- 1Department of Food Science and Biotechnology, Kyungsung University, Busan, South Korea
- 2College of Pharmacy, Kyungpook National University, Daegu, South Korea
- 3Department of Chemical Engineering, Kyung Hee University, Gyeonggi-do, South Korea
A novel oligoalginate lyase from the marine bacterium, Stenotrophomonas maltophilia KJ-2, completely depolymerizes alginate to monomers with generation of oligomer intermediates. In order to analyze whether KJ-2 oligoalginate lyase has endolytic and exolytic alginate lyase activities, five mutants were developed and characterized using homology modeling based on the crystal structure of Alg17c, an exolytic oligoalginate lyase from Saccharophagus degradans 2–40. The important residues of Tyr238, Arg241, Arg418, and Glu644, which were predicted to be located within hydrogen bonding distance of the non-reducing end of alginate during exolytic catalysis, were mutated to Phe, Ala, Ala, and Ala, respectively. Exolytic activities of Arg241Ala, Glu644Ala, and Arg241Ala_Glu644Ala mutants decreased compared to the wild type, indicating that Arg241 and Glu644 are key residues for exolytic catalysis. Interestingly, these mutants produced oligomers as the main product due to inherent endolytic degradation activity. These mutational characterization results showed that KJ-2 oligoalginate lyase possesses catalytic domains for both inherent endolytic and exolytic activities.
Introduction
Oligoalginate lyase is an enzyme belonging to the polysaccharide lyase family. This enzyme degrades the non-reducing end of alginate or oligoalginate and produces unsaturated monomers (Miyake et al., 2003; Ryu and Lee, 2011; Park et al., 2012; Jagtap et al., 2014). Oligoalginate lyase A1-IV of Sphingomonas sp. A1 and oligoalginate lyase atu3025 of Agrobacterium tumefaciens C58 have only exolytic activity and their structures have been classified into the PL-15 family (Hashimoto et al., 2000; Miyake et al., 2003; Ochiai et al., 2010). Alg17c of Saccharophagus degradans 2–40 was characterized as an oligoalginate lyase belonging to the PL-17 family (Kim et al., 2012; Park et al., 2014).
Recently, MJ-3 oligoalginate lyase from Sphingomonas sp. strain MJ-3 was characterized to possess two conserved domains of the polymannuronate specific lyase (AlgL) superfamily and heparinase II/III (Hepa II/III) superfamily. The oligoalginate lyase of MJ-3 showed highly specific alginate-degrading activity and time-dependent degradation profiles that were quite different from other oligoalginate lyases, A1-IV, and atu3025 (Hashimoto et al., 2005; Ochiai et al., 2006; Park et al., 2012; Shin et al., 2015). Generally, exolytic oligoalginate lyases do not generate an oligomer intermediate, because they release monomers one by one from the end of alginate polysaccharide chain. In contrast, MJ-3 oligoalginate lyase simultaneously generate oligomer intermediates, trimer, and dimer together with monomer due to endolytic and exolytic activity. Site-directed mutagenesis indicated the possibility that the oligoalginate lyases of MJ-3 could possess both endolytic and exolytic alginate-degrading activity (Kim et al., 2015).
The cultivation of brown algae is already practiced in several countries and cost-effective large-scale production of brown algae is expected to be implemented in the near future (Roesijadi et al., 2010; Konda et al., 2015). Recently, metabolically engineered microbial cell factories have also been constructed for the production of bioethanol from seaweed biomass (Takeda et al., 2011; Wargacki et al., 2012; Ota et al., 2013; Enquist-Newman et al., 2014; Song et al., 2015). In order to utilize non-fermentable alginate for bioethanol fermentation, alginate monosaccharide is converted to 2-keto-3-deoxygluconate (KDG) by uronate reductase, and then further metabolized to pyruvate by kinase and aldolase via the Entner-Doudoroff pathway (Takase et al., 2010).
As a prerequisite for bioethanol fermentation using metabolically engineered cells, alginate needs to be efficiently degraded into alginate monosaccharide by alginate lyases. Thus, recruitment of various alginate lyases possessing a novel and efficient alginate-degrading activity for alginate saccharification is one of the key factors in successful commercialization of brown seaweed biomass-based biofuels production. For efficient saccharification of alginate, various polyM-, polyG-, polyMG-specific alginate lyases and oligoalginate lyase needs to be used as the biocatalysts. Marine and soil microorganisms are promising resources for identifying alginate lyases (Kim et al., 2011; Zhu and Yin, 2015). Various alginate lyases with different substrate specificity, catalytic action mode, protein structures and their products have been produced from various microbial resources including Pseudomonas aeruginosa, Azotobacter vinelandii, Sphingomonas sp., Agrobacterium tumefaciens, Saccharophagus degradans, Flammeovirga sp., and etc. (Zhu and Yin, 2015; Peng et al., 2018).
In order to identify novel alginate lyases from various microbial strains, we isolated a highly active alginate-degrading marine bacterial strain KJ-2 from salted guts from hairtails (Trichiurus haumela), a traditional Korean food, with alginate as the sole carbon source (Lee et al., 2012; Shin et al., 2015). Based on 16S rDNA analysis, chemotaxonomic and molecular characterization, the isolated strain was designated as Stenotrophomonas maltophilia KJ-2. In the process of recruiting highly efficient alginate-degrading enzyme from S. maltophilia KJ-2, we identified and characterized a polyMG-specific alginate lyase that preferentially degrades the heteropolymeric random sequence of poly-MG block, which was very rarely found in nature (Lee et al., 2012). Moreover, a highly active KJ-2 oligoalginate lyase possessing AlgL- and heparinase II/III-like domains with different alginate lyase activities was identified and used to completely degrade alginate and oligoalginate into monomers. In general, relatively long reaction time is required for oligoalginate lyase with only exolytic lyase activity (Miyake et al., 2003; Ochiai et al., 2010). Compared to exolytic oligoalginate lyase, it is assumed that high degradation activity of KJ-2 oligoalginate lyase might be due to the presence of both endolytic and exolytic degradation activity. In this study, the KJ-2 oligoalginate lyase from S. maltophilia KJ-2 was functionally analyzed using site-specific mutagenesis to prove the presence of the bifunctionality of endolytic and exolytic activity in one enzyme.
Materials and Methods
Homology Modeling and Prediction of the Active Site
The tertiary structures of oligoalginate lyases were homology modeled against the crystal structure of an oligoalginate lyase Alg17c (pdb code: 4OJZ) from Saccharophagus degradans 2–40 by using Swiss Modeler workspace (Arnold et al., 2006). Amino acid residues consisting of the active site and hydrogen bonding with the substrate were predicted and replaced with other amino acids by site-directed mutagenesis. Superimposition and analysis of molecular structures were performed by Chimera 1.10rc (Pettersen et al., 2004).
Site-Specific Mutagenesis of the KJ-2 Oligoalginate Lyase Gene
Site-specific mutagenesis of KJ-2 oligoalginate lyase (KJ-2 Oal) was carried out using a Quickchange II site-directed mutagenesis kit (Stratagene Co., United States) with the primers shown in Table 1. The recombinant pColdI/KJ-2-Oal plasmid containing oligoalginate lyase gene (GenBank Accession No. KR024335; Shin et al., 2015) excluding the signal peptide was used as the template for mutagenesis. To obtain double-site mutants, plasmid DNA containing the Arg241Ala mutant gene was used as a template. For protein expression, purified plasmids containing wild-type and mutated oligoalginate lyase genes were used for transformation of Escherichia coli BL21 (DE3), and the recombinant cells were cultured on a LB agar plate containing 50 μg/ml ampicillin. Recombinant E. coli were cultivated at 37°C for 2–3 h up to an OD600 of 0.4–0.6 in a shaking incubator at 190 rpm. The cells were incubated at 15°C for 30 min and then cultivated at 15°C for 24 h to express the oligoalginate lyase genes by the addition of 1 mM IPTG according to the method developed by Park et al. (2012).
