- 1Grice Marine Lab, Department of Biology, College of Charleston, Charleston, SC, United States
- 2Hollings Marine Lab, Department of Biology, College of Charleston, Charleston, SC, United States
The functional diversity of crustacean hemocyanins is broad, encompassing O2 delivery, innate immune response, metabolite storage, and osmolyte balance, all in a heterogeneous protein structure. As such, the sequence diversity of this class of proteins and its subunit composition are the focus of many studies on crustacean adaptation to environmental challenges. Recent transcriptomic and genomic assemblies on the Pacific whiteleg shrimp Litopenaeus vannamei have identified unique isoforms of hemocyanin including an ancestral β-type subunit thought to be lost in penaeid shrimp. However, it is unknown the degree to which these isoforms are translated as proteins, and their abundances within the hemolymph. The present study uses proteomic approaches to characterize the protein-level abundance and organization of these hemocyanin isoforms within their native oligomeric structures. Fractions of each hemocyanin oligomeric form were purified by size-exclusion high performance liquid chromatography for identification of subunit isoforms using tandem mass spectrometry at < 1% protein false discovery rate. Data are available via ProteomeXchange with identifier PXD014575. Relative abundances of hemocyanin oligomers and monomeric subunits from hemolymph and fractions were also quantified by polyacrylamide gel electrophoresis with and without denaturation for comparison of subunit heterogeneity. Hemocyanin subunits were organized primarily as hexamers (95–99% relative abundance) as opposed to dodecamers. Ten unique hemocyanins predicted by transcriptome and genome assemblies were identified by tandem mass spectrometry in both oligomer fractions including the first protein-level evidence of a β-type subunit in penaeid shrimp. Identified hemocyanins mapped to four genomic scaffolds and had two or three exons. A single small (75 kDa) subunit constituted half or more of all isoforms identified in each oligomer. The dodecameric fraction exhibited greater subunit heterogeneity utilizing a significantly lower ratio (1.04:1) of small subunit to large subunit compared to hexamers (2.05:1). One isoform, XP_027232115.1, appeared to be more dominant within dodecamers than hexamers. The ability to distinguish and quantify hemocyanin isoforms within oligomeric structures will aid future studies linking hemocyanin genes to transcripts to function and physiology as well as offer insight into the evolutionary history of crustaceans.
Introduction
The respiratory pigments of crustaceans, called hemocyanins, have followed a unique evolutionary history diverging from prophenoloxidases to eventually perform functions ranging from oxygen delivery to metabolite storage and innate immunity (Burmester, 2001; Terwilliger, 2007). Understanding the structural and functional plasticity of hemocyanins in marine crustaceans is particularly important as the incidence of low O2 (hypoxic) zones is rapidly increasing, such that approximately 240,000 km2 of coastal waters are unable to support higher forms of aerobic life (Diaz and Rosenberg, 2008). These extreme hypoxic events affect the economics of commercially important shrimp and other crustacean species by decreasing landings, changing harvest timings, and altering the growth and life history of historically captured species (Rabotyagov et al., 2014). However, differences in hemocyanin function and regulation between species may ultimately contribute to varying degrees of tolerance to hypoxia or other environmental challenges (Tommerdahl et al., 2015). Even within species, the regulation of hemocyanin structures can confer functional plasticity as hemocyanin protein domains can be cleaved proteolytically within the hemolymph to yield peptides with anti-fungal, anti-microbial, and anti-viral activity (Destoumieux-Garzón et al., 2001; Lee et al., 2003; Chongsatja et al., 2007).
The diversity of hemocyanin functions stems from evolutionarily derived sequence diversity which is more recently accessible through high-throughput sequencing techniques. A monomeric hemocyanin subunit is composed of roughly 660 amino acids, comprised of three domains (N, M, C) divided at amino acids 180 and 408 (Linzen et al., 1985). The N and C domains are more variable in sequence than the M domain, and are involved in binding carbohydrates (Linzen et al., 1985; Neuteboom et al., 1992). The central M domain is the most conserved region containing six histidine residues which coordinate with two copper atoms to bind O2. The distance between the two copper atoms influences the affinity with which hemocyanin binds O2 (Magnus et al., 1994). Hemocyanin O2 delivery may also be modified by allosteric factors such as lactate, urate, pH, or divalent ions which alter the protein structure to affect O2 binding (Terwilliger, 1998). There have been three major groups of hemocyanin described in the decapod crustaceans: the ancestral and less-understood β type and the more common α and γ types (Markl, 1986). The α type hemocyanin is found in all decapods while β and γ types are absent in many species. Of the α and γ types, seven distinct variants have been described in crustaceans (Burmester, 2001). Burmester calculated a decapod hemocyanin mutation rate, 1.29 × 10–9 substitutions per site per year, to be twice as fast as hemocyanin mutation in chelicerates (horseshoe crabs) and estimated that divergence of α and β type hemocyanins occurred 200 MYA (Burmester, 2001).
Hemocyanins often exist in multiple oligomeric forms each of which may possess distinct functional properties. The overall quaternary structure of hemocyanin in its native form exists in multiples of 6 monomeric subunits such as a 6-mer (hexamer), 12-mer (dodecamer) or higher orders (Burmester, 2002). These higher-ordered hemocyanin oligomers are often held together by linker subunit dimers via ionic or disulfide bonds (Stöcker et al., 1988). Depending on the nature of oligomer linking in each species, hemocyanin oligomers may or may not be dissociated in vitro by the removal of divalent ions, increases in pH, or multiple freeze/thaws (Markl and Decker, 1992; Beltramini et al., 2005). For example, those crabs and prawns in which dodecamer and hexamer usage differ generally express more of the oligomeric form which has a lower affinity for binding O2 (Mangum et al., 1991; Dainese et al., 1998; Molon et al., 2000; Beltramini et al., 2005; Kölsch et al., 2013). Wang et al. (2015) demonstrated that dodecameric hemocyanins in Litopenaeus vannamei can promote phenoloxidase activity absent in hexameric forms alone. Higher-order oligomers consisting of multiple hexamers may also confer distinct agglutination properties on hemocyanin (Pan et al., 2008).
Subunit heterogeneity within a hemocyanin oligomer can also confer functional plasticity in O2 binding or other functions. For example, hemocyanin subunits from the king crab Paralithodes camtschaticae were re-associated in vitro to form homo-hexamers which had lower O2 affinities than their native hetero-hexamer counterparts (Molon et al., 2000). In vivo regulation of hemocyanin subunit heterogeneity was also observed in the Atlantic blue crab, Callinectes sapidus, which increased the O2 affinity of its hemocyanin following 7- or 25-day exposure to hypoxia through shifts in hemocyanin subunit types (deFur et al., 1990). Diversity of both subunit and oligomer composition makes hemocyanin a functionally plastic protein that may play an important role in adaptation to environmental factors such as hypoxia or disease resistance.
Recent studies of L. vannamei hemocyanin have identified a highly expressed small subunit (HcS) and several isoforms of a large subunit (HcL) through transcriptomic and other techniques (Johnson et al., 2015, 2016; Lu et al., 2015; Xu et al., 2015). Recognition of the sequence diversity in this commercially important species has led to tests of function in different scenarios of hypoxia (low O2), hypercapnia (high CO2), or immunological challenge. Seven hemocyanin microarray probes expressed in the hepatopancreas showed significant changes between 24 h hypoxia (Po2 4.0 kPa) and normoxia while two probes were also informative in distinguishing shrimp exposed to hypercapnic hypoxia (Rathburn et al., 2013). In-depth RNA-sequencing efforts by Johnson et al. (2015) showed that all identified isoform transcripts were upregulated (1.65–1.98 fold) after 24 h exposure to hypoxia. However, these analyses also revealed an additional novel β-type hemocyanin transcript which showed patterns of transcriptional regulation that differed from other isoforms in animals exposed to a combination of hypoxia and hypercapnia. Other studies also demonstrated the transcriptional upregulation of hemocyanin isoforms in L. vannamei exposed to white spot syndrome virus (Xu et al., 2015). Further, Lu et al. (2015) cloned and expressed an additional HcL variant; the recombinant protein displayed anti-bacterial properties. While recent efforts to describe the regulation of hemocyanin in response to various challenges have focused on transcriptional regulation, informatics, or artificial over-expression of protein, we are unaware of any studies which examine the natural abundance of each hemocyanin isoform at the protein-level. The present study uses semi-quantitative proteomic approaches to examine all predicted hemocyanin isoforms simultaneously at the protein level and to describe their heterogeneous usage within different oligomeric structures in L. vannamei hemolymph. We report the first protein-level identification of a β-type subunit in penaeid shrimp and demonstrate differential usage of subunit isoforms in native dodecameric and hexameric hemocyanins. These approaches are readily adaptable for developing targeted measurements and testing protein-level regulation of all isoforms in response to environmental challenges.
Materials and Methods
Animal Care and Maintenance
Juvenile L. vannamei shrimp purchased as post-larvae from Shrimp Improvement Systems (Plantation Key, FL, United States) were grown out to approximately 1 g size at the Waddell Mariculture Center (Bluffton, SC, United States) before being transported to Hollings Marine Laboratory for the present study. Shrimp were kept in recirculating tanks with well-aerated, filtered, and UV-treated seawater at 30‰ salinity. Salinity was monitored daily, and ammonia, nitrate, nitrite, and pH were monitored weekly. Water changes of 20% tank volume were performed each week. A ration of commercial shrimp food (Shrimp Grower Semi-Intensive, Zeigler Bros., Inc., Gardners, PA, United States) was supplied daily. A total of eight shrimp were used for characterizing hemocyanin subunit (SDS-PAGE) and oligomer (native PAGE, SE-HPLC) diversity. Hemolymph proteins from these eight shrimps were also fractionated for analysis of subunit diversity by LC-MS/MS, but TIC from fractions from three shrimp were very low and were subsequently omitted.
Hemolymph Extraction
Hemolymph (100–200 μL) was extracted from the base of the 2nd or 3rd walking leg using a 26 ½ gauge needle and immediately extruded into 1.5 mL microcentrifuge tubes to clot on ice. Clot formation occurred within an hour, after which, the hemolymph samples were homogenized with a pestle, and centrifuged for 6 min at 23,000 × g at 4°C. An aliquot of the resulting hemolymph supernatants was immediately used for the determination of hemocyanin concentration while the remaining volumes were stored at −80°C until use.
Hemocyanin and Total Protein Concentration
Fresh hemolymph supernatants were diluted to 1:100 in 10 mM EDTA, 2.5% NaCl, pH 10 to dissociate oligomeric forms. Hemocyanin concentrations were then determined spectrophotometrically by measuring absorbance at 338 nm (OD338) and calculated using the extinction coefficient of Crangon crangon hemocyanin, E1 = 2.83%–1 cm–1 (Nickerson and Van Holde, 1971; Hagerman, 1986). Total protein concentration of each hemolymph sample was determined using the Bradford method with bovine serum albumen dilution standards (Bradford, 1976).
