
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 04 June 2021
Sec. Aquatic Physiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.685429
This article is part of the Research TopicOmics Approaches in Aquatic Nutritional PhysiologyView all 7 articles
Wnt/β-catenin signalling plays an essential role in the immunity of Penaeus vannamei. In this study, the effects of dietary Wnt/β-catenin pathway activator TWS119 on the growth, immunity, and transcriptome response in P. vannamei were investigated. Penaeus vannamei were fed diets with added TWS119 at doses of 0 (T0), 0.25 (T0.25), 1 (T1), 4 (T4), 16 (T16), or 64 mg·kg−1 (T64), respectively. LvGSK3β activity was effectively inhibited in P. vannamei given TWS119. The growth of P. vannamei in the T16 group was significantly improved when compared with the control group. After Vibrio parahaemolyticus infection, the survival rates (SRs) of P. vannamei in all experimental groups except the T64 group were significantly higher than in the T0 group. Compared with the control group, the immune enzymes’ activities in the serum of P. vannamei increased in all the experimental groups, while the malondialdehyde (MDA) contents decreased. Transcriptome analysis identified 5,073 differentially expressed genes (DEGs) for P. vannamei in the T0 and T16 groups. Most of the DEGs are involved in the ribosome pathway, endocytosis, glycerophospholipid metabolism, Wnt signalling, and FoxO =signalling pathways. The majority of the DEGs were from the ribosome pathway, which is also the most significantly enriched pathway. The study confirmed that the growth and immunity status of P. vannamei could improve by increasing dietary TWS119, which probably regulates the activity of the Wnt/β-catenin pathway and may be closely related to ribosome function and energy metabolism.
The white shrimp, Penaeus vannamei, is the most important commercially cultured shrimp species in Asia and around the world (Kuo et al., 2019). However, in recent decades, the P. vannamei aquaculture industry has been threatened by several viral pathogens and Vibrio infections, causing severe economic losses worldwide (Kuo et al., 2019; Zeng et al., 2019). Traditionally, antibiotics are used to treat and prevent bacterial diseases. However, such use of antibiotics has resulted in severe threats to human health and environmental security because of residual antibiotics found in animals and the emergence of resistant bacteria, both of which are not conducive to developing the shrimp industry (Holmström et al., 2003). Thus, it is essential to develop non-toxic, non-polluting, and highly efficient therapeutic agents that can replace antibiotics.
The pathogenicity of shrimp is caused by the complex interaction between the shrimp itself, pathogens, and the environment. One of the fundamental strategies for controlling shrimp diseases is to enhance shrimp’s immunity (Lightner et al., 2005). As a crustacean species, shrimp lacks an antibody-based vertebrate-type adaptive immune system, and innate immune responses play crucial roles in defense against pathogens (Li and Xiang, 2013). Recently, a growing number of signalling pathways have been shown to play roles in the innate immune system of P. vannamei, including Toll, IMD, JAK-STAT, and Wnt/β-catenin pathways (Song et al., 2015; Zhang et al., 2016; Li et al., 2019). Immunostimulants, also known as immune modulators, are substances that activate an animals’ immune system to make them more resistant to microbial infections. Presently, immunostimulants have been considered suitable substitutes for antibiotics in shrimp culture (Apines-Amar and Amar, 2015).
Many kinds of immunostimulants have previously been used in shrimp culture, such as polysaccharides (Dalmo and Bogwald, 2008; Deng et al., 2015), oligosaccharides (Wang and Chen, 2005), vitamins (Liu et al., 2007; Qiao et al., 2011), microbial products (Flores-Miranda et al., 2011; Peraza-Gomez et al., 2014), and medicinal plants (Harikrishnan et al., 2011; Deng et al., 2015). However, the efficacy of immunostimulants is influenced by their timing, delivery, and dosage requirements. Additionally, immunostimulants’ mechanisms of action are challenging to elucidate (Sheng et al., 2014). Interestingly, β-1,3-glucan, lipopolysaccharides (LPS), and peptidoglycan (PG), which are all the pathogen-associated molecular patterns (PAMPs) of the Toll signalling pathway, could improve the immunity status of shrimps and have been reported as effective immunostimulants (Suphantharika et al., 2003; Wang et al., 2003; Rungrassamee et al., 2013). Consideration of this correlation led us to investigate the Wnt/β-catenin pathway, an alternative signalling pathway that may potentially be mitigated to induce immunostimulation.
The Wnt/β-catenin pathway is a well-studied signal transduction pathway involved in a broad range of biological systems, including cell behaviour, cell adhesion, and cell polarity (Gordon and Nusse, 2006). It is highly conserved throughout evolution. When there is no Wnt stimulation, β-catenin is phosphorylated by glycogen synthase kinase 3β (GSK3β), hampering the transport of β-catenin from the cytoplasm to the nucleus, resulting in the Wnt/β-catenin pathway becoming off-state. Clearly, inhibition of GSK3β is an excellent way to activate the Wnt/β-catenin pathway (MacDonald et al., 2009). In humans, the dysregulation of Wnt/β-catenin signalling causes several diseases, especially tumourigenesis (Nusse and Clevers, 2017). Several small molecules have previously been used in medicine as targeting key factors in the Wnt/β-catenin pathway, including GSK3β. 4,6-disubstituted pyridopyrimidine (TWS119) is a potent inhibitor of GSK3β, commonly used as a small-molecule tool for anti-tumour therapy (Ding et al., 2003). In P. vannamei, previous studies have shown that TWS119 effectively inhibits GSK3β, resulting in the activation of the Wnt/β-catenin pathway (Zhang et al., 2020). However, there is no relevant research about the safety evaluation on TWS119 in aquaculture. In the current study, the effect of dietary TWS119 on the growth performance, immunity, and transcriptome responses of P. vannamei was investigated to explore the potential of TWS119 as an immunopotentiator in shrimp.
Experimental diets were designed on the basis of nutrient requirements of shrimp (NRC, 2015). The basal diet was considered as the control diet, and the five experimental diets were supplemented with TWS119 (Med Chem Express, China) at concentrations of 0.25, 1, 4, 16, and 64 mg kg−1, respectively. The ingredients and nutrient composition of the experimental diets are shown in Table 1. Brown fish meal, soybean meal, peanut meal, and corn gluten meal were used as main protein sources. Corn oil, soybean lecithin oil, and fish oil were used as the basal lipid sources and provided a total of 8.06% of crude lipid. Ingredients were ground to a fine powder and sieved through a 178-μm screen. Weighed according to the test formula, mixed step by step, using a twin-screw extruder to make pellets with particle size of 1.0 and 1.5 mm, air-dried, and stored at −20°C until used.
The study protocol was approved by the ethics review board of the Institutional Animal Care and Use Committee of the Guangdong Ocean University. The experiment was conducted in an indoor farming system in the Marine Biological Research Base of Guangdong Ocean University (Zhanjiang, China). Penaeus vannamei were obtained from Guangdong Haixing Agriculture Group Co., Ltd. (Zhanjiang, China) and acclimatizsed to experimental condition for a week while feeding with commercial feed (Haid Group, Guangzhou, China) before commencing the feeding trial. A total of 720 P. vannamei (0.27 ± 0.02 g) were weighed and evenly divided at random into 18 indoor circular fibreglass tanks (0.3 m3). The P. vannamei in the 18 tanks were further divided into six groups (named as group T0, T0.25, T1, T4, T16, and T64, respectively) and hand fed with the corresponding experimental diets to apparent satiation four times daily (07:00, 11:00, 17:00, and 21:00) for 8 weeks, under suitable water salinity (27‰) and temperature (25–27°C) conditions.
At the end of the feeding trial, shrimp were fasted for 24 h before collecting samples. All the shrimp from each tank were counted and measured in weight. After weighing, haemolymph was collected from five randomly selected shrimp from each tank and then stored at 4°C for 12 h. The stored haemolymph samples were centrifuged (3,000 rpm for 10 min at 4°C), and the supernatant was stored at −80°C for the analysis of serum non-specific immune parameters and the enzyme activity of LvGSK3β. The haemolymph of another five shrimp from each tank was placed into a modified anticoagulant-citrate-dextrose (ACD) solution and centrifuged at 3,000 rpm for 5 min at 4°C. The haemocytes were separated and stored in RNA later at −80°C for transcriptome analysis.
