
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 30 July 2021
Sec. Aquatic Physiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.687377
 Peihua Zheng1,2
Peihua Zheng1,2 Xiuxia Zhang2,3
Xiuxia Zhang2,3 Dongmei Wang2,3
Dongmei Wang2,3 Juntao Li2,3
Juntao Li2,3 Zelong Zhang2,3
Zelong Zhang2,3 Yaopeng Lu1,2
Yaopeng Lu1,2 Jianan Xian2,3*
Jianan Xian2,3* Anli Wang1*
Anli Wang1* Lei Wang1*
Lei Wang1*Glutaredoxins (Grxs), small heat-stable oxidoreductases, are key members of the thioredoxin (Trx) superfamily. Recently, an emerging subclass of Grxs with a cysteine residue in the active site was found in shrimps. However, molecular functions of Grx-related proteins in decapods were rarely reported. In this study, a novel full-length Grx 3 (LvGrx 3) complementary DNA (cDNA) was identified in Pacific white shrimp (Litopenaeus vannamei), which had a 975-bp open reading frame (ORF) encoding a polypeptide of 324 amino acids. The nucleic acid sequence of Pacific white shrimp glutaredoxin 3 (LvGrx 3) showed 99.59% identity with genomic DNA (gDNA) sequence and 63.49% coverage. Sequence alignment showed that the amino acid sequence of LvGrx 3 shared 97% identity with black tiger shrimp (Penaeus monodon) Grx 3 and 62% identity with amphipod (Hyalella azteca) Grx 3. LvGrx 3 showed higher expression in the intestine, gill, and hepatopancreas, and lower expression in epithelium and abdominal nerve. In response to ammonia-N stress, LvGrx 3 was significantly upregulated in both the hepatopancreas and gill, and the peak value appeared after 24 h exposure. After lipopolysaccharide (LPS) injection, expression levels of LvGrx 3 in the hepatopancreas were increased in the middle stage, and LvGrx 3 in gill was upregulated in the middle and later periods (24 and 48 h). These results indicate that LvGrx 3 can participate in immune responses against ammonia-N stress and pathogen infection. However, RNA interference (RNAi) assay showed that LvGrx 3 silencing in ammonia-N-challenged shrimp could significantly induce the gene expression of antioxidant enzymes and aggravate the oxidative damage of protein and lipid. These results suggest that LvGrx 3 is involved in regulating the antioxidant system and plays a vital role in defense responses against environmental stress.
Pacific white shrimp (Litopenaeus vannamei) is one of the popular shrimp species cultivated in China (Williams et al., 1996; Chang et al., 2009). In the past two decades, the deteriorating aquaculture environment severely disrupted shrimp farming and caused stress-induced disease outbreaks (Tseng and Chen, 2004; Chiu et al., 2007; Qiu et al., 2011). Change in environmental factors has an important effect on increasing mortality (Mermoud et al., 1998). Environmental factors may induce stress reaction and immune-mediated depression, making shrimps susceptible to pathogens (Horowitz and Horowitz, 2001). Ammonia-N is one of the main environmental factors in the aquaculture system, which is toxic for aquatic animals, especially for shrimps. It can induce oxidative stress in various tissues and organs of shrimp, such as hepatopancreas, hemocytes, and nerve (Kosenko et al., 1997; Liu and Chen, 2004).
In many oxygenic organisms, reactive oxygen species (ROS) are generated as by-products in the process of aerobic metabolism (Wulf, 2002; Balaban et al., 2005). In many events, ROS can provoke severe oxidative stress and damage cellular proteins, lipids, or DNA (Cross et al., 1987; Toren, 2005; Leung and Chan, 2009). Organisms can protect themselves from the toxicity of excess ROS by developing different effective mechanisms (Poyton, 1999; Kondo et al., 2006). Glutaredoxin redox system is an important antioxidant system, which takes part in many physiological and biochemical processes and plays a critical role in defense mechanisms against oxidative stress (Grant, 2001; Lillig et al., 2008). As a key member of the glutaredoxin redox system, Grx is a small and conserved oxidoreductase belonging to the thioredoxin (Trx) superfamily, which is prevalent in both prokaryotes and eukaryotes (Fernandes and Holmgren, 2004; Lillig and Berndt, 2013). Grxs have highly conserved domains, which are exposed to the surface of the protein and accompanied by glutathione (GSH) in the vicinity (Rodríguez-Manzaneque et al., 1999). Grxs mainly use GSH, which serves as a substrate to catalyze the reduction of protein disulfides (Wells et al., 1990). Grxs reduce disulfides via two different mechanisms. Dithiol mechanism requires active site Cys residues, while monothiol mechanism relies on the N-terminal active site cysteine (Bushweller et al., 1992). According to the structure and catalytic mechanism, there are three kinds of Grxs (Vlamis-Gardikas and Holmgren, 2002; Lillig et al., 2005). The first kind of Grx is a classical Grx (9–14 kDa) with two cysteines in their active sites [e.g., Escherichia coli Grx 1, yeast (Saccharomyces cerevisiae) Grx 1 and Grx 2 and human Grxs] (Vlamis-Gardikas and Holmgren, 2002). The second kind of Grx consists of the monothiol Grxs, which is exemplified by the yeast (S. cerevisiae) Grx 3, Grx, 4 and Grx 5, lacking the C-terminal active site cysteine. All the three yeast Grxs have C-G-F-S as the active site motif (Rodríguez-Manzaneque et al., 1999). The third kind of Grx consists of E. coli Grx 2, which is a 24.3 kDa protein with more three-dimensional structural similarities to the glutathione S-transferase (GST) family of protein (Xia et al., 2001). Other proteins belonging to this kind of Grx are human glutathione transferase omega 1 (GSTO1) and chloride intracellular channel 1 (CLIC1) (Vlamis-Gardikas and Holmgren, 2002).
Grxs are of great interest in recent years due to their different and potent functions. Grx 3 participates in the process of cellular defense against oxidative stress, cell growth and proliferation, protection of protein oxidation, biogenesis of iron-sulfur clusters, and iron homeostasis (Li et al., 2007; Haunhorst et al., 2013). However, most researchers care about the species of E. coli (Shi et al., 1999), yeast (Chi et al., 2018), zebrafish (Danio rerio) (Haunhorst et al., 2013), mice, and human (Cheng et al., 2011). However, few studies were reported on decapods. In this study, the full length of Grx 3 was first cloned from Pacific white shrimp, and its molecular characterization was analyzed. In order to investigate the biological function of LvGrx 3, the expression pattern of LvGrx 3 was determined in Pacific white shrimp under ammonia-N stress and lipopolysaccharide (LPS) injection. Besides, LvGrx 3 was silenced to explore its role in Pacific white shrimp after ammonia-N exposure.
Healthy cultured Pacific white shrimps (L. vannamei) (body length of 10.6 ± 1.5 cm; weight of 11.2 ± 2.6 g) were obtained from a local shrimp farm in Haikou, Hainan province, China. The shrimps were maintained in cycling filtered plastic tanks with salinity of 15‰, temperature of 24 ± 2°C, and pH of 7.9–8. A commercial shrimp diet (40% protein, 5% fat, 5% fiber, and 16% ash; Haid Group, Guangzhou, China) was used to feed the shrimps twice daily before starting the experiment.
Hemolymph (400 μl) was extracted from the pericardial sinus of each shrimp with 25 gauge needle and 1.5 ml syringe, absorbing an equal volume of ice-cold shrimp anticoagulant solution (AS, 20.5 g L−1 glucose, 8 g L−1 sodium citrate, and 4.2 g L−1 sodium chloride, pH 7.5) (Xian et al., 2013). Hemocyte was separated by centrifugation at 800 g for 5 min at 4°C, and hemocyte pellets were used for RNA extraction. Tissue samples, such as hepatopancreas, intestine, gill, stomach, pyloric cecum, eyestalk, muscle, hemocytes, scape, epithelium, and abdominal nerve were collected from healthy shrimps and preserved in liquid nitrogen for RNA extraction (Yao et al., 2010).
Based on the previous transcriptome data of Pacific white shrimp, we obtained a partial middle fragment of Grx 3 cDNA using primers, such as Grx3-F1 and Grx3-R1. Semi-nested real-time polymerase chain reaction (RT-PCR) strategy was used to obtain an open reading frame (ORF) sequence near the 3′ end or 5′ end of LvGrx 3. The 3′ ends of Grx 3 were obtained with 3′-Full RACE Core Set with PrimeScript RTase (Takara, Dalian, China), and 5′ ends were obtained with SMART RACE cDNA Amplification Kit (Clontech, Mountain View, CA, United States). For the 3′ end, PCR was performed with gene-specific primer Grx3-3′F1 and 3′ rapid amplification of cDNA ends (RACE) outer primer. The PCR protocol was as follows: 1 cycle of denaturation at 94°C for 3 min, 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min, followed by a 10 min extension at 72°C. The 5′-untranslated regions (UTRs) were amplified with primer Grx3-5′R1 and universal primer mix (UPM). The PCR was carried out using 30 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 3 min. 5′- and 3′-RACE PCR products were gel purified, cloned, and sequenced as described previously (Zheng et al., 2019). All primer sequences are provided in Table 1.
The full-length cDNA of LvGrx 3 was identified using the National Center for Biotechnology Information (NCBI) BLAST search program (http://www.ncbi.nlm.nih.gov/blast). The nucleotide and predicted protein sequences of LvGrx 3 were analyzed using the DNAMAN software and the Expasy search program (http://au.expasy.org/tools/). Multiple sequence alignments of the predicted LvGrx 3 protein with known Grx 3 proteins were analyzed using the ClustalX software.
The phylogenetic tree was constructed using the neighbor-joining (NJ) method and plotted with Molecular Evolutionary Genetics Analysis (MEGA) version 6.0 software. Bootstrap sampling was performed with 1,000 replicates.
In order to assess the expression of LvGrx 3 in different tissues of Pacific white shrimp, quantitative real-time PCR (qRT-PCR) was conducted using the RNA of hepatopancreas, intestine, gill, stomach, pyloric cecum, eyestalk, muscle, hemocytes, scape, epithelium, and abdominal nerve from nine healthy shrimps. The housekeeping gene β-actin was used as an internal control.