Hemocyanin Oligomer Characterization and Fractionation
Size Exclusion HPLC
Hemolymph proteins from L. vannamei were separated using SE-HPLC (7.8 mm × 600 mm BioSep SEC s4000, Phenomenex) using a custom elution buffer mimicking the ionic composition of L. vannamei hemolymph to keep hemocyanin in its native state (10 mM HEPES pH 7.40, 210 mM NaCl, 15 mM CaCl2, 10 mM MgCl2, 8 mM KCl) (Cheng et al., 2002). Hemolymph samples were loaded on a 200 μL injection loop at different amounts for preparative fraction collection (1.5 mg) or analytical quantification (200 μg). Absorbance was monitored at 280 and 338 nm to detect aromatic amino acids and the Cu-O2 complex of hemocyanin respectively. Two eluted peaks absorbing at both 280 and 338 nm were confirmed by LC-MS/MS as hemocyanins and further analyzed. Molecular weights and relative abundances of each peak were estimated based on an external calibrant (Gel Filtration Standard proteins, BioRad) and peak areas were estimated at OD280. To validate the reliability of this method for analyzing crustacean hemocyanin, hemocyanin oligomers from the blue crab, C. sapidus, the brown shrimp Farfantepenaeus aztecus, and the pink shrimp Farfantepenaeus duorarum were also separated. The major protein peaks identified at 280 nm also corresponded with absorbance at 338 nm characteristic of hemocyanin-O2 binding. Chromatograms of C. sapidus hemolymph matched hemocyanin peaks reported by Mangum et al. (1991) and fractions from F. aztecus were further confirmed to be predominantly hemocyanin by LC-MS/MS (data not shown) suggesting that this is a reliable method of separating hemocyanins from crustacean hemolymph.
Oligomer Fraction Collection
Two fractions of column eluate were collected in Protein Lo-Bind tubes (Eppendorf): between 9.0 and 9.7 mL for fraction 1 (here referred to as the dodecamer fraction) and between 10.0 and 11.5 mL for fraction 2 (the hexamer fraction). Due to low abundance of protein in the dodecamer fraction, duplicate injections were collected and combined. Fractions were then re-concentrated using Centricon Ultracel YM-30 (Millipore) spin columns according to the manufacturer’s instructions. Following buffer exchange with mass spectrometry grade water and final re-concentration, each sample had a volume between 50 and 100 μL. Final protein concentrations were measured using the Bradford Assay described above, and samples were frozen at −20°C until use within a few days.
Native PAGE
Hemolymph proteins (5 μg) and purified dodecamer (1 μg) and hexamer hemocyanin (1 μg) fractions were separated on 3–12% Bis-Tris Blue NativePage gels (Life Technologies) with NativeMark Unstained Protein Standard (Life Technologies). All samples were diluted using Ultrapure water and loaded with non-reducing sample buffer (Life Technologies). Following separation, gels were fixed in 40% methanol (v/v), 10% glacial acetic acid (v/v) for 20 min, stained with 0.02% (w/v) Coomassie R-250 in 30% methanol, 10% acetic acid for another 20 min, and de-stained with 8% acetic acid overnight. Molecular weights and relative abundances based on stain intensity were determined using ImageLab software.
Hemocyanin Subunit Characterization
Denaturing PAGE
The subunit compositions of hemolymph supernatant and oligomer fractions were analyzed by SDS-PAGE. Hemolymph proteins (0.5 μg) were combined with LDS Sample Buffer and the NuPAGE reducing agent, then separated on NuPAGE 4–12% Bis-Tris gels (Life Technologies) with Novex Sharp Unstained protein standard (Life Technologies). Gels were stained with 0.1% (w/v) Coomassie R-250 in 40% methanol, 10% acetic acid according to the manufacturer’s protocol (Life Technologies), and de-stained with 8% acetic acid overnight. Gels were imaged and analyzed as indicated above for native gels. Two hemocyanin subunit sizes were consistently seen at approximately 74 and 70 kDa (here referred to as HcL and HcS respectively). The ratios of HcS:HcL were determined from total hemolymph, dodecamer fractions, and hexamer fractions for eight shrimp using repeated measures ANOVA as well as paired t-tests using a Bonferroni correction. All statistical analyses were performed using RStudio (v2.15.1). Preparation of tryptic peptides for LC-MS/MS.
Hemocyanin subunit isoforms and other proteins were identified by LC-MS/MS in dodecamer and hexamer fractions separated by SE-HPLC in five of the same eight shrimps previously described. The three remaining shrimps were omitted due to low TIC between fractions. Fractions collected by SE-HPLC underwent five total centricon washes and five buffer replacements using mass-spectrometry grade water. Following re-concentration of fractions, 5 μg of each sample were brought to 50 μL in100 mM ammonium bicarbonate buffer (pH 8.0). The 50 μL samples were then reduced by adding 100 mM dithiothreitol (DTT) to a final concentration of 5 mM DTT incubated at 60°C for 30 min. After cooling, the reduced samples were alkylated by adding 500 mM iodoacetic acid (IAA) to a final concentration of 15 mM IAA and incubated at room temperature for 30 min in the dark. Trypsin Gold, MS grade (Promega) was added to each sample at an enzyme to protein ratio of 1:25 (w:w). All samples were brought to a total volume of 150 μL with 100 mM ammonium bicarbonate, pH > 7.5, then incubated at 37°C to facilitate trypsin activity.
The trypsin digest was stopped after 18 h by bringing samples to 1 mL volume with 0.1% (v/v) formic acid (FA). Peptides were loaded onto solid phase extraction columns (30 mg Strata-X 33 μ polymeric reverse phase solid phase extraction columns, Phenomenex, Torrance, CA, United States) first conditioned with 1 mL 100% acetonitrile (ACN) followed by 1 mL 0.1% formic acid (v/v). The columns were then washed three times with 1 mL of 0.1% FA, and samples were eluted with 500 μL of 60% ACN (v/v), 0.1% FA into a Protein Lo-bind tube (Eppendorf) and dried by SpeedVac. Dodecamer fraction samples were re-suspended in 30 μL 0.1% FA, and hexamer fraction samples in 80 μL 0.1% FA. Protein concentrations were then estimated using NanoDrop detection (OD280).
LC-MS/MS
The performance of the LC-MS/MS was calibrated by evaluating precursor retention times and fragment ion areas of a standard tryptic digest of β-galactosidase prior to loading samples. Based on NanoDrop estimates of protein concentration, a dilution series of sample peptides was used to determine appropriate loading amounts for each fraction matching TIC between runs. As a result, dodecamer fraction samples were diluted to 0.1 μg μL–1 and hexamer fraction samples were diluted to 0.05 μg μL–1 using 0.1% FA. Samples were then centrifuged at maximum speed for 10 min and 10 μL of dodecamer fraction supernatant and 3 μL of hexamer fraction supernatant was injected onto a 100 μm × 2 cm C18 (100 Å with 5-μm particles) trap column (AcclaimTM PepMap 100; Thermo Fisher Scientific) with mobile phase A [2% ACN (v/v), 0.1% FA], and separated on a 75 μm × 15 cm C18 (300 Å with 3-μm particles) analytical column (AcclaimTM PepMap 100; Thermo Fisher Scientific). Reverse phase separation at 350 nL min–1 was performed with a gradient of 0–50% mobile phase B [95% ACN (v/v), 0.1% FA (v/v)] over 36 min on a 2D + NanoLC system (Eksigent, Dublin, CA, United States). The LC was interfaced to a TripleTOF 5600 Mass Spectrometer (AB Sciex, Foster City, CA, United States) with a nanospray source. Source temperature was 120°C, source voltage was 2,500 V, declustering potential was 100 V, and collision energy was 10 eV. The instrument was run in positive ion instrument dependent acquisition mode with precursor ion scans accumulating for 250 ms with up to 20 candidate ions if precursors were 350–1250 m/z, exceeded 500 counts per second (cps), and had 2 + to 5 + charge. The mass tolerance was 50 mDa and isotopes within 4 Da were excluded. The product ion scans were performed with an accumulation time of 200 ms for TOF masses 100–1500 Da. Collision energy was set to rolling collision energy.
Data were visualized using PeakView (AB Sciex) software and files were converted from.wiff to .mgf format. Mascot (v2.6.2) searches were performed accounting for fixed Carbamidomethyl (C) modifications, variable modifications [oxidation (M), deamidation (NQ), carbamidomethyl (N-term)], a decoy database, 2 maximum missed cleavages, a peptide tolerance of ± 20 ppm, MS/MS tolerance of 0.25 Da, and charges of + 2, + 3, and + 4. Peptides were searched against databases including the representative L. vannamei genome assembly proteins (ASM378908v1; Zhang et al., 2019), a translated L. vannamei transcriptome assembly including five full-length hemocyanins and multiple contig variants (Johnson et al., 2015), and contaminant protein databases. Search results were then imported into Scaffold (4.8.8) software and scored by peptide and protein prophet with legacy grouping. All peptides and proteins were identified at < 1% protein FDR with 2 minimum peptides at < 1% local peptide FDR. Normalized spectral abundance factors were determined using functions embedded in Scaffold. Exponentially modified protein abundance index values were also calculated within Scaffold according to Ishihama et al. (2005). Relative abundances of proteins were calculated as the proportion NSAF or emPAI within each fraction sample and averaged between five replicate shrimps for fractions 1 and 2.
Comparison of Transcriptome and Reference Genomic Hemocyanins
Hemocyanin isoforms HcB, HcS, HcL1-3 predicted from the transcriptome database (Johnson et al., 2015) were aligned to the reference L. vannamei genome (ASM378908v1 Annotation Release 100; Zhang et al., 2019) using tBLASTn. The highest ranking % identity scaffold alignments with over 95% coverage of queries were further examined. Sequences from the top genes from each of the highest ranking scaffold hits were translated and aligned back to the original transcriptomic query sequences using Clustal Omega to compare sequence differences.
Results
Hemocyanin Oligomers
Hemocyanin oligomer and subunit usage was described in eight shrimp using SE-HPLC and native PAGE. Separation of hemolymph proteins by SE-HPLC revealed two main protein peaks which absorbed at OD338 (Figure 1A). These two fractions were confirmed by mass spectrometry to contain primarily hemocyanin (see section “Distinction of Subunit Isoforms by LC-MS/MS”). The dodecamer peak had a molecular weight of approximately 661.3 ± 3.6 (SEM; n = 8) kDa when analyzed by SE-HPLC and 673.1 ± 4.5 (SEM; n = 8) kDa when analyzed by native PAGE. Meanwhile, the hexamer peak was approximately 381.0 ± 3.8 (SEM; n = 8) kDa by SE-HPLC and 402.6 ± 5.6 (SEM; n = 8) kDa by native PAGE. The hexamer accounted for approximately 99.4 ± 0.1% (SEM; n = 8) of the total OD280 peak area of the two hemocyanin peaks, and 95.5 ± 0.4% (SEM; n = 8) of the band intensity of the two oligomers by native PAGE. Purified dodecamer and hexamer fractions separated by native PAGE confirmed that the hexamer fraction excluded the higher molecular weight oligomer (Figure 1B). However, due to the overwhelming amount of hexamer relative to dodecamer, a faint hexamer band was also detected in the dodecamer fraction. Therefore, further analyses of the dodecamer fraction could not necessarily exclude the presence of small amounts of hexamer fraction proteins.
