- 1Department of Biology, Brooklyn College, City University of New York, Brooklyn, NY, United States
- 2Department of Biology, Temple University, Philadelphia, PA, United States
- 3Woods Hole Oceanographic Institution, Woods Hole, MA, United States
Mixotrophic flagellates can comprise significant proportions of plankton biomass in marine ecosystems. Despite the growing recognition of the importance of this ecological strategy, and the identification of major environmental factors controlling phagotrophic behavior (light and nutrients), the physiological and molecular mechanisms underlying mixotrophic behavior are still unclear. In this study, we performed RNA-Seq transcriptomic analysis for two mixotrophic prasinophytes, Micromonas polaris and Pyramimonas tychotreta, under dissolved nutrient regimes that altered their ingestion of bacteria prey. Though the strains examined were polar isolates, both belong to genera with widespread distribution. Our aim was to characterize the transcriptomes of these two non-model phytoflagellates, identify transcripts consistent with phagotrophic activity and assess their differential expression in response to nutrient stress. De novo assembly of the transcriptomes yielded large numbers of novel coding transcripts with no known match within public databases. A summary of the transcripts by Gene Ontology terms showed many expected expression patterns, including genes involved in photosynthetic pathways and enzymes implicated in nutrient uptake pathways. Searches of KEGG databases identified several genes associated with intra-cellular digestive pathways actively transcribed in both prasinophytes. Differential expression analysis showed a larger response in P. tychotreta, where 23,373 genes were up-regulated and 1,752 were down-regulated in the low nutrient treatment when phagotrophy was enhanced. In contrast, in M. polaris, low nutrient treatments resulted in up-regulation of 314 transcripts while down-regulating 371. With respect to phagotrophic-related expression, 37 genes were co-expressed in both P. tychotreta and M. polaris, and although the response was less pronounced in M. polaris, it is consistent with differences in observed ingestion behavior. This study presents the first genomic data for Pyramimonas tychotreta, and also contributes to the limited available data for Micromonas polaris. Furthermore, it provides insight into the presence of genes associated with phagocytosis within the Prasinophyceae and contributes to the understanding of potential target genes required for the construction of a complete model of gene regulation of phagocytic behavior in algae.
Introduction
The traditional view of protistan species is that they can be defined in terms of a single trophic mode, either photo- or hetero-trophic (Azam et al., 1983). However, more recently, a greater recognition of the impact of photosynthetic protists with the ability to ingest prey has been growing (Pomeroy, 1974; Flynn et al., 2012). Ingestion of prey by phytoplankton is found to occur in both freshwater and marine environments, ranging from tropical to polar ecosystems (Boraas et al., 1988; Sanders, 1991; Pålsson and Granéli, 2004; Moorthi et al., 2009; Hartmann et al., 2013). This ecological strategy can account for significant impact with regard to the movement of nutrients and carbon, as well as through substantial reintroduction of immobilized carbon back into the food web (Boraas et al., 1988; Sanders, 2011; Unrein et al., 2014). Mixotrophic behavior has also been demonstrated to satisfy organismal requirements for macro- and micro-nutrient deficiencies, and ingestion behavior is often a response to these limiting conditions (Nygaard and Tobiesen, 1993; Maranger et al., 1998). Mixotrophic phytoflagellates can comprise a considerable proportion of planktonic communities and potentially graze large proportions of bacterial standing stock daily in both freshwater and marine planktonic food webs (Bird and Kalff, 1986; Sanders et al., 1989; Sanders, 2011), accounting for a majority of the bacterial mortality (Hitchman and Jones, 2000; Zubkov and Tarran, 2008; Sanders and Gast, 2012).
Despite the growing recognition of predatory behavior in photosynthetic protists, and the substantial collection of physiological measurements of mixotrophic behavior (e.g., predation rates and conversion efficiencies), the regulatory controls at the genetic level are still poorly understood. This is, in part, a result of the broad spectrum of mixotrophic organisms representing diverse evolutionary lineages, many of which lack established and well-curated model organisms for use in genetic and molecular studies (Sherr and Sherr, 2002). This area is of importance because phagotrophic behavior provides evidence of the mechanisms underlying the endosymbiotic theory, which unites all eukaryotic lineages, and all plastid containing organisms in particular (Burns et al., 2015). Some molecular targets involved in the process of cellular phagotrophy have been previously identified, including receptors associated with phagocytosis, actin filament remodeling and regulatory gene families (Yutin et al., 2009). Liu et al. (2016) also identified genes potentially associated with mixotrophy, however, the details in terms of regulation and biochemical controls need further study.
Within the Archeaplastida, also referred to as the green algae, the Prasinophyta represent an ancient group of plastid containing eukaryotes that are widely distributed in both freshwater and marine environments, and have been found to make up significant portions of the standing biomass in polar microbial communities (Moro et al., 2002; Lovejoy et al., 2007; Li et al., 2009; Gast et al., 2014). Green algae were long thought to be a wholly autotrophic lineage of organisms, but recent observations have shown ingestion behavior in several green algal species, including several Prasinophytes (Maruyama and Kim, 2013; Raven, 2013; McKie-Krisberg and Sanders, 2014; McKie-Krisberg et al., 2015). Within the Prasinophyceae, genomic data is available for a limited number of species, including Ostreococcus tauri, an isolate of Micromonas pusilla and Cymbomonas tetramitiformis (Derelle et al., 2006; Worden et al., 2009; Tirichine and Bowler, 2011; Burns et al., 2015). Of these prasinophytes, aside from C. tetramitiformis, none have been positively identified as mixotrophs. The recent advancements in genomic technologies, particularly RNA-Seq and related computational analysis, provide efficient, cost effective methods to gain a deeper understanding of the changes at the molecular level, particularly in non-model organisms.
In this study, we examine the transcriptional profiles of two polar mixotrophic prasinophytes representing different phytoplankton size fractions (pico- vs. nano-flagellates). We investigate the response at the molecular level, in terms of gene expression, to changes in nutrient availability previously shown to illicit changes in prey ingestion behavior in these two organisms. Our aims were to characterize the transcriptomes of these two non-model phytoflagellates, identify transcripts consistent with phagotrophic activity and assess their differential expression in response to stimulation of mixotrophy due to nutrient limitation. We hypothesize that nutrient availability is a major factor in driving ingestion behavior in polar mixotrophic algae, and that by using a comparative “-omics” approach, we can identify patterns of gene expression common among mixotrophic behavior in microbial eukaryotes. We obtained feeding activity and transcriptomic data simultaneously from an Arctic isolate of Micromonas polaris, and the Antarctic Pyramimonas tychotreta, and identified changes in transcript abundance in response to a reduction in nutrients. By comparing the transcriptomes of the two prasinophytes, we also attempt to identify genes linked to phagotrophic behavior shared by both organisms, which could be potential targets for the evaluation of mixotrophic activity in environmental samples.
Materials and Methods
Culture Origin and Maintenance
Pyramimonas tychotreta (I-9 Pyram) was isolated from the Ross Sea, Antarctica in 1999 and maintained at Woods Hole Oceanographic Institution and at Temple University (http://www.whoi.edu/science/B/protists/). Micromonas isolate (CCMP 2099, Provasoli-Guillard National Center for Marine Algae and Microbiota), recently identified as M. polaris (Simon et al., 2017), was originally isolated from an Arctic polyna. The prasinophytes in the present study differ in terms of cell size (1.2 μm M. polaris vs. 5–7 μm average diameter for P. tychotreta). Experimental cultures of these algae were maintained at 4°C in 32 PSU f/2 + Si (Guillard, 1975; Caron, 1993). The cultures were non-axenic and grown under 14 h light/10 h dark irradiance from 50 W cool-white fluorescent bulbs at ~50 μmol m−2 s−1.
