- 1Key Laboratory of Freshwater Aquatic Genetic Resources, Ministry of Agriculture, Shanghai Ocean University, Shanghai, China
- 2Shanghai Engineering Research Center of Aquaculture, Shanghai Ocean University, Shanghai, China
- 3Shanghai Collaborative Innovation for Aquatic Animal Genetics and Breeding, Shanghai Ocean University, Shanghai, China
Freshwater pearl mussels and marine pearl oysters are major producers of cultured pearls. With the development of pearl farming, pearl bivalve mollusks have attracted significant research attention over the past 25 years. To provide an overview of this research, we conducted a bibliometric analysis of publications from the Web of Science Core Collection database from 1995 to 2020. A network map generated by VOSviewer software was used to evaluate studies of pearl bivalve mollusks in terms of author, country, organization, journal, and keywords. A total of 2,288 publications were obtained, showing an annual increase in the number of papers. Researchers based in China were a major contributor to the field and there was active cooperation among authors. Research focused on mussel and oyster developmental biology, growth, culture mode, molecular biology, and ecological conservation. In recent years, hot topics, such as growth performance and traits, pearl formation and biomineralization, and immune response, have been widely discussed. Genomics, transcriptomics, proteomics, and metabonomics analyses were commonly used to analyze the regulatory mechanism of coding and non-coding genes. The interaction between the environment and farming activities showed the importance of sustainable development. Interdisciplinary research could solve some of the issues facing the pearl bivalve mollusk farming. In conclusion, our findings could serve as another way to understand research trends in pearl bivalve mollusks and contribute to future studies.
Introduction
Cultured pearls are the response of pearl mussels or oysters to a tissue implant (Nagai, 2013). A tissue graft is inserted from a donor mollusk to a recipient mollusk, the mantle tissue of which then secretes nacreous material into the pearl sac, forming a cyst as part of the healing process (Haws and Ellis, 1999; Dan and Ruobo, 2002). This material is known as conchiolin and is formed of calcium carbonate and a fibrous protein (Ma et al., 2005). The growing pearl sac cell secretes nacre, and a pearl results once the nacre fills the growing pearl sac. Pearls can be farmed from two bivalve mollusks: freshwater pearl mussels and marine pearl oysters (Lucas, 2007). Freshwater pearl mussel refers not only to Margaritifera margaritifera but also to other freshwater mussel species that can also form pearls, such as Hyriopsis spp. in Asia and Amblema spp. in North America, both of which belong to the family Unionidae. Marine pearl oysters occur in the genus Pinctada including Pinctada margaritifera and Pinctada maxima which are used for culturing South Sea and Tahitian pearls (Tisdell and Poirine, 2000). Commercial quantities of pearls can also be obtained from some species of the closely related winged oyster genus Pteria (Paul Southgate, 2011). Pearls have been used chiefly for adornment since ancient times (Nagai, 2013; Duncan Peter and Arne, 2019), and pearl powder is often used as a health product in traditional Chinese medicine, having antioxidant and anti-inflammatory effects on cells (Xu et al., 2001; Jian-Ping et al., 2010). Pearl extract is also used in the clinical treatment of various ulcers (Jian-Ping et al., 2010; Velasco, 2011) and wound healing (Li et al., 2013; Chen et al., 2019). In addition, marine pearl oysters are suitable for the restoration of polluted coastal environments (O’Connor and Gifford, 2008) because they have the highest pumping and filtration rates of any bivalve mollusk (Lucas, 2008).
Since Mikimoto succeeded in culturing blister pearls in 1893, pearls have been cultured worldwide by utilizing different bivalve mollusk species (Nagai, 2013), and the pearl farming industry has grown rapidly (Gueguen et al., 2016; Johnston et al., 2019). The black pearl oyster P. margaritifera, found in Tahiti and many other Pacific islands, including the Cook Islands and Fiji, is widely used for the production of cultured pearls (Kishore et al., 2018; Johnston B. et al., 2020; Johnston W. et al., 2020). Freshwater pearl mussels are widely used in various industries and created great wealth in China and Japan (Tisdell and Poirine, 2008; Bai et al., 2014b). Moreover, the industry of pearl farming has become a large part of financial income in some countries or regions (Southgate et al., 2006; Jiddawi, 2008; Johnston et al., 2019). The studies on pearl bivalve mollusks have been generated from various perspectives. Research has focused on a variety of factors relating to the aquaculture of these mollusks, including how pearls form and their color is determined (Chen et al., 2017; Li et al., 2017b), influence of nuclei implantation (Li et al., 2016; Wang et al., 2019), and different aquaculture methods (Muhammad et al., 2017; Ky et al., 2019).
However, pearl bivalve mollusks face multiple threats, not only in aquaculture settings but also in the wild. Red algal tides (Ericson, 2017; Nakayama et al., 2020) and disease (Sakatoku et al., 2018) have resulted in the decrease of specific species, which has also occurred as a result of environmental pollution, caused by water conservancy projects, overfishing, and water contamination (Cosgrove et al., 2016). Thus, in recent years, the global production of pearls has been in decline (Johnston et al., 2019; Mao et al., 2019; Saucedo et al., 2021). According to research released in 2019 by the South China Sea Fisheries Research Institute (SCSFRI) and Australia’s University of the Sunshine Coast (USC), global pearl production had fallen by 60% and output value by 39% over the past decade (Changbo et al., 2019). Therefore, here we review studies on pearl bivalve mollusks to highlight areas for future research on their conservation and further development of pearl aquaculture.
