- 1MARUM – Center for Marine Environmental Sciences, University of Bremen, Bremen, Germany
- 2Department of Physics and Earth Science, Jacobs University Bremen, Bremen, Germany
- 3Department of Life Science and Chemistry, Jacobs University Bremen, Bremen, Germany
Future mining of polymetallic nodules in the Clarion Clipperton Zone (Northeastern Pacific) is expected to affect all benthic ecosystems. The diversity, distribution, and environmental functions of microorganisms inhabiting abyssal sediments are barely understood. To understand consequences of deep-sea mining, experimental in vitro systems needs to be established to test hypotheses on the environmental impact of mining. For this, 40 bacterial strains, belonging to proteobacteria, actinobacteria and firmicutes were isolated from deep-sea sediments and nodules sampled at depths of ≥ 4000 m. Phenotypic characterization revealed a strong inter-species and moderate intra-species variability. Determination of metal minimum inhibitory concentrations indicated the presence of acute manganese-resistant bacteria such as Rhodococcus erythropolis (228.9 mM), Loktanella cinnabarina (57.2 mM), and Dietzia maris (14.3 mM) that might be suitable systems for testing the effects of release of microbes from nodules and their interactions with sediment particles in plumes generated during mining. Comparative genomic analysis indicated the presence of manganese efflux systems relevant for future transcriptomics or proteomics approaches with environmental samples and might serve in paving the way to develop model systems including representative organisms which are currently not cultivable. Monitoring deep-sea mining activity at abyssal depth is a challenge that has to be tackled. We proposed the use of API strips as a fast on-board methodology for bacterial monitoring as an indicator for sediment plume dispersions within the water column.
Introduction
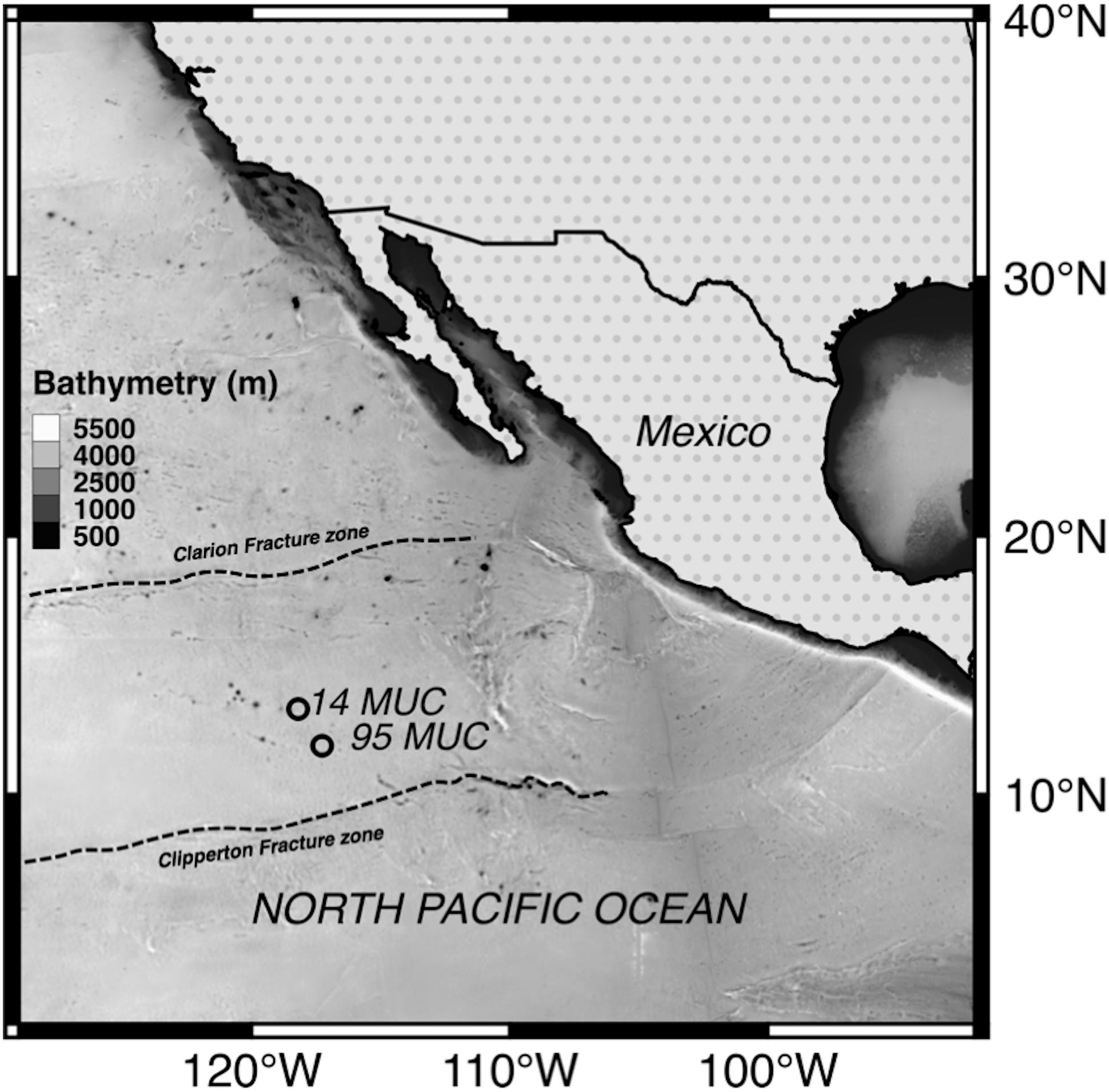
Deep-sea environments are considered the most remote, broad (95% of ocean surface) and least understood ecosystems on Earth (Jørgensen and Boetius, 2007; Smith et al., 2008). Some deep-sea areas, however, contain considerably high amounts of mineral resources, which have recently received increasing attention from governments and private entities. Consequently, the Clarion Clipperton Zone (CCZ; NE equatorial Pacific; Figure 1), with an extent of 4.5 million km2, has been the focus for polymetallic nodule exploration programs.
Polymetallic nodules are marine encrustations rich in precious metals such as manganese (Mn), nickel (Ni), copper (Cu), and cobalt (Co), as well as rare earth elements of both ecological and economical relevance (Hein and Koschinsky, 2013; Fritz, 2016). Lying on the surface of abyssal plains at around 4000 m depth, their genesis is geologically slow and results from diagenetic and hydrogenetic processes that take place over a million years (Graham et al., 2004). Environmental conditions at the CCZ seafloor are characterized by cold temperature (2°C), clay siliceous ooze sediment with an oxygen penetration depth of around 2–3 m, a low sedimentation rate of 0.35 cm kyr–1, and very low organic impact (Mewes et al., 2014).
Nodule harvesting is expected to not only directly impact the mined surface, where the top sediment layer will be removed, but also a much wider area, as a result of the tailing of mined products (sediment plume and nodule debris) into the benthic boundary layer (Oebius et al., 2001; Aleynik et al., 2017; Gillard et al., 2019). This sediment plume, which is rich in manganese-oxide, may lead to potentially high sorption of trace metals in the water column (Koschinsky et al., 2003). To date, nothing is known about the role and interactions of microbial communities within the plume particles and the redeposited sediments that may influence the oxidation state, mobility and flux of metals. Heavy metals are essential elements for the maintenance of cellular functions in microorganisms. However, under elevated concentrations, those elements result in toxicity that is metal- and organism-dependent (Lemire et al., 2013). As such, the anticipated impact of mining activity is very difficult to estimate but will likely affect the entire regional deep-sea ecosystem. The extent to which this ecosystem will be affected is unknown and unpredictable (Ramirez-Llodra et al., 2011).
Although microbial diversity and ecology in most abyssal sediments have barely been investigated (Danovaro et al., 2014), the potential risks that these fragile ecosystems may encounter have forced the scientific community to increase efforts in describing the prokaryotic diversity in the CCZ area over the last few years (Parkes et al., 2014). Culture-independent approaches and the use of next generation sequencing technologies have revealed the high microbial diversity and complexity of bacterial assemblages inhabiting deep-sea sediments (Wu et al., 2013; Corinaldesi, 2015; Shulse et al., 2016; Lindh et al., 2017).
In accompanying the efforts to explore the deep-sea with microbiological approaches and to broaden the understanding of gene activity for heavy metal resistance, a cultivation-based study was conducted herein.
It is hypothesized that isolation and characterization of heavy-metal-resistant and -sensitive bacterial strains from abyssal sediments of the CCZ may help to establish experimental in vitro system to study the dispersal of microorganisms during relocation of material in sediment plumes in dependence of diverse biotic and abiotic environmental factors. Such an in vitro system may complement omics-based microbial analysis, as well as mineralogical and hydrographical analyses, which would therefore shed light on ecosystem alterations caused by mining activities. Our current research aims were to: (1) isolate and characterize of cultivable deep-sea microorganisms, (2) investigate heavy-metal-resistance and sensitivity of these microorganisms, (3) obtain any first insights on potential genes that confer manganese resistance in bacterial isolates.
Materials and Methods
Study Area and Sampling
Abyssal sediments and polymetallic nodules were collected during the RV SONNE cruise SO-240, within the German license area for the exploration of polymetallic nodules in the CCZ (Figure 1). Samples were obtained at two different sites using a multicorer (MUC; samples 14 and 95; Supplementary Table S1 for details). The top water layer and 10 cm of sediment from one multicore (14 MUC) as well as surface nodules (95 MUC) were aseptically sampled, sliced in 1 cm intervals and stored at 4°C in the dark until further analysis in the laboratory.
