
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mar. Sci., 11 April 2018
Sec. Marine Biogeochemistry
Volume 5 - 2018 | https://doi.org/10.3389/fmars.2018.00125
This article is part of the Research TopicBiogeochemistry and Genomics of Silicification and SilicifiersView all 13 articles
The silicified cell walls of diatoms have inspired the interest of researchers for several centuries, and our understanding of their properties and formation has developed in synch with the development of observational and analytical techniques. Over the past 20 years, approaches used to characterize the molecular components involved in cell wall silicification have evolved, and this has provided significant insights into fundamental aspects of silicification, and promises to continue to do so. Diatom cell wall formation is highly dynamic but, apart from microscopic investigations, most previous molecular characterizations have been on completely formed structures, and thus only provide information on a static end point in the process. However, recent studies that monitor when components are made, and how they are transported to the silica deposition vesicle (SDV), indicate that investigation into the true dynamics of the process is possible. Real-time imaging and genetic manipulation offer the promise of elucidating the spatio-temporal dynamics of, and interactions between, components, which will be essential to understand how structure formation is controlled and coordinated. This review is aimed at describing the approaches that have been used to characterize diatom silicification, integrating newer concepts based on results from diverse approaches, and raising questions that still need to be addressed, leveraging the diverse tools and techniques we now have.
The ability of diatoms to make silica-based cell walls has been the subject of fascination for centuries. It started with a microscopic observation by an anonymous English country nobleman in 1703, who observed an object that looked like a chain of regular parallelograms and debated whether it was just crystals of salt, or a plant (Anonymous, 1702). The viewer decided that it was a plant because the parallelograms didn't separate upon agitation, nor did they vary in appearance when dried or subjected to warm water (in an attempt to dissolve the “salt”). Unknowingly, the viewer's confusion captured the essence of diatoms—mineral utilizing plants. It is not clear when it was determined that diatom cell walls are made of silica, but in 1939 a seminal reference characterized the material as silicic acid in a “subcolloidal” state (Rogall, 1939). Identification of the main chemical component of the cell wall spurred investigations into how it was made. These investigations have involved, and been propelled by, diverse approaches including, microscopy, chemistry, biochemistry, material characterization, molecular biology, ‘omics, and transgenic approaches. The results from this work have given us a better understanding of cell wall formation processes, establishing fundamental knowledge which can be used to create models that contextualize current findings and clarify how the process works.
The process of building a mineral-based cell wall inside the cell, then exporting it outside, is a massive event that must involve large numbers of genes and their protein products. The act of building and exocytosing this large structural object in a short time period, synched with cell cycle progression, necessitates substantial physical movements within the cell as well as dedication of a significant proportion of the cell's biosynthetic capacities.
It is nearly two decades since the first characterizations of the biochemical processes and components involved in diatom silicification (Hildebrand et al., 1997; Kröger et al., 1999, 2000, 2002). More recent work has provided insights into how higher order assembly of silica structures might occur (Tesson and Hildebrand, 2010, 2013; Scheffel et al., 2011). Very recent reports describe the identification of novel components involved in higher order processes, the dynamics documented through real-time imaging, and the genetic manipulation of silica structure (Kotzsch et al., 2017; Tesson et al., 2017). The approaches established in these recent works provide practical avenues to not only identify the components involved in silica cell wall formation but to elucidate their interactions and spatio-temporal dynamics. This type of holistic understanding will be necessary to achieve a more complete understanding of cell wall synthesis.
The purpose of this review is to summarize previous work in detail and provide a framework for future work which can take advantage of recent discoveries and tools to explore the dynamic nature of diatom cell wall formation.
Silicon dissolved in an aqueous solution at neutral pH is primarily in the form of silicic acid, Si(OH)4 (Iler, 1979). The solubility of a silicic acid solution is limited to around 2 mM, above that concentration silica (SiO2) begins to polymerize into polymers with a range of lengths, forming an amorphous solid (Iler, 1979; Kley et al., 2014). The surface of forming silica has a net negative charge, and the morphology of the silica that forms varies depending on the solution's ionic strength and pH. In solutions near neutral pH and with low ionic strength, silica polymerizes as a sol consisting of individual particles which are formed due to repulsion between negatively charged particle surfaces. At lower pH or with higher ionic strength, the negative charges are neutralized, leading to aggregation of smaller particles, and in some conditions formation of a gel network. A gel network is desirable for creating a physically robust structure, and is the morphology generally observed in diatom silica, although silica morphology can vary depending on cell wall features (Round et al., 1990; Hildebrand et al., 2006).
Although not the primary focus of this review, the processes of silicic acid transport and stabilization at high concentrations are important aspects of diatom silicification. As a small, uncharged molecule, silicic acid can freely diffuse across membranes. Kinetic data indicate that, under environmentally relevant concentrations, diffusion can be the major mode of uptake (Thamatrakoln and Hildebrand, 2008). Under relatively low silicic acid concentrations, the silicon transporters (SITs) actively facilitate uptake (Hildebrand et al., 1997; Thamatrakoln and Hildebrand, 2008). SITs have been localized to the plasma membrane but it is not clear what intracellular membranes they may be targeted to (Shrestha and Hildebrand, 2015). Silicic acid transport mechanisms within the cell are unknown. Given the diffusibility of silicic acid, a transporter protein may not be required. Several studies (reviewed in Martin-Jezequel et al., 2000) have documented intracellular concentrations of soluble silicic acid substantially greater than its 2 mM saturation limit. How such high concentrations are maintained is still a mystery, but a likely hypothesis is that undescribed organic compounds associate with intracellular silicic acid, preventing polymerization. This hypothesis explains features of silicic acid transport in terms of transport against a potential concentration gradient (the gradient may not actually exist if the chemical form of silicic acid—bound or unbound—differs on two sides of a membrane) as well as high intracellular silicic acid concentrations (Martin-Jezequel et al., 2000).
Thalassiosira pseudonana has developed as the most intensively studied diatom species in regard to silicification, a schematic of its major structural features is shown in Figure 1. Several microscopic approaches, including scanning electron microscopy (SEM), transmission electron microscopy (TEM), and atomic force microscopy (AFM), have sufficiently high resolution to evaluate details of silica polymerization and structure formation in diatoms. The ability to enrich for cells undergoing cell wall formation allows for larger numbers of relevant images to be captured, providing more information on ephemeral phenomena such as intermediate structural stages. A synchronized culturing method was developed for T. pseudonana which enriches for cells making valves (Hildebrand et al., 2007). In this procedure, cells are starved for silicic acid for 24 h, during which time the majority (80%) of the cells arrest in the G1 phase of the cell cycle. Upon silicic acid replenishment, the cell cycle resumes, and cells complete G1, during which time girdle bands are synthesized. Cells then move into S phase succeeded by G2+M where daughter cells are formed within the mother cell. G2+M is followed by valve formation, and ultimately cell separation. The timing of cell cycle events is well-conserved except for the length of G1, which can last 1–4 h after silicic acid replenishment (Hildebrand et al., 2007). Based on findings in yeast, the variability in G1 length could relate to whether enough carbon is available for the cell to complete mitosis (Hall et al., 1998). Despite the variability in the timing of G1, the period of valve synthesis can easily be determined by staining cells with the fluorescent dyes rhodamine 123 or PDMPO (Li et al., 1989; Shimizu et al., 2001; Hildebrand et al., 2007), both of which incorporate into forming silica. Forming valves are visualized by a sharp demarcation of fluorescence at the cleavage area of the cell, and isolation of cells during this time enriches for those making valves. Culture synchronization is not perfect—in addition to less than 100% of all cells arresting in G1, subpopulations of cells can progress through the cell cycle at different rates. However, based on ongoing transcriptomic analysis (Hildebrand et al., in prep), the timing of distinct cell cycle related processes can be defined within 1 h windows.
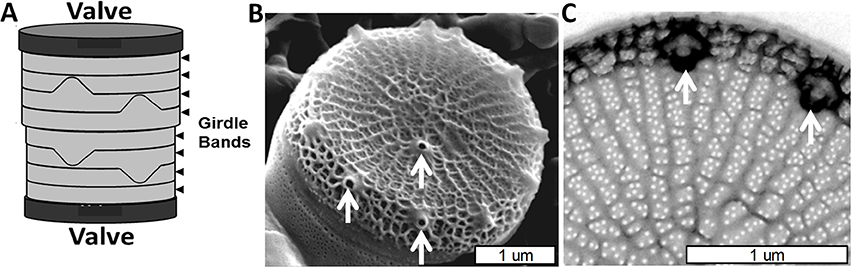
Figure 1. Cell wall structure for T. pseudonana. (A) Schematic of the entire frustule, indicating valves and girdle bands. (B) Scanning electron micrograph (SEM) of the valve and associated first girdle band. (C) Transmission electron micrograph (TEM) of the valve, highlighting the rib and pore structure. White arrows denote a subset of the fultoportulae. From Hildebrand et al. (2006), reproduced with permission.
The valves in T. pseudonana are circular, were measured (Hildebrand et al., 2006) at an average of 3.8 μm (±0.4) diameter, and consist of ribs radiating from an area called the pattern center (where valve formation is initiated) to the rim of the valve (Figure 1B). The valve surfaces are interspersed with numerous 18 nm (±3.1) diameter circular pores positioned between the ribs which cover ~4% of the valve area (Hildebrand et al., 2006). The ribs are not strictly linear, but are consistently spaced (145 nm ± 13 between ribs) and branched to maintain this spacing when the distance between them increases beyond 162 nm (±16). There are also cross connections between the ribs (Figure 1C). The other major feature of the valve is the fultoportulae, which are larger cone shaped openings for secretion of chitin fibrils (Herth, 1979a,b; Round et al., 1990). Fultoportulae are always found on the rim of the valve, and generally a single fultoportula (but occasionally two or none) is found offset from the center of the valve (Figures 1B,C).
As in other diatom species, valve formation in T. pseudonana occurs in distinct steps. The first feature that resembles a valve component is called the base layer, which will form the proximal surface of the completed valve. The base layer is a footprint of the valve which includes the ribs and precursors to the fultoportulae, defining structure in the x and y plane (Figures 2A,B). The structure is very flexible at this stage (Figure 2C and Hildebrand et al., 2007). After base layer formation, the rim of the valve is built up and additional silicification occurs across the surface, eventually resulting in a robust rigid structure (Figure 2D). This process involves thickening the valve via expansion in the z-axis direction toward what will become the distal valve surface (Figure 2E).
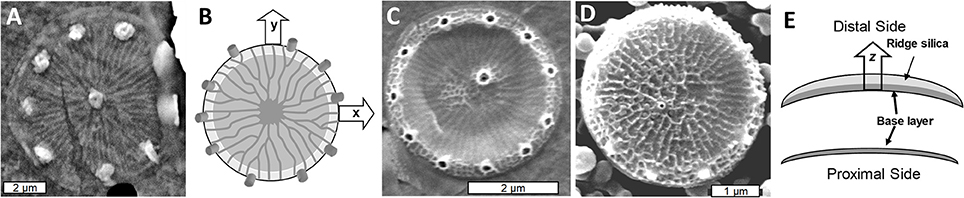
Figure 2. Formation of the T. pseudonana valve. (A) SEM of the base layer. (B) Schematic of x/y axis expansion during base layer formation. (C) Intermediate stage of valve formation showing buildup of the rim, but lack of extensive silicification in the center. (D) Fully formed valve. (E) Cross section schematic of silica deposition in the distal z-axis direction with the lower image representing the initial base layer and the upper representing the fully silicified valve with the final rigid structure. From Hildebrand et al. (2006), reproduced with permission.
The nanoscale morphology of the silica varies between different features and during different stages of the process. The ribs consist of an amalgam of nanoparticulate silica, whereas the area between the ribs is filled with silica characterized by a smooth branched morphology (Figures 3A–C). The nanopores on the surface appear to be formed by objects that obstruct silica polymerization, as they are initially irregular in shape, but become circular as the silica fills in around the proposed obstruction (Figures 3B,C). Such an interpretation is supported by AFM analysis of silica-associated organic material in other diatom species where material associated with pores projects above the base layer (Tesson and Hildebrand, 2013). In T. pseudonana, the distal surface ornamentation is composed of a third silica morphology which is deposited on top of the base layer ribs (Figures 3D,E).
