- 1Experimental Marine Ecology Laboratory, National University of Singapore, Singapore, Singapore
- 2Singapore Centre for Environmental Life Sciences Engineering, Nanyang Technological University, Singapore, Singapore
- 3Centre for Marine Science and Innovation, School of Biological, Earth and Environmental Sciences, University of New South Wales, Sydney, NSW, Australia
- 4Sydney Institute of Marine Science, Mosman, NSW, Australia
- 5The ithree Institute, University of Technology Sydney, Sydney, NSW, Australia
Competitive interactions between corals and macroalgae play an important role in determining benthic community structure on coral reefs. While it is known that macroalgae may negatively affect corals, the relative influence of contact- versus water-mediated macroalgal interactions on corals – such as via an influence on coral-associated microbiomes – is less well understood. Further, the impacts of macroalgae on corals that have persisted in a heavily urbanized reef system have not been explored previously. We examined the effects of the macroalgae Lobophora sp. and Hypnea pannosa on the physiology and microbiome of three Indo-Pacific coral species (Merulina ampliata, Montipora stellata, and Pocillopora acuta) collected from two reefs in Singapore (Pulau Satumu and Kusu Island), and compared how these effects varied between direct contact and water-mediated interactions. Direct contact by Lobophora sp. caused visible tissue bleaching and reduced maximum quantum yield (Fv/Fm) in all three coral species, while direct contact by H. pannosa only led to slight, but significant, suppression of Fv/Fm. No detrimental effects on coral physiology were observed when corals were in close proximity to the macroalgae or when in direct contact with algal mimics. However, both direct contact and water-mediated interactions with Lobophora sp. and H. pannosa altered the prokaryotic community structures in M. stellata. For M. ampliata and P. acuta, the changes in their microbiomes in response to algal treatments were more strongly influenced by the source reefs from which the coral colonies were collected. In particular, coral colonies collected from Kusu Island had proportionately more initial abundances of potentially pathogenic bacteria in their microbiomes than those collected from Pulau Satumu; nevertheless, coral fragments from Kusu Island had the same physiological responses to macroalgal interactions as corals from Pulau Satumu. Overall, our results reveal that, for the species tested, the coral microbiomes were sensitive to both direct contact and water-mediated interactions with macroalgae, while coral physiology was only compromised when in direct contact. Further, the presence of high levels of potentially pathogenic bacteria in some of the coral samples did not lead to the corals being more susceptible to impacts from macroalgae.
Introduction
Competition plays a critical role in shaping the ecology and evolution of species, as well as determining the structure and function of ecological communities (Yodzis, 1978; Hooper et al., 2005). On coral reefs, intense competition for space occurs among sessile benthic organisms, particularly between corals and macroalgae (Miller, 1998; McCook et al., 2001). In recent decades, anthropogenic local stressors such as overfishing and eutrophication, as well as global ocean warming from climate change have resulted in dramatic declines in coral cover and increasing prevalence of macroalgae on degraded reefs (Bellwood et al., 2004; Hughes et al., 2010, 2018). As a consequence, there is a potential for reefs to shift from coral- to macroalgal-dominated systems (Hughes et al., 2010). Macroalgae directly and indirectly impact corals via a suite of physical, chemical, and microbial processes (McCook et al., 2001; Rasher et al., 2011; Barott and Rohwer, 2012). Hence, high macroalgal abundance can reinforce a coral-depauperate state by reducing coral health, growth, reproduction, and recruitment (Tanner, 1995; Box and Mumby, 2007; Evensen et al., 2019; reviewed in McCook et al., 2001 and Birrell et al., 2008). While it is known that macroalgae can negatively affect corals, the relative influence of direct contact and water-mediated interactions with macroalgae are less understood – especially how these interactions may impact coral-associated microbiomes.
Direct macroalgal contact can harm corals through shading, abrasion, and/or production of harmful allelochemicals that cause coral tissue bleaching and/or alter the coral microbiome (River and Edmunds, 2001; Rasher et al., 2011; Morrow et al., 2012). Indirect, water-mediated interactions with macroalgae can also have deleterious effects; however, to what extent this occurs remains unclear. For example, macroalgae can transmit pathogenic bacteria to corals, triggering coral diseases (Nugues et al., 2004). Macroalgae can also exude allelochemicals and compounds (e.g., dissolved organic carbon) into surrounding waters, which has been reported to promote the growth of coral-associated microbes and lead to hypoxic stress in corals (Haas et al., 2011, 2013; Barott and Rohwer, 2012; Jorissen et al., 2016; Roach et al., 2017). Furthermore, advection of macroalgal chemicals and microbes by water flow may indirectly affect the coral microbiome and trigger coral mortality (Barott and Rohwer, 2012). High variability in the competitive abilities among coral and macroalgal species precludes generalizations of the processes involved and the outcomes of these interactions (McCook et al., 2001; Jompa and McCook, 2003).
Corals harbor a range of microorganisms including bacteria, protists, archaea, fungi, and viruses (the “microbiome”) that are critical for maintaining coral health (Rosenberg et al., 2007; Bourne et al., 2016). For example, some coral-associated bacteria assist corals in assimilation of nutrients and prevention of pathogenic infections through antibiotic production (Lesser et al., 2004; Ritchie, 2006; Lema et al., 2012). Coral microbiomes also play important roles in the ecological interactions between corals and other organisms, including macroalgae. Macroalgae can disturb coral microbial communities by transmitting macroalgal-associated microbes and compounds (Smith et al., 2006; Morrow et al., 2012; Thurber et al., 2012), which may reduce the abundance of beneficial bacteria, promote the growth of rare microbial taxa, or vector new, pathogenic microbes (Nugues et al., 2004; Morrow et al., 2011). Such changes in the microbial communities can be detrimental to corals by increasing their susceptibility to pathogens, bleaching, or other environmental stressors (Rosenberg et al., 2007).
Coral reefs in Singapore persist in a heavily disturbed, urbanized reef environment characterized by high sedimentation and nutrient levels (Browne et al., 2015; Heery et al., 2018). As a result, corals in Singapore may be in a reduced health state and therefore more susceptible to stressors including macroalgal competition (Barott and Rohwer, 2012; Barott et al., 2012). Yet, the persistence of corals in Singapore, especially on the reef flats where macroalgae are abundant and herbivory is low (Guest et al., 2016; Bauman et al., 2017; Low et al., 2019), suggests that the corals are potentially adapted to macroalgal competition. In this study, we examined the direct and indirect effects of macroalgae on corals collected from Singapore and their microbial communities. Specifically, we compared how the effect of macroalgae on corals differed between direct contact and indirect, water-mediated interactions between a brown alga Lobophora sp. from a genus previously shown to be allelopathic (Morrow et al., 2011; Evensen et al., 2019; Fong et al., 2019), and a putatively non-allelopathic red alga Hypnea pannosa, on three common coral species in Singapore (Merulina ampliata, Montipora stellata, and Pocillopora acuta).
Materials and Methods
Corals and Macroalgae Collection
This study was conducted over 8 weeks at St John’s Island National Marine Laboratory (SJINML), Singapore from November 2017 to January 2018. Two weeks prior to the experiment, four colonies each of M. ampliata, M. stellata, and P. acuta were collected from the reef flats at two fringing reefs in the Southern Islands of Singapore (Kusu Island: 1°13′32″N, 103°51′35″E; Pulau Satumu: 1°09′39″N, 103°44′26″E). For P. acuta and M. ampliata, two colonies were each collected from Pulau Satumu and Kusu Island, while all four colonies of M. stellata were collected from Pulau Satumu due to lack of suitable colonies at Kusu Island. Samples of two locally abundant macroalgae, Lobophora sp. and H. pannosa, were also collected from Kusu Island on the same day. We chose M. ampliata, M. stellata, and P. acuta as study species because they are abundant on Singapore’s reefs (Huang et al., 2009) and within genera that vary in susceptibility to macroalgal competition (Bonaldo and Hay, 2014). We selected the foliose brown alga Lobophora sp. because this genus is known to be strong competitors against corals (Jompa and McCook, 2002; Nugues and Bak, 2006), while previously studies have shown the corticated mat-forming red alga H. pannosa have negligible impacts on corals (Jompa and McCook, 2003). DNA barcode analysis revealed that the Lobophora sp. studied here is an undescribed species, with its closest sister species being an undescribed Lobophora species from Oman (see Fong et al., 2019).
Coral and macroalgal samples were brought back to SJINML. Seven 4 to 5 cm fragments were obtained from each coral colony and epoxied to small cement tiles (6 × 6 cm). Samples of each macroalgal species were separated into twenty-four equal sized clumps of ∼7 g wet weight. On each cement tile, two 1 cm stainless-steel nails were embedded on the opposite ends so that polyethylene string could be secured over each nail head to hold the macroalgae in place (following Rasher and Hay, 2010). Both corals and algae were allowed to acclimate for 2 weeks in separate holding tanks in the outdoor aquariums before the start of the experiment.
