- 1Fisheries College, Jimei University, Xiamen, China
- 2Key Laboratory of Healthy Mariculture for the East China Sea, Ministry of Agriculture, Xiamen, China
- 3Fujian Engineering Research Center of Aquatic Breeding and Healthy Aquaculture, Xiamen, China
The worsening of soil salinization has become an important factor limiting crop yield and quality. Pyropia haitanensis grows in the intertidal zone and is highly salt tolerant. The response mechanism of organisms exposed to stress is linked to energy metabolism and protein processing. However, the effect of hypersaline stress on the energy metabolism and protein synthesis of the P. haitanensis thallus has not been reported. Therefore, the objective of this study was to apply data-independent acquisition (DIA) technology to analyze the differences in the changes to the abundance of proteins related to energy metabolism and protein processing in P. haitanensis thalli exposed to salinity stress (100%) and semilethal salinity (110%). The examination of energy metabolism in algae under hypersaline stress revealed the decreased abundance of phosphofructokinase inhibits the glycolytic pathway. However, increases in triosephosphate isomerase levels and the enhancement of the pentose phosphate pathway minimize this inhibitory effect, while also providing substrates for the citric acid cycle as well as energy for physiological and biochemical algal reactions under hypersaline stress conditions. Regarding the protein processing related to the endoplasmic reticulum, increases in the abundance of CNX, the transporter COP II, heat shock proteins, protein disulfide isomerase, and other proteins related to endoplasmic reticulum quality control will ensure proteins are synthesized in algae under hypersaline stress. Increases in the levels of endoplasmic reticulum-associated degradation recognition protein BiP and transporter Sec61 as well as the ubiquitination degradation system-related proteins will maintain the dynamic balance between protein folding and clearance in algae. Moreover, algae are tolerant to 100% salinity, but 110% salinity is semilethal to algae. The abundances of citrate synthase, 6-phosphoglucose dehydrogenase, protein disulfide isomerase, and other related proteins increased under 100% salinity stress, but not under 110% salinity stress. Accordingly, in response to 100% salinity stress, P. haitanensis thalli may induce metabolic activities to satisfy the requirements of physiological and biochemical reactions. Furthermore, protein processing and synthesis may be stabilized to ensure necessary stress reactions are completed. The results of this study provide data potentially useful for investigating plant stress resistance mechanisms and for cultivating new salt-tolerant varieties.
Introduction
The degree of soil salinization is increasing worldwide, which is adversely affecting crop quality and yield (Flowers, 2004; Hanin et al., 2016). Therefore, there is an urgent need for the development of new highly saline-tolerant crops, which requires a thorough characterization of the mechanism underlying plant salt tolerance. Pyropia haitanensis is an algal species that grows in the intertidal zone, where it is dehydrated and rehydrated at low and high tides, respectively (Davison and Pearson, 1996; Dittami et al., 2009; Blouin et al., 2011; Brawley et al., 2017). Under water deficit conditions, the water on the surface of the algae gradually evaporates, leading to hypersalinity at the algal surface, which is harmful. However, after rehydrating at high tide, P. haitanensis can return to its normal physiological state, implying P. haitanensis is highly tolerant to hypersalinity (Blouin et al., 2011; Lu et al., 2016). However, P. haitanensis is a relatively primitive multicellular algal species with a special evolutionary status. Accordingly, its stress response mechanism may be different from that of higher plants, unicellular green algae, or blue algae. Therefore, P. haitanensis has become an ideal material for studying the stress tolerance physiology of algae and the salt tolerance mechanism of plants (Blouin et al., 2011; Dittami et al., 2012; Brawley et al., 2017).
Previous studies revealed that the adverse effects of hypersaline conditions on plants can be grouped into the following three categories: osmotic stress, ion toxicity, and oxidative stress (Kirst, 1990; Zhu, 2016; Yang and Guo, 2018). Additionally, plants evolved multi-level response mechanisms and strategies during the long-term natural selection process. Studies have proven that plants can minimize the damage due to ion toxicity through ion transmembrane transport or regional isolation (Munns and Tester, 2008). Moreover, they maintain their osmotic pressure by inducing the synthesis of soluble osmotic substances and stimulate increases in antioxidant enzyme contents or activities to eliminate accumulated reactive oxygen species. The synthesis of secondary metabolites, plant hormone metabolism, and signal transduction pathways are also important for responses to salt stress (Yan et al., 2013; Tang et al., 2015). Some progress has been made regarding the characterization of the salt tolerance mechanism of macroalgae. An earlier investigation on the antioxidant system of Ulva fasciata indicated that exposure to hypersaline stress results in oxidized algae that are damaged because of the accumulation of H2O2, which is eliminated by the significant increase in glutathione reductase activity and glutathione content (Lu et al., 2006). A transcriptomics analysis by Wang et al. (2019) revealed that the P. haitanensis thallus under hypersaline stress is stabilized by accumulating osmotic substances and enhancing antioxidant enzyme activities (Wang et al., 2019). Chen et al. (2019) reported that under hypersaline conditions, P. haitanensis induces the Na+/H+ antiporter and stabilizes the membrane potential by activating the plasma membrane H+-ATPase, thereby maintaining the K+/Na+ homeostasis (Chen et al., 2019).
Research has also confirmed that abiotic stresses (e.g., drought, high temperatures, and salinity) can cause protein misfolding and unfolding (Liu and Howell, 2010). Heat shock proteins (HSPs) help mediate the folding of nascent peptide chains and the repair, degradation, and maintenance of proteins to stabilize intracellular homeostasis, and are important for the correct folding of proteins in organisms exposed to stress. Studies have demonstrated that under high-temperature stress, protein denaturation in P. haitanensis is prevented via the up-regulated expression of HSP70, thereby increasing its resistance to high temperatures (Shi et al., 2017). In a study of the P. haitanensis thallus response to hypersaline stress, many HSP-related genes exhibited up-regulated expression (Wang et al., 2019). Moreover, HSPs are important for endoplasmic reticulum quality control (ERQC), which refers to a system responsible for monitoring protein folding in the endoplasmic reticulum. Its function is to ensure that correctly folded proteins enter the Golgi apparatus. Incorrectly folded proteins remain in the endoplasmic reticulum to be refolded with the help of various molecular chaperones, including HSPs and protein disulfide isomerase, after which they are released from the endoplasmic reticulum. Alternatively, misfolded proteins are ubiquitinated for endoplasmic reticulum-associated degradation (ERAD) by the 26S proteasome to avoid endoplasmic reticulum stress due to the accumulation of misfolded or unfolded proteins, which is crucial for maintaining the steady state of the endoplasmic reticulum (Hegde and Ploegh, 2010). Research on the salt tolerance mechanism of macroalgae has generally focused on the response mechanisms related to the ion balance, osmotic pressure regulation, and active oxygen scavenging. The response mechanism of algae under salt stress is inextricably linked to energy metabolism and protein processing. However, to the best of our knowledge, there are no reports describing the mechanism underlying algal responses to hypersaline stress specifically regarding energy metabolism and endoplasmic reticulum protein processing.
Proteins represent the biochemical machinery and functional processes enabling organisms to respond to changing environmental conditions. Therefore, a systemic analysis of proteins through proteomics-based research is an effective way to elucidate the adaptations of organisms to abiotic stresses. Xu et al. (2016) studied the proteome of P. haitanensis under water deficit stress using 2-DE technology, revealing that P. haitanensis mainly responds by inhibiting photosynthesis and energy metabolism (Xu et al., 2016). Shi et al. (2017) applied iTRAQ technology to study the response of P. haitanensis under high-temperature stress. Their results indicated that at high temperatures, P. haitanensis activates the antioxidant system, glycolytic pathway, molecular chaperones, and other related proteins to establish new stable and balanced metabolic processes (Shi et al., 2017). However, disadvantages of traditional proteomics technology include low protein recognition and inaccurate quantification, which limit its utility for comprehensive proteomics analyses (Schulze and Usadel, 2010). Data-independent acquisition (DIA) proteomics technology uses conventional data-dependent acquisition (DDA) mass spectrometry detection technology to analyze and establish an atlas database, after which a DIA method is used to collect mass spectrometry data, thus enabling qualitative and quantitative analyses of proteins in samples. Unlike traditional DDA technology, DIA technology can produce fragment information for all ions in a sample, which substantially increases the generated data, making it more suitable for detecting proteins in large samples and complex systems (Law and Lim, 2013). Therefore, in this study, we conducted a DIA-based proteomic analysis of the P. haitanensis thallus under diverse hypersaline stresses. We screened key regulatory proteins and explored the response mechanism related to energy metabolism and endoplasmic reticulum protein processing under hypersaline conditions. The resulting data may be relevant for future research on the molecular basis of P. haitanensis salt tolerance and the cultivation of new salt-tolerant plant varieties.
Materials and Methods
Algal Material and Hypersaline Stress Treatment
The P. haitanensis strain Z-61 analyzed in this study was isolated and purified by researchers in the Laboratory of Germplasm Improvements and Applications of Pyropia at Jimei University, Fujian Province, China (Chen et al., 2008; Xu et al., 2014). Every three blades were cultured in a 500-ml aerated flask containing natural seawater and Provasoli’s enrichment solution medium at 21 ± 1°C with a 12-h light: 12-h dark photoperiod (50–60 μmol m–2 s–1 photons). The medium was changed every 2 days. When the Z-61 blades grew to 20 ± 2 cm, they were exposed to hypersaline conditions induced by the addition of a NaCl solution prepared in natural seawater. In a previous study, we observed that P. haitanensis can acclimate to 100% hypersaline stress (HSS 100; approximately 1,700 mM Na+) conditions, whereas 110% hypersaline stress (HSS 110; approximately 1,900 mM Na+) conditions were semilethal for thalli, which can subsequently recover in normal seawater (control; approximately 500 mM Na+) (Chen et al., 2019). Therefore, we selected 100 and 110% as hypersaline stress conditions. The Z-61 blades were harvested after a 4 h salt treatment, immediately frozen in liquid nitrogen, and then stored at −80°C Each treatment was completed with three biological replicates.
Protein Extraction
For each sample, 500 mg was weighed and added to a 1.5-ml centrifuge tube, after which a 5-mm steel bead was added along with 250 μl Lysis Buffer 3 (7 M carbamide, 2 M thiocarbamide, 0.2% SDS, and 20 mM Tris, pH 8.0–8.5), PMSF (1 mM final concentration), and EDTA (2 mM final concentration). The sample was vortexed and then left undisturbed for 5 min before DTT (10 mM final concentration) was added. The solution was oscillated with a tissue grinder for 2 min (frequency = 50 Hz) and then centrifuged (25,000 × g for 20 min at 4°C). The supernatant was collected and after adding DTT (10 mM final concentration), the solution was incubated in a water bath at 56°C for 1 h. The sample was cooled to room temperature before iodoacetamide was added (55 mM final concentration). The resulting solution was incubated in darkness for 45 min, mixed with four volumes of cold acetone, and incubated at −20 °C for 2 h. The previous step was repeated (if necessary) until the supernatant became colorless. The solution was centrifuged (25,000 × g for 20 min at 4°C) and the supernatant was discarded. The pellet was resuspended in 250 μl Lysis Buffer 3. The solution was ultrasonicated to dissolve the precipitated proteins and then centrifuged (25,000 × g for 20 min at 4°C). The supernatant was collected to quantify the protein content.
Bradford Quantification
The protein standard solution (0.2 μg/μl BSA) was added sequentially to a 96-well plate (0, 2, 4, 6, 8, 10, 12, 14, 16, and 18 μl to wells A1 to A10, respectively). Pure water was then added (20, 18, 16, 14, 12, 10, 8, 6, 4, and 2 μl to wells A1 to A10, respectively) along with 180 μl (per well) MAS Bright Blue G-250 Quantitative Working Solution. The optical density at 595 nm (OD595) was measured with a microplate reader, and a linear standard curve was prepared based on the OD595 and protein concentration. The protein solution to be tested was diluted several times, after which 20 μl aliquots were mixed with 180 μl MAS Bright Blue G-250 Quantitative Working Solution and the OD595 was determined. The sample protein concentration was calculated based on the standard curve and the sample OD595.
