- 1Guangdong Provincial Key Laboratory of Marine Biotechnology, Shantou University, Shantou, China
- 2STU-UMT Joint Shellfish Research Laboratory, Shantou University, Shantou, China
- 3Institute of Tropical Aquaculture and Fisheries, Universiti Malaysia Terengganu, Kuala Terengganu, Malaysia
Crustacean culture has been developing rapidly in various parts of the world. Therefore, it is important to understand their reproductive biology. Insulin-like androgenic gland hormone (IAG) secreted from the androgenic gland (AG) is widely accepted as a key regulator of sexual differentiation in male crustaceans. However, recently several sex-related genes (i.e., CFSH, DEAD-box family, Tra-2, Sxl, Dsx, Fem-1, Sox gene family, Foxl2, and Dmrt gene family) have been identified via transcriptomic analysis in crustaceans, indicating that sexual differentiation in crustaceans is more complicated than previously expected. It has been found that several non-coding RNAs (i.e., miRNAs, lncRNAs, and piRNAs) and IAG receptors may be involved in the sexual development of decapods. Identification and study of the regulation mechanism of sex-related genes, non-coding RNAs, and IAG receptors will provide valuable information regarding sexual development in decapods. In this review, the roles of hormonal and genetic factors in both males and females are discussed. In males, crustacean female sex hormone (CFSH), Sxl, Dmrt gene family, Dsx, Sox gene family, GEM, Fem-1, l-GnRH-III, and corazonin play important roles in IAG regulation in the “eyestalk-IAG-testis” endocrine axis. Unlike males, the regulation mechanism and interaction of sexual genes are relatively unknown in females. However, CFSH, IAG, Fem-1, FAMeT, Slo, UCHLs, Erk2, Cdc2, EGFR, Vg, VgR, and VIH seem to play crucial roles during ovarian development. This study summarizes the available information in the field, highlights gaps, and lays the foundations for further studies and a better understanding of the regulatory mechanism of sexual development in decapods.
Introduction
The order Decapoda includes approximately 17,000 species of crayfish, shrimps, crabs, and lobsters (De Grave et al., 2009). Some decapods are economically cultivated and some maintained in domestic aquariums, but some are invasive and are considered as pathogenic vectors. Crustacean culture is a rapidly developing industry, due to the high economic value as a source of animal protein, employment, and foreign exchange gains. Crustaceans production reached 9.4 million tons in 2018 (6.4 and 3 million tons were provided from capture fisheries and aquaculture, respectively), with an estimated sale value of US$ 69.3 billion (FAO, 2020).
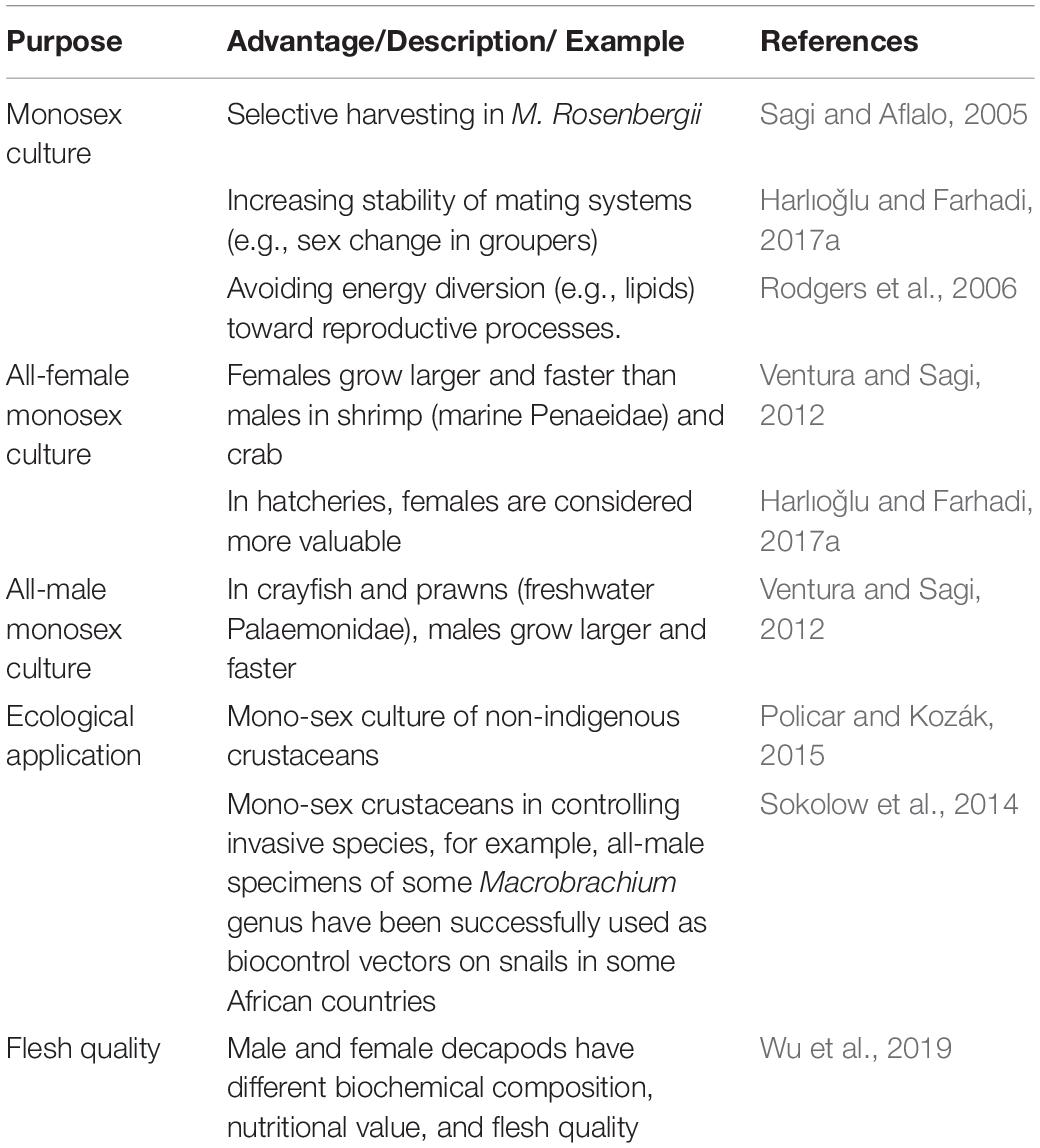
Understanding the reproductive biology and mechanism of sexual differentiation are key issues for sexual manipulation and improving the reproductive efficiency of decapods. Male and female decapods have different growth performances at different growth stages, which is important in artificial selective breeding and farming (Shi X. et al., 2019). For example, in crayfish and prawns (freshwater Palaemonidae), males grow larger and faster, and as such, all-male individual rearing is desired. In contrast, females grow larger and faster than males in shrimps (marine Penaeidae) and crabs (Ventura and Sagi, 2012). In hatcheries, females are considered more valuable than males for increasing the size of the population since males can copulate with more females with no negative effect on the percentage of berried females (Harlıoǧlu and Farhadi, 2017b). Studies showed that male and female decapods have different biochemical composition, nutritional value, and flesh quality (Wu et al., 2019). Another benefit of sexual manipulation is the ecological application of non-reproductive and mono-sex crustaceans in controlling invasive species. For example, all-male specimens of some Macrobrachium genus have been successfully used as biocontrol vectors on snails in some African countries (Sokolow et al., 2014). The advantages of sexual manipulation in decapod crustaceans are summarized in Table 1.
Sexual Development in Decapods
Sex Determination
The sexual development in crustaceans begins with a sex-specific genetic cascade mediated by a chromosomal mechanism of sex determination. In Penaeidae shrimp and prawns a general ZZ/ZW (homogametic male) mechanism seems to exist (Aflalo et al., 2006; Coman et al., 2008; Staelens et al., 2008). In other decapods such as freshwater crayfish (Astacidea) and true crabs (Brachyura), both ZZ/ZW and XY/XX mechanisms occur (Parnes et al., 2003; Cui et al., 2015; Mlinarec et al., 2016). Recently, various genetic techniques have been successfully applied to identify the sex-specific DNA sequences and markers in decapods. Restriction-site associated DNA sequencing (RAD-seq) is based on NGS technology and can discover massive single nucleotide polymorphisms (SNPs) in various species by sequencing parts of the genome at high depth, without a reference genome. RAD-seq has been successfully applied to develop sex-specific markers in several decapod species (Zhang et al., 2018). For example, sex-specific SNP markers and high-density genetic linkage maps showed male homogamety and female heterogamety, presenting reliable genetic evidence for a WZ/ZZ sex-determination system in mud crabs, Scylla tranquebarica, and Scylla serrata (Shi et al., 2018; Waiho et al., 2019b). The discovery of five male-specific SNP markers in crucifix crab (Charybdis feriatus) provided reliable genetic evidence for a XX/XY sex-determination system in C. feriatus (Fang et al., 2020).
The sexual determination process in Drosophila species is controlled through alternative splicing of pre-mRNA of a series of genes involved in the sex determination pathway, including Tra-2, Sxl, and Dsx. A homologue gene of Tra-2 has been identified in Penaeus monodon, however, no sex dimorphism on the expression and alternative splicing patterns was observed (Leelatanawit et al., 2009). Up to now, the sex determination mechanisms in crustacean species are still largely unknown. Kato et al. (2011) identified two Dsx homologues (DapmaDsx1 and DapmaDsx2), which act as a key regulator of the male phenotype in Daphnia magna. However, in contrast to Dsx for Tra no significant difference was detected in the splicing or expression patterns (Kato et al., 2010). In ornate spiny lobster (Panulirus ornatus), Sxl and Tra-2 isoform-encoding transcripts showed broad tissue expression, with no clear bias between males and females, suggesting that Sxl-Tra-Dsx may be unique to Drosophilidae and not conserved among decapods. However, the male-specific iDMY was conserved between the eastern (Sagmariasus verreauxi) and ornate spiny lobsters (P. ornatus), indicating a conserved central function in sex determination (Ventura et al., 2020).
Sexual Differentiation and Maturation
The complex process of sexual differentiation results in sex-specific phenotypic development. Our understanding of sexual differentiation in the decapods far exceeds what is known about sex determination. Neuropeptides from the eyestalk and mandibular organ regulate the sexual development in decapods (Khalaila et al., 2002; Alfaro-Montoya, 2010). Eyestalk is the major neuroendocrine control center and eyestalk ablation influences gonadal development in crustaceans (Fingerman, 1987). The CHHs (crustacean hyperglycemic hormones) secreted from the X-organ have been classified as subfamily I peptides containing a CHH precursor related peptide (CPRP) and the subfamily II peptides lack the CPRP (Jia et al., 2012). The subfamily II peptides including moult inhibiting hormone (MIH), gonad/vitellogenesis-inhibiting hormone (GIH or VIH), and mandibular organ-inhibiting hormone (MOIH) control the moulting and gonadal maturation (Lacombe et al., 1999; Böcking et al., 2002). Serotonin (5-HT), dopamine, and gonadotropin-releasing hormones (l-GnRH-III) are other important factors affecting the sexual development pathway of decapods (Tinikul et al., 2008, 2011; Siangcham et al., 2013).
The insulin-like androgenic gland hormone (IAG) secreted from androgenic gland (AG) is a key regulator in sexual differentiation. Recent studies have shown that IAG itself is controlled by several genes and hormonal factors (Shen et al., 2014; Shi L. et al., 2019; Zhong et al., 2019). The detailed mechanisms and the role of sex-related genes and miRNAs in the IAG regulation are discussed in detail in other sections of the present review. In the present review, we focus on the downstream mechanisms such as novel sex-related genes and miRNAs that play important role in the sexual development process. The present review aims to highlight gaps in knowledge that must be filled to allow further development of sexual manipulation techniques.
Sexual Manipulation in Decapod Crustaceans
Androgenic gland is the key regulator of primary and secondary sexual characters in male decapods. It controls behavior, spermatogenesis, and sexual differentiation by secretion of IAG. AG is associated with the subterminal part of the sperm duct and attached to the posterior vas deferens in male decapods. IAG silencing had adverse effects on sexual development (i.e., spermatogenesis) in male Macrobrachium rosenbergii (Ventura et al., 2009). Silencing accomplished via double-stranded RNA (dsRNA) injection caused considerable sex-related changes such as ovarian activation and testicular degeneration in male Cherax quadricarinatus (Rosen et al., 2010). Similar results were observed in M. rosenbergii and C. quadricarinatus after AG ablation (Barki et al., 2006).
At present, sexual manipulation in decapods only applies through the manipulation of AG. Such as AG ablation and IAG suppression in males, which produce neofemales (phenotypically female but genetically male) in M. rosenbergii (Aflalo et al., 2006; Ventura et al., 2009). Neofemale M. rosenbergii (complete and functional sex reversal) was obtained by long-term IAG silencing at PL30 stage (Ventura et al., 2012). AG implantation into females induced sex reversal in females, called neomales (phenotypically male but genetically female) (Aflalo et al., 2006; Ventura et al., 2009).
Until now, the only successful AG manipulation that led to complete and functional sex reversal has been recorded in M. rosenbergii (Aflalo et al., 2006; Ventura and Sagi, 2012). In Macrobrachium sp., IAG is expressed exclusively and abundantly in the AG rather than in the testis or other tissues (Ventura and Sagi, 2012; Ventura et al., 2012; Ma et al., 2013). However, studies showed that in other decapod groups such as shrimps (Mareddy et al., 2011; Li et al., 2012b), crayfish (Shi X. et al., 2019), and crabs (Zhang et al., 2014; Jiang et al., 2020a) the IAG is expressed not only in the AG, but also in other tissues. The AG-specific expression pattern of IAG could explain why AG manipulation led to complete sex reversal only in M. rosenbergii. The broad tissue expression pattern of IAGs in other decapods indicates that IAGs may have some additional functions.
Recently, several sex-related genes and non-coding RNAs have been identified in decapods, suggesting that sexual differentiation mechanisms in decapods are more complicated than previously expected (Chandler et al., 2018). Further studies are required to identify more sex-related factors such as genes, non-coding RNAs and IAG hormones, which could be involved in the sexual development of decapods. Understating the function of these genetic factors may provide novel insights into the development of new sexual manipulation techniques. Sex-related genes, hormones, and their potential roles in decapods are summarized in Table 2.
Hormonal and Genetic Factors in Male Decapods
Insulin-like Androgenic Gland Hormone
Insulin-like androgenic gland hormones have been isolated from several decapod species including M. rosenbergii (Mr-IAG) (Ventura et al., 2009), C. quadricarinatus (Cq-IAG) (Manor et al., 2007), Portunus pelagicus (Pp-IAG) (Sroyraya et al., 2010), P. monodon (Pem-IAG) (Mareddy et al., 2011), Marsupenaeus japonicus (Maj-IAG) (Banzai et al., 2011), F. chinensis (Fc-IAG1, Fc-IAG2) (Li et al., 2012a), Cherax destructor, Callinectes sapidus (Cas-IAG) (Chung et al., 2011), Scylla paramamosain (Huang et al., 2014), Procambarus clarkii (Shi L. et al., 2019), and Eriocheir sinensis (Liu Z. et al., 2019). There is remarkable structural similarity in encoded amino acids from IAGs and all consist of a signal peptide, a B chain, a C peptide, and an A chain (Chung et al., 2011). Recent studies showed that IAG has a more complex expression pattern and AG is not the only organ for the expression of IAG (Manor et al., 2007; Sroyraya et al., 2010; Mareddy et al., 2011). For example, in C. sapidus IAG is not only expressed in AG but also in the hepatopancreas (Chung et al., 2011). In Fenneropenaeus chinensis, IAG was expressed in both nerve cord and hepatopancreas of female and male individuals at relatively low levels (Li et al., 2012a).