Bibliometrics is the use of statistical methods to analyze books, articles, and other publications (Broadus, 1987). Bibliometric methods were widely used to explore the distribution structure, quantitative relationships, and changes in literature (Narin et al., 1994). It can quantitatively reveal the development history, research focus, and future research direction of academic fields (Narin et al., 1994; Daim et al., 2006). Bibliometrics has been applied in various fields, such as economics (Bonilla et al., 2015), physics (Rinia et al., 1998), ecology (Romanelli et al., 2018), and medicine (Geaney et al., 2015). Thus, the bibliometric analysis of previous pearl bivalve mollusk research could highlight hot topics and potential future research trends and directions.
Materials and Methods
Data Collection
Given that there are two different bivalve mollusks used for pearl production (the freshwater pearl mussel and the marine pearl oyster), the topic search query phrase [TS = (pearl) AND TS = (mussel OR oyster OR bivalve)] in the Web of Science (WOS) search page (TS means topic) was used to compile a bibliography of all publications. The search time ranged from January 1, 1995, to December 23, 2020. The type of document was limited to research article, meeting reports, and reviews. Any irrelevant information was deleted manually.
In total, 2,288 results were selected from the Web of Science Core Collection for the next step. The Statistics plug-in in WOS was utilized to count the number of publications based on authors, organizations, journals, countries, or regions. All information was exported as plain text as a “Full Record and Cited Reference” so that it could be recognized by VOSviewer (Version 1.6.15).
Data Analysis
The VOSviewer software, supported by algorithmic clustering analysis based on bibliometric network data, can be used to visualize existing connections between bibliometric data. With the help of bibliometric maps, the network of relationships on this topic was examined among countries, research institutions, and authors.
For the 2,288 publications, the type of analysis was set as co-authored publication and the units of analysis were set as authors, organizations, and countries (regions included) to provide insights into any partnerships in this research. The minimum number of documents of an author (country or organization) was five. To merge synonyms by setting a thesaurus terms file, “Co-occurrence” and “Author keywords” were chosen to identify keywords to use to explore research hot spots, and the cited source of co-citations was also chosen to build a bibliometric map. The threshold of occurrences of a keyword was 10, whereas the minimum number of occurrences of the cited source of a co-citation was 20.
Results
Publication and Journal Field
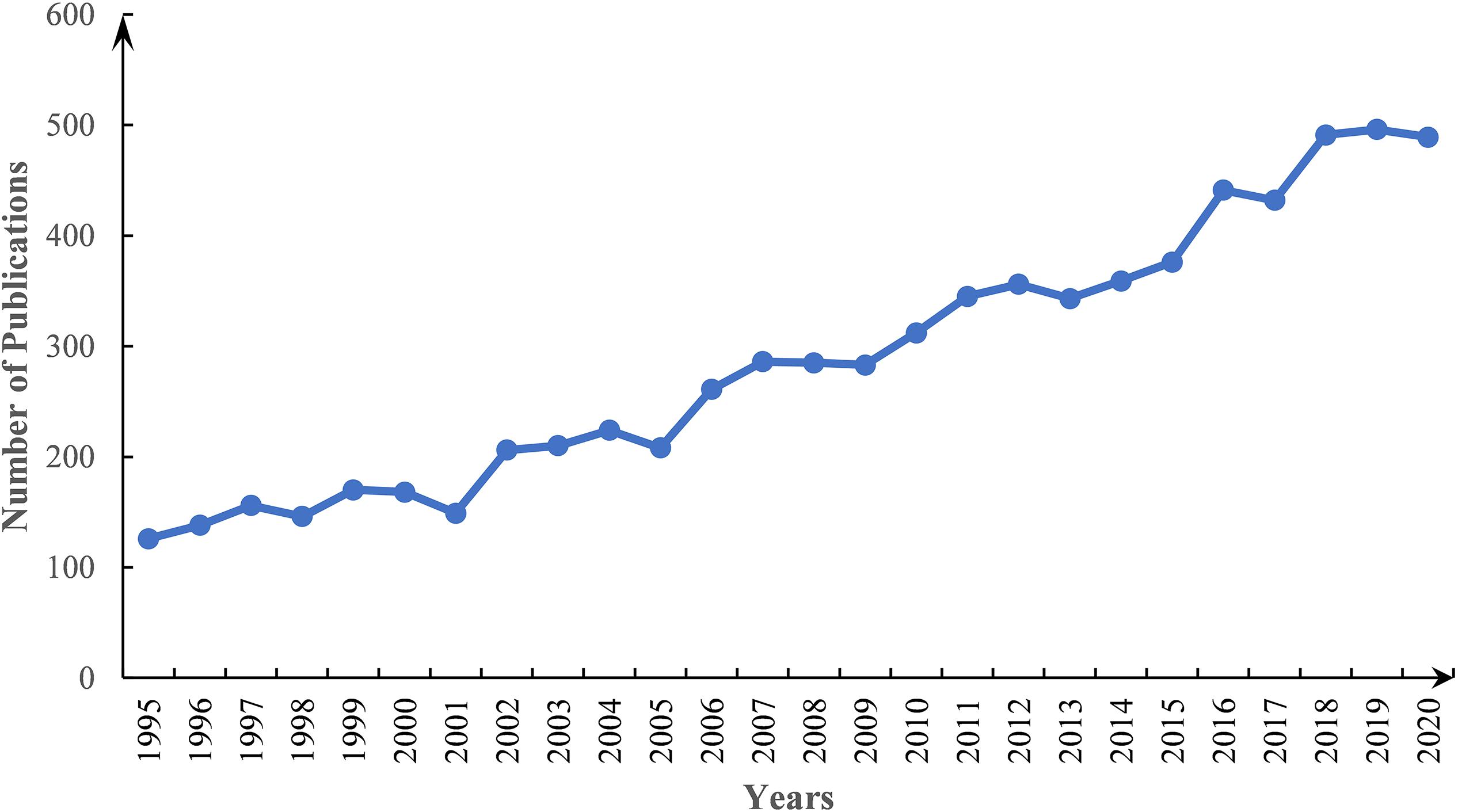
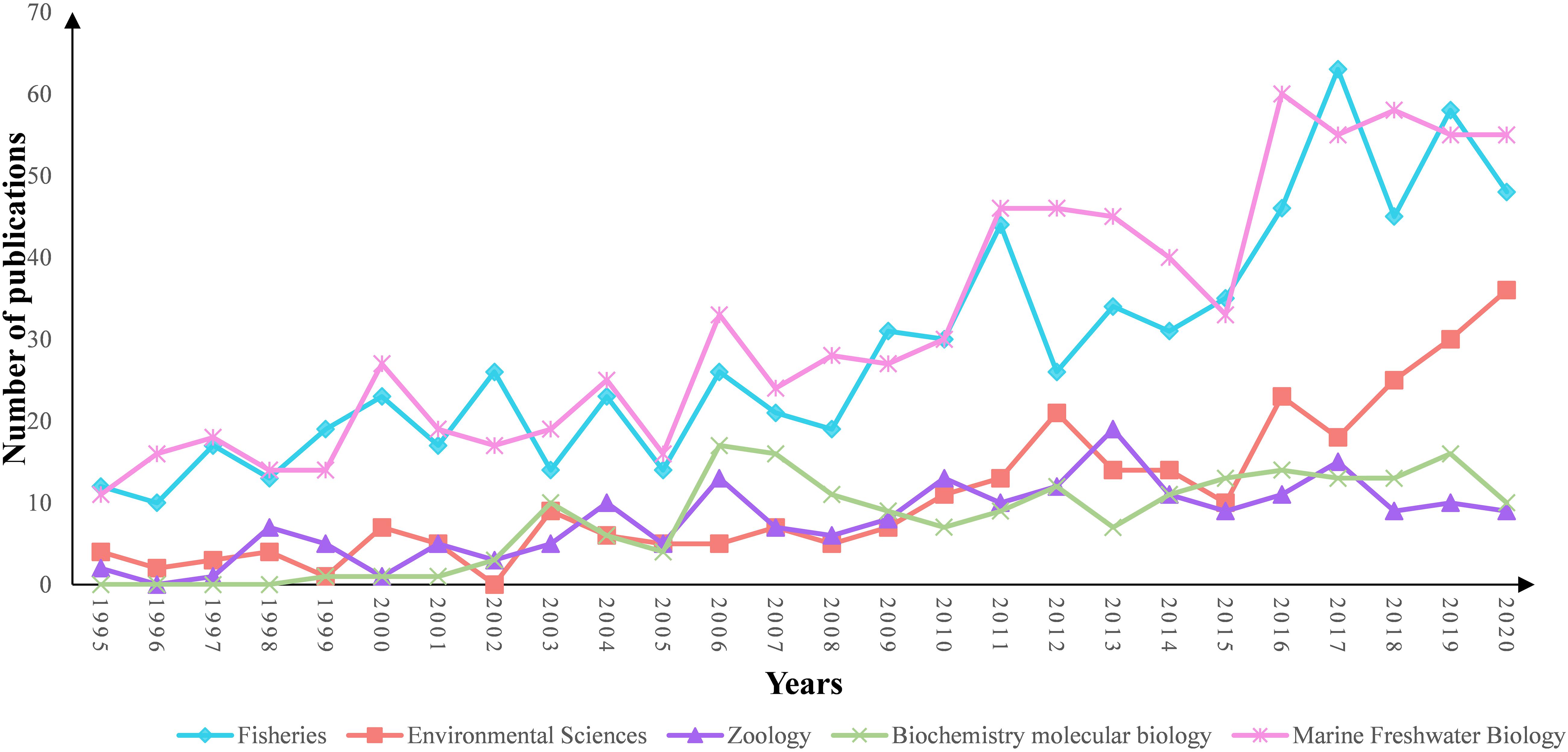
From 1995 to 2020, the number of publications on pearl bivalve mollusks increased, reflecting the increasing interest in these species (Figure 1). The top five subject categories were selected based on the classification of subject categories in Journal Citation Reports (JCR) (Figure 2). The results showed that research in environmental sciences increased gradually, whereas the fisheries and marine freshwater biology research both increased significantly over the study time frame. In total, 2,288 publications were from 548 journals. The top 10 most productive journals accounted for ∼31% of the total publications (Table 1). Among the pearl bivalve mollusks studies, Aquaculture was the most frequent publication, with 180 articles, cited 4,793 times, followed by the Journal of Shellfish Research, with 102 articles, and Fish and Shellfish Immunology, with 99 articles. In addition to these three journals, PLoS ONE and Marine Pollution Bulletin were more frequently cited (>1,100 times) by the 2,288 articles included in the bibliometric analysis.
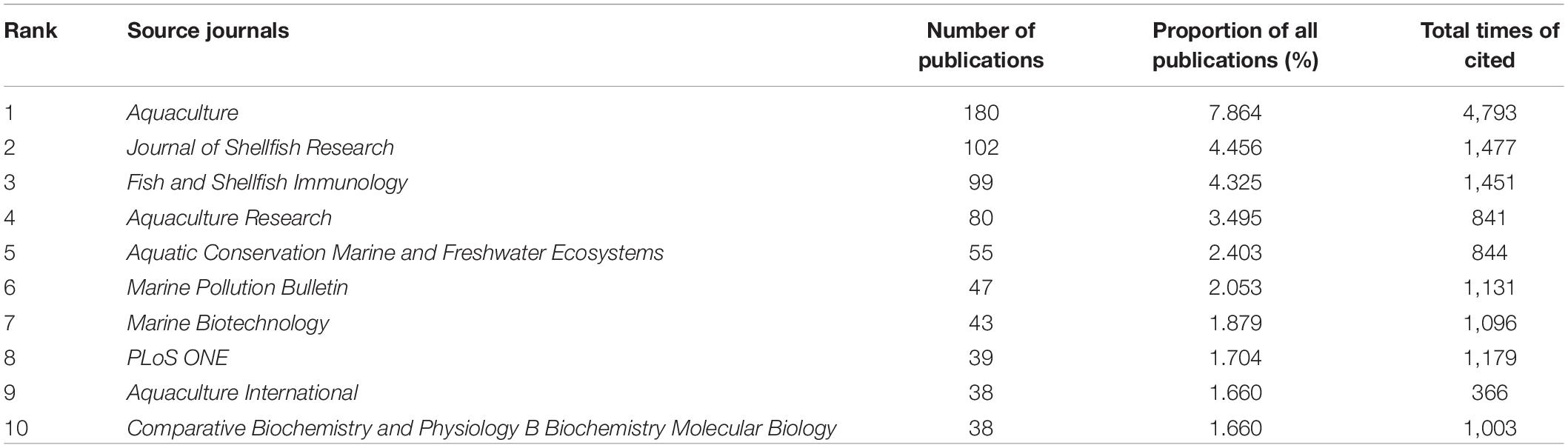
Table 1. Among the studies related with pearl bivalve mollusks as the theme, the numbers of publications, the proportion of all publications, and the total times cited for the top 10 source journals ranked by the number of publications based on Web of Science (WOS).
Countries or Regions
The contribution of the China was especially prominent with the largest number of publications on pearl bivalve mollusks and the highest number of cited articles (Table 2 and Figure 3). Also in the top five countries in terms of publications were Japan, Australia, and France, which all have a long history of pearl bivalve mollusk research. The United States represented the core of academic exchange and cooperation in pearl bivalve mollusks studies from 2010 to 2012, whereas contributions from China increased significantly after 2014. From 2016 onward, articles were published by researchers based in Italy, Poland, New Zealand, Portugal, and Belgium. In Figure 3, points representing a country that were joined to other country points indicated that researchers in those countries or regions worked with researchers from other countries or regions. Thus, this analysis indicated that research on pearl bivalve mollusks is a continuous and evolving research field.
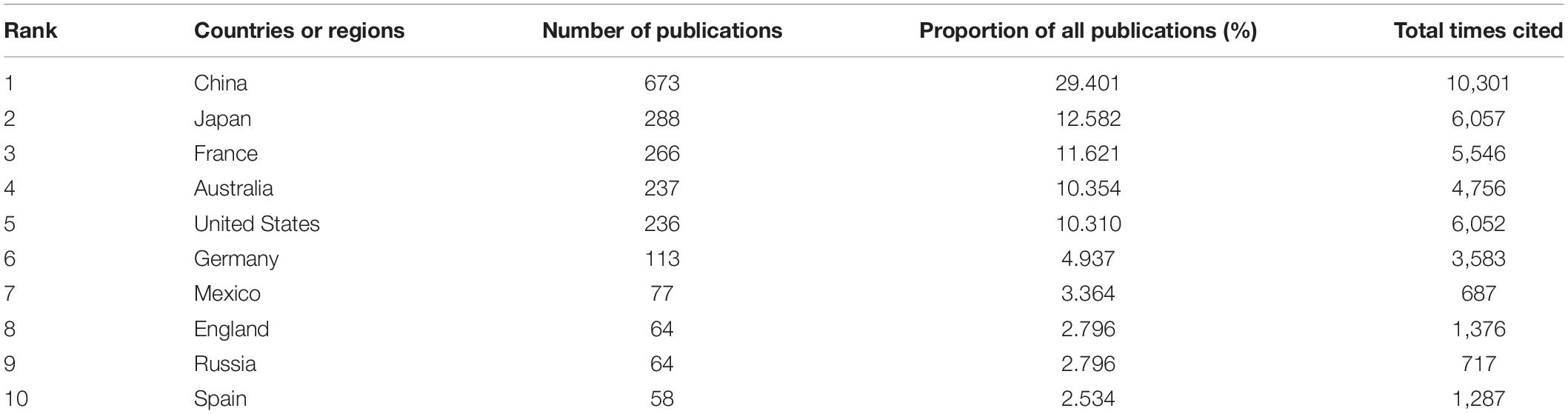
Table 2. Among the studies related with pearl bivalve mollusks as the theme, the numbers of publications, the proportion of all publications, and the total times cited of the top 10 countries or regions ranked by the number of publications based on WOS.
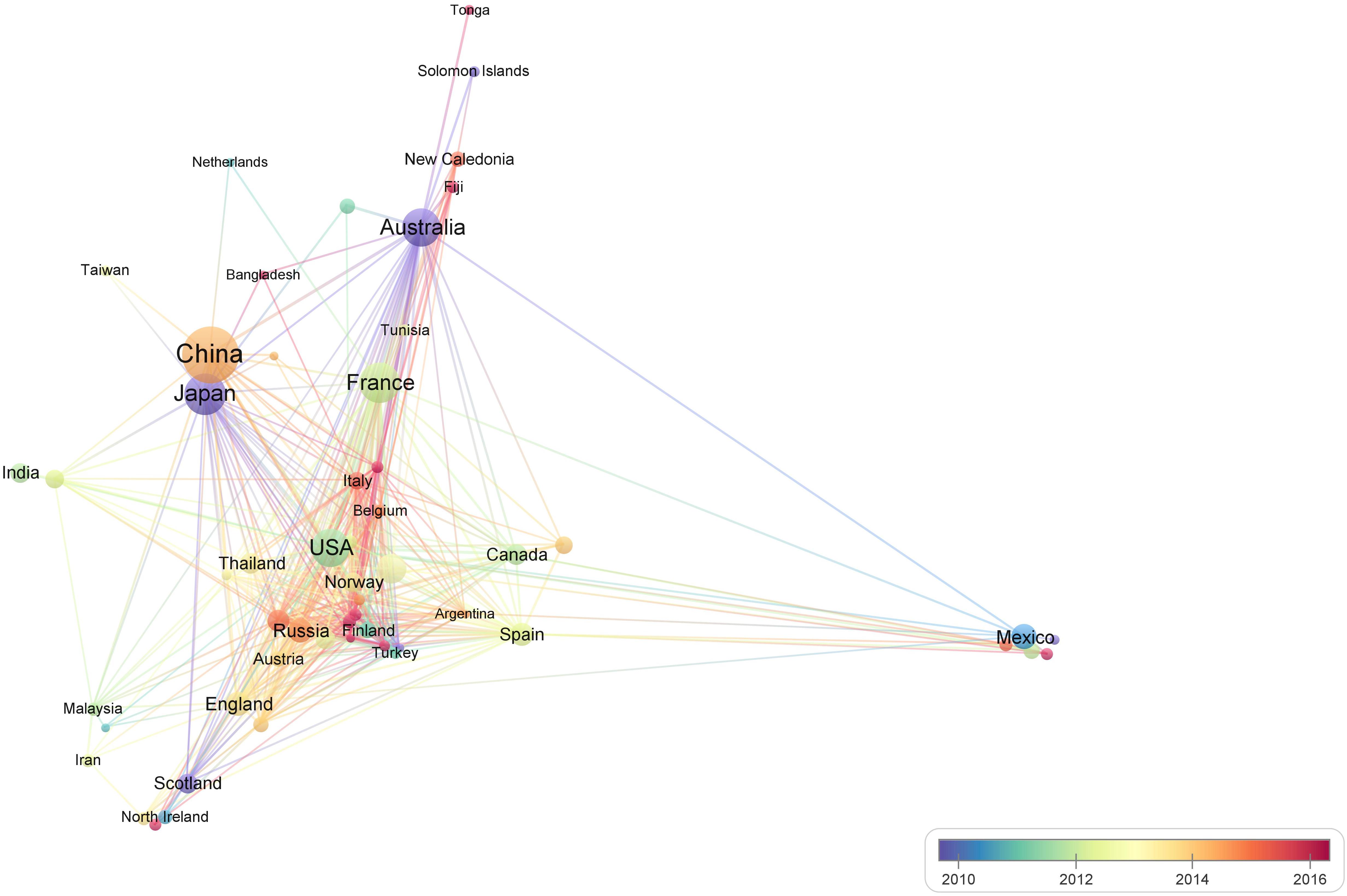
Figure 3. Bibliometric map of the studies of pearl bivalve mollusks as the theme. The size of the dot is proportional to the frequency that a certain country (the region included) appears in the analyzed articles. A line between two dots means that these two countries (regions included) appeared in the same article. The positive correlation between the thicker line and the higher frequency indicated that the two countries (regions included) appeared continually in the same articles. The color stands for the year when a certain country (the region included) most frequently occurred in articles.