Based on the recorded data, growth parameters, including survival rate (SR), specific growth rate (SGR), weight gain rate (WGR), and feed conversion rate (FCR), were calculated using the following formulae:
As previously described (Shi et al., 2017), Vibrio parahaemolyticus were cultured in a Luria-Bertani (LB) agar culture medium at 30°C for 18 h, and the V. parahaemolyticus cells were centrifuged at 5,000 g for 10 min at 4°C, washed with 1 × PBS, and then resuspended as an inoculum with approximately 1 × 106 colony forming units (CFU)·μl−1. A total of 40 P. vannamei from each feed group were chosen to undergo challenge test with V. parahaemolyticus at a dose of 107 CFU·g−1 shrimp. Cumulative mortality was recorded every 4 h and the differences between the groups were analysed using the Mantel-Cox (log-rank χ2 test) method with the software GraphPad Prism.
The enzyme activity of LvGSK3β in the haemolymph was detected using the GSK3β Kinase Activity Quantitative Detection Kit (Genmed, China) as previously described (Zhang et al., 2020). The acid phosphatase (ACP), alkaline phosphatase (AKP), superoxide dismutase (SOD), catalase (CAT), and phenoloxidase (PO) were measured using the ACP assay kit (Cat. No. A060-2), AKP assay kit (Cat. No. A059-2), SOD assay kit (WST-1 method; Cat. No. A001-3), CAT assay kit (Visible light; Cat. No. A007-1), and PO kit (Cat. No. H247), respectively. The content of malondialdehyde (MDA) was determined using the MDA assay kit (TBA method; Cat. No. A003-1). All of these assay kits were bought from the Nanjing Jiancheng Bioengineering Institute (Nanjing, China) and used according to the instructions of the manufacturer.
Based on the results of growth performance analysis and pathogen challenge tests. Total RNA of haemocytes of P. vannamei from T0 and T16 groups were extracted by Trizol method. The RNA concentration was determined by spectrophotometry at OD260 using a SimpliNano (GE Healthcare, United States) and the RNA integrity was examined by electrophoresis on a 1.5% agarose gel. Library construction and sequencing were carried out by Guangzhou GeneDenovo Biotech Co., Ltd. (Guangzhou, China). Briefly, the mRNA was enriched with magnetic beads with Oligo (dT), and the fragmentation buffer was added to the mRNA to break it into short fragments. Using the post-fragment mRNA as a template, the first strand of cDNA was synthesizsed using random hexamers, and dNTPs, buffer, DNA polymerase I, and RNase H were added to synthesize the second strand of cDNA, using Agencourt AMPure XP Beads reagent. The cassette was purified and EB buffer eluted with end-repair, base A, and sequencing linker, and the target fragment was recovered by agarose gel electrophoresis. Finally, PCR was used to complete the whole library preparation work. The PCR product was purified to create the final cDNA libraries. Lastly, the library preparations were sequenced on an Illumina HiSeq™ platform that generated ~200 bp pair-end raw reads.
After removing the raw reads with adaptor, the raw reads all A-base contained, the raw reads containing more than 10%, and the low-quality sequences in which the base number of Q ≤ 20 accounts for more than 50% of the whole read, the filtered reads were mapped to the pacific white shrimp reference genome (NCBI Genome database ID: 10710), using HISAT software (Kim et al., 2015).
As shown in previous report, Fragments Per Kb per Million reads (FPKM) was used to express the relative transcript abundance (Trapnell et al., 2010). Log2(FC) was used as an indicator of the transcriptomic differences between the T16 and T0 group. The results of the statistical test were corrected using false discovery rate (FDR). The unigenes were considered to be differentially expressed genes (DEGs) if the absolute value of log2(FC) was greater than 1 and the FDR was less than 0.05. DEGs were analysed using tools in the Gene Ontology (GO)1 and Kyoto Encyclopedia of Genes and Genomes (KEGG)2 databases.
To validate the data obtained from the transcriptome analysis, a total of eight DEGs, including four upregulated and four downregulated, were selected for quantification of their relative expression by quantitative PCR (qPCR). Using the samples returned after transcriptome sequencing, cDNA samples were prepared using the PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) kits (Takara, Japan). TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) kits (Takara, Japan) were used to perform qPCR analysis in a Roche Light Cycler 480 thermal cycler (Roche Applied Science, Germany). Elongation factor 1α (EF1α, GenBank accession No. GU136229) was used as the internal control. The 2−ΔΔCt method was used to calculate gene expression after normalisation by LvEF1α. The primers used in the qPCR analysis are listed in Table 2.
All results were subjected to one-way ANOVA followed by Duncan’s multiple range tests to determine significant differences among treatment groups using SPSS version 19. The survival percentages post-challenge was calculated by Log-rank Kaplan-Meier analysis. For all results, differences were presented to be significant at p < 0.05 and highly significant at p < 0.01. The data are shown as the mean ± SD.
LvGSK3β activity was assessed to enable evaluation of the effect of TWS119 on the Wnt/β-catenin pathway. As shown in Figure 1, LvGSK3β activity significantly decreased in all the five experimental groups compared to the control group, reaching the lowest level in the T4 group.
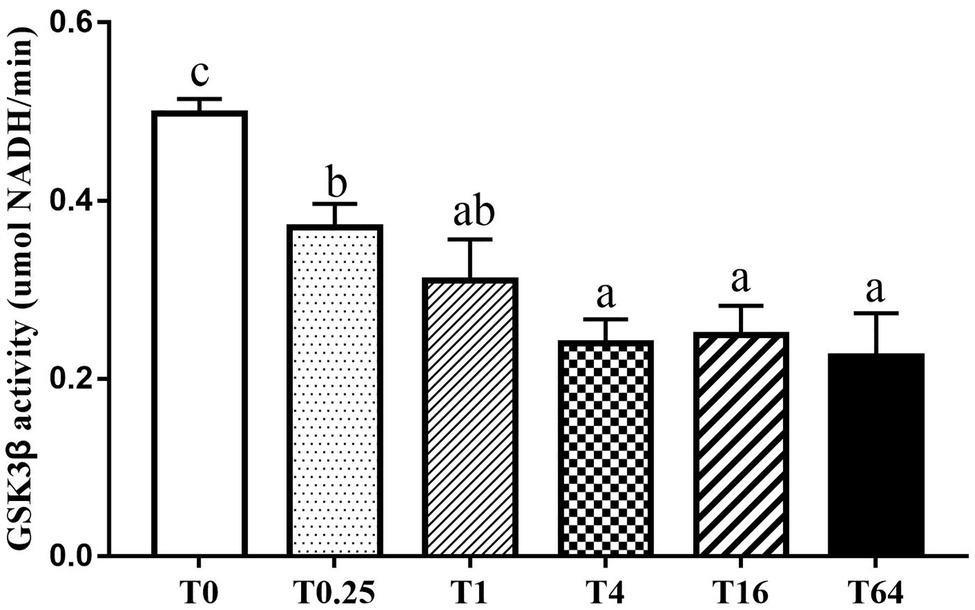
Figure 1. Influence of dietary TWS119 on the enzyme activities of LvGSK3β in P. vannamei. The detection of enzyme activities was performed in triplicate for each sample. Values (means ± SD) with different letter mean significant difference (p < 0.05).
As shown in Table 3, no significant differences were found in the SR of P. vannamei among the different groups (p > 0.05) after the 8-week feeding trial. In the T16 group, the FBW, WGR, and SGR of P. vannamei were significantly higher than those in the T0 group, while the FCR was significantly lower than that in the T0 group (p < 0.05). In the T0.25, T1, T4, and T64 groups, there was no significant difference in FBW, WGR, SGR, and FCR compared to the T0 group.
After V. parahemolyticus infection, the survival rates of P. vannamei in the T0.25, T1, T4, T16, and T64 groups were higher than the T0 group, with a significant difference in the T0.25, T1, T4, and T16 groups. The final survival rates of P. vannamei first increased and then decreased as the additive dosage of TWS119 increased, with the highest level in the T4 group (Figure 2).
