
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 27 August 2021
Sec. Aquatic Physiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.736338
 Fahim Ullah Khan1,2,3†
Fahim Ullah Khan1,2,3† Yueyong Shang1,2†
Yueyong Shang1,2† Xueqing Chang1,2
Xueqing Chang1,2 Hui Kong1,2
Hui Kong1,2 Amina Zuberi3
Amina Zuberi3 James K. H. Fang4
James K. H. Fang4 Wei Liu5
Wei Liu5 Jinxia Peng6
Jinxia Peng6 Xingzhi Zhang6*
Xingzhi Zhang6* Menghong Hu1,2
Menghong Hu1,2 Youji Wang1,2*
Youji Wang1,2*Gut microbiota play a very important role in the health of the host, such as protecting from pathogens and maintaining homeostasis. However, environmental stressors, such as ocean acidification, hypoxia, and warming can affect microbial communities by causing alteration in their structure and relative abundance and by destroying their network. The study aimed to evaluate the combined effects of low pH, low dissolved oxygen (DO) levels, and warming on gut microbiota of the mussel Mytilus coruscus. Mussels were exposed to two pH levels (8.1, 7.7), two DO levels (6, 2 mg L−1), and two temperature levels (20, 30°C) for a total of eight treatments for 30 days. The experiment results showed that ocean acidification, hypoxia, and warming affected the community structure, species richness, and diversity of gut microbiota. The most abundant phyla noted were Proteobacteria, Bacteroidetes, and Firmicutes. Principal coordinate analysis (PCoA) revealed that ocean acidification, hypoxia, and warming change microbial community structure. Low pH, low DO, and increased temperature can cause shifting of microbial communities toward pathogen dominated microbial communities. Linear discriminant analysis effect size (LEfSe) showed that the significantly enriched biomarkers in each group are significantly different at the genus level. Phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) analysis revealed that the gut microbiome of the mussels is associated with many important functions, such as amino acid transport and metabolism, transcription, energy production and conservation, cell wall, membrane and envelope biogenesis, and other functions. This study highlights the complexity of interaction among pH, DO, and temperature in marine organisms and their effects on the gut microbiota and health of marine mussels.
The phenomenon in which pH reduction occurs in seawater due to atmospheric CO2 diffusion into the surface waters of oceans is called ocean acidification (Orr et al., 2005). It is estimated that due to anthropogenic activities ocean pH will decrease ~0.3–0.5 units by the end of this century (Pachauri et al., 2014). It has been reported that ocean acidification has adverse effects on the marine calcified organisms with consequences for species diversity, trophic interaction, and other ecological processes (Pörtner et al., 2014). There is evidence that during the last century, average temperature has increased ~0.7°C, and it will further increase 1.8–4°C by the end of the 21st century (IPCC, 2007). The temperature fluctuations can affect growth, metabolism, and development of marine bivalve mollusks, and in the case of thermal stress conditions, it can also cause increased mortality and decreased growth in the bivalve mussels (Dickinson et al., 2012). Dissolved oxygen (DO) concentration of <2 mg L−1 is considered a hypoxic condition in the aquatic ecosystems (Wu, 2002). The East China Sea has one of the largest coastal hypoxic areas in the world which covers an area of more than 12,000 km2 (Chen C. et al., 2007). Hypoxia can cause reduced ingestion of food and also reduced growth (Welker et al., 2013). Hypoxia can also adversely affect the immune system of bivalves (Monari et al., 2007), leading to an increased risk of pathogen infections in bivalves (Wang et al., 2012, 2014). The digestive physiology and energetic budget of bivalves can be affected by dissolved oxygen concentrations (Wang et al., 2015). The previous studies found that the digestive function and antioxidant response of Mytilus coruscus were affected by hypoxia, acidification, and warming of the ocean (Khan et al., 2020, 2021). It is worth noting that in recent years, more and more scholars have paid attention to the potential combined effects of ocean hypoxia, acidification, and warming on marine life (Sampaio et al., 2021).
It has been proved that microbiota associated with host play a very important and essential role in animal health. Microbiota in the host provide many functions to the host, such as protection from diseases and nutrient processing (Sweet and Bulling, 2017). The gut microbiome is considered an organ contributing to the regulation of host metabolism (Rastelli et al., 2019). The balanced and healthy gut microbiota of animals play an important role to prevent infections and support the host immune system (Kau et al., 2011; Man et al., 2017). The studies have found that gut microbiota have functions in physiology, reproduction, development, energy balance, behavior, immunity, and life history of the host (Semova et al., 2012). Gut microbiota also play an important role in maintaining the homeostasis of the host, and changes in its composition can bring metabolic shifts that may result in host phenotype alterations (Turnbaugh et al., 2006; Visconti et al., 2019). High microbial abundance and diversity have been found in marine invertebrate animals (Olson and Kellogg, 2010). The alterations in the host-microbial communities due to stressful conditions are associated with an increased risk to diseases and poor health conditions because opportunistic pathogens and non-resident microbial species find their way to increase populations within the host (Vezzulli et al., 2013; Lokmer and Wegner, 2015). Mass mortality incidents of pacific oyster (Crassostrea gigas) have been reported in farming areas of Europe (Samain and McCombie, 2008), this loss was attributed to the complex interactions among environmental variables, microbial pathogens, and oysters (Pernet et al., 2014). The marine bivalve mollusks have high microbiome diversity and abundance (Olson and Kellogg, 2010; Pierce and Ward, 2018). The microbiota of invertebrates can be altered partially by fluctuations in the external environment (Tanaka et al., 2004; Beleneva and Zhukova, 2009). The stable host-microbiota associations in the digestive tract of a variety of animals like crustaceans (Freese and Schink, 2011), insects (Robinson et al., 2010), and oysters (Wegner et al., 2013) have been reported.
Various abiotic environmental factors, such as pH, DO, and temperature can affect the mussel microbiota communities (Rubiolo et al., 2019). The structure of the gut microbiome is highly changeable and can be altered as a result of environmental factors (Goodrich et al., 2016; Xie et al., 2016; Bestion et al., 2017). The changes in these environmental factors can directly affect mussel microbiome communities by altering their structure and diversity which will finally affect the ability of the mussel to convert food into energy and other physiological functions (Rubiolo et al., 2019). Low pH value has been reported to increase the growth of Vibrionaceae, and consequently, increased growth of vibrio genus might have adverse effects on mussel growth (Meron et al., 2011). Decreased pH affects bacterial communication, and this process is also called quorum sensing. It is a key process that affects the microbial interactions in the ecosystem, depending on autoinducing peptides (AIPs) in Gram-positive bacteria and acyl homoserine lactones (AHLs) in Gram-negative bacteria (Miller and Bassler, 2001; Manefield and Whiteley, 2007). Both have important ecological and biological functions and regulate symbiosis, competence, virulence, secondary metabolite production, extracellular enzymes, biofilm formation, and bioluminescence in the marine environment (De Kievit, 2009; Chong et al., 2012; Mangwani et al., 2012). Changes in environmental factors such as temperature, would affect microbial community composition either directly or indirectly (Mathai et al., 2020). The increased temperature is a critical factor in regulating the bacteria virulence (Lee et al., 2001; Kimes et al., 2012) and innate immunity of bivalves (Cheng et al., 2004). In marine biotic and abiotic habitats, temperature is an important factor to shape microbiota communities (Fuhrman et al., 2008). Due to global warming, alteration of mutualistic microbial communities to pathogen-dominated communities and increase in infectious diseases in the hosts have already been reported (Ritchie, 2006; Altizer et al., 2013). Few studies have been carried out to reveal the bacterial diversity of mollusks digestive gland (Rubiolo et al., 2018). Previous studies reported that high water temperature altered M. coruscus gut microbiota by favoring the accumulation of opportunistic pathogens in adult mussels (Li et al., 2018). The homeostasis and health controlling mechanisms of gut microbiota in aquatic animals under hypoxic conditions are still unclear (Fan et al., 2020). Especially the combined effects of acidification, warming, and hypoxia on the gut microbiota of the aquatic animals are still not well-known (Wang et al., 2020).
Mytilus coruscus is an ecologically and economically important bivalve species in the coastal waters of Yellow Sea and East China Sea (Dong et al., 2017). The habitats of the economically important bivalve mollusks face the conditions of hypoxia (Diaz and Rosenberg, 2008), thermal stress (Baumann and Doherty, 2013), and ocean acidification (Wallace et al., 2014), which are dangerous for the survival of the bivalves species in recent years (Beck et al., 2011). As these multiple stressors practically exist in the habitat of M. coruscus in the summer season, their effects on the mussels may be synergistic, antagonistic, or additive, it is urgent to understand the complex effects of these multiple stressors (Stevens and Gobler, 2018), and the interactive effects of these stressors need to be clarified (Hu et al., 2015; Sui et al., 2017). Therefore, in this experiment, we elevated the combined effects of ocean acidification, hypoxia, and warming on the gut microbiota of the thick shell mussel M. coruscus, which can help us to understand the physiological responses of the mussels to these stressful conditions and to make plans for better health and protection of marine animals.