Organizations
Chinese Academy of Sciencess published the highest number of articles (Table 3), followed by the French Research Institute for Exploitation of the Sea (Institut Français de Recherche pour l’Exploitation de la Mer; IFREMER), James Cook University, and Tsinghua University. Publication from the Chinese Academy of Sciences was the most cited, followed by those from the IFREMER and James Cook University. The relationship between different organizations was complex, as indicated by the numerous lines connecting to them, representing the tight cooperation between these organizations. IFREMER, Chinese Academy of Sciences, Chinese Academy of Fishery Sciences, and the University of Tokyo were the top four institutions in terms of frequency of cooperation. In Figure 4, the color scheme of yellow, orange, and red indicates that most organizations published articles after 2010.
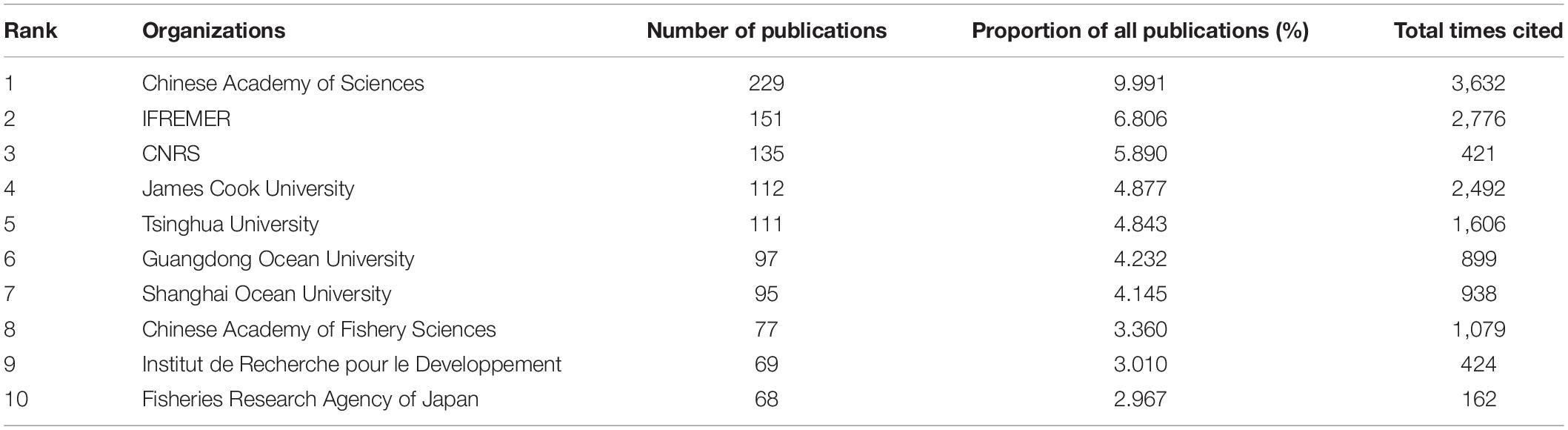
Table 3. Among the studies related with pearl bivalve mollusks as the theme, the numbers of publications, the proportion of all publications, and the total times cited for the top 10 organizations ranked by the number of publications based on WOS.
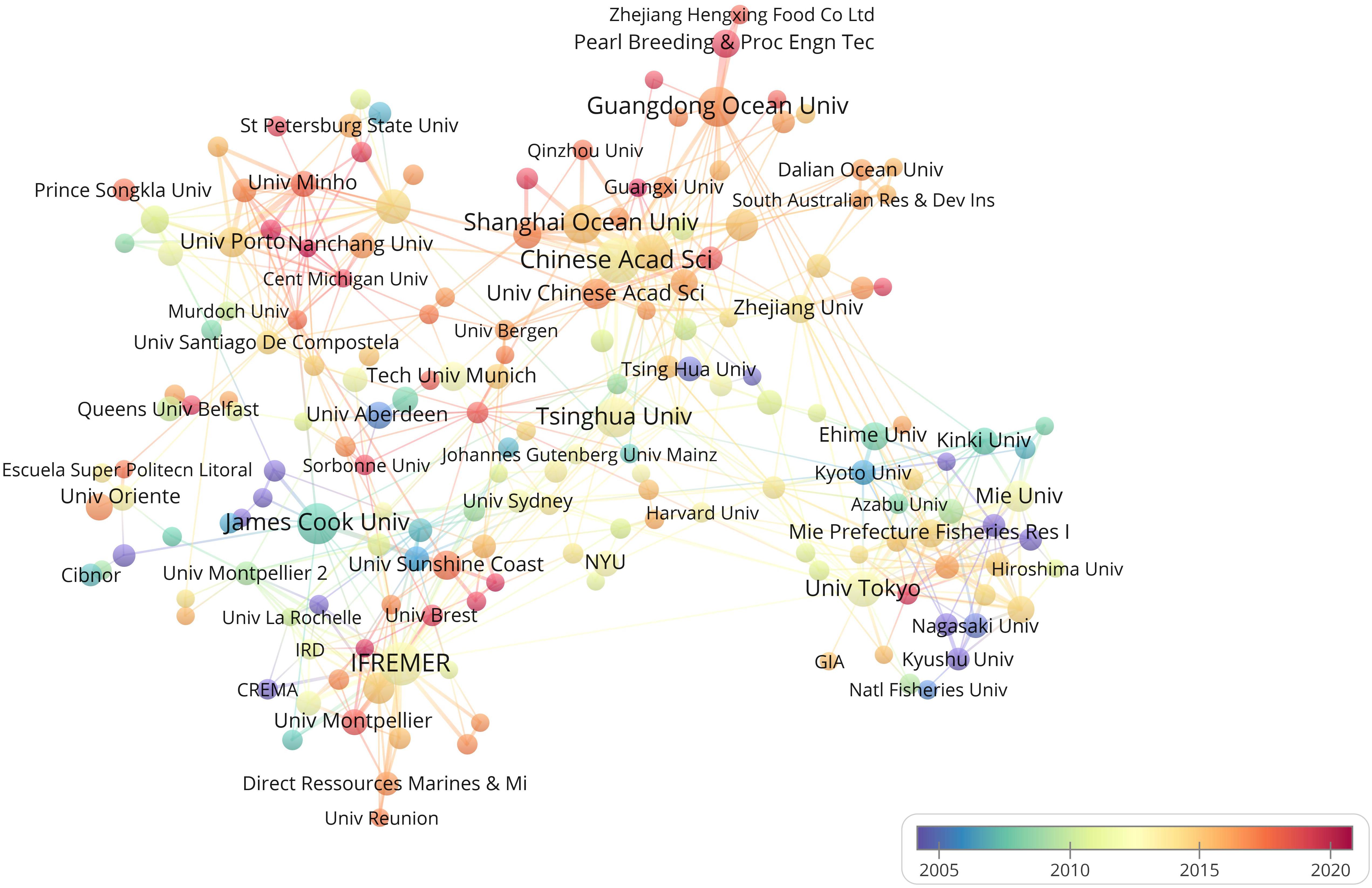
Figure 4. Bibliometric map of the studies of pearl bivalve mollusks as the theme. The size of the dot is proportional to the frequency that an organization appears in the analyzed articles. A line between two dots means that these two organizations appeared in the same article. The positive correlation between the thicker line and the higher frequency indicated that the two organizations appeared continually in the same articles. The color stands for the year when an organization most frequently occurred in articles.
Authors
The top 10 leading authors are listed in Table 4. Analysis of author productivity revealed that a few high-producing authors contributed significantly to the number of publications on pearl bivalve mollusks, especially R. Q. Zhang and L. P. Xie, who had the highest number of publications (∼4% of the total number of publications) and which were also cited most frequently. The main authors contributing to pearl bivalve mollusk research were R. Q. Zhang, L. P. Xie, Y. W. Deng, X. D. Du, G. Le Moullac, P. C. Southgate, and M. X. He (Figure 5). The mode of academic development was more likely to be polycentric, indicating that authors collaborated with others both inside and outside of their research institutions, although collaborations among the main contributors were rare. Except for the studies by R. Q. Zhang and L. P. Xie, most other authors began to publish work only after 2014.
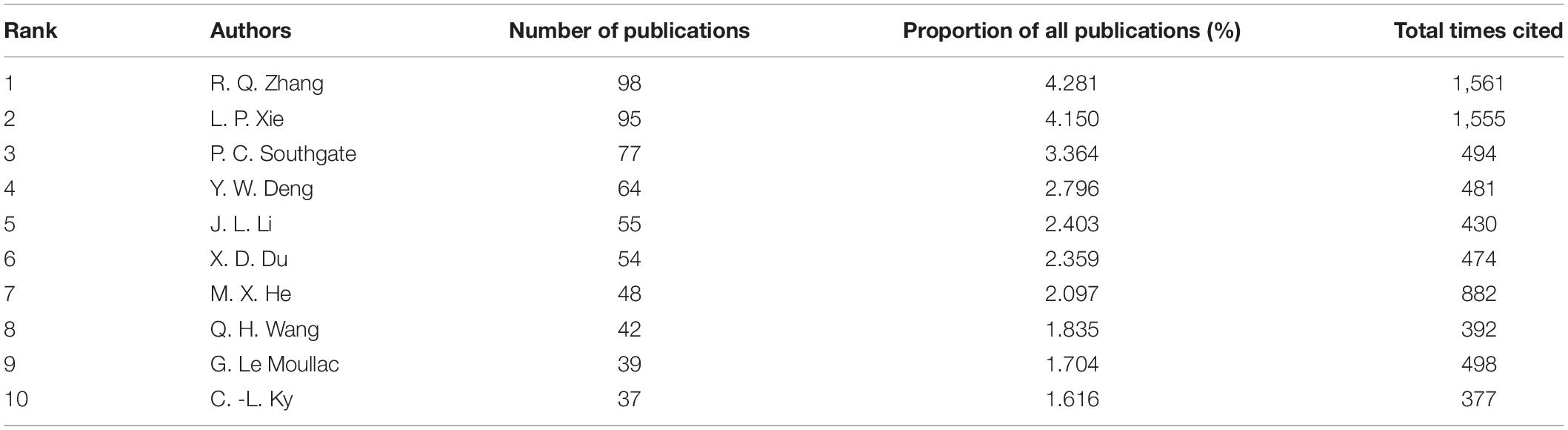
Table 4. Among the studies related with pearl bivalve mollusks as the theme, the numbers of publications, the proportion of all publications, and the total times cited for the top 10 authors ranked by the number of publications based on WOS.
