
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
POLICY AND PRACTICE REVIEWS article
Front. Mar. Sci., 31 May 2021
Sec. Marine Affairs and Policy
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.667274
A correction has been applied to this article in:
Corrigendum: Marine Genetic Resources in Areas Beyond National Jurisdiction: Promoting Marine Scientific Research and Enabling Equitable Benefit Sharing
 Alex D. Rogers1*
Alex D. Rogers1* Amy Baco2
Amy Baco2 Elva Escobar-Briones3
Elva Escobar-Briones3 Duncan Currie4
Duncan Currie4 Kristina Gjerde5
Kristina Gjerde5 Judith Gobin6
Judith Gobin6 Marcel Jaspars7
Marcel Jaspars7 Lisa Levin8
Lisa Levin8 Katrin Linse9
Katrin Linse9 Muriel Rabone10
Muriel Rabone10 Eva Ramirez-Llodra1
Eva Ramirez-Llodra1 Javier Sellanes11
Javier Sellanes11 Timothy M. Shank12
Timothy M. Shank12 Kerry Sink13,14
Kerry Sink13,14 Paul V. R. Snelgrove15
Paul V. R. Snelgrove15 Michelle L. Taylor16
Michelle L. Taylor16 Daniel Wagner17
Daniel Wagner17 Harriet Harden-Davies18
Harriet Harden-Davies18Growing human activity in areas beyond national jurisdiction (ABNJ) is driving increasing impacts on the biodiversity of this vast area of the ocean. As a result, the United Nations General Assembly committed to convening a series of intergovernmental conferences (IGCs) to develop an international legally-binding instrument (ILBI) for the conservation and sustainable use of marine biological diversity of ABNJ [the biodiversity beyond national jurisdiction (BBNJ) agreement] under the United Nations Convention on the Law of the Sea. The BBNJ agreement includes consideration of marine genetic resources (MGR) in ABNJ, including how to share benefits and promote marine scientific research whilst building capacity of developing states in science and technology. Three IGCs have been completed to date with the fourth delayed by the Covid pandemic. This delay has allowed a series of informal dialogues to take place between state parties, which have highlighted a number of areas related to MGR and benefit sharing that require technical guidance from ocean experts. These include: guiding principles on the access and use of MGR from ABNJ; the sharing of knowledge arising from research on MGR in ABNJ; and capacity building and technology transfer for developing states. In this paper, we explain what MGR are, the methods required to collect, study and archive them, including data arising from scientific investigation. We also explore the practical requirements of access by developing countries to scientific cruises, including the sharing of data, as well as participation in research and development on shore whilst promoting rather than hindering marine scientific research. We outline existing infrastructure and shared resources that facilitate access, research, development, and benefit sharing of MGR from ABNJ; and discuss existing gaps. We examine international capacity development and technology transfer schemes that might facilitate or complement non-monetary benefit sharing activities. We end the paper by highlighting what the ILBI can achieve in terms of access, utilization, and benefit sharing of MGR and how we might future-proof the BBNJ Agreement with respect to developments in science and technology.
Areas beyond national jurisdiction (ABNJ) lie outside of the exclusive economic zones of coastal States covering 61% of the ocean surface and 73% of its volume (O’Leary et al., 2019). This vast area, contains a rich biodiversity of marine life representing nearly 4 billion years of evolution, ranging from viruses and bacteria to the largest animal ever to exist on Earth, the blue whale. Because of gaps in the governance framework of ABNJ and increasing threats to its marine biodiversity, the United Nations General Assembly has committed to convene a series of intergovernmental conferences (IGCs) to develop an international legally-binding instrument (ILBI) for the conservation and sustainable use of marine biological diversity of ABNJ (Biodiversity Beyond National Jurisdiction or BBNJ agreement) under UNCLOS (UNGA, 2017; Wright et al., 2018). The BBNJ agreement, focuses on four main areas: Marine Genetic Resources (MGR), area-based management tools, including marine protected areas; environmental impact assessments (EIAs); and capacity building and technology transfer (UNGA, 2015, 2017). This paper focuses on MGR, including the practicalities of equitable access to them and the sharing of benefits arising from their use.
Marine genetic resources include the genetic information marine organisms host enabling them to produce a wide range of biochemicals (Jaspars et al., 2016) that can benefit humankind through applications of biodiscovery of pharmaceutical compounds, cosmetics, food supplements, research tools, and in industrial processes (Blasiak et al., 2020a,b; Harden-Davies, 2020). They also include adaptive solutions found in deep-sea organisms that can also inspire novel materials and structural designs (e.g., light conductance and protective materials; Sundar et al., 2003; Yao et al., 2010). Additional applications of new genetic tools and technologies include tackling invasive species, prevention of biofouling on the hulls of vessels, bioremediation and wildlife management. An understanding of the structure and function of the genetic diversity of ocean life is also crucial for assessing adaptation potential in a changing climate, and for the conservation and sustainable use of marine biodiversity (Supplementary Material 1.0).
Marine genetic resources, including the access and sharing of both monetary and non-monetary benefits (see Supplementary Material 2.0), are complex topics in themselves, as is understanding of the different steps involved in marine scientific research: from exploration at sea, to the analysis and archiving of resulting collections and data; to production of scientific output, publications, decision support tools and technological innovation and development (Rabone et al., 2019). The BBNJ agreement prioritizes promoting, and not inhibiting marine scientific research and innovation; recognizing the benefits of ABNJ to human wellbeing; the key role of science in management and conservation; and in lifting the capacity of all States to support equitable sharing in the benefits of research, technology development, capacity building and innovation (UN, 2019). However, much of the text relating to MGR in the ‘Revised Draft Text of the BBNJ agreement’ (UN, 2019) remains in square brackets, indicating a persisting divergence of views between States on several aspects of this element of the BBNJ negotiations. The issue of MGR has therefore become critical because the BBNJ agreement is a package deal where “nothing is agreed until everything is agreed” (Danilenko, 1993; Wright et al., 2018).
Science and capacity building play prominent roles throughout the provisions on MGR, including in relation to access and sharing benefits from MGR and contributing to the conservation and sustainable use of BBNJ. The United Nations Convention on the Law of the Sea (UNCLOS) gives all States the right to conduct marine scientific research and to use living resources sustainably in ABNJ (Wright et al., 2018; see UNCLOS Articles 143, 238, and 241). These rights are conditional on duties, in accordance with UNCLOS and the general principles of international law, including: the protection and preservation of the marine environment, conservation of high-seas living marine resources, capacity building and technology transfer (Harden-Davies and Gjerde, 2019; Harden-Davies and Snelgrove, 2020). However, because of the remoteness of ABNJ, far from coastal waters and typically at depths well below 200 m, access presents substantial technological challenges, necessitating significant technical, human and financial resources available only to a few States (Bernal and Simcock, 2016; UNESCO, 2017). Requirements for research in ABNJ include: state-of-the-art scientific research vessels equipped with modern sampling tools, including for deep-sea research, and trained personnel in all disciplines of marine sciences and associated technologies (Figure 1; Broggiato et al., 2014; Glover et al., 2016). These vessels, which cost between $25,000 USD (TDI Brooks Proteus) to >$87,500 USD per day (Japan Agency for Marine-Earth Science and Technology’s vessel Yokosuka with submersible Shinkai 6500) excluding fuel costs, are typically operated as publicly funded facilities of developed States, or by offshore industries or philanthropic organizations (e.g., the Schmidt Ocean Institute’s Falkor). Some organizations run research vessels which are affiliated with government institutions (e.g., the 501(c)(3) non-profit organization Ocean Exploration Trust which coordinates with the National Oceanographic and Atmospheric Administration [NOAA]). Research and development involving MGR also require onshore laboratory facilities of varying levels of sophistication, complexity and interaction with industry or government (see below).
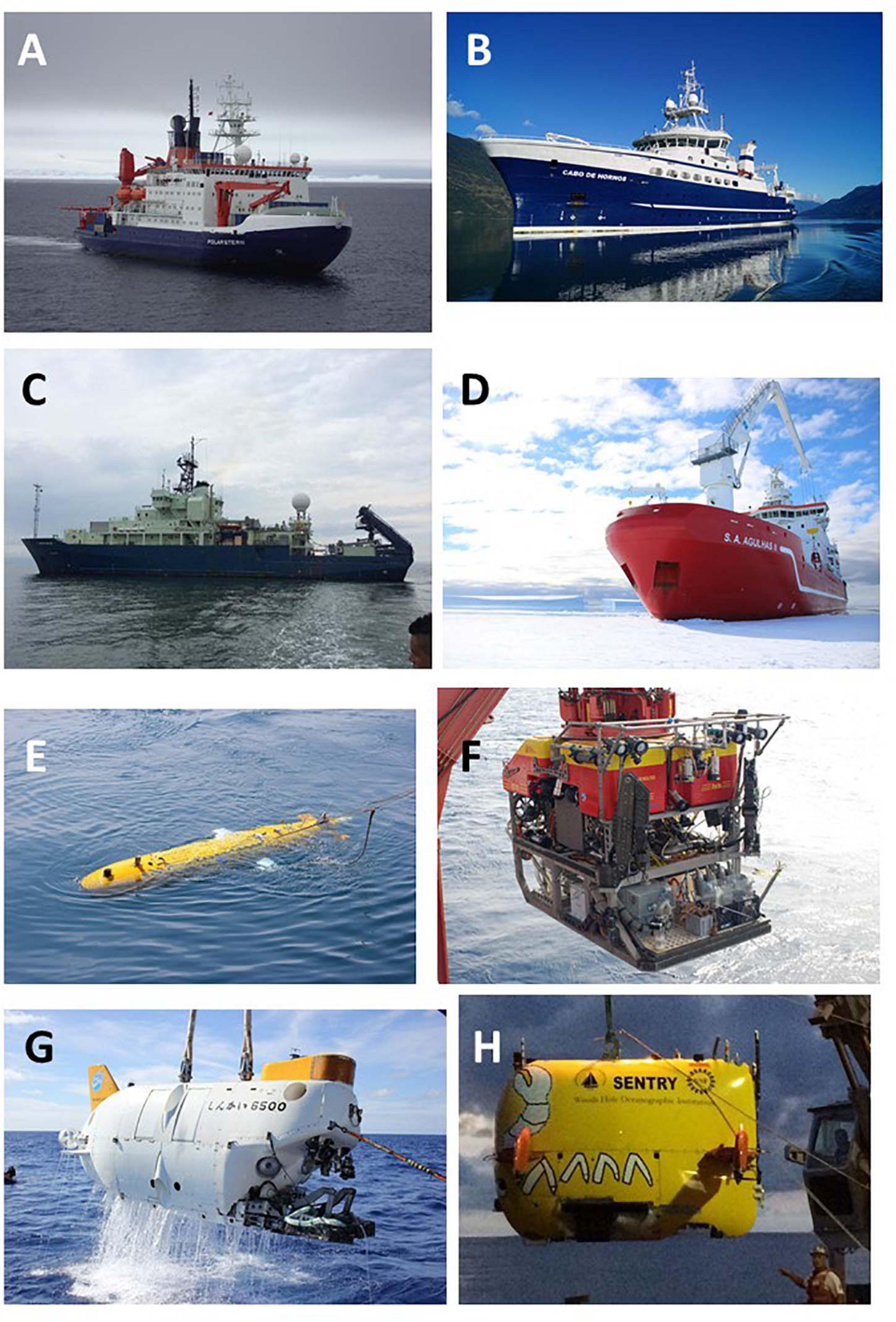
Figure 1. Advanced infrastructure required for marine scientific research in ABNJ. (A) RV Polarstern (Germany; K. Linse), (B) RV Cabo de Hornos (Chile; J. Sellanes), (C) RV Atlantis (United States; T. Shank), (D) SA Agulhas II (South Africa; B. Frinault, University of Oxford), (E) Autosub AUV (United Kingdom; A. D. Rogers), (F) Isis ROV (United Kingdom; A. D. Rogers), (G) Shinkai 6500 submersible (Japan; A. D. Rogers), and (H) Sentry AUV (United States; L. Levin).
Many developing States lack the capacity to access and utilize marine biodiversity and MGR of ABNJ. Even access to collections (e.g., samples of organisms, tissues, and DNA/RNA), data, including digital sequence information (DSI; Rabone et al., 2019; Houssen et al., 2020), or ocean-going vessels may be of limited use if States lack adequately resourced research and educational institutions to acquire, maintain, and update the required infrastructure or cyberinfrastructure, and to strengthen human capacity to assess, analyze, and evaluate such resources (Broggiato et al., 2014; Harden-Davies, 2020).
In this paper, we analyze areas of contention and common misunderstanding in the discourse on access and benefit sharing of MGR in the BBNJ agreement. We intend this analysis to inform ongoing deliberations related to the BBNJ agreement and hope that it will be useful for government representatives as well as supporting experts in ocean law, economics, management, and science. Here, we consider “access1” as access to: (i) specimens or samples of organisms collected from ABNJ, including extracted tissues, DNA, RNA or other extracted biochemicals; (ii) DSI related to organisms collected in ABNJ; (iii) data associated with the collection of samples in ABNJ; (iv) scientific infrastructure, technology and expertise to enable developing States to undertake research and training in research on MGR from ABNJ, including placement of scientists on research vessels and in appropriately resourced laboratories. Our specific objectives are:
(i) To explain MGR and the methods required to explore and collect these resources from the ocean, and access, study and archive them, including data arising from scientific investigation.
