
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 13 May 2021
Sec. Aquatic Physiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.662220
This article is part of the Research TopicThe Cellular Stress Response and Physiological Adaptations of Corals Subjected to Environmental Stressors and PollutantsView all 19 articles
Coral reefs at the northern extent of Florida’s coral reef tract are exposed to many localized anthropogenic influences including controlled freshwater discharges, runoff, upwelling, and seasonal environmental variability. To better understand coral responses to sublethal stressors in nearshore environments, we conducted complementary experiments to assess the impacts of estuarine runoff and temperature stress on local populations of the scleractinian coral species, Montastraea cavernosa, using Tag-Seq global gene expression profiling. In an in situ time series experiment, fate-tracked colonies were sampled during periods of relatively low and high estuarine discharge over 4 years to investigate temporal trends in transcriptional patterns and to identify if coral stress indicators were regulated through time. There was significant transcriptomic variation through time, but patterns did not appear to be attributed to distance from nearby estuarine tidal flux. In an ex situ factorial experiment, clonal replicates of coral genotypes were exposed to temperature (25°C and 30°C) and water (offshore and estuarine discharge, representing typical oceanic conditions and episodic discharge conditions, respectively) treatments to quantify the potential individual and synergistic effects of sublethal stress on coral and algal gene expression. Comparative analyses suggested that corals and their algal symbionts were more responsive to thermal stress than to estuarine discharge, although there was evidence of a synergistic relationship between the two stressors. Strong genotype effects also demonstrated that transcriptomic responses to thermal stress were largely based on coral genotype, indicating the potential for stress resilience among certain members of coral populations from southeast Florida.
Increases in coastal development and changes to watershed use in recent decades have had profound implications for nearshore coral reefs. Coral reef habitats at the northern extent of Florida’s coral reef tract are of particular concern, residing at their latitudinal environmental limits while also increasingly exposed to global and local anthropogenic stressors including land-based sources of pollution (e.g., freshwater runoff, sedimentation), annual temperature flux, and warming sea temperatures (Lirman and Fong, 2007; Gregg, 2013). Within this region, coral reefs inhabit an ecotone transition between subtropical and warm temperate climate zones (Lugo et al., 1999). Reefs in southeast Florida face high levels of both seasonal and episodic environmental variation that combine with anthropogenic impacts to impose multiple stressors on scleractinian corals. Land-based influences (e.g., controlled freshwater discharge and agricultural runoff), as well as natural environmental variability (e.g., seasonal rainfall and upwelling events), impact coastal habitats in the region (Yentsch et al., 2002; Beal et al., 2012; Gregg, 2013).
Two distinct climatological seasons dominate weather patterns along nearshore reefs in southeast Florida: (1) a wet season characterized by increased freshwater discharge and runoff, nutrient over-enrichment, and migration of dark, low-salinity water over coastal zones, and (2) a subsequent dry season during the winter with decreased rainfall and runoff (Beal et al., 2012). This pattern is also influenced by global environmental variations such as the El Niño Southern Oscillation and regional tropical storm intensity and duration (Lapointe et al., 2012). During periods of prolonged rainfall, coastal habitats including nearshore coral reefs are frequently subjected to discharge from a series of natural tributaries and artificial canals which are meant to contain stormwater runoff (Graves et al., 2004; Lapointe et al., 2012). This system controls water levels in Lake Okeechobee and manages a portion of flow from the surrounding watershed via periodic releases through control structures located on major canals (e.g., C-23, C-24, C-44; Figure 1). As a result, the majority of runoff water is drained from urban and agricultural lands into coastal zones, rather than into the Everglades to the south (Perry, 2004). Characteristics of discharge water during high levels of flow from the watershed have been documented to include increased levels of total suspended solids, water color (i.e., colored dissolved organic matter; CDOM), and nutrients such as nitrogen and phosphorous (Collier et al., 2008; Lapointe et al., 2017).
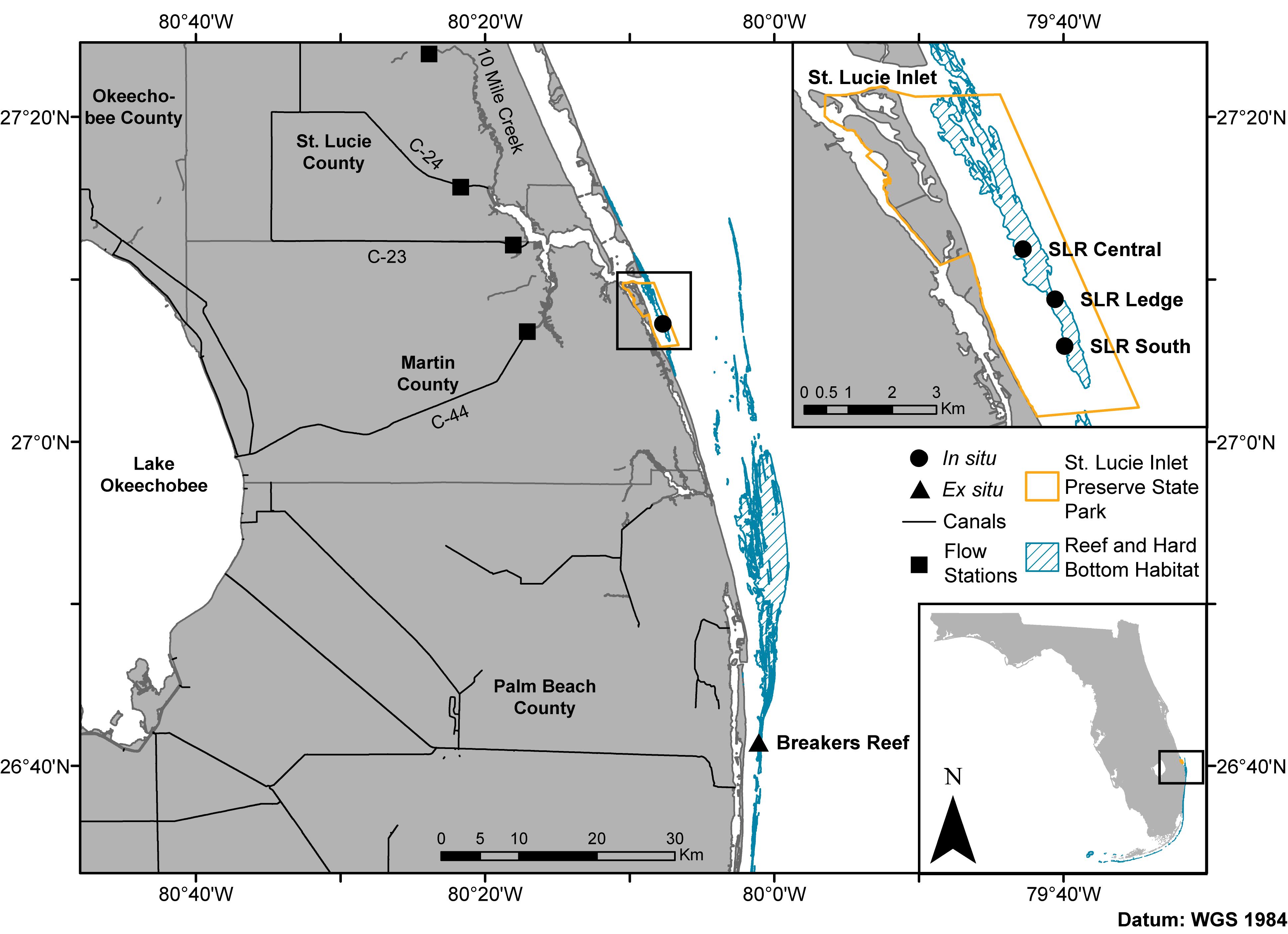
Figure 1. Map of the northern extent of Florida’s coral reef tract including the sampling sites in the in situ experiment (circles: SLR Central, SLR Ledge, and SLR South) and coral collection site for the ex situ experiment (triangle: Breakers Reef). Squares represent canal control structures where discharge flow rates and rainfall (C-44 only) data were obtained from the South Florida Water Management District’s DBHYDRO database. State park shapefile sourced from the Florida Department of Environmental Protection’s Division of Recreation and Parks, reef and hard bottom habitat shapefile sourced from the Florida Fish and Wildlife Conservation Commission-Fish and Wildlife Research Institute, canal shapefile sourced from the South Florida Water Management District.
Restructuring and management of the Okeechobee and St. Lucie watersheds has drastically changed environmental stability through time, whereas water quality can rapidly fluctuate near St. Lucie Inlet at the southern terminus of the Indian River Lagoon (Pickering and Baker, 2015). This region also corresponds with the northernmost portion of Florida’s coral reef tract, St. Lucie Reef (SLR), located within the St. Lucie Inlet Preserve State Park (Figure 1 inset). This nearshore reef environment begins directly outside of the St. Lucie Inlet and continues approximately 7 km to the south (Walker and Gilliam, 2013). St. Lucie Reef is the northern known limit of at least eight scleractinian and octocoral species along the reef tract (Reed, 1982; Beal et al., 2012). Near the inlet, the reef is composed primarily of wormrock (Phragmatopoma lapidosa), with an increase in diversity and abundance of scleractinian corals toward the southern end of the park. The observed gradient of scleractinian diversity is hypothesized to be a result of increasing estuarine influence closer to the inlet (Beal et al., 2012; Shatters, 2017). Despite highly variable conditions, SLR supports a diverse community of 24 species of scleractinian corals, 100 other species of invertebrates, 23 algal species, four sea turtle species, and approximately 250 fish species (Beal et al., 2012). The two dominant scleractinian coral species found at SLR are Pseudodiploria clivosa and Montastraea cavernosa, with the former occurring throughout, and the latter absent from the northern third of the reef nearest the inlet (Walker and Gilliam, 2013). Their persistence on the reef despite highly variable water quality parameters may be in part due to their prolific ability to shed sediments through excessive mucus production and sloughing, heterotrophic feeding, and skeletal morphology characteristics (Bak and Elgershuizen, 1976; Lasker, 1980; Vargas-Ángel et al., 2006, 2007).
There have been ongoing efforts to monitor reef populations and seasonal environmental variability within the state park since 2006 (Beal et al., 2012; Walker and Gilliam, 2013; Gilliam et al., 2018). From 2010 to 2017, quarterly surveys have assessed survival of fate-tracked colonies of the dominant scleractinian species, P. clivosa and M. cavernosa, with occasional tissue sampling events to quantify coral health through physiological, histological, and population genetics approaches. These two species have shown relatively stable coral-algal assemblages over time, suggesting resilient Symbiodiniaceae associations despite environmental and seasonal changes (Klepac et al., 2015). Coral transcriptomic analyses using microarrays, however, have documented an increase in expression of stress response genes during the wet season (Beal et al., 2012). Histological analyses have indicated that these two coral species at SLR are not actively reproductive (Beal et al., 2012), despite evidence of historical population connectivity to other reefs ∼20–50 km to the south (Dodge et al., 2020). Coral skeletons appeared less dense in fate-tracked colonies relative to conspecifics elsewhere along Florida’s coral reef tract, with additional tissue and skeletal loss due to the boring sponges Cliona spp. (Beal et al., 2012). In spite of these stressors, coral mortality was low at SLR from 2010 until Hurricane Irma in 2017 (Walker, 2018) and the recent outbreak of stony coral tissue loss disease (SCTLD) along Florida’s coral reef tract starting in 2014 (reaching the northern regions in 2017) (Walton et al., 2018). Since 2017, monitoring efforts have increased both temporally and by individual fate-tracking to include all remaining scleractinian species on the reef (Voss and Combs, 2018; Walker, 2018). Prior to widespread coral mortality of the two dominant species at SLR due to SCTLD, colonies were remarkably persistent in the face of hypervariable conditions and stress.