Protein Enzymatic Hydrolysis
For each protein sample, 100 μg was diluted with 4 volumes of 50 mM NH4HCO3, after which 2.5 μg trypsin was added for a protein:enzyme ratio of 40:1. Proteins were digested at 37°C for 4 h. Trypsin (Mass Spectrometry grade, Beijing Hualishi Tech. Ltd., Beijing, China) was added again at the same ratio, and the digestion resumed at 37°C for 8 h. The resulting peptides were desalted with a Strata X column and vacuumed to dryness.
High pH RP Separation
Tryptic digest (10 μg) from all samples were combined, and the 200 μg mixture was diluted with 2 mL mobile phase A (5% ACN, pH 9.8) and injected into the Shimadzu LC-20AB HPLC system coupled with a Gemini high pH C18 column (5 μm, 4.6 × 250 mm). Specifically, the peptide sample was added to the column and then eluted at a flow rate of 1 ml min–1 with the following gradient: 5% mobile phase B (95% ACN, pH 9.8) for 10 min, 5 to 35% mobile phase B for 40 min, 35 to 95% mobile phase B for 1 min, mobile phase B for 3 min, and equilibration with 5% mobile phase B for 10 min. The elution peak was monitored at a wavelength of 214 nm and fractions were collected every minute. The collected fractions were combined for a total of 10 fractions, which were then lyophilized.
DDA and DIA Analysis by Nano-LC-MS/MS
The DDA and DIA sample analyses were completed with the Q Exactive HF X mass spectrometer (Thermo Fisher Scientific) coupled with the UltiMate 3000 RSLCnano system (Thermo Fisher Scientific). A nano-LC column (150 μm × 30 cm, 1.8 μm, and 100 Å) was packed in-house to separate peptides at a flow rate of 500 nl/min. For the DDA analysis, peptides were loaded onto a C18 trap column (300 μm × 5 mm and 5 μm; Thermo Fisher Scientific) with buffer A (2% ACN, 0.1% FA) for 5 min, then eluted with a gradient of 5 to 25% buffer B (98% ACN, 0.1% FA) for 155 min, 25 to 30% buffer B for 10 min, and 30 to 80% buffer B for 5 min. The mass spectrometry parameters were as follows: MS scan range 350-1,500 m/z; loop count 30; NCE 28; MS resolution 120,000, maximal injection time (MIT) 50 ms; MS/MS HCD scans with resolution 30,000, MIT 100 ms; dynamic exclusion duration 30 s; isolation window 2.0 m/z; intensity threshold 2.0e4; charge exclusion 1, 7, 8, and >8. For the DIA analysis, the nano-LC system and gradient were the same as those used for the DDA analysis. The DIA mass spectrometry parameters were as follows: full scan range 400–1,250 m/z at resolution 120,000 with MIT 50 ms; DIA isolation window was set to 17 m/z with a loop count of 50 and automatic MIT, scanned at resolution 30,000; step-wise NCE 22.5, 25, and 27.5; AGC target 1e6.
Data Analysis
DDA Data Analysis
MaxQuant1 (Choi et al., 2014) is a free protein identification and quantification software developed by the Max Planck Institutes for high-resolution MS data. This software was used to analyze the DDA data, which served as a spectrum library for the subsequent DIA analysis. The analysis used raw data as input files and set the corresponding parameters and databases for the subsequent identification and quantitative analysis. The identified peptides with a false discovery rate (FDR) ≤ 1% were used to construct the final spectral library.
DIA Data Analysis
The DIA data were analyzed with SpectronautTM (Bruderer et al., 2015), which uses the iRT peptides for calibrating retention times. Additionally, Spectronaut integrates the mProphet scoring algorithm, which accurately reflects the matching level of ion pairs. On the basis of the target-decoy model applicable to SWATH-MS, false positives were controlled with an FDR threshold of 1% to obtain significant quantitative data.
MSstats Differential Analysis
MSstats (Choi et al., 2014) is an R package from the Bioconductor repository. It can be used for evaluating the significant differences in proteins or peptides from diverse samples. Consequently, it has been widely applied for targeted proteomics multiple reaction monitoring (MRM), label-free quantification, and SWATH quantitative experiments. The core algorithm is a linear mixed effect model. The data were preprocessed according to the predefined comparison group, after which the significance test based on the model was completed. Thereafter, differentially abundant proteins were screened based on a fold change (FC) ≥ 1.5 and P < 0.05 as the criteria for determining significant differences. Additionally, the differentially abundant proteins underwent an enrichment analysis. A protein–protein interaction (PPI) network was constructed with the STRING2 and Cytoscape programs.
Validation of Differentially Abundant Proteins With a Multiple Reaction Monitoring Analysis
Samples were extracted and digested as described and then spiked with 50 fmol β-galactosidase to normalize the data. The MRM analyses were completed with a QTRAP 6500 mass spectrometer (SCIEX, Framingham, MA, United States) equipped with the LC-20 AD nano-HPLC system (Shimadzu, Kyoto, Japan). The generated raw data file was integrated with Skyline software. All transitions for each peptide were used for quantification unless interference from the matrix was detected. A sample spiked with β-galactosidase was used for label-free data normalization. We used MSstats with the linear mixed effect model, and the P values were adjusted to control the FDR at a threshold of 0.05. All proteins with at least a 1.5-fold difference in abundance (P < 0.05) were considered significant.
Determination of Organic Acid Contents
After adding 1 ml 80% methanol to 0.5 g P. haitanensis sample, the solution was mixed with magnetic beads and then ultrasonicated at a low temperature for 20 min. The solution was then centrifuged (12,000 rpm for 10 min at 4°C), after which 100 μl supernatant was dried. The dried material was resuspended and thoroughly mixed with 50 μl acetonitrile and 50 μl BSTFA derivatization reagent and then incubated at 80°C for 30 min. After shaking for 1 min, the sample was centrifuged (5,000 rpm for 2 min at 4°C). Finally, 50 μl supernatant was added to a brown bottle and analyzed by GC-MS with the Agilent 7890B gas chromatograph and the Agilent 5977A mass selective detector in the serial SIM detection mode. The mass selective detector ChemStation software was used to process data, and the absolute target compound content in the sample was calculated.
Results
DIA Identification Results
In this study, 2,458 proteins were identified from 19,878 peptide fragments (Supplementary Tables S1–S3). To evaluate the reproducibility of the DIA data, a principal component analysis was conducted involving the three replicates for each salt stress treatment (Figure 1A). The degree of data dispersion between different salt treatments was obvious, with minor differences in the responses to salt stress. The correlations among protein quantities between samples were determined based on Pearson correlation coefficients, which are presented as a heat map (Figure 1B).
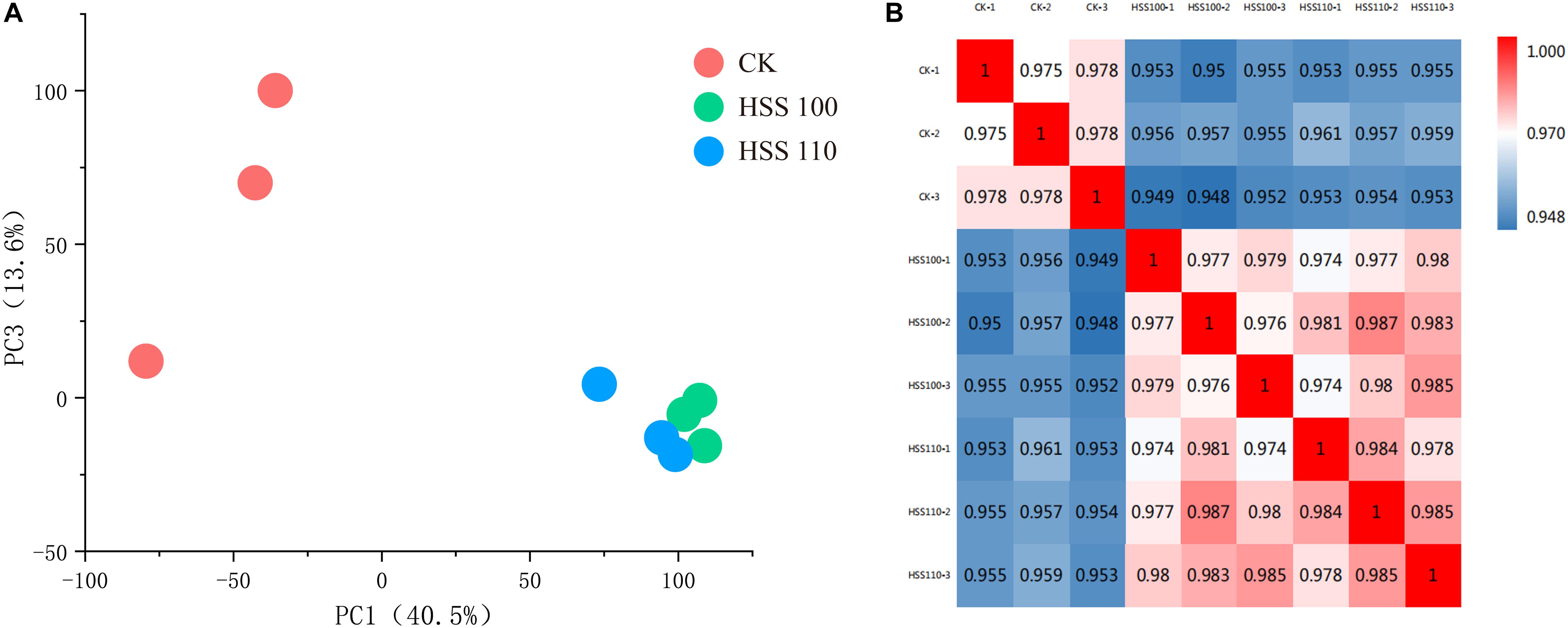
Figure 1. Results of the DIA analysis of P. haitanensis exposed to hypersaline stress for 4 h. (A) Principal component analysis of each sample. (B) Heat map presenting sample correlations.
Analysis of Differentially Abundant Proteins
On the basis of an FC ≥ 1.5 and P value < 0.05, a total of 1,105 differentially abundant proteins were screened, including 1,009 and 823 under the HSS 100 and 110 conditions, respectively (Figure 2A). A subsequent analysis of the differentially abundant proteins under salt stress conditions indicated the abundance of 701 and 140 proteins increased and decreased, respectively, under both stresses (Figures 2B,C). Compared with the effects of the HSS 110 treatment, the HSS 100 condition resulted in more proteins with increased abundance and fewer proteins with decreased abundance. Additionally, the 10 proteins that were the most differentially abundant between the two treatments were statistically analyzed, which revealed the proteins with the largest increases (i.e., hedgehog, proteasome component ECM29, and translation initiation factor 3 subunit C) and decreases (i.e., associated with light-harvesting complex I chlorophyll a/b binding protein 1) in abundance (Table 1).
