- 1Department of Fisheries Science, Faculty of Aquatic Biology, Chonnam National University, Yeosu, South Korea
- 2Department of Fisheries Biology and Genetics, Faculty of Fisheries, Patuakhali Science and Technology University, Patuakhali, Bangladesh
- 3Department of Horticulture, Sunchon National University, Suncheon, South Korea
Pacific abalone (Haliotis discus hannai) is a commercially important high valued molluscan species. Its wild population has decreased in recent years. It is widely cultured in Korea. Traditional breeding programs have been implemented for hatchery production of abalone seeds. To obtain more genetic information for its molecular breeding program, a high-density linkage map and quantitative trait locus (QTL) for three growth-related traits was constructed for Pacific abalone. F1 cross population with two parents were sampled to construct the linkage map using genotyping by sequencing (GBS). A total of 664,630,534 clean reads and 56,686 SNPs were generated and 3,345 segregating SNPs were used to construct a consensus linkage map. The map spanned 1,747.023 cM with 18 linkage groups and an average interval of 0.55 cM. QTL analysis revealed two significant QTL in LG10 on the consensus linkage map of each growth-related trait. Both QTLs were located in the telomere region of the chromosome. Moreover, four potential candidate genes for growth-related traits were identified in the QTL region. Expression analysis revealed that these identified genes are involved in growth regulation of abalone. The newly constructed genetic linkage map, growth-related QTLs and potential candidate genes identified in the present study can be used as valuable genetic resources for marker-assisted selection (MAS) of Pacific abalone in molecular breeding program.
Introduction
Abalones are marine molluscan species distributed worldwide along coastal waters in tropical and temperate regions (Gordon and Cook, 2013). Pacific abalone (Haliotis discus hannai) is the most commercially important invertebrate species in South Korea as well as in South-East Asian countries. Because of its extraordinary food value due to the presence of bioactive molecules and high price, commercial aquaculture of abalone has expanded widely (Park and Kim, 2013; Hossen et al., 2021). It is reared in sea cages and extensive case farming has distributed along small bays and islands of South, South-West and West coasts of Korea (Choi et al., 2015). Commercial-scale production of abalone in Korea began in the 2000s. As of 2015, abalone production reached about 10,090 metric tons. Its production was doubled in the next 4 years, reaching about 20,100 metric tons in 2019 with commercial value of about 536 million USD (KOSTAT, 2020). However, aquaculture production of abalone is facing difficulties due to its low growth rate, summer mortality and frequent outbreaks of infectious diseases. A total of 684 hatcheries are producing abalone seeds in Korea using conventional selective breeding methods (Choi et al., 2015). Recent studies have shown that successful aquaculture mostly depends on molecular breeding for rapid growth, bigger size, disease resistance and higher survival of target species (Wang et al., 2018). Hence, it is essential to ascertain molecular breeding for Pacific abalone.
Growth is the most preferred economic important trait for aquaculture species including abalone. Substantial improvement of abalone seed production has been achieved using conventional phenotype-based selection for growth. However, such method is labor-intensive, time consuming, error prone and influenced by environmental factors (Gjedrem and Baranski, 2009). Molecular breeding using marker-assisted selection (MAS) can increase the accuracy of selection and shorten the operation time (Xu et al., 2017). Economically related traits such as growth, meat quality and immunity are mainly controlled by quantitative trait loci (QTLs). QTLs are defined as chromosomal regions linking single gene or genes clusters (Yue, 2014). With progress and growing application of contemporary biotechnology, marker-assisted selection (MAS) is becoming popular over phenotype-based selection in animal breeding program. Markers linked to QTLs are used in MAS to accelerate the molecular breeding technique that can increase the accuracy of selection and achieve genetic improvement through direct and prompt selection (Feng et al., 2018; Song et al., 2018).
Mapping of QTL is a principal step in molecular breeding program. Genetic linkage map is an essential tool for genomic research as it can provide a basis for QTL mapping (Tian et al., 2015). The availability of a bulk number of molecular markers is a prerequisite to construct a high-density genetic linkage map. Several molecular markers such as express sequence tag (EST), single sequence repeats (SSR), restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), randomly amplified polymorphic DNA (RAPD) and single nucleotide polymorphism (SNP) can be used to construct a genetic linkage map. Among these, SNPs are the most abundant markers located in the genome, and appropriate for broad-scale genotyping and construction of a high-density linkage map (Lindblad-Toh et al., 2000). A number of linkage maps have been reported for Pacific abalone using DNA-based low-density SSR markers (Sekino and Hara, 2007), AFLP markers (Qi et al., 2007) and a combination of AFLP, RAPD, and SSR markers (Liu et al., 2006, 2007). Even so, these linkage maps are limited by the lack of dense polymorphic molecular markers. Recently introduced next-generation sequencing (NGS) technologies are cost-effective, easy, and rapid to detect genome-wide molecular markers (such as SNPs) for construction of a high-density linkage map. Among various NGS technologies, genotype-by-sequencing (GBS) is a new method to perform sequence-based genotyping by preparing libraries of thousands of DNA fragments. Thus, GBS technology provides a large quantity of quality data that can be used to construct a fine map using SNP markers from a reference genome to detect QTLs (Salazar et al., 2019). After the introduction of GBS, QTL mapping using GBS has been performed for several aquaculture species such as snapper (Ashton et al., 2019), Pacific oyster (Li et al., 2018), Pacific bluefin tuna (Uchino et al., 2018) and scallop (Jiao et al., 2014).
In the present study, GBS approach was used for large-scale identification of SNP markers and construction of a SNP-based high-density linkage map for Pacific abalone. Further QTL mapping was performed to detected SNPs associated with growth-related traits. Finally, candidate genes were identified in the QTL region that might regulate growth-related traits of Pacific abalone.
Materials and Methods
Mapping Family
An F1 family of Pacific abalone was produced in May 2019 for genetic mapping. Female and male parents were obtained from two geographically distant zones in Korea: a smaller-sized female (95.64 g; 94.97 mm) from Wando (34°19′12.8″N 126°39′02.6″E) in South coast of Korea and a larger-sized male (115.92 g; 105.21 mm) from Boryeong (36°18′56.3″N 126°26′17.8″E) in West coast of Korea. The parents were maintained in a commercial abalone hatchery, Tou-Jeong Soosan abalone hatchery, Dolsan-eup, Yeosu, Korea. Abalones were reared in recirculating sea water tanks. After reaching at full maturity, induced spawning was performed by heat-induction and UV-irradiation (Uki and Kikuchi, 1984; Leighton, 2008). Briefly, fully mature abalones were exposed to sunlight for one hour in an upside-down orientation and another half an hour in an upside-up orientation. Heat induced abalones were then kept in a spawning tank with UV-irradiated seawater and aeration until spawning. After fertilization and production of abalone larvae, 15,000 progenies were reared under commercial condition at the larval rearing unit with recirculating sea water in Tou-Jeong Soosan abalone hatchery. These larvae were ensured to have optimal grow-out densities.
Husbandry of Abalone Larvae and Monitoring of Water Quality
A 1.5 L min–1 sea water flow rate was maintained continuously in the rearing tanks. Water quality parameters such as temperature, dissolved oxygen, pH and salinity were measured daily with a YSI digital water quality meter (Pro 10102030; Xylem Inc., United States) (Supplementary Table 1). Abalone larvae were fed every day in spring-summer season and every other day in winter season to satiation with commercial abalone feed (Gold Ocean Premium Abalone Feed, JOEUN Corp., Wando, Korea). The rearing tank was cleaned every day in spring-summer season and at every other day interval in winter season by changing water and washing with sea water flash.
Ethics Statement
Animal experiments were conducted in accordance with guidelines of the Institutional Animal Care and Use Committee of Chonnam National University (CNU IACUC) with a permission number CNU IACUC-YS-2020-5. Animals were used according to Article 14th of the Korean Animal Protection Law of the Korean government. Animals were cared for in accordance with Guidelines for Animal Experiments of Chonnam National University.
Sample Collection and Trait Measurement
Twelve months after hatching in May 2020, 96 F1 offspring (48 largest and 48 smallest) were randomly selected and sampled as described previously (Baranski et al., 2008; Li and He, 2014). For linkage analysis, three quantitative growth traits of abalone, i.e., total weight (TW), shell length (SL) and shell width (SW), were measured using an electric balance (SPX223KR, OHAUS, United States) and a digimatic caliper (CD-P20S, Mitutoyo Corp., Japan) to the nearest mg and mm, respectively (Supplementary Table 2). A portion of the nondestructive cephalic tentacle was seized from each juvenile abalone for extraction of genomic DNA (gDNA), washed with phosphate buffered saline (PBS), immediately flash frozen in liquid nitrogen and stored at −80°C until gDNA extraction. After taking cephalic tentacles, juvenile abalones were tagged and returned to a new grow-out tank. Cephalic tentacles of parent abalone were also collected previously during the breeding experiment and stored at –80°C after flash frozen in liquid nitrogen.
