
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 29 May 2018
Sec. Microbial Symbioses
Volume 5 - 2018 | https://doi.org/10.3389/fmars.2018.00188
This article is part of the Research Topic Socio-Ecology of Microbes in a Changing Ocean View all 17 articles
 Anna R. Bramucci1†
Anna R. Bramucci1† Leen Labeeuw1†‡
Leen Labeeuw1†‡ Fabini D. Orata1‡
Fabini D. Orata1‡ Elizabeth M. Ryan2
Elizabeth M. Ryan2 Rex R. Malmstrom2
Rex R. Malmstrom2 Rebecca J. Case1*
Rebecca J. Case1*Marine microbes form host-associated biofilm communities that are shaped by complex interactions between bacteria and their host. The roseobacter Phaeobacter inhibens exploits both symbiotic and pathogenic niches while interacting with its microalgal host Emiliania huxleyi. During co-cultivation over extended periods with E. huxleyi, we show that P. inhibens selectively kills two host cell types, the diploid calcifying strain and the haploid flagellated strain. Meanwhile, various non-calcifying diploid strains are resistant to this pathogen or the pathogen is avirulent to this cell type. This differential pathogenesis has the potential of dramatically altering the composition of E. huxleyi blooms, which are typically dominated by the roseobacter-susceptible calcifying strain. This cell type makes calcite plates, which are an important sink in the marine carbon cycle and forms part of the marine paleobotanic record. P. inhibens kills the haploid cells, which have been proposed as critical to the survival of the algae, as they readily escape both eukaryotic predation and viral infection. Consequently, bacteria such as P. inhibens could influence E. huxleyi's life history by selective pathogenesis, thereby altering the composition of cell types within E. huxleyi populations and its bloom-bust lifestyle.
On a microscopic scale, the marine environment is a heterogeneous mixture of nutrient “hotspots” formed by plankton and marine snow (Azam, 1998). Marine microbes take advantage of this aquatic array of nutrient gradients and “hotspots” by preferentially occupying specific niches (Hunt et al., 2008; Stocker, 2012). Motile microbes capable of directly associating with phytoplankton thereby expose themselves to a continuous stream of metabolites leaked from their algal hosts (Sapp et al., 2007; Geng and Belas, 2010; Sule and Belas, 2013). The concentrations of these metabolites are at their highest at the cell surface and within the phycosphere (Bell and Mitchell, 1972), the area immediately surrounding an algal cell, suggesting that algal-associated bacteria experience the greatest nutrient benefit and most likely to exchange communication and bioactive molecules.
While many bacteria directly benefit from algal metabolites exuding out of leaky or dying algae, some bacteria have fine-tuned their ability to sense and respond to host molecules by expressing symbiotic or pathogenic traits (Miller et al., 2004; Tripp et al., 2008; Seymour et al., 2010; Case et al., 2011; Wang et al., 2014; Amin et al., 2015) or releasing bioactive molecules that in turn alter the host's behavior or survival (Seyedsayamdost et al., 2011b; Labeeuw et al., 2017). Some bacteria engage in symbiotic relationships with their algal host by producing small molecules, such as vitamins or growth hormones, that are beneficial or even required by the alga (Bolch et al., 2011). Other bacteria use bioactive molecules in pathogenesis to coordinate virulence or kill their host (Ashen et al., 1999; Fernandes et al., 2011; Seyedsayamdost et al., 2011b; Gardiner et al., 2017).
The haptophyte Emiliania huxleyi, which dominates coccolithophore blooms (Baumann et al., 2008), has recently been described as a host for bacteria in the marine environment (Seyedsayamdost et al., 2011b; Segev et al., 2016). It can quickly form dense populations (>106 cells·L−1) (Rhodes et al., 1995; Tyrrell and Merico, 2004) over vast expanses of the upper ocean (>250,000 km2) (Holligan et al., 1993). Its bloom-bust life cycle dramatically restructures the marine ecosystem, as it becomes a habitat-forming species for bacteria and viruses in the open ocean. Bloom formation has been associated with a variety of environmental factors (Tyrrell and Merico, 2004), while bloom collapse is frequently attributed to viral infection or microzooplankton grazing (Wolfe et al., 1994, 1997; Wilson et al., 2002). The ubiquity and abundance of E. huxleyi in the oceans, as well as its production of important intermediates in the carbon and sulfur biogeochemical cycles, have made it an important model phytoplankton species.
The E. huxleyi species complex, with an average diameter of 5–7 μm, has three distinct cell types: non-motile non-calcifying cells (N-type, diploid), non-motile coccolith-bearing cells (C-type, diploid), and motile scale-bearing cells (S-type, haploid) (Klaveness, 1972; Rhodes et al., 1995; Frada et al., 2012; von Dassow et al., 2015). E. huxleyi blooms are comprised of complex mixtures of C and S cells (Klaveness, 1972), though the fast growing calcifying C cells make up the majority of the bloom (Bidle et al., 2007; Baumann et al., 2008). It is not presently understood how these three cell types interact or alternate to shape the alga's life history, but each cell type is capable of asexual proliferation (Klaveness, 1972). However, the ways in which these different cell types impact the ecology, evolution, interactions, and metabolites of the E. huxleyi species complex is largely unknown, except for its interaction with viruses (EhVs). EhVs kill both diploid cell types (C and N) while S cells are resistant to infection (Wilson et al., 2002; Frada et al., 2008). Once an EhV infects a diploid cell, it proliferates and produces the viral glycosphingolipids that kill the algal cell (Vardi et al., 2009). Viral infection triggers caspase-like activity in E. huxleyi (associated with an upregulation of algal metacaspases), suggesting that the virus is hijacking algal programmed cell death (PCD) machinery to kill the host (Bidle et al., 2007).
Although viral infection and grazers have been studied (Wolfe et al., 1994; Wilson et al., 2002), the association of bacteria with E. huxleyi is largely unexplored (Seymour et al., 2010; Curson et al., 2011). We have investigated how E. huxleyi interacts with the marine α-proteobacteria Phaeobacter inhibens DSM 17395, previously named Phaeobacter gallaeciensis BS107 (Seyedsayamdost et al., 2011b). P. inhibens is a member of the roseobacter clade that is frequently identified within E. huxleyi blooms (González et al., 2000; Green et al., 2015; Segev et al., 2016). This bacterium produces a number of novel bioactives, including the antibiotic tropodithetic acid (TDA) (Geng et al., 2008; Berger et al., 2011; Thole et al., 2012) and potent algaecides called roseobacticides (Seyedsayamdost et al., 2011b, 2014) and roseochelins (Wang and Seyedsayamdost, 2017). These bioactives might allow P. inhibens to live a duplicitous lifestyle as both a beneficial symbiont and as a pathogen. TDA has been implicated in P. inhibens' chemical defense of various hosts (D'Alvise et al., 2012; Prol García et al., 2013; Beyersmann et al., 2017). Indeed, this bacterium is a symbiont of Ulva australis, chemically defending this ubiquitous seaweed from colonization (Rao et al., 2007). It can also act as a probiotic for turbot cod larvae, protecting it against Vibrio anguillarum infections (Planas et al., 2006). It also produces roseobacticides that could facilitate pathogenesis, as they have specific activity at very low concentrations against certain non-calcifying strains of E. huxleyi, causing these cells to lyse (Seyedsayamdost et al., 2011b). Additionally, a recent report suggested that P. inhibens uses the plant growth hormone indole-3-acetic acid (IAA) to kill calcifying (C-type) E. huxleyi (Segev et al., 2016), although the dynamics of this interaction have yet to be elucidated.
We co-cultured an axenic representative strain of the three E. huxleyi cell types (N diploid, C diploid, and S haploid) with P. inhibens and monitored photosynthetic health, as well as the cell dynamics of both the algae and bacterium. We demonstrate that P. inhibens selectively causes the precipitous population-wide death of aged E. huxleyi coccolith-bearing C cells as well as the motile scale-bearing S cells, while various non-calcifying diploid N populations were not killed. Algal death directly benefits the bacterium, which increases its own population size during its host's decline. Finally we utilized a P. inhibens mutant library to demonstrate that the potent roseobacticides produced by P. inhibens, which were previously shown to kill N cells at nanomolar concentrations (Seyedsayamdost et al., 2011b), are not required for P. inhibens pathogenesis of C or S cells. These findings suggest that alternative, as of yet unknown, pathogenicity factor(s) may play equally critical roles in the pathogenesis of P. inhibens toward E. huxleyi.