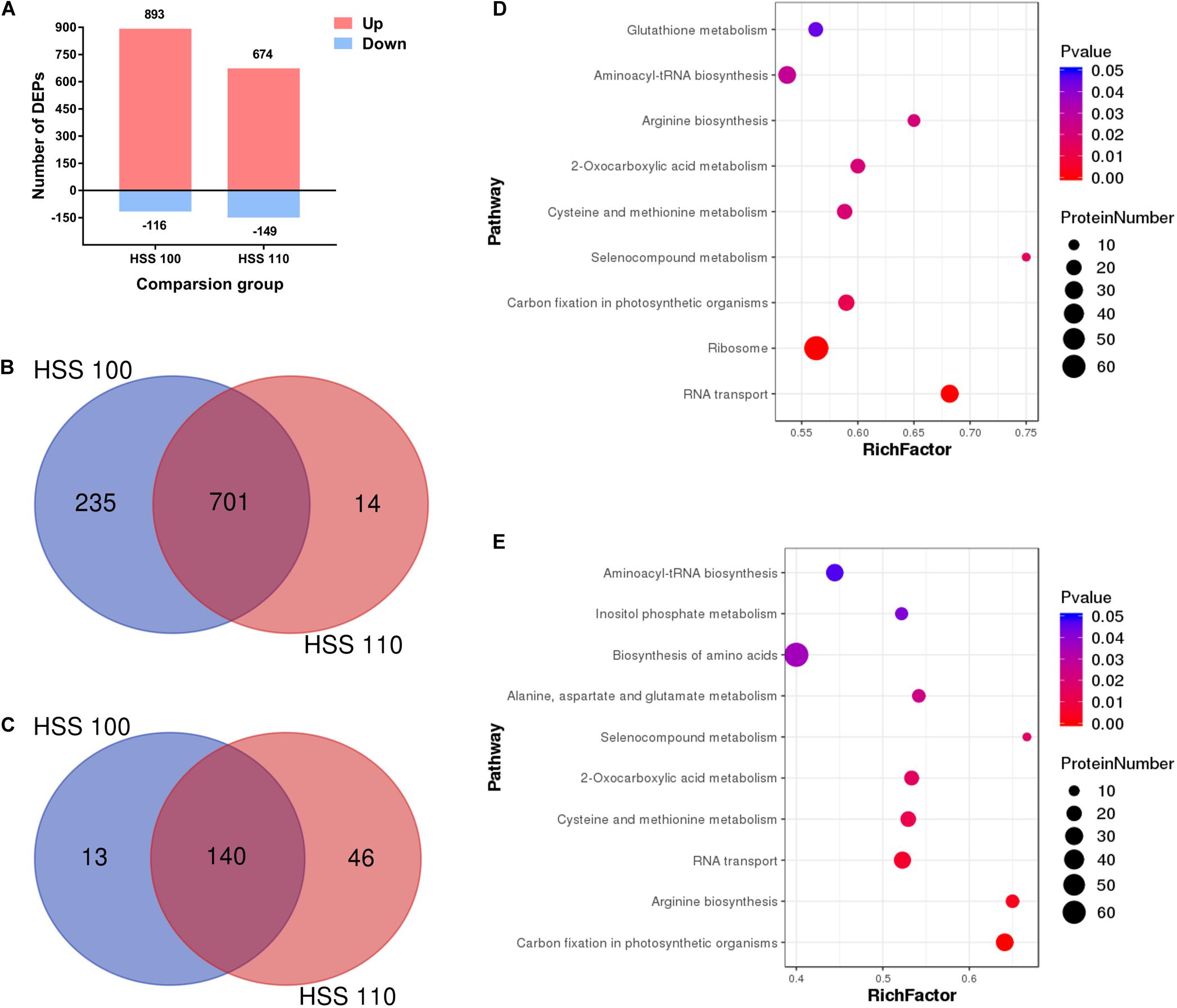
Figure 2. Analysis of differentially abundant proteins in P. haitanensis exposed to hypersaline stress for 4 h. (A) Number of differentially abundant proteins in each comparison. (B) Venn diagram of differentially abundant proteins (increased abundance). (C) Venn diagram of differentially abundant proteins (decreased abundance). (D) KEGG classification of differentially abundant proteins under HSS 100 conditions. (E) KEGG classification of differentially abundant proteins under HSS 110 conditions. FC ≥ 1.5 and P < 0.05.
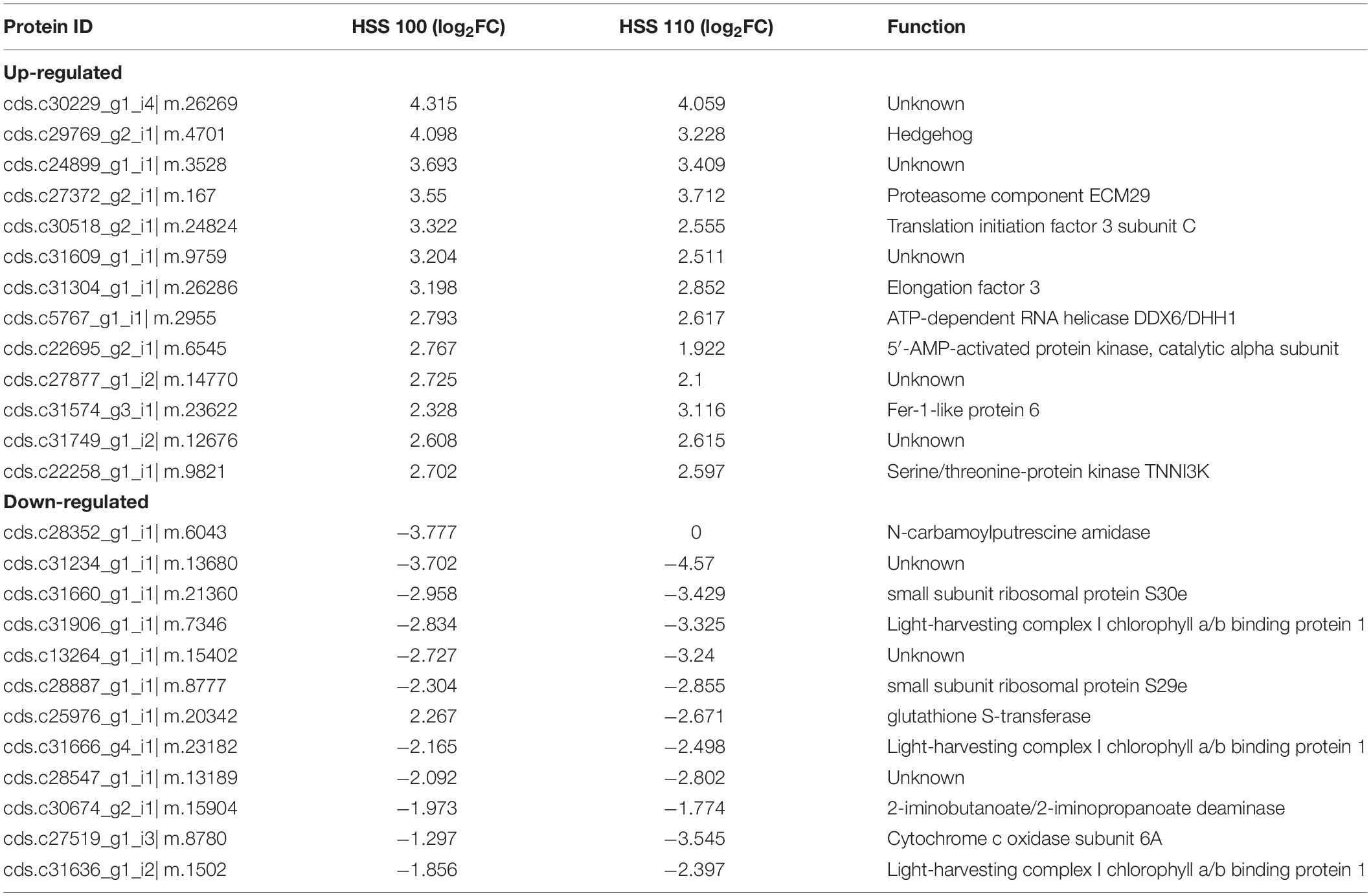
Table 1. Ten most differentially abundant proteins with increased and decreased abundances under HSS 100 and 110 conditions.
To further analyze the key metabolic pathways of P. haitanensis thalli in response to hypersaline stress, proteins differentially abundant under HSS 100 and 110 conditions were analyzed by identifying the associated enhanced KEGG pathways. The differentially abundant proteins under HSS 100 were mainly related to the ribosome, RNA transport, and carbon fixation in photosynthetic organisms (Figures 2D,E), whereas the differentially abundant proteins under HSS 110 were primarily associated with carbon fixation in photosynthetic organisms, RNA transport, and arginine biosynthesis.
Verification of Differentially Abundant Proteins
Twenty differentially abundant proteins were randomly selected from the sequenced proteome database, and their abundance under different hypersaline stress conditions was determined with the MRM method to verify the accuracy of the DIA results (Supplementary Tables S4, S5). The results indicated that the MRM abundance trend was essentially consistent with the DIA results, implying the DIA data were highly accurate.
Effects of Hypersaline Stress on Energy Metabolism
A total of 21 of the differentially abundant proteins in P. haitanensis thalli under hypersaline conditions were associated with the glycolytic (EMP) pathway (Figure 3). For example, the phosphofructokinase and fructose-bisphosphate aldolase levels increased in response to both salt stress treatments. In contrast, the abundances of triosephosphate isomerase (TIM) and pyruvate kinase decreased under both salt stress conditions.
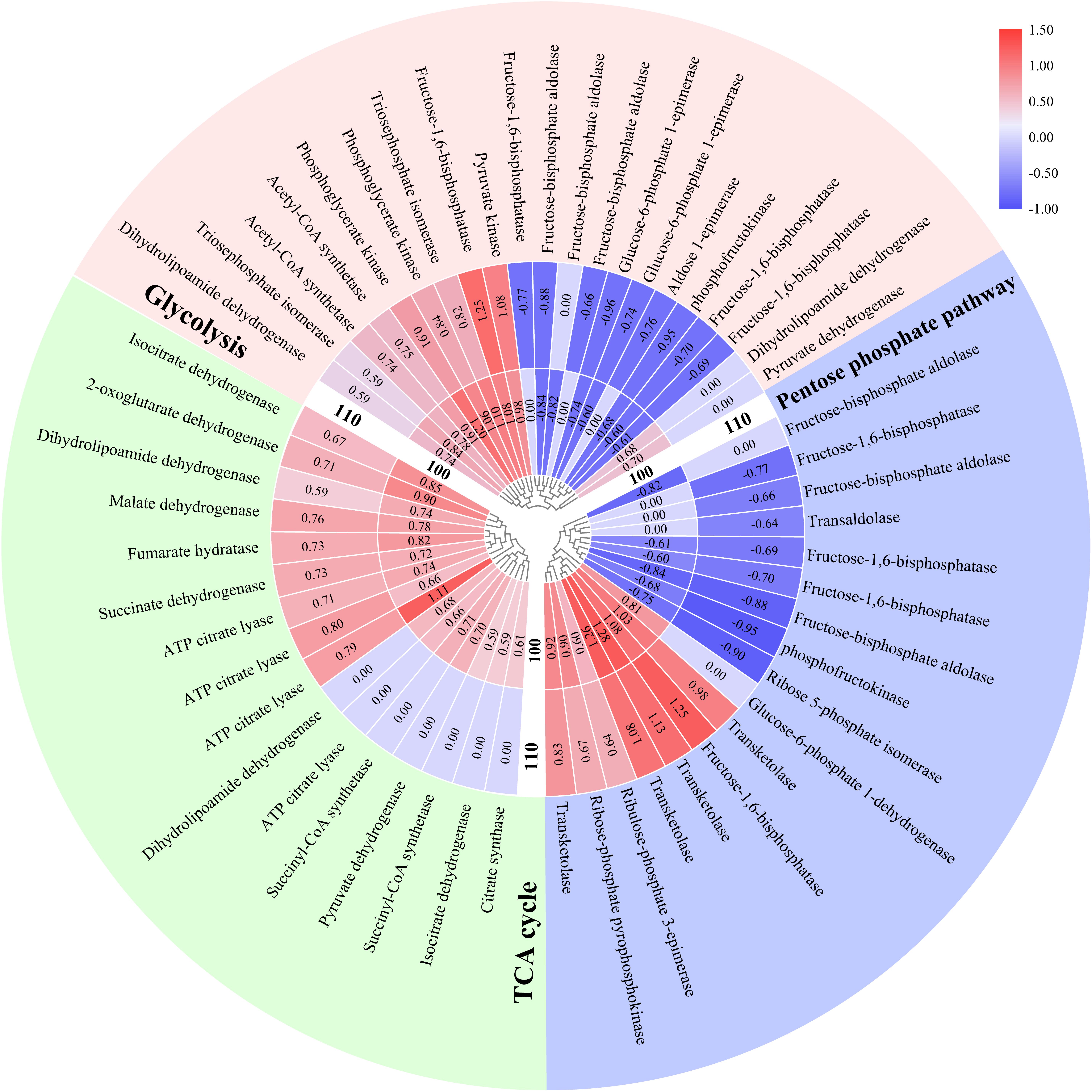
Figure 3. Effects of hypersaline stress on P. haitanensis energy metabolism-related pathways. The heat map was plotted based on log2 fold change (FC) values from the proteomics dataset. The inside and outside tracks represent the HSS 100 and 110 conditions, respectively. FC ≥ 1.5 and P < 0.05.
