- 1Simon F. S. Li Marine Science Laboratory, School of Life Sciences, The Chinese University of Hong Kong, Shatin, Hong Kong SAR, China
- 2School of Biological Sciences, The University of Hong Kong, Pofulam, Hong Kong SAR, China
- 3Graduate School of Integrated Sciences for Life, Hiroshima University, Hiroshima, Japan
Horseshoe crabs provide important ecological services including bioturbation and linking of food web in the shallow waters, but their populations are declining globally, leading to major concerns on conservation of these iconic animals. Baseline information of horseshoe crab ecology, such as their trophic role and food source, is pre-requisite for habitat protection plan and captive restocking program. Trophic ecology of Asian horseshoe crabs is relatively poorly understood and previous studies on their juveniles have suggested that they are selective feeders rather than opportunistic generalists. This study demonstrates a non-invasive approach, using DNA metabarcoding analyses of the nuclear 18S rRNA gene on fecal samples to assess the dietary compositions of Carcinoscorpius rotundicauda and Tachypleus tridentatus juveniles to (1) determine their dietary compositions and trophic roles in their ecosystem, (2) determine any prey selectivity, and (3) distinguish the interspecific dietary differences with potential implications on the habitat requirement and ecological partitioning between these two horseshoe crab species. The results based on relative read abundance (RRA) suggested that oligochaetes were the major prey items for both C. rotundicauda (41.6%) and T. tridentatus (32.4%). Bivalves and crustaceans were second major prey groups for C. rotundicauda (8.6 and 8.4%, respectively). Surprisingly, anthozoans contributed a considerable portion of T. tridentatus’s diet (22.8%), which has never been reported. Furthermore, the major prey groups identified in the fecal samples were not the dominant benthic organisms in the studied area as revealed by environmental DNA (eDNA) analyses on the sediment samples, implying that both species are selective feeders rather than dietary generalists. Significant differences observed in the dietary compositions of the two species might be partially due to the difference in habitat preference between the two species. This study provides new insights into the trophic ecology of the two Asian horseshoe crab species in the estuarine habitat and establishes a new framework for future detailed molecular dietary analyses on all developmental stages of horseshoe crabs around the world, which will allow us to evaluate the food sources needed for the survival of horseshoe crabs and facilitate future conservation strategies without killing the animals.
Introduction
Horseshoe crabs are well known as living fossils with their origin dated back to late Ordovician period in 445 million years ago (Rudkin and Young, 2009). Horseshoe crabs are represented by only four extant species nowadays, including the American horseshoe crab (Limulus polyphemus) in the Atlantic, and three Asian horseshoe crab species (Tachypleus gigas, Tachypleus tridentatus, and Carcinoscorpius rotundicauda) found in the Indo-West Pacific (Sekiguchi and Nakamura, 1979; Vestbo et al., 2018). They play important roles in intertidal communities, for instance they are important biological engineers that facilitate nutrient cycling and ecosystem functioning in the intertidal mudflat by bioturbation (Kraeuter and Fegley, 1994; reviewed by Luo et al., 2020). They also harbor a variety of epibionts and symbionts (Patil and Anil, 2000; Botton, 2009; Tan et al., 2011; Kondo et al., 2018), and their eggs can be the major food sources for different migratory shorebirds (Botton et al., 1994; Karpanty et al., 2006; Michael Haramis et al., 2007; Novcic et al., 2015). The horseshoe crabs are, however, threatened by anthropogenic activities such as habitat destruction, overharvesting, and pollution. Their populations are in a declining trend globally (Rudloe, 1982; Chatterji et al., 1988; Widener and Barlow, 1999; Botton, 2001; Carmichael et al., 2003; Mishra and Mishra, 2011; Wang et al., 2020), in particular, T. tridentatus (commonly known as the tri-spine horseshoe crab) has been listed as an endangered species in the International Union for Conservation of Nature (IUCN) Red List since 2019 (Laurie et al., 2019). Although the population status of C. rotundicauda (commonly known as the mangrove horseshoe crab, or round-tailed horseshoe crab) in the latest IUCN Red List is “data deficient,” prior studies in Hong Kong showed that their populations were threatened and dramatically declined over the past decade (Shin et al., 2009; Morton and Lee, 2010). Thus, major efforts on conservation of these marine iconic taxa should be prompted. To achieve scientifically sounding conservation measurements, detailed baseline information of horseshoe crabs is needed, such as their microhabitats and trophic roles.
Characterizing the dietary compositions of horseshoe crabs is crucial to study their trophic roles in the ecosystem, which is important not only in understanding the estuarine ecology but also in facilitating future conservation and restocking program (Xu et al., 2021). The majority of researches on horseshoe crabs was focused on L. polyphemus, in particular the adults, whilst relatively limited information is known for the feeding habit of the three Asian horseshoe crab species (Luo et al., 2020). There are some studies on the diet of T. tridentatus and C. rotundicauda based on microscopic examinations of gut contents (Zhou and Morton, 2004; Pahang et al., 2012) and stable isotope analyses (Kwan et al., 2015; Fan et al., 2017), yet these studies revealed inconsistent or even contradictory observations and conclusions. Mollusks were most commonly reported food items of adult horseshoe crabs for both Atlantic and Indo-West Pacific species [e.g., L. polyphemus from North America (Botton, 1984; Botton and Ropes, 1989), T. gigas from India (Chatterji et al., 1992), and C. rotundicauda from Malaysia (Pahang et al., 2012)] based on gut content examinations, while polychaetes were also found to be dominant prey for C. rotundicauda from Singapore (Leng and Sivasothi, 2015). The juvenile horseshoe crabs are understudied and the several studies available so far varied in the results. Gut content examination on C. rotundicauda and T. tridentatus suggests that both species had a strong preference on consuming insect larvae (Zhou and Morton, 2004). However, subsequent studies on T. tridentatus juveniles from Hong Kong (Kwan et al., 2015) and C. rotundicauda and T. tridentatus from Pearl Bay in Guangxi (Fan et al., 2017) based on stable isotope analyses suggested that a mixed diet with seagrass contributed to most of the dietary carbon for both species, supplemented with polychaetes, crustaceans, and bivalves. Hence, the findings from various studies are inconsistent, even they were conducted in the same locality [e.g., Pak Nai, Hong Kong (Zhou and Morton, 2004; Kwan et al., 2015)]. Furthermore, gut content examinations (Zhou and Morton, 2004; Pahang et al., 2012) consistently suggested that C. rotundicauda and T. tridentatus are selective feeders, while the stable isotope study showed that the two species are dietary generalists (Kwan et al., 2015; Fan et al., 2017). It remains unclear whether the discrepancies in findings are caused by geographic and temporal variations, differential sensitivity of the adopted techniques and/or among different developmental stages to different food items. Gut content examinations were highly reliable on morphological identification of food residual that is labor demanding and biased toward prey with hard structure, and this approach focused on daily or short-term consumption. On the other hand, the resolution of stable isotopes analyses is low and is sensitive to sample collection size of benthic fauna from the habitat, although it can reflect long-term intake (assimilated items). Thus, a standardized technique with higher resolution that can broadly applied to different geographic settings is undoubtedly needed to allow more accurate comparison among studies and to resolve the major question in trophic roles of horseshoe crabs.
DNA metabarcoding approach represents an accurate and efficient tool for the identification and quantification of consumed species in gut contents and feces (Pompanon et al., 2012), and are increasingly applied on investigating trophic interactions of marine animals (e.g., Berry et al., 2015; Albaina et al., 2016; Jakubavičiūté et al., 2017; van der Reis et al., 2018; Shink et al., 2019; Sonsthagen et al., 2020; van Zinnicq Bergmann et al., 2021). Furthermore, the non-invasive nature of this technique is especially appropriate for studying threatened or endangered species like horseshoe crabs, and will provide important baseline information for biological studies and conservation program. The present study attempts to demonstrate a non-invasive way to determine the diets of horseshoe crabs C. rotundicauda and T. tridentatus from Hong Kong. We used DNA metabarcoding of the nuclear 18S rRNA gene (18S) to (1) determine the dietary compositions and trophic roles of these two horseshoe crab species in their ecosystem, (2) investigate any food selectivity in horseshoe crabs, and (3) evaluate any interspecific dietary differences between C. rotundicauda and T. tridentatus with potential implications on the habitat requirement and ecological partitioning between species. We also evaluate the prey availability in the benthic habitat with the environmental DNA (eDNA) analyses on sediment samples. The results will allow us to evaluate the food sources needed for the survival of horseshoe crabs to facilitate future design of conservation strategies.
