- 1College of Earth, Ocean, and Atmospheric Sciences, Oregon State University, Corvallis, OR, United States
- 2Ocean Networks Canada, University of Victoria, Victoria, BC, Canada
- 3Department of Biology, University of Victoria, Victoria, BC, Canada
- 4Department of Microbiology, College of Science, Oregon State University, Corvallis, OR, United States
Methane seep habitats are widespread chemosynthesis-based ecosystems that span continental margins and interact with surrounding marine systems. With many of these habitats occurring below 200 m, seeps can serve as an important source of nutrients in otherwise food limited deep-sea environments. However, the potential for marauding megafauna to assimilate seep derived nutrition has been difficult to quantify. Here, we provide the first evidence of a commercially harvested species, Chionoecetes tanneri (tanner crab), assimilating chemosynthetic production. Although bulk isotope analysis of C. tanneri tissue indicated no quantifiable incorporation of seep-derived carbon or sulfur (mean δ13C, −18.5‰; mean δ34S, 19.5‰), depletions in 13C (δ13C as light as −38.8‰) were noted in fatty acid (FA) compounds. In addition, diagnostic biomarkers for seep bacteria, including 16:1ω6 and 18:1ω8c FA's, were found to have been assimilated by C. tanneri. Futher supporting a trophic link between the seep and the C. tanneri, seep associated bacteria and archaea were, in certain cases, the dominant taxa in the gut contents of the crabs. This work provides the first insights into a link between seep production and deep-sea ecosystem services, specifically fisheries production. In addition, it reveals a methodological bias that could exist in some trophic studies where bulk isotopes under-represent the role of seep nutrition in the diet of marauding animals.
Introduction
Deep-sea habitats provide a range of ecosystem goods and services to society (Armstrong et al., 2010; Thurber et al., 2014). These habitats are believed to be energy-limited ecosystems, with a slow rain of food from the surface driving community structure across all size classes (Rex et al., 1993; Wei et al., 2010). Microbially facilitated chemosynthetic production in deep-sea habitats, such as methane seeps can produce an abundant supply of food and support dense faunal communities (Levin, 2005; Cordes et al., 2010). However, the role these habitats serve for surrounding ecosystems and society remains largely unknown (Levin et al., 2016). Here we identify the unexpected use of chemosynthetic production by a commercially harvested species, Chionoecetes tanneri (tanner crab), identifying a novel link between seep ecosystems, fisheries, and humans. Through this we discover unexpected methodological biases that could have obscured quantification of export production from seep ecosystems in previous studies.
Methane seeps are increasingly becoming recognized as abundant areas of productivity along margin habitats, with over 1,000 methane seep sites discovered in the past 2 years along the Cascadia subduction zone alone (Bell et al., 2017; Riedel et al., 2018; Seabrook et al., 2018). Non-seep organisms have been observed at seeps, including commercially important species, such as longspine thornyheads (Sebastolobus altivelis; Grupe et al., 2015) and the Patagonian toothfish (Dissostichus eleginoides; Sellanes et al., 2012), however, the nature of these interactions are unclear. In 2012, a dense aggregation of C. tanneri were found inhabiting Clayoquot Slope, a methane seep off the coast of British Columbia. The crabs were observed actively foraging, with the crabs sifting through the sediment at actively bubbling regions, feeding both on and around microbial mats at the site, and in some cases “flipping” off the seafloor from bubble activity (Figures 1a–d; Supplementary Videos 1, 2). In 2014, an additional dense crab aggregation, of various size classes, around the nearby Barkley Canyon seep was also seen (Figure 1c). The observations of active foraging behavior by the C. tanneri specimen suggested that they may be using the seep as a source of trophic support. Chionoecetes tanneri are a commercially harvested species in the NE Pacific (Somerton and Donaldson, 1996; Kruse et al., 2005; Keller et al., 2012) that migrate across the margin (Keller et al., 2012, 2016). As a result, if they are harvesting seep production they could be exporting it to non-seep ecosystems and potentially to humans.
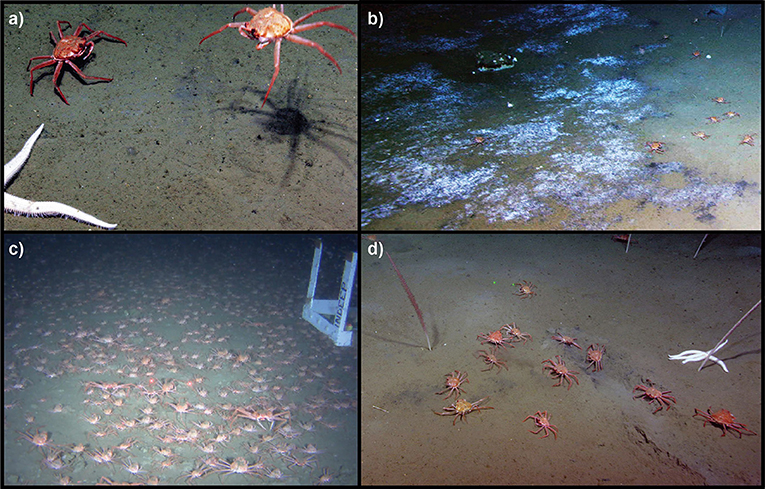
Figure 1. (a,b,d) Chionoecetes tanneri at Clayoquot Slope seep showing active foraging behavior in the sediment amongst bubbles and on microbial mats (a,b,d) and being lifted off the seafloor by methane hydrates building up on its abdomen (a). (c) A migration event of hundreds (estimated) of tanner crabs of multiple size classes at 1000 m depth in the nearby Barkley Canyon, in January 2015. Photos courtesy of Ocean Networks Canada.
Quantification of the transfer of chemosynthetic production to the surrounding ecosystem has often led to enigmatic results, showing little to no use of chemosynthetic production by mobile organisms (MacAvoy et al., 2002, 2008). As microbial production can be difficult to quantify, biomarker analysis, including stable isotope and fatty acid analysis are often employed to measure the transfer of bacterial and archaeal production to higher trophic levels. Stable isotope analysis makes use of incorporations of carbon (C) and sulfur (S) from an organism's diet and can be used to disentangle food webs, if the food sources differ in their stable isotopic composition (Fry and Sherr, 1989; Van Dover, 2007; Boecklen et al., 2011) At seep systems, chemosynthetic bacteria and archaea have unique isotopic signatures compared to non-seep food sources. For example, methanotrophically-derived carbon ranges from −40 to −100‰ in δ13C, and thiotrophically-derived sulfur ranges from −25 to −35‰ δ35S (Levin and Michener, 2002; MacAvoy et al., 2003, 2005; Levin, 2005; Boecklen et al., 2011). This contrasts with carbon and sulfur compounds derived from photosynthesis, which range from −15 to −25‰ δ13C and 18 to 22‰ δ35S, respectively (Fry and Sherr, 1989; Boecklen et al., 2011). To get a more high-resolution view of an organism's diet, fatty acid (FA) analysis can be used, as certain FA compounds as well as branching and double bond patterns can be linked to specific types of producers (e.g., Thurber et al., 2013). Seep associated production can be identified through diagnostic biomarkers for methanotrophy and sulfate reduction, including the bacterial produced 16:1ω6, 16:1ω8, and 18:1ω8 FA's (Bowman et al., 1991; Thurber et al., 2012). The assimilation of photosynthetically produced food can be tracked with the essential FA's FA's20:5ω3 (eicosapentaenoic acid), 22:5ω3 (docosahexaenoic acid), and 20:4ω6 (arachidonic acid) (Copeman et al., 2002; De Troch et al., 2012).
Molecular analysis of stomach contents provides an additional tool within trophic studies, as it allows the identification of what an organism has most recently consumed, including microbial taxa (sensu; Niemann et al., 2013). At seep sites, molecular gut content analysis can identify consumption of bacteria and archaea that dominate biogeochemical cycling at seeps including: sulfate reducing, sulfide oxidizing, and methanotrophic bacteria and methane oxidizing archaea (Knittel and Boetius, 2009; Valentine, 2011). With this method, Niemann et al. (2013) was able to identify a dominance of sulfide-oxidizing Epsilonproteobacteria in the guts of a lithoid crab, linking the microbial community of a microbial mat at a methane seep to the crabs diet. When used in conjunction with fatty acid and stable isotopic analysis, a holistic view of an individual's diet can be obtained. Each of the methods integrate an animals diet over different temporal scales and have the potential to parse out otherwise hard to constrain trophic linkages between highly mobile animals and chemosynthesis-based ecosystems. Here, we use bulk stable isotope, compound specific isotope analysis of Fatty Acid Methyl Esters (FAME), and molecular gut content analysis, to test whether C. tanneri is incorporating chemosynthetic production into their biomass. Further, we compare how each of these three methods reflect the diet of C. tanneri differently.