This study builds on a decade of monitoring efforts to describe, at the molecular level, how corals at SLR respond to environmental variability. Using complementary temporal in situ and manipulative ex situ designs, we sought to quantify transcriptomic variation among individuals exposed to different levels of environmental stress. Recent advancements in RNA sequencing (RNA-seq) and analysis pipelines have enhanced assessments of coral health through whole-transcriptome gene expression profiling, especially in response to stressors (Kenkel et al., 2011, 2013b, 2014; Meyer et al., 2011; Wright et al., 2019). There is a growing body of literature examining transcriptomic responses to thermal stress in corals (Meyer et al., 2011; Kenkel et al., 2013b, 2014; Anderson et al., 2016; Davies et al., 2016; Dixon et al., 2020); though few studies to date have examined transcriptomic variability in corals exposed to estuarine and riverine runoff [Aguilar et al., 2017, 2019; Edge et al., 2013; Mayfield et al., 2013; see non-transcriptomic responses reviewed by Fabricius (2005)], or synergistic effects of multiple stressors therein (Wright et al., 2019). St. Lucie Reef provides an ecologically relevant scenario to investigate the potential synergistic impacts of thermal stress and estuarine discharge on coral health. Through comparative analyses between two experiments, we aimed to identify common responses to thermal and water quality stressors, in order to provide insight into the fundamental metabolic processes that may enhance stress tolerance and coral resilience in marginal reef environments.
The scleractinian coral Montastraea cavernosa was chosen as the study species for both experiments as it is widely distributed throughout the study area, has annotated transcriptomic and genomic references (Kitchen et al., 2015)1, and has temporally stable Symbiodiniaceae communities (Cladocopium spp.) documented for the fate-tracked colonies in the in situ experiment (Klepac et al., 2015). Fifteen M. cavernosa colonies were repeatedly sampled over nine time points spanning periods of relatively low and high estuarine discharge in 2013–2016, resulting in a total of 92 coral tissue fragments collected (Table 1). Variable water visibility and sea state conditions occasionally prevented collection of tissue samples from all colonies at each time point. Sampling occurred at three sites along St. Lucie Reef (SLR Central, SLR Ledge, SLR South; depths ranging from 2.4 to 9.7 m; Table 1 and Figure 1) with increasing distance from the St. Lucie Inlet, which has been shown to approximate a gradient of decreasing exposure to estuarine discharge water (Beal et al., 2012; Shatters, 2017). Cumulative estuarine discharge flow rates measured at the canals C-44, C-23, C-24, and Ten Mile Creek (Figure 1) were obtained from the South Florida Water Management District’s DBHYDRO database (Supplementary Figure 1), where daily flow rates were averaged for 2 weeks leading up to the corresponding sampling time points. Daily rainfall data were obtained from the same database at the C-44 canal control structure. Small tissue fragments (∼5 cm2) were collected from the perimeter of each colony using a hammer and chisel, transported to the surface in separate zip-top bags containing ambient seawater, and preserved in TRIzol reagent within 30 min of collection. Preserved samples were then kept on ice until storage at −80°C prior to RNA extraction and library preparation.
Full details of the experimental apparatus used in the ex situ experiment are described in Shatters (2017). In summary, a two-factor (temperature and water) factorial apparatus was constructed using twelve 150-L glass aquaria across six open-top raceways. Three tank replicates per each of the four experimental treatments were randomly distributed among the raceways, with continuous monitoring of temperature, pH, and ORP with Neptune Systems Apex controllers, daily monitoring of salinity with a refractometer, and a 9-h photoperiod using Hydro Grow Sol LED lights set to comparable PAR levels (∼102 μmol m–2 s–1) found at the coral collection site based on in situ data collection with Onset HOBO data loggers. Temperature treatment levels (control: 25°C; elevated: 30°C) were determined from mean temperatures recorded before and during discharge events, respectively, by Odyssey conductivity and temperature data loggers deployed from October 2013 to January 2015 at the St. Lucie Reef study sites (Shatters, 2017; Figure 1 inset). Raceways were temperature-controlled using immersion heaters and in-line chillers to reflect the two temperature treatments, with three raceways per temperature treatment. Offshore water (∼32 psu) was collected via a subsurface pump from the Florida Oceanographic Society at high tide. The discharge water parameters were similarly determined from salinity data obtained before and during discharge events, where the mean salinity of the estuarine discharge plume during outgoing tides was ∼28 psu. Discharge water was collected via a pump at 0.5 m depth in the inlet during low slack tide, 5 days after flow rates through the canals from Lake Okeechobee reached a flow rate of 99.02 m3 s–1 (Shatters, 2017). The resulting water (19 psu) was then adjusted with offshore water to achieve the treatment salinity of 28 psu, resulting in a ∼17/83% mix between discharge and offshore water, respectively. Both water types were initially filtered (50, 10, then 1 μm) and UV-sterilized, and stored in opaque holding tanks.
Due to state park regulations restricting whole-colony collection, eleven Montastraea cavernosa colonies of 150–175 cm2 in size with no tissue loss, paling, or other visible signs of stress were collected south of SLR at Breakers Reef (26.6908 N, −80.0181 W) at depths of 13–17 m (Figure 1). Temperature data loggers deployed in June 2015 confirmed that ambient temperature was similar between sites (∼28°C) despite difference in depth. Following transport to the lab, colonies were cut with a tile saw into twelve 3 cm × 4 cm colony replicates [genotyped in Dodge et al. (2020)], and randomly positioned in each of the twelve treatment tanks (n = 33 per treatment, n = 132 total; Table 2). The ex situ experiment was conducted over 32 days, with a 7-day recovery/acclimatization period, 5-day ramp to experimental conditions (achieved with daily ∼15% water changes and <1°C temperature adjustments), and 20-day experimental treatment. Coral fragments were fed ∼10 g of brine shrimp per tank every 3–4 days over the course of the experiment. At the end of the treatment period, coral fragments were split into subsamples for gene expression and symbiont/chlorophyll analyses, and preserved in TRIzol/−80°C and dry/−20°C, respectively.
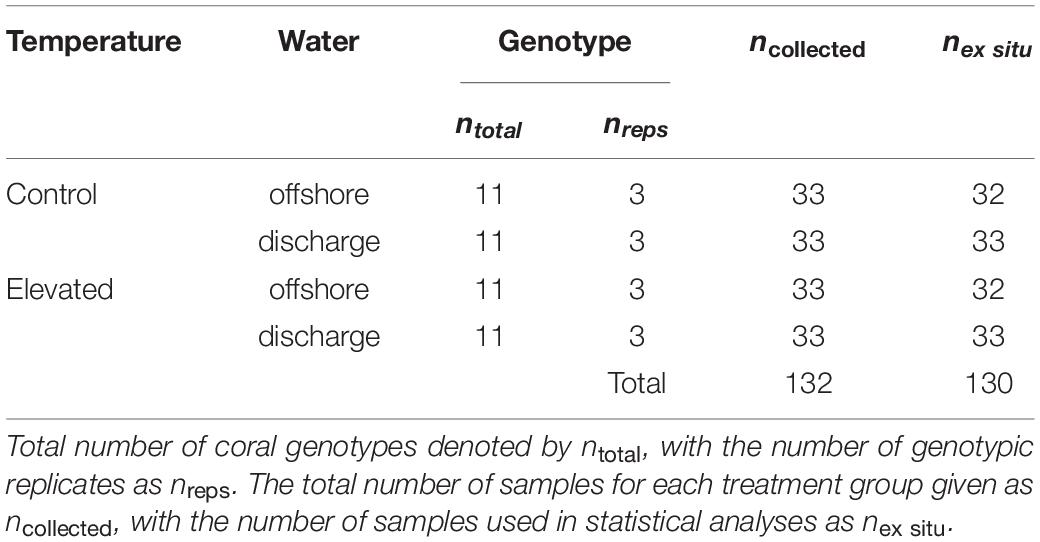
Table 2. Sampling information for temperature and water treatment factors in the ex situ experiment.
Symbiont and chlorophyll metrics, including Symbiodiniaceae density (cells cm–2), areal chlorophylls a and c2 (chl cm–2), cellular chlorophylls a and c2 (chl cell–1), and chlorophyll a:c2 ratio, were quantified from the frozen subsamples as described in Shatters (2017). All statistical analyses were conducted in the R statistical environment (R Core Team, 2019). Symbiont and chlorophyll metrics could not be normalized with data transformation, therefore non-parametric tests were used. Data were first assessed for homogeneity of variance among treatment groups using the betadisper function of the package vegan (Oksanen et al., 2015), utilizing Euclidean distance matrices and 9,999 permutations. Following identification of statistically equal variance (all p > 0.05), univariate PERMANOVAs were conducted using the adonis function in vegan to identify potential effects of experimental treatments (genotype, temperature, water) on symbiont and chlorophyll metrics. Pairwise comparisons were made among single-factor combinations of temperature and water treatments for each of the metrics using the nctp function of the package nparcomp (Konietschke et al., 2015). Multivariate effects were assessed using a three-factor PERMANOVA and pairwise colony tests using the packages vegan and RVAideMemoire (Hervé, 2019), respectively. Both multivariate tests used the same model parameters as indicated before, with FDR p value correction for the pairwise comparisons.
For both experiments, RNA extractions and Tag-Seq library preparation protocols were followed as in Studivan and Voss (2020). In short, total RNA was extracted using a modified phenol-chloroform protocol (Chomczynski and Sacchi, 1987, 2006), DNase I treated using an Ambion kit, and purified using a LiCl precipitation. Only samples with at least 1 μg of total RNA were used for library preparation according to the Tag-Seq protocol first described in Meyer et al. (2011) and modified as in Studivan (2020b). In summary, cDNA libraries were amplified over 22 cycles, dual-barcoded with individual 5′ and 3′ Illumina indices, and equally combined using qPCR into three pools encompassing samples from both experiments. A total of 275 sample pools were generated (in situ: n = 111; ex situ: n = 164), including libraries that were removed for subsequent statistical analyses. Pools were then sequenced at the University of Wisconsin Biotechnology Center on an Illumina HiSeq 2500 with V4 chemistry and 15% PhiX spike-in, generating 1 × 50 bp single-end reads. For all samples across both experiments, 1.30 billion reads were produced, with a mean number of 4.73 ± 0.16 (mean ± SE) million reads per sample.
Raw sequences were first processed separately for each experiment to obtain cleaned, mapped, and quantified reads using pipelines described in a GitHub repository associated with a previous study (Studivan, 2020b; Studivan and Voss, 2020). Cleaned reads were mapped to a concatenated reference transcriptome composed of the coral host M. cavernosa (Kitchen et al., 2015; updated genome-derived transcriptome from https://matzlab.weebly.com/data–code.html), and its dominant algal symbiont identified at SLR (Klepac et al., 2015; Supplementary Data 1), Cladocopium spp. (Davies et al., 2018). Creation and annotation of the combined coral host and algal symbiont reference transcriptome is described in a GitHub repository associated with a previous study (Studivan, 2020a; Studivan and Voss, 2020). Subset datasets were created to remove additional samples not part of the respective in situ and ex situ experimental designs. For the in situ experiment, 49 samples were retained (seven colonies with complete sampling coverage across seven time points) to achieve a balanced design with sufficient replication (Table 1). For the ex situ experiment, 130 samples were retained as part of the design (Table 2). Mapping efficiency to the sequenced samples from both experiments was 17.0 ± 0.3%, resulting in 24,182 isogroups for the in situ experiment (15,712 host and 8,470 symbiont), and 27,933 isogroups for the ex situ experiment (18,210 host and 9,723 symbiont). Low mapping efficiency has been previously shown to be an issue with the updated version of the M. cavernosa transcriptome (Studivan and Voss, 2020), however, improved mapping with the original transcriptome was countered by relatively poor GO and KOG functional annotations. Statistical analyses were therefore conducted using counts generated from the updated coral transcriptome, following the separation of count data into host and symbiont datasets for separate statistical analyses of coral and algal gene expression for each experiment.
Datasets were pre-filtered to remove low-count genes (cumulative count <10 across all samples in a dataset) to decrease processing time, and then tested for outliers using the package arrayQualityMetrics (Kauffmann et al., 2009). Samples violating the distances between sample arrays (Sa) criterion for each of the respective datasets were removed, whereas three host and one symbiont samples were removed from the in situ experiment (Supplementary Figure 2), and three host and five symbiont samples from the ex situ experiment (Supplementary Figure 3). Count data were normalized using the variance stabilized transformation in the package DESeq2 (Love et al., 2014) for data visualization and PERMANOVAs. PERMANOVAs were conducted using vegan with 9,999 permutations for both experiments to identify if model terms were significantly affecting gene expression patterns in either the coral host or algal symbionts. Multivariate differences among samples were visualized using PCoAs of Manhattan distance in vegan and heatmaps of individual gene expression patterns using the package pheatmaps (Kolde, 2019). Tests of differential expression for both experiments were conducted using DESeq2 to identify differentially expressed genes (DEGs) associated with factors (model terms) and among factor levels (contrast statements) using the “reduced” flag on the respective main models. For the in situ experiment, a two-factor main model of time and site was used, with pairwise comparisons between sites and time points. For the ex situ experiment, a three-factor model of coral genotype, temperature, and water was used with interaction terms between the main model factors and a blocking factor (to assess whether tank location had an impact on gene expression patterns). Additionally, pairwise comparisons were made between single-factor levels of temperature and water treatments.