All E. huxleyi strains were obtained from the National Centre for Marine Algae and Microbiota (East Boothbay, ME, USA). This includes coccolith-producing (C-type) strain CCMP3266, scale-bearing haploid (S-type) strain CCMP3268, and non-calcifying diploid (N-type) strains CCMP370, CCMP372, CCMP374, CCMP379, and CCMP2090. The N-type strains are all from unique geographical locations spanning the biogeographical range of all available axenic E. huxleyi cultures (Table 1). They were maintained in L1-Si medium (Guillard and Hargraves, 1993) at 18°C in a diurnal incubator (8:16 h dark-light cycle) with 41.51 ± 11.15 μmol·m−2·s−1 of light during the light period. Algal cultures and medium were checked for bacterial contamination by microscopic observations and by inoculation onto half-strength marine agar (½ MA) (18.7 g Difco Marine Broth 2216 and 9 g NaCl, supplemented with 15 g Difco agar in 1 L) followed by incubation at 30°C for 2 days. E. huxleyi was inoculated with a 10−1 dilution, grown statically for 5 days to 105 cells·mL−1 (early-log) prior to all experiments.
The wild type P. inhibens DSM 17395 (Frank et al., 2014), was maintained at 30°C on ½ MA (see Supplementary Methods for identification of the bacterium). Colonies were then transferred to half-strength marine broth (18.7 g Difco Marine Broth 2216 and 9 g NaCl in 1 L). P. inhibens was grown to stationary phase in a shaking incubator at 30°C, 160 rpm, for 24 h and then subsequently re-cultured in the same conditions prior to experimentation. Transposon mutants of P. inhibens DSM 17395 (Wetmore et al., 2015) were grown under the same conditions with the addition of 200 μg·mL−1 kanamycin (Sigma-Aldrich).
Transposon mutants in TDA biosynthesis have been identified previously as deficient in TDA and roseobacticide production and consequently have white colonies, as opposed to the typical brown colonies of the wild type (Wang et al., 2016). Mutants screened all produced white colonies and were in two genes in the tda gene cluster of a P. inhibens plasmid pPGA1_262: tdaB (PGA1_262p00970) and paaZ2 (PGA1_262p00800); as well as two genes in the paa gene cluster encoded on the chromosome: paaA (PGA1_c04080) and patB (PGA1_c00860) (Wetmore et al., 2015; Wang et al., 2016).
Gene knockout strains of P. inhibens DSM 17395 were obtained from a previously generated library created through molecularly barcoded transposon mutagenesis (Wetmore et al., 2015; Price et al., 2016), and specific mutants were recovered following the approach outlined by Cole et al. (2017). Briefly, the P. inhibens mutant library was spread on MA plates with 50 μg·mL−1 kanamycin and incubated overnight at ~22°C. Colonies were picked using Qpix460 (Molecular Devices), arrayed into 384-well plates containing half-strength YTSS media (2 g yeast, 1.25 g tryptone, and 20 g sea salts in 1 L) with 7.5% glycerol and 50 μg·mL−1 kanamycin, and incubated overnight at ~22°C. Next, 25 nL of each well was collected using an Echo525 liquid handler (Labcyte) as part of a multiplexing strategy involving pooling of rows, columns, and plates. These pools were subject to PCR amplification and sequencing of the molecular barcodes that identified the transposon insertion site of each mutant strain (Wetmore et al., 2015). The well location of each mutant was determined from amplicon sequencing results using an in-house script. Successful interruption of the targeted genes was confirmed by sequencing over the mutation site. Genomic DNA was extracted using GeneJet Genomic DNA Purification Kit (Thermo Scientific). Gene-specific primers were designed, and PCR was performed using the Phusion High-Fidelity DNA polymerase (Thermo Scientific). PCR products were either column- or gel-purified using QIAquick PCR Purification Kit or MinElute Gel Extraction Kit (QIAGEN), respectively. Amplicon sequencing was performed using the Sanger dideoxy method (Applied Biosystems 3730 Genetic Analyzer).
Bacterial-algal co-cultivation was performed as previously described (Bramucci et al., 2015). Briefly, stationary phase bacterial cultures were washed twice by centrifugation and re-suspended in L1-Si medium before further centrifugation and resuspension to the target cell concentration in colony forming units (CFU)·mL−1. E. huxleyi and the bacteria were mixed 1:1 (volume:volume) with a final concentration in the co-culture of 102 CFU·mL−1 bacteria and 105 cells·mL−1 algae, then 1 mL of this co-culture was aliquoted in 48-well plates (Becton Dickinson). The same cell densities of the bacterium and the alga were inoculated as monocultures in L1-Si medium and aliquoted into the microtitre plate. All controls/co-cultures were performed in triplicate. The microtitre plates were incubated in a diurnal incubator (8:16 h dark-light cycle) at 18°C for all experiments.
Co-cultivation of CCMP3266 or CCMP3268 with P. inhibens transposon mutants were performed as described, with control and co-cultures amended with kanamycin to a final concentration of 100 μg·mL−1 to keep selective pressure for the mutants (Wetmore et al., 2015). E. huxleyi was grown with various concentrations of kanamycin (0, 10, 50, 100, 200 μg·mL−1) to determine that 100 μg·mL−1 kanamycin did not adversely affect growth of photosynthetic yield (Supplementary Figure S1). The cell density of mutants was enumerated on ½ MA with and without 200 μg·mL−1 kanamycin throughout the co-cultivation with algae to ensure the transposon mutation had not been lost from P. inhibens.
Roseobacticide B was obtained from Dr. Mohammad Seyedsayamdost (Princeton University, NJ, USA) and dissolved in methanol as previously described (Seyedsayamdost et al., 2011b). The compound was then added to senescent (i.e., cultures were declining after reaching their maximal cell fluorescence) E. huxleyi (CCMP3266 and CCMP3268) at half the maximal inhibitory concentration (IC50) previously identified for the sensitive N strain (Seyedsayamdost et al., 2011b), as well as ten-fold lower and higher (final concentrations of 0.019, 0.19, and 1.9 μM). A final concentration of 2% methanol was added to experimental and control cultures. The samples were incubated in microtiter plates in the same conditions previously used for bacterial co-culture experiments for 24 h before obtaining pulse-amplitude-modulation (PAM) fluorescence measurements.
A PAM fluorometer (WATER-PAM, Heinz Walz) was used to measure chlorophyll fluorescence (Schreiber et al., 1986; Bramucci et al., 2015). All samples were taken at the midpoint of the dark cycle (4 h) and diluted in sterile L1-Si medium to within the detection range of the PAM fluorometer. Samples were maintained at 18°C throughout handling. A dark adaption period of 3 min was determined, after which a saturating pulse was applied and the fluorescence readings were taken in triplicate at intervals of 1 min 30 s to calculate the minimal dark fluorescence (F0), the maximum dark fluorescence (Fm), and the photosystem II (PSII) maximum efficiency (Fv/Fm), Fv/Fm = (Fm − F0)/Fm (Schreiber et al., 1986; Baker, 2008). Triplicate readings of each sample were averaged and the three microtitre wells were treated as replicates to determine the maximum quantum efficiency.
Subsamples from algal controls and co-cultures were fixed for flow cytometry, incubated in the dark for 10 min with 0.15% glutaraldehyde (Sigma-Aldrich), flash-frozen in liquid nitrogen, and stored at −80°C until flow cytometry was performed using a FACSCalibur (Becton Dickinson). A 488 nm laser was used for excitation. Samples were then run using chlorophyll fluorescence (emission = 670 nm) for detection and cell counting (cells·mL−1).
Phaeobacter inhibens population density grown alone and in co-culture was enumerated by counting CFU on ½ MA after 2 days of incubation at 30°C. Five replicate counts from each well were averaged, and the triplicate wells were used as experimental replicates for analysis. Although P. inhibens attaches to E. huxleyi and itself, aggregated cells were not observed microscopically after 5 min of vigorous vortexing.