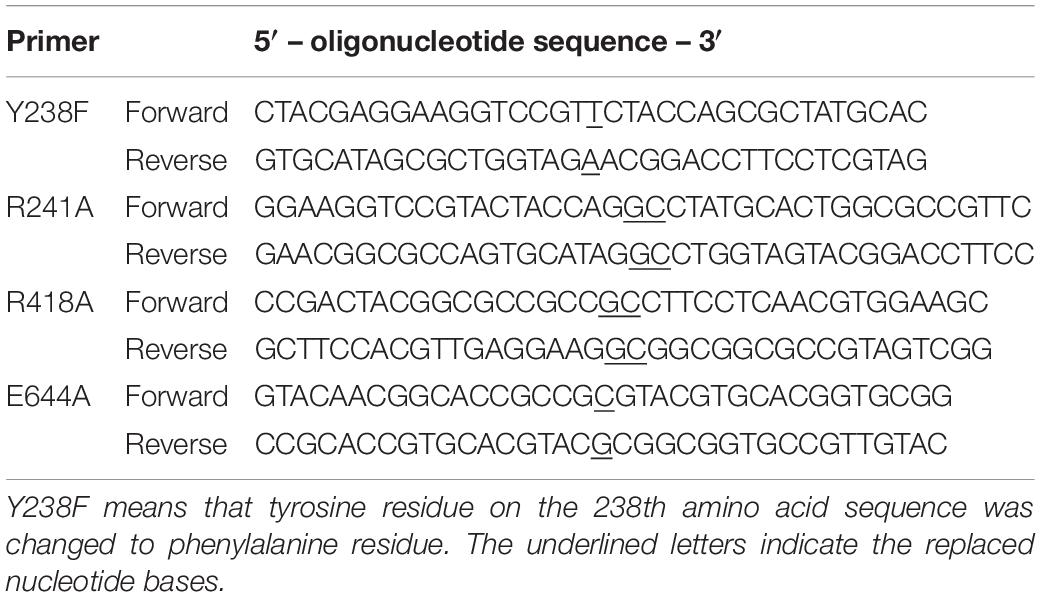
Table 1. Mutation sites and primer sequences for site directed mutagenesis of KJ-2 oligoalginate lyase from Stenotrophomonas maltophilia KJ-2.
To analyze the degradation profile of alginate, an IPTG-induced recombinant cell pellet was collected by centrifugation. To purify the expressed enzyme, the cell pellet was disrupted using an ultrasonicator (Vibra Cell CX400, Sonics & Materials Inc., United States), as described previously (Park et al., 2012). The expressed (His)6-tagged fusion protein was purified using a Ni-Sepharose affinity column (Amersham Biosciences, United States). The imidazole and salt in the elution solution were removed using a HiTrapTM desalting column (Amersham Biosciences, United States), and the protein solution was stored at 4°C. The protein concentration was determined using a BCA protein assay kit (Thermo Scientific, United States). Finally, overexpressed cells were lysed using a BugBuster protein extraction reagent containing lysonase (Novagen, United States) and cell lysates were analyzed by 10% SDS-PAGE electrophoresis and the TBA method.
Enzyme Assay and Analysis of Enzymatic Degradation Products
Enzymatic degradation of alginate was performed in 20 mM phosphate buffer containing 0.2% (w/v) sodium alginate at pH 7.0 and 30°C. The alginate lyase activity was determined using the TBA method (Weissbach and Hurwitz, 1959). The degradation products were analyzed by FPLC and ESI-MS. FPLC equipped with a Superdex peptide 10/100 GL column (GE Healthcare, Sweden) was used with 0.1 M ammonium bicarbonate at a flow rate of 0.5 ml/min. The degraded unsaturated oligomers and monomer were detected by measuring the absorbance at 220 and 235 nm using a UV detector.
For determination of molecular mass, the reaction mixture containing enzyme and the degraded alginate was mixed with methanol (1:1, v/v), filtered through a 0.45-μm cellulose membrane, and then injected into an electrospray-ionization mass spectroscope (6410 Triple Quadrupole LC-MS, Agilent, United States). The mobile phase was 10 mM ammonium acetate:methanol (1:1, v/v). MS was performed in the negative mode with an ion-spray voltage of 4 kV and a source temperature of 350°C.
Results
Homology Modeling and Prediction of Active Site for Exolytic Catalyst
Recently, the KJ-2 oligoalginate lyase gene (KJ-2 Oal, GenBank Accession No. KC430928) was cloned in a pColdI vector, overexpressed and then characterized (Shin et al., 2015). In the previous study, protein alignment analysis was conducted. The amino sequence of KJ-Oal showed very low sequence identity of 9.8 and 8.6% with Sphingomonas sp. A1 (A1-IV) and A. tumefaciens strain C58 (Atu3025), respectively. KJ-Oal had 51.7% sequence identity with oligoalginate lyase of Sphingomonas sp. MJ-3. KJ-2 Oal has two conserved domains, a polymannuronate lyase domain (AlgL superfamily) in the N-terminal region and a heparinase II/III like protein domain (Hepa II/III superfamily) in the C-terminal region (Figure 1).
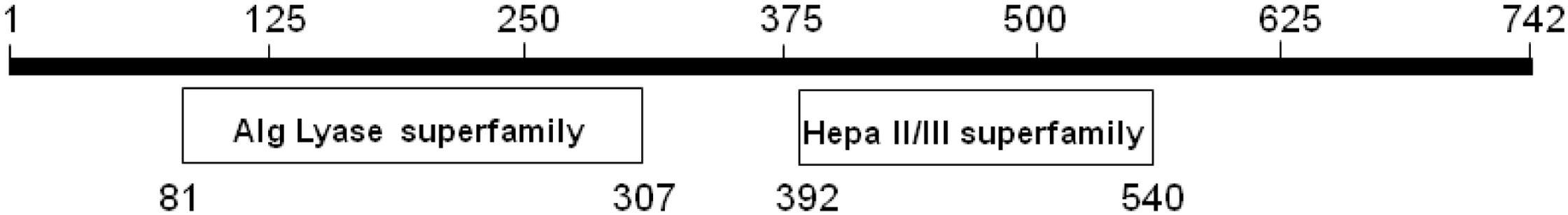
Figure 1. Conserved domains of KJ-2 oligoalginate lyase protein from Stenotrophomonas maltophilia KJ-2. KJ-2 oligoalginate lyase protein sequence was blasted within conserved domain database of NCBI. The number indicates amino acid residues. KJ-2 oligoalginate lyase contains two conserved domains with alginate lyase superfamily and heparinase II/III superfamily.
In order to determine whether KJ-2 Oal possesses a bifunctionality of endolytic and exolytic activity, we tried to mutate key amino acids involved in the exolytic catalysis to remove the exolytic activity and to confirm the existence of inherent endolytic activity. The first step for determination of bifunctionality is to identify important amino acids in exolytic catalysis. Elimination of exolytic activity can open a way to recognize inherent endolytic lyase activity of KJ-2 Oal. The second step is to generate mutants with changing the target amino acid with other one to remove or reduce exolytic lyase activity. The third step is to analyze alginate-degrading products by mutants and compare the degradation pattern with that of wild-type KJ-2 Oal to confirm the presence of inherent endolytic lyase activity of KJ-2 Oal.
For first step, we constructed a homology-model of KJ-2 Oal protein (amino acid residues from 12 to 712) by using the crystal structure of trisaccharide-bound Alg17c oligoalginate lyase from S. degradans 2–40 (Alg17c_Tyr258Ala mutant, PDB code: 4OJZ) as the template. The KJ-2 Oal protein had 50.0% amino acid sequence identity to Alg17c oligoalginate lyase (Alg17c-Oal). When the KJ-2 Oal homology model was superimposed on amino acid residues consisting of the substrate binding pocket, it was strictly superimposed upon the template structure (Figure 2). Alg17c-Oal belonging to the PL17 family has been reported to be a homodimer enzyme, which consists of two domains; imperfect α6/α6-barrel at the N-terminal domain and an antiparallel β-sheet in the C-terminal domain (Park et al., 2014). As shown in Figure 2, Glu644 in the B chain hydrogen bonds with the non-reducing end (ΔM) of the substrate on the A chain. As exolytic oligoalginate lyase cleaves the non-reducing end to an unsaturated monomer by a β-elimination reaction, the amino acid residues forming a hydrogen bond with ΔM are important during catalysis, and thus they were targeted for mutation to remove exolytic activity.
