- 1College of Fisheries, Guangdong Ocean University, Zhanjiang, China
- 2South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 3Key Laboratory of Open-Sea Fishery Development, Ministry of Agriculture and Rural Affairs, Guangzhou, China
- 4CAS Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 5Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
- 6Laboratory for Marine Ecology and Environmental Science, Qingdao National Laboratory for Marine Science and Technology, Chinese Academy of Sciences, Qingdao, China
Although species of Decapterus form important pelagic fisheries in the northern South China Sea, information on their spawning grounds is limited because identification of fish eggs based on their morphology is difficult. We identify eggs of four Decapterus species (D. macrosoma, D. maruadsi, D. macarellus, and D. tabl) with DNA barcodes from the fishery resources surveys in spring and autumn 2018 in Xisha islands, and spring and later summer–autumn 2019 along the continental shelf of the northern South China Sea, and describe egg morphology. Of 1405 fish eggs with obtained cytochrome c oxidase subunit I (COI) sequences, 81 were successfully attributed to four Decapterus species; eggs of each are spherical, have a smooth chorion and narrow perivitelline space, and can be partly differentiated by diameter, melanophore drops on the oil globule, and the notum of the embryo. Seasonal distributions of eggs reveal spawning grounds, with that of D. maruadsi located mainly off the Pearl River estuary in spring; eggs of D. maruadsi rarely co-occur with those of D. macrosoma. Spawning grounds of D. macrosoma are probably further south, where water temperature and salinity are higher. Spawning periods of these four Decapterus species overlap slightly. Spawning habitat of D. maruadsi has been lost, and the spawning season of D. macrosoma has extended. Identification of Decapterus eggs using DNA barcodes can assist with the identification of eggs using traditional morphological approaches. Spatial and temporal information on the distributions of Decapterus eggs can be used for improved conservation of spawning grounds and fisheries management in the northern South China Sea.
Introduction
Pelagic fish play important roles in marine ecosystems, occupy a large proportion of commercially important global fisheries, and are considered increasingly important as a source of protein (Lainez del Pozo, 2013; Food and Agricultural Organization [FAO], 2019). However, being r-strategists, their stocks can vary considerably within and between years and decades, related to changes in climate and oceanography, and physical–biological process at regional and global scales (Checkley et al., 2009; Kawakami et al., 2010; Watson et al., 2018; Ma et al., 2019). Understanding early life stage recruitment is therefore important in fisheries management (Chambers and Trippel, 1997; Fuiman, 2002; Checkley et al., 2009).
Scads in the genus Decapterus are commercially important pelagic fish. Of these, the small Japanese scad D. maruadsi is commercially important in China and, from 1998 to 2006, was the first number catch in the northern South China Sea (SCS). However, the average catch of this species decreased by 28%, relegating it to the third number catch in 2007–2018, with catch varying considerably between years (China Agriculture Press), 1997–2019). While D. maruadsi is caught mainly by pair trawlers, and single bottom trawl and light purse seine nets, two other species (in predominantly subtropical–tropical waters), Shortfin scad D. macrosoma and Mackerel scad D. macarellus, are targeted by light purse seine and light falling nets (Yang et al., 2009; Li et al., 2018). A fourth species, Roughear scad D. tabl, found on the edge of the continental shelf from 200 to 360 m, is uncommon (Froese and Pauly, 2010).
The northern SCS continental shelf is an important spawning and nursery ground for various commercial fishes, such as jack mackerel, sardines, and Japanese scad (Zhang et al., 1985). Recent progress in understanding the biology and ecology of early life history stages and recruitment processes of these pelagic species has been made (Xie and Watanabe, 2007; Kasai et al., 2008; Nishiyama et al., 2014; Sciascia et al., 2018). However, due to difficulties identifying fish eggs and larvae, information on the early life stages of these species, especially their spawning grounds based on ichthyoplankton surveys in the SCS, is limited (Zhang et al., 1985; Zhou et al., 2011; Li et al., 2014; Chen et al., 2018).
Accurate identification of fish eggs based on morphology can be difficult and time-consuming, especially when dealing with samples containing eggs of many species at various stages of development. This is further aggravated by fish adapting to oceanic conditions, changing their reproduction and egg morphology, such as incubation time, egg diameter, pigmentation, and spawning season duration (Neira et al., 2015). Fish egg identification is further complicated by many characters used to distinguish taxa being evident only during later developmental stages, mainly after the sarcomere period, leaving eggs of even unrelated species sometimes morphologically indistinguishable (Shao et al., 2002; Ikeda et al., 2014).
DNA barcoding using molecular markers [e.g., cytochrome c oxidase subunit I (COI)] enables accurate identification of fish eggs and larvae, regardless of developmental stage or morphology (Valdez-Moreno et al., 2010; Frantine-Silva et al., 2015; Hubert et al., 2015). Considerable sequence data for fish taxa are deposited in the BOLD (Barcode of Life Data System1) database; by early 2020, more than 19,000 fish species based on over 260,000 specimens were reported (Ratnasingham and Hebert, 2007). These data enable discrimination of adults and larvae of many fish species.
We use DNA barcodes to discriminate eggs of D. maruadsi, D. macarellus, D. macrosoma, and D. tabl from the northern SCS. We aim to (i) differentiate the eggs of these Decapterus species using DNA barcodes and describe their morphology, (ii) determine the main spawning grounds and reproductive periods of Decapterus species in the northern SCS, and (iii) appraise the value of information on fish reproduction and recruitment in sustainable exploitation and management.
Materials and Methods
Sample Collection
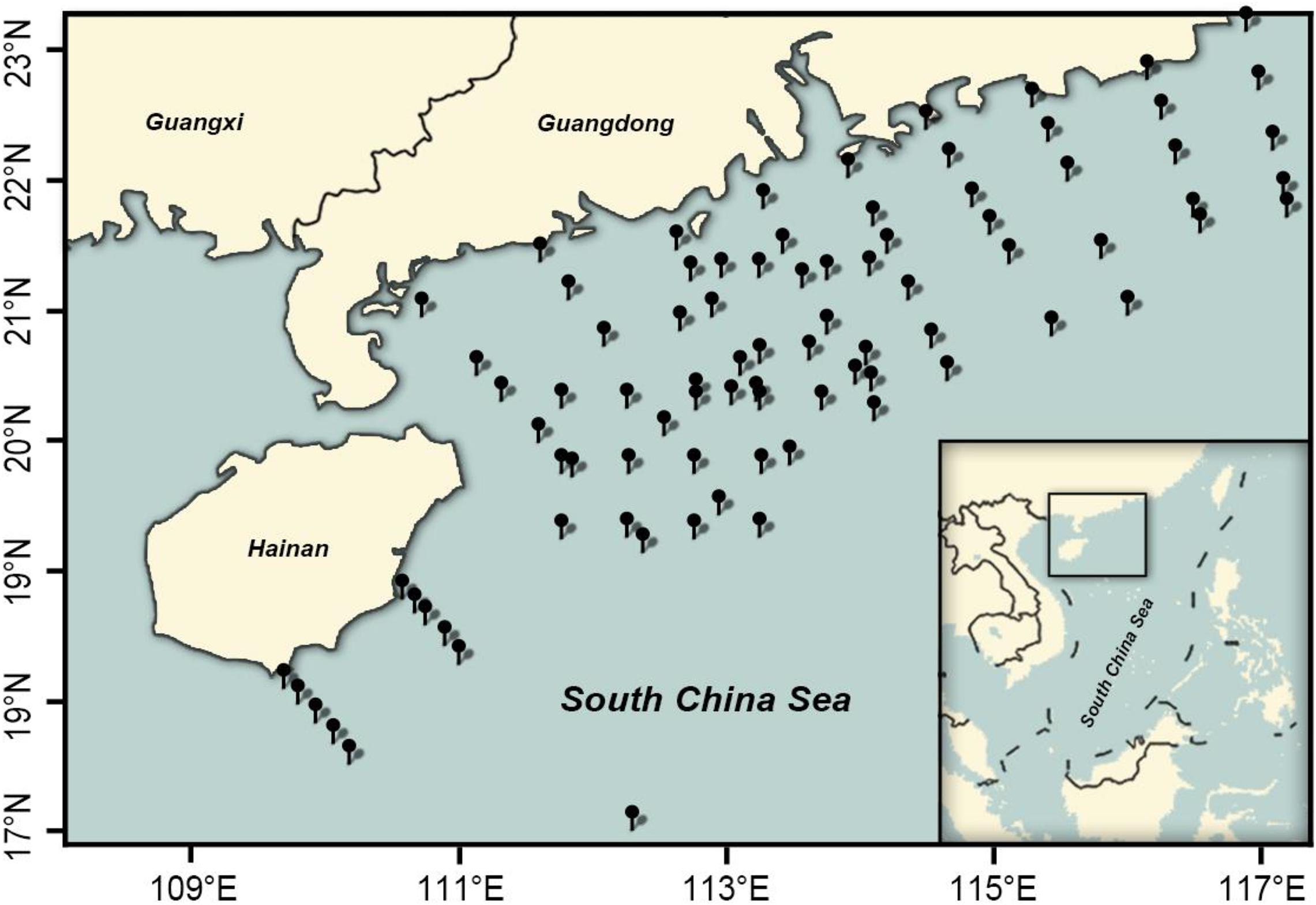
Fish eggs were collected during the austral spring (April–May) and later summer–autumn in the northern SCS and Xisha islands (Figure 1). In all, 85 stations were surveyed, including 84 stations in the Guangdong coastal sea area (19.12–23.15°N, 110.7–117.21°E) in April and September–October 2019 and 1 station off the Xisha islands (16.98°N, 112.28°E) in May and September 2018. Eggs were collected with a 2.7-m-long, 80-cm-diameter zooplankton net, with a 0.505-mm mesh and a cod-end container mesh of 400 μm. Nets were fitted with a General Oceanic flowmeter for estimating filtered water volume. In horizontal trawls, nets were dragged 10–15 min at 1.5–2.2 knots. Samples from each station were fixed in ∼75% ethanol/seawater solution.
