
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Mar. Sci., 16 August 2021
Sec. Ocean Observation
Volume 8 - 2021 | https://doi.org/10.3389/fmars.2021.620368
This article is part of the Research TopicOpen Citizen Science Data and MethodsView all 28 articles
 Eva Turicchia1,2,3,4†‡
Eva Turicchia1,2,3,4†‡ Massimo Ponti2,3,4,5*†‡
Massimo Ponti2,3,4,5*†‡ Gianfranco Rossi3
Gianfranco Rossi3 Martina Milanese3,6
Martina Milanese3,6 Cristina Gioia Di Camillo3,7‡
Cristina Gioia Di Camillo3,7‡ Carlo Cerrano3,4,7,8‡
Carlo Cerrano3,4,7,8‡Since 2001, trained snorkelers, freedivers, and scuba diver volunteers (collectively called EcoDivers) have been recording data on the distribution, abundance, and bathymetric range of 43 selected key marine species along the Mediterranean Sea coasts using the Reef Check Mediterranean Underwater Coastal Environment Monitoring (RCMed U-CEM) protocol. The taxa, including algae, invertebrates, and fishes, were selected by a combination of criteria, including ease of identification and being a key indicator of shifts in the Mediterranean subtidal habitats due to local pressures and climate change. The dataset collected using the RCMed U-CEM protocol is openly accessible across different platforms and allows for various uses. It has proven to be useful for several purposes, such as monitoring the ecological status of Mediterranean coastal environments, assessing the effects of human impacts and management interventions, as well as complementing scientific papers on species distribution and abundance, distribution modeling, and historical series. Also, the commitment of volunteers promotes marine stewardship and environmental awareness in marine conservation. Here, we describe the RCMed U-CEM protocol from training volunteers to recording, delivering, and sharing data, including the quality assurance and control (QA/QC) procedures.
Community-based environmental monitoring is a participatory approach engaging volunteers through citizen science (CS) programs to enhance the ability of decision-makers and non-government organizations to monitor and manage natural resources, track endangered species, and protect biodiversity (Conrad and Hilchey, 2011; Chandler et al., 2017). Therefore, community-based monitoring engages citizen scientists and other stakeholders in the ecosystem-based management of natural heritage, not only aiming to increase the chance of obtaining biodiversity data for conservation purposes but also to raise public awareness and support for environmental protection (Keough and Blahna, 2006; Freiwald et al., 2018; Alexander et al., 2019). Marine citizen science (MCS) may provide a valuable contribution to community-based monitoring in marine environments, given the vastness of the world's oceans and coastlines and the diversity of their habitats, communities, and species (Thiel et al., 2014; Garcia-Soto et al., 2017). Involving millions of people worldwide, MCS programs are becoming increasingly important to conservation science not only by providing monitoring of biodiversity, ecosystem services, and functions but also by influencing and improving the management of marine protected areas and fishery resources (Freiwald et al., 2018). Despite a worldwide increase in program number and extent of MCS (Thiel et al., 2014), the collected information is rarely used for institutional monitoring programs or to inform decision-making processes in marine conservation (Conrad and Hilchey, 2011). This disconnect is partly due to persisting skepticism about the reliability of data collected from volunteers (Burgess et al., 2017) and a co-creation approach in the supply and demand of environmental monitoring data that is still not well-integrated in CS processes (Bonney et al., 2015). However, many studies demonstrate that well-trained citizens can provide valuable data on marine environmental issues and that suitable protocols for volunteer projects can provide results consistent with the methods used by the professional researchers (e.g., Holt et al., 2013; Forrester et al., 2015; Done et al., 2017). Still, there are limits to accessing the MCS data (Thiel et al., 2014), which are not always well-organized and readily available according to the FAIR (findable, accessible, interoperable, and reusable) data principles (sensu Wilkinson et al., 2016).
Here, we describe a well-established MCS protocol i.e., the Reef Check Mediterranean Underwater Coastal Environment Monitoring (RCMed U-CEM) protocol, which is refined and applied since 2001 by trained snorkelers, freedivers, and scuba diver volunteers (hereafter called as EcoDivers) to collect data on the occurrence, distribution, abundance, and bathymetric range of selected key marine species along the Mediterranean Sea coasts. The obtained dataset is openly accessible across different platforms and allows for various uses, such as complementing scientific papers on the species distribution and abundance, aiding distribution modeling, and comparing historical series (Lucrezi et al., 2018 and references therein). The protocol is implemented, and the data are maintained by the non-profit organization Reef Check Italia onlus, collaborating with the other European Reef Check organizations, members of the worldwide Reef Check Foundation, and within the Reef Check Mediterranean Sea network.
Reef Check Mediterranean Underwater Coastal Environment Monitoring (RCMed U-CEM) protocol is intended to collect data on the abundance, and geographical and bathymetric distribution of selected taxa. It requires trained participants (certified EcoDivers) to collect standardized data and send it to the online database. The data are then processed and made available on the various open access sharing platforms.
The 43 target taxa were selected by scientists from a combination of two or more criteria, including ease of identification, inclusion in the international lists of protected species, being sensitive to human impacts, and the effects of climate change occurring in the Mediterranean subtidal habitats (Table 1). Morphologically and ecologically similar species have been included at the genus level or higher taxa (Cerrano et al., 2017). The selected taxa embrace a broad taxonomic range, from algae to invertebrates and vertebrates. Most of them are sessile or sedentary with a limited home range; therefore, they cannot escape the local human disturbances and changes in environmental conditions. In terms of both resistance and resilience of local populations, their sensitivities to pressures were assessed and employed to develop the MedSens biotic index (see Supplementary materials in Turicchia et al., 2021a).
The trained volunteers are involved in the program. Their training is verified before being certified as EcoDivers. Once certified, they can independently apply the RCMed U-CEM protocol. Although they are often passionate naturalists, diving experts, and sometimes marine biologists, participants are not required to have any particular level of scientific background. Of course, they must be sufficiently skilled in snorkeling, freediving, or scuba diving, depending on how the protocol will be applied. Training is based not only on protocol explanation but also on raising awareness of the usefulness and importance of the data collected for the conservation of the Mediterranean Sea's coastal marine habitats. The course syllabus includes knowledge about main marine habitats, field identification of target species, and their ecological role. It also covers a range of geographic localization methods using nautical charts, conspicuous points, and satellite global positioning system (GPS) devices. Training materials encompass an illustrated protocol manual and benefit from a multi-language website1. Teaching methods include the initial lesson, class discussion, full hands-on demonstration, data entry, and final debriefing. Trainers are generally marine biologists, diving instructors, or both, with proven communication skills and experience in applying the protocol. Individual learning assessments are based on an online questionnaire (implemented on the QuestBase platform2), allowing immediate feedback on the ability of the participants to provide the data correctly. Certified EcoDivers are assigned a unique identification code to be used for data entry. They also sign a privacy agreement, compliant with the European general data protection regulation, which allows sharing the collected data on their behalf but leaves each one responsible for the quality of the data they provided.
