- Department of Biology, Memorial University of Newfoundland, St. John’s, NF, Canada
Coral bacterial associates can play important functional roles for the holobiont, such as nitrogen cycling, nutrient processing, and supporting immunity. While bacteria found within the microbiome of corals may benefit the host, they can also be linked to pathogenesis. In the deep-sea, cold-water corals, like their warm shallow-water counterparts, host bacterial communities, but have received little attention due to logistical constraints in sampling. In particular, bacteria associated with surficial mucus of cold-water corals have not yet been investigated. Here, tissue and mucus samples of Paragorgia arborea were collected from three submarine canyons along the continental slope of the Gulf of Maine. Bacterial DNA was extracted from tissue and mucus samples and sequencing of the V6–V8 hypervariable region of the 16S rRNA gene was performed using Illumina MiSeq. The bacterial communities associated with P. arborea compartments (tissue and mucus) and sampling locations (canyon) differed significantly in composition. Proteobacteria, Tenericutes, and Spirochaetes were the dominant phyla across the majority of coral tissue samples, with Gammaproteobacteria and Alphaproteobacteria identified as the largest Proteobacteria contributors across all samples. Operational taxonomic units (OTUs) belonging to the taxa Spirochaeta, Mycoplasma, Flavobacteriaceae, Terasakiellaceae, Campylobacterales, and Rickettsiales were identified as biomarkers (bacterial taxa significantly more abundant in a specific coral microhabitat) of P. arborea tissues, while Paracoccus was a biomarker of P. arborea mucus. Many of the recovered biomarker taxa may be involved in nitrogen cycling. Representatives from several bacterial families (Vibrionaceae, Campylobacteraceae, Rhodobacteraceae, Flavobacteriaceae, and Burkholderiaceae) previously reported in diseased scleractinians, were present in P. arborea as rare bacterial taxa. Characterizing the bacterial associates present in visibly healthy coral colonies provides a benchmark of dominant and rare bacterial groups present in the cold-water coral holobiont. This is the first characterization of bacterial groups associated with P. arborea, examining both tissue- and mucus-specific communities.
Introduction
Corals host a wide range of microbial associates, including bacteria, eukaryotes, archaea, and viruses (Ainsworth et al., 2017). Of these, coral-associated bacteria (herein referred to colloquially as the “microbiome”) may play important roles in host nitrogen metabolism (Grover et al., 2014; Rädecker et al., 2015; Kellogg et al., 2016; Lawler et al., 2016), nutrient cycling (Wild et al., 2004; Naumann et al., 2009), and antibacterial mechanisms (Kelman et al., 1998; Sutherland et al., 2004; Ritchie, 2006). The microbiome typically helps maintain coral health and provides a defense system against disease (Krediet et al., 2013). However, it can also harbor low abundances of pathogens that may become dominant when subjected to environmental stressors; such dysbiosis may lead to coral disease and/or death (Mouchka et al., 2010; Egan and Gardiner, 2016). The coral microbiome likely consists of a combination of commensals, transients, and long-term, stable partners selected by the host (Ainsworth et al., 2015), and is distributed among several anatomical compartments: the skeletal tissue, polyp tissue, and the external surface mucopolysaccharide layer (SML) (Bourne and Munn, 2005; Brown and Bythell, 2005; Sweet et al., 2011; Krediet et al., 2013; Ainsworth et al., 2015).
The SML has been reported as a first line of defense to protect the coral host against pathogens from the surrounding water column (Bythell and Wild, 2011; Sweet et al., 2011; Glasl et al., 2016). Additionally, the mucus is important for particulate feeding and can act as an energy carrier/provider and as a particle trap, as it is sloughed-off into the surrounding waters (Coffroth, 1991; Wild et al., 2008; Bythell and Wild, 2011). In shallow-water scleractinian corals, mucus is subject to diurnal or hourly replacement cycles, and its bacterial biodiversity could be changing with these cycles (Ainsworth et al., 2010; Sweet et al., 2011). The contrast between the more stable tissue/skeletal regions and the frequently renewing mucus compartment can explain differences in bacterial compositions between those coral microhabitats (Bourne and Munn, 2005; Sweet et al., 2011).
Like their warm, shallow-water counterparts, cold-water corals found in the deep-sea (beyond the photic zone to over 2000 m depth) host diverse microbial communities. However, they are difficult to access due to logistical and financial constraints (Kellogg et al., 2016) and therefore little is known about them, and even less about their microbiomes (Holm and Heidelberg, 2016). In particular, the bacterial communities colonizing the SML of corals from deep-sea regions remain unexplored and may differ from those of shallow-water corals due to expected discrepancies in physical conditions within the mucus layer (e.g., the absence of zooxanthellae-linked diel oxygen fluctuations in the SML of corals from deep habitats) and functions (e.g., a greater importance of particulate feeding in the deep sea). This study aims to characterize the bacterial associates of the cold-water alcyonacean coral, Paragorgia arborea (Linnaeus, 1758). P. arborea has been suggested to be a brooding azooxanthellate coral (Roberts et al., 2006; Lacharité and Metaxas, 2013) and is widely distributed in the Northwestern and Northeastern Atlantic, from the Gulf of Maine northward along the eastern coasts of Canada and the Davis Strait, to the continental slopes of Greenland, along the Reykjanes Ridge of Iceland, to the shelf of Norway (Cairns and Bayer, 2005; Mortensen and Buhl-Mortensen, 2005; Buhl-Mortensen et al., 2015; Brooke et al., 2017). P. arborea occupies a wide range of depth (200–1200 m) where temperatures range between 3 and 8°C, serving as a foundation species across a broad geographic area (Buhl-Mortensen et al., 2015). The arborescent morphology of P. arborea provides niches for prey refuge and creates habitats for facultative and obligate deep-sea symbionts (Buhl-Mortensen and Mortensen, 2004; Lacharité and Metaxas, 2013).
The bacterial communities of tropical, cold water, and deep-sea Alcyonacea and gorgonians are typically dominated by Proteobacteria, specifically Gammaproteobacteria (Penn et al., 2006; Bayer et al., 2013; Correa et al., 2013; La Rivière et al., 2013, 2015; Vezzulli et al., 2013; Ransome et al., 2014; Kellogg et al., 2016; Robertson et al., 2016). Additionally, Alphaproteobacteria were identified as notable contributors to the microbial consortia across multiple studies on alcyonaceans and gorgonians (Gray et al., 2011; Bayer et al., 2013; Correa et al., 2013; La Rivière et al., 2013, 2015; Vezzulli et al., 2013; Ransome et al., 2014; Holm and Heidelberg, 2016; Kellogg et al., 2016; Robertson et al., 2016). While Proteobacteria remain largely dominant across alcyonacean microbiomes, in some cases Spirochaetes (Holm and Heidelberg, 2016; Lawler et al., 2016; van de Water et al., 2016) and Tenericutes (Gray et al., 2011; Holm and Heidelberg, 2016) were the most abundant bacterial associates.
In deep-sea ecosystems, food sources are limited and variable and therefore alternate mechanisms are required by benthic invertebrates for metabolic processes (Mueller et al., 2014; Middelburg et al., 2015). Such mechanisms include bacteria-mediated nitrogen cycling, which has been documented in shallow-water corals (Wegley et al., 2007; Rädecker et al., 2015), and more recently in the cold-water scleractinian coral Lophelia pertusa (Middelburg et al., 2015), and postulated for the octocoral Paramuricea placomus (Kellogg et al., 2016) and Anthothelidae corals (Lawler et al., 2016). In these studies, several bacterial species are thought to facilitate nitrogen metabolism, with Spirochaeta implicated in nitrogen fixation (Lawler et al., 2016), Pirellulaceae in nitrification (Kellogg et al., 2016), Kiloniellales and Bacillus spp. in denitrification (Verbaendert et al., 2011; Kellogg et al., 2016; Lawler et al., 2016), Propionibacterium and Oceanospirillales in nitrogen reduction (Kellogg et al., 2016; Lawler et al., 2016), and Campylobacterales in nitrate/nitrite ammonification in a deep-sea octocoral (Kellogg et al., 2016). P. arborea has been observed to occupy the same benthic distribution and topography as L. pertusa (Buhl-Mortensen et al., 2015) and P. placomus (Buhl-Mortensen and Buhl-Mortensen, 2014), and may show similar host–bacterial interactions.
In the deep-sea, corals play a similar role to their shallow water counterparts, providing prey refuge and rugose habitats to many organisms (Hixon and Beets, 1993; Syms and Jones, 2000; Stone, 2006). However, coral ecosystems are threatened by several anthropogenic stressors and there is increasing concern that cold-water corals located along continental shelves are at risk from disturbances such as bottom trawling and oil exploration (Bavestrello et al., 1997; Fosså et al., 2002; Husebø et al., 2002; Roberts et al., 2006; Cordes et al., 2016). As only a few cold-water corals have been studied, there is a knowledge gap regarding these organisms’ bacterial communities and their functional roles in the host. Previous studies have reported variability in bacterial compositions between octocoral genera (Brück et al., 2007), between congeners (Holm and Heidelberg, 2016; Kellogg et al., 2016), between coral microhabitats (Weinbauer et al., 2012), within species and between sampling locations (Gray et al., 2011). In this study, we characterized the bacterial associates of P. arborea across two compartments: (1) skeletal and polyp tissue, and (2) surface mucus, and examined the degree of similarity in bacterial composition across three study locations in the Gulf of Maine (ranging in depth between 411 and 700 m) to explore the effects of relative sampling location proximity and depth on bacterial composition. Due to biological traits observed in P. arborea (i.e., abundant mucus production; Etnoyer et al., 2006, and brooding; Lacharité and Metaxas, 2013), and the previously observed differences in bacterial compositions in tissues and mucus in other coral species, we expect to see between-compartment variation as well as variability among sampling locations. We compared common/dominant bacterial associates, searched for taxa that could be markers of either tissue or mucus due to significant differences in relative abundance, and identified rare bacterial taxa of particular interest. To our knowledge, this work represents the first description of the bacterial associates recovered from tissue and mucus samples of the alcyonacean coral P. arborea.
