
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 21 August 2020
Sec. Deep-Sea Environments and Ecology
Volume 7 - 2020 | https://doi.org/10.3389/fmars.2020.00674
This article is part of the Research Topic Deep-sea Sponge Ecosystems: Knowledge-based Approach Towards Sustainable Management and Conservation View all 29 articles
Deep-sea sponge grounds are underexplored ecosystems that provide numerous goods and services to the functioning of the deep-sea. This study assessed the prokaryotic diversity in embryos, recruits, and adults of Craniella zetlandica and Craniella infrequens, common and abundant representatives of deep-sea sponge grounds in the North Atlantic. Our results reveal that symbiont transmission in the two Craniella sponge species likely occurs vertically, as highly similar microbial consortia have been identified in adults, embryos, and recruits. Moreover, transmission electron microscopy revealed high abundances of sponge-associated microorganisms, among which Chloroflexi (SAR202) were identified as common representatives by amplicon sequencing and fluorescence in situ hybridization (FISH). Equal diversity metrices, a similar overall prokaryotic community composition and a distinct dominance of the phylum Chloroflexi within all life stages are the key findings of our analyses. Information such as presented here provide understanding on the recruitment of deep-sea sponge holobionts which is needed to develop integrated management tools of such vulnerable marine ecosystems.
A fundamental process influencing the fitness of many organisms is the establishment of host-microbial colonization during development (Mortzfeld et al., 2016). Microbial consortia enable the respective animal host to expand its range of nutrition (Nicholson et al., 2012), defense (Flórez et al., 2015), immunity (Eberl, 2010), and development (Koropatnick et al., 2004). Host-microbe interactions may be regulated from early developmental stages and transmitted through generations (reviewed in Rosenberg and Zilber-Rosenberg, 2011; McFall-Ngai et al., 2013). In general, beneficial microbial symbionts are expected to be transmitted vertically (from parents to offspring) in order to assure an establishment of a compatible partnership (Björk et al., 2019 and references therein). Further, probability of vertical transmission is expected to increase with increasing dependence of the host on its microbial partners (Ewald, 1987; Bull et al., 1991; Yamamura, 1993; Douglas, 1994; Thompson, 1994; Herre et al., 1999; Wilkinson, 2001; Sachs et al., 2011). In the marine environment symbioses are common and host-microbe interactions have been assessed at different developmental stages for a variety of model organisms, including cnidarians (Apprill et al., 2009; Mortzfeld et al., 2016), echinoderms (Galac et al., 2016; Carrier et al., 2019), cephalopods (Montgomery and McFall-Ngai, 1994), molluscs (Endow and Ohta, 1990; Wentrup et al., 2014), and annelids (Nussbaumer et al., 2006; Schimak et al., 2015).
Sponges (Porifera) are evolutionarily ancient animals which first appeared between 650 and 700 million years ago (Love et al., 2009; Zumberge et al., 2018). Today, more than 9,000 species (according to the World Porifera Database) are classified into four classes (Demospongiae, Calcarea, Homoscleromorpha, and Hexactinellida) and populate virtually all marine benthic habitats from shallow tropical reefs to the polar seas and the deep ocean (van Soest et al., 2020). Sponges are prominent examples of marine holobionts, consisting of diverse prokaryotic communities with more than 63 prokaryotic phyla detected within sponge hosts (Thomas et al., 2016; Moitinho-Silva et al., 2017a). The prokaryotic communities of sponges are host species-specific and distinct from seawater prokaryotic communities (Hentschel et al., 2002; Thomas et al., 2016). The transmission of microbial sponge symbionts to the next generation occurs vertically or horizontally (De Caralt et al., 2007; Maldonado, 2007; Schmitt et al., 2008; Webster et al., 2010; Gloeckner et al., 2013). Microbial communities have been detected in reproductive stages, such as oocytes, embryos, larvae, and juveniles of various shallow water species (Ereskovsky et al., 2005; Schmitt et al., 2007; Sharp et al., 2007; Gloeckner et al., 2013; Björk et al., 2019) indicating that vertical transmission is an important mechanism for the establishment and maintenance of sponge-microbe associations over evolutionary time scales.
In comparison to sponges from shallow and often warm-water habitats, much less is known about sponges and their microbiomes from deep and cold environments. Sponges are ubiquitous and abundant components of deep-sea benthic communities and can form structurally complex and diverse ecosystems, termed “sponge grounds” (Klitgaard and Tendal, 2004; Xavier et al., 2015; Maldonado et al., 2016). Sponge grounds are known to strongly support biodiversity, either directly by provision of shelter or indirectly via food webs, and further perform a variety of ecosystem functions, among others via participation in bentho-pelagic coupling and biogeochemical cycles (reviewed in Maldonado et al., 2016; Thompson and Fuller, 2020). Deep-sea sponge grounds (syn. “sponge aggregations” or “sponge gardens”) have been flagged as vulnerable marine ecosystems (VMEs) that are currently under pressure through fishing and other human activities (Pham et al., 2019; Murillo et al., 2020). The species Craniella infrequens and Craniella zetlandica are common members of such sponge grounds and have further been recorded in shelf-areas around the globe (Figure 1). Craniella sponges are spherical in shape and grow up to the size of a golf ball and larger. This study aims to describe the prokaryotic community composition and diversity of two Craniella species and their early life stages from the deep North Atlantic Ocean. In particular we sought to address whether prokaryotic community composition, diversity, and dominant microbial phyla differ between different life stages (adults, embryos, and recruits) of the two sponge species. Knowledge about deep-sea sponge holobiont recruitment and plasticity across life stages is crucial to enable, in the long term, an adequate conservation of sponge populations from vulnerable marine ecosystems.
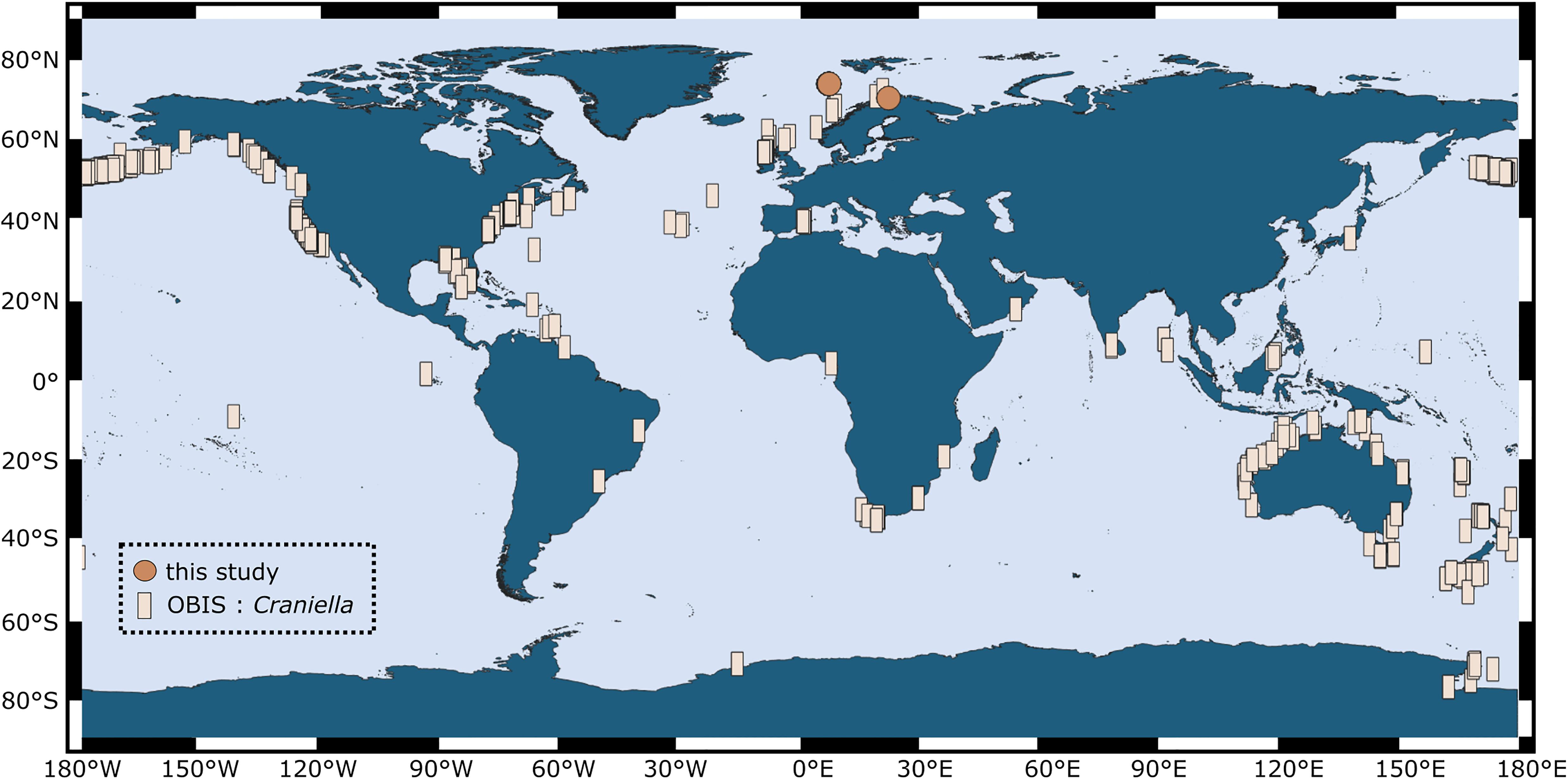
Figure 1. Global distribution of the genus Craniella, as compiled from records in the Ocean Biogeographic Information System (OBIS; squares) in March 2020. Sampling locations of individuals from our study are indicated by points.
