- 1Department of Marine Biology, Institute of Biological Sciences, University of Rostock, Rostock, Germany
- 2Department of Orthopedagogy and Physical Therapy, Ternopil V. Hnatiuk National Pedagogical University, Ternopil, Ukraine
- 3Department of Biological Sciences, Kent State University, Kent, OH, United States
- 4Department of Maritime Systems, Interdisciplinary Faculty, University of Rostock, Rostock, Germany
Coastal environments commonly experience strong oxygen fluctuations. Resulting hypoxia/reoxygenation stress can negatively affect mitochondrial functions, since oxygen deficiency impairs ATP generation, whereas a surge of oxygen causes mitochondrial damage by oxidative stress. Marine intertidal bivalves are adapted to fluctuating oxygen conditions, yet the underlying molecular mechanisms that sustain mitochondrial integrity and function during oxygen fluctuations are not yet well understood. We used targeted mRNA expression analysis to determine the potential involvement of the mitochondrial quality control mechanisms in responses to short-term hypoxia (24 h at <0.01% O2) and subsequent reoxygenation (1.5 h at 21% O2) in two hypoxia-tolerant marine bivalves, the Pacific oysters Crassostrea gigas and the blue mussels Mytilus edulis. We hypothesized that the genes involved in the mitochondrial quality control will be upregulated during hypoxia, and the less hypoxia-tolerant of the two studied species (M. edulis) will show a stronger dependence on transcriptional upregulation of these pathways than C. gigas. To test these hypotheses, mRNA expression of 17 (C. gigas) and 11 (M. edulis) marker genes involved in mitochondrial fusion, fission, proteolysis and mitophagy was analyzed in the digestive gland of M. edulis and C. gigas in normoxia and during hypoxia-reoxygenation (H/R) stress. In the mussels, the mRNA expression of the transcripts related to mitochondrial dynamics and quality control was strongly altered during H/R stress showing a shift toward fission, suppression of fusion, an increase in mitochondrial proteolysis and onset of mitophagy. These changes indicate that H/R stress induces mitochondrial injury in M. edulis requiring upregulation of the protective mechanisms to segregate the dysfunctional mitochondria by fission and degrade the oxidative damaged proteins and/or organelles. Unlike mussels, the transcript levels of all studied genes in the oysters remained at the baseline (normoxic) levels during H/R stress. This muted transcriptional response of C. gigas is in agreement with earlier findings showing better ability to maintain cellular homeostasis and higher resistance to apoptosis during H/R stress in the oysters compared with the mussels. The revealed species-specific differences in the expression of the mitochondrial quality control pathways shed light on the potentially important mechanisms of mitochondrial protection against H/R-induced damage that might contribute to hypoxia tolerance in marine bivalves.
Introduction
Deoxygenation driven by nutrient pollution and warming is a major stressor in estuarine and coastal oceans worldwide (Breitburg et al., 2018, 2019). The deficiency (hypoxia) or lack (anoxia) of oxygen occurs in the estuarine and coastal habitats where oxygen depletion due to respiration outstrips the oxygen influx from diffusion, water mixing and photosynthesis. As a result, these habitats can experience hypoxic periods from a few hours (such as in a diel cycling hypoxia) to several weeks or months in the case of seasonal hypoxia (Diaz and Rosenberg, 1995, 2008). Most benthic metazoans including important ecosystem engineers and keystone species of bivalves, crustaceans and fish require oxygen to survive and complete their life cycles, and oxygen deficiency can lead to a decrease in growth, performance and survival of marine benthos thereby threatening coastal marine ecosystems (Diaz and Rosenberg, 1995). Low motility of benthic marine invertebrates such as bivalves limits their ability to escape local hypoxia, making them reliant on physiological and biochemical adaptations to survive oxygen deficiency (Grieshaber et al., 1994). Metabolic regulation is at the crux of such adaptations due to the direct dependence of the mitochondria (generating >90% of the cellular ATP) on oxygen availability and the key role of these organelles in the energetic and redox balance of the cell. Investigation of mitochondrial responses to fluctuating oxygen conditions is thus important for understanding the fundamental physiological mechanisms setting limits of hypoxic tolerance in marine organisms.
In benthic invertebrates, severe hypoxia (<4% O2) and anoxia results in transition to a metabolically suppressed state, where ATP production and consumption are downregulated and anaerobic pathways are engaged to compensate for insufficient aerobic ATP generation (Hochachka et al., 1996; Hochachka and Lutz, 2001; Storey, 2002). Under these conditions, the mitochondrial respiration strongly declines or ceases (Zwaan et al., 1991; van den Thillart et al., 1992). In hypoxia-sensitive organisms such as terrestrial mammals, mitochondria become depolarized and eventually damaged during hypoxia due to the ATP depletion, acidosis and Ca2+ overload (Piper et al., 2003; Chen et al., 2007; Solaini et al., 2010). Post-hypoxic reoxygenation restores the aerobic ATP synthesis in mammalian mitochondria but may come at a cost due to a surge of the mitochondrial production of reactive oxygen species (ROS) that can damage the cellular proteins, lipids and DNA (Cadenas and Davies, 2000; Paradis et al., 2016). Similar patterns of mitochondrial responses during hypoxia-reoxygenation (H/R) stress involving oxidative damage, collapse of the mitochondrial membrane potential and loss of the OXPHOS capacity has been reported in hypoxia-sensitive marine mollusks such as the bay scallops (Ivanina and Sokolova, 2016; Ivanina et al., 2016). In hypoxia-tolerant organisms such as freshwater turtles, fish and marine intertidal mollusks, mitochondrial respiration, OXPHOS capacity and membrane potential are preserved and the oxidative damage is mitigated during the H/R stress (Galli and Richards, 2014; Sokolova, 2018; Sokolova et al., 2019). These findings imply that maintenance of the mitochondrial integrity through mitochondrial damage sensing and quality control mechanisms might play an important role in mitochondrial tolerance to H/R stress (Berlett and Stadtman, 1997; Cabiscol et al., 2000; Zorov et al., 2014). However, the mitochondrial quality control mechanisms remain poorly understood in hypoxia-tolerant organisms including benthic marine bivalves and require further investigations.
The mitochondrial quality control is well studied in hypoxia-sensitive terrestrial mammals (such as rodents) and encompasses a complex protective network of proteins regulating mitochondrial dynamics (fission and fusion), as well as proteases and mitophagic pathways degrading oxidatively damaged proteins and defective mitochondria (Eisner et al., 2018; Li and Liu, 2018). Under normal conditions, mitochondria tend to form mitochondrial networks by mitochondrial fusion. Fusion of the outer mitochondrial membrane (OMM) is mediated by mitofusins 1 and 2 (Mfn1 and 2), while dynamin-related GTPase OPA1 induces fusion of the inner mitochondrial membrane (IMM) (Chen et al., 2003; Song et al., 2009). Under stress, membrane depolarization activates the metalloendopeptidase OMA1, which degrades OPA1 (Baker et al., 2014; Xiao et al., 2014) and induces mitochondrial fission through interactions of dynamin-related protein DRP1 and its adaptor proteins (the mitochondrial fission factor Mff and fission protein 1 Fis1) (Otera et al., 2010; Losón et al., 2013). Mitochondrial fission removes the damaged mitochondria and facilitates apoptosis under extreme cellular stress (Youle and van der Bliek, 2012), while the hypoxia upregulated protein (HYOU1) supports the mitochondrial repair by suppressing premature hypoxia-induced cell death (Ozawa et al., 1999).
Mechanisms involved in the degradation of the damaged proteins and organelles play an important role in the cellular responses to hypoxia in mammalian models. Protein quality surveillance in the mitochondria is supported by multiple ATP-dependent and –independent proteases including Lon protease and paraplegin (both selectively degrading oxidatively damaged proteins) (Atorino et al., 2003; Venkatesh et al., 2012; Kuo et al., 2015; Pinti et al., 2015; Shanmughapriya et al., 2015; Sepuri et al., 2017), the IMM protease ATP23, and the caseinolytic matrix peptidase chaperone (Osman et al., 2007; Doyle and Wickner, 2008). On DNA level, the helicase Twinkle is responsible for mtDNA maintenance and replication (Spelbrink et al., 2001). Cells can also degrade complete damaged mitochondria through mitophagy to prevent joining of dysfunctional mitochondria to the network. These pathways involve the PTEN-induced kinase 1 (PINK1) which accumulates under the hypoxia and recruits Parkin to tag damaged mitochondria for degradation (Eiyama and Okamoto, 2015). Mitophagic mediators in the PINK1-Parkin pathway are regulated via reversible phosphorylation by a serine/threonine-protein phosphatase PGAM5 (Chen et al., 2016). Additional to the PINK1-Parkin pathway, an alternative mitophagic pathway degrading unhealthy mitochondria is mediated by the mitochondrial eating protein (MIEAP) (Kitamura et al., 2011; Nakamura and Arakawa, 2017).
The important role of mitochondrial quality control mechanisms (involving the regulation of the mitochondrial fission and fusion, proteolysis and mitophagy) in the responses to H/R stress is well established in the hypoxia-sensitive mammalian models such as rodents (Kulek et al., 2020; Wang and Zhou, 2020). Earlier studies also indicate that high activity and upregulated mRNA expression of mitochondrial proteases correlates with elevated hypoxia tolerance in marine bivalves (Ivanina and Sokolova, 2016; Ivanina et al., 2016). However, involvement of the pathways regulating mitochondrial dynamics and quality control in the response to H/R stress have not been extensively studied in hypoxia-tolerant organisms including bivalves. Here, we explored the molecular mechanisms underlying the mitochondrial responses to H/R stress by measuring the mRNA expression of key marker genes in the mitochondrial quality control pathways in the Pacific oyster Crassostrea gigas and the blue mussel Mytilus edulis. The two studied species are commonly exposed to fluctuating oxygen conditions in intertidal, coastal and estuarine habitats and are therefore good models to study the mitochondrial adaptations to hypoxia and reoxygenation stress (Sokolova et al., 2019).
Earlier studies found that the Pacific oyster is more tolerant to abiotic stressors (including hypoxia) than the blue mussel (David et al., 2005; Le Moullac et al., 2007; Sokolov et al., 2019). At the cellular level, higher hypoxia tolerance of C. gigas was associated with the resistance to apoptosis and muted inflammation response indicating less pronounced mitochondrial and cellular damage during the H/R stress in oysters compared with the mussels (Falfushynska et al., 2020a,b). Therefore, we hypothesized that the less hypoxia-tolerant of the two studied species (M. edulis) will more strongly rely on the regulation of the mitochondrial quality control mechanisms to counteract the H/R-induced cellular damage compared with the more tolerant C. gigas that experiences less cellular injury (Falfushynska et al., 2020b) and thus might require less protection from the inducible mitochondrial quality control mechanisms. To test this hypothesis, mRNA expression of 17 (C. gigas) and 11 (M. edulis) key genes in the mitochondrial fusion, fission, proteolysis and mitophagy pathways were analyzed by quantitative RT-PCR in the digestive gland of the two studied species of bivalves exposed to hypoxia and reoxygenation relative to the normoxic controls. We focused on the transcript levels of the following marker genes: for mitochondrial fission and fusion - mfn2 (encoding mitofusin 2), opa1 (mitochondrial dynamin-like 120 kDa protein), dnm1l (dynamin-1-like protein), mff (mitochondrial fission factor), fis1 (mitochondrial fission protein 1); for protein and DNA quality control - tsfm (encoding mitochondrial translation elongation factor Ts), lonp1 (mitochondrial Lon protease), spg7 (paraplegin), oma1 (mitochondrial metalloendopeptidase OMA1), clpB (mitochondrial caseinolytic matrix peptidase chaperone subunit B), atp23 (mitochondrial inner membrane protease ATP23), twnk (mitochondrial twinkle mtDNA helicase); and for mitophagy - mieap (encoding mitochondrial eating protein), hyou1 (hypoxia upregulated protein 1), prkn (parkin), pink1 (PTEN-induced kinase 1), and pgam5 (mitochondrial serine/threonine protein phosphatase PGAM5).
Materials and Methods
Animal Maintenance
Adult oysters [mean shell length ± the standard deviation (SD): 99.3 ± 13.1 mm] were obtained from the low intertidal zone of the German Wadden Sea near List/Sylt (55°01′42″N 8°26′04″E) and transported within 48 h of collection to the University of Rostock, Germany. Adult blue mussels (mean shell length ± SD: 52.5 ± 3.22 mm) were collected at Warnemünde marina “Mittelmole” (54°10′49.4″N 12°05′19.9″E) and brought to the University of Rostock within an hour of collection. All bivalves were transported in coolers lined with seawater-soaked paper towels, and the shells were cleaned from epibionts upon arrival. Two weeks prior to experiments, oysters and mussels were acclimated in recirculated temperature-controlled aquarium systems (Kunststoff-Spranger GmbH, Plauen, Germany) with aerated artificial seawater (ASW) (Tropic Marin®, Wartenberg, Germany) at salinity 33 ± 1 (C. gigas) and 15 ± 1 (M. edulis), respectively, and temperature 15 ± 0.5°C. These salinity and temperature were within natural range of the respective oysters’ and mussels’ habitat conditions. Bivalves were fed ad libitum by continuous addition of a commercial live algal blend (DTs Premium Blend Live Marine Phytoplankton, Coralsands, Mainz Kastel, Germany) according to the manufacturer’s instructions (0.01 ml g–1 fresh mass per day) using an automatic aquarium feeder.
Experimental Exposures
Bivalves (three oysters or six mussels in 2 l of ASW) were exposed to 24 h of severe hypoxia (<0.1% O2) by bubbling the ASW with pure nitrogen (Westfalen AG, Münster, Germany) in air-tight chambers at 15 ± 0.5°C and respective salinity. Oxygen concentration was monitored with an IntellicalTM LDO101 Laboratory Luminescent/Optical Dissolved Oxygen (DO) Sensor (HACH, Loveland, CO, United States). During exposure, bivalves were not fed to prevent bacterial growth in the chambers. It is worth noting that oysters and mussels spontaneously close their shells and cease feeding during hypoxic exposures regardless of the presence of algae in surrounding water. After hypoxia exposure, a subset of bivalves was allowed to recover in normoxic ASW (21% O2) for 1.5 h. The H/R incubation times were chosen based on tidal and diurnal cycles of oxygen fluctuations and previous findings that show that strongest mitochondrial response to reoxygenation occurs within the first hours of recovery (Kurochkin et al., 2009; Richards, 2011; Ivanina and Sokolova, 2016; Andrienko et al., 2017). The control group was maintained in normoxia (21% O2) in recirculated temperature-controlled aquarium systems (10 oysters or 30 mussels per 30 l of seawater) with continuous feeding as described in section “Animal Maintenance.”
