
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 06 September 2021
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.742330
This article is part of the Research Topic Mollusk Breeding and Genetic Improvement View all 25 articles
 Xinghai Zhu1†
Xinghai Zhu1† Junhao Zhang1†
Junhao Zhang1† Xiujiang Hou1
Xiujiang Hou1 Pingping Liu1
Pingping Liu1 Jia Lv1
Jia Lv1 Qiang Xing1,2*
Qiang Xing1,2* Xiaoting Huang1,2
Xiaoting Huang1,2 Jingjie Hu1,3
Jingjie Hu1,3 Zhenmin Bao1,2
Zhenmin Bao1,2Molluscan shell color has consistently drawn attention for its abundant diversity and commercial use in shellfish breeding projects. Recently, two new strains of bay scallop (Argopecten irradians irradians) with different shell colors as marked phenotypic traits have been artificially bred to improve their economic values; however, the inheritance mechanism of their shell pigmentation is still unclear. In this study, a genome-wide association study (GWAS) was conducted to determine the genetic basis of shell color in bay scallops utilizing 29,036 high-quality single-nucleotide polymorphisms (SNPs) derived from 80 purple-red (PP) and 80 black-brown (BP) shell color individuals. The result of the GWAS showed that 469 SNPs (p <1.72E−6) significantly associated with shell color were mainly distributed in chromosome 7. The top three SNPs (i.e., chr7-12764003, chr7-13213864, and chr7-11899306) are located in the genic region of G-protein-coupled receptor-like 101 (GRL101), polyketide synthase 1 (PKS1), and phosphoinositide phospholipase C (PLC1), which have been widely reported to be involved in pigmentation. Successfully, the top three SNPs were verified in another non-breeding bay scallop population. Furthermore, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses obtained 38 GO terms covering 297 genes and aggregating pathways involving 252 annotated genes. Specifically, the expression profiles of the top three identified candidate genes were detected in mantles of PP and BP individuals by real-time quantitative reverse transcription PCR. The significantly higher expression levels of GRL101 (6.43-fold) and PLC1 (6.48-fold) in PP, and PKS1 (12.02-fold) in BP implied that GRL101 and PLC1 potentially functioned in PP shell coloration, and black pigmentation in BP might be principally regulated by PKS1. Our data provide valuable information for deciphering the phenotype differences of shell color in the bay scallop.
The shell of mollusks with diverse forms and alternative colors has always appealed to consumers and scientists (Comfort, 1950). Shell color of marine mollusks, an obvious and marked genetic-based phenotypic trait, has been proven to be closely related to production traits (Cong et al., 2014; Zhang et al., 2016). Therefore, shell color has been widely applied in the selective breeding project of economic shellfish, such as oysters (Xu et al., 2019; Han et al., 2020), mussels (Innes and Leslie, 1977; Li et al., 2014), scallops (Petersen et al., 2012; Ding et al., 2015), clams (Zhang et al., 2018; Nie et al., 2020), and abalones (Liu et al., 2007; Hoang et al., 2017). To reveal the mechanism of shell color formation, several investigations have been conducted and found multiple factors, ranging from environmental factors to inner genetic factors, contributing to the higher variability of shell color in mollusks (Underwood and Creese, 1976; Kraeuter et al., 1984). For example, researches demonstrated that water temperature differences (Heller, 1992), salinity variations (Sokolova and Berger, 2000), and diet sources (Marchais et al., 2017) could affect shell color formation in mollusks. On the aspect of genetic basis, shell color has been demonstrated to be determined by one or a small number of major gene(s) in some shellfish species (Cole, 1975; Palmer, 1985). For instance, experimental crosses of Manila clam, Ruditapes philippinarum, verified that pigmented coloration in two valves is controlled by at least two genes (Peignon et al., 1995). In noble scallop Chlamys nobilis, a one-locus-three-allele model was proposed to elucidate the distribution of four different color variants, and the brown shell color is controlled by a recessive allele distinct from the other colors (Zheng et al., 2013). Investigation in Pacific oyster, Crassostrea gigas, demonstrated that shell pigmentation is controlled by two genetic loci, with one responsible for the secretion of pigments and the other responsible for the distribution mode of pigmentation (Xu et al., 2019). Furthermore, transcriptomes and digital gene expression analysis of four different color clams, Meretrix meretrix, suggested that several potential genes and the Notch pathway played a crucial role in its shell color patterning (Yue et al., 2015). The same method was also exerted in two extreme color phenotypes of Yesso scallop, Patinopecten yessoensis, and 25 significantly differential expression genes were identified for unraveling shell color differences (Sun et al., 2016). These researches provided pivotal loci/genes responsible for the formation of shell coloration and served as candidate markers for mollusks breeding projects.
Biological pigments are considered as the key element of shell color diversity, and the distinct color is perceived because of different types of pigments, such as melanins, porphyrins, tetrapyrroles, and carotenoids, which had been identified in shellfish shells (Comfort, 1951; Stemmer and Nehrke, 2014). Melanins were first derived by oxidation and polymerization of tyrosine in animals or phenolic compounds in lower organisms (D'Iischia et al., 2013). Using the ultraviolet and infrared radiation spectral analysis, melanin was confirmed as the black pigment extracted from the scars where adductor muscle jointed shells in oysters (Hao et al., 2015). Eumelanin and pheomelanin, two forms of melanin, were verified to comprise the dark brown shell pigments with ~76.6 and 23.4% components via spectrophotometry scanning and high-performance liquid chromatography in Yesso scallop (Sun et al., 2017). As for porphyrins, the cyclic structure of tetrapyrroles is often associated with red, brown, or purple shell coloration (Comfort, 1951). Investigation via modern chemical and multimodal spectroscopic techniques of shell pigments in marine snails Clanculus pharaonius and Clanculus margaritarius showed that pink-red dots and lines in shells were caused by porphyrins (Williams et al., 2016). Furthermore, carotenoids were regarded as the principal substance that presented orange adductor muscle and shell color in scallop since their first identification in the adductor muscle of rare orange Yesso scallop variants (Li et al., 2010). Subsequent analysis in reddish-orange and brown shell-color Yesso scallops identified candidate genes involved in carotenoid metabolism during shell color formation (Zhao et al., 2017). A similar result was also reported in noble scallop populations, in which individuals with orange shell color contained dramatically higher carotenoid content compared with the brown ones (Zheng et al., 2010).
The bay scallop, Argopecten irradians irradians, is polymorphic for shell color (purple, orange, yellow, or white) and possesses advantages of fast growth and high production in aquaculture (Shumway and Parsons, 2006; Wang et al., 2007). Selective breeding of bay scallops for better growth and tolerance performance has been consecutively conducted for generations from the 1980s in China (Zheng et al., 2004, 2011; Wang et al., 2017). Specifically, a potential phenotypic correlation between shell color and growth traits has been observed in artificially bred bay scallops (Wang et al., 2020). Consequently, understanding the genetic mechanism of shell color in bay scallops is of theoretical and economic importance. The first comprehensive classification of shell color based on experimental crosses in bay scallop showed that three background colors, six pattern colors, and the diversity of shell colors were under genetic control (Kraeuter et al., 1984; Elek and Adamkewicz, 1990). Subsequent study of shell color inheritance in selfing families found that orange or yellow parents produced both colorful and white progeny with a ratio of ~3:1, implying that shell color in bay scallops was controlled by one locus being dominant. Furthermore, one amplified fragment length polymorphism (AFLP) marker was successfully identified in orange and white shell color bay scallops through genetic linkage analysis (Qin et al., 2007). Considering the large polymorphism of shell color in bay scallop, further genome-wide researches is still needed to identify the comprehensive candidate loci/genes and to explore the potential molecular mechanism.
Genome-wide association study (GWAS) is an accurate and powerful tool to identify novel genetic variants loci underlying complex traits and has been broadly applied in various fields (Zhao et al., 2011; Kim et al., 2012; Sukumaran and Yu, 2014; Ikeda et al., 2018; Ning et al., 2019). Investigations using GWAS have already successfully deciphered genetic variants of coloration traits, in which the identified specific markers could be potentially beneficial for selective breeding. For instance, a strong candidate locus, in arabidopsis pseudo-response regulator 2-like (APRR2) that regulates the accumulation of green pigments, was identified in melon and watermelon via GWAS (Oren et al., 2019). Information of two colors of single-nucleotide polymorphisms (SNPs) in French Saanen goats could be used to remove the undesired colored goats through molecular marker-assisted breeding (Martin et al., 2016). Furthermore, five potential loci related to carotenoid biosynthesis were identified in maize (Zeamays L.); therefore, a greater abundance of carotenoids accompanied by the deeper orange color of the kernel was potentially obtained (Owens et al., 2019). A similar GWAS result has also been concluded in Yesso scallop, in which three candidate genes (i.e., LDLR, FRIS, and FRIY) were detected around the two most significant potential loci and were proven to be involved in carotenoid metabolism in reddish-orange shell individuals (Zhao et al., 2017). These GWAS results not only provide a list of candidate loci/genes to be functionally validated but also offer an efficient approach for deciphering the genetic architecture of complex traits and selecting the breeding strategy.