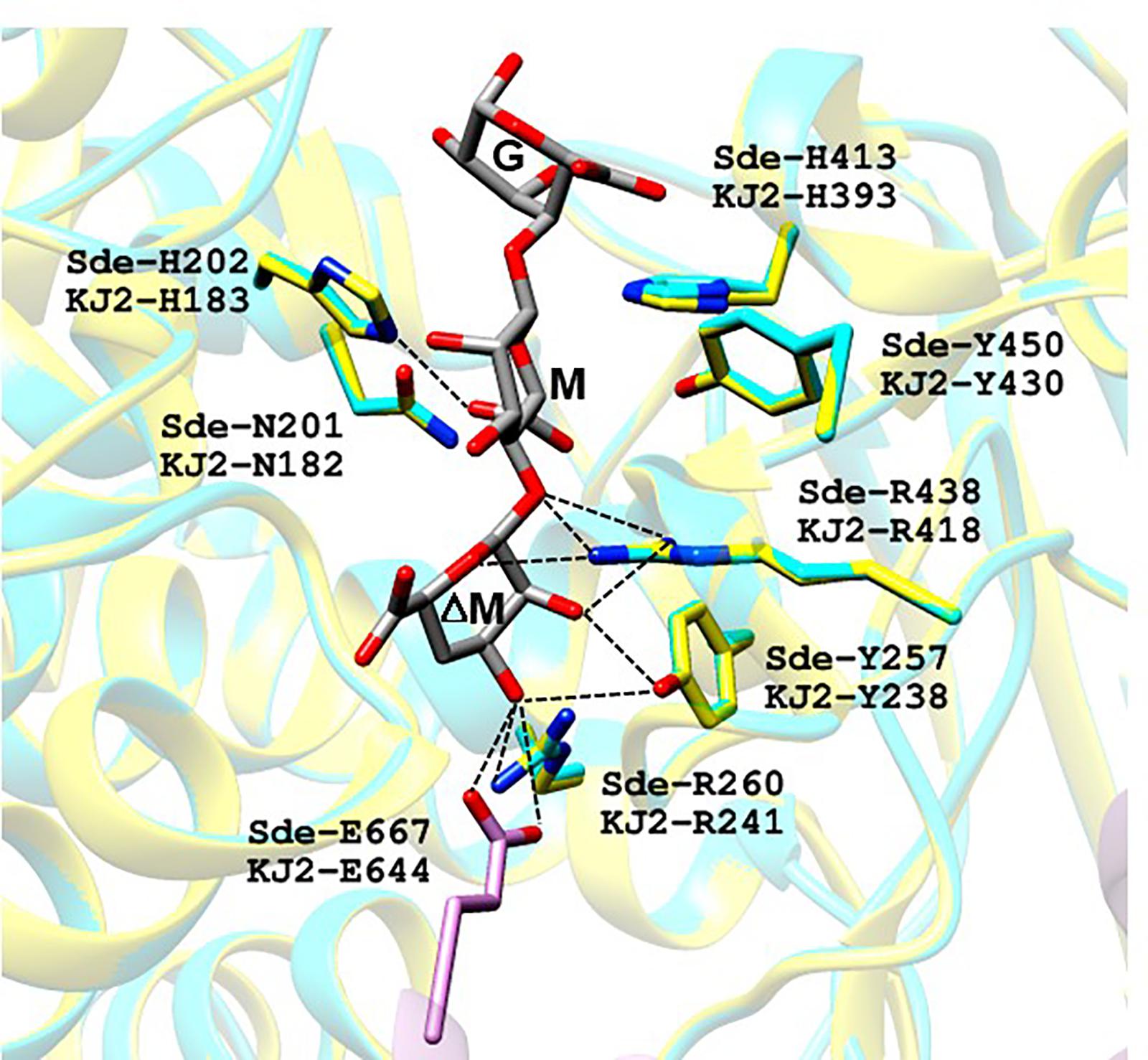
Figure 2. Proposed substrate binding sites of oligoalginate lyases from Stenotrophomonas maltophilia KJ-2. KJ-2 oligoalginate lyase (GenBank Accession No. AGM38186) was homology-modeled based on the crystal structure of Saccharophagus degradans oligoalginate lyase Alg17c (4OJZ.pdb). ΔM, M, and G of trisaccharide indicate unsaturated β-D-mannuronate, α-D-mannuronate, and α-L-guluronate, respectively. KJ-2 oligoalginate lyase is represented as blue, while A chain, and B chain of template were shown as yellow and pink, respectively. Active site residues of both the homology model and template were superimposed well each other. The lable Sde-R260 and KJ-2 R241 means arginine residue at 260th and 241st of template and KJ-2 oligoalginate lyase, respectively. Hydrogen bonding between key amino acids and substrate are shown as dotted lines.
Oligoalginate lyase is an enzyme that finally produces monomers via the β-elimination reaction of glycosidic bond between unsaturated non-reducing uronate and next uronate. The crystal structure of mutant Alg17c (4OJZ.pdb) demonstrated that the bound ΔMMG trisaccharide occupied subsites -1, +1, and +2 with the non-reducing mannuronate occupying the deep cleft of the binding pocket, and then Oal cleaved the bond between -1 (ΔM) and +1 (M) (Park et al., 2014). Analysis of the KJ-2 Oal structure model showed that Arg241 (NH2) and Glu644 (OE1 and OE2 of Glu644 in another chain) were located within the hydrogen bonding distance of O3 (2.9 Å) and O3 (2.8 and 3.2 Å) of non-reducing sugar (ΔM), respectively. Tyr238 (OH) was located adjacent to O2 (2.9 Å) and O3 (3.4 Å) of ΔM, while NH1 and NH2 of Arg418 were near to O2 (2.8 Å) and O5 (3.3 Å) of ΔM, respectively. Therefore, Tyr238, Arg241, Arg418, and Glu644 were predicted to locate within hydrogen bonding distance of the non-reducing end of alginate and were identified as important amino acids in mutational characterization to eliminate exolytic activity and to recognize inherent endolytic lyase activity.
Analysis of Expression and Activity of KJ-2 Oal Mutants Using SDS-PAGE and TBA Method
We performed site-specific mutagenesis for Tyr238Phe, Arg241Ala, Arg418Ala, Glu644Ala, and Arg241Ala_Glu644Ala. The recombinant proteins of KJ-2 Oal mutants were overexpressed and analyzed by 10% SDS-PAGE electrophoresis. The recombinant wild-type and mutated proteins were over expressed (Figure 3A) and purified (Supplementary Figure S1). When 5 μg protein of each recombinant lysate was reacted with 0.2% alginate solution, the wild-type enzyme showed the highest activity, but the Arg418Ala mutant lost almost all alginate degradation activity, as shown in Figure 3B. In comparison to the wild-type KJ-2 Oal, Tyr238Phe, Arg241Ala, Glu644Ala, and Arg241Ala_Glu644Ala double mutant showed decreased activity corresponding to 8.1, 43.1, 77.6 and 61.5% after a 60-min incubation, respectively. Data obtained from kinetic characterization of wild and mutant enzymes are represented in Supplementary Table S1.
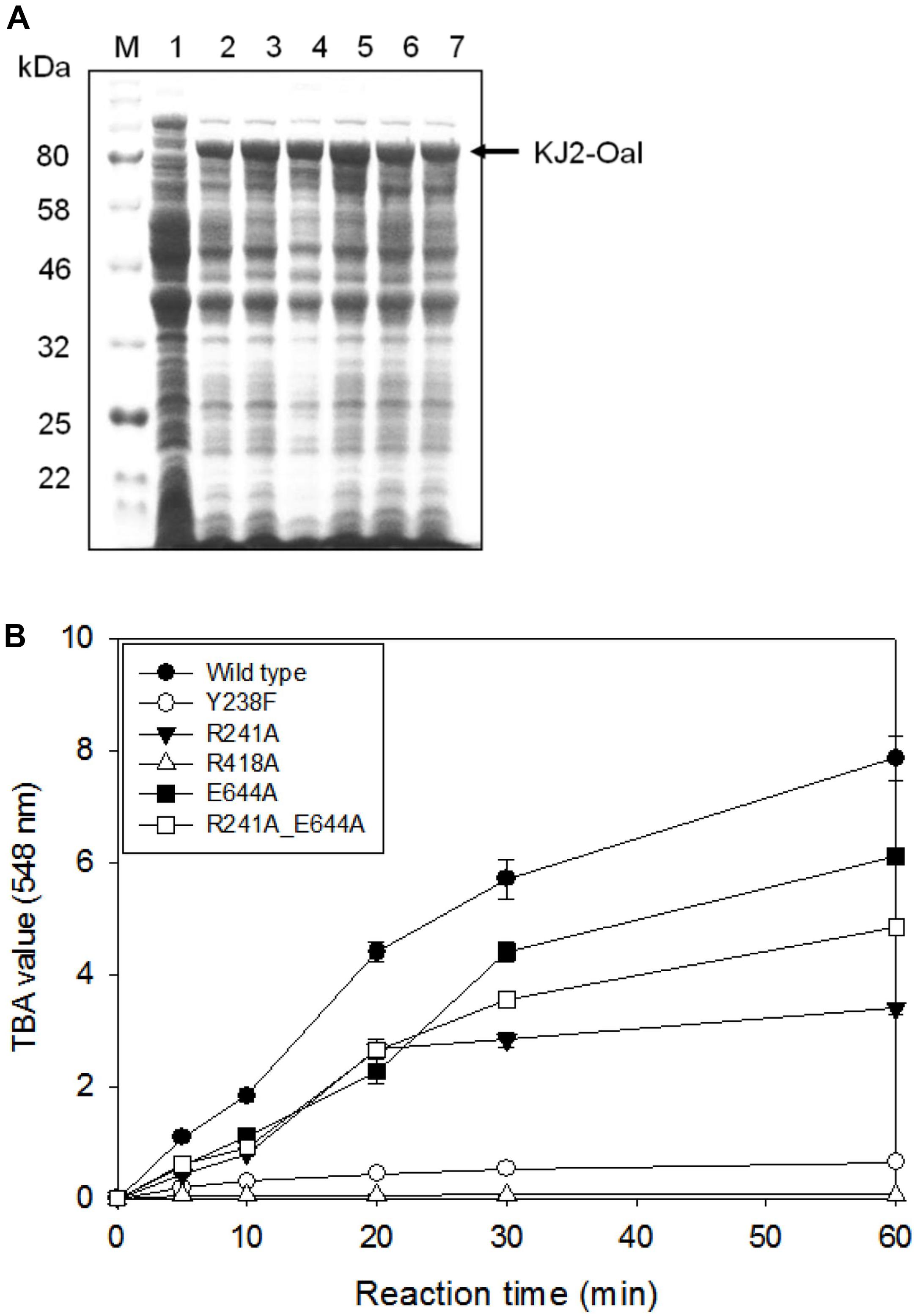
Figure 3. SDS-PAGE analysis and alginate lyase activity of cell lysates harboring KJ-2 oligoalginate lyase mutants. Recombinant proteins were overexpressed by inducing with 1 mM IPTG for 20 h at 15°C and cell pellets were lysed by adding BugBuster solution containing lysonase. (A) 10% SDS-PAGE. Lane 1–7 are cell lysates containing pColdI vector, wild type, Tyr238Phe, Arg241Ala, Arg418Ala, Glu644Ala, and double mutant (Arg241Ala_Glu644Ala), respectively. (B) Alginate lyase activity. Each 5 μg protein of cell lysate was incubated with 0.2% alginate solution for indicated time. Enzyme activity was determined by TBA method.