Mussels were collected from the Gouqi Island located in the Zhejiang province of China. Healthy mussels (shell length: 8.43 ± 0.6 cm; wet weight: 79.58 ± 4.64 g) were selected and cleaned by removing all kinds of attachments adhered to the mussel shell surfaces. After that, mussels were placed in the aquatic tanks (28L), equipped with filtering and circulating water devices. The standard natural seawater conditions of the collection site of mussels were established in all experimental tanks, i.e., pH 8.1 ± 0.1, DO 6 ± 0.1 mg L−1, temperature 20 ± 0.2°C. Before starting the experiment, the mussels were acclimatized for 1 week under the above conditions. Mussels were fed with microalgae Chlorella spp. (2.5 × 104 cells ml−1) twice a day.
In the current experiment, we tested the effects of three major environmental factors (pH, DO, and temperature) on the mussel gut microbiota. Two pH levels (8.1 and 7.7), two DO levels (6 and 2 mg L−1), and two temperature levels (20 and 30°C) were set for the experiment. The pH level 8.1 represents the ongoing mean pH level at the sampling site while pH level 7.7 represents the forecast average pH at the end of 2,100 (Orr et al., 2005; IPCC, 2007), and extreme of present natural variability at the Shengsi island of East China Sea (Shang et al., 2019). At the sampling site of mussels, DO concentrations of <2–3 mg L−1 were reported by Chen C. et al. (2007) and Chen J. H. et al. (2007). DO level ≤ 2 mg L−1 is considered hypoxic level for aquatic organisms (Sui et al., 2017), while 6 mg L−1 is considered as normal DO level. Thus, in our experiment, DO 2 mg L−1 was considered as hypoxic treatment for mussels. In the present experiment, 20°C denotes the mean temperature experienced by the mussels, while 30°C represents the peak temperature level experienced by the mussels at the site of collection (Wu et al., 2016). All factors were used in a full factorial design. A total of 8 treatments were set in the experiment T1 (DO 6 mg L−1 × pH 8.1 × 20°C), T2 (DO 6 mg L−1 × pH 8.1 × 30°C), T3 (DO 6 mg L−1 × pH 7.7 × 20°C), T4 (DO 6 mg L−1 × pH 7.7 × 30°C), T5 (DO 2 mg L−1 × pH 7.7 × 20°C), T6 (DO 2 mg L−1 × pH 7.7 × 30°C), T7 (DO 2 mg L−1 × pH 8.1 × 20°C), and T8 (DO 2 mg L−1 × pH 8.1 × 30°C). The experiment was conducted in triplicates with a density of 20 mussels per tank (28 L). A recirculatory system with filters was used to prevent metabolic wastes accumulation in tanks by the mussels. Before starting the experiment, the mussels were acclimatized gradually to the experimental conditions. DO (from 6 to 2 mg L−1) and pH (from 8.1 to 7.7) were gradually reduced within 2 days, and the temperature was increased step by step from 20 to 30°C in 5 days. The entire experiment lasted for 30 days.
For maintenance of required temperature and DO levels in the experimental tanks, we used electronic thermostats and DO regulators (Loligo systems Apps, Tjele, Denmark), respectively. Low DO level was maintained by injecting N2 directly into tanks using an oxygen regulator linked to a computer. Required experimental low pH level was maintained by introducing pure CO2 directly using WTW pH 3310 connected to pCO2/pH feedback STAT systems (DAQ-M) and SenTix 41 pH electrodes (Loligo Systems Inc.), operated by Cap CTRL software (Loligo Systems Inc.). A Salinometer (S/Mill-E, Atago) was used to measure salinity. Salinity was kept constant at 25 psu. For pH measurement, a portable pH meter (pH-201, MSITECH (Asia-Pacific) Pte. Ltd., Singapore) gauged with the NBS scale was used. The titration method was adopted to determine total alkalinity (AT) values for all treatments. To calculate pCO2, calcite saturation state (Ωca) and aragonite saturation state (Ωar) from AT, pHNBS, temperature, and salinity, CO2sys was used with constants, K1 and K2 (Mehrbach et al., 1973) modified by Millero (2010). The seawater chemical parameters of all treatment groups in the experiment are summarized in Table 1.
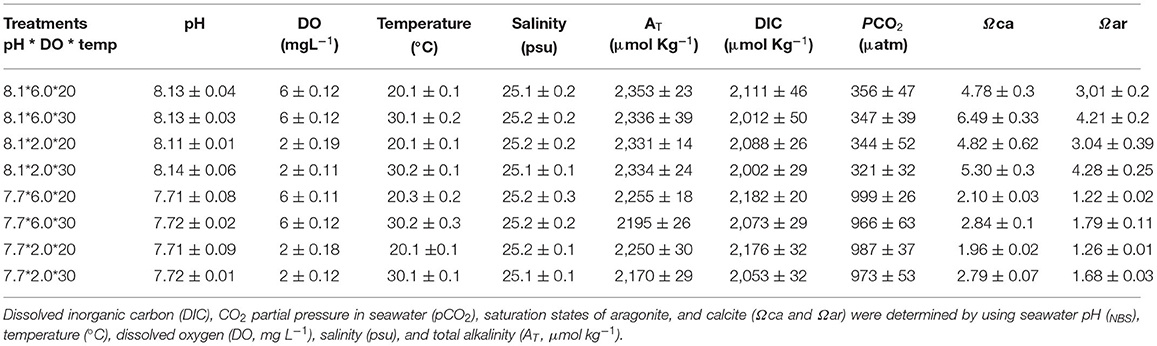
Table 1. Seawater carbonate chemistry variables (mean ± SD, n = 4) during the whole experimental period.
Prior to sampling, mussels were stopped feeding for 12 h. Three mussels per replicate were randomly collected for dissection at the sampling day (day 30). Guts were carefully removed and then cleaned with phosphate buffer (50 mM, pH 7.4), dried using tissue paper, placed in tubes, and kept on ice. For further analysis, the tissues of three mussels from each replicate were pooled representing one sample of the relative treatment and immediately preserved at −80°C. Thus, for each treatment, three samples were collected.
A DNA extraction kit was used for total genomic DNA extraction by following the protocols of the manufacturer. With NanoDrop and agarose gel, the DNA quality and quantity were determined. Dilution of the extracted DNA was done to the concentration of 1 ng μL−1 and for further processing preserved at −20°C. The diluted DNA was used as a template for PCR amplification of bacterial 16S ribosomal ribonucleic acid (rRNA) genes with the barcoded primers and Takara Ex Taq (Takara). For bacterial diversity analysis, V3–V4 variable regions of 16S rRNA genes were amplified with universal primers 343F (5′- TACGGRAGGCAGCAG-3′) and 798R (5′-AGGGTATCTAATCCT-3′) (Nossa et al., 2010).
By using gel electrophoresis, amplicon quality was visualized, purified with AMPure XP beads (Agencourt), and amplified for another round of PCR. After purification with the AMPure XP beads again, a Qubit dsDNA assay kit was used for the final amplicon quantification. For subsequent sequencing, the same amounts of purified amplicon were pooled.
A FASTQ file format was used for raw sequencing data. Using Trimmomatic software, paired-end reads were pre-treated (Bolger et al., 2014) to recognize and break off unclear bases (N). Using the sliding window trimming approach, low-standard sequencing with average quality score <20 was also removed. With the help of Flash software (Reyon et al., 2012) paired-end reads were grouped. The parameters of assembly were: 10 base pairs of minimal overlapping, 200 base pairs of maximum overlapping, and 20% of maximum non-similarity rate. Further denoising of sequences was performed by abandoning reads with ambiguous or homologous sequences or base pairs below 200 bp reads and reads with 75% of bases above Q20 were retained. Reads with chimera were then detected and removed. QIIME software (version 1.8.0) was used to achieve these two steps (Caporaso et al., 2010).
To produce operational taxonomic units (OTUs), clean reads were subjected to the removal and clustering of primer sequences, for this purpose Vsearch software with a 97% similarity cutoff was used (Edgar et al., 2011). With the help of the QIIME package, the representative read of each OTU was selected. Using an RDP classifier (confidence threshold of 70%), all representative reads were annotated and blasted against the Silva database version 123 (Wang et al., 2007). All representative reads were annotated and blasted against the Unite database using blast (Lobo, 2008).