(ii) To identify what is practically required to enable access and benefit sharing of MGR without compromising marine scientific research under the BBNJ agreement and to outline current best practices, risks, potential problems, and their solutions including: pre- and post- research cruise notification; sharing of information and technology regarding sampling at sea; and research and development on-shore including management of data and samples.
(iii) To outline existing infrastructure and shared resources that facilitate access, research and development, and benefit sharing of MGR from ABNJ; and discuss existing gaps.
(iv) To provide illustrative examples of current capacity building and technology transfer that might facilitate or complement non-monetary benefit sharing activities and how the BBNJ agreement could strengthen existing practices or create new avenues and opportunities.
(v) Discuss access, utilization, and benefit sharing of MGR in light of future development of technology and how we might future proof the BBNJ agreement with respect to such developments.
The definition of MGR is complex and the BBNJ draft text includes more than one draft definition (see Supplementary Material 3.0). This is not confined to the BBNJ agreement and discussions are also taking place amongst parties to the Convention on Biological Diversity (CBD) particularly with respect to the inclusion of DSI for access and benefit sharing (Lyal and Zhao, 2020). Here we understand MGR as the genetic material present in all marine life, including the physical genes and gene clusters (DNA and RNA), the information they encode (i.e., DSI), and the products of the genes, including, a wide range of molecules (e.g., enzymes, structural proteins, peptides, secondary metabolites), their derivatives, the biological processes they are a part of (e.g., biosynthetic pathways), and the physical structures they form that offer actual or potential value to humankind (Rabone et al., 2019). Value can include: monetary value in terms of commercial products, but also the concept of ‘inherent’ value’: maintenance or improvement of the resilience and/or adaptability of marine species, including exploited species and ecosystems (Harden-Davies and Gjerde, 2019; Marlow et al., 2019). The definition of MGR must cover the genes of organisms, the information they contain, and the variety of potential uses today, and in the future, to be scientifically valid (Rabone et al., 2019) and for all uses of MGR to benefit humankind.
The use of MGR for biodiscovery may occur through several different pathways. These include (from Broggiato et al., 2014; Blasiak et al., 2020a):
(i) In situ pathway: harvesting biological molecules from preserved tissue samples.
(ii) Ex situ pathway: extracting molecules from breeding, cultivation or culturing of living organisms.
(iii) In vitro pathway: expression of DNA coding for specific proteins in cultured cells (genetically modified organisms) to produce large quantities of a protein which can be purified and used for scientific studies or produced in industrial quantities (e.g., insulin).
(iv) In silico pathway: the use of sequences of nucleic acid (DNA or RNA) stored as digital information to create proteins, molecular processes, biogenic precipitates, innovation, genetically modified organisms and synthetic organisms.
When UNCLOS was originally negotiated many aspects of MGR and the technologies used to identify and develop them for commercial use were not known (Leary, 2018). Furthermore, as we outline in this paper, the ways of collecting, identifying, accessing and using knowledge related to MGR is rapidly advancing. Whilst the majority of MGR identified to date are from coastal or deep waters within exclusive economic zones (Leary, 2018) they also exist, sometimes exclusively, within ABNJ (e.g., Yao et al., 2010). We therefore see it as essential to ensure that any definition of MGR in the BBNJ agreement is clear from a scientific perspective. It is desirable for such a definition to be sufficiently broad to cover the full range of aspects of biology that may be of value to humankind under current scientific practice, including any derivative products, and future technological advances (see Supplementary Material 3.0). We also note that whilst non-monetary benefits can be derived from marine scientific research and discovery of MGR in ABNJ it is only when MGR are actually utilized to generate profit that monetary benefits can be triggered.
For developing States to place personnel on board research vessels in ABNJ for non-monetary benefits such as training, capacity building, or the opportunity to collect samples to assess the presence of MGR, prior cruise notification is required. Such notification guarantees transparency in marine scientific research in ABNJ and confidence for developing States that they can identify opportunities for access and benefit sharing of MGR. This notification will also assist in tracing MGR from sample collection to commercial production, a requirement for sharing monetary benefits. Access to records of past cruises (e.g., cruise reports) can also expedite access to MGR, because they typically contain details of biological material collected and where the material has gone for investigation.
Prior cruise notification has been contentious because of concerns that it could delay or even hamper marine scientific research. Some states have also raised security concerns with respect to advanced cruise notification. Given these discussions, there is a need to streamline the currently fragmented and complex practices in cruise notification (Table 1), combine existing vessel databases and provide accessible information on research vessels (Broggiato et al., 2018; Rabone et al., 2019). Such rationalization of scientific cruise notification at a global scale will not only help enable access for scientists from developing States but will also assist in marine scientific research. It will enhance opportunities for collaboration amongst scientists from all States and might also prevent unnecessary replication of scientific sampling reducing the environmental impact of scientific activities. Given the massive costs of global and ocean-class research vessels, this may also lead to more effective usage of resources through better use of existing cruises and by preventing cruise overlap (Oldham et al., 2014; Collins et al., 2021). In Table 1 (see also Supplementary Material 4.0) we survey the current practices of cruise notification, with examples provided for countries in different geographic regions, as well as for international programs. We note the current context of COVID-related issues with research vessel operations adds further complexity to science cruise planning and may influence some of our observations of current practice.
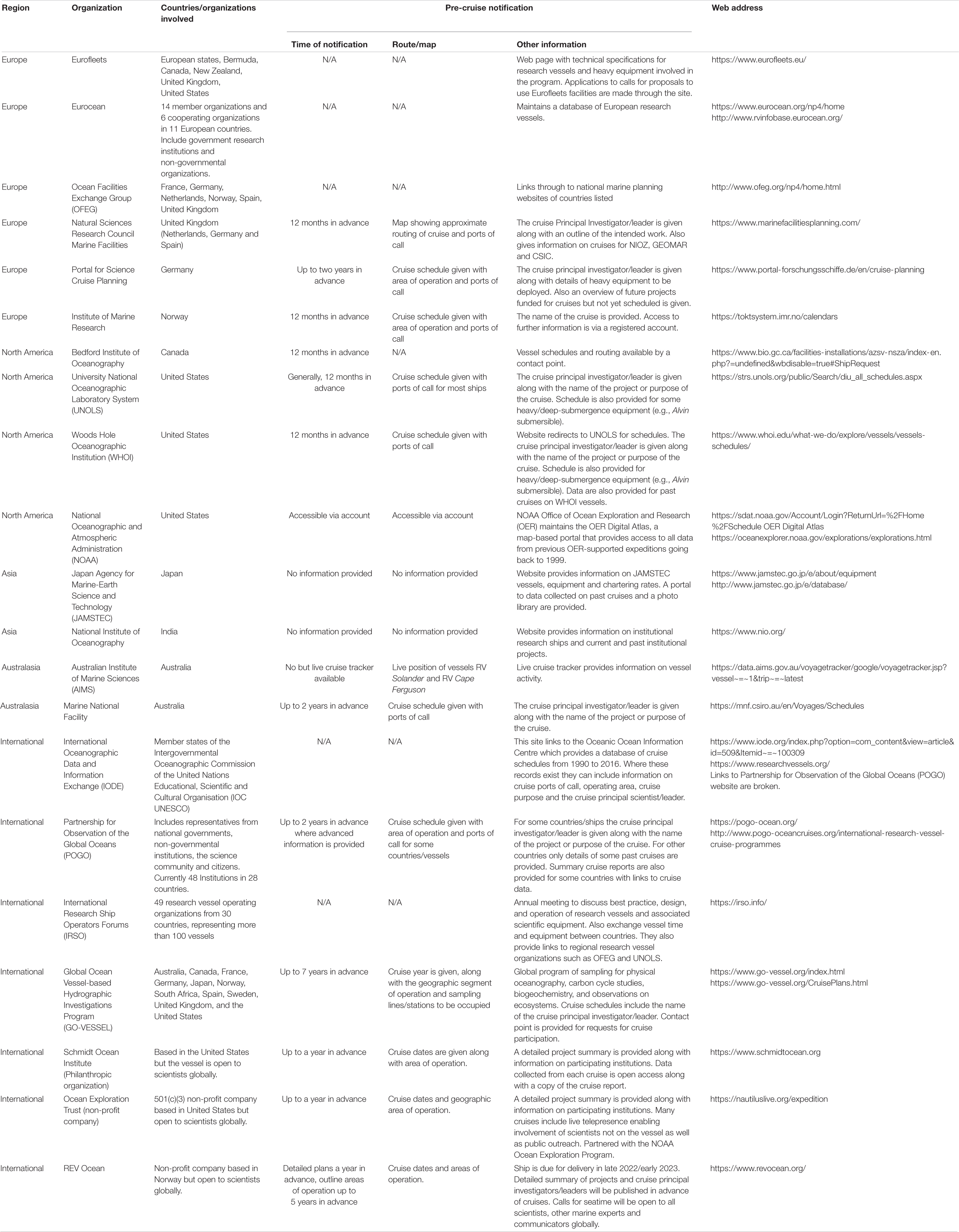
Table 1. Pre-cruise notification practices at national and international level. Organisations which are involved in international cooperation on marine scientific research vessel operations are also included.
Fishing states collect biological samples in ABNJ as a contribution to scientific management of high-seas fisheries by regional fisheries management organizations and these may also provide opportunities for access and benefit sharing for MGR. Industrial activities in ABNJ may necessitate EIAs or other scientific studies (e.g., mineral prospecting) prior to full implementation of projects. This will likely increase following the implementation of the BBNJ agreement as it will require EIA of all activities potentially impacting biodiversity in ABNJ. Such industries include: offshore cables for electricity transmission or telecommunications; renewable energy projects; aquaculture; climate remediation actions including carbon capture and storage; and deep-seabed minerals mining. At present, there is no mechanism for prior notification of such cruises or for archiving of resulting reports, data or samples from such activities with the exception of those related to deep-sea mining which are subject to special requirements by the International Seabed Authority2. While industry cruises specifically for the purposes of investigating MGR are unlikely for economic reasons, it should be noted that companies involved in mineral prospecting for deep-sea mining have already collected biological samples from the seabed and, in some cases, these appear to be available for bioprospecting purposes (e.g., Deep Green, 2020). It is also notable that cruises undertaken with government funding have partnered with private organizations to target MGR for screening for pharmaceutical compounds within the Exclusive Economic Zone of coastal States (e.g., E/V Nautilus Cruise NA124 funded by NOAA3).
Global, ocean and regional-class research vessels, as well as some Remotely-Operated Vehicles (ROVs) and Autonomous Underwater Vehicles (AUVs) are normally used for collection of samples and data from ABNJ (Figure 1). However, other ocean observation systems are used to collect physical oceanographic and geological data (e.g., tsunami warning systems). Increasingly, these autonomous platforms are able to collect biogeochemical and biological data and samples. The major international network which coordinates these platforms is the Global Ocean Observing System (GOOS4). This network includes more than 3,700 Argo, 16 Deep Argo and 275 Bio-Argo floats, surface drifters, fixed sensor platforms, ice buoys, moored buoys and tsunameters. Whilst this infrastructure to date has not collected samples relevant to MGR research, current developments in autonomous platform technology include the ability to collect and sequence DNA (e.g., the Environmental Sample Processor, ESP; Hansen et al., 2020). Similarly, ocean observatories (fixed arrays of sensors, cameras and sampling equipment), including those located at a considerable distance from the coast, in deep water, already sample microorganisms for DNA sequence analyses over time (e.g., Axial Seamount Array; Smith et al., 2018). It is therefore important that access and benefit sharing arrangements incorporated within the BBNJ agreement include the entire range of specimen and data collection technologies currently used in the ocean and which are in development now or in the future. The Deep Ocean Observing Strategy (DOOS) noted the fragmented nature of deep-ocean observations from GoShip, Argo, OceanSITES, observatories and gliders and proposed coordination of sustained observing activities that provide critical context for MGR (Levin et al., 2019). Samples arising from such platforms could also be archived in a similar way to samples from research cruises. This would necessitate a better integration of such observation programs with museums or other institutions that can archive and curate such samples over the long term (see below).
Current practice in advanced cruise notification varies among countries or even among organizations within a country (Table 1). Some countries provide advanced notification of cruises and cruise details (e.g., United Kingdom, United States) while other countries do not or these are not readily discoverable (e.g., China, Japan, Korea, India). Some countries can provide advanced details on cruises through contact with the cruise planning office (e.g., Canada). However, information can vary even within a country (e.g., Germany).
A globally available platform for pre-cruise notification and post-cruise reporting will assist in opening access and expediting benefit sharing for MGR in ABNJ, as well as benefiting marine scientific research. Given the vast size of ABNJ yet to be studied by marine scientists, such a rationalization of global cruise effort will benefit marine science and humanity overall. The United Nations Educational, Scientific and Cultural Organisation’s (UNESCO) Intergovernmental Oceanographic Commission’s (IOC) International Ocean Data Exchange (IODE) Oceanic site offers a suitable platform for notification of science cruises in ABNJ internationally, although this site currently lacks details for cruises beyond 2016 (Table 1). It provides the architecture for pre-cruise and post-cruise notification, possibly with some modifications such as a mechanism to search for cruises by geographic area5, to act as a clearinghouse for announcement of future cruises and for information on past cruises. Ideally, this clearinghouse would be integrated with existing systems for cruise notification rather than a new standalone system. Alternatively, a public private partnership, such as with the Ocean Data Platform (ODP6) might also work well, or integration between IODE, ODP, and other networks such as DOOS.
