
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 02 November 2021
Sec. Aquatic Physiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.760902
Chalcone synthase (CHS) family are plant type III polyketide synthases that participate in the flavonoid synthesis pathway to induce plant resistance to various biotic and abiotic stresses. Zostera marina, a common seagrass, migrated to terrestrial conditions and returned to the sea, achieving the most severe habitat shift of flowering plants. Given the special evolutionary process, we conducted genome-wide, expression and enzyme activity analyses of the ZosmaCHS family to understand its phylogenetic implications. Various duplication modes led to the expansion of 11 CHS homologs in Z. marina. Based on the phylogenetic relationships, ZosmaCHSs were classified into three clades. Further quantitative real time-PCR analyses of the ZosmaCHS homologs showed different light responses and tissue-specific expression, indicating functional diversification of the ZosmaCHSs. Moreover, the ZosmaCHS proteins clustering with the validated chalcone synthases were recombined into prokaryotic expression systems. All the recombinant proteins showed CHS activity to generate naringenin chalcone with varying catalytic efficiencies. ZosmaCHS07 was regarded as the dominant CHS because of its significant light response and the higher catalytic efficiency. Taken together, the disparity of the expression and enzyme activity indicated that sub-functionalization is the primary mechanism of the expansion of the ZosmaCHSs family.
The chalcone synthase (CHS) superfamily mainly participates in the biosynthesis of various plant secondary metabolites. The metabolites include chalcones, stilbenes, phloroglucinols, resorcinols, benzophenones, biphenyls, bibenzyls, chromones, acridones, pyrones, and curcuminoids (Austin and Noel, 2003; Abe and Morita, 2010). These metabolites play key roles in tissue pigmentation, auxin transport, pathogen defense, pollen fertility, and light protection (Dao et al., 2011; Pandith et al., 2016, 2019). CHS (EC 2.3.1.74), the most representative member, carries out the sequential decarboxylative condensation of p-coumaroyl-CoA with three malonyl-CoA molecules. This process generates naringenin chalcone, the starting molecule in the biosynthesis of various flavonoids (Kuo et al., 2019).
Non-CHS genes co-existing in the CHS superfamily are thought to have co-evolved with CHSs during the evolutionary process (Durbin et al., 2000; De Luca and Lauritano, 2020). Most CHS superfamily members have a high sequence similarity, and a conserved catalytic triad of Cys-His-Asn in their active sites. They function as 40–45 kDa protein homodimers (Jiang et al., 2008). However, the enzymes encoded by non-CHS genes differ from CHS on the choice of initial substrates, the number of condensation reactions, or mechanism of the cyclization and aromatization of intermediates (Helariutta et al., 1996; Han et al., 2017).
Most plants retained multiple CHS copies and showed various expression in different tissues. For example, the three grapevine CHS genes mediated the production of different flavonoid compounds in different tissues (Harris et al., 2013). As for the 14 maize CHS genes (ZmCHS01-14), ZmCHS01/02 exhibited constitutive expression in seeds and leaves, while the ZmCHS05/12 primarily expressed in roots (Han et al., 2016). In case of the four CHS genes in Gerbera hybrida (GCHS1-4), only GCHS1 involved in flavonoid biosynthesis (Deng et al., 2014). The tissue specific expression patterns reflect the functional diversification of duplicated CHS genes. Light is one of the most important environmental signals influencing flavonoid biosynthesis in plants. Light intensity and quality (wavelength) can induce the expression of the CHS genes and the synthesis of flavonoid compounds (Zoratti et al., 2014). Exposure to light can increase the higher expression level and anthocyanin, PAs and flavonols content in grape berry skin (Azuma et al., 2012). Positive impact of light on flavonoid accumulation has also been reported in some fruit species and Rosaceae family (Uleberg et al., 2012; Li et al., 2013). Light-activated photoreceptors regulate a core signaling pathway, CONSTITUTIVE PHOTOMORPHOGENIC1/SUPPRESOR OF PHYTOCHROME A-105 (COP1/SPA), to activate the expression of flavonoid pathway genes including CHS (Lau and Deng, 2012). It appears that the accumulation of flavonoids is more sensitive to the blue and UV-light (Zoratti et al., 2014). The anthocyanin concentrations in grape berries were higher in blue light-treated samples than in red light (Kondo et al., 2014).
The availability of numerous sequenced genomes has facilitated the evolutionary studies of the CHS genes family. Extensive genome-wide analyses of CHS genes have been performed in various species, such as Physcomitrella patens (Koduri et al., 2010), Psilotum nudum (Yamazaki et al., 2001), Gerbera hybrida (Deng et al., 2014), Vitis vinifera (Wang et al., 2016), Capsicum annuum (Xu et al., 2015), Salvia miltiorrhiza (Deng et al., 2018), Zea mays (Han et al., 2016), Oryza sativa (Han et al., 2017; Hu et al., 2017) and Solanum melongena (Wu et al., 2020). Most of these studies focused on exploring the identification and diverse expression patterns of CHS homologs. In addition, there are several reports have been studied the enzyme activity of CHS homologs in land plants (Liou et al., 2018). For example, the CHS-D and CHS-E genes of Ipomoea purpurea and the PaCHS in the liverwort Plagiochasma appendiculatum were proved to be the chalcone synthases producing naringenin chalcone (Shiokawa et al., 2000; Yu et al., 2015). However, enzyme properties of the CHSs in seagrass have been scarcely characterized.
Zostera marina, a marine angiosperm, is widely distributed in the northern Pacific and northern Atlantic Oceans (Olsen et al., 2016). It is the main component of seagrass meadows and plays a crucial role in nutrient cycling, sediment stabilization, habitats, and food provision for numerous organisms (Worm et al., 2006; Heck et al., 2008; Hughes et al., 2009). Seagrass migrated to terrestrial conditions approximately 200 million years ago and returned to the sea about 140 million years ago (Les and Waycott, 1997). This migration is regarded as the most severe habitat shift achieved by flowering plants. The process involved significant genomic changes to adapt to its marine lifestyle (Olsen et al., 2016). Given the special evolutionary process of Z. marina, we conducted a genome-wide, expression, and enzyme activity analyses to understand the phylogenetic implications and functional disparity of the ZosmaCHS family.
To identify the CHS family members in Z. marina, CHS profiles (PF00195 and PF02797) downloaded from the Pfam database1 were used as queries to perform BLASTP searches against Z. marina database (Olsen et al., 2016) on Phytozome website2. The gene models of ZosmaCHSs were constructed using the Gene Structure Display Server (GSDS,3) (Hu et al., 2015). The DNAMAN 9 software (LynnonBiosoft, United States) was used to predict the isoelectric points and molecular weights of CHS proteins. Plant Duplicate Gene Database (PlantDGD,4) was used for gene duplication analysis.
