
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mar. Sci., 30 June 2021
Sec. Marine Biology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.673620
This article is part of the Research TopicBioluminescence from Land to the Oceans: Its Role in Evolution, Communication, and EcologyView all 9 articles
Bioluminescence—i.e., the emission of visible light by living organisms—is defined as a biochemical reaction involving, at least, a luciferin substrate, an oxygen derivative, and a specialised luciferase enzyme. In some cases, the enzyme and the substrate are durably associated and form a photoprotein. While this terminology is educatively useful to explain bioluminescence, it gives a false idea that all luminous organisms are using identical or homologous molecular tools to achieve light emission. As usually observed in biology, reality is more complex. To date, at least 11 different luciferins have indeed been discovered, and several non-homologous luciferases lato sensu have been identified which, all together, confirms that bioluminescence emerged independently multiple times during the evolution of living organisms. While some phylogenetically related organisms may use non-homologous luciferases (e.g., at least four convergent luciferases are found in Pancrustacea), it has also been observed that phylogenetically distant organisms may use homologous luciferases (e.g., parallel evolution observed in some cnidarians, tunicates and echinoderms that are sharing a homologous luciferase-based system). The evolution of luciferases then appears puzzling. The present review takes stock of the diversity of known “bioluminescent proteins,” their evolution and potential evolutionary origins. A total of 134 luciferase and photoprotein sequences have been investigated (from 75 species and 11 phyla), and our analyses identified 12 distinct types—defined as a group of homologous bioluminescent proteins. The literature review indicated that genes coding for luciferases and photoproteins have potentially emerged as new genes or have been co-opted from ancestral non-luciferase/photoprotein genes. In this latter case, the homologous gene’s co-options may occur independently in phylogenetically distant organisms.
Bioluminescence, i.e., the biochemical production of visible light by living organisms, is a widespread feature in the tree of life. Bioluminescent species have been found, so far, in at least 700 genera belonging to a large variety of evolutionary lineages (Haddock et al., 2010; Widder, 2010; Lau and Oakley, 2020, for review) such as bacteria, dinoflagellates, arthropods, molluscs, annelids, echinoderms, urochordates, or vertebrates (but only in fishes, e.g., Claes and Mallefet, 2009; Davis et al., 2016). A large number of luminous species inhabit marine habitats (Hastings, 1983; Herring, 1987; Haddock et al., 2010; Widder, 2010) and, for example, about three quarters of the individuals observed in the water column have luminescence capability (Martini and Haddock, 2017).
From a molecular perspective, bioluminescence is the product of the oxidation of a luciferin substrate catalysed by a luciferase enzyme. The electronically excited oxyluciferin emits light as it relaxes to the ground state. In some cases, the luciferase and the luciferin are associated in a single unit, the so-called photoprotein. In photoprotein systems, the substrate/enzyme complex may require additional cofactors to be functionally active (Shimomura, 2012). The general luminescence reaction is ubiquitous in all known luminescent organisms, however, this ability to produce light emerged multiple times independently in the tree of life: more than 94 times according to the recent literature (Hastings, 1983; Wilson and Hastings, 1998; Haddock et al., 2010; Davis et al., 2016; Lau and Oakley, 2020). Several luciferins—i.e., at least 11 different molecules identified so far—and luciferases have indeed been described in a large variety of taxa (Herring, 1987; Haddock et al., 2010; Lau and Oakley, 2020, for review). Around 100 different species have been substantially described using biochemical approaches (Supplementary Table 1). While the majority of investigated luminous species use a luciferase/luciferin system (at least 75 species, Supplementary Table 1), photoproteins have been described in around 25 species (i.e., in cnidarians, ctenophores, annelids, molluscs, crustaceans, echinoderms, and fishes, Supplementary Table 1) (Shimomura, 1986, 2008, 2012).
Luciferases are generally considered as “taxon-specific” (Haddock et al., 2010; Shimomura, 2012). Besides, phylogenetically related organisms may sometimes rely on non-homologous enzymes for photogenesis, supporting the convergent evolution of bioluminescence. Therefore, there is no common luminous ancestor for all bioluminescent species. The luminescent systems have different origins, resulting in highly diverse systems involving different molecular actors, different associated morphological and anatomical structures, and different types of control mechanisms (Haddock et al., 2010). It is strongly suggested that the multi-convergent evolution of bioluminescence demonstrates the existence of intense selective pressures in support of the emergence of bioluminescence mechanisms during organism evolution (Haddock et al., 2010). In that view, the acquisition of the ability to emit light could be seen as an “evolutionary easy process” during evolution (Haddock et al., 2010).
Based on the unpredictable emergence of luminescent species throughout evolution, and the necessity for oxygen in luminescence reactions, it has been speculated that bioluminescence might have evolved to eliminate oxygen or reactive oxygen species from the organism (Timmins et al., 2001; Wilson and Hastings, 2013). Bioluminescence would then be derived from defence mechanisms against free-radicals, i.e., coopted from an oxygen detoxifying mechanism to a light-related communication type (Seliger, 1975). Wilson and Hastings (2013) argue that bioluminescence evolved in response to low oxygen levels during the time between the evolutionary emergence of photosynthesis on earth (the so-called “great oxidation event” that occurred around 2 billion years ago) and the Cambrian explosion (around 500–550 million years ago). According to Wilson and Hastings (2013), all bioluminescence systems “consume” oxygen and could therefore be considered primary oxygen detoxification strategies, with light simply considered a secondary by-product. Bioluminescence would have then acquired a different functional role when antioxidant pathways, such as those involving superoxide dismutases and catalases, became widespread with increasing oxygen levels. As stated by Valiadi and Iglesias-Rodriguez (2013), the hypothesis of Wilson and Hastings is mainly based on the bioluminescence systems of bacteria and fireflies, but it is largely plausible for other bioluminescent organisms. In cell cultures, coelenterazine (i.e., the most common luciferin in the marine environment) has been shown to reduce the death of fibroblasts exposed to oxidative stress (Rees et al., 1998). Coelenterazine is detected not only in luminescent organs but is also found in the digestive tract and hepatopancreas of several luminous and non-luminous decapods, cephalopods and fishes (Shimomura, 1987; Mallefet and Shimomura, 1995; Thomson et al., 1997; Rees et al., 1998; Duchatelet et al., 2019). These observations support an anti-oxidative function of this kind of compound and luciferins might then be antioxidant molecules emitting light as a by-product of their “reactive oxygen scavenging chemical activity” (Rees et al., 1998; Haddock et al., 2010). The presence of common light-emitting luciferins in luminous but also is non-luminous organisms (i.e., ecological notion of luciferin reservoir) led to the hypothesis of the “luciferin dietary acquisition” (Haddock et al., 2010; Shimomura, 2012): luminous organisms acquired their luciferin through their food, and those molecules can transit via the food chain (i.e., a predator can retrieve the luciferin produced by its prey) (demonstrated in some species: Barnes et al., 1973; Warner and Case, 1980; Frank et al., 1984; Thomson et al., 1997; Haddock et al., 2001; Mallefet et al., 2020). The same luciferin can then be found in phylogenetically distant organisms (e.g., coelenterazine is found in at least nine phyla). This “oxygen defence” hypothesis has also been adapted for luciferases that might initially be antioxidative enzymes secondarily co-opted in luciferases (Haddock et al., 2001; Wilson and Hastings, 2013). The hypothesis has the advantage to explain the widespread occurrence of bioluminescence in organisms. However, the general idea that luminescence might have evolved to eliminate oxygen stress is over-simplistic and has been disproved by the discovery of several luciferases which are not homologous to antioxidative enzymes but rather derived, i.e., coopted, from unrelated enzymes (Viviani, 2002; Loening et al., 2006; Müller et al., 2009; Delroisse et al., 2017b).
Understanding the evolution of bioluminescence is challenging because various biological processes were shown to be synergically involved in the emergence of bioluminescence (e.g., substrate dietary acquisition, gene cooption, potential horizontal gene transfers, …) (e.g., Viviani, 2002; Loening et al., 2006; Delroisse et al., 2017b; Bessho-Uehara et al., 2020b). Recently, a remarkable example of dietary enzyme acquisition has been described in the predator fish Parapriacanthus for which not only the luciferin was shown to be recovered from the ostracod prey, but also the functionally active luciferase (Bessho-Uehara et al., 2020b).
Luciferase and photoprotein protein sequences were retrieved from NCBI (Supplementary Table 1). The global dataset was analysed using a sequence-similarity-based clustering approach based on BLASTp e-values and using the CLANS software (Frickey and Lupas, 2004). Based on the CLANS clustering, pairwise sequence identity and similarity were calculated from trimmed multiple sequence alignments of each luciferase/photoprotein subset (defined as a group of potentially homologous luciferases/photoproteins) using SIAS web tool.1
Molecular domain prediction was performed using the Hidden Markov Model of Simple Modular Architecture Research Tool (SMART)2 (Figure 1). Molecular weight values (determined by biochemical approaches, not by in silico analyses) were collected from the literature.
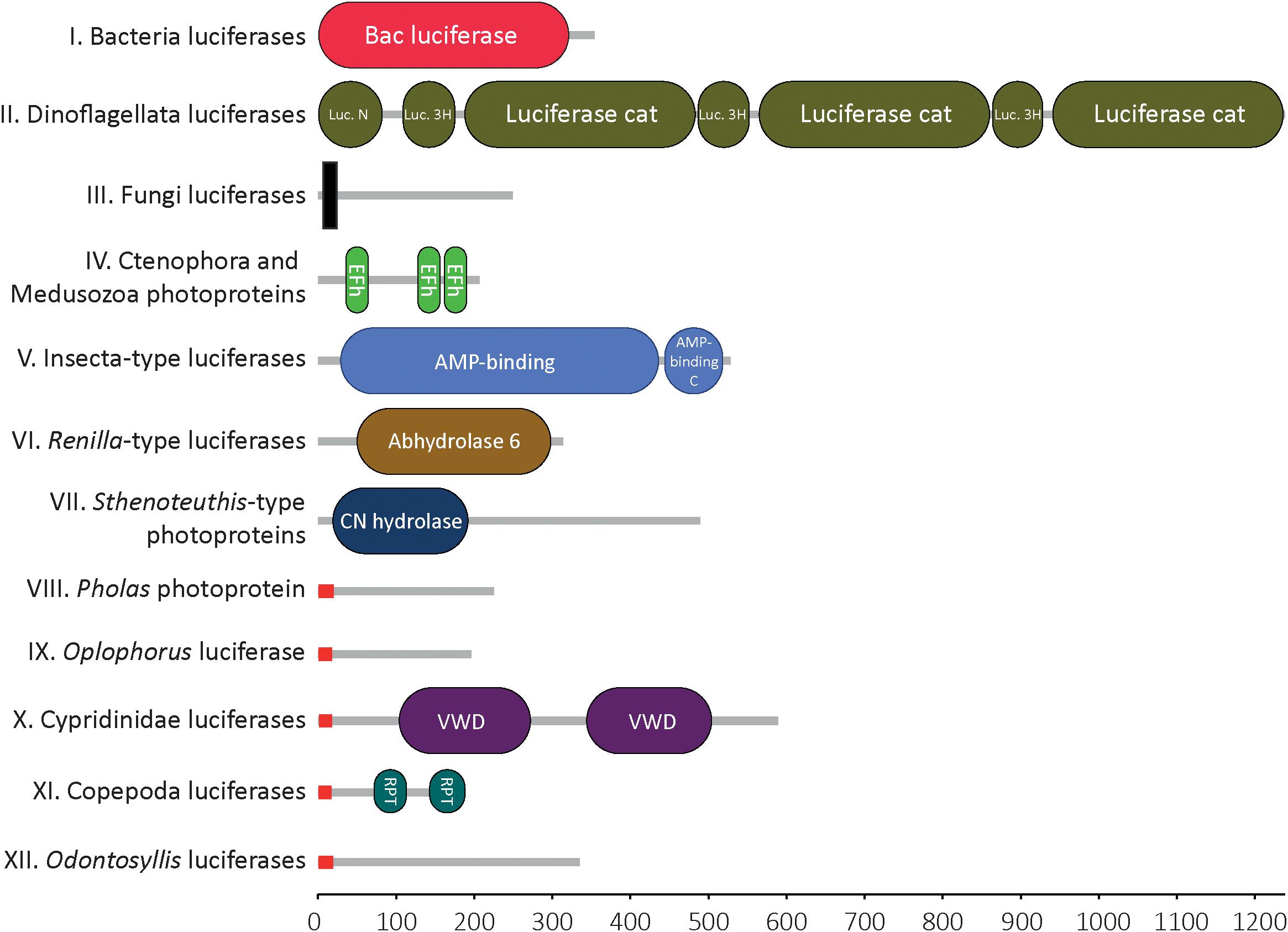
Figure 1. Molecular domains of known bioluminescent proteins. Domain detected within the 12 types of luciferase/photoprotein types using Hidden Markov Model of Simple Modular Architecture Research Tool SMART. For each cluster of luciferases/photoproteins, all available sequences were tested. Only one representative sequence is shown for each cluster. Bacteria-type luciferase: Vibrio fischeri (Q6VFQ4), Dinoflagellata-type luciferase: Pyrocystis lunula (AAL40677), Fungi-type luciferase: Panellus stipticus (BBH43512), Aequora-type photoprotein: Mnemiopsis leidyi (AFK83786), Insecta-type luciferase: Arachnocampa luminosa (AXM90651), Renilla-type luciferase: Renilla reniformis (CAA01908), Sthenoteuthis-type photoprotein: Sthenoteuthis oulalaniensis (BAH89068), Pholas-type photoprotein: Pholas dactylus (CAA10292), Oplophorus-type photoprotein: Oplophorus gracilirostris (BAB13776), Ostracoda-type luciferase: Vargula hilgendorfii (AAB86460), Copepoda-type luciferase: Metridia pacifica (BAY00656), Odontosyllis-type luciferase: Odontosyllis octodentata (BBG43629). The X-axis represents the protein size (i.e., number of amino acids). Signal peptides (in red) were specifically detected in all known secreted luciferases. Transmembrane domain (in black) was detected in the Fungi luciferases, only. Legend: N, N-terminal; Luciferase_3H, luciferase helical bundle domain; Cat, catalytic; Efh, EF-hand domain; C, C-terminal; CN hydrolase, Carbon-nitrogen hydrolase; VWD, Von Willebrand factor type D domain; RPT, internal repeat within the sequence (this does not correspond to a molecular domain per se).