A real-time quantitative PCR (qPCR) using Stratagene Mx3005P (Agilent, Santa Clara, CA, United States) was applied to study the expression of LvGrx 3 in different tissues of Pacific white shrimp. The primers of the target gene were Grx3-RT-F1 and Grx3-RT-R1 (Table 1). The RT-PCR conditions were as follows: 94°C for 3 min to activate the polymerase, followed by 40 cycles of 95°C for 15 s, 58°C for 15 s, and 72°C for 20 s. Dissociation analysis of amplification products was performed at the end of each PCR reaction to confirm that only a PCR product was amplified and detected. The comparative CT method (2−ΔΔCt method) was used to analyze the expression level of LvGrx 3.
Expression analysis of LvGrx 3 in shrimp was tested by ammonia-N stress or LPS injection. Two groups (control and 20 mg L−1 ammonia) were designed in the ammonia-N stress experiment, and the 20 mg L−1 ammonia-N solution was made using NH4Cl and 15‰disinfected seawater (Lin and Chen, 2001; Liu and Chen, 2004). Prior to the experiment, the mean doses of ammonia-N in two groups were tested by 0.01 and 20.45 mg L−1, respectively. Both groups were analyzed in triplicate with 25 shrimps per replicate in plastic tanks with 80 L disinfected seawater (salinity 15‰and temperature of 24 ± 2°C, pH7.9–8), which was aerated continuously using an air stone. The water environment of the plastic tanks was similar to the water environments of the culture ponds where the shrimps were purchased.
For the pathogen infection experiment, healthy shrimps were intramuscularly injected at the third abdominal segment with 2 μg μl−1 LPS or sterile physiological saline solution. LPS (2 mg ml−1, from E. coli 055: B5, Sigma-Aldrich, Shanghai, China) was dissolved in physiological saline solution (0.85% NaCl, Macklin, Shanghai, China) to give a dose of 2 μg μl−1. Shrimps were split into two categories, and each category was set in three replicates with 25 shrimps per replicate. According to a previous study (Xian et al., 2016), the LPS dose was 8 μg g−1 wet weight, and the shrimp in the control group was injected with the same amount of sterile physiological saline solution.
After 0, 3, 6, 12, 24, and 48 h ammonia-N stress or LPS injection, nine healthy shrimps were collected from different groups at each time point. Hemocytes were extracted, separated, and immediately used for RNA extraction. The tissues (gill and hepatopancreas) of each shrimp were dissected quickly and preserved separately in liquid nitrogen and then stored at −80°C before RNA extraction.
The total RNA was extracted from the tissues of shrimps by the TRIzol Reagent (Invitrogen, Carlsbad, CA, United States) following the instructions of the manufacturer. RNA concentration and quality were estimated using a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States), and RNA integrity was verified by 1% agarose gel electrophoresis. First-strand cDNA was generated from total RNA using PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Dalian, China).
production was quantified using the reduction of nitroblue tetrazolium (NBT) to formazan (Zhang et al., 2013). Hepatopancreas samples were homogenized in 10 volumes of ice-cold 0.85% NaCl solution in the ice bath for 2 min. Homogenates were centrifuged at 10,000 g for 15 min at 4°C, and then 20 μl supernatant was stained with 100 μl NBT solution (0.3%) for 30 min at room temperature. The NBT solution was removed and then washed one time with 100% methanol (Sigma-Aldrich, Shanghai, China) and three times with 70% methanol and then air-dried. Formazan was dissolved by adding 120 μl 2 M KOH(Sigma-Aldrich, Shanghai, China) and 140 μl dimethyl sulphoxide (Sigma-Aldrich, Shanghai, China). The optical density at 630 nm was measured using a microplate reader. production was expressed as NBT reduction in 20 μl of supernatant.
The double-stranded RNA (dsRNA) for LvGrx 3 and green fluorescent protein (GFP, as a negative control) gene were synthesized by in vitro transcription using T7 RiboMAX™ Express RNAi System (Promega, Madison, WI, United States). The gene-specific primers of LvGrx 3 and GFP dsRNA are presented in Table 1. The formation of the dsRNAs was quantified by UV spectrophotometry at an absorbance of 260 nm and analyzed by 1% agarose gel electrophoresis. The dsRNAs were stored at −80°C for gene silencing experiments.
RNA interference (RNAi) efficiency was detected by qRT-PCR and semi-quantitative RT-PCR. First, the synthetic dsRNA was dissolved in a protective buffer (10 mM Tris-HCl and 400 mM NaCl, pH 7.5) to make a final concentration of 2.5 μg μl−1. Then, the shrimps were split into two groups and injected with LvGrx 3 (2.5 μg g−1 shrimp) or GFP dsRNA (2.5 μg g−1 shrimp) into each shrimp at the lateral area of the fourth abdominal segment (Kong et al., 2018). The hepatopancreas samples in three shrimps from the test or control group were collected at 0, 12, 24, and 48 h after injection. Then, the total RNA was extracted from the hepatopancreas of shrimps by the method mentioned above. The LvGrx 3 cDNA sequence of hepatopancreas was amplified using specific primers, Grx3-RT-F1 and Grx3-RT-R1, and the β-actingene was used as an internal control. Finally, all the PCR products were identified by 2% agarose gel electrophoresis as a semi-quantitative analysis.
To study the effects of ammonia-N stress on shrimps after LvGrx 3 silencing, shrimps were randomly divided into two groups. One group was injected with Grx 3 dsRNA, and the other group was injected with GFP dsRNA. Then, both groups of shrimps were challenged by ammonia-N (20 mg L−1) after 12 h dsRNA injection, and hepatopancreas samples from nine shrimps in each group were collected at 0, 1.5, 3, 6, 12, and 24 h. These hepatopancreas samples were placed in liquid nitrogen and then stored at −80°C for RNA extraction.
qRT-PCR was performed to analyze the relative expression levels of the antioxidant genes, such as LvGrx 3, LvGrx 2, Trx, GPx, and GST in the shrimp of LvGrx 3-silenced and GFP groups. All gene-specific primer pairs are listed in Table 1.
In this study, the concentrations of protein carbonyls were measured using Protein Carbonyl Test Kit (A087, Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Protein carbonyl proteins were tested by a 2,4-dinitrophenylhydrazine reaction (Levine et al., 1990). Total protein concentrations were measured using Coomassie Brilliant Blue Protein Determination Kit (A045-2, Nanjing Jiancheng Bioengineering Institute, Nanjing, China). All tests were performed in triplicate, measuring the absorbance of each sample at 370 and 595 nm for protein carbonyl and protein contents, respectively.
Under oxidative stress conditions, the MDA content was used to assess the extent of lipid peroxidation by the thiobarbituric acid (TBA) assay. The MDA content in the hepatopancreas of shrimp was carried out using the MDA detection kit (A003-2) purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). The MDA content in lipid hydroperoxide decomposition products could condensate with TBA to produce red compounds with an absorption peak of 532 nm. All data were tested in triplicate, and the mean of the three values was used for statistical analysis.
All data were presented as means ± SD. One-way ANOVA was performed to analyze data, and a multiple comparison (Tukey) test was conducted to compare the significant differences among different treatments using the SPSS 18.0 software (SPSS Inc., Chicago, IL, United States). P < 0.05 was considered statistically significant.
The nucleotide and deduced amino acid sequences of LvGrx 3 are presented in Figure 1. The full-length cDNA for LvGrx 3 of Pacific white shrimp was 1,512 bp, containing a 975-bp ORF, which encodes 324 amino acids. It had 99.59% identity with genomic DNA (gDNA) sequence (GenBank accession no.: XM_027353970.1), and showed 63.49% coverage. It displayed 5.25% identity with LvGrx 2, which was cloned in the previous study (Zheng et al., 2019). The cDNA contained a 5' UTR of 93 nucleotides and a long 3'-UTR of 444 nucleotides, including a stop codon (TAA), a putative polyadenylation consensus signal (AATAAA), and a poly (A) tail. The deduced mature LvGrx 3 protein had a calculated molecular mass of 36 kDa and a pI of 4.93. The GSH binding sites of LvGrx 3 were formed by Lys141, 243, Cys149, 251, Gly150, 252, Phe151, 253, Thr189, 291, and Tyr190, 292.

Figure 1. Nucleotide and deduced amino acid sequences of Grx 3 gene of Pacific white shrimp (Litopenaeus vannamei). The letters in box indicate start codon (ATG), stop codon (TAA), and polyadenylation signal sequence (AATAAA). The catalytic amino acids (C-G-F-S) are shaded in gray. The other cysteine residues, Cys34, Cys45, and Cys136 in LvGrx 3 are underlined.
To investigate the relationship between LvGrx 3 and its homologs, multiple sequence alignment was performed. Blast analysis revealed that LvGrx 3 possessed a Trx domain, two Grx domains, and two active sites of the monothiol Grx CGFS (149–152 and 251–254 aa), indicating that LvGrx 3 is related to the monothiol kind of Grx family. LvGrx 3 shared the highest similarity with Grx 3 from black tiger shrimp (P. monodon) (GenBank accession no.: XP_037789122.1, 97%) and amphipod (H. azteca) (GenBank accession no.: XP_018026525.1, 62% identity). LvGrx 3 showed 61% percent identity with dampwood termite (Zootermopsis nevadensis) (GenBank accession no.: XP_021939072.1), and termite (Cryptotermes secundus) (GenBank accession no.: XP_023701670.1). LvGrx 3 showed 52% identity with brown marmorated stink bug (Halyomorph halys) (GenBank accession no.: XP_014281542.1). Multiple sequence alignment demonstrated that the deduced amino acid sequence of Grx 3 from Pacific white shrimp showed a high similarity in fundamental structure and function with that from other species (Figure 2). The signature monothiol CGFS active site and 12 GSH binding sites were highly conserved in all sequences of Grx 3.
