- 1APEC Climate Center, Busan, South Korea
- 2Marine Climate Change Unit, Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan
- 3ARC Centre of Excellence for Coral Reef Studies, James Cook University, Townsville, QLD, Australia
- 4Department of Biological Sciences, University of Alberta, Edmonton, AB, Canada
Elevated temperature can have detrimental effects on the physiological performance of many marine organisms. However, phenotypic plasticity may enable some populations to maintain their performance under thermal stress. Two longitudinally separated populations of the coral reef fish, Acanthochromis polyacanthus from the Great Barrier Reef have shown differing capacities for thermal plasticity – the southernmost Heron Island population restored aerobic scope within one generation at a higher temperature, whereas the northernmost Palm Island population restored aerobic scope only when two generations were exposed to warmer conditions. We recently discovered an epigenetic signature associated with transgenerational plasticity in the Palm Island population. Here, we aimed to determine if epigenetic changes are also associated with the within-generational plasticity observed in the Heron Island population and, if so, how this epigenetic signature compares to the Palm Island transgenerational epigenome. By sequencing and analyzing the genome-wide DNA methylome of fish reared at control (+0°C) or elevated temperatures (+1.5 and +3°C) since early life, we identified 480 differentially methylated genomic regions and 372 adjacent protein-coding genes associated with within-generational plasticity in the Heron Island population. Functions related to insulin, cardiovascular capacity, development, and heat response were significantly enriched in differentially methylated genes, suggesting that these functions are the core mechanisms for within-generational restoration of aerobic scope. Comparison to the differentially methylated genes identified from F2 Palm Island population revealed little overlap of genes and enriched functions, indicating that distinct genetic toolkits may be used for within- and between-generational plasticity to ocean warming in the same species from different latitudes.
Introduction
Understanding how organisms respond to rapid environmental change is increasingly important as global climate change intensifies (Scheffers et al., 2016; Pecl et al., 2017; IPCC, 2019). For ectothermic animals, changes in environmental temperature have direct effects on physiological performance, due to a lack of internal thermal regulation, making them especially at risk from future warming (Fry and Hart, 1948; Portner and Farrell, 2008; Sunday et al., 2010). Furthermore, for many ectotherms, the projected future warming exceeds the current-day thermal performance range and will likely result in the loss of individual performance and potentially increased rate of mortality (Tewksbury et al., 2008; Hughes et al., 2018). However, ectothermic species may be able to buffer the effects of rising temperature on individual performance through phenotypic plasticity and genetic adaptation over the longer-term (Hoffmann and Sgro, 2011; Munday et al., 2013; Reusch, 2014). Therefore, in order to predict the effect of future warming on ectotherm populations it is necessary to understand the adaptive physiological mechanisms that can reduce the impact of elevated environmental temperature over timescales relevant to the pace of climate change.
The capacity for phenotypic plasticity (i.e., environmentally-induced phenotypic variation) to buffer individual responses to higher temperature is likely to be especially advantageous because, for many species, the climate is changing faster than adaption via natural selection can keep up (Gienapp et al., 2008; Reusch, 2014; Fox et al., 2019). Many short-term experiments have demonstrated negative effects of predicted future temperatures on the physiological performance of fish and other marine ectotherms (Wernberg et al., 2012; Hoey et al., 2016). However, new studies are showing that individual performance can be sustained if environmental change is experienced during critical windows within- and between-generations (Donelson et al., 2018). For example, the intertidal copepod, Tigriopus californicus, showed a plastic shift in adult upper thermal tolerance when they developed at elevated temperature compared to those developed at control temperature throughout the larval stage (Healy et al., 2019). Similarly, the growth rate of juvenile sheepshead minnow is reduced at elevated temperature, but not when parents were also exposed to elevated temperature (Salinas and Munch, 2012). Nevertheless, our ability to project the extent of thermal phenotypic plasticity beyond the few species that have been studied to date is limited. For example, beneficial plasticity does not always occur (Marshall and Uller, 2007; Bautista and Burggren, 2019) and the magnitude of temperature change can be important in inducing phenotypic change (Grenchik et al., 2013; Shama, 2017). The timing of warming in development can also determine whether phenotypic change occurs, and in some cases development at higher temperature during early life stages is critical for phenotypic adjustment (Shama et al., 2014; Le Roy et al., 2017). The diversity of phenotypic responses to environmental change make projecting future biological responses challenging, but an understanding of the underlying mechanisms could enhance our capacity to predict plasticity in the future.
The coral reef damselfish, Acanthochromis polyacanthus, has been extensively studied in terms of within- and between-generational plasticity to elevated temperature. Differing capacities for phenotypic plasticity has been observed between populations of A. polyacanthus from the Great Barrier Reef (GBR) (Donelson and Munday, 2012; Donelson et al., 2012). Specifically, restoration of aerobic physiology was observed within a generation in the Heron Island population (near the cold boundary of the species range on the GBR) when fish developed from early life in elevated temperature conditions (Donelson and Munday, 2012). In contrast, there was only partial restoration of aerobic performance within one generation in the Palm Island population (middle of the species range on the GBR) (Donelson and Munday, 2012; Donelson et al., 2012; Veilleux et al., 2015). Full restoration of aerobic scope only occurred when two generations (parents and offspring) were subjected to warmer conditions. The physiological processes involved were also different between the two populations. The Heron Island fish were able to restore aerobic scope by reducing resting metabolic rates (RMR) when reared from early life at +3°C (within-generational treatment) compared to the present-day control group. Conversely, the Palm Island population that were reared at +3°C slightly reduced their RMR, but not enough to restore aerobic scope (Donelson et al., 2011; Donelson and Munday, 2012). However, when Palm Island fish were reared at +3°C for two generations (transgenerational treatment) or reared at +1.5°C for the first generation and then stepped up to +3°C for the second generation (step treatment), fish possessed enhanced aerobic performance through increased maximum metabolic rates (MMR), with the +3°C transgenerational treatment also showing a reduction in RMR (Donelson et al., 2012; Bernal et al., 2018). This implies that underlying molecular mechanisms for thermal physiological plasticity may differ for these populations, and between within- and between-generational plasticity. Understanding the molecular mechanisms governing thermal plasticity in different populations is important for predicting the response of the species to changing climatic conditions across their geographical range.
Epigenetic regulation, a process that can shape heritable phenotypes without changes in the genetic code and can mediate stable gene expression, may be one important mechanism underlying within- and between-generational phenotypic plasticity (Jablonka and Raz, 2009; Ho and Burggren, 2010; Bonduriansky et al., 2012; Duncan et al., 2014). Although epigenetic mechanisms, such as DNA methylation, have been recognized as putatively playing a role in phenotypic plasticity in the context of climate change (Marsh and Pasqualone, 2014; Munday, 2014; Putnam et al., 2016; Anastasiadi et al., 2017; Torda et al., 2017; Ryu et al., 2018), only a handful of studies have addressed relationships among whole genome DNA methylation changes, physiology, and their related gene functions in marine organisms. For example, warm acclimation for 4 weeks in an adult marine polychaete improved mitochondrial performance accompanied by shift of methylation status at 11% of analyzed CpG sites (Marsh and Pasqualone, 2014). An environmentally sensitive coral species subjected to a 6 week CO2 treatment, which mimics ocean acidification, showed reduced calcification and doubled global methylation level (Putnam et al., 2016). Similarly, sea bass exposed to warm temperatures as larvae showed a distinct global DNA methylation pattern (Anastasiadi et al., 2017). Although these studies identified broad relationships between DNA methylation and phenotype, they were unable to identify detailed mechanisms of DNA methylation and associated gene functions, which can allow a deeper level of understanding about how plasticity is regulated and controlled. This is mainly because previous studies have typically achieved low coverage of methylation detection due to the methods used (e.g., methylation sensitive amplification polymorphism sequencing or colorimetric assessment of the amount of DNA methylation). We previously studied DNA methylation changes and related genes in the Palm Island population of A. polyacanthus with high genome-wide coverage and sequencing depth (Ryu et al., 2018). We found significant changes in methylation among treatments in the F2 generation, and when correlated with the relevant phenotype (adjusted net aerobic scope), we identified genes involved in energy and nutrient homeostasis and cardiovascular functions. This implies these differentially methylated genes play a critical role in between-generational thermal plasticity in the Palm Island population of A. polyacanthus.
In this study, we investigated whether DNA methylation in the Heron Island population of A. polyacanthus was altered following exposure to elevated water temperatures (+1.5 or +3°C) from the early juvenile stage, as a putative mechanism for within-generational thermal plasticity (Donelson and Munday, 2012). We then compared these Heron Island methylome profiles to those from the F2 Palm Island population that exhibited plasticity in transgenerational and step treatments, as well as the within-generational treatment that showed limited plasticity. Identifying the similar or distinct epigenetic mechanisms associated with within- and between-generational plasticity is important to determine if they rely on similar or different molecular and physiological processes. Establishing if there are fundamentally similar or different processes operating in different examples of thermal plasticity is crucial to making predictions how fish populations will respond to climate change across their geographical range.
Materials and Methods
Experimental Design
Recently hatched juveniles of A. polyacanthus were collected from February to March 2009 from the reef slope at Heron Island (23°27′ S, 151°57′ E) on the Great Barrier Reef (Figure 1A). At this location the average water temperature at 6–8 m depth below sea level was 27°C in summer and 21.8°C in winter (from 1999 to 2008 Australian Institute of Marine Science1). Fish were transported to James Cook University’s aquarium facility where 120 juvenile fish (<3 months old) were randomly divided into three seasonally-adjusted temperature treatments modeled off the previous 10 years for the collection location: a present-day temperature (control: +0°C) and two within-generational elevated temperature treatments (+1.5 and +3°C above present-day). Fish were maintained in 60 L replicate tanks for each temperature treatment until maturity (11–14 months of age), as previously described (Donelson and Munday, 2012) (Figure 1B). Sampling of fish for tissue was completed during the Austral summer of 2009–2010 when water temperature for the three treatments were +0: 27°C, +1.5: 28.5°C, and +3: 30°C.