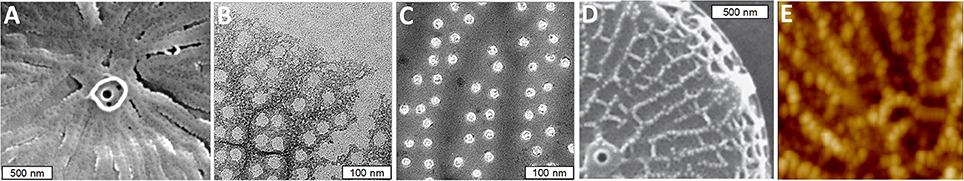
Figure 3. Silica morphology during valve formation in T. pseudonana. (A) SEM of initially formed ribs and a precursor to the central fultoportula. (B) Network structure of initially deposited silica in the rib and pore area. (C) Mature pores, note circularity of the pores compared with (B). (D) SEM of nanoparticles of silica on top of ribs on the distal valve surface. (E) Atomic force micrograph (AFM) of distal surface nanoparticles. From Hildebrand et al. (2006), reproduced with permission.
In summary, there are two distinct major stages in T. pseudonana valve formation, 1) formation of the base layer which defines the valve shape in the x/y plane and major features of ribs, nanopores, and portulae, and 2) expansion in the z-axis direction by additional polymerization of silica to form a rigid structure. The silica morphology of the valve varies depending on the structural feature being made.
Silica structures in diatoms are formed within a membrane-bound compartment called the silica deposition vesicle (SDV) (Drum and Pankratz, 1964; Reimann et al., 1966; Schmid et al., 1981). The membrane surrounding the SDV is called the silicalemma. The SDV has not been isolated, which has complicated characterization of its components. Fortunately, both biochemical (Kröger et al., 1999, 2000, 2002; Wenzl et al., 2008; Scheffel et al., 2011; Kotzsch et al., 2017) and transcript response (Shrestha et al., 2012; Tesson et al., 2017) approaches have proven useful for the identification of components of the SDV.
To be described in more detail below, there are three general classes of proteins known to be associated with the SDV based on biochemical approaches used to purify them and sequence characteristics. A diagram depicting our current understanding of the relative arrangement of components associated with the SDV and the outcomes of various purification procedures is shown in Figure 4. Isolation and detergent cleaning of cell wall silica, followed by dissolution with ammonium fluoride (Figure 4D), isolated soluble silica-associated proteins and long chain polyamines (LCPAs) embedded within the silica (Kröger et al., 1999, 2000, 2002; Poulsen and Kröger, 2004; Wenzl et al., 2008). Isolation of insoluble material resulting from the same cleaning and dissolution procedure (Figure 4D) resulted in characterization of ammonium fluoride insoluble material (AFIM), or the insoluble organic matrix (Scheffel et al., 2011). Most recently, proteins associated with the silicalemma which are extracted by detergent treatment (Figure 4B) have been characterized (Kotzsch et al., 2017; Tesson et al., 2017). To date these are characterized by a single transmembrane domain that spans the silicalemma and have an intralumenal and cytoplasmic portion. Depending on their other associations, such as with the AFIM, such proteins may or may not be extracted solely by detergent (Figure 4D). Harsh acid treatment of diatom cell walls strips away all exposed organic material (Tesson and Hildebrand, 2013), but should leave silica-embedded material intact depending on its degree of exposure (Figure 4C).
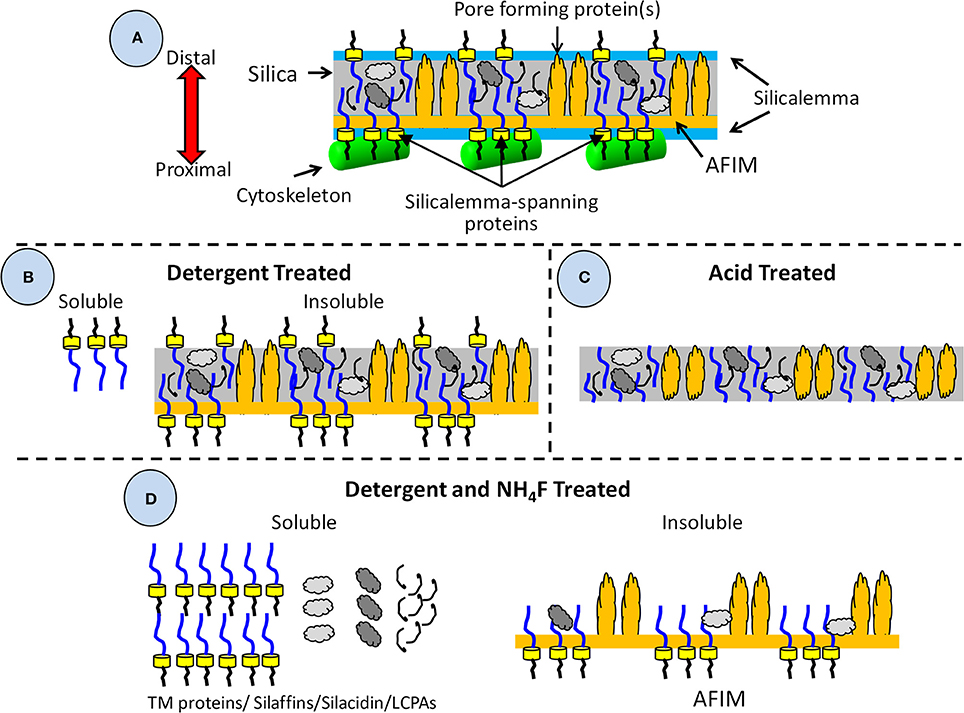
Figure 4. Diagram of valve and associated components detailing their solubility in different detergents and isolation treatments. (A) All components associated with the valve during its formation including those embedded in the silica, or associated with the AFIM or silicalemma, including the cytoskeleton. (B) Components resulting from detergent extraction, which removes the cytoskeleton, silicalemma, and proteins associated with the silicalemma that are not embedded in the silica, leaving a soluble and insoluble fraction. (C) Components resulting from acid treatment, which removes all organic material external to the silica. It is assumed that embedded material is at least partially protected from acid degradation. (D) Components resulting from detergent extraction and dissolution of the silica by ammonium fluoride, resulting in a soluble fraction of silica-embedded and silicalemma components, and an insoluble fraction of the AFIM and any proteins that had become associated via crosslinking to it.
In addition to protein-mediated structure formation, the SDV can also be passively molded (Mann, 1984); diatom silica structures have been observed molded around mitochondria (Pickett-Heaps et al., 1979), compression between daughter cells influences silica structure (van de Meene and Pickett-Heaps, 2002), and microtubules can leave indentations in valve structures (Tesson and Hildebrand, 2010). These observations indicate that the plasticity of the silicalemma can influence shape.
Long chain polyamines (LCPAs) are critical components of the silicification process, and appear to be abundant organic components of diatom silica (Kröger et al., 2000). The LCPAs are found embedded within the silica, and consist of repeating units of N-methyl-propylamine or propylamine attached to ornithine or putrescine which are methylated to different degrees. The number of repeat units varies between species. Incubation of purified LCPAs in a solution of silicic acid rapidly catalyzes the polymerization of silica (Kröger et al., 2000). Altering the ratios of different size classes of LCPAs impacts silica morphology, generally by changing nanoparticle dimensions.
The cytoskeleton plays a significant role in forming diatom silica structures, as evidenced by inhibitor studies and imaging (Chiappino and Volcani, 1977; Schmid, 1980; Blank and Sullivan, 1983; Pickett-Heaps et al., 1990; Pickett-Heaps, 1998a,b; van de Meene and Pickett-Heaps, 2002; Tesson and Hildebrand, 2010). Spatial constraints and imaging support that the cytoskeleton is associated primarily, if not solely, with the proximal surface of the SDV, although protrusions extending through silica structures toward the distal side have been observed (Tesson and Hildebrand, 2010). Microtubules appear to be involved in providing a rigid framework for expansion of the SDV and positioning of components within valves. Additionally, microtubules define a major structure specific to pennate diatom species called the raphe (Pickett-Heaps et al., 1979, 1990). Actin defines the edges of active silica polymerization and both the overall and detailed structure of the silica. In centric diatoms, a ring of actin has been observed that defines the edge of the SDV, this ring expands as the SDV grows (van de Meene and Pickett-Heaps, 2002; Tesson and Hildebrand, 2010). In addition, there is excellent correspondence between the pattern of actin assembly over the valve and silica structure (Tesson and Hildebrand, 2010). These observations suggest that microtubules and actin are responsible for positioning components of the SDV that lead to meso- and micro-scale structural features. Although they are not a direct component of the SDV, they must interact with proteins of the SDV in order to translate their organizational patterns into a corresponding silica structure. A proposed mechanism for this will be discussed below.
There is strong evidence for the involvement of carbohydrate polymers in silicification and structure formation, but their precise roles and contributions are not clear. For that reason, carbohydrate polymers are not included in the diagrams of Figure 4. An additional complication is that different types of carbohydrate polymers are associated with cell wall components in different ways in different diatom species (Tesson and Hildebrand, 2013).
Mannose-6-phosphate was identified to be tightly associated with silica from Stephanopyxis turris, and because its amount increased upon silica dissolution, it was suggested to at least be partially embedded within the silica (Hedrich et al., 2013).
Evidence also indicates an association of chitin with diatom silica structures. Tesson and coworkers (Tesson et al., 2008) performed a solid state NMR analysis of SDS and H2O2 cleaned silica from T. pseudonana which showed the presence of several peaks corresponding to β (1 → 4) N-acetyl glucosamine of chitin. The authors attributed the peaks to contaminating chitin fibrils secreted from the cell, however, the signals were enriched as the material was further purified, which should not be the case with a contaminant. The data suggest an association between chitin and the silica. H2O2 treatment in aqueous solution does not necessarily degrade organic material, so it cannot be concluded whether the chitin was associated with the surface of the silica, or embedded within it.
Durkin et al. (2009) characterized chitin synthase genes from different diatom species, and identified them in species which do not secret chitin fibrils, suggesting another role for chitin. In T. pseudonana they detected chitin using FITC-conjugated wheat germ agglutinin and a chitin binding protein associated with the girdle bands. The staining indicated that chitin was accessible from the outer surface of the girdle bands. No chitin staining was observed in the valves. Previously, a class of highly abundant girdle band associated proteins with chitin binding domains were characterized in T. pseudonana (Davis et al., 2005), consistent with the observation of chitin associated with the girdle bands.
Brunner et al. (2009) used different centrifugation methods to distinguish between secreted chitin fibrils and chitin that might be otherwise associated with the cell. Cell walls were extracted with SDS and EDTA, with no other treatment. Material isolated by flow centrifugation had no visible chitin fibrils associated with it, yet NMR signals characteristic of chitin were seen. NaOH treatment removed NMR signals resulting from amino acid side chains, and enriched the chitin signals, indicating that chitin constituted 25–40% of the organic material associated with the silica. The authors showed images of organic material resulting from dissolution of the silica with HF, which mimicked the overall shape and size of the valves. At the time, the AFIM material had not yet been characterized, and given that the cleaning treatment did not include acid exposure (Figure 4C), the material imaged by Brunner et al., could have been the AFIM associated with T. pseudonana valves (Kotzsch et al., 2016). Considering that Durkin et al. (2009) only observed chitin associated with the girdle bands, it is not clear whether chitin is associated with the valves (the NMR signal could have been from girdle bands), and if so, whether it is a component of the organic material imaged by Brunner et al.