Sox Gene Family
The Sox (Sry-related high-mobility group (HMG) box) family of transcription factors is extensively distributed in different animal groups, and plays crucial roles in development processes including cell pluripotency, neurogenesis, and sex determination. To date, the Sox gene has been isolated from several decapods including Macrobrachium nipponense (Hu et al., 2020), Penaeus vannamei (Peng et al., 2015), Palaemon serratus (González-Castellano et al., 2019), E. sinensis (Liu et al., 2016), and S. paramamosain (Liao et al., 2020; Lin et al., 2020). SoxE1 is involved in testis development in M. nipponense (Hu et al., 2020). In a recent study, Lin et al. (2020) reported that the SoxB2 gene plays a key role during gonad development in the mud crab. Sox gene plays critical roles in sexual dimorphism and participates in the formation of brain nerves and is involved in the regulation of body segments (Liao et al., 2020). In E. sinensis, Sox protein was exclusively localized in the nucleus of spermatid and spermatozoa even at the end of acrosome reaction and was bound to the uncondensed chromatin in the nucleoplasm of mature spermatozoa. SoxB2-1 knockdown leads to an abnormal transformation of the nucleus during spermiogenesis. These results revealed the importance of the Sox gene family in testis development and spermatogenesis in males (Liu et al., 2016; Lin et al., 2020).
Dmrt Gene Family
The Dmrt gene family plays essential roles in sexual differentiation. This gene family has been widely studied in vertebrates, nematodes, and insects, however, limited information is available regarding the molecular function of the Dmrt gene family in decapods (Zhong et al., 2019). To date, Dmrt has been isolated from different crustacean species including E. sinensis (Zhang and Qiu, 2010), the water flea D. magna (Toyota et al., 2013), S. verreauxi (Chandler et al., 2015), F. chinensis (Li et al., 2012b), M. nipponense (Wang et al., 2019), P. serratus (González-Castellano et al., 2019), the giant freshwater prawn M. rosenbergii (Yu et al., 2014; Zhong et al., 2019), and mud crab S. paramamosain (Gao et al., 2014). Targeting the coding region of the Dmrt gene by repetitive injection of dsRNA revealed that Dmrt plays important roles in the spermatogenesis and testicular development of E. sinensis (Ma et al., 2016a). RNAi-targeting of MroDmrt99B in male M. rosenbergii did not affect the expression level of IAG, while MroDmrt11E silencing resulted in a significant reduction in the expression level of IAG (Yu et al., 2014), indicating that IAG is positively regulated by MroDmrt11E. In M. rosenbergii, eyestalk removal significantly increased the expression level of Dsx in the testis and AG. Putative Dsx/Mab-3 binding site was identified on the promoter region of male sexual differentiation effector gene IAG (Yu et al., 2014).
Vasa Gene
Vasa is an important regulatory gene for reproductive system development (Lasko and Ashburner, 1988). In decapod crustaceans, the Vasa gene has been identified in F. chinensis (Zhou et al., 2010) and P. vannamei (Aflalo et al., 2007), M. rosenbergii (Nakkrasae and Damrongphol, 2007), and mud crab S. paramamosain (Wang et al., 2012). Vasa gene can be used as a specific germ cell marker to track the origin and precise route of germ cell migration toward the gonad, and elevates our knowledge on the reproductive and developmental processes of the germline in decapods (Wang et al., 2012). In F. chinensis, the strongest hybridization signal was detected in the spermatogonia and the intensity gradually declined toward the centre of testis tubules indicating that the transcript level of Vasa progressively decreased from spermatogonia to spermatids (Zhou et al., 2010). Vasa RNA was detected in the nucleus and the cytoplasm of the spermatozoa in M. rosenbergii (Nakkrasae and Damrongphol, 2007). It seems that Vasa gene plays various roles in the regulation of spermatogenesis. Further studies are required to investigate the exact role of the Vasa gene in the germ cells of decapods.
Sex-lethal (Sxl)
It has been documented that sex-lethal (Sxl) is involved in the sex determination process of insects. In Drosophila, female embryos expressed a fully functional Sxl protein while males produced a shorter peptide lacking an RNA binding domain. In males, the truncated Sxlm has no regulatory effect, leading to the production of an equally inactive default splice variant of Tra (Mullon et al., 2012). In decapods, Sxl has been isolated from Chinese mitten crab (Shen et al., 2014), redclaw crayfish C. quadricarinatus (Zheng et al., 2019), red swamp crayfish P. clarkii (Shi L. et al., 2019), S. verreauxi (Chandler et al., 2015), M. nipponense (Zhang et al., 2013), and P. vannamei (López-Cuadros et al., 2018). Sxl plays crucial roles in sexual differentiation, embryonic development, and gonad development but not in the sex determination of decapods (Shen et al., 2014; López-Cuadros et al., 2018; Zheng et al., 2019). In E. sinensis, Sxl was expressed in different stages of embryos development. The expression of Sxl had a gradual increase from the cleavage stage to the egg-nauplius stage, however, Sxl rapidly increased in the original zoea stage. The increase of Sxl at the last stages of embryos indicates that Sxl may play a role in embryonic development (Shen et al., 2014). Among the four isoforms of Sxl in P. clarkii, CqSxl3 demonstrated tissue specificity with a higher expression level in the testis than in the ovary. CqSxl4 and CqSxl1 were extensively expressed in various tissues while CqSxl2 was almost undetectable. The transcript levels of CqSxl1/3/4 slowly increased along with embryonic development (Zheng et al., 2019). The transcript pattern of Sxl in females and males show that Sxl may not be sex-specific in decapods (Shen et al., 2014; López-Cuadros et al., 2018; Zheng et al., 2019).
Corazonin
Corazonin (Crz) was first found in insects and is a peptide with the sequence pGlu-Thr-Phe-Gln-Tyr-Ser-Arg-Gly-Trp-Thr-Asn-amide (Nässel, 2002). In crustaceans, Crz has been characterized from Cancer borealis (Li et al., 2003), Carcinus maenas (Ma et al., 2009), Homarus americanus (Ma et al., 2008), and M. rosenbergii (Siangcham et al., 2013). Some studies have shown that Crz is involved in various physiological functions, including stress (Boerjan et al., 2010), heart beat in cockroaches (Veenstra, 1989), pigment production in locusts (Hua et al., 2000), and reduction of silk making in caterpillar (Tanaka et al., 2002).
Bone morphogenetic proteins (BMPs)
It has been demonstrated that bone morphogenetic proteins (BMPs) such as BMPRIB and BMPRII play an important role in the development of ovary in S. paramamosain (Shu et al., 2016). BMP7 may also play a role in the testis development of E. sinensis, probably by promoting the germ cell differentiation/meiotic mitosis and maintaining the self-renewal of spermatogonia or facilitating the successful fertilization (Xu et al., 2018).
Gem-associated Protein 2-like Isoform X1 (GEM)
In M. nipponense, gem-associated protein 2-like isoform X1 (GEM) was highly expressed in the testis and ovary and during larval development indicating that GEM plays important roles in gonad and larval development. In situ hybridization showed that GEM may promote oogenesis and spermatogenesis. RNAi experiment revealed that GEM negatively controls the expression of IAG in M. nipponense (Jin et al., 2019).
Regulatory Mechanism of Sex in Male Decapods
The “eyestalk-AG-testis” endocrine axis is the main regulatory mechanism of sex in male decapods. The X-Organ-Sinus-gland complex (XO-SG) of the eyestalk regulates the activity of AG through the XO-SG-AG-Gonad axis (Rodríguez et al., 2007). Eyestalk ablation causes hyperplasia and hypertrophy in AG cells (Khalaila et al., 2002), elevates IAG expression, and accelerates the testicular development in males (Nagaraju, 2011). XO-SG secretes neurohormones such as GIH, CHH, MIH, and VIH which are suspected to directly regulate the expression of IAG (Li et al., 2015b) and have inhibitory effects on reproduction and sexual development (Suwansa-ard et al., 2015).
Recently, several sexual genes have been identified to be involved in the “eyestalk-IAG-testis” endocrine axis (Guo et al., 2019). It has been revealed that some of these genes are regulated by the eyestalk neurohormones and are located at the upstream of IAG and the sexual differentiation cascade (Shen et al., 2014; Ma et al., 2016a; Shi X. et al., 2019; Zheng et al., 2019; Zhong et al., 2019).
Upstream Mechanisms of Sex in Male Decapods
Crustacean female sex hormone (CFSH) (Liu A. et al., 2018), Sxl (Shi L. et al., 2019; Zheng et al., 2019), Dmrt gene family (Abayed et al., 2019), Dsx (Yu et al., 2014), Sox gene family (Li et al., 2012a; Zhang et al., 2014), GEM (Jin et al., 2019), Fem-1 (Ma et al., 2016b), l-GnRH-III, and Crz (Siangcham et al., 2013) may play key roles in IAG regulation at the upstream of IAG in the “eyestalk-IAG-testis” endocrine axis.
Liu C. et al. (2018) suggested that CFSH is not a female-specific hormone and the expression pattern of CFSH is opposite to the expression pattern of IAG during the development of AG in males. Recombinant CFSH (rCFSH) reduced the expression of IAG in AG by approximately 60% (Liu A. et al., 2018). CFSH from the eyestalk is a negative regulator and inhibits AG development in decapods. Jiang et al. (2020b) found no difference in the expression level of Sox-5 and AP-1 in AG exposed to rCFSH, however, STAT significantly declined, indicating that CFSH controls the expression of STAT, inhibiting its function to control IAG in the mud crab. Further experiment showed that RNAi mediated STAT gene knockdown lowered IAG expression indicating that CFSH regulates IAG by inhibiting STAT. In a study by Jiang et al. (2020b), promoter activity was significantly increased in pGL3-pSp-IAG1 (Sox-5-binding site) and pGL3-pSp-IAG6 (Sox-5-binding site) than in previous plasmids (pGL3-basic and pGL3-pSp-IAG5), indicating that the Sox-5-binding site is probably involved in enhancing this activity, consistent with site-directed mutation (SDM) results of decreased promoter activity in pGL3-pSp-IAG6-Mut (Jiang et al., 2020b).
The Sox gene plays a crucial role in male sexual differentiation and sex determination of vertebrates (Ma K. et al., 2012). In zebrafish, Sox-5 binds to the Dmrt1 promoter (double sex and mab-3-related transcription factor 1) and inhibits Dmrt1 expression that may affect sexual differentiation (Gao et al., 2005). In medaka, Sox-5 knockout reverses female to male by controlling dmrt1bY expression (Schartl et al., 2018). In decapods, 5′ flanking regions for IAG have only been studied in F. chinensis, M. nipponense, and S. paramamosain, which contains a cis-acting element of Sox-5-binding site, indicating that the transcriptional regulation mechanism of IAG may be conserved in decapods (Li et al., 2012a; Zhang et al., 2014). The finding of transcription factor binding sites of Sox in the 5′ flanking region of IAG indicates a regulatory role of Sox at the upstream of IAG.
Sxl is regulated by neuropeptide hormones from the eyestalk (Shen et al., 2014) and acts as the upstream regulator of IAG (Shi L. et al., 2019; Zheng et al., 2019). Sxl transcription in the testis was downregulated 3 days after eyestalk removal while in the ovary it was upregulated 7 days after eyestalk removal (Shen et al., 2014). Sxl silencing significantly decreased the IAG level in C. quadricarinatus and P. clarkii (Shi L. et al., 2019; Zheng et al., 2019). In P. clarkii, no direct relation was observed between the Sxl and IAG and the expression level of Sxl was not affected by IAG-dsRNA injection (Shi L. et al., 2019). In contrast, after IAG silencing, the expression level Sxl1, and Sxl2 decreased significantly in M. rosenbergii (Tan et al., 2020b).
Similarly, Dsx is likely to be negatively regulated by the neuropeptide hormones from the eyestalk and Dsx may be involved in the regulation of IAG in M. rosenbergii (Zhong et al., 2019). Dsx knockdown induced a significant decrease in the expression of IAG (Yu et al., 2014; Li et al., 2018). IAG silencing significantly reduced the expression of two genes, MroiDmrt1b and MroiDmrt1c in M. rosenbergii, thereby indicating their possible roles in the IAG-switch (Abayed et al., 2019; Tan et al., 2020a). Nevertheless, the up-regulation of Dsx at the nauplius stage before the first up-regulation of IAG (Li et al., 2018) indicate that Dsx may act as an upstream regulator of IAG (Yu et al., 2014; Li et al., 2018). Abayed et al. (2019) suggested that Mr-IAG is correlated with MroiDmrt1b and MroiDmrt1c, however, it is difficult to conclude whether the relationship is downstream or upstream.
The Fem-1 gene might be at the upstream of the sex determination cascade when compared with IAG in M. nipponense (Ma et al., 2016b), since IAG was first expressed at the blastula stage (Ma et al., 2013). In contrast to IAG, the Fem-1 showed a high expression level from the unfertilized egg to cleavage, and then the expression declined to a low level from blastula to zoea. Fem-1 probably influences sexual development by interacting with cathepsin L in embryogenesis and vitellogenesis (Ma et al., 2016b). Silencing of IAG results in testicular degeneration and sexual shifts in decapods (Rosen et al., 2010). The insulinase domain protein in Drosophila willistoni may hydrolyze insulin protein in individual development (Clark et al., 2007), indicating that Fem-1 probably controls IAG indirectly and feminizes male-related phenotypes similar to IAG silencing (Ma et al., 2016b).
Crz and l-GnRH-III are other regulatory factors at the upstream of IAG, In M. rosenbergii, Crz has inhibitory effects on the AG through significant decreases in AG size, proliferation of AG cells, and IAG suppression. In contrast, it seems that IAG is positively regulated by l-GnRH-III (Siangcham et al., 2013).
Downstream Mechanisms of Sex in Male Decapods
To better understand the multiple functions of IAG, it is necessary to clarify the mechanisms of how IAG is transported. An insulin family-based signaling system has been proposed based on the structure similarity of IAG to the insulin/IGF family (Sharabi et al., 2016). Several peptide receptors including tyrosine kinase insulin receptor (TKIR), G-protein-coupled receptor (GPCR), insulin receptor (IR), and insulin-like growth factor-binding peptide (IGFBP) have been identified to be the receptor for IAG (Rosen et al., 2010; Aizen et al., 2016; Chandler et al., 2017; Jiang et al., 2020c; Ventura et al., 2020).
IGFBPs act not only as carriers of IGFs to release at the target site, but also function as modulators of IGF availability and activity (Govoni et al., 2005). In vitro study of IGFBP in the red claw crayfish C. quadricarinatus revealed that recombinant Cq-IGFBP protein interacts with the IAG hormone (Rosen et al., 2013). In M. rosenbergii, the highest expression level of IAGBP was detected in AG (Yang et al., 2020). IAGBP-dsRNA significantly reduced the transcript level of IAG in M. rosenbergii and M. nipponense (Li et al., 2015a; Yang et al., 2020). These results indicate that IAGBP probably participates in IAG signaling.