Brightfield images of algal controls and co-cultures were obtained using a 63 × Axio Scope.A1 objective lens (Zeiss), equipped with an Optronics digital camera and PictureFrame software v2.3 (Zeiss). Epifluorescence images were obtained using a 100 × Axio Imager.M2 microscope objective lens (Zeiss), equipped with a monochrome camera (AxioCam 506 mono). Epifluorescence microscopy was also used to assess algal chlorophyll auto-florescence and to visualize algal and bacterial DNA when stained with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) (DNA–DAPI complex: excitation = 364 nm; emission = 454 nm) (Life Technologies). Unfixed E. huxleyi control and co-culture aliquots were stained with DAPI according to manufacturer's instructions (30°C, 20 min) then immediately pelleted by centrifugation (5,000 × g, room temperature, 2 min). Cells were gently washed twice in sterile L1-Si medium and analyzed immediately on the epifluorescence microscope. Images were acquired simultaneously for three different channels and overlaid using Zen 2 Blue Edition software v2 (Zeiss). The differential interference contrast channel was overlaid with 1) algal chlorophyll auto-fluorescence (red: excitation = 610–650 nm; emission = 670–720 nm) and 2) DNA–DAPI complex fluorescence (blue: excitation = 350–400 nm; emission = 417–477 nm).
Flow cytometry data were processed using FlowJo v9.2 (Tree Star Inc.). Quantitative data from all other experiments were processed using SigmaPlot 12.0 (Systat Software). Statistical significance was determined using a one-way ANOVA and Tukey HSD test.
This study involves the bacterium previously identified as P. gallaeciensis BS107 that produces roseobacticides, which have a specific algaecidal effect on E. huxleyi (Seyedsayamdost et al., 2011b); however, there is evidence that there were differences in strains of the bacteria submitted to various repositories (Buddruhs et al., 2013). Therefore, we sequenced the genome of our strain and its identity was confirmed to be P. inhibens DSM 17395 (Frank et al., 2014) based on both 100% average nucleotide identity and percent (in silico) DNA–DNA hybridization to the published DSM 17395 genome (Thole et al., 2012; Supplementary Figure S2).
To determine if P. inhibens has a host cell type preference in its interactions with E. huxleyi, we co-cultured P. inhibens with five axenic N strains from distinct geographical locations, one axenic C strain (CCMP3266), and one axenic S strain (CCMP3268) (Table 1). The C and S strains were both killed by P. inhibens, and all tested N type strains survived, regardless of geographic origin (Table 1). To further investigate the differential pathogenesis of P. inhibens on various E. huxleyi cell types, an axenic N [CCMP2090, derived from the polymicrobial calcifying CCMP1516 (Orata et al., 2016; Zhang et al., 2016)], C (CCMP3266), and S [CCMP3268, derived from a single cell isolation from CCMP3266 (von Dassow et al., 2015)] strains (Figure 1) were investigated in more depth. Each strain was grown alone and in co-culture with P. inhibens for 14 days and monitored for PSII maximum quantum efficiency (Fv/Fm), which is affected by cellular stress and/or loss of functional PSII centers (Figure 2), as well as algal and bacterial cell density (Figure 3).
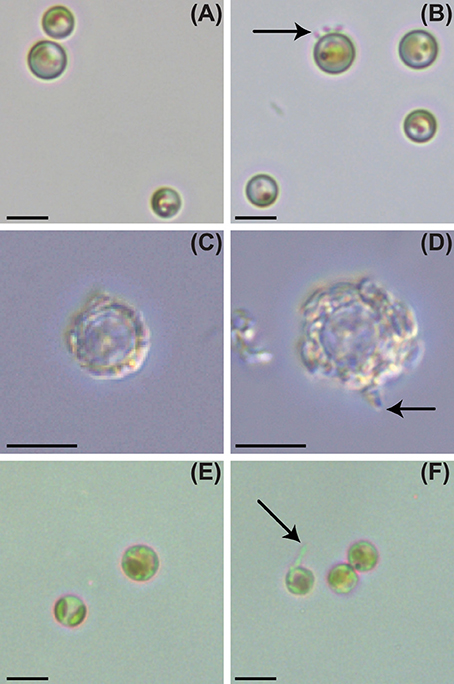
Figure 1. Light and epifluorescence microscopy of P. inhibens attached to non-calcifying diploid N (CCMP2090), diploid calcifying C (CCMP3266), and haploid S (CCMP3268) E. huxleyi strains. Brightfield light microscopy of E. huxleyi, day 6: (A) control CCMP2090, (B) CCMP2090 co-culture with P. inhibens, (C) control CCMP3266, (D) CCMP3266 co-culture with P. inhibens, (E) control CCMP3268, (F) CCMP3268 co-culture with P. inhibens. P. inhibens cells attached to algal host are indicated by black arrows. The scale bar represents 5 μm.
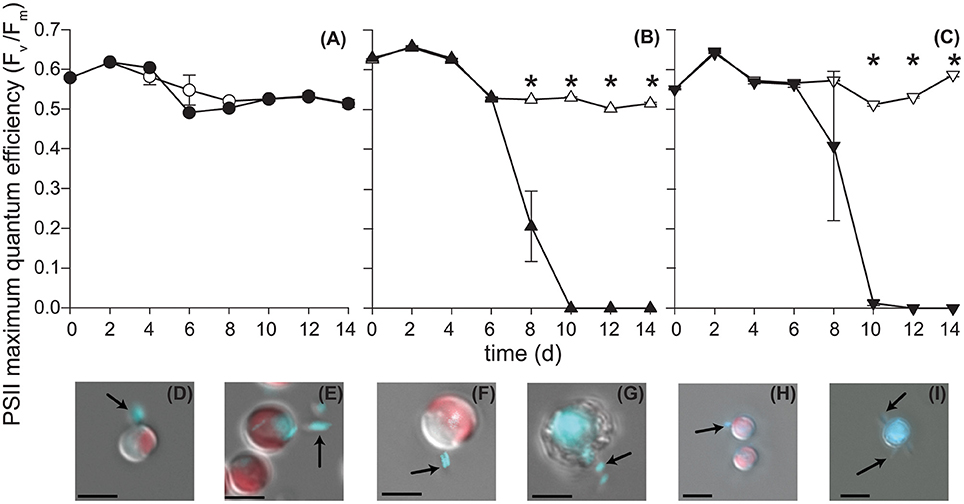
Figure 2. P. inhibens effect on E. huxleyi's photosynthetic health. P. inhibens was co-cultured with non-calcifying diploid N cells (CCMP2090), diploid calcifying C cells (CCMP3266), and haploid S cells (CCMP3268) E. huxleyi. The maximum quantum efficiency (Fv/Fm) is shown for (A) CCMP2090 (circles), (B) CCMP3266 (triangles) and (C) CCMP3268 (upside down triangles). Data for axenic algal cultures are filled in white, whereas data from co-cultured experiments are solid black. Triplicate wells that represent independent experiments were sacrificially sampled at each time point; error bars represent ±1 standard error. An asterisk (*) at a time point indicates that it is significantly different to the control. Statistical significance was determined using a one-way ANOVA and Tukey HSD test. Differential interference contrast images of (D) CCMP2090 co-culture with P. inhibens healthy, day 6 and (E) healthy, day 10, (F) CCMP3266 co-culture with P. inhibens healthy, day 6 and (G) dying, day 10, (H) CCMP3268 co-culture with P. inhibens healthy, day 6 and (I) dying, day 10. Images are shown overlaid with two fluorescent channels: chlorophyll auto-fluorescence (red: excitation 610–650 nm; emission 670–720 nm) and DNA–DAPI complex fluorescence (blue: excitation 350–400 nm; emission 417–477 nm). DAPI-stained P. inhibens cells attached to algal host and non-associated are indicated by black arrows. The scale bar represents 5 μm.
