- 1Department of Biology, Appalachian State University, Boone, NC, United States
- 2CONACyT-Centro de Investigaciones Biológicas del Noroeste (CIBNOR), Laboratorio de Necton y Ecología de Arrecifes, La Paz, Mexico
- 3Department of Biological Sciences, Louisiana State University, Baton Rouge, LA, United States
Most tropical reef corals live at temperatures near 27°C and pH values near 8. Conditions outside of these can stress corals and lead to bleaching, disease, and death. However, some corals can survive in marginal or extreme habitats outside of these ranges. To date there is a paucity of knowledge about the role that associated microbes may play in the acclimation of corals to such extreme habitats. Here, we explore differences in the compositional and functional profile of the microbiomes of the scleractinian coral Porites panamensis living both on and off potentially stressful shallow-water hydrothermal vents. The environment near the vents is extreme, with temperatures exceeding 80°C and pH values below 6. Coral microbiomes under stress often exhibit increased diversity, increased abundance of pathogenic bacteria, and functional profiles that shift toward pathways associated with pathogenic taxa. Samples from along a transect that crossed an arc of hydrothermal vents were sequenced for the 16S rRNA gene (V4 region). On-vent coral microbiomes were distinct from those of off-vent corals, but did not have increased alpha or beta diversity. On-vent samples had a higher relative abundance of the beneficial endosymbiont, Endozoicomonas. On- and off-vent microbiomes did not differ in overall abundance of the endolithic green alga Ostreobium, however, a single ASV, close to O. quekettii was more abundant in on-vent corals. Ostreobium can provide many of the same benefits to corals as zooxanthellae and their symbiosis is better maintained under thermal stress. Surprisingly, on-vent coral microbiomes had fewer microbial taxa that are known to be pathogenic or associated with stress than did off-vent corals. The predicted functional profiles of on-vent microbiomes revealed enrichment of pathways related to aerobic respiration, fermentation and amino acid biosynthesis, but not of virulence-related pathways. Our results suggest that P. panamensis microbiomes have acclimated to the extreme environment of the hydrothermal vent habitat rather than showing signs of stress. These results exemplify the need to focus efforts on examining the mechanisms of resilience, including symbioses with microbiota, in corals living in extreme environments in an effort to design better management strategies for reef-building corals under thermal and pH stress.
Introduction
Reef-building corals are declining globally in response to ocean acidification and thermal stresses associated with rapidly changing seas (Gardner et al., 2003; Carpenter et al., 2008; Toth et al., 2019). Temperature, pH, nutrient levels and light availability all vary over the broad ranges of many coral species, suggesting that historically these species have been able to adapt to differences in their abiotic environments (Matz et al., 2018). What is largely unclear though, is whether reef corals can adapt rapidly enough to keep pace with human-induced climate change, which will ultimately determine their long-term viability (Torda et al., 2017).
While most coral species may struggle to adapt to climate change, some live currently in marginal habitats (high temperatures, low pH, high turbidity) at the periphery of their environmental tolerances. A diverse community of corals has been uncovered thriving in waters with a naturally low pH (∼7.85–7.95) approaching that predicted for ocean waters in 2100 (Shamberger et al., 2014). Several acroporiids have acclimated to warm tidepools (max. temp. ∼35°C) and show potential for long-term adaptation to even more extreme heat stress (Palumbi et al., 2014; Matz et al., 2018). Corals from more challenging and variable habitats generally have higher phenotypic plasticity and can withstand more stress than others (Oliver and Palumbi, 2011; Barshis et al., 2013; Putnam et al., 2016). However, it is not always clear how much of these adaptive responses are owed to the coral host and how much falls to the many coral-associated symbionts (Bourne et al., 2016; van Oppen et al., 2018).
Corals live in a close association with a diverse community of bacteria, archaea, fungi and algae that together with their host constitute the coral holobiont. These microbial symbionts are thought to benefit their host by cycling nutrients (Rohwer et al., 2002) and providing energy (Brown, 1997; Grottoli et al., 2006) and protection from pathogens (Ritchie, 2006). These roles have been best studied for symbiotic microalga (zooxanthellae). Corals that host heat-tolerant strains of zooxanthellae are better able to survive thermal stress (Rowan, 2004; Littman et al., 2010; Oliver and Palumbi, 2011). Some reef-building corals can shuffle less heat-tolerate zooxanthellae types for those that are more heat-tolerant, increasing their ability to survive high temperatures (Baker, 2003; Berkelmans and Van Oppen, 2006).
The role of the prokaryotic members (the microbiome) of the holobiont are not as well known, but corals exposed to experimental stressors experience shifts in the composition of their microbiomes (Thurber et al., 2009; Webster et al., 2016; Grottoli et al., 2018). Coral microbiomes living in thermally variable habitats show signs of microbiome acclimation to high temperatures, with shifts in the microbiome occurring in as little as 20 h after exposure to thermal stress (Ziegler et al., 2017). However, there is still a paucity of knowledge about the composition and functional role of the microbiomes of corals that are surviving in marginal or extreme habitats.
Shallow hydrothermal vents offer an opportunity to examine the role of microbial symbionts in host acclimation to extreme and natural stressful conditions. Environmental conditions around these vents resemble those predicted for ocean waters due to climate change, including high temperatures, low pH (due to high CO2 and CH4), and high concentrations of toxic compounds and nutrients (Bianchi et al., 2012; Dahms et al., 2018; Oprandi et al., 2019). The microbial communities associated with sediments from shallow hydrothermal vents are generally dominated by chemoautotrophic bacteria, especially archaea, and are enriched for sulfur-oxidizing bacteria (Kalanetra et al., 2004; Maugeri et al., 2009; Wang et al., 2015). Composition patterns for the microbial symbionts near- and off-vents are not so clear, although variation in sponge microbiomes appears to be tied to the chemistry of seeps (Coelho et al., 2018). Results for coral microbiomes have been mixed. Meron et al. (2012) found no change in coral microbiome composition along a seep-caused pH gradient, nor any evidence of an increase in pathogenic bacteria. In contrast, Morrow et al. (2015) found distinct microbiomes from corals near vents largely attributed to a decrease in the endosymbiont Endozoicomonas coupled with an increase in Chromohalobacter salexigens, a symbiont capable of hydrogen sulfide production and anaerobic growth in the presence of nitrate. Members of Ostreobium, a endolithic microalga, are heat-tolerant and their presence in the coral skeletal microbiome may ease acclimation of corals to thermal stress by replacing some functions usually provided by the less heat-tolerant zooxanthellae (Fine and Loya, 2002; Del Campo et al., 2017). Overall, the available data for microbiomes of vent-associated corals remains limited despite the insights it could provide about the resilience of corals under climate change conditions.
The hydrothermal vents of Bahía Concepción, Baja California Sur, Mexico, offer an accessible habitat in which to examine changes to microbial communities associated with corals in a natural extreme habitat. These submarine hydrothermal vents occur in shallow waters, some <10 m deep. The water discharged from the vents is acidic (pH = 5.95–6.02) and surrounding sediments rise to 89°C (Prol-Ledesma et al., 2004). Gas released by the vents is >40% CO2 (Prol-Ledesma et al., 2004; Forrest et al., 2005; Dávila-Ramos et al., 2015), over 40 times that of most sea water (Wingenter et al., 2007). Elevated pCO2 hinders calcification and can ultimately lead to reef systems devoid of scleractinians (Hall-Spencer et al., 2008; Milazzo et al., 2015; Camp et al., 2020).
Despite the extreme conditions surrounding the thermal vents in Bahía Concepción, colonies of the reef-building coral Porites panamensis live within 1 m of the vents, where they experience the combined effects of thermal and pH stress (Oporto-Guerrero et al., 2018). P. panamensis produces brooding larvae with limited dispersal potential (Glynn et al., 1994), resulting in modest subdivision of populations within the Gulf of California (Paz-García et al., 2012; Saavedra-Sotelo et al., 2013). The microbiomes of a congener, P. lobata, showed mixed responses to bleaching and temperature stress depending on location (Hadaidi et al., 2017; Rachmawati, 2018; McDevitt-Irwin et al., 2019). A better understanding of the composition and function of the microbial communities in P. panamensis colonies living near the vents could yield insight into whether, and how, prokaryotes help their coral hosts survive in extreme habitats.
Here, we sought to characterize and compare the microbiomes of P. panamensis living near hydrothermal vents to those living in surrounding “normal” conditions. First, we compared the diversity and taxonomic composition of on- and off- vent P. panamensis microbiomes. Next, to gain insight into the possible functional capabilities of the on- and off-vent microbiomes, we completed a metagenomic functional prediction assessment. Lastly, we examined the diversity and abundance of variants of the beneficial endolithic algae, Ostreobium, in both on- and off-vent corals.
Materials and Methods
Sample Collection
Samples were collected from the shallow rocky reefs of La Ventila, Bahía Concepción, Baja California Sur, Mexico (26°41’14.00”N, 111°51’35.30”W) in February 2018, when ambient water temperatures are near their seasonal low. A 50-meter transect that crossed an arc of shallow hydrothermal vents was deployed at a depth of ∼4–5 m. The sediment at each sampled vent ranged from 27 to 66°C, while the average temperature of surrounding sediments was 20°C. Sampled coral colonies were located 1 m or less from the transect line. For all coral, water and sediment samples collected, the substrate temperature was taken at the corresponding location along the transect (see Figure 1 and Supplementary Table 1). Because only a limited number of coral colonies occurred within a meter of the transect, we divided our samples into two groups that, based on substrate temperature, should represent the extremes in vent water influence: samples collected where measured substrate temperatures on the transect were >25°C (“on-vent”) and those collected where substrate temperatures were ≤25°C (“off-vent”). The 25°C line is of no absolute importance – all colonies experience temperatures >30°C in the summer – only as an indicator of relative influence of vent water, which is not only warmer, but more acidic and CO2-rich than off-vent waters.
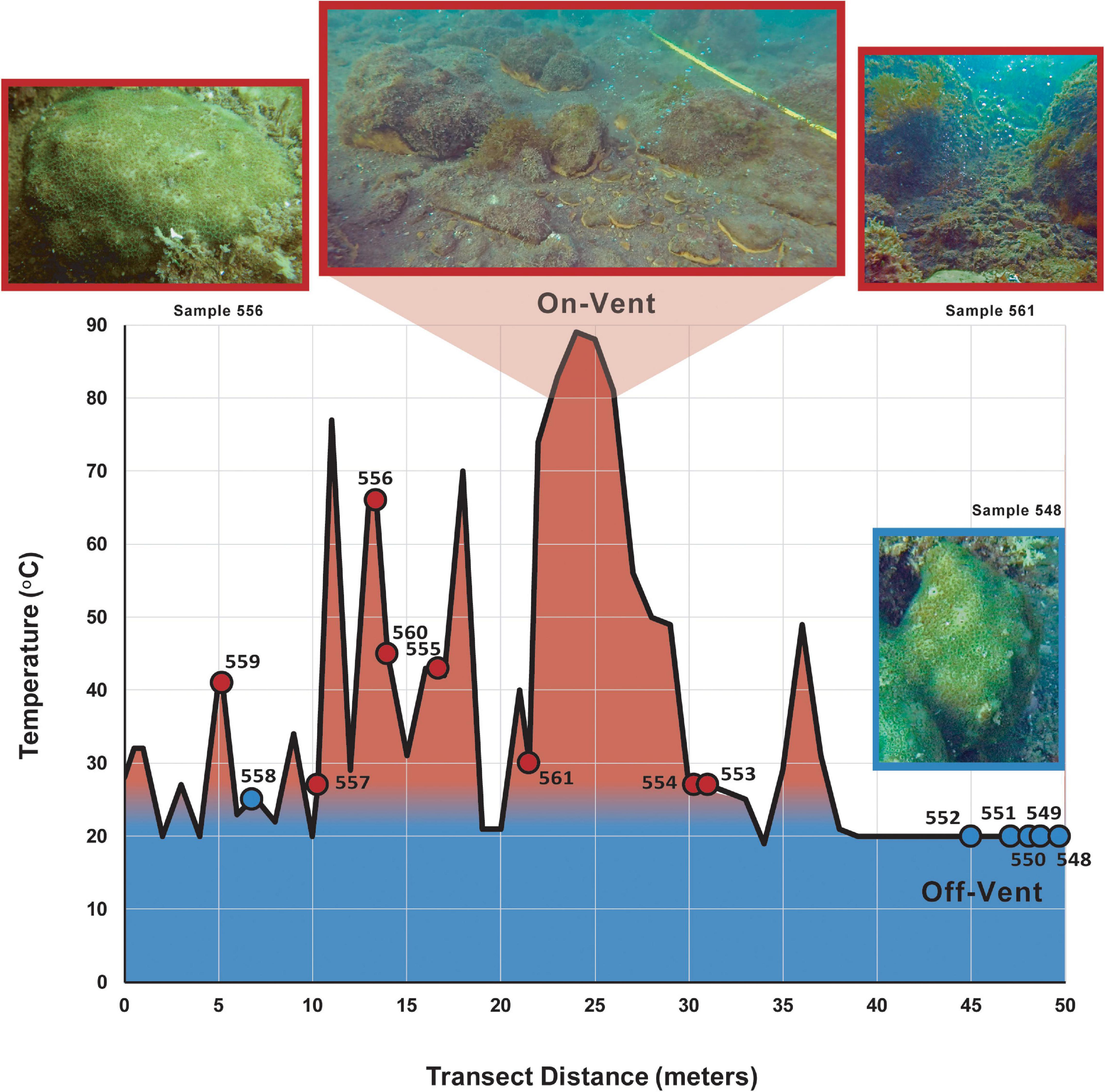
Figure 1. Temperature (red = warmer, blue = cooler) of the reef substrate along the 50 m collection transect. Circles indicate the sediment temperature nearest a sampled on-vent (red) or off-vent (blue) Porites panamensis colony (see Supplementary Table 1).