The surveyed sites are freely chosen by EcoDivers and localized using GPS receivers, nautical charts, or conspicuous points (e.g., mooring buoys in marine protected areas). Geographic coordinates (WGS84) are recorded with ± 6 arc-seconds (i.e., 185 m in latitude) accuracy, the usual distance range explored by divers. Before going snorkeling or diving, each EcoDiver has to choose one or more of the 43 taxa included in the protocol as search targets, according to the expected habitat typology, survey depth, and personal motivations.
EcoDivers can make independent observations along random swim (Hill and Wilkinson, 2004; also called “roving visual census,” sensu Rassweiler et al., 2020). They record the presence/absence of the species and their abundance (using numerical or descriptive classes according to the countability of organisms; see definition in Table 2) and depth range of the searched taxa. Prevailing habitat (chosen from a list; Table 3), estimated underwater visibility, and the presence/abundance of gas bubbles leaching from the seabed are also recorded. Not encountered but actively searched taxa are reported as absent. No data are provided for not searched taxa. The time dedicated to actively search target species (at least 10 minutes) must be recorded. A preset underwater slate, with target species drawings and available with different species selections and languages, helps in the identification and recording tasks (Figures 1, 2).
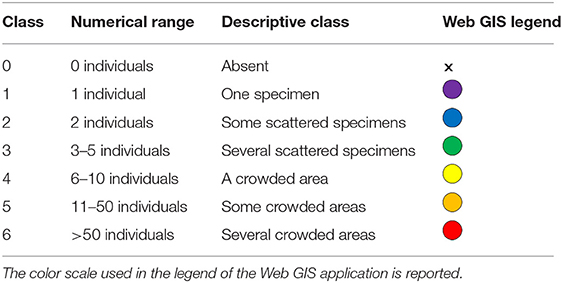
Table 2. Numerical and descriptive abundance classes used to record the abundance of the target species observed during the survey.
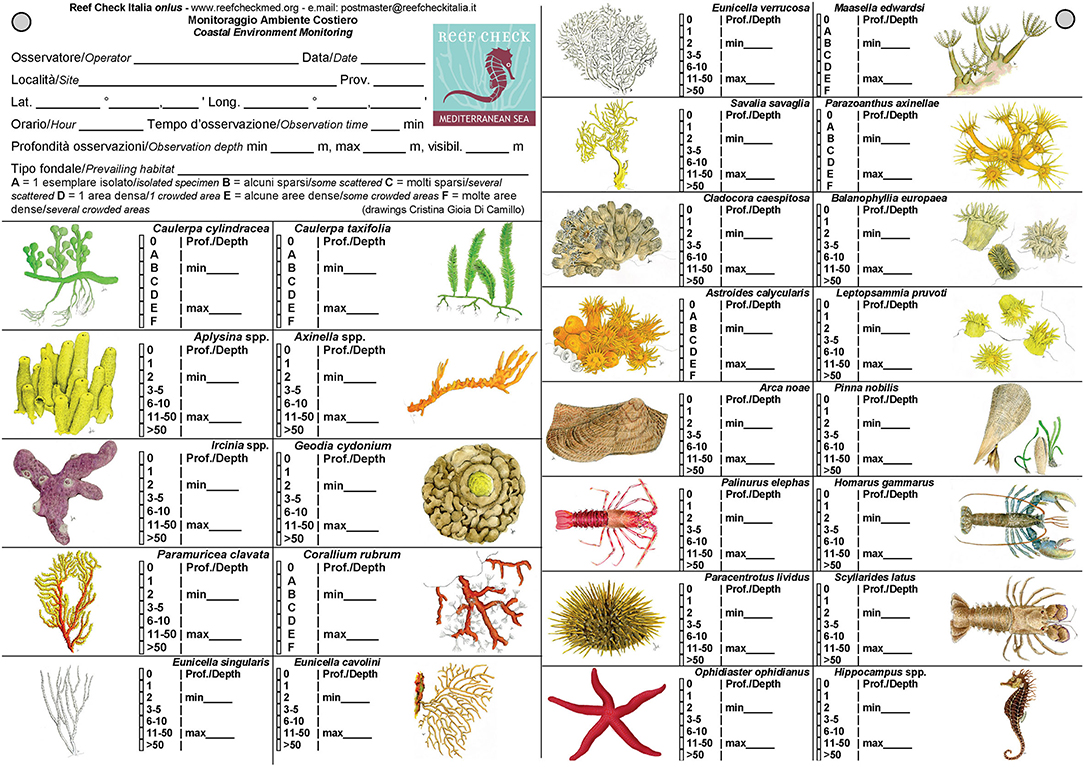
Figure 1. Example of front and back of an underwater slate preset to record data according to the RCMed U-CEM protocol (Italian–English version with a subset of the most common target species on rocky bottoms).
Recorded observations, including absence, site name, geographic coordinates, date and time, underwater visibility, survey depth range (min and max), and observation effort in terms of dedicated time, are uploaded to the online database through an internet form3 or a dedicated multilanguage app for Android smartphones (“Reef Check Med” app) connected to the online database (Figure 3).
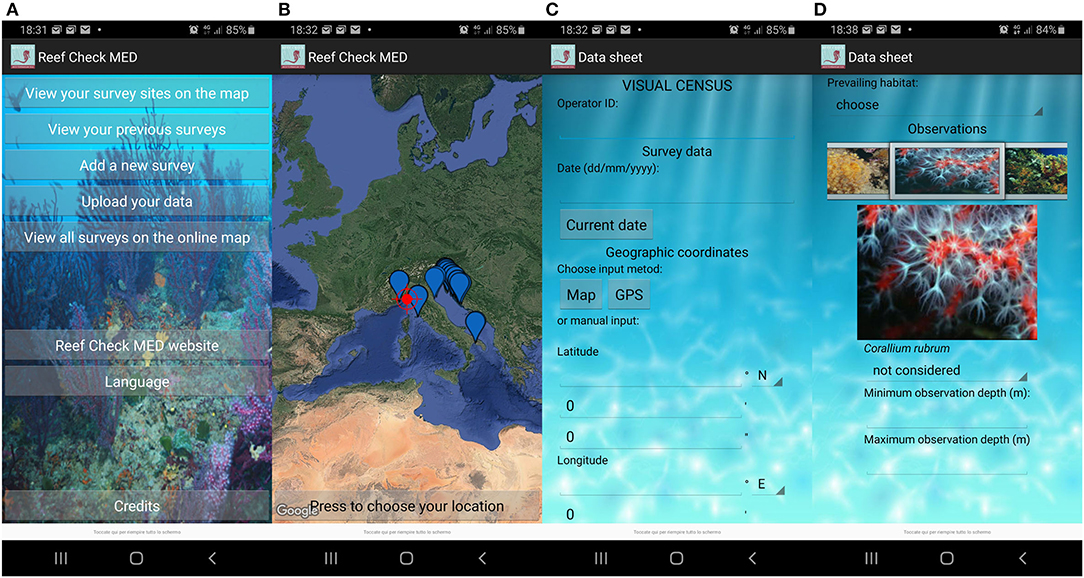
Figure 3. Android smartphone app screenshots: (A) main menu; (B) maps with previously investigated sites; (C) initial part of the data entry form; and (D) final part of the data entry form.