Many sites for cruise notification contained broken links to other addresses reportedly holding cruise information or gaps in information, such as providing schedules for some vessels and not others. This inconsistency suggests a need for increased rigor to meet requirements for advanced cruise notification and for providing details of past cruise activities. This requires consideration of: (a) resource requirements to maintain the information flows and (b) buy-in from national focal points to whom the scientific community would provide the cruise details. Creating a system where it is a requirement that all principal investigators for scientific cruises, both publicly and privately funded, fill out an online form comprising the basic cruise information or relevant data would be an easy means to achieve this.
Where sensitivity exists regarding security for specific research vessel fleets, a system of notification of publicly and privately funded future science projects in ABNJ could be initiated that does not provide specific cruise dates, the vessel and ports of call, and instead outlines the science activity and region of operation. Security concerns regarding advanced cruise notification, however, must be considered in the context of real time positional data provided by many vessels for reasons of maritime safety. Most research vessels can be tracked in real time via their Automated Identification System (AIS) on the Marine Traffic website7 with some exceptions (e.g., some Chinese research vessels).
Scientists have routinely sampled living organisms from the deep ocean since the mid-nineteenth century (e.g., the HMS Challenger expedition 1872–1876). Some technologies have changed little over many decades or even a hundred years or more and sampling still uses over-the-side equipment (e.g., trawls, dredges, water sampling bottles [Niskins], corers), although with greater emphasis on maintaining sample integrity and avoiding contamination. More recently, driven by advances in technology associated with offshore industries, scientists have begun to use submersibles, ROVs, AUVs and autonomous surface vehicles (ASVs) (Baker et al., 2020). These devices can collect samples containing multiple organisms or, with more recent and precise technologies, discrete samples of single animals or even a part of a single animal (Figure 2). These vehicles allow precise sampling and non-intrusive monitoring, reducing impacts on the marine environment. Both benthic landers, deployed for short periods, and long-term observatories may also have technology incorporated to take samples and sensor data over time. Baited traps can also be deployed from the surface or from submersibles or ROVs to collect scavenging animals. Comprehensive coverage of all forms of current biological sampling techniques for the deep ocean is provided in Clark et al. (2016) and general marine sampling in other publications (e.g., Harris et al., 2000; Selvanes et al., 2017).
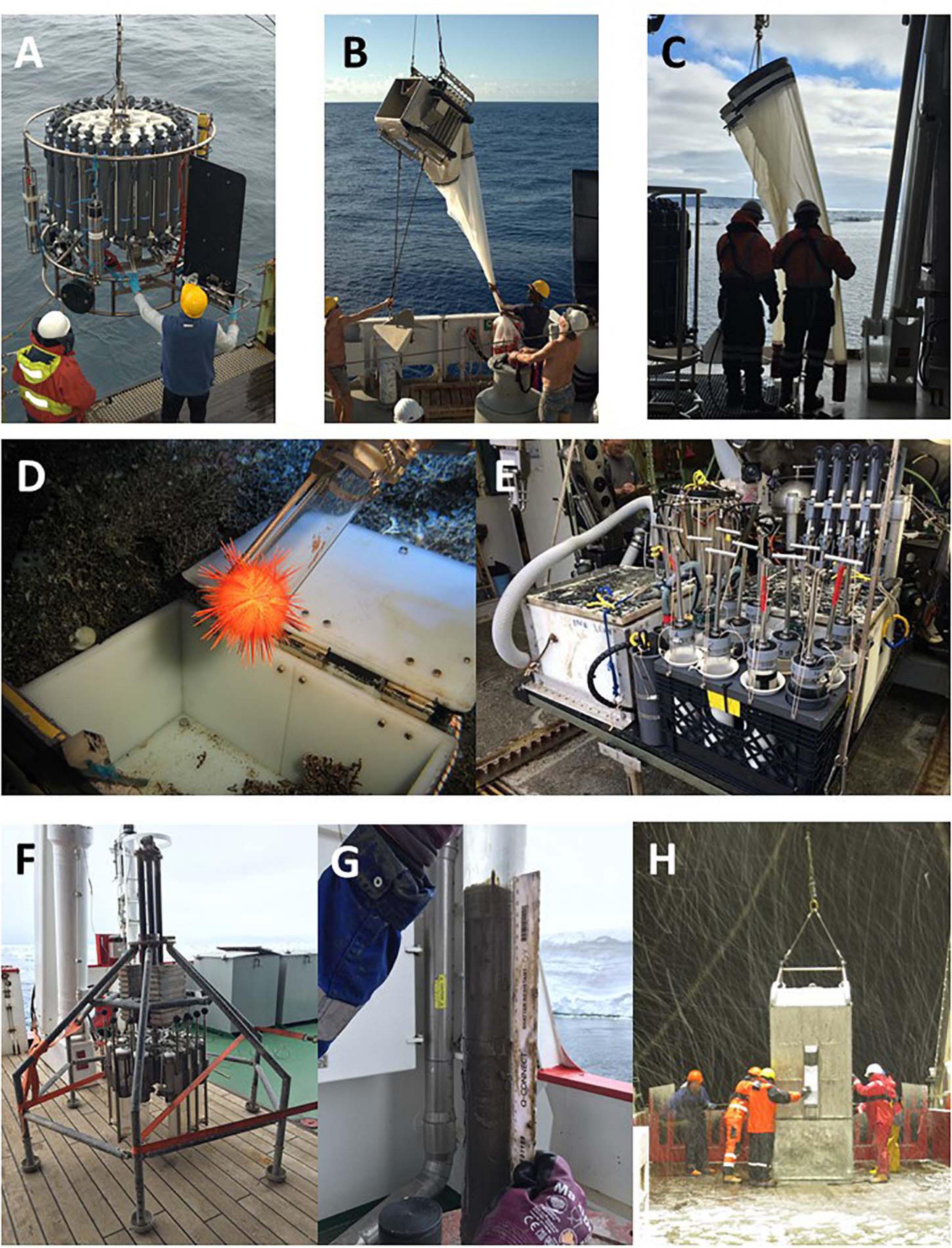
Figure 2. Sampling equipment for marine scientific research. (A) Water sampling rosette with Conductivity, Temperature, and Depth (CTD) sensors for collecting water samples for physical oceanographic measurements, biogeochemistry, and microbiology (A. D. Rogers). (B) Macrozooplankton trawl (A. D. Rogers); a multiple net system which collects samples of small zooplankton. (C) Bongo net for taking vertically hauled samples of plankton (M. L. Taylor). (D) Specimen of the urchin Dermechinus horridus being sampled using an ROV “slurp gun” (A. D. Rogers). (E) Science skid of the submersible Alvin showing push cores and biobox for samples (L. Levin). (F) Multicorer for deployment over the side of a research vessel (M. L. Taylor). (G) Core tube with sample of sediment (M. L. Taylor). (H) Epibenthic sledge, a type of trawl used to take samples of megafauna from the seafloor (M. L. Taylor).
Once collected, samples are processed on board the vessel prior to transport back to land, where further processing and investigation takes place in a laboratory in a university, museum or other research institution (Figures 3, 4 and Supplementary Figure 5). Giving each individual sample a unique identifier post-collection that ties the sample to a specific sampling event along with the associated environmental data represents a critical stage in the collection of samples at sea (Rabone et al., 2019; Figures 3, 4). Such data typically include environmental information such as water depth, temperature, salinity, oxygen, fluorescence (a proxy for chlorophyll concentrations), turbidity, and other parameters all marked by Greenwich Mean Time (GMT) or local time and ship’s and/or vehicle’s navigation information. Data will also include a variety of types and formats. For submersibles, ROVs, and other sampling devices fitted with cameras, data may also include video film and/or photographs of the sampling event. Coupled with multibeam-generated bathymetric maps of sampling locations, these data can provide detailed topographic and geophysical context for where the collected organisms live. Without a unique identifier, tying a sample or specimen to a specific location, water depth, date and time, most forms of subsequent investigation such as taxonomic identification and/or description, ecological studies, EIAs or strategic environmental assessments are impossible or severely compromised. For example, description of a new species requires information on the type locality, the place where the sample was collected. Therefore, best practices widely adopted by marine scientists, already include allocating a unique identifier to samples/specimens and recording environmental data associated with their collection (Glover et al., 2016; Schiaparelli et al., 2016).
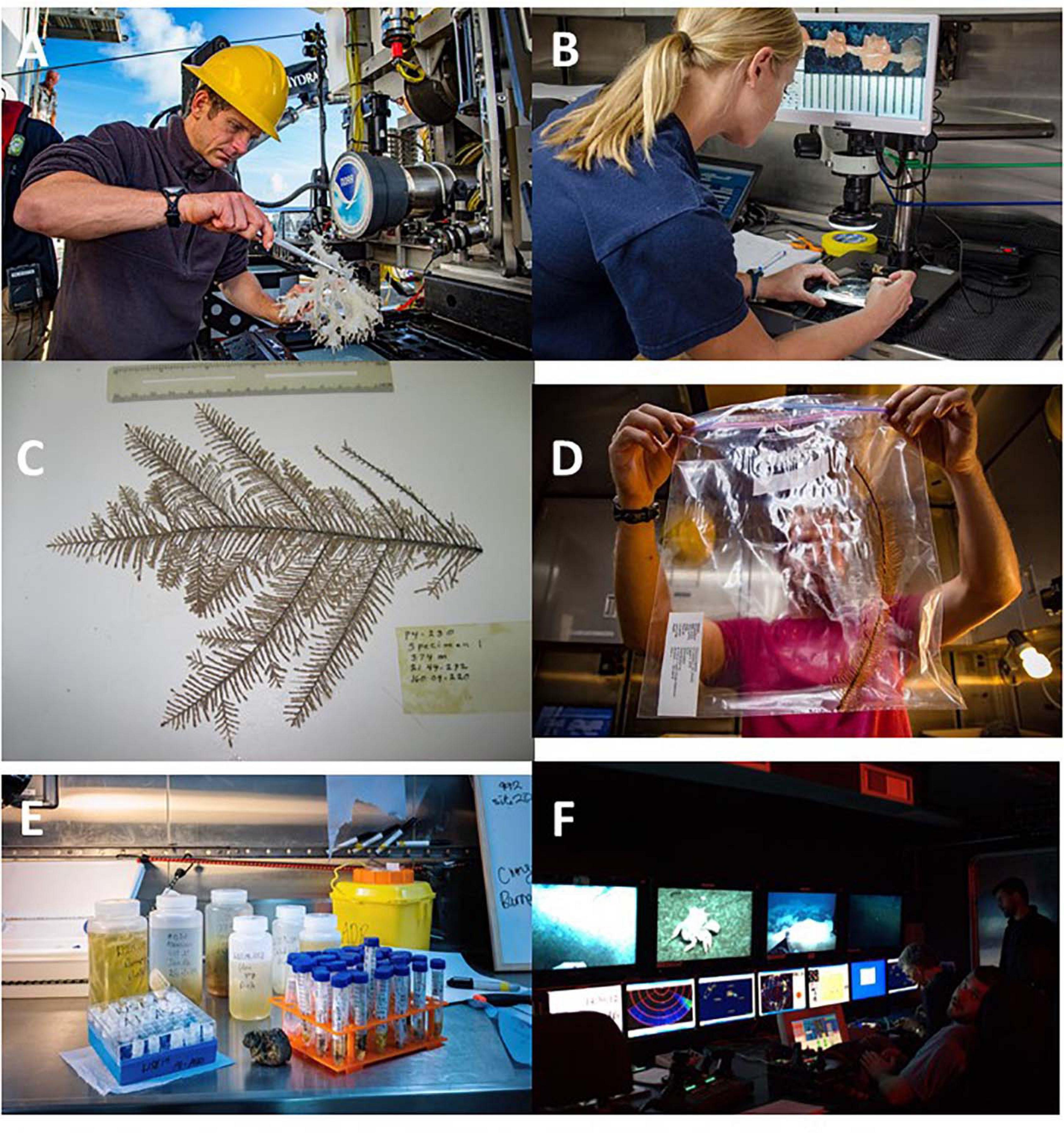
Figure 3. Handling of samples during marine scientific research. (A) Sponge specimen being removed from biobox on ROV science skid (D. Wagner). (B) Specimen of octocoral being photographed in the laboratory on board a ship (D. Wagner). (C) Photograph of black coral specimen showing label with sample details (D. Wagner). (D) Specimen of black coral in plastic bag for preservation with label inserted in bag with sample details (D. Wagner). (E) Samples and sub-samples in containers. Most containers will have sample labels both on the outside of the container and a label inside (A. D. Rogers). (F) ROV control van showing video screens both for navigation and for sampling operations. Sampling operations are recorded so details of the environment where a sample is taken can be retrieved post cruise (A. D. Rogers).