On- and off-vent coral samples (n = 8 and n = 6, respectively) were collected along the transect via SCUBA (Figure 1 and Supplementary Table 1). Samples were taken from Porites panamensis colonies (∼15–20 cm diameter) using a hammer and chisel and placed into a labeled bag. Upon return to the surface, each sample was placed in a sterile tube filled with 95% EtOH and stored at −20°C. Sediment samples were collected using a small shovel and placed in a labeled bag at depth. Upon return to the surface all excess water was drained off, the sample was put in a sterile Whirl-PaK© bag and stored on ice. Water samples were collected in a bleached-rinsed, 100% EtOH sterilized 1-L Nalgene© bottle at depth. At the surface, the water was filtered through a 2 μm glass microfiber prefilter to remove large particles, and then through a 0.22 μm Sterivex filter to collect microbes. Both filters were placed in sterile Whirl-Pak© bags and stored on ice. All sediment and water samples were placed in −80°C for long-term storage.
DNA Extraction
Microbial DNA was extracted from all coral and abiotic samples using the Qiagen DNAEasy PowerSoil kit. Sediment, water, coral and blank samples were extracted following manufacturer protocols. Prior to extraction, coral samples were drained of excess ethanol and rinsed 3× with sterile H20. Each rinsed coral sample was ground up using a sterilized polypropylene pellet pestle. The resulting coral sample was extracted following the protocol of Sunagawa et al. (2010) with two modifications: the initial heated lysis step was extended from 60 min to ∼2–4 h, and an additional centrifugation step was added post-lysis to concentrate coral skeletal fragments for removal. DNA concentrations were quantified using a Qubit2.0 Fluorometer (Life Technologies, Carlsbad, CA, United States) and stored at −20°C.
Amplicon Library Preparation and Sequencing
DNA samples were sent to the Environmental Sample Preparation and Sequencing Facility (ESPSF) at Argonne National Laboratory for amplicon library preparation and sequencing. Briefly, amplicon libraries were generated using a barcoded primer set adapted for the Illumina HiSeq2000 that targeted the 16S rRNA V4 region and modified to reduce sequencing bias of archaeal taxa and SAR11 bacteria (515F-806R; Caporaso et al., 2012; Apprill et al., 2015; Parada et al., 2016). The primers can also amplify coral DNA decreasing bacterial amplicon counts, so to mitigate this issue we utilized the peptide nucleic acid (PNA) clamps and PCR protocols developed and tested in Reigel et al. (2020).
After PCR, the resulting amplicons were quantified, and products were pooled to ensure amplicons were represented in equimolar amounts. The pool was cleaned using AMPure XP Beads (Beckman Coulter) and quantified using a Qubit. The pool was then diluted to 2 nM, denatured and diluted again to 6.75 pM with a 10% PhiX spike for sequencing. Sequence data was generated using a 2 × 251 Illumina HiSeq2000. FASTQ files for P. panamensis are accessioned at the NCBI Sequence Read Archive under BioProject # PRJNA761929.
Sequence Processing and Inference of Amplicon Sequence Variants (ASVs)
Demultiplexed FASTQ files for all coral and abiotic samples were imported to QIIME2 for processing using a modified version of the “Atacama soil microbiome tutorial” bioinformatics pipeline at qiime2.org (Bolyen et al., 2019). Quality control (QC) and ASV clustering were both completed using the QIIME2 wrapper of the “DADA2” package (Callahan et al., 2016, 2017; Glassman and Martiny, 2018; Joos et al., 2020) “denoise-paired” method, which removes low quality (expected error rate >2.0) and chimeric reads (method: “consensus”) and truncates reads. The number of base pairs to truncate from the 3’ end of each read was determined by assessing the interactive quality plot (output by QIIME2) for the point at which read quality consistently drops below 20 (as suggested by QIIME2 developers). Forward reads were truncated to 240 bp and reverse reads to 215 bp. Reads were joined and the resulting sequences aligned by DADA2 to infer unique ASVs. QIIME2 was used to assign taxonomy to each ASV using the Silva rRNA v.132 reference database (Quast et al., 2012). ASVs that did not have at least a phylum-level classification and those identified as mitochondrial were removed from the ASV dataset.
Despite our stringent filtering, ASVs belonging to a genus of green algae, Ostreobium, were not removed. This was a result of an incorrect taxonomic classification of Ostreobium as a cyanobacterium in the Silva database. The identity of all 47 ASVs in our dataset that were designated as Ostreobium by the Silva database were subsequently confirmed by checking their associated sequence reads against the NCBI nucleotide database. All 47 ASVs had matches (>96% identity) to at least one of three NCBI database entries: Ostreobium sp. HV05042 (NCBI ID: KY509314.1), uncultured Ostreobium spp. (NCBI ID: MG011659.1) or Ostreobium quekettii (LT593849.1).
While designed to target bacterial and archaeal DNA, 16S “microbial” primer sets can inadvertently amplify homologs from the mitochondrial and chloroplast DNA of eukaryotes (Hanshew et al., 2013; Reigel et al., 2020). Critically, Marcelino and Verbruggen (2016) found comparable relative abundances of Ostreobium in coral skeletal samples amplified with 16S rRNA primers (one of them the same set as used here) and the tufA gene, a marker commonly used for green algae, suggesting that 16S markers are reliable for amplifying and obtaining accurate relative abundance data for Ostreobium spp. Their 16S rRNA primers did not capture as much Ostreobium diversity as the tufA primers, so our data may underestimate the Ostreobium diversity in P. panamensis microbiomes. However, because there are multiple chloroplasts per Ostreobium cell, the Ostreobium abundance in our dataset are likely inflated compared to that of bacterial and archaeal taxa (hereafter referred to as microbial taxa). Therefore, we chose to perform all diversity analyses for Ostreobium reads independently of those for bacteria and archaea.
Prior to beta diversity analyses for both microbial taxa and Ostreobium, all samples were rarefied in QIIME2 by removing samples below the desired read count and then resampling the remaining reads without replacement. The final ASV non-rarefied and rarefied datasets and accompanying taxonomic information were exported from QIIME2 for analysis of community composition.
Community Analyses
The microbial and Ostreobium ASV abundance tables and taxonomic information were manually formatted for community diversity analyses in the R package “Phyloseq” (McMurdie and Holmes, 2013). Alpha diversity (Chao1, Simpson’s Diversity Index, Shannon’s Index) was calculated using the Phyloseq command estimaterichness. Beta diversity of the microbiomes were examined with Bray-Curtis dissimilarity and ordinated using NMDS. NMDS plots were generated using “ggplot2” (Wickham, 2016). Differences (p < 0.001) between microbiomes of various sampling groups (0.25 < R < 1 suggests dissimilarity between groups) were calculated using the anosim function from the R package “vegan” (Oksanen et al., 2013). Variance in microbial beta diversity within site types was assessed using a homogeneity of dispersion test (permdist) with the QIIME2 command diversity beta-group-significance with 1000 permutations (Anderson, 2001). The Phyloseq function plot_bar was used to plot relative abundance bars for microbial taxa within sample groups. The functions amp_venn, amp_box, and amp_rankabundance from the R package “ampvis2” (Andersen et al., 2018) were used to identify the core microbial ASVs, to calculate and plot the read counts of the most abundant ASVs, and to plot rank abundance curves, respectively. The Phyloseq-formatted ASV and taxonomy was imported into the R package “DESeq2” (Love et al., 2014) and the protocol documented on the Phyloseq website1 was used to identify differentially abundant ASVs between on- and off-vent coral samples (Wald Test; FDR adjusted p-value = 0.05).
Functional Profile Analysis
The QIIME2 wrapper of PICRUSt2 (nsti cutoff = 2 [default], hidden-state prediction (hsp) method) was used to obtain metagenomic functional predictions for the rarefied P. panamensis microbial communities following the protocol recommended by the developers (Langille et al., 20132). PICRUSt2 predicts the functional composition of a microbiome by comparing marker gene data to a reference genome database. The program takes representative 16S rRNA sequences for all ASVs and uses an ancestral-state recognition algorithm to predict the presence of gene families. The resulting gene families are combined to get an estimate of the metagenomic functional capabilities within the data. The QIIME2 wrapper of PICRUSt2 provides an abundance table of KEGG (Kyoto Encyclopedia of Genes and Genomes) orthologs present in each sample. The orthologs are then collapsed into functional categories at two hierarchical levels based on their class-level ontology in the MetaCyc Metabolic Pathway Database (Caspi et al., 2018). The resulting table was imported to R and differentially abundant pathways between on- and off-vent corals were uncovered using “DESeq2” (Wald Test; FDR adjusted p-value = 0.1). The amp_box command from the “ampvis2” package was used to identify and plot the most abundant pathways.
Results
Sequencing Results
Sequencing returned a total of 22,516,603 reads. After standard quality control (QC) measures, 15,161,615 reads remained across all samples. Post-QC, P. panamensis samples retained 3,644,484 microbial reads that clustered into 11,422 unique ASVs and 1,167,759 Ostreobium reads (Supplementary Table 1) in 47 ASVs. The abiotic samples (sediment and water) retained 10,217,235 reads after QC that were clustered into 20,978 unique ASVs (Supplementary Table 1). The two blank samples retained ∼6,600 reads following quality control measures (Supplementary Table 1). The blank sample reads clustered into 27 ASVs (all <924 reads each) and no ASVs with >56 reads were found in the 100 most abundant ASVs of coral, water and sediment. Blank samples were removed from further microbial community analyses.
Microbial Communities of Biotic Versus Abiotic Samples
Post-QC sediment samples retained 2.5× more microbial reads, and water samples 4.3×, than P. panamensis samples (Supplementary Table 1). Due to the disparity in microbial sequence read numbers between P. panamensis and abiotic samples the data were rarefied. While this is atypical for examining alpha diversity, the large difference in reads can impact the results of diversity measures (Weiss et al., 2017). Samples were rarefied to 116,035 reads, which maintained the diversity for coral samples (Supplementary Figure 1) but reduced the diversity for the abiotic samples slightly (Supplementary Figure 2). One off-vent P. panamensis sample (P_551) and one off-vent water sample (WCTRL3) did not meet the minimum read count and were removed from further analyses.
The three types of samples (coral, sediment and water) had distinct microbial assemblages (ANOSIM R: 0.937, p < 0.001; Figure 2A). Microbial composition varied more within sediment and coral samples than within water samples, which clustered tightly (Figure 2A). Sediment microbial microbiomes were more diverse than those of P. panamensis (Chao1; Kruskal-Wallis: chi-squared1 = 4.808, p = 0.029; Figure 2B). Microbial communities in water samples showed no difference in alpha diversity (Chao1) from either sediment or coral communities (Kruskal-Wallis: p = 0.082 and p = 0.924, respectively; Figure 2B). Off-vent sediments had higher overall microbial richness than on-vent sediments (average Chao1 = 7,298 and 1,654, respectively; note bimodal distribution of sediment points in Figure 2B), while the microbial richness between off- and on-vent water was similar (average Chao1 = 836 and 904, respectively). There was no difference in evenness between the three samples types (Inverse Simpson’s Index: p > 0.05, Shannon’s Diversity: p > 0.05; Supplementary Figure 3).
