- 1Guangxi Laboratory on the Study of Coral Reefs in the South China Sea, Coral Reef Research Center of China, School of Marine Sciences, Guangxi University, Nanning, China
- 2Southern Marine Science and Engineering Guangdong Laboratory, Zhuhai, China
The coral-eating crown-of-thorns starfish (COTS; Acanthaster spp.) play a major role in coral reef degradation in the Indo-Pacific region. However, the impacts of environmental factors on the phylogenetic and genetic characteristics of COTS in the northern Indo-Pacific convergence region remains unclear. We used mitochondrial DNA (mtDNA) and microsatellite markers to analyze the phylogenetic relationship, demographic history, genetic diversity and genetic structure of COTS in the South China Sea (SCS) and explored the impact of environmental factors on historical population expansion, genetic differentiation and larval dispersal. There was a clear signature of a population expansion in the SCS using the mtDNA marker. According to microsatellite loci analysis, COTS have high genetic diversity in the SCS. STRUCTURE analysis indicated that COTS in the Pacific Ocean can be divided into four subgroups: the SCS, Western Pacific, Pacific equatorial current affected zone, and Pacific insular atolls populations in the Pacific Ocean. Fst-statistical analysis revealed positive correlations between the Fst values and geographic isolation for all sampling sites. Additionally, there were no clear associations between the Fst values and chlorophyll a concentrations among coral reefs in the SCS; however, there were significant positive associations between the Fst values and particulate organic carbon (POC) concentrations within small geographic distances. These results suggest that COTS underwent historical population expansion after the Last Glacial Maximum, possibly followed by coral population expansion. The genetic structure of COTS populations may have been shaped by distinct nutrient concentrations, particularly those of POC, over small geographic distances. Moreover, ocean currents provide a potential dispersal mechanism for COTS larvae in the SCS. This study demonstrates that environmental and oceanographic factors play important roles in shaping the genetic characteristics and larval dispersal of COTS populations in the northern Indo-Pacific convergence region.
Introduction
Coral reefs are one of the most biologically diverse ecosystems in the oceans (Reaka-Kudla, 1997; Robert et al., 2002; Wilkinson, 2008). However, coral reef ecosystems have been severely threatened by anthropogenic global climate change and this has become especially apparent since the 1970s (Silverstein et al., 2015). Climate change not only leads to drastic declines in coral cover (Hughes et al., 2018; Stuart-Smith et al., 2018), but also appears to promote outbreaks of coral predator crown-of-thorns starfish (COTS; Acanthaster spp; Uthicke et al., 2015a). COTS are the most important biological threat to coral reef ecosystems in the Indo-Pacific region (Pratchett et al., 2017a), and COTS outbreaks not only lead to substantial loss of live coral cover, but also impair the integrity and resilience of reef ecosystems (Death et al., 2012; Kayal et al., 2012; Mellin et al., 2019). In the Indo-Pacific region, at least 246 outbreaks of COTS have been recorded since 1990, which is triple the number of population explosion events reported prior to 1990 (Pratchett et al., 2014). Additionally, coral reefs in the Pacific region experience a high frequency of COTS outbreaks, including the Great Barrier Reef (GBR; Death et al., 2012; Pratchett et al., 2014), and those in Japan (Yamaguchi, 1986; Suzuki et al., 2012; Yasuda, 2018), French Polynesia (Kayal et al., 2012), Guam (Chesher, 1969), and other regions, which has resulted in high levels of coral mortality and reduced live coral cover. Although many COTS outbreak events have been observed, the causes of dramatic COTS population explosions remain equivocal. It is still unclear whether outbreaks start from a single reef or arise simultaneously from separate locations due to limited temporal and spatial resolution of monitoring (Pratchett, 2005).
Crown-of-thorns starfish have high fecundity, and a female COTS can produce 30–70 million eggs (resulting in about 10 million fertilized eggs) per spawning period (Birkeland and Lucas, 1990; Babcock et al., 2016; Caballes et al., 2021; Pratchett et al., 2021). Thus, many studies have suggested that COTS outbreaks can be attributed to higher survival rate during early life stages, and that a small increase in the survival rate of the COTS larvae could lead to rapid increases in population size (Birkeland and Lucas, 1990). Elevated food supply (Brodie et al., 2005), anthropogenic eutrophication of seawater (Lucas, 1973; Birkeland, 1982; Fabricius et al., 2010), reduced predation pressure due to overfishing (Cowan et al., 2016) have been associated with increased larval survival. Notably, the geographical dispersal of COTS plays an important role in the formation of secondary outbreaks of COTS (Pratchett, 2005; Yasuda et al., 2009; Wooldridge and Brodie, 2015; Tusso et al., 2016), given that the duration of the planktonic larval stage (PLS) ranges between 9 and 42 days (Caballes and Pratchett, 2014). Weak or variable longshore currents may promote strong larval retention or limited dispersal, which are likely to give rise to primary outbreaks (Pratchett, 2005; Wooldridge and Brodie, 2015). However, strong directional longshore currents increase the likelihood of inter-reef dispersal, which leads to outbreaks once they become established (Wooldridge and Brodie, 2015). The multiple factors that lead to increased COTS larval survival rates remain unclear, and they may show spatial differences among distinct areas in the Indo-Pacific region. In addition, the dispersal routes and spatial distribution of COTS larvae may also vary between sites (Uthicke et al., 2015b; Yasuda et al., 2015a; Suzuki et al., 2016). Therefore, the knowledge of relationships between environmental-oceanographic factors and population size, demographics and structure of COTS population need to be improved.
The survivorship, settlement, and development of COTS larvae has been difficult to document in the wild, so direct observations have proven ineffective for COTS outbreaks, population demographics and dispersal events at different life stages. Model-based statistics of population genetics have increasingly been used as an indirect and effective method to infer the patterns of historical demographics, genetic diversity, genetic structure and migration routes for COTS, which could aid in understanding the relationship between molecular ecological characteristics of COTS and environmental-oceanographic factors among distinct coral habitats. Vogler et al. (2008) identified four clades of COTS were within the Indo-Pacific regions based on mitochondrial DNA (mtDNA) gene markers, which included Pacific, Red Sea, Southern Indian Ocean and Northern Indian Ocean. These clades were estimated to have diverged 1.95–3.56 Mya in the Pliocene-Early Pleistocene period. Unexpectedly, results from mtDNA studies have shown that genetic differentiation of COTS populations have clear relationships with regions. For instance, Timmers et al. (2012) showed that there are shared mitochondrial haplotypes between the South Central and Northwest Pacific and that their haplotypes do not strictly cluster according to geographic region. Vogler et al. (2013) also found a large geographic cluster in Western Pacific localities, with shared haplotypes across the entire range from the GBR to the Philippines. In addition, studies based on sensitive microsatellite loci makers revealed that COTS populations are genetically differentiated, there is contemporary dispersal among distinct coral reefs, and there are established relationships between ocean currents and maximum larval dispersal distance (Yasuda et al., 2009, 2015b; Tusso et al., 2016; Harrison et al., 2017). A large genetic break has been documented between the Indian Ocean and the Pacific Ocean corresponding to mtDNA study by Vogler et al. (2008), and there is significant genetic differentiation of COTS populations among Southeast Africa, Northwestern Pacific, Palau, the North Central Pacific, GBR, Fiji, and French Polynesia (Yasuda et al., 2009, 2015b). Furthermore, it has also been suggested that western boundary currents played an important role in the dispersal of COTS larvae in the Southeast African, Northwestern Pacific, and GBR oceanic regions (Yasuda et al., 2009). Tusso et al. (2016) also found that COTS populations show significant differentiation among Pacific insular atolls (e.g., Moorea and Johnston Atoll), and there was an effective dispersal pattern and gene flow of the COTS population in the Pacific equatorial current affected zone (e.g., Guam, Kingman reef and Swains; Tusso et al., 2016). However, in previous population genetics studies of COTS, generally conducted in areas within or near the open ocean (Yasuda et al., 2009, 2015b; Vogler et al., 2012, 2013; Tusso et al., 2016), strong ocean currents appear to facilitate effective larval dispersal of marine organisms. There are very few studies about the associations between genetic differentiation and connectivity of COTS populations in northern Indo-Pacific convergence region. Importantly, this region has special geographic barriers (e.g., mainland and archipelago) that may form genetic barriers against the mixing of COTS populations between marginal seas and the open ocean.
The South China Sea (SCS) spans the tropical and subtropical climate zones in the northern Indo-Pacific convergence region, and contains numerous coral reefs and communities across 19 degrees of latitude (Spalding, 2001; Yu, 2012; Chen et al., 2020). The abundant growth of corals in SCS provides favorable habitat for COTS populations (Li et al., 2019). Moreover, the SCS is connected to the Indian Ocean and the Pacific Ocean via several straits, with seawater exchange through the Luzon Strait, Taiwan, and Mindoro (Yu et al., 2010). Some studies have shown that there has been significant genetic differentiation and connectivity between reef organisms in the SCS and the Pacific Ocean (e.g., Symbiodiniaceae), which may be due to large biogeographic barriers and latitudinal environmental factors (Chen et al., 2020). In addition, reef around the Paracel Islands (Xisha Islands) had experienced dramatic population explosions of COTS that destroyed > 95% of live coral cover from 2005 to 2010 (Li et al., 2019). Unexpectedly, in less than 8 years, COTS outbreaks occurred in the Paracel Islands in 2019 again (Li et al., 2019). Nevertheless, the knowledge about genetic structure, connectivity, larval dispersal and its environmental impact factors of COTS in the SCS is still scant.
In this study, mtDNA and microsatellite genetic markers were used to analyze the phylogenetic relationship, demographic history, genetic diversity, genetic structure and differentiation, and recent gene flow of the COTS populations in the SCS. Previously published population genetic datasets of COTS in the Indo-Pacific region were also used for analyses and to draw comparisons (Vogler et al., 2008; Tusso et al., 2016). These analyses improve our understanding of COTS historical population expansion, long-distance larval dispersal, and genetic differentiation of COTS between SCS and other coral habitats.
Materials and Methods
Samples Collection and DNA Extraction
In total, 163 adult COTS with similar sizes (ranging from 210 to 220 mm) were collected by SCUBA diving in June 2018, which ensure that individual samples were approximately the same age (2–3 years old; Pratchett et al., 2014). The sampling sites included five distinct coral reefs in the SCS: First Thomas Shoal (FTS; Xinyi Reef), Passu Keah (PK; Panshiyu), Drummond Island (DI; Jinqing Island), Bombay Reef (BR; Langhua Reef), and Pattle Island (PI; Shanhu Island) (Figure 1 and Table 1). Tube feet of COTS samples were cut off using sterile medical operation scissors. All tube feet were transferred into 50-mL cryogenic vials (Corning, Mexico), 95% ethanol was added, and tube feet samples were stored at –20°C until DNA extraction. DNeasy® Blood and Tissue Kits (Qiagen, Hilden, Germany) were used to extract total DNA from tube feet samples, following the manufacturer’s instructions.