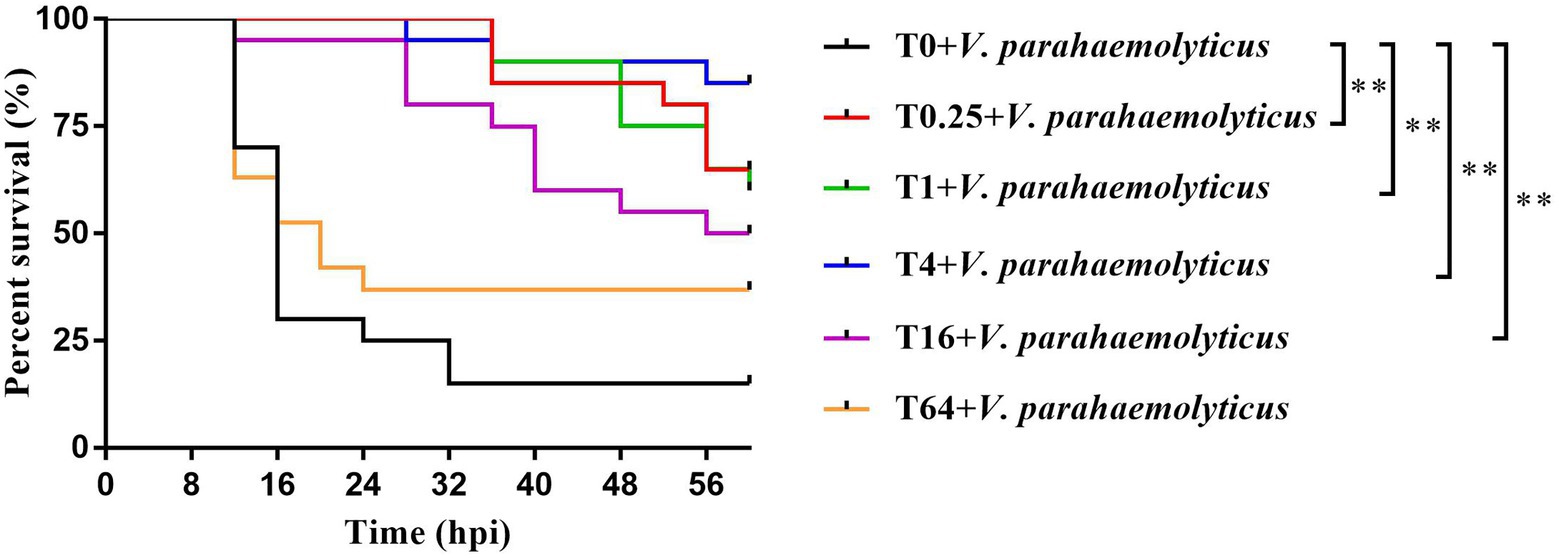
Figure 2. Survival rates of TWS119 fed P. vannamei after Vibrio parahaemolyticus infection. The experiments were performed two times with identical results. Differences in survival levels between treatments were analyzsed by Kaplan-Meier plot (log-rank χ2 test). Significant differences in survival rate were marked with asterisks, ** indicates p < 0.01.
Non-specific immune indices in serum are shown in Figure 3. In general, the immune enzymes ACP, AKP, SOD, CAT, and PO in all the experimental groups had higher activity than the control group, while the MDA contents were lower than the control group. The ACP and CAT activities increased with increasing levels of TWS119 in the diet, reaching the highest levels in the T64 group (Figures 3A,D). The SOD and PO activities increased first and then declined with increasing TWS119 dietary levels (p < 0.05), with the peak in T4 and T1 groups, respectively (Figures 3C,E). The SOD and PO activities in the T1 and T4 groups were significantly higher than the T0 group (p < 0.05). TWS119 addition increased the AKP activity in all experimental groups, with significantly higher levels in the T1, T16, and T64 groups (Figure 3B). As shown in Figure 3F, the T4 and T64 groups’ MDA activities were significantly lower than in the T0 group (p < 0.05).
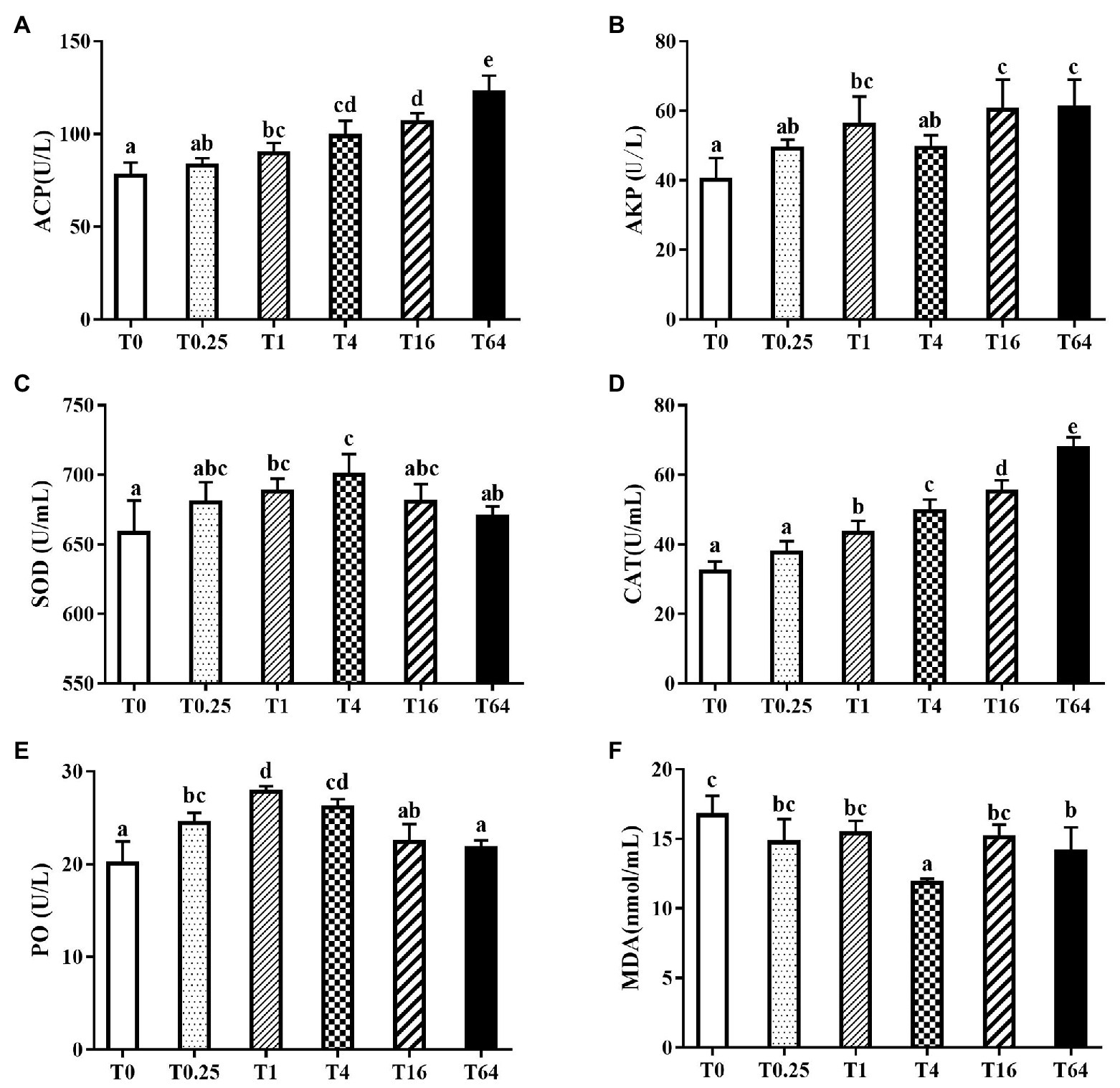
Figure 3. Effects of dietary TWS119 on non-specific immune indices of P. vannamei. The activities of ACP (A), AKP (B), SOD (C), CAT (D) and PO (E), and the content of MDA (F) in the haemolymph were measured. Values (means ± SD) in bars that have the different letters represent significantly different (p < 0.05; Tukey’s test) among treatments (n = 3).