Methods
Study Site and Sampling
Samples were collected from the methane seep habitats at Clayoquot Slope (48°40′15″ N, 126°50′52″ W; depth: 1250 m), a site within the long-term seafloor monitoring sites of Ocean Networks Canada's (ONC) NEPTUNE cabled observatory (Barnes et al., 2010). Sampling occurred during two of ONC's yearly maintenance cruises with ROV Jason aboard R/V TG Thompson (Cruise TN328, Sept 2015), and ROV Hercules aboard E/V Nautilus (Cruise NA071, May-June 2016). Crab traps were deployed at Clayoquot Slope during both of these expeditions to collect C. tanneri. Once the traps were received on deck the C. tanneri specimens (n = 11 for 2015 collection; n = 12 for 2016 collection) were frozen at −80°C until further processing. Push cores were collected during these two expeditions, as well, from seep sites at Clayoquot Slope and the nearby Barkley Canyon, where a mass migration event of C. tanneri was observed in 2014, and non-seep sites at Cascadia Basin within the observatory network (site information in Supplementary Table 1). These cores were vertically sectioned at 0–1, 1–2, 2–3, and 3–5 centimeter intervals and preserved at −80°C until further processing.
Isotope Analysis
Bulk Isotope Analysis
The muscle tissue from the walking legs of the C. tanneri specimens was extracted from the specimen in the laboratory to measure δ13C, δ15N, and δ34S signatures. Walking leg tissues of C. tanneri specimens were dried at 60°C for 48 h. We additionally isolated tissue from the gut wall and gills of a subset of C. tanneri specimens. All tissues were dried at 60°C for 48 h, and acidified with 12 μl of 1 M HCl (Thurber et al., 2010). Sediment samples were freeze dried and treated with 2N phosphoric acid to remove carbonates.
δ13C and δ15N analysis
Dried tissue samples (n = 23; walking leg tissue, guts, and gills) and sediment samples (Clayoquot Slope: 4 cores; Barkley Canyon: 3 cores; Cascadia Basin: 4 cores) were packaged in tin capsules (0.3–0.7 mg dry weight) and analyzed using a Costech (Valencia, CA USA) elemental analyzer interfaced with a continuous flow Micromass (Manchester, UK) Isoprime isotope ratio mass spectrometer (EA-IRMS) for 15N/14N and 13C/12C ratios at Washington State University. Measurements are reported in δ notation [per mil (‰) units] using ovalbumin as a routine standard. Precision for δ13C and δ15N was generally ±0.2 and ±0.4‰. Isotopic ratios are expressed relative to standards (Pee Dee Belemnite for δ13C and atmospheric nitrogen gas for δ15N).
δ34S analysis
Dried walking leg tissue samples (n = 10, 5 from 2015 and 5 from 2016; 3–5 mg dry weight) were packaged in tin capsules for mass spectrometry and sent to UC Davis Stable Isotope Laboratory for δ34S analysis. Samples were measured following their standard procedures on an Elementar vario ISOTOPE cube interfaced to a SerCon 20–22 IRMS (Sercon Ltd., Cheschire UK). The precision for the method employed at this facility is ±0.4‰.
Compound Specific Isotope Analysis
To determine the quantity, type, and source of lipids assimilated into the biomass of C. tanneri, the Fatty Acid Methyl Esters (FAME) and associated C isotope signatures were analyzed. A direct transesterification procedure was performed to extract the FAME from the freeze-dried walking leg tissue of the C. tanneri specimens (n = 23; following Thurber, 2015). FAMEs were quantified and identified using retention times on a Thermo Fisher Scientific TRACE 1310 gas chromatography—flame ionization detector (GC-FID) with a Thermo TR5 (length: 60 m, I.D.: 0.32 mm) column. Identification of compounds was in comparison to known standards (Supelco 37 Component FAME mix, Sigma Aldrich). GC-Mass spectrometer analysis on a Hewlett Packer (HP) 6890 gas chromatography system with a DB1 (length: 30 m, I.D.: 0.25 mm) column and a HP 5973 mass selective detector was used to confirm peak identify. Dimethyl disulfide (DMDS) adducts [derived following (Nichols et al., 1986)] were used to identify mono-unsaturated bond locations and also run on this instrument. Blanks were run with all batches, and all solvents were ACS grade or better. Fatty-Acids (FA) under 14 carbon lengths, and non-FA peaks were not included in analysis.
Compound specific isotope analysis on three selected FAME extracts (selected based on the corresponding gut contents that had high sequencing success) were analyzed with a GC-c-IRMS at the UC Davis Stable Isotope Laboratory on an Agilent 6890 gas chromatograph, coupled to Thermo MAT 253 through a GC-C-III combustion interface on a BPX-70 SGE (length: 60 m, O.D.: 0.25 mm) column. Isotopic concentrations were corrected for the C used in the methanol as part of the transesterification.
Gut Content Analysis
The gut contents of the C. tanneri specimens (n = 11, 2015 collection only) and the sediment cores were sequenced to identify the microbial taxa present at the time of collection. The gut contents were separated from the gut tissue, and DNA was extracted from both the separated gut contents and the gut wall and residue (pieces of the gut tissue that residue from the gut contents remained on). DNA was extracted from sediment cores and gut contents using the DNeasy PowerSoil Kit (Mobio/Qiagen) following manufacturer's instructions. Extracted DNA was PCR-amplified using 515f and 806r (Caporaso et al., 2012; Apprill et al., 2015) primers as in Seabrook et al. (2018). PCR products were sequenced on the Illumina MiSeq platform at the Center for Genome Research and Biocomputing (CGRB) at Oregon State University. Resulting sequences were processed using a custom pipeline (Supplementary Materials) that involved mothur v39, Usearch7 (64 bit), and QIIME v. 1.9.1 (Edgar, 2010; Schloss et al., 2011). Sequences were clustered at 97% Operational Taxonomic Unit (OTU0.03), and assigned by comparison to the Silva v123 database formatted for QIIME (https://www.arb-silva.de/download/archive/qiime/). Aligned sequences were rarefied to the least abundant quality-filtered sequences per sample, after omitting those with uncharacteristically low sequencing success (3690 sequences per sample for this project; resulting in n = 3 gut contents, n = 6 gut wall and residue). The 16S rRNA gene sequences are archived in the National Center for Biotechnology Information public database under BioProject ID PRJNA494176.
Bray-Curtis similarity comparisons on log transformed rarified OTU0.03's were used to generate non-metric multidimensional scaling (NMDS) plots and cluster diagrams which were used to visualize patterns in the community structure. PERmutation Multivariate ANalysis Of VAriance (PERMANOVA; McArdle and Anderson, 2012) was used to identify significant differences between sites and crab gut contents. The taxa responsible for differences among groups and the level of similarity between sites and gut contents were measured using the SIMilarity PERrcentage Analysis (SIMPER; Clarke and Warwick, 2001). All multidimensional analyses were performed using the software package PRIMER v7 with the PERMANOVA+ add on (Clarke and Gorley, 2015).
Results
Bulk Isotopes
Bulk tissue stable isotope results indicated that C. tanneri relied on photosynthetically derived food sources as its main form of nutrition. The δ13C values of the crab tissue ranged between −17.4 and −20.5‰, and δ15N values ranged between 10.7 and 16.1‰ (Figure 2A; Supplementary Table 2). Of the three different tissue types analyzed, the only significant differences were between the δ13C values of the muscle and gill tissues (paired t-test, p = 0.005), and the δ15N values of the gill and gut tissues (paired t-test, p ≤ 0.005). A false discovery rate (0.05) correction was applied to correct for multiple comparisons. There was a significant difference in the bulk 13C of mature vs. juvenile C. tanneri (determined from carapace width; < 8.5 and >8.5 cm; t-test, p = 0.044; Supplementary Table 2; Keller et al., 2016). Clayoquot Slope and Barkley Canyon sediment δ13C values averaged −22.1 ± 0.1 and −22.3 ± 0.2‰, respectively, and Cascadia Basin δ13C values averaged −21.6 ± 0.2‰. Values were averaged across vertical fraction of sediment and then averaged across replicates within site, standard error is reported across replicates within site. δ34S values for the C. tanneri tissues ranged between 17.9 and 20.6‰, reflecting background (non-seep) seawater sulfur (Figure 2B; Supplementary Table 1). There was no indication of the assimilation of matter derived from methanotrophic or thiotrophic sources in the bulk isotopic values measured, as would be indicated by significantly lower isotopic values.
