
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 19 July 2021
Sec. Aquatic Microbiology
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.626207
In the context of global warming, changes in phytoplankton-associated bacterial communities have the potential to change biogeochemical cycling and food webs in marine ecosystems. Skeletonema is a cosmopolitan diatom genus in coastal waters worldwide. Here, we grew a Skeletonema strain with its native bacterial assemblage at different temperatures and examined cell concentrations of Skeletonema sp. and free-living bacteria, dissolved organic carbon (DOC) concentrations of cultures, and the community structure of both free-living and attached bacteria at different culture stages. The results showed that elevated temperature increased the specific growth rates of both Skeletonema and free-living bacteria. Different growth stages had a more pronounced effect on community structure compared with temperatures and different physical states of bacteria. The effects of temperature on the structure of the free-living bacterial community were more pronounced compared with diatom-attached bacteria. Carbon metabolism genes and those for some specific amino acid pathways were found to be positively correlated with elevated temperature, which may have profound implications on the oceanic carbon cycle and the marine microbial loop. Network analysis revealed evidence of enhanced cooperation with an increase in positive interactions among different bacteria at elevated temperature. This may help the whole community to overcome the stress of elevated temperature. We speculate that different bacterial species may build more integrated networks with a modified functional profile of the whole community to cope with elevated temperature. This study contributes to an improved understanding of the response of diatom-associated bacterial communities to elevated temperature.
Human activities have resulted in increasing atmospheric concentrations of greenhouse gases; and as a result, the global ocean temperature has been rising since the mid-20th century. The surface ocean temperature may increase a further 1–3°C by the end of this century (Pörtner et al., 2019). Elevated ocean temperatures may greatly influence oceanic biogeochemical processes and pose threats to marine food webs and ecosystems (Edwards and Richardson, 2004; Sarmento et al., 2010). Observations have confirmed that increased ocean temperatures may greatly affect marine organisms such as microorganisms. Metagenomic data collected across the globe in the Tara Oceans project (Sunagawa et al., 2015) showed that temperature is the main driver of variation in oceanic microbial community structure.
Phytoplankton and heterotrophic bacteria are the two most important groups of microorganisms in the ocean, playing indispensable roles in oceanic ecosystems. Phytoplanktons are important primary producers and play a central role in oceanic food chains. The dissolved organic carbon (DOC) released by marine phytoplankton is an important source of the oceanic DOC reservoir, which can be actively taken up by heterotrophic bacteria in seawater (Thornton, 2014; Moran, 2015). The DOC released by phytoplankton can be transferred to higher trophic levels through the microbial loop, released into CO2, and converted into more refractory carbon (Azam, 1998; Jiao et al., 2010). It has been reported that 2–50% of fixed carbon by phytoplankton is released as DOC into seawater (Thornton, 2014). Meanwhile, bacteria support the growth of phytoplankton by providing vitamins, organic acids, and growth factors (Seymour et al., 2017). The DOC composition and concentration in seawater are substantially determined by the phytoplankton species, growth stages of phytoplankton, and environmental conditions (Norrman and Zweifel, 1996; Barofsky et al., 2009; Thornton, 2014). Different heterotrophic bacterial species also have different capabilities in terms of utilizing specific substances released from phytoplankton (Riemann et al., 2000; Horn, 2011). The transfer of organic carbon from phytoplankton to bacteria may occur through two different routes: via the DOC released by phytoplankton into the surrounding water and its uptake by free-living bacteria, and through bacteria that are physically attached to the phytoplankton taking up the DOC directly from phytoplankton cells (Grossart and Simon, 2007; Mével et al., 2008). Although some recent studies have demonstrated that communities of free-living and attached bacteria were only slightly different, they have generally been considered to have many distinct characteristics (Hu et al., 2020). Compared with free-living cells, attached bacteria generally have stronger capability for involvement in biofilm development, having higher extracellular enzymatic activity and stronger chemotactic behavior (Tout et al., 2015; Dang and Lovell, 2016). Due to their closer physical proximity to the phytoplankton allowing more reliable uptake of substances released by attached bacteria are believed to have stronger physical and metabolic interaction with phytoplankton compared to free-living bacteria (Rooney-Varga et al., 2005; Arandia-Gorostidi et al., 2017). Thus, attached bacteria are thought to gain an ecological advantage compared with their free-living counterparts (Grossart et al., 2007; Arandia-Gorostidi et al., 2017).
Some studies have indicated that elevated temperature has differential effects on free-living and attached bacteria, and that elevated temperature led to a significant increase in cell abundance of attached bacteria and significantly higher DOC incorporation by attached bacteria compared with their free-living counterparts (Arandia-Gorostidi et al., 2017, 2020). However, there is still a lack of studies on the changes in bacterial community structure and functional profile in response to elevated temperature. These could help us better understand the strategies of free-living and attached bacteria coping with elevated temperature.
Diatoms, an important group of phytoplankton, have a complex evolutionary history marked by extensive horizontal gene transfer from bacteria, indicating their possible co-evolution (Armbrust et al., 2004; Bowler et al., 2008; Mock et al., 2017). Skeletonema is a typical centric diatom genus, with a wide distribution from Arctic and Antarctica to tropical waters that can form extensive blooms in the coastal regions (Johansson et al., 2019a). Due to the ecological importance of Skeletonema, Skeletonema marinoi has been proposed as a new genetic model for chain-forming diatoms (Johansson et al., 2019b). It is difficult to separate Skeletonoma from its associated bacteria, which may be because of sensitivity to antibiotics (Johansson et al., 2019b) and potentially close interactions between Skeletonema and heterotrophic bacteria, as described in Synechococcus (Zheng et al., 2018). A previous study has shown pronounced growth enhancement of S. marinoi by heterotrophic bacteria at a lower temperature (8°C) but not at higher temperatures (16 and 24°C) in experiments that involved culturing different bacterial species isolated from S. marinoi culture with non-axenic S. marinoi. However, changes in community composition of native heterotrophic bacteria, which may have potentially close interactions with S. marinoi in the culture, were not investigated (Johansson et al., 2019b). In order to better understand the strategies of bacteria in response to elevated temperature, it is important to explore the effects of elevated temperature on native free-living and attached diatom-associated bacteria.
In this study, we cultured a recently isolated non-axenic Skeletonema sp. at different temperatures, without eliminating native heterotrophic bacteria. (1) We compared the effects of elevated temperatures on the Skeletonema sp. and free-living bacteria in terms of specific growth rate. (2) We attempted to explore the potential impacts of elevated temperature on the carbon fluxes and carbon partitioning in Skeletonema sp. culture by measuring DOC concentrations. (3) We examined the composition of diatom-attached bacteria and free-living bacteria at different stages such as initial growth, early exponential growth, and early decline stage at 20, 25, and 30°C. (4) We then considered the impacts of elevated temperature on the free-living and attached bacteria in the context of community structure, predicted functional profiles, and network configurations. Given the importance of this particular diatom species globally and the potential importance of bacterial interactions, this study aims to contribute to better understanding of the response of the diatom associated-bacterial community to elevated temperatures.
The experimental population of Skeletonema sp. used in this study was isolated from the coastal water of Xiamen, China (24.57°N, 118.12°E), on March 29, 2018 and preserved in Microalgae Culture Center of Diatom Lab, Life Science School, Xiamen University, China (MMDL 506572). There were 8 months of culturing between Skeletonema sp. isolation and the start of the experiment. This Skeletonema sp. strain was maintained in an f/2 + Si medium under low light conditions (20°C, 12 h light: 12 h dark photoperiod with an irradiance of 75 μmol photons m–2 s–1 from LED-light panels). The Skeletonema sp. culture maintained at low light conditions was transferred to standard conditions (20°C, 12 h light: 12 h dark photoperiod with an irradiance of 150 μmol photons m–2 s–1) for a week. The Skeletonema sp. with native bacteria assemblage was then cultured at 20, 25, and 30°C for 3 days. After 3 days of acclimation, Skeletonema sp. was cultured in 1 L polycarbonate bottles containing 700 mL artificial seawater (salinity 35‰) with six replicates at 20, 25, and 30°C with initial concentration 1 × 104 cells/mL. The specific growth rates of Skeletonome sp. and free-living bacteria were calculated, respectively. For the growth rate calculation of Skeletonema sp., first, we identified the exponential phases of cultures at different temperatures. Then, we calculated the specific growth rate using the following formula: μ = (Ln C2 - Ln C1)/(t - t1). μ is the specific growth rate (day–1), and C1 and C2 are the cell concentrations in the exponential phase at times t1 and t2, respectively. Days 0–6 for T30 and T25, days 0–8 for T20 were identified as exponential phases. The specific growth rates of free-living bacteria under different temperatures were calculated based on the bacterial cell concentrations on days 0 and 14.
The Skeletonema sp. cells were counted under a light microscope (Eclipse E100, Nikon, Tokyo, Japan) using a plankton counting chamber every 2 days. After filtration through a 20-μm net, the samples were stained for bacterial cell enumeration. Bacterial cell concentrations were measured every 2 days by flow cytometry (CytoFLEX, Beckman Coulter, Brea, CA, United States) equipped with 488-nm excitation laser and forward scatter, side scatter, and four fluorescence channel (525/40BP, 585/42BP, 690/50BP, and 780/50BP) detectors. 10,000 × SYBR Green I dye was diluted 100 times with 0.22 μm filtered DMSO and stored at 4°C, and 200 μL of each sample was stained with SYBR Green I at 1 × final concentration for 15 min in the dark. At least 30,000 measurements were collected for each sample and analyzed using the CytExpert software. The bacteria were enumerated by plotting side scatter versus green fluorescence, and green fluorescence versus red fluorescence after staining with SYBR green I.
Dissolved organic carbon concentrations were measured on day 4, 8, and 12 during the cultivation, and 20-mL culture from each sample filtered through a 0.2-μm polycarbonate filter (Millipore, Burlington, MA, United States) was used for DOC measurement. The DOC concentrations were measured by high-temperature catalytic oxidation (HTCO) using a Shimadzu TOC-VCPH (Shimadzu, Kyoto, Japan) analyzer with the TOC-control software; and a 7-μm pore size GF/F membrane (25 mm diameter, Whatman, Maidstone, United Kingdom) and 40-mL glass vials (CNW, Dusseldorf, Germany) were combusted in a muffle furnace at 450°C for 4 h. In order to avoid carbon contamination, all glass materials and filters were washed with acid, rinsed with RO-water and Milli-Q water, and pre-combusted as described above. Each sample, 4 mL, was filtered with GF/F filters and diluted to 40 mL with Milli-Q water, and then placed in 40-mL glass vials and stored at −20°C. Before sample measurement, a standard curve was established using potassium hydrogen phthalate and tested with low carbon water and deep seawater. The samples were thawed and acidified with phosphoric acid to adjust the pH to 2–3 before analysis.
