
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 14 January 2019
Sec. Coastal Ocean Processes
Volume 5 - 2018 | https://doi.org/10.3389/fmars.2018.00512
This article is part of the Research Topic Marine Microbiome and Biogeochemical Cycles in Marine Productive Areas View all 13 articles
Global warming is having a great impact on the Arctic region, due to the change of air temperature and precipitation. As a consequence, the glacial ice melts and englacial materials are being transported into the ocean. These substances can constitute a source of nutrients in food webs or, on the contrary, a source of contaminants. In this research seven marine Svalbard glaciers and their tidewater tongues were focused. This survey provides a first attempt comparing microbial communities from coastal and tidewater glaciers that reveal a hitherto unknown microbial diversity. A wider diversity was found in glaciers than in seawater samples. Glacier microorganisms mainly corresponded to the phylum Proteobacteria (48.8%), Bacteroidetes (29.1%) and Cyanobacteria (16.3%) (Figure 3A). Seawater microorganisms belonged to Bacteroidetes (40.3%), Actinobacteria (31.7%) and Proteobacteria (25.4%). Other phyla found such as Firmicutes, Thermi, Gemmatimonadetes, Verrucomicrobia, Nitrospirae, Chloroflexi, Planctomycetes, and Chlamydiae were less abundant. The distribution of microbial communities was affected in different extent by the concentration of nutrients (nitrogen nutrients, dissolved organic carbon and soluble reactive phosphorus) and by environmental parameters such as salinity. Nevertheless, the environmental variables did not influence in the distribution of the microbial communities as much as the concentration of nutrients did. Our results demonstrate an interchange between glacier and coastal microbial populations as well as the presence of some indicator species (i.e., Hymenobacter) as possible sentinels for bacterial transport between glaciers and their downstream seawaters. The consequence of this process could be the alteration of the water composition of the fiords producing serious consequences throughout the marine ecosystem and in the cycling of globally important elements.
Glaciers have been considered authentic biomes (Anesio and Laybourn-Parry, 2012), in which microbial life goes by enduring a hostile environment. Microbial communities associated with glaciers found all over the world, are being increasingly studied. Specifically, these microorganisms have been reported in Polar Arctic (Amato et al., 2007; Hell et al., 2013; Larose et al., 2013) and Antarctic Regions (Foreman et al., 2007) and in high mountains (Garcia-Descalzo et al., 2013; Hotaling et al., 2017). However, glaciers are very different from each other, which influence the life of their inhabitants. The effect of environmental variables, of chemical composition (Møller et al., 2011) and nutrient concentration on microbial communities have been extensively researched (Mindl et al., 2007).
The Svalbard archipelago (74° to 81° N) (Figure 1) comprise an area of 62,248 km2, with more than 2,000 glaciers; and ice covers 60% of its surface (Lang, 2011). Svalbard glaciers are of various kinds (Hagen et al., 1993; Hagen et al., 2003), although most of them are sub-polar or polythermal, sharing the characteristics of the cold and temperate glaciers (Hagen et al., 1993). Some of them have a coastal margin and terminate in a calving front, what establishes important differences with respect to glaciers with inland margin. Coastal glaciers calve icebergs into the ocean (McNabb et al., 2016). These glaciers greatly affect the physical and chemical characteristics of the sea to which they discharge (Garcia-Lopez et al., 2016). In glaciers ending on land, there is continuous permafrost at ice front; while calving glaciers present partly or completely temperate tidewater tongues (Hagen et al., 2003).
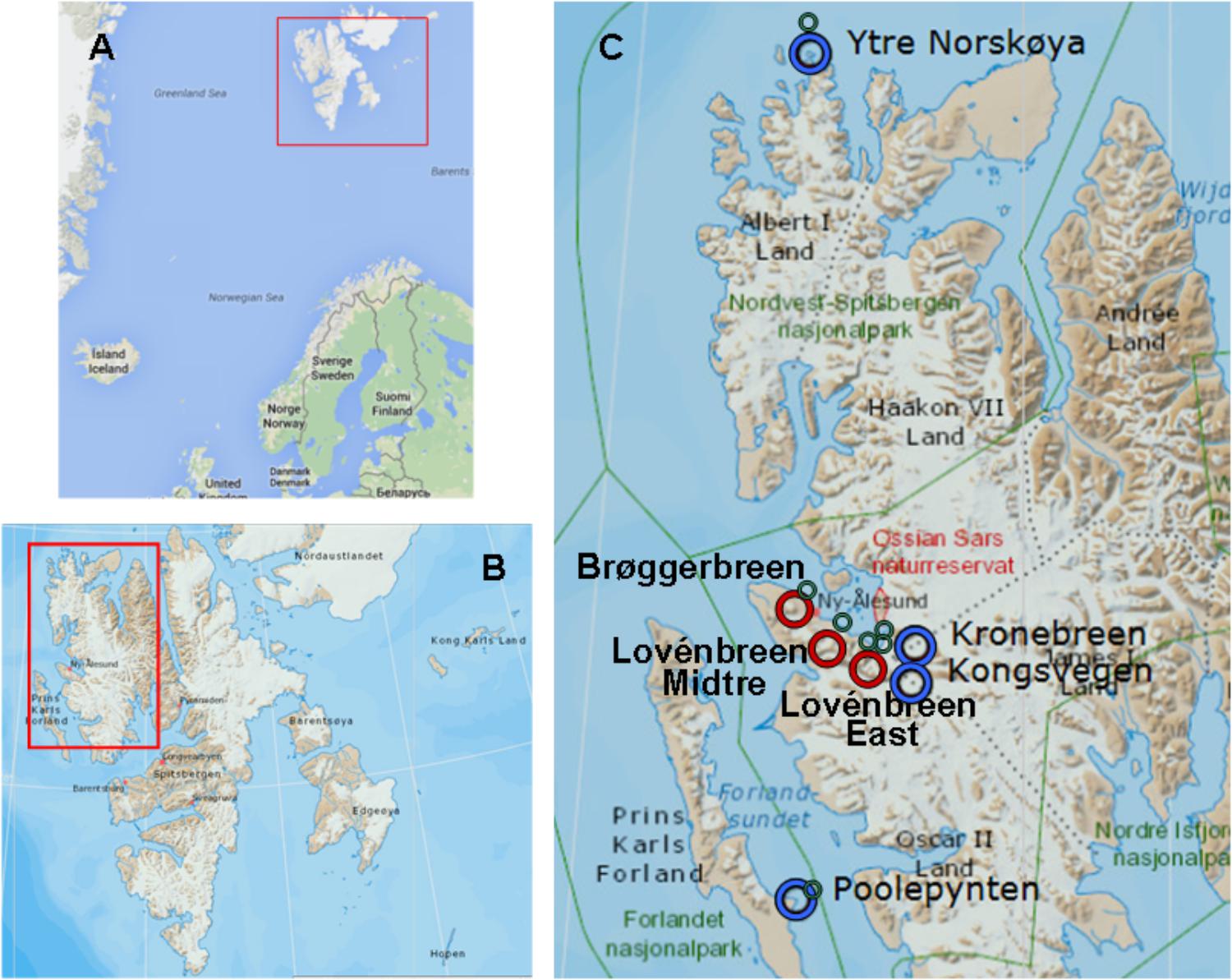
Figure 1. Geographic maps the Arctic Svalbard archipelago. (A) Svalbard archipelago. (B) Spitsbergen and Prins Karls Forland. (C) Map showing the distribution of ice samples collected at marine glaciers (blue rings), inland glaciers (red rings) and tidewaters tongues (green rings). Image sources: Google maps (A) and TopoSvalbard (Copyright Norwegian Polar Institute) (B,C).
Global warming is nowadays one of the most worrying problems. It is having a great impact on the Arctic region, because of the change of air temperature and precipitation (Solomon et al., 2007; Førland et al., 2009). As a consequence, the glacial ice melts and disappears, and microbial communities are being seriously affected (Hallbeck, 2009; Zeng et al., 2013). One of the effects of global warming is the change in the basal temperature of the ice, moving from cold to polythermal; this causes the growth of new microorganisms that are not psychrophiles, thus leading to changes in the diversity (Hell et al., 2013; Nowak and Hodson, 2014). It is essential to know how climate change is shaping the distribution and diversity of microbial communities, as microorganisms participate particularly in the ecology of the Arctic ecosystems. They constitute the basis of trophic networks and they are an essential element in carbon and nutrient cycling (Hoham and Duval, 2001; Garcia-Descalzo et al., 2013).
