
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Mar. Sci. , 10 August 2021
Sec. Marine Evolutionary Biology, Biogeography and Species Diversity
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.687860
This article is part of the Research Topic Benthic Biodiversity of the Indian Ocean View all 14 articles
The deep sea is the largest environment on Earth, comprising important resources of commercial interest. It is composed of a wide variety of ecosystems, which is home to often unique organisms that are yet to be described. The deep-sea is one of the least studied environments, where research is strongly linked to technological access and advances. With the recent advances in the next-generation sequencing and bioinformatics tools, there is an enhanced understanding of microbial diversity and ecological functions in deep sea. Multidisciplinary programs are being undertaken to investigate into microbial communities in diverse marine environments. As compared to other Oceans, the deeper parts of Indian Ocean are still poorly sampled and studied for bacterial, and more so fungal diversity. The studies reporting usage of modern sequencing tools to describe uncultured microbial diversity have seen a rise in numbers in the last decade. In this review, we summarize the important findings of research works carried on bacterial and fungal diversity from the abyssal regions of the Indian Ocean and provide our views on possible future paths.
The abyssal plain (also referred to as the deep-sea) is normally located between 3000 and 6000 m deep in the global ocean. It includes biogenic sediments, ocean waters from the bathypelagic (ranging from 1000 to 4000 m) and abyssopelagic (ranging from 4000 to 6000 m) zones, and occasionally manganese nodules (da Silva et al., 2013; Corinaldesi, 2015). The deep-sea environment plays important roles in nutrient cycling, carbon sequestration and biomass production (Barbier et al., 2014; Danovaro et al., 2014, 2017). Light intensity decreases with depth within this region, and photosynthetic production is arrested due to the absence of sunlight. The temperatures nearing freezing (–1°C to 4°C) and the paucity of organic matter at the seafloor bed are the key features of the deep sea that control benthic productivity and biomass (Jørgensen and Boetius, 2007). Characteristically, the biota of the deep-sea are “food limited,” since it is solely dependent on organic matter input from the photic zone (Danovaro et al., 2017).
Despite the “harsh” conditions, there is a large variety of deep-sea organisms. The diversity of deep-sea benthic communities has intrigued researchers. Over the last two decades, deep-sea explorations have led to the discovery of novel ecosystems and diverse microbes. Gartner et al. (2011) studied the effect of hydrostatic pressure on the microbial communities from the deep sea sediments of Eastern Mediterranean and revealed that Gamma-proteobacteria had selective advantage over others under mimicked nutrient supply in elevated pressure conditions. Marine microbial communities are important for the structure and dynamics of food webs, global biogeochemical cycles, and the remineralization of organic matter (Jørgensen and Boetius, 2007; Parvathi et al., 2020; Wei et al., 2020). The microbial processes occurring along the seafloor are essential in sustaining primary and secondary production in the water column. In deep-sea sediments, the most significant fraction of taxonomic richness and biomass is contributed by the Bacteria and Archaea, which represent around 90% of the total benthic biomass. Multidisciplinary efforts across the globe are undertaken to characterize the microbial communities in different marine environments. For example, the Sorcerer II Global Ocean Sampling expedition (2003–2004) carried out in the Atlantic and Pacific Oceans; the TARA Ocean expedition (2009–2013) carried out in the Atlantic, Pacific and Indian Ocean. Despite this, the deep-sea environment remains one of the most understudied ecosystems on earth.
The total microbial counts in the marine sediments is estimated to be 2.9 × 1029 – 5.4 × 1029 cells, contributing to 3.6% of the entire living biomass on Earth (Kallmeyer et al., 2012; Parkes et al., 2014), and bacterial endospores in the upper kilometer of marine sediment are estimated between 2.5 × 1028 and 1.9 × 1029 (Wörmer et al., 2019). Stratification of the microbial communities is dependent on the depth of the sediment; wherein cell counts are inversely proportional to depth (Inagaki et al., 2003; Parkes et al., 2005; Inagaki et al., 2006; Biddle et al., 2008; Fry et al., 2008; Teske, 2013; Ciobanu et al., 2014; Inagaki et al., 2015; Chen et al., 2017). Additionally, microbial abundance is inversely proportional to the age of marine sediments and is higher in organic-rich anoxic sediments than oligotrophic oxic sediments (Kallmeyer et al., 2012; D’Hondt et al., 2015). Studies carried out on pore-water suggested that the microbial activity in the sub-seafloor environment is extremely low, with means respiration rates ranging between 2.8 × 10–18 and 1.1 × 10–14 moles of electrons per cell per year (D’Hondt et al., 2004; Hoehler and Jørgensen, 2013; D’Hondt et al., 2015).
Although efforts are there to develop newer methods for studying deep sea organisms, the biodiversity in abyssal plains remains one among the least understood (da Silva et al., 2013). In addition, commercial mining of marine mineral resources has regained attention in recent times due to rising global demands and the belief that marine resources would someday replace land-based resources (Molari et al., 2020). The Indian Ocean has attracted attention as a potential mining site for polymetallic nodules; enriched in minerals such as manganese, iron, copper, cobalt, nickel and rare-earth elements (Wegorzewski and Kuhn, 2014). This has increased the curiosity among the scientific community in gaining insights about ‘sedimentary life’ in the Indian Ocean. The Indian Ocean is the third largest ocean, yet it remains among the most underexplored oceans in terms of biodiversity, especially microbial diversity (Hood et al., 2009; Wang et al., 2018). Mining of the deep sea nodules might result in perturbations in the deep-sea environments (Miller et al., 2018); hence, it is important to explore and record the biota present in deep-sea sediments. Among the biota present, microbes are considered important as they are believed to play a key role in organic matter mineralization, thereby affecting the global biogeochemical cycles and carbon sequestration capacity of oceans. This review is aimed at highlighting the bacterial and fungal diversity derived from the abyssal deep-sea environments (including water, hydrothermal vents, polymetallic nodules and sediment) of the Indian Ocean.
The deep sea microbes are difficult to isolate due to poor understanding about their nutritional requirements and lack of novel methods and incubation conditions. Despite this, considerable efforts have been invested in culturing of deep-sea bacteria, mainly to under their taxonomic novelty, physiological adaptations and biotechnological potentials (Zengler et al., 2002).
The first study, by Johnson et al. (1968), investigated the bacterial composition in the sediments of the Indian Ocean. After this, extensive studies have been carried out to gain an understanding about the topography and benthic life in deep-sea sediments of the Central Indian Ocean Basin (CIOB) (Parulekar et al., 1982; Chandramohan et al., 1987; Nath and Mudholkar, 1989; Sharma and Rao, 1992; Sharma and Kodagali, 1993; Ingole et al., 1999; Ingole et al., 2001). The microbial standing stock in the CIOB was measured (Raghukumar et al., 2001) and the results showed a relatively homogenous distribution of bacteria, indicative of possible active microbial processes and a stable ecosystem. The bacterial counts in the sediments were found to be in the range 1010–1011 cfu g–1, higher than those found in sediments of the Pacific and Atlantic Oceans. Furthermore, perturbations in the CIOB stimulated the retrievable bacterial counts from sediments. This resulted in an increase in the culturable bacterial diversity, wherein Acinetobacter and Moraxella species that were dominant before the disturbance were replaced by Acinetobacter, Coryneforms, Enterobacter, Marinococcus, Pseudomonas, and Staphylococcus (LokaBharathi and Nair, 2005). Disturbances in the sediment caused by anthropogenic influence could result in a re-emergence of the buried sedimentary organic material accessible for microbial degradation. This could result in the alteration of bacterial communities therein and thereby impact the ecological process within the ecosystem.
In recent times, researchers have focused on isolation, cultivation and screening of deep-sea microbes for biotechnological applications. Aiming to identify bacteria with potential biotechnological applications, Gawas et al. (2019) isolated 43 heterotrophic bacteria belonging to the phyla Proteobacteria, Actinobacteria, and Firmicutes, from nodule associated sediments of the first generation mining site and preservation reference zone of the CIOB. This is among the first reports of Oceanobacillus (Firmicutes) and Brachybacterium (Actinobacteria) in the CIOB, although these genera have been previously reported from deep-sea sediments of the Pacific and the Atlantic Oceans. Padmanaban et al. (2019) investigated the metabolic potential of bacteria isolated from deep-sea sediments of the Indian Ocean, mainly from the Bay of Bengal and the Andaman Sea. They isolated and screened 34 bacteria for their potential to produce extracellular hydrolases such as caseinase, α-amylase, urease, gelatinase, lipase, DNase and the production of anti-bacterial metabolites against human clinical pathogens. These 34 bacterial isolates were affiliated to Firmicutes, Proteobacteria and Actinobacteria, with Firmicutes and Actinobacteria being the dominant phyla, which was also confirmed through metagenomic studies.
