- 1Evolutionary Genetics Group, Department of Anthropology, University of Zurich, Zurich, Switzerland
- 2Centre for Sustainable Aquatic Ecosystems, Harry Butler Institute, Murdoch University, Murdoch, WA, Australia
- 3School of Biological Sciences, Oceans Institute, University of Western Australia, Crawley, WA, Australia
- 4School of Biological Sciences, University of Bristol, Bristol, United Kingdom
- 5Evolution and Ecology Research Centre, School of Biological Earth and Environmental Science, University of New South Wales, Sydney, NSW, Australia
- 6Curtin Law School, Curtin Business School, Curtin University, Bentley, WA, Australia
Identifying population structure and boundaries among communities of wildlife exposed to anthropogenic threats is key to successful conservation management. Previous studies on the demography, social and spatial structure of Indo-Pacific bottlenose dolphins (Tursiops aduncus) suggested four nearly discrete behavioral communities in Perth metropolitan waters, Western Australia. We investigated the genetic structure of these four communities using highly polymorphic microsatellite markers and part of the hypervariable segment of the mitochondrial control region. Overall, there was no evidence of spatial genetic structure. We found significant, yet very small genetic differentiation between some communities, most likely due to the presence of highly related individuals within these communities. Our findings of high levels of contemporary migration and highly related individuals among communities point toward a panmictic genetic population with continuous gene flow among each of the communities. In species with slow life histories and fission-fusion dynamics, such as Tursiops spp., genetic and socio-spatial structures may reflect different timescales. Thus, despite genetic similarity, each social community should be considered as a distinct ecological unit to be conserved because they are exposed to different anthropogenic threats and occur in different ecological habitats, social structure being as important as genetic information for immediate conservation management. The estuarine community, in particular, is highly vulnerable and appropriate conservation measures are needed in order to maintain its connectivity with the adjacent, semi-enclosed coastal communities.
Introduction
Social structure can play a critical role in the conservation of wildlife as it underpins the demography and behavior of a population and influences processes such as habitat use and dietary specialization (Snijders et al., 2017; Louis et al., 2018). Complex social structure and localized specialization are expected to promote isolation, which may create conservation challenges for communities (i.e., sets of individuals that are behaviorally discrete from neighboring communities and within which most individuals associate with other members of the community, Wells et al., 1987) or populations unable to adapt to rapid environmental changes, notably human impacts in urbanized areas (Barragan-Barrera et al., 2017). Reduced intra-group social cohesion, for instance, appeared to affect the recovery of small populations of killer whales (Parsons et al., 2009; Williams et al., 2009). On the other hand, long-term social affiliations may contribute to natal site fidelity, potentially resulting in reduced gene flow among social groups and thereby shaping genetic variability within and between populations (Krützen et al., 2004; Möller and Beheregaray, 2004; Litz et al., 2012; Pratt et al., 2018).
The low cost of locomotion, high dispersal capabilities and lack of geographic barriers in the marine environment were thought to promote high levels of gene flow, but many taxa reveal surprising structuring even at small spatial scales (e.g., Palumbi, 1992; Taylor and Hellberg, 2003; Sellas et al., 2005). Genetic differentiation among communities and populations can result from recent divergence with no ongoing gene flow or long-term separation with low recurrent gene flow (Nielsen and Wakeley, 2001; Palsbøll et al., 2004). Discriminating between these scenarios has important implications for conservation, as isolated communities or populations may require management measures that are dependent upon their demographic patterns, ecological and social organization, as well as their levels of exposure to anthropogenic stressors (Yannic et al., 2016; Crawford et al., 2018).
Population genetic data have been increasingly used to delineate populations or communities of cetaceans into separate management units for the purposes of meeting conservation objectives (Taylor and Dizon, 1999; Palsbøll et al., 2007). Many of the coastal cetaceans lend themselves to such analyses as they tend to occur as genetically discrete population units, with differentiation being detected at small spatial scales relative to the distances over which they are capable of dispersing (e.g., spinner dolphins Stenella longirostris, Andrews et al., 2010; common dolphins Delphinus delphis, Möller et al., 2011; Bilgmann et al., 2014; Australian snubfin Orcaella heinsohni and humpback dolphins Sousa sahulensis, Brown et al., 2014; bottlenose dolphins Tursiops spp., Hoelzel et al., 1998b; Krützen et al., 2004; Fernández et al., 2011; Ansmann et al., 2012). Population differentiation of these species has been associated with habitat complexity, i.e., estuaries or bays versus open coastline (e.g., Möller et al., 2007), as well as resource or habitat specializations (e.g., Krützen et al., 2014; Louis et al., 2014; Giménez et al., 2018). While geographic isolation-by-distance results in differences between populations (e.g., Allen et al., 2016; Parra et al., 2018), genetic differentiation on particularly small spatial scales has also been attributed to factors pertaining to behavioral variation, including social structure and local philopatry (e.g., Wiszniewski et al., 2009; Andrews et al., 2010; Kopps et al., 2014; Van Cise et al., 2017).
Genetic structure in dolphins of the genus Tursiops is often highly habitat dependent. While there is relatively little differentiation for both nuclear and mitochondrial DNA markers in pelagic bottlenose dolphin populations (e.g., Hoelzel et al., 1998b; Quérouil et al., 2007; Louis et al., 2014), stronger differentiation on markedly smaller spatial scales has been described in coastal populations (Krützen et al., 2004; Sellas et al., 2005; Möller et al., 2007; Rosel et al., 2009; Tezanos-Pinto et al., 2009; Urian et al., 2009; Mirimin et al., 2011). Variation in oceanographic conditions such as salinity and temperature gradients, habitat type, as well as differences in prey preference and foraging specializations, have also been invoked to explain such fine-scale genetic differentiation (Natoli et al., 2005; Kopps et al., 2014; Krützen et al., 2014; Gaspari et al., 2015).
In the Perth metropolitan area, Western Australia, Indo-Pacific bottlenose dolphins (Tursiops aduncus, “bottlenose dolphins” hereafter) inhabit coastal, embayment and estuarine waters (Chabanne et al., 2017a). The population is exposed to year-round anthropogenic activities, including dredging, pile-driving, recreational and commercial shipping and fisheries, and environmental contaminants (Donaldson et al., 2010; Salgado Kent et al., 2012; Paiva et al., 2015; Cannell et al., 2016; Marley et al., 2017). Previous behavioral research recognized four well-defined communities with limited interactions, identified as Gage Roads (GR), Swan Canning Riverpark (SCR), Owen Anchorage (OA), and Cockburn Sound (CS) (Chabanne et al., 2017a,b). Three of these communities form relatively stable, cohesive units with long term residency in the area, occupying coastal embayment and estuarine habitats. The fourth community occurs along open coastline and appears structurally different, being much less socially cohesive and appearing to consist of more transient individuals (Chabanne et al., 2012, 2017a).
We investigated the extent of genetic structure and the degree of gene flow occurring among the four communities in Perth metropolitan waters using several genetic marker systems. We considered the well-documented social and spatial segregation of the four behaviorally defined communities when formulating conservation measures.
Materials and Methods
Genetic Sample Collection
During systematic (Chabanne et al., 2017a) and opportunistic photo-identification surveys, we collected biopsy samples from Indo-Pacific bottlenose dolphins (T. aduncus) in Perth metropolitan waters, Western Australia (Figure 1), between 2007 and 2015 using a PAXARMS remote biopsy system (Krützen et al., 2002). Calves less than 2 years old (based on body length and approximate date of birth) were not sampled. Tissue samples were preserved in saturated NaCl 20% dimethyl sulfoxide (Amos and Hoelzel, 1991) until DNA extraction.
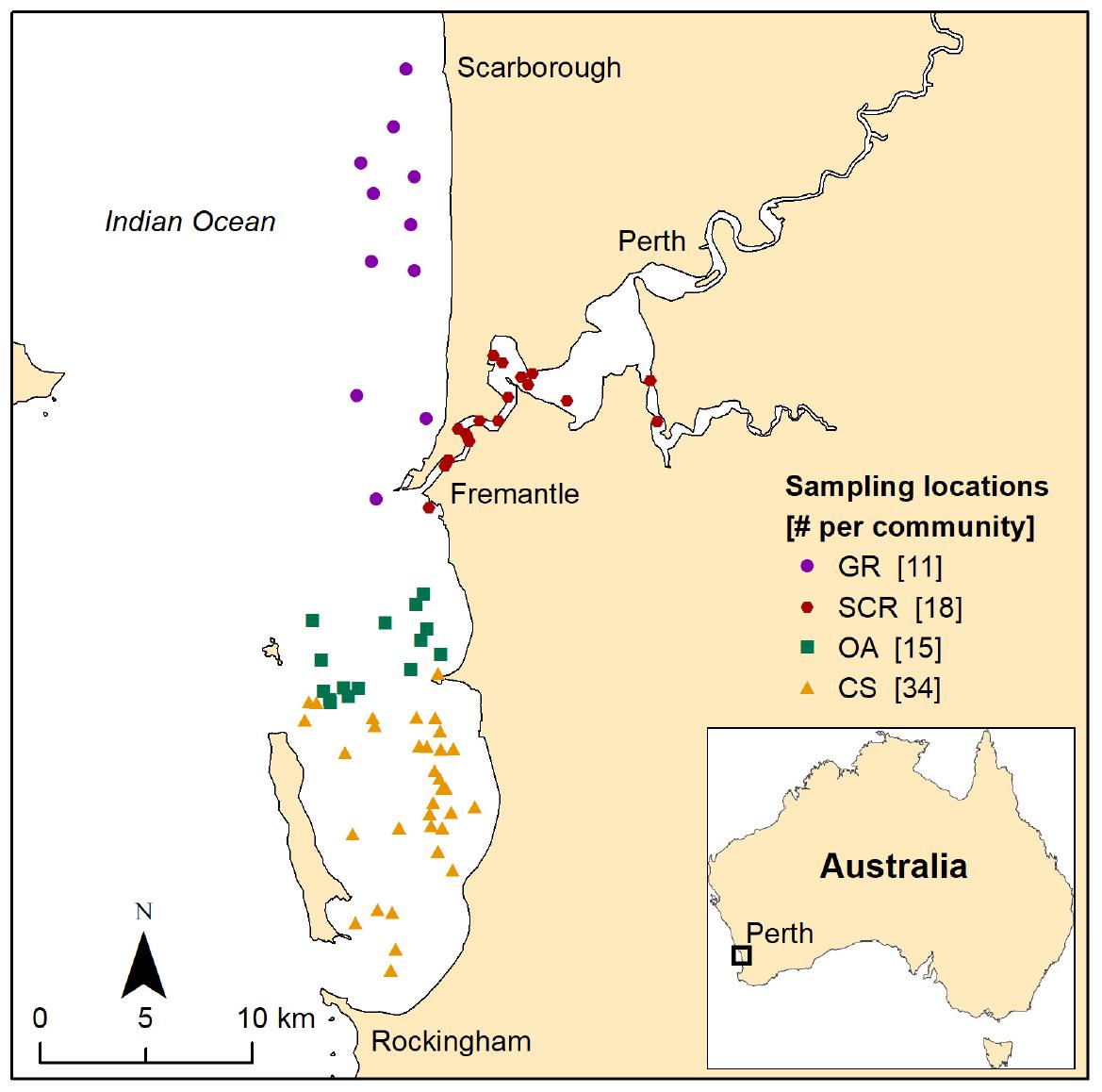
Figure 1. Location of the sampled individuals of bottlenose dolphins associated with one of the four socio-spatial communities in Perth metropolitan waters, Western Australia. GR, Gage Roads; SCR, Swan Canning Riverpark; OA, Owen Anchorage; CS, Cockburn Sound.
