acad. Sci. U.S.A. 103, 14288–14293. doi: 10.1073/pnas.0606291103
Hansen, P. J. (1991). Quantitative importance and trophic role of heterotrophic dinoflagellates in a coastal pelagial food web. Mar. Ecol. Prog. Ser. 73, 253–261. doi: 10.3354/meps073253
Heisler, J., Glibert, P. M., Burkholder, J. M., Anderson, D. M., Cochlan, W., Dennison, W. C., et al. (2008). Eutrophication and harmful algal blooms: a scientific consensus. Harmful Algae 8, 3–13. doi: 10.1016/j.hal.2008.08.006
Hinder, S. L., Hays, G. C., Edwards, M., Roberts, E. C., Walne, A. W., and Gravenor, M. B. (2012). Changes in marine dinoflagellate and diatom abundance under climate change. Nat. Clim. Change 2, 271–275. doi: 10.1038/nclimate1388
Hwang, D. F., and Lu, Y. H. (2000). Influence of environmental and nutritional factors on growth, toxicity, and toxin profile of dinoflagellate Alexandrium minutum. Toxicon 38, 1491–1503. doi: 10.1016/S0041-0101(00)00080-5
Iglesias-Prieto, R., Matta, J. L., Robins, W. A., and Trench, R. K. (1992). Photosynthetic response to elevated temperature in the symbiotic dinoflagellate Symbiodinium microadriaticum in culture. Proc. Natl. Acad. Sci. U.S.A. 89, 10302–10305. doi: 10.1073/pnas.89.21.10302
Inoue, M., Ugajin, A., Kiguchi, O., Yamashita, Y., Komine, M., and Yamakawa, S. (2021). Effects of the Tibetan high and the North Pacific high on the occurrence of hot or cool summers in Japan. Atmosphere 12:307. doi: 10.3390/atmos12030307
Iordachescu, M., and Imai, R. (2008). Trehalose biosynthesis in response to abiotic stresses. J. Integr. Plant Biol. 50, 1223–1229. doi: 10.1111/j.1744-7909.2008.00736.x
Jaeckisch, N., Yang, I., Wohlrab, S., Glöckner, G., Kroymann, J., Vogel, H., et al. (2011). Comparative genomic and transcriptomic characterization of the toxigenic marine dinoflagellate Alexandrium ostenfeldii. PLoS One 6:e28012. doi: 10.1371/journal.pone.0028012
Jang, S. H., and Jeong, H. J. (2020). Spatio-temporal distributions of the newly described mixotrophic dinoflagellate Yihiella yeosuensis (Suessiaceae) in Korean coastal waters and its grazing impact on prey populations. Algae 35, 45–59. doi: 10.4490/algae.2020.35.2.24
Jang, S. H., Jeong, H. J., and Chon, J. K. (2019). De novo transcriptome of the newly described phototrophic dinoflagellate Yihiella yeosuensis: comparison between vegetative cells and cysts. Mar. Biol. 166, 1–21. doi: 10.1007/s00227-019-3554-9
Jang, S. H., Jeong, H. J., Moestrup, Ø, Kang, N. S., Lee, S. Y., Lee, K. H., et al. (2015). Morphological, molecular and ecophysiological characterization of the phototrophic dinoflagellate Biecheleriopsis adriatica from Korean coastal waters. Eur. J. Phycol. 50, 301–317. doi: 10.1080/09670262.2015.1054892
Jeong, H. J., Kang, H. C., Lim, A. S., Jang, S. H., Lee, K., Lee, S. Y., et al. (2021a). Feeding diverse prey as an excellent strategy of mixotrophic dinoflagellates for global dominance. Sci. Adv. 7:eabe4214. doi: 10.1126/sciadv.abe4214
Jeong, H. J., Yoo, Y. D., Lee, K., Kang, H. C., Kim, J. S., and Kim, K. Y. (2021b). Annual carbon retention of a marine-plankton community in the eutrophic Masan Bay, based on daily measurements. Mar. Biol. 168:69. doi: 10.1007/s00227-021-03881-4
Jeong, H. J., Lee, K. H., Yoo, Y. D., Kang, N. S., Song, J. Y., Kim, T. H., et al. (2018). Effects of light intensity, temperature, and salinity on the growth and ingestion rates of the red-tide mixotrophic dinoflagellate Paragymnodinium shiwhaense. Harmful Algae 80, 46–54. doi: 10.1016/j.hal.2018.09.005
Jeong, H. J., Lim, A. S., Franks, P. J., Lee, K. H., Kim, J. H., Kang, N. S., et al. (2015). A hierarchy of conceptual models of red-tide generation: nutrition, behavior, and biological interactions. Harmful Algae 47, 97–115. doi: 10.1016/j.hal.2015.06.004