Experimental Design
To investigate the effects of direct macroalgal contact and indirect water-mediated interactions on coral physiology and microbiome, coral fragments from M. ampliata, M. stellata, and P. acuta were assigned to one of the following seven treatments for 8 weeks: (1) direct contact with Lobophora sp., (2) direct contact with H. pannosa, (3) in close proximity with Lobophora sp. (coral and macroalgae were on separate cement tiles ∼5 cm apart but not in direct contact), (4) in close proximity with H. pannosa, (5) direct contact with mimic Lobophora sp., (6) direct contact with mimic H. pannosa, and (7) without any algal treatment (control). The inert algal mimics were constructed by tying together several semicircles of brown foam paper (mimic Lobophora sp.) and by grouping several lengths of wool yarn (mimic H. pannosa). These mimics served to control for the physical effects caused by macroalgal abrasion as well as any shading effects. Each treatment was replicated four times.
Replicates were placed in individual 3 l plastic tanks, which received independent flow through filtered seawater and were aerated. Tanks were distributed randomly among three large water baths to reduce water temperature fluctuations. To avoid any positional effects, the arrangement of the plastic tanks was changed randomly every week. Minimal variations in water temperature and light levels were recorded among the water baths throughout the experiment (mean ± SE: 28.10 ± 0.08°C, 57.14 ± 8.68 μmol s–1 m–2; HOBO data loggers), and the values were comparable to in situ conditions (Browne et al., 2015). Tanks were cleaned every 3 to 4 days.
Assessment of Coral Physiology
The physiological state of the coral fragments was monitored using image analysis and PAM fluorometry. Previous research indicates that the effects of macroalgae on corals are often localized to areas of physical contact only (Rasher et al., 2011). Therefore, for each coral fragment that was in direct contact with live and mimic algae, we assessed the side of the fragment that was exposed to the algal treatment and also its opposite side away from the exposure. Photographs of coral fragments were taken weekly to quantify the percentage of tissue bleaching using ImageJ photoanalysis software (Schneider et al., 2012). In addition, we examined maximum quantum yield (Fv/Fm) of coral fragments using pulse amplitude modulated fluorometer (Diving-PAM Underwater Fluorometer) every 2 weeks. Fv/Fm is a measure of the quantum efficiency of photosystem II centers in dark-adapted organisms, and many studies have found Fv/Fm a reliable proxy for coral health (Ralph et al., 1999; Fitt et al., 2001). For each side of each coral fragment, three Fv/Fm values were taken and averaged. All PAM measurements were taken between 0400 and 0600 h to ensure adequate dark acclimation in coral samples.
To determine the effects of algal treatments on the percent coral tissue bleaching and Fv/Fm, linear mixed-effect (LME) models were performed using the lme function from the nlme package (Pinheiro et al., 2019) in R v3.6.1 (R Core Team, 2019). Parent colony was added as a random effect to account for the nested data structures, and only data from the final time point (i.e., week 8) were fitted to the models. Models were weighted to allow variance to differ by treatments and corals to ensure homogeneity of variance. Significance tests were based on likelihood ratio tests and changes in Akaike’s Information Criterion (AIC). Post hoc analyses were conducted using the emmeans package with Tukey’s adjusted p-values. To compare the physiological responses between coral fragments sides (i.e., in direct contact with macroalgae and the opposite sides), Wilcoxon signed–rank tests were performed. Assumptions of normality and homoscedasticity were validated by visual inspection of the residual plots for all models (Supplementary Figure 1).
Assessment of Coral Microbiome
To characterize the natural microbiome of the three coral species, a 5 cm fragment was sampled from each individual coral colony at the time the colonies were collected in the field. An additional 5 cm fragment from each colony was sampled 2 weeks after acclimation in the aquarium. At the end of the experiment (and after the physiological testing described above), all coral fragments from the seven treatments were collected for microbial sampling. All samples were immediately frozen in a dry shipper or −80°C freezer. Coral tissue was then removed from the entire fragment using compressed air and subsequently stored at −80°C. DNA was extracted using the Qiagen DNeasy PowerBiofilm Kit and stored at −20°C. PCR was run with 10 μl HotStarTaq Plus Master Mix, 1 μl each of 10 μM forward and reverse primers, 5 μl water, 1 μl 100% DMSO, and 2 μl template DNA (5 ng μl–1). The 515F and 806R primers were used to amplify the V4 region of the 16S rRNA gene (Caporaso et al., 2011). Triplicate PCR reactions were run using the following conditions: an initial denaturation at 95°C for 5 min, followed by 37 cycles of 94°C for 30 s, 53°C for 40 s, and 72°C for 1 min, then a final extension of 10 min at 72°C. For M. stellata samples, no DMSO was used during PCR, and the reaction only ran for 35 cycles. All samples were pooled and cleaned with Agencourt AMPure XP beads and quantified using a Qubit 2.0 fluorometer. All PCR products were quality checked on an Aligent 2200 TapeStation before the samples were delivered to the sequencing facility at the Singapore Centre for Environmental Life Sciences Engineering (Nanyang Technological University, Singapore) for library preparation and amplicon sequencing on an Illumina MiSeq platform.
Amplicon sequence data were processed in Mothur v. 1.39.5 (Schloss et al., 2009) using the corresponding MiSeq SOP analysis pipeline (Kozich et al., 2013). In brief, paired sequences were combined and trimmed, then aligned to the Silva v4 reference database. Sequences that matched within two base pairs were pre-clustered, and chimeric sequences were removed using VSEARCH. Any non-bacterial or non-archaeal sequence reads (i.e., chloroplast, mitochondria, or unknown Eukaryota) were filtered out. Individual treatment replicates were rarefied to the lowest number of sequence reads by coral species to account for variation in sampling depth. Mean relative abundance of the representative taxa for each sample group was calculated. Rarefied data were used to calculate alpha diversity (richness, chao, Shannon evenness, and Inverse Simpson Diversity) and beta diversity. Alpha diversity measures were calculated using Mothur.
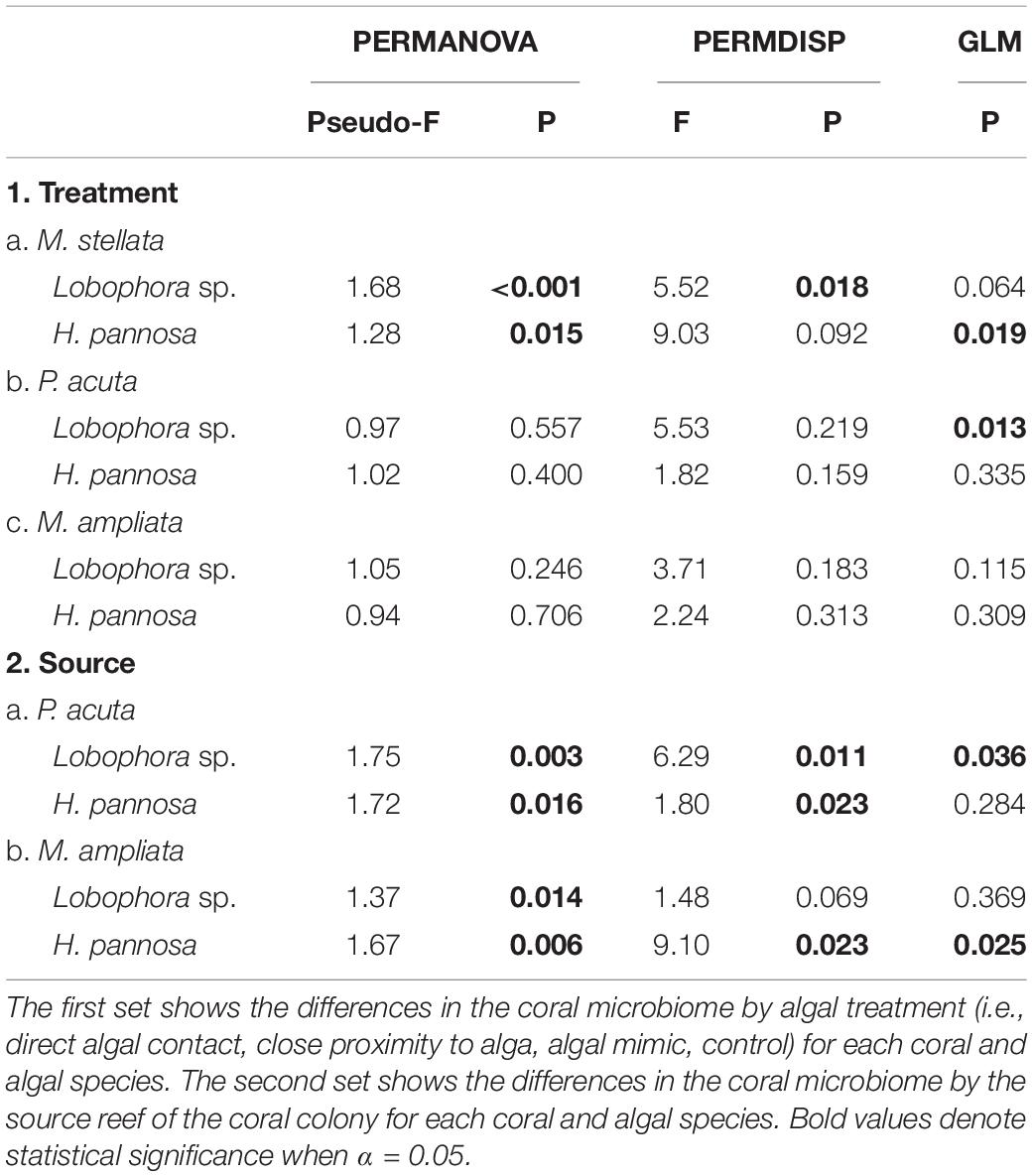
To examine the patterns of coral-associated prokaryotic communities, non-metric multidimensional scaling (nMDS) plots were created using Bray-Curtis distance matrices of square root transformed data with the Vegan package in R v3.4.3. Permutational multivariate analysis of variance (PERMANOVA) and permutational multivariate analysis of dispersion (PERMDISP) were performed using the PERMANOVA+ add-on in PRIMER 7 – both with the controls and with only algal treatments for each coral-algal pairing. PERMANOVA was also performed to compare the response of the prokaryotic community structure between colonies collected from Pulau Satumu and Kusu Island for M. ampliata and P. acuta interactions with Lobophora sp. and H. pannosa. To determine the specific OTUs that were contributing to the differences detected by the PERMANOVA, multivariate generalized linear models (GLMs) with negative binomial distribution were performed using the mvabund package (Wang et al., 2012) with residual plots inspected to ensure fit. Each dataset was subsampled to 500 OTUs for the GLM analysis.