DNA Extraction and Sexing
We extracted genomic DNA from skin samples using the Gentra Puregene Tissue Kit (Qiagen), following the manufacturer’s protocol. We determined the sex of sampled individuals by amplifying parts of the ZFX and SRY genes (Gilson et al., 1998) via polymerase chain reaction (PCR) with 20–25 ng DNA, 0.15 μl of each primer (ZFX forward and reverse and SRY forward and reverse) and standard PCR reagents. The PCR profile consisted of initial denaturation at 95°C for 4 min, followed by 35 cycles of 94°C for 45 s, 50°C for 45 s, and 72°C for 10 min (Natoli et al., 2004; Ansmann et al., 2012). PCR products were separated on agarose gel to determine sex base on length differences.
Genotyping and Validation of Microsatellites
We successfully genotyped 87 non-duplicated samples for 32 microsatellite loci: DIrFCB4, DIrFCB5 (Buchanan et al., 1996), LobsDi_7.1, LobsDi_9, LobsDi_19, LobsDi_21, LobsDi_24, LobsDi_39 (Cassens et al., 2005), SCA9, SCA22, SCA27 (Chen and Yang, 2008), TexVet5, TexVet7 (Rooney et al., 1999), KWM12 (Hoelzel et al., 1998a), EV37 (Valsecchi and Amos, 1996), MK3, MK5, MK6, MK8, MK9 (Krützen et al., 2001), Tur_E12, Tur4_66, Tur4_80, Tur4_98, Tur4_105, Tur4_108, Tur4_111, Tur4_117, Tur4_128, Tur4_132, Tur_142, Tur4_153, and Tur4_162 (Nater et al., 2009). We amplified the loci following the PCR method described in Frère et al. (2010) and in Marfurt (2019). Single stranded PCR products were run on an ABI 3730 DNA Sequencer (Applied Biosystems). We scored microsatellite alleles using GENEIOUS 9.11 (Kearse et al., 2012) with the microsatellite plugin 1.4 (Applied Biosystems). Our genotyping error rate based on 11 individuals scored twice was estimated to be 0.27%, i.e., one incorrect genotype out of 360 scored. Although dolphin calves were not sampled, our study was sufficiently long for some individuals to be sampled when older. All scored individuals were cross-checked for eventual duplicates not initially identified (i.e., misidentification due to change of the dorsal fin). To minimize biases associated with first-order (parent–offspring, full-siblings) related individuals when assessing genetic structure, we removed any known offspring and juveniles from our dataset (n = 9 individuals). Later, we also calculated the relatedness between all possible dyads using the R package “related” (Pew et al., 2014) using the TrioML estimator and tested differences within and between communities using a Mantel test and 104 permutations.
After checking for null alleles and scoring errors due to stuttering or large allele dropout using the software Micro-Checker 2.2.3 (Van Oosterhout et al., 2004), we removed four microsatellite loci (SCA27, TexVet5, MK8, and Tur4_80). No deviation from Hardy-Weinberg equilibrium (HWE) and no evidence of linkage disequilibrium (LD) were detected between the remaining loci and across communities using the Markov chain randomization in the package “genepop” (Rousset, 2008, 2020) with 105 dememorizations, 103 batches, and 104 iterations and the Bonferroni correction (Rice, 1989).
Mitochondrial (mt) DNA Sequencing
We amplified a 412-bp mitochondrial fragment using the primers dlp1.5 (5′-TCA CCC AAA GCT GRA RTT CTA-3′) and dlp5 (5′-CCA TCG WGA TGT CTT ATT TAA GRG GAA-3′) (Baker et al., 1993) and following the PCR conditions described in Bacher et al. (2010). We successfully obtained mtDNA sequences for 73 out of the 78 samples used in the nuclear analyses after manually edited them in GENEIOUS 9.1.
Genetic Diversity
We assessed the genetic diversity based on microsatellite alleles within each of the four communities by calculating the number of alleles (Na), effective number of alleles (Ne), private alleles (NPA), observed (HO) and expected (HE) heterozygosities in GenAlEx (Peakall and Smouse, 2012), and allelic richness (AR) using FSTAT 2.9.3 (Goudet, 2001). We estimated inbreeding coefficient (FIS) for each community separately and pooled as one population in Arlequin 3.5 (Excoffier and Lischer, 2010) testing for significance with 104 permutations. We also evaluated the possibility of inbreeding using INEST 2.2 (Inbreeding/Null Allele Estimation; Chybicki and Burczyk, 2009) and taking into account the estimated null allele frequencies considering null alleles (Chybicki, 2017). For mtDNA, we identified the number of haplotypes (NH), and estimated haplotype (h) and nucleotide (π) divergence using DnaSP 5.10 (Librado and Rozas, 2009).
Genetic Differentiation
We estimated pairwise genetic differentiation of microsatellite alleles (FST) (Weir and Cockerham, 1984) and mtDNA ΦST (Tamura and Nei, 1993) among communities using Arlequin 3.5 (Excoffier and Lischer, 2010). For mtDNA, the choice of the model used was made after computing several nucleotide substitution models in jModelTest 2.1 (Posada, 2008). Although the minimum corrected Akaike Information Criterion (AICc used for small sample size) suggested Hasegawa-Kishino-Yano (Hasegawa et al., 1985) as the best model, this one was unavailable in Arlequin, and therefore we used the Tamura-Nei model (TrN, Tamura and Nei, 1993) as the next best model (ΔAICc < 2). All pairwise comparisons were testing for significance with 104 permutations.
Genetic Population Structure
Given the challenges associated with the inference of the most likely number of distinct genetic clusters (K) in a population (Guillot et al., 2009), we assessed K in Perth metropolitan waters based on the microsatellite loci using four approaches to ensure the robustness of the results: (1) a Bayesian clustering algorithm implemented in STRUCTURE 2.3 (Pritchard et al., 2000), (2) a least-squares optimization approach taking into account spatial autocorrelation based on tessellation (Caye et al., 2016) with the R package “tess3r” (Caye et al., 2017), (3) a discriminant analysis of principal components (DAPC) that allows to maximize the differences between groups while minimizing variation within groups (Jombart et al., 2010) using the package “adegenet” (Jombart and Ahmed, 2011), and (4) a three-dimensional factorial correspondence analysis (FCA) that seeks to identify genetic affinities between individuals and alleles and performed in GENETIX (Belkhir et al., 2004).
In STRUCTURE, we conducted the analysis without and with LOCPRIOR models that assigned samples to their respective socio-spatial community, as defined in Chabanne et al. (2017a). Using a LOCPRIOR model improves clustering when the signal is weak without spuriously inferring structure (Hubisz et al., 2009). We performed the analysis using the admixture model with correlated allele frequencies (Falush et al., 2003), using a burn-in of 106 Markov Chain Monte Carlo (MCMC) steps followed by 107 MCMC steps. We repeated each run 10 times for K varying from one to six (K = 5 and 6 being used to enable calculation of ΔK). The most likely value of K was determined by averaging the log probability LnP(D) among runs for each K value, and selecting the highest mean LnP(D) (Pritchard et al., 2000). Individual genetic cluster assignment estimates (i.e., individual ancestry proportions) were generated for each set for K varying from two to four, using the web service software CLUMPAK (Kopelman et al., 2015). The TESS analysis included the GPS coordinates (latitude and longitude, WGS84) of the sampled individuals as a priori information (Durand et al., 2009). We replicated ten runs for each K value varying from two to six. The most likely number of genetic clusters was identified by plotting the scores of a cross-validation with the best K value corresponding to a plateau before generating a q matrix containing individual admixture coefficients (Frichot et al., 2015). We then performed the DAPC analysis following Jombart and Collins (2015) with the maximum number of clusters set to six. The most likely number of genetic clusters was identified by the lowest Bayesian Information Criterion (BIC) value. Finally, we also assessed the structure with an FCA without a priori community information. The 3D FCA representation was carried out using the R package “scatterplot3d” (Ligges et al., 2018).
Assessment of Gene Flow
Using GENECLASS 2.0 (Piry et al., 2004), we looked for possible first-generation migrants (referred to as the likelihood of migrant detection L) among the communities following the Bayesian computation criteria of Rannala and Mountain (1997). This analysis was based on the likelihood of an individual’s genotype arising from the communities where the individual was sampled given the observed set of allele frequency (LHOME) and the highest likelihood value among all potential source communities (LMAX), including the home community where the individual was sampled, such as L = LHOME/LMAX. Significance of the assignment of individuals was assessed with MCMC resampling of 104 individuals and a threshold of 0.05 (Paetkau, 2004). This analysis was repeated using L = LHOME as the likelihood computation to account for the uncertainty that all source populations were sampled (Piry et al., 2004). We then estimated the effective numbers of migrants per generation between communities based on the Mutual information (SHua) calculated in GenAlEx 6.5 (Peakall and Smouse, 2012). We also used equations 10b and 10c (Sherwin et al., 2006), allowing us to estimate the effective number of individuals exchanged per generation (Nem) with and without Ne known, respectively. Given the small population size here, we estimated Ne as the sum of the samples available for the communities for which the migration rate was calculated.
Results
Genetic Diversity
Autosomal levels of genetic diversity were significantly higher in CS than in other communities, with more than two thirds of the private alleles identified in CS (Table 1). Other communities showed similar genetic diversity (Table 1). For mitochondrial DNA, we found a total of 20 polymorphic sites, defining five haplotypes (Table 1). In contrast to autosomal data, haplotype diversities were significantly lower in CS and SCR.
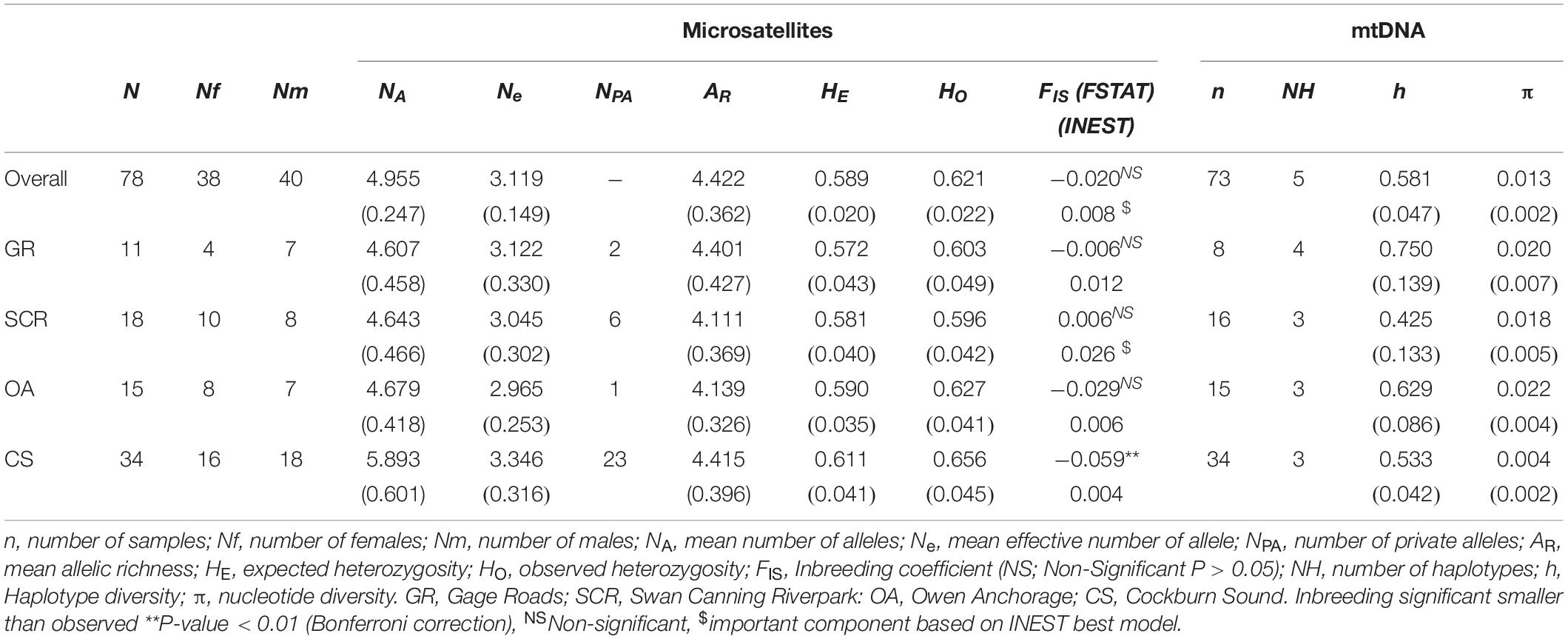
Table 1. Genetic diversity measures (SE) for bottlenose dolphin socio-spatial communities using microsatellite loci (n = 28) and mtDNA.
