
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 19 July 2021
Sec. Aquatic Physiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.705332
This article is part of the Research TopicNew Progresses of Fish Meal Replacement in Aquatic AnimalsView all 16 articles
 Bin Yin1,2,3
Bin Yin1,2,3 Hongyu Liu1,2,3*
Hongyu Liu1,2,3* Beiping Tan1,2,3*
Beiping Tan1,2,3* Xiaohui Dong1,2,3
Xiaohui Dong1,2,3 Shuyan Chi1,2,3
Shuyan Chi1,2,3 Qihui Yang1,2,3
Qihui Yang1,2,3 Shuang Zhang1,2,3
Shuang Zhang1,2,3Different doses of β-conglycinin produce different regulations on the intestinal health of aquatic animals, affecting the absorption of nutrients, indirectly changing water quality. Sodium butyrate (NaB) can effectively alleviate the negative effects caused by high-dose β-conglycinin. We investigated the positive response to low-dose (1.5%, bL) and negative response to high-dose (6.0%, bH) β-conglycinin and supplementation with NaB (6.0% β-conglycinin + 0.13% NaB, bHNaB) in terms of water pollutants, microbiota, transcriptome, and metabolome in hybrid grouper (Epinephelus fuscoguttatus♀ × E. lanceolatus♂). The ammonia nitrogen, nitrite, total nitrogen, and total phosphorus contents were significantly higher in the water from bH than from FMb, bL, and bHNaB. Supplementing with NaB significantly reduced the ammonia nitrogen, nitrite, total nitrogen, and total phosphorus contents. Low-dose β-conglycinin increased the relative abundance of Pelagibacterium, Pediococcus, Staphylococcus, and Lactobacillus and promoted the “ribosome,” “peroxisome proliferator-activated receptor (PPAR) signaling” and “histidine metabolism.” High-dose β-conglycinin increased the relative abundance of pathogenic bacteria Ralstonia and Photobacterium and inhibited the “cell cycle” “PPAR signaling” and “starch and proline metabolism.” NaB supplementation at high-dose β-conglycinin reduced the Ralstonia and Photobacterium abundance and promoted the “cell cycle,” “linoleic acid metabolism,” and “ABC transporters.” Overall, these results reveal differences in the effects of high- and low-dose β-conglycinin, as well as NaB supplementation, on the utilization of proteins, carbohydrates, and lipids and on substance transport and signaling among distal intestinal cells of hybrid grouper. A total of 15 differential metabolite biomarkers were identified: FMb vs. bL contained 10-methylimidazole acetic acid, N-acetyl histamine, urocanic acid, creatinine, glutathione, taurine, nervonic acid, stearic acid, docosanoic acid, and D-serine; FMb vs. bH contained 4-L-fucose, sucrose, α,α-trehalose, and quercetin; and bH vs. bHNaB contained 4-N-acetyl histamine, urocanic acid, creatinine, and S-adenosylhomocysteine, respectively. Our study provides new insights into the regulation of intestinal health by β-conglycinin in aquatic animals and the protective mechanism of NaB.
The hybrid grouper (Epinephelus fuscoguttatus♀ × E. lanceolatus♂) is an economically important fish along the southern coast of China, with a fast growth rate and strong environmental tolerance. In 2017, groupers were officially included in the construction of China’s National Marine Fish Industry Technology System. As a typical carnivorous fish, it usually requires up to about 50% of protein in its feed (Jiang et al., 2016). Fishmeal is a high quality protein source and it has been traditionally used as the main protein source in the aquafeed industry. However, due to the high demand for fishmeal leading to a high price and lack of sources, the farming industry had to find dietary alternative ingredients. The high nitrogen and phosphorus content in the fishmeal feeds is not completely absorbed by hybrid grouper and is most likely to be excreted along with the feces, leading to serious contamination of the farmed water. Currently, the use of soybean meal to replace fish meal can relieve the pressure of fish meal shortage and greatly promote the sustainable development of aquaculture industry (Yang et al., 2011; Smith et al., 2017; Miao et al., 2018). However, owing to low tolerance to soybean meal, excessive intake of soybean meal can lead to varying degrees of intestinal injury (Zhang et al., 2019).
The intestines of aquatic animals are responsible for the dual functions of digestion and absorption. The mucosa of the intestinal tract is the main interface between the internal and external environments of the organism. The intestinal tract is the main site of nutrient exchange and infection or initiation of infection by many pathogens (Jiang et al., 2019) and is highly susceptible to the effects of feed ingredients and living environmental conditions (Duan et al., 2017). Thus, it is essential to maintain the health and stability of the intestinal tracts of aquatic animals. The presence of a large number of microbiota in the intestinal tract is the result of long-term evolution, and they are closely related to the immune function and nutritional requirements of the organism (Yang et al., 2018). Intestinal microbiota can participate in the mediation of multiple metabolic pathways in the host, interacting with host metabolism and signal transduction to form a physiologically connected gut-immune-inflammation axis (Matsumoto et al., 2012). Similarly, intestinal health status could also influence the composition of microorganisms. Therefore, the relationship between intestinal flora and metabolite associations and host intestinal health requires further investigation.
It is accepted that β-conglycinin (7S) of soybean meal is a major factor in the induction of intestinal injury. β-Conglycinin, one of the major antigen proteins in soybean, often causes allergic reactions in young animals, causing inflammatory damage to intestinal epithelial cells accompanied by lipid peroxidation of cell membranes, negatively affecting the function of tight junction protein structures and increasing intestinal epithelial permeability (Zhao et al., 2014). The unbalanced amino acid in soybean meal may lead to increased excretion of nitrogen and phosphorus by fish (Tantikitti et al., 2005), which is not conducive to the healthy and sustainable development of the aquaculture industry. As a widely used additive in livestock and poultry animals, sodium butyrate (NaB) can not only act as a food attractant, but also improve the tight junctions of intestinal epithelium (Huang et al., 2015) and alleviate inflammation by inhibiting nuclear factor kappa B (NF-kB) (Albino et al., 2012), and it also has a positive regulatory effect on the host’s intestinal microbiota (Zou et al., 2019). The active ingredient in NaB is butyric acid, which is a short-chain, volatile fatty acid (O’Hara et al., 2018). In aquatic animals, NaB can stimulate the growth and proliferation of intestinal mucosa and inhibit the proliferation of intestinal pathogenic microorganisms, thus, promoting the growth of fish and enhancing intestinal digestion and antioxidant capacity (Tian et al., 2017; Jesus et al., 2018; Fu et al., 2019). Nevertheless, the physiological mechanisms of intestinal injury and inflammation in hybrid grouper caused by β-conglycinin and the protective effects of NaB on the intestinal tract are not well understood.
In a previous study, we demonstrated that ingestion of high levels of soybean meal by hybrid grouper resulted in intestinal injury (Zhang et al., 2019). The inhibitory effects of β-conglycinin on the growth of fish is usually thought to be due to intestinal inflammation (Zhang et al., 2013); nevertheless, the reports of whether β-conglycinin can negatively affect other physiological functions in fish are rare. In this study, we investigated the effects of β-conglycinin and the protective mechanism of NaB on transcription, intestinal microbiota, and metabolites in the distal intestine of hybrid grouper. Then, transcriptomic, intestinal microbiota, and metabolomic means were combined to model the associations among host metabolism, intestinal microbiota, and aquaculture water pollutants at the system biology level. This will help us understand the metabolic processes of the microflora in the host and their effects on host transcription, as well as their interactions with each other, and search for potential targets for the treatment of soybean meal enteritis.
Healthy juvenile hybrid groupers were purchased from a fish hatchery in East Island (Zhanjiang, China). The juveniles were housed in a concrete pond at the Biological Research Base of Guangdong Ocean University and fed commercial feed for 1 week to acclimatize them to the base environment.
After 1 week, 480 robust and uniformly fit hybrid groupers with an average body weight of 7.70 ± 0.05 g were randomly selected. The experiment was divided into four treatment groups with four replicates of 30 fish each and farmed for 8 weeks. 0.3 m3 fiberglass tanks were used for each replicate breeding indoors. During the experimental period, apparent satiety feeding was carried out twice daily (08:00 and 16:00) with a daily water change of approximately 70% (water flow: 100 L/min). The water temperature was 30.00 ± 1.30°C, salinity was 32.00 ± 2.00, dissolved oxygen was ≥7.00 mg/L, pH was 7.80–8.10, and ammonia nitrogen was <0.09 mg/L.
Four different design approaches to experimental feeds were used in this experiment. Based on the approximate amount of 7S in soybean meal protein (30%) (Ogawa et al., 1991) and the research of soybean meal substitution for fish meal in aquatic animals (Shiu et al., 2015), we set 1.5% 7S as the low dose (equivalent to supplementing 10% dehulled soybean meal to feed) and 6.0% as the high dose group (equivalent to supplementing 40% dehulled soybean meal to feed). Optimum NaB addition level was determined based on the results of studies on aquatic animals (Mirghaed et al., 2019). We used the group with 0.00% 7S and 0.00% NaB as the control group, named FMb group. To the control group, 1.5% of 7S was supplemented as a low-dose 7S addition group, named bL group; to the control group, 6.0% of 7S was supplemented as a high-dose 7S addition group, named bH group; to the control group, both 6.0% of 7S and 0.13% of NaB were supplemented as a repair group, named bHNaB group. Fishmeal, casein, and gelatin were used as the main protein sources for experimental feeds, while fish oil and soy lecithin were used as the main lipid sources. Methionine and lysine were added to the experimental diets at the FM diet level. The diet formulations and amino acid profiles of the four diets are shown in Supplementary Tables 1, 2. Purified 7S was purchased from China Agricultural University (Patent No. 200410029589.4, China). After all the ingredients were thoroughly blended, they were passed through a 380 μm sieve. Then, 30% water (dissolved choline chloride) was added and mixed thoroughly, and a pelletizer was used to produce 2.5 mm diameter pellets. The samples were air-dried at room temperature until the moisture content was close to 10% and then stored at −20°C until use.
Six experimental fish were randomly selected from each treatment at the end of the 8-week breeding experiment. The fish were dissected in a sterile environment using sterile scissors and forceps, and the distal intestine contents were removed and placed in sterile cryopreservation tubes. Following this, the intestines were immediately cleaned with pre-chilled phosphate buffered saline and divided equally into two portions, one for the transcriptomic sample and the other for the metabolomic sample, which were placed in cryopreservation tubes. Each individual fish served as a biological replicate of each microbiota, transcriptome, and metabolome, and corresponded one to one.
The siphon method was used to obtain water samples approximately 10–20 cm from the bottom of each tank for the determination of ammonia nitrogen (AN), nitrite (NIT), total nitrogen (TN), and total phosphorus (TP). One end of the polyvinylchlorid pipe was placed below the water surface, 10–20 cm from the bottom of the tank, and the other end was connected to the sample bottle. Then the air inside the tube was extracted so that the water enters the sample bottle through the tube due to the air pressure difference. The water samples for determining TN and TP were additionally filtered using a filter extractor with a pore size of 0.45 μm. All water samples were temporarily stored at −20°C. According to the Specification for Marine Monitoring-Part 4: Seawater analysis (GB 17378.4-2007) (China, 2007), the hypobromite oxidation method and N-(1-naphthyl)-ethylenediamine dihydrochloride spectrophotometric method were used to determine the contents of NH4+-N and NO2-N, respectively, and the potassium persulfate oxidation method was used to determine the contents of TN and TP.
Trizol (1 mL; TRI Reagent solution, Invitrogen, Carlsbad, CA, United States) was used to extract total RNA from 100–150 mg of the distal intestinal tissue. Electrophoresis of 1% agarose gels and spectrophotometric analysis with a NanoDrop 2000 (260:280 nm) was used to determine the total RNA quality and quantity. Total RNA was reverse-transcribed into cDNA using the PrimerScriptTM RT-PCR kit (TaKaRa, Kusatsu, Japan). SYBR® Premix Ex TaqTM kit was used to perform real-time PCR reactions using the Applied Biosystems 7500 Real-Time PCR System (Life Technologies, Carlsbad, CA, United States). Relative mRNA levels were analyzed using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
The hexad ecyltrimethyl ammonium bromide (CTAB) method was used to extract the total genomic DNA from the distal intestine samples (Griffith and Shaw, 1998). 20 uL of lysozyme was added to 1000 uL of CTAB lysis solution (0.1 M Tris HCl, pH 8.0, 1.4 M NaCl, 0.02 M EDTA, 2% CTAB) and the mixed to be used for adequate lysis of the samples. The lysed solution was centrifuged and 950 uL of the supernatant was taken. The supernatant was again taken and added to a mixture equal in volume to the supernatant [V(phenol):V(chloroform):V(isoamyl alcohol) = 25:24:1], mixed well, and centrifuged at 12,000 rpm for 10 min. The supernatant was again taken and added to a mixture equal in volume to the supernatant [V(chloroform):V(isoamyl alcohol) = 24:1], then mixed and centrifuged at 12,000 rpm for 10 min. The supernatant was then taken and three-fourths of the supernatant volume of isopropanol was added and precipitated at −20°C. Centrifuged the above mixture at 12,000 rpm for 10 min. After pouring out the supernatant, the obtained precipitate was washed using 1 mL of 75% ethanol and the washing was repeated twice. The DNA was dried on the ultra clean bench and was dissolved in 51 uL of double distilled water. Finally, 1 uL of RNase A was added and the RNA was removed by placing it at 37°C for 15 min. The quality and quantity of extracted DNA were examined using agarose gel electrophoresis and Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific), respectively. The V3–V4 region of the bacterial 16S rDNA gene was amplified using a pair of barcoded fusion primers, 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′). All PCR reactions were carried out with 15 μL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs). PCR products were mixed in equi-density ratios and purified using the Qiagen Gel Extraction Kit (Qiagen, Germany). Library quality was assessed using the Qubit@2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on an Illumina NovaSeq platform using TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, United States), and 250 bp paired-end reads were generated. The UCHIME Algorithm1 (Edgar et al., 2011) was used to compare tags with the reference database (Silva database2) to remove chimera sequences (Haas et al., 2011).
The FLASH (v.1.2.73) analysis tool was used to merge paired-end reads (Magoè and Salzberg, 2011). Quality filtering on the raw tags was performed to obtain high-quality clean tags (Bokulich et al., 2013) according to the QIIME (v.1.9.14) (Caporaso et al., 2010) quality controlled process. Uparse software (v7.0.10015) (Edgar, 2013) was used to perform sequence analysis. Sequences with ≥97% similarity were assigned to the same operational taxonomic unit (OTU). Alpha diversity was applied to analyze the complexity of species diversity for a sample through four indices, namely, observed-species, Shannon, Simpson, and Good’s coverage, using the Mothur method (Schloss et al., 2011). QIIME (v.1.7.0) and R software (v.2.15.3) were used to calculate and display all indices in the experimental samples, respectively. Shannon and Simpson indices were used to identify the community diversity. Good’s coverage was used to characterize the sequencing depth. β-diversity analysis was used to determine differences of samples in species complexity. QIIME software (v. 1.9.1) was used to calculate β-diversity on both unweighted and weighted unifrac. And Principal coordinate analysis (PCoA) was used to perform principal coordinates and visualize from complex multidimensional data. WGCNA package, stat packages and ggplot2 package was used to display the PCoA analysis in R software (v. 2.15.3). High-throughput sequencing data in this study are deposited in the NCBI SRA repository, accession number PRJNA733825.