In the last few decades, recently deglaciated areas present in different glacial zones in the world, are available for colonization and primary succession, especially initiated by pioneer microorganisms (Kastovska et al., 2005; Deiglmayr et al., 2006). It has been researched that in glacier forelands, soil microorganisms are essential for plant growth as they play a key role in the nutrient cycling. In this phase, nitrogen, phosphorus, and other nutrients accumulate and facilitate succeeding plant growth (Garcia-Lopez and Cid, 2017; Kim M. et al., 2017). Similarly, in seawater, these microorganisms could also contribute to the development and evolution of marine trophic webs.
The role of microorganisms in polar environments is essential (Kirchman et al., 2009). They are responsible for carrying out key functions in the ecosystems. For instance, phosphorus is a major growth-limiting nutrient, which can be made biologically available by a group of heterotrophic microorganisms (Khan et al., 2009). Other important nutrients such as natural polysaccharides from plants and algae (cellulose, and laminarin) are degraded by some bacteria such as Cytophaga and a few Clostridium species (Lopez-Ramirez et al., 2015). The denitrification or reduction of NO3- to N2, NO, or N2O, is the main means by which N2 and N2O are biologically formed. Nitrification (the oxidation of NH3 to NO3-) is a main process in oxic environments, and it is carried out by the nitrifying bacteria (Madigan et al., 2012). Sulfate-reducing bacteria are also a highly diverse group. Many reduced sulfur compounds are used as electron donors by sulfur bacteria. The most common sulfur compounds used as electron donors are H2S, S0, SO32-. Hydrogen sulfide H2S is oxidized by chemolithotrophic microorganisms to sulfur S0 and sulfate SO42-. SO42- can then be reduced to H2S by activities of the sulfate-reducing bacteria (organisms that consume organic carbon) and this reduction closes the biogeochemical sulfur cycle while regenerating CO2 (Madigan et al., 2012).
With the aim of studying whether a process of colonization analogous to what happens in soils can be initiated in the coastal glaciers, the microbial populations of seven glaciers and their corresponding discharge seawaters were studied. The studied glaciers were: Lovénbreen Midtre (LVM), Lovénbreen Austre (LBE), Kongsvegen (KGV), Kronebreen (KNB), Brøggerbreen (BGN), Ytre Norskøya (YTRE) (Hamiltonbukta), and Poolepynten (POOL) (Archibald Geikiebreen) (Figure 2). Of these seven glaciers, two are more distant (YTRE and POOL) and the other five form a glacial circus. One of them (BGN) is close to a populated area, which can also condition the microbial communities. In addition, some are marine and others are terrestrial, and each has its particular physico-chemical characteristics. The study of such microorganisms involved the sequencing of the 16S rRNA genes to identify bacteria. The chemical composition of ice and seawater was also one of the main goals of study. Once the microorganisms in the samples were identified, their distribution and relationship with the chemical composition of glacial ice and seawater were determined.

Figure 2. Images of the seven Svalbard glaciers. (A) Lovénbreen Midtre, Sherdahlfjellet (LVM); (B) Lovénbreen Austre, Slatlofjellet (LBE); (C) Kongsvegen (KGV); (D) Kronebreen (KNB); (E) Brøggerbreen (BGN); (F) Ytre Norskøya, Hamiltonbukta (YTRE); (G) Poolepynten, Archibald Geikiebreen (POOL). GPS Coordinates: Kongsvegen: 78°51′44″N 12°32′29″E; Kronebreen 78°52′23″N 12°39′14″E; Ytre Norskøya: 79°47′11″N 11°44′14″E; Poolepynten: 78°27′29″N 11°32′55″E; Lovénbreen Midtre: 78°53′12″N 12°2′57″E; Lovénbreen Austre: 78°53′51″N 11°49′15″E; Brøggerbreen: 78°54′49″N 11°43′42″ E.
Two types of samples were obtained, during summer 2014, from seven Svalbard glaciers and their corresponding nearest tidewater (Figures 1, 2). The glaciers considered in the study were: Lovénbreen Midtre, Sherdahlfjellet (LVM); Lovénbreen Austre, Slatlofjellet (LBE); Kongsvegen (KGV); Kronebreen (KNB); Brøggerbreen (BGN); Ytre Norskøya, Hamiltonbukta (YTRE); Poolepynten, Archibald Geikiebreen (POOL). Four glaciers, KGV, KNB, YTRE and POOL, present all or part of the glacier front in the sea, and are termed “coastal glaciers”; the rest are termed “inland glaciers.” Samples were named as: CG, glacial ice samples from coastal glaciers (KGV, KNB, YTRE, and POOL); IG, glacial ice samples from inland glaciers (LVM, LBE, and BGN); CT, seawater samples from the tidewater area closest to a coastal glacier; and IT, seawater samples from the tidewater area closest to an inland glacier. GPS coordinates of the sampling areas are detailed in Table 1.
Glacial ice samples were obtained, by removing 20–30 cm of thick debris and by drilling with a Mark II Kovacs Core System. Each ice core of 9 cm × 3 m was cut into three parts to obtain three sampling replicates. Glacial ice samples (a total of 21) were wrapped in sterile plastic bags, transported in ice and stored at -20°C until analyzing in the laboratory at the Kings Bay Research Station (Ny Ålesund, Svalbard). Then, ice samples were decontaminated following a surface decontamination and melting procedure described in previous studies (Garcia-Descalzo et al., 2013). In a UV-irradiated laminar flow hood, each section of block ice was removed from -20°C and soaked in ice-cold 95% ethanol for 1 min, followed by extensive rinsing with 0.22 μm-filtered MilliQ water, effectively ablating the exterior 2-cm shell of ice samples. The decontaminated interior ice was thawed in a sterile plastic bag at 4°C and used for analyses.
Tidewater samples (3 sampling replicates, a total of 21) were obtained at the seaside, in the nearest point from each glacier. They were collected in Niskin bottles at a depth of 2 m as in Zaikova et al. (2009), transported and immediately filtered in the laboratory.
Both, the meltwater and tidewater samples were individually filtered through filters with pores of 0.22 μm attached to a vacuum pump in a flow hood, previously sterilized with ethanol. Both filters and water were stored at -20°C until use in the laboratory at the Centro de Astrobiología (Madrid, Spain). To control for laboratory contamination, 500 ml of MilliQ water were subjected to identical analytical procedures.
Basic measurements of physical and chemical parameters of meltwater from sampling sites were made with a temperature-calibrated pH, conductivity, and salinity meter (WTW, Weilheim, Germany). Assays for dissolved nitrogen and phosphorus (NH4+, NO2-, NO3-, TDN, SRP, and DOC) were performed by ion chromatographic method using suppressed conductivity detection (Garcia-Descalzo et al., 2013) and by HPLC-Size Exclusion Chromatography with UV and On-Line DOC Detection (Her et al., 2002) in a 861 Advance Compact IC system (Metrohm AG, Herisau, Switzerland). Chromatograms were recorded using the Metrohm IC Net 2.3 SR6 software.
Genomic DNA from each sampling replicate (500 ml) was extracted by using the DNA Isolation PowerWater kit (MO BIO Laboratory, Inc.). Extraction procedures were identical for all samples. DNA concentration was determined using a Nanodrop 2000p.
Then, Illumina sequencing was applied to each sampling replicate. Purified DNA was quantified and 1ng of input DNA was used in a first PCR of 20 cycles with Q5® Hot Start High-Fidelity DNA Polymerase (New England Biolabs) in the presence of 100 nM primers of the regions V3 and V4 of the 16S rRNA gene. Primers sequences were: (V3-V4) 341F, 5′-ACACTGACGACATGGTTCTACACCTACGGGNGGCWGCAG-3′ and (V3-V4) 805R, 5′-TACGGTAGCAGAGACTTGGTCTGACTACHVGGGTATCTAATCC-3′ (Herlemann et al., 2011). After the first PCR, a second PCR of 15 cycles was carried out with Q5® Hot Start High-Fidelity DNA Polymerase (New England Biolabs) in the presence of 400 nM of primers 5′-AATGATACGGCGACCACCGAGATCTACACTGACGACATGGTTCTACA-3′ and 5′-CAAGCAGAAGACGGCATACGAGAT-[10 nucleotides barcode]-TACGGTAGCAGAGACTTGGTCT-3′ of the Access Array Barcode Library for Illumina Sequencers (Fluidigm). Amplicons were validated and quantified by Bioanalyzer and an equimolecular pool was purified using AMPure beads and titrated by quantitative PCR using the Kapa-SYBR FAST qPCR kit for LightCycler 480 and a reference standard for quantification. The pool of amplicons were denatured prior to be seeded on a flowcell at a density of 10 pM, where clusters were formed and sequenced using a MiSeq Reagent Kit v3, in a 2 × 300 pair-end sequencing run on a MiSeq sequencer to obtain 100000 reads per sample approximately.
