
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 30 July 2020
Sec. Global Change and the Future Ocean
Volume 7 - 2020 | https://doi.org/10.3389/fmars.2020.00605
Elevated CO2 levels predicted to occur by the end of the century can affect the physiology and behavior of marine fishes. For one important survival mechanism, the response to chemical alarm cues from conspecifics, substantial among-individual variation in the extent of behavioral impairment when exposed to elevated CO2 has been observed in previous studies. Whole brain transcriptomic data has further emphasized the importance of parental phenotypic variation in the response of juvenile fish to elevated CO2. In this study, we investigate the genome-wide proteomic responses of this variation in the brain of 5-week old spiny damselfish, Acanthochromis polyacanthus. We compared the accumulation of proteins in the brains of juvenile A. polyacanthus from two different parental behavioral phenotypes (sensitive and tolerant) that had been experimentally exposed to short-term, long-term and inter-generational elevated CO2. Our results show differential accumulation of key proteins related to stress response and epigenetic markers with elevated CO2 exposure. Proteins related to neurological development and glucose metabolism were also differentially accumulated particularly in the long-term developmental treatment, which might be critical for juvenile development. By contrast, exposure to elevated CO2 in the parental generation resulted in only three differentially accumulated proteins in the offspring, revealing potential for inter-generational acclimation. Lastly, we found a distinct proteomic pattern in juveniles due to the behavioral sensitivity of parents to elevated CO2, even though the behavior of the juvenile fish was impaired regardless of parental phenotype. Our data shows that developing juveniles are affected in their brain protein accumulation by elevated CO2, but the effect varies with the length of exposure as well as due to variation of parental phenotypes in the population.
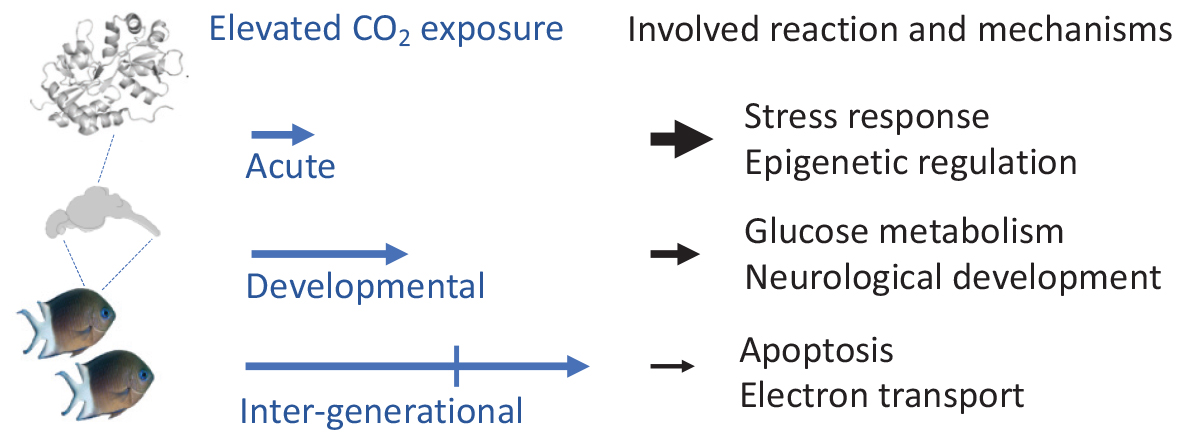
Graphical Abstract | Functional proteome response to varying length of elevated CO2 exposure in fish brains.
Rising CO2 levels in the ocean are anticipated to affect the physiology and behavior of marine organisms, with possible impacts on the structure and function of marine ecosystems (Wittmann and Pörtner, 2013; Cattano et al., 2018; Pörtner et al., 2019). Exposure to elevated CO2 has shown to alter important behaviors, such as navigation, habitat selection, learning and predator-prey interactions in some fish (Nagelkerken and Munday, 2016; Munday et al., 2019; Paula et al., 2019; Williams et al., 2019) and invertebrates (Watson et al., 2013; Jellison et al., 2016). For example, exposure to elevated CO2 impairs the natural avoidance behavior of chemical alarm cues (CAC) in some fishes (Ferrari et al., 2011; Chivers et al., 2014; Welch et al., 2014; Ou et al., 2015; Welch and Munday, 2017; Laubenstein et al., 2019). Chemical alarm cues are compounds released from the skin of injured fishes, thereby providing a reliable cue of immediate predation threat (Ferrari et al., 2009). Most fishes respond to the presence of CAC from conspecifics by making behavioral decisions that reduce the risk of predation, such as reducing activity and seeking shelter (Ferrari et al., 2009). However, when exposed to elevated CO2, some fish fail to exhibit behaviors that reduce their risk of predation. This impaired behavior is hypothesized to be caused by the self-amplifying alteration in the chloride-bicarbonate current in the GABAA receptor in the brain, following acid-base regulation in reaction to elevated environmental CO2 (Nilsson et al., 2012; Chivers et al., 2014; Heuer et al., 2016; Schunter et al., 2019).
Studying the response to a change in environmental condition on the molecular level of an organisms can provide insight into the broad response of a tissue to this change. Some molecular processes might change to acclimate to the new condition and prevent the need of a change on the physiological, behavioral or whole organism level. On the other hand, some molecular patterns will be the underlying processes to the whole organisms’ physiological changes in response to the environmental alteration (Madeira et al., 2017). Transcriptomics can aid in understanding this molecular reaction whereas the protein response can elucidate possible post-translational modifications that have a more direct effect on cellular responses (López, 2007; Tang et al., 2015).
Previous brain transcriptomic studies on five-month-old spiny damselfish A. polyacanthus showed differential gene expression in GABA related pathways especially during short-term elevated CO2 exposure (Schunter et al., 2018). However, when elevated CO2 exposure was prolonged across generations, the transcriptional signal differs and shows an overexpression of genes related to sugar and energy production in the brain (Schunter et al., 2016), whereas the behavioral impairments persist (Welch et al., 2014). There is also large variation in the sensitivity of individual A. polyacanthus to moderately high CO2 levels (∼700 μatm), with a part of the population exhibiting some resistance to behavioral impairment (“tolerant” individuals) whereas others are behaviourally impaired when exposed to elevated CO2 (“sensitive” individuals) (Schunter et al., 2016). This variation in sensitivity to behavioral effects of elevated CO2 has a heritable component as fathers and their offspring show similar sensitivities after short-term exposure to elevated CO2 (Welch and Munday, 2017).
The protein signal of the tolerant and sensitive parental behavioral phenotype in the offspring has been previously investigated in five-month-old spiny damsel fish, by comparing control versus inter-generational CO2 exposed fish (Schunter et al., 2016). However, the early juvenile developmental period is one of the most critical life stages in fish (DePasquale et al., 2015) and environmental conditions could have strong impacts during this early life stage. Therefore, here we focus on investigating the proteomic responses of five-week-old juvenile A. polyacanthus exposed to elevated CO2 (750 μatm) for different periods of time. In our experimental design we use the variation in the olfactory response behavior of adult fish to study the influence of parental phenotype on brain protein accumulation of small juveniles. Further, these developing offspring of spiny damselfish from behaviourally tolerant and sensitive parents were exposed to different exposure lengths of elevated CO2: 1) Acute (5-days of elevated CO2), Developmental (5 weeks of elevated CO2 since hatching) and Inter-generational (previous parental exposure and 5 weeks of elevated CO2). The short-term acute treatment allows to tease apart mechanisms involved in the acclimation process, whereas developmental and in particular inter-generational exposure are more relevant on ecological timescales. We investigated the molecular pathways involved in the reaction to the three different CO2 treatments by analyzing the differential accumulation of the brain proteome of 5-week old developing A. polyacanthus.