All figures presented in the manuscript were edited using Adobe Illustrator 2020 (v24.3.0).
To illustrate the homology status of known bioluminescent proteins, luciferase and photoprotein sequences were retrieved from public databases and analysed using a sequence-similarity-based clustering approach based on BLASTp E-values. Sequence similarity searching is typically performed using BLAST. It is the most widely used and most reliable strategy for characterising newly determined sequences. Sequence similarity searches can identify “homologous” proteins by detecting excess similarity corresponding to the statistically significant similarity that reflects common ancestry (Pearson, 2013).
In total, 134 sequences of luciferases and photoproteins (from 75 species and 11 phyla), were collected in the context of the present review. In parallel, we generated the list of known bioluminescent proteins, including those for which no sequences are available, yet (Supplementary Table 1). To our knowledge, it is the most complete repertoire of known luciferases and photoproteins (however, the idea of generating an exhaustive set is certainly utopian).
Our analyses highlighted the presence of 12 distinct types of bioluminescent proteins, defined as clusters of homologous bioluminescent proteins based on an E-value threshold of 1e–10 (Figure 2). In the following sections, we will discuss each of these photoprotein/luciferase clusters from an evolutionary perspective.
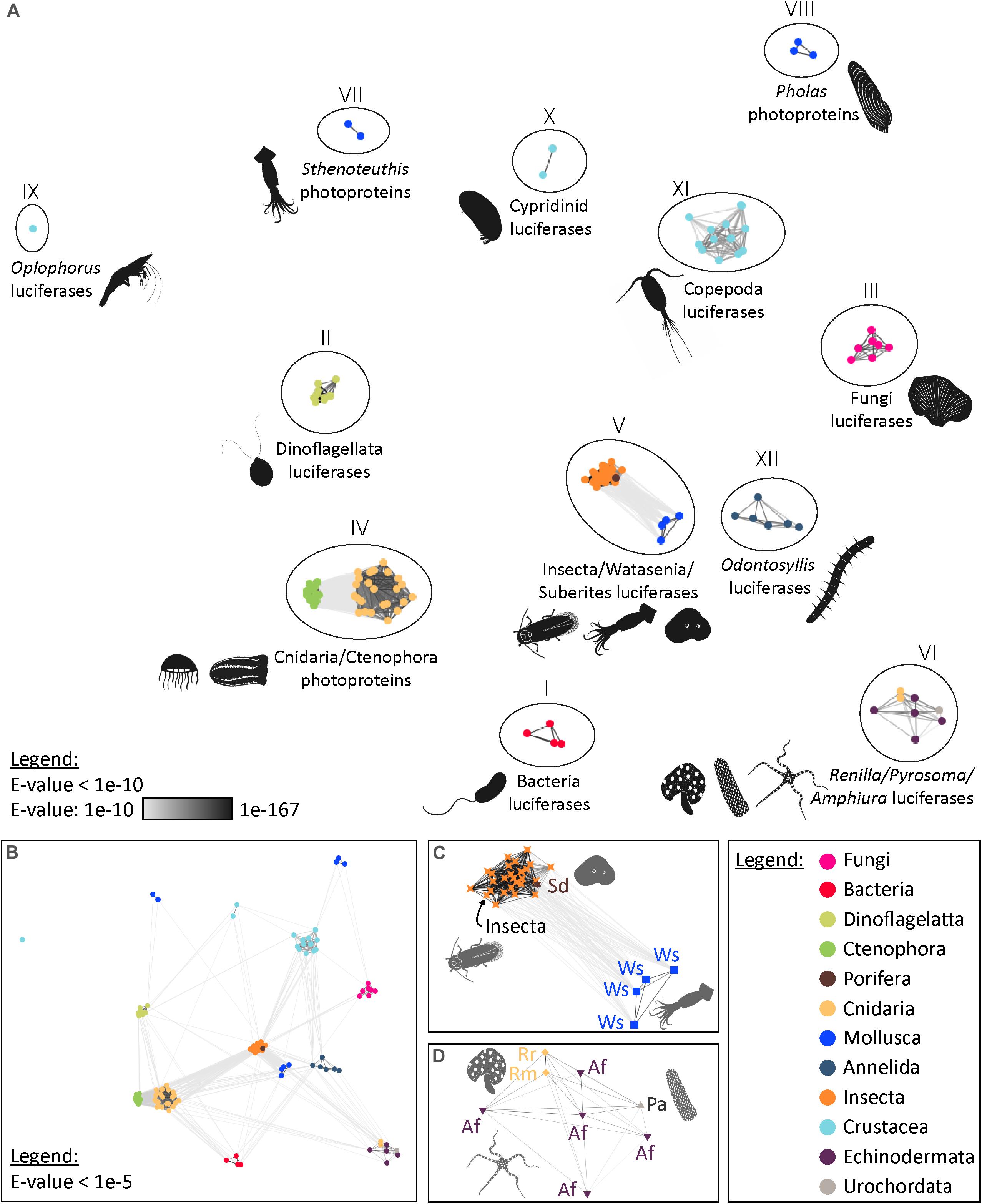
Figure 2. Visualisation of the similarity of known bioluminescent proteins. (A) Sequence-similarity-based clustering approach based on BLASTp e-values (CLANS software) using known luciferases and photoproteins (E-value threshold of 1e–10). (B) Same analysis with an E-value threshold of 1e–5. (C) Focus on the Insecta-type luciferase cluster (Group V) that also contains luciferases of Watasenia scintillans and Suberites domuncula (E-value threshold of 1e–10). (D) Focus on the Renilla-type luciferase cluster (Group VI) containing Pyrosoma atlanticum and Amphiura filiformis candidate luciferases (E-value threshold of 1e–10). The names and accession numbers of the sequences are referenced in the Supplementary Table 1.
Bioluminescent bacteria (i.e., terrestrial or aquatic) are all distributed into seven genera belonging to three families of Gammaproteobacteria: Vibrionaceae, Shewanellaceae and Enterobacteriaceae (Brodl et al., 2018), and at least 28 luminous marine bacteria have been described (e.g., Vibrio harveyi, Photobacterium sp.) (Tanet et al., 2020). Symbiotic associations with luminescent bacteria have also been described in teleost fish and squids (Dunlap and Kita-Tsukamoto, 2006) and suggested as hypotheses, in other organisms (e.g., Mackie and Bone, 1978; Taylor et al., 1983). These last hypotheses (i.e., luminescence produced by symbiotic bacteria) were recently challenged in sharks (Duchatelet et al., 2020) and urochordates (Tessler et al., 2020). Multiple bacteria species have been identified in bioluminescent symbiotic associations: Aliivibrio fischeri, Aliivibrio logei, Photobacterium leiognathi, Photobacterium phosphoreum, Photobacterium kishitanii, Photobacterium mandapamemsis, Candidatus Enterovibrio luxaltus, Candidatus Enterovibrio escacola, Candidatus Photodesmus katoptron, Candidatus Photodesmus blepharon, etc. (Boettcher and Ruby, 1990; Dunlap and Kita-Tsukamoto, 2006; Ast et al., 2007; Dunlap et al., 2007; Kaeding et al., 2007; Hendry et al., 2014, 2018; Freed et al., 2019) and some studies suggested that unidentified species could also be involved (Haygood and Distel, 1993). The bacterial luciferase is described as a flavin-dependent monooxygenase and is composed of two different but homologous subunits: the α subunit is the catalytic core, and the β subunit is crucially required for maintaining the catalytic function of the α subunit. In all described bacterial luminescence cases, the luminous reaction involved the oxidation of both a long chain aldehyde and a reduced flavin mononucleotide (FMNH2) into the corresponding carboxylic acid and the oxidised flavin mononucleotide (FMN). The reaction is catalysed by the heterodimeric enzyme and involves oxygen. Tetradecanal is possibly the natural aldehyde substrate of bacterial luciferases but other aldehydes may also be used (Ulitzur and Hastings, 1979). The product of the oxidation of the reduced riboflavin phosphate (FMNH2), the FMN-4a-hydroxide—in an excited state—reverts to its basic state when it emits light. FMN-4a-hydroxide is then generally considered to be the luciferin, as it is the light emitter (Kurfürst et al., 1984; Lei et al., 2004).
Up to now, all investigated bacteria are sharing a common type of luciferase (observed as the “Bacteria luciferases (Group I)” cluster in our meta-analyses, see Figure 2). It is interesting to note that similar bacterial luciferase-like encoding genes have been detected in Archaea (identity superior to 45%) and Fungi (identity superior to 35%), although more functional information are missing for these groups. Also, no homologous sequences have been found in metazoans (J.D. personal observations).
Dinoflagellates (Dinophyceae) are the most commonly encountered luminescent organisms in coastal environments, and at least 69 luminous species have been discovered (Sweeney, 2012; Marcinko et al., 2013; Valiadi and Iglesias-Rodriguez, 2013). Marine bioluminescence of dinoflagellates is stimulated by hydrodynamic turbulence generated by predators or waves (Latz et al., 1994, 2004; Rohr et al., 1998; Latz and Rohr, 1999, 2005).
The dinoflagellate luciferin is thought to be derived from chlorophyll and has a very similar structure (Dunlap et al., 1981; Topalov and Kishi, 2001). A modified form of this luciferin is also found in herbivorous euphausiid shrimps, indicating a probable dietary link for the luciferin acquisition (Shimomura, 1980, 1995).
The dinoflagellate luciferase contains three homologous domains, and each domain—of approximately 46kDa—is known to be enzymatically active and to participate in the bioluminescence reaction (Li et al., 1997; Fajardo et al., 2020). The crystal structure of one of the domains (D3) in its inactive form was solved by Schultz et al. (2005). All luminous dinoflagellates investigated until now are sharing a homologous luciferase type [Dinoflagellata luciferase (Group II) cluster in Figure 2]. Noctiluca scintillans, considered as a “primitive dinoflagellate species” (non-photosynthetic dinoflagellate), has only one enzymatically active luciferase domain potentially corresponding to the gene structure of the ancestral luciferase of dinoflagellates (Valiadi and Iglesias-Rodriguez, 2013).
In dinoflagellates, the luminous organelles—the scintillons—also contain a luciferin binding protein that protects the luciferin from oxidation by the luciferase at physiological pH. While the luciferases are highly conserved in all investigated dinoflagellate species, this is not the case for the luciferin binding proteins, which are all homologous but also appear to be highly variable in sequence and structure (Fajardo et al., 2020, for review).
The evolutionary origin of dinoflagellate luciferase remains elusive, and no exact homologous sequences have been detected in non-dinoflagellate organisms. However, a structural similarity has been found with fatty-acid-binding proteins (FABPs) (lipocalin family) present in metazoans (Schultz et al., 2005).
Approximately 100 fungi species from the order Agaricales emit light using a standard luciferase-luciferin system (Oliveira et al., 2012; Kotlobay et al., 2018). Although fungal bioluminescence’s ecological role is not fully understood, there is evidence that it might be used to attract spore-dispersing insects (Oliveira et al., 2015).
Fungal bioluminescence is known to utilise molecular oxygen, a reduced luciferin recently identified as 3-hydroxyhispidin and a luciferase (Airth and McElroy, 1959; Oliveira and Stevani, 2009; Purtov et al., 2015; Kotlobay et al., 2018).
Our analyses confirm that all investigated luminous Fungi share a common luciferase type (Group III, Figure 2). Unlike all other described bioluminescent proteins, Fungi luciferases are characterised by the presence of a transmembrane domain (Figure 1). No homologs of these enzymes have yet been found and they most likely represent a novel family of protein (Kotlobay et al., 2018).
Bioluminescence is well represented in ctenophores and cnidarians. More than 90% of known planktonic genera of ctenophores can produce light while no luminous benthic species have been identified (Haddock and Case, 1995). In cnidarians, bioluminescence is found in both benthic and planktonic species. Luminous hydrozoans include both hydromedusae (e.g., the species Aequorea victoria in which GFP was discovered, Prasher et al., 1985, 1992; Shimomura, 2005) and siphonophores (e.g., 91% of known planktonic siphonophore genera are luminous) (Haddock et al., 2010). Two orders of scyphozoans contain luminous members: the Coronatae (e.g., Atolla sp., Periphylla sp.) and the Semaeostomeae (e.g., Pelagia noctiluca, Phacellophora sp., Poralia sp.) (Haddock and Case, 1999; Haddock et al., 2010).