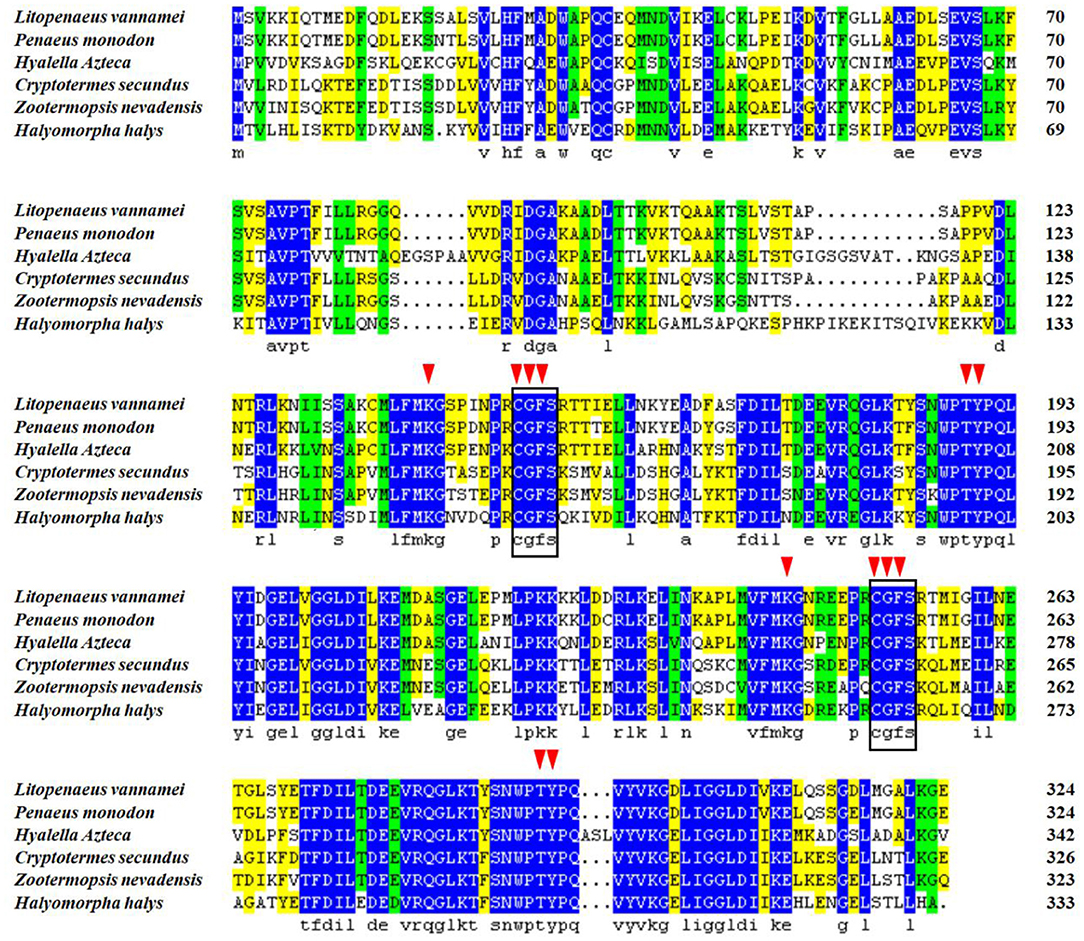
Figure 2. Multiple sequence alignment of Grx 3 in Pacific white shrimp (Litopenaeus vannamei). The blue region indicates that the sequences share the same amino acid residue. Gaps are indicated by spots. A square line box shows the special catalytic residues, and red triangle arrows represent 12 GSH binding sites.
In order to construct a phylogenetic tree, 15 sequences of Grx 3 and five sequences of Grx 2 were selected from 18 different species (Figure 3). Grx 3 sequences of 15 species used for the phylogenetic tree were black tiger shrimp (GenBank accession no: XP_037789122.1), amphipod (GenBank accession no: XP_018026525.1), termite (GenBank accession no: XP_023701670.1), German cockroach (Blattella germanica) (GenBank accession no: PSN36174.1), nematode (Caenorhabditis elegans) (GenBank accession no: NP_001023756.1), fungus (S. cerevisiae) (GenBank accession no: GFP69319.1), goldfish (Carassius auratus) (GenBank accession no: XP_026078075.1), zebrafish (Danio rerio) (GenBank accession no: NP_001005950.1), rohu (Labeo rohita) (GenBank accession no: RXN07040.1), lesser kestrel (Falco naumanni) (GenBank accession no: XP_040461937.1), red junglefowl (Gallus gallus) (GenBank accession no: NP_001264313.1), human (Homo sapiens) (GenBank accession no.: AAH05289.1), brown rat (Rattus norvegicus) (GenBank accession no: AAH86381.1), and house mouse (Mus musculus) (GenBank accession no: NP_075629.2). The Grx 2 sequences of the five species used for the phylogenetic tree were Pacific white shrimp (GenBank accession no: AYO52065.1), black tiger shrimp (GenBank accession no: XP_037791880.1), Chinese mitten crab (Eriocheir sinensis) (GenBank accession no: AXM05418.1), snow crab (Chionoecetes opilio) (GenBank accession no: KAG0711752.1), and gazami crab (Portunus trituberculatus) (GenBank accession no: MPC96022.1).
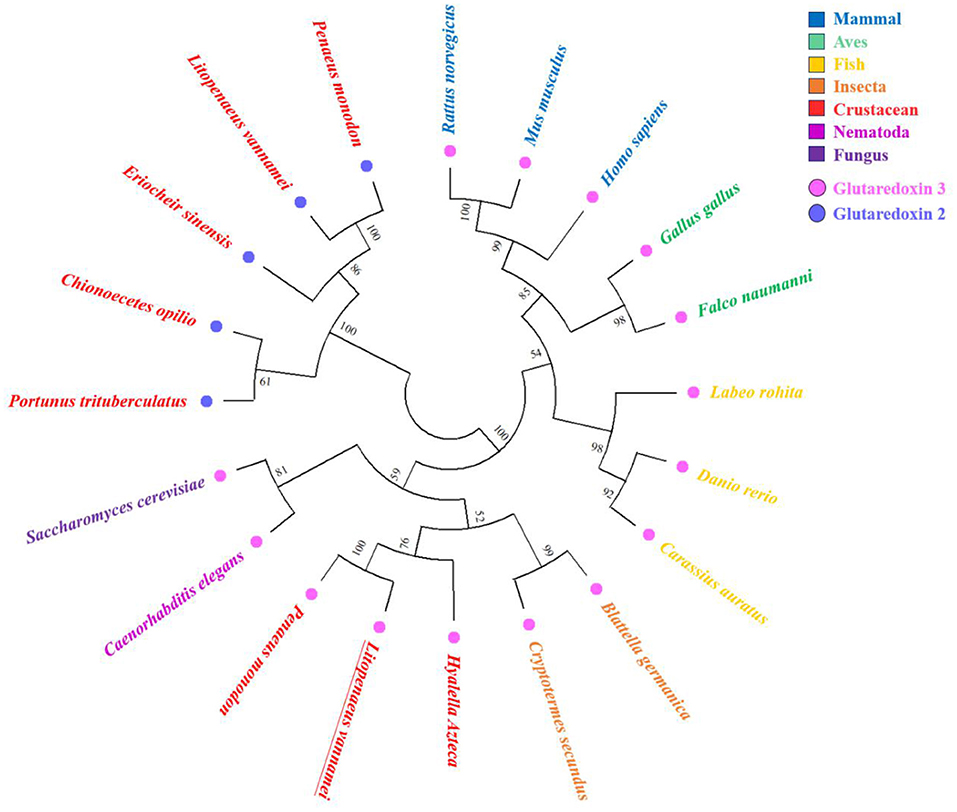
Figure 3. Phylogenetic tree using 15 sequences of Grx 3 and five sequences of Grx 2 from various species. Amino acids sequences are aligned using the ClustalX software, and the tree is constructed with the neighbor-joining (NJ) method in Molecular Evolutionary Genetics Analysis (MEGA) 4. The numbers at the forks indicate the bootstrap values. The corresponding species in different colors are presented on the right side of the figure. The pink dots indicate Grx 3, and the blue dots indicate Grx 2. LvGrx 3 is underlined in the figure.
The phylogenetic tree was mainly divided into two large branches, namely, Grx 2 of the crustaceans and Grx 3 from different classifications. The Grx 3 members were subdivided into two large groups. A group was originated from the vertebrate, and the other group was originated from invertebrate and fungus. The vertebrate group mainly contained mammal, aves, and fish. The invertebrate group included insects, crustaceans, and nematodes. The Grx 3 from the fungus was closer to the invertebrate group. LvGrx 3 formed a large group with fungus Grx 3, which is the typical example of the monothiol Grxs. Therefore, LvGrx 3 was proved to be a member of the Grx 3 family. Finally, LvGrx 3 shared the highest similarity with black tiger shrimp Grx 3 and formed a group with black tiger shrimp Grx 3 in the phylogenetic tree.
The results of qRT-PCR for tissue distribution analysis showed that LvGrx 3 could be detected in all the examined tissues of Pacific white shrimp. LvGrx 3 showed a significant expression (P < 0.05) in intestine, gill, and hepatopancreas compared with hemocytes. LvGrx 3 expression was low in epithelium and abdominal nerve (Figure 4).
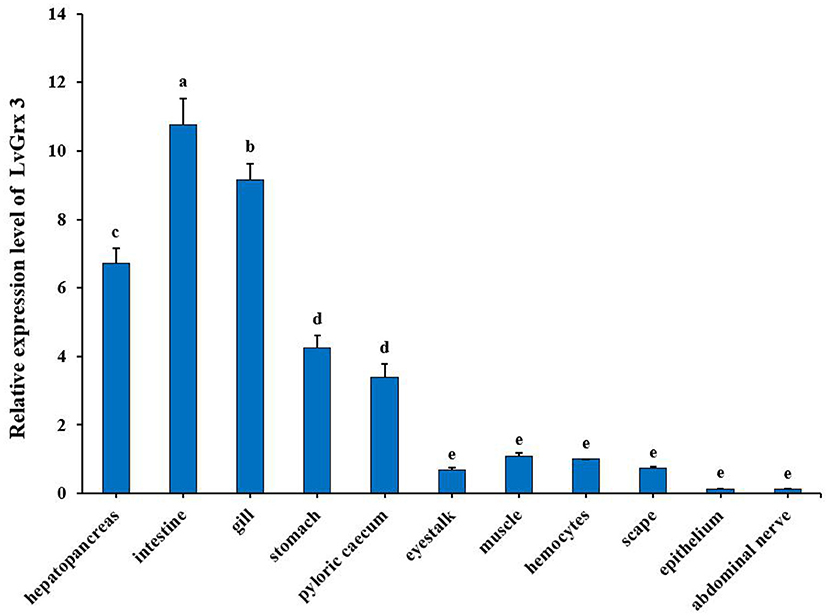
Figure 4. Relative expression of Grx 3 in different tissues of Pacific white shrimp (Litopenaeus vannamei), such as hepatopancreas, intestine, gill, stomach, pyloric cecum, eyestalk, muscle, hemocytes, scape, epithelium, and abdominal nerve (P < 0.05). The calculated data in the same group with different letters are significantly different (P < 0.05).