A total of 16 differentially abundant proteins in the P. haitanensis thalli under hypersaline stress were related to the TCA cycle, with both salt stress treatments increasing the protein abundances (Figure 3). Additionally, the abundances of citrate synthase and pyruvate dehydrogenase increased only in response to the HSS 100 conditions.
Seventeen differentially abundant proteins were associated with the pentose phosphate pathway (PPP) (Figure 3), including transketolase, whose abundance increased under both salt stresses. In contrast, the ribose 5-phosphate isomerase and fructose-1,6-bisphosphatase levels decreased following both salt stress treatments. However, the abundance of glucose-6-phosphate dehydrogenase increased only under HSS 100 conditions.
Effects of Hypersaline Stress on Protein Synthesis
In this study, the HSS 100 and 110 conditions increased the abundances of many molecular chaperones, such as HSP70 and protein disulfide isomerase (Figure 4A), which can help proteins fold correctly. The HSP70 and protein disulfide isomerase levels increased more under the HSS 100 conditions than under the HSS 110 conditions.
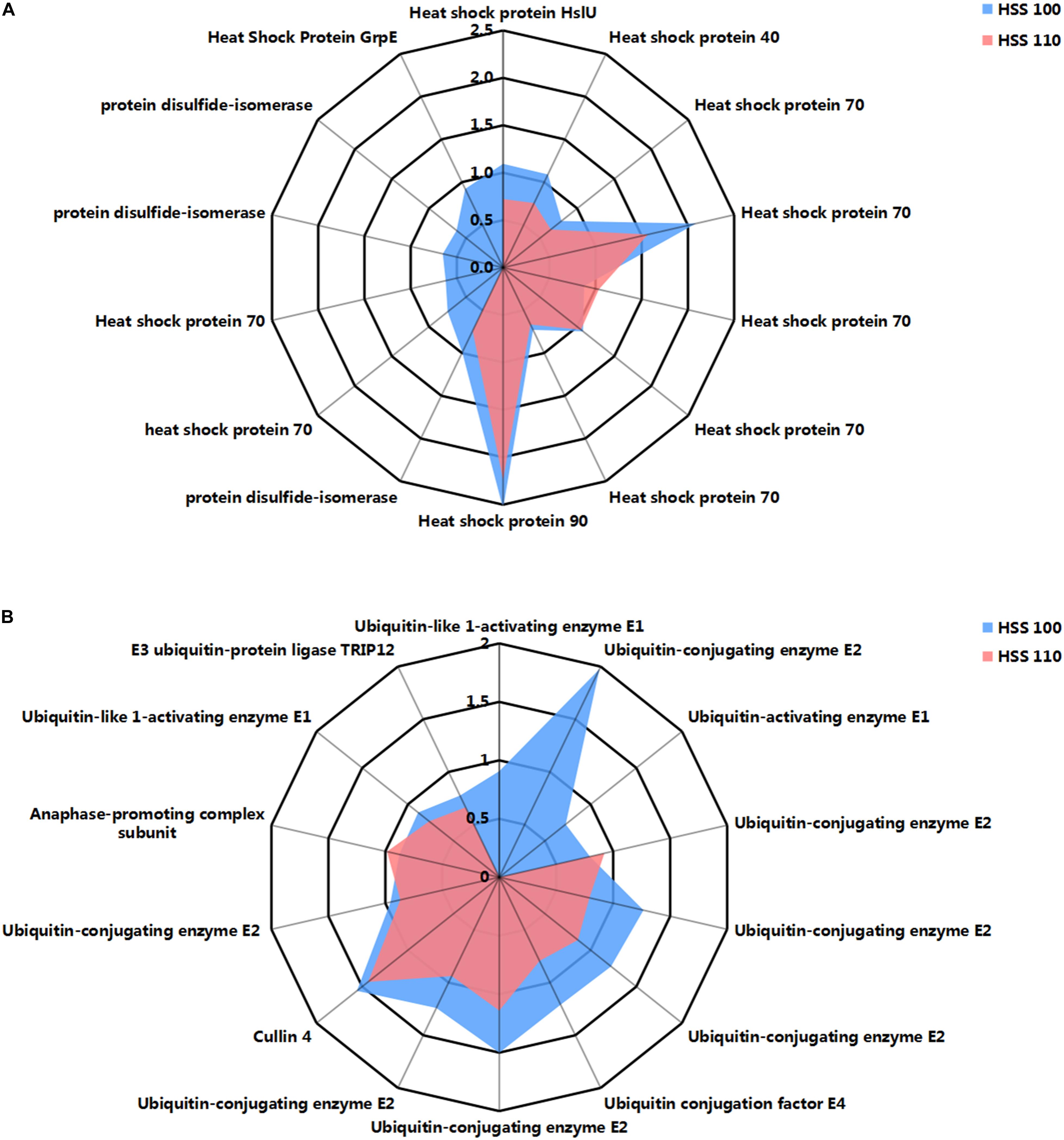
Figure 4. Effects of hypersaline stress on the protein processing-related pathways of P. haitanensis. (A) Differentially abundant molecular chaperone-related proteins. (B) Differentially abundant ubiquitin-mediated proteolysis-related proteins. Radar maps were plotted based on log2 fold change (FC) values from the proteomics dataset. Blue and red correspond to HSS 100 and 110 conditions, respectively. FC ≥ 1.5 and P < 0.05.
Ubiquitin-mediated protein degradation is a highly specific and efficient process that identifies the useless and abnormal proteins accumulated in cells for a subsequent elimination. This process is critical for maintaining normal physiological functions in cells. In this study, the abundances of a considerable number of proteins related to ubiquitin hydrolysis increased in P. haitanensis thalli under HSS 100 and 110 conditions (Figure 4B). More ubiquitin-activating enzyme (E1)- and ubiquitin-conjugating enzyme (E2)-related proteins with increased contents were detected under HSS 100 conditions than under HSS 110 conditions.
Interactions Among Differentially Abundant Proteins
To reveal the functional relationships among differentially abundant proteins, a PPI network was generated with the STRING and Cytoscape programs. The PPI network revealed interactions among 41 of the differentially abundant proteins under hypersaline stress (Figure 5). Specifically, TIM, isocitrate dehydrogenase (ICDH), malate dehydrogenase (MDH), and transketolase (AT2G45290) interacted with 24, 20, 19, and 17 proteins, respectively, implying these proteins are important for P. haitanensis responses to hypersaline stress.
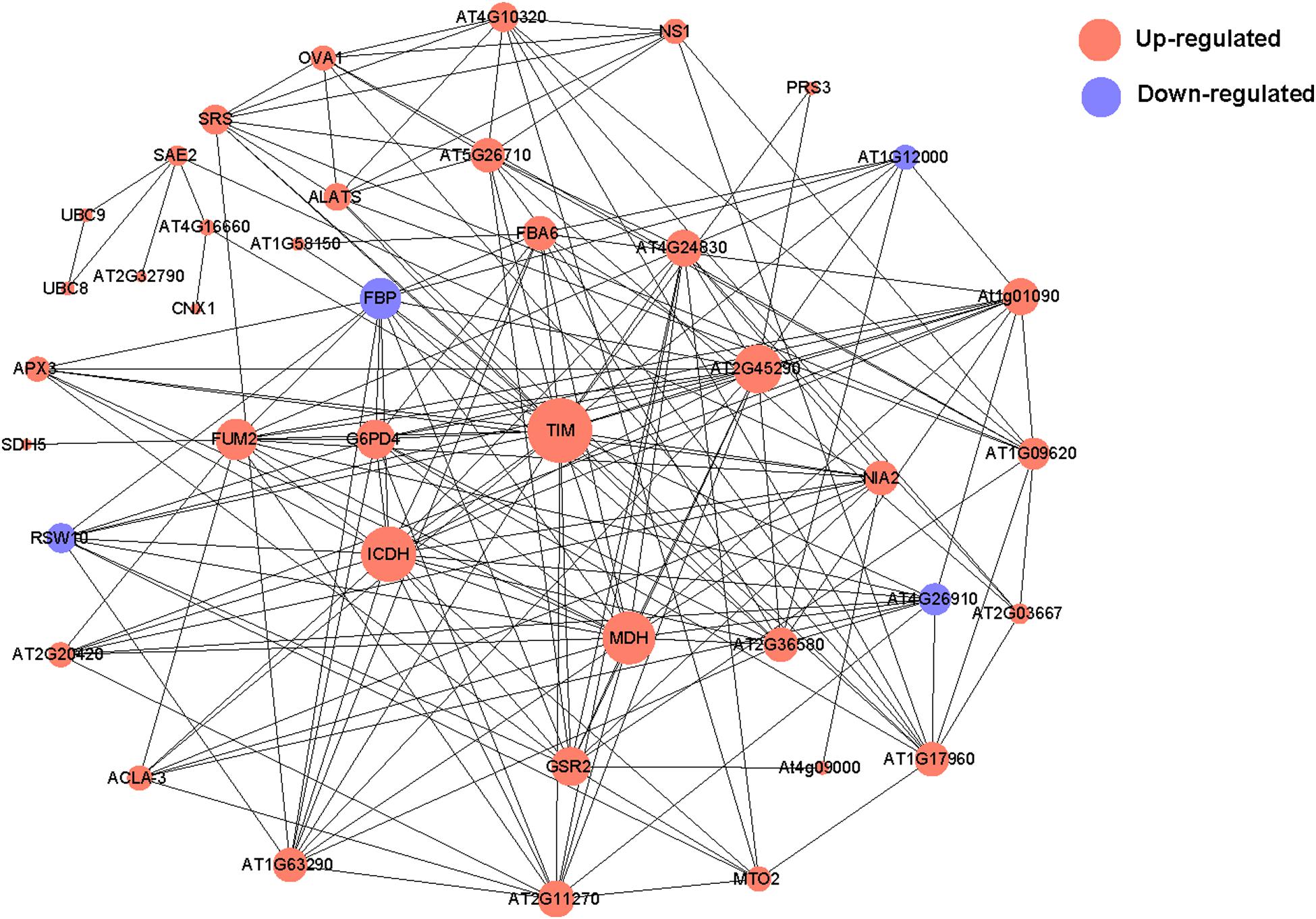
Figure 5. Protein–protein interaction network for P. haitanensis thalli exposed to hypersaline stress for 4 h based on STRING and Cytoscape analyses. Red and blue correspond to increased and decreased abundances, respectively. Circle size represents the number of protein interactions. Minimum required interaction score >0.4. Protein annotations are listed in Supplementary Table S6.
Effects of Hypersaline Stress on Organic Acid Metabolism
The pyruvic acid, citric acid, succinic acid, fumaric acid, and malic acid contents in P. haitanensis under three conditions were determined. Specifically, under the HSS 100 and 110 conditions, the contents of all organic acids increased, with the exception of pyruvic acid. Additionally, the pyruvic acid and citric acid contents in P. haitanensis under hypersaline stress differed significantly from the corresponding contents under normal conditions, whereas the succinic acid content was significantly different from the control level only under HSS 100 conditions (Figure 6).