Fifty-one bacteria capable of degrading polyaromatic hydrocarbons, belonging to 29 genera, were isolated from the deep seawater column (3946 – 4746 m) above the South-West Indian Ridge. The majority of the isolated bacteria were grouped under Proteobacteria (88%), followed by Bacteroides (6%), Actinobateria (4%), and Firmicutes (2%). Of these isolates, 34 isolates showed the ability to use phenanthrene as a sole carbon source, wherein 20 isolates belonged to Alpha-proteobacteria within the genera, Alterierythrobacter, Citricella, Erythrobacter, Kaistia, Lutibacterium, Maricaulis, Martelella, Mesorhizobium, Novosphingobium, Pseudomonas, Phenylobacterium, Roseovarius, Rhodobacter, Salipiger, Stappia, Sphingopyxis, Sphingomonas, Tistrella, and Thalassospira. Within the Gamma-proteobacteria, isolates belonged to the genera Alkaligens, Alkanovorax, Halomonas, Idiomarina, Marinobacter, Pseudoidiomarina, and Pseudomonas. Muricauda and Salegentibacter (Bacteroides), Bacillus (Firmicutes) and Microbacterium (Actinobacteria) also showed phenanthrene-degrading potential (Shao et al., 2015). Moreover, in addition to bioprospecting, isolation of bacterial cultures could yield novel findings with respect to new species within the environment or an undocumented functional role. Yuan et al. (2009) isolated a novel hydrocarbon-degrading bacterium from the deep sea waters of the Indian Ocean and named it Novosphingobium indicum, referring to the Ocean from which the type strain was isolated. Another study reported a unique lineage of Aquificales in the Edmon hydrothermal vent and the Kairei vent field of the Central Indian Ridge (Reysenbach et al., 2002). Bhargavaea cecembensis, a novel Gram-positive, sporulating bacterium, was isolated from sediments of the Chagos-Laccadive ridge system in the Indian Ocean (Manorama et al., 2009). Acuticoccus sedimeni was reported from sediments collected from Indian Ocean from a depth of 2946 m (Lai et al., 2019). This strain was isolated from sediments enriched with poly aromatic hydrocarbons (PAHs) as sole source of carbon and energy. This also shows that the novel strains obtained from deep-sea sediments could have applications in field of bioremediation too like PAH degradation. Cao et al. (2017) reported a novel thermophilic sulfur-reducing bacterium, Desulfurobacteium indicum from sulfide sample collected from a depth at 2771 m from a high temperature hydrothermal vent in the Indian Ocean. Ren et al. (2014) reported a Mn-oxidizing bacterial strain, Fulvimarina manganooxydans from a Fe and Mn-rich hydrothermal plume in the south-west Indian Ocean. Wang J. et al. (2020) reported a novel planctomycete, Gimesia benthica, from the bottom water samples collected with a multi-corer from a depth of 4213 m in Northwest Indian Ocean. Most of the Planctomycetes strains are known for their role in global carbon and nitrogen cycles (Wagner and Horn, 2006). Shivaji et al. (2007) and Bhadra et al. (2008) have reported novel Brevibacterium oceani and Microbacterium indicum species, respectively, from Chagos Trench in Indian Ocean. Qiu et al. (2021) reported a halotolerant Halomonas sedimenti from the deep sea sediment (2699 m) of the Southwest Indian Ocean. Xie et al. (2021) have reported a novel hydrogen and sulfur-oxidizing chemolithoautotroph, Sulfovorum indicum, from deep sea hydrothermal plumes in the Northwestern Indian Ocean. This reiterates the fact that there is a lot more to be explored from the abyssal depths of the Oceans, and it is a very good source of novel bacterial isolates for understanding the diversity and also useful for biotechnological applications.
All the research findings discussed above underline the importance of traditional isolation and cultivation of bacteria, as they allow the possibility of genome sequencing and bioprospecting for biomolecules and industrially relevant processes. Although isolation and culturing of bacteria help us provide holistic information regarding the microorganism, this may not always be feasible due to the stringent nutritional requirements and limited methods available for their isolation. For these reasons, researchers are now focusing on culture-independent methods to gain insights into diversity and ecological roles of deep sea life forms. Additionally, these tools allow for detection of the viable but non-cultivable fraction of the sedimentary microbial community (Polymenakou et al., 2009; Schauer et al., 2010).
Despite technological advances, only 5% of the ocean floors have been scientifically explored. There is a lacuna in understanding the distribution patterns, diversity and ecological roles played by microbes within the infinite seafloor (Corinaldesi, 2015). Describing marine microbial communities is vital to catalog the genetic diversity in a region and understand the functional roles of these organisms in the ecological processes of marine ecosystems. Deep-biosphere bacteria are extremely difficult to cultivate due to their incredibly slow growth rates, hence the number of pure cultures available for in-depth studies does not exceed a few hundred (Jørgensen and Boetius, 2007). Microbial communities interact with each other to exchange nutrients, biochemical products and chemical signals. The presence of this complex community cannot be detected based on traditional cultural methods. Recent developments in molecular fingerprinting techniques have been instrumental in providing new insights into bacterial diversity and their response to environmental heterogeneity (Varliero et al., 2019). The use of molecular methods has made it possible to analyze microbial communities without the conventional isolation and culturing techniques. Molecular analysis using rRNA as a marker began in the early 1970s based on Sanger’s sequencing. This was replaced by the next-generation sequencing (NGS) techniques such as Illumina, Ion Torrent and Roche/454, which are reliable and efficient tools for studying microbial communities. Table 1 summarizes select studies carried out using NGS in the last decade from the abyssal regions of the Indian, Pacific and Atlantic Oceans. This shows that abyssal regions of the Indian Ocean are less explored as compared to the Pacific and Atlantic Oceans.
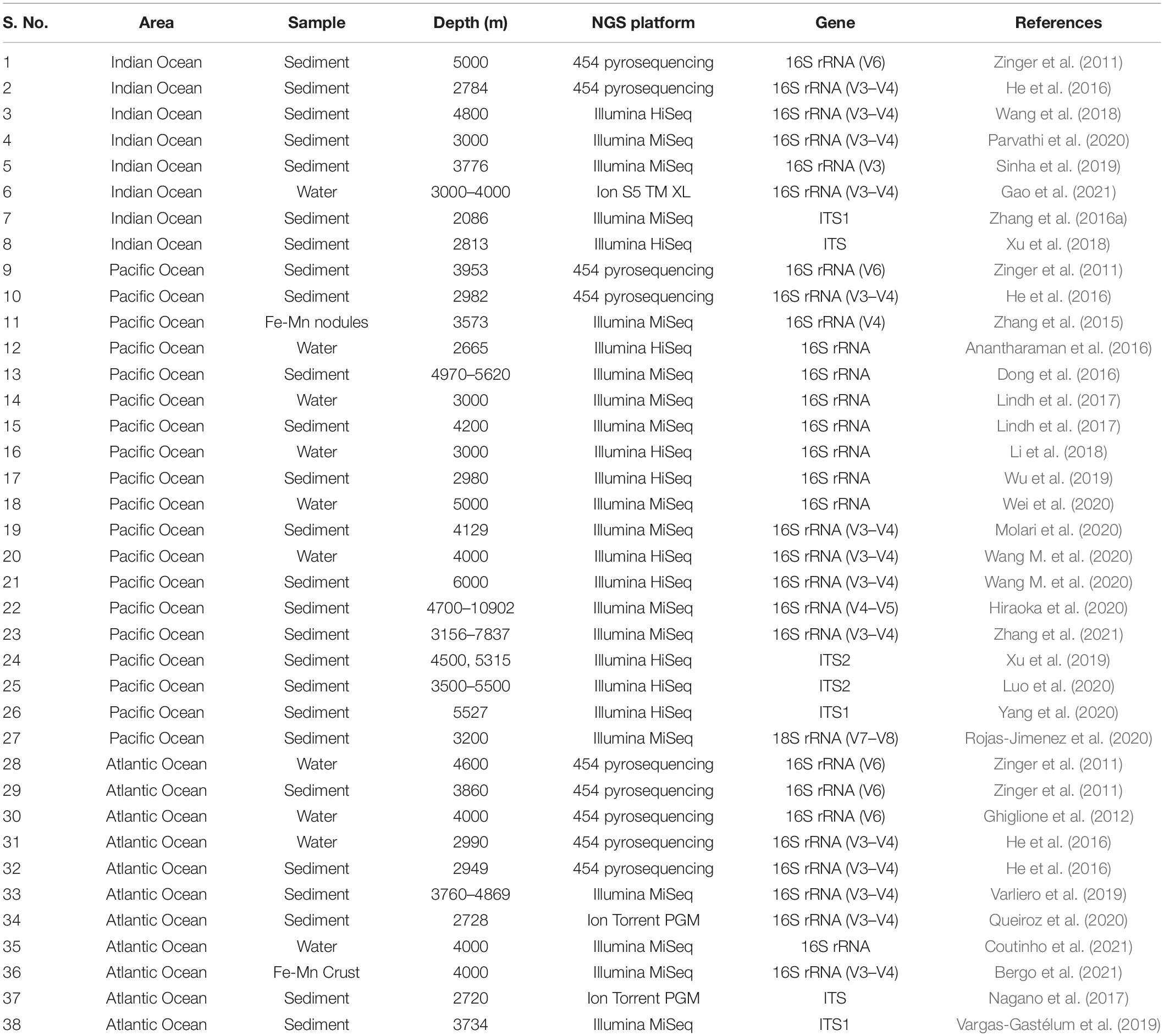
Table 1. Un-cultured bacterial and fungal diversity reported (last 10 years) from the abyssal regions of different Oceans using different next-generation sequencing platforms.