Differentially expressed genes were exported for each of the respective comparisons within experiments and analyzed for larger expression patterns across gene pathways using the package GO-MWU (Wright et al., 2015) for gene ontology (GO) databases, and the package KOGMWU (Dixon et al., 2015; Matz, 2016; Strader et al., 2016) for eukaryotic orthologous groups (KOGs). Both GO and KOG databases contain universal functional group annotations of individual genes based on biological and biochemical functions (Ashburner et al., 2000; Tatusov et al., 2000, 2003; The Gene Ontology Consortium, 2017). GO-MWU identifies GO categories that are significantly enriched by up-regulated or down-regulated genes using Mann–Whitney U tests. The resulting outputs identify significantly enriched GO categories and their relative levels of expression across the three GO divisions (Molecular Function, Biological Processes, and Cellular Components) according to an FDR-corrected p value cutoff of 0.05, or a fold change cutoff of 1. The KOGMWU analysis is similar, although enrichment of functional pathways is compared across universal KOG categories to produce a heatmap (Nordberg et al., 2014).
Prior to weighted gene co-expression network analysis (WGCNA) of the ex situ experiment, host and symbiont datasets were further filtered to remove genes with counts <10 across >90% of samples. Correlational networks were then created blind to experimental design based on gene expression patterns using the package WGCNA (Zhang and Horvath, 2005; Langfelder and Horvath, 2008). In short, groups of genes with similar expression patterns (modules) were first identified across samples, then correlated to experimental factors (coral genotypes, temperature/water treatments, symbiont, and chlorophyll metrics) based on strength of the module-trait associations. The protocol used to conduct WGCNA, as well as scripts for all other analyses described above, can be found in a GitHub repository associated with this study (Studivan, 2021). Soft thresholds were adjusted to fit each particular dataset (11 for host, 4 for symbiont) to achieve a suitable scale-free topology index, with no merging of original modules. Module-associated genes were then exported from the host dataset for GO enrichment analysis.
Pre-filtering of raw counts produced datasets of 8,497 coral host and 1,652 symbiont transcripts across 46 and 48 samples, respectively. Analysis of gene expression patterns across in situ experiment factors (site, time) using PERMANOVA yielded that only time was a significant factor for both host and symbiont datasets (host: R2 = 0.24537, p < 0.0005; symbiont: R2 = 0.27559, p < 0.0001; Table 3). DESeq2 identified 927 host and 50 symbiont differentially expressed genes (DEGs) associated with time, while only 60 host and 30 symbiont DEGs were identified for site, and zero DEGs for the interaction term (Supplementary Table 1). DEGs identified for each of the pairwise comparisons across sites and sampling time points can also be found in Supplementary Table 1. PCoAs and heatmaps of samples through time suggested that transcriptomic variation was primarily attributed to differences from the first two time points (October 2013 and September 2014) to all subsequent time points (Figure 2). There appeared to be limited overlap in the PCoAs between samples collected during periods of relatively high estuarine discharge and rainfall (indicated by cooler colors in Figures 2A,C and Supplementary Figure 1), however, similarities between sample dispersions from consecutive time points (e.g., October 2013 and September 2014) suggest that other environmental factors beyond discharge rates alone may have influenced gene expression patterns in both the coral host and their algal symbionts.
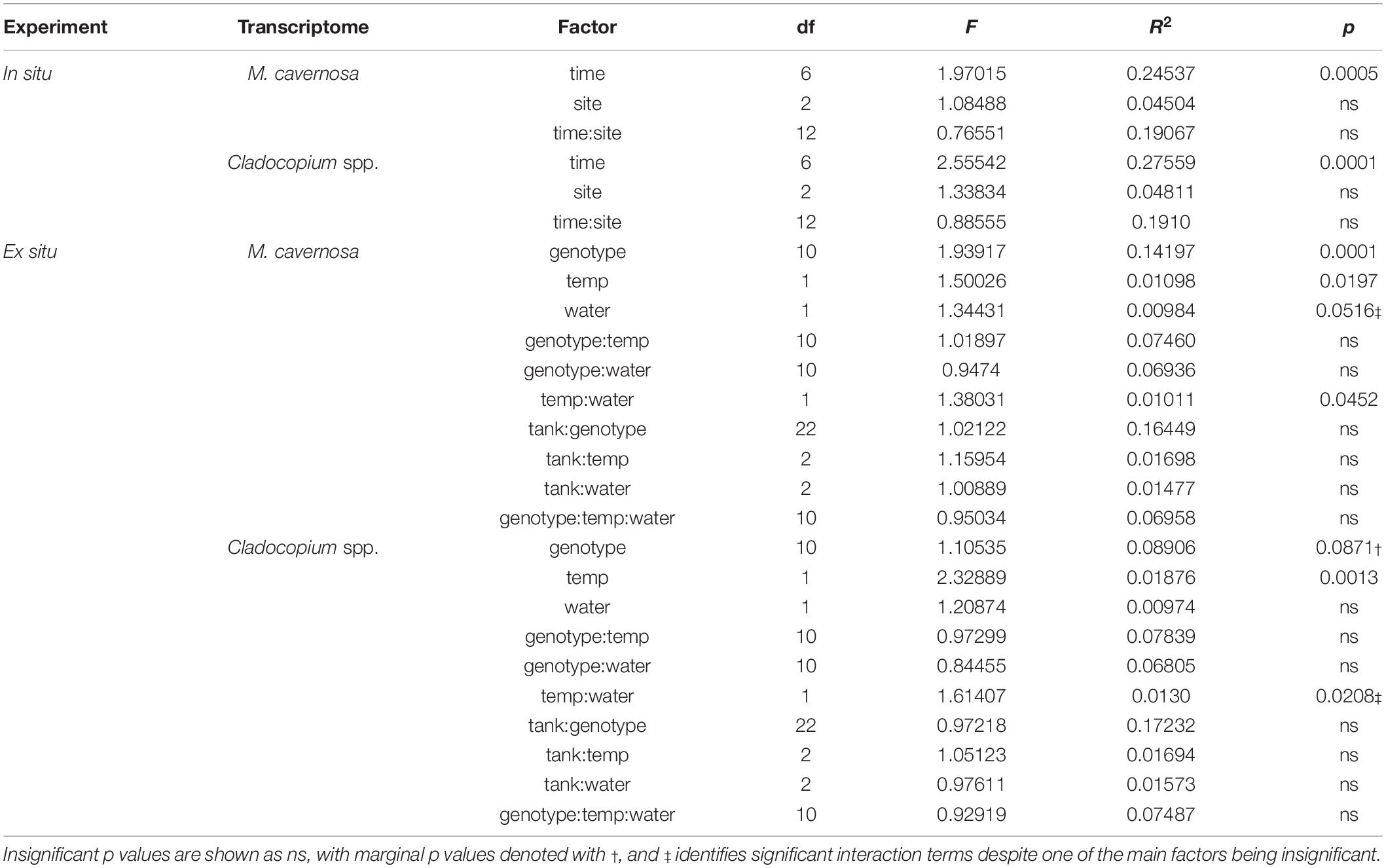
Table 3. PERMANOVA model results for in situ and ex situ experiments, split into coral host (M. cavernosa) and symbiont (Cladocopium spp.) datasets.
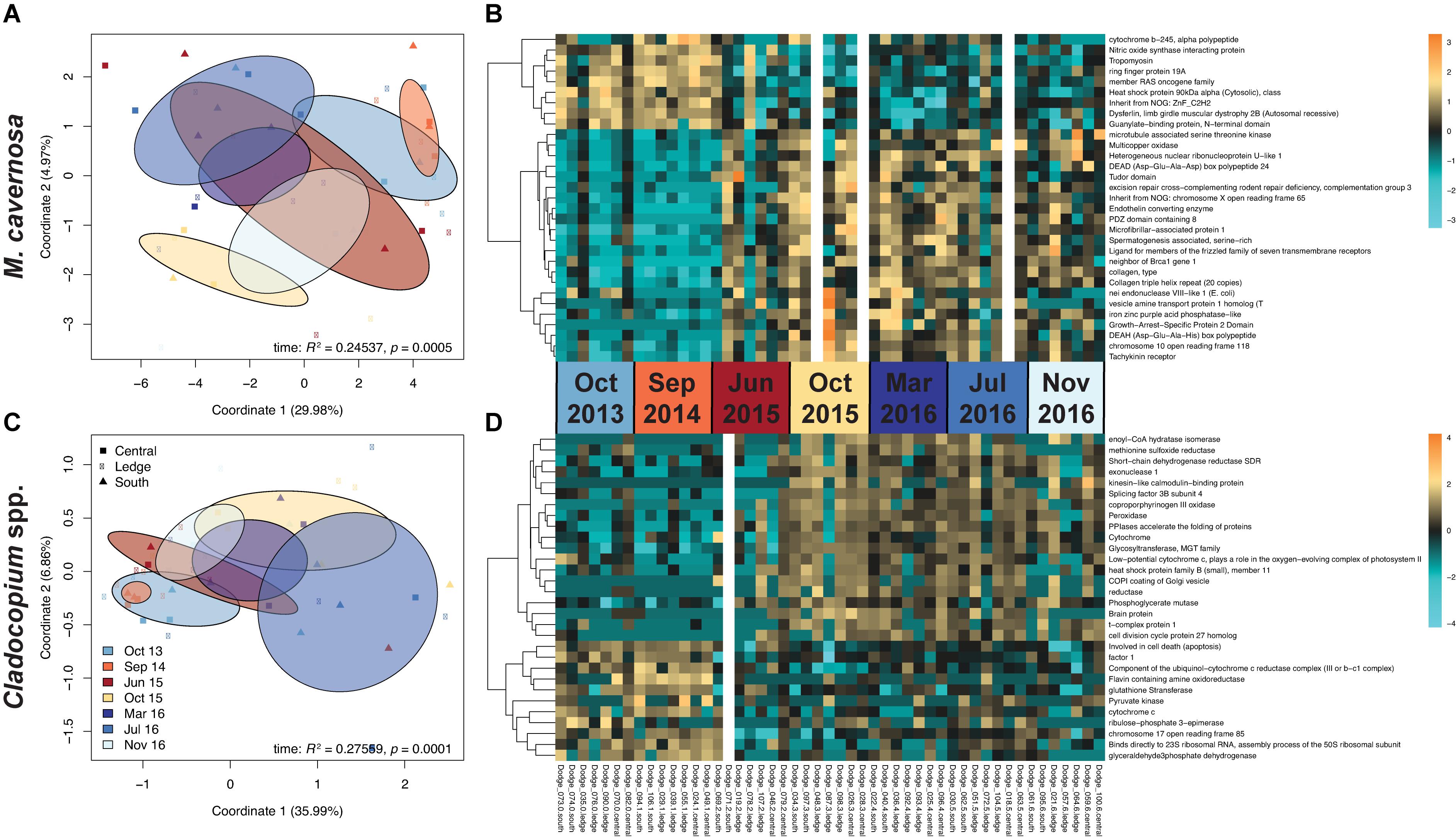
Figure 2. (A,C) PCoAs of coral host and symbiont datasets depicting transcriptomic variation through time from the in situ experiment. Colors correspond to sampling time point, where warmer colors represent periods of relatively low estuarine discharge (South Florida Water Management District DBHYDRO; September 2014: 6.15 m3 s– 1, June 2015: 3.08 m3 s– 1, October 2015: 14.29 m3 s– 1) and cooler colors represent periods of higher discharge (October 2013: 44.99 m3 s– 1, March 2016: 70.52 m3 s– 1, July 2016: 59.37 m3 s– 1, November 2016: 33.31 m3 s– 1). Ellipses denote the dispersion of each time point’s samples, and sites are indicated by point shapes. Test statistics and p values were generated from PERMANOVAs in vegan. (B,D) Heatmaps of the most highly significant DEGs, organized by site within time points using a Benjamini-Hochberg-corrected p value cutoff of 1e– 6 for host and 0.01 for symbiont datasets. Gaps represent outlier samples removed from each dataset. Increasing intensities of gold indicate higher expression of genes in samples, while increasing intensities of turquoise indicate lower expression.