Genetic Differentiation
For autosomal markers, the CS community showed small albeit significant levels of differentiation from all other communities, which was also the case for SCR and OA (Table 2). To verify the accuracy of the significant values of FST reported here, the simulation run in POWSIM indicated that we would be able to detect an FST of 0.018 and above with 100% confidence, 0.010 with 99.40% confidence, and 0.007 with 93.90% confidence, while the type I error probability was lower than the expected rate of 5%. For mtDNA (ΦST), significant differentiation was found between OA and CS only (Table 2).
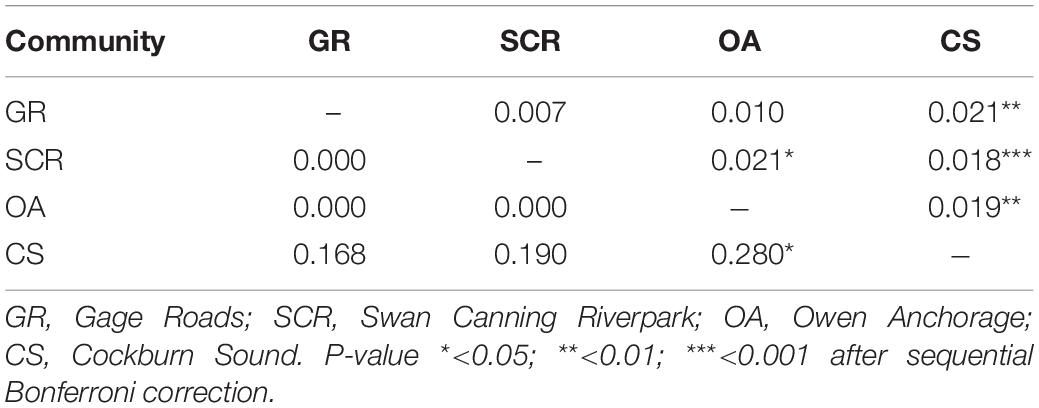
Table 2. Pairwise comparison between the four socio-spatial communities for 28 microsatellite loci (FST, above diagonal) and for mtDNA (ΦST, below diagonal).
Genetic Structure
Based on the highest mean of the estimated posterior probabilities [LnP(D)], the STRUCTURE analyses suggested K = 2 to be the most likely number of genetic clusters, while DAPC analysis suggested no clustering (Supplementary Figure 1). The cross-validation from the TESS method did not exhibit a minimum value or a clear plateau to identify the most likely number of genetic clusters K (Supplementary Figure 1). Both the OA and CS communities appeared to contain individuals belonging to one of the identified clusters or were admixed (Figure 2). Clustering assignment for individuals from GR and SCR contrasted between the results from STRUCTURE and TESS, with the former suggesting that the majority of the individuals were assigned to the same cluster (minimum of 82% and 78% with q > 0.8 in GR and SCR, respectively), while the latter indicated many individuals to be admixed (Figure 2).
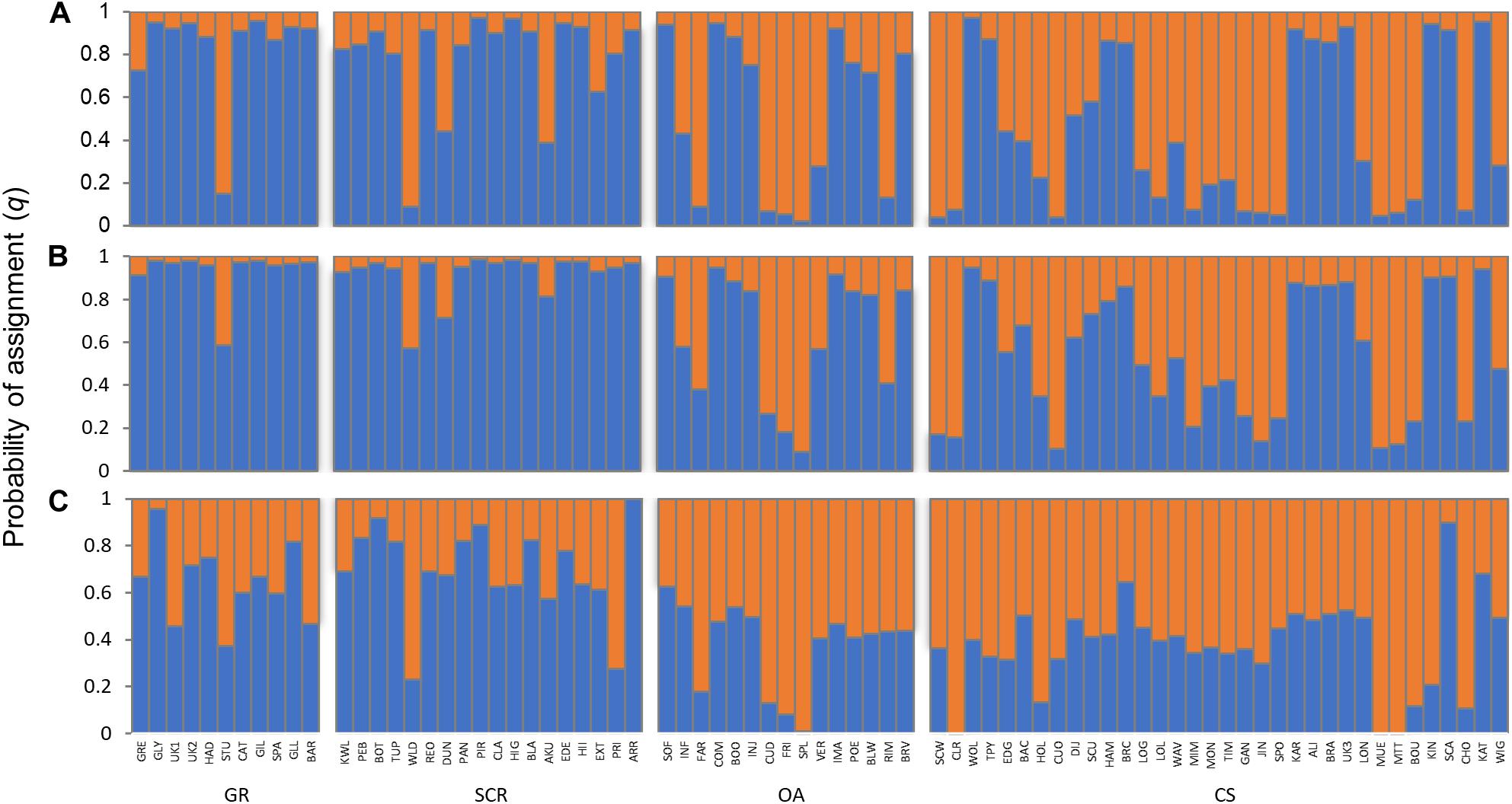
Figure 2. Assignment probabilities of individual bottlenose dolphins based on 28 microsatellite loci and inferred for K = 2 using methods implemented in STRUCTURE (A) without LOCPRIOR, (B) with LOCPRIOR), and (C) in TESS. Each vertical line represents one individual, with the colors indicating the membership proportions (q) to each of the genetic clusters. Individuals are grouped by socio-spatial communities (GR, Gage Roads; SCR, Swan Canning Riverpark; OA, Owen Anchorage; CS, Cockburn Sound).
The 3D FCA showed no clustering (factors 1–3 explaining only 13.98% of the variation, Figure 3) and samples were generally scattered.
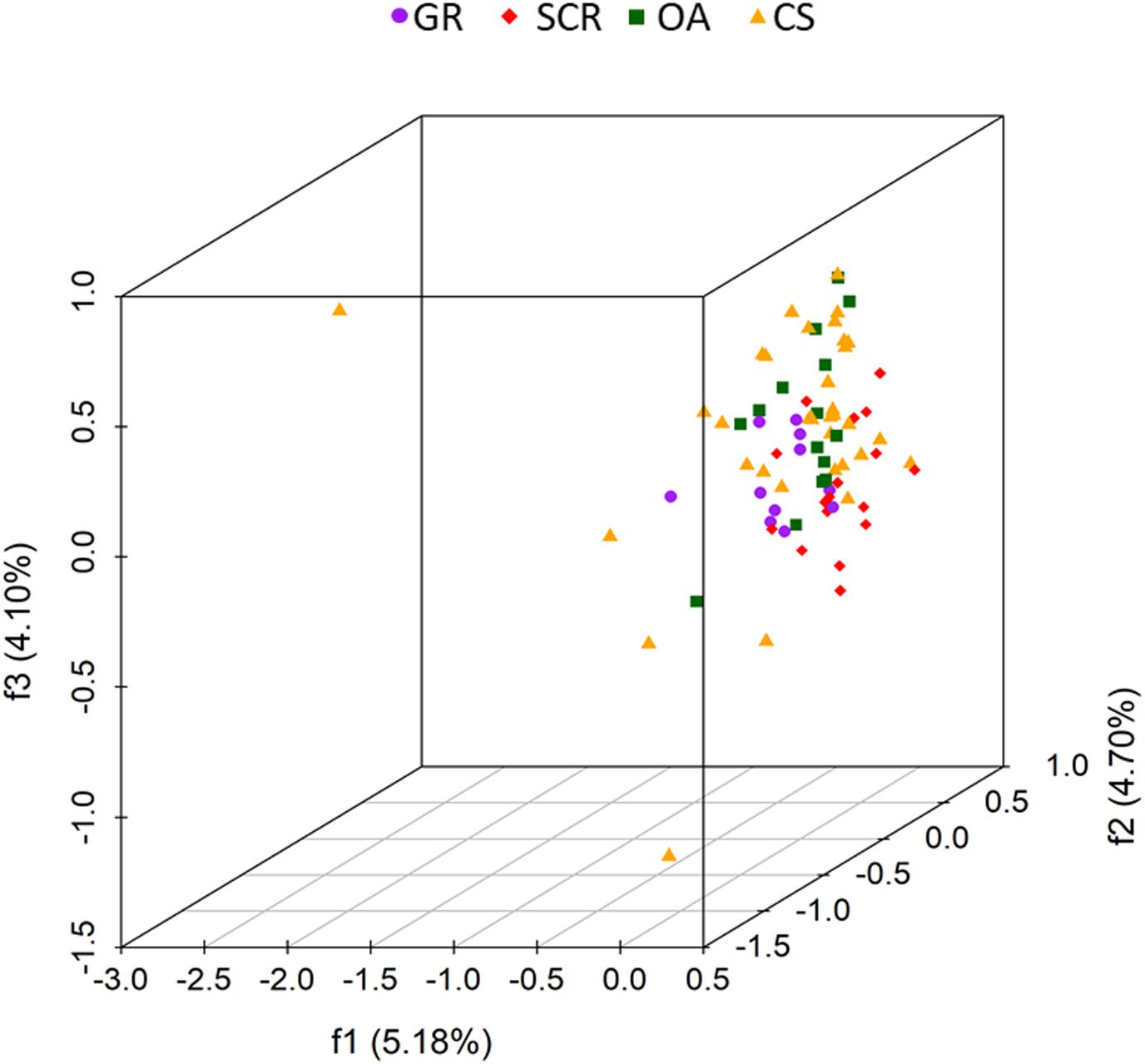
Figure 3. Factorial correspondence analysis (FCA) three-dimensional plots of microsatellite variation of bottlenose dolphins based on 28 microsatellite loci and without a priori information on socio-spatial communities. Each dot represents one individual, with the color/shape indicating its designated socio-spatial community (GR, Gage Roads; SCR, Swan Canning Riverpark; OA, Owen Anchorage; CS, Cockburn Sound). The percentage of variation explained by each of the first three factors are given in parentheses.