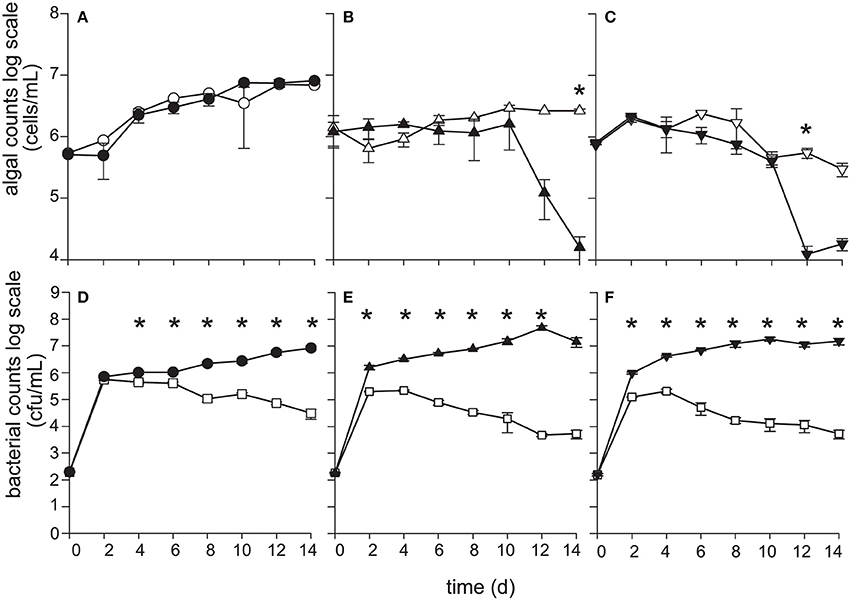
Figure 3. P. inhibens kills diploid calcifying C (CCMP3266), and haploid S (CCMP3268) E. huxleyi while non-calcifying diploid N (CCMP2090) is resistant to P. inhibens pathogenesis. E. huxleyi cell density (cells·mL−1) were enumerated using flow cytometry for (A) CCMP2090 (circles), (B) CCMP3266 (triangles), and (C) CCMP3268 (upside down triangles) for both axenic control (white) and grown in co-culture with P. inhibens (black). P. inhibens cell density (CFU·mL−1) were enumerated for the control in L1-Si medium (white squares) and co-cultured with E. huxleyi: (D) CCMP2090 (black circles) (E) CCMP3266 (black triangles), and (F) CCMP3268 (black upside down triangles). Triplicate wells that represent independent experiments were sacrificially sampled at each time point; error bars represent ±1 standard error. An asterisk (*) at a time point indicates that it is significantly different to the control. Statistical significance was determined using a one-way ANOVA and Tukey HSD test.
When grown alone, all three algal cell types had a stable PSII maximum quantum efficiency (Fv/Fm > 0.5) throughout the experiment, although they entered a senescent stage, represented by reduced fluorescent health after 6–10 days (Figures 2A–C). The algal cultures grown alone did not die in this experiment and have been shown to live for > 60 days in microtitre plates (Bramucci et al., 2015). Both C and S cultures grown alone reached their peak density between 6 and 10 days followed by a characteristic slow decline during senescence (Figures 3B,C), typified by gradual losses of cell numbers (Franklin et al., 2012). It has been suggested that P. inhibens stimulates E. huxleyi's growth rate in the early growth phases (Segev et al., 2016); however, no growth stimulation was detected for the N, C, or S cell types in the co-cultures, with no significant difference between E. huxleyi's cell density for any of the cell types in co-culture or the control in the first 6 days of the experiment (Figure 3).
The N strain grown in co-culture with the bacterial symbiont maintained chlorophyll health, functional PSII systems (Figure 2A), and cell density (Figure 3A) comparable to the axenic cultures throughout the experiment, despite P. inhibens being found attached to its surface (Figures 2D,E). This algal strain, alone or in co-culture with the bacterium, grows to a higher cell density than C- or S-type grown alone from 12 days onwards (P < 0.05) (Figure 3).
In contrast, the C and S cultures grown in co-culture with P. inhibens experienced an accelerated and premature decline in photosystem health (8–10 days) (Figures 2B,C). This population-wide decline is associated with a concurrent decrease in individual chlorophyll content in the algal cells co-cultured with the bacterium (Figures 2F–I). When co-cultured with P. inhibens, C-type cell densities are much lower than the control by 14 days (P < 0.05) compared to the axenic control (Figure 3B). A similar trend is observed for the S-type strain (Figure 3C). All S cells lost their photosynthetic ability in the co-culture with P. inhibens by day 12 with a detectible decrease in photosynthetic health initiated on day 8 (Figure 2C). This timing for rapid decline triggered by a bacterial pathogen coincides closely with the onset of senescence in the algal host.
The rapid decline of PSII efficiency when co-cultured with P. inhibens is intriguing, as it has not been observed in starved senescent E. huxleyi cells (Franklin et al., 2012), but occurs during viral infection and lysis of E. huxleyi cells by EhVs (Bidle et al., 2007). These findings suggest that the algaecidal activity of P. inhibens against its microalgal host, E. huxleyi, is dependent on algal cell type, and that the targeted cell types are different to EhVs which are only known to kill N- and C-type cells (Frada et al., 2008; Mordecai et al., 2017). Similarly, the closely related roseobacter pathogen, Ruegeria sp. R11 (Fernandes et al., 2011) (also known as Nautella italica R11; Vandecandelaere et al., 2009; Rodrigo-Torres et al., 2016), kills C and S cells and not N cells (Mayers et al., 2016). While the mechanism of P. inhibens pathogensis is not known, clues can be found in EhVs, which upregulate metacaspase activity in calcifying diploid cells, inducing alga PCD (Bidle et al., 2007). EhVs induce an autophagy-like PCD event in the algal host (Schatz et al., 2014). Autophagy, or the genetically programed lysosomal degradation of cellular constituents (Kroemer et al., 2009), is a vital part of the host immune response and can infer protection against bacterial pathogens by ensuring the rapid degradation of virulence factors (Cemma and Brumell, 2012). Because this viral-induced PCD response in E. huxleyi results in a loss of PSII function directly before death due to viral lysis (Bidle et al., 2007), it might follow that a similar mechanism is activated by the bacterial pathogen. Environmental stress is a factor in gradual loss of photosynthetic health, but the sudden complete loss of photosynthetic health has only so far been seen in the C cells undergoing viral-induced apoptosis. The activation of algal caspase-like molecules resulting in apoptosis-like PCD has already been described in the pathogenic interaction between Ruegeria sp. R11 and the C- and S-type culture CCMP3266 (Mayers et al., 2016).
This resistance of N-type cells to pathogenic roseobacters has been previously reported for the N-type strain, CCMP2090, cultured with Ruegeria sp. R11 (Mayers et al., 2016). Together, these findings suggest that, although the N cell used in the current study (CCMP2090) was recently derived from a polymicrobial calcifying parent strain (CCMP1516), calcifying and non-calcifying diploid strains have important biological differences, which might for instance confer widespread resistance to roseobacter pathogenesis. There are at least two possible explanations as to why E. huxleyi N cells are widely resistant to P. inhibens pathogenesis, or that P. inhibens does not interact with N cells. The first possibility is that N cells escape the pathogen, similar to the haploid escape from EhVs (Frada et al., 2008; Mordecai et al., 2017). The haploid S-type is thought to be resistant to viral lysis, and it was postulated that the haploid's scaly coverings or that gene loss or mutation might infer viral resistance to the haploid cell type (Frada et al., 2008). Supporting this theory, an increase in culture temperature confers temperature-induced resistance to EhVs by altering the outer sphingolipids of representative calcifying and non-calcifying diploid E. huxleyi strains, impeding viral recognition and infection of target cells (Kendrick et al., 2014). By analogy, it is possible that P. inhibens does not interact with E. huxleyi N cells due to differential recognition and/or attachment to the alga, as was suggested for Ruegeria sp. R11 (Mayers et al., 2016). However, P. inhibens does attach to N cells (Figure 1B) and so N cell resistance, or P. inhibens lack of virulence, must have another mechanism. A second possibility for N cell escape is that there might be genetic differences between C and N cells. There are two prevalent theories as to how N cells are generated: (1) through a series of mutations resulting in, among other differences, a malformed coccolith-forming vesicle (van der Wal et al., 1983) and (2) due to prolonged lab domestication (Zhang et al., 2016). To our knowledge, only one axenic C and S culture exist, and it is commonly reported that coccoliths were lost through subsequent culturing. For example, the polymicrobial CCMP1516 was a C-type culture that recently became an N-type culture; it was a C-type culture when it produced its axenic daughter strain CCMP2090, which is N-type (Zhang et al., 2016). Future genomic effort should focus on sequencing parent (C) and daughter (N) cultures to elucidate if a genetic basis for the phenotypic switch can be identified as it could elucidate resistance mechanisms to the bacterial pathogen and the molecular basis of coccolithogenesis.
