- 1International Research Center for Marine Biosciences, Ministry of Science and Technology, Shanghai Ocean University, Shanghai, China
- 2Key Laboratory of Exploration and Utilization of Aquatic Genetic Resources, Ministry of Education, Shanghai Ocean University, Shanghai, China
- 3Southern Marine Science and Engineering Guangdong Laboratory, Guangzhou, China
- 4Department of Marine Science and Fisheries, College of Agricultural and Marine Sciences, Sultan Qaboos University, Muscat, Oman
- 5Center of Excellence in Marine Biotechnology, Sultan Qaboos University, Muscat, Oman
Bacterial quorum sensing (QS) is a chemical communication that allows bacteria to coordinate their population and gene regulation in response to threshold concentrations of QS signals. Here, we study the effect of QS inhibitor 2(5H)-furanone (FUR) on the formation of Pseudoalteromonas marina ECSMB14103 biofilms and the effect of these biofilms on the settlement of plantigrades of the mussel Mytilus coruscus. FUR was added during biofilm formation of P. marina ECSMB14103 or in the settlement bioassay. FUR at 10–4 M added during biofilm formation significantly (p < 0.05) reduced the settlement of plantigrades, and it was associated with a significant (p < 0.05) reduction of bacterial density. The visualization of individual substances in biofilms shows that β-polysaccharides are the major components in the biofilm matrix. The decreased settlement rate may be attributed to the reduction of the biovolume of α-polysaccharides and β-polysaccharides in biofilms. The transcriptome of biofilms treated with 10–4 M FUR shows 61 genes are differentially expressed. Aspartate kinase and the flagellar assembly protein fliH gene are significantly affected after exposure to 10–4 M FUR (p < 0.05). They are involved in the synthesis of the bacterial cell wall and the mobility of bacteria, respectively. Exposure to 10–4 M FUR also significantly changes the carbapenem biosynthesis pathway, which is regulated by the QS system in bacteria (p < 0.05). These data suggest that FUR directly affects biofilm formation by altering extracellular polymeric substances (EPS) components and metabolic processes, which, in turn, indirectly reduce the settlement of plantigrades. The present study provides new insights into the molecular mechanisms of controlling biofouling.
Introduction
In marine environments, biofilms are the aggregation of microorganisms embedded in extracellular polymeric substances (EPS) on all submerged substrata (Wahl et al., 2012; Dobretsov et al., 2013; Flemming et al., 2016). Biofilms are composed of various inhabitants, including bacteria, diatoms, fungi, and protozoa (Qian et al., 2007; Callow and Callow, 2011). Biofilms are one of the driving forces for mediating habitat selection and the onset of the settlement behavior of various sessile marine organisms (Bao et al., 2007b; Wahl et al., 2012; Wang et al., 2012). Of the different biofilm components, the functions of bacteria and bacterial community composition are crucial for mediating invertebrate larval settlement (Maki et al., 1989; Chung et al., 2010; Hadfield, 2011; Dobretsov and Rittschof, 2020).
Monospecific bacterial biofilms are shown to modulate larval settlement of many marine invertebrates (Huggett et al., 2006; Bao et al., 2007a; Tran and Hadfield, 2011; Nasrolahi et al., 2012). In mussels, those monospecies biofilms can also promote the settlement of Mytilus coruscus (Yang et al., 2013; Li et al., 2014; Peng et al., 2020b). Because many monospecific bacterial biofilms induce a distinct settlement rate at various cell densities, the density-based underlying mechanisms of bacteria warrant further analysis (Yang et al., 2013; Li et al., 2014).
Bacteria have evolved a density-dependent regulatory system named quorum sensing (QS), which allows them to coordinate and reorganize microbial activity, such as biofilm maturation, swarming, luminescence, and virulence factors (Waters and Bassler, 2005; Steinberg et al., 2011). N-acyl homoserine lactones (AHLs) are signal molecules used by many Gram-negative bacteria (Dobretsov et al., 2013). The intracellular regulatory mechanisms of QS are bacterial density-dependent molecular mechanisms through the activation of transcriptional regulators by binding with the diffusible signal molecules at a certain threshold concentration (Antunes et al., 2010). Because bacteria facilitate the larval settlement of many marine invertebrates, disruption of bacterial biofilms can reduce the attractiveness of the substratum to invertebrate larvae (Dobretsov et al., 2007). QS inhibitors have been applied in biofouling and the aquaculture industry to prevent the macrofouling or to control vibriosis (Dobretsov et al., 2011; Galloway et al., 2012; Pande et al., 2013). QS inhibitors are generally believed to put less selective pressure on bacteria and, therefore, reduce the risk of drug-resistant bacteria (Defoirdt et al., 2004, 2010; Rasmussen et al., 2005; Lönn-Stensrud et al., 2007; von Bodman et al., 2008).
The halogenated furanones initially extracted from the red algae Delisea pulchra are shown to act as antifouling compounds or biofilm formation inhibitors (Maximilien et al., 1998; De Nys et al., 2006; Janssens et al., 2008; Dobretsov et al., 2011). It is shown that natural furanones disturb the QS system in many species of microorganisms (Ren et al., 2004; Roy et al., 2010). 2(5H)-Furanone (FUR) is reported to be a QS inhibition compound against various AHLs and inhibit biofilm formation (Ponnusamy et al., 2010). Moreover, FUR reduces bacterial mobility and biofilm formation, interfers with QS activity, and exerts antimicrobial effects against the pathogen Campylobacter jejuni (Castillo et al., 2015). Although the reduction of larval settlement after treatment with QS inhibitors has been described (Dobretsov et al., 2007), the gene regulatory mechanisms remain elusive.
The present study aims to investigate the mode of action of FUR on the formation of monospecies biofilms of Pseudoalteromonas marina ECSMB14103 and the settlement of plantigrades of the mussel M. coruscus. We demonstrate that FUR mediates the settlement of plantigrades indirectly by regulating the biofilm formation (e.g., EPS production). The transcriptome reveals the genetic basis of the modified biofilms exposed to FUR during biofilm formation.
Materials and Methods
Ethics Statement
The mussel M. coruscus is a common aquaculture bivalve species in Zhoushan (Zhejiang Province, China), and it is not endangered. The animal ethics committee of Shanghai Ocean University approved the experimental protocol for mussel acclimation and experimentation (SHOU-DW-2018-013).
Bacterial Strain and Formation of Monospecific Bacterial Biofilms
The bacterial strain P. marina ECSMB14103 was isolated from natural marine biofilms formed in the East China Sea (Peng et al., 2018). The preparation of P. marina ECSMB14103 biofilms was performed as described previously (Yang et al., 2013; Li et al., 2014). P. marina ECSMB14103 was cultured in Zobell 2216E broth at 25°C for 24 h. Then bacteria cultures were centrifuged (1600 g, 15 min). Cell pellets of bacteria were washed, resuspended, and centrifuged three times to remove residual culture media. The cell pellets were resuspended in 50 mL of autoclaved filtered seawater (AFSW), followed by 100 dilutions before filtering through 25 mm Whatman® Nuclepore 0.2 μm filters (Whatman, United Kingdom). Cells were stained by 0.1% acridine orange (AO) (Sigma-Aldrich, St. Louis, MO, United States) for 5 min, and bacterial densities were determined under an Olympus BX51 epifluorescence microscope with excitation/emission wavelengths of 490/519 nm (Yang et al., 2013; Li et al., 2014). Ten random fields of view were counted for each filter. Three biological replicates were conducted for each treatment group.
Preparation of Treated P. marina ECSMB14103 Biofilms
2(5H)-Furanone (FUR, Sigma 283754) was diluted in AFSW to prepare a stock solution of 10–2 M. The stock solution was prepared fresh and used on the same day. The stock solution was then diluted to the desired concentration (10–3 M, 10–4 M, and 10–5 M). The experiment was composed of two treatments: (1) FUR was added during the formation of P. marina ECSMB14103 biofilms, and FUR was not added during the settlement of plantigrades bioassay; (2) the formation of P. marina ECSMB14103 biofilms occurred in the absence of FUR, and FUR was added during the settlement of plantigrades bioassay. Bacterial biofilm formation was conducted in sterile glass Petri dishes containing one piece of sterile glass coverslip (38 mm × 26 mm); 108 cells of P. marina ECSMB14103 were added with FUR (final concentration: 10–3 M, 10–4 M, and 10–5 M) and without FUR (control group). The dishes were incubated at 28°C for 48 h. The control group (CTR) was prepared without adding FUR during or after bacterial biofilm formation. No adding of FUR or bacterial cells in the Petri dish containing a sterile glass slip was set up as the negative control (NC). The unattached bacterial cells were removed by gentle washing three times with 60 mL AFSW. The bacteria firmly attached to the surfaces of glass slips were considered as attached P. marina ECSMB14103 biofilms.