Total RNA from distal intestinal tissues was extracted using the TRIzol Reagent (Life Technologies, United States). Agarose gel electrophoresis, NanoDrop microspectrophotometer, and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States) were used to determine the quality and quantity of the extracted total RNA. Total RNA was enriched using oligo (dT). Sequencing libraries were constructed on high-quality RNA samples using the NEB #7530 kit (New England Biolabs, #E7530) on an Illumina HiSeqTM 2500 by Gene Denovo Biotechnology Co. (Guangzhou, China). Clean reads were filtered using fastp (Chen et al., 2018) (v. 0.18.0) to obtain high-quality reads. The remaining reads were mapped to the reference genome by TopHat2 (Kim et al., 2013) (v. 2.1.1) after rRNA was removed using the short read alignment tool Bowtie2 (Langmead and Salzberg, 2012) (v. 2.2.8). Differentially expressed genes (DEGs) of FMb vs. bL, FMb vs. bH, and bH vs. bHNaB were identified. Each transcript expression level was calculated using the fragments per kilobase of transcript per million mapped reads (FPKM) method. The edgeR package (v. 3.12.1)6 was used for identifying DEGs between two groups. DEGs with a fold change (FC) ≥ 2 and a P-value <0.05 were considered significant. All DEGs were mapped to gene ontology (GO) terms of the GO database7. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed using OmicShare tools8. The GO and KEGG enrichment statistical analyses were set at corrected P-values <0.05 as the threshold for significance. Transcriptome sequencing data in this study are deposited in the NCBI SRA repository, accession number PRJNA735581.
Six distal intestinal tissue sample replicates of hybrid grouper from each treatment were used for metabolomic analysis. 100 mg of intestinal tissue samples ground in liquid nitrogen were placed in Eppendorf tubes and 500 μL of 80% formaldehyde aqueous solution was added. The samples were subjected to vortex shaking, allowed to stand in an ice bath for 5 min, and centrifuged at 15,000 × g at 4°C for 20 min. Mass spectrometry-grade water (liquid chromatograph-mass spectrometer (LC-MS) Grade, Merck, Germany) was added to dilute the formaldehyde content of the sample to a concentration of 53%. The sample was then centrifuged at 15,000 × g at 4°C for another 20 min. The supernatant was collected and detected by LC-MS (Vanquish UHPLC, Thermo Fisher, Germany; Q ExactiveTM HF, Thermo Fisher, Germany). The liquid chromatograph was equipped with a Hypersil Gold column (100 × 2.1 mm, 1.9 μm). In the assay, an equal volume of sample from each experimental sample was mixed as a quality control (QC) sample, and an aqueous formaldehyde solution at 53% concentration was used as a blank sample. Mass spectrometry scans ranged from 100 to 1500 (mass to charge ratio, m/z), and the electrospray ionization source was set up as follows: spray voltage, 3.2 kV; sheath gas flow rate, 40 arb; auxiliary gas flow rate, 10 arb; and capillary temperature, 320°C. Polarity: positive, negative; MS/MS secondary scans were data-dependent.
The offline data (raw) files were imported into Compound Discoverer 3.1 (CD3.1, Thermo Fisher) to perform a simple filtering of retention time, mass to charge ratio, etc. The different samples were then peak-aligned according to a retention time deviation of 0.2 min and a mass deviation of 5 ppm for more accurate identification. Peaks were then extracted based on a set mass deviation of 5 ppm, signal strength deviation of 30%, signal to noise ratio of 3, and minimum signal intensity of 100,000. The peak area was also quantified, and molecular formulae were predicted by ion peaks and fragment ions and compared with mzCloud9, mzVault, and Masslist databases to obtain accurate qualitative and relative quantitative results. The softwares R (R version R-3.4.3), Python (Python 2.7.3 version), and CentOS (CentOS release 6.6) were used for statistical analyses. The transformation was performed using area normalization when the data were not normally distributed. QC and quality assurance were used to determine all data. The KEGG10, HMDB11, and LIPIDMaps12 databases were used to annotate the metabolites. MetaX was used to perform principal component analysis and partial least squares discriminant analysis (PLS-DA). The univariate analysis (t-test) was used to calculate statistical significance (P value). Metabolites with variable importance in projection (VIP) > 1, P-value <0.05, and FC ≥ 2 or ≤ 0.5 were considered to be differential metabolites (DMs). DMs of FMb vs. bL, FMb vs. bH, and bH vs. bHNaB were identified. The metabolites of interest were filtered using Volcano plots based on log2(FC) and −log10(P-value) of metabolites. For clustering heat maps, z-scores of the intensity areas of the DMs were used to normalize the data. The Pheatmap package was used to plot the data in R. The functions of these metabolites and metabolic pathways were studied using the KEGG database13. Metabolic pathway enrichment of the DMs was performed. The pathways were considered to be enriched when ratios were satisfied by x/n > y/N and significantly enriched when P < 0.05.
The correlation between distal intestinal bacteria and water pollutants, distal intestinal DEGs, and DMs was revealed by Spearman correlation analysis using the Cytoscape software coNetplug-in. The P-value and correlation coefficient were not set; ∗ indicated significant differences (P < 0.05), ∗∗ indicated very significant differences (P < 0.01), and ∗∗∗ indicated extremely significant differences (P < 0.001).
Under the premise of variance homogeneity, statistical evaluations of pollutants and RT-PCR data were subjected to one-way analysis of variance followed by Tukey’s multiple range tests to determine significant differences among the four groups. SPSS (v. 22, SPSS Inc., Chicago, IL, United States) was used as described previously (Guo et al., 2017). The results are presented as mean ± standard error.
After 24 h of water exchange, water samples were collected to determine the quality (Figure 1). bH showed a significantly higher ammonia nitrogen concentration than FMb, while the concentration in bL or bHNaB was not significantly different from that in FMb. As for nitrite, significant increases occurred in bH and bHNaB compared with that in FMb and bL, and no significant difference was found between the FMb and bL. TN and TP concentrations showed the same trend as that of ammonia nitrogen, with the concentrations being significantly higher in bH than in FMb, bL, and bHNaB.
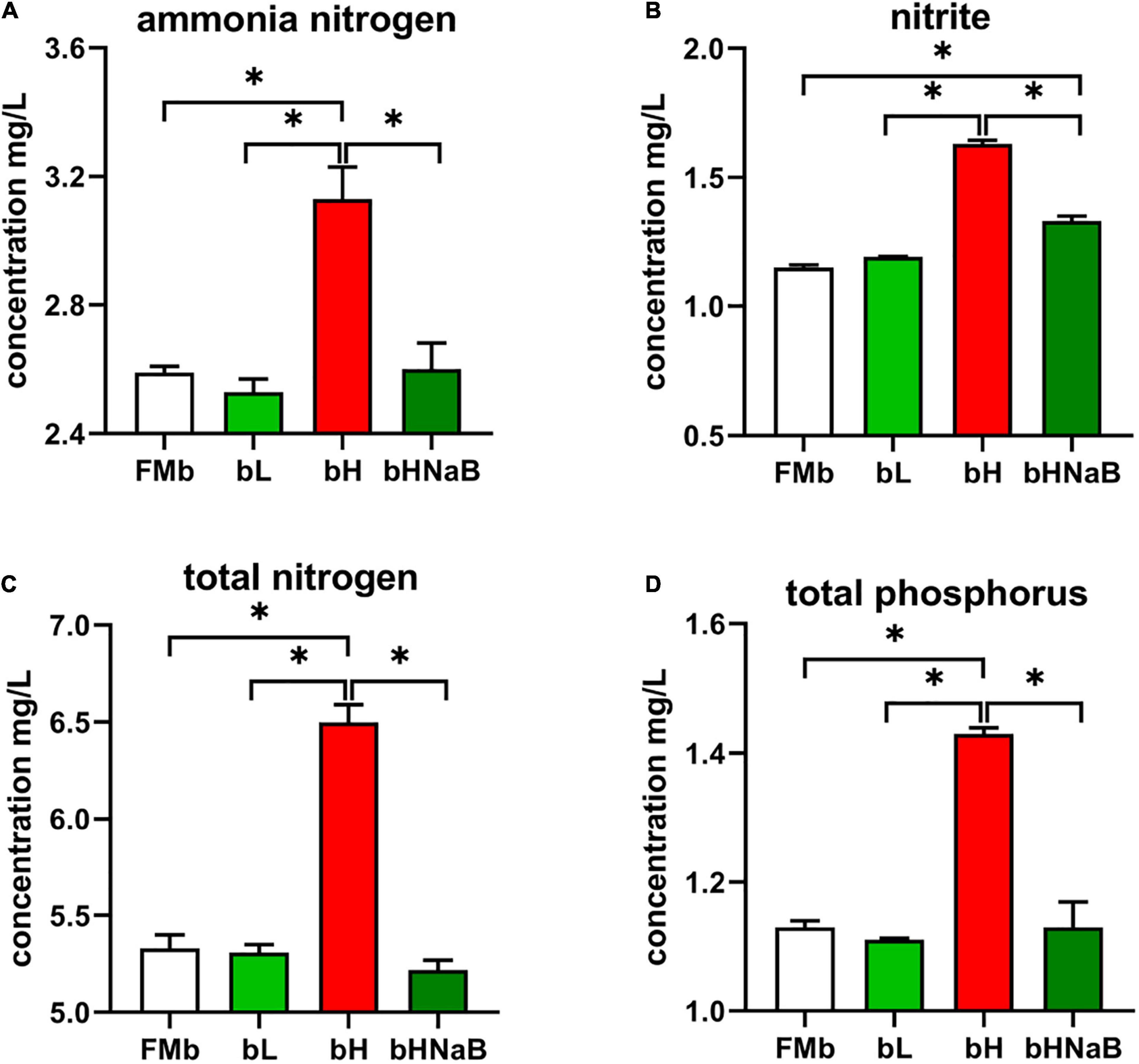
Figure 1. Detection of harmful substances in aquaculture water. The concentrations of (A) ammonia nitrogen, (B) nitrite, (C) total nitrogen concentration, and (D) total phosphorus were determined by Specification for Marine Monitoring-Part 4: Seawater analysis (GB 17378.4-2007). *P < 0.05.
A total of 1,218,101 raw reads were observed in all 16 microbial samples (two samples were excluded from each group). Average numbers of raw reads per treatment were 70,628.50 ± 4793.95, 80,868.00 ± 5197.54, 74,132.00 ± 4256.19, and 78,896.75 ± 5944.74 for FMb, bL, bH, and bHNaB, respectively. After clustering OTUs with 97% consistency, we obtained a total of 11,707 OTUs. The average OTUs in bL (916.75 ± 212.02), bH (561.75 ± 46.00), and bHNaB (352.50 ± 17.59) were lower than those in FMb (1095.75 ± 212.02). In addition, the rarefaction curves indicated that the identification rate of new OTUs gradually decreased as the number of reads per sample increased (Supplementary Figure 1), while the Good’s coverage of all samples exceeded 99%. This indicated that the sequencing results had good accuracy and reproducibility. Compared with FMb and bL, in bH and bHNaB, the Shannon and Simpson indices showed the same significant differences and were decreased, but no difference was found between bH and bHNaB (Supplementary Figure 2). The number of unique OTUs in bL was increased, while that in bH and bHNaB decreased (Figure 2A). Principal coordinate analysis plots of unweighted and weighted UniFrac matrix distances showed that FMb was separated from bL, bH, and bHNaB, while bH and bHNaB were very close to each other (Figures 2B,C).
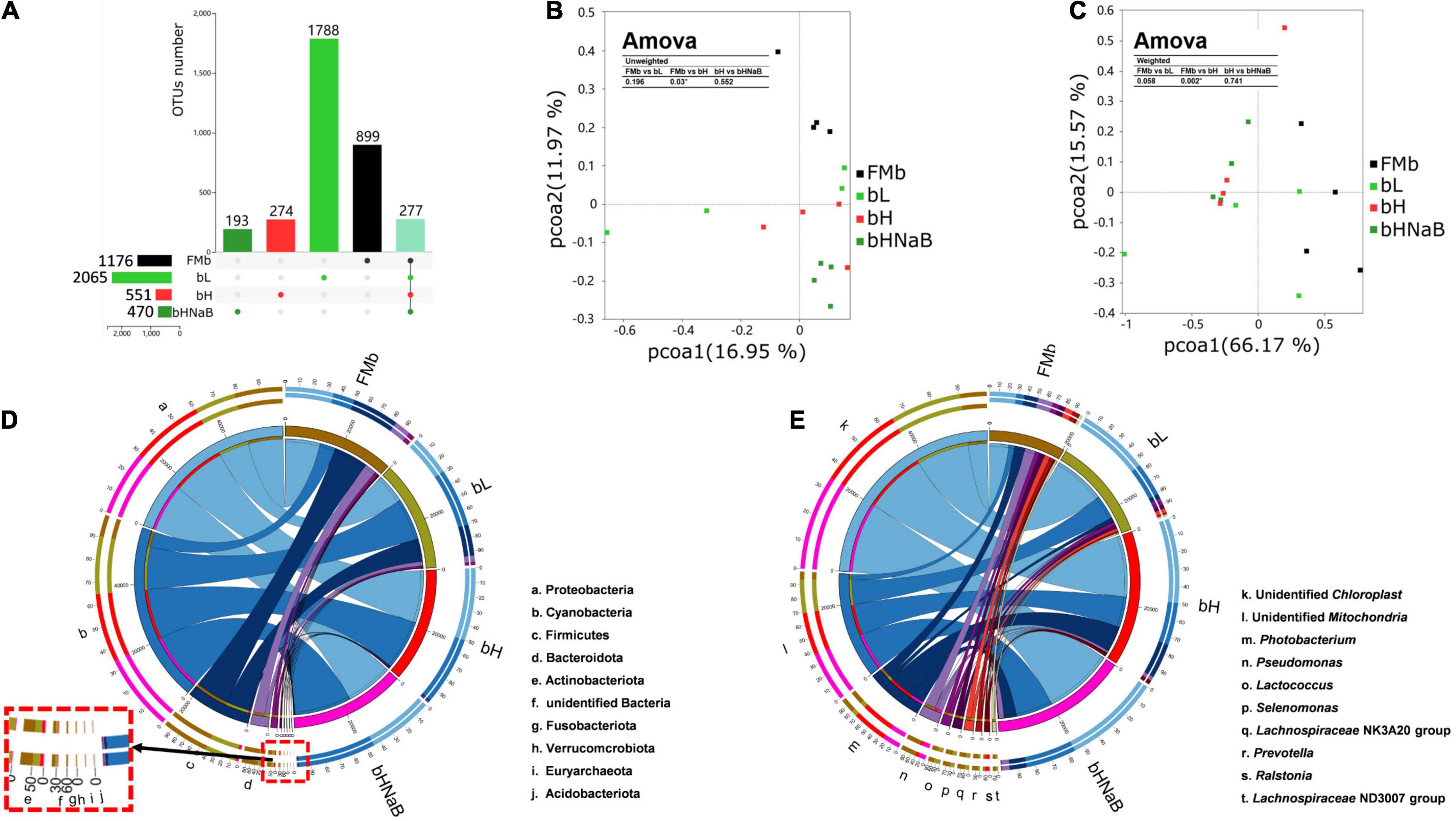
Figure 2. Distal intestinal microbial diversity and composition of hybrid grouper. The numbers of unique and shared OTUs between different treatments indicated by UPSET (A). The horizontal bars indicate the total OTUs for each treatment, the vertical bars indicate the specific OTUs for each treatment, and the rightmost vertical bar indicates the shared OTUs of all treatments. PCoA plots based on unweighted UniFrac metrics (B) and weighted UniFrac metrics (C), and Amova analysis was used to determine the significant differences in distance between two groups. Color code, FMb: #000000; bL: #31CB31; bH: #FF3131; bHNaB: #319831; shared OUTs: 91E1BD. Circos graphs of dominant bacterial phyla (top 10) (D) and genera (top 10) (E) in the distal intestinal. The right semicircle represents the phyla and genera composition of each treatment, and the left semicircle indicates the distribution of each phyla and genera in the different treatments.
At the phylum level, the top 10 phyla in terms of overall relative abundance included Proteobacteria, Cyanobacteria, Firmicutes, Bacteroidetes, and Actinobacteria. The relative abundance of Proteobacteria decreased in bL and increased in bH and bHNaB compared with that in FMb. The relative abundance of Cyanobacteria increased in bL, bH, and bHNaB compared with that in bH. The relative abundance of Firmicutes, Bacteroidetes, and Actinobacteria decreased in all three treatment groups (Figure 2D). At the genus level, the dominant bacteria in FMb included Pseudomonas, Prevotellaceae_UCG-001, Prevotella, Bifidobacterium, and Moraxella. Pelagibacterium, Vibrio, Pediococcus, Staphylococcus, and Lactobacillus showed a higher abundance in bL than in the other three groups. Classification of bacterial relative abundance showed a higher abundance of Ralstonia and Photobacterium in bH than in FMb, while the relative abundance of these genera in bL was also lower than that in FMb. The unidentified_Mitochondria, unidentified_Chloroplast, and Burkholderia-Caballeronia-Paraburkholderia showed higher relative abundance in bHNaB than in bH (Figure 2E).