Experimental Setup
Previous studies have shown both P. tychotreta and M. polaris to ingest particles, and that ingestion rates are influenced by nutrient availability in the culture media (McKie-Krisberg and Sanders, 2014; McKie-Krisberg et al., 2015). Replicate flasks of P. tychotreta (2.5 × 103 cells ml−1) and M. polaris (1.44 × 105 cells ml−1) were grown in either high- or reduced-nutrient conditions for 1 week for transcriptional analysis (n = 4 per treatment/species/time point; total 16 per species) and ingestion experiments (n = 3 per treatment/species/time point; total 12 per species) conducted at the time of whole cell collection for sequencing to confirm differences in feeding. The differences in cell abundances are reflective of the shorter doubling time and smaller size of M. polaris as compared to P. tychotreta. High-nutrient treatments were full strength f/2+Si (i.e., the maintenance media, 882 μM N, 36.2 μM ), while low-nutrient treatments were a 10-fold dilution of f/2 + Si culture media (88.2 μM N, 3.62 μM ) prepared with filter-sterilized seawater. The low-nutrient conditions were previously found to elicit increases in ingestion by these two species (McKie-Krisberg and Sanders, 2014; McKie-Krisberg et al., 2015) and the concentrations of nitrogen and phosphorous fall within the range observed in both the Artic and Antarctic environments (Wheeler et al., 1997; Smith et al., 2003). Illumination and temperature conditions were the same as used for culture maintenance (14 h light/10 h dark irradiance at ~50 μmol m−2 s−1).
RNA Extraction, Transcriptome Assembly and Differential Expression Analysis
Replicate cultures of P. tychotreta (n = 4) and M. polaris (n = 4) that had been incubated under high- and reduced-nutrient conditions known to affect ingestion were collected on 25 mm, 0.2 μm polycarbonate filters, frozen at −80°C, then extracted using the Qiagen Mini RNA kit, with confirmation of nucleic acid abundance and quality assessed by Bioanalyzer (Agilent Technologies, Santa Clara, CA). Replicate samples were sent to the Sulzberger Genome Center at Columbia University for poly-A selection, library construction and sequencing by Illumina Mi-Seq (150 bp paired-end reads, 30 million reads per sample). Raw reads have been deposited in the NCBI Sequence Read Archive under accession number SRP090401.
Overall quality of Illumina raw reads data was assessed using FastQC (Andrews, 2010). Poor quality reads and adapters were removed using Trimmomatic 0.32, with settings for removing seed mismatches = 2, palindrome clip threshold = 10, and a simple clip threshold = 30. Additional settings included removing leading and trailing bases below quality scores of 3 and excluding reads less than 36 bp (Bolger et al., 2014). Prior to assembly, FLASh (version 1.2.11) was used to find overlaps and merge read pairs to increase the quality of the transcript assembly (Magoc and Salzberg, 2011). Following these preparatory steps, all paired-end reads for each species were combined and assembled using Trinity (version 2.3.2), using default assembly settings (Grabherr et al., 2011) (Table 1). Following assembly, bacterial decontamination was performed by Deconseq (version 0.4.3) using the bacterial data base generated by the script designers (Schmieder and Edwards, 2011). Transcriptomes of each species were filtered through CD-hit to identify unique transcripts (Li and Godzik, 2006). This step clustered transcripts that were ≥90% similar. In the absence of sequenced genomes and proper functional annotation of either of these prasinophytes, this was a conservative approach that allowed for the occurrence of differential splicing of transcriptional products, but constrained computational artifacts from the de novo assembly that could not be validated. Transcript abundance from each replicate was determined by sequence-read alignment to transcriptomes with Bowtie2 (Langmead and Salzberg, 2012), followed by count estimation with eXpress (Roberts et al., 2011). Differential expression of M. polaris and P. tychotreta was determined using edgeR according to the experimental design, using the exact test based on quantile-adjusted conditional maximum likelihood (qCML) method (Robinson et al., 2010). Our threshold for differential regulation of Trinity “genes” was defined by transcript differences of at least 2 Log2-fold (4-fold) increase/decrease in expression. Transcript assembly, differential expression analysis and gene annotation were all performed using computational resources available via the Owlsnest Super-Computing Cluster at Temple University, and the CUNY High Performance Computing Center at the CUNY College of Staten Island.
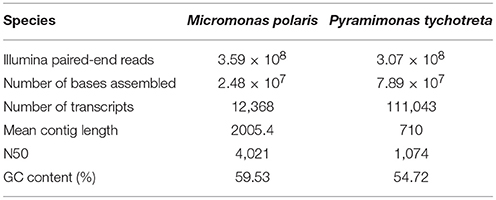
Table 1. Summary statistics from Trinity de novo transcriptome assembly for two polar mixotrophic algae Micromonas polaris and Pyramimonas tychotreta.
Functional Annotation
Functional annotation of assembled transcripts was performed with Trinotate 2.0 (Haas et al., 2013). Searches of several genomic databases, including Uniprot databases (http://www.uniprot.org/) and nr and nt (http://www.ncbi.nlm.nih.gov/) were performed using BLAST utility and employing Transdecoder to identify predicted Open Reading Frames (ORFs). ORFs were also queried using hmmscan, and rnammer for predictions of conserved domains with homology to protein families and domains found in the Pfam database (Lagesen et al., 2007; Eddy, 2011; Finn et al., 2015). Gene Ontology (GO) associated with identified genes obtained from Interproscan searches were grouped according to “Biological Process” (BP), “Cellular Component” (CC) or “Molecular Function” (MF) (Jones et al., 2014). Whole transcriptome Gene Ontologies were summarized with the goSlim function of the GSEABase package, within the R statistical environment using the plant ontology (http://www.plantontology.org) (Gentleman et al., 2004; Morgan et al., 2008; R Core Team, 2013). GO term information from transcript data was integrated into Gene Enrichment analysis. We obtained the ontology pathways that were enriched by the treatment conditions using the runTest function in the topGO package, performing a Fisher Exact test inputting normalized gene expression measurements along with differential expression analysis results (Alexa and Rahnenfuhrer, 2017). Assembled transcripts of both M. polaris and P. tychotreta were submitted to the KAAS-KEGG Automatic Annotation Server provided by the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/tools/kaas/) for the identification of additional enzymes involved in primary nutrient metabolism (Carbon, Nitrogen), as well as phagotrophic related pathways, such as autophagy (Moriya et al., 2007). Additionally, genes found to be involved in phosophorus (P) limitation response in plants (Rouached et al., 2010) were identified by blast searches of M. polaris and P. tychotreta transcriptomes (Supplementary Data Sheet 1).
Ingestion Experiments
Both microspheres and fluorescently-labeled bacteria have been used to demonstrate ingestion behavior (Nygaard and Tobiesen, 1993), but the fluorescent particles employed in the present work were also used in previous studies of phagotrophy of both P. tychotreta and M. polaris, and therefore provide the most suitable comparison to the previous experimental work (McKie-Krisberg and Sanders, 2014; McKie-Krisberg et al., 2015). A total of 1 × 106 microspheres were added to each flask and the T0 replicates (controls) were immediately fixed using the Lugol's/formaldehyde/Na2S2O3 method that prevents egestion (Sherr et al., 1993). The remaining replicates were incubated under the same light and temperature used for culture maintenance for 20 min (Tf), after which they were fixed as for the T0 samples. Previous studies indicate that 20 min is within the period of time when there is a linear uptake of microspheres for both P. tychotreta, M. polaris (McKie-Krisberg et al., McKie-Krisberg and Sanders). Ten milliliter of fixed sample were filtered onto 25 mm Poretics polycarbonate membranes (0.8 μm pore size for M. polaris and 3 μm pore size for P. tychotreta, GE Osmonics, Minnetonka MN, USA) and stained/mounted using VectaShield® with DAPI (Vector Laboratories Inc., Burlingame, CA). Epifluorescence microscopy was used to examine at least 500 cells per filter for P. tychotreta, while 10 fields per filter were counted for M. polaris. Mixotrophic behavior was identified as the ingestion of beads after background correction with the T0 samples.