Culture of Plantigrades
Plantigrades (0.77 ± 0.15 mm in shell length and 0.52 ± 0.09 mm in shell width) were supplied by the Shengsi Service Center of Marine Science and Technology Development (Zhoushan, China) and shipped to Shanghai Ocean University on the same day (Shanghai, China). Plantigrades were cultured in 15-L polycarbonate tanks with filtered seawater (acetate-fiber filter: 1.2 μm pore size) at an initial density of 1 plantigrades mL–1 at 18°C. The algae Platymonas helgolandica var. tsingtaoensis were supplied to the mussel as a food source (1 × 104 cells ml–1). The seawater was renewed every day. Plantigrades were acclimated for 1 week before settlement bioassays.
Enumeration of Bacterial Density
Pseudoalteromonas marina ECSMB14103 biofilms were fixed in 5% formalin solution for a maximum duration of 3 weeks. Three replicate biofilms from each experimental treatment group were carried out. Samples were first washed with AFSW and stained by 0.1% AO for 5 min. Bacterial densities were determined under an Olympus BX51 epifluorescence microscope as previously described (Li et al., 2014). Three biological replicate biofilms were examined for each treatment group, and the biofilm biovolumes were evaluated by 10 random fields of view.
Settlement Bioassays
Settlement bioassays were carried out as previously described (Yang et al., 2013; Li et al., 2014). Ten plantigrades were transferred into individual glass Petri dishes (Ø64 mm × 19 mm height) containing 20 mL of AFSW (with or without FUR) and a P. marina ECSMB14103 biofilm or treated biofilms. The induction rate was evaluated by the percentage of settled plantigrades after 24 h. Plantigrades that attached to the surface of the glass slip by byssal threads were considered “settled.” Nine biological replicates were examined for each treatment group. Assays were performed at 18°C in darkness.
Biofilm Staining Procedure
The 10–4 M FUR-treated biofilms were selected for biofilm staining due to the lowest plantigrade settlement rate for comparing the individual components in the biofilm matrix. The visualization of proteins, lipids, α-polysaccharides, and β-polysaccharides in the P. marina ECSMB14103 biofilms was carried out by following a previously described method (González-Machado et al., 2018; Liang et al., 2020). Four fluorescent dyes, including fluorescein isothiocyanate isomer I (FITC), DiIC18(5) oil, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate (DiD′oil; Invitrogen D307), concanavalin A, tetramethylrhodamine conjugate (ConA-TmR; Invitrogen C860), and calcofluor white M2R (CFW; Sigma 18909) were used for staining the proteins, lipids, α-polysaccharides, and β-polysaccharides, respectively. Four working solutions were prepared in 150 mM NaCl solution: 46.6 μg/ml FITC (stock solution: 2 mg FITC in 100 μL absolute ethanol), 79.4 μg/mL DiD′oil (stock solution: 25 mg DiD′oil in 2.5 ml of absolute ethanol), 944.8 μg/mL ConA-TMR (stock solution: 10 mg ConA-TMR in 2 ml distilled water with 168 mg NaHCO3), and 189 μl/ml CFW. P. marina ECSMB14103 biofilms in glass slides were rinsed with 150 mM NaCl solution and treated with 50 μL working solution for 20 min. The glass slides were rinsed again with 150 mM NaCl solution and allowed to air dry. Image acquisition was performed by confocal laser scanning microscopy (Leica TCS SP8) using the 63 × objective with oil immersion (Liang et al., 2020). For image analysis, the biovolume of proteins, lipids, α-polysaccharides, and β-polysaccharides were calculated using FIJI software (Schindelin et al., 2012). For each staining treatment group, seven replicates were performed.
RNA Isolation, cDNA Library Construction, and Illumina Sequencing
Pseudoalteromonas marina ECSMB14103 biofilms treated with 10–4 M FUR during biofilm formation were further analyzed by Illumina sequencing. The biofilms were scraped from the surface of the glass slides into a sterile 1.5 mL tube. Total RNA was extracted with the RNAiso Plus reagent according to the manufacturer’s instructions (Takara, Japan) and treated with DNAse for removing genomic DNA.
The sequencing libraries were prepared following the TruSeqTM RNA sample preparation Kit from Illumina (San Diego, CA, United States) using 2 μg of total RNA. RNA-seq sequencing was performed with the Illumina HiSeq X Ten (Reuter et al., 2015; Li et al., 2019) with 150 bp pair-end reads at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). Raw sequences have been submitted to the NCBI database under the accession number: SRP226488.
Bioinformatic Analysis
Clean reads were obtained from raw reads by discarding the adapter sequences and removing low-quality sequences and sequences with more than 5% of unknown bases. The clean reads were mapped to the P. marina ECSMB14103 genome (NCBI accession number: CP023558) using the Bowtie2 software (Langmead and Salzberg, 2012). The expression level for each transcript for each gene was calculated by RSEM (Li and Dewey, 2011) using a transcript per million mapped reads (TPM) method. The differentially expressed genes (DEGs) between the two treatment groups were performed by EdgeR (Robinson et al., 2010), and genes with FDR value ≤0.05 and | log2FC| ≥ 2 were assigned as differentially expressed. KOBAS 2.01 was applied to identify significantly enriched KEGG pathways using Fisher’s exact test. Benjamini–Hochberg (BH) multiple testing correction was used for p-value correction. The corrected p-value ≤ 0.05 was considered to be significantly different.
Statistical Analysis
The settlement of plantigrade data expressed in percentages was arcsine-transformed before the normality and homogeneity of variance. The bacterial cell densities and the settlement data were compared by Wilcoxon/Kruskal–Wallis test after being tested for normality (Shapiro–Wilk test) and homogeneity (O’Brien test) using JMPTM software (SAS Institute, Shanghai, China). The analysis of the biovolume of α-polysaccharides, β-polysaccharides, proteins, and lipids were carried out using Student’s t-test. A p-value < 0.05 was considered significantly different.
Results
The Effect of Treated P. marina ECSMB14103 Biofilms on Plantigrade Settlement
The percentage of the settlement of M. coruscus plantigrades on the P. marina ECSMB14103 developed in the absence of FUR exposure (CTR) was 46 ± 6% (Figure 1). In the NC, the percentage of the settlement was 12 ± 4% (Figure 1). Addition of FUR during the settlement assay did not affect the percentage of settlement compared to the CTR group. The biofilms developed in the presence of 10–3 M and 10–4 M FUR significantly reduced the settlement of plantigrades compared to the CTR group (p < 0.05, Figure 1).