The richness index of Chao1 and the diversity indices of Shannon and Simpson were calculated based on OTUs, and the differences of microbial diversity indices were determined by the Kruskal-Wallis test. A principal coordinate analysis (PCoA) was performed to reflect the differences in microbial construction between samples. A linear discriminant analysis of effect size (LEfSe) was performed to assess the difference in the relative abundance of bacteria, and the logarithmic transformation linear discriminant analysis (LDA) score threshold was 3.5. PICRUSt function prediction of the mussel gut microbes is based on the cluster of orthologous groups of proteins (COG) database.
An operational taxonomic unit (OTU) was classified with a sequence similarity of 97%, and 1,878 OTUs were identified from gut samples of the mussels. Through the results of OTUs cluster analysis, it is found that the number of OTUs in each sample is distributed between 165 and 598. Based on the level of phylum taxonomy, the top 15 most abundant bacterial phyla were listed (Figure 1). For the gut microbiome in T1–T8, Proteobacteria was the most abundant phylum (80.9, 89.4, 77.9, 92.7, 81.6, 62.9, 77.4, and 43.3%, respectively), followed by Bacteroidetes (12.2, 7.4, 12.4, 4.1, 10.3, 10.6, 13.8, and 22.5%, respectively), and Firmicutes (3.1, 2.1, 5.5, 2.2, 3.8, 24.8, 5.7, and 22.2%, respectively). In addition, Actinobacteria in the T1, T3, T5, and T8 groups accounted for 1.3, 1.8, 2.9, and 10.1%, respectively. Tenericutes of T1 and T8 accounted for 1.4 and 1.2%, respectively. The abundance of the remaining unmentioned gut microbiota in the experimental groups was <1% (Figure 1).
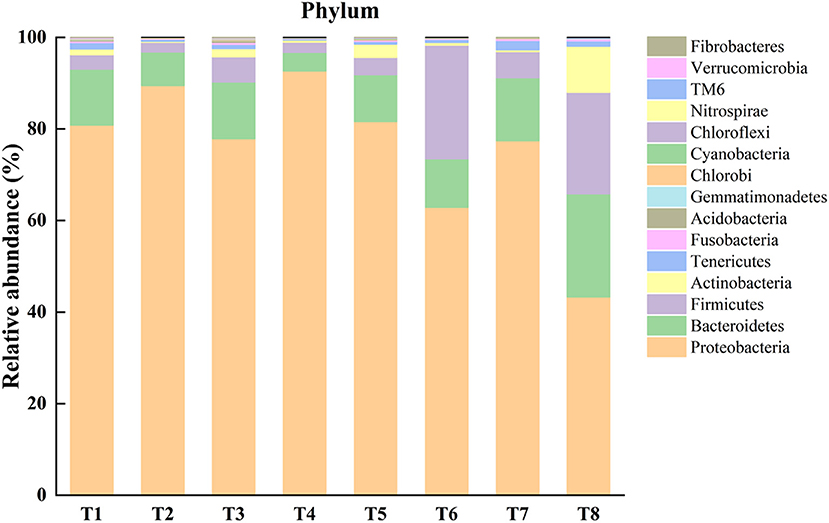
Figure 1. Representing relative abundance of top 15 phyla of microbiota in each sample of different experimental treatments [T1 (DO 6 mg L−1 × pH 8.1 × 20°C), T2 (DO 6 mg L−1 × pH 8.1 × 30°C), T3 (DO 6 mg L−1 × pH 7.7 × 20°C), T4 (DO 6 mg L−1 × pH 7.7 × 30°C), T5 (DO 2 mg L−1 × pH 7.7 × 20°C), T6 (DO 2 mg L−1 × pH 7.7 × 30°C), T7 (DO 2 mg L−1 × pH 8.1 × 20°C), T8 (DO 2 mg L−1 × pH 8.1 × 30°C)] in the gut of Mytilus coruscus.
Alpha diversity metrics, such as Chao 1 (total species abundance index) and Shannon and Simpson indices (species diversity indices) were analyzed for the characterization of microbial diversity in all treatments (Table 2, Figures 2A–C). Temperature has significant effects on T1–T2 (DO 6 mg L−1 × pH 8.1 × 20°C*DO 6 mg L−1 × pH 8.1 × 30°C) groups of Chao1 and Shannon indices (the Kruskal–Wallis test, p < 0.05). Higher temperature (30°C) significantly reduced species richness in T2 group compared with T1 group (20°C). Similarly T3–T4 (DO 6 mg L−1 × pH 7.7 × 20°C*DO 6 mg L−1 × pH 7.7 × 30°C) groups of Chao 1 and Shannon and Simpson indices were significantly affected by temperature. Higher temperature (30°C) significantly decreased the richness (Chao 1 index) and diversity (Shannon and Simpson indices) in T4 group compared to T3 group (20°C). Moreover, T7–T8 (DO 2 mg L−1 × pH 8.1 × 20°C*DO 2 mg L−1 × pH 8.1 × 30°C) groups of Chao 1 and Shannon and Simpson indices were also significantly affected by temperature. Higher temperature (30°C) significantly increased the Chao 1 index and Shannon and Simpson indices in T8 group compared with T7 group (20°C). Only T5–T7 (DO 2 mg L−1 × pH 7.7 × 20°C*DO 2 mg L−1 × pH 8.1 × 20°C) groups of Chao 1 index was significantly affected by pH. Low pH (7.7) significantly increased Chao 1 index in T5 group compared to T7 group (pH 8.1). Chao 1 index of T1–T7 (DO 6 mg L−1 × pH 8.1 × 20°C*DO 2 mg L−1 × pH 8.1 × 20°C) groups, Chao 1 and Shannon and Simpson indices of T2–T8 (DO 6 mg L−1 × pH 8.1 × 30°C*DO 2 mg L−1 × pH 8.1 × 30°C) groups, and Shannon and Simpson indices of T3−T5 (DO 6 mg L−1 × pH 7.7 × 20°C*DO 2 mg L−1 × pH 7.7 × 20°C) groups were significantly affected by hypoxia. Chao 1 index was significantly decreased in T7 group (2 mg L−1) compared with T1 (6 mg L−1). Chao 1 and Shannon and Simpson indices were significantly increased in T8 group (2 mg L−1) compared with T2 group (6 mg L−1). Shannon and Simpson indices were significantly decreased in T5 group (2 mg L−1) compared with T3 group (6 mg L−1) (Table 2, Figures 2A–C). The PCoA analysis revealed that all treatments with different pH, DO, and temperature combinations affected the microbial communities and shaped them by separating various groups from each other. PC1 comprises 38.2% of the overall variance (Figure 2D).
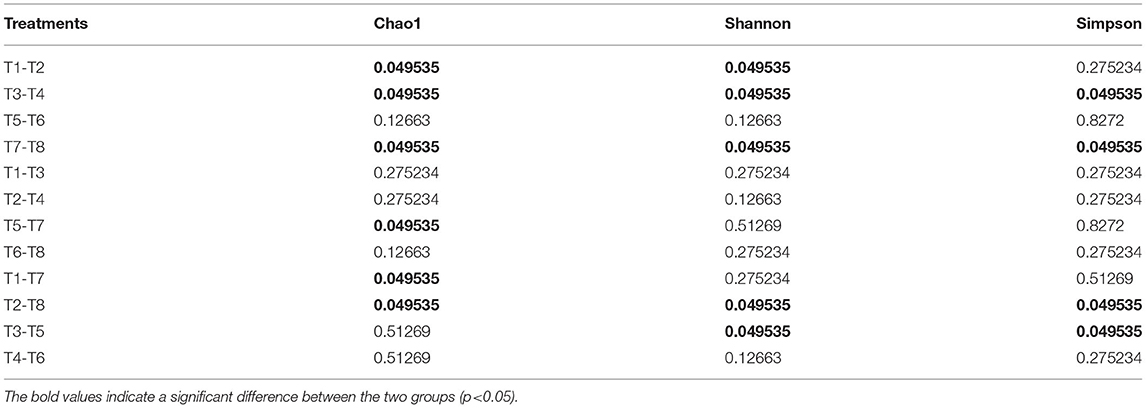
Table 2. The Kruskal–Wallis' test results (p-value) of the Alpha index (Chao1, Shannon, and Simpson) of gut microbiome [T1 (DO 6 mg L−1 × pH 8.1 × 20°C), T2 (DO 6 mg L−1 × pH 8.1 × 30°C), T3 (DO 6 mg L−1 × pH 7.7 × 20°C), T4 (DO 6 mg L−1 × pH 7.7 × 30°C), T5 (DO 2 mg L−1 × pH 7.7 × 20°C), T6 (DO 2 mg L−1 × pH 7.7 × 30°C), T7 (DO 2 mg L−1 × pH 8.1 × 20°C), and T8 (DO 2 mg L−1 × pH 8.1 × 30°C)].