Jeong, H. J., Yoo, Y. D., Kim, J. S., Seong, K. A., Kang, N. S., and Kim, T. H. (2010). Growth, feeding and ecological roles of the mixotrophic and heterotrophic dinoflagellates in marine planktonic food webs. Ocean Sci. J. 45, 65–91. doi: 10.1007/s12601-010-0007-2
Jones, R. J., Ward, S., Amri, A. Y., and Hoegh-Guldberg, O. (2000). Changes in quantum efficiency of Photosystem II of symbiotic dinoflagellates of corals after heat stress, and of bleached corals sampled after the 1998 Great Barrier Reef mass bleaching event. Mar. Freshw. Res. 51, 63–71. doi: 10.1071/MF99100
Kang, H. C., Jeong, H. J., Jang, S. H., and Lee, K. H. (2019a). Feeding by common heterotrophic protists on the phototrophic dinoflagellate Biecheleriopsis adriatica (Suessiaceae) compared to that of other suessioid dinoflagellates. Algae 34, 127–140. doi: 10.4490/algae.2019.34.5.29
Kang, H. C., Jeong, H. J., Ok, J. H., You, J. H., Jang, S. H., Lee, S. Y., et al. (2019b). Spatial and seasonal distributions of the phototrophic dinoflagellate Biecheleriopsis adriatica (Suessiaceae) in Korea: quantification using qPCR. Algae 34, 111–126. doi: 10.4490/algae.2019.34.5.25
Kang, H. C., Jeong, H. J., Kim, S. J., You, J. H., and Ok, J. H. (2018). Differential feeding by common heterotrophic protists on 12 different Alexandrium species. Harmful Algae 78, 106–117. doi: 10.1016/j.hal.2018.08.005
Kang, H. C., Jeong, H. J., Park, S. A., Eom, S. H., Ok, J. H., You, J. H., et al. (2020b). Feeding by the newly described heterotrophic dinoflagellate Gyrodinium jinhaense: comparison with G. dominans and G. moestrupii. Mar. Biol. 167:156. doi: 10.1007/s00227-020-03769-9
Kang, H. C., Jeong, H. J., Lim, A. S., Ok, J. H., You, J. H., Park, S. A., et al. (2020a). Effects of temperature on the growth and ingestion rates of the newly described mixotrophic dinoflagellate Yihiella yeosuensis and its two optimal prey species. Algae 35, 263–275. doi: 10.4490/algae.2020.35.8.20
Kibler, S. R., Litaker, R. W., Holland, W. C., Vandersea, M. W., and Tester, P. A. (2012). Growth of eight Gambierdiscus (Dinophyceae) species: effects of temperature, salinity and irradiance. Harmful Algae 19, 1–14. doi: 10.1016/j.hal.2012.04.007
Kikawada, T., Saito, A., Kanamori, Y., Nakahara, Y., Iwata, K. I., Tanaka, D., et al. (2007). Trehalose transporter 1, a facilitated and high-capacity trehalose transporter, allows exogenous trehalose uptake into cells. Proc. Natl. Acad. Sci. U.S.A. 104, 11585–11590. doi: 10.1073/pnas.0702538104
Kim, B. M., Son, S. W., Min, S. K., Jeong, J. H., Kim, S. J., Zhang, X., et al. (2014). Weakening of the stratospheric polar vortex by Arctic sea-ice loss. Nat. Commun. 5:4646. doi: 10.1038/ncomms5646
Kim, H., Park, H., Wang, H., Yoo, H. Y., Park, J., and Ki, J. S. (2021). Low temperature and cold stress significantly increase saxitoxins (STXs) and expression of STX biosynthesis genes sxtA4 and sxtG in the dinoflagellate Alexandrium catenella. Mar. Drugs 19:291. doi: 10.3390/md19060291
Kim, H. J., and Winge, D. R. (2013). Emerging concepts in the flavinylation of succinate dehydrogenase. Biochim. Biophy. Acta Bioenerg. 1827, 627–636. doi: 10.1016/j.bbabio.2013.01.012
Kohli, G. S., Murray, S. A., Neilan, B. A., Rhodes, L. L., Harwood, D. T., Smith, K. F., et al. (2014). High abundance of the potentially maitotoxic dinoflagellate Gambierdiscus carpenteri in temperate waters of New South Wales, Australia. Harmful Algae 39, 134–145. doi: 10.1016/j.hal.2014.07.007
Korea Hydrographic and Oceanographic Agency. (2021). Ocean Data in Grid Framewokr. Available online at: http://www.khoa.go.kr/oceangrid/gis/category/reference/distribution.do (assessed August 4, 2021).