Materials and Methods
Sample Collection and Study Sites
Paragorgia arborea colony fragments were collected from multiple submarine canyons: Nygren-Heezen Intercanyon (N40°51.96′, W66°32.74′, depth 700 m); Corsair Canyon (N41°21.26′, W66°5.39′, depth 411 m); and Georges Canyon (N41°16.48′, W66°11.59′, depth 423 m), on the continental slope of the Gulf of Maine during a research cruise aboard the National Oceanic and Atmospheric Association (NOAA) Ship Henry B. Bigelow from June 8th to 22nd, 2017. Coral colonies were located and sampled using CSSF-ROPOS (Canadian Scientific Submersible Facility, Remotely Operated Platform for Ocean Sciences), with individual fragments held in separate water-filled chambers until surfacing. Tissue fragments were then dissected from the coral stalk and placed in individual, sterile cryovials. Mucus samples were collected by gently rolling a sterile cotton swab over the exterior of the specimen where mucus was visible. Three tissue replicates, and one mucus swab were collected from individual colonies and frozen (−20°C) until further analysis. At each dive location, a reference water sample was collected from approximately 1 m from the bottom, within proximity of the coral colonies, using a remotely triggered Niskin bottle attached to the ROV. On board, 50 mL aliquots of each water sample were passed through individual 0.22 μm syringe filters (MilliporeSigma, Canada). The syringe filters were frozen at −20°C until further analysis.
Nucleic Acid Extraction
Total DNA was extracted from P. arborea mucus and tissue samples, following the protocol described by Sunagawa et al. (2010) with modifications. Samples of coral tissue and skeleton (between 0.100 and 0.250 g total weight) were washed in 600 μL phosphate-buffered saline (PBS) three times to remove mucus and loosely associated bacteria. Once washed, the samples were flash frozen in liquid nitrogen and crushed using a sterile mortar and pestle. Crushed samples were transferred to tubes included with the PowerViral Environmental RNA/DNA extraction kit (Mo Bio, Carlsbad, CA, United States) containing 0.1 mm glass beads. DNA was extracted from water filters by adding 600 μL PV1 lysis buffer (Mo Bio) into the filter, incubating for 2 min at room temperature, and subsequently purging the filter cartridge using a syringe to collect all lysis buffer. The process was repeated for the reverse side of the syringe filter and once more for the original starting side to ensure maximum PV1 based lysis and recovery. Mucus swabs were placed in microfuge tubes and vortexed with PV1 lysis buffer (Mo Bio) for 10 min. After initial lysis, DNA was extracted according to the manufacturer’s protocol. Prior to high throughput sequencing, the presence of bacterial DNA was confirmed by PCR amplification of 16S rRNA genes using the universal primers ECO8F (Edwards et al., 1989) and 1492R (Stackebrandt and Liesack, 1993), DreamTaq Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, United States), and thermal cycled: 30 cycles of 95°C for 5 min, 95°C for 30 s, 45°C for 30 s, 72°C for 1 min, and final extension of 72°C for 2 min. Extracted DNA was checked for quality and quantity using NanoDropTM 1000 (Thermo Fisher Scientific).
16S rRNA Gene Sequencing and Processing
Coral DNA extracts were outsourced for sequencing at the Centre for Comparative Genomics and Evolutionary Bioinformatics (Dalhousie University, Halifax, Canada). 2 × 300-bp paired-end sequencing of the 16S rRNA gene was performed using Illumina MiSeq v3, with all samples amplified using previously published primers targeting the V6–V8 regions (B969F/B1406R): V6 forward: 5′- CCATCTCATCCCTGCGTGTCTCCGACTCAG and V8 reverse: 5′- CCTATCCCCTGTGTGCCTTGGCAGTCTCAG (Comeau et al., 2011).
Sequence data were processed using the in-house developed SPONS-2 pipeline, as described in Verhoeven and Dufour (2017), with a few modifications. In short, sequences were trimmed to remove both low-quality bases using Trimmomatic version 0.38 (20-base sliding window with a minimum average quality of 15 per base) and short reads (<100 bases) (Bolger et al., 2014). Reads that passed the initial quality check were merged using PEAR version 0.9 (Zhang et al., 2014). Primers were trimmed from merged reads using CutAdapt (maximum error rate 0.2), filtering out reads that lack forward or reverse primers (Martin, 2011). Reads with an average Phred score below 20 were removed as a final quality check prior to defining operational taxonomic units (OTUs) using SWARM version 2.2 (Mahé et al., 2015). The step wherein Swarm defines OTUs was modified so the maximum difference between amplicons (d) was increased from 1 to 3 (decreasing the potential for overestimating defined OTUs). Defined OTUs were analyzed using the RDP naïve Bayesian classifier (Wang et al., 2007) for taxonomic assignment, using the SILVA SSU database (release 132; Quast et al., 2013) with a 51% minimum bootstrap confidence estimate when assigning taxonomy.
Bioinformatics and Statistical Analyses
Microbiome high-throughput sequence (HTS) datasets are compositional (Gloor et al., 2017), and recent concerns have been raised regarding current microbiome analysis methodologies (i.e., normalizing HTS count data by rarefaction or other subsampling methods) (Fernandes et al., 2014; McMurdie and Holmes, 2014). Here, we used a compositional analysis approach to compare the bacterial communities from coral and seawater samples.
Operational taxonomic unit count and taxonomic data were imported in R for analyses (R Development Core Team, 2008). First, filtering was conducted to remove OTUs not classified as bacteria at the Kingdom level. Bacterial alpha diversity in each coral and water sample was then examined through the Hill’s series of diversity indices. Sample count data were square-root transformed, and using the vegan package (Oksanen et al., 2016), we calculated the Hill’s diversity series: the raw number of OTUs (H0), the exponent of Shannon diversity (H1), and the reciprocal of the Simpson’s index (H2). To test for significant differences between sample diversity index values across geographic locations and between anatomical compartments, t-tests were performed in PAST (Hammer et al., 2001).
Before performing beta diversity analysis to compare bacterial community composition across samples, low abundance OTUs were filtered out (minimum proportional abundance: 0.5%) and zero counts were replaced with non-zero calculated values using the count zero multiplicative method from the R packages CoDaSeq and zCompositions (Martín-Fernández et al., 2015; Palarea-Albaladejo and Martín-Fernández, 2015). The zero-adjusted data were then centered log-ratio (clr) transformed using the CoDaSeq package (Gloor and Reid, 2016; Gloor et al., 2017). The clr is scale-invariant, meaning that the same ratio is expected from samples with different read counts (Gloor et al., 2017). A principal component analysis (PCA) was performed by plotting a singular value decomposition of clr transformed values to visualize beta diversity. We also performed hierarchical clustering using the “hclust” command, using a Euclidian distance matrix and agglomeration method “Ward. D2”. Bacterial community composition bar graphs representing phyla contribution (≥1%) and class contribution (≥3%) per sample were produced to visualize trends in the hierarchical cluster analysis dendrogram.
Quantitative analyses were conducted to examine the meta-data factors contributing toward the observed variance in bacterial community structure using the vegan R package “adonis” command for permutational multivariate analysis of variance (PERMANOVA) using a Euclidian distance matrix. A two-way design was used for the PERMANOVA, where location (canyons) and compartment (mucus and tissue) were both considered as fixed factors, and the interaction between the two factors was tested. Lastly, pairwise quantitative [ANOVA-like differential expression (ALDEx), Fernandes et al., 2013] analysis was performed to identify compartment-specific OTUs, or biomarkers. For this, clr-transformed count data were analyzed using differential relative abundance tests generated by 128 Monte Carlo samples sourced from a Dirichlet distribution. This ALDEx analysis was performed using the ALDEx2 package in R (Fernandes et al., 2013, 2014) to generate a list of OTUs (classified according to lowest available taxonomic rank) that possessed a significant association with either tissue or mucus. Here, we consider an OTU to be positively associated with tissue if the effect size is ≥1, and positively associated with mucus if the effect size is <−1.
Results
Alpha and Beta Diversity in Coral Bacterial Assemblages
Rarefaction curve analysis showed that all samples were sequenced sufficiently (i.e., to a read depth considered representative of each sample’s total microbial diversity), as indicated by the plateauing of the OTU count curves (aside from the Corsair Canyon seawater sample, which was omitted from further analysis; Figure 1). Sequence data were deposited in the NCBI short read archive linked to BioProject accession number PRJNA490387. The mucus swabs were amongst the lowest in sequencing depth, and reference seawater samples were among the highest (Table 1). Alpha and beta diversity measures were used to compare diversity trends and similarities between bacterial communities. The reference seawater samples were significantly higher than coral samples in all three Hill’s series diversity indices for bacterial communities [H0, H1, H2, two-sample t-test between coral samples (n = 12) and seawater (n = 2), p < 0.001 for each metric]. There were no significant differences between coral compartments or among geographic locations for any of the Hill’s series diversity indices [H0, H1, H2, two-sample t-test between coral tissue (n = 9) and mucus (n = 3), p > 0.05 for each metric].
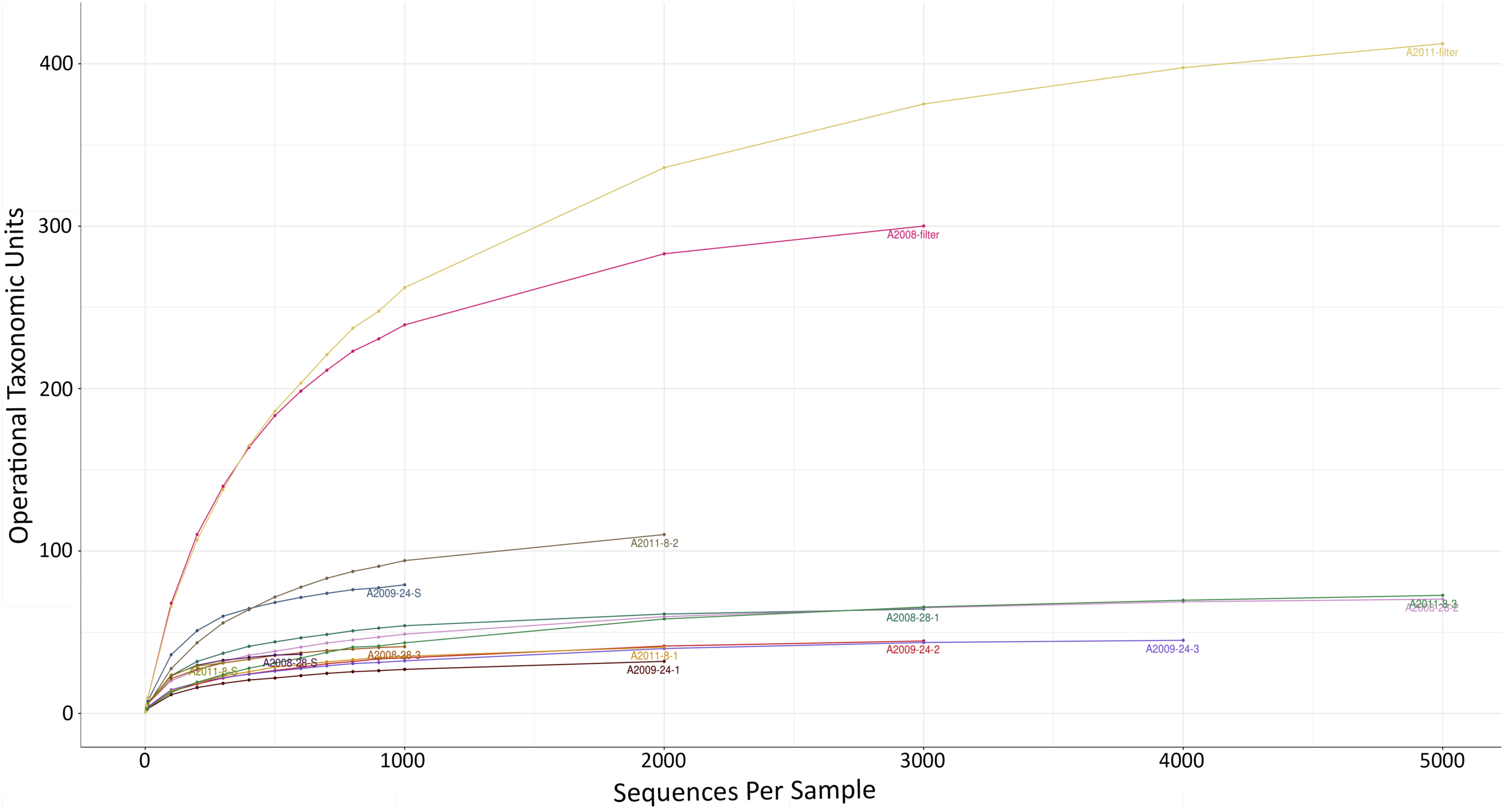
FIGURE 1. Rarefaction curve showing the number of observed bacterial OTUs as a function of the number of sequences. Paragorgia arborea tissue and mucus samples (n = 12) and seawater samples (n = 2). Sample names as in Table 1.