Considering the results that both the growth performance and disease resistance of P. vannamei in T16 group was significantly improved when compared to the T0 group, two cDNA libraries from the mRNA extracted from the haemocytes of the P. vannamei in T0 and T16 groups were sequenced. As Table 4 shows, a total of 86,407,884 original reads were obtained and 86,170,320 clean reads were left after the removal of the reads with a ratio of N greater than 10% and the low-quality reads. Both of the Q20 in T0 and T16 groups were greater than 97.5%. The GC content of the clean reads was 52.63 and 49.82%, respectively. There were 89.83% (74.37 unique mapped) and 88.07% (75.16 unique mapped) of the clean sequences were successfully mapped to the reference genome in T0 and T16 groups, respectively. In addition, the percentages of the clean sequences mapped in exon were about 60.00% both in T0 and T16 groups.
Differentially expressed gene analysis was undertaken for P. vannamei in the T0 and T16 groups. Compared with L.vannamei in the T0 group, a total of 5,073 DEGs in the T16 group were identified, including 3,981 upregulated and 1,092 downregulated DEGs (Figure 4).
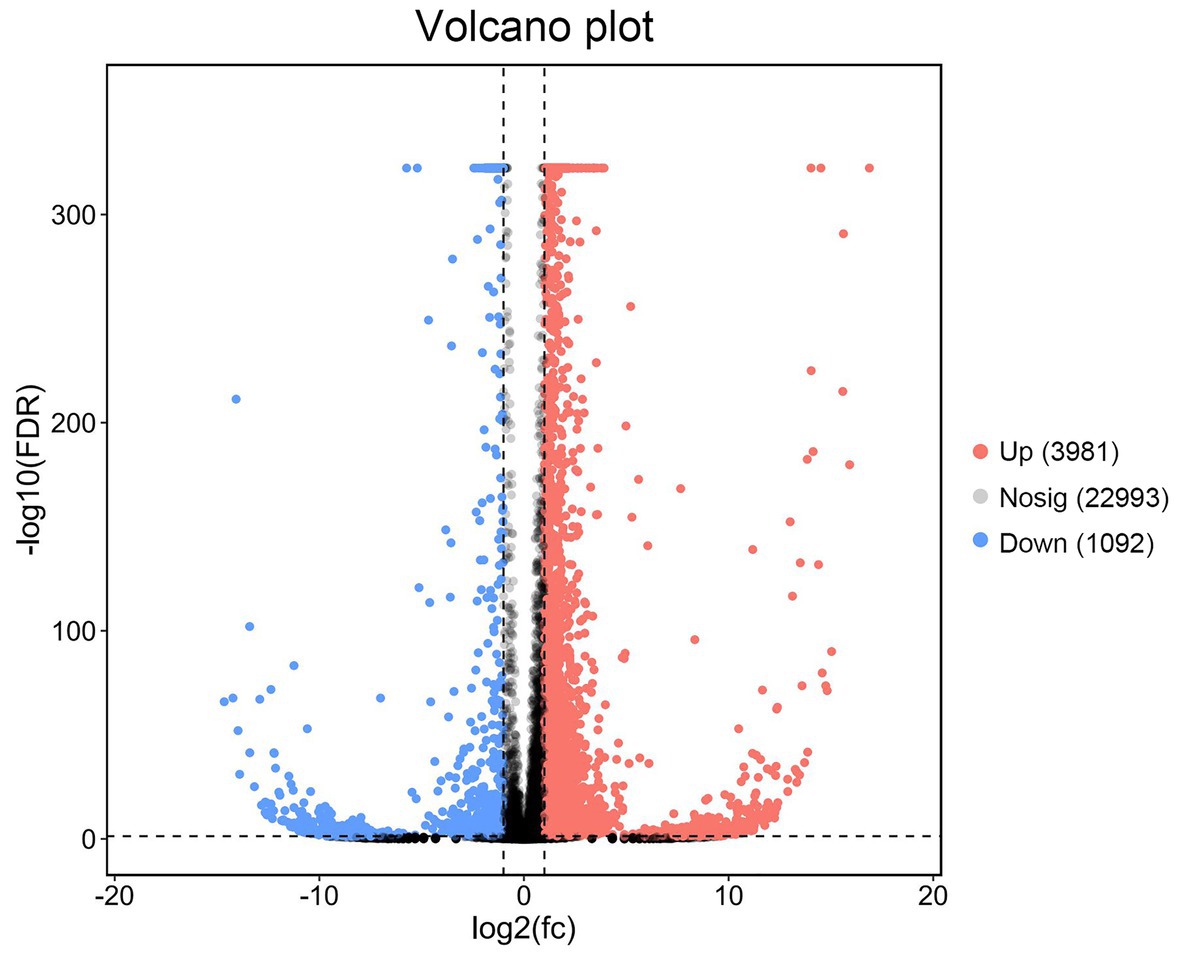
Figure 4. Volcano diagram of differentially expressed genes (DEGs) between TWS119 fed P. vannamei and the control. The X-axis indicates the fold change and the Y-axis indicates the statistical significance of the differences. Red dots represent the significantly upregulated DEGs while blue dots represent the significantly downregulated DEGs (FDR ≤ 0.01 and FC ≥ 2). Gray dots represent the DEGs which are not significantly different.
All the DEGs were mapped to terms in the GO database to evaluate their functions. As shown in Figure 5, 48 significantly (correct p < 0.05) enriched terms were identified and classified into three major functional classes: biological processes, molecular functions, and cellular components. There were 23, 10, and 15 significantly enriched terms classified as biological processes, molecular functions, and cellular components, respectively. The largest number of DEGs was involved in the cellular process, followed by metabolic processes, catalytic activity, single organism processes, and binding.
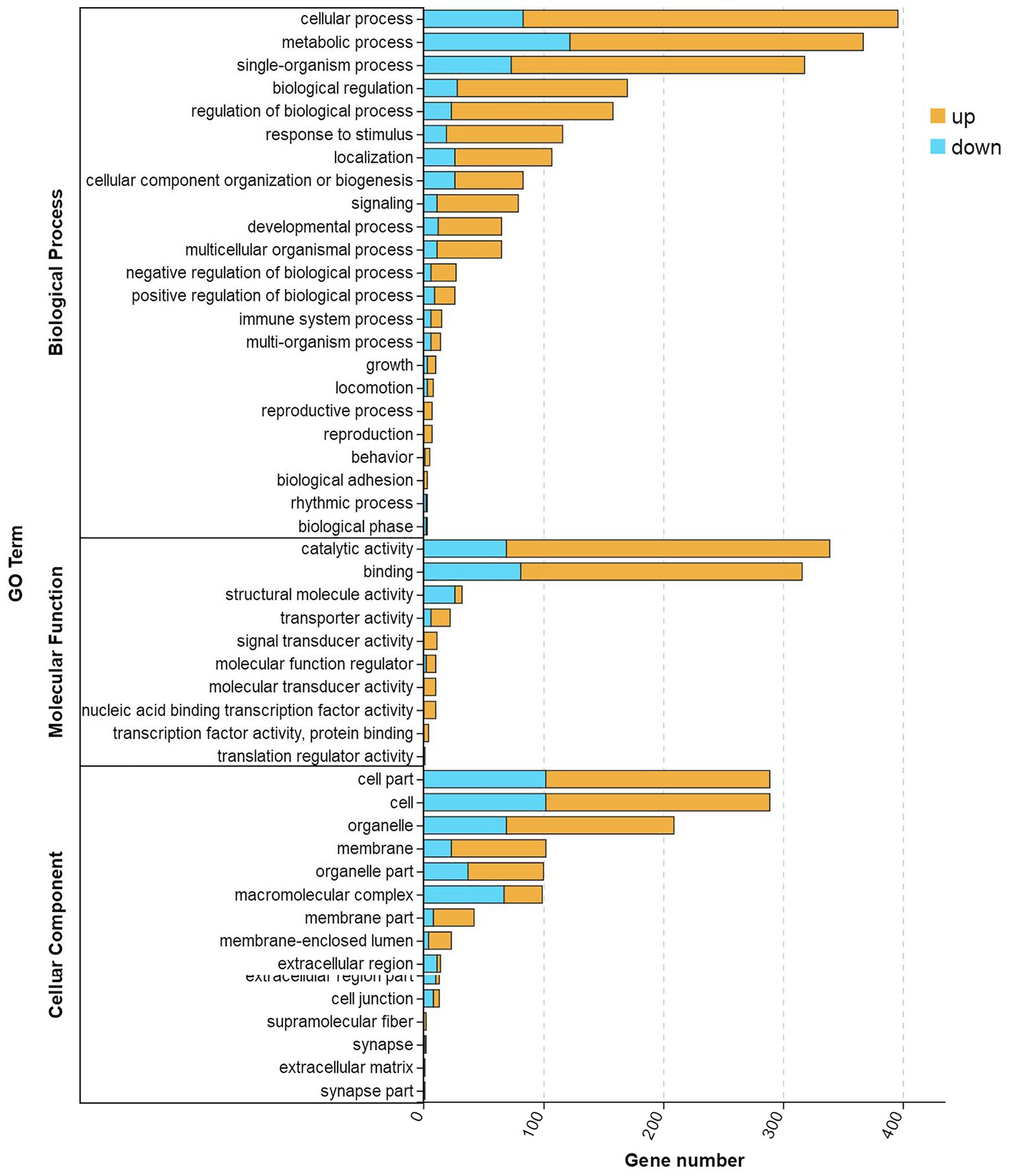
Figure 5. GO enrichment analysis of DEGs in TWS119 fed and control P. vannamei. Three main GO categories: cellular component, molecular function, and biological process. X-axis indicates the number of unigenes and Y-axis indicates GO categories and the subcategories.