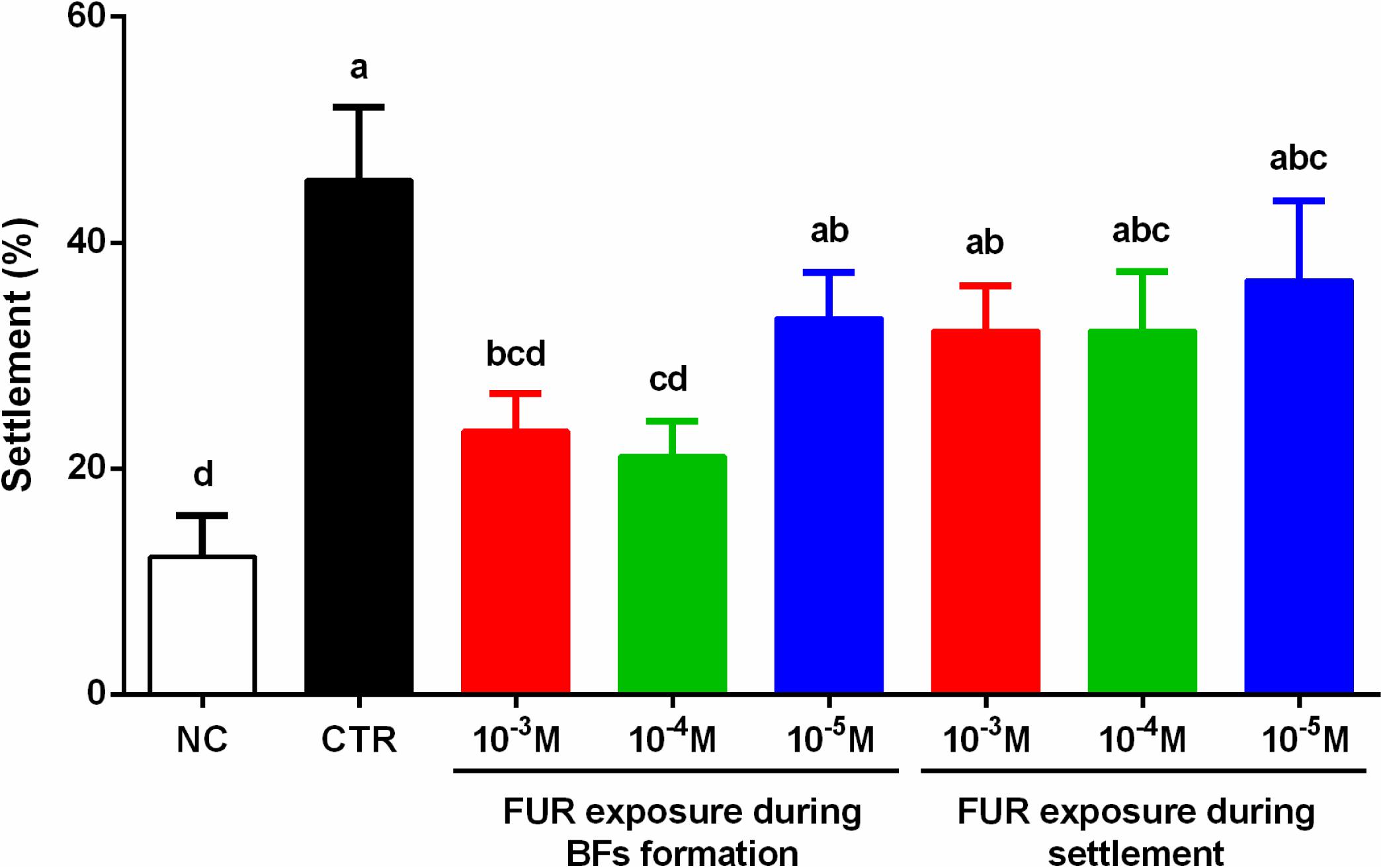
Figure 1. Percentages of the settlement of M. coruscus plantigrades. Significant differences (p < 0.05) are represented by different letters above the columns. Control (CTR), P. marina ECSMB14103 biofilm prepared on a sterile glass slide without adding FUR; negative control (NC), non-biofilm glass slide.
The Effect of FUR on Bacterial Densities and Individual Components in Biofilms
Bacterial densities were significantly increased in the biofilms treated with 10–3 M FUR compared to the CTR biofilms (p < 0.05, Figure 2). Treatment of biofilms with 10–4 M FUR significantly reduced bacterial densities compared to the CTR biofilms (p < 0.05, Figure 2). The individual components, including proteins, lipids, α-polysaccharides, and β-polysaccharides within the EPS matrix of biofilms treated by 10–4 M FUR was also determined (Figure 3). β-polysaccharides were the major components in the biofilm matrix. The 10–4 M FUR treatment significantly reduced the biovolume of the α-polysaccharides and β-polysaccharides relative to the control (CTR) biofilms (p < 0.01 and p < 0.0001, respectively). The biovolume of proteins increased significantly in 10–4 M FUR treated biofilms compared to the CTR biofilms (p < 0.0001). The 10–4 M FUR exposure had no effect on the biovolume of lipids (p > 0.05).
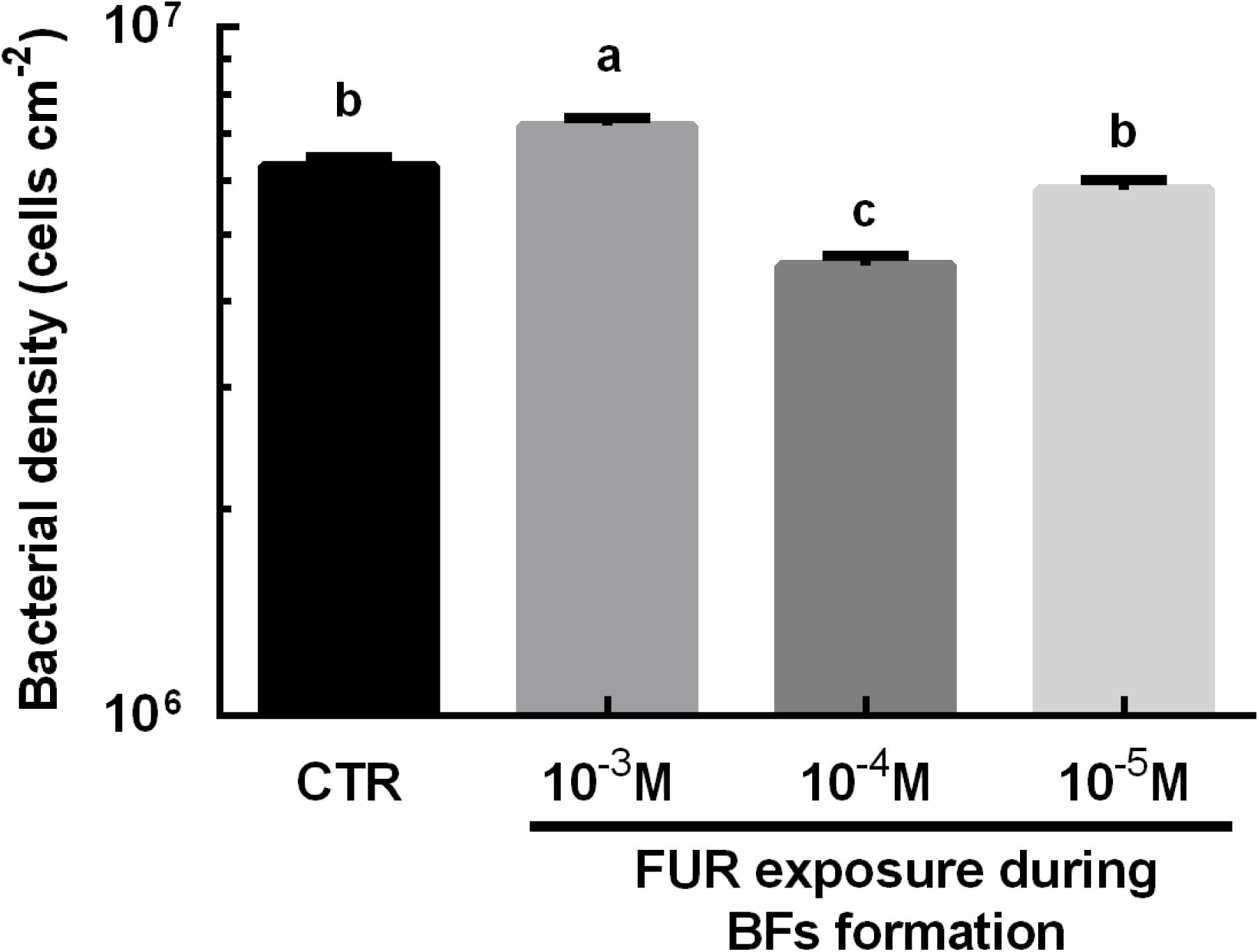
Figure 2. Bacterial densities of FUR-treated biofilms. The results are presented as the mean ± SEM of three biological samples, and each sample was calculated by 10 random fields of view. Control (CTR), P. marina ECSMB14103 biofilm prepared on a sterile glass slide without adding FUR. Significant differences (p < 0.05) are represented by different letters above the columns.
