
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 20 March 2020
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 7 - 2020 | https://doi.org/10.3389/fmars.2020.00117
This article is part of the Research Topic Fish Nutrition, Metabolism and Physiology View all 20 articles
Fatty acyl desaturases 2 (Fads2) and elongases of very-long-chain fatty acid 5 (Elovl5) are two key enzymes involved in the biosynthesis of long-chain polyunsaturated fatty acids (LC-PUFAs), and their activities determine the LC-PUFA biosynthetic ability of teleost. In order to investigate the relation of enzymic activities with fish’s feeding habits and ecological habits, the activities of Fads2 and Elovl5 were compared among six teleosts, namely, freshwater carnivorous mandarin fish (Siniperca chuatsi), freshwater herbivorous grass carp (Ctenopharyngodon idellus), marine carnivorous orange-spotted grouper (Epinephelus coioides), marine herbivorous rabbitfish (Siganus canaliculatus), anadromous Atlantic salmon (Salmo salar), and catadromous Japanese eel (Anguilla japonica). Among them, the enzymatic features of Fads2 and Elovl5 from the last five fish species have been characterized, whereas those of S. chuatsi were unknown. And thus, the coding sequences (CDSs) of S. chuatsi fads2 and elovl5 (elovl5a and elovl5b) were isolated, and their functions were further characterized by heterologous expression in yeast. The results showed that S. chuatsi Fads2 has a monofunctional Δ6 desaturase and that Elovl5a has a higher activity toward C18–C20 PUFAs than has Elovl5b, which showed a noteworthy activity toward C22 PUFAs. The comparison of enzymatic activities among the six teleosts showed that the Δ6 Fad and Elovl5 activities varied markedly among fish species; in particular, the activity of Δ6 Fad in C. idellus, S. canaliculatus, and A. japonica was significantly higher than that in S. chuatsi, S. salar, and E. coioides. For C18 PUFA substrates, A. japonica Elovl5 has a higher elongation than has the other tested fish, and it exhibits a higher activity toward the C20 PUFAs. The results suggest that the Δ6 Fad activity is influenced by both feeding habits and ecological habits, whereas the Elovl5 activity was more affected by the feeding habits. These data enrich our knowledge on LC-PUFA biosynthesis diversity of fatty acid desaturation and elongations in teleosts and provide guidance for the choice of dietary PUFA precursors for farmed fish.
The health benefits of fish consumption are derived from n–3 long-chain polyunsaturated fatty acids (n–3 LC-PUFA), eicosapentaenoic acid (EPA; 20:5n–3) and docosahexaenoic acid (DHA; 22:6n–3). These above-mentioned bioactive molecules are involved in maintaining the normal development of the nervous system (Uauy et al., 2001) and improving in lipid metabolism, inflammatory response, and cardiovascular and neurological health (Delgado-Lista et al., 2012; Awada et al., 2013). Farmed fish are becoming an increasingly important source of LC-PUFA in the human diet, because the wild fishery stocks are declining (FAO, 2014). Fish oils (FOs), rich in digestible energy and n–3 LC-PUFA, are considered as the most important raw materials for aquafeeds (especially for carnivorous fish feed). During the recent two decades, the supplementation of C18 PUFA-rich plant oils [vegetable oils (VOs), devoid of n–3 LC-PUFA] was increased to replace FOs in fish formula feed, because of the limited and increasingly expensive FO resources (Turchini et al., 2011; Lu et al., 2017). Consequently, the increasing supplementation of VO in aquafeeds has resulted in decreasing the levels of n–3 LC-PUFA in farmed fish, especially in marine fish (Hossain, 2011; Sprague et al., 2016).
To efficiently use dietary VO and maximize endogenous n–3 LC-PUFA biosynthesis, much attention has been focused on illuminating the regulation mechanisms of LC-PUFA biosynthesis in farmed fish (Tocher, 2010; Castro et al., 2012; Oboh et al., 2017). Studies conducted in teleosts have demonstrated that the LC-PUFA biosynthesis pathway involves sequential desaturation and elongation steps from C18 PUFA substrates catalyzed by fatty acyl desaturase (Fads) and elongation of very-long-chain fatty acids (Elovl) enzymes, such as Δ6 Fad, Δ5 Fad, Δ4 Fad, Elovl5, Elovl4, and Elovl2 (Li et al., 2010; Castro et al., 2012; Janaranjani et al., 2018; Ferraz et al., 2019). Conventionally, teleosts, like freshwater fish and salmonid species, expressed all necessary desaturase and elongase activities (Castro et al., 2012; Monroig et al., 2013; Fonseca-Madrigal et al., 2014; Kuah et al., 2015; Oboh et al., 2017). However, even with genome sequencing, the Δ5 fad gene has not been found in any marine carnivorous teleost (Leaver et al., 2008), and no Elovl2 has been isolated from any marine fish species (Tocher, 2010). Therefore, marine carnivorous teleost having limited capacity for LC-PUFA biosynthesis probably depends on their genome complement appearing to lack Δ5 Fad and Elovl2 (Monroig et al., 2013).
The extent to which fish species have the LC-PUFA biosynthetic capability varies with species and is associated with not only their complement of fads and elovl genes but also the activities of those key enzymes (Fonseca-Madrigal et al., 2014). Δ6 Fad and Elovl5 have been cloned and characterized from all fish so far investigated including marine carnivorous fishes (Tocher, 2010). However, it remained unclear whether the enzymatic activities of Δ6 Fad and Elovl5 are linked to the habitat, trophic level, and ecology of fish species. To this end, the coding sequences (CDSs) of putative Δ6 Fad and Elovl5 were isolated from the freshwater carnivorous fish, mandarin fish (Siniperca chuatsi), and functionally characterized by heterologous expression in yeast (Saccharomyces cerevisiae). The enzymic activities of Δ6 Fad and Elovl5 among six fish species with different habitats and trophic ecologies, namely, freshwater herbivorous fish grass carp (Ctenopharyngodon idellus), S. chuatsi, marine carnivorous fish orange-spotted grouper (Epinephelus coioides), marine herbivorous fish rabbitfish (Siganus canaliculatus), anadromous fish Atlantic salmon (Salmo salar), and catadromous fish Japanese eel (Anguilla japonica), were determined by heterologous expression in the yeast. The results were expected to identify the relationships between the enzymatic activities of Δ6 Fad and Elovl5 and the habitat, trophic level, and ecology of fish species and to increase our knowledge of the molecular basis of LC-PUFA biosynthesis and its regulation in teleosts.
In order to clone the fads2 and elovl5 CDSs of S. chuatsi, liver samples were collected from three S. chuatsi individuals (about 250 g) brought from the local fish market, after the fish were anesthetized with 0.01% 2-phenoxyethanol. Tissue samples were frozen in liquid nitrogen immediately after collection and stored at –80°C until RNA extraction. The liver tissue of S. chuatsi was sampled, and the total RNA extracted using TRIzol reagent (Invitrogen, United States) was reverse transcribed into cDNA using random primers and an appropriate RT-PCR kit (Invitrogen, United States). The CDS-specific primers of Δ6 fad and elovl5 containing restriction sites BamHI and XbaI were designed on the basis of S. chuatsi Δ6 fad (EU683737) and elovl5 (EU683736) (Table 1). PCR was performed using high-fidelity DNA polymerase (TianGen, Beijing, China). For all genes, PCR consisted of an initial denaturation at 94°C for 4 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min, followed by a final extension at 72°C for 10 min. The resulting PCR fragments were sequenced (Sangon, Shanghai, China).
The recombinant plasmids containing Δ6 fad and elovl5 CDSs of Ctenopharyngodon idellus, Siganus canaliculatus, Epinephelus coioides, S. salar, and Anguilla japonica were constructed and kept in our laboratory (Wang et al., 2014). The Δ6 fad and elovl5 CDSs of C. idellus, S. canaliculatus, E. coioides, S. salar (elovl5a and elovl5b), and A. japonica were cloned from the corresponding recombinant plasmids with CDS-specific primers (Table 1). For all genes, PCR was performed using high-fidelity DNA polymerase (TianGen, Beijing, China) under the following conditions: initial denaturation at 94°C for 4 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min, followed by a final extension at 72°C for 10 min. The PCR fragments were sequenced (Sangon, Shanghai, China).
The amino acid (aa) sequences of the cloned Δ6 Fad and Elovl5 were aligned among the six species by ClustalW2.1 Alignments and similarity matrices were calculated using the EMBOSS Needle Pairwise Sequence Alignment tool.2 Phylogenetic analysis was performed by constructing a tree using the neighbor-joining method (Saitou and Nei, 1987). Confidence in the resulting phylogenetic tree branch topology was measured by bootstrapping through 1,000 iterations.
