- 1School of Life Sciences, University of Essex, Colchester, United Kingdom
- 2Centre for Environment, Fisheries and Aquaculture Science (CEFAS), Lowestoft Laboratory, Lowestoft, United Kingdom
Despite many shipwrecks containing oil there is a paucity of studies investigating their impact on surrounding environments. This study evaluates any potential effect the World War II shipwreck HMS Royal Oak is having on surrounding benthic sediments in Scapa Flow, Scotland. HMS (Her Majesty’s Ship) Royal Oak sank in 1939, subsequently leaked oil in the 1960s and 1990s, and is estimated to still hold 697 tonnes of fuel oil. In this study, sediments were analysed, over a 17.5 cm depth profile, along a 50–950 m cruciform transect away from the shipwreck. Analysis of polycyclic aromatic hydrocarbons (PAHs) revealed low concentrations (205.91 ± 50.15 μg kg–1 of dry sediment), which did not significantly differ with either distance from the shipwreck or sediment depth. PAH concentrations were well below the effects-range low (ERL) for the OSPAR (Oslo/Paris convention for the Protection of the Marine Environment of the North-East Atlantic) maritime area. The average Pyrogenic Index, in sediments around HMS Royal Oak, was 1.06 (±0.34), indicating PAHs were pyrogenic rather than petrogenic. Moreover, analysis of sediment microbiomes revealed no significant differences in bacterial community structure with distance from the shipwreck, with extremely low levels of obligate hydrocarbonoclastic bacteria (OHCB; 0.21% ± 0.54%). Both lines of evidence suggest that sampled sediments are not currently being impacted by petrogenic hydrocarbons and show no long-term impact by previous oil-spills from HMS Royal Oak.
Introduction
In the last two decades, the focus on evaluating the environmental impact from shipwrecks has significantly increased (Landquist et al., 2013), especially those from the two World Wars, which constitute >8,600 shipwrecks in the world’s seas (Monfils et al., 2006). There is particular concern regarding oil pollution from shipwrecks (Michel et al., 2005; Faksness et al., 2015; Amir-Heidari et al., 2019) because they contain an estimated 2.5–20.4 million tonnes of crude oil and petroleum products (Landquist et al., 2017a). Crude oil and its petroleum derivatives can have toxic effects, especially oils with higher proportions of volatile organic compounds (VOCs) and polycyclic aromatic hydrocarbons (PAHs) (IARC, 1987; National Toxicology Program, 2011; Manzetti, 2013). Oil-spill hydrocarbon analysis tends to focus on PAHs due to their persistence, carcinogenic, and mutagenic properties (Lehr and Jerina, 1977).
On the 14th October 1939, 6 weeks after the start of World War II, the British battleship HMS (Her Majesty’s Ship) Royal Oak was torpedoed by a German submarine and sank to the bottom of Scapa Flow (Orkney Islands, Scotland; Figure 1). As many lives were lost, HMS Royal Oak is protected under the Protection of Military Remains Act 1986. The 189 m long and 26,444 tonne battleship lies upside down, approximately 30 m below the surface of the water at its deepest point and 4.9 m at its shallowest point. HMS Royal Oak sank with approximately 3,000 tonnes of fuel oil on board. It is likely that a large fraction of oil was spilled during the incident, however a significant proportion of oil remained on board. Leaking oil was first observed in the 1960s, and then again during the late 1990s (Hill, 2019). Efforts to remove the fuel oil from the vessel’s tanks began in 2006, and by 2010, 1,600 tonnes had been removed (Marine, 2010), though up to 697 tonnes of oil is thought to remain (Hill, 2019). Whilst any visible oil leakage from HMS Royal Oak in Scapa Flow has subsided, it is unknown whether there is a legacy of hydrocarbon contamination in the sediments.
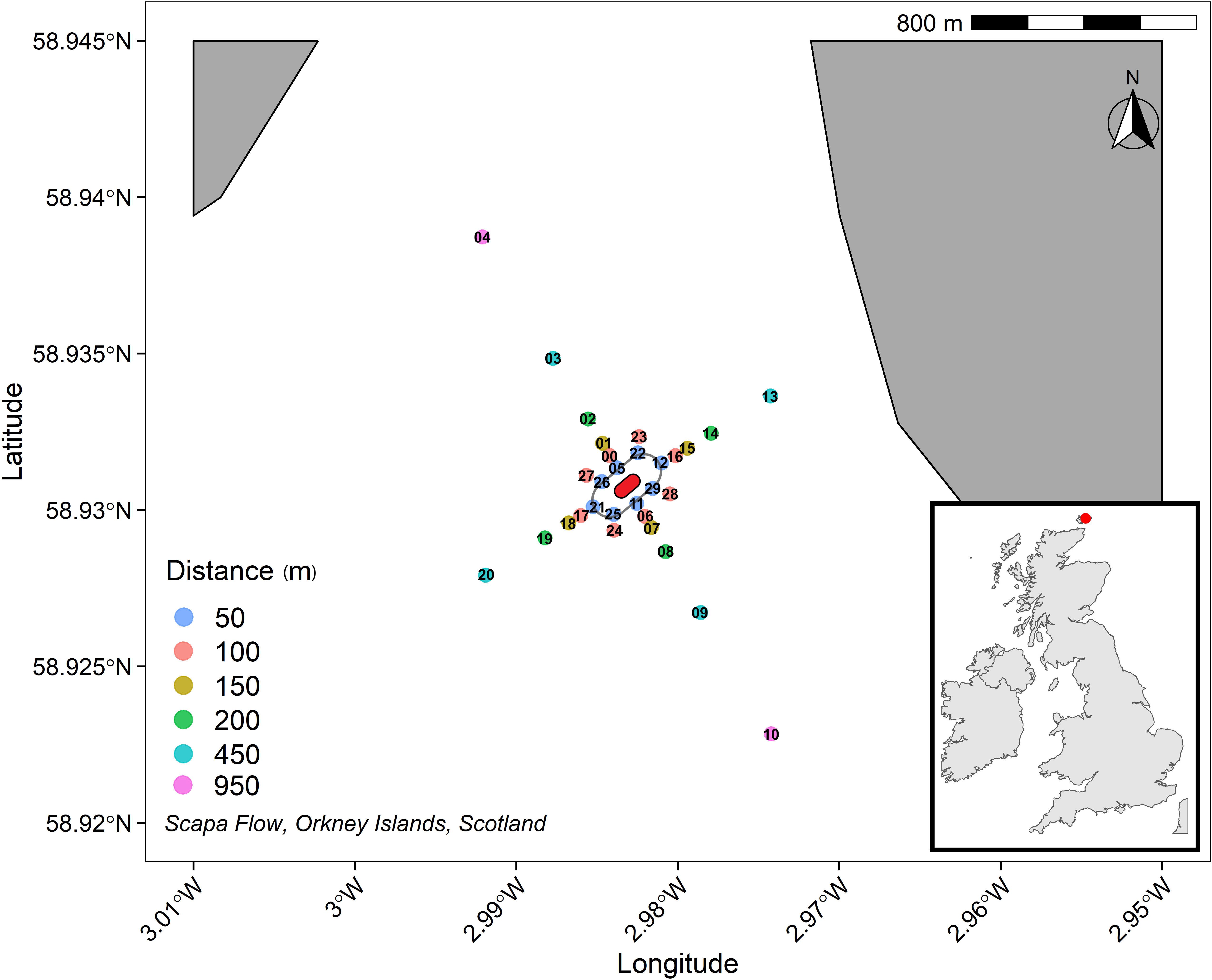
Figure 1. 950 m cruciform transect (where those sediments sampled further away act as controls) centred on HMS Royal Oak shipwreck (approximation, centre, red), labels refer to sample codes as detailed in Supplementary Table 1. A safety boundary (grey oval) around HMS Royal Oak meant sediment sampling commenced 50 m from the shipwreck.
Certain microbes use a range of hydrocarbons, found in crude oil and its derivatives, as a source of carbon and energy. These include the obligate hydrocarbonoclastic bacteria (OHCB), which use hydrocarbons as an almost exclusive source of carbon and energy (Yakimov et al., 2007). Hydrocarbon-degrading bacteria are particularly important responders to oil-pollution as they remove a significant proportion of oil and act as bioindicators of hydrocarbon-pollution (Head et al., 2006; Lozada et al., 2014). Oil pollution will often significantly alter marine microbial community composition, whereby a species richness and diversity decreases as selection for oil-degrading bacteria occurs (McGenity et al., 2012; Thomas et al., 2020). After an oil spill, there is often an initial increase in the abundance of alkane-degrading microbes (e.g., Alcanivorax/Oleispira) followed by growth of PAH-degraders (e.g., Cycloclasticus) (Head et al., 2006; Ribicic et al., 2018). The latter of which, Cycloclasticus (Dyksterhouse et al., 1995; Gutierrez et al., 2013) would be expected to be in high abundance where there are increased levels of petrogenic PAHs. Hamdan et al. (2018) found that World War II and 19th century shipwrecks in the Gulf of Mexico influenced microbial diversity in surface sediments 2 m from the shipwrecks. Sediments around two of the shipwrecks evaluated by Hamdan et al. (2018) had no significant effect on microbial diversity and composition. It was suggested that either the greater depths of these shipwrecks, or the fact they were both impacted by the Macondo oil spill, obscured any potential impacts of the shipwrecks on microbial community composition. Scapa Flow is a large embayment approximately 24 by 13 km, surrounded by the Orkney Islands, situated north east of Scotland, United Kingdom (Figure 1) and was the main British fleet base during both World Wars. Much of the marine microbial research conducted in Scapa Flow has focused on microbial mats in sheltered beaches and photosynthetic bacteria (Herbert, 1985; van Gemerden et al., 1989; Wieland et al., 2003). Despite the large number of wrecks (>150; Muir, 2020) and oil pollution events within Scapa Flow, there is a paucity of data on sediment microbial communities of this region. Indeed, there are few studies investigating how shipwrecks impact indigenous microbial communities or evaluating their actual or potential oil pollution. Most of the current literature researching shipwrecks focuses on archaeological wood (Liu et al., 2018), artefacts (Li et al., 2018), shipwrecks as artificial reefs (Church et al., 2009; Mugge et al., 2019), and microbially induced corrosion (Russell et al., 2004). Analysis of microbial communities in oil-contaminated environments can provide valuable insight into ecosystem function and resilience. Moreover, harnessing such data allows for the refinement of post-incident modelling (Kirby et al., 2018).