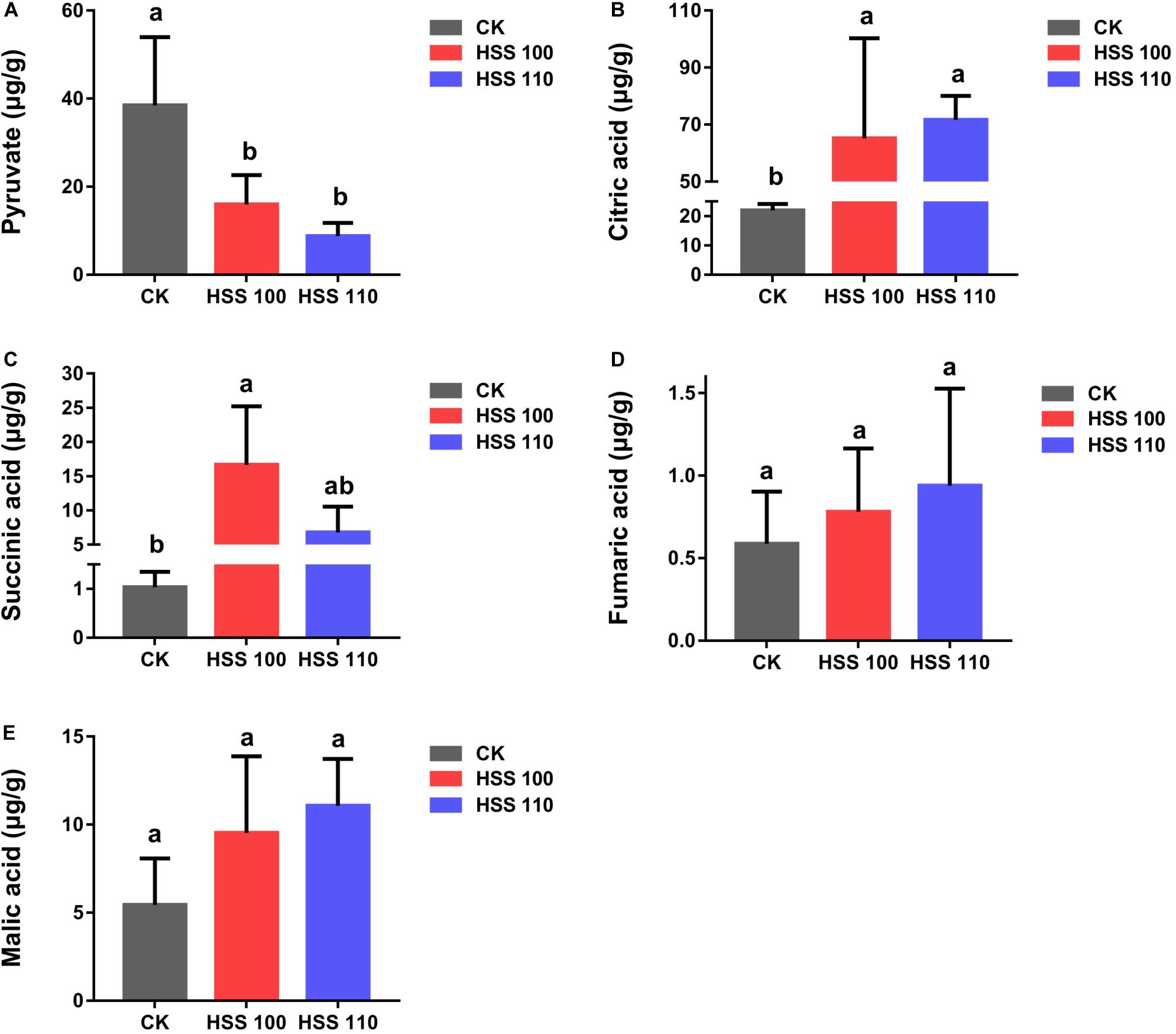
Figure 6. Effects of different conditions on P. haitanensis organic acid contents. (A) Pyruvic acid content. (B) Citric acid content. (C) Succinic acid content. (D) Fumaric acid content. (E) Malic acid content. Data are presented as the mean ± SE for at least three independent biological replicates. Different letters above the bars indicate significant differences at P < 0.05 as determined by a one-way ANOVA followed by Tukey’s test.
Discussion
The recent intensification of soil salinization has seriously affected crop yield and quality, and has increased the urgency of developing salt-tolerant crops (Flowers, 2004; Hanin et al., 2016). Pyropia haitanensis has become an ideal material for studying the mechanism underlying plant responses to saline conditions because of its strong salt tolerance. However, most of the available relevant research has focused on the establishment of a K+/Na+ balance, maintenance of osmotic pressure, and removal of reactive oxygen species in P. haitanensis under hypersaline stress (Chen et al., 2019; Wang et al., 2019). The importance of energy metabolism and protein processing for stress resistance has largely been ignored. Moreover, advances in proteomics-based research have provided researchers with a platform for characterizing the response mechanism of organisms exposed to abiotic stresses. Therefore, in this study, DIA technology was used for a proteome-level analysis of P. haitanensis thalli under different hypersaline stresses to clarify the response mechanism specifically related to energy metabolism and endoplasmic reticulum protein processing. The generated data may be useful for future investigations on the molecular basis of P. haitanensis salt tolerance and for breeding salt-tolerant cultivars of agriculturally important crops.
Energy Metabolism Response Mechanism
Under normal and stress conditions, stable metabolic activities provide organisms with the energy required for life activities. Additionally, whether an organism has a stable energy supply under stress conditions is closely related to its stress resistance. Previous studies revealed that an exposure to hypersaline stress for 4 h significantly decreases the maximum quantum yield (Fv/Fm) of P. haitanensis thalli (Chen et al., 2019; Wang et al., 2019). How P. haitanensis thalli can produce sufficient energy for its physiological activities when photosynthesis is inhibited should be clarified.
Regarding the EMP pathway, phosphofructokinase is the rate-limiting enzyme because of its low catalytic efficiency and irreversible catalytic reaction (Chitlaru and Pick, 1991; Ruan et al., 2009). The decreased abundance of phosphofructokinase observed in the current study will affect the production of fructose 1,6-diphosphate and inhibit the EMP pathway (Figure 3). Moreover, the decrease in the fructose-bisphosphate aldolase level may be related to the limited production of fructose 1,6-diphosphate. Of glyceraldehyde 3-phosphate and dihydroxyacetone phosphate, only glyceraldehyde 3-phosphate is involved in the EMP pathway. Therefore, increases in TIM contents will contribute to the conversion of dihydroxyacetone phosphate (Dorion et al., 2005) to glyceraldehyde 3-phosphate to maintain the dynamic balance between glyceraldehyde 3-phosphate and dihydroxyacetone phosphate in algae and stabilize the EMP pathway (Figure 3). Additionally, the PPI network indicated that TIM can interact with 24 differentially abundant proteins related to energy metabolism and amino acid synthesis identified in this study (Figures 3, 5), suggesting that it is crucial for the P. haitanensis response to hypersaline stress. Increased phosphoglycerate kinase and pyruvate kinase contents will help the intermediate products to fully react to ensure the EMP pathway progresses.
Pyruvate produced by the EMP pathway serves as the substrate for the TCA cycle. Therefore, we examined the TCA cycle-related differentially abundant proteins. Regarding the TCA cycle, pyruvate dehydrogenase first converts pyruvic acid to acetyl coenzyme A (CoA). The formation of citric acid from acetyl-CoA and oxaloacetic acid via a reaction catalyzed by citric acid synthetase represents the beginning of the TCA cycle, in which citric acid synthetase is the rate-limiting enzyme. The upstream substrate activates the downstream enzyme, and the downstream product inhibits the upstream enzyme, which is a characteristic of the TCA cycle. Previous studies on soybean indicated that salt stress increases the expression of genes encoding enzymes such as citrate synthase (Liu et al., 2019). In the current study, the differentially abundant proteins related to the TCA cycle in P. haitanensis increased in abundance under hypersaline stress (Figure 7). Additionally, ICDH and MDH can interact with multiple proteins and are in key regulatory positions of the PPI network (Figure 5). Moreover, we measured the contents of pyruvic acid, citric acid, and other organic acids related to the TCA cycle in P. haitanensis under three conditions. In a previous study on Arabidopsis thaliana, drought stress was revealed to promote the accumulation of the intermediate products of the TCA cycle (Urano et al., 2009), suggesting that it can enhance carbon decomposition to meet the energy needs of the stressed plant. In the present study, we observed that compared with normal conditions, hypersaline stress caused the TCA reaction substrate (pyruvic acid) content to decrease and the reaction product (e.g., citric acid and succinic acid) contents to increase in P. haitanensis (Figure 6). These results indicated that under hypersaline conditions, the TCA cycle of P. haitanensis is induced to produce more energy to ensure physiological and biochemical activities can occur. Additionally, unlike the effects of HSS 110 conditions, the succinic acid content of P. haitanensis under HSS 100 conditions was significantly different from that under normal conditions, which may be closely related to the increased abundance of succinyl-CoA synthetase only under HSS 100 conditions (Figure 7).
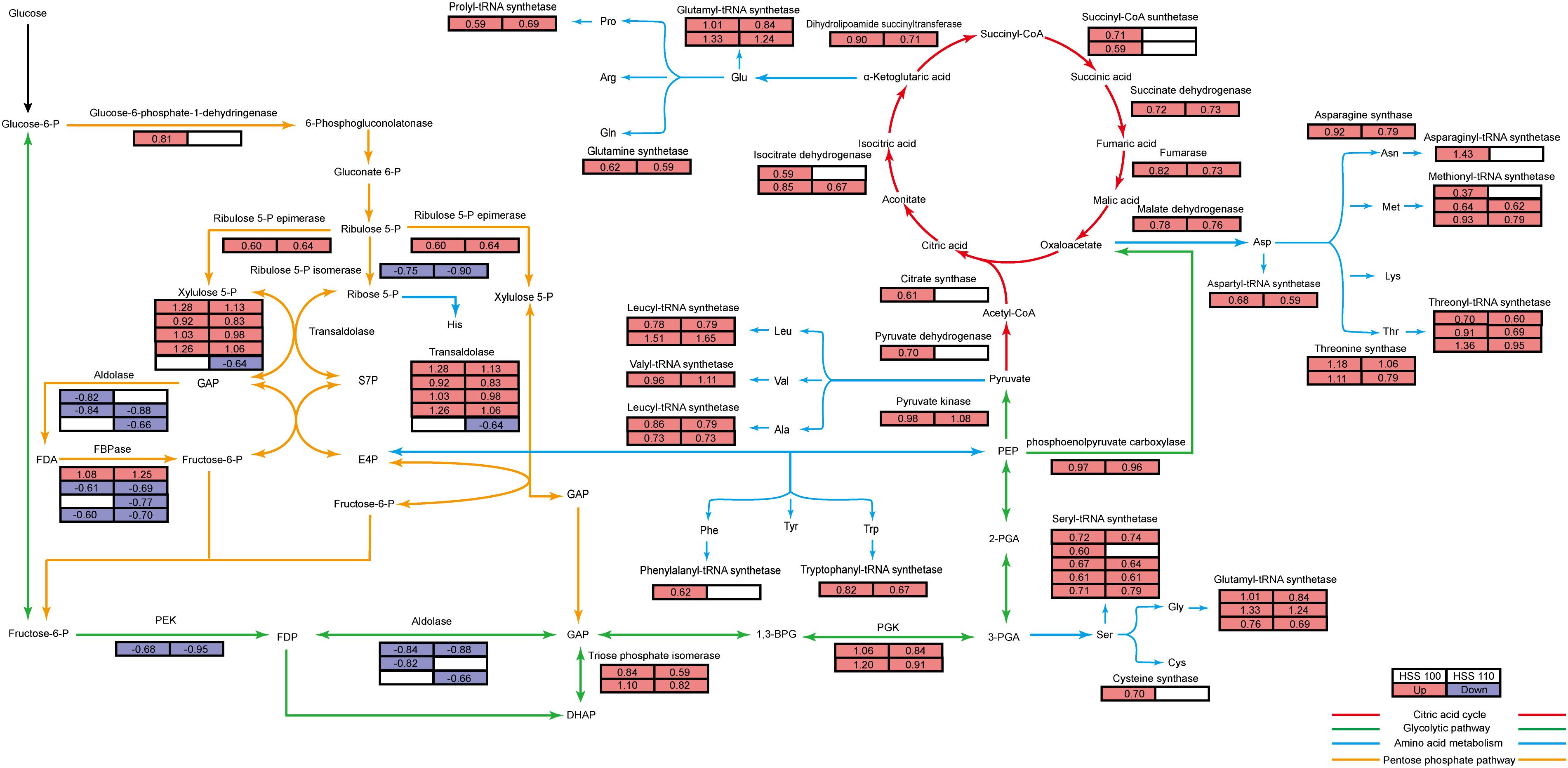
Figure 7. Energy metabolism map of P. haitanensis under hypersaline stress. Green lines represent the glycolytic pathway (i.e., EMP pathway), yellow lines represent the pentose phosphate pathway, red lines represent the TCA cycle, and blue lines represent the amino acid and aminoacyl-tRNA synthesis pathway. The tables on the left and right represent the HSS 100 and 110 conditions, respectively, and the colors reflect the protein content differences relative to the abundance under normal conditions. Red, blue, and white correspond to increased, decreased, and unchanged, respectively. Numbers represent log2 fold change (FC) values. FC ≥ 1.5 and P < 0.05.