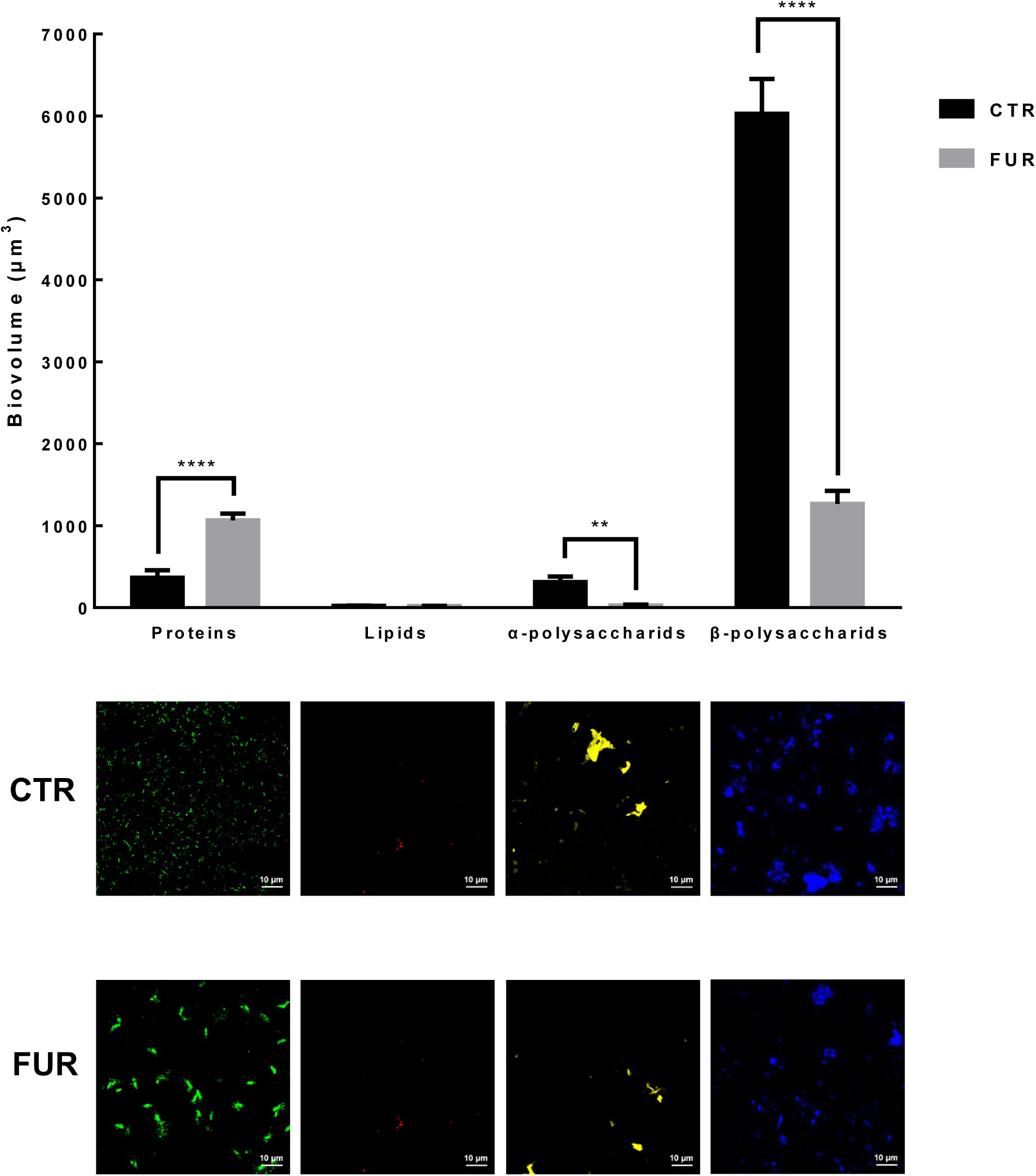
Figure 3. Biovolume of the individual components in biofilms formed by P. marina ECSMB14103. Images of each component are shown. **p < 0.01; **** p < 0.0001. Control (CTR), P. marina ECSMB14103 biofilm prepared on a sterile glass slide without adding FUR; FUR, 104M FUR treated P. marina ECSMB14103 biofilm.
Transcriptomics of P. marina ECSMB14103 Biofilms Exposed to 10–4 M FUR
In order to understand the underlying molecular mechanisms between the CTR group and furanone-treated group during biofilm formation, biofilms treated with 10–4 M FUR were selected for transcriptome sequencing due to the lowest settlement. The transcriptome results revealed 61 DEGs in 10–4 M FUR-treated biofilms in comparison to the CTR biofilms. Among these DEGs, 36 upregulated and 25 downregulated genes were identified. A heat map based on the expression pattern of DEGs was generated, and two replicates exhibited a similar expression pattern (Figure 4).
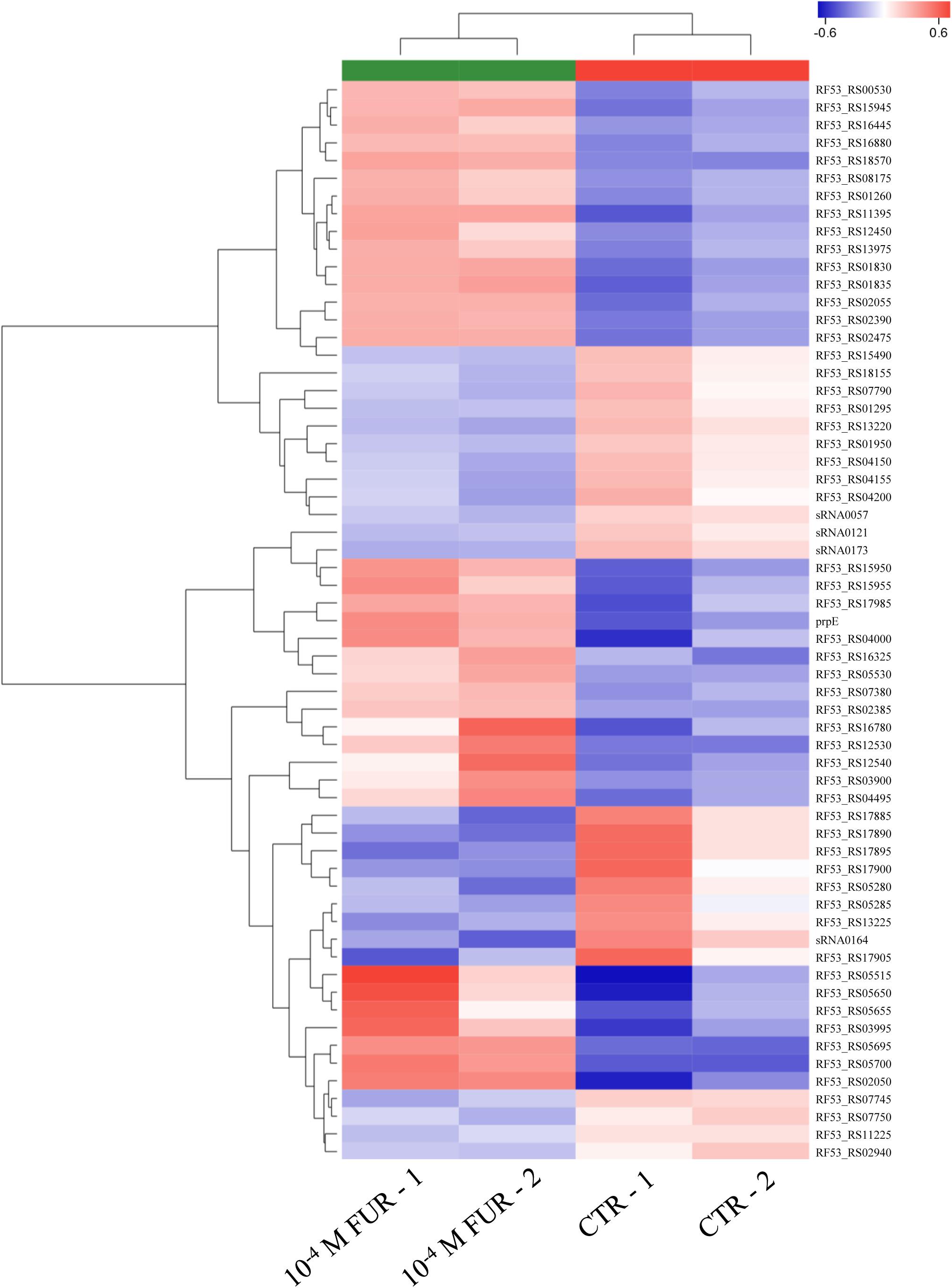
Figure 4. Heat map of the DEGs identified in RNA-seq data in CTR and FUR-treated biofilms are shown. The gene names are shown on the right side of the heat map. The gene clustering is shown on the left side. Control (CTR), P. marina ECSMB14103 biofilm prepared on a sterile glass slide without adding FUR; FUR, 104M FUR treated P. marina ECSMB14103 biofilm.
