
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 08 January 2019
Sec. Marine Ecosystem Ecology
Volume 5 - 2018 | https://doi.org/10.3389/fmars.2018.00502
This article is part of the Research Topic Coralline Algae: Globally Distributed Ecosystem Engineers View all 11 articles
 Suzanne Fredericq1*
Suzanne Fredericq1* Sherry Krayesky-Self1
Sherry Krayesky-Self1 Thomas Sauvage2
Thomas Sauvage2 Joseph Richards1
Joseph Richards1 Ronald Kittle1
Ronald Kittle1 Natalia Arakaki3
Natalia Arakaki3 Emma Hickerson4
Emma Hickerson4 William E. Schmidt1
William E. Schmidt1Rhodoliths are the main hard substrata for the attachment of benthic macroalgae in the NW Gulf of Mexico rubble habitats that are associated with salt domes, unique deep bank habitats at ~50–90 m depth on the continental shelf offshore Louisiana and Texas. With the advent of additional sequencing technologies, methodologies for biodiversity assessments are now rapidly shifting to DNA metabarcoding, i.e., High Throughput Sequencing (HTS) of environmental DNA mixtures with standardized molecular markers, such as 16S V4, for rapid, cost-effective biodiversity measurement. We newly tested 16S V4 metabarcoding on endolithic portions of mesophtic rhodoliths exhibiting low phototroph colonization that revealed a hidden, cryptic algal diversity targeting spores, propagules, and unsuspected life history stages. We explored cryo-SEM as a potentially more informative method than regular SEM to minimize artifacts of sample preparation in the study of endolithic cell inclusions which brought to light a suite of microalgal stages. We were able to differentiate floridean starch from cellular inclusions. We associated the effect of anatomical growth pattern on presence or absence of cellular inclusions in biogenic rhodoliths. Analyses of combined 16S V4 metabarcodes and 16S Sanger sequences of two red algal orders, the Halymeniales and Bonnemaisoniales, increased the established record of diversity in the region. We view rhodoliths as marine biodiversity hotspots that may function as seedbanks, temporary reservoirs for life history stages of ecologically important eukaryotic microalgae, and macroalgae or as refugia for ecosystem resilience following environmental stress.
This paper focuses on rhodoliths from mesophotic rhodolith beds occurring in the northwestern Gulf of Mexico (NWGMx) on the continental shelf offshore Louisiana and Texas (USA) at ~45–90 m depth (Figures 1–3). It is to be viewed as a conceptual primer to address insights and working hypotheses on the dynamic role of rhodoliths as marine biodiversity hotspots and seedbanks for macroalgae and microalgae in maintaining the health of marine ecosystems.
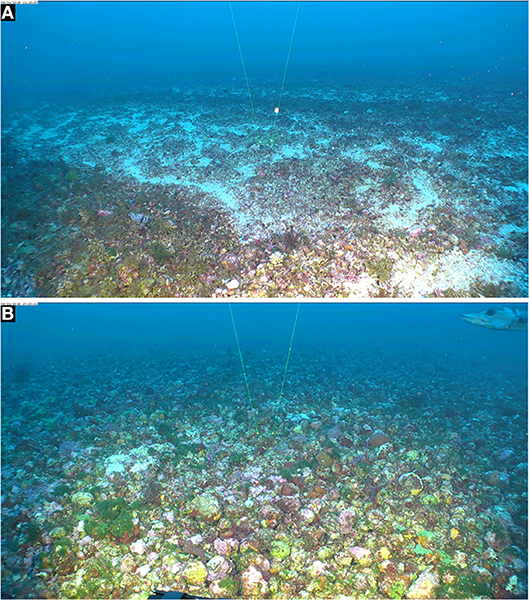
Figure 1. Extensive rhodolith beds in East Flower Garden Bank, FGBNMS. Mohawk ROV photos taken on 24 Sept. 2018. (A) 27°56'22.2288“N, 93°36'1.3068”W, 53 m (Dive 701_0905_111338). (B) 27°54'4.6182“ N; 93°36'40.2732” W, 52.9m (Dive 703_0603_172042). Space between spot lasers = 10 cm.

Figure 2. Rhodoliths form the substratum for attached fleshy and crustose red, brown, and green seaweeds. Mohawk ROV photos taken on 25 Sept. 2018 in West Flower Garden Bank, FGBNMS. (A) 27°52'7.2732“N, 93°51'34.4088”W, 58.8 m depth (Dive 706_0238_122144). The visible macroscopic seaweed is the green alga Codium isthmocladum Vickers (Codiaceae, Bryopsidales). (B) 27°52' 7.1682“N, 93°51'34.185”W, 58.8 m depth (Dive 706_0268_123120). The visible fleshy macroscopic red algal blades are the red alga Anatheca sp. (Areschougiaceae, Gigartinales). Space between spot lasers = 10 cm.

Figure 3. Mound of rhodoliths with attached fleshy and crustose red, brown, and green seaweeds, built by sand tilefish (Malacanthus plumieri Bloch 1786, Malacanthidae). Mohawk ROV photos taken on 24 Sept. 2018 in East Flower Garden Bank (FGBNMS). (A) (27°55'22.1334“N, 93°35'31.8048”W, 51.2 m (Dive 702_0063_131400). (B) Sand tilefish at entry of its rhodolith mound, 27°56'20.7132“N, 93°36'1.1052”W, 51.8 m (Dive 700_0040_075832). Space between spot lasers = 10 cm.
Rhodoliths are unattached, marine, benthic algal nodules of various sizes, and origins that are predominantly accreted by non-geniculate (crustose) coralline red algae (CCA) precipitating CaCO3 within their cell walls (Foster, 2001). Bioturbation or water motion is critical for rhodoliths to grow and remain unburied by sediments, and it also limits fouling by enabling their periodic rotation to allow light exposure on all sides (Steneck, 1986; Hinojosa-Arango et al., 2009). Rhodolith beds are common constituents of modern and fossil marine environments worldwide, especially in clear tropical waters with beds up to 10 m thick (Littler et al., 1985; Amado-Filho et al., 2007, 2012; Pereira-Filho et al., 2011, 2012; Harvey et al., 2017). The taxonomic composition of mesophotic algae (Spalding et al., 2019) associated with rhodolith beds at 45–90 m depth in the NWGMx typically encompasses fleshy and crustose species limited to those low light habitats and includes some taxa from very deep branches of the algal Tree of Life (e.g., the recently erected Class Palmophyllophyceae, Leliaert et al., 2016).
Representative species of three orders of coralline algae (Corallinophycidae) that grow as rhodoliths, i.e., Hapalidiales, Corallinales, and Sporolithales, are found in the NWGMx. On the basis of comparative DNA sequence analysis and anatomy, Richards et al. (2016) recognized eight species of Lithothamnion (Hapalidiales) rhodoliths in the Gulf of Mexico where only a total of three taxa were previously reported. Also discovered were a wealth of other taxa, including, so far, 3 species of Harveylithon, and at least 3 species of Lithophyllum and “Titanoderma” (Corallinales) (Richards et al., 2014, Richards, unpubl. data), as well as two new species of Sporolithon (Sporolithales) (Richards and Fredericq, 2018, and Richards, unpubl. data). Cryptic or pseudocryptic species of CCA (crustose coralline algae) forming rhodoliths abound in the NWGMx because they do not appear that different from one another based on superficial characters and external morphology alone. Likewise, internal morpho-anatomy can be very similar between different species. Overall, it is not unreasonable to assume that the true diversity of rhodoliths species in the NWGMx is much greater than is currently recognized.
Rhodoliths represent an important and understudied component of marine diversity that contributes to major ecosystem functions. They are important ecosystem engineers (Foster et al., 2007; Cavalcanti et al., 2014) providing structurally complex habitats harboring high biodiversity (Nelson et al., 2014; Teigert, 2014; Riosmena-Rodríguez et al., 2017), including many microhabitats for diverse assemblages of algae, invertebrates and other macroscopic taxa (Cabioch, 1969; Hinojosa-Arango and Riosmena-Rodríguez, 2004). For instance, the sand tilefish Malacanthus plumieri (Bloch) further increases habitat complexity provided by the rhodoliths by building mounds that coalesce in more solid structures and may constitute an early successional stage in the formation of large coralline reefs on SW Atlantic tropical shelves (Pereira-Filho et al., 2011). Pereira-Filho et al. (2015) summarize that the purpose of the rhodolith mounds in the Fernando de Noronha Archipelago, Brazil, is yet undetermined and may provide an orientation reference for the tilefish during foraging, a refuge from predation, a nutritional source, and/or are important for social organization.
As primary producers, rhodoliths and their associated macroalgae are crucial components of the photosynthetic community that produces O2, provides food and shelter for invertebrates and vertebrates, and induces settlement and metamorphosis of some invertebrate larvae (Heyward and Negri, 1999; Hadfield and Paul, 2001; Riosmena-Rodríguez and Medina-Lopez, 2011); these processes may also be linked to co-habiting microbial communities. Rhodoliths also fulfill an important function in biogeochemical cycling. For example, they exude organic matter (Smith et al., 2006) that is utilized by co-residing prokaryotes, which in turn cycle key biogeochemical elements necessary for these primary producers and other eukaryotic rhodolith colonizers. Corallines are one of the major producers of dimethylsulfoniopropionate (DMSP) (Kamenos et al., 2008), which, upon being metabolized by algal-associated bacteria, produces volatile compounds such as dimethyl sulfide (DMS) that has direct effects on the global sulfur cycle and global climate change (Burdett et al., 2015; Wang et al., 2018). Yet, integrated baseline data on the rhodoliths' microbiome (microbiota) composition and gene expression in relation to biogeochemical cycling are sorely needed to better understand the functions of these nodules within the marine ecosystem.
CaCO3 provides various advantages such as skeletal strength, protection from grazers and borers, and enhanced survivorship by increasing resistance to wave action (Borowitzka and Larkum, 1987; Foster et al., 2007). The deposition of CaCO3 by rhodoliths and other calcifying marine algae is an essential process in the global carbon cycle (Vecsei, 2004; McCoy and Kamenos, 2015), and rhodoliths are recognized as foremost carbonate builders when they form extensive beds. The modulation of CaCO3 deposition by the corallines and its dissolution is receiving much attention because ocean acidification and rising global sea surface temperatures represent major threats to calcifying marine organisms and their associated microbes (Anthony et al., 2008; Webster et al., 2016). They are highly sensitive to variations in temperature (Adey et al., 2013; Halfar et al., 2013) and are mostly composed of high-Mg biogenic calcite, the most sensitive CaCO3 polymorph to decreases in ocean pH (Kamenos et al., 2013; Webster et al., 2013; Krayesky-Self et al., 2016; Ragazzola et al., 2016). They may be the first indicators of ocean acidification by dissolution (Morse et al., 2006; Andersson et al., 2007; Ragazzola et al., 2012), with ensuing negative effects on a variety of biotic interactions, such as larval settlement reduction of spawning corals (Anthony et al., 2008; Doropoulos et al., 2012; Webster et al., 2013).
Prior to the April 2010 Macondo Well Blowout (04/2010, 28°44'12.01”N, 88°23'13.78”W) and resulting Deepwater Horizon oil spill (DWH) offshore Louisiana (Paris et al., 2012; Rabalais, 2014), mesophotic rhodolith beds occurring throughout the NWGMx, such as Ewing Bank, harbored diversity-rich, lush assemblages of red, green, and brown seaweeds. In contrast, post-DWH, macroalgae in Ewing Bank rhodolith beds in the vicinity of the Macondo Well blowout disappeared, and most rhodoliths themselves appeared bleached, and fully or partially bare of surface macroalgae (Felder et al., 2014; Fredericq et al., 2014), a situation that has persisted in the field as of May 2018, our last expedition to Ewing Bank. These post-DWH impacts appear long-lasting, with little macroalgal growth recovery in the field, for reasons still unknown. Since rhodoliths and their associated macroalgae are ecologically important phototrophs at the base of the oxygen-based food chain, their continued, apparent drastic die-off may have important consequences for the health and recovery of the marine mesophotic bank ecosystem in the NWGMx. The full extent, reasons and ecological consequences of the disappearance of healthy rhodoliths and associated macroalgae on Ewing Bank post-DWH, and the potential link of this decline to consumers is currently unknown. This requires an ecosystem-wide investigation that must include the less visible component of primary producers, such as endolithic algae growing within CaCO3-lined cells of the rhodoliths, resting spores, and carbonate dwellers, heteromorphic life history stages, and their interaction with bacterial communities involved in elemental cycling within the rhodoliths.