Materials and Methods
Sample Collection
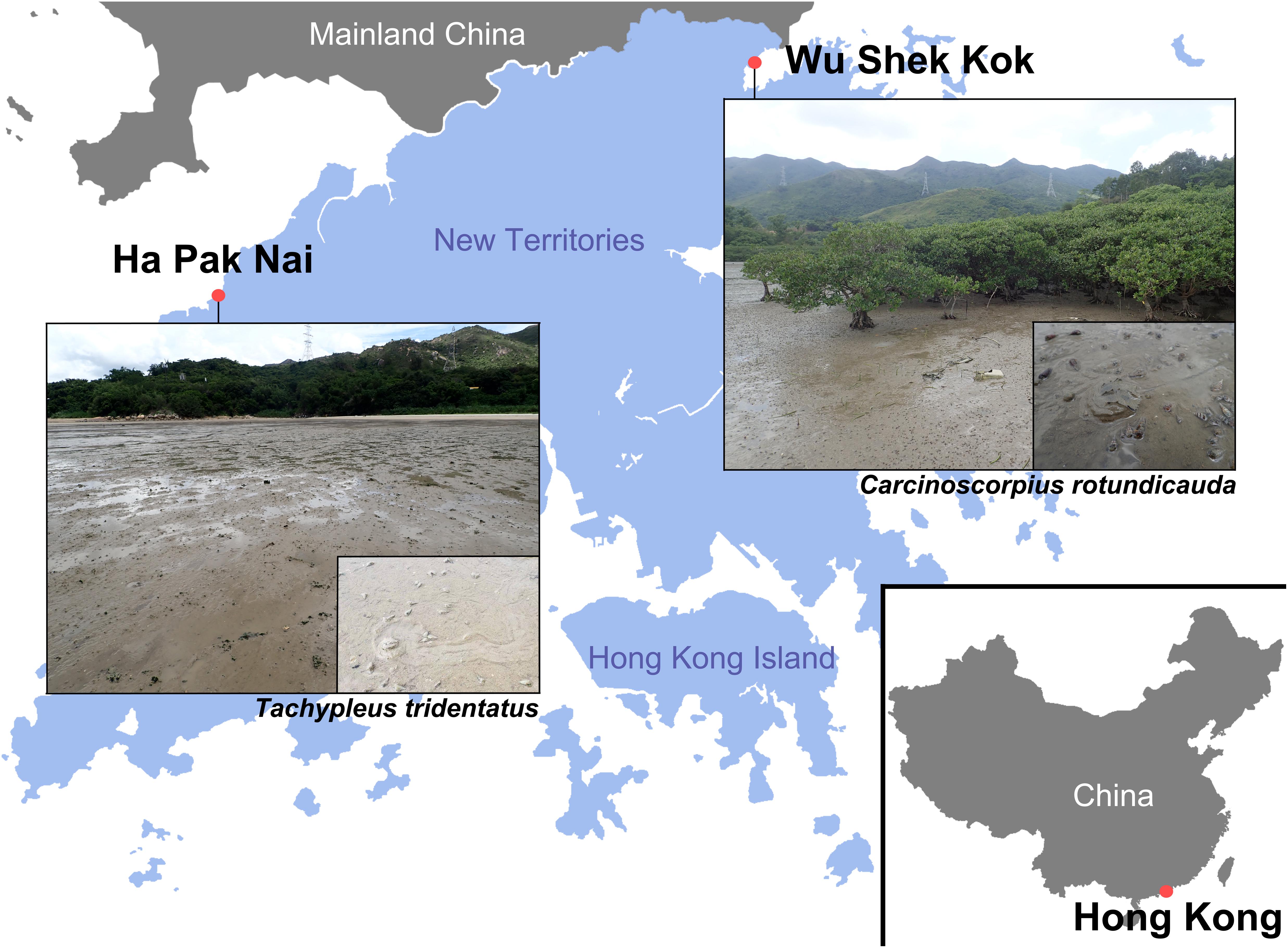
The two Asian horseshoe crab species, C. rotundicauda and T. tridentatus, inhabit different microhabitat and sites in Hong Kong, where C. rotundicauda is usually found in mudflats near mangal habitats whilst T. tridentatus is found in sandier substratum, suggesting potential differential prey preference and/or environmental requirements. Samples of C. rotundicauda were collected from Wu Shek Kok, Sha Tau Kok, Hong Kong (22°32′N, 114°12′E) on September 10 and October 23, 2019, and samples of T. tridentatus were collected from Ha Pak Nai, Yuen Long, Hong Kong (22°25′N, 113°56′E) on September 11 and 24, 2019 (Figure 1). These sampling sites were chosen based on previous study of horseshoe crab populations in Hong Kong of which showed that the largest population of juvenile C. rotundicauda and T. tridentatus could be found in Wu Shek Kok, Sha Tau Kok, and Ha Pak Nai, Yuen Long, respectively. Wu Shek Kok locates on the north-eastern coastline and contains a large patch of dwarf mangrove (Figure 1). This mangrove supported various marine organisms, especially gastropods and bivalves. Ha Pak Nai is a sandy shore in Deep Bay at Northwest Hong Kong. This site is rich in seagrasses that provide nutrients for many marine fauna and oysters were commonly found (Figure 1; Kwan et al., 2016). Horseshoe crabs were collected by hand during low tide and were individually placed in plastic boxes with filtered seawater. They were allowed to stand for 1–3 h until egestion. The feces were collected using a dropper, and preserved in 95% ethanol immediately upon collection and stored in 4°C before DNA extraction. Scaled photos of each horseshoe crab were taken after collection of fecal samples for measurement of six morphometric parameters (i.e., the widths and lengths of prosoma and opisthosoma, paired dorsal compound eye distance and telson length; Supplementary Figure 1) in order to estimate their instar stages based on previous laboratory culture experiment (Chen et al., 2010). The horseshoe crabs were released back into their natural habitats after the feces collection. In each trip, five in situ sediment samples (5 ml in volume each) were collected at the area that horseshoe crabs were collected, to examine the prey availability in the habitat by eDNA analyses. The sediment samples were kept on ice during transportation back to the laboratory and were pooled and mixed well with a spatula to obtain one 25 ml pooled sediment sample in each sampling trip. As a result, a total of four pooled sediment samples were included in the present study (two trips for each of the two sites, Wu Shek Kok and Ha Pak Nai). All pooled sediment samples were stored at −80°C before further treatment.
DNA Extraction, PCR Amplification and Next Generation Sequencing
DNA was extracted from the feces and sediment using QIAamp PowerFecal kit (Qiagen, Hilden, Germany), following the manufacturer’s protocol. The same extraction kit was applied on two different types of samples to minimize discrepancies during DNA extraction and allow more reliable comparisons on metabarcoding data obtained. Five DNA extraction replications were carried out for each pooled sediment sample. The nuclear 18S rRNA gene was examined because this marker can be readily amplified across a broad spectrum of food items from different phyla, including algae which were previously suggested to be a major carbon sources for horseshoe crab species (Kwan et al., 2015; Fan et al., 2017). The V4 region of the 18S (∼450 bp) was amplified using the eukaryotic universal primers 3NDF (5′-GGCAAGTCTGGTGCCAG-3′) and 18S ER1 (5′-GACTACGACG-GTATCTRATCRTCTT-CG-3′) (Bråte et al., 2010). To prevent the amplification of host (horseshoe crab) DNA, a horseshoe crab specific peptide nucleic acid (PNA) probe HSC_18S (5′-NH2-GGCGGTTACTTCCTGGC-Lys-3′) was developed based on the sequence region unique to horseshoe crabs identified from the alignment of 18S sequences of the four horseshoe crab species and other related animals (e.g., spider and scorpion) downloaded from National Center Biotechnology Information (NCBI) GenBank. The PNA probe acted as a blocking primer that differentially bound to the sequence of horseshoe crab and inhibited the amplification.
The PCR amplification of each fecal sample was carried out with a reaction mix containing 7.5 μL of template DNA, 1× PCR buffer, 1 mM dNTPs, 10 nmol of each primer, 20 μg bovine serum albumin (BSA) (New England Biolabs, Ipswich, United States), 12.5 nmol of PNA probe, 0.25 U TaKaRa TaqTM DNA Polymerase (Takara Bio Inc., Shiga, Japan) and sterile distilled water to a total volume of 25 μL. The PCR amplification of each sediment sample followed the same protocol, except no PNA probe was added and template DNA amount was reduced to 2.5 μL. Positive (with horseshoe crab genomic DNA) and negative (with sterile distilled water) controls were included in each batch of PCR reaction to validate the efficiency of PNA probe and reveal any occurrence of false-positive (contamination) and false-negative. The thermal cycle consisted of a 2-min initial denaturation at 94°C, 40 cycles of 15 s at 94°C and 2-min at 63°C, and a finial extension of 3 min at 72°C. Three PCR replicates were done for each sample to obtain a 75 μL PCR product to minimize PCR bias or randomness, and were purified by Gel/PCR DNA fragments kit (Geneaid, Taiwan, China) following the manufacturer’s protocol. All the purified PCR amplicons were sent to commercial company GENEWIZ (Suzhou, China) for high-throughput sequencing using Illumina MiSeq. The purified PCR amplicons were ligated with adaptors incorporated with a 6-bp barcoding region for multiplexing the samples from different locations and species. Preparation of libraries, quality assessment, and paired-end 2 × 300 bp sequencing on Illumina MiSeq platform were conducted by the commercial company.
Bioinformatics and Statistical Analysis
Demultiplex of raw reads were done by GENEWIZ. Paired-end raw reads of each sample were assembled using the standard bioinformatics analysis pipeline of QIIME2 (version: qiime2-2019.10; Bolyen et al., 2019), DADA2 plugin was used for quality and sequence-length filtering, and removal of chimera and singletons. Similarity threshold of 97% was adopted for taxonomic assignment of operational taxonomic units (OTUs). Taxonomic assignment of all the OTUs representative sequences were done by both QIIME-compatible 18S SILVA SSU r119 database (Yilmaz et al., 2014) and NCBI GenBank. Sequences with ≥97% similarity were assigned to class-level, for sequences exhibiting similarity results lower than 97% but higher than 80% were assigned to phylum-level. Sequences were classified as unidentified taxa when exhibiting similarity results lower than 80% or no match was found in both databases. OTUs with less than 1% of the total read in each sample and sequences of horseshoe crabs were removed before statistical analyses. All the common OTUs were assigned into different taxonomic groups based on the classification results.