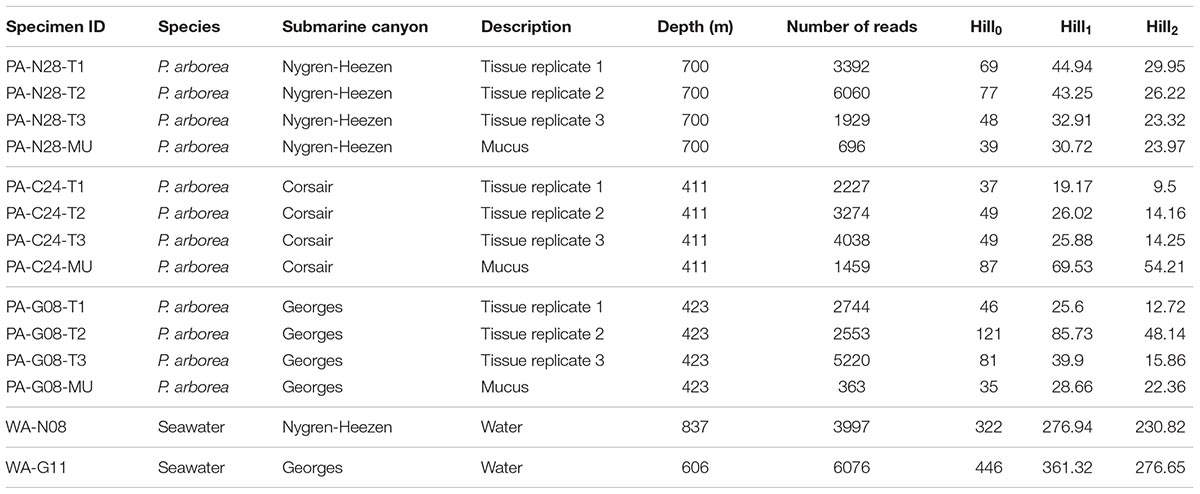
TABLE 1. Sample collection, description, number of reads from processed data, and Hill’s series diversity summary statistics of bacterial communities determined by analysis of 16S rRNA sequence libraries.
Due to the significant differences in bacterial alpha diversity between seawater and coral samples, we chose to produce a PCA ordination using Euclidian distance to further visualize similarities in bacterial composition across samples (Figure 2). The PCA representation of P. arborea tissue, mucus, and surrounding seawater bacterial communities showed some separation between coral samples and the reference seawater along the primary PCA axis (PC1 = 46.2% of the variation in the dataset) (Figure 2). The majority of coral samples clustered together, with some outliers. Differences between coral mucus samples were largely along the secondary PCA axis (PC2 = 16.2% of the variation) (Figure 2). PERMANOVA analysis uncovered a significant difference in bacterial composition between compartments (tissue and mucus) in P. arborea (p = 0.001). Furthermore, sample geographic location (canyon) was a significant explanatory factor for observed differences within P. arborea bacterial composition (p = 0.014). The PERMANOVA analysis showed no significant interaction between compartment and canyon. The PERMANOVA results were cross-checked using beta dispersion analysis, wherein significant results were validated by the PERMANOVA’s assumption of homogeneity of dispersion.
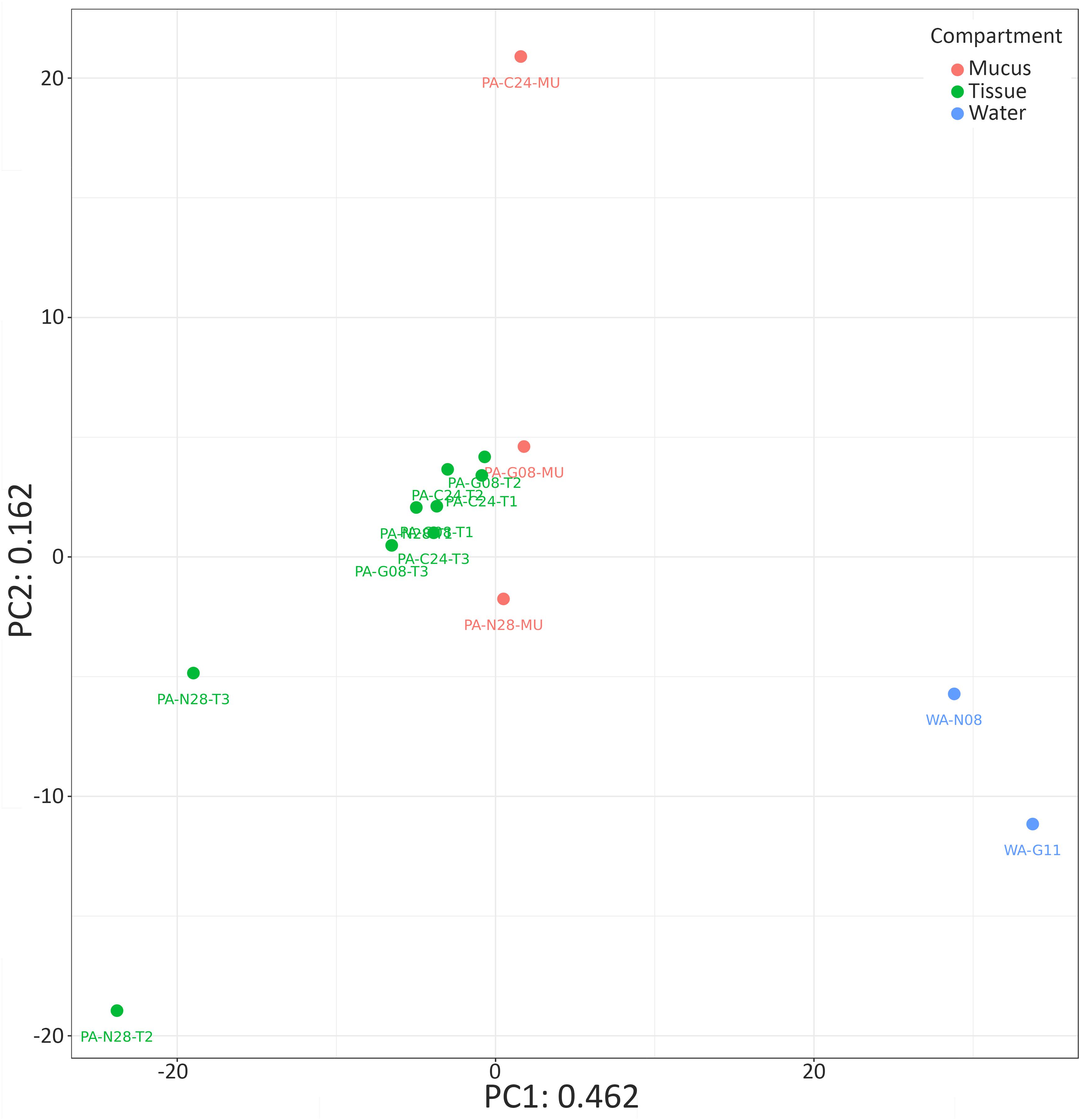
FIGURE 2. Bacterial community similarity for Paragorgia arborea and reference seawater samples. Principal component analysis (PCA) ordination of bacterial communities in P. arborea (tissue samples in green and mucus samples in orange) and seawater samples (blue), developed from a singular value decomposition of centered log-ratio (clr)-transformed compositional data. Sample names as in Table 1.
Bacterial Community Composition Within Coral Samples
The phyla shown in Figure 3A accounted for ∼98% of the bacteria found within the samples. Proteobacteria were present in all coral and seawater samples and were dominant in both seawater samples, in two tissue samples from Nygren-Heezen Intercanyon (PA-N28-T1/T3), and in two mucus samples (PA-G08-MU & PA-N08-MU) (Figure 3A). The third mucus sample (PA-C24-MU) had similar proportions of Verrucomicrobia (∼37%), Proteobacteria (∼27%), and Bacteroidetes (∼26%). Tenericutes dominated (∼68%) all coral tissue samples from Corsair Canyon and Georges Canyon, with Proteobacteria (∼15%) and Spirochaetes (∼7%) as notable contributors. Furthermore, Tenericutes were observed in every coral sample with the exception of one mucus sample (PA-C24-MU) and were absent in the seawater sample. The Nygren-Heezen Intercanyon coral tissue samples appeared more variable in composition and contrasted with samples from Corsair Canyon and Georges Canyon, which showed similar bacterial community composition across tissue samples. Cyanobacteria were present in coral samples, with the exception of two tissue samples (PA-C24-T2 and PA-N28-T2). Actinobacteria were observed in all samples recovered from Nygren-Heezen Intercanyon (including seawater) and were a dominant contributor in one tissue sample from Nygren-Heezen Intercanyon, in all mucus samples, and in the Georges Canyon seawater sample (WA-G11). Lastly, phylum and class Chlamydiae were only present in coral tissue samples from Georges Canyon (∼8%), while Firmicutes were only found in coral mucus from Georges Canyon (∼3%).