Geographic coordinates can be entered either in decimal degrees, degrees and decimal minutes, or degrees, minutes, and seconds, specifying east or west. Users of the smartphone app can benefit from the built-in GPS, remembering to use it near the surveyed site. The website and the app also provide EcoDivers with an online satellite map to retrieve geographic coordinates based on conspicuous points. Finally, the smartphone app also allows to store and review data, even offline, before submitting them to the online database. Additional notes on the characteristics of the site or the presence of anomalous situations such as mortality/disease events of marine organisms, presence of waste, or abandoned fishing nets, can be provided at the end of the form.
Citizen science reliability is a major issue in the acceptance and actual use of data collected by citizen scientists. As such, proper data quality assurance (QA) and data quality control (QC) are essential steps. For the RCMed U-CEM protocol, QA is mainly based on the quality and learning verification of the initial training and the personal responsibility for the provided data. At the end of each training session, a test is carried out to verify the level of competence of each volunteer in the data collection. Only volunteers who provide at least 70% correct answers can be qualified as EcoDivers.
In April 2015, a field test was carried out to verify the ability of the method to discriminate species assemblages among close sites with similar habitats and the ability of the trained volunteers to collect suitable data for this purpose. Ten EcoDivers were divided into three training levels: two participants belonged to the category “professional scientist and trainer” (i.e., marine biologists who are also trainers of EcoDivers), four to “professional scientist” (i.e., marine biologists trained as EcoDiver), and four to “citizen scientist” (i.e., EcoDivers without any academic training in marine sciences). Two dive sites were randomly selected at Gallinara Island, an islet in the Ligurian Sea (NW Mediterranean Sea). At each site, participants independently recorded the presence, abundance, and depth distribution of 20 target taxa selected among the ones in the RCMed U-CEM protocol, along a predefined belt-transect of 100 × 6 m. The dive profile varied from 3 to 30 m in depth. Each participant recorded the data by applying the RCMed U-CEM protocol, except for the constrained path, and entered them into the online database. Afterward, records were extracted from the database, and multivariate species assemblage data were analyzed using principal coordinate analysis (PCoA) based on Bray–Curtis similarities without any transformations. Differences in species assemblage structures (i.e., the combination of species found and their abundance) between the two sites (random factor) and resulting from the observation carried out by participants having three different training levels (fixed factor) were assessed by a two-way crossed permutational non-parametric multivariate ANOVA (PERMANOVA, α = 0.05; Anderson and ter Braak, 2003) under the hypothesis that the assemblage structures differed between sites and these differences were similarly detected independently by the training levels of the observers. The test assesses if there are no assemblage structure differences between the two sites, differences among the assemblage structures detected by the operators with different training levels, or a combination of these two factors (training level and site). The analyses were performed using the software PRIMER v. 6 (Anderson et al., 2008). Patterns of similarities among the observed assemblages are shown in the PCoA ordination plot (Figure 4). In this plot, the distances among the points are inversely proportional to the level of measured similarity in the corresponding assemblage structures. The first two axes of the PCoA explained 34.8% and 28.9% of the total variation. The ordination plot shows some degrees of separation of the points from the two sites but not among the observer training levels. The PERMANOVA test confirmed the pattern showing a significant difference (p < 0.01) in assemblage structure only between sites and not among training levels (Table 4). Even if some minor differences among the single operators were obtained, these represent a random effect related to the accuracy of the method, as occurs in any visual census. However, the method appeared robust enough to distinguish the assemblages between sites with similar habitats, a few 100 m apart. This case study shows that the absence of effects of the training level indicates that the trained citizen scientists can provide the same results as the professional scientists.
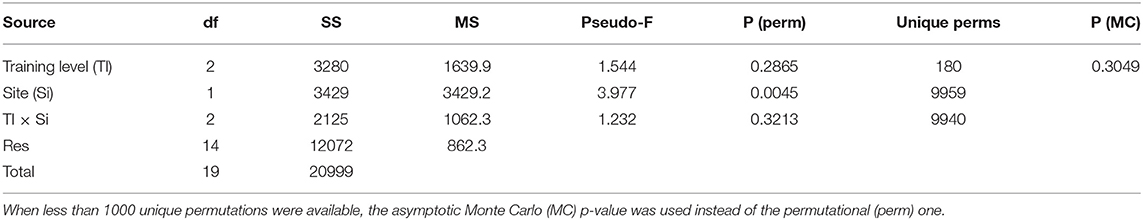
Table 4. Permutational non-parametric multivariate ANOVA (PERMANOVA) test on differences among training levels (Tl, 3 levels, fixed) and between sites (Si, 2 levels, fixed) and their interactions (Tl × Si) (Bray–Curtis similarities abundance data).
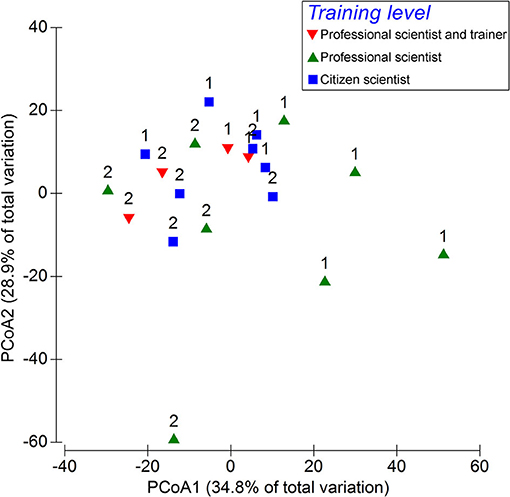
Figure 4. Principal coordinate analysis (PCoA) ordination plot based on Bray–Curtis similarities comparing species assemblages detected by scuba divers with different training levels along the same pathway at two sites (Sites 1 and 2 are indicated by numbers) at Gallinara Island (Ligurian Sea). The training level is indicated with symbols and colors.