Bacterial population dynamics when grown alone and in co-culture were monitored using CFU (Figures 3D–F). Under the experimental conditions used (algal medium L1-Si from seawater, with no additional carbon), P. inhibens was able to grow to a maximum cell density of 105 CFU·mL−1 without E. huxleyi at 2–4 days (Figures 3D–F), after which P. inhibens cell density declined. Prolonged monitoring of P. inhibens grown without a host showed that all bacteria died by 25 days. However, its growth was greatly enhanced by the presence of E. huxleyi, reaching 100–1,000 times higher densities when grown with an algal host (Figures 3D–F).
The growth benefit conveyed to P. inhibens is greater in the short term (14 days and less) when grown with C- and S-type cells of E. huxleyi than the N-type (Figures 3D–F). The bacterial cell density in the C cell co-culture appears to benefit from 2 to 6 days (P < 0.01) and 10 days (P < 0.05) when compared to the N culture (Figures 3D,E). Directly after the C-type culture suffers a decline in both PSII health and numbers of chlorophyll containing cells (day 12), P. inhibens cell density increases 8.5-fold (P < 0.01), compared to those grown in co-culture with N-type, which does not die (day 12) (Figures 3A,B,D,E). P. inhibens co-cultured with S-type also has a significant (P < 0.01) benefit in terms of increased cell counts compared to the N-type co-culture between 4 and 10 days (Figures 3D,F), the time when the algae has reduced PSII maximum quantum efficiency as well as declining cells per mL in the co-culture (Figures 2C, 3C). Additionally, the benefit to P. inhibens persisted after the death of the algal host (12–14 days). In fact, P. inhibens grown with C- and S-type maintained high cell counts until at least 30 days, when both co-cultures had 106 and 107 CFU·mL−1, respectively, despite death of its host. P. inhibens grown in co-culture with N-type maintained high cell counts until at least 30 days (108 CFU·mL−1).
This increased persistence and higher population density of P. inhibens when co-cultured with E. huxleyi is likely the result of nutrients made available by algal exudate (Borchard and Engel, 2012). Phytoplankton constantly leak sugars and oxygen from photosynthesis, as well as other chemicals such as dimethylsulfoniopropionate (DMSP) and amino acids, which are chemoattractants for roseobacters (Mitchell et al., 1985; Baines and Pace, 1991; Miller and Belas, 2004; Miller et al., 2004). Furthermore, DMSP-degrading microbes, such as P. inhibens, benefit directly from being able to efficiently assimilate the sulfur from DMSP directly into bacterial amino acids (Curson et al., 2011). These results demonstrate a population wide benefit for P. inhibens to being grown in co-culture with an algal host, regardless of host cell type. P. inhibens then increases its fecundity and persistence by killing certain cell types of its host, presumably because algal cell lysis could provide it with nutrients (Kolb et al., 2013). This interaction makes P. inhibens an opportunistic pathogen, not a parasite (Seyedsayamdost et al., 2011a), as parasites reduce the health and fecundity of their host and benefit from prolonging the host's life, while P. inhibens causes C- and S-type cells to die prematurely.
The rapid decline in maximum quantum yield and loss of chlorophyll observed in the C- and S-type cultures at 8–10 days (Figure 2) is consistent with the physiological response of E. huxleyi to roseobacticides, which causes cell membrane blebbing, chloroplast loss, and lysis of N-type cells (Seyedsayamdost et al., 2011b). Roseobacticides are produced by P. inhibens when supplied with p-coumaric acid (pCA) (Seyedsayamdost et al., 2011b) and other lignin precursors (Wang and Seyedsayamdost, 2017). pCA is produced by E. huxleyi and hypothesized to be made during senescence (Seyedsayamdost et al., 2011b) where it is an intermediary of the lignin and flavonoid pathways (Labeeuw et al., 2015). P. inhibens thereby produces the algaecidal roseobacticides to precipitously kill a dying host when it ages or is damaged. To determine if roseobacticides were responsible for decreases in quantum yield and chlorophyll loss, roseobacticide B was added to senescent C and S cultures of E. huxleyi on day 9—the day when those E. huxleyi cultures typically decline when grown with P. inhibens—at IC50 of 0.19 μM determined for another N-type strain, CCMP372 (Seyedsayamdost et al., 2011b). Roseobacticide B concentrations 10-fold higher and lower than the IC50 were also tested. Surprisingly, no changes in PSII maximum quantum yield were observed at any roseobacticide concentrations (Figures 4A,B). Furthermore, the characteristic declines in C and S cultures were reproduced when co-cultured with four different P. inhibens mutants whose roseobacticide production genes had been disrupted (Figures 4C,D). These results suggest roseobacticides were not responsible for the death of C- and S-type cells, and that P. inhibens must be producing additional algaecidal compounds or virulence factors to kill E. huxleyi. In addition, non-calcifying diploid strains surviving co-culture with P. inhibens included CCMP372 (Table 1), which were previously shown to be killed by roseobacticides (Seyedsayamdost et al., 2011b).
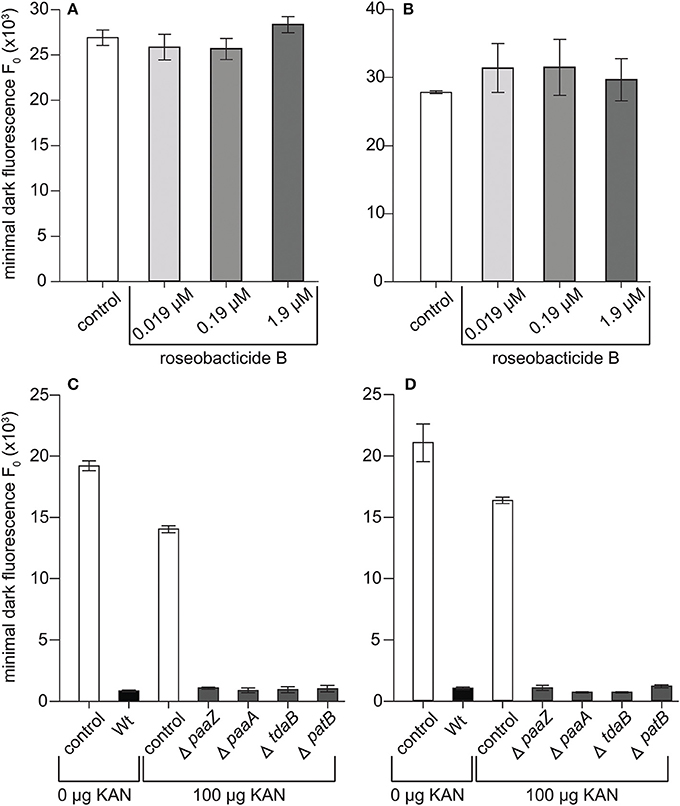
Figure 4. P. inhibens has a roseobacticide-independent mechanism for killing diploid calcifying C cells (CCMP3266) and haploid S cells (CCMP3268). Minimal algal fluorescence of the population (F0) is shown for (A) senescent CCMP3266 cells and (B) senescent CCMP3268 cells cultured with roseobacticide B. Both cell types were incubated for 24 h with methanol (white bar) and with 0.019 μM (lightest gray), 0.19 μM (gray), and 1.9 μM (darkest gray) roseobacticide B, dissolved in methanol. The final concentration of methanol solvent added to control and experimental cultures was 2% of the final volume. P. inhibens transposon mutants deficient in roseobacticide synthesis were co-cultured with (C) CCMP3266 and (D) CCMP3268. Minimal algal fluorescence of the population (F0) was measured on day 10. Triplicate wells that represent independent experiments were sacrificially sampled; error bars represent ±1 standard error.
Phaeobacter inhibens produces many different compounds with roles in microbe-microbe interactions. Segev et al. (2016) has proposed that P. inhibens uses IAA to kill E. huxleyi. This plant auxin is produced by various roseobacters (Fernandes et al., 2011; Wienhausen et al., 2017), including P. inhibens, and can kill the C-type culture, CCMP3266, at high concentrations (1,000 μM) (Labeeuw et al., 2016; Segev et al., 2016). However, Segev et al. (2016) also showed that P. inhibens did not produce IAA at high enough concentrations to kill E. huxleyi CCMP3266 (0.4–10 nM). In addition, E. huxleyi CCMP3266 produces IAA itself as a cell-cell signal at a higher concentration (200 μM) in the presence of tryptophan (Labeeuw et al., 2016). Finally, the N-type strain, CCMP2090, is susceptible to IAA when co-cultured at lower concentrations (10–100 μM), exhibiting morphological responses and reduced health similar to those seen in terrestrial plants (Labeeuw et al., 2016) that we did not observe in the P. inhibens–CCMP2090 co-culture. Taken together, these data show IAA is probably not the bioactive molecule causing E. huxleyi's decline when grown with P. inhibens, and that some other molecule or mechanism is responsible.