Glucose-6-phosphate dehydrogenase is a rate-limiting enzyme in the PPP (Kletzien et al., 1994), and hypersaline stress-induced increases in its content will promote the PPP of P. haitanensis thalli. In the PPP, the non-oxidation stage is characterized by reversible reactions. Thus, increases in transketolase amounts ensure that the alga can satisfy its demand for sugar metabolic intermediates (Figure 3). Our PPI network (Figure 5) indicated that transketolase is very important for the C and N metabolism of algae. Additionally, a previous study demonstrated that the overexpression of the transketolase gene enhances Cucumis sativus L. tolerance to abiotic stresses (Bi et al., 2013). Glucose-6-phosphate dehydrogenase levels also increased only under HSS 100 conditions, indicating that the PPP of P. haitanensis thalli is more responsive to salt stress under HSS 100 conditions than under HSS 110 conditions (Figure 3).
A previous 2-DE investigation of P. haitanensis revealed an increase in the production of energy-related proteins in response to desiccation conditions (Xu et al., 2016). Moreover, Shi et al. (2019) proved that the desiccation tolerance of P. haitanensis is closely related to its energy metabolism. Decreases in phosphofructokinase levels in P. haitanensis thalli under hypersaline stress will generally inhibit the EMP pathway (Figure 7). Increases in the abundance of TIM enhance the conversion of dihydroxyacetone phosphate to glyceraldehyde 3-phosphate to maintain the EMP pathway (Figure 7). Additionally, an induced PPP can provide sufficient intermediate products for the EMP pathway (Georgieva et al., 2017), which may help to explain why phosphoglycerate kinase and pyruvate kinase levels can still increase when phosphofructokinase and aldolase abundances decrease in this pathway (Figure 7). Meanwhile, increases in the phosphoenolpyruvate carboxylase contents result in sufficient oxaloacetic acid for the TCA cycle (Figure 7). The observed increased abundances of citrate synthase, pyruvate dehydrogenase, and 6-phosphoglucose dehydrogenase under HSS 100 conditions suggest that P. haitanensis thalli increase their energy production in response to this stress (Figure 7).
Protein Synthesis Response Mechanism
As the EMP pathway, PPP, and the TCA cycle progress, energy is generated and the intermediate products can be used as carbon chain skeletons for amino acid synthesis (Fernie et al., 2004; Figure 7). The formation of aminoacyl-tRNAs of 20 amino acids in reactions catalyzed by specific enzymes is the first step in protein synthesis. In this study, many of the differentially abundant proteins were related to aminoacyl-tRNA synthesis, and all of these proteins were more abundant after the exposure to hypersaline stress. Thus, hypersalinity may promote protein synthesis in P. haitanensis thalli. More differentially abundant proteins related to aminoacyl-tRNA synthesis were detected under HSS 100 conditions than under HSS 110 conditions (Figure 7).
Our data also revealed the increased abundance of nitrate reductase (NR) (Supplementary Table S7) from the nitrogen assimilation pathway. Increases in NR levels will promote the transformation of nitrates into ammonia in algae (Lam et al., 1996), enhance the nitrogen assimilation rate of P. haitanensis thalli under hypersaline stress, and ensure the synthesis of amino acids and proteins.
Earlier investigations confirmed that stress can increase the probability of protein misfolding or unfolding and that the accumulation of these improperly formed proteins due to inefficient removal will affect the normal physiological activities of organisms, possibly leading to cell death (Liu and Howell, 2010). Therefore, organisms tend to inhibit protein synthesis under stress to avoid the accumulation of misfolded or unfolded proteins (Shi et al., 2017). However, this stress response adversely affects the normal physiological and biochemical reactions of organisms under stress conditions. The mechanism mediating the enhanced protein synthesis in P. haitanensis thalli under hypersaline stress remains unclear. The endoplasmic reticulum processes proteins in eukaryotic cells. Only proteins that have been correctly folded in the endoplasmic reticulum will be released to be transported to the appropriate tissue and organ where their function is required (Ellgaard and Helenius, 2003). Therefore, we analyzed the proteins related to protein processing in the endoplasmic reticulum.
Biotic and abiotic stresses induce protein misfolding and unfolding. The ERQC system recognizes proteins in the endoplasmic reticulum based on interactions with CNX and CRT (Helenius and Aebi, 2004). Correctly folded proteins will be transported to the Golgi apparatus through COP II, while misfolded or unfolded proteins will be refolded via recruited molecular chaperones, such as HSPs and protein disulfide isomerase (Wang et al., 2004). An earlier study indicated the heterologous expression of the rice calnexin gene OsCNX confers drought tolerance to Nicotiana tabacum (Sarwat and Naqvi, 2013). Ozgur et al. (2015) confirmed that oxidative stress enhances CNX expression in A. thaliana. In the current study, the observed increases in the CNX level (Supplementary Table S7) will enhance the recognition of correctly or incorrectly folded proteins in the endoplasmic reticulum, whereas the increased abundance of COP II (Supplementary Table S7) will improve the transport of correctly folded proteins to the Golgi apparatus (Figure 8). Increased HSP and protein disulfide isomerase contents will ensure correct protein folding in the endoplasmic reticulum and prevent protein aggregation (Figure 8). Notably, HSP70 and protein disulfide isomerases were more abundant under HSS 100 conditions than under HSS 110 conditions, implying that of the two salt treatments in this study, protein folding occurs more accurately and efficiently in P. haitanensis thalli under HSS 100 conditions.
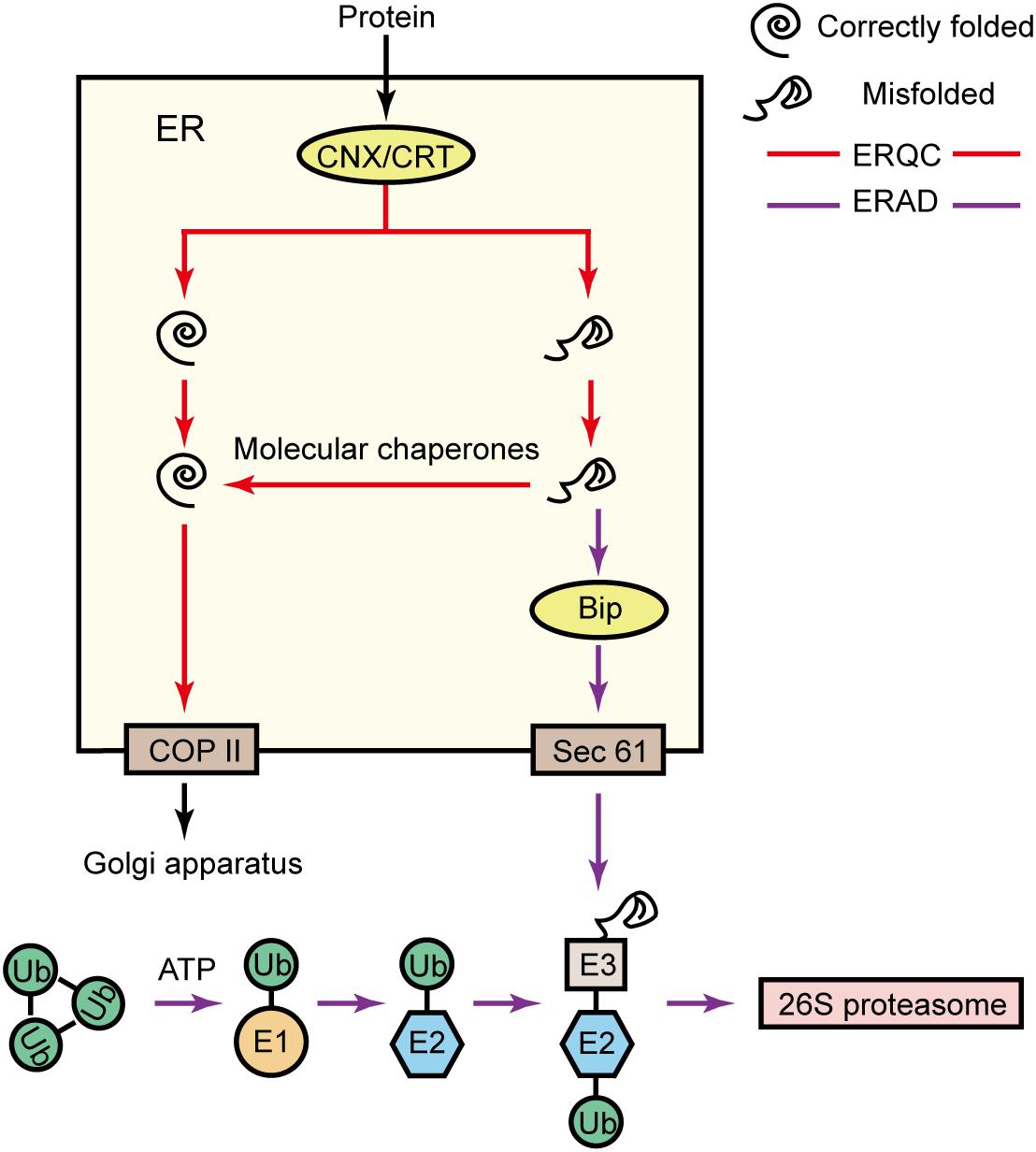
Figure 8. Protein synthesis response mechanism. In the endoplasmic reticulum, the ERQC proteins CNX and CRT detect the folding of proteins. Correctly folded proteins will be transported from COP II to the Golgi apparatus for further processing. However, misfolded proteins are refolded by recruited molecular chaperones. Alternatively, the ERAD system can also identify misfolded proteins via BiP, transport misfolded proteins from Sec61 to the cytoplasm, and mediate ubiquitination-based degradation to prevent the accumulation of misfolded proteins in the endoplasmic reticulum. The ubiquitin-mediated degradation process is as follows: a ubiquitin-activating enzyme (E1) activates a ubiquitin molecule (Ub), with ATP supplying the required energy; the activated Ub is transferred to a ubiquitin-binding enzyme (E2); a ubiquitin ligase (E3) connects Ub-E2 to the target protein; and the ubiquitinated target protein is specifically recognized by the 26S proteasome and degraded (Ruggiano et al., 2014).