Hepatopancreas tissue was sampled on ice after hypoxia, reoxygenation and in normoxic control, immediately shock-frozen in liquid nitrogen and stored at −80°C until further analysis. We have chosen hepatopancreas because it is one of the largest metabolically active organs in bivalves involved in digestion, energy storage and maintenance of energy homeostasis (Gosling, 1992; Kennedy, 1996). Furthermore, earlier studies in bivalves showed strong hypoxia-induced shifts in the hepatopancreas metabolome (Haider et al., 2020) and induction of apoptotic and inflammatory responses in this tissue during H/R stress (Falfushynska et al., 2020a). This makes hepatopancreas a useful tissue to investigate the pathways that maintain the mitochondrial integrity during H/R exposure. Samples sizes were 10 and 6 per treatment group for oysters and mussels, respectively.
Quantitative Real-Time PCR (qRT-PCR)
RNA extraction and cDNA synthesis was conducted as described elsewhere (Falfushynska et al., 2020a). Briefly, 20 to 50 mg hepatopancreas tissue were extracted in RNA Extracol (EURX Ltd.–molecular biology products, Gdansk, Poland) according to manufacturer’s instructions using an automatic homogenizer for 40 s at 6.0 m s–1 (FastPrep-24 MP Biomedicals, Valiant Co. Ltd., Yantai, Shandong, China). To avoid potential cross-reaction with residual DNA, RNA extracts were treated with DNase using the TURBO DNA-free kit (ThemoFisher Scientific, Waltham, MA, United States). RNA with a 280/260 absorbance ratio > 2.0 assessed by NanoVue PlusTM spectrophotometer (GE Healthcare, Chicago, IL, United States) was used for further processing. cDNA was synthesized from 2 μg of total RNA using RevertAid RT Kit (Thermo Fischer Scientific, Waltham, MA, United States). Expression levels were quantified by quantitative PCR (qPCR) using the StepOnePlus Realtime-PCR System instrument (Applied Biosystems, Thermo Fisher Scientific Corp., Foster City, CA, United States) and Biozym Blue S’Green qPCR BlueMix Separate ROX kit (Biozym Scientific GmbH, Hessisch Oldendorf, Germany). Each reaction comprised of 10 μl 2x qPCR master mix with ROX additive, 6.4 μl DNase/RNase-free water, 1.6 μl gene-specific reverse and forward primer mix (each to a final concentration of 0.4 μM) and 2 μl cDNA. For the genes where species-specific annotated transcript sequences were available, gene-specific primers were designed using published gene sequences of M. edulis and C. gigas in the NCBI/GenBank database. This was the case for all C. gigas target genes (except lonp1, opa1, hyou1 and fis1) and for the housekeeping genes from M. edulis. For those Mytilus sequences where no annotated transcripts were found in the NCBI/GenBank, we have used a transcriptome from a closely related species Mytilus galloprovincialis that belongs to the Mytilus edulis species complex (Varvio et al., 1988; Gaitán-Espitia et al., 2016). M. edulis and M. galloprovincialis are closely related, genetically similar and characterized by a high degree of hybridization and introgression in the nature (Gosling and Wilkins, 1981; Varvio et al., 1988; Gardner, 1996; Daguin et al., 2001; Gaitán-Espitia et al., 2016). To identify the candidate regions for primer design in Mytilus, we have aligned M. galloprovincialis sequences with available homologous sequences from multiple molluscan species, including Crassostrea. The regions with the high degree of nucleotide sequence conservation were chosen for primer design, and the qPCR was conducted under stringent conditions to avoid non-specific priming. The relevant fragments of sequence alignments between Crassostrea and M. galloprovincialis are provided in Supplementary Table S1. For lonp1, opa1, hyou1 and fis1 genes, for which no transcript from C. gigas were publicly available at the time of this study, primers were designed using a sequence from a closely related species, Crassostrea virginica (Supplementary Table S2). The identified sequence segments were then used to design gene-specific primers (Table 1). For primer check, the fragments were amplified using the following cycling parameters: 2 min at 95°C for polymerase activation, 40 cycles of 15 s at 95°C and 30 s at 60°C. Only the primers that produced a single product of expected length were used in the following analyses. All 17 target genes were successfully amplified from C. gigas. Six out of 17 target genes (including tsfm, clpB, mieap, atp23, hyou1, and twnk) could not be amplified from Mytilus due to the low quality or insufficient length of the respective transcript sequences available in the NCBI/GenBank.
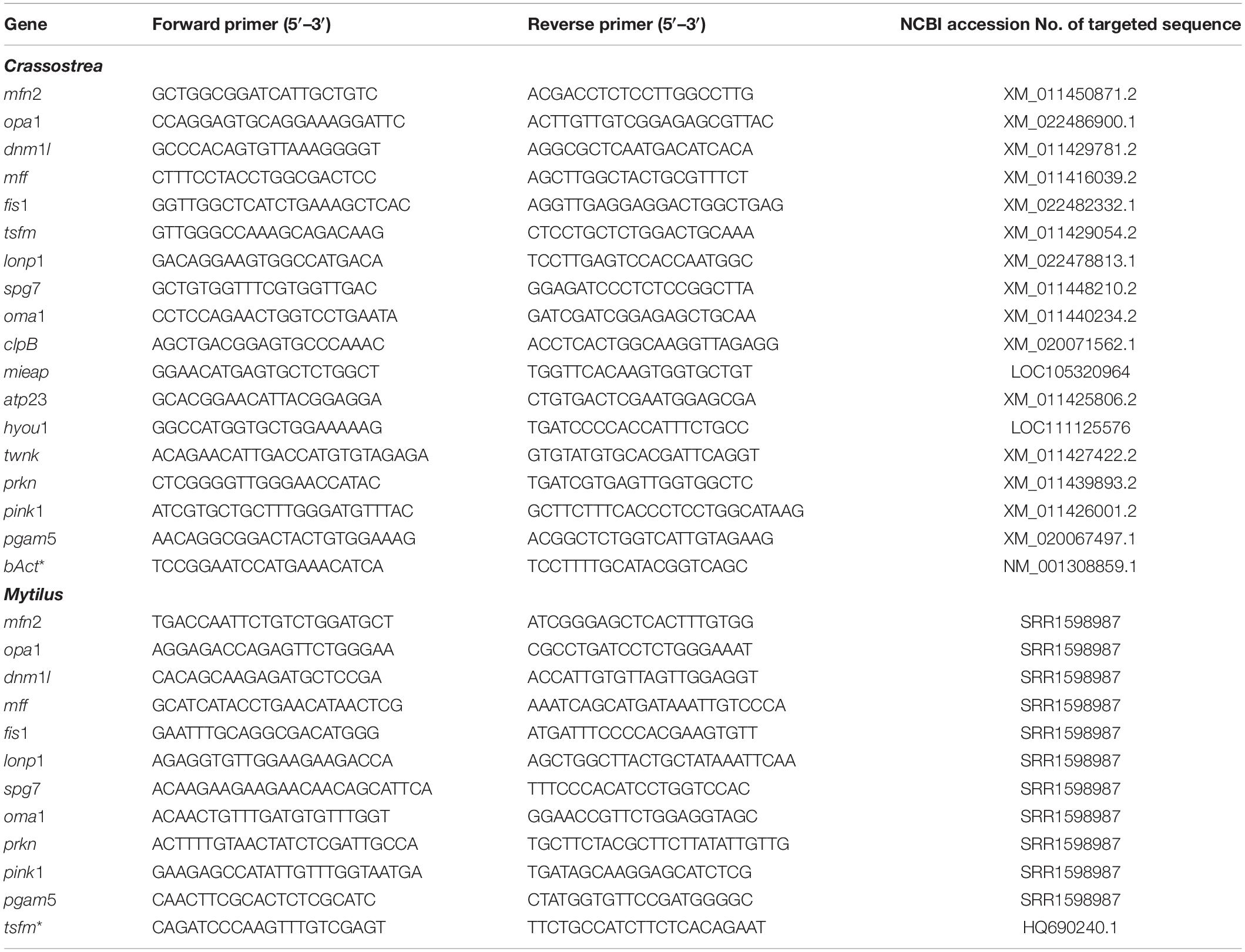
Table 1. Primers used for qRT-PCR for genes of mitochondrial quality control in C. gigas and M. edulis.
To measure mRNA levels of the target and housekeeping genes, the following cycling parameters were used: 2 min at 95°C for polymerase activation, 40 cycles of 15 s at 95°C, 15 s at 57°C (annealing), 20 s at 72°C (extension) and 30 s at a primer specific reading temperature (for acquiring the product signal). For quality control, the melt curve analysis was conducted at the end of each run. To determine the apparent amplification efficiency of the primers and to correct for the possible plate-to-plate variation, a standard dilution series of a single pooled cDNA sample was run on each plate. All measurements were conducted in duplicates. Expression levels of the target samples relative to the reference genes were conducted using the relative standard curve method (Dorak, 2006). Reference genes were chosen as described elsewhere (Falfushynska et al., 2020a). In pilot experiments, we tested five potential housekeeping genes (encoding eukaryotic elongation factor 1, α-tubulin, β-actin, ubiquitin and 18S rRNA) as potential reference genes. Only one out of five tested genes (eukaryotic elongation factor 1 in mussels and actin in oysters) showed no significant change in expression (p > 0.05) with ≤1 difference in mean CT values between different experimental treatments. Therefore, target mRNA expression was normalized to the expression of a reference gene (β-actin βAct for C. gigas and the eukaryotic elongation factor 1 eEF1 tsfm for M. edulis) and the mean CT of the normoxic group as described elsewhere (Dorak, 2006)
Statistics
Data were tested for normal distribution in IBM® SPSS® Statistics v. 25 (IBM Corp., Armonk, NY, United States) using the Shapiro-Wilk test and for homogeneity of variances by the Brown-Forsythe test in SigmaPlot 13 (v. 13.0.0.83, Systat Software Inc., San Jose, CA, United States). Outliers were removed using box-whisker plots in IBM® SPSS® Statistics. Non-normally distributed data were transformed via Box-Cox transformation by Minitab 19 (Minitab LLC., State College, PA, United States). Significant differences between exposures were tested by a one-way ANOVA within each species using SigmaPlot 13. Oxygen regime was used as a fixed factor with three levels (normoxic control, 1 day hypoxia, reoxygenation). Significant differences between the pairs of means were determined using a Tukey’s Honest Significant Differences (HSD) post hoc test. Pearson correlation analysis was carried out by RStudio’s packages “mixOmics” and “factoextra” [v. 1.2.5033 and R v. 3.6.3 (R Core Team, 2020)] with cut off R = 0.6. To reduce the dimensionality of the data, the principal component analysis (PCA) was conducted on transformed data with outliers replaced by group means. PCA and graphs were designed in RStudio. All effects were considered significant at p < 0.05.
Results
Mitochondrial Dynamics: Fusion and Fission
In M. edulis, severe hypoxia suppressed mRNA levels of mfn2 encoding mitofusin-2, which were rapidly restored during reoxygenation (Figure 1A). Transcript levels of opa1 encoding dynamin-like 120 kDa protein did not change during H/R exposure in the mussels (Figure 1C). In C. gigas, tissue levels of mfn2 and opa1 mRNA did not change in response to H/R stress (Figures 1B,D).
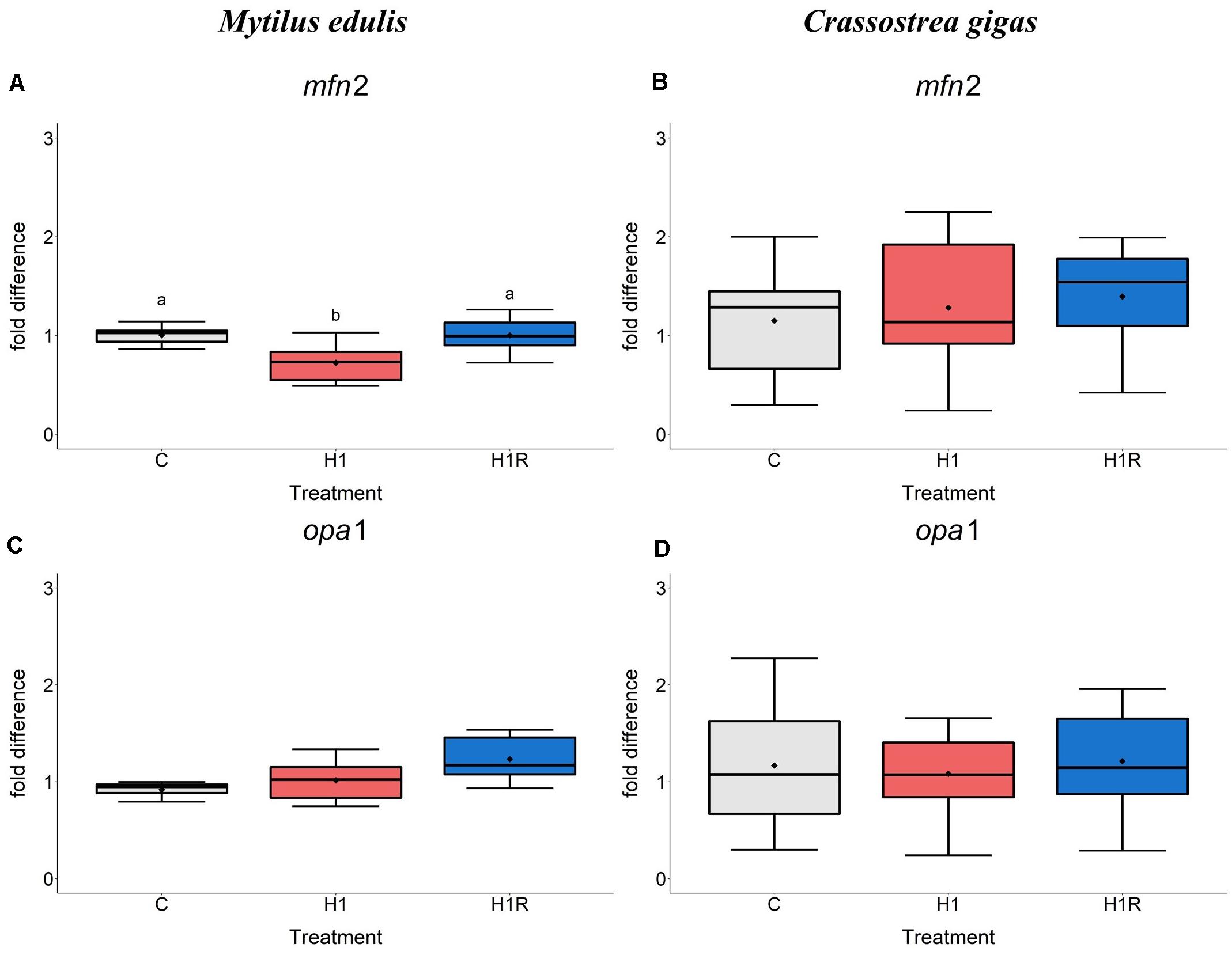
Figure 1. mRNA expression of mitochondrial fusion proteins in M. edulis and C. gigas hepatopancreas in response to H/R stress. (A,C) M. edulis; (B,D) C. gigas; (A,B) mfn1 mRNA; (C,D) opa1 mRNA. Experimental groups: C, control; H1, short-term (1 day) severe hypoxia; H1R, short-term (1 day) severe hypoxia and subsequent 1.5 h reoxygenation. Means are depicted as points within corresponding Box-Whisker-plot. Outliers are shown as asterisk (*). Different letters above plots represent values that are statistical significantly different from each other (p < 0.05). Plots sharing a letter are not significantly different (p > 0.05). N = 6–10.