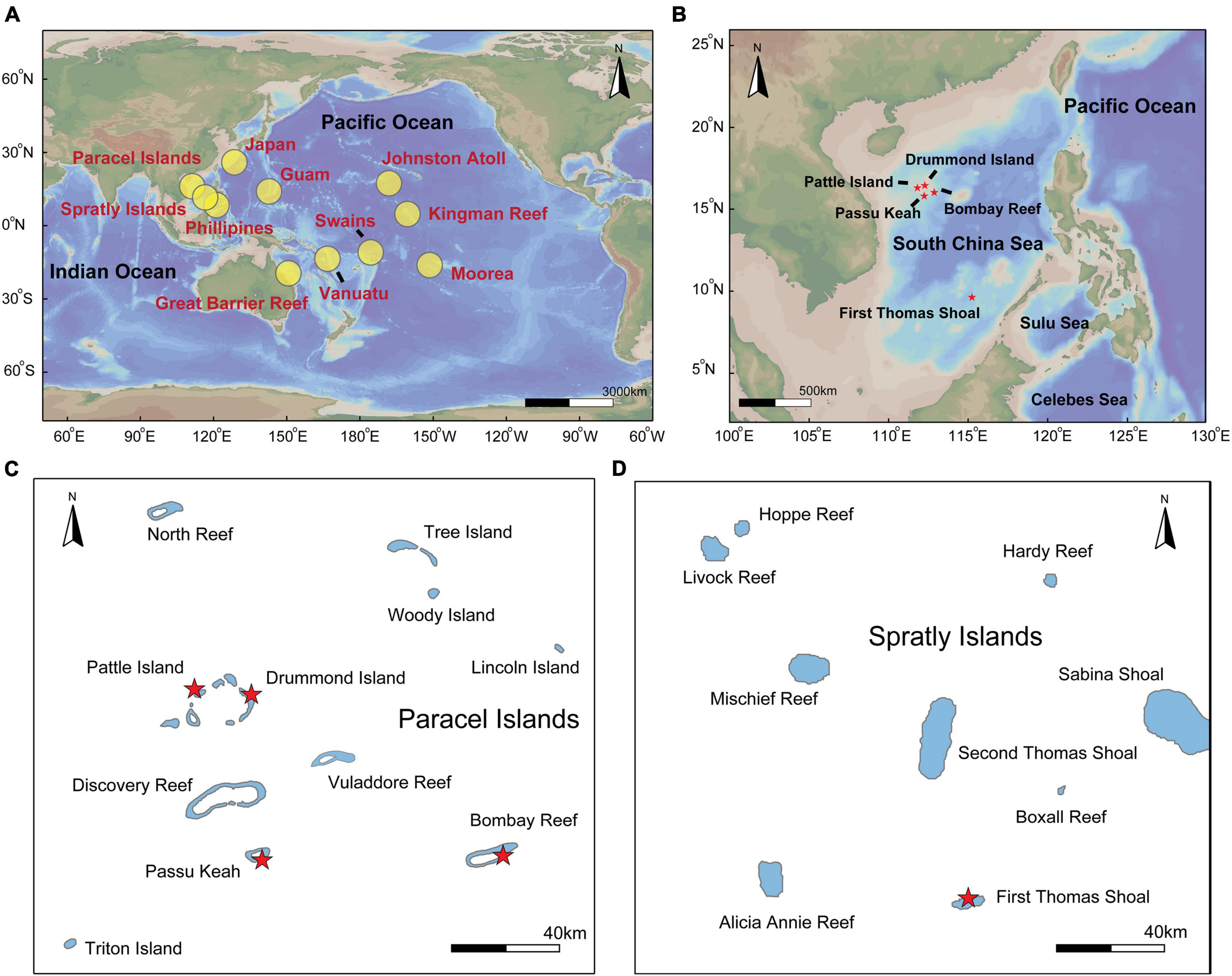
Figure 1. Locations of crown-of-thorns starfish (COTS) sample collections in the Pacific Ocean and South China Sea (SCS). (A) Overview of study locations. Our sample areas which include the Paracel and Spratly Islands within the SCS. COTS microsatellite loci data was retrieved from the Western Pacific (Japan, Philippines, Great Barrier Reef, and Vanuatu), Pacific equatorial currents affected zone (Guam, Kingman Reef, and Swains) and Pacific insular atoll (Johnston Atoll and Moorea; Tusso et al., 2016). These locations were marked by red in the map. (B) The details of sampling locations in the SCS, (C) which including Pattle Island, Drummond Island, Bombay Reef and Passu Keah in the Paracel Islands, (D) also including First Thomas Shoal in Spratly Islands.
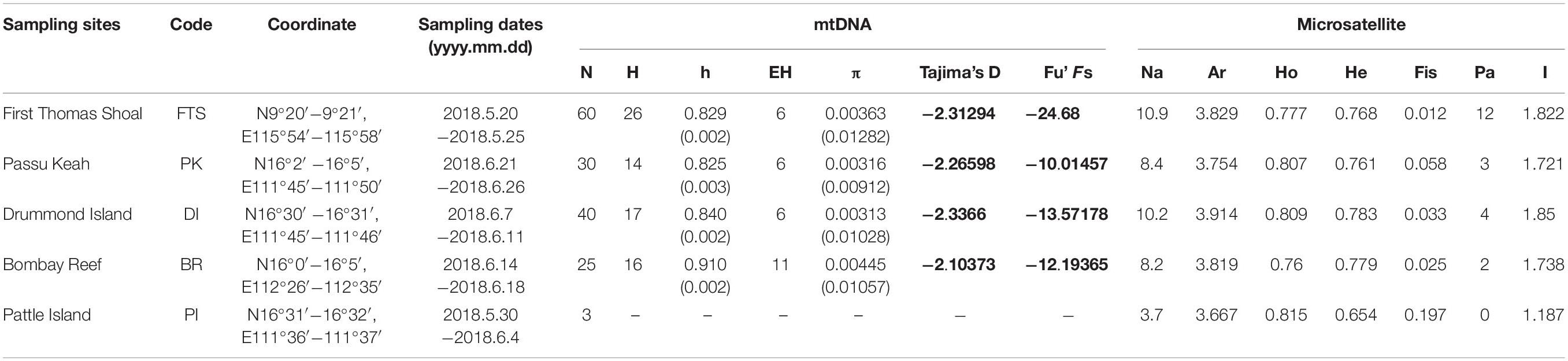
Table 1. Summary information of sampled locations, coordinate, number of samples (N), the number of haplotypes (H), haplotype diversity (h), the number of effective haplotypes (EH), nucleotide diversity (π), Fu’s Fs, Tajima’s D (significant values bold), number of alleles (Na), allelic richness (Ar), observed heterozygosity (Ho), expected heterozygosity (He), inbreeding coefficient (Fis), Private alleles (Pa), and Information index (I) in the South China Sea.
Phylogenetic Tree Construction and Demographic History
A fragment of the mitochondrial cytochrome oxidase subunit I gene (COI) was amplified using COTS_COI_R4734 5′-GCCTGAGCAGGAATGGTTGGAAC-3′ and COTS_COI_ R5433 5′-CGTGGGATATCATTCCAAATCCTGG-3′ (Vogler et al., 2008). PCR was conducted with 25 μL Premix Taq™ (TaKaRa Taq™ Version 2.0 Plus dye, TaKaRa, Dalian, China), ∼75 ng of DNA, 2 μL primer, and ddH2O to a total volume of 50 μL. Based on ABI GeneAmp 9700 thermal cycler, PCR reactions were carried out under the following conditions: 2 min at 94°C, followed by 35 cycles of 94°C for 20 s, 59°C for 45 s, 72°C for 1 min, and a final extension at 72°C for 10 min. Amplification products were directly sequenced on an ABI 3730XL DNA Analyser (Sangon Biotech Company, Shanghai, China). COI sequences were then aligned using the ClustalW algorithm implemented in MEGA 6. Phylogenetic trees of COTS sequences were inferred using maximum likelihood (ML) and neighbor-joining (NJ) methods. ML analyses for sequence data were performed with a K2 + G model. The robustness of the ML and NJ trees was tested with 1000 bootstrap replicates. Novel COI sequences of COTS in the SCS can be accessed in GenBank under accession numbers MW238184-MW238343. In addition, DnaSp6 was used to analyze the number of haplotypes (H), haplotype diversity (h), the number of effective haplotypes [EH; HE = 1/(1 − h)], and nucleotide diversity (π), and NETWORK v10.1.0.0 was used to visualize haplotype variability and clustering based on the median-joining method. To understand the demographic history of COTS in the SCS and investigate signatures of historical population expansions, Fu’s Fs (Fu, 1997) and Tajimas’s D (Tajima, 1989) were calculated using Arlequin v 3.5 (45,000 replicates; Excoffier and Lischer, 2010).
Microsatellite Analysis
Nine polymorphic microsatellite loci were used to analysis COTS genotypes (Supplementary Table 1; Yasuda et al., 2006, 2007; Tusso et al., 2016). Each locus was amplified in 50 μL reaction volumes containing ∼70 ng DNA, 2 μm primer, 24 μL of TaKaRa Premix Taq™ and ddH2O. The PCR were performed under the following conditions: 94°C 2 min, followed by 35 cycles of 94°C for 20 s, 51°C (AyU03), 52°C (Yukina05), 53°C (Hisay01, Maki12), 55°C (Yukina01, Yukina06), 57°C (Aya02) or 58°C (Maki01, Maki03) for 20 s, and a final extension at 72°C for 5 min. ABI 3730XL Genetic Analyzer was used to analyze PCR products at the Sangon Biotech Company (Shanghai, China). Sizing was achieved using LIZ500 internal standards. The GeneMapper v.3.2 was used to identify allele sizes. To explore the genetic differentiation and gene flow of COTS populations between the SCS and the Pacific Ocean, we combined microsatellite data of COTS from the Pacific Ocean (Western Pacific, Pacific equatorial current affected zone, and Pacific insular atolls), which were published by Tusso et al. (2016). The microsatellite genotype data sets have been deposited in Supplementary Dataset 1.
Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium exact tests were conducted by Genepop v4.3.1. Myriad v1.2 software was used to test for the presence of null alleles using Bonferroni-adjusted 95% confidence intervals (Antonio, 2018). The descriptive statistics included the number of alleles (Na), observed heterozygosity (Ho), expected heterozygosity (He), Private alleles (Pa) and Information index (I), which were conducted by GeneAlex v.6.5 (Peakall and Smouse, 2010). Inbreeding coefficient (Fis) was calculated by Genepop v4.3.1, and allelic richness (Ar; with 2000 randomizations) and was used to determine differences in genetic diversity using FSTAT v.2.9.4 (Goudet, 2001). A General Linear Model (GLM) analysis was used to assess differences in genetic diversity with 1000 bootstrap permutations, and the groups of ocean regions was regarded as a fixed factor. The Student–Newman–Keuls (SNK) test was used for post hoc multiple comparisons of significant GLM test results.
STRUCTURE v2.4 was used to analyze the genetic clusters, and Markov chain runs consisted of an initial “burn-in” of 250,000 steps followed by a final 1,000,000 iterations (Evanno et al., 2010). Fifteen independent runs were performed for 1–19 Ks. The most likely value for K was tested by plotting the log probability of data over multiple runs and comparing that with delta K as implemented in Structure Harvester v.0.6.94 (Earl and VonHoldt, 2012), and the results of Ln Pr(X\K) and delta K are shown in the supporting information (Supplementary Figure 1). To choose the appropriate K value the results were averaged using CLUMPP v.1.1.2 (Jakobsson and Rosenberg, 2007), and genetic structure plots were manipulated using DISTRUCT (Rosenberg, 2010). Analyses of molecular variance (AMOVA) were conducted on geographic populations with regional structuring as defined by STRUCTURE and by sampling sites to determine the degree of differentiation, all of which was performed in GeneAlex v.6.5 (Peakall and Smouse, 2010). Genetix software was employed to calculated pairwise Fst values, using Weir and Cockerham genetic distance with 10,000 permutations to determine significance. Correlations between geographic distance and Fst values were tested by Mantel tests performed with IBD software v.1.5.2 (Bohonak, 2002).
Environmental Characteristic Analysis
The genetic differentiation and long-distance dispersal of COTS larvae may be closely related with geographic isolation (Dight et al., 1990; Vogler et al., 2012), nutrient concentrations (Fabricius et al., 2010; Pratchett et al., 2017a,b) and ocean currents (Yasuda et al., 2009; Vogler et al., 2012; Tusso et al., 2016). As the food source of COTS larvae was diverse and complicated (Ayukai, 1994; Okaji et al., 1997; Nakajima et al., 2016; Mellin et al., 2017), the chlorophyll a (Chl a, μg⋅L–1) and particulate organic carbon (POC, μg⋅L––1) concentrations were collected together as proxies for the nutrient concentration, which can represent the phytoplankton biomass and particulate organic matter concentrations in the sampled coral reefs and surrounding sea water, respectively. Aqua-MODIS monthly average Chl a and POC datasets from 2016 to 2020 were obtained from the NASA Giovanni satellite1. Additionally, correlations between Chl a and Fst and between POC and Fst were tested by Mantel tests using IBD software v.1.5.2 (Bohonak, 2002) and multivariate analysis (DistLM) performed with PRIMER 7 (McArdle and Anderson, 2001). In addition, A GLM analysis was used to compare the monthly Chl a and POC concentrations with 1000 bootstrap permutations, and the groups of coral reefs in the SCS was regarded as a fixed factor. The post hoc multiple comparisons of significant GLM results were conducted using SNK tests. The upper-ocean surface current data (depth < 2.5 m) in the SCS during potential COTS breeding periods (May to August; Pratchett et al., 2014, 2017a) from 2016 to 2018 were acquired from the National Marine Science Data Centre, National Science & Technology Resource Sharing Service Platform of China2.
Results
Phylogenetic Analyses and Demographic History of Crown-of-Thorns Starfish Populations
The phylogenetic analysis of COTS mtDNA showed that almost all COI sequences of COTS in the SCS do not show obvious phylogenetic divergence with Pacific populations (Figure 2A). In addition, COTS populations in SCS also have significant phylogenetic divergence with those in the Red Sea, Southern Indian Ocean and Northern Indian Ocean (Figure 2A). For COTS populations within the SCS, there were no significant distinct genetic clusters in the median-joining haplotype (Figure 2B; separated by at least 18 mutation steps; Vogler et al., 2013). This result also showed no clear association between haplotypes and geographic factors, and two dominant haplotypes were widely shared among the populations of the coral reefs in the SCS (Figure 3B). The haplotype diversity in COTS populations in the SCS was low, and only the h of COTS population in BR was >0.1 (h = 0.910; Table 1). Additionally, low nucleotide diversity was determined, and π of all populations was <0.01 in the SCS (Table 1). However, there was a clear signature of a population expansion identified by mismatch distribution analysis (Figure 2C) and the significance of Fu’s Fs and Tajima’s D (Table 1).