Bacterial Extraction and Isolation
Sediments from every core layer were screened for fast growing aerobic bacteria. Aliquots of roughly 300 μg of sediment were resuspended in 1 mL of autoclaved North Sea water. Nodule samples were carefully rinsed several times with the same North Sea water prior to the extraction procedure. Bacterial cells were separated from sediment particles or nodules by three 10-min vortexing intervals (Vortex 2 Genie, Scientific Industries, Bohemia, United States). Samples were centrifuged (Eppendorf 5418R; Eppendorf, Hamburg, Germany) at 750 × g for 10 min at 4°C (Dos Santos Furtado and Casper, 2000). After centrifugation, the supernatant was used for bacterial isolation.
Bacterial strain isolation was conducted using a serial dilution (up to 10–5) plating on a marine broth (MB) agar medium (Sonnenschein et al., 2011). Samples were incubated in the dark at 18°C for 7 days to expedite microbial growth. Colonies with unique morphological features were re-streaked on the MB agar medium. Isolates were maintained on agar plates with regular re-streaking. A suspension of bacterial cells in 30% (v/v) glycerol was prepared to store the bacterial strains for long term at −80°C.
Taxonomic Characterization
The polymerase chain reaction (PCR) was conducted with a re-suspended single bacterial colony as a template in 100 μL of sterile water incubated at 95°C for 10 min prior to reaction. The corresponding 16S rRNA gene was amplified by PCR using the universal primers (5′- AGA GTT TGA TCC TGG CTC AG-3′ and 5′- TAC GGY TAC CTT GTT ACG ACT T-3′; Alfaro-Espinoza and Ullrich, 2014). The PCR reaction mix, with a final volume of 50 μL, consisted of 42.5 μL of target cell suspension (10–100 ng of DNA), 0.5 μL of each primer (50 pmol μL–1), 1 μL dNTP (2 μmol), 5 μL of 10X DreamTaq Green buffer, and 0.5 μL of DreamTaq DNA polymerase (5 U μL–1). All reagents were purchased from Thermo Fisher Scientific (MA, United States). The thermal cycling gradient program was as follows: initial denaturation (5 min at 94°C), subsequent denaturation (32 cycles of 15 s at 94°C), annealing (30 s at 55°C), extension (60 s at 72°C), and a final extension (72°C for 3 min). A negative control containing no DNA extract was conducted to account for any contamination.
The amplification of the 16S rRNA gene was confirmed by agarose [1% (w/v)] gel electrophoresis (120 V, 30 min) and ethidium bromide staining (0.1%). PCR products were purified using a GeneJet PCR Purification Kit (Thermo Fisher Scientific, MA, United States). All 16S amplicons were sequenced by Eurofins Genomics1. All nucleotides sequences were submitted to the NCBI 16S Microbial Database using the Basic Local Alignment Search Tool (BLAST)2 to determine whether they aligned with any closely related organisms. Sequence similarity was set to a threshold value of minimum 99% for a positive match.
Morphological, Biochemical, and Enzymatic Characterization
Colony morphology, bacteria motility, and gram staining were examined using a phase contrast microscope (Axiostar plus, Zeiss). Biochemical characteristics and enzyme activity were determined using the API 20NE and API ZYM kits (BioMérieux, Marcy-l’Étoile, France). The protocol followed the manufacturer’s instructions with the exception of the culture being suspended in autoclaved North Sea water (MacDonell et al., 1982; Kim et al., 2007). The incubation was done at 18°C for 24 h.
Based on the corresponding phenotypical characteristics of the selected bacterial strains, single tests or combinations of tests providing a unique identification were performed in R Core Team (2016), thus providing the most effective identification path for each strain. Tests with results not in accordance with previously reported phenotypes were not considered. Cell or colony morphology was omitted from the analysis.
Minimum Inhibitory Concentration (MIC) of Heavy Metals
The MIC of heavy metal ions for bacteria isolated from deep-sea sediments or nodule surfaces was conducted in triplicate with a twofold dilution assay in 96-well plates (Stahl et al., 2015). The heavy metals tested were: cadmium acetate [Cd(CH3CO2)2⋅2H2O], cobalt chloride (CoCl2⋅H2O), cupric sulfate (CuSO4⋅5H2O), zinc sulfate (ZnSO4⋅7H2O) (Sigma-Aldrich, City, Germany), manganese (II)-sulfate (MnSO4⋅H2O) (Carl Roth, Karlsruhe, Germany) and nickel chloride (NiCl2⋅6H2O) (AppliChem). The concentration of the metal stock solutions (1M) were confirmed by a Cyros Vision inductively coupled plasma optical emission spectrometry (ICP-OES) (SPECTRO Analytical Instruments Inc., Kleve, Germany). Cells were harvested in their exponential growth phase (OD600 = 0.5–1) and adjusted to ∼2.85 × 106 cells mL–1 (OD600 = 0.001). The highest concentration of metal salt used was 0.5 M. Cells were incubated at 18°C for 96 h. The MIC was defined as the lowest concentration of metal salt, which inhibited visible bacterial growth.
Genomic Analysis of Manganese Resistance Related Genes
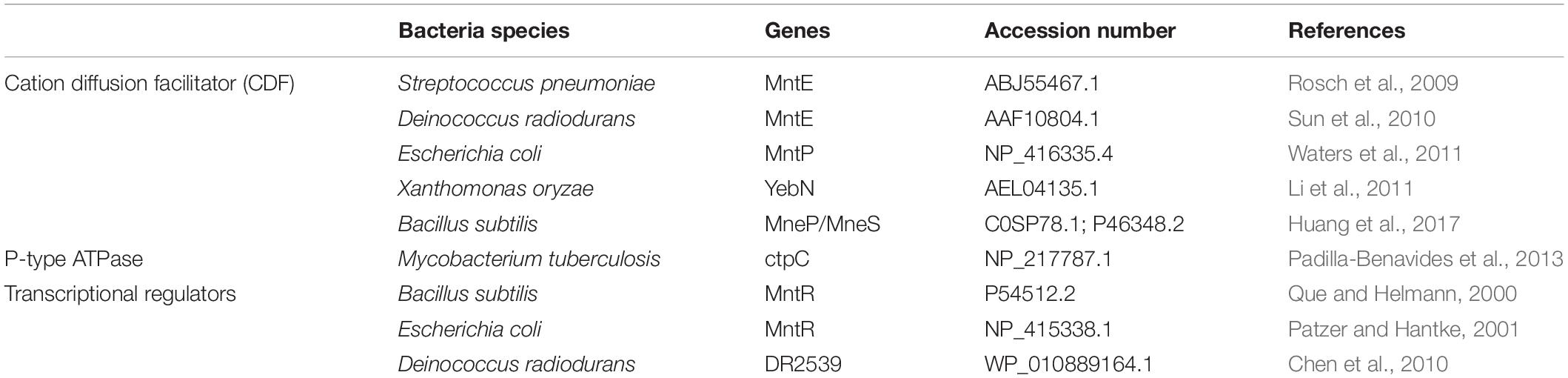
To our knowledge, only a few studies have investigated the differential gene expression mechanisms for Mn (II) efflux systems in several bacterial species over the last decade (Table 1). From the isolated bacterial strains, those showing the highest resistance to Mn as determined by MIC were selected for further analysis.
Based on the availability of genome sequences from the literature, the search tool BlastP (protein-protein) was used to test for the presence of highly similar protein sequences from the tested bacterial species. Protein sequence homology was determined based on the following criteria: minimum sequence coverage of 90%, bit score > 50 and sequence identity > 25% (Pearson, 2014). Protein functional inference was based on the overall similarity, conserved active site domains and residues using the InterPro (Finn et al., 2017) and UniProtKB (Wu et al., 2006) databases. For every protein match, the length of identical DNA sequences was determined using TBlastn (protein-translated nucleotide).