The relative read abundance (RRA) were used as a proxy of (1) proportion of intake by the horseshoe crabs for fecal samples and (2) proportion of prey availability in the habitats for sediment samples, while frequencies of occurrence (%FOO) of different comestibles were calculated to reflect the prevalence of ingested taxa among individuals. Non-metric multi-dimensional scaling (NMDS) and permutational multivariate analysis of variance (PerMANOVA) were performed in RStudio (RStudio Team, 2020) based on the Bray–Curtis dissimilarity matric, using the “ordinate”, “adonis”, and “ggplot2” functions from R packages, to visualize and to examine the differences in (1) dietary compositions between two species, (2) dietary preferences among instar stages, (3) prey availability between two study sites, and (4) dietary compositions and the prey availability in their habitat at the taxonomic group level. The unidentified taxa were excluded in both NMDS and PerMANOVA analyses, as the sequences could be originated from a wide range of organisms that was inappropriate to categorize as a single group. NMDS and PerMANOVA were performed on both RRA and %FOO metrics. Two matrixes were retained for the NMDS: (1) square-root transformed data of RRA or %FOO and (2) sampling sites, instar stages, and sample types.
Results
Illumina Sequencing
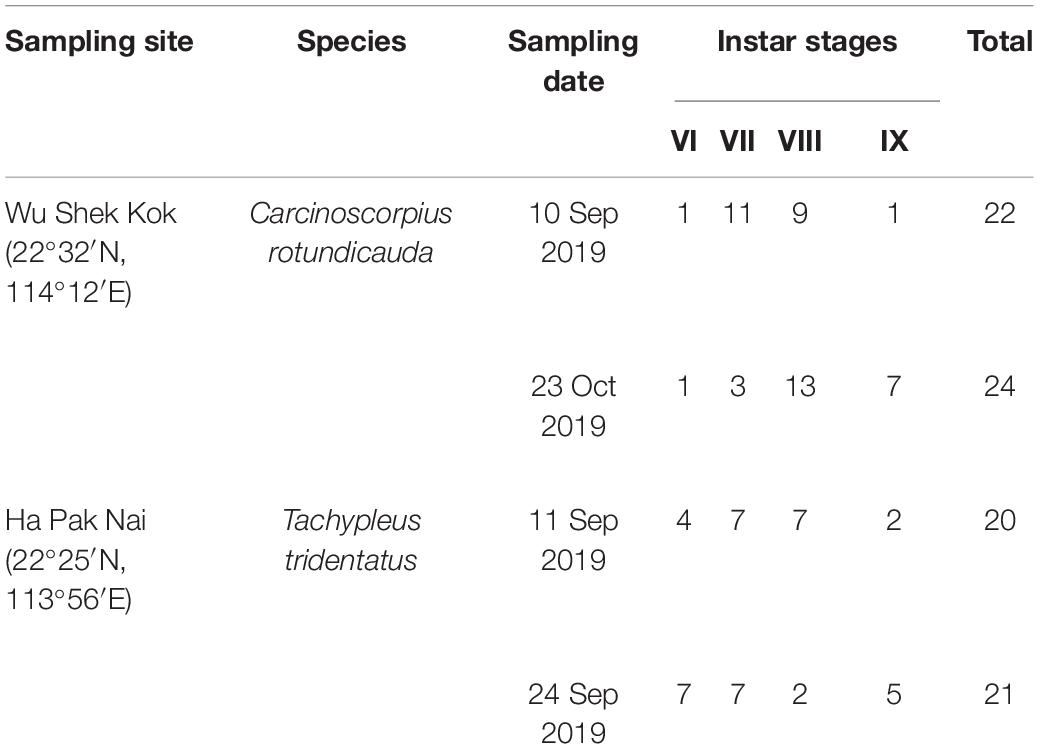
PCR amplifications of the 18S were succeeded in a total 107 DNA samples extracted from 87 fecal samples (46 from C. rotundicauda and 41 from T. tridentatus) and 20 sediment samples (5 replicates from each of the 4 pooled sediment samples). The 87 horseshoe crabs of both species ranged from 6th to 9th instars according to Chen et al. (2010; Table 1). A total number of 2,164,120 filtered reads (mean = 20,225 ± 7,675) were obtained from 107 PCR products, including 942,756 reads from 46 C. rotundicauda fecal samples (mean = 20,495 ± 3,880); 1,011,472 reads from 41 T. tridentatus fecal samples (mean = 24,670 ± 7,413); 114,834 reads from 10 Wu Shek Kok sediment samples (mean = 11,483 ± 5,129); and 94,929 reads from 10 Ha Pak Nai sediment samples (mean = 9,493 ± 6,710). All paired FASTQ files are available in NCBI BioProject (PRJNA758222).
The final OTUs table consisted of 401 OTUs from 21 phyla, assigned from the 107 samples. A total of 160 OTUs (16 phyla) were detected in C. rotundicauda fecal samples; 114 OTUs (14 phyla) were detected in T. tridentatus fecal samples; 62 OTUs (6 phyla) were detected in Wu Shek Kok sediment samples; and 129 OTUs (16 phyla) were detected in Ha Pak Nai sediment samples. A total of 23 OTUs were common in the fecal samples of both species; 16 OTUs were unique in Wu Shek Kok sediment and C. rotundicauda fecal samples; 15 OTUs were unique in Ha Pak Nai sediment and T. tridentatus fecal samples; no OTU was common only in both sediment samples from Wu Shek Kok and Ha Pak Nai; no OTU was shared among all the samples (Figure 2). Furthermore, three OTUs were unique in fecal samples of both species and Wu Shek Kok sediment samples; two OTUs were unique in C. rotundicauda fecal samples and both sediment samples from Wu Shek Kok and Ha Pak Nai, and no OTU was unique in other relations. Of the total number of OTUs detected, 360 could be assigned down to the class or subclass level, and all of the OTUs were assigned to 14 taxonomic groups based on the classification results (Supplementary Table 1). The sequencing depth was sufficient enough to detect the OTUs occurring in each sample, as reflected by the plateau in rarefaction curves (Supplementary Figure 2).
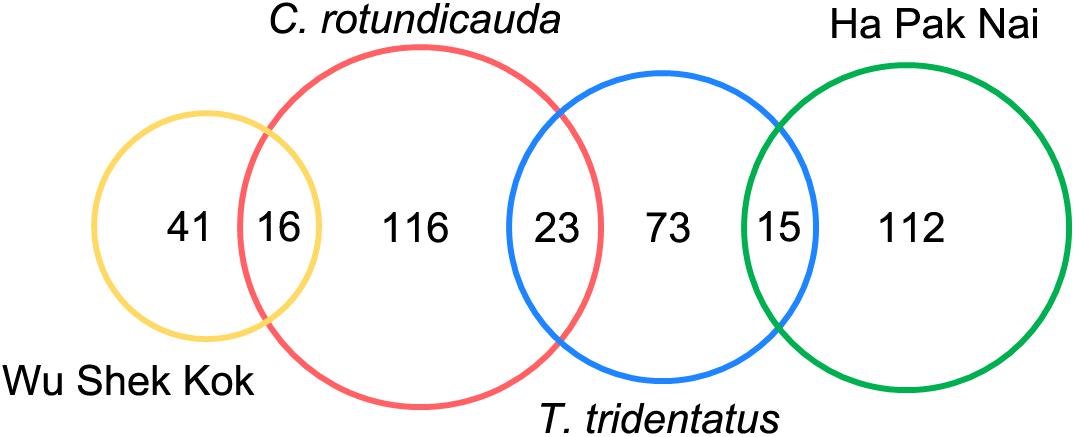
Figure 2. Operational taxonomic units (OTUs) Venn diagram showing the OTUs detected and shared in each group of samples. “Ha Pak Nai” and “Wu Shek Kwok” denote to the sediment samples from the corresponding sites. There were three OTUs shared by the fecal samples of the two species and Wu Shek Kok sediment samples, and two OTUs were shared by C. rotundicauda fecal samples and sediment samples from Wu Shek Kok and Ha Pak Nai. These five OTUs were not shown in the Venn diagram for the ease of reading.