Adult A. polyacanthus pairs (n = 20) were collected as described in Welch and Munday (2017) and exposed to elevated CO2 (754 ± 92 μatm), consistent with average atmospheric CO2 predicted by the end of the century according to the RCP6 emissions trajectory. After a seven-day elevated CO2 exposure, the reaction toward chemical alarm cues (CAC) in adults was tested as described in Welch and Munday (2017) to define the parental phenotype to be either tolerant (T; less than 30% of the trial time was spent in CAC) or sensitive (S; more than 70% of the trial time in CAC) (Figure 1). Individuals of the same behavioral phenotype (T or S) were paired into breeding pairs and either left at control condition (414 ± 46 μatm) or placed into elevated CO2 (754 ± 92 μatm) for three months until breeding started. In this study we used offspring from two breeding pairs from each parental phenotype (Tolerant and Sensitive) within each of the two parental CO2 treatments (Control or elevated CO2). Offspring clutches from each breeding pair were then placed into different experimental conditions resulting in a total of four treatment groups for each parental behavioral phenotype (T and S) (Figure 1): (1) Parents and offspring were kept at a CO2 level of 414 ± 46 μatm (Control treatment); (2) Parents and offspring were kept at control condition, and offspring were exposed to elevated CO2 (754 ± 92 μatm) for five days before dissection (Acute treatment); (3) Parents were kept at control condition and offspring were exposed to elevated CO2 immediately after hatching for their entire developmental period (Developmental treatment); and (4) both parents as well as offspring were exposed to elevated CO2 for the entire duration of the study (Inter-generational treatment). Developmental and inter-generational exposures of elevated CO2 are relevant on an ecological timescale of ocean acidification in the wild. The short-term acute treatment was included to distinguish more immediate physiological responses to elevated CO2 and to compare with the developmental treatment in order to identify which are specific developmental affects that depend on exposure to elevated CO2 from early life. After five weeks juvenile fish were tested for their reaction to CAC as described in Welch and Munday (2017) with a sample number of 15–20 individual offspring for each parental behavioral phenotype (T and S; 7–10 per family line) within each of the four CO2 treatment (N = 120 fish sampled in total). Differences in the percentage of time juveniles spent in CAC was tested by pair-wise comparisons using Tukey’s post hoc tests in R. Due to the limited sample number that can be included in the proteomic analysis six offspring per treatment and phenotype were randomly selected (without prior knowledge of the outcomes of behavioral tests) and dissected for protein accumulation analysis. This means that three offspring from each of the tolerant and sensitive group coming from two family lines (Figure 1) were euthanized and the brain was dissected out and snapped frozen in liquid nitrogen and then stored in −80°C for further processing. This randomly chosen subset of samples represent the molecular reaction to each of the environmental CO2 treatments.
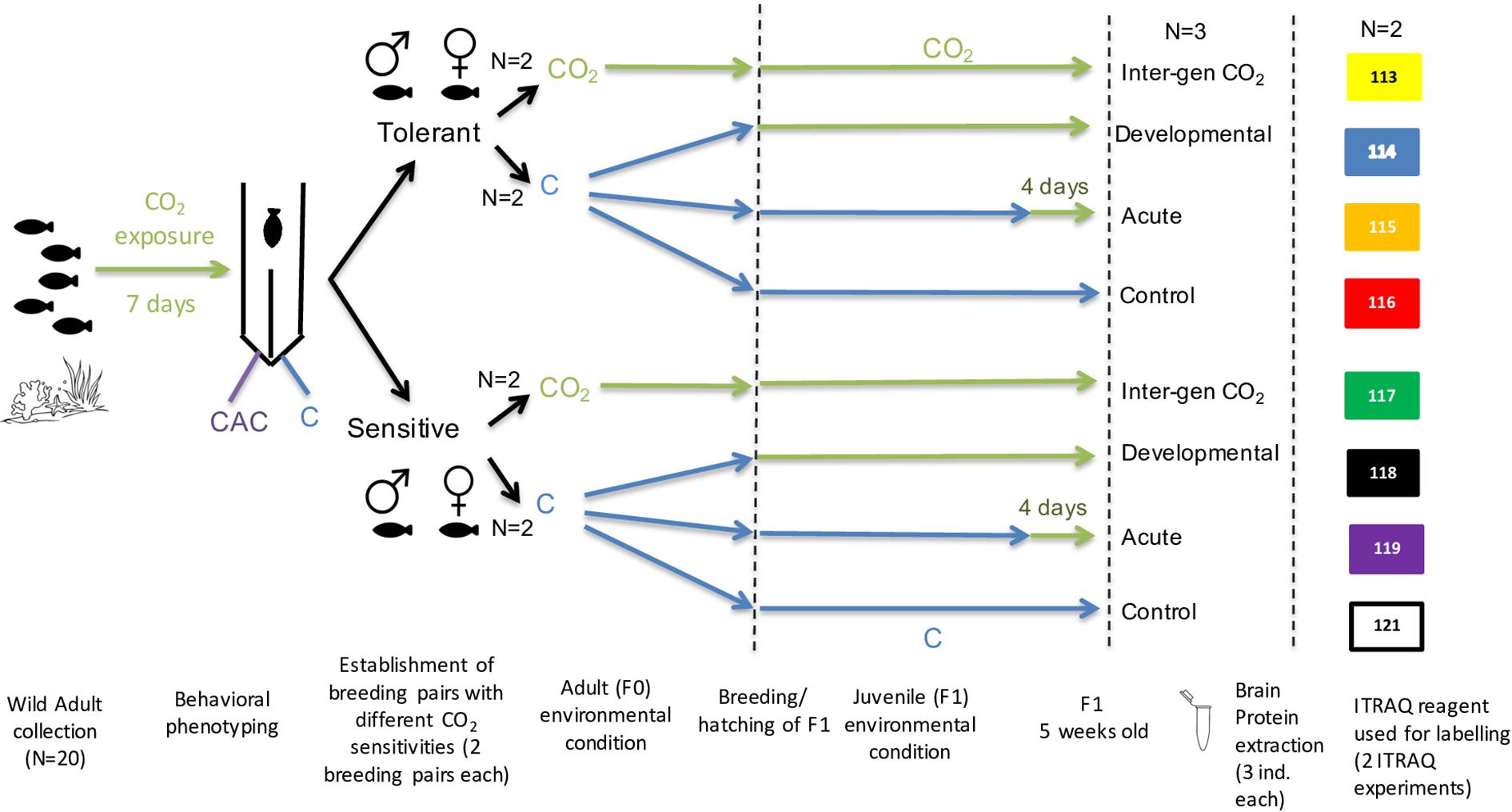
Figure 1. Schematic of experimental design from wild adult fish collection, behavioural testing, environmental CO2 exposure treatments and Proteome iTraq experimental design.
Proteins from whole brain tissue were extracted along with DNA and RNA with a Qiagen AllPrep DNA/RNA Mini Kit. The flow through at the RNA purification in the manufacturer’s instruction was kept and placed on ice for protein extraction. 3.5 μl of Halt protease and phosphatase inhibitor cocktail 100X, Thermo Fisher Scientific, was added, vortexed and split equally into two tubes and placed on ice for 15 min. Then 1000 μl of ice-cold acetone were added and placed on ice for 30 min for protein precipitation. Samples were then spun at full speed in a centrifuge at 4°C for 10 min at full speed of 16,873 RCF. The supernatant of acetone was discarded then without disturbing the protein pellet and the samples were left to dry for 15 min and then stored at −80°C.
Dried protein pellets extracted from fish brains were resuspended in 8 M urea lysis buffer with the help of a cooled stick sonicator for ten seconds at an amplitude of 20 (Fisher Scientific Q55) while kept on ice. Protein concentrations were measured using a 2-D Quant kit (GE Healthcare, United Kingdom). ITRAQ labeling allows for the relative comparison of protein accumulation across a maximum of eight labeled samples. To increase the number of biological samples and get a more representative picture we used three biological replicates of the same treatment and parental phenotype and pooled them at equal concentrations to a final volume of 100 μg. The pools of protein samples were then diluted 1:7 with 50 mM triethylammonium bicarbonate and digested using trypsin (Promega, United States) at an enzyme:protein ratio of 1:40 and left overnight at 37°C. Triflouroacetic acid at 2% was used to inactivate the digestion and peptides were desalted using Sep-Pak C18 cartridges (100 mg, Water Corporation, United States). Each sample (of three pooled biological replicates) was then labeled with iTRAQ Reagents-8plex reagents (Applied Biosystems) and incubated for 60 min. All eight samples (four CO2 treatments for two parental phenotypes) were then mixed together and dried by SpeedVac (Chandramouli et al., 2015). To further increase the representation of biological replicates to six per treatment and parental phenotype (for a total of 48 biological samples) we used two family lines and produced two complete iTraq experiments, each with eight treatment samples (of three pooled biological samples) for a total of 16 treatment samples.