Tesson and Hildebrand characterized insoluble organic material associated with valves and girdle bands from a variety of diatom species, and included staining and carbohydrate quantitation approaches to characterize associated carbohydrate polymers (Tesson and Hildebrand, 2013). Two types of polymer were identified. One was a DAPI-stainable (polyanionic) material that was mannan-rich and found in all species. Staining only occurred after SDS treatment, suggesting that the material was masked in intact cells. The other was calcofluor-stainable and enriched in glucose, and thus was likely a β-1,3 glucan called callose previously identified in diatom cell walls (Waterkeyn and Bienfait, 1987). Although the DAPI-stained material was always immediately adjacent to the silica, the location and accessibility of the DAPI- and calcofluor-stained materials relative to cell wall components varied depending on the species, suggesting distinct roles (Tesson and Hildebrand, 2013). In two species that had calcofluor-stained material associated with forming valves, treatment with mycafungin (a specific beta 1–3 glucan synthesis inhibitor) dramatically altered silica structure morphology, indicating a strong influence on the structure formation process, but not on silica polymerization.
In summary, at least some form of carbohydrate polymer is involved in controlling the morphology of forming silica, but more work is needed to clarify the specific role(s) of carbohydrate polymers in diatom cell wall synthesis.
The first proteins isolated from purified dissolved silica were the silaffins (Kröger et al., 1999). These were initially isolated from Cylindrotheca fusiformis, and consisted of peptides 15–22 amino acids in length with repeated sequence enriched in lysine and serine. The lysines were post translationally modified by methylation or addition of polyamine units, and the serines were phosphorylated (Kröger et al., 1999, 2002). Purified silaffins rapidly catalyzed polymerization of silica from a solution of silicic acid, and the morphology of the silica varied depending on the mixture of silaffins used (Kröger et al., 1999, 2002). Analysis of the gene encoding silaffin indicated a single polypeptide that was proteolytically processed into the shorter peptides, with proteolytic cleavage occurring after an RXL amino acid sequence (Kröger et al., 1999). A survey of silaffins in different diatom species revealed that most were larger polypeptides—in contrast to the smaller peptides in C. fusiformis (Kröger et al., 2000). A detailed characterization of silaffins in T. pseudonana (Poulsen and Kröger, 2004) provided key insights into properties of the larger silaffins. Three proteins were identified, TpSil 1/2 H, TpSil 1/2 L, and TpSil 3. TpSil 3 was a single polypeptide, and TpSil 1 and 2 were very similar proteins encoded by two different genes. TpSil 1/2 were synthesized as single proteins that were proteolytically cleaved into the H and L forms. The two forms had distinct influences over silica formation. In contrast to the C. fusiformis silaffins, none of the T. pseudonana silaffins had silica forming activity in vitro in the absence of LCPAs. In the presence of LCPAs, the H form of TpSil 1/2 initially promoted silica formation, then inhibited it as its concentration was increased, whereas the L form continued to promote silica formation as its concentration was increased. TpSil 3 also promoted silica formation at lower concentrations and inhibited it at higher. These results led to the distinction between “catalytic” silaffins that had a continuous stimulatory effect on silica formation, and “regulatory” silaffins that could both promote and inhibit silica formation depending on the protein concentration. The activities were attributed to differences in amino acid composition, and more specifically, posttranslational modification to amino acids. TpSil 1/2 H was enriched in hydroxyamino acids, which were likely glycosylated as evidenced by substantially higher carbohydrate content in TpSil 1/2 H relative to TpSil 1/2 L (Poulsen and Kröger, 2004). It had been demonstrated with a particular silaffin from C. fusiformis, natSil2, that carbohydrate and sulfate moieties inhibited polyamine-dependent silica formation, whereas protein-bound phosphate groups promoted it (Poulsen et al., 2003). TpSil 1/2 L lacked those particular inhibiting posttranslationally-added groups, and solely promoted silica formation due to the predominance of posttranslationally-added phosphate groups. TpSil 3 contained both types of side chain modifications, and displayed an intermediate ability to promote or repress silica formation.
Characterization of the various silaffins demonstrated that posttranslational modifications have a major influence on silica formation activity. Thus, although amino acid sequence comparisons are a valid way to assess the ability of proteins to control silica formation, it should be kept in mind that it is modification of the amino acids that are largely responsible.
Silica polymerization in vitro by LCPAs required the presence of polyanions in the form of phosphate or pyrophosphate (Kröger et al., 2000), which prompted a search for polyanionic organic material embedded within diatom silica that could serve a similar function. This resulted in the discovery of silacidin, a protein enriched in serine and the acidic amino acids aspartic and glutamic acid (Wenzl et al., 2008). In the presence of LCPAs and in the absence of phosphate, silacidin rapidly promoted silica formation in vitro, and was thus regarded as an excellent candidate for performing the anion role in vivo. Silacidin was encoded by a single open reading frame, but the protein was proteolytically cleaved after RXL sequences into smaller repeated peptides of 29–31 amino acids.
The described characterizations provided fundamental insights into the role of soluble silica-associated material. In vitro silica precipitation experiments have demonstrated a relationship between the size of the catalytic organic material and how ordered the resulting silica morphology was. The short silaffin peptides isolated from C. fusiformis silaffins precipitate silica spheres, with no higher order organization (Kröger et al., 1999). Similarly, LCPAs (Kröger et al., 2000), fully processed silacidins (Wenzl et al., 2008) and TpSil 1/2 L from T. pseudonana (Poulsen and Kröger, 2004) generate silica structures in vitro that lack higher order - they are agglomerations of randomly organized fused silica particles. Only in the presence of higher molecular weight silaffins do higher order structures form (Poulsen and Kröger, 2004). We propose that the smaller organic components are responsible for generating “filler” silica which can assume structure either through interaction with other peptides or by confinement within the SDV. Small molecular size would facilitate this by enabling freer diffusion and more degrees of freedom. Expansion of T. pseudonana valves in the z-axis direction could be an example of this filling process.
The silaffins lacked amino acid sequence conservation, and no clear homologs were found in genome searches, which prompted the examination of genes encoding proteins with similar amino acid composition, and not necessarily sequence, to the silaffins (Scheffel et al., 2011). A search was made of the T. pseudonana genome for proteins with an ER signal sequence (consistent with targeting to the SDV) and ≥18% serine and ≥10% lysine residues. Eighty-nine genes were identified, and a subset of six genes encoded proteins with conserved repetitive features, which were called cingulins. There were two types of cingulins, the “W” cingulins and the “Y” cingulins, which were named based on enrichment of their sequences for either tryptophan (W) or tyrosine (Y). GFP tagging of the cingulins indicated that they were exclusively associated with girdle bands (Scheffel et al., 2011). Subsequent characterization of insoluble material resulting from dissolution of cleaned silica revealed that the proteins were associated with “microrings” that precisely mimicked the girdle band dimensions and structure. The microring material was termed AFIM, or the insoluble organic matrix (Figure 4). It was proposed that the insolubility of the organic matrix was due to covalent cross-links between its protein components, which may include O-phosphoester bonds and O-glycosidic bonds (Kotzsch et al., 2016). Initial characterizations in T. pseudonana found the AFIM associated only with the girdle bands and not valves, although a valve associated AFIM was found in Coscinodiscus wailesii (Scheffel et al., 2011). Subsequent work identified a valve-like AFIM in T. pseudonana (Kotzsch et al., 2016). An extensive microscopic study of the insoluble organic matrix in a variety of diatom species (Tesson and Hildebrand, 2013) determined that the material was fibrous. Direct imaging showed that the material was associated exclusively with the proximal surface of the valves. The material could be DAPI-stained only after SDS treatment, which is consistent with it being obscured by the silicalemma, leading to the model for structural arrangement shown in Figure 4. The AFIM precisely mimicked valve features (Figure 5). Given its location and how it precisely mimics the proximal surface, it is reasonable to hypothesize that the AFIM constitutes an organized pattern for base layer formation. Imaging of the matrix in the location of pores identified projections of organic material (Figures 5D,E) around which silicification occurred. Comparison of AFM imaging of the AFIM with an SEM of the valve of S. turris shows that the pores in the valve are occluded by this material (Figures 5E,F). These observations support the hypotheses derived from observations of forming pores in Figures 3B,C. A consequence of this is that pores are not entirely open channels, but are at least lined, if not occluded, by organic material. The proteins that constitute these projections could impart a level of control over what passes through the pores. If so, this prompts a significant re-framing of how we think about transport processes of diatoms.
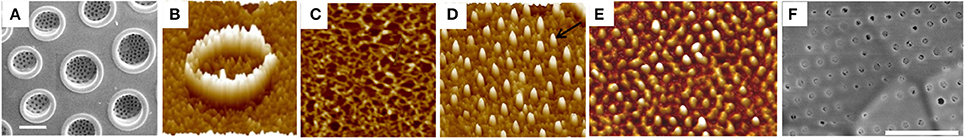
Figure 5. Characterization of the AFIM, or insoluble organic matrix from Coscinodiscus radiatus. (A) SEM of Proximal valve surface, highlighting the large openings of the foramina. Scale bar = 5 μm. (B) AFM of region of foramina in the AFIM. (C) Fibrous structure of AFIM from proximal surface. (D) AFM of AFIM from a girdle band, showing projections that define pores. (E) AFM of AFIM from the valve of Stephanopyxis turris. (F) SEM of proximal valve surface of S. turris. Scale bar = 1 μm. Scan sizes are (B) = 1 μm, (C) = 1.2 μm, (D) = 2 μm, (E) = 3.6 μm. From Tesson and Hildebrand (2013), reproduced under the Creative Commons license.
An elegant nuclear magnetic resonance (NMR) study substantiated the presence of an organic layer associated with the surface of silica in S. turris (Jantschke et al., 2015). Dynamic nuclear polarization solid state NMR on SDS/EDTA cleaned silica (which would not remove the AFIM—Figure 4B) identified an estimated 3 nm thick surface layer, which corresponded to direct measurements using AFM (Tesson and Hildebrand, 2013), enriched in protein and carbohydrate. Predominant amino acids were Asp, Gly, Ser, Glu, and Ala, as confirmed by both NMR and MS analyses. The bulk silica was enriched in LCPAs, and there was excellent agreement between the estimated thickness of the silica structure by NMR and the actual measured thickness. Several lines of evidence supported an abundance of β-strand protein secondary structure and random coils, which the authors proposed would help to establish compactness as well as intermolecular networking. These features correspond well with other studies mentioned previously related to the AFIM.
Biochemical and proteomic analysis of the AFIM from T. pseudonana identified seven proteins called “SiMats” which generally had amino acid sequence characteristics of known silicification proteins (Kotzsch et al., 2016). Most lysine residues in the identified AFIM proteins were post-translationally modified to contain short polyamine chains. SiMat1 was localized to the girdle band AFIM.
Two silaffins initially identified as being associated with the soluble fraction of cleaned dissolved silica, and one other protein later associated with the silicalemma, were also found associated with the insoluble fraction. A GFP fusion of TpSil3 was incorporated into the AFIM, although its abundance was lower than the cingulins (Scheffel et al., 2011). Mass spectrometry characterization of the T. pseudonana AFIM also identified a peptide from the C-terminal L form of TpSil 1/2 (Kotzsch et al., 2016). Although not noted in previous work, the extreme C-term of TpSil 1/2 has a transmembrane domain (Table S1) as predicted by TMHMM v.2.0 (Sonnhammer et al., 1998), so the L form could be associated with the silicalemma. Another explanation for the predominance of TpSil 1/2 and 3 in the soluble fraction, with a lesser amount in the insoluble fraction, could be incorporation of the silaffins in the latter (Figure 4D) by the proposed cross-linking phenomenon (Kotzsch et al., 2016). Described in more detail in the following section, a silicalemma-associated protein called Sin1 was initially isolated from the AFIM and named SiMat7 (Kotzsch et al., 2016).