Previous studies have revealed differences in the composition and structure of the bacterial taxa of the distal intestine of hybrid groupers from different treatment groups. To identify the key bacterial taxa with significant differences, we used LEfSe to analyze the differences in the abundance of taxa among the four groups. LEfSe analysis of all the samples revealed that 34 taxa with significantly different taxa information among the four groups were found at the phylum, class, order, family, genus, and species levels. Among them, the relative abundance of 31 bacterial taxa, including Firmicutes, Clostridia, Bacteroidota, Bacteroidia, Bacteroidales, Lachnospiraceae, Lachnospirales, Bacilli, Lactobacillales, and Negativicutes, in FMb was significantly higher than that in the other groups (P < 0.05). The relative abundance of one bacterial taxon, Lactobacillaceae, in bL was significantly higher than that in the other groups (P < 0.05). The relative abundance of two bacterial taxa, Ralstonia and Ralstonia pickettii, in the bH group was significantly higher than that in the other groups (P < 0.05). No discriminant taxa were found in bHNaB (Supplementary Figures 3A,B).
After transcriptome sequencing analysis of 16 distal intestine samples from the four groups (two samples were excluded from each group), a total of 527,803,196 raw reads and 79,170,479,400 raw bases were obtained, from which 526,786,184 clean reads and 78,684,777,954 clean bases were obtained after filtering and QC.
A total of 1878 DEGs (1231 upregulated and 647 downregulated) in FMb vs. bL, 1455 (702 upregulated and 753 downregulated) in FMb vs. bH, and 802 (570 upregulated and 232 downregulated) in bH vs. bHNaB were identified (Supplementary Figures 4, 5).
After all the DEGs (P < 0.05) were annotated by GO enrichment analysis, the dominant GO terms of the three comparison groups were almost the same. The dominant sub-categories in biological process were “single-organism process,” “cellular process,” and “metabolic process” (Figures 3A,D–F); the dominant sub-categories in molecular function were “binding” and “catalytic activity” (Figures 3B,G–I); and the dominant sub-categories in cellular components were “cell,” “cell part,” and “organelle” (Figures 3C,J–L). After significant enrichment analysis, in FMb vs. bL, DEGs were significantly enriched in “cellular component disassembly,” “multi-organism process,” and “protein complex subunit organization” for biological process; “nucleic acid binding,” “protein binding,” and “binding” for molecular function; and “organelle part,” “organelle,” and “intracellular organelle” for cellular component. In FMb vs. bH, DEGs were significantly enriched in “cell cycle,” “cell cycle process,” and “immune system process” for biological process; “tubulin binding,” “cytoskeletal protein binding,” and “nucleoside-triphosphatase activity” for molecular function; and “intrinsic component of membrane,” “membrane part,” and “intracellular non-membrane-bounded organelle” for cellular component. In bH vs. bHNaB, DEGs were significantly enriched in “regulation of protein stability,” “organonitrogen compound catabolic process,” and “cell cycle” for biological process; “protein binding,” “oxidoreductase activity,” and “RNA binding” for molecular function; and “organelle,” “intracellular organelle,” and “intracellular part” for cellular component. According to the KEGG enrichment analysis, DEGs were enriched in six KEGG A classes: “organismal system,” “metabolism,” “human diseases,” “cellular processes,” “genetic information processing,” and “environmental information processing” (Figure 4). Nineteen of 283 pathways were significantly enriched in FMb vs. bL compared with those in FMb; the enriched pathways of bL were “ribosome,” “protein digestion and absorption,” “Salmonella infection,” and “peroxisome proliferator-activated receptor (PPAR) signaling” (Figures 4A,D). Twenty-six of 261 pathways were significantly enriched in FMb vs. bH compared with those in FMb; the enriched pathways of bH were “cell cycle,” “cytokine–cytokine receptor interaction,” “PPAR signaling,” and “ferroptosis” (Figures 4B,E). Twenty of 198 pathways were significantly enriched in bH vs. bHNaB (P < 0.05) compared with those in bH; the enriched pathways of the bHNaB were “protein processing in endoplasmic reticulum,” “cell cycle,” and “linoleic acid metabolism” (Figures 4C,F).
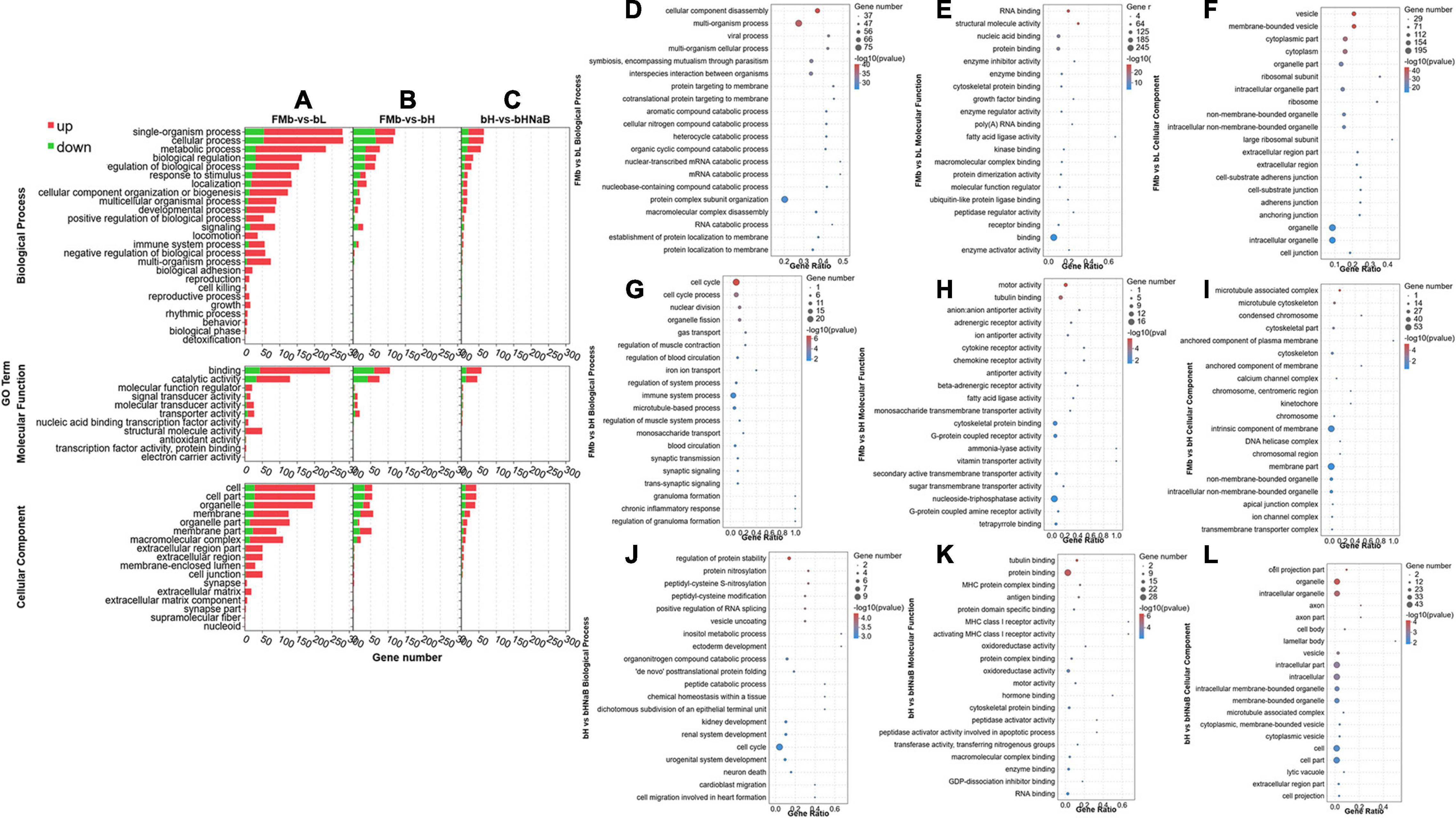
Figure 3. GO classification and enrichment analysis of the DEGs in the distal intestine of hybrid grouper. GO terms of FMb vs. bL, FMb vs. bH and bH vs. bHNaB. Distribution of the top 20 (level-2) GO terms significantly enriched in the categories of biological process, molecular function, and cellular component in FMb vs. bL (A,D–F), FMb vs. bH (B,G–I), and bH vs. bHNaB (C,J–L).
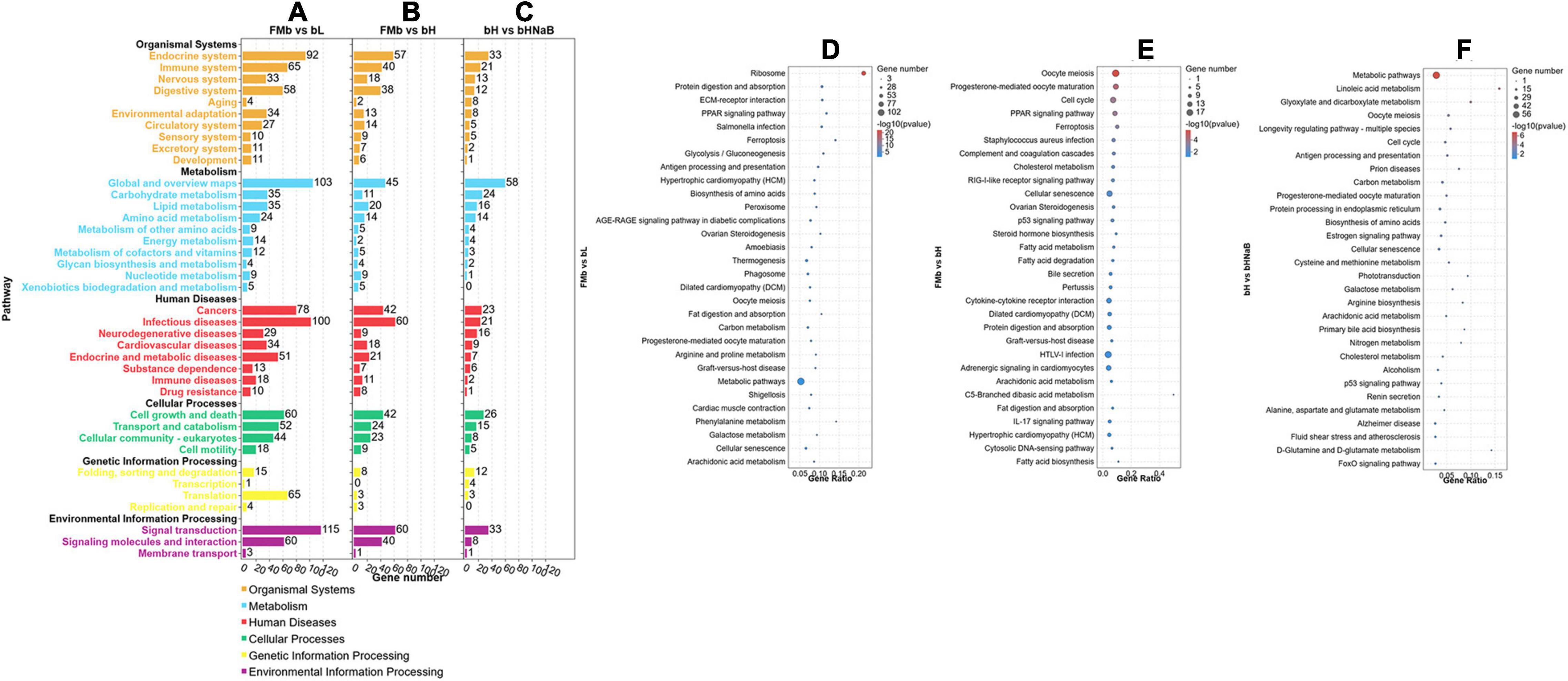
Figure 4. KEGG pathway classification and enrichment analysis of the DEGs in the distal intestine of hybrid grouper. KEGG classification of all DEGs (A–C). Top 30 enriched of KEGG pathways of FMb vs. bL (D), FMb vs. bH (E) and bH vs. bHNaB (F).
To verify the accuracy of the present transcriptome results, we selected 24 genes related to three aspects, namely, intestinal tight junctions, amino acid transporters, and inflammatory factors, and validated the results using RT-PCR (Supplementary Figure 6). The primers used in the experiment (Supplementary Table 3) were designed based on the full-length intestinal sequence of hybrid grouper (Zhang, 2020). Overall, the RT-PCR results were generally consistent with the trend of the transcriptome results, indicating that the transcriptome sequencing results were relatively accurate.
We used metabolome analysis to explore the differences in the distal intestinal metabolite profiles of different comparison objects. 24 distal intestine samples were analyzed by LC-MS in both positive and negative ion modes, and the LC-MS spectra are shown in Supplementary Figure 7 in positive (A) and negative (B) ions. Score plots of the PLS-DA were performed to verify the DMs in different comparison objects, and a multivariate analysis was performed to validate Supplementary Figure 8. The samples in each comparison object were clearly separated in both positive and negative ions, and the samples in each group were within the 95% confidence interval (Supplementary Figures 8A1,A3,B1,B3,C1,C3), indicating that different doses of 7S and NaB produced significant effects on the distal intestinal metabolic profile of hybrid grouper. Meanwhile, the R2 value was greater than the Q2 value, and the intercept of the Q2 regression line with the Y-axis was less than zero, which indicated that the PLS-DA model in this experiment was not over-fitted and that the model was stable and reliable (Supplementary Figures 8A2,A4,B2,B4,C2,C4).
Next, we performed DM analysis for the two groups in each comparison object using a VIP threshold of 1 and an FC threshold of 1.5 (P < 0.05). Subsequently, hierarchical clustering analysis was performed on the DMs obtained for each group to derive the differences in metabolic expression patterns between and within the two groups of one comparison object. For FMb vs. bL, 161 positively ionized DMs (122 significantly upregulated and 39 significantly downregulated) (Figures 5A1, 6A) and 84 negatively ionized DMs (50 significantly upregulated and 34 significantly downregulated) (Figures 5A2, 6B) were screened. For FMb vs. bH, 154 positively ionized DMs (111 significantly upregulated and 43 significantly downregulated) (Figures 5B1, 7A) and 100 negatively ionized DMs (53 significantly upregulated and 47 significantly downregulated) (Figures 5B2, 7B) were screened. For bH vs. bHNaB, 89 positively ionized DMs (47 significantly upregulated and 42 significantly downregulated) (Figures 5C1, 8A) and 55 negatively ionized DMs (38 significantly upregulated and 17 significantly downregulated) (Figures 5C2, 8B) were screened.