Results
Bacterial Extraction and Isolation
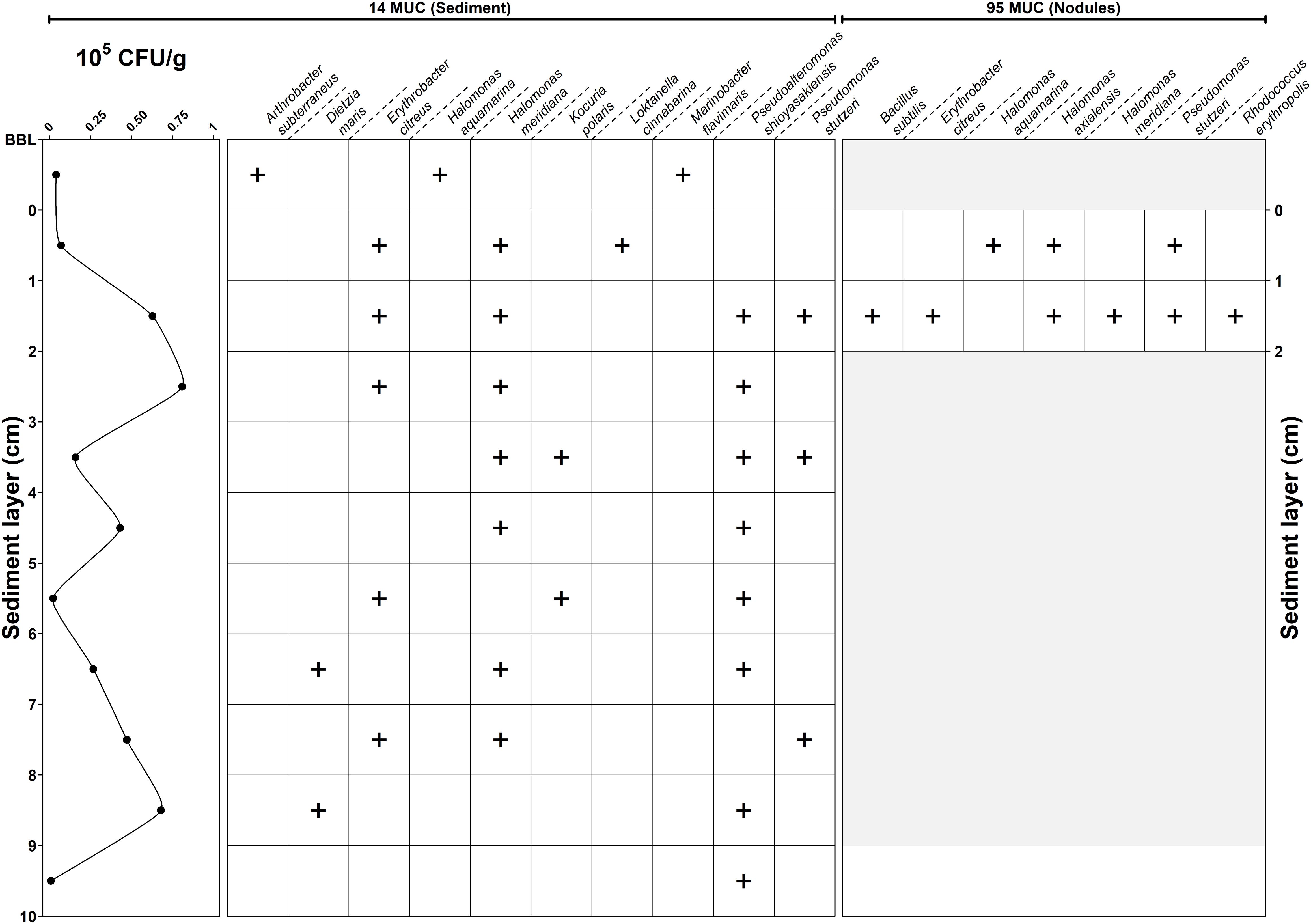
The overall abundance of cultivable bacteria throughout the first 10 cm of sediment core 14 MUC delineates a multi-modal distribution pattern (Figure 2). Three peaks of elevated concentrations were observed at 2–3 cm (0.81 × 105 CFU g–1), 4–5 cm (0.43 × 105 CFU g–1), and 8–9 cm (0.68 × 105 CFU g–1) depth.
In total, 40 bacterial strains were isolated based on their distinguishing morphological characteristics. The amount of isolated strains per depth varied between one (10 cm depth) and six (nodule surface) strains. Species location revealed different trends of spatial distribution (Figure 2). The bottom boundary layer (BBL) representing the water in contact with the sediment core contained bacteria that were not present in the sediment (e.g., Arthrobacter subterraneus, Marinobacter flavimaris). Inside the sediment core, bacterial distribution occurred in three patterns: (1) widely spread (e.g., Pseudoalteromonas shioyasakiensis), (2) confined at certain depth (e.g., Dietzia maris, Kocuria polaris), or (3) present at distinctive depth layers (e.g., Erythrobacter citreus, Halomonas meridiana).
Taxonomic Characterization
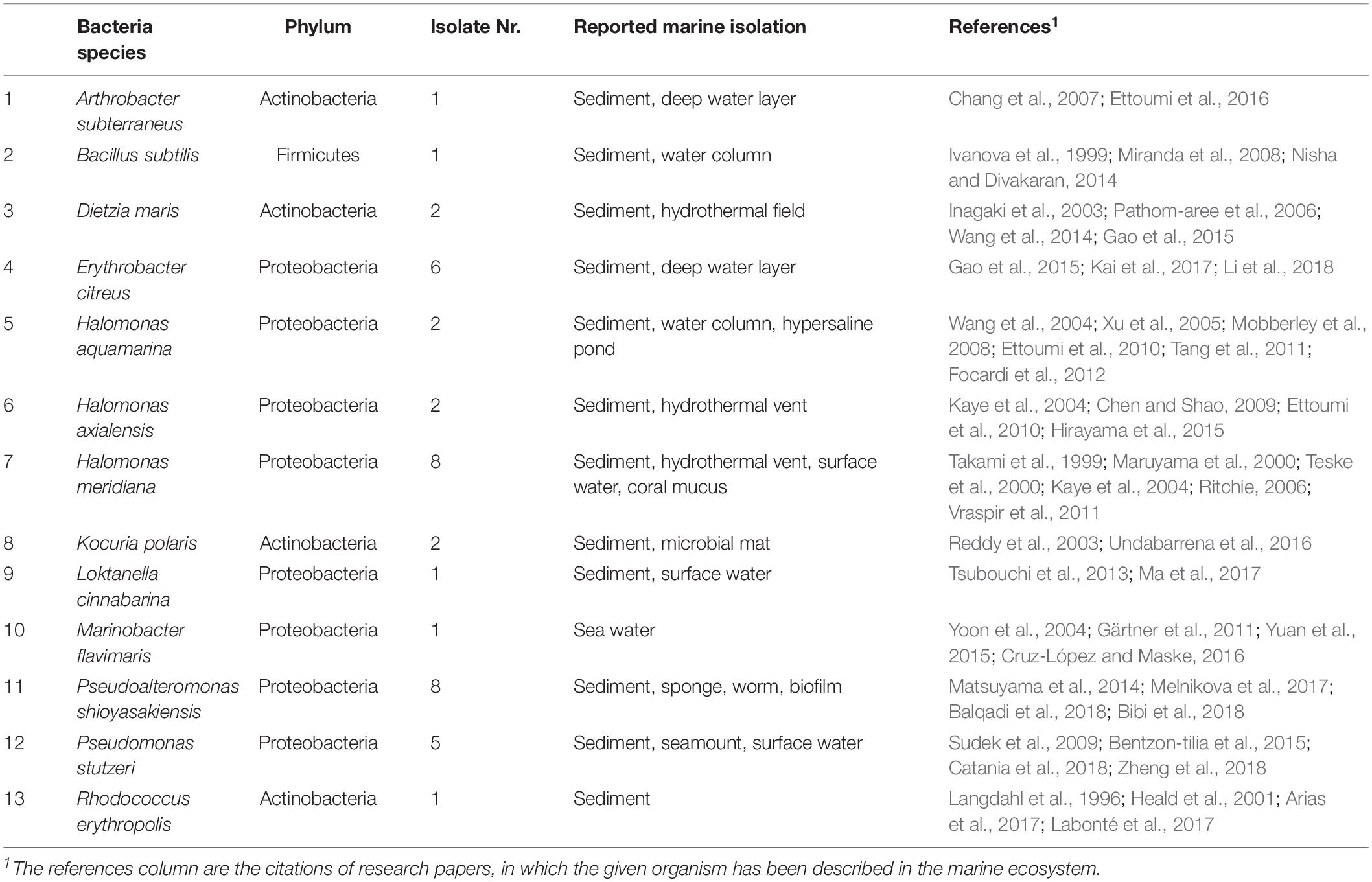
BLASTN analysis results (Supplementary Table S2) based on partial 16S rRNA gene sequences indicated that the 40 bacterial isolates belonged to three distinctive phylogenetical groups (83% proteobacteria, 15% actinobacteria, and 3% firmicutes) and could be classified into 13 species (listed in Table 2). Sequences with similarities of > 99% to sequences from taxonomically closely related species were deposited in GenBank3 under the accession numbers MK254646-MK254685, demonstrating that all bacterial isolates from this study resembled species previously isolated from the marine environment and the marine benthos; the sole exception is M. flavimaris, which was previously only reported for bulk seawater.