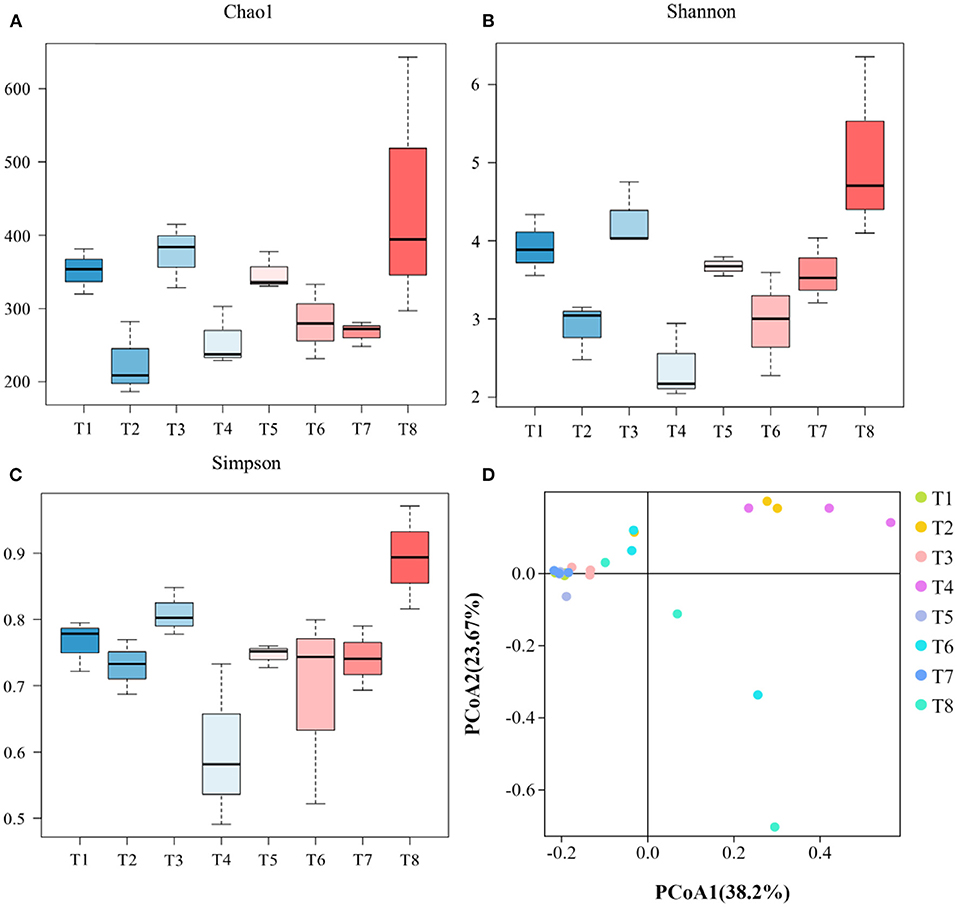
Figure 2. Representing microbial diversity indices of Chao 1 (A), Shannon (B), and Simpson (C) and principal coordinate analysis of gut microbiome (D). [T1 (DO 6 mg L−1 × pH 8.1 × 20°C), T2 (DO 6 mg L−1 × pH 8.1 × 30°C), T3 (DO 6 mg L−1 × pH 7.7 × 20°C), T4 (DO 6 mg L−1 × pH 7.7 × 30°C), T5 (DO 2 mg L−1 × pH 7.7 × 20°C), T6 (DO 2 mg L−1 × pH 7.7 × 30°C), T7 (DO 2 mg L−1 × pH 8.1 × 20°C), T8 (DO 2 mg L−1 × pH 8.1 × 30°C)].
The LEfSe was conducted to determine taxonomic differences in relative abundance of bacteria and unique biomarkers in all experimental treatments. Mucispirillum is the bacterial genus level biomarker in T1, distinguished this group from all the other groups. Acinetobacter is genus level biomarker in T3 that is unique to this group. Endozoicomonas, Kistimonas genera distinguished T4 from all other groups. Pseudomonas is the unique genus of T5. Polaribacter and Candidatus−Rhabdochlamydia are the unique genera of T6. Pseudorhodobacter and Mycoplasma are the special genera of T7. Similarly, Atopostipes and Flammeovirga are the special genera of T8 (Figure 3).
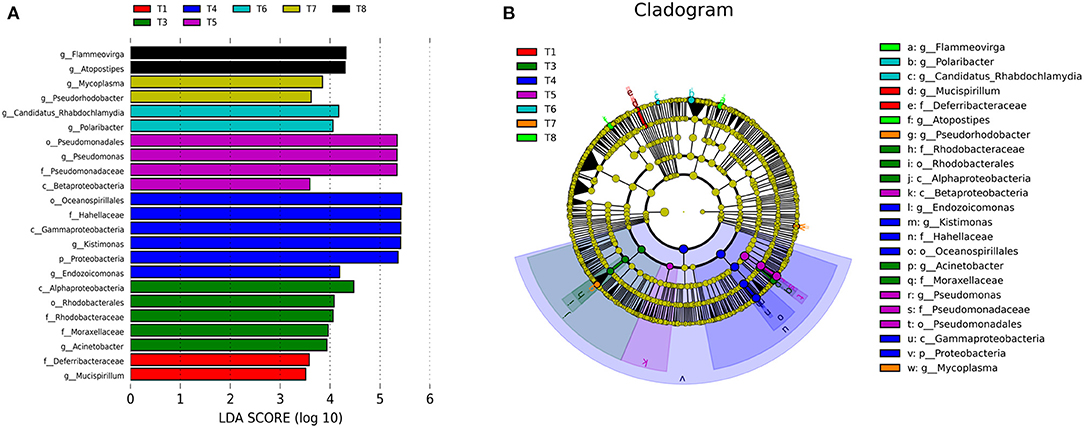
Figure 3. (A) Showing LEfSe analysis of gut microbiota of various experimental groups of mussels Mytilus coruscus. Bar chart showing the log transformed LDA scores of bacterial taxa identified by LEfSe analysis. (B) Cladogram represents the phylogenetic relationships of bacterial taxa revealed by LEfSe. [T1 (DO 6 mg L−1 × pH 8.1 × 20°C), T2 (DO 6 mg L−1 × pH 8.1 × 30°C), T3 (DO 6 mg L−1 × pH 7.7 × 20°C), T4 (DO 6 mg L−1 × pH 7.7 × 30°C), T5 (DO 2 mg L−1 × pH 7.7 × 20°C), T6 (DO 2 mg L−1 × pH 7.7 × 30°C), T7 (DO 2 mg L−1 × pH 8.1 × 20°C), and T8 (DO 2 mg L−1 × pH 8.1 × 30°C)].
Based on the results of gut microbial sequencing and the clusters of orthologous groups of proteins (COG) database, PICRUSt predicted the functions of T1–T8 mussels gut microbial community. The results showed that among the mussels of all treatment groups, 20 functions such as amino acid transport and metabolism, transcription, energy production and conversion, cell wall/membrane/envelope biogenesis, translation, ribosomal structure and biogenrsis, signal transduction mechanisms, inorganic ion/carbohydrate/coenzyme/lipid/ nucleotide transport and metabolism, and other functions account for a higher proportion (>1%) (Figure 4).
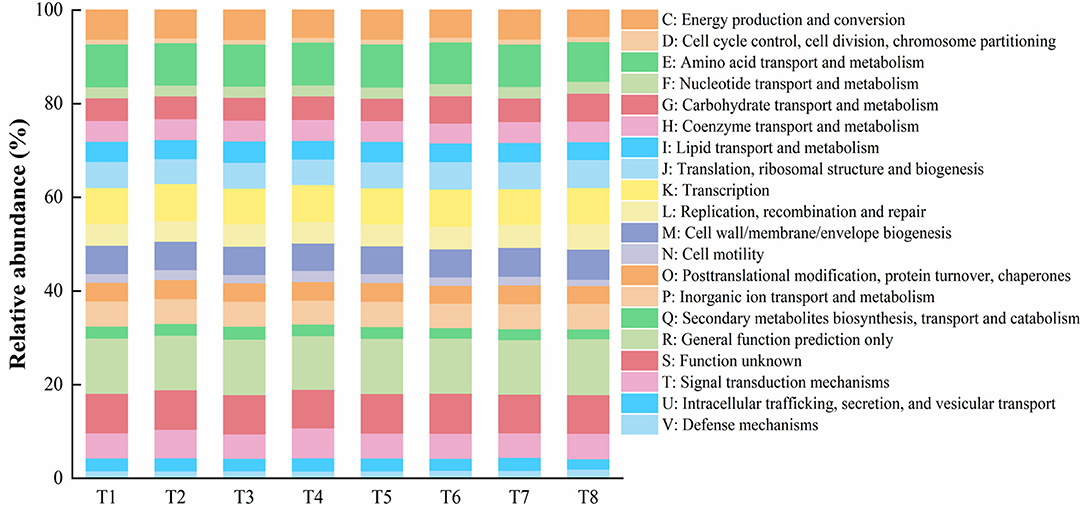
Figure 4. Representing gut microbiota presumptive functions and their relative abundances of the mussel Mytilus coruscus in different experimental treatments. [T1 (DO 6 mg L−1 × pH 8.1 × 20°C), T2 (DO 6 mg L−1 × pH 8.1 × 30°C), T3 (DO 6 mg L−1 × pH 7.7 × 20°C), T4 (DO 6 mg L−1 × pH 7.7 × 30°C), T5 (DO 2 mg L−1 × pH 7.7 × 20°C), T6 (DO 2 mg L−1 × pH 7.7 × 30°C), T7 (DO 2 mg L−1 × pH 8.1 × 20°C), and T8 (DO 2 mg L−1 × pH 8.1 × 30°C)].