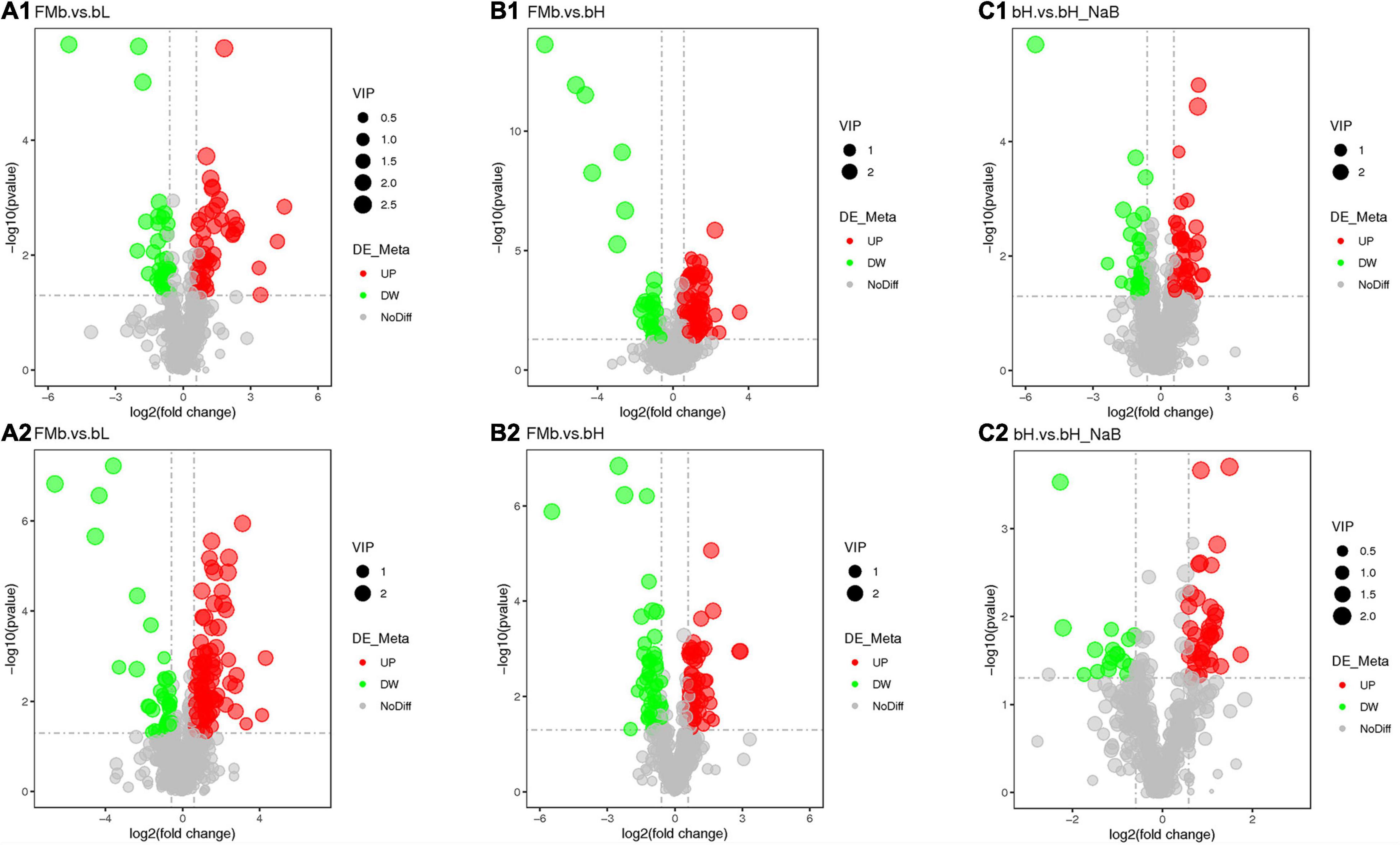
Figure 5. Volcano plots of the DEGs in the distal intestine of hybrid grouper at different comparison objects. FMb vs. bL: positive (A1) and negative (A2) ions; FMb vs. bH: positive (B1) and negative (B2) ions; bH vs. bHNaB: positive (C1) and negative (C2) ions.
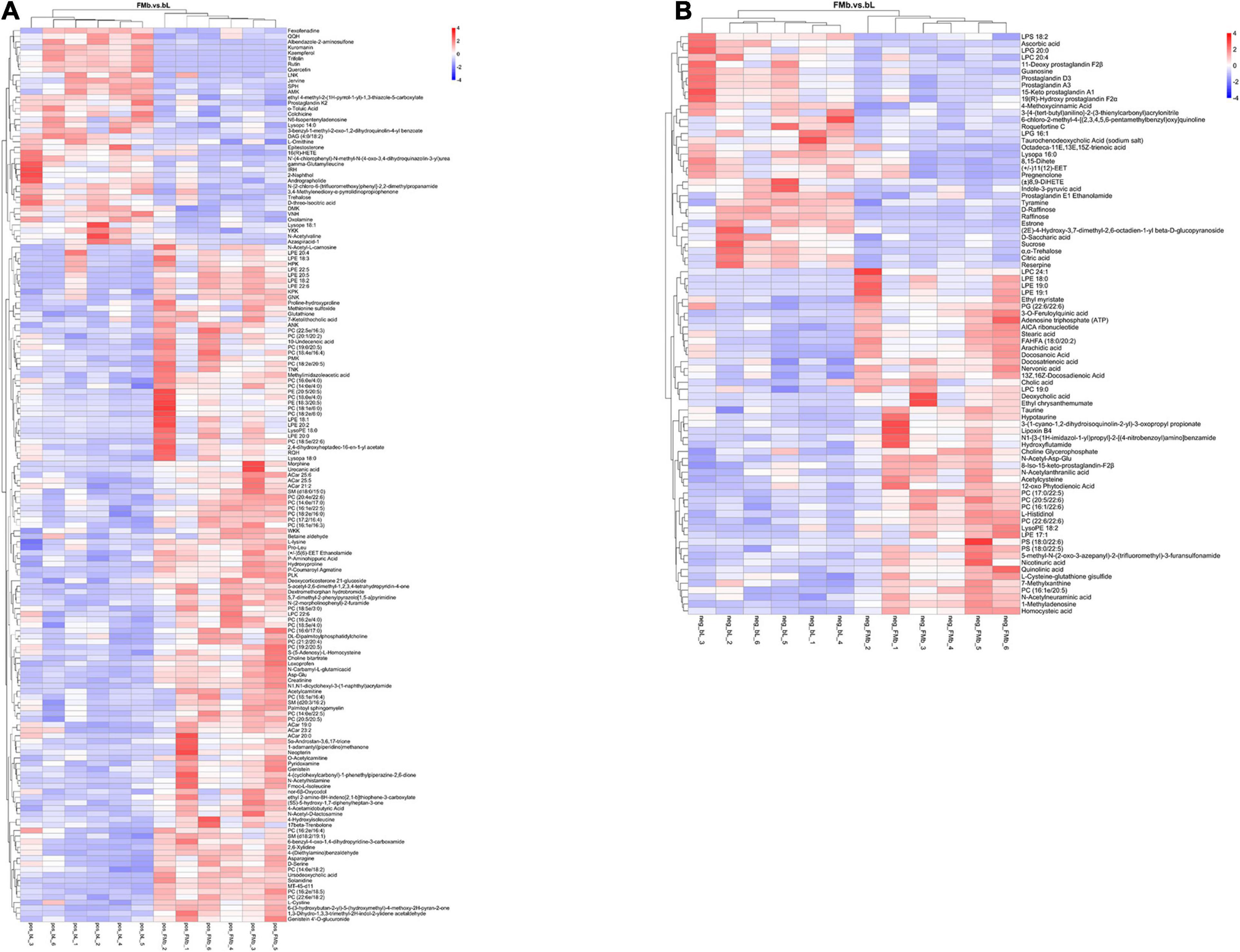
Figure 6. Hierarchical clustering analysis of the DMs in FMb vs. bL: positive (A) and negative (B) ions. Red and blue indicate that the DMs were upregulated and downregulated, respectively.
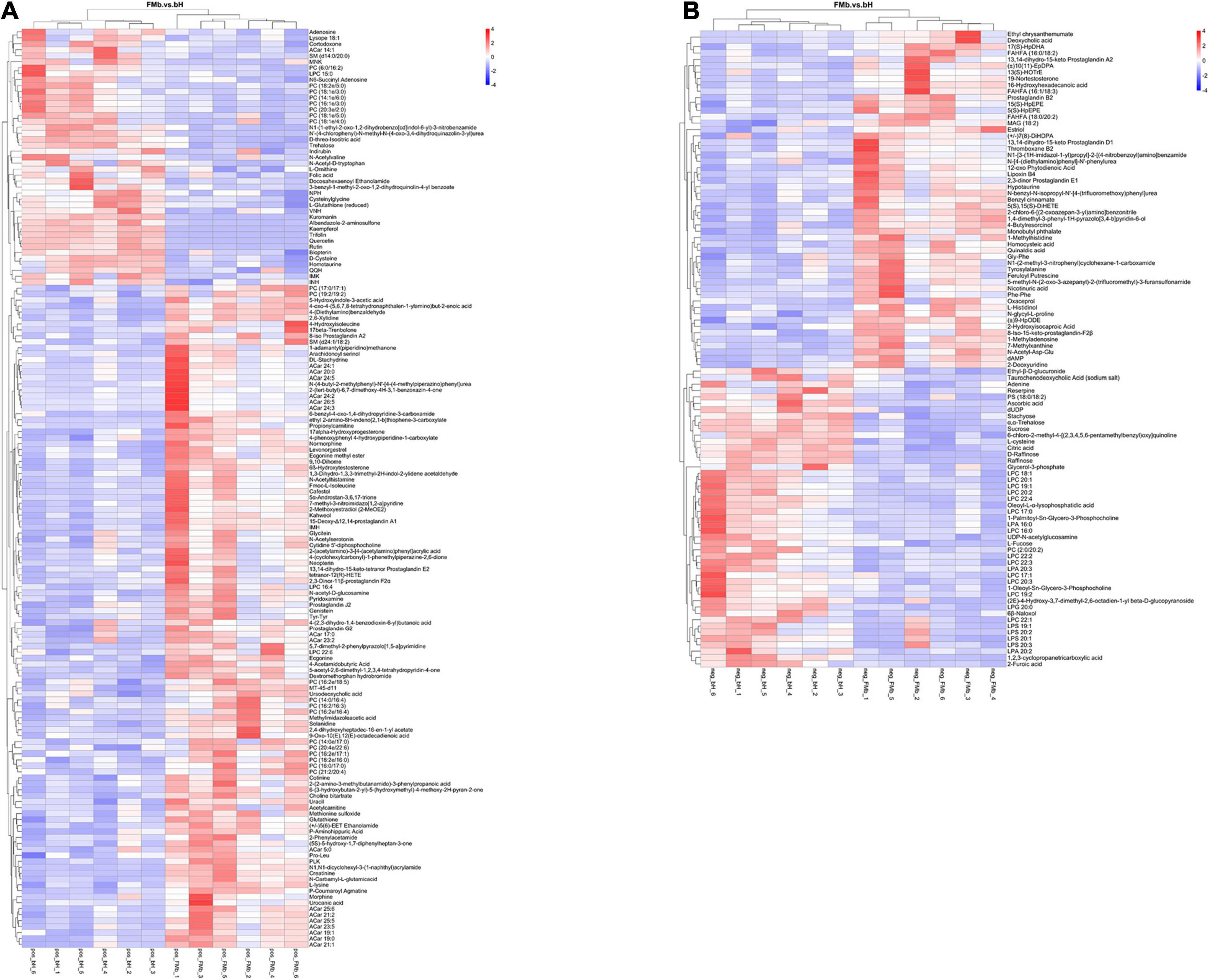
Figure 7. Hierarchical clustering analysis of the DMs in FMb vs. bH: positive (A) and negative (B) ions. Red and blue indicate that the DMs were upregulated and downregulated, respectively.
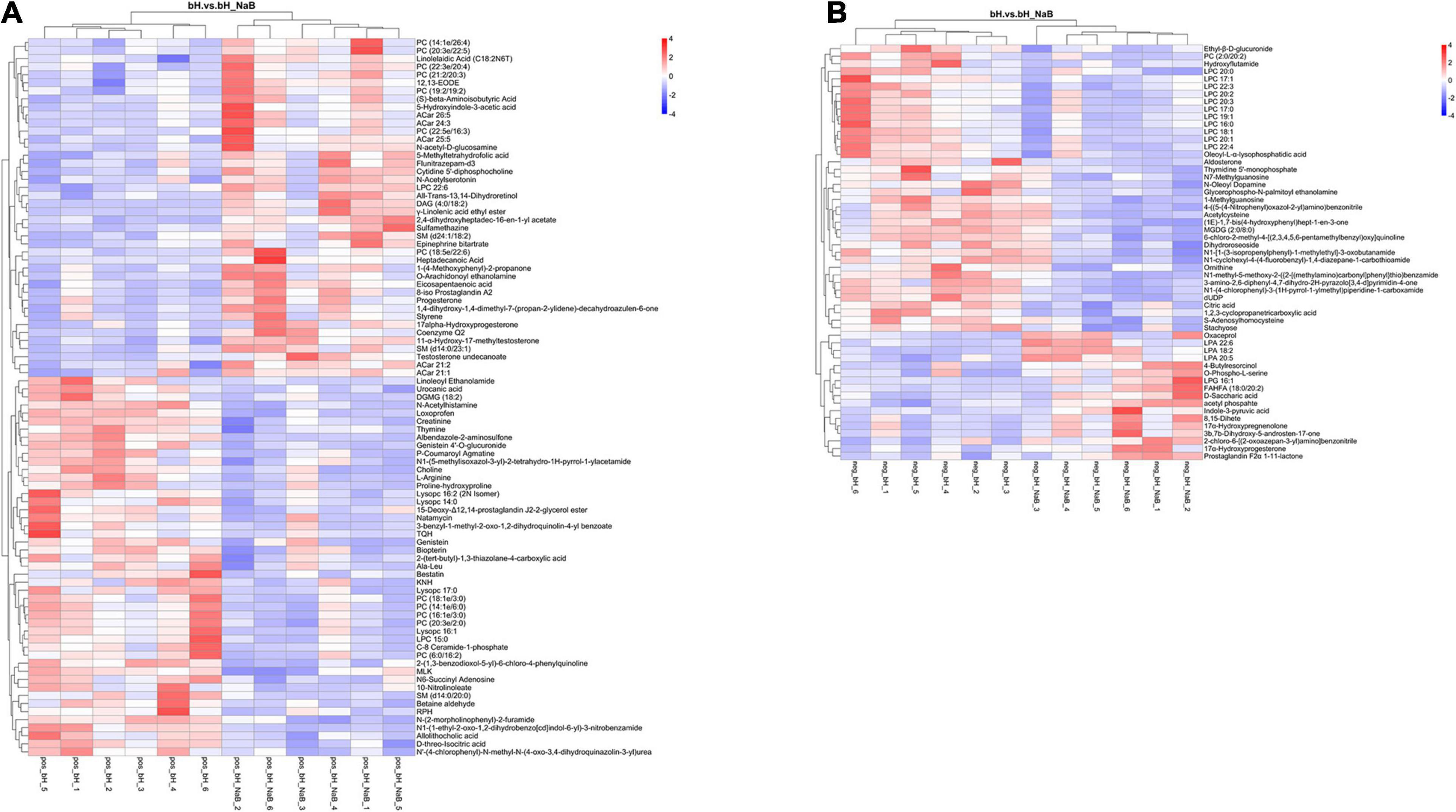
Figure 8. Hierarchical clustering analysis of the DMs in bH vs. bHNaB: positive (A) and negative (B) ions. Red and blue indicate that the DMs were upregulated and downregulated, respectively.
Furthermore, the DMs were analyzed using KEGG annotation. In the FMb vs. bL comparison object, for the positive ionization data, 11 DMs were enriched in 19 pathways, and the “histidine metabolism,” “arginine and proline metabolism,” “cysteine and methionine metabolism,” and “glycine, serine, and threonine metabolism” pathways were enriched (Figure 9A1); for the negative ionization data, 46 DMs were enriched in 31 pathways, and the “biosynthesis of unsaturated fatty acids,” “primary bile acid biosynthesis,” “fatty acid biosynthesis,” and “taurine and hypotaurine metabolism” pathways were enriched (Figure 9A2). In the FMb vs. bH comparison object, for the positive ionization data, 70 DMs were enriched in 36 pathways, and the “cysteine and methionine metabolism,” “ferroptosis,” and “AMP-activated protein kinase (AMPK) signaling” pathways were enriched (Figure 9B1); for the negative ionization data, 37 DMs were enriched in 25 pathways, and the “galactose metabolism,” “starch and sucrose metabolism,” and “amino sugar and nucleotide sugar metabolism” pathways were enriched (Figure 9B2). In the bH vs. bHNaB comparison object, for the positive ionization data, 53 DMs were enriched in 33 pathways, and the “histidine metabolism,” “glycine, serine and threonine metabolism,” “arginine and proline metabolism,” and “ATP-binding cassette (ABC) transporters” pathways were enriched (Figure 9C1); for the negative ionization data, 35 DMs were enriched in 19 pathways, and the “biosynthesis of amino acids” pathway was enriched (Figure 9C2).
