- 1MOE Key Laboratory of Marine Genetics and Breeding, College of Marine Life Sciences, Ocean University of China, Qingdao, China
- 2Laboratory for Marine Fisheries Science and Food Production Processes, Pilot Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Laboratory of Tropical Marine Germplasm Resources and Breeding Engineering, Sanya Oceanographic Institution, Ocean University of China, Sanya, China
Flavin-containing monooxygenase (FMO) is one of the most prominent xenobiotic metabolic enzymes. It can catalyze the conversion of heteroatom-containing chemicals to polar, readily excretable metabolites and is considered an efficient detoxification system for xenobiotics. Bivalves can accumulate paralytic shellfish toxins (PSTs) produced by dinoflagellates, especially during outbreaks of harmful algal blooms. Exploring FMO genes in bivalves may contribute to a better understanding of the adaptation of these species and the mechanisms of PSTs bioavailability. Therefore, through genome screening, we examined the expansion of FMO genes in two scallops (Patinopecten yessoensis and Chlamys farreri) and found a new subfamily (FMO_like). Our expression analyses revealed that, in both scallops, members of the FMO_N-oxide and FMO_like subfamilies were mainly expressed from the D-stage larvae to juveniles, whereas the FMO_GS-OX subfamily genes were mainly expressed at and prior to the trochophore stage. In adult organs, higher expressions of FMOs were observed in the kidney and hepatopancreas than in other organs. After exposure to PST-producing algae, expression changes in FMOs occurred in hepatopancreas and kidney of both scallops, with more members being up-regulated in hepatopancreas than in kidney for Alexandrium catenella exposure, while more up-regulated FMOs were found in kidney than in hepatopancreas of C. farreri exposed to A. minutum. Our findings suggest the adaptive functional diversity of scallop FMO genes in coping with the toxicity of PST-producing algae.
Introduction
Organisms have developed extensive molecular mechanisms to protect against the toxicity of various xenobiotic compounds, producing a variety of enzymes and proteins in response to xenobiotic exposure (Li et al., 2007; Raghunath et al., 2018). Flavin-containing monooxygenase (FMO), a prominent family of monooxygenases in eukaryotes (Naumann et al., 2002; Huijbers et al., 2014), is a major contributor to oxidative xenobiotic metabolism and is as important as cytochrome P450 (CYP450) (Manikandan and Nagini, 2018). FMO was first discovered in the 1960s in hepatic microsomes in pigs as an NADPH- and O2-requiring activity that converted N, N0-dimethylaniline to N-oxide (Ziegler and Pettit, 1964). To date, FMO homologs have been found in a range of organisms, including bacteria (Chen et al., 2011), fungi (Suh et al., 2000), marine invertebrates (Boutet et al., 2004), insects (Naumann et al., 2002; Sehlmeyer et al., 2010), and teleost fish (Peters et al., 1995; Larsen and Schlenk, 2001). FMO genes have been studied widely in mammals that possess five FMO genes (e.g., human FMO1-FMO5) (Cashman and Zhang, 2006). In unicellular eukaryotic organisms, such as yeast, there is only one FMO isoform (Eswaramoorthy et al., 2006), and in insects, there are two [e.g., in Drosophila melanogaster (Scharf et al., 2004)] or three [e.g., FMO1-FMO3 in lepidoptera (Sehlmeyer et al., 2010)]. Conversely, plants have various FMO genes. For example, there are 29 FMO genes in Arabidopsis thaliana (Schlaich, 2007).
FMOs utilize a tightly bound FAD prosthetic group, NAD(P)H, and molecular oxygen to create monooxygenate or otherwise oxidize substrates, producing water and NAD(P)+ as byproducts (Poulsen and Ziegler, 1995; Krueger and Williams, 2005). Generally, FMOs act on a large variety of nitrogen-, sulfur-, selenium-, or iodine-containing compounds, but carbon, phosphorus, and other elements are also amenable to FMO-mediated oxidation (Başaran and Can Eke, 2017). FMOs transfer hydroxyl groups to these substrates, rendering them more polar and more readily excretable (Cashman, 1995), and thus are thought to participate in the detoxification of foodstuffs and other xenobiotics (Hodgson et al., 1995). In humans, substantial evidence has shown that FMOs play an important role in the detoxification of tertiary amines [e.g., trimethylamine (Cholerton and Smith, 1991) and (S)-nicotine (Park et al., 1993)]. In mice, one FMO is involved in the detoxification pathway of l-methyl-4-phenyl-l,2,3,6-tetrahydropyridine (Cashman, 1989; Chiba et al., 1990; Di-Monte et al., 1991). A role for detoxification of xenobiotics by FMOs has also been observed for insects, including the European cinnabar moth (Tyria jacobaea), which utilizes senecionine N-oxygenase, an FMO, to efficiently N-oxidize pyrrolizidine alkaloids (PAs), which are toxic to unspecialized insects and vertebrates, thereby allowing larvae to feed on PA-containing plants (Naumann et al., 2002). A relatively high level of FMO activity has also been observed in guinea pigs, which can also efficiently detoxify PAs (Miranda et al., 1991). In bivalves, which are continuously exposed to non-biological contaminants (e.g., industry, agriculture, and urban effluents) (Boutet et al., 2003), FMOs may function in a tissue-specific toxin-responsive manner with differences in FMO expression between tissues in the stress response induced by hydrocarbons in Pacific oysters (Crassostrea gigas), revealing that FMO was upregulated after exposure to hydrocarbons in the digestive gland on day 7, while it was not expressed in the gill (Boutet et al., 2004).
In addition to non-biological contaminants (Boutet et al., 2003), bivalves are also exposed to a variety of biological contaminants. As filter-feeding organisms that mainly feed on microalgae, bivalves can accumulate large amounts of paralytic shellfish toxins (PSTs) produced by dinoflagellates, especially during the outbreak of harmful algal blooms (Andres et al., 2019). PSTs are a class of potent neurotoxins that reversibly block voltage-gated sodium channels in the cell membrane to prevent the occurrence of neuronal signals, eventually leading to muscle paralysis (Boullot et al., 2017). The contamination of bivalves by PSTs negatively impacts sanitary quality and often leads to the closure of related industries (Farabegoli et al., 2018). Although bivalves showed high PSTs resistance mainly because of PST-resistant amino acids in the voltage-gated sodium channel (Li et al., 2017), PSTs accumulation changes bivalve behavior and physiology (Shin et al., 2018). Many studies on the response of bivalves to PSTs stress found that CYP450 and GST, which are related to detoxification, were significantly upregulated in bivalves after exposure to PST-producing algae (Garcia-Lagunas et al., 2013; Lou et al., 2020), indicating that the detoxification process might be a strategy bivalves employ to cope with the effects of PST accumulation (Garcia-Lagunas et al., 2013). However, whether FMO genes are involved in this process remains unknown.
Yesso scallops (Patinopecten yessoensis) and Zhikong scallops (Chlamys farreri) are important aquaculture species in the two main sea areas (yellow sea and Bohai sea) in China (Li et al., 2017; Wang et al., 2017; Yang et al., 2019), which are both economically important, and the two species have been reported to accumulate PSTs (Li et al., 2017; Hu et al., 2019; Liu et al., 2020). Therefore, they are widely used to study PST accumulation and transformation (Hu et al., 2019; Xun et al., 2020). Previous studies have indicated that the hepatopancreas and kidney are the most toxic organs in scallops exposed to PSTs (Li et al., 2017). In this study, FMO genes were identified and characterized, and their responses were systematically assessed in the hepatopancreas and kidney after exposure to two different PST-producing dinoflagellates (Alexandrium catenella and Alexandrium minutum) in two scallops (P. yessoensis and C. farreri). We found a new subfamily of FMOs that has not been identified previously and indicated the involvement of bivalve FMOs in the response to the accumulation of PSTs. Our results will contribute to a better understanding of the functional diversity and adaptive evolution of FMO genes in bivalves in coping with toxic algae.
Materials and Methods
Genome-Wide Identification and Characterization of FMO Genes in Scallops
Candidate FMO genes have been identified in the genome (Li et al., 2017; Wang et al., 2017) and transcriptome (Hou et al., 2011; Wang et al., 2013) sequences of both P. yessoensis and C. farreri. The orthologous FMO sequences of some representative species, including humans (Homo sapiens), frogs (Xenopus laevis), zebrafish (Danio rerio), mice (Mus musculus), fruit flies (Drosophila melanogaster), Pacific oysters (C. gigas), and nematodes (Caenorhabditis elegans), from the NCBI1, and Uniprot2 databases were downloaded and used as queries to perform whole-genome and transcriptome-based blast with an e-value threshold of 1E-05. Then, the coding sequences were translated into amino acid sequences using an ORF Finder3, and the predicted FMO proteins were aligned to KEGG4, Uniprot (see text footnote 2), and the non-redundant database of the NCBI (see text footnote 1) using BLASTP (e-value set: 1E-05) to confirm the completeness and accuracy of the identified FMOs. The translated amino acid sequences were further analyzed using SMART tools (Schultz et al., 1998) and subjected to the NCBI Conserved Domains Database5 to identify the conserved domains. Finally, the physicochemical characteristics of the identified proteins, including the number of amino acids, molecular weight, and theoretical isoelectric point, were calculated using the ProtParam tool6.