Two major rhodolith categories can be found in the NWGMx, i.e., biogenic and autogenic rhodoliths. Biogenic rhodoliths (Figures 4A–C,F, 5) are formed by the non-geniculate crustose coralline algae (CAA) themselves, e.g., Lithothamnion spp. (Hapalidiales) and Sporolithon sinusmexicanum (Sporolithales). In contrast, autogenic rhodoliths (Figures 4D–F) are derived from already existing calcium carbonate rubble established by differential erosion processes of the caprock (Gore, 1992), with the rubble becoming secondarily covered by various encrusting and fleshy algae (Felder et al., 2014; Fredericq et al., 2014; Richards et al., 2014, 2016; Krayesky-Self et al., 2017; Schmidt et al., 2017). We view the autogenic rhodoliths as a specific type of nucleated rhodoliths (sensu Freiwald and Henrich, 1994) in which the core derives from calcium carbonate rubble as opposed to other materials. These two categories of rhodoliths co-inhabit the same rhodolith beds but the internal (endolithic) microbiome of each category differs with regard to the number and diversity of taxa (biogenic: Krayesky-Self et al., 2017, and autogenic: Sauvage et al., 2016). Even though corallines belong in the Supergroup Plants (Yang et al., 2016), the calcium carbonate-encrusted cell lumina are hard and “stony,” and the endophytic (“inside plant”) nature of algal inclusions can be interpreted as endolithic (“inside stone”).
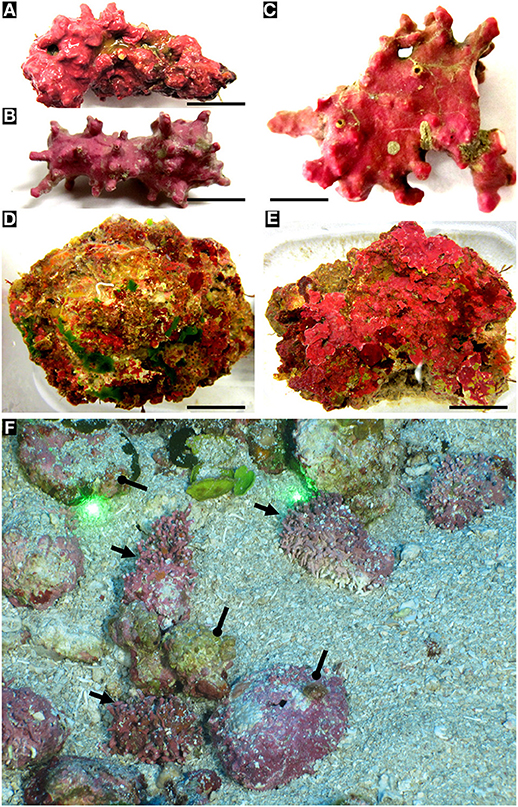
Figure 4. Biogenic and autogenic rhodoliths. (A) biogenic rhodoliths. (A,B) Lithothamnion spp. (Hapalidiales). Scale bars: 0.8 cm; 0.5 cm. (C) Sporolithon sinumexicanum (Sporolithales) collected from the central cavity of a sponge. Scale bar: 0.5 cm. (D,E) Examples of NWGMx autogenic rhodoliths. Scale bars: 1.5 cm; 2 cm. (F) Mohawk ROV photo taken on 24 Sept. 2018 of biogenic (arrows) and autogenic rhodoliths (circle pointers) in East Flower Garden Bank (FGBNMS), 27°55' 21.9612“N, 93°35'31.6674”W, 50.9 depth (Dive 702_0007_131247). Space between spot lasers = 10 cm.
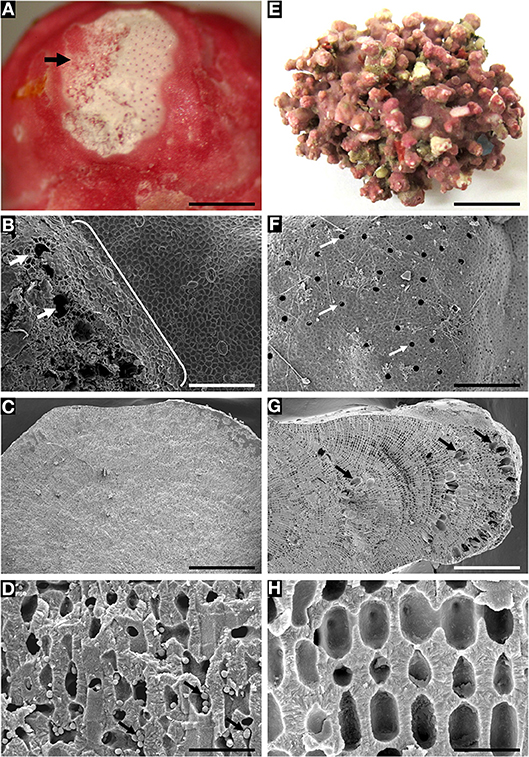
Figure 5. Rhodolith species with sloughed tetrasporangial sori and epithallial layers showing endolithic cells (A–D), and species with non-sloughed tetrasporangial sori (E–H) lacking endolithic cells. (A–D) Sporolithon sinusmexicanum (A,C,D = specimen LAF6970B from Dry Tortugas vicinity [24° 31.494'N, 83°19.793'W, 69 m]; B = specimen LAF6956A from Sackett Bank, LA [28° 38.0'N, 89°33.028'W, 69 m]). (A) Thallus surface showing tetrasporangial sorus (arrow) in the process of sloughing. Scale bar = 500 μm. (B) Disintegrating tetrasporangial sorus showing exposed tetrasporangial compartments (arrows) and sloughing epithallial cells (bracket). Scale bar = 160 μm. (C) Longitudinal fracture of rhodolith protuberance showing lack of buried tetrasporangial compartments. Scale bar = 720 μm. (D) Perithallial cells with putative endolithic cell stages (arrows). Scale bar = 32 μm. (E–H) Sporolithon sp. nov. (LAF7260). (E) Thallus habit. Scale bar = 1.1 cm. (F) Thallus surface with tetrasporangial sorus showing lack of sloughing and pores of intact tetrasporangial compartments. Scale bar = 176 μm. (G) Longitudinal fracture of rhodolith protuberance showing layers of unshed and buried tetrasporangial compartments (arrows). Scale bar = 400 μm. (H) Perithallial cells lacking endolithic cell stages. Scale bar = 22 μm.
Post-DWH observations that prompted us to explore algal diversity within and on the surface of rhodoliths include the fact that (1) Bare, denuded, and “apparently dead” rhodoliths collected at Ewing and Sackett Banks offshore Louisiana were brought back to the laboratory and placed into 75 liter microcosms. After a few weeks, diverse macroalgal growth started to emerge from the rhodoliths' surface, reflecting macroalgal community present prior to the DWH oil spill (Arakaki et al., 2014; Felder et al., 2014; Fredericq et al., 2014), with many taxa reaching sexual maturity and completing their life life cycle. (2) Presence of algal propagules and spores, bacteria, fungal hyphae, and diatoms, dinoflagellates, were shown on the surface (epilithic) or inside (endolithic) autogenic rhodoliths with SEM and epifluorescence microscopy (Felder et al., 2014; Fredericq et al., 2014). Furthermore, SEM, TEM, and Fluorescence microscopy documented previously unrecognized benthic life history stages of bloom-forming microalgae such as the dinoflagellate Prorocentrum lima and the haptophyte Ochrosphaera verrucosa (Krayesky-Self et al., 2017) residing endolithically inside calcium carbonate-lined cell lumina of biogenic CCA rhodoliths (Lithothamnion sp., Hapalidiales). (3) Metabarcoding (amplicon environmental sequencing) of endolithic DNAs from within an Ewing Bank autogenic rhodolith with degenerate primers for plastid tuf A (elongation factor EF-Ttu) (Sauvage et al., 2016) recovered a wide microbiotal diversity of photosynthetic prokaryotic (cyanobacteria) and eukaryotic algae, including red algae (Florideophyceae, Rhodophyta), green algae (Ulvophyceae, Chlorophyta), and Haptophyta. In addition, sequencing of observed single cell inclusion within live CCA cells of biogenic rhodoliths also confirmed their microalgal identity (Krayesky-Self et al., 2017) and remains understudied.
In this paper we expanded upon our previous work by testing 16S V4 for recovering phototrophic diversity of mesophotic rhodoliths exhibiting low phototroph colonization. We also tested cryo-SEM as a potentially a more informative method than regular SEM to minimize artifacts of sample preparation in the study endolithic cell inclusions, and utilized staining techniques to differentiate floridean starch from cellular inclusions. We associated the effect of anatomical growth pattern on presence or absence of cellular inclusions in biogenic rhodoliths.
Rhodolith beds offshore Louisiana and Texas in the NW Gulf of Mexico (NWGMx, Supplementary Figure S1) are associated with salt domes (diapirs), unique mesophotic bank habitats on the continental shelf (Rezak et al., 1985; Felder and Camp, 2009) that are peculiar to that part of the NW Gulf, at ~45–90 m depth (Figures 1, 2). By virtue of their geological history, especially in that they often surmount salt domes where strata trap hydrocarbons, many of the rhodolith habitats are located in or immediately adjacent to areas of intensive oil and gas exploration, production, and transportation (Felder and Camp, 2009). The composite sedimentary overlayer of CaCO3, gypsum, and anhydrite above a salt dome is geologically known as caprock. Anaerobic bacteria obtain the carbon necessary to reduce anhydrite to limestone from petroleum hydrocarbons accumulating in pockets along the edge of the salt dome banks (Gore, 1992), and it is the caprock slopes and peaks that are covered by rhodolith (algal nodule) beds (Minnery et al., 1985; Minnery, 1990; Fredericq et al., 2009, 2014; Felder et al., 2014; Richards et al., 2014, 2016). Offshore Louisiana and Texas (Figures 1, 2), the majority of benthic fleshy, erect, decumbent and crust-forming red, green, and brown seaweeds grow attached to the surface of rhodoliths (Figures 3, 4). In the NWGMX, the continuous and extended rhodolith bed facies may become spatially discontinuous by sandy or silty patches forming scattered mounds of rhodoliths (Figure 3) built by sand tilefish (Malacanthus plumieri Bloch) (Rezak et al., 1985; Tom Bright, pers. comm.; also shown in multiple Flower Garden Banks National Marine Sanctuary (FGBNMS) annotations, photographs, and videos at FGBNMS, a true ecosystem engineer!
Extensive rhodolith beds occur also at the edge of thriving coral reefs in the FGBNMS on the outer continental shelf about 170 km south of the Texas-Louisiana border (Scanlon et al., 2003; Gardner and Beaudoin, 2005; Slowey et al., 2008) but these rhodolith beds (Figures 1–3) are only found below the hermatypic coral zone. These coral reefs are an important natural laboratory for interdisciplinary studies, and we contend the same holds true for their associated rhodolith beds. The FGBNMS has a web page that shows a series of mapped deep banks throughout Louisiana and Texas, with a discussion of continental slope and carbonate bank sedimentary processes, high-resolution seismic stratigraphy, and physical properties of marine sediments in the Gulf of Mexico (http://sanctuaries.noaa.gov/about/pdfs/se_gom.pdf). Hickerson et al. (2008) provided an in-depth habitat characterization scheme that identifies the CCA zone which includes the rhodolith (algal nodule) zone.