The DEGs were mapped to the KEGG database to define their metabolic pathway for understanding the biological function of the DEGs further. The DEGs were classified into five categories and 15 KEGG metabolic pathways (Figure 5). The 34 DEGs were divided into a metabolic category, including amino acid metabolism (8 DEGs), biosynthesis of other secondary metabolites (3 DEGs), carbohydrate metabolism (9 DEGs), energy metabolism (5 DEGs), lipid metabolism (3 DEGs), metabolism of cofactors and vitamins (2 DEGs), metabolism of other amino acids (1 DEG), nucleotide metabolism (2 DEGs), and xenobiotic biodegradation and metabolism (1 DEG) (Figure 5). The remaining six pathways contained 1 DEG each (Figure 6). KEGG pathway enrichment analysis showed that propanoate metabolism, carbapenem biosynthesis, arginine and proline metabolism, and methane metabolism were significantly enriched in the 10–4 M FUR exposure biofilms compared to the CTR biofilms (Figure 6).
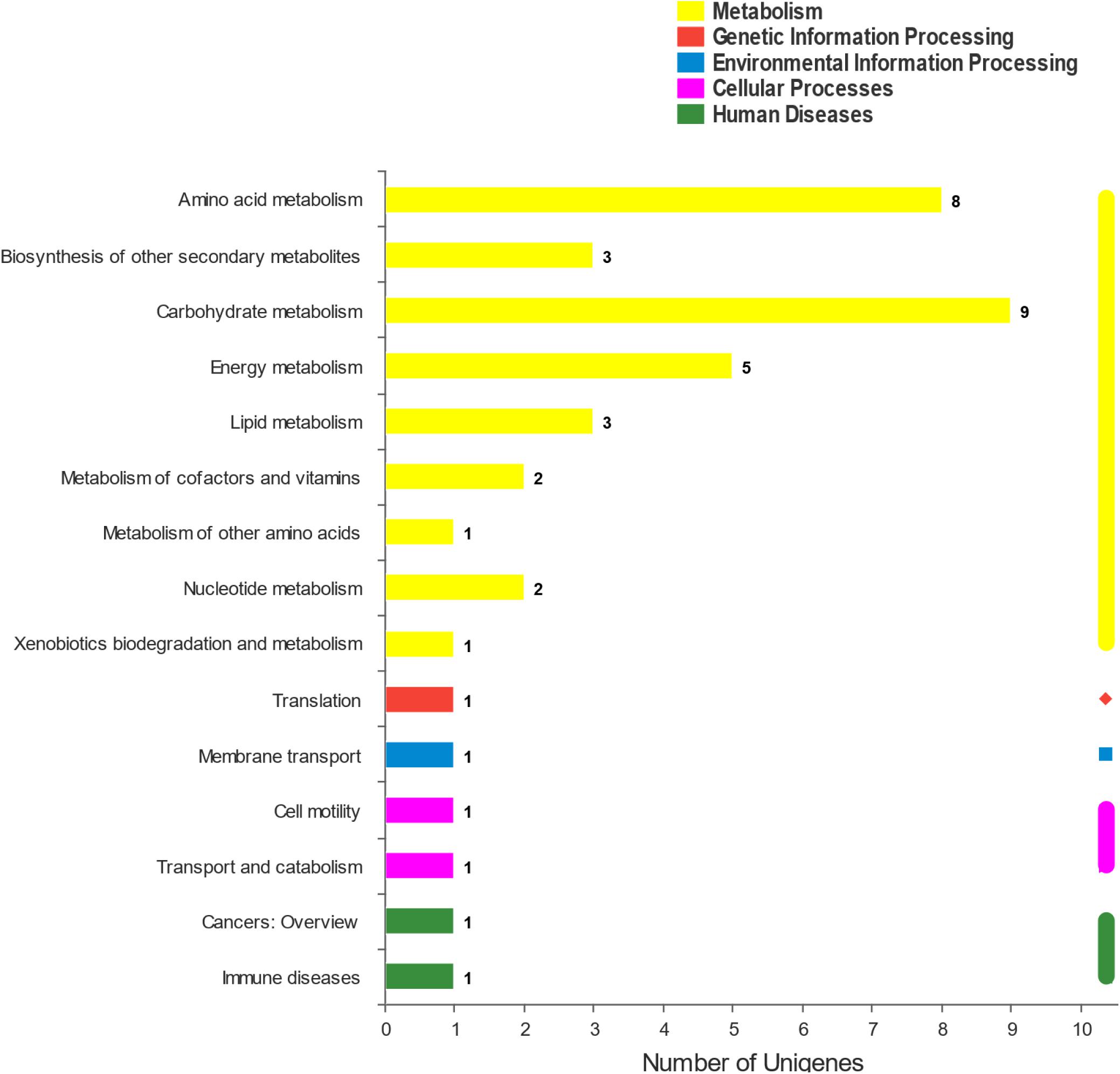
Figure 5. KEGG pathway classification of DEGs between CTR and FUR exposure biofilms. The horizontal coordinate represents the number of DGEs annotated to each subcategory. Control (CTR), P. marina ECSMB14103 biofilm prepared on a sterile glass slide without adding FUR; FUR, 104M FUR-treated P. marina ECSMB14103 biofilm.
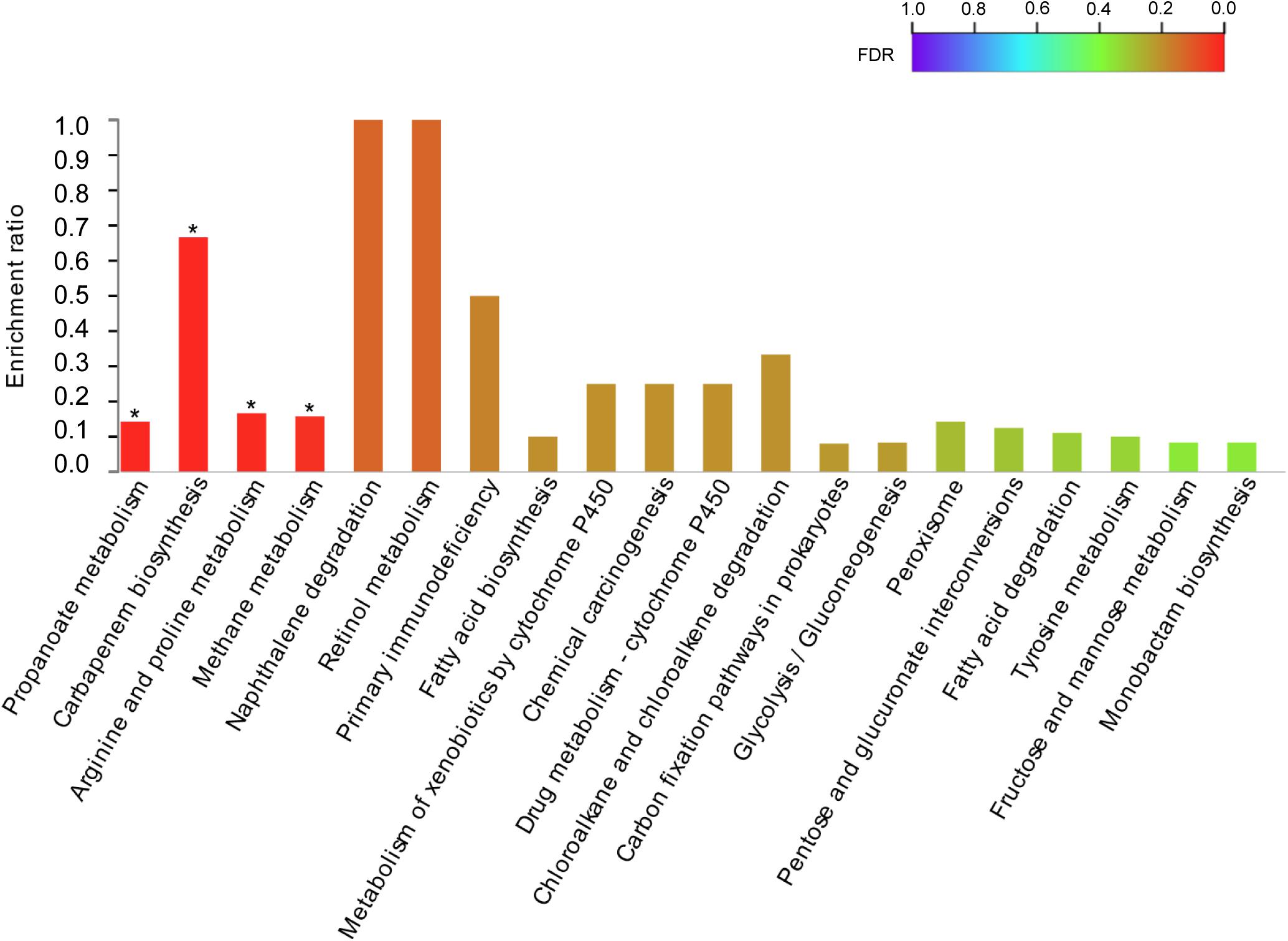
Figure 6. KEGG pathway enrichment analysis of DEGs between CTR and FUR exposure biofilms. *p < 0.05. Control (CTR), P. marina ECSMB14103 biofilm prepared on a sterile glass slide without adding FUR; FUR, 104M FUR-treated P. marina ECSMB14103 biofilm.