As data are sent to the online database, EcoDivers can see their survey counter increasing. That represents an important feedback that allows everyone to verify that the system has accepted the data submission. Moreover, it provides immediate public recognition for the volunteers' work. However, the collected data are not made public immediately but only after undergoing the quality control (QC).
The user interface is designed in such a way to facilitate data entry; however, typos in manual entry are difficult to prevent. For this purpose, both web and app data entry forms are equipped with automatic checks that prevent oversights and common errors. Some fields are mandatory; some are preset (all species are initially set to “not considered”) or contain limits in the values that can be entered (for example, for geographic coordinates). Yet, this cannot completely avoid all possible errors. Post-entry QC is based on automatic procedures (e.g., consistency among survey and observation depth ranges, check for the possible exchange of the max and min depths) and manual checks (e.g., matching between the site name and geographic coordinates). Automatic algorithms are applied during data extraction from the online database using specific queries and procedures implemented in R (R Core Team, 2019), ending with the automatic creation of shapefiles (ESRI, 1998; Stabler, 2013). The shapefiles are closely inspected by the Reef Check Med operators and then validated. In the case of inconsistencies that cannot be resolved uniquely by the operators, the EcoDivers who provided the data are contacted by email to ask for further information. If the problem cannot be solved, the indicated data are definitively discarded.
An online interactive peer review represents a further step in the QC of the data. To this end, as soon as the data are published online, EcoDivers are informed and invited to check the data in their favorite areas and report any possible doubts or inconsistencies through an online form. Based on their reports, the data are then re-analyzed and, where possible, the errors are fixed in the subsequent data publication.
Data that has passed the QC are made public through the various platforms. The most updated data are made freely available on a web-based geographic information system (Web GIS) built-up using the QGIS Cloud free platform4 This platform allows the visualization of all data on the Bing Aerial base-map (Figure 5). The user can choose to visualize the survey points, displayed as yellow dots, or the data inherent single target species distribution and abundance, displayed as dots colored according to an abundance scale or as a black cross in case of species not found (Table 2). With the pointer, it is possible to query the data stored for each point, including geographic coordinates in decimal degrees, survey date and depth, and the EcoDiver's name, according to their informed consent. The QGIS Cloud platform provides users with a full-screen version and a smartphone version. Moreover, being the platform an Open Geospatial Consortium compliant web services allows the display of the maps via the Web Map Service (WMS) or downloading the data via the web feature service (WFS). Data are distributed under the international Creative Common license (CC BY 4.0), which allows for free sharing and adaptation, giving appropriate credit to the Reef Check Mediterranean network.
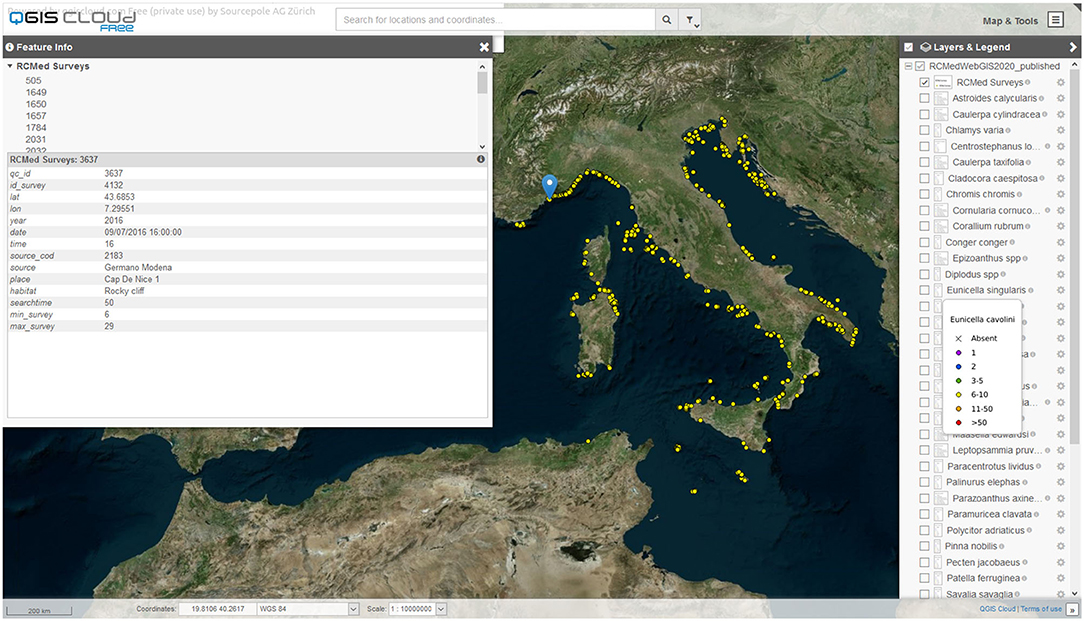
Figure 5. Web GIS screenshot shows some data interrogation options: on the left, the data of a single survey and on the right, the list of species layers that can be activated with the relative symbol legends, which display the abundance classes (see Table 2).
Following the FAIR principles, a Darwin Core (Wieczorek et al., 2012) compliant version of the whole dataset is available at the Biology data portal of the European Marine Observation and Data Network (EMODnet; Miguez et al., 2019) and redistributed under the Ocean Biodiversity Information System (OBIS) networks (including EurOBIS, MedOBIS; Costello and Vanden Berghe, 2006 and references therein), the European infrastructure on biodiversity and ecosystem research (LifeWatch; Basset and Los, 2012), and the Global Biodiversity Information Facility (GBIF; Flemons et al., 2007).
As of December 2020, the dataset consisted of 50,255 records (including absence records) unevenly distributed among 43 taxa in the Mediterranean Sea collected in 4,898 single survey events from 2001 and carried out by 692 EcoDivers (Ponti et al., 2021; Turicchia et al., 2021b). The data comes from Croatia, France, Greece, Italy, Spain, and Tunisia, covering part of the following ecoregions (sensu Spalding et al., 2007): Western Mediterranean (52.3% of the surveys), Adriatic Sea (42.2%), Ionian Sea (4.9%), Alboran Sea (0.2%), Aegean Sea (0.2%), and Tunisian Plateau/Gulf of Sidra (0.2%). The possibility to focus on a few target species during the underwater surveys ensures a higher accuracy of the data collection: EcoDivers select the species based on confidence (thereby reducing identification errors), personal interest (increasing satisfaction), or because some species are more charismatic than others (KrŽelj et al., 2020). Although never assessed, a reduced number of species to consider may reduce psychological stress during the surveys; however, this generates skewed distribution efforts among the searched taxa and surveyed coastal habitats. Indeed, the most-searched taxa are common species like seabreams (Diplodus spp.) and the edible sea urchin (Paracentrotus lividus), the noble pen shell (Pinna nobilis), the red coral (Corallium rubrum), and sea fans (Paramuricea clavata and Eunicella cavolini). Less conspicuous but highly concerning species, such as invasive algae in the genus Caulerpa, are also frequently surveyed (Figure 6A). As expected, the most investigated habitats are those most attractive to divers, namely, rocky coasts, cliffs, and wrecks (Figure 6B).