The Pfam database and Simple Modular Architecture Research Tool (SMART,5) were used to predict the conserved domains of the ZosmaCHS proteins. The online MEME program6 was used to identify the conserved motifs of the CHS sequences. The parameters set as following: any number of repetitions, maximum of 10 motifs, and optimum motif width of 6–50 amino acid residues.
The neighbor-joining method and maximum likelihood method with a 1,000 times bootstrap value were used to construct phylogenetic tree by multiple sequence alignment of the protein sequences of 11 ZosmaCHSs in MEGA X software. The accession numbers and resources of all CHSs used for the tree were also provided in Supplementary Table 6. All CHSs can be found in Uniprot database7. Default settings were used for the other parameters.
The PlantCare website8 was used to analyze the putative promoter sequences of ZosmaCHSs. All the predicted cis-elements except the TATA-box and CAAT-box were visualized using the TBtools software following the author’s instructions (Chen et al., 2020).
Zostera marina with intact rhizome-systems was collected during the growing season from 3 m deep sub-tidal seagrass beds in Rongcheng (37° 91′N, 120° 73′E), Shandong Province, China. Samples were then cultured in seawater-aquarium that was continuously aerated and renewed daily. The plants were pre-cultivated with a 10/14 h (light/dark) photoperiod under minimum saturation light intensity (100 μmol photons m–2s–1) at 15°C for 3 days before experimentation. The roots, leaves, flowers, stems, and rhizomes were collected during the plant’s flowering stage for tissue-specific expression analysis using quantitative real time-PCR (qRT-PCR). The leaves were further exposed to blue, red and white light at the intensity of 300 μmol photons m–2s–1, after dark-adaptation overnight. Sampling was done at 0, 1, and 3 h for the qRT-PCR. Blue light was the dominant spectral components in the ecological niche of Z. marina, while the red light was at low intensity (Olsen et al., 2016).
Total RNA was extracted from the five tissues as well as the leaves exposed to different light treatments using RNAiso Plus (Takara, Japan). RNA quality was determined through agarose gel electrophoresis and NanoQuant (TECAN, Switzerland). Subsequently, 1 μg of the total RNA was reverse transcribed to cDNA using HiScript® II 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) following the manufacturer’s instructions. The qRT-PCR assays were performed on a Bio-Rad CFX96 Real-Time PCR System using AceQ Universal SYBR qPCR Master Mix (Vazyme). Primer sequences used in qRT-PCR are listed in Supplementary Table 1. The housekeeping gene gapdh of Z. marina was used as the internal control. The qRT-PCR program was set as follows: 95°C for 10 s, followed by 40 cycles of 56°C for 10 s and 72°C for 30 s. Each reaction was performed in three biological replicates. The relative expression level of each gene was calculated using the 2–Δ Δ CT method. Heatmaps were constructed based on the transformed log2 (2–ΔΔCT + 1) values using the TBtools software.
The open reading frames of the five CHS genes (ZosmaCHS01, ZosmaCHS02, ZosmaCHS07, ZosmaCHS08, and ZosmaCHS11) were amplified using KOD -Plus- Neo DNA Polymerase (TOYOBO, Japan). Primer sequences with restriction enzymes (EcoRI and BamHI) cutting sites are listed in Supplementary Table 2. The amplified products were purified using a FastPure Gel DNA Extraction Mini Kit (Vazyme, Nanjing, China), cloned into pEASY®-Blunt Simple Cloning Vector (TransGen Biotech, Beijing, China), and then transformed into T1 competent cell for sequencing to select sequences without the base mutations and deletions. Plastids with correct CHS sequences, which was extracted by TIANprep Mini Plasmid Kit (TIANGEN, Beijing, China), and pET-28a (+) vector were cut using EcoRI and BamHI restriction enzymes (Invitrogen, Carlsbad, CA, United States), purified using a FastPure Gel DNA Extraction Mini Kit, and then linked by T4 DNA ligase (Invitrogen, Carlsbad, CA, United States). The pET-28a (+)-CHS was transformed into Transetta (DE3) Chemically Competent Cell. Until their OD600 to 0.4–0.5, the transformant cultures were added with isopropyl β-D-thiogalactopyranoside (IPTG) to a final concentration of 1 mmol L–1, and incubated at 16°C overnight. The heterologous expression of the CHS was then examined using SDS-PAGE. The following buffers were used for purification of the ZosmaCHSs: lysis buffer (50 mM Na2HPO4, 0.3 M NaCl, pH = 8.0), washing buffer (50 mM NaH2PO4, 0.3 M NaCl, 10 mM imidazole pH = 8.0), and elution buffer (50 mM NaH2PO4, 0.3 M NaCl, 250 mM imidazole pH = 8.0). The bacterial cells were centrifugation, resuspended in lysis buffer, and broken by ultrasonication for 30 min on ice. The proteins were loaded on High-Affinity Ni-NTA Resin (GenScript Biotech, Nanjing, China), washed by washing buffer, eluted by elution buffer and further examined using SDS-PAGE.
The enzyme activities of the five purified CHS proteins were examined using a CHS enzyme activity kit (GENMED Scientifics Inc., United States) following the manufacturer’s instructions. Their absorbance was measured at 412 nm using a multifunctional enzyme-labeled instrument (Tecan, Switzerland). Each reaction was performed in three technical replicates. The enzyme activity of ZosmaCHS represented the test enzyme activity subtracted the activity of the empty buffer.
Data were analyzed using one-way ANOVA and Tukey’s tests on SPSS 22.0. P < 0.05 indicated significant differences between groups.
Eleven full-length CHS homologs (ZosmaCHS01-11) corresponding to the Pfam CHS family were identified and described (Table 1). Among them, ZosmaCHS09 was the smallest protein with 23 kDa, while the remaining were about 43 kDa. The isoelectric point (pI) of the proteins ranged between 6.25 and 7.58. Most CHS genes had multiple exons, of which ZosmaCHS02, 04, 07, 08, and 09 had two exons, ZosmaCHS03 had three exons, ZosmaCHS10 had four exons, while ZosmaCHS01, 05, 06, and 11 had one exon (Figure 1A).