Mesophotic rhodolith collections used in this study were collected at Ewing Bank (vicinity of 28°05.7′N, 91°01.2′W) during August 2008 (Pre-DWH), October 2013, and May 2018 (Post-DWH) cruises aboard the R/V Pelican. Rhodoliths were retrieved using an Hourglass-design box dredge (Joyce and Williams, 1969) with minimum tows (usually 10 min or less) at depths ranging from 50–110 m. Water samples and environmental data were collected in situ using a CTD/Rosette system with sensors and Niskin bottles aboard ship. Samples were initially stored on-site by location in containers filled with seawater collected in situ from the same depth and site of the sampled rhodoliths using the onboard CTD water sampling rosette. Samples were kept aerated aboard ship for the duration of the trip (2–4 days) and transferred into microcosms, filled with in situ collected seawater, located in our laboratory at UL Lafayette. Subsamples of rhodoliths were also immediately upon retrieval placed in Ziploc bags filled with the desiccant silica gel for long-term preservation for subsequent DNA analysis and microscopy studies. Pre-and post-DWH-collected rhodoliths from Ewing Bank are housed at the University of Louisiana at Lafayette Herbarium (LAF). Images and samples collected by a Mohawk ROV of rhodolith beds in the West and East Flower Garden Banks were taken at ~55 m depth aboard the R/V Manta in September 2018. An extensive library of images collected during ROV surveys since 2001 throughout the reefs and banks of the northwestern Gulf of Mexico are held by the Flower Garden Banks National Marine Sanctuary, and will be assessed in the future.
Metabarcoding of 16S ribosomal RNA (rRNA) gene with the V4 region (Caporaso et al., 2011) was conducted on three pre- and three post-DWH-collected autogenic rhodoliths from Ewing Bank selected from previous collections desiccated in silica gel (see above). 16S V4 was selected to explore bacterial profiles from these rhodoliths as well as test its viability as an easily amplifiable marker to recover eukaryotic algae in mesophotic rhodolith samples exhibiting relatively low density of endolithic phototrophs. Moreover, our laboratory has previously used the 16S marker in phylogenetic studies of red algae via Sanger sequencing (Olsen et al., 2004; Hommersand et al., 2006; Rodríguez-Prieto et al., 2013). Succinctly, the sampling of endolithic communities consisted in drilling non-encrusted patches (with a drill, n = 5–6 patches per rhodolith) adjacent to CCA or peyssonnelioid crusts, i.e., “bare” substratum with a sterile 1.6 mm (1/16”) bit (Sauvage et al., 2016). DNA extraction was then carried with a PowerSoil DNA Isolation kit (MO BIO Laboratories, Carlsbad, CA, USA. DNA extracts were shipped to MRDNA, where they were amplified with the HotStarTaq Plus Master Mix Kit (Qiagen, USA) with indexed primer for the 16S rRNA V4 region (515F and 806R, Caporaso et al., 2011). Paired-end reads (2 × 250 bp) of the V4 amplicons were generated on the Illumina MiSeq Next-Generation Sequencing platform at MRDNA (www.mrdnalab.com, Shallowater, TX, USA).
Demultiplexed FASTQ files (NCBI SRA accession PRJNA508570) were processed with the DADA2 pipeline (Callahan et al., 2016) to produce RDP-annotated (Cole et al., 2014) 16S V4 OTUs denoised and devoid of chimeras. OTUs as “Chloroplast” and Cyanobacteria were extracted for further taxonomic investigation. We first merge this OTU file with the PhytoREF (Decelle et al., 2015) and built an exploratory tree to visualize without ambiguity the different classes retrieved from the rhodoliths (note that percent identity thresholds to be used with 16S for accurate classification have not been worked out for phototrophs, and thus the tree method provided a much less ambiguous alternative). We then used TREE2FASTA (Sauvage et al., 2018) to rapidly segregate OTUs belonging to the Rhodophyta to a separate file. These were then used for BLASTn search against Genbank's nt to find their closest matches and retrieve contextual sequences for tree building and determine their best identification (ordinal level) (see below and results).
We generated plastid 16S Sanger sequences of pertinent species of Halymeniaceae (Halymeniales) and Bonnemaisoniales, including many taxa from throughout the Gulf of Mexico, following the primer and sequencing protocols listed in Hommersand et al. (2006).
OTUs, Genbank matches, and Sanger sequences (see Supplementary Table S1) were merged, multiple-aligned and run with RAxML (Stamatakis, 2014) with a GTR+ G model of evolution rooted with early-branching Rhodophyta to display Florideophycean taxa found within the sampled endolithic communities (the best ML tree out of 1,000 restarts was kept and branch support assessed with 1,000 bootstrap replicates, full tree not shown). To showcase the phylogenetic representation of OTUs present within endolithic communities or as epilithic spores, we display trees for the Halymeniales, and Bonnemaisoniales.
SEM was used to document coralline morpho-anatomy and endolithic life history stages inside perithallial cells of biogenic rhodoliths. Portions of biogenic rhodoliths from silica gel-dried specimens were removed using a single-edged razor blade and forceps, or live rhodoliths of interest were placed into a folded sheet of paper and underwent short bursts of directed force with a hammer and resulting fragments subsequently preserved in silica gel. Vertical fractures were performed on crustose portions whereas protuberances were sectioned longitudinally using a new razor blade for each fracture. Sections were mounted using conductive adhesive tape or liquid graphite and coated with 5–16.5 nm of gold. Specimens were viewed using a Hitachi S-3000N SEM at a voltage of 15 or 20 kV, or a JEOL field emission scanning electron microscope (FESEM), both housed in the Microscopy Center at UL Lafayette (Richards et al., 2016; Krayesky-Self et al., 2017).
Live rhodoliths for the Cryo-SEM study were taken from the microcosm and immediately placed in liquid nitrogen for 1–2 min. Portions were moved to a Cryo-modified stub that held small amounts of liquid nitrogen. The Cryo-SEM stub was placed into a Hitachi S-3000N SEM at a voltage of 15V. Backscatter electrons were collected for viewing and imaging purposes. The backscatter electron detector was set to either composition mode or to topographic mode. Sublimation was allowed to occur for 2 h under a high vacuum in some cases. Each sublimated sample was sputter-coated with 10–13 nm gold and viewed on a JEOL SEM microscope following the procedure of Pesacreta and Hasenstein (2018).
Microcosm rhodoliths collected from Ewing Bank in May 2018 (Figures 8A,B) and 2013 (Figure 8C) were cut into small sections using a new razor blade for each sample. The hand sections were done on a glass slide covered by seawater. Seawater was then removed using a narrow-tipped transfer pipette. A 2% iodine solution (Carolina Biological Supply, CAT#869093) was added to the sections to test for the presence of floridean starch, and allowed to react for 5 min. The same samples were then rinsed with seawater, viewed with an Olympus SZX7 microscope and photographed using the True Chrome IIS digital system.
Sporolithon sinusmexicanum, a biogenic rhodolith species that sloughs its tetrasporangial sori and epithallial layers, showed putative endolithic cell stages within perithallial cell lumina (Figures 5A–D). In contrast, Sporolithon sp. nov., a species with non-sloughed tetrasporangial sori and epithallial layers, lacked endolithic cell stages (Figures 5E–H) within perithallial cell lumina. At sites where tetrasporangial sori underwent sloughing from the rhodolith surface (Figure 5B), exposed openings into the rhodolith interior were observed that are larger than the pores of the tetrasporangial compartments which are not sloughed (Figure 5F).
SEM documented a variety of presumed microalgae residing endolithically inside calcium carbonate-lined cell lumina of biogenic Lithothamnion (Hapalidiales) rhodoliths (Figures 6A–I). The identity of many of these microalgal stages still needs to be confirmed. Micrographs reveal rhodolith surfaces (epithallus) with underlying perithallus enclosing endolithic cellular inclusions within their calcium carbonate-enclosed cell lumina (Figures 6, 7). Often, free-living microalgae were also seen on the rhodolith surface (Figure 6C). Burrowing taxa were also observed (Figure 6F). Perithallial cells can also be filled with aragonite crystals (Figure 6G) next to perithallial cells containing endolithic cells. There is a great variety of shape and surface ornamentation on endolithic cells (Figures 6H,I). Some of the endolithic cells reside in perithallial fusion cells (Figure 6H)
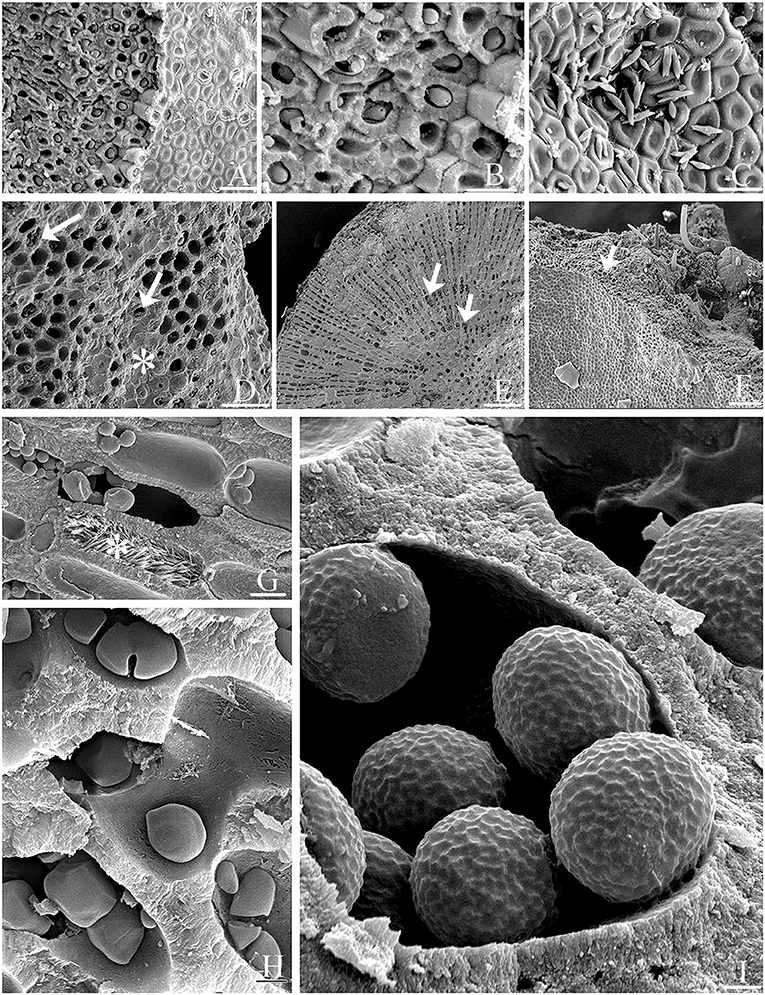
Figure 6. Epilithic microalgae and various endolithic cellular inclusions of unknown taxonomic identity in biogenic rhodoliths. (A) Biogenic rhodolith surface (epithallus, at right) revealing underlying perithallus (at left) with endolithic cellular inclusions within calcium carbonate-enclosed cell lumina. Scale bar = 10 μm. (B) Close-up of (A) Scale bar = 20 μm. (C) Free-living microalgae on rhodolith surface Scale bar = 14 μm. (D) Surface cells (asterisk), and underlying exposed perithallial cells. Epithallial cells (asterisk) have been partly removed exposing endolithic cellular inclusions (arrows) in some perithallial cells. Scale bar = 5 μm. (E) Transverse view of rhodolith nodule showing endolithic cellular inclusions (arrows) within perithallial cells. Scale bar = 200 μm. (F) Surface (epithallus, at left) and fractured perithallus (in center, arrow) with cellular inclusions, and with burrowing taxa (at top). Scale bar = 300 μm. (G) Perithallial cells filled with aragonite crystals (asterisk), or endolithic cells. Scale bar = 20 μm. (H) Perithallial cells filled with endolithic cells. Some endolithic cells reside in perithallial fusion cells. Scale bar = 0.5 μm. (I) Endolithic cells showing surface ornamentation within perithallial cells. Scale bar = 0.7 μm.