Marine bacteria, being metabolically diverse, significantly impact the nutrient composition and energy flow in both: the sediments as well as the overlaying water column; thus, governing the critical biogeochemical cycles (Arrigo, 2005; Fennel et al., 2005; Emerson et al., 2010). Distinct biological patterns have been revealed in marine environments due to the advances in sequencing techniques. For example, Proteobacteria is found to be dominant in the Atlantic Ocean sediments and the deeper depths of the Arctic and Pacific Oceans (Fuhrman et al., 1993; Schauer et al., 2010). Seasonal changes have been reported in the Pacific Ocean, with Bacteroidetes being more abundant than Proteobacteria in winter (Suh et al., 2014). Compared to other Oceans, studies with respect to the Indian Ocean are in their infancy (Wang et al., 2016).
The first culture-independent molecular study in the Indian Ocean was carried out in the Edmon hydrothermal vent (3300 m) on the Central Indian Ridge (Hoek et al., 2003). Sequencing of 150 clones of the amplified 16S rDNA revealed the incidence of 26 unique bacterial phylotypes. The majority of the clones were affiliated with the Epsilon-proteobacteria. Among the Epsilon-proteobacteria, more than 40% of the phylotypes were closely related to Nautilia lithotrophica, a thermophilic chemoautotrophic sulfur reducer isolated from deep-sea vents; while the majority of the remaining sequences clustered with uncultured clones. Epsilon-proteobacteria displays wide metabolic diversity, but most are hydrogen- and sulfur-oxidizing chemolithoautotrophs. They play significant roles in carbon and sulfur cycles and are also dominant in the active hydrothermal vents in the Pacific and Atlantic Oceans. One of the 150 clones analyzed was identified as Hydrogenobacter thermolithotrophum, which is the first report of the incidence of this genus in deep-sea hydrothermal environments.
Studies on sediments obtained from the Afanasiy- Nikitin seamount in the Equatorial East Indian Ocean revealed a unique bacterial diversity depending on the depth of the sediment (Khandeparker et al., 2014). Sixty-four clones obtained from the top surface sediments classified into Firmicutes (63%) and Gamma-proteobacteria (37%). Within the Firmicutes, Bacillus was dominant, followed by Dolosigranulum. Pseudomonas, and Shigella or in some cases Escherichia were prominent among the Gamma-proteobacteria. The 58 clones obtained from deeper sediments (200 m) were dominated by the Gamma-proteobacteria (66%) represented by Pseudomonas and Enterobacteriaceae, followed by Beta-proteobacteria (34%) represented by Limnobacter and Burkholderiales. Hence, it can be said that bacterial communities are stratified based on the depth of the sediment. Gamma-proteobacteria members are metabolically versatile and abundant in various environments, including the cold deep ocean, hydrothermal region, nodule province, polluted continental area, and water column. Gamma-proteobacteria is known to mediate sulfide reduction and oxidation, which is one of the most important microbial chemosynthetic pathways in deep-sea hydrothermal ecosystems.
Li et al. (2014) examined the microbial diversity inhabiting the exterior (black Fe-Mn oxides) as well as the interior (white carbonates) of carbonate sediments of the Southwest Indian Ridge (SWIR). Analyses of the 16S rRNA gene of the 145 clones indicated that diverse bacteria are associated with the exterior and interior of the sediments. Ninety-three phylotypes were obtained from the clone analysis, among which a large majority belonged to the Proteobacteria (alpha, beta, gamma, and delta), followed by the Acidobacteria (22.07%), Actinobacteria and Bacteroidetes (6.9%); and a small fraction classified as Chloroflexi, Deferribacteres, Nitrospirales, Planctomycete, Verrucomicrobia, and uncultured taxonomic groups WS3. Nitrospira, which plays a role in nitrogen cycling in the conversion of nitrites to nitrates, is widespread in marine environments and has been reported from metal-rich sediments of Pacific Ocean. Deferribacteres and Nitrospirales are known metal reducers and could play a role in the formation of metal oxides. These bacterial groups detected are common in deep-sea sediments and participate in various biochemical processes such as recycling nutrients (carbon, nitrogen, sulfur) and trace metals (iron and manganese).
Another study conducted in the SWIR, analyzed the bacterial communities within inactive hydrothermal vents (Zhang et al., 2016b). Samples were collected at two different sites for comparison using the Illumina MiSeq2500 platform, and it was found that the Proteobacteria and Bacteroidetes were dominant at both locations. The major classes in the two samples were α-proteobacteria, followed by Gamma-proteobacteria, Sphingobacteria, Beta-proteobacteria, and Flavobacteria. Alpha- and Gamma-proteobacteria are known to mediate sulfide reduction and oxidation. Sphingobacteria and Flavobacteria are known degraders of biopolymers in sedimentary organic matter. Thus, the bacterial community identified could be involved in nitrogen and sulfur cycling and metal metabolism, suggesting that they may play important ecological roles in inactive deep-sea hydrothermal vents (Zhang et al., 2016b). Furthermore, the diversity and abundance varied between the two samples isolated from the two adjacent sites (E 50.9277°, S 37.6251°, and E 50.9643°, S 37.6174°), suggesting that the ambient environmental parameters probably influence the formation of microbial communities, even within the same oceanic ridge. Environmental conditions are considered to have a strong influence on microbial biogeography (Li et al., 2009). Several studies in Pacific and Atlantic Oceans have also previously shown that spatial distances affect microbial diversity (Papke et al., 2003; Whitaker et al., 2003; Martiny et al., 2006; Ramette and Tiedje, 2007; Schauer et al., 2010; Zinger et al., 2011).
Bacterial diversity from rare earth elements-rich sediment (4800 m) in the Indian Ocean was characterized using the Illumina HiSeq platform (Wang et al., 2018). The results revealed the occurrence of 49 different phyla, among which the most abundant bacteria were Proteobacteria, with Gamma-proteobacteria being present in all sections of the core; followed by Firmicutes (27.95%), Actinobacteria (5.45%), Bacteroidetes (4.03%), Cyanobacteria (1.71%), and Chloroflexi (0.68%). The major genera included, Lactobacillus, Profundibacterium, Shigella, Escherichia, Pseudoalteromonas, Vibrio, Propionibacterium, Alteromonas, Enterobacter, Sphingomonas, and Staphylococcus. The results also revealed that the microbial diversity at this site was less sensitive to changes in the vertical depth within the sediment core; however, the abundance of bacteria changed with the vertical depth profile. Pseudomonas and Alteromonas, which are predicted to play a role in building matrix outside cells to induce or control mineralization, suggest that these bacteria possibly play a role in the formation of polymetallic nodules in the Indian Ocean. Another study using the Illumina MiSeq platform was conducted on sediments from the Bay of Bengal region of the Indian Ocean also showed that bacterial diversity within the deep-sea sediments is quite similar (Parvathi et al., 2020). Proteobacteria dominated the sediments, followed by Firmicutes, Cyanobacteria, Bacteroidetes, Actinobacteria, Chloroflexi, and Planctomycetes. Alpha-proteobacteria was the most abundant class among Proteobacteria, followed by Gamma-proteobacteria and Delta-proteobacteria, and are known to mediate sulfide reduction and oxidation. Planctomycetes have a suggestive role in methane oxidation. Wolbachia belonging to α-proteobacteria was found to be the dominant genus in the deep-sea. Functional analysis revealed that dehalogenation activities were higher in Wolbachia dominated sediments, hinting at the role of these bacteria in biogeochemical cycles of chlorine, iodine, bromine and halogenated carbon substrates.
