
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 12 October 2021
Sec. Marine Molecular Biology and Ecology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.753222
This article is part of the Research Topic Algal Symbiotic Relationships in Freshwater and Marine Environments View all 5 articles
 Young Mok Heo1
Young Mok Heo1 Seung-Yoon Oh2
Seung-Yoon Oh2 Kyeongwon Kim3
Kyeongwon Kim3 Sang-Il Han3
Sang-Il Han3 Sun Lul Kwon3
Sun Lul Kwon3 Yeonjae Yoo3
Yeonjae Yoo3 Dongjun Kim3
Dongjun Kim3 Jong Seong Khim4
Jong Seong Khim4 Seunghyun Kang1
Seunghyun Kang1 Hanbyul Lee5*
Hanbyul Lee5* Jae-Jin Kim3*
Jae-Jin Kim3*The whole genome and transcriptome analyses were performed for prediction of the ecological characteristics of Arthrinium and the genes involved in gentisyl alcohol biosynthesis. Whole genome sequences of A. koreanum KUC21332 and A. saccharicola KUC21221 were analyzed, and the genes involved in interspecies interaction, carbohydrate-active enzymes, and secondary metabolites were investigated. Three of the seven genes associated with interspecies interactions shared by four Arthrinium spp. were involved in pathogenesis. A. koreanum and A. saccharicola exhibit the enzyme profiles similar to those observed in plant pathogens and endophytes rather than saprobes. Furthermore, six of the seven metabolites of known clusters identified in the genomes of the four Arthrinium spp. are associated with plant virulence. These results indicate that Arthrinium spp. are potentially pathogenic to plants. Subsequently, different conditions for gentisyl alcohol production in A. koreanum were established, and mRNA extracted from cultures of each condition was subjected to RNA-Seq to analyze the differentially-expressed genes. The gentisyl alcohol biosynthetic pathway and related biosynthetic gene clusters were identified, and gentisyl alcohol biosynthesis was significantly downregulated in the mannitol-supplemented group where remarkably low antioxidant activity was observed. These results indicate that gentisyl alcohol production in algicolous Arthrinium spp. is influenced by mannitol. It was suggested that the algicolous Arthrinium spp. form a symbiotic relationship that provides antioxidants when the photosynthetic activity of brown algae decreases in exchange for receiving mannitol. This is the first study to analyze the lifestyle of marine algicolous Arthrinium spp. at the molecular level and suggests a symbiotic mechanism with brown algae. It also improves the understanding of fungal secondary metabolite production via identification of the gentisyl alcohol biosynthetic gene clusters in Arthrinium spp.
Arthrinium spp. are filamentous ascomycetes that have been isolated from various environments such as bamboo, sedges, soil, seaweeds, egg masses of sailfin sandfish, corals, and beach sand (Elissawy et al., 2017; Heo et al., 2018; Wang et al., 2018; Pintos et al., 2019). They have been reported to thrive as saprophytes, endophytes, and pathogens. Among saprophytes, few species (ex. A. phaeospermum) have been identified as wood-decaying fungi (Astiti and Suprapta, 2012). In contrast, many Arthrinium spp. (A. arundinis, A. garethjonesii, A. hydei, A. hyphopodii, A. neosubglobosa, A. phaeospermum, A. sacchari, A. saccharicola, etc.) have been isolated from the inner tissue of a variety of hosts including bamboos, candle bush (Senna alata), seeds of white leadtree (Leucaena leucocephala), yacon (Smallanthus sonchifolius), cock’s-foot (Dactylis glomerata), Malabar nut (Adhatoda vasica), Japanese sedge (Carex konomugi Ohwi), and brown algae (Sargassum fulvellum) (Sánchez Márquez et al., 2007; Khan et al., 2009; Maehara et al., 2010; Ramos et al., 2010; Shamsi et al., 2013; Shen et al., 2014; Lezcano et al., 2015; Dai et al., 2016; Pansanit and Pripdeevech, 2018; Suradkar and Hande, 2018). A. arundinis was isolated from a lichen (Cladonia sp.) (Wang et al., 2017). Moreover, the pathogenicity of at least four Arthrinium spp. (A. arundinis, A. phaeospermum, A. sacchari, and A. xenocordella) has been reported in various hosts, including wheat (damping-off), barley (kernel blight), bamboos (blight, brown culm streak, culm rot, and foot rot), rosemary (leaf spot), olive trees (leaf necrosis), aloe (flower malformation), and legumes (fruit blight) (Khan and Sullia, 1980; Suxuan et al., 1999; Mavragani et al., 2007; Li et al., 2013; Piccolo et al., 2013; Bagherabadi et al., 2014; Chen K. et al., 2014; Aiello et al., 2018; Jiang et al., 2018). Therefore, Arthrinium spp. may be endophytic or pathogenic rather than saprotrophic.
In general, symbiosis of endophytes is based on conferring competitive advantages to their hosts and there are various possibilities for the types of benefits provided by Arthrinium to the specific host (Rudgers et al., 2004). First, it has been observed that A. phaeospermum produces gibberellin, a plant growth-promoting compound, and promote mycorrhizal formation in pink rock-rose (Khan et al., 2009; Sabella et al., 2015). Additionally, A. arundinis accelerates strawberry seed germination by promoting their coat destruction (Guttridge et al., 1984). These Arthrinium spp. benefit host plant growth. On the contrary, at least five Arthrinium spp. (A. arundinis, A. aureum, A. phaeospermum, A. saccharicola, and A. serenense) exhibit antibiotic activity against over 39 microorganisms (Oka et al., 1993; Alfatafta et al., 1994; Vijayakumar et al., 1996; Aissaoui et al., 1999, 2001; Sato et al., 2000; Calvo et al., 2005; Miao et al., 2006; Bloor, 2008; Ramos et al., 2010; Heo et al., 2018; Pansanit and Pripdeevech, 2018; Hinterdobler and Schinnerl, 2019). Among them, the antibiotics apiosporamide, arthrichitin and arthrinic acid, and griseofulvin and terpestacin were isolated and identified from A. arundinis, A. phaeospermum, and an unidentified Arthrinium species, respectively (Oka et al., 1993; Alfatafta et al., 1994; Vijayakumar et al., 1996; Bloor, 2008; Elissawy et al., 2017). Thus, it is speculated that few Arthrinium spp. help the host defend against harmful microorganisms by producing antimicrobial compounds. Additionally, various roles of Arthrinium spp. in symbiosis have been suggested and verified. However, their relationship with brown algae and sponges, which are their major hosts in the marine environment, has not been extensively studied, perhaps due to the difficulty of conducting empirical experiments using marine organisms. Marine algicolous species have been proposed to help maintain the redox equilibrium in the host by producing antioxidants to remove excess reactive oxygen species (ROS), which are generated by the absorption of ultraviolet (UV) radiation by dissolved organic matter in seawater (Mopper and Kieber, 2000). A previous study has confirmed that most marine Arthrinium spp., including A. saccharicola and A. koreanum isolated from gulfweeds (Sargassum fulvellum) and egg masses of sailfin sandfish (Arctoscopus japonicus) that spawn on them, demonstrate high antioxidant activity and produce gentisyl alcohol (Heo et al., 2018). Therefore, the physiological and ecological characteristics of Arthrinium were investigated at the molecular level and genes related to gentisyl alcohol biosynthesis were investigated by analyzing the genome and transcriptome. This is the first comparative genome analysis of the genus Arthrinium.
Two algicolous Arthrinium species, A. saccharicola KUC21221 and A. koreanum KUC21332, were obtained from the Korea University Culture (KUC) collection. Arthrinium species found in the egg mass of A. japonicus are known to have originated from an adjacent brown alga, as A. japonicus spawns on S. fulvellum, one of the major endophytic hosts of Arthrinium spp. (Heo et al., 2018). For genome sequencing, they were cultured in 1-L Erlenmeyer flasks containing 500 mL potato dextrose broth for 7 days at 25°C in dark. Genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, United States) according to the manufacturer’s instructions with slight modifications (Kohler et al., 2011). To improve the accuracy of genome annotation, total RNA was extracted from the same cultures using the RNeasy Plant Mini Kit (Valencia, CA, United States) according to the manufacturer’s instructions. The DNA and RNA qualities were determined using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States) with a DNA 1000 chip.
Arthrinium koreanum KUC21332 was inoculated on malt extract agar (MEA, Bacto, Sparks, MD, United States). The fungus was cultured for 7 days, and three agar plugs with mycelium were used as the inocula. To assess the difference in gentisyl alcohol production according to the nitrogen source [peptone, sodium glutamate, KNO3, and (NH4)2SO4], the fungus was cultured in 250-mL Erlenmeyer flasks containing 50 mL medium (40 g glucose, 10 g nitrogen source, 0.5 g MgSO4, 0.5 g KH2PO4, and 0.5 g K2HPO4 in a liter of distilled water) for 10 days in dark. To determine the carbon source (glucose, sucrose, maltose, soluble starch, and mannitol) and to perform RNA-Seq, it was cultured in the medium (40 g carbon source, 10 g sodium glutamate, 0.5 g MgSO4, 0.5 g KH2PO4, and 0.5 g K2HPO4 in a liter of distilled water) for 3 days in dark. Total RNA was extracted in the same way as described above.
A DNA library with approximately 20-kb fragment sizes was constructed and the WGS was acquired using the PacBio Sequel platform at Macrogen Co., Ltd., (Seoul, South Korea). Meanwhile, a single molecule real-time (SMRT) library was constructed and sequenced with a single SMRT cell. The reads were trimmed, corrected, and filtered. Data on high-quality and error-corrected Illumina reads were used as input for the program Proovread v2.14.0 to correct the potential sequencing errors in the PacBio long reads (Hackl et al., 2014). Assembler Falcon was used for de novo assembly of the corrected PacBio reads (Chin et al., 2016). The assemblies were finalized and manually corrected after polishing using the paired-end Illumina reads and Pilon v1.21 (Walker et al., 2014).