Phylogenetic and Evolutionary Analysis of FMO Genes
The amino acid sequences of FMOs from selected organisms, including humans (H. sapiens), original chickens (Gallus gallus), frogs (X. laevis), zebrafish (D. rerio), amphioxus (Branchiostoma floridae), sea urchins (Strongylocentrotus purpuratus), freshwater snails (Biomphalaria glabrata and Pomacea canaliculata), and pacific oysters (C. gigas), were obtained from the NCBI database (Supplementary Table 1), and were combined with the FMOs identified in scallops to construct a phylogenetic tree. A phylogenetic analysis was conducted using MEGA7 (Kumar et al., 2016) to construct a neighbor-joining tree using the p-distance and maximum likelihood methods. We eliminated positions containing gaps and missing data, and tested the robustness of the results via a reanalysis of 1,000 bootstrap replicates.
To measure the selective pressure in the individual codons of the amino acid sequences, we applied codon-based site models in the PAML 4.9 package (Yang, 2007) with a maximum likelihood ratio test, which estimates the non-synonymous (dN) and synonymous (dS) substitution ratios (dN/dS or ω) variation among the codons. The codon-based site model assumes that different branches of the phylogenetic tree are under the same selection pressure. However, different amino acid positions experience different selection pressures, indicating that different positions of the same sequence have different ω values. Different ω values indicate different selective pressures, in which 1, < 1, and > 1 represent neutral evolution, purifying selection/negative selection, or positive selection, respectively. In our study, three pairs (M0 vs. M3, M1a vs. M2a, and M7 vs. M8) of models were applied. Model M0 (one ratio) assumes that all sites have a constant ω ratio, whereas model M3 (discrete) assumes that there are three classes of ω. The null model, M1a (neutral), allows two classes of codon sites for ω, wherein 0 < ω < 1 and ω = 1, whereas the alternative model, M2a (selection), allows one additional site class with ω > 1. The M7 (beta) model assumes that ω (0 < ω < 1) at all sites belongs to the matrix (0, 1) and fits a beta distribution with 10 categories, whereas the alternative model, M8, also has a category with ω > 1. If model M8 (M2a) fitted the data significantly better than model M7 (M1a), we applied a Bayes Empirical Bayes estimation method in the PAML 4.9 package (Yang, 2007) to identify the sites estimated to be under positive selection.
Expression Profiles of FMO Genes in Developmental Stages and Adult Organs/Tissues of Scallops
The expression profiles of FMOs in P. yessoensis (PyFMOs) and C. farreri (CfFMOs) during the developmental stages and in adult tissues were analyzed and represented in the form of RPKM (reads/kilobase of exon model/million mapped reads) based on the RNA-Seq data from previous studies (Li et al., 2017; Wang et al., 2017). The developmental stages include zygotes, 2–8 cell stage, blastula, gastrula, trochophore, D-stage larva, umbo early larva, umbo middle larva, umbo post larva, creeping larva, and juvenile scallop (Supplementary Table 2) and the adult tissues include the mantle, gill, gonad, kidney, hepatopancreas, smooth muscle, striated muscle, and foot. The body size of the sampled scallops were summarized in Supplementary Table 3. The heat map of the expression levels was drawn using custom R scripts to visualize the expression patterns of FMOs in P. yessoensis and C. farreri.
Expression Regulation of FMO Genes in Scallops After Exposure to PST-Producing Dinoflagellates
Scallops were previously subjected to PST-producing dinoflagellates, in which C. farreri was fed with A. catenella (ACDH strain) and A. minutum (AM-1 strain) (Li et al., 2017), and P. yessoensis was fed only A. catenella (ACDH strain). Briefly, two-year-old P. yessoensis and C. farreri were collected from Zhangzidao Group Co., Ltd. (Dalian, Liaoning Province, China) and Xunshan Group Co., Ltd. (Rongcheng, Shandong Province, China), respectively. Individual scallops were acclimated by feeding non-toxic algae (Isocrydid galbana) in pre-filtered seawater (30 spu; pH = 7.9) at 12–13°C for 3 weeks in our laboratory. The two toxic strains of dinoflagellates, A. catenella (ACDH strain) and A. minutum (AM-1 strain), which have been confirmed to produce PSTs (analogs C1 and C2 in ACDH, and GTXs, mainly GTX1-4 in AM-1), were cultured independently (Navarro et al., 2006) at 20°C under a light-dark cycle with 14 h light and 10 h dark (light intensity of 6,000 lx) in 10 L Erlenmeyer flasks (Guillard and Ryther, 1962), and collected when the cell density reached 3 × 105 cells/mL in the exponential growth phase. The acclimated scallops were individually cultured in a flat-bottom beaker with pre-filtered seawater (30 spu; pH = 7.9) at 12–13°C, and half-volume of the seawater were changed every day. A total of 3 L of dinoflagellate cells were fed to each acclimated scallop once a day at a density of 2,000 cells/mL. The exposure experiments lasted for 15 days, and scallop tissues were sampled at six time points, including days 0 (samples collected immediately before feeding A. catenella and A. minutum), 1, 3, 5, 10, and 15, in which three scallops were randomly selected at each time point. For each individual scallop, the total RNA was extracted from the hepatopancreas and kidney using the traditional phenol-chloroform method, following the guidelines described by Puissant and Houdebine (1990), and RNA-Seq libraries were constructed using the NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, United States) according to the manufacturer’s instructions, and then subjected to the Illumina X-ten platform with the mode of paired-end 150 bp. The expression levels of all FMOs were normalized and represented in the form of RPKM. The differentially expressed genes between the exposure and control groups were determined using the R package edgeR (Robinson et al., 2010) with p < 0.05 and | log2 FC| > 1. TBtools (Chen et al., 2020) was used to draw heat maps with the log2FC values.
Results and Discussion
Identification and Characterization of FMO Genes in Chlamys farreri and Patinopecten yessoensis
Through genome and transcriptome mining, 15 and 13 FMO genes were identified in P. yessoensis and C. farreri, respectively. Based on the phylogenetic analysis, the FMOs were classified into three subfamilies (Figure 1), including FMO_N-oxide, FMO_GS-OX, and FMO_like, wherein the gene numbers of these subfamilies were 8, 4, and 3 in P. yessoensis, and 7, 3, and 3 in C. farreri, respectively (Table 1). FMO_N-oxide was the only FMO subfamily that was present in both mammals and other animals, but FMO_GS-OX was not found in mammals, probably because of gene loss. Expansion of the two subfamilies was observed in mollusks, including scallops, when compared with vertebrates (Figure 1 and Table 1). Note that the FMO_like subfamily, which was identified for the first time in this study, was only found in scallops. As it was absent in other mollusks, including bivalve oysters, FMO_like might have evolved independently from the other two FMO subfamilies in scallops. A previous study characterized the long-term evolution of this FMO family and found that this family was strongly selected for purification at the protein level (Hao et al., 2009). Coastal marine organisms are continuously exposed to a wide variety of contaminants from industrial, agricultural, and urban effluents (Boutet et al., 2004). Thus, the diversity of FMO members in scallops may be a defense mechanism to cope with different chemical pollutants in the marine environment.
