
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 22 October 2020
Sec. Aquatic Microbiology
Volume 7 - 2020 | https://doi.org/10.3389/fmars.2020.585970
This article is part of the Research Topic Microbial Communities of Coastal Eutrophic Systems View all 10 articles
Microbial communities are important indicators of aquatic ecosystem health, responsible for functional roles sustaining the ecosystem such as nutrient cycling, as well as environmental problems. Urbanized estuaries are vulnerable intersections between terrestrial and marine ecosystems and are susceptible to pressure and perturbation from both natural processes and human activities over time. The response by microbial communities toward changes in the environment should be closely monitored and studied. In this study, microbial communities in brackish intertidal sediments, sampled seasonally over a year along the coast of Yangtze River estuary, were analyzed using 16S rRNA gene sequencing. Potential compositional and functional changes in microbial communities resulting from temporal variation and associated physiochemical gradients in the environment were studied and notable patterns were observed over time. Summer season was recorded with the lowest microbial community α-diversity. For β-diversity, spatiotemporal differentiation in community structures was recorded with seasonal clustering. Proteobacteria was recorded as the most dominant phylum among all sampling sites throughout the year but its relative abundance showed no temporal changes. Instead, relative abundances of phyla Actinobacteria, Chloroflexi, and Verrucomicrobia were found to differ significantly over time, generally favoring the wet Spring and Summer. In contrast, temporal variation was observed for the class Deltaproteobacteria and some of the corresponding sulfate/sulfur-reducing bacterial genera, favoring the dry Autumn and Winter. However, there were no major temporal changes in ecological functions contributed by microbial communities throughout the study period. While results from redundancy analysis showed that the subtle changes in pH in the environment had the greatest impact on microbial community composition, the variation it explained remained relatively low. Alternatively, precipitation, distinguishing the dry and wet seasons of the subtropical monsoon climate zone, was suggested to be another potential key factor influencing microbial community composition and their relative abundances. The present study provides new data and insights on the impact of temporal variation on microbial community composition and ecological function in an urbanized estuarine ecosystem.
Microbes are important components of estuarine ecosystems responsible for driving many biogeochemical processes, ranging from regulatory functions for the cycling of nutrients, such as nitrogen, phosphorus, and sulfur (Falkowski et al., 2008), to biodegradation and decomposition of pollutants (Xie et al., 2016). However, the role and performance of microbial communities are susceptible to impact from changing environments (Suh et al., 2015).
Estuarine intertidal mudflats are known interactive zones between terrestrial and marine ecosystems. Intertidal sediment conditions can constantly change due to different natural processes such as freshwater input, terrestrial runoff, and incoming tides, as well as anthropogenic impact (Guo et al., 2018). This variability has resulted in microbial communities in intertidal sediments displaying richer composition and overall biodiversity, compared to marine ecosystems (Wang et al., 2012). In particular, the surface sediment layer has the highest microbial activity; the sensitive microbial communities from surface sediments can also be indicators of ecological health (Duarte et al., 2012; Kallmeyer et al., 2012). The status of the active and diverse microbial communities in estuarine ecosystems has great economic and ecological significance (Grizzetti et al., 2019).
In terms of environmental changes, temporal variation such as the succession of seasons can be an important factor, leading to differences in tides and precipitation. Subsequently, the amount of dissolved oxygen in surface sediments are often dynamic (Murphy et al., 2011; Peterson et al., 2013). Oxic-anoxic conditions have knock-on effects on the nutrient conditions of the environment and potential respiration rates of microbes (Karhu et al., 2014). Seasonal changes may have notable effects on microbial community composition patterns and corresponding ecological functions due to microbial adaptation (Gilbert et al., 2012; Aguirre et al., 2017). For marine ecosystems, previous long-term monitoring study showed that microbial communities can display seasonal patterns, and the data can be used to predict the effects microbial communities have on geochemical cycles (Giovannoni and Vergin, 2012). For urbanized estuarine ecosystems, spatiotemporal dynamics of microbial communities can be important indicators of water quality and ecological conditions, with human health consequences (Kaestli et al., 2017). Therefore, microbial ecology studies on community-level responses to temporal variations have important implications. They can provide beneficial data to help evaluate relevant long-term impact imposed on an ecosystem, and to identify patterns in microbial community diversity and ecological functions, especially for an ever-changing environment.
The urbanized Yangtze River estuary receives freshwater from Yangtze River and its tributaries and discharges to the East China Sea. With anthropogenic input including nutrients (Liu et al., 2003) and pollutants (Shi et al., 2014), Yangtze River estuary is a complex hydrological environment with rich fisheries resources and great economic significance (Kindong et al., 2020). Yangtze River is also a source of drinking water for nearby cities. The importance of Yangtze River estuary is well established, but to our knowledge, there are very limited field monitoring data available on the potential temporal or seasonal patterns of microbial communities in the estuarine intertidal mudflats, as previous studies have tended to focus on spatial differences or specific groups of bacteria (Guo et al., 2018; Niu et al., 2018; Xu et al., 2018). The advances in sequencing methods such as high throughput 16S rRNA gene sequencing have facilitated microbial studies on a community-level, allowing research on community composition, phylogeny, and ecology (Dyksma et al., 2016; Fan et al., 2016), as opposed to previous species-level approach. In the present study, microbial communities in the Yangtze River estuary, obtained along the brackish boundary region approaching the East China Sea, were characterized. Microbial communities were sampled seasonally over the course of one year with the aim to monitor and identify potential patterns and shifts in microbial community composition and corresponding ecological functions with changing seasons. Environmental factors which may be responsible for potential differences were evaluated to determine their correlation and contribution to compositional and functional changes. Results will help to inform and allow for timely decision making to better manage the valuable ecosystem. This study contributes to the understanding of microbial communities in coastal waters and will be a useful reference for future studies to evaluate impact on estuarine ecosystems.
The Yangtze River estuary is the mouth of an urbanized waterway, carrying significant volumes of freshwater from Yangtze, subsidiary rivers, and lakes to the East China Sea. The Yangtze River is located in the subtropical monsoon climate zone, with warm, humid summer seasons during May to August, and cool, dry winter seasons during December to February, with an annual temperature range between 6 to 28°C, averaging at 16°C. The annual average precipitation is around 1057 mm, recorded mainly during the Summer season between May to October (Shen, 2020).
In this study, estuarine sediment samples were collected at seasonal intervals on April 12th (Spring), July 23rd (Summer), November 13th (Autumn) of 2016 and January 10th (Winter) of 2017. Five sampling sites were selected along the Yangtze River estuary (Figure 1), including Wusongkou (WSK, 31°24′ N, 121°30′ E), Shidongkou (SDK, 31°28′ N, 121°23′ E), Liuhekou (LHK, 31°31′ N, 121°17′ E), Qiyakou (QYK, 31°36′ N, 121°14′ E) and Xupu (XP, 31°45′ N, 120°57′ E) (Supplementary Figures 1–5). Sampling sites were coastal brackish intertidal mudflats. Notably, a wastewater treatment plant outlet is located near the SDK sampling site. For each season, intertidal sediments for all five sampling sites were collected on the same day. Dates and time were chosen according to the tidal table to allow efficient sample collection during low tide period for all sites.
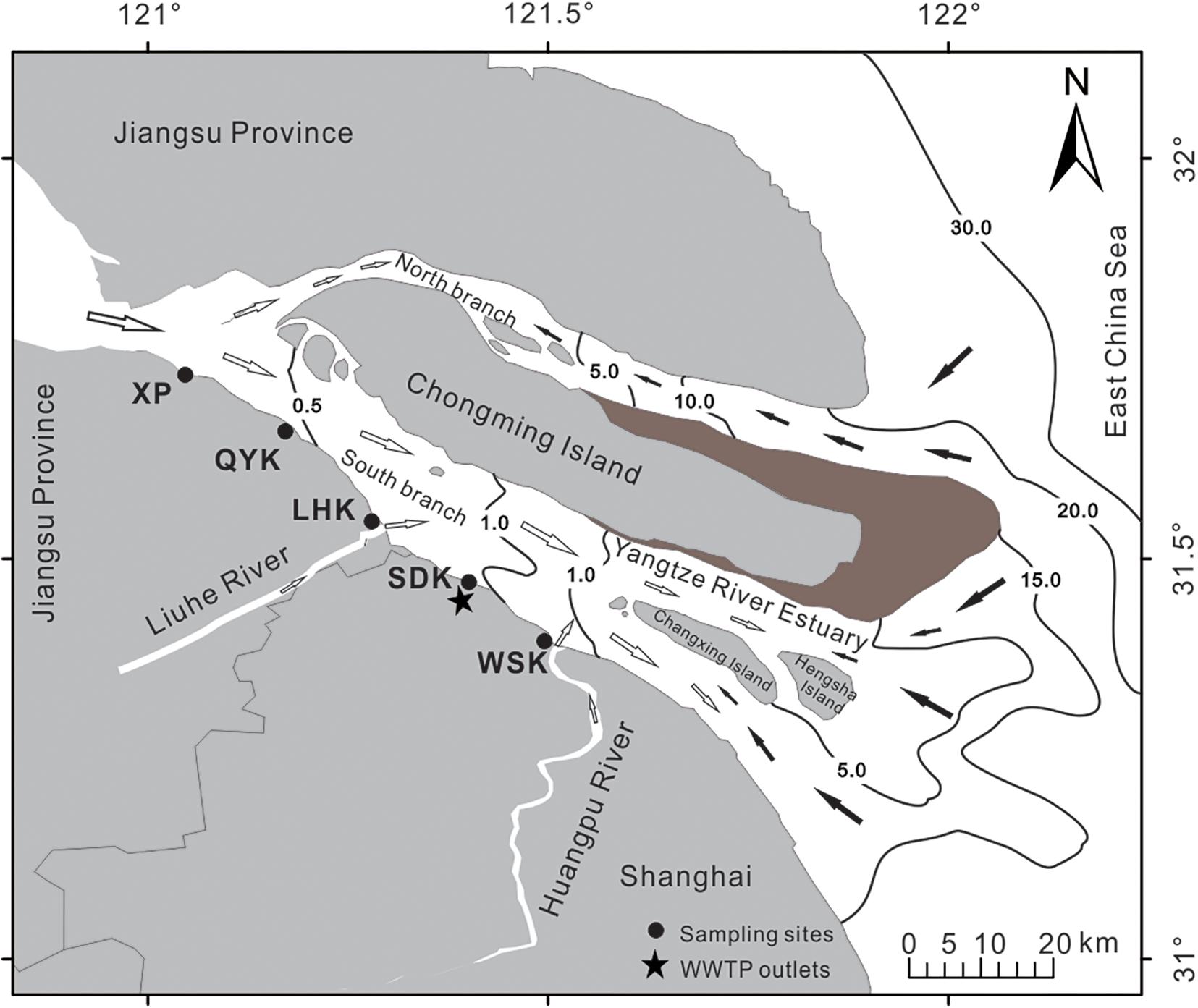
Figure 1. Map of the locations of sampling sites along the Yangtze River estuary tidal mudflats. WSK = Wusongkou; SDK = Shidongkou; LHK = Liuhekou; QYK = Qiyakou; XP = Xupu. Black and white arrows indicate direction of saltwater and freshwater, respectively. Numbers on black lines represent salinity gradients. The black star indicates the outlet of Shidongkou wastewater treatment plant.
A sterile polyethylene shovel was used for sample collection. Above surface sediments (0 - 5 cm) were removed using the shovel before collection of sediment samples (5 - 7 cm). At each site, samples were collected in triplicates and pooled together as one representative sample for subsequent analytical purposes. Over the year, a total of 60 samples were collected and pooled into 20 samples. Samples collected were placed in individual sterile plastic bags in an ice box and transported back to laboratory on the same day. At the laboratory, samples collected were separated into two portions. A portion was stored at 4°C for the characterization of physicochemical properties of the sediments. The other portion was stored at −80°C for DNA extraction. All samples were prepared and handled within seven days after collection.