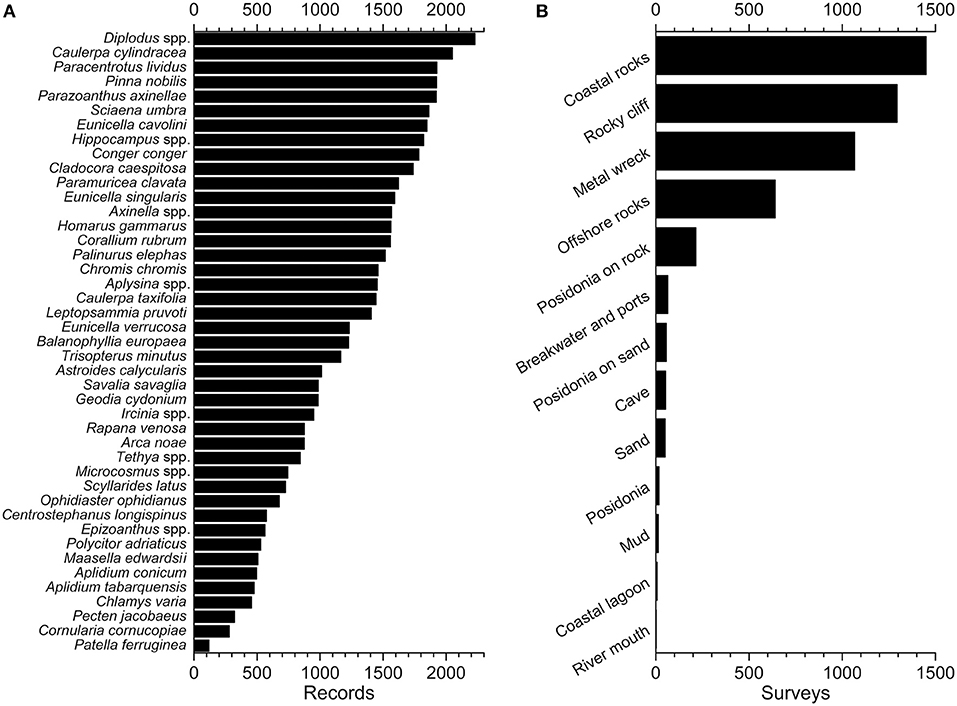
Figure 6. Dataset contents (last access October 18th, 2020): (A) number of records (including absences) for each target species and (B) number of surveys carried out at each habitat.
Over the years the dataset has been successfully used to complement studies on the spatial and the temporal distribution of key marine species such as the habitat-forming corals in the central-eastern Mediterranean (Özalp and Alparslan, 2016; Di Camillo et al., 2018), the pink sea-fan Eunicella verrucosa (Chimienti, 2020), as well as of rare and/or endangered species like the gold coral Savalia savaglia (Giusti et al., 2015), the zooxanthellate soft coral Maasella edwardsii (Özalp and Ateş, 2015), and the sponge Geodia cydonium (Turicchia et al., 2013). These data can also help in tracking mass mortality events and assess the possible effects of climate change (Pairaud et al., 2014; Ponti et al., 2018; Turicchia et al., 2018; Garrabou et al., 2019) and the invasion of the non-indigenous species Caulerpa taxifolia and Caulerpa cylindracea (Montefalcone et al., 2015; Cerrano et al., 2017). Moreover, the dataset can be used in assessing the protected and sensitive species richness within the marine protected areas (Turicchia et al., 2015, 2016; Cerrano et al., 2017), and in offering an effective monitoring tool for the Mediterranean subtidal rocky coastal habitats through the MedSens biotic index (Turicchia et al., 2021a).
The RCMed U-CEM protocol integrates the Reef Check family of protocols, which already includes those for the California coasts (Gillett et al., 2012) and tropical coral reefs around the world (Hodgson, 2001) extending its range to the Mediterranean Sea. These standard protocols aim to report the presence and the abundance of key species and to assess and monitor over time the ecological status of the investigated sites based on the relative abundance of selected indicator species. They are optimized for the habitats and operating conditions for which they are intended. In tropical reefs and California, standard transects are made in shallow waters under the supervision of a scientist and/or a team leader, while in the Mediterranean, volunteers independently apply a roving visual census that allows them to explore from shallow to deep habitats (see further comparisons in Table 5). RCMed U-CEM is not the only protocol that can be adopted by snorkelers and divers to report marine species in the Mediterranean Sea; however, the aims and methods applied differ widely. Among those listed by Earp and Liconti (2020), the most popular and internationally applied alternatives in the Mediterranean Sea are: REEF, Sea Watchers, and iNaturalist. Their main features are summarized in Table 5 for comparison. REEF, started in 1993 with the fish visual census in the tropical western Atlantic, was one of the forerunners in involving volunteer divers (Pattengill-Semmens and Semmens, 2003). Over the years, the protocol has been extended to various regions of the world, and since 2014, it also includes the Eastern Atlantic and the Mediterranean Sea. As RCMed U-CEM, REEF is based on roving visual census carried out by trained volunteers. While the main focus of REEF's program is marine fish, they also survey selected invertebrates and algae in the temperate water regions. In addition to the different lists of considered species, the major difference between the two protocols lies in the less explicit and not always precise location of the sites (based on hierarchical area codes and not geographic coordinates), the lack of information on the bathymetric distribution of species, and the reduced findability, accessibility, and interoperability of the REEF data. Sea Watchers, launched in Spain in 2009 as “Observadores del Mar,” includes several thematic sub-projects (e.g., massive mortalities of corals, death of pen shells, alien fishes, invasive algae, decapod crustaceans, sharks and rays, and zooxanthellate scleractinian; see Mariani et al., 2018); they are based on visual observations supplemented by photographs that are sent to experts for identification and analysis. Involved experts assess and archive the observations, possibly integrating with additional information interactively requested to the participants. Data collected from some sub-projects are already distributed by EMODnet or other data portals. iNaturalist, which began in 2008, is the widest social network of nature enthusiasts that share and cross-validate photographic observations and it is one of the more FAIR initiatives worldwide (Bowser et al., 2014). Unfortunately, despite initiatives on marine organisms are multiplying within it (Reef Check Italia has launched two calls for contributions on Mediterranean nudibranchs and corals), most of the collected data concerns terrestrial species. In addition, many of the marine species' records lack important additional data such as observation depth or surveyed habitat. Notably, except for RCMed U-CEM, none of the other initiatives mentioned here for the Mediterranean Sea provide information on the absence of key species, a “dark diversity” (sensu Partel et al., 2011) usually neglected but potentially very useful in assessing biological integrity, ecological status, and the effects of global changes.