All of these findings portray the importance of diversity studies within an environment. Bacteria are highly adaptive and sensitive to environmental perturbations. The bacterial community structure differs depending on the features of the environment, the nutrient profiles, the vertical zonation, and the interior and exterior of sediments obtained from the same ecological niche. Therefore, cataloging the diversity of bacteria is of paramount importance to fully understand the processes taking place in a particular environment. Unfortunately, the Indian Ocean is yet to be explored, and a large portion of the Indian Ocean remains under-explored and under-sampled (Figure 1).
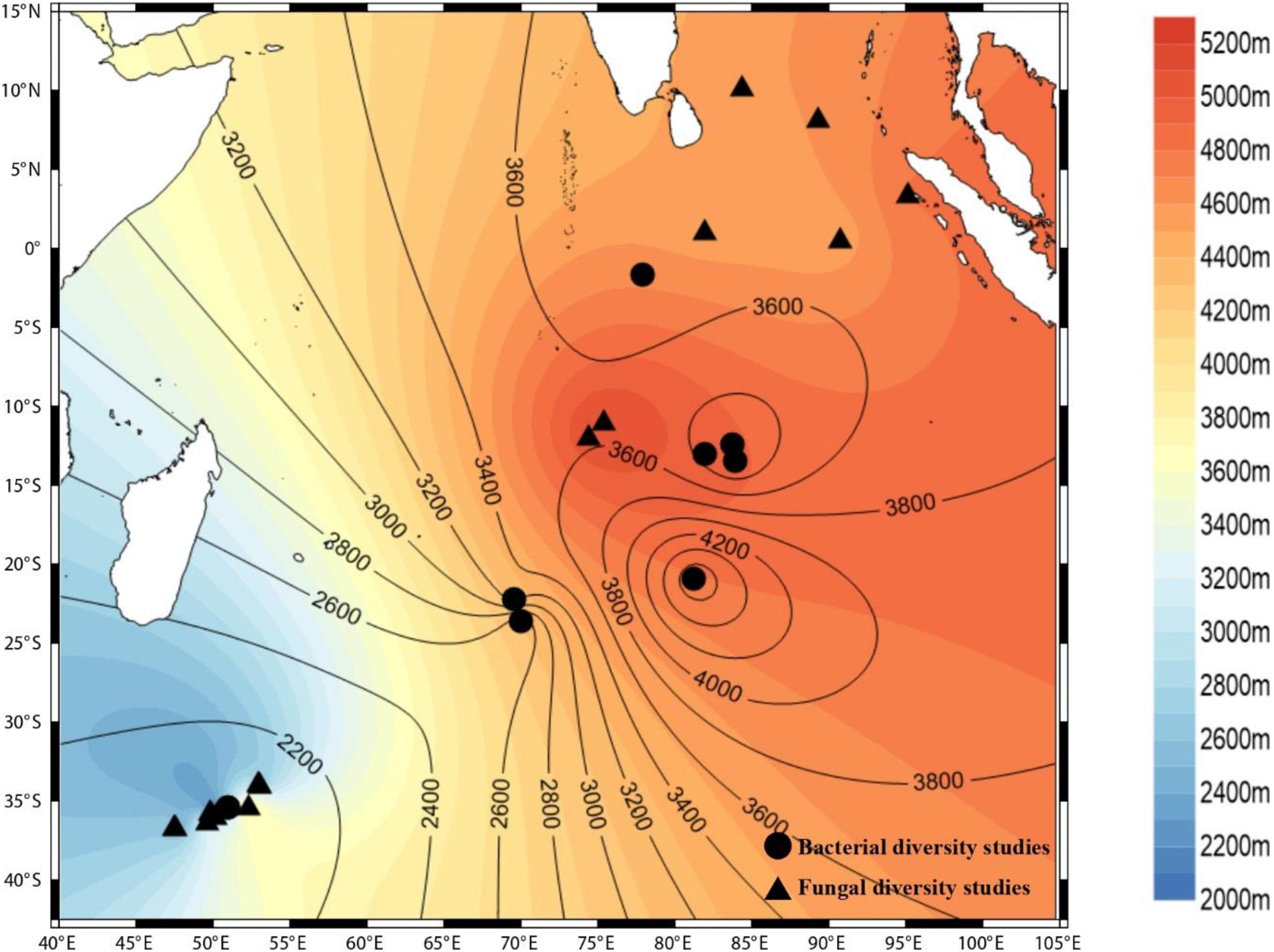
Figure 1. Culture-independent studies carried out at various locations in the Indian Ocean. The circles depict the bacterial diversity studies; the triangles depict the fungal diversity studies. (Surfer, Golden software LLC).
Compared to the exploration of bacterial diversity in the Indian Ocean, fungal diversity studies are still in their infancy. The first report on the isolation of deep-sea fungi from a depth of 4450 m from the Atlantic Ocean was published by Roth et al. (1964). Although the role and diversity of deep-sea fungi remains largely ambiguous, their ecological significance is being recognized. Numerous studies have reported isolation of fungal isolates from deep-sea environments including hydrothermal vents and the Mariana Trench (Takami et al., 1997; Gadanho and Sampaio, 2005; Nagahama et al., 2006, 2008). Fungal diversity in the deep-sea realm has been investigated by both traditional culturing and modern culture-independent methods.
The ability of fungi to adapt/thrive under deep-sea conditions in the Indian Ocean was first discussed by Raghukumar and Raghukumar (1998). Wherein two isolates, Aspergillus ustus and Graphium sp. obtained from calcareous shells in the sediments from the Arabian Sea and the Bay of Bengal, demonstrated conidial germination and hyphal growth at conditions mimicking the deep-sea environment, i.e., high pressures and low temperature. Following this, another study, based on culturing, reported the presence of fungi at a depth of 5904 m in the Chagos trench of the Indian Ocean (Raghukumar et al., 2004). They comprised non-sporulating as well as sporulating forms, identified as Aspergillus sydowii, and present up to 370 m depth of the core subsection mbsf. A total of 181 fungal cultures were isolated from 5000 m depth in the CIOB, most of which were terrestrial sporulating species belonging to Aspergillus, Penicillium, and Cladosporium genera (Damare et al., 2006). The presence of Aspergillus terreus in the sediments was confirmed in these studies using immunofluorescence techniques. Singh et al. (2010) isolated 16 filamentous and 12 yeast cultures, belonging to the Ascomycota and Basidiomycota, from sediments of the CIOB at depths of 5000 m. This was the first report of isolation of filamentous fungi (Capronia, Exophiala, Sagenomella, and Tilletiopsis) from deep-sea sediments, showing that the Indian Ocean basin is a host to a unique diversity of fungi.
The first culture-independent report on fungal diversity from the Indian Ocean sediments was published by Singh et al. (2011). A total of 39 OTUs, belonging to 32 fungal taxa, majorly the Ascomycota and Basidiomycota, were retrieved from 5000 m depth. Within the Ascomycota, members of Sordariomycetes, Dothideomycetes, and Saccharomycetes were identified. Members belonging to Tremellomycetes, Microbotryomycetes, and Ustilaginomycetes were identified within the Basidiomycota. This study revealed heterogeneity with respect to the fungal diversity in deep-sea sediments. In a comparative study using culture-dependent and culture-independent techniques, it was found that the fungal diversity altered vertically with the increase in the depth of the sediment (Singh et al., 2012a,b). Furthermore, 19 fungi belonging to 12 genera were obtained via culture-based methods, wherein a higher diversity was obtained (42 unique OTUs) using sequencing techniques. This was also confirmed in another study in the Eastern Indian Ocean, wherein 45 OTUs were obtained using sequencing approaches compared to 20 culturable fungal phylotypes (Zhang et al., 2014). The culturable fraction was dominated by filamentous fungi, including Aspergillus, Penicillium, Simplicillium, Cladosporium, and Phoma. Moreover, six of the twenty culturable fungal isolates and 20 of the 45 OTUs were first time reports from deep-sea sediments. A more significant fraction of fungal phylotypes was recovered using targeted environmental sequencing than the conventional culturable techniques from deep-sea sediments of hydrothermal vents in the Southwest Indian Ridge (Xu et al., 2018). In this study, 79 fungal taxa were identified from sediments, whereas isolates belonging to only 14 taxa could be cultivated. In a recent study, 42 OTUs and 10 different fungal phylotypes belonging majorly to Aspergillus, Penicillium, Ophiocordyceps, and Phoma could be recovered (Tang et al., 2020). All the findings discussed here suggest that sediments from the abyssal regions of the Indian Ocean are dominated by Ascomycota and Basidiomycota. Fungi detected in deep-sea sediments are similar to those present in the terrestrial environment, indicating the possibility of connectivity between these two environments, mediated by either aerial dispersal or terrestrial runoff.