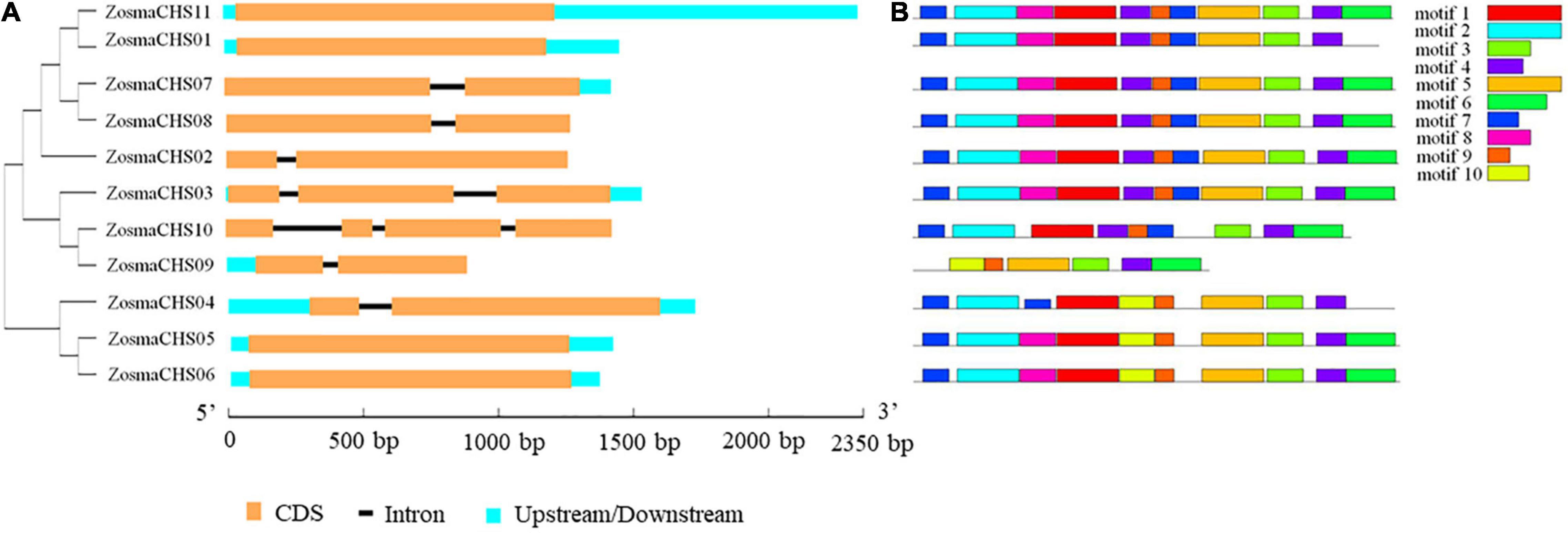
Figure 1. Gene structure and motifs compositions of ZosmaCHS proteins. (A) Gene model of CHS genes in Z. marina. Blue boxes indicate untranslated regions, black lines indicate introns and yellow boxes indicate exons. (B) The motif compositions of ZosmaCHS proteins. The motifs 1–10 are displayed in different colored boxes. The sequence information for each motif is shown in Supplementary Table 4.
Ten of the 11 CHS homologs harbored integral chalcone/stilbene synthases N-terminal (Chal_sti_synt_N) and chalcone/stilbene synthases C-terminal (Chal_sti_synt_C) domain represented by motif 1, 2, 7, and 9, and motif 3, 4 and 6, respectively (Figure 1B). ZosmaCHS09, had only part of the two domains with the absence of motif 1, 2, and 7. Moreover, ZosmaCHSs within clades I and III shared similar motif composition. The duplicated pairs ZosmaCHS05/06 and ZosmaCHS 07/08 showed highly similar motif distribution, indicating that their protein architecture is conserved in duplicated homologs.
Five widely accepted duplication modes (whole genome, tandem, proximal, transposed, and dispersed duplication) were systematically scanned among the ZosmaCHS genes to explore the evolution clues of the CHS gene family. The gene pairs ZosmaCHS05/06, ZosmaCHS07/08, and ZosmaCHS01/11 originated from tandem, proximal, and transposed duplication, respectively. Both ZosmaCHS09 and 10 formed dispersed duplication pairs with ZosmaCHS03. To further elucidate the evolutionary trend of duplicated ZosmaCHS genes, the ratios of the number of non-synonymous substitutions per non-synonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks) (Ka/Ks) were calculated between the duplication pairs. Most duplicated pairs had a Ka/Ks ratio of less than one, indicating that the ZosmaCHS gene family could have undergone strong purifying selective pressure during evolution (Supplementary Table 3).
All the ZosmaCHS proteins contained conserved catalytic triad of Cys-His-Asn and the CHS superfamily-specific Pro386 residues (Supplementary Figure 1). All the CHS members retained the residue Phe 220 responsible for CoA binding, except ZosmaCHS09. Phe 271, which is also essential for CoA binding, varied amongst the ZosmaCHS proteins. It was not retained in ZosmaCHS10, converted to alanine in ZosmaCHS04-06 and leucine in ZosmaCHS03 and 09. However, it was conserved in the five ZosmaCHSs (ZosmaCHS01, ZosmaCHS02, ZosmaCHS07, ZosmaCHS08, and ZosmaCHS11).
The phylogenetic relationships between Z. marina and other CHS genes using the maximum likelihood (Figure 2) and neighbor-joining method (Supplementary Figure 2) exhibited similar phylogenetic topology. It revealed that 11 ZosmaCHSs clustered into three clades. Notably, ZosmaCHS01, 02, 07, 08, and 11 clustered with the validated chalcone synthases in land plants (clade I), indicating that these CHS genes could have CHS activity to generate naringenin chalcone. ZosmaCHS03, 09, and 10 clustered with Hydrangea macrophylla coumaroyl triacetic acid lactone synthase (CTAS) and Rheum palmatum benzalacetone synthase (BAS) (clade II). ZosmaCHS04-06 formed clade III, with no CHS homologs in other plants.