The labeled pool of peptides was fractionated by strong cation exchange chromatography (SCX)26 using an Accela 1250 LC system (Thermo Scientific, United States). We obtained a total of 15 peptide fractions and then desalted, dried and resuspended them in 20 μL of LC-MS sample buffer (97% H20, 3% ACN, 0.1% formic acid). The samples were analyzed through three technical replicates using a Q Exactive HF mass spectrometer (Thermo Scientific, Germany) coupled with an UltiMateTM 3000 UHPLC (Thermo Scientific). Introduction of the sample into mass spectrometer was done through a Nanospray Flex (Thermo Scientific) with an electrospray potential of 1.5 kV and the ion transfer tube temperature was set at 160°C. The Q Exactive was set to perform data acquisition in the positive ion mode. A full MS scan (350–1400 m/z range) was acquired in the Orbitrap at a resolution of 60,000 (at 200 m/z) in a profile mode, a maximum ion accumulation time of 100 ms and a target value of 3 × e6. Charge state screening for precursor ion was activated. The ten most intense ions above a 2e4 threshold and carrying multiple charges were selected for fragmentation using higher energy collision dissociation (HCD) and the resolution was set as 15000. Dynamic exclusion for HCD fragmentation was 20 s. Other setting for fragment ions included a maximum ion accumulation time of 100 ms, a target value of 1 × e5, a normalized collision energy at 28%, and isolation width of 1.8.
Mascot generic format (mgf) files was used to convert the raw MS data in the Proteome Discoverer 1.4 software (Thermo Scientific) and submitted to MASCOT v2.4 (Matrix Sciences Ltd., United Kingdom) for database search against an A. polyacanthus brain protein dataset developed in-house from the genome assembly (Schunter et al., 2016). The mass tolerance was set to 10 ppm for precursors, and 0.5 Da for the MS/MS fragment ion with a maximum allowance of one missed cleavage. Variable modifications included 8-plex iTRAQ at tyrosine and oxidation at methionine. The fixed modifications were set to methylethanethiosulfonate at cysteine and lysine, and 8-plex iTRAQ at N-terminal. The MASCOT result files were processed using Scaffold v4.1.1 (Proteome Software Inc., United States) software for validation of peptide and protein identifications with a threshold of 95% using the Prophet algorithm with Scaffold delta-mass correction. iTRAQ label-based quantitation of the identified proteins was performed using the Scaffold Q+ algorithm. The intensities of all labeled peptides were normalized across all runs (Zhang et al., 2010) and individual quantitative data acquired in each run were normalized using the i-Tracker algorithm (Shadforth et al., 2005; Tian et al., 2011). Peptide intensity was normalized within the assigned protein. The reference channel was normalized to produce a 1:1 fold change, and the iTRAQ ratios were then transformed to a log scale. P-values were calculated using a paired t-test with significance level of 0.05. We allowed for missing data in one of the three technical replicates and accepted differential accumulation at a fold-change level of 1.5 (consistent over technical replicates).
We performed two different iTraq experiments including two family lines for differential protein accumulation among treatments. The final number of proteins varied between 2,100 to 2,300 with a total of 1,862 proteins overlapping among the two iTraq experiments/family lines. Protein quantification was only considered valid if minimum two technical measurements were valid. As we are measuring relative protein accumulation between samples, we used average values of technical replicates and normalized these against the control replicate for each biological replicate and protein. Each labeled iTraq replicate was compared to the seven other iTraq replicates (each including 3 biological pooled samples) for all commonly measured proteins. Significant differential accumulation was accepted at a fold change level of 1.5 between two replicates. Differential accumulation for each treatment comparison was consolidated among both iTraq experiments and was called when both family lines (represented by the two iTraq experiments) showed differential accumulation for the particular treatment comparison. In cases where only one of the two families showed differential accumulation but the other technical replicates in the family have a trend of up/down regulation, the overall average of all technical replicates within the two families was considered. Hence, the sum of relative protein accumulation values of all technical replicates was divided by the number of valid technical replicates. The proteins were considered to be differentially accumulated the average of the technical replicates showed 1.5-fold change in accumulation difference. Functional annotation of detected proteins was performed on the whole list of proteins obtained by using the in-house A. polyacanthus protein database and annotated with gene ontology terms in Blast2GO using an FDR significance cutoff of 0.05 (Conesa et al., 2005).
The behavioral response of 5-week-old A. polyacanthus to CAC was variable among individuals within the same CO2 treatment (Figure 2). Nevertheless, there were significant differences among CO2 treatments and parental phenotypes. At control conditions, the average percent of time spent by offspring of tolerant and sensitive parent in CAC was 14.9 and 20.3%, respectively. In the acute elevated CO2 treatment, offspring of sensitive parents (W = 647, P ≤ 0.001), but not tolerant parents (W = 870.5, P = 0.243) spent significantly more time in CAC compared with controls (Figure 2). For both parental phenotypes, the developmentally and the inter-generationally elevated CO2 treated offspring showed a significant difference in behavior when compared with the offspring in control condition (T Dev: W = 155 and P ≤ 0.001; T Inter: W = 334 and P ≤ 0.001; S Dev: W = 833.5 and P ≤ (0.001; S Inter: W = 673 and P ≤ 0.001). When comparing directly between parental phenotypes, only the acute elevated CO2 treatment resulted in a significant difference with more time spent in CAC for the offspring of sensitive parents (W = 563.5, P < 0.001).
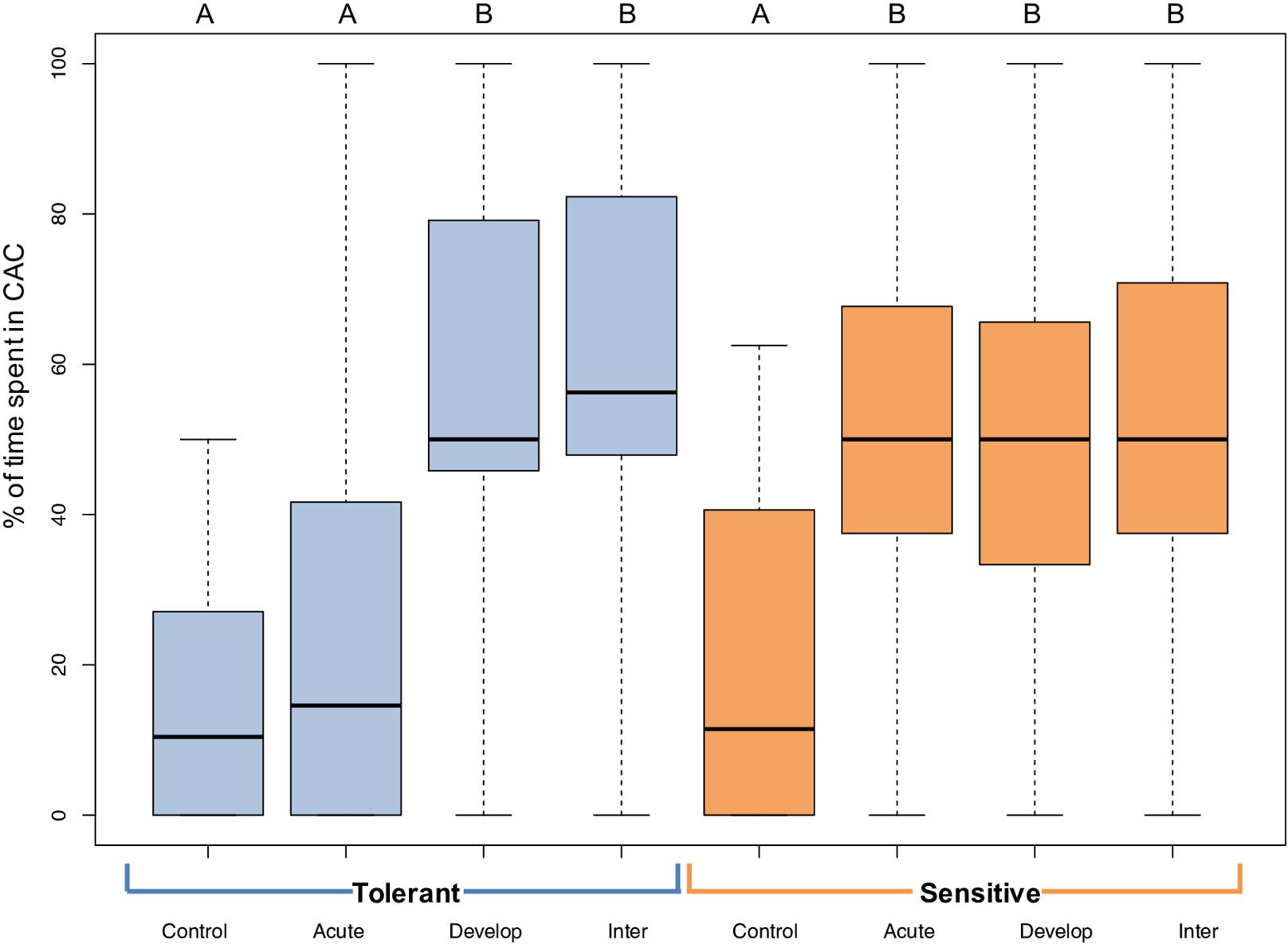
Figure 2. Behavioral responses to chemical alarm cues (CAC) of 5-week-old juvenile A. polyacanthus exposed to different CO2 treatments. Offspring of tolerant parental phenotype are in blue and sensitive in orange. Control, Acute, Develop, and Inter are the four different environmental CO2 exposures. The thick black line represents the median, the lower line of the colored box is the first quartile and the top line of the box represents the third quartile; whiskers show minimum and maximum values. Different letters represent significant differences between treatments (Tukey, P < 0.05). N = 15–20 for each of the eight treatments - phenotypes combinations.