Sample Photographs
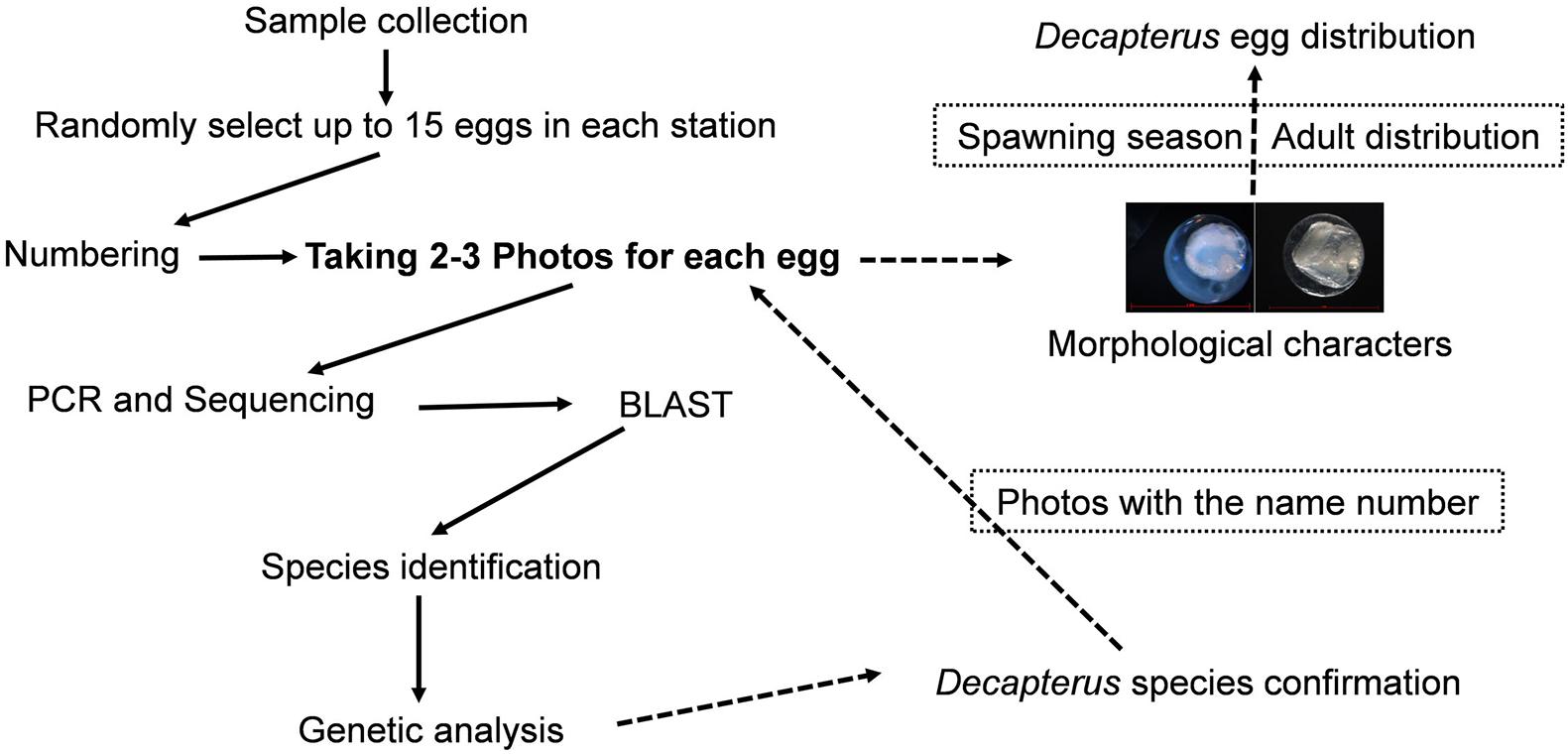
All eggs from each station were sorted beneath a stereomicroscope in a laboratory; up to 15 eggs were randomly selected (if more than 15 eggs at a station, or all eggs if less) and photographed. Alcohol-preserved eggs were rehydrated in hydrogen peroxide for ∼8 min for cleaning and to better measure egg diameter (to the nearest 0.001 mm). Eggs were photographed using a Zeiss microscope (Axioplan 2 imaging E) and numbered, and their DNA was subsequently extracted (Figure 2).
DNA Extraction and PCR Amplification
Each numbered egg was firstly put in a cleaned centrifuge tube, dried in the air and volatilized alcohol, and then quickly punctured by a fine needle within 50 s. Total genomic DNA was then extracted using an Axygen Genomic DNA Miniprep Kit (Axygen, Shanghai, China). COI sequences (∼648 bp) were amplified using the universal primers FishF1 and FishR1 (Ward et al., 2005). The polymerase chain reaction (PCR) contained 20 μl of Tsingke TM Master Mix, 1 μl of each primer (10 pmol), 1–10 μl of template DNA and 8–17 μl of ddH2O (total template DNA + ddH2O = 18 μl), to make a total volume of 40 μl. PCR conditions were 94°C for 3 min, 35 cycles at 94°C for 30 s, 51°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 10 min. Amplification products of PCR reactions were purified using 1% low-melting agarose by electrophoresis, and sequenced bidirectionally on an ABI 3730 XL DNA system following manufacturer protocols. High-quality DNA sequences were edited with MEGA v6.0 (Tamura et al., 2013).
Sequence Analysis
Egg sequences were initially differentiated by Blast search in BOLD (the Barcode of life Data System, BOLD)2. Sequences exceeding 98% similarity and 2% divergence threshold with the nearest neighbor were tagged with species name following the criteria of Hubert et al. (2015). The DNA barcode library under the project “South China Sea Fishes” in BOLD comprised six locally and proximally occurring Decapterus species; it was used in combination with BOLD to confirm egg species when aforementioned criteria were not met, using Selar crumenophthalmus as an outgroup in tree-based delimitation (Jaafar et al., 2012; Chang et al., 2017; Hou et al., 2018). Available Decapterus COI sequences were downloaded from Genbank3 (8 February 2020), combined with our Decapterus egg sequences and submitted to statistical parsimony networks for haplotype network construction (Templeton et al., 1992) using the default 95% connection limit in TCS 1.2 (Clement et al., 2000).
Egg Morphology
Images of identified Decapterus eggs were used to describe their morphology following Shao et al. (2001) and Ikeda et al. (2014).
Egg Distribution
Catch rates of Decapterus eggs for each station are presented as numbers of eggs per 100 m3 of filtered water (flowmeter data). Depth (D), surface sea temperature (SST), and salinity (S) were measured and analyzed under the guidance of “Specification of Oceanographic Investigation” (GB12763-2007). The distribution and egg catch rates at stations with Decapterus spp. were plotted using Golden Software Surfer (version 15.0, Golden Software Inc., Golden, CO, United States).