To improve the accuracy of genome annotation, mRNA in the samples was sequenced. The RNA-Seq library was constructed and Illumina HiSeq 4000 sequencing was performed at Macrogen Co., Ltd., (Seoul, South Korea). Trimmed and corrected RNA-Seq reads were aligned to the reference genome using HISAT2 v2.1.0 (Kim et al., 2019). The genes were predicted using Augustus v3.3.3, BRAKER v2.1.5, GenMark-ES v4.61, and GlimmerM v2.5.1 (Majoros et al., 2003; Stanke et al., 2006, 2008; Ter-Hovhannisyan et al., 2008; Hoff et al., 2016, 2019). The final consensus gene model was constructed using all predictions using EVidenceModeler v1.1.1 (Haas et al., 2008). Repeats were masked using RepeatMasker v4.1.0 and RepeatModeler v2.01. Annotation completeness was evaluated using BUSCO v4.1.2 on fungi_odb10, ascomycota_odb10, and sordariomyceta_odb10 gene sets (Seppey et al., 2019). Orthologous protein families with the reference genome datasets were identified using Orthofinder v2.4.0 with default setting except for an inflation parameter (I = 2.5) (Emms and Kelly, 2019).
The proteins were annotated by predicting functional domains from Pfam using InterProScan (Hunter et al., 2009; El-Gebali et al., 2019). To further facilitate functional interpretation, proteins were aligned to the non-redundant database of NCBI2. Gene ontology (GO) terms were mapped using InterProScan v5.29-68.0 (Gene Ontology Consortium, 2012; Jones et al., 2014). Carbohydrate-active enzymes (CAZymes) were analyzed using dbCAN2 (hmmer) (Potter et al., 2018; Zhang et al., 2018). Gene clusters related to secondary metabolism were analyzed using antiSMASH Fungi v5.1.2, and secondary metabolite regions were identified using strictness “relaxed” (Blin et al., 2019).
To obtain fungal extracts, the fungal cultures were filtered with 0.45 μm syringe filters (Chromafil PE- 20/15 MS, Macherey-Nagel, Düren, Germany) and extracted with 50 mL of ethyl acetate for 24 h. The ethyl acetate layer was collected and dried at 35°C using a rotary evaporator, and the obtained extracts were stored at −18°C until use.
The 2,2-diphenyl-1-picrylhydrazyl (DPPH, Sigma-Aldrich Inc., St. Louis, MO, United States) was dissolved in 80% methanol at 150 μM. The 198 μL of DPPH solution and 22 μL of the fungal extracts (10 mg/mL DMSO) was mixed in each well in a 96-well plate. The plate was allowed to reach a steady state for 30 min at room temperature in dark. The absorbance was measured at 540 nm using a microplate reader (SunriseTM, Tecan Group Ltd., Port Melbourne, VIC, Australia).
To analyze differentially-expressed genes (DEGs), RNA-Seq libraries were constructed using the Illumina TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, United States) and were sequenced on the Illumina NovaSeq 6000 platform at Macrogen Co., Ltd., (Seoul, South Korea). Quality check of sequenced reads was performed using the FastQC v0.11.73.
The RNA-Seq data were analyzed using the “new Tuxedo” pipeline according to a published protocol (Pertea et al., 2016). Briefly, the raw reads were quality filtered using the program Trimmomatic v0.39 and aligned to each genomic DNA reference obtained in this study using HISAT2 v2.1.0 (Bolger et al., 2014; Kim et al., 2019). Data on the transcripts and their expression levels were assembled and estimated using the StringTie v1.3.4 (Kovaka et al., 2019). The DEG analysis was conducted using the DESeq2 (Love et al., 2014). To reduce systematic bias that could affect biological meaning in comparison between samples, the size factor was estimated using count data and normalized using median of ratios method. Between the experimental groups, there was no significant difference (p = 0.564) in the expression level of pyruvate kinase (GO:0004743), which catalyzes the final step of glycolysis, indicating that the difference in secondary metabolism was independent of the amount of available energy (Abdel Fattah et al., 2010; Zhang et al., 2016).
Statistical analyses were performed with R version 3.5.3 (R Development Core Team, 2013). Non-metric multidimensional scaling (NMDS) and permutational multivariate analysis of variance (adonis2) were performed using the package “vegan” (Oksanen et al., 2013).
In the present study, the genomes of two marine Arthrinium spp., A. koreanum KUC21332 and A. saccharicola KUC21221, isolated from the inner tissue of a brown alga Sargassum fulvellum and egg mass of A. japonicus, respectively, were analyzed. A. koreanum demonstrates a genome size of 48.75 Mbp (GC content: 52.09%), including 14,381 gene models, and A. saccharicola exhibits a 55.08-Mbp genome (GC content: 50.07%) with 14,773 models (Table 1). Both genomes are larger than those of A. phaeospermum AP-Z13 (48.45 Mbp) and A. arundinis NRRL 25634 (47.67 Mbp), with A. saccharicola genome being the largest among the four species. The number of gene models of both species is similar to A. phaeospermum (14,055 models), while that of A. arundinis NRRL 25634 is higher (16,992 models). The genome assembly completeness of both species ranges from 96.3–98.8%.
The four Arthrinium spp. shared 10,001 genes (Figure 1A). A. koreanum shared more genes (615) with A. arundinis compared to the genes shared with A. saccharicola (212) and A. phaeospermum (146). A. saccharicola shared more orthogroups (672) with A. phaeospermum compared to those shared with A. arundinis (208) and A. koreanum (212). Compositions of shared and specific genes were analyzed by gene ontology (GO) categories and have been illustrated in Figure 1B. The genes that only existed in A. koreanum (circle XII) were unidentified. Approximately half (mean 51.2%) of the genes were involved in biological process (BP), and 37.0 and 11.5% were involved in molecular function (MP) and cellular component (CC), respectively. Metabolic process (GO:0008152), cellular process (GO:0009987), and localization (GO:0051179) accounted for 48.6, 29.0, and 13.1% of the total genes in BP, respectively. Additionally, there were seven genes involved in interspecies interactions between organisms (GO:0044419), all of which were common in at least two Arthrinium spp. Four of them were shared by all four Arthrinium spp., of which two were involved in the defense response to gram-negative bacteria (GO:0050829), one was responsible for anti-bacterial defense response (GO:0042742), and the other were involved in pathogenesis (GO:0009405). One gene shared by A. arundinis, A. koreanum, and A. phaeospermum was involved in the DNA restriction-modification system (GO:0009307). One gene shared by A. arundinis, A. saccharicola, and A. phaeospermum, and the other shared by A. koreanum and A. saccharicola, were involved in pathogenesis (GO:0009405). In the case of CC, cellular anatomical entity (GO:0110165), protein-containing complex (GO:0032991), and intercellular (GO:0005622) accounted for 84.6, 7.8, and 7.2% of the total, respectively. In the case of MF, catalytic activity (GO:0003824) and binding (GO:0005488) accounted for 46.9 and 40.1% of the total genes, respectively, followed by transporter activity (GO:0005215) and molecular function regulator (GO:0098772), accounting for 7.5 and 3.6%, respectively.
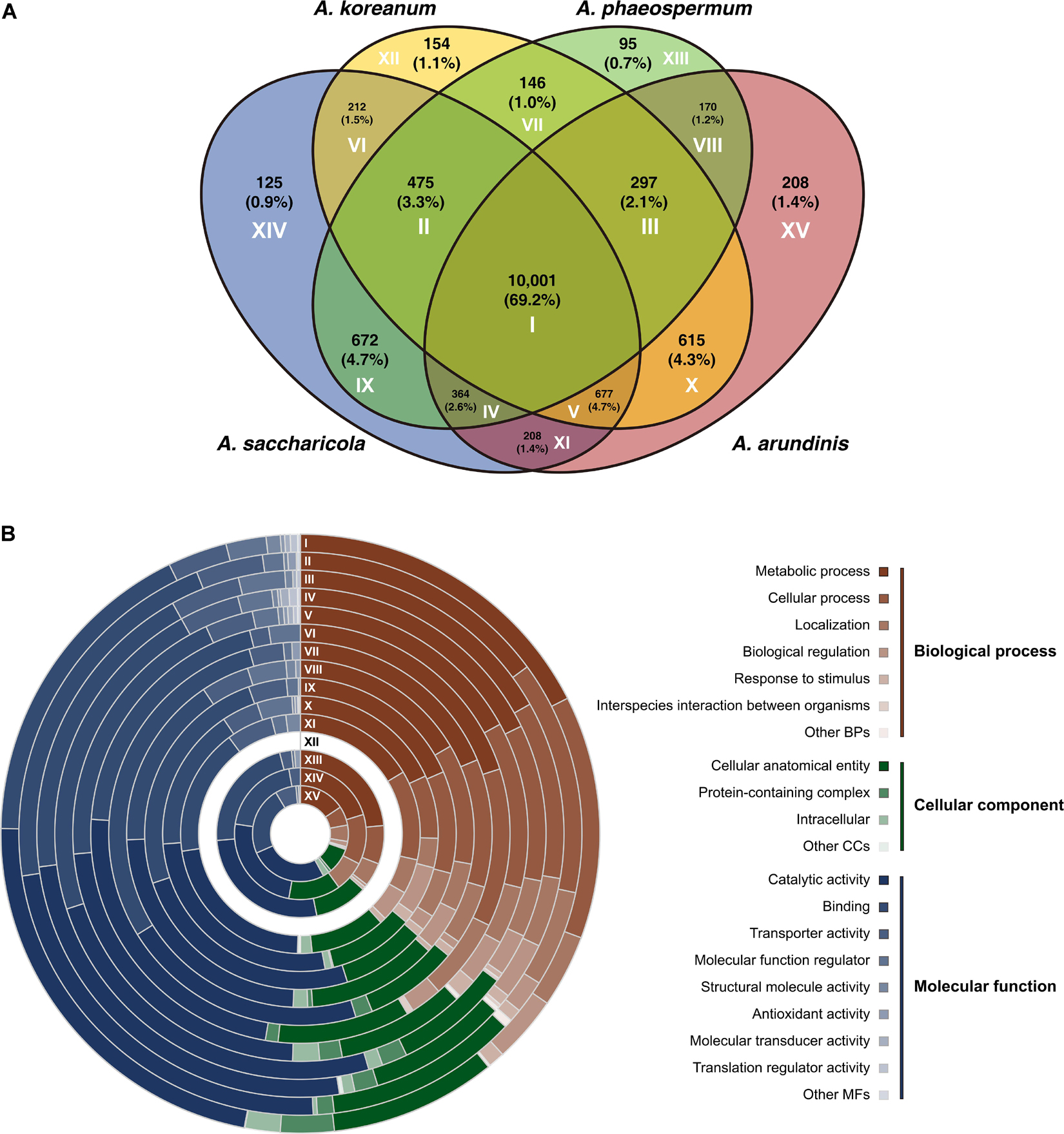
Figure 1. Genome comparison between Arthrinium koreanum, A. saccharicola, A. phaeospermum, and A. arundinis. (A) Venn diagram showing the number of shared and specific gene orthologs. (B) Compositions of gene families according to gene ontology categories. Circles I-XV correspond to areas with same Roman numerals in panel (A).