Since site did not have a significant impact on gene expression, subsequent analyses were conducted among sampling time points, combining samples across sites. Analysis of gene ontology (GO) categories enriched for the coral host dataset over sampling time points identified two main groups of pairwise time comparisons. From the first two time points (October 2013 and September 2014) to subsequent time points, all but one GO category in the biological process (BP) division demonstrated reduced expression in the later versus earlier time point (Supplementary Figure 4). Of these, several categories were consistently represented in pairwise time comparisons, including: “RNA catabolic process,” “nuclear-transcribed mRNA catabolic process, nonsense-mediated decay,” “protein localization to endoplasmic reticulum,” and “protein-containing complex disassembly.” Between time points after October 2015, the majority of GO categories were instead over-expressed in later versus earlier time points, including several of the aforementioned categories (Supplementary Figure 4). This indicated a shift in expression of gene pathways through time, primarily occurring between 2013 and 2015–2016 time points, and between 2015 and 2016 time points. For the symbiont dataset, only one GO category in the BP division was significantly enriched in pairwise comparisons of time points (“negative regulation of metabolic process,” comparing March 2016 versus June 2015).
Analysis of eukaryotic orthologous groups (KOGs) identified only one KOG class in the host dataset with relatively strong variation through time, where “nuclear structure” was negatively enriched between the first two time points and subsequent time points, and represented by higher-expressed genes between 2015 and 2016 time points (Supplementary Figure 5). Other KOG classes, including “cell motility,” “nucleotide transport and metabolism,” and “extracellular structures” demonstrated mild variation in expression through time. For the symbiont dataset, however, the majority of the KOG classes contained under-expressed genes in later versus earlier time points, with the exception of “extracellular structures” that showed variation through time. This KOG class was more highly-expressed in 2015–2016 versus 2013–2014 time points, but less-expressed in 2016 versus 2015 time points (Supplementary Figure 5).
Coral genotype and temperature treatments were significant factors affecting all symbiont and chlorophyll metrics, with the exception of cellular chlorophylls a and c2 for temperature (Supplementary Table 2). Only areal chlorophyll a was marginally affected (p < 0.0612) by the water treatment; all other metrics were non-significant. Multivariate pairwise differences among coral genotypes are found in Supplementary Table 3. Significant pairwise p values between single-factor combinations of temperature and water treatments were only found for comparisons between control and elevated temperature treatments for symbiont density and both areal chlorophyll metrics. Symbiont density and areal chlorophylls a and c2 were significantly reduced in both elevated temperature treatments, with no significant pairwise differences between offshore and discharge water treatments, respectively (Supplementary Figure 6). The ratio of chlorophylls a:c2 and cellular chlorophylls a and c2 did not vary significantly among treatments.
Pre-filtering of raw counts resulted in 11,248 coral host and 1,834 symbiont transcripts across 127 and 125 samples, respectively. PERMANOVA results for the ex situ experiment indicated that genotype and temperature were significant factors, while water was marginally significant for the host dataset, and there was a significant temperature:water interaction (genotype: R2 = 0.14197, p < 0.0001; temperature: R2 = 0.01098, p < 0.0197; water: R2 = 0.00984, p < 0.0516; temperature:water: R2 = 0.01011, p < 0.0452; Table 3). For the symbiont dataset, genotype was marginally significant while temperature significantly affected algal gene expression (genotype: R2 = 0.08906, p < 0.0871; temperature: R2 = 0.01876, p < 0.0013; Table 3). DESeq2 identified 1,450 host and 18 symbiont differentially expressed genes (DEGs) associated with genotype, 1,575 [725 up, 850 down; (elevated/control)] host and 25 (8 up, 17 down) symbiont DEGs for temperature, while only 45 [13 up, 32 down; (discharge/offshore)] host and 0 symbiont DEGs for water, and 35 host and 0 symbiont DEGs for temperature:water (Supplementary Table 1). For both datasets, the highest number of DEGs were associated with pairwise treatment comparisons of elevated discharge over control offshore samples, with 269 (74 up, 195 down) host and 33 (6 up, 27 down) symbiont genes (Figure 3). Corals appeared to respond more strongly to estuarine discharge than did the symbionts across all treatments. DEGs associated with each of the remaining interaction terms and pairwise comparisons between treatments are found in Supplementary Table 1. PCoAs demonstrated that there was stronger genotypic variation in the coral host than in the symbiont dataset, as well as evidence of a synergistic effect of temperature and water treatments for both datasets (Figure 4). Each of the temperature and water treatments alone were relatively close to one another in the ordination space, with a somewhat distinct sample spread for the combined treatment (elevated discharge).
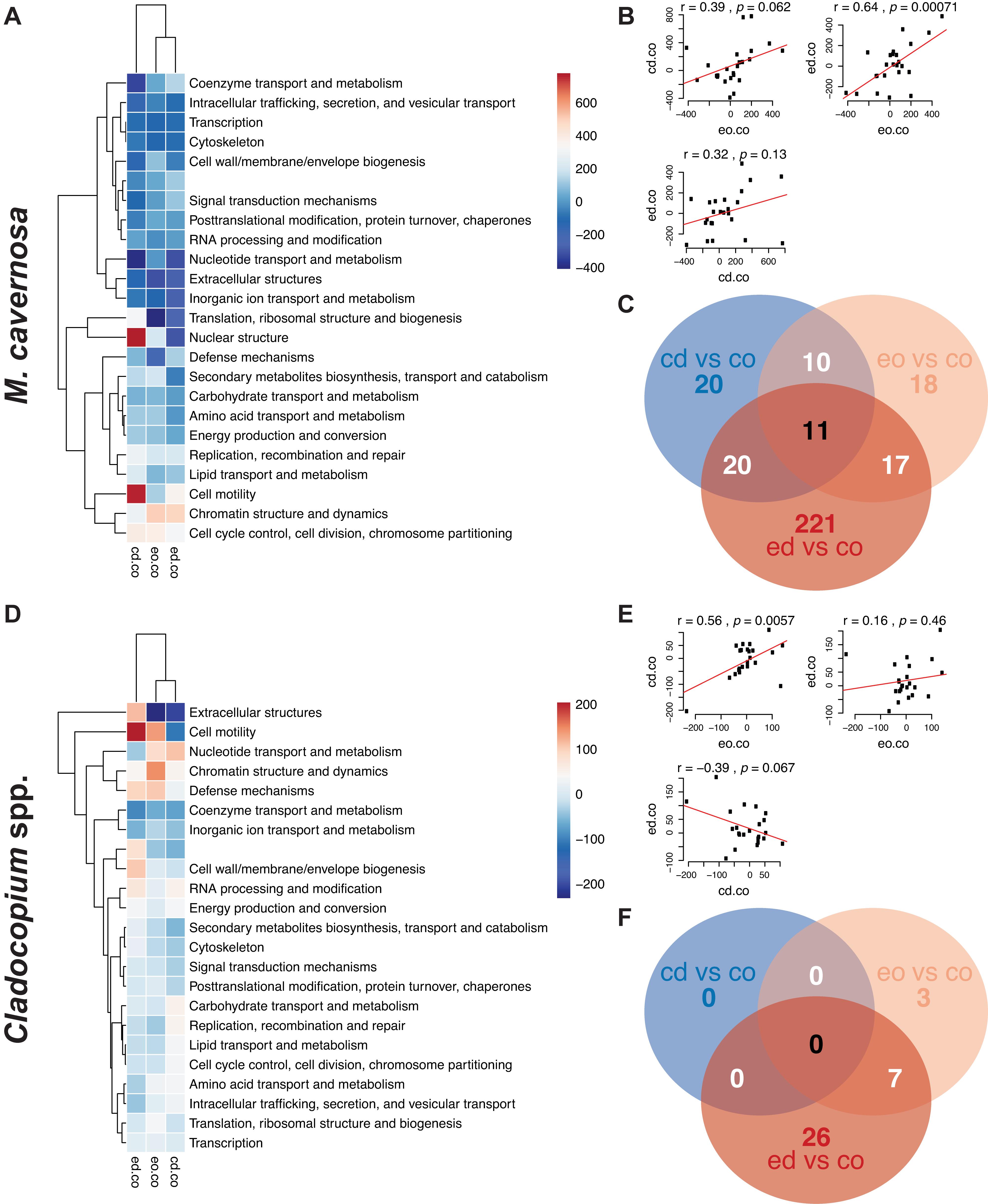
Figure 3. (A,D) Heatmaps of eukaryotic orthologous groups (KOGs) among pairwise comparisons of water + temperature factors for the ex situ experiment, split into coral host (M. cavernosa) and symbiont (Cladocopium spp.) datasets. Increasing intensities of red/blue indicate enrichment of KOGs more-expressed/less-expressed in treatments (cd, control discharge; eo, elevated offshore; ed, elevated discharge) versus control offshore (co) samples. Color values represent ranks generated from Mann–Whitney U tests. Hierarchical networks on both axes display the similarities among pairwise comparisons (x-axis) and KOG classes (y-axis). (B,E) Panels of correlation analyses between pairwise comparison combinations. (C,F) Venn diagram displaying the number of DEGs associated with each pairwise treatment comparison.
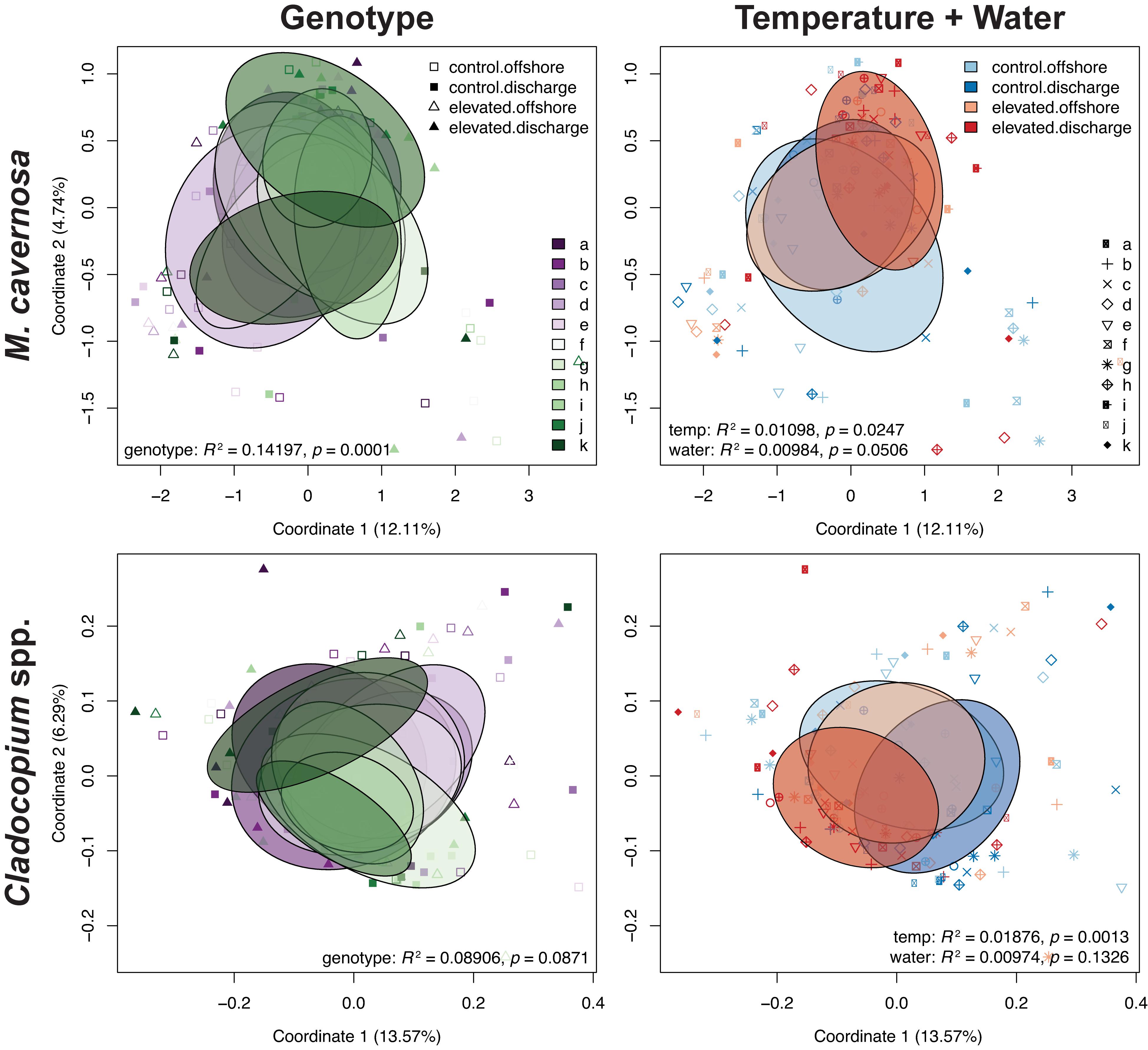
Figure 4. PCoAs of coral host and symbiont datasets depicting transcriptomic variation for coral genotypes and temperature + water treatments from the ex situ experiment. Ellipses denote the dispersion of each factor’s samples. Test statistics and p values generated from PERMANOVAs in vegan.