A modelling approach (Landquist et al., 2013, 2014, 2017a,b) allows the potential for oil leaks from shipwrecks to be predicted. However, in situ sampling of the environment around a shipwreck is ultimately needed to quantify and evaluate whether such shipwrecks pose a threat to the surrounding environment. Furthermore, analyses of in situ microbial communities can provide data that are valuable in the design of post-incident monitoring guidelines (Kirby et al., 2018). This study evaluates historic impact of oil contamination from the shipwreck HMS Royal Oak on the surrounding benthic microbial communities. The objective of this study was to determine whether fuel oil from HMS Royal Oak displays elevated concentrations in surrounding benthic sediments, by determining both the concentration and source of sediment PAHs over a 950 m radius of the shipwreck (where those sediments sampled further away act as controls). Furthermore, 16S rRNA gene qPCR and amplicon libraries were used to determine any effects of the shipwreck on sediment bacterial community composition, and to quantify any hydrocarbon-degrading or sentinel bacteria associated with oil pollution.
Materials and Methods
Sampling Campaign
Sediment cores were sampled using an OSIL (Ocean Scientific International Ltd.) extended box corer from 30 sites located on a cruciform transect (four transects: North East, South East, South West, and North West) centred around HMS Royal Oak shipwreck, employed to capture the dispersion trajectory of any sediment contaminants. Samples were taken 50, 100, 150, 200, 450, and 950 m from the shipwreck along the transects (Scapa Flow, Scotland, United Kingdom; 58°55′50″N, 02°58′58″W″), between the 3rd and 6th November 2019 (Figure 1). No other shipwrecks or scuttling were observed, or are known to be located, within the sampled transect area. HMS Royal Oak is protected under the Protection of Military Remains Act 1986; therefore a 50 m safety boundary was placed around the shipwreck. Transects were to 950 m, allowing for sufficient sampling coverage and comparison between those closer to the shipwreck and control sediments further away (i.e., 450–950 m). At each of the 30 sites, samples were taken from 2.5, 7.5, 12.5, and 17.5 cm sediment depth. See Supplementary Table 1 for full sample details. The box corer and all subsampling equipment were pentane-rinsed between sites to prevent cross-station contamination. A CTD (Conductivity, Temperature, and Depth) sensor was deployed at each sampling site; no significant temporal or spatial differences were recorded regarding temperature (10.35°C) or salinity (35.27 PSU). Sediments within the survey area comprised poorly sorted fine muddy sands. Scapa Flow experiences limited tidal and/or wind-generated water flows. Sediment Profile Imagery (SPI) provided accompanying evidence of this (Supplementary Figure 4) and revealed that oxidised sediments were limited to the surficial region (0 to ∼2.5 to 5 cm), with clear anoxia below. There were no signs of oil contamination during the survey, either visible on the sea surface or in the sediment samples collected.
Polycyclic Aromatic Hydrocarbon Analysis
Homogenised sediment samples were extracted using alkaline saponification prior to filtering and subsequent liquid-liquid extraction into pentane (Kelly et al., 2000). Samples were passed through an alumina chromatography column to remove residual polar compounds and concentrated to 1 ml. Quantification for PAHs was performed using surrogate standards and five calibrations levels (range 25–500 ng ml–1). Surrogate and recovery standards were formed of deuterated PAHs [naphthalened8, acenaphthylened8, anthracened10, dibenzothiophened8, pyrened10, benzo(a)anthracened12, benzo(a)pyrened12, and dibenz(a,h)anthracened14], and were added to each sample prior to quantification on an Agilent 6890 Gas Chromatography system coupled with an Agilent 5975 Mass Spectrometer with Triple-Axis detector, operating at 70 eV in positive-ion mode, using conditions previously described by Kelly et al. (2000). For quality control, a calibration check [(CRM) HS-6] and a blank were analysed with each sample batch (∼four samples), the limit of detection was 0.1 μg kg–1 dry weight sediment for each PAH compound/class, and the extraction efficiency was measured at 99%.
A suite of alkylated and parent PAHs, including compounds with both petrogenic and combustion sources, were then determined. This suite comprised naphthalene and the C1-C3 methylnaphthalenes, acenaphthylene, acenaphthene, fluorene, phenanthrene, anthracene, C1-C2 methylphenanthrenes/anthracenes, dibenzothiophene and C1-C3 methyldibenzothiophenes, fluoranthene, pyrene, C1 methylpyrenes, benz[a]anthracene, chrysene, C1 methylchrysene, 1,2-benzodiphenylene sulphide, benzo[b]fluoranthene, benzo[k]fluoranthene, benzo[e]pyrene, benzo[a]pyrene, perylene, indeno[1,2,3-cd]pyrene, benzo[ghi]perylene and dibenz[a,h]anthracene. The % oil, within a sample, is calculated by the proportion of low molecular weight (2− and 3-ring) and alkylated (C1-C3 methylnaphthalenes, C1-C2 methylphenanthrenes/anthracenes, C1-C3 methyldibenzothiophenes, C1 methylpyrenes, and C1 methylchrysene) PAHs. The % combustion is the remaining proportion of PAHs within the sample.
qPCR Analysis of Bacterial 16S rRNA Genes
DNA was extracted from 0.25 g of thawed sediment samples with a DNeasy PowerSoil Kit (Qiagen), according to the manufacturer’s instructions. The primers used for quantification of bacterial 16S rRNA were 341f – CCTACGGGNGGCWGCAG and 785r – GACTACHVGGGTATCTAATCC (Klindworth et al., 2013). qPCR reactions were performed using a CFX384TM Real-Time PCR Detection System (BioRad) using reagents, cycle conditions, and standards as previously described (McKew and Smith, 2015; Tatti et al., 2016). Inspection of standard curves showed that all assays produced satisfactory efficiency (74%) and R2 values (>0.99).
Amplicon Sequencing and Bioinformatics
Amplicon libraries were prepared, as per Illumina instructions by a 25-cycle PCR. PCR primers were the same as those used for qPCR but flanked with Illumina overhang sequences. A unique combination of Nextera XT v2 Indices (Illumina) were added to PCR products from each sample, via an 8-cycle PCR. PCR products were quantified using Quant-iT PicoGreen dsDNA Assay Kit (TheromFisher Scientific) and pooled in equimolar concentrations. Quantification of the amplicon libraries was determined via NEBNext® Library Quant Kit for Illumina (New England BioLabs Inc.), prior to sequencing on the Illumina MiSeq® platform, using a MiSeq® 600 cycle v3 reagent kit and 20% PhiX sequencing control standard. Raw sequence data have been submitted to the European Nucleotide Archive database under accession number PRJEB37440. Sequence output from the Illumina MiSeq platform were analysed within BioLinux (Field et al., 2006), using a bioinformatics pipeline as described by Dumbrell et al. (2016). Forward sequence reads were quality trimmed using Sickle (Joshi and Fass, 2011) prior to error correction within SPAdes (Nurk et al., 2013) using the BayesHammer algorithm (Nikolenko et al., 2013). The quality filter and error corrected sequence reads were dereplicated, sorted by abundance, and clustered into OTUs (Operational Taxonomic Units) at the 97% level via VSEARCH (Rognes et al., 2016). Singleton OTUs were discarded, as well as chimeras using reference based chimera checking with UCHIME (Edgar et al., 2011). Taxonomic assignment was conducted with RDP Classifier (Wang et al., 2007). Non-locus-specific, or artefactual, OTUs were discarded prior to statistical analyses, along with any OTUs that had <70% identity with any sequence in the RDP database.
Statistical Analysis
The average sequence reads per sediment sample was 65,239 (range from 25,232 to 146,401); prior to community analysis, sequence data were rarefied to the lowest library sequence value. Data were first tested for normality (Shapiro-Wilk’s test), those data which were normally distributed were tested for significance with ANOVAs or appropriate linear models. Non-normally distributed data were analysed using appropriate GLMs (Generalised Linear Models) as follows. The relative abundance of OTUs or genera in relation depth, distance, or transect were modelled using multivariate negative binomial GLMs (Wang et al., 2010). Here, the number of sequences in each library was accounted for using an offset term, as described previously (Alzarhani et al., 2019). The abundance of bacterial 16S rRNA gene copies was modelled using negative binomial GLMs. The concentration of PAHs, and the difference between means and threshold values, was also modelled using negative binomial GLMs (Venables and Ripley, 2002). The significance of model terms was assessed via likelihood ratio tests. Correlations were performed using Spearman rank correlation with an alpha of 0.05. The Ecological Index of Hydrocarbon Exposure (EIHE; Lozada et al., 2014) was calculated using the script available at the ecolFudge GitHub page1 (Clark, 2019) and EIHE values modelled using poisson GLMs. The EIHE aims to establish hydrocarbon exposure in an environmental sample by calculating the proportion of 16S rRNA sequences within a bacterial community assigned to bacterial genera with hydrocarbon-biodegradation potential; the genera are detailed in the paper by Lozada et al. (2014). It should be noted that samples from the compared studies (Figure 5D) come from aerobic surface sediments and thus only surface sediments from this study were used in the comparison.
All statistical analyses were carried out in R3.6.1 (R Development Core Team, 2011) using a variety of packages available through the references (Venables and Ripley, 2002; Hope, 2013; Wilke, 2015, 2020; Becker et al., 2016; Auguie, 2017; Oksanen et al., 2019; Hvitfeldt, 2020; Kassambara, 2020; Lenth, 2020; Pedersen, 2020). All plots were constructed using the “ggplot2” (Bodenhofer et al., 2011) and “patchwork” (Pedersen, 2019) R packages.
