- 1Department of Marine Biology, Leon H. Charney School of Marine Sciences, University of Haifa, Haifa, Israel
- 2Morris Kahn Marine Research Station, University of Haifa, Haifa, Israel
- 3Bioinformatics Core Unit, University of Haifa, Haifa, Israel
- 4Moses Strauss Department of Marine Geosciences, Leon H. Charney School of Marine Sciences, University of Haifa, Haifa, Israel
Amino acids compound-specific nitrogen stable isotope analysis (AA-CSIA) is an emerging tool in ecology for understanding trophic system dynamics. While it has been successfully used for several independent studies across a range of environments and study locations, researchers have encountered calculation issues for determining trophic position values. Most studies introduce modifications to the constants of trophic position equation calculations, but then fail to account for the equation variations when comparing across separate research studies. The broad acceptance of this approach is anchored in an underlying presumption that no addition of the exogenous nitrogen atom occurs in the different methods; and therefore, such variations should not affect the outcome. In this paper, we evaluate the use of the EZfaast amino acid derivatization kit (chloroformate) and compare it to the isotopic results of two other derivatization methods. We highlight new considerations for working with AA-CSIA that might account for some of the variations in the results and lead researchers to modify constants in the equation. This study concludes that developing unique constants per derivatization method is required to have more accurate cross-study comparisons of trophic positions.
Introduction
Traditional methods for calculating the trophic position include stomach content analysis, bulk δ15N stable isotope analysis and, more recently, amino acid compound-specific nitrogen isotopic analysis (AA-CSIA). Stomach content analysis provides information on an individual’s prey taxonomy and their relative importance within the food web. However, because it represents only a snapshot in time, this method exhibits bias from a myriad of factors including the proportion of identifiable dietary items, significant numbers of “empty stomach” samples in top predator collections, and varying digestibility of different prey species (i.e., residence time in the stomach). Therefore, when using the stomach content approach, large numbers of samples are needed to correctly evaluate the trophic position (Rindorf and Lewy, 2004), which is both labor-intensive and sometimes impossible. Bulk δ15N of whole organisms and their tissues has been used in several ecological studies as an alternative, or in tandem, to assess the trophic position and nitrogen flow in the food web (Yoshii et al., 1999; Post, 2002; Logan and Dodge, 2013; Vander Zanden et al., 2013). This approach maintains the observed relationship between rising δ15N (2–4‰) values and higher trophic positions (Minagawa and Wada, 1984). However, the increase in δ15N is not constant under all conditions and, therefore, it is influenced by the food sources, stressors, consumer physiology, and natural δ15N of the surrounding environment. Constraining the nitrogen isotopic baseline, or isotopic composition of primary producers at the base of an ecosystem, can be complicated, difficult, or impossible in certain environments, and therefore most studies rely on the relative values between samples within the same system (Popp et al., 2007). Thus, researchers were led to seek new methods to determine the trophic position using δ15N, while obtaining robust estimates of the trophic position in consumers without knowing the isotopic baselines.
McClelland and Montoya (2002) were the first to examine AA-CSIA for establishing a trophic position using nitrogen isotope. The research investigated the relationship between lab-cultivated phytoplankton and its consumer, zooplankton. They discovered that the “non-essential” AA glutamic-acid (also known as “trophic” AA) becomes “heavier” (richer in 15N) compared to the bulk tissue. The “essential” AA phenylalanine (also known as “source” AA) is inert in terms of trophic position and is not affected by the organism’s position on the food chain. Therefore, it records the δ15N signature of the primary producer in the particular food web in question. Both the isotopic baseline and fractionated information are retained in the sample and enables the calculation of the trophic position independently of the surrounding values. In order to confirm the applicability of this approach to ecosystem-level studies (rather than species-specific), later studies tested several different macroalgae, phytoplankton, zooplankton gastropods, and fish in the natural environment and lab (Chikaraishi et al., 2007, 2009). It was concluded that due to the different traits of the AAs (glutamic-acid and phenylalanine), the trophic position can be readily calculated TPGlu/Phe = ((δ15NGlu−δ15NPhe−β)/TDFAA) + 1 without the need to directly measure the primary producer’s δ15N (Chikaraishi et al., 2009; Steffan et al., 2013).
The constant, β, is the difference between the δ15N values of glutamic-acid and phenylalanine in primary producers (trophic position 1). The trophic discrimination factor (TDFAA) is the average δ15N enrichment between the trophic AA (glutamic-acid) relative to source AAs (phenylalanine) per trophic position. When calculating the trophic position based on AA-CSIA, the “β” and “TDFAA” are broadly constant but mostly dependent on the nitrogen source, physiology, and diet (Chikaraishi et al., 2010; McMahon and McCarthy, 2016). Chikaraishi et al. (2009) calculated marine environment values as β = 3.4 ± 0.9‰, while terrestrial environment values for β in C3 and C4 plants were -8.4 ± 1.6 and -0.4 ± 1.7‰, respectively (Chikaraishi et al., 2010). As for TDFAA, it was thought to be 7.6 ± 1.7‰ for all environments (Chikaraishi et al., 2009, 2010). However, further studies have found that this is not always accurate. Bradley et al. (2015) recalculated the “TDFAA” based on a variety of teleost from various trophic positions and found it to be inaccurate in the higher trophic positions, and instead established a value of 5.7 ± 0.3‰. Nielsen et al. (2015) performed similar work and produced calculations of 6.6 ± 1.7‰. McMahon and McCarthy (2016) concluded that the variability is higher (0–10‰) and dependent on a range of variables such as nitrogen excretion, diet, and trophic position, based on a thorough review of the literature.