Sponges of different developmental stages (adults, embryos, recruits) were collected during two research cruises onboard the Norwegian research vessel RV G.O.Sars (“GS2017110” and “GS2018108”) to the North Atlantic Ocean (Schulz Bank seamount at 73.8°N, 7.5°E; and Stjernsund at 70.3°N, 22.5°E) in July 2017 and August 2018. Samples were collected by remotely operated vehicle (ROV AEgir, University of Bergen) or Agassiz trawl at an average depth of 600 m below surface. Tissue chunks of adult sponges and recruits were subsampled onboard ship with sterile scalpels from the mesohyl, rinsed, flash-frozen and stored at −80°C for molecular analyses. Additional pieces of tissue were subsampled with biopsy punches and preserved onboard ship for microscopy. Embryos were carefully picked out of the parent sponges (Figure 2) with sterile spring steel forceps using a stereomicroscope. After rinsing, 10 whole embryos were each pooled per adult sponge to account for the small biomass, fixed in RNAlaterTM Stabilizing Solution (Thermo Fisher Scientific, Waltham, MA, United States), stored at 4°C overnight, transferred to −20°C on the next day and subsequently to −80°C. In addition, intact embryos were preserved onboard ship for microscopy. In terms of sample numbers, we sampled eight adult C. infrequens sponges, of which four were brooding embryos, and nine C. infrequens recruits. During the cruise we further sampled eight adult C. zetlandica sponges of which one was brooding embryos. In addition to our cruise activities, we sampled nine C. zetlandica recruits in September 2018 from an aquarium system (Bergen, Norway). The respective parent sponges had been maintained since summer 2017 in 45 L tanks with sea-water flow through (1 L min–1) pumped from the adjacent fjord at 105 m depth below surface. Seawater was treated with a 20 μm drum filter prior to flowing through the holding tanks. Under these conditions spawning was documented frequently and the new recruits had settled in the tanks. C. zetlandica recruits from the aquarium system were smaller (and thus most likely younger) than the in situ sampled C. infrequens recruits and therefore flash-frozen as a whole. In our case the new sponge recruits had the following sizes: 1 cm mean diameter for C. infrequens; and 0.3 cm maximal diameter for C. zetlandica. Adult sponges of both sponge species had the size of a fist. In summary, for this study in total 39 sponge individuals of the two sponge species were collected: C. zetlandica (eight adults, of which one was brooding embryos, and nine recruits) and C. infrequens (eight adults, of which four were brooding embryos, and nine recruits). In addition to the sponge samples, we also sampled 18 seawater microbial communities during the two cruises (at all sponge sampling locations) and from the aquarium system as environmental reference samples. Two liters of seawater sample were filtered onto polyvinylidene fluoride (PVDF) filter membranes (with 0.22 μm pore size; Merck Millipore) and stored at −80°C.
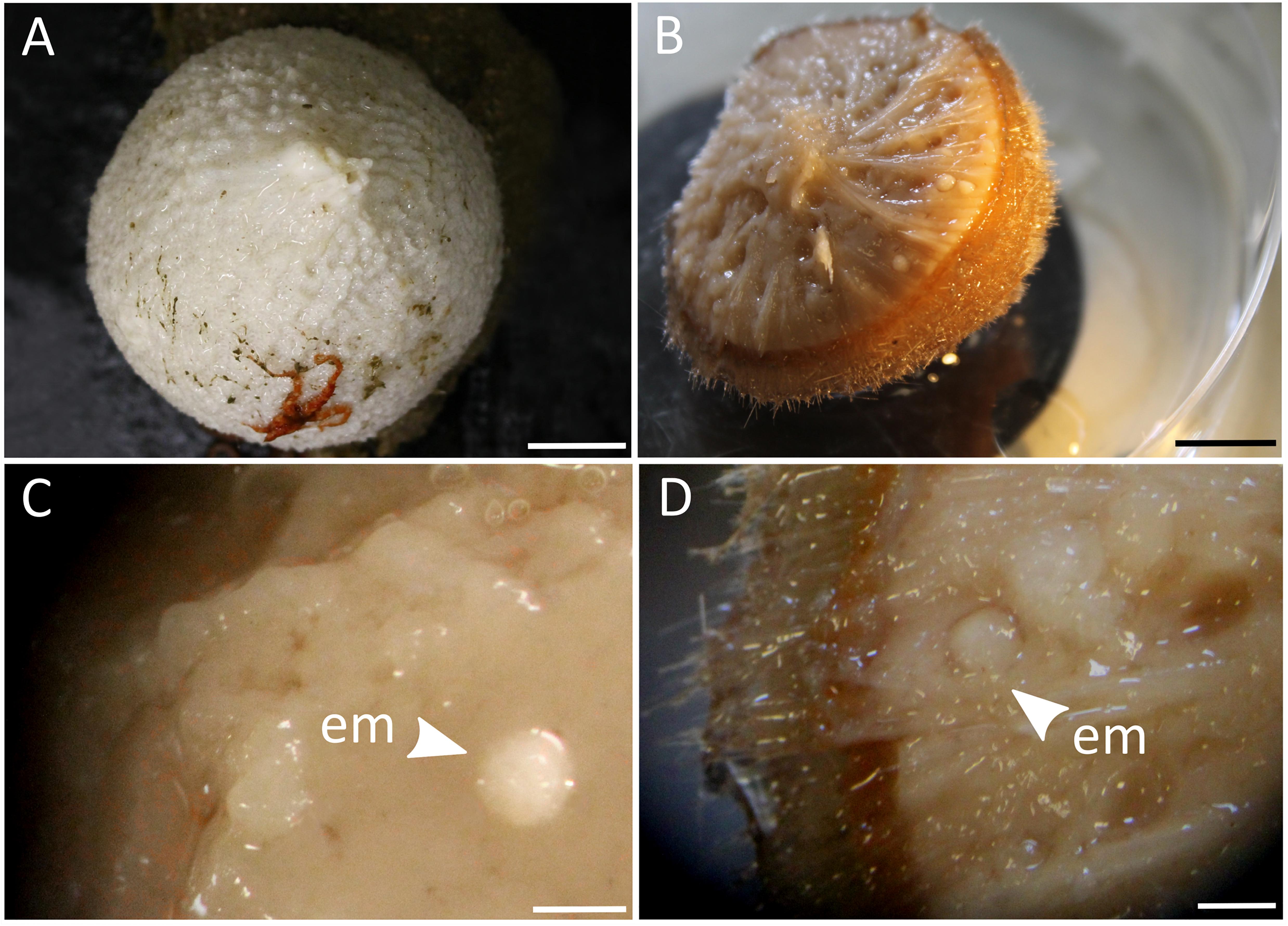
Figure 2. (A) Photograph of an adult C. zetlandica individual with an attached red brittle star at the bottom. Scale bar = 1 cm. (B) Photograph of an adult C. infrequens individual that was cut in half, showing numerous embryos inside the sponge tissue. Scale bar 1 cm. (C) Stereomicroscopic magnification of C. zetlandica tissue. em, embryo. Scale bar = 1 mm. (D) Stereomicroscopic magnification of C. infrequens tissue. em, embryo(s) brooding in the mesohyl. Scale bar = 1 mm.
DNA extraction was performed on either ∼ 0.25 g of tissue or bulk of 10 pooled embryos using the DNeasy Power Soil kit (Qiagen, Venlo, Netherlands). For seawater samples DNA was extracted of one half of a PVDF filter membrane. Subsequent DNA quantity and quality controls were performed with a NanoDrop spectrophotometer and by polymerase chain reaction with universal 16S primers (27F-1429R; Weisburg et al., 1991), followed by gel electrophoresis. For amplicon sequencing, the V3-V4 variable regions of the 16S rRNA gene were amplified in a one-step PCR with the primer pair 341F-806R (dual barcoding approach; Kozich et al., 2013). The primer pair has the following nucleotide sequences: 5′-CCTACGGGAGGCAGCAG-3′ (Muyzer et al., 1993) and 5′-GGACTACHVGGGTWTCTAAT-3′ (Caporaso et al., 2011). Verification of PCR-products was performed with gel electrophoresis, followed by normalization and pooling of samples. Sequencing was conducted on a MiSeq platform (MiSeqFGx, Illumina, San Diego, CA, United States) using v3 chemistry. Zero mismatches were allowed in the barcode sequence for demultiplexing.
Raw reads were quality-checked and processed within the QIIME2 environment (version 2018.11; Bolyen et al., 2019). Based on forward reads (truncated to 270 nt), Amplicon Sequence Variants (ASVs) were generated using the DADA2 algorithm (python-based; Callahan et al., 2016). Data were analyzed based on ASVs instead of classical operational taxonomic units (OTUs), in order to resolve fine-scale community patterns and to use a state-of-the-art method which profits from the error correction model conducted by the DADA2 algorithm. Singletons and chimeric sequences were removed during this process.