Krueger, T., Becker, S., Pontasch, S., Dove, S., Hoegh-Guldberg, O., Leggat, W., et al. (2014). Antioxidant plasticity and thermal sensitivity in four types of Symbiodinium sp. J. Phycol. 50, 1035–1047. doi: 10.1111/jpy.12232
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., et al. (2018). Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580. doi: 10.1016/j.cub.2018.07.008
Landsberg, J. H. (2002). The effects of harmful algal blooms on aquatic organisms. Rev. Fish. Sci. 10, 113–390. doi: 10.1080/20026491051695
Lauritano, C., De Luca, D., Ferrarini, A., Avanzato, C., Minio, A., Esposito, F., et al. (2017). De novo transcriptome of the cosmopolitan dinoflagellate Amphidinium carterae to identify enzymes with biotechnological potential. Sci. Rep. 7:11701. doi: 10.1038/s41598-017-12092-1
Lee, H. D., Min, K. H., Bae, J. H., and Cha, D. H. (2020). Characteristics and comparison of 2016 and 2018 heat wave in Korea. Atmos. Korean Meteorol. Soc. 30, 209–220. doi: 10.14191/Atmos.2020.30.1.001
Lee, T. F., Zhai, J., and Meyers, B. C. (2010). Conservation and divergence in eukaryotic DNA methylation. Proc. Natl. Acad. Sci.U.S.A. 107, 9027–9028. doi: 10.1073/pnas.1005440107
Leggat, W., Seneca, F., Wasmund, K., Ukani, L., Yellowlees, D., and Ainsworth, T. D. (2011). Differential responses of the coral host and their algal symbiont to thermal stress. PLoS One 6:e26687. doi: 10.1371/journal.pone.0026687
Levin, R. A., Beltran, V. H., Hill, R., Kjelleberg, S., McDougald, D., Steinberg, P. D., et al. (2016). Sex, scavengers, and chaperones: transcriptome secrets of divergent Symbiodinium thermal tolerances. Mol. Biol. Evol. 33, 2201–2215. doi: 10.1093/molbev/msw119
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12:323. doi: 10.1186/1471-2105-12-323
Li, T., Chen, X., and Lin, S. (2021). Physiological and transcriptomic responses to N-deficiency and ammonium: nitrate shift in Fugacium kawagutii (Symbiodiniaceae). Sci. Total Environ. 753:141906. doi: 10.1016/j.scitotenv.2020.141906
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Li, Y., Liew, Y. J., Cui, G., Cziesielski, M. J., Zahran, N., Michell, C. T., et al. (2018). DNA methylation regulates transcriptional homeostasis of algal endosymbiosis in the coral model Aiptasia. Sci. Adv. 4:eaat2142. doi: 10.1126/sciadv.aat2142
Lim, A. S., Jeong, H. J., Ok, J. H., You, J. H., Kang, H. C., and Kim, S. J. (2019). Effects of light intensity and temperature on growth and ingestion rates of the mixotrophic dinoflagellate Alexandrium pohangense. Mar. Biol. 166:98. doi: 10.1007/s00227-019-3546-9
Lin, S., Yu, L., and Zhang, H. (2019). Transcriptomic responses to thermal stress and varied phosphorus conditions in Fugacium kawagutii. Microorganisms 7:96. doi: 10.3390/microorganisms7040096