The median-joining network using mtDNA revealed no clear clustering based on socio-spatial communities with most of the haplotypes being shared between communities, despite showing two main ancestral groups of haplotypes separated by at least 15 mutational steps (Figure 4). However, most samples were represented in one matriline group with haplotype H3 found in the majority of the samples (57, 75, 53, and 50% for GR, SCR, OA, and CS, respectively). The low level of nucleotide diversity found in CS (0.004 and Table 1) is consistent with all but one sample (WOL with H1) having haplotypes from the same ancestral matriline group (H2 and H3, Figure 4) and separated by a maximum number of mutations (n = 17).
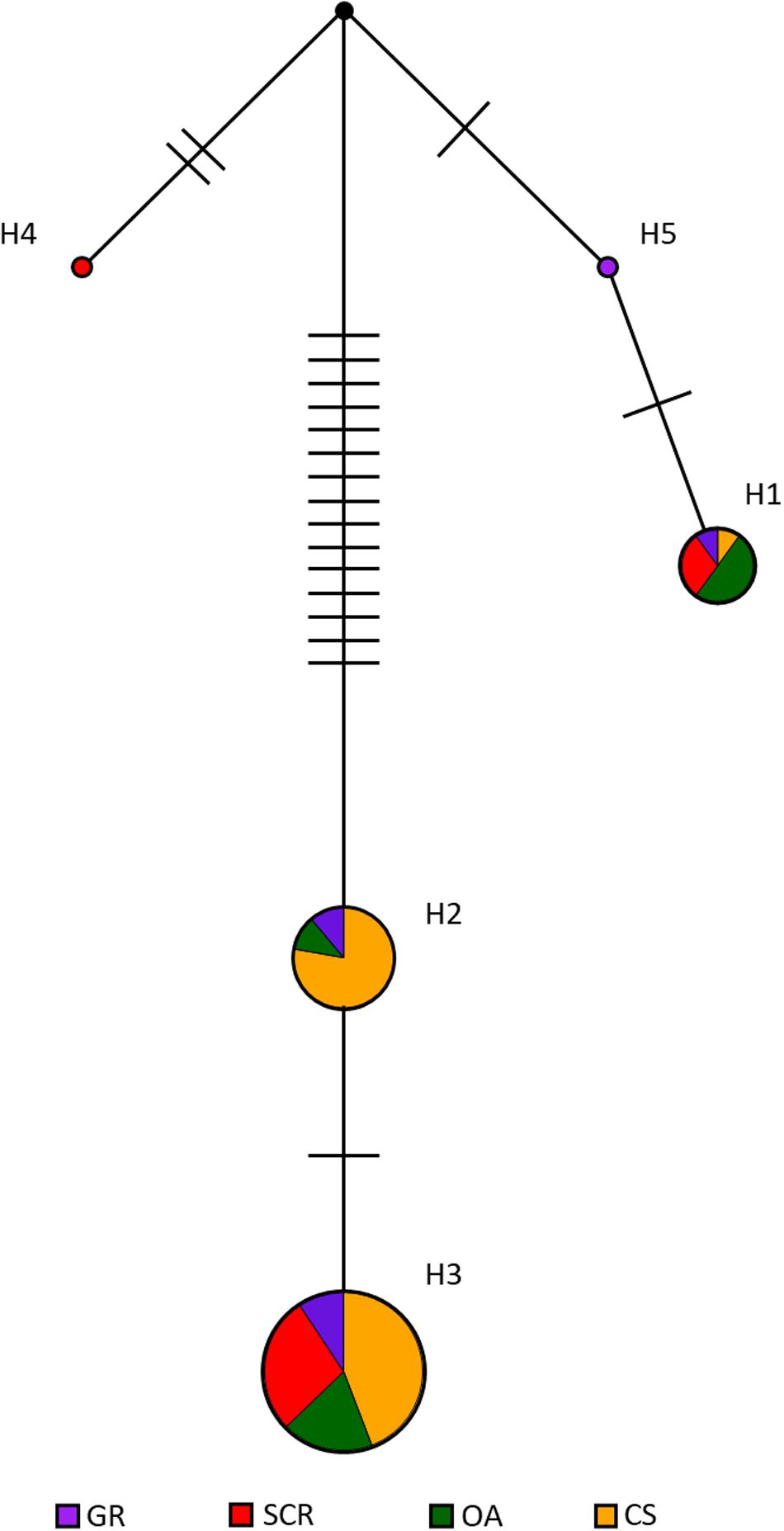
Figure 4. Median-joining network of mtDNA control region haplotypes in bottlenose dolphins in Perth metropolitan waters. The size of the circles is proportional to the total number of individuals carrying that haplotype. Different colors denote the four different sampled communities: GR, Gage Roads; SCR, Swan Canning Riverpark; OA, Owen Anchorage; CS, Cockburn Sound. The number of mutational events between each haplotype is indicated by hash marks. The black circle indicates intermediate haplotypes not found in our samples.
Contemporary Dispersal
The assignment/exclusion test in GENECLASS2 indicated that only 59% of the individuals were correctly assigned to their respective socio-spatial communities and detected 17 first-generation migrants in all directions, except from CS to SCR (Supplementary Table 1). Based on the estimated mutual information (SHua, Supplementary Table 2), we estimated the effective number of individuals exchanged per generation (Nem) varying from 2.33 to 4.97 and from 4.90 to 8.69 when Ne was estimated based on the number of samples available for each community pair (Table 3).
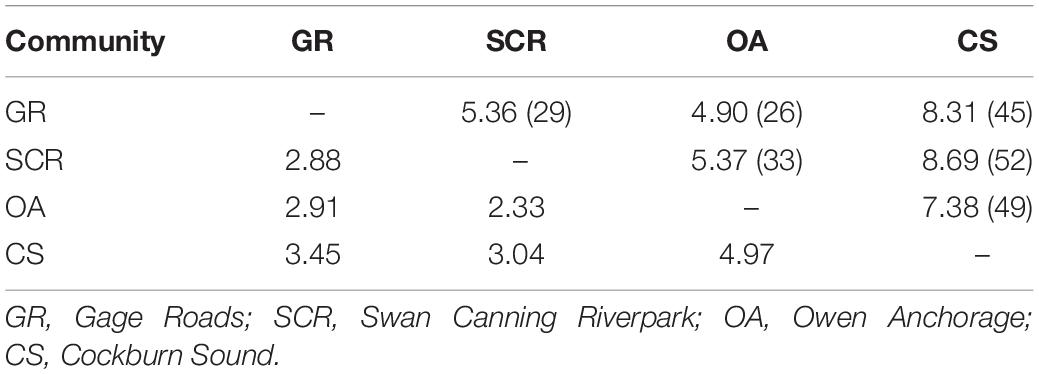
Table 3. Effective number of individuals exchanged per generation (Nem) between communities estimated without knowledge of Ne [equation 10c from Sherwin et al. (2006), estimates below the diagonal] and with our estimate of Ne as the sum of the paired communities [equation 10b from Sherwin et al. (2006), estimates above the diagonal with Ne between brackets].
Relatedness
Comparison of intra- and inter-community relatedness indicated significantly higher values within communities than between communities (Mantel test, r = 0.1327, and P-value < 0.001). Notably, individuals related at the first-order level (parent-offspring, full-siblings; for the purpose of this study defined by having a pairwise relatedness value of 0.3811 or larger, the lowest value observed from a known mother-calf pair in this study) were found primarily within communities rather than between (Figure 5). Half-siblings (r > 0.25) were roughly equally represented within and between communities with 4.6 and 2.4% of the pairs, respectively (Figure 5). Removing at least one individual from each first-order pair had no bearing on genetic differentiation and genetic structure analyses (Supplementary Table 3).
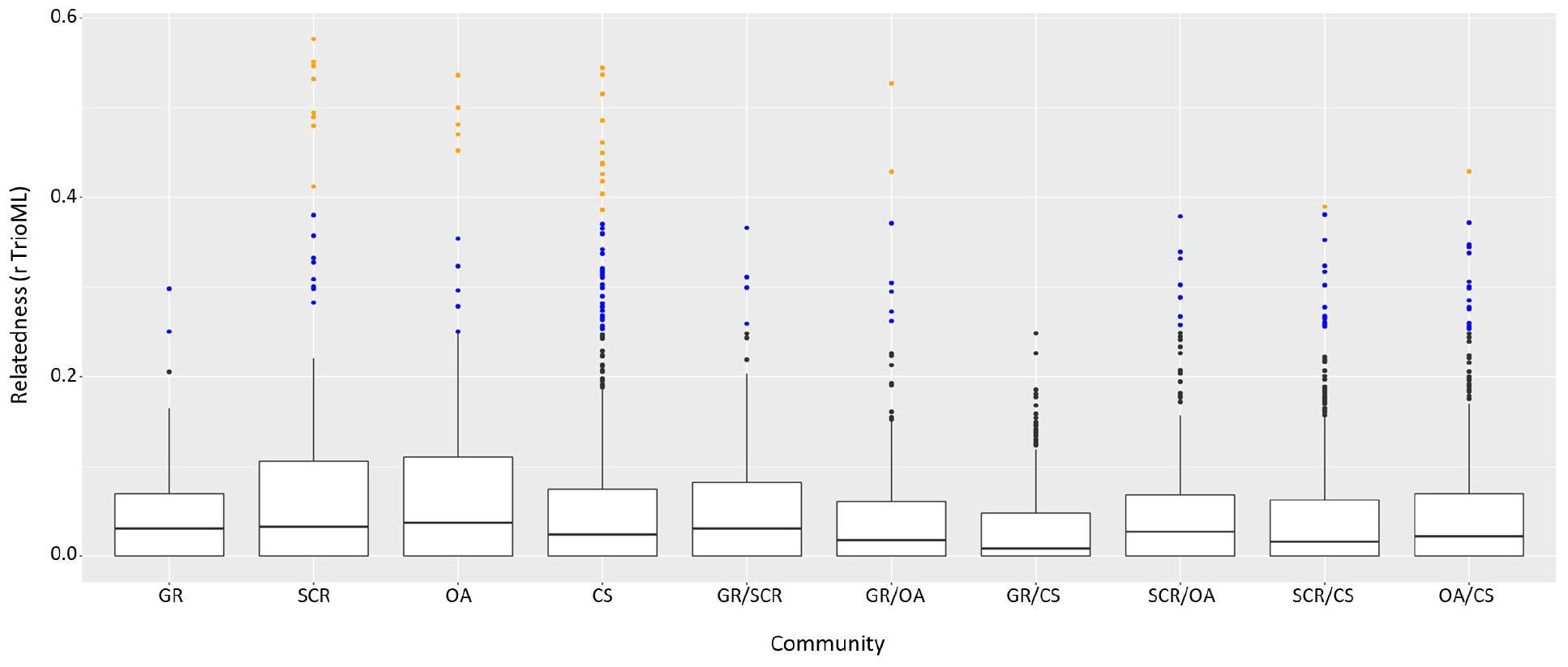
Figure 5. Pairwise relatedness among and between communities using the triadic likelihood (TrioML) estimator. Outliers in orange represent first-order pairwise defined with r > 0.3811 (percentage of those pairs respective to the number of pairs available are noted above the figure). Outliers in blue represent half-sibling order pairwise (r > 0.25).
Discussion
Overall, we found evidence for one panmictic bottlenose dolphin population with continuous gene flow. Analyses of both microsatellites and mtDNA markers revealed no conclusive structure, despite using various clustering algorithms that can produce different solutions due to differences in the underlying models and prior assumptions (Francois and Durand, 2010), and only low nuclear differentiation (FST < 0.021) among the four socio-spatial communities. This may be due to the presence of highly related individuals in our data, mirroring family structures present within each community (Latch et al., 2006; Anderson and Dunham, 2008; Pritchard et al., 2010). The post hoc removal of highly related individuals leading to even lower levels of differentiation (results in Supplementary Table 3) supports this hypothesis.