Phylogenetic trees were calculated based on the resulting ASVs via the FastTree2 plugin. Taxonomic classification of representative ASVs was achieved with the help of a primer-specific trained Naive Bayes taxonomic classifier, based on the Silva 132 99% OTUs 16S database (Quast et al., 2013). Taxonomic filtering was applied on the dataset, meaning that mitochondria, chloroplasts and unassigned sequences were removed. Further, a sampling depth of 3,900 was applied to standardize the number of reads across samples. Diversity metrices were calculated, such as Shannon indices (alpha diversity) and weighted UniFrac distances (beta diversity). Non-metric multidimensional scaling (nMDS) was conducted to evaluate sample separation in ordination space (based on weighted UniFrac distances). We performed PERMANOVAs (999 permutations) on weighted UniFrac distances, and Tukey Posthoc tests on Shannon indices following ANOVA type III tests, to assess if the prokaryotic community differed significantly between sample groups. To evaluate further on the differential abundance of microbial phyla present in aquarium sponges (i.e., C. zetlandica recruits) in comparison to in situ sampled sponges (and in aquarium water vs in situ water), we ran the Linear Discriminant Analysis (LDA) Effect Size (LEfSe) algorithm (Segata et al., 2011). Visualizations were created using R (version 3.0.2 with the ggplot2 package loaded; R Development Core Team, 2008; Wickham, 2016) and Inkscape (version 0.92.4; Harrington and the Inkscape Development Team, 2005). The ternary plot was created using the Ternary plot maker web tool (Blom, 20191).
In order to provide an independent proof-of-concept accompanying our 16S amplicon sequencing results we performed qualitative microscopic analyses. These included light microscopy, transmission electron microscopy (TEM), and fluorescence in situ hybridization of the most dominant microbial phylum detected by amplicon sequencing. Especially for the embryo-based DNA libraries we aimed at confirming by microscopy that they contained sequences which were indeed present within the embryos and not merely contaminants.
Tissue samples for light microscopy and TEM were fixed in a cocktail of glutaraldehyde (2.5%) with sodium chloride (0.34 M) and phosphate buffered saline (PBS; 0.4 M). Samples were stored at 4°C. Back on land, samples were washed 3x with buffer (at 4°C; 15 min per washing step), post-fixed in 2% osmiumtetroxide for 2 h and afterward washed three more times with buffer (at 4°C; 15 min per washing step). Subsequently partial dehydration was conducted with an ascending ethanol series, from 30% EtOH (2 × 15 min) to 50% EtOH (1 × 15 min), to 70% EtOH (2 × 15 min, including an overnight storage at 4°C), to 90% EtOH (1 × 15 min) up to 100% EtOH (2 × 15 min). After dehydration, the samples were gradually infiltrated with LR-White resin (at room temperature). The infiltration series covered the following steps: from 1 × 1 h 2:1 EtOH:LR-White, to 1 × 1 h 1:1 EtOH:LR-White, to 1 × 1 h 1:2 EtOH:LR-White, up to 2 × 2 h pure LR-White. Samples were transferred to gelatine embedding capsules filled with fresh LR-White resin and subsequently incubated for polymerization at 57°C for two days. The resulting resin blocks were manually pre-trimmed. Semithin sections (0.5 μm) were cut for light microscopy using an ultramicrotome (Reichert-Jung ULTRACUT E; Leica Camera AG, Wetzlar, Germany) which was equipped with a diamond knife (DIATOME, Biel, Switzerland). After staining with a Richardson solution (after Mulisch and Welsch, 2015), semi-thin sections were mounted onto SuperFrost Ultra Plus® microscopy slides and visualized with the help of an Axio Observer.Z1 microscope (Zeiss, Göttingen, Germany).
For TEM, ultrathin sections (70 nm) were cut with the ultramicrotome, mounted on pioloform coated copper grids (75 mesh) and contrasted with uranyl acetate (20 min incubation and subsequent washing steps) as well as with Reynold’s lead citrate (3 min incubation, followed by washing steps). Visualization of the ultra-thin sections was performed on a Tecnai G2 Spirit Bio Twin transmission electron microscope (FEI Company, Hillsboro, United States) at an acceleration voltage of 80 kV.
Sponge tissue samples were fixed overnight (at 4°C) onboard with 4% paraformaldehyde (PFA) in 1xPBS (pH was adjusted to approximately 6.9 with the help of NaOH and HCl). For preparation of all FISH solutions, we used filtered (0.22 μm membrane) diethylpyrocarbonate (DEPC)-treated water (Thermo Fisher Scientific, Waltham, United States) to inactivate RNase enzymes, and performed autoclaving for sterilization of buffers and solvents onboard the ship. After fixation with PFA, five washing steps (3 min) were performed with 1xPBS (4°C). Subsequently quenching of free aldehyde groups was conducted by incubation with glycine (50 mM) in 1xPBS for 15 min (room temperature). The quenching step was followed by five additional washing steps (3 min) in DEPC water (at 4°C). Dehydration was performed onboard in a ascending ethanol series (molecular grade, RNase free), covering the following steps: 30% EtOH in 1xPBS (2 × 15 min at 4°C), 50% EtOH in 1xPBS (2 × 30 min). Samples were subsequently stored at −20°C. Back on land, dehydration was continued in following steps: 50% EtOH in MilliQ water (1 × 15 min), 60% EtOH in MilliQ water (2 × 15 min), 70% EtOH in MilliQ water (2 × 15 min), 90% EtOH in MilliQ water (1 × 15 min) up to 100% EtOH (2 × 15 min). The samples were gradually infiltrated with LR-White resin, polymerized, pre-trimmed and cut into semithin sections (0.5 μm) with an ultramicrotome, in the same manner as described in section 2.3. As embryos were embedded in whole, cutting with the microtome was run for a substantial time, until the central part of the embryo was reached. At the area with widest diameter, sections were received and fixed onto two separate SuperFrost Ultra Plus® microscopy slides. While one half of the slides was directly stained with a Richardson solution and mounted using Biomount medium (Plano, Germany; see description in section “Light Microscopy and TEM”). The other half was only fixed onto microscopy slides, without any staining or covering in Biomount medium. Following the procedures described in Bayer et al. (2018), these latter samples were afterward labeled with Chloroflexi clade-specific probes. In particular the following probes were used: Cal825 (5′-[Cy3]-ACACCGCCCACACCTCGT-[Cy3]-3′; E. coli binding positions 825 to 843) for Caldilineae; Ana1005 (5′-[Alexa Fluor 647]-TCCGCTTTCGCTTCCGTA-[Alexa Fluor 647]-3′; E. coli binding positions 1005 to 1023) for Anaerolineae; and SAR202-104 (5′-[Alexa Fluor 488]-GTTACTCAGCCGTCTGCC-[Alexa Fluor 488]-3′; E. coli binding positions 104 to 122) for the SAR202 group. A double labeling approach at 5′ and 3′ ends was performed for all probes (Sigma-Aldrich, Germany).
Hybridization was performed in Sylgard chambers (GEOMAR in-house production) which were located inside an equilibrated humid chamber (at 46°C for 3 h). The hybridization buffer consisted of 900 mM NaCl, 20 mM Tris-HCl (pH = 7.4), 10% formamide, 0.01% sodium dodecyl sulfate and 20% dextransulfate. All Chloroflexi clade-specific probes were co-hybridized, in addition counterstaining of bacterial nucleic acids and sponge cell nuclei was performed with pre-warmed DAPI in hybridization buffer (1 ng/μL), with an incubation time of 20 min at 46°C. After hybridization, the slides were incubated in pre-warmed wash buffer (450 mM NaCl, 20 mM Tris–HCl, 0.01% sodium dodecyl sulfate) at 48°C for 25 min. The washing step was repeated twice. Subsequently, the slides were carefully rinsed with ice cold molecular grade water, dried and mounted in Mowiol medium (Mowiol® 40–88, Kuraray Europe GmbH, Tokyo, Japan). The same procedure was conducted for a negative control for non-specific binding (i.e., hybridization buffer without Chloroflexi-specific probes, but with an oligonucleotide complementary to the probe EUB338 (NON-338-probe; Wallner et al., 1993), to prevent falling into the trap of false-positive results. Fluorescence signals at the four specific wavelength ranges were detected using an Axio Observer.Z1 microscope equipped with AxioCam 506 and Zen 2 software (version 2.0.0.0; Zeiss Germany).
In this study, the prokaryotic community composition of 39 Craniella sponge individuals was analyzed. A visual overview of the denoising stats is compiled in Supplementary Figure S1. The complete dataset after quality filtering consisted of 614,928 reads, which is 82% of the initial input read count. On average each sample had 15,767 reads, but as several samples contained less reads, a sampling depth of 3,900 was applied on the complete dataset for normalization. Four adult C. infrequens individuals and one adult C. zetlandica individual were found brooding embryos inside their tissue (Figure 2). For each of these brooding individuals, ten embryos had been picked and pooled. In C. infrequens, the embryos were more numerous compared to C. zetlandica and were scattered throughout the mesohyl of the parental sponge. Prokaryotic diversity (as estimated by Shannon index) was similar between the developmental stages within each sponge species (Figure 3 and Supplementary Table S1). Other alpha diversity indices (Pielou’s eveness, Faith’s PD and number of observed ASVs) led to a similar result (Supplementary Figure S2). Average Shannon indices were slighly lower in C. zetlandica than in C. infrequens, despite not statistically significantly (Figure 3 and Supplementary Table S1). The prokaryotic community compositions of both sponge species were significantly different from seawater (Figure 4A and Supplementary Table S2). Further, C. infrequens and C. zetlandica displayed species-specific microbiomes, as shown by their distinct clustering based on weighted UniFrac distances and on PERMANOVA analyses (Figure 4B, with some exceptions; Supplementary Table S2). Overall C. zetlandica samples showed a slightly higher variability than C. infrequens samples (please note in this regard that while C. infrequens specimens were all sampled at the same geographic location, C. zetlandica specimens analyzed in this study originate from different geographic locations).