Los, D. A., Mironov, K. S., and Allakhverdiev, S. I. (2013). Regulatory role of membrane fluidity in gene expression and physiological functions. Photosynth. Res. 116, 489–509. doi: 10.1007/s11120-013-9823-4
Los, D. A., and Murata, N. (2004). Membrane fluidity and its roles in the perception of environmental signals. Biochim. Biophys. Acta 1666, 142–157. doi: 10.1016/j.bbamem.2004.08.002
Lowe, C. D., Mello, L. V., Samatar, N., Martin, L. E., Montagnes, D. J., and Watts, P. C. (2011). The transcriptome of the novel dinoflagellate Oxyrrhis marina (Alveolata: Dinophyceae): response to salinity examined by 454 sequencing. BMC Genom. 12:519. doi: 10.1186/1471-2164-12-519
Lu, W., Tang, X., Huo, Y., Xu, R., Qi, S., Huang, J., et al. (2012). Identification and characterization of fructose 1, 6-bisphosphate aldolase genes in Arabidopsis reveal a gene family with diverse responses to abiotic stresses. Gene 503, 65–74. doi: 10.1016/j.gene.2012.04.042
Luo, Z., Yang, W., Xu, B., Zheng, B., and Gu, H. (2015). Morphology, ultrastructure, and phylogeny of Protodinium simplex and Biecheleriopsis cf. adriatica (Dinophyceae) from the China Sea. Nova Hedwigia 101, 251–268. doi: 10.1127/nova_hedwigia/2015/0268
Maciver, S. K. (1996). Myosin II function in non-muscle cells. Bioessays 18, 179–182. doi: 10.1002/bies.950180304
Mansoorabadi, S. O., Thibodeaux, C. J., and Liu, H. W. (2007). The diverse roles of flavin coenzymes nature’s most versatile thespians. J. Org. Chem. 72, 6329–6342. doi: 10.1021/jo0703092
Matsubara, T., Nagasoe, S., Yamasaki, Y., Shikata, T., Shimasaki, Y., Oshima, Y., et al. (2007). Effects of temperature, salinity, and irradiance on the growth of the dinoflagellate Akashiwo sanguinea. J. Exp. Mar. Biol. Ecol. 342, 226–230. doi: 10.1016/j.jembe.2006.09.013
McClanahan, T. R., Ateweberhan, M., Muhando, C. A., Maina, J., and Mohammed, M. S. (2007). Effects of climate and seawater temperature variation on coral bleaching and mortality. Ecol. Monogr. 77, 503–525. doi: 10.1890/06-1182.1
McClymont, E. L., Sosdian, S. M., Rosell-Melé, A., and Rosenthal, Y. (2013). Pleistocene sea-surface temperature evolution: early cooling, delayed glacial intensification, and implications for the mid-Pleistocene climate transition. Earth Sci. Rev. 123, 173–193. doi: 10.1016/j.earscirev.2013.04.006
Middelbeek, J., Clark, K., Venselaar, H., Huynen, M. A., and Van Leeuwen, F. N. (2010). The alpha-kinase family: an exceptional branch on the protein kinase tree. Cell. Mol. Life Sci. 67, 875–890. doi: 10.1007/s00018-009-0215-z
Minarik, P., Tomaskova, N., Kollarova, M., and Antalik, M. (2002). Malate dehydrogenases-structure and function. Gen. Physiol. Biophy. 21, 257–266.