This study aimed to identify significant SNP makers associated with shell color (purple-red and black-brown) in two new strains of bay scallops by performing GWAS, with the aid of whole-genome sequence databases of A. irradians irradians (unpublished data). The top three SNPs that dominantly control the shell color formation were further analyzed in their located candidate genes and then verified in another population. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted to reveal the potential molecular mechanism in bay scallop shell coloration. Finally, we investigated the expression levels of the top three candidate genes in bay scallop mantles to detect their vital roles in shell coloration and to provide valuable information for the efficient selective breeding of shell color strains in bay scallop.
Two strains of healthy bay scallops with nearly 9-month-old, namely, the purple-red shell color population (PP) (N > 500) and the black-brown shell color population (BP) (N > 500), were used as samples in 2016. The shell color trait of both strains is genetically stable due to the genomic selection of four consecutive generations from 2012 to 2015 by our group (Figure 1). Samples were collected from artificial scallop-rearing substrates at Qingdao Jinshatan Fishery Group Co. Qingdao (35°97′23 N, 120°27′28 E, Shandong Province). Subsequently, we randomly selected scallop samples (N = 80) from each group based on their shell color. Gills were removed from each scallop according to the procedure of Mao et al. (2013) and then preserved in 100% ethanol at −20°C before genetic analysis. Our experiments were conducted according to the guidelines and regulations established by the Ocean University of China and the local government.
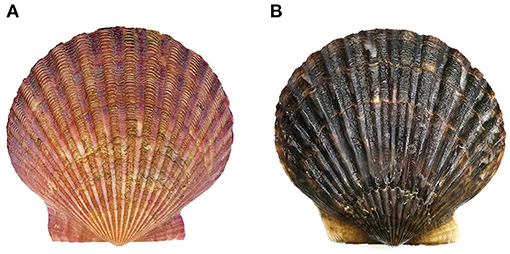
Figure 1. (A) The purple-red shell color (PP) and (B) black-brown shell color (BP) bay scallops Argopecten irradians.
Genomic DNA was extracted from the gill using a standard phenol-trichloromethane method (Sambrook et al., 1989). Subsequently, Miso-RAD libraries were constructed by using 80 PP and 80 BP individuals, following the protocol developed by Wang et al. (2016). On summarizing, genomic DNA of the 160 selected bay scallop individuals was digested with BsaXI (New England BioLabs, MA, USA, Cat. no. R0609) at 37°C for 45 min. Adaptor ligation, PCR amplification, digestion, and ligation were followed and after which every five DNA fragments were set as one group (barcoding and pooling). MinElute PCR Purification Kit (Qiagen, Hilden, Germany, Cat. no. 28004) was used to purify each product that concatenated five tags from five samples after performing amplification for two times. The 32 libraries were then pooled to run double-end sequencing on the Illumina Hiseq2000 system (San Diego, USA).
Raw reads were first preprocessed to eliminate paired-end (PE) reads with ambiguous base calls (N), long homopolymer regions (>10 bp), or excessive low-quality bases (>20% of bases with quality score <10) to obtain high-quality reads. PEAR software (Zhang J. et al., 2014) was exerted to assemble the forward and reverse reads. The merged reads containing five tags were divided into single-tag datasets by using homemade Perl script, and via SOAP software (Li et al., 2009), each tag was subsequently aligned to the chromosome-level reference bay scallop genome (unpublished data) that was constructed by our group. The RADtyping software (Fu et al., 2013) was employed in the genotyping analysis of sequencing data. The selective rules of markers included polymorphism, sufficient genotype rate (with available genotype over 80% progenies), and minor allele frequency (MAF) higher than 0.05. Then, the qualified markers were used for downstream analysis.
To reduce the false-positive rate, the population genomic structure analysis was performed to provide a population membership matrix through Admixture software 1.3 (Alexander et al., 2009), and a kinship matrix was estimated by TASSEL 5.0 (Bradbury et al., 2007). Association analysis between genotypes and shell colors was conducted using PLINK software 1.9 (Chang et al., 2015) by employing the logistic regression model with these two covariates to detect the associate SNPs. The statistic p-value for each SNP was calculated and summarized in ascending order. The threshold p-value for genome-wide significance was calculated using the Bonferroni correction based on the number of qualified makers (Johnson et al., 2010). When an SNP scored less than the significance cutoff (1.72E−6), the sequence of each maker was extracted and used as a query to exert BLASTN (Altschul et al., 1997) to search the draft genome of A. irradians irradians (unpublished data) that were annotated by the protein database NR. In addition, the Manhattan plot was drawn by an R package named “ggplot,” and these potential SNPs (p <1.00E−5) were captured to conduct bioinformatics analysis, including GO function annotation and KEGG pathway enrichment analyses (http://www.omicshare.com/tools).
To further evaluate the accuracy of GWAS, the top three SNPs (p-value <1.00E−17) were selected to verify in the 30 purple-red shell color samples and 30 black-brown shell color samples collected from another separate bay scallop population (N > 500) in the sea area of Huangdao District, Qingdao, Shandong Province, in 2018. Striated muscles of the above 60 individuals were separately sampled and preserved at −80°C for subsequent analysis. The primer sequences are provided in Table 1. The PCR of fragments containing the above three SNPs was carried out as follows: 2 μl of template DNA (50 ng/μl), 15 μl of Q5 High-Fidelity 2 × Master Mix (New England BioLabs, MA, USA, Cat. no. M0492S), 0.5 μl (10 pmol/μl) of each forward and reverse primer, and sterile water up to 30 μl volume was added for amplifying. The related parameters in the PCR system for three primers were pre-denaturing under 98°C for 30 s, then denaturing under 98°C for 10 s, annealing under 56°C for 15 s, and extending under 72°C for 10 s, respectively. After 28 cycles, the sample was extended under 72°C for 2 min. The PCR products were sequenced using the Sanger method by Sangon Biotech (Shanghai, China). Sequences amplified by the primers were compared among the 60 scallops (30 PP and 30 BP, respectively) using the ClustalW2 multiple alignment program (http://www.ebi.ac.uk/Tools/msa/clustalw2/). When a mutation was detected, comparisons of genotype frequencies between the PP and BP were performed using Fisher's exact test to identify the shell color-associated mutation. p-values < 0.05 were considered statistically significant.
To compare the differences in the expression levels of genes which are the top three SNPs annotated between PP and BP, 30 PP and 30 BP individuals from another non-breeding bay scallop population (N > 500) were randomly recruited, and the mantle was sampled in the sea area of Huangdao District, Qingdao, Shandong Province, in 2020. Total RNA of the sampled mantles was isolated following the method described by Hu et al. (2006) and then digested with DNaseI (Takara, Shiga, Japan). The concentration and purity of the RNA were determined using a Nanovue Plus spectrophotometer (GE Healthcare, Piscataway, NJ, USA), and the RNA integrity was assessed by agarose gel electrophoresis. The first-strand cDNA was synthesized according to the protocol of the manufacturer using Moloney murine leukemia virus (MMLV) reverse transcriptase (Thermo, Wilmington, USA) in a 20 μl volume with 2 μg of each total RNA sample as the template and 0.5 μg of oligo (dT)18 (Takara Biotechnology, Liaoning, China) as the primer. The mixture was denatured at 65°C for 5 min and then chilled immediately on ice. After adding the reverse transcriptase, reaction buffer, and dNTPs, cDNA was amplified under the following conditions: 42°C for 90 min and 72°C 10 min. The cDNA was stored at −20°C and diluted to 5 ng/μl for use as the template in the real-time quantitative reverse transcription PCR (qRT-PCR). Data from the qRT-PCR were obtained and the gene expression level is shown as the fold change. The statistical analysis of the data was performed with SPSS software (version 21.0) via an independent sample t-test. Differences were considered significant at p < 0.05.
In total, sequencing of 32 Miso-RAD libraries produced 447,286,098 reads (GenBank: PRJNA689862), and the average sequencing depth was 22.32 × with 2,795,538 reads per individual. After quality control, we obtained 219,383,262 high-quality reads, and an average of 42.49% of high-quality reads in each individual was uniquely mapped to the bay scallop genome (unpublished data) by SOAP. Eventually, a total of 211,728 SNPs were genotyped in 160 scallop samples (80 PP and 80 BP) via RADtyping.