Transcript levels of mitochondrial fission mediators dnm1l, mff and fis1 were significantly higher in the hepatopancreas of the mussels exposed to hypoxia than the control group (Figures 2A,C,F). Reoxygenation led to a further significant increase in the mRNA expression levels of these genes. Levels of mff mRNA were ∼4-fold and 6-fold elevated in hypoxia and reoxygenation, respectively, compared to control. Expression of fis1 mRNA was ∼2.5- to 3-fold increased in hypoxia and reoxygenation compared to normoxia. The dnm1l gene was ∼1.5- to 2-fold elevated by hypoxia and reoxygenation in the mussels compared to normoxic control. In C. gigas, transcript levels of the mitochondrial fission factors dnm1l, mff, tsfm and fis1 were not significantly affected by hypoxia and reoxygenation (Figures 2B,D,E,G).
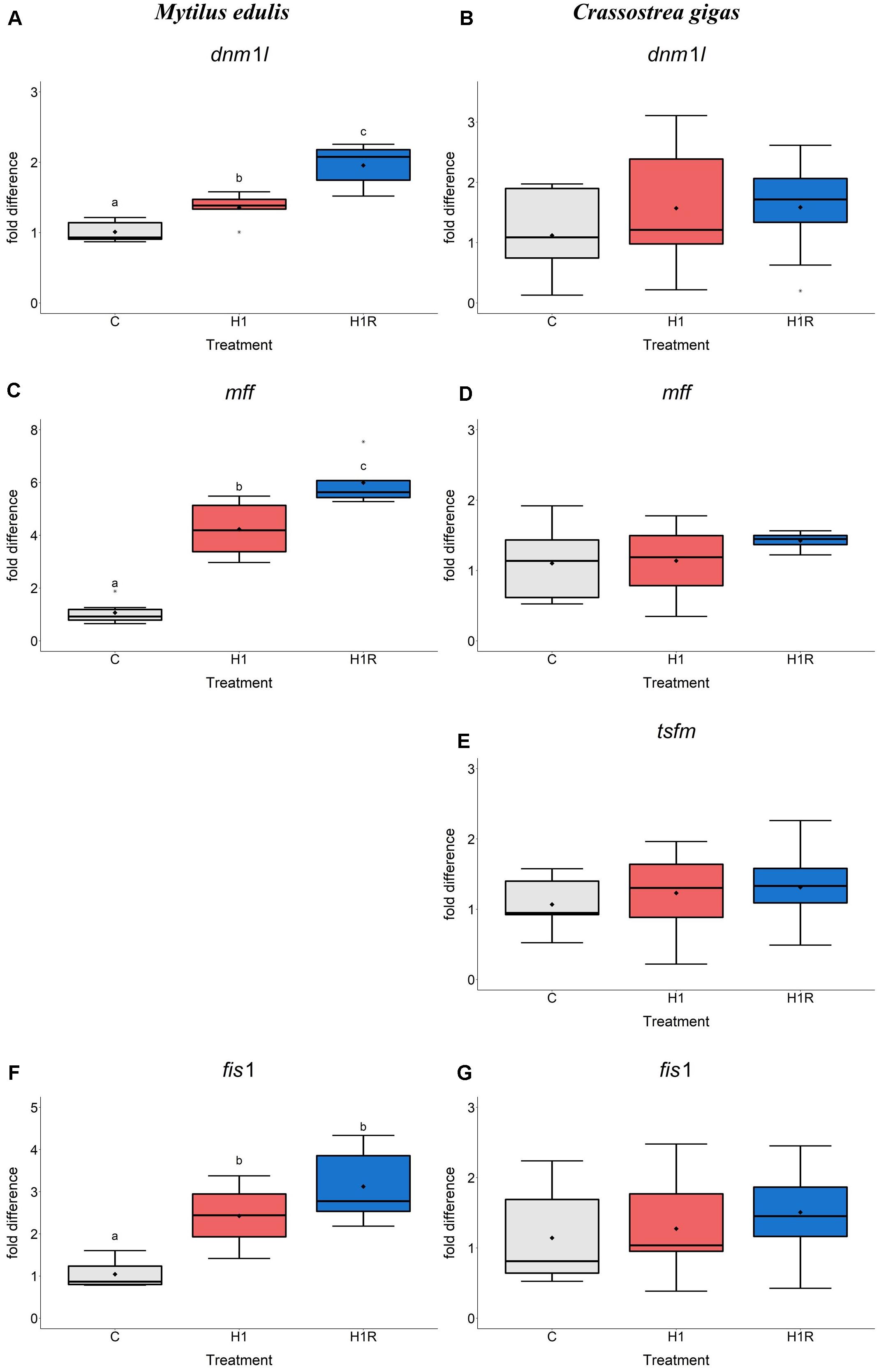
Figure 2. mRNA expression of mitochondrial fission proteins in M. edulis and C. gigas hepatopancreas in response to H/R stress. (A,C,F) M. edulis; (B,D,E,G) C. gigas; (A,B) dnm1l mRNA; (C,D) mff mRNA; (E) tsfm mRNA; (F,G) fis1 mRNA. Experimental groups: C, control; H1, short-term (1 day) severe hypoxia; H1R, short-term (1 day) severe hypoxia and subsequent 1.5 h reoxygenation. Means are depicted as points within corresponding Box-Whisker-plot. Outliers are shown as asterisk (*). Different letters above plots represent values that are statistical significantly different from each other (p < 0.05). Plots sharing a letter are not significantly different (p > 0.05). N = 6–10.
Mitochondrial Protein and DNA Quality Control
Transcript levels of the genes encoding mitochondrial proteases involved in the protein quality control including lonp1 and spg7 were not affected by hypoxia and reoxygenation in M. edulis (Figures 3A,C). In contrast, transcript levels of metalloendopeptidase OMA1 (oma1) increased during hypoxia and reoxygenation by ∼1.5-fold and ∼1.7-fold, respectively, compared to the normoxic controls (Figure 3E).
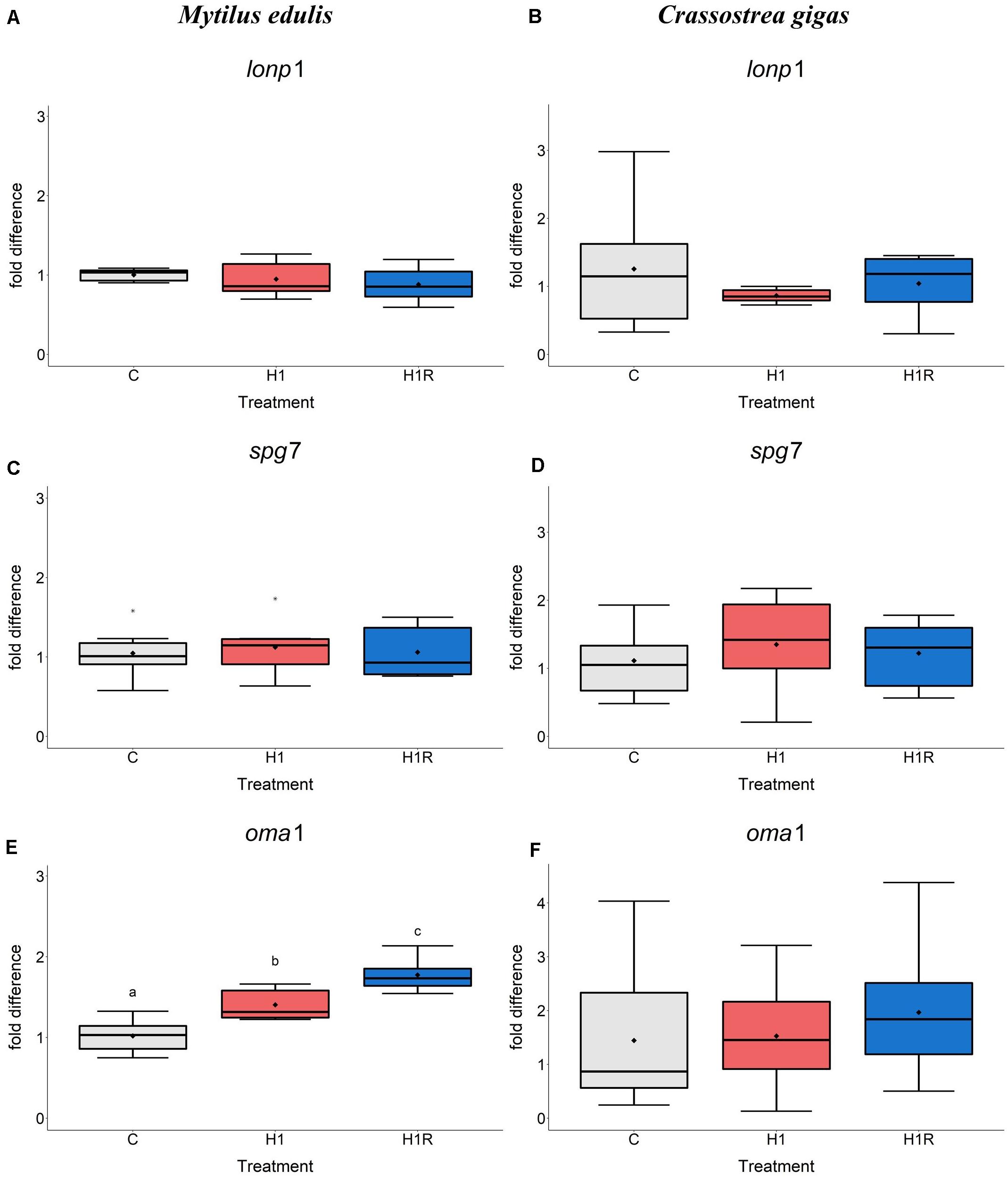
Figure 3. mRNA expression of mitochondrial protein quality control and proteolysis proteins in M. edulis and C. gigas hepatopancreas in response to H/R stress. (A,C,E) M. edulis; (B,D,F) C. gigas; (A,B) lonp1 mRNA; (C,D) spg7 mRNA, (E,F) oma1 mRNA. Experimental groups: C, control; H1, short-term (1 day) severe hypoxia; H1R, short-term (1 day) severe hypoxia and subsequent 1.5 h reoxygenation. Means are depicted as points within corresponding Box-Whisker-plot. Outliers are shown as asterisk (*). Different letters above plots represent values that are statistical significantly different from each other (p < 0.05). Plots sharing a letter are not significantly different (p > 0.05). N = 6–10.
In C. gigas, the mRNA level of the genes involved in mitochondrial protein quality control including lonp1, spg7, oma1 (Figures 3B,D,F) as well as four additional genes (not measured in M. edulis due to the lack of specific primers) including clpB, mieap, atp23 and hyou1 did not change in response to H/R stress (Figures 4A–D). Transcript levels of the mitochondrial helicase TWINKLE (twnk) responsible for mtDNA maintenance, remained unchanged in C. gigas throughout experimental exposures (Figure 4E) (no data are available for M. edulis due to the lack of specific primers).