Discussion
An earlier study demonstrates that QS blockers modify the marine bacteria communities and indirectly affect the larval attachment of the polychaete Hydroides elegans and the bryozoan Bugula neritina (Dobretsov et al., 2007). In the present study, we investigate the extracellular substances in P. marina ECSMB14103 biofilms and transcriptome analysis of biofilms in response to FUR, which allows for understanding how these changes impact the biofilm formation on a molecular level and, subsequently, the settlement of M. coruscus plantigrades. Our data also reveal an indirect effect of exposure to FUR on the inhibition of the settlement of plantigrades.
The mode of action of FUR exposure is a crucial factor for determining the settlement of plantigrades. Addition of FUR during the settlement assay did not affect the settlement of plantigrades, but the FUR exposure during biofilm formation significantly decreased the settlement of plantigrades. These results suggest that the inducing activity of biofilms for the settlement of plantigrades depends on a properly functioning biofilm, which formed in the absence of FUR during biofilm formation. These observations are in agreement with the previous study, which shows that the furanone added during biofilm formation inhibits the larval attachment of H. elegans and B. neritina (Dobretsov et al., 2007). It is further demonstrated that exposure to furanone significantly modifies the bacterial density and bacterial communities in multispecies bacterial biofilms (Dobretsov et al., 2007). Halogenated furanones of D. pulchra inhibit the swarming motility of biofilm through QS regulated gene expression (Rasmussen et al., 2000). Biofilm formation can also be effectively inhibited by many synthetic brominated furanones (He et al., 2012; Kayumov et al., 2015).
Previous studies suggest that the cell density of bacteria is positively correlated with the percentage of larval settlement (Huang and Hadfield, 2003; Wang et al., 2012). In this study, the highest bacterial density was found in the 10–3 M FUR-treated group, but the inducing activity of the biofilm was significantly reduced. In addition, the bacterial density in FUR treated groups shows a threshold effect. This may suggest that FUR does not affect the growth of P. marina, which is similar to previous reports that QS inhibitors effectively influence the pathogenicity rather than bacterial growth (Rasmussen and Givskov, 2006; Kalia et al., 2014). Generally, because bacterial cells account for only 2–5% of the biofilm mass (Kristensen et al., 2008), the EPS of biofilm is involved in regulating larval settlement (Lau et al., 2003; Qian et al., 2007; Peng et al., 2020a). The EPS of biofilms consists of many organic substances. The main constituents of EPS are the polysaccharides, proteins, and lipids, which responsible for maintaining the architecture and activity of the biofilm (González-Machado et al., 2018). A previous study shows that polysaccharides rather than proteins in the EPS matrix function as larval settlement cues in H. elegans (Lam et al., 2005). The observed reduction in the biovolume of both α-polysaccharides and β-polysaccharides in biofilms exposed to 10–4 M FUR when compared to control biofilms may explain the low settlement rate. Although proteins in EPS did not affect the larval settlement of H. elegans (Lam et al., 2005), those extracellular proteins are necessary for maintaining the structure of EPS, providing nutrition, and releasing the cells (Flemming and Wingender, 2010).
A previous study shows that FUR inhibits biofilm formation and has a negative effect on a wide range of AHLs (Ponnusamy et al., 2010). It is recently reported that FUR interfers with the QS system and affects the motility and biofilm formation in C. jejuni (Castillo et al., 2015). Although FUR has no effect on the QS-dependent processes in Bacillus subtilis, FUR affected the cellular metabolism in B. subtilis (Kayumov et al., 2015). We hypothesized that exposure to FUR may impair the metabolic processes in P. marina ECSMB14103 during biofilm formation. Transcriptome data shows that 61 genes are differentially expressed after exposure to 10–4 M FUR. The majority of DEG-encoding proteins are involved in the metabolism process (Figure 5). Glutamate 5-kinase (downregulated; RF53_RS13220) and glutamate-5-semialdehyde dehydrogenase (downregulated; RF53_RS13225) and aspartate kinase (upregulated; RF53_RS12450) involved in the biosynthesis of other secondary metabolites are significantly affected in the FUR-treatment biofilms compared to the CTR biofilms. Aspartate kinase is important for the synthesis of an intermediate compound, diaminopimelic acid, which is involved in lysine biosynthesis, and this compound is crucial for the synthesis of the cell wall in most bacteria (Yoshida et al., 2007). Although it has been proposed that the aspartate kinase gene (lysC) might act in biofilm formation (Mueller et al., 2007), there is not much evidence to support that. We found that flagellar assembly protein fliH (RF53_RS02940) participating in cell motility was significantly downregulated in FUR-exposed P. marina biofilms relative to the CTR biofilms. Our recent study also demonstrates that a flagellin synthetic protein gene fliP deletion of P. marina contributes to the loss of the flagella structure and motility (Liang et al., 2020). The flagellum is critical for the mobility of bacteria, which is crucial for the initial surface attachment, subsequent biofilm formation and virulence (O’Toole and Kolter, 1998; Watnick et al., 2001; Lemon et al., 2007; Liang et al., 2020). Similarly, exposure to furanone inhibits gene expression involved in flagellar biosynthesis and metabolism in Salmonella enterica Serovar Typhimurium (Janssens et al., 2008).
In the present study, propanoate metabolism, carbapenem biosynthesis, arginine and proline metabolism, and methane metabolism are significantly affected in response to FUR exposure. Carbapenems are naturally produced metabolites acting as potent β-lactam antibiotics with a broad spectrum of antibiotic activity against both Gram-positive and Gram-negative bacteria (McGowan et al., 1999). It is demonstrated that the production of the secondary metabolite, carbapenem, is coordinately governed by a QS system (Thomson et al., 2000). Taken together, our data indicates that the alteration of metabolism in P. marina ECSMB14103 is impaired in the presence of FUR.