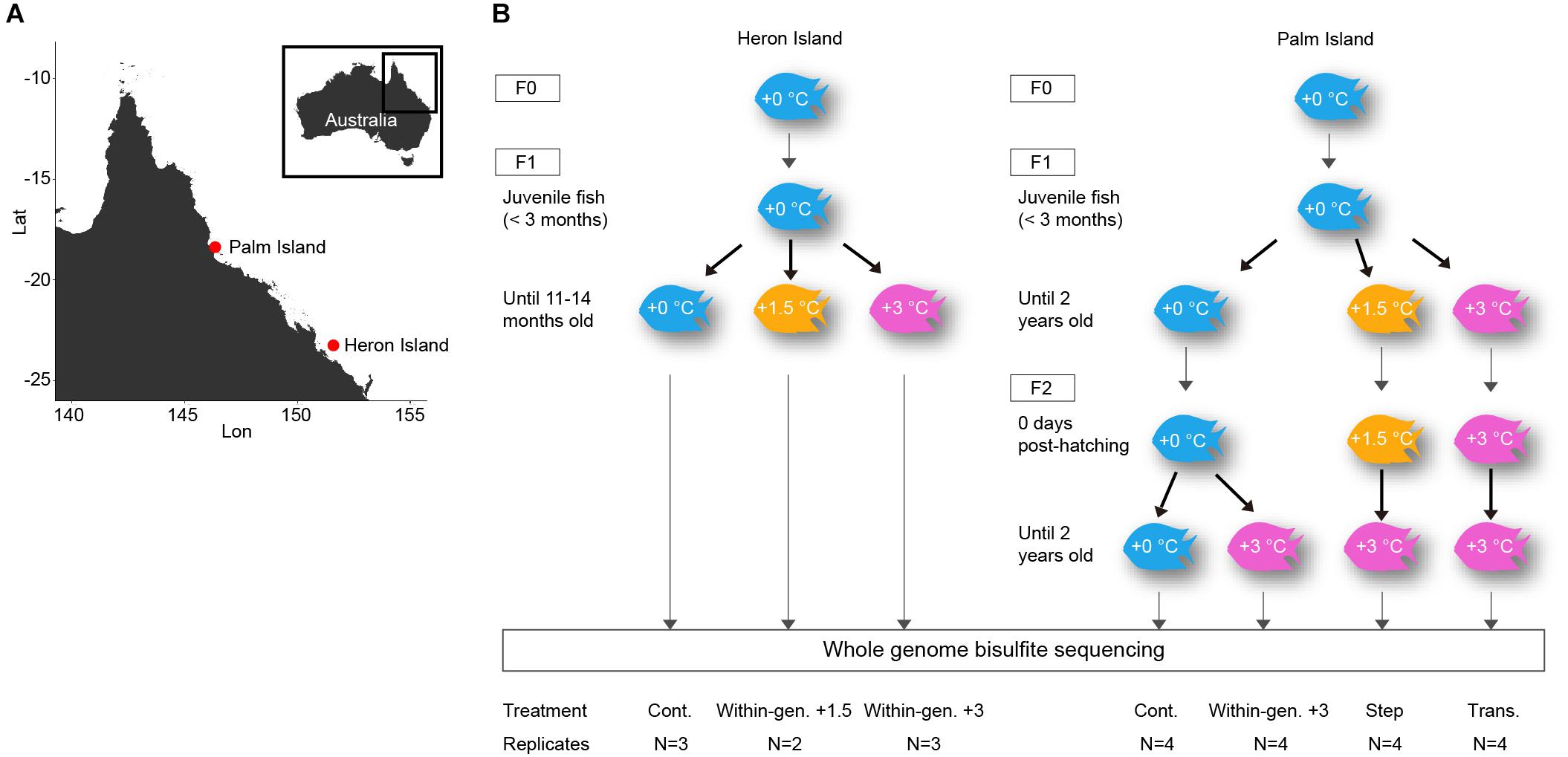
Figure 1. Experimental design to analyze DNA methylation related to thermal plasticity. (A) Sampling location of two populations of A. polyacanthus on the Great Barrier Reef, Australia. (B) Scheme of the fish rearing experiment to study within-generational plasticity (left) and transgenerational plasticity (right). The treatment names and the number of fish samples per treatment (biological replicates) are indicated below.
Liver was the target tissue because it is a major metabolic organ. Liver is an important long-term storage site for excess energy as glycogen, allowing glucose release to the blood and provision of fuel to the body as required (Wasserman, 2009). Liver also synthesizes bile and heme for fat absorption and oxygen delivery, respectively (Campbell, 2006). Adult fish from each treatment were sampled and whole livers immediately dissected, snap-frozen in liquid nitrogen, and then stored at −80°C until further processing. To avoid sex-related epigenome variations we used only liver tissue from male fish.
Nucleotide Extraction and Sequencing
Liver genomic DNA (gDNA) of 8 fish (3, 2, and 3 fish from control, within-generational +1.5°C, and within-generational +3°C groups, respectively) was extracted by homogenizing 100 mg of frozen liver tissue in 700 μL CTAB (hexadecyltrimethylammonium bromide) buffer. After adding proteinase K, the homogenate was incubated overnight at 55°C and then RNase treated for 10 min at room temperature. A standard chloroform/isoamyl alcohol extraction and ethanol precipitation method (Sambrook and Russell, 2001) was used. DNA quality and quantity were checked using NanoDrop Spectrophotometer absorbance readings (Invitrogen, Mulgrave, VIC, Australia) and on a 0.8% agarose gel.
gDNA was bisulfite-converted using Methyl-MaxiSeqTM Kit and sequenced using llumina HiSeq1500/2500 system by Zymo Research (Irvine, CA, United States) as described previously (Ryu et al., 2018). Briefly, 500 ng of genomic DNA was treated with dsDNA ShearaseTM Plus (Zymo Research), end-blunted, 3′ A-extended, and purified by DNA Clean & ConcentratorTM –5 kit (Zymo Research). DNA fragments were then ligated with adapters and filled in. EZ DNA Methylation–Lightning kit (Zymo Research) was used for bisulfite treatment and DNA fragments were sequenced on Illumina HiSeq1500/2500. All procedures were conducted according to the manufacturer’s instructions. 1.26 ∼ 1.37 × 109 reads of 101 bp were generated for each sample (Supplementary Table 1). The methylomic reads for this study were generated together with those from our previous work (Ryu et al., 2018). The quality of these methylome datasets were validated using targeted bisulfite sequencing assays in the same study.
Analysis of Whole Genome DNA Methylation Patterns
Draft genome assembly and gene models for A. polyacanthus from Heron Island were generated as described in our previous work (Veilleux et al., 2018). The assembled genome consisted of 16,609 genomic scaffolds ranging from 500 ∼ 3,215,819 bp with N50 of 527,731 bp. Base coverage was 129X on average and assembly completeness measured by CEGMA (Parra et al., 2007) was 99.19%. 23,464 gene models were identified based on ab initio and A. polyacanthus transcriptome-based gene prediction by Maker2 pipeline (Holt and Yandell, 2011).
Adapters and low quality bases in reads from the whole genome bisulfite sequencing (WGBS) were trimmed by Trim Galore v0.4 (Krueger, 2015). Trimmed reads were mapped to A. polyacanthus genome sequences from Heron Island using Bismark v0.13.1 (Krueger and Andrews, 2011) with the ‘–non-_directional’ option. Mapping efficiency was 67.17 ∼ 71.23% per sample. The bisulfite conversion rate is often used as a metric to assess the efficiency of bisulfite treatment and is calculated as numT/(numT +numC) × 100, where numC and numT represent the number of times that cytosines and thymines are observed, respectively at CHG and CHH sites. We used methylKit v.1.2.0 (Akalin et al., 2012) to identify differentially methylated regions (DMRs) as follows. Per base coverage of methylomic reads on the genome was 43 ∼53 × calculated by ‘getCoverageStats’ function (Supplementary Table 2). The ‘methRead’ function scanned the methylome mapping information and kept cytosines that were covered by a minimum of 10 reads. Highly covered bases (>99.9%) were excluded to remove PCR bias by ‘filterByCoverage’ function. Variances across entire samples were normalized by ‘normalizeCoverage’ functions. The genome was broadly categorized as eight functional units (CpG island, CpG shore, promoter, 5′ untranslated region (UTR), exon, introns, 3′ UTR, and repeats), following our previous study (Ryu et al., 2018). Methylation status of individual cytosines in each genomic region were summed per sample by the ‘regionCounts’ function. Statistical significance of differential methylation between samples was calculated using the Chi-squared test of the ‘calculateDiffMeth’ function. Multiple testing correction was performed by the Benjamini & Hochberg (BH) method of the ‘p.adjust’ function from p-values from all pairwise comparisons. DMRs for three methylation contexts (CpG, CHH, CHG; H = A, C, T) were identified using the two thresholds: adjusted p-value ≤ 0.05 and ≥ 33.3% methylation difference between two treatments. The adjacent genes to each DMR on the same scaffold were assigned using the ‘bedtools closest’ v2.23 (Quinlan, 2014).
The ‘heatmap.2’ function of the ‘gplots’ package (Warnes et al., 2016) was used to draw heatmaps. For multidimensional scaling analysis, dissimilarity between samples was calculated as ‘1 – Spearman correlation coefficient’ and confidence ellipsoid of each sample was obtained using the ‘bootmds’ function of the ‘smacof’ package (De Leeuw and Mair, 2011) with the setting of alpha = 0.05 and 1,000 bootstrap replications. Statistical significance of separation between experimental groups was assessed by ind.ctest function of the ‘ICSNP’ R package (Nordhausen et al., 2015). p-Values were adjusted for multiple testing correction using the BH method.
Functional Analysis
Homologs of A. polyacanthus proteins were identified by a BLASTP search against the NCBI non-redundant protein database with e-value of 10–4 as a threshold (Altschul et al., 1990). Gene Ontology (GO) terms of homologous proteins were then assigned to A. polyacanthus proteins using the NCBI gene2go.gz file2 and custom script. GO terms were considered when associated with more than two genes. Only Biological Process among the three GO categories (Biological Process, Molecular Function, and Cellular Component) were analyzed to focus on the biological outcome of each gene rather than a specific molecular activity (Molecular Function) or a location relative to cellular structures (Cellular Component) of the gene (Gene Ontology Consortium, 2008). Statistical significance of GO enrichment in DMR genes from all groups or between experimental groups were calculated by cumulative hypergeometric test and adjusted by the BH method using the custom R script respectively.
Comparison of Differential Methylation Between Within- and Between-Generational Treatments
To allow comparison of DNA methylation profiles for fish exposed to elevated temperature within- and between-generations, we used the methylome dataset from our previous study (Ryu et al., 2018). This dataset was generated from F2 A. polyacanthus from the Palm Island population (central Great Barrier Reef, Australia; 18°37′ S, 146°30′ E) that had been reared at control and elevated temperatures for two generations (Figure 1A). Briefly, F1 offspring (<3 months old) of wild-caught adult fish were split between three temperature treatments: a present-day control for the collection location +0°C (mean winter: 23.2°C and summer: 28.5°C) as well as +1.5 and +3°C above present-day (seasonally cycling temperature treatments). Fish were reared in treatment conditions in 40–60 L aquaria until 2 years of age when fish were mature. At this time pairs of fish from unrelated families were randomly mated (Donelson et al., 2012 for more details). F2 offspring from the three F1 parental treatments were divided between two F2 developmental conditions, +0 and +3°C immediately after hatching and remained in treatment until 2 years of age. This produced four F2 treatment groups: control (F1: +0°C, F2: +0°C), within-generational (F1: +0°C, F2: +3°C), step (F1: +1.5°C, F2: +3°C), and transgenerational (F1: +3°C, F2: +3°C) (Figure 1B).
To minimize the effect of sex on the downstream analysis, two males and two females were selected for each Palm Island population treatment. DNA extraction from adult liver and WGBS was conducted as written above for Heron Island fish. The methylomic reads were aligned to the genome assembly from Palm Island fish. Each sample has 0.95 ∼ 1.36 × 109 reads of 101 bp and yielded 34 ∼ 62 × coverage depth after alignment to the genome sequences. The same computational methods used to identify DMRs in Heron Island populations were used except for one parameter: to remove the influence of unbalanced sex ratio in this population, the information of males and females was included as the ‘covariates’ parameter in the ‘calculateDiffMeth’ function of methylKit (Akalin et al., 2012). DMRs were identified using the same thresholds (adjusted p-value ≤ 0.05 and ≥ 33.3% methylation difference between two treatments), which resulted in 445 CpG DMR genes.