KEGG pathway enrichment analysis reported 2,034 DEGs significantly enriched, associated with 310 different signalling pathways classified into 41 subcategories. All the signalling pathways were subordinated to six different KEGG categories – metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems, and human diseases. Additionally, 926 of the significantly enriched DEGs were related to metabolism, accounting for more than 45% (Figure 6). As shown in Figure 7, six of the top 20 pathways were related to metabolism, and six pathways were related to environmental information processing and cellular processes. Most of the DEGs are involved in the ribosome pathway (ko03010), endocytosis (ko04144), glycerophospholipid metabolism (ko00564), Wnt signalling pathway (ko04310), and FoxO signalling pathway (ko04068). The ribosome pathway (ko03010) accounted for the most DEGs and was also the most significantly enriched pathway. Another three significantly enriched pathways were the pathways involved in nicotinate and nicotinamide metabolism (ko00760), the phosphatidylinositol signalling system (ko04070), and Kaposi’s sarcoma-associated herpesvirus infection (ko05167).
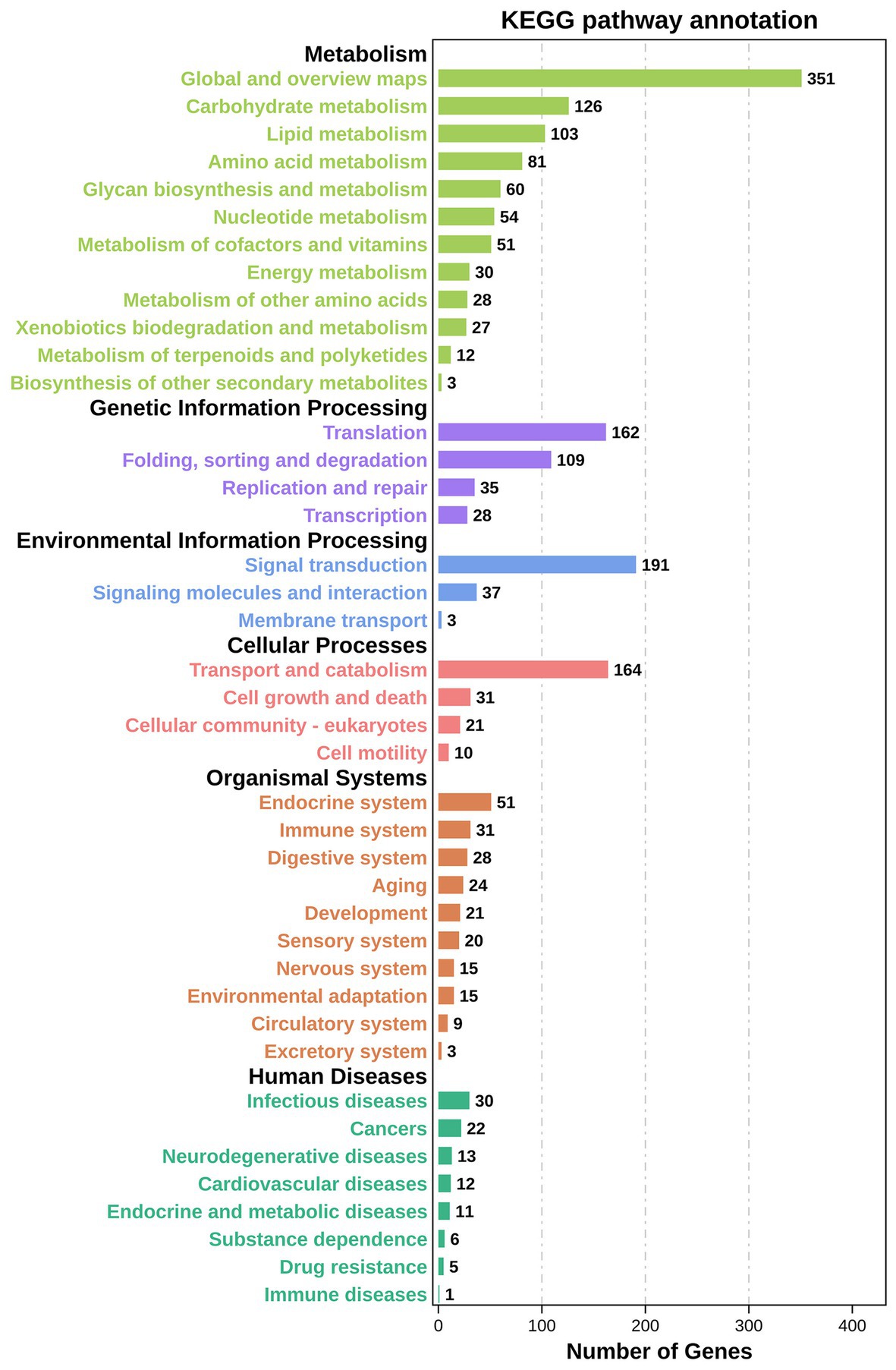
Figure 6. KEGG enrichment analyses of DEGs in TWS119 fed and control P. vannamei. Highly expressed biological pathways represented in the transcriptome retrieved from the KEGG database. DEGs were assigned to six special KEGG pathways, including organismal systems, metabolism, genetic information processing and environmental information processing, cellular processes, and human diseases.