Results
Under high nutrient treatment conditions, a low level of particle ingestion was observed for M. polaris, and undetectable for P. tychotreta. However, in the low nutrient treatments, the percent of cells that ingested microspheres increased by 69% for M. polaris and 31% for P. tychotreta. These observations are consistent with previous studies where grazing in both species was affected by concentrations of nutrients in the culture media, with P. tychotreta showing a particularly distinct response (McKie-Krisberg and Sanders, 2014; McKie-Krisberg et al., 2015).
Transcriptome Assembly and Functional Annotation
The de novo transcriptome assembly of RNA-Seq data identified ~12,000 and ~110,000 unique transcripts for M. polaris and P. tychotreta, respectively (Table 1). Current available genomic data for two isolates of Micromonas (CCMP1545 v3.0, and RCC299 v.3, Phytozome) include annotation of 10,660 and 10,103 genes, respectively (Goodstein et al., 2012). Therefore, even with our conservative approach, the transcriptome for M. polaris likely includes differential splicing products. The large transcriptome size of P. tychotreta was associated with a smaller N50 value, which may be due to a large portion of short length contigs (unique, consensus regions of RNA produced as a result of the de novo assembly) (Table 1). No genomic data is currently available for P. tychotreta, but annotation of the related prasinophyte, Cymbomonas tetramitiformis (Order Pyramimondales) was found to have almost 4 times the number of genes (~37,000 genes) compared to the published Micromonas pusilla genomes (Burns et al., 2015).
The functional annotation of the transcriptomes for each species revealed a large number of novel transcripts that had no matches to previously identified genomic information in available databases. Functional annotation identified 64% of transcripts in M. polaris, while in P. tychotreta, only ~24% of transcripts matched previously identified proteins in the SwissProt database, indicating a large proportion of the transcripts represented potential novel sequences (Figure 1). Regardless of the relatively small percentage of identified transcripts, 2.7 × 103 transcripts were matched to existing accessions and functional annotation via the KEGG database. Looking in detail at differential expression results for both M. polaris and P. tychotreta, we observed changes in mechanisms associated with acquisition for both light and nutrient uptake pathways (Supplementary Table 1). Not surprisingly, proteins involved in photosynthesis represented a large number of the GO terms associated with Biological Processes in both M. polaris and P. tychotreta (Figure 2, Supplementary Data Sheet 2). These include Light Harvesting Complex proteins (LHC) and Ribuose-1,5-Bisphosphate Carboxylase/Oxygenase (RUBISCO) (Supplementary Table 1).
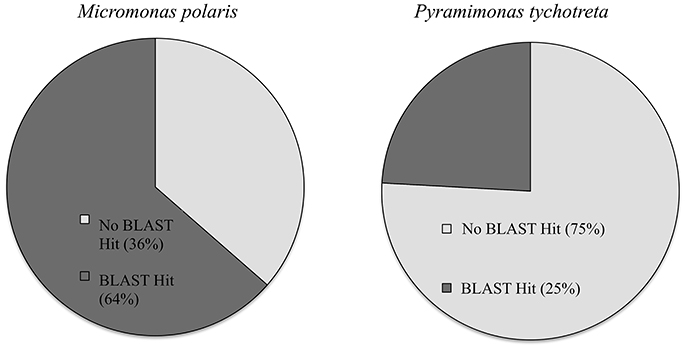
Figure 1. Percentage of transcripts with BLAST matches in Uniprot database in Micromonas polaris and Pyramimonas tychotreta.
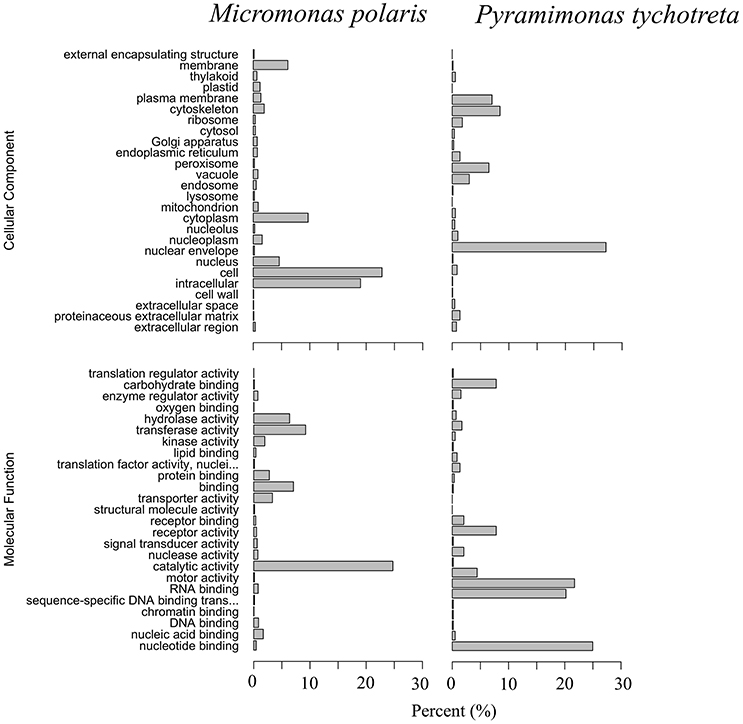
Figure 2. Gene Ontology (GO) summaries of M. polaris and P. tychotreta transcriptomes as a percentage of total identified transcripts. GO terms were grouped by Molecular Function and Cellular Component.
Overall, the effect of lower concentrations of dissolved nutrients was less pronounced in M. polaris, resulting in fewer changes in gene expression as compared to P. tychotreta. In cultures of M. polaris, lower nutrient concentrations resulted in 314 up-regulated and 371 down-regulated, at least a Log2-fold change of 2 (Figure 2). This response is less, but still comparable to, earlier studies of a tropical isolate of Micromonas (CCMP2709), which found up-regulation of 738 out of 960 differentially-expressed transcripts under of the effect of phosphorus limitation (Whitney and Lomas, 2016). In low-nutrient treatments of P. tychotreta, we found 23,373 up-regulated and 1,752 down-regulated at a 2 Log2-fold change) (Figure 3). Reduction in nutrient availability resulted in up-regulation of genes related to carbon metabolism, and to some extent nitrogen metabolism, in both prasinophytes, although the response was more pronounced in cultures of P. tychotreta (Figures 4,5). Differential expression of genes associated with phosphorus limitation was greater in cultures of P. tychotreta, but M. polaris appears to constitutively express these genes, as they did not respond to the experimental conditions (Figures 3–5).