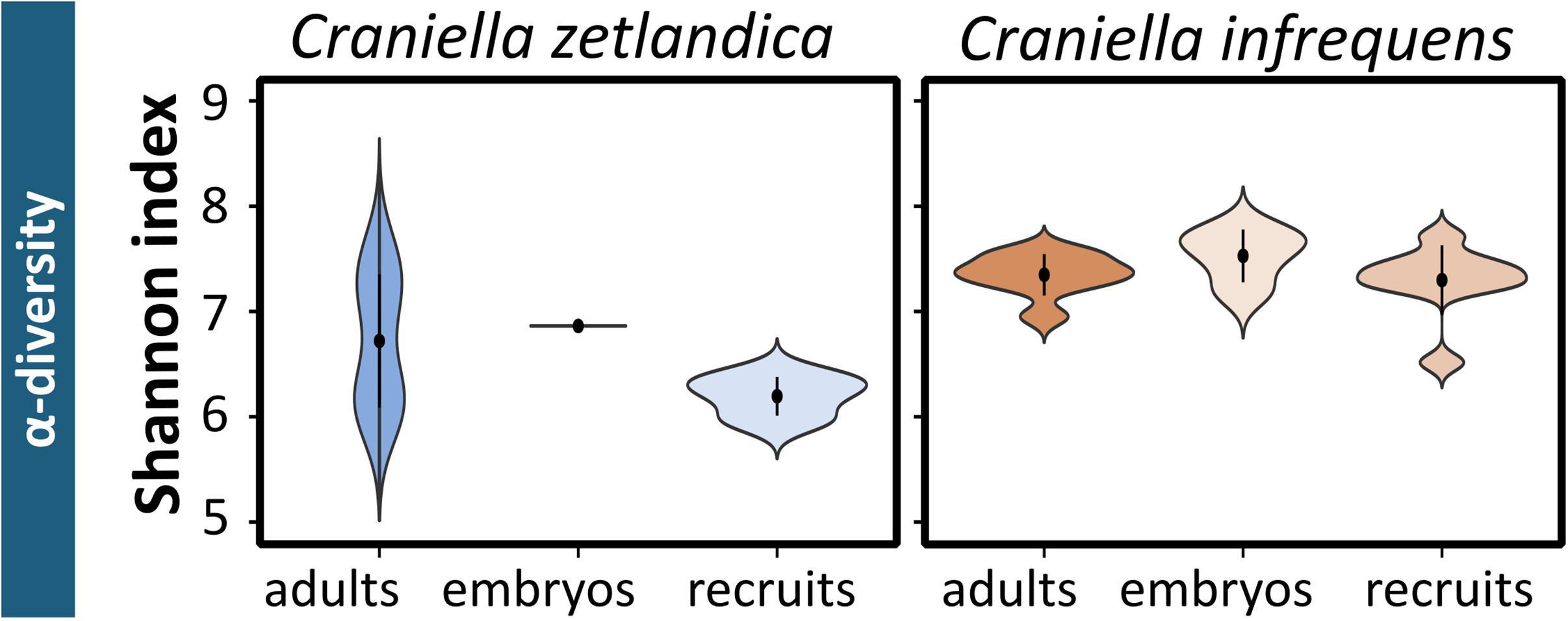
Figure 3. Prokaryotic alpha-diversity for the sampled Craniella species. Shannon indices calculated for the different developmental stages of C. zetlandica and C. infrequens.
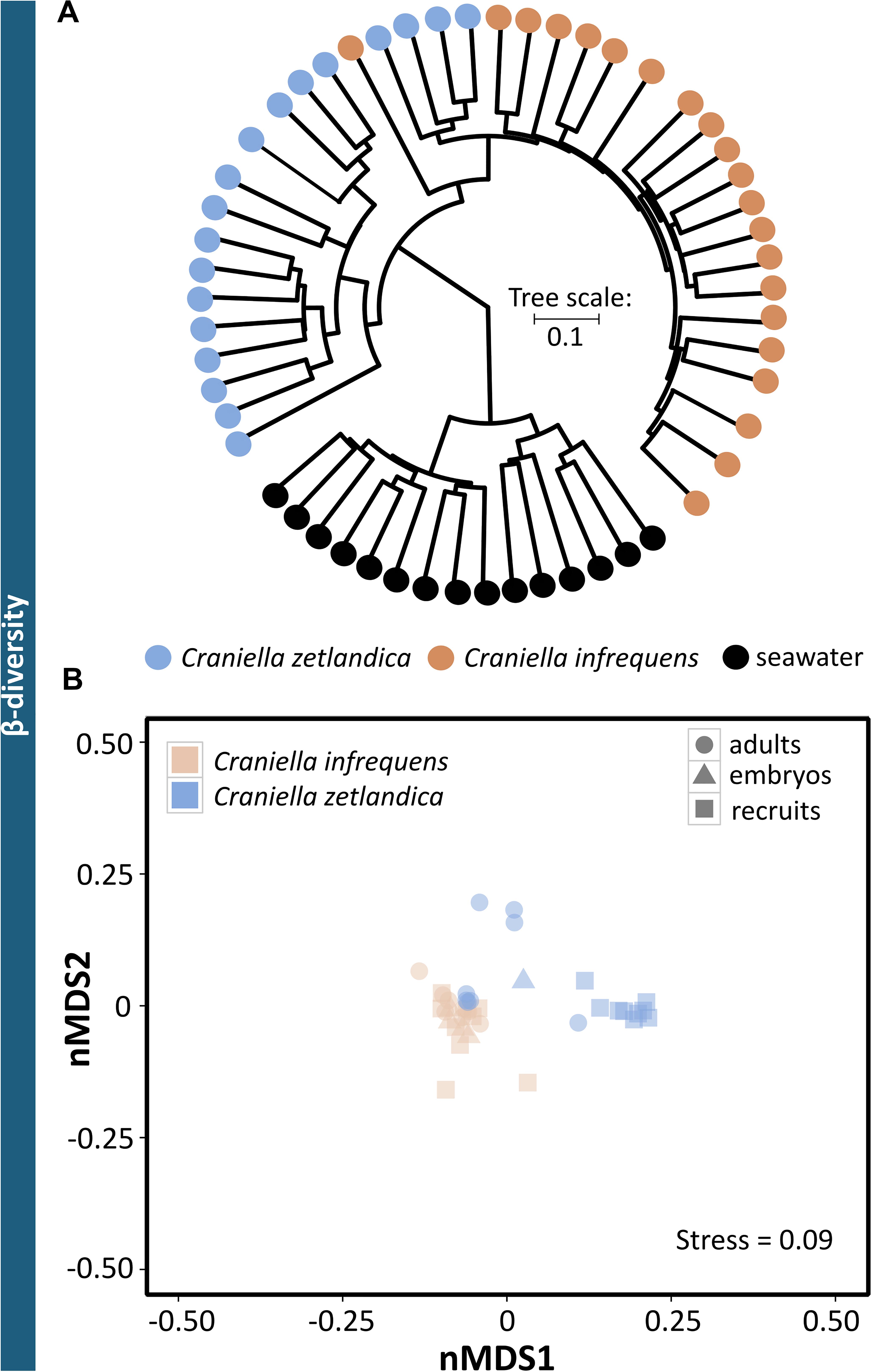
Figure 4. Prokaryotic beta-diversity across the sampled Craniella species and reference samples. (A) Clustering dendrogram based on weighted UniFrac distances for all samples of seawater and both sponge species. (B) Non-metric multidimensional scaling (nMDS) plot based on weighted UniFrac distances. The sponge species C. zetlandica and C. infrequens are marked by different colors, the different developmental stages (adult, recruit, and embryo) are indicated by different symbols.
With respect to the prokaryotic community composition, most bacterial phyla were shared across all life stages, and were not particularly enriched in any specific life stage (Figure 5A). Chloroflexi were the most abundant prokaryotic phylum in both sponge species and across all life stages and SAR202 was the most abundant clade, constituting on average 73% of all recorded Chloroflexi (Table 1). C. zetlandica recruits obtained from the aquarium system were a noticeable exception that were dominated by Proteobacteria and significantly enriched in this phylum compared to in situ sampled sponges (Figure 5B and Supplementary Figure S3A). Proteobacteria were however not significantly enriched in the aquarium water compared to in situ sampled seawater (Supplementary Figures S3B C). Neither were Chloroflexi significantly depleted in the aquarium water compared to in situ sampled seawater, although Chloroflexi were significantly enriched in the in situ sampled sponges compared to the aquarium sampled sponges. A total of 390 Chloroflexi ASVs were detected in the 39 Craniella samples, of which 171 were present in ≥50% of the samples per category (adult, embryo, recruit) (Figure 6A). Of these 171 common Chloroflexi ASVs, 16 were present in all life stages and both species, while being absent from seawater reference samples. These 16 ASVs are particularly abundant and account for 20–40% of the total Chloroflexi prokaryotic community in each sponge sample. Seven ASVs of the 16 ASVs belong to the SAR202 clade, while the remainder belongs to the classes JG30-KF-CM66 (4 ASVs), Anaerolineae (2 ASVs), TK17 (2 ASVs), and the order S085 (1 ASV).