Physicochemical properties of sediment samples were measured either in situ or in the laboratory. Salinity of pore water was measured in situ using a portable salinity meter (Model 30, YSI Incorporated, United States). Similarly, pH of pore water was measured in situ using a portable pH meter (SIN-PH170, Hangzhou Sinomeasure Automation, China). Sediment samples were then transported to the laboratory and dried at 50°C before measurement of grain size and nutrient concentrations. The grain size of sediments was analyzed using a laser granulometer (LS 13320, Beckman Coulter Corporation, United States). Total nitrogen (TN) concentration in sediments was measured using an elemental analyzer (Vario EL III, Elementar, Germany). Total organic carbon (TOC) concentration in sediments was determined using the K2Cr2O7 redox titration method. Total phosphorus (TP) concentration in sediments was determined using the Murphy-Riley modified solution method (Murphy and Riley, 1962).
DNA extraction and isolation from sediment samples were carried out using the E.Z.N.A.TM Soil DNA Kit (D5625-01, Omega Bio-tek, Inc., United States). Extracted DNA were measured through quantitation assays with a Qubit 2.0 Fluorometer (Invitrogen, United States) for subsequent PCR. Primers used for the amplification of bacterial 16S rRNA gene were 314F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′), targeting the V3–V4 hypervariable regions (Herlemann et al., 2011). A 30 μL PCR was carried out, containing: 15 μL of 2 × Taq Master Mix, 1 μL of Bar-PCR primer F (10 μM), 1 μL of Primer R (10 μM), 20 μg of DNA template, and deionized water. PCR was conducted under the following cycling conditions: initial denaturation at 94°C for 3 min; followed by 5 cycles of amplification in which each cycle consisted of denaturation at 94°C for 30 s, primer annealing at 45°C for 20 s and extension at 65°C for 30 s; followed by 20 cycles of 94°C for 20 s, 55°C for 20 s and 72°C for 30 s; followed by final extension at 72°C for 5 min. Products of the first PCR were used as the template for subsequent PCR, with the addition of primers allowing for IlluminaTM Bridge Amplification. Second round of PCR was conducted under the following cycling conditions: initial denaturation at 95°C for 3 min; followed by 5 cycles of amplification in which each cycle consisted of denaturation at 94°C for 30 s, primer annealing at 55°C for 20 s and extension at 72°C for 30 s; followed by final extension at 72°C for 5 min. Amplicon products were quantified using Qubit 2.0 Fluorometer. Finally, for each sample, 10 ng (20 pmol) of the product were paired-end sequenced using the IlluminaTM MiSeq sequencing platform (Illumina, United States), conducted at Sangon Biotech (Shanghai, China). Sequences generated from the samples used in this study have been deposited in the Sequence Read Archive (SRA) database of the NCBI. Accession numbers for the 20 samples collected from the Yangtze River estuary are SRX 3851127 - 3851146.
Sequencing data were analyzed using the data processing tool Quantitative Insights Into Microbial Ecology 2 (QIIME2) (Bolyen et al., 2019). After importing raw sequencing data (FASTQ) into QIIME2, sequence reads from different samples were demultiplexed according to the barcode sequences. The bacterial paired-end reads were then merged using the VSEARCH pipeline (Rognes et al., 2016). After filtering chimeric and short sequences, reads were clustered into OTUs using closed-reference OTU picking based on a 98% similarity cutoff (Edgar et al., 2011; Rognes et al., 2016).
Classifying OTUs into taxonomy can allow further analysis of the microbial community composition at different taxonomic ranks (Lan et al., 2012). Representative sequences from each OTU were assigned to taxonomy according to the SILVA r138.1 reference database using QIIME2 at > 93% confidence (Quast et al., 2013). The relative abundances of bacterial communities were studied at different taxonomic levels, such as phylum, class and genus. At each level, taxa were considered dominant if the average relative abundance was above 1%.
An array of α- and β-diversity measures were calculated using QIIME2. The α-diversity of bacterial communities was analyzed using ACE, Chao1 and Shannon’s indices, based on the assigned OTUs. Rarefaction curve for random sampling was established to assess species richness and diversity between samples (Supplementary Figure 6). The differences in α-diversity between microbial communities under the influence of different environmental factors were evaluated using Tukey’s HSD test followed by Pearson’s correlation.
The analysis of β-diversity was conducted through an unweighted UniFrac analysis, as it is sensitive toward rare species of low abundance and is more representative of microbial community diversity in the environment. Based on the UniFrac analysis, a principal coordinate analysis (PCoA) was used to compare microbial communities (Lozupone and Knight, 2005), using the QIIME2 pipeline. Differences between microbial communities were analyzed through Permutational Multivariate Analysis of Variance (PERMANOVA, Adonis test) using QIIME2 (Anderson, 2014).
Microbial ecological functions were annotated using Functional Annotation of Prokaryotic Taxa (FAPROTAX) based on OTU classification (Louca et al., 2016). FAPROTAX is one of the common predictive methods that utilizes 16S rRNA gene sequencing, extrapolating microbial taxa into putative functions based on literature and cultured microorganism database (Kumar et al., 2019).
Differences in physicochemical properties between collected samples were analyzed using one-way ANOVA and Tukey’s HSD test. Differences in relative abundances of microbial community compositions were analyzed using the Welch’s t-test (White et al., 2009) and Statistical analysis of metagenomics profiles (STAMP) tool (Parks et al., 2014).
Principal component analysis (PCA) and redundancy analysis (RDA) were then carried out using Canoco5 to investigate the relationships between environmental factors, microbial community composition and abundance, and ecological functions (Ter Braak and Šmilauer, 2002). PCA can reflect spatial and temporal variation in microbial communities. Based on PCA, RDA can reveal the impact of different environmental factors on the composition and abundance of microbial communities, and determine whether they contribute significantly to changes (Sun et al., 2018). The used data were log (x + 1) transformed prior to the RDA.
The correlation between environmental factors, relative abundances of bacterial taxa, and their contribution toward ecological functions were analyzed using Spearman’s correlation. Statistical analyses were carried out using the SPSS Statistics Version 22.0 (IBM Corp., United States) software. Differences were considered significant when P < 0.05.
Physicochemical properties of estuarine sediments were summarized in Table 1. The salinity of pore water from sampled areas were generally low, ranging between 0.1–0.4. The pH values ranged between 7.20–7.82 and displayed no significant temporal or spatial differences. Salinity, pH, and grain size of sediments in intertidal mudflats of the Yangtze River estuary showed low variance throughout the year, whereas nutrient concentrations showed greater variability. The 20 samples were further separated into four seasonal groups to identify seasonal patterns. Results showed that only TOC concentrations displayed a significant temporal change from Spring to Summer (P < 0.05). It is suspected that anthropogenic sources of wastewater input and Liuhe River may have an impact on the environmental conditions of downstream sampling sites along Yangtze River estuary, such as WSK and SDK. By grouping the collected data based on location, results revealed that the TN concentrations of WSK and SDK region were significantly higher than its counterpart from LHK, QYK and XP region (P = 0.03). Similar patterns were found for TP, but the difference was statistically insignificant (P = 0.07).
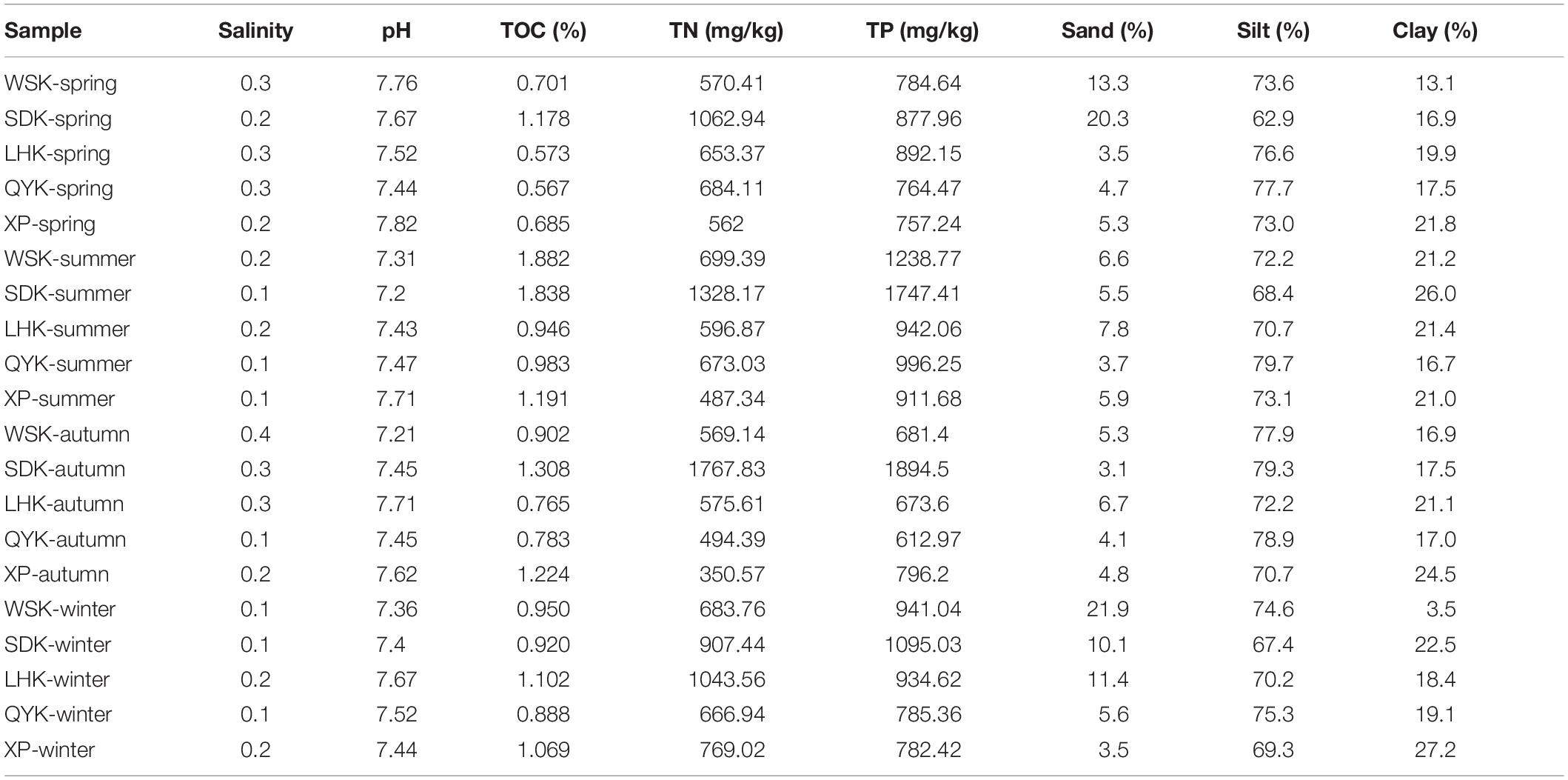
Table 1. Physicochemical properties of intertidal sediments along the Yangtze River estuary at different seasons.
The differences in α-diversity of microbial communities with changing seasons were calculated. A high Good’s coverage value showed that the samples could be considered representative of the intertidal sediments. Overall, the structure of microbial communities was relatively stable. When samples were grouped according to sampling season, the Shannon index showed no significant change in overall diversity across all four seasons. However, during Summer, the OTUs in microbial communities were significantly lower compared to other seasons (Tukey’s test, P < 0.05). Furthermore, the Chao1 index revealed that richness of microbial communities can show significant temporal differences, with Spring having the highest richness, followed by Winter, Autumn and Summer (Tukey’s test, P < 0.05). Notably, only the difference between Winter and Autumn was insignificant (Tukey’s test, P = 0.801). The α-diversity indices of the samples are summarized in Table 2.