All investigated ctenophores and cnidarians use coelenterazine as their light-emitting substrate. All investigated ctenophores and most investigated medusozoans (Cnidaria) species use calcium-activated photoproteins. The specific case of Periphylla sp., however, will be discussed in the section “Other Groups of Luciferases or Photoproteins” as the species has been shown to use a luciferase system. In addition, the case of the anthozoans appears to be different, as well, and will be tackled in the section “Renilla-Type Luciferases (Group VI), a Luciferase Type also Found in the Brittle Star Amphiura filiformis and, Possibly, in the Tunicate Pyrosoma atlanticum” as the octocorals Renilla sp., and most probably other sea pansies and sea pens (Bessho-Uehara et al., 2020a), are using a different and non-homologous luciferase system.
Ctenophore photoproteins share around 20–25% sequence identity and around 40–45% sequence similarity with known hydromedusan cnidarian photoproteins. Mnemiopsis photoproteins share 85–91% sequence identity with other ctenophore photoproteins (i.e., Beroe and Bolinopsis). Within all hydromedusan photoproteins, there is around 60–94% sequence identity (Schnitzler et al., 2012).
Ctenophoran and cnidarian photoproteins are functionally related to coelenterazine-binding proteins from Renilla, sarcoplasmic calcium-binding protein from the marine worm Nereis diversicolor and calmodulin proteins (Schnitzler et al., 2012).
Schnitzler et al. (2012) proposed a metazoan-wide phylogeny for the “Aequora-type photoprotein” (i.e., Ctenophora and Medusozoa Photoproteins) gene family. They identified photoprotein-like genes in non-luminescent taxa (i.e., the poriferan Amphimedon and the cnidarian Nematostella), and demonstrated that the gene family likely arose at the base of the Metazoa (Schnitzler et al., 2012). Light-emitting calcium-binding photoproteins (i.e., functional photoproteins) may have evolved independently from a homologous gene found in ctenophores, cnidarians, and non-luminous sponges (Prasher et al., 1985; Tsuji et al., 1995). The emergence of “light-emitting photoproteins” in cnidarians and ctenophores could then appear as an example of parallel evolution of conserved and homologous genes.
All luminous insects [i.e., Coleoptera with around 2,300 luminous species (Li et al., 2021); Diptera with a lower specific diversity but not yet exhaustively evaluated to the best of our knowledge; Collembola and Hemiptera (Harvey, 1952; McElroy et al., 1974)] share a unique homologous luciferase-type, and the insect proto-luciferase is known to derive from Acyl-CoA ligase enzymes that have a primary metabolic function (Viviani, 2002). These enzymes are members of the ANL superfamily of adenylating enzymes. This superfamily consists of various enzymes, in addition to the Insecta-type luciferase, such as long-chain fatty acid Co-A ligases and acetyl-CoA synthetases as well as other closely related synthetases and a plant auxin-responsive promoter family. The name ANL derives from three subfamilies—Acyl-CoA synthetases, the NRPS adenylation domains, and the Luciferase enzymes. Members of this superfamily catalyse the initial adenylation of a carboxylate to form an acyl-AMP intermediate, followed by a second partial reaction, most commonly forming a thioester (Gulick, 2009). While the homology between all insect luciferase genes is apparent (Figure 2), the evolutionary origin of bioluminescence in this group still appears very complicated and recent studies supported independent emergences of bioluminescence and parallel evolution of luciferases in fireflies, click beetles and Diptera (Fallon et al., 2018; Watkins et al., 2018). Several authors suggested that the high abundance of ancestral gene duplications in this gene family, and as a result the associated closely related enzymatic activities, served as “raw materials for the selection of new adaptive catalytic functions” (Weng, 2014; Fallon et al., 2018).
Generally, the insect luciferin-luciferase system uses the firefly-type luciferin and requires ATP as a cofactor. Interestingly, the use of a distinct bioluminescent system was demonstrated for Arachnocampa flava and Orfelia fultoni (Viviani et al., 2002), two phylogenetically related dipteran species from Australia and North America, respectively. While both species produce light via an insect-type luciferase, A. flava is using a different type of luciferin compared to the other insects (Watkins et al., 2018).
Surprisingly, homologous luciferases to the Insecta-type luciferase were also proposed in the marine squid Watasenia scintillans (Cephalopod, Mollusca) and the sponge Suberites domuncula (Porifera) (Figure 2). The firefly squid W. scintillans emits intense blue bioluminescence from photophores located at the tip of two of its arms. Within the photophore, luciferases are specifically organised in microcrystals, and these proteins are catalysing the bioluminescent reaction using the coelenterazine disulfate luciferin and ATP (Tsuji, 1985; Hamanaka et al., 2011). The involvement of ATP in the luminescence reaction was, however, questioned (Teranishi and Shimomura, 2008; Shimomura, 2012). Gimenez et al. (2016) identified potential Watasenia luciferases (i.e., several related proteins were pinpointed: wsluc1–3) that are sharing around 20% sequence identity with firefly luciferases (Gimenez et al., 2016). While the authors demonstrated that the expression profile of the predicted luciferases is matching with the luminous patterns, additional functional studies would be necessary to confirm the bioactivity of the predicted luciferase.
Müller et al. (2009) suggested that an Insecta-type luciferase (acetyl-CoA synthetase) was involved in the bioluminescence of the sponge Suberites domuncula (Wiens et al., 2010) but this hypothesis remains very speculative. The authors showed that tissue extracts produce light that was detected using sensitive films in the dark. Then, the luciferase-like protein was immunodetected within the tissue. Finally, the recombinant sponge luciferase-like protein produced in Escherichia coli has been shown to emit light, but no information was given as to the actual amount of light produced. The authors suggested that the marine sponge Suberites may be using a firefly luciferase homolog and a luciferin similar to the firefly luciferin which raises many unanswered questions. Additional data would be required to test the presumed involvement of the insect-type luciferase in ecologically relevant light emission in the poriferan Suberites domuncula. The records of luminescence in Porifera are extremely limited and the clear status of intrinsic bioluminescence in these organisms still needs to be confirmed. Martini et al. (2020) recently published the most documented observation of bioluminescence in a deep-sea sponge paving the way to a better understanding of bioluminescence in these organisms. These authors suggested that an undescribed carnivorous sponge species (Cladorhizidae) is using a coelenterazine-dependent bioluminescence (Martini et al., 2020).
From the above examples, it appears probable that enzyme of the ANL superfamily have independently evolved in distant species to produce light using unrelated substrates (Gimenez et al., 2016). It may then represent a striking example of parallel evolution.
The luminescence of the sea pansy Renilla reniformis, a shallow-water soft coral (octocoral) that displays blue-green bioluminescence upon mechanical stimulation, has been intensively studied since the luciferase has been cloned and sequenced in 1991 (Lorenz et al., 1991). The Renilla luciferase enzyme catalyses coelenterazine oxidation leading to bioluminescence. The Renilla luciferase shows a characteristic alpha/betahydrolase fold (Marchler-Bauer et al., 2003). It is found to have a high level of tertiary structure similarity and to be homologous to bacterial haloalkane dehalogenases which are primarily hydrolase enzymes cleaving a carbon-halogen bond in halogenated compounds (Hynková et al., 1999; Loening et al., 2006). Horizontal gene transfers that are known to play critical roles in the evolutionary acquisition of novel traits in eukaryotes (Boto, 2014), have been suspected to explain the high similarity of the Renilla luciferase compared to bacterial haloalkane dehalogenases (Loening et al., 2006; Delroisse et al., 2017b).
Several other luminous anthozoans are found within the octocorals (Alcyonaria) in shallow sandy bottoms (e.g., Ptilosarcus, Pennatula), and in the deep sea (e.g., Stylatula, Halipterus, Anthomastus). A recent work performed on diverse sea pen species strongly suggests the use of a conserved luciferase throughout the lineage (families Isididae, Alcyoniidae, Umbellulidae, Funiculinidae, Kophobelemnidae, and Protoptilidae) (Bessho-Uehara et al., 2020a). In parallel, bioluminescent species have also been found in hexacorals (e.g., bamboo corals), but the biochemistry of their bioluminescence remains unexplored.
A Renilla-type luciferase was recently identified in the brittle star Amphiura filiformis (Echinodermata, Ophiuroidea). In this species, which emits a blue luminescence at the level of the arm spines (Delroisse et al., 2017a), coelenterazine is the luciferin and is acquired via a dietary pathway (Mallefet et al., 2020). The predicted A. filiformis luciferase, highly similar to the Renilla luciferase (up to 47% of identity, up to 69% of similarity) (Figure 2), would constitute the unique example of luciferase described so far in echinoderms. Amphiura luciferase has been detected specifically in the animal’s spine photocytes, which constitutes a strong indication of its photogenesis implication (Delroisse et al., 2014, 2017a, b). However, given the expression of Renilla luciferase-like proteins in non-luminous echinoderms, this hypothesis must be confirmed by the recombinant expression of the A. filiformis protein sequence to verify its luciferase activity.
It has been suggested that the haloalkane-dehalogenase function constitutes the metazoan ancestral state, which shifted to luciferase in cnidarians (lineage of Renilla) and brittle stars (lineage of A. filiformis) (Loening et al., 2006; Delroisse et al., 2017b). Haloalkane dehalogenases were presumably co-opted in luciferases in these two specific lineages. In A. filiformis, the apparent late duplications of luciferase-like genes could suggest the co-occurrence of both functions (Delroisse et al., 2017b). Renilla sp. and A. filiformis would then possess similar and homologous luciferases to catalyse the photogenous reaction. Delroisse et al. (2017b) hypothesised that a co-emergence happened between these two luminous systems using the same compounds under similar environmental pressure. The ecological similarities between the Renilla (Renilla mulleri, Renilla reniformis, and potentially luminous sea-pens in general) and A. filiformis, such as the benthic position on loose sediment and the suspension-feeding strategy, would presumably permit to acquire coelenterazine from planktonic organisms in a “dietary way.” The predation pressure would positively select the emergence of the bioluminescence function endowing these slow-moving organisms with an efficient anti-predation strategy.
Similar to what was observed in the brittle star A. filiformis, a Renilla-like luciferase has also been found in the luminous tunicate Pyrosoma atlanticum (Figure 2). Immunodetections of the luciferase have been performed within the Pyrosoma tissues. In parallel, in vitro expression and functional testing of the protein qualitatively confirmed the enzyme bioactivity (Tessler et al., 2020). However, the hypothesis of the Renilla-like luciferase involved in the bioluminescence of P. atlanticum has recently been questioned (Berger et al., 2021).
The flying squid Sthenoteuthis oualaniensis is characterised by a light organ on its mantle. The light organ contains thousands of small granules, in which a photoprotein exists as the active form (Kuse, 2014; for review). The species emits light using the oxidation of the dehydro-coelenterazine luciferin substrate by the so-called symplectin photoprotein enzyme (Fujii et al., 2002). The 60-kDa symplectin photoprotein was extracted and characterised (Fujii et al., 2002). Sequence analyses revealed no sequence similarity to known bioluminescent proteins (Figure 2) but a significant similarity to the carbon-nitrogen hydrolase domain found in mammalian biotinidase and vanin (pantetheinase) (Fujii et al., 2002).
Francis et al. (2017) recently explored the phylogenetic distribution of these enzymes, grouped in the symplectin/pantetheinase protein family, in metazoans. These authors suggested that symplectins may have multiple functions including hydrolase activity (Francis et al., 2017). Both dehydro-coelenterazine and symplectin-like photoprotein were recently demonstrated to be responsible for the light emission of the Humboldt squid, Dosidicus gigas, one of the largest cephalopods on Earth (Francis et al., 2017; Galeazzo et al., 2019).
Pholas dactylus, the common glowing piddock, is a famous luminescent organism because Dubois, who discovered the general luciferin-luciferase reaction back in the 19th century, was specifically working on this species (Dubois, 1889). The biochemistry of the Pholas bioluminescence was studied in depth in the 1970s (Henry and Michelson, 1973; Michelson, 1978). The term Pholasin was initially dedicated to the luciferin before Pholasin was confirmed to be a photoprotein (Henry and Michelson, 1973; Knight and Campbell, 1987; Kuse, 2020).
Dunstan et al. (2000) cloned the gene coding for the Pholasin apoprotein. These authors compared the amino acid sequence with known proteins present in the public databases and more specifically with the sequences of other cloned bioluminescent proteins (available at that time). A small region of similarity was found between the recombinant protein and the putative luciferin-binding sites of Vargula luciferase and Renilla luciferin-binding protein. However, these sites are very small and do not inform on the potential homology status of these proteins. Our analyses indicated that the Pholas photoprotein have no clear homology with other known bioluminescent proteins [Figure 2, three protein sequences, predicted from different clones sequenced by Dunstan et al. (2000) are presented but these identical protein sequences most probably correspond to a unique gene].
This cluster only contains the luciferase of the deep-sea shrimp Oplophorus gracilirostris. O. gracilirostris secretes a luminous blue cloud from the basal part of its antennae when disturbed (Shimomura et al., 1978). Similar behaviours are observed in various luminescent decapod shrimps including the genera Heterocarpus, Systellaspis, and Acanthephyra (Harvey, 1952). However, the involvement of an Oplophorus-type luciferase in the light emission has only been confirmed in O. gracilirostris.
The Oplophorus luciferase catalyses the oxidation of coelenterazine. The enzyme consists of two subunits (19 and 35 kDa), but the smaller subunit is the only one to have a catalytic activity while the 35 kDa protein is thought to have a role in the stabilisation of the catalytic unit. The 19 KDa protein of Oplophorus luciferase is the smallest known catalytic component having a luciferase function (Inouye et al., 2000).
Oplophorus luciferase presents no homology with other known bioluminescent proteins (Figure 2).