Results
Fish Egg Identification by DNA Barcode
A total of 81 Decapterus eggs were screened from 1,405 successfully amplified samples (referable to 120 fish species). All sequences of Decapterus eggs were of high quality, with no evidence of insertions/deletions, heterozygous sites, or stop codons; each has been uploaded to NCBI (Genbank accession numbers: MT609955–MT610035). The 81 sequences were 619 nucleotides in length after alignment and trimming of noisy sites; a BOLD Blast search identified four candidate species belonging to four species of genus Decapterus (based on 98% similarity and 2% of genetic divergence among species); all egg sequences reached a match of similarity 98–100%, with matching results for the nearest neighbor ranging similarity 90.78–100%. Only four sequences fitted the criterion and were identified as D. tabl or D. macarellus; the remaining 77 sequences were 98–100%, similar to both the best match and nearest neighbor, with the genetic divergence from the nearest neighbor less than 2% (indicating that 95% of the samples could not be unambiguously assigned to species directly using Blast in Bold; Supporting information S1). Thus, haplotype analysis and genetic distance were conducted for the downloaded COI sequences of genus Decapterus from the Bold system and Genbank, to detect if there is haplotype sharing with low genetic divergence among them (Supplementary Table S2). Five groups, i.e., D. macrosoma and D. maruadsi, D. kurroides, and D. maruadsi, etc., were detected to likely share the COI haplotype but with high genetic divergence (>2%), which indicated that they are not really sharing haplotypes but with wrong identification or mismatch species name among the sequences (Figure 3 and Supplementary Table S3). The tree-based method was then used to delimit the fish eggs with local DNA barcode library, which revealed six species clusters in genus Decapterus supported by bootstrap values of 100%, attributing 61 sequences to D. macrosoma, 16 sequences to D. maruadsi, 2 sequences to D. macarellus, and 2 sequences to D. tabl (Figure 4).
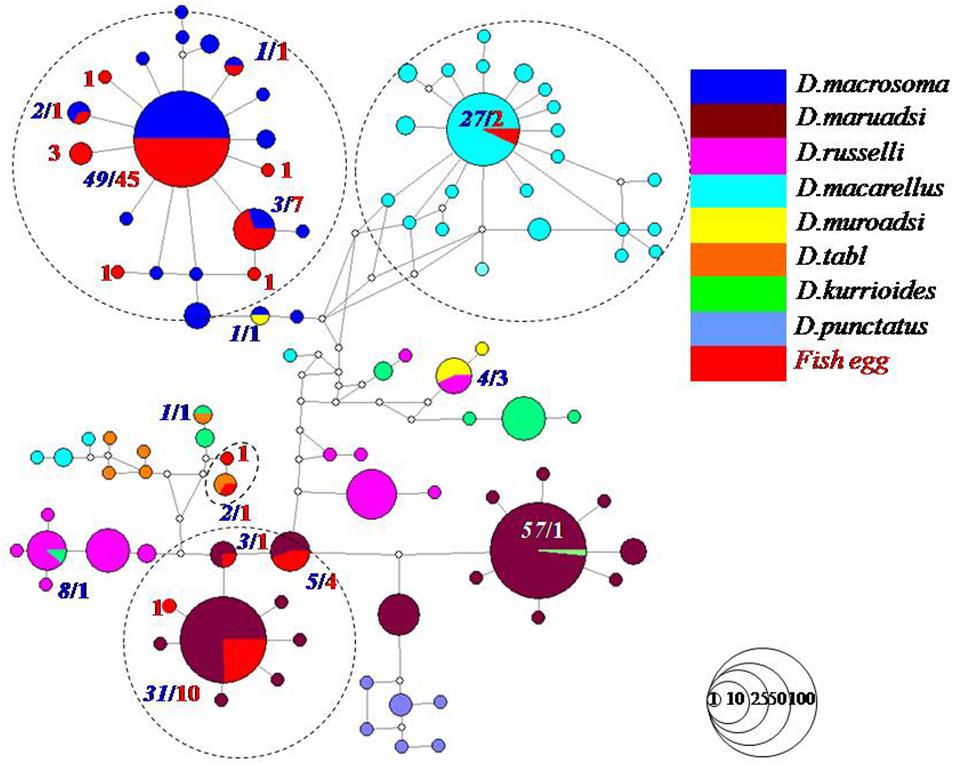
Figure 3. Decapterus spp. egg haplotype network analysis combined with downloaded sequences from NCBI. Red numbers in bold and italic indicated fish egg sequences, and blue and white numbers in bold and italic indicated downloaded fish sequences. Circles represent haplotypes, and each circle is proportional to the number of occurrences of the corresponding haplotype. The dotted circles contained the eggs and the downloaded sequences of four Decapterus fish species.
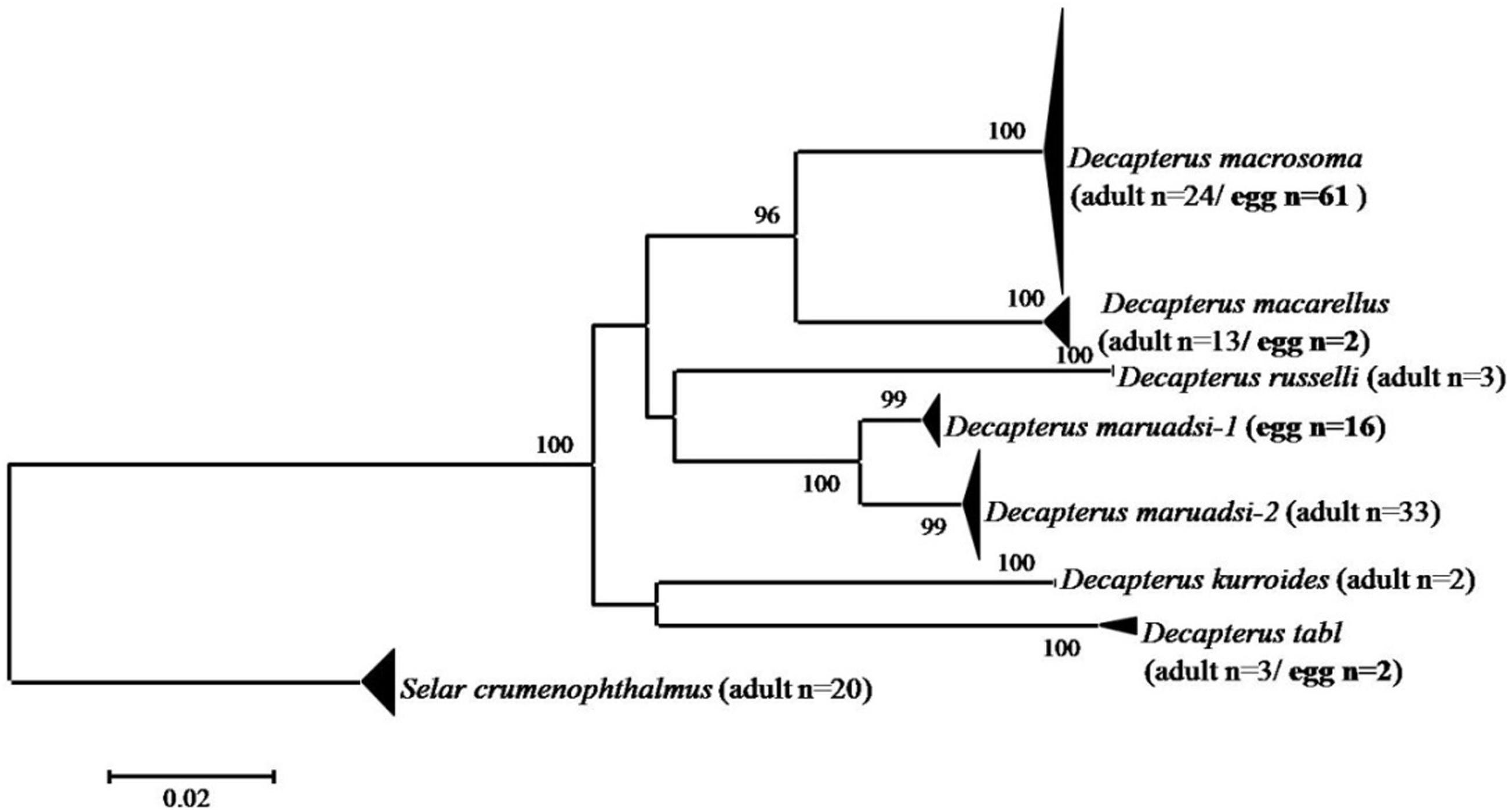
Figure 4. Neighbor-joining tree of fish egg sequences combined with local DNA barcode library of Decapterus species. The numbers at the nodes are bootstrap values based on 1,000 replications.