A 100-mL culture was taken from each sample on day 4, 8, and 14. Two replicates from each treatment were combined into one replicate with 200 mL in order to get enough material for bacteria DNA extraction. Sequential size fractionated filtration (2- and 0.2-μm polycarbonate filters) with peristaltic pump was performed to filter the culture collected from each combined replicate. The samples collected with 0.2-μm and 2-μm polycarbonate filters (Millipore, Burlington, MA, United States) were washed with phosphate-buffered saline and then centrifuged at 9,600 g to obtain cell pellets. The bacteria collected with 0.2- and 2-μm polycarbonate filters were considered as free-living bacteria and attached bacteria, respectively. A previously described DNA extraction protocol (Francis et al., 2005) was utilized with some modifications. One milliliter of hot (∼70°C) lysis buffer (100 mM Tris, 40 mM EDTA, 100 mM NaCl, 1% SDS) with 10-μL lysozyme (100 mg/mL) was used to wash the samples from filters, and 5-μL RNase A (10 mg/mL) was then added and incubated at 37°C for 5 min. Proteinase K 20 L was added and mixed gently. GB solution 220 μL was added from the bacteria DNA extraction kit (Tiangen DP302, Tiangen, Sichuan, China) and the instructions in the kit were followed for DNA extraction. Primers were 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′), targeting the V3–V4 hypervariable regions of the bacterial 16S rRNA gene. The PCR amplification conditions were as follows: initial denaturation at 95°C for 3 min, 27 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s extension at 72°C for 45 s, and then final extension at 72°C for 10 min. PCR products were purified using a AxyPrepDNA gel extraction kit (Axygen, Union City, CA, United States) from 2% agarose gel after electrophoresis. DNA library was constructed following the MiSeq Reagent Kit preparation guide (Illumina, San Diego, CA, United States).
The sequencing was conducted using an Illumina MiSeq PE300 platform (Shanghai Majorbio Bio-pharm Technology Co. Ltd., Shanghai, China) after purification and quantification of PCR products. The sequencing data have been uploaded in NCBI (project ID: PRJNA706479). Raw fastq sequences were quality-filtered by fastp (v0.19.6) and merged by FLASH (v1.2.7) before analysis. The filtered reads were imported to QIIME 2 (released on 2020/11/1), and DADA2 (released on 2020/11/1) was used to de-noise sequences, resulting in high-resolution amplicon sequence variants (ASVs). For taxonomic classification, we used the SILVA database1 with 97% identity. Sample 4_30_3_2 was considered as abnormal based on rank-abundance curve and beta analysis compared with other samples. In the following analysis, chloroplast sequences and sequences from 4_30_3_2 were removed. Alpha diversity was estimated using Mothur 1.30. Beta diversity was analyzed with QIIME 2. The aligned ASVs were used to construct a phylogenetic tree using the neighbor joining algorithm with 1,000 bootstraps in IQ-TREE. Jaccard distance matrixes were used to calculate beta diversity and were visualized by non-metric multidimensional scaling (NMDS) analysis. We used redundancy analysis (RDA) to attribute the causes of variation within the bacterial communities, such as bacterial fractions (free-living or attached bacteria), temperatures, and cultivation stages. Beta diversity linear regression analysis with different variables by Bray–Curtis distance was carried out in R. We performed linear discriminant analysis effect size (LEfSe) analysis to identify taxa differentially abundant cultured in different samples.2 First, a non-parametric factorial Kruskal–Wallis sum-rank test was performed to detect features with significant differential abundance. Second, linear discriminant analysis (LDA) was conducted to calculate the effect size of each feature. Alpha-value for the factorial Kruskal–Wallis test among classes = 0.05; Alpha value for the pairwise Wilcoxon test between subclasses = 0.05; non-negative threshold on the logarithmic LDA score for discriminative features = 2. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) (Douglas et al., 2020) was used to predict the functional profiles of bacterial communities on the basis of the bacterial community. Spearman correlation analysis was performed to detect the correlation between variables and bacterial taxa or functional profiles.
In order to investigate the potential differential effects of temperatures on bacteria network interaction of free-living and attached bacteria regardless of cultivation stages, the sequencing data from different temperatures were combined and analyzed. The sequences were divided into six groups: 02t20 (free-living bacteria at 20°C), 02t25 (free-living bacteria at 25°C), 02t30 (free-living bacteria at 30°C), 2t20 (attached bacteria at 20°C), 2t25 (attached bacteria at 25°C), and 2t30 (attached bacteria at 30°C). Ecological network construction and analyses were performed based on the relative abundance of ASVs in different samples with three biological replicates. The similarity matrices of the relative abundance of ASVs of different groups were created using Spearman correlation coefficient across time points with biological replicates by a random matrix theory (RMT)-based approach in R studio. The “psych” and “qvalue” packages were used to calculate r-value, p-value, and q-value. Cut-off values were determined according to | r| > 0.6, p < 0.01, and q < 0.05 to construct network structure. The bacteria network interaction was then visualized using Cytoscape. Topological property parameters of interaction networks were derived from Cytoscape.
One-way ANOVA was performed for testing the significant differences between different temperature treatments. The Turkey test was performed when the variance was homogeneous, while the unmet test was performed when the variance was uneven. Statistical significance was determined at the level of p-value less than 0.05. Two-way ANOVA was performed for DOC concentration interaction analysis (different temperatures and incubation stages were used as two factors). To detect the statistical differences between groups of alpha diversity index, we performed non-parametric Kruskal–Wallis rank tests.
As shown in Figure 1A, significant differences among cell abundances of Skeletonema sp. at different time points were detected. At 30°C, the cell abundance of Skeletonema sp. reached a peak earlier with the lowest maximum cell abundance. In contrast, the maximum cell abundance of Skeletonema sp. was highest with a postponed cell abundance peak at 2°C. At 30°C, Skeletonema sp. cell concentration began to decrease rapidly after reaching the maximum value on day 6 [(13.633 ± 1.975) × 104 cells/mL], while the cell concentration at 25°C began to decline rapidly after reaching the maximum value on day 8 [(17.05 ± 2.884) × 104 cells/mL]. In contrast, the cell concentration of culture at 20°C reached a maximum value on day 8 [(23.217 ± 2.221) × 104 cells/mL] and gradually decreased. No significant differences between maximum cell abundance at 25 and at 30°C were detected. However, significant differences between maximum cell abundance at 20 and 30°C (p = 0.005), and 20 and 25°C (p = 0.04) were detected. In a word, elevating temperature from 20 to 25°C significantly accelerated the onset of Skeletonema sp. cell abundance peak and decreased the maximum cell abundance. The specific growth rate of Skeletonema sp. was 0.793 ± 0.106 (day–1) at 30°C,0.723 ± 0.035 (day–1) at 25°C, and 0.619 ± 0.044 (day–1) at 20°C. The specific growth rate of Skeletonema sp. at 30°C was significantly higher than that at 20°C (p = 0.049) (Figure 1B). The growth rate ratio of Skeletonema sp. at 30/20°C was 1.31. The free-living bacteria cell abundance showed an upward trend during the cultivation at different temperatures. On day 6, the free-living bacteria cell abundance at 25°C was significantly higher than at 20 and 30°C. On day 12 and day 14, significant higher free-living bacteria cell abundances were observed at 30°C compared with at 25 and 20°C (Figure 1C). The specific growth rate of free-living bacteria was 0.412 ± 0.02 (day–1) at 30°C,0.356 ± 0.03 (day–1) at 25°C, and 0.343 ± 0.003 (day–1) at 20°C (Figure 1D). The specific growth rate of free-living bacteria at 30°C was significantly higher than at 20 (p = 0.014) and 25°C (p = 0.019). No significant differences between the growth rate of free-living bacteria at 20 and 25°C were detected. The growth rate ratio of free-living bacteria at 30/20°C was 1.12.
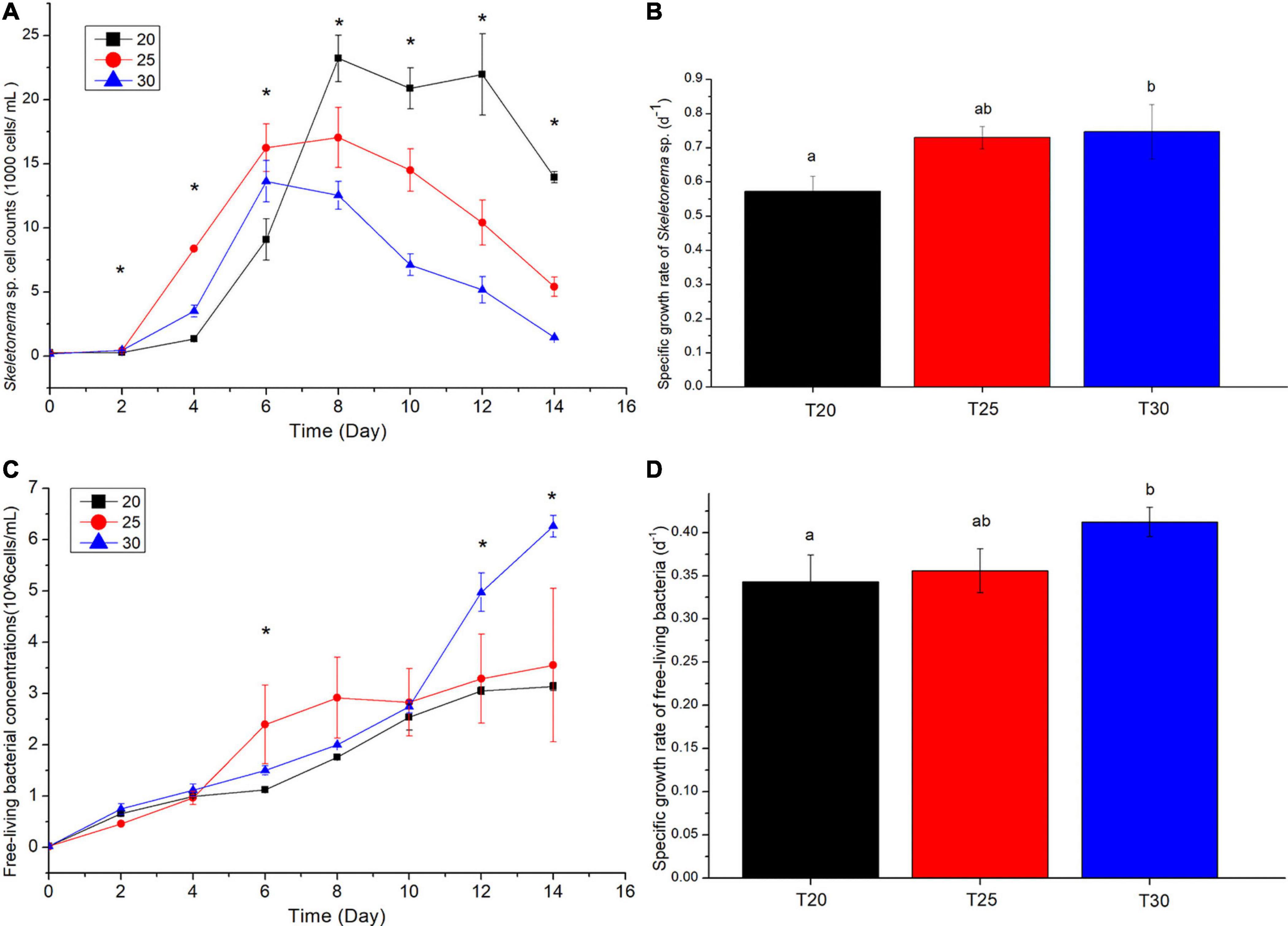
Figure 1. (A) Growth curve and (B) specific growth rate of Skeletonema sp. at 20 (T20), 25 (T25), and 30°C (T30). The (C) growth curve and (D) specific growth rate of free-living bacteria in the culture at 20 (T20), 25 (T25), and 30°C (T30). *Indicates significant difference above all the three temperatures (p < 0.05). Values are the means ± SD, n = 6 (six replicate cultures). Different letters indicate significant difference between different treatments.