Results and Discussion
Sediment PAHs Originate From Pyrogenic Sources
The concentration of PAHs (containing 2− to 6-ring parent PAHs and alkylated derivatives (C1-C3)-naphthalenes, (C1-C2)-phenanthrenes, (C1-C3)-dibenzothiophenes, and (C1)-pyrenes) in all sediment samples, around HMS Royal Oak shipwreck, ranged from 121 to 348 μg kg–1 of dry sediment (Figure 2). There were no significant differences (p > 0.05) in PAH concentrations between core depths (2.5, 7.5, 12.5, and 17.5 cm) or over distance (50–950 m) from the shipwreck. When data from transects were pooled there was no correlation between PAH concentration and distance from the wreck (R2 = 0.00019 p > 0.05) nor with sediment depth (R2 = 0.036 p > 0.05) (see Supplementary Figure 1). In comparing PAH concentrations, sediments near a World War II cargo ship, Nordvard (30 m depth near the Norwegian harbour of Moss), contained PAH concentrations ranging from 3,000 to 25,000 μg kg–1 of dry sediment (Ndungu et al., 2017). PAH concentrations in sediments around HMS Royal Oak were well within typical levels of PAHs found in United Kingdom coastal sediments. For example, muddy sediments around Scotland (Woodhead et al., 1999), contained PAHs from 75 to 1,212 μg kg–1 of dry sediment. Additionally, the observed PAH concentrations within sediments around HMS Royal Oak were well below the effects-range low (ERL) and within background concentration (BC) levels (3,340 and 212 μg kg–1 of dry sediment for ERL and BC, respectively) for sediments in the OSPAR (Oslo/Paris convention for the Protection of the Marine Environment of the North-East Atlantic) maritime area (OSPAR Commission, 2009). Therefore, the observed PAH concentrations of sediments around HMS Royal Oak would seldom cause negative effects in marine organisms (OSPAR Commission, 2017).
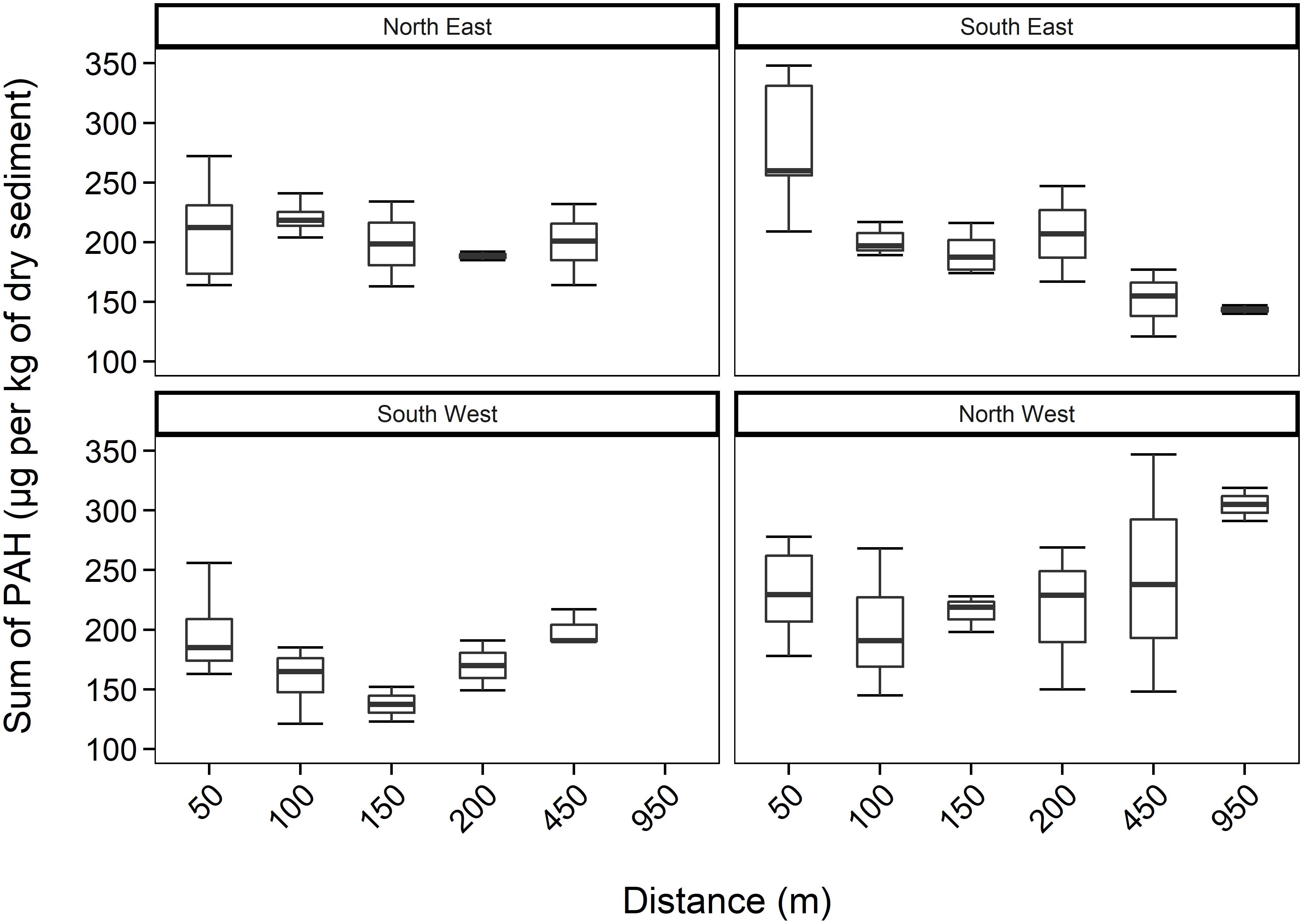
Figure 2. Total PAH concentrations (mean ± SE, n = 2) within sediments, including all depths, over 950 m transects (where those sediments sampled further away act as controls) in four different directions from HMS Royal Oak shipwreck; located in Scapa Flow, Scotland, United Kingdom, November 2019.
As well as evaluating the concentration of environmental PAHs it is also important to determine their origin. Despite PAH concentrations being below the ERL and within BC for the area, evaluating the source of PAHs is especially important considering the negative relationship between PAH concentration and distance from the shipwreck in the South East direction. PAHs generally enter the environment through three major routes: pyrogenic, petrogenic, or biogenic (Soclo et al., 2000). Pyrogenic PAHs come from the combustion of organic substances at high temperature and low oxygen concentrations, such as industrial thermal cracking and the incomplete combustion of petroleum in motor engines, or from forest fires. Petrogenic PAHs are formed over millions of years at relatively lower temperatures during crude oil maturation and enter the sea from natural seeps or spills and leaks from oil extraction, transportation or storage, or shipwrecks. Finally, biogenic PAHs originate from specific algae and bacteria, or from the degradation of vegetative material (Abdel-Shafy and Mansour, 2016). The evaluation of PAH ratios can provide an accurate identification of the PAH source (Zakaria et al., 2002; Wang and Fingas, 2003; Hussain et al., 2016). Biogenic PAHs are identified by a number of hydrocarbon bioindicators, including a unique n-alkane distribution with an increased abundance of odd carbon-number alkanes over even carbon-number alkanes (Peters et al., 2004; Wang et al., 2009) as well as an increased abundance of PAH perylene (Venkatesan, 1988). Petrogenic PAHs are distinct from pyrogenic PAHs in their chemical composition, with a dominance of C1 to C4 alkylated naphthalenes, phenanthrenes, dibenzothiophenes, fluorenes, and chrysenes (Wang and Fingas, 2005). Furthermore, the abundance of LMW (low molecular weight) 2− to 3-ring PAHs is generally much higher than HMW (high molecular weight) 4− to 6-ring PAHs in petrogenic PAHs. Such analysis can determine whether sediment PAHs originated naturally or from anthropogenic inputs (i.e., fuel oil from shipwrecks) and is important as petrogenic PAHs have increased bioavailability to organisms, especially in filter feeders (Baumard et al., 1998).
Generally, a proportion of LMW (2− and 3-ring) and alkylated (C1–C3 methylnaphthalenes, C1–C2 methylphenanthrenes/anthracenes, C1–C3 methyldibenzothiophenes, C1 methylpyrenes, and C1 methylchrysene) PAHs to total PAHs above 70% would indicate that the PAHs are petrogenic (Law et al., 1999). Our data imply that sediment PAHs around HMS Royal Oak were pyrogenic, as the proportion of LMW and alkylated PAHs to total PAHs was significantly below the 70% threshold (coef. 4.02, z 285.5, p < 0.001); with an overall average of 55% (± 5%) (Figure 3 and see Supplementary Table 1). Additionally, the proportion of perylene to total 5-ringed aromatics averaged 4% (±0.6%; Figure 3 and Supplementary Tables 1, 3); significantly below the 10% threshold (coef. 1.40, z 26.89, p < 0.001) that is indicative of pyrogenic PAHs (Pereira et al., 1999; Lima et al., 2003; Wang et al., 2009). An increased abundance (i.e., >10%) of perylene compared to other 5-ringed aromatics is indicative of biogenic PAHs; as perylene is produced by early diagenesis mechanisms on organic matter within sediments (Peters et al., 2004; Wang et al., 2014). Only one sample (RO27, 12.5 cm depth) met the 70% threshold (% LMW/alkylated PAHS of total PAHs) for petrogenic PAHs but had a perylene to 5-ringed aromatics ratio of only 4.76%. There are many other PAH ratios that can distinguish pyrogenic from petrogenic PAHs. Widely used ratios include anthracene/(anthracene + phenanthrene) (Baumard et al., 1998; Qiao et al., 2006), benzo[a]anthracene/[benzo(a)anthracene + chrysene] (Yunker et al., 2002; Akyüz and Çabuk, 2010), fluoranthene/(fluoranthene + pyrene) (Guinan et al., 2001; De La Torre-Roche et al., 2009), and indeno[1,2,3-cd]pyrene/[indeno(1,2,3-cd)pyrene + benzo(ghi)perylene] (Yunker et al., 2002). All four ratios were calculated for PAH data from sediments sampled around HMS Royal Oak; all PAH ratio confirmed that sediment PAHs were pyrogenic, with no significant difference with transect distance, direction, or over sediment depth (see Supplementary Table 2).