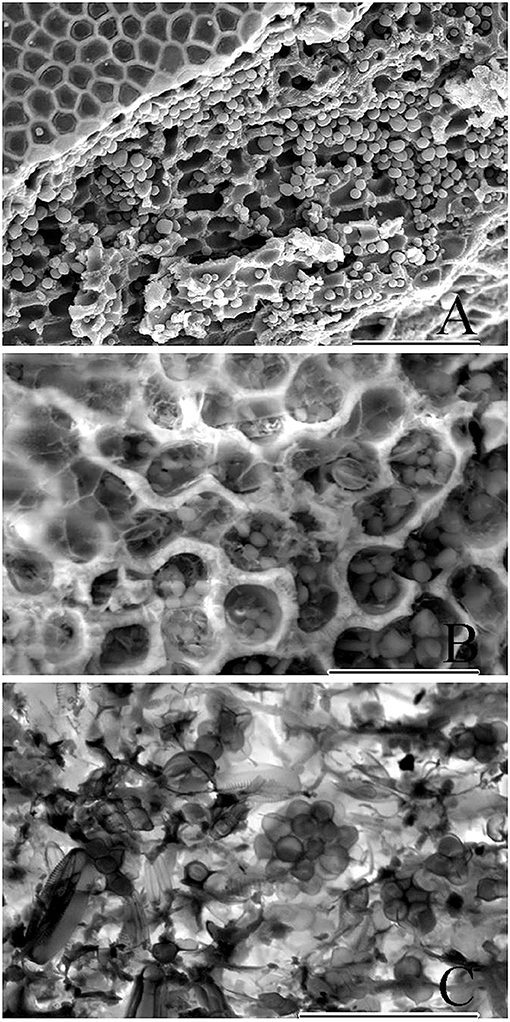
Figure 7. Cryo-SEM micrographs showing copious amounts of endolithic cells. (A) Epithallus (at left) of a biogenic rhodolith, and clusters of endolithic cells viewed following sublimation and gold-coating. Scale bar = 100 μm. (B) Exposed perithalial cells enclosing endolithic cells. Micrograph produced using backscatter electrons collected with the detector mode set to topographic. Scale bar = 30 μm. (C) Micrograph showing clusters of endolithic cells, produced using backscatter electrons with the detector set to composition mode. Scale bar = 30 μm.
Cryo-SEM micrographs (Figures 7A–C) showed copious amounts of clusters of endolithic cells within perithallial cells and validate the use of this method as a better approach when live material is available to better document endolithic cells. In samples that have been frozen, sublimation, and gold-coating bypass traditional fixation and dehydration protocols (Figure 7A) resulting in micrographs that include larger numbers of endolithic cells since they were not lost during the fixation and dehydration process. Furthermore, frozen samples can be viewed using backscatter electrons collected with the detector mode set to topographic (Figure 7B) or composition mode (Figure 7C). The composition mode is sensitive to variance in atomic number contrast, while topographic mode is sensitive to contrast between depths.
When rhodolith sections were exposed to iodine (Figure 8), floridean starch granules inside some rhodolith cells turned purple, indicating that these represented the floridean starch intrinsic to the coralline cells. Some areas contained brownish cellular inclusions but tested negative for floridean starch, whereas other regions contained a mixture of starch and cellular inclusions (Figures 8A–C, box).
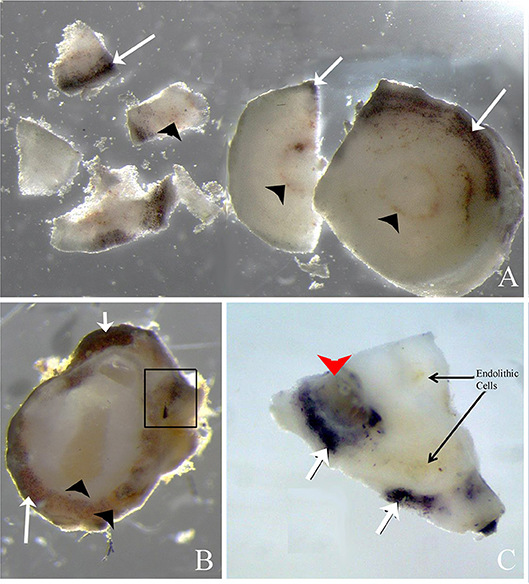
Figure 8. Rhodolith sections exposed to 2% iodine solution. (A,B) Cross sections of rhodolith protuberance that has been exposed to iodine resulting in purple staining of floridean starch (arrows) present in the coralline cells. Brownish endolithic cells (arrowheads) do not stain purple after exposure to iodine. Some areas contain perithalial cells with and without purple stain, indicating that endolithic cells may have consumed floridean starch (in box, B). (C) Periclinal section of rhodolith nodule showing darkly staining floridean starch, brownish endolithic cells, and areas where both floridean starch and endolithic cells co-occur (arrowhead).
Metabarcoding of 16S V4 rRNA recovered a large proportion of bacterial taxa (heterotrophs) as compared to phototrophs (Cyanobacteria and eukaryotic algae) in terms of richness (number of OTUs per phylum) and abundance (total read number per phylum) (see C/C annotation for “Cyanobacteria/Chloroplast” on Figures 9A,B bar graphs) (Table 1). The most abundant prokaryotes consisted of 10 phyla of Bacteria and 1 phylum of Archaea (Thaumarchaeota). Sequencing depth strongly varied (see total read number per sample, Figure 9B and Table 1) with ENV14 (pre-DWH) and ENV10 (post-DWH) being the most sampled. Among these two samples, ENV10 exhibited higher richness (i.e., more OTUs) than ENV14 in spite of the latter larger sequencing depth (compare bar height in Figures 9A,B. Because of their more adequate sampling (sequencing depth), ENV10 and ENV14 may reflect phyletic profiles of the endolithic community within a mesophotic rhodolith more accurately as compared to the other samples, which mostly harbored the most abundant bacterial phyla (Proteobacteria, Firmicutes, Actinobacteria). A total of 55 phototrophic OTUs were found aside 1,321 heterotrophic OTUs in 4 phyla (Cyanophyta, Chlorophyta, Rhodophyta, Ochrophyta) (Figures 9C,D). Phototrophic OTUs represented <5% of total OTUs per sample (range 0.8 to 4.7%), and <4% of total read abundance per sample (range 0.2 to 3.4%) (Table 1, Figure 9). As for heterotrophic profiles, the post-DWH ENV10 exhibited greater richness and abundance than the pre-DWH ENV14 and may highly likely represent an exceptionally rich endolithic community rather than an effect of the DWH-spill since other samples (ENV11 and ENV12) exhibited very few phototrophs comparable to other pre-DWH samples (ENV04 and ENV13). Overall, the phototrophic OTU profiles may be extremely skewed because of the unequal sequencing depth of the samples and their relative low proportion in 16S data.
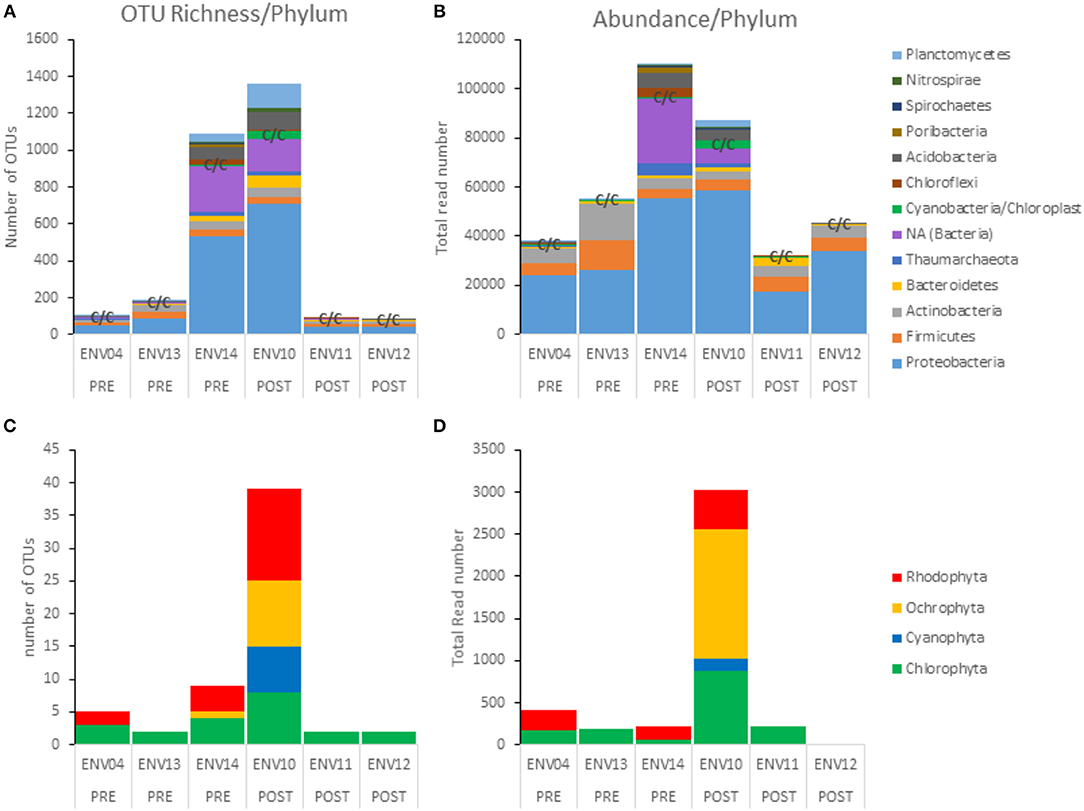
Figure 9. Phototroph richness (OTU number) and abundance (read counts) retrieved with 16S V4 rDNA from pre- and post-DWH rhodolith endolithic communities. (A,B) All phyla. (C,D) Phototrophic phyla only. Note the relatively small number of OTUs and abundance of phototrophic OTUs (C/C for Cyanobacteria/Chloroplast) as compared to heterotrophic phyla.
Using tree methods with PhytoREF coupled with Genbank Blastn search (not shown), phototrophic OTUs obtained from the rhodoliths CaCO3 could be identified in seven classes. These included “Bacillariophyceae” (5 OTUs), Cyanophyceae (7 OTUs), Florideophyceae (20 OTUs), Pedinophyceae (1 OTU), Phaeophyceae (4 OTUs), Pinguiophyceae (1 OTU), and Ulvophyceae (16 OTUs). Interestingly, the Ulvophyceae and the Florideophyceae were the diversity-richest classes. The Ulvophyceae included primarily euendoliths, such as Ostreobium and related unresolved Bryopsidales with 16S V4. The Florideophyceae represented spores or alternative filamentous life stages of otherwise epilithic/epiphytic fleshy macroalgae from several orders, i.e., the Bonnemaisoniales, Gracilariales, Gelidales, Gigartinales, Dumontiaceae-complex, Halymeniales, Nemaliales, and Sebdeniales. A few crustose members of Florideophytes, e.g., Corallinophycidae, and Peyssonneliales were also detected representing filaments or contaminants introduced from accidental drilling during sample preparation prior to DNA extraction. The Phaeophyceae represented primarily members of the order Ectocarpales. Phylogenetic trees showcasing Halymeniales and Bonnemaisoniales (Figure 10) and retrieved OTUs belonging to these orders showed 100% identity to a previously unrecorded genus for the Gulf of Mexico, Reticulocaulis (Naccariaceae, Bonnemaisoniales), and a Halymenia sp. (Halymeniaceae, Halymeniales) previously sequenced in our laboratory rom thalli collected during collecting cruises.