As stated previously, the cytoskeleton has a substantial influence over silica structure, and in particular actin assembly patterns have been correlated with silica structures (Tesson and Hildebrand, 2010). Actin assembles outside of the SDV; therefore, its assembly pattern must be transmitted into the SDV lumen for it to influence silica patterning. Robinson and Sullivan (1987) proposed that silicalemma-spanning proteins could serve as intermediates that translate cytoskeletal assembly patterns into similar silica structures. Such proteins would be predicted to have a transmembrane domain, a cytoplasmic portion that could interact with the cytoskeleton (either directly or indirectly), and an intralumenal portion that would either have characteristics of silicification proteins, or domains that would facilitate interactions with such proteins or LCPAs (Figure 6). They also could have different characteristics depending on which region of membrane they are in. In the proximal silicalemma, they could interact with the AFIM. These interactions would not be possible from the distal membrane unless they extended completely across the SDV lumen (Figure 6), if the interactions do not occur one would surmise that this subset of silicalemma associated proteins might be extractable by detergent. Examination of genes upregulated during valve formation in synchronized cultures of T. pseudonana resulted in the identification of a multiple silicalemma-associated candidate proteins with predicted ER targeting, a single transmembrane segment, and sequence features characteristic of silica-associated proteins (Tesson et al., 2017). One protein, called SAP1 (Silicalemma Associated Protein), was used to identify two other proteins (SAP2 and 3) with similar characteristics in the T. pseudonana genome. These proteins constitute a gene family in diatoms, and are found mainly in other centric diatom species. They have a longer N-terminal portion which is enriched in serine, and which in SAP3 was associated with silica in the SDV lumen. The SAPs also have a shorter C-terminal portion which contains a conserved, but undefined, sequence domain found in the other SAPs. The SAPs have the structural organization predicted by Robinson and Sullivan, but the C-terminal portion does not match any known cytoskeleton-interacting sequence. It is possible that (1) the conserved sequence is a novel cytoskeleton-interacting sequence, (2) the SAPs interact indirectly with the cytoskeleton via other proteins, or (3) these particular proteins have no interaction with the cytoskeleton. SAP1 and SAP3, when tagged with GFP on their C-termini, were seen associated with forming silica structures in both the girdle bands and valves. In addition, SAP3 was associated with a relatively large intracellular vesicle, and an increase in GFP fluorescence over time during silicon starvation suggested that the protein accumulated there (Tesson et al., 2017). The C-terminal GFP tagged proteins were never observed in the completed cell wall, and evidence was presented that proteolytic cleavage to remove the C-terminus of SAP3 occurred. If the C-terminus was involved in interacting with other proteins, this cleavage could cease the interaction. One possibility in this scenario is that interactions with the cytoskeleton must be terminated in order for the valve to be exocytosed. Tagging of SAP3 in the N-terminal region with GFP indicated that this portion was embedded in the silica. Transcript knockdown of SAP1 and 3 using antisense and RNAi approaches generated distinct phenotypic changes in silica structure. These were the first demonstrations of genetic manipulation of silica structure in a diatom. SAP3 knockdowns displayed a lack of decoration of the distal surface where silica particles are normally found on top of the ribs (Figure 7). SAP1 knockdowns displayed aberrations in proximal surface silicification separately or in addition to alterations in the location of their pattern forming centers (Figure 7), which is the characteristic location of valve formation initiation. It is known that the pattern forming center is positioned by microtubules (Pickett-Heaps et al., 1990), therefore, it is possible that SAP1 knockdown interferes with the interactions of SDV-associated proteins and microtubules. These results demonstrate that manipulation of a single gene can generate a consistent alteration in silica structure. This indicates that the role of specific proteins can be probed using genetic manipulation approaches.
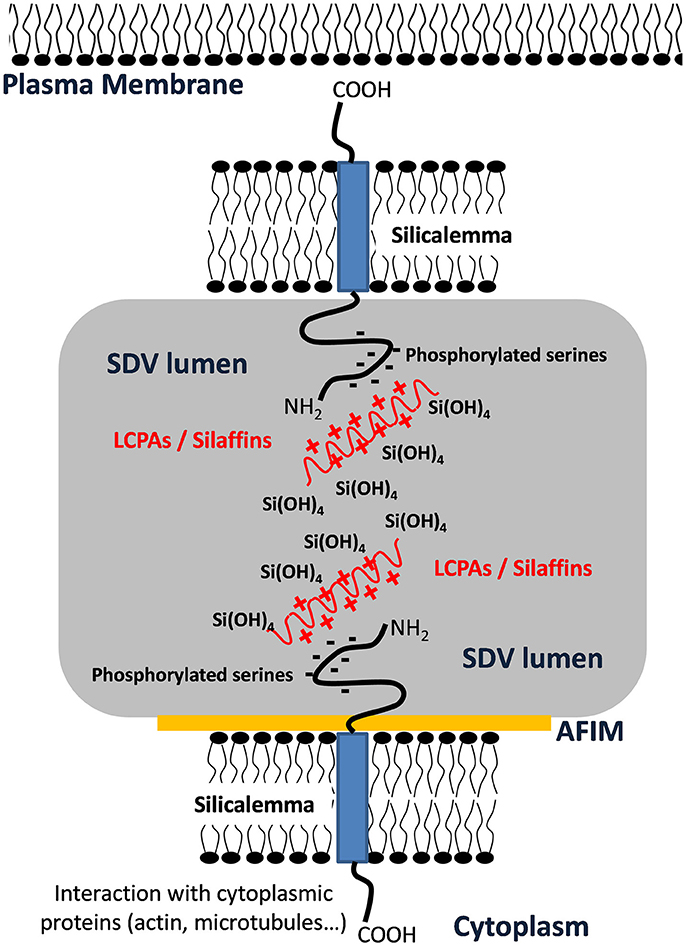
Figure 6. Schematic representation of transmembrane protein organization into the SDV membrane showing putative interactions of phosphorylated serine with Long Chain Polyamines (LCPAs) or silaffins inside the SDV lumen and interaction with cytoplasmic proteins via the cytoplasmic domain. This figure was modified from Tesson et al. (2017) and is based on a model by Robinson and Sullivan (1987).
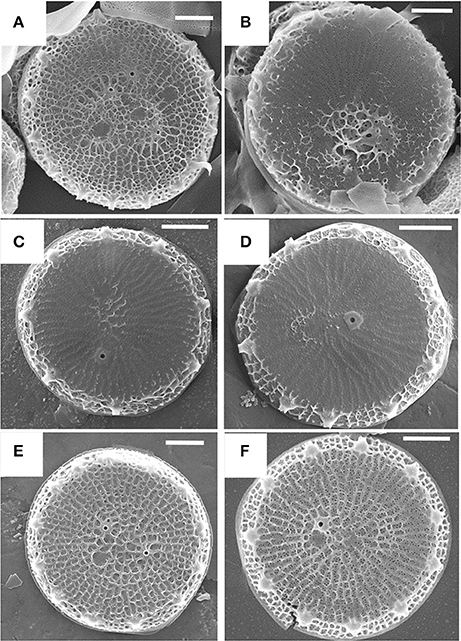
Figure 7. Alterations in T. pseudonana valve structure resulting from antisense knockdowns of TpSAP1 (A,B) and TpSAP3 (C,D). Wild-type controls are (E,F). From Tesson et al. (2017), reproduced under the Creative Commons license.
A distinct silicalemma-associated protein, called Silicanin-1 (Sin1) has also been characterized (Kotzsch et al., 2017), which was the same protein characterized as the AFIM-associated SiMat7 (Kotzsch et al., 2016). Similar to the SAPs, Sin1 has a long intralumenal N-terminal region, a single transmembrane segment, and a short cytoplasmic sequence. The N-terminal region was shown to be embedded within the silica and the C-terminal region appeared to be proteolytically released. Sin1 was found associated with both girdle bands and valves. The dynamics of Sin1 localization over time were determined, showing that the protein was associated with the plasma membrane as well as an intracellular vesicle (similar in appearance to that found with SAP3). Sin1 was recruited to the SDV during silica formation, and then dispersed into the plasma membrane afterwards. Unique targeting signals must be involved to accomplish this. No direct function of the protein in valve formation was demonstrated, but it was shown that recombinantly-expressed protein assembled into clusters in a pH-dependent manner—at lower pH (similar to the SDV lumen), clusters formed, and at higher pH, they disaggregated. pH-dependent aggregation was also demonstrated for the cingulins (Kotzsch et al., 2016).
For Sin1, antibody accessibility experiments indicated that <20% of the protein was accessible in the silica relative to the AFIM (Kotzsch et al., 2016), meaning that most of the protein was embedded in the silica. As such, SDS extraction would not be expected to produce the highest yields, although it would isolate protein localized to other cellular locations such as the plasma membrane or an intracellular vesicle. A similar situation may exist for the SAPs; SAP3 was found embedded in the silica (Tesson et al., 2017) but also could be extracted with detergent only. In this case the most abundant source of the protein could have been from the intracellular vesicle.
One hypothesis regarding the role of silica- or silicalemma-associated proteins is that if they are exclusively localized to one particular macroscale structure (valve or girdle band) they could be involved in formation of the specific features of that structure, but if they are localized to different structures, they may be involved in more general silicification processes. Following this line of reasoning, because SAP3 was associated with both valves and girdle bands, both structures contain nanoparticulate silica on their distal surfaces (Hildebrand et al., 2006), and knockdown of SAP3 prevented formation of the distal valve nanoparticulate silica, perhaps SAP3 performs a general function of facilitating distal nanoparticulate silica formation. Similarly, although a specific function for Sin1 has not been defined, we would propose that it plays a similar role in valve and girdle band silicification. In contrast, the cingulins have only been identified associated with girdle bands, suggesting that they are involved in the meso- or micro-scale processes specifically associated with shaping the girdle bands.
Because formation of diatom cell wall structures is intimately associated with the cell cycle, it may seem logical that the induction of transcripts for silicification proteins relate to the formation of particular structures at particular times in the cell cycle. TpSil3 served as the first marker for valve formation, because it was specifically upregulated during that period (Frigeri et al., 2006). However, analysis of transcript expression patterns obtained from synchronized cultures of T. pseudonana (Shrestha et al., 2012) indicated that known and proposed genes encoding proteins involved in silicification have distinct expression patterns and maxima (Hildebrand and Lerch, 2015), which do not necessarily relate to the timing of formation of structures. A recent microarray analysis confirms this conclusion (Brembu et al., 2017). As detailed in the following, there appear to be four major types of responses, which cover all major stages of the cell cycle.
In Figure 8, we examine a whole genome microarray response (Shrestha et al., 2012) of three classes of genes encoding proteins peripherally involved in silicification, the silicon transporters (SITs), chitin synthases, and aminopropyltransferases (APTs) and a spermidine synthase that may be involved in polyamine synthesis.
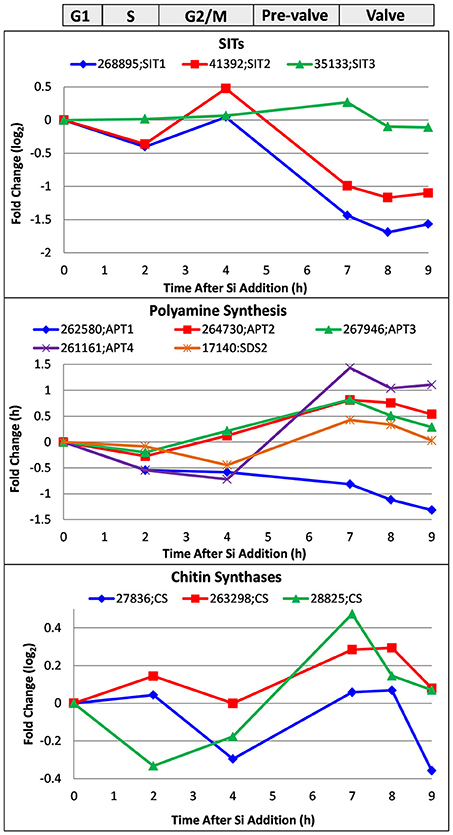
Figure 8. Whole genome microarray analysis of transcript level changes of proteins peripherally involved in T. pseudonana silicification. Log base 2 fold changes relative to 0 h are plotted. Transcripts were measured in a synchronized culture where cell cycle progression began after silicic addition to a silicon-starved culture. Approximate timing of different cell cycle stages is shown at top. Data are from Shrestha et al. (2012).