A total of 211,728 SNP markers obtained from 160 bay scallop individuals were exerted to quality control performed by PLINK software. After quality filtering (call rate > 80% and MAF > 5%), a set of 29,036 qualified SNPs were successfully assigned to the 16 chromosomes of bay scallop and used for further genome-wide analysis (Figure 2). Genetic clustering illustrated that the admixed ancestry of PP and BP was shown for K = 2, with kinship coefficients varying from 0 to 0.02 between PP and BP individuals, then the corresponding components were brought as covariates. After conducting Bonferroni correction and 0.05 cutoff (p <1.72E−6), we detected 469 SNPs significantly associated with shell color, and 285 of these 27 bp tags could be directly annotated in the bay scallop genome. Furthermore, the –log10 (p-value) values of the top eight SNPs with a significant association of shell color were higher than 15, and these top eight SNPs were mainly distributed in chromosome 7 (Table 2). Specifically, the top eight loci were separately located at the genic regions of GRL101 (G-protein-coupled receptor-like 101), PKS1 (polyketide synthase 1), PLC1 (phosphoinositide phospholipase C), GLRA1 (glycine receptor subunit alpha-1-like), COL4 (zinc finger protein constants-like 4), CL1 (cathepsin L1-like), and gene LOC110449017 (not yet characterized), as well as at an intergenic region (where SNP Chr13-253787 anchored).
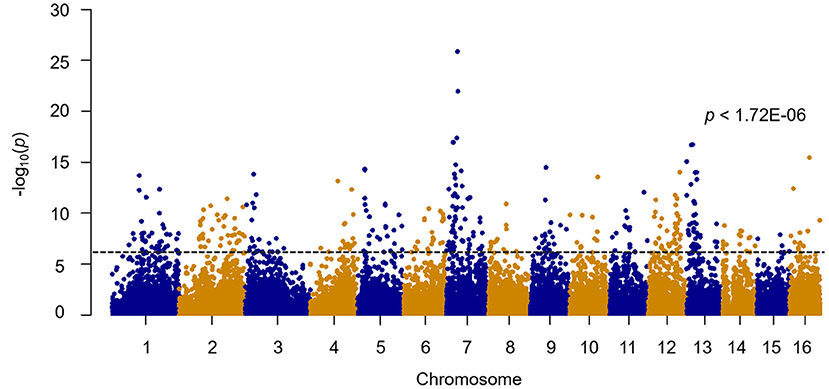
Figure 2. Genome-wide association study (GWAS) of shell color with 29,036 single-nucleotide polymorphisms (SNPs) in bay scallops (p < 1.72E−06).
Among these, the top three loci were distributed in the genic region of chromosome 7 (Figure 3). More specifically, SNP chr7-12764003 was mapped to location 41 bp in the coding sequence (CDS) 7 of GRL101, and SNP chr7-13213864 was cited in position 1,347 bp in CDS 5 of PKS1. For chr7-11899306, an SNP was located at position 14,136 bp in the first intron of PCL1. Subsequently, the top three SNPs were selected to be verified by sequencing the PCR products using the Sanger method by Sangon Biotech (Shanghai, China).
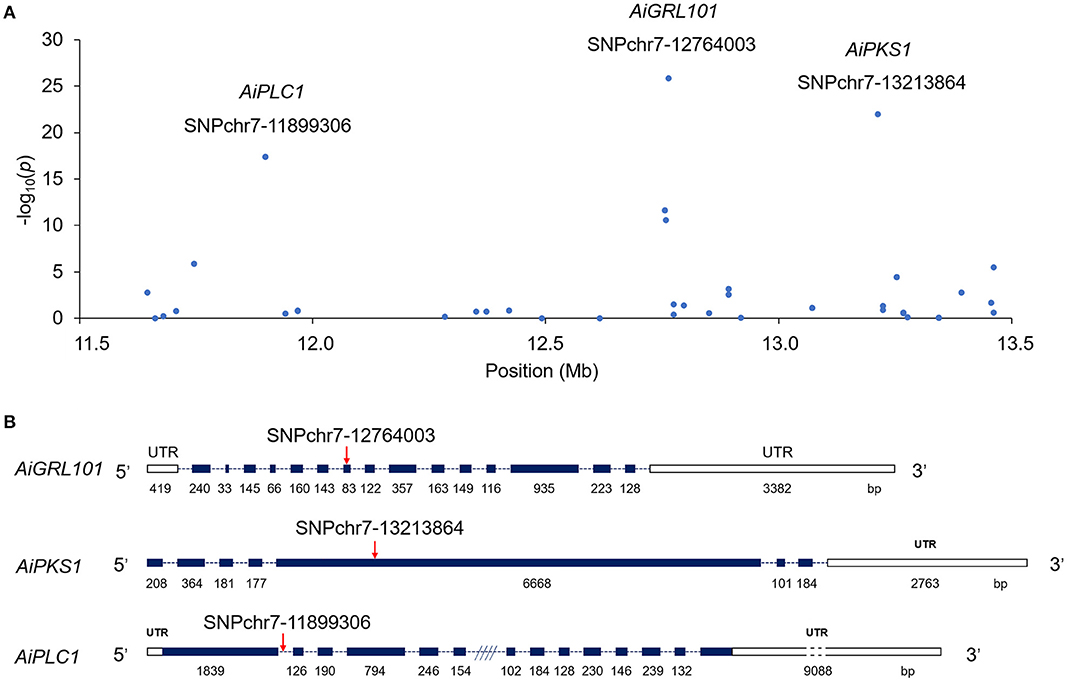
Figure 3. A few neighboring SNP loci on chromosome 7 are independently associated with shell color in the bay scallop. (A) The genic structure of AiGRL101, AiPKS1, and AiPLC1. (B) The blank boxes indicate the 5′UTRs and 3′UTRs (untranslated region); the dark blue boxes represent coding sequence (CDS); the dotted lines indicate the introns; and the red arrows indicate the verified SNPs in responding genes. The CDS are shown relative to their lengths and the SNPs are marked in the accurate positions in the genic sequences.
All the top three SNPs provided by the GWAS results were successfully verified by Sanger sequencing in another separate bay scallop population. The allele and genotype frequency of these three SNP markers are shown in Table 3. For SNP chr7-12764003, dimorphic alleles could be observed, with AA and GG presenting in PP and BP, respectively. As for SNP chr7-13213864, the genotype of AA (100%) was only observed in PP, while another two genotypes of TT and AT were present in BP, with frequencies of 76.67 and 23.33%, respectively. Besides, the genotype is varied in SNP chr7-11899306 (for CC, TT, CT; 76.67, 0, 23.33% in PP and 6.67, 63.33, 30% in BP, respectively). Comparison between PP and BP individuals showed that genotype frequencies of the above three SNP markers were significantly different (p < 0.05). These results support that the verified SNPs might be involved in shell color formation of bay scallop PP and BP.
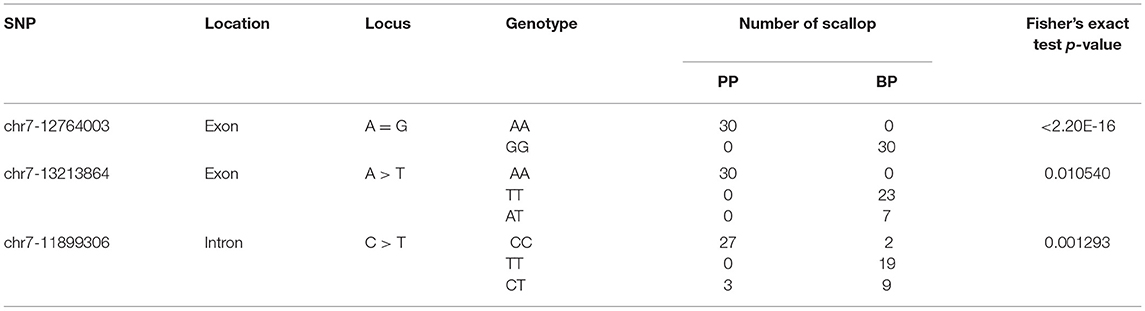
Table 3. Comparison of genotype frequencies of the top three SNPs between PP (N = 30) and BP (N = 30) bay scallops.