Sequencing data were analyzed with the Base Space platform. All DNA sequences obtained have been deposited in NCBI Short Read Archive (SRA).
A rarefaction analysis was performed using Analytic Rarefaction 1.3 software1 (Tipper, 1979).
Statistical differences on the number of sequences and number of OTUs were studied by ANOVA test. Data of sequences and OTUs are media values of three sampling replicates. Significant differences between two types of samples were identified using Student’s t-test. All of the statistical analyses were performed using GraphPad Prism version 7.00 (GraphPad Software, La Jolla, CA, United States)2.
Effects of the type of sample, nutrient concentrations and environmental variables on the bacterial community composition were investigated by a combination of multivariate statistical analysis developed with CANOCO version 5 software (Microcomputer Power, Ithaca, NY, United States) (Jongman et al., 1995; Ter Braak, 1995). For statistical analysis, Monte Carlo permutation tests with 500 permutations were used.
Most of the physico-chemical characteristics of glacier samples were very analogous to each other (Tables 1, 2). These samples differed mainly on their salinity, since the samples from coastal glaciers had a much higher salinity than those from inland glaciers.
Nevertheless, it was observed that seawater samples corresponding to the two types of glaciers were very different in terms of their physico-chemical characteristics. IT samples are combined with the runoff waters dragged from glaciers located inland. These waters transport inorganic and organic matter from the snow or soil they pass through in their way to the sea. However, as coastal glaciers emit icebergs directly into the sea, waters of these tidewater tongues (CT samples) contain less organic matter and less inorganic salts in solution. Their different physico-chemical characteristics can affect the microbial populations these samples contain.
Soluble nitrogen nutrients were analyzed in the forms of NH4+, NO2-, NO3-, and TDN. The occurrence of the different forms of ammonia depends on pH.
In an aqueous medium, ammonia (NH3) and ammonium (NH4+) ions take part in an equilibrium reaction. Ammonia reacts as a base, raising the pH by generating OH- ions. This equilibrium depends on pH and temperature. At a lower pH, the concentration of NH3 is lower (Weiner, 2008). At the observed pH of between 4 and 5, about 95% of ammonia is in the cationic form ammonium (NH4+). Highly variable contents were obtained for each glacier (Table 2). Samples from KNB showed the highest amounts of NH4+, NO2-, and NO3- while the highest TDN values were found in LVM, LBE and KNB. The soluble reactive phosphorous (SRP) ranged between 0.51 μM in Brøggerbreen Vestre and 1.55 μM in LBE. No differences with respect to these parameters could be established between samples from coastal or land margin neither between ice or seawater samples.
In glaciers, the movement of carbon from the atmosphere to the rocks and moraines begins with snow precipitation. Atmospheric carbon combines with water to form carbonic acid that falls to the glacier surface. The acid dissolves rocks and releases several ions such as calcium, magnesium, potassium, or sodium. Calving glaciers transport these ions (Zeebe and Caldeira, 2008), carbon particles and organic matter contained in ice to their downstreaming tidewater (Sipler et al., 2017). Storage and release of organic carbon from glaciers have been extensively studied (Hood et al., 2015); and it has been calculated that climate change contributes to this release approximately 13% of the annual flux of glacier dissolved organic carbon as a result of glacier mass loss. In coastal glacier samples, DOC concentrations were very similar to DOC values in their downstreaming seawater samples. But DOC concentrations were significantly higher in seawater samples than they were in inland glacier samples. These amounts are comparable to previously described DOC concentrations for glaciers and ice sheets (Hood et al., 2015), and for Arctic seawater (Benner et al., 2005). All results about the chemical composition of ice and seawater will be discussed in relationship to the structure of microbial communities in glaciers.
In order to identify the microbial communities that inhabit the glacial and seawater samples, 16S rRNA gene sequencing was performed by Illumina MiSeq. A total of 695307 sequences were obtained (Supplementary Table S1). The correspondence of sequences to OTUs was assessed considering a 97% identity threshold. According to Supplementary Figure S1, corresponding to the seven glacier samples and seven tidewater samples, graphs tended to an asymptotic behavior. At 3% sequence divergence rarefaction curves reached saturation, indicating that the surveying attempt covered almost the full extent of taxonomic diversity at this genetic distance.
Statistical analyses of microbial abundance were only run on OTUs that were present in two or more sampling point. Glacier microorganisms mainly corresponded to the phylum Proteobacteria (48.8%), Bacteroidetes (29.1%) and Cyanobacteria (16.3%) (Figure 3A). Seawater microorganisms belonged to Bacteroidetes (40.3%), Actinobacteria (31.7%), and Proteobacteria (25.4%). Other phyla found such as Firmicutes, Thermi, Gemmatimonadetes, Verrucomicrobia, Nitrospirae, Chloroflexi, Planctomycetes, and Chlamydiae were less abundant. The total number of sequences found in glaciers and seawater was significantly different (∗∗∗p < 0.0001) (Figure 1A). When abundances were compared at the phylum level, significant differences were also observed for Cyanobacteria, Proteobacteria, and Actinobacteria (∗∗∗p < 0.0001). It has been published that these bacterial phylum constitute some of the typically most well-represented bacterial groups in glaciers (Lutz et al., 2015) and marine habitats (Michaud et al., 2014) respectively.
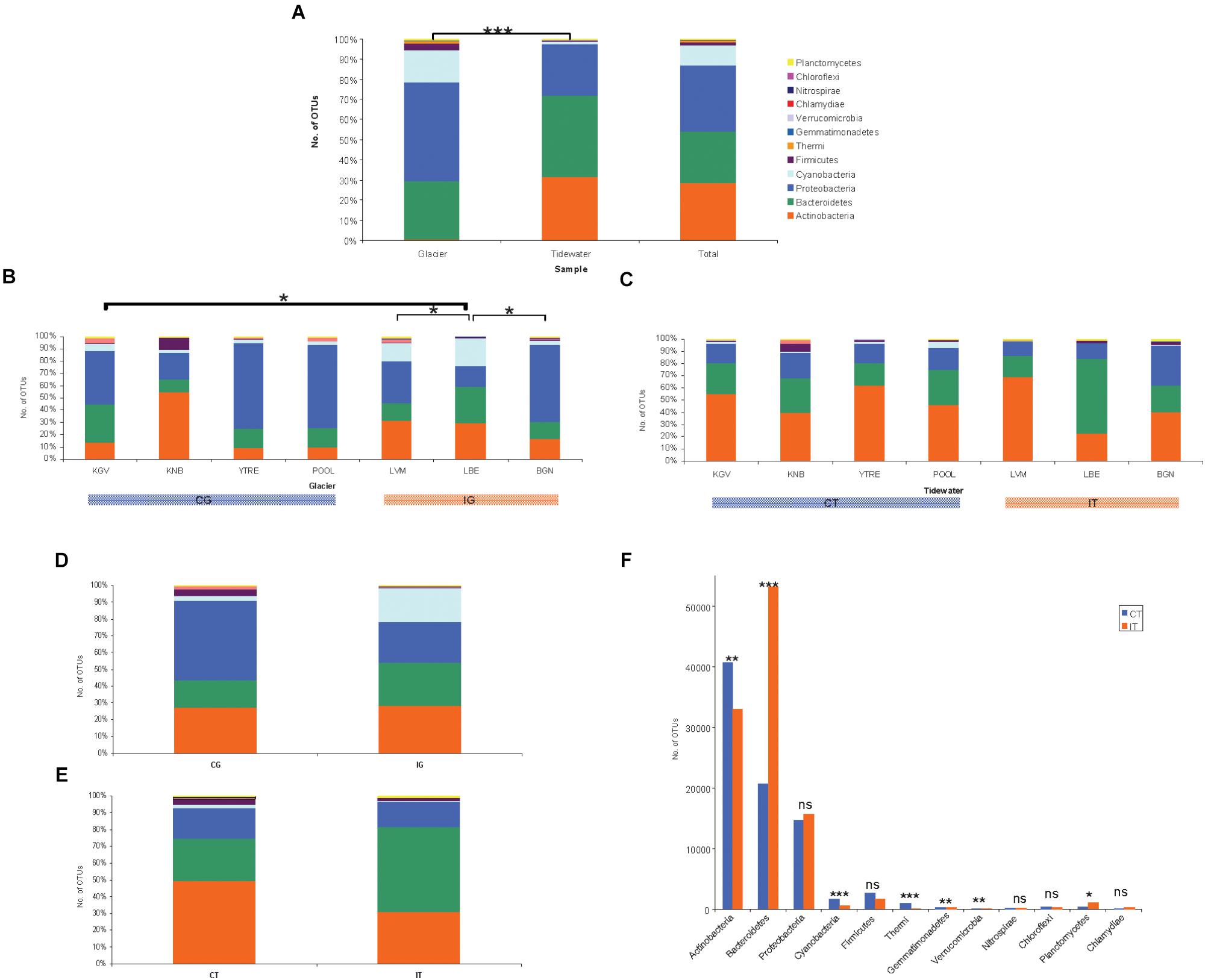