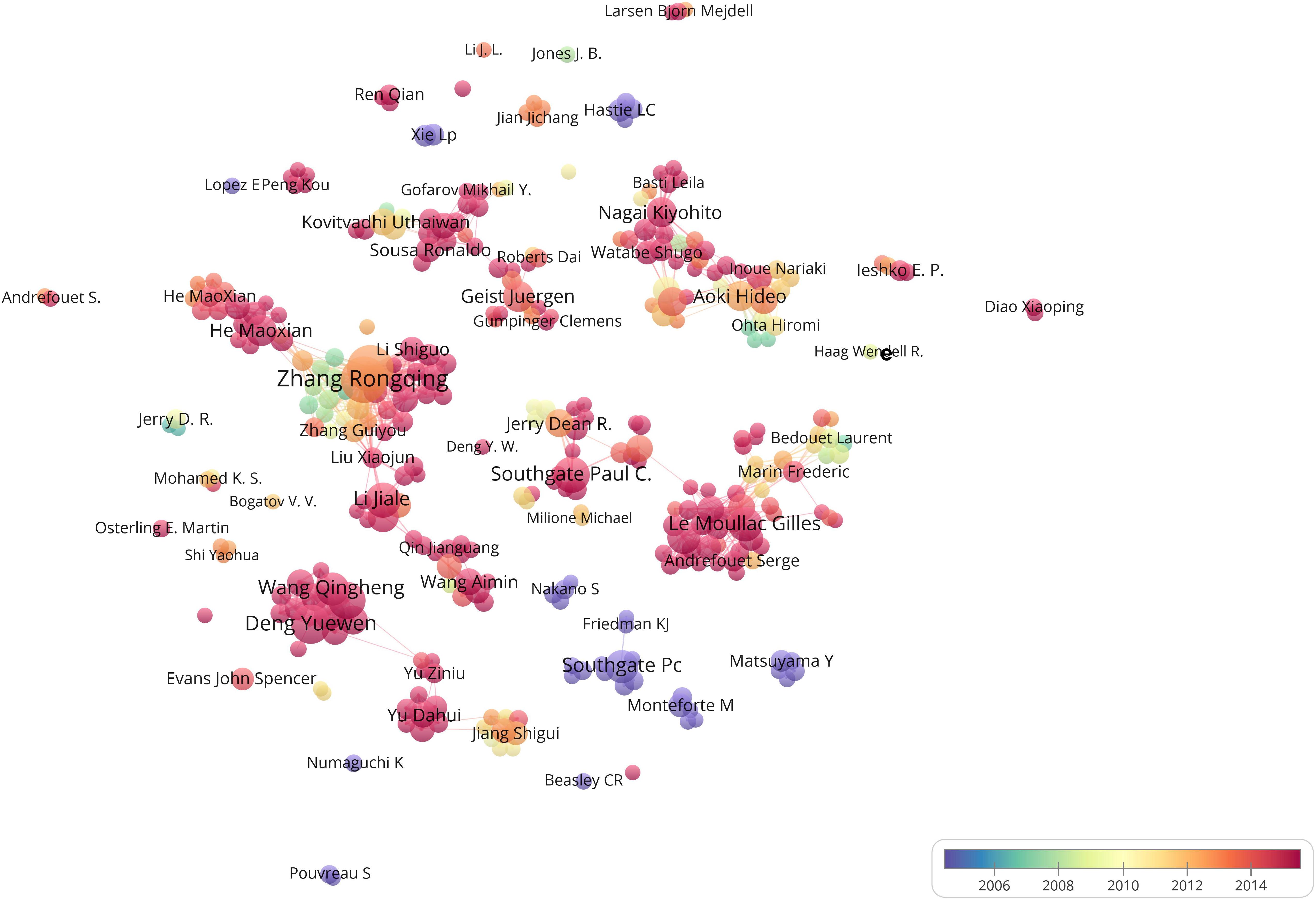
Figure 5. Bibliometric map of the studies of pearl bivalve mollusks as the theme. The size of the dot is proportional to the frequency that a certain author appears in the analyzed articles. A line between two dots means that these two authors appeared in the same article. The positive correlation between the thicker line and the higher frequency indicated that the two authors appeared continually in the same articles. The color stands for the year when a certain author most frequently occurred in articles.
Cited Journals
Aquaculture, Fish and Shellfish Immunology, Marine Pollution Bulletin, Hydrobiologia, and Nature formed the major cores in terms of cited journals (Figure 6). Each cluster is represented by a different color and can be connected with other clusters. Thus, these journals represent the main publication sources for pearl bivalve mollusk research.
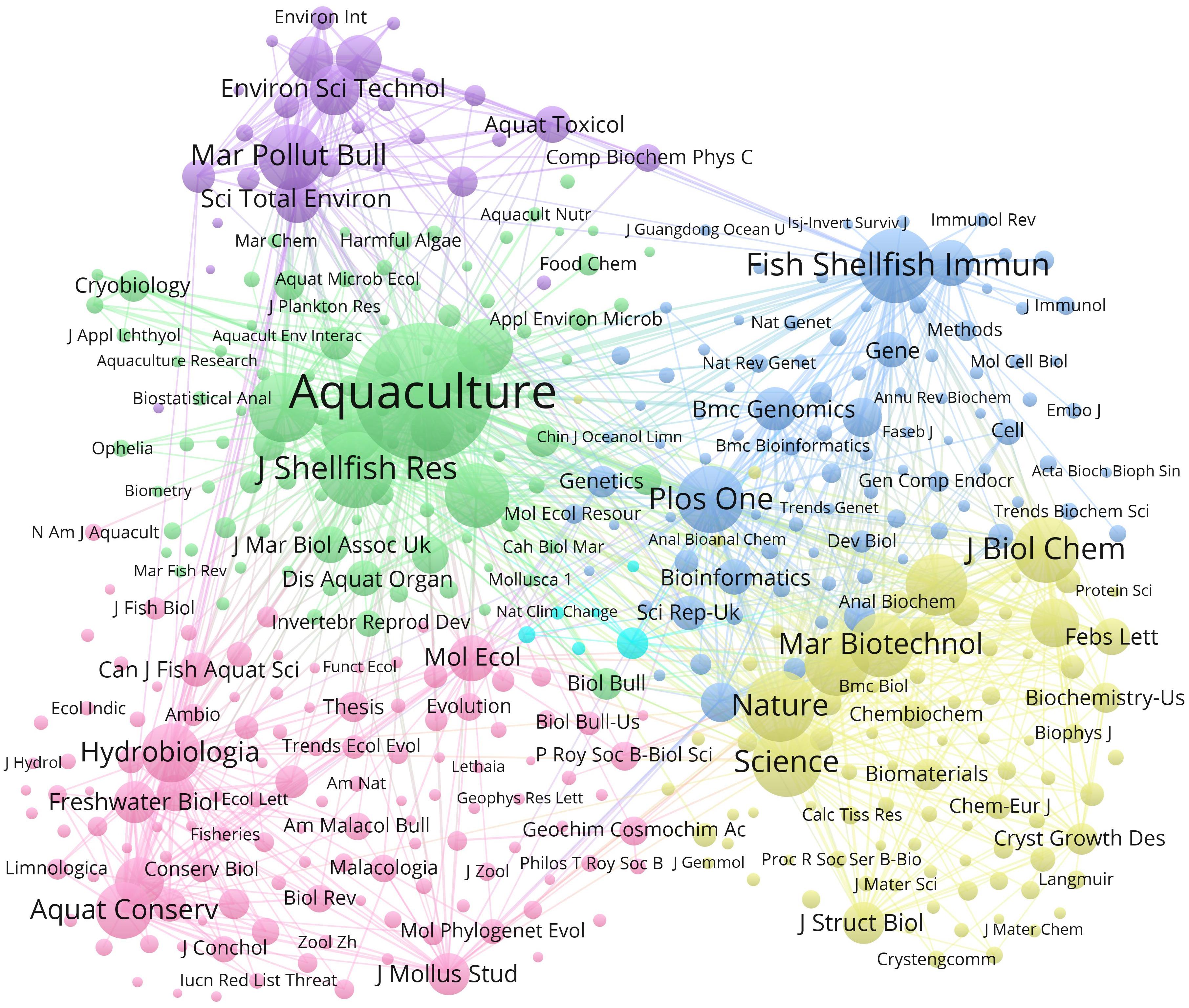
Figure 6. Bibliometric map of the studies of pearl bivalve mollusks as the theme. The size of the dot is proportional to the frequency that a certain co-citation appears in the analyzed articles. A line between two dots means that these two co-citations appeared in the same article. The positive correlation between the thicker line and the higher frequency indicated that the two co-citations appeared continually in the same articles.
Hot Keywords
Obviously, the phrase “pearl oyster” occurs the most because it was the name of the study object (Figure 7). Around 2008, “spat,” “culture,” “P. maxima,” “gametogenesis,” and “larvae” were the significant keywords, suggesting that researchers were mainly focused on the development of oyster larvae especially for silverlip pearl oyster during this period. Around 2010, the growth of Pacific pearl oysters and blacklip pearl oysters was the most popular topic, followed by “growth,” “P. margaritifera,” and “Crassostrea gigas” becoming the focus of pearl bivalve mollusk research. Hot keywords that emerged around 2012 were “conservation,” “temperature,” “survival,” and “Pinctada fucata,” indicating that researchers began to focus on environmental problems, such as global warming and marine pollution which affect the survival of Akoya pearl oysters. In 2014, the focus transferred to “biomineralization,” “prismatic layer,” “nacreous layer,” “family,” and “shell formation,” which means the specific formation of pearls was notable. The hot keywords changed to “quality,” “gene expression,” “identification,” “molecular-cloning,” and “Hyriopsis cumingii” around 2016. In recent years, researchers have focused on using advanced molecular techniques to improve the quality of pearls especially for specific species of freshwater pearl mussels.
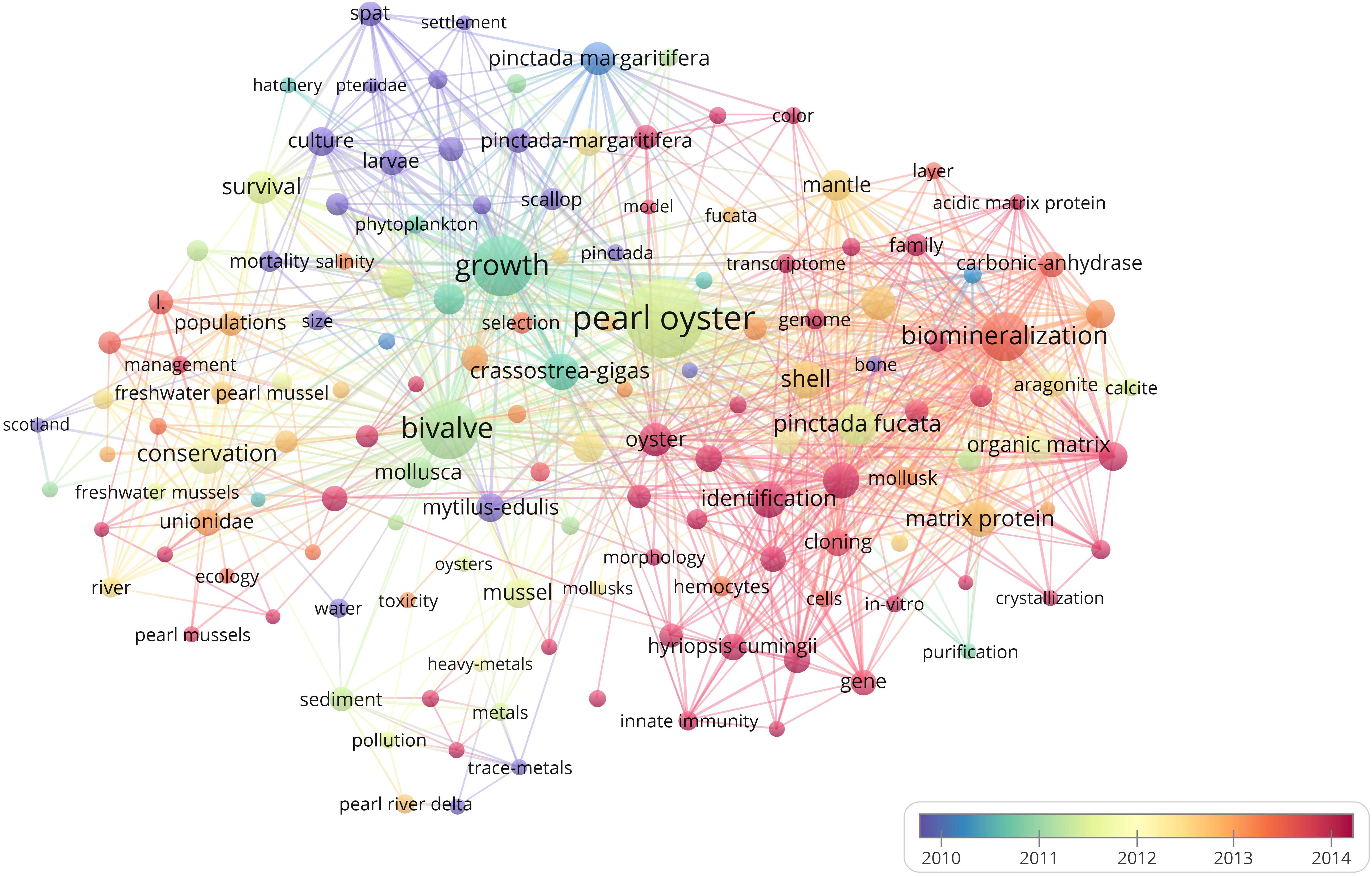
Figure 7. Bibliometric map of the studies of pearl bivalve mollusks as the theme. The size of the dot is proportional to the frequency that a certain keyword appears in the analyzed articles. A line between two dots means that these two keywords appeared in the same article. The positive correlation between the thicker line and the higher frequency indicated that the two keywords appeared continually in the same articles. The color stands for the year when a certain keyword most frequently occurred in articles.