Phenotypic Characterization
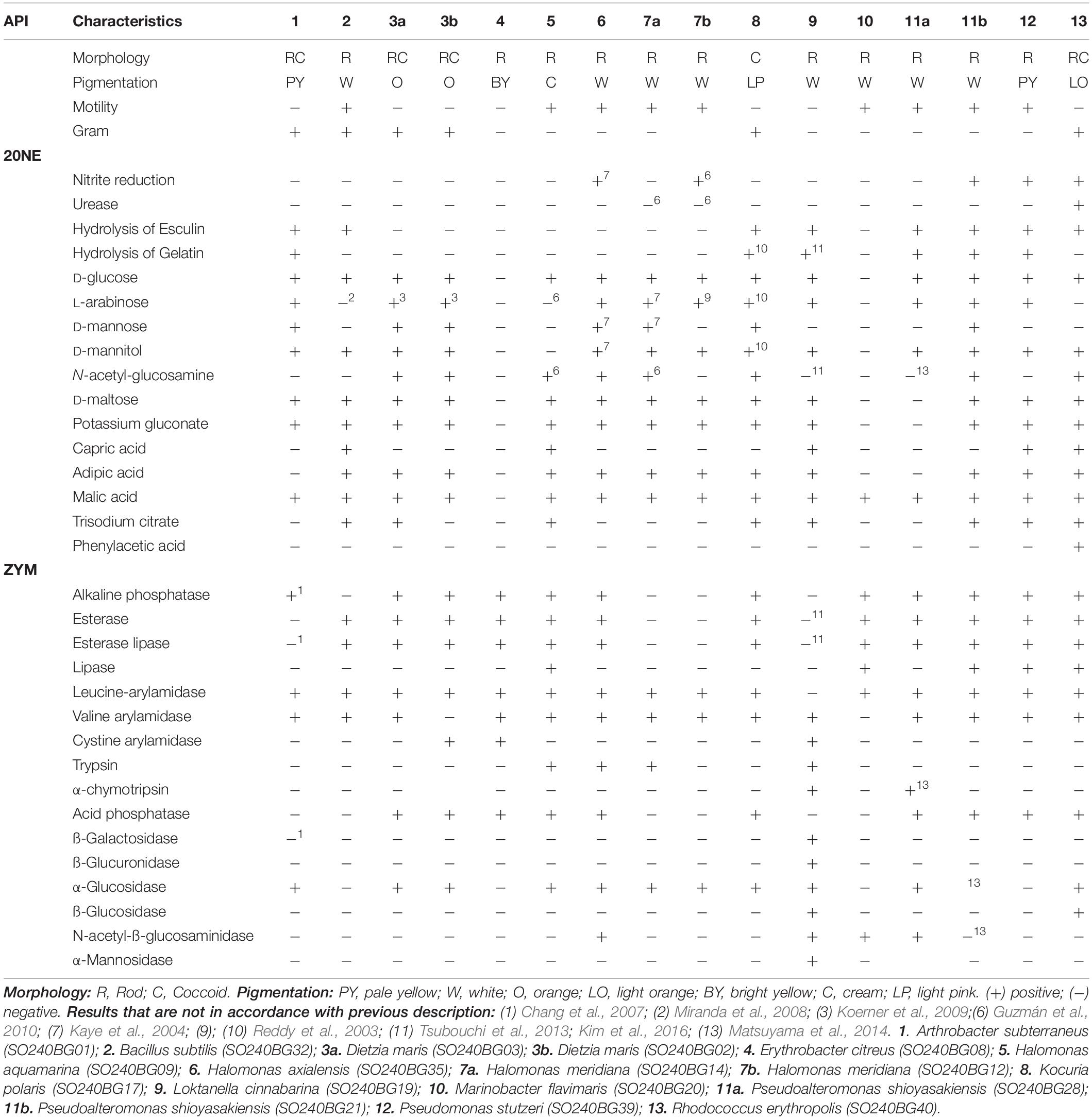
Based on the isolation depth profile and 16S rDNA sequence similarities, 16 bacterial strains were selected for further phenotypic characterization. Most strains were capable of using diverse substrates as sole carbon sources and synthesized a wide spectrum of hydrolytic enzymes (Table 3). The following tests were negative for all isolated bacterial strains: indole production, glucose fermentation, arginine dihydrolase, α-galactosidase, and α-fucosidase. After 24 h of incubation, E. citreus did not show any visible growth for any of the tested sole carbon sources. A similar result was observed for M. flavimaris, which could, however, utilize malic acid as its only carbon source. In the case of D. maris and H. meridiana, for which two strains each were isolated, biochemical analyses indicated high phenotypic similarity. In clear contrast, the two isolates of P. shioyasakiensis displayed substantial biochemical and enzymatic variability.
The applied identification path indicated that a combination of two phenotypical parameters allowed for the identification of 11 of the 13 species; for example, Rhodococcus erythropolis was distinguishable by its nitrate reduction and uptake of L-arabinose as its carbon source (Supplementary Table S3).
Determination of Minimal Inhibitory Concentrations (MIC) of Heavy Metals
The multi-metal resistance of the 16 bacterial strains of interest in liquid medium was tested by determining the minimal inhibitory concentrations (MIC) following a two-fold dilution technique approach. High MIC values indicate high tolerance of the bacterial isolate toward the metal and vice-versa. Metal tolerance appeared to be heterogeneous; the results are listed in Table 4.
The overall level of metal toxicity increased in the order of Mn2+ < Cu2+ < Ni2+ < Zn2+ < Co2+ < Cd2+, corresponding to an average MIC of 20.6, 4.8, 4.3, 2.5, 1.1, and 0.8 mM, respectively. As reported for the phenotypic characterization above, inter-species variability was observed for D. maris, H. meridiana, and P. shioyasakiensis. The highest metal tolerance was found using manganese salt with D. maris (MIC of 14.3 mM), Loktanella cinnabarina (MIC of 57.2 mM) and R. erythropolis (MIC of 228.9 mM). K. polaris also exhibited higher metal tolerance to nickel salt (12.1 mM) as compared to the other tested bacterial strains.
Genomic Analysis of Manganese Resistance Related Genes
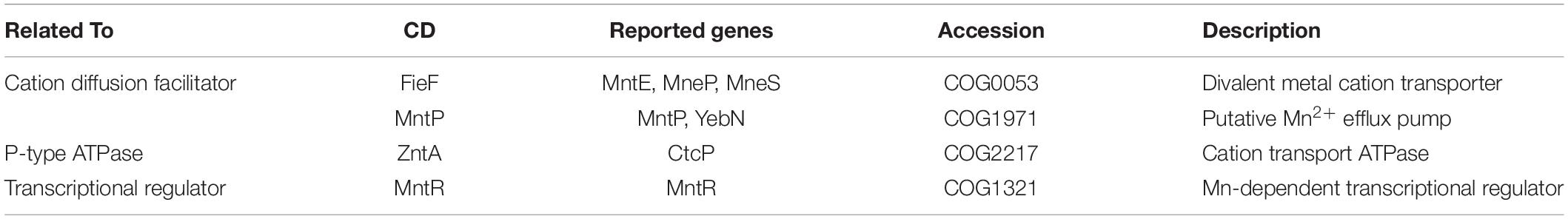
As the deep-sea environment in focus is characterized by manganese nodules, the majority of isolated bacterial strains showed an elevated tolerance to Mn. Consequently, previously reported amino acid sequences of Mn efflux systems, P-type ATPase and Mn resistance-associated transcriptional regulators from relevant marine bacterial species were compared by sequence alignment (data not shown) and their functional conserved domains (CD) identified (Table 5). Protein similarity and domain architecture resulted in a functional classification of two cation diffusion facilitator family proteins (FieF and MntP), one P-type ATPase (ZntA) and one transcriptional regulator (MntR).
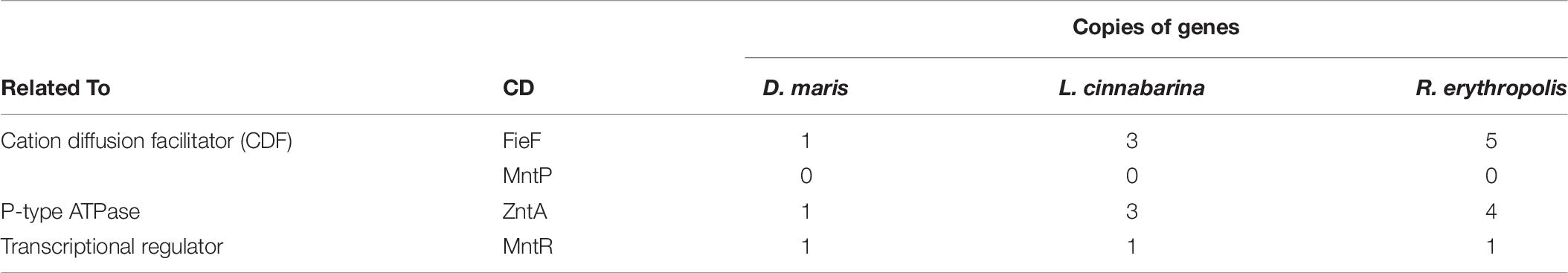
The genomic analysis of D. maris, L. cinnabarina, and R. erythropolis, which exhibited the highest Mn2+ resistance, was conducted using Blast, InterPro, and UniProtKB to give first hints on possible mechanisms of their metal tolerance. The published genomes of D. maris (LVFF00000000), L. cinnabarina (BATB00000000), and R. erythropolis (MDCH00000000) were retrieved from the Genbank database. The numbers of putative gene copies involved in Mn tolerance are presented in Table 6. Details of sequence identification are provided in Supplementary Table S4. Surprisingly, none of the investigated genomes possessed any copies of the putative Mn2+ efflux pump MntP. In contrast, the divalent metal transporter FieF was found in one or more copies of the genomes of D. maris, L. cinnabarina, and R. erythropolis. A similar situation was observed for the P-type ATPase, ZntA. Finally, only one copy of the transcriptional regulator MntR was found per genome investigated.
Discussion
The removal of surface sediment layers and subsequent dispersion of sediment plumes during a deep-sea mining operation is expected to disturb the benthic ecosystem to an unknown extent. The main aim of this study was to provide a pilot study for the development of an in vitro system containing metal-resistant and metal-sensitive bacterial organisms derived from deep-sea sediments. In the future, those organisms will allow a better assessment of heavy metal resistance, bacterial behavior and bacterial dispersion during deep-sea mining in order to optimize comprehensive environmental monitoring in preparation for and during mining activities.
It must be admitted that in vitro cultivation of sub-seafloor microbial community representatives only reveals a very small fraction (less than 0.1%) of the total bacterial diversity (Hondt et al., 2004). However, our study is one of the first (Wang et al., 2018) in which cultivated deep-sea sediment bacteria from the CCZ were characterized in the laboratory in combination with assessment of their metals resistance and related gene repertoire. Comparable studies have been done in other deep-sea ecosystems such as hydrothermal vent or abyssal plain from different ocean in the world (Farias et al., 2015; Zhang et al., 2015).
Consequently, future studies on the development of a respective model system will complement ecosystem-wide ‘omics’ studies (metagenomics and metatranscriptomics) and prokaryotic taxonomic diversity studies to ultimately provide a better understanding of the role of microbes and their interactions in the abyssal plain of the CCZ ecosystems during disruptive anthropogenic processes.