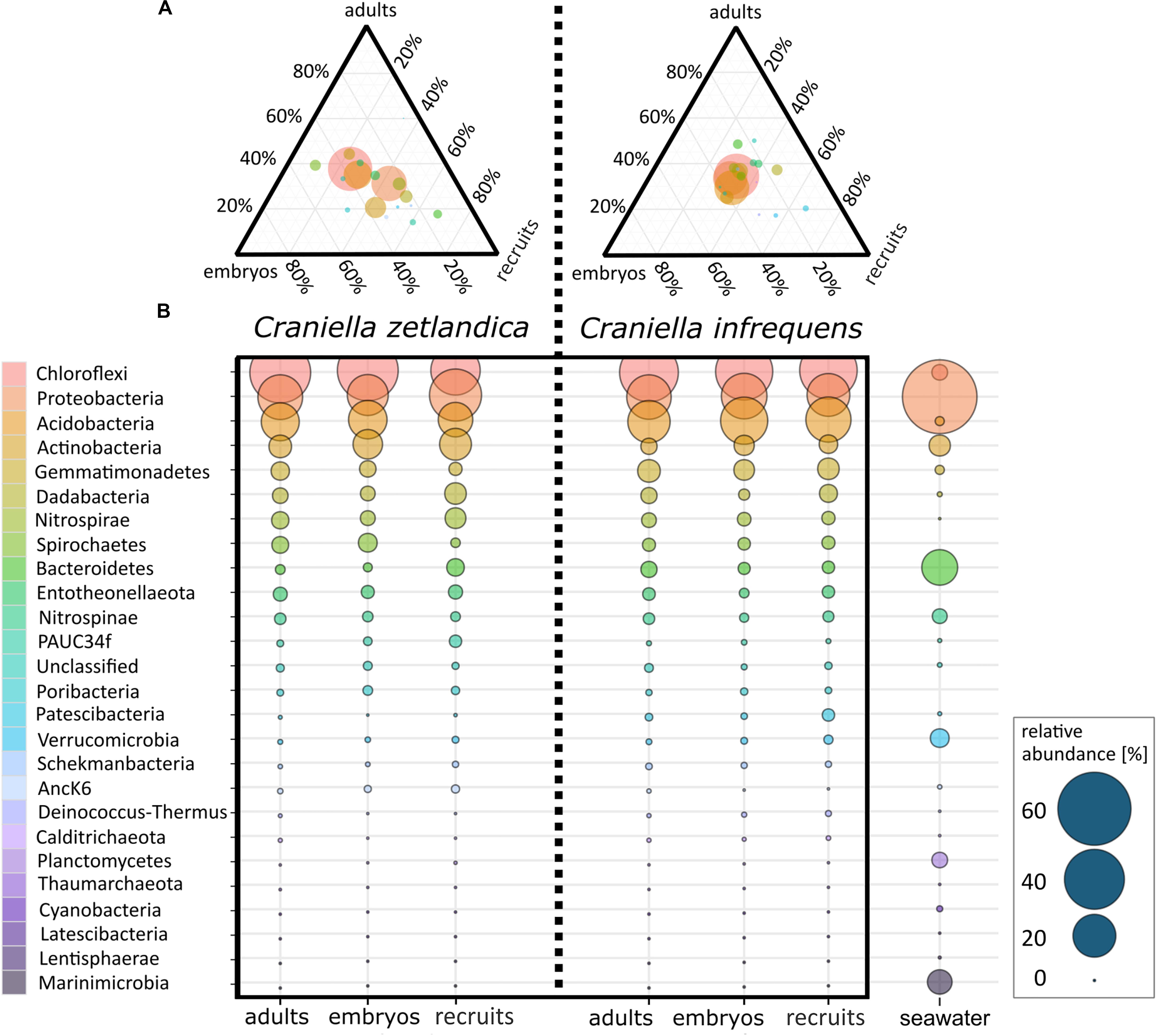
Figure 5. (A) Ternary plot of average prokaryotic phyla distributions for embryos, recruits, and adults of C. zetlandica and C. infrequens. Colors of phyla are the same in both figures: (A,B). Circle size indicates relative abundance of respective phyla. (B) Average prokaryotic community composition of C. zetlandica and C. infrequens developmental stages as well as seawater. Microbial phyla are sorted in descending order by mean relative abundances. Circle size indicates relative abundances (for exact relative abundance values consider the scale presented at the right bottom).
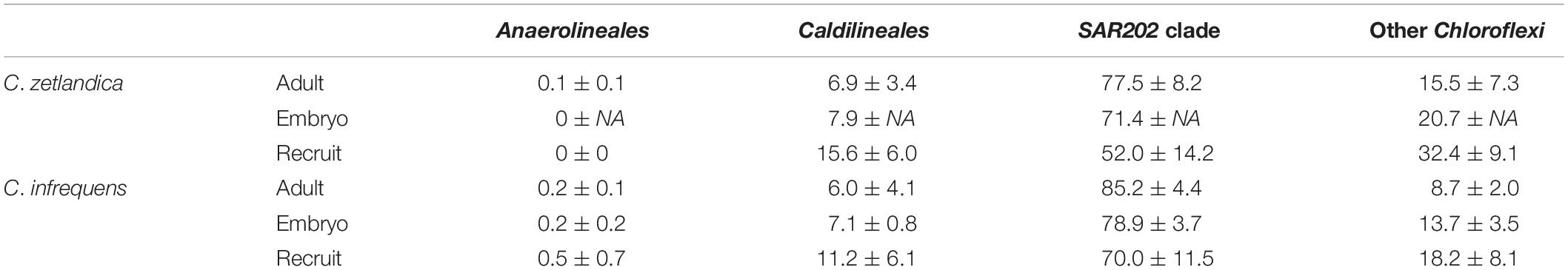
Table 1. Average relative abundances of Chloroflexi (Anaerolinae, Caldilineae, SAR202, others) as determined by 16S amplicon sequencing. mean, and standard deviation per group are indicated.
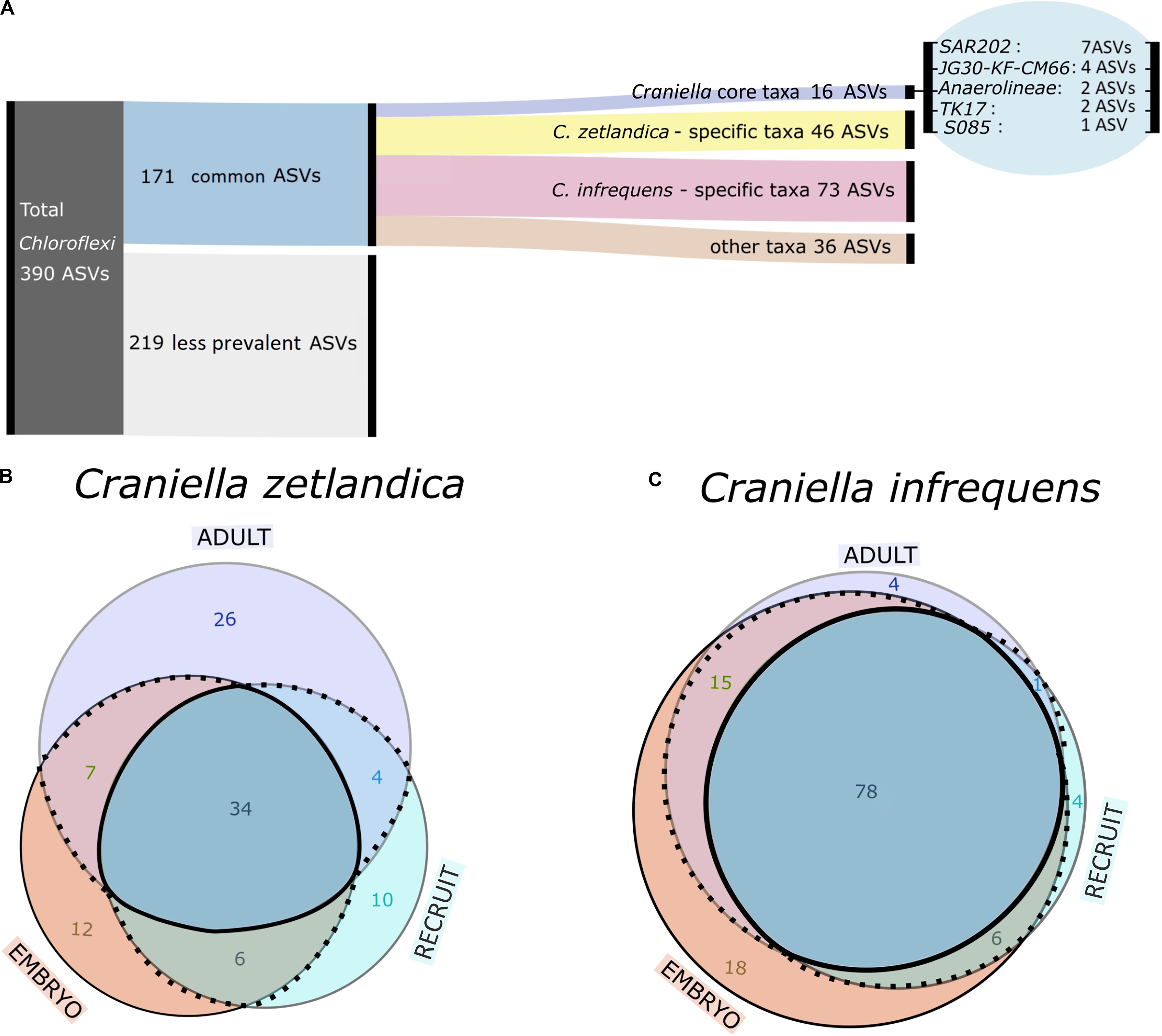
Figure 6. (A) Alluvial diagram showing grouping scheme of Chloroflexi ASVs. (B,C) Area proportional Venn-diagram showing counts of common Chloroflexi ASVs being shared or specific across different life stages for (B) C. zetlandica and (C) C. infrequens.