Over 1,800 proteins were detected and commonly accumulated among the two-family lines. We first considered the offspring of the two parental phenotypes separately to understand the reaction to the different CO2 treatments against the respective control individuals. We detected between 0 and 40 differentially accumulated proteins among the pair-wise comparisons of different elevated CO2 exposures (Figure 3). Individuals exposed to acute elevated CO2 for five days exhibited a total of 17 and 40 differentially accumulated brain proteins in comparison to the control group for the sensitive and tolerant parental group, respectively (Figure 3). Fifteen proteins were found commonly differentially accumulated in both groups, representing 37.5% and 88% in tolerant and sensitive groups, respectively, showing a common response to acute exposure to elevated CO2 (Table 1). Fourteen out of the 15 proteins were accumulated at lower levels in the acute treatment than in the control condition. One of these proteins was B-cell receptor-associated protein 31-like isoform 1 (BCAP31), which is involved in the expression of MHC class I protein and the negative regulation of GABAergic synapses density (Elmer and McAllister, 2012). Three down-regulated proteins are involved in histone modifications. This includes the non-histone chromosomal protein HMG-14A-like isoform X2 (HMGN2) which is involved in phosphorylation inhibition of nucleosomal histones H3 and H2A; chromobox protein homolog 1-like (CBX1) which is associated with histone binding; and prothymosin alpha-B-like (PTMAB) which plays a role in histone methylation. Expression of two histone proteins (H1 and H2) were also differentially accumulated in the short-term elevated CO2 treatment.
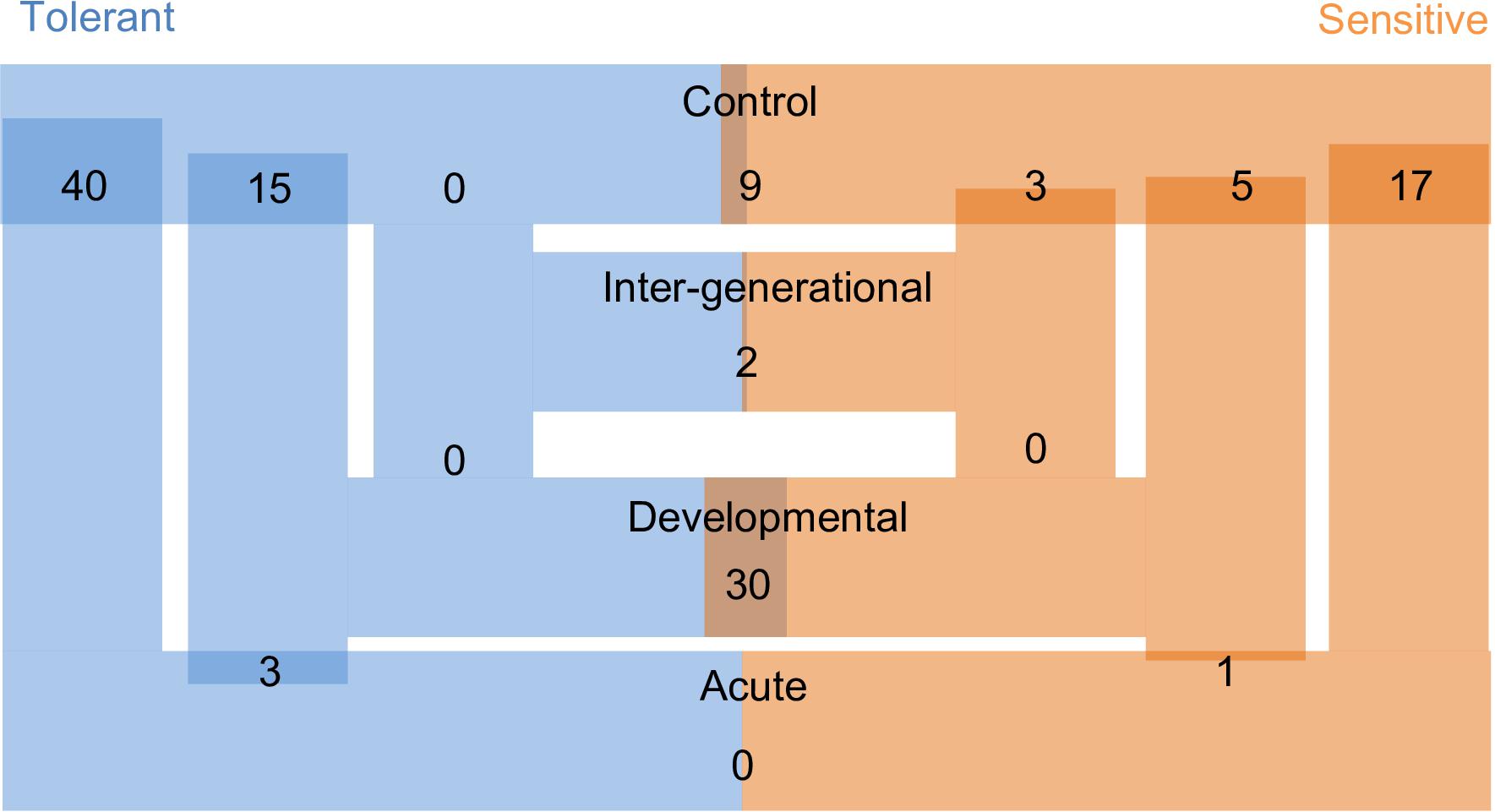
Figure 3. Number of differentially accumulated proteins (DAPs) across all comparisons between CO2 treatments and parental phenotypes. Larger overlap of bars represent a higher number of DAPs. ‘Tolerant’ and ‘Sensitive’ refers to the behavioral parental phenotypes. Control, Acute, Developmental and Inter-generational refer to each of the experimental CO2 conditions (Figure 1).