Expression profiles of LvGrx 3 in the hepatopancreas and gill of shrimp under ammonia-N stress are shown in Figure 5. In hepatopancreas, LvGrx 3 transcript level was increased at 12–48 h under ammonia-N stress (P < 0.05). After 24 h exposure, LvGrx 3 expression in the ammonia-N group was 2.44-fold higher than that of the control group. In gill, LvGrx 3 expression level was upregulated at 6–48 h exposure (P < 0.05), and the peak value was found after 24 h exposure, which was 8.23-fold of that in the control group.
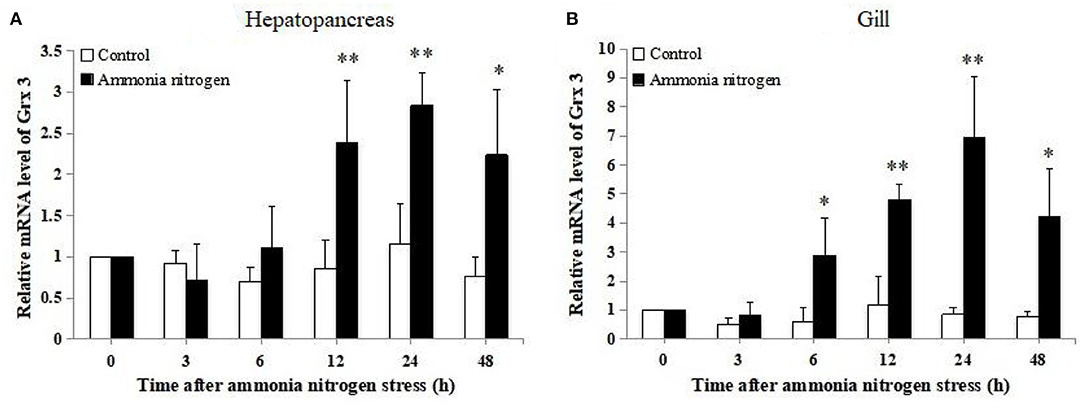
Figure 5. Analysis of Grx 3 expression in (A) hepatopancreas and (B) gill of ammonia-N stress and the control groups by real-time polymerase chain reaction (RT-PCR) at 0, 3, 6, 12, 24, and 48 h post-stress (n = 9). Statistical significance was calculated using Statistical Package for the Social Sciences (SPSS) 18.0 (*P < 0.05; **P < 0.01; ***P < 0.001).
The mRNA transcript levels of LvGrx 3 of hepatopancreas and gill in LPS-injected shrimps are presented in Figure 6. The transcriptional level of LvGrx 3 in hepatopancreas was upregulated after 24 h LPS injection. The transcriptional level of LvGrx 3 reached the highest level (P < 0.05) and then fell back to the level of the control group after 48 h (P > 0.05). LvGrx 3 expression level in gill was significantly increased after 12–48 h injection (P < 0.05), and the highest level was observed after 12 h.
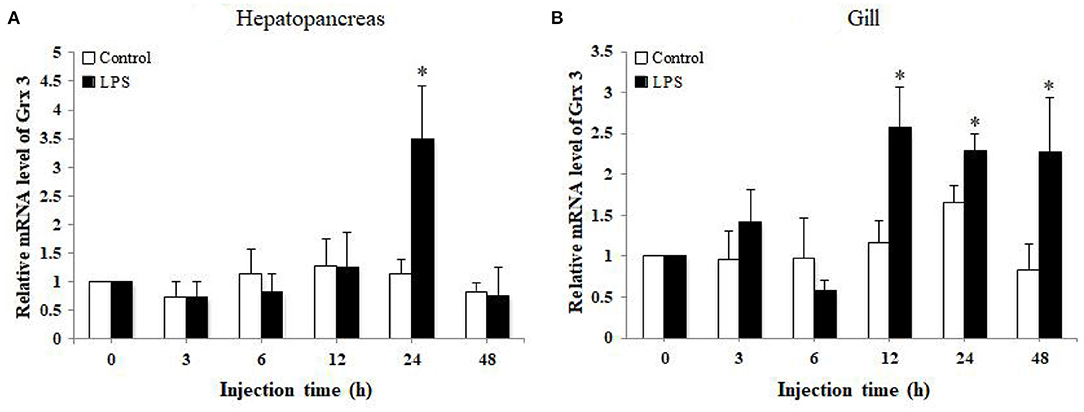
Figure 6. Analysis of Grx 3 expression in (A) hepatopancreas and (B) gill of LPS-injected and control groups by quantitative real-time polymerase chain reaction (qRT-PCR) at 0, 3, 6, 12, 24, and 48 h post injection (n = 9). Statistical significance was calculated using Statistical Package for the Social Sciences (SPSS) 18.0 (*P < 0.05; **P < 0.01).
The production in the hepatopancreas and gill of shrimps under ammonia-N stress is shown in Figure 7. production in hepatopancreas significantly increased at 6 and 24 h after ammonia-N stress (P < 0.05) and reached the peak at 6 h. In gill, production increased after 12–48 hexposure (P < 0.05), and the highest value was observed after 12 h exposure.
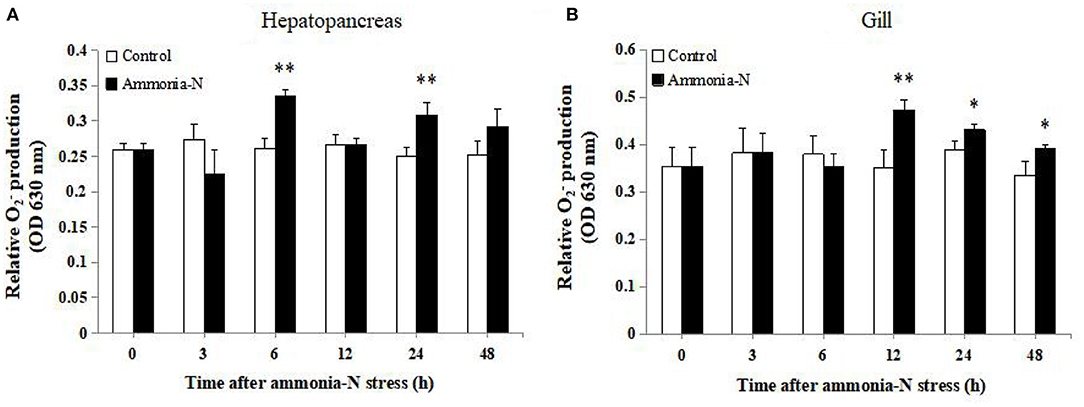
Figure 7. Analysis of production in (A) hepatopancreas and (B) gill of ammonia-N stress and control groups at 0, 3, 6, 12, 24, and 48 h post stress (n = 9). Statistical significance was calculated using Statistical Package for the Social Sciences (SPSS) 18.0 (*P < 0.05; **P < 0.01; ***P < 0.001).
As shown in Figure 8A, compared with the control group, LPS stimulation could significantly enhance production in hepatopancreas after 6, 12, and 24 h exposure (P < 0.05). At the same time, a significant increase in production of gill was observed after 3–12 h LPS injection (P < 0.05), and the peak level was detected after 3 h injection (Figure 8B).
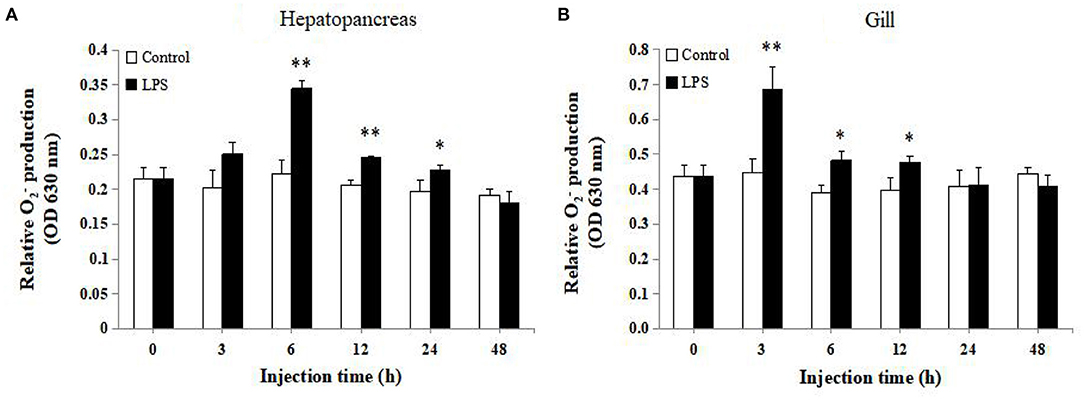
Figure 8. Analysis of production in (A) hepatopancreas and (B) gill of LPS-injected and control groups at 0, 3, 6, 12, 24, and 48 h post injection (n = 9). Statistical significance was calculated using Statistical Package for the Social Sciences (SPSS) 18.0 (*P < 0.05; **P < 0.01; ***P < 0.001).