Tryptophan, the precursor to IAA, was also found to be lethal at the high concentration (1,000 μM) required to produce sufficient IAA to kill E. huxleyi (Labeeuw et al., 2016). Tryptophan was also shown to be released by CCMP3266, and the addition of exogenous tryptophan causes faster killing of CCMP3266 by both P. inhibens (Segev et al., 2016) and another roseobacter, Ruegeria sp. R11 (Labeeuw et al., 2016). This indicates that tryptophan may be a signal or metabolite for the bacteria to become virulent.
Phaeobacter inhibens interaction with E. huxleyi is dependent on the cell type of its algal host. This differential pathogenesis is also observed for EhVs, although the targeted cell types differ. Currently, N-type cells are heavily relied upon for cell biology studies of E. huxleyi because they are readily maintained axenically and avoid many of the problems associated with working with an autofluorescent mineral (i.e., coccoliths). However, our findings suggest that we should not assume that N-type cells will have similar biological interactions to their calcifying counterparts. The ability of P. inhibens to target coccolith-bearing (C) diploid cells and scale-bearing (S) haploid cells, while not killing non-calcifying diploid cells is a unique role, likely representing differentiation from EhVs, which kills both diploid cell types but not the haploid cell type (Frada et al., 2008). We have now shown two roseobacters, P. inhibens and Ruegeria sp. R11, can kill populations of C- and S-type cells (Mayers et al., 2016). This may have wide-reaching implications, as S cells are resistant to EhV lysis, which was postulated as a mechanism for the algal population to regenerate the dominant C cell population following viral-induced bloom collapse (Frada et al., 2008). Given the present findings, it is possible that the roseobacter's rapid pathogenesis of S cells could limit the alga's ability to reseed C cell populations. Additionally, roseobacter killing of C and S populations, while N populations evade bacterial induced death, could have important implications for the marine carbon cycle and formation of the paleobotanical record (Coolen, 2011), as well as the overall distribution of E. huxleyi blooms.
AB, LL, and RC conceived of the experiments. AB and LL carried out the experiments. FO carried out the whole-genome sequencing and analysis. ER and RM constructed the P. inhibens mutants. AB, LL, FO, and RC drafted the manuscript. All authors have read and approved the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Dr. Mohammad Seyedsayamdost (Princeton University) for providing the roseobacticide B and Albert Rosana (University of Alberta) for assistance with sequence confirmation of the barcoded transposon mutants. This work was supported by the Natural Sciences and Engineering Research Council of Canada (grant 402105) to RC. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under Contract No. DE-AC02-05CH11231.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00188/full#supplementary-material
Amin, S. A., Hmelo, L. R., van Tol, H. M., Durham, B. P., Carlson, L. T., Heal, K. R., et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101. doi: 10.1038/nature14488
Ashen, J. B., Cohen, J. D., and Goff, L. J. (1999). GC-SIM-MS detection and quantification of free indole-3-acetic acid in bacterial galls on the marine alga Prionitis lanceolata (Rhodophyta). J. Phycol. 35, 493–500. doi: 10.1046/j.1529-8817.1999.3530493.x
Azam, F. (1998). Microbial control of oceanic carbon flux: the plot thickens. Science 280, 694–696. doi: 10.1126/science.280.5364.694
Baines, S. B., and Pace, M. L. (1991). The production of dissolved organic matter by phytoplankton and its importance to bacteria: patterns across marine and freshwater systems. Limnol. Oceanogr. 36, 1078–1090. doi: 10.4319/lo.1991.36.6.1078
Baker, N. R. (2008). Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59, 89–113. doi: 10.1146/annurev.arplant.59.032607.092759
Baumann, K., Boeckel, B., and Cepek, M. (2008). Spatial distribution of living coccolithophores along an east- west transect in the subtropical South Atlantic. J. Nannoplankt. Res. 30, 9–21.
Bell, W., and Mitchell, R. (1972). Chemotactic and growth responses of marine bacteria to algal extracellular products. Mar. Biol. Lab. 143, 265–277. doi: 10.2307/1540052
Berger, M., Neumann, A., Schulz, S., Simon, M., and Brinkhoff, T. (2011). Tropodithietic acid production in Phaeobacter gallaeciensis is regulated by N-acyl homoserine lactone-mediated quorum sensing. J. Bacteriol. 193, 6576–6585. doi: 10.1128/JB.05818-11
Beyersmann, P. G., Tomasch, J., Son, K., Stocker, R., Göker, M., Wagner-Döbler, I., et al. (2017). Dual function of tropodithietic acid as antibiotic and signaling molecule in global gene regulation of the probiotic bacterium Phaeobacter inhibens. Sci. Rep. 7, 1–9. doi: 10.1038/s41598-017-00784-7
Bidle, K. D., Haramaty, L., Barcelose Ramos, J., and Falkowski, P. (2007). Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi. Proc. Natl. Acad. Sci. U.S.A. 104, 6049–6054. doi: 10.1073/pnas.0701240104
Bolch, C. J., Subramanian, T. A., and Green, D. H. (2011). The toxic dinoflagellate Gymnodinium catenatum (Dinophyceae) requires marine bacteria for growth. J. Phycol. 47, 1009–1022. doi: 10.1111/j.1529-8817.2011.01043.x
Borchard, C., and Engel, A. (2012). Organic matter exudation by Emiliania huxleyi under simulated future ocean conditions. Biogeosciences 9, 3405–3423. doi: 10.5194/bg-9-3405-2012
Bramucci, A. R., Labeeuw, L., Mayers, T. J., Saby, J. A., and Case, R. J. (2015). A small volume bioassay to assess bacterial/phytoplankton co-culture using WATER-Pulse-Amplitude-Modulated (WATER-PAM) fluorometry. J. Vis. Exp. 97:e52455. doi: 10.3791/52455
Buddruhs, N., Pradella, S., Göker, M., Päuker, O., Pukall, R., Spröer, C., et al. (2013). Molecular and phenotypic analyses reveal the non-identity of the Phaeobacter gallaeciensis type strain deposits CIP 105210T and DSM 17395. Int. J. Syst. Evol. Microbiol. 63, 4340–4349. doi: 10.1099/ijs.0.053900-0
Case, R. J., Longford, S. R., Campbell, A. H., Low, A., Tujula, N., Steinberg, P. D., et al. (2011). Temperature induced bacterial virulence and bleaching disease in a chemically defended marine macroalga. Environ. Microbiol. 13, 529–537. doi: 10.1111/j.1462-2920.2010.02356.x
Cemma, M., and Brumell, J. H. (2012). Interactions of pathogenic bacteria with autophagy systems. Curr. Biol. 22, R540–R545. doi: 10.1016/j.cub.2012.06.001
Cole, B. J., Feltcher, M. E., Waters, R. J., Wetmore, K. M., Mucyn, T. S., Ryan, E. M., et al. (2017). Genome-wide identification of bacterial plant colonization genes. PLoS Biol. 15:e2002860. doi: 10.1371/journal.pbio.2002860
Coolen, M. J. (2011). 7000 years of Emiliania huxleyi viruses in the Black Sea. Science 333, 451–452. doi: 10.1126/science.1200072
Curson, A. R., Todd, J. D., Sullivan, M. J., and Johnston, A. W. (2011). Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat. Rev. Microbiol. 9, 849–859. doi: 10.1038/nrmicro2653
D'Alvise, P. W., Lillebø, S., Prol-Garcia, M. J., Wergeland, H. I., Nielsen, K. F., Bergh, Ø., et al. (2012). Phaeobacter gallaeciensis reduces Vibrio anguillarum in cultures of microalgae and rotifers, and prevents vibriosis in cod larvae. PLoS ONE 7:e43996. doi: 10.1371/journal.pone.0043996