Additionally, the genes encoding CAZymes and plant cell wall degrading enzymes (PCWDEs) and compare to those of other ascomycetes exhibiting various lifestyles. Five, eight, and twelve species were selected for dark septate endophytes (DSEs), plant pathogens, and saprobes, respectively, by referring to the Mycocosm group of the Joint Genome Institute and previous studies (Supplementary Table 1; Galagan et al., 2003; Dean et al., 2005; Güldener et al., 2006; Hane et al., 2007; Van Den Berg et al., 2008; Ellwood et al., 2010; Andersen et al., 2011; Rouxel et al., 2011; Gianoulis et al., 2012; Ohm et al., 2012; O’Connell et al., 2012; Grigoriev et al., 2013; Koike et al., 2013; Traeger et al., 2013; Nordberg et al., 2014; Xu et al., 2015; David et al., 2016; Jourdier et al., 2017; Knapp et al., 2018). DSE is a paraphyletic group (Sordariomycetes, Pezizomycetes, Dothideomycetes, Leotiomycetes, Eurotiomycetes, etc.) of endophytic fungi belonging to Ascomycota, and was selected because it represented endophytic ascomycetes. The sequence data of A. arundinis NRRL 25634, Entoleuca mammata CFL468, Pestalotiopsis sp. NC0098, and Truncatella angustata MPI-SDFR-AT-0073 were produced by the United States Department of Energy Joint Genome Institute4 in collaboration with the user community.
The Arthrinium spp. had a total of 601-626 CAZymes, of which 307–323, 96–104, 9–11, 62–68, 11–14, and 113–117 were associated with glycoside hydrolase (GH), glycosyltransferase (GT), polysaccharide lyase (PL), carbohydrate esterase (CE), carbohydrate-binding module (CBM), and auxiliary activity (AA), respectively. The average number of CAZyme by lifestyle was 713, 553, and 397 for DSE, plant pathogen, and saprobe, respectively. Arthrinium spp. had 127–134 of PCWDEs, while DSE, plant pathogen, and saprobe had an average of 168, 130, and 65 PCWDEs, respectively. In the NMDS plot generated using the CAZyme profile of these species, Arthrinium-DSE-plant pathogen was clustered together, and the saprobe was widely distributed at a distance from this cluster. Based on the permutational multivariate analysis of variance, significant dissimilarities were observed between clusters according to lifestyles, and A. koreanum and A. saccharicola, whose lifestyles have not been identified, showed significant dissimilarity only with the saprobe (Figure 2). The NMDS plot generated using the PCWDE profile showed similar results, except that the dissimilarity between the DSE and the plant pathogen cluster was not significant. Further, among PCWDEs, GH53, GH127, PL4, and CE12 were significant predictors in the dimension 2 direction, and the representative enzymes of these CAZyme families are arabinogalactan endo-β-1,4-galactanase (EC 3.2.1.89), non-reducing end β-L-arabinofuranosidase (EC 3.2.1.185), rhamnogalacturonan endolyase (EC 4.2.2.23), and acetylxylan esterase (EC 3.1.1.72), respectively.
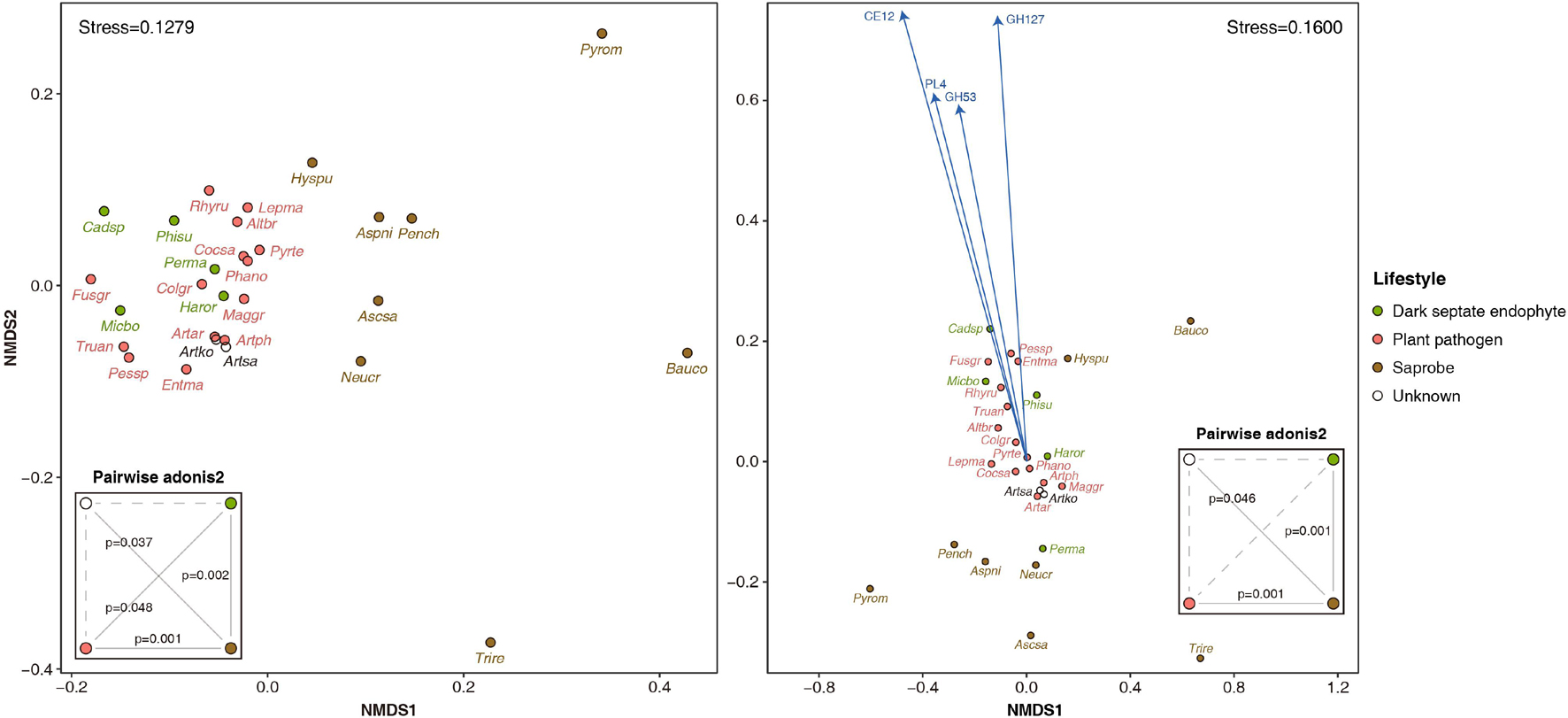
Figure 2. Non-metric multidimensional scaling plot generated using profiles of carbohydrate active enzymes (CAZymes, left) and plant cell wall degrading enzymes (PCWDEs, right) of four Arthrinium spp. and other 25 ascomycetes with different lifestyles. Fitted variables are scaled by Pearson correlation coefficient (p < 0.05). Dissimilarities between CAZyme and PCWDE profiles by lifestyle based on permutational multivariate analysis of variance were summarized in the small boxes titled pairwise adonis2. The solid and dotted lines in the pairwise adonis2 boxes indicate significant (p < 0.05) and insignificant (p > 0.05), respectively. Altbr, Alternaria brassicicola; Artar, Arthrinium arundinis; Artko, A. koreanum; Artph, A. phaeospermum; Artsa, A. saccharicola; Ascsa, Ascocoryne sarcoides; Aspni, Aspergillus niger; Bauco, Baudoinia compniacensis; Cadsp, Cadophora sp.; Cocsa, Cochliobolus sativus; Colgr, Colletotrichum graminicola; Entma, Entoleuca mammata; Fusgr, Fusarium graminearum; Haror, Harpophora oryzae; Hyspu, Hysterium pulicare; Lepma, Leptosphaeria maculans; Maggr, Magnaporthe grisea; Micbo, Microdochium bolleyi; Neucr, Neurospora crassa; Pench, Penicillium chrysogenum; Perma, Periconia macrospinosa; Pessp, Pestalotiopsis sp.; Phano, Phaeosphaeria nodorum; Phisu, Phialocephala subalpina; Pyrte, Pyrenophora teres; Pyrom, Pyronema omphalodes; Rhyru, Rhytidhysteron rufulum; Trire, Trichoderma reesei; Truan, Truncatella angustata.