The disconnect between genetic and behavioral structures is to some extent unexpected. Other studies comparing communities of T. aduncus (e.g., Möller et al., 2007; Wiszniewski et al., 2010) and its congener T. truncatus (e.g., Mirimin et al., 2011; De Los Ángeles Bayas-Rea et al., 2018) detected fine-scale genetic structure at a similar geographic scale. The presence of genetic structure may indicate limited gene flow between communities, perhaps due to behavioral specializations or environmental discontinuities (Wiszniewski et al., 2010). The permanent dispersal described here, with the detection of first-generation migrants and high migration rates among communities, could reduce inbreeding within communities (Mills and Allendorf, 1996; Perrin and Mazalov, 1999; Faubet et al., 2007).
The low travel costs associated with short distances between the communities in Perth metropolitan waters provide the potential for reproduction outside social groups (Connor, 2000) without the necessity for individuals to permanently leave them. Killer whales, for example, live in permanent social groups from which males temporarily emigrate to mate with females from other groups (Pilot et al., 2010; Martien et al., 2019). In our study, some SCR resident males were observed herding reproductive females from OA into SCR (data not published), with females returning to their community core area afterward, a behavior recorded elsewhere for this species (Tsai and Mann, 2013; Wallen et al., 2016) and supported by the presence of a few highly related pairs of individuals from different communities.
Theory suggests that animals forming social groups may favor close relatives (Hamilton, 1964a,b). This appears to occur among dolphins of Perth metropolitan waters, as we found relatedness levels to be significantly higher within than between communities. Additionally, we found highly related individuals that were members of different communities. Such relatedness structure appears to be reflected by the long lifespan and slow life history leading to highly overlapping generations in bottlenose dolphins (Wells and Scott, 2002), as well as fission-fusion dynamics (Connor et al., 2000). Indeed, the maximum estimate of residency time (i.e., site fidelity) was 18 years (Chabanne et al., 2017a), a period considerably less than maximum life span in this species (i.e., more than 40 years; Wang and Yang, 2009). Individuals may switch communities in response to significant events (e.g., loss of a social partner, a new resource becoming available). Thus, the lack of genetic structure among social communities may mirror past and present fission-fusion dynamics among the communities as they overlap on the edge of their home range.
While genetic parameters do not match socio-spatial structure in this study, other factors need to be considered for management purposes. Over the last two decades, anthropogenic impacts were variable between communities (Chabanne et al., 2017a). Differences in habitat quality and population density commonly influence dispersal between populations, with areas characterized by lower-quality habitat and lower-density requiring an influx of dispersing individuals from higher-quality and higher-density areas for populations to persist (Pulliam, 1988; Figueira and Crowder, 2006; Liggins et al., 2013; Draheim et al., 2016; Sundqvist et al., 2016). Within Perth metropolitan waters, environmental changes associated with anthropogenic activities varied among the sites, requiring site-specific management strategies. Indeed, a gradual decline of seagrass and fish communities have been observed for the last three decades in SCR, in addition to problems common in other urban estuaries, such as eutrophication, deoxygenation, algal blooms, and the presence of per- and polyfluorinated alkyl substances (PFAS), a group of synthetic industrial chemicals (Deeley and Paling, 1998; Nice, 2009; Kilminster and Forbes, 2014; Valesini et al., 2017). In CS, important loss of seagrass coverage occurred between the 1960s and 1990s, with no substantial recovery of seagrasses on the eastern shelf (Kendrick et al., 2002) as a result of remaining problems associated with nutrient influx from groundwater plumes and contaminated sediments (Fraser and Kendrick, 2017). Also, changes in fish diversity were found along a gradient north to south (Sampey et al., 2011), indicating variation in suitable fish habitat that could result in heterogeneous dolphin distribution. In comparison, seagrass coverage in OA has not changed or even increased (Kendrick et al., 2000) despite dredging occurring year-round since 1972, suggesting a better habitat supporting a broad assemblage of prey species for dolphins. Ongoing, past and future environmental impacts (i.e., development of an Outer Harbor in CS) described above suggest that habitat and oceanographic characteristics, physical barriers, and dietary specializations are all conditions that may lead to discontinuous ecological communities, potentially representing unique ecological units to conserve even in the absence of genetic structure (Giménez et al., 2018). In this context, future research may disentangle the socio-spatial structure of bottlenose dolphins in Perth metropolitan waters.
Finally, none of these communities are safe from reductions in genetic diversity should stochastic events lead to a bottleneck (Storz, 1999; Leffler et al., 2012; Vachon et al., 2018). The low haplotype diversity found in CS could be suggestive of the occurrence of an historical bottleneck event or selection (Rand, 1996). Such scenarios are often challenging to evaluate because of the difficulty in assessing small sample sizes or when there is low effective population size pre-bottleneck (Bjorklund, 2003; Peery et al., 2012; Subramanian, 2016). The SCR lost almost 1/3 of its individuals (Chabanne et al., 2012) in 2009 from an outbreak of the cetacean morbillivirus (CeMV), which has also affected several other dolphin populations worldwide (Holyoake et al., 2010; Stone et al., 2011; West et al., 2012; Casalone et al., 2014; Stephens et al., 2014; Kemper et al., 2016). After the event, there was a noteworthy decrease in mtDNA haplotypes from at least seven before 2010 (Holyoake et al., 2010) to only three in the SCR (this study), with a potential loss of haplotypes at Perth population scale, i.e., only five defined in this study, while Manlik et al. (2019) described 10 haplotypes using a subsample of the Perth population. Mortalities caused by viral pandemics, environmental pollution or human activities have already negatively affected population size and genetic diversity in other species of dolphin (Pichler and Baker, 2000; Krützen et al., 2018). Although more work is required to better understand possible selection processes associated with resistance to CeMV (Batley et al., 2019), the reduction in genetic diversity exacerbated by the negative effects of genetic drift in small populations and potential inbreeding, and thus lower resilience to stochastic processes, poses an extinction risk to some cetacean populations (Oremus et al., 2015).
Conclusion
Our study suggests that the present socio-spatial structure of Indo-Pacific bottlenose dolphins in Perth metropolitan waters does not reflect a genetic structure defined by a clear separation between communities. Managers might then consider that, should any of these communities (CS, GR, SCR, and/or OA) become extinct, the locality of the community would be repopulated by members of one or more of the adjacent communities. However, the recovery of dolphin abundance to original numbers or the re-colonization of available habitats is likely to be hampered if the cause of the community decline and local extinction in the first place is not correctly identified and managed accordingly (Irwin and Würsig, 2004; Nichols et al., 2007). Therefore, each community should still be considered a distinct ecological unit to conserve based on the available information on anthropogenic stressors and that presented here, as well as in Chabanne et al. (2017a).
The lack of fine-scale genetic structure should not lead to the conclusion that no population structure exists [e.g., Louis et al. (2018) and Giménez et al. (2018)]. The discrete social and spatial parameters, the long-term residency of the communities and the higher relatedness found within than between communities still support community-specific management actions (Chabanne et al., 2017a). Other extrinsic factors such as resource availability, habitat, or foraging specializations have significant explanatory power of socio-spatial structure in dolphin populations (Krützen et al., 2004; Möller et al., 2007; Ansmann et al., 2012; Kopps et al., 2014; Giménez et al., 2018; Louis et al., 2018). Any ecological differences between communities must therefore be considered and, in some circumstances, may be as informative as genetic differences (Taylor, 2005; Giménez et al., 2018; Louis et al., 2018). Until further research can be done to better understand the consequences of high intra-community relatedness, and despite the communities appearing to belong to a single panmictic population, the different anthropogenic threats support the current socio-spatial division for practical conservation management. With increased environmental stochasticity, even comparatively large dolphin populations have been subject to significant negative impacts on demographic and vital rates (e.g., Wild et al., 2019). In particular, the estuarine SCR community in Perth waters appears highly vulnerable to rapid environmental change and appropriate conservation measures are needed, in addition to maintaining connectivity between it and the coastal communities.
Data Availability Statement
Microsatellite genotypes are available in the figshare repository, doi: 10.6084/m9.figshare.13078823. Haplotype sequences were deposited into GenBank under Accession numbers MW221830 to MW221834.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee, Murdoch University.
Author Contributions
DC conceived and designed the study and processed the data. DC and SA collected the data. DC analyzed the data with advice from MK and WS. DC wrote the manuscript, with contributions to drafting, review and editorial input from MK, SA, HF, and WS. All authors contributed to the article and approved the submitted version.
Funding
Funding for this research was primarily provided by the Swan River Trust, with additional contributions from Fremantle Ports and the University of Zurich. DC acknowledges the Australian Postgraduate Award from Murdoch University (Ph.D.) and the Swiss Government Excellence Scholarship (Postdoctoral Fellowship).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the Whadjuk People of the Noongar Nation as the traditional custodians of the lands and waters on which this research took place, including the Derbal Nara, Derbal Yerrigan, and Djarlgarra. We would like to thank Celine Frere, Benthan Littleford-Colquhoun, Livia Gerber, and Eliane Escher for contributions to genetic laboratory work. We thank Lars Bejder for his contribution during DC’s Ph.D. We also thank many field assistants, without whom this research would not have been possible. We would also like to thank the Fremantle Sailing Club for logistical support. Field research was conducted under the conditions of licenses, authorities, and permits from the Western Australia Department of Biodiversity, Conservation and Attractions (SF005997, SF006538, SF007046, SF007596, SF008480, SF009119, SF009734, and SF0101223) and the Murdoch University Animal Ethics Committee (W2076/07, W2307/10, W2342/10, and R2649/14).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.617540/full#supplementary-material
Footnotes
References
Allen, S. J., Bryant, K. A., Kraus, R. H., Loneragan, N. R., Kopps, A. M., Brown, A. M., et al. (2016). Genetic isolation between coastal and fishery-impacted, offshore bottlenose dolphin (Tursiops spp.) populations. Mol. Ecol. 25, 2735–2753. doi: 10.1111/mec.13622
Amos, W., and Hoelzel, A. R. (1991). “Long term preservation of whale skin for DNA analysis,” in Genetic Ecology of Whales and Dolphins. Report of the International Whaling Commission Special Issue 13, ed. A. R. Hoelzel (Cambridge: International Whaling Commission), 99–104.
Anderson, E. C., and Dunham, K. K. (2008). The influence of family groups on inferences made with the program structure. Mol. Ecol. Resour. 8, 1219–1229. doi: 10.1111/j.1755-0998.2008.02355.x
Andrews, K. R., Karczmarski, L., Au, W. W. L., Rickards, S. H., Vanderlip, C. A., Bowen, B. W., et al. (2010). Rolling stones and stable homes: social structure, habitat diversity and population genetics of the Hawaiian spinner dolphin (Stenella longirostris). Mol. Ecol. 19, 732–748. doi: 10.1111/j.1365-294x.2010.04521.x
Ansmann, I. C., Parra, G. J., Lanyon, J. M., and Seddon, J. M. (2012). Fine-scale genetic population structure in a mobile marine mammal: inshore bottlenose dolphins in Moreton Bay, Australia. Mol. Ecol. 21, 4472–4485. doi: 10.1111/j.1365-294x.2012.05722.x
Bacher, K., Allen, S., Lindholm, A. K., Bejder, L., and Krützen, M. (2010). Genes or culture: are mitochondrial genes associated with tool use in bottlenose dolphins (Tursiops sp.)? Behav. Genet. 40, 706–714. doi: 10.1007/s10519-010-9375-8
Baker, C. S., Perry, A., Bannister, J. L., Weinrich, M. T., Abernethy, R. B., Calambokidis, J., et al. (1993). Abundant mitochondrial DNA variation and world-wide population structure in humpback whales. Proc. Natl. Acad. Sci. U.S.A. 90, 8239–8243. doi: 10.1073/pnas.90.17.8239
Barragan-Barrera, D. C., May-Collado, L. J., Tezanos-Pinto, G., Islas-Villanueva, V., Correa-Cardenas, C. A., and Caballero, S. (2017). High genetic structure and low mitochondrial diversity in bottlenose dolphins of the Archipelago of Bocas del Toro, Panama: a population at risk? PLoS One 12:e0189370. doi: 10.1371/journal.pone.0189370
Batley, K. C., Sandoval-Castillo, J., Kemper, C. M., Attard, C. R. M., Zanardo, N., Tomo, I., et al. (2019). Genome-wide association study of an unusual dolphin mortality event reveals candidate genes for susceptibility and resistance to cetacean Morbillivirus. Evol. Appl. 12, 718–732. doi: 10.1111/eva.12747
Belkhir, K., Goudet, J., Chikhi, L., and Bonhomme, F. (2004). Genetix, Logiciel Sous WindowsTM Pour la Génétique des Populations, ver. 4.05. Available online at: http://www.genetix.univ-montp2.fr/genetix/genetix.htm (accessed April 3, 2020).