We next explored whether the 171 common Chloroflexi ASVs are species-specific or developmental stage-specific (Figures 6B,C). We determined 46 ASVs as being C. zetlandica specific (absent from C. infrequens), and 73 ASVs as being C. infrequens specific (absent from C. zetlandica). For C. zetlandica 26 ASVs were exclusively occurring in adults, 12 in embryos, and 10 in recruits. For C. infrequens four ASVs were occurring only in adults, 18 in embryos, and four in recruits. Interestingly, one Chloroflexi-ASV, occurred only in embryos of both sponge species. This ASV was affiliated with the Chloroflexi-class TK17, that could only be classified to class-level. Upon blasting against a sponge microbiome reference database (Moitinho-Silva et al., 2017a), this particular ASV was found in 134 sponges samples, of which 88 samples were identified as Geodia barretti. In summary, the major fraction of the detected Chloroflexi ASVs were sponge species-specific but not developmental-stage specific. Further information on the relative abundances of the ASV groups is presented in Supplementary Figure S4.
Light microscopy revealed that bacterial cells were numerous inside C. infrequens. In the embryos, the microbial cells were restricted to a narrow band at the peripheral rim (Figures 7A,B), while the interior was devoid of cellular material or bacterial morphotypes. An artifact resulting from cutting through the possibly more fluidic embryo interior can however not be excluded. Fluorescence in situ hybridization confirmed high abundances of Chloroflexi, in particular SAR202, within the “bacterial band” at the embryo rim (Figure 7C). The negative control for non-specific binding was negative (Supplementary Figures S5I–L). Bacterial cells were further found to be abundant and widely distributed in the mesohyl in adult C. infrequens (Supplementary Figure S6A). FISH confirmed high abundances of SAR202 also for adult individuals (Supplementary Figure S6B). Microbial cells within the embryos displayed diverse morphologies and were found in dividing stages (see Figure 7D for examples).
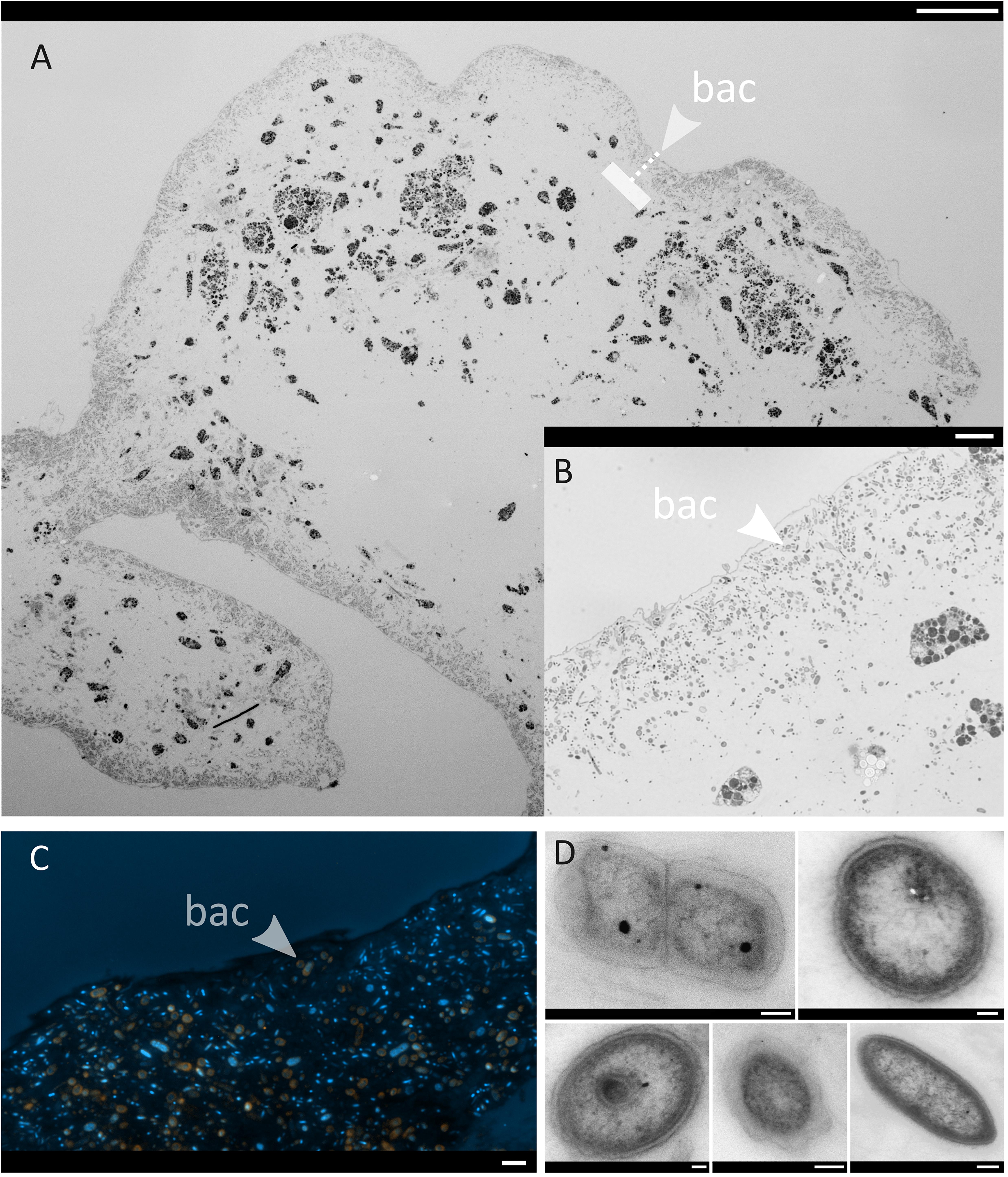
Figure 7. Micrographs of one representative C. infrequens embryo (n = 3 inspected). Arrows point from the outside to the inside. bac = band of bacterial cells (A) A whole C. infrequens embryo stained with Richardson solution. Scale bar = 100 μm. The dashed line indicates the band with highest bacterial densities (B) Brightfield microscopy of the bacterial band, scale bar = 10 μm. (C) Fluorescence in situ hybridization of the bacterial band. Merged channels of SAR202 signals (orange) and the DAPI counter stain (blue) are shown, scale bar = 2 μm. Micrographs of isolated hybridization signals for the specific probes (Anaerolinae, Caldilineae, and SAR202) are included in Supplementary Figures S5A–D. (D) TEM close-up of microbial cells located within the bacterial band, scale bars = 200 nm.
For C. zetlandica we observed an in situ (stress-induced) release of ripe propagules, visible by ROV live-footage in August 2018, during the sampling campaign “GS2018108” in Stjernsund (Supplementary Video S1: doi.pangaea.de/10.1594/PANGAEA.918218). We suspect a similar timing of spawning in summer for those sponges maintained in the aquarium system in Bergen, as high numbers of settled recruits were found in October 2018 (Supplementary Figure S7). These observations point to a synchronized reproductive cycle in aquarium and in situ thriving C. zetlandica with a spawning in late summer.
Deep-sea sponge grounds are underexplored ecosystems that provide numerous goods and services to the functioning of the deep-sea (Maldonado et al., 2016), such as the provision of food, habitat and refugia for commercially relevant fish species. The need to preserve these vulnerable marine ecosystems (VMEs) is thus contradictory to the high demand for socio-economic processes such as fishing in North Atlantic waters. In this study we assessed the prokaryotic diversity of two adult deep-sea sponge species of the genus Craniella and their developmental stages. These sponges are prominent although understudied members of deep-sea sponge grounds and have further been recorded in shelf-areas around the globe. While there is one report on the Proteobacteria-dominated microbiome of Craniella australiensis, which is a shallow water species in the Indo-Pacific and South China Sea (Anbuchezhian and Li, 2014), the microbiome of C. infrequens and C. zetlandica have—to our knowledge—not been characterized to date.