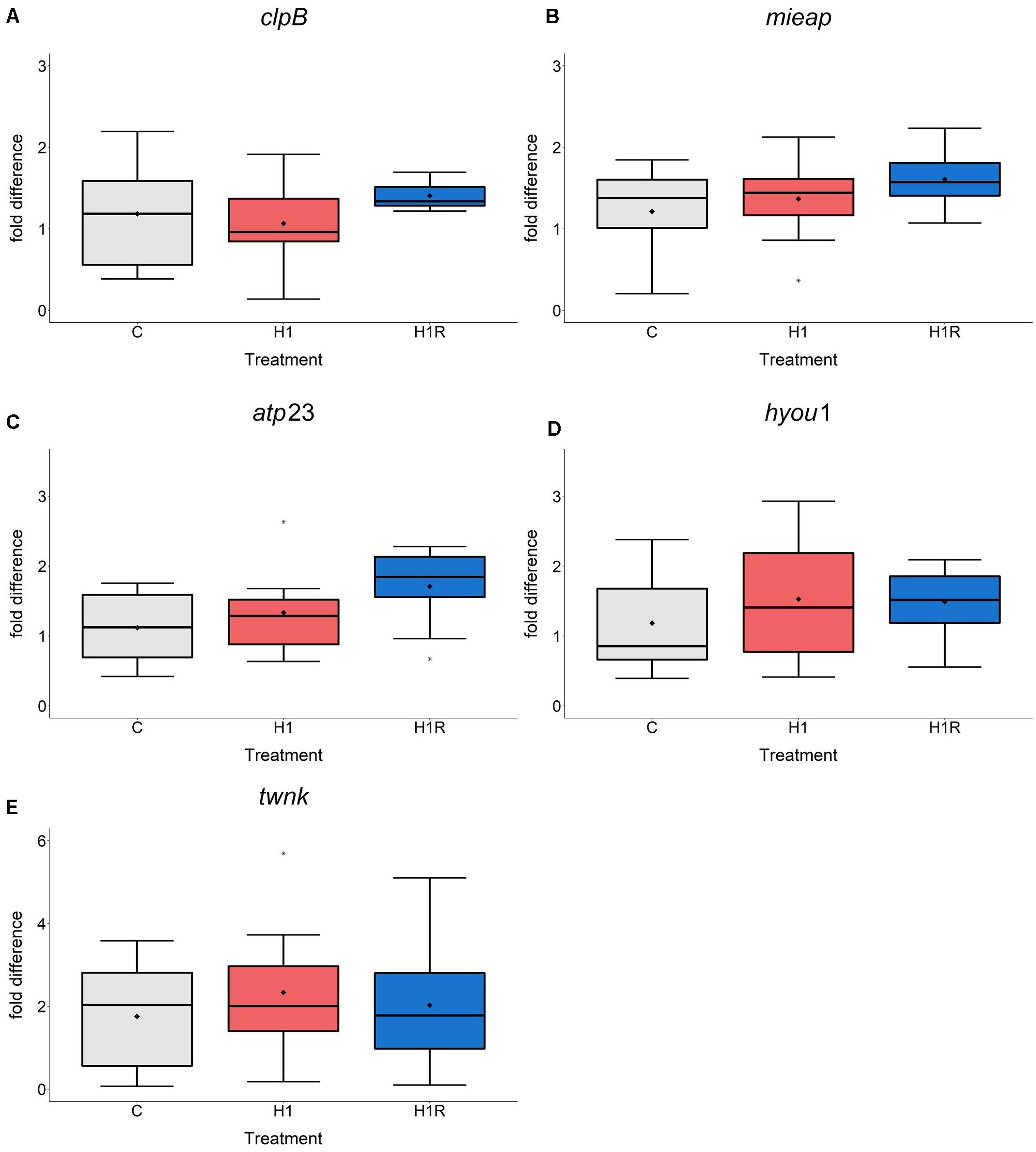
Figure 4. mRNA expression of mitochondrial quality control proteolysis and mtDNA maintenance in C. gigas hepatopancreas in response to H/R stress. (A–D) C. gigas; (A) clpB mRNA; (B) mieap mRNA; (C) atp23 mRNA; (D) hyou1 mRNA; (E) twnk mRNA. Experimental groups: C, control; H1, short-term (1 day) severe hypoxia; H1R, short-term (1 day) severe hypoxia and subsequent 1.5 h reoxygenation. Means are depicted as points within corresponding Box-Whisker-plot. Outliers are shown as asterisk (*). Different letters above plots represent values that are statistical significantly different from each other (p < 0.05). Plots sharing a letter are not significantly different (p > 0.05). N = 6–10.
Mitophagy
In M. edulis, hypoxia induced a ∼2-fold higher expression of prkn mRNA (encoding a mitophagy mediator Parkin) compared to normoxic condition (Figure 5A). The levels of prkn remained elevated during reoxygenation in the mussels (Figure 5A). Expression of its co-mediator pink1 did not change with hypoxia, while reoxygenation led to a ∼2.5-fold increase of pink1 expression compared to normoxic controls (Figure 5C). Transcript levels of the protein phosphatase PGAM5 showed a decreasing trend with hypoxia, followed by a significant increase during reoxygenation (Figure 5E). In C. gigas, the expression pattern of the studied mitophagy genes did not show pronounced effects of hypoxia and subsequent reoxygenation (Figures 5B,D,F).
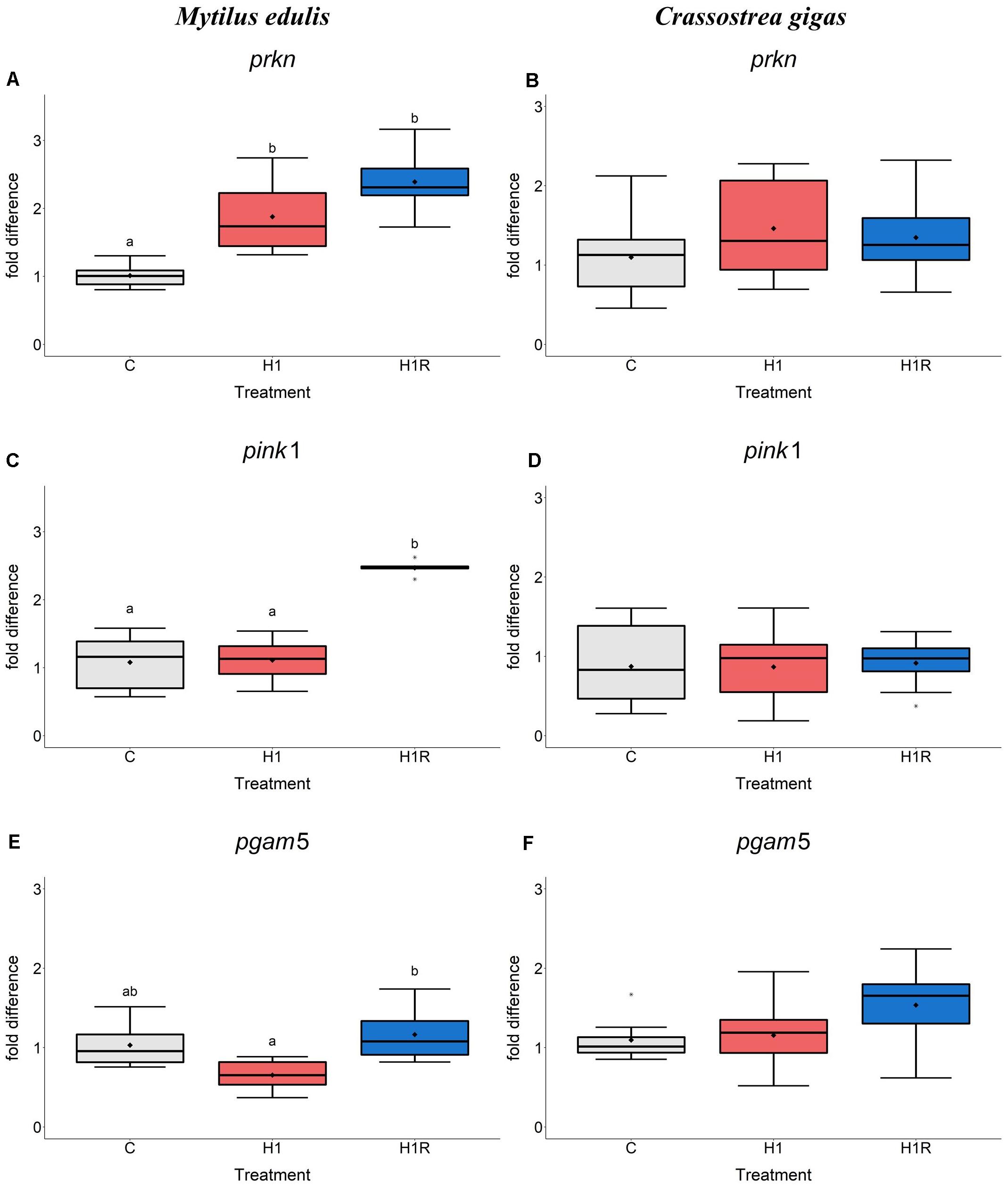
Figure 5. mRNA expression of mitophagy proteins in M. edulis and C. gigas hepatopancreas in response to H/R stress. (A,C,E) M. edulis; (B,D,F) C. gigas; (A,B) pink1 mRNA; (C,D) prkn mRNA, (E,F) pgam5 mRNA. Experimental groups: C, control; H1, short-term (1 day) severe hypoxia; H1R, short-term (1 day) severe hypoxia and subsequent 1.5 h reoxygenation. Means are depicted as points within corresponding Box-Whisker-plot. Outliers are shown as asterisk (*). Different letters above plots represent values that are statistical significantly different from each other (p < 0.05). Plots sharing a letter are not significantly different (p > 0.05). N = 6–10.
Data Integration
The principal component analysis in M. edulis hepatopancreas revealed two principal components explaining 47.2% (PC1) and 19.1% (PC2) of the data variation (Figure 6C and Supplementary Table S3). The 1st component showed high positive loadings for genes of mitochondrial fission (dnm1l, mff, fis1) and fusion (opa1) and mitophagy (prkn, pink1) (Figure 6D and Supplementary Table S4). The 2nd principal component had high positive loadings of genes of mitochondrial quality control (spg7, lonp1) and a strong negative loading of mfn2, mediator of mitochondrial fusion.
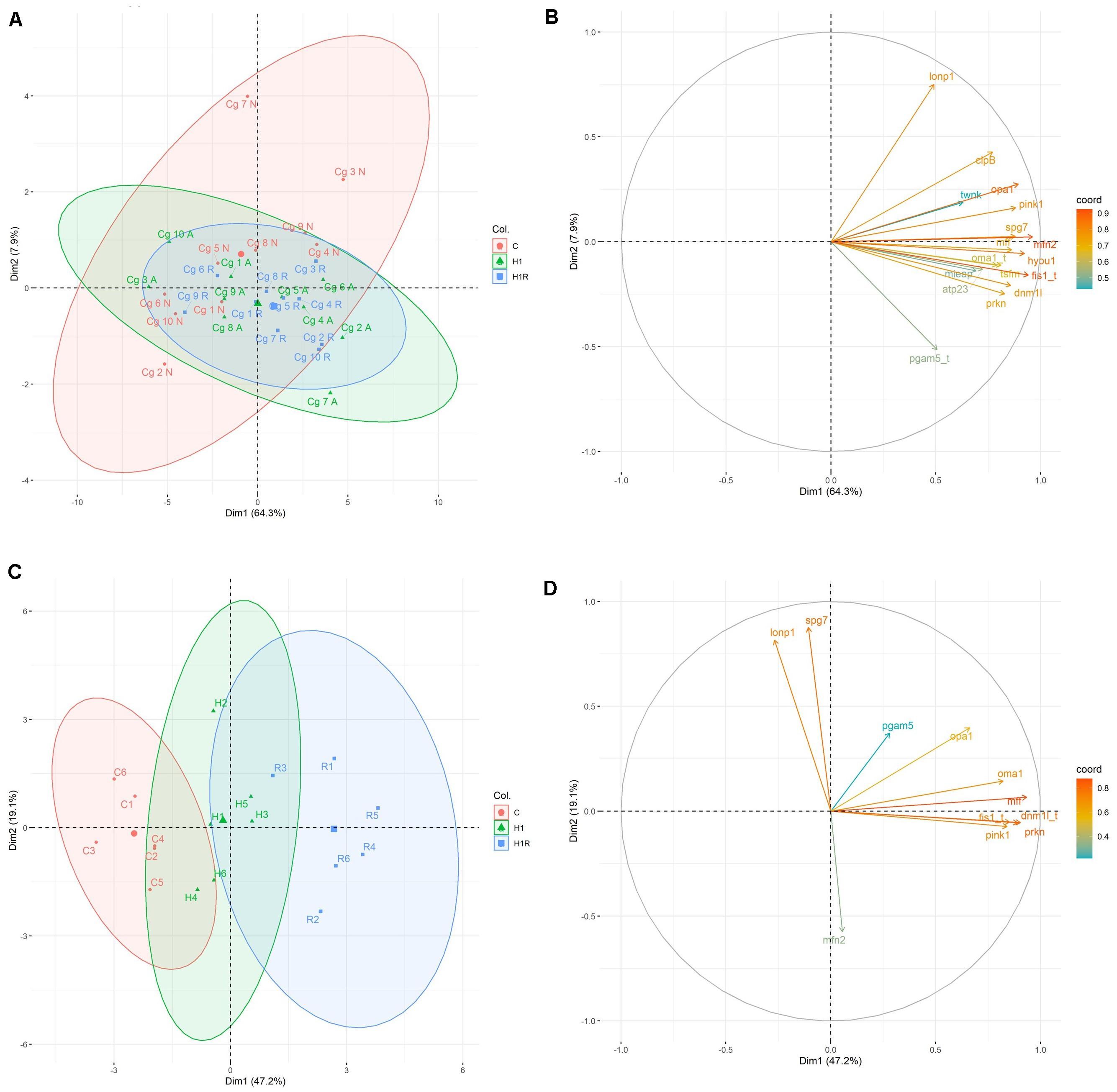
Figure 6. PCA score plots and variable loading plots based on the studied biomarkers from M. edulis and C. gigas during H/R stress. (A,B) C. gigas; (C,D) M. edulis; (A,C) PCA score plot, position of the individual measurements from different experimental group within a coordinate system of first and second principal component. Horizontal axis depicts PC1 and vertical axis PC2. Colored ellipse includes all samples within 95% confidence interval. (B,D) Variable loading plot, visualization of weight distribution of different biomarkers on first and second principal component based on correlations between variables and dimensions. Close vectors define positive correlation between corresponding biomarkers. Coloring of vectors visualize coordinates. Horizontal axis depicts PC1 and vertical axis PC2. Gene name with appended “t” depicts transformed data to ensure normal distribution. Experimental groups: C, control, red; H1, short-term (1 day) severe hypoxia, green; H1R, short-term (1 day) severe hypoxia and subsequent 1.5 h reoxygenation, blue. Gene abbreviations: mfn2, mitofusin 2; opa1, mitochondrial dynamin-like 120 kDa protein; dnm1l, dynamin-1-like protein; mff, mitochondrial fission factor; fis1, mitochondrial fission protein; tsfm, mitochondrial translation elongation factor Ts; lonp1, mitochondrial Lon protease; spg7, paraplegin; oma1, mitochondrial metalloendopeptidase OMA1; clpB, mitochondrial caseinolytic matrix peptidase chaperone subunit B protein homolog; mieap, mitochondrial eating protein; atp23, mitochondrial inner membrane protease ATP23; hyou1, hypoxia upregulated protein 1; twnk, mitochondrial twinkle mtDNA helicase; prkn, E3 ubiquitin-protein ligase parkin; pink1, PTEN-induced kinase 1; pgam5, mitochondrial serine/threonine protein phosphatase PGAM5; βAct, beta Actin
In M. edulis, control and reoxygenation group were clearly separated along the 1st principal component (Figure 6C). Hypoxia and to greater degree reoxygenation induced a shift toward more positive values of the 1st principal component compared to the normoxic control reflecting upregulation of fission genes mff, fis1 and dnm1l and a gene encoding a mitochondrial protease oma1. All experimental treatment groups showed a high variance along the 2nd principal component axis.