Figure 3. Microbial communities composition. Number of counts aggregates at OTU level of three sampling replicates from glacial ice and tidewater samples. The values represent means of three replicates from each glacier. (A) Comparison of bacterial abundance between samples from glacier ice and tidewater. Significant differences were observed by Student’s t-test for Cyanobacteria, Proteobacteria, and Actinobacteria (∗∗∗p < 0.0001). (B) Comparison of bacterial abundance among samples from glaciers was analyzed by ANOVA and Newman–Keuls Multiple Comparison Test. The number of OTUs was significantly different to between LBE and BGN, LVM and KGV (∗p < 0.05). (C) Comparison of bacterial abundance among seawater samples by ANOVA demonstrated significant differences (∗∗∗p < 0.0001). The Newman–Keuls Multiple Comparison Test revealed that the number of OTUs in LBE was significantly different to the rest seawater samples in almost all pairs (∗p < 0.001). (D) Comparison of bacterial total abundance between samples from coastal glaciers (CG) and inland glaciers (IG). Differences determined by Student’s t-test were not significant. (E) Comparison of bacterial total abundance between samples from the tidewater area closest to a coastal glacier (CT) and samples from the tidewater area closest to an inland glacier (IT). Differences determined by Student’s t-test were not significant. (F) Comparison of bacterial abundance at the phylum level between samples from the tidewater area closest to a coastal glacier (CT) and samples from the tidewater area closest to an inland glacier (IT) by Student’s t-test. Differences in the number of OTUs of were very significant for Bacteroidetes, Cyanobacteria and Thermi (∗∗∗p < 0.0001), moderately significant for actinobacteria, Gemmatimonadetes and Verrucomicrobia (∗∗p < 0.001), and little significant for and (∗p < 0.01) Planctomycetes.
Several researches (Michaud et al., 2014) have described the input of marine microorganisms and organic particles which could be transported to glaciers by atmospheric deposition. We observe that, even in the inland glaciers, an important influence from marine environment seems to exist in both directions, from the glaciers to the sea and vice versa. The presence of bacterial genus characteristic of a given glacier could be sentinels for tracking this transport.
The best example of sentinel microorganism in this study would be the OTUs identified as Hymenobacter. Hymenobacter is the most abundant genus found in this study due to its great presence in the inland glacier LBE. In spite of being the glacier farthest from the coast of all considered in this study, a great influence in its tidewater is observed, because it is also the most abundant OTU in its corresponding tidewater. Currently, about 50 species of the genus Hymenobacter are known (Sun et al., 2018). Some of them are psychotrophs from Arctic and Antarctic environments and high altitudes at Qinghai-Tibet Plateau (Zhang et al., 2008), but they have also been isolated from deep sea water samples (Sun et al., 2018) and from Arctic marine sediments (Kim M.C. et al., 2017).
Other sentinel microorganisms from the exchange of materials from the glaciers to the seawaters could be numerous OTUs common to the sampling points. Among them, the most abundant OTUs belong to the genus Aquaspirillum, Streptomyces, Polaromonas, Candidatus Pelagibacter, Pseudonocardia, Polaribacter, Salinibacterium, Paracoccus, and Streptosporangium.
When of the number of OTUs among all glaciers was analyzed by ANOVA it was little significant. The Newman–Keuls Multiple Comparison Test demonstrated that the most different glacier in terms of the number of OTUs, was LBE, which is significantly different to BGN, LVM, and KGV (∗p < 0.05) (Figure 3B). The number of sequences found in LBE-CG samples was more abundant than the number of sequences in any of the rest glaciers.
In the seawater samples, the number of OTUs was also significantly different by ANOVA (∗∗∗p < 0.0001) (Figure 3C). When comparing the number of OTUs using the Newman–Keuls Multiple Comparison Test, LBE samples were significantly different to the rest seawater samples in almost all pairs (∗p < 0.001), except for Proteobacteria and Firmicutes. The number of sequences found in LBE-CT samples also was more abundant than the number of sequences in any of the other seawater samples.
Regarding the number of sequences corresponding to the most abundant genus (Supplementary Table S2), the highest abundance of sequences belonged to Hymenobacter, both in glaciers and seawater. The abundance of the genus Hymenobacter could be a consequence of its broad presence in the LBE glacier. LBE is an inner glacier but its flux of water can transport this bacterium that is also the most abundant in the seawater samples (LBE-CT). However, in the other glaciers, the highest number of sequences corresponded to Pseudomonas, Polaromonas and Aquaspirillum (Supplementary Table S2). In seawater samples, in addition to Hymenobacter, the most abundant OTUs belonged to Pelagibacter, Salinibacterium and Polaribacter (Supplementary Table S2).
When comparing by Student’s t-test the number of OTUs found in the CG samples with the number of OTUs in the IG samples, the differences were not significant (Figure 3D). Neither were they when comparing the number of OTUs between the CT and IT samples (Figure 3E).
Microbial diversity in glaciers was very similar (Figure 3B). The most significant difference was a greater abundance of Actinobacteria in KNB as well as of Cyanobacteria in LVM and LBE.
Microbial diversity among seawater samples (Figure 3C) was only higher for Cyanobacteria at POOL, Bacteroidetes at LBE and Firmicutes at KNB. The higher and specific abundance of Firmicutes at KNB glacier and its downstreaming tidewater could be also a consequence of the microbial interchange between both populations.
In addition, OTUs have been found in glaciers that corresponded with typical marine microorganisms such as Pelagibacter (Giovannoni, 2017), Polaribacter (Staley and Gosink, 1999), and Roseospira (Kalyan Chakravarthy et al., 2007) or vice versa, OTUs of terrestrial microorganisms as Acidisoma (Belova et al., 2009) or Propionibacterium (Koussémon et al., 2001) were found in seawater samples.
A relevant result was the significant difference found between microbial populations inhabiting CT and IT by Student’s t-test (Figure 3F). The difference in the number of OTUs was very significant for Bacteroidetes, Cyanobacteria, and Thermi (∗∗∗p < 0.0001), moderately significant for Actinobacteria, Gemmatimonadetes, and Verrucomicrobia (∗∗p < 0.001), and little significant for (∗p < 0.01) Planctomycetes.
In order to determine the distribution of microbial communities and how they are affected by environmental variables and by the concentration of nutrients in the environment, several multivariate statistical analysis of community data were carried out with.
Detrended Correspondence Analysis with the relative abundances of the microbial phyla from tidewater samples was performed. This analysis clustered tidewater samples into two groups (Figure 4 and Supplementary Table S3): seawater samples corresponding to inland glaciers (samples IT) and seawater samples corresponding to coastal glaciers (CT). These two microbial populations presented an equivalent number of species (Figure 3C), but their microbial composition in the tidewater region were different, although they are very close geographically and their seawaters can be mixed easily and quickly by wave effect.