Decapterus Egg Morphology
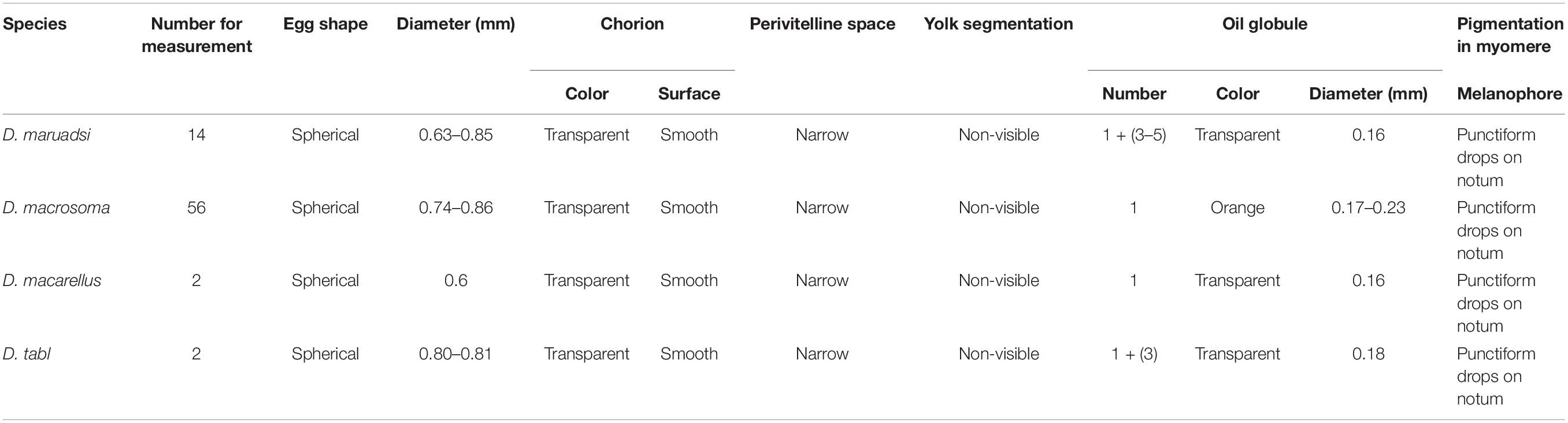
Eggs of all four Decapterus species are spherical, with a smooth chorion and diameters ranging 0.60–0.86 mm; the smallest is D. maruadsi (n = 16; mean ± SE = 0.75 ± 0.06), and the largest is D. macarellus (n = 2; mean ± SE = 0.81 ± 0.01) (Figure 5). All eggs have a narrow perivitelline space, with that of D. maruadsi widest after the myotome stage (Figure 6). Yolk segmentation was not observed in alcohol-preserved eggs.
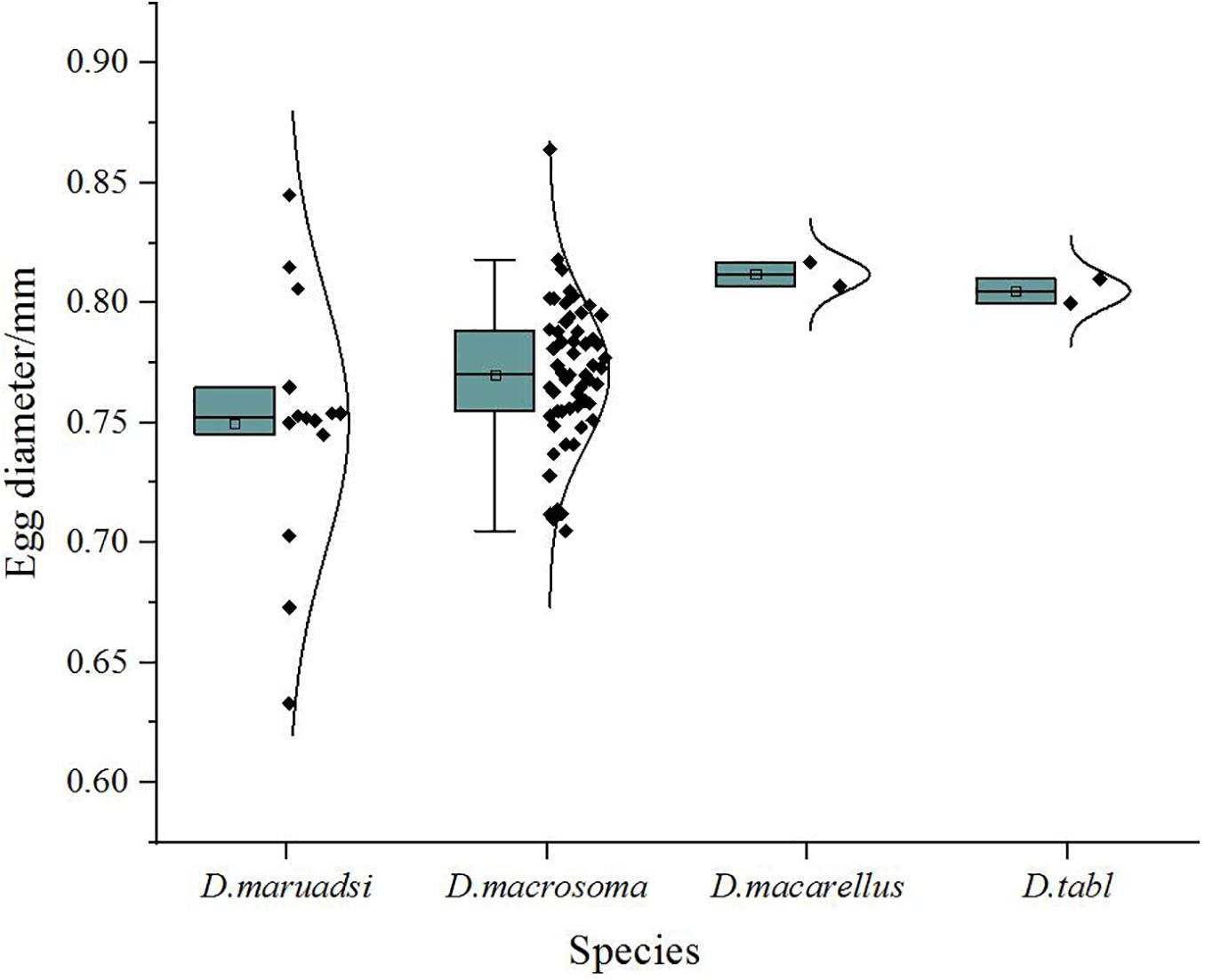
Figure 5. Egg diameter (mm) in Decapterus species, northern South China Sea. Whisker plots depict median, mean, 1st and 3rd quartiles, and ranges (minima, maxima).
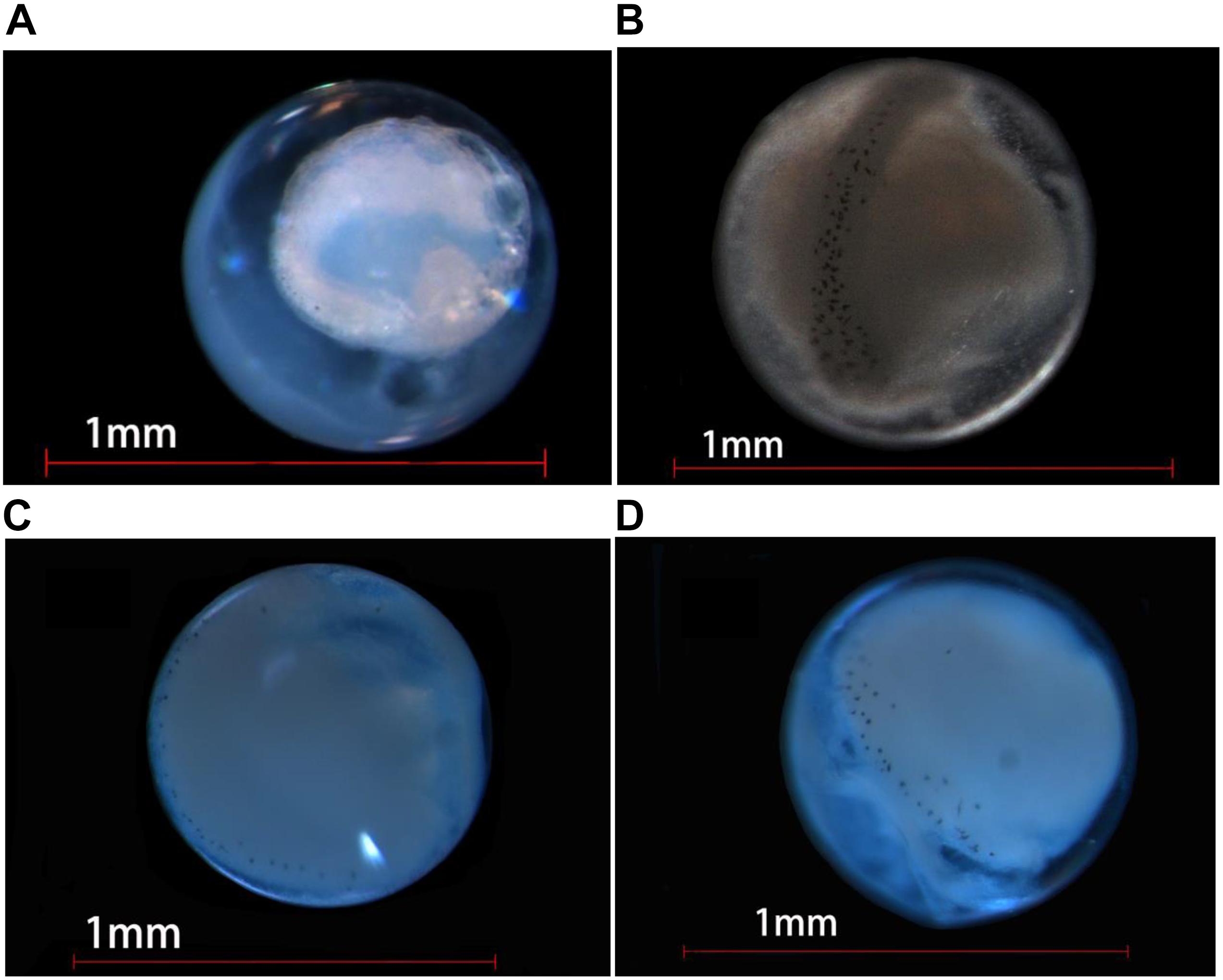
Figure 6. Photographs of eggs of four Decapterus species delimited by COI. (A) D. maruadsi, (B) D. macrosoma, (C) D. macarellus, and (D) D. tabl.