Dietary Compositions of Carcinoscorpius rotundicauda and Tachypleus tridentatus
The results from the two sampling days for each of the two species were largely consistent so the data from the two trips were analyzed together since the sample size in the present study was not sufficient for revealing any temporal pattern. The analyses of RRA showed that oligochaetes were the major prey group for both of the two horseshoe crab species in Hong Kong, which contributed to the largest proportion of reads in both C. rotundicauda (41.6%) and T. tridentatus (32.4%) (Figures 3A, 4). These results also matched with the percentage of frequencies of occurrence (%FOO; Figure 5, showing that oligochaetes could be recovered in 91.3% of C. rotundicauda fecal samples and 82.9% of T. tridentatus fecal samples. Bivalves and crustaceans were other prey groups that contributed to a relatively high proportion of reads (8.6 and 8.4%, respectively) in the diet of C. rotundicauda, where bivalves were recovered in 21.7% of C. rotundicauda individuals and crustaceans were found in 52.2%. Surprisingly, a relatively high proportion of anthozoans (22.8% of RRA) was detected in the diet of T. tridentatus, and it was found in 65.9% of individuals analyzed. This resulted in a significant difference between the dietary compositions of C. rotundicauda and T. tridentatus, both in term of RRA and %FOO (p < 0.001, Table 2; NMDS, Figure 6). One noteworthy observation was that the difference of diet compositions between different instar stages is significant in C. rotundicauda (RRA and %FOO: p < 0.01) but not in T. tridentatus (RRA and %FOO: p < 0.5, Table 2 and Figure 3B). Additionally, SAR (stramenopiles, alveolates, and rhizarians) contributed 24.4% of reads in C. rotundicauda and 13.3% in T. tridentatus (Figures 3A, 4), where SAR was recovered in 91.3% of C. rotundicauda diet and 73.2% of T. tridentatus (Figure 5). However, these were probably a result of incidental consumption or secondary predation, rather than both species were actively feed on SAR. Apart from SAR, unidentified taxa contributed 1.2% of reads in C. rotundicauda and 13.3% in T. tridentatus (Figures 3A, 4), where unidentified taxa were recovered in 13% of C. rotundicauda diet and 78% of T. tridentatus (Figure 5).
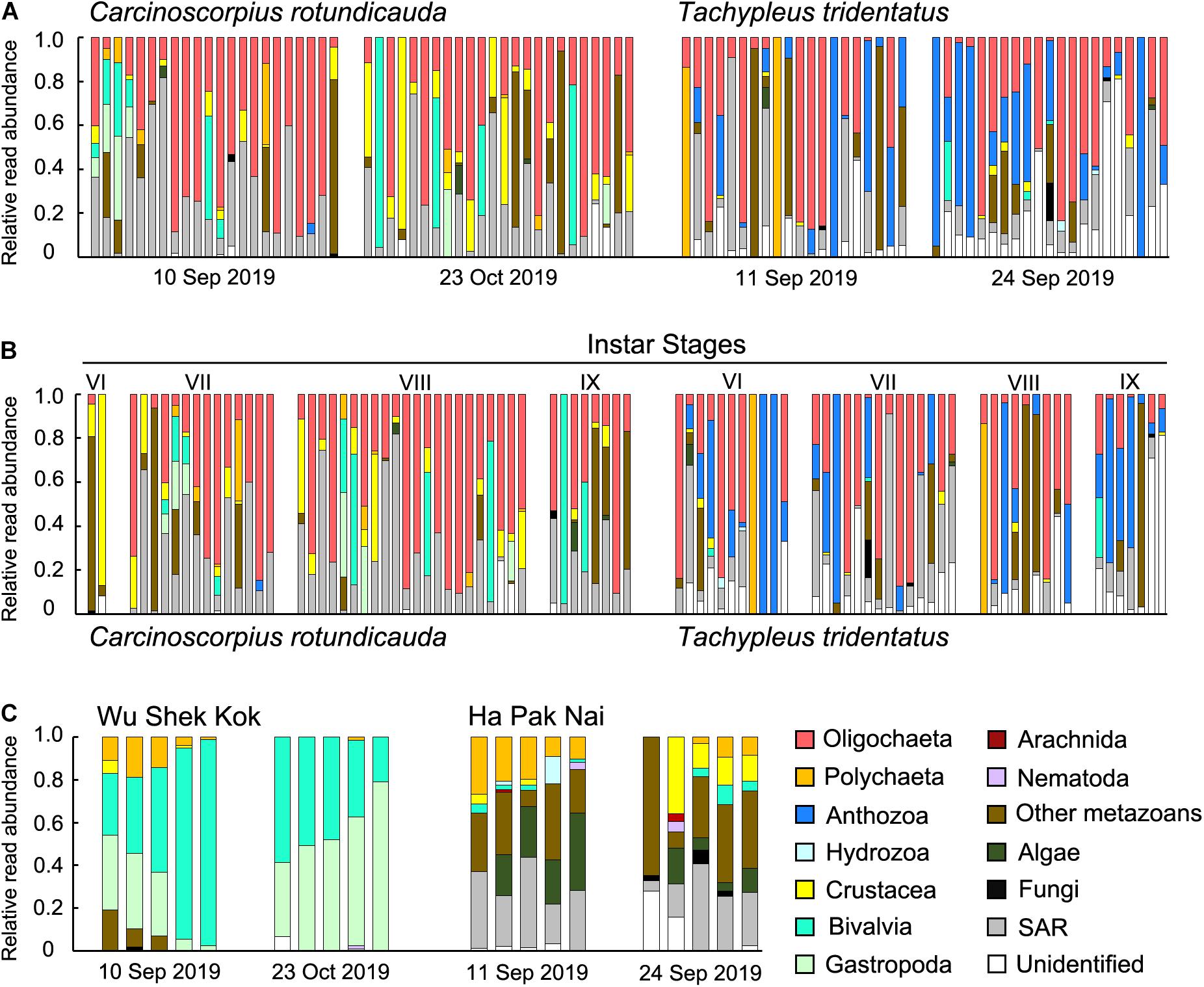
Figure 3. Relative read abundance (RRA) of 14 taxonomic groups detected in each sample of (A) C. rotundicauda and T. tridentatus fecal samples in each sampling trip; (B) C. rotundicauda and T. tridentatus fecal samples ranked by instar stages; and (C) sediment samples in each sampling trip. Each bar represents one individual sample. The taxonomic groups “SAR” includes the OTUs from phyla stramenopile, alveolata, and rhizaria; “other metazoans” includes the OTUs from phyla platyhelminthes, gastrotricha, cnidaria, kinorhyncha, and rotifera.
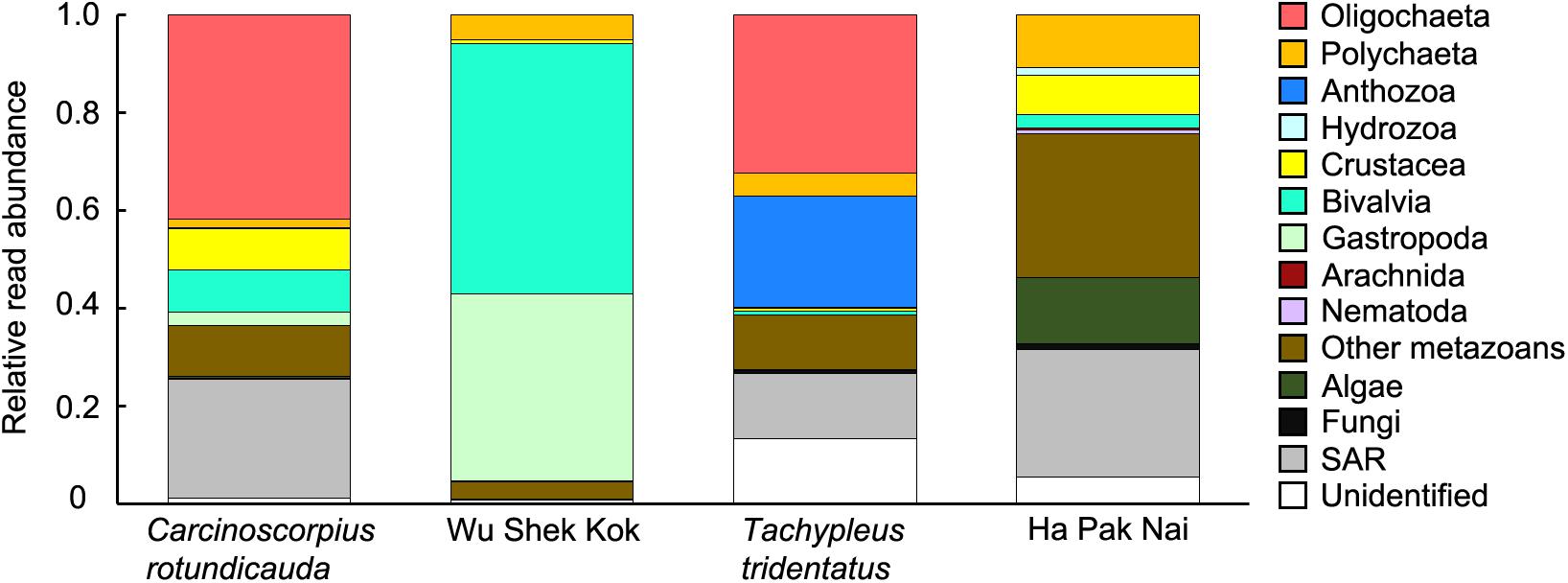
Figure 4. Average relative read abundance (RRA) of 14 taxonomic groups detected in different types of samples.