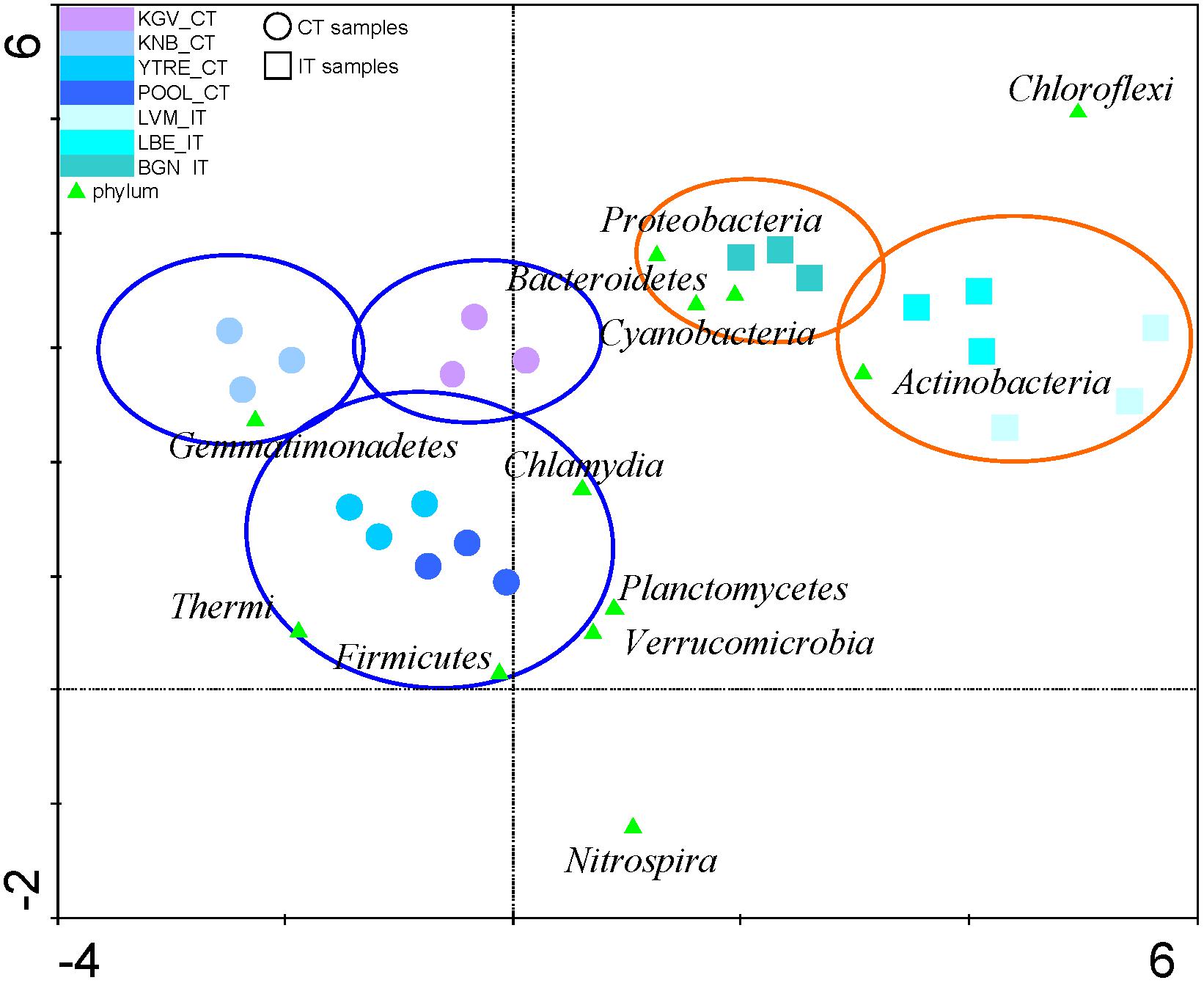
Figure 4. Detrended Correspondence Analysis. The relative abundances of the microbial phyla from tidewater samples were compared to find their gradient. The diagram displays triangles that represent phyla. Sampling sites are represented by different shapes (∘: CT samples and  : IT samples). Venn diagrams cluster and discriminate sampling points in different groups (glacier orange diagrams and tidewater blue diagrams). The axes are scaled in standard deviation units. CT, samples from the tidewater area closest to a coastal glacier; IT, samples from the tidewater area closest to an inland glacier.
: IT samples). Venn diagrams cluster and discriminate sampling points in different groups (glacier orange diagrams and tidewater blue diagrams). The axes are scaled in standard deviation units. CT, samples from the tidewater area closest to a coastal glacier; IT, samples from the tidewater area closest to an inland glacier.
The CCA of all phyla and of the most abundant OTUs in each sampling site demonstrated a significant correlation of the genus Hymenobacter with NO2-, NO3- and NH4+ (Figure 5 and Supplementary Table S3). Other abundant genus such as Pseudomonas and Polaribacter were correlated with levels of all nitrogen species (NO2-, NO3-, NH4+ and TDN). OTUs identified as Salibacterium were correlated to DOC, on the contrary other OTUs such as Polaromonas presented a negative correlation with these compounds.
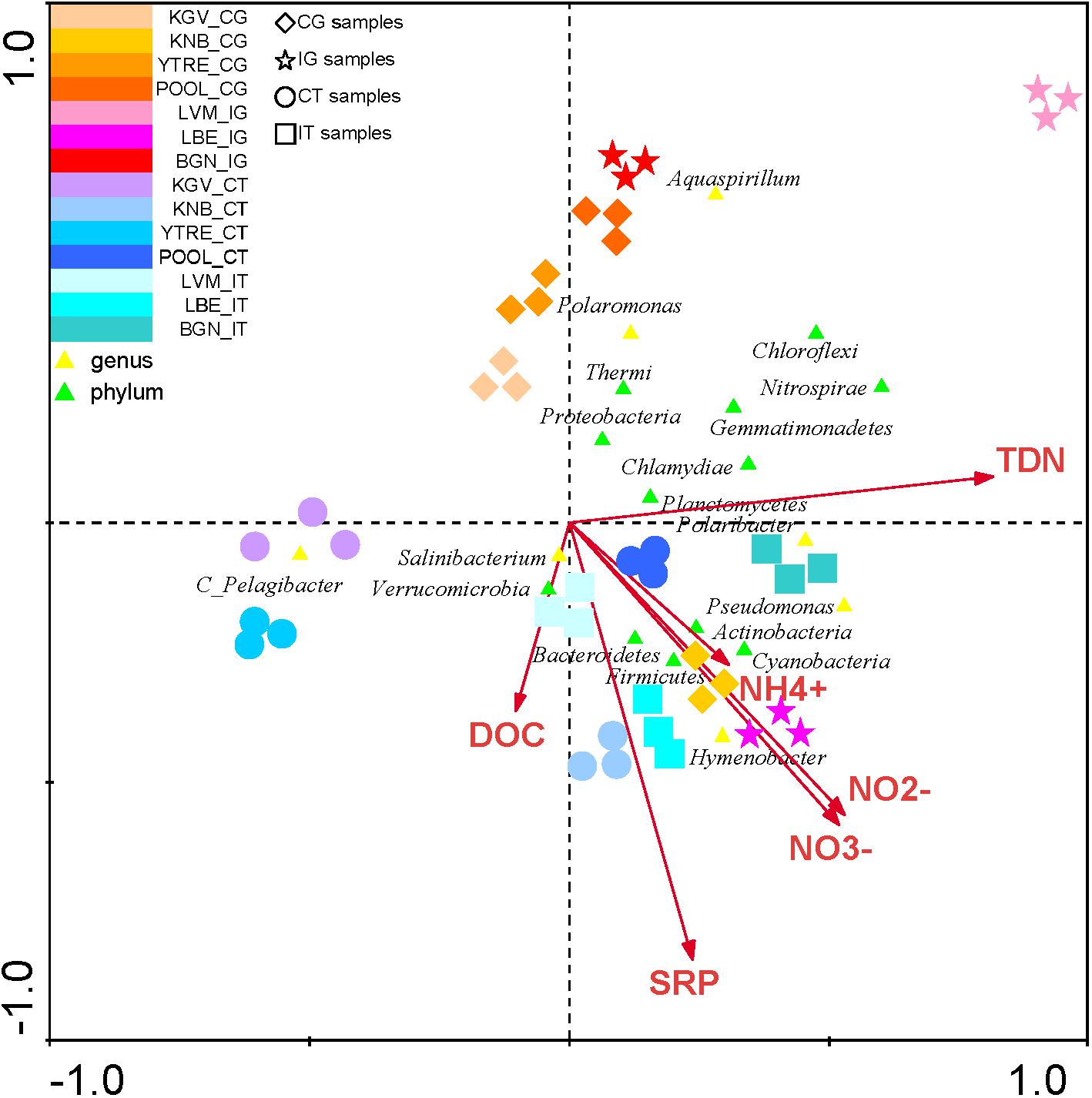
Figure 5. Canonical Correspondence Analysis of the microbial phyla and of the main genus in each sampling site with respect to the concentration of nutrients. The analysis of the bacterial communities with respect to concentration of NH4+, NO2-, NO3-, TDN, SRP, and DOC. The diagram displays triangles that represent taxonomic groups. Sampling sites are represented by different shapes (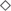 : CG samples, ∗: IG samples, ∘: CT samples, and
: CG samples, ∗: IG samples, ∘: CT samples, and  : IT samples). Arrows symbolize dissolved nitrogen and phosphorus concentrations. The axes are scaled in standard deviation units. CT, samples from the tidewater area closest to a coastal glacier; CG, samples from coastal glaciers; IG, samples from inland glaciers; IT, samples from the tidewater area closest to an inland glacier.