Aizen et al. (2016) suggested that IAG hormone in S. verreauxi is regulating phosphorylation through a tyrosine kinase insulin receptor (Sv-TKIR). The Sv-TKIR receptor signaling pathway is mediated through the MAPK/ERK pathway and IAG activates this signaling pathway in a dose-dependent manner (Aizen et al., 2016). This activation confirms the existence of receptors in the testis which can bind to AG ligands, coinciding with the testis regression in response to IAG silencing (Rosen et al., 2010; Aizen et al., 2016).
Insulin receptors (IRs) belong to the large class of tyrosine kinase receptors. IRs play important roles in signaling pathways, which mediate insulin and insulin-like peptides (ILP) signaling (Jin Chan and Steiner, 2000). The expression of IR in M. rosenbergii (Mr-IR) was detected in most tissues in both males and females, including the AG and gonads (Sharabi et al., 2016). Mr-IR silencing resulted in hypertrophy of AG and increased the expression level of Mr-IAG, with an unusual mass of immature spermatozoa cells in the distal vas deferens. RNAi experiments revealed that MrIR functions as a receptor for MrIAG; and the knockdown of MrIR, using both siRNA and dsRNA approaches, induced sex reversal (neofemales) in M. rosenbergii (Tan et al., 2020a). Mr-IR could also regulate Mr-IAG expression, possibly through cross-talk with eyestalk-borne neuropeptide (s) or potentially via auto/paracrine feedback with the AG itself (Sharabi et al., 2016). The regulatory mechanism of the eyestalk-IAG-testis endocrine axis, sex-related genes, hormones, and miRNAs are presented in Figure 1.
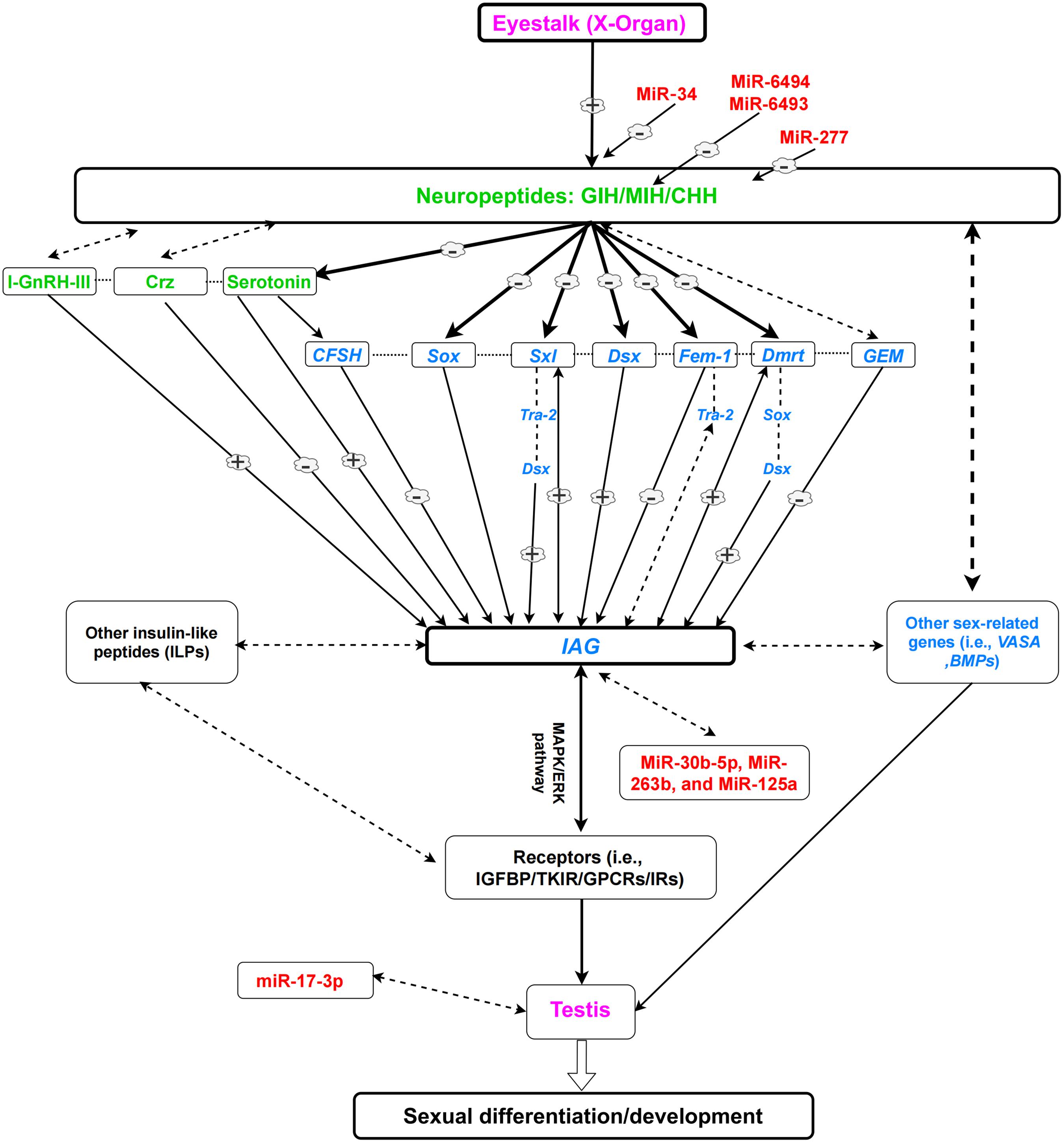
Figure 1. A simplified view of the eyestalk-IAG-testis endocrine axis and its role in the sexual development of male decapods, based on previous literature. Dotted lines indicate possible regulations which are not proved. (–) Shows downregulation. (+) Shows upregulation. Neuropeptides and hormones (green); sex-related genes (blue); miRNAs (red); organs (purple). Note: Every species (or group of species) is unique and could have variable regulatory networks.
Hormonal and Genetic Factors in Female Decapods
CFSH
CFSH has been identified in several decapod species including C. sapidus (Zmora and Chung, 2014), S. verreauxi (Ventura et al., 2014), C. quadricarinatus (Nguyen et al., 2016), P. clarkii (Veenstra, 2015), Chorismus antarcticus (Toullec et al., 2017), M. japonicus (Kotaka and Ohira, 2018), S. paramamosain (Liu A. et al., 2018), and M. rosenbergii (Thongbuakaew et al., 2019). It has been reported that CFSH plays an important role in the development of reproductive phenotypes of female blue crab (C. sapidus) (Zmora and Chung, 2014). The expression pattern of CFSH has been evaluated in several decapod species. In C. sapidus and S. verreauxi, CFSH was exclusively expressed in the eyestalk (Ventura et al., 2014; Zmora and Chung, 2014). In P. clarkii CFSH had the highest expression level in the ovary (Veenstra, 2015). In M. rosenbergii, the expression of CFSH was not exclusive to the eyestalk, but also expressed in several tissues including the CNS, gonads (testis and ovaries), hepatopancreas, stomach, gill, heart, and muscle (Thongbuakaew et al., 2019). However, in some species including shrimps (M. japonicus), prawns (M. rosenbergii), lobsters (S. verreauxi), and crayfish (P. clarkii) CFSH is expressed in both females and males at roughly the same level (Ventura et al., 2014; Veenstra, 2015; Kotaka and Ohira, 2018; Thongbuakaew et al., 2019). These results indicate that CFSH might regulate sexual development only in some crab species and is not a female sex hormone in other decapods.
Fem-1 Gene Family
Fem-1 gene family includes three members, Fem-1a, Fem-1b, and Fem-1c (Ventura-Holman et al., 2003). It has been demonstrated that the sex determination pathway in the roundworm Caenorhabditis elegans is controlled by Fem-1, together with Fem-2 and Fem-3 (Doniach and Hodgkin, 1984). Fem-1 is known to be required for sexual development, involving both somatic and germline sex determination together with other Fem genes (Fem-2 and Fem-3) in C. elegans. Fem-1 negatively regulates Tra-1, which is a key gene for sexual development in females (Doniach and Hodgkin, 1984; Mehra et al., 1999). To date, Fem-1 has been isolated from several decapod species including P. serratus (González-Castellano et al., 2019), E. sinensis (Song et al., 2015), P. vannamei (Galindo-Torres et al., 2019), M. nipponense (Ma et al., 2016b, a), S. paramamosain (Gao et al., 2014), and P. monodon (Robinson et al., 2014). Based on the high expression of Fem-1 in early embryonic development it might be maternal genes in decapods (Song et al., 2015; Ma et al., 2016b). The high expression level of Fem-1 in unfertilized eggs and embryos at the cleavage stage and the low expression level from blastula to zoea in M. nipponense, indicate that Fem-1 in early embryos is maternal. The expression of Fem-1 significantly increased in the larvae, an important stage of sexual differentiation. Fem-1 protein can be potentially interactive with cathepsin L and proteins containing the domains of insulinase, ankyrin or ubiquitin (Ma et al., 2016b). Galindo-Torres et al. (2019) suggested that a negative post-transcriptional regulation controls the expression of Fem-1 in female gonads, which allows the occurrence of sexual differentiation in female P. vannamei. The expression of Fem-1 in spermatogonia indicates it’s also requirment for differentiation of the male gonad (Galindo-Torres et al., 2019). In E. sinensis, the expression pattern of Fem-1 shows a certain degree of sexually dimorphic expression in some tissues. The higher expression level of Fem-1 consistently detected in the testis, ovary, hepatopancreas, and muscles shows its potential role in the last stages of gonadal development (Song et al., 2015).
Tra-2
Transformer-2 (Tra-2) plays important roles in the regulation of pre-mRNA splicing and a pivotal downstream component of the genetic somatic sex determination pathway in Drosophila (Salvemini et al., 2009). In decapods, Tra-2 has been isolated from P. monodon (Leelatanawit et al., 2009), F. chinensis (Li et al., 2012b), P. serratus (González-Castellano et al., 2019), M. nipponense (Zhang et al., 2013), S. verreauxi (Chandler et al., 2015), and E. sinensis (Luo et al., 2017). It has been reported that Tra-2 plays key roles in sexual differentiation and embryonic development of M. nipponense (Zhang et al., 2013). In F. chinensis, Tra-2c expression levels suddenly increased at the mysis stage. In addition, FcTra-2c showed a significantly higher expression level in females than in the male. Tra-2 might therefore be involved in the sex determination of female F. chinensis. Luo et al. (2017) isolated four Tra-2 isoforms, designated as EsTra-2a, EsTra-2b, EsTra-2c, and EsTra-2d from E. sinensis. Remarkably, EsTra-2a had a higher expression level in somatic tissues of males, while EsTra-2c had a higher expression level in the ovary.
Ubiquitin C-Terminal Hydrolases (UCHLs)
Ubiquitin C-terminal hydrolases (UCHLs) are involved in several physiological processes. However, little information is available regarding the role of UCHLs in gonadal development of crustaceans. UCHLs have been identified only in S. paramamosain (Han et al., 2018). Han et al. (2018) reported that the expression level of UCHL3 and UCHL5 in the ovary was significantly higher than in other tissues which provides basic data indicating that UCHL3 and UCHL5 may play important roles in the ovarian development of S. paramamosain.
Epidermal Growth Factor Receptor (EGFR)
Epidermal growth factor receptor (EGFR) is a member of the tyrosine kinase receptor superfamily found in many species. EGFR is a pleiotropic glycoprotein that plays a role in regulating cell proliferation, migration, and differentiation. In mud crab, EGFR is mainly localized in the oocyte perinuclear region with notably obvious signals. EGFR might promote ovarian development by stimulating the expression of VgR and Cyclin B (Lu et al., 2020). The role of EGFR in M. rosenbergii seems to be mostly related to growth control. EGFR-dsRNA injection resulted in delay in the appearance of a male secondary sexual characteristic, growth reduction, and affected eye formation. It seems that Mr-IAG is probably not a ligand of Mr-EGFR (Sharabi et al., 2013).
Regulatory Mechanism of Sex in Female Decapods
Upstream Mechanisms of Sex in Female Decapods
The upstream regulation mechanisms in female decapods include the role of several neurohormones and neuropeptides such as methyl farnesoate (MF), ecdysone, ecdysone receptor (EcR), CHH, GIH, MIH, 5-HT, DA, VIH, octopamine, and red pigment concentrating hormone (RPCH). The major source of these neuropeptides is the X-organ located in the eyestalk ganglia. These neuropeptides involved in carbohydrate and lipid metabolism, moulting, and ovarian development (Böcking et al., 2002; Zhou et al., 2019). The RPCH is not only involved in the regulation of pigmentation but also the regulation of ovarian maturation in crustaceans (Fingerman, 1997). EcR and farnesoic acid O-methyltransferase (FAMeT) are important genes and have a high expression in the eyestalk (Gong et al., 2015). FAMeT is an important rate-limiting enzyme for the conversion from Farnesoic acid (FA) to methyl farnesoate (MF) in the final stage of MF synthesis (Holford et al., 2004). MF is synthesized from the mandibular organ. MF plays important roles in ovarian development (Rodríguez et al., 2002). Based on the high structural similarity, MF is considered to be the homologue of juvenile hormones (JH) from insects (Xie et al., 2015). Localization studies indicated that FAMeT is co-localized in the neurosecretory cells that produce CHH and MIH. Higher levels of FAMeT mRNA transcript and protein levels were reported along with higher levels of CHH and GIH (Zhou et al., 2019). MF biosynthesis is regulated by secreted neuropeptides from the X-organ which regulates the activity of FAMeT and thereby vitellogenesis (Gunawardene et al., 2002).
Downstream Mechanisms of Sex in Female Decapods
CFSH, IAG, Vg, UCHLs, Erk2, SUMO, 14-3-3 zeta, Slo, CKS1, Cdc2, MAPK1, FAMeT, Wnt4, and EGFR probably play important roles in the sexual development of females. Unlike males, the downstream mechanisms in females are relatively unknown. Except for CFSH and IAG, the regulation mechanism of other genes is unknown and information is limited to the molecular characterization and expression profile.
The bilateral eyestalk ablation and silencing of CFSH altered reproductive structures (i.e., sharper abdomens, absent or misplaced gonopores, fewer and shorter setae on pleopods) and impaired mating in female C. sapidus (Zmora and Chung, 2014). It seems that CFSH plays an important role in sexual differentiation rather than sex determination because the absence of CFSH neither resulted in sex reversal nor ovarian degeneration (Zmora and Chung, 2014). The expression level of CFSH changed at different moulting stages and the highest expression level was detected in the pre-moult stage. Moreover, the expression of CFSH increased at the beginning of gonopore development in females and decreased in males, indicating that the expression of CFSH affects the formation of gonopores in S. paramamosain (Jiang et al., 2020a). CFSH may also participate in the regulation of sexual differentiation of early juveniles (Jiang et al., 2020a).
The role of IAG is not limited to males, IAG may play an inhibitory role in ovarian development in female decapods. The expression level of IAG is maintained at low levels during the undeveloped stage (stage I) to late vitellogenic stage (stage IV) and then increased at the mature stage (stage V), indicating that IAG participates in inhibiting oocyte growth and vitellogenesis (Huang et al., 2014). In C. sapidus, ovarian development and season affected the expression level of both IAG and insulin-like peptide binding protein (ILPBP) in the ovary. These results indicated that ovarina IAG and ILPBP may be involved in the ovarian development (Huang et al., 2017).