In order to elucidate the role of LvGrx 3 in shrimps under ammonia-N stress, RNAi assay was conducted using LvGrx 3 gene-specific dsRNA. dsRNA (700 bp) of LvGrx 3 and GFP genes were synthesized, and their expression patterns were determined after dsRNA injection (Figure 9A). The silencing efficiency of dsLvGrx 3 was evaluated by qRT-PCR. The LvGrx 3 expression levels in dsLvGrx 3-injected shrimps were decreased to <50% of the original level at 12, 24, and 48 h post-injection. In contrast, LvGrx 3 transcription remained constant in shrimps injected with dsGFP (Figure 9B). After semi-quantitative RT-PCR analysis, a similar expression trend was observed in dsLvGrx 3 and dsGFP-injected shrimps (Figure 9C).
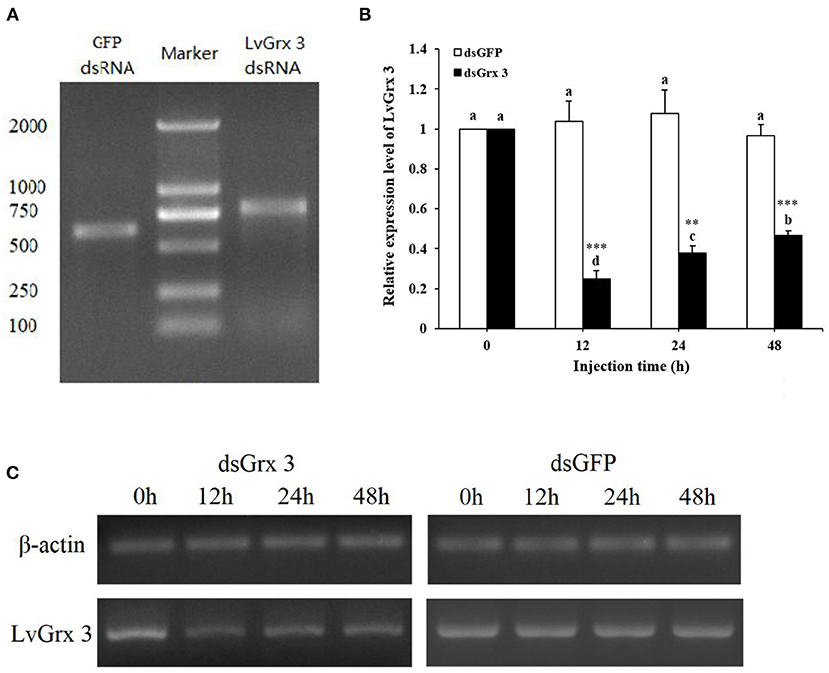
Figure 9. Quality of dsRNA synthesized in vitro and silencing efficiency of LvGrx 3 dsRNA injection. (A) Detection of purified dsRNA of GFP and LvGrx 3 genes by agarose gel electrophoresis. Lane 1, LvGrx 3 dsRNA; Lane 2, DNA marker (DL 2000, Takara, Dalian, China); Lane 3, GFP dsRNA. (B) Efficiency of dsRNA knock down of LvGrx 3 expression levels. Hepatopancreas samples from each shrimp were collected after LvGrx 3 or GFP dsRNA injection for quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis. Data are expressed as means ± standard deviation (SD) relative to the untreated group (0 h). Significant differences between dsGrx 3 and dsGFP-injected shrimps at the same exposure time are indicated with asterisks (*P < 0.05; **P < 0.01; ***P < 0.001). The calculated data in the same group with different letters are significantly different (P < 0.05). (C) LvGrx 3 expression in dsGFP and dsGrx 3-injected shrimps by semi-quantitative real time-polymerase chain reaction (RT-PCR). The β-actin gene was used as a control.
The relative expression level of LvGrx 3 is shown in Figure 10A. In the dsGFP-injected group, no significant change in LvGrx 3 expression was observed at 12 h injection (pre-exposure, 0 h). However, after 6–24 h ammonia-N exposure, LvGrx 3 of dsGFP-injected shrimps were significantly increased and showed a peak at 24 h (P < 0.05). In dsGrx 3-injected groups, the LvGrx 3 transcription was significantly inhibited at 12 h injection (P < 0.05). Although it was slightly upregulated during 1.5–24 h ammonia-N stress, the expression level of LvGrx 3 was lower than that under pre-injection (P < 0.05). The LvGrx 3-interfered shrimps had a significantly lower LvGrx 3 transcription than the dsGFP group before (0 h) ammonia-N stress and at all sampling times (P < 0.05).
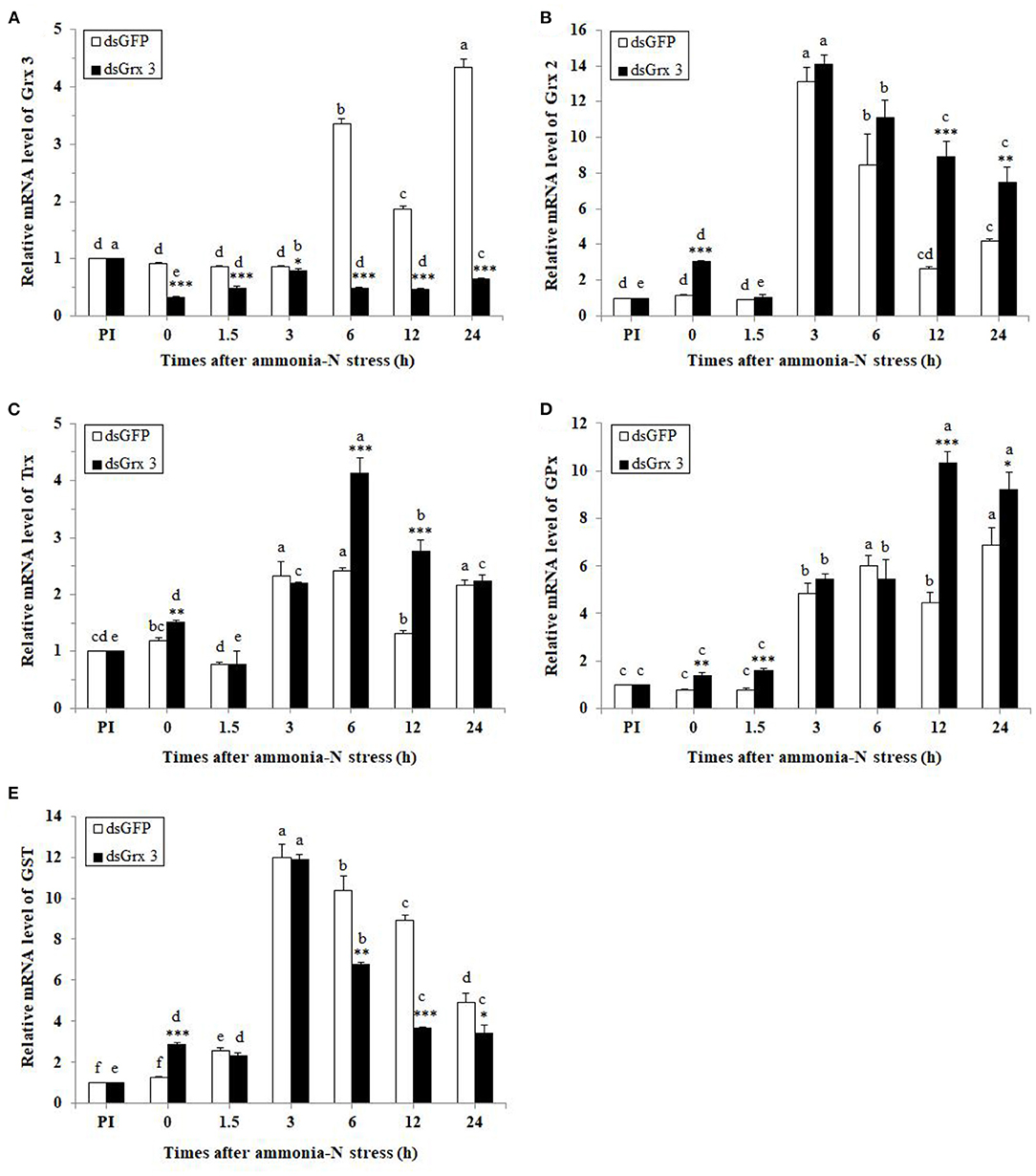
Figure 10. Expression profiles of five genes [LvGrx 3 (A), LvGrx 2 (B), Trx (C), GPx (D), and GST (E)] in the hepatopancreas of dsGrx 3-injected shrimp were collected after exposure to ammonia-N (dsGFP-injected shrimp was used as the control) (n = 9). PI, pre-injection of dsRNA. Significant differences between dsGrx 3 and dsGFP-injected shrimps at the same exposure time are indicated with asterisks (*P < 0.05; **P < 0.01; ***P < 0.001). The calculated data in the same group with different letters are significantly different (P < 0.05).
The relative level of LvGrx 2 is shown in Figure 10B. LvGrx 2 transcriptional levels in dsGFP-injected shrimps were significantly upregulated after 3, 6, and 24 h ammonia-N exposure (P < 0.05), while those in LvGrx 3-interfered shrimps were increased after 0 and 3–24 h (P < 0.05) with a peak at 3 h. Compared with the dsGFP-injected shrimps, transcriptional levels of LvGrx 2 in dsGrx 3-injected shrimps were higher than those of LvGrx 3 exposure after 0, 12, and 24 h (P < 0.05).
The relative level of Trx is shown in Figure 10C. Trx mRNA levels in dsGFP-injected shrimps were significantly enhanced after 3 to 24 h exposure (P < 0.05). In dsGrx 3-injected shrimps, a sharp increase in the expression of Trx was found after 0 and 3–24 h ammonia-N stress (P < 0.05). Compared with dsGFP-injected shrimps, transcriptional levels of Trx in LvGrx 3-interfered shrimps were relatively higher after 0, 6, and 12h exposure (P < 0.05).
The relative level of GPx is shown in Figure 10D. GPxmRNA levels were increased by ammonia-N exposure after 3 h and at all test times (P < 0.05) in both dsGFP and LvGrx 3-suppressed shrimps. Compared with dsGFP-injected shrimps, GPxmRNA expression was enhanced at 0–1.5 and 12–24 h (P < 0.05) in dsGrx 3-injected shrimps.