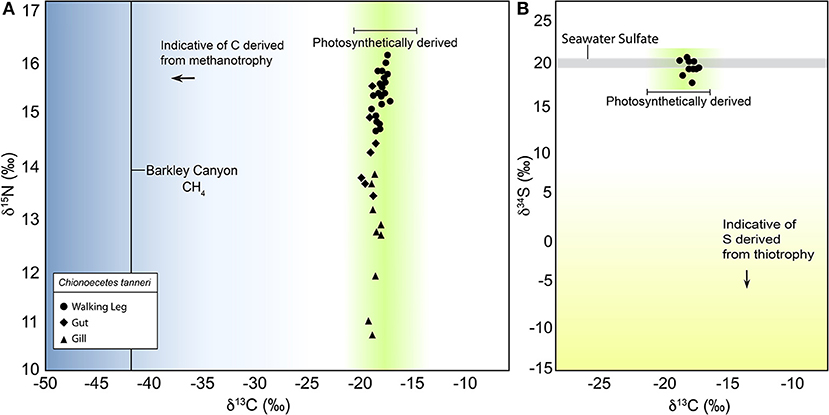
Figure 2. Bulk stable isotope signatures of the different C. tanneri tissue analyzed. In (A) δ13C δ15N indicating the values of C. tanneri in relation to expected values of photosynthetic production. The black line indicates the δ13C value of methane at the nearby Barkley Canyon methane seep, and the blue gradient indicates the range that methane derived carbon would be expected in. (B) δ13C in relation to δ34S for C. tanneri where the gray line indicates the δ34S range for background (non-seep) seawater sulfate, the yellow gradient indicates the range that seep derived sulfide compounds would be expected in. In both (A,B) the green gradient is the range photosynthetically derived carbon is expected in.
Compound Specific Isotopes and FAME Identification
Diagnostic biomarkers for seep bacteria were found in all three crabs, including 16:1ω6 and 18:1ω8c (aerobic methanotrophs), and 16:1ω5 and 18:1ω7c (sulfate reducing bacteria). These later FAs are not specific to sulfate reducing bacteria but are common biomarkers for sulfate reducing bacteria. Essential FA's 20:5ω3 (eicosapentaenoic acid), 22:5ω3 (docosahexaenoic acid), and 20:4ω6 (arachidonic acid) were very abundant in all three crabs measured and had δ13C values associated with photosynthetic sources. The three crab specimens analyzed had fatty acids that were 13C depleted relative to photosynthetic sources (Figure 3). In crab 3, the fatty acids that showed 13C depletion were 16:1ω6 (δ13C = −38.8‰), 18:3 FA's (δ13C = −38.6 and −34.7‰), 19:0 (δ13C = −33.1‰), and 19:1ω12 (δ13C = −33.7‰). Additionally, in crab 7, the 17:1ω6 FA was depleted (δ13C = −33.2‰). The more depleted δ13C values were not found across all specimen studied, indicating that this depletion is not due to isotopic fractionation during synthesis of the FA within the organisms, but its isotopic composition is sourced from the diet.
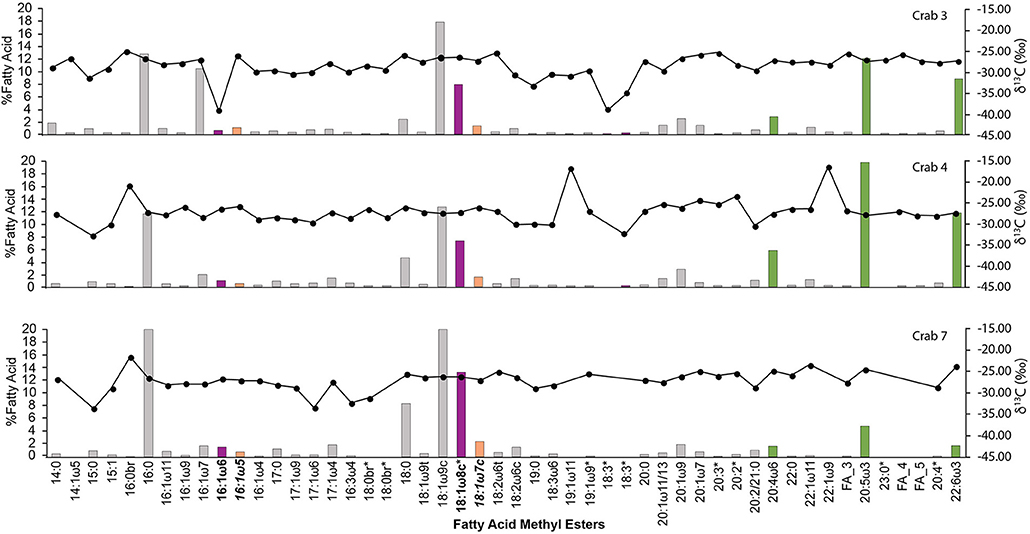
Figure 3. The fatty acid (FA) distribution and carbon isotope composition of FAs extracted from C. tanneri muscle tissue. Left y-axis and bars represent percentage of FA, while right axis and points are the isotopic composition. Pink bars with FAs in bold have been associated with methanotrophic bacteria, orange bars with FAs in italic bold have been associated with sulfate reducing bacteria, and green bars are essential fatty acids linked to photosynthetic sources. Asterisks represent instance where FA compounds were inferred from mass spectra.
Gut Content Analysis
Seep-associated bacteria and archaea were found in the gut contents of C. tanneri specimen (Figures 4A,B). Seep associated bacterial taxa comprised >80% of the stomach contents in crab 7 and were found in smaller numbers (~3%) in crabs 3, 2, and 9 as well. These bacterial taxa included: sulfate reducing Desulfobulbus, Desulfocapsa, Seep-SRBs 1, 2, and 4; methanotrophic Methylococcales; and sulfide oxidizing Sulfurovum and Thiotrichales (mat-forming). Anaerobic methane oxidizing archaea (ANME) comprised up to 100% of the archaeal taxa found in the guts of the C. tanneri specimen. The seep-associated bacteria and archaea that were found in the C. tanneri gut contents were found in the sediment from both the Barkley Canyon and Clayoquot Slope methane seep sites, but were rare or absent in the Cascadia Basin non-seep sites (Seabrook et al., 2018; Supplementary Tables 5, 6). The seep associated bacteria and archaea that were found in C. tanneri guts comprised 4.34% of all sequences found in the seep sediment, while only comprising 0.05% of the sequences found in the non-seep sediment. Two non-seep associated bacterial clades, Mycoplasmatales and Entomoplasmatales, were also dominant in the crab gut contents and gut walls with Mycoplasmatales abundant in all but Crab 4's gut microbiome; in this crab Entomoplasmatales was instead the dominant bacterial group. These groups were very common in the gut contents but rare or absent in the seep sediment. SIMPER analysis revealed that these two taxonomic groups were dominant drivers in the observed dissimilarity between the microbial community in the C. tanneri guts and the seep and non-seep sediments (responsible for up to 1.16% of observed dissimilarity between the C. tanneri gut and sediment samples; Supplementary Tables 7–10).