After a widely used and less-stringent threshold p-value <1.00E−5 (Marigorta et al., 2018), we obtained 713 significant SNPs annotated in 374 unigenes. Subsequently, GO function annotation and KEGG pathway enrichment were applied to excavate the potential association between these genes and shell color. Results displayed that 38 GO terms were classified into the biological process (BP), cellular component (CC), and molecular function (MF) under level 2 covering 297 genes (Figure 4). In detail, genes related to the cellular process (105 genes), single-organism process (94 genes), and metabolic process (68 genes) showed remarkable terms in BP. Genes related to the membrane (53 genes) displayed the notable term in CC. Genes associated with binding (166 genes) and catalytic activity (80 genes) were highly presented in MF (Figure 4). In terms of KEGG pathway enrichment analysis, a total of 252 genes were acquired annotations and involved in 44 pathways (Figure 5). The above candidate genes were chiefly distributed in metabolism (69 genes), genetic information processing (15 genes), environmental information process (55 genes), cellular process (38 genes), organismal systems (90 genes), and human disease (97 genes) (Figure 5). Specifically, signal transduction (37 genes), global and overview maps (29 genes), infectious diseases (23 genes), cancers (21 genes), and endocrine system (19 genes) were mainly presented. Overall, the result indicated that the extracted GO terms and KEGG pathway are closely related to cellular process, membrane, signaling, and binding, and these possible molecular mechanisms may attribute to the shell color formation.
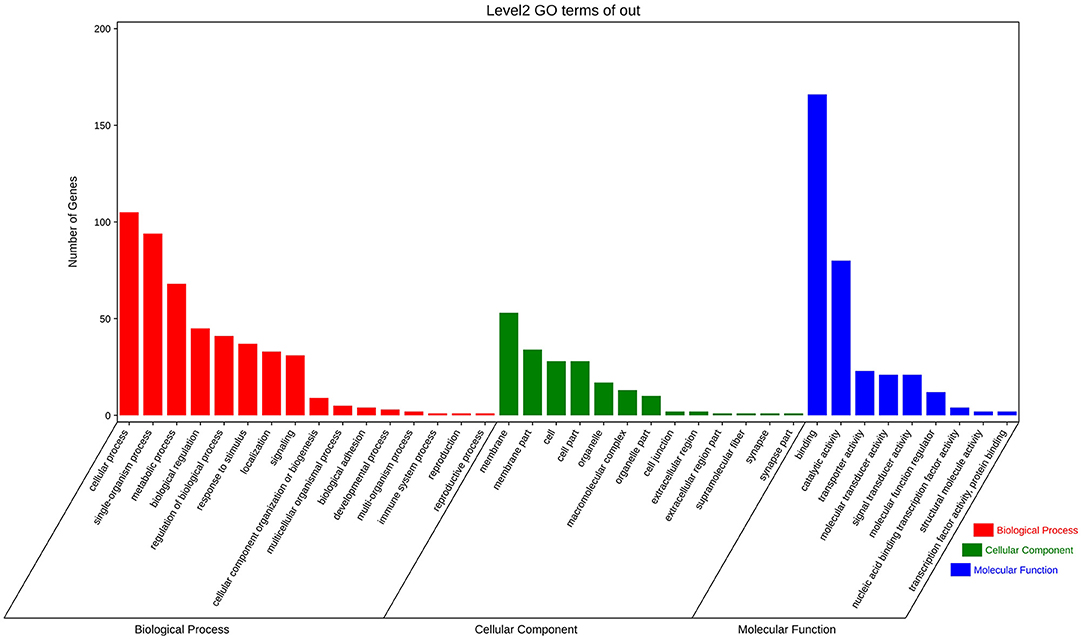
Figure 4. The summary of Gene Ontology (GO) function annotation analysis for candidate genes that comprised significant SNPs (p < 1.00E−5). Each term describes the function of the gene cluster, and the length of colored bars represents the difference of the number of genes.

Figure 5. The summary of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis performed for biological process, cellular component, and molecular function. Functional annotation of candidate selective genes that comprised significant SNPs (p < 1.00E−5).
Annotation results showed that the top three SNPs were located in three genes, including GRL101 (chr7-12764003), PKS1 (chr7-13213864), and PLC1 (chr7-11899306), which were further selected for comparing their expression differences in mantles between PP and BP individuals. As shown in Figure 6, the expression profiles of the three target genes were ubiquitous in both PP and BP individuals but exhibited distinct expression patterns. Specifically, an extremely significant higher (6.43-fold, p < 0.01) expression level of GRL101 was detected in PP than that in BP. A similar phenomenon was also observed in PLC1, which displayed a prominently increasing expression level in PP (6.48-fold, p < 0.05) compared with BP. On the contrary, the expression of PSK1 was notably correlated with black-brown shell color rather than purple-red shell color (12.02-fold of BP compared with PP). Overall, in the bay scallop, the significantly higher expression profiles of GRL101 and PLC1 were detected in PP, implying that they may play a pivotal role in purple-red shell formation. Besides, PSK1 is principally expressed in BP instead of PP, suggesting that it may involve black-brown pigmentation during shell formation.
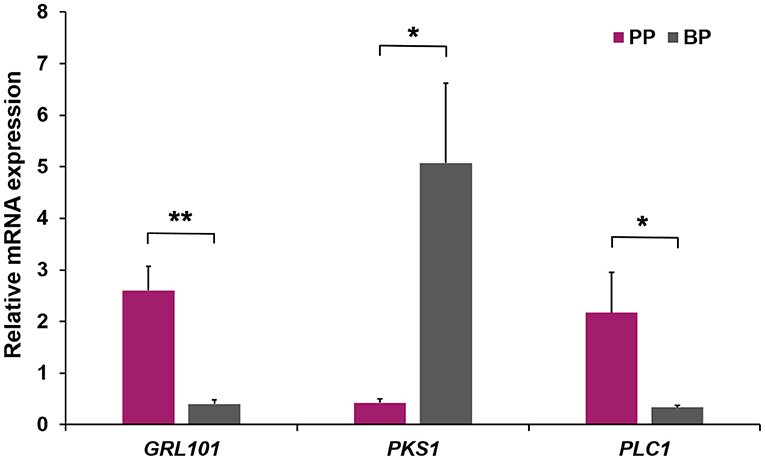
Figure 6. The relative expression levels of GRL101, PKS1, and PLC1 in the mantle of PP and BP bay scallops. Three replicates were performed for each adult mantle, and three technical replicates were conducted for each PCR. The comparison of the expression levels of GRL101, PKS1, and PLC1 in the mantle of PP and BP bay scallops was performed using an independent sample t-test. * and ** indicate statistically significant differences (p < 0.05 and p < 0.01, respectively).
Aquaculture breeding researches have confirmed that color trait in most aquaculture species is heritable, including red/white flesh color in salmon (Iwamoto et al., 1990), carapace/hepatopancreas color in crab (Li Q. et al., 2019), black/white shell color in oyster (Xu et al., 2019), white/orange in scallop adductor muscle (Li X. et al., 2019), etc. Particularly, the bay scallop is polymorphic for the background color of the shell and presents a highly variable distribution of overlying pigments (Clarke, 1965). Although previous investigations showed one or several major genes contributed to shell color diversity of bay scallop (Adamkewicz and Castagna, 1988; Qin et al., 2007; Teng et al., 2018), the precise loci/genes for specific color difference and the mechanism underneath shell color formation in bay scallop are elusive. With rapid development in biotechnology, constantly upgraded methods for studying shell color polymorphisms in bay scallop would dredge out specific genes behind target trait. GWAS has been proven to be an efficient approach to elucidate the genetic variations related to complex phenotypes and to identify the pivotal candidate genes of target economic traits, thereby providing referential makers for the selective breeding projects (Luo et al., 2012). By this method, 469 SNPs that displayed statistically significant differences between PP and BP were identified in our research. As the Manhattan plot displayed, the top value of –log10 (p-value) reached nearly 26, which is much more stringent than 5.8 (p <1.72E−06) that referred to Bonferroni correction, demonstrating that our result is reliable (false positives are rare). Intriguingly, most of the significant SNPs associated with shell color were primarily allocated in chromosome 7 of bay scallop. With the similarity to that in Yesso scallop, the major SNPs associated with shell color were located in chromosome 11 (Zhao et al., 2017), and almost all the significant SNPs associated with carotenoid coloration in scallop adductor muscle were distributed in chromosome 8 (Li X. et al., 2019). Based on the fact that most qualitative traits are typically determined by one or several pivotal loci/genes, we speculated that color variants in scallop were potentially regulated by limited candidate loci/genes gathered closely in association with the genomic region(s) of the specific linkage group. In addition, the tandem or proximal (in a nearby chromosomal region but not adjacent) locations may not only facilitate coordinate expressions of candidate genes but also guarantee efficient expression regulations by pivotal loci.
The top three SNPs with a p-value <1.00E−17 were successfully verified in another separate population via Sanger sequencing. The extremely significant (p < 0.01) or significant (p < 0.05) differences of the three selected loci between PP and BP suggested that these SNPs might be responsible for shell color formation and potentially used as selective breeding markers in shell color-specific projects of bay scallop. In addition, the top eight SNPs (p <1.00E−15) were principally and separately located in genes (exon or intron) except on locus (SNP chr13-253787) located in intergenic regions. A similar phenomenon has also been concluded in the shell color of the Yesso scallop (Zhao et al., 2017). The above results revealed that color traits in scallops may be principally controlled by the coding regions (where SNPs are located) of most critical candidate genes (Cargill et al., 1999) and meanwhile regulated through crucial SNPs within introns or intergenic regions via transcriptional regulation (Sturm et al., 2008; Visser et al., 2014).