Oil globules occurred in the rear position of each species and were numerous in D. maruadsi and D. tabl. Some D. maruadsi eggs had a single large oil globule (0.16 mm) and three to five smaller oil globules (0.030–0.039 mm), with their number dependent on developmental stage. Eggs of D. tabl had one large oil globule (0.18 mm) and three small oil globules (0.065–0.118 mm). Oil globules in D. macrosoma are orange colored, while in the other three species, they are transparent. Melanophore drops occurred on the large oil globule of both D. maruadsi and particularly D. macrosoma; no melanophore drops were found in D. macarellus or D. tabl. The myotome stage was pigmented in each species. Melanophore drops were widely scattered on the embryo notum from the cephalosome to the tail sarcomere, more densely in D. macrosoma (Table 1 and Figure 6).
Seasonal Distribution of Decapterus Eggs
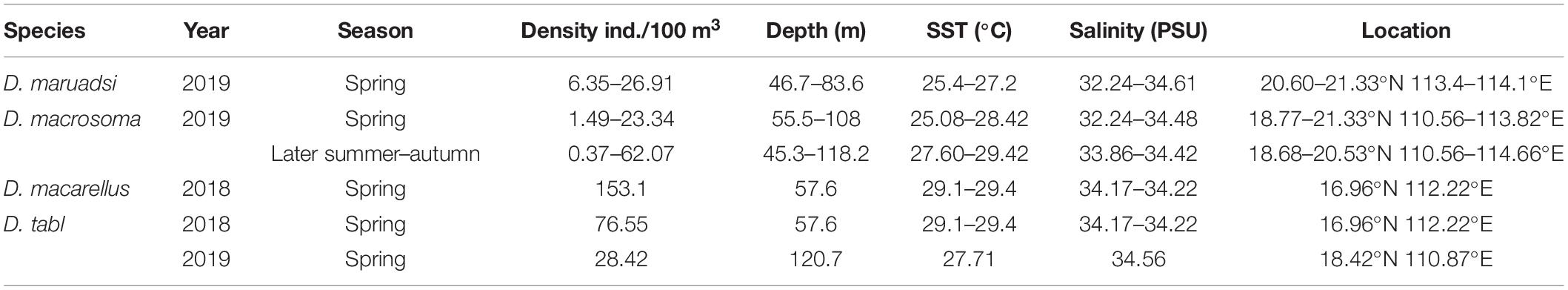
Eggs were collected at 85 stations, with those of Decapterus at 9 stations in both spring and autumn. Eggs of D. maruadsi occurred at four stations concentrated off the Pearl River Estuary in spring, at 46.7–83.6 m, SST 25.4–27.2°C, and salinity 32.24–34.61 (Table 2 and Figure 7A). Eggs of D. maruadsi were not found in autumn. Eggs of D. macrosoma occurred at four stations from 55.5 to 108.0 m in spring, at SST 25.08–28.42°C and salinity 32.24–34.48. In autumn, eggs of D. macrosoma occurred at nine stations from 45.3 to 118.2 m from the Pearl River Estuary to south of Hainan Island (Figure 7B) at SST 27.60–29.42°C and salinity 33.86–34.42. Eggs of D. macarellus occurred at one station in spring of 2018, and eggs of D. tabl occurred at one station in spring of 2018 and one station in spring of 2019 (Table 2 and Figure 7A).
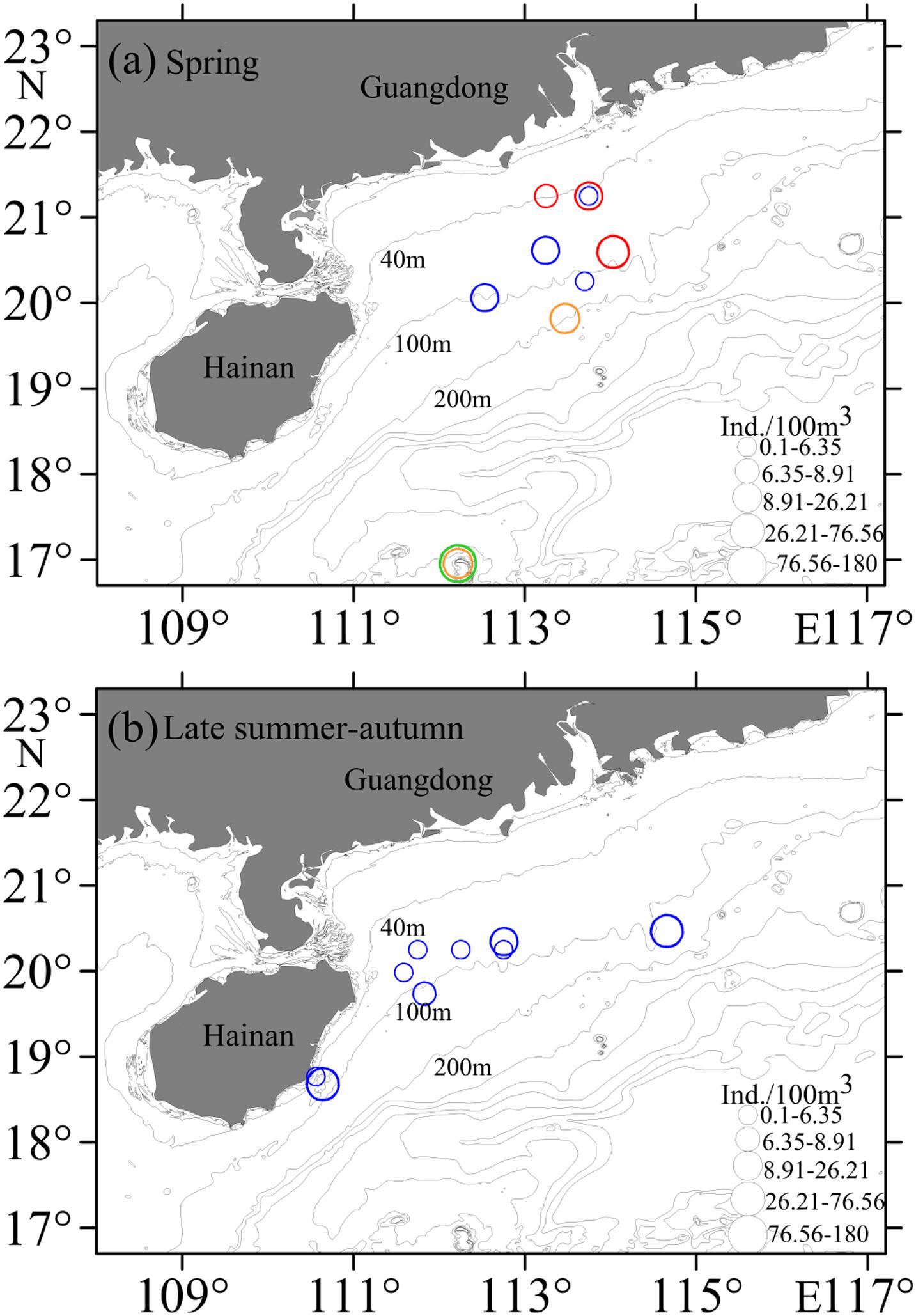
Figure 7. Horizontal distributions of Decapterus eggs in the northern South China Sea. (a) Spring; (b) Late summer-autumn. Symbols represent catch rates per 100 m3. Red circles indicate D. maruadsi, blue circles indicate D. macrosoma, green circles indicate D. macarellus, and orange circles indicate D. tabl.