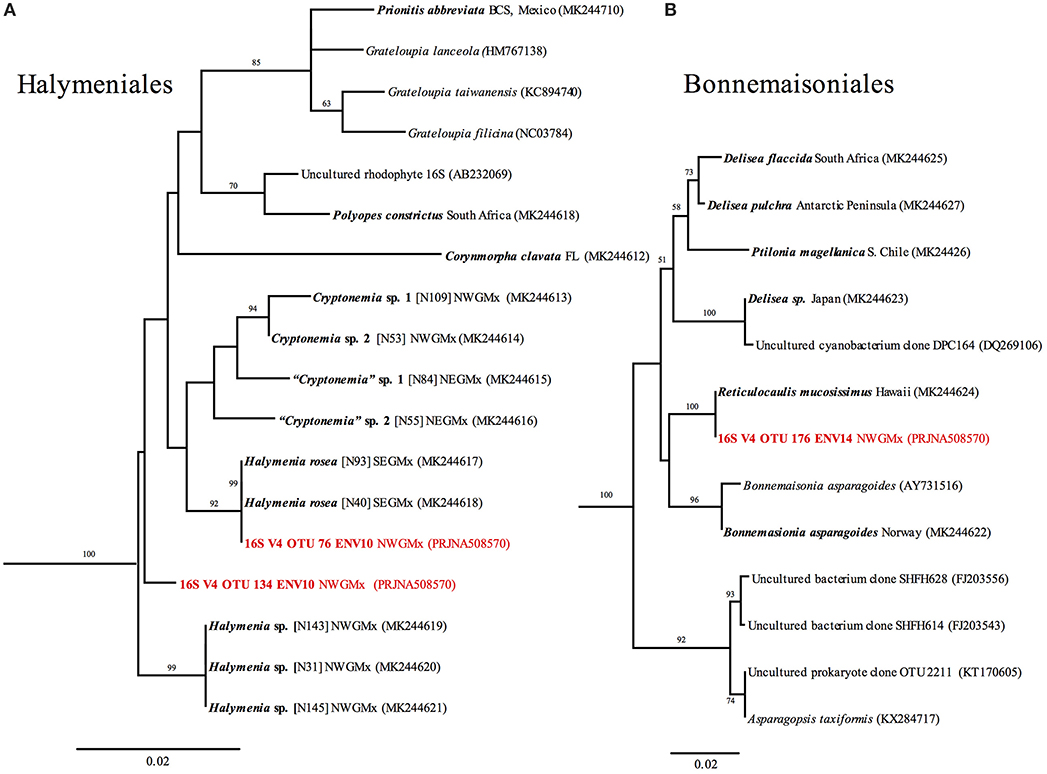
Figure 10. RaxML trees of (A) Halymeniaceae and (B) Bonnemaisoniales based on 16S sequences, and 16S V4 metabarcodes (in red). Newly generated sequences are shown in bold. Branch numbers indicate bootstrap values >50 out of 1,000 replicates.
Based on our previous research, the fact that post-DWH-collected macroalgae were not conspicuously visible at Ewing Bank in situ at their corresponding pre-DWH-collecting depths, sites and dates, but subsequently appeared in microcosms, suggested that rhodoliths play an important role as seedbank reservoirs of dormant microscopic stages of macroalgae (Fredericq et al., 2014; Sauvage et al., 2016; Krayesky-Self et al., 2017) to persist through adverse environmental conditions. Hitherto unknown cryptic, microscopic life history stages (e.g., small sporophytes) of macroalgae that are part of the rhodolith microbiota can now be linked with their macroscopic thalli using environmental sequencing. Our previous metabarcoding investigations of tuf A, which included a rhodolith from the Gulf of Mexico (ENV14, Sauvage et al., 2016) and newly presented 16S V4 data (herein), coupled with the above observations of newly emerging taxa in our microcosms, reveal that the interior of rhodoliths contained previously unknown life history stages of macroalgae, some of which we described taxonomically earlier (Arakaki et al., 2014). Additional linking of microscopic sporophytic stages with their macroscopic female/male gametophytes using multi-marker metabarcoding, Sanger sequencing, and various microscopy tools are needed to better ascertain the true biodiversity of the NWGMx's phototrophic component. Other calcareous substrata are well-known to be essential for life cycle completion of some seaweeds in which the heteromorphic stage of the sporophyte individual (in which meiosis occurs) is often a small crust, disk or aggregation of creeping filaments that does not resemble the larger gametophyte individual (e.g., Drew, 1949; Guiry, 1990; Hawkes, 1990; Hommersand and Fredericq, 1990).
Many planktonic microalgae, such as dinoflagellates (Dale, 1983; Steidinger and Garcés, 2006; Steidinger, 2010; Bravo and Figueroa, 2014), are known to possess benthic stages (i.e., cysts). Our previous work on rhodolith-associated microalgae (Krayesky-Self et al., 2017) demonstrated that common coastal bloom-forming microalgae may also be endolithically associated within biogenic rhodolith cells for part of their life cycle and we hypothesized that this phenomenon is common and widespread in the marine environment. For instance, our previous tuf A metabarcoding analysis pointed out that there was a large proportion of Ochrosphaera verrucosa Schussnig sequences originating from endolithic cells inside calcareous substrata (Sauvage et al., 2016). Krayesky-Self et al. (2017) were able to extract this species from Ewing Bank rhodoliths using single cell genome amplification. O. verrucosa is a common haptophyte in coastal waters worldwide and a taxon of which all the stages in its proposed haplo-diplontic cycle have not yet been observed (Fresnel and Probert, 2005) and may have been overlooked in the endolithic niche. Here, 16S V4 retrieved limited microalgae that we attribute to inadequate sequencing depth. In ENV10, an autogenic rhodolith showing the most phototrophic diversity, we retrieved an OTU identified as a Pinguiophyceae (Pinguiococcus pyrenoidosus), whose cyst is unknown (Andersen et al., 2002), and like O. verrucosa, may also occur in the endolithic niche. With deeper sequencing on a larger number of samples, we expect that more microalgae will be documented. Overall, we contend that numerous other microalgae use the calcareous niche for part of their life cycle and that the discovery of unsuspected endolithic stages may reveal important implications for understanding and predicting the onset of phytoplankton blooms, including those that form harmful algal blooms (HABs, Faust and Gulledge, 2002). Further research may help resolve incomplete life histories and enable one to link previously unknown endolithic stages of bloom-forming microalgae with their free-living stages, a current “hot” topic of great interest to HAB research (e.g., Steidinger and Garcés, 2006; Steidinger, 2010).
Our observations of thalli newly emerging in laboratory microcosms revealed that rhodoliths housed previously unknown life history stages of red (e.g., Halarachnion louisianensis, Arakaki et al., 2014; Schmitzia sp., unpubl. data), brown (e.g.,“Syringoderma” floridana, Camacho et al., 2018, 2019), and green algae (Sauvage et al., 2016). However, when excluding the rhodolith substratum, the culturing of these taxa prevented their life cycle completion, thus corroborating the critical importance of CaCO3 substrata in the life cycle of many macroalgae.
Using 16S V4 metabarcoding, we were able to link the taxonomic identity of the “invisible,” cryptic parts of a macroalga inside a rhodolith with their corresponding “visible” macroscopic thalli through Sanger sequencing of plastid 16S-generated reference sequences from the NWGMx. Whereas 16S V4 is already an established metabarcode for all prokaryotes, we will continue to show that it can resolve phototrophic diversity (i.e., cyanobacteria, eukaryotes) as well. The macroalgal component will thus vastly improve with our preliminary 16S reference database and greatly increase our understanding of general phototrophic biodiversity in rhodolith beds and other marine communities. Our research answers the call by Yoon et al. (2016) that efforts should be made to update 23S and 16S sequences in databases. A 16S framework will allow the scientific community at large to re-examine their 16S datasets and accurately resolve their phototrophic component (e.g.,. 16S sequences labeled “uncultured cyanobacterium” in Genbank actually refer to specific species of Delisea and Asparagopsis (Bonnemaisoniales, Figure 10B). Thus, grossly unexplored biodiversity, new species discovery, and information relevant at both the barcode and phylogenetic level will improve with 16S V4 metabarcoding, as with other metabarcodes, and will greatly contribute to viewing both biogenic and autogenic rhodoliths as overlooked major marine diversity hotspots.
The point of entry into the interior of biogenic rhodoliths (those fully accreted by corallines as opposed to secondarily colonizing nodules, i.e., autogenic rhodoliths) and later exit of life history stages of micro- and macroalgae are currently unresolved and purely speculative at this time. Corallines have the capacity to slough off their external (epithallial) cell layers (Wegeberg and Pueschel, 2002), including parts of their conceptacles (reproductive structures), that can become replenished with a unique type of intercalary meristem. It is possible that the roundish cellular inclusions (= microalgal life history stages) present within individual calcium carbonate-lined coralline cell lumina may passively become surrounded by new coralline surface cell layer growth, or perhaps enter/exit via the large pit connections that approximate the cell width of the endolithic stages, or via cell fusions (Krayesky-Self et al., 2017). There is no consensus about the nature of such cellular inclusions that have either been referred to as floridean starch (Viola et al., 2001), chloroplasts (Kunkel, 2005-2013), or bacteria (Garbary and Veltkamp, 1980; Kazmierczak and Iryu, 1999; Roh and Sim, 2012). The hypothesis that these inclusions were floridean starch was quickly rejected when iodine starch reaction turned negative for the inclusions (Figure 8). Likewise, the large size of the cellular inclusions excluded the concentric structures from being chloroplasts or bacteria (O'Reilly et al., 2012). Because we observed some coralline cells containing starch grains, and others which did not, we speculate that some of the cellular inclusions in biogenic rhodoliths are heterotrophic, consuming starch granules within perithallial cells of a rhodolith for their required maintenance. It is interesting that, from what we have observed so far, the biogenic rhodoliths that include the cellular inclusions also slough off their tetrasporangial sori and surface layers, e.g., Sporolithon sinusmexicanum (Figures 5A–D). In contrast, the species that does not slough off tetrasporangial sori layers does not include cellular inclusions, e.g., Sporolithon sp. nov. (Figures 5E–H). The larger openings formed by sloughing off of the tetrasporangial sori may provide a larger and more direct route for life history stages of microalgae to enter the rhodolith. Observing larger sample sizes of rhodoliths in future studies may shed light on whether this phenomenon is consistent throughout the populations of these two species and if similar phenomena occur in other species.
Our Cryo-SEM studies have shown us that the number of endolithic cells (Figure 7) is greater than revealed by traditional SEM. Furthermore, the variation of the surface ornamentation of the endolithic cells is more variable than previously observed with SEM and FESEM (Krayesky-Self et al., 2017).
We speculate that the majority of microalgal endolithic populations (aside from euendolithic taxa such as Ostreobium) within biogenic rhodoliths may not be permanent residents of these rhodoliths but are transient life history stages (potentially resting stages) that may form blooms once released in the water column from the rhodolith's interior following abrasion or sloughing off of the coralline's surface cell layers. Alternatively, micro- and macro-borers could also be responsible for releasing these cellular inclusions by opening burrows to the water column. This is a fascinating aspect of bentho-planktonic coupling of microalgal species and their association with CCA that we hope will get resolved in the future. Euendolithic taxa and filaments of alternate life stages of macroalgae may possibly enter biogenic rhodoliths via such mechansism. For comparison, in autogenic rhodoliths (those secondarily colonized from initially inert CaCO3), patches of “open” substratum adjacent to encrusting red algae may function as a direct port of entry to the endolithic niche.