Genotyping-by-Sequencing (GBS) Library Preparation, Sequencing and SNP Genotyping
Extraction of Genomic DNA
Genomic DNA of each F1 progeny and parent abalone were extracted from cephalic tentacle using the CTAB (Cetyl trimethylammonium bromide) method as describe previously (Winnepenninckx, 1993; Arseneau et al., 2017) with slight modifications. Briefly, samples were cryogenically ground to fine power in a bead beater. After adding 200 μL of CTAB extraction buffer (0.1 m Tris−HCl pH 8.0, 20 mm EDTA pH 8.0, 1.4 M NaCl, 2% CTAB w/v) containing 73 μL 2-mercaptoethanol to each 100 mg homogenized tissue, the mixture was vortexed thoroughly. The homogenate was then incubated at 65°C in a water bath for 1 h and then centrifuged at 14,000 g for 5 min. The supernatant was then transferred to a new tube. After adding 5 μL of RNase A, the mixture was incubated at 37°C for 20 min. An equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) was added to the mixture. After vortexing for 5 s, the mixture was incubated at room temperature for 5 min and then centrifuged at 13,000 g for 5 min to separate aqueous phases. The clear aqueous upper phase was then transferred to a new tube. After adding 200 μL of chloroform/isoamyl alcohol (24:1), previous steps were repeated and the upper clear aqueous phase was transferred to a new tube. Later, DNA samples were precipitated by adding 2.5 volumes of ice-cold 100% ethanol and incubated at −70°C for 30 min. After the incubation, samples were centrifuged at 13,000 g for 30 min at −4°C. The supernatant was then decanted without disturbing the pellet. The pellet was subsequently washed with 500 μL ice-cold 70% ethanol. Ethanol was decanted and residual ethanol was removed by air-drying. Finally, DNA was dissolved in 20 μL TE buffer (10 mM Tris, pH 8, 1 mM EDTA). The integrity of each DNA sample was evaluated by 1.2% agarose gel electrophoresis and concentration of the DNA was determined with a Nano Photometer spectrophotometer (IMPLEN, United States). The ratio of absorbance at 260/280 nm was used to assess the purity of DNA.
Construction of GBS Library and Sequencing
Construction of GBS library and Illumina HiSeq X sequencing were performed at SEEDERS, Korea using genomic DNA of each female and male parents and 90 progenies. A GBS library was constructed following the protocol described previously (Mascher et al., 2013) by double digestion with two restriction enzymes: PstI and MspI. Chronological steps of GBS library construction included adaptor annealing, DNA digestion with PstI and MspI, adaptor ligation, sample pooling, DNA purification and multiplexed PCR. The quality of the constructed library was analyzed through a quality control check and agarose gel electrophoresis. Pooled GBS library was then sequenced using a HiSeq X platform (Illumina, Inc., United States) by the paired-end read method.
Sequence Data Analysis
Raw sequences were demultiplexed into 96 samples according to barcode sequences. Barcode and adapter sequences were removed using Cutadapt (version 1.8.3) software (Martin, 2011). Low quality sequences were trimmed using DynamicTrim and LengthSort programs of SolexaQA (v.1.13) package (Cox et al., 2010). For DynamicTrim, a phred score ≥20 was used as the criterion. For LengthSort, a read length of ≥25 pb was applied. DynamicTrim removed low-quality bases at either ends of short reads based on the phred score to carry out a purification process for high-quality clean reads. LengthSort removed the reads, from which excess bases were then cut with DynamicTrim.
Raw SNP Detection and Generation of SNP Matrix
Cleaned sequences were aligned to the reference genome sequence of Pacific abalone consisting of 80,032 scaffolds (Supplementary Table 3) using a Burrows-Wheeler Aligner (BWA) program version 0.6.1-r104 (Li and Durbin, 2009). A BAM format file was generated to detect raw SNP (In/Del) using default values with the following options: a seed length (-l) of 30, a maximum difference in the seed (-k) of 1, number of threads (-t) of 16, a mismatch penalty (-M) of six, a gap open penalty (-O) of 15, and a gap extension penalty (-E) of eight. Raw SNPs (In/Del) were detected and consensus sequences were extracted using SAMTools version 0.1.16 (Li et al., 2009). Prior to raw SNP detection, SNP validation was carried out using an in-house script of SEEDERS (Kim et al., 2014). To compare SNPs among analytic targets, an integrated SNP matrix was produced for samples. Based on respective coordinates, SNPs were classified as homozygous (SNP read rate ≥ 90%), heterozygous (40% ≤ SNP read rate ≤ 60%) and ambiguous (20% ≤ SNP read rate ≤ 40% or 60% < SNP read rate < 90%).
SNP Filtering, Genotyping, and Construction of Genetic Linkage Map
Genetic linkage maps were constructed using the JoinMap 4.1 program (Van Ooijen, 2006) with population type cross-pollination (CP) (Grattapaglia and Sederoff, 1994) under the condition of logarithm of odds (LOD) of 6.0 or higher with a maximum distance of 30 cM. To construct an integrated map of male and female parents, all segregating markers that showed polymorphism in at least one parent were used in the JoinMap configuration for CP mode (lmxll, nnxnp, hkxhk). Ratios of marker segregation were calculated using Chi-square test. Markers that satisfied the expected Mendelian segregation ratio were included for mapping. Filtered markers contained different segregation types, including < nn x np >, < lm x ll > and < hk x hk >. Markers showing significantly distorted segregation (p < 0.05) were excluded from map construction. Markers were grouped with a minimum logarithm of odds (LOD) score of 6.0 and a recombination frequency of 0.45. A regression mapping algorithm was used to build the linkage map. Map distances were calculated in centiMorgans (cM) according to the Kosambi mapping function (Kosambi, 2016). For all other calculations, default settings were used.
QTL Mapping
QTL analysis was performed using R/qtl (Broman et al., 2003). Initially, an interval mapping (IM) was performed to identify potential QTLs. Later, a multiple QTL model (MQM) using the stepwiseqtl function was performed for forward and backward selections with a check for interactions to identify multiple QTLs. Based on IM results, the maximum number of QTLs to search for each trait was set to be three. Significance thresholds were determined using 1,000 permutations (p < 0.05). Results from QTL analysis were used to construct a QTL map, and their positions were used in a default model.
Identification of Potential Candidate Genes in the QTL Region
To identify potential candidate genes associated with growth was investigated in the QTL region. Genes encoded in the four scaffolds regions located in LG10 were BLAST searched against the reference genome sequence of Pacific abalone (Nam et al., 2017) and sequences of the NCBI non-redundant Molluscan database.
Quantitative Real Time PCR (qRT-PCR) Analysis of Candidate Genes
The mRNA expression of candidate genes in fast growth (49.38 ± 3.36 g) and slow growth (14.02 ± 3.27 g) abalone groups were investigated by qRT-PCR. The experimental abalones were two years old and obtained from an independent family having ten individuals in each group. Total weight (TW) was measured to the nearest mg and, shell length (SL) and shell width (SW) were measured to the nearest mm (Supplementary Table 4). From each abalone, tissue samples such as cephalic tentacle (CT), epipodial tentacle (ET), muscle (MUS), mantle (MNT), heart (HRT), gill (GIL), hepatopancreas/digestive gland (DG), ovary (Ov), and testis (Te) were collected, snap frozen in liquid nitrogen, and stored at –80°C until total RNA extraction. A separate qRT-PCR analysis of candidate genes was also conducted for parent abalones and presented in the Supplementary Figure 1.
Total RNA Extraction and cDNA Synthesis
Total RNAs were extracted from all sampled tissues using an ISOSPIN Cell & Tissue RNA kit (Nippon Gene, Tokyo, Japan). First-strand cDNAs were synthesized from total RNAs using and oligo (dT) primer (OdT) (Sigma) and a Superscript® III First-Strand cDNA synthesis kit (Invitrogen, United States). All steps of RNA extraction and cDNA synthesis were conducted following the manufacturer’s instructions.
qRT-PCR Analysis
Quantitative real-time PCR (qRT-PCR) assay was carried out using 2x qPCRBIO SyGreen Mix Lo-Rox kit (PCR Biosystems, United Kingdom) on a LightCycler® 96 System (Roche, Germany) as described previously (Sukhan et al., 2021). Briefly, qRT-PCR was executed using 1 μL of cDNA template in a total reaction volume of 20 μL containing 1 μL (10 pmol) of each forward and reverse primer (Supplementary Table 5), 10 μL SyGreen Mix, and 7 μL ultra-pure water. Triplicate reactions were performed for target and reference genes for each tissue sample. PCR amplification conditions were: pre-incubation at 95°C for 3 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. At the end of each cycle, a fluorescence reading was recorded for quantification. Specific amplification of each cDNA sample was verified by melting curve analysis and gel electrophoresis of the qRT-PCR product. Relative gene expression was quantified based on cycle threshold using the 2–ΔΔCT method (Livak and Schmittgen, 2001). Housekeeping β-actin gene (GenBank accession no. AY380809) was used as an internal reference to normalize the gene expression. Relative mRNA expression levels were analyzed statistically and expressed as mean ± SEM (standard error of the mean). Changes in relative mRNA expression levels between slow and fast growth groups of abalone were analyzed by Student’s t-test. Statistical significance was set at p < 0.05. All data were analyzed, and graphs were generated using GraphPad Prism 5.1 software1.
Results
Values of Phenotypic Traits
A total of 90 Pacific abalone F1 progenies were used to generate the linkage map. Average values of total weight, shell length and shell width were 15.58 ± 6.80 g, 49.30 ± 9.50 mm and 33.36 ± 6.21 mm, respectively. These growth traits were strongly correlated with each other (r = 0.968–0.990; p < 0.01 for all traits). Shell length and shell width showed the highest correlation (r = 0.990) (Supplementary Table 6).