At 20 and 25°C, DOC concentrations gradually increased along the increase of cell concentrations of Skeletonema sp. and free-living bacteria in the culture (Figure 2). At 20°C, DOC concentration increased from 0.49 mMC on day 4 to 0.52 mMC on day 8 and 0.97 mMC on day 14. At 25°C, DOC concentration increased from 0.41 mMC on day 4 to 0.52 mMC on day 8 and 0.66 mMC on day 14. In contrast, the DOC concentration at 30°C increased from 0.57 mMC on day 4 to 1.02 mMC on day 8 but decreased from day 8 to day 14 (0.77 mMC). On day 4 and day 14, no significant differences in DOC concentrations were detected among the three different temperatures. On day 8, the DOC concentration at 30°C was significantly higher than that at 20 and 25°C (p = 0.009). Different temperatures and cultivation stages showed interaction with each other in DOC concentration (two-way ANOVA, p = 0.027) (Supplementary Table 1).
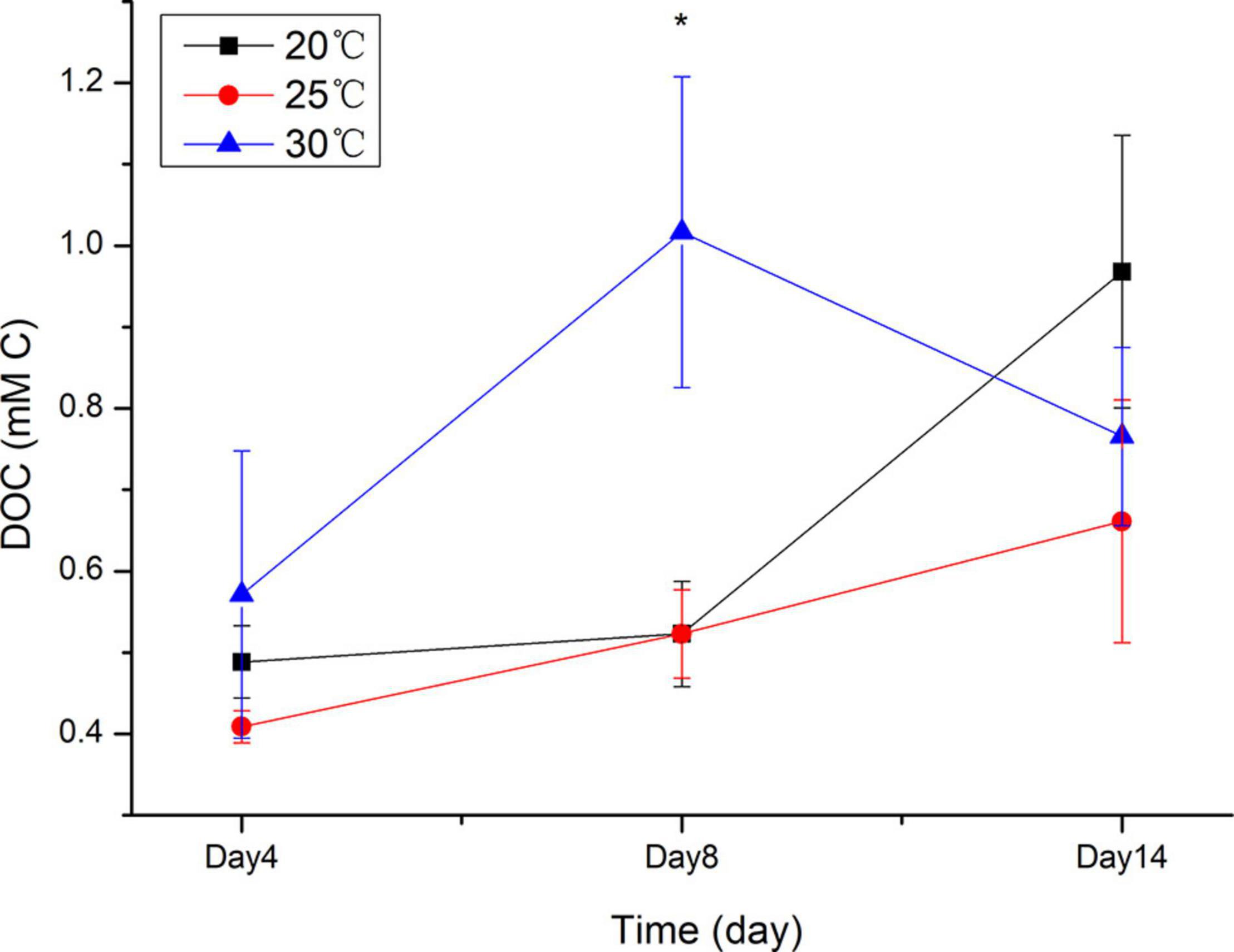
Figure 2. DOC concentrations on days 4, 8, and 14 at different temperatures in the Skeletonema sp. culture. Values are the means ± SD, n = 6 (six replicate cultures). *Indicates significant difference between different treatments (p < 0.05).
Sequences obtained were 3,130,097 after optimization with average length in 449.86 bp. From these, 666 ASVs, 76 species, 72 genera, 60 families, 39 orders, and 14 classes mainly Proteobacteria were obtained after the elimination of erroneous sequences (Supplementary Table 2 and Figures 1, 2). The alpha diversity indexed by Chao, Sobs, Ace, Shannon, Coverage, Simpsoneven, Shannoneven, and Simpson of different samples were demonstrated in Supplementary Table 3. Supplementary Figure 3 shows the beta diversity of Skeletonema sp. associated free-living and attached bacteria at different time points and growth stages by NMDS analysis. Two major genera of bacteria associated with Skeletonema sp. were Alteromonas and Marinobacterium in this study (Figure 3 and Supplementary Figure 2). Free-living and attached bacterial communities were not significantly distinct from each other overall, based on total bacterial community composition (Figure 3 and Supplementary Figure 2). However, some genera showed preferences for free-living or attached states. Vibrio and Pseudomonas had significantly higher relative abundances in the attached bacteria category than free-living bacteria compared with other genera. In contrast, Colwelliaceae showed significantly higher relative abundance among the free-living bacteria (Figure 4).
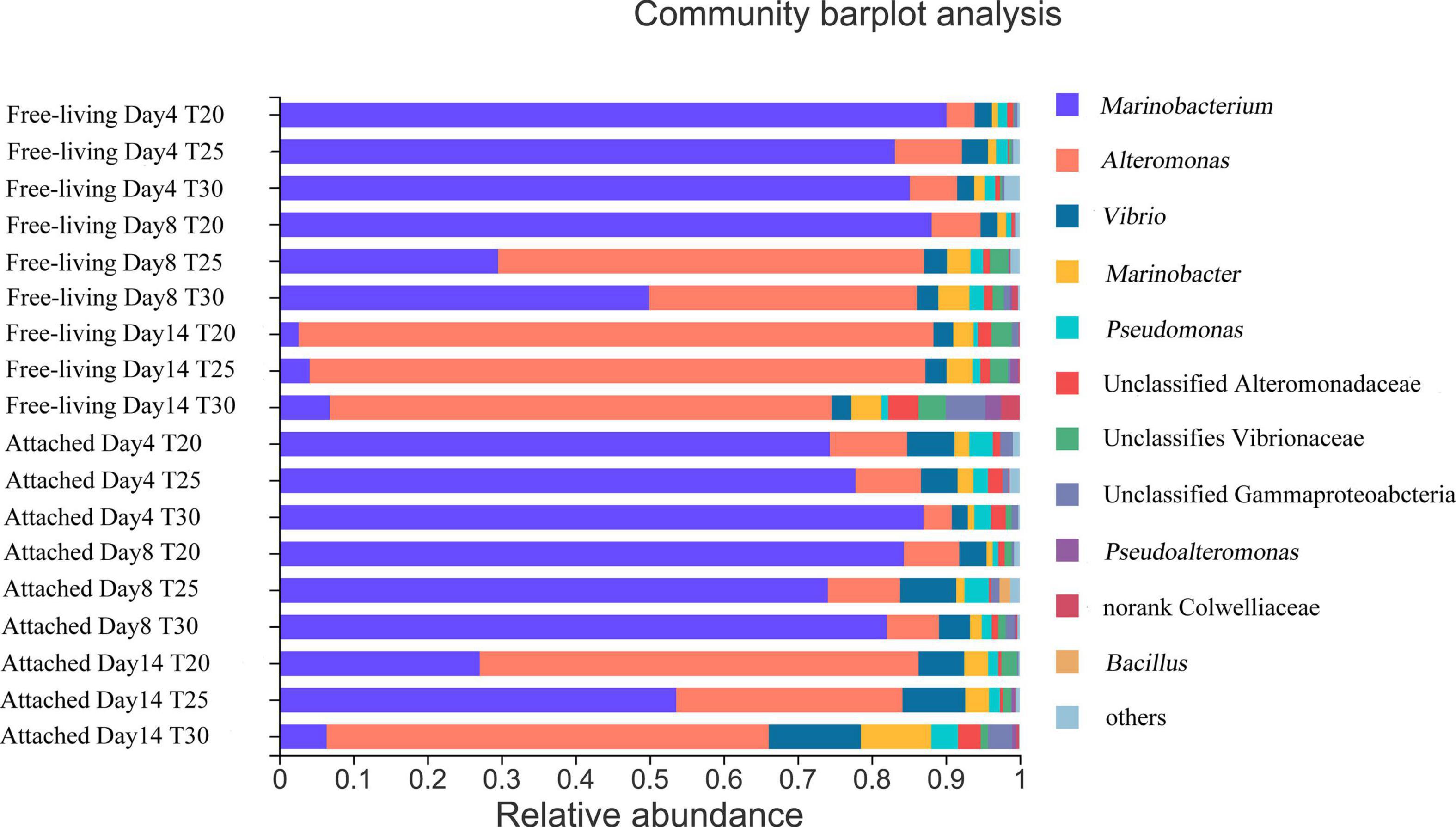
Figure 3. Bacterial composition of the microbial consortium of samples at different time points (days 4, 8, and 14) and at different temperatures (T20, T25, and T30) of free-living and attached bacteria with average value from three replicates for each treatment in genus level.