The analysis of biosynthetic gene clusters (BGCs) of the four Arthrinium spp. showed that the highest number of BGCs was found in A. arundinis (84), followed by A. koreanum (76), and A. phaeospermum (69) and A. saccharicola (69) (Supplementary Figure 1). The secondary metabolite backbone genes consisted of 23–28 type I polyketide synthase (T1PKS) genes, 16–21 non-ribosomal peptide synthetase (NRPS) genes, 13–18 terpene genes, 0–3 indole genes, and 1–2 type III polyketide synthase (T3PKS) genes. Six BGCs were annotated with 100% similarity in the four Arthrinium spp. NRPS and two T1PKS BGCs, known to produce dimethylcoprogen, alternapyrone and ACR toxin I, respectively, were observed in all four species, and another two T1PKS BGCs, known to produce (R)-mellein and alternariol, were commonly found in A. koreanum and A. arundinis (Fujii et al., 2005; Izumi et al., 2012; Chen et al., 2013; Chooi et al., 2015a, b). Additionally, a T1PKS BGC, known to produce pyranonigrin E, was observed in A. phaeospermum (Andersen et al., 2011).
Arthrinium koreanum KUC21332 and A. saccharicola KUC21221, the gentisyl alcohol-producing species, commonly harbor 6-MSA BGCs (42.3 and 42.5 kb, respectively), which show 40% similarity with patulin BGC, while A. phaeospermum AP-Z13 does not harbor one (Figure 3). The nine genes that they commonly harbor in their BGCs are GsaA-GsaI. GsaH is orthologous to PatK, the backbone gene encoding 6-methylsalisylic acid (6MSA) synthase that catalyzes the first step of patulin biosynthesis (conversion of acetyl-CoA and malonyl-CoA to 6MSA); GsaE is orthologous to PatG encoding 6MSA decarboxylase that catalyzes the second step (conversion of 6MSA to m-cresol); GsaD is orthologous to PatH encoding m-cresol methyl hydroxylase that catalyzes the third step (conversion of m-cresol to m-hydroxybenzyl alcohol); GsaF is orthologous to PatI encoding m-hydroxybenzyl alcohol hydroxylase that catalyzes the fourth step (conversion of m-hydroxybenzyl alcohol to gentisyl alcohol); GsaA is orthologous to PatL encoding C6 transcription factor that activates gene expression; GsaB and GsaG are orthologous to PatO and PatJ encoding isoamyl alcohol oxidase and a putative dioxygenase, respectively, that catalyze the sixth step (conversion of gentisaldehyde to isoepoxydon); and GsaC and GsaI have been predicted to encode major facilitator superfamily (MFS) transporter (77.98% identical to EKG18982.1) and aldehyde reductase (73.39% identical to KAF6798856.1), respectively (Li et al., 2019). A. arundinis NRRL 25634 also harbors a 6-MSA BGC (44.9 kb) similar to patulin BGC (46% similarity), with two additional genes named GsaJ and GsaK, orthologous to PatM and PatC encoding ATP-binding cassette (ABC) transporter and MFS transporter, respectively; PatM and PatC were proposed to be involved in the extracellular patulin secretion (Li et al., 2019). In the other two Arthrinium spp., GsaC may replace the function of PatC, and the function of GsaI in the BGC should be further studied.
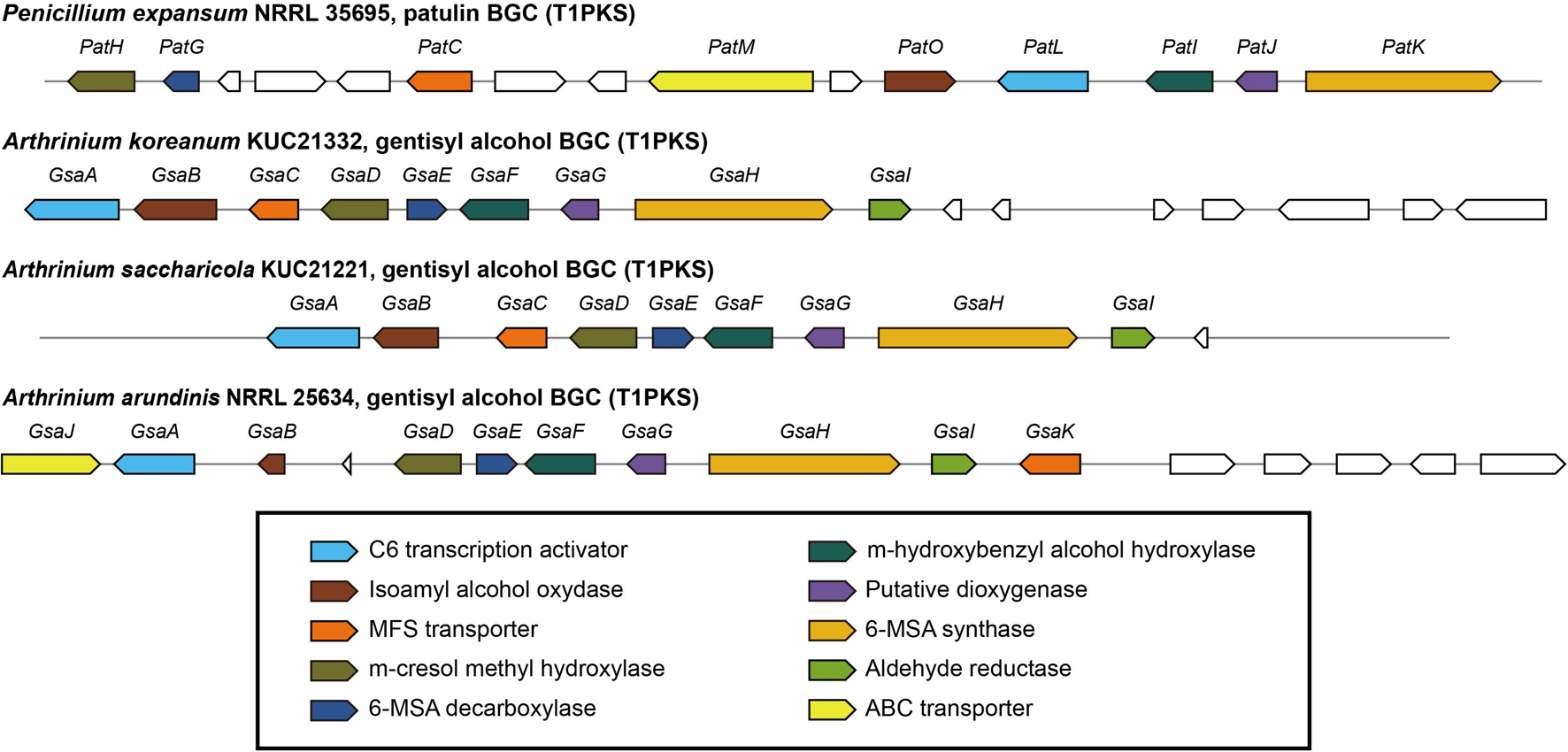
Figure 3. Predicted gentisyl alcohol biosynthetic gene clusters (BGCs) of Arthrinium spp. matched to the patulin BGC of Penicillium expansum. Transcription direction is indicated by arrowheads.
By measuring the DPPH radical-scavenging activity of A. koreanum KUC21332 extract using different organic (peptone and sodium glutamate) and inorganic nitrogen compounds (KNO3 and (NH4)2SO4) as a nitrogen source, the highest activity (90.6–98.3%) was observed with sodium glutamate (Figure 4A). The DPPH radical-scavenging activity was also determined using a monosaccharide (glucose), disaccharides (sucrose and maltose), a polysaccharide (soluble starch), and a sugar alcohol (mannitol) as the carbon source and sodium glutamate as the nitrogen source. As the activities rapidly increased within 2 days in the nitrogen source test, the carbon source test was conducted using 3-day cultures. The highest activity (79.2%) was observed with glucose, which was approximately 25 times higher than the lowest activity (3.2%) observed with mannitol (Figure 4B). To verify whether the Gsa BGC was responsible for gentisyl alcohol production, DEG analysis was performed with the transcriptomes from the mannitol- and glucose-supplemented cultures. The expression of GsaA, GsaC, GsaG, and GsaH was found to be significantly lower in the mannitol-supplemented group compared to that in the glucose-supplemented group. The fold-change values of the four genes in the glucose-/mannitol-supplemented group were 2.03, 27.28, 8.53, and 5.10 (q < 0.001 for all; the Wald test with Benjamini–Hochberg correction), respectively.