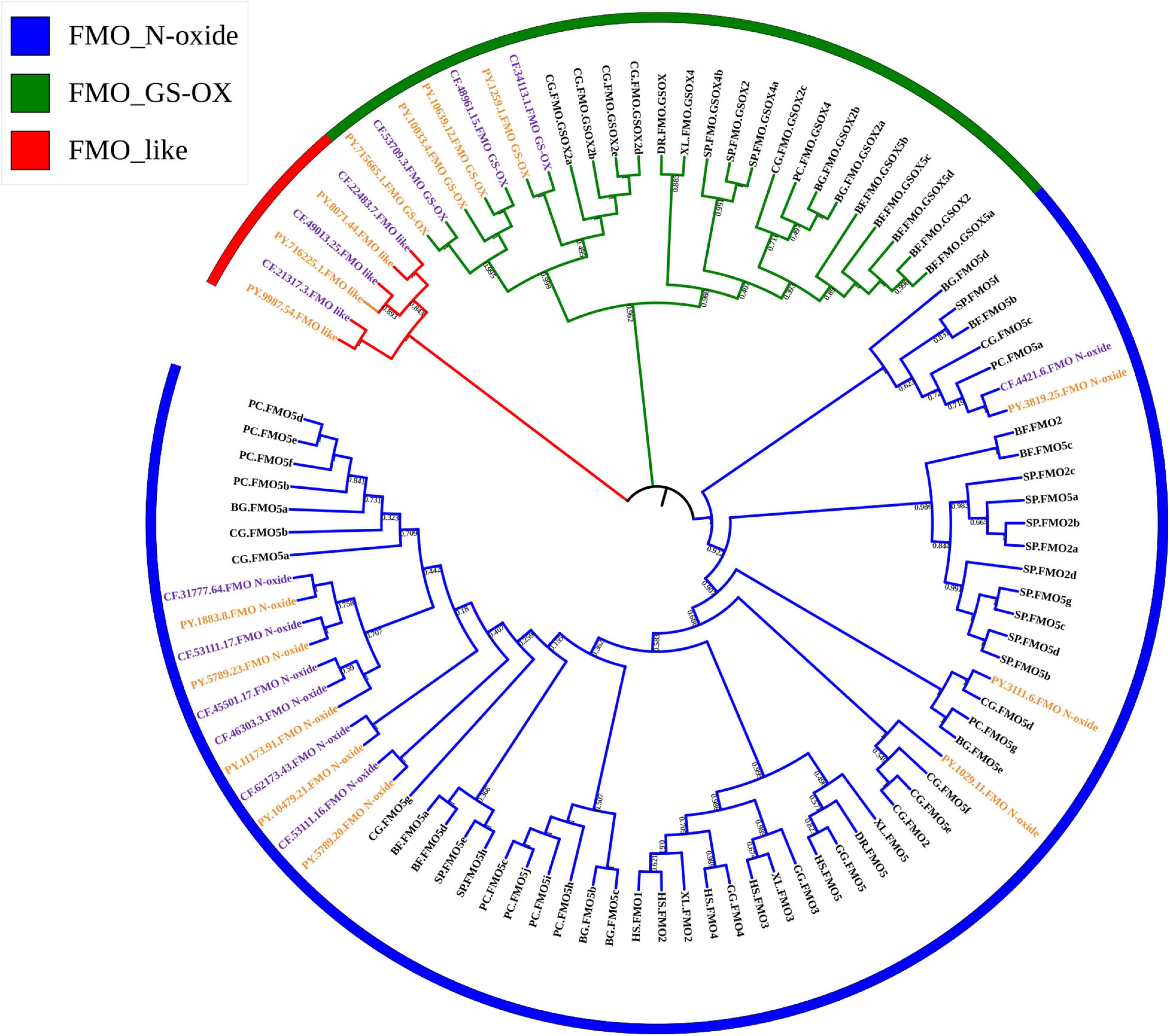
Figure 1. Phylogenetic tree of flavin-containing monooxygenase (FMO) amino acid sequences of selected organisms. The trees were constructed using the neighbor-joining (NJ) method with bootstrapping of 1,000 pseudo replicates. HS, Homo sapiens; GG, Gallus; XL, Xenopus laevis; DR, Danio rerio; BF, Branchiostoma floridae; SP, Strongylocentrotus purpuratus; BG, Biomphalaria glabrata; PC, Pomacea canaliculata; CG, Crassostrea gigas; CF, Chlamys farreri; PY, Patinopecten yessoensis.
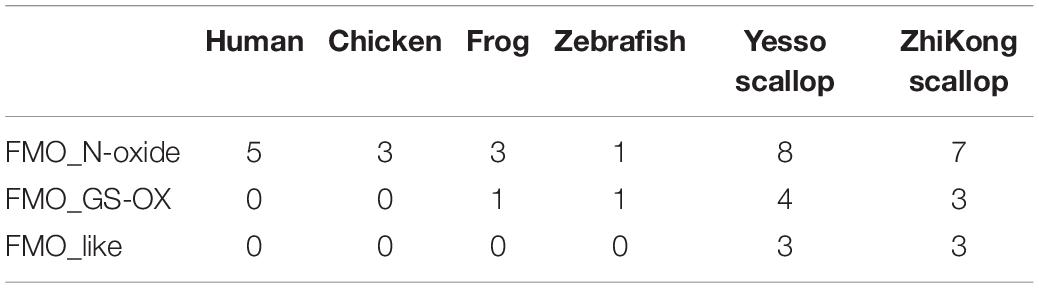
Table 1. Copy numbers of flavin-containing monooxygenase (FMO) subfamilies among selected vertebrate and scallop genomes.
The basic information regarding the PyFMOs and CfFMOs, including gene name, length of coding sequence (CDS), length of amino acid sequence, exon number, isoelectric point, molecular weight, and SwissProt annotation, is summarized in Supplementary Table 4. The CDS lengths of PyFMOs and CfFMOs varied from 1,224 to 1,758 bp and 1,287 to 3,084 bp, respectively, encoding 407 to 585 and 428 to 1,027 amino acids, respectively (Supplementary Table 4). The predicted molecular weight of the FMO proteins ranged from 49.00 to 65.90 kDa with PIs ranging from 5.40 to 9.11 in P. yessoensis and 49.20–117.00 kDa with PIs from 6.44 to 9.21 in C. farreri (Supplementary Table 4). Various exon/intron organization patterns of FMO genes are present in P. yessoensis and C. farreri (Figure 2), wherein the number of exons ranged from 3 to 10, and 3 to 12, respectively (Figure 2 and Supplementary Table 4).
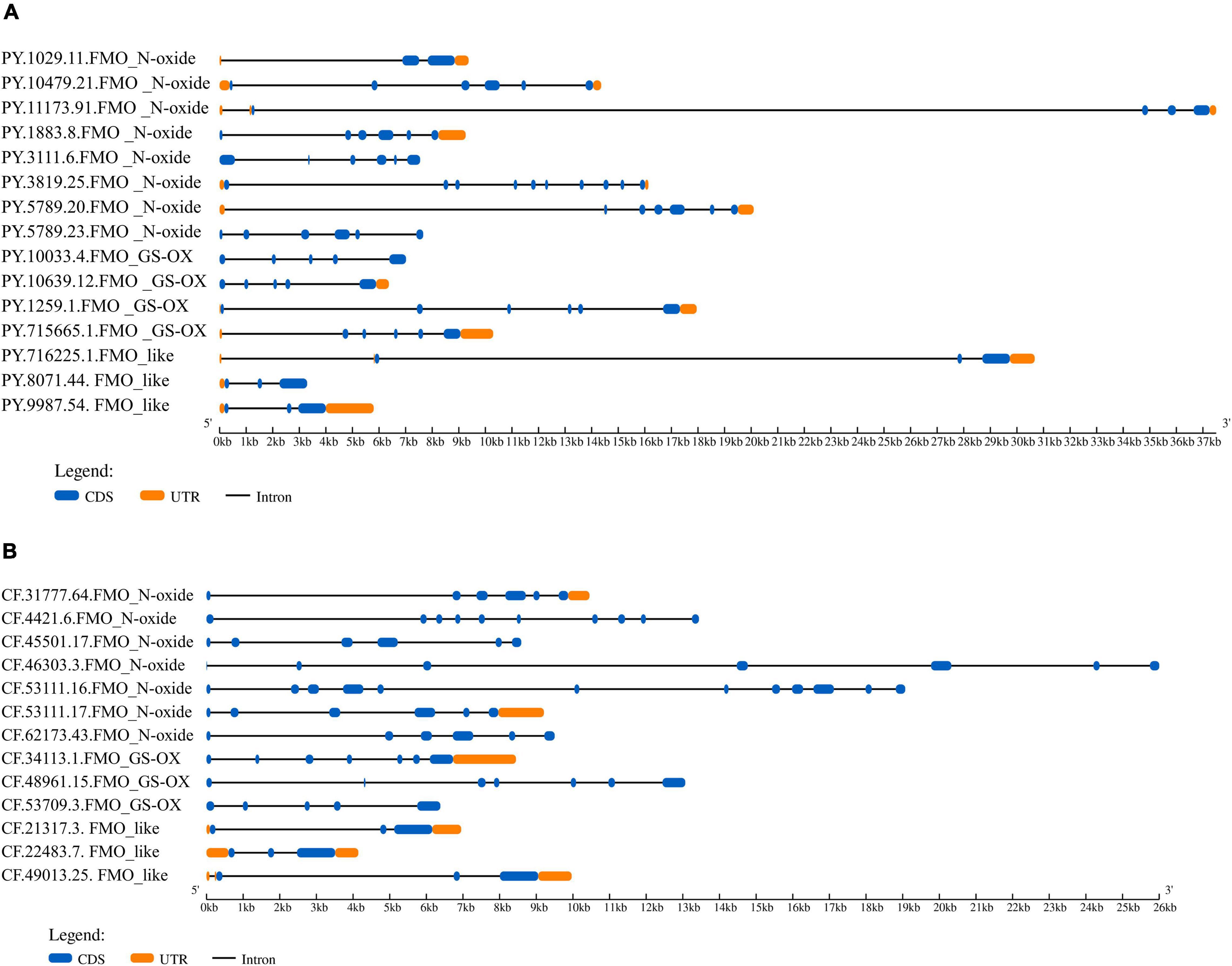
Figure 2. The structure of flavin-containing monooxygenase (FMO) genes in panel (A) Patinopecten yessoensis and (B) and Chlamys farreri. The blue boxes indicate the coding sequence (CDS). The orange boxes indicate UTR (untranslated region) sequence. The solid black lines represent intron sequences.