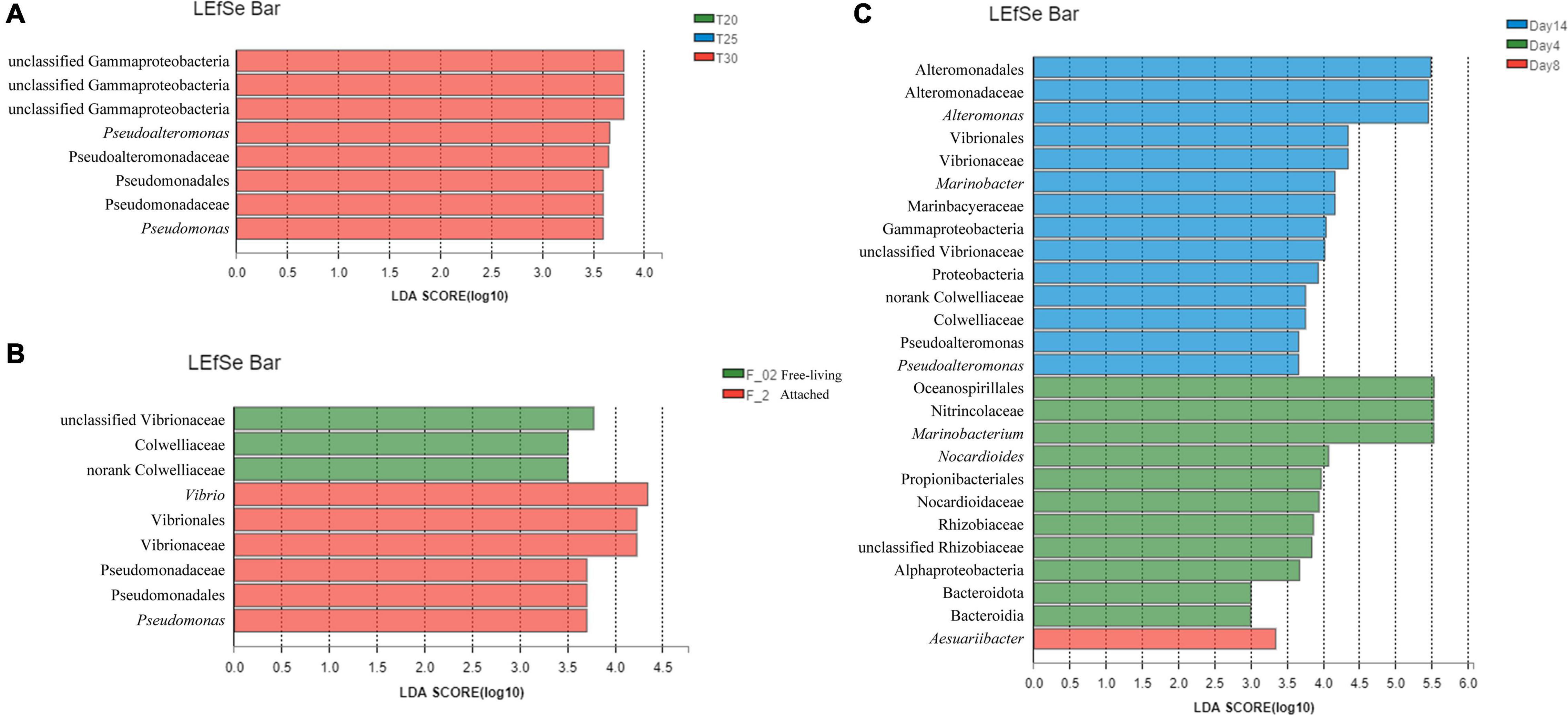
Figure 4. Taxa differentially abundant at (A) different temperatures in (B) free-living and attached bacteria, and at (C) different culturing time points based on the LEfSe analysis.
Cultivation stage had a strong impact on bacterial community composition for both free-living and attached bacteria. From day 4 to 14, the relative proportion of Marinobacterium decreased while Alteromonas increased in both free-living bacteria and attached bacteria (Figure 3 and Supplementary Figure 4). A significant shift in bacterial community composition occurred on day 14 at the degradation phase compared with the other two time points. On day 4, significantly higher relative proportions of Mariobacterium (LDA > 5.5) and Nocardioides (LDA > 4) were observed based on the LEfSe analysis (Figure 4). On day 14, Alteromonas (LDA > 4.5), Marinobacter (LDA > 4), and Pseudoalteromonas (LDA > 3.5) showed significantly higher relative abundances compared with other time points (Figure 4). Regardless of culturing temperatures and physical states of bacteria, the alpha diversity of bacterial community was significantly higher on day 14 compared with day 4 based on Shannon, Shannoneven, Simpsoneven indexes, indicating community diversity and evenness. However, the Simpson index indicating community diversity was significantly higher on day 4 compared with that on day 14 (Supplementary Table 3).
This study showed that elevated temperature influenced the bacterial community. Regardless of cultivation stages and physical states of bacteria, the alpha diversity of bacterial community was significantly higher at 30°C compared with that at 20°C based on Chao, Sobs, Shannon indexes, indicating community richness and diversity (Supplementary Table 3). Although the samples cultured at different temperatures did not form clearly separated clusters (Supplementary Figure 3), the LEfSe analysis showed that Pseudoalteromonas (LDA > 3.5) and Pseudomonas (LDA > 3.5) had significant higher relative abundances at 30°C compared with the samples cultured at 20 and 25°C (Figure 4).
Cultivation stage shows the strongest effect on the bacterial community, while temperature demonstrates the weakest effects, as illustrated in Figure 5. Linear regression analysis of the bacterial community beta diversity demonstrated that cultivation time was negatively correlated with beta diversity (PcoA R2 = 0.600, p = 0.000001). Temperature and physical states did not have a significant positive or negative linear correlation with the beta diversity of the bacterial community. However, some bacterial taxa showed differential relative abundances under different temperatures. Pseudoalteromonas showed significantly higher relative abundance at elevated temperature in both free-living and attached bacteria (Figures 4, 6). Of the attached bacteria, only Pseudoalteromonas showed a positive correlation with elevated temperature. Of the free-living bacteria, Pseudoalteromonas, Colwelliaceae, Marinobacter, and Pseudomonas showed a positive correlation with elevated temperature (Figure 6). In the free-living bacteria, the relative abundances of Alteromonas, Marinobacter, Vibrionaceae, Alteromonadaceace, Colwelliaceae, and Pseudoalteromonas were higher on day 14. In attached bacteria, the relative proportions of Marinobacter, Alteromonas, Vibrio, and Pseudoalteromonas were significantly higher on day 14 (Figure 6). On day 8, Marinobacter showed significantly higher relative abundances (p = 0.0004) at higher temperature in free-living bacteria, but no bacterial taxa were significantly influenced among the attached bacteria (Supplementary Figure 4).
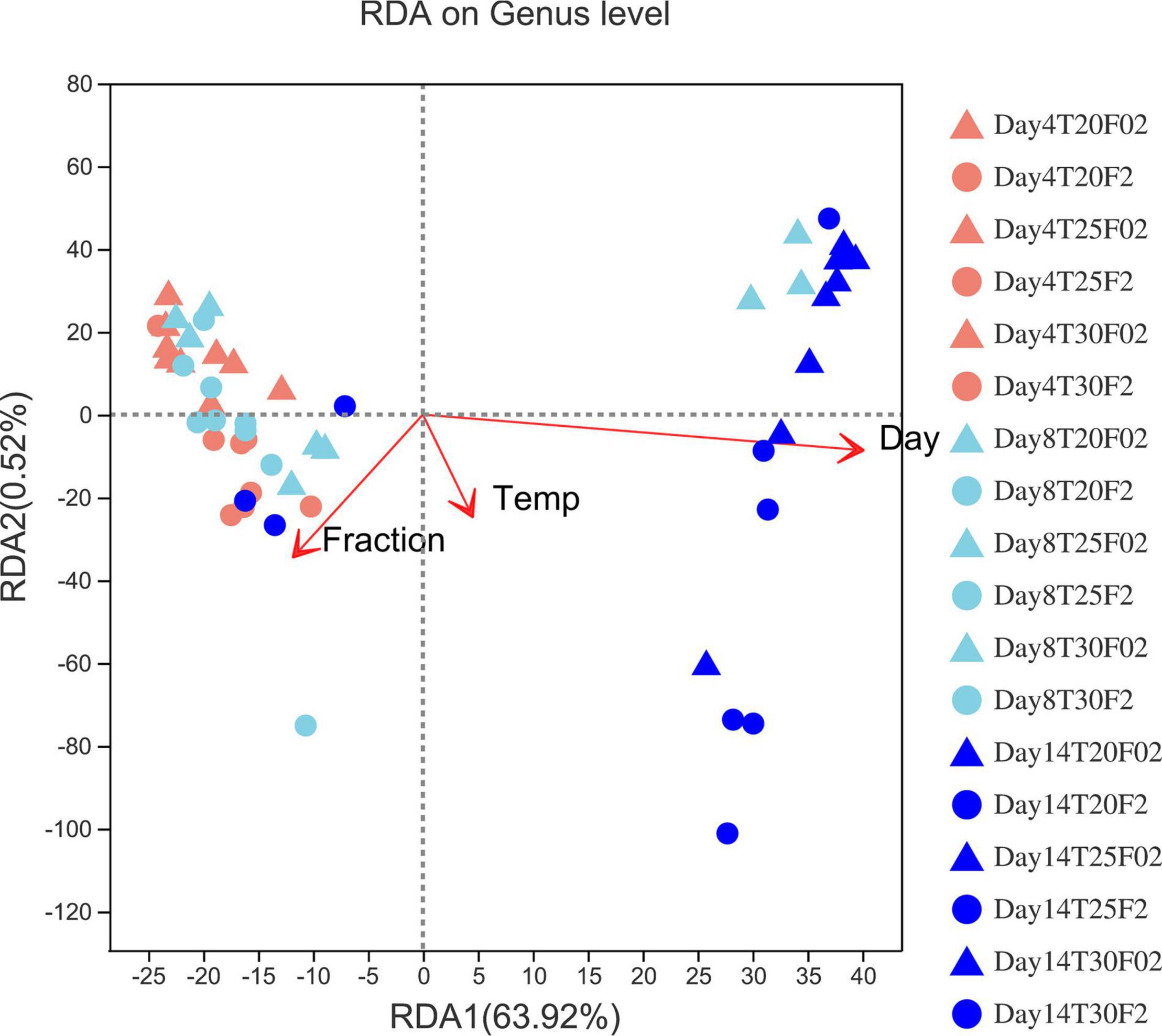
Figure 5. Redundancy analysis (RDA) of microbial communities and environmental variables such as fraction, temperature, and sampling day (red arrows). The lengths of arrows indicate the magnitude of variance to which that variable could be explained. Smaller angles between two variables indicate stronger correlations between them.