Fernandes, N., Case, R. J., Longford, S. R., Seyedsayamdost, M. R., Steinberg, P. D., Kjelleberg, S., et al. (2011). Genomes and virulence factors of novel bacterial pathogens causing bleaching disease in the marine red alga Delisea pulchra. PLoS ONE 6:e27387. doi: 10.1371/journal.pone.0027387
Frada, M. J., Bidle, K. D., Probert, I., and de Vargas, C. (2012). In situ survey of life cycle phases of the coccolithophore Emiliania huxleyi (Haptophyta). Environ. Microbiol. 14, 1558–1569. doi: 10.1111/j.1462-2920.2012.02745.x
Frada, M., Probert, I., Allen, M. J., Wilson, W. H., and de Vargas, C. (2008). The “Cheshire Cat” escape strategy of the coccolithophore Emiliania huxleyi in response to viral infection. Proc. Natl. Acad. Sci. U.S.A. 105, 15944–15949. doi: 10.1073/pnas.0807707105
Frank, O., Pradella, S., Rohde, M., Scheuner, C., Klenk, H.-P., Göker, M., et al. (2014). Complete genome sequence of the Phaeobacter gallaeciensis type strain CIP 105210T (= DSM 26640T = BS107T). Stand. Genomic Sci. 9, 914–932. doi: 10.4056/sigs.5179110
Franklin, D. J., Airs, R. L., Fernandes, M., Bell, T. G., Bongaerts, R. J., Berges, J. A., et al. (2012). Identification of senescence and death in Emiliania huxleyi and Thalassiosira pseudonana: cell staining, chlorophyll alterations, and dimethylsulfoniopropionate (DMSP) metabolism. Limnol. Oceanogr. 57, 305–317. doi: 10.4319/lo.2012.57.1.0305
Gardiner, M., Bournazos, A. M., Maturana-Martinez, C., Zhong, L., and Egan, S. (2017). Exoproteome analysis of the seaweed pathogen Nautella italica R11 reveals temperature-dependent regulation of RTX-like proteins. Front. Microbiol. 8:1203. doi: 10.3389/fmicb.2017.01203
Geng, H., and Belas, R. (2010). Molecular mechanisms underlying roseobacter-phytoplankton symbioses. Curr. Opin. Biotechnol. 21, 332–338. doi: 10.1016/j.copbio.2010.03.013
Geng, H., Bruhn, J. B., Nielsen, K. F., Gram, L., and Belas, R. (2008). Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl. Environ. Microbiol. 74, 1535–1545. doi: 10.1128/AEM.02339-07
González, J. M., Simó, R., Massana, R., Covert, J. S., Casamayor, E. O., Pedrós-Alió, C., et al. (2000). Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66, 4237–4246. doi: 10.1128/AEM.66.10.4237-4246.2000
Green, D. H., Echavarri-Bravo, V., Brennan, D., and Hart, M. C. (2015). Bacterial diversity associated with the coccolithophorid algae Emiliania huxleyi and Coccolithus pelagicus f. braarudii. Biomed. Res. Int. 2015:194540. doi: 10.1155/2015/194540
Guillard, R. R. L., and Hargraves, P. E. (1993). Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia 32, 234–236. doi: 10.2216/i0031-8884-32-3-234.1
Holligan, P. M., Fernández, E., Aiken, J., Balch, W. M., Boyd, P., Burkill, P. H., et al. (1993). A biogeochemical study of the coccolithophore, Emiliania huxleyi, in the North Atlantic. Global Biogeochem. Cycles 7, 879–900. doi: 10.1029/93GB01731
Hunt, D. E., David, L. A., Gevers, D., Preheim, S. P., Alm, E. J., and Polz, M. F. (2008). Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320, 1081–1085. doi: 10.1126/science.1157890
Kendrick, B. J., DiTullio, G. R., Cyronak, T. J., Fulton, J. M., Van Mooy, B. A. S., and Bidle, K. D. (2014). Temperature-induced viral resistance in Emiliania huxleyi (Prymnesiophyceae). PLoS ONE 9:e112134. doi: 10.1371/journal.pone.0112134
Klaveness, D. (1972). Coccolithus huxleyi (Lohm.) Kamptn. II. The flagellate cell, aberrant cell types, vegetative propagation and life cycles. Br. Phycol. J. 7, 309–318. doi: 10.1080/00071617200650321
Kolb, A., Strom, S., Center, S., and Anacortes, W. (2013). An inducible antipredatory defense in haploid cells of the marine microalga Emiliania huxleyi (Prymnesiophyceae). Limnol. Oceanogr. 58, 932–944. doi: 10.4319/lo.2013.58.3.0932
Kroemer, G., Galluzzi, L., Vandenabeele, P., Abrams, J., Alnemri, E. S., Baehrecke, E. H., et al. (2009). Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 16, 3–11. doi: 10.1038/cdd.2008.150
Labeeuw, L., Bramucci, A. R., and Case, R. J. (2017). “Bioactive small molecules mediate microalgal-bacterial interactions,” in Systems Biology of Marine Ecosystems, eds M. Kumar and P. J. Ralph (Cham: Springer International Publishing), 279–300.
Labeeuw, L., Khey, J., Bramucci, A. R., Atwal, H., De La Mata, A. P., Harynuk, J., et al. (2016). Indole-3-acetic acid is produced by Emiliania huxleyi coccolith-bearing cells and triggers a physiological response in bald cells. Front. Microbiol. 7:828. doi: 10.3389/fmicb.2016.00828
Labeeuw, L., Martone, P. T., Boucher, Y., and Case, R. J. (2015). Ancient origin of the biosynthesis of lignin precursors. Biol. Direct 10:23. doi: 10.1186/s13062-015-0052-y
Mayers, T. J., Bramucci, A. R., Yakimovich, K. M., and Case, R. J. (2016). A bacterial pathogen displaying temperature-enhanced virulence of the microalga Emiliania huxleyi. Front. Microbiol. 7:892. doi: 10.3389/fmicb.2016.00892
Miller, T. R., and Belas, R. (2004). Dimethylsulfoniopropionate metabolism by Pfiesteria-associated Roseobacter spp. Appl. Environ. Microbiol. 70, 3383–3391. doi: 10.1128/AEM.70.6.3383-3391.2004
Miller, T. R., Hnilicka, K., Dziedzic, A., Desplats, P., and Belas, R. (2004). Chemotaxis of Silicibacter sp. strain TM1040 toward dinoflagellate products. Appl. Environ. Microbiol. 70, 4692–4701. doi: 10.1128/AEM.70.8.4692-4701.2004
Mitchell, J. G., Okubo, A., and Fuhrman, J. A. (1985). Microzones surrounding phytoplankton form the basis for a stratified marine microbial ecosystem. Nature 316, 58–59. doi: 10.1038/316058a0
Mordecai, G., Verret, F., Highfield, A., and Schroeder, D. (2017). Schrödinger's Cheshire Cat: Are haploid Emiliania huxleyi cells resistant to viral infection or not? Viruses 9:51. doi: 10.3390/v9030051
Orata, F. D., Rosana, A. R., Xu, Y., Simkus, D. N., Bramucci, A. R., Boucher, Y., et al. (2016). Draft genome sequences of four bacterial strains isolated from a polymicrobial culture of naked (N-type) Emiliania huxleyi CCMP1516. Genome Announc. 4, e00674–e00616. doi: 10.1128/genomeA.00674-16
Planas, M., Perez-Lorenzo, M., Hjelm, M., Gram, L., Fiksdal, I. U., Bergh, O., et al. (2006). Probiotic effect in vivo of Roseobacter strain 27-4 against Vibrio (Listonella) anguillarum infections in turbot (Scophthalmus maximus L.) larvae. Aquaculture 255, 323–333. doi: 10.1016/j.aquaculture.2005.11.039
Price, M. N., Wetmore, K. M., Deutschbauer, A. M., and Arkin, A. P. (2016). A comparison of the costs and benefits of bacterial gene expression. PLoS ONE 11:e0164314. doi: 10.1371/journal.pone.0164314
Prol García, M. J., D'Alvise, P. W., and Gram, L. (2013). Disruption of cell-to-cell signaling does not abolish the antagonism of Phaeobacter gallaeciensis toward the fish pathogen Vibrio anguillarum in algal systems. Appl. Environ. Microbiol. 79, 5414–5417. doi: 10.1128/AEM.01436-13
Rao, D., Webb, J. S., Holmström, C., Case, R. J., Low, A., Steinberg, P., et al. (2007). Low densities of epiphytic bacteria from the marine alga Ulva australis inhibit settlement of fouling organisms. Appl. Environ. Microbiol. 73, 7844–7852. doi: 10.1128/AEM.01543-07
Rhodes, L. L., Peake, B. M., MacKenzie, A. L., Simon, M., and Edwards, A. R. (1995). Coccolithophores Gephyrocapsa oceanica and Emiliania huxleyi (Prymnesiophyceae = Haptophyceae) in New Zealand's coastal waters: characteristics of blooms and growth in laboratory culture. New Zeal. J. Mar. Freshw. Res. 29, 345–357.