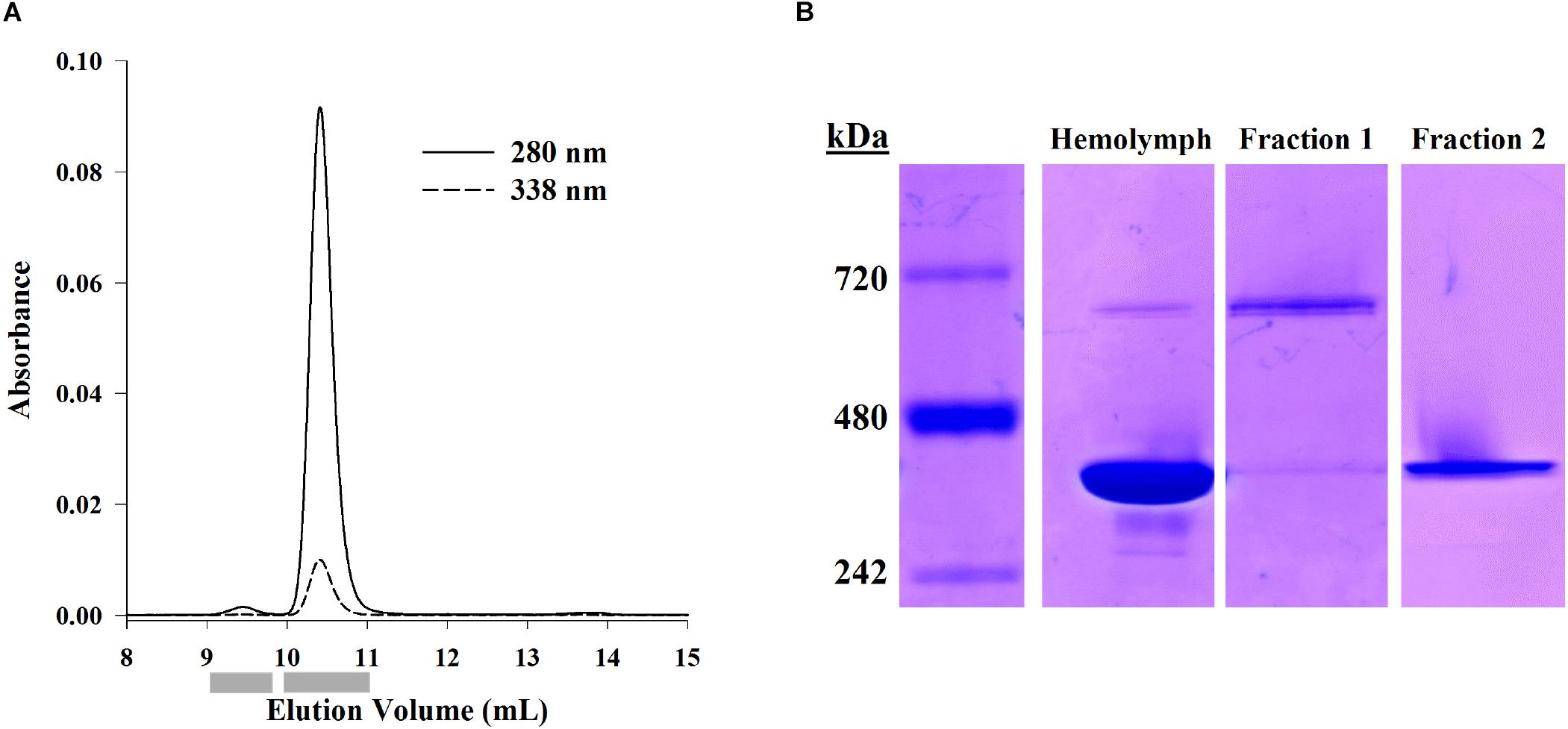
Figure 1. Separation of hemocyanin oligomer forms using SE-HPLC or native PAGE. (A) Representative SE-HPLC chromatogram (OD280 and OD338) of L. vannamei hemolymph proteins showing two peaks at 9.5 mL (dodecamer) and 10.4 mL (hexamer). Gray bars indicate fraction collection windows. (B) Proteins from hemolymph, dodecamer fractions, and hexamer fractions separated by native PAGE (Bis-Tris 3–12%) and visualized with Coomassie-R250 staining show hexameric (403 kDa) and dodecameric (673 kDa) hemocyanin in hemolymph. The hexamer fraction is exclusively hexamer while the dodecamer fraction shows some hexamer as well.
Hemocyanin Subunits
Denatured hemolymph proteins and hemocyanin oligomers, separated by denaturing PAGE and visualized by Coomassie R-250 staining, consisted of two main hemocyanin bands: the large subunit (HcL) at 73.7 ± 0.3 (SEM; n = 8) kDa and the small subunit (HcS) at 70.4 ± 0.3 (SEM; n = 8) kDa (Figure 2A). Significant differences in HcS:HcL ratios between shrimp-matched hemolymph dodecamer, and hexamer fractions were found by repeated measures ANOVA (F2,7 = 94.8, p = 7.3 × 10–9; Figure 2B). Total hemolymph and the hexamer fraction ratios of HcS:HcL were 1.83 ± 0.09 and 2.05 ± 0.08 respectively while the dodecamer fraction had a HcS:HcL ratio of 1.04 ± 0.04. All these ratios were significantly different from each other based on Bonferroni-corrected paired t-test comparisons (p = 0.0002, 0.0459, 0.00001 for respective comparisons between hemolymph/dodecamer, hemolymph/hexamer, dodecamer/hexamer subunits).
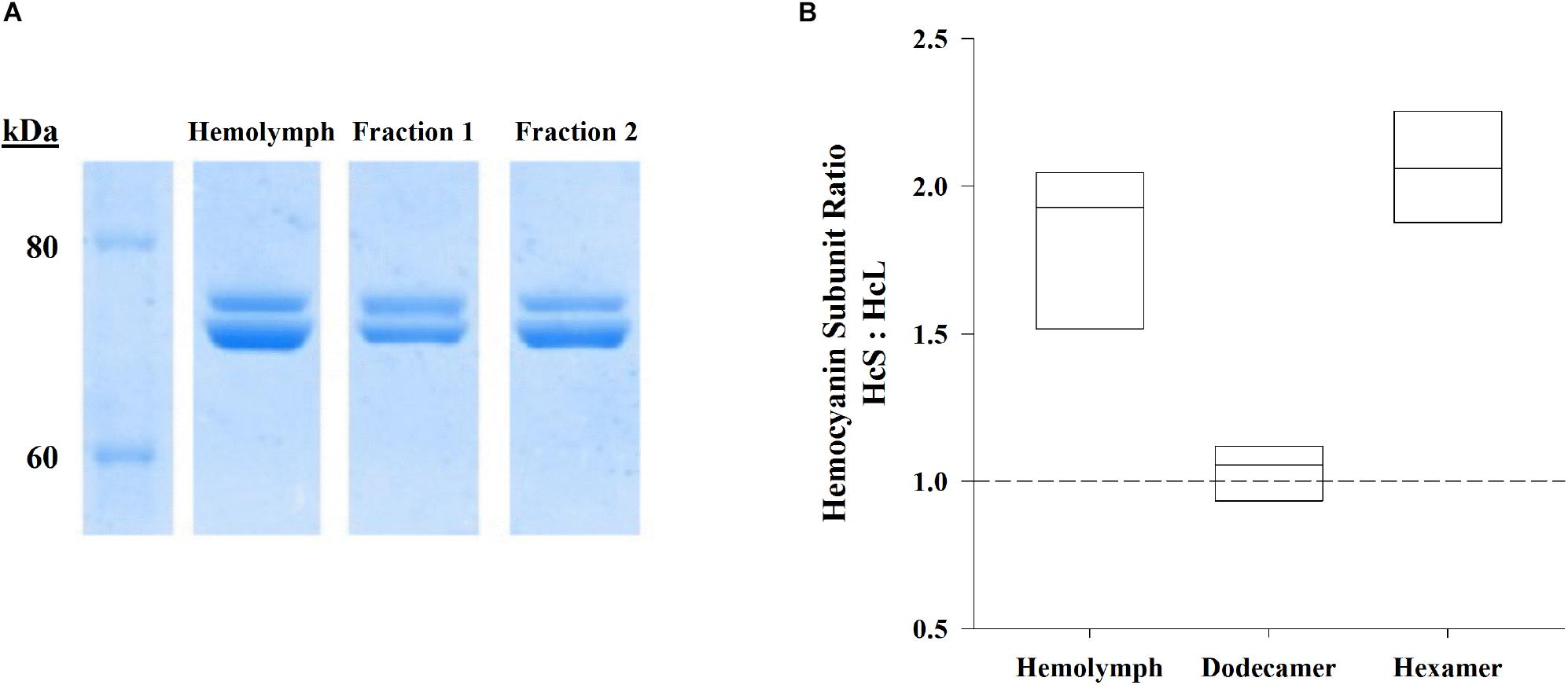
Figure 2. Hemolymph samples and their shrimp-matched dodecamer and hexamer fractions (n = 8) separated by SE-HPLC (BioSep SEC s4000, Phenomenex) were further separated by denaturing PAGE. (A) Representative SDS-PAGE (Bis-Tris 4–12%) gel visualized by Coomassie R-250 staining shows hemocyanin subunits HcL (top) and HcS (bottom) for hemolymph, dodecamer fractions, and hexamer fractions against Novex Unstained Protein Standard (Life Technologies). (B) Ratios of HcS:HcL were significantly different for hemolymph hemocyanins as well as their corresponding fractionated hemocyanin oligomers (repeated measures ANOVA, F2,7 = 94.8, p = 7.3 × 10–9). Boxes indicate interquartile ranges and median HcS:HcL ratios (solid lines). The dashed reference line indicates a 1:1 ratio.