Moestrup, Ø, Lindberg, K., and Daugbjerg, N. (2009). Studies on woloszynskioid dinoflagellates V. ultrastructure of Biecheleriopsis gen. nov., with description of Biecheleriopsis adriatica sp. nov. Phycol. Res. 57, 221–237. doi: 10.1111/j.1440-1835.2009.00541.x
Morey, J. S., Monroe, E. A., Kinney, A. L., Beal, M., Johnson, J. G., Hitchcock, G. L., et al. (2011). Transcriptomic response of the red tide dinoflagellate, Karenia brevis, to nitrogen and phosphorus depletion and addition. BMC Genomics 12:346. doi: 10.1186/1471-2164-12-346
Morton, S. L., Norris, D. R., and Bomber, J. W. (1992). Effect of temperature, salinity and light intensity on the growth and seasonality of toxic dinoflagellates associated with ciguatera. J. Exp. Mar. Biol. Ecol. 157, 79–90. doi: 10.1016/0022-0981(92)90076-M
Muffat, J., and Walker, D. W. (2010). Apolipoprotein D: an overview of its role in aging and age-related diseases. Cell Cycle 9, 269–273. doi: 10.4161/cc.9.2.10433
Murata, N., and Los, D. A. (1997). Membrane fluidity and temperature perception. Plant Physiol. 115:875. doi: 10.1104/pp.115.3.875
Murray, S. A., Suggett, D. J., Doblin, M. A., Kohli, G. S., Seymour, J. R., Fabris, M., et al. (2016). Unravelling the functional genetics of dinoflagellates: a review of approaches and opportunities. Perspect. Phycol. 3, 37–52. doi: 10.1127/pip/2016/0039
Musrati, R. A., Kollarova, M., Mernik, N., and Mikulasova, D. (1998). Malate dehydrogenase: distribution, function and properties. Gen. Physiol. Biophys. 17, 193–210.
Nagasoe, S., Shikata, T., Yamasaki, Y., Matsubara, T., Shimasaki, Y., Oshima, Y., et al. (2010). Effects of nutrients on growth of the red-tide dinoflagellate Gyrodinium instriatum Freudenthal et Lee and a possible link to blooms of this species. Hydrobiologia 651, 225–238. doi: 10.1007/s10750-010-0301-0
Naydenov, M., Baev, V., Apostolova, E., Gospodinova, N., Sablok, G., Gozmanova, M., et al. (2015). High-temperature effect on genes engaged in DNA methylation and affected by DNA methylation in Arabidopsis. Plant Physiol. Biochem. 87, 102–108. doi: 10.1016/j.plaphy.2014.12.022
Neustupa, J., St’astny, J., and Hodac, L. (2008). Temperature-related phenotypic plasticity in the green microalga Micrasterias rotata. Aquat. Microb. Ecol. 51, 77–86. doi: 10.3354/ame01184
Ok, J. H., Jeong, H. J., Lim, A. S., You, J. H., Kang, H. C., Kim, S. J., et al. (2019). Effects of light and temperature on the growth of Takayama helix (Dinophyceae): mixotrophy as a survival strategy against photoinhibition. J. Phycol. 55, 1181–1195. doi: 10.1111/jpy.12907
Pospelova, V., de Vernal, A., and Pedersen, T. F. (2008). Distribution of dinoflagellate cysts in surface sediments from the northeastern Pacific Ocean (43–25°N) in relation to sea-surface temperature, salinity, productivity and coastal upwelling. Mar. Micropaleontol. 68, 21–48. doi: 10.1016/j.marmicro.2008.01.008
Rajan, V. B. V., and D’Silva, P. (2009). Arabidopsis thaliana J-class heat shock proteins: cellular stress sensors. Funct. Integr. Genom. 9, 433–446. doi: 10.1007/s10142-009-0132-0
Raven, J. A., and Geider, R. J. (1988). Temperature and algal growth. New Phytol. 110, 441–461. doi: 10.1111/j.1469-8137.1988.tb00282.x
Rosic, N. N., Pernice, M., Rodriguez-Lanetty, M., and Hoegh-Guldberg, O. (2011). Validation of housekeeping genes for gene expression studies in Symbiodinium exposed to thermal and light stress. Mar. Biotechnol. 13, 355–365. doi: 10.1007/s10126-010-9308-9
Sakamoto, S., Lim, W. A., Lu, D., Dai, X., Orlova, T., and Iwataki, M. (2021). Harmful algal blooms and associated fisheries damage in East Asia: current status and trends in China, Japan, Korea and Russia. Harmful Algae 102:101787. doi: 10.1016/j.hal.2020.101787