Under aerobic and nutrient-rich conditions, the composition of our cultivatable samples was dominated by proteobacteria over actinobacteria and much less firmicutes. The presence of representatives of the genera Halomonas (35%), Pseudomonas, (13%), and Pseudoalteromonas (20%) in our samples is in agreement with previously reported predominant isolate groups from other deep-sea sediment locations (Xu et al., 2005; Kobayashi et al., 2008; da Silva et al., 2013; Parkes et al., 2014). This result is not surprising, as those genera are among the most cultivatable ones from the marine environment (Giovannoni and Rappé, 2000). Even if those genera were certainly not the major microbial players in benthic microbial communities, their detection and simple “on-board” monitoring could help to better understand the dispersion and microbial dissemination processes that occur during deep-sea mining activities.
The vertical distribution of species revealed a marked difference between the surface water (e.g., M. flavimaris, A. subterraneus) and the sediment microbial communities. Another interesting bacterium is R. erythropolis, which was isolated from the nodule’s surface. A similar relation was previously reported in prokaryotic diversity studies conducted in the Clarion Clipperton Zone (Shulse et al., 2016; Lindh et al., 2017), where there were distinct microbial populations within the sediments, nodules and ambient water. The species richness and biomass of marine sediments are primarily related to organic degradation rates (electron donor diversity) and trace metal elements like Mn, Fe, and Co, which act as micronutrients (Gillan et al., 2012; Walsh et al., 2016). Unfortunately, as of yet, no geochemical record has been retrieved from the sediment cores investigated here.
Heterotrophic bacteria are dominant players in the remineralization of organic material and carbon cycling in deep-sea environments (Lochte, 1992). The availability, composition and distribution of organic substrates in the sediment are directly related to the bacterial production and diversity of hydrolytic enzymes (Boetius, 1995; Hoppe et al., 2002). The phenotypic characterization of the 16 herein isolated bacterial strains revealed a strong inter-species and moderate intra-species variability. Surprisingly, our data for the two P. shioyasakiensis isolates displayed numerous inconsistencies in comparison to each other and the literature data (Matsuyama et al., 2014). Either this observation may indicate that both strains evolved independently over time, which is unlikely, or that the 16S rRNA molecular marker (99.93% similarity) was not sufficiently sensitive to differentiate two closely related species.
All isolated bacterial strains were able to express at least three hydrolytic and proteolytic enzymes (e.g., H. meridiana), with a maximum of 10 such enzymes in L. cinnabarina. The most commonly detected enzyme activities were leucine-arylamidase (94%), valine-arylamidase (88%), alkaline phosphatase (75%), esterase (75%), esterase-lipase (75%), α-glucosidase (69%), and acid phosphatase (63%). The revealed ratios for the occurrence of such enzymes was characteristic of a typical activity spectrum for marine sediment bacteria (Boetius, 1995; Turley, 2000; Arnosti, 2014; Li et al., 2017; Liu et al., 2018). The vertical distribution did not indicate any specific trends in enzymes utilization. This observation could be explained by the lack of information from the uncultivated bacteria, yet might also reveal a rich diversity within the micro-environment, in which species are adapted to certain ecological niches.
The results of the API strip analysis conducted here provide only very limited information on the enzymatic activity spectra and are not reflective for their intensities under deep-sea conditions, such as low temperatures and high pressure. For instance, exposure to higher hydrostatic pressure (>100 bar) might limit microbial growth, disrupt protein homeostasis, and conformational change in ribosomes structure (Gayán et al., 2017). Reaching environmental conditions for experimental in vitro study is crucial but rarely feasible in the case of deep-sea conditions (e.g., the hydrostatic pressure of min 400 bar). However, our results are relevant for the establishment of an in vitro system and therefore enhance the general knowledge of degradation processes for organic matter in the deep sea.
The use of fast-growing organisms combined with API strip assays is an inexpensive and reliable tool, which can easily be implemented on research vessels at sea for impact assessment studies during deep-sea mining activities. Partial enrichment media, designed based on the results of this study (Supplementary Table S3) could be used to select for potential indicator organisms related to either sediment (i.e., D. maris, P. shioyasakiensis), nodules (i.e., R. erythropolis) or water environments (i.e., M. flavimaris), with the aim of monitoring the plume propagation over distance and time. Such on-board studies could be easily further combined with metal tolerance or sensitivity assessments using MIC determination, as shown in our current study.
Multi-metal tolerance of isolated bacteria was increased in the order of Mn2+ < Cu2+ < Ni2+ < Zn2+ < Co2+ < Cd2+. All strains delineate a similar trend in their metal tolerance with the exception of the strongest tolerance level from K. polaris (Ni2+: 12.1 mM), D. maris (Mn2+: 14.3 mM), L. cinnabarina (Mn2+: 57,2 mM), and R. erythropolis (Mn2+: 228.9 mM). The intra-species variabilities observed for D. maris and H. meridiana might suggest evolution and adaptation of those strains to cope with higher metal concentrations that are related directly to their micro-environment. It is important to note that the general response to a combined effect of cold temperature and higher hydrostatic pressure on heavy metal resistance may vary between organism but also from the metal tested (Brown et al., 2017).
As the main metal constituent of polymetallic nodules (Hein and Koschinsky, 2013), Mn2+ was selected to reveal the potential resistance pathway using a simple genomic analysis of the most tolerant bacterial strains: D. maris, L. cinnabarina, and R. erythropolis. The results here suggest that all strains possess at least one gene copy with high sequence similarity to previously described cation diffusion facilitator (FieF), P-type ATPase, ZntA, and the transcriptional regulator, MntR. Such gene copies might be indicative of additional functions or denote a redundancy, which would indicate a need of these efflux systems in the corresponding environment. Interestingly, R. erythropolis, which was isolated on the nodule’s surface, exhibits the highest resistance to manganese salt and additionally has a remarkable sequence redundancy of homologous Mn2+ efflux systems. Furthermore, higher Mn resistance did not lead to other higher metal resistances, which might imply the specificity of those efflux systems.
Conclusion
This study has suggested a microbial cultivation-based approach for the broadening of our knowledge of deep-sea microorganisms. In total, 13 fast-growing bacterial species were identified, from which one to three potential organisms could be selected in future studies. Intra-species variabilities were not only found in phenotypic profiles but also in heavy metal tolerance although the taxonomic marker 16s rRNA sequences were almost identical. We propose the use of API strips and partial-enrichment media that can easily be implemented on-board for a rapid and inexpensive monitoring of deep-sea mining plume dispersion using microbial dissemination analyses. In this context, a heavy-metal resistance analysis provides a new scope for future research on Mn2+ resistance pathways and their role in microbial dispersion from anthropogenic impacts on deep-sea environments. Our genomic analysis indicated the presence of a potential efflux system(s), which could be subject to future transcriptomics or proteomics investigations.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
BG, LT, and MU contributed to the conception and design of the study. BG acquired, analyzed, and organized the database. DC performed the statistical analysis. BG wrote the first draft of the manuscript. All authors contributed to the manuscript revision and read and approved the submitted version.
Funding
BG was supported by the JPI Oceans2 project “EcoMining-DEU – Ecological Aspects of Deep-Sea Mining.” This work was also supported by the HGF Young Investigator Group SeaPump “Seasonal and regional food web interactions with the biological pump”: VH-NG-1000 and by the OceanLab of Jacobs University. This study was also co-financed by the BGR Hannover.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the scientific staff of the BGR Hannover, crew members of the Sonne 240 and particularly Annemiek Vink for providing samples. The authors are grateful to Charlotte Kleint and Song Wang for proofreading our manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00462/full#supplementary-material
Footnotes
- ^ www.Eurofinsgenomics.eu
- ^ http://www.ncbi.nlm.nih.gov/blast
- ^ https://www.ncbi.nlm.nih.gov/genbank/
References
Aleynik, D., Inall, M. E., Dale, A., and Vink, A. (2017). Impact of remotely generated eddies on plume dispersion at abyssal mining sites in the Pacific. Sci. Rep. 7, 1–14. doi: 10.1038/s41598-017-16912-2
Alfaro-Espinoza, G., and Ullrich, M. S. (2014). Marinobacterium mangrovicola sp. nov., a marine nitrogen-fixing bacterium isolated from mangrove roots of Rhizophora mangle. Int. J. Syst. Evol. Microbiol. 64, 3988–3993. doi: 10.1099/ijs.0.067462-0
Arias, D., Cisternas, L. A., and Rivas, M. (2017). Biomineralization of calcium and magnesium crystals from seawater by halotolerant bacteria isolated from Atacama Salar (Chile). Desalination 405, 1–9. doi: 10.1016/j.desal.2016.11.027
Arnosti, C. (2014). Patterns of microbially driven carbon cycling in the ocean: links between extracellular enzymes and microbial communities. Adv. Oceanogr. 2014, 1–12. doi: 10.1155/2014/706082
Balqadi, A. A., Salama, A. J., and Satheesh, S. (2018). Microfouling development on artificial substrates deployed in the central Red Sea. Oceanologia 60, 219–231. doi: 10.1016/j.oceano.2017.10.006
Bentzon-tilia, M., Severin, I., Hansen, L. H., and Riemann, L. (2015). Genomics and ecophysiology of heterotrophic nitrogen-fixing bacteria isolated from estuarine surface water. mBio 6, 1–11. doi: 10.1128/mBio.00929-15.Editor
Bibi, F., Alvi, S. A., Al-sofyani, A., Yasir, M., Kensarah, E. A., and Azhar, E. I. (2018). Two marine sponges-associated cultivable bacteria: diversity and biological activities. Genet. Mol. Res. 17:gmr16039910. doi: 10.4238/gmr16039910
Boetius, A. (1995). Microbial hydrolytic enzyme activities in deep-sea sediments. Helgoländer Meeresuntersuchungen 49, 177–187. doi: 10.1007/BF02368348
Brown, A., Thatje, S., and Hauton, C. (2017). The effects of temperature and hydrostatic pressure on metal toxicity: insights into toxicity in the deep sea. Environ. Sci. Technol. 51, 10222–10231. doi: 10.1021/acs.est.7b02988
Catania, V., Cappello, S., Di Giorgi, V., Santisi, S., Di Maria, R., Mazzola, A., et al. (2018). Microbial communities of polluted sub-surface marine sediments. Mar. Pollut. Bull. 131, 396–406. doi: 10.1016/j.marpolbul.2018.04.015
Chang, H.-W., Bae, J.-W., Nam, Y.-D., Kwon, H.-Y., Park, J. R., Shin, K.-S., et al. (2007). Arthrobacter subterraneus sp. nov., isolated from deep subsurface water of the South Coast of Korea. J. Microbiol. Biotechnol. 17, 1875–1879.