In order to use AA-CSIA, proteins must be extracted from the samples, broken into their separate AAs, separate the different AA, and only then calculate for their isotopic value. The amino acids in a sample (e.g., muscle) are extracted using a hydrolysis procedure that breaks the peptide bonds of the constituent proteins. The hydrolysis is generally conducted with 6 to 12 M HCl at 100°–150°C for 1 h to 1 day (Ohkouchi et al., 2017). A GC column is used to separate AA, however, in their natural state AA does not react with the GC columns and they require derivatization to reduce polarity and increase their volatility. In this process, the polar carboxyl (-COOH), amino (-NH2), and hydroxyl (-OH) groups are neutralized by replacing active hydrogen atoms with non-polar moieties. Although there is a variety of derivatization methods, the most commonly used for AA-CSIA are the N-Acetyl-n-propyl (NAP), Trifluoroacetic anhydride (TFAA), and chloroformate (Ohkouchi et al., 2017). Walsh et al. (2014) tested the chloroformate derivatization in the context of AA-CSIA and recommended the use of this method for its simple aqueous derivatization, speed of preparation, good chromatographic resolution, and suitability for the analysis of a range of biological samples. They also mention the commercially available kit from Phenomenex (EZfaast). In contrast to the carbon analysis correction for the addition of exogenous carbon atoms in the derivatization process (Walsh et al., 2014), in nitrogen there is no need, that is since no nitrogen atoms are added in the process. Although there is no need to account for added atom there is still a need for normalization of the results through calibration. Compound specific calibration that has a wide range of isotopic values that include the expected values is the most suitable for that purpose (Reinnicke et al., 2012; Yarnes and Herszage, 2017). Therefore, it can be thought that using different methods will not influence the final result and there is no limitation of using the different methods.
In this study, we test the influence of different derivatization methods and sampling location on the AA isotopic ratio and trophic position in situ, in order to determine the influence of these factors on the calculation of trophic position and a better understanding of the used protocols.
Materials and Methods
Sample Collection
For comparison between sites, we used samples of fish and algae from the Eastern Mediterranean, Western Mediterranean, and the Red Sea, as well as samples (fish only) from the Indian Ocean. For the comparison between derivatization methods, we used strictly herbivorous fish (Siganus rivulatus and S. luridus) for their discrete and known trophic position (TP = 2.1 ± 0.1) (Woodland, 1990).
Sample Preparation
All collected samples were immediately frozen and then lyophilized at the lab prior to hydrolyzation. Approximately 1.5 mg of fish muscle (between the dorsal fin and the head) and 3–5 mg of algae was acid hydrolyzed in 1 ml of 6 nmol HCl at 150°C for 75 min (Cowie and Hedges, 1992) within a nitrogen atmosphere inside a 4 ml glass vial with PTFE cap. Samples were cooled to room temperature and algae samples were filtered through a 0.22 μ PTFE filter to remove all undissolved particles. The HCl was evaporated under a gentle stream of nitrogen and neutralized twice with 1 ml of ultra-pure water (also evaporated). For chloroformate derivatization, we used the EZfaast amino acid analysis kit, slightly modified by replacing reagent 6 with dichloromethane (DCM) as a solvent. For comparison, herbivorous fish samples were also derivatized following the Metges et al. (1996) protocol for N-Acetyl-n-propyl (NAP)-amino acid derivatization. The third approach from Silfer et al. (1991) used Trifluoroacetic anhydride (TFAA) for the acylation. With all methods, the samples were derivatized the same day of analysis with no more than six samples a day.
For all methods, we injected 1.5 μl in a splitless mode at 250°C. Helium was used as a carrier gas at a constant flow of 1.5 ml/min for the chloroformate and N-Acetyl-n-propyl; for the TFAA we used 1.1 ml/min. The chloroformate amino acids were separated on a Zebron ZB-50 column (30 m, 0.25 mm, and 0.25 μm) in a Thermo Scientific Trace 1300 Gas chromatographer (GC). Conditions were set to optimize peak separation for the desired amino acids as follows: Initial temperature 110°C ramped to 240°C at 8°C per min and then ramped to 320°C at 20°C per min and held for 2.5 min (Supplementary Figure 1A). The TFAA derivatized amino acid were separated on the Thermo Scientific TraceGOLD TG-1MS column (30 m, 0.25 mm, and 0.25 μm) in a Thermo Scientific Trace 1300 Gas chromatographer (GC). Conditions were set to optimize peak separation for the desired amino acids as follows: Initial temperature 75°C held for 1 min ramped to 90°C at 7.5°C per min, held for 1.5 min, ramped to 160°C at 7°C per min, held for 3.5 min, ramped to 320°C at 25°C per min, and held for 2 min (Supplementary Figure 1B). The N-Acetyl-n-propyl (NAP)-amino acid was separated on the Thermo Scientific TraceGOLD TG-5MS column (30 m, 0.25 mm, and 0.25 μm) in a Thermo Scientific Trace 1300 Gas chromatographer (GC). Conditions were set to optimize peak separation for the desired amino acids as follows: Initial temperature 75°C ramped to 130°C at 4°C per min, held for 2 min, ramped to 180°C at 5°C per min, held for 2 min, ramped to 320°C at 20°C per min, and held for 1 min (Supplementary Figure 1C). The separated amino acids were split on a MicroChannel Device into two lines, one toward Thermo Scientific ISQ quadruple for amino acid identification and the second toward the Thermo Scientific Delta V Advantage for N2 isotope analysis. The ISQ conditions were set to transfer line 310°C, ion source 240°C, and scanned in the range 43–450 m/z mass range. To define the isotopic ratio of nitrogen, the separated amino acids were combusted in a Thermo scientific GC isolink II at 1,000°C. Before entering to Delta V for the N2 analysis, the sample went through a liquid nitrogen cold trap to freeze all other gases. Delta V peak detection were set as follow: start slope 0.2 mV/s, end slope 0.4 mV/s, peak minimum height 50 mV, peak resolution 50%, and maximum peak width 100 s. A triplicate was injected from each sample.