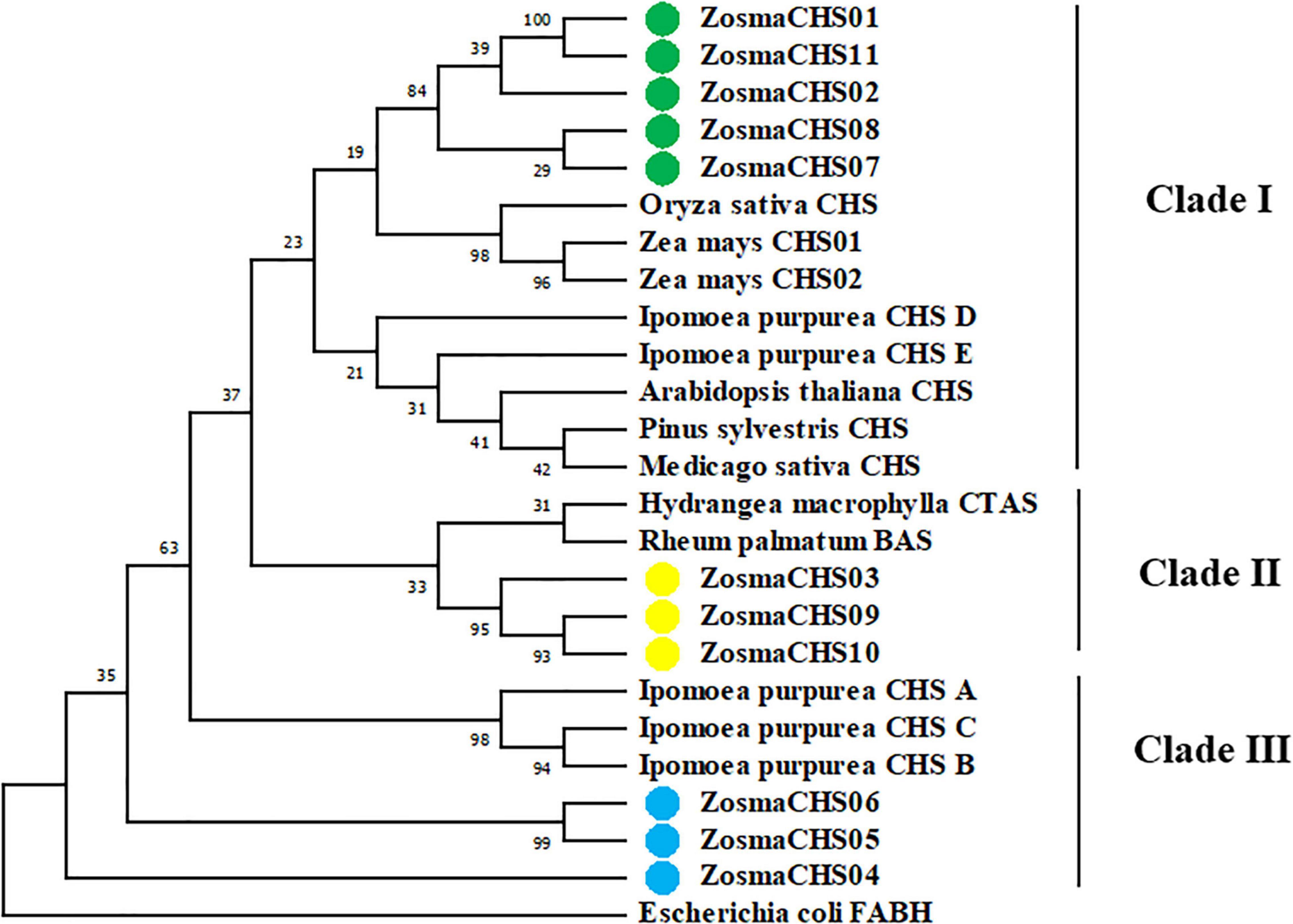
Figure 2. Phylogenetic relationships of the ZosmaCHSs and CHS homologs from other plants using maximum likelihood method. Green, yellow and blue cycles indicate the ZosmaCHSs in clade I, II, and III, respectively.
The putative cis-elements located 2.0 kb upstream of the start codon (ATG) were analyzed to investigate the potential regulatory mechanisms of ZosmaCHSs during different stress responses. Four types of cis-elements were enriched in the promoter region of the ZosmaCHS genes (Figure 3). Numerous light-related elements, including Box 4, G-box, and the GATA-, GT1-, and TCT- motifs, were detected. Box 4 was the most abundant light-responsive element. Methyl jasmonate (MeJA) responsive elements, including CGTCA- and TGACG-motif, were the common cis-acting elements indicating their involvement in regulating the MeJA-pathway. Besides the ubiquitous elements, the MYB, MYC and MBS signature sequences were in the promoter region of ZosmaCHSs, contributing to the plant tolerance to various stresses.
Cluster analysis of the upstream sequences of ZosmaCHSs (Figure 3 and Supplementary Table 5) further revealed that the promoter regions of the 11 ZosmaCHS genes clustered into three groups. TCT-motif, GA-motif, and MRE related to light response and TCA-element related to salicylic acid responsiveness were in group I (ZosmaCHS05-07 and 09) and II (ZosmaCHS02, 04 and 10), indicating that the genes could be regulated by light stress and the salicylic acid pathway. Nevertheless, circadian elements were in group III (ZosmaCHS01, 03, 08, and 11), suggesting the photoperiod had effects on regulating these CHS genes. The functional diversification of ZosmaCHSs was attribute to differences in the promoter region.
Blue light was the dominant spectral components in the ecological niche of Z. marina, while the red light was at low intensity (Olsen et al., 2016). The expression patterns of all the 11 ZosmaCHS genes were investigated to confirm whether the expression of ZosmaCHS genes was influenced by different light qualities. The ZosmaCHSs were highly expressed in red light than in other lights. Moreover, ZosmaCHS02 was predominantly induced at a relatively early stage (after 1 h treatment), while ZosmaCHS01, 07, 08, and 11, were significantly and continuously up-regulated and peaked at 3 h after treatment (Figure 4A). These ZosmaCHS genes were predicted to produce naringenin chalcone. However, most ZosmaCHSs clustering with non-CHS genes were slightly up-regulated in red light or hardly induced. The duplication pairs ZosmaCHS07/08, ZosmaCHS03/09, and ZosmaCHS03/10 exhibited different expression patterns, implying the subfunctionalization of the CHS family.