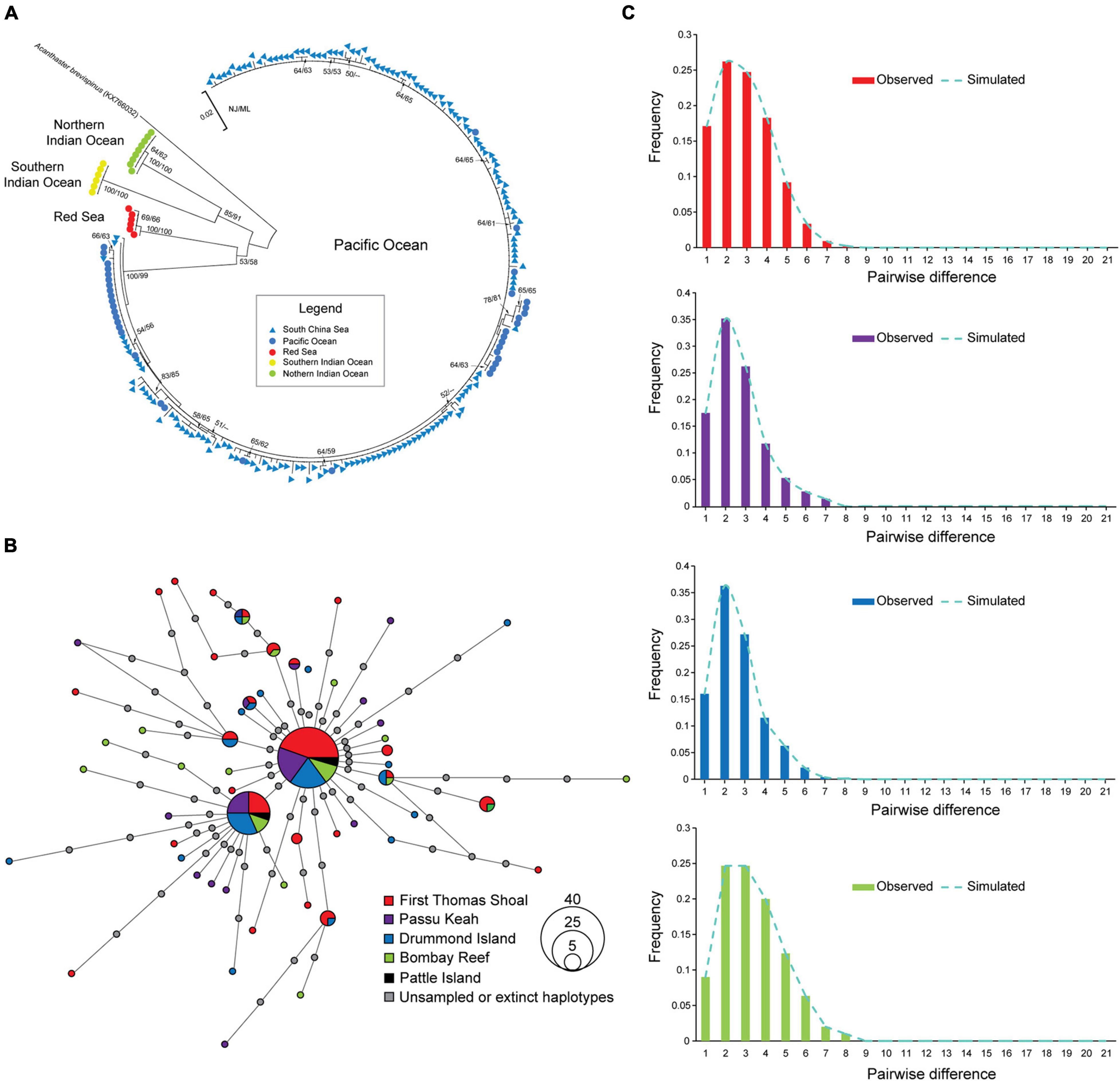
Figure 2. Phylogenetic relationship, haplotype network and historical demography of crown-of-thorns starfish (COTS) in the South China Sea (SCS). (A) Neighbor joining (NJ) tree of mitochondrial cytochrome oxidase subunit I gene (COI) of COTS was presented. (B) COI sequence based on median-joining algorithm. Colors represent geographic COTS populations including First Thomas Shoal (FTS), Passu Keah (PK), Drummond Island (DI), and Bombay Reef (BR) in the SCS, and gray circles represent extinct or unsampled haplotypes. Circle size is proportional to the haplotype frequency. (C) Pairwise mismatch distribution for COTS populations in FTS, PK, DI, and BR in the SCS.
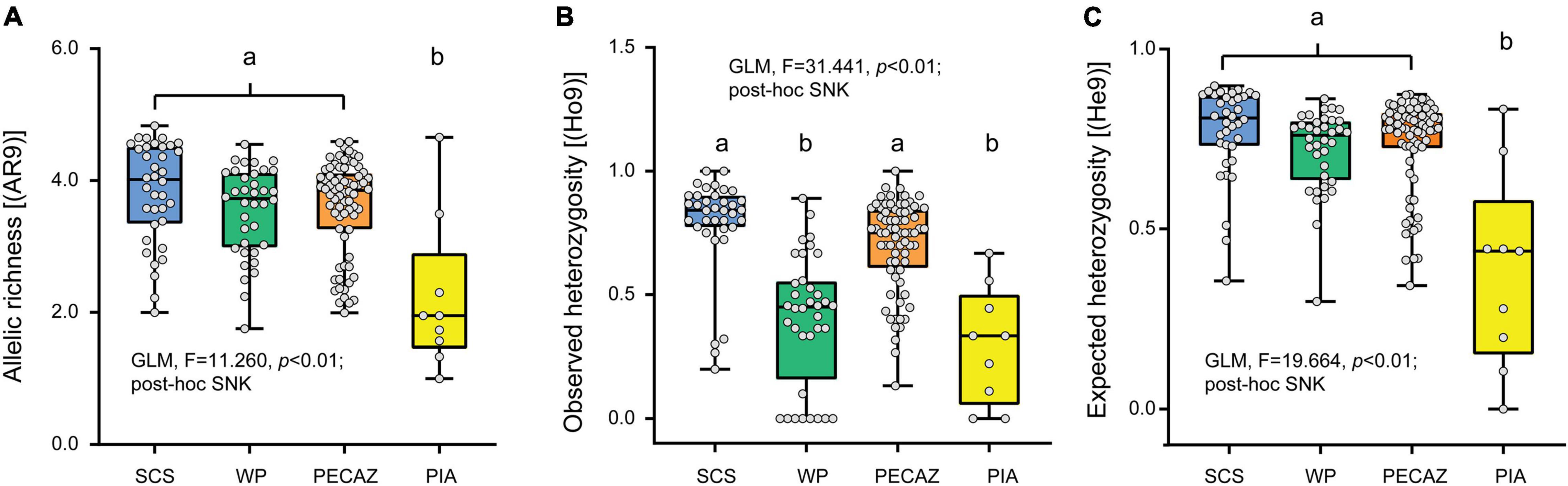
Figure 3. Genetic diversity of crown-of-thorns starfish (COTS) populations in the Pacific Ocean. (A) Allelic richness [AR(9)], (B) observed heterozygosity [Ho(9)], and (C) expected heterozygosity [He(9)] rarefied to a sample size of four regions and 9 genes per regions. SCS mean South China Sea, WP mean Western Pacific, PECAZ mean Pacific equatorial currents affected zone and PIA mean Pacific insular atoll. Genetic diversity differences across regions were analyzed by General Linear Model (GLM) analysis, and post hoc multiple comparisons of significant GLM results were conducted by Student-Newman-Keuls (SNK) test. Lowercase letters indicate post hoc comparisons (SNK) (e.g., a, b) with the significant factors among regions.
Hardy–Weinberg Equilibrium and Linkage Disequilibrium
For microsatellite loci analysis, based on the multi-locus test for five COTS populations in the SCS, most microsatellite loci were at or near HWE after the Bonferroni correction test (p < 0.05). In addition, there were some deviations in COTS populations from the SCS; for example, Aya02 and Yukina06 deviated from HWE in the FTS, PK, and BR. However, almost all of these loci exhibited HWE in the COTS populations of the SCS. Moreover, none of the pairwise comparisons between loci within COTS populations from the SCS and the Pacific Ocean exhibited significant evidence of linkage disequilibrium after Bonferroni correction. Therefore, these loci were included in the data set for subsequent analyses.
Genetic Diversity
The results of the genetic diversity analysis using the microsatellite gene marker were not consistent with those of the mtDNA sequences analysis. COTS populations in the SCS, Western Pacific, and Pacific equatorial current affected zone had higher average allelic richness than that in the Pacific insular atolls (GLM; F = 11.260, p < 0.01; Figure 3A). The average allelic richness was highest in the SCS (Armean = 3.829; SD = 0.763), followed by the Pacific equatorial current affected zone (Armean = 3.603; SD = 0.716), Western Pacific (Armean = 3.538; SD = 0.675), and Pacific insular atolls (Armean = 2.478; SD = 1.002). The COTS population in DI had the highest average allelic richness (Armean = 3.914; SD = 0.803), and the lowest average allelic richness was in the Johnston Atoll COTs population (Armean = 2.220; SD = 1.153). In addition, the observed heterozygosity of COTS populations in the SCS and Pacific equatorial current affected zone were higher than those in Western Pacific and Pacific insular atolls (GLM; F = 31.441, p < 0.01; Figure 3B). However, the changing rule of the expected heterozygosity was consistent with average allelic richness among distinct regions, and the COTS populations in the SCS, Western Pacific, and Pacific equatorial current affected zone regions had higher expected heterozygosity than that in the Pacific insular atolls (GLM; F = 19.664, p < 0.01; Figure 3C). It is worth noting that the COTS populations in the SCS have relatively lower inbreeding coefficients (Fis) and higher private alleles (Pa) than that in other regions (Table 1). Accordingly, COTS populations in the SCS, Western Pacific, and Pacific equatorial current affected zone have higher genetic diversity than that in the Pacific insular atolls.
Genetic Differentiation and Structure
The analysis by STRUCTURE identified two peaks (K = 2 and K = 6) as the most likely number of COTS genetic clusters based on estimates of delta K and LnP(K) (Figures 4A,B and Supplementary Figure 1). At K = 2, the Pacific equatorial current affected zone formed a separate cluster from the other COTS samples (Figure 4A). At K = 6, COTS populations from the SCS, Western Pacific, Pacific equatorial current affected zone, and Pacific insular atolls comprised separate cohesive genetic units (Figure 4B). COTs populations in the Pacific equatorial current affected zone had admixed genotypes, and two main clusters (orange and dark blue) were identified in samples from Guam, Kingman Reef and Swains. The samples from Japan, the Philippines, the GBR and Vanuatu belonged mostly to the same inferred population (green), which was located in the Western Pacific. An additional genetic cluster (bright yellow) was found at low frequencies but with high individual membership probabilities in the Johnston Atoll (Figure 4B). Interestingly, COTS samples from the SCS formed two separate genetic clusters, namely the First Thomas Shoal-Passu Keah cluster (FTS-PK; blue) and the Drummond-Bombay Reef cluster (DI-BR; gray; Figure 4B). In addition, COTS populations of PI and Moorea had complex compositions of genetic clusters, which may be associated with insufficient sample sizes. Thus, microsatellite loci data sets from these two locations were not used to calculate Fst values.