If misfolded or unfolded proteins in the endoplasmic reticulum cannot be eliminated in a timely fashion, the resulting stress on the organelle will be minimized via ERAD to maintain the internal stability of the endoplasmic reticulum (Yoshida, 2007). Specifically, ERAD is mainly divided into the following four steps: (1) identification of misfolded proteins; (2) transport of misfolded proteins to the cytoplasm; (3) ubiquitin modification catalyzed by a specific ubiquitin-activating enzyme (E1), a ubiquitin-binding enzyme (E2), and a ubiquitin ligase (E3); (4) degradation of the misfolded protein by the 26S proteasome (Ruggiano et al., 2014; Figure 8). Previous research proved that the overexpression of the BiP gene improves the tolerance of tobacco to osmotic stress (Alvim et al., 2001). In the current study, we detected a salt-induced increase in the contents of BiP, which contributes to ERAD substrate recognition (Supplementary Table S7), Sec61, which is involved in ERAD substrate transport (Supplementary Table S7), and many E1-, E2-, and E3-related proteins. Increases in the abundances of these proteins enhance the ability of the endoplasmic reticulum to efficiently remove misfolded or unfolded proteins in P. haitanensis thalli exposed to hypersaline stress, thereby minimizing the damage due to endoplasmic reticulum stress. Additionally, the E1- and E2-related proteins were more abundant under HSS 100 conditions than under HSS 110 conditions. Therefore, P. haitanensis thalli can likely eliminate misfolded or unfolded proteins and prevent stress-induced endoplasmic reticulum damages better at the lower of the two salt concentrations tested in this study.
In general, the intermediate products of the EMP pathway, the PPP, and the TCA cycle provide sufficient carbon skeletons for amino acid synthesis under hypersaline stress. Moreover, an increase in the nitrogen assimilation rate also ensures amino acids and proteins are synthesized under stress conditions. Under these conditions of enhanced protein synthesis, the increased abundance of endoplasmic reticulum proteins involved in detecting and transporting developing proteins helps to maintain protein synthesis under salt stress. Moreover, increases in the contents of HSPs and proteins related to the ubiquitin-mediated protein degradation system stabilize the dynamic balance between protein folding and removal in algae. This ultimately ensures proteins are synthesized even at high salt concentrations, enabling P. haitanensis thalli to complete physiological and biochemical reactions under hypersaline conditions.
14-3-3 Protein
As an important regulatory protein, the 14-3-3 protein is widely involved in plant physiological processes, including metabolism, hormone signal transduction, stomatal regulation, and responses to abiotic and biotic stresses (Lozano-Durán and Robatzek, 2015; Cotelle and Leonhardt, 2016). Studies have proven that the 14-3-3 protein plays an important role in plant responses to salt stress (Wu and Seliskar, 1998; Maathuis and Amtmann, 1999; Very and Sentenac, 2003). Under normal conditions, PKS5 kinase activity is high, resulting in the phosphorylation of the plasma membrane H+-ATPase AHA2, which inhibits the enzyme activity. After being stimulated by salt stress, the 14-3-3 protein can recognize and decode Ca+ signals induced by salt stress and enhance its interaction with PKS5. This interaction inhibits PKS5 activity, which decreases the phosphorylation of AHA2 to increase the plasma membrane H+-ATPase activity. This increased activity provides the driving force for SOS1 and is important for maintaining the K+/Na+ balance in organisms exposed to salt stress (Yang et al., 2019). Additionally, the 14-3-3 protein can also participate in the abscisic acid (ABA) signal transduction pathway and regulate ascorbic acid peroxidase (APX) activity. This protein is also important for plant stress resistance (Zhang et al., 1997; Jia et al., 2002; Badawi et al., 2004; Sun et al., 2015). In the current study, although an increase in the 14-3-3 protein abundance was noted, the abundance of related proteins, such as the plasma membrane H+-ATPase and APX, also increased, as did the ABA level (Supplementary Table S7). Nevertheless, our findings imply that the 14-3-3 protein is an important contributor to P. haitanensis responses to hypersaline stress.
Conclusion
Under hypersaline stress conditions, P. haitanensis thalli produce substrates for the TCA cycle by regulating the EMP pathway and the PPP, ultimately ensuring sufficient energy is generated for the required physiological and biochemical reactions. Meanwhile, the ability to refold or remove misfolded and unfolded proteins is enhanced to maintain the synthesis of proteins necessary for physiological and biochemical changes to resist stresses (Figure 9). Moreover, P. haitanensis thalli exhibit better energy metabolism and more efficient protein processing and synthesis under HSS 100 conditions than under HSS 110 conditions. The results of this study may be useful for future investigations of plant stress resistance mechanisms and the development of new salt-tolerant varieties of important crops.
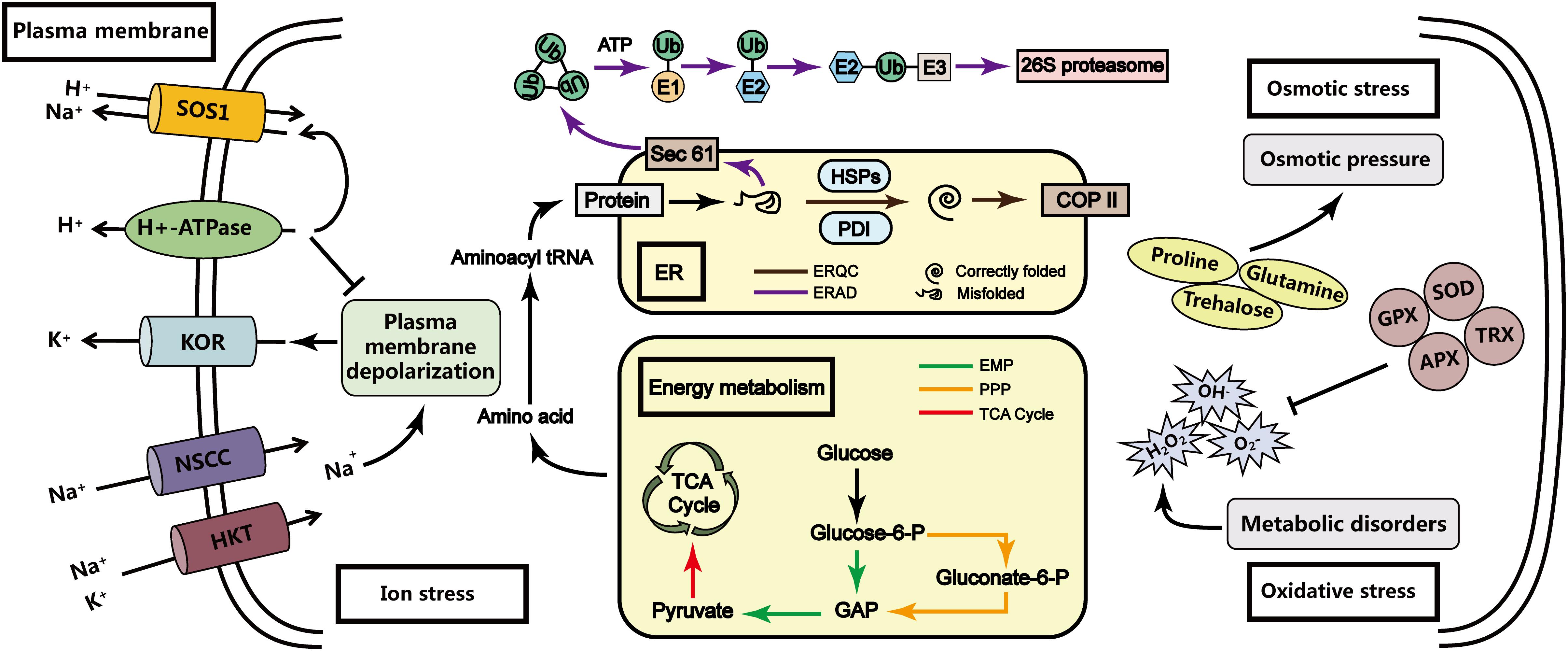
Figure 9. Model of the P. haitanensis response mechanism under hypersaline stress. Previous research indicated that under hypersaline stress, P. haitanensis is primarily subjected to ion stress, osmotic stress, and oxidative stress. However, P. haitanensis can respond to hypersaline stress via the transmembrane transport of Na+ (Na+/H+ antiporter and H+-ATPase), the synthesis or accumulation of available osmotic substances (e.g., proline and trehalose), and increases in the content or activity of antioxidant enzymes (e.g., superoxide dismutase and ascorbate peroxidase) (Chen et al., 2019; Wang et al., 2019). Finally, a new steady-state equilibrium is established to ensure that the algal bodies can survive and perform certain functions under hypersaline stress. Additionally, the P. haitanensis response mechanism under hypersaline conditions is linked to energy metabolism and protein processing. The results of this study imply that under hypersaline stress, P. haitanensis thalli can regulate the EMP pathway and the PPP to provide substrates for the TCA cycle, and enhance the folding and clearing ability of the unfolded proteins. The protein synthesis can provide the energy and components required for physiological and biochemical reactions in P. haitanensis exposed to hypersaline stress.
Data Availability Statement
The raw data generated in this study were deposited in the iProX database (accession number: PXD018664).
Author Contributions
JW and WW conducted the data analysis and wrote the first draft of the manuscript. KX, DJ, and YX participated in the data collection and sample processing. CX and CC contributed to design and interpretation of results. All authors contributed to writing, revising, and approving the submitted version of the manuscript.
Funding
This work was supported by the National Key R&D Program of China (grant numbers: 2018YFD0900100 and 2018YFD0901500), the National Natural Science Foundation of China (grant number: 41806185), and the China’s Agriculture Research System (grant number: CARS-50).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Liwen Bianji of Edanz Editing China (www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00415/full#supplementary-material
TABLE S1 | Peptide summary.
TABLE S2 | Protein summary.
TABLE S3 | Protein identification.
TABLE S4 | Verification of differentially abundant proteins in Pyropia haitanensis under HSS 100 conditions based on multiple reaction monitoring. The relative changes are presented as log2 fold changes.
TABLE S5 | Verification of differentially abundant proteins in Pyropia haitanensis under HSS 110 conditions based on multiple reaction monitoring. The relative changes are presented as log2 fold changes.
TABLE S6 | Protein annotations of the protein–protein interaction network.
TABLE S7 | Differential abundances of related proteins.