GBS Sequencing
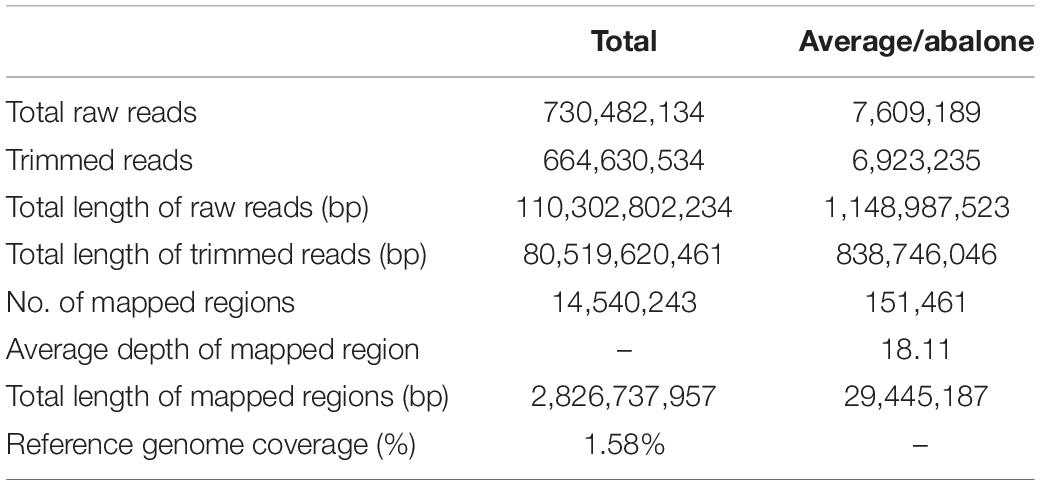
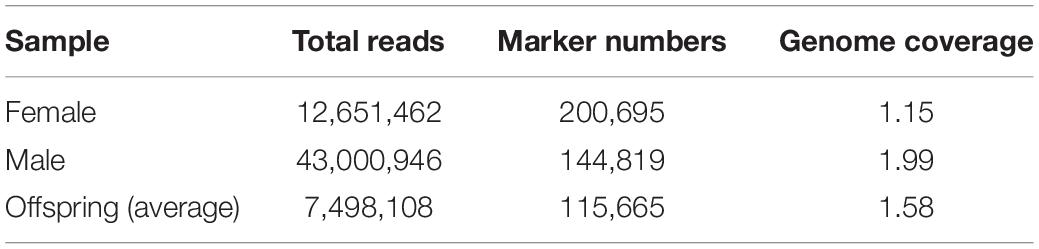
A total of 752,414,780 reads were obtained from 90 progenies (among 96 sample, 6 progenies were discarded because of poor DNA quality) and parents with an average length of 151 bp. The total length was 113,614,631,780 bp with a GC content of 45.25% and an average Q30 ratio of 91.69% (Table 1). A total of 664,630,534 (91.69%) reads passed the quality filter with a total length of 80,519,620,461 bp (Table 2). There were 12,651,462 reads from the female parent and 43,000946 reads from the male parent. Offspring per capita had an average of 7,498,108 reads (Table 3).
SNP Discovery and Genotyping
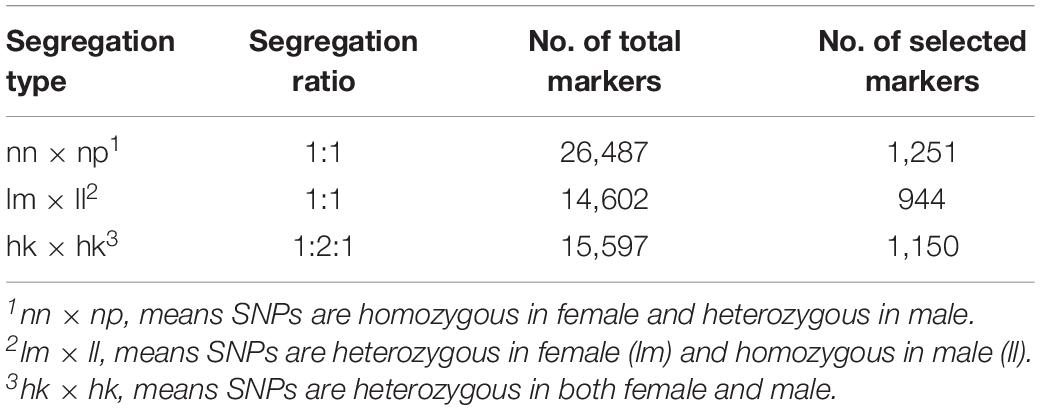
A total number of 815,970 putative SNPs were detected. Among these SNPs, 144,819 and 200,695 putative SNPs were found in the female parent and the male parent, respectively (Table 3). There were 73,025 and 104,357 homozygous SNPs, and 32,525 and 49,280 heterozygous SNPs in the female and male parents, respectively. In the offspring, there were 58,985 homozygous SNPs and 24,120 heterozygous SNPs (Table 4). Numbers of homozygous and heterozygous SNPs in all samples are presented in Figure 1, with parents having more SNPs than the offspring. All sequences containing SNPs are presented in Supplementary Table 7. After initial genotyping, the CP model in JoinMap 4.0 was used to select segregation types nn x np (26,487 SNP markers), lm x ll (14,602 SNP markers) and hk x hk (15,597 SNP markers), resulting in a total of 56,686 SNPs that were polymorphic in at least one parent and one progeny (Table 5).
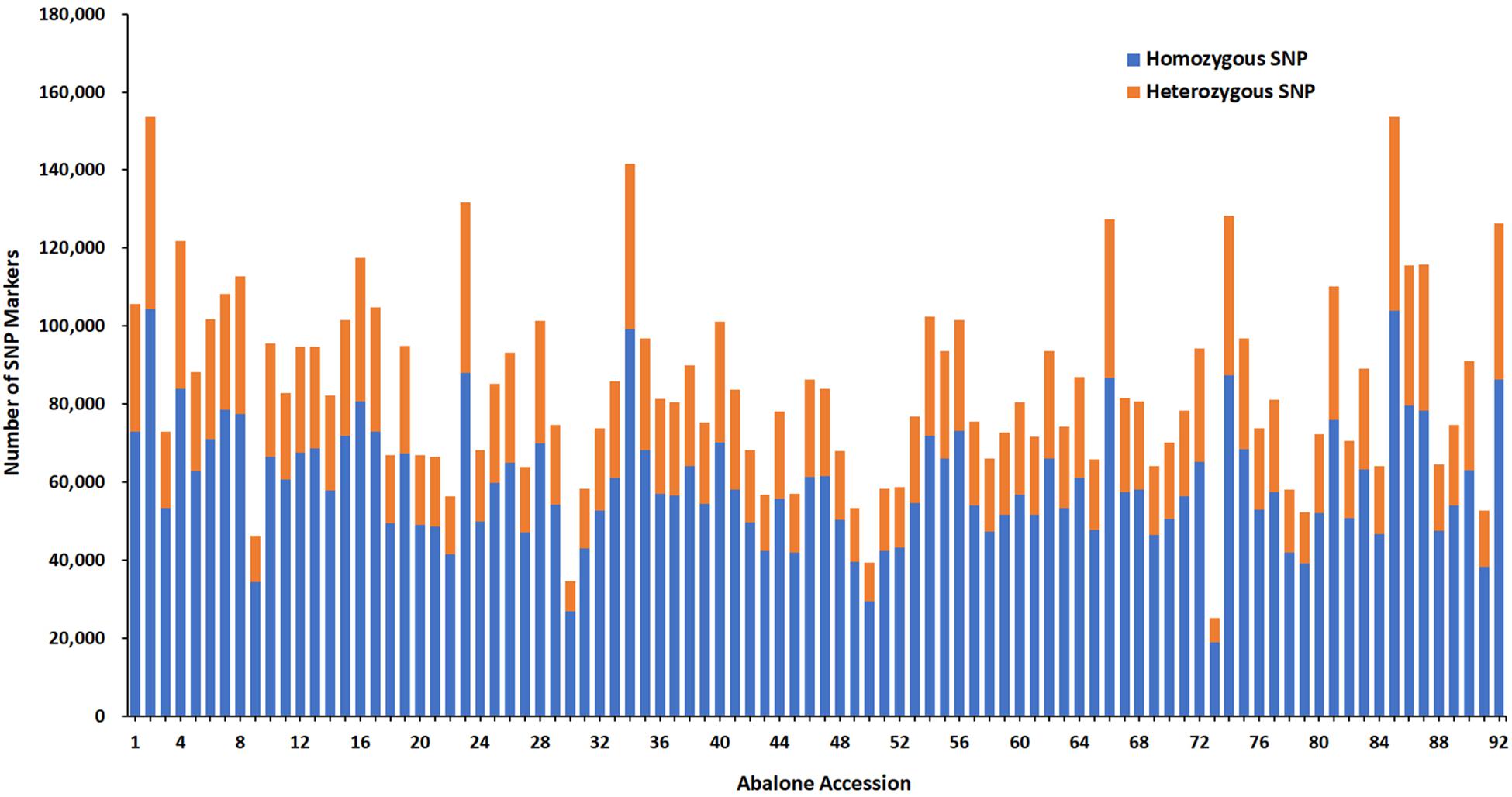
Figure 1. Number of SNP markers for all samples. The x-axis indicates abalone accession including parents and 90 progenies and the y-axis indicates the number of SNP markers.