The application of the RCMed U-CEM protocol may generate a range of direct societal impacts, including a higher public awareness of environmental threats and the involvement of stakeholders (e.g., tourists, divers, and diving centers) in the monitoring and conservation of coastal marine environments (Turicchia et al., 2021a). Therefore, it may enhance the collaboration between coastal management authorities, stakeholders, and researchers, increasing the acceptability of management decisions and enabling more participatory conservation tactics (Markantonatou et al., 2013; de Francesco et al., 2017; Lucrezi et al., 2018).
To obtain and distribute reliable and scientifically sound data, RCMed U-CEM protocol was designed to minimize taxonomic and geolocation errors. However, taxonomic and spatial biases, recognized as major issues in CS projects and biodiversity databases, remain intrinsically unavoidable for this and most CS initiatives (Beck et al., 2014; Troudet et al., 2017). New technologies may help improve the protocol and reduce the possibility of common errors. For example, the widespread use of the smartphone app has already reduced localization errors, thanks to the integrated GPS. The increasing adoption of waterproof cases for smartphones and tablets suggests the possibility of entering data directly during the in-water activity and collecting photos to verify the species identification later using a specifically preconfigured app (Max and Gualdesi, 2013). Advances in image analysis and deep learning algorithms coupled with photo databases can support the development of apps for identifying marine species, as has been the case for identifying plants and terrestrial animals (e.g., the app Seek by iNaturalist; see Waldchen and Mader, 2018 for a review about automatic terrestrial plant identification), and this technology could be implemented on the Reef Check Med app and possibly used directly underwater.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
MP conceived the early version of the protocol, which was refined together with CC, GR, CGDC, and ET. MP and ET wrote the first draft of the manuscript. All authors contributed to and approved the final version of the manuscript.
The preparation of this document was supported by Reef Check Italia Onlus and partially founded by the marine protected area Tavolara—Capo Coda Cavallo under the citizen science project TAVOLARA LAB and by the EMODnet Biology Data Grant (Network Ref. EASME/EMFF/2016/006—Lot No 5—Biology).
MM was employed by company Studio Associato Gaia Snc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank EcoDivers and their trainers. The following marine protected areas supported the training of EcoDivers and promoted data collection: Cabo de Palos, Capo Gallo –Isola delle Femmine, Cinque Terre, Isola di Ustica, Isole Egadi, Isole Tremiti, Miramare, Porto Cesareo, Portofino, Tavolara—Capo Coda Cavallo. The authors are grateful to the reviewers and the editor for their valuable comments and suggestions. This study is part of ET's PhD thesis.
1. ^https://www.reefcheckmed.org
3. ^https://www.reefcheckmed.org/english/underwater-monitoring-protocol/upload-your-data/.
4. ^https://www.reefcheckmed.org/english/underwater-monitoring-protocol/webgis-map/.
Alexander, K. A., Hobday, A. J., Cvitanovic, C., Ogier, E., Nash, K. L., Cottrell, R. S., et al. (2019). Progress in integrating natural and social science in marine ecosystem-based management research. Mar. Freshw. Res. 70, 71–83. doi: 10.1071/MF17248
Anderson, M. J., Gorley, R. N., and Clarke, K. R. (2008). PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods. Plymouth, MA: PRIMER-E Ltd.
Anderson, M. J., and ter Braak, C. J. F. (2003). Permutation tests for multi-factorial analysis of variance. J. Stat. Comput. Sim. 73, 85–113. doi: 10.1080/00949650215733
Basset, A., and Los, W. (2012). Biodiversity e-Science: lifewatch, the European infrastructure on biodiversity and ecosystem research. Plant Biosyst. 146, 780–782. doi: 10.1080/11263504.2012.740091
Beck, J., Boller, M., Erhardt, A., and Schwanghart, W. (2014). Spatial bias in the GBIF database and its effect on modeling species' geographic distributions. Ecol. Inform. 19, 10–15. doi: 10.1016/j.ecoinf.2013.11.002
Bonney, R., Phillips, T. B., Ballard, H. L., and Enck, J. W. (2015). Can citizen science enhance public understanding of science? Public Underst. Sci. 25, 2–16. doi: 10.1177/0963662515607406
Bowser, A., Wiggins, A., Shanley, L., Preece, J., and Henderson, S. (2014). Sharing data while protecting privacy in citizen science. Interactions 21, 70–73. doi: 10.1145/2540032
Burgess, H. K., DeBey, L. B., Froehlich, H. E., Schmidt, N., Theobald, E. J., Ettinger, A. K., et al. (2017). The science of citizen science: exploring barriers to use as a primary research tool. Biol. Conserv. 208, 113–120. doi: 10.1016/j.biocon.2016.05.014
Cerrano, C., Milanese, M., and Ponti, M. (2017). Diving for science—science for diving: volunteer scuba divers support science and conservation in the Mediterranean Sea. Aquat. Conserv. 27, 303–323. doi: 10.1002/aqc.2663
Chandler, M., See, L., Copas, K., Bonde, A. M. Z., López, B. C., Danielsen, F., et al. (2017). Contribution of citizen science towards international biodiversity monitoring. Biol. Conserv. 213(Part B), 280–294. doi: 10.1016/j.biocon.2016.09.004
Chimienti, G. (2020). Vulnerable forests of the pink sea fan Eunicella verrucosa in the Mediterranean Sea. Diversity 12:176. doi: 10.3390/d12050176
Conrad, C. C., and Hilchey, K. G. (2011). A review of citizen science and community-based environmental monitoring: issues and opportunities. Environ. Monit. Assess. 176, 273–291. doi: 10.1007/s10661-010-1582-5
Costello, M. J., and Vanden Berghe, E. (2006). 'Ocean biodiversity informatics': a new era in marine biology research and management. Mar. Ecol. Prog. Ser. 316, 203–214. doi: 10.3354/meps316203
de Francesco, M. C., Cerrano, C., Pica, D., D'Onofrio, D., and Stanisci, A. (2017). Characterization of Teatina coast marine habitats (central Adriatic Sea) toward an integrated coastal management. Oceanography and Fisheries 5:555653. doi: 10.19080/OFOAJ.2017.05.555653
Di Camillo, C. G., Ponti, M., Bavestrello, G., Krzelj, M., and Cerrano, C. (2018). Building a baseline for habitat-forming corals by a multi-source approach, including web ecological knowledge. Biodivers. Conserv. 27, 1257–1276. doi: 10.1007/s10531-017-1492-8
Done, T., Roelfsema, C., Harvey, A., Schuller, L., Hill, J., Schläppy, M.-L., et al. (2017). Reliability and utility of citizen science reef monitoring data collected by Reef Check Australia, 2002–2015. Mar. Pollut. Bull. 117, 148–155. doi: 10.1016/j.marpolbul.2017.01.054
Earp, H. S., and Liconti, A. (2020). Science for the future: the use of citizen science in marine research and conservation, in YOUMARES 9-The Oceans: Our Research, Our Future, ed L.V. Jungblut and S. Bode-Dalby (New York, NY: Springer), 2809.