Transcript behavior for SITs was previously documented (Shrestha and Hildebrand, 2015; Brembu et al., 2017)—SIT1 and 2 are upregulated during silicon starvation, then generally downregulated upon silicon replenishment until an increase in M phase, followed by downregulation thereafter (Figure 8). As suggested, because silicic acid can freely diffuse into the cell, the SITs do not have to substantially contribute to uptake, and appear to play more of a regulatory role over silicic acid metabolism (Shrestha and Hildebrand, 2015). Shrestha and Hildebrand proposed that SIT upregulation during M phase could be due to a need to populate the new daughter cell with SITs rather than a silicic acid requirement.
Various enzymatic steps are involved in polyamine and LCPA synthesis, and key enzymes to elongate the polyamines have fused aminopropyltransferase (APT) and S-adenosylmethionine decarboxylase (SDS) domains (Michael, 2011). Polyamines also contain spermidine and therefore require its synthesis (Michael, 2016). We plot the expression pattern for four APT/ SDS genes and one spermidine synthase in T. pseudonana in Figure 8. Four of the five genes are upregulated during valve formation. Two of the three APTs (Thaps3_267946 and 261161) had predicted ER targeting, which could be consistent with elongation of LCPAs in the steps prior to formation of the SDV or within the SDV itself. The spermidine synthase, Thaps3_17140, also had predicted ER targeting, and was upregulated during valve formation.
As discussed, chitin is closely associated with the cell wall and plays some role in its formation. Three chitin synthases were identified in the genome, and all three were at least somewhat upregulated during valve formation (Figure 8). Although this increase in transcripts occurs when chitin putatively involved in valve formation would be needed, it is also at a time when the fultoportulae should be made. The synthesizing machinery for chitin fibril extrusion is positioned under the fultoportulae, and is likely to be a permanent fixture once in place (Herth, 1979a,b). It is therefore unclear whether upregulation of the synthases during valve formation relates to making chitin for valve formation or positioning the chitin synthesizing machinery for daughter cells.
In Figure 9, we show the transcript response of genes encoding proteins known to be involved in silicification, which includes the silaffins, silacidin, cingulins, SiMat proteins, the SAPs, and Sin1. These encompass proteins embedded within the silica, in the AFIM, and associated with the SDV membrane. The responses fell into four categories. The valve formation response, which was modeled after the established Sil3 pattern (Frigeri et al., 2006), included Sil3, CinW1 and 2, SiMat3 and 5, and SAP1. The S or M phase response (Figure 9) displayed maximum upregulation between 2 and 4 h, and included the Y cingulins, Cin W3, SiMat1, 2, 4, and SiMat7/Sin1. The valve and S/M phase responses have been substantiated in a recent transcriptomic analysis (Brembu et al., 2017). In our analysis, four genes exhibited patterns of either little change related to cell cycle stage, or down regulation during the synchrony (Figure 9), including Sil1 and 4, silacidin, and SAP2. SAP3, which was examined in a distinct RNAseq time course synchrony (Tesson et al., 2017), exhibited a distinct pattern from previously described (Shrestha et al., 2012; Brembu et al., 2017), with maximal upregulation during pre-valve formation, between mitosis and valve formation at 5 h (Figure 9).
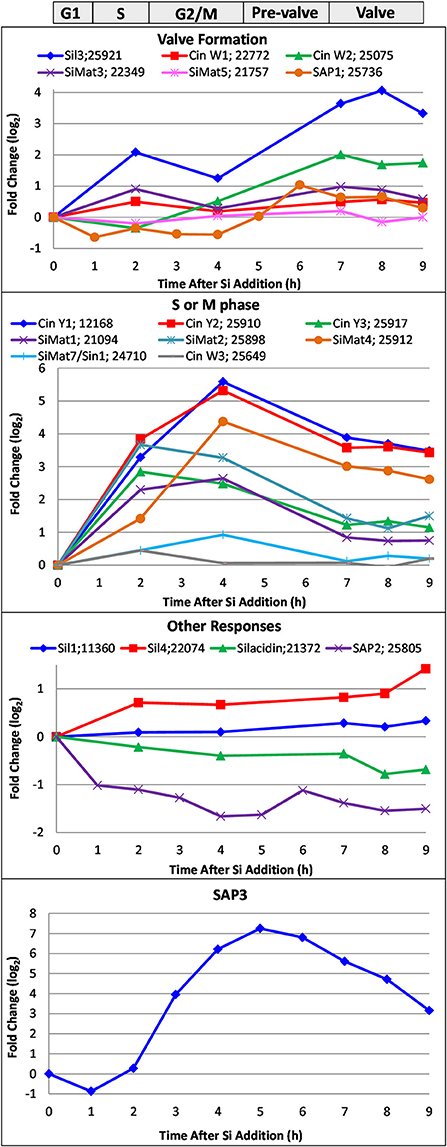
Figure 9. Whole genome microarray analysis of transcript level changes of proteins known to be involved in silicification in T. pseudonana. Log base 2 fold changes relative to 0 h are plotted. Transcripts were measured in a synchronized culture where cell cycle progression began after silicic addition to a silicon-starved culture. Approximate timing of different cell cycle stages are shown at top. Data are from Shrestha et al. (2012).
The amino acid composition of known proteins involved in silicification is shown in Figure 10. The most abundant amino acids (5% or higher) were Ser, Gly, Lys, Asp, Ala, Thr, Glu, and Pro. Since these proteins play different functional roles, we compared the amino acid composition (Table S2) between subsets of proteins, separating them into categories as the silaffins, AFIM associated proteins, and silicalemma-associated proteins (silacidin has a highly distinct amino acid composition and different properties than the silaffins and was therefore excluded). We acknowledge that individual proteins in a given category could also have different functional roles which would influence amino acid composition, hence we do not include any statistical comparison between categories. Differences between the categories were accentuated by comparing ratios of all three and plotting them on a log2 scale (Figure 10). The silaffins were enriched over the other two categories in Ala, Glu, and Met, and depleted in Asn, Cys, Gln, Trp, and Tyr. The silaffins were also depleted in Gly relative to the AFIM proteins. The AFIM proteins were enriched in amino acids depleted in the silaffins and depleted in those enriched in the silaffins. The silicalemma associated proteins were generally enriched in Ala, Arg, Asn, Gln, and His relative to the silaffins, and depleted in those same amino acids relative to the AFIM. Phe, Pro, Ser, Thr, and Val were similarly conserved across all categories. Comparing the averages of the ratios for all amino acids in each pairwise comparison indicates that the AFIM and silicalemma associated proteins had more similar amino acid composition than either compared with the silaffins. In context of the transcript data discussed below these trends may have implications for temporal regulation of different types of proteins based on amino acid based functions.
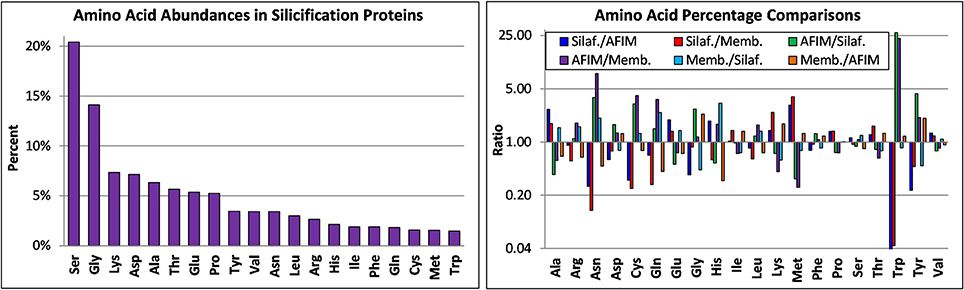
Figure 10. Amino acid composition of silicification proteins based on analysis of their published sequences. Top, amino acid abundances of all proteins, bottom, ratio of amino acid abundances for different classes of proteins, including silaffins (Silaf), those associated with the AFIM (AFIM), and the SDV membrane or silicalemma (Memb).
We examined whether specific amino acids or amino acid motifs were enriched in the known silicification proteins (Table S1). Cysteine was completely absent in 9 of the 20 proteins, but was enriched above the average found in all proteins (1.55%) in 7 of them, specifically in Sil1 H, CinY1, SiMat3-7, and SAP3 (Table S1). Even though Cys is not very highly enriched in Sin1/SiMat7, analysis indicates that 8 cysteines are conserved in a large protein family related to Sin1/SiMat7 in T. pseudonana and Phaeodactylum tricornutum (Brembu et al., 2017), indicating an important functional role. Cysteine is typically involved in disulfide bonding as a means of stabilizing protein tertiary structure. Tryptophan was not highly abundant among the proteins with the exception of the CinW's. SAP1 and 3 were somewhat enriched in Trp. It was proposed that Trp could facilitate aggregation of the CinW's under acidic conditions (Kotzsch et al., 2016). The “KXXK” motif (which includes KXK, KXXK, and KXXXK) has been proposed to promote silicification via posttranslational modification of the lysines and be involved in targeting proteins to the SDV (Poulsen et al., 2013). It is highly enriched in many of the proteins, up to 45% of the total amino acids in Sil1/2 L are KXXK (Table S1). Recent analyses have identified other KXXK motif containing silicification candidate proteins in T. pseudonana (Brembu et al., 2017). If X = Ser, then that side chain could be phosphorylated. If repeated every 4 amino acids in an alpha helical structure, the side chains would consistently orient toward one side, providing a spatial constraint. Analysis of the silaffins and cingulins indicates a prevalence of clusters of KXXKs, with the average spacing between lysines of 4.4 residues. In an alpha helix, that would be equivalent to 1.25 turns between lysines. If in a beta strand structure, which was shown to be enriched in silica associated proteins by NMR in S. turris (Jantschke et al., 2015), side chains repeated on average of 4.4 residues would tend to orient in the opposite directions. These data lend credence to the concept of specific orientation of the side chains in these motifs. The SAPs lack a KXXK motif entirely, and in Sin1 and silacidin it is depleted relative to its average prevalence in all proteins. PT or TP repeats were common in the known silicification proteins, and have been noted in other candidate silicification proteins implicated by transcript response (Brembu et al., 2017). In the context of the threonine being glycosylated, the glycosyl group would be found on the same face of the helix in a repeat, or in a beta strand structure, facing in the same orientation. It has been noted that repeated sequence patterns generates regular arrays of spatial and functional groups, useful for structural packing or for one to one interactions with target molecules (Katti et al., 2000). Glycosylated residues can stimulate silica formation at low concentration, and inhibit it at higher concentration (Poulsen and Kröger, 2004), thus proteins containing PT repeats could regulate the extent of silicification. Sil1/2 H has a high percentage of PT repeats, and it has been shown to regulate the extent of silicification (Poulsen and Kröger, 2004). The CinY's are enriched in PT repeats, also suggesting a regulatory role. Sil3 and 4 were specifically enriched in MS or SM motifs (Table S1), which were largely absent from the other proteins. It is unclear what their effect would be on silicification, although the serine could be phosphorylated.
All of the known silicification proteins have a signal peptide based on Signal P (Bendtsen et al., 2004) prediction, indicative of ER targeting, however six of them actually had predicted periplastid compartment (PPC) targeting because they have a predicted ER signal peptide along with a robust chloroplast targeting prediction (Emanuelsson et al., 1999) lacking the characteristic ASAFAP sequence (specifically lacking an aromatic residue at the F-position—Kilian and Kroth, 2005; Gruber et al., 2007) at the cleavage site between the two targeting sequences. GFP fusions of all of these proteins were experimentally localized to within the silica or silicalemma, therefore we interpret the PPC targeting as a prediction artifact. Interestingly, in an examination of SDV targeting signals, Poulsen et al. (2013) determined that particular sequences targeted TpSil3 to a sub-compartment in the ER, which looked very similar to authentic PPC localization. This suggests that silicification proteins with a PPC targeting motif could be targeted to the SDV by a distinct route than other SDV-associated proteins. Dynamic localization data indicates that Sin1 is targeted to the plasma membrane and then recruited to the SDV (Kotzsch et al., 2017).