A protein structure analysis indicated that all PyFMOs and CfFMOs were composed of three conserved domains (Figure 3). The FAD-binding domain (GXGXXG), which was present in all the species examined, has been shown to be critical for their correct structure, catalytic function (Eswaramoorthy et al., 2006), and the FMO-identifying motif (FxGxxxHxxxY/F) (Eswaramoorthy et al., 2006), which interacts with the flavin of FAD (Figure 3). In addition to these two domains, the NADPH binding motif (GXGXXG/A) was detected at the C-terminal of the scallop FMO_N-oxide subfamily, but not in the other two subfamilies (Figure 3). Such circumstance was also observed in A. thaliana with this motif only detected in some members of all the FMO proteins (Eswaramoorthy et al., 2006), while the mechanism of the lack of this domain in some FMO proteins must be further studied. Identical characteristic sequences of three conserved domains have also been reported in other organisms (Choi et al., 2003; Eswaramoorthy et al., 2006; Schlaich, 2007).
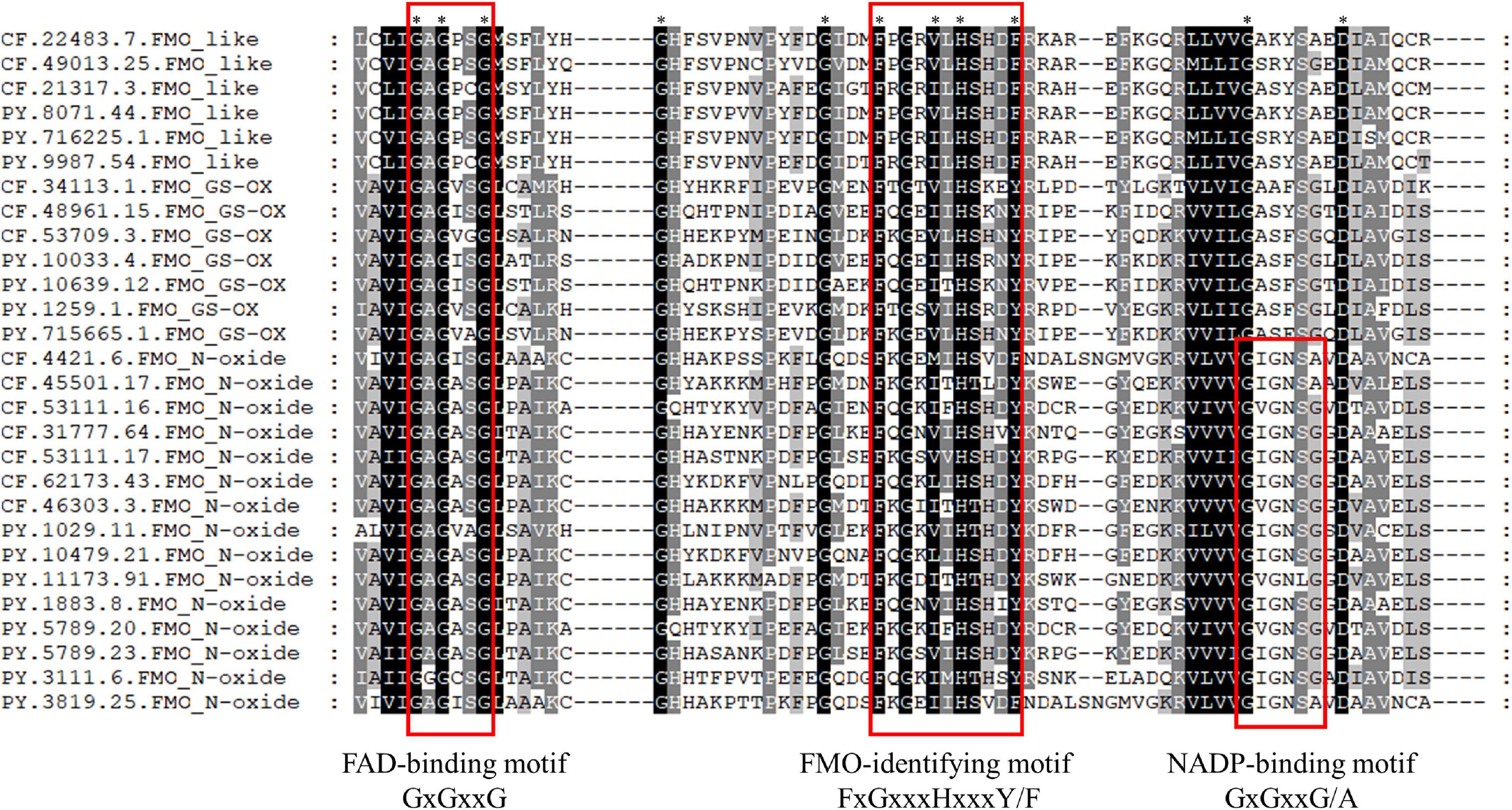
Figure 3. Alignment of the three conserved flavin-containing monooxygenase (FMO) motif sequences of Patinopecten yessoensis and Chlamys farreri scallops. ∗ indicates that all sequences have the same amino acid at the same site, and amino acids that are present in most sequences at the same site are shaded.
Selective Pressure Analysis of the Scallop FMO Genes
Gene replication and expansion by acquiring new genes or modifying existing genes driven by positive selection could provide support for the emergence of new functional genes (Kulmuni et al., 2013), which could possibly help organisms adapt to complex living conditions (Zhang et al., 2012; Ellegren, 2014). To analyze the selective pressure of the expanded scallop FMO genes at the protein level, we used the codon models implemented in PAML to evaluate their ω values in the two subfamilies (FMO_N-oxide and FMO_GS-OX) present in both vertebrates and scallops. We observed that the FMO_N-oxide and FMO_GS-OX subfamilies in P. yessoensis and C. farreri were under variable selection pressure with p-values of 0 (less than 0.05), indicating the rejection of the M0 model (single ratio) and support of the M3 model (discrete). These results demonstrate that P. yessoensis and C. farreri allow for different ω ratios of the two subfamilies. The P-value of the chi-square comparison test between the M7 and M8 models of the two subfamilies was 0 in P. yessoensis (Table 2), indicating the statistical significance of allowing positive selection for M8 in P. yessoensis, while in C. farreri, the P-values were 0.95 and 0.96, displaying no statistical significance (Table 2). These results suggest the presence of positively selected sites in the FMO_N-oxide and FMO_GS-OX subfamilies in P. yessoensis. We further analyzed the sites of the two subfamilies under positive selection (ω > 1) in the M8 model using the Bayes Empirical Bayes approach in PAML and observed 9 and 11 positively selected sites in the FMO_N-oxide and FMO_GS-OX subfamilies, respectively, with a posterior probability > 95% (Table 2). The expansion of FMO genes in scallops may reflect the requirement of a certain selection pressure for such genes as a strategy of molecular sequence evolution, which may be an important way to adapt to the complex and variable marine environment.
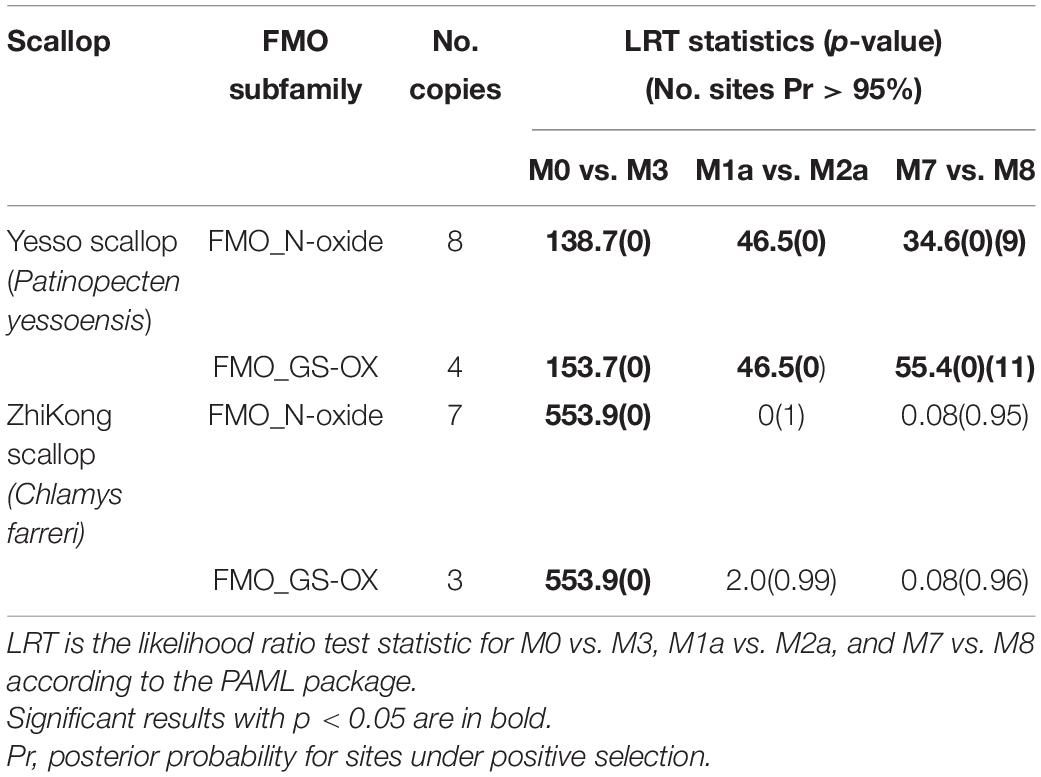
Table 2. Site model analysis for the expanded scallop flavin-containing monooxygenase (FMO) subfamilies.