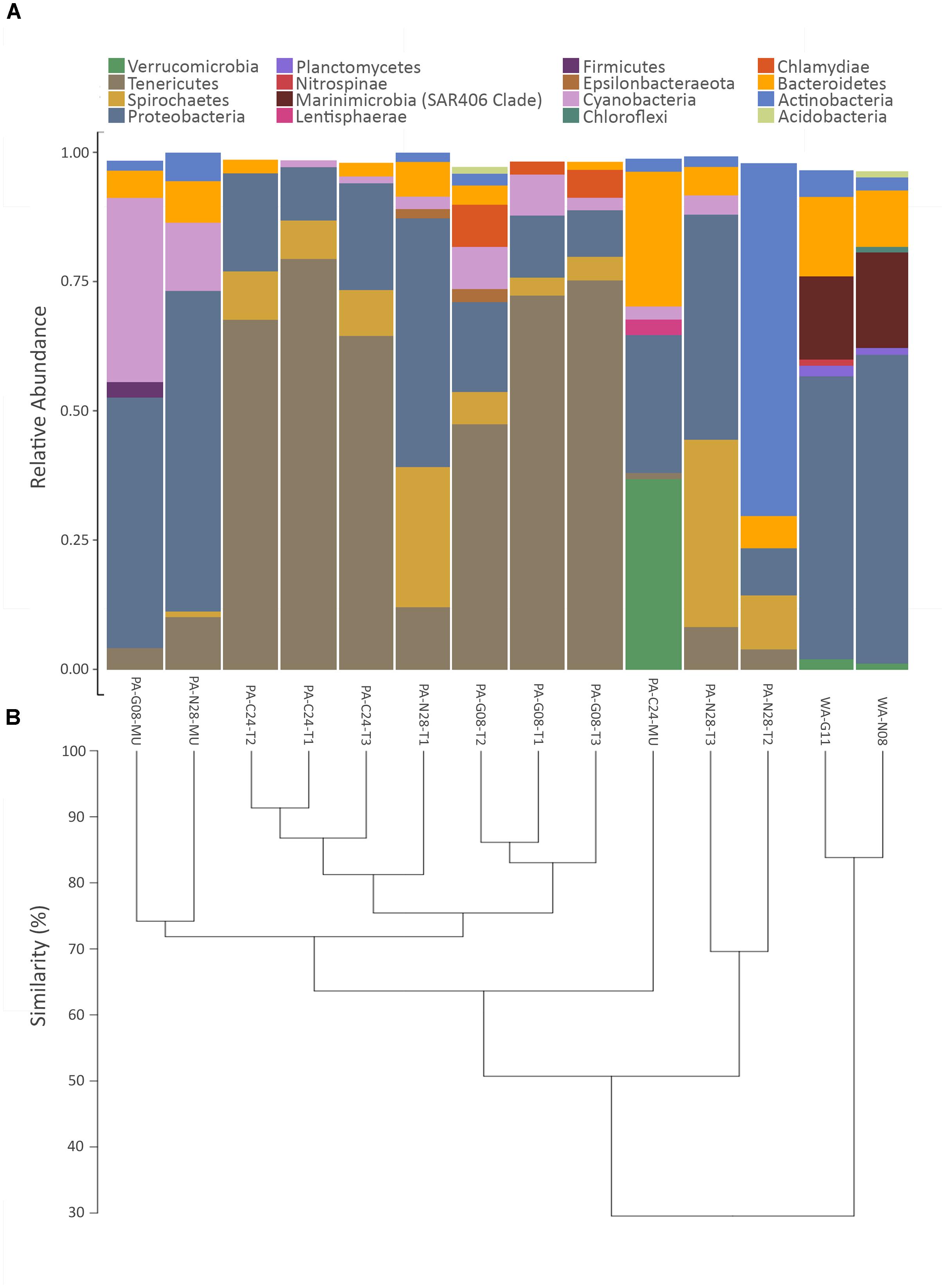
FIGURE 3. Relative abundance of bacterial phyla and hierarchical dendrogram. (A) X-axis represents samples of Paragorgia arborea tissue and mucus, and seawater. Y-axis represents the most dominant bacterial phyla found within samples, with taxa contributing <1% relative abundance excluded; and (B) hierarchical clustering dendrogram for P. arborea and reference seawater samples, developed from a Euclidian distance matrix produced from centered log-ratio (clr)-transformed compositional data. Sample names as in Table 1.
At the class level, bacterial compositions were nearly identical in coral tissue samples from Corsair Canyon, where samples were made up of Mollicutes (∼70%), Alphaproteobacteria (∼11%), Spirochaetes (∼9%), and Gammaproteobacteria (∼6%) (Figure 4). Coral tissue samples from Georges Canyon were also similar in composition, with dominant contributors being Mollicutes (∼65%), Gammaproteobacteria (∼9%), Oxyphotobacteria (∼6%), Chlamydiae (∼5%), and Spirochaetes (∼5%). There was less consistency among tissue samples from the Nygren-Heezen Intercanyon, mainly due to the variation in sample PA-N28-T2. Excluding sample PA-N28-T2, dominated by Actinobacteria (68%), the other samples were very similar in composition, made up of Spirochaetes (∼32%), Gammaproteobacteria (∼28%), Alphaproteobacteria (∼18%), Mollicutes (∼10%), and Bacteroidia (∼6%). Among mucus samples, the one from Corsair Canyon differed from those from Nygren-Heezen Intercanyon and Georges Canyon. Mucus recovered from Corsair Canyon showed a large contribution from Verrucomicrobia (∼37%), and further composed of Bacteroidia (∼26%), Gammaproteobacteria (∼23%), and Alphaproteobacteria (∼4%). The remaining mucus samples were relatively consistent in composition, containing mostly Gammaproteobacteria (∼45%), Oxyphotobacteria (∼24%), Alphaproteobacteria (∼10%), Mollicutes (∼7%), and Bacteroidia (∼7%).
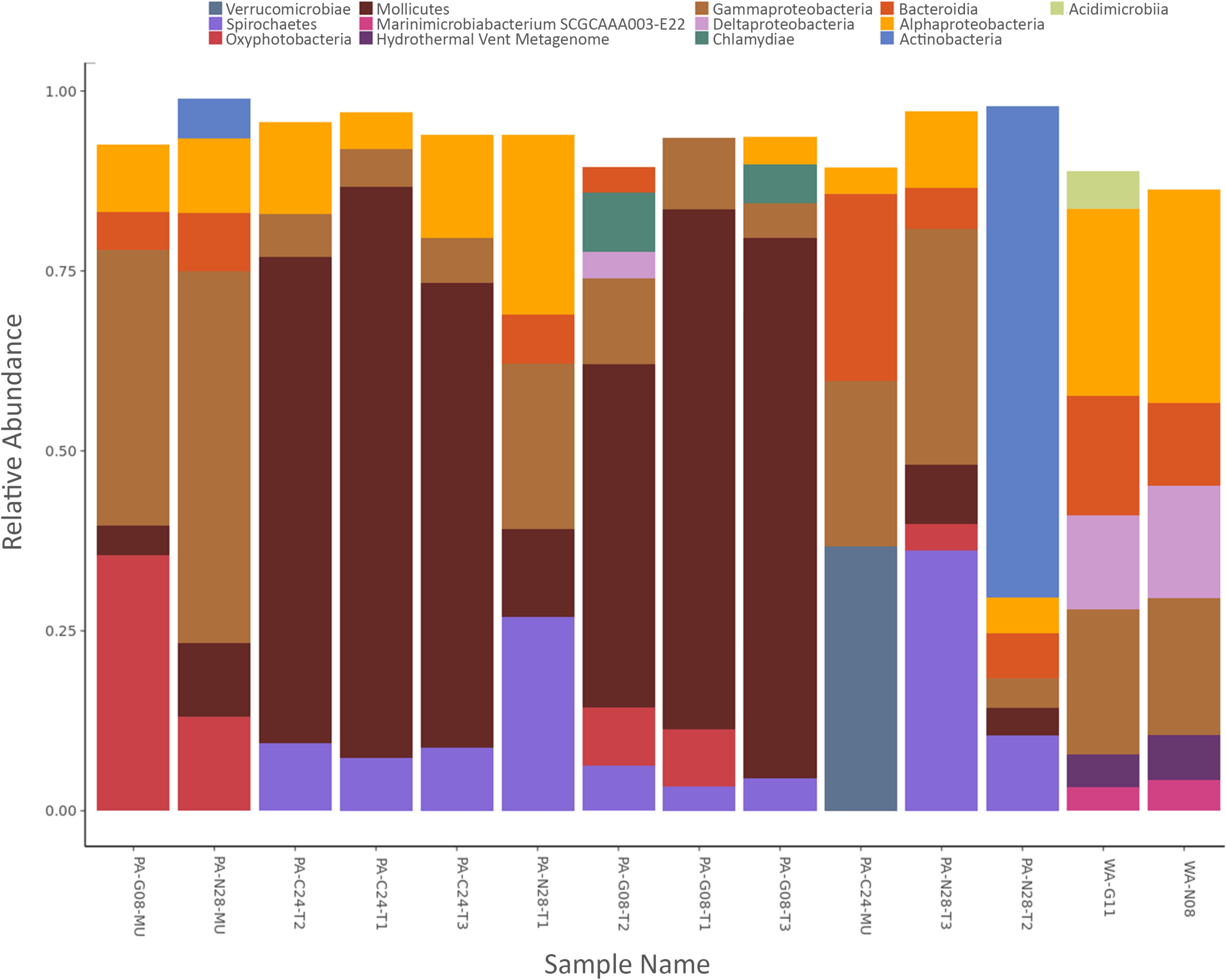
FIGURE 4. Relative abundance of bacterial classes. X-axis represents samples of Paragorgia arborea tissue and mucus, and seawater. Y-axis represents the most dominant bacterial classes found within samples, with taxa contributing <3% relative abundance excluded.
The trends in bacterial composition were observed across all samples in the produced hierarchical dendrogram (Figure 3B). Coral and seawater samples were observed to branch apart at 30% similarity, with the two seawater samples showing similar compositions (∼85% similarity). Coral tissue samples predominantly formed clusters, with groupings of Corsair Canyon samples (∼87% similar) and Georges Canyon samples (∼83% similar) most evident. Two mucus samples (PA-G08-MU and PA-N28-MU) contained relatively similar compositions (∼74%). Coral tissue samples from all three canyons converged at ∼51% similarity.