Bilgmann, K., Parra, G. J., Zanardo, N., Beheregaray, L. B., and Möller, L. M. (2014). Multiple management units of short-beaked common dolphins subject to fisheries bycatch off southern and southeastern Australia. Mar. Ecol. Prog. Ser. 500, 265–279. doi: 10.3354/meps10649
Bjorklund, M. (2003). Test for a population expansion after a drastic reduction in population size using DNA sequence data. Heredity 91, 481–486. doi: 10.1038/sj.hdy.6800309
Brown, A. M., Kopps, A. M., Allen, S. J., Bejder, L., Littleford-Colquhoun, B., Parra, G. J., et al. (2014). Population differentiation and hybridisation of Australian snubfin (Orcaella heinsohni) and Indo-Pacific humpback (Sousa chinensis) dolphins in north-western Australia. PLoS One 9:e101427. doi: 10.1371/journal.pone.0101427
Buchanan, F. C., Friesen, M. K., Littlejohn, R. P., and Clayton, J. W. (1996). Microsatellites from the beluga whale Delphinapterus leucas. Mol. Ecol. 5, 571–575. doi: 10.1111/j.1365-294x.1996.tb00348.x
Cannell, B. L., Campbell, K., Fitzgerald, L., Lewis, J. A., Baran, I. J., and Stephens, N. S. (2016). Anthropogenic trauma is the most prevalent cause of mortality in Little Penguins, Eudyptula minor, in Perth, Western Australia. EMU Austr. Ornithol. 116, 52–61. doi: 10.1071/mu15039
Casalone, C., Mazzariol, S., Pautasso, A., Di Guardo, G., Di Nocera, F., Lucifora, G., et al. (2014). Cetacean strandings in Italy: an unusual mortality event along the Tyrrhenian Sea coast in 2013. Dis. Aquat. Organ. 109, 81–86. doi: 10.3354/dao02726
Cassens, I., Van Waerebeek, K., Best, P. B., Tzika, A., Van Helden, A. L., Crespo, E. A., et al. (2005). Evidence for male dispersal along the coasts but no migration in pelagic waters in dusky dolphins (Lagenorhynchus obscurus). Mol. Ecol. 14, 107–121. doi: 10.1111/j.1365-294x.2004.02407.x
Caye, K., Deist, T. M., Martins, H., Michel, O., and Francois, O. (2016). TESS3: fast inference of spatial population structure and genome scans for selection. Mol. Ecol. Resour. 2, 540–548. doi: 10.1111/1755-0998.12471
Caye, K., Jay, F., and François, O. (2017). Analyzing population genomic data with tess3r. Available online at: https://bcm-uga.github.io/TESS3_encho_sen/articles/main-vignette.html (accessed July 1, 2020).
Chabanne, D., Finn, H., Salgado-Kent, C., and Bejder, L. (2012). Identification of a resident community of bottlenose dolphins (Tursiops aduncus) in the Swan Canning Riverpark, Western Australia, using behavioural information. Pac. Conserv. Biol. 18, 247–262. doi: 10.1071/pc120247
Chabanne, D. B. H., Finn, H., and Bejder, L. (2017a). Identifying the relevant local population for environmental impact assessments of mobile marine fauna. Front. Mar. Sci. 4:148. doi: 10.3389/fmars.2017.00148
Chabanne, D. B. H., Pollock, K. H., Finn, H., and Bejder, L. (2017b). Applying the multistate capture-recapture robust design to characterize metapopulation structure. Methods Ecol. Evol. 8, 1547–1557. doi: 10.1111/2041-210x.12792
Chen, L., and Yang, G. (2008). A set of polymorphic dinucleotide and tetranucleotide microsatellite markers for the Indo-Pacific humpback dolphin (Sousa chinensis) and cross-amplification in other cetacean species. Conserv. Genet. 10, 697–700. doi: 10.1007/s10592-008-9618-x
Chybicki, I. J. (2017). INEST 2.2: The User Manual. Poland: Department of Genetics, Kazimierz Wielki University.
Chybicki, I. J., and Burczyk, J. (2009). Simultaneous estimation of null alleles and inbreeding coefficients. J. Hered. 100, 106–113. doi: 10.1093/jhered/esn088
Connor, R. (2000). “Group living in whales and dolphins,” in Cetacean Societies: Field Studies of Dolphins and Whales, eds J. Mann, R. C. Connor, P. L. Tyack, and H. Whitehead (Chicago, IL: The University of Chicago Press), 199–218.
Connor, R. C., Wells, R. S., Mann, J., and Read, A. J. (2000). “The bottlenose dolphin, social relationship in a fission-fusion society,” in Cetacean Societies, Field Studies of Dolphins and Whales, eds J. Mann, R. C. Connor, P. L. Tyack, and H. Whitehead (Chicago, IL: The University of Chicago Press), 91–126.
Crawford, J. C., Dechen Quinn, A., Williams, D. M., Rudolph, B. A., Scribner, K. T., and Porter, W. F. (2018). Fine-scale spatial genetic structure of deer in a suburban landscape. J. Wildlife Manag. 82, 596–607. doi: 10.1002/jwmg.21417
De Los Ángeles Bayas-Rea, R., Félix, F., and Montufar, R. (2018). Genetic divergence and fine scale population structure of the common bottlenose dolphin (Tursiops truncatus, Montagu) found in the Gulf of Guayaquil, Ecuador. PeerJ 6:e4589. doi: 10.7717/peerj.4589
Deeley, D. M., and Paling, E. I. (1998). “Assessing the ecological health of estuaries in southwest Australia,” in Wetlands for the Future, eds A. J. Mccomb and J. A. Davis (Adelaide: Gleneagles Publishing), 257–271.
Donaldson, R., Finn, H., and Calver, M. (2010). Illegal feeding increases risk of boat-strike and entanglement in bottlenose dolphins in Perth, Western Australia. Pac. Conserv. Biol. 16, 157–161. doi: 10.1071/pc100157
Draheim, H. M., Moore, J. A., Etter, D., Winterstein, S. R., and Scribner, K. T. (2016). Detecting black bear source-sink dynamics using individual-based genetic graphs. Proc. R. Soc. B 283:20161002. doi: 10.1098/rspb.2016.1002
Durand, E., Chen, C., and François, O. (2009). TESS version 2.3 - Reference Manual. Available online at: https://www.scribd.com/document/459462207/manual-pdf (accessed September 15, 2018).
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Falush, D., Stephens, M., and Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587.
Faubet, P., Waples, R. S., and Gaggiotti, O. E. (2007). Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol. Ecol. 16, 1149–1166. doi: 10.1111/j.1365-294x.2007.03218.x
Fernández, R., Santos, M. B., Pierce, G. J., Llavona, Á, López, A., Silva, M. A., et al. (2011). Fine-scale genetic structure of bottlenose dolphins, Tursiops truncatus, in Atlantic coastal waters of the Iberian Peninsula. Hydrobiologia 670, 111–125. doi: 10.1007/s10750-011-0669-5
Figueira, W. F., and Crowder, L. B. (2006). Defining patch contribution in source-sink metapopulations: the importance of including dispersal and its relevance to marine systems. Popul. Ecol. 48, 215–224. doi: 10.1007/s10144-006-0265-0
Francois, O., and Durand, E. (2010). Spatially explicit Bayesian clustering models in population genetics. Mol. Ecol. Resour. 10, 773–784. doi: 10.1111/j.1755-0998.2010.02868.x
Fraser, M. W., and Kendrick, G. A. (2017). Belowground stressors and long-term seagrass declines in a historically degraded seagrass ecosystem after improved water quality. Sci. Rep. 7:14469.
Frère, C. H., Krützen, M., Mann, J., Connor, R. C., Bejder, L., and Sherwin, W. B. (2010). Social and genetic interactions drive fitness variation in a free-living dolphin population. Proc. Natl. Acad. Sci. U.S.A. 107, 19949–19954. doi: 10.1073/pnas.1007997107
Frichot, E., François, O., and O’meara, B. (2015). LEA: an R package for landscape and ecological association studies. Methods Ecol. Evol. 6, 925–929. doi: 10.1111/2041-210x.12382
Gaspari, S., Scheinin, A., Holcer, D., Fortuna, C., Natali, C., Genov, T., et al. (2015). Drivers of population structure of the bottlenose dolphin (Tursiops truncatus) in the eastern Mediterranean Sea. Evol. Biol. 42, 177–190. doi: 10.1007/s11692-015-9309-8
Gilson, A., Syvanen, M., Levine, K., and Banks, J. (1998). Deer gender determination by polymerase chain reaction: validation study and application to tissues, bloodstains, and hair forensic samples from California. Calif. Fish Game 84, 159–169.
Giménez, J., Louis, M., Barón, E., Ramírez, F., Verborgh, P., Gauffier, P., et al. (2018). Towards the identification of ecological management units: a multidisciplinary approach for the effective management of bottlenose dolphins in the southern Iberian Peninsula. Aquat. Conserv. 28, 205–215. doi: 10.1002/aqc.2814
Goudet, J. (2001). FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices (version 2.9.3). Available from http://www.unil.ch/izea/softwares/fstat.html (accessed July 1, 2015).
Guillot, G., Leblois, R., Coulon, A., and Frantz, A. C. (2009). Statistical methods in spatial genetics. Mol. Ecol. 18, 4734–4756.
Hamilton, W. D. (1964a). The genetical evolution of social behaviour. I. J. Theor. Biol. 7, 1–16. doi: 10.1016/0022-5193(64)90038-4
Hamilton, W. D. (1964b). The genetical evolution of social behaviour. II. J. Theor. Biol. 7, 17–52. doi: 10.1016/0022-5193(64)90039-6
Hasegawa, M., Kishino, H., and Yano, T.-A. (1985). Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174. doi: 10.1007/bf02101694
Hoelzel, A. R., Dahlheim, M. E., and Stern, S. J. (1998a). Low genetic variation among killer whales (Orcinus orca) in the Eastern North Pacific and genetic differentiation between foraging specialists. J. Hered. 89, 121–128. doi: 10.1093/jhered/89.2.121
Hoelzel, A. R., Potter, C. W., and Best, P. B. (1998b). Genetic differentiation between parapatric ‘nearshore’ and ‘offshore’ populations of the bottlenose dolphin. Proc. R. Soc. B 265, 1177–1183. doi: 10.1098/rspb.1998.0416
Holyoake, C., Finn, H., Stephens, N., Duignan, P., Salgado, C., Smith, H., et al. (2010). Technical Report on the Bottlenose Dolphin (Tursiops aduncus) Unusual Mortality Event within the Swan Canning Riverpark, June-October 2009. Perth, WA: Murdoch University.
Hubisz, M. J., Falush, D., Stephens, M., and Pritchard, J. K. (2009). Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources 9, 1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x
Irwin, L.-J., and Würsig, B. (2004). A small resident community of bottlenose dolphins, Tursiops truncatus, in Texas: monitoring recommendations. Gulf Mexico Sci. 22, 13–21.
Jombart, T., and Ahmed, I. (2011). Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071. doi: 10.1093/bioinformatics/btr521
Jombart, T., and Collins, C. (2015). A Tutorial for Discriminant Analysis of Principal Components (DAPC) Using Adegenet 2.0.0. Available online at: https://adegenet.r-forge.r-project.org/files/tutorial-dapc.pdf (accessed April 3, 2019).