The interaction of sex-related genes in females is poorly understood. Therefore, further studies are required to study the function and interaction of these genes. The regulatory mechanism of sexual development in female decapods and the roles of sex-related genes, hormones, and miRNAs are presented in Figure 2.
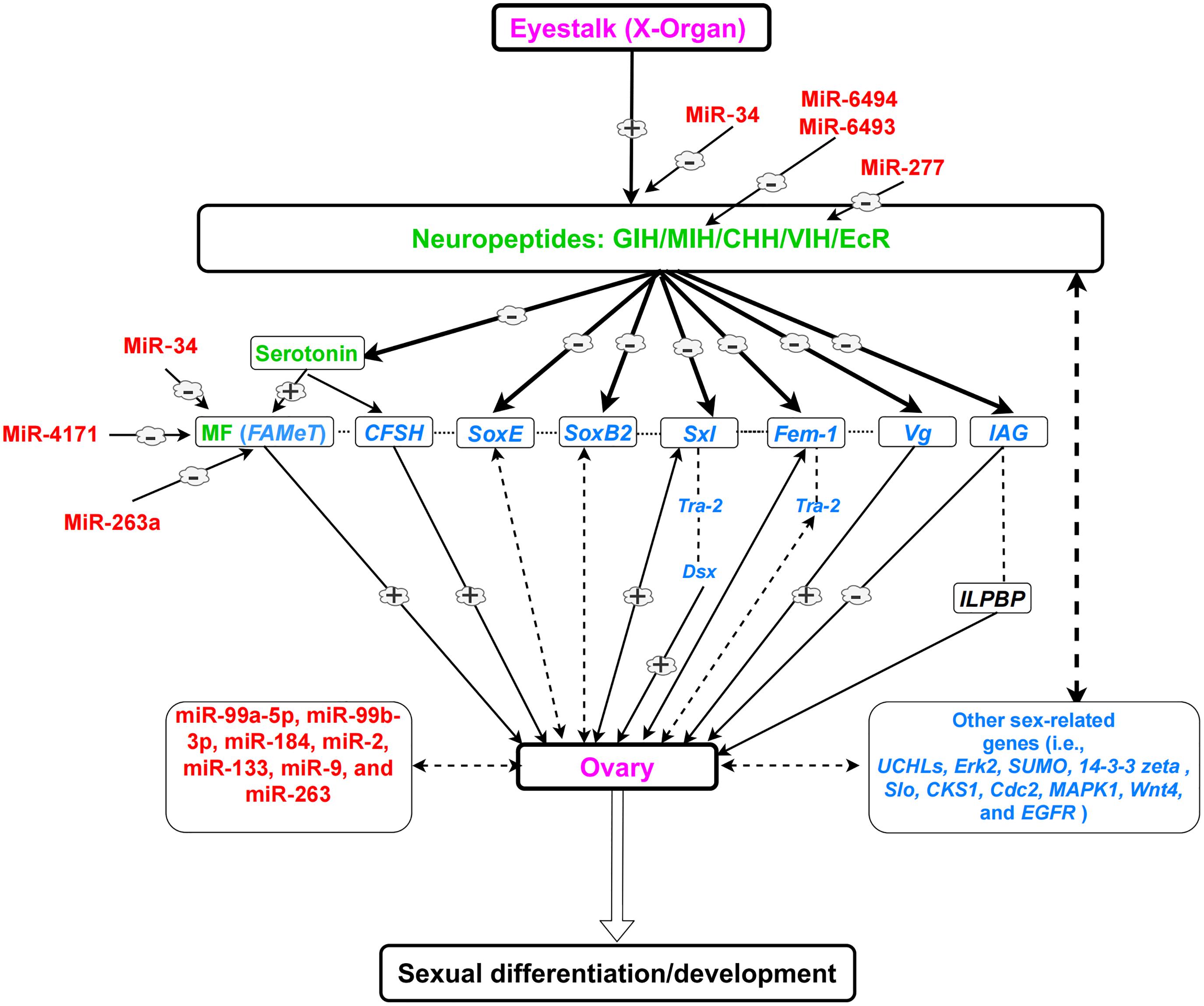
Figure 2. A simplified view of the eyestalk-ovary endocrine axis and its role in sexual development in female decapods, based on previous literature. Dotted lines indicate possible regulations which are not proved. (–) shows downregulation. (+) shows upregulation. Neuropeptides and hormones (green); sex-related genes (blue); miRNAs (red); organs (purple). Note: Every species (or group of species) is unique and could have variable regulatory networks.
Non-Coding RNAs (ncRNAs)
MicroRNAs (miRNAs)
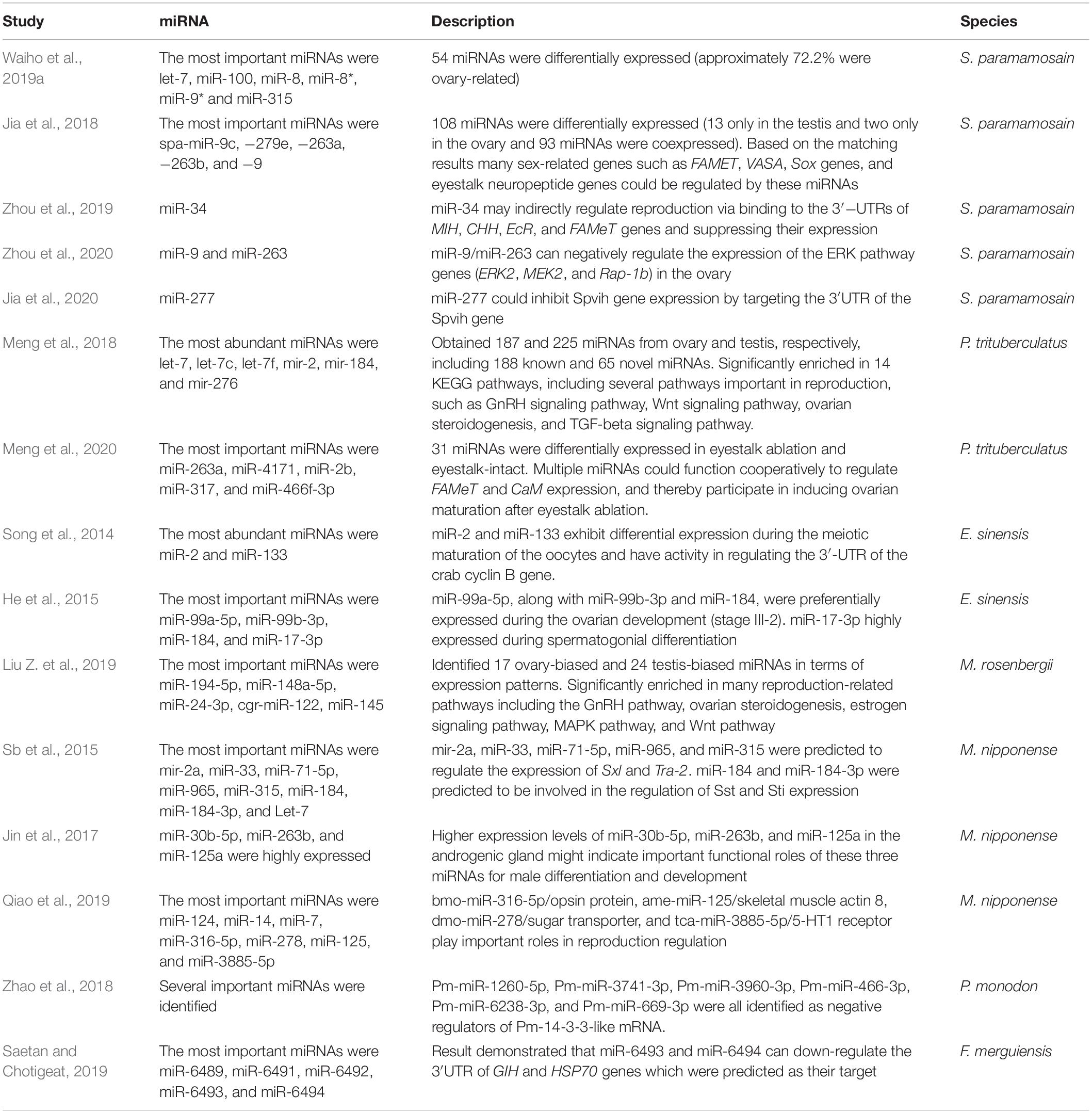
MicroRNAs (miRNAs) are small non-coding RNA with a length of about 22 bp. miRNAs inhibit gene expression through translational repression by binding to the 3′-untranslated regions (UTRs) of the target mRNAs (Christensen et al., 2009). Moreover, one miRNA can control numerous genes, and one gene can be regulated by multiple miRNAs (Jingsheng et al., 2015). There are two main stages for miRNAs generation. The first step is the synthesis of the 70-100 bp precursor-miRNA which has a characteristic hairpin structure generated from a primary miRNA transcript by the Drosha and its RNA-binding cofactor DiGeorge syndrome critical region gene 8 (DGCR8) in the nucleus. In the second step, Exprotin5-RAN-GTP carries the pre-miRNA to the cytoplasm for further processing by Dicer (a RNAse III endonuclease) to form the mature miRNA (Zeng, 2006; Winter et al., 2009). In mammals, miRNAs control gonad development (ovary and testis), induce differentiation and maturation of oocytes and spermatozoa, and influence zygote development (Scherr et al., 2007). In the rat, miR-141-3p inhibits ovarian granulosa by targeting DAPK1 gene and participates in the etiology of polycystic ovary syndrome (Li et al., 2017). It has been reported that miR-143 plays a key role in follicle stimulating hormone (FSH) which induces estradiol production and granulosa cell proliferation via binding to the 3′-UTR of Kirsten rat sarcoma (KRAS) mRNA (Zhang et al., 2017). miRNAs are also essential for ovulation and oocyte maturation in invertebrates (Kadener et al., 2009). In decapods, several sex-related miRNAs have been identified in S. paramamosain (Jia et al., 2018; Waiho et al., 2019a; Zhou et al., 2019, 2020), Portunus trituberculatus (Meng et al., 2018, 2020), E. sinensis (Song et al., 2014; He et al., 2015), M. rosenbergii (Liu X. et al., 2019; Qiao et al., 2019) M. nipponense (Sb et al., 2015; Jin et al., 2017), P. monodon (Zhao et al., 2018), and Fenneropenaeus merguiensis (Saetan and Chotigeat, 2019). The roles of sex-related miRNAs in decapods are summarized in Table 3.
Transcriptomic analysis revealed that miRNAs may affect the sexual development of decapods through several pathways. The most important predicted pathways are ERK, MAPK, necroptosis pathway, GnRH signaling pathway, Wnt signaling pathway, ovarian steroidogenesis, TGF-beta signalling pathway, gluconeogenesis pathway, glycolysis pathway, estrogen signaling pathway, ovarian steroidogenesis, and insulin signaling pathway (He et al., 2015; Jia et al., 2018; Meng et al., 2018; Zhao et al., 2018; Liu X. et al., 2019; Zhou et al., 2019).
In males, miRNAs such as miR-17-3p, let-7c, and miR-21-5p demonstrated different expression levels during different stages of testicular development indicating their probable involvement in spermatogenesis (He et al., 2015). MiR-34 probably regulates the reproductive function via binding to the 3′-UTRs of MIH, EcR, FAMeT, and CHH genes (Zhou et al., 2019).
In females, miR-34 and miR-277 may play a key role in the ovarian development of S. paramamosain. AgomiR-34 reduced the expression level of MIH, CHH, VIH, EcR, RPCH, and FAMeT genes, whereas antagomiR-34 elevates the expression level of these genes (Zhou et al., 2019). Injection of agomiR-277 in female mud crab increased the expression of the Spvtg gene in the ovary and hepatopancreas of mud crab. On the contrary, the expression of Spvih was significantly decreased after injection with agomiR-277, indicating that miR-277 could positively regulate the expression of the Spvtg gene in mud crab indirectly (by negative regulation on VIH) (Jia et al., 2020). It has been demonstrated that miR-133 and miR-2 downregulated cyclin B gene expression during meiotic oocyte maturation in E. sinensis (Song et al., 2014). In addition, the expression level of miR-87 and miR-7 was significantly increased and they were predicted to target cyclin-dependent kinase 2 (Cdk2)7 and cell division cycle4 (Cdc4), which are important for regulation of the G1-to-S-phase transition during the cell cycle (Schnackenberg et al., 2007). Meng et al. (2020) showed that MF synthesis and FAMeT expression are directly regulated by miR-4171 and miR-263a. Also, miR-317, miR-2b, and miR-466f-3p repress the expression of calcium signal transduction (CaM). The eyestalk ablation showed a negative association between miR-263a, miR-4171, and FAMeT and between miR-2b, miR-317, and CaM. Taken together, these results indicate that multiple miRNAs operate cooperatively to regulate the expression of FAMeT and CaM, and thereby participate in inducing ovarian maturation after eyestalk ablation.
PIWI-Interacting RNAs (piRNAs)
PIWI-interacting RNAs (piRNAs) are widely presented in germ cells and are involved in gonad development and gametogenesis. piRNAs play a key role in the gametogenesis processes by silencing transposable elements (Rastetter et al., 2015). piRNAs are longer (26-31 nt) than miRNAs (21-23 nt). However, piRNAs are seldom expressed in somatic cells and show a higher composition present compared to miRNAs during gametogenesis (Aravin et al., 2006). Small RNAs function by guiding a crucial co-factor–an Argonaute/PIWI family of proteins to target RNAs (Hilz et al., 2016). Two major pathways are known for piRNA synthesis: (A) primary biogenesis: piRNAs synthesis from several portions of long single-stranded precursor RNAs transcribed from both uni-directional and bi-directional piRNA clusters in the genomic loci (Mohn et al., 2014), and (B) secondary biogenesis, also known as the ping-pong cycle, selectively amplifies specific piRNAs targeting active transposon (Brennecke et al., 2007). Scarce information is available regarding the role of piRNAs in the sexual development of crustaceans. Waiho et al. (2020) identified gonadal piRNAs of S. paramamosain and out of 115,491 novel piRNAs, 596 were differentially expressed. Furthermore, 389,887 potential piRNA-target genes were predicted. The expression level of four piRNAs and nine genes with high piRNA interactions were validated with the inclusion of additional immature specimens including low-density lipoprotein receptor-related protein 2 (LRP2) that is involved in reproduction and growth, midasin (MDN1) in ribosome biogenesis pathway and gametogenesis, and PRKDC, a DNA repair gene involved in gonad development and differentiation.
Long Non-coding RNAs (lncRNAs)
Long non-coding RNAs (lncRNAs) are non-protein-coding RNAs longer than 200 nt (Rinn and Chang, 2012). lncRNAs are divided into genic and intergenic based on their proximity to the protein-coding genes in a genome (Gibb et al., 2011). Unlike small non-coding RNAs, the annotation and identification of lncRNA are difficult due to their low expression level (Derrien et al., 2012). Similar to protein-coding genes, they are mostly transcribed by RNA polymerase II and can be post-transcriptionally modified by splicing, capping, and polyadenylation (Guttman and Rinn, 2012). lncRNAs usually have a small number of exons and a shorter transcript size (average of 800 nt) compared to protein-coding genes (Derrien et al., 2012). lncRNAs often demonstrate tissue-specific expression patterns and even sometimes are localized in a specific cellular compartment (Ginger et al., 2006). Although several lncRNAs have been identified, the molecular function of only a small number of lncRNAs has been experimentally validated. lncRNAs play an important role in gene regulation in three ways including serving as decoys, scaffolds, and guides. Many lncRNAs serve as decoys and affect the transcription of genes by preventing access to DNA by regulatory proteins (Hung et al., 2011). Some lncRNAs act as scaffolds and bring proteins into a discrete complex. Other lncRNAs regulate different developmental and cellular processes by guiding a specific protein complex to a specific promoter in response to certain molecular signals (Tsai et al., 2010). In addition, lncRNAs regulate several molecular processes such as alternative splicing and mRNA transport (Tripathi et al., 2010).