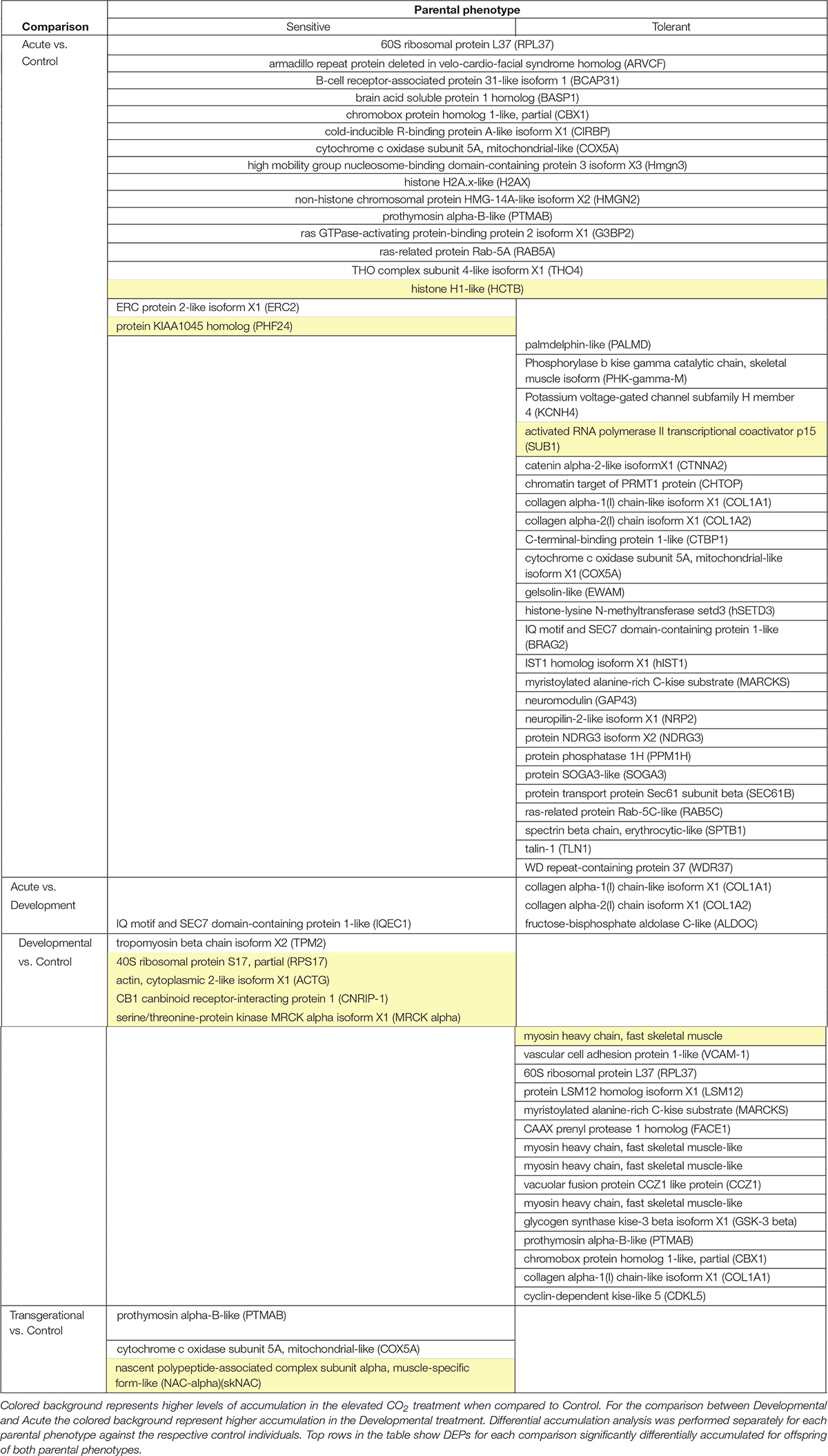
Table 1. Differentially accumulated proteins (DAPs) in the brain of 5-week-old juvenile A. polyacanthus exposed to four different CO2 treatments: Acute (n = 6) vs. Control (n = 6), Developmental (n = 6) vs. Control (n = 6), and Inter-generational (n = 6) vs. Control (n = 6) for each parental phenotype (sensitive and tolerant).
The longer-term developmental elevated CO2 exposure resulted in a less pronounced protein response when compared with the respective control individuals, with fewer differentially accumulated proteins than in the acute CO2 treatment (Figure 3). There was also no overlap in the differential accumulation among the offspring of sensitive and tolerant parents for this CO2 treatment (Table 1). In the offspring of sensitive parents, five proteins were differentially accumulated with one accumulated at a lower level and four at a higher level, including CB1 cannabinoid receptor-interacting protein 1 (CNRIP-1), which is related to synaptic plasticity. Fifteen proteins were differentially accumulated in the offspring of tolerant parents, none of which were differentially accumulated in the offspring of sensitive parents (Figure 3 and Supplementary Table S1). Two down-regulated proteins, vacuolar fusion CCZ1 like protein (CCZ1) and vascular cell adhesion protein (VCAM-1), are functionally related to calcium ion binding and organ development, respectively (Supplementary Table S1). 60S ribosomal protein L37 (RPL37), chromobox protein homolog 1-like (CBX1) and prothymosin alpha-B-like (PTMAB) in the offspring of tolerant parents showed similar down-regulation patterns as seen for fish in the acute CO2 treatment. Further proteins found to be differentially accumulated between the more longer-term developmental treatment and the acute exposure are several collagen alpha chain isoforms (COL1A1 and COL1A2; Table 1) as well as fructose-biphosphate aldolase C (ALDOC) for the offspring of tolerant parents whereas only one protein was differentially accumulated for the offspring of sensitive parents (IQ motif and SEC7 domain-containing protein).
The acute and developmental CO2 treatments reflected the within-generational responses. In the offspring of sensitive parents, 17 proteins were differentially accumulated in the acute CO2 condition while only five were differentially accumulated in the developmental CO2 condition and none overlap between the two comparisons (Figure 3 and Supplementary Table S1). As for the offspring of tolerant parents, five proteins were commonly differentially accumulated for the acute and developmental CO2 condition against the control condition which include functions such as immune response and epigenetic control (RPL37, PTMAB, MARCKS, CBX1, and COL1A1; see Supplementary Table S1).
To determine the effects of parental CO2 exposure on the offspring brain protein accumulation, we compared the inter-generational CO2 treatment with the control condition for both parental groups. No proteins were found to be differentially accumulated in the tolerant group, whereas three proteins were differentially accumulated in the sensitive group (Table 1). The two down-regulated proteins are prothymosin alpha-B-like (PTMAB) and cytochrome c oxidase subunit 5A mitochondrial-like protein (COX5A). The third (up-regulated) protein, nascent polypeptide-associated complex subunit complex alpha muscle specific form-like protein (NAC-alpha), is functionally related to the TBP-class protein binding.
In our experiment we were also able to directly compare differential brain protein accumulation between the offspring of the two parental phenotypes in the same environmental treatment (Figure 3). In control condition, nine proteins differed significantly in accumulation between offspring of sensitive and of tolerant parents of which five were accumulated at lower levels and four at higher levels for the offspring of sensitive parents in comparison to the offspring of tolerant parents (Supplementary Table S2). Both parental phenotypes reacted similarly in the acute elevated CO2 treatment as there was no significant difference in protein accumulation directly between the offspring of the two parental phenotypes for this environmental condition (Figure 3). The similar response between offspring of tolerant and sensitive parents in the acute elevated CO2 treatment is also shown by the previously described overlap of differentially accumulated proteins when comparing between environmental conditions of acute and control CO2 (Table 1). By contrast, 30 proteins were differentially accumulated between the offspring of the two parental phenotypes in the developmental elevated CO2 treatment. The majority of the differentially accumulated proteins (83%) were found at a higher level in the offspring of tolerant parents (Supplementary Table S2). As for the inter-generational CO2 condition, only two proteins were differentially accumulated between the offspring of the two parental phenotypes. The function of one of the proteins is not known. The other protein was AMP deamise 2-like isoform X1 protein (AMPD2), which is functionally related to the cyclic purine nucleotide metabolic process and energy homeostasis.
The different responses on the brain protein level for offspring of both parental phenotypes as well as for the different environmental treatments can be visualized in four recurring differentially accumulated proteins, including ribosomal protein L37 (RPL37), chromobox protein homolog 1-like (CBX1), cytochrome c oxidase subunit 5A (COX5A) and prothymosin alpha-B-like (PTMAB; Figure 4). In the acute elevated CO2 treatment, all four proteins were down-regulated in offspring of both sensitive and tolerant parental phenotype (Figures 4A–D). However, in the developmental elevated CO2 treatment, the proteomic responses of the offspring from the two parental phenotypes diverged. RPL37 and CBX1 were down-regulated in the offspring of tolerant parents, whereas the accumulation of these proteins was at control levels in offspring of sensitive parents (Figures 4A,B). The diverged proteomic responses between the two parental phenotype groups were further detected in the inter-generational elevated CO2 treatment. COX5A and PTMAB were down-regulated in the offspring of sensitive parents (Figures 4C,D), whereas the accumulation of these proteins was at control levels in the offspring of tolerant parents.