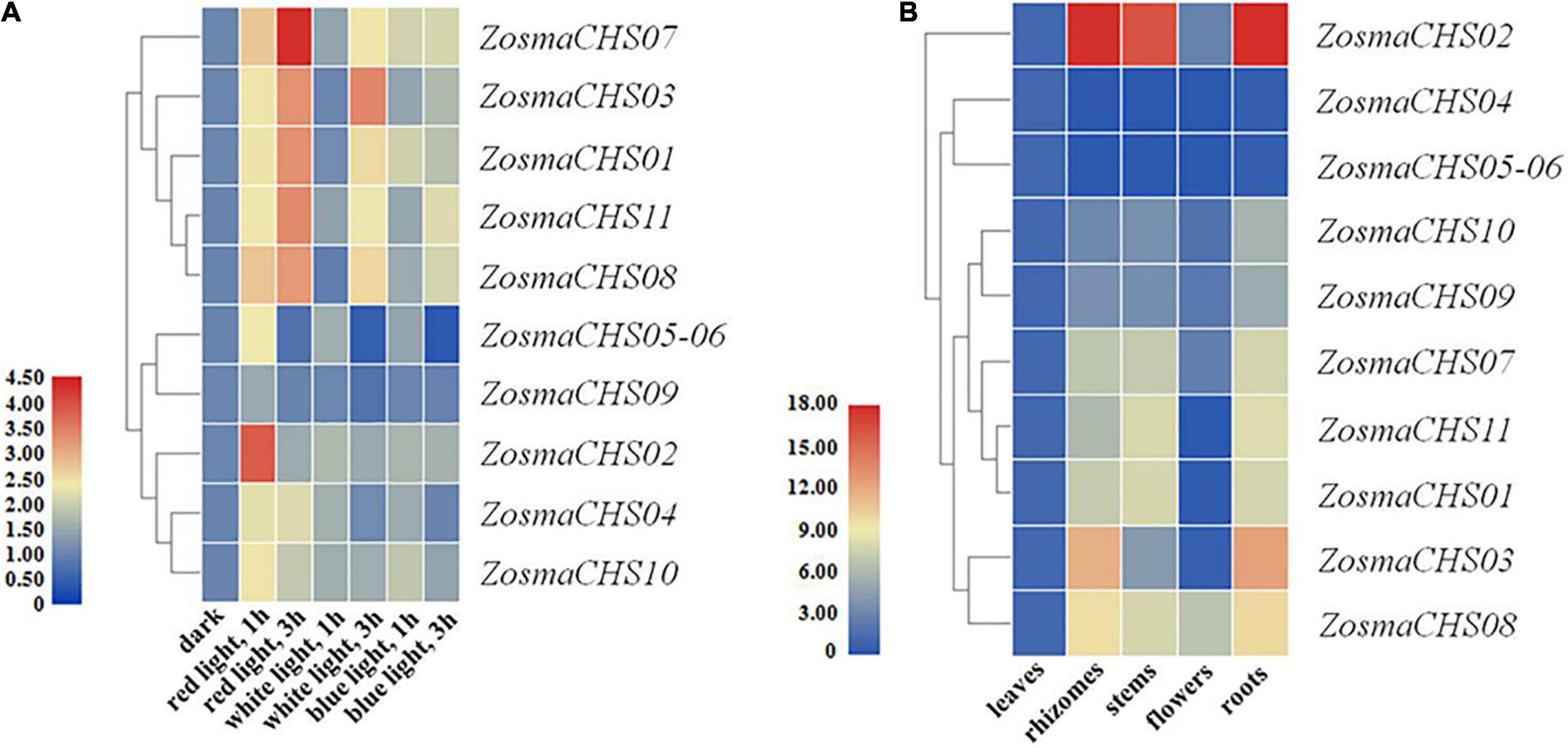
Figure 4. Expression patterns of ZosmaCHS genes. To calculate the relative expression level, the expression of ZosmaCHS gene under dark condition was set as control. Gapdh was used as reference gene. Relative level of expression was transformed by log2 (2–ΔΔCT + 1). ZosmaCHS05-06 represented the sum of ZosmaCHS05 and ZosmaCHS06 expression level because of their high sequence similarity. (A) Expression patterns of ZosmaCHS genes in different light conditions by qRT-PCR. (B) Expression patterns of ZosmaCHS genes in five tissues by qRT-PCR.
The 11 CHS homologs could be detected in all the five tissues, though at varying expression levels (Figure 4B). ZosmaCHS04-06 was highly expressed in the leaves. In contrast, most ZosmaCHSs, including ZosmaCHS01-03, 07-11, showed higher expression levels in rhizomes, stems, and roots than in leaves and flowers, possibly because the CHS was involved in light protection and other biological functions. The diverse tissue-specific expression patterns of ZosmaCHSs suggested the subfunctionalization of the CHS family.
As a crucial step to elucidate biological function of ZosmaCHSs, the proteins which clustered with the validated chalcone synthases were recombined into prokaryotic expression systems to further confirm the ZosmaCHSs producing naringenin chalcone. Five ZosmaCHSs (ZosmaCHS01, 02, 07, 08, and 11) from clade I were heterologously expressed and purified to perform in vitro enzyme activity assays. SDS-PAGE analysis showed that the corresponding protein bands (around 43 kDa) were induced in the IPTG treated sample (Figure 5A and Supplementary Figure 3B), implying that the pET-28a-ZosmaCHSs recombinant plasmid could be successfully induced and expressed in Escherichia coli (E. coli). The purification experiments indicated that His-tag affinity column could effectively enrich the recombinant proteins, while the third time elution buffer, which contained the relatively pure target proteins, could be used in enzyme activity assays. The loss of target band in empty vector control avoided the false positive result (Supplementary Figure 3A). The five proteins showed CHS activity with varying efficiencies (Figure 5B). ZosmaCHS07 had the highest CHS activity and significantly higher enzyme activity than ZosmaCHS08. However, there were no significant differences in the catalytic efficiency in the ZosmaCHS01/11 pair.
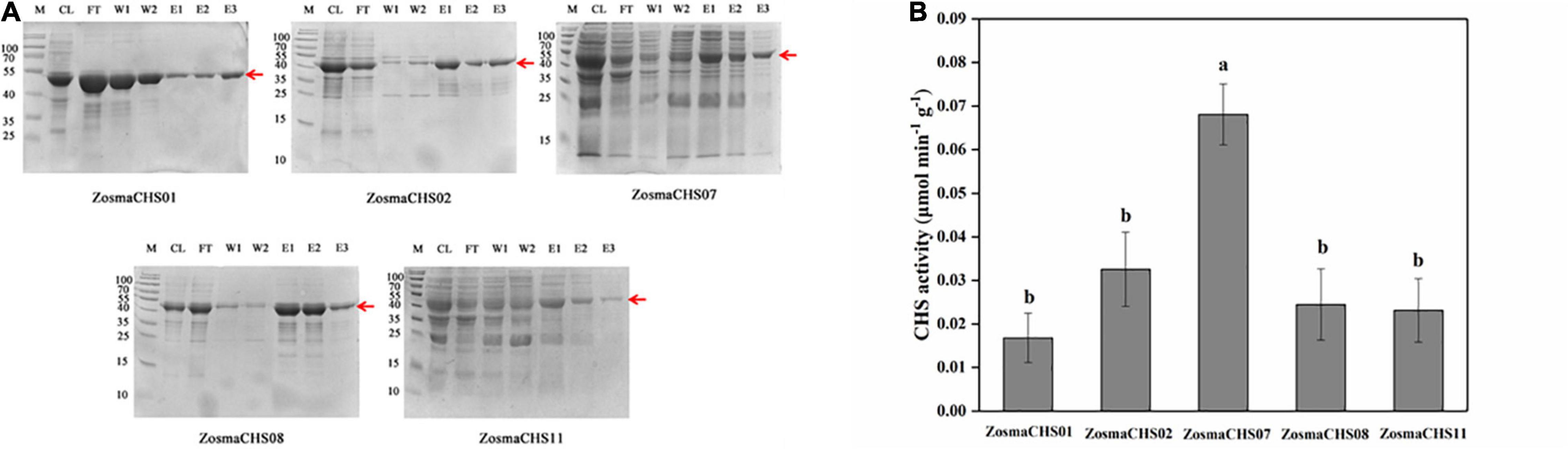
Figure 5. The validation of CHSs in Z. marina. (A) Prokaryotic purification of the five ZosmaCHS proteins. The corresponding protein bands were pointed out by the red arrow at the expected position (around 43 kDa). CL: cell with lysis buffer; FT: flow through buffer; W1: primary washing buffer; W2: secondary washing buffer; E1: primary elution buffer; E2: secondary elution buffer; E3: third time elution buffer. (B) Enzyme activity of ZosmaCHS01, 02, 07, 08, and 11. Different letters indicate significant differences (P < 0.05, One-way ANOVA).