Characteristics of Genetic Mapping
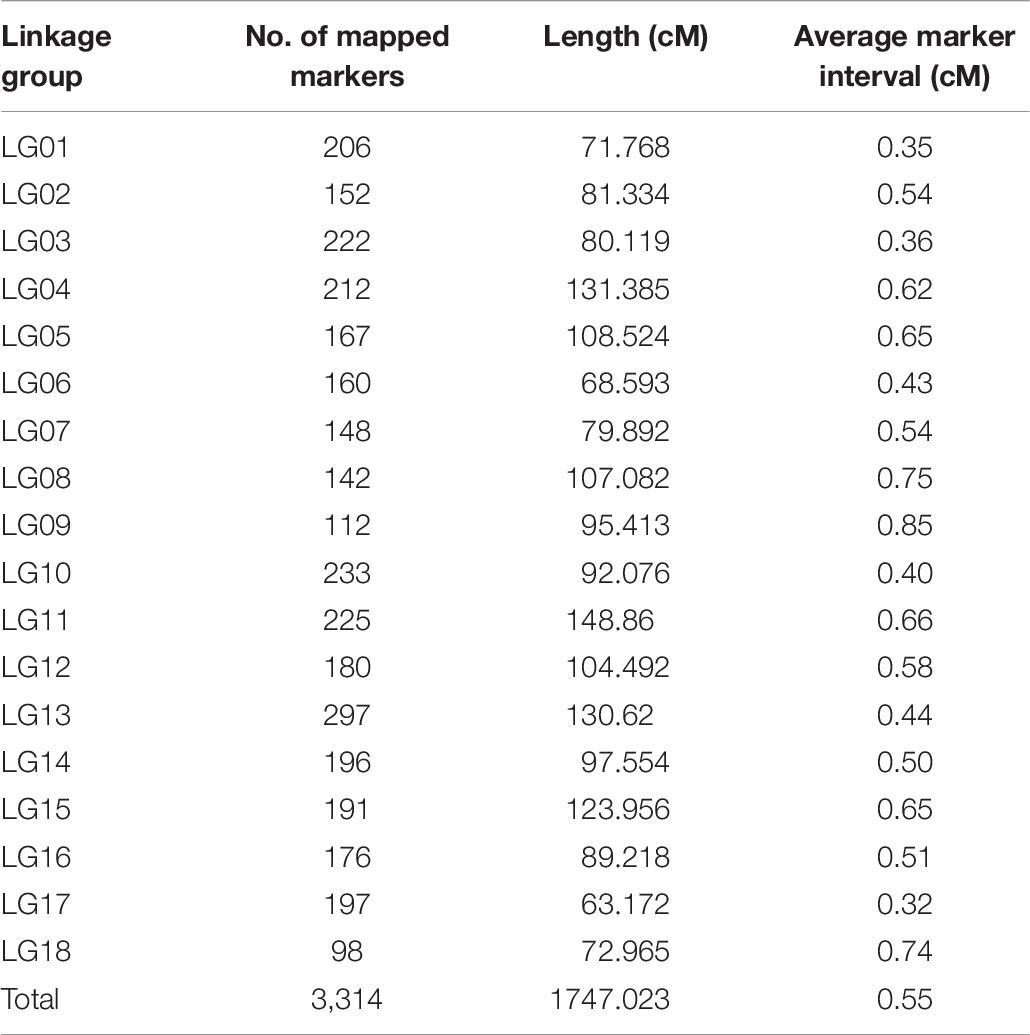
A total of 3,345 markers (1,251 female-specific, 944 male-specific and 1,150 anchor) and phenotypic data of F1 population developed from two geographically distant parents were used to construct a linkage map. A total of 18 linkage groups (LGs) were generated from 3,314 markers (Figure 2). The consensus linkage map had a map distance of 1747.023 cM. The average distance between adjacent markers was 0.55 cM, with the lowest distance of 0.32 cM and the highest distance of 0.85 cM (Table 6). The number of markers per LG varied from 98 to 297, with a mean of 184.11 markers per LG.
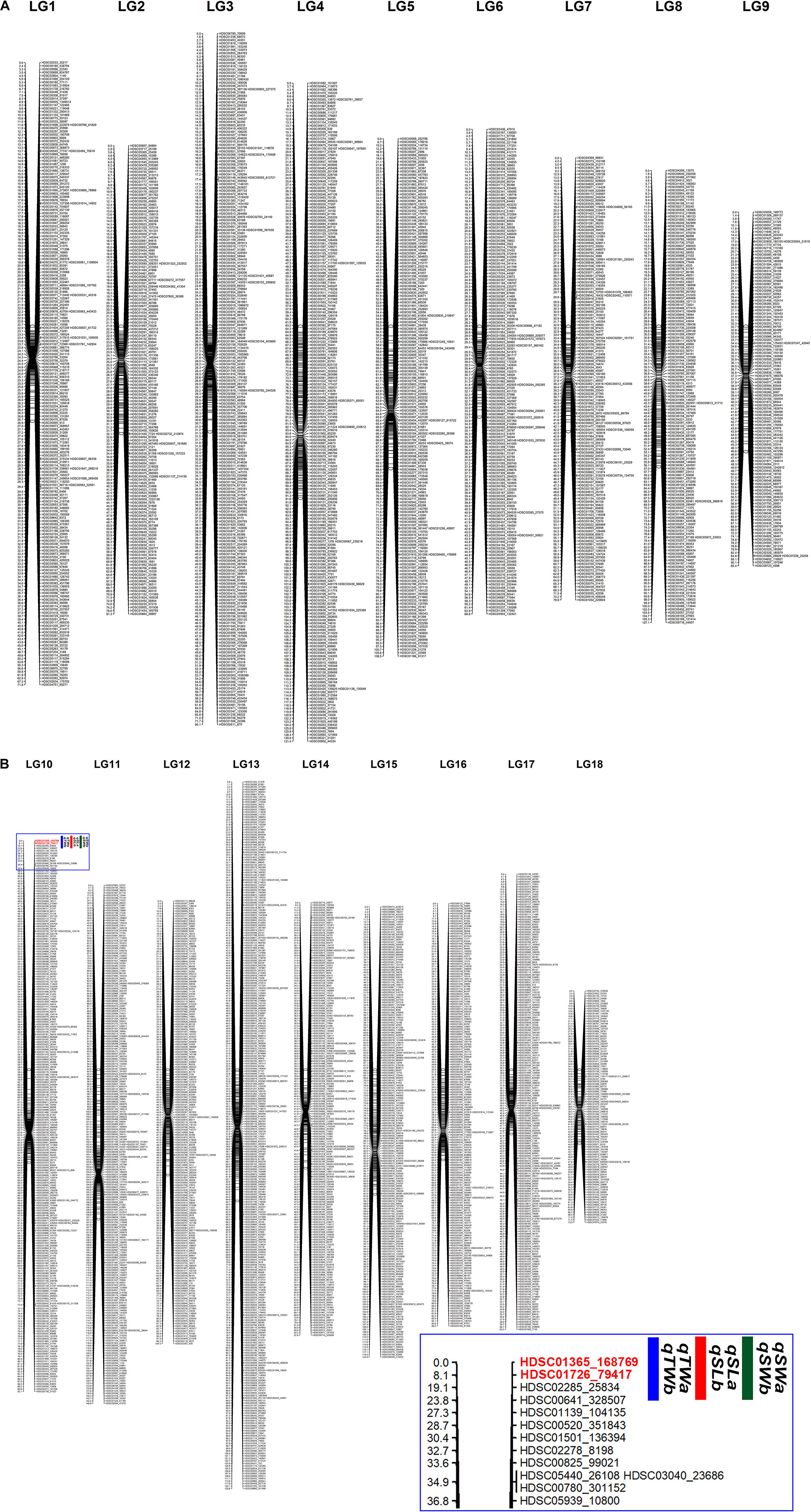
Figure 2. A consensus linkage map of Pacific abalone containing 3,314 effective SNP markers in 18 linkage groups. (A) Linkage group 1 – 9. (B) Linkage group 10 – 18. Inset image shows the zoomed portion of the marked area in the blue box where QTLs were detected.
QTL Analysis
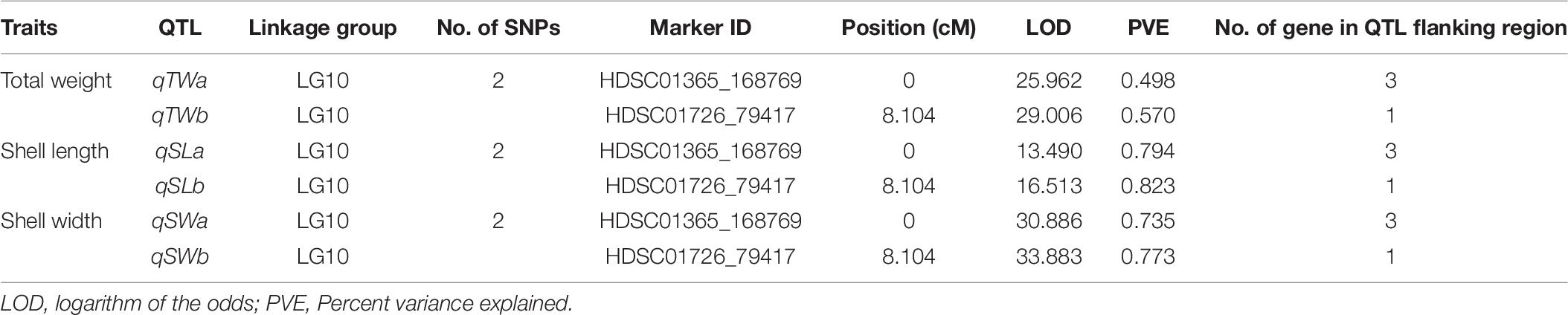
The integrated linkage map with 3,314 effective SNPs in 18 LGs of Pacific abalone was used to detect QTLs for three growth-related traits. Results are presented in Table 7. Two suggestive overlapped QTLs for three growth-related traits were detected in one linkage group (LG 10, Figure 2) at the position of 0 and 8.104 cM in the telomere region of the chromosome. Two SNPs were located on each growth-related trait, with LOD values ranging from 13.490 to 16.513 for shell length, from 30.886 to 33.883 for shell width, and from 25.962 to 29.006 for total weight (Table 7 and Figure 3).