Gene ontology categories across all three divisions (molecular function, MF; biological process, BP; cellular component, CC) were largely positively-enriched in elevated versus control temperature and discharge versus offshore water, respectively (Figure 5). The overall number of significantly enriched GO terms, as well as the number of DEGs within those GO categories, were higher for the temperature treatment compared to the water treatment, although there were several similarities between the two. GO categories “structural constituent of ribosome” (MF), “translational initiation” (BP), “translational elongation” (BP), “RNA catabolic process” (BP), “cellular component disassembly” (BP), and several ribosomal pathways (CC) were elevated in both temperature and water treatments relative to their respective controls. Few GO categories, on the other hand, were under-expressed in treatments [“lysase” (MF), “transferase” (MF), “carboxylic acid catabolic process” (BP), “cell projection part” (CC), and “basolateral plasma membrane” (CC)], although none of these were represented across both temperature and water treatments.
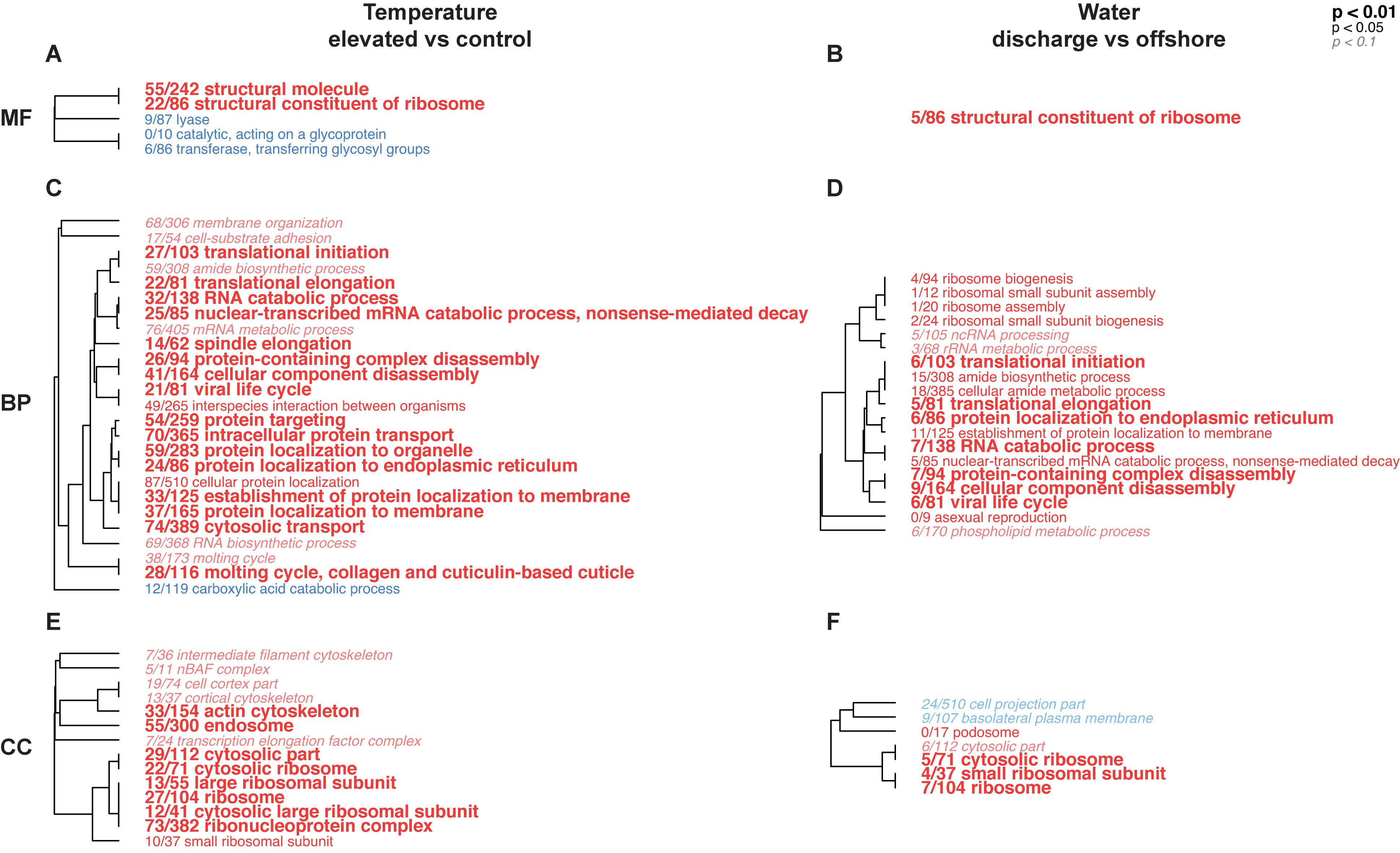
Figure 5. Hierarchical trees representing significantly enriched gene ontology (GO) categories in pairwise comparisons between temperature and water treatments in the coral host dataset for the ex situ experiment. Blue terms represent under-expressed GO categories in (treatment/control), while red terms indicate over-expressed GO categories in (treatment/control). The size and typeface of the font correspond to the p value threshold, where larger, bolded GO terms are more significantly enriched compared to smaller, italicized GO terms, and the number fraction refers to the number of genes represented in each GO category. Annotations are presented for molecular function (MF; A,B), biological process (BP; C,D), and cellular component (CC; E,F) GO divisions.
Eukaryotic orthologous group analyses across main factors identified three main trends: (1) expression patterns for genotype and temperature factors were strongly correlated for both host and symbiont datasets, (2) some KOG classes had an inverse relationship between responses to temperature and water stress in both datasets, and (3) corals and symbionts responded differently to the same factors (Supplementary Figure 7). Indeed, correlation coefficients between genotype and temperature responses were r = 0.99 with highly significant p values in both datasets, suggesting that variation in response to temperature stress was innately tied to coral genotype, and was largely captured by the latter term in statistical models. Several KOG classes, including “cytoskeleton,” “extracellular structures,” and “nuclear structure” were differentially expressed between water and temperature treatments for the coral host (Supplementary Figure 7A). For the symbiont dataset, “extracellular structures” and “coenzyme transport and metabolism” were differentially expressed between water and temperature treatments, while other KOG classes showed generally weaker enrichment in the water treatment compared to the temperature and genotype factors (Supplementary Figure 7D).
When examining pairwise combinations of treatments against the control (control offshore) for the host dataset, only two KOG classes (“nuclear structure” and “cell motility”) were positively enriched in control discharge versus both elevated temperature treatments (Figure 3A). In the symbiont dataset, the same was true of “cell motility,” while “extracellular structures” and “nucleotide transport and metabolism” classes were more-expressed in control discharge and elevated offshore treatments versus elevated discharge samples (Figure 3D). Both of these observations may imply that the coral host responded more strongly to elevated temperatures than estuarine discharge, while algal symbionts demonstrated a synergistic response to elevated temperatures and estuarine discharge combined.
Further filtering of raw gene counts for WGCNA resulted in a dataset of 967 host and 43 symbiont genes. WGCNA clustered genes into five modules for the host dataset and a single module for symbionts; therefore only host data are presented (Supplementary Figure 8; five host genes were unassigned). All of the modules demonstrated significant correlations to individual coral genotypes, while turquoise, yellow, green, and blue modules were also significantly correlated with experimental treatments, suggesting genotype-driven transcriptomic responses to stress. None of the symbiont or chlorophyll metrics were significantly correlated to any of the modules. Variation in module correlation directions between elevated offshore and elevated discharge treatments (turquoise, yellow, and green modules) corroborated a potential synergistic effect of temperature stress and water treatments on coral gene expression patterns. GO-MWU analysis of host modules identified significantly enriched GO categories within blue and brown modules only, with enriched ribosomal and cell division GO categories in the blue module, and immune/stress response and cell component GO terms enriched in the brown module (Supplementary Figure 8).
Temporal analyses of gene expression across three sites at St. Lucie Reef revealed that time was the only significant factor driving transcriptomic patterns for both the coral host and its symbiotic algae. This suggests that despite observations of rapid changes in water quality parameters following discharge events along a gradient with increasing distance from the St. Lucie Inlet (Beal et al., 2012; Pickering and Baker, 2015; Shatters, 2017), corals responded similarly at the transcriptomic level among sites. There did not appear to be a strong relationship between estuarine discharge rates at the time of sampling to either coral or symbiont expression profiles. Taken in the larger context of seasonal variation including rainfall and discharge rates, however, there were observed shifts in expression patterns from the first two time points to all subsequent time points (Figure 2). The wet and dry seasons in 2013–2014 followed the typical pattern described earlier (Beal et al., 2012; Klepac et al., 2015), but the 2015 “wet” season was unusually dry (Shatters, 2017), and was followed by an abnormally rainy 2015–2016 “dry” season (Stockley et al., 2018). Likewise, the number and magnitude of discharge events varied over the course of the study (Supplementary Figure 1). In 2013–2014, there were several larger pulses of discharge (>50 m3 s–1) during the wet season, with relatively low or no discharge events during the dry season. Beginning in 2015, conversely, there were lower (<40 m3 s–1), but consistent, discharges throughout the year, followed by approximately 9 months of discharge at rates in excess of 50 m3 s–1, and in excess of 150 m3 s–1 in 2016 (Supplementary Figure 1; Stockley et al., 2018).
The apparent shifts in gene expression patterns at later versus earlier time points is perhaps the result of covarying environmental factors through time, namely compounding rainfall and discharge flow rates that introduce hyposalinity, nutrient, and elevated temperature stressors to the reef. In the coral host, this is best illustrated by relative expression through time of genes within the “nuclear structure” KOG class. Despite relatively low sequencing coverage associated with the algal symbionts, genes in the “extracellular structures” class were differentially expressed over time (Supplementary Figure 5). Many of the gene ontology (GO) categories that were under-expressed in comparisons between 2015–2016 and initial 2013–2014 time points were then over-expressed in 2016 versus 2015 time points, suggesting variable expression of the same gene pathways in response to different levels of estuarine discharge exposure and hyposalinity stress associated with increased rainfall. Few studies have examined transcriptomic responses to sublethal exposures of estuarine water in natural environments, where differentially expressed stress response genes were linked to increased precipitation and runoff events [Beal et al., 2012; Edge et al., 2013; but see also Wright et al. (2019)].
A recent metanalysis, however, has suggested that transcriptomic responses may be conserved across various stressors, with diverging responses dependent on stress intensity. Dixon et al. (2020) identified a type “A” response to high-intensity bleaching, heat, hyposalinity, or immune challenge stressors where cell death, immune response, and protein degradation pathways were generally up-regulated, and a type “B” response to lower-intensity stress exposures with an inverse relationship to type “A” expression levels. During the time period of our study, no phenotypic changes occurred in the fate-tracked colonies (i.e., bleaching or tissue mortality) to suggest that corals were exposed to high-intensity stress. Expression of gene pathways associated with cell growth [e.g., KOG terms “nuclear structure” in the coral host and “extracellular structures” in the symbionts (Supplementary Figure 5); GO terms “protein localization to endoplasmic reticulum” and several mitosis-related pathways in the coral host (Supplementary Figure 4)] were generally more expressed during later sampling time points when discharge rates and rainfall were higher, compared to earlier time points with less consistent freshwater inputs to the reef (Supplementary Figure 1). Immune GO pathways including “regulation of immune effector process” and “regulation of production of molecular mediator of immune response” were conversely down-regulated during the later sampling time points (Supplementary Figure 4). These patterns would therefore suggest that corals at St. Lucie Reef were exhibiting a more type “B” response following sublethal exposures to estuarine discharge and hyposaline stress.