Around 150 ostracod species from the family Cypridinidae (out of about 300 species) can produce light (Brandão et al., 2015; Morin, 2019). These organisms use bioluminescence for defence or to create courtship displays. All investigated luminous cypridinid ostracods use the same luciferin and homologous enzymes to produce light (Harvey, 1924). It has been proposed that luminous cypridinid ostracods synthesise their luciferin from the amino acids tryptophan, isoleucine, and arginine (Oba et al., 2002). This luciferin, called Vargulin or Cypridina-type luciferin as it was initially found in the ostracods Vargula and Cypridina, is also the one used by the midshipman fish Porichthys sp. A clear dietary link has been established, and fishes are losing their ability to luminesce if they are not fed with luciferin-containing food (Mensinger and Case, 1991). The luminescent fish Parapriacanthus ransonneti obtains its luciferin but also its luciferase enzyme from bioluminescent ostracod preys (Bessho-Uehara et al., 2020a).
In parallel, the ostracod family Halocyprididae also contains a dozen luminous species and five of them—belonging to the same genus—were shown to use a coelenterazine-based system (e.g., Conchoecia pseudodiscophora) (Angels, 1968; Campbell and Herring, 1990; Oba et al., 2004). Their corresponding luciferases are still unknown.
Previous research indicated that cypridinid luciferases evolved independently from other luciferases. These enzymes have specific features such as a signal peptide leading to protein secretion outside of the cell, two Von Willebrand Factor type D domains (VWD), multiple disulphide bonds between conserved cysteines, and post-translational N-linked glycosylation (Figure 1; Nakajima et al., 2004; Oakley, 2005; Inouye and Sahara, 2008; Hunt et al., 2017; Mitani et al., 2017; Tessler et al., 2018; Yasuno et al., 2018).
Cypridinid luciferases have no homology with other known bioluminescent proteins (Figure 2).
Some marine copepods emit a bright blue light using a classical luciferase-luciferin system based on the coelenterazine substrate. It is a case of secreted bioluminescence, and the simple oxidation reaction do not require any additional cofactors. Copepod luciferases are small secreted proteins of around 18-24 kDa. Several copepod luciferase genes were cloned (e.g., Gaussia, Metridia, Pleuromamma, Lucicutia, Heterorhabdus, Heterostylites…) (Takenaka et al., 2008, 2012; Thouand and Marks, 2014). The luciferases from the copepods Gaussia princeps and Metridia longa have been used as bioluminescent reporters in various applications (Thouand and Marks, 2014).
Our analyses confirm that copepod luciferases do not share sequence or structural similarity with other identified bioluminescent proteins, including other coelenterazine-dependent luciferases (e.g., Oplophorus luciferase, Renilla-type luciferases) (Figure 2).
The Group XII cluster only contains the luciferase of the luminous annelid worms from the genus Odontosyllis. The bioluminescent systems of Odontosyllis enopla and Odontosyllis octodentata have been partly characterised and the luciferin of O. enopla has been partially purified, showing that light emission requires the presence of magnesium, molecular oxygen, and crude luciferase (Shimomura et al., 1963; Trainor, 1979; Shimomura, 2012). More recently, the luciferase and luciferin of Odontosyllis umdecimdonta have been characterised, the enzyme was cloned, and it was shown that magnesium ions are not required for the luciferin-luciferase reaction (Mitani et al., 2018, 2019; Schultz et al., 2018; Kotlobay et al., 2019). Odontosyllis luciferases have no homology with other known bioluminescent proteins (Figure 2).
Interestingly, Deheyn and Latz proposed that a photoprotein could be involved in the bioluminescence of the species Odontosyllis phosphorea (Deheyn and Latz, 2009). If this assumption is confirmed, it will imply the presence of two convergent bioluminescent protein types (a luciferase and a photoprotein) in the genus Odontosyllis (Deheyn and Latz, 2009).
There are many luminous organisms in which, although no luciferase sequence is available, crucial biochemical information indicate that the bioluminescent proteins involved do not correspond to any of the 12 luciferase types described above. This suggests that additional groups might be described in the future.
Conversely to the other medusozoans (see section “Ctenophora and Medusozoa Photoproteins (Group IV), the First Described Photoprotein-Type”), Periphylla periphylla has been depicted as using a luciferase rather than a photoprotein for its light emission. Shimomura and Flood (1998) described two types of luciferases catalysing the luminous reaction—i.e., luciferase-L (32 kDa) and luciferase-O (75 kDa) using coelenterazine as substrate-, occurring in the jellyfish marginal exumbrella photocytes and eggs, respectively (Shimomura and Flood, 1998; Shimomura, 2012). The bioluminescent system of Periphylla suggests the emergence of convergent bioluminescent protein in medusozoans, with at least two different systems: the luciferases observed in Periphylla and the photoproteins observed in several Medusozoa species.
In annelids, there is a wide diversity of chemical reactions and kinetics recognised among the different luminous species (Verdes and Gruber, 2017, for review). A 300-kDa heterotrimeric Cu2+ metalloprotein has been identified as the luciferase of the Megascolecidae earthworm Diplocardia longa (Bellisario and Cormier, 1971; Bellisario et al., 1972; Ohtsuka et al., 1976; Oba et al., 2017) and a 65-kDa polynoidin photoprotein is used in Polynoidae scale worms (Nicolas et al., 1982; Bassot, 1987; Bassot and Nicolas, 1995; Plyuscheva and Martin, 2009). The chemistry of bioluminescence in parchment tubeworms has been mainly studied in Chaetopterus variopedatus (Shimomura and Johnson, 1968; Anctil, 1979; Martin and Anctil, 1984; Zinner and Vani, 1986; Shimomura, 2012; Deheyn et al., 2013; Branchini et al., 2014; Rawat and Deheyn, 2016; Mirza et al., 2020). The luminous system includes a photoprotein (Shimomura and Johnson, 1968; Shimomura, 2012). In the holopelagic Tomopteridae, the bioluminescence system is thought to be a membrane-bound photoprotein tightly associated with small particles (Shimomura, 2012; Francis et al., 2014). The independent emergence of multiple types of bioluminescent enzymes is evident in the phylum Annelida (Verdes and Gruber, 2017).
In echinoderms, despite the relatively common occurrence of luminous species, only two ophiuroid species, A. filiformis and Ophiopsila californica have been investigated biochemically. The former luminesces with a luciferin-luciferase system stricto sensu (see above) whereas the latter emits light with a photoprotein system (Mallefet et al., 2012, 2020; Shimomura, 1986, 2012). A high diversity of physiological luminescence control mechanisms has been described in these organisms. Species from the same genus (e.g., O. californica and Ophiopsila aranea) can sometimes exhibit different control mechanisms of the photogenous reaction (Mallefet, 2009 for review).
For a considerable number of studied bioluminescent organisms, the luminous system is partially or totally unknown. For the majority of decapods or bony fishes, for example, only one part of the system (i.e., the luciferin) is known and the luciferases or photoproteins remain unknown (Shimomura, 2012; see Supplementary Table 1). This is also the case for the terrestrial potworms Fridericia heliota (Petushkov et al., 2014; Tsarkova et al., 2016) and Henlea sp. (Petushkov and Rodionova, 2005; Oba et al., 2017), the earthworm Diplocardia longa (Ohtsuka et al., 1976; Rudie et al., 1981), as well as the freshwater limpet, Latia neritoides (Shimomura and Johnson, 1968; Ohmiya et al., 2005); for which the luciferins are identified, and luciferases only partially revealed. Within the terrestrial environment, the biochemistry of bioluminescence of species such as the luminous centipedes (Myriapoda, Chilopoda) and millipedes (Myriapoda, Diplopoda)—such as the millipede Motoxya sp.—have not been investigated to date (Rosenberg and Meyer-Rochow, 2009; Oba et al., 2017). Some studies only rely on cross-reactivity between extracts of closely related, or even phylogenetically distant species, to determine the type of luciferin used (Shimomura, 2012). Most often, no data on the luciferase or photoprotein sequence, activity or specificity is available for species that are difficult to collect and fragile, such as those found in deep oceanic strata. Therefore, efforts are still needed to discover hitherto unknown bioluminescent systems. For instance, lanternfish, dragonfish and viperfish luminous systems were demonstrated to use coelenterazine as luciferin, but no luciferase/photoprotein has been identified to date (Tsuji and Haneda, 1971; Mallefet and Shimomura, 1995; Duchatelet et al., 2019). Similarly, shark bioluminescence systems—while these organisms have been extensively studied these last years (e.g., Claes et al., 2020; Mallefet et al., 2021)—remain totally enigmatic, even though attempts were made to decipher the bioluminescent compound in the lanternshark Etmopterus spinax (Renwart and Mallefet, 2013). Cross-reactivity with known luciferin failed to trigger light production, and preliminary search for luciferase homologues within the available transcriptomic data did not yield any results suggesting the involvement of an unknown bioluminescent system in luminous sharks (Renwart and Mallefet, 2013; Delroisse et al., 2018, 2021).
Bioluminescence evolution is often used as a striking illustration of convergent evolution in life history. While it appears clear that many extant bioluminescent systems have evolved independently on earth, the number of fully characterised bioluminescent proteins is still minimal, and the evolution of these light-emitting luciferases and photoproteins remains mostly enigmatic. While the diversity of luciferins involved in bioluminescent systems is rather well evaluated (Lau and Oakley, 2020, for review), the diversity of bioluminescent proteins is, without a doubt, mostly under-evaluated.
It is essential to clarify that the present review does not illustrate the evolutionary history of bioluminescence because it only focuses on visualising the bioluminescent protein homology across the tree of life. Knowing the luciferase evolutionary history is not enough to explain the global bioluminescence’s evolutionary history. The reality is indeed more complex, and bioluminescence substrates likely possess different evolutionary histories from those of luciferases (as presented by Fallon et al., 2018 in the case of luminous insects). The same remark could also be made for other associated actors such as luciferin binding proteins, for example. As illustrated in Lau and Oakley (2020), understanding how bioluminescence emerged in living organisms requires the investigation of all potential actors of bioluminescence including luciferin biosynthetic pathways or dietary acquisition pathways, luciferases, bioluminescence control, … While bioluminescence can be convergent at one biological level, the convergence may not be found at other levels (Lau and Oakley, 2020).
Multiple types of luciferases emerged convergently in the tree of life (Figure 3). Based on the currently available sequence data, our meta-analyses suggest that at least 12 non-homologous bioluminescent protein types (i.e., three types of photoproteins, nine types of luciferases stricto sensu) appeared independently during evolution. Our analyses confirmed that luciferases/photoproteins appear relatively lineage-specific (e.g., all described luminous bacteria share a common and homologous luciferase type). To cite Lau and Oakley (2020), “most known bioluminescent proteins exhibit wide molecular diversity and are not homologous across distantly related taxa, which suggest that most origins of bioluminescent proteins are the result of convergent, but not parallel, evolution.” However, our analyses also highlighted that, in some cases, a similar system—i.e., homologous enzymes—could be used by phylogenetically distant organisms: Group IV photoproteins are shared by ctenophores and medusozoans, Group V luciferases are shared by insects, the cephalopod Watasenia scintillans and, putatively, the sponge Suberites domuncula; Group VI luciferases are shared by the sea pansy Renilla sp., the tunicate Pyrosoma atlanticum and the brittle star Amphiura filiformis. While enzymes appear to be homologous within all precited Groups (IV, V, VI), it also appears that they have been independently co-opted into luciferases in these distant lineages. In short, in these examples of parallel molecular evolution, the proteins themselves are homologous, but their luciferase function is not. However, as exemplified by Tyler (1988), homology should apply most appropriately to the structural features, not their functions.