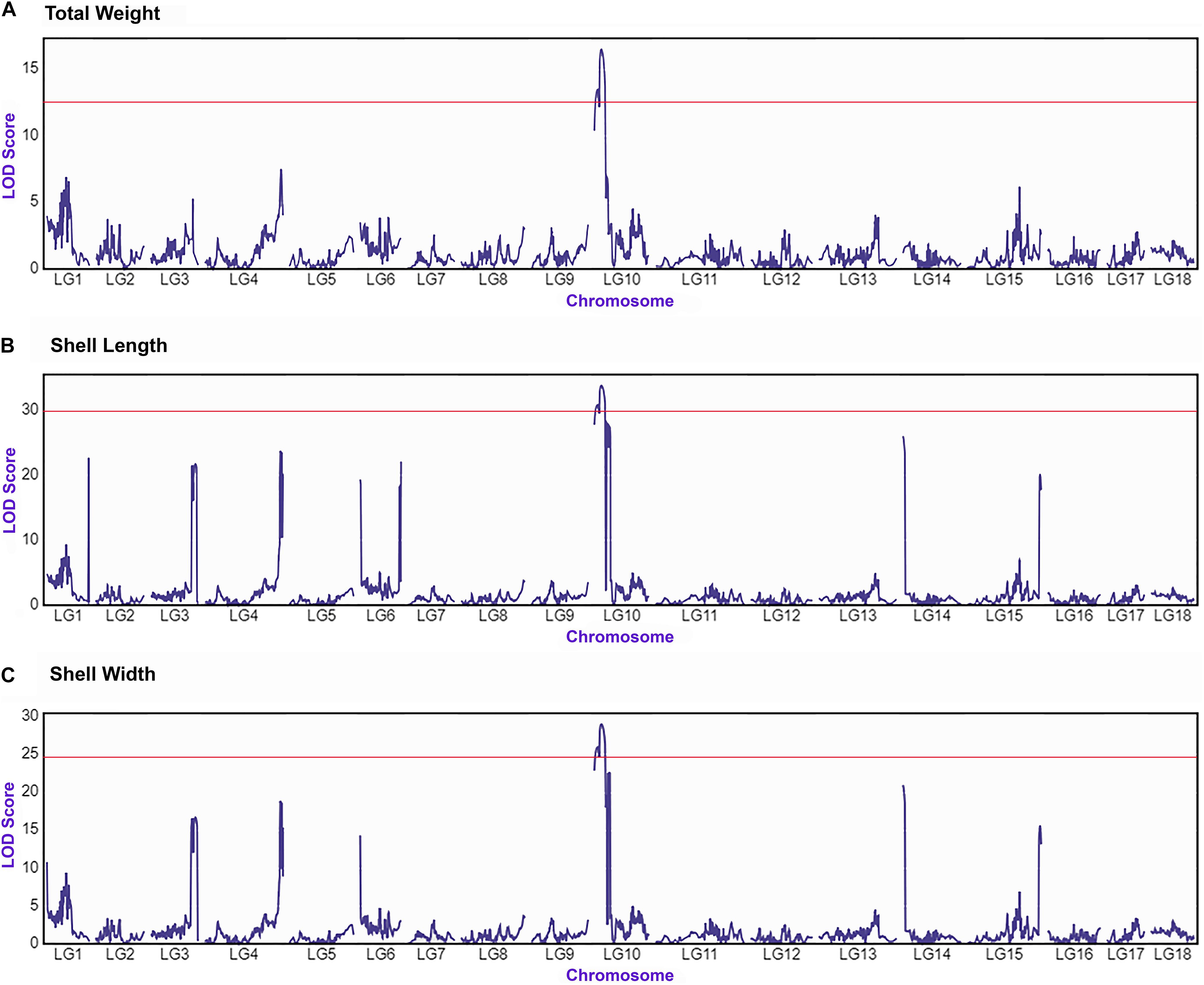
Figure 3. QTL mapping and LOD profile of growth-related traits total weight (A), shell length (B) and shell width (C) of Pacific abalone among 18 linkage groups. The red solid line indicates chromosome-wide significance thresholds.
Identification of Potential Candidate Gene in the QTL Region and Expression Analysis
Four candidate genes were identified encoded on four scaffolds (HDSC01365CG00050, HDSC01365CG00060, HDSC01365CG00070 and HDSC01726CG00010) in the QTL region of LG10. Associated GO categories of these genes included activating signal cointegrator 1 complex subunit (ascc1), ecdysoneless homolog (Ecd), galectin-4 (Gal-4) and calmodulin-B (CaM-B). Details of these identified candidate genes are presented in Table 8.
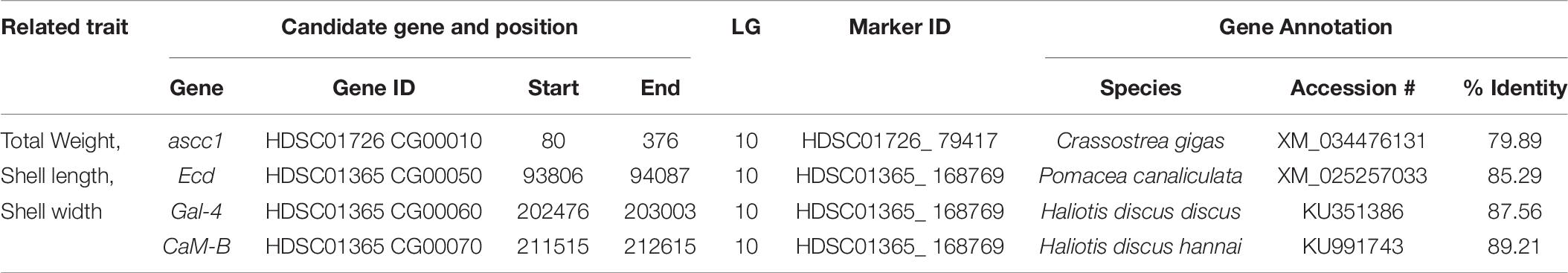
Table 8. Summary of candidate genes identified by QTL mapping of growth-related traits in Pacific abalone.
The mRNA Expression analysis of identified candidate genes in slow and fast growth groups of abalones showed that mRNA expression levels of these genes were significantly upregulated or downregulated in experimental groups of abalones (Figure 4). Expression levels of ascc1 were found to be significantly higher in CT, ET, and MNT of abalones with fast growth than in abalone with slow growth (Figure 4A), whereas expression of Ecd was downregulated in abalone with fast growth, i.e., its expression levels were significantly lower in CT, ET, and MNT of abalone with fast growth than in abalones with slow growth (Figure 4B). mRNA expression levels of Gal-4 gene were significantly higher in ET, MNT, and HRT of abalones with fast growth than in abalones with slow growth (Figure 4C). Expression levels of CaM-B in ET and MNT of abalones with fast growth were also significantly higher than in abalones with slow growth (Figure 4D).
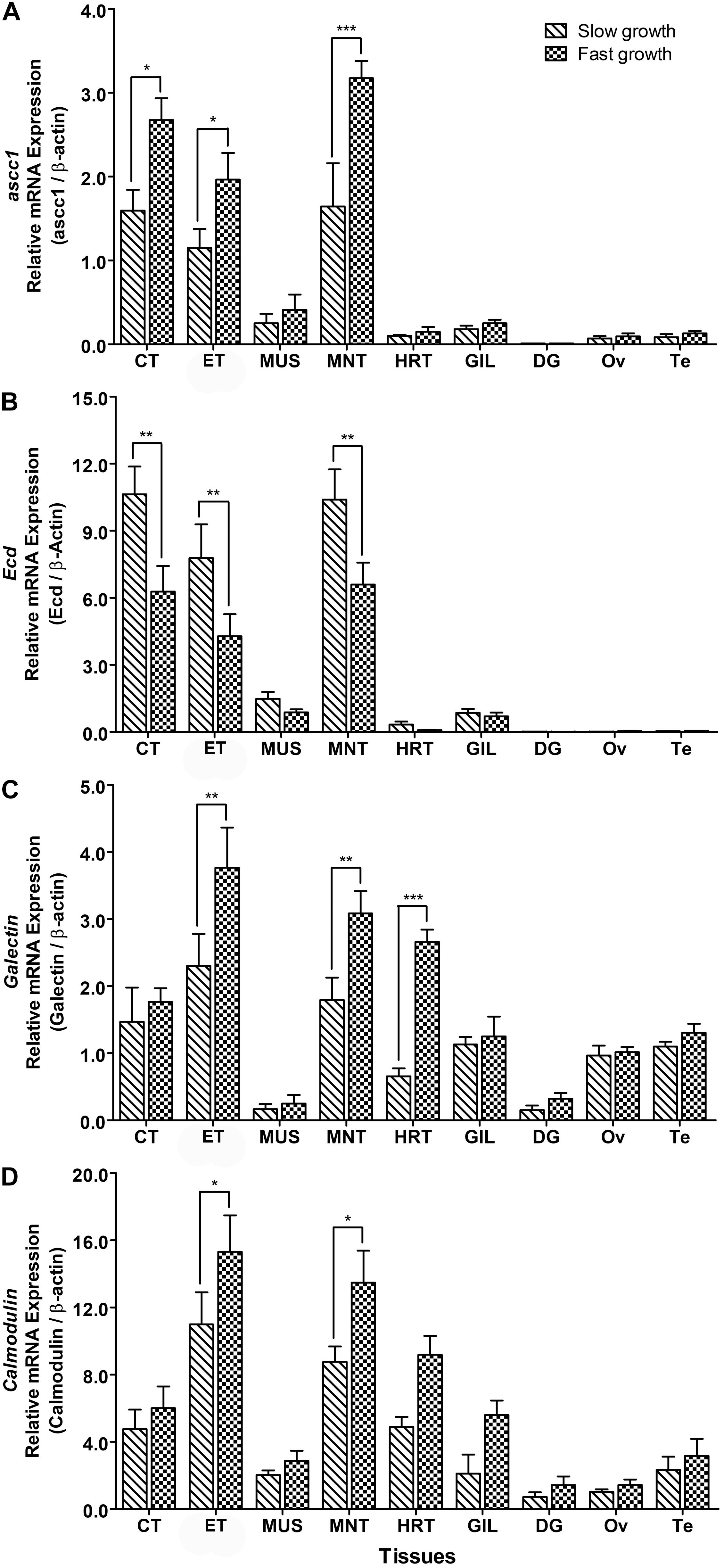
Figure 4. Relative mRNA expression levels of (A) ascc1, (B) Ecd, (C) Gal-4 and (D) CaM-B in different tissues of abalone with slow for fast growth. Asterisks indicated significant differences between slow and fast growth groups (p < 0.05). CT, cephalic tentacle; ET, epipodial tentacle; MUS, muscle; MNT, mantle; HRT, heart; GIL, gill; DG, digestive gland; Ov, ovary; Te, testis.