The concept of identifying conserved domains in silicification protein amino acid sequences will be important moving forward since conserved domains should endow specific functional properties. This is especially important considering that the amino acid composition, and not sequence, appears to be relevant to function in many of the proteins. A conserved domain stands out from such background sequence. The conserved domain identified in the SAPs and its presence in a protein family conserved across diatom species (Tesson et al., 2017) indicates that it has some importance for the function of the proteins. A conserved set of eight cysteines is diagnostic of a Sin1/SiMat7 protein family in T. pseudonana and P. tricornutum (Brembu et al., 2017). The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP) dataset, which contains 650 assembled, functionally annotated marine microbial transcriptomes (Keeling et al., 2014), is a good resource to perform cross-species comparisons of these and other candidate genes (Brembu et al., 2017). Sequences that are conserved across diverse species could either be involved in general aspects of silicification or not involved at all, whereas sequences conserved in a subset of species, for example the Thalassiosirales, could be involved in processes related to formation of specific structures in those species.
Transcript expression patterns, when combined with observations of amino acid composition and targeting, give a more comprehensive picture of silicification protein regulation than any single line of evidence can provide. This data shows interesting trends relating the timing of transcript induction to encoded protein properties and has implications for the trafficking and regulation of silicification related proteins.
A subset of genes involved in silicification are upregulated during either S or M phase, or have expression patterns which do not respond to any canonical cell cycle events (Figure 9). SAP3 transcripts are also induced between M phase and valve formation. We are particularly intrigued by the expression pattern of SAP3 as its transcript peak coincides with the period when we propose that SDV assembly is occurring. This hypothesis is supported by dynamic localization of Sin1, where it is localized to the silicalemma prior to or concurrent with the initiation of silica polymerization (Kotzsch et al., 2017).
The responses described above do not relate to the formation of a particular silica structure. It should be noted that Figure 9 shows changes in transcript levels, not absolute abundance, so lack of induction does not mean a protein is not being expressed. However, transcript induction at a particular time is consistent with an increase in the protein encoded by that transcript (with the caveat that the relation between transcript and protein abundance can vary). Upregulation of silicification related genes when cell wall structures are not being made suggests that encoded proteins are synthesized in advance of when they are needed for cell wall synthesis. The cingulins were localized to the girdle bands (Scheffel et al., 2011), yet transcripts for Cin W1 and 2 have a valve response, and the other cingulins have an S or M phase response (Figure 9). Sil3, SAP1 and 3, and Sin1 were localized to both valves and girdle bands (Scheffel et al., 2011; Kotzsch et al., 2017; Tesson et al., 2017), but have valve-formation, S or M phase, or pre-valve transcript responses. Sil1, the only protein that has been localized exclusively to the valves, had relatively little change in transcript abundance over the cell cycle. These observations are also consistent with intracellular accumulation of silicification proteins prior to their use. SAP3 and Sin1 were directly shown to accumulate in the cell (Kotzsch et al., 2017; Tesson et al., 2017). Intracellular accumulation of GFP-tagged protein was not documented for Sil1/2 and 4 (Poulsen et al., 2013), the cingulins (Scheffel et al., 2011) or SAP1 (Tesson et al., 2017), even though the transcript data suggests that accumulation could occur. These proteins either didn't accumulate prior to use, the GFP tag could have been rapidly removed and degraded, or GFP fluorescence could have been inhibited if the protein was present in an acidic environment (Poulsen et al., 2013).
One possible explanation for the accumulation of silicification proteins is that it would be challenging for the cell to make the large amount of all proteins required to create the cell wall in the short time period allotted. Stockpiling protein that could then be targeted en mass to the SDV would circumvent overloading the synthesis system. Such an approach could also impart a level of regulation over the process if particular proteins can co-exist in storage locations such that they don't catalyze silicification, but then can do so when combined with other separately stored proteins.
Following this concept, a potentially interesting observation stems from categorizing the amino acid composition of proteins according to transcript expression patterns, in which we compare the three expression categories (valve formation, S or M phase, and other) to the mean of all proteins irrespective of category (Figure 11). The “other” category proteins are enriched (at least 1.5-fold log2) in Arg and Glu, and depleted (at least 0.5-fold log2) in Cys, Gly, Trp, and Tyr relative to proteins in the other expression categories. The valve formation and S or M phase response proteins are similar to each other, with exceptions being that the valve formation response group is depleted for Thr and enriched in Trp. The general tendency is that amino acids that have interaction or aggregation properties are enriched or depleted. These include electrostatic (Arg, Glu), covalent (Cys), and hydrophobic (Trp, Tyr) interactions. The average pI of proteins in the different expression categories also differ, with the other category (pIave = 9.13 ± 2.1, n = 4) being basic, and the valve (pIave = 6.52 ± 2.1, n = 6) and S or M phase (pIave = 6.29 ± 1.9, n = 8) categories being slightly acidic. Given the proposed models for electrostatic interactions and pH-dependent aggregation phenomenon demonstrated for silaffins and Sin1 (Kröger et al., 1999; Kotzsch et al., 2016, 2017), this data infers that expression stage-specific biases could relate to functional requirements related to protein-protein interactions at different stages in the process.
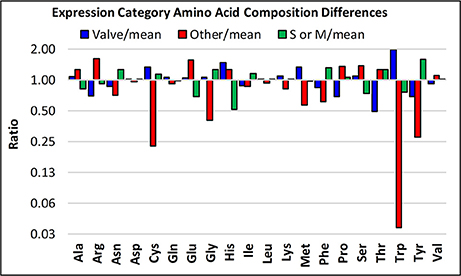
Figure 11. Amino acid enrichments based on expression response category. The ratios of the abundances of particular amino acids are plotted, comparing three of the different transcript expression patterns defined in Figure 9, specifically, valve, S or M phase, and “other”.
Direct observational data indicates that there are multiple means of targeting and trafficking proteins to the SDV. One way is direct targeting to the SDV, apparently through a sub compartment of the ER, as demonstrated in Sil3 (Poulsen et al., 2013). Amino acid substitution experiments (Poulsen et al., 2013) were consistent with posttranslational modification of lysines and serines being essential for silaffin targeting. The former can be modified by methyl groups and polyamine chains, and the latter by phosphate or carbohydrate moieties (Poulsen and Kröger, 2004). An analogy was made (Poulsen and Kröger, 2004) to the targeting of proteins from the trans Golgi network to the lysosome, which involves a post-translational modification to incorporate mannose 6-phosphate. This suggests in this case that modifications to the amino acids, and not the amino acid sequence, determine targeting. A second mode of targeting is through large intracellular vesicles, as seen with GFP fusions of SAP3 and Sin1 (Kotzsch et al., 2017; Tesson et al., 2017). Initial data is consistent with these being the location of protein accumulation (Tesson et al., 2017). Presumably, these vesicles fuse with the SDV during valve formation. The third documented mode of targeting is recruitment from the plasma membrane, as occurs with Sin1. This protein is targeted to both the intracellular vesicle and plasma membrane, and is recruited to the SDV from the plasma membrane during valve formation, then returns to the plasma membrane afterwards (Kotzsch et al., 2017). Because the protein is also localized to both valves and girdle bands, there must be versatility built into the targeting and possibly a repurposing process beyond what is typical for secreted proteins. Although a role in silicification is likely for SDV associated Sin1, it is unclear what role the protein might play in the plasma membrane.
The reason(s) for these diverse targeting and trafficking processes is unclear, but we can return to hypotheses about the need to synthesize and store particular proteins in advance and/or to maintain particular combinations of proteins separately until assembly in the SDV. The system could provide a means of checks and balances, for example, unless requirements (e.g., adequate abundance) for a particular protein are met, then subsequent processes will not occur. This would be especially important in terms of valve formation, where ideally the cell would be able to assess whether it was capable of completing the valve before attempting to make it, as an incompletely formed valve could severely compromise cellular integrity. There is support for this hypothesis in relation to function and expression patterns of some known silicification genes. Silaffin 1/2 and silacidin lack cell cycle specific transcript responses, and silacidin and Sil1/2 L solely promote silica formation (Poulsen and Kröger, 2004; Wenzl et al., 2008). Under depleted silicon conditions, there is increased processing of Sil 1/2 from H to L form and also an increase in the amount of silacidin in the cell wall (Richthammer et al., 2011), consistent with silica formation capability being favored. In contrast, TpSil3 has regulatory activity with regards to silica formation (Poulsen and Kröger, 2004), and is expressed during valve formation. Because of the substantial differences in their timing of maximal expression, TpSil3 may also use a distinct targeting route compared with TpSil1/2 and silacidins. A simplistic view is that under silicon limiting conditions the cell prepares for silica formation by increasing the amount of silicification promoting proteins, but it is not until some level of regulation over silica formation can be imparted that structures such as the valve are made.
Based on the expression and targeting discussion, as well as other data in the literature, we propose a series of events that occur during valve formation in T. pseudonana. These events are delineated by hypothesized checkpoints, events which must be completed in full before cell wall formation can progress. An initial checkpoint is related to the availability of silicic acid. In T. pseudonana, the majority of cells arrest in the G1 phase of the cell cycle if silicic acid levels are insufficient (Brzezinski et al., 1990; Hildebrand et al., 2007). This prevents initiation of the division process. During valve formation, the cytoskeleton appears to be involved in positioning mesoscale structures via microtubules, and patterning the overall surface features by actin (Tesson and Hildebrand, 2010). For this to occur, an interaction between the cytoskeletal components and SDV must exist, and based on the model of Robinson and Sullivan (1987), this is facilitated through the silicalemma associated proteins. Thus, the assembly of the cytoskeleton and the SDV must be coordinated. Additionally, because the AFIM precisely mimics mesoscale silica structure, its constituent proteins must be assembled in the SDV with similar timing. We suggest that the silicalemma associated proteins either appear first, or concurrently with proteins of the AFIM. Either scenario would facilitate cytoskeletal-based positioning of components to organize them for base layer formation. It should be noted that the initial deposition appears to occur prior to when the SDV has reached its final position and dimensions, ion abrasion SEM (IASEM) of an extremely early stage of silica deposition showed a flexible structure with base layer features, which was positioned near one edge of the cell (Hildebrand et al., 2009). Because complete base layer intermediate structures are commonly observed by standard SEM (Hildebrand et al., 2006), we propose that their complete formation, likely mediated in part via actin controlled expansion of the SDV (Tesson and Hildebrand, 2010) and the AFIM (Tesson and Hildebrand, 2013) as previously described, serves as the second checkpoint in valve formation. This would ensure that z-axis expansion was triggered only after x/y axis expansion was complete, ensuring that z-axis structures covered the entirety of the valve.
An important question related to the process of z-axis expansion and the hypothesis that proteins of the SDV are accumulated as precursors, is what prevents silica polymerization at cellular locations other than the SDV, and what stimulates silica formation to occur? For the former, the lack of high enough silicic acid concentrations might be a factor, but because silicic acid freely diffuses across membranes (Thamatrakoln and Hildebrand, 2008), there would be a lack of control over its supply once it entered the cell. We suggest that a shift in pH within the SDV is an important trigger for silicification, and that a lack of pH change in other compartments could prevent silica formation. Data supports the importance of low pH for silica formation; non-postranslationally modified C. fusiformis silaffins had a silica forming optima at pH 7, whereas the native silaffins were optimal at pH 5 (Kröger et al., 1999). Recombinant Sin1, cingulins CinY2 and CinW2 all exhibited silica polymerization and aggregation at low pH (Kotzsch et al., 2016, 2017). Lower pH is also a fundamental chemical characteristic of formation of silica network structures (Iler, 1979), such as those seen in diatoms. Lowering the pH in intracellular compartments typically occurs by proton pumping by the vacuolar H+-ATPase or VHA (Finbow and Harrison, 1997), and we suggest VHA involvement in SDV acidification. This would provide an overall control over the process distinct from the involvement of silicification proteins and LCPAs. Some aspects of structure formation may not be pH dependent, for example, given the distinct stage of base layer formation, and its distinct silica structure compared with the bulk of the valve silica, it could be formed in the absence of a pH shift. Z-axis expansion in which the existing base layer valve structure is filled in could be an example of a pH dependent process.