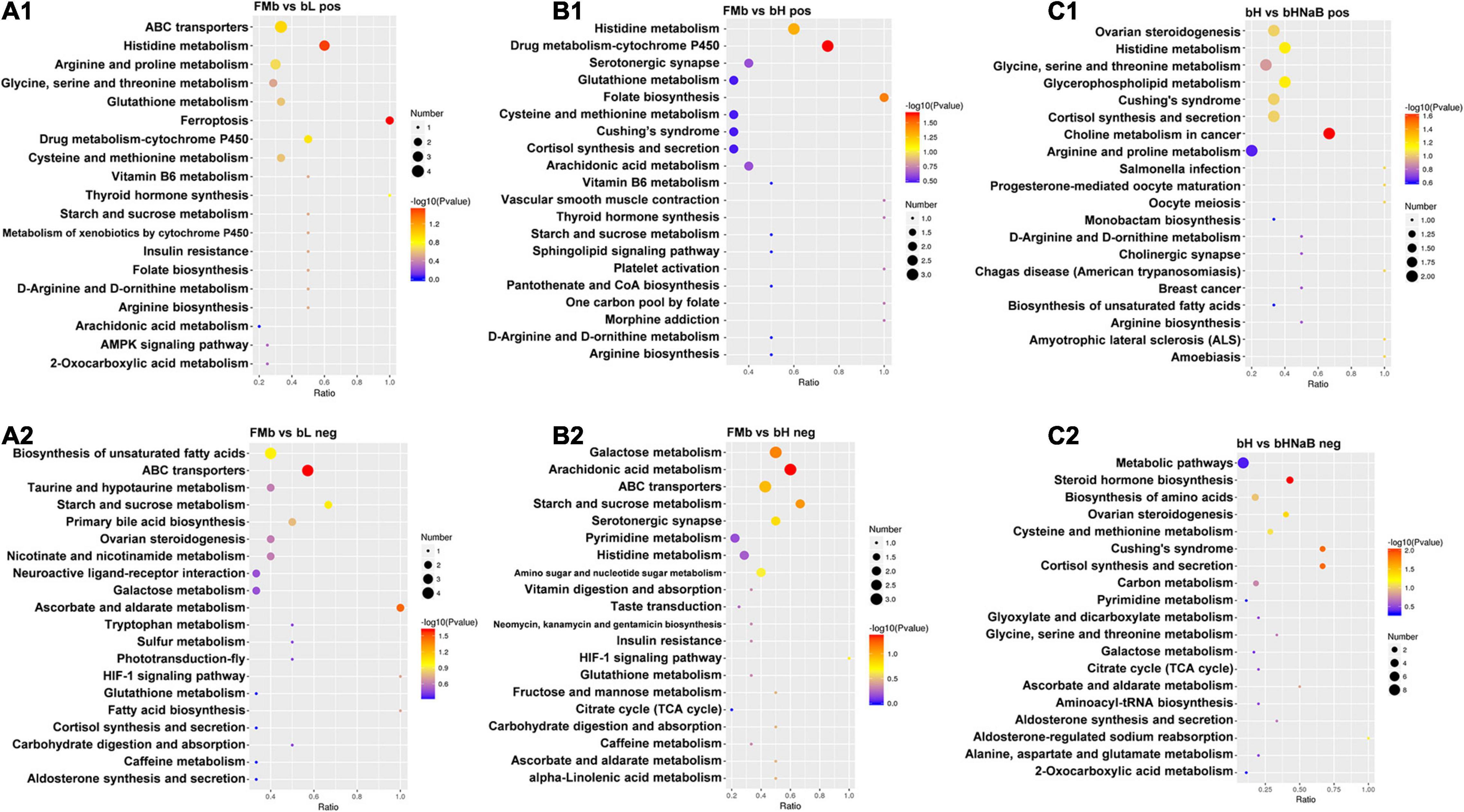
Figure 9. Metabolic pathway enrichment analysis following positive and negative mode ion. DMs were enriched in the different comparison objects. FMb vs. bL: positive (A1) and negative (A2) ions; FMb vs. bH: positive (B1) and negative (B2) ions; bH vs. bHNaB: positive (C1) and negative (C2) ions.
We performed Spearman correlation analysis of the top 30 intestinal genera with pollutants (AN, NIT, TN, and TP), DEGs (Table 1), and DMs (Table 2). In the correlation between intestinal bacteria and pollutants, Ralstonia and Rothia were positively correlated with changes in ammonia nitrogen and Lactococcus, Pediococcus, Pelagibacterium, Anaerovibrio, Prevotella, and Bacillus were negatively correlated with changes in nitrite (Figure 10A). In the correlation between intestinal bacterial and DEGs, Lactococcus, Pediococcus, and Pelagibacterium were positively correlated with changes in ccnf, Numa1, pkb, spc25, and Cks1b genes (Figure 10B). In the correlation between intestinal bacteria and DMs, Lactococcus was negatively correlated with changes in sucrose and α,α-trehalose; Pelagibacterium was negatively correlated with changes in methylimidazole acetic acid; Anaerovibrio was positively correlated with changes in choline, S-adenosylhomocysteine, creatinine, urocanic acid, N-acetyl histamine, and methylimidazole acetic acid and negatively correlated with changes in quercetin, α,α-trehalose, sucrose, L-fucose, and UDP-N-acetylglucosamine; Prevotella was positively correlated with changes in N-acetyl histamine and methylimidazole acetic acid and negatively correlated with changes in raffinose, quercetin, α,α-trehalose, sucrose, and L-fucose; and Bacillus was positively correlated with changes in S-adenosylhomocysteine, urocanic acid, N-acetyl histamine, and methylimidazole acetic acid and negatively correlated with changes in raffinose, quercetin, and α,α-trehalose (Figure 10C). Finally, 10, 4, and 4 potential biomarkers were screened in groups bL, bH, and bHNaB, respectively. ROC analysis showed that the metabolite AUCs all exceeded 0.88 at 95% confidence intervals (Supplementary Figure 9), indicating that the potential biomarkers had good predictive power.
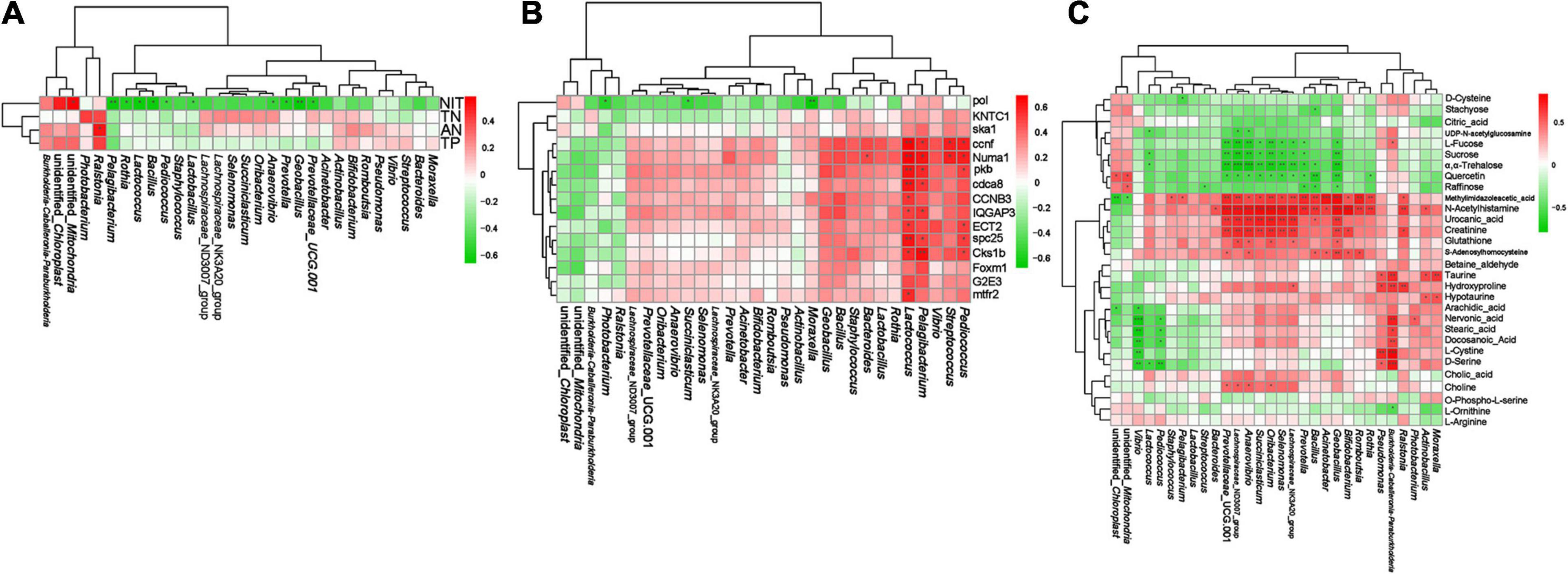
Figure 10. Spearman correlation analysis between distal intestinal bacteria at genus level and water pollutants, DEGs, DMs. The correlation analysis of distal intestinal bacteria and water pollutants (A). The correlation analysis of distal intestinal bacteria and DEGs (B). The correlation analysis of distal intestinal bacteria and DMs (C). Red and green indicate positive and negative correlation, respectively. * significant differences (P < 0.05), ** very significant differences (P < 0.01), *** extremely significant differences (P < 0.001).
The intestinal environment and microbiota interact with food ingested by the host (Wahlström et al., 2016). The intestinal microbiota is quite sensitive to changes in the quality and quantity of food (Wang et al., 2017). In this experiment, we observed that the dominant bacteria were Proteobacteria, Cyanobacteria, Firmicutes, Bacteroidetes, and Actinobacteria, which is consistent with previous results (Li, 2019). The dominant bacteria were influenced by 7S. The process of intestinal microecological dysbiosis is usually accompanied by an increase in Proteobacteria (Shin et al., 2015). This indicates that the intestinal inflammation induced by high-dose 7S in hybrid grouper is closely related to the abundance of Proteobacteria. In addition, the abundance of Proteobacteria and Cyanobacteria were not affected by NaB, and the abundance of Firmicutes, Bacteroidetes, and Actinobacteria decreased further after the addition of NaB compared with that in bH. To further analyze the effects of 7S and NaB on the intestinal microbiota of hybrid grouper, we conducted another assessment at the genus level.
At the genus level, different treatments resulted in different dominant genera in each group. We selected some bacteria that were closely related to the intestinal health of hybrid grouper for analysis. Pelagibacterium, Pediococcus, Staphylococcus, and Lactobacillus became the dominant bacteria in the bL. These bacteria played important roles in maintain intestinal resistance to disease and absorption of nutrients (Uymaz et al., 2009; Jiang et al., 2012; Liu and Yu, 2015; Ortiz-Rivera et al., 2017). Therefore, the growth promotion of hybrid grouper by low doses of 7S may be closely related to the enhanced intestinal resistance of these four dominant bacteria (Supplementary Table 4). The relative abundance of Ralstonia is highly positively correlated with the host inflammatory response (Fu et al., 2017), and a decrease in its relative abundance is effective in alleviating chronic inflammation (Kwon et al., 2018). In addition, increased abundance of Photobacterium may disrupt the intestinal morphology of Litopenaeus vannamei (Tzuc et al., 2014). The increased abundance of Ralstonia and Photobacterium might be the cause of intestinal inflammation in hybrid groupers owing to high doses of 7S. After supplementation with NaB, the abundance of Ralstonia and Photobacterium decreased in bH and became similar to that in FMb and bL. Thus, we speculate that the protective effect of NaB on the distal intestine of hybrid grouper may be achieved by suppressing the abundance of Ralstonia and Photobacterium, which reduces the risk of intestinal inflammation.
To gain further insight into the regulatory mechanism of 7S and the restorative effects of NaB, the distal intestinal tissue was analyzed using RNA sequencing technology. In this study, based on the GO significant enrichment analysis, 7S and NaB have similarities and differences in the regulation of the distal intestine. We selected the top three GO enrichment sub-categories in each category for our analysis. Ribosomes are the site of intracellular protein synthesis (Dauloudet et al., 2020), and ribosomal protein L31 and L3 contribute to the binding and translation of ribosomal subunits (Petrov et al., 2014; Ueta et al., 2017); furthermore, L7 is an important ribosomal component required for translation process (Pettersson and Kurland, 1980) and plays a critical role in the synthesis of host proteins. A low dose of 7S elevated the expression of associated ribosomal proteins, which may imply that a low dose of 7S effectively promoted the synthesis of distal intestine-associated proteins in hybrid grouper. To test this hypothesis, we further analyzed the expression of genes controlling proteins related to intestinal health and observed that the expression of genes controlling collagen (Claudio et al., 2017), cofilin-1 (Wang et al., 2016), calreticulin (Krzysztof et al., 2017), and tubulin (Saegusa et al., 2014) protein synthesis in the distal intestine was significantly increased at low doses of 7S, suggesting improvement in tight junctions and barrier function in the distal intestine. Fructose-bisphosphate A catalyzes fructose 1,6-diphosphate to produce glyceraldehyde 3-phosphate and dihydroxyacetone phosphate (Katebi and Jernigan, 2015). Therefore, the glycolytic capacity of hybrid grouper was also improved to some extent by promoting the expression of fructose-bisphosphate aldolase A.
When high-dose 7S was administered, the intestinal injury became critical. Mitosis plays an important role in maintaining normal growth and development of individuals (Sanz-Gómez et al., 2020). G2/mitotic-specific cyclin-B1 (Xie et al., 2019), B2 (Waesch and Cross, 2002), and B3 (Garrido et al., 2020) are three proteins essential for mitosis, and their deletion or inhibition of expression usually results in varying degrees of cellular damage, consequently affecting cellular function. We observed that these three cyclins were significantly downregulated, and the expression of genes related to cell cycle regulatory proteins and chromosome structure maintenance, such as cyclin-F (D’Angiolella et al., 2012), double-strand break repair protein (Sinha et al., 2020), and structural maintenance of chromosome protein 2 (Heidelberg, 2011) were inhibited. These results suggest that high doses of 7S inhibit the division of hybrid grouper intestinal cells and disrupt their normal functions. In addition, we observed that the expression of genes associated with ion channels, transient receptor potential cation channel subfamily M, and voltage-dependent L-type calcium channel subunit β-4 in intestinal cells was also negatively affected. The function of ion channels in the cell membrane, in addition to regulating the osmotic pressure inside and outside the cell, is maintaining the cell membrane potential (Page et al., 2005; Wu and Cui, 2014), indicating that high-dose 7S also disrupted distal intestinal cell membrane ion channels, thus, affecting the function of regulating ion transport.
To investigate how NaB protects the distal intestine of hybrid groupers, we supplemented NaB after high-dose 7S treatment. The expression of cyclin B1, B2, and B3, which were repressed at high-dose 7S, was significantly increased after NaB supplementation, and the gene expressions related to transcriptional and translational processes, such as RNA polymerase II transcription, eukaryotic translation initiation factor 1A, and transcription factor GATA-4, were also upregulated. This may indicate that NaB effectively protected the process of mitosis in the intestinal cells of hybrid grouper, allowing the intestinal cells to undergo normal division. NaB is a common and effective inhibitor of cell mitosis, which can inhibit the G1 phase of mitosis in mouse fibroblasts (Wintersberger et al., 2010) and also induce G2 blockade of the human breast cancer cell line MDA-MB-231 (Lallemand et al., 1999). However, the addition of NaB to the post-weaned heifer diet can effectively stimulate the mitosis of small intestinal epithelial cells and inhibit their apoptosis, thus, improving intestinal absorption function and promoting the effective absorption and utilization of nutrients (Rice et al., 2019). We speculate that the reason for this difference in mechanism may be species-related and may also be related to direct addition to cells and to feed; NaB may have the same repair mechanism in the intestine of post-weaned heifer and hybrid grouper. Glyceraldehyde 3-phosphate dehydrogenase can catalyze the oxidation (dehydrogenation) and phosphorylation of glyceraldehyde 3-phosphate to produce 1,3-diphosphoglyceric acid, which is the central link in sugar metabolism and, therefore, plays an important role in glycometabolism (Allonso et al., 2015; Zhang et al., 2016). As a member of the glyceraldehyde 3-phosphate dehydrogenase family, chitinase can catalyze the hydrolysis of chitin to produce N-acetylglucosamine (Rosa et al., 2016). Galactosyltransferase can transfer active galactose residues from nucleoside diphosphate galactose to glycosyl receptor molecules (Klohs et al., 2010). The activities of all three enzymes, glyceraldehyde 3-phosphate dehydrogenase, chitinase, and galactosyltransferase, were upregulated after NaB supplementation, suggesting that NaB could also enhance the absorption of glucose in hybrid grouper to some extent by promoting distal intestinal glycometabolism.