Orthology was calculated between Heron and Palm Island populations’ gene models using Inparanoid (O’brien et al., 2005). This resulted in 17,423 ortholog groups with 17,569 and 17,758 genes from each population, respectively.
Results
Measuring Within-Generational Changes in Whole-Genome Methylation
We extracted methylomes from liver samples that were collected from Heron Island populations of A. polyacanthus exposed to control (+0°C) and increased temperatures (+1.5 and +3.0°C) from early life (Figure 1B). This resulted in 841.6 ∼ 884.8 million reads that were uniquely mapped, yielding from 67.2 to 71.2% mapping efficiency (Supplementary Table 2). Methylated cytosines ranged from 61.1 to 63.4% in the CpG context and from 0.77 to 3.68 in CHG and CHH contexts, across samples (Supplementary Table 2). Average bisulfite non-conversion rates were from 0.57 to 2.91% per sample, which is comparable to our previous study (0.84 to 3.46%) (Ryu et al., 2018). Base coverage of methylome reads were ≥ 43 × for the three methylation contexts for all Heron Island samples (Supplementary Table 2).
Differential methylation in CpG, CHH, and CHG contexts was calculated between pairs of treatments and 480, 88, and 3 DMRs were identified, respectively (Table 1). As there were so few CHG DMRs this context was not analyzed further. CpG DMRs were mostly located on exons (22.24%), repeats (21.72%), and introns (20.81%) whilst CHH DMRs were mostly located in repeats (42.66%), introns (16.78%), and CpG shores (13.99%) (Figure 2C and Supplementary Figure 1C, Tables 4, 5).
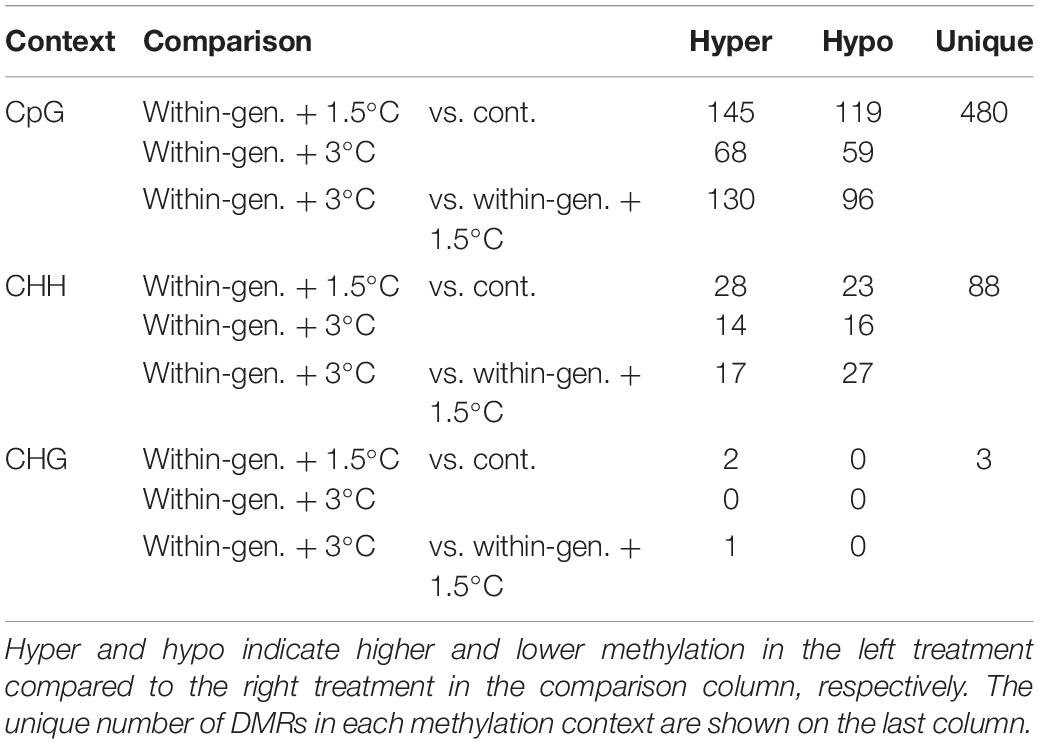
Table 1. The number of differentially methylated regions (DMRs) between experimental groups from the Heron Island population.
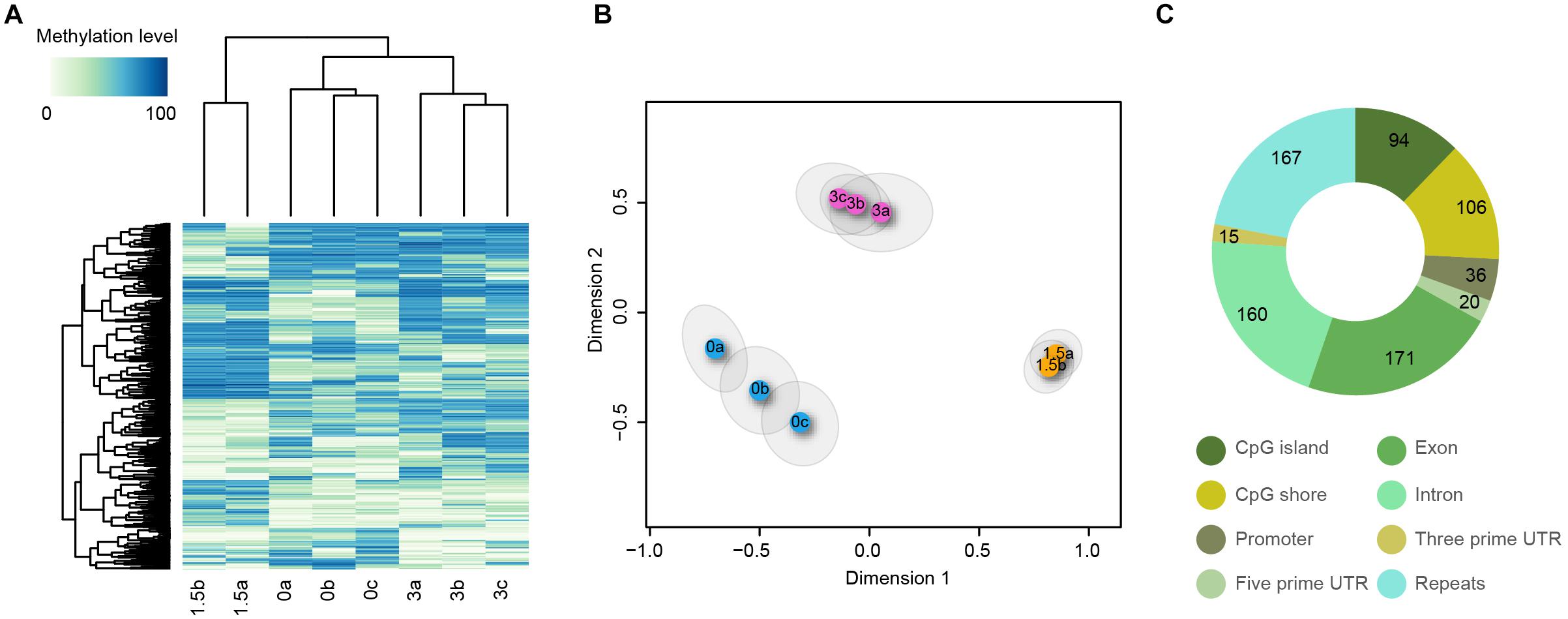
Figure 2. Differential methylation in the CpG context for within-generational plasticity. (A) Heatmap of methylation levels for DMRs is shown with the color key in the upper left corner. Column labels indicate the sample code represented by the combination of the increased temperature (0, 1.5, 3) and individual fish number (a, b, and c). (B) Multidimensional scaling analysis of DMRs. Each circle corresponds to one fish sample and experimental groups are marked by different colors. 95% confidence ellipses from 1,000 bootstrapping are shown for each sample. No overlap of ellipses for CpG DMRs indicates that CpG methylation patterns are significantly different among experimental groups. (C) Distribution of DMRs for CpG contexts across categorized genomic elements.
To determine if within-generational exposure to increased temperatures in the Heron Island population affected global DNA methylation patterns, we performed a multidimensional scaling analysis using the methylation level of DMRs. Individual fish samples were clustered by CpG DMRs and were separated by temperature treatment (Figures 2A,B). Statistical tests also showed clear separation of control samples from the elevated temperature treatments (Supplementary Table 3). These patterns were less evident for the CHH context (Supplementary Figures 1A,B and Table 3), which is similar to our previous observation of methylation patterns in control, within-generational, step, and transgenerational in Palm Island fish (Ryu et al., 2018).
Differentially Methylated Gene Functions for Within-Generational Plasticity
We identified 372 CpG DMR genes (nearest genes to any given CpG DMRs, Supplementary Table 5) in the Heron Island population. GO enrichment analysis of these 372 genes showed 20 enriched terms (adjusted p-value < 0.05) (Supplementary Table 6) mostly related to animal development (e.g., ‘cardiac neural crest cell development involved in heart development,’ ‘central nervous system projection neuron axonogenesis,’ and ‘retina layer formation’) and physiological homeostasis (e.g., ‘cellular chloride ion homeostasis,’ ‘insulin catabolic process,’ and ‘hypotonic response’).
Further investigation of enriched GO terms identified by comparing treatments in a pairwise manner also showed similar but slightly different categories that were treatment-dependent. Most enriched GO terms were related to pathways involved in developmental processes and cardiovascular functions, and the majority of these were found when comparing control to within-generational +1.5°C treated fish (Figure 3). GO terms related to diverse developmental processes, such as ‘bone morphogenesis,’ ‘multicellular organism growth,’ ‘type B pancreatic cell development,’ and ‘neural retina development’ were the most prevalent functions. The enriched terms for cardiovascular physiology included ‘cardiac muscle hypertrophy,’ ‘cardiac myofibril assembly,’ and ‘regulation of heart rate. Nutrient control and heat stress functions were also noteworthy. ‘Insulin catabolic process’ was the most significantly enriched GO term in hypomethylated DMR genes from both elevated temperature groups (both +1.5 and +3°C) compared to control. ‘Glucose homeostasis’ was another enriched term in the comparison between control and within-generational +1.5°C. The GO term ‘regulation of cellular response to heat’ was enriched among DMR genes that were hypomethylated in within-generational +1.5°C fish compared to control.