Comparison of Transcriptome and Reference Genome Hemocyanins
Alignments of hemocyanins from transcriptomic and genomic databases resulted in consensus genes for HcS as XP_027239062.1 and HcB as XP_027236657.1 (Table 1). Identical sequences for transcriptomic HcL1, HcL2 and HcL3 could not be located within the reference genome, but matched most closely to XP_027227689.1 (99% coverage, 93.6% ID), XP_027232108.1 (100% coverage, 98.1% ID), and XP_027232124.1 (100% coverage, 95.9% ID) respectively (Table 1). Unique peptides were identified by LC-MS/MS which distinguish HcL1 and HcL3 from their respective reference genome counterparts and vice versa, leading us to subsequently quantify them as separate isoforms. However, uniquely identified peptides matching XP_027232108.1 and not HcL2 were observed in the present study so only the genomic accession was used for subsequent analysis. Further attempts to locate transcriptomic HcL1-3 within the genome were conducted using tBLASTn separately for each of three assumed exon regions separated at codons for residues 27 and 534 (Table 2) but none of the exon region queries matched any genomic sequences more closely than the accessions listed above. It is thus unknown whether transcriptomic isoforms HcL1 and HcL3 are strain-specific variants of XP_027227689.1 and XP_027232124.1 respectively or coded for by genes unique to the population used in the present study.
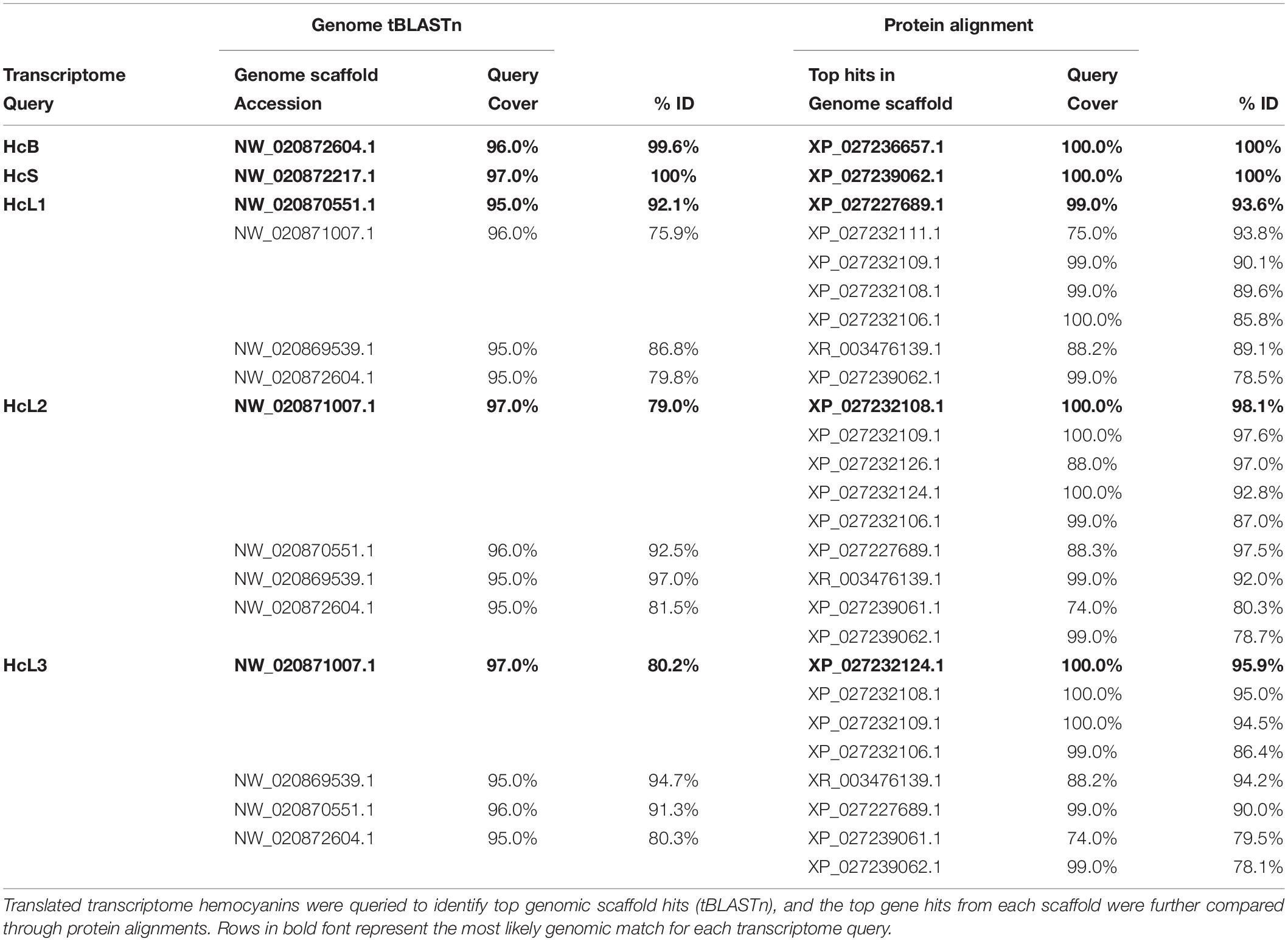
Table 1. Alignments of hemocyanin isoforms identified in L. vannamei transcriptomes (Johnson et al., 2015) to isoforms in the reference genome assembly ASM378908v1 (Zhang et al., 2019).
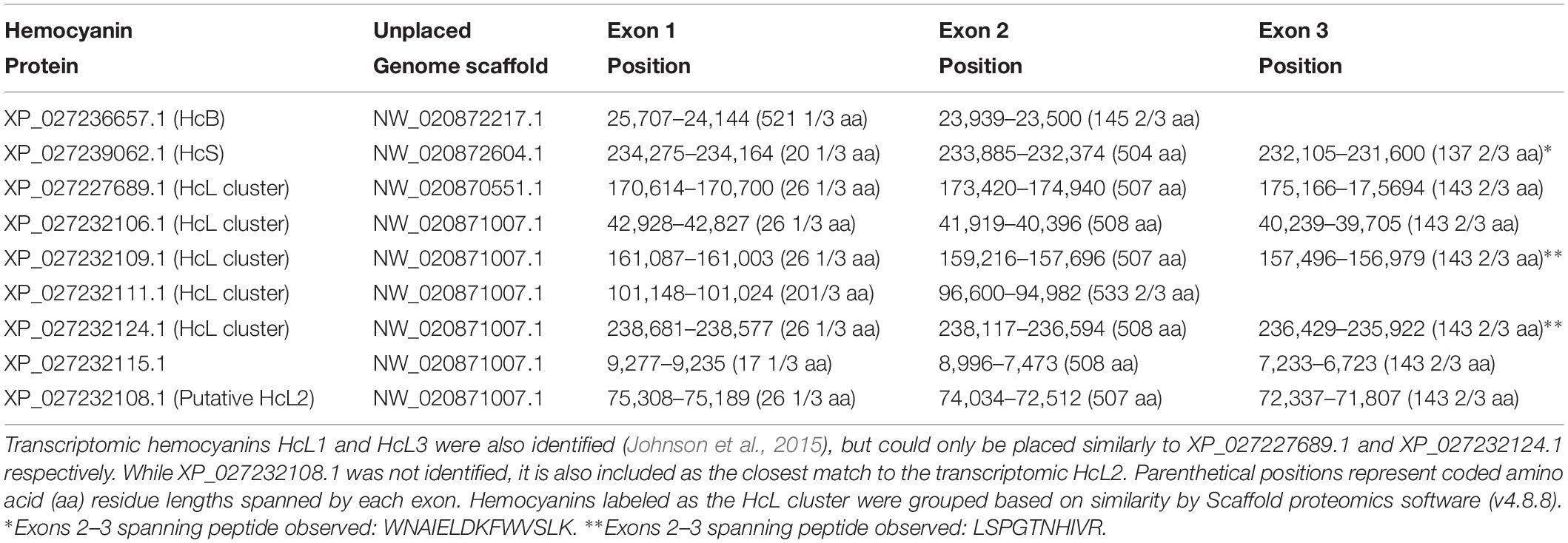
Table 2. Exon positions for L. vannamei hemocyanins identified through LC-MS/MS and their respective positions on unplaced genome scaffolds from the reference genome assembly ASM378908v1 (Zhang et al., 2019).
Further genomic analyses revealed that HcB, HcS, and XP_027227689.1 (putative HcL1 variant) were each found on distinct unplaced genomic scaffolds with the putative HcL1 gene oriented on the complementary strand relative to all other identified isoforms (Table 2). The remaining identified isoforms were all found on a single unplaced genomic scaffold NW_020871007.1. Most hemocyanin isoforms from this scaffold were coded for by three exons with splices in the codons for residues 27 and 534, except for XP_027232115.1 which had its first splice site eight codons earlier, and XP_027232111.1 which contained only two exons spliced at the codon for residue 21. Interestingly, HcB was coded for by only two exons spliced at the codon for residue 522 and lacked the earlier N-domain splice observed in all other isoforms.
Distinction of Subunit Isoforms by LC-MS/MS
Dodecamer and hexamer hemocyanin fractions separated by SE-HPLC were analyzed and identified by LC-MS/MS and the Mascot search engine (<1% local FDR) against a database containing representative L. vannamei genome assembly proteins (ASM378908v1; Zhang et al., 2019) and a translated L. vannamei transcriptome assembly (Johnson et al., 2015). Almost all proteins identified from both fractions were hemocyanins, accounting for over 86% of proteins quantified by NSAF (Figure 3A) and over 96% by emPAI (Figure 3B). Eighteen other non-hemocyanin protein groups were observed at low abundances making up less than 15% (based on NSAF) of the dodecamer fraction and 2% (based on NSAF) of hexamer fraction (Figure 3A). Eight protein groups were exclusive to the dodecamer fraction accounting for < 1% based on emPAI and < 4% based on NSAF in the total fraction. These protein groups included putative cholinesterases and carboxylases, glucan binding proteins, down syndrome cell adhesion molecules, plexins, fibrocystins, muscle M-line assembly proteins, neprilysin 1, and neural cadherins. On the other hand, only two protein groups were exclusively identified in the hexamer fraction accounting for < 0.5% relative abundance by either emPAI or NSAF. These included an uncharacterized protein (XP_27212713.1), and a transferrin like protein (XP_027215079.1) which was previously annotated as a pacifastin heavy chain in the transcriptomic database.