Based on the KEGG pathway analysis, the results have similarities and differences with the results of GO annotation. Under low-dose 7S conditions, the formation of ribosome-associated component proteins was promoted, which in turn enabled better protein synthesis by the host. In addition, two signaling pathways, “protein digestion and absorption” and “PPAR signaling,” which are closely related to the digestion and absorption of two major nutrients, proteins (Yang et al., 2019) and lipids (Calejman et al., 2020), were also positively affected, suggesting that 7S can significantly promote the absorption and metabolism of proteins and lipids in hybrid groupers. Salmonella is a common foodborne pathogen that can enhance bacterial virulence by inhibiting autophagy through the plasmid virulence gene spvB (Li et al., 2016). Salmonella infection in fish can cause disturbances in the intestinal environment and reduce immunity in fish (Wu et al., 2016). A low dose of 7S improved intestinal immunity in hybrid groupers and might be closely related to suppression of the Salmonella infection pathway.
Under high-dose 7S conditions, the results were the opposite of the partial results for low-dose 7S. “PPAR signaling pathway” was significantly negatively affected. Ferroptosis is an impairment of intracellular lipid oxide metabolism, with abnormal metabolism catalyzed by iron ions (Yang and Stockwell, 2016). The promotion of “ferroptosis” pathway further illustrates that high-dose 7S leads to disturbances in host lipid metabolism. Similarly, high doses of 7S significantly inhibited “cell cycle” and “cytokine–cytokine receptor interaction,” two pathways that are closely related in cell division (Sarraf et al., 2019) and intercellular signaling (Leung, 2004).
After NaB supplementation in the high-dose 7S condition, we observed that the “progesterone-mediated oocyte maturation” pathway was promoted, similar to the results of bL, indicating that NaB enhanced the digestion and absorption of proteins in hybrid grouper. In addition, the “cell cycle” pathway, which was inhibited in bH, was activated after NaB supplementation, indicating that NaB could protect the normal division and maintain the normal function of intestinal cells in hybrid grouper. Linoleic acid is a naturally occurring unsaturated fatty acid with many important physiological functions (Smith et al., 2004). It can promote the secretion of immunoglobulins, and thus, enhance the immunity of fish (Huang et al., 2018), and it can improve the lipid metabolism of fish (Makol et al., 2012). Facilitation of the “linoleic acid metabolism” pathway indicates that NaB can effectively improve lipid utilization by hybrid groupers.
To reveal the regulatory mechanism of 7S on the distal intestine of hybrid groupers and the protective effect of NaB in a multidimensional manner, we further utilized the LC-MS technique to analyze the metabolic profile changes in the distal intestine. Low-dose 7S had positive effects on “histidine metabolism,” “arginine and proline metabolism,” “cysteine and methionine metabolism,” and “glycine, serine, and threonine metabolism” in the positive ion mode and positive effects on “biosynthesis of unsaturated fatty acids,” “primary bile acid biosynthesis,” “fatty acid biosynthesis,” and “taurine and hypotaurine metabolism” in the negative ion mode. There are 10 amino acids that fish cannot synthesize in sufficient amount to support maximum growth and must be provided in the diets; these essential amino acids include: methionine, arginine, histidine, isoleucine, leucine, lysine, phenylalanine, threonine, tryptophan and valine (Hua and Bureau, 2019; Nascimento et al., 2020). Proline and glycine are considered as non-essential amino acids, and are involved in key metabolic pathways, they can modulate immune function health, reproduction and growth (Zhao et al., 2015; Li and Wu, 2018). Histidine is involved in the synthesis of hemoglobin in aquatic and terrestrial animals and is important for growth, tissue formation, and repair (Michelato et al., 2017; Khan, 2018). Histidine also inhibits oxidative stress-induced inflammation in human intestinal epithelial cells (Dong et al., 2005). Proline can act as a protective substance for membranes and enzymes and as a free radical scavenger, enhancing the body’s antioxidant capacity (Vunnam et al., 2010; Zabirnyk et al., 2010). The immunomodulatory effect of arginine on fish is mainly achieved through the regulation of endocrine hormones by the arginine/nitric oxide pathway, which plays an important role in the regulation of body immunity and protection of intestinal mucosa function (Zhou, 2011). Methionine can improve the antioxidant system by inhibiting apoptosis, and it can also improve immunity by upregulating the tight junction proteins claudin-b, claudin-c, ZO-1, and ZO-2 in the head, kidney, and spleen (Pan et al., 2016). The addition of glycinin to feed can significantly increase the expression of peptide-transporters 1 (PepT1) in the intestines of grass carp (Cyprinus carpio L.) and promote the absorption of peptides (Ostaszewska et al., 2010). Serine is an important component of the catalytic active site of serine proteases that can significantly enhance host recognition of pathogens (Liu et al., 2017) and improve the cellular immunity of the organism (Ashton-Rickardt, 2009). Threonine deficiency leads to impaired protein synthesis and poor proximal intestinal development in grass carp (Gao et al., 2014), upregulates the expression of the pro-inflammatory factors TNF-α and IL-1β, and downregulates the anti-inflammatory factor TGF-β, resulting in increased intestinal inflammation (Dong et al., 2017). These observations combined with the results of this study suggest that 7S could enhance the antioxidant capacity, immunity, and protein hydrolysate absorption efficiency of the distal intestine by promoting the metabolism of essential amino acids.
Highly unsaturated fatty acids (HUFAs) are essential fatty acids for marine fish because they lack the ability to synthesize HUFAs. HUFAs can prevent inflammation caused by high lipid deposition by inhibiting the activity of enzymes related to lipid deposition (Ma, 2008) and also enhance the innate immunity of grass carp by upregulating the expression of TLR22 and MyD88 (Li, 2013). Bile acids (Bertaggia et al., 2017) and taurine (El-Sayed, 2014) can increase the efficiency of intestinal lipid transport and promote lipid absorption. Consistent with these observations, our results suggest that low-dose 7S can promote lipid utilization, inhibit fat production, and avoid inflammation caused by lipid deposition in hybrid groupers.
However, high-dose 7S negatively affected the distal intestine of hybrid grouper from a different perspective. Our results showed that high-dose 7S had negative effects on “cysteine and methionine metabolism,” “AMPK signaling pathway,” “galactose metabolism,” “starch and sucrose metabolism,” “ABC transporters,” and “amino sugar and nucleotide sugar metabolism” and a positive effect on “ferroptosis.” Quercetin could inhibit oxidative stress and inflammatory responses by regulating the AMPK/SIRT1/NF-κB signaling pathway (Zhang et al., 2020), indicating that high-dose 7S might inhibit the absorption and utilization of carbohydrates in the feed of hybrid grouper, resulting in insufficient energy supply. Ferroptosis is caused by the accumulation of reactive oxygen radicals on membrane lipids because of the failure of glutathione peroxidase activity (Doll et al., 2016). Similar to the transcriptome annotation results, in the metabolome, we also found that the “ferroptosis” was activated by high doses of 7S, which further suggests that high-dose 7S could disrupt the normal function of intestinal cells by inducing ferroptosis. Most ABC transporters are extremely energy-dependent and require the transport of various endogenous substrates and xenobiotics across the lipid bilayer via ATP (Popovic et al., 2010). We hypothesize that the negative effect on cell signaling in hybrid grouper is due to 7S inhibition of the uptake and utilization of carbohydrates, such as sucrose, raffinose, and α,α-trehalose.
After supplementation with NaB in the context of high-dose 7S, the DMs closely related to essential amino acid metabolism in hybrid groupers, such as urocanic acid, betaine aldehyde, choline, creatinine, and L-arginine, were all upregulated. Consequently, the uptake and utilization of protein hydrolysis products by hybrid grouper were improved. One of the reasons why NaB can be utilized in livestock animals is that it can supply energy to intestinal epithelial cells (Huang et al., 2015). ATP is required for the proper functioning of ABC transporters, which suggests that the addition of NaB to the feed of hybrid grouper can effectively supply energy to the intestinal cells and, thus, promote the efficiency of ABC transporters and biosynthesis of amino acids.
The intestinal microbiota co-evolved with the host and plays an important role in host nutrient absorption, metabolism, information transfer, and disease infection (Carlos Magno Da Costa et al., 2015; Jones and Guillemin, 2018; Eckel, 2021; Post et al., 2021). Imbalance of intestinal microbiota can disrupt the intestinal internal environment, which is an important site for nutrient absorption and digestion (Liu et al., 2020). This leads to the excretion of unabsorbed nitrogen and phosphorus from the feed, causing pollution of farm water (Wu et al., 2019). In this study, the changes in the abundance of Ralstonia were significantly positively correlated with the ammonia nitrogen content, indicating that the increase in Ralstonia relative abundance may be the main reason for the high TN content in water. Lactobacillus can enhance the uptake of amino acids mediated by PepT1 in in mice with spontaneous colitis (Chen et al., 2010). Pediococcus is able to secrete proteases that accelerate protein hydrolysis (Afriani et al., 2018). These results indicate that these bacteria might affect the hydrolysis and absorption processes of nitrogen-containing nutrients, such as proteins, in the diet of hybrid grouper. The abundance of Pediococcus, Pelagibacterium, and Lactococcus was significantly positively correlated with the expression of ccnf, Numa1, pkb, spc25, and Cks1b genes associated with cell proliferation, suggesting that these bacteria may play important roles in the proliferation of distal intestinal cells in hybrid grouper. The changes in the abundance of Prevotella were significantly negatively correlated with nitrite content and significantly positively correlated with methylimidazole acetic acid and N-acetyl histamine levels. Prevotella has proteolytic activity, a function similar to that of exopeptidases, with positive effects on protein degradation and utilization of hydrolysis products (Griswold and Mackie, 1997), and plays an important role in carbohydrate utilization (Durb‘An et al., 2013; Aakko et al., 2020), indicating that the decrease in Prevotella abundance caused by high-dose β-conglycinin had a more serious negative effect on the response to nitrogenous nutrients and “carbohydrate utilization and metabolism” in hybrid groupers. The decrease in the abundance of Anaerovibrio was significantly negatively correlated with nitrite content and significantly positively correlated with methylimidazole acetic acid, N-acetyl histamine, urocanic acid, creatinine, glutathione, and S-adenosylhomocysteine levels. Anaerovibrio mainly utilizes lipids in the intestine, yet its abundance is highly correlated with metabolites associated with protein absorption and metabolism, which may be related to hindering the utilization of lipids by Anaerovibrio to produce unsaturated fatty acids (Castagnino et al., 2015), thereby reducing protein absorption by the host. The increase in the abundance of Pelagibacterium was significantly negatively correlated with nitrite level and positively correlated with methylimidazole acetic acid level, indicating that low-dose 7S may promote protein absorption and metabolism in hybrid grouper and, thus, reduce nitrogen emissions. The increase in the abundance of Burkholderia-Caballeronia-Paraburkholderia in bHNaB was significantly positively correlated with taurine, hydroxyproline, nervonic acid, stearic acid, docosanoic acid, L-cystine, and D-serine levels, suggesting that supplementation with NaB significantly improved lipid utilization and biosynthesis in hybrid grouper possibly regulated by Burkholderia-Caballeronia-Paraburkholderia. A number of highly relevant bacteria, genes, and metabolites were selected separately in this study, which could serve as potential biomarkers for evaluating the regulation of 7S and NaB in the intestine of hybrid grouper.
In this study, altered intestinal flora affected aquaculture water quality, host transcription, and metabolism, thus, affecting host health. Two bacteria, Ralstonia and Rothia, had negative effects on water quality, and Lactococcus, Pelagibacterium, Anaerovibrio, Prevotella, and Bacillus had positive effects. Low-dose 7S significantly increased the relative abundance of Pelagibacterium, Pediococcus, and Staphylococcus. High-dose 7S increased the probability of distal intestinal inflammation by increasing the relative abundance of pathogenic bacteria Ralstonia and Photobacterium, while NaB effectively inhibited the relative abundance of pathogenic bacteria Ralstonia and Photobacterium. Gene expression also showed significant differences between the treatments. The pathways involved in the regulation of low-dose 7S include “ribosome,” “protein digestion and absorption,” and “PPAR signaling” to promote protein synthesis and lipid uptake in hybrid grouper; the pathways involved in the regulation of high-dose 7S include “cell cycle,” “cytokine–cytokine receptor interaction,” and “PPAR signaling,” which interfered with the normal nutrient absorption function of intestinal cells in hybrid grouper. The pathways involved in the regulation of NaB supplementation include “protein processing in endoplasmic reticulum,” “cell cycle,” and “linoleic acid metabolism.” Distal intestinal metabolism was observed, and low-dose 7S mainly affected pathways associated with protein absorption, metabolism, and lipid utilization and biosynthesis; high-dose 7S mainly affected pathways associated with carbohydrate utilization and metabolism; and NaB supplementation contributed to protein absorption and metabolism. In addition, 15 DM markers were identified, including UDP-N-acetylglucosamine, L-fucose, sucrose, α,α-trehalose, quercetin, raffinose, methylimidazole acetic acid, N-acetyl histamine, urocanic acid, creatinine, S-adenosylhomocysteine, and choline. Above all, we initially predicted some potential biomarkers associated with water pollutants, host microbiota and genes based on Spearman association analysis, proving that cell proliferation and utilization of three major nutrients (protein, lipid, carbohydrate) might be potential targets for treating the negative effects caused by 7S in hybrid grouper, but further in-depth validation is highly imperative.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal protocol was approved by the Ethics Review Board of Guangdong Ocean University. All procedures were performedaccording to the standards of the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication No. 8023, revised 1978) and relevant Chinese policies.
All of the indicated authors have actively contributed to this study. HL and BT designed the study. BY conducted the study and analyzed the data. XD participated in the interpretation of results. BY wrote the manuscript. QY, SC, and SZ purchased the reagents. HL revised the manuscript. All authors have read and approved the final version of the manuscript.
This work was supported by the National Key R&D Program of China (Grant Number: 2019YFD0900200), the National Natural Science Foundation of China (Grant Number: 31772864), and the Natural Science Foundation of Guangdong Province (Grant Numbers: 2018A030313154 and 2020A1515011129).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Shanghai Menon Animal Nutrition Technology Co., Ltd. (Shanghai, China) for kindly providing the sodium butyrate in this experiment.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.705332/full#supplementary-material
Supplementary Figure 1 | Rarefaction curves of the samples.
Supplementary Figure 2 | Differences in hybrid grouper bacterial diversity and richness.
Supplementary Figure 3 | Differences analysis among the groups. (A) LefSe cladogram. (B) LDA score (3.90) of LEfSe.
Supplementary Figure 4 | The statistics on the number of significantly different genes in different comparison objects.
Supplementary Figure 5 | Volcano plots of the DEGs in the distal intestine of hybrid grouper. (A) FMb vs. bL. (B) FMb vs. bH. (C) bH vs. bHNaB.
Supplementary Figure 6 | Comparison of the RT-PCR and RNA Seq results.
Supplementary Figure 7 | The LC-MS spectra of the distal intestine contents in positive (A) and negative (B) modes.
Supplementary Figure 8 | Partial least squares discriminant analysis (PLS-DA) plot of distal intestinal of the FMb and bL following positive (A1,A2) and negative (A3,A4) ions; FMb and bH following positive (B1,B2) and negative (B3,B4) ions; bH and bHNaB following positive (C1,C2) and negative (C3,C4) ions.