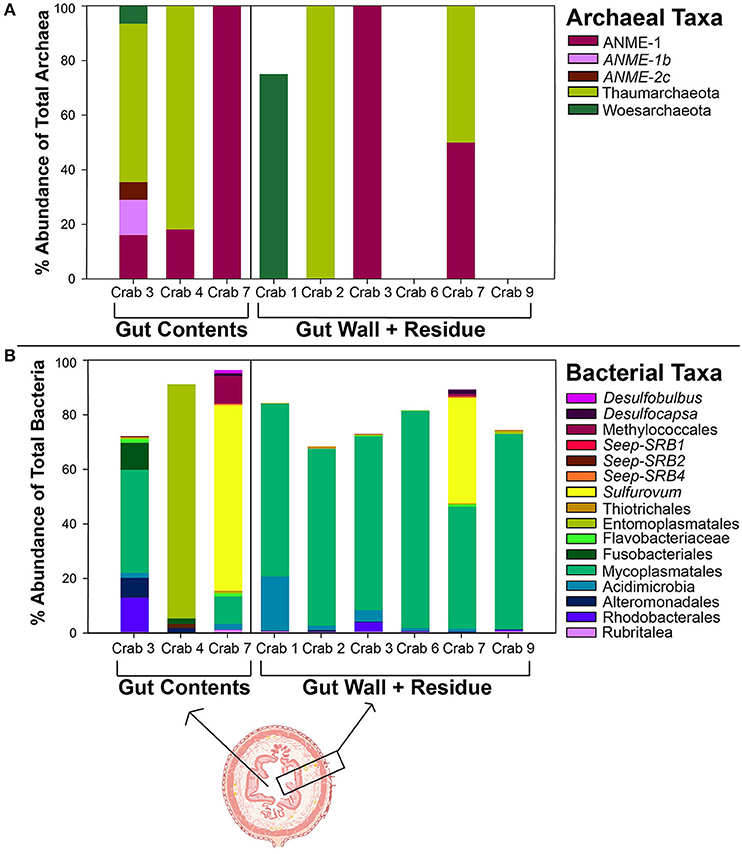
Figure 4. The percent abundance of the (A) archaea and (B) bacteria isolated from the gut contents and gut wall and residue of C. tanneri. In (A) only the five most dominant archaeal groups are shown, and in (B) the 16 most dominant bacterial groups are shown. The order is given for the bacterial and archaeal groups, except in italicized instances when genus is given for clarity.
The seep, non-seep, gut content, and gut wall microbial communities were all distinct from each other (Supplementary Figure 1, PERMANOVA; pseudo-F:8.323, p-value = 0.001). Similarity numbers provided are a result of this PERMANOVA test. The gut wall microbial community across the specimen analyzed were 25.3% similar to each other. The gut wall microbial community shared more similarity with the gut content microbial communities (at 21.9% similarity) than was observed within the gut content microbial community itself (at 16.3% similarity). The dissimilarity among the microbial communities isolated from the gut contents was largely driven by Crab 4's gut community which was dominated by the Entomoplasmatales group. It was possible to compare the microbial community isolated from the gut contents and gut wall in only two of the C. tanneri specimen (Crab 7 and Crab 3). The microbial community isolated from Crab 7's gut contents and gut walls clustered together, sharing >40% similarity, whereas the microbial community isolated from Crab 3's gut content was more distinct from the microbial community associated with the gut wall, sharing only 25% similarity (Supplementary Figure 2).
Discussion and Conclusions
We found multiple lines of evidence that demonstrated that the C. tanneri were grazing upon seep nutrition, although it was not reflected in bulk isotopic measurements. Although the FA signatures revealed a dominance of photosynthetically derived carbon in the crab tissue, there were also multiple bacterial FAs associated with chemosynthetic processes and instances of depletion in 13C associated with methane-derived carbon within particular FAs. The high abundance of seep-associated microbes in some C. tanneri guts provides more resolution to these conflicting results, showing the direct ingestion of seep associated bacteria and archaea. This is strengthened by the increased abundance of these specific seep-associated bacteria and archaea in the seep sediment compared to the non-seep sediment.
While greater sample size could have provided a better quantification of the variability among crabs that occur at these seeps, the samples analyzed provided concrete evidence of methane seep usage by C. tanneri. Although the gut contents from all crabs collected in 2015 were extracted and sequenced, only the gut contents of three specimen and the gut wall and residue of 6 specimen passed our quality control and sequencing depth cut offs. In many cases, amplification was repeatedly attempted on the other individuals with no increased success. Gut contents can be difficult to sequence, especially in trap caught organisms. This is due to a combination of the crabs being trapped for a period of time during which they may have evacuated their gut and not had the ability to feed or digestive enzymes degrading present (Symondson, 2002; King et al., 2008; Liu et al., 2015). Durand et al. (2010) showed that 72 h or less was enough time for the guts of Rimicaris exoculata to be completely cleared of transient bacteria, so the time between some of the C. tanneri specimen entering the traps and on ship preservation may have been long enough for this to occur. Despite sample size limitations, the molecular gut content analysis and compound specific isotopes did reveal the ingestion of seep associated microbes and the assimilation of seep-derived nutrition by some of the C. tanneri specimen. Behavioral observations also indicate a link between C. tanneri and the seep habitat that suggest our findings based on a few specimens are likely common within the species. The persistent dense population of C. tanneri at Clayoquot Slope contrasts with the highly migratory nature of this species (Keller et al., 2012, 2016) and provides an additional indication that C. tanneri use the seep habitat. In addition, C. tanneri are normally separated geographically by size classes and sex yet there were multiple size classes and both sexes present at Clayoquot Slope (Supplementary Table 2). Mass migration events have also been observed near the seep, including a mass migration event at the nearby Barkley Canyon seep (Figure 1c; Doya et al., 2017). The sustained high density of C. tanneri at the Clayoquot Slope seep is coupled with observations of the crabs feeding amongst the seep sediment and microbial mats. Considered with the compound specific isotope and molecular gut content results, it is evident that the crabs are getting some trophic support from the seep site, despite their dominant nutritional pathway appearing to be sourced from photosynthetic-detritus.
The variability within and among the microbial communities isolated from the gut contents and gut walls illustrates natural heterogeneity in the environment, the differences in time since last meal and capture, and potential variability in the resident communities of the C. tanneri gut microbiome. Although we were only able to compare the gut contents and gut walls of two of the specimens, it was interesting that Crab 7 had similarity between the two gut portions while Crab 3 did not. This is potentially a result of Crab 7's gut walls having more gut content residue left on the gut wall. The two non-seep associated bacterial clades that were dominant in the crab gut contents (Mycoplasmatales and Entomoplasmatales) are commensal or pathogenic bacteria found in many animal species including crustaceans and are likely not from the diet (Wang et al., 2004; Ku et al., 2013; Terahara et al., 2017). These two groups were also shown to be two of the biggest drivers of the dissimilarity between the microbial communities in the gut contents and in the sediment from the seep and non-seep sites, supporting that these groups are resident and not transient in the C. tanneri guts. Similarly, Durand et al. (2010) found these two groups to be prominant components of the resident gut microbiome in Rimicaris exoculata specimens, and hypothesized that they are ubiquitous members of crustacean guts. The proportion of Mycoplasmatales and Entomoplasmatales varied among the crabs studied, with Mycoplasmatales dominant in all samples except the gut contents of Crab 4 where Entomoplasmatales comprised ~90% of the bacterial community present. Stark differences between the microbial communities within mobile consumers, such as C. tanneri may be a result of ecosystem and community influence on the composition of animal gut microbiomes. These differences may be an indication of new arrivals vs. longer term site residents that have had more time for genetic exchange to occur between specimens. Similar to the microbial exchange that occurs between human populations, the aggregation of the highly mobile C. tanneri at the Clayoquot Slope seep could be leading to genetic dispersal between formerly distinct communities and habitats (Pehrsson et al., 2016).
The compound specific isotope analysis of FAMEs extracted from C. tanneri muscle tissue revealed mixed signals, but a clear incorporation of specific compounds that are derived from chemosynthetic biomass is evident. Specifically, in the 16:1ω6 and 18:3 FA's (δ13C = −38.8 and −38.6‰, respectively). Although these signatures are not as depleted in 13C as has been noted previously, they are around 10‰ more depleted in 13C than would be expected from photosynthetic sources and are similar to values (i.e., δ13C = −37 to −40‰ δ13C) that have been linked to chemosynthetic bacterial biomass in similar studies (Boshker and Middelburg, 2002; Niemann et al., 2013). In addition the methane that is present at these sites has been previously identified as thermogenic in origin based on its heavy carbon isotopic signature (δ13C = −30 to −43‰; Pohlman et al., 2009, 2011) which would also lead to a heavier δ13C signal from methane-derived carbon. It is important to note that, due to more similar isotopic values between thermogenic methane and photosynthetic production, the ability to easily quantify the role of seeps in the diet of migratory grazers is further hampered at seeps fueled by this heavier methane.