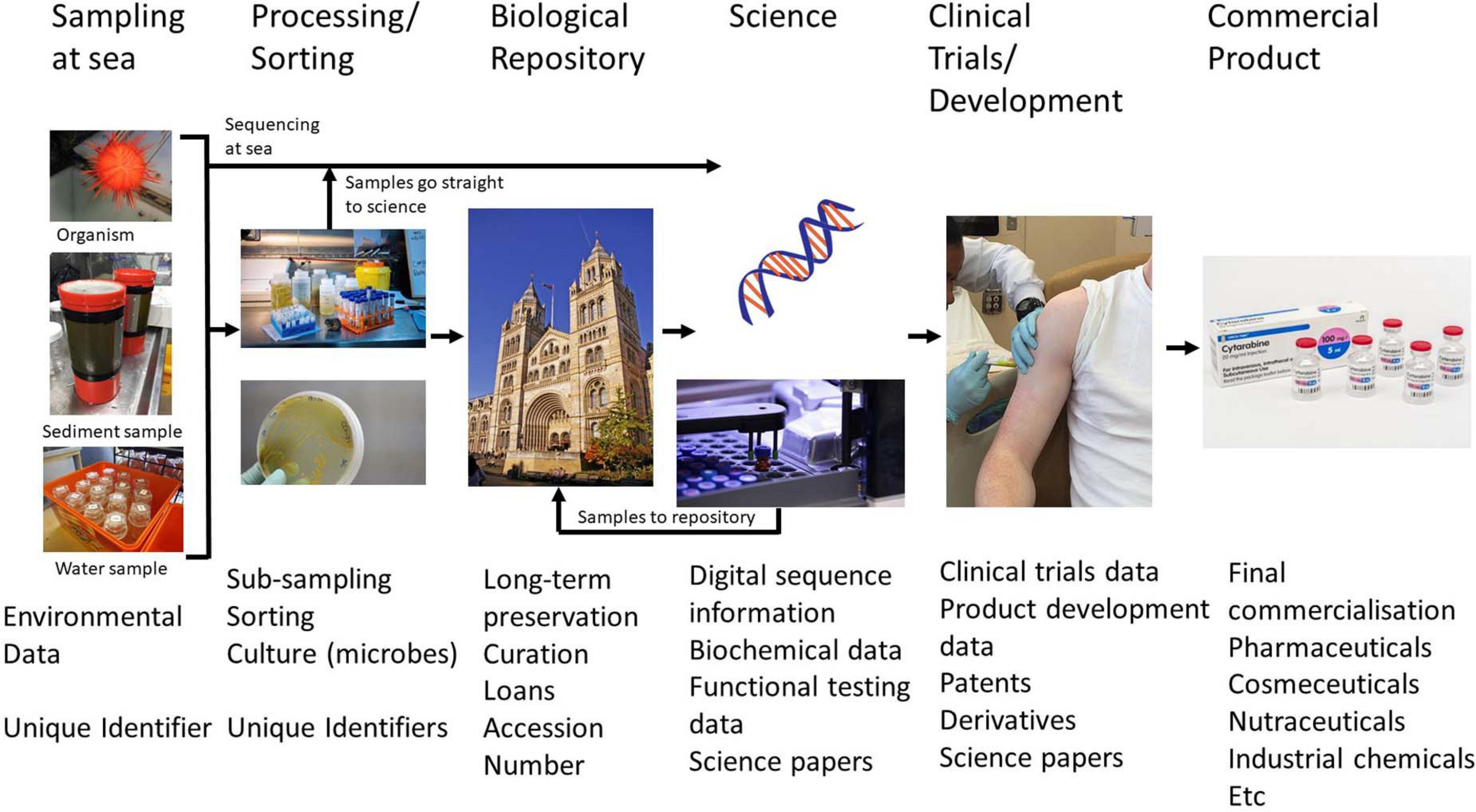
Figure 4. Schematic diagram showing the processes of marine scientific/MGR research from sampling, sample processing, laboratory investigation through to commercial product. Stages where unique identifiers are assigned and data produced are shown. Images are from the following sources: A. D. Rogers; M. L. Taylor; M Jaspers; Christine Matthews (Geograph Project); NIAID (Wikipedia Creative Commons); Pfizer, Netherlands (https://www.pfizer.nl/product/cytarabine-hospira).
Ideally, specimens or cultures in the case of some microbial samples, should be archived following investigation in a curated collection where they can be maintained in the best possible state of preservation for the purposes of the investigation. The form of preservation usually relates to the final intended use (e.g., for taxonomy; see below), sometimes rendering the material of low value for other uses. Also, it is important that the treatment of samples during storage is recorded as this can alter the properties, or, in the case of microbial cultures, the taxonomic composition of samples.
The majority of marine scientific research undertaken in ABNJ that involves sampling biological material is not for purposes of investigation of MGR. Instead it is for understanding patterns of biodiversity, ecology, evolution, and biogeochemical processes in the ocean and includes sampling of microbes, small organisms living on or in the seafloor or the water column to larger individual animals or seaweed (e.g., Sargassum). Conventionally, samples for ecological studies at sea undergo preliminary sorting on the vessel where practical prior to preservation for taxonomic identification, stable isotopic analyses, for genetic/genomic sequencing studies, or other investigations (Figure 3). For individual specimens of megafauna (animals visible in video or photographic images underwater, generally ≥ 2 cm in length; Clark et al., 2016) and sometimes macrofauna (animals collected in sediment cores and retained by a 0.3 mm meshed sieve; Clark et al., 2016), researchers often take photographs of specimens on board using a camera on a stand or mounted on a microscope (Glover et al., 2016; Schiaparelli et al., 2016; Figures 3B,C). This practice captures features that may be lost or obscured during preservation, such as color and fine structural features. Measurements and biomass of specimens may also be taken at this point (Schiaparelli et al., 2016). Photographs may aid subsequent identification or description of the species sampled. The inclusion of a label with the unique identifier assigned to a specimen in the photograph can also help further tie a sample to a sampling location and subsequent laboratory examination (Figures 3C,D). For bulk-processed samples (i.e., mixed-taxa lots with multiple organisms within a net sample or sieved from mud in core samples), some limited sorting into major organismal groups (e.g., copepods, polychete worms etc.) may occur before preservation, and researchers may measure sample biomass or volume (Harris et al., 2000; Schiaparelli et al., 2016). Alternatively, a sample may be bulk fixed for sorting after a cruise. Microbial communities in the water column are often collected using Niskin bottles and seawater filtered with the filters then frozen for subsequent extraction of DNA/RNA (e.g., Biller et al., 2018). Sediment microbial communities, microbial mats and meiofauna (organisms passing through a 0.3 mm sieve and retained on 32–63 μm sieve) can be sampled using cores (Figures 2F,G) and these can likewise be frozen prior to DNA/RNA extraction (e.g., Vonnahme et al., 2020).
Following initial documentation, most samples of organisms are fixed in a tissue fixative, such as ethanol or formalin in buffered seawater, the strength of which varies depending on the taxon (Wilson, 2005; Schiaparelli et al., 2016). Following fixation, specimens are usually transferred to 70–100% ethanol, depending on taxon, for long-term storage (Wilson, 2005; Rainbow, 2009). However, in some cases initial fixation may use 70% ethanol (e.g., sponges; Wilson, 2005) or use formalin for long-term storage (e.g., Scyphozoa or jellyfish; Wilson, 2005; Rainbow, 2009). Conventional methods for fixation and preservation of samples destroy genetic material or make it difficult to extract and sequence, therefore samples or sub-samples for extraction of nucleic acids are generally fixed and preserved in 100% molecular grade ethanol (for DNA extraction and sequencing) or RNALater (for RNA and DNA extraction and sequencing). These samples are also refrigerated or placed in an ultra-cold freezer immediately to minimize degradation of nucleic acids over time. Alternatively, researchers may flash freeze specimens for genomic, transcriptomic or proteomic analyses in liquid nitrogen and then store them in low temperature freezers (–80°C) or in a dry vessel filled with liquid nitrogen (–180°C). While liquid nitrogen storage is ideal for stabilizing RNA for example, small dry shippers typically carried on research cruises can only maintain temperature for a period of 2–3 weeks, therefore storage beyond this point may lead to tissue thawing and subsequent loss. This can be a problem if samples are delayed at customs on route to a laboratory or museum. Split sampling where possible therefore is best-practice, for example, tissue sub-samples from megafauna stored separately in ethanol, RNAlater, and a dry-shipper. Some biological specimens can be dried for preservation, such as skeletons or calcified parts of bodies (e.g., teeth, otoliths, and shells) as well as herbarium specimens of marine algae. These specimens can yield DNA for sequencing studies more easily than formalin-fixed material (Morin et al., 2006; Williamson et al., 2015). It is also notable that the technology for ROVs and autonomous underwater or surface platforms to preserve samples in situ prior to recovery of the vehicle or mooring has been developed (e.g., the ESP; Hansen et al., 2020; see McQuillan and Robidart, 2017 for review of emerging technologies).
Rabone et al. (2019) reviewed current best practice in the archiving of biological specimens and samples for analyses of nucleic acids from ABNJ. Ideally, researchers should deposit biological specimens collected during marine scientific research in the collections of museums or other scientific, research, and educational institutions with adequate infrastructure and funding to manage and curate them long-term. This archiving includes physical specimens of animals, plants, and algae but also culture collections including microbial samples. As stated above, specimens for marine scientific research are generally conserved over the long term in >70% ethanol, or in formalin for some taxa. Frozen samples or sub-samples (usually DNA, isotope or microbiological samples) require storage at low temperatures usually within biorepositories or biobanks (e.g., Strand et al., 2020). Other organisms may be stored as dried specimens or as series of slides showing important structures for taxonomic differentiation. Typically, scientists can borrow specimens internationally from such institutions to aid their investigations, which requires a system for cataloging, labeling, and organizing individual specimens, to facilitate location both for maintenance and for internal and external loans. Curators achieve this objective by assigning a permanent accession (or registration) number for every sample when placed into an institutional collection (Figure 4). International loans are subject to CITES and CBD regulations, which have resulted in significant problems for international collaboration in marine scientific research especially in countries where administrative infrastructure is inadequate. Microbial culture collections may be operated on a commercial or semi-commercial basis. The long-term storage and curation of biological material in institutions is challenging, both technically and in terms of costs but is essential for monetary and non-monetary access and benefit sharing of MGR and marine science in general for ABNJ (see Supplementary Material 5.0).
In practice, for some types of scientific investigation there is not an intention (or requirement) to archive samples over the long term, so they may be destroyed following completion of an investigation. In the deep sea, however, such practices are highly problematic as the deep-sea fauna in most environments are poorly characterized (e.g., samples often include undescribed species), and are collected at immense expense (Rabone et al., 2019). Given the immense cost of deep-sea sampling, this discarding is wasteful and better coordination and practice in making samples available to others with facilities for long-term archiving following initial scientific investigation would improve knowledge on ocean biodiversity as well as other aspects of biology or research on MGR. For instance, even to this day new taxa are described from specimens collected by HMS Challenger close to 150 years ago (e.g., Lemaitre et al., 2018). Reducing the need for collection of biological material also may reduce the environmental impacts of sampling which is important in the deep sea where natural levels of disturbance are low. Existing archiving and infrastructure in museums and some university collections also need improved coordination, for example through distributed collections. Efforts to improve visibility of collections holdings, the discoverability of records at collections level and to improve existing registries/directories of collections, are currently underway at the Global Biodiversity Information System (GBIF) and Biodiversity Information Standards (formally known as the Taxonomic Databases Working Group or TDWG; e.g., GRBio; Schindel et al., 2016). These developments are highly relevant in terms of transparency and discoverability of MGR.
Key to both scientific research and access and benefit sharing is that samples can be tracked for further work (Rabone et al., 2019). In the past, this might be achieved through direct contact with an institution holding specimens or samples, or the researcher who collected the material. At present, the entire process of tracking and tracing samples, linking them to environmental data, as well as even examining specimens is being revolutionized through digitization (Drew et al., 2017; Nelson and Ellis, 2018; Hedrick et al., 2020). This is providing enormous benefits to the scientific community in being able to draw on data on marine biodiversity from institutions holding specimens and samples globally (Drew et al., 2017; Hedrick et al., 2020). For example, if visual examination of a sample is all that is required to identify a sample or for comparative study as part of a new species description, this can also be done through digital 2D or 3D images taken by the holding institution (e.g., Blagoderov et al., 2012).
For purposes of access and benefit sharing for investigation of MGR, the aim is more likely to be to gain access to samples for study. As outlined above, key to both marine scientific research and access and benefit sharing of MGR is that individual specimens or samples are assigned a globally unique identifier (GUID) and that data on the location of sampling as well as any environmental data associated with the sampling event and subsequent processing remains linked to the specimen/sample record. Potential users of specimens, samples or associated data should be able to identify such samples through accessing the cruise report, open databases related to biological collections, records of biological samples from the ocean, nucleic acid and any other forms of data related to MGR, and through searches based on peer-reviewed scientific papers or reports. This means that all of these data sources must be able to communicate through the web so that data on samples are shared and there are many potential routes to identify relevant specimens or samples for access and benefit sharing, whether it be for marine scientific research or investigation of MGR. Key to this interoperability of databases is the usage of data standards, for example MIxS, OBIS-ENV-DATA, Darwin Core, and others (reviewed in Rabone et al., 2019). A key international standard for biological data is Darwin Core (DwC) revolving around a standard file format and standardized information on taxonomy, occurrence and sampling event (Wieczorek et al., 2012). Darwin Core terms are published on the TDWG website, a glossary of standardized terms including those describing the identification of the specimen or samples, geographic origin, the method of sampling, environmental data associated with sampling, identification of sample relationships (e.g., one sample is a sub-sample of another), information related to the specimen or sample (e.g., its biomass), where it is stored and how it is preserved8.