The deep-sea is a largely unexplored area and is attracting attention in recent times due to the presence of polymetallic nodules. The mining of polymetallic nodules found in the Indian Ocean has been viewed as an alternative to terrestrial mining; therefore, it is important to understand the marine life existing in these regions. However, the knowledge about the inhabitants of the deep-sea ecosystem is limited. Furthermore, information on possible effects of anthropogenic disturbances due to deep-sea mining on the ecosystem and its dwellers is yet to be generated. Although newer technology such as submersibles and drilling tools are made available, similar efforts have not yet been made to explore the microbial diversity thoroughly in the Indian Ocean deep-sea.
This review has summarized the important findings on distribution and diversity of bacterial and fungal communities in the abyssal regions of the Indian Ocean. The deep-sea environments of the Indian Ocean possess diverse assemblages of bacteria and fungi, including novel taxa and new records of species that could be endemic to the region (For example, Novosphingobium indicum). The bacterial distribution in the Indian Ocean differs with respect to the depth of the sediment as well as spatial distances between the sampling locations, probably due to the differing environmental conditions. A variety of bacteria, with unique metabolic abilities for biotechnological potentials such as industrial and medical applications (extracellular hydrolases and anti-bacterial metabolites), have been retrieved from the Indian Ocean. Moreover, bacteria (Pseudomonas and Alteromonas) that may play a key role in formation of polymetallic nodules have been detected in sediments from the Indian Ocean. The presence of metal-utilizing bacteria (Acinetobacter) in samples from the Indian Ocean suggest for an environmentally induced adaptation mechanism due to the richness of metals in sediments. Although the third largest Ocean in the world, only a small fraction of the Indian Ocean deep-sea environments has been sampled, most of the high-throughput sequencing studies carried out mainly focus on the water column. Metagenomic and functional gene studies are lacking from the abyssal depths. Therefore, extensive sampling and functional gene characterization and quantification of microbes and their activities in the abyssal regions of the Indian Ocean need to be carried out to build the knowledge with respect to their diversity and the ecological processes they affect.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was a part of PMN-EIA studies funded by the Ministry of Earth Sciences (MoES), Govt. of India (GAP2128). The NB was supported for Fellowship under the same project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the Director of CSIR-National Institute of Oceanography for the support and infrastructure facilities. We are also grateful to the reviewers for their suggestions and constructive comments, which have substantially improved the manuscript. This manuscript is NIO contribution number 6778.
Anantharaman, K., Breier, J. A., and Dick, G. J. (2016). Metagenomic resolution of microbial functions in deep-sea hydrothermal plumes across the Eastern Lau Spreading Center. ISME J. 10, 225–239. doi: 10.1038/ismej.2015.81
Arrigo, K. R. (2005). Marine microorganisms and global nutrient cycles. Nature 437, 349–355. doi: 10.1038/nature04159
Barbier, E. B., Moreno-Mateos, D., Rogers, A. D., Aronson, J., Pendleton, L., Danovaro, R., et al. (2014). Ecology: protect the deep sea. Nat. News 505, 475–477.
Bergo, N. M., Bendia, A. G., Ferreira, J. C. N., Murton, B. J., Brandini, F. P., and Pellizari, V. H. (2021). Microbial diversity of deep-sea ferromanganese crust field in the Rio Grande Rise, Southwestern Atlantic Ocean. Microb. Ecol. [Epub ahead of print].
Bhadra, B., Raghukumar, C., Pindi, P. K., and Shivaji, S. (2008). Brevibacterium oceani sp. nov., isolated from deep-sea sediment of the Chagos Trench, Indian Ocean. Int. J. Syst. Evol. Microbiol. 58, 57–60. doi: 10.1099/ijs.0.64869-0
Biddle, J. F., Fitz-Gibbon, S., Schuster, S. C., Brenchley, J. E., and House, C. H. (2008). Metagenomic signatures of the Peru Margin sub-seafloor biosphere show a genetically distinct environment. Proc. Natl. Acad. Sci. U.S.A. 105, 10583–10588. doi: 10.1073/pnas.0709942105
Cao, J., Birien, T., Gayet, N., Huang, Z., Shao, Z., Jebbar, M., et al. (2017). Desulfurobacterium indicum sp. nov., a thermophilic sulfur-reducing bacterium from the Indian Ocean. Int. J. Syst. Evol. Microbiol. 67, 1665–1668. doi: 10.1099/ijsem.0.001837
Chandramohan, D., Lokabharathi, P. A., Nair, S., and Matondkar, S. G. P. (1987). Bacteriology of ferromanganese nodules from the Indian Ocean. Geomicrob. J. 5, 17–31. doi: 10.1080/01490458709385954
Chen, X., Andersen, T. J., Morono, Y., Inagaki, F., Jørgensen, B. B., and Lever, M. A. (2017). Bioturbation as a key driver behind the dominance of bacteria over archaea in near-surface sediment. Sci. Rep. 7:2400.
Ciobanu, M. C., Burgaud, G., Dufresne, A., Breuker, A., Rédou, V., Maamar, S. B., et al. (2014). Microorganisms persist at record depths in the subseafloor of the Canterbury Basin. ISME J. 8, 1370–1380. doi: 10.1038/ismej.2013.250
Corinaldesi, C. (2015). New perspectives in benthic deep-sea microbial ecology. Font. Mar. Sci. 2:17. doi: 10.3389/fmars.2015.00017
Coutinho, F. H., von Meijenfeldt, F. A. B., Walter, J. M., Haro-Moreno, J. M., Lopéz-Pérez, M., van Verk, M. C., et al. (2021). Ecogenomics and metabolic potential of the South Atlantic Ocean microbiome. Sci. Total Environ. 765:142758. doi: 10.1016/j.scitotenv.2020.142758
D’Hondt, S., Inagaki, F., Zarikian, C. A., Abrams, L. J., Dubois, N., Engelhardt, T., et al. (2015). Presence of oxygen and aerobic communities from the sea floor to basement in deep-sea sediments. Nat. Geosci. 8, 299–304. doi: 10.1038/ngeo2387
D’Hondt, S., Jørgensen, B. B., Miller, D. J., Batzke, A., Blake, R., Cragg, B. A., et al. (2004). Distributions of microbial activities in deep sub-seafloor sediments. Science 306, 2216–2221. doi: 10.1126/science.1101155
da Silva, M. A. C., Cavalett, A., Spinner, A., Rosa, D. C., Jasper, R. B., Quecine, M. C., et al. (2013). Phylogenetic identification of marine bacteria isolated from deep-sea sediments of the eastern South Atlantic Ocean. SpringerPlus 2:127.
Damare, S., Raghukumar, C., and Raghukumar, S. (2006). Fungi in deep-sea sediments of the Central Indian Basin. Deep Sea Res. Part I 53, 14–27. doi: 10.1016/j.dsr.2005.09.005
Danovaro, R., Corinaldesi, C., Dell’Anno, A., and Snelgrove, P. V. (2017). The deep-sea under global change. Curr. Biol. 27, 461–465.