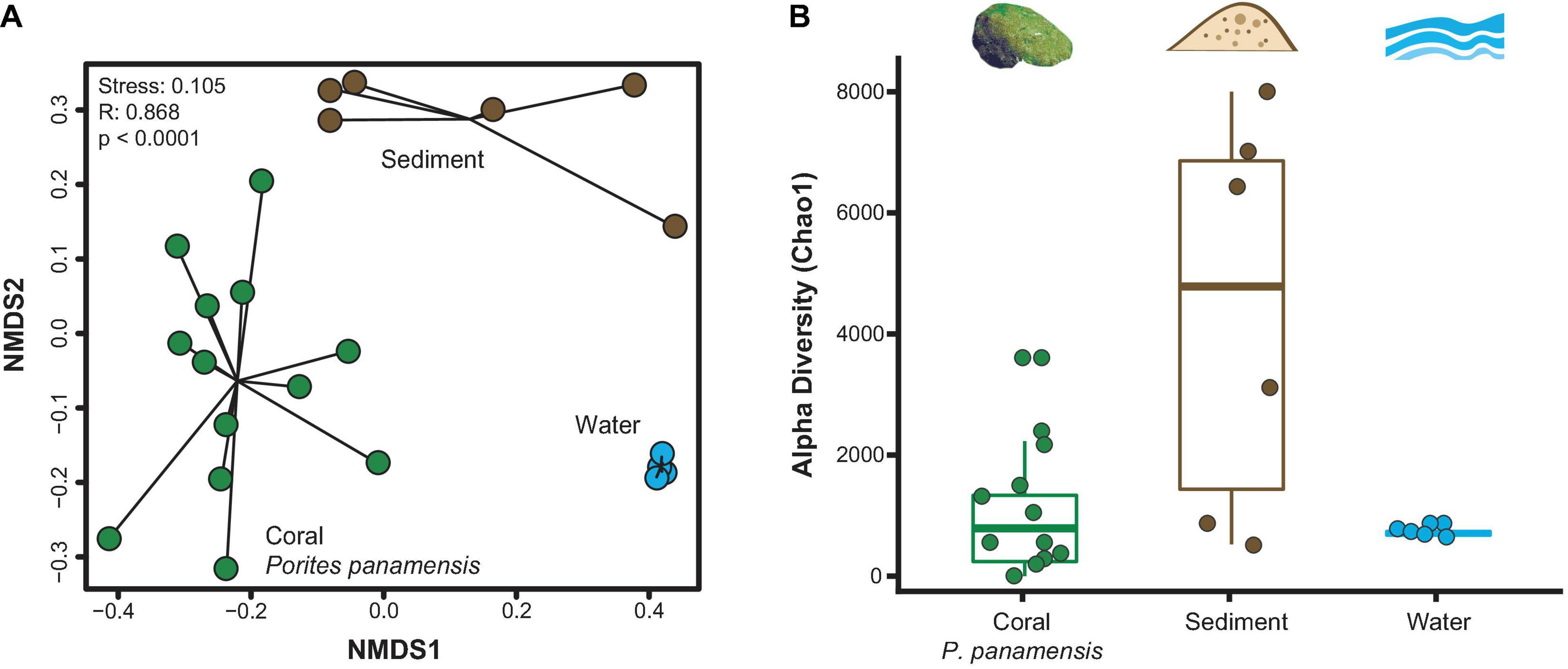
Figure 2. Microbial community diversity between rarefied Porites panamensis (green), sediment (brown) and water (blue) samples. (A) NMDS of beta diversity (Bray-Curtis) with black lines connecting samples of the same type to their centroid. (B) Alpha diversity (Chao1). The lower and upper hinges represent the first and third quartiles. The lower and upper whiskers extend to either the largest or smallest value that is no more than 1.5* the interquartile range.
Porites panamensis Samples: On-Vent vs. Off-Vent Microbial Community Analysis
Neither microbial richness (Chao1) nor evenness (Inverse Simpson’s, and Shannon’s Diversity) differed between on- and off-vent P. panamensis colonies (Kruskal-Wallis: all p > 0.05; Supplementary Figure 4). Beta diversity within off- and on-vent coral microbiomes was similar (permdist: F-value = 0.895, p = 0.683). Initial examinations of the rarefied coral samples showed strong distinctions between off- and on-vent samples (Bray-Curtis: ANOSIM R: 0.228, p < 0.0001; Supplementary Figure 5). A single on-vent sample (P_561) appeared to be an outlier (Supplementary Figure 5) but removing it did not alter overall clustering of site types (ANOSIM R: 0.270, p < 0.0001; Supplementary Figure 5).
The coral microbiomes contained many low abundance ASVs (Supplementary Figure 6), most of which were found in just one colony. When the data were pared to consider just 355 high abundance ASVs (>500 reads), differences between on- and off-vent corals were more defined (ANOSIM R: 0.282, p < 0.0001; Figure 3). Both overall compositional differences (compare Figure 3 and Supplementary Figure 5) and major taxonomic patterns (Supplementary Table 2; compare Figure 4 and Supplementary Figure 7) between site types were retained when the dataset was reduced to only high abundance ASVs, therefore, all further compositional analyses were completed using only high abundance ASVs.
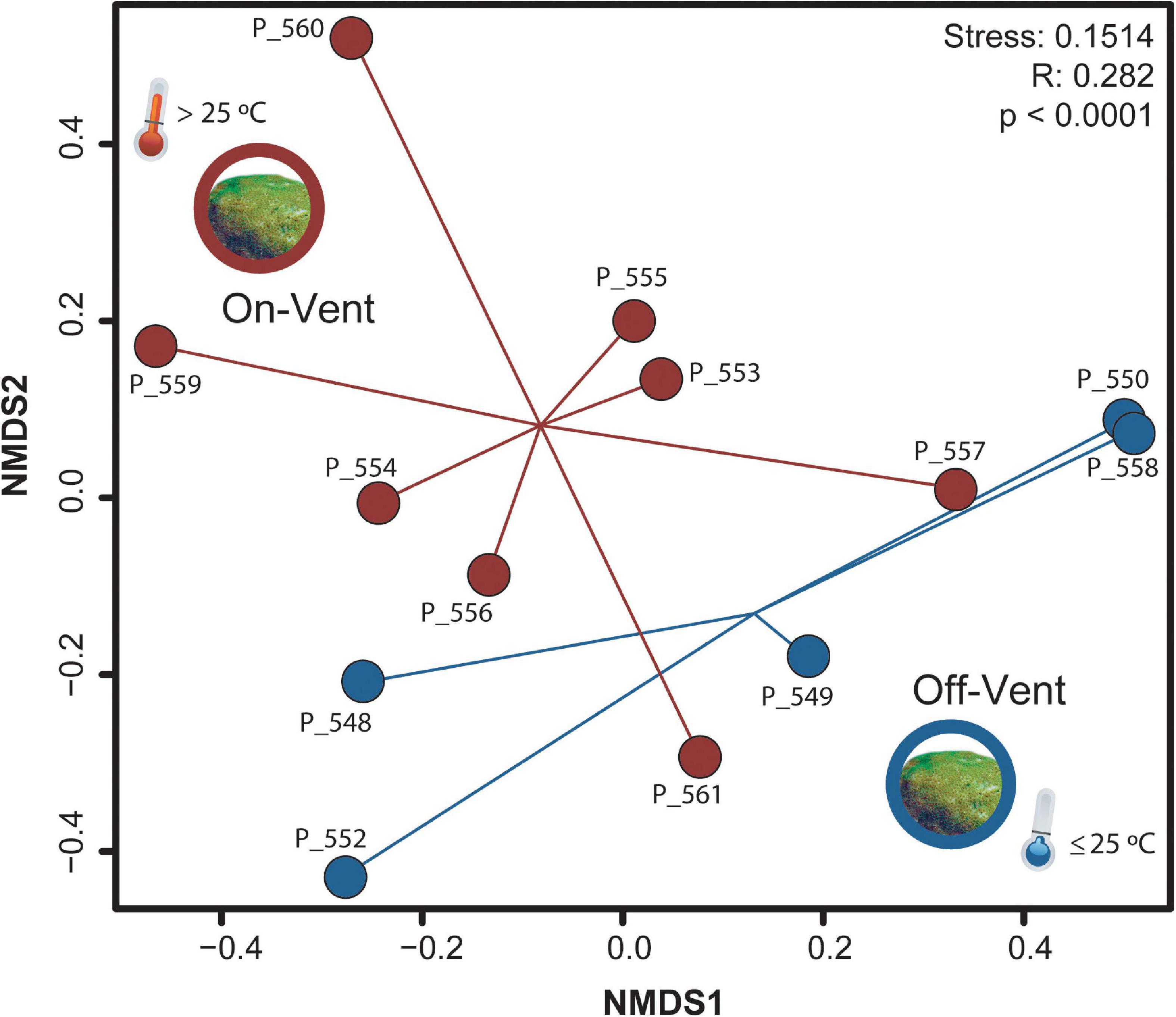
Figure 3. NMDS plot of microbial community beta diversity (Bray-Curtis) of high abundance ASVs (>500 reads) for off- (blue) and on-vent (red) Porites panamensis samples.
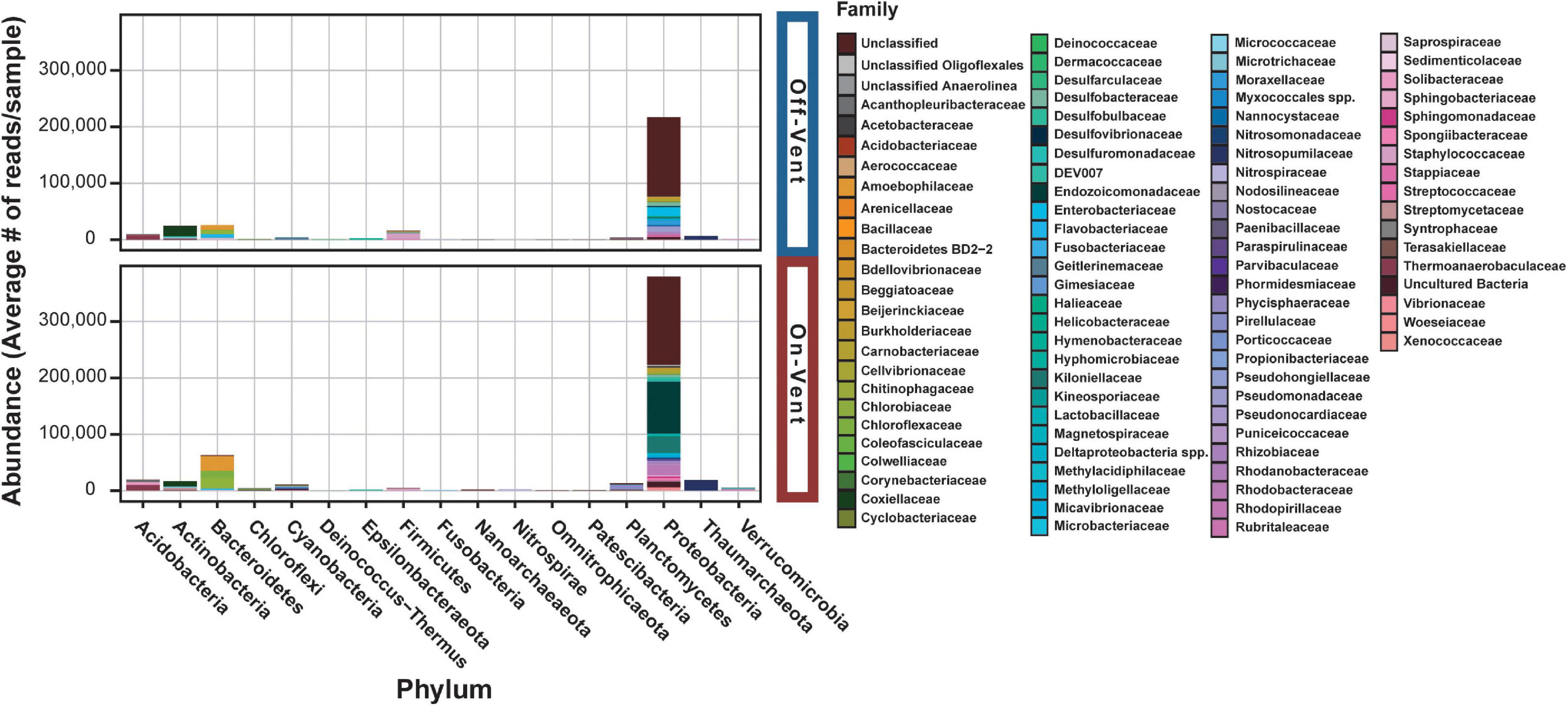
Figure 4. Relative abundance plots of high abundance ASVs (>500 reads) for off- (top) and on-vent (bottom) Porites panamensis samples. ASVs are clustered by phylum designation and colored by family.
Seventeen phyla were represented across the off- and on-vent samples (Figure 4), but two (Proteobacteria and Bacteroidetes) were by far the most abundant (∼80% of all reads in high abundance ASVs). On-vent samples were composed almost entirely of two phyla: Proteobacteria (69.6%) and Bacteroidetes (11.5%). Off-vent microbiomes were similar in composition with Proteobacteria, Bacteroidetes and Actinobacteria being the most abundant taxa (69.8, 8.4, and 8%, respectively). For both off- and on-vent, 219 of the 355 highest abundance ASVs were identified as Proteobacteria but composition within that phylum differed between the two habitat types at the family level (Figure 4 and Supplementary Figure 8).