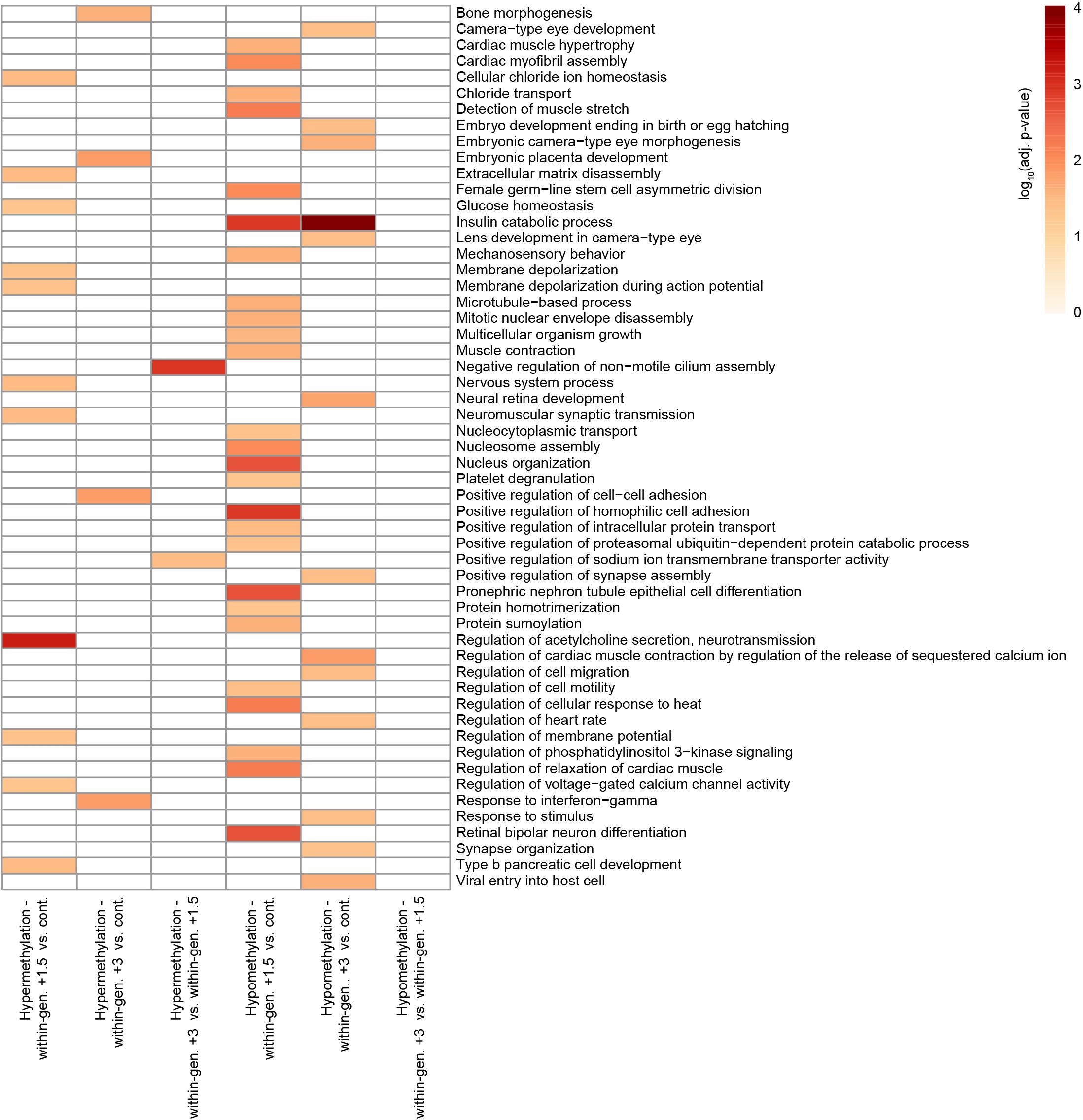
Figure 3. Gene Ontology enrichment of DMR genes related to within-generational plasticity. Significantly enriched GO terms (adjusted p < 0.05) between two experimental groups are listed on the right side of the heatmap. Adjusted p-values are –log10 transformed and colored as indicated on the color key.
This analysis also revealed that functions differed depending on the magnitude of temperature change. The greatest number of enriched GO terms were found when the fish reared in elevated temperatures were compared to control fish, but not when the elevated temperature fish (+1.5 and +3°C groups) were compared to each other (Figure 3). For example, the largest number of enriched terms were found among hypomethylated DMR genes in within-generational +1.5°C fish compared to control fish (26 terms), while only two terms were enriched in within-generational +3°C fish compared to within-generational +1.5°C fish. This suggests that major change of physiological functions occurs when a temperature change is experienced during early life, but the magnitude of change, either +1.5 and +3°C, is relatively unimportant.
Comparison of Within- and Between-Generational Epigenetic Signatures
To compare epigenetic mechanisms between within- and between-generational plasticity, we performed the same GO enrichment analysis on the DNA methylome dataset of the F2 population from Palm Island (Ryu et al., 2018). First, we identified 445 CpG DMR genes by pairwise comparison of WGBS data from the four Palm Island treatments (control, within-generational +3°C, step, and transgenerational) (Supplementary Table 7). Enrichment analysis on all 445 genes in the F2 Palm Island population identified only three significant GO terms (adjusted p-value < 0.05) (Supplementary Table 8), while we identified 20 different GO terms among the 372 CpG DMR genes in the Heron Island population (Supplementary Table 6). Enrichment analysis per treatment pair also showed different patterns with regard to the Heron Island population (Supplementary Figure 2). Five or fewer GO terms were enriched across treatment pairs and there was no overlap with the enriched terms from the Heron Island population.
Because there was no overlap of GO terms between two populations, we examined if there was similarity between within- and between-generational responses at the individual gene level. Among 372 and 445 DMR genes from Heron and Palm Island populations, respectively, only 12 genes were differentially methylated in both populations (Figure 4). These include genes involved in regulation of cell migration (slit2), cell volume homeostasis (znf706), cytoskeleton organization (dtnb), DNA replication (rfc4), immune response (btn1a1), nutrient metabolism (grpr, ren, spon2, and srebf1), sodium channel gating (unc80), and transcriptional regulation (rax2). Interestingly, all 12 genes that were differentially methylated in at least one of the Heron Island within-generational treatments were also differentially methylated in the Palm Island transgenerational or step treatments compared to control or within-generational treatments.
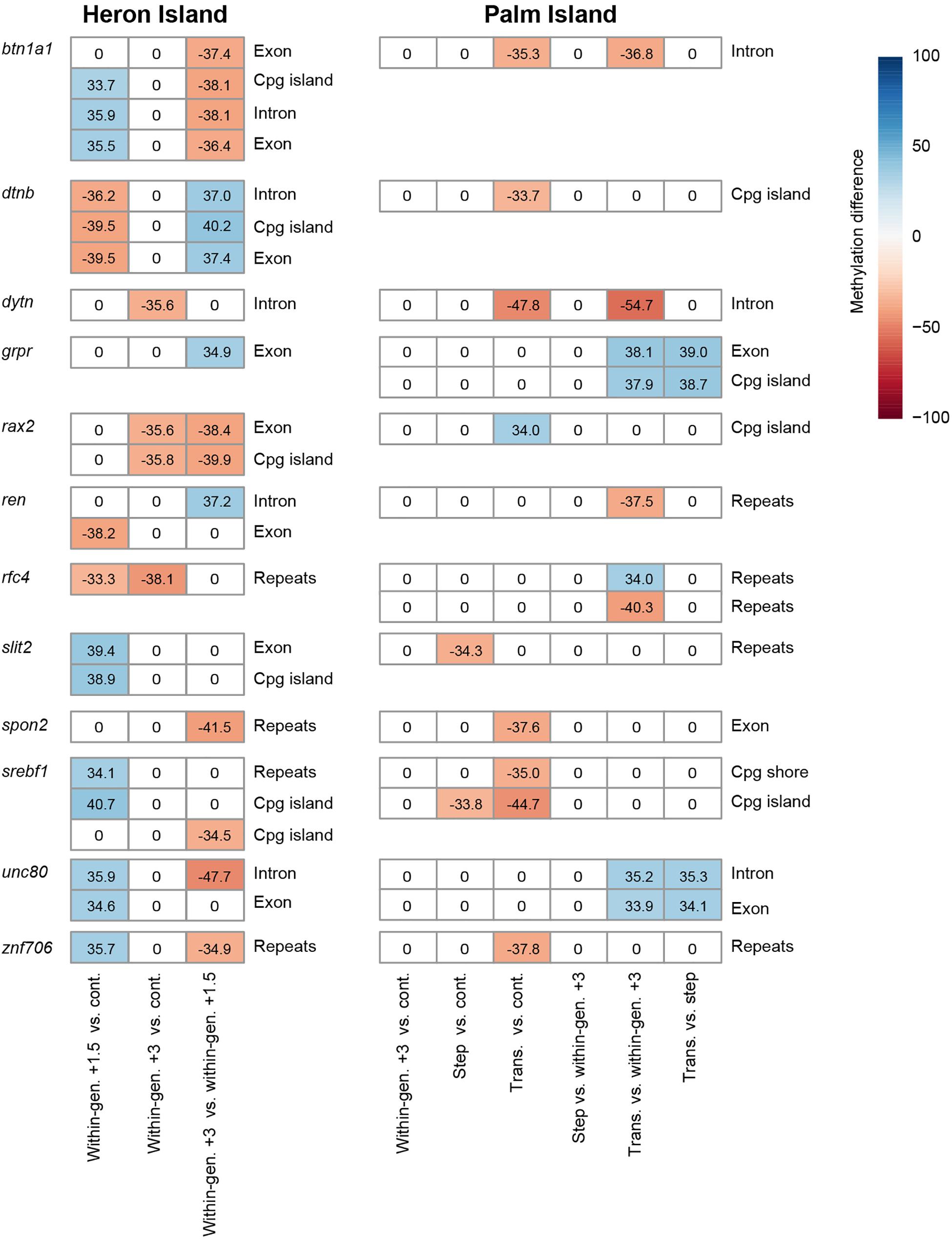
Figure 4. Common DMR genes in Heron and Palm populations. These 12 genes may be involved in both within- and between-generational plasticity to elevated temperatures. Each row and column correspond to one DMR and comparison of two treatments, respectively. The colors and values indicate the percentage differences of methylation level of genomic regions. Methylation differences for non-DMRs are represented as zero.
Discussion
There is ample evidence that environmental experiences during early development can affect phenotypes throughout life; however, the governing cellular mechanisms, especially the role of non-genetic mechanisms in thermal plasticity, are not well-understood. A mechanistic understanding is essential to predict the response of natural populations living in rapidly changing environments. In this study, we investigated if changes in DNA methylation are associated with the capacity for within-generational thermal plasticity of a coral reef fish and compared these methylation patterns to those in a lower latitude population that exhibited between-generational thermal plasticity. In both elevated temperature treatments, the function of DMR genes was related to metabolic control, developmental, cardiovascular, and heat-response functions. However, we found very little overlap between within- and between-generational epigenetic signatures of thermal plasticity from the two populations of the same species; only 3% of the DMR genes from each population were shared.