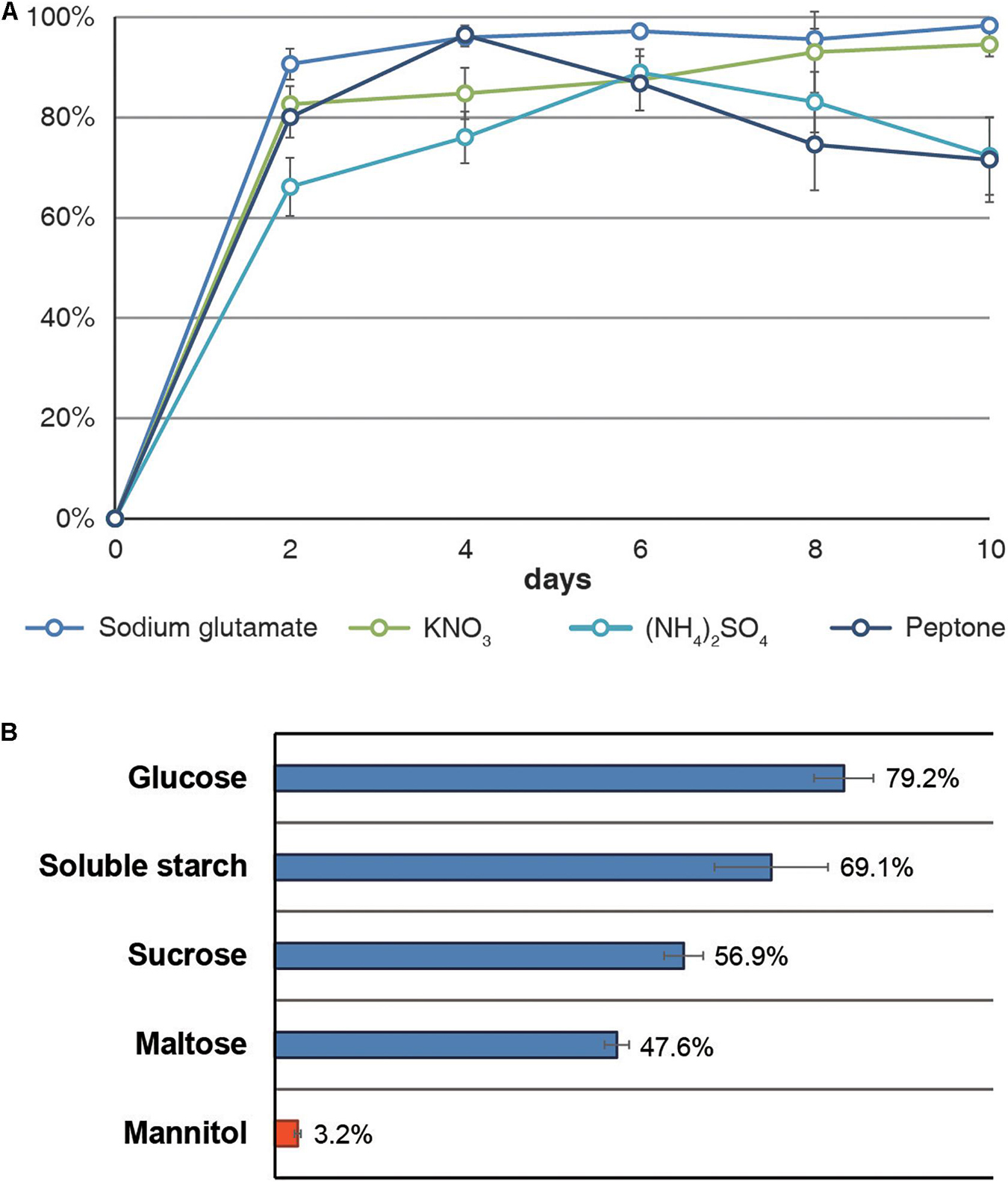
Figure 4. DPPH radical-scavenging activities of Arthrinium koreanum KUC21332 culture extracts. (A) Activities by nitrogen source with glucose as a carbon source. (B) Activities by carbon source after being cultured for 3 days with sodium glutamate as a nitrogen source.
To understand the lifestyle of Arthrinium spp. at the molecular level, the full-length genomes of A. koreanum KUC21332 and A. saccharicola KUC21221 were analyzed and those of A. phaeospermum AP-Z13 and A. arundinis NRRL 25634 were comparatively analyzed. Since few species, including A. arundinis and A. phaeospermum, are plant pathogens and many species have been isolated from the internal tissues of plants or marine algae, genes related to interspecies interaction, CAZymes, and secondary metabolites were analyzed, focusing on pathogenicity and endogenous symbiosis. The seven genes associated with interspecies interaction and shared by the four Arthrinium spp. were involved in pathogenesis and defense processes against bacteria and invading foreign DNA via other biological agents. This suggests that A. arundinis and A. phaeospermum, which are known to be pathogenic to various plants, and A. koreanum and A. saccharicola, can establish abnormal conditions in other organisms. In the NMDS plot generated with CAZyme profiles of other ascomycetes with different lifestyles, A. arundinis and A. phaeospermum were clustered together with plant pathogens, and A. koreanum and A. saccharicola were distributed close to them (Figure 2). The overlap of DSEs and plant pathogens indicates that pathogenesis is an unbalanced symbiosis of endophytes (Kogel et al., 2006). According to the results of permutational multivariate analysis of variance, A. koreanum and A. saccharicola showed CAZyme profiles similar to plant pathogens and DSEs rather than saprobes. Additionally, the total number of CAZymes and PCWDEs in Arthrinium spp. was similar to that of plant pathogens, followed by that of DSEs. These results indicate that their lifestyles are on the mutualism-parasitism-continuum. Furthermore, metabolites of known clusters identified in the genomes of four Arthrinium spp. are associated with virulence, except for pyranonigrin E, which is found only in A. phaeospermum. A previous study has suggested that alternapyrone and dimethylcoprogen play roles in plant pathogenesis (Gluck-Thaler et al., 2020). Dimethylcoprogen, a fungal extracellular siderophore, was reported to be responsible for the pathogenicity of Alternaria alternata to citrus (Chen L. H. et al., 2014). ACR toxin I is a toxin of Alternaria alternata, which is essential for its pathogenicity, and alternariol is a colonization and virulence factor of this fungus during plant infection (Izumi et al., 2012; Wenderoth et al., 2019). (R)-mellein has been identified as a non-host-specific phytotoxin of various plant pathogens, such as Parastagonospora nodorum, Botryosphaeria obtusa, Phoma tracheiphila, Neofusicoccum parvum, and Sphaeropsis sapinea (Chooi et al., 2015a). These results indicate that Arthrinium spp., including A. koreanum and A. saccharicola, are potential plant pathogens. Since most sources of Arthrinium spp. did not show disease symptoms, they were presumed to be opportunistic pathogens that were pathogenic depending on the host condition.
Meanwhile, Arthrinium spp. are reportedly not pathogenic in brown algae, an endogenous host in the marine environment, and no abnormal or detrimental state has been reported in brown algae when Arthrinium spp. were isolated from their internal tissues. Instead, most algicolous Arthrinium strains showed high antioxidant activity, and it was hypothesized that antioxidant production was a strategy of marine Arthrinium spp. in endophytic symbiosis with brown algae (Heo et al., 2018). The antioxidant assay and DEG analysis based on different nutrient sources, including glutamic acid and mannitol, support this hypothesis. Glutamic acid is an accessible nitrogen source for algicolous Arthrinium spp., which is the most abundant amino acid accounting for about 13% of the total protein content of Sargassum spp., the most common endophytic host of Arthrinium spp., and mannitol is the main carbon storage material in brown algae, accounting for 20–30% of the dry weight of brown algae (Reed et al., 1985; Peng et al., 2013). According to the antioxidant assay results, the activity was the highest in the sodium glutamate-supplemented group, but all other groups showed high activity as well (Figure 4A). On the contrary, the activity was markedly different for the carbon source. The activity was barely observed with mannitol as a carbon source and sodium glutamate as a nitrogen source, whereas glucose-, starch-, sucrose-, and maltose-supplemented groups showed high activity (47.6–79.2%) (Figure 4B). Through the DEG analysis, it was confirmed that the low antioxidant activity of the mannitol-supplemented group was due to the downregulation of gentisyl alcohol biosynthesis. These results suggest that gentisyl alcohol production in algicolous Arthrinium spp., such as A. koreanum, is primarily influenced by mannitol rather than glutamic acid. In marine environments, mannitol functions as an osmoprotectant and acts as an antioxidant that prevents peroxidation of cellular components by ROS, which is generated by the absorption of UV rays by dissolved organic matter in seawater (Groisillier et al., 2014). Since the residual mannitol amount in brown algae reflects the photosynthetic activity in brown algae, it is expected that algicolous Arthrinium spp. produce gentisyl alcohol to compensate for the reduced antioxidant activity when the mannitol content decreases at night. Additionally, deterioration of the cellular function or vitality of brown algae due to aging can also cause a situation where algicolous fungi are not sufficiently supplemented with mannitol. Likewise, gentisyl alcohol produced in response to mannitol deficiency in these fungi can help regulate the redox equilibrium of brown algae, when the ability of the algal host to control the redox equilibrium is poor. It is suggested that algicolous Arthrinium spp. establish a symbiotic relationship that enhances the viability of brown algae by providing antioxidants when the photosynthetic activity decreases in exchange for receiving mannitol.
Gentisyl alcohol biosynthetic pathway in Arthrinium spp. was also identified and the related BGCs were reported. The known gentisyl alcohol biosynthetic pathways were reportedly a part of the aculin A and patulin biosynthetic pathways of Aspergillus aculeatus and Penicillium expansum, respectively (Petersen et al., 2015; Li et al., 2019). Both pathways share the reactions of conversion of acetyl-CoA and malonyl-CoA to gentisyl alcohol, an intermediate product. It was expected that the gentisyl alcohol biosynthetic pathway of Arthrinium spp. might consist of genes orthologous to the genes in aculin A and patulin biosynthetic pathways. Indeed, 6-MSA BGCs similar to P. expansum patulin BGCs were observed. Considering that A. koreanum KUC21332 and A. saccharicola KUC21221 produced gentisyl alcohol as a major secondary metabolite instead of patulin, it was expected that the biosynthetic genes responsible for the later steps of patulin biosynthesis were absent. As expected, orthologs of the other six genes (PatA, PatB, PatD, PatE, PatF, and PatN) of patulin BGC were not present in the 6-MSA BGCs of the Arthrinium spp.; PatN, PatF, PatD, and PatE encode isoepoxydon dehydrogenase, neopatulin synthase, alcohol dehydrogenase, and glucose-methanol-choline oxidoreductase (patulin synthase) that catalyze the seventh to final reactions (from isoepoxydon to patulin) (Li et al., 2019). This confirms that enzyme-catalyzed steps after production of gentisyl alcohol in the patulinw biosynthetic pathway of Penicillium are absent in the suggested gentisyl alcohol biosynthetic pathway of Arthrinium (Figure 5). According to the DEG analysis results, GsaA, GsaC, and GsaH expression levels were downregulated. GsaG was neglected as its substrate, gentisaldehyde, cannot be produced because of the absence of responsible genes. GsaA expression downregulation in the mannitol-supplemented group was suspected to be responsible for the suppression of gentisyl alcohol biosynthesis, since GsaA acts as a specific transcription factor in this pathway. GsaH and GsaC expression downregulation decreases 6-MSA and MFS transporter production, respectively, thereby reducing the production and extracellular secretion of gentisyl alcohol. This result is consistent with the results of the antioxidant assay, thus demonstrating that the 6-MSA BGC is responsible for gentisyl alcohol biosynthesis. The expression levels of other genes in the BGC were not significantly different, indicating that GsaC and GsaH were targeted in gentisyl alcohol biosynthesis regulation.