The relative level of GST is shown in Figure 10E. The levels of GST transcript in dsGFP-injected shrimps were upregulated at 1.5–24 h, and the highest level was observed after 3 h exposure (P < 0.05). Similarly, GST transcripts were high in dsGrx 3-injected shrimps throughout the exposure period and peaked at 3 h (P < 0.05). Compared with dsGFP-injected shrimps, GSTs in dsGrx 3-injected shrimps were lower at 6–24 h exposure (P < 0.05).
In dsGFP shrimps, protein carbonyl content gathered rapidly at 3–24 h, and the maximum level was observed at 6 h (P < 0.05). In LvGrx 3-interfered shrimps, its trend was continuously and substantially raised after 6 h, and was higher than the level of PI (P < 0.05). At the same time, the level of protein carbonyl in the dsGFP-injected group was significantly increased at 12 and 24 h after injection with the LvGrx 3 dsRNA (Figure 11A).
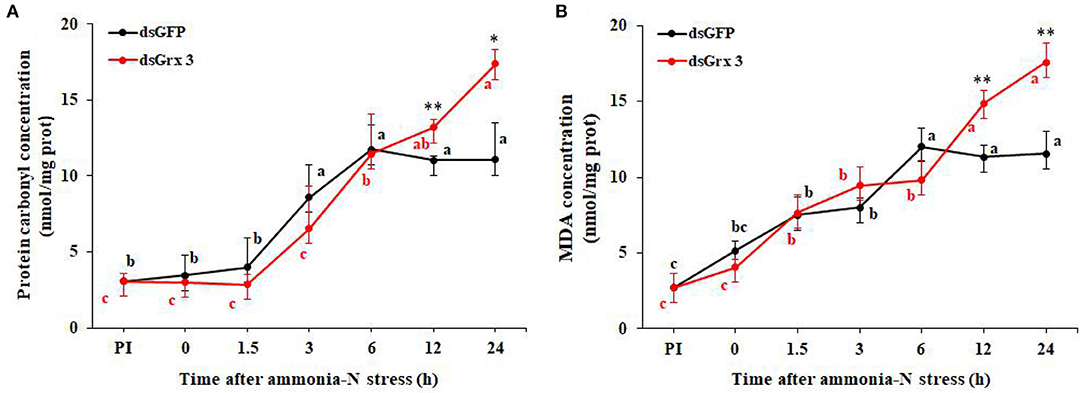
Figure 11. Time course of protein carbonyl (A) and MDA contents (B) in LvGrx 3-interfered shrimps after exposure to ammonia-N (n = 9). PI, pre-injection of dsRNA. Significant differences between dsGrx 3 and dsGFP-injected shrimps at the same exposure time are indicated with asterisks (*P < 0.05; **P < 0.01; ***P < 0.001). The calculated data in the same group with different letters are significantly different (P < 0.05).
As shown in Figure 11B, an obvious increase in the content of MDA was observed at 1.5–24 h in both dsGrx 3 and dsGFP-injected shrimps (P < 0.05). The MDA content increase (P < 0.05) was found in dsGrx 2 shrimps after ammonia-N exposure at 12 and 24 h.
Grx 3 is a small and conserved monothiol Grx, with a typically active Cys-Gly-Phe-Ser motif (Holmgren et al., 1975; Isakov et al., 2000; Vlamis-Gardikas and Holmgren, 2002). In previous studies, Grx 3 of E. coli, yeast, zebrafish, mice, and human was mainly studied (Shi et al., 1999; Cheng et al., 2011; Haunhorst et al., 2013; Chi et al., 2018). However, few studies reported molecular information and functions of Grx 3 in crustaceans. In this study, the full-length ORF of LvGrx 3 was identified, and it had a low homology (5.25% identity) with LvGrx 2, which was cloned in the study (Zheng et al., 2019). The distinct sequence difference in LvGrx 2 and LvGrx 3 genes suggests that this may perform different physiological functions in shrimps. In structure, LvGrx 3 was formed by two Grx-like domains (149CGFS152 and 251CGFS254) in tandem, belonging to monothiol Grx, whereas LvGrx 2 contained dimercap to active site CPYC (Zheng et al., 2019).
The tissue distribution of Grx 3 was studied in some vertebrates. In humans, Grx 3 showed stronger expression in the heart, spleen, testis, thymus, and peripheral blood leukocytes but displayed weaker expression in the lungs, placenta, colon, and small intestine (Witte et al., 2000). Similarly, mouse Grx3 was highly expressed in most tissues and developed organs. It was more robust in testis, brain, kidney, and stomach (Cheng et al., 2011). In zebrafish, Grx3 transcripts were presented in eye, brain, heart, and ventral region by in situ hybridization (Haunhorst et al., 2013). However, there are few reports on the tissue expression of Grx3 mRNA in crustaceans. In this study, LvGrx 3 was similar to the human and mouse Grx 3, which could be detected in all examined tissues. The abundance of Grx3 in different tissues was quite different, indicating that LvGrx 3 had a wide range of biological functions in various tissues and organs. The expression of LvGrx 3 was higher in the intestine, followed by gill and hepatopancreas, and lower in the epithelium and abdominal nerve. The intestinal immune system is an important defense line for crustaceans against environmental stress and pathogen infection. Hepatopancreas is the key organ for metabolism, immunity, and detoxification in crustaceans. Gill is the organ that has a direct contact with the external environment, and ambient biotic or abiotic substances enter into the aquatic animals through the gill, which is the main tissue involved in detoxification after environmental stress in shrimps. The higher expression of LvGrx 3 was detected in intestine, gill, and hepatopancreas, which might imply that this gene took part in innate immunity and detoxification.
Grx 3s participate in the process of cellular defense against oxidative stress, cell growth and proliferation, protection of protein oxidation, biogenesis of iron-sulfur clusters, and iron homeostasis (Li et al., 2007; Haunhorst et al., 2013). In fungus, Grx 3 could form a complex with Grx 4 and the bolA homolog Fra2, and it could assemble a [2Fe-2S] cluster acting as a signal to control the iron regulon in response to cellular iron status (Pujol Carrion et al., 2006; Li et al., 2009; Chi et al., 2018). Grx 3 and Grx 4 play important roles in actin cytoskeleton remodeling and cellular defense. The Trx domain of Grx3 and Grx4 regulates the remodeling of the actin cytoskeleton, whereas the Grx domains regulate the correct redox status of the cells. Both functions are essential for cell survival under oxidative conditions (Pujol Carrion and Torre Ruiz, 2010). However, the depletion of Grx3 impairs the maturation of hemoglobin and the major iron-consuming process during embryonic development in zebrafish. The house mouse Grx 3 was induced during oxidative stress, which indicates that it plays a crucial part in protecting cells against oxidative stress. Besides, the deletion of Grx3 in mice caused early embryonic lethality, which shows that Grx3 is required for efficient cell cycle progression (Cheng et al., 2011). Silencing of human Grx 3 expression in HeLa cells decreased the activities of several cytosolic Fe/S proteins, which could make cells unable to use iron efficiently (Haunhorst et al., 2013). However, the biological and physiological functions of Grx 3 in crustaceans have not been well-characterized. In this study, we determined the expression responses of LvGrx 3 against ammonia-N stress and LPS injection and its roles to participate in the defense system against environmental stress and pathogen infection.
Ammonia-N, a common and severe water pollutant in aquaculture, was derived from nitrogenous waste excretion of aquatic animals and food decomposition. It was proved to be toxic in aquatic animals, especially in decapods, such as Pacific white shrimp, Chinese shrimp (Fenneropenaeus chinensis), and crayfish (Procambarus clarkia) (Lee et al., 2010; Chang et al., 2015; Yao et al., 2016). In previous studies, ammonia-N could induce harmful ROS/reactive nitrogen species (RNS) overproduction and cause oxidative damage to tissues and organs. In order to prevent severe damage, the organism could produce antioxidant substances to resist oxidative stress (Ching et al., 2009; Li et al., 2014, 2016; Cheng et al., 2018). In this study, an upward trend in LvGrx 3 transcript was observed in the hepatopancreas and gill of shrimps after ammonia-N exposure, which was similar to the responses of LvGrx 2 (Zheng et al., 2019). LvGrx 3 and LvGrx 2 play key roles in defense mechanisms against ammonia-N stress by their common functions in redox regulation and antioxidant defense. Although the responses of LvGrx 3 were similar in hepatopancreas and gill, the ammonia-induced expression of LvGrx 3 in gill was higher than that in hepatopancreas. The highest level of LvGrx 3 expression in hepatopancreas was about 2.44-fold compared with the control group, while that was about 8.23-fold compared with the control group in gill. The enhancement of LvGrx 3 transcription in gill could be detected after 6 h stress, while upregulation occurred in hepatopancreas after 12 h exposure, demonstrating that the molecular response of LvGrx 3 in gill was more sensitive than that in hepatopancreas after ammonia-N challenge. These findings suggest that LvGrx 3 plays an important role in defense activities of gill against ammonia-N stress compared with hepatopancreas. Gill is the main channel through which ambient toxic factors, such as ammonia, enter into the aquatic animals (Xian et al., 2014). Therefore, gill is the most vulnerable organ under environmental stress, and the increase in LvGrx 3 expression of gill can protect this organ involved in respiration.
Furthermore, LvGrx 3 expression was knocked down in vivo to verify the defense mechanism against ammonia-N stress. The results confirmed that LvGrx 3 expression could be upregulated in the dsGFP control group after 6 h ammonia-N exposure, while LvGrx 3 expression was decreased in LvGrx 3-suppressed shrimps. Transcriptional levels of three antioxidant enzymes (LvGrx 2, TRx, and GPx) showed upregulation in the middle and later periods of exposure in LvGrx 3-suppressed shrimps, whereas GST expression was increased first and then decreased at the later stage. The results proved that decline of LvGrx 3 expression was linked to the upregulation of LvGrx 2, TRx, and GPx, which suggests that these genes play a vital role in the function of LvGrx 3. Both protein carbonyl and MDA contents in LvGrx 3-suppressed shrimps presented significant upregulation at 12 and 24 h exposure, which indicates that LvGrx 3 silencing increases the degree of oxidative damage in the middle and later stages. These results suggest that LvGrx 3 plays a vital role in antioxidative defense during ammonia-N stress.