Exposure to elevated temperatures (+1.5 and +3°C) from the early juvenile stage in the Heron Island population altered the CpG methylation landscape across the genome compared to controls, in addition to differences between the temperature treatments. This suggests that exposure to an altered thermal environment from early life was sufficient to alter the epigenome, which may be related to the phenotype. The strong differentiation of CpG methylation and limited difference for CHH methylation may be explained by their differing maintenance mechanisms. Once established, CpG methylation is preserved with high accuracy; DNA methyltransferases recognize hemimethylated CpG sites generated during DNA replication to re-establish appropriate methylation patterns. However, a maintenance mechanism such as this is unknown for CHH or CHG contexts in animals (Dyachenko et al., 2010). Previously, we obtained similar results for Palm Island A. polyacanthus methylomes in that only CpG and not CHH or CHG methylation was affected following within-generational, step, and transgenerational exposure to elevated temperatures (Ryu et al., 2018). Together with the results from this study, our data suggests that CpG methylation is the most appropriate context in which to evaluate either within- or between-generational plasticity in reef fish, at least in A. polyacanthus.
Functional analysis of Heron Island DMR genes showed significant enrichment of GO terms related to metabolic control and developmental, cardiovascular, and heat-response functions. In fish, as in other ectotherms, basal metabolic rate increases as ambient temperatures increases (Fry and Hart, 1948; Portner and Farrell, 2008; Clark et al., 2013); hence, maintaining efficient metabolic performance may be critical for normal physiological activities as the oceans warm (Portner, 2010). One metabolic term, ‘insulin catabolic process’ was enriched among hypomethylated genes in both elevated temperature treatments (+1.5 and +3°C) compared to the current-day control temperature. Insulin serves as a master regulator of energy and metabolic functions, by promoting glucose uptake into cells (Rui, 2014). In heat-stressed animals, basal insulin secretion and sensitivity increases in order to compensate for altered intracellular energetics (Victoria Sanz Fernandez et al., 2015). In our study, the ceacam1 (carcinoembryonic antigen-related cell adhesion molecule 1) gene, which is hypomethylated in both elevated temperature treatments, encodes a glycoprotein that controls hepatic insulin sensitivity and lipogenesis by mediating insulin clearance and fatty acid synthase activity (DeAngelis et al., 2008; Russo et al., 2016). ‘Glucose homeostasis’ was enriched in the comparison between +1.5°C and control treatments. An associated ‘glucose homeostasis’ gene was gcgr, which was hypermethylated in +1.5°C fish compared to control fish. Its encoded protein, glucagon receptor, binds to glucagon (a peptide hormone secreted by α cells of the pancreas in response to low blood glucose level) and then promotes the conversion of stored glycogen in liver to glucose and is also involved in gluconeogenesis (Miller and Birnbaum, 2016). The glucagon receptor’s mechanisms, which increase the hepatic glucose production and bloodstream glucose levels, are opposite to insulin action that reduces bloodstream glucose levels. Balanced modulation of glucagon and insulin pathways is critical to maintain stable blood glucose level, which is a major cellular energy source. Therefore, epigenetic control of key hepatic metabolic genes may be an important mechanism for within-generational thermal plasticity.
Many enriched functions in the Heron Island elevated temperature treatments were related to developmental processes such as ‘multicellular organism growth’ and ‘type B pancreatic cell development.’ The selenom and h3f3a genes, which encode selenoprotein M and histone H3.3 respectively, were associated with ‘multicellular organism growth’ and were hypomethylated in +1.5°C fish compared to control fish. Selenium is an essential micronutrient with antioxidant activities and, in chickens, contributes to growth performance and regulation of blood glucose (Zoidis et al., 2018). Selenoprotein M, one of the selenoproteins that chemically incorporate selenium during translation, has regulatory roles in energy metabolism and body weight, identified through a knockout study in mice (Pitts et al., 2013). Moreover, when overexpressed in rats, this protein kept the body in high antioxidant status by altering antioxidant enzyme activity and immune cell composition (Hwang et al., 2008). Thus, epigenetic control of the selenom gene may help fish balance cellular redox state and energy usage during juvenile growth when reared in warmer water, potentially contributing to within-generational plasticity. H3.3, which was hypomethylated in the +1.5°C treatment compared to control, is a histone variant that can replace the canonical histones H3.1 and H3.2, and proper replacement among H3 proteins is the frequent target in the epigenetic regulation for multiple processes such as development, fertilization, and somatic growth (Tang et al., 2015). The wnt5a gene, associated with the GO term ‘type B pancreatic cell development,’ was hypermethylated in +1.5°C fish. Type B pancreatic cells (commonly called β cells) are the predominant cell in the islets of Langerhans, where they synthesize insulin. In addition to its close association to a variety of processes such as cell proliferation, cell fate determination, and cardiovascular diseases (Bhatt and Malgor, 2014), the WNT5A protein has been suggested to be an essential molecule for the formation of pancreatic islets and proliferation of Type B cells in vertebrates (Kim et al., 2005). Differential methylation of genes involved in developmental functions seems counter-intuitive, since we sequenced the epigenome of adult livers; however, recent reports indicate that DNA methylation can reflect the epigenetic history of cellular activities and can be unchanged through development and across tissues (Hon et al., 2013; Huse et al., 2015). Although the possibility that these genes may have additional roles in adult liver cannot be ruled out, DNA methylation of these genes may have been established during early stages of development to maximize the physiological performance in a warmer environment.
Cardiovascular physiology (e.g., enrichment terms ‘cardiac myofibril assembly,’ ‘regulation of heart rate,’ and ‘regulation of relaxation of cardiac muscle’) was also significantly enriched among DMR genes in the Heron Island elevated temperature treatments. Delivering sufficient oxygen to tissues is a limiting factor in the thermal tolerance in fish, as above a critical temperature the cardiovascular system cannot meet the higher demand for oxygen in the tissues (Portner and Farrell, 2008). Thus, enhanced vascular structure may be a central component of thermal plasticity. The titin protein-encoding gene, which was hypomethylated in Heron +1.5°C fish compared to controls, is a key component in the assembly and operation of vertebrate muscles, and is also responsible for the morphogenesis of the vascular network (May et al., 2004). Two genes, prkaca and slc8a1, were hypomethylated in Heron Island +3°C fish compared to controls and were associated with two enriched terms: ‘regulation of heart rate’ and ‘regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion.’ The prkaca gene encodes the catalytic subunit α of protein kinase A that is responsible for post-translational modification to alter various cellular protein activities. Protein kinase A regulates vascular development in vertebrates (Nedvetsky et al., 2016) and mis-regulation of this protein’s activities is related to many cardiovascular diseases (Turnham and Scott, 2016). Moreover, protein kinase A serves as the primary mediator of glucagon’s effect on hepatic metabolism by phosphorylating many enzymes in the glucoregulatory pathway (Miller and Birnbaum, 2016), implying the possible multifunction of this protein in thermal plasticity. SLC8A1 is a sodium/calcium exchanger protein of cardiomyocytes and is responsible for cardiac contraction (Takimoto et al., 2002). This protein also has a putative role in blood pressure regulation according to the transgenic mouse study (Takimoto et al., 2002; Warren et al., 2017). Thus, modulation of DNA methylation of the genes associated with vascular architecture modification in response to altered metabolic demands could be a key characteristic of within-generational thermal plasticity, and has been similarly reported in transgenerational plasticity (Ryu et al., 2018).
Heat stress can induce negative physiological effects, including protein unfolding, collapse of cytoskeleton networks, mis-localization of organelles, reduced number of mitochondria, aberrant RNA splicing, and changes in membrane morphology (Richter et al., 2010). Therefore, minimizing detrimental effects caused by elevated temperatures is critical to maintain normal physiology. In our study, the GO term ‘regulation of cellular response to heat’ was enriched among DMR genes that were hypomethylated in +1.5°C fish compared to control. For example, the nup98 gene associated with this term is found to be associated with heat shock recovery by reactivating the transcription of genes suppressed by the heat shock (Capelson et al., 2010). The alpha-crystallin B chain heat shock protein encoded by the cryab gene prevents aggregation of proteins under a variety of stressful conditions and keeps the structural integrity of heart and muscle tissues under heat stress (Wójtowicz et al., 2015; Yin et al., 2019). Methylation control of these genes may help the fish to adjust to the effects of lifelong exposure to increased temperatures.
Epigenetic Signature Shared Between Plastically Adjusted Within- and Between-Generational Treatments
Fish from Heron Island phenotypically adjusted aerobic scope by reducing RMR without a significant change in MMR (Donelson and Munday, 2012). In contrast, the F2 Palm population did not achieve within-generational plasticity, but was able to do so when both parents and offspring were exposed to increased temperatures, either in the step treatment by decreasing RMR or in the transgenerational treatment by increasing MMR and decreasing RMR (Donelson et al., 2012; Bernal et al., 2018). This implies that there could be different physiological mechanisms and underlying genomic responses for within- and between-generational plasticity of aerobic scope, as well as between populations, following exposure to predicted future temperatures. Indeed, we found that each population might employ a different epigenetic toolkit in order to plastically adjust to warmer conditions. Only 12 DMR genes were common among 372 and 445 DMR genes from Heron and Palm Island fish, respectively. However, these 12 genes might be linked to the phenotypic reductions in RMR of both populations. Independent GO analysis for each population also suggested distinct functions may be employed for within- and between-generational plasticity.
These 12 genes shared by the two populations are involved in various regulatory functions for cell migration (slit2) (Zeng et al., 2018), cell volume homeostasis (znf706) (Dossena et al., 2011), cytoskeleton organization (dtnb) (Veroni et al., 2007), DNA replication (rfc4) (Kim and Brill, 2001), immune response (btn1a1) (Smith et al., 2010), nutrient metabolism (grpr, ren, spon2, and srebf1) (Persson et al., 2002; Ferre and Foufelle, 2010; Zhu et al., 2014; Silva et al., 2017), sodium channel gating (unc80) (Lear et al., 2013), transcriptional regulation (rax2) (Wang et al., 2004), and one had unknown function (dytn).