Analysis of FPLC Profiles and Molecular Mass of Alginate Degradation Products Generated by KJ-2 Oal Mutants to Confirm the Existence of Endolytic Activity
For the analysis of alginate digestion profile, overexpressed recombinant proteins were purified using a Ni-Sepharose affinity column, and then incubated with 0.2% alginate solution. The alginate degradation products in the reaction mixture were analyzed time-dependently by FPLC. As shown in Figure 4, 50 μg of the purified wild-type KJ-2 Oal rapidly produced unsaturated monomers. This FPLC profile shows that KJ-2 Oal is an apparently exolytic alginate lyase that degrades alginate to unsaturated monomer as a final product.
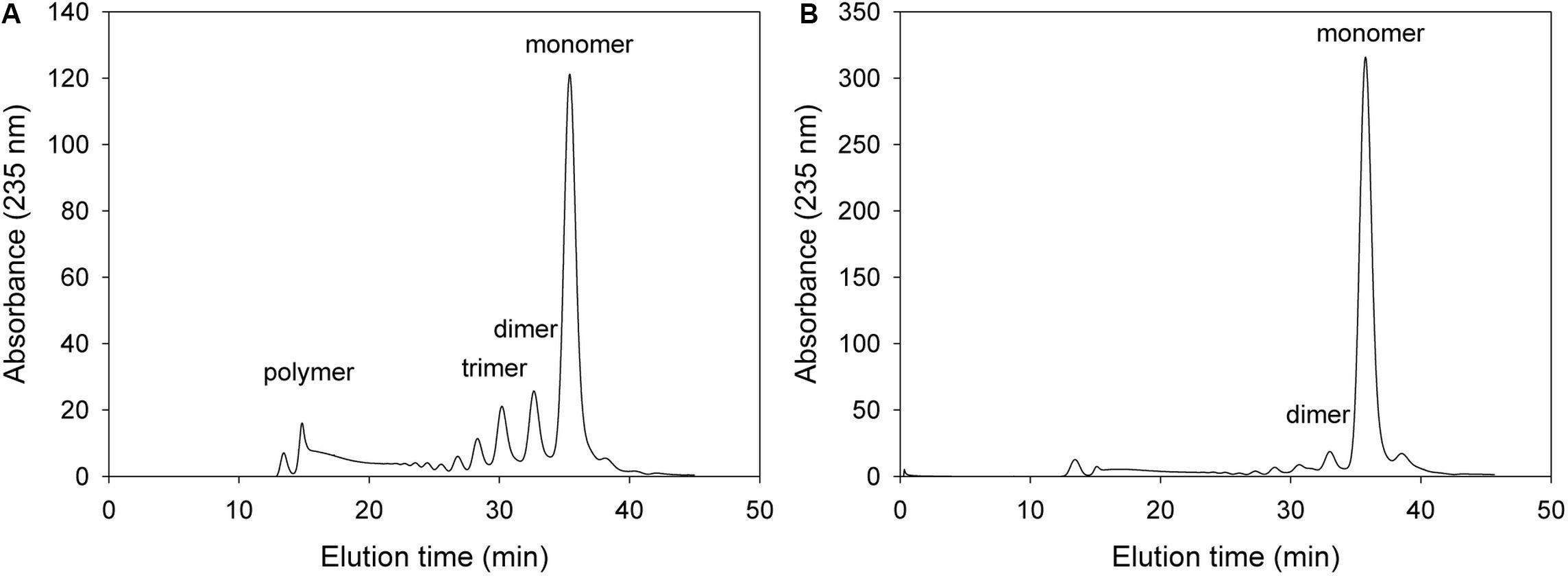
Figure 4. FPLC profile of time-dependent alginate degradation products digested by wild-type KJ-2 oligoalginate lyase. A 50 μg purified enzyme was reacted with 0.2% sodium alginate solution in total reaction volume 1 ml for 10 min (A) and 1 h (B), respectively. Monomer, dimer, trimer, and tetramer were detected at 35.7, 32.8, 30.6, and 35.7 min, respectively.
Firstly, we analyzed alginate degradation pattern for the mutant with changing amino acid that is expected to be not critical for lyase activity but helping exolytic degradation such as Tyr238. In the case of the Tyr238Phe mutant, this mutant initially produced unsaturated monomers and the major product was monomer after 1 h incubation (Figure 5). The FPLC profile of 1 h reaction mixture showed that replacement of phenylalanine instead of tyrosine did not remarkably affect the exolytic activity. Notably, Tyr238 was not an amino acid of the catalytic active site, but the Tyr238Phe mutant showed lower exolytic activity than wild type because the OH of Tyr238 was located within the hydrogen bonding distance of the non-reducing sugar.
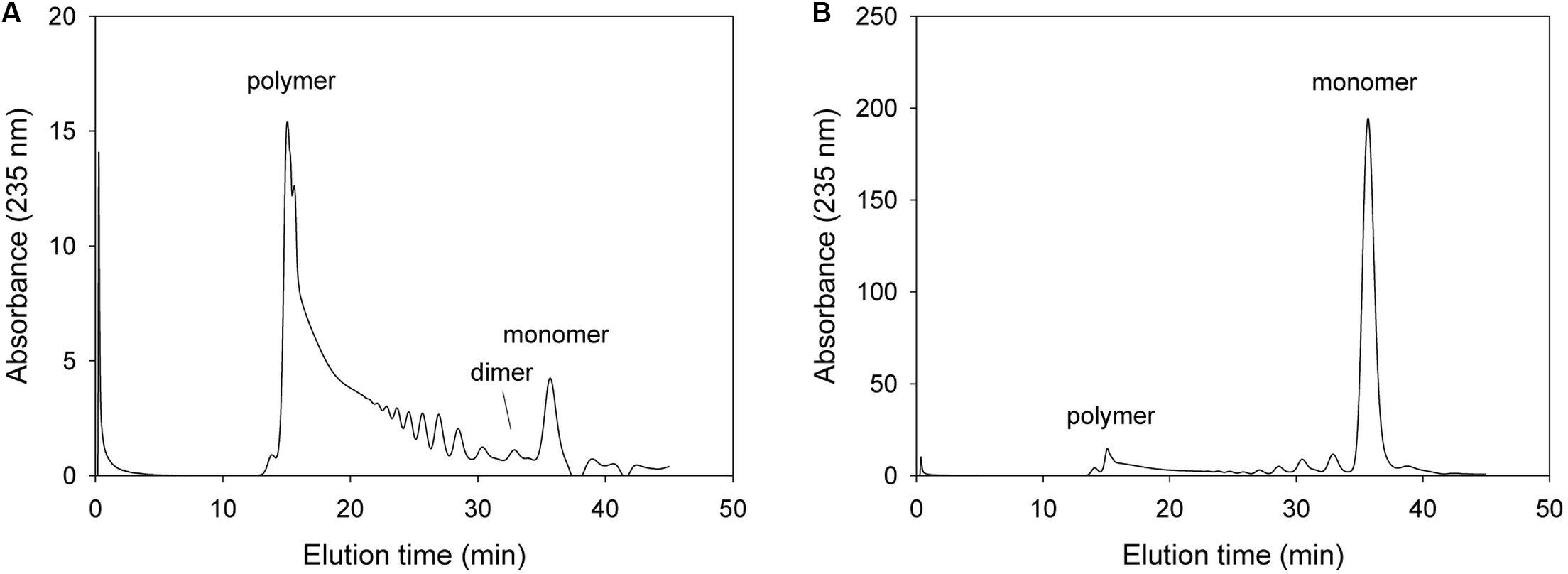
Figure 5. FPLC profile of time-dependent alginate degradation products digested by KJ-2 Tyr238Phe mutant. A 50 μg purified enzyme was reacted with 0.2% sodium alginate solution in total reaction volume 1 ml for 10 min (A) and 1 h (B), respectively. Monomer, dimer, trimer, and tetramer were detected at 35.7, 32.8, 30.6, and 35.7 min, respectively.