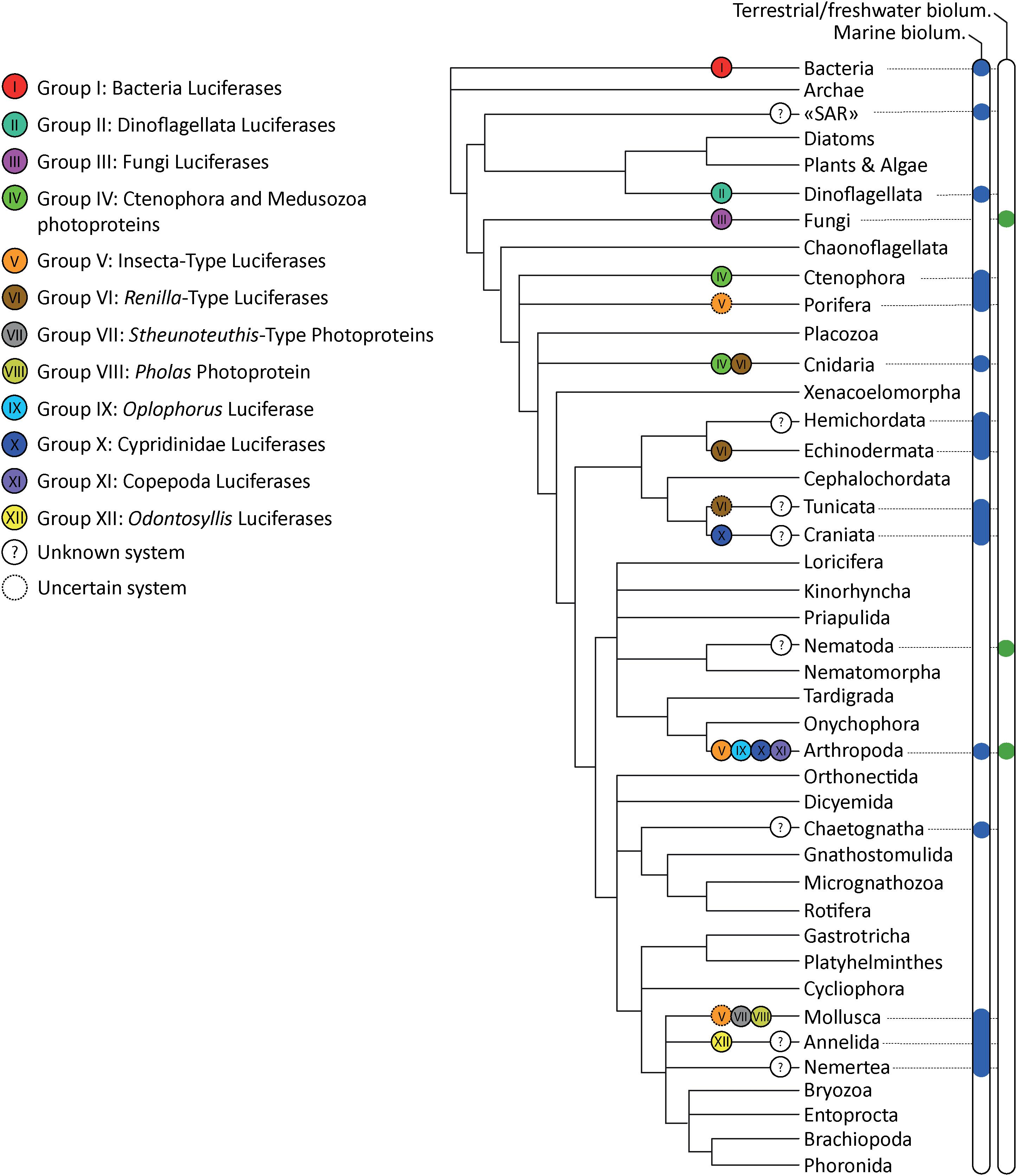
Figure 3. Bioluminescence tree of life annotated with the corresponding bioluminescent protein Group. Distribution of terrestrial and aquatic organisms is based on Lau and Oakley (2020) and Haddock et al. (2010). Phylogenetic tree based on Giribet and Edgecombe (2020).
The evolution of bioluminescence in insects is thought to have emerged from the activity of ancestral fatty acyl-CoA synthetase (ACS) enzymes present in all insects. Beetle luciferases share high sequence identity with these enzymes and often retain the ACS activity (Adams and Miller, 2020). Besides, some ACS enzymes from non-luminous insects can catalyse bioluminescence from synthetic D-luciferin analogues (Mofford et al., 2017; Adams and Miller, 2020).
The annelid polynoidin is also present in non-luminescent scale worms suggesting that bioluminescence might have originated from a non-related mechanism (in this case: quenching of superoxide radicals) (Plyuscheva and Martin, 2009).
The case of the Renilla-type luciferase characterising the brittle star A. filiformis, was investigated in detail based on genomic and transcriptomic data. While the Renilla-type luciferase appears to be specifically expressed in the photocytes in the luminous brittle star, it was also highlighted that similar Renilla-type luciferases are present in non-luminous echinoderms, raising questions about the evolution of bioluminescence in echinoderms. In the sea-urchin Strongylocentrotus purpuratus, a Renilla-type luciferase protein (DspA) was identified as the first biochemically characterised haloalkane dehalogenase of non-microbial origin (Fortova et al., 2013). Nagata et al. (2015) noted homology between the luciferase from R. reniformis and some microbial hydrolases catalysing the removal of halogens from aliphatic hydrocarbons, the so-called haloalkane dehalogenases. Both enzymes share the conserved catalytic triad of residues. Microbial haloalkane dehalogenase (Shingomonas sp.) shares high sequence identity (42%) and similarity (62%) with the sequence of Renilla luciferase. This similarity is somewhat surprising considering that haloalkane dehalogenases are hydrolases and Renilla luciferase is an oxygenase. It seems therefore that the “luciferase-like” proteins could have kept the original microbial function of haloalkane dehalogenases, at least, in sea urchins. Chaloupkova et al. (2019) have recently reconstructed an ancestor of evolutionarily related (but catalytically distinct) haloalkane dehalogenases and Renilla luciferase that has both hydrolase and monooxygenase activities. Fortova et al. (2013) and Delroisse et al. (2017b) reported the absence of light emission after coelenterazine addition to luciferase-like protein extracts from the sea-urchin S. purpuratus and in the sea-star Asterias rubens, which strongly suggest the absence of a luciferase function for the enzymes of these non-luminous species.
Symplectins, from the cephalopod Sthenoteuthis and Dosidicus, which are derived from pantetheinase enzymes, also contain active site residues involved in pantetheinase catalysis suggesting that these photoproteins may have multiple functions including a hydrolase activity (Francis et al., 2017).
These examples of functional shifts indicate that luciferases did not necessarily derive from ancestral oxygenases, and that luciferases may retain the ancestral function and be bifunctional in some cases. Outside the bioluminescence field, this observation has already been reported for oxygenases (Chen et al., 2005). Cooptions of genes non-related to monooxygenases into luciferases are predicted for the Groups IV (Ctenophora/Cnidaria photoproteins), V (Insecta/Watasenia/Suberites luciferases) and VI (Renilla/Pyrosoma/Amphiura luciferases) bioluminescent enzymes that emerged from calmodulin, acyl-CoA ligase and bacterial haloalkane dehalogenase enzymes, respectively. However, determining the ancestral function of a protein is difficult, and these enzymes might have been functioning as oxygenases long before the emergence of their non-oxygenase activity.
Evolution (and natural selection, in particular) often promotes evolutionary innovation by co-opting preexisting genes for new functions, and gene duplication is known to facilitate this process (Hoffmann et al., 2010): citing Hoffmann et al. (2010), “some examples of convergent evolution of protein function provides an impressive demonstration of the ability of natural selection to cobble together complex design solutions by tinkering with different variations of the same basic protein scaffold” (Hoffmann et al., 2010). Here we emphasise that multiple bioluminescent proteins potentially appeared during evolution by the independent emergences of new genes or by the cooption of existing genes with an ancestral function unrelated to bioluminescence (i.e., convergent evolution). In this latter case, cooption might have occurred independently across the tree of life (i.e., parallel evolution) leading to homologous light-emitting systems in non-related luminous organisms. As already suggested for other types of proteins (e.g., Casewell, 2017), our findings suggest that co-option may be an underappreciated process underpinning protein “neofunctionalisation.”
JD performed analyses and wrote the first draft of the manuscript. LD participated in the data collection from the literature. PF and JM supervised the work. All authors participated in discussions and revised the final manuscript.
This work was supported by the F.R.S.-FNRS PDR project “Glow & See” (T.0169.20) awarded to the University of Louvain (Marine Biology Laboratory) and the University of Mons (Biology of Marine Organisms and Biomimetics Laboratory).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study is a contribution from the “Centre Interuniversitaire de Biologie Marine” (CIBIM). This study is the contribution BRC #378 of the Biodiversity Research Center (UCLouvain) from the Earth and Life Institute Biodiversity (ELIB). JD, JM, and PF are, respectively, postdoctoral fellow, Research Associate, and Research Director of the Fund for Scientific Research of Belgium (F.R.S-FNRS). LD is postdoctoral researcher at the University of Louvain. We thank the reviewers as well as Warren Francis and Oba Yuichi for their constructive comments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.673620/full#supplementary-material
Supplementary Table 1 | List of known bioluminescent systems based on biochemical and molecular data (including the sequence references used for the CLANS analysis presented in Figure 2).
Supplementary File 1 | References used to generate the list of known bioluminescent systems based on biochemical and molecular data (Supplementary Table 1).
Adams, S. T. Jr., and Miller, S. C. (2020). Enzymatic promiscuity and the evolution of bioluminescence. FEBS J. 287, 1369–1380. doi: 10.1111/febs.15176
Airth, R. L., and McElroy, W. D. (1959). Light emission from extracts of luminous fungi. J. Bacteriol. 77, 249–250. doi: 10.1128/jb.77.2.249-250.1959
Anctil, M. (1979). The epithelial luminescent system of Chaetopterus variopedatus. Can. J. Zool. 57, 1290–1310. doi: 10.1139/z79-166
Angels, M. V. (1968). Bioluminescence in planktonic halocyprid ostracods. J. Mar. Biol. Ass. U.K. 48, 255–257. doi: 10.1017/S0025315400032562
Ast, J. C., Cleenwerck, I., Engelbeen, K., Urbanczyk, H., Thompson, F. L., De Vos, P., et al. (2007). Photobacterium kishitanii sp. nov., a luminous marine bacterium symbiotic with deep-sea fishes. Int. J. Syst. Evol. Microbiol. 57, 2073–2078. doi: 10.1099/ijs.0.65153-0
Barnes, A. T., Case, J. F., and Tsuji, F. I. (1973). Induction of bioluminescence in a luciferin deficient form of the marine teleost, Porichthys, in response to exogenous luciferin. Comparat. Biochem. Physiol. Part A Physiol. 46, 709–723. doi: 10.1016/0300-9629(73)90123-0
Bassot, J. M. (1987). A transient intracellular coupling explains the facilitation of responses in the bioluminescent system of scale worms. J. Cell Biol. 105, 2235–2243. doi: 10.1083/jcb.105.5.2235
Bassot, J. M., and Nicolas, M. T. (1995). Bioluminescence in scale-worm photosomes: the photoprotein polynoidin is specific for the detection of superoxide radicals. Histochem. Cell Biol. 104, 199–210. doi: 10.1007/bf01835153
Bellisario, R., and Cormier, M. J. (1971). Peroxide-linked bioluminescence catalyzed by a copper-containing, non-heme luciferase isolated from a bioluminescent earthworm. Biochem. Biophys. Res. Commun. 43, 800–805. doi: 10.1016/0006-291x(71)90687-5
Bellisario, R., Spencer, T. E., and Cormier, M. J. (1972). Isolation and properties of luciferase, a non-heme peroxidase, from the bioluminescent earthworm, Diplocardia longa. Biochemistry 11, 2256–2266. doi: 10.1021/bi00762a008
Berger, A., Blackwelder, P., Frank, T., Sutton, T. T., Pruzinsky, N. M., Slayden, N., et al. (2021). Microscopic and genetic characterization of bacterial symbionts with bioluminescent potential in Pyrosoma atlanticum. Front. Mar. Sci. 8:606818. doi: 10.3389/fmars.2021.606818
Bessho-Uehara, M., Francis, W. R., and Haddock, S. H. D. (2020a). Biochemical characterization of diverse deep-sea anthozoan bioluminescence systems. Mar. Biol. 167:114. doi: 10.1007/s00227-020-03706-w
Bessho-Uehara, M., Yamamoto, N., Shigenobu, S., Mori, H., Kuwata, K., and Oba, Y. (2020b). Kleptoprotein bioluminescence: Parapriacanthus fish obtain luciferase from ostracod prey. Sci. Adv. 6:eaax4942. doi: 10.1126/sciadv.aax4942
Boettcher, K. J., and Ruby, E. G. (1990). Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172, 3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990
Boto, L. (2014). Horizontal gene transfer in the acquisition of novel traits by metazoans. Proc. R. Soc. B, Biol. Sci. 281, 20132450. doi: 10.1098/rspb.2013.2450
Branchini, B. R., Behney, C. E., Southworth, T. L., Rawat, R., and Deheyn, D. D. (2014). Chemical analysis of the luminous slime secreted by the marine worm Chaetopterus (Annelida. Polychaeta). Photochem. Photobiol. 90, 247–251. doi: 10.1111/php.12169
Brandão, S. N., Angel, M. V., Karanovic, I., Parker, A., Perrier, V., Sames, B., et al. (2015). World Ostracoda Database. Ostend: Accessed Through: World Register of Marine Species.
Brodl, E., Winkler, A., and Macheroux, P. (2018). Molecular mechanisms of bacterial bioluminescence. Comput. Struct. Biotechnol. J. 16, 551–564. doi: 10.1016/j.csbj.2018.11.003
Campbell, A. K., and Herring, P. J. (1990). Imidazolopyrazine bioluminescence in copepods and other marine organisms. Mar. Biol. 104, 219–225. doi: 10.1007/BF01313261
Casewell, N. R. (2017). Evolution: gene co-option underpins venom protein evolution. Curr. Biol. 27, R647–R649.
Chaloupkova, R., Liskova, V., Toul, M., Markova, K., Sebestova, E., Hernychova, L., et al. (2019). Light-emitting dehalogenases: reconstruction of multifunctional biocatalysts. ACS Catalysis 9, 4810–4823. doi: 10.1021/acscatal.9b01031
Chen, Y. H., Wang, C. C., Greenwell, L., Rix, U., Hoffmeister, D., Vining, L. C., et al. (2005). Functional analyses of oxygenases in jadomycin biosynthesis and identification of JadH as a bifunctional oxygenase/dehydrase. J. Biol. Chem. 280, 22508–22514. doi: 10.1074/jbc.M414229200
Claes, J. M., and Mallefet, J. (2009). “Bioluminescence of sharks: first synthesis,” in Bioluminescence in Focus – A Collection of Illuminating Essays, ed. V. Meyer Rochow (Kerala: Research Signpost), 51–65.