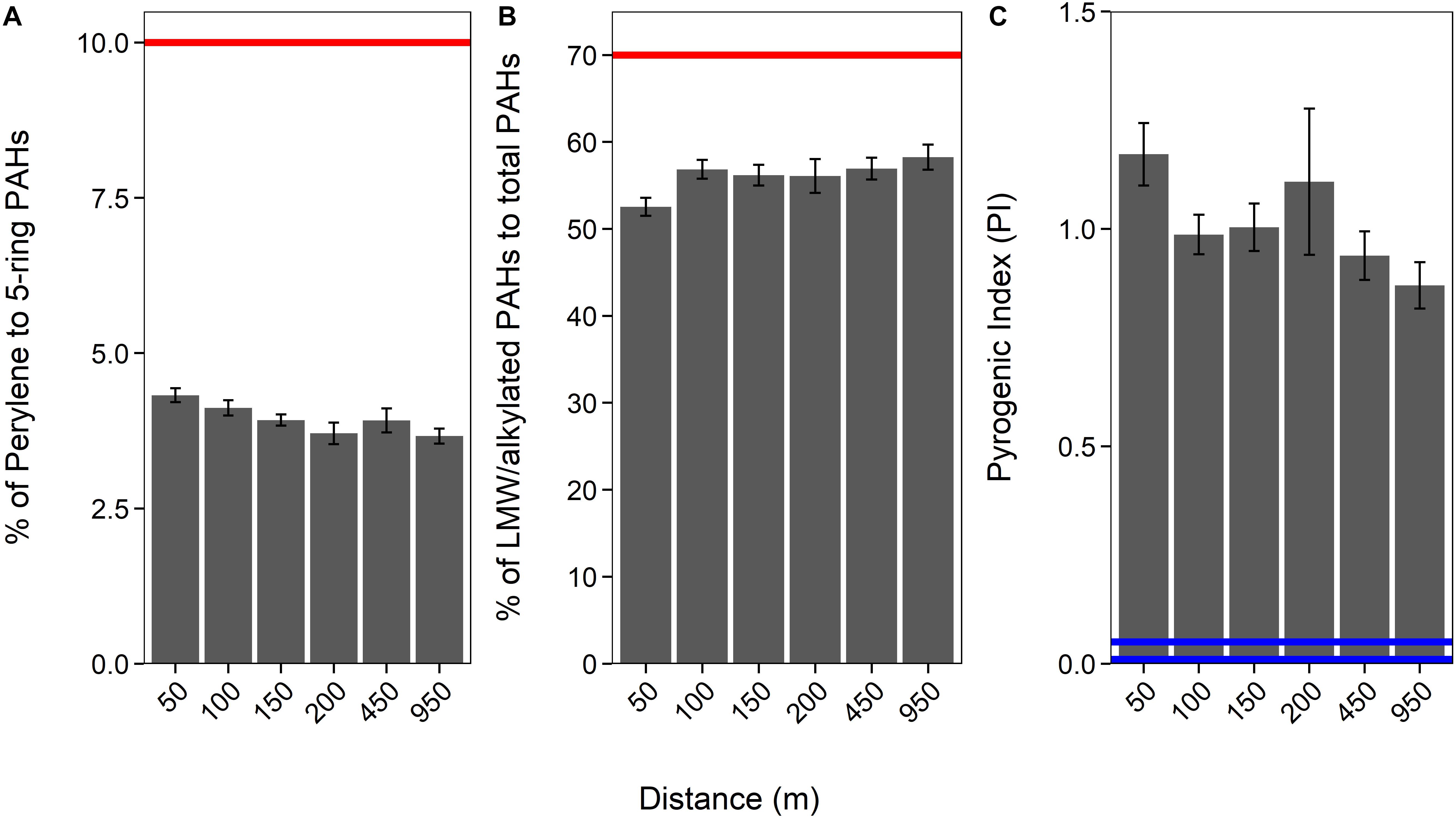
Figure 3. Proportion of perylene to 5-ringed aromatics (A), of LMW/alkylated PAHs to total PAHs (B), and the Pyrogenic Index [PI: Σ(3− to 6-ring EPA priority PAHs) / Σ(5 alkylated PAH series)] over a 950 m cruciform transect (“Distance”; where those sediments sampled further away act as controls), from HMS Royal Oak, Scapa Flow, November 2019. Red lines (A,B) represent the threshold at which PAHs are considered petrogenic. Blue lines (C) represent the points below which PAHs are considered petrogenic (0.01 = lighter refined products and most crude oils and 0.01–0.05 heavy oil and heavy fuel oils).
The accuracy of PAH fingerprinting techniques, such as the above, can be questioned when natural weathering processes and biodegradation may alter PAH distribution patterns. To this regard, Wang et al. (1999) created the “Pyrogenic Index” (PI), which is: Σ(3− to 6-ring EPA priority PAHs) / Σ(5 alkylated PAH series); the exact PAHs are available in the original literature. It is suggested this ratio provides better accuracy and less uncertainty than ratios that use individual or LMW PAHs, as the PI is subject to diminutive fluctuation due to changes in individual PAH concentrations. Long-term natural weathering processes and biodegradation have little impact on the ratio, whereas the ratio is significantly altered by combustion. Lighter refined products and most crude oils (i.e., petrogenic) demonstrate a PI of 0.01 while heavy oils and heavy fuel oils range from 0.01 to 0.05. The PI significantly increases for pyrogenic material, for example diesel soot samples were found to be within a range of 0.80–2.0 (Wang et al., 1999). The average PI, in sediments around HMS Royal Oak, was 1.06 (±0.34; Figure 3), significantly greater than the thresholds for petrogenic PAHs (coef. 1.04, z 30.67, p < 0.001). Therefore, considering the results of all three calculations, and the fact none of these results were significantly different over distance from the shipwreck, it can be assumed PAHs detected within sampled sediments around HMS Royal Oak were pyrogenic and not from oil that has historically leaked from the shipwreck.
Upon analysis of individual transect directions, a moderate negative correlation between PAH concentration and distance from the shipwreck was observed in the South East transect (R2 = −0.54 p < 0.01) whilst a moderate positive correlation between PAH concentration and distance from the shipwreck was observed in the North West transect (R2 = 0.46 p < 0.05). There was no signification relationship between PAH concentration and distance from the shipwreck in either the South West or North East transects. The moderate correlations between PAH concentrations and the South East and North West transect directions were not attributed to the shipwreck as the PAHs were determined to be of pyrogenic origin. Therefore, these correlations are likely to be natural variations in deposition of pyrogenic PAHS driven by water movement. The water is very shallow throughout Scapa Flow and around HMS Royal Oak (18–30 m), and therefore it is likely water movement is wind-driven, with current speeds often being <0.25 m s–1 (Ramsay and Brampton, 2000).
Bacterial 16S rRNA Gene Analysis Reveals Low Levels of Hydrocarbon-Degrading Bacteria
Whilst there was no evidence of petrogenic PAHs by GC-MS analysis, evaluating bacterial community composition provides the ability to highlight any localised effects of previous oil-contamination, and reveal any lasting effects caused by previous oil-contamination. Exposure to hydrocarbons can lead to adaptation within indigenous sediment bacterial communities and increases in hydrocarbon oxidising potential, priming the communities for any current or future oil-contamination (Leahy and Colwell, 1990). For example, increased abundance of known hydrocarbon-degrading bacteria (i.e., Alcanivorax and Cycloclasticus) has been observed in oil-contaminated coastal sediments, many months after hydrocarbons were undetectable (Thomas et al., 2020). Therefore, significantly higher abundance, and a broad diversity, of hydrocarbon-degrading bacteria in sediments close to the shipwreck compared to background levels (i.e., sediments sampled further away from the shipwreck), would indicate that any oil-contamination could be rapidly degraded.
Typically in environments impacted by oil, a large increase in the absolute abundance of Bacteria would be expected due to a selection for hydrocarbon-degrading bacteria, and secondary consumers (Head et al., 2006). For example, bacterial 16S rRNA gene abundance was ∼10-fold higher in coastal oiled sediments impacted by the Deep-Water Horizon oil-well-head blowout from Pensacola Beach, Florida, compared to clean sand samples (Kostka et al., 2011). In sediments across the 950 m transects, around HMS Royal Oak, the mean bacterial 16S rRNA gene abundance was 1.31 × 108 ± 7.04 × 107 copies/g of dry sediment. There were no significant differences (p > 0.05) observed over the 950 m distance from the shipwreck, nor by any transect direction (Supplementary Figures 2A,B), providing additional evidence that sediments localised around the ship, were not currently being contaminated with oil.
There was, however, a significant increase in 16S rRNA gene abundance (1.57-fold; coef. 0.26, t 2.60, p < 0.05) at 2.5 cm depth compared to the deeper sediments at 7.5, 12.5, and 17.5 cm (Supplementary Figure 2A). Differences in 16S rRNA gene bacterial sequence diversity (assigned to OTUs at a 97% similarity threshold) were driven primarily by sediment depth (R2 = 0.15, p < 0.001, n = 4; Figure 4). As sediments transitioned from aerobic to anaerobic beyond 2.5 cm depth, the bacterial community differences between 2.5 cm and the deeper sediments (7.5–17.5 cm) were primarily driven by significant increases in the relative abundance of anaerobic sulphate-reducing bacteria (e.g., Desulfatiglans spp., coef. 1.87, t 7.17, p < 0.001) and significant decreases in the relative abundance of aerobic ammonia-oxidising bacteria (e.g., Nitrosospira spp., coef. −1.72, t −12.18, p < 0.001). Proximity to the shipwreck had no significant effect on bacterial community composition (p > 0.05, n = 4; Supplementary Figure 3), suggesting no evidence of oil-related community changes and adaption within the sediments.