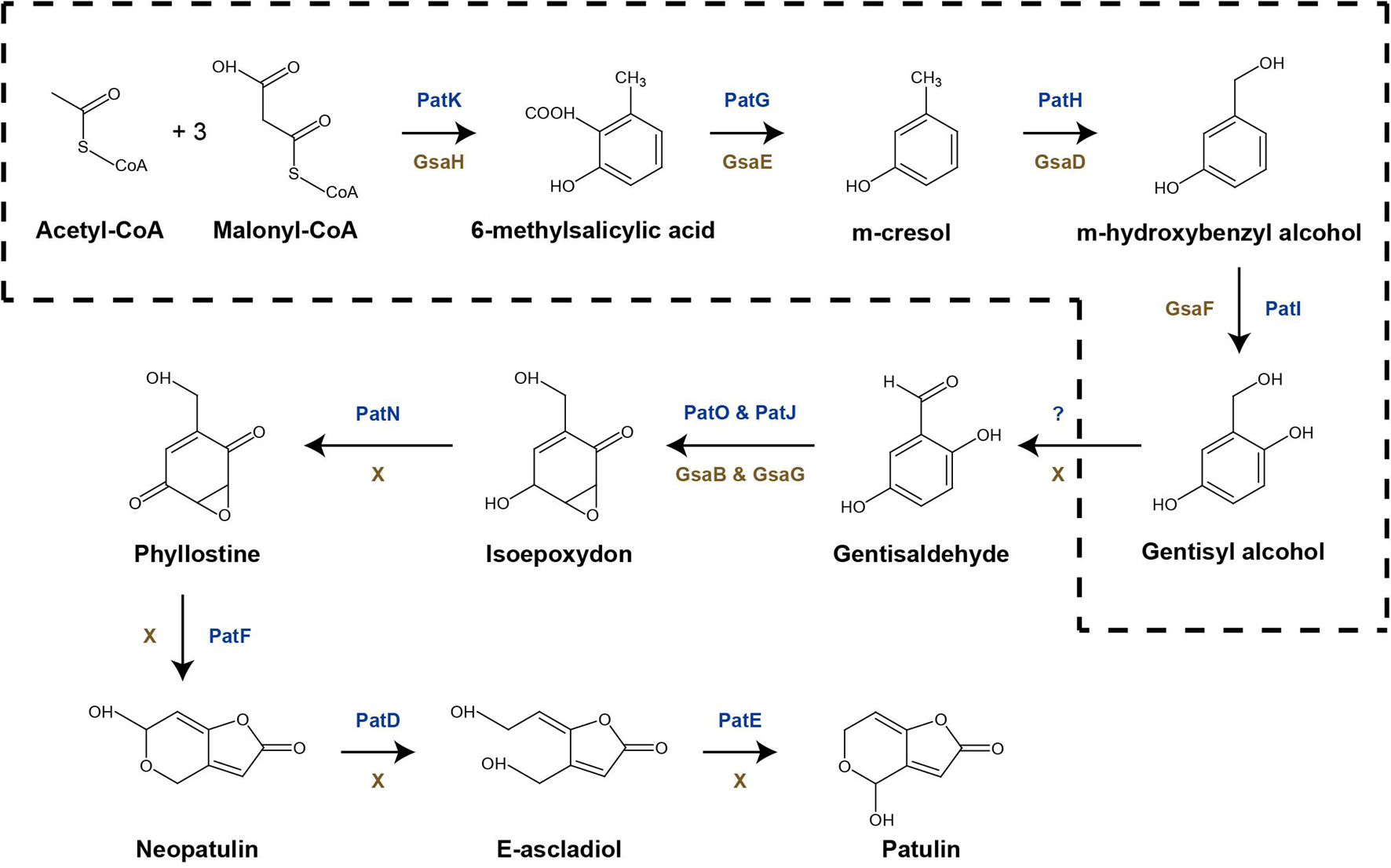
Figure 5. Proposed scheme of gentisyl alcohol biosynthetic pathway in Arthrinium spp. (dotted box) expressed in the patulin biosynthetic pathway.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA692538.
YH and HL: conceptualization. YH, S-YO, and S-IH: methodology. S-YO and S-IH: validation. YH: formal analysis and writing—original draft preparation. KK, SKw, YY, and DK: investigation. J-JK: project administration and resources. S-YO and JK: writing—review and editing. YH and S-YO: visualization. HL, SKa, and J-JK: supervision. JK and J-JK: funding acquisition. All authors have read and agreed to the published version of the manuscript.
This research was supported by National Research Foundation of Korea (NRF) funded by the Korean government (MSIT) (2021R1A2C1011894), the Ministry of Oceans and Fisheries of Korea (MOF) (20170325 and 20210427), and a Korea University Grant.
YH and SKa are employed by COSMAX BTI.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.753222/full#supplementary-material
Abdel Fattah, M., Abdel Ghany, E., Adel, A., Mosallam, D., and Kamal, S. (2010). Glucose-6-phosphate dehydrogenase and red cell pyruvate kinase deficiency in neonatal jaundice cases in egypt. Pediatr. Hematol. Oncol. 27, 262–271.
Aiello, D., Gulisano, S., Gusella, G., Polizzi, G., and Guarnaccia, V. (2018). First report of fruit blight caused by Arthrinium xenocordella on Pistacia vera in Italy. Plant Dis. 102:1853. doi: 10.1094/pdis-02-18-0290-pdn
Aissaoui, H., Agut, M., and Calvo, M. (2001). Effect of the raw extracts of Arthrinium strains (Hyphomycetes, Dematiaceae) on the growth of some deleterious fungi in poultry feed. Microbios 105:103.
Aissaoui, H., Agut, M., and Torras, C. (1999). Effect of the raw extract of Arthrinium strains (Hyphomycetes, Dematiaceae) on the growth of pathogenic bacteria in poultry feed. Microbios 100, 109–116.
Alfatafta, A. A., Gloer, J. B., Scott, J. A., and Malloch, D. (1994). Apiosporamide, a new antifungal agent from the coprophilous fungus Apiospora montagnei. J. Nat. Prod. 57, 1696–1702. doi: 10.1021/np50114a012
Andersen, M. R., Salazar, M. P., Schaap, P. J., Van De Vondervoort, P. J., Culley, D., Thykaer, J., et al. (2011). Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 21, 885–897. doi: 10.1101/gr.112169.110
Astiti, N. P. A., and Suprapta, D. N. (2012). Antifungal activity of teak (Tectona grandis L.f) leaf extract against Arthrinium phaeospermum (Corda) M.B. Ellis, the cause of wood decay on Albizia falcataria (L.) Fosberg. J. ISSAAS 18, 62–69.
Bagherabadi, S., Zafari, D., and Anvar, F. G. (2014). First report of leaf spot caused by Arthrinium arundinis on rosemary in Iran. J. Plant Pathol. 96:4.
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Bloor, S. (2008). Arthrinic acid, a novel antifungal polyhydroxyacid from Arthrinium phaeospermum. J. Antibiot. 61, 515–517.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Calvo, M., Arosemena, L., Agut, M., Adelantado, C., and Aissaoui, H. (2005). Biological control of bayoud in the palm trees by strains of Arthrinium. J. Biol. Sci. 5, 236–238. doi: 10.3923/jbs.2005.236.238
Chen, K., Wu, X.-Q., Huang, M.-X., and Han, Y.-Y. (2014). First report of brown culm streak of Phyllostachys praecox caused by Arthrinium arundinis in Nanjing, China. Plant Dis. 98, 1274–1274. doi: 10.1094/pdis-02-14-0165-pdn
Chen, L. H., Lin, C. H., and Chung, K. R. (2013). A nonribosomal peptide synthetase mediates siderophore production and virulence in the citrus fungal pathogen Alternaria alternata. Mol. Plant Pathol. 14, 497–505. doi: 10.1111/mpp.12021
Chen, L.-H., Yang, S. L., and Chung, K.-R. (2014). Resistance to oxidative stress via regulating siderophore-mediated iron acquisition by the citrus fungal pathogen Alternaria alternata. Microbiology 160, 970–979. doi: 10.1099/mic.0.076182-0
Chin, C.-S., Peluso, P., Sedlazeck, F. J., Nattestad, M., Concepcion, G. T., Clum, A., et al. (2016). Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054. doi: 10.1038/nmeth.4035
Chooi, Y.-H., Krill, C., Barrow, R. A., Chen, S., Trengove, R., Oliver, R. P., et al. (2015a). An in planta-expressed polyketide synthase produces (R)-mellein in the wheat pathogen Parastagonospora nodorum. Appl. Environ. Microbiol. 81, 177–186. doi: 10.1128/aem.02745-14
Chooi, Y.-H., Muria-Gonzalez, M. J., Mead, O. L., and Solomon, P. S. (2015b). SnPKS19 encodes the polyketide synthase for alternariol mycotoxin biosynthesis in the wheat pathogen Parastagonospora nodorum. Appl. Environ. Microbiol. 81, 5309–5317.
Dai, D.-Q., Jiang, H.-B., Tang, L.-Z., and Bhat, D. J. (2016). Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere 7, 1332–1345. doi: 10.5943/mycosphere/7/9/7
David, A. S., Haridas, S., Labutti, K., Lim, J., Lipzen, A., Wang, M., et al. (2016). Draft genome sequence of Microdochium bolleyi, a dark septate fungal endophyte of beach grass. Genome Announc. 4, e270–e216.