Claes, J. M., Delroisse, J., Grace, M. A., Doosey, M. H., Duchatelet, L., and Mallefet, J. (2020). Histological evidence for secretory bioluminescence from pectoral pockets of the American Pocket Shark (Mollisquama mississippiensis). Sci. Rep. 10:18762. doi: 10.1038/s41598-020-75656-8
Davis, M. P., Sparks, J. S., and Smith, W. L. (2016). Repeated and widespread evolution of bioluminescence in marine fishes. PLoS One 11:e0155154. doi: 10.1371/journal.pone.0155154
Deheyn, D. D., Enzor, L. A., Dubowitz, A., Urbach, J. S., and Blair, D. (2013). Optical and physicochemical characterization of the luminous mucous secreted by the marine worm Chaetopterus sp. Physiol. Biochem. Zool. 86, 702–715. doi: 10.1086/673869
Deheyn, D. D., and Latz, M. I. (2009). Internal and secreted bioluminescence of the marine polychaete Odontosyllis phosphorea (Syllidae). Invertebr. Biol. 128, 31–45. doi: 10.1111/j.1744-7410.2008.00149.x
Delroisse, J., Duchatelet, L., Flammang, P., and Mallefet, J. (2018). De novo transcriptome analyses provide insights into opsin-based photoreception in the lanternshark Etmopterus spinax. PLoS One 13:e0209767. doi: 10.1371/journal.pone.0209767
Delroisse, J., Duchatelet, L., Flammang, P., and Mallefet, J. (2021). Photophore distribution and enzymatic diversity within the photogenic integument of the cookie cutter shark Isistius brasiliensis (Chondrichthyes: Dalatiidae). Front. Mar. Sci. 8:627045. doi: 10.3389/fmars.2021.627045
Delroisse, J., Flammang, P., and Mallefet, J. (2014). Marine luciferases: are they really taxon-specific? A putative luciferase evolved by co-option in an echinoderm lineage. Luminescence 29:15.
Delroisse, J., Ullrich-Lüter, E., Blaue, S., Eeckhaut, I., Flammang, P., and Mallefet, J. (2017a). Fine structure of the luminous spines and luciferase detection in the brittle star Amphiura filiformis. Zool. Anz. 269, 1–12. doi: 10.1016/j.jcz.2017.05.001
Delroisse, J., Ullrich-Lüter, E., Blaue, S., Ortega-Martinez, O., Eeckhaut, I., Flammang, P., et al. (2017b). A puzzling homology: a brittle star using a putative cnidarian-type luciferase for bioluminescence. Open Biol. 7:160300. doi: 10.1098/rsob.160300
Dubois, R. (1889). Sur le mécanisme des Fonctions Photodermatique et Photogénique Dans le Siphon du Pholas Dactylus. Paris: Compt. rend. de l’acad. des sciences, 109.
Duchatelet, L., Hermans, C., Duhamel, G., Cherel, Y., Guinet, C., and Mallefet, J. (2019). “Coelenterazine detection in five myctophid species from the kerguelen plateau,” in The Kerguelen Plateau: Marine Ecosystem and Fisheries. Proceedings of the Second Symposium, eds D. Welsford, J. Dell, and G. Duhamel (Tasmania: Australian Antarctic Division, Kingston), 31–41.
Duchatelet, L., Moris, V. C., Tomita, T., Mahillon, J., Sato, K., Behets, C., et al. (2020). The megamouth shark, Megachasma pelagios, is not a luminous species. PLoS One 15:e0242196. doi: 10.1371/journal.pone.0242196
Dunlap, P. V., and Kita-Tsukamoto, K. (2006). “Luminous bacteria,” in The Prokaryotes, eds M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (New York, NY: Springer), doi: 10.1007/0-387-30742-7_27
Dunlap, J. C., Hastings, J. W., and Shimomura, O. (1981). Dinoflagellate luciferin is structurally related to chlorophyll. FEBS Lett. 135, 273–276. doi: 10.1016/0014-5793(81)80799-5
Dunlap, P. V., Ast, J. C., Kimura, S., Fukui, A., Yoshino, T., and Endo, H. (2007). Phylogenetic analysis of host-symbiont specificity and codivergence in bioluminescent symbioses. Cladistics 23, 507–532. doi: 10.1111/j.1096-0031.2007.00157.x
Dunstan, S. L., Sala-Newby, G. B., Fajardo, A. B., Taylor, K. M., and Campbell, A. K. (2000). Cloning and expression of the bioluminescent photoprotein pholasin from the bivalve mollusc Pholas dactylus. J. Biol. Chem. 275, 9403–9409. doi: 10.1074/jbc.275.13.9403
Fajardo, C., De Donato, M., Rodulfo, H., Martinez-Rodriguez, G., Costas, B., Mancera, J. M., et al. (2020). New perspectives related to the bioluminescent system in dinoflagellates: Pyrocystis lunula, a case study. Int. J. Mol. Sci. 21:1784. doi: 10.3390/ijms21051784
Fallon, T. R., Lower, S. E., Chang, C. H., Bessho-Uehara, M., Martin, G. J., Bewick, A. J., et al. (2018). Firefly genomes illuminate parallel origins of bioluminescence in beetles. Elife 7:e36495.
Fortova, A., Sebestova, E., Stepankova, V., Koudelakova, T., Palkova, L., Damborsky, J., et al. (2013). DspA from Strongylocentrotus purpuratus: the first biochemically characterized haloalkane dehalogenase of non-microbial origin. Biochimie 95, 2091–2096. doi: 10.1016/j.biochi.2013.07.025
Francis, W. R., Christianson, L. M., and Haddock, S. H. D. (2017). Symplectin evolved from multiple duplications in bioluminescent squid. PeerJ 5:e3633. doi: 10.7717/peerj.3633
Francis, W. R., Powers, M. L., and Haddock, S. H. (2014). Characterization of an anthraquinone fluor from the bioluminescent, pelagic polychaete Tomopteris. Luminescence 29, 1135–1140. doi: 10.1002/bio.2671
Frank, T. M., Widder, E. A., Latz, M. I., and Case, J. F. (1984). Dietary maintenance of bioluminescence in a deep-sea mysid. J. Exp. Biol. 109:385. doi: 10.1242/jeb.109.1.385
Freed, L. L., Easson, C., Baker, L. J., Fenolio, D., Sutton, T. T., Khan, Y., et al. (2019). Characterization of the microbiome and bioluminescent symbionts across life stages of Ceratioid Anglerfishes of the Gulf of Mexico. FEMS Microbiol. Ecol. 95:fiz146. doi: 10.1093/femsec/fiz146-
Frickey, T., and Lupas, A. (2004). CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20, 3702–3704. doi: 10.1093/bioinformatics/bth444
Fujii, T., Ahn, J. Y., Kuse, M., Mori, H., Matsuda, T., and Isobe, M. (2002). A novel photoprotein from oceanic squid (Symplectoteuthis oualaniensis) with sequence similarity to mammalian carbon-nitrogen hydrolase domains. Biochem. Biophys. Res. Commun. 293, 874–879. doi: 10.1016/S0006-291X(02)00296-6
Galeazzo, G. A., Mirza, J. D., Dorr, F. A., Pinto, E., Stevani, C. V., Lohrmann, K. B., et al. (2019). Characterizing the bioluminescence of the Humboldt squid, Dosidicus gigas (d’Orbigny, 1835): one of the largest luminescent animals in the world. Photochem. Photobiol. 95, 1179–1185. doi: 10.1111/php.13106
Gimenez, G., Metcalf, P., Paterson, N. G., and Sharpe, M. L. (2016). Mass spectrometry analysis and transcriptome sequencing reveal glowing squid crystal proteins are in the same superfamily as firefly luciferase. Sci. Rep. 6:27638. doi: 10.1038/srep27638
Giribet, G., and Edgecombe, G. D. (2020). The Invertebrate Tree of Life. New Jersey, NJ: Princeton University Press.
Gulick, A. M. (2009). Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811–827. doi: 10.1021/cb900156h
Haddock, S. H. D., Moline, M. A., and Case, J. F. (2010). Bioluminescence in the sea. Ann. Rev. Mar. Sci. 2, 443–493. doi: 10.1146/annurev-marine-120308-081028
Haddock, S. H., and Case, J. F. (1995). Not all ctenophores are bioluminescent: Pleurobrachia. The Biological Bulletin 189, 356–362. doi: 10.2307/1542153
Haddock, S. H., and Case, J. F. (1999). Bioluminescence spectra of shallow and deep-sea gelatinous zooplankton: ctenophores, medusae and siphonophores. Mar. Biol. 133, 571–582. doi: 10.1007/s002270050497
Haddock, S. H., Rivers, T. J., and Robison, B. H. (2001). Can coelenterates make coelenterazine? Dietary requirement for luciferin in cnidarian bioluminescence. Proc. Natl Acad. Sci. U.S.A. 98, 11148–11151. doi: 10.1073/pnas.201329798
Hamanaka, T., Michinomae, M., Seidou, M., Miura, K., Inoue, K., and Kito, Y. (2011). Luciferase activity of the intracellular microcrystal of the firefly squid. Watasenia scintillans. FEBS Lett. 585, 2735–2738. doi: 10.1016/j.febslet.2011.07.033
Harvey, E. N. (1924). STUDIES ON BIOLUMINESCENCE: XVI. what determines the color of the light of luminous animals? Am. J. Physiol. Legacy Content 70, 619–623. doi: 10.1152/ajplegacy.1924.70.3.619
Hastings, J. W. (1983). Biological diversity, chemical mechanisms, and the evolutionary origins of bioluminescent systems. J. Mol. Evol. 19, 309–321. doi: 10.1007/BF02101634
Haygood, M. G., and Distel, D. L. (1993). Bioluminescent symbionts of flashlight fishes and deep-sea anglerfishes form unique lineages related to the genus Vibrio. Nature 363, 154–156. doi: 10.1038/363154a0
Hendry, T. A., de Wet, J. R., and Dunlap, P. V. (2014). Genomic signatures of obligate host dependence in the luminous bacterial symbiont of a vertebrate. Environ. Microbiol. 16, 2611–2622. doi: 10.1111/1462-2920.12302
Hendry, T. A., Freed, L. L., Fader, D., Fenolio, D., Sutton, T. T., and Lopez, J. V. (2018). Ongoing transposon-mediated genome reduction in the luminous bacterial symbionts of deep-sea ceratioid anglerfishes. mBio 9, e01033–18. doi: 10.1128/mBio.01033-18
Henry, J. P., and Michelson, A. M. (1973). Studies in bioluminescence: VIII. chemically induced luminescence of pholas dactylus luciferin. Biochimie 55, 75–81.
Herring, P. J. (1987). Systematic distribution of bioluminescence in living organisms. J. Biolum. Chemilum. 1, 147–163. doi: 10.1002/bio.1170010303
Hoffmann, F. G., Opazo, J. C., and Storz, J. F. (2010). Gene cooption and convergent evolution of oxygen transport hemoglobins in jawed and jawless vertebrates. Proc. Natl Acad. Sci. U.S.A. 107, 14274–14279. doi: 10.1073/pnas.1006756107
Hunt, E. A., Moutsiopoulou, A., Broyles, D., Head, T., Dikici, E., Daunert, S., et al. (2017). Expression of a soluble truncated Vargula luciferase in Escherichia coli. Protein Expr. Purif. 132, 68–74. doi: 10.1016/j.pep.2017.01.007
Hynková, K., Nagata, Y., Takagi, M., and Damborský, J. (1999). Identification of the catalytic triad in the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. FEBS letters 446, 177–181.