Conclusion
In conclusion, the present study shows that 2(5H)-Furanone directly affects biofilm formation by altering EPS components and metabolic processes in P. marina ECSMB14103 biofilms and indirectly reduces the mussel settlement. We propose that FUR could be used in the biofouling industry for reducing the initial attachment of micro- or macrofouling. The present study provides new insights into the molecular mechanisms of controlling biofouling. The established method here could be used for screening antifouling compounds in the biofouling industry. More studies are needed to further understand the molecular regulatory mechanisms of FUR on bacterial biofilms.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
Y-FL, XL, and J-LY designed the experiments. XZ and Z-YC performed the experiments. Y-FL, Y-TZ, and D-DF analyzed the data. Y-FL, SD, and J-LY critically reviewed the data and wrote the manuscript. All authors read and approved the manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (2018YFD0900601), National Natural Science Foundation of China (Nos. 31802321, 41876159, and 41476131), Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0402), and the Shanghai Sailing Program (19YF1419500).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Antunes, L. C. M., Ferreira, R. B. R., Buckner, M. M. C., and Finlay, B. B. (2010). Quorum sensing in bacterial virulence. Microbiology 156, 2271–2282. doi: 10.1099/mic.0.038794-0
Bao, W. Y., Satuito, C. G., Yang, J. L., and Kitamura, H. (2007a). Larval metamorphosis of the mussel Mytilus galloprovincialis in response to Alteromonas sp. 1: evidence for two chemical cues? Mar. Biol. 152, 657–666. doi: 10.1007/s00227-007-0720-2
Bao, W. Y., Satuito, C. G., Yang, J. L., and Kitamura, H. (2007b). Larval settlement and metamorphosis of the mussel Mytilus galloprovincialis in response to biofilms. Mar. Biol. 150, 565–574. doi: 10.1007/s00227-006-0383-4
Callow, J. A., and Callow, M. E. (2011). Trends in the development of environmentally friendly fouling-resistant marine coatings. Nat. Commun. 2:244. doi: 10.1038/ncomms1251
Castillo, S., Heredia, N., and García, S. (2015). 2 (5H)-Furanone, epigallocatechin gallate, and a citric-based disinfectant disturb quorum-sensing activity and reduce motility and biofilm formation of Campylobacter jejuni. Folia Microbiol. 60, 89–95. doi: 10.1007/s12223-014-0344-0
Chung, H. C., Lee, O. O., Huang, Y. L., Mok, S. Y., Kolter, R., and Qian, P. Y. (2010). Bacterial community succession and chemical profiles of subtidal biofilms in relation to larval settlement of the polychaete Hydroides elegans. ISME J. 4, 817–828. doi: 10.1038/ismej.2009.157
De Nys, R., Givskov, M., Kumar, N., Kjelleberg, S., and Steinberg, P. (2006). “Furanones,” in Antifouling Compounds, eds N. Fusetani and A. S. Clare (Berlin: Springer), 55–86.
Defoirdt, T., Boon, N., and Bossier, P. (2010). Can bacteria evolve resistance to quorum sensing disruption? PLoS Pathog. 6:e1000989. doi: 10.1371/journal.ppat.1000989
Defoirdt, T., Boon, N., Bossier, P., and Verstraete, W. (2004). Disruption of bacterial quorum sensing: an unexplored strategy to fight infections in aquaculture. Aquaculture 240, 69–88. doi: 10.1016/j.aquaculture.2004.06.031
Dobretsov, S., Abed, R. M., and Teplitski, M. (2013). Mini-review: inhibition of biofouling by marine microorganisms. Biofouling 29, 423–441. doi: 10.1080/08927014.2013.776042
Dobretsov, S., Dahms, H. U., Yili, H., Wahl, M., and Qian, P. Y. (2007). The effect of quorum-sensing blockers on the formation of marine microbial communities and larval attachment. FEMS Microbiol. Ecol. 60, 177–188. doi: 10.1111/j.1574-6941.2007.00285.x
Dobretsov, S., and Rittschof, D. (2020). Love at first taste: induction of larval settlement by marine microbes. Int. J. Mol. Sci. 21:731. doi: 10.3390/ijms21030731
Dobretsov, S., Teplitski, M., Bayer, M., Gunasekera, S., Proksch, P., and Paul, V. J. (2011). Inhibition of marine biofouling by bacterial quorum sensing inhibitors. Biofouling 27, 893–905. doi: 10.1080/08927014.2011.609616
Flemming, H. C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Galloway, W. R., Hodgkinson, J. T., Bowden, S., Welch, M., and Spring, D. R. (2012). Applications of small molecule activators and inhibitors of quorum sensing in Gram-negative bacteria. Trends Microbiol. 20, 449–458. doi: 10.1016/j.tim.2012.06.003
González-Machado, C., Capita, R., Riesco-Pelaez, F., and Alonso-Calleja, C. (2018). Visualization and quantification of the cellular and extracellular components of Salmonella Agona biofilms at different stages of development. PLoS One 13:e0200011. doi: 10.1371/journal.pone.0200011
Hadfield, M. G. (2011). Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Annu. Rev. Mar. Sci. 3, 453–470. doi: 10.1146/annurev-marine-120709-142753
He, Z. Y., Wang, Q., Hu, Y. J., Liang, J. P., Jiang, Y. T., Ma, R., et al. (2012). Use of the quorum sensing inhibitor furanone C-30 to interfere with biofilm formation by Streptococcus mutans and its luxS mutant strain. Int. J. Antimicrob. Agents 40, 30–35. doi: 10.1016/j.ijantimicag.2012.03.016
Huang, S., and Hadfield, M. G. (2003). Composition and density of bacterial biofilms determine larval settlement of the polychaete Hydroides elegans. Mar. Ecol. Prog. Ser. 260, 161–172. doi: 10.3354/meps260161
Huggett, M. J., Williamson, J. E., de Nys, R., Kjelleberg, S., and Steinberg, P. D. (2006). Larval settlement of the common Australian sea urchin Heliocidaris erythrogramma in response to bacteria from the surface of coralline algae. Oecologia 149, 604–619. doi: 10.1007/s00442-006-0470-8
Janssens, J. C. A., Steenackers, H., Robijns, S., Gellens, E., Levin, J., Zhao, H., et al. (2008). Brominated furanones inhibit biofilm formation by Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 74, 6639–6648. doi: 10.1128/AEM.01262-08
Kalia, V. C., Wood, T. K., and Kumar, P. (2014). Evolution of resistance to quorum sensing inhibitors. Microb. Ecol. 68, 13–23. doi: 10.1007/s00248-013-0316-y
Kayumov, A. R., Khakimullina, E. N., Sharafutdinov, I. S., Trizna, E. Y., Latypova, L. Z., Lien, H. T., et al. (2015). Inhibition of biofilm formation in Bacillus subtilis by new halogenated furanones. J. Antibiot. 68, 297–301. doi: 10.1038/ja.2014.143
Kristensen, J. B., Meyer, R. L., Laursen, B. S., Shipovskov, S., Besenbacher, F., and Poulsen, C. H. (2008). Antifouling enzymes and the biochemistry of marine settlement. Biotechnol. Adv. 26, 471–481. doi: 10.1016/j.biotechadv.2008.05.005
Lam, C., Harder, T., and Qian, P. Y. (2005). Induction of larval settlement in the polychaete Hydroides elegans by extracellular polymers of benthic diatoms. Mar. Ecol. Prog. Ser. 286, 145–154. doi: 10.3354/meps286145
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lau, S. C. K., Harder, T., and Qian, P. Y. (2003). Induction of larval settlement in the serpulid polychaete Hydroides elegans (Haswell): role of bacterial extracellular polymers. Biofouling 19, 197–204. doi: 10.1080/08927014.2003.10382982
Lemon, K. P., Higgins, D. E., and Kolter, R. (2007). Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 189, 4418–4424. doi: 10.1128/JB.01967-06
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, G., Shi, M., Zhao, S., Long, Y., and Zhu, Y. (2019). Toxicity response of silkworm intestine to Bacillus cereus SW7-1 pathogen. Sci. Total Environ. 692, 1282–1290. doi: 10.1016/j.scitotenv.2019.07.349
Li, Y. F., Guo, X. P., Yang, J. L., Liang, X., Bao, W. Y., Shen, P. J., et al. (2014). Effects of bacterial biofilms on settlement of plantigrades of the mussel Mytilus coruscus. Aquaculture 433, 434–441. doi: 10.1016/j.aquaculture.2014.06.031
Liang, X., Zhang, X. K., Peng, L. H., Zhu, Y. T., Yoshida, A., Osatomi, K., et al. (2020). The flagellar gene regulates biofilm formation and mussel larval settlement and metamorphosis. Int. J. Mol. Sci. 21:710. doi: 10.3390/ijms21030710
Lönn-Stensrud, J., Petersen, F. C., Benneche, T., and Scheie, A. A. (2007). Synthetic bromated furanone inhibits autoinducer-2-mediated communication and biofilm formation in oral streptococci. Oral Microbiol. Immunol. 22, 340–346. doi: 10.1111/j.1399-302x.2007.00367.x
Maki, J. S., Rittschof, D., Schmidt, A. R., Snyder, A. G., and Mitchell, R. (1989). Factors controlling attachment of bryozoan larvae: a comparison of bacterial films and unfilmed surfaces. Biol. Bull. 177, 295–302. doi: 10.2307/1541944
Maximilien, R., de Nys, R., Holmström, C., Gram, L., Givskov, M., Crass, K., et al. (1998). Chemical mediation of bacterial surface colonisation by secondary metabolites from the red alga Delisea pulchra. Aquat. Microb. Ecol. 15, 233–246. doi: 10.3354/ame015233
McGowan, S. J., Holden, M. T., Bycroft, B. W., and Salmond, G. P. (1999). Molecular genetics of carbapenem antibiotic biosynthesis. Antonie Van Leeuwenhoek 75, 135–141. doi: 10.1023/a:1001768428383
Mueller, R. S., McDougald, D., Cusumano, D., Sodhi, N., Kjelleberg, S., Azam, F., et al. (2007). Vibrio cholerae strains possess multiple strategies for abiotic and biotic surface colonization. J. Bacteriol. 189, 5348–5360. doi: 10.1128/JB.01867-06
Nasrolahi, A., Stratil, S. B., Jacob, K. J., and Wahl, M. (2012). A protective coat of microorganisms on macroalgae: inhibitory effects of bacterial biofilms and epibiotic microbial assemblages on barnacle attachment. FEMS Microbiol. Ecol. 81, 583–595. doi: 10.1111/j.1574-6941.2012.01384.x
O’Toole, G. A., and Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304. doi: 10.1046/j.1365-2958.1998.01062.x
Pande, G. S. J., Scheie, A. A., Benneche, T., Wille, M., Sorgeloos, P., Bossier, P., et al. (2013). Quorum sensing-disrupting compounds protect larvae of the giant freshwater prawn Macrobrachium rosenbergii from Vibrio harveyi infection. Aquaculture 406, 121–124. doi: 10.1016/j.aquaculture.2013.05.015
Peng, L. H., Liang, X., Chang, R. H., Mu, J. Y., Chen, H. E., Yoshida, A., et al. (2020a). A bacterial polysaccharide biosynthesis-related gene inversely regulates larval settlement and metamorphosis of Mytilus coruscus. Biofouling doi: 10.1080/08927014.2020.1807520 [Epub ahead of print].