The Δ6 fad and elovl5 CDS fragments of S. chuatsi were purified and digested with the corresponding restriction endonucleases (New England Biolabs, United Kingdom) and ligated into the yeast episomal plasmid pYES2 (Invitrogen). The recombinant plasmids (pYES2-ScΔ6 fad or pYES2-Scelovl5) were transformed into yeast (strain INVSc1, Invitrogen) using the S.C. Easy Comp Transformation kit (Invitrogen).
The functional characterization of the putative Δ6 Fad and Elovl5 was determined according to the methods we previously described (Li et al., 2010). Briefly, for testing the Δ6 Fad activity, linolenic acid (LNA; 18:3n–3), linoleic acid (LA; 18:2n–6), eicosatetraenoic acid (20:4n–3), dihomo-γ-linolenic acid (20:3n–6), docosapentaenoic acid (DPA; 22:5n–3), and docosatetraenoic acid (DTA; 22:4n–6) were used as substrates; for testing the Elovl5 activity, stearidonic acid (18:4n–3), γ-linolenic acid (18:3n–6), EPA (20:5n–3), arachidonic acid (ARA; 20:4n–6), DPA, and DTA were used as substrates. All the fatty acids were purchased from Cayman Chemicals, Co. (Ann Arbor, MI, United States). The PUFA substrates were added at final concentrations of 0.5 (C18), 0.75 (C20), and 1.0 (C22) mM. After 2 days of culture in SCMMuracil, yeast cells were harvested and washed as described previously (Li et al., 2010; Xie et al., 2014).
To maximize the reliability of results, the comparison of Fads2 and Elovl5 conversion rates among the six species was performed in the same experimental conditions. Briefly, the recombinant plasmids (0.8 μg) containing Δ6 fad and elovl5 CDS of C. idellus, S. canaliculatus, E. coioides, S. salar (elovl5a and elovl5b), and A. japonica were sequenced and transformed into yeast (strain INVSc1, Invitrogen) using the S.C. Easy Comp Transformation kit (Invitrogen, United States). Recombinant yeast solution (100 μl) was coated on the SCMMuraci plate and cultured for 3 days; a single colony was picked and propagated in the SCMMuracil culture. For determining the Δ6 Fad activity, the recombinant yeast was cultured in SCMMuracil and supplemented with C18 PUFA substrates, LNA or LA. For testing the Elovl5 activity, the transgenic yeast was supplemented with one of the PUFA substrates from among the following: 18:3n–6, 18:4n–3, ARA, and EPA. The PUFA substrates were added at final concentrations of 0.5 (C18) and 0.75 (C20) mM. After 2 days (the OD600 of bacterium solution reached 10), recombinant yeast cells were harvested and washed as described previously (Li et al., 2010; Xie et al., 2014).
The total lipid of yeast samples was extracted by homogenization in chloroform/methanol (2:1, v/v) containing 0.01% BHT (Sigma, United States) (Folch et al., 1957). Fatty acid methyl esters (FAMEs) from yeast total lipids were prepared by transesterification with boron trifluoride etherate (ca. 48%, Acros Organics, Morris Township, NJ, United States) as described previously (Li et al., 2010). FAMEs were determined by the gas chromatograph GC2010-plus (Shimadzu, Japan). The parameters were the same as we used before (Li et al., 2010). The conversion rates of genes were calculated as follows: 100 × [product areas/(product area + substrate area)] (Li et al., 2010).
Genes conversion rates were presented as means ± standard error of the mean (n = 3). Differences among the six fish species were tested using one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison. Differences were considered significant at P < 0.05. All analyses were conducted using SPSS v17.0 (SPSS, Inc., Chicago, IL, United States).
The sequence characteristics of Ctenopharyngodon idellus, Siniperca chuatsi, Siganus canaliculatus, Epinephelus coioides, Salmo salar, and Anguilla japonica Δ6 fad CDS was 1,335, 1,338, 1,332, 1,338, 1,365, and 1,335 bp (including stop codons) in length encoding peptides of 444, 445, 443, 445, 454, and 444 amino acids, respectively (Supplementary Figure 1). The Δ6 Fad polypeptides of C. idellus, S. chuatsi, E. coioides, and A. japonica deduced from their CDS obtained in the present study showed 1, 3, 1, and 1 aa differences, respectively, compared with previously published results (Li et al., 2010, 2014; Du et al., 2011; Wang et al., 2014). The polypeptides of S. canaliculatus and S. salar Δ6 Fad obtained in this study showed the same sequences as previously published (Li et al., 2010; Monroig et al., 2010).
The Δ6 Fad polypeptide of C. idellus had 65.86, 68.99, 69.21, 69.59, and 71.91% sequence identities with other Δ6 Fad from S. salar, S. chuatsi, E. coioides, A. japonica, and S. canaliculatus, respectively (Figure 1). Higher identity scores (79.78∼88.54%) were obtained when the E. coioides Δ6 Fad was compared with other Δ6 Fad sequences from S. canaliculatus, S. salar, A. japonica, and S. chuatsi. All polypeptide sequences contained typical conserved features of front-end desaturases including a putative cytochrome b5-like domain, HPGG, and three histidine boxes (HXXXH, HXXHH, and QXXHH) (Hashimoto et al., 2008).

Figure 1. Alignment of the deduced amino acid (aa) sequences of fatty acyl desaturases 2 among Anguilla japonica, Siganus canaliculatus, Ctenopharyngodon idellus, Siniperca chuatsi, Epinephelus coioides, and Salmo salar using ClustalW2. Identical and similar residues are marked with an asterisk and a colon, respectively. The cytochrome-b5-like domain is underlined with a fine line and the heme-binding motifs with a short bold line. The three histidine boxes are highlighted with frames.
Phylogenetic analysis compared all the deduced amino acid sequences of Fads2 along with a variety of Δ4 Fad, Δ5 Fad, and Δ6 Fad desaturases from different fish species (Figure 2). The result showed that teleost Fads2 sequences cluster according to accepted habitat ecology as displayed in the phylogenetic tree, with all the marine fish Fads2 sequences clustered together and more closely related to migration fish Fads2 sequences than freshwater fish species sequences. The salmonid sequences clustered together with the other diadromous fish species, A. japonica. Interestingly, the Nile tilapia and S. chuatsi sequence clustered closer to the marine fish than to the other freshwater fish, clarias leather (Clarias gariepinus), carp, and zebrafish (Danio rerio), which clustered together.
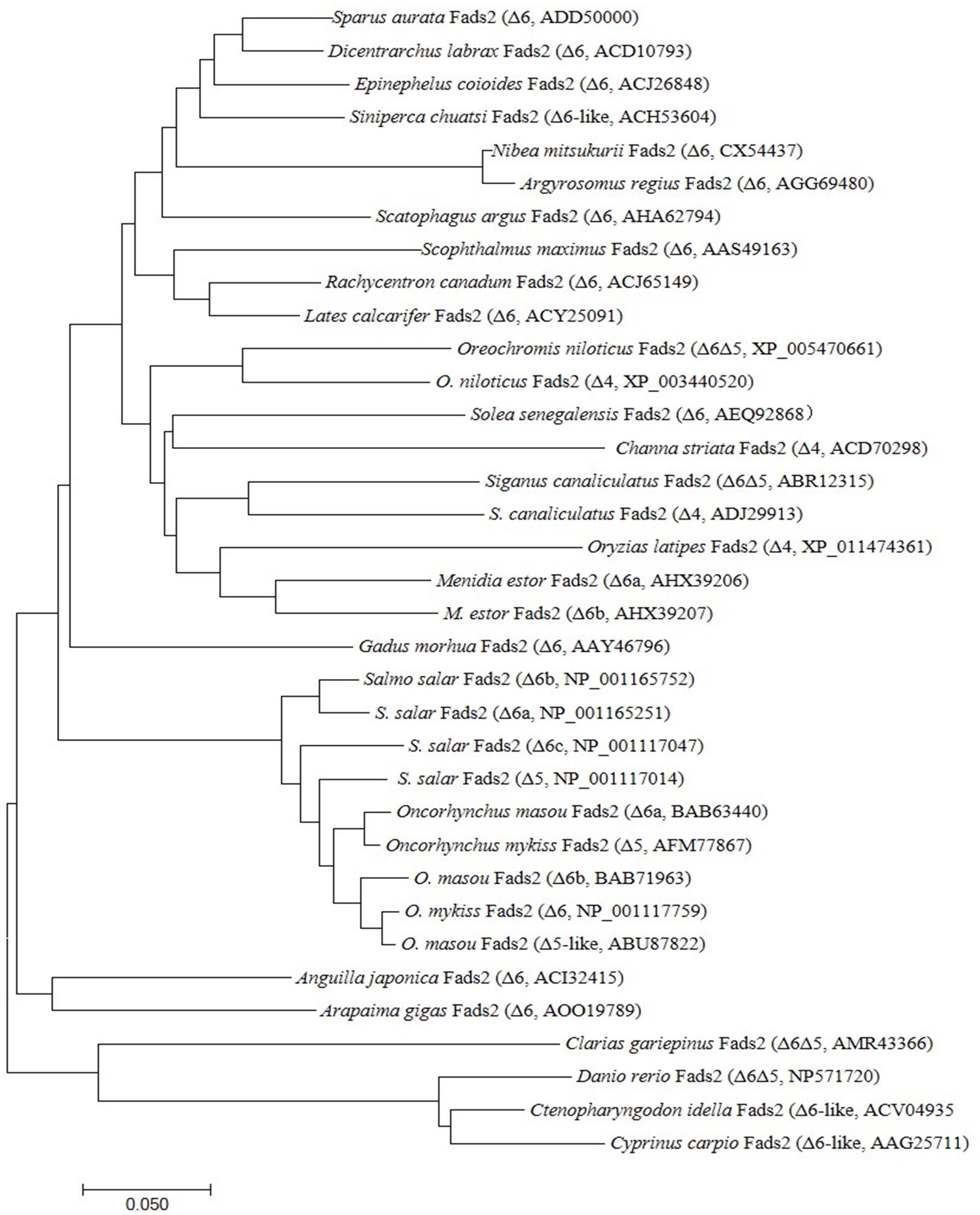
Figure 2. Phylogeny of deduced amino acid (aa) sequences of Fads2 of different fish species. The tree was constructed using the neighbor-joining (N-J) method (Saitou and Nei, 1987) using CLUSTALX and MEGA 7.0. The horizontal branch length is proportional aa substitution rate per site. The numbers represent the frequencies with which the tree topology presented here was replicated after 1,000 bootstrap iterations.