In order to better forecast and understand the physiological responses and other ecological mechanisms of marine organisms in a continuously changing ocean environment due to the combined stress of ocean acidification, hypoxia, and warming, we need multi-stressor exposure experiments. The environmental stressors can act independently or exert synergistic, antagonistic effects on organism performances (Todgham and Stillman, 2013). Very less is known about the variations in microbial compositions in bivalve mollusks (Lokmer and Wegner, 2015). This study is the first report to provide the evidence of the combined effects of ocean acidification, hypoxia, and warming on the gut microbiota of mussels. This study highlights the importance of multi-stressor exposure experiments to reveal the combined effects of three global stressors that practically exist in the oceans at the same time. The intestinal microbiota populations arrange an ecological network via interspecific interactions and rely on the ecological network to regulate its stability and dynamic balance (Coyte et al., 2015). Strong relationships and more sub-modules are the key network properties for effectiveness and strength of the network (Barabasi and Oltvai, 2004; Kitano, 2004).
Bacterial communication is mediated by quorum sensing, which involves the production, release, and detection of autoinducers (small hormone-like molecules). Using these chemical communications, bacteria can detect their environment and respond by changing in population size and/or community structure, which in some instances may be harmful for the host organisms (Waters and Bassler, 2005). The resident microbes by competing with pathogen colonization may contribute to the host protection and therefore play a role in the host immune system (Kamada et al., 2012; Chu and Mazmanian, 2013; Desriac et al., 2014). In the current study, the results also explain such phenomena. Evidence from Goodrich et al. (2016) and Xie et al. (2016) explained that gut microbiota are strongly pliable and changeable under varying environmental conditions, whereas the core gut microbiota remain essentially stable (Nicholson et al., 2012). Bivalve microbiota are highly diverse in nature and are easily affected by environmental conditions, such as water temperature, pH, salinity, DO, nutrients, and infections (Green and Barnes, 2010; Lokmer and Wegner, 2015). In the current study, different pH, DO, and temperature levels affected the relative abundance and community composition of gut microbiota of the M. coruscus among the treatments. The stability of the microbiota communities can be detected in organisms as a result of any ecological or environmental disturbance, which is very important for doing community functions (Shade et al., 2012). In the present experiment results, we did not find any perpetuity of microbial communities among experimental treatments as a result of different pH, DO, and temperature levels. The differences in the relative abundance and community composition of microbiome populations among treatments indicate the unstable state of microbiome populations in the results; it also indicates that environmental conditions can influence the relative abundance and community composition of gut microbiota. The high-level interaction disturbances among the host, environment, and microbiota can destabilize the balance of the community composition and lead to poor states which have drastic consequences for the host. In order to achieve the microbial stability, the host has the capacity for balancing the community composition (Shade et al., 2012; Brown et al., 2013). In the results, the unstable community structure may indicate that the host attempts to stabilize the microbiome community structure and composition and arrange a stable ecological network. However, the current research cannot determine whether the change of bacterial community is due to the physiological changes of mussels caused by the environment or the direct response of bacteria to the environment.
The results showed that dominant microbiota in the gut of the M. coruscus are Proteobacteria, Becteroidetes, and Firmicutes at the phylum level. In addition, the results are similar to Yang et al. (2020), who investigated the effects of ocean acidification and microplastics on the gut microbiota of this species. Similarly, Song et al. (2018) reported that Bacteroidetes and Firmicutes are the two dominant bacteria phyla that colonize the digestive tract of aquatic animals. Firmicutes flourish within mussels because they have capacity to produce spores that show resistance to dehydration and also severe environmental conditions (Sathe et al., 2017). Unstable microbiota community structure in the intestine and microecological disorders can be reflected by intestinal Proteobacteria. Proteobacteria symbiotic bacteria are present in the intestine of healthy animals. Under certain environmental conditions, in the intestine these Proteobacteria may become intestinal microorganisms that are responsible for causing inflammation (Shin et al., 2015). Previous studies reported that microbial communities in Rapana venosa and Achatina fulica are dominated by Tenericutes, Bacteroidetes, and Proteobacteria (Pawar et al., 2014; Yang et al., 2019). The differences in the dominant microflora in the digestive tract of different organisms in these studies and the current study may be due to differences in their diets and different environmental conditions.
Yang et al. (2020) reported that pH variations have no significant effects on the gut microbiota of the M. coruscus. But Zeng et al. (2020) reported that the gut microbiota of moth larva have been significantly affected by dietary pH. In the current experiment results, low pH has no significant effects in most treatments groups. Some authors indicated that high temperature increases heterogeneity of the microbiota communities among individuals (Erwin et al., 2012; Lokmer and Wegner, 2015). Li et al. (2019a) reported that increased temperature affected the microbial diversity in the hemolymph of mussels. The microbiota with lower diversity in unhealthy oysters were found and the diversity was largely affected by heat stress (Lokmer and Wegner, 2015). Hypoxic conditions can disturb the homeostasis of the gut microbiota by quenching important communities and destroying their network structure (Fan et al., 2020). Results of these experiments coincide with the current experimental findings. In current results, we also found that increased temperature, low pH, and low DO have affected the microbiome diversity, relative abundance and community structure, and composition of gut microbiota populations. Probably mussels need to adjust their physiological activities when facing unfavorable environmental conditions and thus the gut microbiota changes.
According to Li et al. (2019a), PCoA analysis revealed that temperature is an important factor affecting the microbial community, 21°C treatments groups were separated from 27°C treatments groups, suggesting that disturbance of microbial community may be a sign of an unhealthy status. Similar results were also reported by Lokmer and Wegner (2015). In the current experiment, the PCoA analysis revealed that all treatments having different pH, DO, and temperature influenced microbial populations and shaped them by separating various groups from each other.
Microbial diversity significantly increased in the gut microbiota of mussels (Mytilus galloprovincialis) by elevated temperature as revealed by Chao1 and Shannon and Simpson indices (Li et al., 2019b). We also observed significantly increased gut microbiota as a result of elevated temperature in some treatments groups as indicated by Chao1 and Shannon and Simpson indices. Li et al. (2018) reported that significant reduction in microbial diversity was noted in live mussels (M. coruscus) gut microbiota as a result of heat stress. In our results, we also found decreased microbial diversity in some groups due to high temperature as revealed by Chao 1 and Shannon and Simpson indices. Similarly, some groups showed decreased microbial diversity due to hypoxia and decreased pH in terms of Chao 1 and Shannon and Simpson indices. High microbial species diversity in healthy animals enhances their ability to tolerate unfavorable environmental conditions (Lozupone et al., 2012). Low microbial diversity highly favors unhealthy status in many organisms (Chang et al., 2008; Green and Barnes, 2010).
The LEfSe analysis in the current study showed that the most unique taxa of all experimental treatments have pathogenic potential. This analysis indicates that acidification, hypoxia, and warming have caused establishment and dominancy of pathogenic taxa in the gut of mussels, as stated by Lokmer and Wegner (2015), in C.gigas environmental stressors such as high temperature favored the shift of bacterial communities toward pathogen-dominated microbiota communities and also enhanced the secondary opportunistic pathogens colonization. Moreover, we found no unique phylum, family, class, order, and genus levels taxa in T2 group compared with other experimental groups. It may be due to that this treatment has favored the environmental conditions for species taxon or sub taxa of phylum, family, class, and order. The deviations from the normal microbiome may indicate pathogenic infections or changes in metabolic processes. This can help us to identify and predict the effects of global change on mussel health (Shade and Handelsman, 2012).