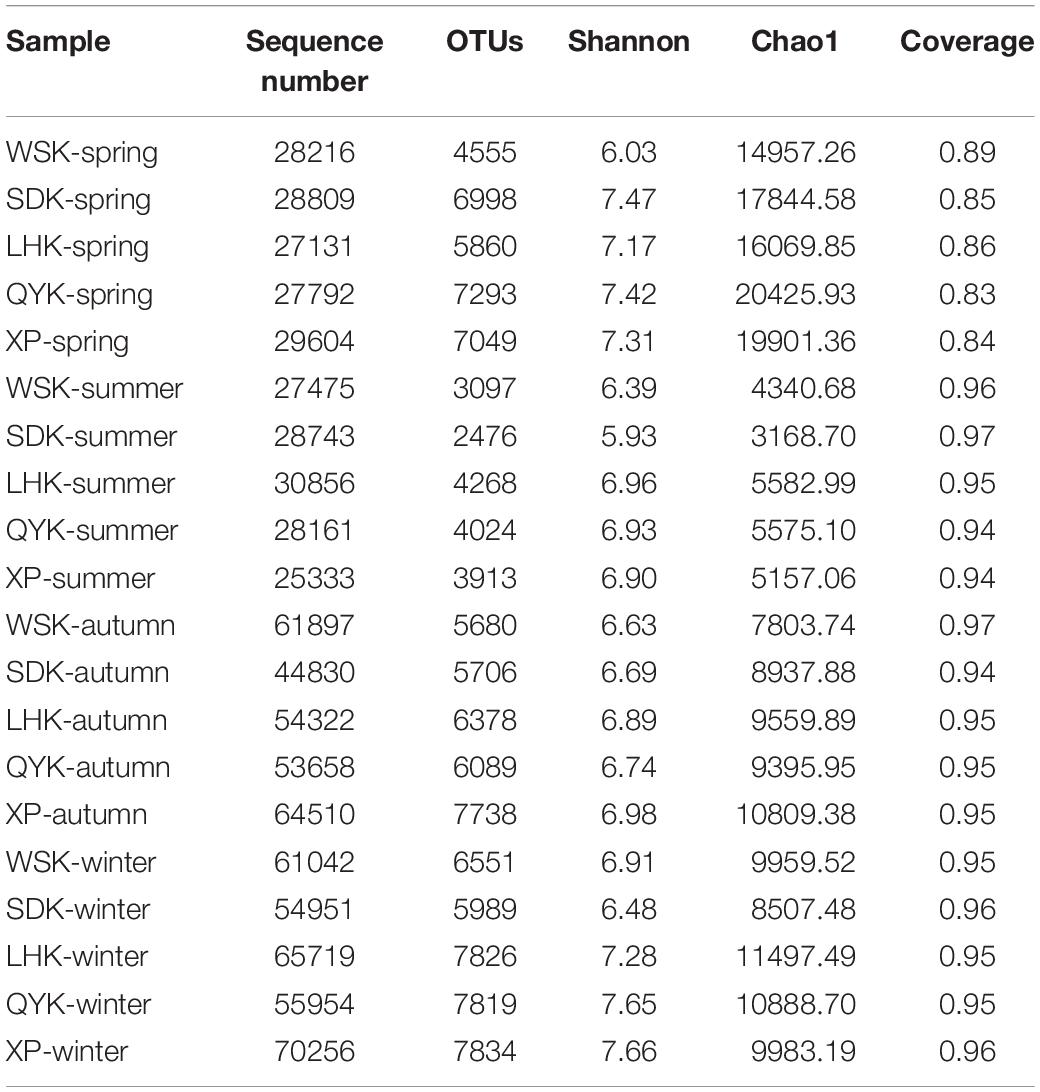
Table 2. Summary of the richness and diversity indices of microbial communities among different seasons.
Pearson’s correlation was used to further investigate the relationship between microbial community richness, diversity, and physicochemical properties of sediments, as summarized in Table 3. Microbial community richness and diversity indices of the 20 samples showed significant positive correlation (R = 0.503, P = 0.024), confirming that as richness increases, diversity is also expected to increase. Among all the samples collected, there is a significant positive correlation between microbial community richness and pH values (R = 0.531, P = 0.016); and a significant negative correlation with TOC (R = −0.633, P = 0.003). Furthermore, microbial community diversity showed a significant negative correlation with TP of the sample (R = −0.465, P = 0.039).
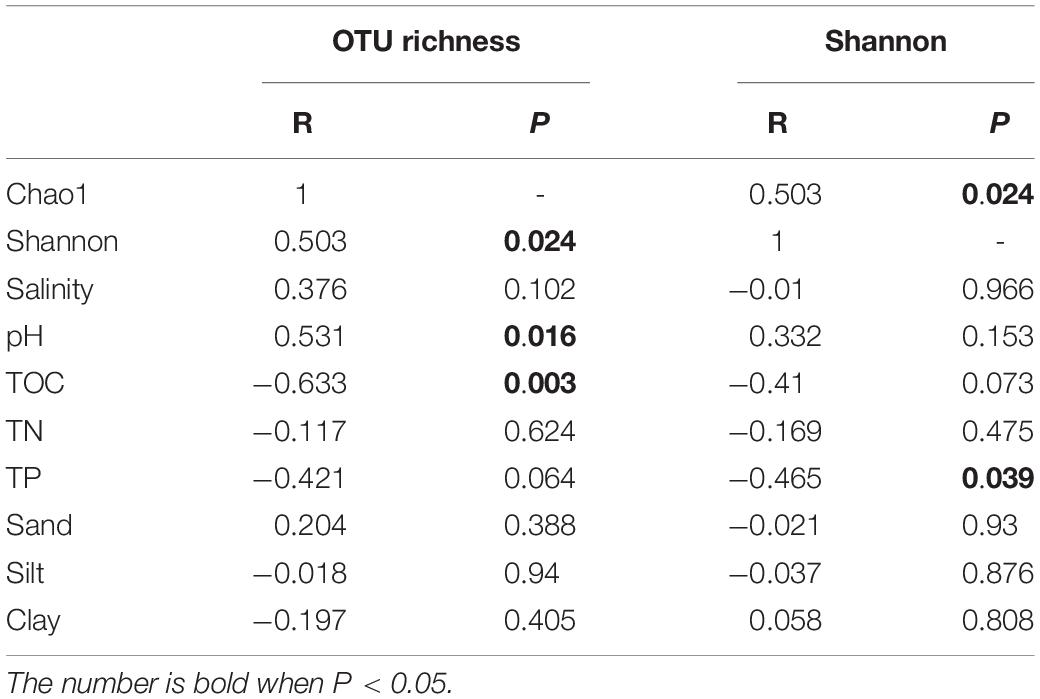
Table 3. Summary of Pearson’s correlation between the richness and diversity of microbial community, and physicochemical properties of sediments.
Significant spatial and/or temporal difference in microbial community structure can be observed between any two samples collected along the Yangtze River estuary, as shown in Supplementary Table 1; a score of 1 is assigned if communities were unrelated with 0% of shared sequences. When samples were grouped based on sampling season to identify seasonal patterns, PCoA results showed that microbial community structures within each seasonal group were relatively similar and clustered together, as shown in Figure 2A. Significant difference was found among the four seasonal groups (PERMANOVA, Adonis test, P = 0.003), reflected by the dispersed groups of colored points. The PCoA results were more visually distinguished when microbial community structures of collected samples were grouped based on wet and dry seasons (Spring and Summer, Autumn and Winter, respectively), as shown in Figure 2B. Microbial community structures from samples collected in Spring and Summer were shown to cluster together; samples collected in Autumn and Winter also showed similar clustering. The two clusters were found to be significantly different (PERMANOVA, Adonis test, P = 0.002), suggesting precipitation as a potential key factor determining microbial community structures.
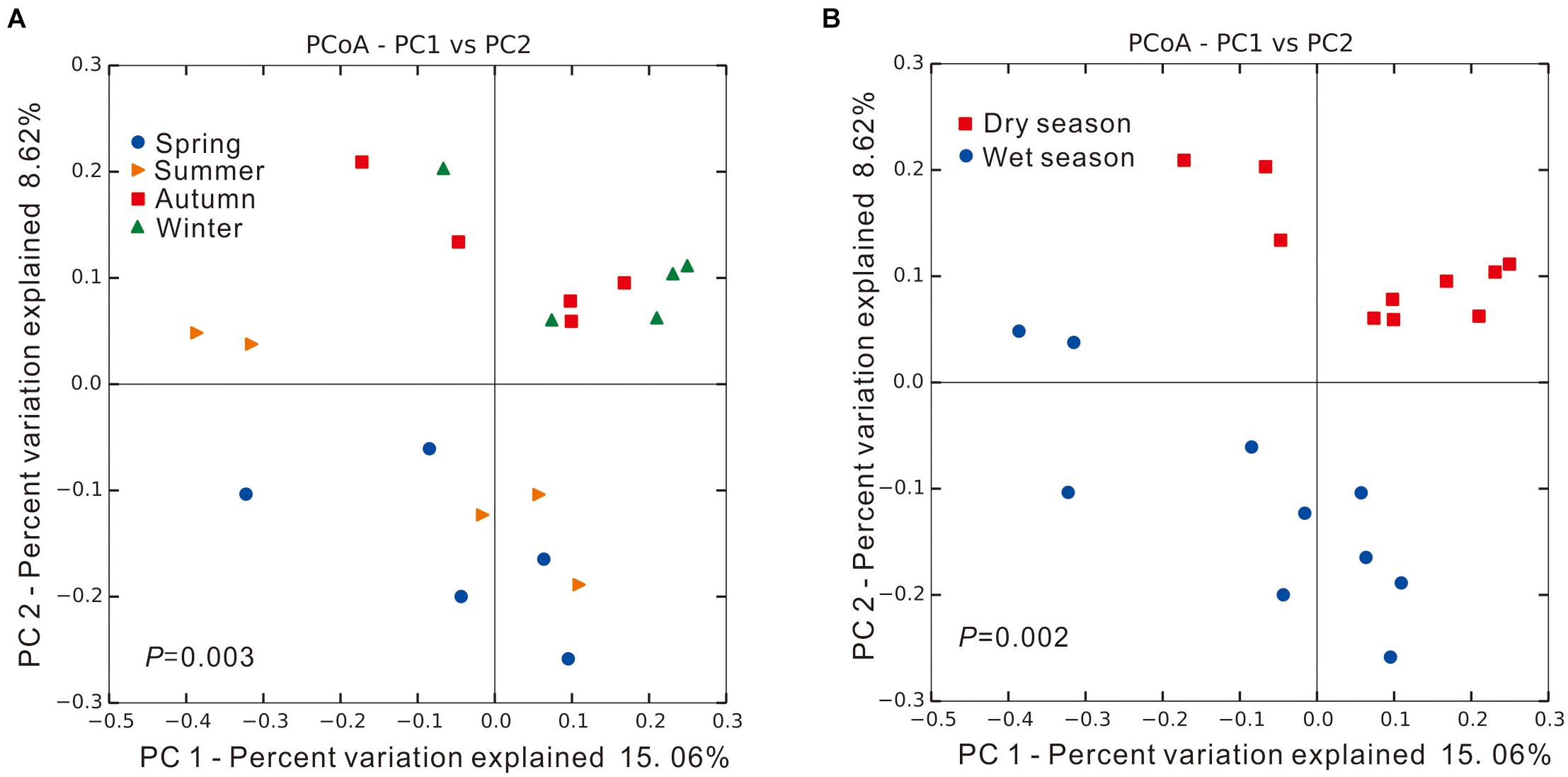
Figure 2. The unweighted UniFrac principal coordinate analysis (PCoA) of microbial communities in intertidal sediments of Yangtze River estuary, among different (A) seasons and (B) precipitation (dry/wet season). Significant difference was identified using Adonis test in PERMANOVA, P < 0.05.