In the present study, the Elovl5 CDS characteristics of C. idellus, S. chuatsi (Elovl5a and Elovl5b), S. canaliculatus, E. coioides, S. salar (Elovl5a and Elovl5b), and A. japonica were 876, 885, 885, 876, 885, 888, 885, and 885 bp (including stop codons) in length encoding peptides of 291, 294, 294, 291 294, 295, 294, and 294 aa, respectively (Supplementary Figure 2). Interestingly, two CDS sequences were cloned in S. chuatsi Elovl5 (Elovl5a and Elovl5b) in the present study, and the Elovl5a sequences showed the same sequence as previously published (EU683736). On the other hand, the S. chuatsi Elovl5b, compared with Elovl5a, showed 7 aa differences. The Elovl5 polypeptides of A. japonica deduced from its CDS showed 3 aa differences compared with those of previously published work (Wang et al., 2014). The obtained Elovl5 polypeptides of C. idellus, S. canaliculatus, E. coioides, and S. salar in this study showed the same sequence as previously published (Du et al., 2011; Monroig et al., 2012; Carmona-Antoñanzas et al., 2013; Li et al., 2016).
The Elovl5 polypeptide of C. idellus had 70.45, 70.75, 73.56, 76.53, and 78.57% sequence identities with other Elovl5 from S. canaliculatus, S. chuatsi, S. salar, E. coioides, and A. japonica, respectively (Figure 3). E. coioides Elovl5 shares aa sequence identities of 76.53–87.76% with Elovl5 from other teleosts, C. idellus, A. japonica, S. canaliculatus, S. salar, and S. chuatsi. The Elovl5 polypeptide sequences contained all the typical structural characteristics, including a conserved histidine box HXXHH, five putative transmembrane domains, and a putative ER retrieval signal (Jakobsson et al., 2006).
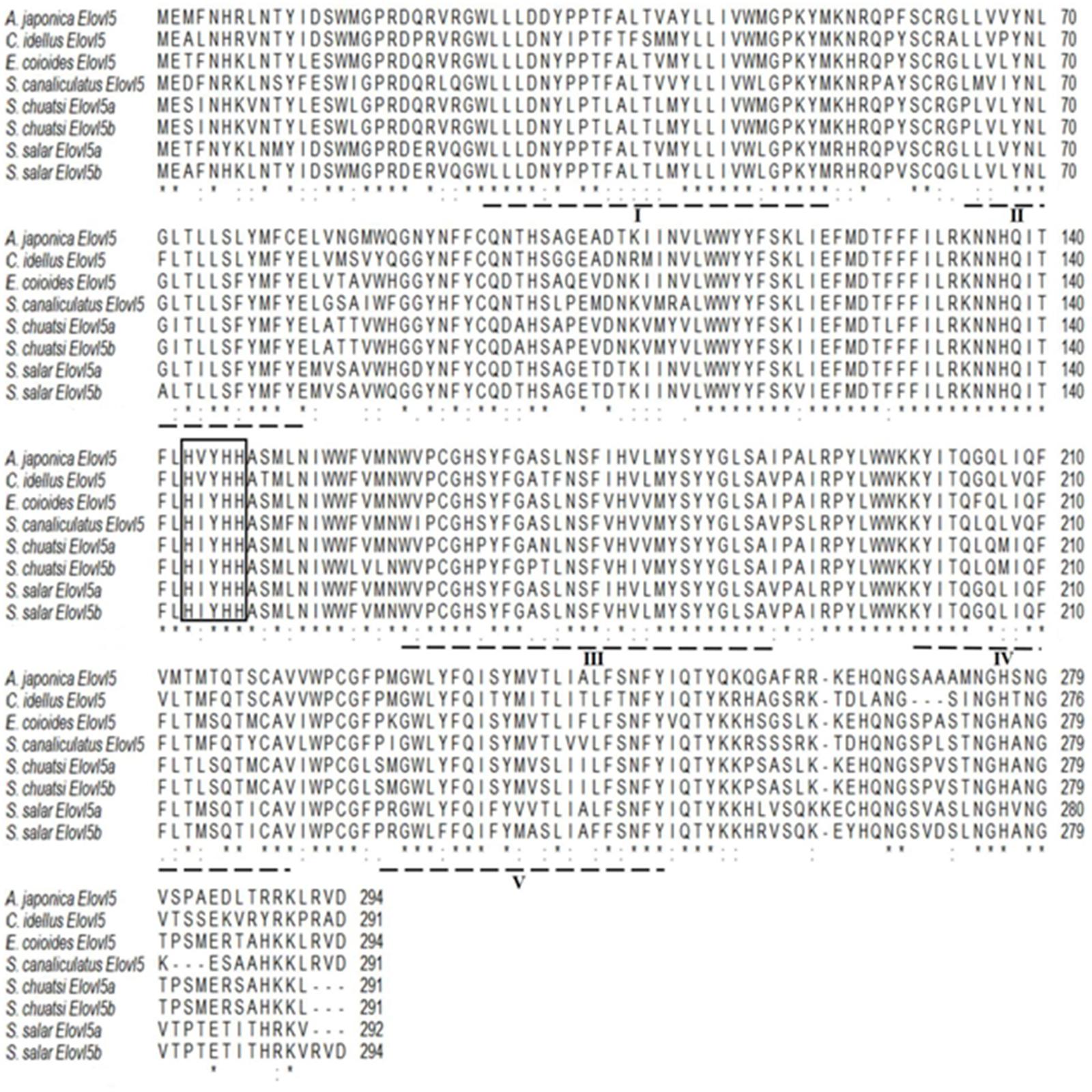
Figure 3. Alignment of the deduced amino acid (aa) sequences of elongases Elovl5 among Anguilla japonica, Siganus canaliculatus, Ctenopharyngodon idellus, Siniperca chuatsi (Elvol5a and Elovl5b), Epinephelus coioides, and Salmo salar (Elvol5a and Elovl5b) using ClustalW2. Identical and similar residues are marked with an asterisk and a colon, respectively. The conserved histidine box HXXHH is highlighted with frames, five putative transmembrane domains are dash-underlined, and the putative ER retrieval signal is solid underlined.
A neighbor-joining phylogenic tree was constructed based on the deduced Elovl5 aa sequences from different species (Figure 4). Our results showed that the phylogenic tree of Elovl5 homologs is almost in accordance with the affinity of those fish. The phylogenic tree of Elovl5 homologs does not really cluster according to habitat ecology as displayed in the phylogenetic tree of teleosts Fads2. The salmonid sequences clustered together with a freshwater fish, Esox lucius, but not with the other diadromous fish species, A. japonica. Interestingly, Oreochromis niloticus and S. chuatsi Elovl5 sequences clustered more closely to the marine fish than to the other freshwater fish.