The microbial families with the highest relative abundance off-vent were Enterobacteriaceae (5.0%; Proteobacteria), Corynebacteriaceae (4.2%; Actinobacteria; Supplementary Table 2). Endozoicomonadaceae was the most abundant family in on-vent corals (11.9%; Proteobacteria), followed by Kiloniellaceae (6.2%; Proteobacteria) and Amoebophilaceae (4.9%; Proteobacteria; Supplementary Table 2). At the family level, 26 of 129 taxonomic families were differentially abundant between off- and on-vent corals (p < 0.05; data not shown), but only two increased in abundance in on-vent samples: Endozoicomonadaceae and an unknown Gammaproteobacteria associated with UBA10353 marine group. Notable decreases in on-vent samples included the members of the sulfate-reducing family Desulfovibrionaceae and an unknown microbe identified only as a member of the phylum Bacteroidetes subgroup BD2-2.
Of the 15 most abundant ASVs in coral microbiomes, two differed in abundance between on- and off-vent corals (Figure 5 and Supplementary Figure 9): a Candidatus Amoebophilus spp. (the fifth most abundant ASV overall) was more common in on-vent samples and Candidatus Blochmannia (12th most abundant) was more common in off-vent colonies (p < 0.05 for both; Figure 5 and Supplementary Figure 9). At finer taxonomic resolution, 83 of the 355 high abundance ASVs (>500 reads) were differentially abundant between the off- and on-vent coral microbiomes (Supplementary Figure 10).
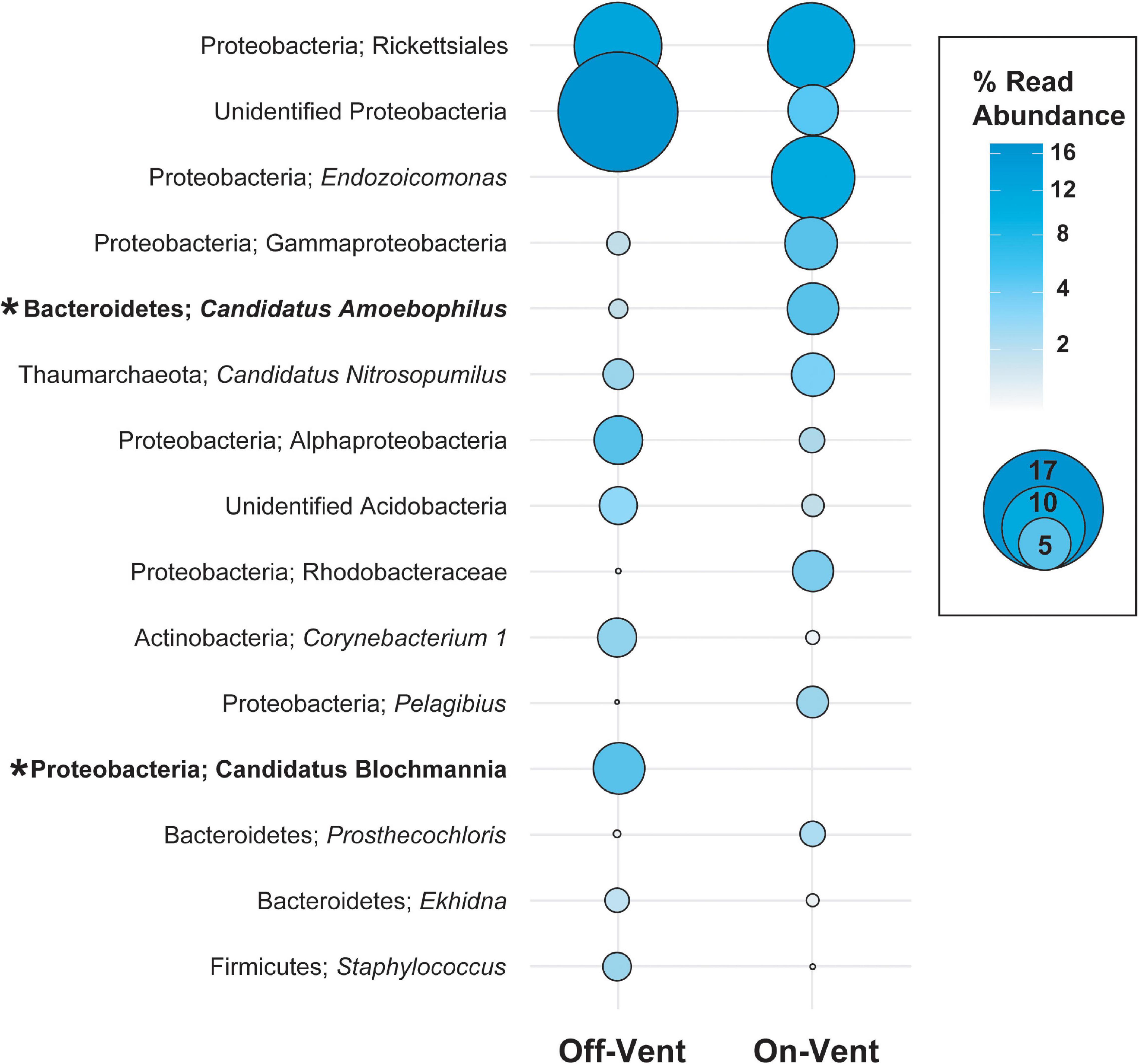
Figure 5. Relative abundance (in descending order) for the ten most abundant high abundance ASVs (>500 reads) in on- and off-vent Porites panamensis samples. Size of the circle indicates the relative abundance of the taxa within each group. Asterisks denote ASVs that were differentially abundant between off- and on-vent colonies.
The core microbial microbiome for all P. panamensis samples was composed of only two ASVs, which accounted for only 1.5% of the reads/sample (Table 1). All on-vent corals contained 4 additional core ASVs, including two distinct archaeal taxa, both identified to the genus level as Candidatus Nitrosopumilus, while off-vent samples only had one additional core ASV, a Helicobacteraceae spp. (Table 1). The most abundant ASV overall was an Endozoicomonas, but it was not part of the 100% core microbiome for on- or off-vent corals (Table 1, Figure 5, and Supplementary Figure 8).
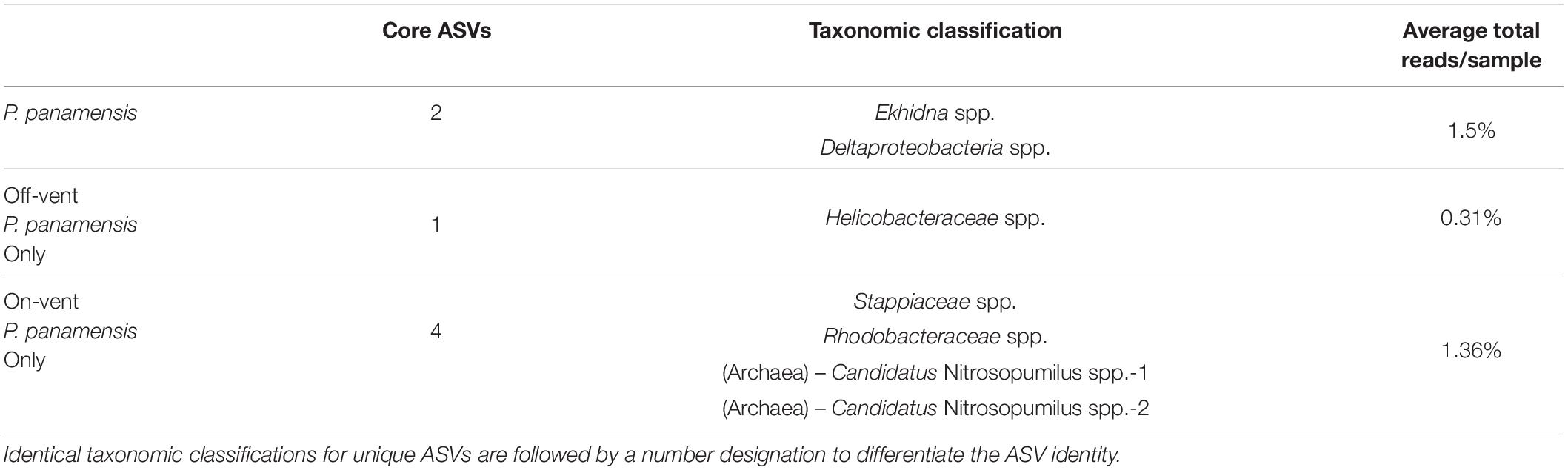
Table 1. The taxonomic classifications and average per sample read abundance of the 100% core high abundance ASVs (>500 reads) for all Porites panamensis corals, off-vent coral only, and on-vent coral only samples.
Microbial Community Functional Analysis
The functional metagenomic analysis of the full ASV dataset for the rarefied microbiome of P. panamensis identified 7,438 KEGG orthologs. The KEGG orthologs were collapsed into 427 MetaCyc Class level pathways, 20 of which were differentially abundant between on- and off-vent samples (Supplementary Figure 9), the majority (17) of which were less abundant in on-vent microbiomes. Of the 20 Class level pathways, 16 of these were related to biosynthesis and degradation of small molecules, and four were involved in glycolysis and fermentation (Supplementary Figure 11). All ten of the most abundant subclass level pathways (most specific pathway designation) were related to biosynthesis of small molecules and energy production including aerobic respiration and fermentation (Figure 6). Of these ten subclass pathways, four were differentially abundant between on- and off-vent P. panamensis microbiomes and all four were more abundant in on-vent samples (Figure 6).
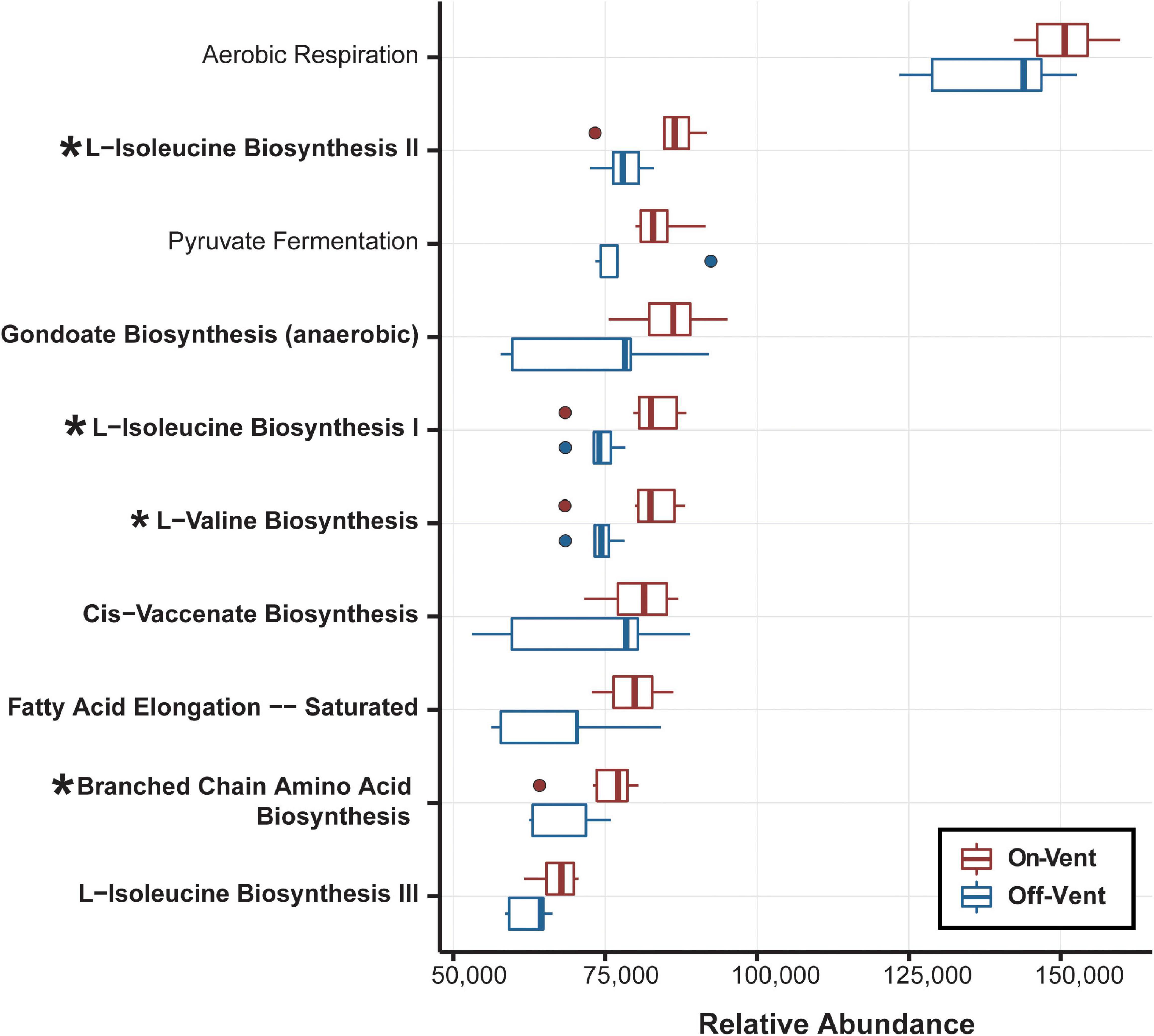
Figure 6. Relative abundance of the ten most abundant biological pathways inferred from the PICRUSt2 analysis. Bolded pathways indicate those involved in biosynthesis of small molecules, macromolecules and cell structure components. Asterisks identify the four pathways that display significant differences.