In decapods, little information is available regarding the sex-related lncRNAs. Yang et al. (2018) provided fundamental resources for further studies on the roles of lncRNAs in decapod sexual development. Several lncRNAs negatively regulated the expression of some genes, indicating their potential role in the sexual differentiation of mud crab. lncRNAs including lncRNA-ncr2, lncRNA-ncr10, and lncRNA-ncr14 suppressed the expression of their target genes (Yang et al., 2018).
Conclusion and Future Perspective
Although relatively adequate information is available regarding molecular mechanisms of sexual development in males, limited information is available in females. Several sex-related genes have been identified to be involved in the “eyestalk-IAG-testis” endocrine axis in males. Most of these genes are negatively regulated by the eyestalk neurohormones and have a regulatory effect on the AG and IAG expression. For example, it has been documented that CFSH, Sox, Dsx, Slx, Dmrt, GEM, and Fem-1 regulate the expression of IAG. IAG expression is negatively regulated by GEM, Fem-1, and CFSH. In contrast, Slx, Dsx, and Dmrt silencing significantly decrease the expression of IAG indicating the existence of positive regulation. IAG, Sox, Dmrt, Dsx, Vasa, BMPs, and Slx exhibit male-biased expression patterns and play crucial roles in testis development and spermatogenesis. In contrast, CFSH, Fem-1, FAMeT, Slo, Tra-2, UCHLs, Erk2, Cdc2, EGFR, Vg, VgR, and VIH have female-biased expression patterns and play important roles during ovarian development.
Also, it has been revealed that several non-coding RNAs (i.e., miRNAs, lncRNAs, and piRNAs) may affect the sexual development of decapods. Studies on non-coding RNAs are limited to some miRNAs and further studies are needed to clarify the roles of other non-coding RNAs (i.e., piRNA and lncRNAs) in the regulation mechanism of sex in decapods. For example, miR-34 indirectly regulates the reproductive function via binding to the 3′-UTRs of MIH, CHH, EcR, and FAMeT genes and suppresses their expression.
Further research is required to (1) identify more sex-related genes and non-coding RNAs; (2) investigate the role and function of sex-related genes and non-coding RNAs; (3) understand the interaction of these sex-related genes together and with non-coding RNAs, (4) understand the role and regulation mechanism of IAG in female decapods, and (5) identify more IAG receptors.
Author Contributions
AF designed the framework of the draft and collected the literature. AF and HM drafted the manuscript. Other authors revised and finalized the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key Research & Development Program of China (No. 2018YFD0900201), National Natural Science Foundation of China (No. 31772837), the Leading Talent Project of Special Support Plan of Guangdong Province (2019TX05N067), the Science and Technology Project of Guangdong Province (2018A050506080), the “Sail Plan” Program for the Introduction of Outstanding Talents of Guangdong Province, China, and the STU Scientific Research Foundation for Talents (No. NTF17006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully thank Prof. Julian D. Reynolds for the English correction of the manuscript.
References
Abayed, F. A. A., Manor, R., Aflalo, E. D., and Sagi, A. (2019). Screening for Dmrt genes from embryo to mature Macrobrachium rosenbergii prawns. Gen. Comp. Endocrinol. 282:113205. doi: 10.1016/j.ygcen.2019.06.009
Aflalo, E. D., Bakhrat, A., Raviv, S., Harari, D., Sagi, A., and Abdu, U. (2007). Characterization of a vasa-like gene from the pacific white shrimp Litopenaeus vannamei and its expression during oogenesis. Mol. Reprod. Dev. 74, 172–177. doi: 10.1002/mrd.20622
Aflalo, E. D., Hoang, T. T. T., Nguyen, V. H., Lam, Q., Nguyen, D. M., Trinh, Q. S., et al. (2006). A novel two-step procedure for mass production of all-male populations of the giant freshwater prawn Macrobrachium rosenbergii. Aquaculture 256, 468–478. doi: 10.1016/j.aquaculture.2006.01.035
Aizen, J., Chandler, J. C., Fitzgibbon, Q. P., Sagi, A., Battaglene, S. C., Elizur, A., et al. (2016). Production of recombinant insulin-like androgenic gland hormones from three decapod species: in vitro testicular phosphorylation and activation of a newly identified tyrosine kinase receptor from the Eastern spiny lobster, Sagmariasus verreauxi. Gen. Comp. Endocrinol. 229, 8–18. doi: 10.1016/j.ygcen.2016.02.013
Alfaro-Montoya, J. (2010). The reproductive conditions of male shrimps, genus Penaeus, sub-genus Litopenaeus (open thelyca penaeoid shrimps): a review. Aquaculture 300, 1–9. doi: 10.1016/j.aquaculture.2009.12.008
Aravin, A., Gaidatzis, D., Pfeffer, S., Lagos-Quintana, M., Landgraf, P., Iovino, N., et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207. doi: 10.1038/nature04916
Banzai, K., Ishizaka, N., Asahina, K., Suitoh, K., Izumi, S., and Ohira, T. (2011). Molecular cloning of a cDNA encoding insulin-like androgenic gland factor from the kuruma prawn Marsupenaeus japonicus and analysis of its expression. Fish. Sci. 77, 329–335. doi: 10.1007/s12562-011-0337-8
Barki, A., Karplus, I., Manor, R., and Sagi, A. (2006). Intersexuality and behavior in crayfish: the de-masculinization effects of androgenic gland ablation. Horm. Behav. 50, 322–331. doi: 10.1016/j.yhbeh.2006.03.017
Böcking, D., Dircksen, H., and Keller, R. (2002). “The crustacean neuropeptides of the CHH/MIH/GIH family: structures and biological activities,” in The CRUSTACEAN Nervous System, ed. K. Wiese (Berlin: Springer), 84–97. doi: 10.1007/978-3-662-04843-6_6
Boerjan, B., Verleyen, P., Huybrechts, J., Schoofs, L., and De Loof, A. (2010). In search for a common denominator for the diverse functions of arthropod corazonin: a role in the physiology of stress? Gen. Comp. Endocrinol. 166, 222–233. doi: 10.1016/j.ygcen.2009.09.004
Brennecke, J., Aravin, A. A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., et al. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103. doi: 10.1016/j.cell.2007.01.043
Buaklin, A., Sittikankaew, K., Phinyo, M., Prasertlux, S., Janpoom, S., Klinbunga, S., et al. (2016). Expression of catechol O-methyltransferase during ovarian development and association between its SNP and reproduction-related parameters of the giant tiger shrimp Penaeus monodon. Aquaculture 465, 245–257. doi: 10.1016/j.aquaculture.2016.09.011
Chandler, J. C., Aizen, J., Elizur, A., Battaglene, S. C., and Ventura, T. (2015). Male sexual development and the androgenic gland: novel insights through the de novo assembled transcriptome of the eastern spiny lobster, Sagmariasus verreauxi. Sex. Dev. 9, 338–354. doi: 10.1159/000443943
Chandler, J. C., Elizur, A., and Ventura, T. (2018). The decapod researcher’s guide to the galaxy of sex determination. Hydrobiologia 825, 61–80. doi: 10.1007/s10750-017-3452-4
Chandler, J. C., Gandhi, N. S., Mancera, R. L., Smith, G., Elizur, A., and Ventura, T. (2017). Understanding insulin endocrinology in decapod crustacea: molecular modelling characterization of an insulin-binding protein and insulin-like peptides in the eastern spiny lobster, Sagmariasus verreauxi. Int. J. Mol. Sci. 18:1832. doi: 10.3390/ijms18091832
Chen, T., Zhang, L., Wong, N., Zhong, M., Ren, C., and Hu, C. (2014). Pacific white shrimp (Litopenaeus vannamei) vitellogenesis-inhibiting hormone (VIH) is predominantly expressed in the brain and negatively regulates hepatopancreatic vitellogenin (VTG) gene expression. Biol. Reprod. 90, 1–10.
Christensen, B. C., Moyer, B. J., Avissar, M., Ouellet, L. G., Plaza, S. L., McClean, M. D., et al. (2009). A let-7 microRNA-binding site polymorphism in the KRAS 3’ UTR is associated with reduced survival in oral cancers. Carcinogenesis 30, 1003–1007. doi: 10.1093/carcin/bgp099
Chung, J. S., Manor, R., and Sagi, A. (2011). Cloning of an insulin-like androgenic gland factor (IAG) from the blue crab, Callinectes sapidus: implications for eyestalk regulation of IAG expression. Gen. Comp. Endocrinol. 173, 4–10. doi: 10.1016/j.ygcen.2011.04.017
Clark, A. G., Eisen, M. B., Smith, D. R., Bergman, C. M., Oliver, B., Markow, T. A., et al. (2007). Evolution of genes and genomes on the Drosophila phylogeny. Nature 450, 203–218. doi: 10.1038/nature06341
Coman, F. E., Sellars, M. J., Norris, B. J., Coman, G. J., and Preston, N. P. (2008). The effects of triploidy on Penaeus (Marsupenaeus) japonicus (Bate) survival, growth and gender when compared to diploid siblings. Aquaculture 276, 50–59. doi: 10.1016/j.aquaculture.2008.01.031
Cui, Z., Hui, M., Liu, Y., Song, C., Li, X., Li, Y., et al. (2015). High-density linkage mapping aided by transcriptomics documents ZW sex determination system in the Chinese mitten crab Eriocheir sinensis. Heredity 115, 206–215. doi: 10.1038/hdy.2015.26
De Grave, S., Pentcheff, N. D., Ahyong, S. T., Chan, T.-Y., Crandall, K. A., Dworschak, P. C., et al. (2009). A classification of living and fossil genera of decapod crustaceans. Raffles Bull. Zool. 21, 1–109. doi: 10.5479/si.00963801.78-2851.1
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. doi: 10.1101/gr.132159.111
Doniach, T., and Hodgkin, J. (1984). A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev. Biol. 106, 223–235. doi: 10.1016/0012-1606(84)90077-0
Fang, S., Zhang, Y., Shi, X., Zheng, H., Li, S., Zhang, Y., et al. (2020). Identification of male-specific SNP markers and development of PCR-based genetic sex identification technique in crucifix crab (Charybdis feriatus) with implication of an XX/XY sex determination system. Genomics 112, 404–411. doi: 10.1016/j.ygeno.2019.03.003
FAO (2020). FAO Yearbook. Fishery and Aquaculture Statistics 2018/FAO Annuaire. Statistiques des Pêches et de L’aquaculture 2018/FAO Anuario. Estadísticas de Pesca y Acuicultura 2018. Rome: FAO
Fingerman, M. (1987). The endocrine mechanisms of crustaceans. J. Crustac. Biol. 7, 1–24. doi: 10.2307/1548622
Fingerman, M. (1997). Roles of neurotransmitters in regulating reproductive hormone release and gonadal maturation in decapod crustaceans. Invertebr. Reprod. Dev. 31, 47–54. doi: 10.1080/07924259.1997.9672562
Galindo-Torres, P., Ventura-López, C., Llera-Herrera, R., and Ibarra, A. M. (2019). A natural antisense transcript of the fem-1 gene was found expressed in female gonads during the characterization, expression profile, and cellular localization of the fem-1 gene in Pacific white shrimp Penaeus vannamei. Gene 706, 19–31. doi: 10.1016/j.gene.2019.04.066
Gao, J., Wang, X., Zou, Z., Jia, X., Wang, Y., and Zhang, Z. (2014). Transcriptome analysis of the differences in gene expression between testis and ovary in green mud crab (Scylla paramamosain). BMC Genomics 15:585. doi: 10.1186/1471-2164-15-585
Gao, S., Zhang, T., Zhou, X., Zhao, Y., Li, Q., Guo, Y., et al. (2005). Molecular cloning, expression of sox5 and its down-regulation of Dmrt1 transcription in zebrafish. J. Exp. Zool. 304, 476–483. doi: 10.1002/jez.b.21053
Gibb, E. A., Brown, C. J., and Lam, W. L. (2011). The functional role of long non-coding RNA in human carcinomas. Mol. Cancer 10:38. doi: 10.1186/1476-4598-10-38
Ginger, M. R., Shore, A. N., Contreras, A., Rijnkels, M., Miller, J., Gonzalez-Rimbau, M. F., et al. (2006). A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc. Natl. Acad. Sci. U.S.A. 103, 5781–5786. doi: 10.1073/pnas.0600745103
Gong, J., Yang, Y., and Ye, H. (2014). Characterization and expression of calcium-activated potassium channels (Slo) in ovary of the mud crab, Scylla paramamosain. Genes Genomics 36, 77–86. doi: 10.1007/s13258-013-0143-z
Gong, J., Ye, H., Xie, Y., Yang, Y., Huang, H., Li, S., et al. (2015). Ecdysone receptor in the mud crab. J. Endocrinol. 224, 273–287. doi: 10.1530/joe-14-0526
González-Castellano, I., Manfrin, C., Pallavicini, A., and Martínez-Lage, A. (2019). De novo gonad transcriptome analysis of the common littoral shrimp Palaemon serratus: novel insights into sex-related genes. BMC Genomics 20:757. doi: 10.1186/s12864-019-6157-4
Govoni, K. E., Baylink, D. J., and Mohan, S. (2005). The multi-functional role of insulin-like growth factor binding proteins in bone. Pediatr. Nephrol. 20, 261–268. doi: 10.1007/s00467-004-1658-y
Gunawardene, Y. I. N. S., Tobe, S. S., Bendena, W. G., Chow, B. K. C., Yagi, K. J., and Chan, S.-M. (2002). Function and cellular localization of farnesoic acid o-methyltransferase (FAMeT) in the shrimp, Metapenaeus ensis. FEBS J. 269, 3587–3595. doi: 10.1046/j.1432-1033.2002.03048.x
Guo, Q., Li, S., Lv, X., Xiang, J., Manor, R., Sagi, A., et al. (2019). Sex-Biased CHHs and Their putative receptor regulate the expression of IAG Gene in the Shrimp Litopenaeus vannamei. Front. Physiol. 10:1525. doi: 10.3389/fphys.2019.01525
Guttman, M., and Rinn, J. L. (2012). Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346. doi: 10.1038/nature10887
Han, K., Dai, Y., Zhang, Z., Zou, Z., and Wang, Y. (2018). Molecular characterization and expression profiles of Sp-uchl3 and Sp-uchl5 during gonad development of Scylla paramamosain. Molecules 23:213. doi: 10.3390/molecules23010213
Han, K., Dai, Y., Zou, Z., Fu, M., Wang, Y., and Zhang, Z. (2012). Molecular characterization and expression profiles of cdc2 and cyclin B during oogenesis and spermatogenesis in green mud crab (Scylla paramamosain). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 163, 292–302. doi: 10.1016/j.cbpb.2012.07.001
Harlıoǧlu, M. M., and Farhadi, A. (2017a). Factors affecting the reproductive efficiency in crayfish: implications for aquaculture. Aquac. Res. 48, 1983–1997. doi: 10.1111/are.13263
Harlıoǧlu, M. M., and Farhadi, A. (2017b). Feminization strategies in crustacean aquaculture. Aquac. Int. 25, 1453–1468. doi: 10.1007/s10499-017-0128-z
He, L., Wang, Y.-L., Li, Q., Yang, H.-D., Duan, Z.-L., and Wang, Q. (2015). Profiling microRNAs in the testis during sexual maturation stages in Eriocheir sinensis. Anim. Reprod. Sci. 162, 52–61. doi: 10.1016/j.anireprosci.2015.09.008
Hilz, S., Modzelewski, A. J., Cohen, P. E., and Grimson, A. (2016). The roles of microRNAs and siRNAs in mammalian spermatogenesis. Development 143, 3061–3073. doi: 10.1242/dev.136721
Holford, K. C., Edwards, K. A., Bendena, W. G., Tobe, S. S., Wang, Z., and Borst, D. W. (2004). Purification and characterization of a mandibular organ protein from the American lobster, Homarus americanus: a putative farnesoic acid O-methyltransferase. Insect Biochem. Mol. Biol. 34, 785–798. doi: 10.1016/j.ibmb.2004.04.003
Hu, Y., Jin, S., Fu, H., Qiao, H., Zhang, W., Jiang, S., et al. (2020). Functional analysis of a SoxE gene in the oriental freshwater prawn, Macrobrachium nipponense by molecular cloning, expression pattern analysis, and in situ hybridization (de Haan, 1849). 3 Biotech 10:10.