Discussion
The present study reported a SNP marker based genetic linkage map that was constructed for Pacific abalone using GBS approach. GBS is an efficient and cost-effective method that could be used to develop SNP markers, construct genetic linkage map and select marker-based complex traits of many species with or without a reference genome (Elshire et al., 2011; Nie et al., 2017; Liu et al., 2020). This is the first report of QTL mapping using GBS to screen high quality and abundant SNP markers for constructing a genetic linkage map for Pacific abalone. Previously, several genetic linkage maps have been constructed for Pacific abalone using SSR markers (Sekino and Hara, 2007), AFLP markers (Qi et al., 2007), and a combination of AFLP, RAPD, and SSR markers (Liu et al., 2006). A growth-related QTL has also been reported for Pacific abalone using a combination of RAPD, AFLP and SSR markers (Liu et al., 2007). SNP markers are most abundant in an organism. They more appropriate than AFLP, RAPD, or SSR markers for genetic studies, especially for constructing a genetic linkage map, identification of QTLs and population genetic analysis (Vignal et al., 2002; Yu et al., 2011; Ren et al., 2016; Yáñez et al., 2016). To date, the development of SNP markers using GBS or RAD sequencing of an organism is simple and quick to construct a genetic linkage map and perform QTL analysis for molluscan aquaculture species with large genomes, such as pearl oyster (Li and He, 2014; Shi et al., 2014) small abalone (Ren et al., 2016), Pacific oyster (Li et al., 2018; Shi et al., 2020), scallop (Jiao et al., 2014) and Manila clam (Nie et al., 2017). In this study, a genetic linkage map was constructed using 3,345 SNP markers that were filtered from 56,686 polymorphic SNP markers using GBS analysis. The consensus linkage map had 18 linkage groups, consistent with the number of haploid chromosomes of Pacific abalone (Arai et al., 1982). The total length of the consensus map was 1747.023 cM with an average inter-location interval of 0.55 cM. This value of inter-location interval was higher than that of Pacific oyster (0.31 cM) (Shi et al., 2020), scallop (0.41 cM) (Jiao et al., 2014), or pearl oyster, Pinctada fucata martensii (0.39 cM) (Shi et al., 2014), but lower than that of small abalone (0.59 cM) (Ren et al., 2016) or pearl oyster, Pinctada fucata (1.14 cM) (Li and He, 2014).
In this study, the target quantitative trait was growth. It was measured as total weight, shell length and shell width (Table 2). As growth is directly related to the production performance of aquaculture species, it is one of primary targets for marker assisted selection in genetic breeding programs. Two outbred geographically distant parents were used to produce F1 population in this experiment. Due to a longer reproductive cycle, it is difficult to produce F2 progenies for molluscan species. Therefore, most genetical mapping and QTL studies have used F1 cross population for molluscan species (Baranski et al., 2006; Liu et al., 2006; Sekino and Hara, 2007; Shi et al., 2010, 2014; Zhan et al., 2012; Vervalle et al., 2013; Jiao et al., 2014; Ren et al., 2016; Li et al., 2018). Two suggestive growth-related QTLs in each quantitative growth trait were detected in the linkage group LG10 of Pacific abalone. Fewer numbers of growth-related QTLs have also been reported for aquaculture species, such as one QTL for Pacific bluefin tuna (Uchino et al., 2018), two QTLs for scallop (Jiao et al., 2014) and three QTLs for kelp grouper (Kessuwan et al., 2016). The two QTLs detected in the present study overlapped for three growth-related traits, suggesting that these growth traits might be controlled by a common region. Overlapping QTL region has also been reported for small abalone (Ren et al., 2016) and Asian seabass (Xia et al., 2014). In the present study, both detected QTLs were located in the telomere of chromosome 10. Since recombination rates are considered to be higher in telomeric region than in centrometric regions of chromosomes (Sakamoto et al., 2000; Anderson et al., 2012), a higher residual heterozygosity level is expected for telomeric regions (Fraslin et al., 2018). Therefore, the present result of the location of QTL in telomere region of chromosome suggests more advanced segregating population and it should be considered as a more robust result.
Beside for detecting genetic markers associated with targeted traits, QTL mapping is also useful for identifying candidate genes that regulate targeted traits (Mackay et al., 2009; Liu et al., 2020). Using BLAST search against the reference genome and sequences in the NCBI non-redundant Molluscan database, four potential candidate genes were identified in scaffolds of the QTL region (Table 8). These genes were mainly involved in signal transduction, regulation of transcription factor and growth. Although whether these genes are directly or indirectly responsible for growth of abalone remains unknown, there are some evidences showing that these genes are involved in the growth of several animal species. For example, Ecd is an evolutionarily conserved protein that plays a crucial role in embryonic development and cell growth (Garen et al., 1997; Kainou et al., 2006). Galectin gene plays an important role in diverse fundamental cellular processes such as growth regulation, lipid raft stabilization, protein apical trafficking, adhesion/migration and mediator release, cell adhesion, and wound healing (López-Lucendo et al., 2004; Cao and Guo, 2016). It has also been reported that galectins contribute to larval development of Pacific abalone (Sandamalika and Lee, 2020), muscle development in Drosophila (Pace et al., 2002) and chick muscle (Cooper and Barondes, 1990). The Gal-3 gene in Siniperca chuatsi was significantly upregulated in groups of fish with higher growth performance, suggesting that protein glycosylation might contribute to the growth performance of S. chuatsi (He et al., 2020). In general, CaM genes and their associated kinases are involved in increased growth and anabolic functions. Thus, their expression levels are expected to be higher in faster growing organisms (Kocmarek et al., 2014). It is evident that CaM genes are involved in regulatory mechanisms related to growth, calcium metabolism, biomineralization and shell formation of Mollusca (Li et al., 2004; Huang et al., 2007; Lim et al., 2018). Genes encoding insulin-like growth factor binding proteins, molluscan insulin related peptide, epidermal growth factor and myostatin are considered to be directly associated with growths of abalone and molluscan species. However, these genes were not detected in the QTL region of Pacific abalone. Results of the present study indicate that mapping of new SNP markers and assemblage of gene information in the QTL region can facilitate the identification of specific regions in the genome that contain growth-related genes. Furthermore, the differential expression patterns of ascc1, Ecd, Gal-4, and Cal-B between slow and fast growth abalones suggest that these genes might be associated with growth regulation of abalone.
In conclusion, QTL mapping using GBS in Pacific abalone revealed two new SNP markers on one linkage group that were significantly associated with growth traits. In addition, four candidate genes were identified on the same LG of these QTLs; which were found to be upregulated or downregulated in abalones with fast growth or slow growth. These genomic resources can be used in selective breeding program of Pacific abalone. These data can also be used as basic data for further genomic investigation of Pacific abalone.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://doi.org/10.5061/dryad.msbcc2fz3.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Chonnam National University (CNU IACUC).
Author Contributions
KK: conceptualization, resources, writing – original draft preparation, supervision, project administration, and funding acquisition. KK, ZS, SH, YC, SK, and MS: methodology. ZS, SH, and YC: data curation. ZS and SH: formal analysis. ZS: software and visualization. KK, ZS, H-JJ, and I-SN: validation. KK, ZS, and I-SN: writing – review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was a component of a project entitled “Development of technology for abalone aquaculture using sperm cryopreservation (Grant No. 2018- 2129)” funded by the Ministry of Oceans and Fisheries, South Korea.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors gratefully acknowledge the support of Dongik Choi (President, Tou-Jeong Soosan abalone hatchery, Dolsan-eup, Yeosu-si, Jeollanam-do, South Korea) for conducting the field experiment in his hatchery facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.713783/full#supplementary-material
Supplementary Figure 1 | Relative mRNA expression levels of (A) ascc1, (B) Ecd, (C) Gal-4 and (D) CaM-B in different tissues of female and male parents of abalone used for generating of F1 progeny of QTL. Asterisks indicated significant differences between slow and fast growth group (p < 0.05). CT, cephalic tentacle; ET, epipodial tentacle; MUS, muscle; MNT, mantle; HRT, heart; GIL, gill; DG, digestive gland; GND, gonad.
Supplementary Table 1 | Water quality parameter of larval rearing tank of F1 progeny.
Supplementary Table 2 | Trait data of 96 F1 progeny.
Supplementary Table 3 | Summary of Reference genome sequence of Pacific abalone.
Supplementary Table 4 | Trait data of slow growth and fast growth abalones used in qRT-PCR expression analysis.
Supplementary Table 5 | List of primers used for quantitative real-time polymerase chain reaction (qRT-PCR) for expression analysis of QTL candidate genes in Pacific abalone with slow and fast growth.
Supplementary Table 6 | Pearson correlation coefficients (r) between growth-related traits measured in F1 progeny of Pacific abalone (p < 0.01).
Supplementary Table 7 | Details of SNPs that used to construct the linkage map.