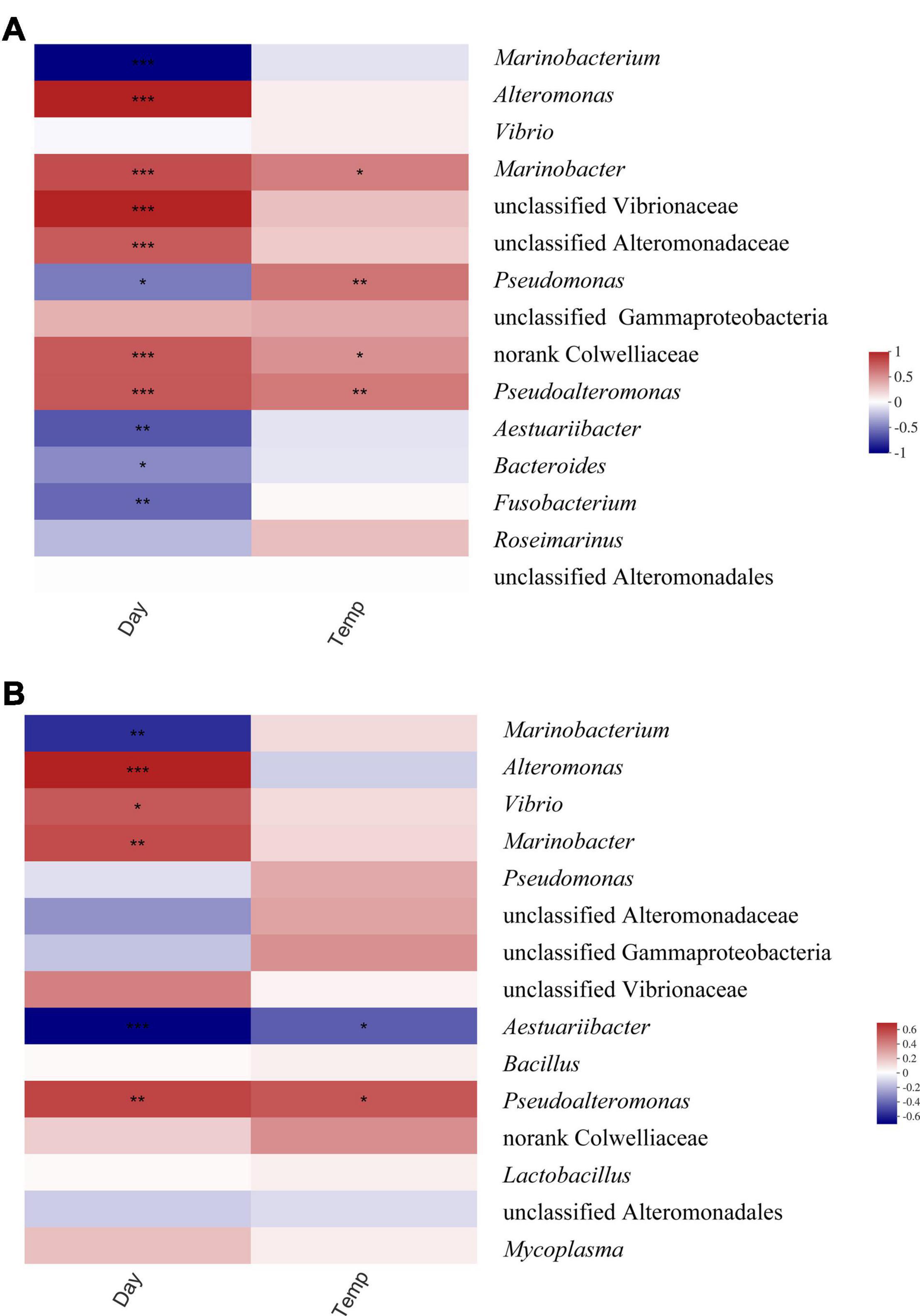
Figure 6. Spearman’s rank correlation coefficients and the corresponding p-values (0.01 < p ≤ 0.05, **0.001 < p ≤ 0.01, ***p ≤ 0.001) were calculated based on correlation of the relative abundance of top 15 abundant taxa and different factors, such as cultivation time (days 4, 8, and 14) and temperatures (T20, T25, and T30) in (A) free-living and (B) attached bacteria.
The functional profiles (KO and metabolic features) of bacterial communities from different samples based on PICRUSt2 analysis are listed in Supplementary Table 4. Many metabolism pathways either positively or negatively correlate with cultivation stages based on Spearman correlation analysis, as shown in Supplementary Table 5. In contrast, far fewer metabolic pathways were correlated with different temperatures and bacterial physical states. ABC-2 type transport system ATP-binding protein (K01990, R = 0.52, p = 7.11E-05), carbon fixation pathways in prokaryotes (Ko00720, R = 0.41, p = 0.002), and carbapenem resistance (M00851, R = 0.5, p = 0.04) were highly correlated with elevated temperature (Supplementary Table 5). Lysosome (ko04142, R = 0.3, p = 0.03) and N-glycan biosynthesis (ko00513, R = 0.3, p = 0.05) were also significantly correlated with elevated temperature (Supplementary Table 5). Amino acid biosynthesis, such as lysine biosynthesis (Ko00300, R = 0.27, p = 0.049), and tyrosine and tryptophan biosynthesis (Ko00400, R = 0.3, p = 0.02), were found to be positively correlated with elevated temperature (Supplementary Table 5). We further carried out a significant difference analysis of carbon fixation metabolism (Ko00720) and lysine (Ko00300), phenylalanine, tyrosine, and tryptophan biosynthesis (Ko00400) at different time points of both free-living and attached bacteria. The results showed that significantly increased carbon fixation metabolism (Ko00720, p = 0.005) was detected at 30°C on day 14 in free-living bacteria (Figure 7) but not in attached bacteria (data not shown). Lysine biosynthesis (ko00300, p = 0.001), and tyrosine and tryptophan biosynthesis (Ko00400, p = 0.002) also showed significantly higher abundances at 30°C on day 14 in free-living bacteria.
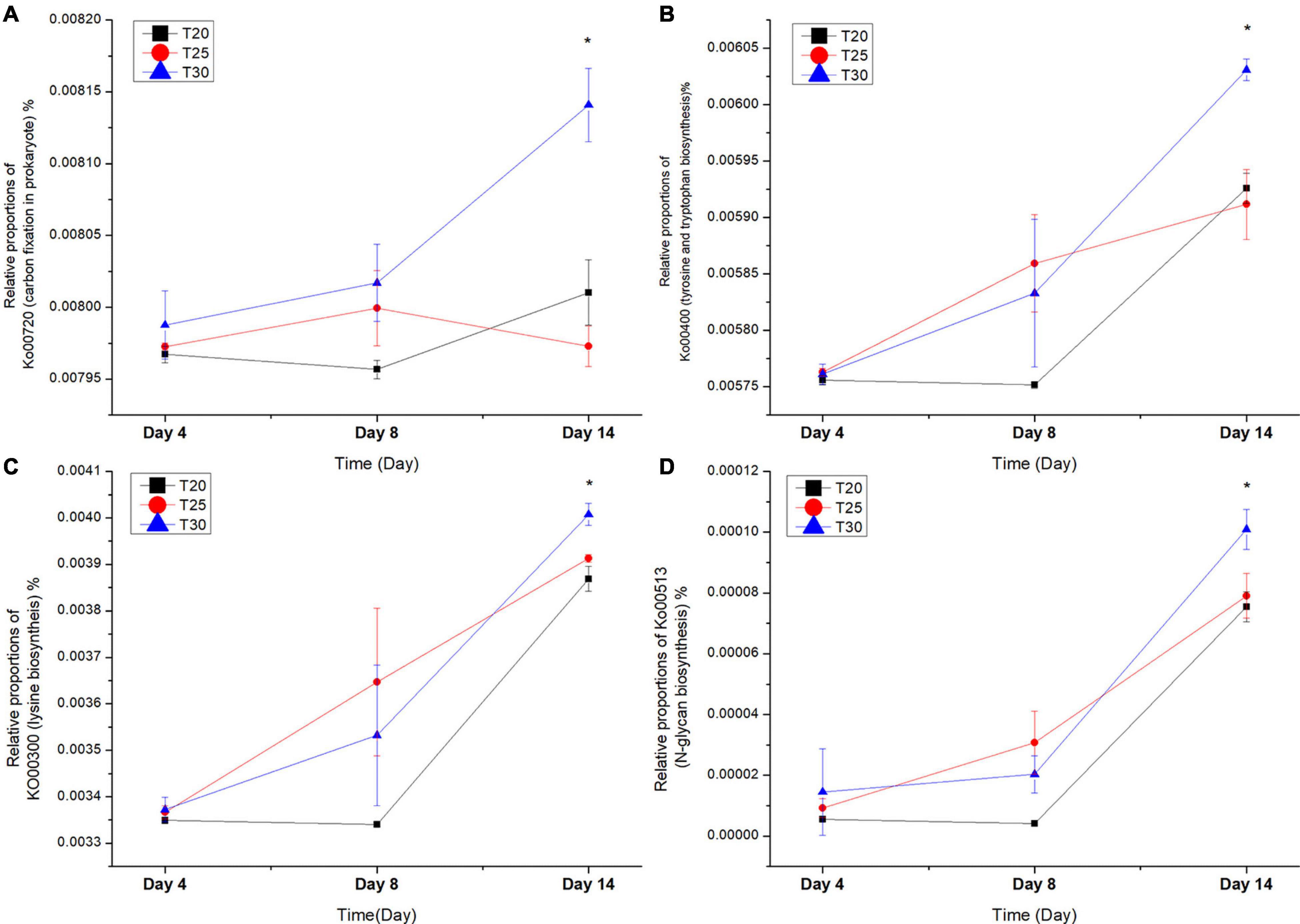
Figure 7. The relative proportions of (A) Ko00720 (carbon fixation pathways in prokaryotes, (B) Ko00400 (phenylalanine, tyrosine and tryptophan biosynthesis), (C) Ko00300 (lysine biosynthesis), (D) Ko00513 (N-glycan biosynthesis at different temperatures and time points). The one-way ANOVA method was used to perform the significance differences analysis. *Indicates significant difference between different treatments (p < 0.05).
The configuration of the network was more stable at elevated temperature, with an increased number of nodes and edges and higher average degrees (Table 1). The number of interactions between different bacteria was dramatically increased as the temperature increased, especially for free-living bacteria. The connections among bacteria, dominated by positive connections, were substantially higher at higher temperatures (Figure 8 and Table 1). For free-living bacteria, 139 nodes with 880 positive and 13 negative connections at 20°C, 153 nodes with 951 positive and three negative connections at 25°C, and 217 nodes, 2,283 positive and 0 negative connections at 30°C were found. For attached bacteria, 134 nodes with 756 positive and a negative connections at 20°C, 147 nodes with 1130 positive and two negative connections at 25°C, and 154 nodes, 1319 positive and three negative links at 30°C were found (Table 1).