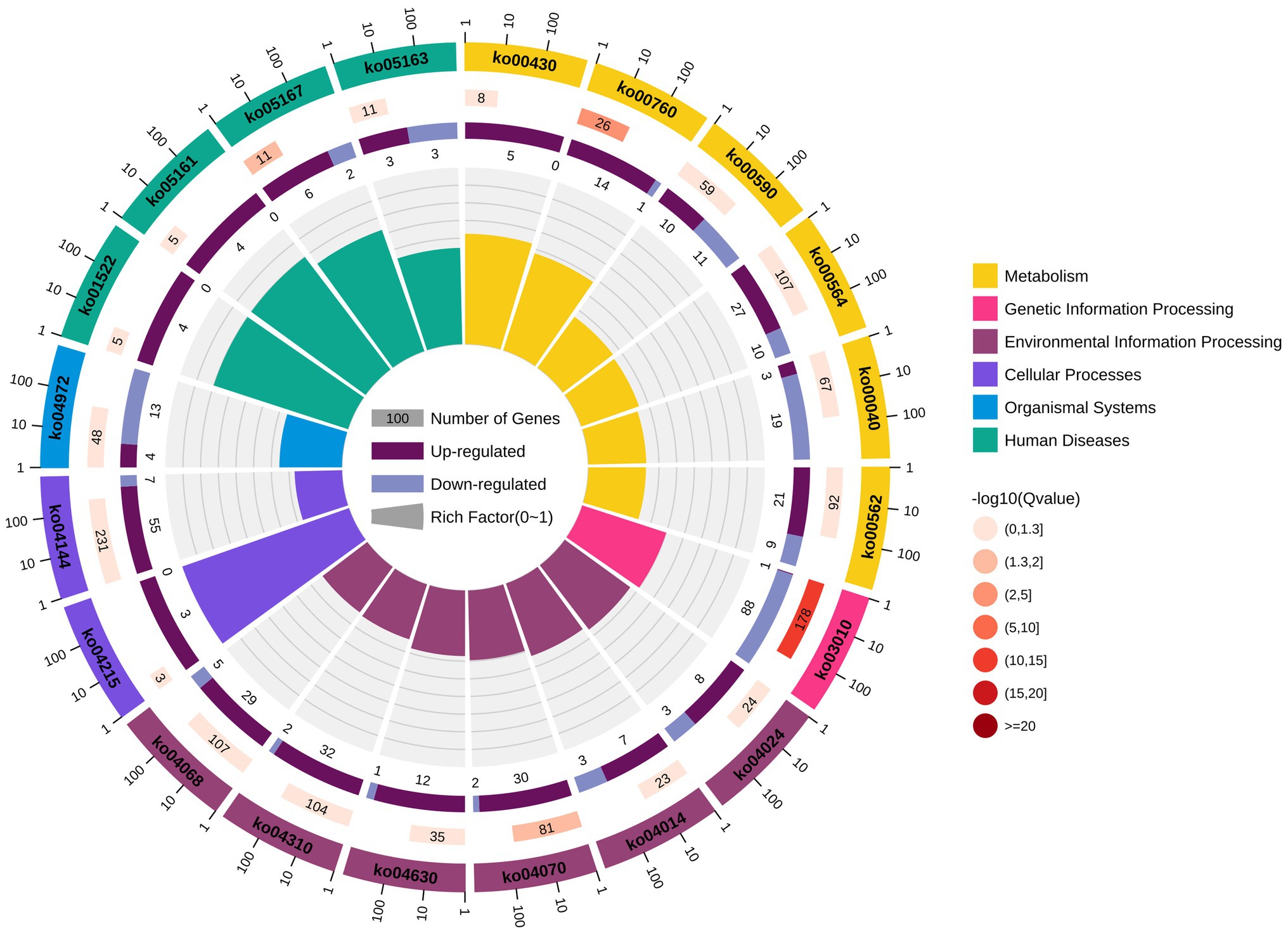
Figure 7. Top 20 pathways of KEGG enrichment analysis. The first lap from outside to inside indicates the top 20 pathways of enrichments, with the coordinate scale of gene numbers outside. The second lap indicates the numbers of the genes in background and p values for each pathway. The more genes, the longer the bar. The smaller the p value, the darker the colour. The third lap indicates the ratio of the upregulated genes (deep purple) and downregulated genes (light purple), with the numbers inside of the bars. The fourth lap indicates the enrichment factor (the ratios of DEGs in background genes) of each pathway. Each cell of background auxiliary line represents 0.1. Different colours in the first and fourth laps represent different classifications of the pathways.
A total of 10 DEGs were randomly selected to further validate the gene expression results obtained from the RNA sequencing data, including five upregulated and five downregulated DEGs. As shown in Figure 8, the 10 selected DEGs’ expression patterns using the qPCR analyses were consistent with the RNA sequencing.
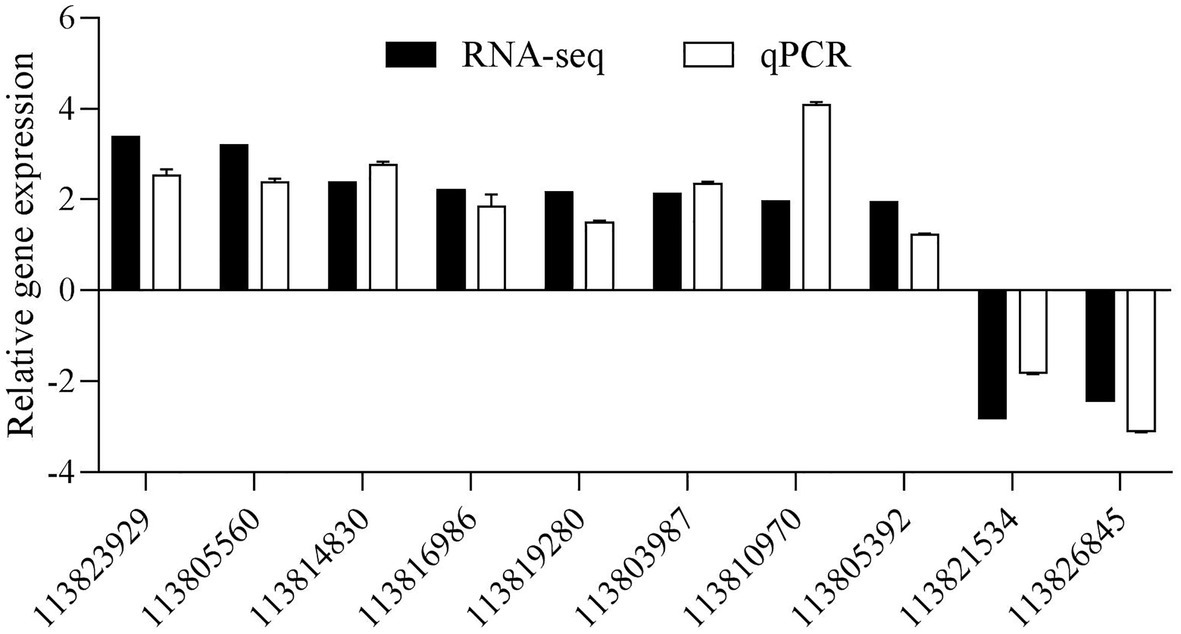
Figure 8. Validation of DEGs by quantitative PCR (qPCR). The detection of gene expression was performed in triplicate for each sample. Expression values were normalizsed to those of LvEF1α using the Livak (2−ΔΔCt) method, and the data were provided as the means ± SD of triplicate assays.
Frequent diseases seriously hinder the healthy development of the shrimp industry. The Wnt/β-catenin pathway has been reported to play a positive role in anti-disease immunity by inhibiting the activity of GSK3β (Zhang et al., 2020). As several PAMPs of the Toll signalling pathway could be used as effective immunostimulants, the current study investigated whether activation of the Wnt/β-catenin pathway could improve shrimp disease resistance. This study showed that dietary GSK3β inhibitor TWS119 could improve the growth performance and immunity of P. vannamei. Also, transcriptome analysis indicates that the improvement in growth performance and immunity of P. vannamei is closely related to ribosome function. All the results provide new insight for utilising immunostimulants in shrimp aquaculture.