Hua, Y.-J., Ishibashi, J., Saito, H., Tawfik, A. I., Sakakibara, M., Tanaka, Y., et al. (2000). Identification of [Arg7] corazonin in the silkworm, Bombyx mori and the cricket, Gryllus bimaculatus, as a factor inducing dark color in an albino strain of the locust, Locusta migratoria. J. Insect Physiol. 46, 853–860. doi: 10.1016/s0022-1910(99)00173-0
Huang, X., Ye, H., and Chung, J. S. (2017). The presence of an insulin-like androgenic gland factor (IAG) and insulin-like peptide binding protein (ILPBP) in the ovary of the blue crab, Callinectes sapidus and their roles in ovarian development. Gen. Comp. Endocrinol. 249, 64–70. doi: 10.1016/j.ygcen.2017.05.001
Huang, X., Ye, H., Huang, H., Yang, Y., and Gong, J. (2014). An insulin-like androgenic gland hormone gene in the mud crab, Scylla paramamosain, extensively expressed and involved in the processes of growth and female reproduction. Gen. Comp. Endocrinol. 204, 229–238. doi: 10.1016/j.ygcen.2014.06.002
Hung, T., Wang, Y., Lin, M. F., Koegel, A. K., Kotake, Y., Grant, G. D., et al. (2011). Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 43, 621–629. doi: 10.1038/ng.848
Jia, C., Hui, L., Cao, W., Lietz, C. B., Jiang, X., Chen, R., et al. (2012). High-definition de novo sequencing of crustacean hyperglycemic hormone (CHH)-family neuropeptides. Mol. Cell. Proteomics 11, 1951–1964. doi: 10.1074/mcp.m112.020537
Jia, X., Chen, Y., Zou, Z., Lin, P., Wang, Y., and Zhang, Z. (2013). Characterization and expression profile of Vitellogenin gene from Scylla paramamosain. Gene 520, 119–130. doi: 10.1016/j.gene.2013.02.035
Jia, X., Fang, Z., Zeng, X., Zhang, X., Zhang, Z., and Wang, Y. (2020). Regulation of VIH by miR-277 in the eyestalk of mud crab Scylla paramamosain. Aquaculture, 534:736254. doi: 10.1016/j.aquaculture.2020.736254
Jia, X., Zhou, M., Zou, Z., Lin, P., Wang, Y., and Zhang, Z. (2018). Identification and comparative analysis of the ovary and testis microRNAome of mud crab Scylla paramamosain. Mol. Reprod. Dev. 85, 519–531.
Jiang, Q., Lu, B., Lin, D., Huang, H., Chen, X., and Ye, H. (2020a). Role of crustacean female sex hormone (CFSH) in sex differentiation in early juvenile mud crabs, Scylla paramamosain. Gen. Comp. Endocrinol. 289:113383. doi: 10.1016/j.ygcen.2019.113383
Jiang, Q., Lu, B., Wang, G., and Ye, H. (2020b). Transcriptional inhibition of Sp-IAG by crustacean female sex hormone in the mud crab, Scylla paramamosain. Int. J. Mol. Sci. 21:5300. doi: 10.3390/ijms21155300
Jiang, Q., Zheng, H., Zheng, L., Wang, Y., Wang, M., Xie, X., et al. (2020c). Molecular characterization of the insulin-like androgenic gland hormone in the swimming crab, Portunus trituberculatus, and its involvement in the insulin signaling system. Front. Endocrinol. 11:585. doi: 10.3389/fendo.2020.00585
Jin, S., Fu, H., Sun, S., Jiang, S., Xiong, Y., Gong, Y., et al. (2017). Integrated analysis of microRNA and mRNA expression profiles during the sex-differentiation sensitive period in oriental river prawn, Macrobrachium nipponense. Sci. Rep. 7:12011.
Jin, S., Hu, Y., Fu, H., Jiang, S., Xiong, Y., Qiao, H., et al. (2019). Potential functions of gem-associated protein 2-Like Isoform X1 in the oriental river prawn Macrobrachium nipponense: cloning, qPCR, In Situ Hybridization, and RNAi Analysis. Int. J. Mol. Sci. 20:3995. doi: 10.3390/ijms20163995
Jin Chan, S., and Steiner, D. F. (2000). Insulin through the ages: phylogeny of a growth promoting and metabolic regulatory hormone. Am. Zool. 40, 213–222. doi: 10.1093/icb/40.2.213
Jingsheng, S., Yibing, W., Jun, X., Siqun, W., Jianguo, W., Feiyan, C., et al. (2015). MicroRNAs are potential prognostic and therapeutic targets in diabetic osteoarthritis. J. Bone Miner. Metab. 33, 1–8. doi: 10.1007/s00774-014-0628-0
Kadener, S., Rodriguez, J., Abruzzi, K. C., Khodor, Y. L., Sugino, K., Marr, M. T., et al. (2009). Genome-wide identification of targets of the drosha–pasha/DGCR8 complex. RNA 15, 537–545. doi: 10.1261/rna.1319309
Kato, Y., Kobayashi, K., Oda, S., Tatarazako, N., Watanabe, H., and Iguchi, T. (2010). Sequence divergence and expression of a transformer gene in the branchiopod crustacean, Daphnia magna. Genomics 95, 160–165. doi: 10.1016/j.ygeno.2009.12.005
Kato, Y., Kobayashi, K., Watanabe, H., and Iguchi, T. (2011). Environmental sex determination in the Branchiopod crustacean Daphnia magna: deep conservation of a Doublesex gene in the sex-determining pathway. PLoS Genet. 7:e1001345. doi: 10.1371/journal.pgen.1001345
Khalaila, I., Manor, R., Weil, S., Granot, Y., Keller, R., and Sagi, A. (2002). The eyestalk–androgenic gland–testis endocrine axis in the crayfish Cherax quadricarinatus. Gen. Comp. Endocrinol. 127, 147–156. doi: 10.1016/s0016-6480(02)00031-x
Kotaka, S., and Ohira, T. (2018). cDNA cloning and in situ localization of a crustacean female sex hormone-like molecule in the kuruma prawn Marsupenaeus japonicus. Fish. Sci. 84, 53–60. doi: 10.1007/s12562-017-1152-7
Lacombe, C., Greve, P., and Martin, G. (1999). Overview on the sub-grouping of the crustacean hyperglycemic hormone family. Neuropeptides 33, 71–80. doi: 10.1054/npep.1999.0016
Lasko, P. F., and Ashburner, M. (1988). The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature 335, 611–617. doi: 10.1038/335611a0
Leelatanawit, R., Sittikankeaw, K., Yocawibun, P., Klinbunga, S., Roytrakul, S., Aoki, T., et al. (2009). Identification, characterization and expression of sex-related genes in testes of the giant tiger shrimp Penaeus monodon. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 152, 66–76. doi: 10.1016/j.cbpa.2008.09.004
Li, D., Xu, D., Xu, Y., Chen, L., Li, C., Dai, X., et al. (2017). MicroRNA-141-3p targets DAPK1 and inhibits apoptosis in rat ovarian granulosa cells. Cell Biochem. Funct. 35, 197–201. doi: 10.1002/cbf.3248
Li, F., Bai, H., Xiong, Y., Fu, H., Jiang, S., Jiang, F., et al. (2015a). Molecular characterization of insulin-like androgenic gland hormone-binding protein gene from the oriental river prawn Macrobrachium nipponense and investigation of its transcriptional relationship with the insulin-like androgenic gland hormone gen. Gen. Comp. Endocrinol. 216, 152–160. doi: 10.1016/j.ygcen.2014.12.007
Li, F., Bai, H., Zhang, W., Fu, H., Jiang, F., Liang, G., et al. (2015b). Cloning of genomic sequences of three crustacean hyperglycemic hormone superfamily genes and elucidation of their roles of regulating insulin-like androgenic gland hormone gene. Gene 561, 68–75. doi: 10.1016/j.gene.2015.02.012
Li, L., Kelley, W. P., Billimoria, C. P., Christie, A. E., Pulver, S. R., and Sweedler, J. V, et al. (2003). Mass spectrometric investigation of the neuropeptide complement and release in the pericardial organs of the crab, Cancer borealis. J. Neurochem. 87, 642–656. doi: 10.1046/j.1471-4159.2003.02031.x
Li, S., Li, F., Sun, Z., and Xiang, J. (2012a). Two spliced variants of insulin-like androgenic gland hormone gene in the Chinese shrimp, Fenneropenaeus chinensis. Gen. Comp. Endocrinol. 177, 246–255. doi: 10.1016/j.ygcen.2012.04.010
Li, S., Li, F., Wen, R., and Xiang, J. (2012b). Identification and characterization of the sex-determiner transformer-2 homologue in Chinese shrimp, Fenneropenaeus chinensis. Sex. Dev. 6, 267–278. doi: 10.1159/000341377
Li, S., Li, F., Yu, K., and Xiang, J. (2018). Identification and characterization of a doublesex gene which regulates the expression of insulin-like androgenic gland hormone in Fenneropenaeus chinensis. Gene 649, 1–7. doi: 10.1016/j.gene.2018.01.043
Li, X., Meng, X., Kong, J., Luo, K., Luan, S., and Cao, B. (2013). Identification, cloning and characterization of an extracellular signal-regulated kinase (ERK) from Chinese shrimp, Fenneropenaeus chinensis. Fish Shellfish Immunol. 35, 1882–1890. doi: 10.1016/j.fsi.2013.09.021
Liao, J., Wan, H., Sun, Y., Zhang, Z., and Wang, Y. (2020). Characterization of gonad differentially expressed SoxB2 genes in mud crab Scylla paramamosain. Gene 740:144507. doi: 10.1016/j.gene.2020.144507
Lin, J., Yuan, Y., Shi, X., Fang, S., Zhang, Y., Guan, M., et al. (2020). Molecular cloning, characterization and expression profiles of a SoxB2 gene related to gonadal development in mud crab (Scylla paramamosain). Invertebr. Reprod. Dev. 62, 1–11.
Liu, A., Liu, J., Liu, F., Huang, Y., Wang, G., and Ye, H. (2018). Crustacean female sex hormone from the mud crab Scylla paramamosain is highly expressed in prepubertal males and inhibits the development of androgenic gland. Front. Physiol. 9:924. doi: 10.3389/fphys.2018.00924
Liu, C., Jia, X., Zou, Z., Wang, X., Wang, Y., and Zhang, Z. (2018). VIH from the mud crab is specifically expressed in the eyestalk and potentially regulated by transactivator of Sox9/Oct4/Oct1. Gen. Comp. Endocrinol. 255, 1–11. doi: 10.1016/j.ygcen.2017.09.018
Liu, X., Luo, B.-Y., Feng, J.-B., Zhou, L.-X., Ma, K.-Y., and Qiu, G.-F. (2019). Identification and profiling of microRNAs during gonadal development in the giant freshwater prawn Macrobrachium rosenbergii. Sci. Rep. 9:2406.
Liu, Y., Hui, M., Cui, Z., Luo, D., Song, C., Li, Y., et al. (2015). Comparative transcriptome analysis reveals sex-biased gene expression in Juvenile Chinese Mitten Crab Eriocheir sinensis. PLoS One 10:e0133068. doi: 10.1371/journal.pone.0133068
Liu, Z., Ma, K., and Qiu, G. (2019). Identification of an insulin-like androgenic gland hormone transcript highly expressed in male precociousness of the Chinese mitten crab Eriocheir sinensis. Aquac. Res. 50, 304–311. doi: 10.1111/are.13899
Liu, Z.-Q., Jiang, X.-H., Qi, H.-Y., Xiong, L.-W., and Qiu, G.-F. (2016). A novel SoxB2 gene is required for maturation of sperm nucleus during spermiogenesis in the Chinese mitten crab, Eriocheir sinensis. Sci. Rep. 6:32139.
López-Cuadros, I., García-Gasca, A., Gomez-Anduro, G., Escobedo-Fregoso, C., Llera-Herrera, R. A., and Ibarra, A. M. (2018). Isolation of the sex-determining gene Sex-lethal (Sxl) in Penaeus (Litopenaeus) vannamei (Boone, 1931) and characterization of its embryogenic, gametogenic, and tissue-specific expression. Gene 668, 33–47. doi: 10.1016/j.gene.2018.05.024
Lu, B., Jiang, Q., Liu, A., Huang, H., and Ye, H. (2020). Stimulatory roles of epidermal growth factor receptor (EGFR) in ovarian development of mud crab Scylla paramamosain. Gen. Comp. Endocrinol. 299:113616. doi: 10.1016/j.ygcen.2020.113616
Luo, D., Liu, Y., Hui, M., Song, C., Liu, H., and Cui, Z. (2017). Molecular characterization and expression profiles of four transformer-2 isoforms in the Chinese mitten crab Eriocheir sinensis. Chinese J. Oceanol. Limnol. 35, 782–791. doi: 10.1007/s00343-017-6071-z
Ma, A., Wang, Y., Zou, Z., Fu, M., Lin, P., and Zhang, Z. (2012). Erk2 in ovarian development of green mud crab Scylla paramamosain. DNA Cell Biol. 31, 1233–1244.