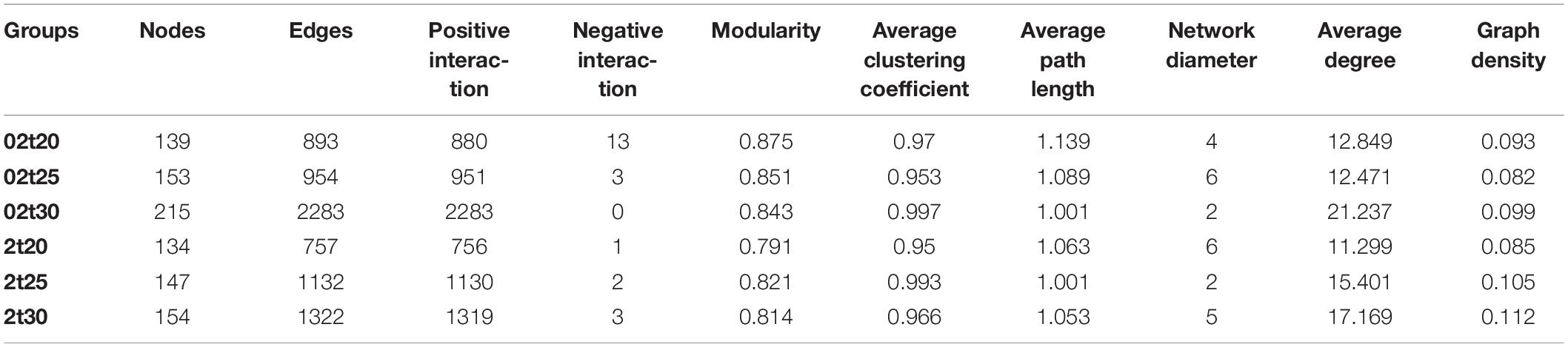
Table 1. Topological properties of interaction networks under different conditions (02t20: free-living bacteria at 20°C; 02t25: free-living bacteria at 25°C; 02t30: free-living bacteria at 30°C; 2t20: attached bacteria at 20°C; 2t25: attached bacteria at 25°C; and 2t30: attached bacteria at 30°C).
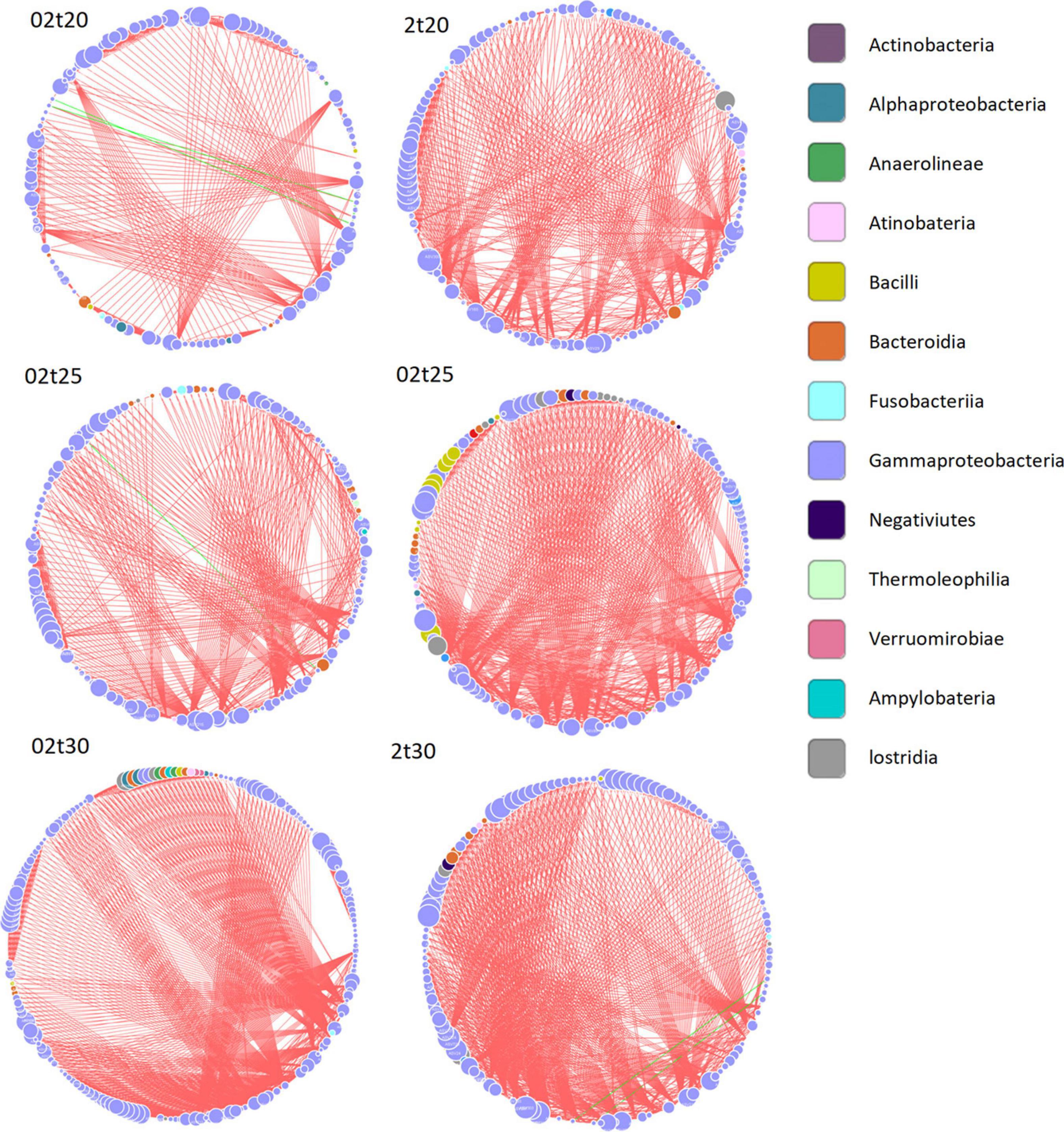
Figure 8. Network analysis of bacterial interaction at different temperatures (02t20: free-living bacteria at 20°C; 02t25: free-living bacteria at 25°C; 02t30: free-living bacteria at 30°C; 2t20: attached bacteria at 20°C; 2t25: attached bacteria at 25°C; and 2t30: attached bacteria at 30°C). Each node represents an ASV. Node color represents taxon on class level. Green lines represent negative correlation, and red lines represent positive correlation between ASVs. The size of node indicates the number of links with other ASVs. The big nodes have more links with other ASVs.
It has been predicted that elevated temperature will increase the metabolic rates across the diversity of living organisms (López-Urrutia and Morán, 2007), and it is generally considered that elevated temperature enhances enzyme activities and metabolic rates within temperature ranges suitable for the growth of both bacteria and phytoplankton (Morán et al., 2015). In this study, elevated temperature significantly increased the growth rate of Skeletonema, shortened the exponential phase, and decreased the maximum cell abundance of Skeletonema sp. (Figure 1). We observed increased growth rate with lower carrying capacity (lower maximum cell abundance) of Skeletonema sp. at elevated temperature, and this is in line with previous studies showing that warming can lead to the lower carrying capacity of organisms (Kaeriyama et al., 2011; Stock et al., 2019). At elevated temperatures, an earlier onset of Skeletonema sp. cell abundance peak was observed in this study, and this was also consistent with a previous mesocosm experiment dominated by diatoms (Engel et al., 2011; von Scheibner et al., 2014). The thermal response of Skeletonema sp. may have also been influenced by the native bacteria assemblage. In this study, the cell abundances of free-living bacteria were significantly higher at higher temperatures at different stages during cultivation, especially at 3°C in the decline stage (Figure 1), which is consistent with previous studies (von Scheibner et al., 2014; Moran, 2015). It has been shown that heterotrophic bacteria are more affected by elevated temperature than phytoplankton (Eppley, 1972; Pomeroy and Wiebe, 2001). However, Skeletonoma sp. was more strongly affected than free-living bacteria in terms of specific growth rate in this study. The abundance of attached bacteria needs to be quantified to fully demonstrate the effects of elevated temperature on bacterial densities.
We found higher relative abundances of Alteromonas at the decline stage in different temperature regimes. Alteromonas, a global marine opportunistic taxon in the phycosphere and commonly found in phytoplankton lab cultures and field samples (López-Pérez et al., 2012, 2013; Shibl et al., 2020), has been known to be good at degrading DOC, and plays vital roles in carbon cycling in the oceanic ecosystem (Pedler et al., 2014). We speculate that a higher relative abundance of Alteromonas may help to consume a large amount of DOC released from Skeletonema sp. resulting in no detectable differences in DOC concentration at the decline stage compared with other stages. In contrast to Alteromonas, Marinobacterium dominated at the early stage in the culture but showed lower relative abundance in the decline stage. Previous studies have shown that DOC compositions in the early stage were distinct from those in the decline stage in Skeletonema culture (Mannino and Harvey, 2002). Alteromonas sp. and Marinobacterium jannaschii have been reported to have distinct capabilities in utilizing small peptides in seawater, which may be correlated with their distinct relative abundances in relation to the cultivation of Skeletonema sp. in this study (Huang et al., 2020). On day 8, significantly higher DOC concentration was detected at 30°C than at the lower temperatures tested (Figure 2), which is consistent with previous studies indicating that increased proportion of primary produced organic matter channeled into DOC at elevated temperature, most notably after nutrient exhaustion (Wohlers et al., 2009; Kim et al., 2011). Considering the interactions of DOM quantity, DOM quality, and bacterial community composition (Engel et al., 2011), the intercorrelations of bacterial community structure, DOM composition, and carbon flow at elevated temperature merit further exploration in the future.
According to the LEfSe analysis (Figure 4), a higher relative abundance of Vibrio was detected in the attached bacteria. Vibrio has been reported to attach to algal cells, as growing algal populations provide additional attachment sites on organic-rich substrates (Takemura et al., 2014). The attached state of Vibrio may be able to benefit from absorbing organic matter from Skeletonema sp. in this study. Pseudoalteromonas and Pseudomonas tended to appear in 30°C culture regardless of physical states compared with 25 and 20°C cultures (Figure 6). The potential algicidal effects of Pseudoalteromonas (Kato et al., 1998; Lee et al., 2000; Lyu et al., 2019) and Pseudomonas (Wang et al., 2005) on Skeletonema costatum and harmful algae species have been reported. Higher relative abundances of Pseudoalteromonas and Pseudomonas under elevated temperature may result in potential higher activity of algicidal activity against Skeletonema sp. reflected by the lower maximum Skeletonema sp. abundance in 30°C compared to lower temperatures (Figure 1) was probably partially due to higher relative abundance of Pseudoalteromonas and Pseudomonas under higher temperature. Pseudoalteromonas also showed significantly higher relative abundances on day 14 in both attached and free-living bacteria (Figure 4). Pseudoalteromonas has been reported to be dominant in the decline stage of S. costatum and Akashiwo sanguinea bloom in the Xiamen sea area (Li et al., 2012). The algicidal activity and capability in metabolizing the organic matrix of diatom of Pseudoalteromonas (Bidle and Azam, 2001; Li et al., 2012) may explain its dominance at the decline stage.