The overlapping differential methylation in genes associated with nutrient delivery and metabolism is not unexpected since these are known to be critical physiological attributes for survival of ectotherms in warming conditions (Portner and Knust, 2007; Portner and Farrell, 2008). The gene ren was differentially methylated in both populations (hypomethylated in Heron Island +1.5°C fish vs. controls, hypermethylated in Heron Island +3°C fish vs. +1.5°C fish, and hypomethylated in Palm Island transgenerational fish. vs. +3°C fish) and is particularly interesting as the encoded protein, RENIN, is the master hormone mediating the volume of extracellular fluid and blood pressure (Persson, 2003). As a component of the renin-angiotensin system, this protein catalyzes angiotensinogen released from the liver, which is eventually converted into angiotensin II, the vasoactive peptide (Persson, 2003). Renin-angiotensin system is also transcriptionally regulated in liver to control glucose and lipid level through hepatic insulin signaling (Silva et al., 2017). We speculate that epigenetic control on this protein may be a key mechanism to improve blood flow and nutrient homeostasis in order to plastically adjust to warmer conditions in A. polyacanthus. The grpr (gastrin-releasing peptide receptor) gene was hypermethylated in Heron Island +3°C fish vs. +1.5°C fish, hypermethylated in Palm Island transgenerational fish vs. +3°C fish, and hypermethylated in Palm Island transgenerational fish vs. step fish. Its encoded product, GRPR, mediates insulin secretion in pancreatic islet cells (Persson et al., 2002) and regulates glucose metabolism by modulating components of the aerobic glycolysis pathway (Rellinger et al., 2015). A recent mice model study suggested that GRPR may play an important role in hepatic ischemia injury (a status characterized by an insufficient supply of oxygen and nutrients), although the underlying molecular mechanism is yet unclear (Guo et al., 2019). SPON2 (spondin-2), whose encoding gene was hypomethylated in Heron Island +3°C fish vs. +1.5°C fish and also hypomethylated in Palm Island transgenerational fish vs. control, controls hepatic lipid metabolism and insulin resistance in rodents (Zhu et al., 2014). Furthermore, SPON2 is known to protect from cardiovascular diseases (Yan et al., 2011) and function in initiation of innate immunity (He et al., 2004) in rodent studies. SREBF1 (sterol regulatory element-binding protein 1), whose encoding gene was differentially methylated in both populations (hypermethylated in Heron Island +1.5°C fish vs. controls, hypomethylated in Heron Island +3°C fish vs. +1.5°C fish, hypomethylated in Palm Island step fish vs. control, and hypomethylated in Palm Island transgenerational fish vs. control), is the master regulator of the lipogenesis in liver in response to environmental signals such as nutrient demand and mediating the transcription of glycolytic and lipogenic genes in insulin- and sterol-dependent manner (Ferre and Foufelle, 2010; Shao and Espenshade, 2012). Epigenetic control on these regulatory genes involved in cell metabolism might be critical for the strict maintenance of homeostatic status at high temperatures.
There is insufficient data available to understand the likely role in thermal plasticity for the other genes that were differentially methylated in both populations (slit2, btn1a1, dtnb, dytn, rax2, rfc4, unc80, and znf706). The slit2 (slit guidance ligand 2) gene was differentially methylated in treatments where temperature was increased +1.5°C within-generations (Heron Island +1.5°C fish vs. controls and Palm Island step fish vs. control). In mammals, the encoded protein, SLIT2, modulates the migration of hepatic stellate cells, the major cell type involved in fibrogenic activity in the liver in chronically abnormal states such as hepatic fibrosis caused by metabolic disorder (Chang et al., 2015; Zeng et al., 2018). The remaining seven genes are not well-studied in relation to stress responses and/or their function in the liver. Although the role of these genes needs to be investigated further in fish liver following within- and between-generation exposure to increased temperatures, our results suggest that they may be useful as epigenetic markers for thermal plasticity.
The epigenetic patterns of within- and between-generational plasticity for the Heron and Palm Island populations of A. polyacanthus could be partially caused by different inheritance mechanisms for DNA methylation. Compared to the epigenomes for within-generational plasticity, which are only transmitted through somatic cell divisions, the epigenomes for between-generational plasticity include transmission through germ and somatic cell division. In mammals, the latter processes include epigenetic reprogramming events during gametogenesis and early embryogenesis, in which DNA methylation marks on the genome are erased and remodeled (Morgan et al., 2005; Messerschmidt et al., 2014). Although not extensively investigated in fish, epigenetic reprogramming differs by fish species. For example, zebrafish did not show global demethylation and recovery, while medaka showed similar reprogramming patterns to mammals (Jiang et al., 2013; Ortega-Recalde et al., 2019; Wang and Bhandari, 2019a, b). If complicated reprogramming event occurs in A. polyacanthus, modifications of epigenetic information during reprogramming (e.g., incomplete erasure and recovery) are possible for Palm Island fish (Kearns et al., 2000; Jacob and Moley, 2005). Furthermore, the exposure of Palm Island fish to increased temperatures for longer time frames compared to Heron Island fish (two vs. one generations, respectively) may increase the chances that the genome could be subjected to modification of DNA methylomes, such as replication-dependent passive demethylation or epigenetic drift that can dilute epigenetic memory (Guo et al., 2014; Messerschmidt et al., 2014; Ciccarone et al., 2018). In future studies, it would be of interest to delve more deeply into the cellular mechanisms of methylation inheritance and reprogramming in A. polyacanthus and other species that show differing capacities for within- and between-generational plasticity.
Conclusion
We performed large scale epigenotyping at the single nucleotide resolution in a reef fish population that can plastically adjust to elevated temperature within a generation. We then compared these epigenetic profiles to those of another population of the same species that failed to plastically adjust within a generation, but which acclimated in the second generation to the same environmental warming. Analysis of genome-wide differential methylation of liver tissue revealed genes involved in nutrient control, developmental processes, cardiovascular functions, and response to heat stress were highly enriched in the population capable of within-generational plasticity compared with the other population. Consequently, these DMR genes might serve as hot spot regions for epigenetic regulation of within-generational plasticity. We also identified 12 shared DMR genes for both types of plasticity, which might serve as part of a general molecular toolkit for thermal plasticity. Exposure to predicted increased temperatures at early life stages seemed to be imprinted in the form of DNA methylation around genes improving oxygen delivery, nutrient usage, and tolerance to heat stress. Our results indicate that DNA methylation may be an important mechanism associated with buffering the effects of elevated temperature in reef fishes, both within- and between-generations.
Data Availability Statement
Short reads from methylome sequencing have been deposited in GenBank under BioProject ID PRJNA348663.
Ethics Statement
This project was carried out in accordance with the recommendations of James Cook University Animal Ethics Committee (JCU Ethics A1233 and A1415).
Author Contributions
JD and PM managed the fish rearing experiments. HV prepared samples for sequencing. TRy extracted genomic DNA for methylome sequencing. TRy, HV, and JD selected fish samples for sequencing. TRy and TRa designed the computational analysis. TRy performed the analysis, interpreted the results, and drafted the manuscript. IJ contributed on the data analysis. TRy, HV, JD, PM, and TRa completed the manuscript.
Funding
This work was supported by the Competitive Research Funds OCRF-2014-CRG3-62140408 from the King Abdullah University of Science and Technology to TRa and PM. TRy acknowledges the support from the APEC Climate Center. PM was supported by the Australian Research Council (ARC) and the ARC Centre of Excellence for Coral Reef Studies. HV and JD were supported by funding from the King Abdullah University of Science and Technology and the ARC Centre of Excellence for Coral Reef Studies.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Eike Joachim Steinig and Clara Ortiz Alvarez (James Cook University) for assisting methylome sequencing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00284/full#supplementary-material
Footnotes
References
Akalin, A., Kormaksson, M., Li, S., Garrett-Bakelman, F. E., Figueroa, M. E., Melnick, A., et al. (2012). methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13:R87. doi: 10.1186/gb-2012-13-10-r87
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Anastasiadi, D., Díaz, N., and Piferrer, F. (2017). Small ocean temperature increases elicit stage-dependent changes in DNA methylation and gene expression in a fish, the European sea bass. Sci. Rep. 7:12401. doi: 10.1038/s41598-017-10861-6
Bautista, N. M., and Burggren, W. W. (2019). Parental stressor exposure simultaneously conveys both adaptive and maladaptive larval phenotypes through epigenetic inheritance in the zebrafish (Danio rerio). J. Exp. Biol. 222:jeb208918. doi: 10.1242/jeb.208918
Bernal, M. A., Donelson, J. M., Veilleux, H. D., Ryu, T., Munday, P. L., and Ravasi, T. (2018). Phenotypic and molecular consequences of stepwise temperature increase across generations in a coral reef fish. Mol. Ecol. 27, 4516–4528. doi: 10.1111/mec.14884
Bhatt, P. M., and Malgor, R. (2014). Wnt5a: a player in the pathogenesis of atherosclerosis and other inflammatory disorders. Atherosclerosis 237, 155–162. doi: 10.1016/j.atherosclerosis.2014.08.027
Bonduriansky, R., Crean, A. J., and Day, T. (2012). The implications of nongenetic inheritance for evolution in changing environments. Evol. Appl. 5, 192–201. doi: 10.1111/j.1752-4571.2011.00213.x
Campbell, I. (2006). Liver: metabolic functions. Anaesth. Intensive Care Med. 7, 51–54. doi: 10.1383/anes.2006.7.2.51
Capelson, M., Liang, Y., Schulte, R., Mair, W., Wagner, U., and Hetzer, M. W. (2010). Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140, 372–383. doi: 10.1016/j.cell.2009.12.054
Chang, J., Lan, T., Li, C., Ji, X., Zheng, L., Gou, H., et al. (2015). Activation of Slit2-Robo1 signaling promotes liver fibrosis. J. Hepatol. 63, 1413–1420. doi: 10.1016/j.jhep.2015.07.033
Ciccarone, F., Tagliatesta, S., Caiafa, P., and Zampieri, M. (2018). DNA methylation dynamics in aging: how far are we from understanding the mechanisms? Mech. Ageing Dev. 174, 3–17. doi: 10.1016/j.mad.2017.12.002
Clark, T. D., Sandblom, E., and Jutfelt, F. (2013). Aerobic scope measurements of fishes in an era of climate change: respirometry, relevance and recommendations. J. Exp. Biol. 216, 2771–2782. doi: 10.1242/jeb.084251
De Leeuw, J., and Mair, P. (2011). Multidimensional Scaling Using Majorization: SMACOF in R. Available online at: http://CRAN.R-project.org/package=smacof (accessed March 3, 2020).