Microbial community function prediction has revealed that intestinal microbiota are associated with carbohydrates, purines, pyrimidines, and amino acid metabolism in the host (Human Microbiome Project Consortium, 2012). The results from PICRUSt analysis predicted that microbiota of the thick shell mussel M. coruscus is mainly associated with amino acid transport and metabolism, transcription, energy production and conversion, cell wall, membrane and envelop biogenesis, translation, ribosomal structure and biogenesis, and other functions. Therefore, further multi-stressor exposure studies are necessary to reveal the potential mechanisms involved in these pathways.
The present study is the first report on the combined effects of three major global environmental problems, i.e., ocean acidification, hypoxia, and warming on the gut microbiome of the mussel M. coruscus. The current study demonstrates that ocean acidification, hypoxia, and warming have effects on the relative abundance, community composition, species richness, and diversity of the gut microbiota communities of mussels. Our results revealed that low pH, low dissolved oxygen (DO), and high temperature favor the proliferation of opportunistic and pathogenic bacteria, and predicted many possible functions of gut microbiota which need further investigations to explore their mechanisms. Further studies are needed for the development of strategic plans to save our marine organisms from the adverse effects of these ongoing global environmental problems.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI [accession: PRJNA744459].
YW, MH, and XZ contributed to the conception and design of the study. FU, YS, XC, and HK performed the experiments. FU and YS performed the statistical analysis. FU, AZ, JF, and WL wrote the first draft of the manuscript. JP, XZ, MH, and YW wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
The authors acknowledged grants from the National Natural Science Foundation of China (31872587). This study was supported by the Open Fund of Guangxi Key Laboratory of Aquatic Genetic Breeding and Healthy Aquaculture. The support from Guangdong South China Sea Key Laboratory of Aquaculture for Aquatic Economic Animals, Guangdong Ocean University (KFKT2019ZD04) was also highly acknowledged.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Altizer, S., Ostfeld, R. S., Johnson, P. T., Kutz, S., and Harvell, C. D. (2013). Climate change and infectious diseases: from evidence to a predictive framework. Science 341, 514–519. doi: 10.1126/science.1239401
Barabasi, A. L., and Oltvai, Z. N. (2004). Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 5, 101–113. doi: 10.1038/nrg1272
Baumann, H., and Doherty, O. (2013). Decadal changes in the world's coastal latitudinal temperature gradients. PLoS ONE 8:e67596. doi: 10.1371/journal.pone.0067596
Beck, M. W., Brumbaugh, R. D., Airoldi, L., Carranza, A., Coen, L. D., Crawford, C., et al. (2011). Oyster reefs at risk and recommendations for conservation, restoration, and management. Bioscience 61, 107–116. doi: 10.1525/bio.2011.61.2.5
Beleneva, I. A., and Zhukova, N. V. (2009). Seasonal dynamics of cell numbers and biodiversity of marine heterotrophic bacteria inhabiting invertebrates and water ecosystems of the Peter the Great Bay, Sea of Japan. Microbiology 78, 369–375. doi: 10.1134/S0026261709030163
Bestion, E., Jacob, S., Zinger, L., Di Gesu, L., Richard, M., White, J., et al. (2017). Climate warming reduces gut microbiota diversity in a vertebrate ectotherm. Nat. Ecol. Evol. 1, 1–3. doi: 10.1038/s41559-017-0161
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brown, E. M., Sadarangani, M., and Finlay, B. B. (2013). The role of the immune system in governing host-microbe interactions in the intestine. Nat. Immunol. 14, 660–667. doi: 10.1038/ni.2611
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chang, J. Y., Antonopoulos, D. A., Kalra, A., Tonelli, A., Khalife, W. T., Schmidt, T. M., et al. (2008). Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197, 435–438. doi: 10.1086/525047
Chen, C., Gong, G. C., and Shiah, F. K. (2007). Hypoxia in the East China Sea: one of the largest coastal low-oxygen areas in the world. Mar. Environ. Res. 64, 399–408. doi: 10.1016/j.marenvres.2007.01.007
Chen, J. H., Mai, K. S., Ma, H. M., Wang, X. J., Deng, D., Liu, X. W., et al. (2007). Effects of dissolved oxygen on survival and immune responses of scallop (Chlamys farreri Jones et Preston). Fish. Shellfish. Immunol. 22, 272–281. doi: 10.1016/j.fsi.2006.06.003
Cheng, W., Hsiao, I. S., Hsu, C. H., and Chen, J. C. (2004). Change in water temperature on the immune response of Taiwan abalone Haliotis diversicolor supertexta and its susceptibility to Vibrio parahaemolyticus. Fish. Shellfish. Immunol. 17, 235–243 doi: 10.1016/j.fsi.2004.03.007
Chong, G., Kimyon, O., Rice, S. A., Kjelleberg, S., and Manefield, M. (2012). The presence and role of bacterial quorum sensing in activated sludge. Microb. Biotechnol. 5, 621–633. doi: 10.1111/j.1751-7915.2012.00348.x
Chu, H., and Mazmanian, S. K. (2013). Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol. 14, 668–675. doi: 10.1038/ni.2635
Coyte, K. Z., Schluter, J., and Foster, K. R. (2015). The ecology of microbiome, network, competition and stability. Science 350, 663–666. doi: 10.1126/science.aad2602
De Kievit, T. R. (2009). Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 11, 279–288. doi: 10.1111/j.1462-2920.2008.01792.x
Desriac, F., Le Chevalier, P., Brillet, B., Leguerinel, I., Thuillier, B., Paillard, C., et al. (2014). Exploring the hologenome concept in marine bivalvia: haemolymph microbiota as a pertinent source of probiotics for aquaculture. FEMS. Microbiol. Lett. 350, 107–116. doi: 10.1111/1574-6968.12308
Diaz, R., and Rosenberg, R. (2008). Spreading dead zones and consequences for marine ecosystems. Science 321, 926–929. doi: 10.1126/science.1156401
Dickinson, G. H., Ivanina, A. V., Matoo, O. B., Pörtner, H. O., Lannig, G., Bock, C., et al. (2012). Interactive effects of salinity and elevated CO2 levels on juvenile eastern oysters, Crassostrea virginica. J. Exp. Biol. 215, 29–43. doi: 10.1242/jeb.061481
Dong, W., Chen, Y., Lu, W., Wu, B., and Qi, P. (2017). Transcriptome analysis of Mytilus coruscus hemocytes in response to Vibrio alginnolyficus infection. Fish. Shellfish. Immun. 70, 560–567. doi: 10.1016/j.fsi.2017.08.034
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Erwin, P. M., Pita, L., Lopez-Legentil, S., and Turon, X. (2012). Stability of sponge-associated bacteria over large seasonal shifts in temperature and irradiance. Appl. Environ. Microbiol. 78, 7358–7368. doi: 10.1128/AEM.02035-12
Fan, S., Li, H., and Zhao, R. (2020). Effects of normoxic and hypoxic conditions on the immune response and gut microbiota of Bostrichthys sinensis. Aquaculture 525:735336. doi: 10.1016/j.aquaculture.2020.735336
Freese, H. M., and Schink, B. (2011). Composition and stability of the microbial community inside the digestive tract of the aquatic crustacean Daphnia magna. Microb. Ecol. 62, 882–894. doi: 10.1007/s00248-011-9886-8
Fuhrman, J. A., Steele, J. A., Hewson, I., Schwalbach, M. S., Brown, M. V., Green, J. L., et al. (2008). A latitudinal diversity gradient in planktonic marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 7774–7778. doi: 10.1073/pnas.0803070105
Goodrich, J. K., Davenport, E. R., Beaumont, M., Jackson, M. A., Knight, R., Ober, C., et al. (2016). Genetic determinants of the gut microbiome in UK twins. Cell Host. Microbe. 19, 731–743. doi: 10.1016/j.chom.2016.04.017
Green, T. J., and Barnes, A. C. (2010). Bacterial diversity of the digestive gland of Sydney rock oysters, Saccostrea glomerata infected with the paramyxean, Marteilia sydneyi. J. Appl. Microbiol. 109, 613–622. doi: 10.1111/j.1365-2672.2010.04687.x
Hu, M. H., Li, L. S., Sui, Y. M., Li, J. L., Wang, Y. J., Lu, W. Q., et al. (2015). Effects of pH and temperature on antioxidant responses of the thick shell mussel Mytilus coruscus. Fish. Shellfish. Immunol. 46, 573–583. doi: 10.1016/j.fsi.2015.07.025
Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
IPCC (2007). “The physical science basis,” in Proceedings of Theontribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change (Cambridge: Cambridge University Press), 996.