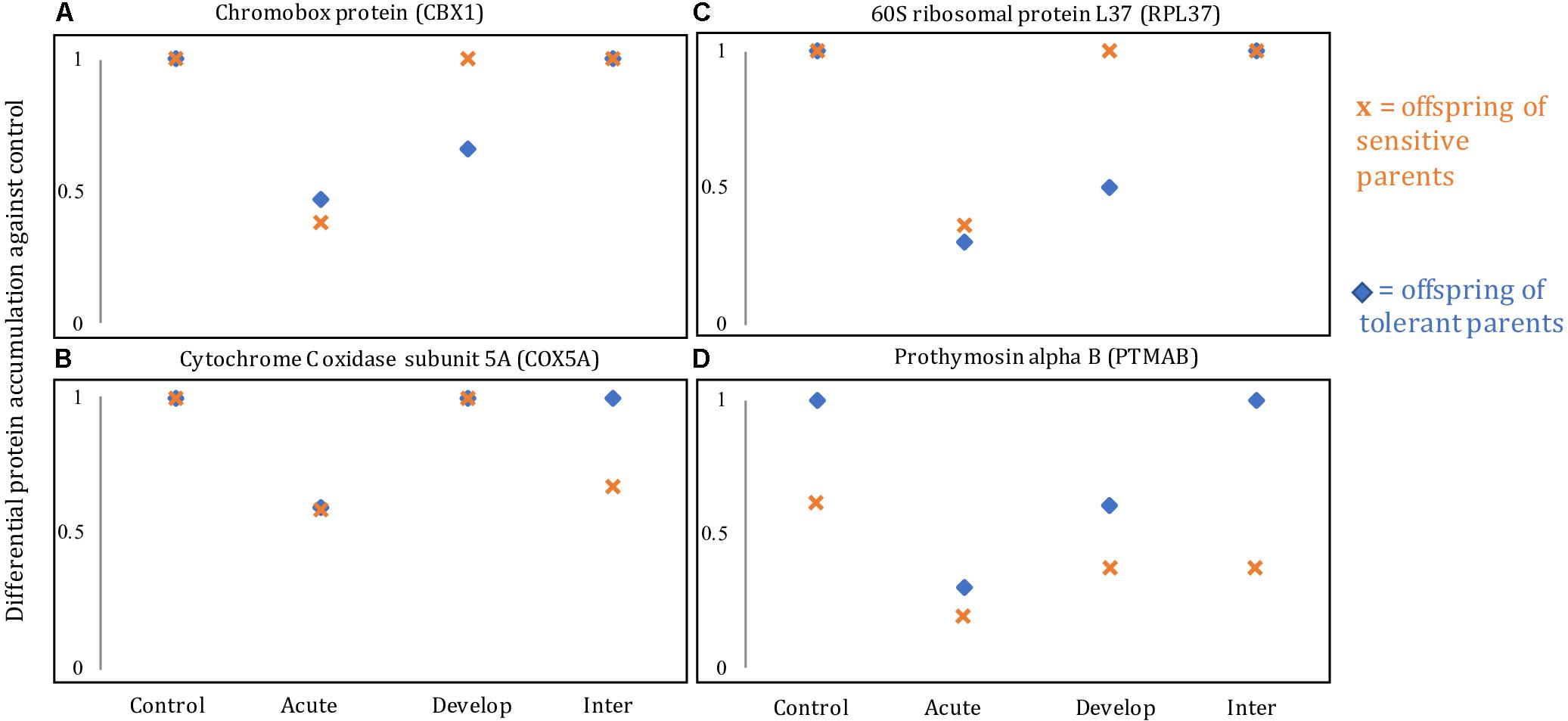
Figure 4. Levels of relative protein accumulation in the offspring of sensitive and tolerant parents of four different proteins: (A) chromobox protein homolog 1-like (CBX1), (B) cytochrome C oxidase subunit 5A, mitochondrial-like (COX5A), (C) 60S ribosomal protein L37 (RPL37), and (D) prothymosin alpha B-like (PTMAB). Control, Acute, Develop, Inter refer to the control, acute, developmental and inter-generational treatment, respectively.
Measuring brain proteome accumulation of organisms exposed to elevated CO2 conditions for different durations, within and between generations, can provide insight into how ocean acidification affects biological processes over relevant timescales. In this study we tested the effects of elevated CO2 treatment and parental phenotype on protein accumulation in the brain of five-week old developing juvenile Acanthochromis polyacanthus. Previous studies have investigated transcriptional effects in the fish brain, but at five months of age, whereas here we particularly focus on the developmental stage of a young juvenile and investigate the protein molecular response. Protein accumulation varied greatly among the different CO2 treatments, as well as between offspring from the two parental phenotypes, revealing treatment-specific responses to ocean acidification conditions in the brain proteome. This is consistent with previous results on the transcriptional level, where responses largely depended on the fish’s life stage and the duration of the exposure (Schunter et al., 2018).
The acute (5 day) elevated CO2 treatment resulted in the largest change in brain protein abundance of all treatments. Acutely treated fish were also the only group with a partial overlap in proteins response between the parental phenotypes. This reveals a large and partially common stress response to an acute change in environment CO2. We found changes in the accumulation of stress-related proteins, for example the cytochrome c oxidase subunit 5A (COX5A) which regulates cell activation and differentiation (Tarasenko et al., 2017). Studies in fish have shown that highly oxidative organs such as the brain depend on cytochrome c oxidase for normal functioning as it serves as a marker for neuronal activity (Wong-Riley, 2012) and Cox deficient fish diplayed neurological abnormalities (Diaz, 2010). Another stress-related response in the acute treatment was the differential regulation of proteins involved in epigenetic regulation. Amelioration of environmental CO2 changes by epigenetic regulation might be seen in our juvenile fish through the downregulation of several proteins linked to histone regulation. Despite the relatively small number of differentially accumulated proteins (maximum of 40 proteins) it represents more than 2% of the recovered proteins with this shot-gun proteomics method. We found differential protein accumulation in histone H1-like, together with PTMAB which binds to linker histone H1 (Karetsou et al., 1998; Zakharova et al., 2011), which also exhibited higher transcriptional levels in brains of five-month-old A. polyacanthus with elevated CO2 exposure (Schunter et al., 2016). The differential regulation of the genome, including silencing, enhancing and splicing of genes, relies heavily on these histone modifiers (Bird, 2002) and has been shown to enable plastic responses to environmentally induced changes (Turner, 2009). The regulation patterns of epigenetic markers at different life stage reveals an important molecular mechanism by which individuals respond to acute environmental change in CO2 conditions.
In contrast, during the longer-term developmental exposure to elevated CO2 the functions of differentially accumulated proteins were related to neurological development in juvenile A. polyacanthus. The CB1 cannabinoid receptor-interacting protein 1 (CNRIP1), for example, is a protein involved in synaptic plasticity and modulates neurotransmission (Howlett et al., 2010) and was differentially accumulated in the offspring of parents that were behaviourally sensitive to elevated CO2. Previous studies on zebrafish revealed that CNRIP1 is highly expressed in the nervous system, particularly in developing fish, and CNRIP1 interacts with the cannabinoid receptor 1, a receptor critical for the functional development of vision and locomotion (Fin et al., 2017; Oltrabella et al., 2017). In the offspring of tolerant parents, we further found differentially accumulated proteins, such as the vascular cell adhesion protein (VCAM-1), to be involved in cellular process and organ development (Kong et al., 2018). One of the few studies on proteomics in fish in the context of ocean acidification found glycolysis to be a key process that is altered with elevated CO2 treatment (Araújo et al., 2018) and for Acanthochromis offspring of tolerant parents glycogen synthase is differentially accumulated in the brain. Between the acute and developmental treatments we also detected differentially accumulated proteins that are involved in glucose metabolism (ALDOC) as well as proteins connected to connective tissue, such as collagen and tropomysin, which is important in brain development as well as brain repair and nerve regeneration (Gregorio et al., 2018) revealing differences among a short-term and longer-term treatment. Our results show that glycolysis as well as some neurological pathways and the structure in the brain could be affected by the differential accumulation of relevant proteins in particular when exposed to elevated CO2 during development.
Few proteins were differentially accumulated when offspring experienced developmental exposure to elevated CO2 in combination with previous exposure of the parents to elevated CO2, revealing an effect of parental exposure on the proteome of the offspring. Only two proteins, prothymosin alpha-B-like (PTMAB) and cytochrome c oxidase subunit 5A (COX5A), were down-regulated independent of parental exposure, but only in the offspring of sensitive parents. Similarly, in the European seabass an immune gene stayed upregulated in the olfactory epithelium despite previous parental exposure to ocean acidification (Mazurais et al., 2020). However, in our study all other differentially accumulated proteins in the within-generational treatments (acute and developmental) were at control levels in the inter-generational treatment. This hints to a non-genetic effect exerted by parents on the offspring through maternal provisioning and/or epigenetic inheritance (Bonduriansky et al., 2012; Salinas et al., 2013). Such parental effects have widely been recognized and has, in some cases, been directly proven to increase offspring’s fitness (Evans et al., 2014). The molecular adjustments in fish exposed to ocean acidification conditions are of lesser magnitude when their parents were also exposed to elevated CO2, both on the transcriptional level (Schunter et al., 2018) as well as seen here on the protein level. Despite this influence of previous parental exposure to elevated CO2 on the proteome, behavioral responses to chemical alarm cues (CAC) remained impaired in the inter-generational treatment. This is consistent with a previous study on the effect of elevated CO2 on behavior, based on a larger sample size, where previous parental treatment revealed no difference in the reaction to CAC (Welch and Munday, 2017). Hence, we see a change in the proteome response of juvenile damselfish with parental exposure, but this is not reflected in the behavioral response. It has to be noted that although the iTraq method allows for shot-gun proteomics in non-model organisms, there are constraints to the methodology, in particular the limitation to compare only eight labeled protein libraries. Therefore, the measured protein response is based on pooled biological samples and represents averages for the environmental treatments and parental phenotypes. Nonetheless, we exhibit inter-generational acclimation on the molecular level in the brains of A. polycanthus, but this does not alleviate the behavioral impairments to ocean acidification.