Bay scallop shell color significance-associated candidate genes, namely, GRL101, PKS1, and PLC1, where the top three SNPs were located, were further selected for comparing their expression differences between PP and BP individuals. The scallop mantle constantly contacts with the shell; therefore, the mantle is considered to manufacture and transport pigments to the shell along the growing edge during shell formation (Fowler et al., 1992). Upon this basis, further expression patterns of the top three candidate genes were characterized in the mantle of PP and BP individuals, respectively. In this study, the mRNA expression level of GRL101 in PP was over six times that in BP (p < 0.05), suggesting GRL101 might function in purple-red shell coloration. As reported, GRL101 belongs to the G-protein-coupled receptors (GPCRs) superfamily (Karnik et al., 2003) that exerts multifunctions, such as a variety of hormones and neurotransmitters regulation (Nilaweera et al., 2006, 2008; Rosenbaum et al., 2009). Specifically, a set number of GPCRs were proven to be involved in pigments formation, known as responding to red-pigment concentrating hormone (RPCH) in Carcinus maenas (Alexander et al., 2018) and interceding pigment granule dispersion in Xenopus laevis melanophores (Teh and Sugden, 2001). Furthermore, GPCRs activated by chromatophores can affect pigment redistribution to alter the color appearance of the animal (Lerner, 1994). In particular, visual pigment rhodopsin (the first order of GPCRs) performs functions on the absorption and conversion of light into chemical signals (Smith, 2010). Based on this, our results implied that GRL101 involved and functioned in the purple-red color shell formation of bay scallop, whereas GRL101 protein was regarded as an orphan receptor performing an uncertain function that was extensively explored in humans (Gene ID: 83550), mice (Gene ID: 245424), sea urchins (Gene ID: 115922752), king scallops (Gene ID: 117318188), and other species. Whether the GRL101 in the bay scallop is also considered an orphan receptor still needs to be further investigated. Coincidently, the majority of GPCRs were coupled to the phosphoinositide signaling pathway where the third candidate gene PLC1 played a pivotal role (Tobin, 1997). Previous researches reported that PLC1 has been classified into an important class of enzymes involved in lipids-related signaling in plants (Rupwate and Rajasekharan, 2012) and acted as signal transducers to generate messengers participating in lipid metabolism in mammals (Essen et al., 1996). Furthermore, the lipid-related genes were proved to be closely associated with carotenoids (Zhang Y. et al., 2014) that contributed to the formation of reddish-orange shells in Yesso scallop (Zhao et al., 2017). Hence, it might then make sense that much higher expression of PLC1 detected in PP individuals leads to the accumulation of carotenoid which in turn resulted in a red-colored shell. Therefore, our results suggest that PLC1 might be related to lipid-associated carotenoid absorption and/or transportation. Collectively, we speculated that GRL101 might perform a common feature of coloration-related GPCRs and activate PLC1 in the phosphoinositide signaling pathway to affect lipid metabolism, including lipid-associated carotenoid regulation, resulting in the purple-red visualization of PP.
Another candidate gene PKS1 belongs to polyketide synthase-related genes (Moriwaki et al., 2004), and the ancient and diversified family members have been well studied in animal fatty acid synthase (Kroken et al., 2003). As reported, plenty of microorganisms employed PKS to produce pigments and other products of intermediate metabolism (Hutchinson, 2003). For example, the biosynthesis of black pigment melanin derived by PKS1 was reported in Colletotrichum lagenarium (Takano et al., 1995) and Aspergillus fumigatus (Heinekamp et al., 2013), respectively. Unlike PKS1 as the pivotal component in melanin biosynthesis in microorganisms, melanin biosynthesis in various tissues of vertebrates was prevailingly affected by tyrosinase genes (Sato et al., 2001). Similarly, melanin biosynthesis was more inclined to be regulated by tyrosinase that contributes to black-brown shell pigmentation in most mollusks (Bai et al., 2013; Sun et al., 2015). However, our study showed that a higher expression level (12.02-fold, p < 0.05) of PKS1 in the black-brown shell individuals was observed than that in purple-red ones; therefore, we speculated that black pigmentation in BP might be regulated by PKS1 as it functioned in microorganisms. Although melanin synthesis in scallop has not been reported yet, the typical biosynthesis pathway (in which tyrosinase is involved) as an alternative method seems worth further study.
Generally, causal SNPs were identified to be strongly associated with target traits, especially the SNPs that were located in the coding region would affect traits directly by changing their encoding amino acid and therefore resulting in the alteration of protein (Seong et al., 2011). However, in this study, the two most significant SNPs, namely, chr7-12764003, and chr7-13213864, were separately located in the CDS of AiGRL101 and AiPSK1, caused synonymous mutations in target amino acids. We speculated that SNPs located in the CDS of AiGRL101 and AiPSK1 might not control the scallop shell coloration directly via mutation of amino acids, but mediate regulating candidate genes by affecting codon bias (Plotkin and Kudla, 2011), translation efficiency (Miyuki and Kentaro, 2013), mRNA secondary structure stability (Chamary and Hurst, 2005), and splicing (Li et al., 2020). Furthermore, linkage disequilibrium (LD) analysis of the three closely arranged SNPs (Figure 2) via MEGA (Version 10.1.8) (Sudhir et al., 2018) showed that LD coefficient r2 between each pair of SNPs was higher than 0.7 (ranged from 0.72 to 0.78), demonstrating that shell color in bay scallop may be regulated by closely linked major SNPs and/or specific interactions among them which potentially affect the expression of candidate genes resulting in shell color formation and differentiation. Further research is needed for investigating the potential molecular mechanism of synonymous mutation of CDS in candidate genes and interactions of target genes in regulating shell coloration of bay scallops.
Furthermore, 713 SNPs (p <1.00E−5) significantly associated with bay scallop shell color were mapped to 374 unigenes, followed by GO function annotation and KEGG pathway enrichment for elucidating their important molecular mechanism (Wang et al., 2019). GO function annotation in this study primarily participated in cellular process, membrane, and binding function, which is similar to previous research that cellular process and binding are the largest group of biological process and molecular function in the transcriptomic analysis of red/white-colored valves of moon scallop (Huang et al., 2015), as well as different shell color lines in Yesso scallop (Ding et al., 2015). Furthermore, transcriptome sequencing of orange shell color bay scallops revealed that differentially expressed genes were mainly annotated in signal transduction, metabolism, human diseases, and organismal systems (Teng et al., 2018) and that is in accordance with our result of KEGG pathway analysis. Taken together, the above results suggest that the shell color of the bay scallop is not only dominated by several loci/genes but also modified by plenty of related minor loci/genes.
We identified SNPs associated with the purple-red and black-brown shell colors bay scallop through GWAS. The genomic regions associated with shell color were mainly located in chromosome 7. The top three SNPs were located in the genic region of GRL101, PKS1, and PLC1 and successfully verified in another separate population. Expression patterns analyses suggested that GRL101 and PLC1 potentially functioned in purple-red shell coloration, and black pigmentation in BP might be principally regulated by PKS1. GO function annotation and KEGG pathway enrichment analyses revealed the possible molecular mechanism of shell color formation. Taken together, this evidence collectively provides valuable information for deciphering the phenotype differences of shell color and helps in the molecular marker-assisted breeding in bay scallops.
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI GenBank (accession number: PRJNA689862).
The animal study was reviewed and approved by Ocean University of China.
QX, JH, and ZB conceived and designed the experiments. XZ, JZ, and XHo collected the samples. QX, XZ, PL, and JL performed the experiments. XZ, QX, JZ, and XHu analyzed the data. QX and XZ wrote the manuscript. All authors have read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China-Shandong Joint Fund (U1706203), the Earmarked Fund for Agriculture Seed Improvement Project of Shandong Province (2020LZGC016), Sanya Yazhou Bay Science and Technology City (SKJC-KJ-2019KY01), and the China Agriculture Research System of MOF and MARA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adamkewicz, L., and Castagna, M. (1988). Genetics of shell color and pattern in the bay scallop Argopecten irradians. J. Heredity 79, 14–17. doi: 10.1093/oxfordjournals.jhered.a110436
Alexander, D. H., Novembre, J., and Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Alexander, J. L., Oliphant, A., Wilcockson, D. C., Audsley, N., Down, R. E., Lafont, R., et al. (2018). Functional characterization and signaling systems of corazonin and red pigment concentrating hormone in the green shore crab, Carcinus maenas. Front. Neurosci. 11:752. doi: 10.3389/fnins.2017.00752
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Bai, Z., Zheng, H., Lin, J., Wang, G., and Li, J. (2013). Comparative analysis of the transcriptome in tissues secreting purple and white nacre in the pearl mussel Hyriopsis cumingii. PLoS ONE 8:e53617. doi: 10.1371/journal.pone.0053617
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Cargill, M., Altshuler, D., Ireland, J., Sklar, P., Ardlie, K., Patil, N., et al. (1999). Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22, 231–238. doi: 10.1038/10290
Chamary, J. V., and Hurst, L. D. (2005). Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol. 6:R75. doi: 10.1186/gb-2005-6-9-r75
Chang, C. C., Chow, C. C., Tellier, L. C., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7. doi: 10.1186/s13742-015-0047-8
Clarke, A. H. (1965). The scallop superspecies Aequipecten irradians (Lamarck). Malacologia 2:161–168.