Dean, R. A., Talbot, N. J., Ebbole, D. J., Farman, M. L., Mitchell, T. K., Orbach, M. J., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986.
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432.
Elissawy, A. M., Ebada, S. S., Ashour, M. L., Øzkaya, F. C., Ebrahim, W., Singab, A. B., et al. (2017). Spiroarthrinols A and B, two novel meroterpenoids isolated from the sponge-derived fungus Arthrinium sp. Phytochem. Lett. 20, 246–251. doi: 10.1016/j.phytol.2017.05.008
Ellwood, S. R., Liu, Z., Syme, R. A., Lai, Z., Hane, J. K., Keiper, F., et al. (2010). A first genome assembly of the barley fungal pathogen Pyrenophora teres f. teres. Genome Biol. 11:R109.
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 1–14. doi: 10.1186/s13059-019-1832-y
Fujii, I., Yoshida, N., Shimomaki, S., Oikawa, H., and Ebizuka, Y. (2005). An iterative type I polyketide synthase PKSN catalyzes synthesis of the decaketide alternapyrone with regio-specific octa-methylation. Chem. Biol. 12, 1301–1309. doi: 10.1016/j.chembiol.2005.09.015
Galagan, J. E., Calvo, S. E., Borkovich, K. A., Selker, E. U., Read, N. D., Jaffe, D., et al. (2003). The genome sequence of the filamentous fungus Neurospora crassa. Nature 422, 859–868.
Gene Ontology Consortium (2012). Gene ontology annotations and resources. Nucleic Acids Res. 41, D530–D535.
Gianoulis, T. A., Griffin, M. A., Spakowicz, D. J., Dunican, B. F., Sboner, A., Sismour, A. M., et al. (2012). Genomic analysis of the hydrocarbon-producing, cellulolytic, endophytic fungus Ascocoryne sarcoides. PLoS Genet. 8:e1002558. doi: 10.1371/journal.pgen.1002558
Gluck-Thaler, E., Haridas, S., Binder, M., Grigoriev, I. V., Crous, P. W., Spatafora, J. W., et al. (2020). The architecture of metabolism maximizes biosynthetic diversity in the largest class of fungi. Mol. Biol. Evol. 37, 2838–2856. doi: 10.1093/molbev/msaa122
Grigoriev, I. V., Nikitin, R., Haridas, S., Kuo, A., Ohm, R., Otillar, R., et al. (2013). MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42, D699–D704.
Groisillier, A., Shao, Z., Michel, G., Goulitquer, S., Bonin, P., Krahulec, S., et al. (2014). Mannitol metabolism in brown algae involves a new phosphatase family. J. Exp. Bot. 65, 559–570. doi: 10.1093/jxb/ert405
Güldener, U., Mannhaupt, G., Münsterkötter, M., Haase, D., Oesterheld, M., Stümpflen, V., et al. (2006). FGDB: a comprehensive fungal genome resource on the plant pathogen Fusarium graminearum. Nucleic Acids Res. 34, D456–D458.
Guttridge, C., Woodley, S. E., and Hunter, T. (1984). Accelerating strawberry seed germination by fungal infection. Anna. Bot. 54, 223–230. doi: 10.1093/oxfordjournals.aob.a086786
Haas, B. J., Salzberg, S. L., Zhu, W., Pertea, M., Allen, J. E., Orvis, J., et al. (2008). Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9:R7.
Hackl, T., Hedrich, R., Schultz, J., and Förster, F. (2014). Proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30, 3004–3011. doi: 10.1093/bioinformatics/btu392
Hane, J. K., Lowe, R. G., Solomon, P. S., Tan, K.-C., Schoch, C. L., Spatafora, J. W., et al. (2007). Dothideomycete–plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum. Plant Cell 19, 3347–3368. doi: 10.1105/tpc.107.052829
Heo, Y. M., Kim, K., Ryu, S. M., Kwon, S. L., Park, M. Y., Kang, J. E., et al. (2018). Diversity and ecology of marine algicolous Arthrinium species as a source of bioactive natural products. Mar. Drugs 16:508. doi: 10.3390/md16120508
Hinterdobler, W., and Schinnerl, J. (2019). Chemical diversity and richness of fungal endophytes from Costa Rican Palicourea and Psychotria species (Rubiaceae). Acta ZooBot. Austria 156, 215–230.
Hoff, K. J., Lange, S., Lomsadze, A., Borodovsky, M., and Stanke, M. (2016). BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32, 767–769. doi: 10.1093/bioinformatics/btv661
Hoff, K. J., Lomsadze, A., Borodovsky, M., and Stanke, M. (2019). Whole-genome annotation with BRAKER. Methods Mol. Biol. 1962, 65–95. doi: 10.1007/978-1-4939-9173-0_5
Hunter, S., Apweiler, R., Attwood, T. K., Bairoch, A., Bateman, A., Binns, D., et al. (2009). InterPro: the integrative protein signature database. Nucleic Acids Res. 37, D211–D215.
Izumi, Y., Ohtani, K., Miyamoto, Y., Masunaka, A., Fukumoto, T., Gomi, K., et al. (2012). A polyketide synthase gene, ACRTS2, is responsible for biosynthesis of host-selective ACR-toxin in the rough lemon pathotype of Alternaria alternata. Mol. Plant-Microbe Inter. 25, 1419–1429. doi: 10.1094/mpmi-06-12-0155-r
Jiang, N., Li, J., and Tian, C. (2018). Arthrinium species associated with bamboo and reed plants in China. Fungal Syst. Evol. 2:1. doi: 10.3897/mycokeys.34.24221
Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., Mcanulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Jourdier, E., Baudry, L., Poggi-Parodi, D., Vicq, Y., Koszul, R., Margeot, A., et al. (2017). Proximity ligation scaffolding and comparison of two Trichoderma reesei strains genomes. Biotechnol. Biofuels 10:151.
Khan, K., and Sullia, S. (1980). Arthrinium phaeospermium var. indicum var. nov., a new market pathogen of cowpea, garden pea and french bean. Acta Bot. Ind. 8, 103–104.
Khan, S. A., Hamayun, M., Kim, H.-Y., Yoon, H.-J., Seo, J.-C., Choo, Y.-S., et al. (2009). A new strain of Arthrinium phaeospermum isolated from Carex kobomugi Ohwi is capable of gibberellin production. Biotechnol. Lett. 31, 283–287. doi: 10.1007/s10529-008-9862-7
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Knapp, D. G., Németh, J. B., Barry, K., Hainaut, M., Henrissat, B., Johnson, J., et al. (2018). Comparative genomics provides insights into the lifestyle and reveals functional heterogeneity of dark septate endophytic fungi. Sci. Rep. 8, 1–13.
Kogel, K.-H., Franken, P., and Hückelhoven, R. (2006). Endophyte or parasite–what decides? Curr. Opin. Plant Biol. 9, 358–363. doi: 10.1016/j.pbi.2006.05.001
Kohler, A., Murat, C., and Costa, M. (2011). High Quality Genomic DNA Extraction using CTAB and Qiagen Genomic-Tip. Champenoux: INRA Nancy Equipe Ecogénomique.
Koike, H., Aerts, A., Labutti, K., Grigoriev, I. V., and Baker, S. E. (2013). Comparative genomics analysis of Trichoderma reesei strains. Ind. Biotechnol. 9, 352–367. doi: 10.1089/ind.2013.0015
Kovaka, S., Zimin, A. V., Pertea, G. M., Razaghi, R., Salzberg, S. L., and Pertea, M. (2019). Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20, 1–13.
Lezcano, J., Alonso, O., and Navarro, M. (2015). Fungal population associated to the germination process of stored seeds of Leucaena leucocephala cv. Peru. Pastures Forages 38, 226–230.
Li, B., Chen, F., Zong, Y., Shang, Y., Zhang, Z., Xu, X., et al. (2019). Dissection of patulin biosynthesis, spatial control and regulation mechanism in Penicillium expansum. Environ. Microbiol. 21, 1124–1139. doi: 10.1111/1462-2920.14542
Li, S., Tang, Y., Fang, X., Qiao, T., Han, S., and Zhu, T. (2020). Whole-genome sequence of Arthrinium phaeospermum, a globally distributed pathogenic fungus. Genomics 112, 919–929.
Li, S.-J., Zhu, T.-H., Zhu, H.-M.-Y., Liang, M., Qiao, T.-M., Han, S., et al. (2013). Purification of protein AP-toxin from Arthrinium phaeospermum causing blight in Bambusa pervariabilis × Dendrocalamopisis grandis and its metabolic effects on four bamboo varieties. Phytopathology 103, 135–145. doi: 10.1094/phyto-07-12-0164-r
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550.
Maehara, S., Simanjuntak, P., Ohashi, K., and Shibuya, H. (2010). Composition of endophytic fungi living in Cinchona ledgeriana (Rubiaceae). J. Nat. Med. 64, 227–230. doi: 10.1007/s11418-009-0380-2
Majoros, W. H., Pertea, M., Antonescu, C., and Salzberg, S. L. (2003). GlimmerM, exonomy and unveil: three ab initio eukaryotic genefinders. Nucleic Acids Res. 31, 3601–3604. doi: 10.1093/nar/gkg527
Mavragani, D., Abdellatif, L., Mcconkey, B., Hamel, C., and Vujanovic, V. (2007). First report of damping-off of durum wheat caused by Arthrinium sacchari in the semi-arid Saskatchewan fields. Plant Dis. 91, 469–469. doi: 10.1094/pdis-91-4-0469a
Miao, L., Kwong, T. F., and Qian, P.-Y. (2006). Effect of culture conditions on mycelial growth, antibacterial activity, and metabolite profiles of the marine-derived fungus Arthrinium cf saccharicola. Appl. Microbiol. Biotechnol. 72, 1063–1073. doi: 10.1007/s00253-006-0376-8
Mopper, K., and Kieber, D. J. (2000). “Marine photochemistry and its impact on carbon cycling,” in The Effects of UV Radiation in the Marine Environment, eds S. D. Mora, S. Demers, and M. Vernet (Cambridge: Cambridge University Press), 101–129. doi: 10.1017/cbo9780511535444.005
Nordberg, H., Cantor, M., Dusheyko, S., Hua, S., Poliakov, A., Shabalov, I., et al. (2014). The genome portal of the department of energy joint genome institute: 2014 updates. Nucleic Acids Res. 42, D26–D31.