Data Analysis and Corrections
Separated amino acids were purchased from Sigma Aldrich and analyzed with an elemental analyzer isotope ratio mass spectrometer (Geological Survey of Israel). To extend the nitrogen isotopic range, two certified amino acids (Alanine + 43.25‰ and Valine + 30.19‰) were purchased from Arndt Schimmelmann (Biogeochemical Laboratories, Indiana University). We used a standard that contains seven amino acids with known isotopic ratios (Alanine, Valine, Leucine, Isoleucine, Methionine, Glutamic acid, and Phenylalanine), with an isotopic range for the nitrogen of −6.69‰ to + 43.25‰. Since nitrogen is not added in the process of derivatization, corrections for nitrogen addition were not required. The standard of amino acids was injected three times after the combustion reactor oxidation phase, and then the standard was injected again three times after a maximum of 18 sample injections to allow for drift correction (Supplementary Table 1). Since AAs differ in the presence of heteroatoms and functional groups, this may lead to different combustion efficiencies and, therefore, variation in drift. To compensate for this drift an average of the standard injection from the beginning and the end of the sequence were used (Reinnicke et al., 2012; Walsh et al., 2014). For each sequence, a correction factor was applied based on the linear regression equation of the ratio between the known AA isotopic ratio and the acquired result for the sequence (Supplementary Figures 1D–F). Stable isotope ratios were expressed in standard δ notation where the standard for nitrogen is atmospheric N2 (air).
Trophic Position Calculation
The trophic position was calculated from the equationTPGlu/Phe = ((δ15NGlu − δ15NPhe − β)/TDFAA) + 1 (Chikaraishi et al., 2009). To examine the influence of the site factor on the trophic position, we re-calculated the values needed for the equation. All samples analyzed were from the eastern Mediterranean Sea. Since Chikaraishi et al. (2009) determined that the marine ecosystem is not dependent on physiological process and independent from cultured/natural or micro/macro alga to calculate β, we used 10 different algae (Supplementary Table 2). For TDFAA calculation, we used 17 herbivorous fish (Siganus rivulatus and S. luridus) (Supplementary Table 3). Calculations yielded the following values: β = -0.36 ± 1.49‰ and TDFAA = 4.54 ± 1.36‰.
Statistical Analyses
Statistical analyses were performed using R v3.2.2 (R Development Core Team, 2015). Because the δ15N data for most parameters were not normally distributed and/or of homogenous variance, they were tested using the non-parametric Kruskal–Wallis test with Dunn as a post hoc analysis. All post hoc p-values were adjusted using the Benjamini Hochberg (BH) correction factor.
Results
Trophic Position of Samples From Different Locations
Samples from different locations (Red Sea, Indian Ocean, and western Mediterranean Sea) were compared for the calculated trophic position against samples from the eastern Mediterranean Sea (Figure 1 and Supplementary Table 4). Trophic position calculations were based on AA-CSIA of nitrogen using the equation we constructed from the eastern Mediterranean Sea samples. We did not find any significant differences between samples (Kruskal-Wallis, p > 0.08).