Kamada, N., Kim, Y. G., Sham, H. P., Vallance, B. A., Puente, J. L., Martens, E. C., et al. (2012). Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329. doi: 10.1126/science.1222195
Kau, A. L., Ahern, P. P., Griffin, N. W., Goodman, A. L., Gordon, J. I., et al. (2011). Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336. doi: 10.1038/nature10213
Khan, F. U., Chen, H., Gu, H. X., Wang, T., Dupont, S., Kong, H., et al. (2021). Antioxidant responses of the mussel Mytilus coruscus co-exposed to ocean acidification, hypoxia and warming. Mar. Pollut. Bull. 162:111869. doi: 10.1016/j.marpolbul.2020.111869
Khan, F. U., Hu, M., Kong, H., Shang, Y., Wang, T., Wang, X. H., et al. (2020). Ocean acidification, hypoxia and warming impair digestive parameters of marine mussels. Chemosphere 256:127096. doi: 10.1016/j.chemosphere.2020.127096
Kimes, N. E., Grim, C. J., Johnson, W. R., Hasan, N. A., Tall, B. D., Kothary, M. H., et al. (2012). Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME. J. 6, 835–846. doi: 10.1038/ismej.2011.154
Lee, K. K., Liu, P. C., Chen, Y. C., and Huang, C. Y. (2001). The implication of ambient temperature with the outbreak of vibriosis in cultured small abalone Haliotis diversicolor supertexta Lischke. J. Therm. Biol. 26, 585–587. doi: 10.1016/S0306-4565(01)00004-3
Li, Y. F., Chen, Y. W., Xu, J. K., Ding, W. Y., Shao, A. Q., Zhu, Y. T., et al. (2019a). Temperature elevation and Vibrio cyclitrophicus infection reduce the diversity of haemolymph microbiome of the mussel Mytilus coruscus. Sci. Rep. 9:16391. doi: 10.1038/s41598-019-52752-y
Li, Y. F., Xu, J. K., Chen, Y. W., Ding, W. Y., Shao, A. Q., Liang, X., et al. (2019b). Characterization of gut microbiome in the mussel Mytilus galloprovincialis in response to thermal Stress. Front. Physiol. 10:1086. doi: 10.3389/fphys.2019.01086
Li, Y. F., Yang, N., Liang, X., Yoshida, A., Osatomi, K., Power, D., et al. (2018). Elevated seawater temperatures decrease microbial diversity in the gut of Mytilus coruscus. Front. Physiol. 9:839. doi: 10.3389/fphys.2018.00839
Lobo, I. (2008). Basic local alignment search tool (BLAST). J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Lokmer, A., and Wegner, K. M. (2015). Hemolymph microbiome of Pacific oysters in response to temperature, temperature stress, and infection. ISME. J. 9, 670–682. doi: 10.1038/ismej.2014.160
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., and Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. doi: 10.1038/nature11550
Man, W. H., de Steenhuijsen Piters, W. A. A., and Bogaert, D. (2017). The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 15, 259–270. doi: 10.1038/nrmicro.2017.14
Manefield, M., and Whiteley, A. S. (2007). Acylated homoserine lactones in the environment: chameleons of bioactivity. Philos. Trans. R. Soc. B 362, 1235–1240. doi: 10.1098/rstb.2007.2048
Mangwani, N., Dash, H. R., Chauhan, A., and Das, S. (2012). Bacterial quorum sensing: functional features and potential applications in biotechnology. J. Mol. Microbiol. Biotechnol. 22, 215–227. doi: 10.1159/000341847
Mathai, P. P., Bertram, J. H., Padhi, S. K., Singh, V., Tolo, I. E., Primus, A., et al. (2020). Influence of environmental stressors on the microbiota of zebra mussels (Dreissena polymorpha). Microb. Ecol. 81, 1042–1053. doi: 10.1007/s00248-020-01642-2
Mehrbach, C., Culberson, C., Hawley, J., and Pytkowicx, R. (1973). Measurement of the apparent dissociation constants of carbonic acid in seawater at atmospheric pressure. Limnol. Oceanogr. 18, 897–907. doi: 10.4319/lo.1973.18.6.0897
Meron, D., Atias, E., Kruh, L. I., Elifantz, H., Minz, D., Fine, M., et al. (2011). The impact of reduced pH on the microbial community of the coral Acropora eurystoma. ISME. J. 5, 51–60. doi: 10.1038/ismej.2010.102
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Millero, F. J. (2010). Carbonate constants for estuarine waters. Mar. Freshw. Res. 61, 139–142. doi: 10.1071/MF09254
Monari, M., Matozzo, V., Foschi, J., Cattani, O., Serrazanetti, G. P., and Marin, M. G. (2007). Effects of high temperatures on the functional responses of haemocytes in the clam chamelea gallina. Fish. Shellfish. Immunol. 22, 98–114. doi: 10.1016/j.fsi.2006.03.016
Nicholson, J. K., Holmes, E., Kinross, J., Burcelin, R., Gibson, G., Jia, W., et al. (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. doi: 10.1126/science.1223813
Nossa, C. W., Oberdorf, W. E., Yang, L., Aas, J. A., Paster, B. J., DeSantis, T. Z., et al. (2010). Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World. J. Gastroenterol. 16:4135. doi: 10.3748/wjg.v16.i33.4135
Olson, J. B., and Kellogg, C. A. (2010). Microbial ecology of corals, sponges, and algae in mesophotic coral environments. FEMS. Microbiol. Ecol. 73, 17–30. doi: 10.1111/j.1574-6941.2010.00862.x
Orr, J., Fabry, V., Aumont, O., Bopp, L., Doney, S., Feely, R., et al. (2005). Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms. Nature 437, 681–686. doi: 10.1038/nature04095
Pachauri, R. K., Allen, M. R., Barros, V. R., Broome, J., Cramer, W., Christ, R., et al. (2014). “Climate change 2014: synthesis report,” in Proceedings of the Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds R. Pachauri, and L. Meyer (Geneva: IPCC).
Pawar, K. D., Dar, M. A., Rajput, B. P., and Kulkarni, G. J. (2014). Enrichment and identification of cellulolytic bacteria from the gastrointestinal tract of giant African snail, Achatina fulica. Appl. Biochem. Biotechnol. 175, 1971–1980. doi: 10.1007/s12010-014-1379-z
Pernet, F., Lagarde, F., Le Gall, P., and D'Orbcastel, E. R. (2014). Associations between farming practices and disease mortality of Pacific oyster Crassostrea gigas in a Mediterranean lagoon. Aquac. Environ. Interact. 5, 99–106. doi: 10.3354/aei00096
Pierce, M. L., and Ward, J. E. (2018). Microbial ecology of the bivalvia, with an emphasis on the family ostreidae. J. Shellfish. Res. 37, 793–806. doi: 10.2983/035.037.0410
Pörtner, H. O., Karl, D. M., Boyd, P. W., et al. (2014). “Climate change 2014: impacts, adaptation, and vulnerability. Part A: global and sectoral aspects,” in Proceedings of the Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (Cambridge: Cambridge University Press), 411–484.
Rastelli, M., Cani, P. D., and Knauf, C. (2019). The gut microbiome influences host endocrine functions. Endocr. Rev. 40, 1271–1284. doi: 10.1210/er.2018-00280
Reyon, D., Maeder, M. L., Khayter, C., Tsai, S. Q., Foley, J. E., Sander, J. D., et al. (2012). FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30, 460–465. doi: 10.1038/nbt.2170
Ritchie, K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Robinson, C. J., Schloss, P., Ramos, Y., Raffa, K., and Handelsman, J. (2010). Robustness of the bacterial community in the cabbage white butterfly larval midgut. Microb. Ecol. 59, 199–211. doi: 10.1007/s00248-009-9595-8
Rubiolo, J. A., Botana, L. M., and Martínez, P. (2019). “Insights into mussel microbiome,” in Microbial Communities in Aquaculture Ecosystems, ed N. Derome (Cham: Springer), 95–120. doi: 10.1007/978-3-030-16190-3_5
Rubiolo, J. A., Lozano-Leon, A., Rodriguez-Souto, R., Rodríguez, N. F., Vieytes, M. R., and Botana, L. M. (2018). The impact of depuration on mussel hepatopancreas bacteriome composition and predicted metagenome. Anton Van Leeuwenhoek 111, 1117–1129. doi: 10.1007/s10482-018-1015-y
Samain, J. F., and McCombie, H. (2008). Summer mortality of Pacific oyster Crassostrea gigas, the Morest project. Editions Quae.