ESRI (1998). ESRI Shapefile Technical Description. West Redlands, CA: Environmental Systems Research Institute, Inc.
Flemons, P., Guralnick, R., Krieger, J., Ranipeta, A., and Neufeld, D. (2007). A web-based GIS tool for exploring the world's biodiversity: The Global Biodiversity Information Facility Mapping and Analysis Portal Application (GBIF-MAPA). Ecol. Inform. 2, 49–60. doi: 10.1016/j.ecoinf.2007.03.004
Forrester, G., Baily, P., Conetta, D., Forrester, L., Kintzing, E., and Jarecki, L. (2015). Comparing monitoring data collected by volunteers and professionals shows that citizen scientists can detect long-term change on coral reefs. J. Nat. Conserv. 24, 1–9. doi: 10.1016/j.jnc.2015.01.002
Freiwald, J., Meyer, R., Caselle, J. E., Blanchette, C. A., Hovel, K., Neilson, D., et al. (2018). Citizen science monitoring of marine protected areas: case studies and recommendations for integration into monitoring programs. Mar. Ecol. 39:e12470. doi: 10.1111/maec.12470
Garcia-Soto, C., van der Meeren, G. I., Busch, J. A., Delany, J., Domegan, C., Dubsky, K., et al. (2017). Advancing citizen science for coastal and ocean research, in Position paper 23 of the European Marine Board, eds V. French, P. Kellett, J. Delany and N. McDonough. (Belgium: Ostend), 112.
Garrabou, J., Gómez-Gras, D., Ledoux, J.-B., Linares, C., Bensoussan, N., López-Sendino, P., et al. (2019). Collaborative database to track mass mortality events in the Mediterranean Sea. Front. Mar. Sci. 6:707. doi: 10.3389/fmars.2019.00707
Gillett, D. J., Pondella, D. J. II., Freiwald, J., Schiff, K. C., Caselle, J. E., Shuman, C., et al. (2012). Comparing volunteer and professionally collected monitoring data from the rocky subtidal reefs of Southern California, USA. Environ. Monit. Assess. 184, 3239–3257. doi: 10.1007/s10661-011-2185-5
Giusti, M., Cerrano, C., Angiolillo, M., Tunesi, L., and Canese, S. (2015). An updated overview of the geographic and bathymetric distribution of Savalia savaglia. Mediterr. Mar. Sci. 16, 128–135. doi: 10.12681/mms.890
Hill, J., and Wilkinson, C. (2004). Methods for Ecological Monitoring of Coral Reefs. A Resource for Managers. Townsville, QD: Australian Institute of Marine Science.
Hodgson, G. (2001). Reef Check: the first step in community-based management. Bull. Mar. Sci. 69, 861–868.
Holt, B. G., Rioja-Nieto, R., Aaron MacNeil, M., Lupton, J., and Rahbek, C. (2013). Comparing diversity data collected using a protocol designed for volunteers with results from a professional alternative. Methods Ecol. Evol. 4, 383–392. doi: 10.1111/2041-210X.12031
Keough, H. L., and Blahna, D. J. (2006). Achieving integrative, collaborative ecosystem management. Conserv. Biol. 20, 1373–1382. doi: 10.1111/j.1523-1739.2006.00445.x
KrŽelj, M., Cerrano, C., and Di Camillo, C. G. (2020). Enhancing diversity knowledge through marine citizen science and social platforms: The case of Hermodice carunculata (Annelida, Polycheta). Diversity 12:311. doi: 10.3390/d12080311
Lucrezi, S., Milanese, M., Palma, M., and Cerrano, C. (2018). Stirring the strategic direction of scuba diving marine Citizen science: a survey of active and potential participants. PLoS ONE 13:e0202484. doi: 10.1371/journal.pone.0202484
Mariani, S., Ocaña Vicente, O., López-Sendino, P., García, M., Ricart, A. M., Garrabou, J., et al. (2018). The zooxanthellate scleractinian coral Oulastrea crispata (Lamarck, 1816), an overlooked newcomer in the Mediterranean Sea? Mediterr. Mar. Sci. 19, 589–597. doi: 10.12681/mms.16986
Markantonatou, V., Meidinger, M., Sano, M., Oikonomou, E., di Carlo, G., Palma, M., et al. (2013). Stakeholder participation and the use of web technology for MPA management. Adv. Oceanogr. Limnol. 4, 260–276. doi: 10.4081/aiol.2013.5347
Max, M. D., and Gualdesi, L. (2013). Using preconfigured tablets underwater to increase data rate recording and improve dive efficiency, in MTS/IEEE Oceans Conference. (San Diego, CA: IEEE).