Spatiotemporal Expression Pattern of Scallop FMO Genes
Previous studies have found stage-dependent expressions of FMOs during early organogenesis in Oryzias latipes that were regulated by developmental factors (Kupsco and Schlenk, 2017), suggesting the involvement of FMOs in early development. In this study, the expression patterns of PyFMO and CfFMO genes during different developmental stages (Figure 4) were analyzed. In general, most members of the FMO_N-oxide and FMO_like subfamilies in the two scallops were expressed during the post-trochophore stages from D-stage larvae to juveniles, while genes of the FMO_GS-OX subfamily were mainly expressed prior to and at the trochophore stage (Figure 4). PyFMO and CfFMO genes showed, to some extent, different stage-specific expressions. For PyFMOs, two members (PY.1259.1 and PY.10033.4) of the FMO_GS-OX subfamily and one member (PY.716225.1) of the FMO_like subfamily were mainly expressed from the zygote to the blastula stages, while two members (PY.1029.11 and PY3111.6) of the FMO_N-oxide subfamily and one member (PY.8071.44) of the FMO_like subfamily were expressed at the trochophore larval stage. The remaining eight members played a major role in post-larval development (Figure 4A). Regarding CfFMOs, one member (CF.45501.17) of the FMO_N-oxide subfamily and one member (CF.48961.15) of the FMO_GS-OX subfamily showed elevated expression at multiple developmental stages, while one member (CF.22483.7) of the FMO_like subfamily and two members (CF.53709.3 and CF.34113.1) of the FMO_GS-OX subfamily were highly expressed at only one or two developmental stages, specifically prior to and at the D-stage (Figure 4B). The remaining eight members were highly expressed in the middle and late umbo stages (Figure 4).
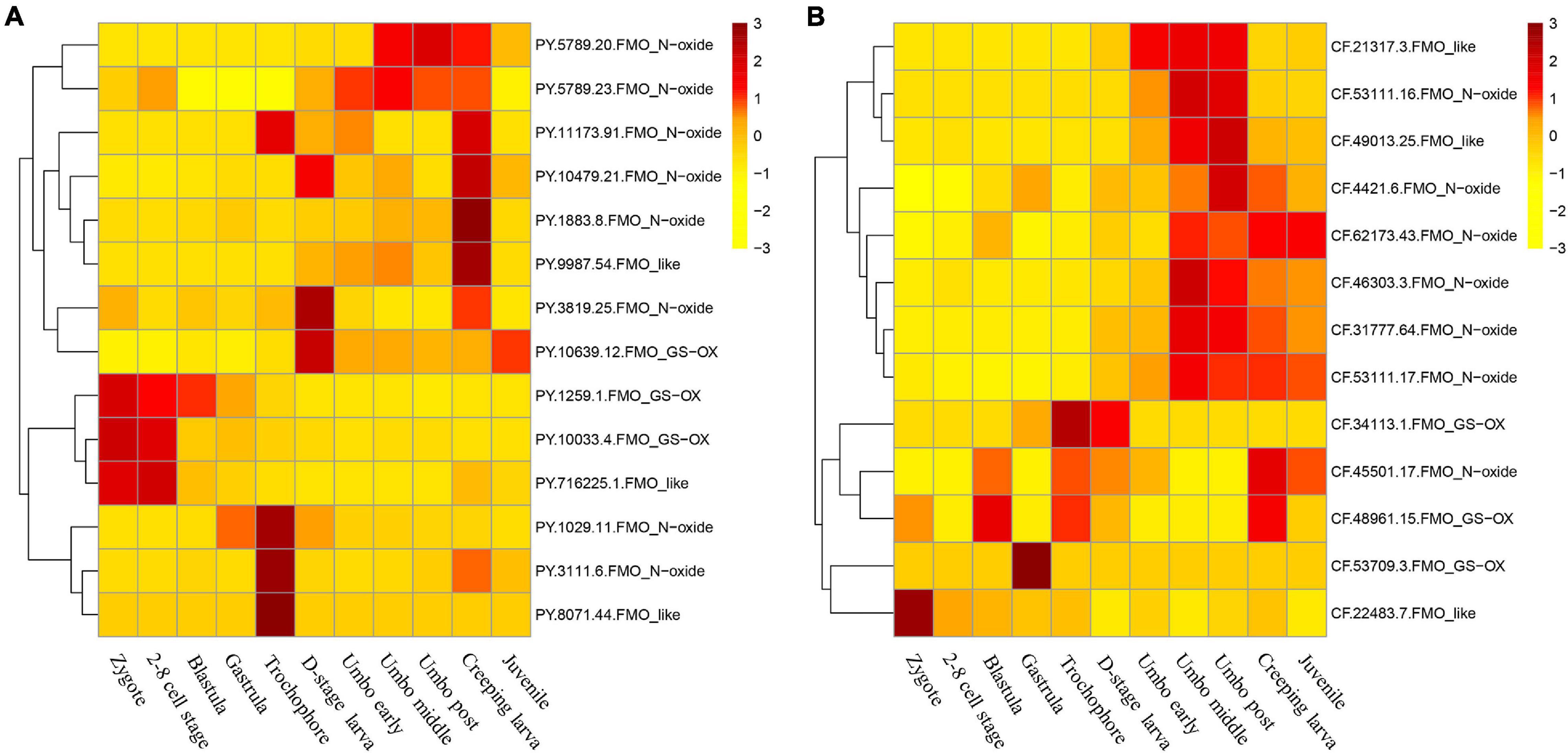
Figure 4. Expression patterns of scallop flavin-containing monooxygenases (FMOs) during the developmental stages of (A) Patinopecten yessoensis and (B) Chlamys farreri (In P. yessoensis, one member (PY.715665.1.FMO_GS-OX) was not expressed in the developmental stages, thus only 14 genes were showed in the heat map).
In adult scallops, higher FMO expressions were detected mostly in the kidney and hepatopancreas, wherein, five and six members of PyFMO, and eight and three members of CfFMO were highly expressed, respectively (Figure 5). Abundant FMO transcripts in the kidney and liver (or hepatopancreas) were also found in many other organisms, including humans (Cashman and Zhang, 2006) and marmosets (Callithrix jacchus) (Uehara et al., 2017). The kidney and hepatopancreas are crucial organs for detoxifying chemicals (Metian et al., 2008; Banni et al., 2010) and removing metabolic waste (Bourioug et al., 2018; Wakashin et al., 2018) in mollusks. Previous studies in scallops have indicated that the hepatopancreas and kidney are two major toxic organs of bivalves (Tian et al., 2010; Li et al., 2017). The scallop FMOs with abundant expression in the kidney and hepatopancreas may participate in similar xenobiotic detoxification processes observed in other organisms (Phillips and Shephard, 2017). In addition, some other FMOs in scallops exhibited high expression in the mantle, gills, and gonads (Figure 5), implying their functions in detoxification in these tissues.
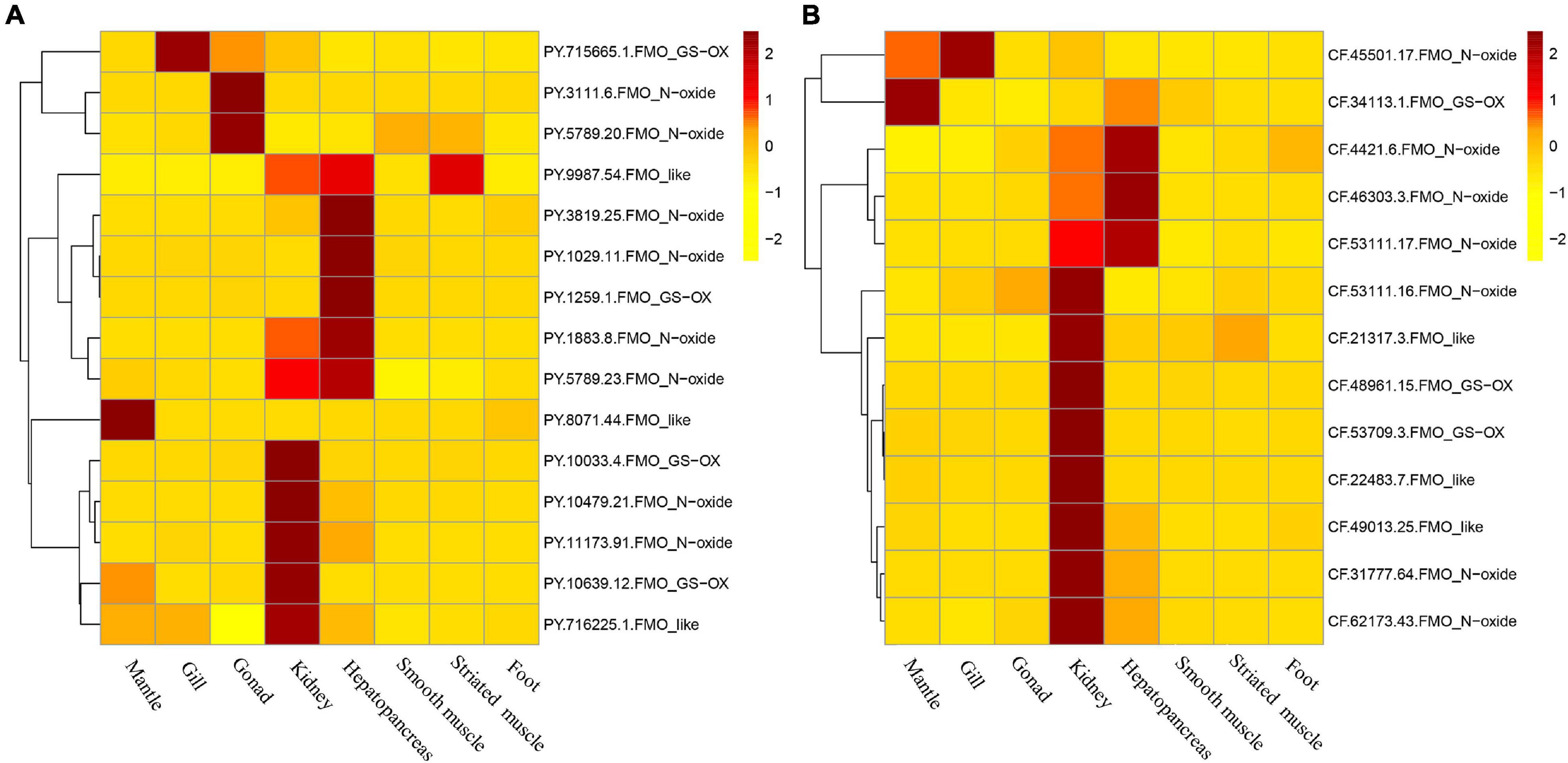
Figure 5. Expression patterns of scallop flavin-containing monooxygenases (FMOs) in adult tissues of (A) Patinopecten yessoensis and (B) and Chlamys farreri.