Results
Macroalgal Effects on Coral Physiology
Interactions with macroalgae had significant effects on coral tissue and Fv/Fm (Table 1). Direct contact with Lobophora sp. caused 12.26 ± 2.11% (mean ± SE) of the coral tissue area to bleach, while direct H. pannosa contact did not cause significant tissue bleaching compared to control corals (Figure 1). Direct macroalgal contact also caused significant suppression of Fv/Fm, from 0.760 ± 0.005 in control corals to 0.529 ± 0.023 in Lobophora sp. treatment and 0.653 ± 0.021 in H. pannosa treatment (Figure 1). Macroalgal impacts were limited to the areas of direct contact (Wilcoxon signed–rank test; p < 0.001; Supplementary Figures 2, 3) and became evident after 4 weeks (Supplementary Figure 4). No detrimental effects on coral physiology were observed when corals were in close proximity (i.e., ∼5 cm apart) to Lobophora sp. and H. pannosa or when in direct contact with algal mimics (Figure 1).
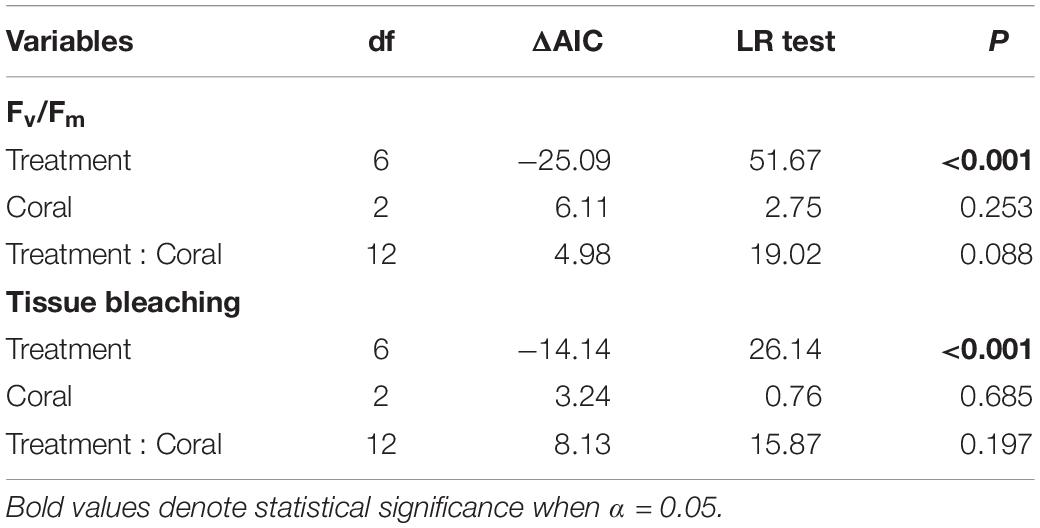
Table 1. Summary of LME models on coral maximum quantum yield (Fv/Fm) and percentage of coral tissue bleaching after 8 weeks of interactions with macroalgae.
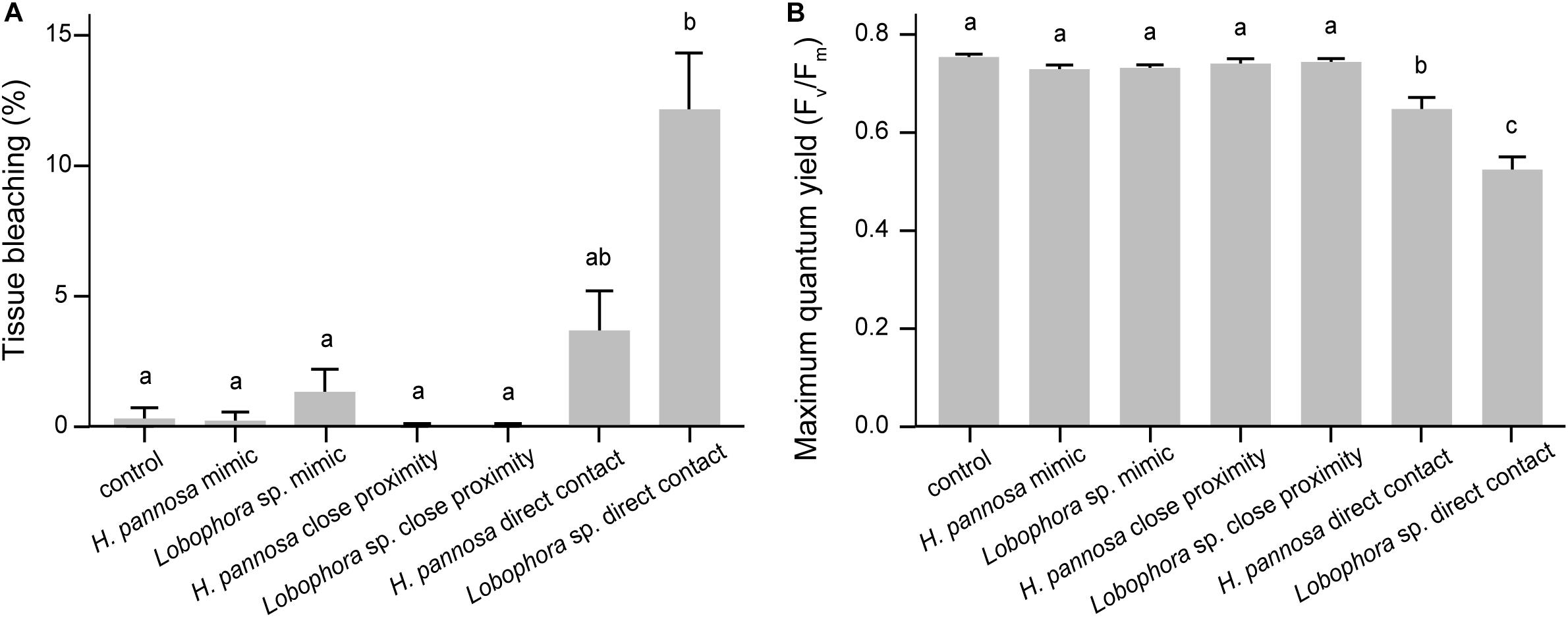
Figure 1. Effects of macroalgal interactions on (A) coral tissue and (B) maximum quantum yield (Fv/Fm) after 8 weeks of experiment (mean ±SE). Letters above bars indicate significant differences among treatments based on Tukey-adjusted comparisons.
Macroalgal Effects on Coral Microbiome
Amplicon sequence data were subsampled by species to 115,813 sequences for M. stellata, 55,399 sequences for P. acuta, and 35,566 sequences for M. ampliata. The dominant bacterial phylum in all coral species was Proteobacteria, particularly Alphaproteobacteria and Gammaproteobacteria (Supplementary Figures 5–7). In general, macroalgal treatments led to higher richness and diversity in the microbiome of all coral species compared to samples collected from the field at the start of the experiment and tank-acclimated controls (Supplementary Tables 1–3). For M. stellata, there was a significant shift in the prokaryotic community structure between the field control samples and the control treatments by the end of the experiment (Pairwise PERMANOVA: t = 2.202, p = 0.007; Supplementary Figure 8). Due to the temporal shift in microbiomes observed throughout the experiment, all subsequent analyses to assess the specific response of the coral microbiome to algal treatment focused on comparisons to the control fragments that were sampled at the same time as those that received algal treatments. For M. ampliata and P. acuta, the temporal shift was less apparent due to the confounding effect of source reef; however, for consistency all coral species were analyzed in the same manner with treatments being compared to experimental controls and not directly to field controls. For each coral species, there were also significant differences in the prokaryotic community structure across all treatments (PERMANOVA: M. stellata Pseudo-F = 2.544, p = 0.001; P. acuta Pseudo-F = 1.549, p = 0.001; M. ampliata Pseudo-F = 1.577, p = 0.001).