Cole, T. J. (1975). Inheritance of juvenile shell colour of the oyster drill Urosalpinx cinerea. Nature 257, 794–795. doi: 10.1038/257794a0
Comfort, A. (1950). Acid-soluble pigments of molluscan shells. 5. Identity of some subsidiary fractions derived from Pinctada vulgaris. Biochem. J. 47, 254–255. doi: 10.1042/bj0470254
Comfort, A. (1951). The pigmentation of molluscan shells. Biol. Rev. 26, 285–301. doi: 10.1111/j.1469-185X.1951.tb01358.x
Cong, R., Kong, L., Yu, H., and Li, Q. (2014). Association between polymorphism in the insulin receptor-related receptor gene and growth traits in the Pacific oyster Crassostrea gigas. Biochem. Syst. Ecol. 54, 144–149. doi: 10.1016/j.bse.2014.02.003
D'Iischia, M., Wakamatsu, K., Napolitano, A., Briganti, S., Garcia-Borron, J. C., Kovacs, D., et al. (2013). Melanins and melanogenesis: methods, standards, protocols. Pigm. Cell Melanoma Res. 26, 616–633. doi: 10.1111/pcmr.12121
Ding, J., Zhao, L., Chang, Y., Zhao, W., Du, Z., and Hao, Z. (2015). Transcriptome sequencing and characterization of Japanese scallop Patinopecten yessoensis from different shell color lines. PLoS ONE 10:e0116406. doi: 10.1371/journal.pone.0116406
Elek, J., and Adamkewicz, S. L. (1990). Polymorphism for shell color in the Atlantic bay scallop Argopecten irradians (Lamarck) (Mollusca: Bivalvia) on Martha's Vineyard island. Am. Malacol. Bull. 7, 117–126.
Essen, L. O., Perisic, O., Cheung, R., Katan, M., and Williams, R. L. (1996). Crystal structure of a mammalian phosphoinositide-specific phospholipase Cδ. Nature 380, 595–602. doi: 10.1038/380595a0
Fowler, D. R., Meinhardt, H., and Prusinkiewicz, P. (1992). Modeling seashells. Acm Siggraph Comput. Graph. 26, 379–387. doi: 10.1145/142920.134096
Fu, X., Dou, J., Mao, J., Su, H., Jiao, W., Zhang, L., et al. (2013). RADtyping: an integrated package for accurate de novo codominant and dominant RAD genotyping in mapping populations. PLoS ONE 8:e79960. doi: 10.1371/journal.pone.0079960
Han, Z., Li, Q., Liu, S., and Kong, L. (2020). Crossbreeding of three different shell color lines in the Pacific oyster reveals high heterosis for survival but low heterosis for growth. Aquaculture 529:735621. doi: 10.1016/j.aquaculture.2020.735621
Hao, S., Hou, X., Wei, L., Li, J., Li, Z., and Wang, X. (2015). Extraction and identification of the pigment in the adductor muscle scar of pacific oyster Crassostrea gigas. PLoS ONE 10:e0142439. doi: 10.1371/journal.pone.0142439
Heinekamp, T., Thywißen, A., Macheleidt, J., Keller, S., Valiante, V., and Brakhage, A. A. (2013). Aspergillus fumigatus melanins: interference with the host endocytosis pathway and impact on virulence. Front. Microbiol. 3:440. doi: 10.3389/fmicb.2012.00440
Heller, J. (1992). Shell colour variation in Bullia digitalis, a sand-dwelling, intertidal whelk (Gastropoda: Prosobranchia). Biol. J. Linn. Soc. 46, 247–258. doi: 10.1111/j.1095-8312.1992.tb00863.x
Hoang, T. H., Stone, D. A., Duong, D. N., Bansemer, M. S., Harris, J. O., and Qin, J. G. (2017). Colour change of greenlip abalone (Haliotis laevigata Donovan) fed formulated diets containing graded levels of dried macroalgae meal. Aquaculture 468, 278–285. doi: 10.1016/j.aquaculture.2016.10.027
Hu, X., Bao, Z., Hu, J., Shao, M., Zhang, L., Bi, K., et al. (2006). Cloning and characterization of tryptophan 2, 3-dioxygenase gene of Zhikong scallop Chlamys farreri (Jones and Preston 1904). Aquac. Res. 37, 1187–1194. doi: 10.1111/j.1365-2109.2006.01546.x
Huang, R., Zheng, Z., Wang, Q., Zhao, X., Deng, Y., Jiao, Y., et al. (2015). Mantle branch-specific RNA sequences of moon scallop Amusium pleuronectes to identify shell color-associated genes. PLoS ONE 10:e0141390. doi: 10.1371/journal.pone.0141390
Hutchinson, C. R. (2003). Polyketide and non-ribosomal peptide synthases: falling together by coming apart. Proc. Nat. Acad. Sci. 100, 3010–3012. doi: 10.1073/pnas.0730689100
Ikeda, M., Takahashi, A., Kamatani, Y., Okahisa, Y., Kunugi, H., Mori, N., et al. (2018). A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol. Psychiatry 23, 639–647. doi: 10.1038/mp.2016.259
Innes, D. J., and Leslie, E. H. (1977). Inheritance of a shell-color polymorphism in the mussel. J. Heredity 68, 203–204. doi: 10.1093/oxfordjournals.jhered.a108814
Iwamoto, R. N., Myers, J. M., and Hershberger, W. K. (1990). Heritability and genetic correlations for flesh coloration in pen-reared coho salmon. Aquaculture 86, 181–190. doi: 10.1016/0044-8486(90)90111-Y
Johnson, R. C., Nelson, G. W., Troyer, J. L., Lautenberger, J. A., Kessing, B. D., Winkler, C. A., et al. (2010). Accounting for multiple comparisons in a genome-wide association study (GWAS). BMC Genomics 11:724. doi: 10.1186/1471-2164-11-724
Karnik, S. S., Gogonea, C., Patil, S., Saad, Y., and Takezako, T. (2003). Activation of G-protein-coupled receptors: a common molecular mechanism. Trends Endocrinol. Metabol. 14, 431–437. doi: 10.1016/j.tem.2003.09.007
Kim, H. C., Lee, J. Y., Sung, H., Choi, J. Y., Park, S. K., Lee, K. M., et al. (2012). A genome-wide association study identifies a breast cancer risk variant in ERBB4 at 2q34: results from the Seoul Breast Cancer Study. Breast Cancer Res. 14:R56. doi: 10.1186/bcr3158
Kraeuter, J., Adamkewicz, L., Castagna, M., Wall, R., and Karney, R. (1984). Rib number and shell color in hybridized subspecies of the Atlantic bay scallop, Argopecten irradians. Nautilus 98, 17–20.