Porites panamensis Samples: On-Vent vs. Off-Vent Ostreobium Community Analysis
Amplicon Sequence Variants closest to Ostreobium quekettii were more abundant in on-vent samples, while ASVs closest to Ostreobium HV05042 were more abundant in off-vent samples, but neither significantly so (p > 0.05). There was no difference in Ostreobium ASV diversity (Chao1) between off- and on-vent samples (Kruskal-Wallis: chi-squared1 = 1.967, p = 0.161; Figure 7A).
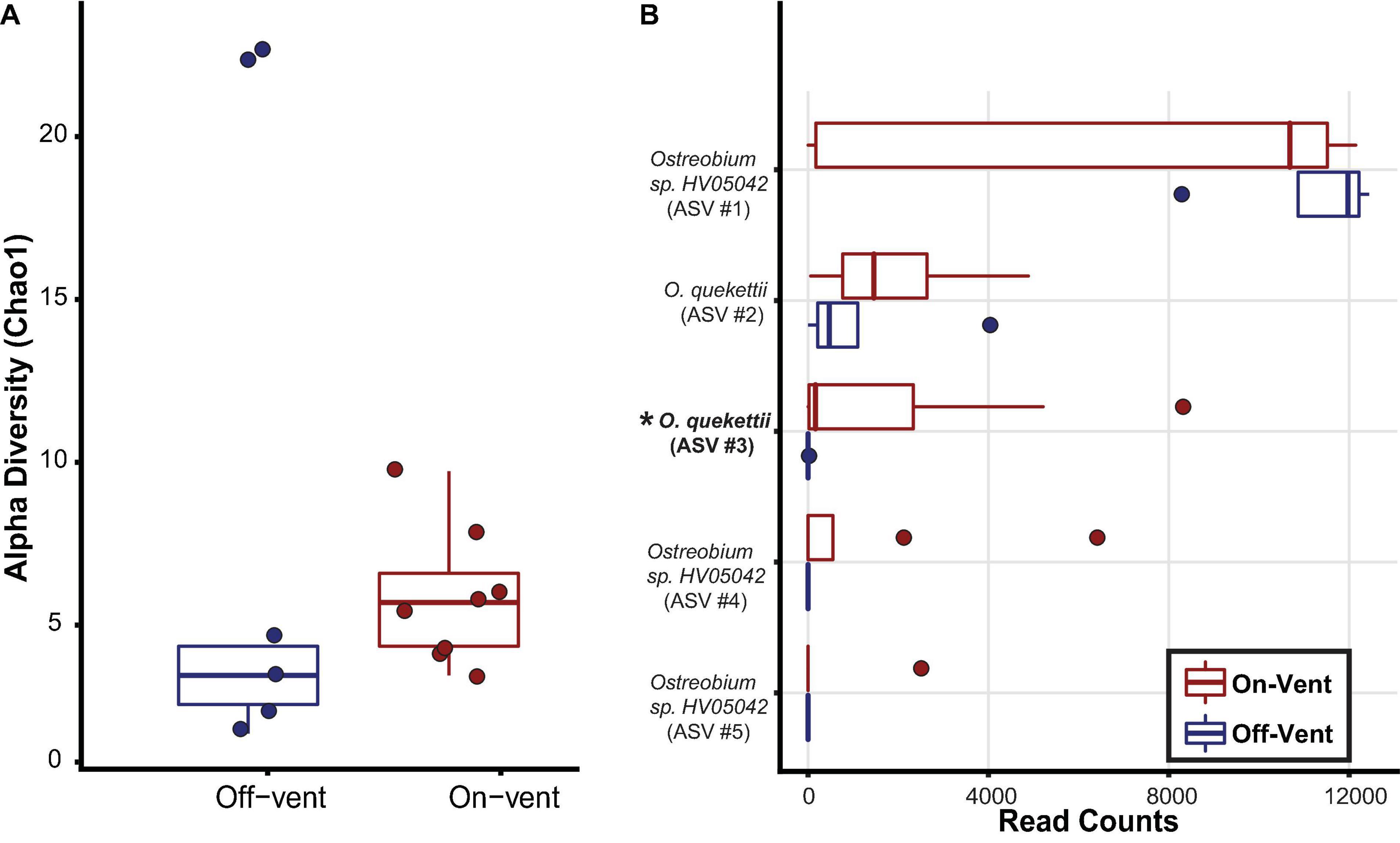
Figure 7. Ostreobium community diversity and abundance for off-(blue) and on-vent (red) Porites panamensis samples. (A) Alpha diversity (Chao1). The lower and upper hinges represent the first and third quartiles. The lower and upper whiskers extend to either the largest or smallest value that is no more than 1.5* the interquartile range. (B) Relative abundance (in descending order) for the five most abundant Ostreobium ASVs in P. panamensis samples. Asterisks and bold lettering denote ASVs that were differentially abundant between off- and on-vent colonies.
To assess beta diversity (Bray-Curtis) Ostreobium reads were rarefied to 12,445 reads per coral sample that clustered into 45 ASVs, of which only five were found in high abundance (>500 reads; Figure 7B). Of the 5 most abundant ASVs, only a single ASV, close to O. quekettii (ASV #3), was differentially abundant between off- and on-vent samples (Kruskal-Wallis: chi-squared1 = 4.647, p = 0.031; Figure 7). Individual Ostreobium ASVs showed high variability in relative abundance among samples within groups (Figure 7B).
Discussion
Differentiation of Biotic and Abiotic Microbial Communities
Microbiomes were distinct between all three sample types and sediment microbiomes were more diverse than those from coral, while coral and water were similar in overall diversity (Figure 2). Marine eukaryote-associated microbiomes rarely match the composition of the local abiotic microbiomes (Sunagawa et al., 2010; Chiarello et al., 2019, although see Rodríguez-Marconi et al., 2015, for an Antarctic sponge exception), even in extreme conditions (Goffredi, 2010; Moreno-Pino et al., 2020).
Microbial Diversity Measures for On-Vent Corals Do Not Show “Symptoms” of Stress
The microbiomes of off- and on-vent colonies of P. panamensis exhibited similar diversity measures (richness, evenness, and beta diversity, Supplementary Figure 3). This was surprising given that conditions near the vents (low pH, high CO2, high temperature) are generally considered stressful to corals and diversity measures of coral microbial communities usually increase under stress (McDevitt-Irwin et al., 2019). This oft-observed higher richness may result from the inability of stressed corals to resist invasion by atypical symbionts, thereby increasing taxon counts (Welsh et al., 2017). This appears to be a common symptom of corals exposed to acute stressors, either experimentally (Meron et al., 2011) or via anthropogenic activity (Morrow et al., 2012; Jessen et al., 2013). Increased beta diversity is expected under stress because the microbiome becomes dysbiotic (Zaneveld et al., 2017). Thus, the host may have difficulty regulating their community structure, as indicated by changes in evenness (McDevitt-Irwin et al., 2019).
On-vent corals live at higher temperatures and lower pH than their off-vent counterparts and if stressed by these conditions, the on-vent coral microbiomes would be expected to show signs of dysbiosis. Beta diversity increases have been seen for corals exposed to high temperatures, increased salinity, and pollution (Klaus et al., 2007; Röthig et al., 2016; Zaneveld et al., 2016; Camp et al., 2020). However, the microbiomes of diverse marine hosts experiencing long-term or natural exposure do not always display increases in alpha (Morrow et al., 2015; Coelho et al., 2018; Camp et al., 2020) or beta diversity (Webster et al., 2016).
Because the microbial per sample read counts for P. panamensis samples were high (>260,000 reads/sample on average; Supplementary Table 1) and the rarefied dataset captured most of the diversity in each sample (Supplementary Figures 1, 2), low power was probably not responsible for the lack of differences between off- and on-vent microbiomes. More likely, the on-vent P. panamensis colonies were not stressed, or rather their microbiomes are acclimated to the stressors associated with vent conditions. We note that coral colonies were sampled in February, when water temperatures are near their annual cool point. While temperature is not the sole potential source of stress from vent water, it may be that colonies experience greater stress during warmer summer months.
On-Vent Coral Microbiomes Are Dominated by Beneficial Endosymbionts
While diversity measures were similar between off- and on-vent P. panamensis microbiomes, their taxonomic compositions were distinct (Figure 3). Proteobacteria and Bacteroidetes dominated microbial phyla for corals in both settings, but both were more abundant on-vent (Figure 4). In addition, the beneficial endolithic alga, Ostreobium, was found in high abundance in both on- and off-vent corals, though a single Ostreobium ASV, close to O. quekettii, was more abundant in on-vent corals than off-vent colonies (Figure 7B).
Ostreobium are endolithic green algae that are well-known coral symbionts. They may benefit their hosts via their role in carbonate dissolution (Verbruggen et al., 2017), their ability to provide photosynthates, and protection from light stress(Fork and Larkum, 1989; Del Campo et al., 2017; Pernice et al., 2020). Carbonate dissolution may be particularly important for on-vent corals because waters of the hydrothermal vents in Bahía Concepción are enriched for CO2 (Dávila-Ramos et al., 2015), which decreases the carbonate ions available for calcification (Eyre et al., 2014).
Coral microbiomes generally lose beneficial endosymbionts under stressful conditions (Bourne et al., 2016; McDevitt-Irwin et al., 2019), however, total Ostreobium abundance did not differ between on- and off-vent corals and the common coral bacterial symbiont Endozoicomonas spp. was more abundant in on-vent P. panamensis than off-vent colonies (Figure 5). Like Ostreobium, Endozoicomonas provide benefits to their host corals, including nutrient cycling, protection from pathogens, carbohydrate transport and sulfur cycling (McDevitt-Irwin et al., 2019), and likely co-evolve with them (Neave et al., 2016). Members of this genus have been consistently found at low abundance in corals stressed by climate change, water pollution, and overfishing (McDevitt-Irwin et al., 2019). In contrast to our findings, Acropora millepora from near a volcanic CO2 seep had a 50% decrease in Endozoicomonas abundance relative to off-seep habitat (Morrow et al., 2015). However, P. panamensis is considered to be a resilient coral species, in contrast to most acroporiids, often found in marginal or even extreme habitats (Halfar et al., 2005; Anlauf et al., 2011; Oporto-Guerrero et al., 2018). Coral-mediated tolerance (via gene expression, genetic adaptation or some other metabolic/genetic mechanism) to stressful conditions may help Porites colonies maintain the symbioses with important microbes such as Endozoicomonas.
Ostreobium, was found in high relative abundance in all corals sampled, however, of the five most abundant Ostreobium ASVs, four were more abundant (one significantly so) in on-vent samples. Unlike shifts in the abundances of Endozoicomonas between treatments or conditions, changes to Ostreobium have not been commonly reported (although see changes with depth in the coral Agaricia undata, Gonzalez-Zapata et al., 2018). Ostreobium inhabit the coral skeleton (Marcelino and Verbruggen, 2016) and while most coral microbiome studies separate coral tissue and mucus from the skeleton before lysing to obtain microbial DNA (e.g., Apprill et al., 2016; Weber et al., 2017), we lysed microbial cells from the entire coral fragment (skeleton, tissue and mucus). This difference in technique may be why Ostreobium is rarely mentioned in comparative microbiome studies from scleractinian corals, though more recently their symbiosis with corals has received more attention (Pernice et al., 2020).
The shift in the abundance of different Ostreobium ASVs in on-vent coral microbiomes may be due to their ability to assume the role of zooxanthellae (Fine and Loya, 2002). Ostreobium can adapt to changes in temperature and their symbiosis with corals does not break down under thermal stress (Fine et al., 2005). In contrast, the symbiosis between corals and zooxanthellae is more tenuous at high temperatures (Brown, 1997). In particular, P. panamensis exposed to high temperatures (28.5–31.1°C) had fewer zooxanthellae in their primary polyps (Anlauf et al., 2011). Although none of the corals sampled in our study displayed any obvious signs of bleaching, we did not measure the abundance of zooxanthellae. If high temperatures depleted zooxanthellae in on-vent corals, this could favor a compensatory rise in Ostreobium abundance to meet the nutritional demands of the coral host. The overall high abundance of Endozoicomonas and Ostreobium, two beneficial coral symbionts, combined with very low differential host and symbiont gene expression between on- and off-vent colonies (Balandra et al., 2021) suggest that the on-vent corals are not stressed and that the coral microbiome in these colonies may have shifted to promote local acclimation.