Bacterial Biomarkers Within Coral Anatomical Compartments
To observe whether any specific bacterial OTUs (biomarkers) were significantly more abundant in either compartment of P. arborea (tissue or mucus), samples were analyzed using an ALDEx analysis using the R package ALDEx2 (Fernandes et al., 2013, 2014). Three OTUS with genus-level assignment were recovered as biomarkers in P. arborea, with two recovered in the tissue and one in the surficial mucus (Figure 5). Two OTUs recovered as significant within the tissues belonged to the genus Spirochaeta of the phylum Spirochaetes and Mycoplasma of the phylum Tenericutes (effect sizes = 4.02 and 2.37, respectively) and were observed as relatively large contributors to the associated bacterial assemblage, particularly in samples from Nygren-Heezen Intercanyon (Supplementary Table S1). The mucus biomarker was identified as Paracoccus marcusii from the phylum Proteobacteria (effect size = −1.47). This biomarker was only recovered in two tissue samples, but was noted in the three mucus samples, comprising ∼1.2–2.4% of the bacterial assemblage (Supplementary Table S1). Four additional OTUs were recovered as significantly associated with tissues, but had no genus level assignment. Those four OTUs belonged to the families Flavobacteriaceae (phylum Bacteroidetes), and Terasakiellaceae (phylum Proteobacteria), and the orders Campylobacterales (phylum Epsilonbacteraeota) and Rickettsiales (phylum Proteobacteria) (effect sizes = 1.72, 1.41, 1.84, and 2.34, respectively). These four OTUs were rarely encountered in mucus and were most abundant in Nygren-Heezen Intercanyon tissue samples (Supplementary Table S1).
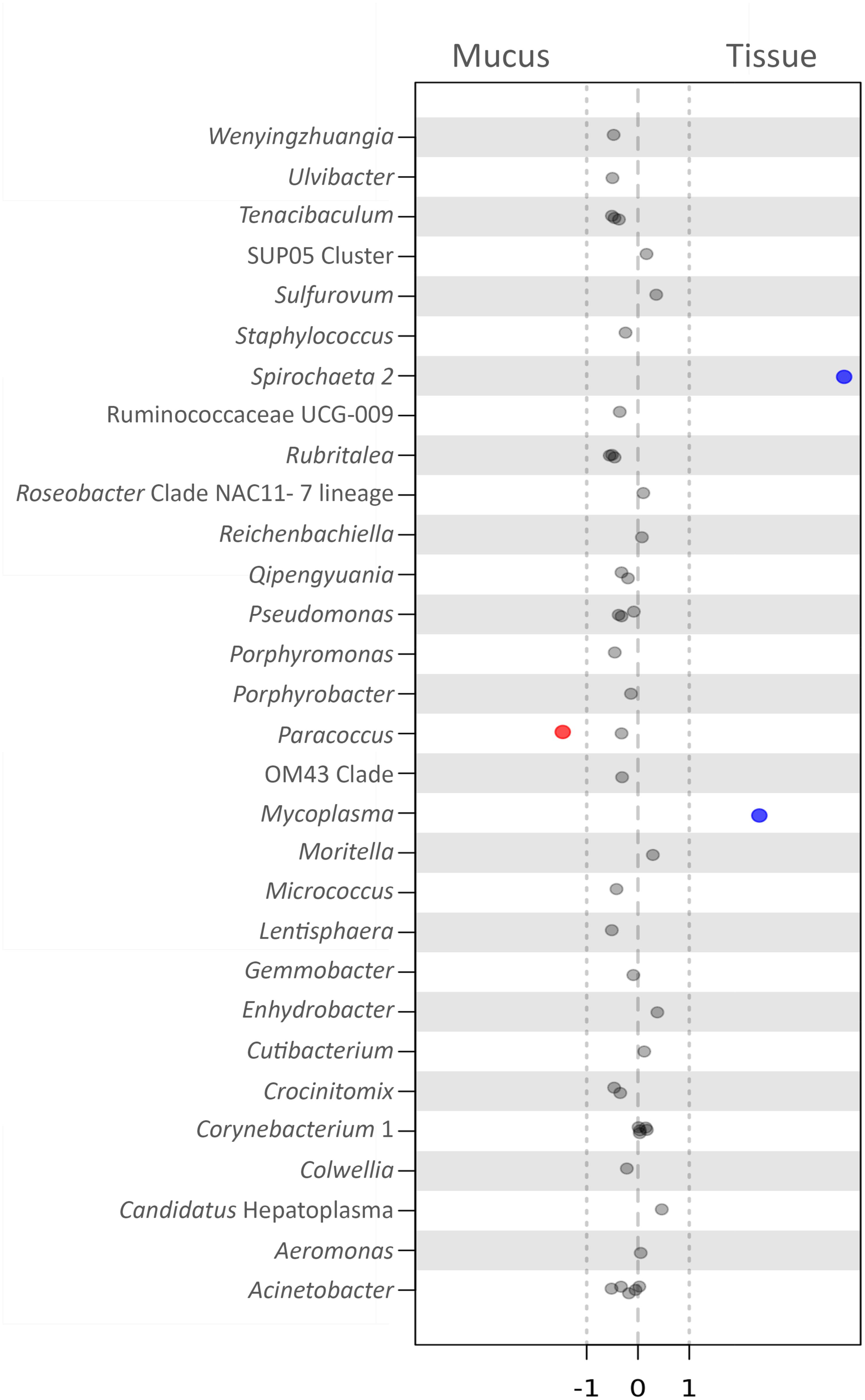
FIGURE 5. Biomarker analysis of bacteria in Paragorgia arborea compartments. Effect size scores for bacterial OTUs, classified at the genus level, defined as biomarkers through the use of ANOVA-Like Differential Expression (ALDEx) analysis (R package ALDEx2). Significance is determined by effect size, comparing tissue to mucus: values >1 represent significant, positive associations with tissue (blue), values <–1 show a negative association with tissue and a significant, positive association with mucus (red), and values between –1 and 1 are close to but not significant. Each dot represents an OTU (within the specified genus) tested as a biomarker. The 30 OTUs (having assigned genera) with effect sizes closest to significance are shown here.
Discussion
This is the first described characterization of the bacterial associates for Paragorgia arborea, a widely distributed cold-water alcyonacean coral that was collected from three submarine canyons off the Gulf of Maine (Nygren-Heezen Intercanyon, Georges Canyon, and Corsair Canyon). Sequencing of a fragment of the 16S rRNA gene allowed us to observe differences in bacterial composition between coral sample locations, and between compartments (tissue and mucus microhabitats). The numbers of reads obtained from extracts of P. arborea tissue samples were much higher than in mucus swabs, suggesting that our mucus sampling methodology could be improved. The low read depth of the water samples was likely due to the small aliquots (50 ml) of seawater at depth available to us on board the research cruise. Coral bacterial associates likely consist of a combination of commensals, long-term, stable partners selected by the host (including potentially intracellular symbionts), bacteria recently ingested and present in the gastro-vascular cavity, and, for mucus samples, either passively adhering bacteria or host-specific taxa (Ainsworth et al., 2015). While we are unable to confidently discriminate among those associate categories in this analysis, we draw upon our comparative analyses and on coral microbiomes described in the literature to suggest some bacterial taxa as potentially important members of the bacterial microbiome of those deep-sea, cold-water corals.
Alpha and Beta Measures of Bacterial Diversity
As observed in similar studies, the corals examined hosted bacterial communities with significantly lower bacterial biodiversity than those in surrounding seawater (Bayer et al., 2013; Holm and Heidelberg, 2016; van de Water et al., 2016). Within P. arborea, there was no significant difference in diversity measures between anatomical compartments (tissue and mucus) and among geographic locations (submarine canyons), suggesting spatial stability in the diversity (as characterized by Hill’s indices) of bacterial communities associated with P. arborea.
The bacterial communities of seawater were ∼70% different in composition (according to hierarchical clustering) from those associated with P. arborea; therefore, we consider that the coral microbiome was predominantly comprised of taxa that were uncommon in surrounding seawater and that may show some specificity for this particular host. Other studies on corals provide evidence for bacterial host-specificity: for example, distinct bacterial populations were observed in three octocorals from the coast of Florida, Leptogorgia minimata, Iciligorgia schrammi, and Swiftia exertia (Brück et al., 2007). Similarly, two octocorals from the Eastern Pacific of the genus Muricea had distinct microbial assemblages, despite co-occurring habitats and similar morphological structure (Holm and Heidelberg, 2016). Even the alcyonacean congeners P. placomus and Paramuricea clavata showed negligible bacterial community similarities (Kellogg et al., 2016). However, another study reported a lack of significant variation between microbiomes in two species of Anthothelidae (Lawler et al., 2016). Based on our study, the bacterial communities associated with P. arborea show some species specificity (given the ∼50% assemblage similarity among samples from different locations) and are dominated by many of the same taxa reported in other cold-water corals. We note the presence of phototrophic bacteria among the recovered taxa (phylum Cyanobacteria and class Oxyphotobacteria), particularly within mucus samples. These bacteria were likely among phytoplankton and other surficial detritus transported to the deep-sea along the canyons and captured by the corals.
The bacterial assemblages in P. arborea samples differed significantly in composition across geographic locations (submarine canyons); substantial variability in bacterial community composition across colonies of a particular coral species has been reported previously (see Hernandez-Agreda et al., 2017). Considering only tissue samples, those from Corsair Canyon (depth = 411 m) and Georges Canyon (depth = 423 m) were roughly 75% similar in bacterial community composition (Figure 3B); although those sites are within relative proximity to one another, they may differ in environmental and biotic factors, which may play a role in the diversity of available microbial associates and explain observed differences. The bacterial communities in coral tissue samples from the more distant and deeper (700 m) Nygren-Heezen Intercanyon showed greater compositional dissimilarity to the Corsair and Georges Canyon samples. Geographical differences in those bacterial assemblages may be driven mostly by commensals and/or more loosely associated or transient bacteria, but could also be influenced by the reproductive behavior of the host. P. arborea has been suggested to be a brooding coral, with fertilization occurring inside or at the surface of the female colony and the larval offspring settling nearby the parent colony (Lacharité and Metaxas, 2013). Some bacteria may be transmitted vertically in both broadcast and brooding corals (Apprill and Rappe, 2011; Ceh et al., 2013), and therefore the greater similarity in bacterial community composition between P. arborea colonies from nearby canyons could be partly linked to the relatively small larval dispersal distances in this coral species. Further research on vertical bacterial transmission in coral microbial communities is needed to shed light on this matter.