DeAngelis, A. M., Heinrich, G., Dai, T., Bowman, T. A., Patel, P. R., Lee, S. J., et al. (2008). Carcinoembryonic antigen-related cell adhesion molecule 1: a link between insulin and lipid metabolism. Diabetes 57, 2296–2303. doi: 10.2337/db08-0379
Donelson, J., Munday, P., McCormick, M., and Pitcher, C. (2012). Rapid transgenerational acclimation of a tropical reef fish to climate change. Nat. Clim. Chang. 2, 30–32. doi: 10.1038/nclimate1323
Donelson, J. M., and Munday, P. L. (2012). Thermal sensitivity does not determine acclimation capacity for a tropical reef fish. J. Anim. Ecol. 81, 1126–1131. doi: 10.1111/j.1365-2656.2012.01982.x
Donelson, J. M., Munday, P. L., McCormick, M. I., and Nilsson, G. E. (2011). Acclimation to predicted ocean warming through developmental plasticity in a tropical reef fish. Glob. Chang. Biol. 17, 1712–1719. doi: 10.1371/journal.pone.0097223
Donelson, J. M., Salinas, S., Munday, P. L., and Shama, L. N. (2018). Transgenerational plasticity and climate change experiments: where do we go from here? Glob. Chang. Biol. 24, 13–34. doi: 10.1111/gcb.13903
Dossena, S., Gandini, R., Tamma, G., Vezzoli, V., Nofziger, C., Tamplenizza, M., et al. (2011). The molecular and functional interaction between ICln and HSPC038 proteins modulates the regulation of cell volume. J. Biol. Chem. 286, 40659–40670. doi: 10.1074/jbc.M111.260430
Duncan, E. J., Gluckman, P. D., and Dearden, P. K. (2014). Epigenetics, plasticity, and evolution: how do we link epigenetic change to phenotype? J. Exp. Zool. B Mol. Dev. Evol. 322, 208–220. doi: 10.1002/jez.b.22571
Dyachenko, O. V., Schevchuk, T. V., Kretzner, L., Buryanov, Y. I., and Smith, S. S. (2010). Human non-CG methylation: are human stem cells plant-like? Epigenetics 5, 569–572. doi: 10.4161/epi.5.7.12702
Ferre, P., and Foufelle, F. (2010). Hepatic steatosis: a role for de novo lipogenesis and the transcription factor SREBP−1c. Diabetes Obes. Metab. 12, 83–92. doi: 10.1111/j.1463-1326.2010.01275.x
Fox, R. J., Donelson, J. M., Schunter, C., Ravasi, T., and Gaitán-Espitia, J. D. (2019). Beyond buying time: the role of plasticity in phenotypic adaptation to rapid environmental change. Philos. Trans. R. Soc. B Biol. 374:20180174. doi: 10.1098/rstb.2018.0174
Fry, F. E. J., and Hart, J. S. (1948). The relation of temperature to oxygen consumption in the goldfish. Biol. Bull. 94, 66–77. doi: 10.2307/1538211
Gene Ontology Consortium, (2008). The gene ontology project in 2008. Nucleic Acids Res. 36, D440–D444. doi: 10.1093/nar/gkm883
Gienapp, P., Teplitsky, C., Alho, J., Mills, J., and Merilä, J. (2008). Climate change and evolution: disentangling environmental and genetic responses. Mol. Ecol. 17, 167–178. doi: 10.1111/j.1365-294X.2007.03413.x
Grenchik, M., Donelson, J., and Munday, P. (2013). Evidence for developmental thermal acclimation in the damselfish, Pomacentrus moluccensis. Coral Reefs 32, 85–90. doi: 10.1007/s00338-012-0949-1
Guo, F., Li, X., Liang, D., Li, T., Zhu, P., Guo, H., et al. (2014). Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15, 447–459. doi: 10.1016/j.stem.2014.08.003
Guo, L., Wu, X., Zhang, Y., Wang, F., Li, J., and Zhu, J. (2019). Protective effects of gastrin−releasing peptide receptor antagonist RC−3095 in an animal model of hepatic ischemia/reperfusion injury. Hepatol. Res. 49, 247–255. doi: 10.1111/hepr.13315
He, Y.-W., Li, H., Zhang, J., Hsu, C.-L., Lin, E., Zhang, N., et al. (2004). The extracellular matrix protein mindin is a pattern-recognition molecule for microbial pathogens. Nat. Immunol. 5, 88–97. doi: 10.1038/ni1021
Healy, T. M., Bock, A. K., and Burton, R. S. (2019). Variation in developmental temperature alters adulthood plasticity of thermal tolerance in Tigriopus californicus. J. Exp. Biol. 222:jeb213405. doi: 10.1242/jeb.213405
Ho, D. H., and Burggren, W. W. (2010). Epigenetics and transgenerational transfer: a physiological perspective. J. Exp. Biol. 213, 3–16. doi: 10.1242/jeb.019752
Hoey, A. S., Howells, E., Johansen, J. L., Hobbs, J.-P. A., Messmer, V., McCowan, D. M., et al. (2016). Recent advances in understanding the effects of climate change on coral reefs. Diversity 8:12. doi: 10.3390/d8020012
Hoffmann, A. A., and Sgro, C. M. (2011). Climate change and evolutionary adaptation. Nature 470, 479–485. doi: 10.1038/nature09670
Holt, C., and Yandell, M. (2011). MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491. doi: 10.1186/1471-2105-12-491
Hon, G. C., Rajagopal, N., Shen, Y., McCleary, D. F., Yue, F., Dang, M. D., et al. (2013). Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 45, 1198–1206. doi: 10.1038/ng.2746
Hughes, T. P., Kerry, J. T., Baird, A. H., Connolly, S. R., Dietzel, A., Eakin, C. M., et al. (2018). Global warming transforms coral reef assemblages. Nature 556, 492–496. doi: 10.1038/s41586-018-0041-2
Huse, S. M., Gruppuso, P. A., Boekelheide, K., and Sanders, J. A. (2015). Patterns of gene expression and DNA methylation in human fetal and adult liver. BMC Genomics 16:981. doi: 10.1186/s12864-015-2066-3
Hwang, D. Y., Sin, J. S., Kim, M. S., Yim, S. Y., Kim, Y. K., Kim, C. K., et al. (2008). Overexpression of human selenoprotein M differentially regulates the concentrations of antioxidants and H2O2, the activity of antioxidant enzymes, and the composition of white blood cells in a transgenic rat. Int. J. Mol. Med. 21, 169–179.
IPCC, (2019). IPCC Special Report on the Ocean and Cryosphere in a Changing Climate. Geneva: Intergovernmental Panel on Climate Change.
Jablonka, E., and Raz, G. (2009). Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84, 131–176. doi: 10.1086/598822
Jacob, S., and Moley, K. H. (2005). Gametes and embryo epigenetic reprogramming affect developmental outcome: implication for assisted reproductive technologies. Pediatr. Res. 58, 437–446. doi: 10.1203/01.PDR.0000179401.17161.D3
Jiang, L., Zhang, J., Wang, J. J., Wang, L., Zhang, L., Li, G., et al. (2013). Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell 153, 773–784. doi: 10.1016/j.cell.2013.04.041
Kearns, M., Preis, J., McDonald, M., Morris, C., and Whitelaw, E. (2000). Complex patterns of inheritance of an imprinted murine transgene suggest incomplete germline erasure. Nucleic Acids Res. 28, 3301–3309. doi: 10.1093/nar/28.17.3301
Kim, H.-S., and Brill, S. J. (2001). Rfc4 interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 3725–3737. doi: 10.1128/mcb.21.11.3725-3737.2001
Kim, H. J., Schleiffarth, J. R., Jessurun, J., Sumanas, S., Petryk, A., Lin, S., et al. (2005). Wnt5 signaling in vertebrate pancreas development. BMC Biol. 3:23. doi: 10.1186/1741-7007-3-23
Krueger, F. (2015). Trim Galore. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed August 10, 2015).
Krueger, F., and Andrews, S. R. (2011). Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572. doi: 10.1093/bioinformatics/btr167
Le Roy, A., Loughland, I., and Seebacher, F. (2017). Differential effects of developmental thermal plasticity across three generations of guppies (Poecilia reticulata): canalization and anticipatory matching. Sci. Rep. 7:4313. doi: 10.1038/s41598-017-03300-z
Lear, B. C., Darrah, E. J., Aldrich, B. T., Gebre, S., Scott, R. L., Nash, H. A., et al. (2013). UNC79 and UNC80, putative auxiliary subunits of the narrow abdomen ion channel, are indispensable for robust circadian locomotor rhythms in Drosophila. PLoS One 8:e78147. doi: 10.1371/journal.pone.0078147
Marsh, A. G., and Pasqualone, A. A. (2014). DNA methylation and temperature stress in an Antarctic polychaete, Spiophanes tcherniai. Front. Physiol. 5:173. doi: 10.3389/fphys.2014.00173
Marshall, D. J., and Uller, T. (2007). When is a maternal effect adaptive? Oikos 116, 1957–1963. doi: 10.1111/j.2007.0030-1299.16203.x
May, S. R., Stewart, N. J., Chang, W., and Peterson, A. S. (2004). A Titin mutation defines roles for circulation in endothelial morphogenesis. Dev. Biol. 270, 31–46. doi: 10.1016/j.ydbio.2004.02.006
Messerschmidt, D. M., Knowles, B. B., and Solter, D. (2014). DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 28, 812–828. doi: 10.1101/gad.234294.113
Miller, R. A., and Birnbaum, M. J. (2016). Glucagon: acute actions on hepatic metabolism. Diabetologia 59, 1376–1381. doi: 10.1007/s00125-016-3955-y
Morgan, H. D., Santos, F., Green, K., Dean, W., and Reik, W. (2005). Epigenetic reprogramming in mammals. Hum. Mol. Genet. 14, R47–R58. doi: 10.1093/hmg/ddi114
Munday, P. L. (2014). Transgenerational acclimation of fishes to climate change and ocean acidification. F1000prime Rep. 6:99.
Munday, P. L., Warner, R. R., Monro, K., Pandolfi, J. M., and Marshall, D. J. (2013). Predicting evolutionary responses to climate change in the sea. Ecol. Lett. 16, 1488–1500. doi: 10.1111/ele.12185
Nedvetsky, P. I., Zhao, X., Mathivet, T., Aspalter, I. M., Stanchi, F., Metzger, R. J., et al. (2016). cAMP-dependent protein kinase A (PKA) regulates angiogenesis by modulating tip cell behavior in a Notch-independent manner. Development 143, 3582–3590. doi: 10.1242/dev.134767
Nordhausen, K., Sirkia, S., Oja, H., and Tyler, D. (2015). ICSNP: Tools for Multivariate Nonparametrics. Available online at: http://CRAN.R-project.org/package=ICSNP (accessed March 10, 2018).
O’brien, K. P., Remm, M., and Sonnhammer, E. L. (2005). Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 33(Suppl. 1), D476–D480.
Ortega-Recalde, O., Day, R. C., Gemmell, N. J., and Hore, T. A. (2019). Zebrafish preserve global germline DNA methylation while sex-linked rDNA is amplified and demethylated during feminisation. Nat. Commun. 10:3053. doi: 10.1038/s41467-019-10894-7
Parra, G., Bradnam, K., and Korf, I. (2007). CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067. doi: 10.1093/bioinformatics/btm071
Pecl, G. T., Araújo, M. B., Bell, J. D., Blanchard, J., Bonebrake, T. C., Chen, I.-C., et al. (2017). Biodiversity redistribution under climate change: impacts on ecosystems and human well-being. Science 355:eaai9214. doi: 10.1126/science.aai9214
Persson, K., Pacini, G., Sundler, F., and Ahren, B. (2002). Islet function phenotype in gastrin-releasing peptide receptor gene-deficient mice. Endocrinology 143, 3717–3726. doi: 10.1210/en.2002-220371
Persson, P. B. (2003). Renin: origin, secretion and synthesis. J. Physiol. 552, 667–671. doi: 10.1113/jphysiol.2003.049890
Pitts, M. W., Reeves, M. A., Hashimoto, A. C., Ogawa, A., Kremer, P., Seale, L. A., et al. (2013). Deletion of selenoprotein M leads to obesity without cognitive deficits. J. Biol. Chem. 288, 26121–26134. doi: 10.1074/jbc.M113.471235
Portner, H. O. (2010). Oxygen- and capacity-limitation of thermal tolerance: a matrix for integrating climate-related stressor effects in marine ecosystems. J. Exp. Biol. 213, 881–893. doi: 10.1242/jeb.037523
Portner, H. O., and Farrell, A. P. (2008). Ecology, physiology and climate change. Science 322, 690–692. doi: 10.1126/science.1163156
Portner, H. O., and Knust, R. (2007). Climate change affects marine fishes through the oxygen limitation of thermal tolerance. Science 315, 95–97. doi: 10.1126/science.1135471
Putnam, H. M., Davidson, J. M., and Gates, R. D. (2016). Ocean acidification influences host DNA methylation and phenotypic plasticity in environmentally susceptible corals. Evol. Appl. 9, 1165–1178. doi: 10.1111/eva.12408
Quinlan, A. R. (2014). BEDTools: the Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinform. 47, 11.12.11–11.12.34. doi: 10.1002/0471250953.bi1112s47
Rellinger, E. J., Romain, C., Choi, S., Qiao, J., and Chung, D. H. (2015). Silencing gastrin−releasing peptide receptor suppresses key regulators of aerobic glycolysis in neuroblastoma cells. Pediatr. Blood Cancer 62, 581–586. doi: 10.1002/pbc.25348
Reusch, T. B. (2014). Climate change in the oceans: evolutionary versus phenotypically plastic responses of marine animals and plants. Evol. Appl. 7, 104–122. doi: 10.1111/eva.12109
Richter, K., Haslbeck, M., and Buchner, J. (2010). The heat shock response: life on the verge of death. Mol. Cell 40, 253–266. doi: 10.1016/j.molcel.2010.10.006
Rui, L. (2014). Energy metabolism in the liver. Compr. Physiol. 4, 177–197. doi: 10.1002/cphy.c130024
Russo, L., Ghadieh, H. E., Ghanem, S. S., Al-Share, Q. Y., Smiley, Z. N., Gatto-Weis, C., et al. (2016). Role for hepatic CEACAM1 in regulating fatty acid metabolism along the adipocyte-hepatocyte axis. J. Lipid Res. 57, 2163–2175. doi: 10.1194/jlr.m072066
Ryu, T., Veilleux, H. D., Donelson, J. M., Munday, P. L., and Ravasi, T. (2018). The epigenetic landscape of transgenerational acclimation to ocean warming. Nat. Clim. Chang. 8, 504–509. doi: 10.1038/s41558-018-0159-0
Salinas, S., and Munch, S. B. (2012). Thermal legacies: transgenerational effects of temperature on growth in a vertebrate. Ecol. Lett. 15, 159–163. doi: 10.1111/j.1461-0248.2011.01721.x
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning : a Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press.