Specific databases relate to specific aspects of specimens and samples collected at sea. For records of the occurrence of marine species sampled in the ocean the main global database is the Ocean Biodiversity Information System (OBIS9), which is a part of the GBIF10. Environmental data associated with collection of samples can also be deposited in a number of databases including, for example, PANGAEA11 (Alberti et al., 2017) and the National Science Foundation (NSF) sponsored Biological and Chemical Oceanography Data Management Office (BCO-DMO) project12. The International Nucleotide Sequence Database Collaboration (INSDC13) provides a gateway to nucleic acid databases such as Genbank14 and the European Nucleotide Archive15. However, at present, the storage of sequence data does not require a recognized scientific name (i.e., species name) or the geographic location from which it was derived. This has led to a proliferation of “dark taxa,” sequences published without reference to valid scientific names, therefore no information on what species they were derived from (Page, 2016). For many organism samples, particularly those found in the open ocean, this situation is partially created by the lag between identification of specimens and genetic/genomic studies that are undertaken upon them. In such cases, specimens are often designated as operational taxonomic units (OTUs) particularly where they have been identified on the basis of DNA sequence information alone. In some cases, the species may be identified or described at a later date, but the original sequence may not be updated with the valid name. It is also common, especially for small organisms, that they are destroyed or become useless during processing for molecular studies.
Environmental DNA (eDNA) approaches, currently becoming more popular for biodiversity surveys, also tend to destroy samples but it is now being recognized that best practice is to submit voucher samples from such studies to museums or other institutions. The Global Genome Biodiversity Network (GGBN16) has developed standard specifications for treatment of such samples within databases (Droege et al., 2016). For microorganisms, because of the vast diversity of bacteria and microbial eukaryotes and the lack of physical characters to separate species, for practical reasons they are often identified as OTUs, so the “dark taxa” issue applies only to macrofauna and megafauna. To resolve this issue, valid, resolvable ‘temporary names’ are needed, used in a consistent way and linked to all key databases (Horton et al., 2021). The Genomic Standards Consortium (GSC17) and the GGBN and OBIS are currently trying to standardize approaches to improved integration of occurrence and sequence data or to provide links to these data in other databases (Rabone et al., 2019). The progress made in recent years illustrates that the scientific community is driving and benefiting from efforts to improve accessibility of samples and data as well as transparency in research.
Research on MGR requires different approaches to that of most marine scientific research in ABNJ. Researchers working on MGR have developed protocols for large-scale treatment of samples for chemical investigation, with the Developmental Therapeutics Programme of the US National Cancer Institute the largest and longest running program (NCI; McCloud, 2010). By 2010, the NCI Natural Product Repository had accumulated extracts from 15,000 marine species including invertebrates and “plants” (presumably algae), mainly from the shallow waters of the Pacific (McCloud, 2010). The protocols developed by the NCI include transport of frozen material to the institute, subsequent homogenization of frozen samples, and extraction of the pulverized tissue using aqueous and organic solvents (McCloud, 2010). The liquid extracts containing polar and non-polar compounds from the original specimens are freeze-dried for long-term storage (McCloud, 2010).
At smaller scales typical of research laboratories, lyophilizing (freeze drying) occurs immediately after collection (Houssen and Jaspars, 2006) or samples are frozen until this process can take place. Alternatively, immersion of specimens in a 50:50 solution of ethanol-water for 24 h prior to discarding the liquid precedes sealing the damp sample in a container for transport back to the laboratory (Houssen and Jaspars, 2006). When samples arrive at the laboratory, they are immersed in methanol. Ideally, collection of 1–2 kg (wet weight) of samples precedes processing (Houssen and Jaspars, 2006). At the laboratory scale, processing of samples can follow that for large-scale preparation, with maceration of tissues in solvent (typically ethanol/methanol and/or dichloromethane; Houssen and Jaspars, 2006). Tissue residues are often repeatedly extracted until they yield no further natural products (Houssen and Jaspars, 2006). Sometimes researchers use a series of solvents of increasing polarity for extraction, typically removing solvents from extracted biochemicals using rotary evaporation (Houssen and Jaspars, 2006; Supplementary Figure 5a). An alternative extraction method uses supercritical fluids, usually carbon dioxide (Houssen and Jaspars, 2006).
Whether samples are collected for large-scale extraction collections or for smaller laboratory-scale screening programs, voucher specimens must be retained for taxonomic identification of the species involved (Houssen and Jaspars, 2006; McCloud, 2010). In the context of bioprospecting, vouchering is additionally relevant as patents require taxonomic information and the archiving of voucher material, and also in terms of assuring traceability, key to ensuring realization of benefit sharing post-commercialization. Furthermore, knowing the identification of the taxon as well as the geographic location from which the sample comes from greatly assists choice of methods for isolation of compounds and their identification as well as understanding their potential properties (Houssen and Jaspars, 2006). Again, a unique specimen identifier can tie compounds to specific samples of organisms and the associated data. At a fundamental level, voucher specimens critically support reproducible science.
The crude extracts from marine organisms contain a complex mixture of compounds, complicating screening and separation (Houssen and Jaspars, 2006). Fractionation is an important initial step in screening for MGR, a process that separates the complex mixtures of compounds in crude extracts into fractions containing fewer compounds. The NCI’s Natural Product Repository has now made publicly available a pre-fractionated library from their collection of >125,000 extractions adding up to 1,000,000 fractions (Thornburg et al., 2018). This, and other fractionated libraries of extracts reduce the complexity of the whole process of high-throughput screening for biological activity (Thornburg et al., 2018). Fractionation of compounds is usually achieved by chromatography, with screening of fractions for biological activity and initial identification of compounds through mass spectrometry (MS) and/or nuclear magnetic resonance (NMR; Houssen and Jaspars, 2006; Wolfender et al., 2019). These expensive technologies require skilled and experienced researchers (Houssen and Jaspars, 2006) emphasizing the point that states without a scientific policy plan that invests in science will be disadvantaged (see Supplementary Material 6.0). Dereplication, the elimination of previously identified compounds with known biological activities, represents an important step in this process. This step avoids expensive and unnecessary work on further purification and screening of previously identified compounds. Dereplication involves comparisons of compounds identified in samples with known compounds detailed in a range of paid-for or freely available databases (Houssen and Jaspars, 2006; van Santen et al., 2019; Wolfender et al., 2019). Importantly, a broad range of information can help to identify natural products, including the taxonomic information of the species, enabling comparisons with natural products isolated from related taxa, as well as providing information on the geographic origin of the samples and genomic information (Wolfender et al., 2019). Following identification of fractions containing novel bioactive compounds, further purification steps lead to a pure compound (Houssen and Jaspars, 2006). Approaches that combine metabolomic and genomic data (see below) also show great promise in the search for novel MGR (Wolfender et al., 2019).
We note that the significant quantities of tissue required for identification of MGR raises questions regarding the impacts of sampling, particularly for smaller and/or rare organisms (the majority of species in the deep sea) and especially if further samples are required for investigation of specific MGR. It is therefore important to consider the sustainability of sampling for MGR in ABNJ, especially where repeated sampling may be required to accumulate sufficient material for investigations. EIAs are therefore likely to be required prior to undertaking such work in ABNJ and it will be important to apply the precautionary approach where impacts on species or ecosystems are uncertain. However, improvements in analytical, spectroscopic and bioassay techniques mean that the whole pipeline can be executed with smaller samples than 1–2 kg wet weight. Additionally, many researchers work on microorganisms obtained from marine macroorganisms, sediment, or other samples and this reduces the need for larger collections. Once compounds of interest are isolated, they can often be synthesized in the laboratory negating the need for further collection of specimens of the organism of origin from the natural environment. These examples further illustrate the trend in the scientific community in coordinating and streamlining research, reducing its environmental impact and increasing its reproducibility.
Rather than profiling of metabolites (natural products), methods are being developed to screen genomic libraries or genomic sequence data to identify gene clusters for compounds that may act against specific drug targets (Trindade et al., 2015; Ziemert et al., 2016; Wolfender et al., 2019). This approach, known as genome mining, is particularly useful for marine bacteria because genes coding for biosynthesis pathways often cluster within the microbial genome (Trindade et al., 2015). Therefore, a single clone or a small number of overlapping clones in a library may capture the entire pathway in terms of encoding genes (Trindade et al., 2015). Polymerase chain reaction (PCR) or southern blotting can identify those clones within a library that contain specific gene clusters (Trindade et al., 2015; Wolfender et al., 2019). Screening for bioactivity can also directly use the genomic library (heterologous expression; Trindade et al., 2015; Ziemert et al., 2016), enabling in silico predictions on the structure of the metabolites for which such gene clusters code (Trindade et al., 2015). Gene mining can be carried out on bacterial genomes or on genomic libraries generated from environmental samples. It is now possible to express the genes identified through mining not only through heterologous expression but also through synthetic biology (Ziemert et al., 2016). Rapid advancements of these technologies explain the importance of considering DSI in the BBNJ agreement with respect to MGR.
Marine scientific research and MGR cross paths in the generation of large-scale genomic (nucleic acid) data from environmental samples. Gene mining enables the search for MGR through nucleic acid data collected for the purposes of describing the genetic diversity and function of organisms in environmental samples. The recent Tara Ocean and Malaspina expeditions illustrate global-scale marine scientific research programs that produced large new datasets on the genome sequences of viruses and a wide range of other microbial organisms from the water column of the ocean, including ABNJ (Karsenti et al., 2011; Duarte, 2015; Pesant et al., 2015; Supplementary Material 7.0). The data sets produced by these expeditions offer significant potential for discovery of novel genes and chemical compounds (i.e., MGR) given minimal exploration of planktonic organisms for their biotechnological potential (Abida et al., 2013). However, such work is still challenging and developments in bioinformatics and other aspects of discovery of MGR through such in silico studies are in progress to realize the full potential of such resources. It is clear, however, that as technology to access the ocean and its interior advances, we can expect large-scale increases in the availability of DSI from a broad range of organisms living in a variety of habitats. This is not only from deep-sea research cruises but also from ocean observing programs such as the GOOS and DOOS. The move toward open access data, as illustrated by Tara Oceans, demonstrates the potential, aided by the rapid development of cheap technology to sequence entire genomes at sea without necessarily even bringing physical samples back to shore (e.g., Oxford Nanopore Technologies18).
While the focus of this paper is not on capacity development per se, it is an important element of the BBNJ agreement, UNCLOS (Harden-Davies and Gjerde, 2019; Harden-Davies and Snelgrove, 2020), other international ocean treaties (Rogers et al., 2020) and initiatives such as the UN Decade of Ocean Science for Sustainable Development. Many different actors including governments, intergovernmental organizations, philanthropic organizations, and academic institutes are trying to address this issue. We therefore give an overview of the topic in the context of the BBNJ agreement, highlighting examples of programs that aim to build capacity of developing States at national and international levels although a thorough treatment will require a separate publication.
Commitments across developed and developing States to cooperate in capacity building and technology exchange are potentially the most effective means to increase equity in marine scientific research. An example of this is the Belem Statement which outlined a strategy for joint prioritization and coproduction of marine science for societal well-being between Europe, South Africa and Brazil (Claassen et al., 2019). The Statement also committed the partners to capacity building, scientific and technology exchange. The importance of such international commitments is in driving national funding agencies to long-term support for capacity building activities for scientists and other technical experts from developing States as well as technology exchange with them.
Capacity building has also been addressed through nationally funded science or aid initiatives. For example, the US NSF Partnerships in International Research and Education (PIRE) program funded collaborative international research projects that emphasized both co-production of science and educational outcomes. PIRE led to collaborative projects on marine science in the coral triangle that included co-produced peer-reviewed publications (Barber et al., 2014). The subsequent USAID Partnerships for Enhanced Engagement in Research (PEER) program was aimed at supporting collaborative research with other countries that furthered the US’s overseas development objectives (Barber et al., 2014). PEER program projects were led by developing country scientists who then co-produced science with US scientists (Barber et al., 2014).
National policy frameworks within developing countries are important for capacity building. An example of this was the Science Without Borders program, which placed more than 100,000 Brazilian students in top-ranking overseas universities from the period 2011 until the end of the program in 2017, including those studying marine science. The policy resulted in Brazil climbing to 11th in the world for science and engineering peer-reviewed papers by 201819.
A range of capacity development initiatives have been sponsored and organized by intergovernmental organizations. These include activities aimed at building technical expertise in the negotiation of, and implementation of international agreements (e.g., the BBNJ agreement) and in sustainable use of marine resources, including in ABNJ (Cincin-Sain et al., 2018). Sponsoring organizations include the CBD, other UN organizations such as the UNESCO IOC and the Global Environment Facility (Cincin-Sain et al., 2018). An example is the IOC’s Ocean Teacher Global Academy20, which provides a web-based training platform that supports classroom training (face-to-face), blended training (combining classroom and distance learning), and online (distance) learning. It is provided through a number of regional centers including in non-English languages and includes basic training in marine science as well as handling of data, data analyses and use of databases.
A number of civil society organizations also provide capacity building opportunities including training in marine scientific research, ocean governance, including the BBNJ agreement, navigation, marine law and maritime security (Cincin-Sain et al., 2018). Examples of sponsoring organizations include Greenpeace, the International Chamber of Shipping, the International Ocean Institute, the Natural Resource Defense Council, the Nippon Foundation, the Sasakawa Peace Foundation, the Tara Expeditions Foundation, the World Wildlife Fund and various academic institutions (Cincin-Sain et al., 2018).