A long-standing debate is whether or not seeps act as oases on continental margins, providing habitat and trophic subsidies to non-seep organisms (MacAvoy et al., 2005; Jørgensen and Boetius, 2007; Guilini et al., 2012). An important aspect of this is whether vagrant species (i.e., non-seep endemic) that are observed at chemosynthesis-based ecosystems consume food produced from either methanotrophic or thiotrophic processes. Bulk isotopes have been used as a one tool to track sources of primary production in consumers, however, isotopic mixing between chemosynthetic and photosynthetic sources can confound results and complicate interpretation of trophic linkages. Seeps are composed of multiple habitats with their own microbial taxa and biomarkers (Levin and Michener, 2002; Boetius and Suess, 2004; Bernardino and Smith, 2010) many of which have non-distinct, or contrasting, bulk SI signatures that reflect the heterogenous nature of seep habitats. This is even clear in the bulk isotopic analysis of sediment presented here, with δ13C values of −22.1 and −22.3‰ that fall within the isotopic range typically indicative of photosynthetic production (Fry and Sherr, 1989; Levin and Michener, 2002). This may be a result of photosynthetic detrital (including refractory) inputs overwhelming the seep signature, microbial fixation pathways that result in heavy isotopic signatures (Hügler and Sievert, 2011), or that thermogenic methane fuels this particular seep. Yet, similar values have been reported in near-seep sediments (e.g., Demopoulos et al., 2018 measured δ13C of −23.3‰) and even seep macrofaunal from within areas of very active seepage often have values that are not depeated in 13C (e.g., Levin and Michener, 2002 measured δ13C of −22.2‰; Levin et al., 2013 measured δ13C as low as −22.6‰; Thurber et al., 2010 measured δ13C as low as −18.5‰). Quantifying the assimilation of chemosynthetic production with bulk isotopes has proved challenging in previous studies, as well. The incorporation of bacterial chemoautotrophic production by grazing gastropods and deposit-feeding sipunculids in the GoM was obscured by more enriched isotopic values that ranged from −32 to −20‰ δ13C and −14 to −1‰ δ 35S due to the mixing with photosynthetic production (MacAvoy et al., 2005). In a key paper, (MacAvoy et al., 2003) found significant input of chemosynthetic production into Sclorasterias cf. tanneri (an Asteroid), Etpatretus sp. (a hag fish), and Rochina crassa (a spider crab) in the GoM using compound specific analysis of fatty acids however this was only reflected in the bulk δ13C of the less mobile sea star S. tanneri, while the more mobile Eptatretus sp. and R. crassa were both enriched in 13C (−20.9 and −18.4‰, respectively). Interestingly, an Eptatretus sp. that was caught in a non-seep environment was slightly more enriched in 13C than the Eptatretus sp. caught at the seep site (by 1‰), potentially reflecting a small signal from seep derived carbon. Similarly, in this study Crab 3, which had the most depleted compound specific δ13C signals, was ~1‰ more depleted in 13C than the other crabs measured; this may also reflect a slight signal from seep derived carbon. Complimenting the results in MacAvoy et al. (2003, 2005), while compound specific isotope analysis of fatty acids and gut content analysis indicated that C. tanneri specimen were assimilating seep derived carbon, the bulk isotope data did not. Our results suggest that bulk isotopes may be under-representing the assimilation of seep production in some fauna.
A contributing factor to the under-representation of seep derived nutrition in bulk isotope data in our study may also be the use of C from different food sources for anabolic vs. catabolic processes, likely driven by the quality vs. quantity of food sources. On margins, various food pools exist including particulate organic matter from surface productivity, organic falls, and localized chemoautotrophic production at seeps and vents (Levin and Dayton, 2009; Levin and Sibuet, 2012; Sweetman et al., 2017). Each of these habitats and food pools contribute to margin ecosystems in a variety of ways and are different in terms of quantity (seep production) and quality (fresh phytodetritus). Previous work has indicated that in habitats with multiple trophic pathways, food quantity and quality can interact to lead to preferential assimilation of high-quality food types, even if it is in lower quantity (Persson et al., 2007; Hessen, 2008; Marcarelli et al., 2011). Mobile consumers have been shown to selectively feed on more high-quality matter when both high-quality and low-quality matter was offered, while sedentary consumers did not show such a discrimination (Cruz-Rivera and Hay, 2008). Detrital matter has been shown to be of higher quality, or more nutritious, than the direct production in habitats, such as coral reefs and kelp forests, and thus often assimilated in a higher proportion than less nutritious direct production (Campanyà-Llovet et al., 2017). The same could be true in methane seep habitats, where the base of production is facilitated by bacteria, which do not produce the essential FAs shown to be critical in a metazoans diet (Copeman et al., 2002; Zuo et al., 2012). The enriched δ13C and δ35S signatures in the bulk isotopes may be reflecting a preferential assimilation of higher quality detrital matter by the C. tanneri specimen studied. Supporting this, we do see a higher abundance of the essential FA's (20:5ω3, 22:5ω3, and 20:4ω6) relative to the bacterial produced 16:1 and 18:1 FA's in the crab tissues. Food quantity and quality effects, such as these may disproportionately affect ecosystem processes and skew food web analysis that is based on those compounds assimilated (i.e., C, N, S isotopes, and FAs; Marcarelli et al., 2011).
Another major factor that could be influencing the diet of species as resolved by biomarker techniques is the temporal resolution of the different techniques employed. Bulk stable isotopes have turnover times of >400 days in some invertebrate heterotrophs (MacAvoy et al., 2001), which could be leading to an underrepresentation of seep nutrition due to the highly mobile and migratory nature of the C. tanneri. Individual fatty acids have differing turnover times, but have been shown to in general be on the order of a few days to a few weeks (Ruess and Chamberlain, 2010). As our analytical techniques increase in power and precision, we are becoming more able to resolve mixed signals in trophic studies that are due to spatial and temporal factors, particularly through the integration of multiple techniques. Analysis of fatty acids allowed us to separate out specific isotope signatures from the bacterial produced FA's and the more abundant essential FA's sourced from photosynthetic production. These compound specific isotopes of FA's can also reveal more short-term assimilation than possible with bulk isotopes due to the shorter turnover times of FA's, potentially allowing us to capture short term use of a new food source by an organism (MacAvoy et al., 2001; Ruess and Chamberlain, 2010; Bec et al., 2011). The analysis of the gut contents of the crabs allowed us to observe the most recent food source consumed by C. tanneri and dietary connections that were missed in the bulk isotopic methods with longer turnover times.
The implications of mobile species like C. tanneri being able to use seep habitats extends beyond the habitat provisions that the crabs derive directly from the seep itself. The use of a seep by highly mobile consumers like C. tanneri could intensify cross-boundary movements of methane sourced carbon and genetic material. This directly contributes to the “sphere of influence” of a seep habitat on the surrounding margin ecosystem (sensus; Levin et al., 2016). The ecological significance of energy transfers between habitats has been highlighted in other systems, such as with the annual returns of anadromous fishes to rivers shown to bring marine-derived nutrients that sustain the productivity of freshwater food webs (Chaloner et al., 2002). Interactions are considered ecologically important when cross-boundary trophic subsidies are assimilated by the consumers in the respective habitats (Carpenter et al., 2005). Here, we show that C. tanneri are assimilating cross-boundary subsides and thus have the potential to be agents in the dispersal of nutrients due to their highly mobile and migratory nature. This could benefit background (non-seep) ecosystems by delivering allochthonous food, as well as distributing genetic material (seep bacteria and archaea in their gut) across the margin and potentially linking disparate seep systems.
Climate change is predicted to reduce the total input of photosynthetic production to the deep sea, with direct impacts on the biomass of deep-sea fauna (Jones et al., 2014; Sweetman et al., 2017). As a result, the use of alternative sources of production may become increasingly important to mitigate impacts of shortages of surface production (Smith et al., 2008; Levin and Sibuet, 2012). In the often food limited deep-sea, the ability for chemosynthetic production to serve as a trophic subsidy for non-seep species, as it has now been shown in the shallow ocean (Higgs et al., 2016), could help support the health and productivity of ocean environments. Thus, the ability for non-seep organisms to use chemosynthetic production as a trophic subsidy could have implications as we seek to spatially manage ecosystems in a way that promotes sustainable and productive ocean environments. This study provides the first evidence of seep derived food sources contributing to the diet of commercially exploited non-seep species, highlighting the potential importance of these environments for providing a provisioning ecosystem service. More robust analysis on the trends presented herein will aid in further understanding cross-boundary assimilations of chemosynthetic production. However, those studies will be faced with the challenges of dealing with the methodological limitations we highlight, which may preclude the application of mixing models to quantify the role of seeps in margin ecosystems, among other things. The use of multiple methods, as presented here, may prove critical to parsing out discrete signals from mixed settings and understanding complex food webs with both photosynthetic and chemosynthetic food sources. While we may still debate whether seeps act as oases in the deep, this study makes it clear that certain lines of evidence could under-represent the role of seeps and highlights the potential for seeps to act as critical oases to non-seep endemic fauna in the future. In the case of C. tanneri, seep production may serve as a trophic subsidy that could help to buffer the impact of climate change on a commercially harvested marine species.