Supplementary Figure 9 | ROC analysis for discrimination among the groups for the potential biomarker metabolites in positive and negative modes.
Supplementary Table 1 | Composition of the diets.
Supplementary Table 2 | Amino acid profile (%) of the diets used in the experiment.
Supplementary Table 3 | Primers sequences used for real-time quantitative PCR.
Supplementary Table 4 | Growth parameters and feed utilization of juvenile hybrid grouper fed the experimental diets for 8 weeks.
Aakko, J., Pietil, S., Toivonen, R., Rokka, A., Mokkala, K., Laitinen, K., et al. (2020). A carbohydrate-active enzyme (CAZy) profile links successful metabolic specialization of Prevotella to its abundance in gut microbiota. Sci. Rep. 10:12411.
Afriani, Arnim, Marlida, Y., and Yuherman (2018). Isolation and characterization of lactic acid bacteria proteases from Bekasam for use as a beef tenderizer. Pak. J. Nutr. 17, 361–367. doi: 10.3923/pjn.2018.361.367
Albino, M. R., Souza, C. L. D., Damiani, T. C., Alberto, R. H., Silva, V. F., Fontana, V. M., et al. (2012). Sodium butyrate decreases the activation of NF-κB reducing inflammation and oxidative damage in the kidney of rats subjected to contrast-induced nephropathy. Nephrol. Dial. Transplant. 27, 3136–3140. doi: 10.1093/ndt/gfr807
Allonso, D., Andrade, I. S., Conde, J. N., Coelho, D. R., Rocha, D. C. P., Silva, M. L. D., et al. (2015). Dengue virus NS1 protein modulates cellular energy metabolism by increasing glyceraldehyde-3-phosphate dehydrogenase activity. J. Virol. 89, 11871–11883. doi: 10.1128/jvi.01342-15
Ashton-Rickardt, P. G. (2009). Serine protease inhibitors and T lymphocyte immunity. Curr. Immunol. Rev. 5, 187–199. doi: 10.2174/157339509788921256
Bertaggia, E., Jensen, K. K., Castro-Perez, J., Xu, Y., Paolo, G. D., Chan, R. B., et al. (2017). Cyp8b1 ablation prevents western diet-induced weight gain and hepatic steatosis due to impaired fat absorption. Am. J. Physiol. Endocrinol. Metab. 2, E121–E133.
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Calejman, C. M., Trefely, S., Entwisle, S. W., Luciano, A. K., and Guertin, D. A. (2020). mTORC2-AKT signaling to ATP-citrate lyase drives brown adipogenesis and de novo lipogenesis. Nat. Commun. 11, 1–16.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336.
Carlos Magno Da Costa, M., Sandra Bertelli Ribeiro De, C., Gustavo Torres De, S., Cristiano, R., Francisco Carlos Da, G., Maria Anete Santana, V., et al. (2015). Intestinal microbiota as modulators of the immune system and neuroimmune system: impact on the host health and homeostasis. J. Immunol. Res. 2015:931574.
Castagnino, P. S., Messana, J. D., Fiorentini, G., Jesus, R. D., Vito, E. S., Carvalho, I., et al. (2015). Glycerol combined with oils did not limit biohydrogenation of unsaturated fatty acid but reduced methane production in vitro. Anim. Feed Sci. Technol. 201, 14–24. doi: 10.1016/j.anifeedsci.2014.12.004
Chen, H. Q., Shen, T. Y., Zhou, Y. K., Zhang, M., Chu, Z. X., Hang, X. M., et al. (2010). Lactobacillus plantarum consumption increases PepT1-mediated amino acid absorption by enhancing protein kinase C activity in spontaneously colitic mice. J. Nutr. 140, 2201–2206. doi: 10.3945/jn.110.123265
Chen, S. F., Zhou, Y. Q., Chen, Y. R., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890.
China, S. A. O. (2007). ”PRC National Standard”, in: The specification for marine monitoring-Part 4: Seawater analysis. China: State Administration for Market Regulation.
Claudio, F. D., Muglia, C. I., Smaldini, P. L., Delgado, M. L. O., Trejo, F. M., Grigera, J. R., et al. (2017). Use of a collagen membrane to enhance the survival of primary intestinal epithelial cells. J. Cell. Physiol. 232, 2489–2496. doi: 10.1002/jcp.25594
D’Angiolella, V., Donato, V., Forrester, F. M., Jeong, Y.-T., Pellacani, C., Kudo, Y., et al. (2012). Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell 149, 1023–1034. doi: 10.1016/j.cell.2012.03.043
Dauloudet, O., Neri, I., Walter, J. C., Dorignac, J., Geniet, F., and Parmeggiani, A. (2020). Modelling the effect of ribosome mobility on the rate of protein synthesis. Eur. Phys. J. E 44, 60–66.
Doll, S., Proneth, B., Tyurina, Y. Y., Panzilius, E., Kobayashi, S., Ingold, I., et al. (2016). ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 13, 91–98. doi: 10.1038/nchembio.2239
Dong, O. S., Satsu, H., and Shimizu, M. (2005). Histidine inhibits oxidative stress- and TNF-α-induced interleukin-8 secretion in intestinal epithelial cells. FEBS Lett. 579, 4671–4677. doi: 10.1016/j.febslet.2005.07.038
Dong, Y. W., Jiang, W. D., Liu, Y., Wu, P., Jiang, J., Kuang, S. Y., et al. (2017). Threonine deficiency decreased intestinal immunity and aggravated inflammation associated with NF-kappa B and i signalling pathways in juvenile grass carp (Ctenopharyngodon idella) after infection with Aeromonas hydrophila. Br. J. Nutr. 118, 92–108. doi: 10.1017/s0007114517001830
Duan, Y. F., Zhang, Y., Dong, H. B., Wang, Y., Zheng, X. T., and Zhang, J. S. (2017). Effect of dietary Clostridium butyricum on growth, intestine health status and resistance to ammonia stress in Pacific white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 65, 25–33. doi: 10.1016/j.fsi.2017.03.048
Durb‘An, A., Abell‘An, J., Latorre, A., and Moya, A. (2013). Effect of dietary carbohydrate restriction on an obesity-related Prevotella-dominated human faecal microbiota. Metagenomics 2, c1–c4.
Eckel, J. (2021). Intestinal microbiota and host metabolism a complex relationship. Acta Physiol. 232:13638.
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
El-Sayed, A. F. M. (2014). Is dietary taurine supplementation beneficial for farmed fish and shrimp? a comprehensive review. Rev. Aquaculture 6, 241–255. doi: 10.1111/raq.12042
Fu, J. H., Li, G. F., Wu, X. M., and Zang, B. (2019). Sodium butyrate ameliorates intestinal injury and improves survival in a rat model of cecal ligation and puncture-induced sepsis. Inflammation 42, 1276–1286. doi: 10.1007/s10753-019-00987-2
Fu, L. L., Song, J. Y., Wang, C., Fu, S. J., and Wang, Y. B. (2017). Bifidobacterium infantis potentially alleviates shrimp tropomyosin-induced allergy by tolerogenic dendritic cell-dependent induction of regulatory T cells and alterations in gut microbiota. Front. Immunol. 8:1536. doi: 10.3389/fimmu.2017.01536
Gao, Y. J., Yang, H. J., Liu, Y. J., Chen, S. J., Guo, D. Q., Yu, Y. Y., et al. (2014). Effects of graded levels of threonine on growth performance, biochemical parameters and intestine morphology of juvenile grass carp Ctenopharyngodon idella. Aquaculture 42, 113–119. doi: 10.1016/j.aquaculture.2013.12.043
Garrido, D., Bourouh, M., Bonneil, R., Thibault, P., and Archambault, V. (2020). Cyclin B3 activates the anaphase-promoting complex/cyclosome in meiosis and mitosis. PLoS Genet. 16:e1009184. doi: 10.1371/journal.pgen.1009184
Griffith, W. G., and Shaw, D. S. (1998). Polymorphisms in Phytophthora infestans: four mitochondrial haplotypes are detected after PCR amplification of DNA from pure cultures or from host lesions. Appl. Environ. Microbiol. 64, 4007–4014. doi: 10.1128/aem.64.10.4007-4014.1998
Griswold, K. E., and Mackie, R. I. (1997). Degradation of protein and utilization of the hydrolytic products by a predominant ruminal bacterium, Prevotella ruminicola B1(4). J. Dairy Sci. 80:167. doi: 10.3168/jds.s0022-0302(97)75924-1
Guo, X. Z., Ran, C., Zhang, Z., He, S. X., Jin, M., and Zhou, Z. G. (2017). The growth-promoting effect of dietary nucleotides in fish is associated with an intestinal microbiota-mediated reduction in energy expenditure. J. Nutr. 147, 781–788. doi: 10.3945/jn.116.245506
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hua, K., and Bureau, D. P. (2019). Estimating changes in essential amino acid requirements of rainbow trout and Atlantic salmon as a function of body weight or diet composition using a novel factorial requirement model. Aquaculture 513, 734440–734440. doi: 10.1016/j.aquaculture.2019.734440
Huang, C., Song, P. X., Fan, P. X., Hou, C. L., Thancker, P., and Ma, X. (2015). Dietary sodium butyrate decreases postweaning diarrhea by modulating intestinal permeability and changing the bacterial communities in weaned piglets. J. Nutr. 145, 2774–2780. doi: 10.3945/jn.115.217406
Huang, Y. S., Lin, Z. D., Rong, H., Hao, M. L., Zhu, D. S., Li, S. K., et al. (2018). Effects of conjugated linoleic acid on growth, body composition, antioxidant status, lipid metabolism and immunity parameters of juvenile Chu’s croaker, Nibea coibor. Aquac. Res. 49, 546–556. doi: 10.1111/are.13486
Jesus, G. F. A., Pereira, S. A., Owatari, M. S., Nicholas, S., Silva, B. C., Allan, S., et al. (2018). Protected forms of sodium butyrate improve the growth and health of Nile tilapia fingerlings during sexual reversion. Aquaculture 499:S0044848618310305.
Jiang, S., Wu, X., Luo, Y., Wu, M., Lu, S., Jin, Z., et al. (2016). Optimal dietary protein level and protein to energy ratio for hybrid grouper (Epinephelus fuscoguttatus ♀×Epinephelus lanceolatus ♂) juveniles. Aquaculture 465, 28–36. doi: 10.1016/j.aquaculture.2016.08.030
Jiang, X. W., Huo, Y. Q., Cheng, H., Zhang, X. Q., and Wu, Z. M. (2012). Cloning, expression and characterization of a halotolerant esterase from a marine bacterium Pelagibacterium halotolerans B2T. Extremophiles 16, 427–435. doi: 10.1007/s00792-012-0442-3
Jiang, Y., Wang, Y., Zhang, Z., Liao, M., and Chen, G. (2019). Responses of microbial community structure in turbot (Scophthalmus maximus) larval intestine to the regulation of probiotic introduced through live feed. PLoS One 14:e0216590. doi: 10.1371/journal.pone.0216590
Jones, T. A., and Guillemin, K. (2018). Racing to stay put: how resident microbiota stimulate intestinal epithelial cell proliferation. Curr. Pathobiol. Rep. 6, 1–6.
Katebi, A. R., and Jernigan, R. L. (2015). Aldolases utilize different oligomeric states to preserve their functional dynamics. Biochemistry 54, 3543–3554. doi: 10.1021/acs.biochem.5b00042
Khan, M. A. (2018). Histidine requirement of cultivable fish species: a review. Oceanogr. Fish. 8, 1–7.
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, 1–13.
Klohs, W. D., Wilson, J. R., Weiser, M. M., Frankfurt, O., and Bernacki, R. J. (2010). Galactosyltransferase activity and cell growth: uridine diphosphate (UDP) galactose inhibition of murine leukemic. J. Cell. Physiol. 119, 23–28. doi: 10.1002/jcp.1041190105
Krzysztof, G., Maciej, U., Jaroslaw, S., Kedzierska, A. E., Rafal, K., Anna, J., et al. (2017). The novel type 1 fimbriae fimh receptor calreticulin plays a role in Salmonella host specificity. Front. Cell. Infect. Microbiol. 7:326. doi: 10.3389/fcimb.2017.00326
Kwon, M. S., Ki, L. S., Jang, J. Y., Lee, J., Kyeong, P. H., Kim, N. H., et al. (2018). Lactobacillus sakei WIKIM30 ameliorates atopic dermatitis-like skin lesions by inducing regulatory t cells and altering gut microbiota structure in mice. Front. immunol. 9:1905. doi: 10.3389/fimmu.2018.01905
Lallemand, F., Courilleau, D., Fagot, B., Atfi, A., Montagne, M. N., and Mester, J. (1999). Sodium butyrate induces G2 arrest in the human breast cancer cells MDA-MB-231 and renders them competent for DNA rereplication. Exp. Cell Res. 247, 432–440. doi: 10.1006/excr.1998.4370
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Leung, K.-C. (2004). Regulation of cytokine receptor signaling by nuclear hormone receptors: a new paradigm for receptor interaction. DNA Cell Biol. 23, 463–474. doi: 10.1089/1044549041562285
Li, C. (2013). Effects of n-3 HUFA on Growth Performance, Lipid Metabolism, and Health Status in Grass Carp, Ctenopharyngodon Idellus. Xianyang: Northwest A&F University.
Li, P., and Wu, G. Y. (2018). Roles of dietary glycine, proline, and hydroxyproline in collagen synthesis and animal growth. Amino Acids 50, 29–38. doi: 10.1007/s00726-017-2490-6
Li, R. M. (2019). The Effects of Inositol and Zinc in Low-Fishmeal Dietary on Nutritional Physiological Functions for Juvenile L. vannamei and Epinephelus lanceolatu ♂× Epinephelus Fuscoguttatus ♀, Respectively. Zhanjiang: Guangdong Ocean University.
Li, Y. Y., Wang, T., Gao, S., Xu, G. M., Niu, H., Huang, R., et al. (2016). Salmonella plasmid virulence gene spvB enhances bacterial virulence by inhibiting autophagy in a zebrafish infection model. Fish Shellfish Immunol. 49, 252–259. doi: 10.1016/j.fsi.2015.12.033
Liu, F. X., and Yu, B. (2015). Efficient production of reuterin from glycerol by magnetically immobilized Lactobacillus reuteri. Appl. Microbiol. Biotechnol. 99, 4659–4666. doi: 10.1007/s00253-015-6530-4
Liu, H. R., Liu, Y., Song, C. W., and Cui, Z. X. (2017). A chymotrypsin-like serine protease from Portunus trituberculatus involved in pathogen recognition and AMP synthesis but not required for prophenoloxidase activation. Fish Shellfish Immunol. 66, 307–316. doi: 10.1016/j.fsi.2017.05.031
Liu, Q., Long, Y., Li, B., Zhao, L., and Yang, S. (2020). Rice-shrimp culture: a better intestinal microbiota, immune enzymatic activities, and muscle relish of crayfish (Procambarus clarkii) in sichuan province. Appl. Microbiol. Biotechnol. 104, 9413–9420. doi: 10.1007/s00253-020-10797-4
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, J. J. (2008). Effect of Dietary n-3 HUFA on Growth Performance and Lipid Metabolism in Juvenile Black, Sparus macrocephalus. Zhejiang: Zhejiang University.
Magoè, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Makol, A., Torrecillas, S., Caballero, M. J., Fernández-Vaquero, A., and Izquierdo, M. S. (2012). Effect of long term feeding with conjugated linoleic acid (CLA) in growth performance and lipid metabolism of European sea bass (Dicentrarchus labrax). Aquaculture 36, 129–137. doi: 10.1016/j.aquaculture.2012.09.015
Matsumoto, M., Kibe, R., Ooga, T., Aiba, Y., and Benno, Y. (2012). Impact of intestinal microbiota on intestinal luminal metabolome. Sci. Rep. 2:233.
Miao, S. Y., Zhao, C. Z., Zhu, J. Y., Hu, J. T., Dong, X. J., and Sun, L. S. (2018). Dietary soybean meal affects intestinal homoeostasis by altering the microbiota, morphology and inflammatory cytokine gene expression in northern snakehead. Sci. Rep. 8:113.