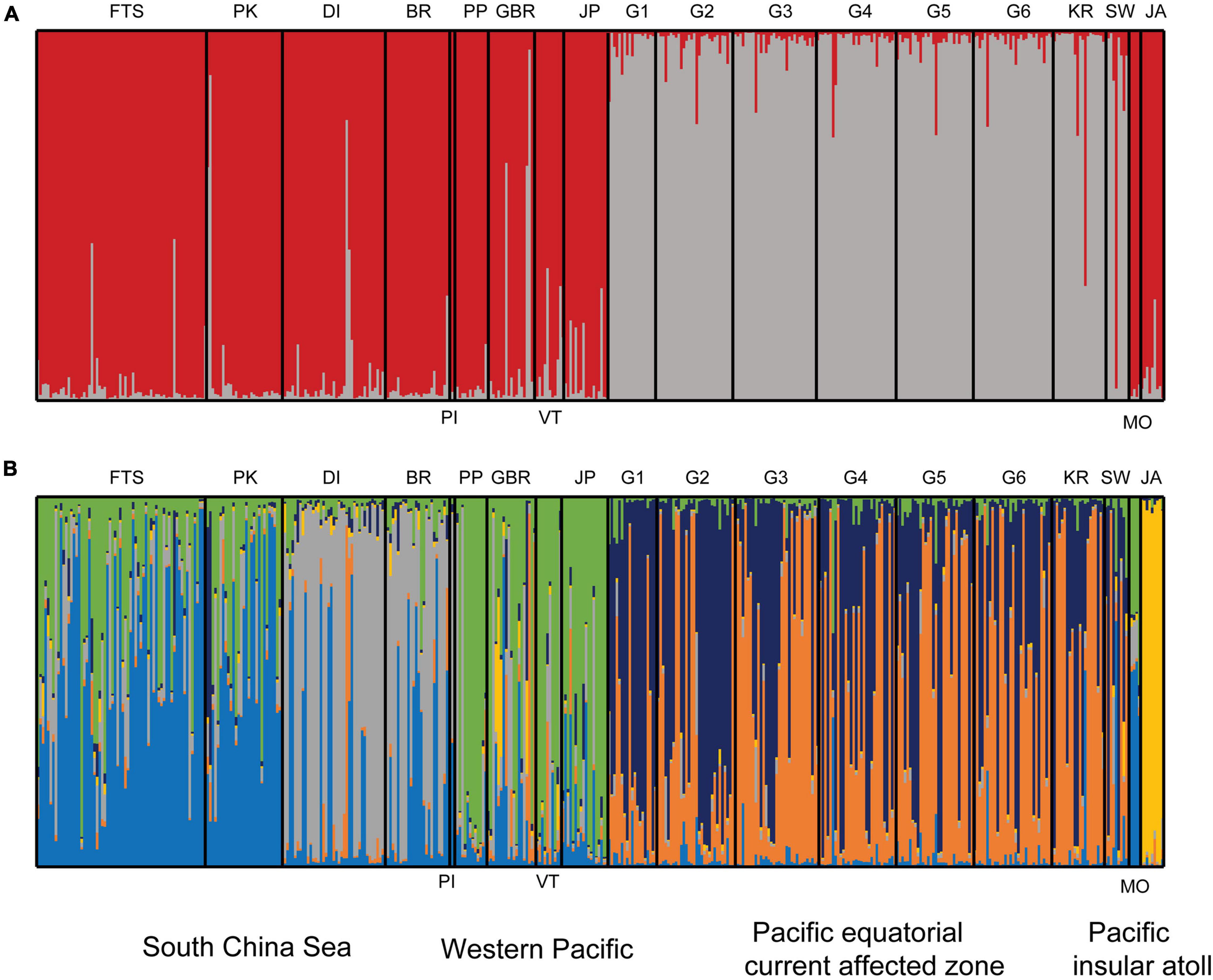
Figure 4. Genetic structure of crown-of-thorns starfish (COTS) in the Pacific Ocean. (A) At the optimal K = 2, COTs sample were assigned to two genetic groups, sampling sites in Pacific equatorial currents affected zone (PECAZ) were significantly distinct from all other locations. (B) A second optimal K = 6, six genotypes were identified in COTS populations. The South China Sea (SCS) mainly has blue genetic cluster in First Thomas Shoal-Passu Keah (FTS and PK) and gray genetic cluster in Drummond Island (DI), Bombay Reef (BR), and Pattle Island (PI); The Western Pacific (WP) mainly has a green genetic cluster, which includes Japan (JP), the Philippines (PP), Great Barrier Reef (GBR), and Vanuatu (VT); the Pacific equatorial currents affected zone (PECAZ) has mixed genotypes (orange and dark blue; G1, Guam-Tipalao; G2, Guam-Uruno; G3, Guam-Taguan; G4, Guam-Notth Haputo; G5, Guam-Tanguisson; G6, Guam-Tagachan; KR, Kingman Reef; SW, Swains), and the Pacific insular atoll (PIA; JA, Johnston Atoll; MO, Moorea) has yellow genetic cluster.
Fst values were significant in 75% of all pairwise comparisons (with 10,000 permutations), with larger values obtained between distinct regions and between more distinct sites (Table 2). There were mainly insignificant genetic differentiation rates among COTS populations within the same region. For instance, FTS and PK in the SCS (Fst = 0.001, FDR, p > 0.05), the Philippines and the GBR in the Western Pacific (Fst = 0.041, FDR, p > 0.05), and some Guam sampling sites and Kingman Reef in the Pacific equatorial current affected zone (Fst values from 0.008 to 0.039, FDR, p > 0.05). AMOVA also found that there were significant genetic differentiation rates among regions (ΦRT = 0.129, p < 0.001, Table 3), among sampling sites (ΦPR = 0.066, p < 0.001, Table 3) and within COTS individuals (ΦPT = 0.187, p < 0.001, Table 3). It was noted that the SCS, Western Pacific, Pacific equatorial current affected zone and Pacific insular atolls yielded highly significant values (percent of variation: 13%), which indicated strong geographical differentiation among the four regions. IBD results also indicated significant positive correlations between the Fst values and geographic distances (Mantel test, Randomization = 10,000, R2 = 0.1347, p = 0.0099; Figure 5A), which was consistent with the results of the AMOVA. It is worth noting that nutrient concentration was significantly distinct among coral habitats within short geographic distances inside the SCS (GLM; Chl a: F = 92.715, p < 0.001; POC: F = 140.043, p < 0.001; Figure 6). Additionally, the results of the Mantel test and DistLM found that there was no significant correlation between the Fst values and Chl a (Mantel test, Randomization = 10,000, R2 = 0.3121, p = 0.1150; DistLM, pseudo-F = 1.815, p = 0.2492; Figure 5B); however, the Fst values were significantly related to POC (Mantel test, Randomization = 10,000, R2 = 0.8670, p = 0.0010; DistLM, pseudo-F = 26.07, p = 0.0070; Figure 5C) within the SCS. Thus, nutrient concentration, especially POC concentration, affect the genetic differentiation of COTS populations among distinct coral habitats, which only had slight geographic isolation from each other.
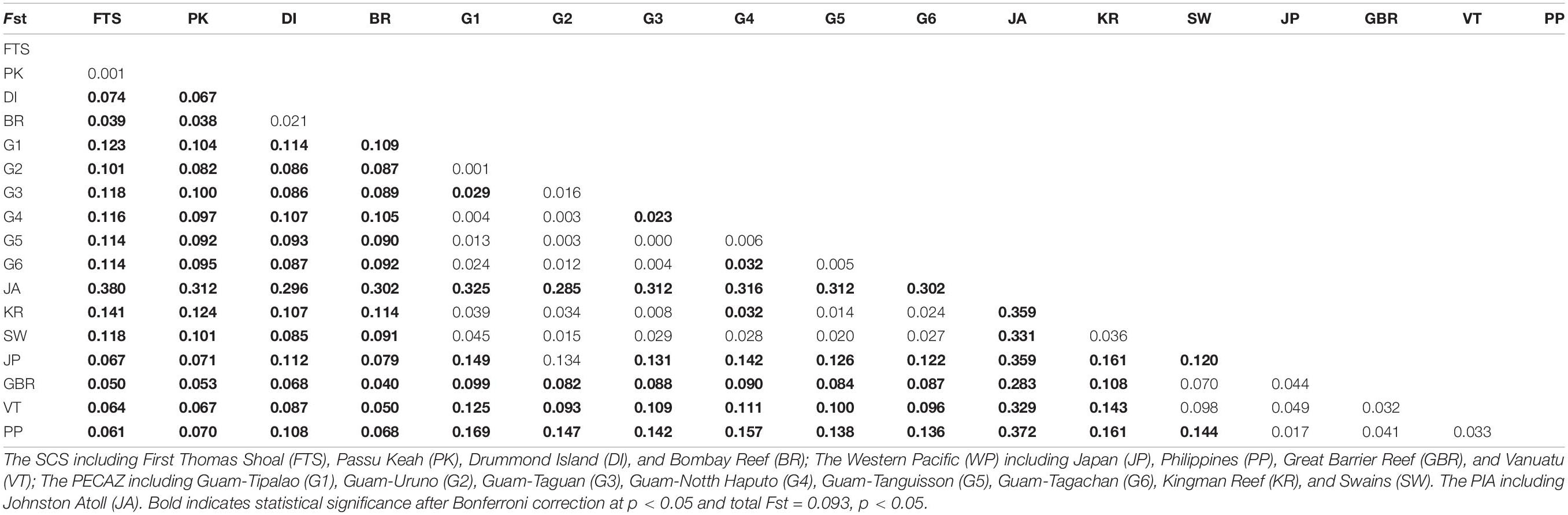
Table 2. Pairwise Fst values for 17 crown-of-thorns starfish populations across the South China Sea (SCS), Western Pacific (WP), Pacific equatorial currents affected zone (PECAZ), and Pacific insular atoll (PIA) in Pacific Ocean.
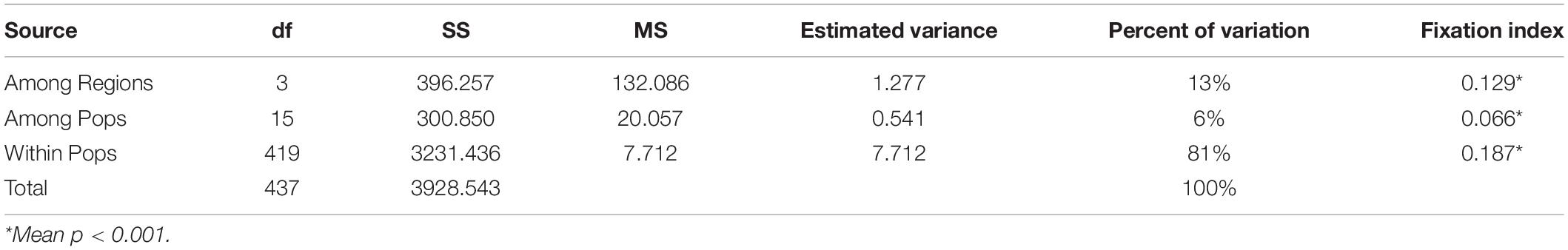
Table 3. Analysis of molecular variance (AMOVA) for crown-of-thorns starfish in the South China Sea and Pacific Ocean from each 17 coral habitats with 4 geographical regional populations (South China Sea, Western Pacific, Pacific equatorial currents affected zone and Pacific insular atoll).
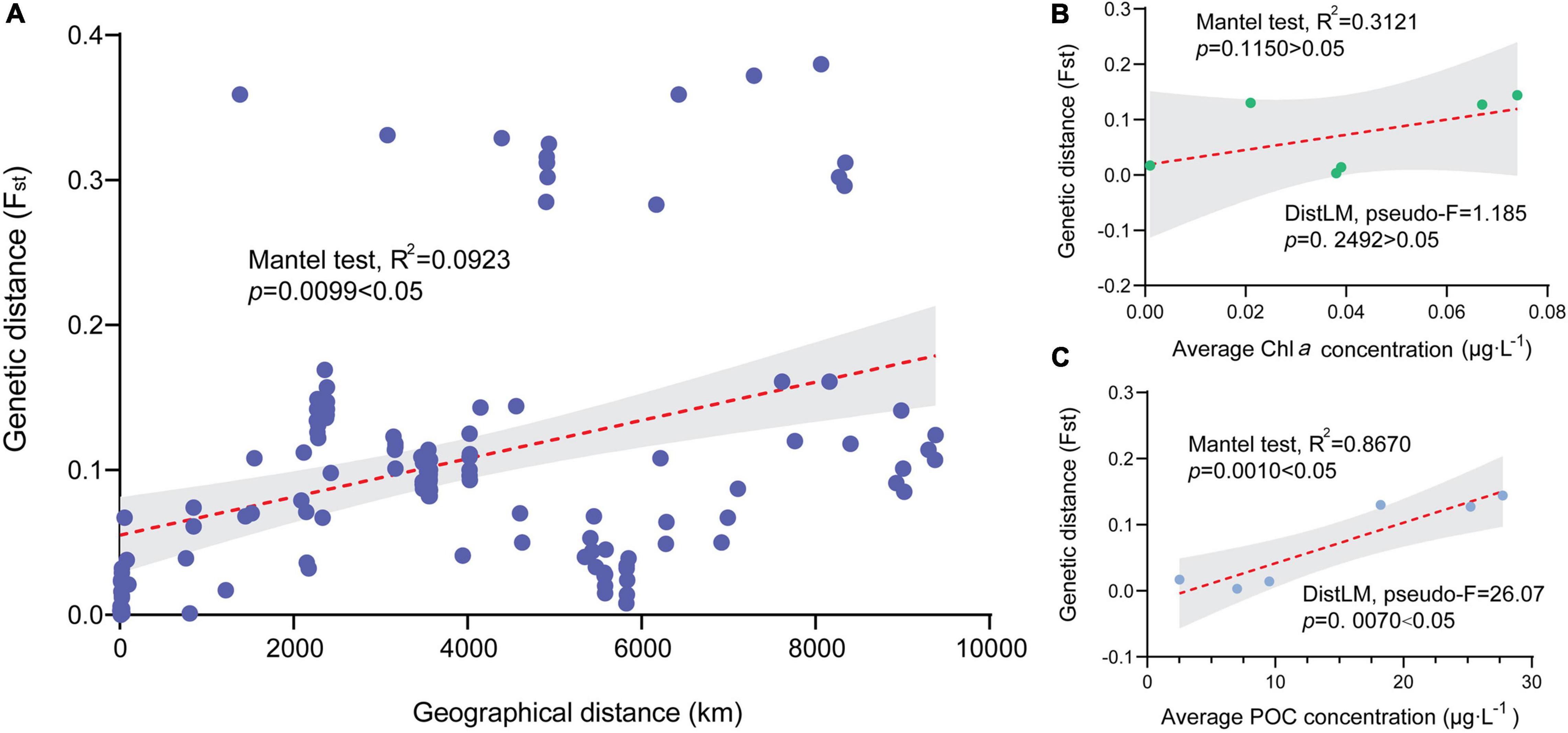
Figure 5. Correlations between genetic differentiation of crown-of-thorns starfish (COTS) populations and geographic and environmental factors. (A) There was a significantly positive relationship between Fst values of COTS populations and geographic distance in the Pacific Ocean (km; Mantel test, R2 = 0.1347, p = 0.0099 < 0.05). (B) There were no significant associations between Fst values of COTS populations and chlorophyll a concentration (μg⋅L–1; Mantel test, R2 = 0.3121, p = 0.1150 < 0.05) in the South China Sea. (C) There was a significantly positive correlation between Fst values of COTS populations and particulate organic carbon concentrations (μg⋅L–1; Mantel test, R2 = 0.8670, p = 0.0010 < 0.05) in the South China Sea.