Discussion
The transfer of hot spots in pearl oyster research fields showed that researchers have continued to focus on developmental biology, growth, culture mode, molecular biology, and ecological conservation of pearl oysters (Figure 7). From 2008 to 2010, researchers focused on the development, growth, and various culture modes of pearl oysters (Dauphin et al., 2008; Furuhashi et al., 2009; Welladsen et al., 2010), whereas in 2012, environmental factors became a focus to enhance the survival of pearl oysters (Gum et al., 2011; Hüning et al., 2013). From 2014 until 2020, researchers were more likely to focus on identifying connections between the different traits of pearl oysters and selected genes to determine the underlying regulatory mechanisms of gene expression by using the integrated application of transcriptomics and metabolomics, which measure the expression levels of genes at different molecular levels (Li and He, 2014; Bai et al., 2015; Lemer et al., 2015; Zheng et al., 2016b; Yang C. et al., 2018).
Cross-Disciplines Technology
Some new interdisciplinary technologies have been applied in traditional pearl farming. Extenics is used to study the possibility of the expansion of a topic and the development of innovation in a formal way (Cai et al., 2013). The essence of extenics is to make all kinds of seemingly contradictory or unsolvable problems solved (Liu et al., 2016). The combination of computer science and extenics brings forth new solutions for many problems across research fields. For example, an extenics-aided innovation system was used to provide a new method for pearl cultivations in China that produced pearls with a high yield but a low value (Yan et al., 2017). With the help of extenics and a mind map, Ye et al. (2019) studied the factors influencing the calcium content, water fluidity, water temperature, algae content, mollusk number, and so on in the cultivation of pearl oyster, providing a complete high-yield breeding program for pearl farmers. Van Wynsberge et al. (2020) used Multiscale Ultrahigh Resolution (MUR) satellite-derived sea surface temperatures (SSTs) to simulate the oyster population and assist the research and management of pearl oysters. In addition, related techniques have been developed to examine pearl nuclei during pearl oyster aquaculture. Digital radiography was used to detect the pearl nucleus by optimizing the voltage, current, and exposure time (Yulianti et al., 2018). Using micro-CT in the live bivalves has advantages in that it is both non-invasive and rapid. Moreover, pearls can be evaluated in terms of their shape, size, smoothness, and dimension (Ding et al., 2021).
Interaction Between Farming and Environment
When discussing pearl farming, it is impossible not to mention the interaction between farming activities and the environment. Following the success of French Polynesia and the Cook Islands in marine pearl oyster aquaculture, other island states in the Pacific have also developed their own pearl mollusk aquacultural setups (Tisdell and Poirine, 2008). Basti et al. (2011) revealed that Heterocapsa circularisquama blooms negatively influence the early life stages of P. fucata. Ocean acidification results in a reduced byssus diameter and amplified byssus nanocavity in P. fucata by altering the abundance and secondary structure of byssal proteins and affecting the metal ion content in distal threads (Li et al., 2017a). Researchers have also highlighted issues with diversity during pearl oyster aquaculture development. For example, Cartier used roving diver census methods to analyze coral reef bommies in a Polynesian lagoon and showed that pearl farming might not negatively alter the number and population structure of fish (Cartier and Carpenter, 2014). Gifford et al. (2004) argued that pearl farms could be regarded as biological filters to solve nutrient enrichment and pollution issues (Gifford et al., 2004). Lacoste et al. (2014) investigated the influence of nutrient regeneration on pearl oyster farming in French Polynesia, revealing that nutrient availability can be enhanced by pearl oyster culture. Thus, these studies did not reveal a significant crisis in marine pearl oyster culture. However, there are more problems associated with freshwater pearl mussel farming. The population numbers of M. margaritifera in Europe had been estimated to have declined >90% by the 1990s (Bauer, 1988), rendering this species highly endangered (Geist, 2010; Gum et al., 2011); therefore, many approaches were taken to protect these mussels, including from aquaculture (Preston et al., 2007; Schmidt and Vandré, 2010) to molecular breeding (Geist et al., 2003; Huff et al., 2004). Given that East Asia represents a significant market for pearls, China and Japan have large market shares of pearl oyster aquacultural outputs (Tisdell and Poirine, 2008). China expanded its pearl farming industry at the expense of the environment which resulted in the decline of both the scale of breeding and pearl yield (Bai et al., 2014b). The 18th National Congress of the Communist Party of China put forward the concept of green development and innovation in 2012. Under these new requirements, the freshwater pearl cultivation industry in China began to transform, resulting in the production of high-quality pearls with fewer environmental impacts.
Hot Research Methods in Gene Expression Regulation
Factors affecting pearl yield and quality can be divided into endogenous factors and exogenous factors, which lead to differences in growth performance, traits, and disease resistance of pearl oysters, prompting researchers to seek solutions to these problems. Important endogenous factors are the molecular mechanisms of growth-related genes and the specific gene expression processes involved in pearl production. In recent years, gene expression regulation as an effective method has attracted extensive attention and in-depth research to improve various indexes of pearl oysters at the genetic level (Adzigbli et al., 2020b). Gene expression regulation technology relates to the control of gene expression during DNA transcription and translation via gene expression regulatory elements, including transcriptional regulation, mRNA regulation, and translation regulation. Genomics (Zenger et al., 2019), transcriptomics (Wei et al., 2017), proteomics (Wang et al., 2019), and metabonomics analyses (Yang et al., 2019) are commonly used to analyze the regulatory mechanism of coding genes and non-coding genes to research hot topics in pearl production, such as growth performance and traits, pearl formation and biomineralization, and immune response (Song et al., 2019; Adzigbli et al., 2020a).
Growth Performance and Traits
As the first comprehensive linkage map for any pearl oyster species, a high-density SNP genetic map for P. maxima not only provides an essential genomic tool facilitating studies investigating the genomic architecture of complex trait variation, but also identifies quantitative trait loci (QTL) for economically important traits in genetic selection programs within the pearl industry (Jones et al., 2013). Shi et al. (2014) generated a high-density genetic linkage map for Pinctada fucata martensii for the first time, which provides a target gene for selective breeding of P. f. martensii to improve pearl production. Results in the study of Li and He (2014) highlighted the effectiveness of restriction site-associated DNA sequencing (RAD-Seq) as a tool for generation of QTL-targeted and genome-wide marker data in P. fucata and its possible utility in marker-assisted selection (MAS). Several growth-related genes have been shown to influence growth in pearl oysters, including insulin-related peptide receptor and the insulin-like growth factor (IGF) system and its components extracellular signal-regulated kinases (ERKs) and mitogen-activated protein kinase 4 (MKK4) (Adzigbli et al., 2020b). Insulin-like growth factor-binding proteins (IGFBPs) are major regulators of IGF activity and regulate the endocrine system in invertebrates. Zhang et al. (2017) use expressed sequence tag (EST) sequences to identify the full-length cDNA of an IGFBP from P. fucata and indicated that Pfigfbp mRNA expression in mantle cells was affected by insulin/IGFs (IGF-I, IGF-II) and that Pfigfbp might be involved in endocrine signaling in P. fucata. In terms of accelerating marker-assisted breeding, whole-genome resequencing (WGR) has the advantages of faster and more accurate detection of recombinant breakpoints and genetic mapping compared with marker-based genotyping. Liu et al. (2020) constructed the first high-density genetic linkage map for P. f. martensii WGR and identified 32 growth-trait-related QTLs; four differentially expressed genes were located on the regions of QTLs related to growth traits. Unsynchronized growth has been observed during pearl culture. Through integrated transcriptome and metabolome analyses, Hao et al. (2019) assessed the metabolic and transcript changes between the fast-growing and slow-growing P. f. martensii groups and determined the mechanism underlying this phenomenon. Pacific Bioscience (PacBio) single-molecular sequencing technologies, an emerging transcriptome sequencing method, enables researchers to understand the complexity and diversity of the molecular mechanisms of the growth of P. f. martensii (Zhang et al., 2020). Using a next-generation sequencing method and RNA-seq technology, Vaihiti et al. performed gonad transcriptome analysis of the hermaphroditic P. margaritifera from several gonadic samples of males and females at different development stages. The specific gene expression profiles identified sex differentiation and potential sex-determining genes in P. margaritifera: pmarg-fem1-like, pmarg-dmrt, and pmarg-foxl2 are potentially involved in the sperm–oocyte switch (Teaniniuraitemoana et al., 2014). Hybridization is documented as an efficient method to improve growth traits in pearl oysters. According to a transcriptome analysis, Yang J. et al. (2018) found that differentially expressed genes in hybrid families and their parental families might be associated with growth heterosis. Mitochondrial (mt) DNA is frequently used in comparative genomic analyses because its gene content and arrangement, as well as mt tRNA secondary structure, provides valuable phylogenetic information. Wu et al. (2012) described the sequence and characterization of the complete mt genome of the silverlip pearl oyster, P. maxima, observing a unique tRNA gene family in this genome and described a novel highly expressed ORF (orfUR1), its potential function, and two possible mechanisms of origin. Piwi-interacting RNAs (piRNAs) are recently discovered small non-coding RNAs, the function of which is repressing transposable element activity. Huang et al. (2019) identified and characterized putative piRNAs by high-throughput sequencing in somatic and gonadal tissues of P. fucata. The discovery that piRNAs are highly expressed in a range of somatic tissues has enriched our understanding of the diversity and function of small RNAs in pearl oyster.