For M. stellata, the coral microbiomes differed significantly among the four macroalgal treatments (i.e., direct algal contact, in close proximity with algae, direct contact with algal mimic, without any algal treatment) for both Lobophora sp. and H. pannosa treatments when examined for each algal species separately (Table 2). Particularly in Lobophora sp. treatments, the nMDS demonstrates how prokaryotic community structures of corals that were in contact or close proximity with the alga were more closely clustered together and distinct from controls and algal mimic treatments (Figure 2A). In contrast, in the H. pannosa treatments, the treatments involving direct contact, either with the algae or algal mimic, were clustered similarly (Figure 2B). For P. acuta and M. ampliata, PERMANOVA analysis revealed no significant differences in prokaryotic community structure among treatment groups for each algal treatment; however, there were significant differences in the response of the coral microbiome to both Lobophora sp. and H. pannosa based on the source reef from which the colony was collected (Table 2). The nMDS plots demonstrate marked separation in the microbiomes of corals collected from Kusu Island and Pulau Satumu (Figure 3).
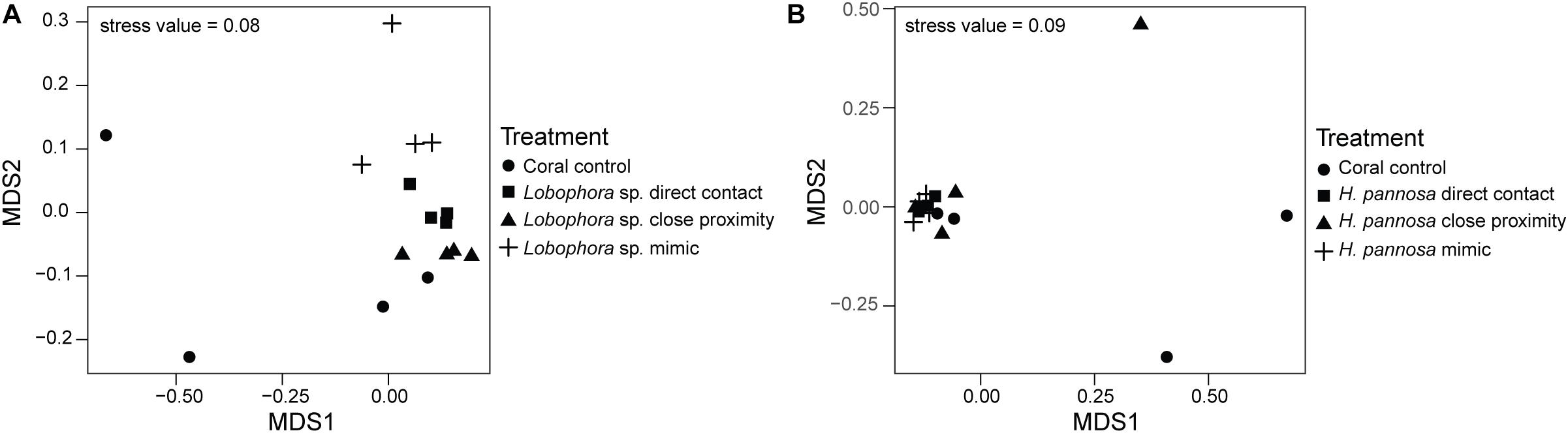
Figure 2. nMDS plots of the prokaryotic community structure of Montipora stellata coral fragments that had interactions with (A) Lobophora sp. and (B) Hypnea pannosa.
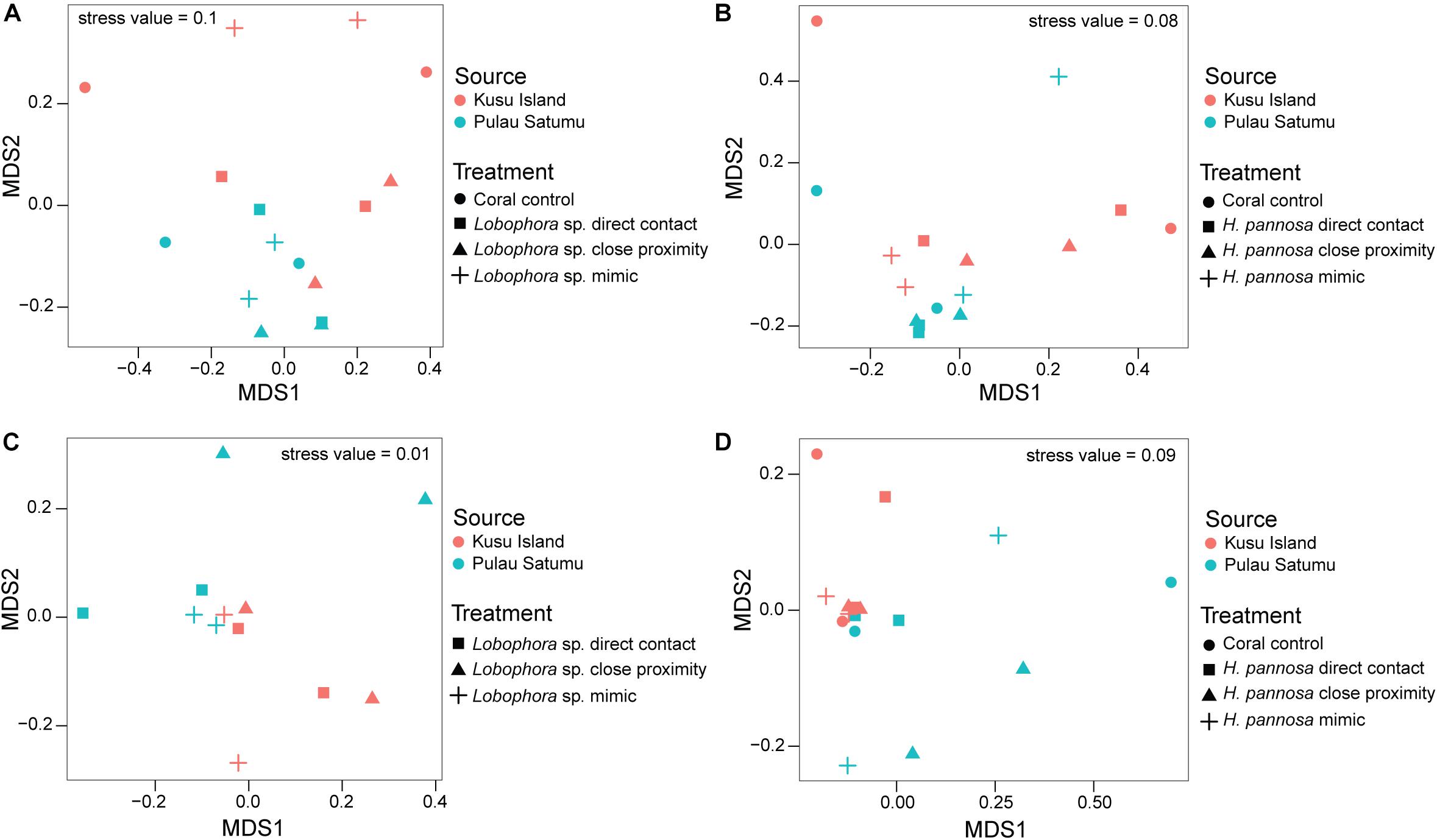
Figure 3. nMDS plots of the prokaryotic community structure of Pocillopora acuta (A,B) and Merulina ampliata (C,D) coral fragments that had interactions with Lobophora sp. (left panels) and Hypnea pannosa (right panels). Color is used to indicate whether corals were collected from either Kusu Island (red) or Pulau Satumu (blue). Control samples were excluded in the nMDS plots for interactions between M. ampliata and Lobophora sp. (see Supplementary Figure 9 for nMDS plot with all samples).
Generalized linear models identified significant differences among treatment groups and source reef consistent with the PERMANOVA results (Table 2). For M. stellata, 18.2% of the OTUs were significantly different in Lobophora sp. treatments while 8.2% of the OTUs were significantly different in H. pannosa treatments (Supplementary Data). For P. acuta and M. ampliata, GLM revealed that significant differences in the coral microbiomes were driven by the colony source reef (Table 2). In the P. acuta treatments, 19.6% of the OTUs were identified as contributing to the significant differences between the colonies from either Kusu Island or Pulau Satumu in response to Lobophora sp. treatments and 18.0% for H. pannosa treatments (Supplementary Data). For M. ampliata, the difference was attributed to 8.8% of the OTUs in the Lobophora sp. treatments and 13.0% for H. pannosa (Supplementary Data). However, when accounting for only the highly abundant OTUs (>1%), the number of significant OTUs between treatment groups ranged from 3 to 11 OTUs (M. stellata: Table 3; P. acuta and M. ampliata: Table 4). Notably in the M. stellata treatments with Lobophora sp., the largest difference between control and algal treatments was due to a high abundance of the coral-associated Endozoicomonas sp. in the control samples (0.175 ± 0.247), compared to a lower relative abundance (0.031 ± 0.021) across all of the algal treatment groups. The single most abundant, significant OTU was a Desulfovibrio sp. found in the P. acuta treatments originating from Kusu Island. Pocillopora acuta was also the only coral species to have Archaea, specifically Thaumarchaeota of the Order Nitrosopumilus, accounting for a large proportion of the difference between groups.