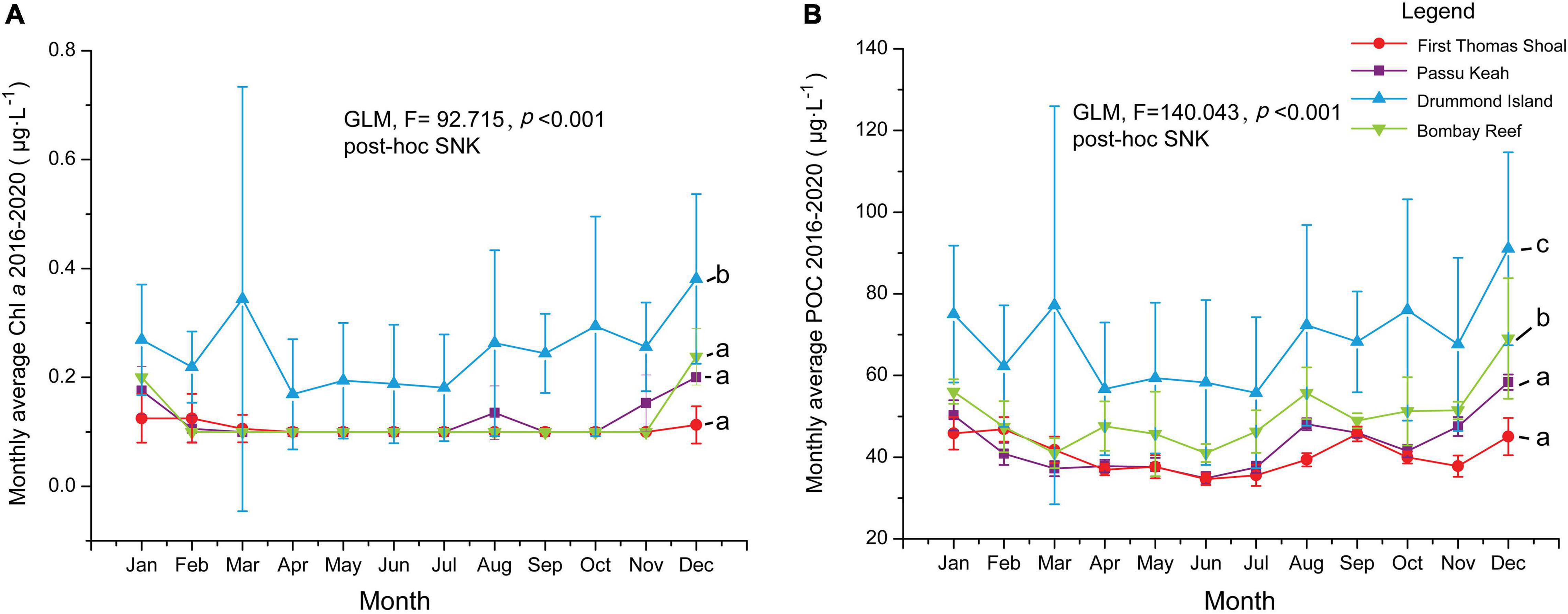
Figure 6. Monthly average of chlorophyll a concentration (Chl a, μg⋅L–1) and particulate organic carbon concentration (POC, μg⋅L–1) from 2016 to 2020 for four coral reefs in the South China Sea (SCS). (A) Monthly average and standard deviations of Chl a in First Thomas Shoal (FTS), Passu Keah (PK), Drummond Island (DI), and Bombay Reef (BR). (B) Monthly average and standard deviations of POC in FTS, PK, DI, and BR. Chl a and POC differences among four coral habitats were analyzed by General Linear Model (GLM). Lowercase letters indicate post hoc comparisons (Student–Newman–Keuls test) (e.g., b, a, a, a) with the significant factors among coral habitats.
Discussion
Historical Population Expansion of Crown-of-Thorns Starfish May Be Associated With Coral Range Expansion After the Last Glacial Maximum in the Pleistocene
The mtDNA marker (4734–5433) for phylogenetic analysis of COTS populations did not belong to the highly variable region of COI fragment (Yasuda et al., 2009; Vogler et al., 2012; Tusso et al., 2016). Thus, this gene marker may be too conservative for detecting haplotype and nucleotide diversity, which likely led to the results of the genetic diversity analysis of COTS based on the mtDNA gene marker not being consistent with those of the microsatellite loci analysis. However, the mtDNA marker could be used to analyze the demographic history and lineage divergence of COTS populations (Vogler et al., 2008, 2013). None of the COTS populations in the SCS showed lineage divergence from that in Pacific Ocean, which may have also diverged in the Pliocene–Early Pleistocene (1.95–3.65 Mya; Vogler et al., 2008). The speciation process of Pacific, Red Sea, and Southern and Northern Indian Ocean COTS groups was likely driven by largely biogeographic barriers between the main oceans due to sea-level changes (Pillans et al., 1998).
Moreover, Tajima’s D, Fu’ Fs, and mismatch distribution indicated that COTS experienced historical population expansion in four coral habitats in the SCS (Table 1 and Figure 2C), which was consistent with the demographic history of COTS populations in the Western Pacific (Vogler et al., 2013). These results may be closely associated with coral population expansion that was driven by sea-level rise after the Last Glacial Maximum (Voris, 2000; Tian et al., 2010; Huang et al., 2018). In the Pleistocene, glacial advances led to a sea-level fall to 120–140 m below present levels in Southeast Asia and Australasia, which led to the bulk of the Sunda and Sahul shelves being largely exposed and formed massive lowland connections between present day islands in this region and adjacent continents (Voris, 2000). Meanwhile, coral habitats in the northern SCS also disappeared during this period (e.g., Weizhou Island, Hainan Island, and Daya Bay) due to land advances (Voris, 2000; Huang et al., 2018), and many marine organisms, including corals, retreated into periodic refugia and subsequently expanded after the end of the glacial age. For example, Huang et al. (2018) found that populations of Porites lutea expanded after the Last Glacial Maximum in the SCS, and re-established populations on Weizhou Island and Hainan Island from the northern SCS as the sea levels rose.
COTS live in coral habitats and feed on scleractinian corals (Glynn, 1988; Rotjan and Lewis, 2008), and consume a maximum of 66–478 cm2 of live coral per day, depending on season and their body size (Glynn, 1973; Keesing and Lucas, 1992). However, densities of 10–15 starfish per hectare (0.001–0.0015 ind./m2) could be sustained in areas with more than 20% coral cover (Keesing and Lucas, 1992). Therefore, re-established coral may provide more food sources for COTS in a larger biogeographical range, and it is reasonable to speculate that the COTS population historical expansion followed coral habitat range expansion. Moreover, COTS generally prefer to prey on Acropora spp. and Pocillopora spp. (Kayal et al., 2012), which were widely distributed in the mature tropical coral reef ecosystems in the SCS, and were rare inshore (Chen et al., 2009; Zhao et al., 2013, 2016; Yu et al., 2019). Birkeland and Lucas (1990) found that variation in the effects of COTS outbreaks in distinct coral habitats of the Pacific may be explained by the relative dominance of Acropora in coral communities. Furthermore, Acropora spp. are consistent among the corals that are worst affected during outbreaks and tend to dominate in the Western and Southern Pacific (Pratchett et al., 2009; Pratchett, 2010). Thus, feeding preferences of COTS may lead to COTS population expansion relying on coral population range expansion. It was found that, as yet, there were no spines or individual COTS in subtropical coral communities in the northern SCS, which are dominated by massive corals, such as Dipsastrea palauensis, Porites lutea, and Dipsastrea halicora (Chen et al., 2009; Liao et al., 2019; Yu et al., 2019).
Furthermore, the large exposure of Sunda and Sahul shelves and the decrease in width of the Balabac Straits (∼12 km) reduced marine organism connectivity between the SCS and surrounding oceans in the glacial period of the Pleistocene (Voris, 2000). However, the Luzon Strait was not closed due to land expansion and was always connected between the SCS and Western Pacific (Voris, 2000), which was also the main seawater exchange channel in the SCS (Yu et al., 2010). Thus, the opening of the Luzon Strait may be the main reason for there being no genetic divergence in the conserved mitochondrial COI fragment of COTS populations between the SCS and the Pacific.
Accordingly, COTS may have undergone historical population expansion after the Last Glacial Maximum in the Pleistocene, which might have been followed by the expansion of coral populations in the SCS. Furthermore, the biogeographical range of COTS population expansion may have been limited by feeding preferences.
Genetic Differentiation of Crown-of-Thorns Starfish Populations May Be Shaped by Distinct Nutrient Concentration in the South China Sea
It is difficult to determine the relationship between environmental factors and the genetic structure of COTS, as COTS cannot be accurately aged (Deaker et al., 2020). However, this problem can be partially solved by measuring the diameter of COTS (Moran, 1988; Pratchett et al., 2014) and selecting an appropriate sampling time (such as a population outbreak period; Pratchett et al., 2009; Saponari et al., 2018). The nutrient concentration analysis results showed that the Chl a concentration in DI was significantly higher than that in the other coral habitats in the SCS (Figure 6A). Additionally, the POC concentration in DI and BR was significantly higher than that in FTS and PK (Figure 6B), which was consistent with the genetic clustering results inferred by STRUCTURE (Figure 4B). Furthermore, the Mantel test indicated that there was a significant correlation between the Fst values and POC concentration in the SCS (Figure 5C), which was consistent with the DistLM analysis results. Thus, the significant genetic differentiation of COTS in distinct coral habitats with slight geographic isolation may be shaped by nutrient concentration, particularly the POC concentration. One adult female COTS can produce approximately 10 million fertilized eggs per spawning (Conand, 1984; Babcock et al., 2016; Caballes et al., 2021; Pratchett et al., 2021), and COTS larvae predominantly feed on mid-sized phytoplankton (e.g., dinoflagellates and pennate diatoms > 5 μm; Ayukai, 1994). However, the larval starvation hypothesis suggests that severe food limitation would lead to high mortality in COTS larvae (Lucas, 1982; Fabricius et al., 2010). Thus, distinct nutrient concentrations may cause stress on COTS larvae among coral habitats, which may lead to genetic differentiation. Pratchett et al. (2017a) combined many studies and suggested that Chl a concentrations between 0.4 and 1.0 μg⋅L–1 may be optimal for COTS larval survival and settlement. Nevertheless, the total food abundance and sources for COTS larvae still cannot be accurately indicated by Chl a concentrations (Ayukai, 1994; Tada, 2003; Mellin et al., 2017; Pratchett et al., 2017a), COTS larvae not only selectively ingest some phytoplankton species (e.g., Chaetoceros sp. and Dunaliella sp.; Ayukai, 1994; Mellin et al., 2017), but can also utilize other nutrients in oligotrophic coral reefs (Olson and Olson, 1989; Hoegh-Guldberg, 1994; Nakajima et al., 2016). These were the main reasons for the lack of a significant correlation between the Chl a concentration and genetic differentiation indices (Fst) of COTS in the SCS.