Expression Regulation of Scallop FMOs After Toxic Dinoflagellates Exposure
To evaluate the possible involvement of scallop FMOs in coping with PST-producing algae, we examined their expression changes after exposure to toxic dinoflagellates in the hepatopancreas and kidney, which are the two most toxic organs with respect to PSTs accumulation (Li et al., 2017). For P. yessoensis exposed to A. catenella, a total of 7 PyFMOs were found to be significantly regulated (| log2FC| ≧ 1, p-value < 0.05) in the hepatopancreas (Figure 6), which is the main organ for PSTs accumulation (Li et al., 2017). Of these, 6 were upregulated, and 2 (PY.11173.91 and PY.8071.44) responded acutely on days 1 and 3, respectively, whereas the other PyFMOs responded chronically on days 5 and 10. For each of the three subfamilies, two upregulated PyFMOs were found (Figure 6). The only down-regulated PyFMO (PY.1883.8) was from the FMO_N-oxide subfamily and was found on day 10 (Figure 6). Meanwhile, in the kidney, wherein PSTs can be transformed into derivatives with higher toxicity (Li et al., 2017; Liu et al., 2020), only two PyFMOs (PY.1259.1 and PY.9987.54) were significantly regulated (Figure 6). Specifically, one (PY.1259.1) was from the FMO_GS-OX subfamily, which was down-regulated on days 3 and 5, and then upregulated on day 10, and one (PY.9987.54) was from the FMO_like subfamily, which was rapidly upregulated on day 1 and showed continuous upregulation throughout the entire exposure experiment, implying its vital role in responding to PST stress (Figure 6). Thus, the PyFMOs regulated in the hepatopancreas and kidney were different.
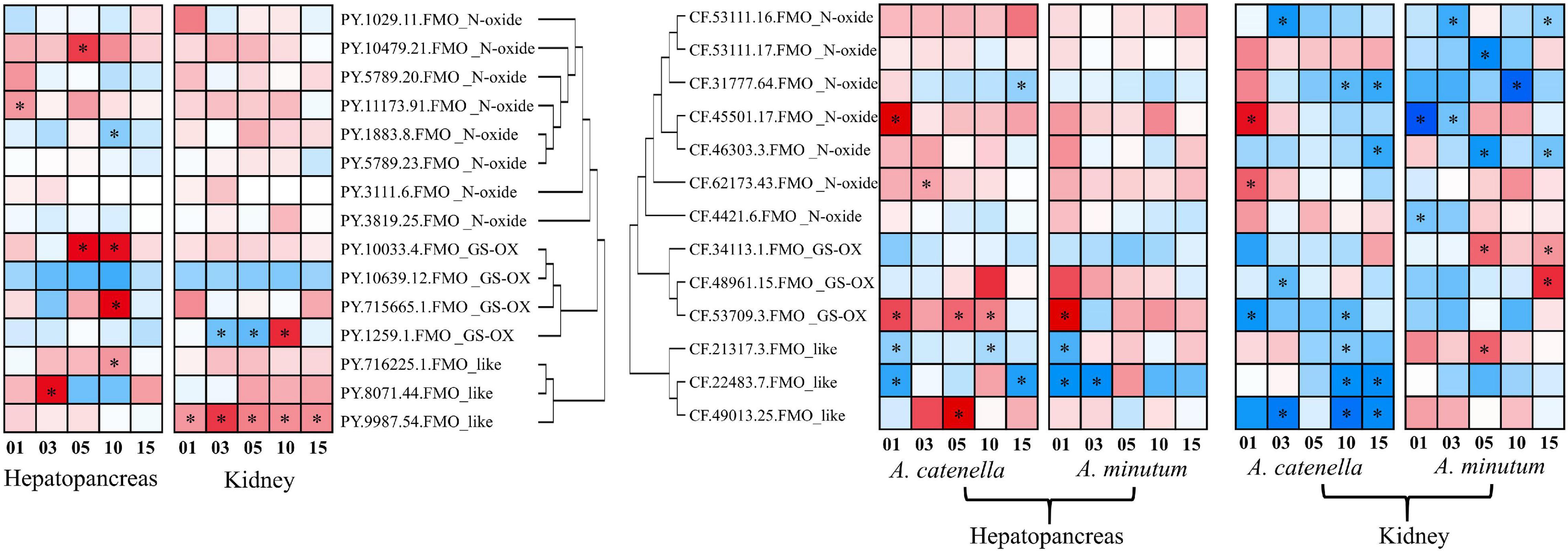
Figure 6. Temporal expressions of scallop flavin-containing monooxygenases (FMOs) in the hepatopancreas and kidney of scallops after Alexandrium catenella and Alexandrium minutum exposure. The heat maps were drawn based on log2 (fold change) values. Exposure time (1, 3, 5, 10, or 15 days) is displayed below the heat map. Significant regulation with | log2FC| > 1 and p < 0.05 is indicated with *.
We further examined the expression changes of FMOs in the hepatopancreas and kidney of C. farreri, another massively cultured scallop in China. In addition to A. catenella, another dinoflagellate, A. minutum was also used to feed C. farreri to assess the response differences of CfFMOs under exposure to different PSTs. Generally, more CfFMOs were significantly regulated in the kidney than in the hepatopancreas (Figure 6). In the hepatopancreas, a total of 7 CfFMOs showed significant responses after exposure to the two toxic dinoflagellates, among which 3 showed similar acute responses with both diets, including 1 (CF.53709.3) upregulated and 2 down-regulated (CF.21317.3 and CF.22483.7). The other four regulated CfFMOs only significantly responded to A. catenella exposure. In particular, two members (CF.45501.17 and CF.62173.43) of the FMO_N-oxide subfamily and one (CF.49013.25) of the FMO_like subfamily were significantly upregulated, respectively, in acute and chronic manners, and one gene (CF.31777.64) of the FMO_N-oxide subfamily was significantly downregulated in a chronic manner on day 15.
In the kidney, all the CfFMOs showed significant responses after exposure to toxic A. catenella or A. minutum, most of which were significantly downregulated and showed dinoflagellate-type dependent responses. In particular, the two upregulated CfFMOs after A. catenella exposure were found on day 1, both of which are from the FMO_N-oxide subfamily. For A. minutum exposure, the three upregulated members were from the other two subfamilies and exhibited chronic responses. These results suggest the PSTs- and organ-dependent responses of CfFMOs. Note that one member (CF.45501.17) showed consistent acute upregulation in both the kidney and hepatopancreas after exposure to A. catenella, which could be a good candidate indicator gene for the accumulation of PSTs.