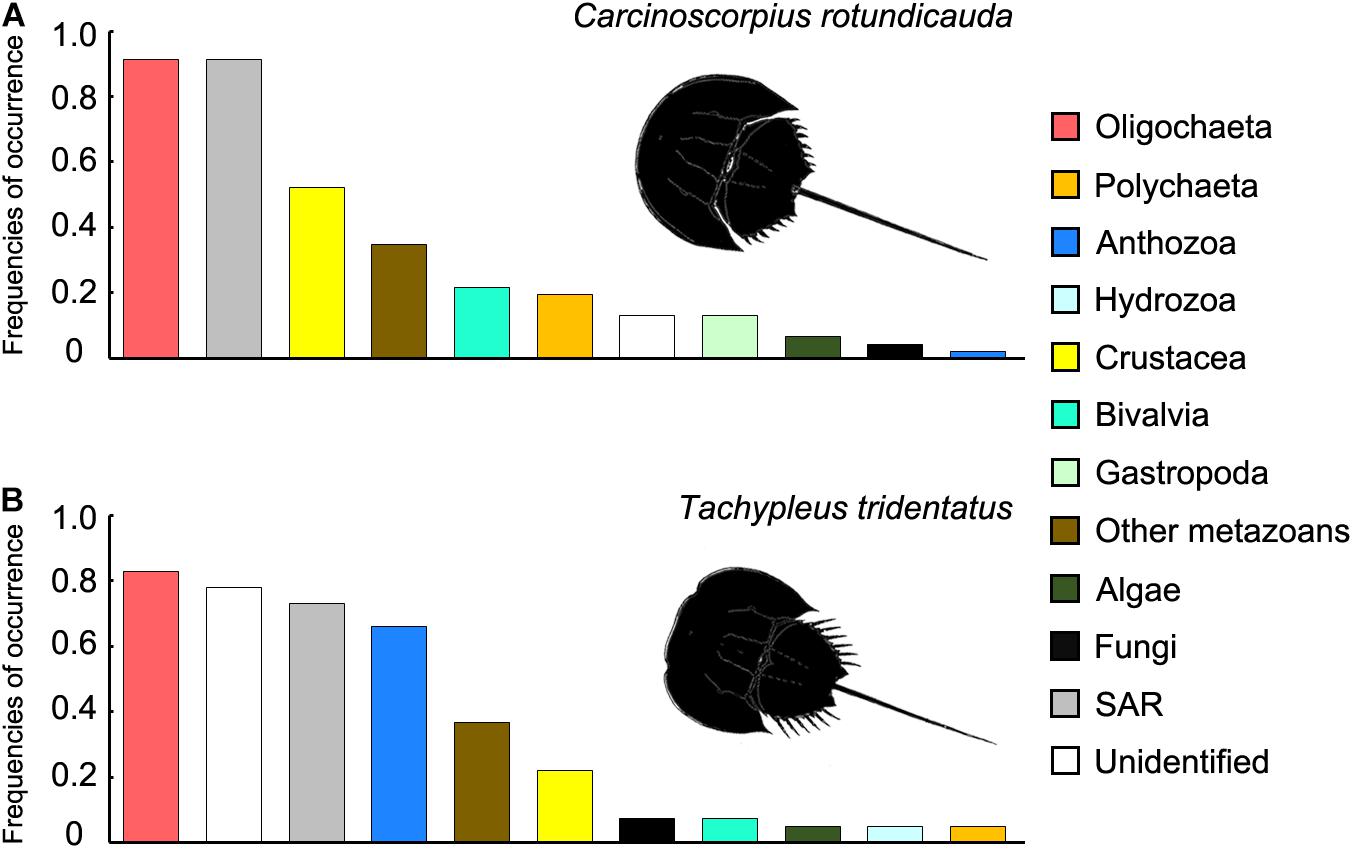
Figure 5. Frequencies of occurrence of 12 taxonomic groups (%FOO ranking) detected in fecal samples of (A) Carcinoscorpius rotundicauda and (B) Tachypleus tridentatus. No OTU from Arachnida and Nematoda was detected in any of the fecal samples and were therefore excluded from this figure.
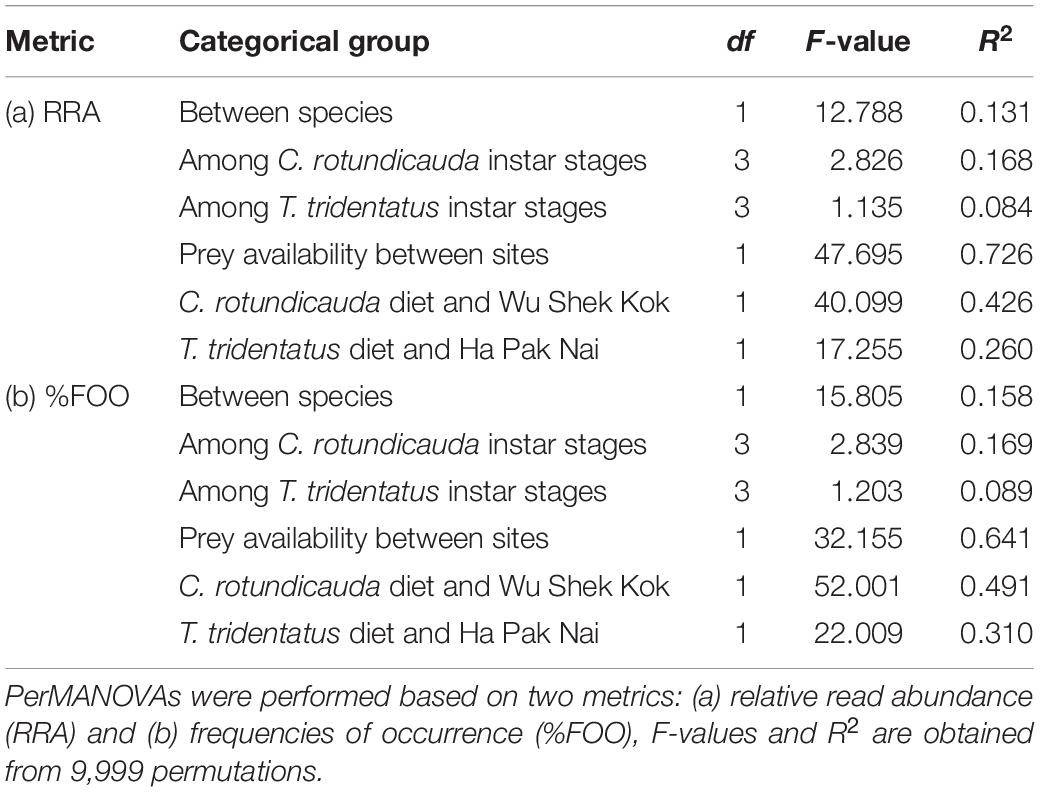
Table 2. The explanatory power of categorical groups on Carcinoscorpius rotundicauda and Tachypleus tridentatus diet.
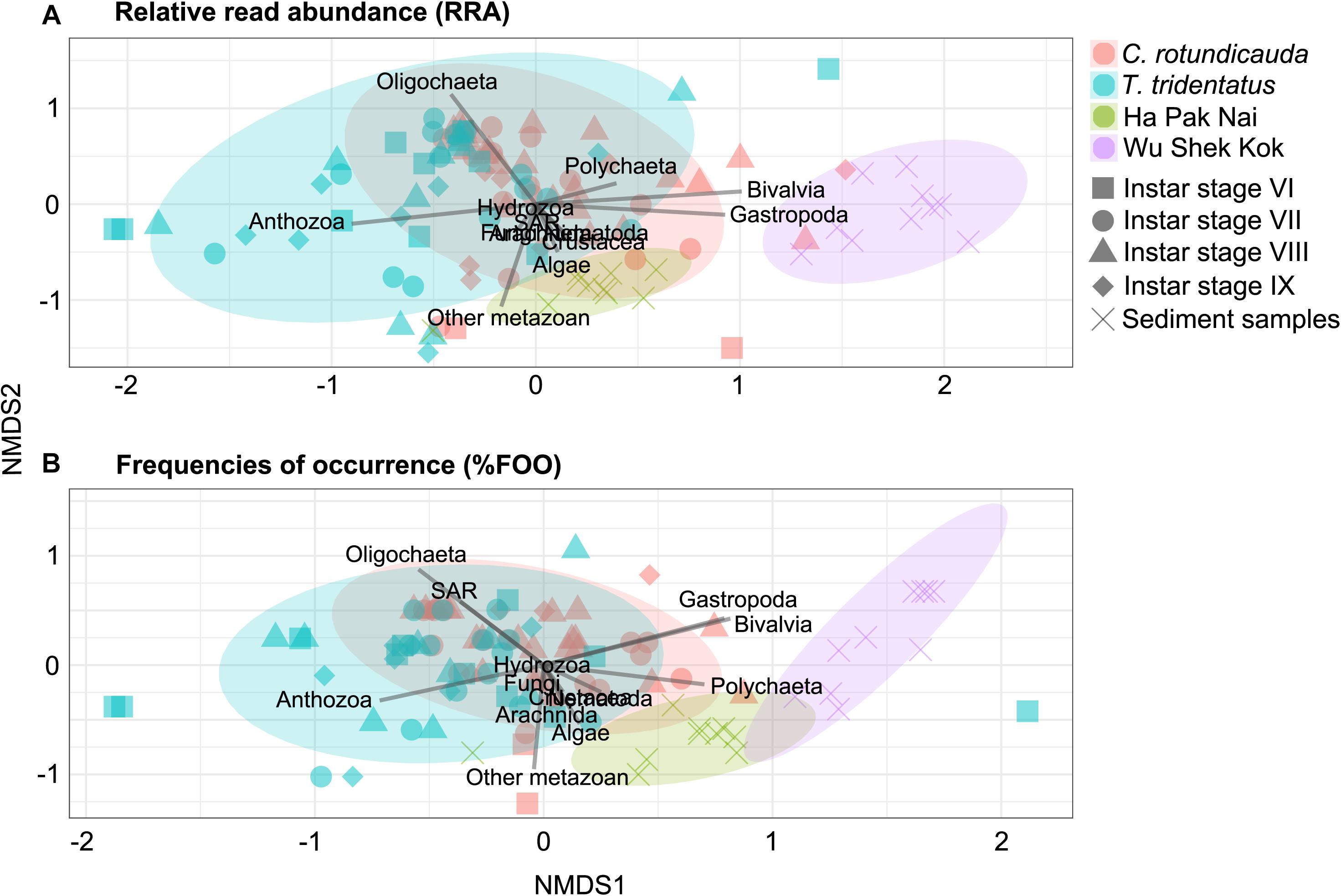
Figure 6. Non-metric multi-dimensional scaling (NMDS) ordinations based on (A) relative read abundance (RRA); (B) frequencies of occurrence (%FOO) of 13 intrinsic variables (i.e., taxonomic groups; indicated by line segments) detected in each group of samples. Each point corresponds to one individual sample with the symbols denote instar stages (for horseshoe crab fecal samples) or sediment samples, while colors represent species or sampling locations.