Footnotes
References
Alvim, F. C., Carolino, S. M., Cascardo, J. C., Nunes, C. C., Martinez, C. A., Otoni, W. C., et al. (2001). Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiol. 126, 1042–1054. doi: 10.1104/pp.126.3.1042
Badawi, G. H., Kawano, N., Yamauchi, Y., Shimada, E., Sasaki, R., Kubo, A., et al. (2004). Over-expression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit. Physiol. Plant 121, 231–238. doi: 10.1111/j.0031-9317.2004.00308.x
Bi, H., Wang, M., Dong, X., and Ai, X. (2013). Cloning and expression analysis of transketolase gene in Cucumis sativus L. Plant Physiol. Bioch. 70, 512–521. doi: 10.1016/j.plaphy.2013.06.017
Blouin, N. A., Brodie, J. A., Grossman, A. C., Xu, P., and Brawley, S. H. (2011). Porphyra : a marine crop shaped by stress. Trends Plant Sci. 16, 29–37. doi: 10.1016/j.tplants.2010.10.004
Brawley, S. H., Blouin, N. A., Ficko-Blean, E., Wheeler, G. L., Lohr, M., Goodson, H. V., et al. (2017). Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae. Rhodophyta). Proc. Natl. Acad. Sci. U.S.A. 114, E6361–E6370. doi: 10.1073/pnas.1703088114
Bruderer, R., Bernhardt, O. M., Gandhi, T., Miladinoviæ, S. M., Cheng, L.-Y., Messner, S., et al. (2015). Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteomics 14, 1400–1410. doi: 10.1074/mcp.M114.044305
Chen, C., Ji, D., Xie, C., Xu, Y., Liang, Y., Zhen, Y., et al. (2008). Preliminary study on selecting the high temperature resistance strains and economic traits of Porphyra haitanensis. Acta Oceanol. Sin. 30, 100–106. doi: 10.1007/s11676-008-0012-9
Chen, T., Wang, W., Xu, K., Xu, Y., Ji, D., Chen, C., et al. (2019). K+ and Na+ transport contribute to K+/Na+ homeostasis in Pyropia haitanensis under hypersaline stress. Algal. Res. 40:101526. doi: 10.1016/j.algal.2019.101526
Chitlaru, E., and Pick, U. (1991). Regulation of glycerol synthesis in response to osmotic changes in Dunaliella. Plant Physiol. 96, 50–60. doi: 10.1104/pp.96.1.50
Choi, M., Chang, C.-Y., Clough, T., Broudy, D., Killeen, T., MacLean, B., et al. (2014). MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526. doi: 10.1093/bioinformatics/btu305
Cotelle, V., and Leonhardt, N. (2016). 14-3-3 proteins in guard cell signaling. Front. Plant Sci. 6:1210. doi: 10.3389/fpls.2015.01210
Davison, I. R., and Pearson, G. A. (1996). Stress tolerance in intertidal seaweeds. J. Phycol. 32, 197–211. doi: 10.1111/j.0022-3646.1996.00197.x
Dittami, S. M., Gravot, A., Goulitquer, S., Rousvoal, S., Peters, A. F., Bouchereau, A., et al. (2012). Towards deciphering dynamic changes and evolutionary mechanisms involved in the adaptation to low salinities in Ectocarpus (brown algae). Plant J. 71, 366–377. doi: 10.1111/j.1365-313X.2012.04982.x
Dittami, S. M., Scornet, D., Petit, J.-L., Ségurens, B., Da Silva, C., Corre, E., et al. (2009). Global expression analysis of the brown alga Ectocarpus siliculosus (Phaeophyceae) reveals large-scale reprogramming of the transcriptome in response to abiotic stress. Genome Biol. 10:R66. doi: 10.1186/gb-2009-10-6-r66
Dorion, S., Jeukens, J., Matton, D. P., and Rivoal, J. (2005). Cloning and characterization of a cytosolic isoform of triosephosphate isomerase developmentally regulated in potato leaves. Plant Sci. 168, 183–194. doi: 10.1080/09670260802573105
Ellgaard, L., and Helenius, A. (2003). Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Bio. 4, 181–191. doi: 10.1016/j.semcdb.2010.03.005
Fernie, A. R., Carrari, F., and Sweetlove, L. J. (2004). Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 7, 254–261. doi: 10.1016/j.pbi.2004.03.007
Flowers, T. (2004). Improving crop salt tolerance. J. Exp. Bot. 55, 307–319. doi: 10.1093/jxb/erh003
Georgieva, K., Dagnon, S., Gesheva, E., Bojilov, D., Mihailova, G., and Doncheva, S. (2017). Antioxidant defense during desiccation of the resurrection plant Haberlea rhodopensis. Plant Physiol. Bioch. 114, 51–59. doi: 10.1016/j.plaphy.2017.02.021
Hanin, M., Ebel, C., Ngom, M., Laplaze, L., and Masmoudi, K. (2016). New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 7:1787. doi: 10.3389/fpls.2016.01787
Hegde, R. S., and Ploegh, H. L. (2010). Quality and quantity control at the endoplasmic reticulum. Curr. Opin. Cell Biol. 22, 437–446. doi: 10.1016/j.ceb.2010.05.005
Helenius, A., and Aebi, M. (2004). Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752
Jia, W., Wang, Y., Zhang, S., and Zhang, J. (2002). Salt-stress-induced ABA accumulation is more sensitively triggered in roots than in shoots. J. Exp. Bot. 53, 2201–2206. doi: 10.1093/jxb/erf079
Kirst, G. (1990). Salinity tolerance of eukaryotic marine algae. Annu. Rev. Plant Biol. 41, 21–53. doi: 10.1146/annurev.pp.41.060190.000321
Kletzien, R., Harris, P., and Foellmi, L. (1994). Glucose-6-phosphate dehydrogenase: a“housekeeping” enzyme subject to tissue-specific regulation by hormones, nutrients, and oxidant stress. FASEB J. 8, 174–181. doi: 10.1096/fasebj.8.2.8119488
Lam, H.-M., Coschigano, K., Oliveira, I., Melo-Oliveira, R., and Coruzzi, G. (1996). The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Biol. 47, 569–593. doi: 10.1146/annurev.arplant.47.1.569
Law, K. P., and Lim, Y. P. (2013). Recent advances in mass spectrometry: data independent analysis and hyper reaction monitoring. Expert Revi. Proteomic. 10, 551–566. doi: 10.1586/14789450.2013.858022
Liu, A., Xiao, Z., Li, M. W., Wong, F. L., Yung, W. S., Ku, Y. S., et al. (2019). Transcriptomic reprogramming in soybean seedlings under salt stress. Plant Cell Environ. 42, 98–114. doi: 10.1111/pce.13186
Liu, J.-X., and Howell, S. H. (2010). Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22, 2930–2942. doi: 10.1105/tpc.110.078154
Lozano-Durán, R., and Robatzek, S. (2015). 14-3-3 proteins in plant-pathogen interactions. Mol. Plant Microbe Int. 28, 511–518. doi: 10.1094/MPMI-10-14-0322-CR
Lu, I.-F., Sung, M.-S., and Lee, T.-M. (2006). Salinity stress and hydrogen peroxide regulation of antioxidant defense system in Ulva fasciata. Mar. Biol. 150, 1–15. doi: 10.1007/s00227-006-0323-3
Lu, X., Huan, L., Gao, S., He, L., and Wang, G. (2016). NADPH from the oxidative pentose phosphate pathway drives the operation of cyclic electron flow around photosystem I in high-intertidal macroalgae under severe salt stress. Physiol. Plant 156, 397–406. doi: 10.1111/ppl.12383
Maathuis, F. J., and Amtmann, A. (1999). K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann. Bot. 84, 123–133. doi: 10.1006/anbo.1999.0912
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Ozgur, R., Uzilday, B., Sekmen, A. H., and Turkan, I. (2015). The effects of induced production of reactive oxygen species in organelles on endoplasmic reticulum stress and on the unfolded protein response in Arabidopsis. Ann. Bot. 116, 541–553. doi: 10.1093/aob/mcv072
Ruan, K., Duan, J., Bai, F., Lemaire, M., Ma, X., and Bai, L. (2009). Function of Dunaliella salina (Dunaliellaceae) enolase and its expression during stress. Eur. J. Phycol. 44, 207–214. doi: 10.1080/09670260802573105
Ruggiano, A., Foresti, O., and Carvalho, P. (2014). ER-associated degradation: protein quality control and beyond. J. Cell Biol. 204, 869–879. doi: 10.1083/jcb.201312042
Sarwat, M., and Naqvi, A. R. (2013). Heterologous expression of rice calnexin (OsCNX) confers drought tolerance in Nicotiana tabacum. Mol. Biol. Rep. 40, 5451–5464. doi: 10.1007/s11033-013-2643-y
Schulze, W. X., and Usadel, B. (2010). Quantitation in mass-spectrometry-based proteomics. Annu. Rev. Plant Biol. 61, 491–516. doi: 10.1146/annurev-arplant-042809-112132
Shi, J., Chen, Y., Xu, Y., Ji, D., Chen, C., and Xie, C. (2017). Differential proteomic analysis by iTRAQ reveals the mechanism of Pyropia haitanensis responding to high temperature stress. Sci. Rep. U. K. 7:44734. doi: 10.1038/srep44734
Shi, J., Wang, W., Lin, Y., Xu, K., Xu, Y., Ji, D., et al. (2019). Insight into transketolase of Pyropia haitanensis under desiccation stress based on integrative analysis of omics and transformation. BMC Plant Biol. 19:475. doi: 10.1186/s12870-019-2076-4
Sun, X., Sun, M., Jia, B., Chen, C., Qin, Z., Yang, K., et al. (2015). A 14-3-3 family protein from wild soybean (Glycine Soja) regulates ABA sensitivity in Arabidopsis. PLos One 10:e0146163. doi: 10.1371/journal.pone.0146163
Tang, X., Mu, X., Shao, H., Wang, H., and Brestic, M. (2015). Global plant-responding mechanisms to salt stress: physiological and molecular levels and implications in biotechnology. Crit. Rev. Biotechnol. 35, 425–437. doi: 10.3109/07388551.2014.889080
Urano, K., Maruyama, K., Ogata, Y., Morishita, Y., Takeda, M., Sakurai, N., et al. (2009). Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 57, 1065–1078. doi: 10.1111/j.1365-313x.2008.03748.x
Very, A.-A., and Sentenac, H. (2003). Molecular mechanisms and regulation of K+ transport in higher plants. Annu. Rev. Plant Biol. 54, 575–603. doi: 10.1146/annurev.arplant.54.031902.134831
Wang, W., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Wang, W., Xu, Y., Chen, T., Xing, L., Xu, K., Ji, D., et al. (2019). Regulatory mechanisms underlying the maintenance of homeostasis in Pyropia haitanensis under hypersaline stress conditions. Sci. Total Environ. 662, 168–179. doi: 10.1016/j.scitotenv.2019.01.214
Wu, J., and Seliskar, D. M. (1998). Salinity adaptation of plasma membrane H+-ATPase in the salt marsh plant Spartina patens : ATP hydrolysis and enzyme kinetics. J. Exp. Bot. 49, 1005–1013. doi: 10.1093/jxb/49.323.1005
Xu, K., Xu, Y., Ji, D., Xie, J., Chen, C., and Xie, C. (2016). Proteomic analysis of the economic seaweed Pyropia haitanensis in response to desiccation. Algal Res. 19, 198–206. doi: 10.1016/j.algal.2016.08.010
Xu, Y., Chen, C., Ji, D., Hang, N., and Xie, C. (2014). Proteomic profile analysis of Pyropia haitanensis in response to high-temperature stress. J. Appl. Phycol. 26, 607–618. doi: 10.1007/s10811-013-0066-8
Yan, K., Shao, H., Shao, C., Chen, P., Zhao, S., Brestic, M., et al. (2013). Physiological adaptive mechanisms of plants grown in saline soil and implications for sustainable saline agriculture in coastal zone. Acta Physiol. Plant 35, 2867–2878. doi: 10.1007/s11738-013-1325-7
Yang, Y., and Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, Z., Wang, C., Xue, Y., Liu, X., Chen, S., Song, C., et al. (2019). Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 10, 1–12. doi: 10.1038/s41467-019-09181-2
Yoshida, H. (2007). ER stress and diseases. FEBS J. 274, 630–658. doi: 10.1111/j.1742-4658.2007.05639.x
Zhang, H., Wang, J., Allen, R. D., and Goodman, H. M. (1997). Cloning and expression of an Arabidopsis gene encoding a putative peroxisomal ascorbate peroxidase. Plant Mol. Biol. 34, 967–971.
Keywords: Pyropia haitanensis, salt stress, energy metabolism, protein processing in the endoplasmic reticulum, DIA
Citation: Wen J, Wang W, Xu K, Ji D, Xu Y, Chen C and Xie C (2020) Comparative Analysis of Proteins Involved in Energy Metabolism and Protein Processing in Pyropia haitanensis at Different Salinity Levels. Front. Mar. Sci. 7:415. doi: 10.3389/fmars.2020.00415
Received: 08 February 2020; Accepted: 12 May 2020;
Published: 10 June 2020.
Edited by:
Susumu Takio, Kumamoto University, JapanCopyright © 2020 Wen, Wang, Xu, Ji, Xu, Chen and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chaotian Xie, Y3R4aWVAam11LmVkdS5jbg==
 Jian Wen
Jian Wen Wenlei Wang
Wenlei Wang Kai Xu
Kai Xu Dehua Ji1,2,3
Dehua Ji1,2,3 Chaotian Xie
Chaotian Xie