The continuing investigation into understanding how diatoms make their silica cell walls has involved a variety of techniques which were state-of-the-art when initially applied. These techniques have not only have improved our understanding of cell wall formation, but in combination, provide different perspectives on the process (Hildebrand and Lerch, 2015). Understanding the outcomes of various cell wall extraction procedures has affected our interpretations of the role of specific organic components in silicification. The logical approach of isolating organic material embedded within detergent-cleaned silica as essential components of the silicification process resulted in the seminal discoveries of the silaffins, LCPAs, and silacidins (Kröger et al., 1999, 2000, 2002; Poulsen and Kröger, 2004; Wenzl et al., 2008). Consistent with our hypotheses about soluble silicification proteins, silica formed by them exhibits nanoscale, but not highly organized meso- or micro-scale order. Later characterization of insoluble organic material resulting from the same isolation procedure identified material (the AFIM) that has a clear relation to higher order structural features (Scheffel et al., 2011; Tesson and Hildebrand, 2013). As discussed, although chitin appears to play an important role in structure formation, to date purification procedures have not enabled distinction between a possible structural arrangement of chitin and the AFIM. To completely remove organic material associated with the surface of the silica, acid treatment is required (Figure 4C)—detergent or peroxide treatment is insufficient. A model that addressed the relation between the cytoskeleton and silica structures formed within the SDV (Robinson and Sullivan, 1987), led to the investigation of silicalemma-associated proteins (currently the SAPs and Sin1), and the genetic manipulation demonstration that they are involved in structure formation (Tesson et al., 2017). It is not clear, however, whether these proteins interact with the cytoskeleton.
Microscopic approaches have provided fundamental insights and will continue to be important in understanding the spatial relationships of organic components and silica structures, these relationships are essential for understanding the function of the organic components. A current limitation in linking molecular entities with ultrastructure is the insufficient resolution of confocal or other optical sectioning microscopy to image fluorescent proteins. The first application of super-resolution fluorescence microscopy on TpSil3 (Gröger et al., 2016) suggests a direction to move toward. In this study, three different fluorescent proteins fused to TpSil3 displayed fluorescence of the protein embedded in silica, with resolution on the order of 25 nm. One issue is that the three proteins exhibited distinct localization patterns in the silica, with the exception of a lack of localization at the fultoportulae, thus it is unclear which protein provides the most accurate assessment of localization, or whether any do. Independent verification using another microscopy technique such as immunogold labeling could clarify that. An unanswered question is whether incorporation of a normally soluble protein (GFP) into silica within the acidic SDV will affect the assembly or localization properties of native silica associated proteins—this also indicates a need for independent localization verification. A combination of localization with genetic manipulation could be especially useful.
We are still in the process of identifying the complete set of proteins involved in silicification, and although significant progress has been made, considering the total number of proteins that must be required, there is more to be done. Both biochemical characterization and transcriptomic analyses have been useful approaches for identifying silicification proteins. Reference transcriptomic datasets may also be good sources for further identification, particularly if efforts are made to identify novel conserved motifs or domains. In this and a previous review (Hildebrand and Lerch, 2015), we identified distinct transcript expression patterns for silicification genes, which expand upon the initial use of only the valve expression pattern (Frigeri et al., 2006; Shrestha et al., 2012) these expression patterns may be useful in further candidate protein identification (Brembu et al., 2017).
In addition to identifying new proteins involved in cell wall formation there are also many unknowns in relatively well-studied areas of this process. Although it has been established that the cytoskeleton is largely responsible for shaping the SDV and positioning structures within it, the dynamics of cytoskeletal movements during structure formation has not been extensively investigated in T. pseudonana. Although not discussed in this review, the process of cell wall exocytosis and the required dynamics of the cytoskeleton and silicalemma associated proteins are also important processes. If SAP3 and Sin1 do associate with in some way with the cytoskeleton or other proteins, proteolytic cleavage of their C-termini could be related to the need to release the SDV after silica formation is complete, enabling exocytosis. The involvement of carbohydrate polymers in silicification is also not well-understood, though they are closely associated with diatom silica, and play a role in structure formation (Tesson et al., 2008; Brunner et al., 2009; Durkin et al., 2009; Hedrich et al., 2013; Tesson and Hildebrand, 2013).
A major emphasis for future work should be better understanding the dynamic processes of cell wall formation. Dynamic imaging approaches have only recently applied to follow the course of events in silicification (Kotzsch et al., 2017). Given the previous discussions, understanding the trafficking processes and timing of key events should provide essential information to understand SDV assembly and associated regulation of its components. Synchronization is a useful approach, but is limited by the fact that even under synchronization individual cells differ in their precise stage of cell wall synthesis—thus details about precise timing are obscured. Imaging single cells progressing through the cell cycle should provide the necessary temporal resolution, thus high-resolution time course imaging on single cells, coupled with co-localization of proteins tagged with different fluorescent markers, is a particularly promising approach.
Given the power of genetically-based approaches to elucidate complex cellular phenomenon such as the cell cycle in yeast, application of genetic manipulation approaches should contribute substantially to clarifying the processes of silicification. Understanding the roles of an individual protein found in a complex assembly of other proteins can be facilitated by selectively removing or decreasing the amount of that protein by knockdown or knockout approaches. One consideration is that the effects of knockdown on structure could be so severe that daughter cells are inviable, and the change in silica structure could not be propagated, and hence, not visualized. In this case, conditional knockdowns could be useful. The most well-developed conditional promoter for diatoms is for nitrate reductase (Poulsen and Kroger, 2005; Poulsen et al., 2006), which is upregulated in the presence of nitrate, and downregulated in ammonium. However, experiments done in our lab (Lerch, unpublished data) indicate that T. pseudonana valve morphology can vary significantly depending on the nitrogen source in the growth medium, which puts a practical limitation on the use of the nitrate reductase promoter in silicification investigations. Identifying another inducible promoter would be a great help. If a suitable inducible promoter was developed, it could be coupled with a synchronized cultivation approach where the knockdown was induced and cells could be isolated after valve formation to evaluate its effect. In this scenario, lack of propagation of daughter cells would not be an issue. Genetic approaches could also improve our understanding of the biochemistry involved by enabling sequence specific alterations in proteins to probe their functions.
Despite an abundant fascination with diatom silica structures, and the growing number of applications which use them, improvement in our understanding of how the structures are made is needed. One reason for this is the lack of precedence for a phenomenon similar to diatom silicification in other biological systems. Each significant discovery in the diatom realm has been novel and required demonstration at a fundamental level. The proteins involved also have unique characteristics, which makes understanding their functions more challenging. Despite the challenges, multiple aspects of cell wall formation have been described through a combination of experimental approaches. We expect that the recent breakthroughs in identifying a new class of SDV associated proteins (Kotzsch et al., 2017; Tesson et al., 2017), the presence of transcriptomic datasets to probe for candidate silicification genes (Shrestha et al., 2012; Keeling et al., 2014; Brembu et al., 2017), dynamic and higher resolution imaging (Gröger et al., 2016; Kotzsch et al., 2017) and genetic manipulation approaches (Tesson et al., 2017) will complement the previously established characterization approaches to allow the field to move forward with a better understanding of the dynamic spatial and temporal protein interactions that are responsible for generating silica structure in a coordinated and controlled manner.
MH assembled an initial draft of the manuscript; MH, SL, and RS were involved in editing and completion.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Diatom silicification research in the Hildebrand lab has been supported by Air Force Office of Scientific Research Multidisciplinary University Research Initiative Grants RF00965521 and FA9550-10-1-0555.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00125/full#supplementary-material
Anonymous (1702). Two letters from a Gentleman in the Country, relating to Mr. Leeuwenhoek's Letter in Transaction, no. 283. Philos. Trans. R. Soc. Lond. B 23, 1494–1501.
Bendtsen, J. D., Nielsen, H., Heijne, G. V., and Brunak, S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. doi: 10.1016/j.jmb.2004.05.028
Blank, G. S., and Sullivan, C. W. (1983). Diatom mineralization of silicic acid. IV. The effect of microtubule inhibitors on silicic acid metabolism in Navicula saprophila. J. Phycol. 19, 39–44
Brembu, T., Skogen Chauton, M., Winge, P., Bones, A. M., and Vadstein, O. (2017). Dynamic responses to silicon in Thalassiosira pseudonana – Identification, characterisation, and classification of signature genes and their corresponding protein motifs. Sci. Rep. 7:4865. doi: 10.1038/s41598-017-04921-0
Brunner, E., Richthammer, P., Ehrlich, H., Paasch, S., Simon, P., Ueberlein, S., et al. (2009). Chitin-based organic networks: an integral part of cell wall biosilica in the diatom Thalassiosira pseudonana. Angew. Chem. Int. Ed. 48, 9724–9727. doi: 10.1002/anie.200905028
Brzezinski, M. A., Olson, R. J., and Chisholm, S. W. (1990). Silicon availability and cell-cycle progression in marine diatoms. Mar. Ecol. Prog. Ser. 67, 83–96. doi: 10.3354/meps067083
Chiappino, M. L., and Volcani, B. E. (1977). Studies on the biochemistry and fine structure of silica shell formation in diatoms. VIII. Sequential cell wall development in the pennate Navicula. Protoplasma 93, 205–221. doi: 10.1007/BF01275654
Davis, A. K., Hildebrand, M., and Palenik, B. (2005). A stress-induced protein associated with the girdle band region of the diatom Thalassiosira pseudonana. J. Phycol. 41, 577–589. doi: 10.1111/j.1529-8817.2005.00076.x
Drum, R. W., and Pankratz, S. H. (1964). Post mitotic fine structure of Gomphonerna parvulum. J. Ultrastruct. Res. 10, 217–223.
Durkin, C. A., Mock, T., and Armbrust, E. V. (2009). Chitin in diatoms and its association with the cell wall. Euk. Cell 8, 1038–1050. doi: 10.1128/EC.00079-09
Emanuelsson, O., Nielsen, H., and Von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. doi: 10.1110/ps.8.5.978
Finbow, M. E., and Harrison, M. A. (1997). The vacuolar H+-ATPase: a universal proton pump of eukaryotes. Biochem. J. 15, 697–712. doi: 10.1042/bj3240697
Frigeri, L. G., Radabaugh, T. R., Haynes, P. A., and Hildebrand, M. (2006). Identification of proteins from a cell wall fraction of the diatom Thalassiosira pseudonana: Insights into silica structure formation. Mol. Cell. Proteomics 5, 182–193. doi: 10.1074/mcp.M500174-MCP200
Gröger, P., Poulsen, N., Klemm, J., Kröger, N., and Schlierf, M. (2016). Establishing super-resolution imaging for proteins in diatom biosilica. Sci. Rep. 6:36824. doi: 10.1038/srep36824
Gruber, A., Vugrinec, S., Hempel, F., Gould, S. B., and Maier, U. G. (2007). Protein targeting into complex diatom plastids: functional characterisation of a specific targeting motif. Plant Mol. Biol. 64, 519–530. doi: 10.1007/s11103-007-9171-x
Hall, D. D., Markwardt, D. D., Parviz, F., and Heideman, W. (1998). Regulation of the Cln3-Cdc28 kinase by cAMP in Saccharomyces cerevisiae. EMBO J. 3, 4370–4378. doi: 10.1093/emboj/17.15.4370
Hedrich, R., Machill, S., and Brunner, E. (2013). Biomineralization in diatoms-phosphorylated saccharides are part of Stephanopyxis turris biosilica. Carb. Res. 365, 52–60. doi: 10.1016/j.carres.2012.11.001
Herth, W. (1979a). Site of beta-chitin fibril formation in centric diatoms. 1. Pores and fibril formation. J. Ultrastruc. Res. 68, 6–15.
Herth, W. (1979b). Site of beta-chitin fibril formation in centric diatoms. 2. Chitin-forming cytoplasmic structures. J. Ultrastruc. Res. 68, 16–27.