Pearson correlation on mRNA expression of M. edulis showed a tight connection between the fission mediators fis1, mff and dnm1l, fusion mediator opa1, mitophagic inducing genes prkn and pink1 and mitochondrial dynamics regulator oma1 (Figures 7C,D).
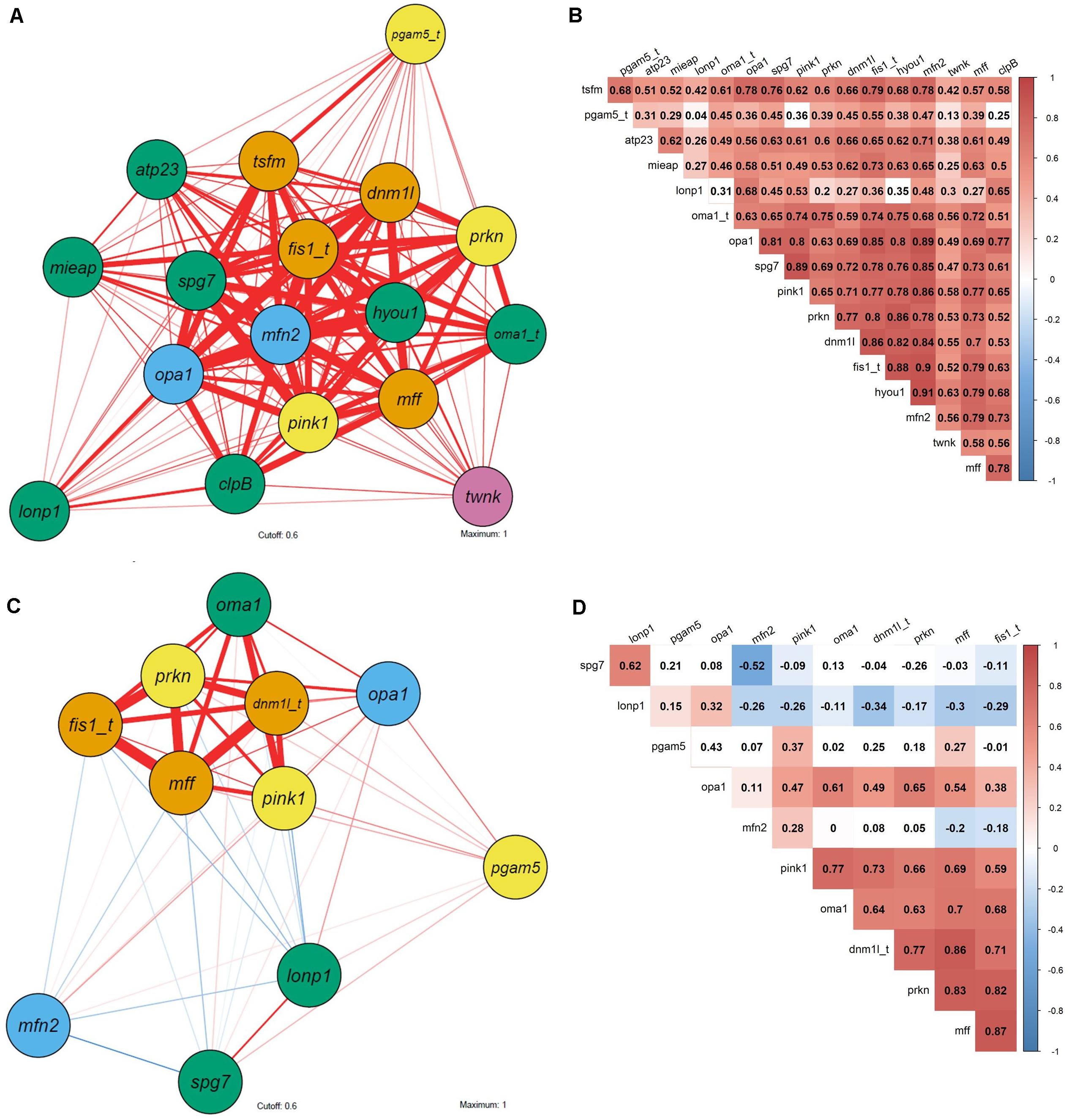
Figure 7. Correlation network and corresponding heatmap of biomarker expression in C. gigas and M. edulis in response to H/R stress. (A,B) C. gigas; (C,D) M. edulis; (A,C) correlation network; (B,D) heatmap. Functional gene groups: orange, mitochondrial fission; light blue, mitochondrial fusion; green, mitochondrial quality control and proteolysis; yellow, mitophagy; pink, mtDNA maintenance. Pearson correlation coefficient: red, positive correlation; blue, negative correlation. Correlation networks are based on Pearson’s correlation with a cutoff of 0.6 and maximum of 1. Non-colored fields of heatmaps and thin shaped correlation network represent non-statistical significant correlations. Thickness of lines represent statistically significant correlations (p < 0.05). Underlying Pearson correlation coefficient are depicted in associated heatmap. Gene name with appended “t” depicts transformed data to ensure normal distribution.
In C. gigas, the 1st and 2nd principal component explained 64.3% and 7.9% of variance, respectively (Figure 6A and Supplementary Table S5). All genes analyzed in this study had high positive loadings on the 1st principal component (>0.05) (Figure 6B and Supplementary Table S6).
Gene transcription of C. gigas from all three experimental treatments showed considerable overlap in the plane of the two first principal components consistent with the lack of change in the mRNA expression profiles of the studied genes in response to the H/R stress. In C. gigas, no apparent correlated gene clusters were found with the tight correlation across all studied genes (Figures 7A,B). These data indicate that the observed variability and correlations in the mRNA expression profiles of C. gigas mostly reflects individual differences between bivalves rather than the effects of the oxygen regime.
Discussion
Short-term severe hypoxia and subsequent reoxygenation had different effects on mRNA expression of mitochondrial quality control in C. gigas and M. edulis (Table 2). In C. gigas, transcript levels were not affected by H/R stress indicating that the mitochondrial injury thresholds needed to induce the protective quality control mechanisms might not have been reached in this hypoxia-tolerant species. In M. edulis, mRNA expression of the transcripts related to mitochondrial dynamics and quality control was strongly regulated during H/R stress showing a shift toward the predominance of mitochondrial fission over fusion and an onset of mitophagy.
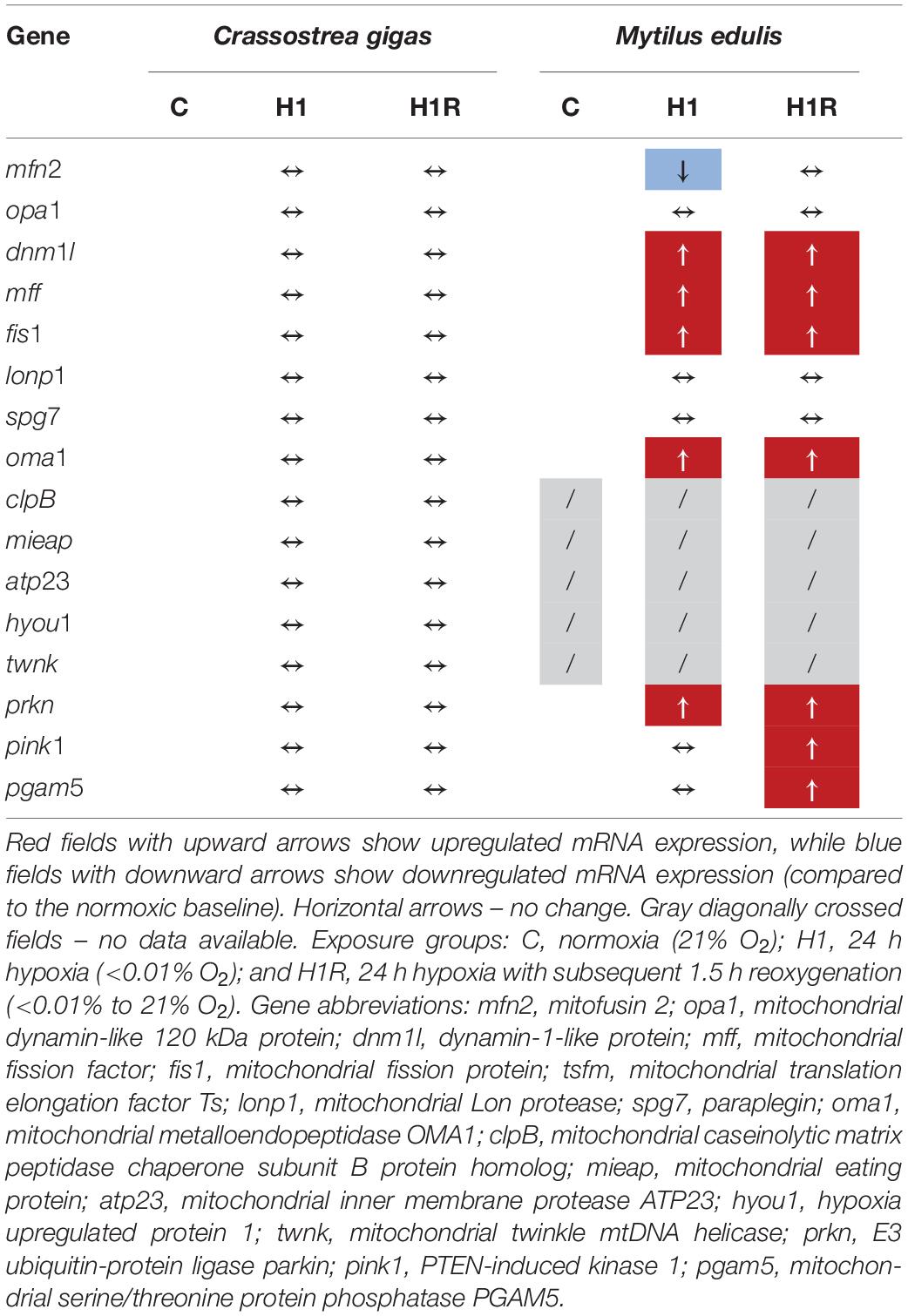
Table 2. Summary of the observed changes in mRNA expression patterns of the mitochondrial quality control genes in Crassostrea gigas and Mytilus edulis.
Effects of H/R Stress on Transcriptional Regulation of Mitochondrial Quality Control in the Mussels
Mitochondrial dynamics (in particular fission and fusion) plays a critical role in maintaining functional mitochondria during stress exposures in animals (Youle and van der Bliek, 2012). In mammalian models, ischemia-reperfusion (I/R) stress regulates transcription and post-translational modifications of fusion and fission proteins reflecting the important role of the mitochondrial dynamics in adjustments to interrupted oxygen supply (Li and Liu, 2018). Cellular stress commonly shifts mitochondrial dynamics toward mitochondrial fission to allow degradation of damaged mitochondria and avoid re-joining of non-functional mitochondria into the mitochondrial network (Chen et al., 2016; Eisner et al., 2018; Sokolova et al., 2019). The GTPase Drp1/Dnm1l mediates mitochondrial fission in response to cellular stress signals such as increase of Ca2 + concentration, ROS production or depolarized mitochondrial membrane as shown during H/R (I/R) stress in mammalian models (Yu et al., 2016; Tian et al., 2017; Yang et al., 2017; Li and Liu, 2018). Drp1/Dnm1l binds to the OMM with the help of the adaptor proteins Mff and Fis1 and initializes division of the membrane (Mears et al., 2011; van der Bliek et al., 2013; Osellame et al., 2016). Drp1/Dnm1l is a transcriptional target of p53 and, in rats, it is regulated at the transcriptional level (Li et al., 2010). The finding of transcriptional upregulation of dnm1l and its two adapter proteins mff and fis1 during H/R stress in M. edulis is consistent with upregulation of the fission pathways possibly as a mechanism to protect mitochondrial integrity by segregation of dysfunctional mitochondria (Twig et al., 2008; Anzell et al., 2018). mRNA expression of the fission-related proteins remained elevated during reoxygenation indicating that 1.5 h reoxygenation might not be sufficient for the full recovery of the mitochondrial fusion-fission balance in M. edulis.
Fusion is an effective strategy to protect against the stress-induced DNA damage, as the damaged mitochondria fuse with the healthy ones and replace the damaged mitochondrial DNA through complementation (Youle and van der Bliek, 2012). In our present study, mRNA levels of a key fusion mediator mfn2 (encoding the mitochondrial Mfn 2 protein) were suppressed during hypoxia in M. edulis indicating downregulation of the mitochondrial fusion pathways. This lack of induction of mfn2 might indicate that mitochondrial DNA damage is not critically involved in the H/R stress responses of the blue mussels. An earlier study in the eastern oysters C. virginica showed an increase in the levels of DNA damage during exposure to 6 days of hypoxia and subsequent reoxygenation in the oysters exposed to a toxic metal cadmium, but not in their counterparts exposed to H/R stress only (Kurochkin et al., 2009). In the mussels M. edulis, hypoxia caused by emersion (72 h) had no effect on the DNA damage assessed by the Comet assay (Singh and Hartl, 2012). In the brown mussels Perna perna, 24 h of emersion-induced hypoxia led to the elevated oxidative DNA damage in the gills but not in the digestive gland (Almeida et al., 2005). In these studies, the oxidative lesions were assessed in the total rather than the mitochondrial DNA. To the best of our knowledge, the effects of H/R stress on the mitochondrial DNA damage have not been studied in marine bivalves. It is worth noting that mitofusin 2 requires energy in the form of GTP to mediate the mitochondrial fusion (Filadi et al., 2018), so that the downregulation of mfn2 in hypoxia-exposed mussels might be a consequence of the metabolic rate depression, a common energy-saving strategy to survive the oxygen deficiency in hypoxia-tolerant mollusks (Hochachka, 1993; Storey, 1998; Storey and Storey, 2004). During reoxygenation mRNA expression of mfn2 returned to the normoxic (baseline) levels in the mussels, which indicates a rapid re-balancing of mitochondrial fusion during post-hypoxic recovery.