The surface mucus layer (SML) of a coral performs multiple functions and hosts a diverse assemblage of microbes (Bythell and Wild, 2011; Sweet et al., 2011). We observed a significant difference in bacterial composition between anatomical compartments of P. arborea and noted compositional differences between mucus samples within a host species. The mucus of shallow-water scleractinian corals is subject to diurnal or hourly replacement cycles, and therefore its bacterial diversity and richness could be constantly changing (Ainsworth et al., 2010; Sweet et al., 2011). In the deep-water scleractinian Lophelia pertusa, bacterial communities within the tissues were more stable than those in mucus, which is continuously being replenished following release in the water column (Wild et al., 2008; Weinbauer et al., 2012). It has been previously documented that P. arborea produces abundant mucus (Etnoyer et al., 2006). We also observed a large quantity of mucus following P. arborea sample collection, and its SML may constitute a perpetually changing microhabitat, as in Lophelia pertusa (Wild et al., 2008). Despite the variation in depth and associated physical parameters between shallow and deep-water habitats, we note that the mucus-derived bacterial composition of P. arborea shares the high diversity and variability of tropical corals.
Community Composition
Based on previous microbiome studies of alcyonaceans, certain bacterial taxa were expected to dominate the bacterial assemblage of P. arborea. In most alcyonaceans examined, Proteobacteria (especially Gammaproteobacteria and Alphaproteobacteria) dominate the bacterial assemblage (Penn et al., 2006; Gray et al., 2011; Bayer et al., 2013; Correa et al., 2013; La Rivière et al., 2013, 2015; Vezzulli et al., 2013; Ransome et al., 2014; Kellogg et al., 2016; Lawler et al., 2016; Robertson et al., 2016). Our results partly agree with these studies: Gammaproteobacteria and Alphaproteobacteria were present in all P. arborea samples, but were not the largest contributor in most samples. In contrast, Tenericutes, or more specifically, Mollicutes were the most dominant across P. arborea tissue samples from two canyons (Corsair and Georges Canyon) and were present in all P. arborea samples, with the exception of mucus sample PA-C24-MU. Tenericutes were reported to be dominant in a few coral species (Kellogg et al., 2009; Gray et al., 2011; Holm and Heidelberg, 2016). The Tenericutes OTUs observed in P. arborea are of the orders Entomoplasmatales and Mycoplasmatales, the latter having been observed in three species of the alcyonacean coral Muricea (Ranzer et al., 2007; Holm and Heidelberg, 2016), in two additional alcyonacean species, Plumarella superba and Cryogorgia koolsae (Gray et al., 2011) and in the scleractinian cold-water coral, Lophelia pertusa (Kellogg et al., 2009). Spirochaetes are dominant bacterial contributors in various alcyonaceans (Holm and Heidelberg, 2016; Lawler et al., 2016; van de Water et al., 2016). Spirochaetes were present in all P. arborea samples, in some cases as major contributors, and have been described as chemoheterotrophic bacteria that thrive in a wide variety of environments (Ludwig et al., 2010). Spirochaetes dominated some Anthothela samples, and were suggested as potential nitrogen fixers (Lawler et al., 2016).
Many of the taxa present at lower abundance may serve crucial roles in coral–bacterial interactions (Ainsworth et al., 2015). Several members of the phylum Firmicutes (orders Bacillales, and Lactobacillales) were observed in extremely low abundance in P. arborea. However, Firmicutes were reported in other alcyonacean microbiome studies (Penn et al., 2006; Brück et al., 2007; Kellogg et al., 2009, 2016; Correa et al., 2013; Lawler et al., 2016). Closek et al. (2014) observed a higher abundance of Firmicutes such as Clostridia in yellow band diseased samples of the scleractinian Orbicella faveolata; our samples showed no evidence of disease. Verrucomicrobia were observed in one P. arborea mucus sample (PA-C24-MU) and in the seawater samples, and were found in low numbers in other alcyonacean microbiome studies (Gray et al., 2011; Correa et al., 2013; Ransome et al., 2014; Kellogg et al., 2016; Lawler et al., 2016). Verrucomicrobiales from our samples are associated with the family Rubritaleaceae. Bacteria from the Rubritaleaceae are carotenoid pigment producers (Rosenberg, 2014), that give a red coloration to colonies; it is uncertain why these bacteria were highly abundant in PA-C24-MU but absent from all other coral samples. Chlamydiae (taxonomic order Chlamydiales) were only observed in tissue samples from Georges Canyon; the role of Chlamydiales in invertebrates is not known. Campylobacterales were found within all P. arborea tissue samples in low abundance, absent in all mucus samples and were part of the Anthothelidae core microbiome (Lawler et al., 2016). Some members of the order Campylobacterales were suggested to contribute to the nitrogen metabolism cycle for nitrate/nitrite ammonification and denitrification (Tiedje, 1988; Hoffmann et al., 1998; Verbaendert et al., 2011); the Campylobacterales observed here belong to the families Sulfurovaceae, and Thiovulaceae.
Comparing rare taxa found in our study to previously described etiological agents in diseased corals and gorgonians, we found Vibrio spp. in one sample of P. arborea (PA-G08-T2) in very low relative abundance (data not shown). Various Vibrio strains may be etiological agents in diseased P. clavata colonies (Bally and Garrabou, 2007; Vezzulli et al., 2013). Additionally, Daniels et al. (2015) found high abundances of mRNA sequences from Vibrionaceae, Campylobacteraceae, Rhodobacteraceae, Flavobacteriaceae, and Burkholderiaceae in diseased O. faveolata coral samples. Representatives from the same bacterial families were found in low abundances in our visually healthy coral samples (data not shown). Further research regarding these bacterial groups is required to understand their functional roles in coral microbiomes, as rare bacterial taxa may be as important as dominant taxa and are typically overlooked (Ainsworth et al., 2017).
Bacterial Biomarkers Within Paragorgia arborea Anatomical Compartments
As previous studies have shown that bacteria found within specific microhabitats may serve particular roles for the host (Bythell and Wild, 2011; Sweet et al., 2011; Glasl et al., 2016), we explored whether any bacterial OTUs were considered statistically significant biomarkers within P. arborea tissues or mucus. Within P. arborea, OTUs from the genera Spirochaeta and Mycoplasma, as well as from the orders Campylobacterales and Rickettsiales and the families Flavobacteriaceae and Terasakiellaceae were biomarkers of tissue, while the genus Paracoccus was a biomarker of mucus. While these OTUs have been identified significantly as biomarkers, their abundance can vary markedly across samples and geographic locations and interpretations of key functional roles for these bacteria remain highly speculative.
The genus Spirochaeta has been observed to dominate Anthothela coral samples and was suggested to be a nitrogen fixer (Lawler et al., 2016). Interestingly, the Spirochaeta 2 OTU identified here as a tissue biomarker comprised up to 36% of the identified bacterial assemblage in a tissue sample from Nygren-Heezen Intercanyon. Mycoplasma strains have been described as pathogens and/or parasites (Rottem, 2003) and were observed in Lophelia pertusa (Kellogg et al., 2009). Mycoplasma were abundant in Muricea coral samples, with specific strains associated with bleached and unbleached coral samples (Ranzer et al., 2007; Holm and Heidelberg, 2016); although P. arborea is azooxanthellate (Roberts et al., 2006), Mycoplasma may nonetheless play some role in P. arborea health. In previous studies, Campylobacterales were suggested to play a role in denitrification (Verbaendert et al., 2011) and nitrate/nitrite ammonification (Hoffmann et al., 1998). Members of the family Terasakiellaceae and certain Paracoccus species were documented to be nitrogen fixers and denitrifiers, respectively, and may be important for alcyonacean nitrogen metabolism (Tiedje, 1988). A microbially mediated nitrogen cycle has been uncovered in the cold-water coral Lophelia pertusa (Middelburg et al., 2015), and may also be present in alcyonacean corals, such as P. arborea, which occupy similar, nutrient-limited environments (Buhl-Mortensen et al., 2015). Notably, we observed the highest proportions of Spirochaeta, Campylobacterales, Terasakiellaceae and Paracoccus, the biomarkers that might be involved in nitrogen cycling, in Nygren-Heezen Intercanyon, the deepest of our three sampling locations. This could signal a greater importance for bacterial associates involved in nitrogen cycling where food limitation could be more pronounced. Further investigations would be required to characterize any bacterial functions and their impact on the coral host.
Conclusion
This study provides the first characterization of bacterial associates for Paragorgia arborea, detailing the microbiome of this deep-sea cold-water coral during a visibly healthy state. While the bacterial communities of this species did not differ significantly in terms of diversity indices between compartments (mucus and tissue) or sampling location (canyon), there were significant compositional differences found among the bacterial assemblages from different compartments and sampling locations. Bacterial communities appeared more stable across colonies in P. arborea tissues than in mucus, and the relative abundance of the more common taxonomic groups tended to fluctuate across samples. In general, the bacterial microbiome of P. arborea was dominated by Tenericutes (orders Entomoplasmatales and Mycoplasmatales), with Spirochaetes, Gammaproteobacteria, and Alphaproteobacteria also making notable contributions to the bacterial assemblages. Bacteria from taxa known to contribute to nitrogen recycling and metabolism were identified as tissue and mucus biomarkers in P. arborea. Representatives of bacterial families previously found in higher abundance in diseased scleractinians (Vibrionaceae, Campylobacteraceae, Rhodobacteraceae, Flavobacteraceae, and Burkholderiaceae; Daniels et al., 2015) were present (but rare) in our coral samples. The work presented here provides baseline microbiome data for P. arborea, a common habitat-forming cold-water coral taxon. Additional research on deep-sea and cold-water coral health and susceptibility to stress is urgently needed for more informed conservation and marine policy planning.
Author Contributions
BW, JV, and SD designed the experiments. JV collected the samples during the research cruise. BW and JV did the extractions and analyzed the data and prepared the figures. BW wrote the paper. JV and SD edited and reviewed drafts of the paper.