Sakamoto, T., and Murata, N. (2002). Regulation of the desaturation of fatty acids and its role in tolerance to cold and salt stress. Curr. Opin. Microbiol. 5, 206–210. doi: 10.1016/S1369-5274(02)00306-5
Saxby, T., Dennison, W. C., and Hoegh-Guldberg, O. (2003). Photosynthetic responses of the coral Montipora digitata to cold temperature stress. Mar. Ecol. Prog. Ser. 248, 85–97. doi: 10.3354/meps248085
Selina, M. S., Morozova, T. V., Vyshkvartsev, D. I., and Orlova, T. Y. (2014). Seasonal dynamics and spatial distribution of epiphytic dinoflagellates in Peter the Great Bay (Sea of Japan) with special emphasis on Ostreopsis species. Harmful Algae 32, 1–10. doi: 10.1016/j.hal.2013.11.005
Sherr, E. B., and Sherr, B. F. (2007). Heterotrophic dinoflagellates: a significant component of microzooplankton biomass and major grazers of diatoms in the sea. Mar. Ecol. Prog. Ser. 352, 187–197. doi: 10.3354/meps07161
Shi, X., Lin, X., Li, L., Li, M., Palenik, B., and Lin, S. (2017). Transcriptomic and microRNAomic profiling reveals multi-faceted mechanisms to cope with phosphate stress in a dinoflagellate. ISME J. 11, 2209–2218. doi: 10.1038/ismej.2017.81
Shumway, S. E. (1990). A review of the effects of algal blooms on shellfish and aquaculture. J. World Aquac. Soc. 21, 65–104. doi: 10.1111/j.1749-7345.1990.tb00529.x
Silflow, C. D., and Lefebvre, P. A. (2001). Assembly and motility of eukaryotic cilia and flagella. Lessons from Chlamydomonas reinhardtii. Plant Physiol. 127, 1500–1507. doi: 10.1104/pp.010807
Sluijs, A., Pross, J., and Brinkhuis, H. (2005). From greenhouse to icehouse; organic-walled dinoflagellate cysts as paleoenvironmental indicators in the Paleogene. Earth Sci. Rev. 68, 281–315. doi: 10.1016/j.earscirev.2004.06.001
Smith, S. J., Edmonds, J., Hartin, C. A., Mundra, A., and Calvin, K. (2015). Near-term acceleration in the rate of temperature change. Nat. Clim. Change 5, 333–336. doi: 10.1038/nclimate2552
Spilling, K., Olli, K., Lehtoranta, J., Kremp, A., Tedesco, L., Tamelander, T., et al. (2018). Shifting diatom: dinoflagellate dominance during spring bloom in the Baltic Sea and its potential effects on biogeochemical cycling. Front. Mar. Sci. 5:327. doi: 10.3389/fmars.2018.00327
Stoecker, D. K., Hansen, P. J., Caron, D. A., and Mitra, A. (2017). Mixotrophy in the marine plankton. Annu. Rev. Mar. Sci. 9, 311–335. doi: 10.1146/annurev-marine-010816-060617
Sun, X. M., Ren, L. J., Zhao, Q. Y., Ji, X. J., and Huang, H. (2018). Microalgae for the production of lipid and carotenoids: a review with focus on stress regulation and adaptation. Biotechnol. Biofuels 11:272. doi: 10.1186/s13068-018-1275-9
Takahashi, K., Sarai, C., and Iwataki, M. (2014). Morphology of two marine woloszynskioid dinoflagellates, Biecheleria brevisulcata sp. nov. and Biecheleriopsis adriatica (Suessiaceae, Dinophyceae), from Japanese coasts. Phycologia 53, 52–65. doi: 10.2216/13-192.1
Taucher, J., Arístegui, J., Bach, L. T., Guan, W., Montero, M. F., Nauendorf, A., et al. (2018). Response of subtropical phytoplankton communities to ocean acidification under oligotrophic conditions and during nutrient fertilization. Front. Mar. Sci. 5:330. doi: 10.3389/fmars.2018.00330
Taylor, F. J. R., Hoppenrath, M., and Saldarriaga, J. F. (2008). Dinoflagellate diversity and distribution. Biodivers. Conserv. 17, 407–418. doi: 10.1007/s10531-007-9258-3
Tong, M., Zhou, Q., David, K. M., Jiang, T., Qi, Y., and Donald, A. M. (2010). Culture techniques and growth characteristics of Dinophysis acuminata and its prey. Chin. J. Oceanol. Limnol. 28, 1230–1239. doi: 10.1007/s00343-010-9960-y
Tsipas, G. (2020). Metagenomic Characterization of Unicellular Eukaryotes in the Urban Thessaloniki Bay. Master’s thesis. Thessaloniki: International Hellenic university.