: IT samples). Arrows symbolize dissolved nitrogen and phosphorus concentrations. The axes are scaled in standard deviation units. CT, samples from the tidewater area closest to a coastal glacier; CG, samples from coastal glaciers; IG, samples from inland glaciers; IT, samples from the tidewater area closest to an inland glacier.
The CCA of was also applied to study the influence of environmental variables such as altitude, temperature, pH and salinity in all phyla and in the most abundant OTUs in each sampling site (Analyses 1–4) (Figure 6 and Supplementary Table S3). The presence of some genus such as Pelagibacter was correlated to salinity, and the presence of the genus Polaromonas and the phylum Thermi were correlated to low temperatures. However, according to the eigenvalues obtained in the analysis 5 (Supplementary Table S3), which are less significant than in the previous analysis, the environmental variables do not influence in the distribution of the microbial communities as much as the nutrients do.
Figure 6. Canonical Correspondence Analysis of the microbial phyla and of the main genus in each sampling site with respect to environmental variables. The analysis of the bacterial communities with respect to both environmental variables (temperature, pH, salinity and altitude) was represented. The diagram displays triangles that represent taxonomic groups. Sampling sites are represented by different shapes (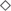 : CG samples, ∗: IG samples, ∘: CT samples and
: CG samples, ∗: IG samples, ∘: CT samples and  : IT samples). Arrows symbolize environmental variables. The axes are scaled in standard deviation units.
: IT samples). Arrows symbolize environmental variables. The axes are scaled in standard deviation units.
In glaciers, organic compounds CH2O (Figure 7) are scarce. In a dark environment such as the englacial area these compounds must be biologically synthesized by CO2 fixation by chemolithotrophs. Even though in most environments the contributions of these organisms to the accumulation of organic matter are trivial compared to that of oxygenic phototrophs, in the englacial ecosystem their small inputs are important. They are degraded biologically to CH4 and CO2.
Figure 7. Overview of the tentative interactions with the biogeochemical cycles in microorganisms from glaciers and tidewater samples. Microorganisms were identified by Illumina sequencing. Some key members of carbon, nitrogen, iron, and sulfur cycles in glacier (left) and tidewater samples (right). They are represented at the genus level.
Some aerobes and facultative anaerobes oxidize CO to produce CO2. The aerobic CO oxidizers described in the scientific literature include members of Proteobacteria, Firmicutes, and Actinobacteria (Park et al., 2003). These organisms can grow chemolithotrophically on CO as the sole carbon and energy source under aerobic conditions and play important roles in global CO biogeochemistry; and it has been assessed that they contribute to CO uptake in soils, plant roots, seas and other natural systems (King, 2006), among which glaciers should be included. In our research we have found the Actinobacteria Streptomyces as well as other aerobic CO oxidizers like Bacillus, Nocardia, and Corynebacterium (Park et al., 2003).
In tidewater samples, much of the primary productivity comes from photosynthesis by Cyanobacteria. Phototrophs and chemolithotrophs, produce new organic carbon from CO2. Besides oxygenic phototrophs (which are the majority), anoxygenic phototrophs are also present. Aerobic anoxygenic phototrophs include bacteria such as Erythrobacter.
Some marine bacteria such as Pelagibacter are ecologically important in the so called “secondary production” because they consume high amounts of organic carbon produced from photosynthesis and are responsible for nutrient regeneration. Thus, these marine bacteria return organic matter to the marine food web that would otherwise be lost because of the inability of larger organisms to use such organic nutrients.
On glacier surface, phototrophs such as cyanobacteria (i.e. Trichodesmium) are important nitrogen-fixing bacteria. In tidewater samples, nitrogen-fixing cyanobacteria were also identified (Figure 7).
The nitrogen cycle is driven by both chemolithotrophic and chemoorganotrophic microorganisms. In the biological utilization of N2, it is reduced to NH3, and then assimilated into organic forms, such as amino acids and nucleotides. Nitrification results from the sequential activities of two physiological groups of organisms: the ammonia-oxidizing bacteria (which oxidize NH3 to nitrite, NO2-), and the nitrite-oxidizing bacteria, which oxidize NO2- to NO3-. Many species of bacteria are nitrifiers. The majority of them belonged to Proteobacteria and Nitrospirae (i.e., Thermodesulfovibrio). The first step is performed both in glacier and tidewater samples by very abundant bacteria such as Hymenobacter and Polaribacter.
The biological redox reactions in the iron cycle include both oxidations and reductions. Some bacteria can use Fe3+ as an electron acceptor in anaerobic respiration, and Fe2+ is oxidized in the chemolithotrophic metabolism. Important Fe3+ reducers as Shewanella, Rhodoferax, and Acidiphilium were identified in glacier samples (Figure 7). Among Fe2+ oxidizers we found Ferrimicrobium. The electron donors that are coupled to iron reduction could be inorganic (sulfur or hydrogen), as in the case of chemolithotrophic acidophiles; or it could be organic (glucose, glycerol), as in the case of heterotrophic acidophiles (e.g., Acidiphilium). In tidewater samples, Shewanella, Rhodoferax, were also identified, as well as some Fe2+ oxidezers such as Gallionella.
In glaciers, several Epsilonproteobacteria (Sulfurospirillum and Sulfurimonas) were identified. These bacteria oxidize sulfide and sulfur as electron donors with either O2 or nitrate as electron acceptors. Some Deltaproteobacteria are specialized in anaerobic metabolisms using oxidized sulfur compounds as electron acceptors. These include organisms such as Desulfovibrio and Desulfuromusa, a group of sulfate-reducing bacteria that reduces sulfate to sulfide, with lactate, pyruvate, or H2 as electron donors. However, in anoxic environments acetate-oxidizing sulfate-reducing bacteria dominate. Acetate-oxidizing sulfate reducers include genera such as Desulfofrigus identified in glacier samples.
Other bacteria, Paracoccus, found both in glacier and tidewater samples (Figure 7), contains a sulfide and thiosulfate oxidation system, named Sox, that is also present in and many other sulfur bacteria both chemolithotrophic and phototrophic. This system oxidizes reduced sulfur compounds.
In tidewater samples, hydrogen sulfide (H2S) is oxidized by phototrophic and chemolithotrophic microorganisms (Thiomonas, Thiomicrospira, and Thioalkalimucrobium) to sulfur (S0) and sulfate (SO42-), the latter being a key nutrient for algae and marine plants.
However, SO42- can be reduced to H2S by activities of the sulfate-reducing bacteria. These organisms consume organic carbon, and this reduction closes the biogeochemical sulfur cycle while regenerating CO2.
Among the phosphorous solubilizing bacterial communities, strains from Pseudomonas, Bacilli, Rhizobium and Enterobacter have been described as phosphate solubilizers (Khan et al., 2009). In glacier samples the presence of microorganisms involved in phosphorus acquisition include strains from bacterial genera Pseudomonas, Mesorhizobium, Azorhizobium, Bradyrhizobium, Bacillus and Azospirillum. In tidewater samples, these same genera except Azospirillum were found.
Glaciers constitute a key link between coasts and their downstreaming tidewaters and are of increasing importance in land-to-ocean fluxes. In published reports about other coastal glaciers such as the Antarctic Wanda Glacier (Pessi et al., 2015), it has been observed that marine microorganisms could colonize glaciers. In our study we have also observed that some marine bacterial OTUs were present in the glaciers. Additionally, we did also observe that OTUs corresponding to specific seawater bacteria were identified in the glacier samples. That is to say, it is possible that part of the microbial diversity found in the seawater samples comes partially from the upstream glaciers. When the glaciers thaw, the inland runoff waters transport more materials than the coastal glaciers do. This progression would alter the seawater composition, by increasing both nutrients and contaminants, or by changing the physicochemical characteristics of water. Over time, microbial populations from CT samples would become more similar to populations from IT samples.