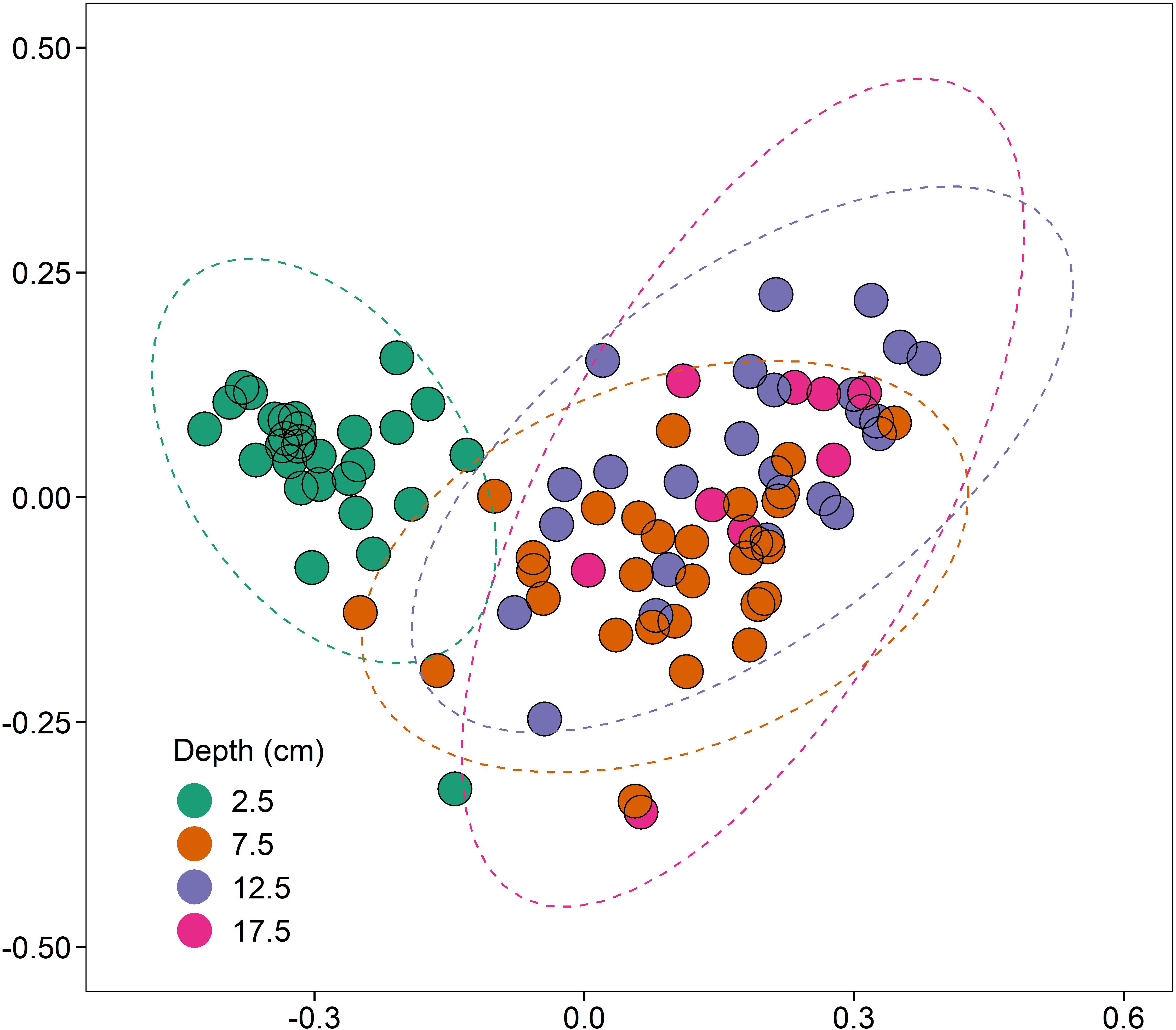
Figure 4. NMDS (non-metric multidimensional scaling) ordination (stress = 0.10), based on clustered bacterial 16S rRNA OTUs at a 97% similarity threshold, displaying the effect of depth (cm) on bacterial community composition (R2 = 0.15, p < 0.001, n = 4).
The Ecological Index of Hydrocarbon Exposure (EIHE) is an index from 0–1, whereby 1 represents 100% of 16S rRNA gene sequences assigned to bacterial genera with hydrocarbon-biodegradation potential (Lozada et al., 2014). The EIHE, within sediments around HMS Royal Oak, was on average 0.008 (± 0.022) over all sediment depths and distances from the shipwreck (Figures 5A–C). There were no significant differences (p > 0.05) in the EIHE between sediment depth, distance from the shipwreck, or direction of the transect (Figures 5A–C). The low index (0.012 ± 0.018) at the surface (2.5 cm) would suggest that the greater abundance of the bacterial 16S rRNA gene was not driven by the presence of hydrocarbon-degrading bacteria. The genera Bacillus, Mycobacterium, Shewanella, Sulfitobacter, and Vibrio made up a large proportion of sequences assigned to the EIHE (87%), in sediments around HMS Royal Oak (see Supplementary Table 4), all of which are genera that primarily contain species that do not degrade hydrocarbons. Compared to other studies of oil-polluted environments, an EIHE of 0.012 (± 0.018) in surface sediments is exceptionally low (see Figure 5D and Supplementary Table 3). Comparatively, a study within the Gulf of Mexico revealed that sediments around the shipwreck of the oil-tanker “Halo” (sunk May 1942) contained PAH concentrations ranging approximately 0–1,400 μg kg–1 of dry sediment respectively, resulting in an EIHE of 0.025 (see Figure 5 and Supplementary Table 3; Hamdan et al., 2018), twofold higher than the EIHE observed in surface sediments around HMS Royal Oak. Additionally, contaminated sediments sampled 5 days after the Agia Zoni II oil-spill, which contained average PAH concentrations of 210,000 μg kg–1 of dry sediment, resulted in an EIHE of 0.52, 42-fold higher than the EIHE observed in surface sediments of the present study (see Figure 5D and Supplementary Table 3; Thomas et al., 2020).
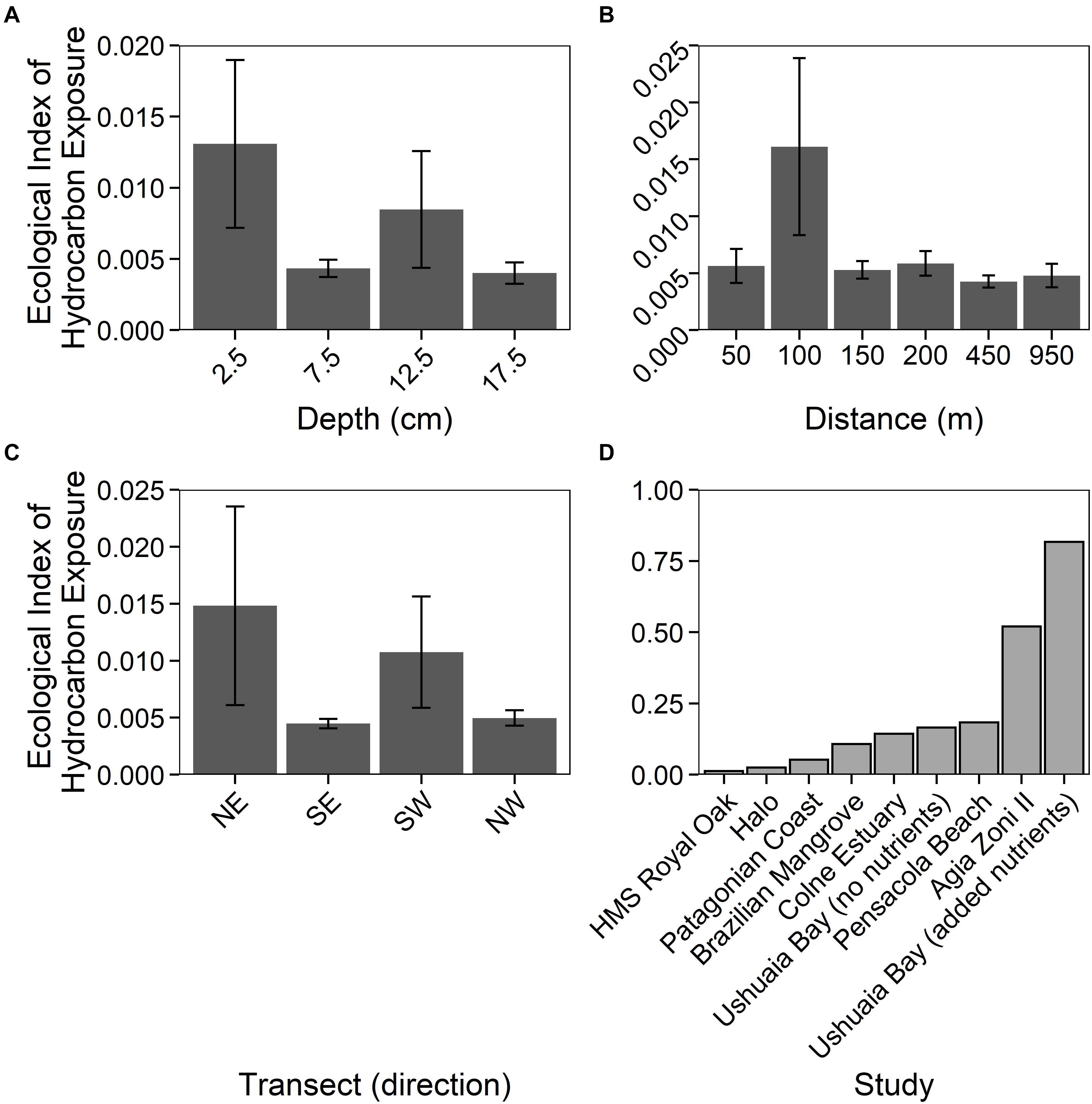
Figure 5. Ecological Index of Hydrocarbon Exposure (EIHE) (ratio up to 1) representing relative abundance of oil-degrading/associated Bacteria, (Lozada et al., 2014) measurements (±SE, n = 4), for sediments sampled around the shipwreck HMS Royal Oak (located in Scapa Flow, Scotland, United Kingdom, November 2019), over a 17.5 cm depth profile (A), a 950 m transect (where sediments further away act as controls) (B), and given the transect direction (C). Additionally, a comparison between the HMS Royal Oak EIHE and other studies investigating oil-polluted environments (D): “HMS Royal Oak” (present study, surface sediments), “Halo” (Hamdan et al., 2018), “Patagonian Coast” (Lozada et al., 2014), “Brazilian Mangrove” (Dos Santos et al., 2011), “Colne Estuary” (Coulon et al., 2012), “Ushuaia Bay (no nutrients)”, “Pensacola Beach” (Kostka et al., 2011), “Agia Zoni II” (Thomas et al., 2020) and “Ushuaia Bay (added nutrients)” (Guibert et al., 2012).