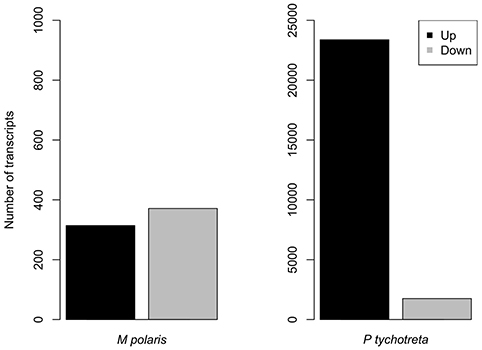
Figure 3. Number or genes differentially expressed in M. polaris and P. tychotreta. The threshold for significant differential expression is defined as a 2 log2 fold-change in expression in response to changes in nutrient condition Note difference in scales.
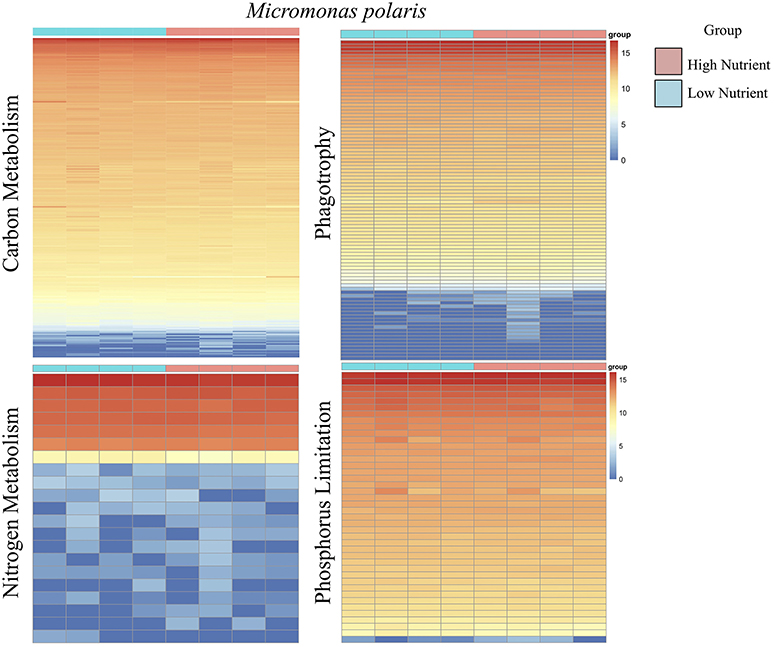
Figure 4. A series of heatmaps for Micromonas polaris comparing gene expression associated with carbon (C) metabolism, nitrogen (N) metabolism, phagotrophy, and phosphorus (P) limitation, in cultures grown under high nutrient vs. low nutrient conditions. Tiles represent log-normalized count estimations of transcripts. Genes for C metabolism, N metabolism, and phagotrophy were identified by KEGG and categories were selected based on BRITE hierarchies, except P limitation genes, where genes were selected based on genes identified by Rouached et al. (2010).
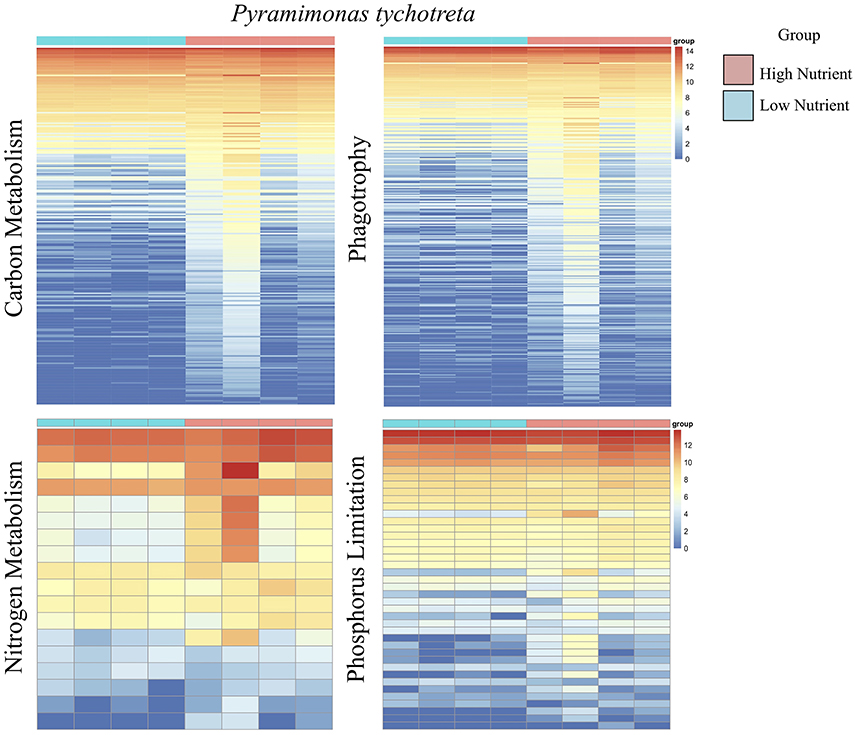
Figure 5. A series of heatmaps for Pyramimonas tychotreta comparing gene expression associated with carbon (C) metabolism, nitrogen (N) metabolism, phagotrophy and phosphorus (P) limitation, in cultures grown under high nutrient vs. low nutrient conditions. Tiles represent log-normalized count estimations of transcripts. Genes for C metabolism, N metabolism, and phagotrophy were identified by KEGG and categories were selected based on BRITE hierarchies, except P limitation genes, where genes were selected based on genes identified by Rouached et al. (2010).
Specific to phagotrophy, low nutrient conditions resulted in up-regulation of 153 genes related to phagotrophic ingestion, as defined by KEGG (Table 2). We also found 37 genes (in some cases, with multiple transcript matches) to be present in both M. polaris and P. tychotreta. These include genes that function in endocytosis (20), formation of phagosome (8), lysosome production (11), and regulation of autophagy (7) (Table 2, Supplementary Table 1). Several transcripts in the transcriptome of both M. polaris and P. tychotreta contained predicted Ankyrin domains (identified by KEGG), which have been suggested to play a role in phagocytosis (Moffat et al., 1996; Grolleau et al., 2003; Nguyen et al., 2014). Many of these genes appear to be found across eukaryotic taxa and not only among prasinophytes (Yutin et al., 2009).