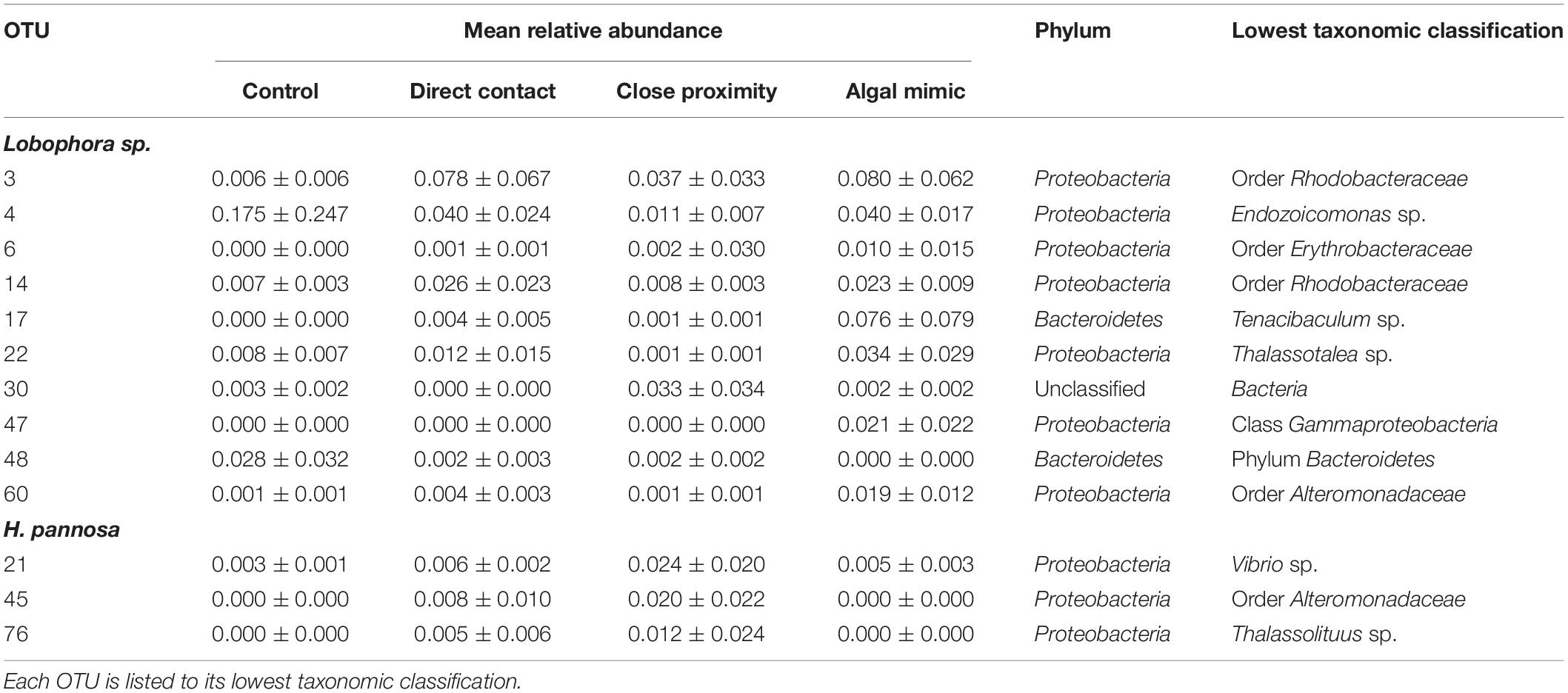
Table 3. Highly abundant (>0.01 mean relative abundance of total community ±SD) OTUs identified as contributing significantly to the differences between treatment groups for Montipora stellata interactions with Lobophora sp. and Hypnea pannosa.
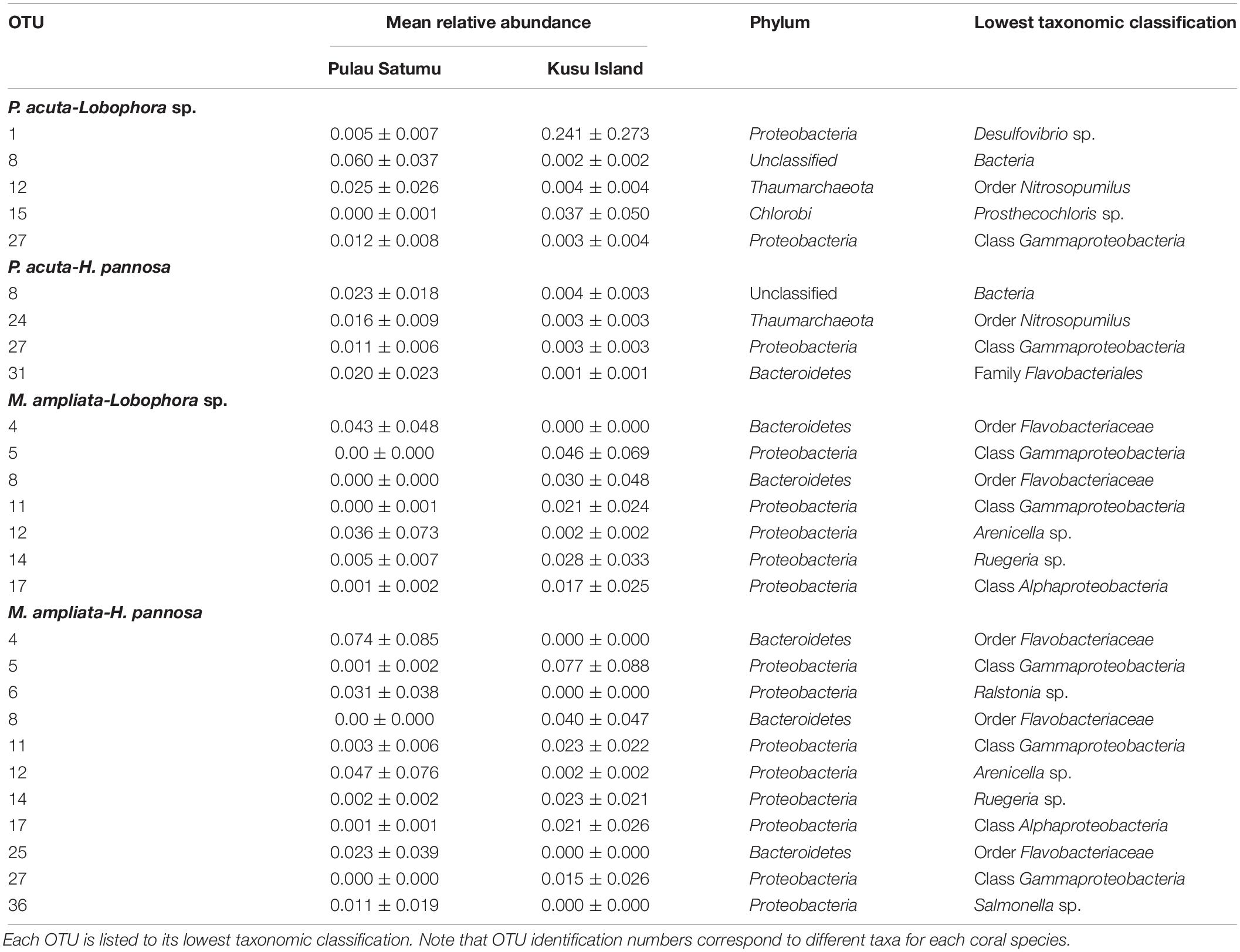
Table 4. Highly abundant (>0.01 mean relative abundance of total community ±SD) OTUs identified as contributing significantly to the differences between Pulau Hantu and Kusu Island for each coral and algal species interaction.
Discussion
Competitive interactions between corals and macroalgae are becoming increasingly pervasive on coral reefs (Hughes et al., 2010; Brown et al., 2018), yet, there remains considerable uncertainties regarding the extent to which macroalgae can impact corals, particularly within heavily disturbed reef systems. While direct macroalgal contact has largely been shown to harm corals, there is limited understanding regarding the water-mediated effects of macroalgae on corals. In this study, we found direct macroalgal contact caused tissue bleaching and reduced Fv/Fm of all three coral species, particularly when in direct contact with Lobophora sp., while macroalgae that were only in close proximity but not touching corals had no effects on coral physiology. Both contact- and water-mediated interactions with Lobophora sp. and H. pannosa altered the coral-associated microbiomes in M. stellata, while the effects on the microbiomes of M. ampliata and P. acuta were more strongly associated with the source reefs from which they were collected rather than algal treatments. Overall, our results reveal that, for the species tested, the coral microbiomes were sensitive to both direct contact and water-mediated interactions with macroalgae, while coral physiology was compromised only when in direct contact with macroalgae.