In subsequent mutation study, we determined the alginate degradation pattern of mutants with changing key amino acids that most probably play an important role in exolytic lyase activity, such as Arg241, Arg418, and Glu644. For these mutation study, we deduced that the mutations with key amino acid of exolytic lyase activity would result in decrease or removal of exolytic activity with showing inherent endolytic lyase activity of KJ-2 Oal. With respect to Arg241, the NH2 of Arg241 was predicted to interact with O3 (2.9 Å) of the non-reducing sugar (ΔM). When 50 μg of the purified Arg241Ala mutant protein was incubated with 0.2% alginate solution, the resulting FPLC profile was different from those of wild-type KJ-2 Oal and Tyr238Phe mutant (Figure 6). The mixture after a 10-min incubation showed many peaks indicating unsaturated oligomers, including pentamer, tetramer, trimer, and dimer that were more abundant than the monomer peak. This is first clue that KJ-2 Oal with Arg241Ala mutation exhibited inherent endolytic activity. The dimer peak was still higher than the other peaks even after the Arg241Ala mutant was incubated for 1 h. The absorbance of the monomer generated by the mutant exolytic activity was much lower than that generated by the wild type. These data showed that arginine at 241, which interacted with O3 of non-reducing end by hydrogen bonding, was an important residue during exolytic catalysis.
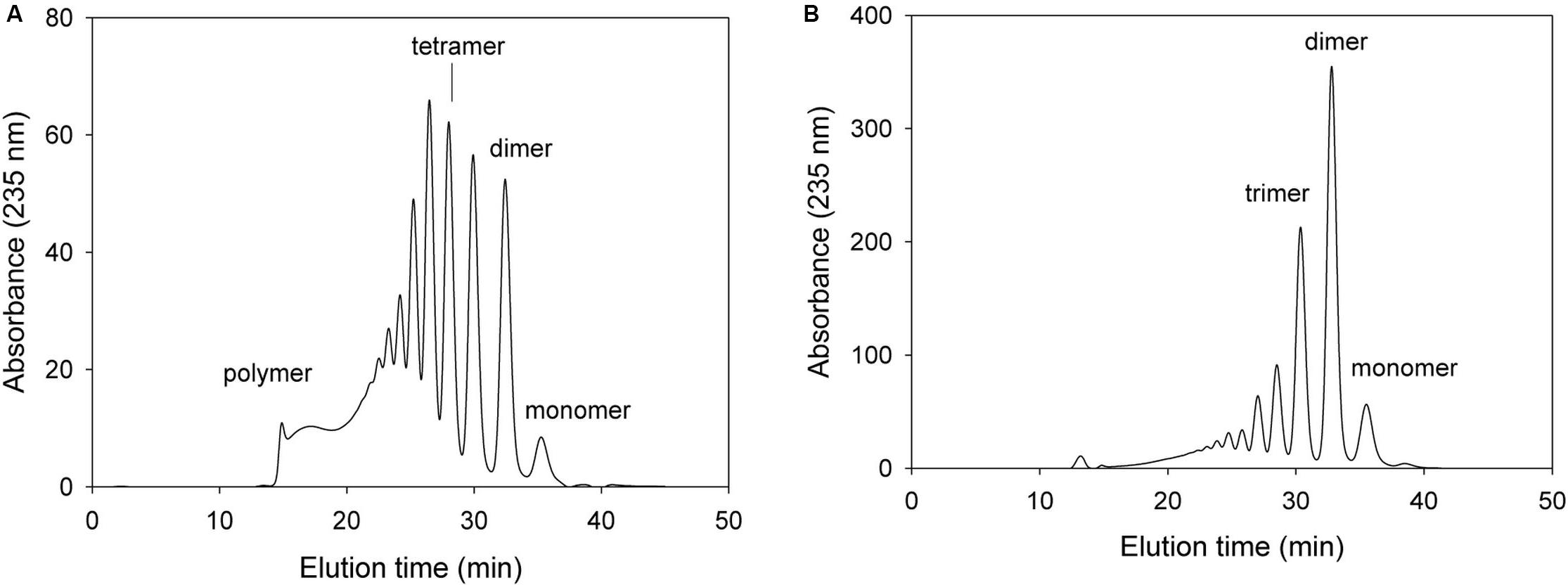
Figure 6. FPLC profile of time-dependent alginate degradation products digested by KJ-2 Arg241Ala mutant. A 50 μg purified enzyme was reacted with 0.2% sodium alginate solution in total reaction volume 1 ml for 10 min (A) and 1 h (B), respectively. Monomer, dimer, trimer, tetramer, and pentamer were detected at 35.7, 32.8, 30.6, 28.5, and 27.2 min, respectively.
The NH1 and NH2 of Arg418 was predicted to interact with O2 and O5 of ΔM, respectively, and also with O4 of M [O4 of β(1–4) glycosidic bond]. When 100 μg of the purified Arg418Ala mutant protein was incubated with 0.2% alginate solution, the absorbance of total degradation products including the monomer was very low (see the scale of Figure 7). Thus, we have demonstrated Arg418 as an important residue for exolytic degradation of alginate.
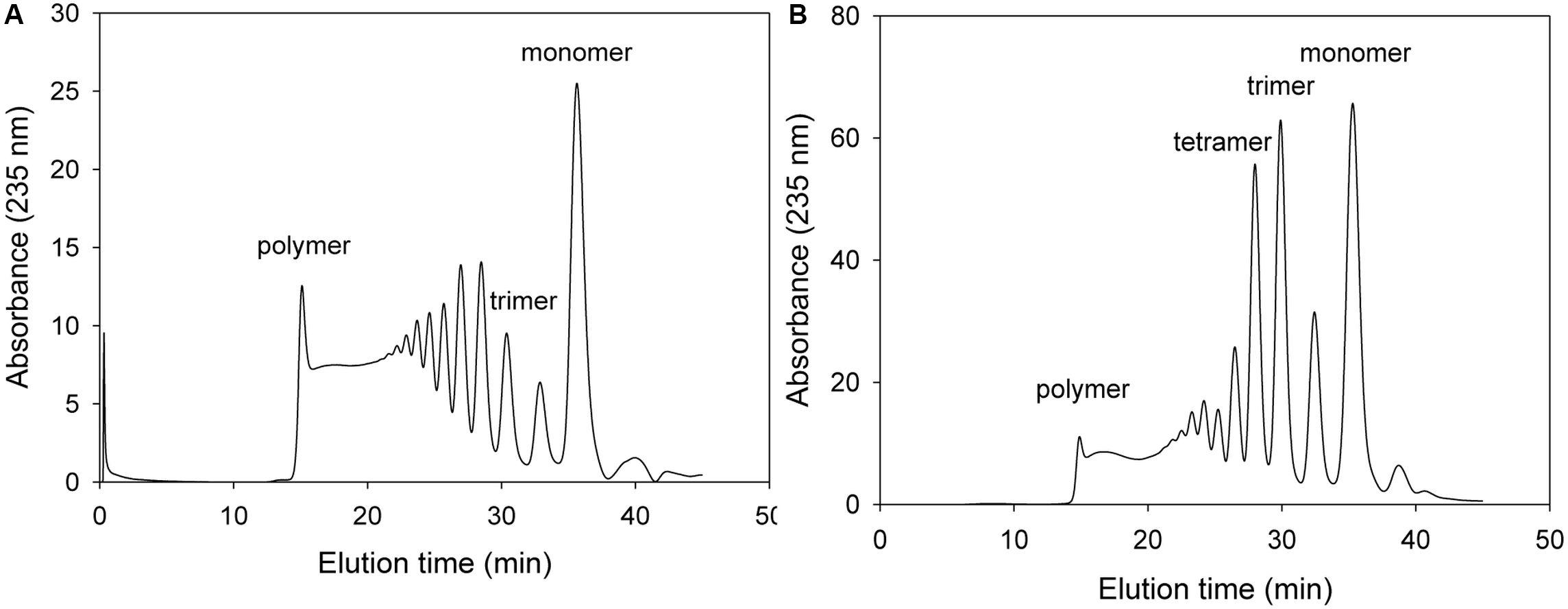
Figure 7. FPLC profile of time-dependent alginate degradation products digested by KJ-2 Arg418Ala mutant. A 100 μg purified enzyme was reacted with 0.2% sodium alginate solution in total reaction volume 1 ml for 10 min (A) and 1 h (B), respectively. Monomer, dimer, trimer, tetramer, and pentamer were detected at 35.7, 32.8, 30.6, 28.5, and 27.2 min, respectively.