While individual sublethal stressors may not cause significant disruptions to the coral-algal symbiosis, consecutive or concurrent stress events that are common in nearshore environments may form a “negative feedback loop” ultimately leading to colony mortality. In particular, exposure to salinity stress and/or sedimentation in estuarine discharge water has been shown to increase susceptibility of corals to diseases (Pollock et al., 2014; Shore-Maggio et al., 2018). While it was perhaps inevitable that SCTLD would reach coral populations in the northern extent of the reef tract, repeated exposure to estuarine discharge may have exacerbated individual corals’ susceptibility to SCTLD infection and contributed to the spread of the disease across additional reefs. Testing this hypothesis requires further manipulative experiments applying multiple stressors experienced by nearshore reefs to observations of SCTLD transmission and progression. Transcriptomic approaches may also increase the ability to recognize early signs of infection following exposure to multiple sublethal stressors. Beyond synergistic effects on coral/symbiont physiology and health, estuarine discharge may have additional ramifications for the survival of coral populations in southeast Florida. For example, exposure to hyposalinity stress can severely limit coral fertilization success (Hédouin et al., 2015). The frequent hyposaline conditions (e.g., 27–28 psu commonly measured at St. Lucie Reef but as low as 19 psu for brief periods) in this region due to estuarine outflow, Lake Okeechobee discharges, and watershed runoff may contribute to the rare occurrence of reproduction in St. Lucie Reef corals (Beal et al., 2012).
In the ex situ factorial experiment, there was clear evidence of transcriptomic responses to thermal stress in both the coral host, and to a lesser extent, their algal symbionts (Supplementary Table 1 and Supplementary Figure 7). It is possible that reduced representation of symbiont transcripts in part explains the limited variation among treatments and lack of significantly enriched GO pathways in the symbiont dataset. In the coral host, several gene pathways were elevated following exposure to the increased temperature treatments, including GO categories for metabolic and catabolic processes, protein translation and localization, and translational elongation, which are part of the metabolic and photosynthetic responses to thermal stress (Murata et al., 2007; Reyes-Bermudez et al., 2009; Seneca and Palumbi, 2015; Davies et al., 2016; Studivan and Voss, 2020). Ribosomal expression was also higher in elevated temperature corals, which is likely associated with an attempt by the corals to return to homeostasis following exposure to stress (Kültz, 2005; DeSalvo et al., 2008; Kenkel et al., 2013b). Several of the GO categories described above, including “protein localization to endoplasmic reticulum,” “nuclear-transcribed mRNA catabolic process,” and protein-containing complex and cellular component disassembly, were positively expressed in the final time points of the in situ experiment (Supplementary Figure 4). The KOG classes “nuclear structure” and “extracellular structures” were also differentially enriched for temperature and water treatments (Supplementary Figure 7), reinforcing the notion that in situ corals at St. Lucie Reef were likely responding to sublethal stress during 2015 and 2016 time points similarly to the artificial stress treatments applied in the ex situ experiment. These similarities also demonstrated that corals in the ex situ experiment were demonstrating a type “B” transcriptomic response to sublethal environmental stress (Dixon et al., 2020).
While exposure to estuarine discharge water alone did not significantly alter coral or symbiont gene expression patterns in the ex situ experiment, many of the same GO categories that were positively enriched with thermal stress were also enriched with exposure to discharge water (Figure 5). This observation concurs with the hypothesis of a conserved stress response among broad taxonomic groups including corals (Wang et al., 2009; Dixon et al., 2020), with variation of genes or expression levels within a general environmental stress response that may be stressor-specific. For example, hyposaline conditions can lead to differential expression of heat shock protein and oxidative stress genes, among others (Seveso et al., 2013; Aguilar et al., 2019). Our data suggest that responses were similar in terms of gene function, but that thermal stress caused stronger responses in both corals and their symbionts. There was also a synergistic effect between temperature and water treatments that could not be explained by elevated temperature or estuarine discharge alone (Figures 3, 4 and Supplementary Figure 6). Previous studies have observed synergistic effects of multiple stressors including temperature, salinity, and nutrient factors, with proteomic indicators of oxidative stress (Dias et al., 2019b), bleaching and reduced growth/condition (Coles and Jokiel, 1978), and reduced photosynthetic performance and/or mortality (Li et al., 2009; Faxneld et al., 2010; Dias et al., 2019a). Exposure to multiple stressors may cumulatively result in more severe impacts than with individual type “B” stressors alone, and can lead to a type “A” stress response (Dixon et al., 2020). In addition to direct effects of multiple stressors on coral health, thermal stress has been shown to impede several species’ ability to cope with enhanced sedimentation (Bessell-Browne et al., 2017), and causes reductions of surface mucopolysaccharide layers (i.e., where mucus is produced) in P. clivosa (Pratte and Richardson, 2014). Such synergistic relationships could have important ramifications for the health of corals in Florida, as nearshore reef environments are often exposed to several stressors simultaneously with tidal fluctuations (Yentsch et al., 2002; Lirman and Fong, 2007; Beal et al., 2012; Gregg, 2013).
Synergistic effects of discharge water and thermal stress treatments were not apparent in the symbiont and chlorophyll metrics; significant differences among treatments were attributed only to elevated temperatures (Supplementary Figure 6). Previous studies examining physiological responses to hyposaline conditions identified reductions in symbiont densities and photosynthetic efficiency (Kerswell and Jones, 2003; Downs et al., 2009; Gardner et al., 2016), although this response may be dependent on the coral species. Siderastrea radians, a common shallow-water species found extensively in nearshore reef environments in Florida, was not affected by hyposalinity stress until its predicted lower tolerance limit of 10 psu (Chartrand et al., 2009; Lirman and Manzello, 2009). Such limits have not been established for M. cavernosa, but it has been hypothesized that salinity and light attenuation are major limiting factors in the distribution of nearshore coral populations (Yentsch et al., 2002; True, 2012), and that inshore environments experience seasonal stress and recovery phases (Haslun et al., 2016). While our ex situ experiment did not examine as many salinity treatments as the aforementioned studies, significant differences were observed between similar salinity levels as tested here. Also, areal chlorophyll metrics differed among treatments whereas cellular chlorophyll metrics did not, implying that decreases in areal chlorophylls a and c2 were largely driven by reduced symbiont density in elevated temperature treatments, rather than a change in the number of symbiont cells within coral symbiosomes.
Variation in host responses to thermal stress was highly linked with coral genotype (Figure 3 and Supplementary Figure 6), suggesting a strong genetic component to thermal stress susceptibility/resilience. A similar relationship has been identified with several coral species (Baums et al., 2013; Kenkel et al., 2013a; Bay and Palumbi, 2014), and host transcriptomic patterns may also be influenced by symbiont genotypes as well (DeSalvo et al., 2010; McGinley et al., 2012; Rocker et al., 2012; Cunning and Baker, 2020). In our study, it appears that M. cavernosa genotypes demonstrated variable susceptibility to thermal stress, as genotype explained an order of magnitude more transcriptomic variation in the coral dataset compared to temperature and water treatments (14.2% versus 1.1% and 1.0%, respectively; Table 3). Additionally, there was a strong correlation of KOG expression between genotype and temperature factors (Supplementary Figure 7). Furthermore, in WGCNA, many of the significant correlations observed between experimental factors and modules of related genes were attributed to individual coral genotypes rather than to temperature or water treatments (Supplementary Figure 8).
In the symbiont dataset, there was also an influence of coral genotype on transcriptomic patterns, despite relatively homogeneous symbiont assemblages across all samples in the ex situ experiment (Supplementary Data 1). The marginally significant p value associated with the genotype factor was likely the direct result of a lack of variation in symbiont among coral genotypes. However, 8.9% of the transcriptomic variation was explained by coral genotype, versus 1.9% explained by the temperature treatment (Table 3). The strong links between coral genotypes and responses to thermal stress in both host and symbiont gene expression patterns may be indicative of resilience among some individuals in the sampled coral populations. Nearshore reef environments in southeast Florida (including Breakers Reef) experience seasonal and freshwater discharge-related environmental fluctuations, notably including changes in temperature at episodic and chronic temporal scales (Yentsch et al., 2002; Beal et al., 2012; Gregg, 2013). An interesting hypothesis is that these corals are particularly resilient to thermal stress given repeated exposure to temperature variation, resulting in a conserved transcriptomic response to sublethal stress events. Additional research is therefore warranted to further elucidate potential relationships between coral-algal genotypes and susceptibility to multiple stressors in this species across populations in Florida and beyond (Cunning and Baker, 2020; Smith et al., 2020).
Few studies to date have examined coral responses to multiple stressors (e.g., reduced salinity, higher nutrients, sedimentation, increased light attenuation) associated with estuarine discharge in coastal zones. Through complementary ex situ and in situ experiments, this study demonstrated that corals and their algal symbionts responded to thermal and estuarine discharge stressors, and that synergistic impacts from these stressors could influence their transcriptomic responses. Compared to a recent metanalysis of general environmental stress responses that generalized two characteristic transcriptomic patterns (Dixon et al., 2020), our results suggest that M. cavernosa displays a type “B” response following exposure to sublethal stress in both their natural environments and following manipulative treatments. Cell growth and protein production/modification pathways were observed to be up-regulated while immune pathways were down-regulated following temperature and hyposaline stress. Nearshore coral populations at the northern extent of Florida’s coral reef tract are commonly exposed to multiple stressors in the form of variable temperature, salinity, and water quality (Yentsch et al., 2002; Lirman and Fong, 2007; Beal et al., 2012; Gregg, 2013) that may combine to negatively affect coral health. Continued examination of marginal populations found in this region may not only identify the molecular mechanisms responsible for coral resilience, but may also identify resilient genotypes for conservation and restoration initiatives. Given that modern coral reef ecosystems, especially those found in coastal environments, are more frequently exposed to multiple stressors, management actions are needed to mitigate potentially synergistic impacts of stressors on the health and resilience of coral reef organisms.
Raw Tag-Seq sequences are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject PRJNA698305, Accession numbers SAMN17712326 through SAMN17712600. Datasets generated from this study, including analysis scripts, are available in a GitHub repository (Studivan, 2021). Pipelines used for transcriptome annotation and bioinformatics followed protocols in previously-released GitHub repositories (Studivan, 2020a,b).
JV, JB, AS, and DD secured funding for this research, designed and performed the research, experiments, and initial data analyses, with field and lab assistance from MS. MS analyzed the data and wrote the manuscript, with significant contributions and revisions by all co-authors. All authors contributed to the article and approved the submitted version.