Two other ASVs from the 100% core of on-vent microbiomes belonged to the archaeal genus Candidatus Nitrosopumilus (Könneke et al., 2005; Table 1). Each of these ASVs was modestly abundant (<1% reads/sample), but their ammonia-oxidizing capabilities may make their presence important to corals living on vents (Feng et al., 2016). In a previous study, corals enriched with nitrogen were better able to withstand thermal stress (Béraud et al., 2013). Shallow hydrothermal vents in Bahía Concepción are typically low in bioavailable nitrogen (Dávila-Ramos et al., 2015) for use by eukaryotic hosts, which could favor nitrifying microbes. Candidatus Nitrosopumilus oxidizes ammonia that is excreted by its host to produce nitrite that can then be transferred to the host (Feng et al., 2016). Thus, these two ammonia-oxidizing ASVs should benefit their on-vent hosts that live in a habitat depleted of available nitrogen and are outside their normal thermal range. These vents have reduced sulfate (SO42–) and methane (CH4) as compared to surrounding waters (Prol-Ledesma et al., 2004) and notable shifts in on-vent microbiomes that correspond with vent water chemistry included decreased abundances of the sulfate-reducing bacterial family, Desulfovibrionaceae, and Bacteroidetes BD2-2, whose members may interact with methanotrophic archaea and sulfate-reducing bacterial taxa (Trembath-Reichert et al., 2016).
On-vent corals also had lower relative abundances of taxa in the order Rickettsiales (phylum: Proteobacteria) and the genus Staphylococcus (phylum: Firmicutes), although these differences were not significant. These intracellular symbionts are widespread among corals and often most abundant in stressed corals (Rosenberg et al., 2007; Chiou et al., 2010; Garcia et al., 2013; Daniels et al., 2015); although see Casas et al., 2004; Godoy-Vitorino et al., 2017). The abundance of Rickettsiales is negatively correlated with coral growth (Shaver et al., 2017) and their genome exhibits pathogenic capabilities (Klinges et al., 2019). Similarly, some Staphylococcus species are known to be opportunistic coral pathogens (Divya et al., 2018), though they can often be in high abundance in healthy coral tissue and may only become pathogenic following stress-related microbiome shifts (Pereira de Castro et al., 2010). If the on-vent environment stressed coral colonies then an increase in potentially pathogenic bacterial taxa such as Staphylococcus and Rickettsiales would have been expected, however, there is evidence that Rickettsiales may not be able to withstand the high temperatures near the vent (Mikolajczyk et al., 1964).
Functional Capabilities of On-Vent Coral Microbiomes Match an Inhospitable Habitat
Twenty functional MetaCyc Class-level pathways showed differences between on- and off-vent microbiomes (Supplementary Figure 11) and most (17) of them decreased in abundance in on-vent P. panamensis samples (Supplementary Figure 11). As a potential response to the energy demands of increased thermal stress, on-vent coral microbiomes were enriched for three Class-level pathways related to energy production and small molecule degradation (largely as a precursor to energy production; Supplementary Figure 11; Dijkstra et al., 2011; Slavov et al., 2014). Ziegler et al. (2017) also found that thermally tolerant A. hyacinthus microbiomes were enriched for metabolic functions. On-vent samples were significantly enriched for the sulfolactate degradation pathway (see Sulfur compound metabolism in Supplementary Figure 9), a metabolic pathway that has been identified in members of the family Rhodobacteraceae (phylum: Proteobacteria; MetaCyc Database; Denger et al., 2009), which were also found in higher abundance in on-vent samples (Figure 5).
While small molecule biosynthesis and sulfur metabolisms were enriched in on-vent coral microbial communities, there was a reduction in genes related to the more traditional energy pathways including glycolysis and fermentation (Supplementary Figure 11). Dombrowski et al. (2018), found that hydrothermal vent sediment microbial communities hosted a more diverse set of metabolisms, including those that utilize methane, hydrogen and sulfate/sulfite, than those of background samples. A similar phenomenon may exist in on-vent coral microbial communities, where the extreme water chemistry (hot, acidic, metallic, low nitrogen, high CO2; Prol-Ledesma et al., 2004) has led to a set of symbionts that are less reliant on traditional energy pathways, but rather can utilize the elements and compounds in vent waters as an energy source.
Of the ten most abundant Subclass-level pathways, all were enriched in on-vent samples, four significantly so (Figure 6). Such a bias for enrichment in the hotter setting may seem unusual, but similar results were found in Acropora hyacinthus (Ziegler et al., 2017). Thus, the enrichment of pathways may represent a signature of acclimation to higher temperatures in coral microbiomes. Both off- and on-vent coral microbiomes were dominated by genes associated with the aerobic respiration Subclass pathway, but on-vent corals more so (Figure 6). Elevated temperatures increase respiration rates in both eukaryotes (Coles and Jokiel, 1977; Strand et al., 2017) and microbial communities (Wan et al., 2007; Dijkstra et al., 2011). Further, the significant differential enrichment of four biosynthesis subclass pathways in on-vent coral microbiomes may be tied to increased energy demands of the warmer waters (Vallon and Spalding, 2009).
Enrichment of energy metabolism pathways can be attributed to stressors like increased upright algal cover, temperature, and pH, but such instances should be accompanied by increases in other stress-related pathways and potentially virulent taxa (Thurber et al., 2009; Zaneveld et al., 2017; Wang et al., 2018). On-vent coral microbiomes were not enriched for typical stress-related pathways such as antimicrobial production, bacterial motility, chemotaxis and invasion (Thurber et al., 2009; Zaneveld et al., 2016) nor did we find any increases in pathogenic taxa like Vibrio spp. (Ben-Haim et al., 2003; Thurber et al., 2009). While the high abundance of genes associated with energy production may be the result of stress, the on-vent microbiomes are missing the virulent pathways and presence of known virulent taxa that should accompany these increases if they were stress-related.
The functional data presented here suggest that on-vent P. panamensis microbiomes are not stressed, however, these insights are based on metagenomic predictions of closely related microbial taxa that have available genomes. A metagenomic or metatranscriptomic study of the microbiomes of on- and off-vent P. panamensis colonies would be needed to confirm these results.
Conclusion
The microbiomes of the coral Porites panamensis living near shallow hydrothermal vents appear well-suited to the high temperature and low pH associated with these extreme conditions and do not show signs of stress. The on-vent coral microbiome is dominated by taxa that are thermally tolerant and capable of providing and cycling nutrients and other benefits to their coral hosts, including the Proteobacteria Endozoicomonas and the endolithic alga Ostreobium. Pathogenic microbial taxa did not increase near vents, as would be expected if coral microbiomes were stressed. Furthermore, the functional pathways in on-vent P. panamensis microbiomes are consistent with the biological requirements for coping at high temperatures, such as increased respiration and energy demand.
Overall, we have seen that vent-associated P. panamensis colonies living at extreme thermal and pH conditions are associated with changes in the composition and functional capabilities of their microbial symbionts, perhaps contributing to their acclimation. These findings exemplify the need to examine microbiota of corals living in natural extreme habitats to determine taxonomic and functional signatures of host duress versus host acclimation to less hospitable conditions.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA761929.
Author Contributions
AR processed the samples, analyzed the data, and wrote the first draft of the manuscript. DP-G and MH performed field work. AR and DP-G developed and edited figures. All authors contributed to the conceptualization of the work, edited, and approved the final version of the manuscript.
Funding
This project was supported by funds provided to MH by the Department of Biological Sciences at Louisiana State University and to DP-G by the Program of Fishery Ecology of CIBNOR (PEP) and Marine BioGenomics Funding.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Andrea Puga, Sofia Mendez, and Julio Arce for field assistance. Logistic support was provided by Eduardo Balart and Noemi Bocanegra of CIBNOR. We thank Anna Lucchesi for laboratory assistance and Erick Balandra for environmental information about the vent sites. We also thank Morgan Kelly and J. Cameron Thrash for valuable feedback on this manuscript. Samples were collected under CONAPESCA permit 0305004228.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.652633/full#supplementary-material
Footnotes
References
Andersen, K. S., Kirkegaard, R. H., Karst, S. M., and Albertsen, M. (2018). ampvis2: an R package to analyse and visualise 16S rRNA amplicon data. bioRxiv [Preprint] doi: 10.1101/299537
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1046/j.1442-9993.2001.01070.x
Anlauf, H., D’Croz, L., and O’Dea, A. (2011). A corrosive concoction: the combined effects of ocean warming and acidification on the early growth of a stony coral are multiplicative. J. Exp. Mar. Bio. Ecol. 397, 13–20. doi: 10.1016/j.jembe.2010.11.009
Apprill, A., Mcnally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Apprill, A., Weber, L. G., and Santoro, A. E. (2016). Distinguishing between microbial habitats unravels ecological complexity in coral microbiomes. mSystems 1:e00143–16. doi: 10.1128/mSystems.00143-16
Baker, A. C. (2003). Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 34, 661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417
Balandra, E., Fernandez Valverde, S., Balart, E. F., Hellberg, M. E., and Paz-García, D. A. (2021) Beyond Marginality: Transcriptomic Insights Between Marginal and Extreme Environments in a Reef-Building Coral. (Unpublished manuscript)
Barshis, D. J., Ladner, J. T., Oliver, T. A., Seneca, F. O., Traylor-Knowles, N., and Palumbi, S. R. (2013). Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. U.S.A. 110, 1387–1392. doi: 10.1073/pnas.1210224110
Ben-Haim, Y., Zicherman-Keren, M., and Rosenberg, E. (2003). Temperature-regulated bleaching and lysis of the coral Pocillopora damicomis by the novel pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 69, 4236–4242. doi: 10.1128/AEM.69.7.4236-4242.2003
Béraud, E., Gevaert, F., Rottier, C., and Ferrier-Pagès, C. (2013). The response of the scleractinian coral Turbinaria reniformis to thermal stress depends on the nitrogen status of the coral holobiont. J. Exp. Biol. 216, 2665–2674. doi: 10.1242/jeb.085183
Berkelmans, R., and Van Oppen, M. J. H. (2006). The role of zooxanthellae in the thermal tolerance of corals: A “nugget of hope” for coral reefs in an era of climate change. Proc. R. Soc. B Biol. Sci. 273, 2305–2312. doi: 10.1098/rspb.2006.3567
Bianchi, C. N., Morri, C., Chiantore, M., Montefalcone, M., Parravicini, V., and Rovere, A. (2012). “Life in the mediterranean sea: a look at habitat changes mediterranean sea biodiversity between the legacy from the past and a future of change,” in Life in the Mediterranean Sea: A Look at Habitat Changes, ed. N. Stambler (Hauppauge, NY: Nova Science).
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Brown, B. E. (1997). Coral bleaching: causes and consequences. Coral Reefs 16, S129–S138. doi: 10.1007/s003380050249
Callahan, B. J., McMurdie, P. J., and Holmes, S. P. (2017). Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643. doi: 10.1038/ismej.2017.119
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583.