The glaciers on the Northwest coast of Svalbard are undergoing a rapid change due to climate change. This effect is important in the coastal glaciers, but it is even more so in the interior glaciers. If the glaciers inland continue to melt, they will increasingly drag a greater amount of materials and sediments in their runoff waters. The consequence of this process will be the possible alteration of the water composition of the fiords. If this effect is maintained over time, the microbial populations of the coasts (CT) could be modified and they would be more similar to the current populations of the IT samples. This process can have serious consequences throughout the marine ecosystem and in the cycling of globally important elements.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was supported by the Spanish Ministerio de Economía y Competitividad (MINECO) (CTM2011-16003-E) and by the Institute of Health Carlos III (PI14/00705). EG-L is recipient of a MINECO Fellowship (PTAT2010-03424).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are indebted to Maria Paz Martín Redondo from the Geology Laboratory of the Centro de Astrobiologia. We thank Christiane Hübner and Christian Zoelly from the Norwegian Sverdrup Station (Ny Ålesund, Svalbard) for their skillful assistance. We also thank the staff of the Norwegian Kings Bay Research Station (Ny Ålesund, Svalbard) for their logistic support (RiS-ID 6845).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00512/full#supplementary-material
BGN, Brøggerbreen glacier; CCA, Canonical Correspondence Analysis; CG, samples from coastal glaciers; CT, samples from the tidewater area closest to a coastal glacier; DCA, Detrended Correspondence Analysis; DOC, Dissolved organic matter; ICP-MS, Inductively coupled plasma-mass spectrometry; IG, samples from inland glaciers; IT, samples from the tidewater area closest to an inland glacier; KGV, Kongsvegen glacier; KNB, Kronebreen glacier; LBE, Lovénbreen Austre glacier; LVM, Lovénbreen Midtre glacier; OTU, operational taxonomic units; PCA, principal components analysis; POOL, Poolepynten; SRP, soluble reactive phosphorus; TDN, total dissolved nitrogen; YTRE, Ytre Norskøya.
Amato, P., Hennebelle, R., Magand, O., Sancelme, M., Delort, A. M., Barbante, C., et al. (2007). Bacterial characterization of the snow cover at Spitzberg, Svalbard. FEMS Microbiol. Ecol. 59, 255–264. doi: 10.1111/j.1574-6941.2006.00198.x
Anesio, A. M., and Laybourn-Parry, J. (2012). Glaciers and ice sheets as a biome. Trends Ecol. Evol. 27, 219–225. doi: 10.1016/j.tree.2011.09.012
Belova, S. E., Pankratov, T. A., Detkova, E. N., Kaparullina, E. N., and Dedysh, S. N. (2009). Acidisoma tundrae gen. nov., sp. nov. and Acidisoma sibiricum sp. nov., two acidophilic, psychrotolerant members of the Alphaproteobacteria from acidic northern wetlands. Int. J. Syst. Evol. Microbiol. 59, 2283–2290. doi: 10.1099/ijs.0.009209-0
Benner, R., Louchouarn, P., and Amon, R. M. W. (2005). Terrigenous dissolved organic matter in the arctic ocean and its transport to surface and deep waters of the north atlantic. Global Biogeochem. Cycles 19:GB2025. doi: 10.1029/2004GB002398
Deiglmayr, K., Philippot, L., Tscherko, D., and Kandeler, E. (2006). Microbial succession of nitrate-reducing bacteria in the rhizosphere of Poa alpina across a glacier foreland in the central alps. Environ. Microbiol. 8, 1600–1612. doi: 10.1111/j.1462-2920.2006.01051.x
Foreman, C. M., Sattler, B., Mikucki, J. A., Porazinska, D. L., and Priscu, J. C. (2007). Metabolic activity and diversity of cryoconites in the taylor valley, Antarctica. J. Geophys. Res. 112:G04S32. doi: 10.1029/2006JG000358
Førland, E. J., Benestad, R. E., Flatøy, F., Hanssen-Bauer, I., Haugen, J. E., Isaksen, K., et al. (2009). Climate Development in North Norway and the Svalbard Region During 1900-2100. Norwegian Polar Institute Report Series No. 128. Tromsø: Norwegian Polar Institute.
Garcia-Descalzo, L., Garcia-Lopez, E., Postigo, M., Baquero, F., Alcazar, A., and Cid, C. (2013). Eukariotic microorganisms in cold enviroments: examples from Pyrenean glaciers. Front. Microbiol. 4:55. doi: 10.3389/fmicb.2013.00055
Garcia-Lopez, E., Alcazar, P., Postigo, M., and Cid, C. (2016). “The effect of climate change on microbial communities from glaciers,” in Glaciers: Formation, Climate Change and Their Effects, ed. N. Doyle (New York, NY: Nova Science Publishers, Inc. ), 71–88.
Garcia-Lopez, E., and Cid, C. (2017). “The role of microbial ecology in glacier retreat analysis,” in Glaciers, ed. W. V. Tangborn (Rijeka: InTech).
Giovannoni, S. J. (2017). SAR11 Bacteria: the most abundant plankton in the oceans. Annu. Rev. Mar. Sci. 9, 231–255. doi: 10.1146/annurev-marine-010814-015934
Hagen, J. O., Kohler, J., Melvold, K., and Winther, J. G. (2003). Glaciers in Svalbard: mass balance, runoff and freshwater flux. Polar Res. 22, 145–159. doi: 10.3402/polar.v22i2.6452
Hagen, J. O., Liestøl, O., Roland, E., and Jørgensen, T. (1993). Glacier Atlas of Svalbard and Jan Mayen. Oslo: Norwegian Polar Institute.
Hallbeck, L. (2009). Microbial Processes in Glaciers and Permafrost. A Literature Study on Microbiology Affecting Groundwater at Ice Sheet Melting. SKB R-09-37. Stockholm: Svensk Kärnbränslehantering AB.
Hell, K., Edwards, A., Zarsky, J., Podmirseg, S. M., Girdwood, S., Pachebat, J. A., et al. (2013). The dynamic bacterial communities of a melting High Arctic glacier snowpack. ISME J. 7, 1814–1826. doi: 10.1038/ismej.2013.51
Her, N., Amy, G., Foss, D., Cho, J., Yoon, Y., and Kosenka, P. (2002). Optimization of method for detecting and characterizing nom by HPLC-size exclusion chromatography with UV and on-line DOC detection. Environ. Sci. Technol. 36, 1069–1076. doi: 10.1021/es015505j
Herlemann, D. P., Labrenz, M., Jurgens, K., Bertilsson, S., Waniek, J. J., and Andersson, A. F. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579. doi: 10.1038/ismej.2011.41
Hoham, R. W., and Duval, B. (2001). “Microbial ecology of snow and freshwater ice with emphasis on snow algae,” in Snow Ecology: An Interdisciplinary Examination of Snow-Covered Ecosystems, eds H. G. Jones, J. W. Pomeroy, D. A. Walker, and R. W. Hoham (Cambridge: Cambridge University Press), 168–228.
Hood, E., Battin, T. J., Fellman, J., O’Neel, S., and Spencer, R. G. (2015). Storage and release of organic carbon from glaciers and ice sheets. Nat. Geosci. 8, 91–96. doi: 10.1038/NGEO2331
Hotaling, S., Hood, E., and Hamilton, T. L. (2017). Microbial ecology of mountain glacier ecosystems: biodiversity, ecological connections and implications of a warming climate. Environ. Microbiol. 19, 2935–2948. doi: 10.1111/1462-2920.13766
Jongman, R. H. G., Ter Braak, C. J. F., and Van Tongeren, O. F. R. (1995). Data Analysis in Community and Landscape Ecology. Cambridge: Cambridge University Press. doi: 10.1017/CBO9780511525575
Kalyan Chakravarthy, S., Srinivas, T. N., Anil Kumar, P., Sasikala, C. H., and Ramana, C. H. V. (2007). Roseospira visakhapatnamensis sp. nov. and Roseospira goensis sp. nov. Int. J. Syst. Evol. Microbiol. 57, 2453–2457. doi: 10.1099/ijs.0.65105-0
Kastovska, K., Elster, J., Stibal, M., and Santruckova, H. (2005). Microbial assemblages in soil microbial succession after glacial retreat in Svalbard (high Arctic). Microb. Ecol. 50, 396–407. doi: 10.1007/s00248-005-0246-4
Khan, A., Jilani, G., Akhtar, M. S., Naqvi, S. M. S., and Rasheed, M. (2009). Phosphorus solubilizing bacteria: occurrence, mechanisms and their role in crop production. J. Agric. Biol. Sci. 1, 48–58.