Ma, K., Qiu, G., Feng, J., and Li, J. (2012). Transcriptome analysis of the oriental river prawn, Macrobrachium nipponense using 454 pyrosequencing for discovery of genes and markers. PLoS One 7:e39727. doi: 10.1371/journal.pone.0039727
Ma, K. Y., Chen, J., Liu, Z. Q., and Qiu, G. F. (2016a). Inhibitory effects of RNAi-mediated knockdown of EsDmrt-like gene on testicular development in the Chinese mitten crab Eriocheir sinensis. Aquaculture 463, 217–223. doi: 10.1016/j.aquaculture.2016.06.003
Ma, K. Y., Lin, J. Y., Guo, S. Z., Chen, Y., Li, J. L., and Qiu, G. F. (2013). Molecular characterization and expression analysis of an insulin-like gene from the androgenic gland of the oriental river prawn, Macrobrachium nipponense. Gen. Comp. Endocrinol. 185, 90–96. doi: 10.1016/j.ygcen.2013.01.018
Ma, K. Y., Liu, Z. Q., Lin, J. Y., Li, J. L., and Qiu, G. F. (2016b). Molecular characterization of a novel ovary-specific gene fem-1 homolog from the oriental river prawn, Macrobrachium nipponense. Gene 575, 244–252. doi: 10.1016/j.gene.2015.08.070
Ma, M., Bors, E. K., Dickinson, E. S., Kwiatkowski, M. A., Sousa, G. L., Henry, R. P., et al. (2009). Characterization of the Carcinus maenas neuropeptidome by mass spectrometry and functional genomics. Gen. Comp. Endocrinol. 161, 320–334. doi: 10.1016/j.ygcen.2009.01.015
Ma, M., Chen, R., Sousa, G. L., Bors, E. K., Kwiatkowski, M. A., Goiney, C. C., et al. (2008). Mass spectral characterization of peptide transmitters/hormones in the nervous system and neuroendocrine organs of the American lobster Homarus americanus. Gen. Comp. Endocrinol. 156, 395–409. doi: 10.1016/j.ygcen.2008.01.009
Manor, R., Weil, S., Oren, S., Glazer, L., Aflalo, E. D., Ventura, T., et al. (2007). Insulin and gender: an insulin-like gene expressed exclusively in the androgenic gland of the male crayfish. Gen. Comp. Endocrinol. 150, 326–336. doi: 10.1016/j.ygcen.2006.09.006
Mareddy, V. R., Rosen, O., Thaggard, H. B., Manor, R., Kuballa, A. V, Aflalo, E. D., et al. (2011). Isolation and characterization of the complete cDNA sequence encoding a putative insulin-like peptide from the androgenic gland of Penaeus monodon. Aquaculture 318, 364–370. doi: 10.1016/j.aquaculture.2011.05.027
Mehra, A., Gaudet, J., Heck, L., Kuwabara, P. E., and Spence, A. M. (1999). Negative regulation of male development in Caenorhabditis elegans by a protein–protein interaction between TRA-2A and FEM-3. Genes Dev. 13, 1453–1463. doi: 10.1101/gad.13.11.1453
Meng, X., Zhang, M., Gao, B., Lv, J., Li, J., and Liu, P. (2020). Integrative proteomic and microRNA analysis: insights into mechanisms of eyestalk ablation-induced ovarian maturation in the swimming crab Portunus trituberculatus. Front. Endocrinol. 11:533. doi: 10.3389/fendo.2020.00533
Meng, X., Zhang, X., Li, J., and Liu, P. (2018). Identification and comparative profiling of ovarian and testicular microRNAs in the swimming crab Portunus trituberculatus. Gene 640, 6–13. doi: 10.1016/j.gene.2017.10.026
Mlinarec, J., Porupski, I., Maguire, I., and Klobučar, G. (2016). Comparative karyotype investigations in the white-clawed crayfish Austropotamobius pallipes (Lereboullet, 1858) species complex and stone crayfish A. torrentium (Schrank, 1803) (Decapoda: Astacidae). J. Crustac. Biol. 36, 87–93.
Mohn, F., Sienski, G., Handler, D., and Brennecke, J. (2014). The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157, 1364–1379. doi: 10.1016/j.cell.2014.04.031
Mullon, C., Pomiankowski, A., and Reuter, M. (2012). Molecular evolution of Drosophila Sex-lethal and related sex determining genes. BMC Evol. Biol. 12:5. doi: 10.1186/1471-2148-12-5
Nagaraju, G. P. C. (2011). Reproductive regulators in decapod crustaceans: an overview. J. Exp. Biol. 214, 3–16. doi: 10.1242/jeb.047183
Nakkrasae, L.-I., and Damrongphol, P. (2007). A vasa-like gene in the giant freshwater prawn, Macrobrachium rosenbergii. Mol. Reprod. Dev. 74, 835–842. doi: 10.1002/mrd.20680
Nässel, D. R. (2002). Neuropeptides in the nervous system of Drosophila and other insects: multiple roles as neuromodulators and neurohormones. Prog. Neurobiol. 68, 1–84. doi: 10.1016/s0301-0082(02)00057-6
Nguyen, T. V., Cummins, S. F., Elizur, A., and Ventura, T. (2016). Transcriptomic characterization and curation of candidate neuropeptides regulating reproduction in the eyestalk ganglia of the Australian crayfish, Cherax quadricarinatus. Sci. Rep. 6:38658.
Parnes, S., Khalaila, I., Hulata, G., and Sagi, A. (2003). Sex determination in crayfish: Are intersex Cherax quadricarinatus (Decapoda, Parastacidae) genetically females? Genet. Res. 82, 107–116. doi: 10.1017/s0016672303006372
Peng, J., Wei, P., Zhang, B., Zhao, Y., Zeng, D., Chen, X., et al. (2015). Gonadal transcriptomic analysis and differentially expressed genes in the testis and ovary of the Pacific white shrimp (Litopenaeus vannamei). BMC Genomics 16:1006. doi: 10.1186/s12864-015-2219-4
Phinyo, M., Visudtiphole, V., Roytrakul, S., and Phaonakrop, N. (2013). Characterization and expression of cell division cycle 2 (Cdc2) mRNA and protein during ovarian development of the giant tiger shrimp Penaeus monodon. Gen. Comp. Endocrinol. 193, 103–111. doi: 10.1016/j.ygcen.2013.07.012
Ponza, P., Yocawibun, P., Sittikankaew, K., and Hiransuchalert, R. (2011). Molecular cloning and expression analysis of the mitogen-activating Protein Kinase 1 (MAPK1) gene and protein during ovarian development of the giant tiger shrimp Penaeus monodon. Mol. Reprod. Dev. 360, 347–360. doi: 10.1002/mrd.21310
Policar, T., and Kozák, P. (2015). “Production and culture of crayfish,” in Crayfish Biology and Culture, eds P. Kozák, Z. Ïuriš, A. Petrusek, M. Buřič, I. Horká, and A. Kouba et al. (Vodňany: University of South Bohemia in České Budějovice), 556.
Qiao, H., Jiang, S., Xiong, Y., Fu, H., Zhang, W., Wang, Y., et al. (2019). Integrated analysis of differentially expressed microRNAs and mRNAs to screen miRNAs and genes related to reproduction in Macrobrachium nipponense. 3 Biotech 9:327.
Qiu, G. F., and Liu, P. (2009). On the role of Cdc2 kinase during meiotic maturation of oocyte in the Chinese mitten crab, Eriocheir sinensis. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 152, 243–248. doi: 10.1016/j.cbpb.2008.12.004
Qiu, L., Zhao, C., Wang, P., Fan, S., Yan, L., Xie, B., et al. (2018). Genomic structure, expression, and functional characterization of checkpoint kinase 1 from Penaeus monodon. PLoS One 13:e0198036. doi: 10.1371/journal.pone.0198036
Rahman, N. M. A., Fu, H., Qiao, H., Jin, S., Bai, H., Zhang, W., et al. (2016). Molecular cloning and expression analysis of Fem1b from oriental river prawn Macrobrachium nipponense. Genet. Mol. Res. 15: gmr.15027950 doi: 10.4238/gmr.15027950
Rastetter, R. H., Smith, C. A., and Wilhelm, D. (2015). The role of non-coding RNAs in male sex determination and differentiation. Reproduction 150, 93–107.
Rinn, J. L., and Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166.
Robinson, N. A., Gopikrishna, G., Baranski, M., Katneni, V. K., Shekhar, M. S., Shanmugakarthik, J., et al. (2014). QTL for white spot syndrome virus resistance and the sex-determining locus in the Indian black tiger shrimp (Penaeus monodon). BMC Genomics 15:732. doi: 10.1186/1471-2164-15-731
Rodgers, L. J., Saoud, P. I., and Rouse, D. B. (2006). The effects of monosex culture and stocking density on survival, growth and yield of redclaw crayfish (Cherax quadricarinatus) in earthen ponds. Aquaculture 259, 164–168. doi: 10.1016/j.aquaculture.2005.11.056
Rodríguez, E. M., Greco, L. S. L., Medesani, D. A., Laufer, H., and Fingerman, M. (2002). Effect of methyl farnesoate, alone and in combination with other hormones, on ovarian growth of the red swamp crayfish, Procambarus clarkii, during vitellogenesis. Gen. Comp. Endocrinol. 125, 34–40. doi: 10.1006/gcen.2001.7724
Rodríguez, E. M., Medesani, D. A., and Fingerman, M. (2007). Endocrine disruption in crustaceans due to pollutants: a review. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 146, 661–671. doi: 10.1016/j.cbpa.2006.04.030
Rosen, O., Manor, R., Weil, S., Gafni, O., Linial, A., Aflalo, E. D., et al. (2010). A sexual shift induced by silencing of a single insulin-like gene in crayfish: ovarian upregulation and testicular degeneration. PLoS One 5:e15281. doi: 10.1371/journal.pone.0015281
Rosen, O., Weil, S., Manor, R., Roth, Z., Khalaila, I., and Sagi, A. (2013). A crayfish insulin-like-binding protein another piece in the androgenic gland insulin-like hormone puzzle is revealed. J. Biol. Chem. 288, 22289–22298.
Rotllant, G., Wade, N. M., Arnold, S. J., Coman, G. J., Preston, N. P., and Glencross, B. D. (2015). Identification of genes involved in reproduction and lipid pathway metabolism in wild and domesticated shrimps. Mar. Genomics 22, 55–61. doi: 10.1016/j.margen.2015.04.001
Saetan, U., and Chotigeat, W. (2019). Identification of ovarian miRNAs in banana shrimp (Fenneropenaeus merguiensis) during ovarian development between non-vitellogenic and vitellogenic shrimp. Aquaculture 511:734240. doi: 10.1016/j.aquaculture.2019.734240
Sagi, A., and Aflalo, E. D. (2005). The androgenic gland and monosex culture of freshwater prawn Macrobrachium rosenbergii (De Man): a biotechnological perspective. Aquac. Res. 36, 231–237. doi: 10.1111/j.1365-2109.2005.01238.x
Saikrithi, P., Balasubramanian, C. P., Otta, S. K., and Tomy, S. (2019). Characterization and expression profile of farnesoic acid O-methyltransferase gene from Indian white shrimp, Penaeus indicus. Comp. Biochem. Physiol. B 232, 79–86. doi: 10.1016/j.cbpb.2019.03.004
Salvemini, M., Robertson, M., Aronson, B., Atkinson, P., Polito, L. C., and Saccone, G. (2009). Ceratitis capitata transformer-2 gene is required to establish and maintain the autoregulation of Cctra, the master gene for female sex determination. Int. J. Dev. Biol. 53, 109–120. doi: 10.1387/ijdb.082681ms
Sb, J., Ht, F., Sf, J., Yw, X., H, Q., Wy, Z., et al. (2015). Identification of androgenic gland microRNA and their target genes to discover sex-related microRNA in the oriental river prawn, Macrobrachium nipponense. Genet. Mol. Res. 14, 18396–18406. doi: 10.4238/2015.december.23.27
Schartl, M., Schories, S., Wakamatsu, Y., Nagao, Y., Hashimoto, H., Bertin, C., et al. (2018). Sox5 is involved in germ-cell regulation and sex determination in medaka following co-option of nested transposable elements. BMC Biol. 16:16. doi: 10.1186/s12915-018-0485-8
Scherr, M., Venturini, L., Battmer, K., Schaller-Schoenitz, M., Schaefer, D., Dallmann, I., et al. (2007). Lentivirus-mediated antagomir expression for specific inhibition of miRNA function. Nucleic Acids Res. 35:e149. doi: 10.1093/nar/gkm971
Schnackenberg, B. J., Palazzo, R. E., and Marzluff, W. F. (2007). Cyclin E/Cdk2 is required for sperm maturation, but not DNA replication, in early sea urchin embryos. Genesis 45, 282–291. doi: 10.1002/dvg.20291
Sharabi, O., Manor, R., Weil, S., Aflalo, E. D., Lezer, Y., Levy, T., et al. (2016). Identification and characterization of an insulin-like receptor involved in crustacean reproduction. Endocrinology 157, 928–941. doi: 10.1210/en.2015-1391
Sharabi, O., Ventura, T., Manor, R., Aflalo, E. D., and Sagi, A. (2013). Epidermal growth factor receptor in the prawn Macrobrachium rosenbergii: function and putative signaling cascade. Endocrinology 154, 3188–3196. doi: 10.1210/en.2013-1259
Shen, H., Hu, Y., and Zhou, X. (2014). Sex-lethal gene of the Chinese mitten crab Eriocheir sinensis: cDNA cloning, induction by eyestalk ablation, and expression of two splice variants in males and females. Dev. Genes Evol. 224, 97–105. doi: 10.1007/s00427-014-0467-y
Shi, L., Han, S., Fei, J., Zhang, L., Ray, J. W., Wang, W., et al. (2019). Molecular characterization and functional study of insulin-like androgenic gland hormone gene in the red swamp crayfish, Procambarus clarkii. Genes 10:645. doi: 10.3390/genes10090645
Shi, X., Lu, J., Wu, Q., Waiho, K., Aweya, J. J., Fazhan, H., et al. (2019). Comparative analysis of growth performance between female and male mud crab Scylla paramamosain crablets: evidences from a four-month successive growth experiment. Aquaculture 505, 351–362. doi: 10.1016/j.aquaculture.2019.02.062
Shi, X., Waiho, K., Li, X., Ikhwanuddin, M., Miao, G., Lin, F., et al. (2018). Female-specific SNP markers provide insights into a WZ/ZZ sex determination system for mud crabs Scylla paramamosain, S. tranquebarica and S. serrata with a rapid method for genetic sex identification. BMC Genomics 19:981. doi: 10.1186/s12864-018-5380-8
Shu, L., Yang, Y., Huang, H., and Ye, H. (2016). A bone morphogenetic protein ligand and receptors in mud crab: a potential role in the ovarian development. Mol. Cell. Endocrinol. 434, 99–107. doi: 10.1016/j.mce.2016.06.023
Siangcham, T., Tinikul, Y., Poljaroen, J., Sroyraya, M., Changklungmoa, N., Phoungpetchara, I., et al. (2013). The effects of serotonin, dopamine, gonadotropin-releasing hormones, and corazonin, on the androgenic gland of the giant freshwater prawn, Macrobrachium rosenbergii. Gen. Comp. Endocrinol. 193, 10–18. doi: 10.1016/j.ygcen.2013.06.028