The ten most dominant bacterial phyla, recorded from all sediment samples collected in this study, in descending order of mean relative abundance, were Proteobacteria (37.46%), Acidobacteria (9.38%), Planctomycetes (8.93%), Chloroflexi (8.89%), Verrucomicrobia (6.82%), Bacteroidetes (6.14%), Firmicutes (3.66%), Actinobacteria (3.24%), Patescibacteria (2.84%), and Spirochaetes (1.26%), as summarized in Figure 3. Proteobacteria, which is commonly found in estuarine sediments, had the highest relative abundance among all phyla reported from sampled microbial communities. The ten dominant bacterial phyla showed varying degree of differences in relative abundance with changing seasons, as summarized in Table 4. When the relative abundances of the ten phyla were compared between seasons and, on a broader scale, precipitation in dry and wet seasons, it is revealed that Actinobacteria, Chloroflexi, Planctomycetes and Verrucomicrobia showed occasional significant differences (Welch’s t-test, P < 0.05). Among the four phyla, relative abundance of Verrucomicrobia displayed more obvious changes with changing seasons and precipitation; relative abundance was notably higher in Spring and, therefore, wet seasons. Alternatively, the Kruskal-Wallis H-test showed that the relative abundances of Actinobacteria, Chloroflexi and Verrucomicrobia were significantly different among the four seasons (P < 0.05). Verrucomicrobia is a methanotrophic phylum found in aquatic sediments and human fecal microbiota (Op, den Camp et al., 2009). Actinobacteria is a much more diverse phylum found in both aquatic and terrestrial ecosystems, known for its economic value in producing metabolites (Barka et al., 2016).
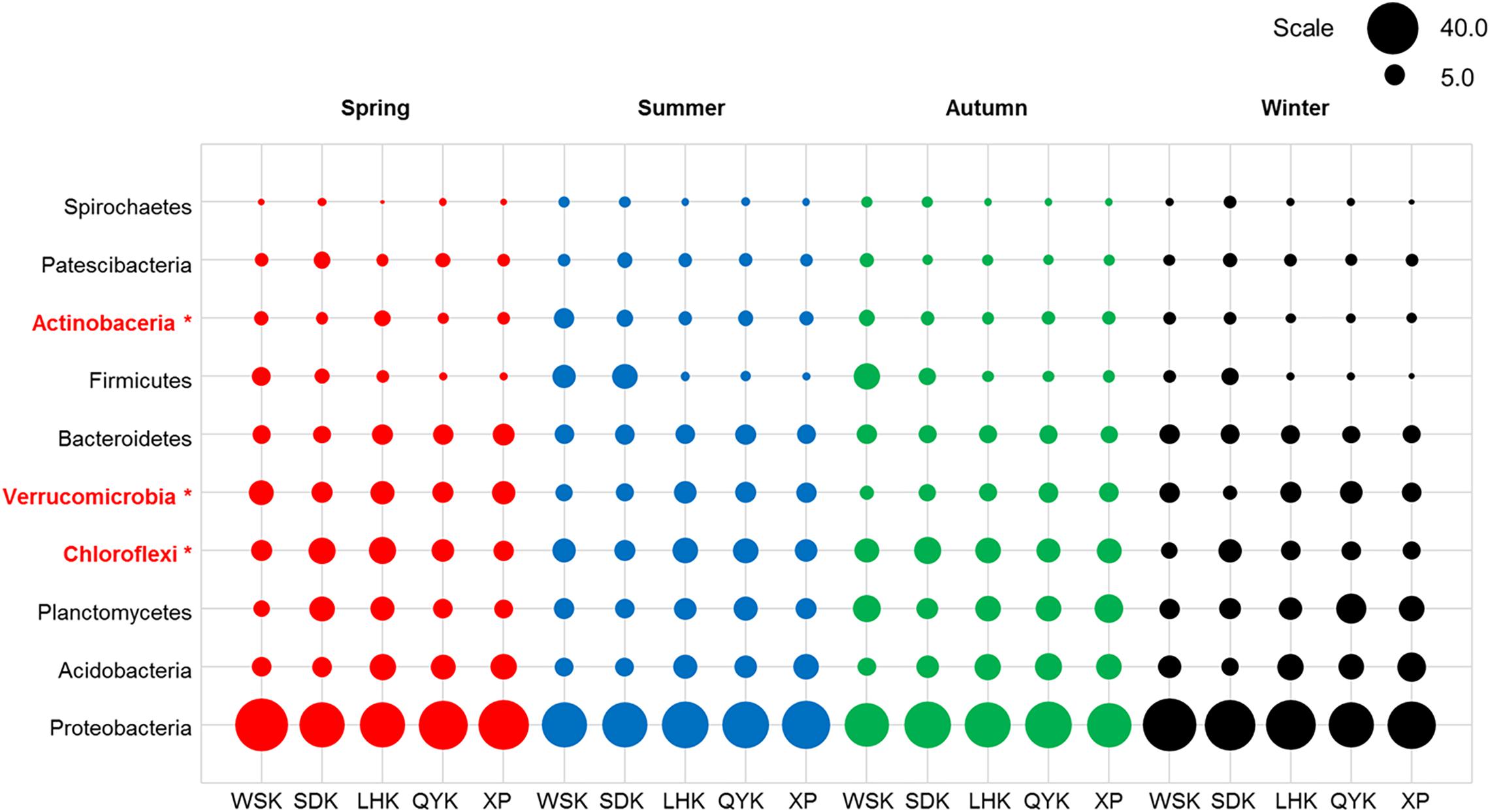
Figure 3. Relative abundances of dominant bacterial phyla in sediments from different sampling sites among various seasons (Red = Spring, Blue = Summer, Green = Autumn, Black = Winter). Asterisks represent a significant difference among the seasons according to Kruskal-Wallis H-test from STAMP pipeline (Kruskal-Wallis H-test, P < 0.05).
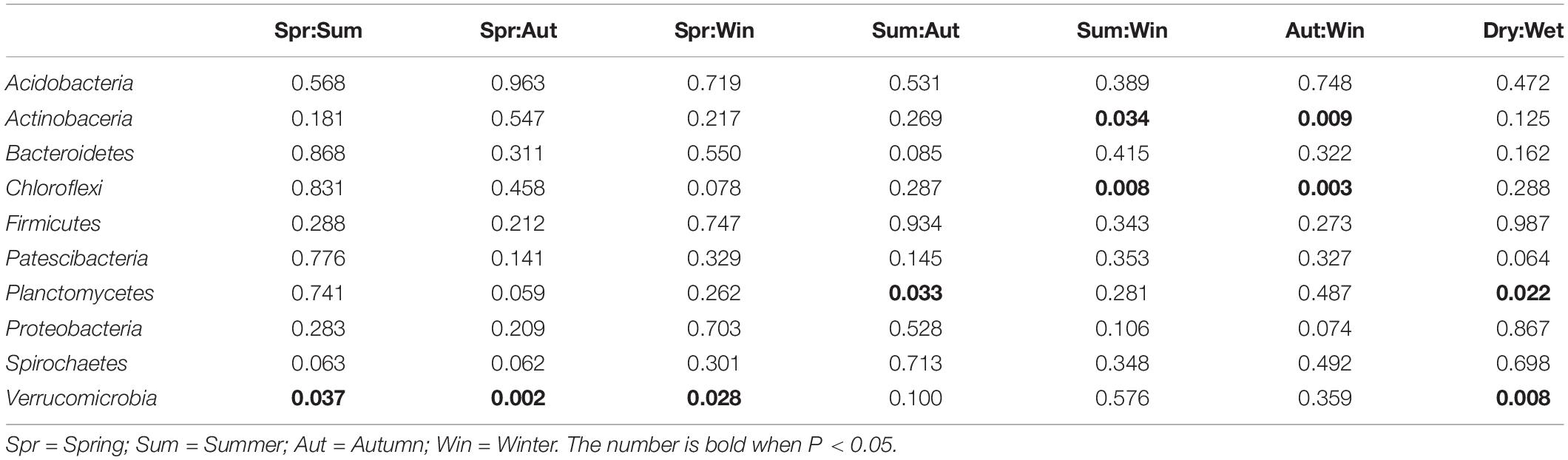
Table 4. Summary of temporal differences in relative abundances of dominant bacterial phyla in microbial communities (Welch’s t-test, P < 0.05).
Bacterial composition at the class level was further investigated to reveal potential temporal variations in microbial communities which may have been masked by the diversity within phyla. As shown in Table 5, the relative abundances of Anaerolineae, Planctomycetacia, and Verrucomicrobiae showed occasional significant differences between particular seasons or precipitation. For Deltaproteobacteria, significant difference was found between Spring and Winter (P < 0.05), favoring Winter. Kruskal-Wallis H-test results (Figure 4) showed that only classes Anaerolineae and Verrucomicrobiae were reported with relative abundances that differed significantly among the four seasons (P < 0.05). Both classes are major subdivisions of their respective phyla Chloroflexi and Verrucomicrobia and are directly responsible for the variation recorded at the phylum level.
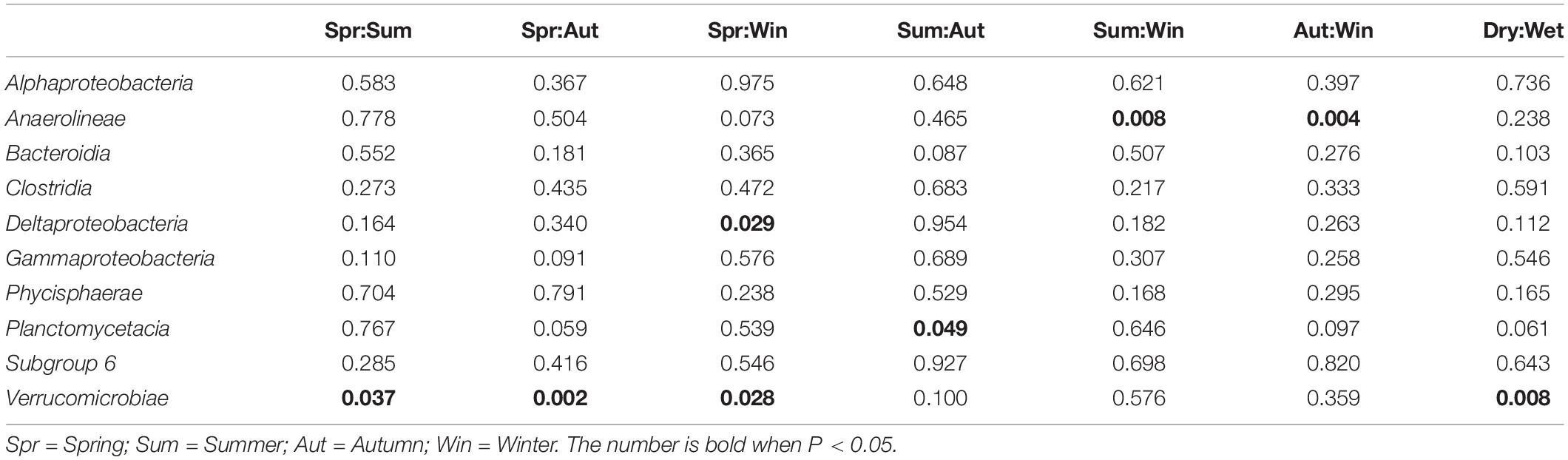
Table 5. Summary of temporal differences in relative abundances of dominant bacterial classes in microbial communities (Welch’s t-test, P < 0.05).