Author Contributions
SS, FDL, and AT conceived and designed the experiments. SS performed the experiments. SS and AT analyzed the data. SS, FDL, and AT contributed reagents, materials, analysis tools. SS, FDL, and AT wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the Canada Foundation for Innovation (Major Science Initiative Fund 30199) for ship and ROV time devoted to the collection of the Chionoecetes tanneri specimen used in this study during two of Ocean Networks Canada-NEPTUNE observatory maintenance cruises (TN328, NA071). We are indebted to the generosity of ONC in providing samples to facilitate this study. We further thank Dr. Miguel Goni, who gave both his time and expertise to help with GC/MS runs and mass-spectra analysis. We also thank our reviewers for their insightful comments and suggestions. A portion of SS time in the preparation of this manuscript was supported by the U.S. National Science Foundation Office of Polar Programs 1642570 and Division of Ocean Sciences 1635913 awards to AT. A portion of this project was supported by the International Network for Scientific Investigation of Deep-Sea Ecosystems (INDEEP) supported by Foundation Total.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00043/full#supplementary-material
Supplementary Videos 1 and 2. Remotely Operated Vehicle video footage of tanner crabs ‘farming’ on methane bubbles at Clayoquot Slope seep (Supplementary Video 1), and an areal overview showing the patched nature of bacterial mats and methane bubbling at Clayoquot Slope seep (Supplementary Video 2). Both video clips have been edited increasing its original frame rate to 150% in some instances. Original footage from ROPOS R1555 dive, on June 3, 2012, is available through Ocean Networks Canada's free data web portal https://data.oceannetworks.ca/SeaTube?resourceTypeId=1000&resourceId=1001&diveId=219&time=2012-06-03T14:00:00.000Z, and time stamp ranges from 15:51:34 to 16:03:02 UTC.
References
Apprill, A., Mcnally, S., Parsons, Rs., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Armstrong, C. W., Foley, N., Tinch, R., and van den Hove, S. (2010). Ecosystem goods and services of the deep sea. Deliv. D6.2 Hermione Proj. 1–68. Available online at: https://www.pik-potsdam.de/news/public-events/archiv/alter-net/former-ss/2010/13.09.2010/van_den_hove/d6-2-final.pdf
Barnes, C. R., Best, M. M., Johnson, F. R., and Pirenne, B. (2010). Final installation and initial operation of the world's first regional cabled ocean observatory (NEPTUNE Canada). Can. Met. Ocean. Soc. 38, 89–96.
Bec, A., Perga, M. E., Koussoroplis, A., Bardoux, G., Desvilettes, C., Bourdier, G., et al. (2011). Assessing the reliability of fatty acid-specific stable isotope analysis for trophic studies. Methods Ecol. Evol. 2, 651–659. doi: 10.1111/j.2041-210X.2011.00111.x
Bell, K. L., Flanders, J., Bowman, A., and Raineault, N. A. (2017). New frontiers in ocean exploration: the E/V nautilus, NOAA ship okeanos explorer, and R/V falkor 2016 field season. Oceanography 30, 1–94. doi: 10.5670/oceanog.2017.supplement.01
Bernardino, A. F., and Smith, C. R. (2010). Community structure of infaunal macrobenthos around vestimentiferan thickets at the San Clemente cold seep, NE Pacific. Mar. Ecol. 31, 608–621. doi: 10.1111/j.1439-0485.2010.00389.x
Boecklen, W. J., Yarnes, C. T., Cook, B. A., and James, A. C. (2011). On the use of stable isotopes in trophic ecology. Annu. Rev. Ecol. Evol. Syst. 42, 411–440. doi: 10.1146/annurev-ecolsys-102209-144726
Boetius, A., and Suess, E. (2004). Hydrate ridge: a natural laboratory for the study of microbial life fueled by methane from near-surface gas hydrates. Chem. Geol. 205, 291–310. doi: 10.1016/j.chemgeo.2003.12.034
Boshker, H. T., and Middelburg, J. J. (2002). Stable isotopes and biomarkers in microbial ecology. FEMS Microbiol. Ecol. 40, 85–95. doi: 10.1111/j.1574-6941.2002.tb00940.x
Bowman, J. P., Skerratt, J. H., Nichols, P. D., and Sly, L. I. (1991). Phospholipid fatty acid and diplopolysaccharide fatty acid signature lipids in methane-utilizing bacteria. FEMS Microbiol. Ecol. 85, 15–22. doi: 10.1111/j.1574-6968.1991.tb04693.x
Campanyà-Llovet, N., Snelgrove, P. V. R., and Parrish, C. C. (2017). Rethinking the importance of food quality in marine benthic food webs. Prog. Oceanogr. 156, 240–251. doi: 10.1016/j.pocean.2017.07.006
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Carpenter, S. R., Cole, J. J., Pace, M. L., Bogert, M., Van, D.e, Bade, D. L., et al. (2005). Ecosystem subsidies: terrestrial support of aquatic food webs from 13 C addition to contrasting lakes. Ecology 86, 2737–2750. doi: 10.1890/04-1282
Chaloner, D. T., Martin, K. M., Wipfli, M. S., Ostrom, P. H., and Lamberti, G. A. (2002). Marine carbon and nitrogen in southeastern Alaska stream food webs: evidence from artificial and natural streams. Can. J. Fish. Aquat. Sci. 59, 1257–1265. doi: 10.1139/f02-084
Clarke, K. R., and Gorley, R. N. (2015). Getting Started with PRIMER v7 Plymouth Routines In Multivariate Ecological Research. Available online at: http://www.primer-e.com
Clarke, K. R., and Warwick, R. M. (2001). A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Mar. Ecol. Prog. Ser. 216, 265–278. doi: 10.3354/meps216265
Copeman, L. A., Parrish, C. C., Brown, J. A., and Harel, M. (2002). Effects of docosahexaenoic, eicosapentaenoic, and arachidonic acids on the early growth, survival, lipid composition and pigmentation of yellowtail flounder (Limanda ferruginea): a live food enrichment experiment. Aquaculture 210, 285–304. doi: 10.1016/S0044-8486(01)00849-3
Cordes, E. E., Cunha, M. R., Galéron, J., Mora, C., Olu-Le Roy, K., Sibuet, M., et al. (2010). The influence of geological, geochemical, and biogenic habitat heterogeneity on seep biodiversity. Mar. Ecol. 31, 51–65. doi: 10.1111/j.1439-0485.2009.00334.x
Cruz-Rivera, E., and Hay, M. E. (2008). Can quantity replace quality ? food choice, compensatory feeding, and fitness of marine mesograzers. Ecology 81, 201–219. doi: 10.1890/0012-9658(2000)081<0201:CQRQFC>2.0.CO;2
De Troch, M., Boeckx, P., Cnudde, C., Van Gansbeke, D., Vanreusel, A., Vincx, M., et al. (2012). Bioconversion of fatty acids at the basis of marine food webs: insights from a compound-specific stable isotope analysis. Mar. Ecol. Prog. Ser. 465, 53–67. doi: 10.3354/meps09920
Demopoulos, A. W. J., Bourque, J. R., Durkin, A., and Cordes, E. (2018). The influence of seep habitats on sediment macrofaunal biodiversity and functional traits. Deep Sea Res. Part I Oceanogr. Res. Pap. 142, 1–17. doi: 10.1016/j.dsr.2018.10.004
Doya, C., Chatzievangelou, D., Bahamon, N., Purser, A., De Leo, F. C., Juniper, S. K., et al. (2017). Seasonal monitoring of deep-sea megabenthos in Barkley Canyon cold seep by internet operated vehicle (IOV). PLoS ONE 12:e0176917. doi: 10.1371/journal.pone.0176917
Durand, L., Zbinden, M., Cueff-Gauchard, V., Duperron, S., Roussel, E. G., Shillito, B., et al. (2010). Microbial diversity associated with the hydrothermal shrimp Rimicaris exoculata gut and occurrence of a resident microbial community. FEMS Microbiol. Ecol. 71, 291–303. doi: 10.1111/j.1574-6941.2009.00806.x
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Fry, B., and Sherr, E. B. (1989). “δ13C Measurements as indicators of carbon flow in marine and freshwater ecosystems,” in Stable Isotopes in Ecological Research, eds. P. W. Rundel, J. R. Ehleringer, and K. A. Nagy (New York, NY: Springer New York), 196–229.