Kim, M., Jung, J. Y., Laffly, D., Kwon, H. Y., and Lee, Y. K. (2017). Shifts in bacterial community structure during succession in a glacier foreland of the High Arctic. FEMS Microbiol. Ecol. 93:fiw213. doi: 10.1093/femsec/fiw213
Kim, M. C., Kim, C. M., Kang, O. C., Zhang, Y., Liu, Z., Wangmu, D., et al. (2017). Hymenobacter rutilus sp. nov., isolated from marine sediment in the Arctic. Int. J. Syst. Evol. Microbiol. 67, 856–861. doi: 10.1099/ijsem.0.001685
King, G. M. (2006). Nitrate-dependent anaerobic carbon monoxide oxidation by aerobic CO-oxidizing bacteria. FEMS Microbiol. Ecol. 56, 1–7. doi: 10.1111/j.1574-6941.2006.00065.x
Kirchman, D. L., Morán, X. A., and Ducklow, H. (2009). Microbial growth in the polar oceans - role of temperature and potential impact of climate change. Nat. Rev. Microbiol. 7, 451–459. doi: 10.1038/nrmicro2115
Koussémon, M., Combet-Blanc, Y., Patel, B. K., Cayol, J. L., Thomas, P., Garcia, J. L., et al. (2001). Propionibacterium microaerophilum sp. nov., a microaerophilic bacterium isolated from olive mill wastewater. Int. J. Syst. Evol. Microbiol. 51, 1373–1382. doi: 10.1099/00207713-51-4-1373
Lang, C. (2011). Modeling of the Surface Mass Balance in Svalbard With the Regional Climate Model Mar Over 1958-2010. Ph.D. thesis, Université de Liège, Liège.
Larose, C., Dommergue, A., and Vogel, T. M. (2013). The dynamic arctic snow pack: an unexplored environment for microbial diversity and activity. Biology 2, 317–330. doi: 10.3390/biology2010317
Lopez-Ramirez, M. P., Sanchez-Lopez, K. B., Sarria-Guzman, Y., Bello-Lopez, J. M., Cano-Garcia, V. L., Ruiz-Valdiviezo, V. M., et al. (2015). Haloalkalophilic cellulose-degrading bacteria isolated from an alkaline saline soil. J. Pure Appl. Microbiol. 9, 2879–2886.
Lutz, S., Anesio, A. M., Edwards, A., and Benning, L. G. (2015). Microbial diversity on icelandic glaciers and ice caps. Front. Microbiol. 6:307. doi: 10.3389/fmicb.2015.00307
Madigan, M. T., Martinko, J. M., Stahl, D. A., and Clark, D. P. (2012). Brock Biology of Microorganisms, 13th Edn. San Francisco, CA: Pearson Education, Inc.
McNabb, R. W., Womble, J. N., Prakash, A., Gens, R., and Haselwimmer, C. E. (2016). Quantification and analysis of icebergs in a tidewater glacier fjord using an object-based approach. PLoS One 11:e0164444. doi: 10.1371/journal.pone.0164444
Michaud, L., Lo Giudice, A., Mysara, M., Monsieurs, P., Raffa, C., Leys, N., et al. (2014). Snow surface microbiome on the high antarctic plateau (DOME C). PLoS One 9:e104505. doi: 10.1371/journal.pone.0104505
Mindl, B., Anesio, A. M., Meirer, K., Hodson, A. J., Laybourn-Parry, J., Sommaruga, R., et al. (2007). Factors influencing bacterial dynamics along a transect from supraglacial runoff to proglacial lakes of a high Arctic glacier. FEMS Microbiol. Ecol. 59, 307–317. doi: 10.1111/j.1574-6941.2006.00262.x
Møller, A. K., Barkay, T., Al-Soud, W. A., Sørensen, S. J., Skov, H., and Kroer, N. (2011). Diversity and characterization of mercury-resistant bacteria in snow, freshwater and sea-ice brine from the high arctic. FEMS Microbiol. Ecol. 75, 390–401. doi: 10.1111/j.1574-6941.2010.01016.x
Nowak, A., and Hodson, A. (2014). Changes in meltwater chemistry over a 20-year period following a termal regime switch from polythermal to cold-based glaciation at austre broggerbreen, Svalbard. Polar Res. 33:22779. doi: 10.3402/polar.v33.22779
Park, S. W., Hwang, E. H., Park, H., Kim, J. A., Heo, J., Lee, K. H., et al. (2003). Growth of mycobacteria on carbon monoxide and methanol. J. Bacteriol. 185, 142–147. doi: 10.1128/JB.185.1.142-147.2003
Pessi, I. S., Osorio-Forero, C., Gálvez, E. J. C., Simões, F. L., Simões, J. C., Junca, H., et al. (2015). Distinct composition signatures of archaeal and bacterial phylotypes in the wanda glacier forefield, Antarctic Peninsula. FEMS Microbiol. Ecol. 91, 1–10. doi: 10.1093/femsec/fiu005
Sipler, R. E., Kellogg, C. T. E., Connelly, T. L., Roberts, Q. N., Yager, P. L., and Bronk, D. A. (2017). Microbial community response to terrestrially derived dissolved organic matter in the coastal Arctic. Front. Microbiol. 8:1018. doi: 10.3389/fmicb.2017.01018
Solomon, S., Qin, D., Manning, M., Chen, M., Marquis, K., Averyt, B., et al. (2007). The Physical Science Basis: Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change, in Climate Change 2007. Cambridge: Cambridge University Press.
Staley, J. T., and Gosink, J. J. (1999). Poles apart: biodiversity and biogeography of sea ice bacteria. Annu. Rev. Microbiol. 53, 189–215. doi: 10.1146/annurev.micro.53.1.189
Sun, J., Xing, M., Wang, W., Dai, F., Liu, J., and Hao, J. (2018). Hymenobacter profundi sp. nov., isolated from deep-sea water. Int. J. Syst. Evol. Microbiol. 68, 947–950. doi: 10.1099/ijsem.0.002621
Ter Braak, C. J. F. (1995). Data Analysis in Community and Landscape Ecology, eds R. H. G. Jongman, C. J. F. Ter Braak, and O. F. R. Van Tongeren. Cambridge: Cambridge University Press.
Tipper, J. C. (1979). Rarefaction and rarefaction -the use and abuse of a method in paleontology. Paleobiology 5:423434. doi: 10.1017/S0094837300016924
Weiner, E. R. (2008). Applications of Environmental Aquatic Chemistry: A Practical Guide, 3rd Edn. Boca Raton, FL: CRC Press. doi: 10.1201/9781420008371
Zaikova, E., Hawley, A., Walsh, D. A., and Hallam, S. J. (2009). Seawater sampling and collection. J. Vis. Exp. 28:e1159. doi: 10.3791/1159
Zeebe, R. E., and Caldeira, K. (2008). Close mass balance of long-term carbon fluxes from ice-core CO2 and ocean chemistry records. Nat. Geosci. 1, 312–315. doi: 10.1038/ngeo185
Zeng, Y. X., Yan, M., Yu, Y., Li, H. R., He, J. F., Sun, K., et al. (2013). Diversity of bacteria in surface ice of austre lovénbreen glacier, Svalbard. Arch. Microbiol. 195, 313–322. doi: 10.1007/s00203-013-0880-z
Keywords: coastal glaciers, next-generation sequencing, food web, Svalbard archipelago, Arctic
Citation: Garcia-Lopez E, Rodriguez-Lorente I, Alcazar P and Cid C (2019) Microbial Communities in Coastal Glaciers and Tidewater Tongues of Svalbard Archipelago, Norway. Front. Mar. Sci. 5:512. doi: 10.3389/fmars.2018.00512
Received: 26 June 2018; Accepted: 21 December 2018;
Published: 14 January 2019.
Edited by:
Alejandro A. Murillo, EMBL Heidelberg, GermanyReviewed by:
Raghab Ray, University of Tokyo, JapanCopyright © 2019 Garcia-Lopez, Rodriguez-Lorente, Alcazar and Cid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cristina Cid, Y2lkc2NAaW50YS5lcw==; Y2lkc2NAY2FiLmludGEtY3NpYy5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.