Coral fragments of M. ampliata, M. stellata, and P. acuta that were in direct contact with Lobophora sp. had visible tissue bleaching and approximately 30% reduction in Fv/Fm, whereas coral fragments that were in direct contact with H. pannosa exhibited a ∼14% reduction. These results are consistent with patterns reported in previous studies showing Lobophora spp. were competitively superior to corals (Rasher and Hay, 2010; Morrow et al., 2012) whereas H. pannosa only caused minor damage to corals (Jompa and McCook, 2003). The damage caused by Lobophora sp. and H. pannosa was only observed in the area of direct contact, providing further evidence of the limited spatial impact of macroalgae on coral health (Rasher et al., 2011; Clements et al., 2018). This is in contrast with previous findings that show macroalgal waterborne compounds (e.g., dissolved organic compounds) could cause coral mortality (Kline et al., 2006; Smith et al., 2006). For instance, Smith et al. (2006) reported that Pocillopora verrucosa fragments placed next to the green alga Dictyosphaeria cavernosa, but not in direct contact, suffered 100% mortality within 2 days. Additionally in our experiment, coral tissue bleaching was only observed after 4 weeks (Supplementary Figure 4), compared to previous studies which have reported more immediate macroalgal impacts within the first few days to a week (e.g., Smith et al., 2006; Andras et al., 2012), or slightly longer period (e.g., 20 days in Rasher et al., 2011). These differences could be due to species-specific variability in coral susceptibility or macroalgal potency among the species tested (McCook et al., 2001). Alternatively, lag effects of macroalgae on coral physiology observed in this study could be related to the highly urbanized nature of Singapore’s reef environment; for instance corals from Singapore may be adapted to algal competition, but additional work is required to clarify this link.
Interactions with macroalgae also caused substantial changes to the coral microbiomes. Macroalgal treatments generally led to higher richness and diversity in the microbiomes of all coral species (Supplementary Tables 1–3), which are consistent with previous research suggesting coral stress can result in higher microbiome beta diversity (McDevitt-Irwin et al., 2017; Zaneveld et al., 2017). For M. stellata, the extent of changes to the microbiome differed between the Lobophora sp. and H. pannosa treatments. Coral microbiomes that were in contact or close proximity to Lobophora sp. were significantly different from the control and algal mimic treatments, indicating that Lobophora sp. was able to impact the coral microbiomes through indirect interactions. The genus Lobophora is known to contain allelopathic chemicals that can harm corals (Vieira et al., 2016; Evensen et al., 2019), including the species in this study (Fong et al., 2019). Crude extracts from Lobophora have also been found to display strong antimicrobial activities (Morrow et al., 2012). Some studies have shown that macroalgae can indirectly impact coral microbiomes that are several centimeters away via water-mediated macroalgal compounds and microbes (Barott and Rohwer, 2012; Morrow et al., 2013; Pratte et al., 2018). For example, Morrow et al. (2013) reported that Halimeda opuntia and Dictyota menstrualis altered bacterial communities associated with Montastraea faveolata that were up to 5 cm away from the direct macroalgal contact zone. Similarly, Pratte et al. (2018) found significant shifts in the microbial communities of Porites sp. 5 cm from turf contact, although there were more pronounced shifts in direct contact zone. While the allelopathic effects of Lobophora sp. has been established (Fong et al., 2019), it remains unclear what role the algal microbiome may have played in the strong response of the coral microbiomes to the presence of Lobophora sp., particularly given that the M. stellata colonies and Lobophora sp. were collected from two different reefs. Future studies should examine the effects of site-specific algal microbial communities on coral microbiomes. In the H. pannosa treatments, M. stellata microbiomes were more similarly affected by the direct contact of live and mimic algae than by the presence of algae nearby, suggesting that the changes in the coral microbiomes were more strongly driven by the effects of physical contact. In contrast to the leathery surface of Lobophora sp., the corticated thallus of H. pannosa might inflict surface abrasions on corals triggering shifts in the coral microbiomes (Mydlarz et al., 2006). Nevertheless, the presence of H. pannosa on coral reefs appears to be less harmful to the coral holobiont than the presence of a strongly allelopathic alga as the impact is restricted to direct interactions.
While Lobophora sp. and H. pannosa induced shifts in the microbial communities of M. ampliata and P. acuta, the source reefs from where the coral colonies were collected (Pulau Satumu and Kusu Island) had a greater effect. The differences in the coral microbiomes between the two reefs were first identified in the field samples collected from Kusu Island, which had proportionately higher abundance of potentially human-associated pathogens in their microbiomes than those collected from Pulau Satumu. For example, the most abundant taxon in the two P. acuta colonies collected from Kusu Island was Salmonella sp. and, in one of the M. ampliata colonies from Kusu Island, the most abundant taxon was Streptococcus sp. Closer proximity to human influences, i.e., the main island of Singapore (∼4 km for Kusu Island versus ∼13 km for Pulau Satumu) might explain why the microbial communities on the corals collected from Kusu Island were dominated by bacterial genera associated with human pathogens (Kaczmarsky et al., 2005; Dinsdale et al., 2008). Differences in environmental conditions among reefs, such as light, nutrients, and water currents, can also have a strong influence on the specificity of coral microbiome (Klaus et al., 2007; Apprill and Rappé, 2011; Pantos et al., 2015). Ng et al. (2019) reported that water motion was significantly greater at Kusu Island than Pulau Satumu. Our results were also consistent with patterns reported by Wainwright et al. (2019), who found that the bacterial communities associated with P. acuta collected from Kusu Island were markedly different from those collected Pulau Satumu. Nevertheless, despite higher levels of potentially pathogenic bacteria, P. acuta and M. ampliata from Kusu Island had the same physiological response to environmental perturbation (interaction with algae) as corals from Pulau Satumu. This is in contrast with the patterns observed in the Line Islands of the Central Pacific, where reefs closer to human influences, which presumably have higher prevalence of pathogens and coral disease, were poorer competitors against algae compared to corals from more pristine habitats (Dinsdale et al., 2008; Barott and Rohwer, 2012; Barott et al., 2012).
Only direct contact with macroalgae initiated a physiological response in the corals, while we found evidence of water-mediated effects of Lobophora sp. and H. pannosa on the microbiome. Coral microbiomes have been shown to be highly dynamic and capable of responding to environmental perturbations, particularly those induced by the presence of macroalgae, and their diffusible compounds (i.e., chemicals and dissolved organic carbon) and microbes (Ainsworth et al., 2010; Bourne et al., 2016). It has been proposed that the coral microbiome may be composed of a small conserved core while the majority of microbes in the microbiome change routinely in response to environmental conditions with little negative effect on the functioning of the coral holobiont (Hernandez-Agreda et al., 2018). While our ability to determine a core microbiome for P. acuta and M. ampliata is limited by the size of the current study, the shared proportion of prokaryotic symbionts for each species within each reef was small, with 31 OTUs shared among the four P. acuta control colonies and 27 OTUs shared among the four M. ampliata control colonies (Supplementary Table 4). Given the variability that exists in their natural microbiomes, it is not surprising to see variability in the response of those microbiomes to the same algal treatments. However, some caution is required when interpreting our results because coral microbiomes were likely different between ex situ and in situ samples, as evidenced in M. stellata fragments, and this might influence the effects of macroalgae on coral microbial communities. Further investigations would be required to (1) compare the impacts of macroalgae on coral microbiomes under different environmental settings, and (2) determine whether a core or resident microbiome is maintained for each of the coral species examined here and the specific symbiotic benefits imparted to the coral from that microbiome.
Overall, this study provides important insights into the impacts of direct contact and water-mediated interactions with macroalgae on corals and their microbiomes. Importantly, our results demonstrate that contact- and water-mediated interactions with Lobophora sp. and H. pannosa altered coral-associated microbiomes in corals, but only direct macroalgal contact resulted in reduced coral health. Notably, the presence of high levels of bacterial genera associated with human pathogens in some of the coral samples did not lead to the corals being more susceptible to macroalgal competition.
Data Availability Statement
The raw sequence data were cataloged in the NCBI Sequence Read Archive under BioProject accession number PRJNA548204.
Author Contributions
All authors contributed to the conception and design of the study, manuscript revision, read, and approved the submitted version. JF carried out the experiment and conducted the coral physiology assessment. LD conducted the coral microbiome work. JF and LD performed the statistical analysis and wrote the first draft of the manuscript.