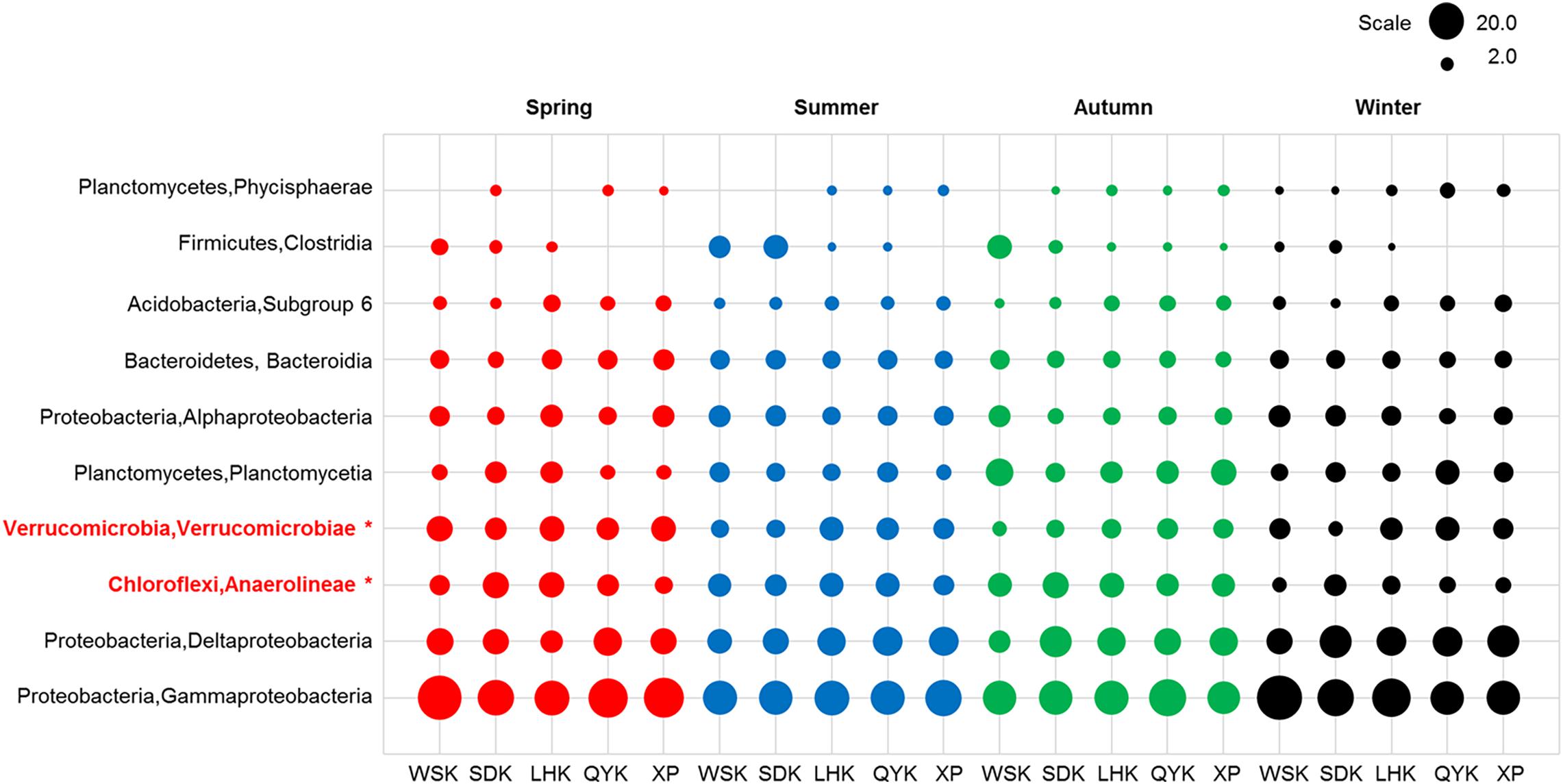
Figure 4. Relative abundances of dominant bacterial classes in sediments from different sampling sites among various seasons (Red = Spring, Blue = Summer, Green = Autumn, Black = Winter). Asterisks represent a significant difference among the seasons according to Kruskal-Wallis H-test from STAMP pipeline (Kruskal-Wallis H-test, P < 0.05).
Deltaproteobacteria is a bacterial class known for its contributions in sulfate/sulfur-reduction. The subset of bacterial genera responsible for sulfate/sulfur-reduction was selected for study. Among the 15 bacterial genera, the relative abundances of Desulfomonile and Desulfovibrio had significant differences between Spring/Autumn and dry/wet seasons, respectively (Table 6). In addition, only the relative abundances of genera Desulfuromonas and Desulfovirga showed similar patterns and had significant differences among the four seasons (Kruskal-Wallis H-test, P < 0.05), favoring the dry seasons, as shown in Figure 5. While Desulfovirga is reportedly sulfate-reducing, Desulfuromonas is characterized as a genus of Gram-negative bacteria capable of converting elemental sulfur into sulfide but does not reduce sulfate.
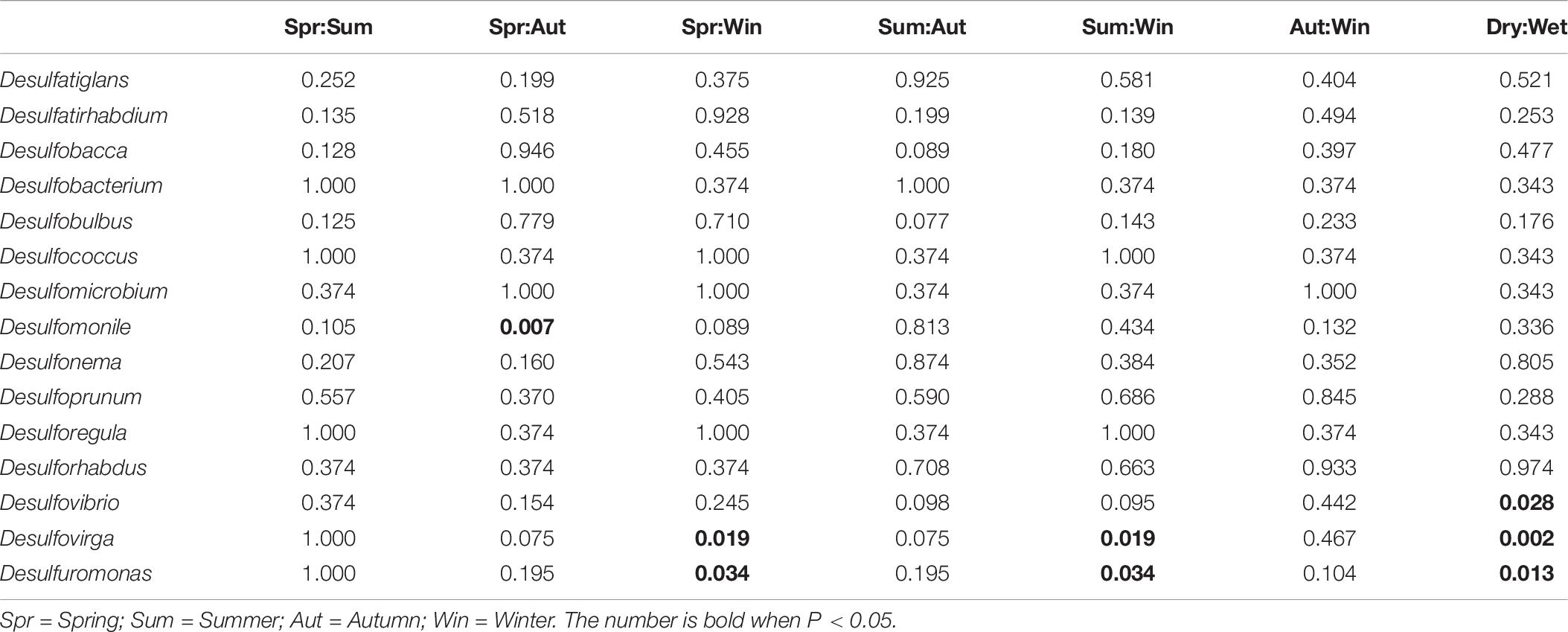
Table 6. Summary of temporal differences in relative abundances of sulfate/sulfur-reducing bacterial genera in microbial communities (Welch’s t-test, P < 0.05).
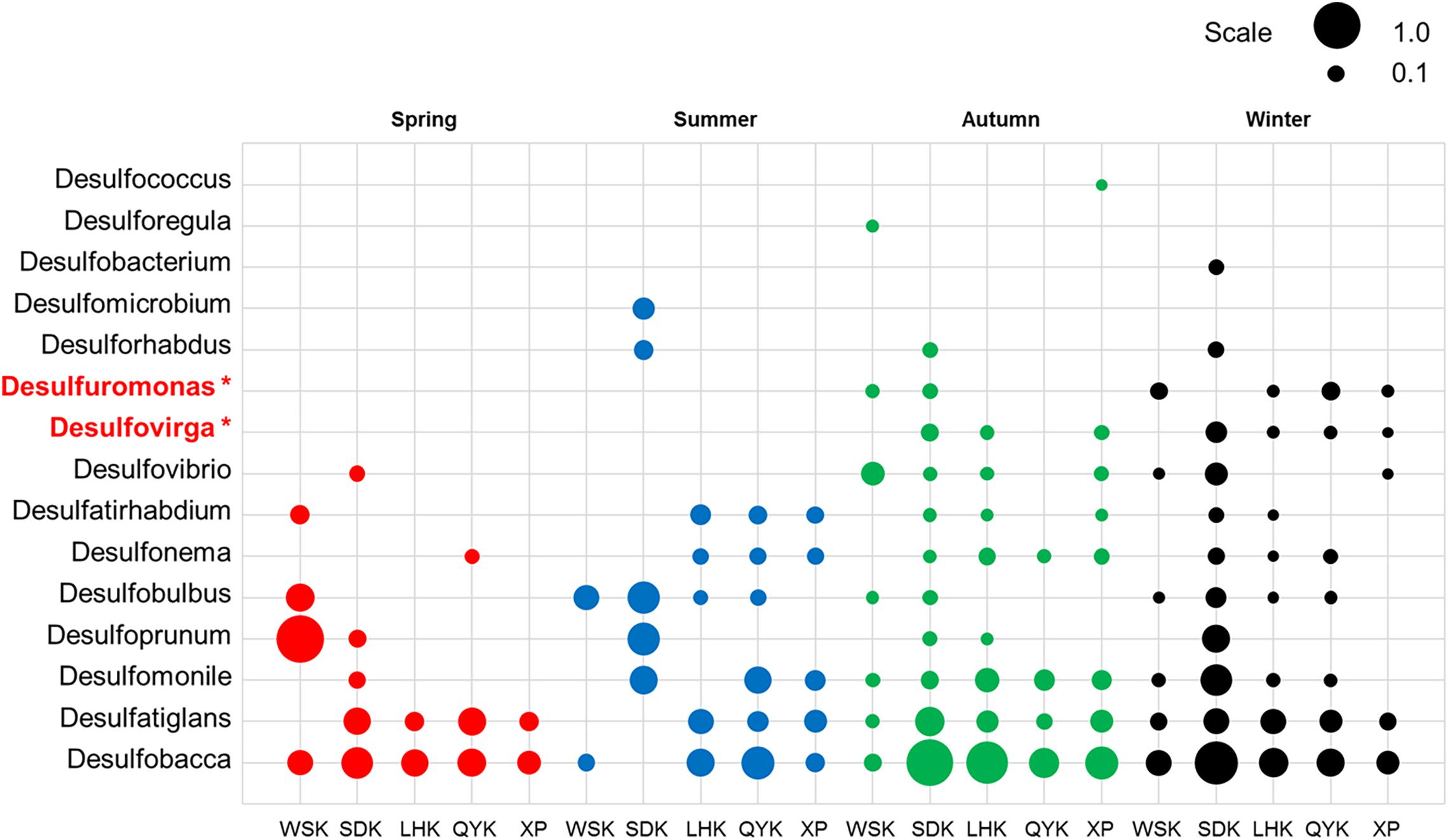
Figure 5. Relative abundances of sulfate/sulfur-reducing bacterial genera in sediments from different sampling sites among various seasons (Red = Spring, Blue = Summer, Green = Autumn, Black = Winter). Asterisks represent a significant difference among the seasons according to Kruskal-Wallis H-test from STAMP pipeline (Kruskal-Wallis H-test, P < 0.05).