Chen, H., Wu, R., Xu, G., Fang, X., Qiu, X., Guo, H., et al. (2010). DR2539 is a novel DtxR-like regulator of Mn/Fe ion homeostasis and antioxidant enzyme in Deinococcus radiodurans. Biochem. Biophys. Res. Commun. 396, 413–418. doi: 10.1016/j.bbrc.2010.04.106
Chen, S., and Shao, Z. (2009). Isolation and diversity analysis of arsenite-resistant bacteria in communities enriched from deep-sea sediments of the Southwest Indian Ocean Ridge. Extremophiles 13, 39–48. doi: 10.1007/s00792-008-0195-1
Corinaldesi, C. (2015). New perspectives in benthic deep-sea microbial ecology. Front. Mar. Sci. 2:17. doi: 10.3389/fmars.2015.00017
Cruz-López, R., and Maske, H. (2016). The vitamin B1 and B12 required by the marine dinoflagellate Lingulodinium polyedrum can be provided by its associated bacterial community in culture. Front. Microbiol. 7:560. doi: 10.3389/fmicb.2016.00560
da Silva, M. A. C., Cavalett, A., Spinner, A., Rosa, D. C., Jasper, R. B., Quecine, M. C., et al. (2013). Phylogenetic identification of marine bacteria isolated from deep-sea sediments of the eastern South Atlantic Ocean. Springerplus 2, 1–10. doi: 10.1186/2193-1801-2-127
Danovaro, R., Snelgrove, P. V. R., and Tyler, P. (2014). Challenging the paradigms of deep-sea ecology. Trends Ecol. Evol. 29, 465–475. doi: 10.1016/j.tree.2014.06.002
Dos Santos Furtado, A. L., and Casper, P. (2000). Different methods for extracting bacteria from freshwater sediment and a simple method to measure bacterial production in sediment samples. J. Microbiol. Methods 41, 249–257. doi: 10.1016/S0167-7012(00)00163-9
Ettoumi, B., Bouhajja, E., Borin, S., Daffonchio, D., Boudabous, A., and Cherif, A. (2010). Gammaproteobacteria occurrence and microdiversity in Tyrrhenian Sea sediments as revealed by cultivation-dependent and -independent approaches. Syst. Appl. Microbiol. 33, 222–231. doi: 10.1016/j.syapm.2010.02.005
Ettoumi, B., Chouchane, H., Guesmi, A., Mahjoubi, M., Brusetti, L., Neifar, M., et al. (2016). Diversity, ecological distribution and biotechnological potential of Actinobacteria inhabiting seamounts and non-seamounts in the Tyrrhenian Sea. Microbiol. Res. 186–187, 71–80. doi: 10.1016/j.micres.2016.03.006
Farias, P., Santo, C. E., Branco, R., Francisco, R., Santos, S., Hansen, L., et al. (2015). Natural hot spots for gain of multiple resistances: arsenic and antibiotic resistances in heterotrophic, aerobic bacteria from marine hydrothermal vent fields. Appl. Environ. Microbiol. 81, 2534–2543. doi: 10.1128/AEM.03240-14
Finn, R. D., Attwood, T. K., Babbitt, P. C., Bateman, A., Bork, P., Bridge, A. J., et al. (2017). InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199. doi: 10.1093/nar/gkw1107
Focardi, S., Pepi, M., Landi, G., Gasperini, S., Ruta, M., Di Biasio, P., et al. (2012). Hexavalent chromium reduction by whole cells and cell free extract of the moderate halophilic bacterial strain Halomonas sp. TA-04. Int. Biodeterior. Biodegrad. 66, 63–70. doi: 10.1016/j.ibiod.2011.11.003
Fritz, J.-S. (2016). Commentary: threatened by mining, polymetallic nodules are required to preserve abyssal epifauna. Front. Mar. Sci. 3:190. doi: 10.1038/srep26808
Gao, X., Gao, W., Cui, Z., Han, B., Yang, P., Sun, C., et al. (2015). Biodiversity and degradation potential of oil-degrading bacteria isolated from deep-sea sediments of South Mid-Atlantic Ridge. Mar. Pollut. Bull. 97, 373–380. doi: 10.1016/j.marpolbul.2015.05.065
Gärtner, A., Blümel, M., Wiese, J., and Imhoff, J. F. (2011). Isolation and characterisation of bacteria from the Eastern Mediterranean deep sea. Antonie van Leeuwenhoek 100, 421–435. doi: 10.1007/s10482-011-9599-5
Gayán, E., Govers, S. K., and Aertsen, A. (2017). Impact of high hydrostatic pressure on bacterial proteostasis. Biophys. Chem. 231, 3–9. doi: 10.1016/j.bpc.2017.03.005
Gillan, D. C., Baeyens, W., Bechara, R., Billon, G., Denis, K., Grosjean, P., et al. (2012). Links between bacterial communities in marine sediments and trace metal geochemistry as measured by in situ DET/DGT approaches. Mar. Pollut. Bull. 64, 353–362. doi: 10.1016/j.marpolbul.2011.11.001
Gillard, B., Purkiani, K., Chatzievangelou, D., Vink, A., Iversen, M. H., and Thomsen, L. (2019). Physical and hydrodynamic properties of deep sea mining-generated, abyssal sediment plumes in the Clarion Clipperton Fracture Zone (eastern-central Pacific). Elem. Sci. Anthr. 7:5. doi: 10.1525/elementa.343
Giovannoni, S. J., and Rappé, M. (2000). “Evolution, diversity, and molecular ecology of marine prokaryotes,” in Microbial Ecology of the Oceans, ed. D. L. Kirchman (Hoboken, NJ: Wiley-Blackwell), 47–84.
Graham, I. J., Dichburn, R. G., and Zondervan, A. (2004). Beryllium isotope dating of ferromanganese nodules and crusts. New Zeal. Sci. Rev. 36, 57–61.
Guzmán, D., Quillaguamán, J., Muñoz, M., and Hatti-Kaul, R. (2010). Halomonas andesensis sp. nov., a moderate halophile isolated from the saline lake Laguna Colorada in Bolivia. Int. J. Syst. Evol. Microbiol. 60, 749–753. doi: 10.1099/ijs.0.014522-0
Heald, S. C., Brandão, P. F. B., Hardicre, R., and Bull, A. T. (2001). Physiology, biochemistry and taxonomy of deep-sea nitrile metabolising Rhodoccus strains. Antonie van Leeuwenhoek 80, 169–183. doi: 10.1023/A:1012227302373
Hein, J. R., and Koschinsky, A. (2013). “Deep-ocean ferromanganese crusts and nodules,” in Treatise on Geochemistry, eds H. D. Holland, K. K. Turekian, 2nd Edn, (Amsterdam: Elsevier Inc.), 273–291. doi: 10.1016/B978-0-08-095975-7.01111-6
Hirayama, H., Abe, M., Miyazaki, J., Sakai, S., Nagano, Y., and Takai, K. (2015). “Data report: cultivation of microorganisms from basaltic rock and sediment cores from the North Pond on the western flank of the Mid-Atlantic Ridge,” in Proceedings of the IODP Expedition 336, (Tokyo: Integrated Ocean Drilling Program Management International, Inc.).
Hondt, S. D., Jørgensen, B. B., Miller, D. J., Batzke, A., Blake, R., Cragg, B. A., et al. (2004). Distributions of microbial activities in deep subseafloor sediments. Science 306, 2216–2222.
Hoppe, H.-G., Arnosti, C., and Herndl, G. F. (2002). “Ecological significance of bacterial enzymes in marine environment,” in Microbial Enzymes in the Environment Activity, Ecology and Application, eds R. G. Burns and R. P. Dick (New York, NY: Marcel Dekker), 73.
Huang, X., Shin, J. H., Pinochet-Barros, A., Su, T. T., and Helmann, J. D. (2017). Bacillus subtilis MntR coordinates the transcriptional regulation of manganese uptake and efflux systems. Mol. Microbiol. 103, 253–268. doi: 10.1111/mmi.13554
Inagaki, F., Suzuki, M., Takai, K., Oida, H., Sakamoto, T., Aoki, K., et al. (2003). Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl. Environ. Microbiol. 69, 7224–7235. doi: 10.1128/AEM.69.12.7224
Ivanova, E. P., Vysotskii, M. V., Svetashev, V. I., Nedashkovskaya, O. I., Gorshkova, N. M., Mikhailov, V. V., et al. (1999). Characterization of Bacillus strains of marine origin. Int. Microbiol. 2, 267–271.