Metabarcoding of 16S V4 rRNA recovered abundant Bacteria, some Archaea and limited phototrophic diversity, i.e., cyanobacteria and eukaryotic algae (Figure 9). Sequencing depth seemed grossly insufficient in order to comprehensively enumerate the biodiversity present in 4 of the samples (ENV04, ENV11, ENV12, ENV13). Nonetheless, while sequencing depth is important, it did not always equate to greater richness (i.e., compare ENV10 vs. ENV14). Overall, based on our analyses, we recommend that exploration of mesophotic rhodolith biodiversity via 16S V4 should be done with depth of >100,000 to 200,000 paired-end reads (possibly more) to stabilize prokaryotic profiles and better access their phototrophic fraction. Indeed, because of the extremely conserved nature of V4's priming region in the chloroplast of algae, which is derived from prokaryotic ancestors, and the inherent overabundance of bacterial taxa in any environmental samples, the latter tend to strongly “mask” phototrophs. For instance in a recent study of phytoplankton communities, the use of 16S for metabarcoding still only resulted in 3–9% of phototrophic reads (Bennke et al., 2018), about twice more than obtained here (<5%). In this context, additional or alternative markers better targeting phototrophic diversity may be considered. For instance, UPA, the Universal Plastid Amplicon (Sherwood and Presting, 2007) should amplify as easily as 16S V4 and retrieve a much larger phototrophic component because of its greater specificity to chloroplasts, but it may not amplify some Chlorophyta that harbor introns in this region (T. Sauvage, pers. obs.). The chloroplast gene tuf A is another alternative to retrieve phototrophs in higher relative abundance (Sauvage et al., 2016). While we had successfully amplified tuf A at UL Lafayette, MRDNA failed to produce PCR products using fusion primers (containing Illumina adapters), except for the ENV14 rhodolith (see Sauvage et al., 2016, sample labeled GM14). We recognize that tuf A, while a very high quality metabarcode for its phylogenetic informativeness across numerous algal phyla, can be more difficult to amplify because of the high degeneracy of the environmental primers. Samples with low phototroph density, such as those of mesophotic rhodoliths here, can be more challenging to optimize and thus successful amplicons should rely on alternative library preparation than fusion primers. Regardless, tuf A and 16S V4 metabarcodes seemed to retrieve the same 3 phyla for ENV14 (see Figure 13b in Sauvage et al., 2016, and Figure 9), but further database curation and referencing of specimens than presently available in the 16S PhytoREF (Decelle et al., 2015) will be needed for finer scale identification of 16S OTUs at lower taxonomic rank (Edgar, 2018).
A display of OTUs' topological relationships with our newly generated Sanger data for reference specimens in the Halymeniales and Bonnemaisoniales (Figure 10) illustrates the power of metabarcoding to track alternate life stages (filaments/spores) of macroalgae. These trees, showcasing these two orders, demonstrated the presence of a previously unknown record of Reticulocaulis mucosissimus (Naccariaceae, Bonnemaisoniales) for the Gulf of Mexico, whose OTU exhibited a 100% identity match with our 16S reference collections from Hawaii. Reticulocaulis started to emerge from the surface of pre-DWH rhodoliths in our laboratory micoscosms. Since we never collected it in the field, it may be either ephemeral, rare, or not expressed in its macroalgal stage in the NWGMx. A Halymenia sp. OTU (Halymeniaceae, Halymeniales), also exhibited 100% identity to previously sequenced specimens in our laboratory on the basis of macroalgae dredged during previous cruises or emerging in our laboratory microcosms. A second OTU represented an unknown member of the Halymeniales that was also present but for which we could not yet produce a matching 16S sequence. Both Halymeniales OTUs originated from ENV10, a post-DWH sample, and while grown thalli were not observed in the field, were present endolithically.
Because of the 2010 DWH oil spill offshore Louisiana (see Supplementary Figure S1 for site of the Deepwater Horizon Macondo Well Blowout) and disappearance of mesophotic macroalgae at Ewing Bank, NWGMx, prompted us to look at rhodoliths endolithically following observation of regeneration of macroalgae in microcosms (Felder et al., 2014; Fredericq et al., 2014; Krayesky-Self et al., 2017), it opened many doors for new research directions and paradigms. One such direction made clear that further research is needed to link the eukaryotic component of the rhodolith holobiont (“total organism”) with its co-occurring prokaryotic component. Great taxon diversity implies great genetic diversity, hence more gene families that will be expressed, and hence more modes of metabolism that will be directly related to biogeochemical cycles, among others. The biogeochemical cycles, in turn, provide the nutrients to support the high taxon diversity of the rhodolith holobiont. If any part of this described sequence is disturbed, the integration of the entire system falls apart and may take years to recover. This is what we believe happened in the NWGMx following the 2010 DWH oil spill: a biogeochemical upheaval post-DWH may have caused a shift in nutrients which led to a change in both the gene expression of the prokaryotic component and a shift in prokaryotic and eukaryotic taxon diversity (i.e., algal die-off) externally, on the surface of the rhodoliths. In contrast, the cryptic microbiotal component inside the rhodolith post-DWH acted as a stable buffer zone resilient to outside anthropogenic disturbances potentially allowing for regeneration. This is the reason macroalgae have emerged from “apparently dead” rhodoliths in our microcosms, while their growth, for reasons still unknown (e.g., herbivory, residual contaminants?), is still suppressed in the field (Ewing Bank). There is an obvious need to continue going back to monitor Ewing Bank to track the fate of the rhodolith beds. Additional research will shed light about ecosystem resilience and the extent rhodoliths play in biogeochemical and biodiversity cycling in their respective ecosystems. To address this, quantitative biogeochemical and transcriptomics data (both metatranscriptomics and qPCR) will need to be compared with the phylogenetic diversity of algal, prokaryotic, viral, and fungal communities and their role in biological succession following a disturbance using statistical models.
To inform us about the entire community composition associated with a rhodolith, deep metagenomic (non-amplicon environmental sequencing) studies will need to be conducted on rhodoliths worldwide. Metagenomes will allow us to recover viruses, fungi, metazoans, and other taxa that were not captured by our selected metabarcodes, but some of which were seen in our micrographs. Metagenomics will also allow us to evaluate the total rhodolith holobiont regardless of metabarcode bias. Cavalcanti et al. (2014) conducted metagenomic studies on rhodoliths from the Abrolhos Bank, Brazil, but the focus was on prokaryotes and they did not differentiate between the rhodolith interior (endolithic) and surface (epilithic) and whether the rhodoliths were biogenic or autogenic. It is especially timely to define the dynamics of the essential biogeochemical cycles associated with rhodolith beds in the NWGMx considering the recent disappearance of conspicuous macroalgae at Ewing Bank since the DWH oil spill (April 2010) with no visible sign of their recovery as of May 2018, our last collecting cruise date.
The fact that phototrophic OTUs could be obtained from within pre- and post-DWH rhodoliths suggests that although currently not visible in the field, seaweeds are still present in the NWGMx as “resting” microscopic stages (e.g., spores) within the rhodolith CaCO3. Additional research on rhodoliths from different NWGMx areas is needed. Further exploration worldwide is also essential to confirm our findings that rhodoliths are marine biodiversity hotspots for unsuspected eukaryotic life and to ascertain whether the rhodolith interior functions as seedbanks for algal stages, as temporary reservoirs for life history stages of algal bloom-forming species, or as refugia for ecosystem resilience following environmental stress. Such studies will allow detection of community changes (including bioinvasions) in the face of possible re-colonization after catastrophic events. The study may have major implications for the prediction of microalgal blooms, shifts in the benthic primary-producer rhodolith community that followed the 2010 Deepwater Horizon Oil Spill in the NW Gulf of Mexico, and the possible effects of global warming and ocean acidification on the calcifying rhodoliths and their microbiota.
SF, TS, SK-S, JR, and WS conceived the study; SF, SK-S, WS, TS, NA, RK, and JR collected the samples; TS, SK-S, JR, WS, and NA conducted the laboratory work; TS, SK-S, and WS performed the data analyses. SF and TS wrote the manuscript with contributions from WS, SK-S, JR, NA, EH, and RK. All authors edited the manuscript before submission.
This work was funded by NSF DEB-1754504.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We greatly acknowledge the support from NSF research grants DEB-0315995, and DEB1754504, and thank the crew of the R/V Pelican for their help with sampling protocols aboard ship and close research collaborators Darryl L. Felder, James N. Norris, Olga Camacho, Dago Venera-Ponton, Daniela Gabriel and D. Wilson Freshwater. We thank the FGBNMS, R/V Manta crew and Jason White and Eric Glidden from the University of North Carolina-Undersea Vehicle Program (WUNCW-UVP) for the ROV-collections and images taken at the East and West Flower Garden Banks. We also thank Brooke Bocklud for her work with the light microscopy, and the UL Lafayette Microscopy Center for their interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00502/full#supplementary-material
Supplementary Figure 1. Map of the Gulf of Mexico showing collection locality data ( ). From left to right: West Flower Garden Bank (WFG), East Flower Garden Bank (EFG), Ewing Bank (EB), Sackett Bank (SB), and vicinity of the Dry Tortugas, Florida (DT), and site of the Deepwater Horizon Macondo Well Blowout (
). From left to right: West Flower Garden Bank (WFG), East Flower Garden Bank (EFG), Ewing Bank (EB), Sackett Bank (SB), and vicinity of the Dry Tortugas, Florida (DT), and site of the Deepwater Horizon Macondo Well Blowout ( ).
).
Supplementary Table 1. Taxon and collection data of plastid 16S reference sequences and GenBank accession numbers (including newly generated sequences shown in boldface) and 16 V4 sequences of Halymeniaceae (Halymeniales) and Bonnemaisoniales. Newly generated 16 V4 OTUs are shown in red (NCBI SRA). SF, Suzanne Fredericq; DWF, Wilson Freshwater; MHH, Max H. Hommersand; JR, Jan Rueness.
Adey, W. J., Halfar, J., and Williams, B. (2013). The coralline genus Clathromorphum Foslie emend. Adey; biological, physiological and ecological factors controlling carbonate production in an Arctic/Subarctic climate archive. Smithsonian Contr. Mar. Sc. 40, 1–83. doi: 10.5479/si.1943667X.40.1
Amado-Filho, G. M., Maneveldt, G., Marins, B., Manso, R. C. C., Pacheco, M. R., and Guimarães, S. P. B. (2007). Structure of rhodolith beds from 4 to 55 meters deep along the southern coast of Espírito Santo State, Brazil. Cienc. Mar. 33, 399–410. doi: 10.7773/cm.v33i4.1148
Amado-Filho, G. M., Moura, R. L., Bastos, A. C., Salgado, L. T., Sumida, P. Y., Guth, A. Z., et al. (2012). Rhodolith beds are major CaCO3 bio-factories in the tropical South West Atlantic. PLoS ONE 7:e35171. doi: 10.1371/journal.pone.0035171
Andersen, R. A., Potter, D., and Bailey, J. C. (2002). Pinguiococcus pyrenoidosus gen. et sp. nov. (Pinguiophyceae), a new marine coccoid alga. Phycol. Res. 50, 57–66. doi: 10.1111/j.1440-1835.2002.tb00136.x
Andersson, A. J., Bates, N. R., and Mackenzie, F. T. (2007). Dissolution of carbonate sediments under rising pCO2 and ocean acidification: observations from Devil's Hole, Bermuda. Aquat. Geochem. 13, 237–264. doi: 10.1007/s10498-007-9018-8
Anthony, K. R., Kline, D. I., Diaz-Pulido, G., Dove, S., and Hoegh-Guldberg, O. (2008). Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc. Natl. Acad. Sci. U.S.A. 105, 17442–17446. doi: 10.1073/pnas.0804478105
Arakaki, N., M., Suzuki, M., and Fredericq, S. (2014). Halarachnion (Furcellariaceae, Rhodophyta), a newly reported genus for the Gulf of Mexico, with the description of H. louisianensis, sp. nov. Phycol. Res. 62, 306–315. doi: 10.1111/pre.12065
Bennke, C. M., Pollehne, F., Müller, A., Hansen, R., Kreikemeyer, B., and Labrenz, M. (2018). The distribution of phytoplankton in the Baltic Sea assessed by a prokaryotic 16S rRNA gene primer system. J. Plankton Res. 40, 244–254. doi: 10.1093/plankt/fby008
Borowitzka, M. A., and Larkum, A. W. D. (1987). Calcification in algae: mechanisms and the role of metabolism. Crit. Rev. Plant Sci. 6, 1–45. doi: 10.1080/07352688709382246
Bravo, I., and Figueroa, R. I. (2014). Towards and ecological understanding of dinoflagellate cyst functions. Microorganisms 2, 11–32. doi: 10.3390/microorganisms2010011
Burdett, J. L., Hatton, A. D., and Kamenos, N. A. (2015). Coralline algae as a globally significant pool of marine dimethylated sulfur. Glob. Biogeochem. Cycl. 29, 1845–1853. doi: 10.1002/2015GB005274
Cabioch, J. (1969). Les fonds de maërl de la baie de Morlaix et leur peuplement végétal. Cah. Biol. Mar. 10, 139–161.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Camacho, O., Fernández-García, C., Vieira, C., Gurgel, C. F. D., Norris, J. N., Freshwater, D. W., et al. (2019). The genus Lobophora (Dictyotales, Phaeophyceae) in the western Atlantic and eastern Pacific oceans with the description of eight new species. J. Phycol.