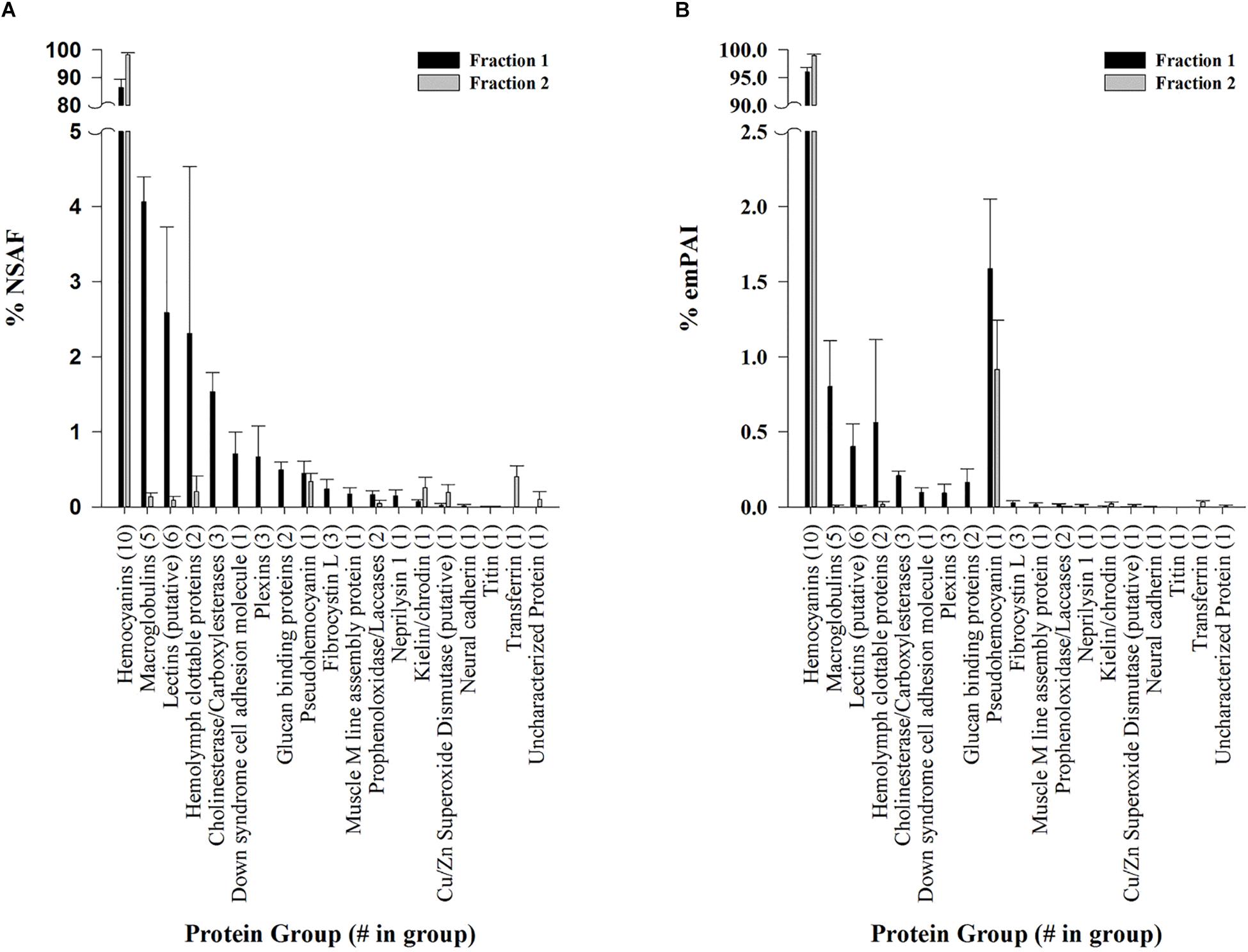
Figure 3. Protein groups identified in L. vannamei hemolymph fractions by LC-MS/MS. Semi-quantitative values are listed as molar fractions (average ± SEM; n = 5) calculated from (A) NSAF and (B) emPAI values summed over all identified proteins. The number of putative protein hits from the reference genome assembly ASM378908v1 (Zhang et al., 2019) or contig hits from the translated L. vannamei transcriptome database (Johnson et al., 2015) are listed in parenthesis for each group. All peptides and proteins were identified at < 1% protein FDR with 2 minimum peptides at < 1% local peptide FDR. Fraction 1 (black bars) and 2 (gray bars) represent dodecamer and hexamer fractions respectively.
Overall, 10 hemocyanin sequences predicted from both RNA sequencing (Johnson et al., 2015) and genome assemblies (Zhang et al., 2019) were identified as distinct isoforms in both fractions (Figure 4). HcS was the single most abundant hemocyanin within each fraction, accounting for 48.25 ± 0.67% and 60.89 ± 0.62% (SEM; n = 5) NSAF of hemocyanins within dodecamer and hexamer fractions, respectively (Figure 4A). Quantification of HcS by emPAI resulted in similar proportions, 42.01 ± 5.39% emPAI in fraction 1 and 53.11 ± 4.97% emPAI in fraction 2 (Figure 4B). In all cases, the transcriptomic version of HcL1 was the next most abundant hemocyanin isoform accounting for 28.54 ± 0.64 and 33.16 ± 0.50% (SEM; n = 5) NSAF of hemocyanins within dodecamer hexamer fractions respectively (Figure 4A). Quantification of HcL1 by emPAI resulted in similar proportions, 19.41 ± 0.57% emPAI in fraction 1 and 28.62 ± 2.81% emPAI in fraction 2 (Figure 4B). The novel β-type hemocyanin subunit (HcB) predicted from transcriptome assemblies was also identified in four of the five shrimp at the protein level (<1% FDR), although the relative abundances were low at 0.02 ± 0.02 and 0.16 ± 0.07% (SEM; n = 5) of NSAF in the dodecamer and hexamer fractions, respectively (Figure 4A). In total, six peptides exclusive and unique to HcB were identified (<1% local FDR), providing the first protein-level confirmation of a β-type hemocyanin in penaeid shrimp (Table 3).
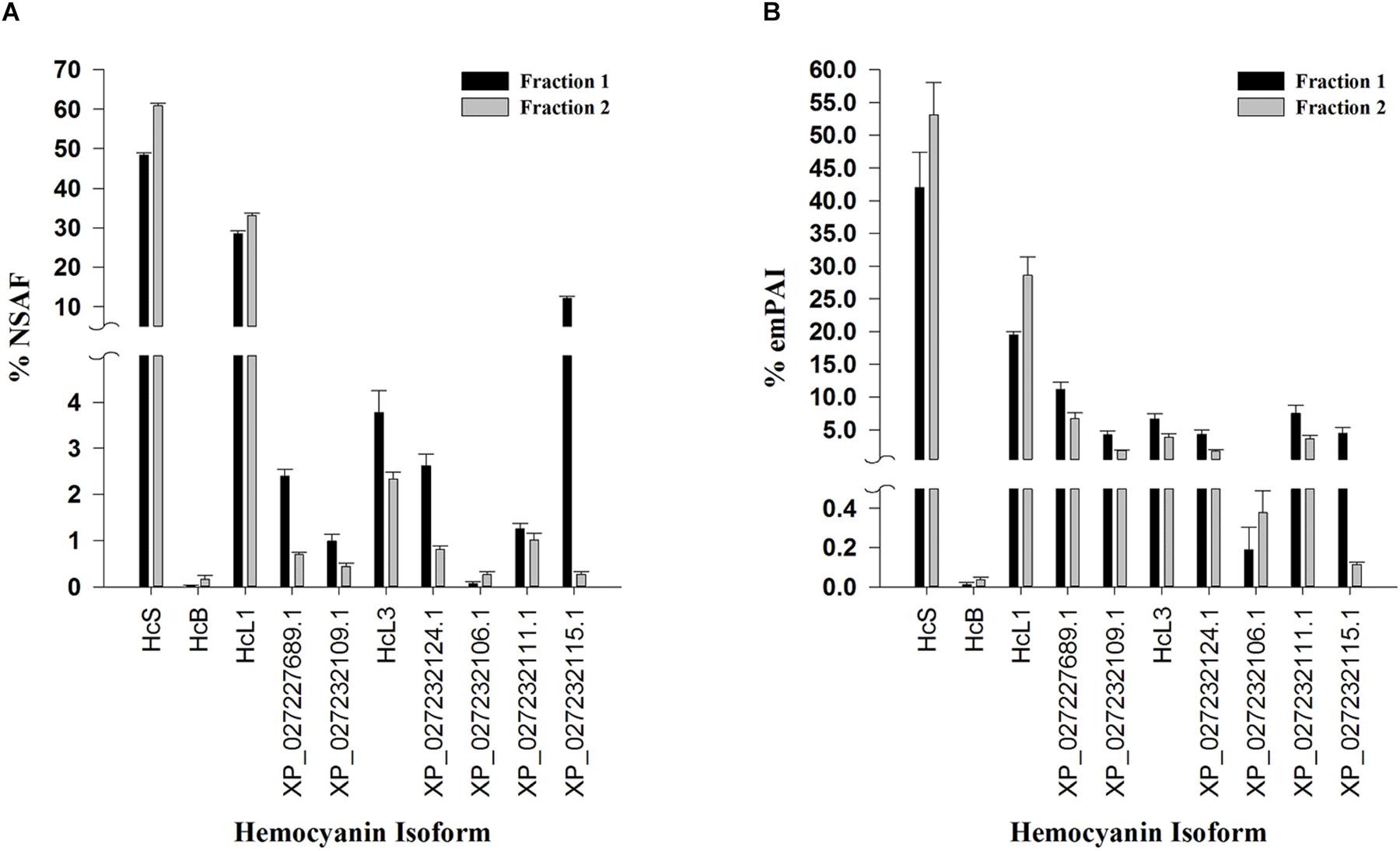
Figure 4. Relative proportion of hemocyanin subunits identified in L. vannamei hemolymph by LC-MS/MS. Semi-quantitative values are listed as molar fractions (average ± SEM; n = 5) calculated from (A) NSAF and (B) emPAI values summed over all identified hemocyanins from the reference genome assembly ASM378908v1 (Zhang et al., 2019) or the translated L. vannamei transcriptome database (Johnson et al., 2015). All peptides and proteins were identified at < 1% protein FDR with 2 minimum peptides at < 1% local peptide FDR. Fraction 1 (black bars) and 2 (gray bars) represent dodecamer and hexamer fractions respectively. HcS (XP_027239062.1) and HcB (XP_027236657.1) refer to small, and β-type hemocyanin isoforms respectively while HcL1 and HcL3 are hemocyanin variants unique to the transcriptome database used.

Table 3. β-type hemocyanin (HcB; XP_027236657.1) peptides identified in Litopenaeus vannamei (n = 5 shrimp denoted by ‘X’) by LC-MS/MS providing 15% predicted protein sequence coverage (<1% protein FDR with 2 minimum peptides at < 1% local peptide FDR).
Seven of the identified hemocyanins contained multiple shared peptides and were clustered together as an HcL group by Scaffold (v4.8.8) software but were treated as distinct genes based on genomic evidence (Table 2). Despite sharing multiple peptides, the predicted molecular weights of the seven isoforms in the HcL cluster also ranged from 64 to 81 kDa. Therefore, when calculating small to large subunit ratios for the purposes of comparison with native PAGE HcS:HcL results, “small” subunits included HcS (75 kDa), HcB (76 kDa), and XP_027232111.1 (64 kDa) while the remaining seven isoforms were counted as ‘large’ subunits (77–81 kDa) to obtain these ratios. This resulted in HcS:HcL ratios of 1.03 ± 0.15% emPAI or 0.98 ± 0.02% NSAF for fraction 1 and 1.45 ± 0.33% emPAI or 1.64 ± 0.03% NSAF for fraction 2.