Jørgensen, B. B., and Boetius, A. (2007). Feast and famine - Microbial life in the deep-sea bed. Nat. Rev. Microbiol. 5, 770–781. doi: 10.1038/nrmicro1745
Kai, W., Peisheng, Y., Rui, M., Wenwen, J., and Zongze, S. (2017). Diversity of culturable bacteria in deep-sea water from the South Atlantic Ocean. Bioengineered 8, 572–584. doi: 10.1080/21655979.2017.1284711
Kaye, J. Z., Márquez, M. C., Ventosa, A., and Baross, J. A. (2004). Halomonas neptunia sp. nov., Halomonas sulfidaeris sp. nov., Halomonas axialensis sp. nov. and Halomonas hydrothermalis sp. nov.: halophilic bacteria isolated from deep-sea hydrothermal-vent environments. Int. J. Syst. Evol. Microbiol. 54, 499–511. doi: 10.1099/ijs.0.02799-0
Kim, H., Choo, Y. J., Song, J., Lee, J. S., Lee, K. C., and Cho, J. C. (2007). Marinobacterium litorale sp. nov. in the order Oceanospirillales. Int. J. Syst. Evol. Microbiol. 57, 1659–1662. doi: 10.1099/ijs.0.64892-0
Kim, K., Srinivasan, S., and Lee, S.-S. (2016). Loktanella aquimaris sp. nov., Isolated from Seawater. Curr. Microbiol. 72, 228–233. doi: 10.1007/s00284-015-0945-0
Kobayashi, T., Koide, O., Mori, K., Shimamura, S., Matsuura, T., Miura, T., et al. (2008). Phylogenetic and enzymatic diversity of deep subseafloor aerobic microorganisms in organics- and methane-rich sediments off Shimokita Peninsula. Extremophiles 12, 519–527. doi: 10.1007/s00792-008-0157-7
Koerner, R. J., Goodfellow, M., and Jones, A. L. (2009). The genus Dietzia: a new home for some known and emerging opportunist pathogens. FEMS Immunol. Med. Microbiol. 55, 296–305. doi: 10.1111/j.1574-695X.2008.00513.x
Koschinsky, A., Winkler, A., and Fritsche, U. (2003). Importance of different types of marine particles for the scavenging of heavy metals in the deep-sea bottom water. Appl. Geochem. 18, 693–710. doi: 10.1016/S0883-2927(02)00161-0
Labonté, J. M., Lever, M. A., Edwards, K. J., and Orcutt, B. N. (2017). Influence of igneous basement on deep sediment microbial diversity on the eastern Juan de Fuca Ridge flank. Front. Microbiol. 8:1434. doi: 10.3389/fmicb.2017.01434
Langdahl, B. R., Bisp, P., and Ingvorsen, K. (1996). Nitrile hydrolysis by Rhodococcus erythropolis BLI, an acetonitrile-tolerant strain isolated from a marine sediment. Microbiology 142, 145–154. doi: 10.1099/13500872-142-1-145
Lemire, J. A., Harrison, J. J., and Turner, R. J. (2013). Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 11, 371–384. doi: 10.1038/nrmicro3028
Li, C., Tao, J., Mao, D., and He, C. (2011). A novel manganese efflux system, YebN, is required for virulence by Xanthomonas oryzae pv. oryzae. PLoS One 6:e21983. doi: 10.1371/journal.pone.0021983
Li, L., Ding, F., Sang, L., Liu, J., Mao, D., Liu, X., et al. (2018). Study on the oxygen reduction reaction catalyzed by a cold-tolerant marine strain phylogenetically related to Erythrobacter citreus. Bioelectrochemistry 119, 51–58. doi: 10.1016/j.bioelechem.2017.08.011
Li, Y., Wu, C., Zhou, M., Wang, E. T., Zhang, Z., Liu, W., et al. (2017). Diversity of cultivable protease-producing bacteria in Laizhou Bay sediments, Bohai Sea, China. Front. Microbiol. 8:405. doi: 10.3389/fmicb.2017.00405
Lindh, M. V., Maillot, B. M., Shulse, C. N., Gooday, A. J., Amon, D. J., Smith, C. R., et al. (2017). From the surface to the deep-sea: bacterial distributions across polymetallic nodule fields in the clarion-clipperton zone of the Pacific Ocean. Front. Microbiol. 8:1696. doi: 10.3389/fmicb.2017.01696
Liu, Q., Fang, J., Li, J., Zhang, L., Xie, B. B., Chen, X. L., et al. (2018). Depth-resolved variations of cultivable bacteria and their extracellular enzymes in the water column of the new britain trench. Front. Microbiol. 9:135. doi: 10.3389/fmicb.2018.00135
Lochte, K. (1992). “Bacterial standing stock and consumption of organic carbon in the benthic boundary layer of the Abyssal North Atlantic,” in Deep-Sea Food Chains and the Global Carbon Cycle, eds G. T. Rowe and V. Pariente (Dordrecht: Springer), 1–10. doi: 10.1007/978-94-011-2452-2_1
Ma, R., Wang, J., Wang, Q., Ma, Z., Li, J., and Chen, L. (2017). Draft genome sequence of Loktanella cinnabarina strain XM1 isolated from coastal surface water. Am. Soc. Microbiol. 5, 4–6. doi: 10.1128/genomeA.00504-17
MacDonell, M. T., Singleton, F. L., and Hood, M. A. (1982). Diluent composition for use of API 20E in characterizing marine and estuarine bacteria. Appl. Environ. Microbiol. 44, 423–427.
Maruyama, A., Honda, D., Yamamoto, H., Kitamura, K., and Higashihara, T. (2000). Phylogenetic analysis of psychrophilic bacteria isolated from the Japan Trench, including a description of the deep-sea species Psychrobacter pacificensis sp. nov. Int. J. Syst. Evol. Microbiol. 50, 835–846. doi: 10.1099/00207713-50-2-835
Matsuyama, H., Sawazaki, K., Minami, H., Kasahara, H., Horikawa, K., and Yumoto, I. (2014). Pseudoalteromonas shioyasakiensis sp. nov., a marine polysaccharide-producing bacterium. Int. J. Syst. Evol. Microbiol. 64, 101–106. doi: 10.1099/ijs.0.055558-0
Melnikova, D. I., Beleneva, I. A., Tyunin, A. P., and Magarlamov, T. Y. (2017). The taxonomic composition, characteristics, and neurotoxic activities of ribbon worm-associated bacteria from the Sea of Japan. Russ. J. Mar. Biol. 43, 383–391. doi: 10.1134/S1063074017050066
Mewes, K., Mogollón, J. M., Picard, A., Rühlemann, C., Kuhn, T., Nöthen, K., et al. (2014). Impact of depositional and biogeochemical processes on small scale variations in nodule abundance in the Clarion-Clipperton Fracture Zone. Deep. Res. Part I Oceanogr. Res. Pap. 91, 125–141. doi: 10.1016/j.dsr.2014.06.001
Miranda, C. A. C., Martins, O. B., and Clementino, M. M. (2008). Species-level identification of Bacillus strains isolates from marine sediments by conventional biochemical, 16S rRNA gene sequencing and inter-tRNA gene sequence lengths analysis. Antonie van Leeuwenhoek 93, 297–304. doi: 10.1007/s10482-007-9204-0
Mobberley, J. M., Authement, R. N., Segall, A. M., and Paul, J. H. (2008). The temperate marine phage HAP-1 of Halomonas aquamarina possesses a linear plasmid-like prophage genome. J. Virol. 82, 6618–6630. doi: 10.1128/JVI.00140-08
Nisha, N. S., and Divakaran, J. (2014). Optimization of alkaline protease production from Bacillus subtilis NS isolated from sea water. Afr. J. Biotechnol. 13, 1707–1713. doi: 10.5897/AJB2014.13652
Oebius, H. U., Becker, H. J., Rolinski, S., and Jankowski, J. A. (2001). Parametrization and evaluation of marine environmental impacts produced by deep-sea manganese nodule mining. Deep. Res. Part II Top. Stud. Oceanogr. 48, 3453–3467. doi: 10.1016/S0967-0645(01)00052-2
Padilla-Benavides, T., Long, J. E., Raimunda, D., Sassetti, C. M., and Argüello, J. M. (2013). A novel P1B-type Mn2+-transporting ATPase is required for secreted protein metallation in mycobacteria. J. Biol. Chem. 288, 11334–11347. doi: 10.1074/jbc.M112.448175
Parkes, R. J., Cragg, B., Roussel, E., Webster, G., Weightman, A., and Sass, H. (2014). A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere: geosphere interactions. Mar. Geol. 352, 409–425. doi: 10.1016/j.margeo.2014.02.009
Pathom-aree, W., Stach, J. E. M., Ward, A. C., Horikoshi, K., Bull, A. T., and Goodfellow, M. (2006). Diversity of actinomycetes isolated from challenger deep sediment (10,898 m) from the Mariana Trench. Extremophiles 10, 181–189. doi: 10.1007/s00792-005-0482-z
Patzer, S. I., and Hantke, K. (2001). Dual repression by Fe 2+ -Fur and Mn 2+ -MntR of the mntH Gene, encoding an NRAMP-Like Mn 2+ transporter in Escherichia coli. J. Bacteriol. 183, 4806–4813. doi: 10.1128/JB.183.16.4806
Pearson, W. R. (2014). An introduction to sequence and series. Int. J. Res. 1, 1286–1292. doi: 10.1002/0471250953.bi0301s42.An
Que, Q., and Helmann, J. D. (2000). Manganese homestasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35, 1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x