The CHS gene family is essential for plant growth and development. They are ubiquitously distributed in various plants, from moss to angiosperms. For instance, 17, 4, 27, 14, 8, 7, 7, and 6 CHS genes have been identified in P. patens, Psilotum nudum (Yamazaki et al., 2001), O. sativa, Z. mays, Pisum sativum, C. annuum, S. melongena, and I. purpurea (Yang et al., 2004), respectively. Most CHS genes in higher plants have one intron at the conserved position (Pandith et al., 2019). Z. marina is composed of 11 full-length medium-sized CHS family genes. Numerous intronless genes were observed in ZosmaCHSs, similar to P. patens (Koduri et al., 2010), a primordial plant, indicating that Z. marina contains more primitive forms of CHS homologs.
There are four duplication types (tandem, proximal, transposed, and dispersed) events in the ZosmaCHS gene family. Tandem and proximal duplication originating from the interruption of ancient tandem duplication are important for plant adaptation to rapidly changing environments (Hanada et al., 2008). Cognizant of this, ZosmaCHS05/06 and ZosmaCHS07/08, which originated from tandem and proximal duplication, respectively, could have contributed to the adaptation of Z. marina in the sea. Most duplicated CHSs had Ka/Ks < 1, indicating that purifying selection dominates the evolution of CHS family.
Phylogenetic analysis of the CHS proteins provided more information on the CHS gene family evolution. The five ZosmaCHSs (ZosmaCHS01, 02, 07, 08, and 11) in clade I clustered with the well-characterized chalcone synthases, which produced naringenin chalcone (Shiokawa et al., 2000). Conversely, ZosmaCHSs in Clade II and III were closely related to the non-CHSs including CTAS, BAS, Ipomoea purpurea CHS A, B, and C (Flores-Sanchez and Verpoorte, 2009; Han et al., 2016). The CoA binding sites (Phe 220 and 271) whose substitution potentially leads to substrate specificity (Deng et al., 2018) were merely conserved in the ZosmaCHSs in clade I. These phenomena implied that the ZosmaCHSs in clade I could be the authentic chalcone synthases. However, an enzyme in the non-CHS cluster is a non-CHS protein, while that in the CHS cluster is not always a CHS, similar to Arachis hypogaea STS and Humulus lupulus VPS (Jiang et al., 2008; Hu et al., 2016). Cognizant of this, it is necessary to confirm the CHS activity of the five proteins in clade I.
Like other plants, the two domains Chal_sti_synt_N and Chal_sti_synt_C are highly conserved in all the ZosmaCHS genes (Han et al., 2016). The motif distribution of the ZosmaCHSs strongly supported the phylogenetic relationships. ZosmaCHS proteins in the same clade have similar motif compositions. Notably, clade II motifs arrangement was an intergradation of clade I and clade III, which probably caused the functional division during the evolution.
The ubiquity of light and MeJA responsive elements in the upstream of the ZosmaCHS genes suggested that light stress and MeJA could significantly regulate the ZosmaCHS family. This finding was consistent with previous studies that reported that plant hormones, photoperiod, light intensity, direction, and quality regulate CHS expressions (Zhang et al., 2017). Cluster analysis of the upstream sequences further showed that some light and salicylic acid response elements were in group I and II, while circadian elements responding to photoperiods were unique to group III. These findings implied that the elements had gene-specific regulation. Unlike land plants, cis-elements associated with pathogen defense were absent in the promoter sequences of ZosmaCHSs. The phenomenon could be Z. marina adaptation to its marine habitat because it lacks stomates which are the main entry points of pests and pathogens in terrestrial plants (Olsen et al., 2016).
Although the 11 ZosmaCHS genes were expressed in all the examined tissues, most genes exhibited variable expression patterns, suggesting the neo- or subfunctionalization of the CHS gene family in Z. marina. The CHS genes in rice, maize and Physcomitrella patens have been reported the various expression patterns in different tissues (Koduri et al., 2010; Han et al., 2017), suggesting its functional diversification during the evolutionary process. Unlike other plants, the transcriptional activation of ZosmaCHS in response to blue light was lower than red light. During the evolutionary process, Z. marina had experienced the respective losses of the ultraviolet photo-receptors (UV resistance locus 8), and most of the blue light photoreceptors (cryptochromes 2, 5 and DASH) (Olsen et al., 2016). The fewer blue light receptors explained the comparatively weak induction of ZosmaCHSs expression in blue light (Olsen et al., 2016), because photoreceptors are involved in light perception in the COP1/SPA-R2R3-MYB pathway to activate CHS transcription (Zhang et al., 2017).
Further enzyme activity assays of the five clade I ZosmaCHSs showed that all these proteins were authentic CHS that could use one p-coumaroyl-CoA molecule and three malonyl-CoA molecules as substrates to produce naringenin chalcone. ZosmaCHS07 was regarded to be the dominant CHS because of its significant light response and higher catalytic efficiency. The ZosmaCHS activities were measured to be 0.0168–0.068 μmol min–1 g–1, which were higher than CHS activity in peanut roots (Zhang et al., 2016). However, it may be caused by the fact that the proteins used in this study were purified proteins rather than total proteins. Moreover, some research conducted the assays of the CHS activities using HPLC (Yu et al., 2015), which was unreasonable to compare activity with ZosmaCHSs.