It should be noted that COTS larvae can utilize nutrients directly associated with POC and dissolved organic carbon. Some studies found that COTS larvae can assimilate and utilize dissolved organic matter and bacteria in the sea water column (Olson and Olson, 1989; Hoegh-Guldberg, 1994), and the large amounts of energy required for developing larvae could be provided by dissolved free amino acids (Hoegh-Guldberg, 1994). However, the concentration of dissolved free amino acids in natural seawater is extremely low, and cannot make a significant contribution to the nutrition of COTS larvae (Ayukai, 1993). Moreover, coral and other reef benthos can release large amounts of mucus aggregates and organism-derived organic matter to reef water, which are closely associated with POC and provide alternative food resource for COTS larvae. Many previous studies also showed that particulate organic matter in reef water can be utilized by COTS larvae (Okaji et al., 1997). An experiment with labeled stable isotope tracers (13C and 15N) also found that COTS larvae can utilize coral mucus particles and associated microorganisms, which may be an additional food resource (Nakajima et al., 2016). Therefore, the impact of Chl a concentration on the genetic structure of COTS larvae in the oligotrophic waters around Paracel and Spratly Islands (Nansha Islands) was weak. But, distinct POC concentration may directly lead to significant genetic differentiations of COTS populations among coral habitats in the SCS, over small geographic distances (e.g., DI-BR genotype and FTS-PK genotype; Figure 4). In addition, because the spatial difference of nutrient availability has a substantial impact on the survival and growth of COTS larvae (Olson, 1987; Okaji et al., 1997; Wolfe et al., 2015), it was speculated that COTS larvae of the FTS-PK genetic cluster may be able to tolerate low nutrient concentrations, whereas DI-BR COTS genotypes are more competitive in coral habitats with higher POC concentration. The low nutrition resistance of COTS larvae may be associated with maternal nutrition (Caballes et al., 2016). Previous studies have shown that the coral cover of Acropora was decreased with the increasing of the latitude in the SCS, and the abundance of Acropora in FTS and PS was higher than that in DI and BR (Zhao et al., 2013, 2016; Li et al., 2019; Liao et al., 2021). Acropora was as preferred coral prey for COTS, which may provide high quality and quantity of food to female COTS in the FTS and PS, which might promote COTS larvae to tolerate low nutrient availability and to progress to more advanced stages faster (Caballes et al., 2016). However, this advantage may diminish under the high POC condition.
There was a weak genetic differentiation in COTS populations between FTS and PK, which implied that COTS larval populations have effective connectivity between two coral habitats with a distance of ∼800 km (Figure 1). In addition, the STRUCTURE results showed that the frequency of the FTS-PK genotype (blue) was also higher than that of the DI-BR (gray) genotype in the COTS populations in the Western Pacific. Thus, the FTS-PK genotype may have a longer PLS, which might be able to achieve long-distance dispersal via ocean currents. It has been found that several COTS larval genotypes have a relatively long PLS period in the Pacific Ocean, and have spread among Guam, Kingman Reef, and Swains through equatorial current systems (Tusso et al., 2016). However, the frequency of the DI-BR genotype was rare except on the Paracel Islands, which suggested that the DI-BR genotype may belong to the local COTS population.
Accordingly, distinct nutrient concentration, especially different POC concentration may lead to genetic differentiation of COTS populations among coral habitats within short geographic distance in the SCS. Also, FTS-PK genotypes may have relative longer PLS and potentially stronger dispersal ability than the DI-BR genotype in the Paracel Islands.
Ocean Currents Provide a Potential Dispersal Mechanism for Crown-of-Thorns Starfish Larvae in the South China Sea
The COTS populations have effective connectivity and gene flow between PK and FTS (Figure 4B), which may benefit from the transport of COTS larvae by ocean currents. The COTS are similar to many reef benthic organisms (e.g., coral and anemones; Kool et al., 2011; Treml et al., 2012), which are almost incapable of migrating between isolated atolls as adults (move < 35 m/day; Chesher, 1969; Keesing and Lucas, 1992; Pratchett et al., 2017c). Thus, the dispersal of COTS was more likely to be completed in the PLS. The shortest geographic distance is about 800 km between PK and FTS, which is a large geographic barrier for COTS larval dispersal. The PLS of COTS lasts between 9 and 42 days (Caballes and Pratchett, 2014), and therefore, larvae were expected to disperse only 10–100 km between reefs without assistance from oceanic currents (Dight et al., 1990). The results of IBD analysis also suggested that large geographic isolation would lead to a genetic break among COTS populations (Figure 5A; Dight et al., 1990; Vogler et al., 2012). For instance, there was significant genetic differentiation between COTS in Southeast Africa and that in East Asia (Yasuda et al., 2009). However, the gene flow inference suggested that COTS populations have effective connectivity between reefs separated by <1000 km (Yasuda et al., 2009). Unexpectedly, it has been found that COTS larvae can spread to remote coral reefs with the assistance of strong ocean currents in the Pacific Ocean (Vogler et al., 2013; Tusso et al., 2016). In addition, the spawning period of COTS is from May to August in the Northern Hemisphere (Pratchett et al., 2014, 2017a), and thus, the ocean currents from May to August were critical to COTS individual dispersal in the SCS. The results of ocean current analysis showed that there were stable currents from the southern part of the Paracel Islands to the Spratly Islands from June to August (current velocity: 0.25–0.4 m/s; average current velocity: 0.33 m/s; Figure 7), which may assist COTS larvae in the Paracel Islands waters to travel to the Spratly Islands within 23 days. Therefore, based on these results, we suggest that some COTS larvae in FTS may have originated from the southern part of the Paracel Islands. Since the ocean current was from south to north in May from the SCS, the migration of COTS larvae from PK to FTS may have started in June.
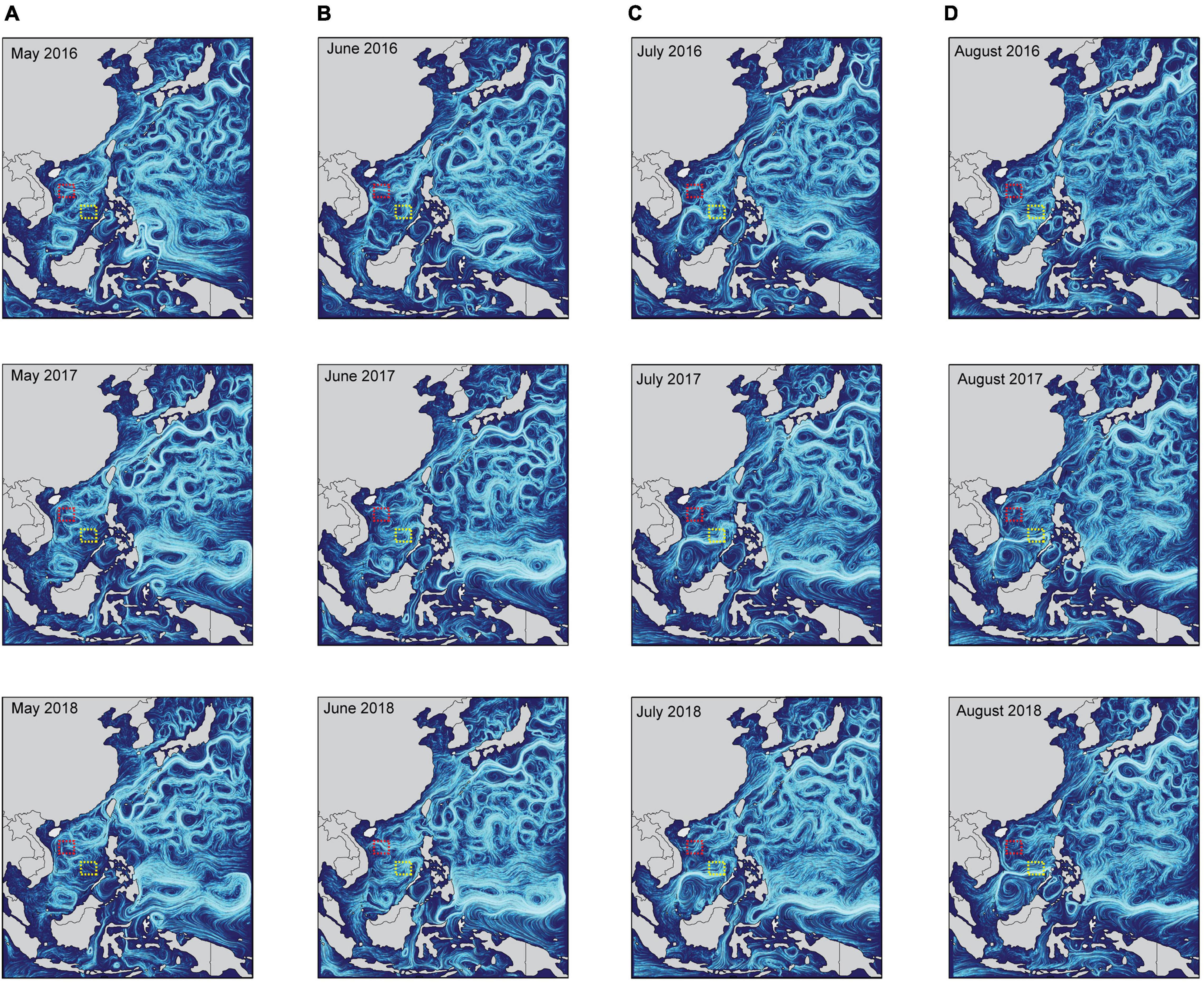
Figure 7. The characteristics of ocean currents from 2016 to 2018 in the South China Sea. The upper-ocean surface current condition (depth < 2.5 m) in (A) May, (B) June, (C) July, and (D) August, which during potential crown-of-thorns starfish breeding periods in the Northern Hemisphere. These data were collected from the National Marine Science Data Center, National Science & Technology Resource Sharing Service Platform of China (http://mds.nmdis.org.cn/). Red and yellow dotted boxes contain sampling sites in the Paracel and Spratly Islands, respectively.
Interestingly, the STRUCTURE results indicated that blue and green genotypes were shared between the FTS-PK and Western Pacific, which implied that COTS populations exhibit recombination and gene exchange between the two regions (Figure 4B). The SCS is a semi-enclosed marginal sea, and the main seawater exchange is through the Luzon Strait to the Pacific Ocean (∼2400 m; Yu et al., 2010). The Taiwan Strait (∼70 m) and the Mindoro Strait (∼400 m) are outflow zones for the SCS (Fang et al., 2003; Su, 2004; Yu et al., 2010). Since the Karimata Strait is shallow (∼29 m), there is negligible exchange of water between the SCS and the Indian Ocean (Yu et al., 2010). Some studies have suggested that reef organisms in the southern SCS have genetic connectivity with the “Coral Triangle,” and Symbiodiniaceae and coral larvae could enter the SCS through the Sulu Sea (Kool et al., 2011; Chen et al., 2020). However, many marine organisms that disperse mainly during the PLS still have large biogeographical barriers between the SCS and the Indo-Pacific region. Notably, recent gene flow and ocean current data revealed that western boundary currents and coastal currents play an important role in COTS larval dispersal in the Pacific Ocean (Dight et al., 1990; Moran et al., 1992; Yasuda et al., 2009). For instance, COTS larvae were transported from Mindoro in central Philippines to northern Luzon Island by the Northwest Luzon Coastal Current (Hu et al., 2000; Yasuda et al., 2009), and the strong Kuroshio Current is likely to continue to transport them to Ryukyu Island in southern Japan (Yasuda et al., 2009; Yasuda, 2018). Some mtDNA and microsatellite data have shown that there was high connectivity of COTS populations between the Philippines and the Western Pacific Ocean (Vogler et al., 2013; Tusso et al., 2016). Thus, the boundary and coastal current systems may help the COTS larval populations in the Western Pacific enter the SCS. Many COTS larvae may have migrated and spread among many coral habitats in a stepping-stone fashion within the SCS (Tusso et al., 2016).
Conclusion
Based on mtDNA sequences, and phylogenetic and demographic history analysis of COTS populations, there was no significant phylogenetic divergence in COTS populations between the SCS and Pacific Ocean. However, there was a clear signature of population expansion in the SCS. These results suggested that COTS may have undergone historical population expansion after the Last Glacial Maximum in the Pleistocene, which might have been followed by the expansion of coral populations in the SCS. Furthermore, the opening of the Luzon Strait may be one of the reasons for the lack of significant genetic divergence in COTS populations between the SCS and the Pacific. In addition, according to the microsatellite loci analysis, the genetic diversity of COTS populations in the SCS was consistent with that in the Western Pacific and Pacific equatorial current affected zone COTS populations, which was higher than that in the Pacific insular atolls. However, the genetic structure of COTS populations in the SCS may have been shaped by distinct nutrient concentrations, particularly different POC concentrations over small geographic distances. FTS-PK genotypes may have stronger dispersal ability than the local DI-BR genotype in the SCS. Moreover, ocean currents provide a potential dispersal mechanism for COTS larvae in the SCS. Stable ocean currents may assist COTS larvae to travel from the Paracel Islands to the Spratly Islands from June to August, and COTS larvae in the Western Pacific may undergo gene exchange with those in the SCS by boundary and coastal current systems. Accordingly, this study demonstrates that environmental and oceanographic factors play important roles in shaping the genetic characteristics and larval dispersal of COTS populations in the northern Indo-Pacific convergence region.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
KY and BC deigned the research. QY, ZL, ZQ, XY, QW, and BH contributed the materials. BC, QW, and BH performed the research. BC analyzed the data and drawn all pictures. BC and KY wrote the manuscript. All the authors reviewed the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 42090041 and 42030502), the Guangxi Scientific Projects (Nos. AD17129063 and AA17204074), and the Innovation Project of Guangxi Graduate Education (No. YCBZ2018006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Zhixian Li for improving the quality of the English writing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.728349/full#supplementary-material
Footnotes
References
Antonio, C. R. (2018). Myriads: P-value-based multiple testing correction. Bioinformatics 2018, 1043–1045. doi: 10.1093/bioinformatics/btx746
Ayukai, T. (1993). “Resource availability to the larvae of the crown-of-thorns starfish in Great Barrier Reef waters,” in The possible causes and consequences of outbreaks of the crown-of-thorns starfish, Vol. 18, eds U. Engelhardt and B. Lassig (Queensland, MD: Great Barrier Reef), 21–35.