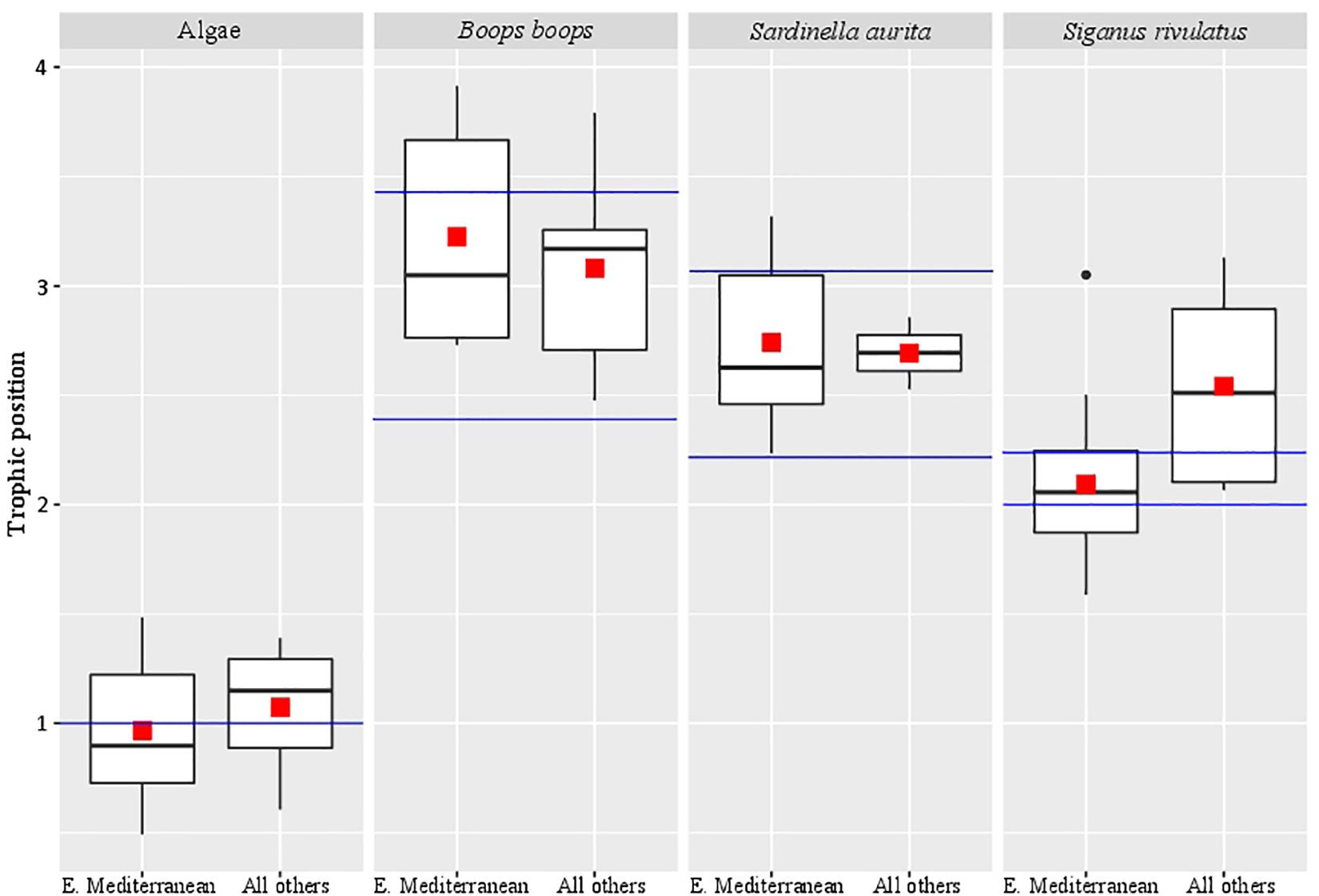
Figure 1. Trophic position of samples from different locations. The red square marks the average trophic position, the black bar in the box marks the median, the black dots are outlier values and the blue bars mark the literature-based trophic position range.
Comparison of the Nitrogen Isotopic Values Between Different Derivatization Methods
S. rivulatus (n = 10) was analyzed using three different methods: chloroformate, NAP (N-Acetyl-n-propyl), and TFAA. We compared Glutamic acid and Phenylalanine δ15N (Figure 2 and Supplementary Table 5), the two most commonly used amino acids for calculation of trophic position (Chikaraishi et al., 2009). A significant difference was observed between methods (Kruskal-Wallis, p < 0.0001). Post hoc analysis reveals a significant difference in Glutamic acid between Chloroformate to NAP (Dunn, BH p = 0.005) and between Chloroformate to TFAA (Dunn, BH p = 0.05). In the Phenylalanine, there was a significant difference between Chloroformate to TFAA (Dunn, BH < 0.0001) and between TFAA to NAP (Dunn, BH = 0.01). There was no consistency in the shift of the isotopic value between the methods and, therefore, no correction factor can be applied.