Pearl Formation and Biomineralization
The study of Takeuchi et al. (2012) provides the first opportunity to obtain insight into a bivalvian mollusk genome. Draft genome of the pearl oyster P. fucata provides a platform for the identification of selection markers and genes for calcification, the knowledge of which has a significant importance in the understanding of the molecular mechanisms involved in pearl biosynthesis and biology of bivalve mollusks. Liu J. et al. (2015) reported a global gene analysis during larval development of P. fucata based on a microarray and proposed a mechanism of how biomineralization-related genes regulate the larval shell formation process. Biomineralization refers to the dynamic physiological processes whereby living organisms elaborate mineralized tissues. Outer mantle epithelial cells (OME) secrete shell matrix proteins (SMPs) which determine the mineralogical and crystallographic properties of the shell. Researchers have identified many novel SMPs by using molecular biology techniques (i.e., gene cloning, in situ hybridization, and immunohistochemistry) coupled with high-throughput sequencing data (genome, proteome, secretome, and transcriptome), as well as confirming their corresponding functions during shell formation (Song et al., 2019). Using liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of proteins extracted from the shells of P. fucata, Liu C. et al. (2015) identified 72 unique SMPs and combined with a draft genome. Bai et al. (2014a) compared the relative expression of seven housekeeping genes across different tissue types in the mantle or pearl sac by quantitative real-time polymerase chain reaction (PCR) and suggested that Ubi, Rpl18, and EF1α are suitable reference genes to facilitate the analysis of gene expressions during shell or pearl formation in the pearl mussel, H. cumingii. Using proteomic analyses combined with genomic and transcriptomic analyses with the addition of RNA-seq data, Zhao et al. (2018) showed that the gene expression profiles and SMPs from larval shells of the pearl oyster, P. fucata, and the Pacific oyster, C. gigas, are almost entirely different from those of adults in both species, which indicates that bivalves have two repertoires of SMP genes to construct larval and adult shells. Blay et al. (2018) analyzed the expression profiles of eight genes encoding nacreous and/or prismatic SMPs in the pearl sac of P. margaritifera during pearl biogenesis and identified three genes implicated in pearl surface quality by multivariate regression tree building. Thus, SHEM9, ASP, and PIF could be used as biomarkers for genetic improvement of P. margaritifera pearl quality. The study of Gao et al. (2016) suggested that the transcription factor Pf-POU3F4 regulates the expression of the matrix protein genes Aspein and Prismalin-14 in pearl oyster. Using high-throughput transcriptome profiling, Take et al. (2019) revealed the genes involved in the development of the pearl sac and pearls, as well as the sequential expression patterns of different SMPs secreted from the pearl sac during pearl formation in P. fucata. miRNAs are a class of non-coding RNA molecules with post-transcriptional regulatory activity in biomineralization. The study of Zheng et al. (2016a) added to the miRNA databases of P. martensii by deep sequencing and indicated pm-miR-2386 and pm-miR-13b as key factors regulating the formation of organic matrix or the differentiation of mineralogic cells during shell formation. Du et al. (2017) sequenced the highly polymorphic genome of the pearl oyster and conducted multi-omic and biochemical studies to study nacre formation, which is formed through a process of matrix-assisted biomineralization, as well as identified a large set of novel proteins participating in matrix–framework formation. Given that color is an indicator of pearl quality, Bai et al. (2013) identified genes involved in nacre coloring through comparative transcriptome analysis of tissues secreting purple and white nacre in the triangle sail mussel, H. cumingii. As nacre color has an effect on pearl color, Chen et al. (2017) studied the effect of tyrosinase on nacre color in H. cumingii (two types of purple-shelled and white-shelled freshwater pearl mussels) by cloning novel tyrosinase protein and tyrosinase-related protein genes (HcTyr and HcTyp-1, respectively) from the mantle. Results indicated that HcTyr and HcTyp-1 are involved in the formation of nacre color in H. cumingii. The albino phenotype of P. margaritifera provides a new way to examine the molecular mechanisms underlying pigmentation. Using whole transcriptome RNA-sequencing analysis, Auffret et al. (2020) found that these genes were involved in five main molecular pathways, tightly linked to known pigmentation pathways: melanogenesis, calcium signaling pathway, Notch signaling pathway, pigment transport, and biomineralization.
Immune Response
Multi-omic studies have documented responses to nuclei insertion operation and diseases and in P. f. martensii, P. maxima, P. margaritifera, and Pteria penguin (Adzigbli et al., 2020a). Bivalve mollusks have evolved an innate immune system with differentially expressed immune-related genes, which relies on the interaction between cellular and humoral factors to prevent them from potential infection because of their frequent exposure to microbial pathogens and environmental perturbations (Xu et al., 2010; Adzigbli et al., 2020b). Matsuyama et al. (2017) used shotgun and 16s rRNA-based metagenomic analysis to identify genes that are present specifically in oysters affected by Akoya oyster disease. ERKs are conserved and related to protein-serine/threonine kinases that participate in the regulation of multiple biological processes. Zhang et al. (2018) cloned and identified a novel ERK from P. fucata (PfERK) that shared significant homology with other mollusk ERK proteins. Results suggested that PfERK was expressed in seven different tissues examined from pearl oyster in all developmental stages and is involved in innate mantle immunity in vivo and in vitro. Proteins in the tumor necrosis factor receptor (TNFR) superfamily have significant roles in inflammation, apoptosis, autoimmunity, and organogenesis. Two TNFR gene homologs (PmTNFR1 and PmTNFR5) were characterized from P. f. martensii, and Wu et al. (2020) found RNAi inhibition of PmTNFR1 and PmTNFR5 gene expression. Results suggested that the NF-κB signaling pathway may be activated by PmTNFR1 and PmTNFR5 and that these genes are closely related to allograft immunity in P. f. martensii. Long non-coding RNAs (lncRNAs) have regulatory roles in exoskeleton formation and immune response. Zheng et al. (2020) found a novel lncRNA, LncMSEN2, from the pearl oyster P. f. martensii, and its sequence was validated via PCR. High expression of LncMSEN2 in mantle tissues and pearl sac detected by quantitative real-time PCR, in situ hybridization experiments, and RNAi experiments indicated that LncMSEN2 may be a novel regulator of the mantle–shell defense system of pearl oyster.
Conclusion and Future Directions
Cultured pearls are mainly produced by freshwater pearl mussels and marine pearl oysters, and the economic importance associated with pearl farming has focused the attention of researchers from various fields on pearl bivalve mollusks. It is precisely because the market demands both the quantity and quality of pearls, which has focused research on these issues over the 25 years. Given that there are many internal and external factors affecting pearl production, researchers have reported corresponding solutions for specific topics, such as different culture modes, color formation, and growth performance of pearl bivalve mollusks. In the meantime, researchers have not ignored other threats to the pearl industry, in terms of environmental pollution and genetic characterization decline. With improvements in technology, genetic regulation technology will continue to be the most fundamental and effective technological method to regulate the character of pearls at the molecular level in the coming decades. However, for the sustainable development of the pearl industry, we need additional collaboration and cooperation among researchers and those involved in the industry to discuss and improve pearl bivalve mollusks research in the future.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
QC performed the bibliometric analysis and drafted the manuscript. WH collected the data and drafted the manuscript. ZB conceived the idea and supported the study. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (31872565), the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-49), and the Program of Shanghai Academic Research Leader (19XD1421500).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adzigbli, L., Hao, R., Jiao, Y., Deng, Y., Du, X., Wang, Q., et al. (2020a). Immune response of pearl oysters to stress and diseases. Rev. Aquac. 12, 513–523. doi: 10.1111/raq.12329
Adzigbli, L., Wang, Z., Zhao, Z., Yang, C., Li, J., and Deng, Y. (2020b). Growth in pearl oysters: a review of genetic and environmental influences. Aquac. Res. 51, 18–28. doi: 10.1111/are.14365
Auffret, P., Le Luyer, J., Koua, M. S., Quillien, V., and Ky, C.-L. (2020). Tracing key genes associated with the Pinctada margaritifera albino phenotype from juvenile to cultured pearl harvest stages using multiple whole transcriptome sequencing. BMC Genomics 21:662. doi: 10.1186/s12864-020-07015-w
Bai, Z., Han, X., Luo, M., Lin, J., Wang, G., and Li, J. (2015). Constructing a microsatellite-based linkage map and identifying QTL for pearl quality traits in triangle pearl mussel (Hyriopsis cumingii). Aquaculture 437, 102–110. doi: 10.1016/j.aquaculture.2014.11.008
Bai, Z., Lin, J., Ma, K., Wang, G., Niu, D., and Li, J. (2014a). Identification of housekeeping genes suitable for gene expression analysis in the pearl mussel, Hyriopsis cumingii, during biomineralization. Mol. Genet. Genomics 289, 717–725. doi: 10.1007/s00438-014-0837-1
Bai, Z., Wang, G., Liu, X., and Li, J. (2014b). The status and development trend of freshwater pearl seed industry in China. J. Shanghai Ocean Univ. 23, 874–881.
Bai, Z., Zheng, H., Lin, J., Wang, G., and Li, J. (2013). Comparative analysis of the transcriptome in tissues secreting purple and white nacre in the pearl mussel Hyriopsis cumingii. PLoS One 8:e53617. doi: 10.1371/journal.pone.0053617
Basti, L., Go, J., Higuchi, K., Nagai, K., and Segawa, S. (2011). Effects of the toxic dinoflagellate Heterocapsa circularisquama on larvae of the pearl oyster Pinctada fucata martensii (Dunker, 1873). J. Shellfish Res. 30, 177–186. doi: 10.2983/035.030.0125
Bauer, G. (1988). Threats to the freshwater pearl mussel Margaritifera margaritifera L. in central Europe. Biol. Conserv. 45, 239–253. doi: 10.1016/0006-3207(88)90056-0
Blay, C., Planes, S., and Ky, C.-L. (2018). Cultured pearl surface quality profiling by the shell matrix protein gene expression in the biomineralised pearl sac tissue of Pinctada margaritifera. Mar. Biotechnol. 20, 490–501. doi: 10.1007/s10126-018-9811-y
Bonilla, C. A., Merigó, J. M., and Torres-Abad, C. (2015). Economics in Latin America: a bibliometric analysis. Scientometrics 105, 1239–1252. doi: 10.1007/s11192-015-1747-7
Cai, W., Yang, C., Smarandache, F., Vladareanu, L., Li, Q., Zou, G., et al. (2013). Extenics and Innovation Methods. Boca Raton, FL: CRC Press.
Cartier, L. E., and Carpenter, K. E. (2014). The influence of pearl oyster farming on reef fish abundance and diversity in Ahe, French Polynesia. Mar. Pollut. Bull. 78, 43–50. doi: 10.1016/j.marpolbul.2013.11.027
Changbo, Z., Paul, C. S., and Ting, L. (2019). “Production of pearls,” in Goods and Services of Marine Bivalves, eds O. Strand, J. G. Ferreira, and J. K. Petersen (Cham: Springer), 73–93.
Chen, X., Liu, X., Bai, Z., Zhao, L., and Li, J. (2017). HcTyr and HcTyp-1 of Hyriopsis cumingii, novel tyrosinase and tyrosinase-related protein genes involved in nacre color formation. Comparative Biochem. Physiol. Part B: Biochem. Mol. Biol. 204, 1–8. doi: 10.1016/j.cbpb.2016.11.005
Chen, X., Peng, L.-H., Chee, S.-S., Shan, Y.-H., Liang, W.-Q., and Gao, J.-Q. (2019). Nanoscaled pearl powder accelerates wound repair and regeneration in vitro and in vivo. Drug Dev. Industrial Pharm. 45, 1009–1016. doi: 10.1080/03639045.2019.1593436
Cosgrove, P., Watt, J., Hastie, L., Sime, I., Shields, D., Cosgrove, C., et al. (2016). The status of the freshwater pearl mussel Margaritifera margaritifera in Scotland: extent of change since 1990s, threats and management implications. Biodiver. Conserv. 25, 2093–2112. doi: 10.1007/s10531-016-1180-0
Daim, T. U., Rueda, G., Martin, H., and Gerdsri, P. (2006). Forecasting emerging technologies: use of bibliometrics and patent analysis. Technol. Forecast. Soc. Change 73, 981–1012. doi: 10.1016/j.techfore.2006.04.004
Dan, H., and Ruobo, G. (2002). Freshwater pearl culture and production in China. Aquac. Asia 7, 6–8.
Dauphin, Y., Ball, A. D., Cotte, M., Cuif, J.-P., Meibom, A., Salomé, M., et al. (2008). Structure and composition of the nacre–prisms transition in the shell of Pinctada margaritifera (Mollusca, Bivalvia). Analytical Bioanalytical Chem. 390, 1659–1669. doi: 10.1007/s00216-008-1860-z
Ding, R., Liu, L., Hu, Z., Wu, A., Zhu, H., Ruan, Q., et al. (2021). In vivo noninvasive and rapid evaluation of pearls in freshwater mussels based on micro-CT. Aquaculture 534:736232. doi: 10.1016/j.aquaculture.2020.736232
Du, X., Fan, G., Jiao, Y., Zhang, H., Guo, X., Huang, R., et al. (2017). The pearl oyster Pinctada fucata martensii genome and multi-omic analyses provide insights into biomineralization. Gigascience 6:gix059. doi: 10.1093/gigascience/gix059
Duncan Peter, F., and Arne, G. (2019). “Shells as collector’s items,” in Goods and Services of Marine Bivalves, eds O. Strand, J. G. Ferreira, and J. K. Petersen (Cham: Springer Nature), 381–411.