Discussion
Building on recent transcriptomic and genomic work revealing hemocyanin diversity in L. vannamei, the present study provided a framework for characterizing and comparing protein-level isoform use through classical and proteomic approaches. Proteomic analyses allowed us to distinguish 10 hemocyanins from a reference genome (Zhang et al., 2019) and transcriptome (Johnson et al., 2015) including the first protein-level identification of a β-type subunit in penaeid shrimp. Hemocyanin variants (HcL1, HcL3) were also identified by peptide spectral matching which were unique to the population-specific transcriptome; otherwise not included in the reference genome. Such an analysis could be readily adapted to a wide variety of studies on hemocyanin modification, hypoxic adaptation, or immune response in crustacean species with assembled transcriptomes. While high throughput RNA sequencing has broadened our understanding of gene diversity and helped to generate hypotheses on organismal responses to challenge, in some cases mRNA levels may only explain approximately 40% of the variation in protein levels (Schwanhausser et al., 2011). Furthermore, protein oligomerization can have significant effects on protein function, so we sought to characterize L. vannamei hemocyanin oligomer and monomer usage to examine their protein-level composition first in non-experimental conditions.
Hemocyanin oligomer use in L. vannamei resembled that of previous studies as well as other penaeid shrimp. Two oligomeric forms of hemocyanin, hexamer and dodecamer, were routinely detected in hemolymph by SE-HPLC or native PAGE with the dominant hexamer representing 95–99% of the two oligomer forms. The dominance of the hexamer was also seen in L. vannamei by Figueroa-Soto et al. (1997) and García-Carreño et al. (2008). Pan et al. (2008) reported a greater abundance of dodecamer relative to hexamer by gel filtration chromatography, but the opposite relative abundance using PAGE in the same study, which was explained as dodecamers dissociating in PAGE conditions. Due to conflicting reports in the literature regarding hemocyanin oligomer abundance in L. vannamei, SE-HPLC and native PAGE conditions were tested in the current study to ensure that dodecamers were not dissociating during separation, and C. sapidus hemocyanin, dominantly dodecameric, was used as a positive control for the detectability of dodecamers (Mangum et al., 1991). Thus we concluded that L. vannamei are like other penaeid shrimp L. setiferus, F. aztecus, F. duorarum, and P. monodon in that they utilize dominantly hexameric rather than dodecameric hemocyanin (Figure 5; Brouwer et al., 1978; Beltramini et al., 2005).
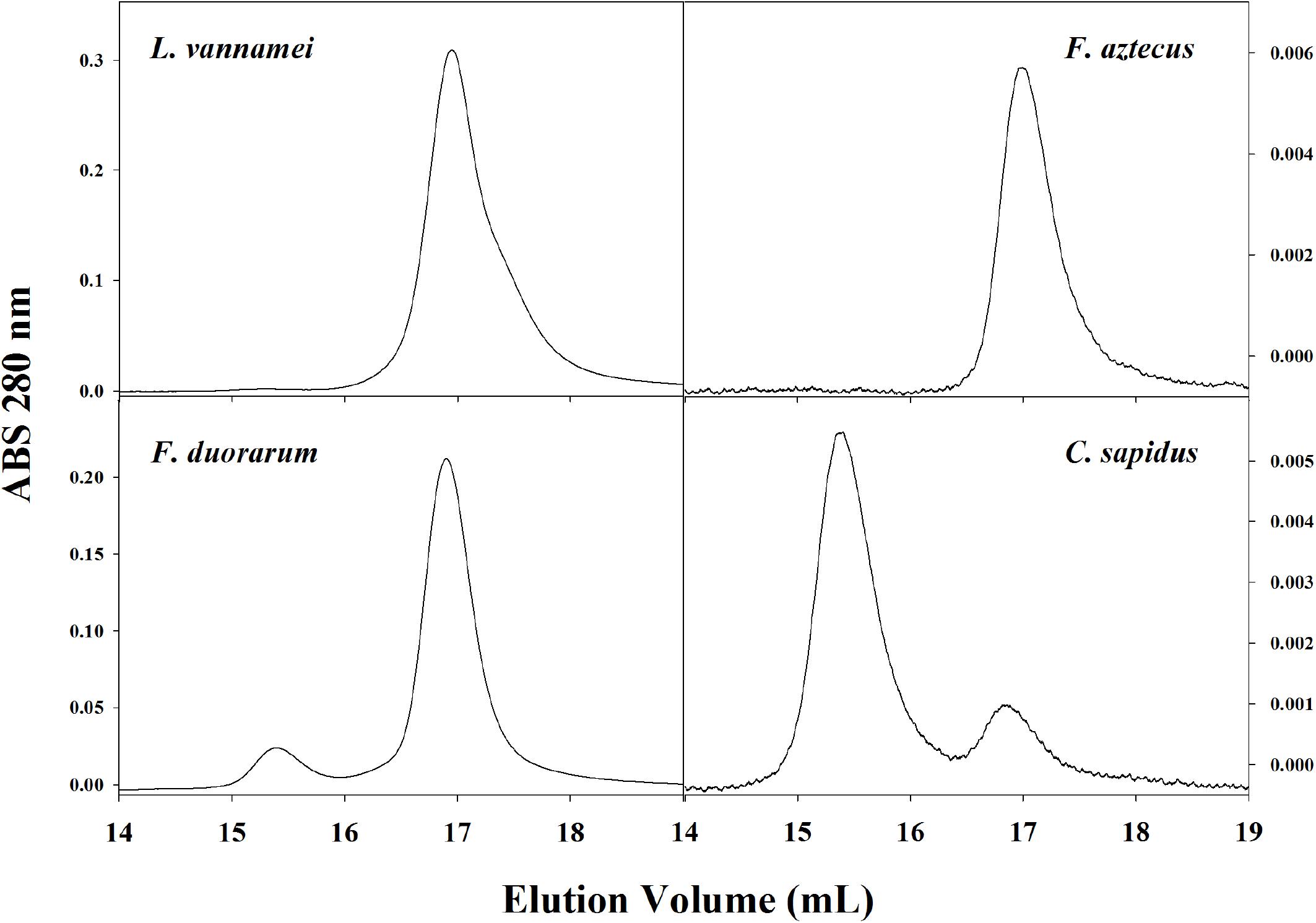
Figure 5. Representative SE-HPLC (2x BioSep SEC-400-5 in series) chromatograms (OD280) of four crustacean species show different usage of hemocyanin oligomers in hemolymph. The dodecameric hemocyanin appears between 15 and 16 mL elution volume and the hexamer appears at approximately 17 mL elution volume.
The molecular weights of oligomers measured in the present study were smaller than expected (673 kDa dodecamer and 403 kDa hexamer by PAGE). Hexameric and dodecameric hemocyanins of crustaceans are generally reported to be 450 and 900 kDa respectively, based on an average monomer size of 75 kDa (Markl and Decker, 1992). Underestimations of size by SE-HPLC and PAGE may be due to the condensed/globular form of oligomers in their native state during separation, which would migrate as physically smaller molecules than when they are denatured or linearized. Hemocyanin amino acid sequences translated in silico have negative charges at a neutral pH as predicted by isoelectric points ranging from 5.3 to 6.01 (ExPASy). Already negatively charged proteins stained with a further negatively charged Coomassie dye may travel faster during PAGE, resulting in underestimations of protein size. Based on a deep literature review of the quaternary structure of hemocyanins, it is unlikely that the hemocyanin oligomers observed in this species deviate from the predicted n × 6-mers characteristic of arthropod hemocyanins. Arthropod hemocyanin oligomeric structures have historically been characterized based on sedimentation coefficients of 5S, 16S, 24S, 34S, and 60S structures corresponding to 1, 6, 12, 24, and 48-mers respectively. These sizes were also confirmed as the only existing oligomeric forms found in native conditions in over 65 arthropod species including penaeid shrimp P. monodon and L. setiferus (Ellerton et al., 1983). The use of x-ray diffraction, small angle x-ray scattering, and cryo-electron microscopy on hemocyanin complexes of many arthropod species also confirmed that the higher order oligomeric hemocyanins form exclusively from the dimerization of 6-mers, and that each 6-mer is composed of a trimer of dimers which, themselves, are unstable when dissociated from the 6-mer (van Holde and Miller, 1982, 1995; Ellerton et al., 1983; Decker et al., 2007; Mičetić et al., 2010). Furthermore, van Holde and Miller (1982) reviewed experiments showing that hexamers can be readily dissociated into monomers following increases in pH or the removal of divalent ions. Following the reversal of these effects, monomers re-associate as hexamers. The consistency of these observations across arthropod taxa led van Holde and Miller (1995) to conclude that arthropod hemocyanin oligomers intermediate of monomers and hexamers do not exist stably in native conditions. To our knowledge, there is only one report which speculates that L. vannamei hemocyanin might function as a dimer or trimer (150 and 230 kDa), but this was only observed in vitro in conditions of low pH and high Ca2+ preceding SDS treatment, and the dimers and trimers further dissociated to monomers upon a second treatment of SDS suggesting an inefficiency in the first SDS exposure (Zhang et al., 2009). While in the present study, neither ultracentrifugation nor crystal structure were examined directly, the assumption was made that the two oligomers observed were hexamers and dodecamers with underestimated sizes due to the in vitro conditions of size determination.
In the present study, SDS-PAGE of hemolymph proteins and purified hemocyanin oligomers revealed only two distinct monomer bands at 70 (HcS) and 74 kDa (HcL), which were also smaller than predicted by translated RNA sequences (75 and 77 kDa). Much like Coomassie, the SDS used in denaturing PAGE acts as a charge shift molecule to further confer charge to the already negatively charged hemocyanin resulting in underestimations of size. The HcS and HcL subunits were also detected by denaturing PAGE in other penaeid shrimp species with minor discrepancies in estimated size. The closely related Atlantic white shrimp, L. setiferus, has 82 and 77 kDa hemocyanin subunits found at a ratio of 1:2.6 in the hemolymph (Brouwer et al., 1978). García-Carreño et al. (2008) report 75 and 82 kDa subunits for L. vannamei while Figueroa-Soto et al. (1997) report 75 and 82 kDa subunits at a 1.5:1 ratio of HcS:HcL in hemolymph. Pan et al. (2008) also examined subunit use in purified hexamers and dodecamers, finding higher abundances of HcL (75 kDa) relative to HcS (73 kDa) in both oligomers. In contrast, in the present study HcS was the most dominant subunit type in hemolymph as detected by denaturing PAGE. Further, the ratio of HcS to HcL varies between purified dodecamers and hexamers. Hexamers have the highest amount of HcS with HcS:HcL ratios approximately 2.05:1 based on PAGE. On the other hand, dodecamers utilize roughly the same amount of HcS and HcL at a ratio of 1.04:1. Total hemolymph HcS:HcL ratios (1.83:1), while statistically distinct, were closer to hexamer ratios, which was expected since hexamers comprise 95–99% of the hemocyanin oligomers within the hemolymph. Heterogeneity of subunit composition among different hemocyanin oligomers is also seen for other crustacean species. Of the six subunit types found in C. sapidus hemocyanin, the presence of type 4 was positively correlated with the proportion of dodecamer while type 5 was negatively correlated with dodecamers (Mangum et al., 1991). Subunits responsible for the inter-hexamer link have been described in higher order hemocyanin oligomers (Stöcker et al., 1988). These linker subunits may be found in variable amounts even within species, correlating with oligomer abundance. Greaves et al. (1992) have shown that C. sapidus sampled from wild populations exhibit up to a fourfold variation in dodecamer/hexamer usage. Apart from the function of linking hexamers to higher order oligomers, such subunit heterogeneity may affect hemocyanin’s ability to deliver oxygen (deFur et al., 1990; Molon et al., 2000; Beltramini et al., 2005).