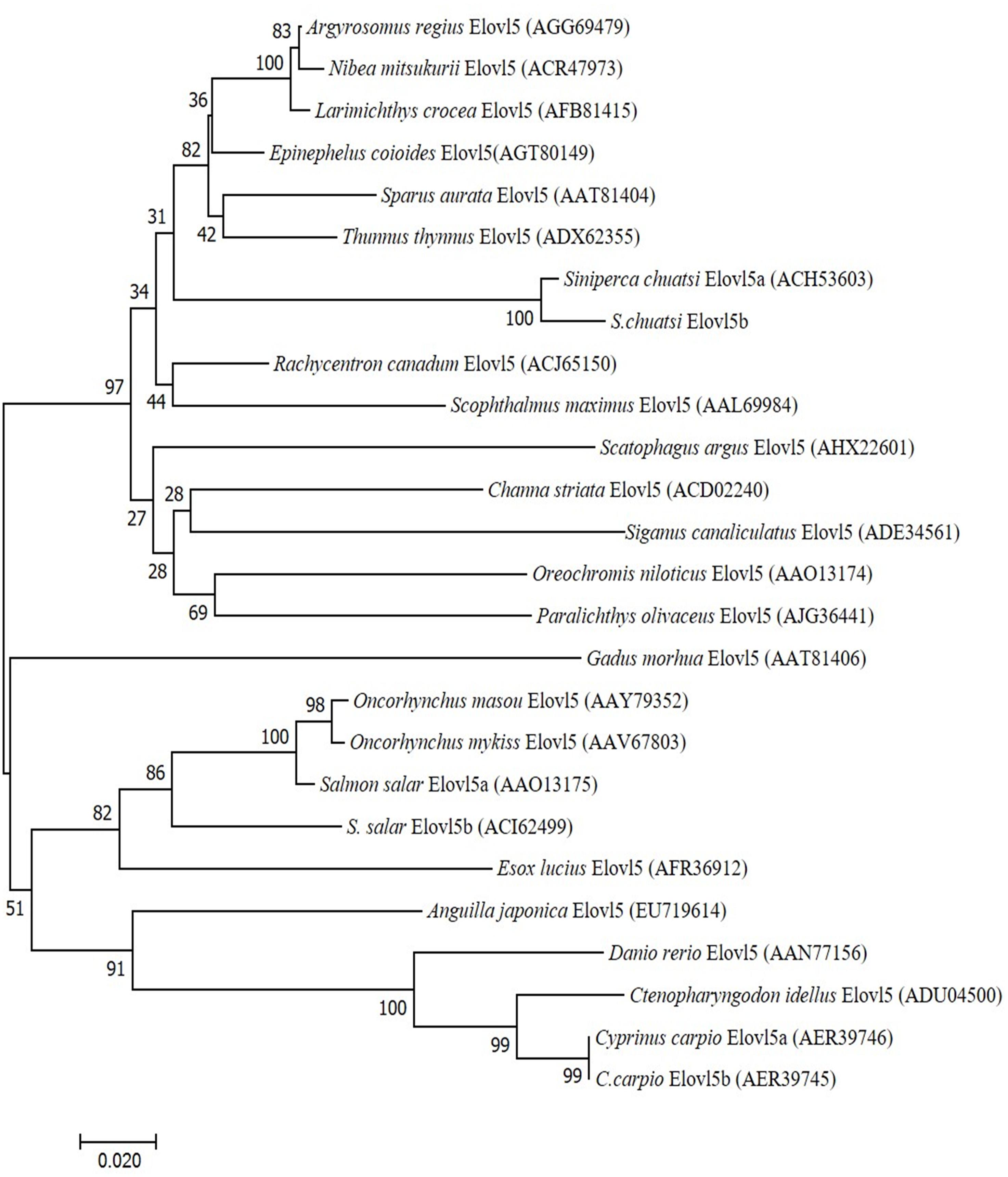
Figure 4. Phylogeny of deduced amino acid (aa) sequences of Elovl5 of different fish species. The tree was constructed using the neighbor-joining (N-J) method (Saitou and Nei, 1987) using CLUSTALX and MEGA 7.0. The horizontal branch length is proportional aa substitution rate per site. The numbers represent the frequencies with which the tree topology presented here was replicated after 1,000 bootstrap iterations.
The functional characteristics of putative Δ6 Fad and Elovl5 isolated from S. chuatsi were determined by heterologous expression in yeast S. cerevisiae grown in the presence of potential FA substrates. The FA profiles of control yeast transformed with the empty pYES2 vector was 16:0 and 16:1 isomers (16:1n–9 and 16:1n–7), 18:0 and 18:1n–9, and any exogenously added PUFA substrate (data not shown).
The FA profile of yeast transformed with S. chuatsi putative Δ6 Fad cDNA additionally showed extra peaks when grown in the presence of 18:2n–6 and 18:3n–3, which corresponded to 18:3n–6 and 18:4n–3, respectively, whereas Δ5 and Δ4 activities were not detected (Figure 5 and Table 2). These data show clearly that the cloned S. chuatsi putative Fads2 had Δ6 Fad specificities. When the S. chuatsi putative Elovl5a/b cDNA was expressed in the yeast cells, evidence of elongation of all fatty acids was observed (Figures 6, 7 and Table 2). Generally, n–3 PUFAs were elongated to a greater extent compared with their corresponding n–6 isomers. Moreover, S. chuatsi Elovl5a had an apparent preference for C18 and C20 over C22 FA substrates. Interestingly, the Elovl5b more effectively converted C22 PUFA substrates than did C18 and C20 substrates.
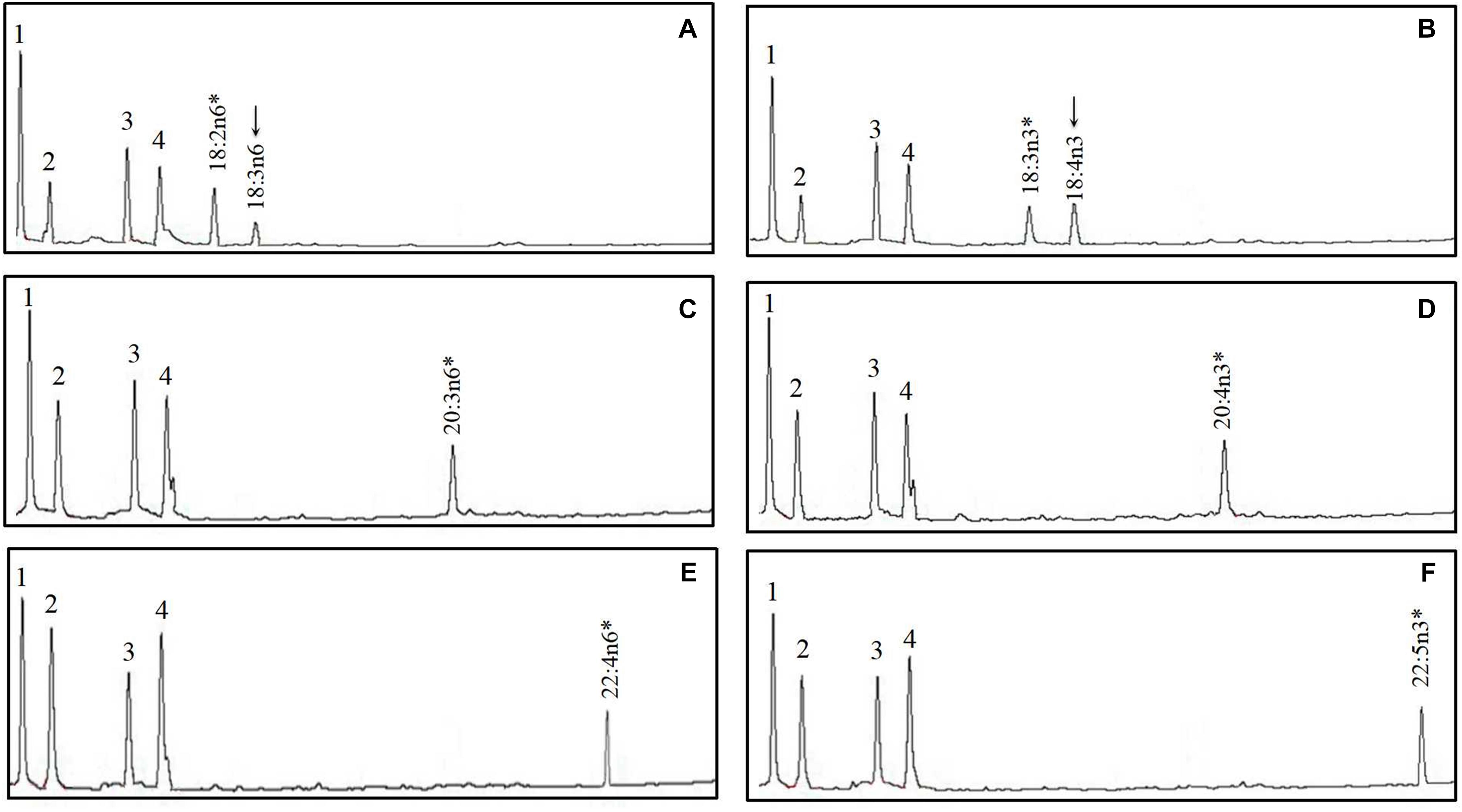
Figure 5. Functional characterization of Fads2 of mandarin fish in Saccharomyces cerevisiae. Fatty acids were extracted from yeast transformed with pYES2 vector containing the putative fatty acid desaturase coding sequence (CDS) inserts from mandarin fish. The added FA substrates (*) were 18:2n–6 (A), 18:3n–3 (B), 20:3n–6 (C), 20:4n–3 (D), 22:4n–6 (E), and 22:5n–3 (F). The first four peaks in (A–F) are the main endogenous fatty acids of S. cerevisiae, namely, 16:0 (1), 16:1n–7 (2), 18:0 (3), and 18:1n–9 (4). Based on retention times, additional peaks (arrowed) were identified as 18:3n–6 (A) and 18:4n–3 (B). Vertical axis, flame ionization detector (FID) response; horizontal axis, retention time.