Gene loss and gain are related to the non-functionalization and sub/neo functionalization, respectively. The varied expression patterns in both light-treated leaves and different tissues, and catalytic efficiency of duplicated genes ZosmaCHS07/08 implied the subfunctionalization of ZosmaCHSs during evolution. However, ZosmaCHS01/11 with a similar pattern could have adopted the Innovation–Amplification–Divergence model. This model suggests that selection pressures promote the mutational improvement of CHS copies and develop side functions to adapt to environmental conditions (Bergthorsson et al., 2007; Copley, 2020).
The 11 CHS genes identified in Z. marina clustered into three clades. All the recombinant CHS proteins in clade I were confirmed to be the authentic CHS producing naringenin chalcone despite their varying catalytic efficiencies with ZosmaCHS07 being the dominant CHS. Moreover, the ZosmaCHSs family exhibited different expression pattern, with those in clade I that showed significant light-induction. Collectively, the varied catalytic efficiency combined with the different expression patterns suggests the sub-functionalization of the CHS family during evolution, which could be viewed as an adaption of Z. marina to its marine habitat.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
MM, MZ, and QZ conceived and designed the experiments, analyzed and interpreted the data. MM conducted the experiments. MZ performed the software. MZ and MM participated in original draft preparation. QZ revised the draft. WZ, MW, and CL participated in the preparation and review of the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the National Natural Science Foundation of China (No. 41376154) and the Yantai Municipal Key Research and Development Project (No. 2019XDHZ096).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank all volunteers who helped with fish collection.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.760902/full#supplementary-material
Abe, I., and Morita, H. (2010). Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat. Prod. Rep. 27, 809–838. doi: 10.1039/b909988n
Austin, M. B., and Noel, J. P. (2003). The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 20, 79–110. doi: 10.1039/b100917f
Azuma, A., Yakushiji, H., Koshita, Y., and Kobayashi, S. (2012). Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta 236, 1067–1080. doi: 10.1007/s00425-012-1650-x
Bergthorsson, U., Andersson, D. I., and Roth, J. R. (2007). Ohno’s dilemma: evolution of new genes under continuous selection. Proc. Natl. Acad. Sci. U. S. A. 104, 17004–17009. doi: 10.1073/pnas.0707158104
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., and Xia, R. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Copley, S. D. (2020). Evolution of new enzymes by gene duplication and divergence. FEBS J. 287, 1262–1283. doi: 10.1111/febs.15299
Dao, T. T., Linthorst, H. J., and Verpoorte, R. (2011). Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 10, 397–412. doi: 10.1007/s11101-011-9211-7
De Luca, D., and Lauritano, C. (2020). In silico identification of type III PKS chalcone and stilbene synthase homologs in marine photosynthetic organisms. Biology 9:110. doi: 10.3390/biology9050110
Deng, X., Bashandy, H., Ainasoja, M., Kontturi, J., Pietiainen, M., Laitinen, R. A. E., et al. (2014). Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida. New Phytol. 201, 1469–1483. doi: 10.1111/nph.12610
Deng, Y., Li, C., Li, H., and Lu, S. (2018). Identification and characterization of flavonoid biosynthetic enzyme genes in salvia miltiorrhiza (Lamiaceae). Molecules 23:1467. doi: 10.3390/molecules23061467
Durbin, M. L., McCaig, B., and Clegg, M. T. (2000). Molecular evolution of the chalcone synthase multigene family in the morning glory genome. Plant Mol. Biol. 42, 79–92. doi: 10.1023/A:1006375904820
Flores-Sanchez, I. J., and Verpoorte, R. (2009). Plant polyketide synthases: a fascinating group of enzymes. Plant Physiol. Biochem. 47, 167–174. doi: 10.1016/j.plaphy.2008.11.005
Han, Y., Cao, Y., Jiang, H., and Ding, T. (2017). Genome-wide dissection of the chalcone synthase gene family in Oryza sativa. Mol. Breed. 37:119. doi: 10.1007/s11032-017-0721-x
Han, Y., Ding, T., Su, B., and Jiang, H. (2016). Genome-wide identification, characterization and expression analysis of the chalcone synthase family in maize. Int. J. Mol. Sci. 17:161. doi: 10.3390/ijms17020161
Hanada, K., Zou, C., Lehti-Shiu, M. D., Shinozaki, K., and Shiu, S. H. (2008). Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 148, 993–1003. doi: 10.1104/pp.108.122457
Harris, N. N., Luczo, J. M., Robinson, S. P., and Walker, A. R. (2013). Transcriptional regulation of the three grapevine chalcone synthase genes and their role in flavonoid synthesis in Shiraz. Aus. J. Grape Wine Res. 19, 221–229. doi: 10.1111/ajgw.12026
Heck, K. L., Carmthers, T. J. B., Duarte, C. M., Hughes, A. R., Kendrick, G., and Williams, O. S. W. (2008). Trophic transfers from seagrass meadows subsidize diverse marine and terrestrial consumers. Ecosystems 11, 1198–1210. doi: 10.1007/s10021-008-9155-y
Helariutta, Y., Kotilainen, M., Elomaa, P., Kalkkinen, N., Bremer, K., Teeri, T. H., et al. (1996). Duplication and functional divergence in the chalcone synthase gene family of Asteraceae: evolution with substrate change and catalytic simplification. Proc. Natl. Acad. Sci. U. S. A. 93, 9033–9038. doi: 10.1073/pnas.93.17.9033
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, L., He, H., Zhu, C., Peng, X., Fu, J., He, X., et al. (2017). Genome-wide identification and phylogenetic analysis of the chalcone synthase gene family in rice. J. Plant Res. 130, 95–105. doi: 10.1007/s10265-016-0871-7
Hu, L., He, H., Zhu, C., Peng, X., and Liu, S. (2016). Genome-wide identification and phylogenetic analysis of the chalcone synthase gene family in rice. J. Plant Res. 130, 1–11.