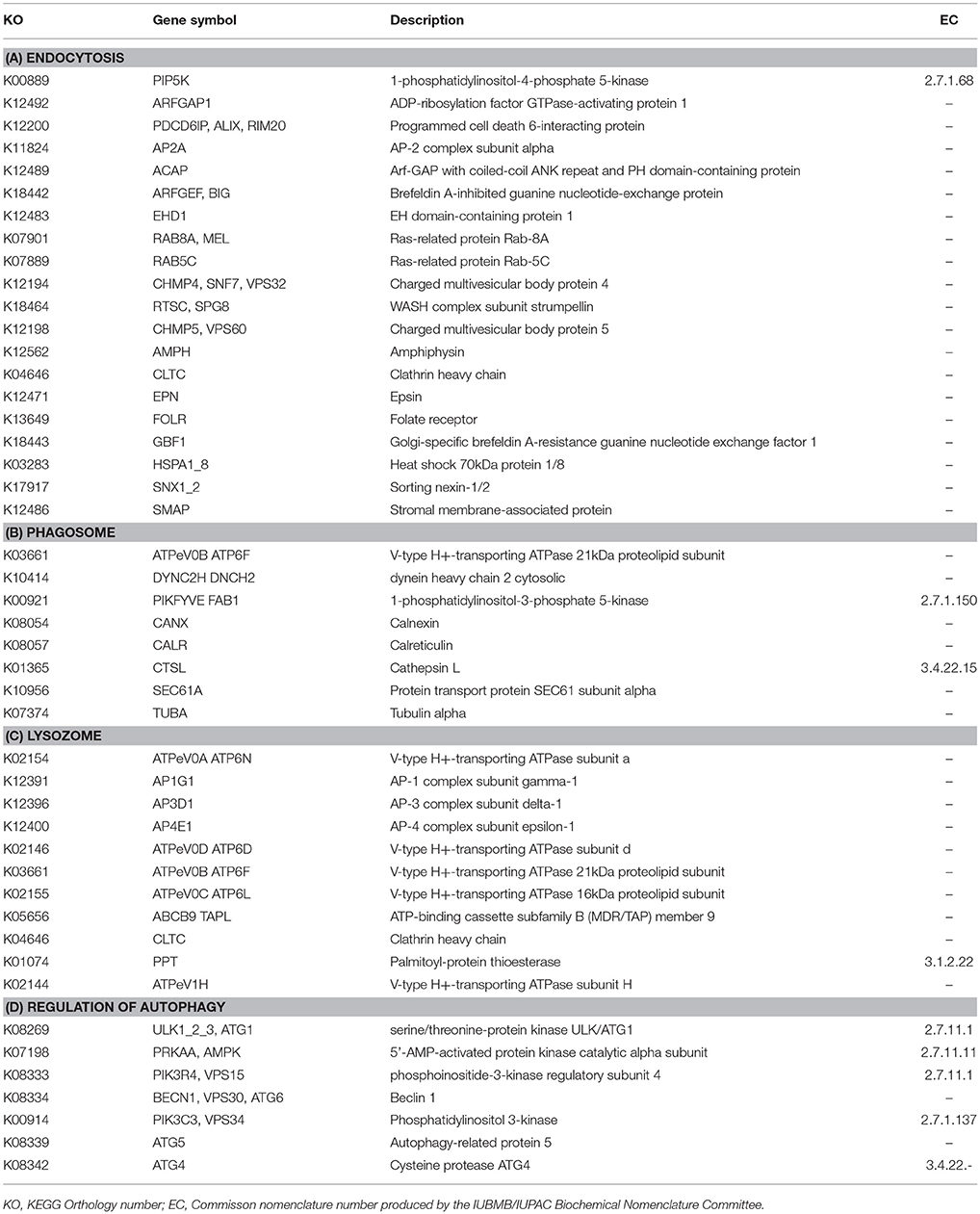
Table 2. Functional annotation of transcripts encoding for enzymes associated with (A) Endocytosis, (B) Phagosome, (C) Lysozome, (D) Regulation of Autophagy, expressed in both M. polaris and P. tychotreta that as identified by the Kyoto Encyclopedia of Genes and Genomes.
Although genes related to phagotrophic processes, as identified by KEGG, were represented in M. polaris, none of these genes were found to be differentially expressed at the defined threshold rate (at least 2 Log2-fold change). The relatively constant level of expression of many of these genes may indicate the ubiquitous, though low, nature of ingestion behavior exhibited by M. polaris. Ingestion by this isolate has been found to be less responsive to media conditions than was the case for P. tychotreta, which was not phagotrophic in nutrient replete media (McKie-Krisberg and Sanders, 2014; McKie-Krisberg et al., 2015) (Figure 4, Supplementary Table 1, Normalized Transcript count data).
Discussion
Light and dissolved nutrients are often considered to be the most important factors affecting phagotrophy in mixotrophs, as these environmental factors represent two significant resource pools required by obligate phototrophs and also used by mixotrophs. The degree to which these factors may induce changes in trophic plasticity (i.e., phagotrophic activity) in a given mixotroph will vary (Stoecker, 1998; Sanders et al., 2001; Liu et al., 2011). Liu et al. (2016) modified light levels when comparing the transcriptomes of three temperate mixotrophs and found significant differences in patterns of gene expression in response to those changes that were consistent with the degree of mixotrophic behavior. We did not include dark treatment here because previous work indicated that P. tychotreta populations decline in dark conditions, and that nutrient concentrations were the major driver of ingestion behavior in this organism (McKie-Krisberg et al., 2015). Both macro- and micro-nutrient concentrations have previously been shown to affect the ingestion rates of these and other mixotrophic phytoflagellates and, in some cases, can dictate whether flagellates will engage in phagotrophy at all (Nygaard and Tobiesen, 1993; Maranger et al., 1998; McKie-Krisberg and Sanders, 2014; McKie-Krisberg et al., 2015). Other factors can also affect ingestion in both mixotrophs and heterotrophs. For example, the differences in cell size between the prasinophytes in the present study (1.2 μm M. polaris vs. 5–7 μm average diameter for P. tychotreta) seems to constrain the number (and size) of bacteria that can be ingested, and prey size has been recognized as a factor limiting ingestion in various single-celled eukaryotes (Fenchel, 1986; Sanders and Gast, 2012; McKie-Krisberg and Sanders, 2014).
M. pusilla (CCMP1545) has a relatively small genome (~20 Mbp, Joint Genome Institute), certainly much smaller than its green algal counterpart, P. tychotreta. Although a sequenced genome of P. tychotreta is not yet available, a recent study of the alga C. tetramitiformis, which is from the same Order, identified a genome size of ~250 Mbp, more than 10 times greater than that of M. pusilla (Worden et al., 2009). The contrasting size of the transcriptomes for M. polaris and P. tychotreta is likely a reflection of the diversity of genome size within the green algal clade. The extensive number of transcripts (over 100 K) found in P. tychotreta suggest additional functions present in this alga, compared to M. polaris, but it may also represent an increase in differential splicing, and/or alternative transcripts. These mechanisms have been shown to exist in other green algae such as Chlamydomonas reinhardtii, as well as in other algal groups including the cryptophytes and chlorarachniophyte algae (Curtis et al., 2012; Blaby et al., 2014). Understanding these types of regulatory mechanisms can only be resolved with additional genome sequencing and annotation efforts, creating a reference wherein the true number of genes and the relation of gene-to-transcript can be determined.
Previous work with marine mixotrophs indicated shifts in gene expression related to both nutrient metabolism and limitation in response to changes in light and nutrient conditions (Koid et al., 2014; Liu et al., 2016; Caron et al., 2017). The results presented here indicate a comparable, though lesser, response noted for a tropical isolate of Micromonas (CCMP2709), which found up-regulation of 738 out of 960 differentially-expressed transcripts under of the effect of phosphorus limitation (Whitney and Lomas, 2016). However, low phosphorus treatments in our present work are 4 times higher than the low nutrient treatments used by Whitney and Lomas (0.5 μM vs. 1.81 μM in our study). Furthermore, Whitney and Lomas (2016) identified the isolate as M. pusilla, but the current nomenclature for this isolate via CCMP, designates CCMP2709 as M. commoda, and was found to be evolutionarily distinct from our M. polaris, which may also contribute to the relative differences in patterns of transcriptional response to changes in nutrients (Worden et al., 2009). In addition, M. polaris may respond to fluctuations in nutrient concentration by employing alternative acquisition strategies, like mixotrophy, which have not been observed in Micromonas sp. (CCMP 2709) to date, and may not be consistent across the genus. In the case of P. tychotreta, we observed significant up regulation of at least one transcript identified as nitrate reductase (NR, EC 1.7.1.1, EC 1.7.1.2, EC 1.7.1.3) as well as ferredoxin-nitrite reductase (EC 1.7.7.1). These results are consistent with previous results of nutrient limitation responses in several non-phagotrophic types of algae from diverse evolutionary lineages, suggesting a basic algal response that is still consistent with a mixotroph that is primarily photosynthetic (Allen, 2003; Berg et al., 2008; Wurch et al., 2011; Dyhrman et al., 2012).