LPS is a highly antigenic and cytotoxic substance, which is found in the outer membrane of Gram-negative bacteria. Recognition of LPS is essential for host antimicrobial defense reactions (Doe et al., 1978; Lorenzon et al., 2002). The cellular toxicity of LPS on hemocytes has been elucidated in shrimps (Lorenzon et al., 1999; Rodríguez-Ramos et al., 2008). The previous study demonstrated that LPS caused ROS/RNS-induced Ca2+-mediated hemocyte apoptosis and subsequent THC decline (Xian et al., 2013). LPS affected the expression levels of antioxidant-related genes in shrimps, such as LvGrx 2, catalase (CAT) and manganese superoxide dismutase (MnSOD) (Ji et al., 2009; Gómez-Anduro et al., 2012; Zheng et al., 2019). In this study, LvGrx 3 transcription was affected by LPS injection. Expression of LvGrx 3 in hepatopancreas was significantly upregulated at the middle stage, while ascension in gill was observed during middle and later periods (24 and 48 h), indicating that LvGrx 3 was involved in the immune response against Gram-negative bacterial infection. Response of LvGrx 3 was more sensitive in gill than that in hepatopancreas after the LPS challenge, but the highest relative expression level of LvGrx 3 was detected in hepatopancreas. The expression profile of LvGrx 3 after LPS stimulation was different from that under ammonia-N stress.
In summary, the full-length LvGrx 3 cDNA was cloned in Pacific white shrimp and identified as a member of the Trx superfamily. LvGrx 3 was observed in all tested tissues of Pacific white shrimp with higher expression in the intestine, gill, and hepatopancreas. The mRNA expression levels of LvGrx 3 in hepatopancreas and gill were induced when challenged by ammonia-N exposure and LPS injection, suggesting that LvGrx 3 plays a critical role in shrimp defense mechanism against environmental stress and pathogen infection. LvGrx 3 silencing in shrimp challenged with ammonia-N significantly induced the gene expressions of some antioxidant enzymes and aggravated the oxidative damages of protein and lipid. These results demonstrate that LvGrx 3 is involved in regulating the antioxidant system and plays a vital role in the defense mechanism against environmental stress.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
PZ, JX, AW, and LW: conceptualization. PZ, XZ, JL, ZZ, YL, and LW: methodology. PZ, XZ, JX, and LW: data curation. PZ: writing—original draft preparation. JX, AW, LW, and DW: writing—review and editing. All authors contributed to the article and approved the submitted version.
This research was supported by Hainan Provincial Natural Science Foundation of China (Grant No. 320RC710), Guangdong Provincial Natural Science Foundation (Grant No. 2017A030313194), Central Public Interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (Grant No. 1630052019013), and National Natural Science Foundation of China (Grant No. 31500326).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Balaban, R. S., Nemoto, S., and Finkel, T. (2005). Mitochondria, oxidants, and aging. Cell 120, 483–495. doi: 10.1016/j.cell.2005.02.001
Bushweller, J. H., Aaslund, F., Wuthrich, K., and Holmgren, A. (1992). Structural and functional characterization of the mutant Escherichia coli glutaredoxin (C14.fwdarw.S) and its mixed disulfide with glutathione. Biochemistry 31, 9288–9293. doi: 10.1021/bi00153a023
Chang, M., Wang, W. N., Wang, A. L., Tian, T. T., Wang, P., Zheng, Y., et al. (2009). Effects of cadmium on respiratory burst, intracellular Ca2+ and DNA damage in thewhite shrimp Litopenaeus vannamei. Comp. Biochem. Physiol. C 149, 581–586. doi: 10.1016/j.cbpc.2008.12.011
Chang, Z. W., Chiang, P. C., Cheng, W., and Chang, C. C. (2015). Impact of ammonia exposure on coagulation in white shrimp, Litopenaeus vannamei. Ecotox. Environ. Safe. 118, 98–102. doi: 10.1016/j.ecoenv.2015.04.019
Cheng, C. H., Guo, Z. X., and Wang, A. L. (2018). Growth performance and protective effect of vitamin E on oxidative stress pufferfish (Takifugu obscurus) following by ammonia stress. Fish Physiol. Biochem. 44, 735–745. doi: 10.1007/s10695-018-0468-2
Cheng, N. H., Zhang, W., Chen, W. Q., Jin, J., Cui, X., Butte, N. F., et al. (2011). Amammalian monothiol glutaredoxin, Grx3, is critical for cell cycle progression during embryogenesis. FEBS J. 278, 2525–2539. doi: 10.1111/j.1742-4658.2011.08178.x
Chi, C. B., Tang, Y. J., Zhang, J., Dai, Y. N., Abdalla, M., Chen, Y., et al. (2018). Structural and biochemical insights into the multiple functions of yeast Grx3. J. Mol. Biol. 430, 1235–1248. doi: 10.1016/j.jmb.2018.02.024
Ching, B., Chew, S. F., Wong, W. P., and Ip, Y. K. (2009). Environmental ammonia exposure induces oxidative stress in gills and brain of Boleophthalmus boddarti (mudskipper). Aquat. Toxicol. 95, 203–212. doi: 10.1016/j.aquatox.2009.09.004
Chiu, C. H., Guu, Y. K., Liu, C. H., Pan, T. M., and Cheng, W. (2007). Immune responses and gene expression in white shrimp, Litopenaeus vannamei, induced by Lactobacillus plantarum. Fish Shellfish Immunol. 23, 364–377. doi: 10.1016/j.fsi.2006.11.010
Cross, C. E., Halliwell, B., Borish, E. T., Pryor, W. A., Ames, B. N., Saul, R. L., et al. (1987). Oxygen radicals and human disease. Ann. Intern. Med. 107, 526–545. doi: 10.7326/0003-4819-107-4-526
Doe, W. F., Yang, S. T., Morrison, D. C., Betz, S. J., and Henson, P. M. (1978). Macrophage stimulation by bacterial lipopolysaccharides. II. Evidence for differentiation signals delivered by lipid A and by a protein rich fraction of lipopolysaccharides. J. Exp. Med. 148, 557–568. doi: 10.1084/jem.148.2.557
Fernandes, A. P., and Holmgren, A. (2004). Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid. Redox Signal 6, 63–74. doi: 10.1089/152308604771978354
Gómez-Anduro, G. A., Ascencio-Valle, F., Peregrino-Uriarte, A. B., Cámpa-Córdova, A., and Yepiz-Plascencia, G. (2012). Cytosolic manganese superoxide dismutase genes from the white shrimp Litopenaeus vannameiare differentially expressed in response to lipopolysaccharides, white spot virus and during ontogeny. Comp. Biochem. Physiol. B 162, 120–125. doi: 10.1016/j.cbpb.2012.03.003
Grant, C. M. (2001). Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol. Microbiol. 39, 533–541. doi: 10.1046/j.1365-2958.2001.02283.x
Haunhorst, P., Hanschmann, E. M., Bräutigam, L., Stehling, O., Hoffmann, B., Mühlenhoff, U., et al. (2013). Crucial function of vertebrate glutaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation. Mol. Biol. Cell 24, 1895–1903. doi: 10.1091/mbc.e12-09-0648
Holmgren, A., Soderberg, B., Eklund, H., and Branden, C. I. (1975). Three-dimensional structure of Escherichia coli thioredoxin-S2 to 2.8 A resolution. P. Natl. Acad.Sci. U.S.A. 72, 2305–2309. doi: 10.1073/pnas.72.6.2305
Horowitz, A., and Horowitz, S. (2001). “Disease control in shrimp aquaculture from a microbial ecology perspective,” in The New Wave, Proceedings of the Special Session on Sustainable Shrimp Culture (Baton Rouge, LA), 199–218.
Isakov, N., Witte, S., and Altman, A. (2000). PICOT-HD: a highly conserved protein domain that is often associated with thioredoxin and glutaredoxin modules. Trends Biochem. Sci. 25, 537–539. doi: 10.1016/S0968-0004(00)01685-6
Ji, P. F., Yao, C. L., and Wang, Z. Y. (2009). Immune response and gene expression in shrimp (Litopenaeus vannamei) hemocytes and hepatopancreas against some pathogen-associated molecular patterns. Fish Shellfish Immunol. 27, 563–570. doi: 10.1016/j.fsi.2009.08.001
Kondo, N., Nakamura, H., Masutani, H., and Yodoi, J. (2006). Redox regulation of human thioredoxin network. Antioxid. Redox Signal. 8, 1881–1890. doi: 10.1089/ars.2006.8.1881
Kong, J. R., Wei, W., Qiao, X. L., Kang, H., Huang, D., Liu, Y., et al. (2018). Identifying the function of lvpi3k during the pathogenic infection of litopenaeus vannamei by Vibrio alginolyticus. Fish Shellfish Immunol. 76, 355–367. doi: 10.1016/j.fsi.2018.03.016
Kosenko, E., Kaminsky, M., Kaminsky, A., Valencia, M., Lee, L., Hermenegildo, C., et al. (1997). Superoxide production and antioxidant enzymes in ammonia intoxication in rats. Free Rad. Res. 27, 637–644. doi: 10.3109/10715769709097867
Lee, B. J., Sis, R. F., Lewis, D. H., and Marks, J. E. (2010). Histology of select organs of the crawfish procambarus clarkii maintained at various temperatures and levels of calcium and ammonia1, 2. J. World Aquacult. Soc. 16, 193–204. doi: 10.1111/j.1749-7345.1985.tb00201.x
Leung, P. S., and Chan, Y. C. (2009). Role of oxidative stress in pancreatic inflammation. Antioxid. Redox Signal. 11, 135–165. doi: 10.1089/ars.2008.2109
Levine, R. L., Garland, D., and Oliver, C. N. (1990). Determination of carbonyl content in oxidatively modified proteins. Method Enzymol. 186, 467–478. doi: 10.1016/0076-6879(90)86141-H
Li, H., Mapolelo, D. T., Dingra, N. N., Naik, S. G., Lees, N. S., Hoffman, B. M., et al. (2009). The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry 48, 9569–9581. doi: 10.1021/bi901182w
Li, M., Gang, F. Y., Gao, Y., and Wu, Q. Y. (2007). Glutaredoxin system and its regulation to the cytosolic thiol-redox status. Acta Biophys. Sin. 23, 343–350.