Very few of these initiatives are targeted at training early career researchers (ECRs) in marine scientific research at sea. From 2016 to 2020 the Tara Expeditions Foundation implemented a program to train early career scientists from developing States both on at sea expeditions and in European laboratories in oceanography and particularly plankton research. The aim was not only to build capacity but also a network of researchers in developing States working collaboratively on plankton studies (Cincin-Sain et al., 2018). Another example was the Training Through Research (TTR) program, proposed by the Moscow State University and supported by UNESCO, the European Science Foundation and IOC. Between 1991 and 2012, the TTR cruises trained undergraduates and postgraduates with hands-on experience in at-sea research, with over 1000 students from 30 nations trained on 18 major cruises. Training ECRs at sea or in the lab is now an important evaluation criterion in most calls for funding of research cruises, including both public- and philanthropic funders (e.g., SOI’s R/V Falkor, REV Ocean). However, the term ECR does not specifically refer to researchers from developing States, so the success of such programs in developing capacity worldwide is unknown. A new international Deep Ocean Training (DOT) program for training at sea is under development within the framework of the Decade of Ocean Science for Sustainable Development (Howell et al., 2020) and is intended to build capacity in deep-sea research for scientists at all levels of career especially those from small island and coastal developing States. In the US, the NOAA Office of Ocean Exploration and Research (OER) has developed an Explorer-in-Training program, where over 120 undergraduate and graduate students have received hands-on experience at sea on deep-water seafloor mapping and exploration since 2009. Both SCOR (Scientific Committee on Ocean Research) and POGO (Partnerships for Observation of the Global Ocean) also manage and promote shipboard training, regional and summer workshops and fellowship programs to expand oceanographic engagement by developing State scientists.
Increased capacity is only achieved if new knowledge is incorporated over the long term by developing States. For example, the Namibian Government’s National Marine Information and Research Centre, recently organized training on benthic ecology and sampling with international experts from DOSI/INDEEP (Deep-Ocean Stewardship Initiative; International Network for Scientific Investigation of Deep-Sea Ecosystems). Subsequently, a new Namibian benthic scientific team was created within the same research institute to conduct coordinated deep-sea benthic observations and sampling during all annual government deep-sea fisheries research cruises, thereby building long-lasting capacity and a comprehensive curated collection of deep-sea benthic fauna for the country. This emphasizes the importance of having institutional support so that scientists can secure long-term employment in roles that allow them to use their talent (Harden-Davies et al., 2020). Supporting and sustaining scientific experts locally, and enabling long-term collaborations are critical to form science networks and secure funding for local infrastructure and investment for research in ABNJ. Capacity building programs for ABNJ can have synergistic impacts on areas within national jurisdiction (Vierros and Harden-Davies, 2020).
Investment in data management infrastructure and training on data management is crucial in an era where we rely increasingly on computational power to archive and analyze the vast amount of complex data being produced at ever increasing rates (Stocks et al., 2016). Digital assets should follow the FAIR principles21 for scientific data management and stewardship: Findable, Accessible, Interoperable, and Reusable (Wilkinson et al., 2016). However, this is a complex topic and a large proportion of scientists and other ocean stakeholders do not have the expertise to correctly archive data, making it available to the wider community. Training on data management is essential at all levels of career development, but particularly for the next generation of ocean experts. This is highly relevant for research on MGR in ABNJ, where vast amounts of data are produced, which need to be easily findable and accessible to ensure sharing of information and knowledge (Leonelli, 2016).
The above examples illustrate that science cooperation is already happening and that capacity building in all States, and at all career levels can happen as a by-product or central focus of collaboration. Yet the fact that there are continuing disparities in science capacity worldwide (IOC-UNESCO, 2020) and imbalances in biodiversity research including for ABNJ (Tolochko and Vadrot, 2021) indicates that the current status quo is not resulting in equitability in research. More effort is needed.
Although assessments of current capacity in marine science are improving (IOC-UNESCO, 2020) it is still extremely difficult to identify where capacity and technology transfer is needed. This requires an international effort to map what the capacity building and technology transfer needs are of developing States. A barrier to achieving this is that the outcomes of capacity building and technology transfer measures are not monitored over the medium to long term and approaches are piecemeal. For example, a cruise opportunity might be viewed as tokenistic, not building lasting capacity if a trainee then has no job to go to in their home country. At worst, such capacity building activities can perpetuate brain drain from developing States. Monitoring and review of capacity building and technology transfer activities will be an important aspect of implementation of the BBNJ agreement. The best examples of capacity building generate equitable partnerships over the long term from which everyone benefits.
Given our overview of aspects of MGR, access and benefit sharing as well as capacity building, we summarize what elements of best practice in marine scientific research are already in place that would assist in the implementation of access and benefit sharing provisions in the BBNJ agreement and what still needs to be developed.
Cruise notification is not only feasible but is already best practice in many States, international organizations and projects. The infrastructure exists under IOC IODE for development of an international system of cruise notification for ABNJ, giving information on past cruises and also current and future cruise opportunities. Current best practice can already address security sensitivities during scientific operations.
Pre-cruise notification is required for aspects of access and benefit sharing. It would also further marine scientific research cooperation through transparency and coordination at regional and global scales. The transparency brought about by pre-cruise notification and post-cruise reporting will also assist in meeting international obligations under Part XIII of UNCLOS, specifically Article 244.
The information that is needed for pre-cruise notification includes:
• The vessel
• The geographic area of operation
• The expedition start and end ports (can be omitted if there are security issues)
• The expedition start and end date, or the general time window in which the cruise is taking place (if there are security issues)
• Types of sampling operations and heavy equipment deployed
• The main scientific objective of the cruise
• The name of the Principal Investigator or Cruise Coordinator
• Contact details for enquiries for cruise participation or access to samples
However, we do not consider that the text of the BBNJ agreement needs to be prescriptive in the details of what is provided in pre-cruise notification. Some of the examples shown in this paper already represent best practice in the provision of this information. Pre-cruise notification should conform to FAIR principles.
Post-cruise reporting is undertaken as best practice for many research vessel operators and is often required by granting agencies. These reports are a record of the work undertaken on board a research vessel, including details of the sampling, data collected, treatment of samples and scientific investigations undertaken. Usually there will also be an indication of where samples are to be worked up following the cruise. Post-cruise reporting also often includes summaries of the operation of scientific equipment as a means of guiding future improvements in practice. Examples of post cruise reports by authors of this paper can be found here:
https://www.bodc.ac.uk/resources/inventories/cruise_inventory/reports/jc042.pdf
https://repository.library.noaa.gov/view/noaa/22706/noaa_22706_DS1.pdf
For the purposes of access and benefit sharing, it is important to be able to link sampled organisms to marine scientific or MGR research, to where they are ultimately archived. Tracking the collection of MGR from ABNJ to the eventual realization of commercial products and benefit sharing requires identification of samples including their origin (Humphries et al., 2021). The assignment of unique identifiers to samples and/or individual specimens is standard best practice for marine science. This includes assignment of unique identifiers to:
• Organisms subsampled from samples of multiple organisms
• Organisms collected from larger organisms on or in which they live (e.g., epizooites or endozooites)
• Samples split and preserved in different ways
• Subsamples of tissue removed from organisms for specific preservation or extraction protocols (e.g., DNA-friendly preservation or DNA extraction)
Labeling protocols should allow assignment of a sample or subsample to an original parent sample or specimen (i.e., they can be traced back to the original sample; Rabone et al., 2019). Usage of global data standards and ontologies together with persistent identifiers (e.g., GUIDs) allows for interoperability between databases facilitating traceability and transparency (Rabone et al., 2019). Such practices are already used by institutions such as large museums in developed countries with global accessibility of data achieved through databases such as OBIS and GBIF. However, the application of such practices at global scale and with not just species but OTUs and environmental samples requires further standardization, and potentially the application of new technologies for traceability. Such careful curation of species, samples and subsamples comes with costs in terms of database infrastructure, the adoption of existing or new technologies and for training and capacity building.
When scientific investigations are complete (including for MGR), samples or specimens are, according to best practice, archived in a museum or other appropriate institution for permanent storage. Such institutions require the resources to ensure that samples are properly curated, and they are accessible to researchers at a national, regional and global level. As part of the process of submitting specimens or samples to such institutions they are usually also assigned an accession number. This is also an identifier and is linked to all the information associated with the original sample, including the original sample identifier. In both marine scientific and MGR research, specimens or samples can be destroyed in which case voucher specimens or samples can be submitted for archiving (the same species, types of samples or subsamples as the destroyed species/samples from the same locality).
Enabling access to samples for marine scientific or MGR research for scientists from developing States requires that information on samples is findable and accessible. Information with respect to samples should therefore be located on databases through the world wide web. Ideally such information should enable the identification of samples, specimens or species and their sampling locations and be linked to data on where they are currently held and information on how to access physical samples. These data should also be linked to environmental and other information collected during sampling events.
There are more than 1,700 databases available through the world wide web which hold data on DNA, RNA and protein sequences, gene expression, small molecules, protein structures and metabolic systems (Imker, 2018; Karger et al., 2019). However, the main repositories of DSI are the European Molecular Biology Laboratory’s European Bioinformatics Institute (EMBL-EBI), the DNA Databank of Japan (DDBJ), and the National Centre for Biotechnology Information (NCBI) in the United States (Karger et al., 2019). Together, these form the International Nucleotide Sequence Database Collaboration (INSDC; Karger et al., 2019). It is not just best practice that DSI is deposited in these databases, but it is often a requirement for submission of peer-reviewed papers to scientific publications that contain such data. All data submitted to DSI databases receive an accession number that is unique to that particular data (e.g., sequence or set of sequences). It is also best practice that all information pertaining to what specimen/sample or species the sequence data have come from as well as information or links to information on its geographic origin is included in submission to these databases. The best way to ensure this happens is by including the GUID of the original specimen/sample in the submission of sequence data to the DSI database. DSI submitted to these databases is freely accessible to the public via the web. However, specialist knowledge is required to understand how to search for, download and use such resources for research on MGR.
Leveling the playing field in marine scientific and MGR research in ABNJ will require concerted and coordinated efforts internationally over extended time to meet the needs of developing States (Harden-Davies and Snelgrove, 2020). Resourcing and training are expensive and so it is important that the needs and aspirations of developing States are identified so that resources are deployed, and partnerships forged to have maximum impact in meeting those needs. The BBNJ agreement would usefully enable discussion and identification of the needs to enable access and benefit sharing, these would likely arise in the following categories:
With respect to MGR this includes all stages of the scientific process from planning research projects, writing proposals for funding, EIA, undertaking research at sea, sample processing, laboratory skills, training in use of advanced laboratory equipment, acquiring and analyzing data, archiving data and samples, preparation and submission of reports, research papers and patents. Outside of postgraduate training (Masters or Ph.D.), there is no single course that provides all of these skills so it is likely that many ECRs or other experts from developing countries will acquire these skills across multiple courses, research placements, practical courses and through coproduction of science and long-term mentoring and networking.
It is important that access to advanced research vessels and deep-submergence technologies, including low-cost but effective sampling and survey tools (e.g., drop cameras) are opened-up for training and research in ABNJ for marine scientific and MGR research. Not only do opportunities need to be created for participation in “at sea” research, but they must be adequately disseminated internationally to enable all to access. A one-stop shop for opportunities could be created through a BBNJ agreement clearing-house mechanism or through a cruise notification system.
Access to adequately resourced advanced laboratories for training and co-production of science and studies on MGR is a critical aspect of access and benefit sharing. Equally, if not more important, is building institutional capacity in developing States for capacity building to have a long-term impact. Scientists trained from developing States need employment opportunities that use their talent. Dedicated personnel are also needed to maintain and operate equipment and local leaders must be empowered to develop lasting capacity and infrastructure.
Growing capacity for scientific research, including studying MGR, requires international effort extended over time. The funding required for such efforts is significant and needs to be recognized as a benefit in the BBNJ agreement. Funding can come through specific training programs, scholarships and many other activities tailored to build capacity in developing States. The BBNJ agreement should be a focal point to coordinate funding and capacity building opportunities as well as a forum for discussing funding priorities and garnering the support and trust of funders.
Research networks, whether formal (e.g., professional societies) or informal (e.g., collaborating scientists) provide a means to identify and access capacity building opportunities through training or coproduction of science. Under the BBNJ agreement, opportunities should be identified to bring together early career researchers (ECRs) from developing States with scientists from developed countries to enable capacity building. This might include sponsorship of international meetings related to marine scientific or MGR research in ABNJ to enable meaningful participation by developing States or designing events to encourage contact with research networks. Some initiatives such as DOSI and the Deep-Sea Biology Society already provide grants for ECRs from developing and small island States to participate in large international symposia. Mentoring, of ECRs by more senior scientists including across disciplines (e.g., bringing science to policy) is a critical function of these networks and can develop long-term collaborations.
Access to data, such as DSI, for research on MGR requires training, access to advanced computing facilities and reliable connection to the internet. Digital information is now integrated into all aspects of scientific research and so should not be a secondary consideration in capacity building activities. Support for such activities needs to be sustained as digital technologies are advancing very quickly, enabling in silico approaches to identification of MGR as well as other activities such as remote participation in scientific cruises.
Development of capacity for marine science, including research on MGR, also requires access to samples. It is therefore important that samples are archived and made available for research in ways that are findable and accessible. Researchers need to be trained in the collection and curation of samples and associated data as an integral part of ocean science. Whilst many samples are held in large institutions in developed States, consideration should be given to the development of regional and national centers for depositing samples in developing States. Examples of these already exist (e.g., South African Institute for Aquatic Biodiversity) and can enable both the development of national or regional programs in developing States and regions, international collaborations and training of ECRs. The BBNJ agreement represents a significant opportunity to convene and coordinate such efforts at international and regional levels.
Translating science to management and policy is an important aspect of training, especially for developing States. It is therefore important that this should be included in capacity building activities, including for non-scientific experts such as those working in law, resource and ecosystem management, governance and other relevant areas.