This study was supported by grants to JV and JB from Florida Sea Grant (R/LR-B-70), EPA (X700D66717), and NOAA Coral Reef Conservation Program (NA16NOS4820052), as well as a donation from the River Branch Foundation and from the Banbury Fund in memory of John and Andreija Robertson to establish the Robertson Coral Reef Research and Conservation Program at Harbor Branch Oceanographic Institute. Additional graduate fellowships and grants from the Women Divers Hall of Fame, PADI Foundation, and Indian River Lagoon Graduate Research Fellowship augmented this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Maureen Williams, Courtney Klepac, Amanda Alker, Jennifer Polinski, Patrick Gardner, Ashley Sproles, Miguel Martini, Lisa Cohen, and Sara Edge for assistance with field activities and sample collection. St. Lucie Inlet Preserve State Park, Martin County representatives, Florida Fish and Wildlife Conservation Commission, and the Florida Department of Environmental Protection, Coral Reef Conservation Program provided critical feedback and support on the design of this study. Coral tissue samples were collected from the St. Lucie Inlet Preserve State Park under a permit from the Florida State Park Service (FL State Park permit number 06261715). Coral colonies were collected from Breakers Reef by permission of FWC. Paul Wills, Richard Baptiste, Peter Stock, Gary Luisi, Susan Laramore, and Jay Adams were integral in the development and construction of the ex situ experimental apparatus. Dennis Hanisak and Kristen Davis from the Indian River Lagoon Observatory at Florida Atlantic University’s Harbor Branch Oceanographic Institute, and Brittany Biber from the Florida Oceanographic Society facilitated collection of estuarine discharge water and seawater. Sequencing services were provided by the University of Wisconsin Biotechnology Center, with exceptional sequencing support from Jeremy Niece and Joshua Hyman, and high-performance computing services were provided by Research Computing Services at Florida Atlantic University. This is contribution 2290 from Harbor Branch Oceanographic Institute at Florida Atlantic University.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.662220/full#supplementary-material
Aguilar, C., Raina, J. B., Fôret, S., Hayward, D. C., Lapeyre, B., Bourne, D. G., et al. (2019). Transcriptomic analysis reveals protein homeostasis breakdown in the coral Acropora millepora during hypo-saline stress. BMC Genomics 20:148. doi: 10.1186/s12864-019-5527-2
Aguilar, C., Raina, J. B., Motti, C. A., Fôret, S., Hayward, D. C., Lapeyre, B., et al. (2017). Transcriptomic analysis of the response of Acropora millepora to hypo-osmotic stress provides insights into DMSP biosynthesis by corals. BMC Genomics 18:612. doi: 10.1186/s12864-017-3959-0
Anderson, D. A., Walz, M. E., Weil, E., Tonellato, P., and Smith, M. C. (2016). RNA-Seq of the Caribbean reef-building coral Orbicella faveolata (Scleractinia–Merulinidae) under bleaching and disease stress expands models of coral innate immunity. PeerJ 4:e1616. doi: 10.7717/peerj.1616
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J., et al. (2000). Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Bak, R. P. M., and Elgershuizen, J. H. B. W. (1976). Patterns of oil-sediment rejection in corals. Mar. Biol. 37, 105–113. doi: 10.1007/BF00389121
Baums, I. B., Devlin-Durante, M. K., Polato, N. R., Xu, D., Giri, S., Altman, N. S., et al. (2013). Genotypic variation influences reproductive success and thermal stress tolerance in the reef building coral. Acropora palmata. Coral Reefs 32, 703–717. doi: 10.1007/s00338-013-1012-6
Bay, R. A., and Palumbi, S. R. (2014). Multilocus adaptation associated with heat resistance in reef-building corals. Curr. Biol. 24, 2952–2956. doi: 10.1016/j.cub.2014.10.044
Beal, J., Voss, J. D., Edge, S., and Cohen, L. (2012). Assessment of Coral Stressors on St. Lucie Reef: Florida’s Northernmost Coral Reef. Washington, DC: US Fish and Wildlife Service, 1–54.
Bessell-Browne, P., Negri, A. P., Fisher, R., Clode, P. L., and Jones, R. (2017). Cumulative impacts: thermally bleached corals have reduced capacity to clear deposited sediment. Sci. Rep. 7:2716. doi: 10.1038/s41598-017-02810-0
Chartrand, K. M., Durako, M. J., and Blum, J. E. (2009). Effect of hyposalinity on the photophysiology of Siderastrea radians. Mar. Biol. 156, 1691–1702. doi: 10.1007/s00227-009-1204-3
Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159.
Chomczynski, P., and Sacchi, N. (2006). The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nat. Protoc. 1, 581–585. doi: 10.1038/nprot.2006.83
Coles, S. L., and Jokiel, P. L. (1978). Synergistic effects of temperature, salinity and light on the hermatypic coral Montipora verrucosa. Mar. Biol. 49, 187–195. doi: 10.1007/BF00391130
Collier, C., Ruzicka, R., Banks, K., Barbieri, L., Beal, J., Bingham, D., et al. (2008). “The state of coral reef ecosystems of Southeast Florida,” in The state of coral reef ecosystems of the United States and Pacific Freely Associated States: 2008, eds J. Waddel and A. Clarke Silver Spring, MD, 131–160.
Cunning, R., and Baker, A. C. (2020). Thermotolerant coral symbionts modulate heat stress- responsive genes in their hosts. Mol. Ecol. 29, 2940–2950. doi: 10.1111/mec.15526
Davies, S. W., Marchetti, A., Ries, J. B., and Castillo, K. D. (2016). Thermal and pCO2 stress elicit divergent transcriptomic responses in a resilient coral. Front. Mar. Sci. 3:112. doi: 10.3389/fmars.2016.00112
Davies, S. W., Ries, J. B., Marchetti, A., and Castillo, K. D. (2018). Symbiodinium functional diversity in the coral Siderastrea siderea is influenced by thermal stress and reef environment, but not ocean acidification. Front. Mar. Sci. 5:150. doi: 10.3389/fmars.2018.00150
DeSalvo, M. K., Sunagawa, S., Fisher, P. L., Voolstra, C. R., Iglesias-Prieto, R., and Medina, M. (2010). Coral host transcriptomic states are correlated with Symbiodinium genotypes. Mol. Ecol. 19, 1174–1186. doi: 10.1111/j.1365-294X.2010.04534.x
DeSalvo, M. K., Voolstra, C. R., Sunagawa, S., Schwarz, J. A., Stillman, J. H., Coffroth, M. A., et al. (2008). Differential gene expression during thermal stress and bleaching in the Caribbean coral Montastraea faveolata. Mol. Ecol. 17, 3952–3971. doi: 10.1111/j.1365-294X.2008.03879.x
Dias, M., Ferreira, A., Gouveia, R., and Vinagre, C. (2019a). Synergistic effects of warming and lower salinity on the asexual reproduction of reef-forming corals. Ecol. Ind. 98, 334–348. doi: 10.1016/j.ecolind.2018.11.011
Dias, M., Madeira, C., Jogee, N., Ferreira, A., Gouveia, R., Cabral, H., et al. (2019b). Oxidative stress on scleractinian coral fragments following exposure to high temperature and low salinity. Ecol. Ind. 107:105586. doi: 10.1016/j.ecolind.2019.105586
Dixon, G., Abbott, E., and Matz, M. V. (2020). Meta-analysis of the general coral stress response: Acropora corals show opposing responses depending on stress intensity. Mol. Ecol. 29, 2855–2870. doi: 10.1111/mec.15535
Dixon, G. B., Davies, S. W., Aglyamova, G. A., Meyer, E., Bay, L. K., and Matz, M. V. (2015). Genomic determinants of coral heat tolerance across latitudes. Science 348, 1460–1462. doi: 10.1126/science.1261224
Dodge, D., Studivan, M. S., Eckert, R., Chei, E., Beal, J., and Voss, J. (2020). Population structure of the scleractinian coral. Montastraea cavernosa, in southeast Florida. Bull. Mar. Sci. 96, 767–782. doi: 10.5343/bms.2019.0074
Downs, C. A., Kramarsky-Winter, E., Woodley, C. M., Downs, A., Winters, G., Loya, Y., et al. (2009). Cellular pathology and histopathology of hypo-salinity exposure on the coral Stylophora pistillata. Sci. Total Environ. 407, 4838–4851. doi: 10.1016/j.scitotenv.2009.05.015
Edge, S. E., Shearer, T. L., Morgan, M. B., and Snell, T. W. (2013). Sub-lethal coral stress: detecting molecular responses of coral populations to environmental conditions over space and time. Aquat. Toxicol. 128–129, 135–146. doi: 10.1016/j.aquatox.2012.11.014
Fabricius, K. E. (2005). Effects of terrestrial runoff on the ecology of corals and coral reefs: review and synthesis. Mar. Pollut. Bull, 50, 125–146. doi: 10.1016/j.marpolbul.2004.11.028
Faxneld, S., Jörgensen, T. L., and Tedengren, M. (2010). Effects of elevated water temperature, reduced salinity and nutrient enrichment on the metabolism of the coral Turbinaria mesenterina. Estuar. Coast. Shelf Sci. 88, 482–487. doi: 10.1016/j.ecss.2010.05.008
Gardner, S. G., Nielsen, D. A., Laczka, O., Shimmon, R., Beltran, V. H., Ralph, P. J., et al. (2016). Dimethylsulfoniopropionate, superoxide dismutase and glutathione as stress response indicators in three corals under short-term hyposalinity stress. Proc. R. Soc. B Biol. Sci. 283, 1–9. doi: 10.1098/rspb.2015.2418
Gilliam, D. S., Hayes, N. K., Ruzicka, R. R., and Colella, M. (2018). Southeast Florida Coral Reef Evaluation and Monitoring Project (SECREMP). Miami, FL: Florida DEP, 1–79.
Graves, G. A., Wan, Y., and Fike, D. L. (2004). Water quality characteristics of storm water from major land use in South Florida. J. Am. Water Resour. Assoc. 40, 1405–1419. doi: 10.1111/j.1752-1688.2004.tb01595.x
Gregg, K. L. (2013). Literature Review and Synthesis of Land-Based Sources of Pollution Affecting Essential Fish Habitats in Southeast Florida. West Palm Beach, FL: NOAA Fisheries Southeast Region, 1–55.
Haslun, J., Hauff, B., Strychar, K., and Cervino, J. (2016). Decoupled seasonal stress as an indication of chronic stress in Montastraea cavernosa and Porites astreoides inhabiting the Florida Reef Tract. Int. J. Mar. Sci. 6, 1–20. doi: 10.5376/ijms.2016.06.0021
Hédouin, L., Pilon, R., and Puisay, A. (2015). Hyposalinity stress compromises the fertilization of gametes more than the survival of coral larvae. Mar. Environ. Res. 104, 1–9. doi: 10.1016/j.marenvres.2014.12.001
Hervé, M. (2019). RVAideMemoire: Testing and Plotting Procedures for Biostatistics. R Package Version 0.9-73.
Kauffmann, A., Gentleman, R., and Huber, W. (2009). arrayQualityMetrics - a bioconductor package for quality assessment of microarray data. Bioinformatics 25, 415–416. doi: 10.1093/bioinformatics/btn647
Kenkel, C. D., Aglyamova, G., Alamaru, A., Bhagooli, R., Capper, R., Cunning, R., et al. (2011). Development of gene expression markers of acute heat-light stress in reef-building corals of the genus Porites. PLoS One 6:e26914. doi: 10.1371/journal.pone.0026914
Kenkel, C. D., Goodbody-Gringley, G., Caillaud, D., Davies, S. W., Bartels, E., and Matz, M. V. (2013a). Evidence for a host role in thermotolerance divergence between populations of the mustard hill coral (Porites astreoides) from different reef environments. Mol. Ecol. 22, 4335–4348. doi: 10.1111/mec.12391
Kenkel, C. D., Meyer, E., and Matz, M. V. (2013b). Gene expression under chronic heat stress in populations of the mustard hill coral (Porites astreoides) from different thermal environments. Mol. Ecol. 22, 4322–4334. doi: 10.1111/mec.12390
Kenkel, C. D., Sheridan, C., Leal, M. C., Bhagooli, R., Castillo, K. D., Kurata, N., et al. (2014). Diagnostic gene expression biomarkers of coral thermal stress. Mol. Ecol. Resour. 14, 667–678. doi: 10.1111/1755-0998.12218
Kerswell, A. A. P., and Jones, R. J. R. (2003). Effects of hypo-osmosis on the coral Stylophora pistillata: nature and cause of “low-salinity bleaching.”. Mar. Ecol. Prog. Ser. 253, 145–154. doi: 10.3354/meps253145
Kitchen, S. A., Crowder, C. M., Poole, A. Z., Weis, V. M., and Meyer, E. (2015). De novo assembly and characterization of four anthozoan (Phylum Cnidaria) transcriptomes. G3 Genes Genomes Genet. 5, 2441–2452. doi: 10.1534/g3.115.020164
Klepac, C. N., Beal, J., Kenkel, C. D., Sproles, A., Polinski, J. M., Williams, M. A., et al. (2015). Seasonal stability of coral-Symbiodinium associations in the subtropical coral habitat of St. Lucie Reef, Florida. Mar. Ecol. Prog. Ser. 532, 137–151. doi: 10.3354/meps11369
Konietschke, F., Placzek, M., Schaarschmidt, F., and Hothorn, L. A. (2015). nparcomp: an R software package for nonparametric multiple comparisons and simultaneous confidence intervals. J. Stat. Softw. 64, 1–17. doi: 10.18637/jss.v064.i09
Kültz, D. (2005). Molecular and evolutionary basis of the cellular stress response. Annu. Rev. Physiol. 67, 225–257. doi: 10.1146/annurev.physiol.67.040403.103635
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Lapointe, B. E., Herren, L. W., and Bedford, B. J. (2012). Effects of hurricanes, land use, and water management on nutrient and microbial pollution: St. Lucie Estuary, Southeast Florida. J. Coast. Res. 28, 1345–1361. doi: 10.2112/JCOASTRES-D-12-00070.1
Lapointe, B. E., Herren, L. W., and Paule, A. L. (2017). Septic systems contribute to nutrient pollution and harmful algal blooms in the St. Lucie Estuary, Southeast Florida, USA. Harmful Algae 70, 1–22. doi: 10.1016/j.hal.2017.09.005
Lasker, H. R. (1980). Sediment rejection by reef corals: the roles of behavior and morphology in Montastrea cavernosa (Linnaeus). J. Exp. Mar. Biol. Ecol. 47, 77–87. doi: 10.1016/0022-0981(80)90139-2
Li, X., Huang, H., Lian, J., Huang, L., and Dong, J. (2009). Effects of the multiple stressors high temperature and reduced salinity on the photosynthesis of the hermatypic coral Galaxea fascicularis. Acta Ecol. Sin. 29, 155–159. doi: 10.1016/j.chnaes.2009.07.002
Lirman, D., and Fong, P. (2007). Is proximity to land-based sources of coral stressors an appropriate measure of risk to coral reefs? An example from the Florida Reef Tract. Mar. Pollut. Bull. 54, 779–791. doi: 10.1016/j.marpolbul.2006.12.014
Lirman, D., and Manzello, D. P. (2009). Patterns of resistance and resilience of the stress-tolerant coral Siderastrea radians (Pallas) to sub-optimal salinity and sediment burial. J. Exp. Mar. Biol. Ecol. 369, 72–77. doi: 10.1016/j.jembe.2008.10.024
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550–571. doi: 10.1186/s13059-014-0550-8
Lugo, A. E., Brown, S. B., Dodson, R., Smith, T. S., and Shugart, H. H. (1999). The Holdridge life zones of the conterminous United States in relation to ecosystem mapping. J. Biogeogr. 26, 1025–1038. doi: 10.1046/j.1365-2699.1999.00329.x
Matz, M. V. (2016). KOGMWU: Functional Summary and Meta-Analysis of Gene Expression Data. R Package Version 1.1.