Ayukai, T. (1994). Ingestion of Ultraplankton by the Planktonic Larvae of the Crown-of-Thorns Starfish, Acanthaster planci. Biol. Bull. 186, 90–100. doi: 10.2307/1542039
Babcock, R. C., Milton, D. A., and Pratchett, M. S. (2016). Relationships between size and reproductive output in the crown-of-thorns starfish. Mar. Biol. 163:5. doi: 10.1007/s00227-016-3009-5
Birkeland, C. (1982). Terrestrial runoff as a cause of outbreaks of Acanthaster. planci (Echinodermata: Asteroidea). Mar. Biol. 69, 175–185. doi: 10.1007/BF00396897
Birkeland, C., and Lucas, J. S. (1990). Acanthaster planci: major management problem of coral reefs. Boca Raton, FL: CRC Press.
Bohonak, A. J. (2002). IBD (Isolation by Distance): a program for analyses of isolation by distance. J. Hered. 93, 153–154. doi: 10.1093/jhered/93.2.153
Brodie, J., Fabricius, K., De’ath, G., and Okaji, K. (2005). Are increased nutrient inputs responsible for more outbreaks of crown-of-thorns starfish? An appraisal of the evidence. Mar. Pollut. Bull. 51, 266–278. doi: 10.1016/j.marpolbul.2004.10.035
Caballes, C. F., and Pratchett, M. S. (2014). “Reproductive biology and early life history of the crown-of-thorns starfish,” in Echinoderms: Ecology, Habitats and Reproductive Biology, ed. E. Whitmore (New York, NY: Nova Science Publishers, Inc), 101–146.
Caballes, C. F., Byrne, M., Messmer, V., and Pratchett, M. S. (2021). Temporal variability in gametogenesis and spawning patterns of crown-of-thorns starfish within the outbreak initiation zone in the northern Great Barrier Reef. Mar. Biol. 168:3. doi: 10.1007/s00227-020-03818-3
Caballes, C. F., Pratchett, M. S., Kerr, A. M., and Rivera-Posada, J. A. (2016). The role of maternal nutrition on oocyte size and quality, with respect to early larval development in the coral-eating starfish, Acanthaster planci. PLoS One 11:e0158007. doi: 10.1371/journal.pone.0158007
Chen, B., Yu, K., Qin, Z., Liang, J., Wang, G., Huang, X., et al. (2020). Dispersal, genetic variation, and symbiont interaction network of heat-tolerant endosymbiont Durusdinium trenchii: Insights into the adaptive potential of coral to climate change. Sci. Total Environ. 723:138026. doi: 10.1016/j.scitotenv.2020.138026
Chen, T., Yu, K., Shi, Q., Li, S., Price, G. J., Wang, R., et al. (2009). Twenty-five years of change in scleractinian coral communities of Daya Bay (northern South China Sea) and its response to the 2008 AD extreme cold climate event. Sci. Bull. 54, 2107–2117. doi: 10.1007/s11434-009-0007-8
Chesher, R. H. (1969). Destruction of Pacific Corals by the Sea Star Acanthaster planci. Science 165, 280–283. doi: 10.1126/science.165.3890.280
Conand, C. (1984). Distribution, reproductive cycle and morphometric relationships of Acanthaster planci (Echinodermata: Asteroidea) in New Caledonia, western tropical Pacific. Proc. Fifth Int. Echinoderm Confer. 1984, 499–506.
Cowan, Z., Dworjanyn, S. A., Caballes, C. F., and Pratchett, M. S. (2016). Predation on crown-of-thorns starfish larvae by damselfishes. Coral Reefs 35, 1253–1262. doi: 10.1007/s00338-016-1491-3
Deaker, D. J., Aguera, A., Lin, H. A., Lawson, C., and Byrne, M. (2020). The hidden army: corallivorous crown-of-thorns seastars can spend years as herbivorous juveniles. Biol. Lett. 16:20190849. doi: 10.1098/rsbl.2019.0849
Death, G., Fabricius, K. E., Sweatman, H., and Puotinen, M. (2012). From the cover: The 27-year decline of coral cover on the Great Barrier Reef and its causes. Proc. Natl. Acad. Sci. U S A. 2012:109. doi: 10.1073/pnas.1208909109
Dight, I. J., Bode, L., and James, M. K. (1990). Modelling the larval dispersal of Acanthaster planci. Coral Reefs 9, 115–123. doi: 10.1007/BF00258224
Earl, D. A., and VonHoldt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Evanno, G., Regnaut, S., and Goudet, J. (2010). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fabricius, K. E., Okaji, K., and Death, G. (2010). Three lines of evidence to link outbreaks of the crown-of-thorns seastar Acanthaster planci to the release of larval food limitation. Coral Reefs 29, 593–605. doi: 10.1007/s00338-010-0628-z
Fang, G., Wei, Z., Choi, B. H., Wang, K., Fang, Y., and Li, W. (2003). Interbasin freshwater, heat and salt transport through the boundaries of the East and South China Seas from a variable-grid global ocean circulation model. Sci. China Earth Sci. 46, 149–161. doi: 10.1360/03yd9014
Fu, Y. (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925.
Glynn, P. W. (1973). Acanthaster: Effect on coral reef growth in Panama. Science 180, 504–506. doi: 10.1126/science.180.4085.504
Glynn, P. W. (1988). Predation on coral reefs: some key processes concepts and research directions. Proc. 6th Int. Coral Reef Sympos. 1, 51–62.
Goudet, J. (2001). FSTAT (version 2.9.3) : A program to estimate and test gene diversities and fixation indices. Burlington, MA: ScienceOpen, Inc.
Harrison, H., Pratchett, M., Messmer, V., Saenz-Agudelo, P., and Berumen, M. (2017). Microsatellites reveal genetic homogeneity among outbreak populations of crown-of-thorns starfish (Acanthaster cf. solaris) on Australia’s Great Barrier Reef. Diversity 9:16. doi: 10.3390/d9010016
Hoegh-Guldberg, O. (1994). Uptake of dissolved organic matter by larval stage of the crown-of-thorns starfish Acanthaster planci. Mar. Biol. 120, 55–63. doi: 10.1007/BF00381942
Hu, J., Kawamura, H., Hong, H., and Qi, Y. (2000). A Review on the Currents in the South China Sea: Seasonal Circulation, South China Sea Warm Current and Kuroshio Intrusion. J. Oceanogr. 56, 607–624. doi: 10.1023/A:1011117531252
Huang, W., Li, M., Yu, K., Wang, Y., Li, J., Liang, J., et al. (2018). Genetic diversity and large-scale connectivity of the scleractinian coral Porites lutea in the South China Sea. Coral Reefs 2018:8. doi: 10.1007/s00338-018-1724-8
Hughes, T. P., Anderson, K. D., Connolly, S. R., Heron, S. F., Kerry, J. T., Lough, J. M., et al. (2018). Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359, 80–83. doi: 10.1126/science.aan8048
Jakobsson, M., and Rosenberg, N. (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801. doi: 10.1093/bioinformatics/btm233
Kayal, M., Vercelloni, J., Lison De Loma, T., Bosserelle, P., Chancerelle, Y., Geoffroy, S., et al. (2012). Predator crown-of-thorns starfish (Acanthaster planci) outbreak, mass mortality of corals, and cascading effects on reef fish and benthic communities. PLoS One 7:e47363. doi: 10.1371/journal.pone.0047363
Keesing, J. K., and Lucas, J. S. (1992). Field measurement of feeding and movement rates of the crown-of-thorns starfish Acanthaster planci (L.). J. Exp. Mar. Biol. Ecol. 156, 89–104. doi: 10.1016/0022-0981(92)90018-6
Kool, J. T., Paris, C. B., Barber, P. H., and Cowen, R. K. (2011). Connectivity and the development of population genetic structure in Indo-West Pacific coral reef communities. Glob. Ecol. Biogeogr. 20, 695–706. doi: 10.1111/j.1466-8238.2010.00637.x
Li, Y., Wu, Z., Liang, J., Chen, S., and Zhao, J. (2019). Analysis on the outbreak period and cause of Acanthaster planci in Xisha Islands in recent 15 years (in Chinese). Chinese Sci. Bull. 64, 3478–3484. doi: 10.1360/TB-2019-0152
Liao, Z., Yu, K., Chen, B., Huang, X., Qin, Z., and Yu, X. (2021). Spatial distribution of benthic algae in the South China Sea: responses to gradually changing environmental factors and ecological impacts on coral communities. Divers. Distrib. 27, 929–943. doi: 10.1111/ddi.13243
Liao, Z., Yu, K., Wang, Y., Huang, X., and Xu, L. (2019). Coral-algal interactions at Weizhou Island in the northern South China Sea: variations by taxa and the exacerbating impact of sediments trapped in turf algae. Peer J 7:e6590. doi: 10.7717/peerj.6590
Lucas, J. S. (1973). Reproductive and larval biology of Acanthaster planci (L.) in Great Barrier Reef waters. Micronesica 9, 197–203.
Lucas, J. S. (1982). Quantitative studies of feeding and nutrition during larval development of the coral reef asteroid Acanthaster planci (L.). J. Exp. Mar. Biol. Ecol. 65, 173–193. doi: 10.1016/0022-0981(82)90043-0
McArdle, B. H., and Anderson, M. J. (2001). Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82, 290–297.