Our results reveal that C. zetlandica and C. infrequens are typical high microbial abundance (HMA) sponges (Gloeckner et al., 2014; Moitinho-Silva et al., 2017b). We base this statement on our observation that both sponge species harbor diverse and dense microbial communities within their tissues, as observed by TEM. This finding is further supported by the observation that the sponge microbiomes are dominated by Chloroflexi (SAR202), which are known indicator taxa of HMA sponges (Moitinho-Silva et al., 2017b).
The microbiomes of C. zetlandica and C. infrequens were host-species specific and distinctly different from seawater which is consistent with previous reports on sponge microbiomes (Easson and Thacker, 2014; Thomas et al., 2016; Steinert et al., 2017). The prokaryotic community composition and alpha diversity were very similar between adults, embryos and recruits within the same sponge species, indicating that entire microbial consortia are very likely vertically transmitted. Vertical transmission has been previously reported for a number of shallow-water HMA sponges (i.e., Schmitt et al., 2007; Björk et al., 2019), but is, to the best of our knowledge, novel in the context of sponge species from sponge grounds. Our results indicate that young deep-sea sponges can already contain a high diversity and density of microorganisms. Despite the observation of very similar prokaryotic community compositions between all examined life stages per species in our study, the recruits of C. zetlandica fell somewhat distant from all other samples in the nMDS plots and were dominated by Proteobacteria. It is noteworthy that these samples were collected in an aquarium system and not in the field. Indeed, previous studies have shown that the maintenance of marine organisms in captivity can affect the relative abundances of some associated microbial taxa (Galand et al., 2018). In addition, microbial variability has been observed to be particularly high in recently settled recruits compared to adult individuals (Schmitt et al., 2007; Busch et al., 2020).
The most dominant microbial phylum in all in situ sampled sponges of both sponge species and across all developmental stages were Chloroflexi. Generally, the probability of vertical symbiont-transmission is assumed to be positively related with the dependence of the host on the microbial partner partners (Ewald, 1987; Bull et al., 1991; Yamamura, 1993; Douglas, 1994; Thompson, 1994; Herre et al., 1999; Wilkinson, 2001; Sachs et al., 2011). Bacteria of the phylum Chloroflexi (in particular SAR202) are abundant and wide-spread in HMA sponges (Moitinho-Silva et al., 2017b; Bayer et al., 2018) and are suggested to play an important function in the degradation of recalcitrant organic matter (Landry et al., 2017; Bayer et al., 2018). We propose that the 16 Craniella core ASVs detected within our study should be considered as sponge symbionts as they were found in high abundances across both species and all developmental stages, while being absent from seawater. Almost half of the 16 Craniella core ASVs belong to the SAR202 clade, making it tempting to speculate about a potential crucial functional role in respect to (recalcitrant) organic matter cycling. Our FISH analyses revealed high abundances of Chloroflexi scattered within the bacterial band at the inner periphery of sponge embryos. We conclude that the transmission of Chloroflexi through the embryonal stages reflects their tight and evolutionarily long-standing integration in the sponge holobiont. Future research efforts should be geared toward understanding the specific functional roles of Chloroflexi symbionts for deep-sea sponges.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA612549.
KBu and UH designed the study, wrote the manuscript, and were responsible for the microbial pipeline. KBu, HR, and EW conducted sampling. KBa and KBu performed fluorescence in situ hybridization. HR led sponge taxonomic analysis. AF was involved in sequencing. KBu performed the data analysis (bioinformatics and visualizations). EW and KBa reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
This study was funded by the European Union’s Horizon 2020 Research and Innovation Program under Grant Agreement No. 679849 (the SponGES project). This document reflects only the authors’ view and the Executive Agency for Small and Medium-sized Enterprises (EASME) is not responsible for any use that may be made of the information it contains.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a past co-authorship with one of the authors HR.
We express our gratitude to HR, coordinator of the H2020-SponGES project on deep-sea sponge grounds of the North Atlantic, deep-sea sponge taxonomist, explorer, mentor, and friend. We thank the crews and scientific parties of RV G.O.Sars cruises “GS2017110” and “GS2018108”. On these cruises the ROV AEgir-team (University of Bergen) enabled sampling of precious samples. Ina Clefsen and Andrea Hethke provided valuable support in the lab back on land. We also thank Heikki Savolainen and Tone Ulvatn for help in aquaria facility management and technical support at the University of Bergen. Heidi Kristina Meyer’s support in the acquisition of video material is much appreciated.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00674/full#supplementary-material
Anbuchezhian, R., and Li, Z. (2014). “Marine sponge Craniella australiensis-associated bacterial,” in Encyclopedia of Metagenomics, ed. K. E. Nelson (Berlin: Springer-Verlag), 319–326. doi: 10.1007/978-1-4899-7475-4_604
Apprill, A., Marlow, H. Q., Martindale, M. Q., and Rappé, M. S. (2009). The onset of microbial associations in the coral Pocillopora meandrina. ISME J. 3, 685–699. doi: 10.1038/ismej.2009.3
Bayer, K., Jahn, M. T., Slaby, B. M., Moitinho-Silva, L., and Hentschel, U. (2018). Marine sponges as Chloroflexi hot spots: genomic insights and high-resolution visualization of an abundant and diverse symbiotic clade. mSystems 3:e00150-18.
Björk, J. R., Díez-Vives, C., Astudillo-García, C., Archie, E. A., and Montoya, J. M. (2019). Vertical transmission of sponge microbiota is inconsistent and unfaithful. Nat Ecol Evol. 3, 1172–1183. doi: 10.1038/s41559-019-0935-x
Blom, J. (2019). Ternary Plot Maker. Available online at: https://www.ternaryplot.com/ (accessed March 1, 2020).
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C., Al Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable, and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857.
Bull, J. J., Molineux, I. J., and Rice, W. R. (1991). Selection of benevolence in a host-parasite system. Evolution 45, 875–882. doi: 10.1111/j.1558-5646.1991.tb04356.x
Busch, K., Beazley, L., Kenchington, E., Whoriskey, F., Slaby, B., and Hentschel, U. (2020). Microbial diversity of the glass sponge Vazella pourtalesii in response to anthropogenic activities. bioRxiv [Preprint]. doi: 10.1101/2020.05.19.102806
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Knight, R., Lozupone, C. A., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Carrier, T. J., Dupont, S., and Reitzel, A. M. (2019). Geographic location and food availability offer differing levels of influence on the bacterial communities associated with larval sea urchins. FEMS Microbiol. Ecol. 95, 1–9.
De Caralt, S., Uriz, M. J., and Wijffels, R. H. (2007). Vertical transmission and successive location of symbiotic bacteria during embryo development and larva formation in Corticium candelabrum (Porifera: Demospongiae). J. Mar. Biol. Assoc. U. K. 87, 1693–1699. doi: 10.1017/s0025315407056846
Easson, C. G., and Thacker, R. W. (2014). Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front. Microbiol. 5:532. doi: 10.3389/fmicb.2014.00532
Eberl, G. (2010). A new vision of immunity: homeostasis of the superorganism. Mucosal. Immunol. 3, 450–460. doi: 10.1038/mi.2010.20
Endow, K., and Ohta, S. (1990). Occurrence of bacteria in the primary oocytes of vesicomyid clam Calyptogena soyoae. Mar. Ecol. Prog. Ser. 64, 309–311. doi: 10.3354/meps064309
Ereskovsky, A. V., Gonobobleva, E., and Vishnyakov, A. (2005). Morphological evidence for vertical transmission of symbiotic bacteria in the viviparous sponge Halisarca dujardini Johnston (Porifera. Demospongiae, Halisarcida). Mar. Biol. 146, 869–875. doi: 10.1007/s00227-004-1489-1
Ewald, P. W. (1987). Transmission modes and evolution of the parasitism-mutualism continuum. Ann. N. Y. Acad. Sci. 503, 295–306. doi: 10.1111/j.1749-6632.1987.tb40616.x
Flórez, L. V., Biedermann, P. H. W., Engl, T., and Kaltenpoth, M. (2015). Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat. Prod. Rep. 32, 904–936. doi: 10.1039/c5np00010f
Galac, M. R., Bosch, I., and Janies, D. A. (2016). Bacterial communities of oceanic sea star (Asteroidea: Echinodermata) larvae. Mar. Biol. 163:162.
Galand, P. E., Chapron, L., Meistertzheim, A.-L., Peru, E., and Lartaud, F. (2018). The effect of captivity on the dynamics of active bacterial communities differs between two deep-sea coral species. Front. Microb. 9:2565. doi: 10.3389/fmicb.2018.02565
Gloeckner, V., Lindquist, N., Schmitt, S., and Hentschel, U. (2013). Ectyoplasia ferox, an experimentally tractable model for vertical microbial transmission in marine sponges. Microb. Ecol. 65, 462–474. doi: 10.1007/s00248-012-0142-7
Gloeckner, V., Wehrl, M., Moitinho-silva, L., Schupp, P., Pawlik, J. R., Lindquist, N. L., et al. (2014). The HMA-LMA dichotomy revisited?: an electron microscopical survey of 56 sponge species. Biol. Bull. 227, 78–88. doi: 10.1086/bblv227n1p78
Harrington, B., and the Inkscape Development Team (2005). Inkscape. Available online at: http://www.inkscape.org/ (accessed February 18, 2019).