Discussion
Molecular Identification by DNA Barcode
The lack of morphological characters in descriptions of ontogenetic stages of fish eggs renders their identification by light microscopy difficult, especially for early stages (Kawakami et al., 2010; Nishiyama et al., 2014). DNA, however, facilitates their identification. We apply DNA barcodes (COI sequences) to identify eggs of Decapterus species and demonstrate the importance of this technique for egg identification, particularly in spawning ground surveys (Lewis et al., 2016; Leyva-Cruz et al., 2016; Ahern et al., 2018).
Disadvantages of using COI sequences for DNA barcodes include sequence alignment, where sequences match multiple species in BOLD: 16 sequences were highly similar (≥99%) to D. maruadsi and D. russelli, and 61 sequences were highly similar (≥99%) to D. macrosoma and D. maruadsi (S1). Identification can be ambiguous, possibly because of hybridization, recent divergence, inadequate taxonomy, or incorrect identification (Meyer and Paulay, 2005; Ivanova et al., 2007). Hybridization is a problem for DNA barcode delimitation for matrilineal inheritance. The two carangids Caranxignobilis and C. sexfasciatus were artificially crossed with C. melampygus, with C. melampygus demonstrated to be the maternal parent of the hybrid (using 16S and COI sequences) (Murakami et al., 2007). Murosaba was even considered to be a natural hybrid between Scomber australasicus and D. muroadsi, but mitochondrial and nuclear markers demonstrated this to be untrue (Yanagimoto and Mahito, 2015). No further evidence proves that hybridization and introgression naturally occur in the Carangidae, especially in Genus Decapterus.
Some closely related species have recently diverged and may be closely genetically related or/and even share identical haplotypes in isolated gene pools, e.g., in tunas (Arnaud et al., 1999; Vinas and Tudela, 2009; Chang et al., 2017; Hou et al., 2018). However, the haplotype sharing may not have been found in Genus Decapterus (Figure 3 and Supplementary Table S3). Due to poor taxonomy, two species may share the same DNA barcode if nominal species come from the same gene pool, and were in fact conspecific (Dahruddin et al., 2017). Homonyms, synonyms, and misidentifications within the reference library could also introduce cases of apparent haplotype sharing or deep intraspecific divergence (Chakraborty et al., 2006; Tzeng et al., 2007; Chen et al., 2020). For Decapterus species, misidentification may be the main reason for the haplotype sharing, according to the different retrieval systems, categories, books, or multiple independent identifications in different oceans (Sun and Chen, 2013). This could be proven by haplotype and genetic divergence analysis in our study (Figure 3).
Morphology and Identification
Photographs can be used to describe egg morphology for species identification and would be a valuable approach in ecological studies combining DNA barcode analysis and morphology (Kawakami et al., 2010; Harada et al., 2015). Eggs of the four Decapterus species were similar in their spherical shape, smooth chorion, narrow perivitelline space after the myotome stage, and in having melanophores distributed in myotome—features that may inadequately separate taxa. Nishiyama et al. (2014) suggested that egg diameter and yolk segmentation may be diagnostic characteristics for differentiation of Trachurus japonicas and D. maruadsi eggs. The average diameter of D. maruadsi eggs was smaller than for the three other Decapterus species, but ranges in diameter overlap (Figure 5); accordingly, egg diameter can be used to differentiate fish eggs to specific taxa but not in species level. We report the egg diameter of D. maruadsi as ranging from 0.63 to 0.85 mm, similar to dimensions reported from China (0.67–0.85 mm, Zhang et al., 1985) and Japan (0.67–0.80 mm, Ikeda et al., 2014). However, our D. macrosma (0.74–0.86 mm) and D. macarellus (0.81–0.82 mm) egg diameters differ from previous measurements reported for these two species (0.6–0.64 mm and 0.6 mm, respectively) by Zhang et al. (1985).
We summarize information on eggs of nine fish species with similar morphology, including the eggs of Parapristipoma trilineatum and Selar crumenopthalmus that occurred simultaneously in the field surveys (Supplementary Table S4; Mito, 1966). Yolk segmentation can be observed in formalin-preserved eggs of T. japonicus, D. maruadsi, P. trilineatum, S. crumenopthalmus, and S. boops, but not for the alcohol-preserved eggs—species whose eggs have similar egg diameter and oil globule diameter. Matuda (1969) and Ikeda et al. (2014) emphasized the importance of spawning seasons for fish egg identification; however, the spawning seasons of these nine species overlap in the northern SCS, so this cannot be used to differentiate eggs either (Matuda, 1969). We even found that the eggs of D. macrosma identified by molecular method co-occurred in the survey of April and August–October, much longer than reported by Zhang et al. (1985). Thus, using morphological characteristics, i.e., egg diameter, oil globule, yolk segmentation, melanophore drops, etc., together with spawning season information can partly differentiate the fish eggs to some special taxa, but cannot differentiate them to species level. This further highlights the importance and benefits of using a DNA barcode method for fish egg species identification and spawning ground surveys in the subtropical tropical waters in the SCS.
Spawning Ground Seasonal Distribution
We report seasonal distribution patterns of Decapterus eggs in the northern SCS. Eggs of D. maruadsi occurred mainly off the Pearl River Estuary in spring, over 40–100 m, identifying this area as a key spawning ground for this species; eggs were not, however, found in autumn. Eggs of D. macrosoma occurred during both spring and autumn, mainly over 45–118 m (Table 2). Eggs of D. macrosoma co-occurred with those of D. maruadsi at one station only in spring, probably indicating a greater spawning area further south where water temperature and salinity were higher (Table 2). Compared with surveys in the 1960s and 1970s, no eggs of D. maruadsi occurred west of the Pearl River estuary or Hailing Islands, indicating that the spawning sea area may have decreased in these two areas (South China Sea Fisheries Research Institute [SCSFRI], 1966, 1979). As one of the last remaining core fishing grounds in the SCS, human activities such as overfishing, aquaculture, and coastal reclamation have already contributed to the collapse of many fish resources and damaged key habitat (Li and Huang, 2008; Darwall and Freyhof, 2016; Amorim et al., 2017; Hamilton et al., 2017). Furthermore, the preferred temperature range of D. maruadsi (<28°C) indicates that a shift in its spawning habitat may be a response to climate change (Kienast et al., 2001; Liu et al., 2008; Wernberg et al., 2016; Cai et al., 2017; Asch and Erisman, 2018; Asch et al., 2019; Sandø et al., 2020). For D. macrosma, Zhang et al. (1985) reported its spawning season to continue from March to July, but we identified 56 eggs by molecular method from eight stations from August to October, indicating that the spawning period of this species has extended.
Fishery Conservation and Management
We demonstrate the value of DNA barcoding for identifying fish eggs and for this technique to assist with identification of fish eggs in more traditional morphological studies. Spawning ground protection and moratoria on fishing activities in the SCS should take into consideration possible losses of D. maruadsi spawning habitat and the extension of the spawning season of D. macrosma. The SCS is a complex environment, with high fish diversity and inconspicuous variation in water temperature, so fish egg morphology, spawning season, spawning ground distribution, and hydrological conditions in this region require further study.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: MT609955–M610035).
Ethics Statement
The animal study was reviewed and approved by the Department of Ocean and Fishery, Guangdong Province.
Author Contributions
GH and HZ analyzed the data and completed the first draft. GH and ZC performed the filed survey. ZC and HZ provided the guidance on data analysis and structure. GH, JW, JZ, and WH conducted the experiments. All authors contributed to the article and approved the submitted version.