Alg17c oligoalginate lyase from S. degradans 2–40 was composed of a homodimer, and dimerization occurred via insertion of an antiparallel β-turn, encompassing residues from Gly660 through Ala672, from the C-terminal β-sheet domain on one monomer into the α6/α6-barrel of the other monomer (Park et al., 2014). When KJ-2 Oal was homology-modeled on the Alg17c co-crystal structure, an antiparallel β-turn (Gly637 through Ala649) was expected adjacent to the α6/α6-barrel of the other chain. The carboxylate (OE1 and OE2) of Glu644 in the B chain (pink color) was predicted to interact with O3 of ΔM in the A chain (yellow color), as shown in Figure 2. Interestingly, when the Glu644Ala mutant of KJ-2 Oal was incubated with 0.2% alginate solution, the FPLC chromatogram showed very low exolytic activity, but high endolytic activity (Figure 8). When the reaction was prolonged to 7 h, the dimer peak of 7 h incubation was higher than that of 1 h, but the monomer peak after 7 h was similar to that of 1 h (data not shown). This clearly supports that KJ-2 Oal possesses inherent endolytic activity. Similarly, the Arg241Ala_Glu644Ala double mutant of KJ-2 Oal also produced large amounts of oligomers (data not shown for the double mutant), which also indicates that KJ-2 Oal possesses endolytic activity. Based on these data, we concluded that KJ-2 Oal possesses inherent endolytic activities together with an exolytic activity. Furthermore, Arg241, and Glu644 of KJ-2 Oal were proven to be important residues for exolytic activity of KJ-2 Oal.
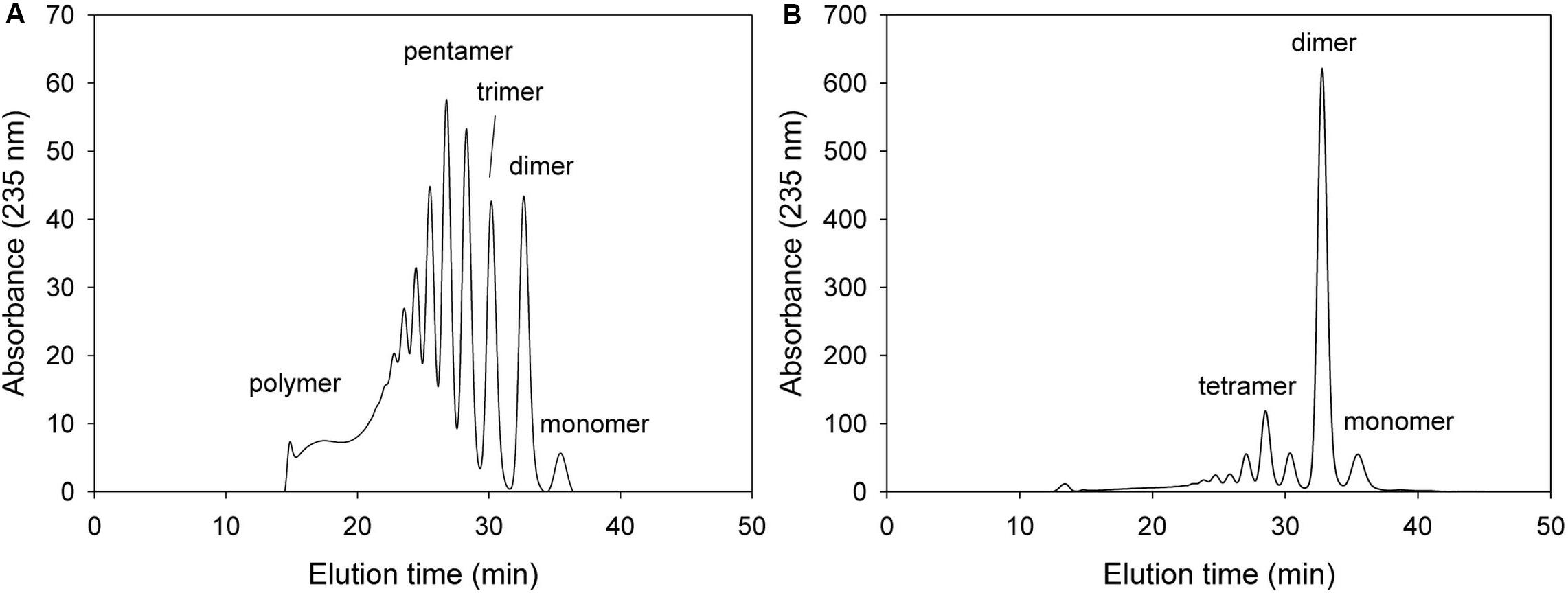
Figure 8. FPLC profile of time-dependent alginate degradation products digested by KJ-2 Glu644Ala mutant. A 50 μg purified enzyme was reacted with 0.2% sodium alginate solution in total reaction volume 1 ml for 10 min (A) and 1 h (B), respectively. Monomer, dimer, trimer, tetramer, and pentamer were detected at 35.7, 32.8, 30.6, 28.5, and 27.2 ml, respectively.
Figure 9 shows the analytical results on alginate degradation products digested by wild-type KJ-2 Oal and Glu644Ala mutant. The degradation products were analyzed by FPLC, LC-MS, and UV/Vis. Herein, we found that the intensity of monomer and dimer via FPLC at 220 nm and 235 nm were quite different from each other (Figures 9A,B). The reason why this discrepancy occurs can be explained by Figure 9E. The dotted and solid lines of Figures 9A,B represent absorbance curves for monomer and dimer, respectively. The absorbance at 235 nm of monomer was lower than that at 220 nm, while the absorbance values at 235 and 220 nm were almost identical for the dimer and oligomer. Thus, FPLC analysis of the monomer could not be applied for quantification of monomer, but only indicates the presence of monomer in the degradation mixture. The intensities of monomer and dimer peaks via FPLC were not linearly proportional to those of LC-MS. Figures 9C,D shows the LC-MS profiles of alginate degradation products by wild-type KJ-2 Oal and Glu644Ala after a 1-h incubation (Figures 9C,D are from the same samples as Figures 9A,B, respectively). The reaction product of wild-type KJ-2 Oal was confirmed to be an unsaturated monomer (m/z = 175.0), which has a molecular weight of 176 Da (Kim et al., 2012). In the case of Glu644Ala, although the FPLC monomer peak was six-fold smaller than the dimer, the LC/MS monomer peak (m/z = 175.0) was almost 60% of the dimer peak (m/z = 351.0). These results also support the fact that FPLC analysis shows the presence of monomer in the degradation mixture, but not in a quantitative manner. Thus, FPLC analysis of alginate degradation products generated by Oal, such as KJ-2 Oal, should be used in qualitative analysis for the existence of a monomer, dimer, and oligomers.
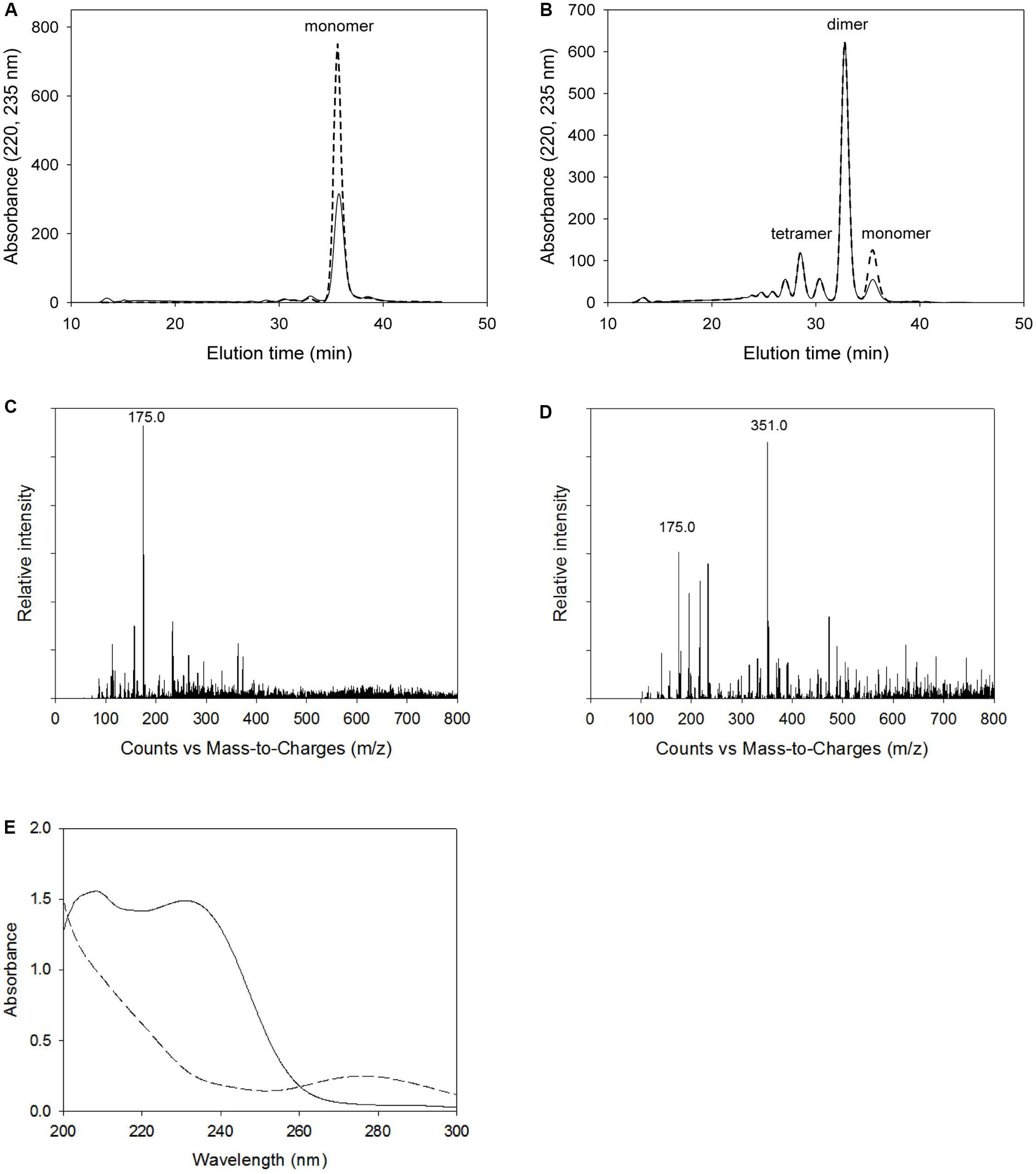
Figure 9. Analysis of alginate degradation products digested by wild-type KJ-2 oligoalginate lyase and Glu644Ala mutant. A 50 μg purified enzyme was reacted with 0.2% sodium alginate solution in total reaction volume 1 ml for 1 h. (A,B) FPLC chromatograms of degradation products by wild type and Glu644Ala mutant. Solid line and dashed line mean the absorbance at 235 nm and 220 nm, respectively. (C,D) LC/MS spectra of degradation products by wild type and Glu644Ala. 175 and 351 indicate m/z main peaks of monomer and dimer, respectively. (E) UV absorption spectra. Absorption spectrum of the alginate degradation products by Glu644Ala mutant (solid line) was compared to that of wild type (dashed line). The degradation products of same samples of (A) and (B) were diluted to five folds of for the determination of UV spectra.