Access and benefit sharing is important in the international governance of biodiversity conservation and management. The bilateral approach to access and benefit sharing within the CBD is not applicable to ABNJ (CBD, 2020). The BBNJ agreement represents an opportunity to clarify definitions and regulations for access and benefit sharing of MGR resources in ABNJ in a way that is fair and equitable and also facilitates marine scientific research. It could increase rigor, efficiency and effectiveness in a number of aspects of marine scientific research including on MGR in ABNJ. This ranges from improving approaches to pre-cruise notification and post-cruise reporting, ensuring such activities are undertaken in an environmentally sustainable manner, to more standardized approaches to identifying and recording samples and associated data as well as permanent archiving of samples and data in ways which are findable and fully accessible (FAIR principles for data).
A requirement for pre-cruise notification for government and privately funded research in ABNJ as well as post-cruise reporting would be consistent with existing best-practice. It is important that any infrastructure or mechanism that is established facilitates easy access to information on cruise notification to enable opportunities for access and benefit sharing – including capacity building. Better coordination internationally would optimize the use of precious samples gathered from some of the most remote ecosystems on Earth.
The BBNJ agreement could promote global community support for international research infrastructure, such as databases, and capacity building efforts, as well as keeping pace with the development of technologies and scientific approaches to ensure that that the intent of the treaty is realized over the medium to long term. Engaging diverse voices from the scientific community worldwide, with the institutional arrangements such as the Conference of the Parties (CoP) and scientific and technical body could help to track progress.
Capacity building is a key aspect of all international treaties related to the ocean, but efforts are still fragmented, and results slow to materialize. Robust provisions within the BBNJ agreement for funding capacity building and technology transfer, for coordination, and for communicating and meeting the needs of developing States would support best-practice approaches. The BBNJ agreement could, crucially, provide opportunities to build scientific and technical capacity in nations where it is not currently possible to engage equitably in research in ABNJ. For example, by supporting training for scientists and other marine experts, building capacity in marine sciences as well as for sustainable management of marine resources and their ecosystems both within ABNJ but also in areas within national jurisdiction. With the right encouragement through equitable, genuine and durable partnerships such as joint programs, this will also provide developing States with the opportunities to support building of scientific infrastructure and institutional capacity and to invest in young scientists, building global capacity in a more equitable way for marine science and ocean management. If successful, the BBNJ agreement would improve governance of the ocean as a whole at local, regional and global levels.
Finally, we note that discussions are on the table for CBD 15th CoP in 2021 both on the definition of MGR, especially with respect to digital sequence information but also on the possibility of a Global Multilateral Benefit Sharing Mechanism (GMBSM; CBD, 2020; Lyal and Zhao, 2020). It would be desirable that the outcomes of discussions relating to the BBNJ agreement were complementary with decisions made at CBD CoP (or thereafter) with respect to these issues.
AR and HH-D developed the concept of the manuscript and lead the writing of the manuscript. AR prepared the figures, table, and Supplementary Materials. MJ contributed sections on analyses of marine genetic resources as well as text and comments on the rest of the manuscript. AB, EE-B, DC, KG, JG, LL, KL, MR, ER-L, JS, TS, KS, PS, MT, and DW contributed equally to text, ideas, citations, and images for figures. All authors contributed to the article and approved the submitted version.
DC was employed by Globe Law, New Zealand. MJ is a founder of, shareholder in, and consultant to GyreOx Therapeutics, a company that uses DSI to engineer enzymes to create complex molecules to treat human disease.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank the Governments of The Kingdom of Belgium, The Principality of Monaco and Costa Rica, as well as The Prince Albert II Monaco Foundation, The Norwegian Nobel Institute, The Nobel Institute, The High Seas Alliance, The Pew Charitable Trusts, Ocean Unite and REV Ocean for supporting the High Seas Treaty Dialogues which have allowed informal discussions between States representatives on the Biodiversity Beyond National Jurisdiction agreement. We would also like to thank the participants of the High Seas Treaty Dialogues for their thoughtful contributions to these discussions which highlighted the need for this paper. We would also like to acknowledge REV Ocean for supporting publication costs for this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.667274/full#supplementary-material
Abida, H., Ruchaud, S., Rios, L., Humeau, A., Probert, I., De Vargas, C., et al. (2013). Bioprospecting marine plankton. Mar. Drugs 11, 4594–4611. doi: 10.3390/md11114594
Alberti, A., Poulain, J., Engelen, S., Labadie, K., Romac, S., Ferrera, I., et al. (2017). Viral to metazoan marine plankton nucleotide sequences from the Tara Oceans expedition. Sci. Data 4:170093.
Baker, M., Ramirez-Llodra, E., and Tyler, P. (2020). “Introduction: evolution of knowledge, exploration, and exploitation of the deep ocean,” in Natural Capital and Exploitation of the Deep Ocean, eds M. Baker, E. Ramirez-Llodra, and P. Tyler (Oxford: Oxford University Press), 1–24. doi: 10.1093/oso/9780198841654.003.0001
Barber, P. H., Ablan-Lagman, M. C. A., Ambariyanto, A., Berlinck, R. G. S., Cahyani, D., and Crandall, E. D. (2014). Advancing biodiversity research in developing countries: the need for changing paradigms. Bull. Mar. Sci. 90, 187–210. doi: 10.5343/bms.2012.1108
Bernal, P., and Simcock, A. (2016). “Marine scientific research,” in The First Global Integrated Marine Assessment: World Ocean Assessment I, eds L. Inniss, A. Simcock, A. Y. Ajawin, A. C. Alcala, P. Bernal, H. P. Calumpong, et al. (Cambridge: Cambridge University Press), 459–470. doi: 10.1017/9781108186148.033
Biller, S. J., Berube, P. M., Dooley, K., Williams, M., Satinsky, B. M., Hackl, T., et al. (2018). Data Descriptor: marine microbial metagenomes sampled across space and time. Sci. Data 5:180176.
Blagoderov, V., Kitching, I. J., Livermore, L., Simonsen, T. J., and Smith, V. S. (2012). No specimen left behind: industrial scale digitization of natural history collections. ZooKeys 209, 133–146. doi: 10.3897/zookeys.209.3178
Blasiak, R., Wynberg, R., Grorud-Colvert, K., Thambisetty, S., Bandarra, N., Canário, A. V. M., et al. (2020a). Blue Paper 4: The Ocean Genome: Conservation and the Fair, Equitable and Sustainable Use of Marine Genetic Resources. Report prepared for the (Norwegian) Prime Minister’s High-Level Panel on a Sustainable Ocean Economy. London: World Resources Institute, 83.
Blasiak, R., Wynberg, R., Grorud-Colvert, K., Thambisetty, S., Bandarra, N., Canario, A. V. M., et al. (2020b). Prospects for conservation and sustainable use of the ocean genome. Nat. Sustain. 3, 588–596.
Broggiato, A., Arnaud-Haond, S., Chiarolla, C., and Greiber, T. (2014). Fair and equitable sharing of benefits from the utilization of marine genetic resources in areas beyond national jurisdiction: bridging the gaps between science and policy. Mar. Policy 49, 176–185. doi: 10.1016/j.marpol.2014.02.012
Broggiato, A., Vanagt, T., Lallier, L. E., Jaspars, M., Burton, G., and Muyldermans, D. (2018). Mare Geneticum: balancing governance of marine genetic resources in international waters. Int. J. Mar. Coast. Law 33, 3–33. doi: 10.1163/15718085-13310030
CBD (2020). Global Multilateral Benefit-Sharing Mechanism (Article 10) Of The Nagoya Protocol Note by the Executive Secretary. Convention on Biological Diversity CBD/SBI/3/15, 11, Available online at: https://www.cbd.int/doc/c/dc43/75f8/c5e5cae2443c63a4e81b04bb/sbi-03-15-en.pdf (accessed July 13, 2020).
Cincin-Sain, B., Vierros, M., Balgos, M., Maxwell, A., Kurz, M., Farmer, T., et al. (2018). Policy Brief on Capacity Development as a Key Aspect of a New International Agreement on Marine Biodiversity Beyond National Jurisdiction (BBNJ). Washington, DC: Global Ocean Forum.
Claassen, M., Zagalo-Pereira, G., Soares-Cordeiro, A. S., Funke, N., and Nortje, K. (2019). Research and innovation cooperation in the South Atlantic Ocean. S. Afr. J. Sci. 115:6114.
Clark, M. R., Consalvey, M., and Rowden, A. A. (2016). Biological Sampling in the Deep Sea. Chichester: Wiley Blackwell, 451.
Collins, J. E., Rabone, M., Vanagt, T., Amon, D. J., Gobin, J., and Huys, I. (2021). Strengthening the global network for sharing of marine biological collections: recommendations for a new agreement for biodiversity beyond national jurisdiction. ICES J. Mar. Sci. 78, 305–314.
Danilenko, G. M. (1993). Law-Making in the International Community. Leiden: Martinus Nijhoff Publishers.
Deep Green. (2020). Compiled Q&As for May 6 and May 20 Webinars With Lead Authors of the Life Cycle Assessment White Paper: Where Should Metals for the Green Transition Come From?. Vancouver, BC: Deep Green, 49.
Drew, J. A., Moreau, C. S., and Stiassny, M. L. J. (2017). Digitization of museum collections holds the potential to enhance researcher diversity. Nat. Ecol. Evol. 1, 1789–1790. doi: 10.1038/s41559-017-0401-6
Droege, G., Barker, K., Seberg, O., Coddington, J., Benson, E., Berendsohn, W. G., et al. (2016). The global genome biodiversity network (GGBN) data standard specification. Database 2016:baw125. doi: 10.1093/database/baw125
Duarte, C. M. (2015). Seafaring in the 21st century: the Malaspina 2010. Circumnavigation expedition. Limnol. Oceanogr. Bull. 24, 11–14. doi: 10.1002/lob.10008
Glover, A. G., Dahlgren, T. G., Wiklund, H., Mohrbeck, I., and Smith, C. R. (2016). An end-to-end DNA taxonomy methodology for benthic biodiversity survey in the Clarion-Clipperton Zone, central Pacific Abyss. J. Mar. Sci. Eng. 4:2. doi: 10.3390/jmse4010002
Hansen, B. K., Jacobsen, M. W., Middelboe, A. L., Preston, C. M., Marin, R. III, Bekkevold, D., et al. (2020). Remote, autonomous real-time monitoring of environmental DNA from commercial fish. Sci. Rep. U. K. 10:13272.
Harden-Davies, H. (2020). “The exploitation of deep-sea biodiversity: components, capacity and conservation,” in Natural Capital and Exploitation of the Deep Ocean, eds M. Baker, E. Ramirez-Llodra, and P. Tyler (Oxford: Oxford University Press), 127–139. doi: 10.1093/oso/9780198841654.003.0007
Harden-Davies, H., and Gjerde, K. (2019). Building scientific and technological capacity: a role for benefit-sharing in the conservation and sustainable use of marine biodiversity beyond national jurisdiction. Ocean Yearbook 33:377. doi: 10.1163/9789004395633_015
Harden-Davies, H., and Snelgrove, P. (2020). Science collaboration for capacity building: advancing technology transfer through a treaty for biodiversity beyond national jurisdiction. Front. Mar. Sci. 7, doi: 10.3389/fmars.2020.00040
Harden-Davies, H., Vierros, M., Gobin, J., Jaspars, M., von der Porten, S., Pouponneau, A., et al. (2020). Science in Small Island Developing States: Capacity Challenges and Options relating to Marine Genetic Resources of Areas Beyond National Jurisdiction. Report for the Alliance of Small Island States. Wollongong, NSW: University of Wollongong, Australia, 103.
Harris, R., Wiebe, P., Lenz, J., Skjoldal, H. R., and Huntley, M. (eds). (2000). ICES Zooplankton Methodology Manual. Cambridge, MA: Academic Press, 684.
Hedrick, B. P., Heberling, J. M., Meineke, E. K., Turner, K. G., Grassa, C. J., Park, D. S., et al. (2020). Digitization and the future of natural history collections. BioScience 70, 243–251.
Horton, T., Marsh, L., Bett, B. J., Gates, A. R., Jones, D. O. B., Benoist, N. M. A., et al. (2021). Recommendations for the standardisation of open taxonomic nomenclature for image-based identifications. Front. Mar. Sci. 8:620702.
Houssen, W. E., and Jaspars, M. (2006). “Isolation of marine natural products,” in Natural Products Isolation, Methods in Biotechnology 20, 2 Edn, eds S. D. Sarker, Z. Latif, and A. I. Grey (Totowa, NJ: Humana Press), 353–390. doi: 10.1385/1-59259-955-9:353
Houssen, W., Rodrigo, S., and Jaspars, M. (2020). Digital Sequence Information on Genetic Resources: Concept, Scope and Current Use. Report to the Secretariat of the Convention on Biological Diversity. CBD/DSI/AHTEG/2020/1/3. Convention on Biological Diversitiy. Ad Hoc Technical Expert Group on Digital Sequence Information on Genetic Resources. Montreal, QC: Convention for Biological Diversity.