Jombart, T., Devillard, S., and Balloux, F. (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11:94. doi: 10.1186/1471-2156-11-94
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kemper, C. M., Tomo, I., Bingham, J., Bastianello, S. S., Wang, J., Gibbs, S. E., et al. (2016). Morbillivirus-associated unusual mortality event in South Australian bottlenose dolphins is largest reported for the Southern Hemisphere. R. Soc. Open Sci. 3:160838. doi: 10.1098/rsos.160838
Kendrick, G. A., Aylward, M. J., Heggeb, B. J., Cambridge, M. L., Hillman, K., Wyllie, A., et al. (2002). Changes in seagrass coverage in Cockburn Sound, Western Australia between 1967 and 1999. Aquat. Bot. 73, 75–87. doi: 10.1016/s0304-3770(02)00005-0
Kendrick, G. A., Hegge, B. J., Wyllie, A., Davidson, A., and Lord, D. A. (2000). Changes in seagrass cover on success and Parmelia Banks, Western Australia Between 1965 and 1995. Estuar. Coast. Shelf Sci. 50, 341–353. doi: 10.1006/ecss.1999.0569
Kilminster, K., and Forbes, V. (2014). Seagrass as an indicator of estuary condition in the Swan-Canning estuary. Water Science Technical Series, report no. 62. Perth, WA: Department of Water, Western Australia.
Kopelman, N. M., Mayzel, J., Jakobsson, M., Rosenberg, N. A., and Mayrose, I. (2015). Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 15, 1179–1191. doi: 10.1111/1755-0998.12387
Kopps, A. M., Ackermann, C. Y., Sherwin, W. B., Allen, S. J., Bejder, L., and Krützen, M. (2014). Cultural transmission of tool use combined with habitat specializations leads to fine-scale genetic structure in bottlenose dolphins. Proc. R. Soc. B 281:20133245. doi: 10.1098/rspb.2013.3245
Krützen, M., Barre, L. M., Moller, L. M., Heithaus, M. R., Simms, C., and Sherwin, W. B. (2002). A biopsy system for small cetaceans: darting success and wound healing in Tursiops spp. Mar. Mamm. Sci. 18, 863–878. doi: 10.1111/j.1748-7692.2002.tb01078.x
Krützen, M., Beasley, I., Ackermann, C. Y., Lieckfeldt, D., Ludwig, A., Ryan, G. E., et al. (2018). Demographic collapse and low genetic diversity of the Irrawaddy dolphin population inhabiting the Mekong River. PLoS One 13:e0189200. doi: 10.1371/journal.pone.0189200
Krützen, M., Kreicker, S., Macleod, C. D., Learmonth, J., Kopps, A. M., Walsham, P., et al. (2014). Cultural transmission of tool use by Indo-Pacific bottlenose dolphins (Tursiops sp.) provides access to a novel foraging niche. Proc. R. Soc. B 281:20140374. doi: 10.1098/rspb.2014.0374
Krützen, M., Sherwin, W. B., Berggrem, P., and Gales, N. (2004). Population structure in an inshore cetacean revealed by microsatellite and mtDNA analysis: bottlenose dolphins (Tursiops sp.) in Shark Bay, Western Australia. Mar. Mamm. Sci. 20, 28–47. doi: 10.1111/j.1748-7692.2004.tb01139.x
Krützen, M., Valsecchi, E., Connor, R. C., and Sherwin, W. B. (2001). Characterization of microsatellite loci in Tursiops aduncus. Mol. Ecol. Notes 1, 170–172. doi: 10.1046/j.1471-8278.2001.00065.x
Latch, E. K., Dharmarajan, G., Glaubitz, J. C., and Rhodes, O. E. (2006). Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv. Genet. 7, 295–302. doi: 10.1007/s10592-005-9098-1
Leffler, E. M., Bullaughey, K., Matute, D. R., Meyer, W. K., Segurel, L., Venkat, A., et al. (2012). Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol. 10:e1001388. doi: 10.1371/journal.pone.1001388
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liggins, L., Treml, E. A., and Riginos, C. (2013). Taking the plunge: an introduction to undertaking seascape genetic studies and using biophysical models. Geogr. Compass 7, 173–196. doi: 10.1111/gec3.12031
Litz, J. A., Hughes, C. R., Garrison, L. P., Fieber, L., and Rosel, P. E. (2012). Genetic structure of common bottlenose dolphins (Tursiops truncatus) inhabiting adjacent South Florida estuaries - Biscayne Bay and Florida Bay. J. Cetacean Res. Manag. 12, 107–117.
Louis, M., Simon-Bouhet, B., Viricel, A., Lucas, T., Gally, F., Cherel, Y., et al. (2018). Evaluating the influence of ecology, sex and kinship on the social structure of resident coastal bottlenose dolphins. Mar. Biol. 165:80.
Louis, M., Viricel, A., Lucas, T., Peltier, H., Alfonsi, E., Berrow, S., et al. (2014). Habitat-driven population structure of bottlenose dolphins, Tursiops truncatus, in the North-East Atlantic. Mol. Ecol. 23, 857–874. doi: 10.1111/mec.12653
Manlik, O., Chabanne, D., Daniel, C., Bejder, L., Allen, S. J., and Sherwin, W. B. (2019). Demography and genetics suggest reversal of dolphin source-sink dynamics, with implications for conservation. Mar. Mamm. Sci. 35, 732–759. doi: 10.1111/mms.12555
Marfurt, S. (2019). Using Social Network Analysis to Investigate Association Patterns and Community Structure of Female Indo-Pacific Bottlenose Dolphins (Tursiops aduncus). Master thesis, University of Zurich, Zurich.
Marley, S. A., Salgado Kent, C. P., and Erbe, C. (2017). Occupancy of bottlenose dolphins (Tursiops aduncus) in relation to vessel traffic, dredging, and environmental variables within a highly urbanised estuary. Hydrobiologia 792, 243–263. doi: 10.1007/s10750-016-3061-7
Martien, K. K., Taylor, B. L., Chivers, S. J., Mahaffy, S. D., Gorgone, A. M., and Baird, R. W. (2019). Fidelity to natal social groups and mating within and between social groups in an endangered false killer whale population. Endang. Spec. Res. 40, 219–230. doi: 10.3354/esr00995
Mills, L. S., and Allendorf, F. W. (1996). The one-migrant per-generation rule in conservation and management. Conserv. Biol. 10, 1509–1518. doi: 10.1046/j.1523-1739.1996.10061509.x
Mirimin, L., Miller, R., Dillane, E., Berrow, S. D., Ingram, S., Cross, T. F., et al. (2011). Fine-scale population genetic structuring of bottlenose dolphins in Irish coastal waters. Anim. Conserv. 14, 342–353. doi: 10.1111/j.1469-1795.2010.00432.x
Möller, L., Valdez, F. P., Allen, S., Bilgmann, K., Corrigan, S., and Beheregaray, L. B. (2011). Fine-scale genetic structure in short-beaked common dolphins (Delphinus delphis) along the East Australian Current. Mar. Biol. 158, 113–126. doi: 10.1007/s00227-010-1546-x
Möller, L. M., and Beheregaray, L. B. (2004). Genetic evidence for sex-biased dispersal in resident bottlenose dolphins (Tursiops aduncus). Mol. Ecol. 13, 1607–1612. doi: 10.1111/j.1365-294x.2004.02137.x
Möller, L. M., Wiszniewski, J., Allen, S. J., and Beheregaray, L. B. (2007). Habitat type promotes rapid and extremely localised genetic differentiation in dolphins. Mar. Freshw. Res. 58, 640–648. doi: 10.1071/mf06218
Nater, A., Kopps, A. M., and Krützen, M. (2009). New polymorphic tetranucleotide microsatellites improve scoring accuracy in the bottlenose dolphin Tursiops aduncus. Mol. Ecol. Resour. 9, 531–534. doi: 10.1111/j.1755-0998.2008.02246.x
Natoli, A., Birkun, A., Aguilar, A., Lopez, A., and Hoelzel, A. R. (2005). Habitat structure and the dispersal of male and female bottlenose dolphins (Tursiops truncatus). Proc. R. Soc. B 272, 1217–1226. doi: 10.1098/rspb.2005.3076
Natoli, A., Peddemors, V. M., and Hoelzel, A. R. (2004). Population structure and speciation in the genus Tursiops based on microsatellite and mitochondrial DNA analyses. J. Evol. Biol. 17, 363–375. doi: 10.1046/j.1420-9101.2003.00672.x
Nice, H. (2009). A baseline study of contaminants in the sediments of the Swan and Canning estuaries. Water Science Technical Series Report No. 6. Perth, WA: Government of Western Australia.
Nichols, C., Herman, J., Gaggiotti, O. E., Dobney, K. M., Parsons, K., and Hoelzel, A. R. (2007). Genetic isolation of a now extinct population of bottlenose dolphins (Tursiops truncatus). Proc. R. Soc. B 274, 1611–1616. doi: 10.1098/rspb.2007.0176
Nielsen, R., and Wakeley, J. (2001). Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics 158, 885–896.
Oremus, M., Garrigue, C., Tezanos-Pinto, G., and Scott Baker, C. (2015). Phylogenetic identification and population differentiation of bottlenose dolphins (Tursiops spp.) in Melanesia, as revealed by mitochondrial DNA. Mar. Mamm. Sci. 31, 1035–1056. doi: 10.1111/mms.12210
Paetkau, D. (2004). Direct real-time estimation of migration rate using assignment methods: a simulation-based exploration of accuracy and power. Mol. Ecol. 13, 55–65. doi: 10.1046/j.1365-294x.2004.02008.x
Paiva, E. G., Salgado Kent, C. P., Gagnon, M. M., Mccauley, R., and Finn, H. (2015). Reduced detection of Indo-Pacific bottlenose dolphins (Tursiops aduncus) in an Inner Harbour channel during pile driving activities. Aquat. Mamm. 41, 455–468. doi: 10.1578/am.41.4.2015.455
Palsbøll, P. J., Berube, M., Aguilar, A., Notarbartolo-Di-Sciara, G., and Nielsen, R. (2004). Discerning between recurrent gene flow and recent divergence under a finite-site mutation model applied to North Atlantic and Mediterranean Sea fin whale (Balaenoptera physalus) populations. Evolution 58, 670–675. doi: 10.1554/02-529
Palsbøll, P. J., Berube, M., and Allendorf, F. W. (2007). Identification of management units using population genetic data. Trends Ecol. Evol. 22, 11–16. doi: 10.1016/j.tree.2006.09.003
Palumbi, S. R. (1992). Marine speciation on a small planet. Trends Ecol. Evol. 7, 114–118. doi: 10.1016/0169-5347(92)90144-z
Parra, G. J., Cagnazzi, D., Jedensjo, M., Ackermann, C., Frère, C., Seddon, J. M., et al. (2018). Low genetic diversity, limited gene flow and widespread genetic bottleneck effects in a threatened dolphin species, the Australian humpback dolphin. Biol. Conserv. 220, 192–200. doi: 10.1016/j.biocon.2017.12.028
Parsons, K. M., Balcomb, K. C., Ford, J. K. B., and Durban, J. W. (2009). The social dynamics of southern resident killer whales and conservation implications for this endangered population. Anim. Behav. 77, 963–971. doi: 10.1016/j.anbehav.2009.01.018
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Peery, M. Z., Kirby, R., Reid, B. N., Stoelting, R., Doucet-Beer, E., Robinson, S., et al. (2012). Reliability of genetic bottleneck tests for detecting recent population declines. Mol. Ecol. 21, 3403–3418. doi: 10.1111/j.1365-294x.2012.05635.x
Perrin, N., and Mazalov, V. (1999). Dispersal and inbreeding avoidance. Am. Nat. 154, 282–292. doi: 10.2307/2463651
Pew, J., Wang, J., Muir, P., and Frasier, T. (2014). related: an R package for analyzing pairwise relatedness data based on codominant molecular markers. Mol. Ecol. Resour. 15, 557–561. doi: 10.1111/1755-0998.12323
Pichler, F. B., and Baker, C. S. (2000). Loss of genetic diversity in the endemic Hector’s dolphin due to fisheries-related mortality. Proc. R. Soc. B 267, 97–102. doi: 10.1098/rspb.2000.0972
Pilot, M., Dahlheim, M. E., and Hoelzel, A. R. (2010). Social cohesion among kin, gene flow without dispersal and the evolution of population genetic structure in the killer whale (Orcinus orca). J. Evol. Biol. 23, 20–31. doi: 10.1111/j.1420-9101.2009.01887.x
Piry, S., Alapetite, A., Cornuet, J. M., Paetkau, D., Baudouin, L., and Estoup, A. (2004). GENECLASS2: a software for genetic assignment and first-generation migrant detection. J. Hered. 95, 536–539. doi: 10.1093/jhered/esh074
Posada, D. (2008). jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Pratt, E. A. L., Beheregaray, L. B., Bilgmann, K., Zanardo, N., Diaz-Aguirre, F., and Möller, L. M. (2018). Hierarchical metapopulation structure in a highly mobile marine predator: the southern Australian coastal bottlenose dolphin (Tursiops cf. australis). Conserv. Genet. 19, 637–654. doi: 10.1007/s10592-017-1043-6
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Pritchard, J. K., Wen, W., and Falush, D. (2010). Documentation for Structure Software: Version 2.3. Chicago, IL: Department of Human Genetics, University of Chicago.