Inouye, S., and Sahara, Y. (2008). Soluble protein expression in E. coli cells using IgG-binding domain of protein A as a solubilizing partner in the cold induced system. Biochem. Biophys. Res. Commun. 376, 448–453. doi: 10.1016/j.bbrc.2008.08.149
Inouye, S., Watanabe, K., Nakamura, H., and Shimomura, O. (2000). Secretional luciferase of the luminous shrimp Oplophorus gracilirostris: cDNA cloning of a novel imidazopyrazinone luciferase. FEBS Lett. 481, 19–25. doi: 10.1016/S0014-5793(00)01963-3
Kaeding, A. J., Ast, J. C., Pearce, M. M., Urbanczyk, H., Kimura, S., Endo, H., et al. (2007). Phylogenetic diversity and cosymbiosis in the bioluminescent symbioses of “Photobacterium mandapamensis”. Appl. Environ. Microbiol. 73, 3173–3182. doi: 10.1128/AEM.02212-06
Knight, J., and Campbell, A. K. (1987). Pholasin—a bioluminescent indicator for detecting activation of single neutrophils. Analyt. Biochem. 160, 139–148. doi: 10.1016/0003-2697(87)90624-5
Kotlobay, A. A., Dubinnyi, M. A., Purtov, K. V., Guglya, E. B., Rodionova, N. S., Petushkov, V. N., et al. (2019). Bioluminescence chemistry of fireworm Odontosyllis. Proc. Natl. Acad. Sci. U.S.A. 116, 18911–18916. doi: 10.1073/pnas.1902095116
Kotlobay, A. A., Sarkisyan, K. S., Mokrushina, Y. A., Marcet-Houben, M., Serebrovskaya, E. O., Markina, N. M., et al. (2018). Genetically encodable bioluminescent system from fungi. Proc. Natl. Acad. Sci. U.S.A. 115, 12728–12732. doi: 10.1073/pnas.1803615115
Kurfürst, M., Ghisla, S., and Hastings, J. W. (1984). Characterization and postulated structure of the primary emitter in the bacterial luciferase reaction. Proc. Natl Acad. Sci. U.S.A. 81, 2990–2994. doi: 10.1073/pnas.81.10.2990
Kuse, M. (2020). “Marine bioluminescence with dehydrocoelenterazine, an imidazopyrazinone compound,” in Topics in Heterocyclic Chemistry (Berlin, Heidelberg: Springer). doi: 10.1007/7081_2020_41
Kuse, M. (2014). Chromophores in photoproteins of a glowing squid and mollusk. Biosci. Biotechnol. Biochem. 78, 731–736. doi: 10.1080/09168451.2014.915724
Latz, M. I., and Rohr, J. (1999). Luminescence response of a red tide dinoflagellate Lingulodinium polyedrum to laminar and turbulent flow. Limnol. Oceanogr. 44, 1423–1435. doi: 10.4319/lo.1999.44.6.1423
Latz, M. I., and Rohr, J. (2005). Glowing with the flow: ecology and applications of flow-stimulated bioluminescence. Opt. Photon. News 16, 40–45. doi: 10.1364/OPN.16.10.000040
Latz, M. I., Case, J. F., and Gran, R. L. (1994). Excitation of bioluminescence by laminar fluid shear associated with simple Couette flow. Limnol. Oceanogr. 39, 1424–1439. doi: 10.4319/lo.1994.39.6.1424
Latz, M. I., Juhl, A. R., Ahmed, A. M., Elghobashi, S. E., and Rohr, J. (2004). Hydrodynamic stimulation of dinoflagellate bioluminescence: a computational and experimental study. J. Exp. Biol. 207, 1941–1951. doi: 10.1242/jeb.00973
Lau, E. S., and Oakley, T. H. (2020). Multi-level convergence of complex traits and the evolution of bioluminescence. Biol. Rev. 96, 673–691. doi: 10.1111/brv.12672
Lei, B., Ding, Q., and Tu, S. C. (2004). Identity of the emitter in the bacterial luciferase luminescence reaction: binding and fluorescence quantum yield studies of 5-decyl-4a-hydroxy-4a, 5-dihydroriboflavin-5 ‘-phosphate as a model. Biochem. 43, 15975–15982. doi: 10.1021/bi0480640
Li, L., Hong, R., and Hastings, J. W. (1997). Three functional luciferase domains in a single polypeptide chain. Proc. Natl Acad. Sci. U.S.A. 94, 8954–8958. doi: 10.1073/pnas.94.17.8954
Li, Y. D., Kundrata, R., Tihelka, E., Liu, Z., Huang, D., and Cai, C. (2021). Cretophengodidae, a new Cretaceous beetle family, sheds light on the evolution of bioluminescence. Proc. R. Soc. B 288:20202730. doi: 10.1098/rspb.2020.2730
Loening, A. M., Fenn, T. D., Wu, A. M., and Gambhir, S. S. (2006). Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 19, 391–400. doi: 10.1093/protein/gzl023
Lorenz, W. W., McCann, R. O., Longiaru, M., and Cormier, M. J. (1991). Isolation and expression of a cDNA encoding Renilla reniformis luciferase. Proc. Natl. Acad. Sci. U.S.A. 88, 4438–4442. doi: 10.1073/pnas.88.10.4438
Mackie, G. O., and Bone, Q. (1978). Luminescence and associated effector activity in Pyrosoma (Tunicata: Pyrosomida). Proc. R. Soc. B 202, 483–495. doi: 10.1098/rspb.1978.0081
Mallefet, J. (2009). “Echinoderm bioluminescence: why, when, and how do so many ophiuroids glow?,” in Bioluminescence in Focus: a Collection of Illuminating Essays, ed. V. B. Rochow (Kerala: Research Signpost), 67–84.
Mallefet, J., and Shimomura, O. (1995). Presence of coelenterazine in mesopelagic fishes from the Strait of Messina. Mar. Biol. 124, 381–385. doi: 10.1007/BF00363911
Mallefet, J., Duchatelet, L., and Coubris, C. (2020). Bioluminescence induction in the ophiuroid Amphiura filiformis (Echinodermata). J. Exp. Biol. 223, jeb218719. doi: 10.1242/jeb.218719
Mallefet, J., Parmentier, B., Mulliez, X., Shimomura, O., and Morsomme, P. (2012). “Characterisation of Amphiura filiformis luciferase (Ophiuroidea, Echinodermata). In Echinoderms in a Changing World,” in Proceedings of the 13th International Echinoderm Conference, January 5-9 2009, (Tasmania), 293.
Mallefet, J., Stevens, D. W., and Duchatelet, L. (2021). Bioluminescence of the largest luminous vertebrate, the kitefin shark, Dalatias licha: first insights and comparative aspects. Front. Mar. Sci. 8:153.
Marchler-Bauer, A., Anderson, J. B., DeWeese-Scott, C., Fedorova, N. D., Geer, L. Y., He, S., et al. (2003). CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31, 383–387. doi: 10.1093/nar/gkg087
Marcinko, C. L., Painter, S. C., Martin, A. P., and Allen, J. T. (2013). A review of the measurement and modelling of dinoflagellate bioluminescence. Prog. Oceanogr. 109, 117–129. doi: 10.1016/j.pocean.2012.10.008
Martin, N., and Anctil, M. (1984). Luminescence control in the tube-worm Chaetopterus variopedatus: role of nerve cord and photogenic gland. Biol. Bull. 166, 583–593. doi: 10.2307/1541164
Martini, S., and Haddock, S. H. D. (2017). Quantification of bioluminescence from the surface to the deep sea demonstrates its predominance as an ecological trait. Sci. Rep. 7:45750. doi: 10.1038/srep45750
Martini, S., Schultz, D. T., Lundsten, L., and Haddock, S. H. (2020). Bioluminescence in an undescribed species of carnivorous sponge (Cladorhizidae) from the deep sea. Front. Mar. Sci. 7:1041. doi: 10.3389/fmars.2020.576476
McElroy, W. D., Seliger, H. H., and DeLuca, M. (1974). “Insect bioluminescence,” in The Physiology of Insecta ed. Morris Rockstein (Miami, FL: Academic Press), 411–460. doi: 10.1016/b978-0-12-591602-8.50015-1
Mensinger, A. F., and Case, J. F. (1991). Bioluminescence maintenance in juvenile Porichthys notatus. Biol. Bull. 181, 181–188. doi: 10.2307/1542501
Michelson, A. M. (1978). Purification and properties of Pholas dactylus luciferin and luciferase. Methods Enzymol. 57, 385–406. doi: 10.1016/0076-6879(78)57037-7
Mirza, J. D., Migotto, A. E., Yampolsky, I. V., de Moraes, G. V., Tsarkova, A. S., and Oliveira, A. G. (2020). Chaetopterus variopedatus bioluminescence: a review of light emission within a species complex. Photochem. Photobiol. 96, 768–778. doi: 10.1111/php.13221
Mitani, Y., Oshima, Y., Mitsuda, N., Tomioka, A., Sukegawa, M., Fujita, M., et al. (2017). Efficient production of glycosylated Cypridina luciferase using plant cells. Protein Expr. Purif. 133, 102–109. doi: 10.1016/j.pep.2017.03.008
Mitani, Y., Yasuno, R., Futahashi, R., Oakley, T. H., and Ohmiya, Y. (2019). Luciferase gene of a Caribbean fireworm (Syllidae) from Puerto Rico. Sci. Rep. 9:13015. doi: 10.1038/s41598-019-49538-7
Mitani, Y., Yasuno, R., Isaka, M., Mitsuda, N., Futahashi, R., Kamagata, Y., et al. (2018). Novel gene encoding a unique luciferase from the fireworm Odontsyllis undecimdonta. Sci. Rep. 8:12789. doi: 10.1038/s41598-018-31086-1
Mofford, D. M., Liebmann, K. L., Sankaran, G. S., Reddy, G. K. K., Reddy, G. R., and Miller, S. C. (2017). Luciferase activity of insect fatty Acyl-CoA synthetases with synthetic luciferins. ACS Chem. Biol. 12, 2946–2951. doi: 10.1021/acschembio.7b00813
Morin, J. G. (2019). Luminaries of the reef: the history of luminescent ostracods and their courtship displays in the Caribbean. J. Crustacean Biol. 39, 227–243. doi: 10.1093/jcbiol/ruz009
Müller, W. E. G., Kasueske, M., Wang, X., Schröder, H. C., Wang, Y., Pisignano, D., et al. (2009). Luciferase a light source for the silica-based optical waveguides (spicules) in the demosponge Suberites domuncula. Cell. Mol. Life Sci. 66:537. doi: 10.1007/s00018-008-8492-5
Nagata, Y., Ohtsubo, Y., and Tsuda, M. (2015). Properties and biotechnological applications of natural and engineered haloalkane dehalogenases. Appl. Microbiol. Biotechnol. 99, 9865–9881. doi: 10.1007/s00253-015-6954-x
Nakajima, Y., Kobayashi, K., Yamagishi, K., Enomoto, T., and Ohmiya, Y. (2004). cDNA cloning and characterization of a secreted luciferase from the luminous Japanese ostracod. Cypridina noctiluca. Biosci. Biotechnol. Biochem. 68, 565–570. doi: 10.1271/bbb.68.565
Nicolas, M. T., Bassot, J. M., and Shimomura, O. (1982). Polynoidin: a membrane photoprotein isolated from the bioluminescent system of scale-worms. Photochem. Photobiol. 35, 201–207. doi: 10.1111/j.1751-1097.1982.tb03832.x
Oakley, T. H. (2005). Myodocopa (Crustacea: Ostracoda) as models for evolutionary studies of light and vision: multiple origins of bioluminescence and extreme sexual dimorphism. Hydrobiologia 538, 179–192. doi: 10.1007/s10750-004-4961-5
Oba, Y., Kato, S. I., Ojika, M., and Inouye, S. (2002). Biosynthesis of luciferin in the sea firefly, Cypridina hilgendorfii: L-tryptophan is a component in Cypridina luciferin. Tetrahedron Lett. 43, 2389–2392. doi: 10.1016/S0040-4039(02)00257-5
Oba, Y., Stevani, C. V., Oliveira, A. G., Tsarkova, A. S., Chepurnykh, T. V., and Yampolsky, I. V. (2017). Selected least studied but not forgotten bioluminescent systems. Photochem. Photobiol. 93, 405–415. doi: 10.1111/php.12704
Oba, Y., Tsuduki, H., Kato, S. I., Ojika, M., and Inouye, S. (2004). Identification of the luciferin–luciferase system and quantification of coelenterazine by mass spectrometry in the deep-sea luminous ostracod Conchoecia pseudodiscophora. ChemBioChem 5, 1495–1499. doi: 10.1002/cbic.200400102
Ohmiya, Y., Kojima, S., Nakamura, M., and Niwa, H. (2005). Bioluminescence in the limpet-like snail, Latia neritoides. Bulletin of the Chemical Society of Japan 78, 1197–1205. doi: 10.1246/bcsj.78.1197
Ohtsuka, H., Rudie, N. G., and Wampler, J. E. (1976). Structural identification and synthesis of luciferin from the bioluminescent earthworm, Diplocardia longa. Biochemistry 15, 1001–1004. doi: 10.1021/bi00650a009
Oliveira, A. G., and Stevani, C. V. (2009). The enzymatic nature of fungal bioluminescence. Photochem. Photobiol. Sci. 8, 1416–1421. doi: 10.1039/B908982A
Oliveira, A. G., Desjardin, D. E., Perry, B. A., and Stevani, C. V. (2012). Evidence that a single bioluminescent system is shared by all known bioluminescent fungal lineages. Photochem. Photobiol. Sci. 11, 848–852. doi: 10.1039/C2PP25032B
Oliveira, A. G., Stevani, C. V., Waldenmaier, H. E., Viviani, V., Emerson, J. M., Loros, J. J., et al. (2015). Circadian control sheds light on fungal bioluminescence. Curr. Biol. 25, 964–968. doi: 10.1016/j.cub.2015.02.021
Pearson, W. R. (2013). An introduction to sequence similarity (“homology”) searching. Curr. Protoc. Bioinformatics 42:Unit3.1. doi: 10.1002/0471250953.bi0301s42
Petushkov, V. N., and Rodionova, N. S. (2005). New types of luminescent systems of soil enchytraeids (Annelida: Clitellata: Oligochaeta: Enchytraeidae). Doklady Biochem. Biophys. 401, 263–266.
Petushkov, V. N., Dubinnyi, M. A., Tsarkova, A. S., Rodionova, N. S., Baranov, M. S., Kublitski, V. S., et al. (2014). A novel type of luciferin from the Siberian luminous earthworm Fridericia heliota: structure elucidation by spectral studies and total synthesis. Angew. Chem. 126, 5672–5674. doi: 10.1002/ange.201400529
Plyuscheva, M., and Martin, D. (2009). On the morphology of elytra as luminescent organs in scale-worms (Polychaeta, Polynoidae). Zoosymposia 2, 379–389. doi: 10.11646/zoosymposia.2.1.26
Prasher, D. C., Eckenrode, V. K., Ward, W. W., Prendergast, F. G., and Cormier, M. J. (1992). Primary structure of the Aequorea victoria green-fluorescent protein. Gene 111, 229–233.
Prasher, D., McCann, R. O., and Cormier, M. J. (1985). Cloning and expression of the cDNA coding for aequorin, a bioluminescent calcium-binding protein. Biochem. Biophys. Res. commun. 126, 1259–1268. doi: 10.1016/0006-291X(85)90321-3
Purtov, K. V., Petushkov, V. N., Baranov, M. S., Mineev, K. S., Rodionova, N. S., Kaskova, Z. M., et al. (2015). The chemical basis of fungal bioluminescence. Angew. Chem. 127, 8242–8246. doi: 10.1002/ange.201501779
Rawat, R., and Deheyn, D. D. (2016). Evidence that ferritin is associated with light production in the mucus of the marine worm Chaetopterus. Sci. Rep 6:36854.