Funding
This study was supported by the National Research Foundation, Prime Minister’s Office, Singapore under the Marine Science Research and Development Programme (MSRDP-P03), Wildlife Reserves Singapore Conservation Fund, and the Ministry of Education, Singapore under its Research Centre of Excellence Program to the Singapore Centre for Environmental Life Sciences Engineering, Nanyang Technological University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Zhiyuan Chen and Jing Jie Teh for their assistance with the experiment. We acknowledge the St. John’s Island National Marine Laboratory, a National Research Infrastructure under the National Research Foundation Singapore, for providing the facility necessary for conducting the research. All research was carried out with permission of the Singapore National Parks Board (Permit no. NP/RP17-046).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00831/full#supplementary-material
References
Ainsworth, T. D., Thurber, R. V., and Gates, R. D. (2010). The future of coral reefs: a microbial perspective. Trends Ecol. Evol. 25, 233–240. doi: 10.1016/j.tree.2009.11.001
Andras, T. D., Alexander, T. S., Gahlena, A., Parry, R. M., Fernandez, F. M., Kubanek, J., et al. (2012). Seaweed allelopathy against coral: surface distribution of a seaweed secondary metabolite by imaging mass spectrometry. J. Chem. Ecol. 38, 1203–1214. doi: 10.1007/s10886-012-0204-9
Apprill, A., and Rappé, M. S. (2011). Response of the microbial community to coral spawning in lagoon and reef flat environments of Hawaii, USA. Aquat. Microb. Ecol. 62, 251–266. doi: 10.3354/ame01471
Barott, K. L., and Rohwer, F. L. (2012). Unseen players shape benthic competition on coral reefs. Trends Microbiol. 20, 621–628. doi: 10.1016/j.tim.2012.08.004
Barott, K. L., Williams, G. J., Vermeij, M. J. A., Harris, J., Smith, J. E., Rohwer, F. L., et al. (2012). Natural history of coral–algae competition across a gradient of human activity in the Line Islands. Mar. Ecol. Prog. Ser. 460, 1–12. doi: 10.3354/meps09874
Bauman, A. G., Hoey, A. S., Dunshea, G., Feary, D. A., Low, J., and Todd, P. A. (2017). Macroalgal browsing on a heavily degraded, urbanized equatorial reef system. Sci. Rep. 7:8352. doi: 10.1038/s41598-017-08873-3
Bellwood, D. R., Hughes, T. P., Folke, C., and Nyström, M. (2004). Confronting the coral reef crisis. Nature 429, 827–833. doi: 10.1038/nature02691
Birrell, C. L., McCook, L. J., Willis, B. L., and Diaz-Pulido, G. A. (2008). Effects of benthic algae on the replenishment of corals and the implications for the resilience of coral reefs. Oceanogr. Mar. Biol. 46, 25–63. doi: 10.1201/9781420065756.ch2
Bonaldo, R. M., and Hay, M. E. (2014). Seaweed-coral interactions: variance in seaweed allelopathy, coral susceptibility, and potential effects on coral resilience. PLoS One 9:e85786. doi: 10.1371/journal.pone.0085786
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Box, S. J., and Mumby, P. J. (2007). Effect of macroalgal competition on growth and survival of juvenile Caribbean corals. Mar. Ecol. Prog. Ser. 342, 139–149. doi: 10.3354/meps342139
Brown, K. T., Bender-Champ, D., Kubicek, A., van der Zande, R., Achlatis, M., Hoegh-Guldberg, O., et al. (2018). The dynamics of coral-algal interactions in space and time on the southern great barrier reef. Front. Mar. Sci. 5:181. doi: 10.3389/fmars.2018.00181
Browne, N. K., Tay, J. K. L., Low, J., Larson, O., and Todd, P. A. (2015). Fluctuations in coral health of four common inshore reef corals in response to seasonal and anthropogenic changes in water quality. Mar. Environ. Res. 105, 39–52. doi: 10.1016/j.marenvres.2015.02.002
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Clements, C. S., Rasher, D. B., Hoey, A. S., Bonito, V. E., and Hay, M. E. (2018). Spatial and temporal limits of coral-macroalgal competition: the negative impacts of macroalgal density, proximity, and history of contact. Mar. Ecol. Prog. Ser. 586, 11–20. doi: 10.3354/meps12410
Dinsdale, E. A., Pantos, O., Smriga, S., Edwards, R. A., Angly, F., Wegley, L., et al. (2008). Microbial ecology of four coral atolls in the Northern Line Islands. PLoS One 3:e1584. doi: 10.1371/journal.pone.0001584
Evensen, N. R., Doropoulos, C., Morrow, K. M., Motti, C. A., and Mumby, P. J. (2019). Inhibition of coral settlement at multiple spatial scales by a pervasive algal competitor. Mar. Ecol. Prog. Ser. 612, 29–42. doi: 10.3354/meps12879
Fitt, W. K., Brown, B. E., Warner, M. E., and Dunne, R. P. (2001). Coral bleaching: interpretation of thermal tolerance limits and thermal thresholds in tropical corals. Coral Reefs 20, 51–65. doi: 10.1007/s003380100146
Fong, J., Lim, Z. W., Bauman, A. G., Valiyaveettil, S., Liao, L. M., Yip, Z. T., et al. (2019). Allelopathic effects of macroalgae on Pocillopora acuta coral larvae. Mar. Environ. Res. 151:104745. doi: 10.1016/j.marenvres.2019.06.007
Guest, J. R., Tun, K., Low, J., Vergés, A., Marzinelli, E. M., Campbell, A. H., et al. (2016). 27 years of benthic and coral community dynamics on turbid, highly urbanised reefs off Singapore. Sci. Rep. 6:36260. doi: 10.1038/srep36260
Haas, A. F., Nelson, C. E., Rohwer, F., Wegley-Kelly, L., Quistad, S. D., Carlson, C. A., et al. (2013). Influence of coral and algal exudates on microbially mediated reef metabolism. PeerJ 1:e108. doi: 10.7717/peerj.108
Haas, A. F., Nelson, C. E., Wegley Kelly, L., Carlson, C. A., Rohwer, F., Leichter, J. J., et al. (2011). Effects of coral reef benthic primary producers on dissolved organic carbon and microbial activity. PLoS One 6:e27973. doi: 10.1371/journal.pone.0027973
Heery, E. C., Hoeksema, B. W., Browne, N. K., Reimer, J. D., Ang, P. O., Huang, D., et al. (2018). Urban coral reefs: degradation and resilience of hard coral assemblages in coastal cities of East and Southeast Asia. Mar. Pollut. Bull. 135, 654–681. doi: 10.1016/j.marpolbul.2018.07.041
Hernandez-Agreda, A., Leggat, W., Bongaerts, P., Herrera, C., and Ainsworth, T. D. (2018). Rethinking the coral microbiome: simplicity exists within a diverse microbial biosphere. mBio 9, e812–e818. doi: 10.1128/mBio.00812-18
Hooper, D. U., Chapin, F. S. III, Ewel, J. J., Hector, A., Inchausti, P., Lavorel, S., et al. (2005). Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol. Monogr. 75, 3–35. doi: 10.1890/04-0922
Huang, D., Tun, K. P. P., Chou, K. M., and Todd, P. A. (2009). An inventory of zooxanthellate scleractinian corals in Singapore, including 33 new records. Raffles Bull. Zool. 22, 69–80.
Hughes, T. P., Anderson, K. D., Connolly, S. R., Heron, S. F., Kerry, J. T., Lough, J. M., et al. (2018). Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 259, 80–83. doi: 10.1126/science.aan8048
Hughes, T. P., Graham, N. A. J., Jackson, J. B. C., Mumby, P. J., and Steneck, R. S. (2010). Rising to the challenge of sustaining coral reef resilience. Trends Ecol. Evol. 25, 633–642. doi: 10.1016/j.tree.2010.07.011
Jompa, J., and McCook, L. J. (2002). Effects of competition and herbivory on interactions between a hard coral and a brown alga. J. Exp. Mar. Biol. Ecol. 271, 25–39. doi: 10.1016/S0022-0981(02)00040-0
Jompa, J., and McCook, L. J. (2003). Coral–algal competition: macroalgae with different properties have different effects on corals. Mar. Ecol. Prog. Ser. 258, 87–95. doi: 10.3354/meps258087
Jorissen, H., Skinner, C., Osinga, R., De Beer, D., and Nugues, M. M. (2016). Evidence for water-mediated mechanisms in coral–algal interactions. Proc. R. Soc. B 283:20161137. doi: 10.1098/rspb.2016.1137
Kaczmarsky, L. T., Draud, M. A., and Williams, E. H. (2005). Is there a relationship between proximity to sewage effluent and the prevalence of coral disease? Caribb. J. Sci. 41, 124–137.