Danovaro, R., Snelgrove, P. V., and Tyler, P. (2014). Challenging the paradigms of deep-sea ecology. Trends Ecol. Evol. 29, 465–475. doi: 10.1016/j.tree.2014.06.002
Dong, Y., Li, J., Zhang, W., Zhang, W., Zhao, Y., Xiao, T., et al. (2016). The detection of magnetotactic bacteria in deep sea sediments from the east Pacific Manganese Nodule Province. Environ. Microbiol. Rep. 8, 239–249. doi: 10.1111/1758-2229.12374
Emerson, D., Fleming, E. J., and McBeth, J. M. (2010). Iron-oxidising bacteria: an environmental and genomic perspective. Annu. Rev. Microbiol. 64, 561–583. doi: 10.1146/annurev.micro.112408.134208
Fennel, K., Follows, M., and Falkowski, P. G. (2005). The co-evolution of the nitrogen, carbon and oxygen cycles in the Proterozoic ocean. Am. J. Sci. 305, 526–545. doi: 10.2475/ajs.305.6-8.526
Fry, J. C., Parkes, R. J., Cragg, B. A., Weightman, A. J., and Webster, G. (2008). Prokaryotic biodiversity and activity in the deep sub-seafloor biosphere. FEMS Microbiol. Ecol. 66, 181–196. doi: 10.1111/j.1574-6941.2008.00566.x
Fuhrman, J. A., McCallum, K., and Davis, A. A. (1993). Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl. Environ. Microbiol. 59, 1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993
Gadanho, M., and Sampaio, J. P. (2005). Occurrence and diversity of yeasts in the mid-Atlantic ridge hydrothermal fields near the Azores Archipelago. Microb. Ecol. 50, 408–417. doi: 10.1007/s00248-005-0195-y
Gao, P., Qu, L., Du, G., Wei, Q., Zhang, X., and Yang, G. (2021). Bacterial and archaeal communities in deep sea waters near the Ninety-east Ridge in Indian Ocean. J. Oceanol. Limnol. 39, 582–597. doi: 10.1007/s00343-020-9343-y
Gartner, A., Blumel, M., Wiese, J., and Imhoff, J. F. (2011). Isolation and characterisation of bacteria from the Eastern Mediterranean deep sea. Antonie Van Leeuwenhoek 100, 421–435. doi: 10.1007/s10482-011-9599-5
Gawas, V. S., Shivaramu, M. S., Damare, S. R., Pujitha, D., Meena, R. M., and Shenoy, B. D. (2019). Diversity and extracellular enzyme activities of heterotrophic bacteria from sediments of the Central Indian Ocean Basin. Sci. Rep. 9:9403.
Ghiglione, J. F., Galand, P. E., Pommier, T., Pedrós-Alió, C., Maas, E. W., Bakker, K., et al. (2012). Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 109, 17633–17638. doi: 10.1073/pnas.1208160109
He, Y., Li, M., Perumal, V., Feng, X., Fang, J., Xie, J., et al. (2016). Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat. Microbiol. 1:16035.
Hiraoka, S., Hirai, M., Matsui, Y., Makabe, A., Minegishi, H., Tsuda, M., et al. (2020). Microbial community and geochemical analyses of trans-trench sediments for understanding the roles of hadal environments. ISME J. 14, 740–756. doi: 10.1038/s41396-019-0564-z
Hoehler, T. M., and Jørgensen, B. B. (2013). Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 11, 83–94. doi: 10.1038/nrmicro2939
Hoek, J., Banta, A., Hubler, F., and Reysenbach, A. L. (2003). Microbial diversity of a sulphide spire located in the Edmond deep-sea hydrothermal vent field on the Central Indian Ridge. Geobiology 1, 119–127. doi: 10.1046/j.1472-4669.2003.00015.x
Hood, R. R., Wiggert, J. D., and Naqvi, S. W. A. (2009). “Indian Ocean research: opportunitiesand challenges,” in Indian Ocean Biogeochemical Processes and Ecological Variability. Geophysical Monograph Series, eds J. D. Wiggert, R. R. Hood, and S. W. A. Naqvi (Washington DC: American Geophysical Union), 409–429. doi: 10.1029/2008gm000714
Inagaki, F., Hinrichs, K. U., Kubo, Y., Bowles, M. W., Heuer, V. B., Hong, W. L., et al. (2015). Exploring deep microbial life in coal-bearing sediment down to~ 2.5 km below the ocean floor. Science 349, 420–424. doi: 10.1126/science.aaa6882
Inagaki, F., Nunoura, T., Nakagawa, S., Teske, A., Lever, M., Lauer, A., et al. (2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. U.S.A. 103, 2815–2820. doi: 10.1073/pnas.0511033103
Inagaki, F., Suzuki, M., Takai, K., Oida, H., Sakamoto, T., Aoki, K., et al. (2003). Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl. Environ. Microbiol. 69, 7224–7235. doi: 10.1128/aem.69.12.7224-7235.2003
Ingole, B. S., Ansari, Z. A., Matondkar, S. G. P., and Rodrigues, N. (1999). “Immediate response of meio and macrobenthos to disturbance caused by a benthic disturber,” in Proceedings of the Third ISOPE Ocean Mining Symposium International Society of Off-Shore and Polar Engineers. Goa.
Ingole, B. S., Ansari, Z. A., Rathod, V., and Rodrigues, N. (2001). Response of deep-sea macrobenthos to a small-scale environmental disturbance. Deep Sea Res. Part II 48, 3401–3410. doi: 10.1016/s0967-0645(01)00048-0
Johnson, R. M., Schwent, R. M., and Press, W. (1968). The characteristics and distribution of marine bacteria isolated from the Indian Ocean. Limnol. Oceanogr. 13, 656–664. doi: 10.4319/lo.1968.13.4.0656
Jørgensen, B. B., and Boetius, A. (2007). Feast and famine—microbial life in the deep-sea bed. Nat. Rev. Microbiol. 5, 770–781. doi: 10.1038/nrmicro1745
Kallmeyer, J., Pockalny, R., Adhikari, R. R., Smith, D. C., and D’Hondt, S. (2012). Global distribution of microbial abundance and biomass in sub-seafloor sediment. Proc. Natl. Acad. Sci. U.S.A. 109, 16213–16216. doi: 10.1073/pnas.1203849109
Khandeparker, R., Meena, R. M., and Deobagkar, D. (2014). Bacterial diversity in deep-sea sediments from Afanasy-Nikitin seamount, equatorial Indian Ocean. Geomicrobiol. J. 31, 942–949. doi: 10.1080/01490451.2014.918214
Lai, Q., Liu, X., Sun, F., and Shao, Z. (2019). Acuticoccus sediminis sp. nov., isolated from deep-sea sediment of the Indian Ocean and proposal of Acuticoccaceae fam. nov. Int. J. Syst. Evol. Microbiol. 69, 1173–1178. doi: 10.1099/ijsem.0.003289
Li, H., Yu, Y., Luo, W., Zeng, Y., and Chen, B. (2009). Bacterial diversity in surface sediments from the Pacific Arctic Ocean. Extremophiles 13, 233–246. doi: 10.1007/s00792-009-0225-7
Li, J., Peng, X., Zhou, H., Li, J., Sun, Z., and Chen, S. (2014). Microbial communities in semi-consolidated carbonate sediments of the Southwest Indian Ridge. J. Microbiol. 52, 111–119. doi: 10.1007/s12275-014-3133-1
Li, Y., Jing, H., Xia, X., Cheung, S., Suzuki, K., and Liu, H. (2018). Metagenomic insights into the microbial community and nutrient cycling in the western subarctic Pacific Ocean. Front. Microbiol. 9:623. doi: 10.3389/fmicb.2018.00623
Lindh, M. V., Maillot, B. M., Shulse, C. N., Gooday, A. J., Amon, D. J., Smith, C. R., et al. (2017). From the surface to the deep-sea: bacterial distributions across polymetallic nodule fields in the clarion-clipperton zone of the Pacific Ocean. Front. Microbiol. 8:1696. doi: 10.3389/fmicb.2017.01696
LokaBharathi, P., and Nair, S. (2005). Rise of the dormant: simulated disturbance improves culturable abundance, diversity, and functions of deep-sea bacteria of Central Indian Ocean Basin. Mar. Georesour. Geotechnol. 23, 419–428. doi: 10.1080/10641190500446805
Luo, Y., Wei, X., Yang, S., Gao, Y. H., and Luo, Z. H. (2020). Fungal diversity in deep-sea sediments from the Magellan seamounts as revealed by a meta-barcoding approach targeting the ITS2 regions. Mycology 11, 214–229. doi: 10.1080/21501203.2020.1799878
Manorama, R., Pindi, P. K., Reddy, G. S. N., and Shivaji, S. (2009). Bhargavaea cecembensis gen. nov., sp. nov., isolated from the Chagos–Laccadive ridge system in the Indian Ocean. Int. J. Sys. Evol. Microbiol. 59, 2618–2623. doi: 10.1099/ijs.0.002691-0
Martiny, J. B. H., Bohannan, B. J., Brown, J. H., Colwell, R. K., Fuhrman, J. A., Green, J. L., et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112. doi: 10.1038/nrmicro1341
Miller, K. A., Thompson, K. F., Johnston, P., and Santillo, D. (2018). An overview of seabed mining including the current state of development, environmental impacts, and knowledge gaps. Front. Mar. Sci. 4:418. doi: 10.3389/fmars.2017.00418
Molari, M., Janssen, F., Vonnahme, T. R., Wenzhöfer, F., and Boetius, A. (2020). The contribution of microbial communities in polymetallic nodules to the diversity of the deep-sea microbiome of the Peru Basin (4130–4198 m depth). Biogeosciences 17, 3203–3222. doi: 10.5194/bg-17-3203-2020
Nagahama, T., Abdel-Wahab, M. A., Nogi, Y., Miyazaki, M., Uematsu, K., Hamamoto, M., et al. (2008). Dipodascus tetrasporeus sp. nov., an ascosporogenous yeast isolated from deep-sea sediments in the Japan Trench. Int. J. Syst. Evol. Microbiol. 58, 1040–1046. doi: 10.1099/ijs.0.65471-0
Nagahama, T., Hamamoto, M., and Horikoshi, K. (2006). Rhodotorula pacifica sp. nov., a novel yeast species from sediment collected on the deep-sea floor of the north-west Pacific Ocean. Int. J. Syst. Evol. Microbiol. 56, 295–299. doi: 10.1099/ijs.0.63584-0
Nagano, Y., Miura, T., Nishi, S., Lima, A. O., Nakayama, C., Pellizari, V. H., et al. (2017). Fungal diversity in deep-sea sediments associated with asphalt seeps at the Sao Paulo Plateau. Deep Sea Res. Part II Top. Stud. Oceanogr. 146, 59–67. doi: 10.1016/j.dsr2.2017.05.012
Nath, B. N., and Mudholkar, A. V. (1989). Early diagenetic processes affecting nutrients in the pore waters of Central Indian Ocean cores. Mar. Geol. 86, 57–66. doi: 10.1016/0025-3227(89)90018-2
Padmanaban, V. P., Verma, P., Gopal, D., Sekar, A. K., and Ramalingam, K. (2019). Phylogenetic identification and metabolic potential of bacteria isolated from deep sea sediments of Bay of Bengal and Andaman Sea. Indian J. Exp. Biol. 57, 561–572.