Funding
This project was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Ship Time Grant 501171-2017 (applicant: A. Metaxas; co-applicants P. Snelgrove, SD, and M. Kienast) and NSERC Discovery Grant RGPIN-2015-06548 to SD. The research cruise was part of a Northeast Regional Deep-Sea Coral Initiative, funded in part by NOAA National Marine Fisheries Service, NOAA Office of Marine and Aviation Operations, and NOAA’s Deep-Sea Coral Research and Technology Program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank chief scientists A. Metaxas (Dalhousie University) and M. Nizinski (National Systematics Laboratory, National Museum of Natural History, Smithsonian Institution), the captain and crew of the NOAA Ship Henry B. Bigelow, and the CSSF-ROPOS team.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00378/full#supplementary-material
References
Ainsworth, T. D., Fordyce, A. J., and Camp, E. F. (2017). The other microeukaryotes of the coral reef microbiome. Trends Microbiol. 25, 980–991. doi: 10.1016/j.tim.2017.06.007
Ainsworth, T. D., Krause, L., Bridge, T., Torda, G., Raina, J. B., Zakrzewski, M., et al. (2015). The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 9, 2261–2274. doi: 10.1038/ismej.2015.39
Ainsworth, T. D., Thurber, R. V., and Gates, R. D. (2010). The future of coral reefs: a microbial perspective. Trends Ecol. Evol. 25, 233–240. doi: 10.1016/j.tree.2009.11.001
Apprill, A., and Rappe, M. S. (2011). Response of the microbial community to coral spawning in lagoon and reef flat environments of Hawaii, USA. Aquat. Microb. Ecol. 62, 251–266. doi: 10.3354/ame01471
Bally, M., and Garrabou, J. (2007). Thermodependent bacterial pathogens and mass mortalities in temperate benthic communities: a new case of emerging disease linked to climate change. Glob. Chang. Biol. 13, 2078–2088. doi: 10.1111/j.1365-2486.2007.01423.x
Bavestrello, G., Cerrano, C., Zanzi, D., and Cattaneo-Vietti, R. (1997). Damage by fishing activities to the gorgonian coral Paramuricea clavata in the Ligurian Sea. Aquat. Conserv. 7, 253–262. doi: 10.1002/(SICI)1099-0755(199709)7:3<253::AID-AQC243>3.0.CO;2-1
Bayer, T., Arif, C., Ferrier-Pagès, C., Zoccola, D., Aranda, M., and Voolstra, C. R. (2013). Bacteria of the genus Endozoicomonas dominate the microbiome of the Mediterranean gorgonian coral Eunicella cavolini. Mar. Ecol. Prog. Ser. 479, 75–84. doi: 10.3354/meps10197
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bourne, D. G., and Munn, C. B. (2005). Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ. Microbiol. 7, 1162–1174. doi: 10.1111/j.1462-2920.2005.00793.x
Brooke, S. D., Watts, M. W., Heil, A. D., Rhode, M., Mienis, F., Duineveld, G. C. A., et al. (2017). Distributions and habitat associations of deep-water corals in Norfolk and Baltimore Canyons, Mid-Atlantic Bight, USA. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 137, 131–147. doi: 10.1016/j.dsr2.2016.05.008
Brown, B. E., and Bythell, J. C. (2005). Perspectives on mucus secretion in reef corals. Mar. Ecol. Prog. Ser. 296, 291–309. doi: 10.3354/meps296291
Brück, T. B., Brück, W. M., Santiago-Vázquez, L. Z., McCarthy, P. J., and Kerr, R. G. (2007). Diversity of the bacterial communities associated with the azooxanthellate deep water octocorals Leptogorgia minimata, Iciligorgia schrammi, and Swiftia exertia. Mar. Biotechnol. 9, 561–576. doi: 10.1007/s10126-007-9009-1
Buhl-Mortensen, L., and Mortensen, P. B. (2004). Symbiosis in deep-water corals. Symbiosis 37, 33–61. doi: 10.1016/j.seares.2013.10.001
Buhl-Mortensen, L., Olafsdottir, S. H., Buhl-Mortensen, P., Burgos, J. M., and Ragnarsson, S. A. (2015). Distribution of nine cold-water coral species (Scleractinia and Gorgonacea) in the cold temperate North Atlantic: effects of bathymetry and hydrography. Hydrobiologia 759, 39–61. doi: 10.1007/s10750-014-2116-x
Buhl-Mortensen, P., and Buhl-Mortensen, L. (2014). Diverse and vulnerable deep-water biotopes in the Hardangerfjord. Mar. Biol. Res. 10, 253–267. doi: 10.1080/17451000.2013.810759
Bythell, J. C., and Wild, C. (2011). Biology and ecology of coral mucus release. J. Exp. Mar. Biol. Ecol. 408, 88–93. doi: 10.1016/j.jembe.2011.07.028
Cairns, S. D., and Bayer, F. M. (2005). New taxa paper a review of the genus Primnoa (Octocorallia: Gorgonacea: Primnoidae), with the description of two new species. Bull. Mar. Sci. 77, 225–256.
Ceh, J., van Keulen, M., and Bourne, D. G. (2013). Intergenerational transfer of specific bacteria in corals and possible implications for offspring fitness. Microb. Ecol. 65, 227–231. doi: 10.1007/s00248-012-0105-z
Closek, C. J., Sunagawa, S., DeSalvo, M. K., Piceno, Y. M., DeSantis, T. Z., Brodie, E. L., et al. (2014). Coral transcriptome and bacterial community profiles reveal distinct Yellow Band Disease states in Orbicella faveolata. ISME J. 8, 1–12. doi: 10.1038/ismej.2014.85
Coffroth, M. A. (1991). Cyclical mucous sheet formation on poritid corals in the San- Blas-Islands, Panama. Mar. Biol. 109, 35–40. doi: 10.1007/BF01320229
Comeau, A. M., Li, W. K., Tremblay, J. É, Carmack, E. C., and Lovejoy, C. (2011). Arctic ocean microbial community structure before and after the 2007 record sea ice minimum. PLoS One 6:27492. doi: 10.1371/journal.pone.0027492
Cordes, E. E., Jones, D. O. B., Schlacher, T. A., Amon, D. J., Bernardino, A. F., Brooke, S., et al. (2016). Environmental impacts of the deep-water oil and gas industry: a review to guide management strategies. Front. Environ. Sci. 4:58. doi: 10.3389/fenvs.2016.00058
Correa, H., Haltli, B., Duque, C., and Kerr, R. (2013). Bacterial communities of the gorgonian octocoral Pseudopterogorgia elisabethae. Microb. Ecol. 66, 972–985. doi: 10.1007/s00248-013-0267-3
Daniels, C. A., Baumgarten, S., Yum, L. K., Mihell, C. T., Bayer, T., Arif, C., et al. (2015). Metatranscriptome analysis of the reef-building coral Orbicella faveolata indicates holobiont response to coral disease. Front. Mar. Sci. 2:62. doi: 10.3389/fmars.2015.00062
Edwards, U., Rogall, T., Blocker, H., Emde, M., and Bottger, E. C. (1989). Isolation and direct complete nucleotide determination of entire genes: characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17, 7843–7853. doi: 10.1093/nar/17.19.7843
Egan, S., and Gardiner, M. (2016). Microbial dysbiosis: rethinking disease in marine ecosystems. Front. Microbiol. 7:991. doi: 10.3389/fmicb.2016.00991
Etnoyer, Peter, J., Cairns, Stephen, D., Sanchez, Juan Armando, et al. (2006). Deep-Sea Coral Collection Protocols. Silver Spring, MD: NOAA, 53.
Fernandes, A. D., Macklaim, J. M., Linn, T., Reid, G., and Gloor, G. B. (2013). ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-seq. PLoS One 8:e67019. doi: 10.1371/journal.pone.0067019
Fernandes, A. D., Reid, J. N. S., Macklaim, J. M., Thomas, A. M., Edgell, D. R., and Gloor, G. B. (2014). Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2, 1–13. doi: 10.1186/2049-2618-2-15
Fosså, J. H., Mortensen, P. B., and Furevik, D. M. (2002). The deepwater coral Lophelia pertusa in Norwegian waters: distribution and fisheries impacts. Hydrobiologia 471, 1–12. doi: 10.1023/A:1016504430684
Glasl, B., Herndl, G. J., and Frade, P. R. (2016). The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 10, 2280–2292. doi: 10.1038/ismej.2016.9
Gloor, G. B., Macklaim, J. M., Pawlowsky-Glahn, V., and Egozcue, J. J. (2017). Microbiome datasets are compositional: and this is not optional. Front. Microbiol. 8:2224. doi: 10.3389/fmicb.2017.02224
Gloor, G. B., and Reid, G. (2016). Compositional analysis: a valid approach to analyze microbiome high-throughput sequencing data. Can. J. Microbiol. 62, 692–703. doi: 10.1139/cjm-2015-0821
Gray, M. A., Stone, R. P., Mclaughlin, M. R., and Kellogg, C. A. (2011). Microbial consortia of gorgonian corals from the Aleutian islands. FEMS Microbiol. Ecol. 76, 109–120. doi: 10.1111/j.1574-6941.2010.01033.x
Grover, R., Ferrier-Pages, C., Maguer, J.-F., Ezzat, L., and Fine, M. (2014). Nitrogen fixation in the mucus of Red Sea corals. J. Exp. Biol. 217, 3962–3963. doi: 10.1242/jeb.111591
Hammer,Ø, Harper, D. A. T., and Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 1–9.
Hernandez-Agreda, A., Gates, R. D., and Ainsworth, T. D. (2017). Defining the core microbiome in coral’s microbial soup. Trends Microbiol. 25, 125–140. doi: 10.1016/j.tim.2016.11.003
Hixon, M. A., and Beets, J. P. (1993). Predation, prey refuges, and the structure of coral-reef fish assemblages. Ecol. Monogr. 63, 77–101. doi: 10.2307/2937124
Hoffmann, T., Frankenberg, N., Marino, M., and Jahn, D. (1998). Ammonification in Bacillus subtilis utilizing dissimilatory nitrite reductase is dependent on resDE. J. Bacteriol. 180, 186–199.