Klaus, J. S., Janse, I., Heikoop, J. M., Sanford, R. A., and Fouke, B. W. (2007). Coral microbial communities, zooxanthellae and mucus along gradients of seawater depth and coastal pollution. Environ. Microbiol. 9, 1291–1305. doi: 10.1111/j.1462-2920.2007.01249.x
Kline, D. I., Kuntz, N. M., Breitbart, M., Knowlton, N., and Rohwer, F. (2006). Role of elevated organic carbon levels and microbial activity in coral mortality. Mar. Ecol. Prog. Ser. 314, 119–125. doi: 10.3354/meps314119
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Lema, K. A., Willis, B. L., and Bourne, D. G. (2012). Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl. Environ. Microbiol. 78, 3136–3144. doi: 10.1128/AEM.07800-11
Lesser, M. P., Mazel, C. H., Gorbunov, M. Y., and Falkowski, P. G. (2004). Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305, 997–1000. doi: 10.1126/science.1099128
Low, J. K. Y., Fong, J., Todd, P. A., Chou, L. M., and Bauman, A. G. (2019). Seasonal variation of Sargassum ilicifolium (Phaeophyceae) growth on equatorial coral reefs. J. Phycol. 55, 289–296. doi: 10.1111/jpy.12818
McCook, L. J., Jompa, J., and Diaz-Pulido, G. (2001). Competition between corals and algae on coral reefs: a review of evidence and mechanisms. Coral Reefs 19, 400–417. doi: 10.1007/s003380000129
McDevitt-Irwin, J. M., Baum, J. K., Garren, M., and Vega Thurber, R. L. (2017). Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 4:262. doi: 10.3389/fmars.2017.00262
Miller, M. W. (1998). Coral/seaweed competition and the control of reef community structure within and between latitudes. Oceanogr. Mar. Biol. 36, 65–96.
Morrow, K. M., Liles, M. R., Paul, V. J., Moss, A. G., and Chadwick, N. E. (2013). Bacterial shifts associated with coral–macroalgal competition in the Caribbean Sea. Mar. Ecol. Prog. Ser. 488, 103–117. doi: 10.3354/meps10394
Morrow, K. M., Paul, V. J., Liles, M. R., and Chadwick, N. E. (2011). Allelochemicals produced by Caribbean macroalgae and Cyanobacteria have species-specific effects on reef coral microorganisms. Coral Reefs 30, 309–320. doi: 10.1007/s00338-011-0747-1
Morrow, K. M., Ritson-Williams, R., Ross, C., Liles, M. R., and Paul, V. J. (2012). Macroalgal extracts induce bacterial assemblage shifts and sublethal tissue stress in Caribbean corals. PLoS One 7:e44859. doi: 10.1371/journal.pone.0044859
Mydlarz, L. D., Jones, L. E., and Harvell, C. D. (2006). Innate immunity, environmental drivers, and disease ecology of marine and freshwater invertebrates. Annu. Rev. Ecol. Evol. Syst. 37, 251–288. doi: 10.1146/annurev.ecolsys.37.091305.110103
Ng, C. S. L., Lim, J. X., Sam, S. Q., Kikuzawa, Y. P., Toh, T. C., Wee, T. W., et al. (2019). Variability in skeletal bulk densities of common hard corals in Southeast Asia. Coral Reefs 38, 1133–1143. doi: 10.1007/s00338-019-01852-2
Nugues, M. M., and Bak, R. P. (2006). Differential competitive abilities between Caribbean coral species and a brown alga: a year of experiments and a long-term perspective. Mar. Ecol. Prog. Ser. 315, 75–86. doi: 10.3354/meps315075
Nugues, M. M., Smith, G. W., Van Hooidonk, R. J., Seabra, M. I., and Bak, R. P. M. (2004). Algal contact as a trigger for coral disease. Ecol. Lett. 7, 919–923. doi: 10.1111/j.1461-0248.2004.00651.x
Pantos, O., Bongaerts, P., Dennis, P. G., Tyson, G. W., and Hoegh-Guldberg, O. (2015). Habitat-specific environmental conditions primarily control the microbiomes of the coral Seriatopora hystrix. ISME J. 9, 1916–1927. doi: 10.1038/ismej.2015.3
Pinheiro, J., Bates, D., DebRoy, S., and Sarkar, D., R Core Team, (2019). Nlme: Linear and Nonlinear Mixed Effects Models. R Package Version 3.1-142.
Pratte, Z. A., Longo, G. O., Burns, A. S., Hay, M. E., and Stewart, F. J. (2018). Contact with turf algae alters the coral microbiome: contact versus systemic impacts. Coral Reefs 37, 1–3. doi: 10.1007/s00338-017-1615-4
R Core Team, (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ralph, P. J., Gademann, R., Larkum, A. W. D., and Schreiber, U. (1999). In situ underwater measurements of photosynthetic activity of coral zooxanthellae and other reef-dwelling dinoflagellate endosymbionts. Mar. Ecol. Prog. Ser. 180, 139–147. doi: 10.3354/meps180139
Rasher, D. B., and Hay, M. E. (2010). Chemically rich seaweeds poison corals when not controlled by herbivores. Proc. Natl. Acad. Sci. U.S.A. 107, 9683–9688. doi: 10.1073/pnas.0912095107
Rasher, D. B., Stout, E. P., Engel, S., Kubanek, J., and Hay, M. E. (2011). Macroalgal terpenes function as allelopathic agents against reef corals. Proc. Natl. Acad. Sci. U.S.A. 108, 17725–17731. doi: 10.1073/pnas.1108628108
Ritchie, K. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
River, G. F., and Edmunds, P. J. (2001). Mechanisms of interaction between macroalgae and scleractinians on a coral reef in Jamaica. J. Exp. Mar. Biol. Ecol. 261, 159–172. doi: 10.1016/S0022-0981(01)00266-0
Roach, T. N., Abieri, M. L., George, E. E., Knowles, B., Naliboff, D. S., Smurthwaite, C. A., et al. (2017). Microbial bioenergetics of coral-algal interactions. PeerJ 5:e3423. doi: 10.7717/peerj.3423
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to imagej: 25 years of image analysis. Nat. Methods 9:671. doi: 10.1038/nmeth.2089
Smith, J. E., Shaw, M., Edwards, R. A., Obura, D., Pantos, O., Sala, E., et al. (2006). Indirect effects of algae on coral: algae-mediated, microbe- induced coral mortality. Ecol. Lett. 9, 835–845. doi: 10.1111/j.1461-0248.2006.00937.x
Tanner, J. E. (1995). Competition between scleractinian corals and macroalgae: an experimental investigation of coral growth, survival and reproduction. J. Exp. Mar. Biol. Ecol. 90, 151–168. doi: 10.1016/0022-0981(95)00027-O
Thurber, R. V., Burkepile, D. R., Correa, A. M. S., Thurber, A. R., Shantz, A. A., Welsh, R., et al. (2012). Macroalgae decrease growth and alter microbial community structure of the reef-building coral, Porites astreoides. PLoS One 7:e44246. doi: 10.1371/journal.pone.0044246
Vieira, C., Engelen, A. H., Guentas, L., Aires, T., Houlbreque, F., Gaubert, J., et al. (2016). Species specificity of bacteria associated to the brown seaweeds Lobophora (Dictyotales, Phaeophyceae) and their potential for induction of rapid coral bleaching in Acropora muricata. Front. Microbiol. 7:316. doi: 10.3389/fmicb.2016.00316
Wainwright, B. J., Afiq-Rosli, L., Zahn, G. L., and Huang, D. (2019). Characterisation of coral-associated bacterial communities in an urbanised marine environment shows strong divergence over small spatial scales. Coral Reefs 38, 1097–1106. doi: 10.1007/s00338-019-01837-1
Wang, Y., Naumann, U., Wright, S. T., and Warton, D. I. (2012). mvabund—an R package for model-based analysis of multivariate abundance data. Methods Ecol. Evol. 3, 471–474. doi: 10.1111/j.2041-210X.2012.00190.x
Yodzis, P. (1978). Competition for Space and the Structure of Ecological Communities. Berlin: Springer.
Keywords: competitive interactions, urbanized reef, Singapore, corals, macroalgae, coral microbiome
Citation: Fong J, Deignan LK, Bauman AG, Steinberg PD, McDougald D and Todd PA (2020) Contact- and Water-Mediated Effects of Macroalgae on the Physiology and Microbiome of Three Indo-Pacific Coral Species. Front. Mar. Sci. 6:831. doi: 10.3389/fmars.2019.00831
Received: 19 September 2019; Accepted: 26 December 2019;
Published: 22 January 2020.
Edited by:
Linda Wegley Kelly, San Diego State University, United StatesReviewed by:
Ty N. F. Roach, University of Hawai’i at Mānoa, United StatesMaggy Nugues, Université de Sciences Lettres de Paris, France
Andrew A. Shantz, The Pennsylvania State University (PSU), United States
Copyright © 2020 Fong, Deignan, Bauman, Steinberg, McDougald and Todd. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jenny Fong, amVubnkuamVubnlAdS5udXMuZWR1
†These authors have contributed equally to this work
 Jenny Fong
Jenny Fong Lindsey K. Deignan
Lindsey K. Deignan Andrew G. Bauman
Andrew G. Bauman Peter D. Steinberg
Peter D. Steinberg Diane McDougald
Diane McDougald Peter A. Todd
Peter A. Todd