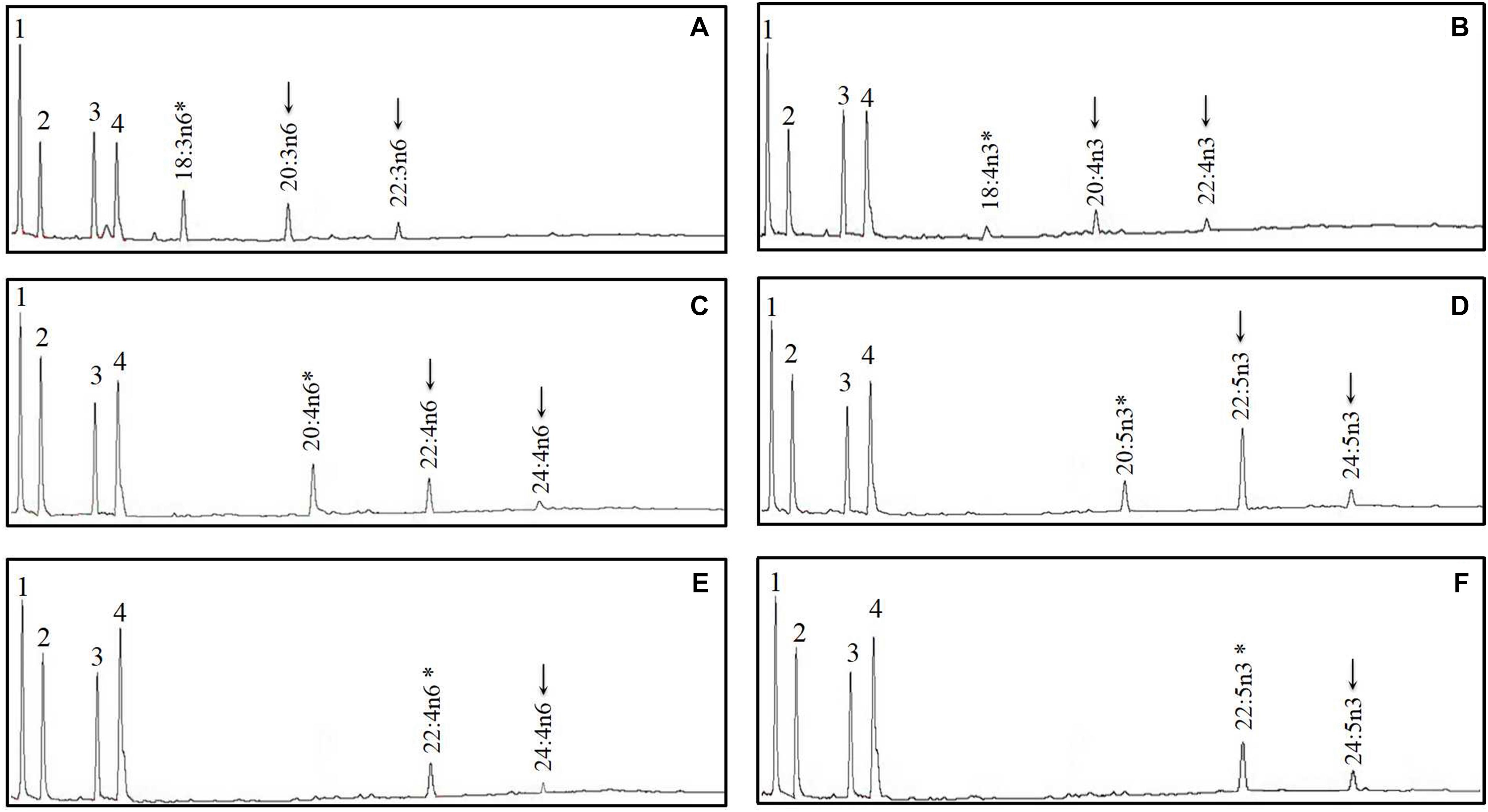
Figure 6. Functional characterization of Elovl5a of mandarin fish in Saccharomyces cerevisiae. Fatty acids were extracted from yeast transformed with pYES2 vector containing the elovl5a CDS inserts from mandarin fish and grown in the presence of polyunsaturated fatty acid (PUFA) substrates 18:3n–6 (A), 18:4n–3 (B), 20:4n–6 (C), 20:5n–3 (D), 22:4n–6 (E), and 22:5n–3 (F). Based on retention times, substrates (*) and their corresponding elongated products (↓) are indicated accordingly. The first four peaks in (A–F) are the main endogenous fatty acids of S. cerevisiae, namely, 16:0 (1), 16:1n–7 (2), 18:0 (3), and 18:1n–9 (4). Vertical axis, FID response; horizontal axis, retention time.
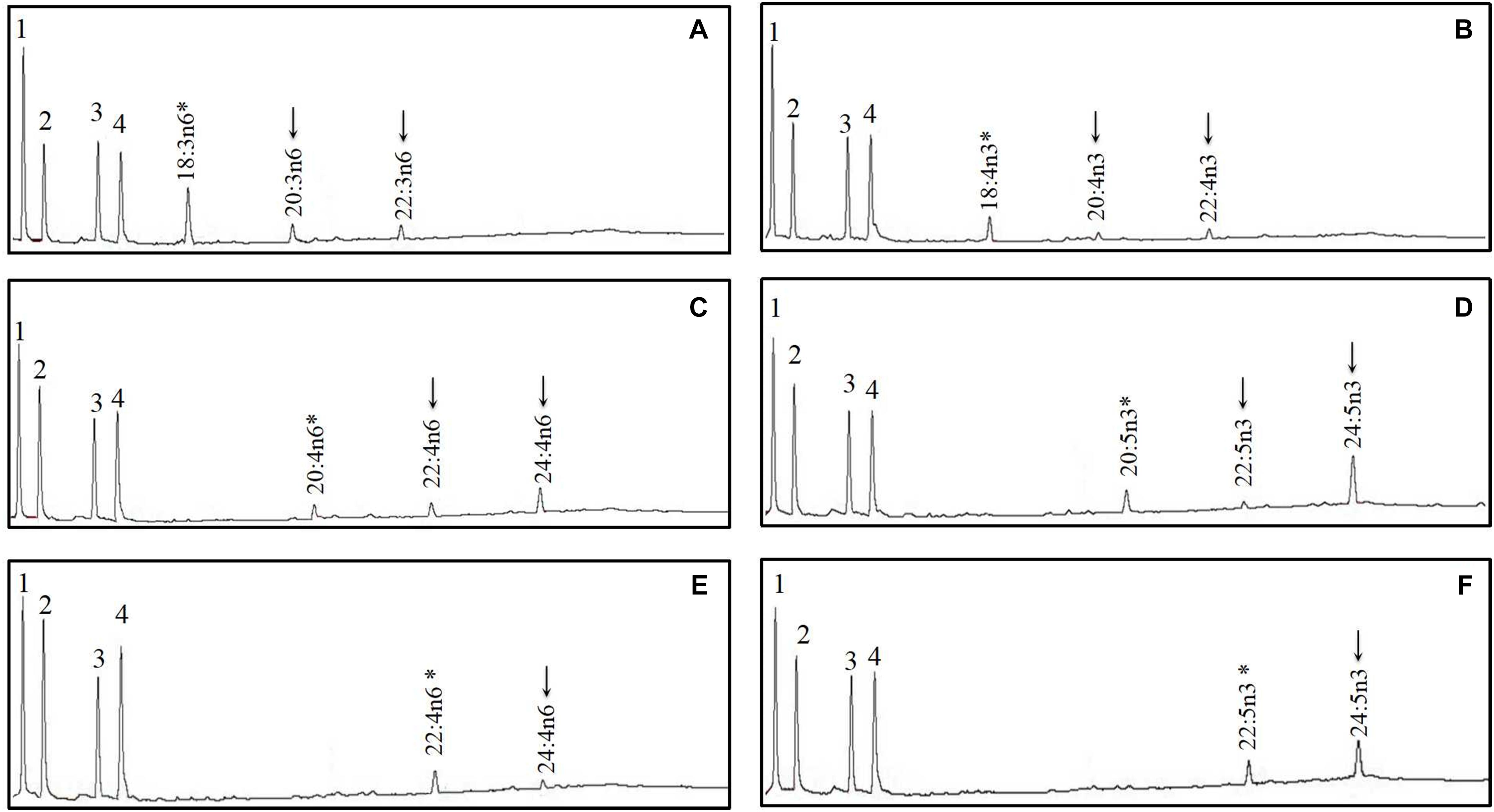
Figure 7. Functional characterization of Elovl5b of mandarin fish in Saccharomyces cerevisiae. Transformed yeast with pYES2 vector containing the elovl5b CDS inserts from mandarin fish grown in the presence of polyunsaturated fatty acid (PUFA) substrates 18:3n–6 (A), 18:4n–3 (B), 20:4n–6 (C), 20:5n–3 (D), 22:4n–6 (E), and 22:5n–3 (F). Based on retention times, substrates (*) and their corresponding elongated products (↓) are indicated accordingly. The first four peaks in panels A–F are the main endogenous fatty acids of S. cerevisiae, namely, 16:0 (1), 16:1n–7 (2), 18:0 (3), and 18:1n–9 (4). Vertical axis, FID response; horizontal axis, retention time.