Only a minority (5 out of 63) of the genera within the EIHE are assigned to the OHCB, a group of widely distributed marine bacteria that use hydrocarbons as an almost exclusive source of carbon and energy (Yakimov et al., 2007). Analysis of OHCB genera revealed only a sporadic and very low abundance of Alcanivorax, Oleispira, Oleibacter, and Thalassolituus sequences, accounting for only 0.002% relative abundance of the bacterial community (see Supplementary Table 4). OHCB are typically present in low numbers in uncontaminated environments but rapidly increase in abundance following oil spills (Head et al., 2006). The OHCB are aerobic and so would not be expected to grow in the anaerobic sediments (7.5–17.5 cm; see Supplementary Figure 4). Whilst many OHCB can use alternative electron acceptors, they cannot degrade oil under anoxic conditions, as activation of hydrocarbons by their oxygenase enzymes requires molecular oxygen (van Beilen et al., 2003). The EIHE does however also include genera that contain known anaerobic hydrocarbon degraders (Lozada et al., 2014), including for example the genera Desulfococcus, Desulfatibacillum, Desulfatiferula, and Desulfobacterium, which were detected only in very low abundance in sampled sediments (Supplementary Table 4). Desulfococcus oleovorans strain Hxd3 was the first described organism capable of anaerobic hydrocarbon-degradation (Aeckersberg et al., 1991) and others have since been described, including alkane-degraders (e.g., Desulfatibacillum alkanivorans strain AK-01; So and Young, 1999). However, there are numerous strains of anaerobic hydrocarbon degraders that are yet to be assigned to any known genus (e.g., see Rabus et al., 2016), such as the PAH-degrading sulphate-reducing Deltaproteobacteria strain NaphS2 (Galushko et al., 1999). Furthermore, certain anaerobic hydrocarbon-degrading bacteria are missing from the EIHE reference list [e.g., Desulfatiglans (Jochum et al., 2018), Desulfosarcina (Watanabe et al., 2017), and Desulfobacula (Rabus et al., 1993)]. These issues, coupled with the fact that in comparison to aerobic hydrocarbon degradation there are many anaerobic hydrocarbon degradation pathways (Widdel et al., 2010; Rabus et al., 2016; Davidova et al., 2018), means that assessments of anaerobic marine environments with the EIHE is much more problematic, and as such, analysis of sediments around HMS Royal Oak could potentially underestimate biosignatures for hydrocarbon presence. To this end, the abundance of missing anaerobic hydrocarbon-degrading sulphate-reducing bacteria was calculated separately and constituted 15% of overall sulphate-reducing bacteria. Whilst these hydrocarbon-degrading sulphate-reducing bacteria may have been utilising buried pyrogenic PAHs there was no significant (p > 0.05) difference in abundance with distance from the shipwreck. The extremely low EIHE, and a low abundance of OHCB and other genera associated with hydrocarbon-degrading (both aerobic and anaerobic), in sediments around HMS Royal Oak, suggest an absence of hydrocarbons required to support the growth of hydrocarbon-degrading bacteria.
Is There Evidence of Historic Oil-Spills, From HMS Royal Oak Shipwreck, Impacting Surrounding Sediment?
The low levels of sediment PAHs around the HMS Royal Oak were deemed to originate from pyrogenic sources. Bacterial community analysis also revealed extremely low abundances of OHCB and other potential aerobic or anaerobic oil-degrading genera both within the EIHE and those not included. Both lines of evidence therefore suggest that the sampled sediments (50–950 m from HMS Royal Oak) show no impacts from current oil leaks. Moreover, there is no evidence of impact by the reported historical oil-spills by HMS Royal Oak. There remains the possibility of more localised pollution within a 50 m radius, but this region was not sampled in this study due to a 50 m safety boundary. However, should this be the case, it would infer that any pollution would be highly localised to the immediate vicinity around the shipwreck, and this study confirms minimal impacts from leaking oil to the wider surrounding environment.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in materials and methods in the main manuscript.
Author Contributions
GT wrote the manuscript/Supplementary Material, data analysis/visualisation, and performed the molecular work. SB, PM, and RB conducted the sampling campaign. CH performed the analytical chemistry. FG, JB, BM, and TM secured funding. All authors reviewed the manuscript, Supplementary Information, figures, tables, contributed to the article, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
The work was funded by the Ministry of Defence Salvage and Marine Operations Wreck Management Programme. We would like to acknowledge and thank the National Environmental Research Council (NERC) (NE/L002582/1), via EnvEast DTP, and the Centre for Environment, Fisheries and Aquaculture Science for access and funding of this work.
Acknowledgments
We would like to thank John Green and Tania Cresswell-Maynard from the University of Essex for providing technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.650139/full#supplementary-material
Footnotes
References
Abdel-Shafy, H. I., and Mansour, M. S. M. (2016). A review on polycyclic aromatic hydrocarbons: source, environmental impact, effect on human health and remediation. Egypt. J. Pet. 25, 107–123. doi: 10.1016/j.ejpe.2015.03.011
Aeckersberg, F., Bak, F., and Widdel, F. (1991). Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch. Microbiol. 156, 5–14. doi: 10.1007/BF00418180
Akyüz, M., and Çabuk, H. (2010). Gas-particle partitioning and seasonal variation of polycyclic aromatic hydrocarbons in the atmosphere of Zonguldak, Turkey. Sci. Total Environ. 408, 5550–5558. doi: 10.1016/j.scitotenv.2010.07.063
Alzarhani, A. K., Clark, D. R., Underwood, G. J. C., Ford, H., Cotton, T. E. A., and Dumbrell, A. J. (2019). Are drivers of root-associated fungal community structure context specific? ISME J. 13, 1330–1344. doi: 10.1038/s41396-019-0350-y
Amir-Heidari, P., Arneborg, L., Lindgren, J. F., Lindhe, A., Rosén, L., Raie, M., et al. (2019). A state-of-the-art model for spatial and stochastic oil spill risk assessment: a case study of oil spill from a shipwreck. Environ. Int. 126, 309–320. doi: 10.1016/j.envint.2019.02.037
Baumard, P., Budzinski, H., Michon, Q., Garrigues, P., Burgeot, T., and Bellocq, J. (1998). Origin and bioavailability of PAHs in the Mediterranean Sea from mussel and sediment records. Estuar. Coast. Shelf Sci. 47, 77–90. doi: 10.1006/ecss.1998.0337
Becker, R. A., Wilks, A. R., Brownrigg, R., Minka, T. P., and Deckmyn, A. (2016). Package “Maps”: Draw Geographical Maps. R Packag. version 2.3-6.
Bodenhofer, U., Kothmeier, A., and Hochreiter, S. (2011). Apcluster: an R package for affinity propagation clustering. Bioinformatics 27, 2463–2464. doi: 10.1093/bioinformatics/btr406
Church, R. A., Warren, D. J., and Irion, J. B. (2009). Analysis of deepwater shipwrecks in the gulf of Mexico artificial reef effect of six world war ii shipwrecks. Oceanography 22, 50–63. doi: 10.5670/oceanog.2009.38
Clark, D. R. (2019). ecolFudge. Available online at: https://github.com/Dave-Clark/ecolFudge (accessed October 1, 2019).
Coulon, F., Chronopoulou, P. M., Fahy, A., Païssé, S., Goñi-Urriza, M., Peperzak, L., et al. (2012). Central role of dynamic tidal biofilms dominated by aerobic hydrocarbonoclastic bacteria and diatoms in the biodegradation of hydrocarbons in coastal mudflats. Appl. Environ. Microbiol. 78, 3638–3648. doi: 10.1128/AEM.00072-12
Davidova, I. A., Marks, C. R., and Suflita, J. M. (2018). “Anaerobic Hydrocarbon-Degrading Deltaproteobacteria,” in Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes, ed. T. J. McGenity (Cham: Springer), 1–38. doi: 10.1007/978-3-319-60053-6_12-1
Davies, A. J., and Evans, J. G. (1980). An analysis of the one-dimensional steady-state glow discharge. J. Phys. D. Appl. Phys. 13. doi: 10.1088/0022-3727/13/9/001
De La Torre-Roche, R. J., Lee, W. Y., and Campos-Díaz, S. I. (2009). Soil-borne polycyclic aromatic hydrocarbons in El Paso, Texas: analysis of a potential problem in the United States/Mexico border region. J. Hazard. Mater. 163, 946–958. doi: 10.1016/j.jhazmat.2008.07.089
Dos Santos, H. F., Cury, J. C., do Carmo, F. L., Dos Santos, A. L., Tiedje, J., van Elsas, J. D., et al. (2011). Mangrove bacterial diversity and the impact of oil contamination revealed by pyrosequencing: bacterial proxies for oil pollution. PLoS One 6:e16943. doi: 10.1371/journal.pone.0016943
Dumbrell, A. J., Ferguson, R. M. W., and Clark, D. R. (2016). “Microbial community analysis by single-amplicon high-throughput next generation sequencing: data analysis – from raw output to ecology,” in Hydrocarbon and Lipid Microbiology Protocols, eds T. J. McGenity, K. N. Timmis, and B. Nogales (Berlin: Springer Protocols Handbooks), 155–206. doi: 10.1007/8623_2016_228
Dyksterhouse, S. E., Gray, J. P., Herwig, R. P., Lara, J. C., and Staley, J. T. (1995). Cycloclasticus pugetii gen. nov., sp. nov., an Aromatic hydrocarbon- degrading bacterium from marine sediments. Int. J. Syst. Bacteriol. 45, 116–123. doi: 10.1099/00207713-45-1-116
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Faksness, L. G., Daling, P., Altin, D., Dolva, H., Fosbæk, B., and Bergstrøm, R. (2015). Relative bioavailability and toxicity of fuel oils leaking from World War II shipwrecks. Mar. Pollut. Bull. 94, 123–130. doi: 10.1016/j.marpolbul.2015.03.002
Field, D., Tiwari, B., Booth, T., Houten, S., Swan, D., Bertrand, N., et al. (2006). Open software for biologists: from famine to feast. Nat. Biotechnol. 24, 801–803. doi: 10.1038/nbt0706-801
Galushko, A., Minz, D., Schink, B., and Widdel, F. (1999). Anaerobic degradation of naphthalene by a pure culture of a novel type of marine sulphate-reducing bacterium. Environ. Microbiol. 1, 415–420. doi: 10.1046/j.1462-2920.1999.00051.x
Guibert, L. M., Loviso, C. L., Marcos, M. S., Commendatore, M. G., Dionisi, H. M., and Lozada, M. (2012). Alkane biodegradation genes from chronically polluted subantarctic coastal sediments and their shifts in response to oil exposure. Microb. Ecol. 64, 605–616. doi: 10.1007/s00248-012-0051-9
Guinan, J., Charlesworth, M., Service, M., and Oliver, T. (2001). Sources and geochemical constraints of polycyclic aromatic hydrocarbons (PAHs) in sediments and mussels of two Northern Irish Sea-loughs. Mar. Pollut. Bull. 42, 1073–1081. doi: 10.1016/S0025-326X(01)00077-7
Gutierrez, T., Singleton, D. R., Berry, D., Yang, T., Aitken, M. D., and Teske, A. (2013). Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP. ISME J. 7, 2091–2104. doi: 10.1038/ismej.2013.98
Hamdan, L. J., Salerno, J. L., Reed, A., Joye, S. B., and Damour, M. (2018). The impact of the deepwater horizon blowout on historic shipwreck-associated sediment microbiomes in the northern Gulf of Mexico. Sci. Rep. 8:9057. doi: 10.1038/s41598-018-27350-z
Head, I. M., Jones, D. M., and Röling, W. F. M. (2006). Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 4, 173–182. doi: 10.1038/nrmicro1348
Herbert, R. A. (1985). Development of mass blooms of photosynthetic bacteria on sheltered beaches in Scapa Flow, Orkney Islands. Proc. R. Soc. Edinburgh. Sect. B. Biol. Sci. 87, 15–25. doi: 10.1017/s0269727000004139
Hill, P. (2019). Managing the Wreck of HMS Royal Oak. Available online at: https://spillcon.com/2019/Resources/Downloads/Speakers/Polly-Hill.pdf (accessed August 10, 2020).