Hughes, A. R., Williams, S. L., Duarte, C. M., Heck, K. L., and Waycott, M. (2009). Associations of concern: declining seagrasses and threatened dependent species. Front. Ecol. Environ. 7, 242–246. doi: 10.1890/080041
Jiang, C., Kim, S. Y., and Suh, D. Y. (2008). Divergent evolution of the thiolase superfamily and chalcone synthase family. Mol. Phylogenet. Evol. 49, 691–701. doi: 10.1016/j.ympev.2008.09.002
Koduri, P. K., Gordon, G. S., Barker, E. I., Colpitts, C. C., Ashton, N. W., and Suh, D. Y. (2010). Genome-wide analysis of the chalcone synthase superfamily genes of Physcomitrella patens. Plant Mol. Biol. 72, 247–263. doi: 10.1007/s11103-009-9565-z
Kondo, S., Tomiyama, H., Rodyoung, A., Okawa, K., Ohara, H., Sugaya, S., et al. (2014). Abscisic acid metabolism and anthocyanin synthesis in grape skin are affected by light emitting diode (LED) irradiation at night. J. Plant Physiol. 171, 823–829. doi: 10.1016/j.jplph.2014.01.001
Kuo, Y. T., Chao, Y. T., Chen, W. C., Shih, M. C., and Chang, S. B. (2019). Segmental and tandem chromosome duplications led to divergent evolution of the chalcone synthase gene family in Phalaenopsis orchids. Annal. Bot. 123, 69–77. doi: 10.1093/aob/mcy136
Lau, O. S., and Deng, X. W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17, 584–593. doi: 10.1016/j.tplants.2012.05.004
Les, D. H., and Waycott, C. M. (1997). Phylogenetic studies in alismatidae, II: evolution of marine angiosperms (Seagrasses) and hydrophily. Syst. Bot. 22, 443–463. doi: 10.2307/2419820
Li, P., Ma, F., and Cheng, L. (2013). Primary and secondary metabolism in the sun-exposed peel and the shaded peel of apple fruit. Physiol. Plant 148, 9–24. doi: 10.1111/j.1399-3054.2012.01692.x
Liou, G., Chiang, Y. C., Wang, Y., and Weng, J. K. (2018). Mechanistic basis for the evolution of chalcone synthase catalytic cysteine reactivity in land plants. J. Biol. Chem. 293, 18601–18612. doi: 10.1074/jbc.RA118.005695
Olsen, J. L., Rouze, P., Verhelst, B., Lin, Y. C., Bayer, T., Collen, J., et al. (2016). The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530, 331–335. doi: 10.1038/nature16548
Pandith, S. A., Dhar, N., Rana, S., Bhat, W. W., Kushwaha, M., Gupta, A. P., et al. (2016). Functional promiscuity of two divergent paralogs of type III plant polyketide synthases. Plant Physiol. 171, 2599–2619. doi: 10.1104/pp.16.00003
Pandith, S. A., Ramazan, S., Khan, M. I., Reshi, Z. A., and Shah, M. A. (2019). Chalcone synthases (CHSs): the symbolic type III polyketide synthases. Planta 251:15. doi: 10.1007/s00425-019-03307-y
Shiokawa, K. I., Inagaki, Y., Morita, H., Hsu, T. J., and Noguchi, H. (2000). The functional expression of the CHS-D and CHS-E genes of the common morning glory (Ipomoea purpurea) in Escherichia coli and characterization of their gene products. Plant Biotechnol. 17, 203–210. doi: 10.5511/plantbiotechnology.17.203
Uleberg, E., Rohloff, J., Jaakola, L., Trost, K., Junttila, O., Haggman, H., et al. (2012). Effects of temperature and photoperiod on yield and chemical composition of northern and southern clones of bilberry (Vaccinium myrtillus L.). J. Agric. Food Chem. 60, 10406–10414. doi: 10.1021/jf302924m
Wang, H., Wang, W., Zhan, J., Yan, A., Sun, L., Zhang, G., et al. (2016). The accumulation and localization of chalcone synthase in grapevine (Vitis vinifera L.). Plant Physiol. Biochem. 106, 165–176. doi: 10.1016/j.plaphy.2016.04.042
Worm, B., Barbier, E. B., Beaumont, N., Duffy, J. E., Folke, C., Halpern, B. S., et al. (2006). Impacts of biodiversity loss on ocean ecosystem services. Science 314, 787–790. doi: 10.1126/science.1132294
Wu, X., Zhang, S., Liu, X., Shang, J., Zhang, A., Zhu, Z., et al. (2020). Chalcone synthase (CHS) family members analysis from eggplant (Solanum melongena L.) in the flavonoid biosynthetic pathway and expression patterns in response to heat stress. PloS One 15:e0226537. doi: 10.1371/journal.pone.0226537
Xu, Wl, Pei, Xl, Jing, Z. G., and Xiong, Zl (2015). Genome-wide identification and expression analysis of chalcone synthase gene in pepper. Genom. Appl. Biol. 34, 1747–1572.
Yamazaki, Y., Suh, D. Y., Sitthithaworn, W., Ishiguro, K., Kobayashi, Y., Shibuya, M., et al. (2001). Diverse chalcone synthase superfamily enzymes from the most primitive vascular plant, Psilotum nudum. Planta 214, 75–84. doi: 10.1007/s004250100586
Yang, J., Gu, H., and Yang, Z. (2004). Likelihood analysis of the chalcone synthase genes suggests the role of positive selection in morning glories (Ipomoea). J. Mol. Evol. 58, 54–63. doi: 10.1007/s00239-003-2525-3
Yu, H.-N., Wang, L., Sun, B., Gao, S., Cheng, A.-X., and Lou, H.-X. (2015). Functional characterization of a chalcone synthase from the liverwort Plagiochasma appendiculatum. Plant Cell Rep. 34, 233–245. doi: 10.1007/s00299-014-1702-8
Zhang, W., Wang, H. W., Wang, X. X., Xie, X. G., Siddikee, M. A., Xu, R. S., et al. (2016). Enhanced nodulation of peanut when co-inoculated with fungal endophyte Phomopsis liquidambari and bradyrhizobium. Plant Physiol. Biochem. 98, 1–11. doi: 10.1016/j.plaphy.2015.11.002
Zhang, X., Abrahan, C., Colquhoun, T. A., and Liu, C. J. (2017). A proteolytic regulator controlling chalcone synthase stability and flavonoid biosynthesis in arabidopsis. Plant Cell 29, 1157–1174. doi: 10.1105/tpc.16.00855
Keywords: Zostera marina, chalcone synthase, expression pattern, evolution, enzyme activity
Citation: Ma M, Zhong M, Zhang Q, Zhao W, Wang M and Luo C (2021) Phylogenetic Implications and Functional Disparity in the Chalcone synthase Gene Family of Common Seagrass Zostera marina. Front. Mar. Sci. 8:760902. doi: 10.3389/fmars.2021.760902
Received: 19 August 2021; Accepted: 14 October 2021;
Published: 02 November 2021.
Edited by:
Xiaotong Wang, Ludong University, ChinaReviewed by:
Zhanru Shao, Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaCopyright © 2021 Ma, Zhong, Zhang, Zhao, Wang and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quansheng Zhang, eXR1cXN6QGhvdG1haWwuY29t
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.