Kroken, S., Glass, N. L., Taylor, J. W., Yoder, O. C., and Turgeon, B. G. (2003). Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Nat. Acad. Sci. 100, 15670–15675. doi: 10.1073/pnas.2532165100
Lerner, M. R. (1994). Tools for investigating functional interactions between ligands and G-protein-coupled receptors. Trends Neurosci. 17, 142–146. doi: 10.1016/0166-2236(94)90087-6
Li, M., Lu, X., Dong, J., Yao, Z., Wu, Y, Rao, H., et al. (2020). A synonymous mutation in exon 39 of FBN1 causes exon skipping leading to Marfan syndrome. Genomics 112, 3856–3861. doi: 10.1016/j.ygeno.2020.06.024
Li, N., Hu, J., Wang, S., Cheng, J., Hu, X., Lu, Z., et al. (2010). Isolation and identification of the main carotenoid pigment from the rare orange muscle of the Yesso scallop. Food Chem. 118, 616–619. doi: 10.1016/j.foodchem.2009.05.043
Li, Q., Liu, Q., Zhang, C., Cheng, Y., and Wu, X. (2019). Can color-related traits in the Chinese mitten crab (Eriocheir sinensis) be improved through quantitative genetic inheritance? Aquaculture 512:734355. doi: 10.1016/j.aquaculture.2019.734355
Li, R., Yu, C., Li, Y., Lam, T. W., Yiu, S. M., Kristiansen, K., et al. (2009). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. doi: 10.1093/bioinformatics/btp336
Li, X., Bai, Z., Luo, H., Wang, G., and Li, J. (2014). Comparative analysis of total carotenoid content in tissues of purple and white inner-shell color pearl mussel, Hyriopsis cumingii. Aquacult. Int. 22, 1577–1585. doi: 10.1007/s10499-014-9766-6
Li, X., Wang, S., Xun, X., Zhang, M., Wang, S., Li, H., et al. (2019). A carotenoid oxygenase is responsible for muscle coloration in scallop. Biochimica et Biophysica Acta (BBA)—Molecul. Cell Biol. Lipids 1864, 966–975. doi: 10.1016/j.bbalip.2019.03.003
Liu, X., Wu, F., and Zhang, G. (2007). Genetic basis of tangerine shell color variation in the Pacific abalone, Haliotis discus hannai. Aquaculture 272:S286. doi: 10.1016/j.aquaculture.2007.07.127
Luo, W., Cheng, D., Chen, S., Wang, L., Li, Y., Ma, X., et al. (2012). Genome-wide association analysis of meat quality traits in a porcine large white × minzhu intercross population. Int. J. Biol. Sci. 8:580. doi: 10.7150/ijbs.3614
Mao, J., Lv, J., Miao, Y., Sun, C., Hu, L., Zhang, R., et al. (2013). Development of a rapid and efficient method for non-lethal DNA sampling and genotyping in scallops. PLoS ONE 8:e68096. doi: 10.1371/journal.pone.0068096
Marchais, V., Jolivet, A., Herv,é, S., Roussel, S., Schöne, B. R., Grall, J., et al. (2017). New tool to elucidate the diet of the ormer Haliotis tuberculata (L.): Digital shell color analysis. Mar. Biol. 164:71. doi: 10.1007/s00227-017-3103-3
Marigorta, U. M., Rodríguez, J. A., Gibson, G., and Navarro, A. (2018). Replicability and prediction: lessons and challenges from GWAS. Trends Gene. 34, 504–517. doi: 10.1016/j.tig.2018.03.005
Martin, P. M., Palhiere, I., Ricard, A., Tosser-Klopp, G., and Rupp, R. (2016). Genome wide association study identifies new loci associated with undesired coat color phenotypes in Saanen goats. PLoS ONE 11:e0152426. doi: 10.1371/journal.pone.0152426
Miyuki, T., and Kentaro, M. (2013). Directed evolution study unveiling key sequence factors that affect translation efficiency in Escherichia coli. J. Biosci. Bioeng. 116, 540–545. doi: 10.1016/j.jbiosc.2013.05.013
Moriwaki, A., Kihara, J., Kobayashi, T., Tokunaga, T., Arase, S., and Honda, Y. (2004). Insertional mutagenesis and characterization of a polyketide synthase gene (PKS1) required for melanin biosynthesis in Bipolaris oryzae. FEMS Microbiol. Lett. 238, 1–8. doi: 10.1111/j.1574-6968.2004.tb09729.x
Nie, H., Jiang, K., Jiang, L., Huo, Z., Ding, J., and Yan, X. (2020). Transcriptome analysis reveals the pigmentation related genes in four different shell color strains of the Manila clam Ruditapes philippinarum. Genomics 112, 2011–2020. doi: 10.1016/j.ygeno.2019.11.013
Nilaweera, K. N., Ozanne, D., Wilson, D., Mercer, J. G., Morgan, P. J., and Barrett, P. (2006). G protein-coupled receptor 101 mRNA expression in the mouse brain: altered expression in the posterior hypothalamus and amygdala by energetic challenges. J. Neuroendocrinol. 19, 34–45. doi: 10.1111/j.1365-2826.2006.01502.x
Nilaweera, K. N., Wilson, D., Bell, L., Mercer, J. G., Morgan, P. J., and Barrett, P. (2008). G protein-coupled receptor 101 mRNA expression in supraoptic and paraventricular nuclei in rat hypothalamus is altered by pregnancy and lactation. Brain Res. 1193, 76–83. doi: 10.1016/j.brainres.2007.11.048
Ning, X., Li, X., Wang, J., Zhang, X., Kong, L., Meng, D., et al. (2019). Genome-wide association study reveals E2F3 as the candidate gene for scallop growth. Aquaculture 511:734216. doi: 10.1016/j.aquaculture.2019.734216
Oren, E., Tzuri, G., Vexler, L., Dafna, A., Meir, A., Faigenboim, A., et al. (2019). The multi-allelic APRR2 gene is associated with fruit pigment accumulation in melon and watermelon. J. Exp. Bot. 70, 3781–3794. doi: 10.1093/jxb/erz182
Owens, B. F., Mathew, D., Diepenbrock, C. H., Tiede, T., Wu, D., Mateos-Hernandez, M., et al. (2019). Genome-wide association study and pathway-level analysis of kernel color in maize. G3: Genes, Genomes, Genetics 9, 1945–1955. doi: 10.1534/g3.119.400040
Palmer, A. R. (1985). Genetic basis of shell variation in Thais emarginata (Prosobranchia, Muricacea). I. Banding in populations from Vancouver Island. The Biological Bulletin 169, 638–651. doi: 10.2307/1541306
Peignon, J. M., Gerard, A., Naciri, Y., Ledu, C., and Phelipot, P. (1995). Analysis of shell colour determinism in the Manila clam Ruditapes philippinarum. Aquat. Living Resour. 8, 181–189. doi: 10.1051/alr:1995015
Petersen, J. L., Baerwald, M. R., Ibarra, A. M., and May, B. (2012). A first-generation linkage map of the Pacific lion-paw scallop (Nodipecten subnodosus): Initial evidence of QTL for size traits and markers linked to orange shell color. Aquaculture 350, 200–209. doi: 10.1016/j.aquaculture.2012.03.039
Plotkin, J. B., and Kudla, G. (2011). Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 12, 32–42. doi: 10.1038/nrg2899
Qin, Y., Liu, X., Zhang, H., Zhang, G., and Guo, X. (2007). Identification and mapping of amplified fragment length polymorphism markers linked to shell color in bay scallop, Argopecten irradians (Lamarck, 1819). Marine Biotechnol. 9, 66–73. doi: 10.1007/s10126-006-6076-7
Rosenbaum, D. M., Rasmussen, S. G., and Kobilka, B. K. (2009). The structure and function of G-protein-coupled receptors. Nature 459, 356–363. doi: 10.1038/nature08144
Rupwate, S. D., and Rajasekharan, R. (2012). Plant phosphoinositide-specific phospholipase C: an insight. Plant Signal. Behav. 7, 1281–1283. doi: 10.4161/psb.21436
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Second Edition. Cold Spring Harbor Laboratory Press.