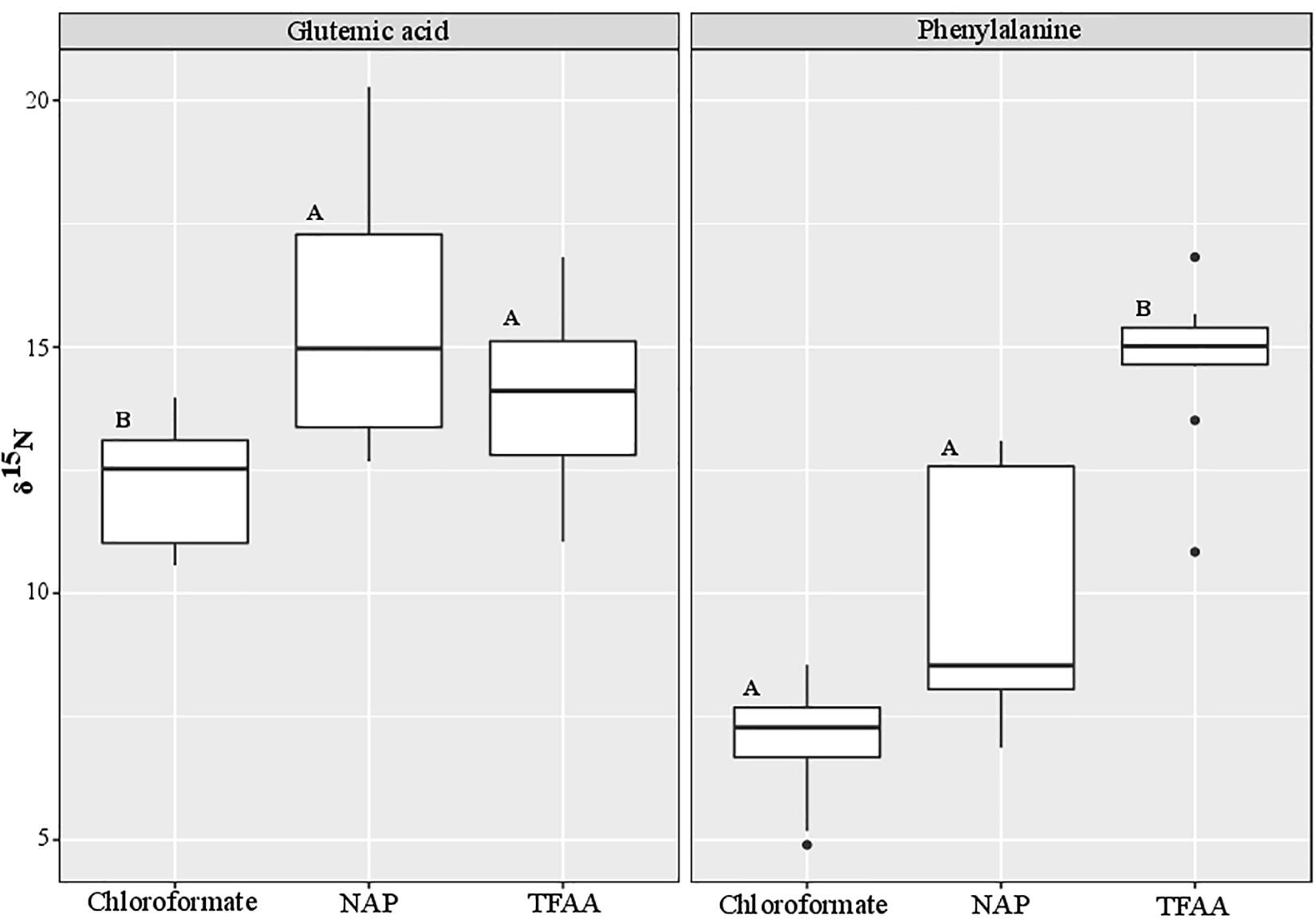
Figure 2. Comparison of the nitrogen isotopic values between different derivatization methods. The black bar in the box marks the median δ15N value and the black dots are outlier values. Statistical significance is indicated in bold letters, and p-values are considered for p < 0.05.
Discussion
In reviewing the literature related to the calculation of the trophic position of teleost using AA-CSIA, it was noted that different studies used different derivatization methods (e.g., Chikaraishi et al., 2009; Bradley et al., 2015; Nuche-Pascual et al., 2018). In all these methods, no correction was made for nitrogen, given that no nitrogen atoms were added in the process. In this study, we used the EZfaast kit, as it is considered to be the easiest, fastest, and safest method to work in GC analysis. This kit was tested previously for the use in AA-CSIA, was considered unsuccessful due to poor separation between amino acids (Philben et al., 2018). However, the likely cause of this poor separation was the use of the Zebron AAA (10 m) column that comes standard with the ezfaast kit and is designed for rapid GC analysis (10 min). This column is too short for this purpose and should be replaced with a longer column that will result in better separation, such as the ZB-50 (30 m). This is especially true in the context of working with nitrogen analysis, where the need for larger sample amounts makes peak separation even more difficult. When measuring organisms with higher trophic position the interrelationship between the δ15N values of glutamic acid and phenylalanine can affect the isotopic values, which in turn may lead to wrong trophic position assignments. Therefore achieving peak separation at the baseline is important. The biological isotopic ratio results from the present analysis did not match any known equations in the literature. This research was concentrated on the Eastern Mediterranean Sea, which is ultraoligotrophic and phosphate-limited, even when compared to classic “blue deserts” such as tropical coral reef environments and mid-ocean gyres in the Pacific Ocean (Krom et al., 2010). Because of these conditions, we must distinguish between the potential effect of the method we are using and the unique influence of local environmental effects. To resolve this, we performed our measurements on samples from the eastern Mediterranean Sea as well as the Red Sea, Indian Ocean, and western Mediterranean Sea (Supplementary Table 4). None of the results matched previously reported values using traditional equations. Therefore, we decided, initially, to form our equation based on samples that are available locally. For calculating β we used ten different algae which produced a β value of -0.36 ± 1.49‰. To calculate the TDFAA, we chose two herbivorous fish species (S. rivulatus and S. luridus) that, based on the literature, have a purely herbivorous diet (Woodland, 1990). From those 17 specimens, we calculated the TDFAA value of 4.54 ± 1.36‰. Using these newly calculated constants, we compared the trophic position of samples from the eastern Mediterranean Sea to samples from the other sources (Red Sea, Indian Ocean, and the Western Mediterranean). We did not find any significant difference between the eastern Mediterranean Sea to the other samples (Figure 1), hence verifying it was not an environmental factor that caused the differential results in trophic position. Also, the trophic position we calculated for Boops boops and Sardinella aurita samples from all locations are in the range that is reported in the literature (Tsikliras et al., 2005; Bode et al., 2006; Madkour, 2012; Mancinelli et al., 2013; Cresson et al., 2014; Albo-Puigserver et al., 2016). Although not significantly different from the Mediterranean Sea, the trophic position measured for the Red Sea samples of S. rivulatus, described as a pure herbivore (TP = 2.1 ± 0.1) were slightly higher (TP = 2.5 ± 0.5) than previously reported (Woodland, 1990). Recent reports documented these fish eating invertebrates such as ctenophores, scyphozoans, and other invertebrates that are part of the alga biome (Bos et al., 2017; Guy-Haim et al., 2017). Therefore, we conclude that although in many aspects the eastern Mediterranean Sea is a unique environment, the measured variations as compared to other places are not large enough (within the error) to change their assigned trophic position (TP = 2.1 ± 0.1 vs. 2.5 ± 0.5) and, therefore, the equation is robust enough to be more broadly applied.