Ericson, K. (2017). Making space for red tide: discolored water and the early twentieth century Bayscape of Japanese pearl cultivation. J. Hist. Biol. 50, 393–423. doi: 10.1007/s10739-016-9443-x
Furuhashi, T., Schwarzinger, C., Miksik, I., Smrz, M., and Beran, A. (2009). Molluscan shell evolution with review of shell calcification hypothesis. Compar. Biochem. Physiol. Part B: Biochem. Mol. Biol. 154, 351–371. doi: 10.1016/j.cbpb.2009.07.011
Gao, J., Chen, Y., Yang, Y., Liang, J., Xie, J., Liu, J., et al. (2016). The transcription factor Pf-POU 3F4 regulates expression of the matrix protein genes Aspein and Prismalin-14 in pearl oyster (Pinctada fucata). FEBS J. 283, 1962–1978. doi: 10.1111/febs.13716
Geaney, F., Scutaru, C., Kelly, C., Glynn, R. W., and Perry, I. J. (2015). Type 2 diabetes research yield, 1951-2012: bibliometrics analysis and density-equalizing mapping. PLoS One 10:e0133009. doi: 10.1371/journal.pone.0133009
Geist, J. (2010). Strategies for the conservation of endangered freshwater pearl mussels (Margaritifera margaritifera L.): a synthesis of conservation genetics and ecology. Hydrobiologia 644, 69–88. doi: 10.1007/s10750-010-0190-2
Geist, J., Rottmann, O., Schröder, W., and Kühn, R. (2003). Development of microsatellite markers for the endangered freshwater pearl mussel Margaritifera margaritifera L.(Bivalvia: Unionoidea). Mol. Ecol. Notes 3, 444–446. doi: 10.1046/j.1471-8286.2003.00476.x
Gifford, S., Dunstan, R., O’Connor, W., Roberts, T., and Toia, R. (2004). Pearl aquaculture—profitable environmental remediation? Sci. Tot. Env. 319, 27–37. doi: 10.1016/S0048-9697(03)00437-6
Gueguen, Y., Gaertner-Mazouni, N., Lo, C., and Le Moullac, G. (2016). Integrated management of pearl culture in French Polynesia in the context of global change: synopsis of existing results. Estuarine Coastal Shelf Sci. 182, 229–234. doi: 10.1016/j.ecss.2016.07.010
Gum, B., Lange, M., and Geist, J. (2011). A critical reflection on the success of rearing and culturing juvenile freshwater mussels with a focus on the endangered freshwater pearl mussel (Margaritifera margaritifera L.). Aquatic Conserv.: Mar. Freshw. Ecosyst. 21, 743–751. doi: 10.1002/aqc.1222
Hao, R., Du, X., Yang, C., Deng, Y., Zheng, Z., and Wang, Q. (2019). Integrated application of transcriptomics and metabolomics provides insights into unsynchronized growth in pearl oyster Pinctada fucata martensii. Sci. Tot. Env. 666, 46–56. doi: 10.1016/j.scitotenv.2019.02.221
Haws, M., and Ellis, S. (1999). Producing Pearls Using the Black-Lip Pearl Oyster (Pinctada Margaritifera)”. CTSA Aquafarmer Information Sheet). Rohini: CTSA.
Huang, S., Ichikawa, Y., Igarashi, Y., Yoshitake, K., Kinoshita, S., Omori, F., et al. (2019). Piwi-interacting RNA (piRNA) expression patterns in pearl oyster (Pinctada fucata) somatic tissues. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-018-36726-0
Huff, S. W., Campbell, D., Gustafson, D. L., Lydeard, C., Altaba, C. R., and Giribet, G. (2004). Investigations into the phylogenetic relationships of freshwater pearl mussels (Bivalvia: Margaritiferidae) based on molecular data: implications for their taxonomy and biogeography. J. Molluscan Stud. 70, 379–388. doi: 10.1093/mollus/70.4.379
Hüning, A. K., Melzner, F., Thomsen, J., Gutowska, M. A., Krämer, L., Frickenhaus, S., et al. (2013). Impacts of seawater acidification on mantle gene expression patterns of the Baltic Sea blue mussel: implications for shell formation and energy metabolism. Mar. Biol. 160, 1845–1861. doi: 10.1007/s00227-012-1930-9
Jian-Ping, D., Jun, C., Yu-Fei, B., Bang-Xing, H., Shang-Bin, G., and Li-Li, J. (2010). Effects of pearl powder extract and its fractions on fibroblast function relevant to wound repair. Pharmaceutical Biol. 48, 122–127. doi: 10.3109/13880200903046211
Jiddawi, N. (2008). Pearl farming in Zanzibar. Secretariat of the Pacific Community (SPC). Pearl Oyster Inform. Bull. 18, 18–18.
Johnston, B., Kishore, P., Vuibeqa, G. B., Hine, D., and Southgate, P. C. (2020). Economic assessment of community-based pearl oyster spat collection and mabé pearl production in the western Pacific. Aquaculture 514:734505. doi: 10.1016/j.aquaculture.2019.734505
Johnston, W., Gordon, S. E., Wingfield, M., Hine, D., and Southgate, P. C. (2020). Economic feasibility of small-scale mabé pearl production in Tonga using the winged pearl oyster, Pteria penguin. Aquac. Rep. 17:100347. doi: 10.1016/j.aqrep.2020.100347
Johnston, W., Hine, D., and Southgate, P. C. (2019). Overview of the development and modern landscape of marine pearl culture in the south Pacific. J. Shellfish Res. 38, 499–518. doi: 10.2983/035.038.0301
Jones, D. B., Jerry, D. R., Khatkar, M. S., Raadsma, H. W., and Zenger, K. R. (2013). A high-density SNP genetic linkage map for the silver-lipped pearl oyster, Pinctada maxima: a valuable resource for gene localisation and marker-assisted selection. BMC Genomics 14:810. doi: 10.1186/1471-2164-14-810
Kishore, P., Vuibeqa, G. B., and Southgate, P. C. (2018). Developing a national spat collection program for pearl oysters in the Fiji Islands supporting pearl industry development and livelihoods. Aquac. Rep. 9, 46–52. doi: 10.1016/j.aqrep.2017.12.004
Ky, C.-L., Broustal, F., Potin, D., and Lo, C. (2019). The pearl oyster (Pinctada margaritifera) aquaculture in French Polynesia and the indirect impact of long-distance transfers and collection-culture site combinations on pearl quality traits. Aquac. Rep. 13:100182. doi: 10.1016/j.aqrep.2019.100182
Lacoste, E., Gueguen, Y., Le Moullac, G., Koua, M. S., and Gaertner-Mazouni, N. (2014). Influence of farmed pearl oysters and associated biofouling communities on nutrient regeneration in lagoons of French Polynesia. Aquac. Env. Interact. 5, 209–219. doi: 10.3354/aei00107
Lemer, S., Saulnier, D., Gueguen, Y., and Planes, S. (2015). Identification of genes associated with shell color in the black-lipped pearl oyster, Pinctada margaritifera. BMC Genomics 16:568. doi: 10.1186/s12864-015-1776-x
Li, Q., Bai, Z., Zhao, L., and Li, J. (2016). Characterization of allograft inflammatory factor-1 in Hyriopsis cumingii and its expression in response to immune challenge and pearl sac formation. Fish Shellfish Immunol. 59, 241–249. doi: 10.1016/j.fsi.2016.10.037
Li, S., Liu, C., Zhan, A., Xie, L., and Zhang, R. (2017a). Influencing mechanism of ocean acidification on byssus performance in the pearl oyster Pinctada fucata. Environ. Sci. Technol. 51, 7696–7706. doi: 10.1021/acs.est.7b02132
Li, S., Liu, Y., Huang, J., Zhan, A., Xie, L., and Zhang, R. (2017b). The receptor genes PfBMPR1B and PfBAMBI are involved in regulating shell biomineralization in the pearl oyster Pinctada fucata. Sci. Rep. 7, 1–15. doi: 10.1038/s41598-017-10011-y
Li, Y., and He, M. (2014). Genetic mapping and QTL analysis of growth-related traits in Pinctada fucata using restriction-site associated DNA sequencing. PLoS One 9:e111707. doi: 10.1371/journal.pone.0111707
Li, Y.-C., Chen, C.-R., and Young, T.-H. (2013). Pearl extract enhances the migratory ability of fibroblasts in a wound healing model. Pharmaceutical Biol. 51, 289–297. doi: 10.3109/13880209.2012.721130
Liu, C., Li, S., Kong, J., Liu, Y., Wang, T., Xie, L., et al. (2015). In-depth proteomic analysis of shell matrix proteins of Pinctada fucata. Sci. Rep. 5, 1–14. doi: 10.1038/srep17269
Liu, J., Yang, D., Liu, S., Li, S., Xu, G., Zheng, G., et al. (2015). Microarray: a global analysis of biomineralization-related gene expression profiles during larval development in the pearl oyster, Pinctada fucata. BMC Genomics 16:325. doi: 10.1186/s12864-015-1524-2
Liu, F., Li, X., and Shi, Y. (2016). Scientific breakthrough study of extenics. Procedia Comput. Sci. 91, 526–531. doi: 10.1016/j.procs.2016.07.134
Liu, H., Zhang, H., Pan, X., Xu, M., Huang, J., and He, M. (2020). A high density genetic map by whole-genome resequencing for QTL fine-mapping and dissecting candidate genes for growth or sex traits in the pearl oyster (Pinctada fucata martensii). Aquaculture 519:734839. doi: 10.1016/j.aquaculture.2019.734839
Lucas, J. S. (2008). “Feeding and metabolism,” in The Pearl Oyster, ed. P. S. J. Lucas (London: Elsevier), 103–130.
Lucas, P. S. J. (2007). “Overview of the Cultured Pearl Industry,” in Pearl Oyster Health Management: A Manual, eds M. G. Bondad-Reantaso, S. E. McGladdery, and F. C. J. Berthe (Rome: Food and Agriculture Organization of the United Nations), 7–16.
Ma, C., Zhang, C., Nie, Y., Xie, L., and Zhang, R. (2005). Extraction and purification of matrix protein from the nacre of pearl oyster Pinetada fueata. Tsinghua Sci. Technol. 10, 499–503. doi: 10.1016/S1007-0214(05)70107-3
Mao, Y., Lin, F., Fang, J., Fang, J., Li, J., and Du, M. (2019). “Bivalve production in China,” in Goods and Services of Marine Bivalves, eds A. C. Smaal, J. G. Ferreira, J. Grant, J. K. Petersen, and O. Strand (Cham: Springer), 51–72.
Matsuyama, T., Yasuike, M., Fujiwara, A., Nakamura, Y., Takano, T., Takeuchi, T., et al. (2017). A Spirochaete is suggested as the causative agent of Akoya oyster disease by metagenomic analysis. PLoS One 12:e0182280. doi: 10.1371/journal.pone.0182280
Muhammad, G., Atsumi, T., and Komaru, A. (2017). Nacre growth and thickness of Akoya pearls from Japanese and Hybrid Pinctada fucata in response to the aquaculture temperature condition in Ago Bay, Japan. Aquaculture 477, 35–42. doi: 10.1016/j.aquaculture.2017.04.032
Nagai, K. (2013). A history of the cultured pearl industry. Zool. Sci. 30, 783–793. doi: 10.2108/zsj.30.783
Nakayama, N., Hamaguchi, M., Yamaguchi, H., Masuda, K., and Fujiwara, M. (2020). Evaluation of a virus-based control method to protect cultured oysters from the harmful dinoflagellate Heterocapsa circularisquama. Aquaculture 529:735625. doi: 10.1016/j.aquaculture.2020.735625
Narin, F., Olivastro, D., and Stevens, K. A. (1994). Bibliometrics/theory, practice and problems. Evaluat. Rev. 18, 65–76. doi: 10.1177/0193841X9401800107
O’Connor, W. A., and Gifford, S. P. (2008). “Environmental impacts of pearl farming,” in The Pearl Oyster, ed. J. S. L. Paul C. Southgate (London: Elsevier), 497–525.