Conclusion
In this study, we performed the first systematic analysis of the FMO gene family in scallops P. yessoensis and C. farreri, and observed the expansion of this family possibly with a bivalve- or scallop-specific gene duplication event. In addition, considerable structural diversity, conserved domains, and motifs were identified, and stage- and tissue-specific expression patterns were revealed. Furthermore, the genes from the expanded subfamily of P. yessoensis may be more inclined toward positive selection and may be involved in lineage-specific adaptation. After exposure to two different toxic Alexandrium species, the regulation patterns of the scallop FMOs presented time-, algal strain-, and organ-dependent expression patterns. These findings will contribute to a better understanding of the evolutionary origin of the FMO gene family and the detoxification strategies of bivalves in response to algal biotoxins in complex marine environments.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XH conceived and designed the study. ML and JS performed the experiments. LK, PL, YL, and HW participated in the data analysis. LK and PL wrote the manuscript. YL and XH revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The project was supported by National Key R&D Project (2018YFD0900604), National Natural Science Foundation of China (U1706203), Taishan Industry Leading Talent Project (tscy2018116), and Sanya Yazhou Bay Science and Technology City (SKJC-KJ-2019KY01).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.732000/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/
- ^ https://www.uniprot.org/
- ^ https://www.ncbi.nlm.nih.gov/orffinder/
- ^ http://www.kegg.jp/
- ^ https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^ http://www.expasy.org/tools/protparam.html
References
Andres, J. K., Yñiguez, A. T., Maister, J. M., Turner, A. D., Olano, D., Mendoza, J., et al. (2019). Paralytic shellfish toxin uptake, assimilation, depuration, and transformation in the Southeast Asian green-lipped mussel (Perna viridis). Toxins 11:468. doi: 10.3390/toxins11080468
Banni, M., Negri, A., Dagnino, A., Jebali, J., Ameur, S., and Boussetta, H. (2010). Acute effects of benzo[a]pyrene on digestive gland enzymatic biomarkers and DNA damage on mussel Mytilus galloprovincialis. Ecotoxicol. Environ. Saf. 73, 842–848. doi: 10.1016/j.ecoenv.2009.12.032
Başaran, R., and Can Eke, B. (2017). Flavin containing monooxygenases and metabolism of xenobiotics. Turk. J. Pharm. Sci. 14, 90–94. doi: 10.4274/tjps.30592
Boullot, F., Castrec, J., Bidault, A., Dantas, N., Payton, L., Perrigault, M., et al. (2017). Molecular characterization of voltage-gated sodium channels and their relations with paralytic shellfish toxin bioaccumulation in the Pacific Oyster Crassostrea gigas. Mar. Drugs 15:21. doi: 10.3390/md15010021
Bourioug, M., Mazzitelli, J. Y., Marty, P., Budzinsky, H., Aleya, L., Bonnafé, E., et al. (2018). Assessment of Lemna minor (duckweed) and Corbicula fluminea (freshwater clam) as potential indicators of contaminated aquatic ecosystems: responses to presence of psychoactive drug mixtures. Environ. Sci. Pollut. Res. Int. 25, 11192–11204. doi: 10.1007/s11356-017-8447-1
Boutet, I., Tanguy, A., and Moraga, D. (2004). Molecular identification and expression of two non-P450 enzymes, monoamine oxidase A and flavin-containing monooxygenase 2, involved in phase I of xenobiotic biotransformation in the Pacific oyster, Crassostrea gigas. Biochim. Biophys. Acta 1679, 29–36. doi: 10.1016/j.bbaexp.2004.04.001
Boutet, I., Tanguy, A., Rousseau, S., Auffret, M., and Moraga, D. (2003). Molecular identification and expression of heat shock cognate 70 (hsc70) and heat shock protein 70 (hsp70) genes in the Pacific Oyster Crassostrea gigas. Cell Stress Chaperones 8, 76–85. doi: 10.1379/1466-126820038<76:miaeoh<2.0.co;2
Cashman, J. R. (1989). Enantioselective N-oxygenation of verapamil by the hepatic flavin-containing monooxygenase. Mol. Pharmacol. 36, 497–503. doi: 10.1111/j.1365-2885.1989.tb00683.x
Cashman, J. R. (1995). Structural and catalytic properties of the mammalian flavin-containing monooxygenase. Chem. Res. Toxicol. 8, 166–181. doi: 10.1021/tx00044a001
Cashman, J. R., and Zhang, J. (2006). Human flavin-containing monooxygenases. Annu. Rev. Pharmacol. Toxicol. 46, 65–100. doi: 10.1146/annurev.pharmtox.46.120604.141043
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Y., Patel, N. A., Crombie, A., Scrivens, J. H., and Murrell, J. C. (2011). Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc. Natl. Acad. Sci. U. S. A. 108, 17791–17796. doi: 10.1073/pnas.1112928108
Chiba, K., Horii, H., Kubota, E., Ishizaki, T., and Kato, Y. (1990). Effects of N-methylmercaptoimidazole on the disposition of MPTP and its metabolites in mice. Eur. J. Pharmacol. 180, 59–67. doi: 10.1016/0014-2999(90)90592-t
Choi, H. S., Kim, J. K., Vho, E. H., Kim, Y. C., Kim, J. I., and Kim, S. W. (2003). A novel flavin-containing monooxygenase from Methylophaga sp. strain SK1 and its indigo synthesis in Escherichia coli. Biochem. Biophys. Res. Commun. 306, 930–936. doi: 10.1016/S0006-291X(03)01087-8
Cholerton, S., and Smith, R. L. (1991). “Human pharmacogenetics of nitrogen oxidations,” in N-Oxidation of Drugs, eds P. Hlavica and L. A. Damani (Berlin: Springer) 107–131.
Di-Monte, D. A., Wu, E. Y., Irwin, I., Delanney, L. E., and Langston, J. W. (1991). Biotransformation of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine in primary cultures of mouse astrocytes. J. Pharmacol. Exp. Ther. 258, 594–600. doi: 10.1016/0160-5402(91)90057-C
Ellegren, H. (2014). Genome sequencing and population genomics in non-model organisms. Trends Ecol. Evol. 29, 51–63. doi: 10.1016/j.tree.2013.09.008
Eswaramoorthy, S., Bonanno, J. B., Burley, S. K., and Swaminathan, S. (2006). Mechanism of action of a flavin-containing monooxygenase. Proc. Natl. Acad. Sci. U. S. A. 103, 9832–9837. doi: 10.1073/pnas.0602398103
Farabegoli, F., Blanco, L., Rodríguez, L. P., Vieites, J. M., and Cabado, A. G. (2018). Phycotoxins in marine shellfish: origin, occurrence and effects on humans. Mar. Drugs 16:188. doi: 10.3390/md16060188
Garcia-Lagunas, N., Romero-Geraldo, R., and Hernandez-Saavedra, N. Y. (2013). Genomics study of the exposure effect of Gymnodinium catenatum, a paralyzing toxin producer, on Crassostrea gigas’ defense system and detoxification genes. PLoS One 8:e72323. doi: 10.1371/journal.pone.0072323
Guillard, R. R. L., and Ryther, J. H. (1962). Studies of marine planktonic diatoms. III Some effects of temperature on respiration of five species. Can. J. Microbiol. 8, 447–453. doi: 10.1139/m62-058
Hao, D., Chen, S. L., Mu, J., and Xiao, P. G. (2009). Molecular phylogeny, long-term evolution, and functional divergence of flavin-containing monooxygenases. Genetica 137, 173–187. doi: 10.1007/s10709-009-9382-y
Hodgson, E., Blake, B. L., Levi, P. E., Mailman, R. B., Lawton, M. P., Philpot, R. M., et al. (1995). “Flavin-containing monooxygenases: substrate specificity and complex metabolic pathways,” in Molecular Aspects of Oxidative Drug Metabolizing Enzymes. Nato ASI Series, eds E. Arinç, J. B. Schenkman, and E. Hodgson (Berlin: Springer) 225–235.