Howell, K. L., Hilário, A., Allcock, A. L., Bailey, D., Baker, M., Clark, M. R., et al. (2020). A blueprint for an inclusive, global deep-sea Ocean Decade field programme. Front. Mar. Sci. 7:584861. doi: 10.3389/fmars.2020.584861
Humphries, F., Rabone, M., and Jaspars, M. (2021). Traceability Approaches for marine genetic resources under the proposed Ocean (BBNJ) Treaty. Front. Mar. Sci. 8:661313. doi: 10.3389/fmars.2021.661313
Imker, H. J. (2018). 25 Years of molecular biology databases: a study of proliferation, impact, and maintenance. Front. Res. Metr. Anal. 3:18. doi: 10.3389/frma.2018.00018
IOC-UNESCO (2020). Global Ocean Science Report 2020–Charting Capacity for Ocean Sustainability, ed. K. Isensee (Paris: UNESCO Publishing), 248.
Jaspars, M., de Pascale, D., Andersen, J. H., Reyes, F., Crawford, A. D., and Ianora, A. (2016). The marine biodiscovery pipeline and ocean medicines of tomorrow. J. Mar. Biol. Assoc. U. K. 96, 151–158. doi: 10.1017/s0025315415002106
Karger, E., du Plessis, P., and Meyer, H. (2019). Digital Sequence Information on Genetic Resources (DSI): An Introductory Guide for African Policymakers and Stakeholders. Deutsche Gesellschaft für Internationale Zusammenarbeit (GIZ) GmbH Bonn and Eschborn, 19.
Karsenti, E., Acinas, S. G., Bork, P., Bowler, C., De Vargas, C., Raes, J., et al. (2011). A holistic approach to marine eco-systems biology. PLoS Biol. 9:e1001177. doi: 10.1371/journal.pbio.1001177
Leary, D. (2018). Marine genetic resources in areas beyond national jurisdiction: do we need to regulate them in a new agreement? Mar. Safe Law J. 5, 22–47.
Lemaitre, R., Rahayu, D. L., and Komai, T. (2018). A revision of “blanket-hermit crabs” of the genus Paguropsis Henderson, 1888, with the description of a new genus and five new species (Crustacea, Anomura, Diogenidae). ZooKeys 752, 17–97.
Leonelli, S. (2016). Data-Centric Biology: A Philosophical Study. Chicago, IL: University of Chicago Press, 288.
Levin, L. A., Bett, B. J., Gates, A. R., Heimbach, P., Howe, B. M., Jannssen, F., et al. (2019). Global observing needs in the deep ocean. Front. Mar. Sci. 6:241.
Lyal, C., and Zhao, F.-W. (2020). EU-China Workshop on ABS and DSI: Access and Benefiting Sharing and Digital Sequence Information, 28-29 May, Virtually. Organised by the EU-China Environment Project. 8. Available online at: https://www.documents.clientearth.org/wp-content/uploads/library/2020-05-29-eu-china-workshop-on-abs-and-dsi-coll-en.pdf (accessed May 14, 2021).
Marlow, J., Harden-Davies, H., Snelgrove, P., Jaspars, M., Blasiak, R., and Wabnitz, C. (2019). The Full Value of Marine Genetic Resources (MGR). Southampton: Deep Ocean Stewardship Council, 4.
McCloud, T. G. (2010). High throughput extraction of plant, marine and fungal specimens for preservation of biologically active molecules. Molecules 15, 4526–4563. doi: 10.3390/molecules15074526
McQuillan, J. S., and Robidart, J. C. (2017). Molecular-biological sensing in aquatic environments: recent developments and emerging capabilities. Curr. Opin. Biotech. 45, 43–50. doi: 10.1016/j.copbio.2016.11.022
Morin, P. A., Leduc, R. G., Robertson, K. M., Hedrick, N. M., Perrin, W. F., Etnier, M., et al. (2006). Genetic analysis of killer whale (Orcinus orca) historical bone and tooth samples to identify western U.S. ecotypes. Mar. Mammal Sci. 22, 897–909. doi: 10.1111/j.1748-7692.2006.00070.x
Nelson, G., and Ellis, S. (2018). The history and impact of digitization and digital data mobilization on biodiversity research. Philos. Trans. R. Soc. B 374:20170391. doi: 10.1098/rstb.2017.0391
O’Leary, B. C., Allen, H. L., Yates, K. L., Page, R. W., Tudhope, A. W., McClean, C., et al. (2019). 30X30 A Blueprint for Ocean Protection: How We Can Protect 30% of Our Oceans by 2030. London: Greenpeace UK, 93.
Oldham, P., Hall, S., Barnes, C., Oldham, C., Cutter, M., Burns, N., et al. (2014). Valuing the Deep: Marine Genetic Resource in Areas Beyond National Jurisdiction Defra Contract MB0128– A Review of Current Knowledge Regarding Marine Genetic Resources and their Current and Projected Economic Value to the UK Economy. Final Report Version One. London: DEFRA.
Page, R. D. M. (2016). DNA barcoding and taxonomy: dark taxa and dark texts. Philos. Trans. R. Sci. B 371:20150334. doi: 10.1098/rstb.2015.0334
Pesant, S., Not, F., Picheral, M., Kandels-Lewis, S., Le Bescot, N., Gorsky, G., et al. (2015). Open science resources for the discovery and analysis of Tara Oceans data. Sci. Data 2:150023.
Rabone, M., Harden-Davies, H., Collins, J. E., Zajderman, S., Appeltans, W., Droege, G., et al. (2019). Access to marine genetic resources (MGR): raising awareness of best-practice through a new agreement for biodiversity beyond national jurisdiction (BBNJ). Front. Mar. Sci. 6:520.
Rainbow, P. S. (2009). Marine biological collections in the 21st century. Zool. Scr. 38(Suppl. 1), 33–40. doi: 10.1111/j.1463-6409.2007.00313.x
Rogers, A., Aburto-Oropeza, O., Appeltans, W., Assis, J., Balance, L. T., Cury, P., et al. (2020). Blue Paper 10: Critical Habitats and Biodiversity: Inventory, Thresholds, and Governance. Report prepared for the (Norwegian) Prime Minister’s High-Level Panel on a Sustainable Ocean Economy. London:World Resources Institute, 85.
Schiaparelli, S., Schnabel, K. E., Richer de Forges, B., and Chan, T.-Y. (2016). “Sorting, recording, preservation and storage of biological samples,” in Biological Sampling in the Deep Sea, eds M. R. Clark, M. Consalvey, and A. A. Rowden (Chichester: Wiley Blackwell), 338–367. doi: 10.1002/9781118332535.ch15
Schindel, D., Miller, S., Trizna, M., Graham, E., and Crane, A. (2016). The global registry of biodiversity repositories: a call for community curation. Biodivers. Data J. 4:e10293. doi: 10.3897/bdj.4.e10293
Selvanes, A. G. V., Devine, J., Jensen, K. H., Hestetun, J. T., Sjøtun, K., and Glenner, H. (2017). Marine Ecological Field Methods: A Guide for Marine Biologists and Fisheries Scientists. Chichester: Wiley-Blackwell, 240.
Smith, L. M., Barth, J. A., Kelley, D. S., Plueddemann, A., Rodero, I., Ulses, G. A., et al. (2018). The Ocean Observatories Initiative. Oceanography 31, 16–35.
Stocks, K., Jacobson-Stout, N., and Shank, T. M. (2016). “Information management strategies for deep-sea biological research,” in Biological Sampling in the Deep Sea, eds M. R. Clark, M. Consalvey, and A. A. Rowden (Chichester: Wiley Blackwell), 368–385. doi: 10.1002/9781118332535.ch16
Strand, J., Thomsen, H., Jensen, J. B., Marcussen, C., Nicolajsen, T. B., Skriver, M. B., et al. (2020). Biobanking in amphibian and reptilian conservation and management: opportunities and challenges. Conserv. Genet. Res. 12, 709–725. doi: 10.1007/s12686-020-01142-y
Sundar, V. C., Yablon, A. D., Grazul, J. L., Ilan, M., and Aizenberg, J. (2003). Fibre-optical features of a glass sponge. Nature 424, 899–900. doi: 10.1038/424899a
Thornburg, C. C., Britt, J. R., Evans, J. R., Akee, R. K., Whitt, J. A., Trinh, S. K., et al. (2018). NCI program for natural product discovery: a publicly-accessible library of natural product fractions for high-throughput screening. ACS Chem. Biol. 13, 2484–2497. doi: 10.1021/acschembio.8b00389
Tolochko, P., and Vadrot, A. B. M. (2021). The usual suspects? Distribution of collaboration capital in marine biodiversity research. Mar. Policy 124:104318. doi: 10.1016/j.marpol.2020.104318
Trindade, M., van Zyl, L. J., Navarro-Fernández, J., and Elrazak, A. A. (2015). Targeted metagenomics as a tool to tap into marine natural product diversity for the discovery and production of drug candidates. Front. Microbiol. 6:890.
UN (2019). Revised Draft text of an Agreement Under the United Nations Convention on the Law of the Sea on the Conservation and Sustainable use of Marine Biological Diversity of Areas Beyond National Jurisdiction. Intergovernmental Conference on an International Legally Binding Instrument Under the United Nations Convention on the Law of the Sea on the Conservation and Sustainable use of Marine Biological Diversity of Areas Beyond National Jurisdiction (Fourth Session, New York, 23 March–3 April 2020). UN doc A/CONF.232/2020/3. 18 November 2019. New York, NY: UN.
UNESCO (2017). Global Ocean Science Report. Paris: United Nations Educational, Scientific and Cultural Organisation, 272.
UNGA (2015). Development of an International Legally Binding Instrument Under the United Nations Convention on the Law of the Sea on the Conservation and Sustainable use of Marine Biological Diversity of Areas Beyond National Jurisdiction. United Nations General Assembly Resolution 69/292. 69th sess, Agenda Item 74(a). UN doc A/RES/69/292. New York, NY: UNGA.
UNGA (2017). International Legally Binding Instrument under the United Nations Convention on the Law of the Sea on the Conservation and Sustainable use of Marine Biological Diversity of Areas Beyond National Jurisdiction. United Nations General Assembly Resolution 72/249. 72nd sess, Agenda Item 77. UN doc A/RES/72/249. New York, NY: UNGA.
van Santen, J. A., Jacob, G., Singh, A. L., Aniebok, V., Balunas, M. J., Bunsko, D., et al. (2019). The Natural Products Atlas: an open access knowledge base for microbial natural products discovery. ACS Central Sci. 5, 1824–1833.
Vierros, M. K., and Harden-Davies, H. (2020). Capacity building and technology transfer for improving governance of marine areas both beyond and within national jurisdiction. Mar. Policy 122:104198.
Vonnahme, T. R., Molari, M., Janssen, F., Wenzhöfer, F., Haeckel, M., Titschack, J., et al. (2020). Effects of a deep-sea mining experiment on seafloor microbial communities and functions after 26 years. Sci. Adv. 6:eaaz5922. doi: 10.1126/sciadv.aaz5922
Wieczorek, J., Bloom, D., Guralnick, R., Blum, S., Döring, M., Giovanni, R., et al. (2012). Darwin Core: an evolving community-developed biodiversity data standard. PLoS One 7:e29715. doi: 10.1371/journal.pone.0029715
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., et al. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 3:160018.
Williamson, C. J., Walker, R. H., Robba, L., Yesson, C., Russell, S., Irvine, L. M., et al. (2015). Toward resolution of species diversity and distribution in the calcified red algal genera Corallina and Ellisolandia (Corallinales, Rhodophyta). Phycologia 54, 2–11. doi: 10.2216/14-024.1
Wilson, R. (2005). Marine Invertebrate Sample Processing Procedures. Melbourne, VIC: Museum of Victoria, 26.
Wolfender, J.-L., Litaudon, M., Touboul, D., and Queiroz, E. F. (2019). Innovative omics-based approaches for prioritisation and targeted isolation of natural products – new strategies for drug discovery. Nat. Prod. Rep. 36:855. doi: 10.1039/c9np00004f
Wright, G., Rochette, J., Gjerde, K., and Seeger, I. (2018). The long and winding road: negotiating a treaty for the conservation and sustainable use of marine biodiversity in areas beyond national jurisdiction. IDDRI Stud. 18:82.
Yao, H., Dao, M., Imholt, T., Huang, J., Wheeler, K., Bonilla, A., et al. (2010). Protection mechanisms of the iron-plated armor of a deep-sea hydrothermal vent gastropod. Proc. Natl. Acad. Sci. U.S.A. 107, 987–992. doi: 10.1073/pnas.0912988107
Keywords: high seas, marine genetic resources, access and benefit sharing, UNCLOS, developing states
Citation: Rogers AD, Baco A, Escobar-Briones E, Currie D, Gjerde K, Gobin J, Jaspars M, Levin L, Linse K, Rabone M, Ramirez-Llodra E, Sellanes J, Shank TM, Sink K, Snelgrove PVR, Taylor ML, Wagner D and Harden-Davies H (2021) Marine Genetic Resources in Areas Beyond National Jurisdiction: Promoting Marine Scientific Research and Enabling Equitable Benefit Sharing. Front. Mar. Sci. 8:667274. doi: 10.3389/fmars.2021.667274
Received: 12 February 2021; Accepted: 29 April 2021;
Published: 31 May 2021.
Edited by:
Elizabeth Mendenhall, University of Rhode Island, United StatesReviewed by:
Nengye Liu, Macquarie University, AustraliaCopyright © 2021 Rogers, Baco, Escobar-Briones, Currie, Gjerde, Gobin, Jaspars, Levin, Linse, Rabone, Ramirez-Llodra, Sellanes, Shank, Sink, Snelgrove, Taylor, Wagner and Harden-Davies. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alex D. Rogers, YWxleC5yb2dlcnNAcmV2b2NlYW4ub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.