Mellin, C., Lugrin, C., Okaji, K., Francis, D., and Uthicke, S. (2017). Selective Feeding and Microalgal Consumption Rates by Crown-Of-Thorns Seastar (Acanthaster cf. solaris) Larvae. Diversity 9:8. doi: 10.3390/d9010008
Mellin, C., Matthews, S. A., Anthony, K. R. N., Brown, S. C., Caley, M. J., Johns, K. A., et al. (2019). Spatial resilience of the Great Barrier Reef under cumulative disturbance impacts. Glob. Chang. Biol. 25, 2431–2445. doi: 10.1111/gcb.14625
Moran, P. J. (1988). The Acanthaster phenomenon. Oceanogr. Mar. Biol. Annu. Rev. 24, 379–480. doi: 10.1080/07328318608068694
Moran, P. J., De’ath, G., Baker, V. J., Bass, D. K., and Thompson, A. A. (1992). Pattern of outbreaks of crown-of-thorns starfish (Acanthaster planci L.) along the Great Barrier Reef since 1966. Mar. Freshwater Res. 43, 555–567. doi: 10.1071/MF9920555
Nakajima, R., Nakatomi, N., Kurihara, H., Fox, M., Smith, J., and Okaji, K. (2016). Crown-of-Thorns Starfish larvae can feed on organic matter released from Corals. Diversity 8:18. doi: 10.3390/d8040018
Okaji, K., Ayukai, T., and Lucas, J. S. (1997). Selective feeding by larvae of the crown-of-thorns starfish, Acanthaster planci (L.). Coral Reefs 16, 47–50. doi: 10.1007/s003380050058
Olson, R. R. (1987). In situ culturing as a test of the larval starvation hypothesis for the crown-of-thorns starfish, Acanthaster planci. Limnol. Oceanogr. 32, 895–904. doi: 10.4319/lo.1987.32.4.0895
Olson, R. R., and Olson, M. H. (1989). Food limitation of planktotrophic marine invertebrate larvae: Does it control recruitment success? Annu. Rev. Ecol. Syst. 20, 225–247. doi: 10.1146/annurev.es.20.110189.001301
Peakall, R., and Smouse, P. (2010). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. 6, 288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Pillans, B., Chappell, J., and Naish, T. R. (1998). A review of the Milankovitch climatic beat: template for Plio–Pleistocene sea-level changes and sequence stratigraphy. Sediment Geol. 122, 5–21. doi: 10.1016/S0037-0738(98)00095-5
Pratchett, M. S. (2005). Dynamics of an outbreak population of Acanthaster planci at Lizard Island, northern Great Barrier Reef (1995–1999). Coral Reefs 24, 453–462. doi: 10.1007/s00338-005-0006-4
Pratchett, M. S. (2010). Changes in coral assemblages during an outbreak of Acanthaster planci at Lizard Island, northern Great Barrier Reef (1995-1999). Coral Reefs 29, 717–725. doi: 10.1007/s00338-010-0602-9
Pratchett, M. S., Caballes, C. F., Rivera-Posada, J. A., and Sweatman, H. P. A. (2014). Limits to understanding and managing outbreaks of crown-of-thorns starfish (Acanthaster spp.). Oceanogr. Mar. Biol. 52, 133–200. doi: 10.1201/b17143-4
Pratchett, M., Caballes, C., Wilmes, J., Matthews, S., Mellin, C., Sweatman, H., et al. (2017a). Thirty Years of Research on Crown-of-Thorns Starfish (1986–2016): Scientific Advances and Emerging Opportunities. Diversity 9:41. doi: 10.3390/d9040041
Pratchett, M. S., Dworjanyn, S., Mos, B., Caballes, C., Thompson, C., and Blowes, S. (2017b). Larval Survivorship and Settlement of Crown-of-Thorns Starfish (Acanthaster cf. solaris) at Varying Algal Cell Densities. Diversity 9:2. doi: 10.3390/d9010002
Pratchett, M. S., Cowan, Z., Nadler, L. E., Caballes, C. F., Hoey, A. S., Messmer, V., et al. (2017c). Body size and substrate type modulate movement by the western Pacific crown-of-thorns starfish, Acanthaster solaris. PLoS One 12:e0180805. doi: 10.1371/journal.pone.0180805
Pratchett, M. S., Nadler, L. E., Burn, D., Lang, B. J., Messmer, V., and Caballes, C. F. (2021). Reproductive investment and fecundity of Pacific crown-of-thorns starfish (Acanthaster cf. solaris) on the Great Barrier Reef. Mar. Biol. 168:87. doi: 10.1007/s00227-021-03897-w
Pratchett, M. S., Schenk, T. J., Baine, M., Syms, C., and Baird, A. H. (2009). Selective coral mortality associated with outbreaks of Acanthaster planci L. in Bootless Bay, Papua New Guinea. Mar. Environ. Res. 67, 230–236. doi: 10.1016/j.marenvres.2009.03.001
Reaka-Kudla, M. L. (1997). “The global biodiversity of coral reefs: A comparison with rain forests,” in Biodiversity II:Understanding and Protecting our Natureal Resources, eds M. L. Reaka-Kudla and E. O. Wilson (Washington, D. C: National Academy Press), 83–108.
Robert, C. M., McClean, C. J., Verson, J. E. N., Hawkins, J. P., Allen, G. R., McAllister, D. E., et al. (2002). Marine biodiversity hotspots and conservation priorities for tropical reefs. Science 295, 1280–1284. doi: 10.1126/science.1067728
Rosenberg, N. A. (2010). DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Resour. 4, 137–138. doi: 10.1046/j.1471-8286.2003.00566.x
Rotjan, R. D., and Lewis, S. M. (2008). Impact of coral predators on tropical reefs. Mar. Ecol. Prog. Ser. 367, 73–91. doi: 10.3354/meps07531
Saponari, L., Montalbetti, E., Galli, P., Strona, G., Seveso, D., Dehnert, I., et al. (2018). Monitoring and assessing a 2-year outbreak of the corallivorous seastar Acanthaster planci in Ari Atoll, Republic of Maldives. Environ. Monitor. Assess. 190:344. doi: 10.1007/s10661
Silverstein, R. N., Cunning, R., and Baker, A. C. (2015). Change in algal symbiont communities after bleaching, not prior heat exposure, increases heat tolerance of reef corals. Glob. Change Biol. 21, 236–249. doi: 10.1111/gcb.12706
Stuart-Smith, R. D., Brown, C. J., Ceccarelli, D. M., and Edgar, G. J. (2018). Ecosystem restructuring along the Great Barrier Reef following mass coral bleaching. Nature 560, 92–96. doi: 10.1038/s41586-018-0359-9
Su, J. (2004). Overview of the South China Sea circulation and its influence on the coastal physical oceanography outside the Pearl River Estuary. Cont. Shelf Res. 24, 1745–1760. doi: 10.1016/j.csr.2004.06.005
Suzuki, G., Kai, S., and Yamashita, H. (2012). Mass stranding of crown-of-thorns starfish. Coral Reefs 31, 821–821. doi: 10.1007/s00338-012-0906-z
Suzuki, G., Yasuda, N., Ikehara, K., Fukuoka, K., Kameda, T., Kai, S., et al. (2016). Detection of a High-Density Brachiolaria-Stage Larval Population of Crown-of-Thorns Sea Star (Acanthaster planci) in Sekisei Lagoon (Okinawa, Japan). Diversity 8:9. doi: 10.3390/d8020009
Tada, K. (2003). Size-fractionated phytoplankton biomass in coral reef waters off Sesoko Island, Okinawa, Japan. J. Plankton Res. 25, 991–997. doi: 10.1093/plankt/25.8.991
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595.
Tian, J., Huang, E., and Pak, D. K. (2010). East Asian winter monsoon variability over the last glacial cycle: Insights from a latitudinal sea-surface temperature gradient across the South China Sea. Palaeogeogr. Palaeocl. 292, 319–324. doi: 10.1016/j.palaeo.2010.04.005
Timmers, M. A., Bird, C. E., Skillings, D. J., Smouse, P. E., and Toonen, R. J. (2012). There’s no place like home: crown-of-thorns outbreaks in the central pacific are regionally derived and independent events. PLoS One 7:e31159. doi: 10.1371/journal.pone.0031159
Treml, E. A., Roberts, J. J., Chao, Y., Halpin, P. N., Possingham, H. P., and Riginos, C. (2012). Reproductive Output and Duration of the Pelagic Larval Stage Determine Seascape-Wide Connectivity of Marine Populations. Integr. Comp. Biol. 52, 525–537. doi: 10.1093/icb/ics101
Tusso, S., Morcinek, K., Vogler, C., Schupp, P. J., Caballes, C. F., Vargas, S., et al. (2016). Genetic structure of the crown-of-thorns seastar in the Pacific Ocean, with focus on Guam. Peer J 4:e1970. doi: 10.7717/peerj.1970
Uthicke, S., Logan, M., Liddy, M., Francis, D., Hardy, N., and Lamare, M. (2015a). Climate change as an unexpected co-factor promoting coral eating seastar (Acanthaster planci) outbreaks. Sci. Rep. 5:sre08402. doi: 10.1038/srep08402
Uthicke, S., Doyle, J., Duggan, S., Yasuda, N., and McKinnon, A. D. (2015b). Outbreak of coral-eating Crown-of-Thorns creates continuous cloud of larvae over 320 km of the Great Barrier Reef. Sci. Rep. 5:sre16885. doi: 10.1038/srep16885
Vogler, C., Benzie, J. A. H., Tenggardjaja, K., Ambariyanto Barber, P. H., and Wörheide, G. (2013). Phylogeography of the crown-of-thorns starfish: genetic structure within the Pacific species. Coral Reefs 32, 515–525. doi: 10.1007/s00338-012-1003-z
Vogler, C., Benzie, J., Barber, P. H., Erdmann, M. V., Ambariyanto Sheppard, C., et al. (2012). Phylogeography of the crown-of-thorns starfish in the Indian Ocean. PLoS One 7:e43499. doi: 10.1371/journal.pone.0043499
Vogler, C., Benzie, J., Lessios, H., Barber, P., and Wörheide, G. (2008). A threat to coral reefs multiplied? Four species of crown-of-thorns starfish. Biol. Lett. 4, 696–699. doi: 10.1098/rsbl.2008.0454
Voris, H. K. (2000). Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. J. Biogeogr. 27, 1153–1167. doi: 10.1046/j.1365-2699.2000.00489.x
Wilkinson, C. A. (2008). Status of coral reefs of the word. Townsville: Australian Institute of Marine Science, 1–304.
Wolfe, K., Graba-Landry, A., Dworjanyn, S. A., and Byrne, M. (2015). Larval starvation to satiation: Influence of nutrient regime on the success of Acanthaster planci. PLoS One 10:1–18. doi: 10.1371/journal.pone
Wooldridge, S. A., and Brodie, J. E. (2015). Environmental triggers for primary outbreaks of crown-of-thorns starfish on the Great Barrier Reef, Australia. Mar. Pollut. Bull. 101, 805–815. doi: 10.1016/j.marpolbul.2015.08.049
Yamaguchi, M. (1986). Acanthaster planci infestations of reefs and coral assemblages in Japan: a retrospective analysis of control efforts. Coral Reefs 5, 23–30. doi: 10.1007/BF00302168
Yasuda, N. (2018). Distribution Expansion and Historical Population Outbreak Patterns of Crown-of-Thorns Starfish, Acanthaster planci sensu lato, in Japan from 1912 to 2015. Coral Reefs World 2018, 125–148. doi: 10.1007/978-981-10-6473-9_9
Yasuda, N., Kajiwara, K., NagaiI, S., Ikehara, K., and Nadaoka, K. (2015a). First report of field sampling and identification of crown-of-thorns starfish larvae. Galaxea 17, 15–16. doi: 10.3755/galaxea.17.15
Yasuda, N., Taquet, C., Nagai, S., Yoshida, T., and Adjeroud, M. (2015b). Genetic connectivity of the coral-eating sea star Acanthaster planci during the severe outbreak of 2006–2009 in the Society Islands, French Polynesia. Mar. Ecol. 36, 668–678. doi: 10.1111/maec.12175
Yasuda, N., Nagai, S., Hamaguchi, M., and Nadaoka, K. (2007). Seven new microsatellite markers for crown-of-thorns starfish Acanthaster planci. Plankton Benthos. Res. 2, 103–106. doi: 10.3800/pbr.2.103
Yasuda, N., Nagai, S., Hamaguchi, M., Lian, C. L., and Nadaoka, K. (2006). Development of microsatellite markers for the crown-of-thorns starfish Acanthaster planci. Mol. Ecol. Notes 6, 141–143. doi: 10.1111/j.1471-8286.2005.01168.x
Yasuda, N., Nagai, S., Hamaguchi, M., Okaji, K., Gérard, K., and Nadaoka, K. (2009). Gene flow of Acanthaster planci (L.) in relation to ocean currents revealed by microsatellite analysis. Mol. Ecol. 18, 1574–1590. doi: 10.1111/j.1365-294X.2009.04133.x
Yu, K. (2012). Coral reefs in the South China Sea: Their response to and records on past environmental changes. Sci. China Earth Sci. 55, 1217–1229. doi: 10.1007/s11430-012-4449-5
Yu, K., Hua, Q., Zhao, J. X., Hodge, E., Fink, D., and Barbetti, M. (2010). Holocene marine 14C reservoir age variability: Evidence from 230Th-dated corals in the South China Sea. Paleoceanogr. Paleocl. 25:3205. doi: 10.1029/2009PA001831
Yu, W., Wang, W., Yu, K., Wang, Y., and Chen, X. (2019). Rapid decline of a relatively high latitude coral assemblage at Weizhou Island, northern South China Sea. Biodivers. Conserv. 28, 3925–3949. doi: 10.1007/s10531-019-01858-w
Zhao, M. X., Yu, K. F., Shi, Q., Chen, T. R., Zhang, H. L., and Chen, T. G. (2013). Coral communities of the remote atoll reefs in the Nansha Islands, southern South China Sea. Environ. Monit. Assess. 185, 7381–7392. doi: 10.1007/s10661-013-3107-5
Keywords: coral reefs, environmental impact, larval dispersal, nutrient concentration, genetic structure, population expansion, crown-of-thorns starfish
Citation: Chen B, Yu K, Yao Q, Liao Z, Qin Z, Yu X, Wu Q and He B (2021) Insights Into the Environmental Impact on Genetic Structure and Larval Dispersal of Crown-of-Thorns Starfish in the South China Sea. Front. Mar. Sci. 8:728349. doi: 10.3389/fmars.2021.728349
Received: 21 June 2021; Accepted: 15 November 2021;
Published: 06 December 2021.
Edited by:
Charles Alan Jacoby, St. Johns River Water Management District, United StatesReviewed by:
Benjamin Mos, Southern Cross University, AustraliaCiemon Frank Caballes, ARC Centre of Excellence for Coral Reef Studies, Australia
Copyright © 2021 Chen, Yu, Yao, Liao, Qin, Yu, Wu and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kefu Yu, a2VmdXl1QHNjc2lvLmFjLmNu
 Biao Chen
Biao Chen Kefu Yu
Kefu Yu Qiucui Yao
Qiucui Yao Zhiheng Liao1
Zhiheng Liao1 Zhenjun Qin
Zhenjun Qin Xiaopeng Yu
Xiaopeng Yu