Rodrigo-Torres, L., Pujalte, M. J., and Arahal, D. R. (2016). Draft genomes of Nautella italica strains CECT 7645T and CECT 7321: two roseobacters with potential pathogenic and biotechnological traits. Mar. Genomics 26, 73–80. doi: 10.1016/j.margen.2016.01.001
Sapp, M., Schwaderer, A. S., Wiltshire, K. H., Hoppe, H. G., Gerdts, G., and Wichels, A. (2007). Species-specific bacterial communities in the phycosphere of microalgae? Microb. Ecol. 53, 683–699. doi: 10.1007/s00248-006-9162-5
Schatz, D., Shemi, A., Rosenwasser, S., Sabanay, H., Wolf, S. G., Ben-Dor, S., et al. (2014). Hijacking of an autophagy-like process is critical for the life cycle of a DNA virus infecting oceanic algal blooms. New Phytol. 204, 854–863. doi: 10.1111/nph.13008
Schreiber, U., Schliwa, U., and Bilger, W. (1986). Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosyn. Res. 10, 51–62. doi: 10.1007/BF00024185
Segev, E., Wyche, T. P., Kim, K. H., Petersen, J., Ellebrandt, C., Vlamakis, H., et al. (2016). Dynamic metabolic exchange governs a marine algal-bacterial interaction. Elife 5:e17473. doi: 10.7554/eLife.17473
Seyedsayamdost, M. R., Carr, G., Kolter, R., and Clardy, J. (2011a). Roseobacticides: small molecule modulators of an algal-bacterial symbiosis. J. Am. Chem. Soc. 133, 18343–18349. doi: 10.1021/ja207172s
Seyedsayamdost, M. R., Case, R. J., Kolter, R., and Clardy, J. (2011b). The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3, 331–335. doi: 10.1038/nchem.1002
Seyedsayamdost, M. R., Wang, R., Kolter, R., and Clardy, J. (2014). Hybrid biosynthesis of roseobacticides from algal and bacterial precursor molecules. J. Am. Chem. Soc. 136, 15150–15153. doi: 10.1021/ja508782y
Seymour, J. R., Simo, R., Ahmed, T., Stocker, R., Simó, R., Ahmed, T., et al. (2010). Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 329, 342–345. doi: 10.1126/science.1188418
Stocker, R. (2012). Marine microbes see a sea of gradients. Science 338, 628–633. doi: 10.1126/science.1208929
Sule, P., and Belas, R. (2013). A novel inducer of Roseobacter motility is also a disruptor of algal symbiosis. J. Bacteriol. 195, 637–646. doi: 10.1128/JB.01777-12
Thole, S., Kalhoefer, D., Voget, S., Berger, M., Engelhardt, T., Liesegang, H., et al. (2012). Phaeobacter gallaeciensis genomes from globally opposite locations reveal high similarity of adaptation to surface life. ISME J. 6, 2229–2244. doi: 10.1038/ismej.2012.62
Tripp, H. J., Kitner, J. B., Schwalbach, M. S., Dacey, J. W. H., Wilhelm, L. J., and Giovannoni, S. J. (2008). SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452, 741–744. doi: 10.1038/nature06776
Tyrrell, T., and Merico, A. (2004). “Emiliania huxleyi: bloom observation and the conditions that induce them,” in Coccolithophores: From Molecular Processes to Global Impact, ed H. R. Thierstein and J. R. Young (Berlin; Heidelberg: Springer), 585–604.
Vandecandelaere, I., Nercessian, O., Segaert, E., Achouak, W., Mollica, A., Faimali, M., et al. (2009). Nautella italica gen. nov., sp. nov., isolated from a marine electroactive biofilm. Int. J. Syst. Evol. Microbiol. 59, 811–817. doi: 10.1099/ijs.0.002683-0
van der Wal, P., de Jong, E. W., Westbroek, P., de Bruijn, W. C., and Mulder-Stapel, A. A. (1983). Ultrastructural polysaccharide localization in calcifying and naked cells of the coccolithophorid Emiliania huxleyi. Protoplasma 118, 157–168. doi: 10.1007/BF01293073
Vardi, A., Van Mooy, B. A. S., Fredricks, H. F., Popendorf, K. J., Ossolinski, J. E., Haramaty, L., et al. (2009). Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Science 326, 861–865. doi: 10.1126/science.1177322
von Dassow, P., John, U., Ogata, H., Probert, I., Bendif, E. M., Kegel, J. U., et al. (2015). Life-cycle modification in open oceans accounts for genome variability in a cosmopolitan phytoplankton. ISME J. 9, 1–13. doi: 10.1038/ismej.2014.221
Wang, H., Tomasch, J., Jarek, M., and Wagner-Döbler, I. (2014). A dual-species co-cultivation system to study the interactions between Roseobacters and dinoflagellates. Front. Microbiol. 5:311. doi: 10.3389/fmicb.2014.00311
Wang, R., Gallant, É., and Seyedsayamdost, R. (2016). Investigation of the genetics and biochemistry of roseobacticide production in the Roseobacter clade bacterium Phaeobacter inhibens. MBio 7:e02118–e02115. doi: 10.1128/mBio.02118-15
Wang, R., and Seyedsayamdost, M. R. (2017). Roseochelin B, an algaecidal natural product synthesized by the roseobacter Phaeobacter inhibens in response to algal sinapic acid. Org. Lett. 19, 5138–5141. doi: 10.1021/acs.orglett.7b02424
Wetmore, K. M., Price, M. N., Waters, R. J., Lamson, J. S., He, J., Hoover, C. A., et al. (2015). Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. MBio 6, e00306–e00315. doi: 10.1128/mBio.00306-15
Wienhausen, G., Noriega-Ortega, B. E., Niggemann, J., Dittmar, T., and Simon, M. (2017). The exometabolome of two model strains of the Roseobacter group: a marketplace of microbial metabolites. Front. Microbiol. 8:1985. doi: 10.3389/fmicb.2017.01985
Wilson, W. H., Tarran, G., and Zubkov, M., V (2002). Virus dynamics in a coccolithophore-dominated bloom in the North Sea. Deep Sea Res. II Top. Stud. Oceanogr. 49, 2951–2963. doi: 10.1016/S0967-0645(02)00065-6
Wolfe, G. V., Sherr, E. B., and Sherr, B. F. (1994). Release and consumption of DMSP from Emiliania huxleyi during grazing by Oxyrrhis marina. Mar. Ecol. Prog. Ser. 111, 111–119. doi: 10.3354/meps111111
Wolfe, G. V., Steinke, M., and Kirst, G. O. (1997). Grazing-activated chemical defense in a unicellular marine alga. Nature 387, 894–897. doi: 10.1038/43168
Keywords: coccolithophore, roseobacter, phytoplankton pathogen, marine pathogens, pathogen ecology, cell type, phytoplankton life history, bacterial–algal interactions
Citation: Bramucci AR, Labeeuw L, Orata FD, Ryan EM, Malmstrom RR and Case RJ (2018) The Bacterial Symbiont Phaeobacter inhibens Shapes the Life History of Its Algal Host Emiliania huxleyi. Front. Mar. Sci. 5:188. doi: 10.3389/fmars.2018.00188
Received: 17 February 2018; Accepted: 09 May 2018;
Published: 29 May 2018.
Edited by:
Matthias Wietz, University of Oldenburg, GermanyReviewed by:
Rurun Wang, Merck, United StatesCopyright © 2018 Bramucci, Labeeuw, Orata, Ryan, Malmstrom and Case. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rebecca J. Case, cmNhc2VAdWFsYmVydGEuY2E=
†These authors have contributed equally to this work.
‡Present Address: Leen Labeeuw, Plant Functional Biology and Climate Change Cluster (C3), University of Technology Sydney, Ultimo, NSW, Australia
Fabini D. Orata, Department of Chemical and Materials Engineering, University of Alberta, Edmonton, AB, Canada
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.