Previous studies have linked behavioral impairments in fish under elevated CO2 conditions to the altered functions of GABA neurotransmitter receptor (Nilsson et al., 2012; Schunter et al., 2019). One previous study in A. polyacanthus found highly up-regulated expression of GABA related genes in acute and developmental exposure to elevated CO2 (Schunter et al., 2018). In our study of five-week-old fish, there were no differentially accumulated proteins related to GABA receptor functions. Our experiment recovered a subset of approximately 2,000 proteins, in which we did detect gamma-aminobutyric acid receptor-associated proteins and aldehyde dehydrogese family 9 member A1-B-like protein, previously found to be differentially expressed on the transcript level in older fish (Schunter et al., 2016); however, in our 5-week old fish brains these proteins were not differentially accumulated. Our behavioral trials showed that after acute CO2 exposure our five-week old fish differed in their behavioral responses to CAC between the two different parental phenotypes. Despite this difference in behavior, these fish exhibited no differentially accumulated proteins, when directly compared, revealing that under short-term elevated CO2 fish of both parental phenotypes exhibit some similarity in their reaction at the brain protein level. For any other CO2 treatment (developmental or inter-generational), we do find distinct protein accumulation patterns between offspring from the different parental phenotypes. Hence in our five-week old fish the signal found in the brain proteome and the behavior does not show a highly concordant pattern. The connection between impaired behavior and protein responses to environmental elevated CO2 therefore remains unclear and it is possible that the brain protein accumulation is not representative of behavioral patterns due to the need for biological sample pooling in this proteomics method or the extended time needed for protein synthesis. Nevertheless, our results show that: (1) developing juveniles are affected in their brain proteins accumulation by near-future elevated CO2 conditions, (2) the impact on protein accumulation depends on the length of exposure to elevated CO2 exposure, with fewer differentially accumulated proteins being exhibited in longer-terms and inter-generational treatments, and (3) variation of parental sensitivity to behavioral effects of elevated CO2 affect the brain proteome of juveniles, especially under developmental exposure to elevated CO2. This variation in molecular response in a juvenile fish reveals a wide repertoire of plastic and inter-generational responses to elevated CO2 which could aid a coral reef fish to adjust to near-future ocean acidification conditions.
The mass spectrometry proteomics data acquired during this experiment have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD020350 and 10.6019/PXD020350.
The animal study was reviewed and approved by James Cook University (JCU) ethics permit A1828.
MW and PM designed and managed the fish rearing experiments. MW performed the fish behavioral phenotyping. CS extracted the proteins, designed the protein accumulation experiment, prepared the i-TRAQ libraries, and identified the proteins. HT and CS analyzed protein accumulation data and interpreted the results. HT, CS, PM, and TR wrote the manuscript and all authors read and approved the final manuscript.
This study was supported by the Australian Research Council (ARC) and the ARC Centre of Excellence for Coral Reef Studies (PM), the Office of Competitive Research Funds OCRF-2014-CRG3-62140408 from the King Abdullah University of Science and Technology (TR, PM, and CS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Marine and Aquaculture Research Facilities Unit (JCU), the Schunter lab members at the Swire Institute of Marine Science (SWIMS), and Biosciences Core Laboratory (KAUST) in particular Huoming Zhang for support and assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00605/full#supplementary-material
Araújo, J. E., Madeira, D., Vitorino, R., Repolho, T., Rosa, R., and Diniz, M. (2018). Negative synergistic impacts of ocean warming and acidification on the survival and proteome of the commercial sea bream, Sparus aurata. J. Sea Res. 139, 50–61. doi: 10.1016/j.seares.2018.06.011
Bird, A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21. doi: 10.1101/gad.947102
Bonduriansky, R., Crean, A. J., and Day, T. (2012). The implications of nongenetic inheritance for evolution in changing environments. Evol. Appl. 5, 192–201. doi: 10.1111/j.1752-4571.2011.00213.x
Cattano, C., Claudet, J., Domenici, P., and Milazzo, M. (2018). Living in a high CO2 world: a global meta-analysis shows multiple trait-mediated fish responses to ocean acidification. Ecol. Monogr. 88, 320–335. doi: 10.1002/ecm.1297
Chandramouli, K. H., Al-Aqeel, S., Ryu, T., Zhang, H., Seridi, L., Ghosheh, Y., et al. (2015). Transcriptome and proteome dynamics in larvae of the barnacle Balanus Amphitrite from the Red Sea. BMC Genomics 16:1063. doi: 10.1186/s12864-015-2262-1
Chivers, D. P., McCormick, M. I., Nilsson, G. E., Munday, P. L., Watson, S.-A., Meekan, M. G., et al. (2014). Impaired learning of predators and lower prey survival under elevated CO 2: a consequence of neurotransmitter interference. Glob. Chang. Biol. 20, 515–522. doi: 10.1111/gcb.12291
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
DePasquale, E., Baumann, H., and Gobler, C. J. (2015). Vulnerability of early life stage Northwest Atlantic forage fish to ocean acidification and low oxygen. J. Mar. Ecol. Prog. Ser. 523, 145–156. doi: 10.3354/meps11142
Diaz, F. (2010). Cytochrome c oxidase deficiency: patients and animal models. Biochim. Biophys. Acta Mol. Basis Dis. 1802, 100–110. doi: 10.1016/j.bbadis.2009.07.013
Elmer, B. M., and McAllister, A. K. (2012). Major histocompatibility complex class I proteins in brain development and plasticity. J. Trends Neurosci. 35, 660–670. doi: 10.1016/j.tins.2012.08.001
Evans, M. L., Wilke, N. F., O’Reilly, P. T., and Fleming, I. A. (2014). Transgenerational effects of parental rearing environment influence the survivorship of captive-born offspring in the wild. Conserv. Lett. 7, 371–379. doi: 10.1111/conl.12092
Ferrari, M. C. O., Dixson, D. L., Munday, P. L., McCormick, M. I., Meekan, M. G., Sih, A., et al. (2011). Intrageneric variation in antipredator responses of coral reef fishes affected by ocean acidification: implications for climate change projections on marine communities. Glob. Chang. Biol. 17, 2980–2986. doi: 10.1111/j.1365-2486.2011.02439.x
Ferrari, M. C. O., Sih, A., and Chivers, D. P. (2009). The paradox of risk allocation: a review and prospectus. Anim. Behav. 78, 579–585. doi: 10.1016/j.anbehav.2009.05.034
Fin, L., Bergamin, G., Steiner, R. A., and Hughes, S. M. (2017). The Cannabinoid Receptor Interacting Proteins 1 of zebrafish are not required for morphological development, viability or fertility. Sci. Rep. 7:4858.