Prey Availability and Selectivity in Horseshoe Crab Diets
Bivalves and gastropods were the most abundance prey groups in Wu Shek Kok (51.3 and 38.3%, respectively) as shown by eDNA analyses on the sediment samples, while other metazoan (including taxa from platyhelminthes, gastrotrichs, cnidarians, kinorhynchs, and rotifers) and SAR contributed most reads in Ha Pak Nai sediment samples (29.4 and 26.0%, respectively; Figures 3C, 4). There was a significant difference in prey availability between the two sampling sites (RRA and %FOO: p < 0.001, Table 2), this result was also reflected by NMDS (Figure 6) and the analysis of RRA (Figures 3C, 4). More importantly, the differences between the dietary composition of C. rotundicauda and the prey availability in Wu Shek Kok were significant (RRA and %FOO: p < 0.001, Table 2). It was the same case between diet composition of T. tridentatus and the prey availability in Ha Pak Nai (RRA and %FOO: p < 0.001, Table 2). Less than 10% of reads from the major prey items of both species (i.e., oligochaetes, crustaceans, and anthozoans) were detected in sediment samples, indicating that both horseshoe crab species are selective feeders.
Discussion
Trophic Role of Horseshoe Crabs and Potential Energy Contribution From Cnidarians
Our DNA metabarcoding results show that juveniles of C. rotundicauda and T. tridentatus are carnivores which mainly consume benthic animals such as oligochaetes, bivalves, and crustaceans. This finding is largely consistent with previous knowledge of prey consumption detected using morphological examinations and stable isotope analyses (Zhou and Morton, 2004; Pahang et al., 2012; Kwan et al., 2015). Oligochaete worms often constitute an important proportion of the meiofauna from estuarine mangrove sediments in subtropical and tropical waters (Erséus, 2002), so it is not surprising that they can serve as major food source for juvenile horseshoe crabs. However, our data did not find any evidence of seagrass as food sources for juveniles of T. tridentatus and C. rotundicauda in Hong Kong and supported previous studies using stable isotope analyses (Kwan et al., 2015; Fan et al., 2017). Although algal reads were detected in the fecal samples of some individuals (5 out of 87 individuals: 3 C. rotundicauda and 2 T. tridentatus), those reads only occupied a small proportion (<0.5%) of the total reads (Figures 3A, 4). Thus, it is probably a result of incidental consumption or secondary consumption, rather than evidence for omnivorous nor herbivorous behaviors for the crabs. Although the possibility of technical artifacts (e.g., differential digestibility and PCR amplification efficiency amongst food items) and/or seasonal variations in dietary composition in resulting failure of recovery of seagrass cannot be ignored, we justified it was unlikely in our study. The animal tissues are generally easier to be digested compared to algal tissues and we could detect a considerable amount of algal DNA from sediment samples collected from Ha Pak Nai, suggesting the technical artifact is not significant in this circumstance. On the other hand, our study was conducted in late summer, which overlapped with the studied period by Kwan et al. (2015) and several species of seagrass were observed in the Ha Pak Nai during our sampling trip, in consistent with the results of eDNA analyses. The carbon values from the seagrasses reported in previous stable isotope studies might be originated from the preys of horseshoe crabs as many marine meiofauna feed on seagrass detritus. Therefore, our data confirmed with previous studies that juveniles of C. rotundicauda and T. tridentatus were carnivores (Zhou and Morton, 2004; Pahang et al., 2012), which also matched with the morphology of their mouth that gnathobases (chitinous bristles) are adapted for holding and shredding the tissues of benthic animal (Wyse, 1971; Hayes and Barber, 1982; Razak and Kassim, 2018).
Remarkably, our molecular approach enabled to detect unexpected interactions between horseshoe crabs and other marine taxa. We showed that the sea anemone was one of the major prey groups of juvenile T. tridentatus, which has never been reported in any other previous studies. Several species of burrowing anemone, including Paracondylactis hertwigi and Edwardsia japonica, are commonly found in soft shores in western water of Hong Kong, including Ha Pak Nai where the juveniles of T. tridentatus were collected in the present study (Morton, 1983). The closest match for the Anthozoa 18S sequences detected in the horseshoe crab metabarcoding data was Edwardsia (>98%), suggesting that the detected anthozoan likely belongs to the family Edwardsiidae which consisted predominantly sediment burrowing anemone. This provides further evidence that the horseshoe crabs preyed on the burrowing anemone in the sandy habitat where the anemone was present. The anemone was only detected in a single individual of C. rotundicauda probably because they are rare in mangal habitat. Similarly, previous dietary studies on other marine taxa also reported that DNA metabarcoding enable us to reveal cryptic predation relationships on jellyfish and other cnidarians [e.g., jellyfish is a major prey group of Adélie penguin (Jarman et al., 2013); black-browed and Campbell albatross selectively feed on jellyfish (McInnes et al., 2017)], which could be easily overlooked by gut microscopic examinations and stable isotope analysis. Thus, our results recapitulate the omitted role of cnidarians in many food webs and provide new insights into the energy flow in the estuarine mudflat ecosystems.
Prey Selectivity and Interspecific Dietary Difference in Horseshoe Crabs
Our results provide further evidence that the two horseshoe crab species are selective feeders as suggested by gut content analyses (Chiu and Morton, 2004; Zhou and Morton, 2004; Pahang et al., 2012). If horseshoe crabs are non-selective opportunistic feeders that consume whatever available in the habitat, we would expect their diet composition would closely mirror the benthic composition in the sediment. Yet comparison of metabarcoding results from the fecal samples and eDNA analyses on sediment showed that oligochaetaes were the major food items for both two species, but the relative abundance of oligochaetes was low in both habitat (<1% of RRA), indicating significant discrepancies between the ingested food and the prey availability in their corresponding habitats. Therefore, our findings are consistent with previous studies that suggest juvenile of both C. rotundicauda and T. tridentatus are selective feeders (Chiu and Morton, 2004; Zhou and Morton, 2004; Pahang et al., 2012), and provide no support to the hypothesis of generalist as proposed by stable isotope analyses (Kwan et al., 2015; Fan et al., 2017). Interestingly, although mollusks are commonly found as major prey for various horseshoe crab species, especially for the adult (e.g., Botton, 1984; Botton and Ropes, 1989; Chatterji et al., 1992; Pahang et al., 2012), we found the consumption of mollusks was uncommon in the juveniles of the two studied species, with only approximately 10% contribution to the diet in C. rotundicauda on average and only observed detected in 20% of the individuals in spite of mollusks were highly abundant in its habitat (approaching 90% in the eDNA analyses), and <10% in T. tridentatus (in term of both RRA and FOO). We anticipate the mouth part of juvenile horseshoe crab is not strong enough to crush the mollusk shells, so they prefer softer preys that are easier to manipulate such as annelid worms and sea anemone as shown here. This may imply a possible shift to more abundant and nutritious mollusks as major prey items once the horseshoe crabs grow up. This hypothesis on dietary change at different life stages remains to be verified in future studies.
Carcinoscorpius rotundicauda and T. tridentatus live in different habitats: C. rotundicauda is more common in muddy habitat in close proximity to mangrove with finer grain size, whilst T. tridentatus is usually found in more exposed bottom with larger grain size. This implies that dietary preference is anticipated to be one of the factors that lead to the separation in habitat between the two species. We found that the two species exhibit overlapping but significantly different diet compositions. Both species consume oligochaetes as the major food source that may reflect conservatism in functional morphology and ecological niche in closely related species. The major difference reported here emphasizes that C. rotundicauda consumes more crustaceans whilst T. tridentatus ingests a larger amount of anthozoans. However, these differences in dietary composition did not correlate with the prey availability as reflected by eDNA analyses, suggesting that establishment of habitat preference may not be based on the prey preference. This implies that the habitat requirement and ecological partitioning of horseshoe crab species may not be directly affected by prey availability, but other physical parameters, for instance sediment grain size is proposed to be an important factor that affect the ecological partitioning of C. rotundicauda and T. tridentatus (Li, 2008). Thus, habitat protection for both mangrove mudflats and sandy shores should be enforced in order to conserve both populations of C. rotundicauda and T. tridentatus.