Li, M., Gong, S., Li, Q., Yuan, L., Meng, F., and Wang, R. (2016). Ammonia toxicity induces glutamine accumulation, oxidative stress and immunosuppression in juvenile yellow catfish Pelteobagrus fulvidraco. Comp. Biochem. Physiol. C 183–184, 1–6. doi: 10.1016/j.cbpc.2016.01.005
Li, M., Yu, N., Qin, J. G., Li, E., Du, Z. Y., and Chen, L. Q. (2014). Effects of ammonia stress, dietary linseed oil and Edwardsiella ictaluri challenge on juvenile darkbarbel catfish Pelteobagrus vachelli. Fish Shellfish Immunol. 38, 158–165. doi: 10.1016/j.fsi.2014.03.015
Lillig, C. H., and Berndt, C. (2013). Glutaredoxins in Thiol/Disulfide Exchange. Antioxid. Redox Signal. 18, 1654–1665. doi: 10.1089/ars.2012.5007
Lillig, C. H., Berndt, C., and Holmgren, A. (2008). Glutaredoxin systems. Biochim. Biophys. Acta. 1780, 1304–1317. doi: 10.1016/j.bbagen.2008.06.003
Lillig, C. H., Berndt, C., Vergnolle, O., Lonn, M. E., Hudemann, C., Bill, E., et al. (2005). Characterization of human glutaredoxin 2 as iron-sulfur protein: a possible role as redox sensor. PNAS 102, 8168–8173. doi: 10.1073/pnas.0500735102
Lin, Y. C., and Chen, J. C. (2001). Acute toxicity of ammonia on Litopenaeus vannamei Boone juveniles at different salinity levels. J. Exp. Mar. Bio. Ecol. 259, 109–119. doi: 10.1016/S0022-0981(01)00227-1
Liu, C. H., and Chen, J. C. (2004). Effect of ammonia on the immune response of white shrimpLitopenaeus vannamei and its susceptibility to Vibrio alginolyticus. Fish Shellfish Immunol. 16, 321–334. doi: 10.1016/S1050-4648(03)00113-X
Lorenzon, S., Guarrini, S. D., Smith, V. J., and Ferrero, E. A. (1999). Effects of LPS injection on circulating haemocytes in crustaceans in vivo. Fish Shellfish Immunol. 9, 31–50. doi: 10.1006/fsim.1998.0168
Lorenzon, S., Pasqual, P., and Ferrero, E. A. (2002). Different bacterial lipopolysaccharides as toxicants and stressors in the shrimp Palaemon elegans. Fish Shellfish Immunol. 13, 27–45. doi: 10.1006/fsim.2001.0379
Mermoud, I., Costa, R., Ferré, O., Goarant, C., and Haffner, P. (1998). 'Syndrome 93' in New Caledonian outdoor rearing ponds of Penaeus stylirostris: History and description of three major outbreaks. Aquaculture 164, 323–335. doi: 10.1016/S0044-8486(98)00197-5
Poyton, R. O. (1999). Models for oxygen sensing in yeast: implications for oxygen-regulated gene expression in higher eucaryotes. Resp. Physiol. 115, 119–133. doi: 10.1016/S0034-5687(99)00028-6
Pujol Carrion, N., Belli, G., Herrero, E., Nogues, A., and Torre Ruiz, M. A. (2006). Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J. Cell Sci. 119, 4554–4564. doi: 10.1242/jcs.03229
Pujol Carrion, N., and Torre Ruiz, M. A. (2010). Glutaredoxins Grx4 and Grx3 of Saccharomyces cerevisiae play a role in actin dynamics through their Trx domains, which contributes to oxidative stress resistance. Appl. Environ. Microb. 76, 7826–7835. doi: 10.1128/AEM.01755-10
Qiu, J., Wang, W. N., Wang, L., Liu, Y. F., and Wang, A. L. (2011). Oxidative stress, DNA damage and osmolality in the Pacific white shrimp, Litopenaeus vannamei exposed to acute low temperature stress. Comp. Biochem. Physiol. C 154, 36–41. doi: 10.1016/j.cbpc.2011.02.007
Rodríguez-Manzaneque, M. T., Ros, J., Cabiscol, E., Sorribas, A., and Herrero, E. (1999). Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol. Cell Biol. 19, 8180–8190. doi: 10.1128/MCB.19.12.8180
Rodríguez-Ramos, T., Espinosa, G., Hernández-López, J., Gollas-Galván, T., Marrero, J., Borrell, Y., et al. (2008). Effects of Echerichia coli lipopolysaccharides and dissolved ammonia on immune response in southern white shrimp Litopenaeus schmitti. Aquaculture 274, 118–125. doi: 10.1016/j.aquaculture.2007.10.049
Shi, J., Vlamis-Gardikas, A., Aslund, F., Holmgren, A., and Rosen, B. P. (1999). Reactivity of glutaredoxins 1, 2, and 3 from Escherichia coli shows that glutaredoxin 2 is the primary hydrogen donor to ArsC-catalyzed arsenate reduction. J. Biol. Chem. 274, 36039–36042. doi: 10.1074/jbc.274.51.36039
Toren, F. (2005). Radical medicine: treating ageing to cure disease. Nat. Rev. Mol. Cell Bio. 6, 971–976. doi: 10.1038/nrm1763
Tseng, I., and Chen, J. C. (2004). The immune response of white shrimp Litopenaeus vannamei and its susceptibility to Vibrio alginolyticus under nitrite stress. Fish Shellfish Immunol. 17, 325–333. doi: 10.1016/j.fsi.2004.04.010
Vlamis-Gardikas, A., and Holmgren, A. (2002). Thioredoxin and glutaredoxin isoforms. Method. Enzymol. 347, 286–296. doi: 10.1016/S0076-6879(02)47028-0
Wells, W. W., Xu, D. P., Yang, Y. F., and Rocque, P. A. (1990). Mammalian thioltransferase (glutaredoxin) and protein disulfide isomerase have dehydroascorbate reductase activity. J. Biol. Chem. 265, 15361–15364. doi: 10.1016/S0021-9258(18)55401-6
Williams, A. S., Davis, D. A., and Arnold, C. R. (1996). Density-Dependent growth and survival of Penaeus setiferus and Penaeus vannamei in a semi-closed recirculating system. J. World Aquacult. Soc. 27, 107–112. doi: 10.1111/j.1749-7345.1996.tb00600.x
Witte, S., Villalba, M., Bi, K., Liu, Y., Isakov, N., and Altman, A. (2000). Inhibition of the c-Jun N-terminal kinase/AP-1 and NF-kappaB pathways by PICOT, a novel protein kinase C-interacting protein with a thioredoxin homology domain. J. Biol. Chem. 275, 1902–1909. doi: 10.1074/jbc.275.3.1902
Wulf, D. (2002). Free radicals in the physiological control of cell function. Physiol. Rev. 82, 47–95. doi: 10.1152/physrev.00018.2001
Xia, B., Vlamis-Gardikas, A., Holmgren, A., Wright, P. E., and Dyson, H. J. (2001). Solution structure of Escherichia coli glutaredoxin-2 shows similarity to mammalian glutathione-s-transferases. J. Mol. Biol. 310, 907–918. doi: 10.1006/jmbi.2001.4721
Xian, J. A., Miao, Y. T., Li, B., Guo, H., and Wang, A. L. (2013). Apoptosis of tiger shrimp (Penaeus monodon) haemocytes induced by Escherichia coli lipopolysaccharide. Comp. Biochem. Physiol. A164, 301–306. doi: 10.1016/j.cbpa.2012.10.008
Xian, J. A., Qian, K., Hui, G., Miao, Y. T., Wang, A. L., and Wang, D. M. (2014). Research progress intoxic effects of ammonia-N on shrimp. Feed Indust. 35, 52–58.
Xian, J. A., Zhang, X. X., Guo, H., Wang, D. M., and Wang, A. L. (2016). Cellular responses of the tiger shrimp Penaeus monodon haemocytes after lipopolysaccharide injection. Fish Shellfish Immunol. 54, 385–390. doi: 10.1016/j.fsi.2016.04.130
Yao, C. L., Ji, P. F., Wang, Z. Y., Li, F. H., and Xiang, J. H. (2010). Molecular cloning and expression of NOS in shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 28, 453–460. doi: 10.1016/j.fsi.2009.12.002
Yao, W. L., He, Y. Y., Liu, P., Li, J., and Wang, Q. Y. (2016). Cloning and expression analysis of the MKK3 gene in Fenneropenaeus chinensis under ammonia-N stress. J. Fish. Sci. China 23, 34–43.
Zhang, S. P., Li, J. F., Wu, X. C., Zhong, W. J., Xian, J. A., Liao, S. A., et al. (2013). Effects of different dietary lipid level on the growth, survival and immune-relating genes expression in Pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 34, 1131–1138. doi: 10.1016/j.fsi.2013.01.016
Keywords: glutaredoxin, Ammonia-N, LPS, stress, shrimp
Citation: Zheng P, Zhang X, Wang D, Li J, Zhang Z, Lu Y, Xian J, Wang A and Wang L (2021) Molecular Characterization and Expression Analysis of a Novel Glutaredoxin 3 Gene in Pacific White Shrimp (Litopenaeus vannamei). Front. Mar. Sci. 8:687377. doi: 10.3389/fmars.2021.687377
Received: 29 March 2021; Accepted: 28 June 2021;
Published: 30 July 2021.
Edited by:
Samad Rahimnejad, University of South Bohemia in Ceské Budějovice, CzechiaReviewed by:
Haoyang Li, Sun Yat-sen University, ChinaCopyright © 2021 Zheng, Zhang, Wang, Li, Zhang, Lu, Xian, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianan Xian, eGlhbi1qYUAxNjMuY29t; Anli Wang, d2FuZ2FubDE5NTdAMTYzLmNvbQ==; Lei Wang, d2FuZ2xlaUBzY251LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.