RDA was conducted to analyze the relationship between relative abundances of dominant bacterial compositions in microbial communities and physicochemical properties of sediments. As shown in Figure 6A, the two RDA axes explained 54.73% (49.13% and 5.60% for axes 1 and 2, respectively) of the variation. Physicochemical properties of sediments explained 62.6% of the variation in microbial compositions. The contribution of individual properties toward variation in microbial composition and abundance is summarized in Supplementary Table 2. Results showed that pH (Explains = 22.0%, F = 5.1, P = 0.016), TOC (Explains = 18.5%, F = 4.1, P = 0.02), and TP (Explains = 17.8%, F = 3.9, P = 0.018) were of significant impact. Among the three factors, pH was found to be the overall most impactful factor on microbial composition and abundance (Explains = 22.0%, F = 5.1, P = 0.016), as shown in Supplementary Table 3.
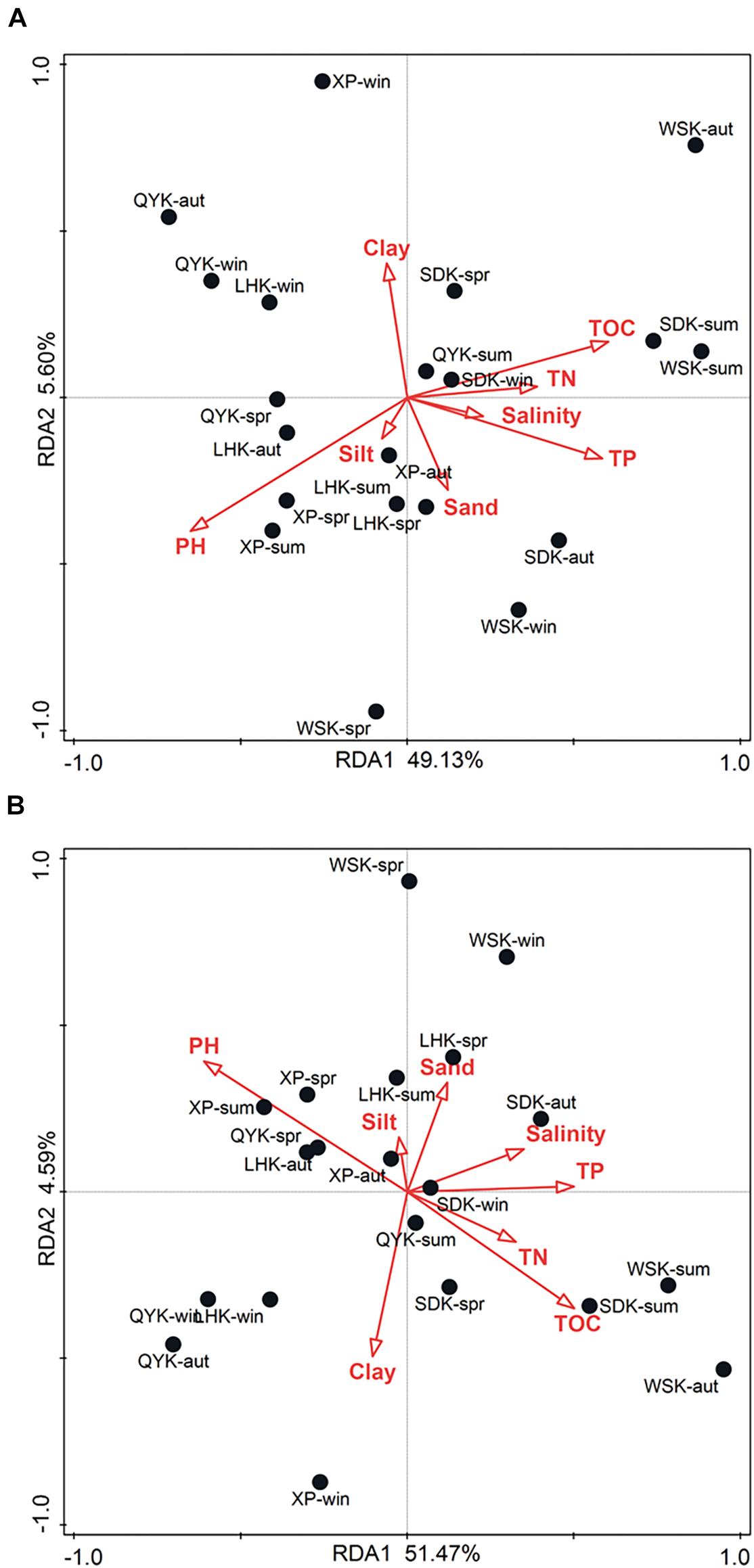
Figure 6. Redundancy analysis (RDA) ordination plot of the composition and abundance of dominant bacterial (A) phyla and (B) classes in intertidal sediments of Yangtze River estuary, in relation to physicochemical properties of sediments.
As shown in Figure 6B, the two RDA axes explained 56.06% (51.47% and 4.59% for axes 1 and 2, respectively) of the variation. Physicochemical properties of sediments explained 62.4% of the variation in microbial composition. The contribution of individual properties toward variation in microbial composition and abundance is summarized in Supplementary Table 4. Similarly, results showed that pH (Explains = 20.1%, F = 4.5, P = 0.024), TOC (Explains = 14.2%, F = 3.0, P = 0.0.04), and TP (Explains = 14.2%, F = 3.0, P = 0.042) were of significant impact. Among the three factors, pH was also determined the overall most impactful factor on microbial composition and abundance (Explains = 20.1%, F = 4.5, P = 0.028), as shown in Supplementary Table 5.
The ecological functions of microbial communities were annotated using FAPROTAX. From the results, approximately 20% (15217) of the total sequence records (75719) were assigned with 82 ecological functions, contributing to a variety of biogeochemical processes. As shown in Figure 7A, the major ecological functions contributed by microbial communities, in descending order of relative contribution, were chemoheterotrophy, aerobic chemoheterotrophy, respiration of sulfur compounds, sulfate respiration, nitrification, aerobic nitrite oxidation, nitrate reduction, nitrate respiration, chlorate reducers, and fermentation.
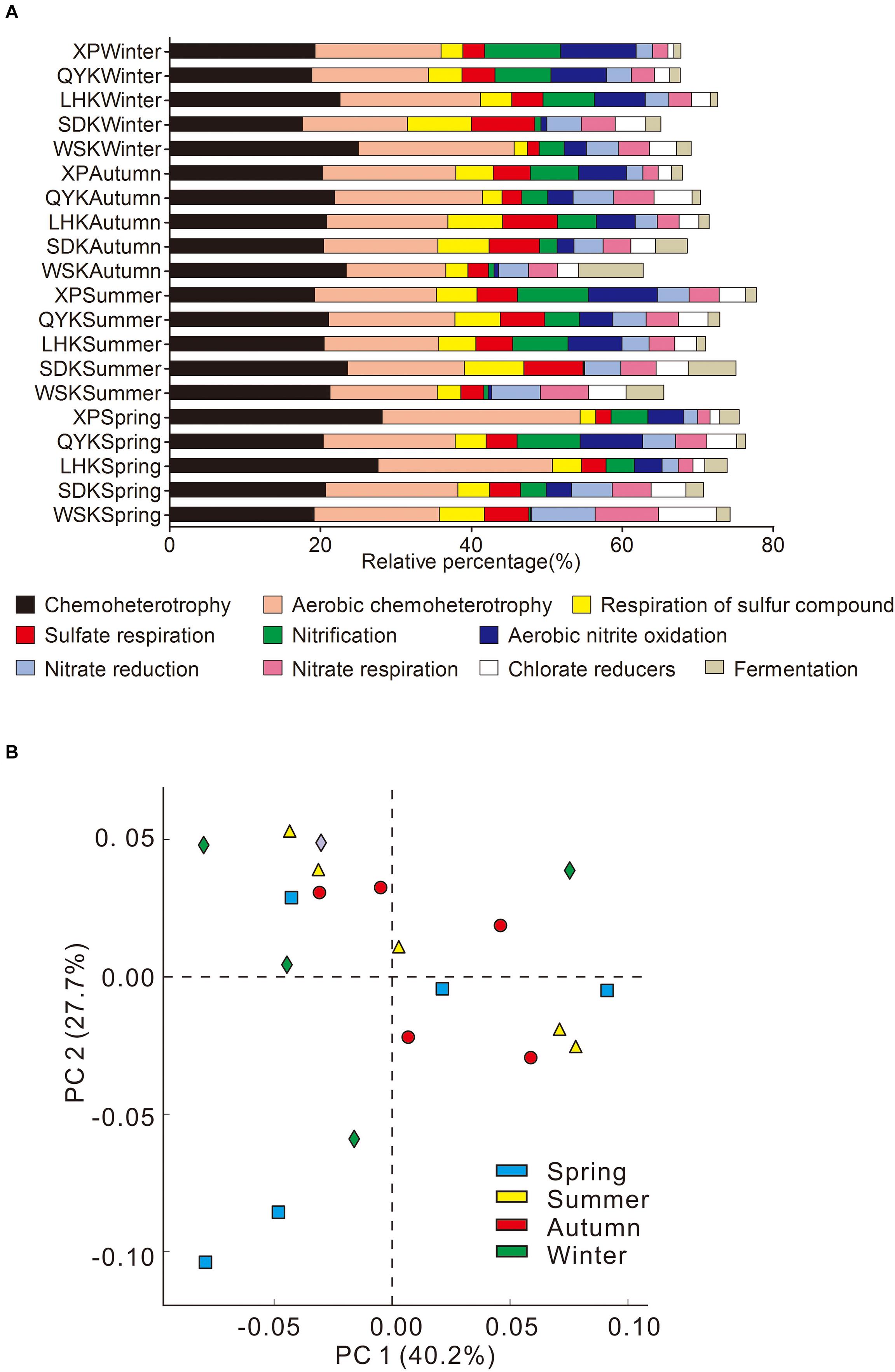
Figure 7. Summary of major ecological functions (A) and principal component analysis (PCA) of ecological functions (B) contributed by microbial communities in the intertidal sediments of Yangtze River estuary, among different seasons (Kruskal-Wallis H-test).
The major ecological functions of microbial communities in studied sampling sites included both aerobic processes, such as aerobic chemoheterotrophy, and anaerobic processes, such as sulfate respiration. PCA was conducted to investigate the temporal variation in ecological functions contributed by microbial communities among all the sampling sites. Results revealed that the ten major ecological functions contributed by microbial communities show no temporal differences (Figure 7B; Kruskal-Wallis H-test). Only nine of the 82 ecological functions assigned showed differences with changing seasons, of which only two ecological functions had an average contribution above 1% (Animal parasites or symbionts, 1.85%; Human pathogens all, 1.57%).