Grupe, B. M., Krach, M. L., Pasulka, A. L., Maloney, J. M., Levin, L. A., and Frieder, C. A. (2015). Methane seep ecosystem functions and services from a recently discovered southern California seep. Mar. Ecol. 36, 91–108. doi: 10.1111/maec.12243
Guilini, K., Levin, L. A., and Vanreusel, A. (2012). Cold seep and oxygen minimum zone associated sources of margin heterogeneity affect benthic assemblages, diversity and nutrition at the Cascadian margin (NE Pacific Ocean). Prog. Oceanogr. 96, 77–92. doi: 10.1016/j.pocean.2011.10.003
Hessen, D. O. (2008). Efficiency, energy and stoichiometry in pelagic food webs; reciprocal roles of food quality and food quantity. Freshw. Rev. 1, 43–57. doi: 10.1608/FRJ-1.1.3
Higgs, N. D., Newton, J., and Attrill, M. J. (2016). Caribbean spiny lobster fishery is underpinned by trophic subsidies from chemosynthetic primary production. Curr. Biol. 26, 3393–3398. doi: 10.1016/j.cub.2016.10.034
Hügler, M., and Sievert, S. M. (2011). Beyond the calvin cycle: autotrophic carbon fixation in the ocean. Ann. Rev. Mar. Sci. 3, 261–289. doi: 10.1146/annurev-marine-120709-142712
Jones, D. O., Yool, A., Wei, C. L., Henson, S. A., Ruhl, H. A., Watson, R. A., et al. (2014). Global reductions in seafloor biomass in response to climate change. Glob. Chang. Biol. 20, 1861–1872. doi: 10.1111/gcb.12480
Jørgensen, B. B., and Boetius, A. (2007). Feast and famine–microbial life in the deep-sea bed. Nat. Rev. Microbiol. 5, 770–781. doi: 10.1038/nrmicro1745
Keller, A. A., Buchanan, J. C., Steiner, E., Draper, D., Chappell, A., Frey, P., et al. (2016). Size at maturity for grooved tanner crab (Chionoecetes tanneri) along the U.S. west coast (Washington to California). Fish. Oceanogr. 25, 292–305. doi: 10.1111/fog.12155
Keller, A. A., Harms, J. H., and Buchanan, J. C. (2012). Distribution, biomass and size of grooved Tanner crabs (Chionoecetes tanneri) from annual bottom trawl surveys (2003-2010) along the U.S. west coast (Washington to California). Deep. Res. Part I Oceanogr. Res. Pap. 67, 44–54. doi: 10.1016/j.dsr.2012.05.002
King, R. A., Read, D. S., Traugott, M., and Symondson, W. O. C. (2008). Molecular analysis of predation: a review of best practice for DNA-based approaches. Mol. Ecol. 17, 947–963. doi: 10.1111/j.1365-294X.2007.03613.x
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334. doi: 10.1146/annurev.micro.61.080706.093130
Kruse, G. H., Gallucci, V. F., Hay, D. E., Perry, R. I., Peterman, R. M., and Shirley, T. C., et al (eds.). (2005). Fisheries assessment and management in data-limited situations. in Proceedings of the Symposium Assessment and Management of New and Developed Fisheries in Data-Limited Situations (Fairbanks), 958. doi: 10.4027/famdis.2005.04
Ku, C., Lo, W. S., Chen, L. L., and Kuo, C. H. (2013). Completegenomesoftwodipteran-associatedspiroplasmas provided insights into the origin, dynamics, and impacts of viral invasion in Spiroplasma. Genome. Biol. Evol. 5, 1151–1164. doi: 10.1093/gbe/evt084
Levin, L. A. (2005). Ecology of cold seep sediments: interactions of fauna with flow, chemistry and microbes. An Annu. Rev. 43, 1–46. doi: 10.1201/9781420037449.ch1
Levin, L. A., Baco, A. R., Bowden, D. A., Colaco, A., Cordes, E. E., Cunha, M. R., et al. (2016). Hydrothermal vents and methane seeps: rethinking the sphere of influence. Front. Mar. Sci. 3:72. doi: 10.3389/fmars.2016.00072
Levin, L. A., and Dayton, P. K. (2009). Ecological theory and continental margins: where shallow meets deep. Trends Ecol. Evol. 24, 606–617. doi: 10.1016/j.tree.2009.04.012
Levin, L. A., and Michener, R. H. (2002). Isotopic evidence for chemosynthesis-based nutrition of macrobenthos: the lightness of being at pacific methane seeps. Limnol. Oceanogr. 47, 1336–1345. doi: 10.4319/lo.2002.47.5.1336
Levin, L. A., and Sibuet, M. (2012). Understanding continental margin biodiversity: a new imperative. Ann. Rev. Mar. Sci. 4, 79–112. doi: 10.1146/annurev-marine-120709-142714
Levin, L. A., Ziebis, W., F., Mendoza, G., Bertics, V. J., and Washington, T., Gonzalez, J., et al. (2013). Ecological release and niche partitioning under stress: lessons from dorvilleid polychaetes in sulfidic sediments at methane seeps. Deep. Res. Part II Top Stud. Oceanogr. 92, 214–233. doi: 10.1016/j.dsr2.2013.02.006
Liu, Y., Zhang, Y., Dong, P., An, R., Xue, C., Ge, Y., et al. (2015). Digestion of nucleic acids starts in the stomach. Sci. Rep. 5, 1–11. doi: 10.1038/srep11936
MacAvoy, S. E., Carney, R. S., Fisher, C. R., and Macko, S. A. (2002). Use of chemosynthetic biomass by large, mobile, benthic predators in the Gulf of Mexico. Mar. Ecol. Prog. Ser. 225, 65–78. doi: 10.3354/meps225065
MacAvoy, S. E., Fisher, C. R., Carney, R. S., and Macko, S. A. (2005). Nutritional associations among fauna at hydrocarbon seep communities in the Gulf of Mexico. Mar. Ecol. Prog. Ser. 292, 51–60. doi: 10.3354/meps292051
MacAvoy, S. E., Macko, S. A., and Carney, R. S. (2003). Links between chemosynthetic production and mobile predators on the Louisiana continental slope: Stable carbon isotopes of specific fatty acids. Chem. Geol. 201, 229–237. doi: 10.1016/S0009-2541(03)00204-3
MacAvoy, S. E., Macko, S. A., and Garman, G. C. (2001). Isotopic turnover in aquatic predators: quantifying the exploitation of migratory prey. Can. J. Fish. Aquat. Sci. 58, 923–932. doi: 10.1139/f01-045
MacAvoy, S. E., Morgan, E., Carney, R. S., and Macko, S. A. (2008). Chemoautotrophic production incorporated by heterotrophs in gulf of mexico hydrocarbon seeps: an examination of mobile benthic predators and seep residents. J. Shellfish Res. 27, 153–161. doi: 10.2983/0730-8000(2008)27<153:CPIBHI>2.0.CO;2
Marcarelli, A. M., Baxter, C. V., Mineau, M. M., and Hall, R. O. (2011). Quantity and quality: unifying food web and ecosystem perspectives on the role of resource subsidies in freshwaters. Ecology 92, 1215–1225. doi: 10.1890/10-2240.1
McArdle, B. H., and Anderson, M. J. (2012). Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82, 290–297. doi: 10.1890/0012-9658(2001)082<0290:FMMTCD>2.0.CO;2
Nichols, P. D., Guckert, J. B., and White, D. C. (1986). Determination of monosaturated fatty acid double-bond position and geometry for microbial monocultures and complex consortia by capillary GC-MS of their dimethyl disulphide adducts. J. Microbiol. Methods 5, 49–55. doi: 10.1016/0167-7012(86)90023-0
Niemann, H., Linke, P., Knittel, K., MacPherson, E., Boetius, A., Brückmann, W., et al. (2013). Methane-carbon flow into the benthic food web at cold seeps–a case study from the costa rica subduction zone. PLoS ONE 8, 4–13. doi: 10.1371/journal.pone.0074894