Sampaio, E., Santos, C., Rosa, I. C., Ferreira, V., Pörtner, H. O., Duarte, C. M., et al. (2021). Impacts of hypoxic events surpass those of future ocean warming and acidification. Nat. Ecol. Evol. 5, 311–321. doi: 10.1038/s41559-020-01370-3
Sathe, P., Laxman, K., Myint, M., Dobretsov, S., Richter, J., and Dutta, J. (2017). Bioinspired nanocoatings for biofouling prevention by photocatalytic redox reactions. Sci. Rep. 7:3624. doi: 10.1038/s41598-017-03636-6
Semova, I., Carten, J. D., Stombaugh, J., Mackey, L. C., Knight, R., Farber, S. A., et al. (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host. Microbe. 12, 277–288. doi: 10.1016/j.chom.2012.08.003
Shade, A., and Handelsman, J. (2012). Beyond the Venn diagram: The hunt for a core microbiome. Environ. Microbiol. 14, 4–12. doi: 10.1111/j.1462-2920.2011.02585.x
Shade, A., Peter, H., Allison, S. D., Baho, D., Berga, M., Bürgmann, H., et al. (2012). Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3:417. doi: 10.3389/fmicb.2012.00417
Shang, Y.Y., Wang, X.H., Kong, H., Huang, W., Hu, M. H., and Wang, Y.J. (2019). Nano-zno impairs anti-predation capacity of marine mussels under seawater acidification. J. Hazard. Mater. 371, 521–528. doi: 10.1016/j.jhazmat.2019.02.072
Shin, N. R., Whon, T. W., and Bae, J. W. (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends. Biotechnol. 33, 496–503. doi: 10.1016/j.tibtech.2015.06.011
Song, H., Yu, Z. L., Yang, M. J., Zhang, T., and Wang, H. Y. (2018). Analysis of microbial abundance and community composition in esophagus and intestinal tract of wild veined rapa whelk (Rapana venosa) by 16S rRNA gene sequencing. J. Gen. Appl. Microbiol. 64, 158–166. doi: 10.2323/jgam.2017.11.003
Stevens, A. M., and Gobler, C. J. (2018). Interactive effects of acidification, hypoxia, and thermal stress on growth, respiration, and survival of four North Atlantic bivalves. Mar. Ecol. Prog. Ser. 604, 143–161. doi: 10.3354/meps12725
Sui, Y. M., Hu, M. H., Shang, Y. Y., Wu, F. L., Huang, X. Z., Dupont, S., et al. (2017). Antioxidant response of the hard shell mussel Mytilus coruscus exposed to reduced pH and oxygen concentration. Ecotox. Environ. Safe 137, 94–102. doi: 10.1016/j.ecoenv.2016.11.023
Sweet, M. J., and Bulling, M. T. (2017). On the importance of the microbiome and pathobiome in coral health and disease. Front. Mar. Sci. 4:9. doi: 10.3389/fmars.2017.00009
Tanaka, R., Ootsubo, M., Sawabe, T., Ezura, Y., and Tajima, K. (2004). Biodiversity and in situ abundance of gut microflora of abalone (Haliotis discus hannai) determined by culture-independent techniques. Aquaculture 241, 453–463. doi: 10.1016/j.aquaculture.2004.08.032
Todgham, A. E., and Stillman, J. H. (2013). Physiological responses to shifts in multiple environmental stressors: relevance in a changing world. Integr. Comp. Biol. 53, 539–544. doi: 10.1093/icb/ict086
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
Vezzulli, L., Pezzati, E., Huete-Stauffer, C., Pruzzo, C., and Cerrano, C. (2013). 16SrDNA pyrosequencing of the Mediterranean gorgonian Paramuricea Clavata reveals a link among alterations in bacterial holobiont members, anthropogenic influence and disease outbreaks. PLoS ONE 8:e67745. doi: 10.1371/journal.pone.0067745
Visconti, A., Le Roy, C. I., Rosa, F., Rossi, N., Martin, T. C., Mohney, R. P., et al. (2019). Interplay between the human gut microbiome and host metabolism. Nat. Commun. 10:4505. doi: 10.1038/s41467-019-12476-z
Wallace, R. B., Baumann, H., Grear, J. S., Aller, R. C., and Gobler, C. J. (2014). Coastal ocean acidification: the other eutrophication problem. Estuar. Coast. Shelf. Sci. 148, 1–13. doi: 10.1016/j.ecss.2014.05.027
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, X., Tang, B., Luo, X., Ke, C., Huang, M., You, W., et al. (2020). Effects of temperature, diet and genotype-induced variations on the gut microbiota of abalone. Aquaculture 524:735269. doi: 10.1016/j.aquaculture.2020.735269
Wang, Y., Hu, M., Li, Q., Li, J., Lin, D., and Lu, W. (2014). Immune toxicity of TiO2 under hypoxia in the green-lipped mussel Perna viridis based on flow cytometric analysis of hemocyte parameters. Sci. Tot. Environ. 470, 791–799. doi: 10.1016/j.scitotenv.2013.09.060
Wang, Y. J., Hu, M. H., Cheung, S. G., Shin, P. K. S., Lu, W. Q., and Li, J. L. (2012). Immune parameter changes of hemocytes in green-lipped mussel Perna viridis exposure to hypoxia and hyposalinity. Aquaculture 356-357, 22–29. doi: 10.1016/j.aquaculture.2012.06.001
Wang, Y. J., Li, L. S., Hu, M. H., and Lu, W. Q. (2015). Physiological energetics of the thick shell mussel Mytilus coruscus exposed to seawater acidification and thermal stress. Sci. Total. Environ. 514, 261–272. doi: 10.1016/j.scitotenv.2015.01.092
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell. Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Wegner, K. M., Volkenborn, N., Peter, H., and Eiler, A. (2013). Disturbance induced decoupling between host genetics and composition of the associated microbiome. BMC. Microbiol. 13:252. doi: 10.1186/1471-2180-13-252
Welker, A. F., Moreira, D. C., Campos, E. G., and Hermes-Lima, M. (2013). Role of redox metabolism for adaptation of aquatic animals to drastic changes in oxygen availability. Comp. Biochem. Physiol Part A: Mol. Integr. Physiol. 165, 384–404. doi: 10.1016/j.cbpa.2013.04.003
Wu, F. L., Lu, W. Q., Shang, Y. Y., Kong, H., Li, L. S., Sui, Y. M., et al. (2016). Combined effects of seawater acidification and high temperature on hemocyte parameters in the thick shell mussel Mytilus coruscus. Fish. Shellfish. Immunol. 56, 554–562. doi: 10.1016/j.fsi.2016.08.012
Wu, R. S. (2002). Hypoxia: from molecular responses to ecosystem responses. Mar. Pollut. Bull. 45, 35–45. doi: 10.1016/S0025-326X(02)00061-9
Xie, H. L., Guo, R. J., Zhong, H.Z, Feng, Q., Lan, Z., et al. (2016). Shotgun metagenomics of 250 adult twins reveals genetic and environmental impacts on the gut microbiome. Cell. Syst. 3, 572–584. doi: 10.1016/j.cels.2016.10.004
Yang, L. G., Lv, L. L., Liu, H. J., Wang, M. R., Sui, Y. M., and Wang, Y. J. (2020). Effects of ocean acidification and microplastics on microflora community composition in the digestive tract of the thick shell mussel Mytilus Coruscus through 16S RNA gene sequencing. Bull. Environ. Contam. Toxicol. 1–10. doi: 10.1007/s00128-020-03022-5
Yang, M. J., Song, H., Sun, L. N., Yu, Z. L., Hu, Z., Wang, X. L., et al. (2019). Effect of temperature on the microflora community composition in the digestive tract of the veined rapa whelk (Rapana venosa) revealed by 16S rRNA gene sequencing. Comp. Biochem Physiol. 29, 145–153. doi: 10.1016/j.cbd.2018.10.006
Keywords: gut microbiota, ocean acidification, hypoxia, warming, 16S rRNA
Citation: Ullah Khan F, Shang Y, Chang X, Kong H, Zuberi A, Fang JKH, Liu W, Peng J, Zhang X, Hu M and Wang Y (2021) Effects of Ocean Acidification, Hypoxia, and Warming on the Gut Microbiota of the Thick Shell Mussel Mytilus coruscus Through 16S rRNA Gene Sequencing. Front. Mar. Sci. 8:736338. doi: 10.3389/fmars.2021.736338
Received: 05 July 2021; Accepted: 28 July 2021;
Published: 27 August 2021.
Edited by:
Jianmin Zhao, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Liqiang Zhao, Guangdong Ocean University, ChinaCopyright © 2021 Ullah Khan, Shang, Chang, Kong, Zuberi, Fang, Liu, Peng, Zhang, Hu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingzhi Zhang, ODE0NDQxNjU2QHFxLmNvbQ==; Youji Wang, eW91aml3YW5nMkBnbWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.