Camacho, O., Sauvage, T., and Fredericq, S. (2018). Taxonomic transfer of syringoderma to microzonia (Syringodermataceae, Syringodermatales), including the new record M. floridana (E.C.Henry) comb. nov. in the Gulf of Mexico. Phycologia 57, 413–421. doi: 10.2216/17-51.1
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS 108 (Suppl. 1), 4516–4522. doi: 10.1073/pnas.1000080107
Cavalcanti, G. S., Gregoracci, G. B., dos Santos, E. O., C. B., Silveira, C. B., Meirelles, P. M., Longo, L., et al. (2014). Physiologic and metagenomic attributes of the rhodoliths forming the largest CaCO3 bed in the South Atlantic Ocean. ISME J. 8, 52–62. doi: 10.1038/ismej.2013.133
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucl. Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Dale, D. (1983). “Dinoflagellate resting cysts: “benthic plankton” in Survival Strategies of the Algae, ed G. A. Fryxell (New York, NY: Cambridge University Press), 69–136.
Decelle, J., Romac, S., Stern, R. F., Bendif, E. M., Zingone, A., Audic, S., et al. (2015). PhytoREF: a reference database of the plastidial 16S rRNA gene of photosynthetic eukaryotes with curated taxonomy. Mol. Ecol. Res. 5, 1435–1445. doi: 10.1111/1755-0998.12401
Doropoulos, C., Ward, S., Diaz-Pulido, G., Hoegh-Guldberg, O., and Mumby, P. J. (2012). Ocean acidification reduces coral recruitment by disrupting intimate larval-algal settlement interactions. Ecol. Lett. 15, 338–346. doi: 10.1111/j.1461-0248.2012.01743.x
Drew, K. M. (1949). Conchocelis-phase in the life-history of Porphyra umbilicalis (L.) Kütz. Nature 164, 748–749. doi: 10.1038/164748a0
Edgar, R. (2018). Taxonomy annotation and guide tree errors in 16S rRNA databases. PeerJ 6:e5030. doi: 10.7717/peerj.5030
Faust, M. A., and Gulledge, R. A. (2002). Identifying harmful marine dinoflagellates. Contr. U.S. Natl. Herbarium 42, 1–144.
Felder, D. L., and Camp, D. K. (Eds) (2009). Gulf of Mexico Origin, Waters and Bioota, Biodiversity. Vol. 1. College Station, TX: A & M University press.
Felder, D. L., Thoma, B. P., Schmidt, W. E., Sauvage, T., Self-Krayesky, S., Chistoserdov, A., et al. (2014). Seaweeds and decapod crustaceans on Gulf deep banks after the Macondo Oil Spill. Bioscience 64, 808–819. doi: 10.1093/biosci/biu119
Foster, M. S. (2001). Rhodoliths: between rocks and soft places—minireview. J. Phycol. 37, 659–667. doi: 10.1046/j.1529-8817.2001.00195.x
Foster, M. S., McConnico, L. M., Lundsten, L., Wadsworth, T., Kimball, T., Brooks, L. B., et al. (2007). Diversity and natural history of a Lithothamnion muelleri-Sargassum horridum community in the Gulf of California. Cienc. Mar. 33, 367–384. doi: 10.7773/cm.v33i4.1174
Fredericq, S., Arakaki, N., Camacho, O., Gabriel, D., Krayesky, D., Self-Krayesky, S., et al. (2014). A dynamic approach to the study of rhodoliths: a case study for the Northwestern Gulf of Mexico. Crypt. Algol. 35, 77–98. doi: 10.7872/crya.v35.iss1.2014.77
Fredericq, S., Cho, T. O., Earle, S. A., Gurgel, C. F., Krayesky, D. M., Mateo Cid, L. E., et al. (2009). “Seaweeds of the Gulf of Mexico,” in Gulf of Mexico: Its Origins, Waters, and Biota. I. Biodiversity, eds D. L. Felder and D. K. Camp (College Station: Texas A&M Univ. Press), 187–259.
Freiwald, A., and Henrich, R. (1994). Reefal coralline algal build-ups within the Arctic Circle: morphology and sedimentary dynamics under extreme environmental seasonality. Sediment 41, 963–984. doi: 10.1111/j.1365-3091.1994.tb01435.x
Fresnel, J., and Probert, I. (2005). The ultrastructure and life cycle of the coastal coccolithophorid Ochrosphaera neapolitana (Prymnesiophyceae). Eur. J. Phycol. 40, 105–122. doi: 10.1080/09670260400024659
Garbary, D., and Veltkamp, C. J. (1980). Observations on Mesophyllum lichenoides (Corallinaceae, Rhodophyta) with the scannning electron microscope. Phycologia 19, 49–53. doi: 10.2216/i0031-8884-19-1-49.1
Gardner, J. V., and Beaudoin, J. (2005). High-resolution multibeam bathymetry and acoustic backscatter of selected northwestern Gulf of Mexico outer shelf banks. Gulf Mexico Sci. 1, 5–29. doi: 10.18785/goms.2301.03
Guiry, M. D. (1990). “Sporangia and spores,” in Biology of the Red Algae, eds K. M. Cole and R. G. Sheath (Cambridge: Cambridge Univ. Press), 347–376.
Hadfield, M. J., and Paul, V. J. (2001). “Natural chemical cues for settlement and metamorphosis of marine invertebrate larvae,” in Marine Chemical Ecology, eds J. B. Clintock and B. J. Baker (Boca Raton: CRC Press), pp. 431–461
Halfar, J., Adey, W. H., Kronz, A., Hetzinger, S., Edinger, E., and Fitzhugh, W. (2013). Arctic sea ice decline archived by multicentury annual-resolution crustose coralline algal proxy. Proc. Natl. Acad. Sci. 110, 19737–19741. doi: 10.1073/pnas.1313775110
Harvey, A. S., Harvey, R. M., and Merton, E. (2017). The distribution, significance and vulnerability of Australian rhodolith beds: a review. Mar. Freshw. Res. 68, 411–428. doi: 10.1071/MF15434
Hawkes, M. W. (1990). “Reproductive strategies,” in Biology of the Red Algae, eds K. M. Cole and R. G. Sheath (Cambridge: Cambridge Univ. Press), 455–476.
Heyward, A. J., and Negri, A. P. (1999). Natural inducers for coral larval metamorphosis. Coral Reefs 18, 273–279. doi: 10.1007/s003380050193
Hickerson, E. L., Schmahl, G. P., Robart, M., Precht, W. E., and Caldow, C. (2008). “The state of coral reef ecosystems of the Flower Garden Banks, Stetson Bank, and other banks in the northwestern Gulf of Mexico,” in: The State of Coral Reef Ecosystems of Flower Garden Banks, 190–218. Available online at: https://nmsflowergarden.blob.core.windows.net/flowergarden-prod/media/archive/document_library/scidocs/stateofcoralfgbnms08.pdf
Hinojosa-Arango, G., Maggs, C. A., and Johnson, M. P. (2009). Like a rolling stone: the mobility of maerl (Corallinaceae) and the neutrality of the associated assemblages. Ecology 90, 517–528. doi: 10.1890/07-2110.1
Hinojosa-Arango, G., and Riosmena-Rodríguez, R. (2004). Influence of rhodolith-forming species and growth-form on associated fauna of rhodolith beds in the central-west Gulf of California, México. Mar. Ecol. 25, 109–127. doi: 10.1111/j.1439-0485.2004.00019.x
Hommersand, M. H., and Fredericq, S. (1990). “Sexual reproduction and cystocarp development,” in Biology of the Red Algae, eds K. M. Cole and R. G. Sheath (Cambridge: Cambridge Univ. Press) 305–45.
Hommersand, M. H., Freshwater, D. W., Lopez Bautista, J., and Fredericq, S. (2006). Proposal of the Euptiloteae Hommersand and Fredericq, Trib. Nov., and transfer of some Southern Hemisphere Ptiloteae to the Callithamnieae (Rhodophyta). J. Phycol. 42, 203–225. doi: 10.1111/j.1529-8817.2006.00175.x
Joyce, A. E., and Williams, J. (1969). Rationale and pertinent data. Mem. Hourglass Cruises 1, 11–50.
Kamenos, N. A., Burdett, H. L., Aloisio, E., Findlay, H. S., Martin, S., Longbone, C., et al. (2013). Coralline algal structure is more sensitive to rate, rather than the magnitude, of ocean acidification. Glob. Change Biol. 19, 3621–3628. doi: 10.1111/gcb.12351
Kamenos, N. A., Strong, S. C., Shenoy, D. M., Wilson, S. T., Hatton, A. D., and Moore, P. G. (2008). Red coralline algae as a source of marine biogenic dimethylsulphonio-propionate. Mar. Ecol. Progr. Ser. 372, 61–66. doi: 10.3354/meps07687
Kazmierczak, J., and Iryu, Y. (1999). Cyanobacterial origin of microcrystalline cements from Pleistocene rhodoliths and coralline algal crusts of Okierabu-jima, Japan. Acta Palaeont. Polon. 44, 117–130.
Krayesky-Self, S., Richards, J. L., Rahmatian, M., and Fredericq, S. (2016). Aragonite infill in overgrown conceptacles of coralline Lithothamnion spp. (Hapalidiaceae, Hapalidiales, Rhodophyta): new insights in biomineralization and phylomineralogy. J. Phycol. 52, 161–173. doi: 10.1111/jpy.12392
Krayesky-Self, S., Schmidt, W. E., Phung, D., Henry, C., Sauvage, T., Camacho, O., et al. (2017). Eukaryotic life inhabits rhodolith-forming coralline algae (Hapalidiales, Rhodophyta), remarkable marine benthic microhabitats. Sc. Rep. 7:45850. doi: 10.1038/srep45850
Kunkel, D. (2005-2013). Dennis Kunkel Microscopy, Inc. Available online at: https://pixels.com/featured/1-coralline-red-alga-dennis-kunkel-microscopysci ence-photo-library.html
Leliaert, F., Tronholm, A., Lemieux, C., Turmel, M., DePriest, M. S., Bhattacharya, D., et al. (2016). Chloroplast phylogenomic analyses reveal the deepest-branching lineage of the Chlorophyta, Palmophyllophyceae class. nov. Sci. Rep. 6:25367. doi: 10.1038/srep25367
Littler, M. M., Littler, D. S., Blair, S. M., and Norris, J. N. (1985). Deepest known plant life discovered on an unchartered seamount. Science 227, 57–59. doi: 10.1126/science.227.4682.57
McCoy, S. J., and Kamenos, N. A. (2015). Coralline algae (Rhodophyta) in a changing world: integrating ecological, physiological, and geochemical responses to global change. J. Phycol. 51, 6–24. doi: 10.1111/jpy.12262
Minnery, G. A. (1990). Crustose coralline algae from the Flower Garden Banks, Northwestern Gulf of Mexico - controls on distribution and growth-morphology. J. Sedim. Petrol. 60, 992–1007.
Minnery, G. A., Rezak, R., and Bright, T. J. (1985). “Chapter 18: Depth zonation and growth form of crustose coralline algae: Flower Garden Banks, northwestern Gulf of Mexico,” in Paleoalgology: Contemporary Research and Applications eds. D. F. Toomey and M. H. Titecki (Berlin; Heidelberg: Springer-Verlag), 238–246.