To further validate the equation (in combination with the technique), we tested ten different samples of S. rivulatus using three different methods [Chloroformate (EZfaast), NAP (N-Acetyl-n-propyl) and TFAA; Supplementary Table 5]. We compared Glutamic acid and Phenylalanine δ15N, the two most widely used amino acids for trophic position calculations (Figure 2). Although nitrogen is not added in any of the derivatization protocols, we still observe differences in the isotopic ratios of nitrogen. Our study is in agreement with a previous study by Hofmann et al. (2003) that found differences between the isotopic values of different analytical methods and, therefore, an additional source must be present that causes these isotopic differences. There is a multitude of possible explanations. We can attribute the difference to the impurity of the AA, specifically from a non-AA matrix in the derivatization process or some fatty acids that can also go through the derivatization process alongside the amino acids (Castro et al., 1997; Goto et al., 2011; Walsh et al., 2014). In addition, under acidic conditions with some of the derivatization methods, glutamic acid can partially be cyclized into Pyroglutamic acid (Castro et al., 1997; Walsh et al., 2014; Ohkouchi et al., 2017). Variations can occur due to the differential reaction of derivatized compounds with the combustion reactor on different conditions, oxidation status of the reactor, or different combustion reactors (CuO/NiO/Pt vs. NiO tube/CuO-NiO) (Reinnicke et al., 2012). There are also differences between AA extraction efficiencies between the methods that may cause different yield which affect the AA pick height (Supplementary Figures 1A–C) and for some AA not to be derivatized (Castro et al., 1997; Goto et al., 2011; Walsh et al., 2014; Ohkouchi et al., 2017). Unfortunately, we could not find any consistency in the shift of the isotopic value between the methods. Therefore, there is no option to apply a correction factor to be able to compare the methods. On the other hand, there is consistency between isotopic ratios within any given protocol. That consistency emphasizes the importance of applying the correct constant of β and TDFAA per specific protocol to conduct interstudy comparisons of the trophic position. Still, it does not solve the problem of comparing the absolute isotopic value between the different methods. Here, we adapted the Ezfaast kit (chloroformate) for quick, safe, and easy analyses of AA-CSIA and provided a robust equation for this protocol that allows for accuracy and precision regardless of the geographic origins of the samples. However, we call for thorough examination of the different protocols and determination of the most accurate protocol for nitrogen isotope studies that will became a standard in that field.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
SM, DT, SE, and ML participated in the design of the study. SM and ES conducted the isotopic analysis. SM, DT, and BG, contributed to the writing and improving of the manuscript. SM and ML participated in the data analysis. All authors contributed and approved the manuscript.
Funding
This research was supported by the Wolfson Foundation, Kahn Foundation, J. Isaacs Charitable Foundation, and the Kadas Charitable Trust.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Drs. Tali Mass, Nir Stern, and Christine Ferrier Pages for their assistance in collecting samples at the various sites.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.561568/full#supplementary-material
Supplementary Figure 1 | Amino acids standards calibration and chromatography. Typical results of derivatized vs. underivatized (elemental analyzer) amino acids standard isotopic values (A)-Chloroformate (Ezfaast), (B)- TFAA, (C)- NAP. Typical isotopic ratio mass spectrometry (IRMS) chromatogram of amino acids standard (D)-Chloroformate (Ezfaast), (E)- TFAA, (F)- NAP.
References
Albo-Puigserver, M., Navarro, J., Coll, M., Layman, C. A., and Palomera, I. (2016). Trophic structure of pelagic species in the northwestern Mediterranean Sea. J. Sea Res. 117, 27–35. doi: 10.1016/j.seares.2016.09.003
Bode, A., Carrera, P., and Porteiro, C. (2006). “Stable nitrogen isotopes reveal weak dependence of trophic position of planktivorous fish on individual size: a consequence of omnivorism and mobility,” in Proceedings of the Radionuclides in the Environment: International Conference on Isotopes in Environmental Studies, (Amsterdam: Elsevier Masson SAS), 281–293.