Preston, S., Keys, A., and Roberts, D. (2007). Culturing freshwater pearl mussel Margaritifera margaritifera: a breakthrough in the conservation of an endangered species. Aquatic Conserv.: Mar. Freshw. Ecosyst. 17, 539–549. doi: 10.1002/aqc.799
Rinia, E. J., Van Leeuwen, T. N., Van Vuren, H. G., and Van Raan, A. F. (1998). Comparative analysis of a set of bibliometric indicators and central peer review criteria: evaluation of condensed matter physics in the Netherlands. Res. Policy 27, 95–107. doi: 10.1016/S0048-7333(98)00026-2
Romanelli, J. P., Fujimoto, J. T., Ferreira, M. D., and Milanez, D. H. (2018). Assessing ecological restoration as a research topic using bibliometric indicators. Ecol. Eng. 120, 311–320. doi: 10.1016/j.ecoleng.2018.06.015
Sakatoku, A., Fujimura, T., Ito, M., Takashima, S., and Isshiki, T. (2018). Newly isolated bacterium Tenacibaculum sp. strain Pbs-1 from diseased pearl oysters is associated with black-spot shell disease. Aquaculture 493, 61–67. doi: 10.1016/j.aquaculture.2018.04.049
Saucedo, P. E., Acosta-Salmón, H., McLaurin-Moreno, D., Castillo-Domínguez, A., Melgar-Valdés, C. E., and Mazón-Suástegui, J. M. (2021). Freshwater pearl culture in Mexico: historic context, present status and future perspectives. Rev. Aquac. doi: 10.1111/raq.12527
Schmidt, C., and Vandré, R. (2010). Ten years of experience in the rearing of young freshwater pearl mussels (Margaritifera margaritifera). Aquatic Conserv. Mar. Freshw. Ecosyst. 20, 735–747. doi: 10.1002/aqc.1150
Shi, Y., Wang, S., Gu, Z., Lv, J., Zhan, X., Yu, C., et al. (2014). High-density single nucleotide polymorphisms linkage and quantitative trait locus mapping of the pearl oyster, Pinctada fucata martensii Dunker. Aquaculture 434, 376–384. doi: 10.1016/j.aquaculture.2014.08.044
Song, X., Liu, Z., Wang, L., and Song, L. (2019). Recent advances of shell matrix proteins and cellular orchestration in marine molluscan shell biomineralization. Front. Mar. Sci. 6:41. doi: 10.3389/fmars.2019.00041
Southgate, P., Rubens, J., Kipanga, M., and Msumi, G. (2006). Pearls from Africa. SPC Pearl Oyster Inform. Bull. 17, 16–17.
Take, S., Igarashi, Y., Yoshitake, K., Asakawa, S., Maeyama, K., Nagai, K., et al. (2019). Gene expression profiles at different stages for formation of pearl sac and pearl in the pearl oyster Pinctada fucata. BMC Genomics 20:240. doi: 10.1186/s12864-019-5579-3
Takeuchi, T., Kawashima, T., Koyanagi, R., Gyoja, F., Tanaka, M., Ikuta, T., et al. (2012). Draft genome of the pearl oyster Pinctada fucata: a platform for understanding bivalve biology. DNA Res. 19, 117–130. doi: 10.1093/dnares/dss005
Teaniniuraitemoana, V., Huvet, A., Levy, P., Klopp, C., Lhuillier, E., Gaertner-Mazouni, N., et al. (2014). Gonad transcriptome analysis of pearl oyster Pinctada margaritifera: identification of potential sex differentiation and sex determining genes. BMC Genomics 15:491. doi: 10.1186/1471-2164-15-491
Tisdell, C., and Poirine, B. (2000). Socio-economics of pearl culture: industry changes and comparisons focusing on Australia and French Polynesia. Secretariat of the Pacific Community. Pearl Oyster Inform. Bull. 14, 21–31.
Tisdell, C., and Poirine, B. (2008). “Economics of pearl farming,” in The Pearl Oyster, ed. J. L. Paul Southgate (London: Elsevier), 473–496.
Van Wynsberge, S., Le Gendre, R., Sangare, N., Aucan, J., Menkes, C., Liao, V., et al. (2020). Monitoring pearl farming lagoon temperature with global high resolution satellite-derived products: an evaluation using Raroia Atoll, French Polynesia. Mar. Pollut. Bull. 160:111576. doi: 10.1016/j.marpolbul.2020.111576
Velasco, M. (2011). Diagnostic and treatment of leg ulcers. Actas Dermo-Sifiliográficas (English Edition) 102, 780–790. doi: 10.1016/j.adengl.2012.01.008
Wang, W., Lei, Q., Liang, H., and He, J. (2019). Towards a better understanding of allograft-induced stress response in the pearl oyster Pinctada fucata martensii: insights from iTRAQ-based comparative proteomic analysis. Fish Shellfish Immunol. 86, 186–195. doi: 10.1016/j.fsi.2018.11.044
Wei, J., Liu, B., Fan, S., Li, H., Chen, M., Zhang, B., et al. (2017). Differentially expressed immune-related genes in hemocytes of the pearl oyster Pinctada fucata against allograft identified by transcriptome analysis. Fish Shellfish Immunol. 62, 247–256. doi: 10.1016/j.fsi.2017.01.025
Welladsen, H. M., Southgate, P. C., and Heimann, K. (2010). The effects of exposure to near-future levels of ocean acidification on shell characteristics of Pinctada fucata (Bivalvia: Pteriidae). Molluscan Res. 30:125.
Wu, X., Li, X., Li, L., and Yu, Z. (2012). A unique tRNA gene family and a novel, highly expressed ORF in the mitochondrial genome of the silver-lip pearl oyster, Pinctada maxima (Bivalvia: Pteriidae). Gene 510, 22–31. doi: 10.1016/j.gene.2012.08.037
Wu, Y., He, J., Yao, G., Liang, H., and Huang, X. (2020). Molecular cloning, characterization, and expression of two TNFRs from the pearl oyster Pinctada fucata martensii. Fish Shellfish Immunol. 98, 147–159. doi: 10.1016/j.fsi.2020.01.010
Xu, H., Huang, K., Gao, Q., Gao, Z., and Han, X. (2001). A study on the prevention and treatment of myopia with nacre on chicks. Pharmacol. Res. 44, 1–6. doi: 10.1006/phrs.2000.0780
Xu, Q., Wang, G., Yuan, H., Chai, Y., and Xiao, Z. (2010). cDNA sequence and expression analysis of an antimicrobial peptide, theromacin, in the triangle-shell pearl mussel Hyriopsis cumingii. Compar. Biochem. Physiol. Part B: Biochem. Mol. Biol. 157, 119–126. doi: 10.1016/j.cbpb.2010.05.010
Yan, S., Fan, R., Chen, Y., Wang, D., Huang, C., Xie, J., et al. (2017). Research on web services-based extenics aided innovation system. Procedia Comput. Sci. 107, 103–110. doi: 10.1016/j.procs.2017.03.064
Yang, C., Du, X., Hao, R., Wang, Q., Deng, Y., and Sun, R. (2019). Effect of vitamin D3 on immunity and antioxidant capacity of pearl oyster Pinctada fucata martensii after transplantation: insights from LC–MS-based metabolomics analysis. Fish Shellfish Immunol. 94, 271–279. doi: 10.1016/j.fsi.2019.09.017
Yang, C., Hao, R., Du, X., Wang, Q., Deng, Y., Sun, R., et al. (2018). GC–TOF/MS-based metabolomics studies on the effect of protein sources in formulated diet for pearl oyster Pinctada fucata martensii. Aquaculture 486, 139–147. doi: 10.1016/j.aquaculture.2017.12.020
Yang, J., Luo, S., Li, J., Zheng, Z., Du, X., and Deng, Y. (2018). Transcriptome analysis of growth heterosis in pearl oyster Pinctada fucata martensii. FEBS Open Bio 8, 1794–1803. doi: 10.1002/2211-5463.12502
Ye, S., Gao, S., Guo, B., Lin, P., Feng, Y., and Hong, W. (2019). “Research into effects of pearl oyster cultivation in freshwater with extensible mind mapping,” in Proceeding of the International Conference on Mechatronics and Intelligent Robotics, (Berlin: Springer), 223–235.
Yulianti, I., Addawiyah, A., and Setiawan, R. (2018). “Optimization of exposure factors for X-ray radiography non-destructive testing of pearl oyster,” in Proceeding of the Journal of Physics: Conference Series, (Bristol: IOP Publishing), 012004.
Zenger, K. R., Khatkar, M. S., Jones, D. B., Khalilisamani, N., Jerry, D. R., and Raadsma, H. W. (2019). Genomic selection in aquaculture: application, limitations and opportunities with special reference to marine shrimp and pearl oysters. Front. Genet. 9:693. doi: 10.3389/fgene.2018.00693
Zhang, H., Ou, Z., Xu, M., Huang, X., Liu, W., Shi, Y., et al. (2018). Molecular cloning and characterization of a putative mitogen-activated protein kinase (erk1/2) gene: involvement in mantle immunity of Pinctada fucata. Fish Shellfish Immunol. 80, 63–70. doi: 10.1016/j.fsi.2018.05.047
Zhang, H., Shi, Y., and He, M. (2017). Molecular identification of an insulin growth factor binding protein (IGFBP) and its potential role in an insulin-like peptide system of the pearl oyster, Pinctada fucata. Comparat. Biochem. Physiol. Part B: Biochem. Mol. Biol. 214, 27–35. doi: 10.1016/j.cbpb.2017.09.003
Zhang, H., Xu, H., Liu, H., Pan, X., Xu, M., Zhang, G., et al. (2020). PacBio single molecule long-read sequencing provides insight into the complexity and diversity of the Pinctada fucata martensii transcriptome. BMC Genomics 21:481. doi: 10.1186/s12864-020-06894-3
Zhao, R., Takeuchi, T., Luo, Y.-J., Ishikawa, A., Kobayashi, T., Koyanagi, R., et al. (2018). Dual gene repertoires for larval and adult shells reveal molecules essential for molluscan shell formation. Mol. Biol. Evolut. 35, 2751–2761. doi: 10.1093/molbev/msy172
Zheng, Z., Huang, R., Tian, R., Jiao, Y., and Du, X. (2016a). Pm-miR-133 hosting in one potential lncRNA regulates RhoA expression in pearl oyster Pinctada martensii. Gene 591, 484–489. doi: 10.1016/j.gene.2016.06.051
Zheng, Z., Jiao, Y., Du, X., Tian, Q., Wang, Q., Huang, R., et al. (2016b). Computational prediction of candidate miRNAs and their potential functions in biomineralization in pearl oyster Pinctada martensii. Saudi J. Biol. Sci. 23, 372–378. doi: 10.1016/j.sjbs.2015.04.001
Keywords: pearl mussel, pearl oyster, aquaculture, bibliometric analysis, VOSviewer
Citation: Cheng Q, Hu W and Bai Z (2021) Research Trends of Development on Pearl Bivalve Mollusks Based on a Bibliometric Network Analysis in the Past 25 Years. Front. Mar. Sci. 8:657263. doi: 10.3389/fmars.2021.657263
Received: 22 January 2021; Accepted: 22 March 2021;
Published: 29 April 2021.
Edited by:
Libin Zhang, Institute of Oceanology (CAS), ChinaReviewed by:
Qingheng Wang, Guangdong Ocean University, ChinaLili Xing, Institute of Oceanology (CAS), China
Copyright © 2021 Cheng, Hu and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyi Bai, enliYWlAc2hvdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Qichao Cheng1†
Qichao Cheng1† Zhiyi Bai
Zhiyi Bai