Morse, J. W., Andersson, A. J., and Mackenzie, F. T. (2006). Initial responses of carbonate-rich shelf sediments to rising atmospheric pCO2 and “ocean acidification”: Role of high Mg-calcites. Geochim. Cosmochim. Acta 70, 5814–3580. doi: 10.1016/j.gca.2006.08.017
Nelson, W., D'Arcino, R., Neill, K., and Farr, T. (2014). Macroalgal diversity associated with rhodolith beds in northern New Zealand. Cryptogam Algol 35, 27–47. doi: 10.7872/crya.v35.iss1.2014.27
Olsen, K. N., Melton, R. D., Yaudes, K. M., Norwood, K. G., and Freshwater, D. W. (2004). Characteristics and utility of plastid-encoded 16S rRNA gene sequence data in phylogenetic studies of red algae. J. North Carolina Acad. Sc. 120, 143–151.
O'Reilly, S., Jurley, S., Coleman, N., Monteys, X., Szpak, M., O'Dwyer, T., et al. (2012). Chemical and physical features of living and non-living maerl rhodoliths. Aquat. Biol. 15, 215–224. doi: 10.3354/ab00431
Paris, C. B., Hénaff, M. L., Aman, Z. M., Subramaniam, A., Helgers, J., Wang, D. P., et al. (2012). Evolution of the Macondo well blowout: simulating the effects of the circulation and synthetic dispersants on the subsea oil transport. Env. Sc. Technol. 46, 13293–13302, doi: 10.1021/es303197h
Pereira-Filho, G. H., Amado-Filho, G. M., de Moura, R. L., Bastos, A. C., Guimarães, S. M. P. B., Salgado, L. T., et al. (2012). Extensive rhodolith beds cover the summits of southwesten Atlantic Ocean seamounts. J. Coast. Res. 28, 261–269. doi: 10.2112/11T-00007.1
Pereira-Filho, G. H., Amado-Filho, G. M., Guimarães, S. M., Moura, R. L., Sumida, P. Y., Abrantes, D. P., et al. (2011). Reef fish and benthic assemblages of the Trindade and Martin Vaz island group, southwestern Atlantic. Braz. J. Oceanogr. 59, 201–212. doi: 10.1590/S1679-87592011000300001
Pereira-Filho, G. H., Veras, P., de, C., Francini-Filho, R. B., Moura, R. L. de, Pinheiro, H. T., Gibran, F. Z., et al. M (2015). Effects of the sand tilefish Malacanthus plumieri on the structure and dynamics of a rhodolith bed in the Fernando de Noronha Archipelago, tropical West Atlantic. MEPS 541, 65–73. doi: 10.3354/meps11569
Pesacreta, T. C., and Hasenstein, K. H. (2018). Tissue accumulation patterns and concentrations of potassium, phosphorus, and carboxyfluorescein translocated from pine seed to the root. Planta 248, 393–407. doi: 10.1007/s00425-018-2897-7
Rabalais, N. (2014). Assessing early looks at biological responses to the Macondo Event. BioScience 64, 757–759. doi: 10.1093/biosci/biu132
Ragazzola, F., Foster, L. C., Form, A., Anderson, P. S. L., Hansteen, T. H., and Fietzke, J. (2012). Ocean acidification weakens the structural integrity of coralline algae. Glob. Change Biol. 18, 2804–2812. doi: 10.1111/j.1365-2486.2012.02756.x
Ragazzola, F., Foster, L. C., Jones, J. J., Scott, T. B., Fietzke, J., Kilburn, M. R., et al. (2016). Impact of high CO2 on the geochemistry of the coralline algae Lithothamnion glaciale. Sci. Rep. 6:20572. doi: 10.1038/srep20572
Rezak, R., Bright, T. J., and McGrail, D. W. (1985). Reefs and Banks of the Northwestern Gulf of Mexico: Their Geological, Biological, and Physical Dynamics. New York,NY: Wiley. 259.
Richards, J. L., and Fredericq, S. (2018). Sporolithon sinusmexicanum sp. nov. (Sporolithales, Rhodophyta): a new rhodolith-forming coralline species from deepwater rhodolith beds in the Gulf of Mexico. Phytotaxa 350, 135–146. doi: 10.11646/phytotaxa.350.2.2
Richards, J. L., Gabrielson, P. W., and Fredericq, S. (2014). New insights into the genus Lithophyllum (Lithophylloideae, Corallinaceae, Corallinales) from offshore the NW Gulf of Mexico. Phytotaxa 190, 162–175. doi: 10.11646/phytotaxa.190.1.11
Richards, J. L., Vieira-Pinto, T., Schmidt, W. E., Sauvage, T., Gabrielson, P. W., Oliveira, M. C., et al. (2016). Molecular and morphological diversity of Lithothamnion spp. rhodoliths (Hapalidiaceae, Hapalidiales) from deepwater rhodolith beds in the northwestern Gulf of Mexico. Phytotaxa 278, 81–114. doi: 10.11646/phytotaxa.278.2.1
Riosmena-Rodríguez, R., and Medina-Lopez, M. (2011). “The role of rhodolith beds in the recruitment of invertebrate species in the Southwestern Gulf of Mexico,” in: All Flesh is Green, Plant-Animal Interrelationships. Cellular Origin, Life in Extreme Habitats and Astrobiology Vol. 16, eds J. Seckbach and Z. Dubinsky (New York, NY: Springer), 417–428.
Riosmena-Rodríguez, R., Nelson, W., and Aguirre, J. (Eds) (2017). Rhodolith/Maërl Beds: A Global Perspective. Switzerland: Springer International Publishing. 368.
Rodríguez-Prieto, C., Freshwater, D. W., and Hommersand, M. H. (2013). Vegetative and reproductive development of Mediterranean Gulsonia nodulosa (Ceramiales, Rhodophyta) and its genetic affinities. Phycologia 52, 357–367. doi: 10.2216/13-132.1
Roh, E., and Sim, J. (2012). “Biomineralization of carbonate minerals: implications for rhodolith formation,” in Proceeding 2nd International Conference on Environment and BioScience Vol 44 (Singapore), 48–53.
Sauvage, T., Plouviez, S., Schmidt, W. E., and Fredericq, S. (2018). TREE2FASTA: a flexible Perl script for batch extraction of FASTA sequences from exploratory phylogenetic trees. BMC Res. Notes 11:164. doi: 10.1186/s13104-018-3268-y
Sauvage, T., Schmidt, W. E., Suda, S., and Fredericq, S. (2016). A metabarcoding framework for facilitated survey of coral reef and rhodolith endolithic communities with tufA. BMC Ecol. 16:8. doi: 10.1186/s12898-016-0068-x
Scanlon, K. M., Ackerman, S. D., and Rozycki, J. E. (2003). Texture, Carbonate Content, and Preliminary Maps of Surficial Sediments, Flower Garden Banks Area, Northwest Gulf of Mexico outer shelf. U.S. Geol. Surv. Open File Report 03–002.
Schmidt, W. E., Lozada-Troche, C., Ballantine, D. L., Arakaki, N., Norris, J. N., Gabriel, D., et al. (2017). Taxonomic transfer of Chrysymenia enteromorpha and C. wrightii to Botryocladia (Rhodymeniaceae, Rhodymeniales, Rhodophyta). Phytotaxa 324, 122–138. doi: 10.11646/phytotaxa.324.2.2
Sherwood, A. R., and Presting, G. G. (2007). Universal primers amplify a 23S rDNA plastid marker in eukaryotic algae and cyanobacteria. J. Phycol. 43, 605–608. doi: 10.1111/j.1529-8817.2007.00341.x
Slowey, N., Holcombe, T., Betts, M., and Bryant, W. (2008). “Habitat islands along the shelf edge of the northwestern Gulf of Mexico,” in A Scientific Forum on the Gulf of Mexico: The Islands in the Stream Concept, Marine Sanctuaries Conservation Series, eds K. B. Ritchie and B. Keller (Silver Spring: NOAA), 19–24.
Smith, J. E., Shaw, M., Edwards, R. A., Obura, D., Pantos, O., Sala, E., et al. (2006). Indirect effects of algae on coral: algae-mediated, microbe-induced coral mortality. Ecol. Lett. 9, 835–845. doi: 10.1111/j.1461-0248.2006.00937.x
Spalding, H. L., Amado-Filho, G. M., Bahia, R. G., Ballantine, D. L., Fredericq, S., Leichter, J. J., et al. (2019). “Macroalgae,” In: Mesophotic Coral Ecosystems, eds Y. Loya, K. A. Puglise, and T. C. L. Bridge (New York, NY: Springer).
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Steidinger, K. A. (2010). Research on the life cycles of harmful algae: A commentary. Deep Sea Res. II 57, 162–165. doi: 10.1016/j.dsr2.2009.09.001
Steidinger, K. A., and Garcés, E. (2006). “Importance of life cycles in the ecology of harmful microalgae,” in: Ecology of Harmful Algae, eds E. Granéli and, J. T. Turner (Berlin: Springer), 37–49.
Steneck, R. S. (1986). The ecology of coralline algal crusts: convergent patterns and adaptive strategies. Ann. Rev. Ecol. Syst. 17, 273–303. doi: 10.1146/annurev.es.17.110186.001421
Teigert, S. (2014). Hollow rhodoliths increase Svalbard's shelf biodiversity. Sci. Rep. 4:6972. doi: 10.1038/srep06972
Vecsei, A. (2004). A new estimate of global reefal carbonate production including the fore-reefs. Global Planet. Change 43, 1–18. doi: 10.1016/j.gloplacha.2003.12.002
Viola, R., Nyvall, P., and Pedersen, M. (2001). The unique features of starch metabolism in red algae. Proc. Biol. Sc. 268, 1417–1422. doi: 10.1098/rspb.2001.1644
Wang, S., Maltrud, M., Elliott, S., Cameron-Smith, O., and Jonko, A. (2018). Influence of dimethyl sulfide on the carbon cycle and biological production. Biogeochem 138, 49–68. doi: 10.1007/s10533-018-0430-5
Webster, N. S., Negri, A. P., Botté, E. S., Laffy, P. W., Flores, F., Nooonan, S., et al. (2016). (2016). Host-associated coral reef microbes respond to the cumulative pressures of ocean warming and ocean acidification. Sci. Rep. 6:19324. doi: 10.1038/srep19324
Webster, N. S., Uthicke, S., Botte, E. S., Flores, F., and Negri, A. P. (2013). Ocean acidification reduces induction of coral settlement by crustose coralline algae. Glob. Change Biol. 19, 303–315. doi: 10.1111/gcb.12008
Wegeberg, S., and Pueschel, C. M. (2002). Epithallial and initial cell fine structure in species of Lithothamnion and Phymatolithon (Corallinales, Rhodophyta). Phycologia 41, 228–244. doi: 10.2216/i0031-8884-41-3-228.1
Yang, E. C., Boo, S. M., Bhattacharya, D., Saunders, G. W., Knoll, A. H., Fredericq, S., et al. (2016). Divergence time estimates and evolution of major lineages in the florideophyte red algae. Sci. Rep. 6:21361. doi: 10.1038/srep21361
Keywords: CCA, coralline algae, Gulf of Mexico, marine biodiversity, mesophotic, metabarcoding, rhodoliths
Citation: Fredericq S, Krayesky-Self S, Sauvage T, Richards J, Kittle R, Arakaki N, Hickerson E and Schmidt WE (2019) The Critical Importance of Rhodoliths in the Life Cycle Completion of Both Macro- and Microalgae, and as Holobionts for the Establishment and Maintenance of Marine Biodiversity. Front. Mar. Sci. 5:502. doi: 10.3389/fmars.2018.00502
Received: 31 October 2018; Accepted: 14 December 2018;
Published: 08 January 2019.
Edited by:
Julio Aguirre, University of Granada, SpainReviewed by:
Christine Maggs, Queen's University Belfast, United KingdomCopyright © 2019 Fredericq, Krayesky-Self, Sauvage, Richards, Kittle, Arakaki, Hickerson and Schmidt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suzanne Fredericq, c2xmOTIwOUBsb3Vpc2lhbmEuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.