Koid et al. (2014) used the EuKaryotic Orthologous Groups (KOG) tool to compare functional categories in four prymnesiophyte algae, including three known mixotrophs, and found that the overall distributions were very similar. However, when data from an additional 37 protistan species were added to their analysis, the effects of both trophic mode and phylogenetic grouping were detected and non-alveolate mixotrophs formed a distinct cluster. The Micromonas strain CCMP2099 used in our study was also included in the broader analysis of Koid et al. (2014). The prasinophytes clustered together, but were distant from other known mixotrophs, so it may be that in this case the clustering more strongly reflected phylogeny than nutritional mode for the mixotrophs. Genome size may also have affected the analysis of Koid et al. (2014), as the three Micromonas strains and Ostreococcus lucimarinus that were included and clustered together are among the smallest eukaryotes with correspondingly small genomes.
Using the genes linked to phagotrophic behavior proposed by the work of Yutin et al. (2009), a recent genomic study of Cymbomonas tetramitiformis identified the presence of enzymes associated with phagotrophic pathways (Burns et al., 2015). Some of those phagotrophy-associated enzymes could also be identified in the current study. However, the majority of these enzymes were described in metazoan cell types, for example, in macrophages of the mouse, Mus musculus. Additionally, many of these associated genes may have multiple functions within a cell, which makes it difficult to determine the direct participation of a specific gene in cell phagocytosis based solely on the presence of the transcript coding for these enzymes. From an evolutionary perspective, dual function in genes creates the opportunity for conservation of these pathways among phyla containing phagotrophic and non-phagotrophic members, such as the Chlorophyceae. Conversely, it has been suggested that feeding behavior in algae may be associated with loss of biosynthetic pathways, which could drive mixotrophs to seek out prey to replace essential biomolecules—just as this loss is associated with obligatory phagotrophy (Swingley et al., 2007; Burns et al., 2015). Also, even for those genes able to be identified, the nucleotide sequence for a particular gene can vary among organisms from different evolutionary lineages. Depending on the limitations of genomic databases available for query, this variation in nucleotide sequence for a gene may limit reliable identification, even when genes are present in the subject organism.
The dearth of well-annotated reference genomes as well as transcriptomic data on a wide distribution of microbial eukaryotes, particularly within the Prasinophyceae, presents a challenge to interpreting these results. Despite this, the more pronounced response of ingestion-related genes in P. tychotreta as compared to M. polaris is consistent with previous work, which indicated that in P. tychotreta grazing activity is induced only under nutrient-limited conditions. In comparison, M. polaris grazing activity was found at low rates when nutrients were replete, and were only slightly further stimulated when nutrients were limited (McKie-Krisberg and Sanders, 2014; McKie-Krisberg et al., 2015). The results presented here also echo those of Liu et al. (2016), in which expression changes in phagotrophy-associated genes (specifically PEP carboxykinase and vacuolar ATPase) reflected the different ingestion behaviors of Prymnesium parvum and Dinobryon. Under dark conditions, the higher expression of PEP carboxykinase by P. parvum indicated that prey became an important source of organic carbon. In contrast, Dinobryon did not increase PEP carboxykinase expression in the dark, and showed a decrease in vacuolar ATPase expression. This suggests that prey digestion is not active in the dark, and is also supported by the fact that Dinobryon prey ingestion dramatically decreased in the dark (Caron et al., 1993). An additional outcome of employing a comparative genomic approach in this study is the identification of 37 phagotrophy related genes that are shared, and expressed, between these two prasinophytes (Supplementary Information). These studies, and that of a mixotrophic oligotrich ciliate by Santoferrara et al. (2014), provide the basis for future multi-species comparative genomic studies of mixotrophic regulation at the genetic level. These efforts will yield greater understanding of whether the mechanisms regulating prey ingestion in mixotrophic protists are conserved across evolutionarily distinct groups.
The present work contributes to the limited, but growing, data set on the transcription dynamics of green algal lineages in contrasting nutrient regimes. We observed responses to reduction in nutrients consistent with predicted responses at the level of gene expression by photosynthetic marine microalgae. These results were obtained using RNA-Seq technology, and despite the presence of bacteria in these non-axenic conditions, we were able to obtain good quality transcriptomic data from target species using a sufficiently conservative bioinformatic analysis pipeline. In addition, we have identified genes in both organisms that are associated with phagotrophic activity, one of the main aims of the study, despite the fact that M. polaris and P. tychotreta are representatives of different phylogenetic Orders with a large difference in genome size. Additionally, several transcripts containing conserved domains associated with phagotrophic ingestion were found in both transcriptomes, further supporting that these processes are operating within otherwise photosynthetic organisms.
Author Contributions
RG conducted grazing experiments and RNA isolation for sequencing. RG and ZM-K conducted the bioinformatic analysis of RNA-seq data including, de novo assembly, functional annotation, and differential expression analysis. ZM-K was the primary writer of the manuscript. RG and RS provided critical revision of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The Owlsnest Super-Computing Cluster at Temple University is funded by a National Science Foundation Grant CNS-09-58854. The CUNY HPCC is operated by the College of Staten Island and funded, in part, by grants from the City of New York, State of New York, CUNY Research Foundation, and National Science Foundation Grants CNS-0958379, CNS-0855217, and ACI 1126113. Support for this work was also supplied by National Science Foundation grants PLR-1341362 (RG), PLR-1603538 (RS), and PLR-1603833 (RG).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00273/full#supplementary-material
References
Alexa, A., and Rahnenfuhrer, J. (2017). topGO: Enrichment Analysis for Gene Ontology. R Package version 2.32.0. doi: 10.18129/B9.bioc.topGo
Allen, J. F. (2003). Cyclic, pseudocyclic and noncyclic photophosphorylation: new links in the chain. Trend Plant Sci. 8, 15–19. doi: 10.1016/S1360-1385(02)00006-7
Andrews, S. (2010). FastQC A Quality Control tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Azam, F., Fenchel, T., Field, J. G., Gray, J. S., Meyer-Reil, L. A., and Thingstad, F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263. doi: 10.3354/meps010257
Berg, G. M., Shrager, J., Glöckner, G., Arrigo, K. R., and Grossman, A. R. (2008). Understanding nitrogen limitation in Aureococcus anophagefferens (Pelagophyceae) through cDNA and qRT-PCR analysis. J. Phycol. 44, 1235–1249. doi: 10.1111/j.1529-8817.2008.00571.x
Bird, D. F., and Kalff, J. (1986). Bacterial grazing by planktonic lake algae. Science 231, 493–495. doi: 10.1126/science.231.4737.493
Blaby, I. K., Blaby-Haas, C. E., Tourasse, N., Hom, E. F., Lopez, D., Aksoy, M., et al. (2014). The Chlamydomonas genome project: a decade on. Trends Plant Sci. 19, 672–680. doi: 10.1016/j.tplants.2014.05.008
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boraas, M. E., Estep, K. W., Johnson, P. W., and Sieburth, J. (1988). Phagotrophic phototrophs: the ecological significance of mixotrophy. J. Protozool. 35, 249–252. doi: 10.1111/j.1550-7408.1988.tb04336.x
Burns, J. A., Paasch, A., Narechania, A., and Kim, E. (2015). Comparative genomics of a bacterivorous green alga reveals evolutionary causalities and consequences of phago-mixotrophic mode of nutrition. Genome Biol. Evol. 7, 3047–3061. doi: 10.1093/gbe/evv144
Caron, D. A. (1993). “Enrichment, isolation, and culture of free-living heterotrophic flagellates,” in Handbook of Methods in Aquatic Microbial Ecology, eds P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (Boca Raton, FL: Lewis Publishers), 77–90.