Gregorio, I., Braghetta, P., Bonaldo, P., and Cescon, M. (2018). Collagen VI in healthy and diseased nervous system. DMM Dis. Model. Mech. 11:dmm032946. doi: 10.1242/dmm.032946
Heuer, R. M., Welch, M. J., Rummer, J. L., Munday, P. L., Grosell, M., Heuer, R. M., et al. (2016). Altered brain ion gradients following compensation for elevated CO2 are linked to behavioural alterations in a coral reef fish. Sci. Rep. 6:33216. doi: 10.1038/srep33216
Howlett, A. C., Blume, L. C., and Dalton, G. D. (2010). CB1 cannabinoid receptors and their associated proteins. Curr. Med. Chem. 17, 1382–1393. doi: 10.2174/092986710790980023
Jellison, B. M., Ninokawa, A. T., Hill, T. M., Sanford, E., and Gaylord, B. (2016). Ocean acidification alters the response of intertidal snails to a key sea star predator. Proc. R. Soc. B Biol. Sci. 283:20160890. doi: 10.1098/rspb.2016.0890
Karetsou, Z., Sandaltzopoulos, R., Frangou-Lazaridis, M., Lai, C.-Y., Tsolas, O., Becker, P. B., et al. (1998). Prothymosin α modulates the interaction of histone H1 with chromatin. Nucleic Acids Res. 26, 3111–3118. doi: 10.1093/nar/26.13.3111
Kong, D.-H., Kim, Y. K., Kim, M. R., Jang, J. H., and Lee, S. (2018). Emerging roles of vascular cell adhesion molecule-1 (VCAM-1) in immunological disorders and cancer. Int. J. Mol. Sci. 19:1057. doi: 10.3390/ijms19041057
Laubenstein, T. D., Rummer, J. L., McCormick, M. I., and Munday, P. L. (2019). A negative correlation between behavioural and physiological performance under ocean acidification and warming. Sci. Rep. 9:4265. doi: 10.1038/s41598-018-36747-9
López, J. (2007). Applications of proteomics in marine ecology. Mar. Ecol. Prog. Ser. 332, 275–279. doi: 10.3354/meps332275
Madeira, D., Araújo, J. E., Vitorino, R., Costa, P. M., Capelo, J. L., Vinagre, C., et al. (2017). Molecular plasticity under ocean warming: proteomics and fitness data provides clues for a better understanding of the thermal tolerance in fish. Front. Physiol. 8:825. doi: 10.3389/fphys.2017.00825
Mazurais, D., Servili, A., Noel, C., Cormier, A., Collet, S., Leseur, R., et al. (2020). Transgenerational regulation of cbln11 gene expression in the olfactory rosette of the European sea bass (Dicentrarchus labrax) exposed to ocean acidification. Mar. Environ. Res. 159:105022. doi: 10.1016/j.marenvres.2020.105022
Munday, P. L., Jarrold, M. D., and Nagelkerken, I. (2019). “Ecological effects of elevated CO2 on marine and freshwater fishes: from individual to community effects,” in Fish Physiology, eds M. Grosell, P. L. Munday, and A. P. Farrell, and C. J. Brauner, (Amsterdam: Elsevier Inc.), 323–368. doi: 10.1016/bs.fp.2019.07.005
Nagelkerken, I., and Munday, P. L. (2016). Animal behaviour shapes the ecological effects of ocean acidification and warming: moving from individual to community-level responses. Glob. Chang. Biol. 22, 974–989. doi: 10.1111/gcb.13167
Nilsson, G. E., Dixson, D. L., Domenici, P., McCormick, M. I., Sørensen, C., Watson, S.-A., et al. (2012). Near-future carbon dioxide levels alter fish behaviour by interfering with neurotransmitter function. Nat. Clim. Chang. 2, 201–204. doi: 10.1038/nclimate1352
Oltrabella, F., Melgoza, A., Nguyen, B., and Guo, S. (2017). Role of the endocannabinoid system in vertebrates: emphasis on the zebrafish model. Dev. Differ. 59, 194–210. doi: 10.1111/dgd.12351
Ou, M., Hamilton, T. J., Eom, J., Lyall, E. M., Gallup, J., Jiang, A., et al. (2015). Responses of pink salmon to CO2-induced aquatic acidification. Nat. Clim. Chang. 5, 950–955. doi: 10.1038/nclimate2694
Paula, J. R., Repolho, T., Pegado, M. R., Thörnqvist, P. O., Bispo, R., Winberg, S., et al. (2019). Neurobiological and behavioural responses of cleaning mutualisms to ocean warming and acidification. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-019-49086-0
Pörtner, H. O., Roberts, D. C., Masson-Delmotte, V., Zhai, P., Tignor, M., Poloczanska, E., et al. (2019). “IPCC, 2019: summary for policymakers,” in Proceedings of the IPCC, 2019: Summary for Policymakers IPCC Special Report on the Ocean and Cryosphere in a Changing Climate, (Geneva: IPCC).
Salinas, S., Brown, S. C., Mangel, M., and Munch, S. B. (2013). Non-genetic inheritance and changing environments. Rockville: ScienceDaily.
Schunter, C., Ravasi, T., Munday, P. L., and Nilsson, G. E. (2019). Neural effects of elevated CO2 in fish may be amplified by a vicious cycle. Conserv. Physiol. 7:coz100. doi: 10.1093/conphys/coz100
Schunter, C., Welch, M. J., Nilsson, G. E., Rummer, J. L., Munday, P. L., and Ravasi, T. (2018). An interplay between plasticity and parental phenotype determines impacts of ocean acidification on a reef fish. Nat. Ecol. Evol. 2, 334–342. doi: 10.1038/s41559-017-0428-8
Schunter, C., Welch, M. J., Ryu, T., Zhang, H., Berumen, M. L., Nilsson, G. E., et al. (2016). Molecular signatures of transgenerational response to ocean acidification in a species of reef fish. Nat. Clim. Chang. 6, 1014–1018. doi: 10.1038/nclimate3087
Shadforth, I. P., Dunkley, T. P. J., Lilley, K. S., and Bessant, C. (2005). i-Tracker: for quantitative proteomics using iTRAQ. BMC Genomics 6:145. doi: 10.1186/1471-2164-6-145
Tang, X., Meng, Q., Gao, J., Zhang, S., Zhang, H., and Zhang, M. (2015). Label-free quantitative analysis of changes in broiler liver proteins under heat stress using SWATH-MS technology. Sci. Rep. 5, 1–15. doi: 10.1038/srep15119
Tarasenko, T. N., Pacheco, S. E., Koenig, M. K., Gomez-Rodriguez, J., Kapnick, S. M., Diaz, F., et al. (2017). Cytochrome c oxidase activity is a metabolic checkpoint that regulates cell fate decisions during T cell activation and differentiation. Cell Metab. 25, 1254.e7–1268.e7.
Tian, L., Wang, M., Li, X., Lam, P. K. S., Wang, M., Wang, D., et al. (2011). Proteomic modification in gills and brains of medaka fish (Oryzias melastigma) after exposure to a sodium channel activator neurotoxin, brevetoxin-1. Aquat. Toxicol. 104, 211–217. doi: 10.1016/j.aquatox.2011.04.019
Turner, B. M. (2009). Epigenetic responses to environmental change and their evolutionary implications. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 364, 3403–3418. doi: 10.1098/rstb.2009.0125
Watson, S.-A., Lefevre, S., McCormick, M. I., Domenici, P., Nilsson, G. E., and Munday, P. L. (2013). Marine mollusc predator-escape behaviour altered by near-future carbon dioxide levels. Proc. R. Soc. London B Biol. Sci. 281:20132377. doi: 10.1098/rspb.2013.2377
Welch, M. J., and Munday, P. L. (2017). Heritability of behavioural tolerance to high CO 2 in a coral reef fish is masked by non-adaptive phenotypic plasticity. Evol. Appl. 10, 682–693. doi: 10.1111/eva.12483
Welch, M. J., Watson, S.-A., Welsh, J. Q., McCormick, M. I., and Munday, P. L. (2014). Effects of elevated CO2 on fish behaviour undiminished by transgenerational acclimation. Nat. Clim. Chang. 4, 1086–1089. doi: 10.1038/nclimate2400
Williams, C. R., Dittman, A. H., McElhany, P., Busch, D. S., Maher, M. T., Bammler, T. K., et al. (2019). Elevated CO 2 impairs olfactory-mediated neural and behavioral responses and gene expression in ocean-phase coho salmon (Oncorhynchus kisutch). Glob. Chang. Biol. 25, 963–977. doi: 10.1111/gcb.14532
Wittmann, A. C., and Pörtner, H.-O. (2013). Sensitivities of extant animal taxa to ocean acidification. Nat. Clim. Chang. 3, 995–1001. doi: 10.1038/nclimate1982
Wong-Riley, M. T. T. (2012). Bigenomic regulation of cytochrome c oxidase in neurons and the tight coupling between neuronal activity and energy metabolism. Adv. Exp. Med. Biol. 748, 283–304. doi: 10.1007/978-1-4614-3573-0_12
Zakharova, N. I., Sokolov, V. V., Suvorova, A. A., Shiau, A.-L., Wu, C.-L., and Evstafieva, A. G. (2011). Prothymosin alpha interacts with C-terminal domain of histone H1 and dissociates p53-histone H1 complex. Mol. Biol. 45:624. doi: 10.1134/s0026893311040157
Keywords: environmental proteomics, climate change, ocean acidification, behavior, tolerance
Citation: Tsang HH, Welch MJ, Munday PL, Ravasi T and Schunter C (2020) Proteomic Responses to Ocean Acidification in the Brain of Juvenile Coral Reef Fish. Front. Mar. Sci. 7:605. doi: 10.3389/fmars.2020.00605
Received: 27 May 2020; Accepted: 30 June 2020;
Published: 30 July 2020.
Edited by:
Christopher Edward Cornwall, Victoria University of Wellington, New ZealandReviewed by:
David Mazurais, Institut français de recherche pour l’exploitation de la mer (IFREMER), FranceCopyright © 2020 Tsang, Welch, Munday, Ravasi and Schunter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Celia Schunter, Y2VsaWFzY2h1bnRlckBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.