Holm, J. B., and Heidelberg, K. B. (2016). Microbiomes of Muricea californica and M. fruticosa: comparative analyses of two co-occurring Eastern Pacific Octocorals. Front. Microbiol. 7:917. doi: 10.3389/fmicb.2016.00917
Husebø,Å, Nøttestad, L., Fosså, J. H., Furevik, D. M., and Jørgensen, S. B. (2002). Distribution and abundance of fish in deep-sea coral habitats. Hydrobiologia 471, 91–99. doi: 10.1023/A:1016549203368
Kellogg, C. A., Lisle, J. T., and Galkiewicz, J. P. (2009). Culture-independent characterization of bacterial communities associated with the cold-water coral Lophelia pertusa in the northeastern Gulf of Mexico. Appl. Environ. Microbiol. 75, 2294–2303. doi: 10.1128/AEM.02357-08
Kellogg, C. A., Ross, S. W., and Brooke, S. D. (2016). Bacterial community diversity of the deep-sea octocoral Paramuricea placomus. PeerJ 4:e2529. doi: 10.7717/peerj.2529
Kelman, D., Kushmaro, A., Loya, Y., Kashman, Y., and Benayahu, Y. (1998). Antimicrobial activity of the Red Sea soft coral Parerythropodium fulvum: reproductive and developmental considerations. Mar. Ecol. Prog. Ser. 169, 87–95. doi: 10.3354/meps169087
Krediet, C. J., Ritchie, K. B., Paul, V. J., and Teplitski, M. (2013). Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc. R. Soc. Lond. B Biol. Sci. 280, 2012–2328. doi: 10.1098/rspb.2012.2328
La Rivière, M., Garrabou, J., and Bally, M. (2015). Evidence for host specificity among dominant bacterial symbionts in temperate gorgonian corals. Coral Reefs 34, 1087–1098. doi: 10.1007/s00338-015-1334-7
La Rivière, M., Roumagnac, M., Garrabou, J., and Bally, M. (2013). Transient shifts in bacterial communities associated with the temperate gorgonian Paramuricea clavata in the Northwestern Mediterranean Sea. PLoS One 8:e57385. doi: 10.1371/journal.pone.0057385
Lacharité, M., and Metaxas, A. (2013). Early life history of deep-water gorgonian corals may limit their abundance. PLoS One 8:65394. doi: 10.1371/journal.pone.0065394
Lawler, S. N., Kellogg, C. A., France, S. C., Clostio, R. W., Brooke, S. D., and Ross, S. W. (2016). Coral-associated bacterial diversity is conserved across two deep-sea Anthothela species. Front. Microbiol. 7:458. doi: 10.3389/fmicb.2016.00458
Ludwig, W., Euzéby, J., and Whitman, W. B. (2010). “Volume 4: the Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes,” in Bergey’s Manual of Systematic Bacteriology, Vol. 949, eds N. R. Krieg, W. Ludwig, W. B. Whitman, B. P. Hedlund, B. J. Paster, J. T. Staley, et al. (New York, NY: Springer), 1–19. doi: 10.1007/978-0-387-68572-4
Mahé, F., Torbjørn, R., Quince, C., de Vargas, C., and Dunthorn, M. (2015). Swarm v2: highly-scalable and high-resolution amplicon clustering. PeerJ 3:e1420. doi: 10.7717/peerj.1420
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10. doi: 10.14806/ej.17.1.200
Martín-Fernández, J. A., Hron, K., Templ, M., Filzmoser, P., and Palarea-Albaladejo, J. (2015). Bayesian-multiplicative treatment of count zeros in compositional data sets. Stat. Model. 15, 134–158. doi: 10.1177/1471082X14535524
McMurdie, P. J., and Holmes, S. (2014). Phyloseq: an r package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Middelburg, J. J., Mueller, C. E., Veuger, B., Larsson, A. I., Form, A., and van Oevelen, D. (2015). Discovery of symbiotic nitrogen fixation and chemoautotrophy in cold-water corals. Sci. Rep. 5:17962. doi: 10.1038/srep17962
Mortensen, P. B., and Buhl-Mortensen, L. (2005). Morphology and growth of the deep-water gorgonians Primnoa resedaeformis and Paragorgia arborea. Mar. Biol. 147, 775–788. doi: 10.1007/s00227-005-1604-y
Mouchka, M. E., Hewson, I., and Harvell, C. D. (2010). Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 50, 662–674. doi: 10.1093/icb/icq061
Mueller, C. E., Larsson, A. I., Veuger, B., Middelburg, J. J., and van Oevelen, D. (2014). Opportunistic feeding on various organic food sources by the cold-water coral Lophelia pertusa. Biogeosciences 11, 123–133. doi: 10.5194/bg-11-123-2014
Naumann, M. S., Richter, C., El-Zibdah, M., and Wild, C. (2009). Coral mucus as an efficient trap for picoplanktonic cyanobacteria: implications for pelagic-benthic coupling in the reef ecosystem. Mar. Ecol. Prog. Ser. 385, 65–76. doi: 10.3354/meps08073
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., et al. (2016). vegan: Community Ecology Package. R Package Version 2.2-0. Available at: http://CRAN.Rproject.org/package=vegan
Palarea-Albaladejo, J., and Martín-Fernández, J. A. (2015). zCompositions — R package for multivariate imputation of left-censored data under a compositional approach. Chemometr. Intel. Lab. Syst. 143, 85–96. doi: 10.1016/j.chemolab.2015.02.019
Penn, K., Wu, D., Eisen, J. A., and Ward, N. (2006). Characterization of bacterial communities associated with deep-sea corals on Gulf of Alaska seamounts. Appl. Environ. Microbiol. 72, 1680–1683. doi: 10.1128/AEM.72.2.1680
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
R Development Core Team (2008). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rädecker, N., Pogoreutz, C., Voolstra, C. R., Wiedenmann, J., and Wild, C. (2015). Nitrogen cycling in corals: the key to understanding holobiont functioning? Trends Microbiol. 23, 490–497. doi: 10.1016/j.tim.2015.03.008
Ransome, E., Rowley, S. J., Thomas, S., Tait, K., and Munn, C. B. (2014). Disturbance to conserved bacterial communities in the cold-water gorgonian coral Eunicella verrucosa. FEMS Microbiol. Ecol. 90, 404–416. doi: 10.1111/1574-6941.12398
Ranzer, L. K., Restrepo, P. F., and Kerr, R. G. (2007). Data from: Microbial Community Profiles of Bleached and Wild-Type Muricea elongata. NCBI PopSet 134140623. Available at: http://www.ncbi.nlm.nih.gov/popset/134140623?report=genbank
Ritchie, K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Roberts, J. M., Wheeler, A. J., and Friewald, A. (2006). Reefs of the deep: the biology and geology of cold-water coral ecosystems. Science 312, 543–547. doi: 10.1126/science.1119861
Robertson, V., Haltli, B., McCauley, E., Overy, D., and Kerr, R. (2016). Highly variable bacterial communities associated with the octocoral Antillogorgia elisabethae. Microorganisms 4, 1–23. doi: 10.3390/microorganisms4030023
Rosenberg, E. (2014). “The family Rubritaleaceae,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), 861–862. doi: 10.1007/978-3-642-38954-2
Rottem, S. (2003). Interaction of mycoplasmas with host cells. Physiol. Rev. 83, 417–432. doi: 10.1152/physrev.00030.2002
Stackebrandt, E., and Liesack, W. (1993). “Nucleic acids and classification,” in Handbook of New Bacterial Systematics, eds M. Goodfellow and A. G. O’Donnell (London: Academic Press), 152–189.
Stone, R. P. (2006). Coral habitat in the Aleutian Islands of Alaska: depth distribution, fine-scale species associations, and fisheries interactions. Coral Reefs 25, 229–238. doi: 10.1007/s00338-006-0091-z
Sunagawa, S., Woodley, C. M., and Medina, M. (2010). Threatened corals provide underexplored microbial habitats. PLoS One 5:9554. doi: 10.1371/journal.pone.0009554
Sutherland, K. P., Porter, J. W., and Torres, C. (2004). Disease and immunity in Caribbean and Indo-Pacific zooxanthellate corals. Mar. Ecol. Prog. Ser. 266, 273–302. doi: 10.3354/meps266273
Sweet, M. J., Croquer, A., and Bythell, J. C. (2011). Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30, 39–52. doi: 10.1007/s00338-010-0695-1
Syms, C., and Jones, G. P. (2000). Disturbance, habitat structure, and the dynamics of a coral-reef fish community. Ecology 81, 2714–2729. doi: 10.1890/0012-9658 (2000)081[2714:DHSATD]2.0.CO;2
Tiedje, J. (1988). “Ecology of denitrification and dissimilatory nitrate reduction to ammonium,” in Biology of Anaerobic Microorganisms, ed. A. Zehnder (New York, NY: John Wiley and Sons),179–244.
van de Water, J. A., Melkonian, R., Junca, H., Voolstra, C. R., Reynaud, S., Allemand, D., et al. (2016). Spirochaetes dominate the microbial community associated with the red coral Corallium rubrum on a broad geographic scale. Sci. Rep. 6:27277. doi: 10.1038/srep27277
Verbaendert, I., Boon, N., De Vos, P., and Heylen, K. (2011). Denitrification is a common feature among members of the genus Bacillus. Syst. Appl. Microbiol. 34, 385–391. doi: 10.1016/j.syapm.2011.02.003
Verhoeven, J. T. P., and Dufour, S. C. (2017). Microbiomes of the Arctic carnivorous sponges Chondrocladia grandis and Cladorhiza oxeata suggest a specific, but differential involvement of bacterial associates. Arct. Sci. 4, 186–204. doi: 10.1139/AS-2017-0015
Vezzulli, L., Pezzati, E., Huete-Stauffer, C., Pruzzo, C., and Cerrano, C. (2013). 16SrDNA pyrosequencing of the Mediterranean gorgonian Paramuricea clavata reveals a link among alterations in bacterial holobiont members, anthropogenic influence and disease outbreaks. PLoS One 8:67745. doi: 10.1371/journal.pone.0067745
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wegley, L., Edwards, R., Rodriguez-Brito, B., Liu, H., and Rower, F. (2007). Metagenomics analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9, 2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x
Weinbauer, M. G., Ogier, J., and Maier, C. (2012). Microbial abundance in the coelenteron and mucus of the cold-water coral Lophelia pertusa and in bottom water of the reef environment. Aquat. Biol. 16, 209–216. doi: 10.3354/ab00443
Wild, C., Huettel, M., Klueter, A., and Kremb, S. G. (2004). Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature 428, 66–70. doi: 10.1038/nature02333.1
Wild, C., Mayr, C., Wehrman, L., Schöttner, S., Naumann, M., Hoffman, F., et al. (2008). Organic matter release by cold water corals and its implication for fauna-microbe interaction. Mar. Ecol. Prog. Ser. 372, 67–75. doi: 10.3354/meps07724
Keywords: cold-water corals, Alcyonacea, bacterial diversity, microbiome, mucus, high-throughput sequencing, 16S rRNA
Citation: Weiler BA, Verhoeven JTP and Dufour SC (2018) Bacterial Communities in Tissues and Surficial Mucus of the Cold-Water Coral Paragorgia arborea. Front. Mar. Sci. 5:378. doi: 10.3389/fmars.2018.00378
Received: 30 April 2018; Accepted: 27 September 2018;
Published: 18 October 2018.
Edited by:
Michael Sweet, University of Derby, United KingdomReviewed by:
Aldo Cróquer, Simón Bolívar University, VenezuelaBryan Wilson, University of Bergen, Norway
Copyright © 2018 Weiler, Verhoeven and Dufour. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bradley A. Weiler, YmF3ZWlsZXJAbXVuLmNh; YnJhZHdlaWxlci5vbkBnbWFpbC5jb20=
†Lead author
 Bradley A. Weiler
Bradley A. Weiler Joost T. P. Verhoeven
Joost T. P. Verhoeven Suzanne C. Dufour
Suzanne C. Dufour