Mayfield, A. B., Fan, T.-Y., and Chen, C.-S. (2013). Real-time PCR-based gene expression analysis in the model reef-building coral Pocillopora damicornis: insight from a salinity stress study. Platax 10, 1–29.
McGinley, M. P., Aschaffenburg, M. D., Pettay, D. T., Smith, R. T., LaJeunesse, T. C., and Warner, M. E. (2012). Transcriptional response of two core photosystem genes in Symbiodinium spp. exposed to thermal stress. PLoS One 7:e50439. doi: 10.1371/journal.pone.0050439
Meyer, E., Aglyamova, G. V., and Matz, M. V. (2011). Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Mol. Ecol. 20, 3599–3616. doi: 10.1111/j.1365-294X.2011.05205.x
Murata, N., Takahashi, S., Nishiyama, Y., and Allakhverdiev, S. I. (2007). Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta Bioenerget. 1767, 414–421. doi: 10.1016/j.bbabio.2006.11.019
Nordberg, H., Cantor, M., Dusheyko, S., Hua, S., Poliakov, A., Shabalov, I., et al. (2014). The genome portal of the department of energy joint genome institute: 2014 updates. Nucleic Acids Res. 42, D26–D31. doi: 10.1093/nar/gkt1069
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R., et al. (2015). vegan: Community Ecology Package. R Package Version 2.0-10.
Perry, W. (2004). Elements of South Florida’s comprehensive everglades restoration plan. Ecotoxicology 13, 185–193. doi: 10.1023/B:ECTX.0000023564.10311.4a
Pickering, N., and Baker, E. S. (2015). Watershed Scale Planning to Reduce Land-Based Sources of Pollution (LBSP) for the Protection of Coral Reefs in Southeast Florida. West Palm Beach, FL: NOAA Fisheries Southeast Region, 1–121.
Pollock, F. J., Lamb, J. B., Field, S. N., Heron, S. F., Schaffelke, B., Shedrawi, G., et al. (2014). Sediment and turbidity associated with offshore dredging increase coral disease prevalence on nearby reefs. PLoS One 9:102498. doi: 10.1371/journal.pone.0102498
Pratte, Z. A., and Richardson, L. L. (2014). Impacts of temperature increase and acidification on thickness of the surface mucopolysaccharide layer of the Caribbean coral Diploria spp. Coral Reefs 33, 487–496. doi: 10.1007/s00338-013-1115-0
R Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Reed, J. K. (1982). A Proposal for the Nearshore Reefs Adjacent to St. Lucie Inlet, Florida, as a part of an Atlantic Hardbottom Sanctuary. Fort Pierce, FL: Harbor Branch Oceanographic Institute, 1–24.
Reyes-Bermudez, A., DeSalvo, M. K., Voolstra, C. R., Sunagawa, S., Szmant, A. M., Iglesias-Prieto, R., et al. (2009). Gene expression microarray analysis encompassing metamorphosis and the onset of calcification in the scleractinian coral Montastraea faveolata. Mar. Genomics 2, 149–159. doi: 10.1016/j.margen.2009.07.002
Rocker, M. M., Willis, B. L., and Bay, L. K. (2012). “Thermal stress-related gene expression in corals with different Symbiodinium types,” in Proceedings of the 12th International Coral Reef Symposium, Vol. 9, Cairns, 1–5.
Seneca, F. O., and Palumbi, S. R. (2015). The role of transcriptome resilience in resistance of corals to bleaching. Mol. Ecol. 24, 1467–1484. doi: 10.1111/mec.13125
Seveso, D., Montano, S., Strona, G., Orlandi, I., Galli, P., and Vai, M. (2013). Exploring the effect of salinity changes on the levels of Hsp60 in the tropical coral Seriatopora caliendrum. Mar. Environ. Res. 90, 96–103. doi: 10.1016/j.marenvres.2013.06.002
Shatters, A. (2017). Effects of St. Lucie Estuarine Discharge Water and Thermal Stress on the Coral Montastraea cavernosa. MSc thesis. Boca Raton, FL: Biological Sciences Florida Atlantic University, 1–91.
Shore-Maggio, A., Aeby, G. S., and Callahan, S. M. (2018). Influence of salinity and sedimentation on Vibrio infection of the Hawaiian coral Montipora capitata. Dis. Aquat. Organ. 128, 63–71. doi: 10.3354/dao03213
Smith, E. G., Gurskaya, A., Hume, B. C. C., Voolstra, C. R., Todd, P. A., Bauman, A. G., et al. (2020). Low Symbiodiniaceae diversity in a turbid marginal reef environment. Coral Reefs 39, 545–553. doi: 10.1007/s00338-020-01956-0
Stockley, N., Sullivan, J. M., Hanisak, M. D., and McFarland, M. N. (2018). “Using observation networks to examine the impact of Lake Okeechobee discharges on the St. Lucie Estuary, Florida,” in Proceedings of SPIE, Ocean Sensing and Monitoring X (Orlando, FL), 1063109. doi: 10.1117/12.2309797
Strader, M. E., Aglyamova, G. V., and Matz, M. V. (2016). Red fluorescence in coral larvae is associated with a diapause-like state. Mol. Ecol. 25, 559–569. doi: 10.1111/mec.13488
Studivan, M. S. (2020a). mstudiva/Mcav-Cladocopium-Annotated-Transcriptome: Final Montastraea cavernosa/Cladocopium spp. transcriptome assembly (Version 2.0.0). Geneva: Zenodo, doi: 10.5281/zenodo.3728261
Studivan, M. S. (2020b). mstudiva/Transcriptional-Plasticity-Mesophotic-Mcav: Final Tag-Seq Scripts for Analysis of Mesophotic Montastraea Cavernosa (Version 2.0.0). Geneva: Zenodo, doi: 10.5281/zenodo.3728281
Studivan, M. S. (2021). mstudiva/Thermal-Stress-Estuarine-Discharge-Mcav: Thermal Stress and Estuarine Discharge Effects on Montastraea Cavernosa Gene Expression (Version 1.1.0). Geneva: Zenodo. doi: 10.5281/zenodo.4643062
Studivan, M. S., and Voss, J. D. (2020). Transcriptomic plasticity of mesophotic corals among natural populations and transplants of Montastraea cavernosa in the Gulf of Mexico and Belize. Mol. Ecol. 29, 2399–2415. doi: 10.1111/mec.15495
Tatusov, R. L., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Kiryutin, B., Koonin, E. V., et al. (2003). The COG database: an updated vesion includes eukaryotes. BMC Bioinformatics 4:41. doi: 10.1186/1471-2105-4-41
Tatusov, R. L., Galperin, M. Y., Natale, D. A., and Koonin, E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28, 33–36. doi: 10.1093/nar/28.1.33
The Gene Ontology Consortium (2017). Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 45, D331–D338. doi: 10.1093/nar/gkw1108
True, J. D. (2012). “Salinity as a structuring force for near shore coral communities,” in Proceedings of the 12th International Coral Reef Symposium, Vol. 9, Cairns, e13.
Vargas-Ángel, B., Peters, E. C., Kramarsky-Winter, E., Gilliam, D. S., and Dodge, R. E. (2007). Cellular reactions to sedimentation and temperature stress in the Caribbean coral Montastraea cavernosa. J. Invertebr. Pathol. 95, 140–145. doi: 10.1016/j.jip.2007.01.003
Vargas-Ángel, B., Riegl, B., Gilliam, D. S., and Dodge, R. E. (2006). “An experimental histopathological rating scale of sedimentation stress in the Caribbean coral Montastraea cavernosa,” in Proceedings of the 10th International Coral Reef Symposium, Okinawa, 1168–1173.
Voss, J. D., and Combs, I. (2018). Fate Tracking, Molecular Investigation, and Amputation Assessment of Tissue Loss Disease on Corals in the Northern Florida Reef Tract. Miami, FL: Florida DEP, 1–22.
Walker, B. K. (2018). Southeast Florida Reef-Wide Post-Irma Coral Disease Surveys. Miami, FL: Florida DEP, 1–37.
Walker, B. K., and Gilliam, D. S. (2013). Determining the extent and characterizing coral reef habitats of the northern latitudes of the Florida Reef Tract (Martin County). PLoS One 8:e80439. doi: 10.1371/journal.pone.0080439
Walton, C. J., Hayes, N. K., and Gilliam, D. S. (2018). Impacts of a regional, multi-year, multi-species coral disease outbreak in Southeast Florida. Front. Mar. Sci. 5:323. doi: 10.3389/fmars.2018.00323
Wang, P., Bouwman, F. G., and Mariman, E. C. M. (2009). Generally detected proteins in comparative proteomics - a matter of cellular stress response? Proteomics 9, 2955–2966. doi: 10.1002/pmic.200800826
Wright, R. M., Aglyamova, G. V., Meyer, E., and Matz, M. V. (2015). Gene expression associated with white syndromes in a reef building coral. Acropora hyacinthus. BMC Genomics 16:371. doi: 10.1186/s12864-015-1540-2
Wright, R. M., Correa, A. M. S., Quigley, L. A., Santiago-Vázquez, L. Z., Shamberger, K. E. F., and Davies, S. W. (2019). Gene expression of endangered coral (Orbicella spp.) in the Flower Garden Banks National Marine Sanctuary after Hurricane Harvey. Front. Mar. Sci. 6:672. doi: 10.3389/fmars.2019.00672
Yentsch, C. S., Yentsch, C. M., Cullen, J. J., Lapointe, B., Phinney, D. A., and Yentsch, S. W. (2002). Sunlight and water transparency: cornerstones in coral research. J. Exp. Mar. Biol. Ecol. 268, 171–183. doi: 10.1016/S0022-0981(01)00379-3
Keywords: Tag-Seq, marginal coral reefs, Florida’s coral reef tract, Florida reef tract, freshwater runoff, sublethal stress, St. Lucie Reef, Symbiodiniaceae
Citation: Studivan MS, Shatters A, Dodge DL, Beal JL and Voss JD (2021) Synergistic Effects of Thermal Stress and Estuarine Discharge on Transcriptomic Variation of Montastraea cavernosa Corals in Southeast Florida. Front. Mar. Sci. 8:662220. doi: 10.3389/fmars.2021.662220
Received: 31 January 2021; Accepted: 06 April 2021;
Published: 13 May 2021.
Edited by:
Davide Seveso, University of Milano-Bicocca, ItalyReviewed by:
Mikhail V. Matz, University of Texas at Austin, United StatesCopyright © 2021 Studivan, Shatters, Dodge, Beal and Voss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael S. Studivan, c3R1ZGl2YW5tc0BnbWFpbC5jb20=; Joshua D. Voss, anZvc3MyQGZhdS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.