The enzymatic activity of Δ6 Fad and Elovl5 was determined by heterologous expression in yeast S. cerevisiae, grown in the presence of a variety of fatty acid substrates including Δ6 Fad substrates (18:2n–6 and 18:3n–3) and Elovl5 substrates (18:3n–6, 18:4n–3, ARA and EPA) (Tables 3, 4). The enzymatic activity of all Δ6 Fad were more active toward the n–3 substrate with 79.23, 68.22, 76.92, 52.61, 76.71, and 68.02% of 18:3n–3 being converted to 18:4n–3 in the case of C. idellus, S. chuatsi, S. canaliculatus, E. coioides, S. salar, and A. japonica, respectively (Table 3 and Supplementary Figure 3). The conversion of 18:3n–3 and 18:2n–6 in C. idellus, S. canaliculatus, and A. japonica was significantly higher than that in S. chuatsi, S. salar, and E. coioides. In comparison, the lowest conversion of 18:3n–3 and 18:2n–6 was shown in E. coioides Δ6 Fad (Supplementary Figure 4).
For the six teleosts, the conversion rates of Elovl5 toward fatty acid substrates are shown in Table 4 and Supplementary Figures 5–8. Generally, high elongations were obtained with n–3 compared with n–6 PUFAs. For C18 PUFA substrates (Supplementary Figures 5, 6), A. japonica Elovl5 had higher elongations than those in other fish species but exhibited lower activities toward the C20 PUFAs (Supplementary Figures 7, 8). Particularly interesting was the difference in the conversion function observed between S. chuatsi Elovl5a and Elovl5b, but the conversion function of S. salar Elovl5a and Elovl5b showed no difference.
The present study provided evidence for the existence of both fads2 and elovl5 encoding cDNAs and demonstrated their role in the LC-PUFA biosynthesis of the freshwater carnivorous Siniperca chuatsi. The aa sequence of S. chuatsi Fads2 possesses all the main structural features common for Fads protein family members, including N-terminal cytochrome-b5-like domains, the heme-binding motif HPGG, three histidine boxes, and four predicted transmembrane domains (Hashimoto et al., 2008). Phylogenetic analysis revealed that S. chuatsi Fads2 shares high homology with other teleosts, and the Fads2 of representatives basically cluster together by their habitats. However, S. chuatsi Fads2 is not closely related to that of the other freshwater fish species but rather to the marine carnivorous fish species (Figure 2). Consistently, the S. chuatsi Fads2, like the majority of Fads2 isolated from marine carnivorous fish (Tocher et al., 2006; Morais et al., 2012), has a monofunctional Δ6 desaturase and does not appear to possess Δ5 or Δ4 activities as in striped snakehead (Kuah et al., 2015). Attempts have been made to search another fads2 gene from S. chuatsi genome data, but these have so far been unsuccessful. Although the S. chuatsi seems more like a marine fish without LC-PUFA biosynthesis ability, a feeding trial demonstrated that total replacement of dietary FOs with alternative VO has no negative impact on the growth performance and health of mandarin fish juvenile, which indirectly suggested this species could bioconvert C18 PUFA to their corresponding LC-PUFA (Sankian et al., 2019). Collectively, these findings highlight that teleosts have an adaptive plasticity and diversity of LC-PUFA biosynthesis mechanism (Fonseca-Madrigal et al., 2014).
In the present study, two elovl5 CDSs (elovl5a and elovl5b) were identified in S. chuatsi, the likes of which have been seen in Atlantic salmon and common carp (Cyprinus carpio var. Jian) (Morais et al., 2009; Ren et al., 2012). Analysis of the deduced aa sequences of S. chuatsi Elovl5a and Elovl5b showed that they both have all the typical characteristic features of the predicted transmembrane domains, the histidine box, and the canonical C-terminal ER retrieval signal (Jakobsson et al., 2006). Phylogenetic analysis showed that the Elovl5 homologs are in accordance with the order of fish but not with their feeding habits and habitat. All the Elovl5 sequences of Pereiformes, Salmoniformes, and Cypriniformes clustered together (Figure 4). A similar phylogenetic grouping was observed previously for the sequences of elongases from a range of teleosts (Agaba et al., 2005). The results of the functional characterization revealed that the capability of S. chuatsi Elovl5a and Elovl5b, similar to the other Elovl5 homologs, exhibits an effective ability to elongate both C18 and C20 PUFA and displays a preference to elongate n–3 PUFA substrates compared with n–6 PUFA substrates (Monroig et al., 2011; Kuah et al., 2015; Xie et al., 2016; Janaranjani et al., 2018; Ferraz et al., 2019). Interestingly, S. chuatsi Elovl5a has a higher activity toward C18–C20 FAs than has Elovl5b, whereas Elovl5b showed a noteworthy activity toward C22 FAs (60.47% conversion of 22:5n–3) as the Elovl2 does (Morais et al., 2009; Gregory and James, 2014; Oboh et al., 2016). As for most fish species, S. chuatsi elovl2 cDNA has not been isolated successfully (data not shown), whereas in these teleost, the conversion of C22 FAs by other elongases, such as Elovl5 and Elovl4, potentially compensates for the absence of Elovl2 in DHA biosynthesis (Morais et al., 2009; Wang et al., 2014; Xie et al., 2016).
To date, fads2 cDNAs have been identified in numerous fish species, and all tested Fads2 enzymes showed the ability to operate as Δ6 Fad, which is likely the primary function of Fads2 for teleosts (Castro et al., 2016; Janaranjani et al., 2018; Ferraz et al., 2019). In the present study, the Δ6 Fad activity of six tested fish species was performed in the same yeast expression system, which showed high efficiency, with conversion rates of 18:2n–6 and 18:3n–3 ranging at 30.38–60.6 and 52.61–79.23%, respectively. However, the activities of desaturases varied markedly among these species. The conversion efficiency of Δ6 Fad in marine carnivorous fish Epinephelus coioides is significantly lower than that in other fish species. Previous studies reported that the conversion rates of Δ6 Fad in grouper, rabbitfish, salmon, and eel were 4.4–9.78% (Li et al., 2014), 35–59% (Li et al., 2010), 25–47% (Monroig et al., 2010), and 20.7–60.8% (Wang et al., 2014), respectively. Although these data of genes conversion rates in the present study are somewhat different from those in the previous studies; the order of genes conversion rates in grouper, rabbitfish, salmon, and eel is consistent in the present study and previous works. Consistently, nutritional trials have shown that VO can satisfy the essential fatty acid (EFA) requirements of Ctenopharyngodon idellus, Siganus canaliculatus, Anguilla japonica, S. chuatsi, and Salmo salar; and their Δ6 fad gene expression was increased by dietary VO (rich in C18 PUFA) (Takeuchi et al., 1980; Monroig et al., 2011; Lei et al., 2017; Xie et al., 2018; Sankian et al., 2019), whereas E. coioides Δ6 Fad had a low enzymatic activity in converting LNA and LA because of the deficiency of binding site for the stimulatory protein 1 (Sp1) in its promoter (Li et al., 2014; Xie et al., 2018). Among the freshwater species, herbivorous C. idellus has a higher Δ6 Fad activity than has carnivorous S. chuatsi, and the similar effect of feeding habit on the desaturase’s activity was also exhibited between the marine species. Among the carnivorous fish species, the highest Δ6 Fad activity was detected in the catadromous A. japonica, followed by freshwater and anadromous species, and the lowest in marine species, whereas the influence of habitats on the Δ6 Fad activity was little in the herbivorous fish species. Those results suggested that the Δ6 Fad activity of fish is under the influence of both feeding habits and habitats. Furthermore, the Δ8 activity of Fads2 varied notably among the different fish species, and a higher Δ8 capability was detected in marine fish compared with freshwater/diadromous species (Monroig et al., 2011). On the other hand, those results confirmed that the functions and capabilities of teleost Fads2 have diversified remarkably as a result of environmental factors including habitat, trophic level, and ecology (Castro et al., 2016).