Caron, D. A., Alexander, H., Allen, A. E., Archibald, J. M., Armbrust, E. V., Bachy, C., et al. (2017). Probing the evolution, ecology and physiology of marine protists using transcriptomics. Nat. Rev. Microbiol. 15, 6–20. doi: 10.1038/nrmicro.2016.160
Caron, D. A., Sanders, R. W., Lim, E. L., Marrasé, C., Amaral, L. A., Whitney, S., et al. (1993). Light-dependent phagotrophy in the freshwater mixotrophic chrysophyte Dinobryon cylindricum. Microb. Ecol. 25, 93–111. doi: 10.1007/BF00182132
Curtis, B. A., Tanifuji, G., Burki, F., Gruber, A., Irimia, M., Maruyama, S., et al. (2012). Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492, 59–65. doi: 10.1038/nature11681
Derelle, E., Ferraz, C., Rombauts, S., Rouzé, P., Worden, A. Z., Robbens, S., et al. (2006). Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. U.S.A. 103, 11647–11652. doi: 10.1073/pnas.0604795103
Dyhrman, S. T., Jenkins, B. D., Rynearson, T. A., Saito, M. A., Mercier, M. L., Alexander, H., et al. (2012). The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS ONE 7:e33768. doi: 10.1371/journal.pone.0033768
Eddy, S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. doi: 10.1371/journal.pcbi.1002195
Fenchel, T. (1986). “Protozoan filter feeding,” in Progress in Protistology, eds J. O. Corliss and D. J. Patterson (Bristol: Biopress, Ltd.), 65–113.
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2015). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Flynn, K. J., Stoecker, D. K., Mitra, A., Raven, J. A., Glibert, P. M., Hansen, P. J., et al. (2012). Misuse of the phytoplankton–zooplankton dichotomy: the need to assign organisms as mixotrophs within plankton functional types. J. Plankton Res. 35, 3–11. doi: 10.1093/plankt/fbs062
Gast, R. J., McKie-Krisberg, Z. M., Fay, S. A., Rose, J. M., and Sanders, R. W. (2014). Antarctic mixotrophic protist abundances by microscopy and molecular methods. FEMS Microbiol. Ecol. 89, 388–401. doi: 10.1111/1574-6941.12334
Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B., Dettling, M., Dudoit, S., et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. doi: 10.1186/gb-2004-5-10-r80
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Grolleau, A., Misek, D. E., Kuick, R., Hanash, S., and Mulé, J. J. (2003). Inducible expression of macrophage receptor Marco by dendritic cells following phagocytic uptake of dead cells uncovered by oligonucleotide arrays. J. Immunol. 171, 2879–2888. doi: 10.4049/jimmunol.171.6.2879
Guillard, R. R. L. (1975). “Culture of phytoplankton for feeding marine invertebrates,” in Culture of Marine Invertebrate Animals, eds W. L. Smith and M. H. Chanley (Boston, MA: Springer), 29–60.
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Hartmann, M., Zubkov, M. V., Scanlan, D. J., and Lepère, C. (2013). In situ interactions between photosynthetic picoeukaryotes and bacterioplankton in the Atlantic Ocean: evidence for mixotrophy. Environ. Microbiol. Rep. 5, 835–840. doi: 10.1111/1758-2229.12084
Hitchman, R. B., and Jones, H. L. (2000). The role of mixotrophic protists in the population dynamics of the microbial food web in a small artificial pond. Freshw. Biol. 43, 231–241. doi: 10.1046/j.1365-2427.2000.00541.x
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Koid, A. E., Liu, Z., Terrado, R., Jones, A. C., Caron, D. A., and Heidelberg, K. B. (2014). Comparative transcriptome analysis of four prymnesiophyte algae. PLoS ONE 9:e97801. doi: 10.1371/journal.pone.0097801
Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H.-H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Li, W. K., McLaughlin, F. A., Lovejoy, C., and Carmack, E. C. (2009). Smallest algae thrive as the Arctic Ocean freshens. Science 326, 539–539. doi: 10.1126/science.1179798
Liu, J., Huang, J., Sun, Z., Zhong, Y., Jiang, Y., and Chen, F. (2011). Differential lipid and fatty acid profiles of photoautotrophic and heterotrophic Chlorella zofingiensis: assessment of algal oils for biodiesel production. Bioresour. Technol. 102, 106–110. doi: 10.1016/j.biortech.2010.06.017
Liu, Z., Campbell, V., Heidelberg, K. B., and Caron, D. A. (2016). Gene expression characterizes different nutritional strategies among three mixotrophic protists. FEMS Microbiol. Ecol. 92:fiw106. doi: 10.1093/femsec/fiw106
Lovejoy, C., Vincent, W. F., Bonilla, S., Roy, S., Martineau, M.-J., Terrado, R., et al. (2007). Distribution, phylogeny, and growth of cold-adapted picoprasinophytes in arctic seas. J. Phycol. 43, 78–89. doi: 10.1111/j.1529-8817.2006.00310.x
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Maranger, R., Bird, D. F., and Price, N. M. (1998). Iron acquisition by photosynthetic marine phytoplankton from ingested bacteria. Nature 396, 248–251. doi: 10.1038/24352
Maruyama, S., and Kim, E. (2013). A modern descendant of early green algal phagotrophs. Curr. Biol. 23, 1081–1084. doi: 10.1016/j.cub.2013.04.063
McKie-Krisberg, Z. M., Gast, R. J., and Sanders, R. W. (2015). Physiological responses of three species of antarctic mixotrophic phytoflagellates to changes in light and dissolved nutrients. Microb. Ecol. 70, 21–29. doi: 10.1007/s00248-014-0543-x
McKie-Krisberg, Z. M., and Sanders, R. W. (2014). Phagotrophy by the picoeukaryotic green alga Micromonas: implications for Arctic Oceans. ISME J. 8, 1953–1961. doi: 10.1038/ismej.2014.16
Moffat, F. L., Han, T., Li, Z. M., Peck, M. D., Falk, R. E., Spalding, P. B., et al. (1996). Involvement of CD44 and the cytoskeletal linker protein ankyrin in human neutrophil bacterial phagocytosis. J. Cell. Physiol. 168, 638–647. doi: 10.1002/(SICI)1097-4652(199609)168:3< 638::AID-JCP16>3.0.CO;2-V
Moorthi, S. D., Caron, D. A., Gast, R. J., and Sanders, R. W. (2009). Mixotrophy: a widespread and important ecological strategy for planktonic and sea-ice nanoflagellates in the Ross Sea, Antarctica. Aquat. Microb. Ecol. 54, 269–277. doi: 10.3354/ame01276
Morgan, M., Falcon, S., and Gentleman, R. (2008). GSEABase: Gene Set Enrichment Data Structures and Methods. R Package Version 1.42.0. doi: 10.18129/B9.bioc.GSEABase
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. doi: 10.1093/nar/gkm321
Moro, I., La Rocca, N., Valle, L. D., Moschin, E., Negrisolo, E., and Andreoli, C. (2002). Pyramimonas australis sp. nov.(Prasinophyceae, Chlorophyta) from Antarctica: fine structure and molecular phylogeny. Eur. J. Phycol. 37, 103–114. doi: 10.1017/S0967026201003493
Nguyen, M. T., Liu, M., and Thomas, T. (2014). Ankyrin-repeat proteins from sponge symbionts modulate amoebal phagocytosis. Mol. Ecol. 23, 1635–1645. doi: 10.1111/mec.12384