The present study characterized intertidal sediments from the Yangtze River estuary and evaluated the effects of temporal variation on inhabiting microbial communities. Nutrient concentrations in sediments as well as α- and β-diversity of microbial communities showed some notable variation which may be attributed to seasonal differences, with summer conditions being reportedly least favorable for microbial diversity. The Summer season was reported with the lowest pH levels, and pH correlated positively with microbial community richness. At the phylum level, relative abundances of the dominant Proteobacteria did not show significant temporal variation. Instead, abundance of Actinobacteria, Chloroflexi, and Verrucomicrobia were reported to differ more significantly, favoring the wet seasons. Most of these phylum-level changes can be associated with similar patterns observed for a major subdivision at the class-level, such as Anaerolineae and Verrucomicrobiae. Notably, Deltaproteobacteria, a class known for containing most of the sulfate/sulfur-reducing bacteria, showed a significant difference in relative abundance between Spring and Winter, favoring Winter. Upon further investigation, sulfur-reducing bacteria at the genus level were also found to favor Autumn and Winter, hence the dry seasons. However, such difference in sediment conditions, microbial diversity, and relative abundance did not translate to corresponding ecological functions showing significant temporal changes. Non-etheless, patterns recorded in the present study should not be overlooked. The composition of microbial communities was found to correlate with pH, TOC, and TP of the environment, but rainfall is proposed to be another important factor to consider, as discussed in the following sections.
Summer seasons of the subtropical monsoon climate zone is characterized by high precipitation. Rainfall and subsequent run-off can be expected to erode and carry surface sediments, exposing sedimentary microbial community to further environmental perturbation. In this study, microbial communities sampled during the Summer season were reportedly of lower richness, despite having no significant difference in sediment physicochemical properties from succeeding seasons. Hence, it is suspected that the summer rainfall may be an unaccounted factor imposing an impact on microbial community diversity. However, throughout the year, species richness and diversity of sampled communities remained to be positively correlated. This meant that differences in microbial community structures were minor; for Yangtze River estuary, overall microbial diversity has displayed a degree of stability. The Summer season may have limited microbial species richness but did not fundamentally impact the community structures observed, reflecting the community’s ability to adapt to changes in the environment (Shade et al., 2012).
In terms of β-diversity of microbial communities, clustering was observed for samples within the same season. The observation was much more distinct between dry and wet seasons, which may be further evidence supporting precipitation as an important factor (Figure 2). With global warming being an imminent issue, it is expected that rate of hydrological cycles will be enhanced, affecting precipitation on a global scale (Park et al., 2016). Previous study by Ren et al. (2018) showed that a reduction in precipitation had no effect on the formation of microbial community. However, this reduction lowered the total sedimentary biomass and bacterial abundance; total organic carbon and respiration rate were also affected by the reduced rainfall, and the two observations were positively correlated (Ren et al., 2018). A lack of rainfall can lead to lowered available water in soil, preventing carbon and nitrogen to be effectively utilized by microbial community and ultimately lowering the biomass (Schrama and Bardgett, 2016). However, some bacterial taxa within microbial communities can withstand the pressure imposed from a loss of water, such as Gram-positive bacteria and Actinobacteria, so the realistic impact on biomass and composition in the field environment may vary. In this study, microbial community diversity under conditions of high precipitation showed decreasing trends, and vice versa. An explanation for this is suggested to be the increase in precipitation causing lowered sediment pH, frequent runoffs, and constant changes in surface sedimentary conditions, which may be costly for the overall proliferation of microbial communities. Furthermore, as tides carry nutrients into intertidal sediments, there is no obvious lack in sediment carbon or nitrogen, even in dry seasons, which mitigates the possibility of a loss in biomass due to nutrient deprivation. Consequently, the present findings highlighted the importance of precipitation for microbial community diversity and provided new insights toward understanding the response of microbial communities to precipitation in a complex environment.
In terms of bacterial abundance, Proteobacteria is the most dominant and diverse phylum, consisting of bacterial taxa which play important ecological roles in the cycling of carbon, nitrogen, and sulfur (Kersters et al., 2006). The metabolic functional diversity of Proteobacteria at lower taxonomic ranks meant that close to 40% of the microbial communities can adapt to different environmental conditions, especially in complex estuarine ecosystems. The adaptability toward oxic and anoxic environments may be an important trait for sedimentary bacteria, as oxygen levels in sediments can vary with vertical depth. In general, as vertical depth increases, sedimentary oxygen content decreases and form hypoxic layer or even anaerobic layer. In the present study, surface sediments were sampled, which should be considered the aerobic layer. Considering that all sediment samples were collected at the same vertical depth of the intertidal mudflats, oxygen content may not be a significant variable for this study. However, it is important to consider the implications that may be present. Since the relative abundance of phylum Verrucomicrobia showed significant differences with change in precipitation, it may be indicative of its adaptability toward the dynamic changes in oxygen. Verrucomicrobia (6.82%) is a dominant bacterial taxon in the Yangtze River estuary. However, very few studies on intertidal sedimentary microbial communities have reported it to be high in relative abundance (Zhang et al., 2014; Obi et al., 2016). In the study of Yu et al. (2012), Verrucomicrobia was reported as a major bacterial taxon (relative abundance of 0.21%). However, its relative contribution toward community abundance was only 1/192 than that of Proteobacteria (relative abundance of 40.5%). Verrucomicrobia is more commonly found in terrestrial environments and soil. Study by Bergmann et al. (2011) showed that among 181 soil samples, 180 were found to contain Verrucomicrobia, with its average abundance reaching 23%. Similarly, in marine environments, study showed that over 98% of the 506 collected terrestrial and marine environmental samples contained Verrucomicrobia. In fact, Verrucomicrobia was also common in marine environments with a reported average abundance of 2%, and a relative abundance of 1.4% in sediments (Freitas et al., 2012). In this study, Yangtze River estuary intertidal sediments were shown to support a rich community of Verrucomicrobia, with the relative abundances of Verrucomicrobia and its subtaxa showing significant seasonal differences, especially between dry and wet seasons. A speculation would be precipitation causing water to carry terrestrial soil with relatively rich Verrucomicrobia content to move to estuarine sediments, leading to its widespread presence. The results can be important to address current gap in knowledge regarding patterns displayed by Verrucomicrobia in response to an ever-changing estuarine environment.
Microbial communities in the Yangtze River estuary displayed diverse ecological functions, including aerobic chemosynthesis and nitrification, as well as anaerobic processes such as sulfate reduction and silver nitrate reduction. Previous study showed that sediments of intertidal mudflats at a depth range between 0–20 cm can be affected by periodic high tides, subsequently affecting microbial community composition and ecological functions (Zheng et al., 2016). Metabolic processes, both aerobic and anaerobic, of microbial communities in surface sediments tend to be more diverse and variable. However, in the present study, ecological function contributed by microbial communities in the field were shown to be stable and consistent, with insignificant temporal variations. For instance, the majority of sulfate/sulfur-reducing bacteria genus were recorded to favor Autumn and Winter in terms of relative abundances, yet the overall contributions by the functional group did not differ significantly, as seen for sulfate respiration (Figure 7). This suggests microbial communities in the Yangtze River estuary to be robust and resilient to changes in environment, instead of being vulnerable to constant perturbation. Investigating functional changes at the gene level e.g., sulfur metabolism gene dsrA can be beneficial for establishing a connection between bacterial community structure and corresponding ecological functions.
A notable factor that is not evaluated in the present study is the dissolved oxygen level in sediments. Periodic changes in tides is a key example of perturbation associated with temporal variation as it affects the utilization of sedimentary oxygen, which determines the composition and relative abundances of aerobes, facultative anaerobes, and anaerobes, in sedimentary microbial communities. Oxygen content and microbial community structure were implied to not follow any direct correlation according to previous study, which showed that aerobic methane-oxidizing bacteria have highest relative abundance and activity in surface sediments of depth range 2–3 cm, despite recording no oxygen content (Rahalkar et al., 2009). At slightly deeper (3–4 cm) sediments, relative abundance of methane-oxidizing bacteria remained at a noticeable level. However, methane-oxidizing activities were not recorded. This partly illustrates the historical problem of measuring oxygen content through instruments which were inaccurate and should be deemed inappropriate as a reference to distinguish aerobic and anaerobic bacterial communities. In addition, sedimentary oxygen may be under dynamic changes, meaning in situ measurements may not be sufficient to reflect the formation process of microbial communities under long-term selective pressure. In this study, intertidal sediment samples were collected at 5–7 cm range from surface. Although the oxygen content in sediments were not characterized, a recent study implied that intertidal sediments of 0 - 10 cm deep tend to contain a concentration of dissolved oxygen and do not form obvious oxic or anoxic layers (Wang et al., 2013). From the results of ecological functional predictions, the contributions of aerobic chemosynthesis regulated by microbial communities showed no spatial or seasonal changes. This may be evidence indicating the functional effectiveness of microbial communities under perceived perturbation.
Compared to temporal variations and potential adaptabilities displayed by the microbial communities, the significant differences in physicochemical properties resulting from different sampling sites may be more impactful for microbial communities. The heavy skew in nutrient concentrations in the SDK site made it a notable potential outlier among the five sites, suspected to be associated with input from Liuhe River. In addition, RDA had determined pH to be the most significant factor explaining the compositional differences. Although pH did not show significant spatiotemporal difference, it is possible that microbial communities were very sensitive to subtle changes. While it may be associated with anthropogenic sources, no convincing conclusion can be drawn at the current stage due to sample size at each sampling site. Spatial variation in microbial communities in estuarine ecosystems is an equally important research direction with ecological implications in human impacted areas, which can further the current understanding.
Overall, temporal variation has notable effects on intertidal sediment conditions and microbial community structure and diversity. This is thought to be related to rainfall conditions in the subtropical monsoon climate zones. At three months sampling intervals, patterns in microbial taxa were identified with respect to the environmental conditions of the intertidal sediments. Seasonality may be an explanation for the present observations but further investigations with rigorous sample collection will be required to provide confirmation. It is also shown that microbial communities and their contributions to ecological functions can be relatively robust and resilient to environmental changes. However, it should be noted that only approximately 20% of the microbial communities were functionally assigned in this study, which is an expected outcome due to limitations in analytical methods and the nature of predictive tools such as FAPROTAX. The drawback of mapping taxa inferred from short marker genes to manually constructed databases based on literature and cultured samples lies in the uncertainty with both data input and output. For 16S rRNA sequencing, results can be unclassified or identified at low specificity (e.g., to order level). Despite the relatively high taxonomic depth, the current database in use still reported 16% of the genus-level taxa to be unclassified. For output, the functional database of FAPROTAX is non-exhaustive, meaning that without frequent updates, new discoveries in the field are omitted, and assumptions have to be made. Nevertheless, the present study provided new temporal data and insights to help understand responses from microbial communities under the changing environment of the urbanized Yangtze River estuary.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
JY performed data curation, methodology, investigation, formal analysis, and writing – original draft. LL performed writing – original draft and writing – reviewing and editing. JC performed conceptualization, resources, supervision, funding acquisition, data curation, formal analysis, writing – original draft, and writing – reviewing and editing. All authors contributed to the article and approved the submitted version.
This work was supported by the Hong Kong Branch of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (SMSEGL20SC01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.585970/full#supplementary-material
Aguirre, M., Abad, D., Albaina, A., Cralle, L., Goni-Urriza, M. S., Estonba, A., et al. (2017). Unraveling the environmental and anthropogenic drivers of bacterial community changes in the estuary of bilbao and its tributaries. PLoS One 12:e0178755. doi: 10.1371/journal.pone.0178755
Anderson, M. J. (2014). Permutational multivariate analysis of variance (PERMANOVA). Wiley Statsref: Stat. Ref. Online 1–15. doi: 10.1002/9781118445112.stat07841
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Klenk, H. P., et al. (2016). Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/mmbr.00019-15
Bergmann, G. T., Bates, S. T., Eilers, K. G., Lauber, C. L., Caporaso, J. G., Walters, W. A., et al. (2011). The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol. Biochem. 43, 1450–1455. doi: 10.1016/j.soilbio.2011.03.012
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857.