Hemocyanin subunit diversity and organization within oligomers is more complex than previously described by denaturing PAGE. From transcriptome and genome assembly databases, the use of LC-MS/MS allowed us to identify 10 distinct hemocyanins coded for by individual genes from four unplaced genomic scaffolds including two variants that uniquely matched the transcriptomic database and not the reference genome (Figure 4 and Table 1). Despite the increase in resolvable hemocyanins relative to SDS-PAGE, HcS was still the single most abundant protein identified in each fraction by LC-MS/MS in our study at approximately 48 and 61% NSAF of dodecamer and hexamer fractions respectively. When including two other lower abundance, similarly sized isoforms (64 and 76 kDa), the ratios of small to large (77–81 kDa) subunits were 0.98:1 and 1.64:1 for dodecameric and hexameric hemocyanins respectively. These ratios were comparable to those determined by denaturing PAGE with dodecameric and hexameric HcS:HcL ratios of 1.0:1 and 2.1:1. Regardless of either PAGE or LC-MS/MS method, the dodecamer fraction contained a larger relative amount of HcL compared to the hexamer fraction representing a greater heterogeneity of subunit usage within the HcL cluster. More noticeably, XP_027232115.1, which did not cluster with HcS, HcL, or HcB, increased from 0.27 ± 0.05% NSAF in hexamers to 12.10 ± 0.51% NSAF (>40 fold%NSAF) in dodecamers suggesting that it may play a more pronounced role in dodecamers (Figure 4A). Patterns of higher subunit heterogeneity with higher order hemocyanin oligomers have also been documented in many other species and attributed to the greater number of 6-mer interacting subunits (van Holde and Miller, 1982). It has also been suggested that dodecameric hemocyanins have agglutination properties not observed in hexamers which may contribute to immune function (Pan et al., 2008). However, the disproportionate abundances of certain isoforms in dodecamers must be examined more specifically.
The present study provided the first protein-level evidence of a β-type hemocyanin in penaeid shrimp as demonstrated by the identification of six exclusive, unique peptides matching HcB transcript assembly at < 1% FDR (Table 3). The ancestral protein of the α and γ type hemocyanin subunits diverged from the original β-type hemocyanin subunit type approximately 520 MYA (Markl et al., 1986; Marxen et al., 2013). The α and γ types commonly found in decapods then split from each other 400 MYA. However, hemocyanin evolution throughout arthropod lineages are complicated by gene duplications or loses. The only report to our knowledge of a β-type hemocyanin in decapods was for the bamboo shrimp Atyopsis moluccensis, but the β-type could not be identified at the protein level by mass spectrometry in the same study (Marxen et al., 2013). Meanwhile, there were no reports of the β-type hemocyanin in the Penaeidae family of decapods. While we report protein evidence for HcB in the hemolymph of the penaeid shrimp L. vannamei, the subunit contributed only 0.16 ± 0.07% (SEM; n = 5) NSAF of the hemocyanin subunits with full-length references within the hexamer fraction and 0.02 ± 0.02% (SEM; n = 5) NSAF in dodecamer fraction suggesting a reduced or specialized function based on relative abundance. Tommerdahl et al. (2015) determined hemocyanin concentrations in aquacultured L. vannamei (same stock source as present study) hemolymph to be 0.103 ± 0.0023 g ml–1 which would equate to approximately 144 μg ml–1 or 1.92 μM of HcB in hemolymph assuming a simplified common subunit weight of 75 kDa and primarily hexameric hemocyanin. Further studies are required to determine whether this relatively small amount of HcB is functionally relevant, but the protein-level presence of HcB in penaeid shrimp has implications for arthropod evolution.
For the purposes of future functional analyses of hemocyanins, it is no longer informative to refer to two classes of Hc in L. vannamei (small and large) when there are at least 10 distinct isoforms coded by separate genes with potentially different functions (Table 2). Furthermore, the transcriptome of shrimp originating from the same stock source as the present study revealed additional variants similar to those in the reference genome (Kehai No. 1 variety, Hainan, China; Zhang et al., 2019), but not identical. Since our stock population was a captive population, it is likely that the conditions of breeding and domestication over many generations has accumulated many divergent polymorphisms. Following the assembly of a reference genome, Zhang et al. (2019) re-sequenced genomes from wild populations in Mexico as well as aquacultured broodstocks from Ecuador, China, United States, and Thailand ranging from 4 to over 20 generations of breeding. Re-sequenced L. vannamei from these different sources exhibited 19 SNPs kb–1 on average with over 200,000 non-synonymous SNPs in coding regions, and selectively divergent genes related to biosynthesis and immune function (Zhang et al., 2019). The transcriptome of shrimp used in the present study suggest that HcL1-3 differ from the reference genome by as much as 43, 13, and 28 amino acids respectively, and several peptides in HcL1 and HcL3 not found in the reference genome were identified (Table 1).
In light of the heterogeneity of hemocyanin usage characterized in different oligomers in the present study, future experiments should not only examine how these isoforms are regulated in different conditions, but how each isoform functions within the oligomeric structures. Mass spectrometry labeling techniques such as isobaric tags for relative and absolute quantification (iTRAQ) or the use of parallel reaction monitoring with synthesized internal standards will allow for quantitative comparisons of hemocyanin isoforms such as HcB relative to all others in different treatment conditions applicable to hypoxia tolerance, metabolism, and disease resistance. Experiments manipulating subunit heterogeneity and relative abundance within oligomers can also be carried out to prove the functions of different isoforms. Finally, the protein-level examination of hemocyanins evolved under many generations of breeding can provide insights into the relationships between polymorphisms, protein abundance, physiology, and adaptation. Since hemocyanins serve many functions from O2 delivery to carbohydrate storage and innate immunity, the characterization of specific protein isoforms within native oligomers offers using LC-MS/MS is a strong tool building upon information from both genomic and transcriptomic data.
Data Availability Statement
The mass spectrometry proteomics datasets generated for this study can be found in the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) (https://www.ebi.ac.uk/pride/archive/) partner repository with the dataset identifier PXD014575.
Ethics Statement
This study is exempt from ethical approval procedures since the study animal (Pacific whiteleg shrimp) is neither a vertebrate nor considered a higher invertebrate (e.g., cephalopod).
Author Contributions
All authors participated in the design of the study, and read and approved the final manuscript. JW fractionated and characterized the hemocyanin oligomers and subunits. MJ and JW performed the proteomic analysis. JW prepared the initial draft of the manuscript with the help of KB.
Funding
This project was funded by the National Science Foundation (IOS-1147008 to Louis Elwood Burnett and KB) and Sigma Xi Grants In Aid of Research (G20141015697244 to JW for the purchase of an HPLC column).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Jillian Johnson for the translation of RNA-sequencing contigs used in our peptide search database as well as Dr. Philip Sobolesky and Alison Bland for their support in the laboratory. This is Grice Marine Lab contribution number 537.
Abbreviations
emPAI, exponentially modified protein abundance index; FDR, false discovery rate; HcB, hemocyanin β-type subunit; HcL, hemocyanin large subunit; HcS, hemocyanin small subunit; iTRAQ, isobaric tag for relative and absolute quantitation; LC-MS/MS, liquid chromatography tandem mass spectrometry; MYA, million years ago; NSAF, normalized spectral abundance factor; PAGE, polyacrylamide gel electrophoresis SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; SE-HPLC, size exclusion high performance liquid chromatography; TIC, total ion current; TOF, time of flight.
References
Beltramini, M., Colangelo, N., Giomi, F., Bubacco, L., Di Muro, P., Hellmann, N., et al. (2005). Quaternary structure and functional properties of Penaeus monodon hemocyanin. FEBS J. 272, 2060–2075. doi: 10.1111/j.1742-4658.2005.04634.x
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Brouwer, M., Bonaventura, C., and Bonaventura, J. (1978). Analysis of the effect of three different allosteric ligands on oxygen binding by hemocyanin of the shrimp, Penaeus setiferus. Biochemistry 17, 2148–2154. doi: 10.1021/bi00604a019
Burmester, T. (2001). Molecular evolution of the arthropod hemocyanin superfamily. Mol. Biol. Evol. 18, 184–195. doi: 10.1093/oxfordjournals.molbev.a003792
Burmester, T. (2002). Origin and evolution of arthropod hemocyanins and related proteins. J. Comp. Physiol. B 172, 95–107. doi: 10.1007/s00360-001-0247-7
Cheng, W., Liu, C. H., Yan, D. F., and Chen, J. C. (2002). Hemolymph oxyhemocyanin, protein, osmolality and electrolyte levels of whiteleg shrimp Litopenaeus vannamei in relation to size and molt stage. Aquaculture 211, 325–339. doi: 10.1016/S0044-8486(01)00768-2
Chongsatja, P., Bourchookarn, A., Lo, C. F., Thongboonkerd, V., and Krittanai, C. (2007). Proteomic analysis of differentially expressed proteins in Penaeus vannamei hemocytes upon Taura syndrome virus infection. Proteomics 7, 3592–3601. doi: 10.1002/pmic.200700281
Dainese, E., Di Muro, P., Beltramini, M., Salvato, B., and Decker, H. (1998). Subunits composition and allosteric control in Carcinus aestuarii hemocyanin. Eur. J. Biochem. 256, 350–358. doi: 10.1046/j.1432-1327.1998.2560350.x
Decker, H., Hellman, N., Jaenicke, E., Lieb, B., Meissner, U., and Markl, J. (2007). Minireview: recent progress in hemocyanin research. Integr. Comp. Biol. 47, 631–644. doi: 10.1093/icb/icm063
deFur, P. L., Mangum, C. P., and Reese, J. E. (1990). Respiratory responses of the blue crab Callinectes sapidus to long-term hypoxia. Biol. Bull. 178, 46–54. doi: 10.2307/1541536
Destoumieux-Garzón, D., Saulnier, D., Garnier, J., Jouffrey, C., Bulet, P., and Bachère, E. (2001). Crustacean immunity. antifungal peptides are generated from the C terminus of shrimp hemocyanin in response to microbial challenge. J. Biol. Chem. 276, 47070–47077. doi: 10.1074/jbc.M103817200
Diaz, R. J., and Rosenberg, R. (2008). Spreading dead zones and consequences for marine ecosystems. Science 321, 926–929. doi: 10.1126/science.1156401
Ellerton, H. D., Ellerton, N. F., and Robinson, H. A. (1983). “Hemocyanins – A current perspective,” in Prog. Biophys. Molec. Biol, eds D. Noble, and T. L. Blundell, (Oxford: Pergamon Press) 143–247.