The transcript levels of opa1 (encoding the mitochondrial dynamin-like GTPase OPA1) were not affected by H/R stress in M. edulis. The mitochondrial OPA1 is involved in the regulation of the mitochondrial fusion and the cross-talk between fusion and fission (Cipolat et al., 2004). In mammals, OPA1 activity is regulated transcriptionally (Ryu et al., 2015; Zheng et al., 2019) as well as post-translationally through proteolytic cleavage by the mitochondrial metalloprotease OMA1 (Chen et al., 2016). Mitochondrial stress such as a decrease in the mitochondrial membrane potential or ATP deficiency stimulates the proteolytic processing of OPA1 generating short isoforms unable to support mitochondrial fusion (Anand et al., 2014; Chen et al., 2016; Ali and McStay, 2018). The posttranslational regulation of OPA1 allows for rapid suppression of mitochondrial fusion and mitochondrial fragmentation but might not be reflected in the changes in mRNA expression (Ali and McStay, 2018). Nevertheless, opa1 was observed to be transcriptionally regulated during formation of immune cells in mice (Ryu et al., 2015), while disorders related to impaired mitochondrial functions were associated with transcriptional regulation of opa1 (Zheng et al., 2019). The observed stability of opa1 mRNA in the mussels might indicate the lack of involvement of OPA1 in the mitochondrial response to the H/R stress in M. edulis. Alternatively, if post-translational regulation plays a predominant role in the OPA1 activation in the mussels, activation of OPA1 might not be reflected at the transcript level. Further studies are needed to distinguish between these alternative explanations.
In M. edulis, H/R stress led to a strong upregulation of oma1 mRNA encoding the mitochondrial metalloprotease OMA1. As discussed above, OMA1 mediates the proteolytic cleavage of fusion mediator OPA1 during stress thereby suppressing mitochondrial fusion (Anand et al., 2014). OMA1-dependent degradation of OPA1 is thus essential for the selective removal of defective mitochondrial fragments and prevention of incorporation of those into the mitochondrial network (Head et al., 2009; Baker et al., 2014). Consequently, elevated oma1 expression during hypoxia and subsequent reoxygenation in the mussels might reflect the overall shift in the mitochondrial fusion-fission dynamics toward the predominance of fission (Ali and McStay, 2018) and is consistent with the transcriptional upregulation of the fission regulating proteins dnm1l, mff and fis1 and suppressed expression of a fusion regulator mfn2 in M. edulis during the H/R stress. Furthermore, OMA1 mediates an ATP-independent proteolytic breakdown of misfolded inner membrane proteins and is complementary to the action of m-AAA proteases (Käser et al., 2003). OMA1 activity is strongly upregulated in response to various stresses in metazoans, plants and yeast (Bohovych et al., 2014; Rainbolt et al., 2015; Migdal et al., 2017) and is required for stabilization of the mitochondrial respiratory supercomplexes and maintenance of the mitochondrial respiration (Bohovych et al., 2015; Richter et al., 2015). Therefore, transcriptional upregulation of oma1 during H/R stress in the mussels might assist in the degradation of the damaged mitochondrial proteins and protection of the mitochondrial integrity and function.
Our study shows that unlike oma1, two other studied mitochondrial proteases (including the mitochondrial Lon protease and paraplegin, encoded by lonp1 and spg7, respectively) are not transcriptionally upregulated during H/R stress in M. edulis. Lon protease is highly conserved throughout organisms and essential for mitochondrial proteostasis (Pinti et al., 2015). It is activated by ROS, protein carbonylation and lipid peroxidation (Kuo et al., 2015) and specifically targets the oxidatively damaged proteins for ATP dependent degradation (Bota and Davies, 2002; Bayot et al., 2010). Lon protease activity is regulated transcriptionally and post-translationally suggesting a fine tuning by several mechanisms, although stronger regulatory patterns are commonly seen at the protein level (Pinti et al., 2015). Earlier studies in bivalves showed that Lon protease mRNA expression was upregulated during H/R stress in the hypoxia-sensitive scallops Argopecten irradians but not in the hypoxia-tolerant hard shell clams Mercenaria mercenaria (Ivanina et al., 2016). Upregulation of lonp1 transcripts in the scallops was associated with accumulation of oxidative lesions (i.e., lipid and protein peroxidation products) in the mitochondria, whereas in the hard shell clams the mitochondrial oxidative stress markers remained at the baseline (normoxic) levels (Ivanina et al., 2016). Upregulation of lonp1 mRNA expression was also found in the Eastern oyster C. virginica during simultaneous exposure to high temperature and a toxic metal cadmium (Sanni et al., 2008), a combination of stressors known to cause strong oxidative stress in oysters (Bagwe et al., 2015). Taken together, these findings indicate that transcriptional upregulation of lonp1 in bivalves might occur when the oxidative stress reaches a species-specific threshold in the mitochondria, and the lack of transcriptional upregulation of lonp1 mRNA levels during H/R stress in M. edulis might indicate that this threshold has not been reached under the conditions of our present study. Similar to lonp1, transcript levels of spg7 gene encoding a mitochondrial metalloprotease paraplegin did not change during H/R stress in the mussels. To the best of our knowledge, expression of spg7 has not been studied in other marine bivalves. In model organisms including mammals and yeast, paraplegin is regulated by mitochondrial stress signals such as Ca2+ accumulation and elevated ROS levels (Ishihara et al., 2006; Shanmughapriya et al., 2015). If similar ROS-dependent mechanisms exist in bivalves, the lack of spg7 (as well as lonp1) transcriptional responses to H/R stress might be explained by the relatively low levels of oxidative stress insufficient to trigger transcriptional upregulation of these proteases. Further studies of the ROS production, oxidative injury and expression of mitochondrial proteases in bivalve mitochondria during exposures to different stressors are required to test this hypothesis and shed light on the thresholds of mitochondrial injury required for induction of these protective mechanisms.
Mitophagy is a selective form of autophagy to degrade damaged mitochondria (Pickles et al., 2018). Based on the studies in the model organisms, the PINK1-Parkin pathways plays a key role in the stress-induced mitophagy (Jin et al., 2010). In healthy mitochondria, the constitutively expressed PINK1 is translocated to mitochondria in a membrane-potential-dependent manner, cleaved by proteases and released to cytosol for further degradation. Membrane depolarization in dysfunctional mitochondria causes stabilization and dimerization of PINK1 on mitochondrial surface (Ding and Yin, 2012). Bound PINK1 recruits and phosphorylates parkin (Dagda et al., 2009; Eiyama and Okamoto, 2015), which tags damaged mitochondria for further degradation by mitophagy (Narendra and Youle, 2011). Activity of the protein phosphatase PGAM5 also contributes to the activation of mitophagy (Chen et al., 2014; Wu et al., 2014) and promotes the mitochondrial fission by activating the Drp1/Dnm1l protein (Wang et al., 2012). In our present study, hypoxia exposure induced elevated mRNA expression of prkn in M. edulis, while the transcript levels of the associated key genes pink1 and pgam5 rose during reoxygenation mitochondria. This expression pattern indicates an onset of mitophagy during reoxygenation as the PINK1-parkin pathway is increasingly required to tag dysfunctional mitochondria caused by oxidative stress. Transcriptional upregulation of PGAM5 also indicates activation of the cell death pathways (Wang et al., 2012) during reoxygenation in M. edulis, consistent with the earlier findings of upregulated apoptosis in this species during H/R stress (Falfushynska et al., 2020a).
The correlation network analysis showed strong correlation of mRNA expression of all three fission mediators, the PINK1-parkin pathway regulators and oma1, underpinning the common regulatory mechanism for these pathways. This is also reflected by the clear separation of mRNA expression profiles of the mussels exposed to hypoxia and (especially) reoxygenation from the normoxic control groups along the 1st principal component axis in the PCA associated with the high loadings of the fission-mitophagy-related transcripts. An earlier study showed a similar major shift of the mRNA expression profiles of the pro-apoptotic and pro-inflammatory genes in M. edulis exposed to the short-term H/R stress (Falfushynska et al., 2020a). The metabolic profiling in a related study showed onset of anaerobiosis, downregulation of gluconeogenesis and a shifted balance between protein synthesis and breakdown in M. edulis indicating early onset of severe stress response during H/R exposure (Haider et al., 2020).
Muted Transcriptional Response of the Mitochondrial Dynamics and Quality Control Genes During H/R Stress in C. gigas
Unlike M. edulis, C. gigas did not change its mRNA expression in any of the 17 studied genes in response to 24 h hypoxia and subsequent 1.5 h reoxygenation. Furthermore, the overlap of gene transcription found by PCA and the apparent tight correlation across all studied genes supports a conclusion of the lack of a transcriptional shift in the studied pathways during H/R stress in the Pacific oyster. A similar transcriptional stability was found for lonp1 expression after 6 days of hypoxia and 1–12 h of reoxygenation in the eastern oyster C. virginica (Ivanina et al., 2010). Previously, metabolic and mRNA expression profiling on apoptotic and pro-inflammatory genes in C. gigas revealed no changes in response to short-term hypoxia (Falfushynska et al., 2020a; Haider et al., 2020). Only 6 days exposure to hypoxia led to a small shift in metabolite profiles indicating metabolic rate depression, maintenance of balanced FAA/metabolite pool and effective detoxification by urea and purine cycles. Furthermore, maintenance of steady activity levels of mitoproteases during the H/R stress was earlier reported in C. gigas (Ivanina and Sokolova, 2016). Together with the muted mRNA expression regulation of mitochondrial quality control genes and mitochondrial dynamics, these findings indicate that 24 h of severe hypoxia might not be sufficient to induce strong stress markers in C. gigas. However, short-term hypoxia (24 h, <1% O2) and subsequent reoxygenation affected a reversible phosphorylation pattern of mitochondrial quality control proteins such as PGAM5 in C. gigas (Sokolov et al., 2019). This indicates that despite the lack of a strong transcriptional induction of the key genes in the mitochondrial quality control pathways, the activity of these pathways might be modulated by posttranslational protein modifications (PTM) in C. gigas. This hypothesis requires further investigation as the role of the PTM in the regulation of the mitochondrial function of marine bivalves has not been extensively studied (Sokolov et al., 2019; Falfushynska et al., 2020b). Notably, a muted transcriptional response to short-term hypoxia and subsequent reoxygenation has been earlier shown for other important pathways involved in cellular bioenergetics of the oysters. Thus, hypoxia induced genes (especially respiratory chain genes) were regulated in C. gigas only after 7 days of exposure to 30% O2 saturation, with no apparent changes at the earlier exposures (David et al., 2005). In the eastern oyster C. virginica, key marker genes in the mitochondrial electron chain and Krebs’ cycle, as well as the cytosolic glycolytic genes showed stable expression during 6 days of severe hypoxia (< 1% O2) and subsequent reoxygenation (Ivanina et al., 2010). Like other hypoxia-tolerant mollusks, the Eastern and Pacific oyster rely on metabolic rate depression to cope with oxygen deficiency (Storey and Storey, 2004) and use a reversible global transcription repression to arrest energy consuming transcription (Larade and Storey, 2002). Only specific transcription factors seem to be upregulated to allow transcription of certain hypoxic genes (David et al., 2005). These factors might contribute to the muted transcriptional response during H/R stress in the oysters. Overall, the hypoxia tolerant Pacific oyster seems to maintain its mitochondrial integrity and functions by reversible enzyme regulations sufficient for fast recovery in reoxygenation, which might contribute to the high stress tolerance of this species adapted to the habitat conditions of daily immersion and emersion (Sussarellu et al., 2013). Further investigations in a broader comparative framework (involving multiple species of bivalves with different levels of hypoxia tolerance) are needed to determine whether the hypoxia-tolerant mitochondrial phenotypes generally correlate with the transcriptional stability and stronger dependence on post-translational regulatory mechanisms during H/R stress in marine bivalves.
Conclusion
Our study showed that differential transcriptional regulation of the mitochondrial quality control pathways is involved in the molecular response to H/R stress in the blue mussels M. edulis but not in the oysters C. gigas. In the mussels, the H/R stress induced the pathways related to the mitochondrial fission, mitophagy and ATP-independent mitochondrial proteolysis. Interestingly, the transcripts of the proteins encoding ATP-dependent mitochondrial quality control proteins (such as mitofusin 2, Lon protease and paraplegin) remained stable or downregulated during hypoxia, possibly as a consequence of the metabolic rate depression and an associated decrease in the ATP turnover rates. In C. gigas, no clear transcriptional response of the mitochondrial quality control pathways was found during H/R stress, with all studied transcripts maintained at control (normoxic) levels. This might indicate that unlike M. edulis, other mechanisms (such as post-transcriptional and post-translational regulation of the respective pathways) are involved in the regulation of the mitochondrial integrity in oysters. Alternatively, it is possible that the level of the mitochondrial damage in oysters did not reach the threshold required to trigger the respective transcriptional activation of the mitochondrial quality control pathways. The latter explanation appears plausible, given the relative stability of the metabolomics profile and the transcript levels of other stress genes (such as those involved in apoptosis and inflammation) during H/R stress in oysters compared with the mussels (Falfushynska et al., 2020a; Haider et al., 2020). Our present study and earlier published research (Falfushynska et al., 2020a; Haider et al., 2020) also indicate that the mRNA expression of the stress-related genes may not be a good biomarker of hypoxia-induced stress in an extremely tolerant species such as C. gigas. It is worth noting that the natural habitat conditions differ with regard to salinity (ocean vs. brackish, respectively) and the tidal height (intertidal vs. subtidal, respectively) between the two studied populations of the oysters and the mussels. Further studies are needed to determine whether the established links between the mRNA expression profiles of the stress-related mitochondrial genes and hypoxia tolerance reflect the species-specific differences between the oysters and the mussels, or some other aspects of the local adaptation to the divergent habitats in which these organisms were collected.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Zenodo: doi: 10.5281/zenodo.4266408.