Sato, S., Tanaka, M., Miura, H., Takeuchi, T., Yamamoto, H., Ikeo, K., et al. (2001). Functional conservation of the promoter regions of vertebrate tyrosinase genes. J. Investigat. Dermatol. Sympo. Proceed. 6, 10–18. doi: 10.1046/j.0022-202x.2001.00008.x
Seong, J., Dong, S. S., Park, K. D., Lee, H. K., and Hong, S. K. (2011). Identification and analysis of MC4R polymorphisms and their association with economic traits of Korean cattle (Hanwoo). Mol. Biol. Rep. 39, 3597–3601. doi: 10.1007/s11033-011-1133-3
Shumway, S. E., and Parsons, J. (2006). Scallops: biology, ecology and aquaculture new edition. Developm. Aquacult. Fisher. Sci. 35. doi: 10.1016/0022-0981(92)90183-B
Smith, S. O. (2010). Structure and activation of the visual pigment rhodopsin. Annu. Rev. Biophys. 39, 309–328. doi: 10.1146/annurev-biophys-101209-104901
Sokolova, I. M., and Berger, V. J. (2000). Physiological variation related to shell colour polymorphism in White Sea Littorina saxatilis. J. Exp. Mar. Biol. Ecol. 245, 1–23. doi: 10.1016/S0022-0981(99)00132-X
Stemmer, K., and Nehrke, G. (2014). The distribution of polyenes in the shell of Arctica islandica from North Atlantic localities: a confocal Raman microscopy study. J. Molluscan Stud. 80, 365–370. doi: 10.1093/mollus/eyu033
Sturm, R. A., Duffy, D. L., Zhao, Z. Z., Leite, F. P., Stark, M. S., Hayward, N. K., et al. (2008). A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am. J. Hum. Genetics 82, 424–431. doi: 10.1016/j.ajhg.2007.11.005
Sudhir, K., Glen, S., Li, M., Christina, K., and Koichiro, T. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Sukumaran, S., and Yu, J. (2014). Association mapping of genetic resources: achievements and future perspectives. Genom. Plant Genetic Resourc. 1, 207–235. doi: 10.1007/978-94-007-7572-5_9
Sun, X., Liu, Z., Zhou, L., Wu, B., Dong, Y., and Yang, A. (2016). Integration of next generation sequencing and EPR analysis to uncover molecular mechanism underlying shell color variation in scallops. PLoS ONE 11:e0161876. doi: 10.1371/journal.pone.0161876
Sun, X., Wu, B., Zhou, L., Liu, Z., Dong, Y., and Yang, A. (2017). Isolation and characterization of melanin pigment from Yesso scallop Patinopecten yessoensis. J. Ocean Univ. China 16, 279–284. doi: 10.1007/s11802-017-3162-6
Sun, X., Yang, A., Wu, B., Zhou, L., and Liu, Z. (2015). Characterization of the mantle transcriptome of yesso scallop (Patinopecten yessoensis): identification of genes potentially involved in biomineralization and pigmentation. PLoS ONE 10:e0122967. doi: 10.1371/journal.pone.0122967
Takano, Y., Kubo, Y., Shimizu, K., Mise, K., Okuno, T., and Furusawa, I. (1995). Structural analysis of PKS1, a polyketide synthase gene involved in melanin biosynthesis in Colletotrichum lagenarium. Molecul. Gene. Genet. MGG 249, 162–167. doi: 10.1007/BF00290362
Teh, M. T., and Sugden, D. (2001). An endogenous 5-HT7 receptor mediates pigment granule dispersion in Xenopus laevis melanophores. Br. J. Pharmacol. 132, 1799–1808. doi: 10.1038/sj.bjp.0703988
Teng, W., Cong, R., Que, H., and Zhang, G. (2018). De novo transcriptome sequencing reveals candidate genes involved in orange shell coloration of bay scallop Argopecten irradians. J. Oceanol. Limnol. 36, 1408–1416. doi: 10.1007/s00343-018-7063-3
Tobin, A. B. (1997). Phosphorylation of phospholipase C-coupled receptors. Pharmacol. Therapeut. 75, 135–151. doi: 10.1016/S0163-7258(97)00053-3
Underwood, A. J., and Creese, R. G. (1976). Observations on the biology of the trochid gastropod Austrocochlea constricta (Lamarck) (Prosobranchia). II. The effects of available food on shell-banding pattern. J. Experim. Marine Biol. Ecol. 23, 229–240. doi: 10.1016/0022-0981(76)90022-8
Visser, M., Palstra, R. J., and Kayser, M. (2014). Human skin color is influenced by an intergenic DNA polymorphism regulating transcription of the nearby BNC2 pigmentation gene. Hum. Mol. Genet. 23, 5750–5762. doi: 10.1093/hmg/ddu289
Wang, C., Liu, B., Liu, X., Ma, B., Zhao, Y., Zhao, X., et al. (2017). Selection of a new scallop strain, the Bohai Red, from the hybrid between the bay scallop and the Peruvian scallop. Aquaculture 479, 250–255. doi: 10.1016/j.aquaculture.2017.05.045
Wang, L., Zhang, H., Song, L., and Guo, X. (2007). Loss of allele diversity in introduced populations of the hermaphroditic bay scallop Argopecten irradians. Aquaculture 271, 252–259. doi: 10.1016/j.aquaculture.2007.06.020
Wang, S., Liu, P., Lv, J., Li, Y., Cheng, T., Zhang, L., et al. (2016). Serial sequencing of isolength RAD tags for cost-efficient genome-wide profiling of genetic and epigenetic variations. Nat. Protoc. 11, 2189–2200. doi: 10.1038/nprot.2016.133
Wang, X., Ding, S., Yin, D., Song, J., and Chang, Y. (2020). Response to selection for growth in the second generation of two shell color lines of the bay scallop Argopecten irradians. Aquaculture 528:735536. doi: 10.1016/j.aquaculture.2020.735536
Wang, Y., Liu, L., and Chen, Z. (2019). Transcriptome profiling of cervical cancer cells acquired resistance to cisplatin by deep sequencing. Artific. Cells Nanomed. Biotechnol. 47, 2820–2829. doi: 10.1080/21691401.2019.1637882
Williams, S. T., Ito, S., Wakamatsu, K., Goral, T., Edwards, N. P., Wogelius, R. A., et al. (2016). Identification of shell colour pigments in marine snails Clanculus pharaonius and C. margaritarius (Trochoidea; Gastropoda). PLOS ONE 11, e0156664. doi: 10.1371/journal.pone.0156664
Xu, C., Li, Q., Yu, H., Liu, S., Kong, L., and Chong, J. (2019). Inheritance of shell pigmentation in Pacific oyster Crassostrea gigas. Aquaculture 512:734249. doi: 10.1016/j.aquaculture.2019.734249
Yue, X., Nie, Q., Xiao, G., and Liu, B. (2015). Transcriptome analysis of shell color-related genes in the clam Meretrix. Marine Biotechnol. 17, 364–374. doi: 10.1007/s10126-015-9625-0
Zhang, G., Zhang, W., Ye, R., Fang, A., Ren, G., Zheng, R., et al. (2016). Analysis of selective breeding of nacre color in two strains of Hyriopsis cumingii Lea based on the cielab colorspace. J. Shellfish Res. 35, 225–229. doi: 10.2983/035.035.0124
Zhang, J., Kobert, K., Flouri, T., and Stamatakis, A. (2014). PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620. doi: 10.1093/bioinformatics/btt593
Zhang, S., Wang, H., Yu, J., Jiang, F., Yue, X., and Liu, B. (2018). Identification of a gene encoding microphthalmia-associated transcription factor and its association with shell color in the clam Meretrix petechialis. Comparat. Biochem. Physiol. Part B: Biochem. Molecul. Biol. 225, 75–83. doi: 10.1016/j.cbpb.2018.04.007
Zhang, Y., Zhang, L., Sun, J., Qiu, J., Hu, X., Hu, J., et al. (2014). Proteomic analysis identifies proteins related to carotenoid accumulation in Yesso scallop (Patinopecten yessoensis). Food Chem. 147, 111–116. doi: 10.1016/j.foodchem.2013.09.078
Zhao, J., Gupta, S., Seielstad, M., Liu, J., and Thalamuthu, A. (2011). Pathway-based analysis using reduced gene subsets in genome-wide association studies. BMC Bioinform. 12:17. doi: 10.1186/1471-2105-12-17
Zhao, L., Li, Y., Li, Y., Yu, J., Liao, H., Wang, S., et al. (2017). A genome-wide association study identifies the genomic region associated with shell color in Yesso scallop, Patinopecten yessoensis. Marine Biotechnol. 19, 301–309. doi: 10.1007/s10126-017-9751-y
Zheng, H., Liu, H., Zhang, T., Wang, S., Sun, Z., Liu, W., et al. (2010). Total carotenoid differences in scallop tissues of Chlamys nobilis (Bivalve: Pectinidae) with regard to gender and shell colour. Food Chem. 122, 1164–1167. doi: 10.1016/j.foodchem.2010.03.109
Zheng, H., Xu, F., and Zhang, G. (2011). Crosses between two subspecies of bay scallop Argopecten irradians and heterosis for yield traits at harvest. Aquac. Res. 42, 602–612. doi: 10.1111/j.1365-2109.2010.02657.x
Zheng, H., Zhang, G., Liu, X., Zhang, F., and Guo, X. (2004). Different responses to selection in two stocks of the bay scallop, Argopecten irradians Lamarck (1819). J. Exp. Mar. Biol. Ecol. 313, 213–223. doi: 10.1016/j.jembe.2004.04.015
Keywords: genome-wide association study, Argopecten irradians irradians, shell color, single nucleotide polymorphism, candidate genes
Citation: Zhu X, Zhang J, Hou X, Liu P, Lv J, Xing Q, Huang X, Hu J and Bao Z (2021) A Genome-Wide Association Study Identifies Candidate Genes Associated With Shell Color in Bay Scallop Argopecten irradians irradians. Front. Mar. Sci. 8:742330. doi: 10.3389/fmars.2021.742330
Received: 16 July 2021; Accepted: 11 August 2021;
Published: 06 September 2021.
Edited by:
Yuehuan Zhang, South China Sea Institute of Oceanology, Chinese Academy of Sciences, ChinaCopyright © 2021 Zhu, Zhang, Hou, Liu, Lv, Xing, Huang, Hu and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Xing, cWlhbmd4aW5nQG91Yy5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.