Ramirez-Llodra, E., Tyler, P. A., Baker, M. C., Bergstad, O. A., Clark, M. R., Escobar, E., et al. (2011). Man and the last great wilderness: human impact on the deep sea. PLoS One 6:e22588. doi: 10.1371/journal.pone.0022588
R Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available at: https://www.R-project.org/
Reddy, G. S. N., Prakash, J. S. S., Prabahar, V., Matsumoto, G. I., Stackebrandt, E., and Shivaji, S. (2003). Kocuria polaris sp. nov., an orange-pigmented psychrophilic bacterium isolated from an Antarctic cyanobacterial mat sample. Int. J. Syst. Evol. Microbiol. 53, 183–187. doi: 10.1099/ijs.0.02336-0
Ritchie, K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Rosch, J. W., Gao, G., Ridout, G., Wang, Y. D., and Tuomanen, E. I. (2009). Role of the manganese efflux system mntE for signalling and pathogenesis in Streptococcus pneumoniae. Mol. Microbiol. 72, 12–25. doi: 10.1111/j.1365-2958.2009.06638.x
Shulse, C. N., Maillot, B., Smith, C. R., and Church, M. J. (2016). Polymetallic nodules, sediments, and deep waters in the equatorial North Pacific exhibit highly diverse and distinct bacterial, archaeal, and microeukaryotic communities. Microbiol. Open 6, 1–16. doi: 10.1002/mbo3.428
Smith, C. R., De Leo, F. C., Bernardino, A. F., Sweetman, A. K., and Arbizu, P. M. (2008). Abyssal food limitation, ecosystem structure and climate change. Trends Ecol. Evol. 23, 518–528. doi: 10.1016/j.tree.2008.05.002
Sonnenschein, E. C., Gärdes, A., Seebah, S., Torres-Monroy, I., Grossart, H. P., and Ullrich, M. S. (2011). Development of a genetic system for Marinobacter adhaerens HP15 involved in marine aggregate formation by interacting with diatom cells. J. Microbiol. Methods 87, 176–183. doi: 10.1016/j.mimet.2011.08.008
Stahl, A., Pletzer, D., Mehmood, A., and Ullrich, M. S. (2015). Marinobacter adhaerens HP15 harbors two CzcCBA efflux pumps involved in zinc detoxification. Antonie van Leeuwenhoek 108, 649–658. doi: 10.1007/s10482-015-0520-5
Sudek, L. A., Templeton, A. S., Tebo, B. M., and Staudigel, H. (2009). Microbial ecology of Fe (hydr)oxide mats and basaltic rock from Vailulu’u Seamount, American Samoa. Geomicrobiol. J. 26, 581–596. doi: 10.1080/01490450903263400
Sun, H., Xu, G., Zhan, H., Chen, H., Sun, Z., Tian, B., et al. (2010). Identification and evaluation of the role of the manganese efflux protein in Deinococcus radiodurans. BMC Microbiol. 10:319. doi: 10.1186/1471-2180-10-319
Takami, H., Kobata, K., Nagahama, T., Kobayashi, H., Inoue, A., and Horikoshi, K. (1999). Biodiversity in deep-sea sites located near the south part of Japan. Extremophiles 3, 97–102. doi: 10.1007/s007920050104
Tang, J., Zheng, A. P., Bromfield, E. S. P., Zhu, J., Li, S. C., Wang, S. Q., et al. (2011). 16S rRNA gene sequence analysis of halophilic and halotolerant bacteria isolated from a hypersaline pond in Sichuan, China. Ann. Microbiol. 61, 375–381. doi: 10.1007/s13213-010-0137-x
Teske, A., Brinkhoff, T., Muyzer, G., Moser, D. P., Rethmeier, J., and Jannasch, H. W. (2000). Diversity of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents. Appl. Environ. Microbiol. 66, 3125–3133. doi: 10.1128/AEM.66.8.3125-3133.2000
Tsubouchi, T., Shimane, Y., Mori, K., Miyazaki, M., Tame, A., Uematsu, K., et al. (2013). Loktanella cinnabarina sp. nov., isolated from a deep subseafloor sediment, and emended description of the genus Loktanella. Int. J. Syst. Evol. Microbiol. 63, 1390–1395. doi: 10.1099/ijs.0.043174-0
Turley, C. (2000). Bacteria in the cold deep-sea benthic boundary layer and sediment-water interface of the NE Atlantic. FEMS Microbiol. Ecol. 33, 89–99. doi: 10.1111/j.1574-6941.2000.tb00731.x
Undabarrena, A., Beltrametti, F., Claverias, F. P., Gonzalez, M., Moore, E. R. B., Seeger, M., et al. (2016). Exploring the diversity and antimicrobial potential of marine actinobacteria from the comau fjord in Northern Patagonia, Chile. Front. Microbiol. 7:1135. doi: 10.3389/fmicb.2016.01135
Vraspir, J. M., Holt, P. D., and Butler, A. (2011). Identification of new members within suites of amphiphilic marine siderophores. BioMetals 24, 85–92. doi: 10.1007/s10534-010-9378-1
Walsh, E. A., Kirkpatrick, J. B., Pockalny, R., Sauvage, J., Spivack, A. J., Murray, R. W., et al. (2016). Relationship of bacterial richness to organic degradation rate and sediment age in subseafloor sediment. Appl. Environ. Microbiol. 82, 4994–4999. doi: 10.1128/AEM.00809-16
Wang, F., Wang, P., Chen, M., and Xiao, X. (2004). Isolation of extremophiles with the detection and retrieval of Shewanella strains in deep-sea sediments from the west Pacific. Extremophiles 8, 165–168. doi: 10.1007/s00792-003-0365-0
Wang, W., Cai, B., and Shao, Z. (2014). Oil degradation and biosurfactant production by the deep sea bacterium Dietzia maris As-13-3. Front. Microbiol. 5:711. doi: 10.3389/fmicb.2014.00711
Wang, X., Lin, D., Jing, X., Zhu, S., Yang, J., and Chen, J. (2018). Complete genome sequence of the highly Mn(II) tolerant Staphylococcus sp. AntiMn-1 isolated from deep-sea sediment in the Clarion-Clipperton Zone. J. Biotechnol. 266, 34–38. doi: 10.1016/j.jbiotec.2017.12.004
Waters, L. S., Sandoval, M., and Storz, G. (2011). The Escherichia coli MntR miniregulon includes genes encoding a small protein and an efflux pump required for manganese homeostasis. J. Bacteriol. 193, 5887–5897. doi: 10.1128/JB.05872-11
Wu, C. H., Apweiler, R., Bairoch, A., Natale, D. A., Barker, W. C., Boeckmann, B., et al. (2006). The universal protein resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 34, D187–D191. doi: 10.1093/nar/gkj161
Wu, Y. H., Liao, L., Wang, C. S., Ma, W. L., Meng, F. X., Wu, M., et al. (2013). A comparison of microbial communities in deep-sea polymetallic nodules and the surrounding sediments in the Pacific Ocean. Deep. Res. Part I Oceanogr. Res. Pap. 79, 40–49. doi: 10.1016/j.dsr.2013.05.004
Xu, M., Wang, P., Wang, F., and Xiao, X. (2005). Microbial diversity at a deep-sea station of the Pacific nodule province. Biodivers. Conserv. 14, 3363–3380. doi: 10.1007/s10531-004-0544-z
Yoon, J. H., Yeo, S. H., Kim, I. G., and Oh, T. K. (2004). Marinobacter flavimaris sp. nov. and Marinobacter daepoensis sp. nov., slightly halophilic organisms isolated from sea water of the Yellow Sea in Korea. Int. J. Syst. Evol. Microbiol. 54, 1799–1803. doi: 10.1099/ijs.0.63151-0
Yuan, J., Lai, Q., Sun, F., Zheng, T., and Shao, Z. (2015). The diversity of PAH-degrading bacteria in a deep-sea water column above the southwest Indian ridge. Front. Microbiol. 6:853. doi: 10.3389/fmicb.2015.00853
Zhang, D. C., Liu, Y. X., and Li, X. Z. (2015). Characterization of bacterial diversity associated with deep sea ferromanganese nodules from the South China Sea. J. Microbiol. 53, 598–605. doi: 10.1007/s12275-015-5217-y
Keywords: deep-sea mining, CCZ, sediment, bacteria, heavy metal, API strips
Citation: Gillard B, Chatzievangelou D, Thomsen L and Ullrich MS (2019) Heavy-Metal-Resistant Microorganisms in Deep-Sea Sediments Disturbed by Mining Activity: An Application Toward the Development of Experimental in vitro Systems. Front. Mar. Sci. 6:462. doi: 10.3389/fmars.2019.00462
Received: 12 February 2019; Accepted: 10 July 2019;
Published: 23 July 2019.
Edited by:
Daniela Zeppilli, Institut Français de Recherche pour l’Exploitation de la Mer (IFREMER), FranceReviewed by:
Alastair Brown, University of Southampton, United KingdomXueWei Xu, State Oceanic Administration, China
Copyright © 2019 Gillard, Chatzievangelou, Thomsen and Ullrich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benjamin Gillard, Yi5naWxsYXJkQGphY29icy11bml2ZXJzaXR5LmRl
 Benjamin Gillard
Benjamin Gillard Damianos Chatzievangelou
Damianos Chatzievangelou Laurenz Thomsen
Laurenz Thomsen Matthias S. Ullrich
Matthias S. Ullrich