Valledor, L., Furuhashi, T., Hanak, A. M., and Weckwerth, W. (2013). Systemic cold stress adaptation of Chlamydomonas reinhardtii. Mol. Cell. Proteomics 12, 2032–2047. doi: 10.1074/mcp.M112.026765
Veron, J. E., Hoegh-Guldberg, O., Lenton, T. M., Lough, J. M., Obura, D. O., Pearce-Kelly, P. A. U. L., et al. (2009). The coral reef crisis: The critical importance of< 350 ppm CO2. Mar. Pollut. Bull. 58, 1428–1436. doi: 10.1016/j.marpolbul.2009.09.009
Wang, H., Kim, H., and Ki, J. S. (2021). Transcriptomic identification and expression analysis of cold shock domain protein (CSP) genes in the marine dinoflagellate Prorocentrum minimum. J. Appl. Phycol. 33, 843–854. doi: 10.1007/s10811-020-02332-9
Wang, W., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Wuitchik, D. M. (2018). Temperature Modulation of Biological Clocks in a Reef-Building Coral. Master’s thesis. Calgary, AB: University of Calgary.
Xu, N., Duan, S., Li, A., Zhang, C., Cai, Z., and Hu, Z. (2010). Effects of temperature, salinity and irradiance on the growth of the harmful dinoflagellate Prorocentrum donghaiense Lu. Harmful Algae 9, 13–17. doi: 10.1016/j.hal.2009.06.002
Xu, Y., Richlen, M. L., Liefer, J. D., Robertson, A., Kulis, D., Smith, T. B., et al. (2016). Influence of environmental variables on Gambierdiscus spp. (Dinophyceae) growth and distribution. PLoS One 11:e0153197. doi: 10.1371/journal.pone.0153197
Ye, J., Fang, L., Zheng, H., Zhang, Y., Chen, J., Zhang, Z., et al. (2006). WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34(suppl 2), W293–W297. doi: 10.1093/nar/gkl031
You, J. H., Jeong, H. J., Lim, A. S., Ok, J. H., and Kang, H. C. (2020). Effects of irradiance and temperature on the growth and feeding of the obligate mixotrophic dinoflagellate Gymnodinium smaydae. Mar. Biol. 167:64. doi: 10.1007/s00227-020-3678-y
Zhang, C., Chen, G., Wang, Y., Guo, C., and Zhou, J. (2018). Physiological and molecular responses of Prorocentrum donghaiense to dissolved inorganic phosphorus limitation. Mar. Pollut. Bull. 129, 562–572. doi: 10.1016/j.marpolbul.2017.10.031
Keywords: cold wave, global warming, heat wave, marine ecosystem, phytoplankton, red tide, temperature
Citation: Kang HC, Jeong HJ, Park SA, Ok JH, You JH, Eom SH, Park EC, Jang SH and Lee SY (2021) Comparative Transcriptome Analysis of the Phototrophic Dinoflagellate Biecheleriopsis adriatica Grown Under Optimal Temperature and Cold and Heat Stress. Front. Mar. Sci. 8:761095. doi: 10.3389/fmars.2021.761095
Received: 19 August 2021; Accepted: 25 October 2021;
Published: 16 November 2021.
Edited by:
Irene Wagner-Doebler, Helmholtz Center for Infection Research, Helmholtz Association of German Research Centers (HZ), GermanyReviewed by:
Shauna Murray, University of Technology Sydney, AustraliaDavid Morse, Université de Montréal, Canada
Copyright © 2021 Kang, Jeong, Park, Ok, You, Eom, Park, Jang and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hae Jin Jeong, aGpqZW9uZ0BzbnUuYWMua3I=
 Hee Chang Kang
Hee Chang Kang Hae Jin Jeong
Hae Jin Jeong Sang Ah Park
Sang Ah Park Jin Hee Ok
Jin Hee Ok Ji Hyun You
Ji Hyun You Se Hee Eom
Se Hee Eom Eun Chong Park
Eun Chong Park Se Hyeon Jang
Se Hyeon Jang Sung Yeon Lee
Sung Yeon Lee