Footnotes
References
Anderson, J. L., Rodríguez Marí, A., Braasch, I., Amores, A., Hohenlohe, P., Batzel, P., et al. (2012). Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics. PLoS One 7:e40701. doi: 10.1371/journal.pone.0040701
Arai, K., Tsubaki, H., Ishitani, Y., and Fujino, K. (1982). Chromosomes of Haliotis discus hannai INO and H. discus REEVE. Nippon Suisan Gakk. 48, 1689–1691. doi: 10.2331/suisan.48.1689
Arseneau, J. R., Steeves, R., and Laflamme, M. (2017). Modified low-salt CTAB extraction of high-quality DNA from contaminant-rich tissues. Mol. Ecol. Resour. 17, 686–693. doi: 10.1111/1755-0998.12616
Ashton, D. T., Ritchie, P. A., and Wellenreuther, M. (2019). High-density linkage map and QTLs for growth in snapper (Chrysophrys auratus). G3 (Bethesda) 9, 1027–1035. doi: 10.1534/g3.118.200905
Baranski, M., Loughnan, S., Austin, C. M., and Robinson, N. (2006). A microsatellite linkage map of the blacklip abalone, Haliotis rubra. Anim Genet. 37, 563–570. doi: 10.1111/j.1365-2052.2006.01531.x
Baranski, M., Rourke, M., Loughnan, S., Hayes, B., Austin, C., and Robinson, N. (2008). Detection of QTL for growth rate in the blacklip abalone (Haliotis rubra Leach) using selective DNA pooling. Anim Genet. 39, 606–614. doi: 10.1111/j.1365-2052.2008.01773.x
Broman, K. W., Wu, H., Sen, S., and Churchill, G. A. (2003). R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890. doi: 10.1093/bioinformatics/btg112
Cao, Z. Q., and Guo, X. L. (2016). The role of galectin-4 in physiology and diseases. Protein Cell 7, 314–324. doi: 10.1007/s13238-016-0262-9
Choi, K. S., Kim, B. H., and Lim, H. K. (2015). Current status of abalone aquaculture in Korea. In: Korean Society of Fisheries and Aquatic Science (KOSFAS) International Conference 2015 proceedings. Available Online at: http://www.dbpia.co.kr/journal/articleDetail?nodeId=NODE06399684 [Accessed April 20, 2021]
Cooper, D. N., and Barondes, S. H. (1990). Evidence for export of a muscle lectin from cytosol to extracellular matrix and for a novel secretory mechanism. J. Cell Bio. 110, 1681–1691. doi: 10.1083/jcb.110.5.1681
Cox, M. P., Peterson, D. A., and Biggs, P. J. (2010). SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. Bioinformatics 11:485. doi: 10.1186/1471-2105-11-485
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple Genotyp-ing-by-Sequencing (GBS) approach for high diversity species. PLoS One 6:e19379. doi: 10.1371/journal.pone.0019379
Feng, X., Yu, X., Fu, B., Wang, X., Liu, H., Pang, M., et al. (2018). A high-resolution genetic linkage map and QTL fine mapping for growth-related traits and sex in the Yangtze River common carp (Cyprinus carpio haematopterus). BMC Genom. 19:230. doi: 10.1186/s12864-018-4613-1
Fraslin, C., Dechamp, N., Bernard, M., Krieg, F., Hervet, C., Guyomard, R., et al. (2018). Quantitative trait loci for resistance to Flavobacterium psychrophilum in rainbow trout: effect of the mode of infection and evidence of epistatic interactions. Genet. Sel. Evol. 50:60. doi: 10.1186/s12711-018-0431-9
Garen, A., Kauvar, L., and Lepesant, J. A. (1997). Roles of ecdysone in Drosophila development. Proc. Natl. Acad. Sci. 74, 5099–5103. doi: 10.1073/pnas.74.11.5099
Gjedrem, T., and Baranski, M. (2009). “The Success of Selective Breeding in Aquaculture”, in Selective Breeding in Aquaculture: An Introduction. Reviews: Methods and Technologies in Fish Biology and Fisheries. Dordrecht, Netherlands: Springer, 221.
Gordon, H. R., and Cook, P. A. (2013). World abalone supply, markets and pricing: 2011 update. J. Shellfish Res. 32, 5–7. doi: 10.2983/35.032.0102
Grattapaglia, D., and Sederoff, R. (1994). Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137, 1121–1137.
He, S., Li, L., Lv, L. Y., Cai, W. J., Dou, Y. Q., Li, J., et al. (2020). Mandarin fish (Sinipercidae) genomes provide insights into innate predatory feeding. Commun. Biol. 3:361. doi: 10.1038/s42003-020-1094-y
Hossen, S., Sukhan, Z. P., Cho, Y., and Kho, K. H. (2021). Effects of cryopreservation on gene expression and post thaw sperm quality of Pacific abalone, Haliotis discus hannai. Front. Mar. Sci. 8:652390. doi: 10.3389/fmars.2021.652390
Huang, J., Zhang, C., Ma, Z., Xie, L., and Zhang, R. (2007). A novel extracellular EF-hand protein involved in the shell formation of pearl oyster. Biochim. Biophys. Acta. 1770, 1037–1044. doi: 10.1016/j.bbagen.2007.03.006
Jiao, W., Fu, X., Dou, J., Li, H., Su, H., Mao, J., et al. (2014). High-resolution linkage and quantitative trait locus mapping aided by genome survey sequencing: building up an integrative genomic framework for a bivalve mollusc. DNA Res. 21, 85–101. doi: 10.1093/dnares/dst043
Kainou, T., Shinzato, T., Sasaki, K., Mitsui, Y., Giga-Hama, Y., Kumagai, H., et al. (2006). Spsgt1, a new essential gene of Schizosaccharomyces pombe, is involved in carbohydrate metabolism. Yeast 23, 35–53. doi: 10.1002/yea.1336
Kessuwan, K., Kubota, S., Liu, Q., Sano, M., Okamoto, N., Sakamoto, T., et al. (2016). Detection of growth-related quantitative trait loci and high-resolution genetic linkage maps using simple sequence repeat markers in the kelp grouper (Epinephelus bruneus). Mar. Biotechnol. 18, 57–84. doi: 10.1007/s10126-015-9673-5
Kim, J. E., Oh, S. K., Lee, J. H., Lee, B. M., and Jo, S. H. (2014). Genome-wide SNP calling using next generation sequencing data in tomato. Mol. Cells 37, 36–42. doi: 10.14348/molcells.2014.2241
Kocmarek, A. L., Ferguson, M. M., and Danzmann, R. G. (2014). Differential gene expression in small and large rainbow trout derived from two seasonal spawning groups. BMC Genom. 15:57. doi: 10.1186/1471-2164-15-57
Kosambi, D. D. (2016). “The Estimation of Map Distances from Recombination Values,” in Ramaswamy R, ed. D. D. Kosambi (New Delhi, India: Springer), 125–130.
KOSTAT (Korea Statistics) (2020). Statistic database for fisheries production. Available online at: http://www.kostat.go.kr [Accessed April 20, 2021]
Leighton, P. (2008). “Abalone hatchery manual,” in Aquaculture Explained, Aquaculture Technical Section, eds G. Robinson and N. McGowan (Dun Laoghaire, Ireland: Aquaculture Development Division, Bord Iascaigh Mhara), 77.