The Wnt/β-catenin pathway controls many biological phenomena throughout most animals’ development and adult life (MacDonald et al., 2009). As an activator of the Wnt/β-catenin pathway, TWS119 has been used in the treatment of oncogenesis in humans, including breast cancer (Vijay et al., 2019), colon cancer (Chen et al., 2017), and lung cancer (Tang et al., 2018). Additionally, TWS119 drives microglial anti-inflammatory activation and facilitates neurological recovery following experimental stroke in rats (Song et al., 2019). Previous studies have shown that injected TWS119 could improve the anti-disease immunity of P. vannamei (Zhang et al., 2020), and a similar effect on shrimp’s immunity status was found in this study. The activity of GSK3β in P. vannamei could be effectively inhibited by dietary TWS119 and the outcome of anti-V. parahemolyticus infection in TWS119 fed P. vannamei was significantly improved. Also, dietary TWS119 improves the non-specific immune indices of P. vannamei. Besides, the mortality after V. parahemolyticus infection vary greatly although there was no significant difference in LvGSK3β activity among T4, T16, and 64 group. Correlating to the reduced activities of SOD and PO, it seemed that the disease-resistant abilities of P. vannamei fed with high dose of TWS119 were interfered by other immune effects, such as antioxidant response. Surprisingly, dietary TWS119 has no adverse effect on P. vannamei growth, and TWS119 could significantly improve the growth performance of P. vannamei at the dose of 16 mg·kg−1, suggesting that there was no perceived security issues of dietary TWS119 on P. vannamei. However, the price of TWS119 at $15/mg seems not acceptable for large-scale use in aquaculture. In any case, the data showed that TWS119 could be a feed additive to enhance shrimp’s growth and autoimmunity, indicating the possibility of moderate doses of TWS119 being an immunopotentiator.
Transcriptome sequencing technology is an important platform for transcriptome research in non-model organisms (Hahn et al., 2009; Ma et al., 2014). The mechanism of TWS119 on the growth and immunity of P. vannamei was investigated using comparative transcriptome analyses performed on control and TWS119-fed P. vannamei groups. GO analysis showed that DEGs were mainly enriched in the subcategories, such as cell, organelle, catalytic activity, binding, single-organism, and cellular processes. The results suggest TWS119 may influence the body-associated biological functions by regulating the expression of these differential genes, leading to changes in growth performance and disease immunity in P. vannamei. As a pathway-based classification of orthologous genes, KEGG enrichment analysis could provide helpful information for gene function prediction (Kanehisa and Goto, 2000). In this study, KEGG analysis showed that there were 34 DEGs significantly enriched in the Wnt signalling pathway. The results infer that TWS119 is the activator of the Wnt/β-catenin pathway through targeting GSK3β and further suggests that dietary TWS119 could have a practical, positive impact on P. vannamei. More than 45% of the significantly enriched DEGs were related to metabolism, including carbohydrate, lipid, and amino acid metabolism, the three sources of a body’s energy (Samat et al., 2020). Furthermore, the ribosome pathways accounted for the largest number of DEGs and were also the most significantly enriched pathway. The ribosome has recently transitioned from being viewed as a passive, indiscriminate machine to a more dynamic macromolecular complex with specializsed roles in the cell (Genuth and Maria, 2018). The ribosome’s primary function is to transform the genetic code into an amino acid sequence and construct protein polymers from amino acid monomers (Moore, 2012). Protein is one of the main energy sources of aquatic animals. Correlated with the data that WGR, SGR, and survival rates after V. parahemolyticus infection of P. vannamei fed with TWS119 at the dose of 16 mg·kg−1 were significantly improved when compared with the control, it is not surprising that the energy metabolic signalling pathways in TWS119 fed P. vannamei were activated, leading to better growth and anti-disease immunity. Activated energy metabolism could provide bigger source for the accumulation of body protein and more energy to stand up to pathogen invasion in P. vannamei. Additionally, the endocytosis signalling pathway, FoxO signalling pathway, and signalling pathway related to Kaposi’s sarcoma-associated herpesvirus infection were closely related to immunity (Cossart and Helenius, 2014; Mohd et al., 2017; Kawaguchi et al., 2018) and also contain many significantly enriched DEGs. These results concur with the overall improved immunity of TWS119 fed P. vannamei.
In this study, the growth and disease resistance of P. vannamei in T16 group was significantly improved, and the comparative transcriptome analysis showed that the ribosome pathway possessed the largest number of DEGs and was also the most significantly enriched pathway. Besides, most of the significantly enriched pathways were related to energy metabolism. All these results suggest that dietary supplementation with TWS119 at the dose of 16 mg·kg−1 could improve the growth and immunity of P. vannamei and is correlated with the function of ribosomes and energy metabolism. However, the specific mechanism for such beneficial outcomes requires further investigation. This study’s findings provide new insights for immunostimulants exploitation in aquaculture as regulators of the critical immune signalling pathways.
The data presented in the study are deposited to the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra) with the accession no. PRJNA723808.
SZ and BT conceived and designed the experiments. CH, SS, LZ, and LS collected samples and performed the experiments. CH analysed the data as well as wrote the paper. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key R&D Program of China (grant no. 2019YFD0900200), the National Natural Science Foundation of China (grant no. 32072988), and the General Program of Natural Science Foundation of Guangdong Province, China (grant no. 2020A1515010319 and 2018A030313963).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Apines-Amar, J. M. S., and Amar, E. C. (2015). “Chapter 3. Use of immunostimulants in shrimp culture: an update” in Biotechnological Advances in Shrimp Health Management in the Philippines. eds. C. M. A. Caipang, M. B. I. Bacano-Maningas, and F. F. Fagutao (Kerala, FL: Research Signpost Press), 45–71.
Chen, Y. Q., Lu, Z., Aldarouish, M., Zhou, Z. H., and Wang, L. X. (2017). Wnt pathway activator tws119 enhances the proliferation and cytolytic activity of human γδt cells against colon cancer. Exp. Cell Res. 362, 63–71. doi: 10.1016/j.yexcr.2017.11.003
Cossart, P., and Helenius, A. (2014). Endocytosis of viruses and bacteria. Cold Spring Harb. Perspect. Biol. 6:a016972. doi: 10.1101/cshperspect.a016972
Dalmo, R. A., and Bogwald, J. (2008). Beta-glucans as conductors of immune symphonies. Fish Shellfish Immunol. 25, 384–396. doi: 10.1016/j.fsi.2008.04.008
Deng, B., Wang, Z. P., Tao, W. J., Li, W. F., Wang, C., Wang, M. Q., et al. (2015). Effects of polysaccharides from mycelia of cordyceps sinensis on growth performance, immunity and antioxidant indicators of the white shrimp Litopenaeus vannamei. Aquac. Nutr. 21, 173–179. doi: 10.1111/anu.12147
Ding, S., Wu, T. Y., Brinker, A., Peters, E. C., Hur, W., and Gray, N. S. (2003). Synthetic small molecules that control stem cell fate. Proc. Natl. Acad. Sci. 100, 7632–7637. doi: 10.1073/pnas.0732087100
Flores-Miranda, M. D. C., Luna-González, A., Campa-Córdova, Á. I., González-Ocampo, H. A., Fierro-Coronado, J. A., and Partida-Arangure, B. O. (2011). Microbial immunostimulants reduce mortality in white leg shrimp (Litopenaeus vannamei) challenged with Vibrio sinaloensis strains. Aquaculture 320, 51–55. doi: 10.1016/j.aquaculture.2011.08.005
Genuth, N. R., and Maria, B. (2018). The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol. Cell 71, 364–374. doi: 10.1016/j.molcel.2018.07.018
Gordon, M. D., and Nusse, R. (2006). Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J. Biol. Chem. 281, 22429–22433. doi: 10.1074/jbc.R600015200
Hahn, D. A., Ragland, G. J., Shoemaker, D. W. D., and Denlinger, D. L. (2009). Gene discovery using massively parallel pyrosequencing to develop ESTs for the flesh fly Sarcophaga crassipalpis. BMC Genomics 10:234. doi: 10.1186/1471-2164-10-234
Harikrishnan, R., Balasundaram, C., and Heo, M. S. (2011). Impact of plant products on innate and adaptive immune system of cultured finfish and shellfish. Aquaculture 317, 1–15. doi: 10.1016/j.aquaculture.2011.03.039
Holmström, K., Grslund, S., Wahlström, A., Poungshompoo, S., Bengtsson, B. E., and Kautsky, N. (2003). Antibiotic use in shrimp farming and implications for environmental impacts and human health. Int. J. Food Sci. Technol. 38, 255–266. doi: 10.1046/j.1365-2621.2003.00671.x
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kawaguchi, Y., Mori, Y., and Kimura, H. (2018). Pathological features of kaposi’s sarcoma-associated herpesvirus infection. Adv. Exp. Med. Biol. 16, 357–376. doi: 10.1007/978-981-10-7230-7_16
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kuo, H. W., Chang, C. C., and Cheng, W. (2019). Tyramine’s modulation of immune resistance functions in Litopenaeus vannamei and its signal pathway. Dev. Comp. Immunol. 96, 68–76. doi: 10.1016/j.dci.2019.01.008
Li, C., Wang, S., and He, J. (2019). The two NF-κB pathways regulating bacterial and WSSV infection of shrimp. Front. Immunol. 10:1785. doi: 10.3389/fimmu.2019.01785
Li, F. H., and Xiang, J. H. (2013). Recent advances in researches on the innate immunity of shrimp in China. Dev. Comp. Immunol. 39, 11–26. doi: 10.1016/j.dci.2012.03.016
Lightner, D. V., Poulos, B. T., Tang-Nelson, K. F., Pantoja, C. R., Nunan, L. M., Navarro, S. A., et al. (2005). “Application of molecular diagnostic methods to penaeid shrimp diseases: advances of the past 10 years for control of viral diseases in farmed shrimp” in New Diagnostic Technology: Applications in Animal Health and Biologics Controls. ed. P. Vannier (Saint-Malo, FL: International Alliance for Biological Standardization) 126, 117–122.