Hou, R., Bao, Z., Wang, S., Su, H., Li, Y., Du, H., et al. (2011). Transcriptome sequencing and de novo analysis for Yesso scallop (Patinopecten yessoensis) using 454 GS FLX. PLoS One 6:e21560. doi: 10.1371/journal.pone.0021560
Hu, B., Li, M., Yu, X., Xun, X., Lu, W., Li, X., et al. (2019). Diverse expression regulation of Hsp70 genes in scallops after exposure to toxic Alexandrium dinoflagellates. Chemosphere 234, 62–69. doi: 10.1016/j.chemosphere.2019.06.034
Huijbers, M. M., Montersino, S., Westphal, A. H., Tischler, D., and van Berkel, W. J. (2014). Flavin dependent monooxygenases. Arch. Biochem. Biophys. 544, 2–17. doi: 10.1016/j.abb.2013.12.005
Krueger, S. K., and Williams, D. E. (2005). Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol. Ther. 106, 357–387. doi: 10.1016/j.pharmthera.2005.01.001
Kulmuni, J., Wurm, Y., and Pamilo, P. (2013). Comparative genomics of chemosensory protein genes reveals rapid evolution and positive selection in ant-specific duplicates. Heredity (Edinb) 110, 538–547. doi: 10.1038/hdy.2012.122
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kupsco, A., and Schlenk, D. (2017). Developmental expression and regulation of flavin-containing monooxygenase by the unfolded protein response in Japanese medaka (Oryzias latipes). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 191, 7–13. doi: 10.1016/j.cbpc.2016.09.003
Larsen, B. K., and Schlenk, D. (2001). Effect of salinity on flavin-containing monooxygenase expression and activity in rainbow trout (Oncorhynchus mykiss). J. Comp. Physiol. B. 171, 421–429. doi: 10.1007/s003600100192
Li, X., Schuler, M. A., and Berenbaum, M. R. (2007). Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 52, 231–253. doi: 10.1146/annurev.ento.51.110104.151104
Li, Y., Sun, X., Hu, X., Xun, X., Zhang, J., Guo, X., et al. (2017). Scallop genome reveals molecular adaptations to semi-sessile life and neurotoxins. Nat. Commun. 8:1721. doi: 10.1038/s41467-017-01927-0
Liu, Y., Kong, F. Z., Xun, X. G., Dai, L., Geng, H. X., Hu, X. L., et al. (2020). Biokinetics and biotransformation of paralytic shellfish toxins in different tissues of Yesso scallops, Patinopecten yessoensis. Chemosphere 261:128063. doi: 10.1016/j.chemosphere.2020.128063
Lou, J., Cheng, J., Xun, X., Li, X., Zhang, X., Li, T., et al. (2020). Glutathione S-transferase genes in scallops and their diverse expression patterns after exposure to PST-producing dinoflagellates. Mar. Life Sci. Technol. 2, 252–261. doi: 10.1007/s42995-020-00050-2
Manikandan, P., and Nagini, S. (2018). Cytochrome P450 structure, function and clinical significance: a Review. Curr. Drug. Targets 19, 38–54. doi: 10.2174/1389450118666170125144557
Metian, M., Warnau, M., Cosson, R. P., Oberhänsli, F., and Bustamante, P. (2008). Bioaccumulation and detoxification processes of Hg in the king scallop Pecten maximus: field and laboratory investigations. Aquat. Toxicol. 90, 204–213. doi: 10.1016/j.aquatox.2008.08.014
Miranda, C. L., Chung, W., Reed, R. E., Zhao, X., Henderson, M. C., Wang, J. L., et al. (1991). Flavin-containing monooxygenase: a major detoxifying enzyme for the pyrrolizidine alkaloid senecionine in guinea pig tissues. Biochem. Biophys. Res. Commun. 178, 546–552. doi: 10.1016/0006-291x(91)90142-t
Naumann, C., Hartmann, T., and Ober, D. (2002). Evolutionary recruitment of a flavin-dependent monooxygenase for the detoxification of host plant-acquired pyrrolizidine alkaloids in the alkaloid-defended arctiid moth Tyria jacobaeae. Proc. Natl. Acad. Sci. U. S. A. 99, 6085–6090. doi: 10.1073/pnas.082674499
Navarro, J., Muñoz, M., and Contreras, A. (2006). Temperature as a factor regulating growth and toxin content in the dinofagellate Alexandrium catenella. Harmful Algae 5, 762–769. doi: 10.1016/j.hal.2006.04.001
Park, S. B., Jacob, P. III., Benowitz, N. L., and Cashman, J. R. (1993). Stereoselective metabolism of (S)-(-)-nicotine in humans: formation of trans-(S)-(-)-nicotine N-1’-oxide. Chem. Res. Toxicol. 6, 880–888. doi: 10.1021/tx00036a019
Peters, L. D., Livingstone, D. R., Shenin-Johnson, S., Hines, R. N., and Schlenk, D. (1995). Characterization of hepatic flavin monooxygenase from the marine teleost turbot (Scophthalmus maximus L.). Xenobiotica 25, 121–131. doi: 10.3109/00498259509061838
Phillips, I. R., and Shephard, E. A. (2017). Drug metabolism by flavin-containing monooxygenases of human and mouse. Expert. Opin. Drug Metab. Toxicol. 13, 167–181. doi: 10.1080/17425255.2017.1239718
Poulsen, L. L., and Ziegler, D. M. (1995). Multisubstrate flavin-containing monooxygenases: applications of mechanism to specificity. Chem. Biol. Interact. 96, 57–73. doi: 10.1016/0009-2797(94)03583-t
Puissant, C., and Houdebine, L. M. (1990). An improvement of the single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. BioTechniques 8, 148–149. doi: 10.1038/nbt0290-148
Raghunath, A., Sundarraj, K., Nagarajan, R., Arfuso, F., Bian, J., Kumar, A. P., et al. (2018). Antioxidant response elements: discovery, classes, regulation and potential applications. Redox. Biol. 17, 297–314. doi: 10.1016/j.redox.2018.05.002
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Scharf, M. E., Scharf, D. W., Bennett, G. W., and Pittendrigh, B. R. (2004). Catalytic activity and expression of two flavin-containing monooxygenases from Drosophila melanogaster. Arch. Insect. Biochem. Physiol. 57, 28–39. doi: 10.1002/arch.20012
Schlaich, N. L. (2007). Flavin-containing monooxygenases in plants: looking beyond detox. Trends Plant Sci. 12, 412–418. doi: 10.1016/j.tplants.2007.08.009
Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95, 5857–5864. doi: 10.1073/pnas.95.11.5857
Sehlmeyer, S., Wang, L., Langel, D., Heckel, D. G., Mohagheghi, H., Petschenka, G., et al. (2010). Flavin-dependent monooxygenases as a detoxification mechanism in insects: new insights from the arctiids (lepidoptera). PLoS One 5:e10435. doi: 10.1371/journal.pone.0010435
Shin, C., Jo, H., Kim, S. H., and Kang, G. J. (2018). Exposure assessment to paralytic shellfish toxins through the shellfish consumption in Korea. Food Res. Int. 108, 274–279. doi: 10.1016/j.foodres.2018.03.061
Suh, J. K., Poulsen, L. L., Ziegler, D. M., and Robertus, J. D. (2000). Redox regulation of yeast flavin-containing monooxygenase. Arch. Biochem. Biophys. 381, 317–322. doi: 10.1006/abbi.2000.1965
Tian, H., Gao, C., Wang, Z., Sun, P., Fan, S., and Zhu, M. (2010). Comparative study on in vitro transformation of paralytic shellfish poisoning (PSP) toxins in different shellfish tissues. Acta Oceanol. Sin. 29, 120–126. doi: 10.1007/s13131-010-0015-1
Uehara, S., Shimizu, M., Uno, Y., Inoue, T., Sasaki, E., and Yamazaki, H. (2017). Marmoset flavin-containing monooxygenase 3 in the liveris a major benzydamine and sulindac sulfide oxygenase. Drug Metab. Dispos. 45, 497–500. doi: 10.1124/dmd.117.075184
Wakashin, H., Seo, E., and Seo, Y. (2018). Accumulation and excretion of manganese ion in the kidney of Mytilus galloprovincialis. J. Exp. Biol. 221:jeb185439. doi: 10.1242/jeb.185439
Wang, S., Hou, R., Bao, Z., Du, H., He, Y., Su, H., et al. (2013). Transcriptome sequencing of Zhikong scallop (Chlamys farreri) and comparative transcriptomic analysis with Yesso scallop (Patinopecten yessoensis). PLoS One 8:e63927. doi: 10.1371/journal.pone.0063927
Wang, S., Zhang, J., Jiao, W., Li, J., Xun, X., Sun, Y., et al. (2017). Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 1:120. doi: 10.1038/s41559-017-0120
Xun, X., Cheng, J., Wang, J., Li, Y., Li, X., Li, M., et al. (2020). Solute carriers in scallop genome: Gene expansion and expression regulation after exposure to toxic dinoflagellate. Chemosphere, 241:124968. doi: 10.1016/j.chemosphere.2019.124968
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yang, Z., Li, X., Liao, H., Hu, L., Peng, C., Wang, S., et al. (2019). A Molecular Cytogenetic Map of Scallop (Patinopecten yessoensis). Mar. Biotechnol. 21, 731–742. doi: 10.1007/s10126-019-09918-6
Zhang, G., Fang, X., Guo, X., Li, L., Luo, R., Xu, F., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. doi: 10.1038/nature11413
Keywords: scallop, flavin-containing monooxygenase, paralytic shellfish toxins exposure, transcriptional responses, detoxification
Citation: Kong L, Liu P, Li M, Wang H, Shi J, Hu J, Li Y and Hu X (2021) Transcriptional Responses of Flavin-Containing Monooxygenase Genes in Scallops Exposed to PST-Producing Dinoflagellates Implying Their Involvements in Detoxification. Front. Mar. Sci. 8:732000. doi: 10.3389/fmars.2021.732000
Received: 29 June 2021; Accepted: 09 August 2021;
Published: 10 September 2021.
Edited by:
Lingling Wang, Dalian Ocean University, ChinaReviewed by:
Jitao Li, Chinese Academy of Fishery Sciences (CAFS), ChinaZhenhua Ma, Chinese Academy of Fishery Sciences (CAFS), China
Copyright © 2021 Kong, Liu, Li, Wang, Shi, Hu, Li and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yueru Li, eXpsMDA2N0BvdXRsb29rLmNvbQ==; Xiaoli Hu, aHhsNzA3QG91Yy5lZHUuY24=
†These authors have contributed equally to this work
 Lingling Kong
Lingling Kong Pingping Liu
Pingping Liu Moli Li1
Moli Li1 Jiaoxia Shi
Jiaoxia Shi Xiaoli Hu
Xiaoli Hu