Miguez, B. M., Novellino, A., Vinci, M., Claus, S., Calewaert, J. B., Vallius, H., et al. (2019). The European marine observation and data network (EMODnet): visions and roles of the gateway to marine data in Europe. Front. Mar. Sci. 6:313. doi: 10.3389/fmars.2019.00313
Montefalcone, M., Morri, C., Parravicini, V., and Bianchi, C. (2015). A tale of two invaders: Divergent spreading kinetics of the alien green algae Caulerpa taxifolia and Caulerpa cylindracea. Biol. Invasions 17, 2717–2728. doi: 10.1007/s10530-015-0908-1
Özalp, H. B., and Alparslan, M. (2016). Scleractinian diversity in the Dardanelles and Marmara Sea (Turkey): morphology, ecology and distributional patterns. Oceanol. Hydrobiol. Stud. 45, 259–285. doi: 10.1515/ohs-2016-0023
Özalp, H. B., and Ateş, A.S. (2015). The occurrence of the Mediterranean soft coral Maasella edwardsi (Cnidaria, Anthozoa, Alcyonacea) in the Aegean Sea coasts of Turkey. Marine Biodiv. Rec. 8:e55. doi: 10.1017/S1755267215000329
Pairaud, I., Bensoussan, N., Garreau, P., Faure, V., and Garrabou, J. (2014). Impacts of climate change on coastal benthic ecosystems: assessing the current risk of mortality outbreaks associated with thermal stress in NW Mediterranean coastal areas. Ocean Dynam. 64, 103–115. doi: 10.1007/s10236-013-0661-x
Partel, M., Szava-Kovats, R., and Zobel, M. (2011). Dark diversity: shedding light on absent species. Trends Ecol. Evol. 26, 124–128. doi: 10.1016/j.tree.2010.12.004
Pattengill-Semmens, C. V., and Semmens, B. X. (2003). Conservation and management applications of the reef volunteer fish monitoring program. Environ. Monit. Assess. 81, 43–50. doi: 10.1023/A:1021300302208
Ponti, M., Turicchia, E., Ferro, F., Cerrano, C., and Abbiati, M. (2018). The understorey of gorgonian forests in mesophotic temperate reefs. Aquat. Conserv. 28, 1153–1166. doi: 10.1002/aqc.2928
Ponti, M., Turicchia, E., Rossi, G., and Cerrano, C. (2021). Reef Check Med— key Mediterranean marine species 2001-2020. Dataset maintained by Reef Check Italia onlus, EMODnet Biology data portal. doi: 10.14284/468
R Core Team (2019). R: A Language and Environment for Statistical Computing Version 3.6.1 [Online]. Available online at: https://www.r-project.org/ (accessed December 1, 2019).
Rassweiler, A., Dubel, A. K., Hernan, G., Kushner, D. J., Caselle, J. E., Sprague, J. L., et al. (2020). Roving divers surveying fish in fixed areas capture similar patterns in biogeography but different estimates of density when compared with belt transects. Front. Mar. Sci. 7:272. doi: 10.3389/fmars.2020.00272
Spalding, M. D., Fox, H. E., Allen, G. R., Davidson, N., Ferdaña, Z. A., Finlayson, M. A. X., et al. (2007). Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. Bioscience 57, 573–583. doi: 10.1641/B570707
Stabler, B. (2013). shapefiles: An R Package for Read and write ESRI Shapefiles [Online]. Available online at: https://CRAN.R-project.org/package=shapefiles (accessed December 1, 2019).
Thiel, M., PennaDíaz, M., LunaJorquera, G., Salas, S., Sellanes, J., and Stotz, W. (2014). Citizen scientists and marine research: Volunteer participants, their contributions, and projection for the future. Oceanogr. Mar. Biol., Annu. Rev. 52, 257–314. doi: 10.1201/b17143-6
Troudet, J., Grandcolas, P., Blin, A., Vignes-Lebbe, R., and Legendre, F. (2017). Taxonomic bias in biodiversity data and societal preferences. Sci. Rep. 7:9048. doi: 10.1038/s41598-017-09084-6
Turicchia, E., Abbiati, M., Sweet, M., and Ponti, M. (2018). Mass mortality hits gorgonian forests at Montecristo Island. Dis. Aquat. Org. 131, 79–85. doi: 10.3354/dao03284
Turicchia, E., Cerrano, C., Abbiati, M., and Ponti, M. (2016). From citizen sciences to environmental quality assessment: The Portofino MPA case study, in Rapport du 41e Congrès de la Commission Internationale pour l'Exploration Scientifique de la mer Méditerranée. (Kiel: CIESM), 12–16.
Turicchia, E., Cerrano, C., Ghetta, M., Abbiati, M., and Ponti, M. (2021a). MedSens index: the bridge between marine citizen science and coastal management. Ecol. Indic. 122:107296. doi: 10.1016/j.ecolind.2020.107296
Turicchia, E., Poli, D., Abbiati, M., and Ponti, M. (2013). Abundance, size, and growth rate of Geodia cydonium (Demospongiae: Geodiidae) in the northern Adriatic temperate biogenic reefs, in Rapport du 40e Congrès de la Commission Internationale pour l'Exploration Scientifique de la mer Méditerranée, (Marseille, France: CIESM).
Turicchia, E., Ponti, M., Abbiati, M., and Cerrano, C. (2015). Citizen science as a tool for the environmental quality assessment of Mediterranean Marine Protected Areas, in Proceedings of the MMMPA/ CIESM International Joint Conference on Mediterranean marine Protected Areas: Integrated Management as a Response to ecosystem threats. eds C. Cerrano, Y. Henocque, K. Hogg, P. Moschella and M. Ponti, (Ancona: CIESM).
Turicchia, E., Ponti, M., Rossi, G., and Cerrano, C. (2021b). The Reef Check Med dataset on key Mediterranean marine species 2001-2020. Front. Mar. Sci. under review.
Waldchen, J., and Mader, P. (2018). Plant species identification using computer vision techniques: A systematic literature review. Archiv. Comput. Methods Eng. 25, 507–543. doi: 10.1007/s11831-016-9206-z
Wieczorek, J., Bloom, D., Guralnick, R., Blum, S., Doring, M., Giovanni, R., et al. (2012). Darwin Core: an evolving community-developed biodiversity data standard. PLoS ONE 7:8. doi: 10.1371/journal.pone.0029715
Keywords: marine citizen science, indicator species, marine protected areas, coastal zone management, monitoring, climate change, human impacts, Mediterranean Sea
Citation: Turicchia E, Ponti M, Rossi G, Milanese M, Di Camillo CG and Cerrano C (2021) The Reef Check Mediterranean Underwater Coastal Environment Monitoring Protocol. Front. Mar. Sci. 8:620368. doi: 10.3389/fmars.2021.620368
Received: 22 October 2020; Accepted: 16 July 2021;
Published: 16 August 2021.
Edited by:
Sven Schade, European Commission, Joint Research Centre (JRC), ItalyReviewed by:
Ioannis Giovos, iSea Environmental Organisation for the Preservation of the Aquatic Ecosystems, GreeceCopyright © 2021 Turicchia, Ponti, Rossi, Milanese, Di Camillo and Cerrano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Massimo Ponti, bWFzc2ltby5wb250aUB1bmliby5pdA==
‡ORCID:Eva Turicchia orcid.org/0000-0002-8952-9028
Massimo Ponti orcid.org/0000-0002-6521-1330
Cristina Gioia Di Camillo orcid.org/0000-0002-4031-8158
Carlo Cerrano orcid.org/0000-0001-9580-5546
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.