Discussion
In this study, we characterized and proved the bifunctionality of the novel oligoalginate lyase of S. maltophilia KJ-2 possessing two conserved domains, an AlgL domain in the N-terminal region and a heparinase-like protein domain in the C-terminal region, by using site-specific mutagenesis (Pettersen et al., 2004; Kim et al., 2015; Shin et al., 2015). In previous studies, the AlgL domain showed endolytic alginate lyase activity, while the heparinase II/III domain showed relatively more exolytic alginate lyase activity (Park et al., 2012; Shin et al., 2015). However, these studies did not analyze the role of key amino acid residues.
Previously, Park et al. (2014) reported that site-directed mutation in some residues of the exolytic oligoalginate lyase, Alg17c from S. degradans, affected exolytic activity. Among the mutants, Tyr258Ala and Tyr450Ala, which function as a general acid and base catalyst, respectively, completely lost its activity. However, in that study, due to using alginate trisaccharides as the substrate, only exolytic activity could be observed. Thus, we used alginate as the substrate instead of trisaccharides, and we mutated key residues of Tyr238, Arg241, Arg418, and Glu644 that were expected to be located within hydrogen bonding distance of the non-reducing end of alginate during exolytic catalysis (Ochiai et al., 2010; Jagtap et al., 2014). These key residues correspond to the template (Alg17c) residues (Tyr257, Arg260, Arg438, and Glu667, respectively) that were previously suggested to be involved in substrate binding (Park et al., 2014). The Arg241Ala and Arg418Ala mutants showed reduced activity compared with the wild-type, which corresponds to the result of Park et al. (2014) that the activities of Arg260Ala and Arg438Ala mutants of Alg17c were significantly decreased. Furthermore, the exolytic activities of Arg241Ala, Glu644Ala, and Arg241Ala_Glu644Ala mutants sharply decreased, indicating that Arg241 and Glu644 play a key role in the exolytic activity of KJ-2 Oal. However, these mutants generated much more oligomers, trimer, and dimer than the wild-type KJ-2 Oal on the basis of same reaction time, which indicates that those mutants have endolytic activity. Because we used alginate as the substrate in this study, the endolytic activity, which could not be observed due to large exolytic activity of wild-type KJ-2 Oal, was detected. As the endolytic activity was observed after removing the exolytic activity, wild-type KJ-2 Oal was proved to possess both intrinsic endolytic and exolytic activities. This is one of the most important results of this study.
We have an evidence demonstrating that the endolytic activity of wild-type KJ-2 Oal is not a novel activity caused by mutation, but an inherent activity of wild-type KJ-2 Oal. In our previous study, when Arg236 of Sphingomonas sp. MJ-3 oligoalginate lyase (MJ-3 Oal) was replaced with alanine, the overexpressed MJ-3 Oal protein was proteolytically cleaved into 32.0 and 47.6 kDa fragments. Each fragment possesses an AlgL domain in the N-terminal and a heparinase II/III-like domain in the C-terminal, which were expected to exhibit endolytic and exolytic activity, respectively. However, neither could degrade trimer into monomer, that is, both fragments showed low endolytic activity despite only 32.0 kDa fragment contained the AlgL domain (Park et al., 2012; Kim et al., 2015). These results indicated that both two domains possess an endolytic activity. Previously, the DNA fragments of the N-terminal AlgL domain (amino acid residues 1 to 282), and C-terminal heparinase II/III-like domain (amino acid residues 283 to 641) of the MJ-3 Oal were independently subcloned, expressed and purified, and then reacted with alginate. When the reaction products were analyzed by LC-MS, both fragments demonstrated a strong dimer peak (m/z = 351, intensity = 12400–3800), while the monomer peak (m/z = 175, intensity = 300–1000) was very low (data not shown). These results also support that both fragments possess an endolytic activity.
In another previous study, we subcloned the AlgL-like or heparinase II/III-like domain of the KJ-2 Oal, and then expressed the recombinant protein for comparison with the wild-type KJ-2 Oal (Shin et al., 2015). In contrast to the MJ-3 Oal, the AlgL-like recombinant protein and heparinase II/III-like recombinant protein exhibited endolytic and exolytic activities, respectively. Therefore, it seems that endolytic and exolytic activity exist separately in each fragment. However, according to the results of the present study, the Arg241Ala mutant, which contains the AlgL domain residue and was expected to exhibit endolytic activity, showed reduced exolytic activity (Supplementary Figure S2). Consequently, we identified that the endolytic activity of KJ-2 Oal existed not only in the AlgL-like domain, but also together with the heparinase II/III-like domain. That is, from the results in this study, we confirmed that KJ-2 Oal has inherent endolytic activity based on site-directed mutagenesis.
Besides the key residues mentioned above, even though we did not check other residue mutations, we can obtain some information about those residues from several references. In Figure 2, His183 and Tyr430 residues, which correspond to the template (Alg17c) residues His202 and Tyr450, respectively, are related to reaction chemistry (Park et al., 2014). Asn182, His183, His393, and Tyr430, which correspond to the MJ-3 Oal residues Asn177, His178, His389, and Tyr426, respectively, are involved in active sites (Park et al., 2012; Kim et al., 2015). However, although these residue mutants showed a decreased activity, alginate was entirely degraded into monomer upon prolonged reaction time. Among those residues, His389 and Tyr 426 of MJ-3 Oal are mentioned as key amino acids, which are involved in binding site of unsaturated heparin dimer (Shaya et al., 2006; Park et al., 2012).
Two important issues have not been addressed. First, other important residues for maintaining exolytic activity may exist, in addition to Arg241 and Glu644. This is because only the monomer was generated when large amounts of mutant enzymes with low exolytic activity, such as Arg241Ala and Glu644Ala mutants, were used for extended reaction time. In addition, it is unclear what amino acids in KJ-2 Oal are involved in endolytic catalysis of KJ-2 Oal. These important issues will be investigated in near future.
In conclusion, functional characterization of KJ-2 Oal using site-specific mutation revealed a bifunctional mode of action possessing both endolytic and exolytic degradation activity. Recruitment of a novel bifunctional alginate lyase, such as KJ-2 Oal, will offer more efficient alginate degradation without the need for many alginate lyases with endolytic and exolytic activity.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
YC, HaK, and MK conducted experiments and analysis. EL and HeK coordinated the study and finalized the manuscript. All authors contributed to improve the manuscript and approved the submission.