Bos, A. R., Cruz-Rivera, E., and Sanad, A. M. (2017). Herbivorous fishes Siganus rivulatus (Siganidae) and Zebrasoma desjardinii (Acanthuridae) feed on Ctenophora and Scyphozoa in the Red Sea. Mar. Biodivers. 47, 243–246. doi: 10.1007/s12526-016-0454-9
Bradley, C. J., Wallsgrove, N. J., Choy, C. A., Drazen, J. C., Hetherington, E. D., Hoen, D. K., et al. (2015). Trophic position estimates of marine teleosts using amino acid compound specific isotopic analysis. Limnol. Oceanogr. Methods 13, 476–493. doi: 10.1002/lom3.10041
Castro, R. M., Carbo, M. T. D., Martinez, V. P., Adelantado, J. V. G., and Reig, F. B. (1997). Study of binding media in works of art by gas chromatographic analysis of amino acids and fatty acids derivatized with ethyl chloroformate. J. Chromatogr. A 778, 373–381. doi: 10.1016/S0021-9673(97)00284-7
Chikaraishi, Y., Kashiyama, Y., Ogawa, N. O., Kitazato, H., and Ohkouchi, N. (2007). Metabolic control of nitrogen isotope composition of amino acids in macroalgae and gastropods: implications for aquatic food web studies. Mar. Ecol. Prog. Ser. 342, 85–90.
Chikaraishi, Y., Ogawa, N. O., Kashiyama, Y., Takano, Y., Suga, H., Tomitani, A., et al. (2009). Determination of aquatic food-web structure based on compound-specific nitrogen isotopic composition of amino acids. Limnol. Oceanogr. Methodes 7, 740–750.
Chikaraishi, Y., Ogawa, N. O., and Ohkouchi, N. (2010). Further evaluation of the trophic level estimation based on nitrogen isotopic composition of amino acids. Earth Life Isot. 6, 37–51.
Cowie, G. L., and Hedges, J. I. (1992). Improved amino acid quantification in environmental samples: charge-matched recovery standards and reduced analysis time. Mar. Chem. 37, 223–238. doi: 10.1016/0304-4203(92)90079-P
Cresson, P., Ruitton, S., Ourgaud, M., and Harmelin-Vivien, M. (2014). Contrasting perception of fish trophic level from stomach content and stable isotope analyses: a Mediterranean artificial reef experience. J. Exp. Mar. Bio. Ecol. 452, 54–62. doi: 10.1016/j.jembe.2013.11.014
Goto, A., Korenaga, T., and Chikaraishi, Y. (2011). Methyl and ethyl chloroformate derivatizations for compound-specific stable isotope analysis (CSIA) of fatty acids(Advances in molecular and stable isotope studies among the organic geochemical and related communities (Part II)). Res. Org. Geochem. 27, 91–95. doi: 10.20612/rog.27.0_91
Guy-Haim, T., Hyams-Kaphzan, O., Yeruham, E., Almogi-Labin, A., and Carlton, J. T. (2017). A novel marine bioinvasion vector: Ichthyochory, live passage through fish. Limnol. Oceanogr. Lett. 2, 81–90. doi: 10.1002/lol2.10039
Hofmann, D., Gehre, M., and Jung, K. (2003). Sample preparation techniques for the determination of natural 15N/14N variations in amino acids by gas chromatography-combustion-isotope ratio mass spectrometry (GC-C-IRMS). Isot. Environ. Health Stud. 39, 233–244. doi: 10.1080/1025601031000147630
Krom, M. D., Emeis, K.-C., and Van Cappellen, P. (2010). Why is the Eastern Mediterranean phosphorus limited? Prog. Oceanogr. 85, 236–244. doi: 10.1016/j.pocean.2010.03.003
Logan, J. M., and Dodge, K. L. (2013). Comment on “stable isotopes challenge the perception of ocean sunfish Mola mola as obligate jellyfish predators”. J. Fish Biol. 82, 1–9. doi: 10.1111/j.1095-8649.2012.03432.x
Madkour, F. F. (2012). Feeding ecology of the round Sardinella, aurita in the Egyptian Mediterranean waters. J. Environ. Sci. Eng. 2, 83–92.
Mancinelli, G., Vizzini, S., Mazzola, A., Maci, S., and Basset, A. (2013). Cross-validation of δ15N and FishBase estimates of fish trophic position in a Mediterranean lagoon: the importance of the isotopic baseline. Estuar. Coast. Shelf Sci. 135, 77–85. doi: 10.1016/j.ecss.2013.04.004
McClelland, J., and Montoya, J. (2002). Trophic relationships and the nitrogen isotopic composition of amino acids in plankton. Ecology 83, 2173–2180.
McMahon, K. W., and McCarthy, M. D. (2016). Embracing variability in amino acid δ15N fractionation: mechanisms, implications, and applications for trophic ecology. Ecosphere 7:e01511. doi: 10.1002/ecs2.1511
Metges, C. C., Petzke, K. J., and Hennig, U. (1996). Gas chromatography/combustion/isotope ratio mass spectrometric comparison of N-acetyl- and N-pivaloyl amino acid esters to measure 15N isotopic abundances in physiological samples: a pilot study on amino acid synthesis in the upper gastro-intestinal tract. J. Mass Spectrom. 31, 367–376.
Minagawa, M., and Wada, E. (1984). Stepwise enrichment of 15 N along food chains: further evidence and the relation between δ 15 N and animal age. Geochim. Cosmochim. Acta 48, 1135–1140.