Liu, Y., Wang, W. N., Wang, A. L., Wang, J. M., and Sun, R. Y. (2007). Effects of dietary vitamin E supplementation on antioxidant enzyme activities in Litopenaeus vannamei (Boone, 1931) exposed to acute salinity changes. Aquaculture 265, 351–358. doi: 10.1016/j.aquaculture.2007.02.010
Ma, D. Y., Yang, H. S., Sun, L. N., and Chen, M. Y. (2014). Transcription profiling using RNA-Seq demonstrates expression differences in the body walls of juvenile albino and normal sea cucumbers Apostichopus japonicus. Chin. J. Oceanol. Limnol. 32, 34–46. doi: 10.1007/s00343-014-3041-6
MacDonald, B. T., Tamai, K., and He, X. (2009). Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26. doi: 10.1016/j.devcel.2009.06.016
Mohd, F., Wang, H., Uma, G., Little, P. J., Xu, J., and Zheng, W. (2017). Foxo signaling pathways as therapeutic targets in cancer. Int. J. Biol. Sci. 13, 815–827. doi: 10.7150/ijbs.20052
Moore, P. B. (2012). How should we think about the ribosome? Annu. Rev. Biophys. 41, 1–19. doi: 10.1146/annurev-biophys-050511-102314
Nusse, R., and Clevers, H. (2017). Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999. doi: 10.1016/j.cell.2017.05.016
Peraza-Gomez, V., Luna-Gonzalez, A., Manuel Gonzalez-Prieto, J., Fierro-Coronado, A., and Gonzalez-Ocampo, H. A. (2014). Protective effect of microbial immunostimulants and antiviral plants against WSSV in Litopenaeus vannamei cultured under laboratory conditions. Aquaculture 420, 160–164. doi: 10.1016/j.aquaculture.2013.10.044
Qiao, J., Du, Z., Zhang, Y., Du, H., Guo, L., Zhong, M., et al. (2011). Proteomic identification of the related immune-enhancing proteins in shrimp Litopenaeus vannamei stimulated with vitamin C and Chinese herbs. Fish Shellfish Immunol. 31, 736–745. doi: 10.1016/j.fsi.2011.07.005
Rungrassamee, W., Maibunkaew, S., Karoonuthaisiri, N., and Jiravanichpaisal, P. (2013). Application of bacterial lipopolysaccharide to improve survival of the black tiger shrimp after Vibrio harveyi exposure. Dev. Comp. Immunol. 41, 257–262. doi: 10.1016/j.dci.2013.05.021
Samat, N. A., Yusoff, F. M., Rasdi, N. W., and Karim, M. (2020). Enhancement of live food nutritional status with essential nutrients for improving aquatic animal health: a review. Animals 10:2457. doi: 10.3390/ani10122457
Sheng, P. C., Yin, W. L., Yao, J. Y., Xu, Y., Lin, F., et al. (2014). Application status and research progress of immune enhancer in shrimp. J. Agric. Catastrophology 4, 31–35. doi: 10.19383/j.cnki.nyzhyj.2014.01.009
Shi, L. L., Chan, S. M., Li, C. Z., and Zhang, S. (2017). Identification and characterization of a laccase from Litopenaeus vannamei involved in anti-bacterial host defense. Fish Shellfish Immunol. 66, 1–10. doi: 10.1016/j.fsi.2017.04.026
Song, D. G., Zhang, X. J., Chen, J. M., Liu, X. X., Xue, J., Zhang, L., et al. (2019). Wnt canonical pathway activator TWS119 drives microglial anti-inflammatory activation and facilitates neurological recovery following experimental stroke. J. Neuroinflammation 16:256. doi: 10.1186/s12974-019-1660-8
Song, X., Zhang, Z. J., Wang, S., Li, H. Y., Zuo, H., Xu, X. P., et al. (2015). A Janus kinase in the JAK/STAT signaling pathway from Litopenaeus vannamei is involved in antiviral immune response. Fish Shellfish Immunol. 44, 662–673. doi: 10.1016/j.fsi.2015.03.031
Suphantharika, M., Khunrae, P., Thanardkit, P., and Verduyn, C. (2003). Preparation of spent brewer’s yeast beta-glucans with a potential application as an immunostimulant for black tiger shrimp, Penaeus monodon. Bioresour. Technol. 88, 55–60. doi: 10.1016/S0960-8524(02)00257-2
Tang, Y. Y., Sheng, S. Y., Lu, C. G., Zhang, Y. Q., Zou, J. Y., Lei, Y. Y., et al. (2018). Effects of glycogen synthase kinase-3β inhibitor tws119 on proliferation and cytokine production of tils from human lung cancer. J. Immunother. 41, 319–328. doi: 10.1097/CJI.0000000000000234
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Vijay, G. V., Zhao, N., Hollander, P. D., Toneff, M. J., Joseph, R., Pietila, M., et al. (2019). Gsk3β regulates epithelial-mesenchymal transition and cancer stem cell properties in triple-negative breast cancer. Breast Cancer Res. 21, 1–14. doi: 10.1186/s13058-019-1125-0
Wang, S. H., and Chen, J. C. (2005). The protective effect of chitin and chitosan against Vibrio alginolyticus in white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 19, 191–204. doi: 10.1016/j.fsi.2004.11.003
Wang, X., Huang, J., and Song, X. L. (2003). The application of immunostimulants-peptidoglycan to the culture of shrimp. Mar. Fish. Res. 24, 69–74.
Zeng, D., Peng, M., Yang, Q., Yang, C. L., Liao, Z. P., Li, Q. Y., et al. (2019). The Penaeus stylirostris densovirus capsid protein interacts with the Litopenaeus vannamei BCCIP protein. Fish Shellfish Immunol. 88, 198–206. doi: 10.1016/j.fsi.2019.02.057
Zhang, S., Shi, L. L., Lv, K., Li, H. Y., Wang, S., He, J. G., et al. (2016). Cloning, identification and functional analysis of a b-catenin homologue from pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 54, 411–418. doi: 10.1016/j.fsi.2016.03.162
Keywords:Penaeus vannamei, TWS119, growth performance, immunity, transcriptome analysis
Citation: Hou C, Song S, Zhu L, Shi L, Tan B and Zhang S (2021) Growth, Immunity, and Transcriptome Response to Dietary Wnt/β-Catenin Pathway Activator TWS119 in Penaeus vannamei. Front. Mar. Sci. 8:685429. doi: 10.3389/fmars.2021.685429
Received: 25 March 2021; Accepted: 12 April 2021;
Published: 04 June 2021.
Edited by:
Erchao Li, Hainan University, ChinaReviewed by:
Hongwei Shan, Ocean University of China, ChinaCopyright © 2021 Hou, Song, Zhu, Shi, Tan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuang Zhang, enNodWFuZ0BnZG91LmVkdS5jbg==; Beiping Tan, YnB0YW5AMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.