Author Contributions
JS and HF conducted the experimental exposures and tissue collections with C. gigas, and M. edulis, respectively. mRNA extraction, cDNA synthesis and quantitative real-time PCR were done by JS and HF, and statistical analysis and data integration was done by JS. HF and JS designed primers according to transcripts provided by bioinformatical analysis from HP. JS, IS, and HP wrote the first draft of the manuscript, and all author contributed to the data interpretation and revisions of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Deutsche Forschungsgemeinschaft within the project “MitoBOX: the mitochondrial basis of hypoxia tolerance in marine mollusks” (grant number 415984732). HP was supported by the NIH grant R21AG064479-01 and a Brain Health Research Institute Pilot Award from Kent State University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mathias Wegner of the Alfred-Wegener-Institute List/Sylt, Germany, for supply and shipping of oysters. We also thank Eugene Sokolov for training and helpful advice in molecular methods. The metadata of the project have been uploaded to an open repository Zenodo (doi: 10.5281/zenodo.4266408).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.604411/full#supplementary-material
References
Ali, S., and McStay, G. P. (2018). Regulation of mitochondrial dynamics by proteolytic processing and protein turnover. Antioxidants 7, 1–15. doi: 10.3390/antiox7010015
Almeida, E. A., Bainy, A. C. D., Dafre, A. L., Gomes, O. F., Medeiros, M. H. G., and Di Mascio, P. (2005). Oxidative stress in digestive gland and gill of the brown mussel (Perna perna) exposed to air and re-submersed. J. Exp. Mar. Biol. Ecol. 318, 21–30. doi: 10.1016/j.jembe.2004.12.007
Anand, R., Wai, T., Baker, M. J., Kladt, N., Schauss, A. C., Rugarli, E., et al. (2014). The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 204, 919–929. doi: 10.1083/jcb.201308006
Andrienko, T. N., Pasdois, P., Pereira, G. C., Ovens, M. J., and Halestrap, A. P. (2017). The role of succinate and ROS in reperfusion injury - A critical appraisal. J. Mol. Cell Cardiol. 110, 1–14. doi: 10.1016/j.yjmcc.2017.06.016
Anzell, A. R., Maizy, R., Przyklenk, K., and Sanderson, T. H. (2018). Mitochondrial quality control and disease: insights into ischemia-reperfusion injury. Mol. Neurobiol. 55, 2547–2564. doi: 10.1007/s12035-017-0503-9
Atorino, L., Silvestri, L., Koppen, M., Cassina, L., Ballabio, A., Marconi, R., et al. (2003). Loss of m-AAA protease in mitochondria causes complex I deficiency and increased sensitivity to oxidative stress in hereditary spastic paraplegia. J. Cell Biol. 163, 777–787. doi: 10.1083/jcb.200304112
Bagwe, R., Beniash, E., and Sokolova, I. M. (2015). Effects of cadmium exposure on critical temperatures of aerobic metabolism in eastern oysters Crassostrea virginica (Gmelin, 1791). Aquat. Toxicol. 167, 77–89. doi: 10.1016/j.aquatox.2015.07.012
Baker, M. J., Lampe, P. A., Stojanovski, D., Korwitz, A., Anand, R., Tatsuta, T., et al. (2014). Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J. 33, 578–593. doi: 10.1002/embj.201386474
Bayot, A., Gareil, M., Rogowska-Wrzesinska, A., Roepstorff, P., Friguet, B., and Bulteau, A. -L. (2010). Identification of novel oxidized protein substrates and physiological partners of the mitochondrial ATP-dependent Lon-like Protease Pim1. J. Biol. Chem. 285, 11445–11457. doi: 10.1074/jbc.M109.065425
Berlett, B. S., and Stadtman, E. R. (1997). Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 272, 20313–20316. doi: 10.1074/jbc.272.33.20313
Bohovych, I., Donaldson, G., Christianson, S., Zahayko, N., and Khalimonchuk, O. (2014). Stress-triggered activation of the metalloprotease Oma1 involves Its C-terminal region and is important for mitochondrial stress protection in Yeast. J. Biol. Chem. 289, 13259–13272. doi: 10.1074/jbc.M113.542910
Bohovych, I., Fernandez, M. R., Rahn, J. J., Stackley, K. D., Bestman, J. E., Anandhan, A., et al. (2015). Metalloprotease OMA1 fine-tunes mitochondrial bioenergetic function and respiratory supercomplex stability. Sci. Rep. 5:13989. doi: 10.1038/srep13989
Bota, D. A., and Davies, K. J. A. (2002). Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell Biol. 4, 674–680. doi: 10.1038/ncb836
Breitburg, D., Baumann, H., Sokolova, I., and Frieder, C. (2019). “6. Multiple stressors -forces that combine to worsen deoxygenation and its effects,” in Ocean Deoxygenation: Everyone’s Problem. Causes, Impacts, Consequences and Solutions, eds. D. D. A. Laffoley and J. M. Baxter (Gland: IUCN), 225–247.
Breitburg, D., Levin, L. A., Oschlies, A., Grégoire, M., Chavez, F. P., Conley, D. J., et al. (2018). Declining oxygen in the global ocean and coastal waters. Science 359, 1–11. doi: 10.1126/science.aam7240
Cabiscol, E., Piulats, E., Echave, P., Herrero, E., and Ros, J. (2000). Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J. Biol. Chem. 275, 27393–27398. doi: 10.1074/jbc.M003140200
Cadenas, E., and Davies, K. J. A. (2000). Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 29, 222–230.
Chen, G., Han, Z., Feng, D., Chen, Y., Chen, L., Wu, H., et al. (2014). A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell 54, 362–377. doi: 10.1016/j.molcel.2014.02.034
Chen, H., Detmer, S. A., Ewald, A. J., Griffin, E. E., Fraser, S. E., and Chan, D. C. (2003). Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189–200. doi: 10.1083/jcb.200211046
Chen, M., Chen, Z., Wang, Y., Tan, Z., Zhu, C., Li, Y., et al. (2016). Mitophagy receptor FUNDC1 regulates mitochondrial dynamics and mitophagy. Autophagy 12, 689–702. doi: 10.1080/15548627.2016.1151580
Chen, Q., Camara, A. K. S., Stowe, D. F., Hoppel, C. L., and Lesnefsky, E. J. (2007). Modulation of electron transport protects cardiac mitochondria and decreases myocardial injury during ischemia and reperfusion. Am. J. Physiol. Cell Physiol. 292, C137-C147. doi: 10.1152/ajpcell.00270.2006
Cipolat, S., Brito, O. M., de Dal Zilio, B., and Scorrano, L. (2004). OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. U.S.A. 101, 15927–15932. doi: 10.1073/pnas.0407043101
Dagda, R. K., Cherra, S. J. III., Kulich, S. M., Tandon, A., Park, D., and Chu, C. T. (2009). Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 284, 13843–13855. doi: 10.1074/jbc.M808515200
Daguin, C., Bonhomme, F., and Borsa, P. (2001). The zone of sympatry and hybridization of Mytilus edulis and M. galloprovincialis, as described by intron length polymorphism at locus mac-1. Heredity 86, 342–354. doi: 10.1046/j.1365-2540.2001.00832.x
David, E., Tanguy, A., Pichavant, K., and Moraga, D. (2005). Response of the Pacific oyster Crassostrea gigas to hypoxia exposure under experimental conditions. FEBS J. 272, 5635–5652. doi: 10.1111/j.1742-4658.2005.04960.x
Diaz, R. J., and Rosenberg, R. (1995). Marine benthic hypoxia: a review of its ecological effects and the behavioural responses of benthic macrofauna. Oceanogr. Mar. Biol. 33, 245–303.
Diaz, R. J., and Rosenberg, R. (2008). Spreading dead zones and consequences for marine Ecosystems. Science 321, 926–929. doi: 10.1126/science.1156401
Ding, W. -X., and Yin, X. -M. (2012). Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol. Chem. 393, 547–564. doi: 10.1515/hsz-2012-0119
Doyle, S. M., and Wickner, S. (2008). Hsp104 and ClpB: protein disaggregating machines. Trends Biochem. Sci. 34, 40–48. doi: 10.1016/j.tibs.2008.09.010
Eisner, V., Picard, M., and Hajnóczky, G. (2018). Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat. Cell Biol. 20, 755–765. doi: 10.1038/s41556-018-0133-0
Eiyama, A., and Okamoto, K. (2015). PINK1/Parkin-mediated mitophagy in mammalian cells. Curr. Opin. Cell Biol. 33, 95–101. doi: 10.1016/j.ceb.2015.01.002
Falfushynska, H. I., Piontkivska, H., and Sokolova, I. M. (2020a). Effects of intermittent hypoxia on cell survival and inflammatory responses in the intertidal marine bivalves Mytilus edulis and Crassostrea gigas. J. Exp. Biol. 223:jeb217026. doi: 10.1242/jeb.217026
Falfushynska, H. I., Sokolov, E., Piontkivska, H., and Sokolova, I. M. (2020b). The role of reversible protein phosphorylation in regulation of the mitochondrial electron transport system during hypoxia and reoxygenation stress in marine bivalves. Front. Mar. Sci. 7:467. doi: 10.3389/fmars.2020.00467
Filadi, R., Pendin, D., and Pizzo, P. (2018). Mitofusin 2: from functions to disease. Cell Death Dis. 9, 1–13. doi: 10.1038/s41419-017-0023-6
Gaitán-Espitia, J. D., Quintero-Galvis, J. F., Mesas, A., and D’Elía, G. (2016). Mitogenomics of southern hemisphere blue mussels (Bivalvia: Pteriomorphia): insights into the evolutionary characteristics of the Mytilus edulis complex. Sci. Rep. 6:26853. doi: 10.1038/srep26853
Galli, G. L. J., and Richards, J. G. (2014). Mitochondria from anoxia-tolerant animals reveal common strategies to survive without oxygen. J. Comp. Physiol. B 184, 285–302. doi: 10.1007/s00360-014-0806-3
Gardner, J. P. A. (1996). The Mytilus edulis species complex in southwest England: effects of hybridization and introgression upon interlocus associations and morphometric variation. Mar. Biol. 125, 385–399. doi: 10.1007/bf00346319
Gosling, E. (ed.) (1992). The Mussel Mytilus: Ecology, Physiology, Genetics and Culture. Amsterdam: Elsevier.
Gosling, E. M., and Wilkins, N. P. (1981). Ecological genetics of the mussels Mytilus edulis and M. galloprovincialis on irish coasts. Mar. Ecol. Prog. Ser. 4, 221–227. doi: 10.3354/meps004221
Grieshaber, M. K., Hardewig, I., Kreutzer, U., and Pörtner, H. -O. (1994). Physiological and metabolic responses to hypoxia in invertebrates. Rev. Physiol. Biochem. Pharmacol. 125, 43–147. doi: 10.1007/bfb0030909
Haider, F., Falfushynska, H. I., Timm, S., and Sokolova, I. M. (2020). Effects of hypoxia and reoxygenation on intermediary metabolite homeostasis of marine bivalves Mytilus edulis and Crassostrea gigas. Comp. Biochem. Phys. A Mol. Integr. Physiol. 242:110657. doi: 10.1016/j.cbpa.2020.110657
Head, B., Griparic, L., Amiri, M., Gandre-Babbe, S., and van der Bliek, A. M. (2009). Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 187, 959–966. doi: 10.1083/jcb.200906083
Hochachka, P. W. (1993). Surviving Hypoxia: Mechanisms of Control and Adaptation. Boca Raton, FL: CRC Press.
Hochachka, P. W., Buck, L. T., Doll, C. J., and Land, S. C. (1996). Unifying theroy of hypoxia tolerance: molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proc. Natl. Acad. Sci. U.S.A. 93, 9493–9498. doi: 10.1073/pnas.93.18.9493
Hochachka, P. W., and Lutz, P. L. (2001). Mechanism, origin, and evolution of anoxia tolerance in animals. Comp. Biochem. Phys. B 130, 435–459. doi: 10.1016/s1096-4959(01)00408-0
Ishihara, N., Fujita, Y., Oka, T., and Mihara, K. (2006). Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 25, 2966–2977. doi: 10.1038/sj.emboj.7601184
Ivanina, A. V., Nesmelova, I., Leamy, L., Sokolov, E. P., and Sokolova, I. M. (2016). Intermittent hypoxia leads to functional reorganization of mitochondria and affects cellular bioenergetics in marine molluscs. J. Exp. Biol. 219, 1659–1674. doi: 10.1242/jeb.134700
Ivanina, A. V., Sokolov, E. P., and Sokolova, I. M. (2010). Effects of cadmium on anaerobic energy metabolism and mRNA expression during air exposure and recovery of an intertidal mollusk Crassostrea virginica. Aquat. Toxicol. 99, 330–342. doi: 10.1016/j.aquatox.2010.05.013
Ivanina, A. V., and Sokolova, I. M. (2016). Effects of intermittent hypoxia on oxidative stress and protein degradation in molluscan mitochondria. J. Exp. Biol. 219, 3794–3802. doi: 10.1242/jeb.146209
Jin, S. M., Lazarou, M., Wang, C., Kane, L. A., Narendra, D. P., and Youle, R. J. (2010). Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191, 933–942. doi: 10.1083/jcb.201008084
Käser, M., Kambacheld, M., Kisters-Woike, B., and Langer, T. (2003). Oma1, a Novel Membrane-bound Metallopeptidase in Mitochondria with Activities Overlapping with the m-AAA Protease. J. Biol. Chem. 278, 46414–46423. doi: 10.1074/jbc.M305584200
Kennedy, V. S. (1996). The Eastern Oyster: Crassostrea virginica. College Park, MD: Maryland Sea Grant College.