In line with previous studies, the community composition of free-living and attached bacterial communities was distinct at different growth stages of Skeletonema sp. (Acinas et al., 1999; Grossart et al., 2005). However, the free-living bacteria and attached bacteria did not show distinct community patterns in this study. Contrasting findings on attached and free-living bacterial communities have been reported (Acinas et al., 1999; Dang and Lovell, 2016; Hu et al., 2020). One of the underlying reasons might be that the filtering method we used did not separate free-living and attached bacteria very well. The similarity between the community of free-living and attached bacteria was also possibly due to high nutrient and algal biomass with high bacterial substrate availability in the lab cultures (Hollibaugh et al., 2000), allowing the same bacterial taxa to exist in both the attached and free-living statuses with different physiological status and activities (Cooksey and Wigglesworth-Cooksey, 1995).
Previous studies using natural microbial assemblies in coastal seawater have indicated more pronounced impacts of elevated temperature on phytoplankton-attached bacteria in terms of cell abundance, carbon uptake, and enzyme activities compared with free-living bacteria. Better access to newly fixed DOC by phytoplankton and increased chemotaxis ability of attached bacteria compared with free-living counterparts were considered as major factors (Arandia-Gorostidi et al., 2017, 2020). In this study, the effects of temperature on free-living bacteria were more pronounced than those on attached bacteria in terms of bacterial community composition, a result that differs from previous studies (Arandia-Gorostidi et al., 2017, 2020; Figure 6). Correlation analysis based on the PICRUSt2 predictions showed that carbon metabolism was significantly correlated with elevated temperature. Further significant difference analysis indicated that carbon metabolism was significantly enhanced at elevated temperature in free-living bacteria but not in attached bacteria during the decline phase on day 14. However, more active organic carbon consumption by free-living bacteria at higher elevated temperature did not result in lower DOC concentration in the culture on day 14 (Figure 2). We speculate that increased rate of both DOC release and consumption at elevated temperature finally resulted in no significant changes in DOC concentration in the decline phase. Besides carbon metabolism, significant enhancement of several specific amino acid metabolisms at higher elevated temperature was detected (Figure 7 and Supplementary Table 5), which may reflect the changes in the amino acid composition of bacteria (Kreil and Ouzounis, 2001; Venev and Zeldovich, 2018). The potential changes in amino acid composition in bacteria at elevated temperature may result in changes in adaptation to different environment conditions (Cabello-Yeves and Rodriguez-Valera, 2019) and would have a profound influence on the microbial loop and oceanic carbon cycle (Repeta, 2015). N-glycans have been reported to play important roles in dealing with heat shock (Cain et al., 2019) and to affect nitrate reductase activity and chemotaxis in pathogenic bacteria (Nothaft and Szymanski, 2019). The results implied that N-glycans may also be vital for marine non-pathogenic bacteria in response to elevated temperature. Significant positive correlation of ABC-2 type transport system ATP-binding protein with elevated temperature was detected in our study. ABC transporters are considered as highly efficient translocation systems with a remarkable diversity of functions (Lewis et al., 2012). The underlying mechanisms of effects of elevated temperature on ABC transporter in marine bacteria remain to be further investigated.
In the future, further metatranscriptomic data and related metabolite measurements may better decipher the changes in the whole bacterial community in response to elevated temperature.
In this study, the interactions, mainly positive, among different bacterial species were significantly enhanced at 30°C (Figure 8), which implied that the enhanced cooperation between different bacterial species with increased positive interactions and more stable network structure may help the whole community to overcome the stress brought by the elevated temperature as indicated in a previous study (Zhang et al., 2020). Previous studies have also shown that bacterial interaction networks were modified by abiotic stresses (Lin et al., 2018; Ratzke et al., 2020). Interestingly, the enhanced interactions in the bacterial community at elevated temperature were more pronounced in attached bacteria compared with free-living bacteria, which implied that elevated temperatures may have a greater impact on the attached bacterial community. The changes in functions and interactions of the bacterial community in response to elevated temperatures might influence energy transfer and element cycling, a topic which deserves further investigation.
In conclusion, we demonstrated that the effects of temperature on the structure of the free-living bacterial community were more pronounced compared with diatom-attached bacteria. Furthermore, the results showed that different bacterial species may build a more integrated network with a modified functional profile of the whole community to cope with elevated temperature. More comprehensive studies on the relationship between bacterial community structure, community functional profiles based on metatranscriptomics data, and element cycling at elevated temperature are needed to better decipher the responses of diatom-associated bacterial community to ocean warming.
The data generated in this manuscript can be found in NCBI (project ID: PRJNA706479).
XL conceived and designed the experiments. SC performed the experiments. ZD, XL, PZ, XZ, and QL analyzed data. XL wrote the manuscript. JA revised the manuscript. All authors reviewed the manuscript.
This study was supported by the National Natural Science Foundation of China (Grant No: 42076128), the National Key Research and Development Program of China (Grant No: 2016YFA0601302), and Fujian Provincial Research Institutes of Basic Research and Public Service Special Operations (2019R1007-2).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors appreciate the assistance of Xiangqi Yi in data analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.626207/full#supplementary-material
Acinas, S. G., Antón, J., and Rodríguez-Valera, F. (1999). Diversity of free-living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl. Environ. Microbiol. 65, 514–522. doi: 10.1128/aem.65.2.514-522.1999
Arandia-Gorostidi, N., Alonso-Sáez, L., Stryhanyuk, H., Richnow, H. H., Morán, X. A. G., and Musat, N. (2020). Warming the phycosphere: differential effect of temperature on the use of diatom-derived carbon by two copiotrophic bacterial taxa. Environ. Microbiol. 22, 1381–1396. doi: 10.1111/1462-2920.14954
Arandia-Gorostidi, N., Weber, P. K., Alonso-Sáez, L., Morán, X. A. G., and Mayali, X. (2017). Elevated temperature increases carbon and nitrogen fluxes between phytoplankton and heterotrophic bacteria through physical attachment. ISME J. 11, 641–650. doi: 10.1038/ismej.2016.156
Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. doi: 10.1126/science.1101156
Azam, F. (1998). Microbial control of oceanic carbon flux: the plot thickens. Science 280, 694–696. doi: 10.1126/science.280.5364.694
Barofsky, A., Vidoudez, C., and Pohnert, G. (2009). Metabolic profiling reveals growth stage variability in diatom exudates. Limnol. Oceanogr. Methods 7, 382–390. doi: 10.4319/lom.2009.7.382
Bidle, K. D., and Azam, F. (2001). Bacterial control of silicon regeneration from diatom detritus: significance of bacterial ectohydrolases and species identity. Limnol. Oceanogr. 46, 1606–1623. doi: 10.4319/lo.2001.46.7.1606
Bowler, C., Allen, A. E., Badger, J. H., Grimwood, J., Jabbari, K., Kuo, A., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. doi: 10.1038/nature07410
Cabello-Yeves, P. J., and Rodriguez-Valera, F. (2019). Marine-freshwater prokaryotic transitions require extensive changes in the proteome. bioRxiv [Preprint]. doi: 10.1101/544619
Cain, J. A., Dale, A. L., Niewold, P., Klare, W. P., Man, L., White, M. Y., et al. (2019). Proteomics reveals multiple phenotypes associated with n-linked glycosylation in Campylobacter jejuni. Mol. Cell. Proteomics 18, 715–734. doi: 10.1074/mcp.RA118.001199
Cooksey, K. E., and Wigglesworth-Cooksey, B. (1995). Adhesion of bacteria and diatoms to surfaces in the sea: a review. Aquat. Microb. Ecol. 9, 87–96. doi: 10.3354/ame009087
Dang, H., and Lovell, C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/mmbr.00037-15
Douglas, G. M., Mafei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 669–688. doi: 10.1038/s41587-020-0548-6
Edwards, M., and Richardson, A. J. (2004). Impact of climate change on marine pelagic phenology and trophic mismatch. Nature 363, 2002–2005.