Michelato, M., Zaminhan, M., Boscolo, W. R., Nogaroto, V., Vicari, M., Artoni, R. F., et al. (2017). Dietary histidine requirement of Nile tilapia juveniles based on growth performance, expression of muscle-growth-related genes and haematological responses. Aquaculture 467, 63–70. doi: 10.1016/j.aquaculture.2016.06.038
Mirghaed, A. T., Yarahmadi, P., Soltani, M., Paknejad, H., and Hoseini, S. M. (2019). Dietary sodium butyrate (Butirex C4) supplementation modulates intestinal transcriptomic responses and augments disease resistance of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunl. 92, 621–628. doi: 10.1016/j.fsi.2019.06.046
Nascimento, T. M. T. D., Mansano, C. F. M., Peres, H., Rodrigues, F. H. F., Khan, K. U., Romaneli, R. S., et al. (2020). Determination of the optimum dietary essential amino acid profile for growing phase of Nile tilapia by deletion method. Aquaculture 523:735204. doi: 10.1016/j.aquaculture.2020.735204
Ogawa, T., Bando, N., Tsuji, H., Okajima, H., Nishikawa, K., Sasaoka, K., et al. (1991). Investigation of the IgE-binding proteins in soybeans by immunoblotting with the sera of the soybean-sensitive patients with atopic dermatitis. J. Nutr. Sci. Vitaminol. 37, 555–565. doi: 10.3177/jnsv.37.555
O’Hara, E., Kelly, A., Mccabe, M., and Kenny, D. (2018). Effect of a butyrate-fortified milk replacer on gastrointestinal microbiota and products of fermentation in artificially reared dairy calves at weaning. Sci. Rep. 8:14901.
Ortiz-Rivera, Y., Sanchez-Vega, R., Gutierrez-Mendez, N., Leon-Felix, J., Acosta-Muniz, C., and Sepulveda, D. R. (2017). Production of reuterin in a fermented milk product by Lactobacillus reuteri: inhibition of pathogens, spoilage microorganisms, and lactic acid bacteria. J. Dairy Sci. 100, 4258–4268. doi: 10.3168/jds.2016-11534
Ostaszewska, T., Dabrowski, K., Kamaszewski, M., Grochowski, P., Verri, T., Rzepkowska, M. G., et al. (2010). The effect of plant protein-based diet supplemented with dipeptide or free amino acids on digestive tract morphology and PepT1 and PepT2 expressions in common carp (Cyprinus carpio L.). Comp. Biochem. Physiol., Part A 157, 158–169. doi: 10.1016/j.cbpa.2010.06.162
Page, A. J., Brierley, S. M., Martin, C. M., Price, M. P., and Blackshaw, L. A. (2005). Different contributions of ASIC channels 1a, 2, and 3 in gastrointestinal mechanosensory function. Gut 54, 1408–1415. doi: 10.1136/gut.2005.071084
Pan, F. Y., Feng, L., Jiang, W. D., Jiang, J., Wu, P., Kuang, S. Y., et al. (2016). Methionine hydroxy analogue enhanced fish immunity via modulation of NF-kappa B, TOR, MLCK, MAPKs and Nrf2 signaling in young grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 56, 208–228. doi: 10.1016/j.fsi.2016.07.020
Petrov, A., Meskauskas, A., and Dinman, J. D. (2014). Ribosomal protein L3: influence on ribosome structure and function. RNA Biol. 1, 58–64. doi: 10.4161/rna.1.1.957
Pettersson, I., and Kurland, C. G. (1980). Ribosomal protein L7/L12 is required for optimal translation. PNAS 77, 4007–4010. doi: 10.1073/pnas.77.7.4007
Popovic, M., Zaja, R., Loncar, J., and Smital, T. (2010). A novel ABC transporter: the first insight into zebrafish (Danio rerio) ABCH1. Mar. Environ. Res. 69, S11–S13.
Post, S. V. D., Birchenough, G. M. H., and Held, J. M. (2021). NOX1-dependent redox signaling potentiates colonic stem cell proliferation to adapt to the intestinal microbiota by linking EGFR and TLR activation. Cell Rep. 35:108959.
Rice, E. M., Aragona, K., Moreland, S. C., and Erickson, P. S. (2019). Supplementation of sodium butyrate to postweaned heifer diets: effects on growth performance, nutrient digestibility, and health. J. Dairy Sci. 102, 3121–3130. doi: 10.3168/jds.2018-15525
Rosa, M. D., Brundo, V. M., and Malaguarnera, L. (2016). New insights on chitinases immunologic activities. World J. Immunol. 6:96. doi: 10.5411/wji.v6.i2.96
Saegusa, K., Sato, M., Sato, K., Nakajima-Shimada, J., Harada, A., and Sato, K. (2014). Caenorhabditis elegans chaperonin CCT/TRiC is required for actin and tubulin biogenesis and microvillus formation in intestinal epithelial cells. Mol. Biol. Cell 25, 3095–3104. doi: 10.1091/mbc.e13-09-0530
Sanz-Gómez, N., Pedro, I. D., Ortigosa, B., Santamaria, D., and Gandarillas, A. (2020). Squamous differentiation requires G2/mitosis slippage to avoid apoptosis. Cell Death. Differ. 27, 2451–2467. doi: 10.1038/s41418-020-0515-2
Sarraf, S. A., Sideris, D. P., Giagtzoglou, N., Ni, L. N., and Pickrell, A. M. (2019). PINK1/Parkin influences cell cycle by sequestering TBK1 at damaged mitochondria, inhibiting mitosis. Cell Rep. 29, 225.e5–235.e5.
Schloss, P. D., Gevers, D., and Westcott, S. L. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. doi: 10.1371/journal.pone.0027310
Shin, N. R., Whon, T. W., and Bae, J. W. (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503. doi: 10.1016/j.tibtech.2015.06.011
Shiu, Y. L., Hsieh, S. L., Guei, W. C., Tsai, Y. T., Chiu, C. H., and Liu, C. H. (2015). Using Bacillus subtilis E20-fermented soybean meal as replacement for fish meal in the diet of orange: potted grouper (Epinephelus coioides, Hamilton). Aquac. Res. 46, 1–14. doi: 10.1016/j.aquaculture.2015.10.020
Sinha, A. K., Possoz, C., and Leach, D. R. F. (2020). The roles of bacterial DNA double-strand break repair proteins in chromosomal DNA replication. FEMS Microbiol. Rev. 44, 351–368. doi: 10.1093/femsre/fuaa009
Smith, A. A., Dumas, A., Yossa, R., Overturf, K. E., and Bureau, D. P. (2017). Effects of soybean meal and high-protein sunflower meal on growth performance, feed utilization, gut health and gene expression in Arctic charr (Salvelinus alpinus) at the grow-out stage. Aquac. Nutr. 481, 191–201. doi: 10.1016/j.aquaculture.2017.08.038
Smith, D. M., Hunter, B. J., Allan, G. L., Roberts, D. C. K., Booth, M. A., and Glencross, B. D. (2004). Essential fatty acids in the diet of silver perch (Bidyanus bidyanus): effect of linolenic and linoleic acid on growth and survival. Aquaculture 236, 377–390. doi: 10.1016/j.aquaculture.2003.10.021
Tantikitti, C., Sangpong, W., and Chiavareesajja, S. J. A. (2005). Effects of defatted soybean protein levels on growth performance and nitrogen and phosphorus excretion in Asian seabass (Lates calcarifer). Aquaculture 248, 41–50. doi: 10.1016/j.aquaculture.2005.04.027
Tian, L., Zhou, X. Q., Jiang, W. D., Liu, Y., Wu, P., Jiang, J., et al. (2017). Sodium butyrate improved intestinal immune function associated with NF-kappa B and p38MAPK signalling pathways in young grass carp (Ctenopharyngodon idella). Fish Shellfish Immun. 66:548. doi: 10.1016/j.fsi.2017.05.049
Tzuc, J. T., Escalante, D. R., Herrera, R. R., Cortés, G. G., and Ortiz, M. L. A. (2014). Microbiota from Litopenaeus vannamei: digestive tract microbial community of Pacific white shrimp (Litopenaeus vannamei). Springer Plus 3:280. doi: 10.1186/2193-1801-3-280
Ueta, M., Wada, C., Bessho, Y., Maeda, M., and Wada, A. (2017). Ribosomal protein L31 in Escherichia coli contributes to ribosome subunit association and translation, whereas short L31 cleaved by protease 7 reduces both activities. Genes Cells 22, 452–471. doi: 10.1111/gtc.12488
Uymaz, B., Şimşek, Ö, Akkoç, N., and Akçelik, H. A. (2009). In vitro characterization of probiotic properties ofPediococcus pentosaceus BH105 isolated from human faeces. Ann. Microbiol. 59, 485–491. doi: 10.1007/bf03175135
Vunnam, S., Juvvadi, P., and Merrifield, R. B. (2010). Synthesis and antibacterial action of cecropin and proline-arginine-rich peptides from pig intestine. J. Rept. Res. 49, 59–66. doi: 10.1111/j.1399-3011.1997.tb01121.x
Waesch, R., and Cross, F. R. (2002). APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature 418, 556–562. doi: 10.1038/nature00856
Wahlström, A., Sayin, Sama, I., Marschall, H. U., and Bäckhed, F. (2016). Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24, 41–50. doi: 10.1016/j.cmet.2016.05.005
Wang, D., Naydenov, N. G., Feygin, A., Baranwal, S., Kuemmerle, J. F., and Ivanov, A. I. (2016). Actin-depolymerizing factor and cofilin-1 have unique and overlapping functions in regulating intestinal epithelial junctions and mucosal inflammation. Am. J. Pathol. 26, 844–858. doi: 10.1016/j.ajpath.2015.11.023
Wang, Y. R., Wang, L., Zhang, C. X., and Song, K. (2017). Effects of substituting fishmeal with soybean meal on growth performance and intestinal morphology in orange-spotted grouper (Epinephelus coioides). Aquac. Rep. 5, 52–57. doi: 10.1016/j.aqrep.2016.12.005
Wintersberger, E., Mudrak, I., and Wintersberger, U. (2010). Butyrate inhibits mouse fibroblasts at a control point in the G1 phase. J. Cell. Biochem. 21, 239–247. doi: 10.1002/jcb.240210306
Wu, D., and Cui, J. (2014). Ion channels in the cell membrane: structure, function, and modeling. J. Physiol. 554(Pt 2), 245–247.
Wu, P., Huo, P., Wang, Y., Dong, Y., and Wu, X. (2019). Effluent containing Rubrivivax gelatinosus improving the yield, digestion system, intestinal microbiota of crucian carp. Aquaculture 514:734418. doi: 10.1016/j.aquaculture.2019.734418
Wu, S. Y., Wang, L. D., Li, J. L., Xu, G. M., He, M. L., Li, Y. Y., et al. (2016). Salmonella spv locus suppresses host innate immune responses to bacterial infection. Fish Shellfish Immunol. 58, 387–396. doi: 10.1016/j.fsi.2016.09.042
Xie, X. H., Lin, W. Z., Zheng, W. L., Chen, T., Yang, H. T., Sun, L. J., et al. (2019). Downregulation of G2/mitotic-specific cyclinB1 triggers autophagy via AMPK-ULK1-dependent signal pathway in nasopharyngeal carcinoma cells. Cell Death Dis. 10:94.
Yang, P., Hu, H., Liu, Y., Li, Y., Ai, Q., Xu, W., et al. (2018). Dietary stachyose altered the intestinal microbiota profile and improved the intestinal mucosal barrier function of juvenile turbot, Scophthalmus maximus L. Aquaculture 486, 98–106. doi: 10.1016/j.aquaculture.2017.12.014
Yang, W. S., and Stockwell, B. R. (2016). Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 26, 165–176. doi: 10.1016/j.tcb.2015.10.014
Yang, W. X., Shi, W. Z., Zhou, S. N., Qu, Y. H., and Wang, Z. H. (2019). Research on the changes of water-soluble flavor substances in grass carp during steaming. J. Food Biochem. 43:e12993.
Yang, Y. H., Wang, Y. Y., Lu, Y., and Li, Q. Z. (2011). Effect of replacing fish meal with soybean meal on growth, feed utilization and nitrogen and phosphorus excretion on rainbow trout (Oncorhynchus mykiss). Aquacult. Int. 19, 405–419. doi: 10.1007/s10499-010-9359-y
Zabirnyk, O., Liu, W., Khalil, S., Sharma, A., and Phang, J. M. (2010). Oxidized low-density lipoproteins upregulate proline oxidase to initiate ROS-dependent autophagy. Carcinogenesis 31, 446–454. doi: 10.1093/carcin/bgp299
Zhang, C. X., Wang, Z. Y., Li, Z., Shen, Q. W., Chen, L. J., Gao, L. L., et al. (2016). Phosphoproteomic profiling of myofibrillar and sarcoplasmic proteins of muscle in response to salting. Food Sci. Biotechnol. 25, 993–1001. doi: 10.1007/s10068-016-0161-0
Zhang, F. W., Feng, J., Zhang, J. Y., Kang, X., and Qian, D. (2020). Quercetin modulates AMPK/SIRT1/NF-κB signaling to inhibit inflammatory/oxidative stress responses in diabetic high fat diet-induced atherosclerosis in the rat carotid artery. Exp. Ther. Med. 20:280.
Zhang, J. X., Guo, L. Y., Feng, L., Jiang, W. D., Kuang, S. Y., Liu, Y., et al. (2013). Soybean β-conglycinin induces inflammation and oxidation and causes dysfunction of intestinal digestion and absorption in fish. PLoS One 8:e58115. doi: 10.1371/journal.pone.0058115
Zhang, W. (2020). Study on the Differential Mechanism of Intestinal Mucosal Barrier Damage in Epinephelus fuscoguttatus♀×E. lanceolatus ♂ Caused by Three Kinds of Soybean Protein. Zhanjiang: Guangdong Ocean University.
Zhang, W., Tan, B. P., Ye, G. L., Wang, J. X., and Zhang, H. T. (2019). Identification of potential biomarkers for soybean meal-induced enteritis in juvenile pearl gentian grouper, Epinephelus lanceolatus ♂× Epinephelus fuscoguttatus ♀. Aquaculture 512:734337. doi: 10.1016/j.aquaculture.2019.734337
Zhao, X. L., Han, Y., Ren, S. T., Ma, Y. M., Li, H., and Peng, X. X. (2015). L-proline increases survival of tilapias infected by Streptococcus agalactiae in higher water temperature. Fish Shellfish Immunol. 44, 33–42. doi: 10.1016/j.fsi.2015.01.025
Zhao, Y., Qin, G. X., Han, R., Wang, J., Zhang, X. D., and Liu, D. D. (2014). β-Conglycinin reduces the tight junction occludin and ZO-1 expression in IPEC-J2. Int. J. Mol. Sci. 15, 1915–1926. doi: 10.3390/ijms15021915
Zhou, F. (2011). Studies on the Effect of Dietary Lysine and Arginine on the Growth of Juvenile Black Seabream (Acanthopagrus schlegelii) and Their Antagonistic Effect Mechanism. Hangzhou: Zhejiang University.
Keywords: hybrid grouper (Epinephelus fuscoguttatus♀ × E. lanceolatus♂), β-conglycinin, sodium butyrate, intestinal microbiota, transcriptome, metabolome
Citation: Yin B, Liu H, Tan B, Dong X, Chi S, Yang Q and Zhang S (2021) Dynamic Alterations of the Distal Intestinal Microbiota, Transcriptome, and Metabolome of Hybrid Grouper by β -Conglycinin With Reconciliations by Sodium Butyrate in Feed. Front. Mar. Sci. 8:705332. doi: 10.3389/fmars.2021.705332
Received: 05 May 2021; Accepted: 23 June 2021;
Published: 19 July 2021.
Edited by:
Ikram Belghit, Norwegian Institute of Marine Research (IMR), NorwayReviewed by:
Fotini Kokou, Wageningen University and Research, NetherlandsCopyright © 2021 Yin, Liu, Tan, Dong, Chi, Yang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyu Liu, bGl1aHl1QGdkb3UuZWR1LmNu; Beiping Tan, YnB0YW5AMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.