Papke, R. T., Ramsing, N. B., Bateson, M. M., and Ward, D. M. (2003). Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 5, 650–659. doi: 10.1046/j.1462-2920.2003.00460.x
Parkes, R. J., Cragg, B., Roussel, E., Webster, G., Weightman, A., and Sass, H. (2014). A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere: geosphere interactions. Mar. Geol. 352, 409–425. doi: 10.1016/j.margeo.2014.02.009
Parkes, R. J., Webster, G., Cragg, B. A., Weightman, A. J., Newberry, C. J., Ferdelman, T. G., et al. (2005). Deep sub-seafloor prokaryotes stimulated at interfaces over geological time. Nature 436, 390–394. doi: 10.1038/nature03796
Parulekar, A. H., Harkantra, S. N., Ansari, Z. A., and Matondkar, S. P. (1982). Abyssal benthos of the central Indian Ocean. Deep Sea Res. 12, 1531–1537. doi: 10.1016/0198-0149(82)90041-3
Parvathi, A., Jasna, V., Aswathy, V. K., Aparna, S., Nathan, V. K., and Jyothibabu, R. (2020). Dominance of Wolbachia sp. in the deep-sea sediment bacterial metataxonomic sequencing analysis in the Bay of Bengal, Indian Ocean. Genomics 112, 1030–1041. doi: 10.1016/j.ygeno.2019.06.019
Polymenakou, P. N., Lampadariou, N., Mandalakis, M., and Tselepides, A. (2009). Phylogenetic diversity of sediment bacteria from the southern Cretan margin, Eastern Mediterranean Sea. Syst. Appl. Microbiol. 32, 17–26. doi: 10.1016/j.syapm.2008.09.006
Qiu, X., Yu, L., Cao, X., Wu, H., Xu, G., and Tang, X. (2021). Halomonas sedimenti sp. nov., a halotolerant bacterium isolated from deep-sea sediment of the Southwest Indian Ocean. Curr. Microbiol. 78, 1662–1669. doi: 10.1007/s00284-021-02425-9
Queiroz, L. L., Bendia, A. G., Duarte, R. T. D., das Graças, D. A., da Silva, A. L. D. C., Nakayama, C. R., et al. (2020). Bacterial diversity in deep-sea sediments under influence of asphalt seep at the São Paulo Plateau. Antonie Van Leeuwenhoek 113, 707–717. doi: 10.1007/s10482-020-01384-8
Raghukumar, C., and Raghukumar, S. (1998). Barotolerance of fungi isolated from deep-sea sediments of the Indian Ocean. Aquat. Microb. Ecol. 15, 153–163. doi: 10.3354/ame015153
Raghukumar, C., Raghukumar, S., Sheelu, G., Gupta, S. M., Nath, B. N., and Rao, B. R. (2004). Buried in time: culturable fungi in a deep-sea sediment core from the Chagos Trench, Indian Ocean. Deep Sea Res. Part I 51, 1759–1768. doi: 10.1016/j.dsr.2004.08.002
Raghukumar, C., Sheelu, G., Loka Bharathi, P. A., Nair, S., and Mohandass, C. (2001). Microbial biomass and organic nutrients in the deep-sea sediments of the Central Indian Ocean Basin. Mar. Georesour. Geotechnol. 19, 1–16. doi: 10.1080/106411901300062385
Ramette, A., and Tiedje, J. M. (2007). Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb. Ecol. 53, 197–207. doi: 10.1007/s00248-005-5010-2
Ren, F., Zhang, L., Song, L., Xu, S., Xi, L., Huang, L., et al. (2014). Fulvimarina manganoxydans sp. nov., isolated from a deep-sea hydrothermal plume in the south-west Indian Ocean. Int. J. Syst. Evol. Microbiol. 64, 2920–2925. doi: 10.1099/ijs.0.060558-0
Reysenbach, A. L., Gotz, D., Banta, A., Jeanthon, C., and Fouquet, Y. (2002). Expanding the distribution of the Aquificales to the deep-sea vents on Mid-Atlantic Ridge and Central Indian Ridge. Cah. Biol. Mar. 43, 425–428.
Rojas-Jimenez, K., Grossart, H. P., Cordes, E., and Cortés, J. (2020). Fungal communities in sediments along a depth gradient in the Eastern Tropical Pacific. Front. Microbiol. 11:2851. doi: 10.3389/fmicb.2020.575207
Roth, F. J. Jr., Orpurt, P. A., and Ahearn, D. G. (1964). Occurrence and distribution of fungi in a subtropical marine environment. Can. J. Bot. 42, 375–383. doi: 10.1139/b64-037
Schauer, R., Bienhold, C., Ramette, A., and Harder, J. (2010). Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J. 4, 159–170. doi: 10.1038/ismej.2009.106
Shao, Z., Yuan, J., Lai, Q., and Zheng, T. (2015). The diversity of PAH-degrading bacteria in a deep-sea water column above the Southwest Indian Ridge. Front. Microbiol. 6:853. doi: 10.3389/fmicb.2015.00853
Sharma, R., and Kodagali, V. N. (1993). Influence of seabed topography on the distribution of manganese nodules and associated features in the Central Indian Basin: a study based on photographic observations. Mar. Geol. 110, 153–162. doi: 10.1016/0025-3227(93)90111-8
Sharma, R., and Rao, A. S. (1992). Geological factors associated with megabenthic activity in the Central Indian Basin. Deep Sea Res. Part I 39, 705–713. doi: 10.1016/0198-0149(92)90096-c
Shivaji, S., Bhadra, B., Rao, R. S., Chaturvedi, P., Pindi, P. K., and Raghukumar, C. (2007). Microbacterium indicum sp. nov., isolated from a deep-sea sediment sample from the Chagos Trench, Indian Ocean. Int. J. Syst. Evol. Microbiol. 57, 1819–1822. doi: 10.1099/ijs.0.64782-0
Singh, P., Raghukumar, C., Meena, R. M., Verma, P., and Shouche, Y. (2012a). Fungal diversity in deep-sea sediments revealed by culture-dependent and culture-independent approaches. Fungal Ecol. 5, 543–553. doi: 10.1016/j.funeco.2012.01.001
Singh, P., Raghukumar, C., Verma, P., and Shouche, Y. (2010). Phylogenetic diversity of culturable fungi from the deep-sea sediments of the Central Indian Basin and their growth characteristics. Fungal Divers. 40, 89–102. doi: 10.1007/s13225-009-0009-5
Singh, P., Raghukumar, C., Verma, P., and Shouche, Y. (2011). Fungal community analysis in the deep-sea sediments of the Central Indian Basin by culture-independent approach. Microb. Ecol. 61, 507–517. doi: 10.1007/s00248-010-9765-8
Singh, P., Raghukumar, C., Verma, P., and Shouche, Y. (2012b). Assessment of fungal diversity in deep-sea sediments by multiple primer approach. World J. Microbiol. Biotechnol. 28, 659–667. doi: 10.1007/s11274-011-0859-3
Sinha, R. K., Krishnan, K. P., Thomas, F. A., Binish, M. B., Mohan, M., and Kurian, P. J. (2019). Polyphasic approach revealed complex bacterial community structure and function in deep sea sediment of ultra-slow spreading Southwest Indian Ridge. Ecol. Indic. 96, 40–51. doi: 10.1016/j.ecolind.2018.08.063
Suh, S. S., Park, M., Hwang, J., Lee, S., Chung, Y., and Lee, T. K. (2014). Distinct patterns of marine bacterial communities in the South and North Pacific Oceans. J. Microbiol. 52, 834–841. doi: 10.1007/s12275-014-4287-6
Takami, H., Inoue, A., Fuji, F., and Horikoshi, K. (1997). Microbial flora in the deepest sea mud of the Mariana Trench. FEMS Microbiol. Lett. 152, 279–285. doi: 10.1111/j.1574-6968.1997.tb10440.x