Application of DNA Metabarcoding on Horseshoe Crabs in Future Ecological and Conservation Studies
This study demonstrated how DNA metabarcoding approach could be adopted to provide new insights into the feeding habit of horseshoe crabs, and hence to improve our knowledge on estuarine trophic ecology and species conservation. Admittedly, the sample size of the current study was relatively small but the non-invasive protocol by using fecal sample collection enables a standardized and accurate investigation that could be easily scaled up to verify current finding and answer further biological questions without the need of sacrificing the threatened animals. The PNA blocking primer developed in this study could also be applied for the other horseshoe crab species that are not included here. Therefore, we would anticipate the protocol can be more widely applied to conduct larger spatial and temporal studies. Furthermore, some evidences of dietary difference among instars were observed in C. rotundicauda, but not T. tridentatus. However, this may also be an artifact of small sample size analyzed for each instar stage in the present study, and more individuals from different instar stages are required for the evaluation of the intra-specific and inter-specific feeding habit to enhance our understanding on these iconic animals of high conservation value.
Future optimization on the protocol will focus on minimizing amplification bias, which is widely criticized as one of the major sources of error in translating the metabarcoding data into the dietary compositions (Alberdi et al., 2018). The inclusion of mitochondrial marker, in particular the cytochrome c oxidase subunit I (COI) that is considered as a barcoding gene for metazoans, can help to reduce the bias arisen from PCR amplification and provide better taxonomic resolution on the identities of prey below family level that cannot be achieved by the more conserved 18S. As aforementioned, we adopted the more conservative 18S marker in the present pilot study because we attempted to maximize the prey items that can be recovered and to test the hypothesis whether seagrass represents one of the main food items for horseshoe crabs. Based on the current data, we show that horseshoe crabs are likely to be selective carnivores, and thus future research will focus on more detailed prey identities down to species level to have a better resolution on the prey composition data. Accordingly, a local inventory sequence database for different fauna will also be necessary, which is not only applicable to horseshoe crabs, but also facilitate the metabarcoding dietary studies in other endangered and ecologically/economically important species. We believe that this technique could provide new directions for us to understand of the trophic relationships between organisms, and therefore facilitate future design of conservation strategies.
Data Availability Statement
The data presented in the study are deposited in the NCBI BioProject repository, accession number PRJNA758222.
Author Contributions
KW, SO, and LMT designed the study. BL, KW, and LMT conducted the field collection. BL carried out lab work and data analysis, and drafted the manuscript. SS and LMT provided the consumables. All authors revised and gave final approval for publication and agreed to be held accountable for the work performed therein.
Funding
This study was supported by grants from the direct grant from The Chinese University of Hong Kong and the Hong Kong Branch of Southern Marine Science and Technology Guangdong Laboratory (Guangzhou), China to LMT.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank K. C. Cheung, K. L. Hui, and T. W. Tse (CUHK) for their help during field sample collection and T. W. Ho (CUHK) for the advice in the development of PNA blocking primer.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.752806/full#supplementary-material
References
Albaina, A., Aguirre, M., Abad, D., Santos, M., and Estonba, A. (2016). 18S rRNA V9 metabarcoding for diet characterization: a critical evaluation with two sympatric zooplanktivorous fish species. Ecol. Evol. 6, 1809–1824. doi: 10.1002/ece3.1986
Alberdi, A., Aizpurua, O., Bohmann, K., Gopalakrishnan, S., Lynggard, C., Nielsen, M., et al. (2018). Promises and pitfalls of using high-throughput sequencing for diet analyses. Mol. Ecol. Resources 19, 327–348. doi: 10.1111/1755-0998.12960
Berry, O., Bulman, C., Bunce, M., Coghlan, M., Murray, D. C., and Ward, R. D. (2015). Comparison of morphological and DNA metabarcoding analyses of diets in exploited marine fishes. Mar. Ecol. Progr. Ser. 540, 167–181. doi: 10.3354/meps11524
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Botton, M. L. (1984). Diet and food preferences of the adult horseshoe crab Limulus polyphemus in Delaware Bay, New Jersey, USA. Mar. Biol. 81, 199–207. doi: 10.1007/BF00393118
Botton, M. L. (2001). “The conservation of horseshoe crabs: what can we learn from the Japanese experience?” in Limulus in the Limelight, ed. J. T. Tanacredi (Amsterdam: Kluwer Academic), 41–51.
Botton, M. L. (2009). “The ecological importance of horseshoe crabs in estuarine and coastal communities: a review and speculative summary,” in Biology and Conservation of Horseshoe Crabs, eds J. T. Tanacredi, M. L. Botton, and D. R. Smith (Berlin: Springer), 45–63.
Botton, M. L., and Ropes, J. W. (1989). Feeding ecology of horseshoe crabs on the continental shelf, New Jersey to North Carolina. Bull. Mar. Sci. 45, 637–647.
Botton, M. L., Loveland, R. E., and Jacobsen, T. R. (1994). Site selection by migratory shorebirds in Delaware Bay, and its relationship to beach characteristics and abundance of horseshoe crab (Limulus polyphemus) eggs. Auk 111, 605–616. doi: 10.1093/auk/111.3.605
Bråte, J., Logares, R., Berney, C., Ree, D. K., Klaveness, D., Jakobsen, K. S., et al. (2010). Freshwater Perkinsea and marine-freshwater colonizations revealed by pyrosequencing and phylogeny of environmental rDNA. ISME J. 4, 1144–1153. doi: 10.1038/ismej.2010.39
Carmichael, R. H., Rutecki, D., and Valiela, I. (2003). Abundance and population structure of the Atlantic horseshoe crab Limulus polyphemus in Pleasant Bay, Cape Cod. Mar. Ecol. Progr. Ser. 246, 225–239. doi: 10.3354/meps246225
Chatterji, A., Mishra, J. K., and Parulekar, A. H. (1992). Feeding behaviour and food selection in the horseshoe crab, Tachypleus gigas (Müller). Hydrobiologia 246, 41–48. doi: 10.1007/BF00005621
Chatterji, A., Vijaykumar, R., and Parulekar, A. H. (1988). Growth and morphometric characteristic in the horseshoe crab, Carcinoscorpius rotundicauda (Latreiile) from Canning (West Bengal), India. Pak. J. Sci. Industrial Res. 33, 352–355.
Chen, Y., Lau, C. W., Cheung, S. G., Ke, C. H., and Shin, P. K. (2010). Enhanced growth of juvenile Tachypleus tridentatus (Chelicerata: Xiphosura) in the laboratory: a step towards population restocking for conservation of the species. Aquat. Biol. 11, 37–46. doi: 10.3354/ab00289
Chiu, H. M., and Morton, B. (2004). The behaviour of juvenile horseshoe crabs, Tachypleus tridentatus (Xiphosura), on a nursery beach at Shui Hau Wan, Hong Kong. Hydrobiologia 523, 29–35. doi: 10.1023/B:HYDR.0000033085.71861.63
Fan, L. F., Chen, C. P., Yang, M. C., Qiu, G., Liao, Y. Y., and Hsieh, H. L. (2017). Ontogenetic changes in dietary carbon sources and trophic position of two co-occurring horseshoe crab species in southwestern China. Aquat. Biol. 26, 15–26. doi: 10.3354/ab00670
Hayes, W. F., and Barber, S. B. (1982). Peripheral synapses in Limulus chemoreceptors. Comp. Biochem. Physiol. A Comp. Physiol. 72, 287–293. doi: 10.1016/0300-9629(82)90221-3
Jakubavičiūté, E., Bergström, U., Eklöf, J. S., Haenel, Q., and Bourlat, S. J. (2017). DNA metabarcoding reveals diverse diet of the three-spined stickleback in a coastal ecosystem. PLoS One 12:e0186929. doi: 10.1371/journal.pone.0186929
Jarman, S. N., McInnes, J. C., Faux, C., Polanowski, A. M., Marthick, J., Deagle, B. E., et al. (2013). Adélie penguin population diet monitoring by analysis of food DNA in scats. PLoS One 8:e82227. doi: 10.1371/journal.pone.0082227
Karpanty, S. M., Fraser, J. D., Berkson, J., Niles, L. J., Dey, A., and Smith, E. P. (2006). Horseshoe crab eggs determine red knot distribution in Delaware Bay. J. Wildlife Manag. 70, 1704–1710.
Kondo, Y., Hirano, K., Kashimoto, K.-I., Yonetami, M., Fuji, T., and Ohtsuka, S. (2018). Biological notes on Tachypleus tridentatus (Leach, 1819) in Hiroshima Prefecture, Japan: two species of epibiotic organisms found on adults, and the carcass of a juvenile presumably eaten by a bird. Bull. Setouchi Field Sci. Center Graduate School Integr. Sci. Life Hiroshima Univ. 16, 1–5. doi: 10.15027/46553
Kraeuter, J. N., and Fegley, S. R. (1994). Vertical disturbance of sediments by horseshoe crabs (Limulus polyphemus) during their spawning season. Estuaries 17, 288–294. doi: 10.2307/1352578
Kwan, B. K., Cheung, S. G., and Shin, P. K. (2015). A dual stable isotope study for diet composition of juvenile Chinese horseshoe crab Tachypleus tridentatus (Xiphosura) on a seagrass-covered intertidal mudflat. Mar. Biol. 162, 1137–1143. doi: 10.1007/s00227-015-2647-3
Kwan, B. K., Hsieh, H. L., Cheung, S. G., and Shin, P. K. (2016). Present population and habitat status of potentially threatened Asian horseshoe crabs Tachypleus tridentatus and Carcinoscorpius rotundicauda in Hong Kong: a proposal for marine protected areas. Biodiv. Conserv. 25, 673–692. doi: 10.1007/s10531-016-1084-z
Laurie, K., Chen, C. P., Cheung, S. G., Do, V., Hsieh, H., John, A., et al. (2019). Tachypleus tridentatus (errata version published in 2019). IUCN Red List Threatened Species 2019:e.T21309A149768986. doi: 10.2305/IUCN.UK.2019-1.RLTS.T21309A149768986.en
Leng, G. J. M., and Sivasothi, N. (2015). “Diet of mangrove horseshoe crab, Carcinoscorpius rotundicauda in Mandai Kechil, Singapore,” in Paper presented at the 3rd International Workshop on the Science and Conservation of Horseshoe Crabs, Nagasaki.