Camp, E. F., Suggett, D. J., Pogoreutz, C., Nitschke, M. R., Houlbreque, F., Hume, B. C. C., et al. (2020). Corals exhibit distinct patterns of microbial reorganisation to thrive in an extreme inshore environment. Coral Reefs 39, 701–716. doi: 10.1007/s00338-019-01889-3
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Carpenter, K. E., Abrar, M., Aeby, G., Aronson, R. B., Banks, S., Bruckner, A., et al. (2008). One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science 321, 560–563. doi: 10.1126/science.1159196
Casas, V., Kline, D. I., Wegley, L., Yu, Y., Breitbart, M., and Rohwer, F. (2004). Widespread association of a Rickettsiales-like bacterium with reef-building corals. Environ. Microbiol. 6, 1137–1148. doi: 10.1111/j.1462-2920.2004.00647.x
Caspi, R., Billington, R., Fulcher, C. A., Keseler, I. M., Kothari, A., Krummenacker, M., et al. (2018). The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 46, D633–D639. doi: 10.1093/nar/gkx935
Chiarello, M., Paz-Vinas, I., Veyssière, C., Santoul, F., Loot, G., Ferriol, J., et al. (2019). Environmental conditions and neutral processes shape the skin microbiome of European catfish (Silurus glanis) populations of Southwestern France. Environ. Microbiol. Rep. 11, 605–614. doi: 10.1111/1758-2229.12774
Chiou, S. F., Kuo, J., Wong, T. Y., Fan, T. Y., Tew, K. S., and Liu, J. K. (2010). Analysis of the coral associated bacterial community structures in healthy and diseased corals from off-shore of southern Taiwan. J. Environ. Sci. Health B 45, 408–415. doi: 10.1080/03601231003800032
Coelho, F. J. R. C., Cleary, D. F. R., Gomes, N. C. M., Pólonia, A. R. M., Huang, Y. M., Liu, L. L., et al. (2018). Sponge prokaryote communities in taiwanese coral reef and shallow hydrothermal vent ecosystems. Microb. Ecol. 75, 239–254. doi: 10.1007/s00248-017-1023-x
Coles, S. L., and Jokiel, P. L. (1977). Effects of temperature on photosynthesis and respiration in hermatypic corals. Mar. Biol. 43, 209–216. doi: 10.1007/BF00402313
Dahms, H. U., Schizas, N. V., James, R. A., Wang, L., and Hwang, J. S. (2018). Marine hydrothermal vents as templates for global change scenarios. Hydrobiologia 818, 1–10. doi: 10.1007/s10750-018-3598-8
Daniels, C. A., Baumgarten, S., Yum, L. K., Michell, C. T., Bayer, T., Arif, C., et al. (2015). Metatranscriptome analysis of the reef-building coral Orbicella faveolata indicates holobiont response to coral disease. Front. Mar. Sci. 2:62. doi: 10.3389/fmars.2015.00062
Dávila-Ramos, S., Estradas-Romero, A., Prol-Ledesma, R. M., and Juárez-López, K. (2015). Bacterial populations (first record) at two shallow hydrothermal vents of the mexican pacific west coast. Geomicrobiol. J. 32, 657–665. doi: 10.1080/01490451.2014.980526
Del Campo, J., Pombert, J. F., Šlapeta, J., Larkum, A., and Keeling, P. J. (2017). The “other” coral symbiont: Ostreobium diversity and distribution. ISME J. 11, 296–299. doi: 10.1038/ismej.2016.101
Denger, K., Mayer, J., Buhmann, M., Weinitschke, S., Smits, T. H. M., and Cook, A. M. (2009). Bifurcated degradative pathway of 3-sulfolactate in Roseovarius nubinhibens ISM via sulfoacetaldehyde acetyltransferase and (S)-cysteate sulfolyase. J. Bacteriol. 191, 5648–5656. doi: 10.1128/JB.00569-09
Dijkstra, P., Thomas, S. C., Heinrich, P. L., Koch, G. W., Schwartz, E., and Hungate, B. A. (2011). Effect of temperature on metabolic activity of intact microbial communities: evidence for altered metabolic pathway activity but not for increased maintenance respiration and reduced carbon use efficiency. Soil Biol. Biochem. 43, 2023–2031. doi: 10.1016/j.soilbio.2011.05.018
Divya, S., Thinesh, T., Seghal Kiran, G., Hassan, S., and Selvin, J. (2018). Emergence of a multi host biofilm forming opportunistic pathogen Staphylococcus sciuri D26 in coral Favites abdita. Microb. Pathog. 120, 204–212. doi: 10.1016/j.micpath.2018.04.037
Dombrowski, N., Teske, A. P., and Baker, B. J. (2018). Expansive microbial metabolic versatility and biodiversity in dynamic guaymas basin hydrothermal sediments. Nat. Commun. 9:4999. doi: 10.1038/s41467-018-07418-0
Eyre, B. D., Andersson, A. J., and Cyronak, T. (2014). Benthic coral reef calcium carbonate dissolution in an acidifying ocean. Nat. Clim. Chang. 4, 969–976. doi: 10.1038/nclimate2380
Feng, G., Sun, W., Zhang, F., Karthik, L., and Li, Z. (2016). Inhabitancy of active Nitrosopumilus-like ammonia-oxidizing archaea and Nitrospira nitrite-oxidizing bacteria in the sponge Theonella swinhoei. Sci. Rep. 6:24966. doi: 10.1038/srep24966
Fine, M., and Loya, Y. (2002). Endolithic algae: an alternative source of photoassimilates during coral bleaching. Proc. R. Soc. London. Ser. B Biol. Sci. 269, 1205–1210. doi: 10.1098/rspb.2002.1983
Fine, M., Meroz-Fine, E., and Hoegh-Guldberg, O. (2005). Tolerance of endolithic algae to elevated temperature and light in the coral Montipora monasteriata from the southern Great Barrier Reef. J. Exp. Biol. 208, 75–81. doi: 10.1242/jeb.01381
Fork, D. C., and Larkum, A. W. D. (1989). Light harvesting in the green alga Ostreobium sp., a coral symbiont adapted to extreme shade. Mar. Biol. 103, 381–385. doi: 10.1007/BF00397273
Forrest, M. J., Ledesma-Vázquez, J., Ussler, W., Kulongoski, J. T., Hilton, D. R., and Greene, H. G. (2005). Gas geochemistry of a shallow submarine hydrothermal vent associated with the El Requesón fault zone, Bahía Concepción, Baja California Sur, México. Chem. Geol. 224, 82–95. doi: 10.1016/j.chemgeo.2005.07.015
Garcia, G. D., Gregoracci, G. B., Santos, E., de, O., Meirelles, P. M., Silva, G. G. Z., et al. (2013). Metagenomic analysis of healthy and white plague-affected Mussismilia braziliensis corals. Microb. Ecol. 65, 1076–1086. doi: 10.1007/s00248-012-0161-4
Gardner, T. A., Côté, I. M., Gill, J. A., Grant, A., and Watkinson, A. R. (2003). Long-term region-wide declines in Caribbean corals. Science 301, 958–960. doi: 10.1126/science.1086050
Glassman, S. I., and Martiny, J. B. H. (2018). Broadscale ecological patterns are robust to use of exact sequence variants versus operational taxonomic units. mSphere 3:e00148–18. doi: 10.1128/msphere.00148-18
Glynn, P. W., Colley, S. B., Eakin, C. M., Smith, D. B., Cortés, J., Gassman, N. J., et al. (1994). Reef coral reproduction in the eastern Pacific: Costa Rica, Panamá, and Galápagos Islands (Ecuador). II. Poritidae. Mar. Biol. 118, 191–208. doi: 10.1007/BF00349785
Godoy-Vitorino, F., Ruiz-Diaz, C. P., Rivera-Seda, A., Ramírez-Lugo, J. S., and Toledo-Hernández, C. (2017). The microbial biosphere of the coral Acropora cervicornis in Northeastern Puerto Rico. PeerJ 5:e3717. doi: 10.7717/peerj.3717
Goffredi, S. K. (2010). Indigenous ectosymbiotic bacteria associated with diverse hydrothermal vent invertebrates. Environ. Microbiol. Rep. 2, 479–488. doi: 10.1111/j.1758-2229.2010.00136.x
Gonzalez-Zapata, F. L., Gómez-Osorio, S., and Sánchez, J. A. (2018). Conspicuous endolithic algal associations in a mesophotic reef-building coral. Coral Reefs 37, 705–709. doi: 10.1007/s00338-018-1695-9
Grottoli, A. G., Dalcin Martins, P., Wilkins, M. J., Johnston, M. D., Warner, M. E., Cai, W.-J., et al. (2018). Coral physiology and microbiome dynamics under combined warming and ocean acidification. PLoS One 13:e0191156. doi: 10.1371/journal.pone.0191156
Grottoli, A. G., Rodrigues, L. J., and Palardy, J. E. (2006). Heterotrophic plasticity and resilience in bleached corals. Nature 440, 1186–1189. doi: 10.1038/nature04565
Hadaidi, G., Röthig, T., Yum, L. K., Ziegler, M., Arif, C., Roder, C., et al. (2017). Stable mucus-associated bacterial communities in bleached and healthy corals of Porites lobata from the Arabian Seas. Sci. Rep. 7:45362. doi: 10.1038/srep45362
Halfar, J., Godinez-Orta, L., Riegl, B., Valdez-Holguin, J. E., and Borges, J. M. (2005). Living on the edge: high-latitude Porites carbonate production under temperate eutrophic conditions. Coral Reefs 24, 582–592. doi: 10.1007/s00338-005-0029-x
Hall-Spencer, J. M., Rodolfo-Metalpa, R., Martin, S., Ransome, E., Fine, M., Turner, S. M., et al. (2008). Volcanic carbon dioxide vents show ecosystem effects of ocean acidification. Nature 454, 96–99. doi: 10.1038/nature07051
Hanshew, A. S., Mason, C. J., Raffa, K. F., and Currie, C. R. (2013). Minimization of chloroplast contamination in 16S rRNA gene pyrosequencing of insect herbivore bacterial communities. J. Microbiol. Methods 95, 149–155. doi: 10.1016/j.mimet.2013.08.007
Jessen, C., Villa Lizcano, J. F., Bayer, T., Roder, C., Aranda, M., Wild, C., et al. (2013). In-situ effects of eutrophication and overfishing on physiology and bacterial diversity of the Red Sea coral Acropora hemprichii. PLoS One 8:e62091. doi: 10.1371/journal.pone.0062091
Joos, L., Beirinckx, S., Haegeman, A., Debode, J., Vandecasteele, B., Baeyen, S., et al. (2020). Daring to be differential: metabarcoding analysis of soil and plant-related microbial communities using amplicon sequence variants and operational taxonomical units. BMC Genomics 21:733. doi: 10.1186/s12864-020-07126-4
Kalanetra, K. M., Huston, S. L., and Nelson, D. C. (2004). Novel, attached, sulfur-oxidizing bacteria at shallow hydrothermal vents possess vacuoles not involved in respiratory nitrate accumulation. Appl. Environ. Microbiol. 70, 7487–7496. doi: 10.1128/AEM.70.12.7487-7496.2004
Klaus, J. S., Janse, I., Heikoop, J. M., Sanford, R. A., and Fouke, B. W. (2007). Coral microbial communities, zooxanthellae and mucus along gradients of seawater depth and coastal pollution. Environ. Microbiol. 9, 1291–1305. doi: 10.1111/j.1462-2920.2007.01249.x
Klinges, J. G., Rosales, S. M., McMinds, R., Shaver, E. C., Shantz, A. A., Peters, E. C., et al. (2019). Phylogenetic, genomic, and biogeographic characterization of a novel and ubiquitous marine invertebrate-associated Rickettsiales parasite, Candidatus Aquarickettsia rohweri, gen. nov., sp. nov. ISME J. 13, 2938–2953. doi: 10.1038/s41396-019-0482-0
Könneke, M., Bernhard, A. E., De La Torre, J. R., Walker, C. B., Waterbury, J. B., and Stahl, D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Littman, R. A., Bourne, D. G., and Willis, B. L. (2010). Responses of coral-associated bacterial communities to heat stress differ with Symbiodinium type on the same coral host. Mol. Ecol. 19, 1978–1990. doi: 10.1111/j.1365-294X.2010.04620.x
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Marcelino, V. R., and Verbruggen, H. (2016). Multi-marker metabarcoding of coral skeletons reveals a rich microbiome and diverse evolutionary origins of endolithic algae. Sci. Rep. 6:31508. doi: 10.1038/srep31508
Matz, M. V., Treml, E. A., Aglyamova, G. V., and Bay, L. K. (2018). Potential and limits for rapid genetic adaptation to warming in a great barrier reef coral. PLoS Genet. 14:e1007220. doi: 10.1371/journal.pgen.1007220
Maugeri, T. L., Lentini, V., Gugliandolo, C., Italiano, F., Cousin, S., and Stackebrandt, E. (2009). Bacterial and archaeal populations at two shallow hydrothermal vents off Panarea Island (Eolian Islands. Italy). Extremophiles 13, 199–212. doi: 10.1007/s00792-008-0210-6
McDevitt-Irwin, J. M., Garren, M., McMinds, R., Vega Thurber, R., and Baum, J. K. (2019). Variable interaction outcomes of local disturbance and El Niño-induced heat stress on coral microbiome alpha and beta diversity. Coral Reefs 38, 331–345. doi: 10.1007/s00338-019-01779-8
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R Package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Meron, D., Atias, E., Iasur Kruh, L., Elifantz, H., Minz, D., Fine, M., et al. (2011). The impact of reduced pH on the microbial community of the coral Acropora eurystoma. ISME J. 5, 51–60. doi: 10.1038/ismej.2010.102
Meron, D., Rodolfo-Metalpa, R., Cunning, R., Baker, A. C., Fine, M., and Banin, E. (2012). Changes in coral microbial communities in response to a natural pH gradient. ISME J. 6, 1775–1785. doi: 10.1038/ismej.2012.19
Mikolajczyk, E., Frygin, C., and Wojciechowski, E. (1964). Thermal inactivation of the toxic and haemolytic activity of typhus fever rickettsiae. Med. Dosw. Mikrobiol. 16, 217–226.