Hildebrand, M., Frigeri, L., and Davis, A. K. (2007). Synchronized growth of Thalassiosira pseudonana (Bacillariophyceae) provides novel insights into cell wall synthesis processes in relation to the cell cycle. J. Phycol. 43, 730–740. doi: 10.1111/j.1529-8817.2007.00361.x
Hildebrand, M., Kim, S., Shi, D., Scott, K., and Subramaniam, S. (2009). 3D imaging of diatoms with ion-abrasion scanning electron microscopy. J. Struct. Biol. 166, 316–328. doi: 10.1016/j.jsb.2009.02.014
Hildebrand, M., and Lerch, S. J. L. (2015). Diatom silica biomineralization: parallel development of approaches and understanding. Semin. Cell Dev. Biol. 46, 27–35. doi: 10.1016/j.semcdb.2015.06.007
Hildebrand, M., Volcani, B. E., Gassmann, W., and Schroeder, J. I. (1997). A gene family of silicon transporters. Nature 385, 688–689.
Hildebrand, M., York, E., Kelz, J. I., Davis, A. K., Frigeri, L. G., Allison, D. P., et al. (2006). Nano-scale control of silica morphology and three-dimensional structure during diatom cell wall formation. J. Mater. Res. 21, 2689–2698. doi: 10.1557/jmr.2006.0333
Iler, R. K. (1979). The Chemistry of Silica: Solubility, Polymerization, Colloid and Surface Properties, and Biochemistry. New York: Wiley-Interscience.
Jantschke, A., Koers, E., Mance, D., Weingarth, M., Brunner, E., and Baldus, M. (2015). Insight into the supramolecular architecture of intact diatom biosilica from DNP-supported solid-state NMR spectroscopy. Angew. Chem. Int. Ed. 54, 15069–15073. doi: 10.1002/anie.201507327
Katti, M. V., Sami-Subbu, R., Ranjekar, P. K., and Gupta, V. S. (2000). Amino acid repeat patterns in protein sequences: their diversity and structural-functional implications. Prot. Sci. 9, 1203–1209. doi: 10.1110/ps.9.6.1203
Keeling, P. J., Burki, F., Wilcox, H. M., Allam, B., Allen, E. E., Amaral-Zettler, L. A., et al. (2014). The marine microbial eukaryote transcriptome sequencing project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12:e1001889. doi: 10.1371/journal.pbio.1001889
Kilian, O., and Kroth, P. G. (2005). Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 41, 175–183. doi: 10.1111/j.1365-313X.2004.02294.x
Kley, M., Kempter, A., Boyko, V., and Huber, K. (2014). Mechanistic studies of silica polymerization from supersaturated aqueous solutions by means of time-resolved light scattering. Langmuir 30, 12664–12674. doi: 10.1021/la502730y
Kotzsch, A., Groeger, P., Pawolski, D., Bomans, P. H. H., Sommerdijk, N. A. J. M., Schlierf, M., et al. (2017). Silicanin-1 is a conserved diatom membrane protein involved in silica biomineralization. BMC Biol. 15:65. doi: 10.1186/s12915-017-0400-8
Kotzsch, A., Pawolski, D., Milentyev, A., Shevchenko, A., Scheffel, A., Poulsen, N., et al. (2016). Biochemical composition and assembly of biosilica-associated insoluble organic matrices from the diatom Thalassiosira pseudonana. J. Biol. Chem. 291, 4982–4997. doi: 10.1074/jbc.M115.706440
Kröger, N., Deutzmann, R., Bergsdorf, C., and Sumper, M. (2000). Species-specific polyamines from diatoms control silica morphology. Proc. Natl. Acad. Sci. U.S.A. 97, 14133–14138. doi: 10.1073/pnas.260496497
Kröger, N., Deutzmann, R., and Sumper, M. (1999). Polycationic peptides from diatom biosilica that direct silica nanosphere formation. Science 286, 1129–1132. doi: 10.1126/science.286.5442.1129
Kröger, N., Lorenz, S., Brunner, E., and Sumper, M. (2002). Self-assembly of highly phosphorylated silaffins and their function in biosilica morphogenesis. Science 298, 584–586. doi: 10.1126/science.1076221
Li, C.-W., Chu, S., and Lee, M. (1989). Characterizing the silicon deposition vesicle of diatoms. Protoplasma 151, 158–163.
Mann, D. G. (1984). “An ontogenetic approach to diatom systematics,” in Proceedings of the Seventh International Diatom Symposium, ed D. G. Mann (Koenigstein: Koeltz), 113.
Martin-Jezequel, V., Hildebrand, M., and Brzezinski, M. A. (2000). Silicon metabolism in diatoms: Implications for growth. J. Phycol. 36, 821–840. doi: 10.1046/j.1529-8817.2000.00019.x
Michael, A. J. (2011). Molecular machines encoded by bacterially-derived multi-domain gene fusions that potentially synthesize, N-methylate and transfer long chain polyamines in diatoms FEBS Lett. 585, 2627–2634. doi: 10.1016/j.febslet.2011.07.038
Michael, A. J. (2016). Polyamines in Eukaryotes, bacteria and archea. J. Biol. Chem. 291, 14896–14903. doi: 10.1074/jbc.R116.734780
Pickett-Heaps, J. (1998a). Cell division and morphogenesis of the centric diatom Chaetoceros decipiens (Bacillariophyceae) I. Living cells. J. Phycol. 34, 989–994.
Pickett-Heaps, J. (1998b). Cell division and morphogenesis of the centric diatom Chaetoceros decipiens (Bacillariophyceae) II. Electron microscopy and a new paradigm for tip growth. J. Phycol. 34, 995–1004.
Pickett-Heaps, J. D., Tippit, D. H., and Andreozzi, J. A. (1979). Cell division in the pennate diatom Pinullaria, I. V. Valve morphogenesis. Biol. Cell 35, 199–206.
Pickett-Heaps, J., Schmid, A.-M. M., and Edgar, L. A. (1990). The cell biology of diatom valve formation. Prog. Phycol. Res. 7, 1–168.
Poulsen, N., Chesley, P. M., and Kroger, N. (2006). Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae). J. Phycol. 42, 1059–1065. doi: 10.1111/j.1529-8817.2006.00269.x
Poulsen, N., and Kroger, N. (2005). A new molecular tool for transgenic diatoms - Control of mRNA and protein biosynthesis by an inducible promoter-terminator cassette. FEBS J. 272, 3413–3423. doi: 10.1111/j.1742-4658.2005.04760.x
Poulsen, N., and Kröger, N. (2004). Silica morphogenesis by alternative processing of silaffins in the diatom Thalassiosira pseudonana. J. Biol. Chem. 279, 42993–42999. doi: 10.1074/jbc.M407734200
Poulsen, N., Scheffel, A., Sheppard, V. C., Chesley, P. M., and Kröger, K. (2013). Pentalysine clusters mediate silica targeting of silaffins in Thalassiosira pseudonana. J. Biol. Chem. 288, 20100–20109. doi: 10.1074/jbc.M113.469379
Poulsen, N., Sumper, M., and Kroger, N. (2003). Biosilica formation in diatoms: characterization of native silaffin-2 and its role in silica morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 12075–12080. doi: 10.1073/pnas.2035131100
Reimann, B. E. F., Volcani, B. E., and Lewin, J. C. (1966). Studies on the biochemistry and fine structure of silica shell formation in diatoms. II. The structure of the cell wall of Navicula pelliculosa (Breb.) Hilse. J. Phycol. 2, 74–84.
Richthammer, P., Bçrmel, M., Brunner, E., and van Pee, K. H. (2011). Biomineralization in diatoms: the role of silacidins. ChemBioChem 12, 1362–1366. doi: 10.1002/cbic.201000775
Robinson, D. H., and Sullivan, C. W. (1987). How do diatoms make silicon biominerals? TIBS 12, 151–154.
Round, F. E., Crawford, R. M., and Mann, D. G. (1990). The Diatoms: Biology and Morphology of the Genera. Cambridge: Cambridge University Press.
Scheffel, A., Poulsen, N., Shian, S., and Kröger, N. (2011). Nanopatterned protein microrings from a diatom that direct silica morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 3175–3180. doi: 10.1073/pnas.1012842108
Schmid, A. M., Borowitzka, M. A., and Volcani, B. E. (1981). “Morphogenesis and biochemistry of diatom cell walls,” in Cell Biology Monographs: Cytomorphogenesis in Plants, Vol. 8, ed O. Kiermayer (Wien: Springer-Verlag), 63.
Schmid, A.-M. M. (1980). Valve morphogenesis in diatoms: a pattern-related filamentous system in pennates and the effect of APM, colchicine, and osmotic pressure. Nova Hedwiga 33, 811–847
Shimizu, K., Del Amo, Y., Brzezinski, M. A., Stucky, G. D., and Morse, D. E. (2001). A novel fluorescent silica tracer for biological silicification studies. Chem. Biol. 8, 1051–1060. doi: 10.1016/S1074-5521(01)00072-2
Shrestha, R. P., and Hildebrand, M. (2015). Evidence for a regulatory role of diatom silicon transporters in cellular silicon responses. Euk. Cell 14, 29–40. doi: 10.1128/EC.00209-14
Shrestha, R. P., Tesson, B., Norden-Krichmar, T., Federowicz, S., Hildebrand, M., and Allen, A. E. (2012). Whole transcriptome analysis of the silicon response of the diatom Thalassiosira pseudonana. BMC Genomics 13:499. doi: 10.1186/1471-2164-13-499
Sonnhammer, E. L. L., von Heijne, G., and Krogh, A. (1998). “A hidden Markov model for predicting transmembrane helices in protein sequences,” in Proc. Sixth Int. Conf. on Intelligent Systems for Molecular Biology, eds J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen (Palo Alto, CA: AAAI Press), 175–182.
Tesson, B., and Hildebrand, M. (2010). Extensive and intimate association of the cytoskeleton with forming silica in diatoms: control over patterning on the meso- and micro-scale. PLoS ONE 5:e14300. doi: 10.1371/journal.pone.0014300
Tesson, B., and Hildebrand, M. (2013). Characterization and localization of insoluble organic matrices associated with diatom cell walls: insight into their roles during cell wall formation. PLOS ONE 8:e61675. doi: 10.1371/journal.pone.0061675
Tesson, B., Lerch, S. J. L., and Hildebrand, M. (2017). Characterization of a new protein family associated with the silica deposition vesicle membrane enables genetic manipulation of diatom silica. Sci. Rep. 7:13457. doi: 10.1038/s41598-017-13613-8
Tesson, B., Masse, S., Laurent, G., Maquet, J., and Livage, J. (2008). Contribution of multi-nuclear solid state NMR to the characterization of the Thalassiosira pseudonana diatom cell wall. Anal. Bioanal. Chem. 390, 1889–1898. doi: 10.1007/s00216-008-1908-0
Thamatrakoln, K., and Hildebrand, M. (2008). Silicon uptake in diatoms revisited: a model for saturable and nonsaturable uptake kinetics and the role of silicon transporters. Plant Physiol. 146, 1397–1407. doi: 10.1104/pp.107.107094
van de Meene, A. M. L., and Pickett-Heaps, J. D. (2002). Valve morphogenesis in the centric diatom Proboscia alata Sundstrom. J. Phycol. 38, 351–363. doi: 10.1046/j.1529-8817.2002.01124.x
Waterkeyn, L., and Bienfait, A. (1987). Localization and function of beta 1,3-glucans (callose and chrysolaminarin) in Pinnularia genus (Diatoms). Cellule 74, 199–226.
Keywords: diatom, silicification, AFIM, silica, trafficking, cell wall synthesis, T. pseudonana
Citation: Hildebrand M, Lerch SJL and Shrestha RP (2018) Understanding Diatom Cell Wall Silicification—Moving Forward. Front. Mar. Sci. 5:125. doi: 10.3389/fmars.2018.00125
Received: 13 December 2017; Accepted: 23 March 2018;
Published: 11 April 2018.
Edited by:
Stephen B. Baines, Stony Brook University, United StatesReviewed by:
Mark Brzezinski, University of California, Santa Barbara, United StatesCopyright © 2018 Hildebrand, Lerch and Shrestha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark Hildebrand, bWhpbGRlYnJhbmRAdWNzZC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.