Funding
This study was financially supported by the Natural Science Foundation of China (No. 31702347), the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDB42000000), the Special Fund for Economic Development of Marine Economy of Guangdong Province (No. GDME-2018E004), the start-up project of Guangdong Ocean University (No. R19006), and Youth Innovation Promotion Association CAS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank colleagues for their generous support of the study, including Bo Feng and Xuefeng Wang. We thank all staff in research vessels Zhongyuke 301 and Beiyu 60011 and students who helped during the surveys and dealt with the samples in the lab. We also thank the editor and reviewers for their constructive comments on our present work. We thank Dr. Steve O’Shea for editing the English text of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.590564/full#supplementary-material
Footnotes
- ^ http://www.barcodinglife.org and http://www.boldsystems.org
- ^ http://www.boldsystems.org/
- ^ https://www.ncbi.nlm.nih.gov/genbank/
References
Ahern, A. L. M., Gómez-Gutiérrez, J., Aburto-Oropeza, O., Saldierna-Martínez, R. J., Johnson, A. F., Harada, A. E., et al. (2018). DNA sequencing of fish eggs and larvae reveals high species diversity and seasonal changes in spawning activity in the southeastern Gulf of California. Mar. Eco. Prog. Ser. 592, 159–179. doi: 10.3354/meps12446
Amorim, E., Ramos, S., Elliott, M., Franco, A., and Bordalo, A. A. (2017). Habitat loss and gain: influence on habitat attractiveness for estuarine fish communities. Estuar. Coast. Shelf. S. 197, 244–257. doi: 10.1016/j.ecss.2017.08.043
Arnaud, S., Bonhomme, F., and Borsa, P. (1999). Mitochondrial DNA analysis of the genetic relationships among populations of scad mackerel (Decapterus macarellus, D. macrosoma and D. russelli) in South-East Asia. Mar. Biol. 135, 699–707. doi: 10.1007/s002270050671
Asch, R. G., and Erisman, B. (2018). Spawning aggregations act as a bottleneck influencing climate change impacts on a critically endangered reef fish. Divers. Distrib. 24, 1712–1728. doi: 10.1111/ddi.12809
Asch, R. G., Stock, C. A., and Sarmiento, J. L. (2019). Climate change impacts on mismatches between phytoplankton blooms and fish spawning phenology. Glob. Chang. Biol. 25, 2544–2559. doi: 10.1111/gcb.14650
Cai, R., Guo, H., Fu, D., Yan, X., and Tan, H. (2017). “Response and adaptation to climate change in the South China Sea and coral sea,” in Climate Change Adaptation in Pacific Countries. Climate Change Management, ed. W. Leal Filho (Cham: Springer), 163–176. doi: 10.1007/978-3-319-50094-2_10
Chakraborty, A., Aranishi, F., and Iwatsuki, Y. (2006). Genetic differentiation of Trichiurus japonicus and T. lepturus (Perciformes: Trichiuridae) based on mitochondrial DNA analysis. Zool. S. 45:419.
Chambers, R. C., and Trippel, E. A. (1997). “Early life history and recruitment: legacy and challenges,” in Early Life History and Recruitment in Fish Populations, Vol. 21, eds R. C. Chambers and T. Edward (Dordrecht: Springer), 515–549. doi: 10.1007/978-94-009-1439-1
Chang, C. H., Shao, K. T., Lin, H. Y., Chiu, Y. C., Lee, M. Y., Liu, S. H., et al. (2017). DNA barcodes of the native ray-finned fishes in Taiwan. Mol. Ecol. Resour. 17, 796–805. doi: 10.1111/1755-0998.12601
Checkley, D., Alheit, J., Oozeki, Y., and Roy, C. (2009). Climate Change and Small Pelagic Fish. Cambridge: Cambridge University Press.
Chen, L. C., Lan, K. W., Chang, Y., and Chen, W. Y. (2018). Summer assemblages and biodiversity of larval fish associated with hydrography in the Northern South China Sea. Mar. Coast. Fish. 10, 467–480. doi: 10.1002/mcf2.10037
Chen, Z., Song, N., Zou, J., Qin, Y., Ma, L., and Gao, T. (2020). Identification of Species in Genus Platycephalus from Seas of China. J. Ocean China 19, 417–427. doi: 10.1007/s11802-020-4158-1
China Agriculture Press (1997-*2019). China Fishery Statistical Yearbook in 1997-2019. Beijing: China Agriculture Press.
Clement, M., Posada, D., and Crandall, K. A. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x
Dahruddin, H., Hutama, A., Busson, F., Sauri, S., Hanner, R., Keith, P., et al. (2017). Revisiting the ichthyodiversity of Java and Bali through DNA barcodes: taxonomic coverage, identification accuracy, cryptic diversity and identification of exotic species. Mol. Ecol. Resour. 17, 288–299. doi: 10.1111/1755-0998.12528
Darwall, W. R., and Freyhof, J. (2016). “Conservation of freshwater fishes,” in Lost Fishes, Who is Counting? The Extent of the Threat to Freshwater Fish Biodiversity, eds G. P. Closs, M. Krkosek, and J. D. Olden (Cambridge: Cambridge University Press), 1–36. doi: 10.1017/CBO9781139627085.002
Food and Agricultural Organization [FAO] (2019). FAO Yearbook: Fishery and Aquaculture Statistics. Rome: FAO.
Frantine-Silva, W., Sofia, S., Orsi, M., and Almeida, F. (2015). DNA barcoding of freshwater ichthyoplankton in the Neotropics as a tool for ecological monitoring. Mol. Ecol. Resour. 15, 1226–1237. doi: 10.1111/1755-0998.12385
Froese, R., and Pauly, D. (2010). FishBase: Fisheries Centre. Vancouver: University of British Columbia.
Fuiman, L. (2002). “Special considerations of fish eggs and larvae,” in Fishery Science, The Unique Contributions of Early Life Stages, eds L. A. Fuiman and R. G. Werner (Hoboken, NJ: Blackwell Science).
Hamilton, R. J., Almany, G. R., Brown, C. J., Pita, J., Peterson, N. A., and Choat, J. H. (2017). Logging degrades nursery habitat for an iconic coral reef fish. Biolo. Conser. 210, 273–280. doi: 10.1016/j.biocon.2017.04.024
Harada, A. E., Lindgren, E. A., Hermsmeier, M. C., Rogowski, P. A., Terrill, E., and Burton, R. S. (2015). Monitoring spawning activity in a southern California marine protected area using molecular identification of fish eggs. PLoS One 10:e0134647. doi: 10.1371/journal.pone.0134647
Hou, G., Chen, W. T., Lu, H. S., Cheng, F., and Xie, S. G. (2018). Developing a DNA barcode library for perciform fishes in the South China Sea: species identification, accuracy and cryptic diversity. Mol. Ecol. Resour. 18, 137–146. doi: 10.1111/1755-0998.12718
Hubert, N., Espiau, B., Meyer, C., and Planes, S. (2015). Identifying the ichthyoplankton of a coral reef using DNA barcodes. Mol. Ecol. Resour. 15, 57–67. doi: 10.1111/1755-0998.12293
Ikeda, T., Hirai, A., Tabata, S., Onishi, Y., and Mito, S. (2014). “Key to fish eggs in Japan,” in An Atlas of the Early Stage Fishes In Japan, 2nd Edn, ed. M. Okiyama (Kanagawa: Tokai University Press), 1–108.
Ivanova, N. V., Zemlak, T. S., Hanner, R. H., and Hebert, P. D. (2007). Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Resour. 7, 544–548. doi: 10.1111/j.1471-8286.2007.01748.x
Jaafar, T. N. A. M., Taylor, I., Nor, S. A. M., de Bruyn, M., and Carvalho, G. R. (2012). DNA barcoding reveals cryptic diversity within commercially exploited Indo-Malay Carangidae (Teleosteii: Perciformes). PLoS One 7:e49623. doi: 10.1371/journal.pone.0049623
Kasai, A., Komatsu, K., Sassa, C., and Konishi, Y. (2008). Transport and survival processes of eggs and larvae of jack mackerel Trachurus japonicus in the East China Sea. Fish. Sci. 74, 8–18. doi: 10.1111/j.1444-2906.2007.01491.x
Kawakami, T., Aoyama, J., and Tsukamoto, K. (2010). Morphology of pelagic fish eggs identified using mitochondrial DNA and their distribution in waters west of the Mariana Islands. Environ. Biol. Fish. 87, 221–235. doi: 10.1007/s10641-010-9592-2
Kienast, M., Steinke, S., Stattegger, K., and Calvert, S. (2001). Synchronous tropical South China Sea SST change and Greenland warming during deglaciation. Science 291, 2132–2134. doi: 10.1126/science.1057131
Lainez del Pozo, D. (2013). Potential Contribution of Small Pelagic Fish to Food Security Within The Asia-Pacific Region. Singapore: Asia-Pacific Economic Cooperation.
Lewis, L. A., Richardson, D. E., Zakharov, E. V., and Hanner, R. (2016). Integrating DNA barcoding of fish eggs into ichthyoplankton monitoring programs. Fish. Bull. 114, 153–165. doi: 10.7755/FB.114.2.3
Leyva-Cruz, E., Vásquez-Yeomans, L., Carrillo, L., and Valdez-Moreno, M. (2016). Identifying pelagic fish eggs in the southeast Yucatan Peninsula using DNA barcodes. Genome 59, 1117–1129. doi: 10.1139/gen-2015-0151
Li, B., and Huang, G. (2008). Ecology effect and countermeasure of urbanization in Pearl River Estuary. Mar. Environ. Sci. 27, 543–546.