Milazzo, M., Rodolfo-Metalpa, R., Chan, V. B. S., Fine, M., Alessi, C., Thiyagarajan, V., et al. (2015). Ocean acidification impairs vermetid reef recruitment. Sci. Rep. 4:4189. doi: 10.1038/srep04189
Moreno-Pino, M., Cristi, A., Gillooly, J. F., and Trefault, N. (2020). Characterizing the microbiomes of Antarctic sponges: a functional metagenomic approach. Sci. Rep. 10:645. doi: 10.1038/s41598-020-57464-2
Morrow, K. M., Bourne, D. G., Humphrey, C., Botté, E. S., Laffy, P., Zaneveld, J., et al. (2015). Natural volcanic CO2 seeps reveal future trajectories for host–microbial associations in corals and sponges. ISME J. 9, 894–908. doi: 10.1038/ismej.2014.188
Morrow, K. M., Moss, A. G., Chadwick, N. E., and Liles, M. R. (2012). Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability. Appl. Environ. Microbiol. 78, 6438–6449. doi: 10.1128/AEM.01162-12
Neave, M. J., Apprill, A., Ferrier-Pagès, C., and Voolstra, C. R. (2016). Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 100, 8315–8324. doi: 10.1007/s00253-016-7777-0
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P., O’Hara, R. B., et al. (2013). Vegan: Community Ecology Package. R Package Version. 2.0-10. CRAN.
Oliver, T. A., and Palumbi, S. R. (2011). Do fluctuating temperature environments elevate coral thermal tolerance? Coral Reefs 30, 429–440. doi: 10.1007/s00338-011-0721-y
Oporto-Guerrero, T., Reyes-Bonilla, H., and Ladah, L. B. (2018). Presence of the reef-building coral, Porites panamensis, in a shallow hydrothermal field in the Gulf of California. Mar. Biodivers. 48, 703–708. doi: 10.1007/s12526-016-0546-6
Oprandi, A., Montefalcone, M., Morri, C., Benelli, F., and Bianchi, C. N. (2019). Water circulation, and not ocean acidification, affects coral recruitment and survival at shallow hydrothermal vents. Estuar. Coast. Shelf Sci. 217, 158–164. doi: 10.1016/j.ecss.2018.11.017
Palumbi, S. R., Barshis, D. J., Traylor-Knowles, N., and Bay, R. A. (2014). Mechanisms of reef coral resistance to future climate change. Science 344, 895–898. doi: 10.1126/science.1251336
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Paz-García, D. A., Chávez-Romo, H. E., Correa-Sandoval, F., Reyes-Bonilla, H., López-Pérez, A., Medina-Rosas, P., et al. (2012). Genetic connectivity patterns of corals Pocillopora damicornis and Porites panamensis (Anthozoa: Scleractinia) along the west coast of Mexico. Pacific Sci. 66, 43–61. doi: 10.2984/66.1.3
Pereira de Castro, A., Dias Araújo, Jr S, M Reis, A. M., Moura, R. L., Francini-Filho, R. B., and Pappas, Jr G, et al. (2010). Bacterial community associated with healthy and diseased reef coral Mussismilia hispida from eastern Brazil. Microb. Ecol. 59, 658–667. doi: 10.1007/S00248-0
Pernice, M., Raina, J. B., Rädecker, N., Cárdenas, A., Pogoreutz, C., and Voolstra, C. R. (2020). Down to the bone: the role of overlooked endolithic microbiomes in reef coral health. ISME J. 14, 325–334. doi: 10.1038/s41396-019-0548-z
Prol-Ledesma, R. M., Canet, C., Torres-Vera, M. A., Forrest, M. J., and Armienta, M. A. (2004). Vent fluid chemistry in Bahía Concepción coastal submarine hydrothermal system, Baja California Sur, Mexico. J. Volcanol. Geotherm. Res. 137, 311–328. doi: 10.1016/j.jvolgeores.2004.06.003
Putnam, H. M., Davidson, J. M., and Gates, R. D. (2016). Ocean acidification influences host DNA methylation and phenotypic plasticity in environmentally susceptible corals. Evol. Appl. 9, 1165–1178. doi: 10.1111/eva.12408
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rachmawati, R. (2018). Differential Responses of Coral-Associated Microbiomes to Elevated Temperatures Across the Indonesian Archipelago at Species, Local and Regional Scales. Available online at: https://escholarship.org/uc/item/79t0h8b0 (accessed July 20, 2020).
Reigel, A. M., Owens, S. M., and Hellberg, M. E. (2020). Reducing host DNA contamination in 16S rRNA gene surveys of anthozoan microbiomes using PNA clamps. Coral Reefs 39, 1817–1827. doi: 10.1007/s00338-020-02006-5
Ritchie, K. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Rodríguez-Marconi, S., De la Iglesia, R., Díez, B., Fonseca, C. A., Hajdu, E., and Trefault, N. (2015). Characterization of bacterial, archaeal and eukaryote symbionts from Antarctic sponges reveals a high diversity at a three-domain level and a particular signature for this ecosystem. PLoS One 10:e0138837. doi: 10.1371/journal.pone.0138837
Rohwer, F., Seguritan, V., Azam, F., and Knowlton, N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Röthig, T., Ochsenkühn, M. A., Roik, A., van der Merwe, R., and Voolstra, C. R. (2016). Long-term salinity tolerance is accompanied by major restructuring of the coral bacterial microbiome. Mol. Ecol. 25, 1308–1323. doi: 10.1111/mec.13567
Rowan, R. (2004). Thermal adaptation in reef coral symbionts. Nature 430, 742–742. doi: 10.1038/430742a
Saavedra-Sotelo, N. C., Calderon-Aguilera, L. E., Reyes-Bonilla, H., Paz-García, D. A., López-Pérez, R. A., Cupul-Magaña, A., et al. (2013). Testing the genetic predictions of a biogeographical model in a dominant endemic Eastern Pacific coral (Porites panamensis) using a genetic seascape approach. Ecol. Evol. 3, 4070–4091. doi: 10.1002/ece3.734
Shamberger, K. E. F., Cohen, A. L., Golbuu, Y., McCorkle, D. C., Lentz, S. J., and Barkley, H. C. (2014). Diverse coral communities in naturally acidified waters of a Western Pacific reef. Geophys. Res. Lett. 41, 499–504. doi: 10.1002/2013GL058489
Shaver, E. C., Shantz, A. A., McMinds, R., Burkepile, D. E., Vega Thurber, R. L., and Silliman, B. R. (2017). Effects of predation and nutrient enrichment on the success and microbiome of a foundational coral. Ecology 98, 830–839. doi: 10.1002/ecy.1709
Slavov, N., Budnik, B. A., Schwab, D., Airoldi, E. M., and van Oudenaarden, A. (2014). Constant growth rate can be supported by decreasing energy flux and increasing aerobic glycolysis. Cell Rep. 7, 705–714. doi: 10.1016/j.celrep.2014.03.057
Strand, R., Whalan, S., Webster, N. S., Kutti, T., Fang, J. K. H., Luter, H. M., et al. (2017). The response of a boreal deep-sea sponge holobiont to acute thermal stress. Sci. Rep. 7:1660. doi: 10.1038/s41598-017-01091-x
Sunagawa, S., Woodley, C. M., and Medina, M. (2010). Threatened corals provide underexplored microbial habitats. PLoS One 5:e9554. doi: 10.1371/journal.pone.0009554
Thurber, R. V., Willner-Hall, D., Rodriguez-Mueller, B., Desnues, C., Edwards, R. A., Angly, F., et al. (2009). Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 11, 2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x
Torda, G., Donelson, J. M., Aranda, M., Barshis, D. J., Bay, L., Berumen, M. L., et al. (2017). Rapid adaptive responses to climate change in corals. Nat. Clim. Chang. 7, 627–636. doi: 10.1038/nclimate3374
Toth, L. T., Stathakopoulos, A., Kuffner, I. B., Ruzicka, R. R., Colella, M. A., and Shinn, E. A. (2019). The unprecedented loss of Florida’s reef−building corals and the emergence of a novel coral−reef assemblage. Ecology 100:e02781. doi: 10.1002/ecy.2781
Trembath-Reichert, E., Case, D. H., and Orphan, V. J. (2016). Characterization of microbial associations with methanotrophic archaea and sulfate-reducing bacteria through statistical comparison of nested Magneto-FISH enrichments. PeerJ 4:e1913. doi: 10.7717/peerj.1913
Vallon, O., and Spalding, M. H. (2009). Chapter 4 - Amino Acid Metabolism,” in The Chlamydomonas Sourcebook (2nd Edn) eds E. H. Harris, D. B. Stern, and G. B. Witman (London: Academic Press), 115–158.
van Oppen, M. J. H., Bongaerts, P., Frade, P., Peplow, L. M., Boyd, S. E., Nim, H. T., et al. (2018). Adaptation to reef habitats through selection on the coral animal and its associated microbiome. Mol. Ecol. 27, 2956–2971. doi: 10.1111/mec.14763
Verbruggen, H., Marcelino, V. R., Guiry, M. D., Cremen, M. C. M., and Jackson, C. J. (2017). Phylogenetic position of the coral symbiont Ostreobium (Ulvophyceae) inferred from chloroplast genome data. J. Phycol. 53, 790–803. doi: 10.1111/jpy.12540
Wan, S., Norby, R. J., Ledford, J., and Weltzin, J. F. (2007). Responses of soil respiration to elevated CO2, air warming, and changing soil water availability in a model old-field grassland. Glob. Chang. Biol. 13, 2411–2424. doi: 10.1111/j.1365-2486.2007.01433.x
Wang, L., Cheung, M. K., Kwan, H. S., Hwang, J.-S., and Wong, C. K. (2015). Microbial diversity in shallow-water hydrothermal sediments of Kueishan Island, Taiwan as revealed by pyrosequencing. J. Basic Microbiol. 55, 1308–1318. doi: 10.1002/jobm.201400811
Wang, L., Shantz, A. A., Payet, J. P., Sharpton, T. J., Foster, A., Burkepile, D. E., et al. (2018). Corals and their microbiomes are differentially affected by exposure to elevated nutrients and a natural thermal anomaly. Front. Mar. Sci. 5:101. doi: 10.3389/fmars.2018.00101
Weber, L., DeForce, E., and Apprill, A. (2017). Optimization of DNA extraction for advancing coral microbiota investigations. Microbiome 5:18. doi: 10.1186/s40168-017-0229-y
Webster, N. S., Negri, A. P., Botté, E. S., Laffy, P. W., Flores, F., Noonan, S., et al. (2016). Host-associated coral reef microbes respond to the cumulative pressures of ocean warming and ocean acidification. Sci. Rep. 6:19324. doi: 10.1038/srep19324
Weiss, S., Xu, Z. Z., Peddada, S., Amir, A., Bittinger, K., Gonzalez, A., et al. (2017). Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5:27. doi: 10.1186/s40168-017-0237-y
Welsh, R. M., Rosales, S. M., Zaneveld, J. R., Payet, J. P., McMinds, R., Hubbs, S. L., et al. (2017). Alien vs. predator: bacterial challenge alters coral microbiomes unless controlled by Halobacteriovorax predators. PeerJ 5:e3315. doi: 10.7717/peerj.3315
Wingenter, O. W., Haase, K. B., Zeigler, M., Blake, D. R., Rowland, F. S., Sive, B. C., et al. (2007). Unexpected consequences of increasing CO2 and ocean acidity on marine production of DMS and CH2ClI: potential climate impacts. Geophys. Res. Lett. 34:L05710. doi: 10.1029/2006GL028139
Zaneveld, J. R., Burkepile, D. E., Shantz, A. A., Pritchard, C. E., McMinds, R., Payet, J. P., et al. (2016). Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat. Commun. 7:11833. doi: 10.1038/ncomms11833
Zaneveld, J. R., McMinds, R., and Vega Thurber, R. (2017). Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2:17121. doi: 10.1038/nmicrobiol.2017.121
Keywords: coral microbiome, Ostreobium, hydrothermal vents, Gulf of California, coral, symbioses, Porites
Citation: Reigel AM, Paz-García DA and Hellberg ME (2021) Microbiome of a Reef-Building Coral Displays Signs of Acclimation to a Stressful Shallow Hydrothermal Vent Habitat. Front. Mar. Sci. 8:652633. doi: 10.3389/fmars.2021.652633
Received: 12 January 2021; Accepted: 09 August 2021;
Published: 15 September 2021.
Edited by:
M. Pilar Francino, Fundación para el Fomento de la Investigación Sanitaria y Biomédica de la Comunitat Valenciana (FISABIO), SpainReviewed by:
Xiubao Li, Hainan University, ChinaYvonne Vallès, The University of the West Indies, Barbados
Copyright © 2021 Reigel, Paz-García and Hellberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael E. Hellberg, bWhlbGxiZUBsc3UuZWR1
 Alicia M. Reigel
Alicia M. Reigel David A. Paz-García
David A. Paz-García Michael E. Hellberg
Michael E. Hellberg