Besides Fads2, the above-mentioned environmental factors have also been suggested as potential drivers modulating the elongation capabilities of teleosts (Agaba et al., 2005; Carmona-Antoñanzas et al., 2013; Wang et al., 2014; Janaranjani et al., 2018). In a comparison of Elovl5 activities on C18–C22 PUFA substrates among seven fish species, Elovl5 activities were more likely to elongate n–3 substrates than n–6 substrates, with the exception of the Atlantic cod (Gadus morhua, Gadiformes) elongase, which was more active toward the n–6 homologs (Agaba et al., 2005). Simiarly, Elovl5 activities of all fish from different ecological backgrounds were high toward n–3 PUFAs, whereas the S. chuatsi Elovl5b exhibited a preference for n–6 substrates. The pattern of activities on different PUFAs substrates showed that the Elovl5 from A. japonica and S. salar exhibited a rank order of C18 > C20, which was very similar to those of most fish species (Castro et al., 2016; Xie et al., 2016; Janaranjani et al., 2018; Ferraz et al., 2019). The Elovl5 from S. chuatsi and E. coioides showed a similar activity with C18 and C20 PUFAs. Interestingly, herbivorous C. idellus and S. canaliculatus elongases were less active toward C18 substrates than were the other elongases, but they displayed higher activities toward the C20 PUFAs (Monroig et al., 2012), which might be linked to the abundance of C18 PUFAs and limited C20 PUFAs in their food web. In general, the trophic level and ecology have a bigger impact on the Elovl5 activity of teleosts than the habitat.
The present study demonstrated S. chuatsi Fads2 with Δ6 Fad capabilities, and its Elovl5a showed a preference toward n–3 C18–C20 PUFAs, whereas the Elovl5b showed substrate specificity toward C22 PUFAs. Furthermore, the desaturation and elongation capabilities of Δ6 Fad and Elovl5 were compared among six fish species from different ecological backgrounds, which indicated that the Δ6 Fad activity of fish is under the double influence of feeding habits and habitats, whereas the Elovl5 activity of teleosts was affected more by the trophic level and ecology. Those differences in the functional competences of the Δ6 Fad and Elovl5 from different fish species may contribute to the different LC-PUFA biosynthesis abilities of the species. These results increase our knowledge of the molecular basis of LC-PUFA biosynthesis and its regulation in teleosts and provide guidance on choosing suitable dietary PUFA precursors for those farmed fish species.
This study has a drawback. The present study compared the conversion rates of Fads2 and Elovl5 among the six teleosts under the same in vitro conditions, which is not enough to explain the difference of Fads2 and Elovl5 activities in vivo. The enzymatic activities are related to the enzymatic kinetics, such as the Michaelis constant (Km) and maximum velocity of the reaction (Vmax). Therefore, more studies are needed to fully investigate the enzymatic kinetics of Fads2 and Elovl5.
All datasets generated for this study are included in the article/Supplementary Material.
The animal study was reviewed and approved by the statement to confirm that all experimental protocols were approved by the Guangdong Provincial Department of Science and Technology on the use and care of experimental animals. The study was reviewed and approved by the Ethics Committee of Animal Experiments of South China Agricultural University.
DX and YL wrote the manuscript. SW, CY, and YL designed the study. JY and ML conducted the experiments. All authors read and approved the final version of the manuscript.
This work was financially supported by the National Natural Science Foundation of China (31873040) and National Key R&D Program of China (2018YFD0900400).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00117/full#supplementary-material
Agaba, M. K., Tocher, D. R., Zheng, X., Dickson, C. A., Dick, J. R., and Teale, A. J. (2005). Cloning and functional characterisation of polyunsaturated fatty acid elongases of marine and freshwater teleost fish. Comp. Biochem. Phys. B. 142, 342–352. doi: 10.1016/j.cbpb.2005.08.005
Awada, M., Meynier, A., Soulage, C. O., Hadji, L., Géloën, A., Viau, M., et al. (2013). n-3 PUFA added to high-fat diets affect differently adiposity and inflammation when carried by phospholipids or triacylglycerols in mice. Nutr. Metab. 10, 23. doi: 10.1186/1743-7075-10-23
Carmona-Antoñanzas, G., Tocher, D. R., Taggart, J. B., and Leaver, M. J. (2013). An evolutionary perspective on Elovl5 fatty acid elongase: comparison of Northern pike and duplicated paralogs from Atlantic salmon. BMC Evol. Biol. 13:85. doi: 10.1186/1471-2148-13-85
Castro, L. F. C., Monroig, O., Leaver, M. J., Wilson, J., Cunha, I., and Tocher, D. R. (2012). Functional desaturase Fads1 (D5) and Fads2 (D6) orthologues evolved before the origin of jawed. PloS One 7:e31950. doi: 10.1371/journal.pone.0031950
Castro, L. F. C., Tocher, D. R., and Monroig, O. (2016). Long-chain polyunsaturated fatty acid biosynthesis in chordates: insights into the evolution of Fads and Elovl gene repertoire. Prog. Lipid Res. 62, 25–40. doi: 10.1016/j.plipres.2016.01.001
Delgado-Lista, J., Perez-Martinez, P., Lopez-Miranda, J., and Perez-Jimenez, F. (2012). Long chain omega-3 fatty acids and cardiovascular disease: a systematic review. Brit. J. Nutr. 107, S201–S213.
Du, J., Liang, X., Cheng, J., Zhu, T., Zhu, Z., and Yu, Y. (2011). Molecular cloning and analyzing of fatty acid desaturase and elongase genes in grass carp (Ctenopharyngodon idellus). Chin. J. Jinan Univ. 32, 513–520.
FAO (2014). The State of World Fisheries and Aquaculture 2014. Rome, Italy: FAO Fisheries and Aquaculture Department.
Ferraz, R. B., Kabeya, N., Lopes-Marques, M., Machado, A. M., Ribeiro, R. A., Salaro, A. L., et al. (2019). A complete enzymatic capacity for long-chain polyunsaturated fatty acid biosynthesis is present in the Amazonian teleost tambaqui. Colossoma macropomum. Comp. Biochem. Phys. B. 227, 90–97. doi: 10.1016/j.cbpb.2018.09.003
Folch, J., Lees, M., and Stanley, G. S. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509.
Fonseca-Madrigal, J., Navarro, J. C., Hontoria, F., Tocher, D. R., Martínez-Palacios, C. A., and Monroig, Ó (2014). Diversification of substrate specificities in teleostei fads2: characterization of Δ4 and Δ6Δ5 desaturases of Chirostomaestor. J. Lipid Res. 55, 1408–1419. doi: 10.1194/jlr.M049791
Gregory, M. K., and James, M. J. (2014). Elovl5 and Elovl2 differ in selectivity for elongation of omega-3 docosapentaenoic acid. BBA Mol. Cell Biol. Lipids 1841, 1656–1660. doi: 10.1016/j.bbalip.2014.10.001
Hashimoto, K., Yoshizawa, A. C., Okuda, S., Kuma, K., Goto, S., and Kanehisa, M. (2008). The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J. Lipid Res. 49, 183–191. doi: 10.1194/jlr.m700377-jlr200
Hossain, M. A. (2011). Fish as source of n-3 polyunsaturated fatty acids (PUFAs), which one is better-farmed or wild. Adv. J. Food Sci. Technol. 3, 455–466.