Sokolow, S. H., Lafferty, K. D., and Kuris, A. M. (2014). Regulation of laboratory populations of snails (Biomphalaria and Bulinus spp.) by river prawns, Macrobrachium spp. (Decapoda, Palaemonidae): implications for control of schistosomiasis. Acta Trop. 132, 64–74. doi: 10.1016/j.actatropica.2013.12.013
Song, C., Cui, Z., Hui, M., Liu, Y., and Li, Y. (2015). Molecular characterization and expression profile of three Fem-1 genes in Eriocheir sinensis provide a new insight into crab sex-determining mechanism. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 189, 6–14. doi: 10.1016/j.cbpb.2015.07.003
Song, Y.-N., Shi, L.-L., Liu, Z.-Q., and Qiu, G.-F. (2014). Global analysis of the ovarian microRNA transcriptome: implication for miR-2 and miR-133 regulation of oocyte meiosis in the Chinese mitten crab, Eriocheir sinensis (Crustacea:Decapoda). BMC Genomics 15:547. doi: 10.1186/1471-2164-15-547
Sroyraya, M., Chotwiwatthanakun, C., Stewart, M. J., Soonklang, N., Kornthong, N., Phoungpetchara, I., et al. (2010). Bilateral eyestalk ablation of the blue swimmer crab, Portunus pelagicus, produces hypertrophy of the androgenic gland and an increase of cells producing insulin-like androgenic gland hormone. Tissue Cell 42, 293–300. doi: 10.1016/j.tice.2010.07.003
Staelens, J., Rombaut, D., Vercauteren, I., Argue, B., Benzie, J., and Vuylsteke, M. (2008). High-density linkage maps and sex-linked markers for the black tiger shrimp (Penaeus monodon). Genetics 179, 917–925. doi: 10.1534/genetics.107.080150
Suwansa-ard, S., Thongbuakaew, T., Wang, T., Zhao, M., Elizur, A., Hanna, P. J., et al. (2015). In silico neuropeptidome of female Macrobrachium rosenbergii based on transcriptome and peptide mining of ryestalk, central nervous system and ovary. PLoS One 10:e0123848. doi: 10.1371/journal.pone.0123848
Tan, K., Li, Y., Zhou, M., and Wang, W. (2020a). siRNA knockdown of MrIR induces sex reversal in Macrobrachium rosenbergii. Aquaculture 523:735172. doi: 10.1016/j.aquaculture.2020.735172
Tan, K., Zhou, M., Jiang, H., Jiang, D., Li, Y., and Wang, W. (2020b). siRNA-mediated MrIAG silencing induces sex reversal in Macrobrachium rosenbergii. Mar. Biotechnol. 22, 456–466. doi: 10.1007/s10126-020-09965-4
Tanaka, Y., Hua, Y.-J., Roller, L., and Tanaka, S. (2002). Corazonin reduces the spinning rate in the silkworm, Bombyx mori. J. Insect Physiol. 48, 707–714. doi: 10.1016/s0022-1910(02)00094-x
Thongbuakaew, T., Suwansa-ard, S., Sretarugsa, P., Sobhon, P., and Cummins, S. F. (2019). Identification and characterization of a crustacean female sex hormone in the giant freshwater prawn, Macrobrachium rosenbergii. Aquaculture 507, 56–68. doi: 10.1016/j.aquaculture.2019.04.002
Tinikul, Y., Mercier, A. J., Soonklang, N., and Sobhon, P. (2008). Changes in the levels of serotonin and dopamine in the central nervous system and ovary, and their possible roles in the ovarian development in the giant freshwater prawn, Macrobrachium rosenbergii. Gen. Comp. Endocrinol. 158, 250–258. doi: 10.1016/j.ygcen.2008.07.009
Tinikul, Y., Poljaroen, J., Kornthong, N., Chotwiwatthanakun, C., Anuracpreeda, P., Poomtong, T., et al. (2011). Distribution and changes of serotonin and dopamine levels in the central nervous system and ovary of the Pacific white shrimp, Litopenaeus vannamei, during ovarian maturation cycle. Cell Tissue Res. 345, 103–124. doi: 10.1007/s00441-011-1176-8
Toullec, J. -Y., Corre, E., Mandon, P., Gonzalez-Aravena, M., Ollivaux, C., and Lee, C. -Y. (2017). Characterization of the neuropeptidome of a southern ocean decapod, the antarctic shrimp Chorismus antarcticus: focusing on a new decapod ITP-like peptide belonging to the CHH peptide family. Gen. Comp. Endocrinol. 252, 60–78. doi: 10.1016/j.ygcen.2017.07.015
Toyota, K., Kato, Y., Sato, M., Sugiura, N., Miyagawa, S., Miyakawa, H., et al. (2013). Molecular cloning of doublesex genes of four cladocera (water flea) species. BMC Genomics 14:239. doi: 10.1186/1471-2164-14-239
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938. doi: 10.1016/j.molcel.2010.08.011
Tsai, M. -C., Manor, O., Wan, Y., Mosammaparast, N., Wang, J. K., Lan, F., et al. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. doi: 10.1126/science.1192002
Veenstra, J. A. (1989). Isolation and structure of corazonin, a cardioactive peptide from the American cockroach. FEBS Lett. 250, 231–234. doi: 10.1016/0014-5793(89)80727-6
Veenstra, J. A. (2015). The power of next-generation sequencing as illustrated by the neuropeptidome of the crayfish Procambarus clarkii. Gen. Comp. Endocrinol. 224, 84–95. doi: 10.1016/j.ygcen.2015.06.013
Ventura, T., Chandler, J. C., Nguyen, T. V, Hyde, C. J., Elizur, A., Fitzgibbon, Q. P., et al. (2020). Multi-tissue transcriptome analysis identifies key sexual development-related genes of the ornate spiny lobster (Panulirus ornatus). Genes 11:1150. doi: 10.3390/genes11101150
Ventura, T., Cummins, S. F., Fitzgibbon, Q., Battaglene, S., and Elizur, A. (2014). Analysis of the central nervous system transcriptome of the eastern rock lobster Sagmariasus verreauxi reveals its putative neuropeptidome. PLoS One 9:e97323. doi: 10.1371/journal.pone.0097323
Ventura, T., Manor, R., Aflalo, E. D., Weil, S., Raviv, S., Glazer, L., et al. (2009). Temporal silencing of an androgenic gland-specific insulin-like gene affecting phenotypical gender differences and spermatogenesis. Endocrinology 150, 1278–1286. doi: 10.1210/en.2008-0906
Ventura, T., Manor, R., Aflalo, E. D., Weil, S., Rosen, O., and Sagi, A. (2012). Timing sexual differentiation: full functional sex reversal achieved through silencing of a single insulin-like gene in the prawn, Macrobrachium rosenbergii. Biol. Reprod. 86:90.
Ventura, T., and Sagi, A. (2012). The insulin-like androgenic gland hormone in crustaceans: from a single gene silencing to a wide array of sexual manipulation-based biotechnologies. Biotechnol. Adv. 30, 1543–1550. doi: 10.1016/j.biotechadv.2012.04.008
Ventura-Holman, T., Lu, D., Si, X., Izevbigie, E. B., and Maher, J. F. (2003). The Fem1c genes: conserved members of the Fem1 gene family in vertebrates. Gene 314, 133–139. doi: 10.1016/s0378-1119(03)00712-1
Waiho, K., Fazhan, H., Zhang, Y., Li, S., Zhang, Y., Zheng, H., et al. (2020). Comparative profiling of ovarian and testicular piRNAs in the mud crab Scylla paramamosain. Genomics 112, 323–331. doi: 10.1016/j.ygeno.2019.02.012
Waiho, K., Fazhan, H., Zhang, Y., Zhang, Y., Li, S., Zheng, H., et al. (2019a). Gonadal microRNA expression profiles and their potential role in sex differentiation and gonadal maturation of mud crab Scylla paramamosain. Mar. Biotechnol. 21, 320–334. doi: 10.1007/s10126-019-09882-1
Waiho, K., Shi, X., Fazhan, H., Li, S., Zhang, Y., Zheng, H., et al. (2019b). High-density genetic linkage maps provide novel insights into ZW/ZZ sex determination system and growth performance in mud crab (Scylla paramamosain). Front. Genet. 10:298. doi: 10.3389/fgene.2019.00298
Wang, Y., Chen, Y., Han, K., Zou, Z., and Zhang, Z. (2012). A vasa gene from green mud crab Scylla paramamosain and its expression during gonadal development and gametogenesis. Mol. Biol. Rep. 39, 4327–4335. doi: 10.1007/s11033-011-1220-5
Wang, Y., Jin, S., Fu, H., Qiao, H., Sun, S., Zhang, W., et al. (2019). Identification and characterization of the Dmrt11e gene in the oriental river prawn Macrobrachium nipponense. Int. J. Mol. Sci. 20:1734. doi: 10.3390/ijms20071734
Winter, J., Jung, S., Keller, S., Gregory, R. I., and Diederichs, S. (2009). Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 11, 228–234. doi: 10.1038/ncb0309-228
Wu, Q., Shi, X., Fang, S., Xie, Z., Guan, M., Li, S., et al. (2019). Different biochemical composition and nutritional value attribute to salinity and rearing period in male and female mud crab Scylla paramamosain. Aquaculture 513:734417. doi: 10.1016/j.aquaculture.2019.734417
Xiaowei, W., Jie, G., Ziping, Z., Zhihua, Z., Xiwei, J., Peng, L., et al. (2013). The full length cDNA cloning and expression of 14-3-3.ZETA. gene in green mud crab (Scylla paramamosain). J. Shanghai Ocean Univ. 22, 496–501.
Xie, X., Zhu, D., Li, Y., Qiu, X., Cui, X., and Tang, J. (2015). Hemolymph levels of methyl farnesoate during ovarian development of the swimming crab Portunus trituberculatus, and its relation to transcript levels of HMG-CoA reductase and farnesoic acid O-methyltransferase. Biol. Bull. 228, 118–124. doi: 10.1086/bblv228n2p118
Xu, Y. -R., Wang, G. -Y., Zhou, Y. -C., and Yang, W. -X. (2018). The characterization and potential roles of bone morphogenetic protein 7 during spermatogenesis in Chinese mitten crab Eriocheir sinensis. Gene 673, 119–129. doi: 10.1016/j.gene.2018.06.020
Yang, G., Lu, Z., Qin, Z., Zhao, L., Pan, G., Shen, H., et al. (2020). Insight into the regulatory relationships between the insulin-like androgenic gland hormone gene and the insulin-llike androgenic gland hormone-binding protein gene in giant freshwater prawns (Macrobrachium rosenbergii). Int. J. Mol. Sci. 21:4207. doi: 10.3390/ijms21124207
Yang, X., Ikhwanuddin, M., Li, X., Lin, F., Wu, Q., Zhang, Y., et al. (2018). Comparative transcriptome analysis provides insights into differentially expressed genes and long non-Coding RNAs between ovary and testis of the mud crab (Scylla paramamosain). Mar. Biotechnol. 20, 20–34. doi: 10.1007/s10126-017-9784-2
Yu, Y. Q., Ma, W. M., Zeng, Q. G., Qian, Y. Q., Yang, J. S., and Yang, W. J. (2014). Molecular cloning and sexually dimorphic expression of two Dmrt genes in the giant freshwater prawn, Macrobrachium rosenbergii. Agric. Res. 3, 181–191. doi: 10.1007/s40003-014-0106-x
Zeng, Y. (2006). Principles of micro-RNA production and maturation. Oncogene 25, 6156–6162. doi: 10.1038/sj.onc.1209908
Zhang, E. -F., and Qiu, G.-F. (2010). A novel Dmrt gene is specifically expressed in the testis of Chinese mitten crab, Eriocheir sinensis. Dev. Genes Evol. 220, 151–159. doi: 10.1007/s00427-010-0336-2
Zhang, L., Zhang, X., Zhang, X., Lu, Y., Li, L., and Cui, S. (2017). MiRNA-143 mediates the proliferative signaling pathway of FSH and regulates estradiol production. J. Endocrinol. 234, 1–14. doi: 10.1530/joe-16-0488
Zhang, Y., Fu, H., Qiao, H., Jin, S., Jiang, S., Xiong, Y., et al. (2013). Molecular cloning and expression analysis of transformer-2 gene during development in Macrobrachium nipponense (de Haan 1849). J. World Aquac. Soc. 44, 338–349. doi: 10.1111/jwas.12039
Zhang, Y., Miao, G., Wu, Q., Lin, F., You, C., Wang, S., et al. (2018). Transcriptome sequencing and molecular markers discovery in the gonads of Portunus sanguinolentus. Sci. Data 5:180131.
Zhang, Y., Qiao, K., Wang, S., Peng, H., Shan, Z., and Wang, K. (2014). Molecular identification of a new androgenic gland-specific insulin-like gene from the mud crab, Scylla paramamosain. Aquaculture 433, 325–334. doi: 10.1016/j.aquaculture.2014.06.033
Zhao, C., Fan, S., and Qiu, L. (2018). Identification of microRNAs and their target genes associated with ovarian development in black tiger shrimp (Penaeus monodon) using high-throughput sequencing. Sci. Rep. 8:11602.
Zheng, J., Cheng, S., Jia, Y., Gu, Z., Li, F., Chi, M., et al. (2019). Molecular identification and expression profiles of four splice variants of Sex-lethal gene in Cherax quadricarinatus. Comp. Biochem. Physiol. B 234, 26–33. doi: 10.1016/j.cbpb.2019.05.002
Zhong, P., Zhou, T., Zhang, Y., Chen, Y., Yi, J., Lin, W., et al. (2019). Potential involvement of a DMRT family member (Mr-Dsx) in the regulation of sexual differentiation and moulting in the giant river prawn Macrobrachium rosenbergii. Aquac. Res. 50, 3037–3049. doi: 10.1111/are.14262
Zhou, M., Jia, X., Wan, H., Wang, S., Zhang, X., Zhang, Z., et al. (2019). miR-34 regulates reproduction by inhibiting the expression of MIH, CHH, EcR, and FAMeT genes in mud crab Scylla paramamosain. Mol. Reprod. Dev. 86, 122–131. doi: 10.1002/mrd.23063
Zhou, M., Jia, X., Wan, H., Wang, S., Zhang, X., Zhang, Z., et al. (2020). miR-9 and miR-263 regulate the key genes of the ERK pathway in the ovary of mud crab Scylla paramamosain. Mar. Biotechnol. 22, 594–606. doi: 10.1007/s10126-020-09981-4
Zhou, Q., Shao, M., Qin, Z., Kyoung, H. K., and Zhang, Z. (2010). Cloning, characterization, and expression analysis of the DEAD-box family genes, Fc-vasa and Fc-PL10a, in Chinese shrimp (Fenneropenaeus chinensis). Chinese J. Oceanol. Limnol. 28, 37–45. doi: 10.1007/s00343-010-9231-y
Keywords: decapods, gametogenesis, sex-related genes/non-coding RNAs, sexual differentiation, regulatory mechanism
Citation: Farhadi A, Cui W, Zheng H, Li S, Zhang Y, Ikhwanuddin M and Ma H (2021) The Regulatory Mechanism of Sexual Development in Decapod Crustaceans. Front. Mar. Sci. 8:679687. doi: 10.3389/fmars.2021.679687
Received: 12 March 2021; Accepted: 12 April 2021;
Published: 29 April 2021.
Edited by:
Shengming Sun, Shanghai Ocean University, ChinaReviewed by:
Huan Wang, Ningbo University, ChinaGuangzhen Jiang, Nanjing Agricultural University, China
Shihao Li, Institute of Oceanology (CAS), China
Copyright © 2021 Farhadi, Cui, Zheng, Li, Zhang, Ikhwanuddin and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyu Ma, bWFoeUBzdHUuZWR1LmNu
 Ardavan Farhadi
Ardavan Farhadi Wenxiao Cui1,2
Wenxiao Cui1,2 Huaiping Zheng
Huaiping Zheng Shengkang Li
Shengkang Li Yueling Zhang
Yueling Zhang Mhd Ikhwanuddin
Mhd Ikhwanuddin Hongyu Ma
Hongyu Ma