Duarte, B., Freitas, J., and Caçador, I. (2012). Sediment microbial activities and physic-chemistry as progress indicators of salt marsh restoration processes. Ecol. Indicators 19, 231–239. doi: 10.1016/j.ecolind.2011.07.014
Dyksma, S., Bischof, K., Fuchs, B. M., Hoffmann, K., Meier, D., Meyerdierks, A., et al. (2016). Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J. 10, 1939–1953. doi: 10.1038/ismej.2015.257
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Falkowski, P. G., Fenchel, T., and Delong, E. F. (2008). The microbial engines that drive Earth’s biogeochemical cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
Fan, M., Lin, Y., Huo, H., Liu, Y., Zhao, L., Wang, E., et al. (2016). Microbial communities in riparian soils of a settling pond for mine drainage treatment. Water Res. 96, 198–207. doi: 10.1016/j.watres.2016.03.061
Freitas, S., Hatosy, S., Fuhrman, J. A., Huse, S. M., Welch, D. B. M., Sogin, M. L., et al. (2012). Global distribution and diversity of marine Verrucomicrobia. ISME J. 6, 1499–1505. doi: 10.1038/ismej.2012.3
Gilbert, J. A., Steele, J. A., Caporaso, J. G., Steinbrück, L., Reeder, J., Temperton, B., et al. (2012). Defining seasonal marine microbial community dynamics. ISME J. 6, 298–308. doi: 10.1038/ismej.2011.107
Giovannoni, S. J., and Vergin, K. L. (2012). Seasonality in ocean microbial communities. Science 335, 671–676. doi: 10.1126/science.1198078
Grizzetti, B., Liquete, C., Pistocchi, A., Vigiak, O., Zulian, G., Bouraoui, F., et al. (2019). Relationship between ecological condition and ecosystem services in European rivers, lakes and coastal waters. Sci. Total Environ. 671, 452–465. doi: 10.1016/j.scitotenv.2019.03.155
Guo, X. P., Lu, D. P., Niu, Z. S., Feng, J. N., Chen, Y. R., Tou, F. Y., et al. (2018). Bacterial community structure in response to environmental impacts in the intertidal sediments along the Yangtze Estuary. China. Mar. Pollut. Bull. 126, 141–149. doi: 10.1016/j.marpolbul.2017.11.003
Herlemann, D. P., Labrenz, M., Jürgens, K., Bertilsson, S., Waniek, J. J., and Andersson, A. F. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579. doi: 10.1038/ismej.2011.41
Kaestli, M., Skillington, A., Kennedy, K., Majid, M., Williams, D., McGuinness, K., et al. (2017). Spatial and temporal microbial patterns in a tropical macrotidal estuary subject to urbanization. Front. Microbiol. 8:1313. doi: 10.3389/fmicb.2017.01313
Kallmeyer, J., Pockalny, R., Adhikari, R. R., Smith, D. C., and D’Hondt, S. (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. 109, 16213–16216. doi: 10.1073/pnas.1203849109
Karhu, K., Auffret, M. D., Dungait, J. A., Hopkins, D. W., Prosser, J. I., Singh, B. K., et al. (2014). Temperature sensitivity of soil respiration rates enhanced by microbial community response. Nature 513, 81–84. doi: 10.1038/nature13604
Kersters, K., De Vos, P., Gillis, M., Swings, J., Vandamme, P., and Stackebrandt, E. (2006). “Introduction to the Proteobacteria,” in The Prokaryotes: a Handbook on the Biology of Bacteria, eds M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (New York, NY: Springer), 3–37.
Kindong, R., Wu, J., Gao, C., Dai, L., Tian, S., Dai, X., et al. (2020). Seasonal changes in fish diversity, density, biomass, and assemblage alongside environmental variables in the Yangtze River Estuary. Environ. Sci. Poll. Res. 27, 25461–25474. doi: 10.1007/s11356-020-08674-8
Kumar, A., Ng, D. H., Wu, Y., and Cao, B. (2019). Microbial community composition and putative biogeochemical functions in the sediment and water of tropical granite quarry lakes. Microbial. Ecol. 77, 1–11. doi: 10.1007/s00248-018-1204-2
Lan, Y., Wang, Q., Cole, J. R., and Rosen, G. L. (2012). Using the RDP classifier to predict taxonomic novelty and reduce the search space for finding novel organisms. PLoS One 7:e32491. doi: 10.1371/journal.pone.0032491
Liu, S. M., Zhang, J., Chen, H. T., Wu, Y., Xiong, H., and Zhang, Z. F. (2003). Nutrients in the Changjiang and its tributaries. Biogeochemistry 62, 1–18.
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/aem.71.12.8228-8235.2005
Murphy, R. R., Kemp, W. M., and Ball, W. P. (2011). Long-term trends in Chesapeake Bay seasonal hypoxia, stratification, and nutrient loading. Estuaries Coasts 34, 1293–1309. doi: 10.1007/s12237-011-9413-7
Murphy, J., and Riley, J. P. (1962). A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta, 27, 31–36. doi: 10.1016/S0003-2670(00)88444-5
Niu, Z. S., Pan, H., Guo, X. P., Lu, D. P., Feng, J. N., Chen, Y. R., et al. (2018). Sulphate-reducing bacteria (SRB) in the Yangtze Estuary sediments: abundance, distribution and implications for the bioavailibility of metals. Sci. Total Environ. 634, 296–304. doi: 10.1016/j.scitotenv.2018.03.345
Obi, C. C., Adebusoye, S. A., Ugoji, E. O., Ilori, M. O., Amund, O. O., and Hickey, W. J. (2016). Microbial communities in sediments of Lagos Lagoon, Nigeria: elucidation of community structure and potential impacts of contamination by municipal and industrial wastes. Front. Microbiol. 7:1213. doi: 10.3389/fmicb.2016.01213
Op, den Camp, H. J., Islam, T., Stott, M. B., Harhangi, H. R., Hynes, A., et al. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1, 293–306. doi: 10.1111/j.1758-2229.2009.00022.x
Park, J. Y., Bader, J., and Matei, D. (2016). Anthropogenic Mediterranean warming essential driver for present and future Sahel rainfall. Nat. Climate Change 6, 941–945. doi: 10.1038/nclimate3065
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Peterson, J. O., Morgan, C. A., Peterson, W. T., and Lorenzo, E. D. (2013). Seasonal and interannual variation in the extent of hypoxia in the northern California Current from 1998–2012. Limnol. Oceanography 58, 2279–2292. doi: 10.4319/lo.2013.58.6.2279
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 41, D590–D596.
Rahalkar, M., Deutzmann, J., Schink, B., and Bussmann, I. (2009). Abundance and activity of methanotrophic bacteria in littoral and profundal sediments of Lake Constance (Germany). Appl. Environ. Microbiol. 75, 119–126. doi: 10.1128/aem.01350-08
Ren, C., Chen, J., Lu, X., Doughty, R., Zhao, F., Zhong, Z., et al. (2018). Responses of soil total microbial biomass and community compositions to rainfall reductions. Soil Biol. Biochem. 116, 4–10. doi: 10.1016/j.soilbio.2017.09.028
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Schrama, M., and Bardgett, R. D. (2016). Grassland invasibility varies with drought effects on soil functioning. J. Ecol. 104, 1250–1258. doi: 10.1111/1365-2745.12606
Shade, A., Peter, H., Allison, S. D., Baho, D., Berga, M., Bürgmann, H., et al. (2012). Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3:417. doi: 10.3389/fmicb.2012.00417
Shen, Z. (ed.) (2020). Studies of the Biogeochemistry of Typical Estuaries, and Bays in China. Berlin: Springer.
Shi, J., Liu, X., Chen, Q., and Zhang, H. (2014). Spatial and seasonal distributions of estrogens and bisphenol a in the Yangtze River Estuary and the adjacent East China Sea. Chemosphere 111, 336–343. doi: 10.1016/j.chemosphere.2014.04.046
Suh, S. S., Park, M., Hwang, J., Kil, E. J., Jung, S. W., Lee, S., et al. (2015). Seasonal dynamics of marine microbial community in the South Sea of Korea. PLoS One 10:e0131633. doi: 10.1371/journal.pone.0131633
Sun, Z., Brittain, J. E., Sokolova, E., Thygesen, H., Saltveit, S. J., Rauch, S., et al. (2018). Aquatic biodiversity in sedimentation ponds receiving road runoff–What are the key drivers? Sci. Total Environ. 610, 1527–1535. doi: 10.1016/j.scitotenv.2017.06.080
Ter Braak, C. J., and Šmilauer, P. (2002). CANOCO Reference Manual and CanoDraw for Windows user’s guide: Software for Canonical Community Ordination (version 4.5). www.canoco.com
Wang, L., Liu, L., Zheng, B., Zhu, Y., and Wang, X. (2013). Analysis of the bacterial community in the two typical intertidal sediments of Bohai Bay, China by pyrosequencing. Mar. Poll. Bull. 72, 181–187. doi: 10.1016/j.marpolbul.2013.04.005
Wang, Y., Sheng, H. F., He, Y., Wu, J. Y., Jiang, Y. X., Tam, N. F. Y., et al. (2012). Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl. Environ. Microbiol. 78, 8264–8271. doi: 10.1128/aem.01821-12
White, J. R., Nagarajan, N., and Pop, M. (2009). Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 5:e1000352. doi: 10.1371/journal.pcbi.1000352
Xie, Y., Wang, J., Wu, Y., Ren, C., Song, C., Yang, J., et al. (2016). Using in situ bacterial communities to monitor contaminants in river sediments. Environ. Poll. 212, 348–357. doi: 10.1016/j.envpol.2016.01.031
Xu, Z., Woodhouse, J. N., Te, S. H., Gin, K. Y. H., He, Y., Xu, C., et al. (2018). Seasonal variation in the bacterial community composition of a large estuarine reservoir and response to cyanobacterial proliferation. Chemosphere 202, 576–585. doi: 10.1016/j.chemosphere.2018.03.037
Yu, Y., Wang, H., Liu, J., Wang, Q., Shen, T., Guo, W., et al. (2012). Shifts in microbial community function and structure along the successional gradient of coastal wetlands in Yellow River Estuary. Eur. J. Soil Biol. 49, 12–21. doi: 10.1016/j.ejsobi.2011.08.006
Zhang, W., Bougouffa, S., Wang, Y., Lee, O. O., Yang, J., Chan, C., et al. (2014). Toward understanding the dynamics of microbial communities in an estuarine system. PloS One 9:e94449. doi: 10.1371/journal.pone.0094449
Keywords: Yangtze River estuary, intertidal sediments, microbial community, community composition, seasonal dynamics, ecological functions
Citation: Yi J, Lo LSH and Cheng J (2020) Dynamics of Microbial Community Structure and Ecological Functions in Estuarine Intertidal Sediments. Front. Mar. Sci. 7:585970. doi: 10.3389/fmars.2020.585970
Received: 22 July 2020; Accepted: 30 September 2020;
Published: 22 October 2020.
Edited by:
Maria Moustaka-Gouni, Aristotle University of Thessaloniki, GreeceReviewed by:
Alma Parada, Stanford University, United StatesCopyright © 2020 Yi, Lo and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinping Cheng, amluY2hlbmdAdXN0Lmhr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.