Engel, A., Händel, N., Wohlers, J., Lunau, M., Grossart, H. P., Sommer, U., et al. (2011). Effects of sea surface warming on the production and composition of dissolved organic matter during phytoplankton blooms: results from a mesocosm study. J. Plankton Res. 33, 357–372. doi: 10.1093/plankt/fbq122
Francis, C. A., Roberts, K. J., Beman, J. M., Santoro, A. E., and Oakley, B. B. (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U.S.A. 102, 14683–14688. doi: 10.1073/pnas.0506625102
Grossart, H., Levold, F., Allgaier, M., Simon, M., and Brinkhoff, T. (2005). Marine diatom species harbour distinct bacterial communities. Environ. Microbiol. 7, 860–873. doi: 10.1111/j.1462-2920.2005.00759.x
Grossart, H., and Simon, M. (2007). Interactions of planktonic algae and bacteria: effects on algal growth and organic matter dynamics. Aquat. Microb. Ecol. 47, 163–176. doi: 10.3354/ame047163
Grossart, H., Tang, K. W., Kiørboe, T., and Ploug, H. (2007). Comparison of cell-specific activity between free-living and attached bacteria using isolates and natural assemblages. FEMS Microbiol. Lett. 266, 194–200. doi: 10.1111/j.1574-6968.2006.00520.x
Hollibaugh, J. T., Wong, P. S., and Murrell, M. C. (2000). Similarity of particle-associated and free-living bacterial communities in northern San Francisco Bay, California. Aquat. Microb. Ecol. 21, 103–114. doi: 10.3354/ame021103
Horn, K. (2011). Alga-derived substrates select for distinct betaproteobacterial lineages and contribute to niche separation in limnohabitans strains. Appl Environ Microbiol. 77, 7307–7315. doi: 10.1128/AEM.05107-11
Hu, Y., Xie, G., Jiang, X., Shao, K., Tang, X., and Gao, G. (2020). The relationships between the free-living and particle-attached bacterial communities in response to elevated eutrophication. Front. Microbiol. 11:423. doi: 10.3389/fmicb.2020.00423
Huang, F., Pan, L., He, Z., Zhang, M., and Zhang, M. (2020). Identification, interactions, nitrogen removal pathways and performances of culturable heterotrophic nitrification-aerobic denitrification bacteria from mariculture water by using cell culture and metagenomics. Sci. Total Environ. 732:139268. doi: 10.1016/j.scitotenv.2020.139268
Jiao, N., Herndl, G. J., Hansell, D. A., Benner, R., Kattner, G., Wilhelm, S. W., et al. (2010). Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 8, 593–599. doi: 10.1038/nrmicro2386
Johansson, O. N., Töpel, M., Pinder, M. I. M., Kourtchenko, O., Blomberg, A., Godhe, A., et al. (2019a). Skeletonema marinoi as a new genetic model for marine chain-forming diatoms. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-019-41085-5
Johansson, O. N., Pinder, M. I. M., Ohlsson, F., Egardt, J., Töpel, M., and Clarke, A. K. (2019b). Friends with benefits: exploring the phycosphere of the marine diatom Skeletonema marinoi. Front. Microbiol. 10:1828. doi: 10.3389/fmicb.2019.01828
Kaeriyama, H., Katsuki, E., Otsubo, M., Yamada, M., Ichimi, K., Tada, K., et al. (2011). Effects of temperature and irradiance on growth of strains belonging to seven Skeletonema species isolated from Dokai Bay, southern Japan. Eur. J. Phycol. 46, 113–124. doi: 10.1080/09670262.2011.565128
Kato, J., Amie, J., Murata, Y., Kuroda, A., Mitsutani, A., and Ohtake, H. (1998). Development of a genetic transformation system for an alga-lysing bacterium. Appl. Environ. Microbiol. 64, 2061–2064. doi: 10.1128/aem.64.6.2061-2064.1998
Kim, J. M., Lee, K., Shin, K., Yang, E. J., Engel, A., Karl, D. M., et al. (2011). Shifts in biogenic carbon flow from particulate to dissolved forms under high carbon dioxide and warm ocean conditions. Geophys. Res. Lett. 38, 1–5. doi: 10.1029/2011GL047346
Kreil, D. P., and Ouzounis, C. A. (2001). Identification of thermophilic species by the amino acid compositions deduced from their genomes. Nucleic Acids Res. 29, 1608–1615. doi: 10.1093/nar/29.7.1608
Lee, S. O., Kato, J., Takiguchi, N., Kuroda, A., Ikeda, T., Mitsutani, A., et al. (2000). Involvement of an extracellular protease in algicidal activity of the marine bacterium Pseudoalteromonas sp. strain A28. Appl. Environ. Microbiol. 66, 4334–4339. doi: 10.1128/AEM.66.10.4334-4339.2000
Lewis, V. G., Ween, M. P., and McDevitt, C. A. (2012). The role of ATP-binding cassette transporters in bacterial pathogenicity. Protoplasma 249, 919–942. doi: 10.1007/s00709-011-0360-8
Li, Y., Yang, C., Li, D., Tia, Y., and Zheng, T. (2012). Dynamics of bacterial community during the bloom caused by Skeletonema costatum and Akashiwo sanguinea in Xiamen sea area. Acta Microbiol. Sin. 52, 1268–1281. doi: 10.1021/cen-v027n032.p2282
Lin, X., Huang, R., Li, Y., Li, F., Wu, Y., Hutchins, D. A., et al. (2018). Interactive network configuration maintains bacterioplankton community structure under elevated CO2 in a eutrophic coastal mesocosm experiment. Biogeosciences 15, 551–565. doi: 10.5194/bg-15-551-2018
López-Pérez, M., Gonzaga, A., Martin-Cuadrado, A. B., Onyshchenko, O., Ghavidel, A., Ghai, R., et al. (2012). Genomes of surface isolates of Alteromonas macleodii: the life of a widespread marine opportunistic copiotroph. Sci. Rep. 2, 1–11. doi: 10.1038/srep00696
López-Pérez, M., Gonzaga, A., and Rodriguez-Valera, F. (2013). Genomic diversity of “deep ecotype” Alteromonas macleodii isolates: evidence for pan-mediterranean clonal frames. Genome Biol. Evol. 5, 1220–1232. doi: 10.1093/gbe/evt089
López-Urrutia, Á, and Morán, X. A. G. (2007). Resource limitation of bacterial production distorts the temperature dependence of oceanic carbon cycling. Ecology 88, 817–822. doi: 10.1890/06-1641
Lyu, Y. H., Zhou, Y. X., Li, Y., Zhou, J., and Xu, Y. X. (2019). Optimized culturing conditions for an algicidal bacterium Pseudoalteromonas sp. SP48 on harmful algal blooms caused by Alexandrium tamarense. Microbiologyopen 8, 1–10. doi: 10.1002/mbo3.803
Mannino, A., and Harvey, H. (2002). Biochemical composition of dissolved organic matter released during experimental diatom blooms. NASA Rep. [Preprint].
Mével, G., Vernet, M., Goutx, M., and Ghiglione, J. F. (2008). Seasonal to hour variation scales in abundance and production of total and particle-attached bacteria in the open NW Mediterranean Sea (0-1000 m). Biogeosciences 5, 1573–1586. doi: 10.5194/bg-5-1573-2008
Mock, T., Otillar, R. P., Strauss, J., McMullan, M., Paajanen, P., Schmutz, J., et al. (2017). Evolutionary genomics of the cold-Adapted diatom Fragilariopsis cylindrus. Nature 541, 536–540. doi: 10.1038/nature20803
Morán, X. A. G., Alonso-Sáez, L., Nogueira, E., Ducklow, H. W., González, N., and López-Urrutia, Á, et al. (2015). More, smaller bacteria in response to ocean’s warming? Proc. R. Soc. B Biol. Sci. 282:20150371. doi: 10.1098/rspb.2015.0371
Norrman, B., and Zweifel, U. L. (1996). Long-term decomposition of DOC from experimental diatom blooms. Limnol. Oceanogr. 41, 1344–1347. doi: 10.4319/lo.1996.41.6.1344
Nothaft, H., and Szymanski, C. M. (2019). New discoveries in bacterial N-glycosylation to expand the synthetic biology toolbox. Curr. Opin. Chem. Biol. 53, 16–24. doi: 10.1016/j.cbpa.2019.05.032
Pedler, B. E., Aluwihare, L. I., and Azam, F. (2014). Single bacterial strain capable of significant contribution to carbon cycling in the surface ocean. Proc. Natl. Acad. Sci. U.S.A. 111, 7202–7207. doi: 10.1073/pnas.1401887111
Pomeroy, L. R., and Wiebe, W. J. (2001). Temperature and substrates as interactive limiting factors for marine heterotrophic bacteria. Aquat. Microb. Ecol. 23, 187–204. doi: 10.3354/ame023187
Pörtner, H.-O., Roberts, D. C., Masson-Delmotte, V., Zhai, P., Tignor, M., Poloczanska, E., et al. (2019). IPCC Special Report on the Ocean and Cryosphere in a Changing Climate. Geneva: IPCC.
Ratzke, C., Barrere, J., and Gore, J. (2020). Strength of species interactions determines biodiversity and stability in microbial communities. Nat. Ecol. Evol. 4, 376–383. doi: 10.1038/s41559-020-1099-4
Repeta, D. J. (2015). “Chemical characterization and cycling of dissolved organic matter,” in Biogeochemistry of Marine Dissolved Organic Matter, eds D. A. Hansell and C. A. Carlson (Cambridge, MA: Academic Press), 21–63. doi: 10.1016/B978-0-12-405940-5.00002-9
Riemann, L., Steward, G. F., and Azam, F. (2000). Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66, 578–587. doi: 10.1128/aem.66.2.578-587.2000
Rooney-Varga, J. N., Giewat, M. W., Savin, M. C., Sood, S., Legresley, M., and Martin, J. L. (2005). Links between phytoplankton and bacterial community dynamics in a coastal marine environment. Microb. Ecol. 49, 163–175. doi: 10.1007/s00248-003-1057-0
Sarmento, H., Montoya, J. M., Vázquez-Domínguez, E., Vaqué, D., and Gasol, J. M. (2010). Warming effects on marine microbial food web processes: how far can we go when it comes to predictions? Philos. Trans. R. Soc. B Biol. Sci. 365, 2137–2149. doi: 10.1098/rstb.2010.0045
Seymour, J. R., Amin, S. A., Raina, J. B., and Stocker, R. (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2:17065. doi: 10.1038/nmicrobiol.2017.65
Shibl, A. A., Isaac, A., Ochsenkühn, M. A., Cárdenas, A., Fei, C., Behringer, G., et al. (2020). Diatom modulation of select bacteria through use of two unique secondary metabolites. Proc. Natl. Acad. Sci. U.S.A. 117, 27445–27455. doi: 10.1073/pnas.2012088117
Stock, W., Vanelslander, B., Rüdiger, F., Sabbe, K., Vyverman, W., and Karsten, U. (2019). Thermal niche differentiation in the benthic diatom cylindrotheca closterium (Bacillariophyceae) complex. Front. Microbiol. 10:1395. doi: 10.3389/fmicb.2019.01395
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., et al. (2015). Structure and function of the global ocean microbiome. Science 348, 1–10. doi: 10.1126/science.1261359
Takemura, A. F., Chien, D. M., and Polz, M. F. (2014). Associations and dynamics of vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 5:38. doi: 10.3389/fmicb.2014.00038
Thornton, D. C. O. (2014). Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Eur. J. Phycol. 49, 20–46. doi: 10.1080/09670262.2013.875596
Tout, J., Jeffries, T. C., Petrou, K., Tyson, G. W., Webster, N. S., Garren, M., et al. (2015). Chemotaxis by natural populations of coral reef bacteria. ISME J. 9, 1764–1777. doi: 10.1038/ismej.2014.261
Venev, S. V., and Zeldovich, K. B. (2018). Thermophilic adaptation in prokaryotes is constrained by metabolic costs of proteostasis. Mol. Biol. Evol. 35, 211–224. doi: 10.1093/molbev/msx282
von Scheibner, M., Dörge, P., Biermann, A., Sommer, U., Hoppe, H. G., and Jürgens, K. (2014). Impact of warming on phyto-bacterioplankton coupling and bacterial community composition in experimental mesocosms. Environ. Microbiol. 16, 718–733. doi: 10.1111/1462-2920.12195
Wang, X., Gong, L., Liang, S., Han, X., Zhu, C., and Li, Y. (2005). Algicidal activity of rhamnolipid biosurfactants produced by Pseudomonas aeruginosa. Harmful Algae 4, 433–443. doi: 10.1016/j.hal.2004.06.001
Wohlers, J., Engel, A., Zöllner, E., Breithaupt, P., Jürgens, K., Hoppe, H. G., et al. (2009). Changes in biogenic carbon flow in response to sea surface warming. Proc. Natl. Acad. Sci. U.S.A. 106, 7067–7072. doi: 10.1073/pnas.0812743106
Zhang, Z., van Kleunen, M., Becks, L., and Thakur, M. P. (2020). Towards a general understanding of bacterial interactions. Trends Microbiol. 28, 1–3. doi: 10.1016/j.tim.2020.05.010
Keywords: global warming, Skeletonema, free-living bacteria, attached bacteria, interaction network
Citation: Deng Z, Chen S, Zhang P, Zhang X, Adams JM, Luo Q and Lin X (2021) Dynamics of Free-Living and Attached Bacterial Assemblages in Skeletonema sp. Diatom Cultures at Elevated Temperatures. Front. Mar. Sci. 8:626207. doi: 10.3389/fmars.2021.626207
Received: 05 November 2020; Accepted: 21 June 2021;
Published: 19 July 2021.
Edited by:
Susana Agusti, King Abdullah University of Science and Technology, Saudi ArabiaReviewed by:
Willem Stock, Ghent University, BelgiumCopyright © 2021 Deng, Chen, Zhang, Zhang, Adams, Luo and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Lin, eGlubGludWxtQHhtdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.