Figueroa-Soto, C. G., de la Barca, A. M. C., Vazquez-Moreno, L., Higuera-Ciapara, I., and Yepiz-Plascencia, G. (1997). Purification of hemocyanin from white shrimp (penaeus vannamei boone) by immobilized metal affinity chromatography. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 117, 203–208. doi: 10.1016/S0305-0491(96)00321-5
García-Carreño, F. L., Cota, K., and Navarrete del Toro, M. A. (2008). Phenoloxidase activity of hemocyanin in whiteleg shrimp Penaeus vannamei: conversion, characterization of catalytic properties, and role in postmortem melanosis. J. Agric. Food. Chem. 56, 6454–6459. doi: 10.1021/jf800839x
Greaves, J., Rainer, J. S., and Mangum, C. P. (1992). Size-exclusion high performance liquid chromatography of the dodecameric and hexameric forms of hemocyanin from Callinectes sapidus. Mar. Biol. 113, 33–36. doi: 10.1007/BF00367635
Hagerman, L. (1986). Haemocyanin concentration in the shrimp crangon crangon (L.) after exposure to moderate hypoxia. Comp. Biochem. Physiol. A Physiol. 85, 721–724. doi: 10.1016/0300-9629(86)90283-5
Ishihama, Y., Oda, Y., Tabata, T., Sato, T., Nagasu, T., Rappsilber, J., et al. (2005). Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272. doi: 10.1074/mcp.M500061-MCP200
Johnson, J. G., Burnett, L. E., and Burnett, K. G. (2016). Uncovering hemocyanin subunit heterogenetiy in penaeid shrimp using RNA-Seq. Integr. Comp. Biol. 56, 1080–1091. doi: 10.1093/icb/icw088
Johnson, J. G., Paul, M. R., Kniffin, C. D., Anderson, P. E., Burnett, L. E., and Burnett, K. G. (2015). High CO2 alters the hypoxia response of the Pacific whiteleg shrimp (Litopenaeus vannamei) transcriptome including known and novel hemocyanin isoforms. Physiol. Genom. 47, 548–558. doi: 10.1152/physiolgenomics.00031.2015
Kölsch, A., Hörnemann, J., Wengenroth, C., and Hellmann, N. (2013). Differential regulation of hexameric and dodecameric hemocyanin from a. leptodactylus. BBA Proteins Proteomics 1834, 1853–1859. doi: 10.1016/j.bbapap.2013.02.037
Lee, S. Y., Lee, B. L., and Söderhäll, K. (2003). Processing of an antibacterial peptide from hemocyanin of the freshwater crayfish Pacifastacus leniusculus. J. Biol. Chem. 278, 7927–7933. doi: 10.1074/jbc.M209239200
Linzen, B., Soeter, N. M., Riggs, A. F., Schnieder, H. J., and Schartau, W. (1985). The structure of arthropod hemocyanins. Science 229, 519–524. doi: 10.1126/science.4023698
Lu, X., Lu, H., Guo, L., Zhang, Z., Zhao, X., Zhong, M., et al. (2015). Cloning and characterization of a novel hemocyanin variant LvHMCV4 from shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 46, 398–405. doi: 10.1016/j.fsi.2015.06.022
Magnus, K. A., Ton-That, H., and Carpenter, J. E. (1994). Recent structural work on the oxygen transport protein hemocyanin. Chem. Rev. 94, 727–735. doi: 10.1021/cr00027a009
Mangum, C. P., Greaves, J., and Rainer, J. S. (1991). Oligomer composition and oxygen binding of the hemocyanin of the blue crab Callinectes sapidus. Biol. Bull. 181, 453–458. doi: 10.2307/1542365
Markl, J. (1986). Evolution and function of structurally diverse subunits in the respiratory protein hemocyanin from arthropods. Biol. Bull. 171, 90–115. doi: 10.2307/1541909
Markl, J., and Decker, H. (1992). “Molecular structure of the arthropod hemocyanins,” in Blood and Tissue Oxygen Carriers, ed Mangum, C. P. (Berlin: Springer), 325–376. doi: 10.1007/978-3-642-76418-9-12
Markl, J., Stöcker, W., Runzler, R., and Precht, E. (1986). “Immunological correspondences between the hemocyanin subunits of 86 arthropods: evolution of a multigene protein family,” in Invertebrate Oxygen Carriers, ed Linzen, B. (Berlin: Springer), 281–292.
Marxen, J., Pick, C., Kwiatkowski, M., and Burmester, T. (2013). Molecular characterization and evolution of haemocyanin from the two freshwater shrimps Caridina multidentata (Stimpson, 1860) and atyopsis moluccensis (De Haan, 1849). J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 183, 613–624. doi: 10.1007/s00360-013-0740-9
Mičetić, I., Losasso, C., Di Muro, P., Tognon, G., Benedetti, P., and Beltramini, M. (2010). Solution structures of 2 x 6-meric and 4 x 6-meric hemocyanins of crustaceans Carcinus aestruarii, squilla mantis, and upogebia pusilla. J. Struct. Biol. 171, 1–10. doi: 10.1016/j.jsb.2010.03.012
Molon, A., Di Muro, P., Bubacco, L., Vasilyev, V., Salvato, B., Beltramini, M., et al. (2000). Molecular heterogeneity of the hemocyanin isolated from the king crab paralithodes camtschaticae. Eur. J. Biochem. 267, 7046–7057. doi: 10.1046/j.1432-1327.2000.01803.x
Neuteboom, B., Jekel, P. A., and Beintema, J. J. (1992). Primary structure of hemocyanin subunit c from Panulirus interruptus. Eur. J. Biochem. 206, 243–249. doi: 10.1111/j.1432-1033.1992.tb16922.x
Nickerson, K. W., and Van Holde, K. E. (1971). A comparison of molluscan and arthropod hemocyanin-I. circular dichroism and absorption spectra. Comp. Biochem. Physiol. B 39, 855–872. doi: 10.1016/0305-0491(71)90109-X
Pan, J., Zhang, Y., Wang, S., and Peng, X. (2008). Dodecamer is required for agglutination of Litopenaeus vannamei hemocyanin with bacterial cells and red blood cells. Mar. Biotechnol. 10, 645–652. doi: 10.1007/s10126-008-9115-8
Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450. doi: 10.1093/nar/gky1106
Rabotyagov, S. S., Kling, C. L., Gassman, P. W., Rabalais, N. N., and Turner, R. E. (2014). The economics of dead zones: causes, impacts, policy challenges, and a model of the gulf of mexico hypoxic zone. Rev. Environ. Econ. Policy 8, 58–79. doi: 10.1093/reep/ret024
Rathburn, C. K., Sharp, N. J., Ryan, J. C., Nealy, M., Cook, M., Chapman, R. W., et al. (2013). Transcriptomic responses of juvenile Pacific whiteleg shrimp, Litopenaeus vannamei, to hypoxia and hypercapnic hypoxia. Physiol. Genomics 45, 794–807. doi: 10.1152/physiolgenomics.00043.2013
Schwanhausser, B., Busse, D., Li, N., Dittmar, G., Schuchhardt, J., Wolf, J., et al. (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. doi: 10.1038/nature10098
Stöcker, W., Raeder, U., Bijholt, M. M. C., Wichertjes, T., Van Bruggen, E. F. J., and Markl, J. (1988). The quaternary structure of four crustacean two-hexameric hemocyanins: immunocorrelation, stoichiometry, reassembly and topology of individual subunits. J. Comp. Physiol. B 158, 271–289. doi: 10.1007/BF00695326
Terwilliger, N. (1998). Functional adaptations of oxygen-transport proteins. J. Exp. Biol. 201, 1085–1098.
Terwilliger, N. (2007). Hemocyanins and the immune response: defense against the dark arts. Integr. Comp. Biol. 47, 662–665. doi: 10.1093/icb/icm039
Tommerdahl, A. P., Burnett, K. G., and Burnett, L. E. (2015). Respiratory properties of hemocyanin from wild and aquacultured penaeid shrimp and the effects of chronic exposure to hypoxia. Biol. Bull. 228, 242–252. doi: 10.1086/BBLv228n3p242
van Holde, K. E., and Miller, K. I. (1982). Haemocyanins. Q. Rev. Biophys. 15, 1–129. doi: 10.1017/s0033583500002705
van Holde, K. E., Miller, K. I. (1995). “Hemocyanins,” in Advances in Protein Chemistry, ed C. B. Anfinsen, F. M. Richards, J. T. Edsall, and D. S. Eisenberg, (San Diego, CA: Academic Press), 1–81.
Wang, K. Z., Wen, Y. L., Ye, Z. C., Wu, H. G., and Pan, J. Y. (2015). Hemocyanin-derived phenoloxidase activity is dependent on dodecameric structure in shrimp Litopenaeus vannamei. Arch. Biol. Sci. 67, 19–23. doi: 10.2298/ABS141103001W
Xu, J., Ruan, L., Li, Z., Yu, X., Li, S., Shi, H., et al. (2015). Characterization of four hemocyanin isoforms in Litopenaeus vannamei. Acta. Oceanol. Sin 34, 36–44. doi: 10.1007/s13131-015-0588-9
Zhang, X., Yuan, J., Sun, Y., Li, S., Gao, Y., Yu, Y., et al. (2019). Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 10:356. doi: 10.1038/s41467-018-08197-4
Keywords: hemocyanin, crustacean, subunit heterogeneity, mass spectrometry, proteomics
Citation: Wang J, Janech MG and Burnett KG (2019) Protein-Level Evidence of Novel β-Type Hemocyanin and Heterogeneous Subunit Usage in the Pacific Whiteleg Shrimp, Litopenaeus vannamei. Front. Mar. Sci. 6:687. doi: 10.3389/fmars.2019.00687
Received: 13 May 2019; Accepted: 25 October 2019;
Published: 07 November 2019.
Edited by:
Francesca Carella, University of Naples Federico II, ItalyReviewed by:
Fan Wang, Shantou University, ChinaRogerio R. Sotelo-Mundo, Centro de Investigación en Alimentación y Desarrollo (CIAD), Mexico
Copyright © 2019 Wang, Janech and Burnett. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jason Wang, amFzb253YW5nMTAzQGdtYWlsLmNvbQ==
†Present address: Jason Wang, Department of Biological Sciences, University of Southern California, Los Angeles, CA, United States
 Jason Wang
Jason Wang Michael Gregory Janech
Michael Gregory Janech Karen Gray Burnett
Karen Gray Burnett