Li, C., Wang, J., Song, K., Meng, J., Xu, F., Li, L., et al. (2018). Construction of a high-density genetic map and fine QTL mapping for growth and nutritional traits of Crassostrea gigas. BMC Genom. 19:626. doi: 10.1186/s12864-018-4996-z
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, S., Xie, L., Zhang, C., Zhang, Y., Gu, M., and Zhang, R. (2004). Cloning and expression of a pivotal calcium metabolism regulator: calmodulin involved in shell formation from pearl oyster (Pinctada fucata). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 138, 235–243. doi: 10.1016/j.cbpc.2004.03.012
Li, Y., and He, M. (2014). Genetic mapping and QTL analysis of growth-related traits in Pinctada fucata using restriction-site associated DNA sequencing. PLoS One 9:e111707. doi: 10.1371/journal.pone.0111707
Lim, H. K., Lee, J. K., Kim, G. D., and Jeong, T. H. (2018). Multiple calmodulin genes of the Pacific abalone, Haliotis discus hannai (Mollusca: Vetigastropoda: Haliotidae). Anim. Cells Syst. 22, 341–351. doi: 10.1080/19768354.2018.1509126
Lindblad-Toh, K., Winchester, E., Daly, M. J., Wang, D. G., Hirschhorn, J. N., Laviolette, J. P., et al. (2000). Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat. Genet. 24, 381–386. doi: 10.1038/74215
Liu, F., Zhan, W., Xie, Q., Chen, H., Lou, B., and Xu, W. (2020). A first genetic linage map construction and QTL mapping for growth traits in Larimichthys polyactis. Sci. Rep. 10, 1–9. doi: 10.1038/s41598-020-68592-0
Liu, X., Liu, X., Guo, X., Gao, Q., Zhao, H., and Zhang, G. (2006). A preliminary genetic linkage map of the Pacific abalone Haliotis discus hannai Ino. Mar. Biotechnol. 8, 386–397. doi: 10.1007/s10126-005-6133-7
Liu, X., Liu, X., and Zhang, G. (2007). Identification of quantitative trait loci for growth-related traits in the Pacific abalone Haliotis discus hannai Ino. Aquac. Res. 38, 789–797. doi: 10.1111/j.1365-2109.2006.01647.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
López-Lucendo, M. F., Solís, D., André, S., Hirabayashi, J., Kasai, K., Kaltner, H., et al. (2004). Growth-regulatory human galectin-1: crystallographic characterisation of the structural changes induced by single-site mutations and their impact on the thermodynamics of ligand binding. J. Mol. Biol. 343, 957–970. doi: 10.1016/j.jmb.2004.08.078
Mackay, T. F., Stone, E. A., and Ayroles, J. F. (2009). The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10, 565–577. doi: 10.1038/nrg2612
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/ej.17.1.200
Mascher, M., Wu, S., Amand, P. S., Stein, N., and Poland, J. (2013). Application of genotyping-by-sequencing on semiconductor sequencing platforms: a comparison of genetic and reference-based marker ordering in barley. PLoS One 8:e76925. doi: 10.1371/journal.pone.0076925
Nam, B. H., Kwak, W., Kim, Y. O., Kim, D. G., Kong, H. J., Kim, W. J., et al. (2017). Genome sequence of Pacific abalone (Haliotis discus hannai): The first draft genome in family Haliotidae. GigaScience 6, 1–8. doi: 10.1093/gigascience/gix014
Nie, H., Yan, X., Huo, Z., Jiang, L., Chen, P., Liu, H., et al. (2017). Construction of a high-density genetic map and quantitative trait locus mapping in the manila clam Ruditapes philippinarum. Sci. Rep. 7, 1–9. doi: 10.1038/s41598-017-00246-0
Pace, K. E., Lebestky, T., Hummel, T., Arnoux, P., Kwan, K., and Baum, L. G. (2002). Characterization of a Novel Drosophila melanogaster galectin: Expression in developing immune, neural, and muscle tissues. J. Biol. Chem. 277, 13091–13098. doi: 10.1074/jbc.M112105200
Park, C. J., and Kim, S. Y. (2013). Abalone aquaculture in Korea. J. Shellfish Res. 32, 17–19. doi: 10.2983/035.032.0104
Qi, L., Yanhong, X., Ruihai, Y., and Akihiro, K. (2007). An AFLP genetic linkage map of pacific abalone (Haliotis discus hannai). J. Ocean Univ. China 6, 259–267. doi: 10.1007/s11802-007-0259-3
Ren, P., Peng, W., You, W., Huang, Z., Guo, Q., Chen, N., et al. (2016). Genetic mapping and quantitative trait loci analysis of growth-related traits in the small abalone Haliotis diversicolor using restriction-site-associated DNA sequencing. Aquaculture 454, 163–170. doi: 10.1016/j.aquaculture.2015.12.026
Sakamoto, T., Danzmann, R. G., Gharbi, K., Howard, P., Ozaki, A., Khoo, S. K., et al. (2000). A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155, 1331–1345.
Salazar, J., Pacheco, I., Shinya, P., Ruiz, D., Martínez-Gómez, P., and Infante, R. (2019). Genotypic characterization of an F1 Japanese plum progeny through genotyping by sequencing (GBS) and preliminary quantitative trait locus (QTL) analysis for important agronomic traits. Acta Hortic. 1260, 49–58. doi: 10.17660/ActaHortic.2019.1260.10
Sandamalika, W. G., and Lee, J. (2020). Quadruple domain-containing galectin from marine invertebrate disk abalone (Haliotis discus discus): Molecular perspectives in early development, immune expression, and potent antiviral responses. Fish Shellfish Immunol. 106, 920–929. doi: 10.1016/j.fsi.2020.09.017
Sekino, M., and Hara, M. (2007). Linkage maps for the Pacific abalone (genus Haliotis) based on microsatellite DNA markers. Genetics 175, 945–958. doi: 10.1534/genetics.106.065839
Shi, R., Li, C., Qi, H., Liu, S., Wang, W., Li, L., et al. (2020). Construction of a high-resolution genetic map of Crassostrea gigas: QTL mapping and GWAS applications revealed candidate genes controlling nutritional traits. Aquaculture 527:735427. doi: 10.1016/j.aquaculture.2020.735427
Shi, Y., Guo, X., Zhifeng, G. U., Wang, A., and Wang, Y. (2010). Preliminary genetic linkage map of the abalone Haliotis diversicolor Reeve. Chin. J. Oceanol. Limnol. 28, 549–557. doi: 10.1007/s00343-010-9026-1
Shi, Y., Wang, S., Gu, Z., Lv, J., Zhan, X., Yu, C., et al. (2014). High-density single nucleotide polymorphisms linkage and quantitative trait locus mapping of the pearl oyster, Pinctada fucata martensii Dunker. Aquaculture 434, 376–384. doi: 10.1016/j.aquaculture.2014.08.044
Song, J., Li, Q., Yu, Y., Wan, S., Han, L., and Du, S. (2018). Mapping genetic loci for quantitative traits of golden shell color, mineral element contents, and growth-related traits in Pacific oyster (Crassostrea gigas). Mar. Biotechnol. 20, 666–675. doi: 10.1007/s10126-018-9837-1
Sukhan, Z. P., Sharker, M. R., Cho, Y., Hossen, S., Choi, K. S., and Kho, K. H. (2021). Thermal stress affects gonadal maturation by regulating GnRH, GnRH receptor, APGWamide, and serotonin receptor gene expression in male pacific abalone, Haliotis discus hannai during breeding season. Front. Mar. Sci. 8:664426. doi: 10.3389/fmars.2021.664426
Tian, M., Li, Y., Jing, J., Mu, C., Du, H., Dou, J., et al. (2015). Construction of a high-density genetic map and quantitative trait locus mapping in the sea cucumber Apostichopus japonicus. Sci. Rep. 5:14852. doi: 10.1038/srep14852
Uchino, T., Hosoda, E., Nakamura, Y., Yasuike, M., Mekuchi, M., Sekino, M., et al. (2018). Genotyping-by-sequencing for construction of a new genetic linkage map and QTL analysis of growth-related traits in Pacific bluefin tuna. Aquac. Res. 49, 1293–1301. doi: 10.1111/are.13584
Uki, N., and Kikuchi, S. (1984). Regulation of maturation and spawning of an abalone, Haliotis (Gastropoda) by external environmental factors. Aquaculture 29, 247–261. doi: 10.1016/0044-8486(84)90270-9
Van Ooijen, J. W. (2006). JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations. Wageningen: Kyazma BV.
Vervalle, J., Hepple, J. A., Jansen, S., Plessis, J. D., Wang, P., Rhode, C., et al. (2013). Integrated linkage map of Haliotis midae linnaeus based on microsatellite and SNP markers. J. Shellfish Res. 32, 89–103. doi: 10.2983/035.032.0115
Vignal, A., Milan, D., SanCristobal, M., and Eggen, A. (2002). A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. 34, 275–305. doi: 10.1186/1297-9686-34-3-275
Wang, X., Fu, B., Yu, X., Qu, C., Zhang, Q., and Tong, J. (2018). Fine mapping of growth-related quantitative trait loci in Yellow River carp (Cyprinus carpio haematoperus). Aquaculture 484, 277–285. doi: 10.1016/j.aquaculture.2017.11.016
Winnepenninckx, B. (1993). Extraction of high molecular weight DNA from molluscs. Trends Genet. 9:407. doi: 10.1016/0168-9525(93)90102-N
Xia, J. H., Lin, G., He, X., Yunping, B., Liu, P., Liu, F., et al. (2014). Mapping quantitative trait loci for omega-3 fatty acids in Asian seabass. Mar. Biotechnol. 16, 1–9. doi: 10.1007/s10126-013-9524-1
Xu, Y., Li, P., Zou, C., Lu, Y., Xie, C., Zhang, X., et al. (2017). Enhancing genetic gain in the era of molecular breeding. J. Exp. Bot. 68, 2641–2666. doi: 10.1093/jxb/erx135
Yáñez, J. M., Naswa, S., López, M. E., Bassini, L., Correa, K., Gilbey, J., et al. (2016). Genomewide single nucleotide polymorphism discovery in Atlantic salmon (Salmo salar): Validation in wild and farmed American and European populations. Mol. Ecol. Resour. 16, 1002–1011. doi: 10.1111/1755-0998.12503
Yu, H., Xie, W., Wang, J., Xing, Y., Xu, C., Li, X., et al. (2011). Gains in QTL detection using an ultra-high density SNP map based on population sequencing relative to traditional RFLP/SSR markers. PloS One 6:e17595. doi: 10.1371/journal.pone.0017595
Yue, G. H. (2014). Recent advances of genome mapping and marker-assisted selection in aquaculture. Fish Fish. 15, 376–396. doi: 10.1111/faf.12020
Keywords: QTL, SNP marker, GBS, candidate gene, growth-related trait, Pacific abalone, Haliotis discus hannai
Citation: Kho KH, Sukhan ZP, Hossen S, Cho Y, Kim SC, Sharker MR, Jung HJ and Nou IS (2021) Construction of a Genetic Linkage Map Based on SNP Markers, QTL Mapping and Detection of Candidate Genes of Growth-Related Traits in Pacific Abalone Using Genotyping-by-Sequencing. Front. Mar. Sci. 8:713783. doi: 10.3389/fmars.2021.713783
Received: 24 May 2021; Accepted: 06 July 2021;
Published: 27 August 2021.
Edited by:
Hao Chen, Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Jitao Li, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences (CAFS), ChinaXiaoli Hu, Ocean University of China, China
Copyright © 2021 Kho, Sukhan, Hossen, Cho, Kim, Sharker, Jung and Nou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kang Hee Kho, a2toQGNob25uYW0uYWMua3I=
 Kang Hee Kho
Kang Hee Kho Zahid Parvez Sukhan
Zahid Parvez Sukhan Shaharior Hossen
Shaharior Hossen Yusin Cho
Yusin Cho Soo Cheol Kim1
Soo Cheol Kim1 Md. Rajib Sharker
Md. Rajib Sharker Ill-Sup Nou
Ill-Sup Nou