Pehrsson, E. C., Tsukayama, P., Patel, S., Mejía-Bautista, M., Sosa-Soto, G., Navarrete, K. M., et al. (2016). Interconnected microbiomes and resistomes in low-income human habitats. Nature 533, 212–216. doi: 10.1038/nature17672
Persson, J., Brett, M. T., Vrede, T., and Ravet, J. L. (2007). Food quantity and quality regulation of trophic transfer between primary producers and a keystone grazer (Daphnia) in pelagic freshwater food webs. Oikos 116, 1152–1163. doi: 10.1111/j.0030-1299.2007.15639.x
Pohlman, J. W., Bauer, J. E., Waite, W. F., Osburn, C. L., and Chapman, N. R. (2011). Methane hydrate-bearing seeps as a source of aged dissolved organic carbon to the oceans. Nat. Geosci. 4:37–41. doi: 10.1038/ngeo1016
Pohlman, J. W., Kaneko, M., Heuer, V. B., Coffin, R. B., and Whiticar, M. (2009). Methane sources and production in the northern Cascadia margin gas hydrate system. Earth Planet. Sci. Lett. 287, 504–512. doi: 10.1016/j.epsl.2009.08.037
Rex, M. A., Stuart, C. T., Hessler, R. R., Allen, J. A., Sanders, H. L., and Wilson, G. D. F. (1993). Global-scale latitudinal patterns of species diversity in the deep sea benthos. Nature 365, 636–638. doi: 10.1038/365636a0
Riedel, M., Scherwath, M., Römer, M., Veloso, M., Heesemann, M., and Spence, G. D. (2018). Distributed natural gas venting offshore along the Cascadia margin. Nat. Commun. 9:3264. doi: 10.1038/s41467-018-05736-x
Ruess, L., and Chamberlain, P. M. (2010). The fat that matters: soil food web analysis using fatty acids and their carbon stable isotope signature. Soil Biol. Biochem. 42, 1898–1910. doi: 10.1016/j.soilbio.2010.07.020
Schloss, P. D., Gevers, D., and Westcott, S. L. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16s rRNA-based studies. PLoS ONE 6:e27310. doi: 10.1371/journal.pone.0027310
Seabrook, S. C., De Leo, F., Baumberger, T., Raineault, N., and Thurber, A. R. (2018). Heterogeneity of methane seep biomes in the Northeast Pacific. Deep. Res. Part II Top. Stud. Oceanogr. 150, 195–209. doi: 10.1016/j.dsr2.2017.10.016
Sellanes, J., Pedraza, M. J., and Zapata Hernandez, G. (2012). Las areas de filtracion de metano constituyen zonas de agregación del bacalao de profundidad (Dissostichus eleginoides) frente a Chile central. Lat. Am. J. Aquat. Res. 40, 980–991. doi: 10.3856/vol40-issue4-fulltext-14
Smith, C. R., De Leo, F. C., Bernardino, A. F., Sweetman, A. K., and Arbizu, P. M. (2008). Abyssal food limitation, ecosystem structure and climate change. Trends Ecol. Evol. 23, 518–528. doi: 10.1016/j.tree.2008.05.002
Somerton, D. A., and Donaldson, W. (1996). Contribution to the biology of the grooved and triangle Tanner crabs, Chionoecetes tanneri and C. angulatus, in the eastern Bering Sea. Fish. Bull. 94, 348–357.
Sweetman, A. K., Thurber, A. R., Smith, C. R., Levin, L. A., Mora, C., Wei, C.-L., et al. (2017). Major impacts of climate change on deep-sea benthic ecosystems. Elem. Sci. Anth. 5:4. doi: 10.1525/elementa.203
Symondson, W. O. (2002). Molecular identification of prey in predator diets. Mol. Ecol. 11, 627–641. doi: 10.1046/j.1365-294X.2002.01471.x
Terahara, N., Tulum, I., and Miyata, M. (2017). Transformation of crustacean pathogenic bacterium Spiroplasma eriocheiris and expression of yellow fluorescent protein. Biochem. Biophys. Res. Commun. 487, 488–493. doi: 10.1016/j.bbrc.2017.03.144
Thurber, A. R. (2015). Diet-dependent incorporation of biomarkers: Implications for food-web studies using stable isotope and fatty acid analyses with special application to chemosynthetic environments. Mar. Ecol. 36, 1–17. doi: 10.1111/maec.12192
Thurber, A. R., Kröger, K., Neira, C., Wiklund, H., and Levin, L. A. (2010). Stable isotope signatures and methane use by New Zealand cold seep benthos. Mar. Geol. 272, 260–269. doi: 10.1016/j.margeo.2009.06.001
Thurber, A. R., Levin, L. A., Orphan, V. J., and Marlow, J. J. (2012). Archaea in metazoan diets: implications for food webs and biogeochemical cycling. ISME J. 6, 1602–1612. doi: 10.1038/ismej.2012.16
Thurber, A. R., Levin, L. A., Rowden, A. A., Sommer, S., Linke, P., and Kröger, K. (2013). Microbes, macrofauna, and methane: a novel seep community fueled by aerobic methanotrophy. Limnol. Oceanogr. 58, 1640–1656. doi: 10.4319/lo.2013.58.5.1640
Thurber, A. R., Sweetman, A. K., Narayanaswamy, B. E., Jones, D. O. B., Ingels, J., and Hansman, R. L. (2014). Ecosystem function and services provided by the deep sea. Biogeosciences 11, 3941–3963. doi: 10.5194/bg-11-3941-2014
Valentine, D. L. (2011). Emerging topics in marine methane biogeochemistry. Ann. Rev. Mar. Sci. 3, 147–171. doi: 10.1146/annurev-marine-120709-142734
Van Dover, C. L. (2007). Stable isotope studies in marine chemoautotrophically based ecosystems: an update. Stable Isot. Ecol. Environ. Sci. 2, 202–237. doi: 10.1002/9780470691854.ch8
Wang, Y., Stingl, U., Anton-Erxleben, F., Geisler, S., Brune, A., and Zimmer, M. (2004). “Candidatus Hepatoplasma crinochetorum,” a new, stalk-forming lineage of mollicutes colonizing the midgut glands of a terrestrial isopod. Appl. Environ. Microbiol. 70, 6166–6172. doi: 10.1128/AEM.70.10.6166-6172.2004
Wei, C. L., Rowe, G. T., Fain Hubbard, G., Scheltema, A. H., Wilson, G. D. F., Petrescu, I., et al. (2010). Bathymetric zonation of deep-sea macrofauna in relation to export of surface phytoplankton production. Mar. Ecol. Prog. Ser. 399, 1–14. doi: 10.3354/meps08388
Zuo, R., Ai, Q., Mai, K., Xu, W., Wang, J., Xu, H., et al. (2012). Effects of dietary docosahexaenoic to eicosapentaenoic acid ratio (DHA/EPA) on growth, non-specific immunity, expression of some immune related genes and disease resistance of large yellow croaker (Larmichthys crocea) following natural infestation of paras. Aquaculture 334–337, 101–109. doi: 10.1016/j.aquaculture.2011.12.045
Keywords: stable isotopes, fisheries, methane seep, microbiology, compound specific, fatty acid, chemosynthesis, trophic subsidy
Citation: Seabrook S, De Leo FC and Thurber AR (2019) Flipping for Food: The Use of a Methane Seep by Tanner Crabs (Chionoecetes tanneri). Front. Mar. Sci. 6:43. doi: 10.3389/fmars.2019.00043
Received: 27 October 2018; Accepted: 28 January 2019;
Published: 19 February 2019.
Edited by:
Ana Colaço, Instituto de Investigação Marinha (IMAR), PortugalReviewed by:
Lucia Bongiorni, Istituto di Scienze Marine (ISMAR), ItalyPeter Deines, University of Kiel, Germany
Copyright © 2019 Seabrook, De Leo and Thurber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah Seabrook, c2VhYnJvb3NAb3JlZ29uc3RhdGUuZWR1
 Sarah Seabrook
Sarah Seabrook Fabio C. De Leo
Fabio C. De Leo Andrew R. Thurber
Andrew R. Thurber