Li, H. Y. (2008). The conservation of horseshoe crabs in Hong Kong. Ph.D. thesis. Hong Kong: University of Hong Kong.
Luo, Z., Miao, F., Hu, M., and Wang, Y. (2020). Research development on horseshoe crab: a 30-year bibliometric analysis. Front. Mar. Sci. 7:41. doi: 10.3389/fmars.2020.00041
McInnes, J. C., Alderman, R., Lea, M. A., Raymond, B., Deagle, B. E., and Phillips, R. A., et al. (2017). High occurrence of jellyfish predation by black-browed and Campbell albatross identified by DNA metabarcoding. Mol. Ecol. 26, 4831–4845. doi: 10.1038/s41598-018-27509-8
Michael Haramis, G., Link, W. A., Osenton, P. C., Carter, D. B., Weber, R. G., Clark, N. A., et al. (2007). Stable isotope and pen feeding trial studies confirm the value of horseshoe crab Limulus polyphemus eggs to spring migrant shorebirds in Delaware Bay. Journal of Avian Biology 38, 367–376. doi: 10.1111/j.0908-8857.2007.03898.x
Mishra, J. K., and Mishra, A. (2011). “Anthropogenic effect on the horseshoe crab breeding ground and impact on their population along the North-East Coast of India,” in International workshop on scientific conservation of Asian horseshoe crabs, (Hong Kong: City University of Hong Kong), 13–16.
Morton, B. (1983). “Soft Shores – Protected flats,” in The Sea Shore Ecology of Hong Kong, ed. J. E. Morton (Hong Kong: Hong Kong University Press), 146–170.
Morton, B., and Lee, C. N. (2010). Spatial and temporal distributions of juvenile horseshoe crabs (Arthropoda: Chelicerata) approaching extirpation along the northwestern shoreline of the New Territories of Hong Kong SAR, China. J. Nat. Hist. 45, 227–251. doi: 10.1080/00222933.2010.522263
Novcic, I., Mizrahi, D. S., Veit, R. R., and Symondson, W. O. (2015). Molecular analysis of the value of Horseshoe Crab eggs to migrating shorebirds. Avian Biol. Res. 8, 210–220. doi: 10.3184/175815515X14455290976316
Pahang, K. T. P. N., John, B. A., Kamaruzzaman, B., Jalal, K., and Zaleha, K. (2012). Feeding ecology and food preferences of Carcinoscorpius rotundicauda collected from the Pahang nesting grounds. Sains Malays. 41, 855–861.
Patil, J. S., and Anil, A. C. (2000). Epibiotic community of the horseshoe crab Tachypleus gigas. Mar. Biol. 136, 699–713. doi: 10.1007/s002270050730
Pompanon, F., Geagle, B. E., Symondson, W. O. C., Brown, D. S., Jarman, S. N., and Taberlet, P. (2012). Who is eating what: diet assessment using next generation sequencing. Mol. Ecol. 21, 1931–1950. doi: 10.1111/j.1365-294X.2011.05403.x
Razak, M. R. M., and Kassim, Z. (2018). Feeding mechanisms of adult tropical horseshoe crab, Tachypleus gigas toward feeds’ conditions. ASM Sci. J. 11, 76–85.
Rudkin, D. M., and Young, G. A. (2009). “Horseshoe crabs – an ancient ancestry revealed,” in Biology and Conservation of Horseshoe Crabs, eds J. T. Tanacredi, M. L. Botton, and D. R. Smith (Berlin: Springer), 25–44.
Rudloe, A. (1982). “Man’s influence as an environmental threat to Limulus,” in Physiology and biology of horseshoe crabs. Studies on normal and environmentally stressed animals, eds J. Bonaventura, C. Bonaventura, and S. Tesh (New York: A. R. Liss), 297–300.
Sekiguchi, K., and Nakamura, K. (1979). “Ecology of the extant horseshoe crabs,” in Biomedical Applications of the Horseshoe Crab (Limulidae), ed. E. Cohen (New York: A. R. Liss), 37–45.
Shin, P. K. S., Li, H. Y., and Cheung, S. G. (2009). “Horseshoe crabs in Hong Kong: current population status and human exploitation,” in Biology and Conservation of Horseshoe Crabs, eds J. T. Tanacredi, M. L. Botton, and D. R. Smith (New York: Springer), 347–360.
Shink, K. G., Sutton, T. M., Murphy, J. M., and López, J. A. (2019). Utilizing DNA metabarcoding to characterize the diet of marine-phase Arctic lamprey (Lethenteron camtschaticum) in the eastern Bering Sea. Can. J. Fish. Aquat. Sci. 76, 1993–2002. doi: 10.1139/cjfas-2018-0299
Sonsthagen, S. A., Jay, C. V., Cornman, R. S., Fischbach, A. S., Grebmeier, J. M., and Talbot, S. L. (2020). DNA metabarcoding of feces to infer summer diet of Pacific walruses. Mar. Mammal Sci. 36, 1196–1211. doi: 10.1111/mms.12717
Tan, A. N., Christianus, A., and Satar, M. A. (2011). Epibiont infestation on horseshoe crab Tachypleus gigas (Müller) at Pantai Balok in Peninsular Malaysia. Our Nat. 9, 9–15. doi: 10.3126/on.v9i1.5725
van der Reis, A. L., Laroche, O., Jeffs, A. G., and Lavery, S. D. (2018). Preliminary analysis of New Zealand scampi (Metanephrops challengeri) diet using metabarcoding. PeerJ. 6:e5641. doi: 10.7717/peerj.5641
van Zinnicq Bergmann, M. P., Postaire, B. D., Gastrich, K., Heithaus, M. R., Hoopes, L. A., Lyons, K., et al. (2021). Elucidating shark diets with DNA metabarcoding from cloacal swabs. Mol. Ecol. Res. 21, 1056–1067. doi: 10.1111/1755-0998.13315
Vestbo, S., Obst, M., Quevedo Fernandez, F. J., Intanai, I., and Funch, P. (2018). Present and potential future distributions of Asian horseshoe crabs determine areas for conservation. Front. Mar. Sci. 5:164. doi: 10.3389/fmars.2018.00164
Wang, C. C., Kwan, K. Y., Shin, P. K., Cheung, S. G., Itaya, S., Iwasaki, Y., et al. (2020). Future of Asian horseshoe crab conservation under explicit baseline gaps: a global perspective. Global Ecol. Conserv. 24:e01373. doi: 10.1016/j.gecco.2020.e01373
Widener, J. W., and Barlow, R. B. (1999). Decline of a horseshoe crab population on Cape Cod. Biol. Bull. 197, 300–302. doi: 10.2307/1542664
Wyse, G. A. (1971). Receptor organization and function in Limulus chelae. Z. Vgl. Physiol. 73, 249–273.
Xu, P., Bai, H., Xie, X., Wang, C. C., Huang, X., Wang, X., et al. (2021). Tri-spine horseshoe crab aquaculture, ranching and stock enhancement: perspectives and challenges. Front. Mar. Sci. 8:50. doi: 10.3389/fmars.2021.608155
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Keywords: diet, conservation, horseshoe crabs, Carcinoscorpius rotundicauda, Tachypleus tridentatus, Xiphosura, trophic ecology
Citation: Lee BY, Wakabayashi K, Sin SYW, Ohtsuka S and Tsang LM (2021) DNA Metabarcoding Revealed Interspecific Dietary Difference and Prey Selectivity in Juvenile Horseshoe Crabs Carcinoscorpius rotundicauda and Tachypleus tridentatus From Hong Kong. Front. Mar. Sci. 8:752806. doi: 10.3389/fmars.2021.752806
Received: 03 August 2021; Accepted: 13 September 2021;
Published: 14 October 2021.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Daniel Garcia-Souto, University of Vigo, SpainSonglin Liu, South China Sea Institute of Oceanology, Chinese Academy of Sciences (CAS), China
Copyright © 2021 Lee, Wakabayashi, Sin, Ohtsuka and Tsang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Ming Tsang, bG10c2FuZ0BjdWhrLmVkdS5oaw==
†ORCID: Simon Yung Wa Sin, orcid.org/0000-0003-2484-2897
 Bo Yee Lee
Bo Yee Lee Kaori Wakabayashi
Kaori Wakabayashi Simon Yung Wa Sin
Simon Yung Wa Sin Susumu Ohtsuka
Susumu Ohtsuka Ling Ming Tsang
Ling Ming Tsang