Nygaard, K., and Tobiesen, A. (1993). Bacterivory in algae: a survival strategy during nutrient limitation. Limnol. Oceanogr. 38, 273–279. doi: 10.4319/lo.1993.38.2.0273
Pålsson, C., and Granéli, W. (2004). Nutrient limitation of autotrophic and mixotrophic phytoplankton in a temperate and tropical humic lake gradient. J. Plankton Res. 26, 1005–1014. doi: 10.1093/plankt/fbh089
Raven, J. A. (2013). Cells inside cells: symbiosis and continuing phagotrophy. Curr. Biol. 23, R530–R531. doi: 10.1016/j.cub.2013.05.006
R Core Team (2013). R: A Language and Environment for Statistical Computing. Available online at: http://www.R-project.org/
Roberts, A., Trapnell, C., Donaghey, J., Rinn, J. L., and Pachter, L. (2011). Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12:R22. doi: 10.1186/gb-2011-12-3-r22
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rouached, H., Arpat, A. B., and Poirier, Y. (2010). Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol. Plant 3, 288–299. doi: 10.1093/mp/ssp120
Sanders, R. W. (1991). Mixotrophic protists in marine and freshwater ecosystems. J. Protozool. 38, 76–81. doi: 10.1111/j.1550-7408.1991.tb04805.x
Sanders, R. W. (2011). Alternative nutritional strategies in protists: symposium introduction and a review of freshwater protists that combine photosynthesis and heterotrophy. J. Eukaryot. Microbiol. 58, 181–184. doi: 10.1111/j.1550-7408.2011.00543.x
Sanders, R. W., Caron, D. A., Davidson, J. M., Dennett, M. R., and Moran, D. M. (2001). Nutrient acquisition and population growth of a mixotrophic alga in axenic and bacterized cultures. Microb. Ecol. 42, 513–523. doi: 10.1007/s00248-001-1024-6
Sanders, R. W., and Gast, R. J. (2012). Bacterivory by phototrophic picoplankton and nanoplankton in Arctic waters. FEMS Microbiol. Ecol. 82, 242–253. doi: 10.1111/j.1574-6941.2011.01253.x
Sanders, R. W., Porter, K. G., Bennett, S. J., and DeBiase, A. E. (1989). Seasonal patterns of bacterivory by flagellates, ciliates, rotifers, and cladocerans in a freshwater planktonic community. Limnol. Oceanogr. 34, 673–687. doi: 10.4319/lo.1989.34.4.0673
Santoferrara, L. F., Guida, S., Zhang, H., and McManus, G. B. (2014). De novo transcriptomes of a mixotrophic and a heterotrophic ciliate from marine plankton. PLoS ONE 9:e101418. doi: 10.1371/journal.pone.0101418
Schmieder, R., and Edwards, R. (2011). Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE 6:e17288. doi: 10.1371/journal.pone.0017288
Sherr, E. B., Caron, D. A., and Sherr, B. F. (1993). “Staining of heterotrophic protists for visualization via epifluorescence microscopy,” in Handbook of Methods in Aquatic Microbial Ecology, eds P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (New York, NY: Lewis Publishers), 213–227.
Sherr, E. B., and Sherr, B. F. (2002). Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek 81, 293–308. doi: 10.1023/A:1020591307260
Simon, N., Foulon, E., Grulois, D., Six, C., Desdevises, Y., Latimier, M., et al. (2017). Revision of the genus Micromonas Manton et Parke (Chlorophyta, Mamiellophyceae), of the type species M. pusilla (Butcher) Manton and Parke and of the species M. commoda van Baren, Bachy and Worden and description of two new species based on the genetic and phenotypic characterization of cultured isolates. Protist 168, 612–635. doi: 10.1016/j.protis.2017.09.002
Smith, W. O., Dinniman, M. S., Klinck, J. M., and Hofmann, E. (2003). Biogeochemical climatologies in the Ross Sea, Antarctica: seasonal patterns of nutrients and biomass. Deep Sea Res. Part II Top. Stud. Oceanogr. 50, 3083–3101. doi: 10.1016/j.dsr2.2003.07.010
Stoecker, D. K. (1998). Conceptual models of mixotrophy in planktonic protists and some ecological and evolutionary implications. Eur. J. Protistol. 34, 281–290. doi: 10.1016/S0932-4739(98)80055-2
Swingley, W. D., Sadekar, S., Mastrian, S. D., Matthies, H. J., Hao, J., Ramos, H., et al. (2007). The complete genome sequence of Roseobacter denitrificans reveals a mixotrophic rather than photosynthetic metabolism. J. Bacteriol. 189, 683–690. doi: 10.1128/JB.01390-06
Tirichine, L., and Bowler, C. (2011). Decoding algal genomes: tracing back the history of photosynthetic life on Earth. Plant. J. 66, 45–57. doi: 10.1111/j.1365-313X.2011.04540.x
Unrein, F., Gasol, J. M., Not, F., Forn, I., and Massana, R. (2014). Mixotrophic haptophytes are key bacterial grazers in oligotrophic coastal waters. ISME J. 8, 164–176. doi: 10.1038/ismej.2013.132
Wheeler, P. A., Watkins, J. M., and Hansing, R. L. (1997). Nutrients, organic carbon and organic nitrogen in the upper water column of the Arctic Ocean: implications for the sources of dissolved organic carbon. Deep Sea Res. Part II Top. Stud. Oceanogr. 44, 1571–1592. doi: 10.1016/S0967-0645(97)00051-9
Whitney, L. P., and Lomas, M. W. (2016). Growth on ATP elicits a P-stress response in the picoeukaryote Micromonas pusilla. PLoS ONE 11:e0155158. doi: 10.1371/journal.pone.0155158
Worden, A. Z., Lee, J. H., Mock, T., Rouzé, P., Simmons, M. P., Aerts, A. L., et al. (2009). Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324, 268–272. doi: 10.1126/science.1167222
Wurch, L. L., Bertrand, E. M., Saito, M. A., Van Mooy, B. A., and Dyhrman, S. T. (2011). Proteome changes driven by phosphorus deficiency and recovery in the brown tide-forming alga Aureococcus anophagefferens. PLoS ONE 6:e28949. doi: 10.1371/journal.pone.0028949
Yutin, N., Wolf, M. Y., Wolf, Y. I., and Koonin, E. V. (2009). The origins of phagocytosis and eukaryogenesis. Biol. Direct 4, 6150–6154. doi: 10.1186/1745-6150-4-9
Keywords: mixotrophy, Pyramimonas, Micromonas, RNA-Seq, transcriptomics, phagotrophy
Citation: McKie-Krisberg ZM, Sanders RW and Gast RJ (2018) Evaluation of Mixotrophy-Associated Gene Expression in Two Species of Polar Marine Algae. Front. Mar. Sci. 5:273. doi: 10.3389/fmars.2018.00273
Received: 01 March 2018; Accepted: 18 July 2018;
Published: 14 August 2018.
Edited by:
Holly V. Moeller, University of California, Santa Barbara, United StatesReviewed by:
Jelena Godrijan, Bigelow Laboratory For Ocean Sciences, United StatesSuzanne Jane Painting, Centre for Environment, Fisheries and Aquaculture Science (CEFAS), United Kingdom
Copyright © 2018 McKie-Krisberg, Sanders and Gast. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zaid M. McKie-Krisberg, WmFpZC5ta0Bicm9va2x5bi5jdW55LmVkdQ==
 Zaid M. McKie-Krisberg
Zaid M. McKie-Krisberg Robert W. Sanders
Robert W. Sanders Rebecca J. Gast
Rebecca J. Gast