O’Connell, R. J., Thon, M. R., Hacquard, S., Amyotte, S. G., Kleemann, J., Torres, M. F., et al. (2012). Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 44, 1060–1065.
Ohm, R. A., Feau, N., Henrissat, B., Schoch, C. L., Horwitz, B. A., Barry, K. W., et al. (2012). Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Path. 9:e1003037. doi: 10.1371/journal.ppat.1003037
Oka, M., Iimura, S., Tenmyo, O., Sawada, Y., Sugawara, M., Ohkusa, N., et al. (1993). Terpestacin, a new syncytium formation inhibitor from Arthrinium sp. J. Antibiot. 46, 367–373. doi: 10.7164/antibiotics.46.367
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’hara, R., et al. (2013). Package ‘Vegan’. Community Ecology Package, Version 2.
Pansanit, A., and Pripdeevech, P. (2018). Antibacterial secondary metabolites from an endophytic fungus, Arthrinium sp. MFLUCC16-1053 isolated from Zingiber cassumunar. Mycology 9, 264–272. doi: 10.1080/21501203.2018.1481154
Peng, Y., Xie, E., Zheng, K., Fredimoses, M., Yang, X., Zhou, X., et al. (2013). Nutritional and chemical composition and antiviral activity of cultivated seaweed Sargassum naozhouense Tseng et Lu. Mar. Drugs 11, 20–32. doi: 10.3390/md11010020
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11:1650. doi: 10.1038/nprot.2016.095
Petersen, L. M., Holm, D. K., Gotfredsen, C. H., Mortensen, U. H., and Larsen, T. O. (2015). Investigation of a 6-MSA synthase gene cluster in Aspergillus aculeatus Reveals 6-MSA-derived Aculinic Acid, Aculins A–B and Epi-Aculin A. Chembiochem 16, 2200–2204. doi: 10.1002/cbic.201500210
Piccolo, S. L., Mondello, V., Giambra, S., Conigliaro, G., Torta, L., and Burruano, S. (2013). Arthrinium phaeospermum, Phoma cladoniicola and Ulocladium consortiale, New Olive Pathogens in Italy. J. Phytopathol. 162, 258–263.
Pintos, Ã, Alvarado, P., Planas, J., and Jarling, R. (2019). Six new species of Arthrinium from Europe and notes about A. caricicola and other species found in Carex spp. hosts. Mycokeys 49, 15–48. doi: 10.3897/mycokeys.49.32115
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
R Development Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ramos, H. P., Braun, G. H., Pupo, M. T., and Said, S. (2010). Antimicrobial activity from endophytic fungi Arthrinium state of Apiospora montagnei Sacc. and Papulaspora immersa. Braz. Arch. Biol. Technol. 53, 629–632. doi: 10.1590/S1516-89132010000300017
Reed, R., Davison, I., Chudek, J., and Foster, R. (1985). The osmotic role of mannitol in the Phaeophyta: an appraisal. Phycologia 24, 35–47. doi: 10.2216/i0031-8884-24-1-35.1
Rouxel, T., Grandaubert, J., Hane, J. K., Hoede, C., Van De Wouw, A. P., Couloux, A., et al. (2011). Effector diversification within compartments of the Leptosphaeria maculans genome affected by repeat-induced point mutations. Nat. Commun. 2, 1–10. doi: 10.1038/ncomms1189
Rudgers, J. A., Koslow, J. M., and Clay, K. (2004). Endophytic fungi alter relationships between diversity and ecosystem properties. Ecol. Lett. 7, 42–51. doi: 10.1046/j.1461-0248.2003.00543.x
Sabella, E., Nutricati, E., Aprile, A., Miceli, A., Sorce, C., Lorenzi, R., et al. (2015). Arthrinium phaeospermum isolated from Tuber borchii ascomata: the first evidence for a “mycorrhization helper fungus”? Mycol. Prog. 14:59. doi: 10.1007/s11557-015-1083-6
Sánchez Márquez, M., Bills, G. F., and Zabalgogeazcoa, I. (2007). The endophytic mycobiota of the grass Dactylis glomerata. Fungal Divers. 27, 171–195.
Sato, J., Goto, K., Nanjo, F., Kawai, S., and Murata, K. (2000). Antifungal activity of plant extracts against Arthrinium sacchari and Chaetomium funicola. J. Biosci. Bioeng. 90, 442–446. doi: 10.1016/S1389-1723(01)80016-5
Seppey, M., Manni, M., and Zdobnov, E. M. (2019). “BUSCO: assessing genome assembly and annotation completeness,” in Gene Prediction, ed. M. Kollmar (New York, NY: Springer), 227–245. doi: 10.1007/978-1-4939-9173-0_14
Shamsi, S., Chowdhury, P., and Naher, N. (2013). Mycoflora associated with the leaves of Senna alata (L.) Roxb. J. Bangladesh Acad. Sci. 37, 249–252. doi: 10.3329/jbas.v37i2.17568
Shen, X.-Y., Cheng, Y.-L., Cai, C.-J., Fan, L., Gao, J., and Hou, C.-L. (2014). Diversity and antimicrobial activity of culturable endophytic fungi isolated from moso bamboo seeds. PLoS One 9:e95838. doi: 10.1371/journal.pone.0095838
Stanke, M., Diekhans, M., Baertsch, R., and Haussler, D. (2008). Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644. doi: 10.1093/bioinformatics/btn013
Stanke, M., Schöffmann, O., Morgenstern, B., and Waack, S. (2006). Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7:62. doi: 10.1186/1471-2105-7-62
Suradkar, K., and Hande, D. (2018). Diversity of fungal endophytes isolated from Adathoda vasica from melghat forest. Aayushi Int. Interdiscip. Res. J. 5:206.
Suxuan, Z., Ning, Z., Zhenyun, C., Qinhua, C., and Weide, Y. (1999). Studies on the integrated control of moso bamboo foot rot. Sci. Silv. Sin. 2, 684–699.
Ter-Hovhannisyan, V., Lomsadze, A., Chernoff, Y. O., and Borodovsky, M. (2008). Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 18, 1979–1990. doi: 10.1101/gr.081612.108
Traeger, S., Altegoer, F., Freitag, M., Gabaldon, T., Kempken, F., Kumar, A., et al. (2013). The genome and development-dependent transcriptomes of Pyronema confluens: a window into fungal evolution. PLoS Genet. 9:e1003820. doi: 10.1371/journal.pgen.1003820
Van Den Berg, M. A., Albang, R., Albermann, K., Badger, J. H., Daran, J.-M., Driessen, A. J., et al. (2008). Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat. Biotechnol. 26, 1161–1168. doi: 10.1038/nbt.1498
Vijayakumar, E., Roy, K., Chatterjee, S., Deshmukh, S., Ganguli, B., Fehlhaber, H.-W., et al. (1996). Arthrichitin. a new cell wall active metabolite from Arthrinium phaeospermum. J. Organ. Chem. 61, 6591–6593. doi: 10.1021/jo960769n
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS one 9:e112963. doi: 10.1371/journal.pone.0112963
Wang, H., Umeokoli, B. O., Eze, P., Heering, C., Janiak, C., Müller, W. E., et al. (2017). Secondary metabolites of the lichen-associated fungus Apiospora montagnei. Tetrahedron Lett. 58, 1702–1705. doi: 10.1016/j.tetlet.2017.03.052
Wang, M., Tan, X.-M., Liu, F., and Cai, L. (2018). Eight new Arthrinium species from China. Mycokeys 34, 1–24.
Wenderoth, M., Garganese, F., Schmidt-Heydt, M., Soukup, S. T., Ippolito, A., Sanzani, S. M., et al. (2019). Alternariol as virulence and colonization factor of Alternaria alternata during plant infection. Mol. Microbiol. 112, 131–146. doi: 10.1111/mmi.14258
Xu, X.-H., Wang, C., Li, S.-X., Su, Z.-Z., Zhou, H.-N., Mao, L.-J., et al. (2015). Friend or foe: differential responses of rice to invasion by mutualistic or pathogenic fungi revealed by RNAseq and metabolite profiling. Sci. Rep. 5:13624. doi: 10.1038/srep13624
Zhang, H., Yohe, T., Huang, L., Entwistle, S., Wu, P., Yang, Z., et al. (2018). dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101. doi: 10.1093/nar/gky418
Keywords: algicolous fungi, biosynthetic gene cluster, comparative genomics, gentisyl alcohol, whole genome sequence
Citation: Heo YM, Oh S-Y, Kim K, Han S-I, Kwon SL, Yoo Y, Kim D, Khim JS, Kang S, Lee H and Kim J-J (2021) Comparative Genomics and Transcriptomics Depict Marine Algicolous Arthrinium Species as Endosymbionts That Help Regulate Oxidative Stress in Brown Algae. Front. Mar. Sci. 8:753222. doi: 10.3389/fmars.2021.753222
Received: 04 August 2021; Accepted: 23 September 2021;
Published: 12 October 2021.
Edited by:
Hao Chen, Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Yonghong Liu, South China Sea Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaCopyright © 2021 Heo, Oh, Kim, Han, Kwon, Yoo, Kim, Khim, Kang, Lee and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanbyul Lee, aGJsZWU5NUBrb3ByaS5yZS5rcg==; Jae-Jin Kim, amFlLWppbmtpbUBrb3JlYS5hYy5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.