Tang, X., Yu, L., Xu, W., Zhang, X., Xu, X., Wang, Q., et al. (2020). Fungal diversity of deep-sea sediments in Mid-Oceanic Ridge area of the East Pacific and the South Indian Oceans. Bot. Mar. 63, 183–196. doi: 10.1515/bot-2018-0112
Teske, A. (2013). Marine deep sediment microbial communities. Prokaryotes 123–138. doi: 10.1007/978-3-642-30123-0_42
Vargas-Gastélum, L., Chong-Robles, J., Lago-Lestón, A., Darcy, J. L., Amend, A. S., and Riquelme, M. (2019). Targeted ITS1 sequencing unravels the mycodiversity of deep-sea sediments from the Gulf of Mexico. Environ. Microbiol. 21, 4046–4061. doi: 10.1111/1462-2920.14754
Varliero, G., Bienhold, C., Schmid, F., Boetius, A., and Molari, M. (2019). Microbial diversity and connectivity in deep-sea sediments of the South Atlantic polar front. Front. Microbiol. 10:665. doi: 10.3389/fmicb.2019.00665
Wagner, M., and Horn, M. (2006). The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 17, 241–249. doi: 10.1016/j.copbio.2006.05.005
Wang, J., Kan, J., Borecki, L., Zhang, X., Wang, D., and Sun, J. (2016). A snapshot on spatial and vertical distribution of bacterial communities in the eastern Indian Ocean. Acta Oceanol. Sin. 35, 85–93. doi: 10.1007/s13131-016-0871-4
Wang, J., Ruan, C. J., Song, L., Li, A., Zhu, Y. X., Zheng, X. W., et al. (2020). Gimesia benthica sp. nov., a planctomycete isolated from a deep-sea water sample of the Northwest Indian Ocean. Int. J. Syst. Evol. Microbiol. 70, 4384–4389. doi: 10.1099/ijsem.0.004301
Wang, M., Ma, Y., Feng, C., Cai, L., and Li, W. (2020). Diversity of pelagic and benthic bacterial assemblages in the Western Pacific Ocean. Front. Microbiol. 11:1730. doi: 10.3389/fmicb.2020.01730
Wang, S., Yu, M., Wei, J., Huang, M., Shi, X., and Chen, H. (2018). Microbial community composition and diversity in the Indian Ocean deep sea REY-rich muds. PLoS One 13:e0208230. doi: 10.1371/journal.pone.0208230
Wegorzewski, A. V., and Kuhn, T. (2014). The influence of suboxic diagenesis on the formation of manganese nodules in the Clarion Clipperton nodule belt of the Pacific Ocean. Mar. Geol. 357, 123–138. doi: 10.1016/j.margeo.2014.07.004
Wei, Z. F., Li, W. L., Huang, J. M., and Wang, Y. (2020). Metagenomic studies of SAR202 bacteria at the full-ocean depth in the Mariana Trench. Deep Sea Res Part I 165:103396. doi: 10.1016/j.dsr.2020.103396
Whitaker, R. J., Grogan, D. W., and Taylor, J. W. (2003). Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301, 976–978. doi: 10.1126/science.1086909
Wörmer, L., Hoshino, T., Bowles, M. W., Viehweger, B., Adhikari, R. R., Xiao, N., et al. (2019). Microbial dormancy in the marine subsurface: Global endospore abundance and response to burial. Sci. Adv. 5:eaav1024. doi: 10.1126/sciadv.aav1024
Wu, J., Hong, Y., Chang, X., Jiao, L., Li, Y., Liu, X., et al. (2019). Unexpectedly high diversity of anammox bacteria detected in deep-sea surface sediments of the South China Sea. FEMS Microbiol. Ecol. 95:fiz013.
Xie, S., Wang, S., Li, D., Shao, Z., Lai, Q., Wang, Y., et al. (2021). Sulfurovum indicum sp. nov., a novel hydrogen-and sulfur-oxidizing chemolithoautotroph isolated from a deep-sea hydrothermal plume in the Northwestern Indian Ocean. Int. J. Syst. Evol. Microbiol. 71:004748.
Xu, W., Gao, Y. H., Gong, L. F., Li, M., Pang, K. L., and Luo, Z. H. (2019). Fungal diversity in the deep-sea hadal sediments of the Yap Trench by cultivation and high throughput sequencing methods based on ITS rRNA gene. Deep Sea Res. Part I 145, 125–136. doi: 10.1016/j.dsr.2019.02.001
Xu, W., Gong, L. F., Pang, K. L., and Luo, Z. H. (2018). Fungal diversity in deep-sea sediments of a hydrothermal vent system in the Southwest Indian Ridge. Deep Sea Res. Part I 131, 16–26. doi: 10.1016/j.dsr.2017.11.001
Yang, S., Xu, W., Gao, Y., Chen, X., and Luo, Z. H. (2020). Fungal diversity in deep-sea sediments from Magellan seamounts environment of the western Pacific revealed by high-throughput Illumina sequencing. J. Microbiol. 58, 841–852. doi: 10.1007/s12275-020-0198-x
Yuan, J., Lai, Q., Zheng, T., and Shao, Z. (2009). Novosphingobium indicum sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from a deep-sea environment. Int. J. Sys. Evol. Microbiol. 59, 2084–2088. doi: 10.1099/ijs.0.002873-0
Zengler, K., Toledo, G., Rappé, M., Elkins, J., Mathur, E. J., Short, J. M., et al. (2002). Cultivating the uncultured. Proc. Natl. Acad. Sci. U.S.A. 99, 15681–15686.
Zhang, C., Liu, Q., Li, X., Wang, M., Liu, X., Yang, J., et al. (2021). Spatial patterns and co-occurrence networks of microbial communities related to environmental heterogeneity in deep-sea surface sediments around Yap Trench, Western Pacific Ocean. Sci. Total Environ. 759:143799. doi: 10.1016/j.scitotenv.2020.143799
Zhang, D. C., Liu, Y. X., and Li, X. Z. (2015). Characterization of bacterial diversity associated with deep sea ferromanganese nodules from the South China Sea. J. Microbiol. 53, 598–605. doi: 10.1007/s12275-015-5217-y
Zhang, L., Kang, M., Huang, Y., and Yang, L. (2016a). Fungal communities from the calcareous deep-sea sediments in the Southwest India Ridge revealed by Illumina sequencing technology. World J. Microbiol. Biotechnol. 32:78.
Zhang, L., Kang, M., Xu, J., Xu, J., Shuai, Y., Zhou, X., et al. (2016b). Bacterial and archaeal communities in the deep-sea sediments of inactive hydrothermal vents in the Southwest India Ridge. Sci. Rep. 6:25982.
Zhang, X. Y., Tang, G. L., Xu, X. Y., Nong, X. H., and Qi, S. H. (2014). Insights into deep-sea sediment fungal communities from the East Indian Ocean using targeted environmental sequencing combined with traditional cultivation. PLoS One 9:e109118. doi: 10.1371/journal.pone.0109118
Keywords: benthic, bioinformatics, deep sea, metagenomic, next-generation sequencing
Citation: Barnes NM, Damare SR and Shenoy BD (2021) Bacterial and Fungal Diversity in Sediment and Water Column From the Abyssal Regions of the Indian Ocean. Front. Mar. Sci. 8:687860. doi: 10.3389/fmars.2021.687860
Received: 30 March 2021; Accepted: 14 July 2021;
Published: 10 August 2021.
Edited by:
De-Chao Zhang, Institute of Oceanology (CAS), ChinaReviewed by:
Eugenio Rastelli, Marche Polytechnic University, ItalyCopyright © 2021 Barnes, Damare and Shenoy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Belle Damodara Shenoy, YmVsbGVzaGVub3lAbmlvLm9yZw==
†ORCID: Samir R. Damare, orcid.org/0000-0002-4201-4844
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.