Nielsen, J. M., Popp, B. N., and Winder, M. (2015). Meta-analysis of amino acid stable nitrogen isotope ratios for estimating trophic position in marine organisms. Oecologia 178, 631–642. doi: 10.1007/s00442-015-3305-7
Nuche-Pascual, M. T., Lazo, J. P., Ruiz-Cooley, R. I., and Herzka, S. Z. (2018). Amino acid-specific δ15N trophic enrichment factors in fish fed with formulated diets varying in protein quantity and quality. Ecol. Evol. 8, 9192–9217. doi: 10.1002/ece3.4295
Ohkouchi, N., Chikaraishi, Y., Close, H. G., Fry, B., Larsen, T., Madigan, D. J., et al. (2017). Advances in the application of amino acid nitrogen isotopic analysis in ecological and biogeochemical studies. Organ. Geochem. 113, 150–174. doi: 10.1016/j.orggeochem.2017.07.009
Philben, M., Billings, S. A., Edwards, K. A., Podrebarac, F. A., van Biesen, G., and Ziegler, S. E. (2018). Amino acid δ15N indicates lack of N isotope fractionation during soil organic nitrogen decomposition. Biogeochemistry 138, 69–83. doi: 10.1007/s10533-018-0429-y
Popp, B. N., Graham, B. S., Olson, R. J., Hannides, C. C. S., Lott, M. J., López-Ibarra, G. A., et al. (2007). Insight into the trophic ecology of yellowfin tuna, Thunnus albacares, from compound-specific nitrogen isotope analysis of proteinaceous amino acids. Terrestrial Ecol. 1, 173–190. doi: 10.1016/S1936-7961(07)01012-3
Post, D. (2002). Using stable isotopes to estimate trophic position: models, methods, and assumptions. Ecology 83, 703–718.
Reinnicke, S., Juchelka, D., Steinbeiss, S., Meyer, A., Hilkert, A., and Elsner, M. (2012). Gas chromatography/isotope ratio mass spectrometry of recalcitrant target compounds: performance of different combustion reactors and strategies for standardization. Rapid Commun. Mass Spectrom. 26, 1053–1060. doi: 10.1002/rcm.6199
Rindorf, A., and Lewy, P. (2004). Bias in estimating food consumption of fish by stomach-content analysis. Can. J. Fish. Aquat. Sci. 61, 2487–2498. doi: 10.1139/f04-200
Silfer, J., Engel, M., Macko, S., and Jumeau, E. (1991). Stable carbon isotope analysis of amino acid enantiomers by conventional isotope ratio mass spectrometry and combined gas chromatography/isotope ratio mass. Anal. Chem. 63, 370–374.
Steffan, S. A., Chikaraishi, Y., Horton, D. R., Ohkouchi, N., Singleton, M. E., Miliczky, E., et al. (2013). Trophic hierarchies illuminated via amino acid isotopic analysis. PLoS One 8:e76152. doi: 10.1371/journal.pone.0076152
Tsikliras, A., Torre, M., and Stergiou, K. (2005). Feeding habits and trophic level of round Sardinella (Sardinella aurita) in the northeastern Mediterranean (Aegean Sea, Greece). J. Biol. Res. 3, 67–75.
Vander Zanden, H., Arthur, K., Bolten, A., Popp, B., Lagueux, C., Harrison, E., et al. (2013). Trophic ecology of a green turtle breeding population. Mar. Ecol. Prog. Ser. 476, 237–249. doi: 10.3354/meps10185
Walsh, R. G., He, S., and Yarnes, C. T. (2014). Compound-specific δ13C and δ15N analysis of amino acids: a rapid, chloroformate-based method for ecological studies. Rapid Commun. Mass Spectrom. 28, 96–108. doi: 10.1002/rcm.6761
Woodland, D. J. (1990). Revision of the fish family Siganidae with descriptions of two new species and comments on distribution and biology. Indo Pac. Fishes 19:136.
Yarnes, C. T., and Herszage, J. (2017). The relative influence of derivatization and normalization procedures on the compound-specific stable isotope analysis of nitrogen in amino acids. Rapid Commun. Mass Spectrom. 31, 693–704. doi: 10.1002/rcm.7832
Keywords: trophic discrimination factor, AA-CSIA, eastern Mediterranean Sea, food web, nitrogen isotopic composition, calibration
Citation: Martinez S, Lalzar M, Shemesh E, Einbinder S, Goodman Tchernov B and Tchernov D (2020) Effect of Different Derivatization Protocols on the Calculation of Trophic Position Using Amino Acids Compound-Specific Stable Isotopes. Front. Mar. Sci. 7:561568. doi: 10.3389/fmars.2020.561568
Received: 13 May 2020; Accepted: 23 November 2020;
Published: 15 December 2020.
Edited by:
Noga Stambler, Bar-Ilan University, IsraelReviewed by:
Naoto F. Ishikawa, Japan Agency for Marine-Earth Science and Technology (JAMSTEC), JapanHenry P. Schwarcz, McMaster University, Canada
Copyright © 2020 Martinez, Lalzar, Shemesh, Einbinder, Goodman Tchernov and Tchernov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephane Martinez, c3RlcGhhbmUubWFydGluZXpAZ21haWwuY29t
 Stephane Martinez
Stephane Martinez Maya Lalzar
Maya Lalzar Eli Shemesh1
Eli Shemesh1 Dan Tchernov
Dan Tchernov