Rees, J. F., De Wergifosse, B., Noiset, O., Dubuisson, M., Janssens, B., and Thompson, E. M. (1998). The origins of marine bioluminescence: turning oxygen defence mechanisms into deep-sea communication tools. J. Exp. Biol. 201, 1211–1221. doi: 10.1242/jeb.201.8.1211
Renwart, M., and Mallefet, J. (2013). First study of the chemistry of the luminous system in a deep-sea shark, Etmopterus spinax Linnaeus, 1758 (Chondrichthyes: Etmopteridae). J. Exp. Mar. Biol. Ecol. 448, 214–219. doi: 10.1016/j.jembe.2013.07.010
Rohr, J., Latz, M. I., Fallon, S., Nauen, J. C., and Hendricks, E. (1998). Experimental approaches towards interpreting dolphin-stimulated bioluminescence. J. Exp. Biol. 201, 1447–1460. doi: 10.1242/jeb.201.9.1447
Rosenberg, J., and Meyer-Rochow, V. B. (2009). ). Luminescent Myriapoda: a Brief Review. Bioluminescence in Focus—a Collection of Illuminating Essays. Trivandrum: Research Signpost, 139–146.
Rudie, N. G., Mulkerrin, M. G., and Wampler, J. E. (1981). Earthworm bioluminescence: characterization of high specific activity Diplocardia longa luciferase and the reaction it catalyzes. Biochemistry 20, 344–350. doi: 10.1021/bi00505a018
Schnitzler, C. E., Pang, K., Powers, M. L., Reitzel, A. M., Ryan, J. F., Simmons, D., et al. (2012). Genomic organization, evolution, and expression of photoprotein and opsin genes in Mnemiopsis leidyi: a new view of ctenophore photocytes. BMC Biol. 10:107. doi: 10.1186/1741-7007-10-107
Schultz, D. T., Kotlobay, A. A., Ziganshin, R., Bannikov, A., Markina, N. M., Chepurnyh, T. V., et al. (2018). Luciferase of the Japanese syllid polychaete Odontosyllis umdecimdonta. Biochem. Biophys. Res. Commun. 502, 318–323. doi: 10.1016/j.bbrc.2018.05.135
Schultz, L. W., Liu, L., Cegielski, M., and Hastings, J. W. (2005). Crystal structure of a pH-regulated luciferase catalyzing the bioluminescent oxidation of an open tetrapyrrole. Proc. Natl Acad. Sci. U.S.A. 102, 1378–1383. doi: 10.1073/pnas.0409335102
Seliger, H. H. (1975). The origin of bioluminescence. Photochem. Photobiol. 21, 355–361. doi: 10.1111/j.1751-1097.1975.tb06684.x
Shimomura, O. (1980). Chlorophyll-derived bile pigment in bioluminescent euphausiids. FEBS Letters 116, 203–206. doi: 10.1016/0014-5793(80)80644-2
Shimomura, O. (1986). Bioluminescence of the brittle star Ophiopsila californica. Photochem. Photobiol. 44, 671–674. doi: 10.1111/j.1751-1097.1986.tb04724.x
Shimomura, O. (1987). Presence of coelenterazine in non-bioluminescent marine organisms. Comparat. Biochem. Physiol. Part B 86, 361–363. doi: 10.1016/0305-0491(87)90306-3
Shimomura, O. (1995). The roles of the two highly unstable components F and P involved in the bioluminescence of euphausiid shrimps. J. Biolumin. Chemilumin. 10, 91–101. doi: 10.1002/bio.1170100205
Shimomura, O. (2005). The discovery of aequorin and green fluorescent protein. J. Microsc. 217, 3–15. doi: 10.1111/j.0022-2720.2005.01441.x
Shimomura, O. (2008). The Photoproteins. Protein Science Encyclopedia. Weinheim, Germany: A.R. Fersht. 1–23. Available online at: https://onlinelibrary.wiley.com/doi/abs/10.1002/9783527610754.mp12
Shimomura, O. (2012). Bioluminescence: Chemical Principles and Methods. Singapore: World Scientific.
Shimomura, O., and Flood, P. R. (1998). Luciferase of the scyphozoan medusa Periphylla periphylla. Biol. Bull. 194, 244–252. doi: 10.2307/1543094
Shimomura, O., and Johnson, F. H. (1968). Chaetopterus photoprotein: crystallization and cofactor requirements for bioluminescence. Science 159, 1239–1240. doi: 10.1126/science.159.3820.1239
Shimomura, O., Johnson, F. H., and Saiga, Y. (1963). Partial purification and properties of the Odontosyllis luminescence system. J. Cell. Comp. Physiol. 61, 275–292. doi: 10.1002/jcp.1030610309
Shimomura, O., Masugi, T., Johnson, F. H., and Haneda, Y. (1978). Properties and reaction mechanism of the bioluminescence system of the deep-sea shrimp Oplophorus gracilorostris. Biochemistry 17, 994–998. doi: 10.1021/bi00599a008
Sweeney, B. M. (2012). “The bioluminescence of dinoflagellates,” in Biochemistry and Physiology of Protozoa, eds M. Levendowsky and S. H. Hutner (New York, NY: Academic Press), 287–306. doi: 10.1016/b978-0-12-444601-4.50016-3
Takenaka, Y., Masuda, H., Yamaguchi, A., Nishikawa, S., Shigeri, Y., Yoshida, Y., et al. (2008). Two forms of secreted and thermostable luciferases from the marine copepod crustacean. Metridia pacifica. Gene 425, 28–35. doi: 10.1016/j.gene.2008.07.041
Takenaka, Y., Yamaguchi, A., Tsuruoka, N., Torimura, M., Gojobori, T., and Shigeri, Y. (2012). Evolution of bioluminescence in marine planktonic copepods. Mol. Biol. Evol. 29, 1669–1681. doi: 10.1093/molbev/mss009
Tanet, L., Martini, S., Casalot, L., and Tamburini, C. (2020). Reviews and syntheses: Bacterial bioluminescence–ecology and impact in the biological carbon pump. Biogeosciences 17, 3757–3778. doi: 10.5194/bg-17-3757-2020
Taylor, L. R., Compagno, L. J., and Struhsaker, P. J. (1983). Megamouth–a new species, genus and family of lamnoid shark (Megachasma pelagios, family Megachasmidae) from the Hawaiian Islands. Proc. Calif. Acad. Sci. 43, 87–110.
Teranishi, K., and Shimomura, O. (2008). Bioluminescence of the arm light organs of the luminous squid Watasenia scintillans. Biochimica Biophysica Acta (BBA) 1780, 784–792. doi: 10.1016/j.bbagen.2008.01.016
Tessler, M., Gaffney, J. P., Crawford, J. M., Trautman, E., Gujarati, N. A., Alatalo, P., et al. (2018). Luciferin production and luciferase transcription in the bioluminescent copepod Metridia lucens. PeerJ 6:e5506. doi: 10.7717/peerj.5506
Tessler, M., Gaffney, J. P., Oliveira, A. G., Guarnaccia, A., Dobi, K. C., Gujarati, N. A., et al. (2020). A putative chordate luciferase from a cosmopolitan tunicate indicates convergent bioluminescence evolution across phyla. Sci. Rep. 10:17724. doi: 10.1038/s41598-020-73446-w
Thomson, C. M., Herring, P. J., and Campbell, A. K. (1997). The widespread occurrence and tissue distribution of the imidazolopyrazine luciferins. J. Biolumin. Chemilumin. 12, 87–91. doi: 10.1002/(sici)1099-1271(199703/04)12:2<87::aid-bio438>3.0.co;2-8
Thouand, G., and Marks, R. (eds) (2014). Bioluminescence: Fundamentals and Applications in Biotechnology, Vol. 2. Berlin: Springer.
Timmins, G. S., Jackson, S. K., and Swartz, H. M. (2001). The evolution of bioluminescent oxygen consumption as an ancient oxygen detoxification mechanism. J. Mol. Evol. 52, 321–332. doi: 10.1007/s002390010162
Topalov, G., and Kishi, Y. (2001). Chlorophyll catabolism leading to the skeleton of dinoflagellate and krill luciferins: hypothesis and model studies. Angewan. Chem. Int. Ed. 40, 3892–3894. doi: 10.1002/1521-3773(20011015)40:20<3892::aid-anie3892>3.0.co;2-h
Trainor, G. L. (1979). Studies on the Odontosyllis Bioluminescence System, Ph.D. Dissertation. Cambridge, MA: Harvard University.
Tsarkova, A. S., Kaskova, Z. M., and Yampolsky, I. V. (2016). A tale of two luciferins: fungal and earthworm new bioluminescent systems. Acc. Chem. Res. 49, 2372–2380. doi: 10.1021/acs.accounts.6b00322
Tsuji, F. I. (1985). ATP-dependent bioluminescence in the firefly squid. Watasenia scintillans. Acc. Chem. Res. Proc. Natl. Acad. Sci. 82, 4629–4632. doi: 10.1073/pnas.82.14.4629
Tsuji, F. I., and Haneda, Y. (1971). Luminescent system in a myctophid fish. Diaphus elucens Brauer. Nature 233, 623–624. doi: 10.1038/233623a0
Tsuji, F. I., Ohmiya, Y., Fagan, T. F., Toh, H., and Inouye, S. (1995). Molecular evolution of the Ca2+-binding photoproteins of the hydrozoa. Photochem. Photobiol. 62, 657–661. doi: 10.1111/j.1751-1097.1995.tb08713.x
Tyler, S. (1988). The role of function in determination of homology and convergence-examples from invertebrates adhesive organs. Fortsch. Zool. 36, 331–347.
Ulitzur, S., and Hastings, J. W. (1979). Evidence for tetradecanal as the natural aldehyde in bacterial bioluminescence. Proc. Natl. Acad. Sci.U.S.A. 76, 265–267. doi: 10.1073/pnas.76.1.265
Valiadi, M., and Iglesias-Rodriguez, D. (2013). Understanding bioluminescence in dinoflagellates—how far have we come? Microorganisms 1, 3–25. doi: 10.3390/microorganisms1010003
Verdes, A., and Gruber, D. F. (2017). Glowing worms: biological, chemical, and functional diversity of bioluminescent annelids. Integr. Comp. Biol. 57, 18–32. doi: 10.1093/icb/icx017
Viviani, V. R. (2002). The origin, diversity, and structure function relationships of insect luciferases. Cell. Mol. Life Sci. 59, 1833–1850. doi: 10.1007/PL00012509
Viviani, V. R., Hastings, J. W., and Wilson, T. (2002). Two bioluminescent Diptera: the north american Orfelia fultoni and the australian Arachnocampa flava. Similar niche, different bioluminescence systems. Photochem. Photobiol. 75, 22–27. doi: 10.1562/0031-865520020750022TBDTNA2.0.CO2
Warner, J. A., and Case, J. F. (1980). The zoogeography and dietary induction of bioluminescence in the midshipman fish, Porichthys notatus. Biol. Bull. 159, 231–246. doi: 10.2307/1541021
Watkins, O. C., Sharpe, M. L., Perry, N. B., and Krause, K. L. (2018). New Zealand glowworm (Arachnocampa luminosa) bioluminescence is produced by a firefly-like luciferase but an entirely new luciferin. Sci. Rep. 8:3278.
Weng, J. K. (2014). The evolutionary paths towards complexity: a metabolic perspective. New Phytol. 201, 1141–1149. doi: 10.1111/nph.12416
Widder, E. A. (2010). Bioluminescence in the ocean: origins of biological, chemical, and ecological diversity. Science 328, 704–708. doi: 10.1126/science.1174269
Wiens, M., Wang, X., Unger, A., Schröder, H. C., Grebenjuk, V. A., Pisignano, D., et al. (2010). Flashing light signaling circuit in sponges: endogenous light generation after tissue ablation in Suberites domuncula. J. Cell. Biochem. 111, 1377–1389. doi: 10.1002/jcb.22866
Wilson, T., and Hastings, J. (2013). Bioluminescence: Living Lights, Lights for Living. Cambridge, MA: Harvard University Press, 185. doi: 10.1002/bio.2543
Wilson, T., and Hastings, J. W. (1998). Bioluminescence. Annu. Rev. Cell Dev. Biol. 14, 197–230. doi: 10.1146/annurev.cellbio.14.1.197
Yasuno, R., Mitani, Y., and Ohmiya, Y. (2018). Effects of N-glycosylation deletions on Cypridina luciferase activity. Photochem. Photobiol. 94, 338–342. doi: 10.1111/php.12847
Keywords: molecular evolution, photoproteins, photobiology, convergence, bioluminescence
Citation: Delroisse J, Duchatelet L, Flammang P and Mallefet J (2021) Leaving the Dark Side? Insights Into the Evolution of Luciferases. Front. Mar. Sci. 8:673620. doi: 10.3389/fmars.2021.673620
Received: 01 March 2021; Accepted: 17 May 2021;
Published: 30 June 2021.
Edited by:
Cassius Vinicius Stevani, University of São Paulo, BrazilReviewed by:
Anderson Garbuglio Oliveira, University of São Paulo, BrazilCopyright © 2021 Delroisse, Duchatelet, Flammang and Mallefet. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jérôme Delroisse, SmVyb21lLkRlbHJvaXNzZUB1bW9ucy5hYy5iZQ==; Jérôme Mallefet, SmVyb21lLk1hbGxlZmV0QHVjbG91dmFpbi5iZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.