Hope, R. M. (2013). Rmisc: Ryan Miscellaneous. Available online at: https://cran.r-project.org/package=Rmisc (accessed August 10, 2020).
Hussain, J., Zhao, Z., Pang, Y., Xia, L., Hussain, I., and Jiang, X. (2016). Effects of different water seasons on the residual characteristics and ecological risk of polycyclic aromatic hydrocarbons in sediments from changdang lake, China. J. Chem. 2016:8545816. doi: 10.1155/2016/8545816
Hvitfeldt, E. (2020). paletteer: Comprehensive Collection of Color Palettes. R package version 1.2.0.
IARC (1987). Overall evaluations of carcinogenicity: an updating of IARC Monographs volumes 1 to 42. IARC Monogr. Eval. Carcinog. Risks Hum. Suppl. 7, 1–440.
Jochum, L. M., Schreiber, L., Marshall, I. P. G., Jørgensen, B. B., Schramm, A., and Kjeldsen, K. U. (2018). Single-cell genomics reveals a diverse metabolic potential of uncultivated Desulfatiglans-related deltaproteobacteria widely distributed in marine sediment. Front. Microbiol. 9:2038. doi: 10.3389/fmicb.2018.02038
Joshi, N., and Fass, J. (2011). Sickle: A Sliding-Window, Adaptive, Quality-based Trimming Tool for FastQ Files (Version 1.33) [Software]. Available at https://github.com/najoshi/sickle
Kelly, C. A., Law, R. J., and Emerson, H. S. (2000). Methods for analysis for hydrocarbons and polycyclic aromatic hydrocarbons (PAH) in marine samples. Aquat. Environ. Prot. Anal. Methods 12:18.
Kirby, M. F., Brant, J., Moore, J., and Lincoln, S. (2018). PREMIAM – Pollution Response in Emergencies – Marine Impact Assessment and Monitoring: Post-Incident Monitoring Guidelines, 2nd Edn. Lowestoft: Cefas. Science Series Technical Report.
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. doi: 10.1093/nar/gks808
Kostka, J. E., Prakash, O., Overholt, W. A., Green, S. J., Freyer, G., Canion, A., et al. (2011). Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the deepwater horizon oil spill. Appl. Environ. Microbiol. 77, 7962–7974. doi: 10.1128/AEM.05402-11
Landquist, H., Hassellöv, I. M., Rosén, L., Lindgren, J. F., and Dahllöf, I. (2013). Evaluating the needs of risk assessment methods of potentially polluting shipwrecks. J. Environ. Manage. 119, 85–92. doi: 10.1016/j.jenvman.2012.12.036
Landquist, H., Norrman, J., Lindhe, A., Norberg, T., Hassellöv, I. M., Lindgren, J. F., et al. (2017a). Expert elicitation for deriving input data for probabilistic risk assessment of shipwrecks. Mar. Pollut. Bull. 125, 399–415. doi: 10.1016/j.marpolbul.2017.09.043
Landquist, H., Rosén, L., Lindhe, A., Norberg, T., and Hassellöv, I. M. (2017b). Bayesian updating in a fault tree model for shipwreck risk assessment. Sci. Total Environ. 590–591, 80–91. doi: 10.1016/j.scitotenv.2017.03.033
Landquist, H., Rosén, L., Lindhe, A., Norberg, T., Hassellöv, I. M., Lindgren, J. F., et al. (2014). A fault tree model to assess probability of contaminant discharge from shipwrecks. Mar. Pollut. Bull. 88, 239–248. doi: 10.1016/j.marpolbul.2014.08.037
Law, R. J., Kelly, C. A., and Nicholson, M. D. (1999). Polycyclic aromatic hydrocarbons (PAH) in shellfish affected by the Sea Empress oil spill in Wales in 1996. Polycycl. Aromat. Compd. 17, 229–239. doi: 10.1080/10406639908020618
Leahy, J. G., and Colwell, R. R. (1990). Microbial degradation of hydrocarbons in the environment. Microbiol. Rev. 54, 305–315. doi: 10.1128/mmbr.54.3.305-315.1990
Lehr, R. E., and Jerina, D. M. (1977). Metabolic activations of polycyclic hydrocarbons - Structure-activity relationships. Arch. Toxicol. 39, 1–6. doi: 10.1007/BF00343269
Lenth, R. (2020). emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 1.4.7.
Li, Q., Cao, L., Wang, W., Tan, H., Jin, T., Wang, G., et al. (2018). Analysis of the bacterial communities in the waterlogged wooden cultural relics of the Xiaobaijiao No. 1 shipwreck via high-throughput sequencing technology. Holzforschung 72, 609–619. doi: 10.1515/hf-2017-0132
Lima, A. L. C., Eglinton, T. I., and Reddy, C. M. (2003). High-resolution record of pyrogenic polycyclic aromatic hydrocarbon deposition during the 20th century. Environ. Sci. Technol. 37, 53–61. doi: 10.1021/es025895p
Liu, Z., Fu, T., Hu, C., Shen, D., Macchioni, N., Sozzi, L., et al. (2018). Microbial community analysis and biodeterioration of waterlogged archaeological wood from the Nanhai No. 1 shipwreck during storage. Sci. Rep. 8:7170. doi: 10.1038/s41598-018-25484-8
Lozada, M., Marcos, M. S., Commendatore, M. G., Gil, M. N., and Dionisi, H. M. (2014). the bacterial community structure of hydrocarbon-polluted marine environments as the basis for the definition of an ecological index of hydrocarbon exposure. Microbes Environ. 29, 269–276. doi: 10.1264/jsme2.me14028
Manzetti, S. (2013). Polycyclic aromatic hydrocarbons in the environment: environmental fate and transformation. Polycycl. Aromat. Compd. 33, 311–330. doi: 10.1080/10406638.2013.781042
Marine, B. (2010). Royal Oak Oil Removal Programme. Available online at: https://www.briggsmarine.com/projects/royal-oak-removal-programme/ (accessed August 10, 2020).
McGenity, T. J., Folwell, B. D., McKew, B. A., and Sanni, G. O. (2012). Marine crude-oil biodegradation: a central role for interspecies interactions. Aquat. Biosyst. 8:10. doi: 10.1186/2046-9063-8-10
McKew, B. A., and Smith, C. J. (2015). “Real-Time PCR approaches for analysis of hydrocarbon-degrading bacterial communities,” in Hydrocarbon and Lipid Microbiology Protocols, eds T. J. McGenity, K. N. Timmis, and B. N. Fernandez (Berlin: Springer Protocols Handbooks).
Michel, J., Gilbert, T., Etkin, D. S., Urban, R., Waldron, J., and Blocksidge, C. T. (2005). Potentially polluting wrecks in Marine waters. 2005 Int. Oil Spill Conf. IOSC 2005, 11590–11629.
Monfils, R., Gilbert, T., and Nawadra, S. (2006). Sunken WWII shipwrecks of the Pacific and East Asia: the need for regional collaboration to address the potential marine pollution threat. Ocean Coast. Manag. 49, 779–788. doi: 10.1016/j.ocecoaman.2006.06.011
Mugge, R. L., Brock, M. L., Salerno, J. L., Damour, M., Church, R. A., Lee, J. S., et al. (2019). Deep-sea biofilms, historic shipwreck preservation and the Deepwater Horizon spill. Front. Mar. Sci. 6:48. doi: 10.3389/fmars.2019.00048
Muir, T. (2020). Scapa Flow Historic Wreck Site. Available online at: http://www.scapaflowwrecks.com/wrecks/other/ (accessed August 10, 2020).
Ndungu, K., Beylich, B. A., Staalstrøm, A., Øxnevad, S., Berge, J. A., Braaten, H. F. V., et al. (2017). Petroleum oil and mercury pollution from shipwrecks in Norwegian coastal waters. Sci. Total Environ. 593–594, 624–633. doi: 10.1016/j.scitotenv.2017.03.213
Nikolenko, S. I., Korobeynikov, A. I., and Alekseyev, M. A. (2013). BayesHammer: bayesian clustering for error correction in single-cell sequencing. BMC Genomics 14:S7. doi: 10.1186/1471-2164-14-S1-S7
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A., Korobeynikov, A., Lapidus, A., et al. (2013). “Assembling genomes and mini-metagenomes from highly chimeric reads,” in Research in Computational Molecular BiologyLecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), eds M. Deng, R. Jiang, F. Sun, and X. Zhang (Berlin: Springer), 158–170.
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). vegan: Community Ecology Package. R package version 2.5.6. 254.
OSPAR Commission (2009). Agreement on CEMP Assessment Criteria for the QSR 2010. Oslo: OSPAR Commission.
OSPAR Commission (2017). Status and Trends in the Concentrations of Polycyclic Aromatic Hydrocarbons (PAHs) in Sediment. Oslo: OSPAR Commission.