Scheffers, B. R., De Meester, L., Bridge, T. C., Hoffmann, A. A., Pandolfi, J. M., Corlett, R. T., et al. (2016). The broad footprint of climate change from genes to biomes to people. Science 354:aaf7671. doi: 10.1126/science.aaf7671
Shama, L. N. (2017). The mean and variance of climate change in the oceans: hidden evolutionary potential under stochastic environmental variability in marine sticklebacks. Sci. Rep. 7:8889. doi: 10.1038/s41598-017-07140-9
Shama, L. N., Strobel, A., Mark, F. C., and Wegner, K. M. (2014). Transgenerational plasticity in marine sticklebacks: maternal effects mediate impacts of a warming ocean. Funct. Ecol. 28, 1482–1493. doi: 10.1111/1365-2435.12280
Shao, W., and Espenshade, P. J. (2012). Expanding roles for SREBP in metabolism. Cell Metab. 16, 414–419. doi: 10.1016/j.cmet.2012.09.002
Silva, A. C. S. E., Miranda, A. S., Rocha, N. P., and Teixeira, A. L. (2017). Renin angiotensin system in liver diseases: friend or foe? World J. Gastroenterol. 23, 3396–3406. doi: 10.3748/wjg.v23.i19.3396
Smith, I. A., Knezevic, B. R., Ammann, J. U., Rhodes, D. A., Aw, D., Palmer, D. B., et al. (2010). BTN1A1, the mammary gland butyrophilin, and BTN2A2 are both inhibitors of T cell activation. J. Immunol. 184, 3514–3525. doi: 10.4049/jimmunol.0900416
Sunday, J. M., Bates, A. E., and Dulvy, N. K. (2010). Global analysis of thermal tolerance and latitude in ectotherms. Proc. R. Soc. B Biol. Sci. 278, 1823–1830. doi: 10.1098/rspb.2010.1295
Takimoto, E., Yao, A., Toko, H., Takano, H., Shimoyama, M., Sonoda, M., et al. (2002). Sodium calcium exchanger plays a key role in alteration of cardiac function in response to pressure overload. FASEB J. 16, 373–378. doi: 10.1096/fj.01-0735com
Tang, M. C., Jacobs, S. A., Mattiske, D. M., Soh, Y. M., Graham, A. N., Tran, A., et al. (2015). Contribution of the two genes encoding histone variant h3. 3 to viability and fertility in mice. PLoS Genet. 11:e1004964. doi: 10.1371/journal.pgen.1004964
Tewksbury, J. J., Huey, R. B., and Deutsch, C. A. (2008). Putting the heat on tropical animals. Science 320, 1296–1297. doi: 10.1126/science.1159328
Torda, G., Donelson, J. M., Aranda, M., Barshis, D. J., Bay, L., Berumen, M. L., et al. (2017). Rapid adaptive responses to climate change in corals. Nat. Clim. Chang. 7, 627–636. doi: 10.1038/nclimate3374
Turnham, R. E., and Scott, J. D. (2016). Protein kinase A catalytic subunit isoform PRKACA; history, function and physiology. Gene 577, 101–108. doi: 10.1016/j.gene.2015.11.052
Veilleux, H. D., Ryu, T., Donelson, J. M., Munday, P., and Ravasi, T. (2018). Molecular response to extreme summer temperatures differs between two genetically differentiated populations of a coral reef fish. Front. Mar. Sci. 5:349. doi: 10.3389/fmars.2018.00349
Veilleux, H. D., Ryu, T., Donelson, J. M., van Herwerden, L., Seridi, L., Ghosheh, Y., et al. (2015). Molecular processes of transgenerational acclimation to a warming ocean. Nat. Clim. Chang. 5, 1074–1078. doi: 10.1038/nclimate2724
Veroni, C., Grasso, M., Macchia, G., Ramoni, C., Ceccarini, M., Petrucci, T. C., et al. (2007). β−dystrobrevin, a kinesin−binding receptor, interacts with the extracellular matrix components pancortins. J. Neurosci. Res. 85, 2631–2639. doi: 10.1002/jnr.21186
Victoria Sanz Fernandez, M., Johnson, J. S., Abuajamieh, M., Stoakes, S. K., Seibert, J. T., Cox, L., et al. (2015). Effects of heat stress on carbohydrate and lipid metabolism in growing pigs. Physiol. Rep. 3:e12315. doi: 10.14814/phy2.12315
Wang, Q.-I., Chen, S., Esumi, N., Swain, P. K., Haines, H. S., Peng, G., et al. (2004). QRX, a novel homeobox gene, modulates photoreceptor gene expression. Hum. Mol. Genet. 13, 1025–1040. doi: 10.1093/hmg/ddh117
Wang, X., and Bhandari, R. K. (2019a). DNA methylation dynamics during epigenetic reprogramming of medaka embryo. Epigenetics 14, 611–622. doi: 10.1080/15592294.2019.1605816
Wang, X., and Bhandari, R. K. (2019b). The dynamics of DNA methylation during epigenetic reprogramming of primordial germ cells in medaka (Oryzias latipes). Epigenetics 15, 483–498. doi: 10.1080/15592294.2019.1695341
Warnes, G. R., Bolker, B., Bonebakker, L., Gentleman, R., Liaw, W. H. A., Lumley, T., et al. (2016). gplots: Various R Programming Tools for Plotting Data. Available online at: http://CRAN.R-project.org/package=gplots (accessed February 25, 2020).
Warren, H. R., Evangelou, E., Cabrera, C. P., Gao, H., Ren, M., Mifsud, B., et al. (2017). Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat. Genet. 49, 403–415. doi: 10.1038/ng.3768
Wasserman, D. H. (2009). Four grams of glucose. Am. J. Physiol. Endocrinol. Metab. 296:E11. doi: 10.1152/ajpendo.90563.2008
Wernberg, T., Smale, D. A., and Thomsen, M. S. (2012). A decade of climate change experiments on marine organisms: procedures, patterns and problems. Glob. Chang. Biol. 18, 1491–1498. doi: 10.1111/j.1365-2486.2012.02656.x
Wójtowicz, I., Jabłońska, J., Zmojdzian, M., Taghli-Lamallem, O., Renaud, Y., Junion, G., et al. (2015). Drosophila small heat shock protein CryAB ensures structural integrity of developing muscles, and proper muscle and heart performance. Development 142, 994–1005. doi: 10.1242/dev.115352
Yan, L., Wei, X., Tang, Q.-Z., Feng, J., Zhang, Y., Liu, C., et al. (2011). Cardiac-specific mindin overexpression attenuates cardiac hypertrophy via blocking AKT/GSK3β and TGF-β1–Smad signalling. Cardiovasc. Res. 92, 85–94. doi: 10.1093/cvr/cvr159
Yin, B., Tang, S., Xu, J., Sun, J., Zhang, X., Li, Y., et al. (2019). CRYAB protects cardiomyocytes against heat stress by preventing caspase-mediated apoptosis and reducing F-actin aggregation. Cell Stress Chaperones 24, 59–68. doi: 10.1007/s12192-018-0941-y
Zeng, Z., Wu, Y., Cao, Y., Yuan, Z., Zhang, Y., Zhang, D. Y., et al. (2018). Slit2-Robo2 signaling modulates the fibrogenic activity and migration of hepatic stellate cells. Life Sci. 203, 39–47. doi: 10.1016/j.lfs.2018.04.017
Zhu, L. H., Wang, A., Luo, P., Wang, X., Jiang, D. S., Deng, W., et al. (2014). Mindin/Spondin 2 inhibits hepatic steatosis, insulin resistance, and obesity via interaction with peroxisome proliferator-activated receptor alpha in mice. J. Hepatol. 60, 1046–1054. doi: 10.1016/j.jhep.2014.01.011
Keywords: climate change, ocean warming, coral reef fish, epigenetics, DNA methylation, phenotypic plasticity
Citation: Ryu T, Veilleux HD, Munday PL, Jung I, Donelson JM and Ravasi T (2020) An Epigenetic Signature for Within-Generational Plasticity of a Reef Fish to Ocean Warming. Front. Mar. Sci. 7:284. doi: 10.3389/fmars.2020.00284
Received: 16 January 2020; Accepted: 07 April 2020;
Published: 30 April 2020.
Edited by:
Jose M. Eirin-Lopez, Florida International University, United StatesReviewed by:
Lisa N. S. Shama, Alfred Wegener Institute Helmholtz Centre for Polar and Marine Research (AWI), GermanyAdam Michael Reitzel, University of North Carolina at Charlotte, United States
Copyright © 2020 Ryu, Veilleux, Munday, Jung, Donelson and Ravasi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Taewoo Ryu, dGFld29vLnJ5dUBvaXN0Lmpw; ZGFrdGVybDc5QGdtYWlsLmNvbQ==; Jennifer M. Donelson, amVubmlmZXIuZG9uZWxzb25AamN1LmVkdS5hdQ==; Timothy Ravasi, dGltb3RoeS5yYXZhc2lAb2lzdC5qcA==
 Taewoo Ryu
Taewoo Ryu Heather D. Veilleux3,4
Heather D. Veilleux3,4 Philip L. Munday
Philip L. Munday Jennifer M. Donelson
Jennifer M. Donelson Timothy Ravasi
Timothy Ravasi