Pulliam, R. (1988). Sources, sinks, and population regulation. Am. Natur. 132, 652–661. doi: 10.1086/284880
Quérouil, S., Silva, M. A., Freitas, L., Prieto, R., Magalhães, S., Dinis, A., et al. (2007). High gene flow in oceanic bottlenose dolphins (Tursiops truncatus) of the North Atlantic. Conserv. Genet. 8, 1405–1419. doi: 10.1007/s10592-007-9291-5
Rand, D. M. (1996). Neutrality tests of molecular markers and the connection between DNA polymorphism, demography, and conservation biology. Conserv. Biol. 10, 665–671. doi: 10.1046/j.1523-1739.1996.10020665.x
Rannala, B., and Mountain, J. L. (1997). Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. U.S.A. 94, 9197–9201. doi: 10.1073/pnas.94.17.9197
Rice, W. R. (1989). Analyzing tables of statistical tests. Evolution 43, 223–225. doi: 10.2307/2409177
Rooney, A. P., Merritt, D. B., and Derr, J. N. (1999). Microsatellite diversity in captive bottlenose dolphins (Tursiops truncatus). J. Hered. 90, 228–253. doi: 10.1093/jhered/90.1.228
Rosel, P. E., Hansen, L., and Hohn, A. A. (2009). Restricted dispersal in a continuously distributed marine species: common bottlenose dolphins Tursiops truncatus in coastal waters of the western North Atlantic. Mol. Ecol. 18, 5030–5045. doi: 10.1111/j.1365-294x.2009.04413.x
Rousset, F. (2008). GENEPOP’007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Salgado Kent, C. P., Mccauley, R. D., Parnum, I. M., and Gavrilov, A. N. (2012). Underwater noise sources in Fremantle inner harbour: dolphins, pile driving and traffic. Proc. Acoust. Soc. Austr. 1–7.
Sampey, A., Fromont, J., and Johnston, D. J. (2011). Demersal and epibenthic fauna in a temperate marine embayment, Cockburn Sound, Western Australia: determination of key indicator species. J. R. Soc. West. Austr. 94, 1–18.
Sellas, A. B., Wells, R. S., and Rosel, P. E. (2005). Mitochondrial and nuclear DNA analyses reveal fine scale geographic structure in bottlenose dolphins (Tursiops truncatus) in the Gulf of Mexico. Conserv. Genet. 6, 715–728. doi: 10.1007/s10592-005-9031-7
Sherwin, W. B., Jabot, F., Rush, R., and Rossetto, M. (2006). Measurement of biological information with applications from genes to landscapes. Mol. Ecol. 15, 2857–2869. doi: 10.1111/j.1365-294x.2006.02992.x
Snijders, L., Blumstein, D. T., Stanley, C. R., and Franks, D. W. (2017). Animal social network theory can help wildlifeconservation. Trends Ecol. Evol. 32, 567–577. doi: 10.1016/j.tree.2017.05.005
Stephens, N., Duignan, P. J., Wang, J., Bingham, J., Finn, H., Bejder, L., et al. (2014). Cetacean morbillivirus in coastal Indo-Pacific bottlenose dolphins, Western Australia. Emerg. Infect. Dis. J. 20, 666–670.
Stone, B. M., Blyde, D. J., Saliki, J. T., Blas-Machado, U., Bingham, J., Hyatt, A., et al. (2011). Fatal cetacean morbillivirus infection in an Australian offshore bottlenose dolphin (Tursiops truncatus). Aust. Vet. J. 89, 452–457. doi: 10.1111/j.1751-0813.2011.00849.x
Storz, J. F. (1999). Genetic consequences of mammalian social structure. J. Mammal. 80, 553–569. doi: 10.2307/1383301
Subramanian, S. (2016). The effects of sample size on population genomic analyses: implications for the tests of neutrality. BMC Genom. 17:123. doi: 10.1186/s12864-016-2441-8
Sundqvist, L., Keenan, K., Zackrisson, M., Prodohl, P., and Kleinhans, D. (2016). Directional genetic differentiation and relative migration. Ecol. Evolut. 6, 3461–3475. doi: 10.1002/ece3.2096
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biolo. Ecol. 10, 512–526.
Taylor, B. L. (2005). “Identifying units to conserve,” in Marine Mammal Research: Conservation Beyond Crisis, eds J. E. Reynolds, W. F. Perrin, R. R. Reeves, S. Montgomery, and T. J. Ragen (Baltimore, MD: The Johns Hopkins University Press), 149–164.
Taylor, B. L., and Dizon, A. E. (1999). First policy then science: why a management unit based solely on genetic criteria cannot work. Mol. Ecol. 8, S11–S16.
Taylor, M. S., and Hellberg, M. E. (2003). Genetic evidence for local retention of pelagic larvar in a Caribbean reef fish. Science 299, 107–109. doi: 10.1126/science.1079365
Tezanos-Pinto, G., Baker, C. S., Russell, K., Martien, K., Baird, R. W., Hutt, A., et al. (2009). A worldwide perspective on the population structure and genetic diversity of bottlenose dolphins (Tursiops truncatus) in New Zealand. J. Hered. 100, 11–24. doi: 10.1093/jhered/esn039
Tsai, Y. J. J., and Mann, J. (2013). Dispersal, philopatry, and the role of fission-fusion dynamics in bottlenose dolphins. Mar. Mamm. Sci. 29, 261–279. doi: 10.1111/j.1748-7692.2011.00559.x
Urian, K. W., Hofmann, S., Wells, R. S., and Read, A. J. (2009). Fine-scale population structure of bottlenose dolphins (Tursiops truncatus) in Tampa Bay, Florida. Mar. Mamm. Sci. 25, 619–638. doi: 10.1111/j.1748-7692.2009.00284.x
Vachon, F., Whitehead, H., and Frasier, T. R. (2018). What factors shape genetic diversity in cetaceans? Ecol. Evol. 8, 1554–1572. doi: 10.1002/ece3.3727
Valesini, F. J., Cottingham, A., Hallett, C. S., and Clarke, K. R. (2017). Interdecadal changes in the community, population and individual levels of the fish fauna of an extensively modified estuary. J. Fish Biol. 90, 1734–1767. doi: 10.1111/jfb.13263
Valsecchi, E., and Amos, W. (1996). Microsatellite markers for the study of cetacean populations. Mol. Ecol. 5, 151–156. doi: 10.1111/j.1365-294x.1996.tb00301.x
Van Cise, A. M., Martien, K. K., Mahaffy, S. D., Baird, R. W., Webster, D. L., Fowler, J. H., et al. (2017). Familial social structure and socially driven genetic differentiation in Hawaiian short-finned pilot whales. Mol. Ecol. 26, 6730–6741. doi: 10.1111/mec.14397
Van Oosterhout, C. V., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Wallen, M. M., Patterson, E. M., Krzyszczyk, E., and Mann, J. (2016). The ecological costs to females in a system with allied sexual coercion. Anim. Behav. 115, 227–236. doi: 10.1016/j.anbehav.2016.02.018
Wang, J. Y., and Yang, S.-C. (2009). “Indo-Pacific bottlenose dolphin (Tursiops aduncus),” in Encyclopedia of Marine Mammals, 2nd Edn, eds W. F. Perrin, B. Würsig, and J. G. M. Thewissen (San Diego, CA: Acadenic Press), 602–607.
Weir, B. S., and Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x
Wells, R. S., and Scott, M. D. (2002). “Bottlenose dolphins,” in The Encyclopedia of Marine Mammals, eds W. F. Perrin, B. Würsig, and J. G. M. Thewissen (New York, NY: Academic Press), 122–128.
Wells, R. S., Scott, M. D., Irvine, A. B., and Genoways, H. (1987). “The social structure of free-ranging bottlenose dolphins,” in Current Mammalogy, ed. H. H. Genoways (New York, NY: Plenum), 247–305. doi: 10.1007/978-1-4757-9909-5_7
West, K. L., Sanchez, S., Rotstein, D., Robertson, K. M., Dennison, S., Levine, G., et al. (2012). A Longman’s beaked whale (Indopacetus pacificus) strands in Maui, Hawaii, with first case of morbillivirus in the central Pacific. Mar. Mamm. Sci. 29, 767–776.
Wild, S., Krützen, M., Rankin, R. W., Hoppitt, W. J. E., Gerber, L., and Allen, S. J. (2019). Long-term decline in survival and reproduction of dolphins following a marine heatwave. Curr. Biol. 29, R239–R240.
Williams, R., Lusseau, D., and Hammond, P. S. (2009). The role of social aggregations and protected areas in killer whale conservation: the mixed blessing of critical habitat. Biol. Conserv. 142, 709–719. doi: 10.1016/j.biocon.2008.12.004
Wiszniewski, J., Allen, S. J., and Moller, L. M. (2009). Social cohesion in a hierarchically structured embayment population of Indo-Pacific bottlenose dolphins. Anim. Behav. 77, 1449–1457. doi: 10.1016/j.anbehav.2009.02.025
Wiszniewski, J., Beheregaray, L. B., Allen, S. J., Moller, L. M., and Möller, L. M. (2010). Environmental and social influences on the genetic structure of bottlenose dolphins (Tursiops aduncus) in Southeastern Australia. Conserv. Genet. 11, 1405–1419. doi: 10.1007/s10592-009-9968-z
Keywords: bottlenose dolphins, population structure, microsatellites, mtDNA, gene flow, conservation, management units, relatedness
Citation: Chabanne DBH, Allen SJ, Sherwin WB, Finn H and Krützen M (2021) Inconsistency Between Socio-Spatial and Genetic Structure in a Coastal Dolphin Population. Front. Mar. Sci. 7:617540. doi: 10.3389/fmars.2020.617540
Received: 15 October 2020; Accepted: 17 December 2020;
Published: 14 January 2021.
Edited by:
Jeremy Kiszka, Florida International University, United StatesReviewed by:
Marie Louis, University of Copenhagen, DenmarkG. Renee Albertson, Oregon State University, United States
Copyright © 2021 Chabanne, Allen, Sherwin, Finn and Krützen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Delphine B. H. Chabanne, ZGVscGhpbmUuY2hhYmFubmVAdXpoLmNo; ZGVscGhpbmUuY2hhYmFubmVAbXVyZG9jaC5lZHUuYXU=
 Delphine B. H. Chabanne
Delphine B. H. Chabanne Simon J. Allen
Simon J. Allen William Bruce Sherwin
William Bruce Sherwin Hugh Finn
Hugh Finn Michael Krützen
Michael Krützen