Peng, L. H., Liang, X., Guo, X. P., Yoshida, A., Osatomi, K., and Yang, J. L. (2018). Complete genome of Pseudoalteromonas marina ECSMB14103, a mussel settlement-inducing bacterium isolated from the East China Sea. Mar. Genomics 41, 46–49. doi: 10.1016/j.margen.2018.04.001
Peng, L. H., Liang, X., Xu, J. K., Dobretsov, S., and Yan, J. L. (2020b). Monospecific biofilms of Pseudoalteromonas promote larval settlement and metamorphosis of Mytilus coruscus. Sci. Rep. 10:2577.
Ponnusamy, K., Paul, D., Kim, Y. S., and Kweon, J. H. (2010). 2 (5H)-Furanone: a prospective strategy for biofouling-control in membrane biofilm bacteria by quorum sensing inhibition. Braz. J. Microbiol. 41, 227–234. doi: 10.1590/S1517-83822010000100032
Qian, P. Y., Lau, S. C. K., Dahms, H. U., Dobretsov, S., and Harder, T. (2007). Marine biofilms as mediators of colonization by marine macroorganisms: implications for antifouling and aquaculture. Mar. Biotechnol. 9, 399–410. doi: 10.1007/s10126-007-9001-9
Rasmussen, T. B., Bjarnsholt, T., Skindersoe, M. E., Hentzer, M., Kristoffersen, P., Köte, M., et al. (2005). Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 187, 1799–1814. doi: 10.1128/JB.187.5.1799-1814.2005
Rasmussen, T. B., and Givskov, M. (2006). Quorum sensing inhibitors: a bargain of effects. Microbiology 152, 895–904. doi: 10.1099/mic.0.28601-0
Rasmussen, T. B., Manefield, M., Andersen, J. B., Eberl, L., Anthoni, U., Christophersen, C., et al. (2000). How Delisea pulchra furanones affect quorum sensing and swarming motility in Serratia liquefaciens MG1. Microbiology 146, 3237–3244. doi: 10.1099/00221287-146-12-3237
Ren, D., Bedzyk, L. A., Ye, R. W., Thomas, S. M., and Wood, T. K. (2004). Differential gene expression shows natural brominated furanones interfere with the autoinducer-2 bacterial signaling system of Escherichia coli. Biotechnol. Bioeng. 88, 630–642. doi: 10.1002/bit.20259
Reuter, J. A., Spacek, D., and Snyder, M. P. (2015). High-throughput sequencing technologies. Mol. Cell 58, 586–597. doi: 10.1016/j.molcel.2015.05.004
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Roy, V., Fernandes, R., Tsao, C. Y., and Bentley, W. E. (2010). Cross species quorum quenching using a native AI-2 processing enzyme. ACS Chem. Biol. 5, 223–232. doi: 10.1021/cb9002738
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Steinberg, P. D., Rice, S. A., Campbell, A. H., McDougald, D., and Harder, T. (2011). Interfaces between bacterial and eukaryotic “neuroecology”. Integr. Comp. Biol. 51, 794–806. doi: 10.1093/icb/icr115
Thomson, N. R., Crow, M. A., McGowan, S. J., Cox, A., and Salmond, G. P. C. (2000). Biosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing control. Mol. Microbiol. 36, 539–556. doi: 10.1046/j.1365-2958.2000.01872.x
Tran, C., and Hadfield, M. G. (2011). Larvae of Pocillopora damicornis (Anthozoa) settle and metamorphose in response to surface-biofilm bacteria. Mar. Ecol. Prog. Ser. 433, 85–96. doi: 10.3354/meps09192
von Bodman, S. B., Willey, J. M., and Diggle, S. P. (2008). Cell-cell communication in bacteria: united we stand. J. Bacteriol. 190, 4377–4391. doi: 10.1128/JB.00486-08
Wahl, M., Goecke, F., Labes, A., Dobretsov, S., and Weinberger, F. (2012). The second skin: ecological role of epibiotic biofilms on marine organisms. Front. Microbiol. 3:292. doi: 10.3389/fmicb.2012.00292
Wang, C., Bao, W. Y., Gu, Z. Q., Li, Y. F., Liang, X., Ling, Y., et al. (2012). Larval settlement and metamorphosis of the mussel Mytilus coruscus in response to natural biofilms. Biofouling 28, 249–256. doi: 10.1080/08927014.2012.671303
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Watnick, P. I., Lauriano, C. M., Klose, K. E., Croal, L., and Kolter, R. (2001). The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39, 223–235. doi: 10.1046/j.1365-2958.2001.02195.x
Yang, J. L., Shen, P. J., Liang, X., Li, Y. F., Bao, W. Y., and Li, J. L. (2013). Larval settlement and metamorphosis of the mussel Mytilus coruscus in response to monospecific bacterial biofilms. Biofouling 29, 247–259. doi: 10.1080/08927014.2013.764412
Keywords: RNA-sequencing, furanone, marine bacterial biofilm, biofilm matrix, plantigrade settlement, Mytilus coruscus
Citation: Li Y-F, Zhu X, Cheng Z-Y, Liang X, Zhu Y-T, Feng D-D, Dobretsov S and Yang J-L (2020) 2(5H)-Furanone Disrupts Bacterial Biofilm Formation and Indirectly Reduces the Settlement of Plantigrades of the Mussel Mytilus coruscus. Front. Mar. Sci. 7:564075. doi: 10.3389/fmars.2020.564075
Received: 22 May 2020; Accepted: 27 August 2020;
Published: 24 September 2020.
Edited by:
Russell T. Hill, University of Maryland, Baltimore County, United StatesReviewed by:
Satheesh Sathianeson, King Abdulaziz University, Saudi ArabiaRubén León, Dentaid S.L., Spain
Copyright © 2020 Li, Zhu, Cheng, Liang, Zhu, Feng, Dobretsov and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sergey Dobretsov, c2VyZ2V5QHNxdS5lZHUub20=; Jin-Long Yang, amx5YW5nQHNob3UuZWR1LmNu
†These authors have contributed equally to this work
 Yi-Feng Li
Yi-Feng Li Xin Zhu
Xin Zhu Zhi-Yang Cheng
Zhi-Yang Cheng Xiao Liang
Xiao Liang You-Ting Zhu
You-Ting Zhu Dan-Dan Feng
Dan-Dan Feng Sergey Dobretsov
Sergey Dobretsov Jin-Long Yang
Jin-Long Yang