Li, K., Yin, J., Huang, L., and Lin, Z. (2014). Seasonal variations in diversity and abundance of surface ichthyoplankton in the northern South China Sea. Acta Oceanol. Sin. 33, 145–154. doi: 10.1007/s13131-014-0533-3
Li, S., Zuozhi, C., Peng, Z., Jie, L., Huanhuan, W., and Jiaxing, H. (2018). Catch composition and spatial-temporal distribution of catch rate of light falling-net fishing in central and southern South China Sea fishing ground in 2017, South China. Fish. Sci. 14, 11–20.
Liu, Y.-J., Yan, J.-Y., and Song, Y.-L. (2008). Features of climate change over the Xishan Island over the South China Sea in Recent 50 years. Sci. Geogr. Sin. 6, 804–808. doi: 10.13249/j.cnki.sgs.2008.06.804
Ma, S., Cheng, J., Li, J., Liu, Y., Wan, R., and Tian, Y. (2019). Interannual to decadal variability in the catches of small pelagic fishes from China Seas and its responses to climatic regime shifts. Deep Sea Res. Pt. II 159, 112–129. doi: 10.1016/j.dsr2.2018.10.005
Matuda, S. (1969). The studies on fish eggs and larvae occurred in the Nansei regional waters of Japan—1. Species occurred and there seasonal variation. Bull. Nansei. Reg. Fish. Res. Lab. 2, 49–83.
Meyer, C. P., and Paulay, G. (2005). DNA barcoding: error rates based on comprehensive sampling. PLoS. Biol. 3:e422. doi: 10.1371/journal.pbio.0030422
Mito, S. (1966). “Fish eggs and larvae,” in Illustrated Encyclopedia of the Marine Plankton of Japan, Vol. 7, ed. S. Motoda (Tokyo: SoyoSha), 1–74.
Murakami, K., James, S. A., Randall, J. E., and Suzumoto, A. Y. (2007). Two hybrids of carangid fishes of the genus Caranx, C. ignobilis × C. melampygus and C. melampygus × C. sexfasciatus, from the Hawaiian Islands. Zool. S. 46:186.
Neira, F. J., Perry, R. A., Burridge, C. P., Lyle, J. M., and Keane, J. P. (2015). Molecular discrimination of shelf-spawned eggs of two co-occurring Trachurus spp.(Carangidae) in southeastern Australia: a key step to future egg-based biomass estimates. ICES J. Mar. Sci. 72, 614–624. doi: 10.1093/icesjms/fsu151
Nishiyama, M., Saito, M., Sanada, Y., Onoue, S., Takasuka, A., and Oozeki, Y. (2014). Revisiting morphological identification of Japanese jack mackerel Trachurus japonicus eggs preserved in formalin. Fish. Sci. 80, 517–529. doi: 10.1007/s12562-014-0732-z
Ratnasingham, S., and Hebert, P. D. (2007). Bold: the barcode of life data system. Mol. Ecol. Resour. 7, 355–364. doi: 10.1111/j.1471-8286.2007.01678.x
Sandø, A. B., Johansen, G. O., Aglen, A., Stiansen, J. E., and Renner, A. H. (2020). Climate change and new potential spawning sites for Northeast Arctic cod. Front. Mar. Sci. 7:28. doi: 10.3389/fmars.2020.00028
Sciascia, R., Berta, M., Carlson, D. F., Griffa, A., Panfili, M., La Mesa, M., et al. (2018). Linking sardine recruitment in coastal areas to ocean currents using surface drifters and HF radar: a case study in the Gulf of Manfredonia. Adriatic Sea Ocean. Sci. 14:1461. doi: 10.5194/os-14-1461-2018
Shao, K.-T., Chen, K.-C., and Wu, J.-H. (2002). Identification of marine fish eggs in Taiwan using light microscopy, scanning electric microscopy and mtDNA sequencing. Mar. Freshw. Res. 53, 355–365. doi: 10.1071/MF01141
Shao, K.-T., Yang, J.-S., Chen, K.-C., and Lee, Y.-S. (2001). An Idenfification Guide of Marine Fish Eggs From Taiwan. Institude of Zoologe Academia Sinica, Taibei: Taiwan Power Company.
South China Sea Fisheries Research Institute [SCSFRI] (1966). The investigation report of fishery resources by bottom trawl survey in Northern South China Sea (East of Hainan Island). SCSFRI 3, 117–121.
South China Sea Fisheries Research Institute [SCSFRI] (1979). The investigation report of fishery resources by bottom trawl survey off the continental shelf in Northern South China Sea. SCSFRI 271–274.
Sun, D. R., and Chen, Z. (2013). Fish categories books in the South China Sea. London: Maritime Press.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Templeton, A. R., Crandall, K. A., and Sing, C. F. (1992). A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132, 619–633.
Tzeng, C., Chen, C., and Chiu, T. (2007). Analysis of morphometry and mitochondrial DNA sequences from two Trichiurus species in waters of the western North Pacific: taxonomic assessment and population structure. Fish J. Biol. 70, 165–176. doi: 10.1111/j.1095-8649.2007.01368.x
Valdez-Moreno, M., Vásquez-Yeomans, L., Elías-Gutiérrez, M., Ivanova, N. V., and Hebert, P. D. (2010). Using DNA barcodes to connect adults and early life stages of marine fishes from the Yucatan Peninsula, Mexico: potential in fisheries management. Mar. Freshw. Res. 61, 655–671. doi: 10.1071/MF09222
Vinas, J., and Tudela, S. (2009). A validated methodology for genetic identification of tuna species (genus Thunnus). PLoS One 4:e7606. doi: 10.1071/MF09222
Ward, R. D., Zemlak, T. S., Innes, B. H., Last, P. R., and Hebert, P. D. (2005). DNA barcoding Australia’s fish species. Philos. Soc. Trans. R. B. 360, 1847–1857. doi: 10.2307/30040931
Watson, S. A., Allan, B. J., McQueen, D. E., Nicol, S., Parsons, D. M., Pether, S. M., et al. (2018). Ocean warming has a greater effect than acidification on the early life history development and swimming performance of a large circumglobal pelagic fish. Glob. Chang. Biol. 24, 4368–4385. doi: 10.1111/gcb.14290
Wernberg, T., Bennett, S., Babcock, R. C., De Bettignies, T., Cure, K., Depczynski, M., et al. (2016). Climate-driven regime shift of a temperate marine ecosystem. Science 353, 169–172. doi: 10.1126/science.aad8745
Xie, S., and Watanabe, Y. (2007). Transport-determined early growth and development of jack mackerel Trachurus japonicus juveniles immigrating into Sagami Bay. Jpn. Mar. Freshw. Res. 58, 1048–1055. doi: 10.1071/MF06165
Yanagimoto, T., and Mahito, M. (2015). Species identification of Murosaba that was considered natural hybridization between Spotted mackerel Scomber australasicus and Brownstriped mackerel Decapterus muroadsi inferred from DNA analysis. DNA Ident. 7, 45–50.
Yang, L., Zhang, X., Tan, Y., and Zhang, P. (2009). Analysis on the catch composition of light-purse seine in the northern South China Sea. S. China Fish. Sci. 5, 65–70. doi: 10.3969/j.issn.1673-2227.2009.06.012
Zhang, R., Lu, S., Zhao, C., Chen, L., Zang, Z., and Zhang, X. (1985). Fish Eggs and Larvae in the Offshore Waters of China. Shanghai: Shanghai Scientific and Technological Press.
Keywords: fish eggs, DNA barcode, morphology, Decapterus, spawning ground, northern South China Sea
Citation: Hou G, Wang J, Chen Z, Zhou J, Huang W and Zhang H (2020) Molecular and Morphological Identification and Seasonal Distribution of Eggs of Four Decapterus Fish Species in the Northern South China Sea: A Key to Conservation of Spawning Ground. Front. Mar. Sci. 7:590564. doi: 10.3389/fmars.2020.590564
Received: 01 August 2020; Accepted: 21 October 2020;
Published: 12 November 2020.
Edited by:
Zhiqiang Han, Zhejiang Ocean University, ChinaReviewed by:
Binbin Shan, South China Sea Fisheries Research Institute (CAFS), ChinaTakashi Yanagimoto, Japan Fisheries Research and Education Agency, Japan
Linlin Zhao, First Institute of Oceanography, Ministry of Natural Resources, China
Copyright © 2020 Hou, Wang, Chen, Zhou, Huang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zuozhi Chen, Y2hlbnp1b3poaUBzY3NmcmkuYWMuY24=; Hui Zhang, emhhbmdodWlAcWRpby5hYy5jbg==
 Gang Hou
Gang Hou Jinrun Wang
Jinrun Wang Zuozhi Chen
Zuozhi Chen Jinlong Zhou
Jinlong Zhou Wangsu Huang1
Wangsu Huang1 Hui Zhang
Hui Zhang