Jakobsson, A., Westerberg, R., and Jacobsson, A. (2006). Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog. Lipid. Res. 45, 237–249. doi: 10.1016/j.plipres.2006.01.004
Janaranjani, M., Mah, M. Q., Kuah, M. K., Fadhilah, N., Hing, S. R., Han, W. Y., et al. (2018). Capacity for eicosapentaenoic acid and arachidonic acid biosynthesis in silver barb (Barbonymus gonionotus): functional characterisation of a Δ6/Δ8/Δ5 Fads2 desaturase and Elovl5 elongase. Aquaculture 497, 469–486. doi: 10.1016/j.aquaculture.2018.08.019
Kuah, M. K., Jaya-Ram, A., and Shu-Chien, A. C. (2015). The capacity for long-chain polyunsaturated fatty acid synthesis in a carnivorous vertebrate: functional characterisation and nutritional regulation of a Fads2 fatty acyl desaturase with Δ4 activity and an Elovl5 elongase in striped snakehead (Channa striata). BBA ol. Cell Biol. Lipids 1851, 248–260. doi: 10.1016/j.bbalip.2014.12.012
Leaver, M. J., Bautista, J. M., Björnsson, B. T., Jönsson, E., Krey, G., Tocher, D. R., et al. (2008). Towards fish lipid nutrigenomics: current state and prospects for fin-fish aquaculture. Rev. Fish. Sci. 16, 73–94. doi: 10.1080/10641260802325278
Lei, C., Tian, J., Ji, H., Chen, L., and Du, Z. (2017). Dietary α-linolenic acid affects lipid metabolism and tissue fatty acid profile and induces apoptosis in intraperitoneal adipose tissue of juvenile grass carp (Ctenopharyngodon idella). Aquacul. Nutr. 23, 160–170. doi: 10.1111/anu.12377
Li, S., Mai, K., Xu, W., Yuan, Y., Zhang, Y., and Ai, Q. (2014). Characterization, mRNA expression and regulation of Δ6 fatty acyl desaturase (FADS2) by dietary n-3 long chain polyunsaturated fatty acid (LC-PUFA) levels in grouper larvae (Epinephelus coioides). Aquaculture 434, 212–219. doi: 10.1016/j.aquaculture.2014.08.009
Li, S., Yuan, Y., Wang, T., Xiang, X., Li, Y., Du, J., et al. (2016). Molecular cloning, functional characterization and nutritional regulation of the putative elongase Elovl5 in the orange-spotted grouper (Epinephelus coioides). PloS One 11:e0150544. doi: 10.1371/journal.pone.0150544
Li, Y., Monroig, Ó, Zhang, L., Wang, S., Zheng, X., Dick, J. R., et al. (2010). Vertebrate fatty acyl desaturase with Delta4 activity. Proc. Natl. Acad. Sci. U.S.A. 107, 16840–16845. doi: 10.1073/pnas.1008429107
Lu, K., Wang, L., Zhang, D., Liu, W., and Xu, W. (2017). Berberine attenuates oxidative stress and hepatocytes apoptosis via protecting mitochondria in blunt snout bream Megalobrama amblycephala fed high-fat diets. Fish Physiol. Biochem. 43, 65–76. doi: 10.1007/s10695-016-0268-5
Monroig, Ó, Li, Y., and Tocher, D. R. (2011). Delta-8 desaturation activity varies among fatty acyl desaturases of teleost fish: high activity in delta-6 desaturases of marine species. Comp. Biochem. Phys. B 159, 206–213. doi: 10.1016/j.cbpb.2011.04.007
Monroig, Ó, Tocher, D. R., Hontoria, F., and Navarro, J. C. (2013). Functional characterisation of a fads2 fatty acyl desaturase with Δ6/Δ8 activity and an elovl5 with C16, C18 and C20 elongase activity in the anadromous teleost meagre (Argyrosomus regius). Aquaculture 412-413, 14–22. doi: 10.1016/j.aquaculture.2013.06.032
Monroig, Ó, Wang, S., Zhang, L., You, C., Tocher, D. R., and Li, Y. (2012). Elongation of long-chain fatty acids in rabbitfish Siganus canaliculatus: cloning, functional characterisation and tissue distribution of Elovl5- and Elovl4-like elongases. Aquaculture 350–353, 63–70. doi: 10.1016/j.aquaculture.2012.04.017
Monroig, Ó, Zheng, X., Morais, S., Leaver, M. J., Taggart, J. B., and Tocher, D. R. (2010). Multiple genes for functionalΔ 6 fatty acyl desaturases (Fad) in Atlantic salmon (Salmo salar L.): gene and cDNA characterization, functional expression, tissue distribution and nutritional regulation. BBA Mol. Cell Biol. Lipids 1801, 1072–1081. doi: 10.1016/j.bbalip.2010.04.007
Morais, S., Castanheira, F., Martinez-Rubio, L., Conceição, L. E., and Tocher, D. R. (2012). Long chain polyunsaturated fatty acid synthesis in a marine vertebrate: ontogenetic and nutritional regulation of a fatty acyl desaturase with Δ4 activity. BBA- Mol. Cell Biol. Lipids 1821, 660–671. doi: 10.1016/j.bbalip.2011.12.011
Morais, S., Monroig, Ó, Zheng, X., Leaver, M. J., and Tocher, D. R. (2009). Highly unsaturated fatty acid synthesis in Atlantic salmon: characterization of ELOVL5- and ELOVL2-like elongases. Mar. Biotechnol. 11, 627–639. doi: 10.1007/s10126-009-9179-0
Oboh, A., Betancor, M. B., Tocher, D. R., and Monroig, Ó (2016). Biosynthesis of long-chain polyunsaturated fatty acids in the African catfish Clarias gariepinus: molecular cloning and functional characterisation of fatty acyl desaturase (fads2) and elongase (elovl2) cDNAs7. Aquaculture 462, 70–79. doi: 10.1016/j.aquaculture.2016.05.018
Oboh, A., Kabeya, N., Carmona-Antoñanzas, G., Castro, L. F. C., Dick, J. R., Tocher, D. R., et al. (2017). Two alternative pathways for docosahexaenoic acid (DHA, 22: 6n-3) biosynthesis are widespread among teleost fish. Sci. Rep.UK 7, 3889. doi: 10.1038/s41598-017-04288-2
Ren, H., Yu, J., Xu, P., and Tang, Y. (2012). Influence of dietary fatty acids on muscle fatty acid composition and expression levels of Δ6 desaturase-like and Elovl5-like elongase in common carp (Cyprinus carpio var. Jian). Comp. Biochem. Phys. B. 163, 184–192. doi: 10.1016/j.cbpb.2012.05.016
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sankian, Z., Khosravi, S., Kim, Y. O., and Lee, S. M. (2019). Total replacement of dietary fish oil with alternative lipid sources in a practical diet for mandarin fish. Siniperca scherzeri, juveniles. Fish Aquat. Sci. 22, 8.
Sprague, M., Dick, J. R., and Tocher, D. R. (2016). Impact of sustainable feeds on omega-3 long-chain fatty acid levels in farmed Atlantic salmon, 2006-2015. Sci. Rep. UK 6, 21892. doi: 10.1038/srep21892
Takeuchi, T., Arai, S., Watanabe, T., and Shimma, Y. (1980). Requirement of eel Anguilla japonica for essential fatty acids. Bull. Japane. Soc. Sci. Fishe. 46, 345–353. doi: 10.2331/suisan.46.345
Tocher, D. R. (2010). Fatty acid requirements in ontogeny of marine and freshwater fish. Aquac. Res. 41, 717–732. doi: 10.1111/j.1365-2109.2008.02150.x
Tocher, D. R., Zheng, X., Schlechtriem, C., Hastings, N., Dick, J. R., and Teale, A. J. (2006). Highly unsaturated fatty acid synthesis in marine fish: cloning, functional characterization, and nutritional regulation of fatty acyl Δ6 desaturase of Atlantic cod (Gadus morhua L.). Lipids 41, 1003–1016. doi: 10.1007/s11745-006-5051-4
Turchini, G. M., Francis, D. S., Senadheera, S. P. S. D., Thanuthong, T., and De Silva, S. S. (2011). Fish oil replacement with different vegetable oils in Murray cod: evidence of an “omega-3 sparing effect” by other dietary fatty acids. Aquaculture 315, 250–259. doi: 10.1016/j.aquaculture.2011.02.016
Uauy, R., Hoffman, D. R., Peirano, P., Birch, D. G., and Birch, E. (2001). Essential fatty acids in visual and brain development. Lipids 36, 885–895.
Wang, S., Monroig, Ó, Tang, G., Zhang, L., You, C., Tocher, D. R., et al. (2014). Investigating long-chain polyunsaturated fatty acid biosynthesis in teleost fish: functional characterization of fatty acyl desaturase (Fads2) and Elovl5 elongase in the catadromous species, Japanese eel Anguilla japonica. Aquaculture 434, 57–65. doi: 10.1016/j.aquaculture.2014.07.016
Xie, D., Chen, F., Lin, S., Wang, S., You, C., and Monroig, Ó, et al. (2014). Cloning, functional characterization and nutritional regulation of Δ6 fatty acyl desaturase in the herbivorous euryhaline teleost Scatophagus argus. Plos One 9:e90200. doi: 10.1371/journal.pone.0090200
Xie, D., Chen, F., Lin, S., You, C., Wang, S., Zhang, Q., et al. (2016). Long-chain polyunsaturated fatty acid biosynthesis in the euryhaline herbivorous teleost Scatophagus argus: functional characterization, tissue expression and nutritional regulation of two fatty acyl elongases. Comp. Biochem. Phys. B 198, 37–45. doi: 10.1016/j.cbpb.2016.03.009
Keywords: Siniperca chuatsi, Δ6 Fad, Elovl5, long-chain polyunsaturated fatty acid, teleosts
Citation: Xie D, Ye J, Lu M, Wang S, You C and Li Y (2020) Comparsion of Activities of Fatty Acyl Desaturases and Elongases Among Six Teleosts With Different Feeding and Ecological Habits. Front. Mar. Sci. 7:117. doi: 10.3389/fmars.2020.00117
Received: 16 January 2020; Accepted: 13 February 2020;
Published: 20 March 2020.
Edited by:
Kang-le Lu, Jimei University, ChinaCopyright © 2020 Xie, Ye, Lu, Wang, You and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanyou Li, eXlsaTE2QHNjYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.