Hentschel, U., Hopke, J., Horn, M., Friedrich, A. B., Wagner, M., Hacker, J., et al. (2002). Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68, 4431–4440. doi: 10.1128/AEM.68.9.4431-4440.2002
Herre, E. A., Knowlton, N., Mueller, U. G., and Rehner, S. A. (1999). The evolution of mutualisms: exploring the paths between conflict and cooperation. Trends Ecol. Evol. 14, 49–53. doi: 10.1016/s0169-5347(98)01529-8
Klitgaard, A. B., and Tendal, O. S. (2004). Distribution and species composition of mass occurrences of large-sized sponges in the northeast Atlantic. Prog. Oceanogr. 61, 57–98. doi: 10.1016/j.pocean.2004.06.002
Koropatnick, T. A., Engle, J. T., Apicella, M. A., Stabb, E. V., Goldman, W. E., and Mcfall-ngai, M. J. (2004). Microbial factor-mediated development in a host-bacterial mutualism. Science 306, 1186–1189.
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the Miseq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/aem.01043-13
Landry, Z., Swa, B. K., Herndl, G. J., Stepanauskas, R., and Giovannoni, S. J. (2017). SAR202 genomes from the dark ocean predict pathways for the oxidation of recalcitrant dissolved organic matter. mBio 8, e00413-17.
Love, G. D., Grosjean, E., Stalvies, C., Fike, D. A., Grotzinger, J. P., Bradley, A. S., et al. (2009). Fossil steroids record the appearance of Demospongiae during the Cryogenian period. Nature 457, 718–721. doi: 10.1038/nature07673
Maldonado, M. (2007). Intergenerational transmission of symbiotic bacteria in oviparous and viviparous demosponges, with emphasis on intracytoplasmically-compartmented bacterial types. J. Mar. Biol. Assoc. U. K. 87, 1701–1713. doi: 10.1017/s0025315407058080
Maldonado, M., Aguilar, R., Bannister, R. J., Bell, J. J., Conway, K. W., Dayton, P. K., et al. (2016). “Sponge grounds as key marine habitats: a synthetic review of types, structure, functional roles, and conservation concerns,” in Marine Animal Forests, eds S. Rossi, L. Bramanti, A. Gori, and C. Orejas (Berlin: Springer-Verlag), 1–39. doi: 10.1007/978-3-319-17001-5_24-1
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C. G., Carey, H. V., Domazet-Lošo, T., and Douglas, A. E. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236.
Moitinho-Silva, L., Nielsen, S., Amir, A., Gonzalez, A., Ackermann, G. L., Cerrano, C., et al. (2017a). The sponge microbiome project. Gigascience 6:gix077.
Moitinho-Silva, L., Steinert, G., Nielsen, S., Hardoim, C. C. P., Wu, Y. C., McCormack, G. P., et al. (2017b). Predicting the HMA-LMA status in marine sponges by machine learning. Front. Microbiol. 8:752. doi: 10.3389/fmicb.2017.00752
Montgomery, M. K., and McFall-Ngai, M. (1994). Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development 120, 1719–1729.
Mortzfeld, B. M., Urbanski, S., Reitzel, A. M., Künzel, S., Technau, U., and Fraune, S. (2016). Response of bacterial colonization in Nematostella vectensis to development, environment and biogeography. Environ. Microbiol. 18, 1764–1781. doi: 10.1111/1462-2920.12926
Murillo, F. J., Kenchington, E., Koen-Alonso, M., Guijarro, J., Kenchington, T. J., Sacau, M., et al. (2020). Mapping benthic ecological diversity and interactions with bottom-contact fishing on the Flemish Cap (northwest Atlantic). Ecol. Indic. 112:106135. doi: 10.1016/j.ecolind.2020.106135
Muyzer, G., Waal, E. C. D. E., and Uitierlinden, A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 1–6.
Nicholson, J. K., Holmes, E., Kinross, J., Burcelin, R., Gibson, G., Jia, W., et al. (2012). Metabolic Interactions. Science 108, 1262–1268.
Nussbaumer, A. D., Fisher, C. R., and Bright, M. (2006). Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature 441, 345–348. doi: 10.1038/nature04793
Pham, C. K., Murillo, F. J., Lirette, C., Maldonado, M., Colaço, A., Ottaviani, D., et al. (2019). Removal of deep-sea sponges by bottom trawling in the Flemish Cap area: conservation, ecology and economic assessment. Sci. Rep. 9:15843.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596.
R Development Core Team (2008). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rosenberg, E., and Zilber-Rosenberg, I. (2011). Symbiosis and development: the hologenome concept. Birth Defects Res. Part C Embryo Today Rev. 93, 56–66. doi: 10.1002/bdrc.20196
Sachs, J. L., Skophammer, R. G., and Regus, J. U. (2011). Evolutionary transitions in bacterial symbiosis. Proc. Natl. Acad. Sci. U.S.A. 108, 10800–10807. doi: 10.1073/pnas.1100304108
Schimak, M. P., Schoffelen, N. J., Gruhl, A., and Dubilier, N. (2015). Transmission of Bacterial Symbionts in the Gutless Oligochaete Olavius Algarvensis. Bremen: University of Bremen, 161–199.
Schmitt, S., Angermeier, H., Schiller, R., Lindquist, N., and Hentschel, U. (2008). Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl. Environ. Microbiol. 74, 7694–7708. doi: 10.1128/aem.00878-08
Schmitt, S., Weisz, J. B., Lindquist, N., and Hentschel, U. (2007). Vertical transmission of a phylogenetically complex microbial consortium in the viviparous sponge Ircinia felix. Appl. Environ. Microbiol. 73, 2067–2078. doi: 10.1128/aem.01944-06
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60.
Sharp, K. H., Eam, B., John Faulkner, D., and Haygood, M. G. (2007). Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl. Environ. Microbiol. 73, 622–629. doi: 10.1128/aem.01493-06
Steinert, G., Rohde, S., Janussen, D., Blaurock, C., and Schupp, P. J. (2017). Host-specific assembly of sponge-associated prokaryotes at high taxonomic ranks. Sci. Rep. 7, 1–9.
Thomas, T., Moitinho-Silva, L., Lurgi, M., Björk, J. R., Easson, C., Astudillo-García, C., et al. (2016). Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7:11870.
Thompson, J. N. (1994). The Coevolutionary Process. Chicago, IL: The University of Chicago Press, 1–383.
Thompson, T., and Fuller, S. D. (2020). Technical Measures and Environmental Risk Assessments for Deep-Sea Sponge Conservation. Rome: FAO, 1–30.
van Soest, R. W. M., Boury-Esnault, N., Hooper, J. N. A., Rützler, K., de Voogd, N. J., Alvarez, B., et al. (2020). World Porifera Database. Accessed at: http://www.marinespecies.org/porifera (accessed February 06, 2020).
Wallner, G., Amann, R., and Beisker, W. (1993). Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14, 136–143. doi: 10.1002/cyto.990140205
Webster, N. S., Taylor, M. W., Behnam, F., Lücker, S., Rattei, T., Whalan, S., et al. (2010). Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. 12, 2070–2082.
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Wentrup, C., Wendeberg, A., Schimak, M., Borowski, C., and Dubilier, N. (2014). Forever competent: deep-sea bivalves are colonized by their chemosynthetic symbionts throughout their lifetime. Environ. Microbiol. 16, 3699–3713. doi: 10.1111/1462-2920.12597
Wickham, H. (2016). ggplot2: Elegant graphics for Data Analysis. New York, NY: Springer-Verlag, 1–213.
Wilkinson, D. M. (2001). Horizontally acquired mutualisms, an unsolved problem in ecology?? Oikos 92, 377–384. doi: 10.1034/j.1600-0706.2001.920222.x
Xavier, J. R., Cárdenas, P., Cristobo, J., Van Soest, R., and Rapp, H. T. (2015). Systematics and biodiversity of deep-sea sponges of the Atlanto-Mediterranean region. J. Mar. Biol. Assoc. U. K. 95, 1285–1286.
Yamamura, N. (1993). Vertical transmission and evolution of mutualism from parasitism. Theor. Popul. Biol. 44, 95–109. doi: 10.1006/tpbi.1993.1020
Keywords: vulnerable marine ecosystems, sponges, Craniella, Chloroflexi, symbiosis, early life stages, amplicon sequencing, fluorescence in situ hybridization
Citation: Busch K, Wurz E, Rapp HT, Bayer K, Franke A and Hentschel U (2020) Chloroflexi Dominate the Deep-Sea Golf Ball Sponges Craniella zetlandica and Craniella infrequens Throughout Different Life Stages. Front. Mar. Sci. 7:674. doi: 10.3389/fmars.2020.00674
Received: 23 March 2020; Accepted: 23 July 2020;
Published: 21 August 2020.
Edited by:
Joana R. Xavier, University of Porto, PortugalReviewed by:
Cristiane Cassiolato Pires Hardoim, São Paulo State University, BrazilCopyright © 2020 Busch, Wurz, Rapp, Bayer, Franke and Hentschel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ute Hentschel, dWhlbnRzY2hlbEBnZW9tYXIuZGU=
†Deceased March 07, 2020
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.