Kitamura, N., Nakamura, Y., Miyamoto, Y., Miyamoto, T., Kabu, K., Yoshida, M., et al. (2011). Mieap, a p53-inducible protein, controls mitochondrial quality by repairing or eliminating unhealthy mitochondria. PLoS One 6:e16060. doi: 10.1371/journal.pone.0016060
Kulek, A. R., Anzell, A. R., Wider, J. M., Sanderson, T. H., and Przyklenk, K. (2020). Mitochondrial quality control: role in cardiac models of lethal ischemia-reperfusion injury. Cells 9:214. doi: 10.3390/cells9010214
Kuo, C. -Y., Chiu, Y. -C., Lee, A. Y.-L., and Hwang, T. -L. (2015). Mitochondrial Lon protease controls ROS-dependent apoptosis in cardiomyocyte under hypoxia. Mitochondrion 23, 7–16. doi: 10.1016/j.mito.2015.04.004
Kurochkin, I. O., Ivanina, A. V., Eilers, S., Downs, C. A., May, L. A., and Sokolova, I. M. (2009). Cadmium affects metabolic responses to prolonged anoxia and reoxygenation in eastern oysters (Crassostrea virginica). Am. J. Physiol. Regul. Integr. Comp. Physiol. 297, R1262-R1272. doi: 10.1152/ajpregu.00324.2009
Larade, K., and Storey, K. B. (2002). “A Profile of the Metabolic Responses to Anoxia in Marine Invertebrates,” in Sensing, Signaling and Cell Adaptation, eds K. B. Storey and J. M. Storey (Amsterdam: Elsevier Science), 27–46. doi: 10.1016/s1568-1254(02)80005-5
Le Moullac, G., Quéau, I., Le Souchu, P., Pouvreau, S., Moal, J., Le René Coz, J., et al. (2007). Metabolic adjustments in the oyster Crassostrea gigas according to oxygen level and temperature. Mar. Biol. Res. 3, 357–366. doi: 10.1080/17451000701635128
Li, J., Donath, S., Li, Y., Qin, D., Prabhakar, B. S., and Li, P. (2010). miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 6:e1000795. doi: 10.1371/journal.pgen.1000795
Li, Y., and Liu, X. (2018). Novel insights into the role of mitochondrial fusion and fission in cardiomyocyte apoptosis induced by ischemia/reperfusion. J. Cell Physiol. 233, 5589–5597. doi: 10.1002/jcp.26522
Losón, O. C., Song, Z., Chen, H., and Chan, D. C. (2013). Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 24, 659–667. doi: 10.1091/mbc.E12-10-0721
Mears, J. A., Lackner, L. L., Fang, S., Ingerman, E., Nunnari, J., and Hinshaw, J. E. (2011). Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 18, 20–26. doi: 10.1038/nsmb.1949
Migdal, I., Skibior-Blaszczyk, R., Heidorn-Czarna, M., Kolodziejczak, M., Garbiec, A., and Janska, H. (2017). AtOMA1 affects the OXPHOS system and plant growth in contrast to other newly identified ATP-independent proteases in Arabidopsis mitochondria. Front. Plant Sci. 8:1543. doi: 10.3389/fpls.2017.01543
Nakamura, Y., and Arakawa, H. (2017). Discovery of Mieap-regulated mitochondrial quality control as a new function of tumor suppressor p53. Cancer Sci. 108, 809–817. doi: 10.1111/cas.13208
Narendra, D. P., and Youle, R. J. (2011). Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. Antioxid Redox. Signal. 14, 1929–1938. doi: 10.1089/ars.2010.3799
Osellame, L. D., Singh, A. P., Stroud, D. A., Palmer, C. S., Stojanovski, D., Ramachandran, R., et al. (2016). Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J. Cell Sci. 129, 2170–2181. doi: 10.1242/jcs.185165
Osman, C., Wilmes, C., Tatsuta, T., and Langer, T. (2007). Prohibitins interact genetically with Atp23, a novel processing peptidase and chaperone for the F1Fo-ATP synthase. Mol. Biol. Cell 18, 627–635. doi: 10.1091/mbc.e06-09-0839
Otera, H., Wang, C., Cleland, M. M., Setoguchi, K., Yokota, S., Youle, R. J., et al. (2010). Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191, 1141–1158. doi: 10.1083/jcb.201007152
Ozawa, K., Kuwabara, K., Tamatani, M., Takatsuji, K., Tsukamoto, Y., Kaneda, S., et al. (1999). 150-kDa Oxygen-regulated protein (ORP150) suppresses hypoxia-induced Apoptotic Cell Death∗. J. Biol. Chem. 274, 6397–6404. doi: 10.1074/jbc.274.10.6397
Paradis, S., Charles, A. -L., Meyer, A., Lejay, A., Scholey, J. W., Chakfé, N., et al. (2016). Chronology of mitochondrial and cellular events during skeletal muscle ischemia-reperfusion. Am. J. Physiol. Cell Physiol. 310, C968-C982. doi: 10.1152/ajpcell.00356.2015
Pickles, S., Vigié, P., and Youle, R. J. (2018). Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170-R185. doi: 10.1016/j.cub.2018.01.004
Pinti, M., Gibellini, L., Liu, Y., Xu, S., Lu, B., and Cossarizza, A. (2015). Mitochondrial Lon protease at the crossroads of oxidative stress, ageing and cancer. Cell Mol. Life Sci. 72, 4807–4824. doi: 10.1007/s00018-015-2039-3
Piper, H. M., Meuter, K., and Schäfer, C. (2003). Cellular mechanisms of ischemia-reperfusion injury. Ann. Thorac. Surg. 75, S644-S648. doi: 10.1016/S0003-4975(02)04686-6
R Core Team (2020). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rainbolt, T. K., Saunders, J. M., and Wiseman, R. L. (2015). YME1L degradation reduces mitochondrial proteolytic capacity during oxidative stress. EMBO Rep. 16, 97–106. doi: 10.15252/embr.201438976
Richards, J. G. (2011). Physiological, behavioral and biochemical adaptations of intertidal fishes to hypoxia. J. Exp. Biol. 214, 191–199. doi: 10.1242/jeb.047951
Richter, V., Singh, A. P., Kvansakul, M., Ryan, M. T., and Osellame, L. D. (2015). Splitting up the powerhouse: structural insights into the mechanism of mitochondrial fission. Cell Mol. Life Sci. 72, 3695–3707. doi: 10.1007/s00018-015-1950-y
Ryu, S. -W., Han, E. C., Yoon, J., and Choi, C. (2015). The mitochondrial fusion-related proteins Mfn2 and OPA1 are transcriptionally induced during differentiation of bone marrow progenitors to immature dendritic cells. Mol. Cells 38, 89–94. doi: 10.14348/molcells.2015.2285
Sanni, B., Williams, K., Sokolov, E. P., and Sokolova, I. M. (2008). Effects of acclimation temperature and cadmium exposure on mitochondrial aconitase and LON protease from a model marine ectotherm, Crassostrea virginica. Comp. Biochem. Phys. C Toxicol. Pharmacol. 147, 101–112. doi: 10.1016/j.cbpc.2007.08.005
Sepuri, N. B. V., Angireddy, R., Srinivasan, S., Guha, M., Spear, J., Lu, B., et al. (2017). Mitochondrial LON protease-dependent degradation of cytochrome c oxidase subunits under hypoxia and myocardial ischemia. Biochim. Biophys. Acta Bioenerg. 1858, 519–528. doi: 10.1016/j.bbabio.2017.04.003
Shanmughapriya, S., Rajan, S., Hoffman, N. E., Higgins, A. M., Tomar, D., Nemani, N., et al. (2015). SPG7 is an essential and conserved component of the mitochondrial permeability transition pore. Mol. Cell 60, 47–62. doi: 10.1016/j.molcel.2015.08.009
Singh, R., and Hartl, M. G. (2012). Fluctuating estuarine conditions are not confounding factors for the Comet assay assessment of DNA damage in the mussel Mytilus edulis. Ecotoxicology 21, 1998–2003. doi: 10.1007/s10646-012-0934-z
Sokolov, E. P., Markert, S., Hinzke, T., Hirschfeld, C., Becher, D., Ponsuksili, S., et al. (2019). Effects of hypoxia-reoxygenation stress on mitochondrial proteome and bioenergetics of the hypoxia-tolerant marine bivalve Crassostrea gigas. J. Prot. 194, 99–111. doi: 10.1016/j.jprot.2018.12.009
Sokolova, I. M. (2018). Mitochondrial adaptations to variable environments and their role in animals’ stress tolerance. Integr. Comp. Biol. 58, 519–531. doi: 10.1093/icb/icy017
Sokolova, I. M., Sokolov, E. P., and Haider, F. (2019). Mitochondrial mechanisms underlying tolerance to fluctuating oxygen conditions: lessons from hypoxia-tolerant organisms. Integr. Comp. Biol. 59, 938–952. doi: 10.1093/icb/icz047
Solaini, G., Baracca, A., Lenaz, G., and Sgarbi, G. (2010). Hypoxia and mitochondrial oxidative metabolism. Biochim. Biophys. Acta 1797, 1171–1177. doi: 10.1016/j.bbabio.2010.02.011
Song, Z., Ghochani, M., McCaffery, J. M., Frey, T. G., and Chan, D. C. (2009). Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol. Biol. Cell 20, 3525–3532. doi: 10.1091/mbc.e09-03-0252
Spelbrink, J. N., Li, F. Y., Tiranti, V., Nikali, K., Yuan, Q. P., Tariq, M., et al. (2001). Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 28, 223–231. doi: 10.1038/90058
Storey, K. B. (1998). Survival under stress: molecular mechanisms of metabolic rate depression in animals. South Afr. J. Zool. 33, 55–64. doi: 10.1080/02541858.1998.11448454
Storey, K. B. (2002). Life in the slow lane: molecular mechanisms of estivation. Comp. Biochem. Phys. A Mol. Integr. Physiol. 133, 733–754. doi: 10.1016/S1095-6433(02)00206-4
Storey, K. B., and Storey, J. M. (2004). Metabolic rate depression in animals: transcriptional and translational controls. Biol. Rev. Camp Philos. Soc. 79, 207–233. doi: 10.1017/s1464793103006195
Sussarellu, R., Dudognon, T., Fabioux, C., Soudant, P., Moraga, D., and Kraffe, E. (2013). Rapid mitochondrial adjustments in response to short-term hypoxia and re-oxygenation in the Pacific oyster, Crassostrea gigas. J. Exp. Biol. 216, 1561–1569. doi: 10.1242/jeb.075879
Tian, L., Neuber-Hess, M., Mewburn, J., Dasgupta, A., Dunham-Snary, K., Wu, D., et al. (2017). Ischemia-induced Drp1 and Fis1-mediated mitochondrial fission and right ventricular dysfunction in pulmonary hypertension. J. Mol. Med. 95, 381–393. doi: 10.1007/s00109-017-1522-8
Twig, G., Elorza, A., Molina, A. J. A., Mohamed, H., Wikstrom, J. D., Walzer, G., et al. (2008). Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446. doi: 10.1038/sj.emboj.7601963
van den Thillart, G., van Lieshout, G., Storey, K., Cortesi, P., Zwaan, A., and de (1992). Influence of long-term hypoxia on the energy metabolism of the haemoglobin-containing bivalve Scapharca inaequivalvis: critical O2 levels for metabolic depression. J. Comp. Physiol. B 162, 297–304. doi: 10.1007/bf00260756
van der Bliek, A. M., Shen, Q., and Kawajiri, S. (2013). Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 5:a011072. doi: 10.1101/cshperspect.a011072
Varvio, S. -L., Koehn, R. K., and Väinölä, R. (1988). Evolutionary genetics of the Mytilus edulis complex in the North Atlantic region. Mar. Biol. 98, 51–60. doi: 10.1007/BF00392658
Venkatesh, S., Lee, J., Singh, K., Lee, I., and Suzuki, C. K. (2012). Multitasking in the mitochondrion by the ATP-dependent Lon protease. Biochim. Biophys. Acta 1823, 56–66. doi: 10.1016/j.bbamcr.2011.11.003
Wang, J., and Zhou, H. (2020). Mitochondrial quality control mechanisms as molecular targets in cardiac ischemia–reperfusion injury. Acta Pharm. Sin. B 10, 1866–1879. doi: 10.1016/j.apsb.2020.03.004
Wang, Z., Jiang, H., Chen, S., Du, F., and Wang, X. (2012). The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 148, 228–243. doi: 10.1016/j.cell.2011.11.030
Wu, H., Xue, D., Chen, G., Han, Z., Huang, L., Zhu, C., et al. (2014). The BCL2L1 and PGAM5 axis defines hypoxia-induced receptor-mediated mitophagy. Autophagy 10, 1712–1725. doi: 10.4161/auto.29568
Xiao, X., Hu, Y., Quirós, P. M., Wei, Q., López-Otín, C., and Dong, Z. (2014). OMA1 mediates OPA1 proteolysis and mitochondrial fragmentation in experimental models of ischemic kidney injury. Am. J. Physiol. Renal Physiol. 306, F1318-F1326. doi: 10.1152/ajprenal.00036.2014
Yang, Y., Zhao, L., and Ma, J. (2017). Penehyclidine hydrochloride preconditioning provides cardiac protection in a rat model of myocardial ischemia/reperfusion injury via the mechanism of mitochondrial dynamics mechanism. Eur. J. Pharmacol. 813, 130–139. doi: 10.1016/j.ejphar.2017.07.031
Youle, R. J., and van der Bliek, A. M. (2012). Mitochondrial fission, fusion, and stress. Science 337, 1062–1065. doi: 10.1126/science.1219855
Yu, J., Wu, J., Xie, P., Maimaitili, Y., Wang, J., Xia, Z., et al. (2016). Sevoflurane postconditioning attenuates cardiomyocyte hypoxia/reoxygenation injury via restoring mitochondrial morphology. PeerJ. 4:e2659. doi: 10.7717/peerj.2659
Zheng, J., Croteau, D. L., Bohr, V. A., and Akbari, M. (2019). Diminished OPA1 expression and impaired mitochondrial morphology and homeostasis in Aprataxin-deficient cells. Nucl. Acids Res. 47, 4086–4110. doi: 10.1093/nar/gkz083
Zorov, D. B., Juhaszova, M., and Sollott, S. J. (2014). Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 94, 909–950. doi: 10.1152/physrev.00026.2013
Keywords: mitochondrial fusion and fission, quantitative real time PCR (qPCR), mRNA expression, mollusca, proteolysis, mitophagy
Citation: Steffen JBM, Falfushynska HI, Piontkivska H and Sokolova IM (2020) Molecular Biomarkers of the Mitochondrial Quality Control Are Differently Affected by Hypoxia-Reoxygenation Stress in Marine Bivalves Crassostrea gigas and Mytilus edulis. Front. Mar. Sci. 7:604411. doi: 10.3389/fmars.2020.604411
Received: 09 September 2020; Accepted: 11 November 2020;
Published: 01 December 2020.
Edited by:
Nina Bednarsek, Southern California Coastal Water Research Project, United StatesReviewed by:
Anwesha Ghosh, Indian Institute of Science Education and Research Kolkata, IndiaDanielle Ferraz Mello, Duke University, United States
Katherine Silliman, Auburn University, United States
Copyright © 2020 Steffen, Falfushynska, Piontkivska and Sokolova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Inna M. Sokolova, aW5uYS5zb2tvbG92YUB1bmktcm9zdG9jay5kZQ==
 Jennifer B. M. Steffen
Jennifer B. M. Steffen Halina I. Falfushynska
Halina I. Falfushynska Helen Piontkivska
Helen Piontkivska Inna M. Sokolova
Inna M. Sokolova