Funding
This study was funded by Ministry of Oceans and Fisheries, Republic of Korea. This research was also supported by the C1 Gas Refinery Program through the National Research Foundation of Korea (NRF) and funded by the Ministry of Science and ICT (2015M3D3A1A01064882).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00420/full#supplementary-material
References
Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22, 195–201. doi: 10.1093/bioinformatics/bti770
Enquist-Newman, M., Faust, A. M., Bravo, D. D., Santos, C. N., Raisner, R. M., Hanel, A., et al. (2014). Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature 505, 239–243. doi: 10.1038/nature12771
Hashimoto, W., Miyake, O., Momma, K., Kawai, S., and Murata, K. (2000). Molecular identification of oligoalginate lyase of Sphingomonas sp. Strain A1 as one of the enzymes required for complete depolymerization of alginate. J. Bacteriol. 182, 4572–4577. doi: 10.1128/jb.182.16.4572-4577.2000
Hashimoto, W., Miyake, O., Ochiai, A., and Murata, K. (2005). Molecular identification of Sphingomonas sp. A1 alginate lyase (A1-IV’) as a member of novel polysaccharide lyase family 15 and implications in alginate lyase evolution. J. Biosci. Bioeng. 99, 48–54. doi: 10.1263/jbb.99.48
Jagtap, S. S., Hehemann, J. H., Polz, M. F., Lee, J. K., and Zhao, H. (2014). Comparative biochemical characterization of three exolytic oligoalginate lyases from Vibrio splendidus reveals complementary substrate scope, temperature, and pH adaptations. Appl. Environ. Microbiol. 80, 4207–4214. doi: 10.1128/AEM.01285-14
Kim, H., Chung, J., Wang, D., Lee, J., Woo, H., Choi, I., et al. (2012). Depolymerization of alginate into a monomeric sugar acid using Alg17C, an exo-oligoalginate lyase cloned from Saccharophagus degradans 2-40. Appl. Microbiol. Biotech. 93, 2233–2239. doi: 10.1007/s00253-012-3882-x
Kim, H. S., Chu, Y. J., Park, C.-H., Lee, E. Y., and Kim, H. S. (2015). Site-directed mutagenesis-based functional analysis and characterization of endolytic lyase activity of N- and C-terminal domains of a novel oligoalginate lyase from Sphingomonas sp. MJ-3 possessing exolytic lyase activity in the intact enzyme. Mar. Biotechnol. 17, 782–792. doi: 10.1007/s10126-015-9658-4
Kim, H. S., Lee, C.-G., and Lee, E. Y. (2011). Alginate lyase: structure, property and application. Biotechnol. Bioprocess Eng. 16, 843–851. doi: 10.1007/s12257-011-0352-8
Konda, N. B. S. N. M., Singh, S., Blake, A., Simmons, B. A., and Klein-Marcuschamer, D. (2015). An investigation on the economic feasibility of macroalgae as a potential feedstock for biorefineries. Bioenerg. Res. 8, 1046–1056. doi: 10.1007/s12155-015-9594-1
Lee, S. I., Choi, S. H., Lee, E. Y., and Kim, H. S. (2012). Molecular cloning, purification, and characterization of a novel polyMG-specific alginate lyase responsible for alginate MG block degradation in Stenotrophomonas maltophilia KJ-2. Appl. Microbiol. Biotechnol. 95, 1643–1653. doi: 10.1007/s00253-012-4266-y
Miyake, O., Hashimoto, W., and Murata, K. (2003). An exotype alginate lyase in Sphingomonas sp. A1: overexpression in Escherichia coli, purification, and characterization of alginate lyase IV (A1-IV). Protein. Expr. Purif. 29, 3–41.
Ochiai, A., Hashimoto, W., and Murata, K. (2006). A biosystem for alginate metabolism in Agrobacterium tumefaciens strain C58: molecular identification of Atu3025 as an exotype family PL-15 alginate lyase. Res. Microbiol. 157, 642–649. doi: 10.1016/j.resmic.2006.02.006
Ochiai, A., Yamasaki, M., Mikami, B., Hashimoto, W., and Murata, K. (2010). Crystal structure of exotype alginate lyase Atu3025 from Agrobacterium tumefaciens. J. Biol. Chem. 285, 24519–24528. doi: 10.1074/jbc.M110.125450
Ota, A., Kawai, S., Oda, H., Iohara, K., and Murata, K. (2013). Production of ethanol from mannitol by the yeast strain Saccharomyces paradoxus NBRC 0259. J. Biosci. Bioeng. 116, 327–332. doi: 10.1016/j.jbiosc.2013.03.018
Park, D., Jagtap, S., and Nair, S. K. (2014). Structure of a PL17 family alginate lyase demonstrates functional similarities among exotype depolymerases. J. Biol. Chem. 289, 8645–8655. doi: 10.1074/jbc.M113.531111
Park, H. H., Kam, N., Lee, E. Y., and Kim, H. S. (2012). Cloning and characterization of a novel oligoalginate lyase from a newly isolated bacterium Sphingomonas sp, MJ-3. Mar. Biotechnol. 14, 189–202. doi: 10.1007/s10126-011-9402-7
Peng, C., Wang, Q., Lu, D., Han, W., and Li, F. (2018). A novel bifunctional endolytic alginate lyase with variable alginate-degrading modes and versatile monosaccharide-producing properties. Front. Microbiol. 9:167. doi: 10.3389/fmicb.2018.00167
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E., et al. (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Roesijadi, G., Jones, S. B., Snowden-Swan, L. J., and Zhu, Y. (2010). Macroalgae as a Biomass Feedstock: a Preliminary Analysis. The US Department of Energy under Contract DE-AC05–76RL01830. PNNL-19944. Richland, WA: Pacific Northwest National Laboratory.
Ryu, M., and Lee, E. Y. (2011). Saccharification of alginate by using exolytic oligoalginate lyase from marine bacterium Sphingomonas sp. MJ-3. J. Ind. Eng. Chem. 17, 853–858. doi: 10.1016/j.jiec.2011.08.001
Shaya, D., Tocilj, A., Li, Y., Myette, J., Venkataraman, G., Sasisekharan, R., et al. (2006). Crystal structure of heparinase II from Pedobacter heparinus and its complex with a disaccharide product. J. Biol. Chem. 281, 15525–15535. doi: 10.1074/jbc.m512055200
Shin, J. W., Lee, O. K., Park, H. H., Kim, H. S., and Lee, E. Y. (2015). Molecular characterization of a novel oligoalginate lyase consisting of AlgL and heparinase II/III-like domains from Stenotrophomonas maltophilia KJ-2 and its application to alginate saccharification. Kor. J. Chem. Eng. 32, 917–924. doi: 10.1007/s11814-014-0282-1
Song, M., Pham, H. D., Seon, J., and Woo, H. C. (2015). Marine brown algae: a conundrum answer for sustainable biofuels production. Renew. Sust. Energ. Rev. 50, 782–792. doi: 10.1016/j.rser.2015.05.021
Takase, R., Ochiai, A., Mikami, B., Hashimoto, W., and Murata, K. (2010). Molecular identification of unsaturated uronate reductase prerequisite for alginate metabolism in Sphingomonas sp, A1. Biochim. Biophys. Acta 1804, 1925–1936. doi: 10.1016/j.bbapap.2010.05.010
Takeda, H., Yoneyama, F., Kawai, S., Hashimoto, W., and Murata, K. (2011). Bioethanol production from marine biomass alginate by metabolically engineered bacteria. Energ. Environ. Sci. 4, 2575–2581.
Wargacki, A. J., Leonard, E., Win, M. N., Regitsky, D. D., Santos, C. N., Kim, P. B., et al. (2012). An engineered microbial platform for direct biofuel production from brown macroalgae. Science 335, 308–313. doi: 10.1126/science.1214547
Weissbach, A., and Hurwitz, J. (1959). The formation of 2-keto-3-deoxyheptonic acid in extracts of Escherichia. J. Biol. Chem. 234, 705–709.
Keywords: alginate, endolytic activity, exolytic activity, oligoalginate lyase, Stenotrophomonas maltophilia KJ-2
Citation: Chu YJ, Kim HS, Kim MS, Lee EY and Kim HS (2020) Functional Characterization of a Novel Oligoalginate Lyase of Stenotrophomonas maltophilia KJ-2 Using Site-Specific Mutation Reveals Bifunctional Mode of Action, Possessing Both Endolytic and Exolytic Degradation Activity Toward Alginate in Seaweed Biomass. Front. Mar. Sci. 7:420. doi: 10.3389/fmars.2020.00420
Received: 09 March 2020; Accepted: 13 May 2020;
Published: 03 June 2020.
Edited by:
Marlen Ines Vasquez, Cyprus University of Technology, CyprusReviewed by:
Sujit Jagtap, University of Illinois at Urbana-Champaign, United StatesLuigi Mandrich, Istituto di Biochimica delle Proteine (IBP), Italy
Copyright © 2020 Chu, Kim, Kim, Lee and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eun Yeol Lee, ZXVueWxlZUBraHUuYWMua3I=
 Yoo Jung Chu1
Yoo Jung Chu1 Eun Yeol Lee
Eun Yeol Lee