Pedersen, T. L. (2019). patchwork: The Composer of Plots. Cran. Available online at: https://cran.r-project.org/package=patchwork (accessed August 10, 2020).
Pedersen, T. L. (2020). ggraph: An Implementation of Grammar of Graphics for Graphs and Networks. R Package Version 2.0.3.
Pereira, W. E., Hostettler, F. D., Luoma, S. N., Van Geen, A., Fuller, C. C., and Anima, R. J. (1999). Sedimentary record of anthropogenic and biogenic polycyclic aromatic hydrocarbons in San Francisco Bay. California. Mar. Chem. 64, 99–113. doi: 10.1016/S0304-4203(98)00087-5
Peters, K. E., Walters, C. C., and Moldowan, J. M. (2004). The Biomarker Guide. Cambridge: Cambridge University Press. doi: 10.1017/cbo9780511524868
Qiao, M., Wang, C., Huang, S., Wang, D., and Wang, Z. (2006). Composition, sources, and potential toxicological significance of PAHs in the surface sediments of the Meiliang Bay, Taihu Lake, China. Environ. Int. 32, 28–33. doi: 10.1016/j.envint.2005.04.005
R Development Core Team (2011). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. doi: 10.1007/978-3-540-74686-7
Rabus, R., Boll, M., Heider, J., Meckenstock, R. U., Buckel, W., Einsle, O., et al. (2016). Anaerobic microbial degradation of hydrocarbons: from enzymatic reactions to the environment. J. Mol. Microbiol. Biotechnol. 26, 5–28. doi: 10.1159/000443997
Rabus, R., Nordhaus, R., Ludwig, W., and Widdel, F. (1993). Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl. Environ. Microbiol. 59, 1444–1451. doi: 10.1128/aem.59.5.1444-1451.1993
Ramsay, D. L., and Brampton, A. H. (2000). Coastal Cells in Scotland: Cell 10 - Orkney. Scottish Natural Heritage Research Survey and Monitoring Report No. 151. Scotland: Scottish Natural Heritage.
Ribicic, D., Netzer, R., Hazen, T. C., Techtmann, S. M., Drabløs, F., and Brakstad, O. G. (2018). Microbial community and metagenome dynamics during biodegradation of dispersed oil reveals potential key-players in cold Norwegian seawater. Mar. Pollut. Bull. 129, 370–378. doi: 10.1016/j.marpolbul.2018.02.034
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ. 4:e2584. doi: 10.7717/peerj.2584
Russell, M. A., Murphy, L. E., Johnson, D. L., Foecke, T. J., Morris, P. J., and Mitchell, R. (2004). Science for stewardship: multidisciplinary research on USS Arizona. Mar. Technol. Soc. J. 38, 54–63. doi: 10.4031/002533204787511255
So, C. M., and Young, L. Y. (1999). Isolation and characterization of a sulfate-reducing bacterium that anaerobically degrades alkanes. Appl. Environ. Microbiol. 65, 2969–2976. doi: 10.1128/aem.65.7.2969-2976.1999
Soclo, H. H., Garrigues, P., and Ewald, M. (2000). Origin of polycyclic aromatic hydrocarbons (PAHs) in coastal marine sediments: case studies in Cotonou (Benin) and Aquitaine (France) Areas. Mar. Pollut. Bull. 40, 387–396. doi: 10.1016/S0025-326X(99)00200-3
Tatti, E., McKew, B. A., Whitby, C., and Smith, C. J. (2016). Simultaneous DNA-RNA extraction from coastal sediments and quantification of 16S rRNA genes and transcripts by real-time PCR. J. Vis. Exp. 112:54067. doi: 10.3791/54067
Thomas, G. E., Cameron, T. C., Campo, P., Clark, D. R., Coulon, F., Gregson, B. H., et al. (2020). Bacterial community legacy effects following the agia zoni II Oil-Spill, Greece. Front. Microbiol. 11:1706. doi: 10.3389/fmicb.2020.01706
van Beilen, J. B., Li, Z., Duetz, W. A., Smits, T. H. M., and Witholt, B. (2003). Diversité des systèmes alcane hydroxylase dans l’environnement. Oil Gas Sci. Technol. 58, 427–440. doi: 10.2516/ogst:2003026
van Gemerden, H., Tughan, C. S., de Wit, R., and Herbert, R. A. (1989). Laminated microbial ecosystems on sheltered beaches in Scapa Flow, Orkney Islands. FEMS Microbiol. Lett. 62, 87–101. doi: 10.1016/0378-1097(89)90018-9
Venables, W. N., and Ripley, B. D. (2002). Modern Applied Statistics with S, 4th Edn. Berlin: Springer-Verlag. doi: 10.2307/2685660
Venkatesan, M. I. (1988). Occurrence and possible sources of perylene in marine sediments - a review. Mar. Chem. 25, 1–27. doi: 10.1016/0304-4203(88)90011-4
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, W., Wang, L., and Shao, Z. (2010). Diversity and abundance of oil-degrading bacteria and alkane hydroxylase (alkB) genes in the subtropical seawater of xiamen Island. Microb. Ecol. 60, 429–439. doi: 10.1007/s00248-010-9724-4
Wang, Z., and Fingas, M. (2005). “Oil and petroleum product fingerprinting analysis by gas chromatographic techniques,” in Chromatographic Analysis of the Environment, eds D. A. Lambropoulou and L. M. L. Nollet (Boca Raton, FL: Taylor and Francis), 1027–1101.
Wang, Z., and Fingas, M. F. (2003). Development of oil hydrocarbon fingerprinting and identification techniques. Marine Pollut. Bull. 47, 423–452. doi: 10.1016/S0025-326X(03)00215-7
Wang, Z., Fingas, M., Shu, Y. Y., Sigouin, L., Landriault, M., Lambert, P., et al. (1999). Quantitative characterization of PAHs in burn residue and soot samples and differentiation of pyrogenic PAH1 from petrogenic PAHs - The 1994 mobile burn study. Environ. Sci. Technol. 33, 3100–3109. doi: 10.1021/es990031y
Wang, Z., Yang, C., Kelly-Hooper, F., Hollebone, B. P., Peng, X., Brown, C. E., et al. (2009). Forensic differentiation of biogenic organic compounds from petroleum hydrocarbons in biogenic and petrogenic compounds cross-contaminated soils and sediments. J. Chromatogr. A 1216, 1174–1191. doi: 10.1016/j.chroma.2008.12.036
Wang, Z., Yang, C., Parrott, J. L., Frank, R. A., Yang, Z., Brown, C. E., et al. (2014). Forensic source differentiation of petrogenic, pyrogenic, and biogenic hydrocarbons in Canadian oil sands environmental samples. J. Hazard. Mater. 271, 166–177. doi: 10.1016/j.jhazmat.2014.02.021
Watanabe, M., Higashioka, Y., Kojima, H., and Fukui, M. (2017). Desulfosarcina widdelii sp. nov. and Desulfosarcina alkanivorans sp. nov., hydrocarbon-degrading sulfate-reducing bacteria isolated from marine sediment and emended description of the genus Desulfosarcina. Int. J. Syst. Evol. Microbiol. 67, 2994–2997. doi: 10.1099/ijsem.0.002062
Widdel, F., Knittel, K., and Galushko, A. (2010). “Anaerobic Hydrocarbon-degrading microorganisms: an overview,” in Handbook of Hydrocarbon and Lipid Microbiology, 1997–2021, ed. K. N. Timmis (Berlin: Springer).
Wieland, A., Kühl, M., McGowan, L., Fourçans, A., Duran, R., Caumette, P., et al. (2003). Microbial mats on the orkney islands revisited: microenvironment and microbial community composition. Microb. Ecol. 46, 371–390. doi: 10.1007/s00248-002-0108-2
Wilke, C. O. (2015). Cowplot: Streamlined Plot Theme and Plot Annotations for ggplot2. R Packag. Version 0.5.0.
Wilke, C. O. (2020). ggtext: Improved Text Rendering Support for “ggplot2”. R package version 0.1.0.
Woodhead, R. J., Law, R. J., and Matthiessen, P. (1999). Polycyclic aromatic hydrocarbons in surface sediments around England and Wales, and their possible biological significance. Mar. Pollut. Bull. 38, 773–790. doi: 10.1016/S0025-326X(99)00039-9
Yakimov, M. M., Timmis, K. N., and Golyshin, P. N. (2007). Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 18, 257–266. doi: 10.1016/j.copbio.2007.04.006
Yunker, M. B., Macdonald, R. W., Vingarzan, R., Mitchell, R. H., Goyette, D., and Sylvestre, S. (2002). PAHs in the Fraser River basin: a critical appraisal of PAH ratios as indicators of PAH source and composition. Org. Geochem. 33, 489–515. doi: 10.1016/S0146-6380(02)00002-5
Keywords: HMS Royal Oak, oil spill, PAH, Scapa Flow, OHCB, sediment, shipwreck
Citation: Thomas GE, Bolam SG, Brant JL, Brash R, Goodsir F, Hynes C, McGenity TJ, McIlwaine PSO and McKew BA (2021) Evaluation of Polycyclic Aromatic Hydrocarbon Pollution From the HMS Royal Oak Shipwreck and Effects on Sediment Microbial Community Structure. Front. Mar. Sci. 8:650139. doi: 10.3389/fmars.2021.650139
Received: 06 January 2021; Accepted: 16 March 2021;
Published: 07 April 2021.
Edited by:
Xiutang Yuan, Yantai Institute of Coastal Zone Research (CAS), ChinaReviewed by:
Xiaolong Yang, National Marine Environmental Monitoring Center, ChinaAndrés Hugo Arias, CONICET Instituto Argentino de Oceanografía (IADO), Argentina
Copyright © 2021 Thomas, Bolam, Brant, Brash, Goodsir, Hynes, McGenity, McIlwaine and McKew. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Boyd A McKew, Ym95ZC5tY2tld0Blc3NleC5hYy51aw==
 Gareth E. Thomas
Gareth E. Thomas Stefan G. Bolam
Stefan G. Bolam Jan L. Brant
Jan L. Brant Rodney Brash2
Rodney Brash2 Clare Hynes
Clare Hynes Terry J. McGenity
Terry J. McGenity Boyd A. McKew
Boyd A. McKew